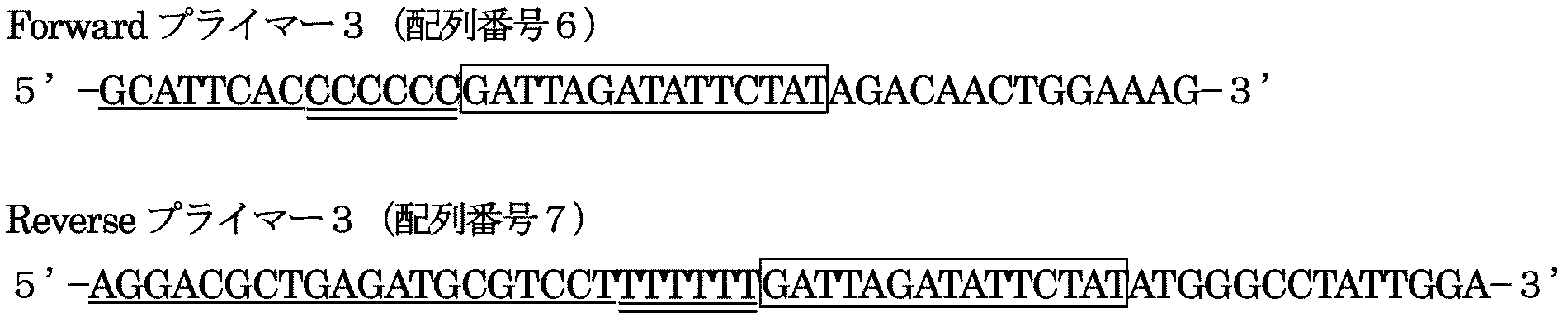WO2013035875A1 - プライマーセット及びそれを用いた標的核酸配列の増幅方法並びに変異核酸の検出方法 - Google Patents
プライマーセット及びそれを用いた標的核酸配列の増幅方法並びに変異核酸の検出方法 Download PDFInfo
- Publication number
- WO2013035875A1 WO2013035875A1 PCT/JP2012/072997 JP2012072997W WO2013035875A1 WO 2013035875 A1 WO2013035875 A1 WO 2013035875A1 JP 2012072997 W JP2012072997 W JP 2012072997W WO 2013035875 A1 WO2013035875 A1 WO 2013035875A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- nucleic acid
- primer
- sequence
- stranded
- reaction
- Prior art date
Links
Images
Classifications
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07H—SUGARS; DERIVATIVES THEREOF; NUCLEOSIDES; NUCLEOTIDES; NUCLEIC ACIDS
- C07H21/00—Compounds containing two or more mononucleotide units having separate phosphate or polyphosphate groups linked by saccharide radicals of nucleoside groups, e.g. nucleic acids
- C07H21/04—Compounds containing two or more mononucleotide units having separate phosphate or polyphosphate groups linked by saccharide radicals of nucleoside groups, e.g. nucleic acids with deoxyribosyl as saccharide radical
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
- C12Q1/6844—Nucleic acid amplification reactions
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
- C12Q1/6844—Nucleic acid amplification reactions
- C12Q1/6853—Nucleic acid amplification reactions using modified primers or templates
Definitions
- the present invention relates to a primer set, a target nucleic acid sequence amplification method using the primer set, and a mutant nucleic acid detection method.
- Amplification of the target gene is mainly performed by an enzymatic method using a DNA polymerase.
- enzymatic methods include polymerase chain reaction (PCR method; US Pat. No. 4,683,195 (Patent Document 1), US Pat. No. 4,683,202 (Patent Document 2) and US Pat. Patent No. 4800159 (Patent Document 3)), reverse transcription PCR method combining RT and reverse transcriptase reaction (RT-PCR method; Trends in Biotechnology 10, pp146-152, 1992 (non-patented) There is literature 1)).
- These methods consist of three steps: dissociation (denaturation) of a double-stranded nucleic acid serving as a template into a single-stranded nucleic acid, annealing of a primer into a single-stranded nucleic acid, and synthesis of a complementary strand from the primer (extension).
- dissociation denaturation
- primer primer into a single-stranded nucleic acid
- synthesis of a complementary strand from the primer extension
- the target gene can be amplified from DNA or RNA.
- Patent Document 4 discloses a ligase chain reaction method (LCR method), in which two-step temperature cycling is performed using a heat-resistant DNA ligase.
- LCR method ligase chain reaction method
- a known gene sequence is amplified by performing a reaction (repeated reaction of heating and cooling).
- it is necessary to use an expensive thermal cycler capable of performing strict temperature control over time in a wide temperature range.
- thermal cycler capable of performing strict temperature control over time in a wide temperature range.
- these reactions are performed under two to three temperature conditions, it takes time to adjust to each reaction temperature, and as the number of cycles increases, the time required for the reaction increases.
- a nucleic acid amplification method that can be carried out in an isothermal state.
- examples of such a method include a strand displacement amplification (SDA) method described in Japanese Patent Publication No. 7-114718 (Patent Document 5), a self-sustained sequence replication (3SR) method, Nucleic acid sequence amplification (NASBA) method described in Japanese Patent No. 2650159 (Patent Document 6), TMA (transcription-mediated amplification) method, Japanese Patent No. 2710159 (Patent Document 7) Qbeta replicase method described in US Pat. No.
- SDA strand displacement amplification
- 3SR self-sustained sequence replication
- NASBA Nucleic acid sequence amplification
- TMA transcription-mediated amplification
- Patent Document 7 Qbeta replicase method described in US Pat. No.
- Patent Document 8 5,824,517
- Patent Document 9 WO99 / 09211
- Patent Document 10 WO95 / 25180
- WO00 / 28082 LAMP method (Loop-Mediated Isothermal Amplification) described
- ICAN method Isothermal and Chimeric primer-initiated Amplification of Nucleic acids
- Patent Document 12 Japanese Patent No. 3897805 Examples thereof include the SmartAmp2 method described in (Patent Document 13). All of the reactions involved in these isothermal nucleic acid amplification methods proceed simultaneously in a reaction mixture maintained at a constant temperature.
- the LAMP method is an isothermal amplification method that requires two turnback primers (TP) and two outer primers (OP). For this reason, four types of primers are required, and the total number of genome recognition sites is six.
- FIG. 11 shows an example of the LAMP method. In the figure, two OPs are omitted and only two TPs are shown. As shown in the figure, TP has a sequence hybridizing to the target nucleic acid sequence on the 3 ′ side and a sequence complementary to the primer extension strand on the 5 ′ side.
- one TP has a sequence (A ′) complementary to the sequence (A) of the target nucleic acid sequence on the 3 ′ side, and primer extension on the 5 ′ side. It has a sequence (M) complementary to the strand sequence (M ′).
- a ′ complementary to the sequence (A) of the target nucleic acid sequence on the 3 ′ side
- M complementary to the strand sequence (M ′).
- FIG. 12 shows an example of the SmartAmp2 method. As shown in the figure, in the SmartAmp2 method, TP is used as one primer and FP is used as the other primer.
- the FP has a sequence (B ′) complementary to the sequence (B) of the target nucleic acid sequence on the 3 ′ side, and the sequences FF complementary to each other on the 5 ′ side. It has a folded sequence containing 'on a single strand. Since the SmartAmp2 method uses TP and FP, there are three genome recognition sites, and FP does not turn back. For this reason, in addition to the advantage that the amplification speed is high and the specificity is high, the primer design is easy and the amplification region can be shortened.
- the SmartAmp2 method has various advantages and is a practical method.
- the SmartAmp2 method uses TP, there are limitations in further reducing the number of genome recognition sites and shortening the amplification distance.
- the present invention provides a primer set used in an isothermal amplification method that has few genome recognition sites and can shorten the amplification distance, an isothermal amplification method using the primer set, and a method for detecting a nucleic acid sequence mutation. Objective.
- the primer set of the present invention is a primer set used in a method for isothermal amplification of a target nucleic acid sequence, wherein the primer set includes a first primer and a second primer, 3 'side contains a sequence (A') capable of hybridizing to the 3 'side sequence (A) of the target nucleic acid sequence, and the second primer is 3' side and the extended strand of the first primer Or a sequence (B ′) that can hybridize to the sequence (B) on the 3 ′ side of the complementary strand of the target nucleic acid sequence, and the first primer and the second primer are It is characterized by containing substantially the same sequence (C).
- the isothermal amplification method of the present invention is an isothermal amplification method in which a target nucleic acid sequence is isothermally amplified using a primer set, wherein the primer set of the present invention is used as the primer set.
- the method for detecting a mutation in a nucleic acid sequence of the present invention is a method for detecting a mutation in a nucleic acid sequence in a nucleic acid sample by an isothermal amplification method using a primer set, wherein the primer set of the present invention is used as the primer set.
- the primer set uses a nucleic acid sequence having the mutation or a nucleic acid sequence not having the mutation as a target nucleic acid sequence, and the nucleotide residue related to the mutation is a complementary sequence (A) of the first primer or a second primer A primer set designed to be included in the complementary sequence (B), wherein an isothermal amplification reaction is performed with the primer set in the presence of the nucleic acid sample.
- the primer set of the present invention there are two genome recognition sites, and since TP is not used, primer design is easy and the amplification region sequence can be shortened. For this reason, by using the primer set of the present invention, it is possible to amplify even a short sequence such as microRNA which could not be amplified by the conventional method.
- the present invention provides a primer set and an isothermal amplification method that are completely different from SmartAmp2 developed by the present inventors.
- FIG. 1 is a diagram showing an example of the primer set of the present invention.
- FIG. 2 is a diagram showing another example of the primer set of the present invention.
- FIG. 3 is a diagram showing still another example of the primer set of the present invention.
- FIG. 4 is a schematic diagram showing an example of the amplification reaction of the primer set of the present invention.
- FIG. 5A is a schematic diagram showing an example of a reaction mechanism in the nucleic acid synthesis method of the present invention.
- FIG. 5B is a schematic diagram illustrating an example of a reaction mechanism in the reaction step subsequent to FIG. 5A.
- FIG. 6 is a schematic diagram showing an example of a reaction mechanism of an extended strand exchange reaction based on nick-passing extension.
- FIG. 1 is a diagram showing an example of the primer set of the present invention.
- FIG. 2 is a diagram showing another example of the primer set of the present invention.
- FIG. 3 is a diagram showing still another example of the primer set of the present invention.
- FIG. 7A is a schematic diagram showing another example of the reaction mechanism in the nucleic acid synthesis method of the present invention.
- FIG. 7B is a schematic diagram illustrating an example of a reaction mechanism in the reaction step subsequent to FIG. 7A.
- FIG. 8A is a schematic diagram showing still another example of the reaction mechanism in the nucleic acid synthesis method of the present invention.
- FIG. 8B is a schematic diagram illustrating an example of a reaction mechanism in the reaction step subsequent to FIG. 8A.
- FIG. 9A is a schematic diagram showing still another example of the reaction mechanism in the nucleic acid synthesis method of the present invention.
- FIG. 9B is a schematic diagram illustrating an example of a reaction mechanism in the reaction step subsequent to FIG. 9A.
- FIG. 9C is a schematic diagram illustrating another example of the reaction mechanism in the reaction step subsequent to FIG. 9A.
- FIG. 10 is a diagram showing an example of the first primer in the primer set of the present invention.
- FIG. 11 is a diagram illustrating an example of the LAMP method.
- FIG. 12 is a diagram illustrating an example of the SmartAmp2 method.
- FIG. 13 is a graph showing a fluorescence amplification curve when the primer set of Example 1 (Forward primer 1 and Reverse primer 1) is used.
- FIG. 14 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in the fluorescence signal was observed in FIG.
- FIG. 10 is a diagram showing an example of the first primer in the primer set of the present invention.
- FIG. 11 is a diagram illustrating an example of the LAMP method.
- FIG. 12 is a diagram illustrating an example of the SmartAmp2 method.
- FIG. 13 is a graph showing a fluorescence a
- FIG. 15 is a graph showing a fluorescence amplification curve when the primer set of Example 1 (Forward primer 2 and Reverse primer 2) is used.
- FIG. 16 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in the fluorescence signal was recognized in FIG.
- FIG. 17 is a graph showing a fluorescence amplification curve when the primer set of Example 2 (Forward primer 3 and Reverse primer 3) is used.
- FIG. 18 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in fluorescence signal was observed in FIG.
- FIG. 19 is a graph showing a fluorescence amplification curve when the primer set of Example 3 (Forward primer 4 and Reverse primer 4) is used.
- FIG. 20 is a graph showing a fluorescence amplification curve when only the Boost primer 1 of Example 3 is used.
- FIG. 21 is a graph showing a fluorescence amplification curve when only the Boost primer 2 of Example 3 is used.
- FIG. 22 is a graph showing a fluorescence amplification curve when Boost primers 1 and 2 of Example 3 are used.
- FIG. 23 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in the fluorescence signal was observed in FIGS.
- FIG. 24 is a graph showing a fluorescence amplification curve when the primer set of Example 4 (Forward primer 5 and Reverse primer 5) is used.
- FIG. 25 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in fluorescence signal was observed in FIG.
- FIG. 26 is a schematic diagram showing the amplification reaction of the primer set of Example 4.
- FIG. 27 is a graph showing a fluorescence amplification curve when the primer set of Example 5 (Forward primer 6 and Reverse primer 6) was used.
- FIG. 28 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in fluorescence signal was observed in FIG.
- FIG. 29 is a schematic diagram showing the amplification reaction of the primer set of Example 5.
- FIG. 30 is a graph showing a fluorescence amplification curve when the primer set of Example 6 (Forward primer 7 and Reverse primer 7) was used.
- FIG. 31 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in fluorescence signal was observed in FIG.
- FIG. 32 is a graph showing fluorescence amplification curves when the primer set of Example 7 (Forward primer 8 and Reverse primer 6) was used.
- FIG. 33 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in fluorescence signal was observed in FIG.
- FIG. 34 is a graph showing fluorescence amplification curves when the primer set of Example 8 (Forward primer 5 and Reverse primer 8) was used.
- FIG. 35 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in fluorescence signal was observed in FIG.
- FIG. 36 is a schematic diagram showing the amplification reaction of the primer set of Example 8.
- the first primer and the second primer each contain substantially the same sequence (C) on the 5 ′ side as described above.
- the phrase “substantially identical” between the two sequences (C) means that the two sequences (C) can each hybridize to the complementary sequence of the other primer.
- the two sequences (C) may be completely identical to each other (full match), or may be sequences (mismatches) in which some bases are different.
- one of the two sequences (C) may be a sequence generated by at least one of base substitution, insertion and deletion with respect to the other sequence (C).
- the number of bases in which substitution, insertion or deletion has occurred is preferably 2/10 or less of the total number of bases in the two sequences (C), It is more preferable that it is 1/10 or less. Further, it is particularly preferable that the two sequences (C) are completely identical (that is, the number of bases causing substitution, insertion or deletion is zero).
- a folded sequence comprising two sequences hybridizing to each other on the same strand ( DD ') may be included.
- the primer set of the present invention includes a folded sequence (DD ′) containing two sequences hybridizing to each other on the same strand on the 5 ′ side of the sequence (C) in the first primer.
- the second primer further comprises a folded sequence (EE ′) containing two sequences hybridizing to each other on the same strand on the 5 ′ side of the sequence (C).
- D ′) and the array (EE ′) may be different from each other.
- the primer set of the present invention further includes a third primer, and the third primer is the target nucleic acid sequence, a complementary sequence of the target nucleic acid sequence, or an extended strand of the first primer or an extension of the second primer. It may be an embodiment that hybridizes to a strand and the hybridization of the third primer does not compete with the first primer and the second primer.
- Another aspect of the present invention is a first method of isothermally synthesizing a double-stranded nucleic acid composed of a single-stranded nucleic acid in which the order of at least two different sequences is repeated at least twice and a nucleic acid complementary to the single-stranded nucleic acid.
- the method for synthesizing a nucleic acid according to the above may be the first nucleic acid synthesis method characterized by including the following steps (A1) to (A6).
- (A3) A step of continuously continuing the extension reaction of the primer that has progressed to the 5 ′ end of the 5 ′ side stem sequence from the 5 ′ end of the 5 ′ side stem sequence to the 3 ′ end of the folded sequence.
- (A4) The extension reaction of the primer that has proceeded to the 3 ′ end of the folded sequence of (A3) is continuously continued again toward the 5 ′ end of the 5′-side stem sequence.
- (A5) A step of stopping the primer extension reaction of (A4) at the 5 ′ end of the 5′-side stem sequence.
- (A6) A step of extending the 3 ′ end of the folded sequence of the single-stranded template nucleic acid using the primer extended strand that has become a single strand in (A4) as a template.
- step (A3) and step (A4) may be repeated at least twice.
- the single-stranded template nucleic acid provided in the step (A1) is the primer set of the present invention, and only the first primer has the sequence (C).
- a single-stranded template nucleic acid formed by an isothermal amplification reaction using a primer set containing the folded sequence (DD ′) on the 5 ′ side of the primer, and hybridizing to the loop in the step (A2) May be the first primer including the folded sequence (DD ′).
- the single-stranded template nucleic acid provided in the step (A1) further hybridizes to the 5 ′ end of the 5 ′ stem loop sequence 2
- a single-stranded template nucleic acid in which folded sequences containing two sequences on the same strand are linked may include the following step (A3-2) instead of the step (A3).
- A3-2) The extension reaction of the primer that has progressed to the 5 ′ end of the 5′-side stem sequence is directly performed without using the folded sequence linked to the 5′-end of the 5′-side stem sequence. The process of continuing continuously from the 5 'end of the' side stem sequence to the 3 'end of the folded sequence.
- the steps (A3-2) and (A4) may be repeated at least twice.
- the first nucleic acid synthesis method of the present invention includes the step (A3-2) instead of the step (A3), the single-stranded template nucleic acid provided in the step (A1) One formed by an isothermal amplification method using the primer set of the present invention comprising the first primer containing the folded sequence (DD ′) and the second primer containing the folded sequence (EE ′).
- the primer that is a double-stranded template nucleic acid and hybridizes to the loop in the step (A2) may be the first primer or the second primer of the primer set of the present invention.
- Still another aspect of the present invention is a method for amplifying a nucleic acid, comprising a single-stranded nucleic acid in which the order of at least two different sequences is repeated at least twice and a nucleic acid complementary to the single-stranded nucleic acid.
- a nucleic acid synthesis method comprising synthesizing a double-stranded nucleic acid isothermally, wherein the nucleic acid synthesis step is performed by the first nucleic acid synthesis method of the present invention. There may be.
- Still another aspect of the present invention provides isothermal synthesis of a single-stranded nucleic acid in which the sequence of at least two different sequences is repeated at least twice and a nucleic acid complementary to the single-stranded nucleic acid.
- a second nucleic acid synthesis method comprising: a first reaction step including the following steps (B1) to (B3); and a second reaction including the following steps (C1) to (C3): It may be a second nucleic acid synthesis method characterized by including at least one of the steps.
- (B1) consisting of a single-stranded nucleic acid containing a folded sequence containing two sequences that hybridize to each other on the same strand in a region containing the 3 ′ end, and a single-stranded nucleic acid complementary to the single-stranded nucleic acid Providing two duplexes in a state in which the orientations of the sequences are opposite to each other.
- (B2) From the 3 ′ end of the double-stranded sequence of the single-stranded nucleic acid of one of the two double-stranded strands of (B1), use the complementary single-stranded nucleic acid of the other double-stranded as a template A partial displacement in which a strand displacement extension reaction occurs, and a complementary single-stranded nucleic acid of the other double strand is hybridized to a part of the extended strand of the single-stranded nucleic acid of the one double strand Forming.
- a complete duplex is formed from the 3 ′ end of the complementary single-stranded nucleic acid by an extension reaction using the single-stranded nucleic acid as a template.
- Process. (C1) It consists of a single-stranded nucleic acid containing a folded sequence containing two sequences that hybridize to each other on the same strand in a region containing the 3 ′ end, and a single-stranded nucleic acid complementary to the single-stranded nucleic acid. Providing a single duplex.
- the double strands of the step (B1) and the step (C1) are further connected to the 5 ′ side of the sequence (C) in at least one of the first primer and the second primer of the primer set of the present invention. It may be a duplex formed by an isothermal amplification reaction using a primer set containing a folded sequence (DD ′) containing two sequences that hybridize to each other on the same strand.
- DD ′ folded sequence
- the duplexes of the steps (B1) and (C1) are further hybridized to each other on the 5 ′ side of the sequence (C) in the first primer of the primer set of the present invention 2
- a folded sequence (DD ′) containing two sequences on the same strand, and in the second primer, two sequences hybridizing to each other are further arranged on the same strand on the 5 ′ side of the sequence (C).
- the sequence (DD ′) and the sequence (EE ′) are formed by isothermal amplification reaction using primer sets having different sequences. It may be a heavy chain.
- Yet another aspect of the present invention is a method for amplifying a nucleic acid, comprising a single-stranded nucleic acid in which the order of at least two different sequences is repeated at least twice, and a nucleic acid complementary to the single-stranded nucleic acid.
- a nucleic acid synthesis method comprising synthesizing a double-stranded nucleic acid isothermally, wherein the nucleic acid synthesis step is carried out by the second nucleic acid synthesis method of the present invention. There may be.
- target nucleic acid or “target nucleic acid sequence” means not only the nucleic acid to be amplified or the sequence itself, but also a complementary sequence or a nucleic acid having the sequence.
- hybridize means that a part of the primer according to the present invention hybridizes to a target nucleic acid under stringent conditions and does not hybridize to a nucleic acid molecule other than the target nucleic acid.
- Stringent conditions can be determined, for example, depending on the melting temperature Tm (° C.) of the duplex of the primer according to the present invention and its complementary strand, the salt concentration of the hybridization solution, etc. You can refer to Sambrook, E. F. Frisch, T. Maniatis; Molecular Cloning 2nd edition, Cold Spring Harbor Laboratory (1989), etc.
- the primer can be specifically hybridized to the target nucleic acid.
- a primer that hybridizes to a target nucleic acid comprises a sequence of all or part of a nucleic acid molecule complementary to the target nucleic acid.
- FIG. 1 shows an example of the primer set of the present invention.
- the primer set of this example is a primer set used for the isothermal amplification method of the target nucleic acid sequence 4, and the primer set includes a first primer 1F and a second primer 1R
- Primer 1F includes a sequence (A ′) capable of hybridizing to the 3 ′ side sequence (A) on the 3 ′ side
- the second primer 1R is arranged on the 3 ′ side to A sequence (B ′) capable of hybridizing to an extended strand of a primer or a sequence (B ′) on the 3 ′ side of a complementary strand of the target nucleic acid sequence 4
- the first primer 1F and the second primer 1R are each The same sequence (C) is included on the 5 ′ side of each other.
- FIG. 4 shows a schematic diagram of an example of the amplification reaction of the primer set of this example.
- the same parts as those in FIG. 4 are identical to FIG. 4 in this figure.
- the first primer 1F and the second primer 1R are hybridized to the target nucleic acid sequence 4 to cause an extension reaction.
- the first primer hybridizes to the extended strand of the first primer by hybridization with the second primer, or the first primer hybridizes to the extended strand of the second primer.
- Extension to form a double-stranded intermediate As shown in (c) and (c ′) in the figure, the double-stranded intermediate has a stem-loop structure by intramolecular hybridization when it becomes a single strand by a dynamic equilibrium reaction.
- the first primer or the second primer hybridizes to the loop of the single-stranded intermediate of the stem-loop structure to form an extended strand, whereby two strands shown in (d) and (d ′) of FIG. Chain intermediates are formed.
- the double-stranded intermediates in FIGS. 4 (d) and (d ′) are the same as the double-stranded intermediate in FIG. 2 (b). It becomes an intermediate of this chain.
- the number of bases of the sequences (A ′) and (B ′) that hybridize with the target nucleic acid sequence is not particularly limited, for example, 3 to 100 bases, preferably Is 10 to 60 bases, more preferably 15 to 50 bases. Further, the number of bases of the same sequence (C) of the first primer and the second primer of the primer set of the present invention is not particularly limited, for example, 3 to 100 bases, preferably 10 to 60 bases, more preferably 15-50 bases.
- DD folded sequence
- the total length of the folded sequence (DD ′) is not particularly limited, and is, for example, 3 to 100 bases, preferably 4 to 60 bases, more preferably 5 to 50 bases.
- the number of bases of any one of the sequences complementary to each other of the folded sequence (DD ′) is not particularly limited, and is, for example, 1 to 50 bases, preferably 1 to 30 bases, more preferably 1 to 20 bases.
- An intervening sequence may exist between the complementary sequences of the folded sequence (DD ′).
- the number of bases in the intervening sequence is, for example, 1 to 50 bases, preferably 1 to 20, and more preferably 1 to 10.
- a part of the folded sequence may form a part of the same sequence (C) of the first primer and the second primer.
- the first primer 3F includes a folded sequence (DD ′) that includes two sequences that hybridize to each other on the same strand on the 5 ′ side of the sequence (C).
- the second primer 3R further includes a folded sequence (EE ′) containing two sequences hybridizing to each other on the same strand on the 5 ′ side of the sequence (C). ′) And the sequence (EE ′) are different from each other.
- a single-stranded nucleic acid in which the order of at least two different sequences is repeated at least twice and a nucleic acid complementary to the single-stranded nucleic acid is used.
- Single-stranded nucleic acid is synthesized isothermally.
- Examples of the reaction mechanism of the nucleic acid synthesis method of the present invention include a first synthesis reaction shown in FIGS. 5A to 7B described later and a second synthesis reaction shown in FIGS. 8A to 9C described later. .
- the first synthesis reaction includes the steps (A1) to (A6), or, as described above, may include the step (A3-2) instead of the step (A3).
- the second synthesis reaction includes at least one of a first reaction step including the steps (B1) to (B3) and a second reaction step including the steps (C1) to (C3).
- the amplification reaction using the primer set of the present invention may include a synthesis reaction other than the first synthesis reaction and the second synthesis reaction.
- the amplification reaction using the primer set of the present invention preferably includes at least one of the first synthesis reaction and the second synthesis reaction, but includes the first synthesis reaction and the second synthesis reaction. You don't have to leave.
- the first synthesis reaction is an extended strand exchange reaction based on nick-passing extension.
- an example of the first synthesis reaction will be described with reference to FIGS. 5A to 7B.
- FIGS. 5A and 5B show an example of the first synthesis reaction.
- This reaction is a reaction using the same primer set as in FIG. 2 in which only one primer contains a folded sequence. That is, the primer set includes a first primer 2F including a folded sequence (DD ′) on the 5 ′ side of the sequence (C), and a second primer 2R not including the folded sequence.
- DD ′ folded sequence
- C 5 ′ side of the sequence
- second primer 2R not including the folded sequence.
- 5A and 5B the same components as those in FIG. 2 are denoted by the same reference numerals.
- an ⁇ intermediate is provided (step (A1)).
- this ⁇ intermediate is composed of a 3 ′ stem sequence (C ′) including a 3 ′ end and a 5 ′ stem sequence (C) including a 5 ′ end.
- the step (A1) (FIGS. 5A (a) to (g)) will be specifically described. That is, first, as shown in FIG. 5A (a), the first primer 2F hybridizes to the sequence (A) of the target nucleic acid sequence. Furthermore, as shown in FIG. 5B, the first strand 2F is extended, whereby the first strand is extended. Next, as shown in FIG. 5C, the extended strand of the first primer 2F has the same sequence (A ′) as the first primer 2F, and the sequence (A ′) is released from the sequence (A) of the target nucleic acid sequence due to fluctuation. A primer having a sequence undergoes strand displacement hybridization (SDH) to the sequence (A) of the target nucleic acid sequence and further undergoes an extension reaction, whereby the extended strand (first strand) of the first primer 2F is released.
- SDH strand displacement hybridization
- the second primer 2R is hybridized and extended to the released first strand, whereby the extended strand of the second primer 2R (first 2 strands) are formed.
- the second strand includes a sequence (DD′-C′-A) complementary to the sequence (A′-CDD ′) of the first primer, as shown in FIG. .
- the extended strand of the second primer 2R has the same sequence (B ′) as the second primer 2R, because the sequence (B ′) is released from the sequence (B) of the target nucleic acid sequence due to fluctuation.
- a primer having a sequence undergoes strand displacement hybridization (SDH) to the sequence (B) of the first strand, and further undergoes an extension reaction, whereby the second strand is released. Then, as shown in FIG. 5G, the released second strand sequences (C) and (C ′) self-hybridize to form a single-stranded template nucleic acid having an ⁇ -like structure. This is the aforementioned ⁇ intermediate.
- the sequence (A) in the loop of the ⁇ intermediate has the same sequence as the first primer 2F.
- the primer is hybridized, and the primer is extended toward the 5 ′ end of the 5 ′ stem sequence (C) of the ⁇ intermediate (step (A2)).
- this extension reaction reaches the 5′-side stem sequence (C)
- the extension reaction using the 5′-side stem sequence (C) as a template is accompanied by a strand displacement reaction.
- the nick formed from the 5 ′ end of the 5 ′ stem sequence (C) (Tail sequence) and the 3 ′ end of the hook sequence (DD ′) is used for extension.
- a haploid amplicon the single-stranded template nucleic acid
- the extension reaction of the primer proceeding to the 5 ′ end of the 5′-side stem sequence (C) is performed on the 5′-side stem sequence (C).
- the process continues continuously from the 5 ′ end to the 3 ′ end of the folded sequence (DD ′) (step (A3)).
- the extension reaction of the primer proceeding to the 3 ′ end of the folded sequence (DD ′) in the same figure (i) step (A3)). Is continued continuously toward the 5 ′ end of the 5′-side stem sequence (C), and the primer extension reaction continues, so that FIGS.
- the primer extension strand hybridized with the single-stranded template nucleic acid ( ⁇ intermediate) formed in (A2) step) is made into a single strand by a strand displacement reaction ((A4) step). Then, the primer extension reaction in the step (A4) is stopped at the 5 ′ end of the 5′-side stem sequence (C) shown in FIG. 5B (l) (step (A5)).
- the primer extension strand becomes a diploid la in which two amplicon sequences (the single-stranded template nucleic acid sequences) are linked in the forward direction (tandem). Furthermore, as shown in FIG.
- the single-stranded template nucleic acid ( ⁇ intermediate) of the single-stranded template nucleic acid ( ⁇ intermediate) is formed using the primer extension strand (tandem diploid la) that has become single-stranded in the step (A4).
- the 3 ′ end of the folded sequence (DD ′) is extended (step (A6)).
- a strand complementary to the primer extension strand (tandem diploid la) is formed, and a complete double strand ma composed of the primer extension strand and a strand complementary thereto is formed.
- 5A and 5B show an example in which a tandem diploid is formed by performing the steps (A3) and (A4) only once.
- steps (A3) and (A4) by repeating the steps (A3) and (A4) at least twice, a tandem chain of triploid or higher can be formed. That is, after the step (A4), the process returns to the step (A3).
- steps (A3) and (A4) are repeated at least twice, the process proceeds to the steps (A5) and (A6). You may form a tandem chain more than a diploid.
- Fig. 6 shows the case where the nick transfer reaction occurs and does not occur.
- (a) of the figure in the ⁇ intermediate, when the 5 ′ end of the 5 ′ stem and the 3 ′ end of the folded sequence are close to each other to form a nick, overcome the nick. Primer extension reaction occurs.
- FIG. 4B in the ⁇ intermediate, when the 5 ′ end of the 5 ′ stem is separated from the 3 ′ end of the folded sequence and a nick is not formed, the primer is extended. The reaction stops at the 5 'end of the 5' stem.
- FIGS. 7A and 7B show another example of the first synthesis reaction.
- This reaction is a reaction using a primer set similar to that shown in FIG. 3 in which both primers contain folded sequences. That is, the primer set includes a first primer 3F including a folded sequence (DD ′) on the 5 ′ side of the sequence (C), and a folded sequence (EE ′) on the 5 ′ side of the sequence (C).
- a second primer 3R. 7A and 7B, the same components as those in FIG. 3 are denoted by the same reference numerals.
- an ⁇ intermediate is provided (step (A1)).
- this ⁇ intermediate has a loop sequence consisting of a 3 ′ stem sequence (C ′) containing a 3 ′ end and a 5 ′ stem sequence (C) containing a 5 ′ end.
- Two sequences (D) and (D) that hybridize with each other at the 3 ′ end of the 3 ′ stem (C ′) of the single-stranded template nucleic acid of the stem-loop structure linked via (AB ′) A single-stranded template nucleic acid in which folded sequences (DD ') containing') in the same strand are linked.
- 5A (g) is different from the ⁇ intermediate shown in FIG. 5A in that the folded arrays (EE ′) included in the shape are connected. Except for including the folded array (EE ′), it is the same as the ⁇ intermediate of FIG. 5A (g).
- FIGS. 7A (a) to (g) In the step (A1) (FIGS. 7A (a) to (g)), FIGS. 7A (a) to (c) (hybridization of the first primer to the template nucleic acid, extension reaction, and extension of the first primer)
- the release of the chain (first chain) is the same as in FIGS. 5A (a) to (c).
- the sequence of the first primer 3F in FIG. 7A is the same as the first primer 2F in FIG. 5A.
- the second primer 3R including the folded sequence (EE ′) is used instead of the second primer 2R not including the folded sequence.
- a single-stranded template nucleic acid ( ⁇ intermediate) shown in FIG. 7A (g) is formed in the same manner as 5A (d) to (g).
- the sequence (A) in the loop of the ⁇ intermediate has the same sequence as the first primer 3F.
- the primer is hybridized, and the primer is extended toward the 5 ′ end of the 5 ′ stem sequence (C) of the ⁇ intermediate (step (A2)).
- this extension reaction reaches the 5′-side stem sequence (C)
- the extension reaction using the 5′-side stem sequence (C) as a template is accompanied by a strand displacement reaction.
- the primer extension reaction may proceed from the middle of the 5′-side stem sequence to the 3 ′ end of the folded sequence, or may be linked to the 5 ′ end of the 5′-side stem.
- the primer extension reaction may proceed in the middle of the folded sequence or from the 5 ′ end of the folded sequence to the 3 ′ end of the folded sequence linked to the 3 ′ stem sequence.
- the extension reaction is continuously continued again toward the 5 ′ end of the 5′-side stem sequence (C), and the primer extension reaction of the primer continues, FIG.
- Step (A4) The primer extension reaction in the step (A4) is stopped at the 5 ′ end of the 5′-side stem sequence (C) shown in FIG. 7B (l) (step (A5)).
- the reaction for converting the primer extension strand into a single strand by a strand displacement reaction is stopped at the 5 ′ end of the 5′-side stem sequence (C), but the extension reaction itself is performed in the 5′-side stem sequence (C ) To the 5 ′ end of the folded sequence (EE ′) of the ⁇ intermediate.
- the primer extension strand becomes a (tandem) diploid in which two amplicon sequences are linked in the forward direction (the upper strand in FIG. 7B (m) or (n)).
- FIGS. 7B (m) and (n) the folded sequence of the single-stranded template nucleic acid ( ⁇ intermediate) (see FIG.
- DD ′ is extended at the 3 ′ end (step (A6)).
- a strand complementary to the primer extension strand (the lower strand in the figure) is formed, and the primer extension strand and the strand complementary thereto are completely formed. Double strands are formed.
- the continuous primer extension reaction from the 5 ′ end of the 5 ′ side stem to the 3 ′ end of the folded sequence (DD ′) occurs by the same mechanism as the nick transfer reaction described above.
- FIG. 7A and 7B show an example in which a tandem diploid is formed by performing the steps (A3-2) and (A4) only once.
- steps (A3-2) and (A4) by repeating the steps (A3-2) and (A4) at least twice, a tandem chain of triploid or higher can be formed. That is, after the step (A4), the process returns to the step (A3-2), and when the step (A3-2) and the step (A4) are repeated at least twice, the step (A5) and the step (A6) Proceeding to tandem chains more than triploid may be formed.
- 8A and 8B show an example of the second synthesis reaction.
- This reaction is a reaction using the same primer set as in FIG. 2 in which only one primer contains a folded sequence. That is, the primer set includes a first primer 2F including a folded sequence (DD ′) on the 5 ′ side of the sequence (C), and a second primer 2R not including the folded sequence.
- DD ′ folded sequence
- B primer 2R not including the folded sequence.
- one of the complementary double strands (haploid amplicons) formed according to FIGS. 8A (a) to (i) is provided as shown in FIG. 8B (j) (step (C1)), Alternatively, as shown in FIG. 8B (j ′), two are provided in a state where the orientations of the arrays are opposite to each other (step (B1)).
- 8A (a) to (g) (formation of ⁇ intermediate) is exactly the same as FIG. 5A (a) to (g) (the first synthesis reaction).
- a primer having the same sequence as the first primer 2F is hybridized to the sequence (A) in the loop of the ⁇ intermediate (single-stranded template nucleic acid), The primer is extended toward the 5 ′ end of the 5 ′ stem sequence (C) of the ⁇ intermediate. This extension reaction was stopped at the 5 ′ end of the 5 ′ stem sequence (C), and as shown in FIG. 5 (i), two complementary strands consisting of the ⁇ intermediate and the extended strand of the primer. A chain (haploid amplicon) is formed. As shown, this amplicon includes a folded sequence (DD ′) containing two sequences (D) and (D ′) that hybridize to each other in the region containing the 3 ′ end on the same strand.
- FIG. 8B From the amplicon of FIG. 8A (i) (identical to FIG. 8B (i)), as shown in FIG. 8B, the ⁇ intermediate or its complementary strand was linked in the forward direction in two reaction paths ( A tandem amplification product is formed.
- One reaction pathway is a pathway in which strand displacement hybridization occurs in the molecule of the double-stranded amplicon (FIG. 8B (i)), as shown in FIGS. 8B (j) to (l).
- the other reaction pathway is a pathway in which strand displacement hybridization occurs between two molecules of the double-stranded amplicon, as shown in FIGS. 8B (j ′) to (l ′). That is, first, one amplicon is provided as shown in FIG.
- step (C1) the double-stranded amplicon is either in the molecule of the amplicon as shown in (j) to (k) of the figure, or as shown in (j ′) to (k ′) of the figure.
- the 5 ′ end sequence (C) hybridizes to the sequence (C ′) of the other strand (Tail substitution).
- this Tail substitution occurs, folding of the folded sequence (DD ′) at each end occurs as shown in FIGS.
- the strand displacement extension reaction takes place from the 3 ′ end to the 5 ′ end of the complementary single-stranded nucleic acid using the complementary single-stranded nucleic acid as a template, and a part of the extended strand of the single-stranded nucleic acid This corresponds to the step ((C2) step) of forming a partial double strand ((m) in the figure) in which the complementary single-stranded nucleic acids are hybridized.
- the reaction from FIG. 8B (l ′) to FIG. 8 (m) shows the folded sequence of the single-stranded nucleic acid of one double strand of the two double strands (B1) (FIG. 8B (j ′)).
- a strand displacement extension reaction occurs using the complementary single-stranded nucleic acid of the other double strand as a template, and the single-stranded nucleic acid of one double strand is extended.
- step (B2) of forming a partial duplex (FIG. (M)) in which a complementary single-stranded nucleic acid of the other duplex is hybridized to a part of the strand.
- FOG. (M) partial duplex
- an extension reaction using the opposite strand as a template occurs from the 3 ′ end portion of the complementary strand of the newly synthesized extension strand.
- a double strand in which the amplicons are linked in the forward direction (tandem) is formed ((o) in the figure).
- the reaction from FIG. 8B (n) to (o) is carried out by using the complementary single-stranded nucleic acid (FIG. 8) in the partial double strand (FIG. 8 (m)) of the step (B2) or (C2).
- a step of forming a complete duplex (FIG. (O)) from the 3 ′ end of (m) upper strand) by an extension reaction using the single-stranded nucleic acid as a template (FIG. (N)) (B3) Step or (C3) Step).
- FIGS. 9A to 9C show another example of the second synthesis reaction.
- This reaction is a reaction using a primer set similar to that shown in FIG. 3 in which both primers contain folded sequences. That is, the primer set includes a first primer 3F including a folded sequence (DD ′) on the 5 ′ side of the sequence (C), and a folded sequence (EE ′) on the 5 ′ side of the sequence (C).
- the primer set includes a first primer 3F including a folded sequence (DD ′) on the 5 ′ side of the sequence (C), and a folded sequence (EE ′) on the 5 ′ side of the sequence (C).
- DD ′ folded sequence
- EE ′ folded sequence
- FIGS. 9A (a) to (i) are provided as shown in FIG. 9C (j) (step (C1)), Alternatively, as shown in FIG. 9C (j ′), two are provided in a state where the orientations of the arrays are opposite to each other (step (B1)).
- 9A (a) to (g) formation of ⁇ intermediate
- FIG. 7A (a) to (g) first synthesis reaction).
- a primer having the same sequence as the first primer 3F is hybridized to the sequence (A) in the loop of the ⁇ intermediate (single-stranded template nucleic acid), The primer is extended toward the 5 ′ end of the 5 ′ stem sequence (C) of the ⁇ intermediate. This extension reaction was stopped at the 5 ′ end of the folding sequence (EE ′) linked to the 5 ′ end of the 5 ′ stem sequence (C), and as shown in FIG. 9A (i), the ⁇ A complementary double strand (haploid amplicon) consisting of the intermediate and the extended strand of the primer is formed.
- this amplicon includes a folded sequence (DD ′) containing two sequences (D) and (D ′) that hybridize to each other in the region containing the 3 ′ end on the same strand. It is a double strand consisting of a single-stranded nucleic acid (lower strand in FIG. 9A (i)) and a single-stranded nucleic acid complementary to the single-stranded nucleic acid (upper strand in FIG. 9). Even when the second primer 3R becomes a reaction starting point, an amplicon is formed in the same reaction process.
- FIG. 9B two reaction routes shown in either FIG. 9B or 9C can be considered.
- one reaction route is a route using a turn having a similar palindromic sequence.
- this is a route when the double-stranded amplicon folding sequences (DD ′) and (EE ′) of FIG. 9A (i) are similar palindromic sequences.
- FIG. 9B (p) hetero-folds (DD ′) and (EE ′) having the similar palindromic sequence between the double-stranded amplicons of two molecules are obtained. , Hybridize to each other.
- the other reaction pathway is a pathway in which recombination hybridization occurs between the double-stranded amplicon sequence (C) and its complementary sequence (C ′) (Tail sequence), as shown in FIG. 9C.
- This reaction path is further divided into two reaction paths as shown in FIG. That is, as shown in FIGS. 9C (j) to (l), one reaction pathway is an intramolecular structure of the double-stranded amplicon (same as FIG. 9A (i)) represented by FIG. 9C (i). This is the pathway through which strand displacement hybridization occurs.
- the other reaction pathway is a pathway in which strand displacement hybridization occurs between two molecules of the double-stranded amplicon, as shown in FIGS. 9C (j ′) to (l ′).
- one amplicon is provided as shown in FIG. 9C (j) (step (C1)), or the arrangement directions are opposite to each other as shown in FIG. 9C (j ′). Two are provided in the state (step (B1)).
- the double-stranded amplicon is either in the molecule of the amplicon as shown in (j) to (k) of the figure, or as shown in (j ′) to (k ′) of the figure.
- the sequence (C) hybridizes to the sequence (C ′) of the other strand (Tail substitution).
- this Tail substitution occurs, folding of each terminal folding sequence (DD ′) or (EE ′) occurs, as shown in FIGS.
- the strand displacement extension reaction takes place from the 3 ′ end to the 5 ′ end of the complementary single-stranded nucleic acid using the complementary single-stranded nucleic acid as a template, and a part of the extended strand of the single-stranded nucleic acid This corresponds to the step ((C2) step) of forming a partial double strand ((m) in the figure) in which the complementary single-stranded nucleic acids are hybridized.
- the reaction from FIG. 9C (l ′) to FIG. 9 (m) shows the folded sequence of the single-stranded nucleic acid of one double strand of the two double strands (B1) (FIG. 9C (j ′)).
- a strand displacement extension reaction occurs using the complementary single-stranded nucleic acid of the other double strand as a template, and the single-stranded nucleic acid of one double strand is extended.
- step (B2) of forming a partial duplex (FIG. (M)) in which a complementary single-stranded nucleic acid of the other duplex is hybridized to a part of the strand.
- step (B2) of forming a partial duplex (FIG. (M)) in which a complementary single-stranded nucleic acid of the other duplex is hybridized to a part of the strand.
- these extension reactions are not switchback extension using the 3 ′ end folding sequence (DD ′) or (EE ′) as its template.
- a recombinant extension reaction occurs using the opposite strand as a template from the 3 ′ folding site of the released folding sequence (EE ′).
- an amplification product is formed in which the heterofolds at the junction of the amplicons form a hammerhead structure ((o) in the figure).
- the reaction from FIG. 9B (n) to (o) is carried out by using the complementary single-stranded nucleic acid (FIG. 9B) in the partial duplex (FIG. 9 (m)) of the step (B2) or (C2).
- a step of forming a complete duplex (FIG. (O)) from the 3 ′ end of (m) upper strand) by an extension reaction using the single-stranded nucleic acid as a template (FIG. (N)) (B3) Step or (C3) Step).
- the first and second synthesis reactions in the nucleic acid synthesis method of the present invention have been described above.
- the isothermal amplification method using the primer set of the present invention may or may not include these synthesis reactions of the present invention.
- the first primer may have an intervening sequence (G) between the sequence (A ′) and the sequence (C).
- the length of the intervening sequence (G) is, for example, 1 to 30 bases, preferably 1 to 20 bases, more preferably 1 to 10 bases.
- the first primer may have an intervening sequence (H) between the sequence (C) and the folded sequence (DD ′).
- the length of the intervening sequence (H) is, for example, 1 to 30 bases, preferably 1 to 20 bases, more preferably 1 to 10 bases.
- the primer set according to the present invention may include a third primer in addition to the first primer and the second primer.
- the third primer hybridizes to the target nucleic acid sequence or its complementary sequence, and does not compete with other primers for hybridization to the target nucleic acid sequence or its complementary sequence.
- not competing means that the primer does not interfere with the provision of a complementary strand synthesis starting point by hybridizing to the target nucleic acid.
- the third primer is a single primer when the amplification product by amplification of the target nucleic acid is partially in a single-stranded state. It can be annealed to a target sequence present in the chain portion. As a result, a new complementary strand synthesis starting point is provided in the target nucleic acid sequence in the amplification product, and an extension reaction takes place therefrom, so that the nucleic acid amplification reaction is performed more rapidly.
- the third primer is not necessarily limited to one type, and two or more types of third primers may be used simultaneously in order to improve the speed and specificity of the nucleic acid amplification reaction. These third primers typically have a different sequence from the first primer and the second primer, but may hybridize to partially overlapping regions as long as they do not compete with these primers.
- the chain length of the third primer is preferably 2 to 100 bases, more preferably 5 to 50 bases, and even more preferably 7 to 30 bases.
- the third primer is mainly intended to assist the nucleic acid amplification reaction by the first primer and the second primer more rapidly. Therefore, the third primer preferably has a Tm lower than the Tm of each 3 'end of the first primer and the second primer. Further, the addition amount of the third primer to the amplification reaction solution is preferably smaller than the addition amounts of the first primer and the second primer.
- the third primer examples include a primer having a structure capable of forming a loop as described in WO 02/24902 and giving a starting point for complementary strand synthesis to the loop portion.
- the present invention is not limited to this. That is, as long as it is within the target nucleic acid sequence, it may provide a complementary strand synthesis origin at any site.
- Primers included in the primer set according to the present invention are composed of deoxynucleotides and / or ribonucleotides.
- ribonucleotide (sometimes simply referred to as “N”) refers to ribonucleotide triphosphate such as ATP, UTP, CTP, GTP, and the like.
- ribonucleotides include these derivatives, such as ribonucleotides ( ⁇ -thio-ribonucleotides) in which the oxygen atom of the phosphate group at the ⁇ -position is replaced with a sulfur atom.
- the primer includes an oligonucleotide primer composed of unmodified deoxynucleotides and / or modified deoxynucleotides, and an oligonucleotide primer composed of unmodified ribonucleotides and / or modified ribonucleotides, unmodified deoxynucleotides and / or Alternatively, chimeric oligonucleotide primers containing modified deoxynucleotides and unmodified ribonucleotides and / or modified ribonucleotides are also included.
- the primers contained in the primer set according to the present invention can be synthesized by any method that can be used for the synthesis of oligonucleotides, such as the phosphate triester method, the H-phosphonate method, the thiophosphonate method, and the like.
- the primer can be easily obtained by, for example, synthesizing by a phosphoramidite method using a DNA synthesizer type 394 manufactured by ABI (Applied Biosystem Inc.).
- the template nucleic acid or nucleic acid sample containing the target nucleic acid sequence used in the nucleic acid amplification reaction may be either DNA or RNA, and may be double-stranded or single-stranded.
- DNA includes any of cDNA, genomic DNA, and synthetic DNA.
- RNA includes all RNA, mRNA, rRNA, siRNA, hnRNA, microRNA, and synthetic RNA.
- These nucleic acids are prepared from, for example, blood, tissues, cells, biological samples such as animals and plants, or microorganism-derived samples or virus-derived samples separated from biological samples, food, soil, waste water, etc. be able to.
- the isolation of the template nucleic acid or nucleic acid sample can be carried out by any method, and examples thereof include a method using dissolution treatment with a surfactant, sonication, shaking and stirring using glass beads, and a French press.
- a surfactant for example, phenol extraction, chromatography, ion exchange, gel electrophoresis, density-dependent centrifugation, and the like.
- the template nucleic acid or the nucleic acid sample is a double-stranded nucleic acid such as genomic DNA or PCR fragment isolated by the above method, or a cDNA prepared by reverse transcription from total RNA or mRNA. Any single stranded nucleic acid can be used. In the case of the above double-stranded nucleic acid, it can be used more optimally by carrying out a denaturing step to make it a single strand.
- the enzyme used in the reverse transcription reaction is not particularly limited as long as it has cDNA synthesis activity using RNA as a template.
- avian myeloblastosis virus-derived reverse transcriptase AMV RTase
- rous-related virus 2 Reverse transcriptases RAV-2 RTase
- Moloney murine leukemia virus-derived reverse transcriptase MMLV RTase
- other reverse transcriptases avian myeloblastosis virus-derived reverse transcriptase
- a DNA polymerase having reverse transcription activity can also be used.
- an enzyme having reverse transcription activity at a high temperature is optimal.
- a DNA polymerase derived from Thermus bacterium (Tth DNA polymerase, etc.), a DNA polymerase derived from Bacillus bacterium, or the like can be used.
- a particularly preferable enzyme for example, as a DNA polymerase derived from a thermophilic Bacillus bacterium, B. st-derived DNA polymerase (Bst DNA polymerase); Examples include ca-derived DNA polymerase (Bca DNA polymerase), such as BcaBEST DNA polymerase and Bca (exo-) DNA polymerase.
- Bca DNA polymerase does not require manganese ions for the reaction, and can synthesize cDNA while suppressing secondary structure formation of template RNA under high temperature conditions.
- the primer nucleic acid can be denatured into a single strand to obtain a primer for the template nucleic acid. Annealing can also be performed efficiently. Increasing the temperature to about 95 ° C is a preferred nucleic acid denaturation method. As another method, it is possible to denature by raising the pH, but in this case, it is necessary to lower the pH in order to hybridize the primer to the target nucleic acid.
- the polymerase used in the nucleic acid amplification reaction only needs to have a strand displacement activity (strand displacement ability), and any of normal temperature, intermediate temperature, and heat resistance can be suitably used. Further, this polymerase may be either a natural body or a mutant with artificial mutation. Examples of such a polymerase include DNA polymerase. Furthermore, it is preferable that the DNA polymerase has substantially no 5 ' ⁇ 3' exonuclease activity. Examples of such DNA polymerase include Bacillus stearothermophilus (hereinafter referred to as “B.st”), Bacillus caldotenax (hereinafter referred to as “B.ca”), and the like.
- B.st Bacillus stearothermophilus
- B.ca Bacillus caldotenax
- thermophilic Bacillus genus DNA polymerase Klenow fragment of E. coli DNA polymerase I, and the like.
- DNA polymerases used in the nucleic acid amplification reaction Vent DNA polymerase, Vent (Exo-) DNA polymerase, DeepVent DNA polymerase, DeepVent (Exo-) DNA polymerase, ⁇ 29 phage DNA polymerase, MS-2 phage DNA polymerase, Z -Taq DNA polymerase, Pfu DNA polymerase, Pfu turbo DNA polymerase, KOD DNA polymerase, 9 ° Nm DNA polymerase, Thermonarter DNA polymerase, etc.
- a DNA polymerase having reverse transcription activity for example, BcaBEST DNA polymerase, Bca (exo-) DNA polymerase, etc.
- reverse transcription reaction from total RNA or mRNA and cDNA as a template.
- the DNA polymerase reaction can be carried out with one kind of polymerase.
- reagents used in the nucleic acid amplification reaction include, for example, catalysts such as magnesium chloride, magnesium acetate, magnesium sulfate, substrates such as dNTP mix, Tris-HCl buffer, tricine buffer, sodium phosphate buffer, potassium phosphate buffer, etc. Can be used. Furthermore, additives such as dimethyl sulfoxide and betaine (N, N, N-trimethylglycine), acidic substances described in International Publication No. 99/54455 pamphlet, cation complexes, and the like may be used.
- catalysts such as magnesium chloride, magnesium acetate, magnesium sulfate
- substrates such as dNTP mix, Tris-HCl buffer, tricine buffer, sodium phosphate buffer, potassium phosphate buffer, etc.
- additives such as dimethyl sulfoxide and betaine (N, N, N-trimethylglycine), acidic substances described in International Publication No. 99/54455 pamphlet, cation complexes,
- a melting temperature adjusting agent can be added to the reaction solution in order to increase the nucleic acid amplification efficiency.
- the melting temperature (Tm) of a nucleic acid is generally determined by the specific nucleotide sequence of the double stranded portion in the nucleic acid. By adding a melting temperature adjusting agent to the reaction solution, this melting temperature can be changed. Therefore, under a certain temperature, the intensity of double strand formation in the nucleic acid can be adjusted.
- a general melting temperature adjusting agent has an effect of lowering the melting temperature.
- a melting temperature adjusting agent By adding such a melting temperature adjusting agent, it is possible to lower the melting temperature of the double-stranded forming part between two nucleic acids, in other words, to reduce the strength of the double-stranded formation. It becomes. Therefore, when such a melting temperature adjusting agent is added to the reaction solution in the nucleic acid amplification reaction, it is efficiently used in a GC-rich nucleic acid region that forms a strong double strand or a region that forms a complex secondary structure. The double-stranded portion can be made into a single strand, and this makes it easy for the next primer to hybridize to the target region after the extension reaction with the primer is completed, so that the nucleic acid amplification efficiency can be increased.
- the melting temperature adjusting agent used in the present invention and its concentration in the reaction solution are appropriately selected by those skilled in the art in consideration of other reaction conditions that affect the hybridization conditions, such as salt concentration, reaction temperature, etc.
- the melting temperature adjusting agent is not particularly limited, but is preferably dimethyl sulfoxide (DMSO), betaine, formamide or glycerol, or any combination thereof, more preferably dimethyl sulfoxide (DMSO). .
- an enzyme stabilizer can be added to the reaction solution in the nucleic acid amplification reaction. Thereby, since the enzyme in the reaction solution is stabilized, the amplification efficiency of the nucleic acid can be increased.
- the enzyme stabilizer used in the present invention may be any known in the art, such as glycerol, bovine serum albumin, saccharides, etc., and is not particularly limited.
- a reagent for enhancing the heat resistance of an enzyme such as DNA polymerase or reverse transcriptase can be added to the reaction solution as an enzyme stabilizer.
- an enzyme stabilizer As a result, the enzyme in the reaction solution is stabilized, so that the nucleic acid synthesis efficiency and amplification efficiency can be increased.
- a reagent may be any known in the art and is not particularly limited, but is preferably a saccharide, more preferably a mono- or oligosaccharide, more preferably trehalose, sorbitol or mannitol, or these It is set as the mixture of 2 or more types.
- the nucleic acid amplification reaction using the primer set according to the present invention can be performed isothermally.
- “isothermal” refers to maintaining an approximately constant temperature condition such that the enzyme and the primer can substantially function.
- the “substantially constant temperature condition” means that not only the set temperature is accurately maintained but also a temperature change that does not impair the substantial functions of the enzyme and the primer is allowed. To do.
- the nucleic acid amplification reaction under a constant temperature condition can be carried out by keeping the temperature at which the activity of the enzyme used can be maintained.
- the reaction temperature is preferably set to a temperature near or below the melting temperature (Tm) of the primer.
- Tm melting temperature
- the level of stringency is preferably set in consideration of the melting temperature (Tm) of the primer. Therefore, this temperature is preferably about 20 ° C. to about 75 ° C., more preferably about 35 ° C. to about 65 ° C.
- the amplification reaction is repeated until either the enzyme is inactivated or one of the reagents including the primer is used up.
- nucleic acid containing a non-natural nucleotide can be used as a template nucleic acid (target nucleic acid sequence).
- non-natural nucleotide means a nucleotide that contains a base other than the base (adenine, guanine, cytosine, and thymine or uracil) contained in a natural nucleotide, and that can be incorporated into a nucleic acid sequence. Examples thereof include xanthosines, diaminopyrimidines, isoG, isoC (Proc. Natl. Acad. Sci. USA 92, 6329-6333, 1995).
- a nucleic acid amplification enzyme having no heat resistance is used for amplification of a target nucleic acid containing a non-natural nucleotide.
- the nucleic acid amplification reaction can be performed at, for example, an isothermal temperature of about 50 ° C., the possibility that the nucleic acid amplification enzyme (DNA polymerase or the like) is inactivated is low as compared with the conventional PCR method. Therefore, the nucleic acid amplification reaction using the primer set according to the present invention is also effective for amplification of a target nucleic acid containing a non-natural nucleotide in which a nucleic acid amplification enzyme having no heat resistance is used.
- the enzyme used to amplify a nucleic acid containing a non-natural nucleotide is not particularly limited as long as it can amplify such a target nucleic acid, but particularly from the viewpoint of uptake efficiency, Y188L / E478Q mutant HIV I reverse transcription Enzymes, AMV reverse transcriptase, Klenow fragment of DNA polymerase, 9 ° N DNA polymerase, HotTub DNA polymerase, etc. are suitable (Michael Sismour 1 et al., Biochemistry 42, No. 28, 8598, 2003 / US Pat. No. 6,617,106) Statement, Michael J. Lutz et al., Bioorganic & Medical Chemistry letters 8, 1149-1152, 1998, etc.). Furthermore, a substance that improves the heat resistance of the nucleic acid amplification enzyme, such as trehalose, can also be added to the reaction solution, whereby the target nucleic acid containing unnatural nucleotides can be more efficiently amplified.
- the presence of the amplification product obtained by the nucleic acid amplification method according to the present invention can be detected by many various methods.
- One method is the detection of amplification products of a specific size by general gel electrophoresis. In this method, for example, it can be detected by a fluorescent substance such as ethidium bromide or cyber green.
- a labeled probe having a label such as biotin and hybridizing it to the amplification product.
- Biotin can be detected by binding to fluorescently labeled avidin, avidin bound to an enzyme such as peroxidase, and the like.
- a method of visually observing the white turbidity of the reaction solution by utilizing the fact that pyrophosphoric acid is bonded to magnesium in the reaction solution to cause white precipitation of magnesium pyrophosphate.
- a metal indicator whose color tone changes depending on the magnesium ion concentration (for example, Eriochrome Black T, Hydroxy Naphthol Blue, etc.) is added to the reaction solution, and the color change of the reaction solution is visually observed. This makes it possible to detect the presence or absence of amplification.
- Calcein or the like it is possible to visually observe the increase in fluorescence accompanying the amplification reaction, so that the amplification product can be detected in real time.
- the presence of the amplification product obtained by the nucleic acid amplification method according to the present invention can also be detected by observing the aggregation of the solid phase carrier resulting from the generation of the amplification product.
- at least one primer contained in the primer set according to the present invention comprises a solid phase carrier or a site capable of binding to the solid phase carrier.
- the solid phase carrier or the site capable of binding to the solid phase carrier may be introduced into any part such as the 3 ′ end portion, 5 ′ end portion, central region of the primer, but preferably the 5 ′ end portion. It is assumed that it was introduced.
- the substrate used in the nucleic acid amplification reaction may comprise a solid phase carrier or a site capable of binding to the solid phase carrier.
- the solid phase carrier used in the present invention is a carrier that is insoluble in the reaction solution used for the nucleic acid amplification reaction, or a property from the liquid phase to the solid phase (gel phase) or from the solid phase (gel phase) to the liquid phase before and after amplification.
- Any phase transition carrier that changes can be used.
- Preferred solid phase carriers include water-insoluble organic polymer carriers, water-insoluble inorganic polymer carriers, synthetic polymer carriers, phase transition carriers, metal colloids, magnetic particles, and the like, and further solvent-insoluble organic polymer carriers. , Solvent-insoluble inorganic polymer carriers, solvent-soluble polymer carriers, gel polymer carriers and the like.
- examples of the water-insoluble organic polymer include silicon-containing materials such as porous silica, porous glass, diatomaceous earth, and celite, and cross-linked polysaccharides such as nitrocellulose, hydroxyapatite, agarose, dextran, cellulose, and carboxymethylcellulose. , Cross-linked protein such as methylated albumin, gelatin, collagen, casein, gel particles, dye sol and the like.
- examples of the water-insoluble inorganic polymer include aluminum oxide, titanium oxide, and ceramic particles.
- Examples of the synthetic polymer include polystyrene, poly (meth) acrylate, polyvinyl alcohol, polyacrylonitrile or a copolymer thereof, styrene-styrene sulfonic acid copolymer, vinyl acetate-acrylic acid ester copolymer, and the like.
- Examples of the metal colloid include gold colloid.
- Examples of magnetic particles include magnetic iron oxide beads, monodispersed magnetic iron oxide finely pulverized particles, superparamagnetic particles (Japanese Patent Publication No. 4-501959), and superparamagnetic oxidation covered with a polymerizable silane coating. Examples thereof include magnetically responsive particles having iron (Japanese Patent Publication No.
- the magnetized solid phase carrier can be easily separated from solid and liquid using magnetic force.
- the shape of the solid support include particles, membranes, fibers, and filters. Particles are particularly preferred as the shape of the solid support, and the surface thereof may be either porous or non-porous.
- Particularly preferred solid phase carriers include latex in which a synthetic polymer carrier is uniformly dispersed in water, metal colloid particles such as gold colloid, magnetic particles such as magnet beads, and the like.
- the immobilization of a primer or a substrate to a solid phase carrier can be performed by a method known to those skilled in the art, and may be a method using either physical bonding or chemical bonding. Immobilization of a primer or a substrate to a solid phase carrier is generally performed by, for example, combining a substance capable of labeling an oligonucleotide such as a primer or a probe with a solid phase carrier to which a substance capable of binding to the substance is bound. be able to.
- a combination of substances used for such purpose those well known in the art can be used, for example, a combination of biotin and avidin or streptavidin, a combination of an antigen and an antibody capable of binding thereto, a ligand, And a combination of two nucleic acids which hybridize with each other.
- a primer or a substrate labeled with biotin is bound to a solid phase carrier whose surface is coated with avidin or streptavidin, whereby the primer or the substrate can be immobilized on the solid phase carrier.
- the antigen include haptens such as FITC, DIG, and DNP.
- antibodies that can bind to these include antibodies such as anti-FITC antibody, anti-DIG antibody, and anti-DNP antibody.
- these antibodies may be either monoclonal antibodies or polyclonal antibodies.
- the combination of biotin and streptavidin has high specificity and good binding efficiency, so that these combinations are particularly preferable. Labeling substances such as biotin, hapten, and ligand may be used alone or in a plurality of combinations if necessary, using known means (Japanese Patent Laid-Open Nos. 59-93099, 59-148798, and No. 59-204200), the primer can be introduced at the 5 ′ end of the primer.
- the site (or group) capable of binding to the solid phase carrier used in the present invention can be selected according to the above-described method used for immobilization of the primer or the substrate to the solid phase carrier, and thus the solid phase carrier. Any of those that allow physical binding to or chemical binding to each other may be used, but specific binding is preferable.
- the site capable of binding to such a solid phase carrier include biotin, avidin, streptavidin, antigen, antibody, ligand, receptor, nucleic acid, protein and the like, and preferably biotin or streptavidin. More preferably, it is biotin.
- the solid phase carrier used in this case may include a binding partner of the site contained in the primer or the substrate, if necessary.
- a binding partner is present in a form capable of binding to the site contained in the primer or the substrate, preferably present on the surface of the solid support, more preferably the solid support. It was assumed that it was applied on the surface of
- a primer set according to the present invention is prepared for each of a plurality of target nucleic acids, the plurality of primer sets are immobilized on a solid phase carrier in a form that can be distinguished from each other, and these immobilizations are performed. Nucleic acid amplification reaction is performed using the optimized primer set. This makes it possible to simultaneously amplify a plurality of target nucleic acids and detect the amplification products for each of them in a distinguishable form. Detection of the amplification product can be performed using an intercalator or the like.
- solid phase carriers that can be used for such purposes include not only the above-mentioned planar solid phase carriers but also bead surfaces that can be distinguished from each other (US Pat. No. 6,046,807 and US Pat. No. 6,057,107). ), A quasi-flat plate carrier (Japanese Patent Laid-Open No. 2000-245460) produced by bundling a fibrous carrier in which each primer set is solid-phased and then cutting into thin pieces. Things can be used.
- the amplified fragment obtained by the nucleic acid amplification method according to the present invention is composed of ordinary bases, it can be subcloned into an appropriate vector using the restriction enzyme site inside the amplified fragment after amplification. Furthermore, the amplified fragment can be treated with a restriction enzyme like RFLP, and can be widely used in the field of genetic testing. Further, the amplified fragment can be generated as containing an RNA polymerase promoter sequence, which makes it possible to synthesize RNA directly from the amplified fragment. The RNA synthesized in this way can also be used as an RNA probe, siRNA or the like.
- a base labeled with biotin or a fluorescent substance can be used as a substrate in place of ordinary dNTP, whereby a DNA probe labeled with biotin or a fluorescent substance can be used. It is also possible to prepare. Furthermore, the presence or absence of an amplification product can be confirmed through some structure such as biotin or a labeling substance.
- the primer included in the primer set according to the present invention can include a restriction enzyme recognition site, thereby improving the efficiency of nucleic acid amplification. That is, since a nick is generated in the amplification product by the restriction enzyme corresponding to the restriction enzyme recognition site in the primer, it is possible to cause a strand displacement type complementary strand synthesis reaction using this nick as a starting point for synthesis.
- This method is basically based on the principle of the SDA method described as the prior art.
- the primer included in the primer set according to the present invention may include an RNA polymerase promoter sequence, thereby improving the efficiency of nucleic acid amplification.
- This method is basically based on the principle of the NASBA method described as the prior art.
- the primer set according to the present invention can include an “outer primer” used in the LAMP method or the SDA method, thereby improving the efficiency of nucleic acid amplification.
- an “outer primer” used in the LAMP method or the SDA method, thereby improving the efficiency of nucleic acid amplification.
- the outer primer a primer capable of providing a complementary strand synthesis origin at a portion located outside the target nucleic acid sequence on the template nucleic acid can be used.
- nucleic acid amplification method By the nucleic acid amplification method according to the present invention, a single-stranded nucleic acid to be immobilized on a DNA chip, a single-stranded DNA probe for determining a base sequence, a megaprimer for a long-chain PCR method, etc. can be easily and rapidly prepared. Can do. Further, according to the nucleic acid amplification method of the present invention, it is possible to selectively amplify only the sense sequence or the antisense sequence of the target nucleic acid depending on the purpose.
- the single-stranded nucleic acid prepared by the nucleic acid amplification method according to the present invention can be used as a DNA fragment for immobilization on a DNA chip. That is, the nucleic acid amplification method according to the present invention can be applied to a method for preparing a DNA chain for immobilization in DNA chip production. It is also possible to prepare a DNA chip by preliminarily fixing the 5 'end of the primer on the DNA chip and performing nucleic acid amplification on the chip. In addition, by adding a fluorescently labeled probe that hybridizes to the amplification product in advance to the reaction solution before performing the nucleic acid amplification, the amplification product can be detected in real time while performing nucleic acid amplification on the DNA chip. It becomes possible.
- the primer set can be designed so that the mutation site is included in the sequence (A) or the sequence (B), thereby confirming the presence or absence of the amplification product. It is possible to detect (determine) the presence or absence.
- the presence of the amplification product after the nucleic acid amplification reaction indicates the presence of the mutation. Absence or reduction of indicates an absence of mutation.
- the presence of the amplification product after the nucleic acid amplification reaction indicates the absence of the mutation, A decrease indicates the presence of the mutation.
- “decrease in amplification product” means that the amount of amplification product obtained is reduced compared to the amount of amplification product obtained when the target nucleic acid sequence is present in the nucleic acid sample. To do.
- mutation means that there is a base (base pair in the case of double-stranded nucleic acid) different from the control nucleic acid sequence in the nucleic acid sequence.
- mutation includes base deletion, insertion, addition and substitution.
- control nucleic acid refers to a standard base sequence, for example, a standard genotype, with respect to a specific base sequence, both a wild type and a normal type.
- test nucleic acid means a nucleic acid to be examined whether or not it has a base (mutation) different from the reference nucleic acid in the mutation detection method according to the present invention, in other words, in a nucleic acid sample.
- a nucleic acid sample Means a nucleic acid having the same sequence as that of the control nucleic acid except for the base related to the mutation.
- mutant base or “mutation nucleotide residue” means a base or nucleotide residue present at the mutation site in the nucleic acid, and therefore included in the mutation site of the control nucleic acid.
- the gene of the patient suspected of having the mutation is a test nucleic acid
- the gene of a healthy person corresponding to this gene is a control nucleic acid.
- test nucleic acid and control nucleic acid may be a natural product-derived nucleic acid or an artificially synthesized nucleic acid.
- nucleic acid as used in the present invention means a polynucleotide comprising any unmodified nucleotide and / or modified nucleotide.
- the test nucleic acid and the control nucleic acid are typically DNA such as cDNA, genomic DNA, and synthetic DNA, or RNA such as mRNA, total RNA, hnRNA, siRNA, and synthetic RNA.
- polynucleotide used in the present invention includes, for convenience, polynucleotides and oligonucleotides, and artificially synthesized nucleic acids such as peptide nucleic acids, morpholino nucleic acids, methylphosphonate nucleic acids, S-oligonucleic acids, and the like. Shall be.
- the test nucleic acid can be freely selected by the tester. Furthermore, when performing detection, these nucleic acids may be mixed.
- a nucleic acid sample containing a test nucleic acid can be obtained from a test subject, for example, a human or non-human animal.
- the nucleic acid can be extracted from a sample such as a desired tissue, organ and cell from the subject by a method known in the art, and if necessary, the size of the nucleic acid fragment can be extracted after the extraction.
- conditions such as purification purity can be adjusted to an appropriate state.
- the test nucleic acid in the nucleic acid sample may be further amplified by performing an amplification reaction by a general polymerase chain reaction (PCR) or the like.
- PCR general polymerase chain reaction
- the test nucleic acid and the control nucleic acid may be single-stranded or double-stranded.
- double-stranded nucleic acid used in the present invention means any of double-stranded DNA, double-stranded RNA, and DNA / RNA.
- the double-stranded nucleic acid may be used as it is as a nucleic acid sample, or may be amplified with a vector such as a phage or a plasmid.
- Example 1 when the folding sequence (DD ′) is added to one of the first primer and the second primer, and the folding sequence (DD ′) is added to the first primer, When the folded sequence (EE ′) was added to the second primer, the target nucleic acid sequence was isothermally amplified using two types of primer sets.
- a forward primer (Forward primer 1: SEQ ID NO: 1 or Forward primer 2: SEQ ID NO: 2) and a reverse primer ( A reaction solution was prepared by adding Reverse primer 1: SEQ ID NO: 3 or Reverse primer 2: SEQ ID NO: 4) at a concentration of 2 ⁇ M each.
- Reverse primer 1 SEQ ID NO: 3 or Reverse primer 2: SEQ ID NO: 4
- the underlined portion indicates the folded sequence
- the enclosed portion indicates the common sequence of the Forward and Reverse primers.
- plasmid DNA As a template DNA, 2 ⁇ 10 4 copies of plasmid DNA having the partial sequence of the cDNA of the RNA genome N2 segment of influenza A (H3N2) (SEQ ID NO: 5) was added to the reaction solution, and the real-time PCR apparatus MX3000p (manufactured by Agilent) was added. The reaction was carried out at a constant temperature of 60 ° C. for 60 minutes, and the nucleic acid amplification activity was examined by obtaining a fluorescence amplification curve through a FAM filter.
- FIG. 13 shows the fluorescence amplification curves ( ⁇ ) for the case where Forward Primer 1 (SEQ ID NO: 1) and Reverse Primer 1 (SEQ ID NO: 3) are used, with and without template DNA ( ⁇ ). The meanings of and ⁇ are the same in the following examples).
- the increase in the fluorescence signal was remarkably observed in the template DNA added.
- FIG. 14 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in the fluorescence signal was recognized in FIG. As shown in FIG. 14, a periodic band pattern extending from a short chain to a long chain was observed, which revealed that isothermal nucleic acid amplification occurred.
- FIG. 15 shows a fluorescence amplification curve when Forward primer 2 (SEQ ID NO: 2) and Reverse primer 2 (SEQ ID NO: 4) are used.
- FIG. 16 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in the fluorescence signal was recognized in FIG.
- a periodic band pattern extending from a short chain to a long chain was observed.
- the pattern of the band period is different, and it was revealed that isothermal nucleic acid amplification having an amplification pattern different from that in the above result 1 occurred.
- Example 2 In this example, an intervening sequence (H) was inserted between the folded sequence (DD ′) or (EE ′) and the common sequence (C) in both the first primer and the second primer. In some cases, the target nucleic acid sequence was amplified isothermally. Further, the intervening sequence (H) was made different between the first primer and the second primer.
- the reaction composition liquid other than the primer was the same as in Example 1 described above. That is, in a liquid having a volume of 25 ⁇ L, final concentrations of 1.4 mM dNTP, 5% DMSO, 20 mM Tris-HCl (pH 8.0), 30 mM potassium acetate, 10 mM sodium sulfate, 8 mM magnesium sulfate, 0.1% Tween 20, 1 / 100,000 diluted SYBR Green I (Takara Shuzo Co., Ltd.) and 12 unit Aac DNA polymerase solution, the primers are Forward primer (Forward primer 3: SEQ ID NO: 6) of the sequence shown below, and Reverse primer (Reverse primer 3: A reaction solution was prepared by adding SEQ ID NO: 7) at a concentration of 2 ⁇ M each.
- the underlined (single underlined) part is a folded sequence
- the enclosed part is a common sequence of Forward and Reverse primers
- the double underlined part is
- Example 2 Similarly to Example 1 described above, 2 ⁇ 10 4 copies of plasmid DNA having the partial sequence of the cDNA of the RNA genome N2 segment of influenza A (H3N2) (SEQ ID NO: 5) as template DNA were added to the reaction solution.
- the nucleic acid amplification activity was examined by performing a reaction at a constant temperature of 60 ° C. for 90 minutes with a real-time PCR apparatus MX3000p (manufactured by Agilent) and obtaining a fluorescence amplification curve through a FAM filter.
- FIG. 17 shows a fluorescence amplification curve when Forward primer 3 (SEQ ID NO: 6) and Reverse primer 3 (SEQ ID NO: 7) are used. As shown in FIG. 17, the increase in the fluorescence signal was remarkably observed in the case where the template DNA was added.
- FIG. 18 is a photograph showing an agarose gel electrophoresis result of the reaction solution in which an increase in fluorescence signal was observed in FIG. As shown in FIG. 18, two periodic band patterns ranging from a short chain to a long chain were observed, and it was revealed that isothermal nucleic acid amplification occurred in the reaction solution.
- Example 3 Effect of third primer
- the effect of the third primer designed complementarily between the annealing regions of the first primer and the second primer in the target nucleic acid to improve the isothermal amplification reaction in the present invention is shown.
- both of the first primer and the second primer used in this example are the intervening sequence (H) between the folded sequence (DD ′) or (EE ′) and the common sequence (C). What inserted was used.
- the reaction composition solution other than the primer was the same as in Example 1 described above. That is, in a liquid having a volume of 25 ⁇ L, final concentrations of 1.4 mM dNTP, 5% DMSO, 20 mM Tris-HCl (pH 8.0), 30 mM potassium acetate, 10 mM sodium sulfate, 8 mM magnesium sulfate, 0.1% Tween 20, 1 / 100,000 diluted SYBR Green I (Takara Shuzo) and 12 unit Aac DNA polymerase.
- Primers were added at a concentration of 2 ⁇ M each using a forward primer (Forward primer 4: SEQ ID NO: 8) having the sequence shown below as a first primer and a reverse primer (Reverse primer 4: SEQ ID NO: 9) as a second primer.
- the following Boost primer (Boost Primer 1 or 2: SEQ ID NO: 10 or 11) was added as a triple primer at a concentration of 0.66 ⁇ M as a reaction solution.
- the underlined (single underlined) part is the folded sequence
- the enclosed part is the common sequence of the Forward and Reverse primers
- the double underlined part is the intervening sequence.
- human genomic DNA Human Genomic DNA, Male; Cat # G1471 manufactured by Promega
- a partial sequence SEQ ID NO: 12
- Nucleic acid amplification activity is obtained by adding 10 ng of the human genomic DNA to the reaction solution, performing a reaction at a constant temperature of 60 ° C. for 100 minutes with a real-time PCR apparatus MX3000p (manufactured by Agilent), and obtaining a fluorescence amplification curve through a FAM filter.
- FIG. 19 shows a fluorescence amplification curve when Forward primer 4 (SEQ ID NO: 8) and Reverse primer 4 (SEQ ID NO: 9) were used as the first and second primers.
- 20 to 22 show the results of fluorescence amplification curves when a third primer is further added to the first and second primers. Only Boost primer 1 is used in the reaction shown in FIG. 20, and only Boost primer 2 is used in the reaction shown in FIG.
- FIG. 22 shows the result of adding both Boost primers 1 and 2 in the reaction of FIG.
- FIG. 19 shows the results of agarose gel electrophoresis of the reaction solution in which an increase in fluorescence signal was observed in FIGS.
- Example 4 the isothermal nucleic acid amplification method was carried out in a final concentration of 1.4 mM dNTP, 5% DMSO, 20 mM Tris-HCl (pH 8.0), 10 mM potassium chloride, 10 mM ammonium sulfate, 8 mM magnesium sulfate in 25 ⁇ L of liquid.
- plasmid DNA having the cDNA partial sequence (SEQ ID NO: 15) of the influenza B RNA genome MP segment was added as a template DNA so as to be 6 ⁇ 10 3 copies in the reaction solution. Also, for amplification from short template DNA, it corresponds to the annealing sequence of Forward primer 5 and Reverse primer 5 and the cDNA partial sequence of influenza B RNA genome MP segment sandwiched between them (SEQ ID NO: 15, underlined)
- the 52-mer oligo DNA was used as a template DNA, and the template DNA was added to the reaction solution so as to be 6 ⁇ 10 3 copies.
- the isothermal nucleic acid amplification reaction was performed by incubation at a constant temperature of 60 ° C. for 90 minutes with a real-time PCR apparatus MX3000p (manufactured by Agilent), and the presence or absence of nucleic acid amplification was examined by obtaining a fluorescence amplification curve through a FAM filter. .
- FIG. 24 shows a fluorescence amplification curve in the case of using Forward primer 5 (SEQ ID NO: 13) and Reverse primer 5 (SEQ ID NO: 14).
- FIG. 25 shows the result of subjecting the reaction solution in the amplification reaction in FIG. 24 to agarose gel electrophoresis.
- a periodic band pattern extending from the short chain to the long chain was observed in the reaction sample containing the template DNA, which revealed that isothermal nucleic acid amplification occurred.
- FIG. 26 shows the result of determining the base sequence of each DNA for the bands indicated by arrows 1 and 2 in FIG. 25A.
- a base sequence of 105 bp was obtained from the DNA corresponding to arrow 1 and a base sequence of 202 bp was obtained from the DNA corresponding to arrow 2, which almost coincided with the DNA fragment length observed by agarose electrophoresis.
- the DNA corresponding to the arrow 1 in FIG. 25A had an amplicon sequence sandwiched between the forward and reverse primers from the template DNA (FIG. 26A).
- the DNA corresponding to the arrow 2 has a sequence in which two amplicon sequences are ligated in the forward direction (tandem), and the amplicon ligation site contains only one primer folding sequence. (FIG. 26B).
- Example 5 the specificity of the primer to the target sequence in the isothermal nucleic acid amplification method of the present invention was examined.
- final concentrations of 1.4 mM dNTP, 5% DMSO, 20 mM Tris-HCl (pH 8.0), 10 mM potassium chloride, 10 mM ammonium sulfate, 8 mM magnesium sulfate, 0.1% Tween 20, 1 / Forward primer 6 (SEQ ID NO: 16) and Reverse primer 6 (SEQ ID NO: 17) shown below were added to a solution containing 100,000 diluted SYBR Green I (Takara Shuzo) and 6 unit Aac DNA polymerase (Danaform), respectively. Added at a concentration of 2.5 ⁇ M.
- the underlined portion indicates the folded sequence, and the enclosed portion indicates the common sequence of the Forward and Reverse primers.
- plasmid DNA having the cDNA partial sequence (SEQ ID NO: 15) of the influenza B RNA genome MP segment was added as a template DNA so as to be 6 ⁇ 10 3 copies in the reaction solution. Further, with the template DNA, was prepared per reaction 20ng of human genomic DNA (Human Genomic DNA, Male, from Promega, 10 3 copies equivalent) and the reaction solution was added.
- FIG. 27 shows fluorescence amplification when Forward Primer 6 (SEQ ID NO: 16) and Reverse Primer 6 (SEQ ID NO: 17) are used as primers, FluB cDNA plasmid containing the target sequence is added as template DNA, and human genomic DNA is added. A curve is shown. As a result, in both the case where the template DNA was added (white circle) and the case where the human genomic DNA was further added (gray square), the fluorescence signal was compared with the case where the template DNA was not added as a control experiment (gray diamond). The increase was noticeable.
- FIG. 28A shows the result of subjecting the reaction solution in the amplification reaction in FIG. 27 to agarose gel electrophoresis. As can be seen from the figure, a periodic band pattern from short to long is observed in the reaction sample containing the template DNA of the FluB cDNA plasmid, and the reaction solution contains no template DNA or only human genomic DNA. In, no significant amplification pattern was observed. Therefore, it was revealed that the primers used in this example performed isothermal nucleic acid amplification and specifically recognized the FluB cDNA plasmid as a template DNA.
- FIG. 29 shows the result of determining the base sequence of each DNA with respect to the band indicated by the arrow in FIG. 28B.
- a base sequence of 105 bp was obtained from the DNA corresponding to the arrow, which almost coincided with the DNA fragment length observed by agarose electrophoresis.
- the DNA indicated by the arrow in FIG. 28B has an amplicon sequence sandwiched between the template DNA and the Forward and Reverse primers, and the sequence corresponding to the 3 ′ end of both primers. In the sandwiched section, it was confirmed that the FluB continuous from the annealing region contains a specific sequence. Therefore, the specificity of the target sequence recognition of the primer used in this example was proved also from the primary sequence structure analysis of the amplification product.
- Example 6 it was examined whether the isothermal nucleic acid amplification method according to the present invention can be used for direct isothermal nucleic acid amplification from a reverse transcription reaction solution using RNA as a template. From the plasmid DNA containing the partial sequence of the FluB cDNA used in Example 4, FluB cDNA linear primers with the T7 promoter sequence added using the following FluB cDNA amplification primers (SEQ ID NOs: 18 and 19) DNA was prepared by PCR.
- RNA obtained by acidic phenol extraction from the reaction solution was used as a template RNA for the reverse transcription reaction in this example.
- Reverse transcription reaction solution 25 ⁇ L (50 mM Tris-HCl [pH 8.3], 75 mM potassium chloride, 3 mM magnesium chloride, 10 mM DTT, 200 unit M-MLV Reverse transcriptase [deletion mutant, RNaseH] (-, Manufactured by Promega Corporation), a primer for reverse transcription [SEQ ID NO: 20]), and reacted at 42 ° C. for 1 hour. Then, in order to inactivate M-MLV Reverse transcriptase, it hold
- Reverse transcription primer (SEQ ID NO: 20) 18-mer 5'-TGGACGTCTTCTCCTTTT-3 '
- FIG. 30 shows a reaction in which a 1/4000 dilution of the reverse transcription reaction solution prepared in this example was added in an isothermal amplification reaction using Forward primer 5 (SEQ ID NO: 13) and Reverse primer 6 (SEQ ID NO: 14). It is the result compared with the case (Gray square) which added FluB cDNA plasmid as template DNA like a sample (white circle) and Example 4. As a result, although there was a delay in the rise time of the fluorescence signal compared to the plasmid template, the reaction sample with the reverse transcription reaction solution added no template DNA as a control experiment (black triangle).
- FIG. 31 shows the result of subjecting the reaction solution in the amplification reaction in FIG. 30 to agarose gel electrophoresis. As a result, even in the reaction sample to which the reverse transcription reaction solution was added, a periodic band pattern extending from the short chain to the long chain was observed in the same pattern as when using template DNA, and the same isothermal nucleic acid amplification occurred. It became clear that
- Example 7 the behavior of the primer structure used in the isothermal nucleic acid amplification method of the present invention was examined when only one primer was removed from the folded sequence located at the 5 ′ end.
- a solution containing diluted SYBR Green I (Takara Shuzo) and 6 unit Aac DNA polymerase (Danaform) Forward primer 8 (SEQ ID NO: 21) shown below and Reverse primer 6 (sequence) used in Example 5 were used.
- Forward primer 8 is a sequence obtained by removing the folded sequence (16 base) at the 5 ′ end of Forward primer 6 (SEQ ID NO: 16). Similar sequence.
- plasmid DNA having the cDNA partial sequence (SEQ ID NO: 15) of the influenza B RNA genome MP segment was added as a template DNA so as to be 6 ⁇ 10 3 copies in the reaction solution. Further, with the template DNA, was prepared per reaction 20ng of human genomic DNA (Human Genomic DNA, Male, from Promega, 10 3 copies equivalent) and the reaction solution was added.
- FIG. 32 shows FluB cDNA containing the target sequence using Forward primer 8 (SEQ ID NO: 21) containing no folding sequence at the 5 ′ end and Reverse primer 6 (SEQ ID NO: 17) containing the folding sequence at the 5 ′ end as primers.
- a fluorescence amplification curve was shown when the plasmid was added as template DNA and human genomic DNA was added.
- the fluorescence signal was compared with the case where the template DNA was not added as a control experiment (gray diamond). The increase was noticeable.
- FIG. 33 shows the result of subjecting the reaction solution in the amplification reaction in FIG. 32 to agarose gel electrophoresis. As a result, a band pattern extending from short chain to long chain was observed in the reaction sample containing the template DNA of FluB cDNA plasmid, and a remarkable amplification pattern was observed in the reaction solution containing no template DNA or only human genomic DNA. There wasn't. Therefore, it was revealed that isothermal nucleic acid amplification with respect to the target template DNA was achieved even when one of the primers did not contain the 5 ′ end.
- Example 8 the isothermal nucleic acid amplification method was carried out in a final concentration of 1.4 mM dNTP, 5% DMSO, 20 mM Tris-HCl (pH 8.0), 10 mM potassium chloride, 10 mM ammonium sulfate, 8 mM magnesium sulfate in 25 ⁇ L of liquid.
- Forward primer 5 (SEQ ID NO: 13) used in Example 4 was added to a solution containing 0.1% Tween 20, 1/10000 diluted SYBR Green I (Takara Shuzo), and 6 unit Aac DNA polymerase (Danaform).
- a reaction solution was prepared by adding Reverse Primer 8 (SEQ ID NO: 22) at a concentration of 2.5 ⁇ M.
- the box indicates the common sequence of the Forward and Reverse primers, and the Reverse primer 8 is the same as the sequence obtained by removing the 5 'terminal folding sequence (13base) of the Reverse primer 5 (SEQ ID NO: 14). Is an array of
- plasmid DNA having the cDNA partial sequence (SEQ ID NO: 15) of the influenza B RNA genome MP segment was added as a template DNA so as to be 6 ⁇ 10 3 copies in the reaction solution.
- the isothermal nucleic acid amplification reaction using the above reaction solution, electrophoresis of amplification reaction products, and sequence analysis of amplification products were performed in the same manner as in Example 4.
- FIG. 34 shows FluB cDNA containing the target sequence using Forward primer 5 (SEQ ID NO: 13) containing a folded sequence at the 5 ′ end and Reverse primer 8 (SEQ ID NO: 22) containing no folded sequence at the 5 ′ end as primers.
- the fluorescence amplification curve when a plasmid was added as a template DNA is shown.
- a plasmid containing FluB cDNA was used as the template DNA (white circle), an increase in the fluorescence signal was significantly observed as compared with the case where no template DNA was added as a control experiment (black triangle).
- FIG. 35 shows the result of subjecting the reaction solution in the amplification reaction in FIG. 34 to agarose gel electrophoresis. As a result, a periodic band pattern extending from the short chain to the long chain was observed in the reaction sample containing the template DNA, and it was revealed that isothermal nucleic acid amplification occurred.
- FIG. 36 shows the result of determining the base sequence of each DNA with respect to the bands indicated by arrows 1 and 2 in FIG.
- a base sequence of 92 bp was obtained from the DNA corresponding to arrow 1 and a base sequence of 186 bp was obtained from the DNA corresponding to arrow 2, which almost coincided with the DNA fragment length observed by agarose electrophoresis.
- the DNA corresponding to arrow 1 in FIG. 35 has a sequence in which the amplicon sequence sandwiched between the forward and reverse primers was copied from the template DNA. (FIG. 36A).
- the DNA corresponding to the arrow 2 has a sequence in which two amplicon sequences are linked in the forward direction (tandem) via two bases of GT (guanine, thymine) (FIG. 36B). .
Landscapes
- Chemical & Material Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Organic Chemistry (AREA)
- Health & Medical Sciences (AREA)
- Engineering & Computer Science (AREA)
- Molecular Biology (AREA)
- Biochemistry (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Zoology (AREA)
- Wood Science & Technology (AREA)
- General Health & Medical Sciences (AREA)
- Biotechnology (AREA)
- Genetics & Genomics (AREA)
- Biophysics (AREA)
- Microbiology (AREA)
- Immunology (AREA)
- Analytical Chemistry (AREA)
- Bioinformatics & Cheminformatics (AREA)
- General Engineering & Computer Science (AREA)
- Physics & Mathematics (AREA)
- Chemical Kinetics & Catalysis (AREA)
- Measuring Or Testing Involving Enzymes Or Micro-Organisms (AREA)
Abstract
Description
(A1) 3´末端を含む3´側ステム配列と、5´末端を含む5´側ステム配列とが、ループ配列を介して連結したステムループ構造の一本鎖鋳型核酸であって、前記3´側ステムの3´末端に、相互にハイブリダイズする2つの配列を同一鎖上に含む折り返し配列が連結した一本鎖鋳型核酸を提供する工程。
(A2) 前記一本鎖鋳型核酸の前記ループにプライマーをハイブリダイズさせ、前記プライマーを前記5´側ステム配列の5´末端に向かって伸長させる工程。
(A3) 前記5´側ステム配列の5´末端まで進んだ前記プライマーの伸長反応を、前記5´側ステム配列の5´末端から前記折り返し配列の3´末端にかけて連続して続行する工程。
(A4) 前記(A3)の前記折り返し配列の3´末端に進んだ前記プライマーの伸長反応を、再び、前記5´側ステム配列の5´末端に向かって連続して続行させ、かつ、続行する前記プライマーの伸長反応により、前記(A2)で形成された一本鎖鋳型核酸にハイブリダイズしているプライマー伸長鎖を鎖置換反応により一本鎖にする工程。
(A5) 前記(A4)のプライマー伸長反応を、前記5´側ステム配列の5´末端で停止する工程。
(A6) 前記(A4)で一本鎖になったプライマー伸長鎖を鋳型として前記一本鎖鋳型核酸の前記折り返し配列の3´末端を伸長させる工程。
(A3-2) 前記5´側ステム配列の5´末端まで進んだ前記プライマーの伸長反応を、前記5´側ステム配列の5´末端に連結した前記折り返し配列を介することなく、直接、前記5´側ステム配列の5´末端から前記折り返し配列の3´末端にかけて連続して続行する工程。
(B1) 3´末端を含む領域に、相互にハイブリダイズする2つの配列を同一鎖上に含む折り返し配列を含む一本鎖核酸と前記一本鎖核酸と相補的な一本鎖核酸とからなる二重鎖を、配列の向きが互いに逆方向になる状態で、二つ提供する工程。
(B2) 前記(B1)の二つの二重鎖の一方の二重鎖の前記一本鎖核酸の折り返し配列の3´末端から、他方の二重鎖の前記相補的な一本鎖核酸を鋳型として鎖置換伸長反応が起こり、前記一方の二重鎖の前記一本鎖核酸の伸長鎖の一部に前記他方の二重鎖の相補的な一本鎖核酸がハイブリダイズした部分的二重鎖を形成する工程。
(B3) 前記工程(B2)の部分的二重鎖において、前記相補的な一本鎖核酸の3´末端から、前記一本鎖核酸を鋳型とした伸長反応により、完全二重鎖を形成する工程。
(C1) 3´末端を含む領域に、相互にハイブリダイズする2つの配列を同一鎖上に含む折り返し配列を含む一本鎖核酸と前記一本鎖核酸と相補的な一本鎖核酸とからなる二重鎖を一つ提供する工程。
(C2) 前記(C1)の二重鎖の前記一本鎖核酸の折り返し配列の3´末端から、前記相補的な一本鎖核酸を鋳型とし、前記相補的な一本鎖核酸の3´末端から5´末端にかけて鎖置換伸長反応が起こり、前記一本鎖核酸の伸長鎖の一部に前記相補的な一本鎖核酸がハイブリダイズした部分的二重鎖を形成する工程。
(C3) 前記工程(C2)の部分的二重鎖において、前記相補的な一本鎖核酸の3´末端から、前記一本鎖核酸を鋳型とした伸長反応により、完全二重鎖を形成する工程。
本実施例では、第一プライマー及び第二プライマーのいずれか一方に折返し配列(D-D´)が付加されている場合、及び、第一プライマーに折返し配列(D-D´)が付加され、第二プライマーに折返し配列(E-E´)が付加されている場合の、二種類のプライマーセットを用いて標的核酸配列を等温で増幅した。
401 bp
5’-CCAGGAGTCAGAATGCGTTTGTATCAATGGAACTTGTACAGTAGTAATGACTGATGGGAGTGCTTCAGGAAAAGCTGATACTAAAATACTATTCATTGAGGAGGGGAAAATCGTTCATACTAGCACATTGTCAGGAAGTGCTCAGCATGTCGAGGAGTGCTCCTGCTATCCTCGATATCCTGGTGTCAGATGTGTCTGCAGAGACAACTGGAAAGGCTCCAATAGGCCCATCGTAGATATAAACATAAAGGATCATAGCATTGTTTCCAGTTATGTGTGTTCAGGACTTGTTGGAGACACACCCAGAAAAAACGACAGCTCCAGCAGTAGCCATTGTTTGGATCCTAACAATGAAGAAGGTGGTCATGGAGTGAAAGGCTGGGCCTTTGATGATGGAAATG-3’
図13は、Forwardプライマー1(配列番号1)およびReverseプライマー1(配列番号3)を用いた場合の、鋳型DNAを加えたもの(○)と加えないもの(●)それぞれの蛍光増幅曲線(○と●の意味は以下の実施例において同じ)を示す。図示のように、鋳型DNAを加えたものにおいて、蛍光シグナルの増加が顕著に認められた。また、図14は、図13で蛍光シグナルの増加が認められた反応溶液のアガロースゲル電気泳動結果を示す写真である。図14に示すように、短鎖から長鎖に渡る周期的なバンドパターンが観察され、等温核酸増幅が起きていることが明らかとなった。
図15は、Forwardプライマー2(配列番号2)およびReverseプライマー2(配列番号4)を用いた場合の蛍光増幅曲線を示す。図15に示すように、前記結果1と同様に、鋳型DNAを加えたものにおいて、蛍光シグナルの増加が顕著に認められた。また、図16は、図15で蛍光シグナルの増加が認められた反応溶液のアガロースゲル電気泳動結果を示す写真である。図16に示すように、短鎖から長鎖に渡る周期的なバンドパターンが観察された。ただし、図14におけるバンドパターンと比較すると、バンド周期のパターンが異なっており、前記結果1の場合と異なる増幅パターンを有する等温核酸増幅が起きていることが明らかとなった。
本実施例では、第一プライマー及び第二プライマーの双方において、折返し配列(D-D´)又は(E-E´)と共通配列(C)との間に、介在配列(H)を挿入した場合で、標的核酸配列を等温で増幅反応した。また、第一プライマー及び第二プライマーにおいて、介在配列(H)は異なるようにした。
図17は、Forwardプライマー3(配列番号6)、およびReverseプライマー3(配列番号7)を用いた場合の蛍光増幅曲線を示す。図17に示すように、鋳型DNAを加えたものにおいて、蛍光シグナルの増加が顕著に認められた。また図18は、図17で蛍光シグナルの増加が認められた反応溶液のアガロースゲル電気泳動結果を示す写真である。図18に示すように、短鎖から長鎖に渡る周期的な2本ずつのバンドパターンが観察され、等温核酸増幅が反応溶液中で起きていることが明らかとなった。
本実施例では、標的核酸において第一プライマー及び第二プライマーのアニーリング領域間に相補的に設計した第三プライマーが、本発明における等温増幅反応を向上させる効果について示す。
401 bp
5’-TTTCATGATTGAATTTTGTAAGGTATTTTGAAATAATTTTTCATATAAAGGTGAGTTTGTATTAAAAGGTACTGGTGGAGTATTTGATAGTGTATTAACCTTATGTGTGACATGTTCTAATATAGTCACATTTTCATTATTTTTATTATAAGGCCTGCTGAAAATGACTGAATATAAACTTGTGGTAGTTGGAGCTGGTGGCGTAGGCAAGAGTGCCTTGACGATACAGCTAATTCAGAATCATTTTGTGGACGAATATGATCCAACAATAGAGGTAAATCTTGTTTTAATATGCATATTACTGGTGCAGGACCATTCTTTGATACAGATAAAGGTTTCTCTGACCATTTTCATGAGTACTTATTACAAGATAATTATGCTGAAAGTTAAGTTATCTGAAA-3’
図19は、第一および第二プライマーとしてForwardプライマー4(配列番号8)およびReverseプライマー4(配列番号9)を用いた場合の蛍光増幅曲線を示した。図20~22は、第一および第二プライマーに更に第三プライマーを加えた場合の蛍光増幅曲線の結果であり、図20の反応ではBoostプライマー1のみを、図21の反応ではBoostプライマー2のみを、図22の反応では、Boostプライマー1および2の両方を添加した結果である。
本実施例における等温核酸増幅法は、25μLの液体中に、最終濃度として1.4mM dNTP、5% DMSO、20 mM Tris-HCl(pH 8.0)、10mM塩化カリウム、10mM硫酸アンモニウム、8mM硫酸マグネシウム、0.1% Tween20、1/100000希釈SYBR Green I(宝酒造社製)、および6unit Aac DNAポリメラーゼ(ダナフォーム社製)を含む溶液に、以下に示す配列のForwardプライマー5(配列番号13)、およびReverseプライマー5(配列番号14)を各2.5μMの濃度で加えたものを反応溶液とした。尚、下記各プライマーにおいて、下線部は折り返し配列、囲部はForward及びReverseプライマーの共通配列を示した。
740 bp
5’-AGCAGAAGCACGCACTTTCTTAAAATGTCGCTGTTTGGAGACACAATTGCCTACCTGCTTTCATTGACAGAAGATGGAGAAGGCAAAGCAGAACTAGCAGAAAAATTACACTGTTGGTTTGGTGGGAAAGAATTTGACCTAGACTCTGCCTTGGAATGGATAAAAAACAAAAGATGCTTAACTGATATACAAAAAGCACTAATTGGTGCCTCTATATGCTTTTTAAAACCCAAAGACCAGGAAAGAAAAAGAAGATTCATCACAGAGCCCTTATCAGGAATGGGAACAACAGCAACAAAAAAGAAAGGCCTGATTCTGGCTGAGAGAAAAATGAGAAGATGTGTGAGCTTTCATGAAGCATTTGAAATAGCAGAAGGCCATGAAAGCTCAGCGCTACTATACTGTCTCATGGTCATGTACCTGAATCCTGGAAATTATTCAATGCAAGTAAAACTAGGAACGCTCTGTGCTTTATGCGAGAAACAAGCATCACATTCACACAGGGCTCATAGCAGAGCAGCGAGATCTTCAGTGCCTGGAGTGAGACGAGAAATGCAGATGGTCTCAGCTATGAACACAGCAAAAACAATGAATGGAATGGGAAAAGGAGAAGACGTCCAAAAGCTGGCAGAAGAGCTGCAAAGCAACATTGGAGTGCTGAGATCTCTTGGGGCAAGTCAAAAGAATGGGGAAGGGATTGCAAAGGATGTAATGGAAGTGCTAAAGCAGAGCTCCATGGG-3’
図24では、Forwardプライマー5(配列番号13)およびReverseプライマー5(配列番号14)を用いた場合の蛍光増幅曲線を示した。この結果、鋳型DNAとしてFluB cDNAを含むプラスミドを用いた場合(白丸)、および両プライマーのアニーリング配列とそれらに挟まれたFluBのターゲット領域のみで構成される52-merの一本鎖オリゴDNAを用いた場合(灰色四角)のどちらにおいても、対照実験として鋳型DNAを加えなかった場合(黒三角)と比べ、蛍光シグナルの増加が顕著に認められた。また図25は、図24での増幅反応における反応溶液をアガロースゲル電気泳動に供した結果である。同図からわかるように、短鎖から長鎖に渡る周期的なバンドパターンが鋳型DNAを含む反応標品で観察され、等温核酸増幅が起きていることが明らかとなった。また、鋳型DNAが長鎖二本鎖となるプラスミドを供した場合と短鎖一本鎖オリゴDNAを供した場合ではバンドパターンの違いは認められず、同一の増幅反応が起きている事が明らかとなった。
本実施例では、本発明の等温核酸増幅法におけるプライマーの標的配列への特異性について検討した。25μLの液体中に、最終濃度として1.4mM dNTP、5% DMSO、20mM Tris-HCl(pH 8.0)、10mM塩化カリウム、10 mM硫酸アンモニウム、8 mM硫酸マグネシウム、0.1% Tween20、1/100000希釈SYBR Green I(宝酒造社製)、および6unit Aac DNAポリメラーゼ(ダナフォーム社製)を含む溶液に、以下に示すForwardプライマー6(配列番号16)、およびReverseプライマー6(配列番号17)を各2.5μMの濃度で加えた。尚、下記各プライマーにおいて、下線部は折り返し配列、囲部はForward及びReverseプライマーの共通配列を示した。
図27は、Forwardプライマー6(配列番号16)およびReverseプライマー6(配列番号17)をプライマーとして用い、その標的配列を含むFluB cDNAプラスミドを鋳型DNAとして加えると共に、ヒトゲノムDNAを添加した場合の蛍光増幅曲線を示した。この結果、鋳型DNAを加えたもの(白丸)、およびヒトゲノムDNAを更に添加した場合(灰色四角)のどちらにおいても、対照実験として鋳型DNAを加えなかった場合(灰色菱形)と比べ、蛍光シグナルの増加が顕著に認められた。また、ヒトゲノムDNAのみを鋳型として加えた場合(黒三角)では蛍光シグナルの増加は認められなかった。また図28Aは、図27での増幅反応における反応溶液をアガロースゲル電気泳動に供した結果である。同図からわかるように、短鎖から長鎖に渡る周期的なバンドパターンが、FluB cDNAプラスミドの鋳型DNAを含む反応標品で観察され、鋳型DNAを含まないもしくはヒトゲノムDNAのみを含んだ反応溶液では、顕著な増幅パターンは認められなかった。よって、本実施例において使用したプライマーは等温核酸増幅を果たし、FluB cDNAプラスミドを鋳型DNAとして特異的に認識していることが明らかとなった。
本実施例では、本発明における等温核酸増幅法により、RNAを鋳型とした逆転写反応溶液からの直接的な等温核酸増幅が実施可能かについて検討した。実施例4で使用したFluB cDNAの部分配列を含むプラスミドDNAから、以下に示すFluB cDNA増幅用プライマー(配列番号18および19)を用い、T7プロモータ配列を付加したFluB cDNAの直鎖状二本鎖DNAをPCR法により作成した。このPCR産物とCUGA T7 RNAポリメラーゼ(ニッポンジーンテク社製)を用いてin vitro転写反応を行ない、反応溶液から酸性フェノール抽出法によって得られたRNAを本実施例における逆転写反応の鋳型RNAとした。
39-mer
5’-TAATACGACTCACTATAGGGAGCAGAAGCACGCACTTTC-3’
T7/FluB cDNA増幅用Reverseプライマー7(配列番号19)
20-mer
5’-CCCATGGAGCTCTGCTTTAG-3’
18-mer
5’-TGGACGTCTTCTCCTTTT-3’
図30は、Forwardプライマー5(配列番号13)およびReverseプライマー6(配列番号14)を用いた等温増幅反応において、本実施例にて作成した逆転写反応液の1/4000希釈液を添加した反応標品(白丸)、および実施例4と同様、FluB cDNAプラスミドを鋳型DNAとして添加した場合(灰色四角)とを比較した結果である。結果、プラスミドを鋳型としたものに比べ、蛍光シグナルの立ち上がり時間に遅延はあるものの、逆転写反応液を添加した反応標品においても、対照実験として鋳型DNAを加えなかった場合(黒三角)と比べ、蛍光シグナルの増加が顕著に認められた。また図31は、図30での増幅反応における反応溶液をアガロースゲル電気泳動に供した結果である。結果、短鎖から長鎖に渡る周期的なバンドパターンが逆転写反応液を添加した反応標品においても、鋳型DNAを用いた場合と同様のパターンが観察され、同一の等温核酸増幅が起きていることが明らかとなった。
本実施例では、本発明における等温核酸増幅法に供するプライマーの構造において、5’末端に位置する折り返し配列を、片方のプライマーのみ除去した場合の挙動について検討した。容積25μLの液体中に、最終濃度として1.4mM dNTP、5% DMSO、20mM Tris-HCl(pH 8.0)、10mM 塩化カリウム、10mM 硫酸アンモニウム、8mM硫酸マグネシウム、0.1% Tween20、1/100000希釈SYBR Green I(宝酒造社製)、および6unit Aac DNAポリメラーゼ(ダナフォーム社製)を含む溶液に、以下に示すForwardプライマー8(配列番号21)、および実施例5で使用したReverseプライマー6(配列番号17)を各2.5μMの濃度で加えた。尚、下記プライマーにおいて、囲部はForward及びReverseプライマーの共通配列を示しており、Forwardプライマー8は、Forwardプライマー6(配列番号16)の5’末端の折り返し配列(16 base)を除去した配列と同様の配列である。
図32は、5’末端に折り返し配列を含まないForwardプライマー8(配列番号21)および5’末端に折り返し配列を含むReverseプライマー6(配列番号17)をプライマーとして用い、その標的配列を含むFluB cDNAプラスミドを鋳型DNAとして加えると共に、ヒトゲノムDNAを添加した場合の蛍光増幅曲線を示した。この結果、鋳型DNAを加えたもの(白丸)、およびヒトゲノムDNAを更に添加した場合(灰色四角)のどちらにおいても、対照実験として鋳型DNAを加えなかった場合(灰色菱形)と比べ、蛍光シグナルの増加が顕著に認められた。また、ヒトゲノムDNAのみを鋳型DNAのみに加えた場合(黒三角)では蛍光シグナルの増加は認められなかった。また図33は、図32での増幅反応における反応溶液をアガロースゲル電気泳動に供した結果である。結果、短鎖から長鎖に渡るバンドパターンが、FluB cDNAプラスミドの鋳型DNA含む反応標品で観察され、鋳型DNAを含まないもしくはヒトゲノムDNAのみを含んだ反応溶液では、顕著な増幅パターンは認められなかった。よって、プライマーの片方が5’末端を含まない場合においても、標的の鋳型DNAに対する等温核酸増幅を果たしていることが明らかとなった。但し、増幅産物として得られたバンドパターンを実施例5の結果(図28)と比較すると、異なるバンドパターンが得られており、片側のプライマー5’末端折り返し配列を除去する事により、異なる増幅産物が得られる事が明らかとなった。
本実施例における等温核酸増幅法は、25μLの液体中に、最終濃度として1.4mM dNTP、5% DMSO、20mM Tris-HCl(pH 8.0)、10mM塩化カリウム、10mM硫酸アンモニウム、8mM硫酸マグネシウム、0.1% Tween20、1/100000希釈SYBR Green I(宝酒造社製)、および6unit Aac DNAポリメラーゼ(ダナフォーム社製)を含む溶液に、実施例4で使用したForwardプライマー5(配列番号13)、およびReverseプライマー8(配列番号22)を各2.5μMの濃度で加えたものを反応溶液とした。尚、下記プライマーにおいて、囲部はForward及びReverseプライマーの共通配列を示しており、Reverseプライマー8は、Reverseプライマー5(配列番号14)の5’末端の折り返し配列(13base)を除去した配列と同様の配列である。
図34は、5’末端に折り返し配列を含むForwardプライマー5(配列番号13)および5’末端に折り返し配列を含まないReverseプライマー8(配列番号22)をプライマーとして用い、その標的配列を含むFluB cDNAプラスミドを鋳型DNAとして加えた場合の蛍光増幅曲線を示した。この結果、鋳型DNAとしてFluB cDNAを含むプラスミドを用いた場合(白丸)において、対照実験として鋳型DNAを加えなかった場合(黒三角)と比べ、蛍光シグナルの増加が顕著に認められた。また図35は、図34での増幅反応における反応溶液をアガロースゲル電気泳動に供した結果である。結果、短鎖から長鎖に渡る周期的なバンドパターンが鋳型DNA含む反応標品で観察され、等温核酸増幅が起きていることが明らかとなった。
Claims (16)
- 標的核酸配列の等温増幅方法に用いるプライマーセットであって、
前記プライマーセットが、第一プライマー及び第二プライマーを含み、
前記第一プライマーが、3´側に、前記標的核酸配列の3´側の配列(A)にハイブリダイズ可能な配列(A´)を含み、
前記第二プライマーが、3´側に、前記第一プライマーの伸長鎖又は前記標的核酸配列の相補鎖の3´側の配列(B)にハイブリダイズ可能な配列(B´)を含み、
さらに、前記第一プライマー及び第二プライマーが、それぞれの5´側に、互いに実質的に同一の配列(C)を含むことを特徴とするプライマーセット。 - 前記第一プライマー及び第二プライマーの少なくとも一方において、前記配列(C)の5´側に、さらに、相互にハイブリダイズする2つの配列を同一鎖上に含む折返し配列(D-D´)を含むことを特徴とする請求項1記載のプライマーセット。
- 前記第一プライマーにおいて、前記配列(C)の5´側に、さらに、相互にハイブリダイズする2つの配列を同一鎖上に含む折返し配列(D-D´)を含み、
前記第二プライマーにおいて、前記配列(C)の5´側に、さらに、相互にハイブリダイズする2つの配列を同一鎖上に含む折返し配列(E-E´)を含み、
前記配列(D-D´)及び前記配列(E-E´)が、互いに異なる配列であることを特徴とする請求項1記載のプライマーセット。 - さらに、第三プライマーを含み、前記第三プライマーは、前記標的核酸配列、前記標的核酸配列の相補配列、又は、前記第一プライマーの伸長鎖若しくは前記第二プライマーの伸長鎖にハイブリダイズし、かつ、前記第三プライマーのハイブリダイゼーションが、前記第一プライマー及び第二プライマーと競合しないことを特徴とする請求項1から3のいずれか一項に記載のプライマーセット。
- プライマーセットを用いて標的核酸配列を等温で増幅する等温増幅方法であって、前記プライマーセットとして、請求項1から4のいずれか一項に記載のプライマーセットを用いることを特徴とする方法。
- プライマーセットを用いた等温増幅方法により核酸試料中の核酸配列における変異を検出する方法であって、
前記プライマーセットとして、請求項1から4のいずれか一項に記載のプライマーセットを用い、
前記プライマーセットが、前記変異を有する核酸配列又は前記変異を有さない核酸配列を標的核酸配列とし、前記変異に係るヌクレオチド残基が、第一プライマーの相補配列(A)又は第二プライマーの相補配列(B)に含まれるように設計されたプライマーセットであり、
前記核酸試料の存在下、前記プライマーセットによる等温増幅反応を行うことを特徴とする方法。 - 少なくとも二つの異なる配列の順序が少なくとも二回繰り返す一本鎖核酸及び前記一本鎖核酸と相補的な核酸から構成される二本鎖核酸を等温で合成する核酸の合成方法であって、
下記の(A1)~(A6)の各工程を含むことを特徴とする核酸の合成方法。
(A1) 3´末端を含む3´側ステム配列と、5´末端を含む5´側ステム配列とが、ループ配列を介して連結したステムループ構造の一本鎖鋳型核酸であって、前記3´側ステムの3´末端に、相互にハイブリダイズする2つの配列を同一鎖上に含む折り返し配列が連結した一本鎖鋳型核酸を提供する工程。
(A2) 前記一本鎖鋳型核酸の前記ループにプライマーをハイブリダイズさせ、前記プライマーを前記5´側ステム配列の5´末端に向かって伸長させる工程。
(A3) 前記5´側ステム配列の5´末端まで進んだ前記プライマーの伸長反応を、前記5´側ステム配列の5´末端から前記折り返し配列の3´末端にかけて連続して続行する工程。
(A4) 前記(A3)の前記折り返し配列の3´末端に進んだ前記プライマーの伸長反応を、再び、前記5´側ステム配列の5´末端に向かって連続して続行させ、かつ、続行する前記プライマーの伸長反応により、前記(A2)で形成された一本鎖鋳型核酸にハイブリダイズしているプライマー伸長鎖を鎖置換反応により一本鎖にする工程。
(A5) 前記(A4)のプライマー伸長反応を、前記5´側ステム配列の5´末端で停止する工程。
(A6) 前記(A4)で一本鎖になったプライマー伸長鎖を鋳型として前記一本鎖鋳型核酸の前記折り返し配列の3´末端を伸長させる工程。 - (A3)工程及び(A4)工程を少なくとも2回繰り返すことを特徴とする請求項7記載の核酸の合成方法。
- 前記(A1)工程で提供される前記一本鎖鋳型核酸が、請求項2記載のプライマーセットであって、第一プライマーにのみ前記折り返し配列(D-D´)を含むプライマーセットを用いた等温増幅反応により形成された一本鎖鋳型核酸であり、
前記(A2)工程で前記ループにハイブリダイズする前記プライマーが、前記折り返し配列(D-D´)を含む第一プライマーである
ことを特徴とする請求項7または8に記載の核酸の合成方法。 - 前記(A1)工程で提供される前記一本鎖鋳型核酸が、さらに、前記5´側ステムループ配列の5´末端に、相互にハイブリダイズする2つの配列を同一鎖上に含む折り返し配列が連結した一本鎖鋳型核酸であり、
前記(A3)工程に代えて、下記(A3-2)工程を含むことを特徴とする請求項7記載の核酸の合成方法。
(A3-2) 前記5´側ステム配列の5´末端まで進んだ前記プライマーの伸長反応を、前記5´側ステム配列の5´末端に連結した前記折り返し配列を介することなく、直接、前記5´側ステム配列の5´末端から前記折り返し配列の3´末端にかけて連続して続行する工程。 - (A3-2)工程及び(A4)工程を少なくとも2回繰り返すことを特徴とする請求項10記載の核酸の合成方法。
- 前記(A1)工程で提供される前記一本鎖鋳型核酸が、請求項3のプライマーセットを用いた等温増幅法で形成された一本鎖鋳型核酸であり、
前記(A2)工程で前記ループにハイブリダイズする前記プライマーが、請求項3のプライマーセットの第一プライマー又は第二プライマーである
ことを特徴とする請求項10又は11に記載の核酸の合成方法。 - 核酸の増幅方法であって、少なくとも二つの異なる配列の順序が少なくとも二回繰り返す一本鎖核酸及び前記一本鎖核酸と相補的な核酸から構成される二本鎖核酸を等温で合成する核酸の合成工程を含み、前記核酸の合成工程が、請求項7から12のいずれか一項に記載の核酸の合成方法により実施されることを特徴とする核酸の増幅方法。
- 少なくとも二つの異なる配列の順序が少なくとも二回繰り返す一本鎖核酸及び前記一本鎖核酸と相補的な核酸から構成される二本鎖核酸を等温で合成する核酸の合成方法であって、下記の(B1)~(B3)の各工程を含む第1の反応工程及び下記の(C1)~(C3)の各工程を含む第2の反応工程の少なくとも一方を含むことを特徴とする核酸の合成方法。
(B1) 3´末端を含む領域に、相互にハイブリダイズする2つの配列を同一鎖上に含む折り返し配列を含む一本鎖核酸と前記一本鎖核酸と相補的な一本鎖核酸とからなる二重鎖を、配列の向きが互いに逆方向になる状態で、二つ提供する工程。
(B2) 前記(B1)の二つの二重鎖の一方の二重鎖の前記一本鎖核酸の折り返し配列の3´末端から、他方の二重鎖の前記相補的な一本鎖核酸を鋳型として鎖置換伸長反応が起こり、前記一方の二重鎖の前記一本鎖核酸の伸長鎖の一部に前記他方の二重鎖の相補的な一本鎖核酸がハイブリダイズした部分的二重鎖を形成する工程。
(B3) 前記工程(B2)の部分的二重鎖において、前記相補的な一本鎖核酸の3´末端から、前記一本鎖核酸を鋳型とした伸長反応により、完全二重鎖を形成する工程。
(C1) 3´末端を含む領域に、相互にハイブリダイズする2つの配列を同一鎖上に含む折り返し配列を含む一本鎖核酸と前記一本鎖核酸と相補的な一本鎖核酸とからなる二重鎖を一つ提供する工程。
(C2) 前記(C1)の二重鎖の前記一本鎖核酸の折り返し配列の3´末端から、前記相補的な一本鎖核酸を鋳型とし、前記相補的な一本鎖核酸の3´末端から5´末端にかけて鎖置換伸長反応が起こり、前記一本鎖核酸の伸長鎖の一部に前記相補的な一本鎖核酸がハイブリダイズした部分的二重鎖を形成する工程。
(C3) 前記工程(C2)の部分的二重鎖において、前記相補的な一本鎖核酸の3´末端から、前記一本鎖核酸を鋳型とした伸長反応により、完全二重鎖を形成する工程。 - 前記(B1)工程及び前記(C1)工程の二重鎖が、請求項2または3に記載のプライマーセットを用いた等温増幅反応により形成された二重鎖であることを特徴とする請求項14記載の核酸の合成方法。
- 核酸の増幅方法であって、少なくとも二つの異なる配列の順序が少なくとも二回繰り返す一本鎖核酸及び前記一本鎖核酸と相補的な核酸から構成される二本鎖核酸を等温で合成する核酸の合成工程を含み、前記核酸の合成工程が、請求項14または15に記載の核酸の合成方法により実施されることを特徴とする核酸の増幅方法。
Priority Applications (3)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| JP2012541264A JP5299981B1 (ja) | 2011-09-08 | 2012-09-07 | プライマーセット及びそれを用いた標的核酸配列の増幅方法並びに変異核酸の検出方法 |
| EP12829819.7A EP2746395B1 (en) | 2011-09-08 | 2012-09-07 | Primer set, method for amplifying target nucleic acid sequence using same, and method for detecting mutated nucleic acid using same |
| US14/343,511 US9586987B2 (en) | 2011-09-08 | 2012-09-07 | Primer set for isothermal amplication of a target nucleic acid sequence |
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| JP2011196597 | 2011-09-08 | ||
| JP2011-196597 | 2011-09-08 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2013035875A1 true WO2013035875A1 (ja) | 2013-03-14 |
Family
ID=47832310
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/JP2012/072997 WO2013035875A1 (ja) | 2011-09-08 | 2012-09-07 | プライマーセット及びそれを用いた標的核酸配列の増幅方法並びに変異核酸の検出方法 |
Country Status (5)
| Country | Link |
|---|---|
| US (1) | US9586987B2 (ja) |
| EP (1) | EP2746395B1 (ja) |
| JP (2) | JP5299981B1 (ja) |
| TW (2) | TWI619722B (ja) |
| WO (1) | WO2013035875A1 (ja) |
Cited By (8)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US9416387B2 (en) | 2013-03-15 | 2016-08-16 | Theranos, Inc. | Nucleic acid amplification |
| US9551027B2 (en) | 2013-03-15 | 2017-01-24 | Theranos, Inc. | Nucleic acid amplification |
| EP3071713A4 (en) * | 2013-11-22 | 2017-08-02 | Theranos, Inc. | Nucleic Acid Amplification |
| US9916428B2 (en) | 2013-09-06 | 2018-03-13 | Theranos Ip Company, Llc | Systems and methods for detecting infectious diseases |
| US10450595B2 (en) | 2013-03-15 | 2019-10-22 | Theranos Ip Company, Llc | Nucleic acid amplification |
| US11254960B2 (en) | 2013-03-15 | 2022-02-22 | Labrador Diagnostics Llc | Nucleic acid amplification |
| WO2023100486A1 (ja) * | 2021-12-02 | 2023-06-08 | 株式会社デンソー | 分析方法 |
| WO2023100898A1 (ja) * | 2021-12-02 | 2023-06-08 | 株式会社デンソー | 結合物質及び分析方法 |
Families Citing this family (27)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CA3138078C (en) | 2007-10-02 | 2024-02-13 | Labrador Diagnostics Llc | Modular point-of-care devices and uses thereof |
| US8691509B2 (en) | 2009-04-02 | 2014-04-08 | Fluidigm Corporation | Multi-primer amplification method for barcoding of target nucleic acids |
| CN106290160A (zh) | 2011-01-21 | 2017-01-04 | 提拉诺斯公司 | 样品使用最大化的系统和方法 |
| SG194745A1 (en) | 2011-05-20 | 2013-12-30 | Fluidigm Corp | Nucleic acid encoding reactions |
| US20140170735A1 (en) | 2011-09-25 | 2014-06-19 | Elizabeth A. Holmes | Systems and methods for multi-analysis |
| US9632102B2 (en) | 2011-09-25 | 2017-04-25 | Theranos, Inc. | Systems and methods for multi-purpose analysis |
| US8475739B2 (en) | 2011-09-25 | 2013-07-02 | Theranos, Inc. | Systems and methods for fluid handling |
| US9268915B2 (en) | 2011-09-25 | 2016-02-23 | Theranos, Inc. | Systems and methods for diagnosis or treatment |
| US9619627B2 (en) | 2011-09-25 | 2017-04-11 | Theranos, Inc. | Systems and methods for collecting and transmitting assay results |
| US9664702B2 (en) | 2011-09-25 | 2017-05-30 | Theranos, Inc. | Fluid handling apparatus and configurations |
| US9250229B2 (en) | 2011-09-25 | 2016-02-02 | Theranos, Inc. | Systems and methods for multi-analysis |
| US10012664B2 (en) | 2011-09-25 | 2018-07-03 | Theranos Ip Company, Llc | Systems and methods for fluid and component handling |
| US9810704B2 (en) | 2013-02-18 | 2017-11-07 | Theranos, Inc. | Systems and methods for multi-analysis |
| MX2016002797A (es) | 2013-09-06 | 2016-05-26 | Theranos Inc | Dispositivos, sistemas, metodos y equipos para recibir un hisopo. |
| US10767222B2 (en) | 2013-12-11 | 2020-09-08 | Accuragen Holdings Limited | Compositions and methods for detecting rare sequence variants |
| US11859246B2 (en) | 2013-12-11 | 2024-01-02 | Accuragen Holdings Limited | Methods and compositions for enrichment of amplification products |
| US11286519B2 (en) | 2013-12-11 | 2022-03-29 | Accuragen Holdings Limited | Methods and compositions for enrichment of amplification products |
| US20170335382A1 (en) * | 2014-11-10 | 2017-11-23 | University Of Pittsburgh - Of The Commonwealth System Of Higher Education | Isothermal Amplification Assay for the Detection of Short Nucleic Acid Sequences |
| CN114807323A (zh) * | 2015-10-09 | 2022-07-29 | 安可济控股有限公司 | 用于富集扩增产物的方法及组合物 |
| WO2017106777A1 (en) * | 2015-12-16 | 2017-06-22 | Fluidigm Corporation | High-level multiplex amplification |
| EP3458586B1 (en) | 2016-05-16 | 2022-12-28 | Accuragen Holdings Limited | Method of improved sequencing by strand identification |
| EP3464592A4 (en) * | 2016-05-24 | 2020-05-13 | Atila Biosystems, Inc. | OMEGA AMPLIFICATION |
| WO2018035170A1 (en) * | 2016-08-15 | 2018-02-22 | Accuragen Holdings Limited | Compositions and methods for detecting rare sequence variants |
| EP3287528A1 (de) | 2016-08-25 | 2018-02-28 | AGCT GmbH | Verfahren zur amplifikation von nukleinsäuren und kit zu dessen durchführung |
| JP7055691B2 (ja) | 2017-07-11 | 2022-04-18 | 株式会社東芝 | 短鎖核酸伸長用プライマーセット、アッセイキット、短鎖核酸伸長方法、増幅方法及び検出方法 |
| US11203782B2 (en) | 2018-03-29 | 2021-12-21 | Accuragen Holdings Limited | Compositions and methods comprising asymmetric barcoding |
| US12049665B2 (en) | 2018-06-12 | 2024-07-30 | Accuragen Holdings Limited | Methods and compositions for forming ligation products |
Citations (24)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| JPS5993099A (ja) | 1983-10-31 | 1984-05-29 | Wakunaga Seiyaku Kk | オリゴヌクレオチド誘導体およびその製造法 |
| JPS59148798A (ja) | 1983-02-14 | 1984-08-25 | Wakunaga Seiyaku Kk | ビオチンヌクレオチド誘導体 |
| JPS59204200A (ja) | 1983-04-28 | 1984-11-19 | Wakunaga Seiyaku Kk | 2,4―ジニトロフェニルヌクレオチド誘導体 |
| US4683195A (en) | 1986-01-30 | 1987-07-28 | Cetus Corporation | Process for amplifying, detecting, and/or-cloning nucleic acid sequences |
| US4683202A (en) | 1985-03-28 | 1987-07-28 | Cetus Corporation | Process for amplifying nucleic acid sequences |
| US4800159A (en) | 1986-02-07 | 1989-01-24 | Cetus Corporation | Process for amplifying, detecting, and/or cloning nucleic acid sequences |
| EP0320308A2 (en) | 1987-12-11 | 1989-06-14 | Abbott Laboratories | Method for detecting a target nucleic acid sequence |
| JPH04501959A (ja) | 1988-11-21 | 1992-04-09 | ダイナル・エイ・エス | 核酸プローブ |
| JPH076986B2 (ja) | 1983-05-12 | 1995-01-30 | チバ コーニング ダイアグノスティクス コーポレーション | リゲートの濃度測定方法 |
| WO1995025180A1 (en) | 1994-03-16 | 1995-09-21 | Gen-Probe Incorporated | Isothermal strand displacement nucleic acid amplification |
| JPH07114718B2 (ja) | 1991-01-31 | 1995-12-13 | ベクトン・ディッキンソン・アンド・カンパニー | 鎖置換型増幅法 |
| JP2650159B2 (ja) | 1988-02-24 | 1997-09-03 | アクゾ・ノベル・エヌ・ベー | 核酸増幅方法 |
| JP2710159B2 (ja) | 1986-04-16 | 1998-02-10 | ザ・ソーク・インステチュート・フォー・バイオロジカル・スタディース | 複製型rnaリポーター・システム |
| US5824517A (en) | 1995-07-24 | 1998-10-20 | Bio Merieux | Method for amplifying nucleic acid sequences by strand displacement using DNA/RNA chimeric primers |
| WO1999009211A1 (en) | 1997-08-13 | 1999-02-25 | Tepnel Medical Limited | Amplification of nucleic acids |
| WO1999054455A1 (en) | 1998-04-23 | 1999-10-28 | Takara Shuzo Co., Ltd. | Method for synthesizing dna |
| US6046807A (en) | 1998-05-14 | 2000-04-04 | Luminex Corporation | Diode laser based measurement apparatus |
| US6057107A (en) | 1995-10-11 | 2000-05-02 | Luminex Corporation | Methods and compositions for flow cytometric determination of DNA sequences |
| WO2000028082A1 (fr) | 1998-11-09 | 2000-05-18 | Eiken Kagaku Kabushiki Kaisha | Procede de synthese d'acide nucleique |
| JP2000245460A (ja) | 1999-03-05 | 2000-09-12 | Mitsubishi Rayon Co Ltd | 核酸固定化中空繊維並びに核酸固定化中空繊維配列体及びその薄片 |
| WO2002016639A1 (fr) | 2000-08-23 | 2002-02-28 | Takara Bio Inc. | Procede d'amplification d'acide nucleique |
| WO2002024902A1 (fr) | 2000-09-19 | 2002-03-28 | Eiken Kagaku Kabushiki Kaisha | Procede permettant de synthetiser un polynucleotide |
| US6617106B1 (en) | 1990-10-09 | 2003-09-09 | Steven Albert Benner | Methods for preparing oligonucleotides containing non-standard nucleotides |
| WO2005063977A1 (ja) * | 2003-12-25 | 2005-07-14 | Riken | 核酸の増幅法およびこれを利用した変異核酸の検出法 |
Family Cites Families (8)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US4605735A (en) | 1983-02-14 | 1986-08-12 | Wakunaga Seiyaku Kabushiki Kaisha | Oligonucleotide derivatives |
| JPH076986A (ja) | 1993-06-16 | 1995-01-10 | Nippondenso Co Ltd | 半導体基板研削方法 |
| JPH07114718A (ja) | 1993-10-18 | 1995-05-02 | Tanashin Denki Co | テープレコーダの回転ヘッド装置 |
| FR2721945B1 (fr) | 1994-07-04 | 1996-10-18 | David Fabrice | Accroissement genique, un procede d'amplicication genique isotherme et ses applications |
| US5882856A (en) * | 1995-06-07 | 1999-03-16 | Genzyme Corporation | Universal primer sequence for multiplex DNA amplification |
| US6117635A (en) | 1996-07-16 | 2000-09-12 | Intergen Company | Nucleic acid amplification oligonucleotides with molecular energy transfer labels and methods based thereon |
| ATE431848T1 (de) | 1999-03-05 | 2009-06-15 | Mitsubishi Rayon Co | Microarray mit einer biologischen substanz |
| GB0703996D0 (en) * | 2007-03-01 | 2007-04-11 | Oxitec Ltd | Nucleic acid detection |
-
2012
- 2012-09-07 US US14/343,511 patent/US9586987B2/en not_active Expired - Fee Related
- 2012-09-07 WO PCT/JP2012/072997 patent/WO2013035875A1/ja active Application Filing
- 2012-09-07 JP JP2012541264A patent/JP5299981B1/ja not_active Expired - Fee Related
- 2012-09-07 EP EP12829819.7A patent/EP2746395B1/en not_active Not-in-force
- 2012-09-10 TW TW105127301A patent/TWI619722B/zh not_active IP Right Cessation
- 2012-09-10 TW TW101132990A patent/TWI583695B/zh not_active IP Right Cessation
-
2013
- 2013-04-26 JP JP2013094786A patent/JP2013143966A/ja active Pending
Patent Citations (28)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| JPS59148798A (ja) | 1983-02-14 | 1984-08-25 | Wakunaga Seiyaku Kk | ビオチンヌクレオチド誘導体 |
| JPS59204200A (ja) | 1983-04-28 | 1984-11-19 | Wakunaga Seiyaku Kk | 2,4―ジニトロフェニルヌクレオチド誘導体 |
| JPH076986B2 (ja) | 1983-05-12 | 1995-01-30 | チバ コーニング ダイアグノスティクス コーポレーション | リゲートの濃度測定方法 |
| JPS5993099A (ja) | 1983-10-31 | 1984-05-29 | Wakunaga Seiyaku Kk | オリゴヌクレオチド誘導体およびその製造法 |
| US4683202A (en) | 1985-03-28 | 1987-07-28 | Cetus Corporation | Process for amplifying nucleic acid sequences |
| US4683202B1 (ja) | 1985-03-28 | 1990-11-27 | Cetus Corp | |
| US4683195A (en) | 1986-01-30 | 1987-07-28 | Cetus Corporation | Process for amplifying, detecting, and/or-cloning nucleic acid sequences |
| US4683195B1 (ja) | 1986-01-30 | 1990-11-27 | Cetus Corp | |
| US4800159A (en) | 1986-02-07 | 1989-01-24 | Cetus Corporation | Process for amplifying, detecting, and/or cloning nucleic acid sequences |
| JP2710159B2 (ja) | 1986-04-16 | 1998-02-10 | ザ・ソーク・インステチュート・フォー・バイオロジカル・スタディース | 複製型rnaリポーター・システム |
| EP0320308A2 (en) | 1987-12-11 | 1989-06-14 | Abbott Laboratories | Method for detecting a target nucleic acid sequence |
| JP2650159B2 (ja) | 1988-02-24 | 1997-09-03 | アクゾ・ノベル・エヌ・ベー | 核酸増幅方法 |
| JPH04501959A (ja) | 1988-11-21 | 1992-04-09 | ダイナル・エイ・エス | 核酸プローブ |
| US6617106B1 (en) | 1990-10-09 | 2003-09-09 | Steven Albert Benner | Methods for preparing oligonucleotides containing non-standard nucleotides |
| JPH07114718B2 (ja) | 1991-01-31 | 1995-12-13 | ベクトン・ディッキンソン・アンド・カンパニー | 鎖置換型増幅法 |
| WO1995025180A1 (en) | 1994-03-16 | 1995-09-21 | Gen-Probe Incorporated | Isothermal strand displacement nucleic acid amplification |
| US5824517A (en) | 1995-07-24 | 1998-10-20 | Bio Merieux | Method for amplifying nucleic acid sequences by strand displacement using DNA/RNA chimeric primers |
| US6057107A (en) | 1995-10-11 | 2000-05-02 | Luminex Corporation | Methods and compositions for flow cytometric determination of DNA sequences |
| WO1999009211A1 (en) | 1997-08-13 | 1999-02-25 | Tepnel Medical Limited | Amplification of nucleic acids |
| WO1999054455A1 (en) | 1998-04-23 | 1999-10-28 | Takara Shuzo Co., Ltd. | Method for synthesizing dna |
| US6046807A (en) | 1998-05-14 | 2000-04-04 | Luminex Corporation | Diode laser based measurement apparatus |
| JP3313358B2 (ja) * | 1998-11-09 | 2002-08-12 | 栄研化学株式会社 | 核酸の合成方法 |
| WO2000028082A1 (fr) | 1998-11-09 | 2000-05-18 | Eiken Kagaku Kabushiki Kaisha | Procede de synthese d'acide nucleique |
| JP2000245460A (ja) | 1999-03-05 | 2000-09-12 | Mitsubishi Rayon Co Ltd | 核酸固定化中空繊維並びに核酸固定化中空繊維配列体及びその薄片 |
| WO2002016639A1 (fr) | 2000-08-23 | 2002-02-28 | Takara Bio Inc. | Procede d'amplification d'acide nucleique |
| WO2002024902A1 (fr) | 2000-09-19 | 2002-03-28 | Eiken Kagaku Kabushiki Kaisha | Procede permettant de synthetiser un polynucleotide |
| WO2005063977A1 (ja) * | 2003-12-25 | 2005-07-14 | Riken | 核酸の増幅法およびこれを利用した変異核酸の検出法 |
| JP3897805B2 (ja) | 2003-12-25 | 2007-03-28 | 独立行政法人理化学研究所 | 核酸の増幅法およびこれを利用した変異核酸の検出法 |
Non-Patent Citations (6)
| Title |
|---|
| J. SAMBROOK; E. F. FRISCH; T. MANIATIS: "Molecular Cloning 2nd edition", 1989, COLD SPRING HARBOR LABORATORY |
| KIMURA Y. ET AL.: "Optimization of turn-back primers in isothermal amplification", NUCLEIC ACIDS RES., vol. 39, no. 9, February 2011 (2011-02-01), pages E59, XP032107525 * |
| MICHAEL J. LUTZ ET AL., BIOORGANIC & MEDICAL CHEMISTRY LETTERS, vol. 8, 1998, pages 1149 - 1152 |
| MICHAEL SISMOUR 1 ET AL., BIOCHEMISTRY, vol. 42, no. 28, 2003, pages 8598 |
| PROC. NATL. ACAD. SCI. USA, vol. 92, 1995, pages 6329 - 6333 |
| TRENDS IN BIOTECHNOLOGY, vol. 10, 1992, pages 146 - 152 |
Cited By (17)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US10745745B2 (en) | 2013-03-15 | 2020-08-18 | Labrador Diagnostics Llc | Nucleic acid amplification |
| US9551027B2 (en) | 2013-03-15 | 2017-01-24 | Theranos, Inc. | Nucleic acid amplification |
| US11649487B2 (en) | 2013-03-15 | 2023-05-16 | Labrador Diagnostics Llc | Nucleic acid amplification |
| US9725760B2 (en) | 2013-03-15 | 2017-08-08 | Theranos, Inc. | Nucleic acid amplification |
| US9416387B2 (en) | 2013-03-15 | 2016-08-16 | Theranos, Inc. | Nucleic acid amplification |
| US10017809B2 (en) | 2013-03-15 | 2018-07-10 | Theranos Ip Company, Llc | Nucleic acid amplification |
| US10131939B2 (en) | 2013-03-15 | 2018-11-20 | Theranos Ip Company, Llc | Nucleic acid amplification |
| US11603558B2 (en) | 2013-03-15 | 2023-03-14 | Labrador Diagnostics Llc | Nucleic acid amplification |
| US10450595B2 (en) | 2013-03-15 | 2019-10-22 | Theranos Ip Company, Llc | Nucleic acid amplification |
| US11254960B2 (en) | 2013-03-15 | 2022-02-22 | Labrador Diagnostics Llc | Nucleic acid amplification |
| US9916428B2 (en) | 2013-09-06 | 2018-03-13 | Theranos Ip Company, Llc | Systems and methods for detecting infectious diseases |
| US10522245B2 (en) | 2013-09-06 | 2019-12-31 | Theranos Ip Company, Llc | Systems and methods for detecting infectious diseases |
| US10283217B2 (en) | 2013-09-06 | 2019-05-07 | Theranos Ip Company, Llc | Systems and methods for detecting infectious diseases |
| EP3957741A1 (en) * | 2013-11-22 | 2022-02-23 | Labrador Diagnostics LLC | Nucleic acid amplification |
| EP3071713A4 (en) * | 2013-11-22 | 2017-08-02 | Theranos, Inc. | Nucleic Acid Amplification |
| WO2023100486A1 (ja) * | 2021-12-02 | 2023-06-08 | 株式会社デンソー | 分析方法 |
| WO2023100898A1 (ja) * | 2021-12-02 | 2023-06-08 | 株式会社デンソー | 結合物質及び分析方法 |
Also Published As
| Publication number | Publication date |
|---|---|
| JP2013143966A (ja) | 2013-07-25 |
| TWI619722B (zh) | 2018-04-01 |
| EP2746395B1 (en) | 2017-11-29 |
| TWI583695B (zh) | 2017-05-21 |
| US20140295447A1 (en) | 2014-10-02 |
| EP2746395A1 (en) | 2014-06-25 |
| EP2746395A4 (en) | 2015-08-19 |
| JPWO2013035875A1 (ja) | 2015-03-23 |
| TW201317250A (zh) | 2013-05-01 |
| US9586987B2 (en) | 2017-03-07 |
| TW201641511A (zh) | 2016-12-01 |
| JP5299981B1 (ja) | 2013-09-25 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| JP5299981B1 (ja) | プライマーセット及びそれを用いた標的核酸配列の増幅方法並びに変異核酸の検出方法 | |
| JP3897805B2 (ja) | 核酸の増幅法およびこれを利用した変異核酸の検出法 | |
| JP5789307B2 (ja) | マルチプレックス配列決定反応における核酸鋳型の完全性および同定を維持するための方法 | |
| JP3867926B2 (ja) | 核酸の増幅法 | |
| US9315860B2 (en) | Conjugates of nucleotides and method for the application thereof | |
| JP3942627B2 (ja) | 変異核酸の検出法 | |
| JP7054398B2 (ja) | 改変されたマルチプレックスおよびマルチステップ増幅反応ならびにそのための試薬 | |
| JP2008029333A (ja) | 新規遺伝子増幅法に用いられるプライマー | |
| JP2008161165A (ja) | 競合オリゴヌクレオチドを用いた遺伝子検出法 | |
| JPWO2002090538A1 (ja) | 核酸を合成する方法 | |
| JP4942160B2 (ja) | RecAタンパク質を利用した核酸の等温増幅法 | |
| JP2005160387A (ja) | 核酸の増幅法および核酸増幅用プライマーセット | |
| JP5618227B2 (ja) | 核酸の増幅方法および遺伝子変異の検出方法 | |
| WO2021132596A1 (ja) | プライマーセット及びそれを用いて標的核酸を検出する方法 | |
| EP4105327A1 (en) | Method for synthesizing nucleic acid under constant temperature conditions, kit, and application | |
| WO2006051991A1 (ja) | 核酸の増幅および検出方法 | |
| JP2008161164A (ja) | 人工ミスマッチ核酸を含むプライマーを用いた遺伝子検出法 | |
| JP2008048603A (ja) | 核酸の増幅および検出方法 | |
| WO2018132939A1 (zh) | 一种恒温条件下合成核酸的方法 | |
| JP2005102502A (ja) | 一本鎖目的核酸断片の増幅方法 |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| ENP | Entry into the national phase |
Ref document number: 2012541264 Country of ref document: JP Kind code of ref document: A |
|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 12829819 Country of ref document: EP Kind code of ref document: A1 |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| REEP | Request for entry into the european phase |
Ref document number: 2012829819 Country of ref document: EP |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 14343511 Country of ref document: US |