EP2493496B1 - Use of gstp1 - Google Patents
Use of gstp1 Download PDFInfo
- Publication number
- EP2493496B1 EP2493496B1 EP10775689.2A EP10775689A EP2493496B1 EP 2493496 B1 EP2493496 B1 EP 2493496B1 EP 10775689 A EP10775689 A EP 10775689A EP 2493496 B1 EP2493496 B1 EP 2493496B1
- Authority
- EP
- European Patent Office
- Prior art keywords
- gstp1
- cardiomyopathy
- cmp
- traf2
- patients
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Not-in-force
Links
- 102100030943 Glutathione S-transferase P Human genes 0.000 claims description 368
- 101710112368 Glutathione S-transferase P 1 Proteins 0.000 claims description 366
- 208000031229 Cardiomyopathies Diseases 0.000 claims description 152
- 206010056370 Congestive cardiomyopathy Diseases 0.000 claims description 79
- 201000010046 Dilated cardiomyopathy Diseases 0.000 claims description 78
- 210000002966 serum Anatomy 0.000 claims description 61
- 238000011282 treatment Methods 0.000 claims description 48
- 230000002107 myocardial effect Effects 0.000 claims description 31
- 108020004999 messenger RNA Proteins 0.000 claims description 26
- 238000000034 method Methods 0.000 claims description 19
- 239000003814 drug Substances 0.000 claims description 18
- 239000000523 sample Substances 0.000 claims description 15
- 201000010099 disease Diseases 0.000 claims description 12
- 208000037265 diseases, disorders, signs and symptoms Diseases 0.000 claims description 12
- 239000012472 biological sample Substances 0.000 claims description 10
- 238000001574 biopsy Methods 0.000 claims description 10
- 239000000872 buffer Substances 0.000 claims description 10
- 238000001990 intravenous administration Methods 0.000 claims description 10
- 230000002265 prevention Effects 0.000 claims description 10
- 229940079593 drug Drugs 0.000 claims description 9
- 238000003745 diagnosis Methods 0.000 claims description 8
- 206010020871 hypertrophic cardiomyopathy Diseases 0.000 claims description 8
- 239000000203 mixture Substances 0.000 claims description 6
- JKMHFZQWWAIEOD-UHFFFAOYSA-N 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid Chemical compound OCC[NH+]1CCN(CCS([O-])(=O)=O)CC1 JKMHFZQWWAIEOD-UHFFFAOYSA-N 0.000 claims description 4
- 206010048858 Ischaemic cardiomyopathy Diseases 0.000 claims description 4
- 206010038748 Restrictive cardiomyopathy Diseases 0.000 claims description 4
- 210000004369 blood Anatomy 0.000 claims description 4
- 239000008280 blood Substances 0.000 claims description 4
- 238000000338 in vitro Methods 0.000 claims description 4
- 206010016654 Fibrosis Diseases 0.000 claims description 3
- 239000007995 HEPES buffer Substances 0.000 claims description 3
- 239000003795 chemical substances by application Substances 0.000 claims description 3
- 201000011304 dilated cardiomyopathy 1A Diseases 0.000 claims description 3
- 239000003937 drug carrier Substances 0.000 claims description 3
- 230000004761 fibrosis Effects 0.000 claims description 3
- 208000019622 heart disease Diseases 0.000 claims description 3
- 239000008363 phosphate buffer Substances 0.000 claims description 3
- 210000002826 placenta Anatomy 0.000 claims description 3
- 210000002381 plasma Anatomy 0.000 claims description 3
- QKNYBSVHEMOAJP-UHFFFAOYSA-N 2-amino-2-(hydroxymethyl)propane-1,3-diol;hydron;chloride Chemical compound Cl.OCC(N)(CO)CO QKNYBSVHEMOAJP-UHFFFAOYSA-N 0.000 claims description 2
- 201000010053 Alcoholic Cardiomyopathy Diseases 0.000 claims description 2
- 206010007509 Cardiac amyloidosis Diseases 0.000 claims description 2
- 206010007637 Cardiomyopathy alcoholic Diseases 0.000 claims description 2
- 208000035473 Communicable disease Diseases 0.000 claims description 2
- 206010018641 Gouty tophus Diseases 0.000 claims description 2
- 208000030852 Parasitic disease Diseases 0.000 claims description 2
- 206010013023 diphtheria Diseases 0.000 claims description 2
- 206010014663 endocardial fibroelastosis Diseases 0.000 claims description 2
- 206010014665 endocarditis Diseases 0.000 claims description 2
- 230000002327 eosinophilic effect Effects 0.000 claims description 2
- 208000014471 histiocytoid cardiomyopathy Diseases 0.000 claims description 2
- 230000002458 infectious effect Effects 0.000 claims description 2
- 208000030159 metabolic disease Diseases 0.000 claims description 2
- 235000016709 nutrition Nutrition 0.000 claims description 2
- 208000019180 nutritional disease Diseases 0.000 claims description 2
- 239000008194 pharmaceutical composition Substances 0.000 claims description 2
- 231100000399 thyrotoxic Toxicity 0.000 claims description 2
- 230000001897 thyrotoxic effect Effects 0.000 claims description 2
- 230000000302 ischemic effect Effects 0.000 description 66
- 230000014509 gene expression Effects 0.000 description 65
- 210000005003 heart tissue Anatomy 0.000 description 60
- 108090000623 proteins and genes Proteins 0.000 description 59
- 102000004169 proteins and genes Human genes 0.000 description 58
- 102000002574 p38 Mitogen-Activated Protein Kinases Human genes 0.000 description 51
- 108010068338 p38 Mitogen-Activated Protein Kinases Proteins 0.000 description 51
- 102100036836 Natriuretic peptides B Human genes 0.000 description 50
- 101710187802 Natriuretic peptides B Proteins 0.000 description 50
- 108060008682 Tumor Necrosis Factor Proteins 0.000 description 50
- MZOFCQQQCNRIBI-VMXHOPILSA-N (3s)-4-[[(2s)-1-[[(2s)-1-[[(1s)-1-carboxy-2-hydroxyethyl]amino]-4-methyl-1-oxopentan-2-yl]amino]-5-(diaminomethylideneamino)-1-oxopentan-2-yl]amino]-3-[[2-[[(2s)-2,6-diaminohexanoyl]amino]acetyl]amino]-4-oxobutanoic acid Chemical compound OC[C@@H](C(O)=O)NC(=O)[C@H](CC(C)C)NC(=O)[C@H](CCCN=C(N)N)NC(=O)[C@H](CC(O)=O)NC(=O)CNC(=O)[C@@H](N)CCCCN MZOFCQQQCNRIBI-VMXHOPILSA-N 0.000 description 48
- 102000000852 Tumor Necrosis Factor-alpha Human genes 0.000 description 48
- 108010055717 JNK Mitogen-Activated Protein Kinases Proteins 0.000 description 33
- 241001465754 Metazoa Species 0.000 description 31
- 208000010125 myocardial infarction Diseases 0.000 description 31
- 206010019280 Heart failures Diseases 0.000 description 30
- 230000000747 cardiac effect Effects 0.000 description 29
- 238000001262 western blot Methods 0.000 description 27
- 238000004458 analytical method Methods 0.000 description 25
- 239000013615 primer Substances 0.000 description 24
- 208000031225 myocardial ischemia Diseases 0.000 description 21
- 108020004414 DNA Proteins 0.000 description 19
- 101000950695 Homo sapiens Mitogen-activated protein kinase 8 Proteins 0.000 description 19
- 102100037808 Mitogen-activated protein kinase 8 Human genes 0.000 description 19
- 241000700159 Rattus Species 0.000 description 19
- 230000004913 activation Effects 0.000 description 18
- 238000007912 intraperitoneal administration Methods 0.000 description 18
- 210000001519 tissue Anatomy 0.000 description 18
- 210000004165 myocardium Anatomy 0.000 description 16
- 230000000694 effects Effects 0.000 description 15
- 230000006870 function Effects 0.000 description 15
- 238000003752 polymerase chain reaction Methods 0.000 description 15
- 230000035945 sensitivity Effects 0.000 description 15
- 230000002829 reductive effect Effects 0.000 description 14
- 230000002861 ventricular Effects 0.000 description 14
- 230000002757 inflammatory effect Effects 0.000 description 13
- 238000011002 quantification Methods 0.000 description 13
- 208000009525 Myocarditis Diseases 0.000 description 12
- 230000006907 apoptotic process Effects 0.000 description 11
- 230000003993 interaction Effects 0.000 description 11
- 239000006166 lysate Substances 0.000 description 11
- 238000005259 measurement Methods 0.000 description 11
- 230000036593 pulmonary vascular resistance Effects 0.000 description 11
- 102000004127 Cytokines Human genes 0.000 description 10
- 108090000695 Cytokines Proteins 0.000 description 10
- 238000002965 ELISA Methods 0.000 description 10
- 230000037396 body weight Effects 0.000 description 10
- 210000004027 cell Anatomy 0.000 description 10
- 230000009918 complex formation Effects 0.000 description 10
- 238000002474 experimental method Methods 0.000 description 10
- RWSXRVCMGQZWBV-WDSKDSINSA-N glutathione Chemical compound OC(=O)[C@@H](N)CCC(=O)N[C@@H](CS)C(=O)NCC(O)=O RWSXRVCMGQZWBV-WDSKDSINSA-N 0.000 description 10
- 230000001404 mediated effect Effects 0.000 description 10
- 238000003498 protein array Methods 0.000 description 10
- 230000011664 signaling Effects 0.000 description 10
- 206010061216 Infarction Diseases 0.000 description 9
- 206010061218 Inflammation Diseases 0.000 description 9
- 230000004054 inflammatory process Effects 0.000 description 9
- 102000043136 MAP kinase family Human genes 0.000 description 8
- 108091054455 MAP kinase family Proteins 0.000 description 8
- 238000003556 assay Methods 0.000 description 8
- DDRJAANPRJIHGJ-UHFFFAOYSA-N creatinine Chemical compound CN1CC(=O)NC1=N DDRJAANPRJIHGJ-UHFFFAOYSA-N 0.000 description 8
- 230000003247 decreasing effect Effects 0.000 description 8
- 230000000004 hemodynamic effect Effects 0.000 description 8
- 238000012360 testing method Methods 0.000 description 8
- 108091032973 (ribonucleotides)n+m Proteins 0.000 description 7
- UUUHXMGGBIUAPW-UHFFFAOYSA-N 1-[1-[2-[[5-amino-2-[[1-[5-(diaminomethylideneamino)-2-[[1-[3-(1h-indol-3-yl)-2-[(5-oxopyrrolidine-2-carbonyl)amino]propanoyl]pyrrolidine-2-carbonyl]amino]pentanoyl]pyrrolidine-2-carbonyl]amino]-5-oxopentanoyl]amino]-3-methylpentanoyl]pyrrolidine-2-carbon Chemical compound C1CCC(C(=O)N2C(CCC2)C(O)=O)N1C(=O)C(C(C)CC)NC(=O)C(CCC(N)=O)NC(=O)C1CCCN1C(=O)C(CCCN=C(N)N)NC(=O)C1CCCN1C(=O)C(CC=1C2=CC=CC=C2NC=1)NC(=O)C1CCC(=O)N1 UUUHXMGGBIUAPW-UHFFFAOYSA-N 0.000 description 7
- 102000004270 Peptidyl-Dipeptidase A Human genes 0.000 description 7
- 108090000882 Peptidyl-Dipeptidase A Proteins 0.000 description 7
- 230000003110 anti-inflammatory effect Effects 0.000 description 7
- 230000027455 binding Effects 0.000 description 7
- 239000002299 complementary DNA Substances 0.000 description 7
- 230000001419 dependent effect Effects 0.000 description 7
- 238000002592 echocardiography Methods 0.000 description 7
- 230000007574 infarction Effects 0.000 description 7
- 208000028867 ischemia Diseases 0.000 description 7
- 238000011068 loading method Methods 0.000 description 7
- 150000007523 nucleic acids Chemical group 0.000 description 7
- 230000000770 proinflammatory effect Effects 0.000 description 7
- 238000007634 remodeling Methods 0.000 description 7
- 238000010186 staining Methods 0.000 description 7
- 238000007619 statistical method Methods 0.000 description 7
- YBJHBAHKTGYVGT-ZKWXMUAHSA-N (+)-Biotin Chemical compound N1C(=O)N[C@@H]2[C@H](CCCCC(=O)O)SC[C@@H]21 YBJHBAHKTGYVGT-ZKWXMUAHSA-N 0.000 description 6
- 108010070675 Glutathione transferase Proteins 0.000 description 6
- PEDCQBHIVMGVHV-UHFFFAOYSA-N Glycerine Chemical compound OCC(O)CO PEDCQBHIVMGVHV-UHFFFAOYSA-N 0.000 description 6
- 206010020772 Hypertension Diseases 0.000 description 6
- 206010028980 Neoplasm Diseases 0.000 description 6
- 230000009286 beneficial effect Effects 0.000 description 6
- 210000004351 coronary vessel Anatomy 0.000 description 6
- 238000001114 immunoprecipitation Methods 0.000 description 6
- 230000006698 induction Effects 0.000 description 6
- 238000004519 manufacturing process Methods 0.000 description 6
- 239000003550 marker Substances 0.000 description 6
- 230000037361 pathway Effects 0.000 description 6
- 230000000638 stimulation Effects 0.000 description 6
- 108010014303 DNA-directed DNA polymerase Proteins 0.000 description 5
- 102000016928 DNA-directed DNA polymerase Human genes 0.000 description 5
- 102000005720 Glutathione transferase Human genes 0.000 description 5
- 101001010139 Homo sapiens Glutathione S-transferase P Proteins 0.000 description 5
- 102000003945 NF-kappa B Human genes 0.000 description 5
- 108010057466 NF-kappa B Proteins 0.000 description 5
- 102000015736 beta 2-Microglobulin Human genes 0.000 description 5
- 108010081355 beta 2-Microglobulin Proteins 0.000 description 5
- 230000000295 complement effect Effects 0.000 description 5
- 238000001514 detection method Methods 0.000 description 5
- 229960003180 glutathione Drugs 0.000 description 5
- 238000011534 incubation Methods 0.000 description 5
- 230000005764 inhibitory process Effects 0.000 description 5
- 239000007924 injection Substances 0.000 description 5
- 238000002347 injection Methods 0.000 description 5
- 238000012544 monitoring process Methods 0.000 description 5
- 239000002773 nucleotide Substances 0.000 description 5
- 125000003729 nucleotide group Chemical group 0.000 description 5
- 238000001543 one-way ANOVA Methods 0.000 description 5
- 230000007170 pathology Effects 0.000 description 5
- 238000003753 real-time PCR Methods 0.000 description 5
- 230000001105 regulatory effect Effects 0.000 description 5
- 238000012216 screening Methods 0.000 description 5
- XLYOFNOQVPJJNP-UHFFFAOYSA-N water Chemical compound O XLYOFNOQVPJJNP-UHFFFAOYSA-N 0.000 description 5
- IJGRMHOSHXDMSA-UHFFFAOYSA-N Atomic nitrogen Chemical compound N#N IJGRMHOSHXDMSA-UHFFFAOYSA-N 0.000 description 4
- 108010024636 Glutathione Proteins 0.000 description 4
- 102000000588 Interleukin-2 Human genes 0.000 description 4
- 108010002350 Interleukin-2 Proteins 0.000 description 4
- 238000011529 RT qPCR Methods 0.000 description 4
- FAPWRFPIFSIZLT-UHFFFAOYSA-M Sodium chloride Chemical compound [Na+].[Cl-] FAPWRFPIFSIZLT-UHFFFAOYSA-M 0.000 description 4
- 102100033733 Tumor necrosis factor receptor superfamily member 1B Human genes 0.000 description 4
- 101710187830 Tumor necrosis factor receptor superfamily member 1B Proteins 0.000 description 4
- 230000001154 acute effect Effects 0.000 description 4
- 206010000891 acute myocardial infarction Diseases 0.000 description 4
- 150000001413 amino acids Chemical group 0.000 description 4
- 238000003491 array Methods 0.000 description 4
- 230000004872 arterial blood pressure Effects 0.000 description 4
- 239000002876 beta blocker Substances 0.000 description 4
- 229940097320 beta blocking agent Drugs 0.000 description 4
- 229940109239 creatinine Drugs 0.000 description 4
- 238000007405 data analysis Methods 0.000 description 4
- 238000011161 development Methods 0.000 description 4
- 230000018109 developmental process Effects 0.000 description 4
- 238000011156 evaluation Methods 0.000 description 4
- 230000004217 heart function Effects 0.000 description 4
- 230000007246 mechanism Effects 0.000 description 4
- 238000002493 microarray Methods 0.000 description 4
- 239000000825 pharmaceutical preparation Substances 0.000 description 4
- YBYRMVIVWMBXKQ-UHFFFAOYSA-N phenylmethanesulfonyl fluoride Chemical compound FS(=O)(=O)CC1=CC=CC=C1 YBYRMVIVWMBXKQ-UHFFFAOYSA-N 0.000 description 4
- -1 polypropylene Polymers 0.000 description 4
- 210000001147 pulmonary artery Anatomy 0.000 description 4
- 230000002685 pulmonary effect Effects 0.000 description 4
- 238000000746 purification Methods 0.000 description 4
- 238000011552 rat model Methods 0.000 description 4
- 230000009467 reduction Effects 0.000 description 4
- 230000004044 response Effects 0.000 description 4
- 239000000243 solution Substances 0.000 description 4
- 238000001356 surgical procedure Methods 0.000 description 4
- 230000001225 therapeutic effect Effects 0.000 description 4
- 238000002560 therapeutic procedure Methods 0.000 description 4
- QTBSBXVTEAMEQO-UHFFFAOYSA-N Acetic acid Chemical compound CC(O)=O QTBSBXVTEAMEQO-UHFFFAOYSA-N 0.000 description 3
- 108700028369 Alleles Proteins 0.000 description 3
- QGZKDVFQNNGYKY-UHFFFAOYSA-N Ammonia Chemical compound N QGZKDVFQNNGYKY-UHFFFAOYSA-N 0.000 description 3
- KCXVZYZYPLLWCC-UHFFFAOYSA-N EDTA Chemical compound OC(=O)CN(CC(O)=O)CCN(CC(O)=O)CC(O)=O KCXVZYZYPLLWCC-UHFFFAOYSA-N 0.000 description 3
- 102100022086 GRB2-related adapter protein 2 Human genes 0.000 description 3
- WQZGKKKJIJFFOK-GASJEMHNSA-N Glucose Natural products OC[C@H]1OC(O)[C@H](O)[C@@H](O)[C@@H]1O WQZGKKKJIJFFOK-GASJEMHNSA-N 0.000 description 3
- 108010001336 Horseradish Peroxidase Proteins 0.000 description 3
- 102000013691 Interleukin-17 Human genes 0.000 description 3
- 108050003558 Interleukin-17 Proteins 0.000 description 3
- 241000124008 Mammalia Species 0.000 description 3
- 238000002944 PCR assay Methods 0.000 description 3
- 101710181935 Phosphate-binding protein PstS 1 Proteins 0.000 description 3
- 108091000080 Phosphotransferase Proteins 0.000 description 3
- 238000012288 TUNEL assay Methods 0.000 description 3
- 238000010162 Tukey test Methods 0.000 description 3
- 206010067584 Type 1 diabetes mellitus Diseases 0.000 description 3
- 238000000540 analysis of variance Methods 0.000 description 3
- 230000000692 anti-sense effect Effects 0.000 description 3
- 239000011324 bead Substances 0.000 description 3
- 230000033228 biological regulation Effects 0.000 description 3
- 239000000090 biomarker Substances 0.000 description 3
- 230000015572 biosynthetic process Effects 0.000 description 3
- 229960002685 biotin Drugs 0.000 description 3
- 235000020958 biotin Nutrition 0.000 description 3
- 239000011616 biotin Substances 0.000 description 3
- 230000003197 catalytic effect Effects 0.000 description 3
- 238000006243 chemical reaction Methods 0.000 description 3
- 238000000546 chi-square test Methods 0.000 description 3
- 230000001684 chronic effect Effects 0.000 description 3
- 238000013211 curve analysis Methods 0.000 description 3
- 238000013461 design Methods 0.000 description 3
- LOKCTEFSRHRXRJ-UHFFFAOYSA-I dipotassium trisodium dihydrogen phosphate hydrogen phosphate dichloride Chemical compound P(=O)(O)(O)[O-].[K+].P(=O)(O)([O-])[O-].[Na+].[Na+].[Cl-].[K+].[Cl-].[Na+] LOKCTEFSRHRXRJ-UHFFFAOYSA-I 0.000 description 3
- 239000008103 glucose Substances 0.000 description 3
- 101150008380 gstp1 gene Proteins 0.000 description 3
- 230000036541 health Effects 0.000 description 3
- 239000012133 immunoprecipitate Substances 0.000 description 3
- 239000004615 ingredient Substances 0.000 description 3
- 239000003112 inhibitor Substances 0.000 description 3
- 230000003834 intracellular effect Effects 0.000 description 3
- 239000007928 intraperitoneal injection Substances 0.000 description 3
- 239000003446 ligand Substances 0.000 description 3
- 238000007477 logistic regression Methods 0.000 description 3
- 238000010841 mRNA extraction Methods 0.000 description 3
- 238000000329 molecular dynamics simulation Methods 0.000 description 3
- 230000005914 myocardial expression Effects 0.000 description 3
- 108020004707 nucleic acids Proteins 0.000 description 3
- 102000039446 nucleic acids Human genes 0.000 description 3
- 239000002953 phosphate buffered saline Substances 0.000 description 3
- 102000020233 phosphotransferase Human genes 0.000 description 3
- IHEHEFLXQFOQJO-UHFFFAOYSA-N piritramide Chemical compound C1CC(C(=O)N)(N2CCCCC2)CCN1CCC(C#N)(C=1C=CC=CC=1)C1=CC=CC=C1 IHEHEFLXQFOQJO-UHFFFAOYSA-N 0.000 description 3
- 239000000047 product Substances 0.000 description 3
- 230000002285 radioactive effect Effects 0.000 description 3
- 238000003127 radioimmunoassay Methods 0.000 description 3
- 239000000758 substrate Substances 0.000 description 3
- 208000024891 symptom Diseases 0.000 description 3
- 230000007838 tissue remodeling Effects 0.000 description 3
- 238000002054 transplantation Methods 0.000 description 3
- 210000004881 tumor cell Anatomy 0.000 description 3
- 208000001072 type 2 diabetes mellitus Diseases 0.000 description 3
- 239000002023 wood Substances 0.000 description 3
- 229920000936 Agarose Polymers 0.000 description 2
- NLXLAEXVIDQMFP-UHFFFAOYSA-N Ammonia chloride Chemical compound [NH4+].[Cl-] NLXLAEXVIDQMFP-UHFFFAOYSA-N 0.000 description 2
- 102000004506 Blood Proteins Human genes 0.000 description 2
- 108010017384 Blood Proteins Proteins 0.000 description 2
- HEDRZPFGACZZDS-UHFFFAOYSA-N Chloroform Chemical compound ClC(Cl)Cl HEDRZPFGACZZDS-UHFFFAOYSA-N 0.000 description 2
- 102000008186 Collagen Human genes 0.000 description 2
- 108010035532 Collagen Proteins 0.000 description 2
- 241000252212 Danio rerio Species 0.000 description 2
- AOJJSUZBOXZQNB-TZSSRYMLSA-N Doxorubicin Chemical compound O([C@H]1C[C@@](O)(CC=2C(O)=C3C(=O)C=4C=CC=C(C=4C(=O)C3=C(O)C=21)OC)C(=O)CO)[C@H]1C[C@H](N)[C@H](O)[C@H](C)O1 AOJJSUZBOXZQNB-TZSSRYMLSA-N 0.000 description 2
- 206010014824 Endotoxic shock Diseases 0.000 description 2
- 102000004190 Enzymes Human genes 0.000 description 2
- 108090000790 Enzymes Proteins 0.000 description 2
- 241000283086 Equidae Species 0.000 description 2
- 108700039887 Essential Genes Proteins 0.000 description 2
- WSFSSNUMVMOOMR-UHFFFAOYSA-N Formaldehyde Chemical compound O=C WSFSSNUMVMOOMR-UHFFFAOYSA-N 0.000 description 2
- DHCLVCXQIBBOPH-UHFFFAOYSA-N Glycerol 2-phosphate Chemical compound OCC(CO)OP(O)(O)=O DHCLVCXQIBBOPH-UHFFFAOYSA-N 0.000 description 2
- OWXMKDGYPWMGEB-UHFFFAOYSA-N HEPPS Chemical compound OCCN1CCN(CCCS(O)(=O)=O)CC1 OWXMKDGYPWMGEB-UHFFFAOYSA-N 0.000 description 2
- 101100400545 Homo sapiens MATN1 gene Proteins 0.000 description 2
- 102100034343 Integrase Human genes 0.000 description 2
- 108010044467 Isoenzymes Proteins 0.000 description 2
- TWRXJAOTZQYOKJ-UHFFFAOYSA-L Magnesium chloride Chemical compound [Mg+2].[Cl-].[Cl-] TWRXJAOTZQYOKJ-UHFFFAOYSA-L 0.000 description 2
- 206010048849 Myocardial haemorrhage Diseases 0.000 description 2
- 206010028604 Myocardial rupture Diseases 0.000 description 2
- 108091028043 Nucleic acid sequence Proteins 0.000 description 2
- 241000282577 Pan troglodytes Species 0.000 description 2
- 239000004743 Polypropylene Substances 0.000 description 2
- 108010092799 RNA-directed DNA polymerase Proteins 0.000 description 2
- 206010040070 Septic Shock Diseases 0.000 description 2
- 108010090804 Streptavidin Proteins 0.000 description 2
- 102000004887 Transforming Growth Factor beta Human genes 0.000 description 2
- 108090001012 Transforming Growth Factor beta Proteins 0.000 description 2
- 239000007983 Tris buffer Substances 0.000 description 2
- 108060008683 Tumor Necrosis Factor Receptor Proteins 0.000 description 2
- 102100033732 Tumor necrosis factor receptor superfamily member 1A Human genes 0.000 description 2
- 101710187743 Tumor necrosis factor receptor superfamily member 1A Proteins 0.000 description 2
- 230000009471 action Effects 0.000 description 2
- 206010069351 acute lung injury Diseases 0.000 description 2
- 238000003915 air pollution Methods 0.000 description 2
- 208000026935 allergic disease Diseases 0.000 description 2
- 230000003321 amplification Effects 0.000 description 2
- 238000002583 angiography Methods 0.000 description 2
- 239000002333 angiotensin II receptor antagonist Substances 0.000 description 2
- 238000010171 animal model Methods 0.000 description 2
- 230000001640 apoptogenic effect Effects 0.000 description 2
- 208000006673 asthma Diseases 0.000 description 2
- 230000000740 bleeding effect Effects 0.000 description 2
- BVKZGUZCCUSVTD-UHFFFAOYSA-N carbonic acid Chemical compound OC(O)=O BVKZGUZCCUSVTD-UHFFFAOYSA-N 0.000 description 2
- 230000008859 change Effects 0.000 description 2
- 238000000749 co-immunoprecipitation Methods 0.000 description 2
- 229920001436 collagen Polymers 0.000 description 2
- 210000003748 coronary sinus Anatomy 0.000 description 2
- 230000007423 decrease Effects 0.000 description 2
- 206010012601 diabetes mellitus Diseases 0.000 description 2
- 230000003205 diastolic effect Effects 0.000 description 2
- VHJLVAABSRFDPM-QWWZWVQMSA-N dithiothreitol Chemical compound SC[C@@H](O)[C@H](O)CS VHJLVAABSRFDPM-QWWZWVQMSA-N 0.000 description 2
- 230000003828 downregulation Effects 0.000 description 2
- 238000001962 electrophoresis Methods 0.000 description 2
- 230000007717 exclusion Effects 0.000 description 2
- 239000007850 fluorescent dye Substances 0.000 description 2
- 239000012634 fragment Substances 0.000 description 2
- 238000003500 gene array Methods 0.000 description 2
- 238000009396 hybridization Methods 0.000 description 2
- 230000008105 immune reaction Effects 0.000 description 2
- 238000003365 immunocytochemistry Methods 0.000 description 2
- 230000001771 impaired effect Effects 0.000 description 2
- WHXMKTBCFHIYNQ-SECBINFHSA-N levosimendan Chemical compound C[C@@H]1CC(=O)NN=C1C1=CC=C(NN=C(C#N)C#N)C=C1 WHXMKTBCFHIYNQ-SECBINFHSA-N 0.000 description 2
- 229960000692 levosimendan Drugs 0.000 description 2
- 239000007788 liquid Substances 0.000 description 2
- 239000012139 lysis buffer Substances 0.000 description 2
- 230000036210 malignancy Effects 0.000 description 2
- 238000002483 medication Methods 0.000 description 2
- 238000010208 microarray analysis Methods 0.000 description 2
- 238000013425 morphometry Methods 0.000 description 2
- 229910052757 nitrogen Inorganic materials 0.000 description 2
- 238000003199 nucleic acid amplification method Methods 0.000 description 2
- 230000008506 pathogenesis Effects 0.000 description 2
- 230000001575 pathological effect Effects 0.000 description 2
- 206010034674 peritonitis Diseases 0.000 description 2
- 229920001155 polypropylene Polymers 0.000 description 2
- 238000002360 preparation method Methods 0.000 description 2
- 230000008569 process Effects 0.000 description 2
- 102000004196 processed proteins & peptides Human genes 0.000 description 2
- 108090000765 processed proteins & peptides Proteins 0.000 description 2
- 102000005962 receptors Human genes 0.000 description 2
- 108020003175 receptors Proteins 0.000 description 2
- 238000003757 reverse transcription PCR Methods 0.000 description 2
- 150000003839 salts Chemical class 0.000 description 2
- 239000011780 sodium chloride Substances 0.000 description 2
- 238000002415 sodium dodecyl sulfate polyacrylamide gel electrophoresis Methods 0.000 description 2
- UCSJYZPVAKXKNQ-HZYVHMACSA-N streptomycin Chemical compound CN[C@H]1[C@H](O)[C@@H](O)[C@H](CO)O[C@H]1O[C@@H]1[C@](C=O)(O)[C@H](C)O[C@H]1O[C@@H]1[C@@H](NC(N)=N)[C@H](O)[C@@H](NC(N)=N)[C@H](O)[C@H]1O UCSJYZPVAKXKNQ-HZYVHMACSA-N 0.000 description 2
- 230000006354 stress signaling Effects 0.000 description 2
- 230000004083 survival effect Effects 0.000 description 2
- 230000008685 targeting Effects 0.000 description 2
- ZRKFYGHZFMAOKI-QMGMOQQFSA-N tgfbeta Chemical compound C([C@H](NC(=O)[C@H](C(C)C)NC(=O)CNC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](CC(N)=O)NC(=O)[C@H](CC(C)C)NC(=O)[C@H]([C@@H](C)O)NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H]([C@@H](C)O)NC(=O)[C@H](CC(C)C)NC(=O)CNC(=O)[C@H](C)NC(=O)[C@H](CO)NC(=O)[C@H](CCC(N)=O)NC(=O)[C@@H](NC(=O)[C@H](C)NC(=O)[C@H](C)NC(=O)[C@@H](NC(=O)[C@H](CC(C)C)NC(=O)[C@@H](N)CCSC)C(C)C)[C@@H](C)CC)C(=O)N[C@@H]([C@@H](C)O)C(=O)N[C@@H](C(C)C)C(=O)N[C@@H](CC=1C=CC=CC=1)C(=O)N[C@@H](C)C(=O)N1[C@@H](CCC1)C(=O)N[C@@H]([C@@H](C)O)C(=O)N[C@@H](CC(N)=O)C(=O)N[C@@H](CCC(O)=O)C(=O)N[C@@H](C)C(=O)N[C@@H](CC=1C=CC=CC=1)C(=O)N[C@@H](CCCNC(N)=N)C(=O)N[C@@H](C)C(=O)N[C@@H](CC(C)C)C(=O)N1[C@@H](CCC1)C(=O)N1[C@@H](CCC1)C(=O)N[C@@H](CCCNC(N)=N)C(=O)N[C@@H](CCC(O)=O)C(=O)N[C@@H](CCCNC(N)=N)C(=O)N[C@@H](CO)C(=O)N[C@@H](CCCNC(N)=N)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](CC(C)C)C(O)=O)C1=CC=C(O)C=C1 ZRKFYGHZFMAOKI-QMGMOQQFSA-N 0.000 description 2
- LENZDBCJOHFCAS-UHFFFAOYSA-N tris Chemical compound OCC(N)(CO)CO LENZDBCJOHFCAS-UHFFFAOYSA-N 0.000 description 2
- 102000003390 tumor necrosis factor Human genes 0.000 description 2
- 102000003298 tumor necrosis factor receptor Human genes 0.000 description 2
- 208000003663 ventricular fibrillation Diseases 0.000 description 2
- HJTAZXHBEBIQQX-UHFFFAOYSA-N 1,5-bis(chloromethyl)naphthalene Chemical compound C1=CC=C2C(CCl)=CC=CC2=C1CCl HJTAZXHBEBIQQX-UHFFFAOYSA-N 0.000 description 1
- OWEGMIWEEQEYGQ-UHFFFAOYSA-N 100676-05-9 Natural products OC1C(O)C(O)C(CO)OC1OCC1C(O)C(O)C(O)C(OC2C(OC(O)C(O)C2O)CO)O1 OWEGMIWEEQEYGQ-UHFFFAOYSA-N 0.000 description 1
- SXGZJKUKBWWHRA-UHFFFAOYSA-N 2-(N-morpholiniumyl)ethanesulfonate Chemical compound [O-]S(=O)(=O)CC[NH+]1CCOCC1 SXGZJKUKBWWHRA-UHFFFAOYSA-N 0.000 description 1
- BUOYTFVLNZIELF-UHFFFAOYSA-N 2-phenyl-1h-indole-4,6-dicarboximidamide Chemical compound N1C2=CC(C(=N)N)=CC(C(N)=N)=C2C=C1C1=CC=CC=C1 BUOYTFVLNZIELF-UHFFFAOYSA-N 0.000 description 1
- FWBHETKCLVMNFS-UHFFFAOYSA-N 4',6-Diamino-2-phenylindol Chemical compound C1=CC(C(=N)N)=CC=C1C1=CC2=CC=C(C(N)=N)C=C2N1 FWBHETKCLVMNFS-UHFFFAOYSA-N 0.000 description 1
- 239000005541 ACE inhibitor Substances 0.000 description 1
- 206010067484 Adverse reaction Diseases 0.000 description 1
- 206010001935 American trypanosomiasis Diseases 0.000 description 1
- ITPDYQOUSLNIHG-UHFFFAOYSA-N Amiodarone hydrochloride Chemical compound [Cl-].CCCCC=1OC2=CC=CC=C2C=1C(=O)C1=CC(I)=C(OCC[NH+](CC)CC)C(I)=C1 ITPDYQOUSLNIHG-UHFFFAOYSA-N 0.000 description 1
- 206010002383 Angina Pectoris Diseases 0.000 description 1
- 241000615866 Antho Species 0.000 description 1
- 241000713838 Avian myeloblastosis virus Species 0.000 description 1
- 108090001008 Avidin Proteins 0.000 description 1
- 208000035143 Bacterial infection Diseases 0.000 description 1
- BVKZGUZCCUSVTD-UHFFFAOYSA-M Bicarbonate Chemical compound OC([O-])=O BVKZGUZCCUSVTD-UHFFFAOYSA-M 0.000 description 1
- 108091003079 Bovine Serum Albumin Proteins 0.000 description 1
- 241000244203 Caenorhabditis elegans Species 0.000 description 1
- 241000282472 Canis lupus familiaris Species 0.000 description 1
- CURLTUGMZLYLDI-UHFFFAOYSA-N Carbon dioxide Chemical compound O=C=O CURLTUGMZLYLDI-UHFFFAOYSA-N 0.000 description 1
- 208000020446 Cardiac disease Diseases 0.000 description 1
- 206010007572 Cardiac hypertrophy Diseases 0.000 description 1
- 208000006029 Cardiomegaly Diseases 0.000 description 1
- 208000024699 Chagas disease Diseases 0.000 description 1
- CMSMOCZEIVJLDB-UHFFFAOYSA-N Cyclophosphamide Chemical compound ClCCN(CCCl)P1(=O)NCCCO1 CMSMOCZEIVJLDB-UHFFFAOYSA-N 0.000 description 1
- 102000053602 DNA Human genes 0.000 description 1
- 239000003155 DNA primer Substances 0.000 description 1
- VYZAHLCBVHPDDF-UHFFFAOYSA-N Dinitrochlorobenzene Chemical compound [O-][N+](=O)C1=CC=C(Cl)C([N+]([O-])=O)=C1 VYZAHLCBVHPDDF-UHFFFAOYSA-N 0.000 description 1
- 239000006144 Dulbecco’s modified Eagle's medium Substances 0.000 description 1
- 241000196324 Embryophyta Species 0.000 description 1
- 241000588724 Escherichia coli Species 0.000 description 1
- 208000000461 Esophageal Neoplasms Diseases 0.000 description 1
- 241000282326 Felis catus Species 0.000 description 1
- 101710195403 Glutathione S-transferase P Proteins 0.000 description 1
- 101710112365 Glutathione S-transferase P 2 Proteins 0.000 description 1
- 239000007996 HEPPS buffer Substances 0.000 description 1
- 241000238631 Hexapoda Species 0.000 description 1
- 241000282412 Homo Species 0.000 description 1
- 101001033249 Homo sapiens Interleukin-1 beta Proteins 0.000 description 1
- 101100537863 Homo sapiens TRAF2 gene Proteins 0.000 description 1
- 101001050288 Homo sapiens Transcription factor Jun Proteins 0.000 description 1
- 108090000144 Human Proteins Proteins 0.000 description 1
- 102000003839 Human Proteins Human genes 0.000 description 1
- 206010020880 Hypertrophy Diseases 0.000 description 1
- DGAQECJNVWCQMB-PUAWFVPOSA-M Ilexoside XXIX Chemical compound C[C@@H]1CC[C@@]2(CC[C@@]3(C(=CC[C@H]4[C@]3(CC[C@@H]5[C@@]4(CC[C@@H](C5(C)C)OS(=O)(=O)[O-])C)C)[C@@H]2[C@]1(C)O)C)C(=O)O[C@H]6[C@@H]([C@H]([C@@H]([C@H](O6)CO)O)O)O.[Na+] DGAQECJNVWCQMB-PUAWFVPOSA-M 0.000 description 1
- 238000012404 In vitro experiment Methods 0.000 description 1
- 102000000589 Interleukin-1 Human genes 0.000 description 1
- 108010002352 Interleukin-1 Proteins 0.000 description 1
- 102100039065 Interleukin-1 beta Human genes 0.000 description 1
- 102000015696 Interleukins Human genes 0.000 description 1
- 108010063738 Interleukins Proteins 0.000 description 1
- PIWKPBJCKXDKJR-UHFFFAOYSA-N Isoflurane Chemical compound FC(F)OC(Cl)C(F)(F)F PIWKPBJCKXDKJR-UHFFFAOYSA-N 0.000 description 1
- 102000019145 JUN kinase activity proteins Human genes 0.000 description 1
- 208000008839 Kidney Neoplasms Diseases 0.000 description 1
- GUBGYTABKSRVRQ-PICCSMPSSA-N Maltose Natural products O[C@@H]1[C@@H](O)[C@H](O)[C@@H](CO)O[C@@H]1O[C@@H]1[C@@H](CO)OC(O)[C@H](O)[C@H]1O GUBGYTABKSRVRQ-PICCSMPSSA-N 0.000 description 1
- 102000002274 Matrix Metalloproteinases Human genes 0.000 description 1
- 108010000684 Matrix Metalloproteinases Proteins 0.000 description 1
- 208000003445 Mouth Neoplasms Diseases 0.000 description 1
- 108700005084 Multigene Family Proteins 0.000 description 1
- 208000005647 Mumps Diseases 0.000 description 1
- 241000699666 Mus <mouse, genus> Species 0.000 description 1
- 241000699660 Mus musculus Species 0.000 description 1
- 241000699670 Mus sp. Species 0.000 description 1
- 206010028615 Myocarditis septic Diseases 0.000 description 1
- 108020001621 Natriuretic Peptide Proteins 0.000 description 1
- 102000004571 Natriuretic peptide Human genes 0.000 description 1
- 244000061176 Nicotiana tabacum Species 0.000 description 1
- 235000002637 Nicotiana tabacum Nutrition 0.000 description 1
- 102100029438 Nitric oxide synthase, inducible Human genes 0.000 description 1
- 101710089543 Nitric oxide synthase, inducible Proteins 0.000 description 1
- 239000000020 Nitrocellulose Substances 0.000 description 1
- 108091092724 Noncoding DNA Proteins 0.000 description 1
- 108091034117 Oligonucleotide Proteins 0.000 description 1
- 108700020796 Oncogene Proteins 0.000 description 1
- BPQQTUXANYXVAA-UHFFFAOYSA-N Orthosilicate Chemical compound [O-][Si]([O-])([O-])[O-] BPQQTUXANYXVAA-UHFFFAOYSA-N 0.000 description 1
- 206010061535 Ovarian neoplasm Diseases 0.000 description 1
- 239000012807 PCR reagent Substances 0.000 description 1
- 208000002193 Pain Diseases 0.000 description 1
- 229930182555 Penicillin Natural products 0.000 description 1
- JGSARLDLIJGVTE-MBNYWOFBSA-N Penicillin G Chemical compound N([C@H]1[C@H]2SC([C@@H](N2C1=O)C(O)=O)(C)C)C(=O)CC1=CC=CC=C1 JGSARLDLIJGVTE-MBNYWOFBSA-N 0.000 description 1
- 108091005804 Peptidases Proteins 0.000 description 1
- WCUXLLCKKVVCTQ-UHFFFAOYSA-M Potassium chloride Chemical compound [Cl-].[K+] WCUXLLCKKVVCTQ-UHFFFAOYSA-M 0.000 description 1
- 102100038280 Prostaglandin G/H synthase 2 Human genes 0.000 description 1
- 108050003267 Prostaglandin G/H synthase 2 Proteins 0.000 description 1
- 206010060862 Prostate cancer Diseases 0.000 description 1
- 208000000236 Prostatic Neoplasms Diseases 0.000 description 1
- 239000004365 Protease Substances 0.000 description 1
- 229940124158 Protease/peptidase inhibitor Drugs 0.000 description 1
- 108010029485 Protein Isoforms Proteins 0.000 description 1
- 102000001708 Protein Isoforms Human genes 0.000 description 1
- 102100037486 Reverse transcriptase/ribonuclease H Human genes 0.000 description 1
- 102000006382 Ribonucleases Human genes 0.000 description 1
- 108010083644 Ribonucleases Proteins 0.000 description 1
- 240000004808 Saccharomyces cerevisiae Species 0.000 description 1
- 229920005654 Sephadex Polymers 0.000 description 1
- 239000012507 Sephadex™ Substances 0.000 description 1
- 238000012352 Spearman correlation analysis Methods 0.000 description 1
- 238000003646 Spearman's rank correlation coefficient Methods 0.000 description 1
- 208000005718 Stomach Neoplasms Diseases 0.000 description 1
- 238000000692 Student's t-test Methods 0.000 description 1
- 229930006000 Sucrose Natural products 0.000 description 1
- CZMRCDWAGMRECN-UGDNZRGBSA-N Sucrose Chemical compound O[C@H]1[C@H](O)[C@@H](CO)O[C@@]1(CO)O[C@@H]1[C@H](O)[C@@H](O)[C@H](O)[C@@H](CO)O1 CZMRCDWAGMRECN-UGDNZRGBSA-N 0.000 description 1
- 101150113594 TRAF2 gene Proteins 0.000 description 1
- 108010006785 Taq Polymerase Proteins 0.000 description 1
- 208000024313 Testicular Neoplasms Diseases 0.000 description 1
- FOCVUCIESVLUNU-UHFFFAOYSA-N Thiotepa Chemical compound C1CN1P(N1CC1)(=S)N1CC1 FOCVUCIESVLUNU-UHFFFAOYSA-N 0.000 description 1
- 201000005485 Toxoplasmosis Diseases 0.000 description 1
- 102100023132 Transcription factor Jun Human genes 0.000 description 1
- 229920004890 Triton X-100 Polymers 0.000 description 1
- 239000013504 Triton X-100 Substances 0.000 description 1
- 102000009524 Vascular Endothelial Growth Factor A Human genes 0.000 description 1
- 108010073929 Vascular Endothelial Growth Factor A Proteins 0.000 description 1
- 208000027418 Wounds and injury Diseases 0.000 description 1
- JLCPHMBAVCMARE-UHFFFAOYSA-N [3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-[[3-[[3-[[3-[[3-[[3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-hydroxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methyl [5-(6-aminopurin-9-yl)-2-(hydroxymethyl)oxolan-3-yl] hydrogen phosphate Polymers Cc1cn(C2CC(OP(O)(=O)OCC3OC(CC3OP(O)(=O)OCC3OC(CC3O)n3cnc4c3nc(N)[nH]c4=O)n3cnc4c3nc(N)[nH]c4=O)C(COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3CO)n3cnc4c(N)ncnc34)n3ccc(N)nc3=O)n3cnc4c(N)ncnc34)n3ccc(N)nc3=O)n3ccc(N)nc3=O)n3ccc(N)nc3=O)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cc(C)c(=O)[nH]c3=O)n3cc(C)c(=O)[nH]c3=O)n3ccc(N)nc3=O)n3cc(C)c(=O)[nH]c3=O)n3cnc4c3nc(N)[nH]c4=O)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)O2)c(=O)[nH]c1=O JLCPHMBAVCMARE-UHFFFAOYSA-N 0.000 description 1
- 239000008351 acetate buffer Substances 0.000 description 1
- 230000002378 acidificating effect Effects 0.000 description 1
- 201000005180 acute myocarditis Diseases 0.000 description 1
- 229940009456 adriamycin Drugs 0.000 description 1
- 230000002411 adverse Effects 0.000 description 1
- 230000006838 adverse reaction Effects 0.000 description 1
- 238000001261 affinity purification Methods 0.000 description 1
- 229960005260 amiodarone Drugs 0.000 description 1
- 229910021529 ammonia Inorganic materials 0.000 description 1
- 230000033115 angiogenesis Effects 0.000 description 1
- 229940044094 angiotensin-converting-enzyme inhibitor Drugs 0.000 description 1
- 238000000137 annealing Methods 0.000 description 1
- 230000000259 anti-tumor effect Effects 0.000 description 1
- 239000000427 antigen Substances 0.000 description 1
- 108091007433 antigens Proteins 0.000 description 1
- 102000036639 antigens Human genes 0.000 description 1
- 210000001765 aortic valve Anatomy 0.000 description 1
- 238000013459 approach Methods 0.000 description 1
- GOLCXWYRSKYTSP-UHFFFAOYSA-N arsenic trioxide Inorganic materials O1[As]2O[As]1O2 GOLCXWYRSKYTSP-UHFFFAOYSA-N 0.000 description 1
- 229960002594 arsenic trioxide Drugs 0.000 description 1
- 239000012298 atmosphere Substances 0.000 description 1
- QVGXLLKOCUKJST-UHFFFAOYSA-N atomic oxygen Chemical compound [O] QVGXLLKOCUKJST-UHFFFAOYSA-N 0.000 description 1
- 230000035578 autophosphorylation Effects 0.000 description 1
- 230000001580 bacterial effect Effects 0.000 description 1
- 208000022362 bacterial infectious disease Diseases 0.000 description 1
- 208000028535 bacterial myocarditis Diseases 0.000 description 1
- 230000008901 benefit Effects 0.000 description 1
- WQZGKKKJIJFFOK-VFUOTHLCSA-N beta-D-glucose Chemical compound OC[C@H]1O[C@@H](O)[C@H](O)[C@@H](O)[C@@H]1O WQZGKKKJIJFFOK-VFUOTHLCSA-N 0.000 description 1
- GUBGYTABKSRVRQ-QUYVBRFLSA-N beta-maltose Chemical compound OC[C@H]1O[C@H](O[C@H]2[C@H](O)[C@@H](O)[C@H](O)O[C@@H]2CO)[C@H](O)[C@@H](O)[C@@H]1O GUBGYTABKSRVRQ-QUYVBRFLSA-N 0.000 description 1
- 230000036772 blood pressure Effects 0.000 description 1
- 210000001124 body fluid Anatomy 0.000 description 1
- 239000010839 body fluid Substances 0.000 description 1
- 238000009395 breeding Methods 0.000 description 1
- 230000001488 breeding effect Effects 0.000 description 1
- 238000010804 cDNA synthesis Methods 0.000 description 1
- 201000011510 cancer Diseases 0.000 description 1
- 210000005242 cardiac chamber Anatomy 0.000 description 1
- 210000004413 cardiac myocyte Anatomy 0.000 description 1
- 230000001364 causal effect Effects 0.000 description 1
- 238000004113 cell culture Methods 0.000 description 1
- 230000030833 cell death Effects 0.000 description 1
- 230000006192 cellular response to oxidative stress Effects 0.000 description 1
- 239000001913 cellulose Substances 0.000 description 1
- 229920002678 cellulose Polymers 0.000 description 1
- 239000002738 chelating agent Substances 0.000 description 1
- 239000003153 chemical reaction reagent Substances 0.000 description 1
- 229960004630 chlorambucil Drugs 0.000 description 1
- JCKYGMPEJWAADB-UHFFFAOYSA-N chlorambucil Chemical compound OC(=O)CCCC1=CC=C(N(CCCl)CCCl)C=C1 JCKYGMPEJWAADB-UHFFFAOYSA-N 0.000 description 1
- 239000003593 chromogenic compound Substances 0.000 description 1
- 229960004316 cisplatin Drugs 0.000 description 1
- DQLATGHUWYMOKM-UHFFFAOYSA-L cisplatin Chemical compound N[Pt](N)(Cl)Cl DQLATGHUWYMOKM-UHFFFAOYSA-L 0.000 description 1
- 238000003759 clinical diagnosis Methods 0.000 description 1
- 210000001072 colon Anatomy 0.000 description 1
- 208000029742 colonic neoplasm Diseases 0.000 description 1
- 208000029078 coronary artery disease Diseases 0.000 description 1
- 229960004397 cyclophosphamide Drugs 0.000 description 1
- 229940127089 cytotoxic agent Drugs 0.000 description 1
- 231100000599 cytotoxic agent Toxicity 0.000 description 1
- 239000002254 cytotoxic agent Substances 0.000 description 1
- RGWHQCVHVJXOKC-SHYZEUOFSA-J dCTP(4-) Chemical compound O=C1N=C(N)C=CN1[C@@H]1O[C@H](COP([O-])(=O)OP([O-])(=O)OP([O-])([O-])=O)[C@@H](O)C1 RGWHQCVHVJXOKC-SHYZEUOFSA-J 0.000 description 1
- 230000006378 damage Effects 0.000 description 1
- 230000034994 death Effects 0.000 description 1
- 239000000104 diagnostic biomarker Substances 0.000 description 1
- 230000035487 diastolic blood pressure Effects 0.000 description 1
- 235000013681 dietary sucrose Nutrition 0.000 description 1
- 230000004069 differentiation Effects 0.000 description 1
- 230000010339 dilation Effects 0.000 description 1
- XEYBRNLFEZDVAW-ARSRFYASSA-N dinoprostone Chemical compound CCCCC[C@H](O)\C=C\[C@H]1[C@H](O)CC(=O)[C@@H]1C\C=C/CCCC(O)=O XEYBRNLFEZDVAW-ARSRFYASSA-N 0.000 description 1
- 229960002986 dinoprostone Drugs 0.000 description 1
- BNIILDVGGAEEIG-UHFFFAOYSA-L disodium hydrogen phosphate Chemical compound [Na+].[Na+].OP([O-])([O-])=O BNIILDVGGAEEIG-UHFFFAOYSA-L 0.000 description 1
- 229910000397 disodium phosphate Inorganic materials 0.000 description 1
- 238000010494 dissociation reaction Methods 0.000 description 1
- 230000005593 dissociations Effects 0.000 description 1
- 238000009826 distribution Methods 0.000 description 1
- 239000002552 dosage form Substances 0.000 description 1
- 239000003596 drug target Substances 0.000 description 1
- 239000000975 dye Substances 0.000 description 1
- 230000002526 effect on cardiovascular system Effects 0.000 description 1
- 239000012039 electrophile Substances 0.000 description 1
- 238000005538 encapsulation Methods 0.000 description 1
- 239000003623 enhancer Substances 0.000 description 1
- 239000002702 enteric coating Substances 0.000 description 1
- 238000009505 enteric coating Methods 0.000 description 1
- 210000003238 esophagus Anatomy 0.000 description 1
- AVOLMBLBETYQHX-UHFFFAOYSA-N etacrynic acid Chemical compound CCC(=C)C(=O)C1=CC=C(OCC(O)=O)C(Cl)=C1Cl AVOLMBLBETYQHX-UHFFFAOYSA-N 0.000 description 1
- 229960003199 etacrynic acid Drugs 0.000 description 1
- VJJPUSNTGOMMGY-MRVIYFEKSA-N etoposide Chemical compound COC1=C(O)C(OC)=CC([C@@H]2C3=CC=4OCOC=4C=C3[C@@H](O[C@H]3[C@@H]([C@@H](O)[C@@H]4O[C@H](C)OC[C@H]4O3)O)[C@@H]3[C@@H]2C(OC3)=O)=C1 VJJPUSNTGOMMGY-MRVIYFEKSA-N 0.000 description 1
- 229960005420 etoposide Drugs 0.000 description 1
- 230000005713 exacerbation Effects 0.000 description 1
- 239000013604 expression vector Substances 0.000 description 1
- 239000000284 extract Substances 0.000 description 1
- 239000012894 fetal calf serum Substances 0.000 description 1
- 238000000799 fluorescence microscopy Methods 0.000 description 1
- 238000001943 fluorescence-activated cell sorting Methods 0.000 description 1
- 238000004108 freeze drying Methods 0.000 description 1
- 238000007710 freezing Methods 0.000 description 1
- 230000008014 freezing Effects 0.000 description 1
- 208000035474 group of disease Diseases 0.000 description 1
- 244000144993 groups of animals Species 0.000 description 1
- 210000002064 heart cell Anatomy 0.000 description 1
- 230000002209 hydrophobic effect Effects 0.000 description 1
- 230000006607 hypermethylation Effects 0.000 description 1
- 230000001631 hypertensive effect Effects 0.000 description 1
- 238000010874 in vitro model Methods 0.000 description 1
- 238000001727 in vivo Methods 0.000 description 1
- 238000011065 in-situ storage Methods 0.000 description 1
- 238000009776 industrial production Methods 0.000 description 1
- 208000015181 infectious disease Diseases 0.000 description 1
- 230000001524 infective effect Effects 0.000 description 1
- 230000003960 inflammatory cascade Effects 0.000 description 1
- 230000028709 inflammatory response Effects 0.000 description 1
- 208000014674 injury Diseases 0.000 description 1
- 230000000297 inotrophic effect Effects 0.000 description 1
- 229940047122 interleukins Drugs 0.000 description 1
- 238000011835 investigation Methods 0.000 description 1
- 229960002725 isoflurane Drugs 0.000 description 1
- 210000003734 kidney Anatomy 0.000 description 1
- 230000003907 kidney function Effects 0.000 description 1
- 210000004185 liver Anatomy 0.000 description 1
- 210000004072 lung Anatomy 0.000 description 1
- 208000020816 lung neoplasm Diseases 0.000 description 1
- 210000002540 macrophage Anatomy 0.000 description 1
- 229910001629 magnesium chloride Inorganic materials 0.000 description 1
- 238000002595 magnetic resonance imaging Methods 0.000 description 1
- 230000003211 malignant effect Effects 0.000 description 1
- 210000004962 mammalian cell Anatomy 0.000 description 1
- 238000007726 management method Methods 0.000 description 1
- 239000000463 material Substances 0.000 description 1
- 238000000816 matrix-assisted laser desorption--ionisation Methods 0.000 description 1
- 239000002609 medium Substances 0.000 description 1
- 239000012528 membrane Substances 0.000 description 1
- 229910052751 metal Inorganic materials 0.000 description 1
- 239000002184 metal Substances 0.000 description 1
- 239000002480 mineral oil Substances 0.000 description 1
- 235000010446 mineral oil Nutrition 0.000 description 1
- 210000004115 mitral valve Anatomy 0.000 description 1
- 239000003068 molecular probe Substances 0.000 description 1
- 230000003562 morphometric effect Effects 0.000 description 1
- 210000000214 mouth Anatomy 0.000 description 1
- 208000010805 mumps infectious disease Diseases 0.000 description 1
- 210000004898 n-terminal fragment Anatomy 0.000 description 1
- 239000000692 natriuretic peptide Substances 0.000 description 1
- 239000013642 negative control Substances 0.000 description 1
- 230000001613 neoplastic effect Effects 0.000 description 1
- 230000002182 neurohumoral effect Effects 0.000 description 1
- 230000007935 neutral effect Effects 0.000 description 1
- 229920001220 nitrocellulos Polymers 0.000 description 1
- 229910000069 nitrogen hydride Inorganic materials 0.000 description 1
- 238000012316 non-parametric ANOVA Methods 0.000 description 1
- 230000036963 noncompetitive effect Effects 0.000 description 1
- 238000010606 normalization Methods 0.000 description 1
- 238000011330 nucleic acid test Methods 0.000 description 1
- 230000000269 nucleophilic effect Effects 0.000 description 1
- 230000000771 oncological effect Effects 0.000 description 1
- 210000001672 ovary Anatomy 0.000 description 1
- 230000002018 overexpression Effects 0.000 description 1
- 239000001301 oxygen Substances 0.000 description 1
- 229910052760 oxygen Inorganic materials 0.000 description 1
- 230000007310 pathophysiology Effects 0.000 description 1
- 229940049954 penicillin Drugs 0.000 description 1
- 239000000137 peptide hydrolase inhibitor Substances 0.000 description 1
- 230000002093 peripheral effect Effects 0.000 description 1
- 150000002978 peroxides Chemical class 0.000 description 1
- 238000001050 pharmacotherapy Methods 0.000 description 1
- 239000002504 physiological saline solution Substances 0.000 description 1
- 229960001286 piritramide Drugs 0.000 description 1
- 231100000614 poison Toxicity 0.000 description 1
- 229920001184 polypeptide Polymers 0.000 description 1
- 239000001267 polyvinylpyrrolidone Substances 0.000 description 1
- 235000013855 polyvinylpyrrolidone Nutrition 0.000 description 1
- 229920000036 polyvinylpyrrolidone Polymers 0.000 description 1
- 239000013641 positive control Substances 0.000 description 1
- 230000034190 positive regulation of NF-kappaB transcription factor activity Effects 0.000 description 1
- 239000001103 potassium chloride Substances 0.000 description 1
- 239000002244 precipitate Substances 0.000 description 1
- 230000035935 pregnancy Effects 0.000 description 1
- 230000003449 preventive effect Effects 0.000 description 1
- 230000037452 priming Effects 0.000 description 1
- 108010008064 pro-brain natriuretic peptide (1-76) Proteins 0.000 description 1
- 230000000750 progressive effect Effects 0.000 description 1
- 230000008741 proinflammatory signaling process Effects 0.000 description 1
- XEYBRNLFEZDVAW-UHFFFAOYSA-N prostaglandin E2 Natural products CCCCCC(O)C=CC1C(O)CC(=O)C1CC=CCCCC(O)=O XEYBRNLFEZDVAW-UHFFFAOYSA-N 0.000 description 1
- 210000002307 prostate Anatomy 0.000 description 1
- 238000000159 protein binding assay Methods 0.000 description 1
- 239000003531 protein hydrolysate Substances 0.000 description 1
- 239000011535 reaction buffer Substances 0.000 description 1
- 230000000306 recurrent effect Effects 0.000 description 1
- 210000002345 respiratory system Anatomy 0.000 description 1
- 230000000717 retained effect Effects 0.000 description 1
- 238000010839 reverse transcription Methods 0.000 description 1
- 201000007529 rheumatic myocarditis Diseases 0.000 description 1
- 230000000630 rising effect Effects 0.000 description 1
- 229940069575 rompun Drugs 0.000 description 1
- 238000005070 sampling Methods 0.000 description 1
- 238000003118 sandwich ELISA Methods 0.000 description 1
- 201000000306 sarcoidosis Diseases 0.000 description 1
- 201000006832 septic myocarditis Diseases 0.000 description 1
- 230000019491 signal transduction Effects 0.000 description 1
- 239000000779 smoke Substances 0.000 description 1
- 239000011734 sodium Substances 0.000 description 1
- 229910052708 sodium Inorganic materials 0.000 description 1
- AJPJDKMHJJGVTQ-UHFFFAOYSA-M sodium dihydrogen phosphate Chemical compound [Na+].OP(O)([O-])=O AJPJDKMHJJGVTQ-UHFFFAOYSA-M 0.000 description 1
- FQENQNTWSFEDLI-UHFFFAOYSA-J sodium diphosphate Chemical compound [Na+].[Na+].[Na+].[Na+].[O-]P([O-])(=O)OP([O-])([O-])=O FQENQNTWSFEDLI-UHFFFAOYSA-J 0.000 description 1
- 229910000162 sodium phosphate Inorganic materials 0.000 description 1
- 229940048086 sodium pyrophosphate Drugs 0.000 description 1
- 238000013222 sprague-dawley male rat Methods 0.000 description 1
- 238000012453 sprague-dawley rat model Methods 0.000 description 1
- 239000003381 stabilizer Substances 0.000 description 1
- 238000010561 standard procedure Methods 0.000 description 1
- 238000007655 standard test method Methods 0.000 description 1
- 239000008223 sterile water Substances 0.000 description 1
- 210000002784 stomach Anatomy 0.000 description 1
- 229960005322 streptomycin Drugs 0.000 description 1
- 239000000126 substance Substances 0.000 description 1
- 229960004793 sucrose Drugs 0.000 description 1
- 229910052717 sulfur Inorganic materials 0.000 description 1
- 125000004434 sulfur atom Chemical group 0.000 description 1
- 238000011477 surgical intervention Methods 0.000 description 1
- 230000009885 systemic effect Effects 0.000 description 1
- 230000035488 systolic blood pressure Effects 0.000 description 1
- 238000012353 t test Methods 0.000 description 1
- 235000019818 tetrasodium diphosphate Nutrition 0.000 description 1
- 239000001577 tetrasodium phosphonato phosphate Substances 0.000 description 1
- 229960001196 thiotepa Drugs 0.000 description 1
- 230000017423 tissue regeneration Effects 0.000 description 1
- 239000003440 toxic substance Substances 0.000 description 1
- 238000013518 transcription Methods 0.000 description 1
- 230000035897 transcription Effects 0.000 description 1
- 230000002103 transcriptional effect Effects 0.000 description 1
- 238000012546 transfer Methods 0.000 description 1
- 238000011269 treatment regimen Methods 0.000 description 1
- IHIXIJGXTJIKRB-UHFFFAOYSA-N trisodium vanadate Chemical compound [Na+].[Na+].[Na+].[O-][V]([O-])([O-])=O IHIXIJGXTJIKRB-UHFFFAOYSA-N 0.000 description 1
- 239000000439 tumor marker Substances 0.000 description 1
- 208000035408 type 1 diabetes mellitus 1 Diseases 0.000 description 1
- 235000015112 vegetable and seed oil Nutrition 0.000 description 1
- 239000008158 vegetable oil Substances 0.000 description 1
- 239000003981 vehicle Substances 0.000 description 1
- 230000035899 viability Effects 0.000 description 1
- 230000003612 virological effect Effects 0.000 description 1
- QYEFBJRXKKSABU-UHFFFAOYSA-N xylazine hydrochloride Chemical compound Cl.CC1=CC=CC(C)=C1NC1=NCCCS1 QYEFBJRXKKSABU-UHFFFAOYSA-N 0.000 description 1
Images
Classifications
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K38/00—Medicinal preparations containing peptides
- A61K38/16—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- A61K38/43—Enzymes; Proenzymes; Derivatives thereof
- A61K38/45—Transferases (2)
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P9/00—Drugs for disorders of the cardiovascular system
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P9/00—Drugs for disorders of the cardiovascular system
- A61P9/04—Inotropic agents, i.e. stimulants of cardiac contraction; Drugs for heart failure
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P9/00—Drugs for disorders of the cardiovascular system
- A61P9/10—Drugs for disorders of the cardiovascular system for treating ischaemic or atherosclerotic diseases, e.g. antianginal drugs, coronary vasodilators, drugs for myocardial infarction, retinopathy, cerebrovascula insufficiency, renal arteriosclerosis
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P9/00—Drugs for disorders of the cardiovascular system
- A61P9/12—Antihypertensives
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
- C12Q1/6876—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes
- C12Q1/6883—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for diseases caused by alterations of genetic material
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q2600/00—Oligonucleotides characterized by their use
- C12Q2600/158—Expression markers
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N2333/00—Assays involving biological materials from specific organisms or of a specific nature
- G01N2333/90—Enzymes; Proenzymes
- G01N2333/91—Transferases (2.)
- G01N2333/9116—Transferases (2.) transferring alkyl or aryl groups other than methyl groups (2.5)
- G01N2333/91165—Transferases (2.) transferring alkyl or aryl groups other than methyl groups (2.5) general (2.5.1)
- G01N2333/91171—Transferases (2.) transferring alkyl or aryl groups other than methyl groups (2.5) general (2.5.1) with definite EC number (2.5.1.-)
- G01N2333/91177—Glutathione transferases (2.5.1.18)
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N2800/00—Detection or diagnosis of diseases
- G01N2800/32—Cardiovascular disorders
- G01N2800/325—Heart failure or cardiac arrest, e.g. cardiomyopathy, congestive heart failure
-
- Y—GENERAL TAGGING OF NEW TECHNOLOGICAL DEVELOPMENTS; GENERAL TAGGING OF CROSS-SECTIONAL TECHNOLOGIES SPANNING OVER SEVERAL SECTIONS OF THE IPC; TECHNICAL SUBJECTS COVERED BY FORMER USPC CROSS-REFERENCE ART COLLECTIONS [XRACs] AND DIGESTS
- Y02—TECHNOLOGIES OR APPLICATIONS FOR MITIGATION OR ADAPTATION AGAINST CLIMATE CHANGE
- Y02A—TECHNOLOGIES FOR ADAPTATION TO CLIMATE CHANGE
- Y02A50/00—TECHNOLOGIES FOR ADAPTATION TO CLIMATE CHANGE in human health protection, e.g. against extreme weather
- Y02A50/30—Against vector-borne diseases, e.g. mosquito-borne, fly-borne, tick-borne or waterborne diseases whose impact is exacerbated by climate change
Definitions
- the present invention relates to the use of Glutathione-S-transferase P1.
- GSTs The glutathione S-transferases (GSTs) are a multigene family of isozymes that catalyze the nucleophilic attack of the sulfur atom of glutathione (GSH) on electrophilic groups of substrate molecules. GSTs are known as detoxicating enzymes which conjugate toxic substances with GSH to become more water-soluble products which can be metabolised in the liver and finally excreted out of the body. On the basis of the amino acid sequence, the mammalian GSTs are divided into six classes: alpha, mu, omega, pi, theta and zeta.
- glutathione S-transferase P1 (GSTP1; GST-pi, GSTP1-1) is the most prevalent one in mammalian cells.
- GSTP1 has also generated interest as a neoplastic marker because it is overexpressed in a variety of different human malignancies, including lung, colon, stomach, esophagus, mouth, kidney, ovary and testicular cancers.
- Most prostate cancer types contain decreased levels of GSTP1 polypeptide relative to normal prostate tissue due to hypermethylation of the GSTP1 promoter resulting in decreased transcription of GSTP1 ( US 2009/0186360 A1 ).
- GSTP1 is determinant in cellular response to oxidative stress and protects tumor cells from apoptosis elicited by a variety of cytotoxic agents, such as H 2 O 2 , UV, cisplatin, adriamycin, etoposide, thiotepa, chlorambucil, ethacrynic acid and arsenic trioxide.
- cytotoxic agents such as H 2 O 2 , UV, cisplatin, adriamycin, etoposide, thiotepa, chlorambucil, ethacrynic acid and arsenic trioxide.
- GSTP1 is therefore considered as a promising target in tumour medicine, both as a target for drug action and as a tumor marker.
- GSTP1 also participates in the regulation of stress signalling and protects cells against apoptosis by the mechanisms related to its non-catalytic and ligand-binding activities. For example, GSTP1 prevents LPS-induced excessive production of pro-inflammatory factors and plays an anti-inflammatory role in response to LPS. GSTP1 expression, both at transcriptional and translational levels, is up-regulated by LPS stimulation. GSTP1 also functions as an endogenous inhibitor of JNK (c-Jun NH 2 -terminal kinase), via interaction with the C-terminus. It could also be demonstrated that exogenous (recombinant) GSTP1 protein could be delivered into macrophages and suppressed iNOS and COX-2 expression in cells. Furthermore, intraperitoneally administered GSTP1 protein to mice decreased mortality of endotoxic shock significantly and inhibited acute lung injury and peritonitis ( Luo et al., Mol. Immunol. 46 (2009), 848-857 ).
- EP 1 500 709 A1 suggests GSTP1 as one of 22 inflammation markers.
- Cardiomyopathies are a heterogeneous group of diseases, including the dilative (DCM), the most frequent entity among cardiomyopathies, and ischemic (ICM) forms.
- DCM dilative
- ICM ischemic
- results of the last decade's work clearly provide evidence that inflammation and enhanced release of pro-inflammatory cytokines, especially tumor necrosis factor (TNF)- ⁇ might play a role in the pathogenesis of CMP.
- TNF tumor necrosis factor
- IHD The pathology of IHD is immediately related to the development of CMP, patients currently receive medications such as beta-blockers or angiotensin converting enzyme (ACE)-blockers with the goal to ameliorate the patient's symptoms and to prevent or delay the development of (ischemic) CMP due to IHD. It is therefore common to use therapies for CMP also for IHD.
- therapies for CMP also for IHD.
- patients who develop CMP based on IHD in particular end-stage CMP, have currently only limited therapeutic options (e.g. cardiac transplantation or the use of ventricular assist devices, which can be in turn offered only to a limited number of patients).
- ischemic heart diseases also called coronary heart diseases that can result in myocardial infarction
- elevated blood pressure-induced CMP and volume-induced CMP especially elevated blood pressure-induced CMP and volume-induced CMP.
- Cardiomyopathies result in recurrent exacerbation leading to hospitalization or death. Therefore, also close surveillance and thorough clinical tests are required to achieve the optimal tailored cardiomyopathy treatment in these patients, associated with tremendous health care costs.
- a biomarker that could identify the evolution of cardiomyopathy and help to optimize treatment monitoring and outcome is therefore highly desirable. Ideally, such a test should be sensitive, specific, none-invasive, quick and cheap.
- BNP brain-type natriuretic peptide
- proBNP inactive N-terminal fragment
- circulating BNP and proBNP levels are dramatically affected by renal function and are age- and gender-dependent. In addition, it remains unknown whether proBNP could be beneficial in diagnosis of preserved as opposed to reduced ejection fraction (EF) CMP.
- the invention provides Glutathione S-transferase P1 (GSTP1) for use in the prevention or treatment of cardiomyopathies (CMPs).
- CMPs cardiomyopathies
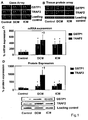
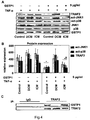
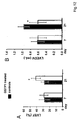
- GSTP1 is beneficial in CMP treatment.
- GSTP1 has been described as a relevant protein in cancer medicine (e.g. US 5,427,917 A , US 5,552,277 A and WO 98/21359 A1 ) and in allergic diseases, such as asthma, especially in connection with traffic-related air pollution (e.g. Melen et al., Env. Health Persp. 116 (2008), 1077-1084 ).
- GSTP1 has also been suggested to decrease mortality of endotoxic shock and inhibition of acute lung injury and peritonitis ( Luo et al., Mol. Immunol. 46 (2009), 848-875 ).
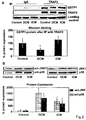
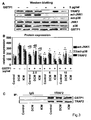
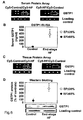
- GSTP1 was reported to interact with TRAF2 (tumor necrosis factor receptor-associated factor 2) as a novel ligand-binding function of GSTP1 in regulating kinase signalling ( Wu et al., Oncogene 25 (2006), 5787-5800 ).
- GSTP1 is specifically upregulated in CMP patients as compared to controls. It was further found that recombinant GSTP1 mediated GSTP1-TRAF2 association, decreased pro-inflammatory JNK1 and p38 activation and TRAF2 expression dependent on its dose and the underlying form of CMP. This enabled the provision of a new therapy strategy for CMP by administration of GSTP1. Since GSTP1 ameliorates TNF- ⁇ -mediated activation of pro-inflammatory JNK1/p38 by association to TRAF2, GSTP1 is not only beneficial in CMP treatment but also for the prevention of CMP, especially in risk patients, such as hypertensive patients.
- the present invention provides the use of GSTP1 for the manufacture of a medicament for the prevention or treatment of CMP.
- the treatment of CMP according to the present invention involves the administration of an effective amount of GSTP1 to an individual in need of such treatment, e.g. human CMP patients.
- an individual in need of such treatment e.g. human CMP patients.
- patients currently receive medications such as beta-blockers or angiotensin converting enzyme (ACE)-blockers with the goal to ameliorate the patient's symptoms and to prevent or delay the development of (ischemic) CMP due to IHD.
- ACE angiotensin converting enzyme
- CMPs can be caused by myocardial infarction, can be idiopathic dilative or be caused by hypertension. All these forms can be treated with GSTP1 according to the present invention.
- ischemic CMPs e.g. caused by myocardial infarction can be treated with GSTP1
- non-ischemic forms can be treated (e.g. idiopathic dilated CMPs or hypertension- or volume-induced CMPs). Since in all these CMPs, inflammatory pathways are involved, this makes them eligible for treatment with GSTP1 according to the present invention (even rare forms of CMP, if such inflammatory pathways are involved).
- preferred CMPs are dilated cardiomyopathy, especially congestive cardiomyopathy; obstructive hypertrophic cardiomyopathy, especially hypertrophic subaortic stenosis; other hypertrophic cardiomyopathies, especially nonobstructive hypertrophic cardiomyopathy; endomyocardial (eosinophilic) disease, especially endomyocardial (tropical) fibrosis or Löffler's endocarditis; endocardial fibroelastosis, especially congenital cardiomyopathy; other restrictive cardiomyopathies, especially constrictive cardiomyopathy NOS (not otherwise specified (according to ICD-10)); alcoholic cardiomyopathy, cardiomyopathies due to drugs and other external agents, unspecified cardiomyopathies, especially cardiomyopathy (primary)(secondary) NOS; cardiomyopathy in infectious and parasitic diseases, especially cardiomyopathy in diphtheria; cardiomyopathy in metabolic diseases, especially cardiac amyloidosis; cardiomyopathy in nutritional diseases, especially nutritional diseases, especially nutritional diseases,
- CMPs to be treated (or prevented) according to the present invention are CMPs caused by ischemia, especially by myocardial infarction; by hypertension, or by myocarditis.
- GSTP1 treatment according to the present invention is therefore preferably applied in patients with acute myocarditis, preferably infective myocarditis, especially septic myocarditis; isolated myocarditis, myocarditis in a bacterial disease, especially diphtheritic, gonococcal, meningococcal, syphilitic or tuberculous myocarditis; myocarditis in a viral disease, especially influenzal myocarditis or mumps myocarditis; acute or chronic myocarditis in Chagas' disease, myocarditis in toxoplasmosis, rheumatoid myocarditis or sarcoid myocarditis.
- the preferred indications according to the present invention are therefore those listed under Chapter IX of the ICD-10 (I11-I15, especially I21, I22 and I23; I20-I25, especially I21 and I22; I40-I43, especially I42; I50-I57, especially I50 and I57).
- the main focus of the present invention is the treatment of human patients; however, based on the present findings, it is clear that also animal (mammal) CMP in which inflammation is involved can be successfully treated by GSTP1 according to the present invention.
- This may be important for farm or zoo animals (especially breeding farm animals, such as (former) racing horses) or pets, for example dogs, cats and horses.
- the present invention is also useable for the prevention of CMP it is clear that the primary focus of the present invention lies in the treatment of human CMP patients who have already been diagnosed of CMP or which have a high risk of developing CMP, for example patients with hypertension (e.g. Stage 2 patients with systolic pressure of 160 mm Hg or more and/or with diastolic pressure of 100 mm Hg or more).
- Another preferred embodiment of the preventive aspect of the present invention is for angina pectoris patients, whereby myocardial infarction might or might not have occurred in this set of patients; and heart insufficiency or heart failure ( Mc Murray et al., Lancet 365 (2005), 1877-1889 ) may or may not be present.
- heart insufficiency or heart failure Mc Murray et al., Lancet 365 (2005), 1877-1889
- Preferred routes of administration of the GSTP1 containing medicament according to the present invention are parenteral routes, preferably intraperitoneal or intravenous administration, intravenous administration being specifically preferred.
- Intravenous administration can be performed e.g. via bolus injection or by continuous intravenous delivery over a longer time period (e.g. 30 min to 6 h, especially 1 to 3 h).
- administration directly to the heart in-tramyocardial injection
- GSTP1 activity can be directly and in high concentrations delivered to the area of need, e.g. to an infarct region.
- Another preferred route of administration is administration to the coronary sinus (e.g. via a catheter inserted into the coronary sinus).
- GSTP1 Oral or mucosal routes (although described for GSTP1 in principle; see e.g. US 5,976,528 A ) are less preferred, since GSTP1 is a protein and for this route specific protection measures (enteric coating, encapsulation, etc.) are necessary as well as a significant optimisation with respect to galenic manufacture.
- Preferred dosages for administration are dosages of 0.001 to 100 mg GSTP1/kg, preferably 0.01 to 10 mg GSTP1/kg, especially 0.1 to 1 mg GSTP1/kg, to a human individual, preferably via intravenous administration; or dosages of 0.1 to 10000 U GSTP1/kg, preferably 1 to 1000 U GSTP1/kg, especially 10 to 100 U GSTP1/kg, to a human individual, preferably via intravenous administration.
- These are preferred daily dosages for intravenous application (“kg” means kg body weight of the person to be treated). During surgery, these dosages may also be applied, either as a whole or even more than one dosage; or only part of the dosage (having in mind that the GSTP1 can directly be applied during surgery in the area needed).
- Ready-to-use daily dosage forms (e.g. for a 80 kg and/or a 60 kg human individual) can be pre-manufactured and can be made storage-stable by lyophilisation or freezing (and keeping at e.g. -20°C). The dosage can then be fully or only partially consumed (e.g. based on the body weight of the person to be treated).
- the human sequence (Seq. ID. No. 1) is listed as “GSTP1_HUMAN, P09211" in the UniProtKB/SwissProt database (HGNC: 4638, Entrez Gene: 2950).
- variants exist for this human protein (homozygous as well as heterozygous), the most important variants are a Ile to Val exchange at position 105 and a Ala to Val exchange at position 114 (Melen et al., 2008).
- Other variants include a Gly to Glu exchange at position 78, a Thr to Ser exchange at position 110, a Gly to Glu exchange at position 139, a Asp to Tyr exchange at position 147 and a Asp to His exchange at position 158.
- SNPs are known without affecting the amino acid sequence.
- the CMP patient to be treated with GSTP1 according to the present invention has such an allele which results in a (heterozygous or homozygous) amino acid exchange compared to SEQ.ID.No. 1, it may be recommendable to administer to such a patient the allelic form of this protein.
- This is specifically preferred for patients who are homozygous for this allele, e.g. patients with homozygous 105Val or 114Val allele (who could then receive the 105Val or 114Val GSTP1 variant, respectively).
- the risk of developing adverse reactions, especially adverse immune reactions (which is always a topic for proteinaceous medicaments), is kept low.
- any ingredients in the pharmaceutical preparation containing GSTP1 according to the present invention eliciting or boosting an immune reaction in the patient should be kept at minimum if possible.
- Orthologs from mammals are known, e.g. from chimpanzee (Pan troglodytes; NCBI accessions: 745954, XM_001152516.1, XP_001152516.1) and mouse (Mus musculus; NCBI accessions: 148701, NM_013541.11, NP_038569.11, AK0791445, BC0020485), but also from non-mammals, such as zebrafish (Danio rerio) and worm (Caenorhabditis elegans).
- the present invention also relates to a pharmaceutical composition
- a pharmaceutical composition comprising GSTP1, preferably human recombinant GSTP1 or human placenta GSTP1, and a pharmaceutically acceptable carrier for the prevention or treatment of cardiomyopathies or ischemic heart diseases.
- the composition according to the present invention contains 1 to 100000 U GSTP1, preferably 10 to 10000 U GSTP1, especially 10 to 1000 U GSTP1.
- a unit of GSTP1 activity conjugates 1.0 micro-mole of 1-chloro-2,4-dinitrobenzene with reduced glutathione per minute at pH 6.5 at 25°C.
- a preferred composition according to the present invention also contains a buffer.
- Typical buffer systems are the carbonic acid/HCO 3 system (pH 6,2 to 8,6; neutral), carbonic acid/silicate buffer (pH 5,0 to 6,2; weakly acidic), acetic acid/acetate buffer (pH 3,7 to 5,7), phosphate buffer (NaH 2 PO 4 + Na 2 HPO 4 ; pH 5,4 to 7,8), ammonia buffer (NH 3 + H 2 O + NH 4 Cl; pH 8,2 to 10,2), TRIS/HCl (Tris(hydroxymethyl)-aminomethane; pH 7,2 to 9,0), HEPES (4-(2-Hydroxyethyl)-1-piperazinethanesulfonic acid; pH 6,8 to 8,2), HEPPS (4-(2-Hydroxyethyl)-piperazine-1-propane sulfonic acid; pH 7,3 to 8,7) or MES (2-(N-Morpholino)ethanesulfonic
- Preferred buffers according to the present invention is a phosphate buffer, a Tris-HCl buffer or a HEPES buffer, with a pH of 5.5 to 9.0, preferably of 6.0 to 8.5, especially of 6.5 to 8.0. Buffer concentrations can preferably be adjusted to 1 mM to 1 M, especially 10 mM to 0.5 M.
- Other preferred additional ingredients include stabilisers, chelators, salts, etc., such as glycerol, glucose, saccharose, maltose, EDTA or similar substances, NaCl, KCl, NH 4 Cl and similar salts usable in pharmaceutical preparations. Of course, all ingredients in a pharmaceutical preparation have to be of pharmaceutical grade quality.
- Pharmaceutically acceptable carriers preferably used are physiological saline, vegetable oils, mineral oil, aqueous sodium caroboxymethyl cellulose or aqueous polyvinylpyrrolidone; however, also sterile water can be used.
- GSTP1 to be used according to the present invention can be provided by various ways. As mentioned above, GSTP1 from human placenta is one of the preferred natural sources of GSTP1. Other preferred sources are cultures of GSTP1 (over-)expressing cells. GSTP1 preparations are also commercially available. The preferred industrial production route according to the present invention is, however, recombinant production of GSTP1. Recombinant production of GSTP1 is well established, as well as its purification from such sources (e.g. Luo et al., 2009, Wu et al., 2006; WO 98/21359 A1 ; US 5,976,528 ; etc.). Expression vectors (e.g.
- WO 98/21359 A1 , Part D (pages 40 to 51) containing the GSTP1 coding sequences are transferred in suitable host cells (such as COS, HEK 293, HeLa, VERO, W138, BHK or MDCK cells, but also yeast, plant, insect and bacterial cells (S. cerevisiae, E. coli, etc.).
- GSTP1 is then expressed and purified according to standard methods. Recombinant GSTP1 production may also use variant forms of GSTP1 with additional sequences which facilitate purification, such as metal chelating peptides, protein A domains, affinity purification tags, etc. (e.g. US 5,976,528 A , column 16).
- Such variants can - after purification or in the course of purification - be removed e.g. via cleavable linker sequences (protease, entereokinase, etc.).
- Purified batch preparations of GSTP1 may contain e.g. 1 to 200 U GSTP1 activity per mg total protein.
- Preferred pharmaceutical GSTP1 compositions contain 5 to 200 U/mg protein, especially 10 to 150 U/mg protein.
- the pharmaceutical preparation according to the present invention is always provided as an appropriately labelled product which is sterile and useable for administration to human patients according to the present invention.
- the present invention relates to GSTP1 as a diagnostic target for CMP.
- the present invention provides the use of GSTP1 for the diagnosis of CMP.
- GSTP1 is a known and established marker in tumor diagnosis, it was surprising that GSTP1 is a biomarker for CMP which is even superior to the current "gold standard", pro-BNP, with respect to sensitivity and specificity.
- the cardiomyopathies referred to above for therapeutical use of GSTP1 can also be diagnosed according to the present invention.
- the present invention therefore relates to a method for diagnosing CMP, especially the indications mentioned above, by determining the amount of GSTP1 or GSTP1 mRNA in a biological sample and comparing this amount to the GSTP1 or GSTP1 mRNA amount in a biological sample with a defined and known CMP status, especially from a healthy human individual or a patient.
- This comparison can be done practically, i.e. in that a real sample is tested in the same manner as the biological sample (side-by-side), it can also be a virtual comparison so that the determined value is compared with a known value of a healthy or diseased sample.
- Preferred techniques for determining the amount of GSTP1 include antibody-liked techniques, such as ELISA, optionally combined with electrophoresis techniques or radio immunoassay (RIA). Electrophoresis techniques may also be combined with MALDI techniques.
- GSTP1 mRNA may be determined, e.g. by quantitative real time RT-PCR.
- the preferred diagnosis target is a human patient who is suspected to have CMP, a human patient who is at risk for developing CMP or a human patient who has CMP (who has to be monitored for progression of the disease).
- Preferred biological samples according to the present invention include human blood, plasma or serum samples or human myocardial biopsy samples. These samples can also be tested by routine methods.
- a high concentration of GSTP1 in serum is indicative of CMP.
- serum concentrations of GSTP1 are below 50 ng/ml or, preferably, even below 10 ng/ml.
- a CMP diseased status can be diagnosed at a serum level at 50 ng/ml or above, preferably at 100 ng/ml or above. Severe forms of CMP can be diagnosed at 200 ng/ml or above. Of course, these levels always depend on the technique used for assaying the GSTP1 protein.
- CMP diseased status can be diagnosed if the serum level of GSTP1 is elevated by at least the factor 2, preferably by at least the factor 5, compared to a healthy status, especially the healthy status of the same patient. Severe forms could be diagnosed, if the serum GSTP1 level is elevated by at least the factor 10, especially by at least the factor 20, compared to a healthy status, especially the healthy status of the same patient.
- determining the amount of GSTP1 or GSTP1 mRNA in a sample can be performed within a very short time by standard test methods, such as ELISA or PCR.
- standard test methods such as ELISA or PCR.
- the present invention Based on the DNA and amino acid sequence according to SEQ.ID.NO.2 and SEQ.ID.NO.1 (and the data base entries mentioned above for the GSTP1 gene and the GSTP1 protein), the present invention also provides e.g. a PCR assay for identification of S. aureus infections or a sandwich ELISA assay.
- GSTP1 can be caught out of a biological sample with a GSTP1-specific antibody and detected and quantified with a second antibody directed against a different epitope of GSTP1 or an antibody being specific for the previous binding event.
- the present invention therefore also provides a kit for determining the amount of GSTP1 mRNA in a least two synthetic nucleic acid sequences for amplifying at least one nucleic acid sequence encoding GSTP1 or parts thereof, wherein at least of one of the synthetic nucleic acid sequences in the kit is selected from the group consisting of: (a) a suitable primer pair, the primers being a nucleic acid sequence comprising 10-30 consecutive nucleotides of at least one of the following: (i) the GSTP1 mRNA according to SEQ.ID.NO.2 or the complementary sequence thereof; or (ii) from the 5' or 3' noncoding region of the GSTP1 gene (e.g.
- primers being usable for polymerising a nucleotide sequence located between the primers on the GSTP1 gene or its 5'- or 3'-region (said polymerizable GSTP1 nucleotide sequence being PCR detectable and preferably up to 1000 bp, more preferred up to 500 bp, even more preferred up to 300bp in length; on the other hand, the primers may also be selected to polymerise (with PCR) the whole GSTP1 m-RNA); and (b) suitable reagents for a polymerase chain reaction (PCR).
- PCR polymerase chain reaction
- the kit further comprises: (c) instructions for use and (d) optionally, a positive and/or negative control for determining and/or quantification of GSTP1 mRNA, especially a control nucleic acid encoding GSTP1 or a fragment thereof.
- PCR reaction buffer examples include PCR reaction buffer, Mg 2+ (e.g., MgCl 2 ), dNTPs, DNA polymerases (such as reverse transcriptases and thermostable DNA polymerases (e.g., Taq-related DNA polymerases and Pfu-related DNA polymerases)), RNase, PCR reaction enhancers or inhibitors, PCR reaction monitoring agents (e.g., double-stranded DNA dye (such as SYBRTM Green), TaqManTM probes, molecular beacons, and Scorpion-sTM), and PCR-grade water.
- DNA polymerases such as reverse transcriptases and thermostable DNA polymerases (e.g., Taq-related DNA polymerases and Pfu-related DNA polymerases)
- RNase e.g., RNase
- PCR reaction enhancers or inhibitors e.g., RNase, PCR reaction enhancers or inhibitors
- PCR reaction monitoring agents e.g., double
- PCR polymerase chain reaction
- a PCR assay may use a heat-stable polymerase and two about 10 to 30 base primers: one complementary to the (+)-strand at one end of the sequence to be amplified, and the other complementary to the (-)-strand at the other end.
- the newly-synthesized biological sample comprising: (a) a primer set comprising at DNA strands can subsequently serve as additional templates for the same primer sequences, successive rounds of primer annealing, strand elongation, and dissociation may produce rapid and highly-specific amplification of the desired sequence.
- PCR also may be used to detect the existence of a defined sequence in a GSTP1 m-RNA containing sample.
- a typical PCR assay according to the present invention might start - optionally after reverse transcription of the mRNA into DNA - with two synthetic oligonucleotide primers which are specifically and complementarily binding to two regions of the target DNA or its complementary strand, respectively, encoding GSTP1 or its 5'- and/or 3' region (one for each strand) that is to be amplified. These may be added to the target DNA (that needs not be pure) in the presence of excess deoxynucleotides (dNTPs) and a thermostable DNA polymerase (e.g., Taq polymerase).
- dNTPs deoxynucleotides
- Taq polymerase e.g., Taq polymerase
- the target DNA may be repeatedly denatured (about 80-100°C, e.g. 90°C), annealed to the primers (typically at 40-65°C), and a daughter strand may be extended from the primers (typically at 65-80°C, e.g. 72°C).
- PCR is specifically suitable in the clinical diagnostics according to the present invention.
- These tests may preferably employ labels which are also suitable for quantification, such as biotin, fluorescent molecules, radioactive molecules, chromogenic substrates, chemiluminescence markers, and the like.
- labels which are also suitable for quantification, such as biotin, fluorescent molecules, radioactive molecules, chromogenic substrates, chemiluminescence markers, and the like.
- the methods for biotinylating nucleic acids are well known in the art, as are methods for introducing fluorescent molecules and radioactive molecules into oligonucleotides and nucleotides.
- Detection methods are well known for fluorescent, radioactive, chemiluminescent, chromogenic labels, as well as other commonly used labels. Briefly, chemiluminescence can be identified and quantitated most directly by their emission wavelengths and intensity.
- biotin When biotin is employed, it is detected by avidin, streptavidin or the like, which is conjugated to a detectable marker, such as an enzyme (e.g. horseradish peroxidase).
- a detectable marker such as an enzyme (e.g. horseradish peroxidase).
- Steptavidin binds with high affinity to biotin, unbound streptavidin is washed away, and the presence of horseradish peroxidase enzyme is then detected using a luminescence-emission substrate in the presence of peroxide and appropriate buffers.
- Determining the amount of GSTP1 in a sample preferably includes ELISA, RIA and FACS. Detection and quantification of GSTP1 by using anti GSTP1 antibodies is well established in the art (especially in tumor medicine) and can be readily applied for the purpose of diagnosing CMP according to the present invention (e.g. WO 98/21359 A1 ("F. Antibodies”); US 5,976,528 A , US 5,427,917 A ). Normal or standard values for GSTP1 expression are established by combining body fluids or cell extracts taken from normal mammalian subjects, preferably human, with antibody to GSTP1 under conditions suitable for complex formation. The amount of standard complex formation may be quantified by various methods, preferably by photometric means.
- Figure 1 shows increased GSTP1 and TRAF2 in DCM and ICM patients.
- A Representative gene array images show increased hybridization to GSTP1 and TRAF2 array sequences (arrows) of labelled cDNA probes.
- B Representative protein array images demonstrating enhanced Cy5 (red) staining for cardiac GSTP1 and TRAF2 proteins in DCM and ICM patients compared to the Cy3 (green) staining of controls.
- C Quantification of myocardial GSTP1 and TRAF2 mRNA expressions by real time RT-PCR.
- GSTP1 (*P ⁇ 0.0001) and TRAF2 (P ⁇ 0.0001) mRNA expression is significantly up-regulated in heart failure patients compared to controls.
- FIG. 1 Representative Western blot images and quantification of myocardial GSTP1 and TRAF2 protein expressions corrected for protein loading control levels.
- FIG. 2 shows GSTP1-TRAF2 interaction and activation of JNK and p38 in DCM and ICM.
- A Representative Western blot images and quantification of myocardial GSTP1-TRAF2 complex formation in DCM, ICM and controls. LV myocardial lysates were immunoprecipitated (IP) and precipitates were analyzed on Western blotts. Significantly lower GSTP1-TRAF2 association was found in DCM compared to ICM and controls (*P ⁇ 0.008).
- FIG. 3 shows that GSTP1 modulates cardiac TRAF2-JNK1/p38 system in cardiac tissue cultures.
- A Representative Western blot images of TRAF2, GSTP1, active JNK and active p38 protein expressions in cardiac tissue cultures treated with 5 ⁇ g/mL recombinant GSTP1.
- B Quantification of TRAF2, active JNKs and active p38 protein expressions in DCM, ICM and control cardiac tissue cultures treated with recombinant GSTP1 (2.5, 5, 10 ⁇ g/ml) for 24 h.
- FIG. 4 shows that TNF- ⁇ abrogates sensitivity of TRAF2-JNK1/p38 cascade to GSTP1 in DCM cardiac cultures.
- A Representative Western blot images and quantification
- B Representative Western blot images and quantification
- FIG. 5 shows that higher GSTP1 concentrations rescue TRAF2-mediated JNK1/p38 downregulation following TNF- ⁇ treatment.
- A Representative Western blot images and quantifications (B) of DCM, ICM and control cardiac tissue cultures treated with TNF- ⁇ (50 ng/ml) and GSTP1 (10 ⁇ g/ml).
- B The results demonstrate that DCM cardiac tissue cultures treated with GSTP1 at 10 ⁇ g/mL and TNF- ⁇ express markedly reduced JNK and p38 activation as well as reduced TRAF2 protein expression (*P ⁇ 0.0001) as compared to TNF- ⁇ -treated controls.
- C The immunoprecipitates of DCM cardiac tissue lysates demonstrate significant (P ⁇ 0.035) increase of GSTP1-TRAF2 association at 10 ⁇ g/ml GSTP1 as compared to 5 mg/ml GSTP1.
- Figure 6 shows that serum and cardiac GSTP1 is associated with CMP.
- D Representative Western blotting images and quantification of myocardial GSTP1 protein expression corrected for protein loading control levels. GSTP1 protein levels were significantly elevated in end-stage HF patients with EF ⁇ 35% versus control cardiac graft tissue (*P ⁇ 0.001).
- Figure 7 shows serum GSTP1 and proBNP association with ejection fraction (EF) and the correlation between GSTP1 and proBNP.
- EF ejection fraction
- Figure 8 shows serum GSTP1 and proBNP correlation to cardiac function.
- Figure 9 shows that GSTP1 inhibits the inflammatory cytokines in a rat acute MI model.
- A Immunoprecipitation with Western blotting revealed no differences in GSTP1-TRAF2 complex formation between GSTP1-treated and control animals;
- B GSTP1 decreased activated JNK1 protein expression levels as analyzed by Western blotting;
- C GSTP1 inhibits the myocardial tissue mRNA expression of several inflammatory cytokines. *; p ⁇ 0.01.
- FIG. 10 shows that GSTP1 ameliorates inflammation in the rat failing heart.
- GSTP1-treated animals had significantly higher levels of complex formation between GSTP1-TRAF2, GSTP1-JNK1 and GSTP1-p38, as shown by immunoprecipitation and Western blot analyses;
- the protein expression of activated JNK1, p38 and NF- ⁇ B is significantly lower in GSTP1-treated animals as compared to controls as shown by Western blot analyses;
- GST-P1 inhibits the mRNA expression of inflammatory cytokines TGF-B, IL-1B, IL-2 and IL-17; and increases the mRNA expression of anti-inflammatory cytokine IL-10 in failing myocardium as compared to controls. *; p ⁇ 0.001.
- Figure 11 shows that GSTP1 attenuates myocardial remodeling in a rat ischemia-induced model of heart failure.
- A Goldner trichrome collagen staining reveals significantly lower tissue remodeling in the heart of GSTP1-treated animals as compared to controls;
- B left ventricular (infarct) wall thickness is significantly larger in GSTP1-treated rats compared to controls;
- GSTP1-treated rats had a significantly lower apoptotic index as compared to controls. *; p ⁇ 0.01.
- Figure 12 shows that GSTP1 improves the left ventricular function in a rat model of ischemia-induced heart failure.
- A GSTP1-treated rats had significantly higher left ventricular ejection fraction at 21 days post-myocardial infarct;
- B the left ventricular end diastolic volume is significantly greater in controls as compared to GSTP1-treated animals. *; p ⁇ 0.05.
- ACE angiotensin converting enzyme
- BMI body mass index
- BSA body surface area
- DCM dilated cardiomyopathy
- ICM ischemic cardiomyopathy
- LVEF left ventricular ejection fraction
- PAP pulmonary artery pressure
- PCWP pulmonary capillary wedge pressure
- PVR pulmonary vascular resistance.
- Poly(A+)-RNA was isolated with the Oligotex-dT kit (Quiagen, Valencia CA) and first-strand cDNA synthesis was performed with avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI) on 2 ⁇ g poly(A+) RNA. The RNA strand within the DNA-RNA duplex was degraded and products were purified on a Sephadex G-50 spun-column (Pharmacia, Uppsala, Sweden).
- first-strand cDNA was used to generate [ ⁇ -32P]dCTP-labeled second-strand cDNA for the cDNA arrays (GEArray Q Human Apoptosis Gene Array, SuperArray Bioscience, Frederick, MD) as described ( Shufer et al., Circulation 108 (2003), 1585-1591 ). Pooled cDNA hybridization signals were quantified using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
- the protein array was performed on antibody Microarray 500 (Clontech, Mountain View, CA; 507 proteins) as described ( Aharinejad et al., Circulation 120 (2009a), 11 Supp1:S198-205 ).
- the proteins in each sample were labeled with 2 different fluorescent dyes and incubated on microarray antibody-coated slides.
- the protein fluorescent signals of both slides were detected by the GenePix 4000B scanner (Molecular Dynamics, Sunnyvale, CA).
- the quantification of the signal was performed by calculating an internally normalized ratio (INR), yielding the abundance of an antigen in CMP sample relative to that in control samples by using the automated Microarray Analysis Workbook (http://bioinfo.clontech.com). Proteins with INR values outside the threshold interval were considered differentially expressed.
- Human cardiac tissue was lysed in the lysis buffer containing 20 mM Tris (pH 7.5), 135 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 2 mM dithiothreitol (DTT), 25 mM ⁇ -glycerophosphate, 2 mM sodium pyrophosphate, 10% glycerol, 1% Triton X-100, 1 mM sodium orthovanadate, 10 mM NaF and 1 mM phenylmethylsulfonyl fluoride (PMSF) supplemented with complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA) at 4 C as described (Aharinejad et al., 2005).
- Tris pH 7.5
- EDTA ethylenediaminetetraacetic acid
- DTT dithiothreitol
- PMSF phenylmethylsulfonyl fluoride
- Lysates were centrifuged (15 000 g) at 4 C for 15 min.
- GSTP1-TRAF2 complexes formation proteins 500 ⁇ g were immunoprecipitated with TRAF2 antibody (0.5 ⁇ g, BD Pharmingen, San Diego, CA).
- TRAF2 antibody 0.5 ⁇ g, BD Pharmingen, San Diego, CA.
- the precleared Protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) were incubated with immunocomplexes for another 2 h and washed four times with the lysis buffer.
- the immunoprecipitates were further subjected on SDS-PAGE and presided with Western blotting analysis using anti-GSTP1 monoclonal antibody (Bethyl, Montgomery, TX).
- Protein lysates 50 ⁇ g/lane were separated by SDS-PAGE (10%) prior to electrophoretic transfer onto a Nitrocellulose membrane (Bio-Rad, Hercules, CA) as described (Aharinejad et al., 2005).
- the blots were incubated with primary human monoclonal anti-GSTP1 antibody (Bethyl, Montgomery, TX), human monoclonal anti-TRAF2 (BD Pharmingen, San Diego, CA), human polyclonal phospho-p38 (Promega, Madison, WI), human polyclonal phosphor-JNK (Santa Cruz Biotechnology, Santa Cruz, CA), human polyclonal anti-JNK (Santa Cruz Biotechnology, Santa Cruz, CA), human polyclonal anti-p38 (Santa Cruz Biotechnology, Santa Cruz, CA), prior to incubation with horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences, Piscataway, NJ).
- primary human monoclonal anti-GSTP1 antibody Bethyl, Montgomery, TX
- human monoclonal anti-TRAF2 BD Pharmingen, San Diego, CA
- human polyclonal phospho-p38 Promega, Madison, WI
- Protein loading was assessed by Ponceau S staining and immunode-tection was performed by chemiluminescence (Supersig-nal-West-Pico, Pierce, Rockford, IL). Bands were quantified by ImageQuant software and specific protein signals were normalized to loading controls and expressed as arbitrary units. The average value of three measurements in each sample was used for data analysis.
- the screening tissue profiling arrays identified higher cardiac GSTP1 and TRAF2 gene as well as protein expression levels in randomly selected patients with both DCM and ICM compared to the control individuals ( Figures 1A, 1B ).
- the myocardial expression of GSTP1 and TRAF2 was then prospectively examined by real time RT-PCR and Western blotting in the study cohort. These analyses indicated significantly elevated myocardial mRNA expression levels in DCM and ICM for both GSTP1 (P ⁇ 0.0001) and TRAF2 (P ⁇ 0.0001) as compared to controls ( Figure 1C ).
- GSTP1-treated cardiac tissue lysates were immunoprecipitated with TRAF2 antibody and immunopellets were subjected to Western blotting.
- TNF- ⁇ abrogates the sensitivity of TRAF2/JNK/p38 cascade to GST-P1 in DCM
- TRAF2 cardiac tissue protein expression in DCM was approximately two times higher as compared to ICM and controls and considering the results described above, we tested the effect of higher GSTP1 concentration following TNF- ⁇ treatment.
- the results demonstrate that DCM cardiac tissue cultures treated with GSTP1 at 10 ⁇ g/mL and TNF- ⁇ at 50 ng/mL express markedly reduced JNK and p38 activation as well as reduced TRAF2 protein expression (P ⁇ 0.0001; Figures 5A, 5B ).
- the immunoprecipitates of DCM cardiac tissue lysates demonstrate significant (P ⁇ 0.035) increase of GSTP1-TRAF2 association at 10 ⁇ g/ml as compared to 5 mg/ml GSTP1.
- TRAF2 is a central regulator of TNF- ⁇ signalling that mediates MAPK activation.
- GSTP1 can abrogate MAPK activation through association with TRAF2 and consequently suppress TNF- ⁇ signalling.
- the present data show significant elevation of active JNK and p38 in DCM myocardium compared to ICM and controls.
- Serum GSTP1 is a sensitive marker of CMP
- the study cohort comprised 141 end-stage CMP patients scheduled for cardiac transplantation and 20 patients with pre-served EF undergoing conventional isolated aortic or mitral valve surgery. All patients underwent echocardiography, coronary artery angiography, and right-heart catheterization for evaluation of ventricular function and standard hemodynamic parameters by two independent cardiologists. The case history, clinical test results, and treatment were documented, coded and blinded to the investigators of serum samples.
- the study patients were subdivided in groups based on their left ventricular EF assessed by echocardiography as follows: EF>52%; EF 52-43%; EF 42-33%, EF 32-23% and EF ⁇ 22% as described ( Lee et al., Circulation 119 (2009) 3070-3077 ).
- LV anterior left ventricular wall
- the protein fluorescent signals of both slides were detected by the GenePix 4000B scanner (Molecular Dynamics, Sunnyvale, CA).
- the quantification of the signal was performed by calculating an internally normalized ratio (INR) using the automated Microarray Analysis Workbook (http://bioinfo.clontech.com).
- Enzyme linked immunosorbent assay for GSTP1 (HEPKITTM-Pi, Biotrin International Ltd., Dublin, Ireland) was performed according to the manufacturer's protocol. The substrate reaction was quantified spectrophotometrically by using a 96-well automated microplate reader (Anthos, Salzburg, Austria) at 450 nm. N-terminal proBNP was measured in undiluted serum automatically by a chemiluminescent noncompetitive ELISA (Roche Diagnostics, Indianapolis, IN) on a Roche Elecsys 2010 analyzer.
- Real-time RT-PCR was performed on a LightCycler instrument (Roche) as described (Aharinejad et al., 2009b, Abraham et al, 2000).
- the primer sequences were sense/antisense: GSTP1: 5'-CCAAAGGTGGTGAGCTTCAT-3'/5'-TCTACCCAGCATGGAGGAAC-3'; and ⁇ 2-microglobulin: 5'-GATGAGTATGCCTGCCGTGTG-3'/5'-CAATCCAAATGCG-GCATCT-3'.
- mRNA expression levels of GSTP1 was normalized to the ⁇ 2-microglobulin signal as a housekeeping gene (Aharinejad et al., 2009b, Abraham et al, 2000). The average value of three PCR measurements in each sample was used for data analysis.
- Tissue lysates were prepared (Aharinejad et al., 2009b) and GSTP1 expression was analyzed by Western blotting using primary human monoclonal anti-GSTP1 antibody (Bethyl, Montgomery, TX). Protein bands were quantified by ImageQuant software and specific protein signals were normalized to loading controls. The average value of three measurements in each sample was used for data analysis.
- GSTP1 and proBNP serum concentrations were compared between patient groups by analysis of variance (one-way ANOVA; Tukey's test).
- PrS Spearman's rank correlation coefficients
- Univariate logistic regression was used to describe the utility of GSTP1 and proBNP as predictors of EF.
- the corresponding receiver operating characteristic curve (ROC) analyses were used to find optimal cut-off levels. Sensitivity and specificity of GSTP1 and proBNP cut-offs were calculated by table analysis. All statistical analyses were performed using SAS system for Windows, version 9.1.3 and the Enterprise Guide, version 4.1 (SAS Institute, Inc., Cary, NC). Statistical significance was set at P ⁇ 0.05. Results are expressed as mean ⁇ standard deviation.
- FIG. 6A shows the array images of GSTP1 and indicates that its serum protein levels are increased in CMP patients as compared to healthy volunteers.
- serum GSTP1 concentration was determined in the same patient cohort selected for screening analyses by GSTP1-specific ELISA. These results show that serum GSTP1 concentrations were significantly upregulated in end-stage HF patients as compared to the control samples ( Figure 6B , P ⁇ 0.001).
- tissue protein profiling was initiated using the LV myocardial samples of the same end-stage HF patients analysed by serum arrays and used LV myocardial biopsies of donated hearts as controls.
- the results of tissue profiling indicate elevated GSTP1 expression levels in myocardium of end-stage HF patients as compared to controls ( Figure 6C ).
- Western blotting analyses were performed on cardiac tissues and found significantly upregulated GSTP1 protein expression levels in patients with end-stage HF as compared to controls (P ⁇ 0.001; Figure 6D ).
- Serum GSTP1 concentrations specifically diagnosed CMP with significant association to EF independently from demographical and clinical characteristics in our patient cohort.
- markedly higher correlation coefficients were shown for serum GSTP1 association to EF as compared to proBNP.
- serum GSTP1 shows better diagnostic power in CMP patients with EF ⁇ 22% as compared to proBNP. More importantly, GSTP1 diagnosed EF ⁇ 42% whereas proBNP failed.
- proBNP is recommended to be mainly used for exclusion of CMP with normal EF in patients with symptoms attributed to CMP.
- gender and older age are associated with higher proBNP levels ( Costello-Boerrigter er al., J. Am. Coll. Cardiol. 47 (2006), 345-353 )
- the proposed diagnostic properties of proBNP might be too unspecific to differentiate CMP with preserved EF in elderly patients.
- proBNP can predict EF ⁇ 30% with a sensitivity and specificity of 90% and 71%, respectively in a CMP population with EF ⁇ 45%.
- Other studies reported that plasma proBNP detect EF ⁇ 28% with 77% sensitivity and 69% specificity in a patients cohort with EF ⁇ 50%, and it was found that proBNP can predict EF ⁇ 40% with an area under the curve of 0.69.
- proBNP had 97% sensitivity but only 26% specificity to diagnose EF ⁇ 22% with an area under the curve of 0.62.
- the lower specificity of proBNP in the present study could be explained by the fact that our cut-off level was selected to identify lower EFs as compared to other studies.
- GSTP1 is a sensitive, specific, cheap and quickly measurable serum marker superior to proBNP in CMP. Importantly, GSTP1 discriminates between preserved and reduced EF with a high sensitivity and specificity and can therefore serve as a novel tool in guiding of clinical trials in CMP patients.
- the model of ligation of the anterior branch of the left coronary artery in the rat has been selected to further prove the principle of the present invention.
- IHD is induced following the ligation of the said coronary artery branch, and over time the animals will subsequently develop CMP. Therefore, the model is best suited to show the efficacy of the suggested invention for prevention or treatment of both IHD and CMP.
- mice Male, Sprague Dawley rats (Harlan, Borchen, Germany) will be coded for experiments. Due to expected hemodynamic compromise, ventricular fibrillation, myocardial rupture and bleeding following myocardial infarction (MI), the number of surviving animals is expected to be about 60-70%. Therefore, in each group with myocardial infarction, a total of 18 animals will be included.
- MI myocardial infarction
- Rats will be anesthetized with intraperitoneal injection of Ketasol (100 mg/kg body weight) and Rompun (10 mg/kg body weight), intubated tracheally using a 14-gauge catheter and ventilated with 1.5% Isofluran at 55 cycles per minute (tidal volume: 2.5 ml) before undergoing a left lateral thoracotomy.
- the anterior branch of the left coronary artery (LCA) will be either ligated with a 7-0 polypropylene snare (Ethicon, Somerville, NJ) or left intact in sham procedure.
- LCA left coronary artery
- LV left ventricular
- mice with comparable base-line cardiac function will be coded and assigned to groups 1-8 as shown in Table 4.
- animals with comparable base-line cardiac function will be coded and assigned to groups 1-8 as shown in Table 4.
- animals with comparable base-line cardiac function will be coded and assigned to groups 1-8 as shown in Table 4.
- animals will receive immediately after MI induction and on the first day post MI 0.6 mg/100 g body weight Piritramid (Dipidolor ® in 5% glucose solution) s.c. Further, animals will receive Piritramid via their drink water for 3 days at 0.6 mg/100 g body weight (250 ml water plus 20 ml 5% glucose solution) and at days 4-7 post MI at 0.3 mg/100 g body weight.
- Piritramid Dipidolor ® in 5% glucose solution
- treatment On d7, immediately after MI induction, treatment will start in groups 1-4 as follows. Group 1 will receive daily intraperi-toneal injections of Ringer's solution for two weeks (control), while groups 2-4 will receive daily injections of recombinant GSTP1 at a dose of 10, 20 and 40 mg/body weight for two weeks, respectively. On d21 the treatment will start in groups 5-8 as follows. Group 5 will receive daily intraperitoneal injections of Ringer's solution for two weeks (control), while groups 6-8 will receive daily injections of recombinant GSTP1 at a dose of 10, 20 and 40 mg/body weight for two weeks, respectively. Group 9 includes 10 animals with sham procedure.
- Table 4 Groups of animals and their treatment Group Treatment Sacrifice No. of animals Application and dose 1 Ringer d88 18 i.p. 2 GSTP1 d88 18 i.p., 10 mg/kg bw, 2 weeks 3 GSTP1 d88 18 i.p., 20 mg/kg bw, 2 weeks 4 GSTP1 d88 18 i.p., 40 mg/kg bw, 2 weeks 5 Ringer d88 18 i.p.
- LV ejection fraction (EF), LV end systolic volume (ESV), LV end diastolic volume (EDV) using echocardiography 2.
- Heart-to-body weight ratio 3. Histology and immunocytochemistry for infarct size, infarct wall thickness and fibrosis as well as angiogenesis, inflammatory proteins including p38, june kinase and interleukins (only examples are mentioned for inflammatory pathways) 4.
- Signaling pathways 5.
- Promoter binding assays 6. TUNEL-assay for apoptosis evaluation 7.
- LVEDV LV end-diastolic
- LVESV end-systolic
- LVEF LVEDV-LVESV/LVEDV
- ventricles were cross-sectioned at the midpoint of their long axis, before they were either frozen or immersion-fixed in formalin. Paraffin-embedded specimens were used for H&E, Tunnel assay as well as Goldner trichrome collagen staining. TUNEL assays (In situ Cell Death Detection Kit) were performed according to the manufacturer's protocol (Roche Molecular Biochemicals, Basel, Switzerland) in triplicate. Slides were counterstained with 4,6-diamidino-2-phenylindole (DAPI, Molecular Probes, Eugene, OR) and embedded in AF1 antifadent (Citifluor, Sheffield, UK). Digital images were obtained by fluorescence microscopy (Nikon, Melville, NY). Morphometry was carried out as described (Aharinejad et al., 2008).
- the primer sequences were sense/antisense: GSTP1: 5'-GGCAACTGAAGCCTTTTGAG-3'/5'-TCATGGATCAGCAGCAAGTC-3'; tumor necrosis factor receptor-associated factor 2 (TRAF2): 5'-GCAGAAG-GTCTTGGAGATGG-3'/5'-GGTGGAGCAGCATTAAGGTC-3'; and ⁇ 2-microglobulin: 5'-GATGAGTATGCCTGCCGTGTG-3'/5'-CAATCCAAATGCG-GCATCT-3'. mRNA expression were calculated and plotted for treated animals as percentage relative to that in controls.
- TRF2 tumor necrosis factor receptor-associated factor 2
- ⁇ 2-microglobulin 5'-GATGAGTATGCCTGCCGTGTG-3'/5'-CAATCCAAATGCG-GCATCT-3'.
- the blots were incubated with primary human monoclonal GSTP1 (Bethyl) and TRAF2 (BD Pharmingen) or polyclonal phospho-p38 (Promega, Madison, WI), phospho-JNK1, JNK1 and p38 (Santa Cruz Biotechnology) antibodies prior to incubation with secondary antibodies (Amersham Biosciences, Piscataway, NJ). All experiments were performed in triplicate and expression levels were corrected for control loading protein.
- GSTP1 ameliorates the myocardial overexpression of inflammatory cytokines following acute MI
- GSTP1 ameliorates the myocardial expression of pro-inflammatory cytokines through TRAF2-regulated NF-kB signaling in the failing myocardium
- GSTP1-treated rats had significantly increased formation of GST-P1-TRAF2, GSTP1-JNK1 and GSTP1-p38 complexes in their failing cardiac tissue at 3 weeks post-MI as compared to controls (p ⁇ 0.001; Figure 10A ).
- GSTP1-treated rats had significantly reduced protein expression of activated JNK1 and p38 in their failing myocardial tissue as compared to controls (p ⁇ 0.001; Figure 10B ).
- GSTP1-treated rats had lower cardiac tissue mRNA expressions of TGF- ⁇ , IL-1 ⁇ , IL-2 and IL-17 as compared to controls ( P ⁇ 0.001; Figure 10C ).
- GSTP1 ameliorates remodeling in rat failing myocardium
- TNF- ⁇ The role of the inflammatory mediators in the patho-physiology of heart failure has gained a growing interest over the last two decades. Different experimental studies have described the negative inotropic effect of TNF- ⁇ on the LV function. Moreover, it has been reported that TNF- ⁇ promotes LV remodeling, cardiac hypertrophy and progressive cardiomyocytes loss through apoptosis. The growing evidence on the pathological role of inflammatory mediators in the setting of heart failure have resulted in a series of multicenter clinical trials that intended to target TNF- ⁇ in heart failure patients. However, the results of these trials were discouraging. One explanation of these results is that the low physiological levels of TNF- ⁇ might be important for cardiac tissue remodeling and repair.
- TRAF2 is a central regulator of TNF- ⁇ signaling that mediates MAPK activation.
- TRAF2 results in activation of NF- ⁇ B.
- GSTP1 is an important negative regulator of TNF- ⁇ -induced signaling by forming interactions with TRAF2.
- GSTP1 could inhibit MAPK activation in malignant cell lines indirectly through its interaction with TRAF2 or directly through its interaction with JNK1 and P38.
- GSTP1 was not only able to suppress the pro-inflammatory mediators like; IL-1, IL-2, JNK1, P38 and NF- ⁇ B but also could increase the expression of the anti-inflammatory cytokine IL-10.
- the results according to the present invention revealed that GSTP1 protects the cardiac tissue against apoptosis in the failing heart. In concert with this, it has been shown in oncological settings that GSTP1 attenuates the autophosphorylation of TRAF2-enhanced apoptosis signal-regulating kinasel (ASK1) and thus inhibits TRAF2-ASK1-induced cell apoptosis by suppressing the interaction of TRAF2 and ASK1.
- ASK1 TRAF2-enhanced apoptosis signal-regulating kinasel
- the present invention provides a novel function of GSTP1 in modulating TNF- ⁇ /TRAF2 elicited JNK1/p38 activation in heart failure.
- the selective inhibition of TNF- ⁇ /TRAF2-mediated inflammatory signaling by GSTP1 was due to the present animal model shown to be a beneficial treatment for patients with cardiomyopathies or ischemic heart diseases, especially for patients with myocardial infarction and heart failure.
Landscapes
- Health & Medical Sciences (AREA)
- Chemical & Material Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Engineering & Computer Science (AREA)
- Organic Chemistry (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- General Health & Medical Sciences (AREA)
- Medicinal Chemistry (AREA)
- Pharmacology & Pharmacy (AREA)
- Animal Behavior & Ethology (AREA)
- Public Health (AREA)
- Veterinary Medicine (AREA)
- Cardiology (AREA)
- Genetics & Genomics (AREA)
- Immunology (AREA)
- Wood Science & Technology (AREA)
- Zoology (AREA)
- Analytical Chemistry (AREA)
- Nuclear Medicine, Radiotherapy & Molecular Imaging (AREA)
- General Chemical & Material Sciences (AREA)
- Heart & Thoracic Surgery (AREA)
- Chemical Kinetics & Catalysis (AREA)
- Microbiology (AREA)
- General Engineering & Computer Science (AREA)
- Biophysics (AREA)
- Biotechnology (AREA)
- Physics & Mathematics (AREA)
- Epidemiology (AREA)
- Molecular Biology (AREA)
- Gastroenterology & Hepatology (AREA)
- Pathology (AREA)
- Biochemistry (AREA)
- Urology & Nephrology (AREA)
- Vascular Medicine (AREA)
- Hospice & Palliative Care (AREA)
- Medicines That Contain Protein Lipid Enzymes And Other Medicines (AREA)
- Investigating Or Analysing Biological Materials (AREA)
- Medicinal Preparation (AREA)
Priority Applications (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| EP16186303.0A EP3120865B1 (en) | 2009-10-30 | 2010-10-28 | Gstp1 for the treatment of ischemic heart diseases |
| EP10775689.2A EP2493496B1 (en) | 2009-10-30 | 2010-10-28 | Use of gstp1 |
Applications Claiming Priority (3)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| EP09174692 | 2009-10-30 | ||
| PCT/AT2010/000408 WO2011050379A1 (en) | 2009-10-30 | 2010-10-28 | Use of gstp1 |
| EP10775689.2A EP2493496B1 (en) | 2009-10-30 | 2010-10-28 | Use of gstp1 |
Related Child Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| EP16186303.0A Division EP3120865B1 (en) | 2009-10-30 | 2010-10-28 | Gstp1 for the treatment of ischemic heart diseases |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| EP2493496A1 EP2493496A1 (en) | 2012-09-05 |
| EP2493496B1 true EP2493496B1 (en) | 2016-08-31 |
Family
ID=42282291
Family Applications (2)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| EP10775689.2A Not-in-force EP2493496B1 (en) | 2009-10-30 | 2010-10-28 | Use of gstp1 |
| EP16186303.0A Not-in-force EP3120865B1 (en) | 2009-10-30 | 2010-10-28 | Gstp1 for the treatment of ischemic heart diseases |
Family Applications After (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| EP16186303.0A Not-in-force EP3120865B1 (en) | 2009-10-30 | 2010-10-28 | Gstp1 for the treatment of ischemic heart diseases |
Country Status (7)
| Country | Link |
|---|---|
| US (2) | US8758745B2 (pl) |
| EP (2) | EP2493496B1 (pl) |
| JP (1) | JP5863660B2 (pl) |
| CN (1) | CN102573892B (pl) |
| ES (1) | ES2630654T3 (pl) |
| PL (1) | PL2493496T3 (pl) |
| WO (1) | WO2011050379A1 (pl) |
Families Citing this family (7)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN109529046B (zh) * | 2018-11-09 | 2020-10-27 | 北京大学 | 一种靶向线粒体的自组装蛋白质纳米颗粒的制备与应用 |
| US20220362326A1 (en) * | 2019-09-12 | 2022-11-17 | The University Of British Columbia | Inhibiting zd17-jnk interaction as a therapy for acute myocardial infarction |
| MX2022009880A (es) * | 2020-02-14 | 2022-08-22 | Galaxy Ccro Inc | Dispositivos y metodos para tratar la isquemia y sindromes de dificultad respiratoria aguda. |
| CN112472821B (zh) * | 2020-11-19 | 2022-11-18 | 北京大学 | 一种蛋白质纳米颗粒用于制备抗vsv病毒产品的用途 |
| CN113265423B (zh) * | 2021-05-25 | 2022-08-23 | 上海家化联合股份有限公司 | 含人gstp1 are的荧光素酶报告基因的细胞株的构建及应用 |
| CN116139117A (zh) * | 2022-05-31 | 2023-05-23 | 西南医科大学附属医院 | 衣康酸及其衍生物在防治阿霉素诱导的心脏毒性中的应用 |
| CN115808529B (zh) * | 2022-11-24 | 2023-05-30 | 哈尔滨医科大学 | 一种蛋白质pars2在制备诊断酒精性心肌病的试剂中的应用及试剂盒 |
Family Cites Families (8)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US5427917A (en) | 1988-02-01 | 1995-06-27 | Teijin Limited | Method of determining human acid glutathione S-transferase, reagent, therefor, kit therefor, method of diagnosing cancer in digestive organs, and monoclonal antibody for use therein |
| US5552277A (en) | 1994-07-19 | 1996-09-03 | The Johns Hopkins University School Of Medicine | Genetic diagnosis of prostate cancer |
| US5968737A (en) | 1996-11-12 | 1999-10-19 | The University Of Mississippi | Method of identifying inhibitors of glutathione S-transferase (GST) gene expression |
| US5817497A (en) | 1996-11-26 | 1998-10-06 | Incyte Pharmaceuticals | Glutathione s-transferase |
| EP1500709A1 (en) | 2003-07-21 | 2005-01-26 | Academisch Ziekenhuis bij de Universiteit van Amsterdam | Means and methods for detecting a risk of infarction related to atherosclerosis |
| US20060154261A1 (en) * | 2005-01-07 | 2006-07-13 | Andrew Saxon | Method for determining susceptibility of an individual to allergen induced hypersensitivity |
| CA2613204A1 (en) | 2005-06-29 | 2007-01-11 | Rules-Based Medicine, Inc. | Methods and kits for the diagnosis of acute coronary syndrome |
| US20090186360A1 (en) | 2008-01-22 | 2009-07-23 | Abhijit Mazumder | Detection of GSTP1 hypermethylation in prostate cancer |
-
2010
- 2010-10-28 EP EP10775689.2A patent/EP2493496B1/en not_active Not-in-force
- 2010-10-28 CN CN201080044454.9A patent/CN102573892B/zh not_active Expired - Fee Related
- 2010-10-28 WO PCT/AT2010/000408 patent/WO2011050379A1/en not_active Ceased
- 2010-10-28 US US13/505,078 patent/US8758745B2/en not_active Expired - Fee Related
- 2010-10-28 EP EP16186303.0A patent/EP3120865B1/en not_active Not-in-force
- 2010-10-28 PL PL10775689T patent/PL2493496T3/pl unknown
- 2010-10-28 JP JP2012535539A patent/JP5863660B2/ja not_active Expired - Fee Related
- 2010-10-28 ES ES10775689.2T patent/ES2630654T3/es active Active
-
2014
- 2014-05-13 US US14/275,923 patent/US9375460B2/en not_active Expired - Fee Related
Also Published As
| Publication number | Publication date |
|---|---|
| EP2493496A1 (en) | 2012-09-05 |
| US9375460B2 (en) | 2016-06-28 |
| ES2630654T3 (es) | 2017-08-22 |
| JP2013508426A (ja) | 2013-03-07 |
| US20140302002A1 (en) | 2014-10-09 |
| EP3120865B1 (en) | 2018-06-06 |
| CN102573892A (zh) | 2012-07-11 |
| EP3120865A1 (en) | 2017-01-25 |
| US8758745B2 (en) | 2014-06-24 |
| JP5863660B2 (ja) | 2016-02-16 |
| PL2493496T3 (pl) | 2017-03-31 |
| WO2011050379A9 (en) | 2011-09-29 |
| US20120213761A1 (en) | 2012-08-23 |
| CN102573892B (zh) | 2015-07-29 |
| WO2011050379A1 (en) | 2011-05-05 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| US9375460B2 (en) | Use of GSTP1 | |
| Stretti et al. | A year in heart failure: an update of recent findings | |
| JP2018023392A (ja) | 肝線維症に関連する遺伝的多型、その検出方法および使用 | |
| US10765739B2 (en) | Use of MIF and MIF pathway agonists | |
| US8052960B2 (en) | Netrin-1 as a biomarker of injury and disease | |
| JP2008515394A (ja) | 心臓の圧負荷関連遺伝子 | |
| US8965708B2 (en) | Method for the treatment of acquired lymphedema | |
| US11654134B2 (en) | Cyclocreatine phosphate: a novel bioenergetic therapy to prevent and treat ischemia-induced and aging-related cardiovascular and neurodegenerative diseases | |
| Hirose et al. | Nicorandil prevents Gαq-induced progressive heart failure and ventricular arrhythmias in transgenic mice | |
| RU2626670C2 (ru) | Способ прогнозирования эффективности терапии больных сахарным диабетом 2 типа | |
| US20220389074A1 (en) | Compositions and uses thereof for treating, prognosing and diagnosing pulmonary hypertension | |
| US8293481B2 (en) | Biomarkers for chronic vascular dysfunction | |
| WO2020097210A1 (en) | Personalized treatment for cardiomyopathy and heart failure by measuring renin activity, pro-renin, pro-renin, receptor levels in blood | |
| US20240175863A1 (en) | Compositions and methods for monitoring enpp1 activity | |
| KR102317737B1 (ko) | 순환 미토콘드리아 dna를 이용한 당뇨병 또는 당뇨 합병증 진단용 조성물 및 진단방법 | |
| Hsieh et al. | Plumbagin improves pulmonary vascular remodeling in PAH via miR-21-5p/MMP/TIMP regulation, with diagnostic implications for cardiac function | |
| US8623601B2 (en) | Methods of diagnosing cancer | |
| US20100034835A1 (en) | Use of inhibitors of leukotriene b4 receptor blt2 for treating asthma | |
| Hassan et al. | Association of cathepsin D (CTSD) polymorphism and serum levels with myocardial infarction risk in type-2 diabetes mellitus | |
| Holst et al. | Proteomic Analysis in Valvular Cardiomyopathy: Aortic Regurgitation vs. Aortic Stenosis. Cells 2023, 12, 878 | |
| Essandoh | The Role of Tsg101 in the Development of Physiological Cardiac Hypertrophy and Cardio-Protection from Endotoxin-Induced Cardiac Dysfunction | |
| Elwing et al. | Idiopathic and Familial Pulmonary Arterial Hypertension | |
| KR20200112069A (ko) | 골 질환의 예방 또는 치료를 위한 Jmjd2b 억제제의 용도 | |
| Hamiltona et al. | Combination of Angiotensin II and LNg-Nitroarginine methyl ester exacerbates mitochondrial dysfunction and oxidative stress to cause heart failure. 2 | |
| Makino et al. | Articles in PresS. Am J Physiol Heart Circ Physiol (October 23, 2009). doi: 10.1152/ajpheart. 00421.2009 |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| PUAI | Public reference made under article 153(3) epc to a published international application that has entered the european phase |
Free format text: ORIGINAL CODE: 0009012 |
|
| 17P | Request for examination filed |
Effective date: 20120330 |
|
| AK | Designated contracting states |
Kind code of ref document: A1 Designated state(s): AL AT BE BG CH CY CZ DE DK EE ES FI FR GB GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL PT RO RS SE SI SK SM TR |
|
| DAX | Request for extension of the european patent (deleted) | ||
| 17Q | First examination report despatched |
Effective date: 20151015 |
|
| GRAP | Despatch of communication of intention to grant a patent |
Free format text: ORIGINAL CODE: EPIDOSNIGR1 |
|
| INTG | Intention to grant announced |
Effective date: 20160331 |
|
| GRAS | Grant fee paid |
Free format text: ORIGINAL CODE: EPIDOSNIGR3 |
|
| GRAA | (expected) grant |
Free format text: ORIGINAL CODE: 0009210 |
|
| AK | Designated contracting states |
Kind code of ref document: B1 Designated state(s): AL AT BE BG CH CY CZ DE DK EE ES FI FR GB GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL PT RO RS SE SI SK SM TR |
|
| REG | Reference to a national code |
Ref country code: CH Ref legal event code: EP Ref country code: GB Ref legal event code: FG4D |
|
| REG | Reference to a national code |
Ref country code: IE Ref legal event code: FG4D |
|
| REG | Reference to a national code |
Ref country code: DE Ref legal event code: R096 Ref document number: 602010036055 Country of ref document: DE |
|
| REG | Reference to a national code |
Ref country code: AT Ref legal event code: REF Ref document number: 824387 Country of ref document: AT Kind code of ref document: T Effective date: 20161015 |
|
| REG | Reference to a national code |
Ref country code: FR Ref legal event code: PLFP Year of fee payment: 7 |
|
| REG | Reference to a national code |
Ref country code: NL Ref legal event code: FP |
|
| REG | Reference to a national code |
Ref country code: SE Ref legal event code: TRGR |
|
| REG | Reference to a national code |
Ref country code: LT Ref legal event code: MG4D |
|
| REG | Reference to a national code |
Ref country code: AT Ref legal event code: MK05 Ref document number: 824387 Country of ref document: AT Kind code of ref document: T Effective date: 20160831 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: RS Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: LT Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: NO Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20161130 Ref country code: FI Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: HR Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: GR Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20161201 Ref country code: AT Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: LV Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: RO Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: EE Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: SK Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: CZ Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: PT Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20170102 Ref country code: SM Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: DK Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: BG Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20161130 |
|
| REG | Reference to a national code |
Ref country code: DE Ref legal event code: R097 Ref document number: 602010036055 Country of ref document: DE |
|
| PLBE | No opposition filed within time limit |
Free format text: ORIGINAL CODE: 0009261 |
|
| STAA | Information on the status of an ep patent application or granted ep patent |
Free format text: STATUS: NO OPPOSITION FILED WITHIN TIME LIMIT |
|
| REG | Reference to a national code |
Ref country code: IE Ref legal event code: MM4A |
|
| 26N | No opposition filed |
Effective date: 20170601 |
|
| REG | Reference to a national code |
Ref country code: ES Ref legal event code: FG2A Ref document number: 2630654 Country of ref document: ES Kind code of ref document: T3 Effective date: 20170822 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: SI Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: LU Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20161028 |
|
| REG | Reference to a national code |
Ref country code: FR Ref legal event code: PLFP Year of fee payment: 8 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: IE Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20161028 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: FR Payment date: 20171012 Year of fee payment: 8 Ref country code: DE Payment date: 20171027 Year of fee payment: 8 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: BE Payment date: 20171023 Year of fee payment: 8 Ref country code: NL Payment date: 20171018 Year of fee payment: 8 Ref country code: PL Payment date: 20171025 Year of fee payment: 8 Ref country code: SE Payment date: 20171012 Year of fee payment: 8 Ref country code: GB Payment date: 20171026 Year of fee payment: 8 Ref country code: IT Payment date: 20171020 Year of fee payment: 8 Ref country code: CH Payment date: 20171030 Year of fee payment: 8 Ref country code: ES Payment date: 20171103 Year of fee payment: 8 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: CY Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: HU Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT; INVALID AB INITIO Effective date: 20101028 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: TR Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: IS Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: MT Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20161031 Ref country code: MC Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 Ref country code: MK Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: AL Free format text: LAPSE BECAUSE OF FAILURE TO SUBMIT A TRANSLATION OF THE DESCRIPTION OR TO PAY THE FEE WITHIN THE PRESCRIBED TIME-LIMIT Effective date: 20160831 |
|
| REG | Reference to a national code |
Ref country code: DE Ref legal event code: R119 Ref document number: 602010036055 Country of ref document: DE |
|
| REG | Reference to a national code |
Ref country code: SE Ref legal event code: EUG |
|
| REG | Reference to a national code |
Ref country code: CH Ref legal event code: PL |
|
| REG | Reference to a national code |
Ref country code: NL Ref legal event code: MM Effective date: 20181101 |
|
| GBPC | Gb: european patent ceased through non-payment of renewal fee |
Effective date: 20181028 |
|
| REG | Reference to a national code |
Ref country code: BE Ref legal event code: FP Effective date: 20161128 Ref country code: BE Ref legal event code: MM Effective date: 20181031 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: SE Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20181029 Ref country code: DE Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20190501 Ref country code: NL Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20181101 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: CH Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20181031 Ref country code: FR Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20181031 Ref country code: ES Free format text: THE PATENT HAS BEEN ANNULLED BY A DECISION OF A NATIONAL AUTHORITY Effective date: 20160831 Ref country code: LI Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20181031 Ref country code: BE Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20181031 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: IT Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20181028 Ref country code: GB Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20181028 |
|
| REG | Reference to a national code |
Ref country code: ES Ref legal event code: FD2A Effective date: 20191204 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: ES Free format text: THE PATENT HAS BEEN ANNULLED BY A DECISION OF A NATIONAL AUTHORITY Effective date: 20181029 |
|
| PG25 | Lapsed in a contracting state [announced via postgrant information from national office to epo] |
Ref country code: PL Free format text: LAPSE BECAUSE OF NON-PAYMENT OF DUE FEES Effective date: 20181028 |