CN106029687B - HTLV衣壳抗原p24的可溶性和免疫反应性变体 - Google Patents
HTLV衣壳抗原p24的可溶性和免疫反应性变体 Download PDFInfo
- Publication number
- CN106029687B CN106029687B CN201580010787.2A CN201580010787A CN106029687B CN 106029687 B CN106029687 B CN 106029687B CN 201580010787 A CN201580010787 A CN 201580010787A CN 106029687 B CN106029687 B CN 106029687B
- Authority
- CN
- China
- Prior art keywords
- ala
- leu
- gly
- gln
- val
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Active
Links
Images
Classifications
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N33/00—Investigating or analysing materials by specific methods not covered by groups G01N1/00 - G01N31/00
- G01N33/48—Biological material, e.g. blood, urine; Haemocytometers
- G01N33/50—Chemical analysis of biological material, e.g. blood, urine; Testing involving biospecific ligand binding methods; Immunological testing
- G01N33/53—Immunoassay; Biospecific binding assay; Materials therefor
- G01N33/569—Immunoassay; Biospecific binding assay; Materials therefor for microorganisms, e.g. protozoa, bacteria, viruses
- G01N33/56983—Viruses
- G01N33/56988—HIV or HTLV
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K14/00—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- C07K14/005—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from viruses
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K14/00—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- C07K14/005—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from viruses
- C07K14/08—RNA viruses
- C07K14/15—Retroviridae, e.g. bovine leukaemia virus, feline leukaemia virus human T-cell leukaemia-lymphoma virus
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N33/00—Investigating or analysing materials by specific methods not covered by groups G01N1/00 - G01N31/00
- G01N33/48—Biological material, e.g. blood, urine; Haemocytometers
- G01N33/50—Chemical analysis of biological material, e.g. blood, urine; Testing involving biospecific ligand binding methods; Immunological testing
- G01N33/53—Immunoassay; Biospecific binding assay; Materials therefor
- G01N33/569—Immunoassay; Biospecific binding assay; Materials therefor for microorganisms, e.g. protozoa, bacteria, viruses
- G01N33/56983—Viruses
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2319/00—Fusion polypeptide
- C07K2319/35—Fusion polypeptide containing a fusion for enhanced stability/folding during expression, e.g. fusions with chaperones or thioredoxin
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2740/00—Reverse transcribing RNA viruses
- C12N2740/00011—Details
- C12N2740/10011—Retroviridae
- C12N2740/14011—Deltaretrovirus, e.g. bovine leukeamia virus
- C12N2740/14022—New viral proteins or individual genes, new structural or functional aspects of known viral proteins or genes
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2740/00—Reverse transcribing RNA viruses
- C12N2740/00011—Details
- C12N2740/10011—Retroviridae
- C12N2740/14011—Deltaretrovirus, e.g. bovine leukeamia virus
- C12N2740/14031—Uses of virus other than therapeutic or vaccine, e.g. disinfectant
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2740/00—Reverse transcribing RNA viruses
- C12N2740/00011—Details
- C12N2740/10011—Retroviridae
- C12N2740/14011—Deltaretrovirus, e.g. bovine leukeamia virus
- C12N2740/14051—Methods of production or purification of viral material
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N2333/00—Assays involving biological materials from specific organisms or of a specific nature
- G01N2333/005—Assays involving biological materials from specific organisms or of a specific nature from viruses
- G01N2333/08—RNA viruses
- G01N2333/15—Retroviridae, e.g. bovine leukaemia virus, feline leukaemia virus, feline leukaemia virus, human T-cell leukaemia-lymphoma virus
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N2469/00—Immunoassays for the detection of microorganisms
- G01N2469/20—Detection of antibodies in sample from host which are directed against antigens from microorganisms
-
- Y—GENERAL TAGGING OF NEW TECHNOLOGICAL DEVELOPMENTS; GENERAL TAGGING OF CROSS-SECTIONAL TECHNOLOGIES SPANNING OVER SEVERAL SECTIONS OF THE IPC; TECHNICAL SUBJECTS COVERED BY FORMER USPC CROSS-REFERENCE ART COLLECTIONS [XRACs] AND DIGESTS
- Y02—TECHNOLOGIES OR APPLICATIONS FOR MITIGATION OR ADAPTATION AGAINST CLIMATE CHANGE
- Y02A—TECHNOLOGIES FOR ADAPTATION TO CLIMATE CHANGE
- Y02A50/00—TECHNOLOGIES FOR ADAPTATION TO CLIMATE CHANGE in human health protection, e.g. against extreme weather
- Y02A50/30—Against vector-borne diseases, e.g. mosquito-borne, fly-borne, tick-borne or waterborne diseases whose impact is exacerbated by climate change
Landscapes
- Health & Medical Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Chemical & Material Sciences (AREA)
- Virology (AREA)
- Immunology (AREA)
- Engineering & Computer Science (AREA)
- Molecular Biology (AREA)
- Hematology (AREA)
- Biomedical Technology (AREA)
- Urology & Nephrology (AREA)
- Biochemistry (AREA)
- Medicinal Chemistry (AREA)
- General Health & Medical Sciences (AREA)
- Organic Chemistry (AREA)
- Food Science & Technology (AREA)
- Tropical Medicine & Parasitology (AREA)
- Physics & Mathematics (AREA)
- Analytical Chemistry (AREA)
- Cell Biology (AREA)
- Biotechnology (AREA)
- General Physics & Mathematics (AREA)
- Pathology (AREA)
- Microbiology (AREA)
- Gastroenterology & Hepatology (AREA)
- Biophysics (AREA)
- Genetics & Genomics (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- AIDS & HIV (AREA)
- Oncology (AREA)
- Peptides Or Proteins (AREA)
- Preparation Of Compounds By Using Micro-Organisms (AREA)
Abstract
本发明涉及可以融合至分子伴侣的可溶性和抗原性HTLV p24变体及其在诊断应用诸如用于检测分离的生物样品中的针对HTLV‑I或HTLV‑II的抗体的免疫测定中的用途。具体而言,本发明涉及包含p24的N‑或C‑末端结构域且缺乏其它结构域的可溶性HTLV‑I或HTLV‑II p24抗原。而且,本发明涵盖编码这些HTLV‑I和‑II融合抗原的重组DNA分子以及使用表达载体和用这样的表达载体转化的宿主细胞的它们的重组产生。此外,本发明聚焦于这些HTLV p24抗原与HTLV gp21抗原的组合物和用于使用本发明的抗原检测HTLV抗体的免疫测定方法。还包括HTLV p24抗原在体外诊断测定中的用途以及包含所述HTLV抗原的用于检测抗‑HTLV‑的抗体的试剂盒。
Description
人T-细胞嗜淋巴细胞病毒(HTLV)I型(HTLV-I)是1980年在人中发现的第一个逆转录病毒。其为T-细胞白血病和/或淋巴瘤和HTLV相关的脊髓病(最终导致热带痉挛性轻截瘫的严重脱髓鞘病况)的病原体。在HTLV-I的无症状携带者中,发展这些致命和难治愈的疾病的累积终生风险总计为~ 5 %。HTLV-I主要感染CD4-阳性T-细胞。其也被称为成人T-细胞淋巴瘤病毒1型。HTLV-II与HTLV-I共享约70%基因同源性(转化为在蛋白水平的80-95%结构相似性)。HTLV-II的致病潜力还没有完全阐明,但HTLV-II被认为是输血的风险标志物,因为其主要在全世界的静脉内吸毒者中发现(Vandamme等人, Evolutionary strategies ofhuman T-cell lymphotropic virus type II, Gene 261 (2000) 171-180)。两种病毒在全球传播,但HTLV-I的流行率在南日本(Kyushu、Shikoku和Okinawa)、撒哈拉以南非洲、加勒比(牙买加和海地)和南美洲的热点地区中是最高的。
HTLV-I/II的主要传播模式是通过性接触、输血、共用注射针头和通过哺乳的母婴传播。当与其它感染性疾病相比时,HTLV感染之后的血清转化期长。窗口期,即,其内无法检测到针对病毒的抗体的感染后的时间段可以范围为数周至数月。
在日本于1986年、在美国和加拿大于1988/1989年、在法国于1991年和在几个欧洲和南美国家在1991年之后首次引入针对HTLV的献血者筛选。至今还没有出现用于诊断HTLV感染的金标准。已经在过去几年中引入了基于重组和/或合成肽抗原的几种免疫测定法。
用于检测抗-HTLV-抗体的市售免疫测定法通常使用源自病毒包膜的多肽(gp46表面蛋白和gp21跨膜蛋白)或源自gag编码的p24衣壳蛋白的多肽。
由于长血清转化时间,一旦抗体在感染后的早期阶段出现即检测甚至非常少量的抗体是重要的。因此,开发适当的抗原用于高度敏感的免疫测定是必需的。理所当然的是,期望闭合感染和检测之间的诊断间隙,以防止病毒的意外传播和繁殖。
已经知道一段时间的是,HTLV-感染后,针对gag蛋白的抗体在血清转化早期出现。具体而言,gag编码的衣壳抗原p24是体液免疫应答的优选早期目标(Manns等人, Blood(1991) 77: 896-905)。迄今为止,p24衣壳蛋白的肽和重组变体已被用作免疫测定中的抗原。借助这些抗原,已经以高精确度和令人满意的灵敏度检测到G型的抗-p24免疫球蛋白。然而,这种类型的p24衣壳抗原无法结合和检测M型的免疫球蛋白。由于IgM分子通常在血清转化期间在IgG分子之前出现,所以我们推理,以其被IgM识别和结合的方式修饰重组的p24衣壳抗原应当是值得的。总之,我们想知道是否可能通过定制和工程改造p24衣壳抗原而改进抗-p24免疫球蛋白检测的灵敏度。具体而言,我们正在寻求设计能够与IgM分子相互作用并检测IgM分子的p24变体。
因此,本发明的基本问题是开发用于检测针对HTLV-I和HTLV-II的抗体的免疫测定法,其克服了迄今为止可用的免疫测定法的有限的血清转化灵敏度。
通过如权利要求中指定的本发明解决了该问题。
发明概述:
本发明涉及融合至分子伴侣的可溶性HTLV p24抗原及其在诊断应用诸如用于检测分离的生物样品中的针对HTLV-I或HTLV-II的抗体的免疫测定中的用途。具体而言,本发明涉及包含p24序列的N-或C-末端结构域的可溶性HTLV-I或HTLV-II p24抗原片段,其中所述HTLV p24抗原片段可以融合至分子伴侣。而且,本发明涵盖编码这些HTLV-I和-II融合抗原的重组DNA分子以及使用表达载体和用这样的表达载体转化的宿主细胞的它们的重组产生。此外,本发明聚焦于这些HTLV p24抗原中的几种的组合物和用于使用本发明的抗原检测HTLV抗体的免疫测定方法。还包括HTLV p24抗原在体外诊断测定中的用途以及包含所述HTLV p24抗原的用于检测抗-HTLV-的抗体的试剂盒。
公开的氨基酸序列的说明:
SEQ ID NO.1: p24/HTLV-I (146-344)// P10274, 199个氨基酸残基
显示如从SwissProt数据库ID P10274检索的HTLV-I p24序列(来自人T-细胞白血病病毒1,毒株Japan ATK-1亚型A的146-344的146-344 Gag-Pro多蛋白)。编号是指未成熟多蛋白前体(aa 1-130的序列是指基质蛋白p19)。注意,已经省略来自aa 131-145的N-末端15个氨基酸残基(富含脯氨酸的序列)。
SEQ ID NO.2: p24 NTD (146-260)/HTLV-I, 115个氨基酸残基
显示HTLV-I p24的来自氨基酸146-260的N-末端结构域(对于氨基酸位置的编号,还参见SEQ ID NO.1)。注意,一个位置被标记为X(加下划线),这意味着天然序列的半胱氨酸残基可以被替代为丙氨酸或丝氨酸(X = C、A或S)。
SEQ ID NO.3: p24 CTD (261-344)/HTLV-I, 84个氨基酸残基
显示HTLV-I p24的来自氨基酸残基261-344的C-末端结构域(对于氨基酸位置的编号,还参见SEQ ID NO.1)。注意,两个位置被标记为X(加下划线),这意味着天然序列的半胱氨酸残基可以被替代为丙氨酸或丝氨酸(X = C、A或S)。
SEQ ID NO.4: p24 (146-344)/HTLV-I, 199个氨基酸残基
显示就长度和位置而言类似于SEQ ID NO.1的HTLV-I p24序列。然而,三个氨基酸位置显示X(加下划线),这意味着在这些位置中,天然存在的半胱氨酸(根据前体多肽序列编号的位置编号193、311和332)可以被取代为丙氨酸或丝氨酸(X = C、A或S)。
SEQ ID NO.5: p24/HTLV-II (152-350)//P03353, 199个氨基酸残基
显示如从SwissProt数据库ID P03353检索的HTLV-II p24序列(来自人T-细胞白血病病毒2的152-3350 Gag-Pro多蛋白)。编号是指未成熟多蛋白前体(aa 1-136的序列是指基质蛋白p19)。注意,已经省略来自aa 137-151的N-末端15个氨基酸(富含脯氨酸的序列)。
SEQ ID NO.6: p24 NTD (152-266)/HTLV-II, 115个氨基酸残基
显示HTLV-II p24的来自氨基酸152-266的N-末端结构域(对于氨基酸位置的编号,还参见SEQ ID NO.5)。注意,一个位置被标记为X(加下划线),这意味着天然序列的半胱氨酸残基可以被替代为丙氨酸或丝氨酸(X = C、A或S)。
SEQ ID NO.7: p24 CTD (267-350)/HTLV-II, 84个氨基酸残基
显示HTLV-II p24的来自氨基酸267-350的C-末端结构域(对于氨基酸位置的编号,还参见SEQ ID NO.5)。注意,三个位置被标记为X(加下划线),这意味着天然序列的半胱氨酸残基可以被替代为丙氨酸或丝氨酸(X = C、A或S)。
SEQ ID NO.8: p24 (152-350)/HTLV-II, 199个氨基酸残基
显示就长度和位置而言类似于SEQ ID NO.5的HTLV-II p24序列。四个氨基酸位置显示X(加下划线),这意味着在这些位置中,天然存在的半胱氨酸(根据前体多肽序列编号的位置编号199、281、317和338)可以被取代为丙氨酸或丝氨酸(X = C、A或S)。
以下氨基酸序列(SEQ ID NO.9-16和18-24)显示如实施例部分中使用的HTLV-I或HTLV-II p24(完全或部分)序列的融合序列。EcSlyD、EcFkpA和EcSkp的蛋白命名中的两个字母Ec表明来自大肠杆菌的蛋白序列来源。各蛋白在其C-末端具有六-组氨酸标签,其用于促进蛋白纯化和重折叠。
SEQ ID NO.25: gp21/HTLV-1 (339-446)// P14075, 108个氨基酸残基
显示根据SwissProt条目ID P14075的包膜糖蛋白gp21(源自env多蛋白前体)的氨基酸残基号339-446。完整多蛋白前体包含:人T-细胞白血病病毒I(分离株Caribbean HS-35亚型A)的表面蛋白(=糖蛋白46,gp46)和跨膜蛋白(=糖蛋白21 gp21)。注意,三个残基被标记为X(加下划线),这意味着天然序列的半胱氨酸残基可以被替代为丙氨酸或丝氨酸(X= C、A或S)。
HTLV gp21也可以有利地应用为增强溶解性的分子伴侣融合多肽,如例如SEQ IDNO.26和27中所示。
附图说明:
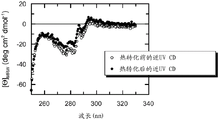
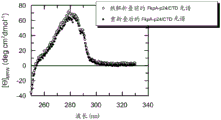
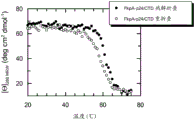
图1显示Skp-p24/CTD (267-350), SEQ ID NO.22的近UV CD光谱。
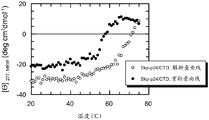
图2显示Skp-p24/CTD (SEQ ID NO.22)的熔化曲线。通过在277 nm的近UV CD光谱监测热诱导的解折叠和重折叠。
图3显示FkpA-p24/CTD (267-350), SEQ ID NO.21的近UV CD光谱。
图4显示在热诱导的解折叠/重折叠循环之后,完全恢复天然FkpA-p24/CTD分子的近UV CD信号。
发明详述
HTLV p24是用于检测抗-HTLV抗体的重要抗原。p24衣壳蛋白在本领域中长时间已知,并且已经在用于检测抗-HTLV抗体的免疫测定中使用(Manns等人, Blood (1991) 77:896-905)。用于检测IgG和IgM分子两者的免疫测定需要一组抗原,其不仅被IgG分子、而且被IgM分子识别和结合。IgM分子通常存在于HTLV感染后的血清转化的早期阶段。多价IgM分子的结合关键取决于高抗原表位密度。因此,设计用于特异性检测IgM分子的抗原具有和展示这样的高表位密度是必要的。
生成具有高表位密度的IgM检测模块的常规方法是借助化学交联聚合单体抗原。存在大量同双功能和异双功能交联剂,其可以以很大优势使用,并且是本领域中众所周知的。然而,在用于用作血清学测定中的指示剂(specifiers)的抗原的化学诱导的聚合中存在一些严重的缺点。例如,将交联剂部分插入抗原可以通过干扰天然样构象或通过掩蔽关键表位而损害抗原性。此外,非天然三级接触的引入可以干扰蛋白折叠/解折叠的可逆性,并且此外,其可以是干扰问题的来源,所述干扰问题必须通过免疫测定混合物中的抗干扰策略来克服。
生成IgM检测模块的一种更近的技术是将目标抗原融合至寡聚分子伴侣,由此为抗原赋予高表位密度。该技术的优点在于其高可重复性和寡聚分子伴侣融合伴侣的三重功能:首先,分子伴侣增强宿主细胞中的融合多肽的表达率,其次,分子伴侣促进目标抗原的重折叠过程并增强其整体溶解度,以及第三,其将目标抗原可重复地组装成有序寡聚结构。
欧洲专利申请公开号EP1982993A2公开了用于使用融合至寡聚分子伴侣的抗原早期检测病原体诸如人巨细胞病毒的真实原发感染的方法和工具。然而,该出版物没有提到HTLV感染的检测。
我们用HTLV p24的全长版本的初始尝试已经揭示,该蛋白当融合至作为分子伴侣的EcSlyD-EcSlyD或EcFkpA时表现出高溶解度。然而,当将p24融合至三聚Skp分子伴侣时,其溶解度是有限的。不言自明的是,所有化合物的溶解度是异质免疫测定应用的关键特征。免疫测定中的蛋白性成分的聚集过程通常导致信号的损失(由于表位的损失)和特异性的损失(由于标记的抗原聚集体与固相的非特异性结合)。我们观察到,来自HTLV的全长p24 -当融合至寡聚分子伴侣EcSkp时 - 显示在环境温度在生理缓冲液中聚集的倾向。因此,从某种程度上在灵敏IgM免疫测定中的简单且直接的应用中排除全长p24变体。
不再聚焦于p24的全长版本,我们现在试图设计截短的、但构象折叠的p24的片段。换言之,我们试图使用蛋白结构域而非全长p24蛋白作为用于抗原开发的基础。蛋白结构域是蛋白结构内的自主折叠实体,即,不依赖于蛋白在其折叠中的其它部分或区域的蛋白结构域。迄今为止,已经阐明了许多天然蛋白结构域,其大小范围为~ 40个氨基酸残基(WW结构域)至超过300个氨基酸残基。也已经表明,可以从scratch设计非常小、但稳定的蛋白结构域:已经显示具有23-28个氨基酸序列的片段长度的人工多肽序列协同地折叠,并且具有蛋白结构域的特有特征(Struthers, M. D.等人., Design of a monomeric 23-residuepolypeptide with defined tertiary structure, Science (1996) 271 (5247) 342-345; Dahiyat, B. I. & Mayo, S.L., De novo protein design: fully automatedsequence selection, Science (1997) 278 (5335) 82-87; Dahiyat, B.I.等人, De novo protein design: towards fully automated protein design, J. Mol. Biol.(1997) 273 (4) 789-796)。从理论考虑和实验证据,假定蛋白结构域的最小长度要求为约25个氨基酸残基(Porter L. L. & Rose, G. D., A thermodynamic definition ofprotein domains, PNAS (2012) 109 (24), 9420-9425)。
在Molecular Biology (1999) Aug. 13; 291(2):491-505的杂志中,Khorasanizadeh等人呈现了衣壳蛋白p24的NMR结构,并且揭示了该蛋白的结构域拓扑学。根据该工作,来自HTLV-I的p24主要是螺旋的,并且由两个良好分离的结构域组成,即,p24包含两个明确定义的自主折叠单元。N-末端结构域(NTD)具有螺旋1-7,而C-末端结构域(CTD)包含螺旋8-12。我们想知道在大肠杆菌中单独表达两种结构域是否是可行的,我们是否能够获得可溶和抗原形式的寡聚分子伴侣多肽融合体。
Khorasanizadeh等人没有提到p24的抗原特性(例如,B-细胞表位)和NMR-表征的HTLV衣壳蛋白的任何诊断应用。从p24衣壳抗原的仅仅三维溶液结构无法预测其抗原性是否主要位于N-末端结构域(NTD)或C-末端结构域(CTD)中,或者其B-细胞表位是否在整个分子均匀散布。
令人惊讶的是,我们能够表达与分子伴侣模块诸如SlyD、FkpA和Skp融合的分离的HTLV p24结构域NTD和CTD。如实施例部分中可见,所有这些构建体都可以被纯化至均质,它们是充分可溶的,并且我们能够在自动免疫测定分析仪中使用抗-HTLV阳性人血清评价它们的抗原性。结果是相当明确的:抗原性对于两个结构域相当高,并且对于C-结构域(CTD)甚至略高。引人注目的是,NTD可以被鉴定为相对于空白值不确定,当与CTD相比时,其显著增加。CTD表现出优异的信号动力学,因为其对于阳性血清生成高信号,并且对于阴性血清生成非常低的信号。这是令人惊讶的,因为推测CTD具有p24衣壳组装所需的天然二聚化基序。凭借其天然寡聚化行为,我们已经推理CTD会表现出比NTD的聚集倾向显著更高的聚集倾向。
当我们用兔抗-HTLV血清学转换血清(没有商购的人HTLV血清转化实验对象组,所以我们必须回到人工兔模型)评价p24 CTD和NTD时,我们发现在DAGS测定的两侧使用分子伴侣诱导的寡聚p24变体大大增强了免疫测定的灵敏度。与单体p24变体相比,用寡聚p24变体对血清转化样品的识别好得多。
简言之,来自HTLV-I和HTLV-II的p24的C-结构域被鉴定为具有高抗原性和高溶解度的p24片段。当融合至分子伴侣诸如SlyD、FkpA或Skp时,p24 CTD保持可溶,稳定,并且非常适合于检测通常在HTLV感染后存在于血清转化的早期阶段中的IgM分子。因此,具体而言,p24 CTD的寡聚FkpA和Skp融合变体可用于增强HTLV-免疫测定的灵敏度。
我们已经开发了来自HTLV的衣壳蛋白p24的变体,其比全长p24分子更可溶且聚集倾向显著更低。以抗原性为代价,溶解度和稳定性得到改善 - 然而,新开发的p24变体有希望作为HTLV免疫测定中的抗原,因为它们在大肠杆菌中大量过表达,容易经由固定化的金属螯合层析法(IMAC)纯化和重折叠,表现出令人满意的稳定特性,并且可以用于可靠地检测人血清中的抗-HTLV抗体(据推测与GP21(来自HTLV的另一种免疫显性蛋白)的胞外结构域组合)。至关重要的是,例如,FkpA-p24/CTD和Skp-p24/CTD融合蛋白形成具有足以检测IgM分子的表位密度的天然寡聚体。因为我们旨在开发用于总免疫球蛋白检测(即IgG和IgM的检测)的免疫测定法,所以寡聚种类FkpA-p24/CTD和Skp-p24/CTD可以有利地用作DAGS形式的两侧的指示剂(例如FkpA-p24/CTD-生物素和Skp-p24/CTD-钌)。初步数据表明,寡聚p24变体的使用确保竞争剂测定法无法匹配的优异的血清转化灵敏度。
因此,本发明涉及分别包含N-末端结构域且缺乏C-末端结构域或包含全长HTLVp24多肽的C-末端结构域且缺乏N-末端结构域的可溶性HTLV p24抗原。根据本发明,p24抗原可以融合至分子伴侣。还包括这些HTLV p24抗原在诊断应用诸如用于检测分离的生物样品中的针对HTLV-I或HTLV-II的抗体的免疫测定中的用途。术语“HTLV”意指“人T-细胞嗜淋巴细胞病毒”。除非特别标记为HTLV-I或HTLV-II,否则术语HTLV是指两种病毒类型两者。
根据本发明,所述抗原仅包含完整HTLV p24抗原的特定结构域,诸如N-末端结构域(NTD)或C-末端结构域(CTD)。优选地,所述抗原包含HTLV-I p24的SEQ ID NO.2的N-末端结构域或SEQ ID NO.3的C-末端结构域。对于HTLV-II抗原,融合抗原优选包含SEQ ID NO.6的N-末端结构域或SEQ ID. NO. 7的C-末端结构域。在一个进一步优选模式中,如果N-末端结构域是所述抗原的部分,则C-末端结构域缺失,反之亦然。
具体而言,本发明涉及可溶性HTLV p24抗原,其包含如SEQ ID NO.2 (p24 NTDHTLV-I)或SEQ ID NO.6 (p24 NTD HTLV-II)中指定的HTLV p24的N-末端结构域(NTD),其中所述HTLV p24抗原缺乏如SEQ ID NO.3 (p24 CTD HTLV-I)和SEQ ID NO.7 (p24 CTDHTLV-II)中指定的C-末端结构域(CTD)。
此外,本发明涉及可溶性HTLV p24抗原,其包含如SEQ ID NO.3 (p24 CTD HTLV-I)或SEQ ID NO.7 (p24 CDT HTLV-II)中指定的HTLV p24的C-末端结构域,其中所述HTLVp24抗原缺乏如SEQ ID NO.2 (p24 NTD HTLV-I)和SEQ ID NO.6 (p24 NTD HTLV-II)中指定的N-末端结构域。
术语HTLV p24抗原也包括变体。HTLV p24变体可以由本领域技术人员通过保守或同源取代公开的氨基酸序列(诸如例如用丙氨酸或丝氨酸取代半胱氨酸)而容易地生成。在该上下文中的术语“变体”还涉及与所述蛋白基本上类似的蛋白或蛋白片段(即,多肽或肽)。例如,修饰诸如C-或N-末端截短1至10个氨基酸在请求保护的HTLV p24抗原的范围内。具体而言, 变体可以是与最普遍的蛋白同种型相比显示氨基酸交换、缺失或插入的同种型。在一个实施方案中,这样基本上类似的蛋白与蛋白最普遍的同种型具有至少80%,在另一个实施方案中至少85%或至少90%, 在又另一个实施方案中至少95%的序列相似性。术语“变体”还涉及翻译后修饰的蛋白诸如糖基化或磷酸化的蛋白。根据本发明,变体分类为HTLV p24抗原变体,只要维持体外诊断免疫测定法中的免疫反应性,即变体仍然能够结合并检测分离的样品中存在的抗-HTLV p24抗体。“变体”也是这样的蛋白或抗原,其已经例如通过将标记物或载体部分共价或非共价附接至蛋白或抗原而被修饰。可能的标记物是放射性的、荧光的、化学发光的、 电化学发光的、酶或其它例如像异羟基洋地黄毒苷或生物素。这些标记物是本领域技术人员已知的。
本发明的HTLV p24抗原是可溶的,稳定的和免疫反应的,即,它们适合作为抗原用于免疫学测定法中使用。这意味着根据本发明的抗原在生理缓冲剂条件下, 例如在没有添加去污剂的情况下在环境温度在磷酸盐缓冲剂系统中是可溶的。所述抗原也能够结合对于HTLV p24特异性的抗体(如例如分离的样品诸如人血清中存在的抗- p24抗体)或被其识别和结合。
根据本发明的HTLV p24抗原可以与分子伴侣融合。如本发明中所使用的术语“融合蛋白”、“融合多肽”或“融合抗原”是指这样的蛋白,其包含至少一个对应于HTLV p24多肽的蛋白部分和至少一个充当融合伴侣的作用的源自分子伴侣的蛋白部分。
分子伴侣,被称为经典的助折叠物,是辅助其它蛋白的折叠和结构完整性的维持的蛋白。助折叠物的实例详细描述于WO 03/000877中。根据本发明,肽基脯氨酰异构酶类别的分子伴侣诸如FKBP家族的分子伴侣可用于与 HTLV p24抗原变体融合。适合作为融合伴侣的FKBP分子伴侣的实例是FkpA、SlyD和SlpA。适合作为HTLV p24的融合伴侣的其它分子伴侣是Skp,来自大肠杆菌的周质的三聚分子伴侣,其不属于FKBP家族。并不总是必须使用分子伴侣的完整序列。也可以使用仍然具有所需能力和功能的分子伴侣的功能片段(所谓的能够结合的模块)(参照WO 98/13496)。
根据本发明的一个进一步实施方案,FKBP分子伴侣(诸如例如大肠杆菌SlyD、SlpA或FkpA)中的至少一个或至少两个模块被用作用于表达HTLV p24抗原的融合部分。分子伴侣Skp同样可用作融合伴侣。两个FKBP-分子伴侣结构域的融合导致所得融合多肽的溶解性提高。融合部分可位于HTLV p24抗原的N-末端或C-末端或两端(夹心样)
优选地,根据本发明的HTLV p24抗原融合至寡聚分子伴侣。寡聚分子伴侣是这样的分子伴侣,其天然形成二聚体、三聚体或更高阶多聚体,使得通过特异性非共价相互作用组装多个单体亚基。优选的寡聚分子伴侣是FkpA和Skp。
特别优选的是融合至选自SEQ ID NO.9至16和18至24的分子伴侣的可溶性HTLVp24抗原。
可以借助重组DNA技术生成和制备根据本发明的HTLV p24抗 原。因此本发明的另一个方面是编码如上进一步定义的HTLV p24抗原及其变体的重组DNA分子。
术语“重组DNA分子”是指这样的分子,其通过组合两个以其它方式分离的DNA序列区段而制成,所述组合由通过遗传工程改造技术或通过化学合成人工操作多核苷酸的分离的区段而完成。这样做时,可以将具有期望功能的多核苷酸区段接合在一起以生成期望的功能组合。用于在原核或低等或高等真核宿主细胞中表达蛋白的重组DNA技术是本领域中众所周知的。它们已经例如由Sambrook等人, (1989, Molecular Cloning: A LaboratoryManual)描述。
根据本发明的重组DNA分子也可以含有这样的序列,所述序列编码在HTLV p24抗原和融合部分之间和还在几个融合部分之间的10至100个氨基酸残基的接头肽。这样的接头序列可以例如携带蛋白水解切割位点。
本发明的一个进一步方面是表达载体,其包含可操作连接的根据本发明的重组DNA分子,即编码HTLV p24抗原和任选肽基脯氨酰异构酶分子伴侣诸如FKBP-分子伴侣的重组DNA分子,其中FKBP-分子伴侣选自FkpA、 SlyD和SlpA。在一个替代实施方案中,所述重组DNA分子编码包含HTLV p24抗原和Skp的融合蛋白。根据本领域众所周知的方法,包含根据本发明的重组 DNA的表达载体可用于在无细胞翻译系统中表达HTLV p24抗原或可用于转化宿主细胞用于表达HTLV p24抗原。本发明的另一个方面因此涉及用根据本发明的表达载体转化的宿主细胞。在本发明的一个实施方案中,在大肠杆菌细胞中产生重组HTLV p24抗原。
一个额外方面是用于产生根据本发明的可溶的、稳定的和免疫反应性的HTLV p24抗原。所述p24抗原可以作为含有HTLV p24抗原和分子伴侣的融合蛋白产生。优选地,使用分子伴侣,诸如Skp或肽基脯氨酰异构酶类别分子伴侣如FKBP分子伴侣。在本发明的一个进一步实施方案中,所述FKBP分子伴侣选自SlyD、FkpA和SlpA。
该方法包括以下步骤
a) 培养用含有编码HTLV p24抗原的基因的上述表达载体转化的宿主细胞
b) 表达编码所述HTLV p24抗原的基因
c) 纯化所述HTLV p24抗原。
任选地,作为额外的步骤d),需要实施功能性增溶,使得借助本领域已知的重折叠技术使HTLV p24抗原成为可溶和免疫反应性的构象。
本发明的一个额外方面涉及用于检测分离的人样品中的抗-HTLV抗体的方法,其中使用根据本发明的HTLV p24抗原作为抗体的结合伴侣。本发明因此涵盖用于检测分离的样品中的对于HTLV特异性的抗体的方法,所述方法包括
a) 通过混合体液样品与根据本发明的HTLV p24抗原而形成免疫反应混合物
b) 维持所述免疫反应混合物一定时间段,所述时间段足以允许所述体液样品中存在的针对所述HTLV p24抗原的抗体与所述HTLV p24抗原免疫反应,以形成免疫反应产物;和
c) 检测所述免疫反应产物中的任一种的存在和/或浓度。
在一个进一步方面,所述方法适合于检测IgG和IgM亚类的HTLV抗体。
用于检测抗体的免疫测定法是本领域中众所周知的,并且用于实施这样的测定法和实际应用的方法和程序也是本领域中众所周知的。根据本发明的HTLV p24抗原可用于不依赖于使用的标记物并且不依赖于检测模式(例如,放射性同位素测定,酶免疫测定,电化学发光测定等)或测定原理(例如测试条测定、夹心测定、间接测试概念或均质测定等)而改进用于检测抗-HTLV抗体的测定法。专家已知的所有生物学液体都可以用作用于检测抗-HTLV抗体的分离的样品。通常使用的样品是体液如全血、血清、血浆、尿或唾液。
本发明的一个进一步实施方案是根据所谓的双抗原夹心概念(DAGS)进行的用于检测分离的样品中的抗-HTLV抗体的免疫测定法。有时,该测定概念也称为双抗原桥概念,因为两个抗原通过抗体分析物桥接。在这样的测定法中,需要和利用抗体用其两个(IgG、IgE)、四个(IgA)或十个(IgM)互补位结合给定抗原的至少两个不同分子的能力。
更详细地,通过孵育含有抗-HTLV抗体的样品与两种不同的HTLV p24抗原,即,第一(“固相”)HTLV p24抗原和第二HTLV p24(“检测”) 抗原,实施根据双抗原桥形式的用于测定抗-HTLV抗体的免疫测定法,其中所述抗原各自与所述-HTLV抗体特异性结合。第一抗原可以直接或间接结合至固相并且通常携带效应基团,所述效应基团是生物亲和力(bioaffine)结合对(如例如生物素和抗生物素蛋白)的部分。例如, 如果第一抗原缀合至生物素,则用抗生物素蛋白或链霉抗生物素蛋白包被固相。第二抗原携带标记物。因此,形成包含第一抗原、样品抗体和第二抗原的免疫反应混合物。在向所述抗原添加样品之前,或在形成免疫反应混合物之后,添加第一抗原可以结合的固相。维持该免疫反应混合物一定时间段,所述时间段足以允许所述体液样品中的针对所述HTLV p24抗原的抗-HTLV抗体与所述HTLV p24抗原免疫反应,以形成免疫反应产物。下一步是分离步骤,其中将液相与固相分离。最后,在所述固相或液相或两者中检测所述免疫反应产物中的任一种的存在。
在所述DAGS免疫测定中,“固相抗原”和“检测抗原”的基础结构是基本上相同的。在双抗原桥测定中,也可能使用相似、但不同的HTLV p24 抗原,所述HTLV p24抗原是免疫学交叉反应性的。进行这样的测定的本质要求是相关的一个或多个表位存在于两个抗原上。根据本发明,可能对每个HTLV p24抗原使用不同的融合部分(例如,融合至固相侧上的HTLV p24的SlyD和融合至检测侧上的HTLV p24的FkpA p24),因为这样的变异显著减轻非特异性结合的问题,因此降低假阳性结果的风险。
优选地,在所述DAGS免疫测定中,应用不对称形式,其组合融合至FkpA的HTLV p24和融合至Skp的HTLV p24抗原。更优选地,在固相侧使用融合至FkpA的HTLV p24,并且在检测侧应用融合至Skp的HTLV p24,但它也可能具有反向排列,即在固相侧上使用融合至Skp的HTLV p24抗原,并且在检测侧上使用融合至FkpA的HTLV p24。最优选地,HTLV p24 FkpA融合蛋白携带用于附接至已经用链霉抗生物素蛋白或抗生物素蛋白包被的固相的生物素部分,并且HTLV p24 Skp融合蛋白携带电化学发光标记物诸如钌络合物。在相反排列的情况下,p24 Skp融合蛋白携带生物素,且p24 FkpA携带所述标记物。
本发明的一个进一步实施方案因此是根据双抗原桥概念的免疫测定法,其中使用根据本发明的第一HTLV p24抗原,和根据本发明的第二HTLV p24抗原。
本发明进一步涉及HTLV p24的至少一种抗原在用于检测抗-HTLV抗体的诊断测试中的用途。
本发明的额外主题是用于检测针对HTLV的抗体的试剂盒,所述试剂盒,除了用于免疫测定法的通常的测试添加剂以外,含有适合于特异性结合待测定的HTLV抗体并且如果必要可能携带标记物以及其它通常的添加剂的根据本发明的HTLV p24抗原的至少一种抗原。
具体而言,所述试剂盒含有根据SEQ ID NO.9至16和18至24中任一种的HTLV p24抗原。
此外,以上定义的试剂盒含有对照和标准溶液以及一种或多种溶液中的试剂与如本领域普通技术人员所使用的通常的添加剂、缓冲剂、盐、去污剂等,连同使用说明书。
另一个实施方案是包含根据本发明的可溶性HTLV p24抗原和HTLV env抗原、优选包含SEQ ID NO.25的gp21的HTLV抗原组合物。术语“组合物”是指作为个别不同分子存在于混合物中的分开表达的多肽。术语组合物排除在单个多肽链具有p24和gp21片段的蛋白。
优选的是包含HTLV p24的C-末端结构域的组合物,特别优选的是包含根据SEQ IDNO.3(缺乏SEQ ID NO.2)的HTLV-I p24抗原和/或根据SEQ ID NO.7 (缺乏SEQ ID NO.6)的HTLV-II p24抗原和HTLV gp21的组合物。例如,在所述组合物中,可以存在包含SEQ IDNO.25、26或27中任一种的 HTLV gp21序列。对于根据DAGS形式的免疫测定中的应用,所述组合物包含两种形式的每种HTLV抗原,所述两种形式即,使得能够将抗原附接至固相的形式(例如,可以结合至用链霉抗生物素蛋白包被的表面的生物素化的抗原),和使得能够检测在样品中存在的HTLV抗体和应用的HTLV抗原之间的免疫复合物的标记的形式。
本发明还涉及根据本发明的HTLV p24抗原在用于检测抗-HTLV抗体的体外诊断测试中的用途。
本发明通过实施例进一步举例说明。
实施例1
p24衣壳融合多肽的克隆和纯化
表达盒的克隆
基于Novagen (Madison, WI, USA)的pET24a表达质粒,基本上如所述(Scholz,C.等人, J. Mol. Biol. (2005) 345, 1229-1241)获得编码来自HTLV-I和HTLV-II的p24融合蛋白的表达盒。从SwissProt数据库检索来自HTLV-I和HTLV-II的p24抗原的序列(分别为SwissProt ID P10274和P03353)。编码来自HTLV-I的p24衣壳抗原aa 146-344(编号是指Gag-Pro多聚蛋白前体)(在成熟衣壳蛋白的N-末端缺乏富含脯氨酸的15个氨基酸)与同框融合至N-末端的富含甘氨酸的接头区域的合成基因购自Medigenomix (Martinsried,Germany)。将p24在位置193、311和332的半胱氨酸残基改变为丙氨酸残基以防止不想要的副作用诸如氧化或分子间二硫桥接。BamHI和XhoI限制性位点分别在p24-编码区的5’和3’末端。编码经由富含甘氨酸的接头区连接的两个EcSlyD单位(SwissProt登录号P0A9K9的残基1-165)和涵盖C-末端的进一步接头区的部分的进一步合成基因同样购自Medigenomix。NdeI和BamHI限制性位点分别在该盒的5’和3’末端。设计基因和限制性位点以使得能够通过简单连接同框融合分子伴侣部分EcSlyD-EcSlyD和p24抗原部分。为了避免意外的重组过程且为了增加大肠杆菌宿主中的表达盒的遗传稳定性,简并编码EcSlyD单位的核苷酸序列,同样简并编码延伸的接头区的核苷酸序列,即,使用不同的密码子组合编码相同的氨基酸序列。
用NdeI和XhoI消化pET24a载体并插入包含与HTLV-I p24 (146-344)同框融合的串联-SlyD的盒。因此构建包含多杀性巴氏杆菌SlyD (1-156, SwissProt ID Q9CKP2)、大肠杆菌Skp (21-161, SwissProt ID P0AEU7)或大肠杆菌FkpA (26-270, SwissProt IDP45523)的表达盒,以及包含来自HTLV-II的p24和p24片段(SwissProt ID P03353)的表达盒。如同来自HTLV-I的p24,将来自HTLV-II的p24的位置199、281、317和338(再次,编号是指前体Gag-Pro多蛋白)的真正半胱氨酸残基改变为丙氨酸残基,以防止不希望的副作用诸如氧化或分子间二硫键桥接。所有重组融合多肽变体含有C-末端六组氨酸标签以便于Ni-NTA辅助的纯化和重折叠。使用QuikChange (Stratagene, La Jolla, CA, USA)和标准PCR 技术以在各自的表达盒中生成点突变、缺失、插入和延伸变体或限制性位点。
下图显示具有与其N-端末端同框融合的两个SlyD分子伴侣单位的N-末端截短的HTLV-I p24抗原146-344的方案。为了表示SlyD融合伴侣的大肠杆菌来源,已经将描绘的融合多肽命名为EcSlyD-EcSlyD-p24 (146-344)。
将所得质粒的插入物测序并发现其编码期望的融合蛋白。来自HTLV-I和HTLV-II的p24融合多肽的完整氨基酸序列显示于SEQ ID NO.9至16和18至24中。接头L的氨基酸序列显示于SEQ ID NO.17中。
包含来自HTLV-I和HTLV-II的p24和p24变体的融合蛋白的纯化
通过使用几乎相同的方案纯化所有p24融合蛋白变体。在37℃在加有卡那霉素(30µg/ml)的LB培养基中使携带特别的pET24a表达质粒的大肠杆菌BL21 (DE3)细胞生长至1.5的OD600,并且通过添加1 mM异丙基-β-D-硫代半乳糖苷诱导胞质溶胶过表达。诱导后三小时,通过离心(20min,5000g)收获细胞,冷冻并储存于-20℃。为了细胞裂解,将冷冻的沉淀在冰冷的50 mM磷酸钠pH 8.0、7.0 M GdmCl、5mM咪唑中重悬浮,并且在冰上搅拌悬浮液2小时以完成细胞裂解。在离心和过滤(0.45μm/0.2μm)之后,将粗裂解物应用于用包括5.0 mMTCEP的裂解缓冲液平衡的Ni-NTA柱上。对于各目标蛋白定制随后的洗涤步骤并且范围为5至15 mM咪唑(在50 mM磷酸钠pH 8.0, 7.0 M GdmCl, 5.0 mM TCEP中)。至少应用10-15体积的洗脱缓冲液。然后,通过50mM磷酸钾pH 8.0、100 mM KCl、10 mM咪唑、5.0 mM TCEP替换GdmCl溶液以诱导基质结合的蛋白的构象重折叠。为了避免共纯化蛋白酶的重活化,在重折叠缓冲液中包括蛋白酶抑制剂混合物(Complete®无EDTA, Roche)。在过夜反应中应用总共15-20柱体积的重折叠缓冲液。然后,通过用3-5柱体积50mM磷酸钾pH 8.0、100 mM KCl、10mM咪唑洗涤而去除TCEP和Complete®无EDTA抑制剂混合物。随后,咪唑浓度-仍然在50mM磷酸钾pH 8.0、100 mM KCl中-升高至20 - 80 mM(取决于各目标蛋白)以去除非特异结合的蛋白污染物。然后通过在相同缓冲液中的500 mM咪唑洗脱天然蛋白。通过Tricine-SDS-PAGE评价含有蛋白的级分的纯度并合并所述级分。最后,对蛋白进行大小排阻层析(Superdex HiLoad, Amersham Pharmacia),合并含有蛋白的级分并在Amicon cell(YM10)将其浓缩至10-20 mg/ml。
在偶联的纯化和重折叠方案之后,可以从1 g大肠杆菌湿细胞获得约10-30 mg的蛋白产量,这取决于各目标蛋白。
实施例2
光谱学测量
用Uvikon XL双光束分光光度计进行蛋白浓度测量。通过使用由Pace (1995),Protein Sci.4, 2411-2423描述的程序测定摩尔消光系数(ε280)。表1中指明了用于独特的融合多肽的摩尔消光系数(ε M280)。
表 1:本研究中使用的p24融合变体的蛋白参数。所有参数都是指各自的蛋白单体。
融合多肽变体的氨基酸序列显示示于SEQ ID NO.9至16和18至24。
实施例3
生物素和钌部分与融合蛋白的偶联
分别用N-羟基-琥珀酰亚胺活化的生物素和钌标记物分子以10-30 mg/ml的蛋白浓度修饰融合多肽的赖氨酸ε-氨基基团。标记物/蛋白比率从2:1至5:1(mol:mol)变化,这取决于各融合蛋白。反应缓冲液是150 mM磷酸钾pH 8.0、100 mM KCl、0.5 mM EDTA。在室温实施反应15分钟并通过添加缓冲的L-赖氨酸至10 mM的最终浓度而停止反应。为了避免标记物的水解失活,在干燥的DMSO(无水品质, Merck, 德国)中制备各储备溶液。研究的所有融合蛋白均良好耐受反应缓冲液中多至25%的DMSO浓度。在偶联反应之后,通过将粗蛋白缀合物通过凝胶过滤柱(Superdex 200 HiLoad)去除未反应的游离标记物。
实施例4
HTLV免疫测定中不同的p24衣壳抗原变体的免疫反应性(即,抗原性)
在自动化Elecsys® 2010和cobas e 411分析仪(Roche Diagnostics GmbH)中评价HTLV p24衣壳抗原的多肽融合变体的免疫反应性(抗原性)。Elecsys®是Roche集团的注册商标。以双抗原夹心形式实施测量。
Elecsys® 2010和cobas e 411中的信号检测基于电化学发光。将生物素-缀合物(即捕获抗原)固定在链霉抗生物素蛋白包被的磁珠的表面上,而检测-抗原具有络合的钌阳离子(在氧化还原态2+和3+之间转换)作为信号传导部分。在特异性免疫球蛋白分析物存在的情况下,生色钌络合物与固相桥接并在铂电极激发之后在620 nm发光。信号输出为相对光单位。
以双抗原夹心(DAGS)免疫测定形式评价重组p24衣壳抗原融合多肽。为此,重组HTLV-I衣壳抗原p24分别用作生物素缀合物和钌缀合物,以检测人血清中的抗p24抗体。
p24是HTLV的免疫显性抗原之一,并且p24的可溶性变体 - 如本专利申请中所公开 - 是用于检测HTLV感染的非常有价值的工具。在所有测量中,在反应缓冲液中以大过量(~ 10 µg/ml)实施化学聚合和未标记的EcSlyD-EcSlyD、EcFkpA和EcSkp作为抗干扰物质以避免经由分子伴侣融合单位的免疫学交叉反应。
具体而言,在本研究中详细检查来自HTLV-I的三种p24变体,即全长p24 (146-344,编号是指Gag-Pro多蛋白前体,参见SEQ ID NO.1和5)、p24 N-末端结构域(p24/NTD,146-260)和p24 C-末端结构域(p24/CTD, 261-344)。为了检测抗-p24 IgG分子,在R1(试剂缓冲液1)和R2(试剂缓冲液2)中分别使用EcSlyD-EcSlyD-p24-生物素和EcSlyD-EcSlyD-p24-钌。为了检测抗-p24 IgM和IgG分子两者,在R1(试剂缓冲液1)和R2(试剂缓冲液2)中分别使用EcFkpA-p24-生物素和EcSkp-p24-钌。R1和R2中的抗原缀合物的浓度分别各自为100ng/ml。N-末端截短的成熟p24 (146-344)用作生物素-侧的EcSlyD-EcSlyD融合多肽和钌侧的EcFkpA融合多肽。
不幸的是,人HTLV血清转化实验对象组 – 其为用于开发改进的体外诊断测定法的不可缺少的工具 – 是不可商购的。为了评价HTLV感染的非常早期阶段中的不同的p24变体的抗原特性,我们必须回到充当血清转化模型的兔血清。为此,用纯化和灭活的HTLV-I和–II病毒裂解物(购自Zeptometrix, New York, USA)和完全弗氏佐剂免疫新西兰白兔来诱导免疫应答(2次免疫,1周时间间隔)。我们意识到,当人中真正HTLV感染之后体液免疫应答的模式可能不同于由兔的病毒裂解物接种触发的免疫应答。然而,人工诱导的兔血清转化是我们可得的最好模拟物。
在第一个实验中,在前述DAGS免疫测定设置中用抗-HTLV-阴性人血清评价单体p24 CTD (p24, 261-344),以知道背景信号。不可避免的系统固有的信号为约500个计数。低背景信号表明各自钌缀合物的高溶解度和通常良性的物理化学特性。我们可以从表2推断,单体p24 CTD的物理化学特性是优异的(第1列)。这同样适用于寡聚p24 CTD(第2列):FkpA-p24(261-344)-生物素和Skp-p24(261-344)-钌,当用作DAGS形式中的抗原对,导致阴性人血清的~ 1100个计数的背景信号,其明确指出良好的溶解特性。然而,第一眼变得明显的是,p24 CTD的单体和寡聚形式在其检测血清转化实验对象组中的抗-HTLV-抗体(且尤其是IgM分子)的能力方面强烈不同,如表2中所示。更近查看血清转化K5645,我们发现,单体p24 CTD在第18天几乎没有检测为阳性(1558个计数),而使用寡聚p24 CTD在第14天已经揭示为明显阳性的(8232个计数),并且导致在第18天高达50118个计数的信号。我们看到用血清转化实验对象组K5646、K5647和K5648相同的景象:寡聚p24 CTD变体在更早时产生更高的信号,因此保证在早期检测血清转化中的抗-p24抗体的优异灵敏度。原则上,该情况与p24的N-末端结构域(NTD)(其涵盖氨基酸残基146-260(编号是指Gag-Pro多蛋白前体))类似。如同CTD,p24 NTD的寡聚形式更好地适合于检测血清转化的早期阶段中出现的抗体(即,M型的免疫球蛋白),这特别用血清转化实验对象组K5647和K5648例举(表2,第3和4列)。然而,当与CTD p24相比时,寡聚NTD p24的背景信号显著增加。此外,p24的C-末端结构域的抗原性似乎大大超过N-末端结构域的抗原性。总之,p24的寡聚C-末端结构域具有显著的物理化学特性和优异的抗原特性,使其成为HTLV血清学的有吸引力的候选。其在早期IgM检测中的灵敏度方面明显优于全长p24(146-344,Gag-pro多蛋白前体的编号)。由于全长p24 (146-344)的Skp融合多肽因为其显著趋于聚集而不可得,所以我们被局限于p24全长版本的SlyD-SlyD和FkpA融合多肽。当在DAGS形式的生物素侧使用单体SlyD-SlyD-p24(146-344)且在DAGS形式的钌侧使用寡聚FkpA-p24 (146-344)时,其结果是相当明确的:全长p24在血清转化实验对象组的晚期阶段得到优异的信号,但其在早期检测中完全失败(表2,第5列)。寡聚CTD p24和NTD p24两者均优于单体全长变体,提供了良好的证据证明灵敏的早期检测主要取决于使用的p24片段的表位密度。提供完整的p24序列作为整体以获得优异的血清转化灵敏度似乎不是强制性的。相反,p24的N和 C-结构域中的表位足以保证HTLV感染的早期阶段中的IgM分子的灵敏和可靠的检测 - 条件是这些表位以寡聚形式提供。凭借其优异的溶解度(如低背景信号中所反映)及其显著的抗原性,来自HTLV-I的p24的C-末端结构域有希望作为HTLV免疫测定中的非常有价值的成分。这有点出乎意料:由于p24衣壳抗原的C-结构域据推测参与p24寡聚化(Khorasanizadeh等人, J. Mol. Biol. (1999)291, 491-505),我们推理,分离的C-结构域可能可能倾向于聚集,至少其应当比N-结构域更难以处理。而且,我们的预期是,在很大程度上隐藏于成熟衣壳颗粒中的p24 C-结构域可能比充分可及的N-结构域具有更少的免疫显性表位。我们惊讶的是,事实相反。实际上,p24的N-结构域也非常适合作为用于HTLV-免疫测定的抗原,尽管其在溶解度和抗原性方面似乎劣于C-末端结构域。表2显示来自HTLV-I的p24变体的结果。我们对于来自HTLV-II的相应的p24变体发现了几乎相同的结果。这与我们的预期一致,由于来自HTLV-I和HTLV-II的p24的氨基酸序列共享84%同一性和93%同源性。来自HTLV-II的p24的相应序列为152-266(N-结构域,NTD)、267-350(C-结构域,CTD)和152-350(成熟全长p24),还参见SEQ ID NO.5-8。
表 2:早期HTLV感染中的寡聚p24变体的优异的免疫反应性(兔血清转化实验对象组中的增加的灵敏度)。
实施例5
不对称双抗原夹心形式的寡聚分子伴侣载体蛋白的组合。
基本上如实施例4中所述进行免疫测定。寡聚分子伴侣诸如FkpA和Skp可以有利地用作融合伴侣以实现其各自的客户抗原的功能寡聚化。FkpA或Skp与目标蛋白(即,其客户或客座抗原)的任何可以得到良好定义的寡聚的融合多肽,其适合于在免疫测定中检测IgM分子。此处,我们解决了这样的问题:当以DAGS(双抗原夹心)形式使用时,是否存在Skp-X和FkpA-X融合多肽的特别优选的组合。(注:“X”通常是指任何目标蛋白或抗原。) 换言之,我们解决了这样的问题:在(捕获)生物素侧、而不是在(信号传导)钌侧使用FkpA-X融合多肽是否是可建议的。相反,我们想知道在(信号传导)钌侧、而不是在(捕获)生物素侧使用Skp-X融合多肽是否是可建议的。
在第一个实验中,如实施例3中所述制备纯化的重组EcSkp和EcFkpA的生物素和钌缀合物。即,借助N-羟基-琥珀酰亚胺-活化的生物素标记物生物素化纯化的分子伴侣(即,没有任何融合的目标序列的裸分子伴侣)。同样,借助N-羟基-琥珀酰亚胺-活化的钌标记物产生钌化的分子伴侣(即,没有任何融合的目标序列的裸分子伴侣)。
然后,以不同的浓度在生物素和钌侧用EcSkp进行对称DAGS。作为样品,使用抗-HTLV阴性人血清的合并物,并且一式两份实施测量。从表3中的数据显而易见,当在DAGS形式的生物素和钌侧上使用非常相同的寡聚分子伴侣(此处:EcSkp)(甚至以非常低的浓度)时,背景信号是相当高的。背景信号以剂量依赖的方式随着缀合物浓度强烈增加。因此,当使用寡聚融合伴侣诸如来自大肠杆菌的三聚分子伴侣Skp时,对称DAGS形式似乎不是可行的选项。对于二聚分子伴侣FkpA,已经发现了类似的结果。
表 3:在DAGS免疫测定的两侧使用非常相同的寡聚分子伴侣(对称DAGS形式的寡聚载体蛋白)
然而,当我们以不对称DAGS形式组合EcSkp和EcFkpA时,景象变得完全不同(参见下表4)。无论分子伴侣的组合,当我们在捕获和信号传导侧使用不同的分子伴侣时,背景信号显著降低。例如,在各250 ng/ml的缀合物浓度,对称(即,Skp-Bi/Skp-Ru) DAGS形式中的背景信号为约21,000个计数。在非常相同的缀合物浓度,不对称DAGS形式中的背景信号分别急剧降低至2,700个计数(Skp-Bi/FkpA-Ru)和860个计数(FkpA-Bi/Skp-Ru)。第一眼不言自明的是,在DAGS免疫测定中的确存在FkpA和Skp的优选组合:在DAGS免疫测定中使用FkpA作为生物素缀合物和Skp作为钌缀合物是可建议的,并且得出对于FkpA-X和Skp-X融合多肽也是如此的结论是合理的。
表 4:在DAGS免疫测定的两侧使用不同的寡聚分子伴侣(不对称DAGS形式的寡聚载体蛋白)
实施例6
Skp-p24/CTD (267-350)和FkpA-p24/CTD (267-350)的CD-检测的热诱导的解折
叠
用具有恒温室固定器的Jasco-720分光偏振计记录近-UV CD光谱,并将其转化为平均残余椭圆率。缓冲液为150 mM磷酸钾pH 8.0、100 mM KCl、0.5 mM EDTA。路径长度为0.2 cm,蛋白浓度为218 µM(是指Skp-p24单体)或147.5 µM(是指FkpA-p24单体)。测量范围为250 - 330 nm,频带宽度为1.0 nm,扫描速度为20 nm/min(分辨率为0.5 nm),且响应为1s。为了改善信噪比,九次测量光谱,并将其平均化。
圆二色光谱(CD)是选择以评价蛋白的二级和三级结构的方法。芳香族区域(250-330 nm)中的椭圆率报道蛋白内的三级接触(即,经常折叠蛋白的球状结构),并且被认为是天然样折叠(构象)的指纹区域。
分别监测Skp-p24/CTD(267-350)和FkpA-p24/CTD(267-350)、SEQ ID NO.22和21的近UV CD光谱以解决这样的问题:作为纯化过程中的关键步骤的基质偶联的重折叠程序之后,融合蛋白是否采用有序构象。答案是相当明确的:Skp-p24/CTD(参见图1)和FkpA-p24/CTD(参见图3)两者的近UV CD信号明确报道了各融合多肽的有序三级结构。显然,Skp-p24/CTD和FkpA-p24/CTD的芳香族残基被嵌于亲脂性蛋白核心中,并且因此经历不对称环境,其强烈指向各自的融合构建体内的载体和目标蛋白组分的天然样构象。Skp-p24/CTD的近UV CD光谱表现出在282和277 nm具有最大值的阴性信号(图1)。FkpA-p24/CTD的近UV CD光谱表现出在280 nm具有最大值的阳性信号(图3)。
为了解决Skp-p24/CTD和FkpA-p24/CTD的热诱导的解折叠是否是可逆的问题,分别在277和280 nm的检测波长的近UV区域中监测熔化曲线。温度范围为20-75℃,频带宽度为2.0 nm,温度斜率为1℃/min,且响应为2 s。
在分别对应于Skp-p24/CTD和FkpA-p24/CTD的最大信号振幅的277和280 nm监测热诱导的解折叠。加热后,稳定融合多肽分子的天然构象的非共价接触变得松散,并且最终分解。对于Skp-p24/CTD,该热诱导的解折叠(如在277 nm所监测)反映于CD信号的增加,如图2中所示。Skp-p24/CTD明显直至55℃保留其天然样折叠及其三聚结构。解折叠的开始在55℃和60℃之间。在70℃,分子是完全解折叠,如通过图2中的熔化曲线所判断。引人注目的是,当将蛋白溶液冷却至20℃时,CD信号得到恢复(图1、2)。然而,重折叠曲线的滞后是显著的,并且可能指向解折叠和重折叠的不同途径。令人惊讶的是,复杂三聚融合蛋白诸如Skp-p24/CTD的热诱导的解折叠 - 至少部分地 – 是可逆过程。已经预期,Skp-p24/CTD,在热诱导的解折叠并解离成单体亚基之后,将在升高温度诸如75℃非常迅速且定量地聚集。然而,我们发现,当蛋白溶液被冷却至20℃时,Skp-p24/CTD显然能够重新采用其天然样构象。的确,在热诱导的解折叠之前和之后监测的近UV CD光谱几乎重叠(参见图1)。总之,Skp-p24/CTD具有稳健的折叠特性,其对于具有这样程度的复杂性的分子是优异的,并且对于免疫测定中使用的抗原是高度期望的。我们对于FkpA-p24/CTD发现了非常类似的结果:就像Skp-p24/CTD,FkpA-p24/CTD在近UV区域(250-330 nm,在280 nm信号最大)中表现出显著的CD信号,指向基质偶联的重折叠过程之后的充分有序的构象(图3)。当分子解折叠并失去其三级结构时,FkpA-p24/CTD的CD信号强烈降低(图4)。借助热转变,我们观察到,FkpA-p24/CTD的确在直至55℃的温度保留其天然样构象。解折叠的开始-如通过在280nm的近UV CD光谱所监测-为60℃左右,并且在70℃,FkpA-p24/CTD被完全解折叠。值得注意的是,在热解折叠/重折叠循环之后,完全恢复天然FkpA-p24/CTD分子的CD信号(图4)。如图3中所举例说明,解折叠/重折叠循环之后的FkpA-p24/CTD的CD光谱几乎完美重叠。
总之,Skp-p24/CTD和FkpA-p24/CTD具有非常稳健的折叠特性,其对于具有这样程度的复杂性的分子是优异的,并且对于充当免疫测定中的抗原成分(即,指示剂)的融合多肽是高度期望的。
序列表
<110> Roche Diagnostics GmbH and Hoffmann-La Roche AG
<120> HTLV衣壳抗原p24的可溶性和免疫反应性变体
<130> P32015 WO-IR
<160> 27
<170> PatentIn version 3.5
<210> 1
<211> 199
<212> PRT
<213> 人T-细胞嗜淋巴细胞病毒1型
<400> 1
Gln Met Lys Asp Leu Gln Ala Ile Lys Gln Glu Val Ser Gln Ala Ala
1 5 10 15
Pro Gly Ser Pro Gln Phe Met Gln Thr Ile Arg Leu Ala Val Gln Gln
20 25 30
Phe Asp Pro Thr Ala Lys Asp Leu Gln Asp Leu Leu Gln Tyr Leu Cys
35 40 45
Ser Ser Leu Val Ala Ser Leu His His Gln Gln Leu Asp Ser Leu Ile
50 55 60
Ser Glu Ala Glu Thr Arg Gly Ile Thr Gly Tyr Asn Pro Leu Ala Gly
65 70 75 80
Pro Leu Arg Val Gln Ala Asn Asn Pro Gln Gln Gln Gly Leu Arg Arg
85 90 95
Glu Tyr Gln Gln Leu Trp Leu Ala Ala Phe Ala Ala Leu Pro Gly Ser
100 105 110
Ala Lys Asp Pro Ser Trp Ala Ser Ile Leu Gln Gly Leu Glu Glu Pro
115 120 125
Tyr His Ala Phe Val Glu Arg Leu Asn Ile Ala Leu Asp Asn Gly Leu
130 135 140
Pro Glu Gly Thr Pro Lys Asp Pro Ile Leu Arg Ser Leu Ala Tyr Ser
145 150 155 160
Asn Ala Asn Lys Glu Cys Gln Lys Leu Leu Gln Ala Arg Gly His Thr
165 170 175
Asn Ser Pro Leu Gly Asp Met Leu Arg Ala Cys Gln Thr Trp Thr Pro
180 185 190
Lys Asp Lys Thr Lys Val Leu
195
<210> 2
<211> 115
<212> PRT
<213> 人工
<220>
<223> 如天然序列,但X=C或A或S
<220>
<221> MISC_FEATURE
<223> Xaa = Cys或Ala或Ser
<400> 2
Gln Met Lys Asp Leu Gln Ala Ile Lys Gln Glu Val Ser Gln Ala Ala
1 5 10 15
Pro Gly Ser Pro Gln Phe Met Gln Thr Ile Arg Leu Ala Val Gln Gln
20 25 30
Phe Asp Pro Thr Ala Lys Asp Leu Gln Asp Leu Leu Gln Tyr Leu Xaa
35 40 45
Ser Ser Leu Val Ala Ser Leu His His Gln Gln Leu Asp Ser Leu Ile
50 55 60
Ser Glu Ala Glu Thr Arg Gly Ile Thr Gly Tyr Asn Pro Leu Ala Gly
65 70 75 80
Pro Leu Arg Val Gln Ala Asn Asn Pro Gln Gln Gln Gly Leu Arg Arg
85 90 95
Glu Tyr Gln Gln Leu Trp Leu Ala Ala Phe Ala Ala Leu Pro Gly Ser
100 105 110
Ala Lys Asp
115
<210> 3
<211> 84
<212> PRT
<213> 人工
<220>
<223> 如天然序列,但X=A或C或S
<220>
<221> MISC_FEATURE
<223> Xaa = Cys或Ala或Ser
<400> 3
Pro Ser Trp Ala Ser Ile Leu Gln Gly Leu Glu Glu Pro Tyr His Ala
1 5 10 15
Phe Val Glu Arg Leu Asn Ile Ala Leu Asp Asn Gly Leu Pro Glu Gly
20 25 30
Thr Pro Lys Asp Pro Ile Leu Arg Ser Leu Ala Tyr Ser Asn Ala Asn
35 40 45
Lys Glu Xaa Gln Lys Leu Leu Gln Ala Arg Gly His Thr Asn Ser Pro
50 55 60
Leu Gly Asp Met Leu Arg Ala Xaa Gln Thr Trp Thr Pro Lys Asp Lys
65 70 75 80
Thr Lys Val Leu
<210> 4
<211> 199
<212> PRT
<213> 人工
<220>
<223> 如天然序列,但X=C或A或S
<220>
<221> MISC_FEATURE
<223> Xaa = Cys或Ala或Ser
<220>
<400> 4
Gln Met Lys Asp Leu Gln Ala Ile Lys Gln Glu Val Ser Gln Ala Ala
1 5 10 15
Pro Gly Ser Pro Gln Phe Met Gln Thr Ile Arg Leu Ala Val Gln Gln
20 25 30
Phe Asp Pro Thr Ala Lys Asp Leu Gln Asp Leu Leu Gln Tyr Leu Xaa
35 40 45
Ser Ser Leu Val Ala Ser Leu His His Gln Gln Leu Asp Ser Leu Ile
50 55 60
Ser Glu Ala Glu Thr Arg Gly Ile Thr Gly Tyr Asn Pro Leu Ala Gly
65 70 75 80
Pro Leu Arg Val Gln Ala Asn Asn Pro Gln Gln Gln Gly Leu Arg Arg
85 90 95
Glu Tyr Gln Gln Leu Trp Leu Ala Ala Phe Ala Ala Leu Pro Gly Ser
100 105 110
Ala Lys Asp Pro Ser Trp Ala Ser Ile Leu Gln Gly Leu Glu Glu Pro
115 120 125
Tyr His Ala Phe Val Glu Arg Leu Asn Ile Ala Leu Asp Asn Gly Leu
130 135 140
Pro Glu Gly Thr Pro Lys Asp Pro Ile Leu Arg Ser Leu Ala Tyr Ser
145 150 155 160
Asn Ala Asn Lys Glu Xaa Gln Lys Leu Leu Gln Ala Arg Gly His Thr
165 170 175
Asn Ser Pro Leu Gly Asp Met Leu Arg Ala Xaa Gln Thr Trp Thr Pro
180 185 190
Lys Asp Lys Thr Lys Val Leu
195
<210> 5
<211> 199
<212> PRT
<213> 人T-细胞嗜淋巴细胞病毒2型
<400> 5
Gln Met Lys Asp Leu Gln Ala Ile Lys Gln Glu Val Ser Ser Ser Ala
1 5 10 15
Leu Gly Ser Pro Gln Phe Met Gln Thr Leu Arg Leu Ala Val Gln Gln
20 25 30
Phe Asp Pro Thr Ala Lys Asp Leu Gln Asp Leu Leu Gln Tyr Leu Cys
35 40 45
Ser Ser Leu Val Val Ser Leu His His Gln Gln Leu Asn Thr Leu Ile
50 55 60
Thr Glu Ala Glu Thr Arg Gly Met Thr Gly Tyr Asn Pro Met Ala Gly
65 70 75 80
Pro Leu Arg Met Gln Ala Asn Asn Pro Ala Gln Gln Gly Leu Arg Arg
85 90 95
Glu Tyr Gln Asn Leu Trp Leu Ala Ala Phe Ser Thr Leu Pro Gly Asn
100 105 110
Thr Arg Asp Pro Ser Trp Ala Ala Ile Leu Gln Gly Leu Glu Glu Pro
115 120 125
Tyr Cys Ala Phe Val Glu Arg Leu Asn Val Ala Leu Asp Asn Gly Leu
130 135 140
Pro Glu Gly Thr Pro Lys Glu Pro Ile Leu Arg Ser Leu Ala Tyr Ser
145 150 155 160
Asn Ala Asn Lys Glu Cys Gln Lys Ile Leu Gln Ala Arg Gly His Thr
165 170 175
Asn Ser Pro Leu Gly Glu Met Leu Arg Thr Cys Gln Ala Trp Thr Pro
180 185 190
Lys Asp Lys Thr Lys Val Leu
195
<210> 6
<211> 115
<212> PRT
<213> 人工
<220>
<223> 如天然序列,但X=C或A或S
<220>
<221> MISC_FEATURE
<223> Xaa = Cys或Ala或Ser
<400> 6
Gln Met Lys Asp Leu Gln Ala Ile Lys Gln Glu Val Ser Ser Ser Ala
1 5 10 15
Leu Gly Ser Pro Gln Phe Met Gln Thr Leu Arg Leu Ala Val Gln Gln
20 25 30
Phe Asp Pro Thr Ala Lys Asp Leu Gln Asp Leu Leu Gln Tyr Leu Xaa
35 40 45
Ser Ser Leu Val Val Ser Leu His His Gln Gln Leu Asn Thr Leu Ile
50 55 60
Thr Glu Ala Glu Thr Arg Gly Met Thr Gly Tyr Asn Pro Met Ala Gly
65 70 75 80
Pro Leu Arg Met Gln Ala Asn Asn Pro Ala Gln Gln Gly Leu Arg Arg
85 90 95
Glu Tyr Gln Asn Leu Trp Leu Ala Ala Phe Ser Thr Leu Pro Gly Asn
100 105 110
Thr Arg Asp
115
<210> 7
<211> 84
<212> PRT
<213> 人工
<220>
<223> 如天然序列,但X=C或A或S
<220>
<221> MISC_FEATURE
<223> Xaa = Cys或Ala或Ser
<400> 7
Pro Ser Trp Ala Ala Ile Leu Gln Gly Leu Glu Glu Pro Tyr Xaa Ala
1 5 10 15
Phe Val Glu Arg Leu Asn Val Ala Leu Asp Asn Gly Leu Pro Glu Gly
20 25 30
Thr Pro Lys Glu Pro Ile Leu Arg Ser Leu Ala Tyr Ser Asn Ala Asn
35 40 45
Lys Glu Xaa Gln Lys Ile Leu Gln Ala Arg Gly His Thr Asn Ser Pro
50 55 60
Leu Gly Glu Met Leu Arg Thr Xaa Gln Ala Trp Thr Pro Lys Asp Lys
65 70 75 80
Thr Lys Val Leu
<210> 8
<211> 199
<212> PRT
<213> 人工
<220>
<223> 如天然序列,但X=C或A或S
<220>
<221> MISC_FEATURE
<223> Xaa = Cys或Ala或Ser
<400> 8
Gln Met Lys Asp Leu Gln Ala Ile Lys Gln Glu Val Ser Ser Ser Ala
1 5 10 15
Leu Gly Ser Pro Gln Phe Met Gln Thr Leu Arg Leu Ala Val Gln Gln
20 25 30
Phe Asp Pro Thr Ala Lys Asp Leu Gln Asp Leu Leu Gln Tyr Leu Xaa
35 40 45
Ser Ser Leu Val Val Ser Leu His His Gln Gln Leu Asn Thr Leu Ile
50 55 60
Thr Glu Ala Glu Thr Arg Gly Met Thr Gly Tyr Asn Pro Met Ala Gly
65 70 75 80
Pro Leu Arg Met Gln Ala Asn Asn Pro Ala Gln Gln Gly Leu Arg Arg
85 90 95
Glu Tyr Gln Asn Leu Trp Leu Ala Ala Phe Ser Thr Leu Pro Gly Asn
100 105 110
Thr Arg Asp Pro Ser Trp Ala Ala Ile Leu Gln Gly Leu Glu Glu Pro
115 120 125
Tyr Xaa Ala Phe Val Glu Arg Leu Asn Val Ala Leu Asp Asn Gly Leu
130 135 140
Pro Glu Gly Thr Pro Lys Glu Pro Ile Leu Arg Ser Leu Ala Tyr Ser
145 150 155 160
Asn Ala Asn Lys Glu Xaa Gln Lys Ile Leu Gln Ala Arg Gly His Thr
165 170 175
Asn Ser Pro Leu Gly Glu Met Leu Arg Thr Xaa Gln Ala Trp Thr Pro
180 185 190
Lys Asp Lys Thr Lys Val Leu
195
<210> 9
<211> 582
<212> PRT
<213> 人工
<220>
<223> 融合蛋白
<400> 9
Met Lys Val Ala Lys Asp Leu Val Val Ser Leu Ala Tyr Gln Val Arg
1 5 10 15
Thr Glu Asp Gly Val Leu Val Asp Glu Ser Pro Val Ser Ala Pro Leu
20 25 30
Asp Tyr Leu His Gly His Gly Ser Leu Ile Ser Gly Leu Glu Thr Ala
35 40 45
Leu Glu Gly His Glu Val Gly Asp Lys Phe Asp Val Ala Val Gly Ala
50 55 60
Asn Asp Ala Tyr Gly Gln Tyr Asp Glu Asn Leu Val Gln Arg Val Pro
65 70 75 80
Lys Asp Val Phe Met Gly Val Asp Glu Leu Gln Val Gly Met Arg Phe
85 90 95
Leu Ala Glu Thr Asp Gln Gly Pro Val Pro Val Glu Ile Thr Ala Val
100 105 110
Glu Asp Asp His Val Val Val Asp Gly Asn His Met Leu Ala Gly Gln
115 120 125
Asn Leu Lys Phe Asn Val Glu Val Val Ala Ile Arg Glu Ala Thr Glu
130 135 140
Glu Glu Leu Ala His Gly His Val His Gly Ala His Asp His His His
145 150 155 160
Asp His Asp His Asp Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly
165 170 175
Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Lys Val Ala Lys
180 185 190
Asp Leu Val Val Ser Leu Ala Tyr Gln Val Arg Thr Glu Asp Gly Val
195 200 205
Leu Val Asp Glu Ser Pro Val Ser Ala Pro Leu Asp Tyr Leu His Gly
210 215 220
His Gly Ser Leu Ile Ser Gly Leu Glu Thr Ala Leu Glu Gly His Glu
225 230 235 240
Val Gly Asp Lys Phe Asp Val Ala Val Gly Ala Asn Asp Ala Tyr Gly
245 250 255
Gln Tyr Asp Glu Asn Leu Val Gln Arg Val Pro Lys Asp Val Phe Met
260 265 270
Gly Val Asp Glu Leu Gln Val Gly Met Arg Phe Leu Ala Glu Thr Asp
275 280 285
Gln Gly Pro Val Pro Val Glu Ile Thr Ala Val Glu Asp Asp His Val
290 295 300
Val Val Asp Gly Asn His Met Leu Ala Gly Gln Asn Leu Lys Phe Asn
305 310 315 320
Val Glu Val Val Ala Ile Arg Glu Ala Thr Glu Glu Glu Leu Ala His
325 330 335
Gly His Val His Gly Ala His Asp His His His Asp His Asp His Asp
340 345 350
Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser
355 360 365
Gly Gly Gly Ser Gly Gly Gly Gln Met Lys Asp Leu Gln Ala Ile Lys
370 375 380
Gln Glu Val Ser Gln Ala Ala Pro Gly Ser Pro Gln Phe Met Gln Thr
385 390 395 400
Ile Arg Leu Ala Val Gln Gln Phe Asp Pro Thr Ala Lys Asp Leu Gln
405 410 415
Asp Leu Leu Gln Tyr Leu Ala Ser Ser Leu Val Ala Ser Leu His His
420 425 430
Gln Gln Leu Asp Ser Leu Ile Ser Glu Ala Glu Thr Arg Gly Ile Thr
435 440 445
Gly Tyr Asn Pro Leu Ala Gly Pro Leu Arg Val Gln Ala Asn Asn Pro
450 455 460
Gln Gln Gln Gly Leu Arg Arg Glu Tyr Gln Gln Leu Trp Leu Ala Ala
465 470 475 480
Phe Ala Ala Leu Pro Gly Ser Ala Lys Asp Pro Ser Trp Ala Ser Ile
485 490 495
Leu Gln Gly Leu Glu Glu Pro Tyr His Ala Phe Val Glu Arg Leu Asn
500 505 510
Ile Ala Leu Asp Asn Gly Leu Pro Glu Gly Thr Pro Lys Asp Pro Ile
515 520 525
Leu Arg Ser Leu Ala Tyr Ser Asn Ala Asn Lys Glu Ala Gln Lys Leu
530 535 540
Leu Gln Ala Arg Gly His Thr Asn Ser Pro Leu Gly Asp Met Leu Arg
545 550 555 560
Ala Ala Gln Thr Trp Thr Pro Lys Asp Lys Thr Lys Val Leu Leu Glu
565 570 575
His His His His His His
580
<210> 10
<211> 476
<212> PRT
<213> 人工
<220>
<223> 融合蛋白
<400> 10
Met Ala Glu Ala Ala Lys Pro Ala Thr Thr Ala Asp Ser Lys Ala Ala
1 5 10 15
Phe Lys Asn Asp Asp Gln Lys Ser Ala Tyr Ala Leu Gly Ala Ser Leu
20 25 30
Gly Arg Tyr Met Glu Asn Ser Leu Lys Glu Gln Glu Lys Leu Gly Ile
35 40 45
Lys Leu Asp Lys Asp Gln Leu Ile Ala Gly Val Gln Asp Ala Phe Ala
50 55 60
Asp Lys Ser Lys Leu Ser Asp Gln Glu Ile Glu Gln Thr Leu Gln Ala
65 70 75 80
Phe Glu Ala Arg Val Lys Ser Ser Ala Gln Ala Lys Met Glu Lys Asp
85 90 95
Ala Ala Asp Asn Glu Ala Lys Gly Lys Glu Tyr Arg Glu Lys Phe Ala
100 105 110
Lys Glu Lys Gly Val Lys Thr Ser Ser Thr Gly Leu Val Tyr Gln Val
115 120 125
Val Glu Ala Gly Lys Gly Glu Ala Pro Lys Asp Ser Asp Thr Val Val
130 135 140
Val Asn Tyr Lys Gly Thr Leu Ile Asp Gly Lys Glu Phe Asp Asn Ser
145 150 155 160
Tyr Thr Arg Gly Glu Pro Leu Ser Phe Arg Leu Asp Gly Val Ile Pro
165 170 175
Gly Trp Thr Glu Gly Leu Lys Asn Ile Lys Lys Gly Gly Lys Ile Lys
180 185 190
Leu Val Ile Pro Pro Glu Leu Ala Tyr Gly Lys Ala Gly Val Pro Gly
195 200 205
Ile Pro Pro Asn Ser Thr Leu Val Phe Asp Val Glu Leu Leu Asp Val
210 215 220
Lys Pro Ala Pro Lys Ala Asp Ala Lys Pro Glu Ala Asp Ala Lys Ala
225 230 235 240
Ala Asp Ser Ala Lys Lys Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
245 250 255
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Gln Met Lys
260 265 270
Asp Leu Gln Ala Ile Lys Gln Glu Val Ser Gln Ala Ala Pro Gly Ser
275 280 285
Pro Gln Phe Met Gln Thr Ile Arg Leu Ala Val Gln Gln Phe Asp Pro
290 295 300
Thr Ala Lys Asp Leu Gln Asp Leu Leu Gln Tyr Leu Ala Ser Ser Leu
305 310 315 320
Val Ala Ser Leu His His Gln Gln Leu Asp Ser Leu Ile Ser Glu Ala
325 330 335
Glu Thr Arg Gly Ile Thr Gly Tyr Asn Pro Leu Ala Gly Pro Leu Arg
340 345 350
Val Gln Ala Asn Asn Pro Gln Gln Gln Gly Leu Arg Arg Glu Tyr Gln
355 360 365
Gln Leu Trp Leu Ala Ala Phe Ala Ala Leu Pro Gly Ser Ala Lys Asp
370 375 380
Pro Ser Trp Ala Ser Ile Leu Gln Gly Leu Glu Glu Pro Tyr His Ala
385 390 395 400
Phe Val Glu Arg Leu Asn Ile Ala Leu Asp Asn Gly Leu Pro Glu Gly
405 410 415
Thr Pro Lys Asp Pro Ile Leu Arg Ser Leu Ala Tyr Ser Asn Ala Asn
420 425 430
Lys Glu Ala Gln Lys Leu Leu Gln Ala Arg Gly His Thr Asn Ser Pro
435 440 445
Leu Gly Asp Met Leu Arg Ala Ala Gln Thr Trp Thr Pro Lys Asp Lys
450 455 460
Thr Lys Val Leu Leu Glu His His His His His His
465 470 475
<210> 11
<211> 469
<212> PRT
<213> 人工
<220>
<223> 融合蛋白
<400> 11
Met Lys Val Ala Lys Asp Leu Val Val Ser Leu Ala Tyr Gln Val Arg
1 5 10 15
Thr Glu Asp Gly Val Leu Val Asp Glu Ser Pro Val Ser Ala Pro Leu
20 25 30
Asp Tyr Leu His Gly His Gly Ser Leu Ile Ser Gly Leu Glu Thr Ala
35 40 45
Leu Glu Gly His Glu Val Gly Asp Lys Phe Asp Val Ala Val Gly Ala
50 55 60
Asn Asp Ala Tyr Gly Gln Tyr Asp Glu Asn Leu Val Gln Arg Val Pro
65 70 75 80
Lys Asp Val Phe Met Gly Val Asp Glu Leu Gln Val Gly Met Arg Phe
85 90 95
Leu Ala Glu Thr Asp Gln Gly Pro Val Pro Val Glu Ile Thr Ala Val
100 105 110
Glu Asp Asp His Val Val Val Asp Gly Asn His Met Leu Ala Gly Gln
115 120 125
Asn Leu Lys Phe Asn Val Glu Val Val Ala Ile Arg Glu Ala Thr Glu
130 135 140
Glu Glu Leu Ala His Gly His Val His Gly Ala His Asp His His His
145 150 155 160
Asp His Asp His Asp Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly
165 170 175
Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Lys Val Ala Lys
180 185 190
Asp Leu Val Val Ser Leu Ala Tyr Gln Val Arg Thr Glu Asp Gly Val
195 200 205
Leu Val Asp Glu Ser Pro Val Ser Ala Pro Leu Asp Tyr Leu His Gly
210 215 220
His Gly Ser Leu Ile Ser Gly Leu Glu Thr Ala Leu Glu Gly His Glu
225 230 235 240
Val Gly Asp Lys Phe Asp Val Ala Val Gly Ala Asn Asp Ala Tyr Gly
245 250 255
Gln Tyr Asp Glu Asn Leu Val Gln Arg Val Pro Lys Asp Val Phe Met
260 265 270
Gly Val Asp Glu Leu Gln Val Gly Met Arg Phe Leu Ala Glu Thr Asp
275 280 285
Gln Gly Pro Val Pro Val Glu Ile Thr Ala Val Glu Asp Asp His Val
290 295 300
Val Val Asp Gly Asn His Met Leu Ala Gly Gln Asn Leu Lys Phe Asn
305 310 315 320
Val Glu Val Val Ala Ile Arg Glu Ala Thr Glu Glu Glu Leu Ala His
325 330 335
Gly His Val His Gly Ala His Asp His His His Asp His Asp His Asp
340 345 350
Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser
355 360 365
Gly Gly Gly Ser Gly Gly Gly Ala Lys Asp Pro Ser Trp Ala Ser Ile
370 375 380
Leu Gln Gly Leu Glu Glu Pro Tyr His Ala Phe Val Glu Arg Leu Asn
385 390 395 400
Ile Ala Leu Asp Asn Gly Leu Pro Glu Gly Thr Pro Lys Asp Pro Ile
405 410 415
Leu Arg Ser Leu Ala Tyr Ser Asn Ala Asn Lys Glu Ala Gln Lys Leu
420 425 430
Leu Gln Ala Arg Gly His Thr Asn Ser Pro Leu Gly Asp Met Leu Arg
435 440 445
Ala Ala Gln Thr Trp Thr Pro Lys Asp Lys Thr Lys Val Leu Glu His
450 455 460
His His His His His
465
<210> 12
<211> 363
<212> PRT
<213> 人工
<220>
<223> 融合蛋白
<400> 12
Met Ala Glu Ala Ala Lys Pro Ala Thr Thr Ala Asp Ser Lys Ala Ala
1 5 10 15
Phe Lys Asn Asp Asp Gln Lys Ser Ala Tyr Ala Leu Gly Ala Ser Leu
20 25 30
Gly Arg Tyr Met Glu Asn Ser Leu Lys Glu Gln Glu Lys Leu Gly Ile
35 40 45
Lys Leu Asp Lys Asp Gln Leu Ile Ala Gly Val Gln Asp Ala Phe Ala
50 55 60
Asp Lys Ser Lys Leu Ser Asp Gln Glu Ile Glu Gln Thr Leu Gln Ala
65 70 75 80
Phe Glu Ala Arg Val Lys Ser Ser Ala Gln Ala Lys Met Glu Lys Asp
85 90 95
Ala Ala Asp Asn Glu Ala Lys Gly Lys Glu Tyr Arg Glu Lys Phe Ala
100 105 110
Lys Glu Lys Gly Val Lys Thr Ser Ser Thr Gly Leu Val Tyr Gln Val
115 120 125
Val Glu Ala Gly Lys Gly Glu Ala Pro Lys Asp Ser Asp Thr Val Val
130 135 140
Val Asn Tyr Lys Gly Thr Leu Ile Asp Gly Lys Glu Phe Asp Asn Ser
145 150 155 160
Tyr Thr Arg Gly Glu Pro Leu Ser Phe Arg Leu Asp Gly Val Ile Pro
165 170 175
Gly Trp Thr Glu Gly Leu Lys Asn Ile Lys Lys Gly Gly Lys Ile Lys
180 185 190
Leu Val Ile Pro Pro Glu Leu Ala Tyr Gly Lys Ala Gly Val Pro Gly
195 200 205
Ile Pro Pro Asn Ser Thr Leu Val Phe Asp Val Glu Leu Leu Asp Val
210 215 220
Lys Pro Ala Pro Lys Ala Asp Ala Lys Pro Glu Ala Asp Ala Lys Ala
225 230 235 240
Ala Asp Ser Ala Lys Lys Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
245 250 255
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ala Lys Asp
260 265 270
Pro Ser Trp Ala Ser Ile Leu Gln Gly Leu Glu Glu Pro Tyr His Ala
275 280 285
Phe Val Glu Arg Leu Asn Ile Ala Leu Asp Asn Gly Leu Pro Glu Gly
290 295 300
Thr Pro Lys Asp Pro Ile Leu Arg Ser Leu Ala Tyr Ser Asn Ala Asn
305 310 315 320
Lys Glu Ala Gln Lys Leu Leu Gln Ala Arg Gly His Thr Asn Ser Pro
325 330 335
Leu Gly Asp Met Leu Arg Ala Ala Gln Thr Trp Thr Pro Lys Asp Lys
340 345 350
Thr Lys Val Leu Glu His His His His His His
355 360
<210> 13
<211> 259
<212> PRT
<213> 人工
<220>
<223> 融合蛋白
<400> 13
Met Ala Asp Lys Ile Ala Ile Val Asn Met Gly Ser Leu Phe Gln Gln
1 5 10 15
Val Ala Gln Lys Thr Gly Val Ser Asn Thr Leu Glu Asn Glu Phe Arg
20 25 30
Gly Arg Ala Ser Glu Leu Gln Arg Met Glu Thr Asp Leu Gln Ala Lys
35 40 45
Met Lys Lys Leu Gln Ser Met Lys Ala Gly Ser Asp Arg Thr Lys Leu
50 55 60
Glu Lys Asp Val Met Ala Gln Arg Gln Thr Phe Ala Gln Lys Ala Gln
65 70 75 80
Ala Phe Glu Gln Asp Arg Ala Arg Arg Ser Asn Glu Glu Arg Gly Lys
85 90 95
Leu Val Thr Arg Ile Gln Thr Ala Val Lys Ser Val Ala Asn Ser Gln
100 105 110
Asp Ile Asp Leu Val Val Asp Ala Asn Ala Val Ala Tyr Asn Ser Ser
115 120 125
Asp Val Lys Asp Ile Thr Ala Asp Val Leu Lys Gln Val Lys Gly Gly
130 135 140
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
145 150 155 160
Gly Ser Gly Gly Gly Ala Lys Asp Pro Ser Trp Ala Ser Ile Leu Gln
165 170 175
Gly Leu Glu Glu Pro Tyr His Ala Phe Val Glu Arg Leu Asn Ile Ala
180 185 190
Leu Asp Asn Gly Leu Pro Glu Gly Thr Pro Lys Asp Pro Ile Leu Arg
195 200 205
Ser Leu Ala Tyr Ser Asn Ala Asn Lys Glu Ala Gln Lys Leu Leu Gln
210 215 220
Ala Arg Gly His Thr Asn Ser Pro Leu Gly Asp Met Leu Arg Ala Ala
225 230 235 240
Gln Thr Trp Thr Pro Lys Asp Lys Thr Lys Val Leu Glu His His His
245 250 255
His His His
<210> 14
<211> 498
<212> PRT
<213> 人工
<220>
<223> 融合蛋白
<400> 14
Met Lys Val Ala Lys Asp Leu Val Val Ser Leu Ala Tyr Gln Val Arg
1 5 10 15
Thr Glu Asp Gly Val Leu Val Asp Glu Ser Pro Val Ser Ala Pro Leu
20 25 30
Asp Tyr Leu His Gly His Gly Ser Leu Ile Ser Gly Leu Glu Thr Ala
35 40 45
Leu Glu Gly His Glu Val Gly Asp Lys Phe Asp Val Ala Val Gly Ala
50 55 60
Asn Asp Ala Tyr Gly Gln Tyr Asp Glu Asn Leu Val Gln Arg Val Pro
65 70 75 80
Lys Asp Val Phe Met Gly Val Asp Glu Leu Gln Val Gly Met Arg Phe
85 90 95
Leu Ala Glu Thr Asp Gln Gly Pro Val Pro Val Glu Ile Thr Ala Val
100 105 110
Glu Asp Asp His Val Val Val Asp Gly Asn His Met Leu Ala Gly Gln
115 120 125
Asn Leu Lys Phe Asn Val Glu Val Val Ala Ile Arg Glu Ala Thr Glu
130 135 140
Glu Glu Leu Ala His Gly His Val His Gly Ala His Asp His His His
145 150 155 160
Asp His Asp His Asp Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly
165 170 175
Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Lys Val Ala Lys
180 185 190
Asp Leu Val Val Ser Leu Ala Tyr Gln Val Arg Thr Glu Asp Gly Val
195 200 205
Leu Val Asp Glu Ser Pro Val Ser Ala Pro Leu Asp Tyr Leu His Gly
210 215 220
His Gly Ser Leu Ile Ser Gly Leu Glu Thr Ala Leu Glu Gly His Glu
225 230 235 240
Val Gly Asp Lys Phe Asp Val Ala Val Gly Ala Asn Asp Ala Tyr Gly
245 250 255
Gln Tyr Asp Glu Asn Leu Val Gln Arg Val Pro Lys Asp Val Phe Met
260 265 270
Gly Val Asp Glu Leu Gln Val Gly Met Arg Phe Leu Ala Glu Thr Asp
275 280 285
Gln Gly Pro Val Pro Val Glu Ile Thr Ala Val Glu Asp Asp His Val
290 295 300
Val Val Asp Gly Asn His Met Leu Ala Gly Gln Asn Leu Lys Phe Asn
305 310 315 320
Val Glu Val Val Ala Ile Arg Glu Ala Thr Glu Glu Glu Leu Ala His
325 330 335
Gly His Val His Gly Ala His Asp His His His Asp His Asp His Asp
340 345 350
Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser
355 360 365
Gly Gly Gly Ser Gly Gly Gly Gln Met Lys Asp Leu Gln Ala Ile Lys
370 375 380
Gln Glu Val Ser Gln Ala Ala Pro Gly Ser Pro Gln Phe Met Gln Thr
385 390 395 400
Ile Arg Leu Ala Val Gln Gln Phe Asp Pro Thr Ala Lys Asp Leu Gln
405 410 415
Asp Leu Leu Gln Tyr Leu Ala Ser Ser Leu Val Ala Ser Leu His His
420 425 430
Gln Gln Leu Asp Ser Leu Ile Ser Glu Ala Glu Thr Arg Gly Ile Thr
435 440 445
Gly Tyr Asn Pro Leu Ala Gly Pro Leu Arg Val Gln Ala Asn Asn Pro
450 455 460
Gln Gln Gln Gly Leu Arg Arg Glu Tyr Gln Gln Leu Trp Leu Ala Ala
465 470 475 480
Phe Ala Ala Leu Pro Gly Ser Ala Lys Asp Leu Glu His His His His
485 490 495
His His
<210> 15
<211> 392
<212> PRT
<213> 人工
<220>
<223> 融合蛋白
<400> 15
Met Ala Glu Ala Ala Lys Pro Ala Thr Thr Ala Asp Ser Lys Ala Ala
1 5 10 15
Phe Lys Asn Asp Asp Gln Lys Ser Ala Tyr Ala Leu Gly Ala Ser Leu
20 25 30
Gly Arg Tyr Met Glu Asn Ser Leu Lys Glu Gln Glu Lys Leu Gly Ile
35 40 45
Lys Leu Asp Lys Asp Gln Leu Ile Ala Gly Val Gln Asp Ala Phe Ala
50 55 60
Asp Lys Ser Lys Leu Ser Asp Gln Glu Ile Glu Gln Thr Leu Gln Ala
65 70 75 80
Phe Glu Ala Arg Val Lys Ser Ser Ala Gln Ala Lys Met Glu Lys Asp
85 90 95
Ala Ala Asp Asn Glu Ala Lys Gly Lys Glu Tyr Arg Glu Lys Phe Ala
100 105 110
Lys Glu Lys Gly Val Lys Thr Ser Ser Thr Gly Leu Val Tyr Gln Val
115 120 125
Val Glu Ala Gly Lys Gly Glu Ala Pro Lys Asp Ser Asp Thr Val Val
130 135 140
Val Asn Tyr Lys Gly Thr Leu Ile Asp Gly Lys Glu Phe Asp Asn Ser
145 150 155 160
Tyr Thr Arg Gly Glu Pro Leu Ser Phe Arg Leu Asp Gly Val Ile Pro
165 170 175
Gly Trp Thr Glu Gly Leu Lys Asn Ile Lys Lys Gly Gly Lys Ile Lys
180 185 190
Leu Val Ile Pro Pro Glu Leu Ala Tyr Gly Lys Ala Gly Val Pro Gly
195 200 205
Ile Pro Pro Asn Ser Thr Leu Val Phe Asp Val Glu Leu Leu Asp Val
210 215 220
Lys Pro Ala Pro Lys Ala Asp Ala Lys Pro Glu Ala Asp Ala Lys Ala
225 230 235 240
Ala Asp Ser Ala Lys Lys Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
245 250 255
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Gln Met Lys
260 265 270
Asp Leu Gln Ala Ile Lys Gln Glu Val Ser Gln Ala Ala Pro Gly Ser
275 280 285
Pro Gln Phe Met Gln Thr Ile Arg Leu Ala Val Gln Gln Phe Asp Pro
290 295 300
Thr Ala Lys Asp Leu Gln Asp Leu Leu Gln Tyr Leu Ala Ser Ser Leu
305 310 315 320
Val Ala Ser Leu His His Gln Gln Leu Asp Ser Leu Ile Ser Glu Ala
325 330 335
Glu Thr Arg Gly Ile Thr Gly Tyr Asn Pro Leu Ala Gly Pro Leu Arg
340 345 350
Val Gln Ala Asn Asn Pro Gln Gln Gln Gly Leu Arg Arg Glu Tyr Gln
355 360 365
Gln Leu Trp Leu Ala Ala Phe Ala Ala Leu Pro Gly Ser Ala Lys Asp
370 375 380
Leu Glu His His His His His His
385 390
<210> 16
<211> 288
<212> PRT
<213> 人工
<220>
<223> 融合蛋白
<400> 16
Met Ala Asp Lys Ile Ala Ile Val Asn Met Gly Ser Leu Phe Gln Gln
1 5 10 15
Val Ala Gln Lys Thr Gly Val Ser Asn Thr Leu Glu Asn Glu Phe Arg
20 25 30
Gly Arg Ala Ser Glu Leu Gln Arg Met Glu Thr Asp Leu Gln Ala Lys
35 40 45
Met Lys Lys Leu Gln Ser Met Lys Ala Gly Ser Asp Arg Thr Lys Leu
50 55 60
Glu Lys Asp Val Met Ala Gln Arg Gln Thr Phe Ala Gln Lys Ala Gln
65 70 75 80
Ala Phe Glu Gln Asp Arg Ala Arg Arg Ser Asn Glu Glu Arg Gly Lys
85 90 95
Leu Val Thr Arg Ile Gln Thr Ala Val Lys Ser Val Ala Asn Ser Gln
100 105 110
Asp Ile Asp Leu Val Val Asp Ala Asn Ala Val Ala Tyr Asn Ser Ser
115 120 125
Asp Val Lys Asp Ile Thr Ala Asp Val Leu Lys Gln Val Lys Gly Gly
130 135 140
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
145 150 155 160
Gly Ser Gly Gly Gly Gln Met Lys Asp Leu Gln Ala Ile Lys Gln Glu
165 170 175
Val Ser Gln Ala Ala Pro Gly Ser Pro Gln Phe Met Gln Thr Ile Arg
180 185 190
Leu Ala Val Gln Gln Phe Asp Pro Thr Ala Lys Asp Leu Gln Asp Leu
195 200 205
Leu Gln Tyr Leu Ala Ser Ser Leu Val Ala Ser Leu His His Gln Gln
210 215 220
Leu Asp Ser Leu Ile Ser Glu Ala Glu Thr Arg Gly Ile Thr Gly Tyr
225 230 235 240
Asn Pro Leu Ala Gly Pro Leu Arg Val Gln Ala Asn Asn Pro Gln Gln
245 250 255
Gln Gly Leu Arg Arg Glu Tyr Gln Gln Leu Trp Leu Ala Ala Phe Ala
260 265 270
Ala Leu Pro Gly Ser Ala Lys Asp Leu Glu His His His His His His
275 280 285
<210> 17
<211> 23
<212> PRT
<213> 人工
<220>
<223> 接头
<400> 17
Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser
1 5 10 15
Gly Gly Gly Ser Gly Gly Gly
20
<210> 18
<211> 582
<212> PRT
<213> 人工
<220>
<223> EcSlyD-EcSlyD-p24(152-350)/HTLV-II融合蛋白
<400> 18
Met Lys Val Ala Lys Asp Leu Val Val Ser Leu Ala Tyr Gln Val Arg
1 5 10 15
Thr Glu Asp Gly Val Leu Val Asp Glu Ser Pro Val Ser Ala Pro Leu
20 25 30
Asp Tyr Leu His Gly His Gly Ser Leu Ile Ser Gly Leu Glu Thr Ala
35 40 45
Leu Glu Gly His Glu Val Gly Asp Lys Phe Asp Val Ala Val Gly Ala
50 55 60
Asn Asp Ala Tyr Gly Gln Tyr Asp Glu Asn Leu Val Gln Arg Val Pro
65 70 75 80
Lys Asp Val Phe Met Gly Val Asp Glu Leu Gln Val Gly Met Arg Phe
85 90 95
Leu Ala Glu Thr Asp Gln Gly Pro Val Pro Val Glu Ile Thr Ala Val
100 105 110
Glu Asp Asp His Val Val Val Asp Gly Asn His Met Leu Ala Gly Gln
115 120 125
Asn Leu Lys Phe Asn Val Glu Val Val Ala Ile Arg Glu Ala Thr Glu
130 135 140
Glu Glu Leu Ala His Gly His Val His Gly Ala His Asp His His His
145 150 155 160
Asp His Asp His Asp Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly
165 170 175
Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Lys Val Ala Lys
180 185 190
Asp Leu Val Val Ser Leu Ala Tyr Gln Val Arg Thr Glu Asp Gly Val
195 200 205
Leu Val Asp Glu Ser Pro Val Ser Ala Pro Leu Asp Tyr Leu His Gly
210 215 220
His Gly Ser Leu Ile Ser Gly Leu Glu Thr Ala Leu Glu Gly His Glu
225 230 235 240
Val Gly Asp Lys Phe Asp Val Ala Val Gly Ala Asn Asp Ala Tyr Gly
245 250 255
Gln Tyr Asp Glu Asn Leu Val Gln Arg Val Pro Lys Asp Val Phe Met
260 265 270
Gly Val Asp Glu Leu Gln Val Gly Met Arg Phe Leu Ala Glu Thr Asp
275 280 285
Gln Gly Pro Val Pro Val Glu Ile Thr Ala Val Glu Asp Asp His Val
290 295 300
Val Val Asp Gly Asn His Met Leu Ala Gly Gln Asn Leu Lys Phe Asn
305 310 315 320
Val Glu Val Val Ala Ile Arg Glu Ala Thr Glu Glu Glu Leu Ala His
325 330 335
Gly His Val His Gly Ala His Asp His His His Asp His Asp His Asp
340 345 350
Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser
355 360 365
Gly Gly Gly Ser Gly Gly Gly Gln Met Lys Asp Leu Gln Ala Ile Lys
370 375 380
Gln Glu Val Ser Ser Ser Ala Leu Gly Ser Pro Gln Phe Met Gln Thr
385 390 395 400
Leu Arg Leu Ala Val Gln Gln Phe Asp Pro Thr Ala Lys Asp Leu Gln
405 410 415
Asp Leu Leu Gln Tyr Leu Ala Ser Ser Leu Val Val Ser Leu His His
420 425 430
Gln Gln Leu Asn Thr Leu Ile Thr Glu Ala Glu Thr Arg Gly Met Thr
435 440 445
Gly Tyr Asn Pro Met Ala Gly Pro Leu Arg Met Gln Ala Asn Asn Pro
450 455 460
Ala Gln Gln Gly Leu Arg Arg Glu Tyr Gln Asn Leu Trp Leu Ala Ala
465 470 475 480
Phe Ser Thr Leu Pro Gly Asn Thr Arg Asp Pro Ser Trp Ala Ala Ile
485 490 495
Leu Gln Gly Leu Glu Glu Pro Tyr Ala Ala Phe Val Glu Arg Leu Asn
500 505 510
Val Ala Leu Asp Asn Gly Leu Pro Glu Gly Thr Pro Lys Glu Pro Ile
515 520 525
Leu Arg Ser Leu Ala Tyr Ser Asn Ala Asn Lys Glu Ala Gln Lys Ile
530 535 540
Leu Gln Ala Arg Gly His Thr Asn Ser Pro Leu Gly Glu Met Leu Arg
545 550 555 560
Thr Ala Gln Ala Trp Thr Pro Lys Asp Lys Thr Lys Val Leu Leu Glu
565 570 575
His His His His His His
580
<210> 19
<211> 476
<212> PRT
<213> 人工
<220>
<223> EcFkpA-p24(152-350)/HTLV-II融合蛋白
<400> 19
Met Ala Glu Ala Ala Lys Pro Ala Thr Thr Ala Asp Ser Lys Ala Ala
1 5 10 15
Phe Lys Asn Asp Asp Gln Lys Ser Ala Tyr Ala Leu Gly Ala Ser Leu
20 25 30
Gly Arg Tyr Met Glu Asn Ser Leu Lys Glu Gln Glu Lys Leu Gly Ile
35 40 45
Lys Leu Asp Lys Asp Gln Leu Ile Ala Gly Val Gln Asp Ala Phe Ala
50 55 60
Asp Lys Ser Lys Leu Ser Asp Gln Glu Ile Glu Gln Thr Leu Gln Ala
65 70 75 80
Phe Glu Ala Arg Val Lys Ser Ser Ala Gln Ala Lys Met Glu Lys Asp
85 90 95
Ala Ala Asp Asn Glu Ala Lys Gly Lys Glu Tyr Arg Glu Lys Phe Ala
100 105 110
Lys Glu Lys Gly Val Lys Thr Ser Ser Thr Gly Leu Val Tyr Gln Val
115 120 125
Val Glu Ala Gly Lys Gly Glu Ala Pro Lys Asp Ser Asp Thr Val Val
130 135 140
Val Asn Tyr Lys Gly Thr Leu Ile Asp Gly Lys Glu Phe Asp Asn Ser
145 150 155 160
Tyr Thr Arg Gly Glu Pro Leu Ser Phe Arg Leu Asp Gly Val Ile Pro
165 170 175
Gly Trp Thr Glu Gly Leu Lys Asn Ile Lys Lys Gly Gly Lys Ile Lys
180 185 190
Leu Val Ile Pro Pro Glu Leu Ala Tyr Gly Lys Ala Gly Val Pro Gly
195 200 205
Ile Pro Pro Asn Ser Thr Leu Val Phe Asp Val Glu Leu Leu Asp Val
210 215 220
Lys Pro Ala Pro Lys Ala Asp Ala Lys Pro Glu Ala Asp Ala Lys Ala
225 230 235 240
Ala Asp Ser Ala Lys Lys Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
245 250 255
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Gln Met Lys
260 265 270
Asp Leu Gln Ala Ile Lys Gln Glu Val Ser Ser Ser Ala Leu Gly Ser
275 280 285
Pro Gln Phe Met Gln Thr Leu Arg Leu Ala Val Gln Gln Phe Asp Pro
290 295 300
Thr Ala Lys Asp Leu Gln Asp Leu Leu Gln Tyr Leu Ala Ser Ser Leu
305 310 315 320
Val Val Ser Leu His His Gln Gln Leu Asn Thr Leu Ile Thr Glu Ala
325 330 335
Glu Thr Arg Gly Met Thr Gly Tyr Asn Pro Met Ala Gly Pro Leu Arg
340 345 350
Met Gln Ala Asn Asn Pro Ala Gln Gln Gly Leu Arg Arg Glu Tyr Gln
355 360 365
Asn Leu Trp Leu Ala Ala Phe Ser Thr Leu Pro Gly Asn Thr Arg Asp
370 375 380
Pro Ser Trp Ala Ala Ile Leu Gln Gly Leu Glu Glu Pro Tyr Ala Ala
385 390 395 400
Phe Val Glu Arg Leu Asn Val Ala Leu Asp Asn Gly Leu Pro Glu Gly
405 410 415
Thr Pro Lys Glu Pro Ile Leu Arg Ser Leu Ala Tyr Ser Asn Ala Asn
420 425 430
Lys Glu Ala Gln Lys Ile Leu Gln Ala Arg Gly His Thr Asn Ser Pro
435 440 445
Leu Gly Glu Met Leu Arg Thr Ala Gln Ala Trp Thr Pro Lys Asp Lys
450 455 460
Thr Lys Val Leu Leu Glu His His His His His His
465 470 475
<210> 20
<211> 372
<212> PRT
<213> 人工
<220>
<223> EcSkp-p24(152-350)/HTLV-II融合蛋白
<400> 20
Met Ala Asp Lys Ile Ala Ile Val Asn Met Gly Ser Leu Phe Gln Gln
1 5 10 15
Val Ala Gln Lys Thr Gly Val Ser Asn Thr Leu Glu Asn Glu Phe Arg
20 25 30
Gly Arg Ala Ser Glu Leu Gln Arg Met Glu Thr Asp Leu Gln Ala Lys
35 40 45
Met Lys Lys Leu Gln Ser Met Lys Ala Gly Ser Asp Arg Thr Lys Leu
50 55 60
Glu Lys Asp Val Met Ala Gln Arg Gln Thr Phe Ala Gln Lys Ala Gln
65 70 75 80
Ala Phe Glu Gln Asp Arg Ala Arg Arg Ser Asn Glu Glu Arg Gly Lys
85 90 95
Leu Val Thr Arg Ile Gln Thr Ala Val Lys Ser Val Ala Asn Ser Gln
100 105 110
Asp Ile Asp Leu Val Val Asp Ala Asn Ala Val Ala Tyr Asn Ser Ser
115 120 125
Asp Val Lys Asp Ile Thr Ala Asp Val Leu Lys Gln Val Lys Gly Gly
130 135 140
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
145 150 155 160
Gly Ser Gly Gly Gly Gln Met Lys Asp Leu Gln Ala Ile Lys Gln Glu
165 170 175
Val Ser Ser Ser Ala Leu Gly Ser Pro Gln Phe Met Gln Thr Leu Arg
180 185 190
Leu Ala Val Gln Gln Phe Asp Pro Thr Ala Lys Asp Leu Gln Asp Leu
195 200 205
Leu Gln Tyr Leu Ala Ser Ser Leu Val Val Ser Leu His His Gln Gln
210 215 220
Leu Asn Thr Leu Ile Thr Glu Ala Glu Thr Arg Gly Met Thr Gly Tyr
225 230 235 240
Asn Pro Met Ala Gly Pro Leu Arg Met Gln Ala Asn Asn Pro Ala Gln
245 250 255
Gln Gly Leu Arg Arg Glu Tyr Gln Asn Leu Trp Leu Ala Ala Phe Ser
260 265 270
Thr Leu Pro Gly Asn Thr Arg Asp Pro Ser Trp Ala Ala Ile Leu Gln
275 280 285
Gly Leu Glu Glu Pro Tyr Ala Ala Phe Val Glu Arg Leu Asn Val Ala
290 295 300
Leu Asp Asn Gly Leu Pro Glu Gly Thr Pro Lys Glu Pro Ile Leu Arg
305 310 315 320
Ser Leu Ala Tyr Ser Asn Ala Asn Lys Glu Ala Gln Lys Ile Leu Gln
325 330 335
Ala Arg Gly His Thr Asn Ser Pro Leu Gly Glu Met Leu Arg Thr Ala
340 345 350
Gln Ala Trp Thr Pro Lys Asp Lys Thr Lys Val Leu Leu Glu His His
355 360 365
His His His His
370
<210> 21
<211> 361
<212> PRT
<213> 人工
<220>
<223> EcFkpA-p24/CTD(267-350)/HTLV-II融合蛋白
<400> 21
Met Ala Glu Ala Ala Lys Pro Ala Thr Thr Ala Asp Ser Lys Ala Ala
1 5 10 15
Phe Lys Asn Asp Asp Gln Lys Ser Ala Tyr Ala Leu Gly Ala Ser Leu
20 25 30
Gly Arg Tyr Met Glu Asn Ser Leu Lys Glu Gln Glu Lys Leu Gly Ile
35 40 45
Lys Leu Asp Lys Asp Gln Leu Ile Ala Gly Val Gln Asp Ala Phe Ala
50 55 60
Asp Lys Ser Lys Leu Ser Asp Gln Glu Ile Glu Gln Thr Leu Gln Ala
65 70 75 80
Phe Glu Ala Arg Val Lys Ser Ser Ala Gln Ala Lys Met Glu Lys Asp
85 90 95
Ala Ala Asp Asn Glu Ala Lys Gly Lys Glu Tyr Arg Glu Lys Phe Ala
100 105 110
Lys Glu Lys Gly Val Lys Thr Ser Ser Thr Gly Leu Val Tyr Gln Val
115 120 125
Val Glu Ala Gly Lys Gly Glu Ala Pro Lys Asp Ser Asp Thr Val Val
130 135 140
Val Asn Tyr Lys Gly Thr Leu Ile Asp Gly Lys Glu Phe Asp Asn Ser
145 150 155 160
Tyr Thr Arg Gly Glu Pro Leu Ser Phe Arg Leu Asp Gly Val Ile Pro
165 170 175
Gly Trp Thr Glu Gly Leu Lys Asn Ile Lys Lys Gly Gly Lys Ile Lys
180 185 190
Leu Val Ile Pro Pro Glu Leu Ala Tyr Gly Lys Ala Gly Val Pro Gly
195 200 205
Ile Pro Pro Asn Ser Thr Leu Val Phe Asp Val Glu Leu Leu Asp Val
210 215 220
Lys Pro Ala Pro Lys Ala Asp Ala Lys Pro Glu Ala Asp Ala Lys Ala
225 230 235 240
Ala Asp Ser Ala Lys Lys Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
245 250 255
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Pro Ser Trp
260 265 270
Ala Ala Ile Leu Gln Gly Leu Glu Glu Pro Tyr Ala Ala Phe Val Glu
275 280 285
Arg Leu Asn Val Ala Leu Asp Asn Gly Leu Pro Glu Gly Thr Pro Lys
290 295 300
Glu Pro Ile Leu Arg Ser Leu Ala Tyr Ser Asn Ala Asn Lys Glu Ala
305 310 315 320
Gln Lys Ile Leu Gln Ala Arg Gly His Thr Asn Ser Pro Leu Gly Glu
325 330 335
Met Leu Arg Thr Ala Gln Ala Trp Thr Pro Lys Asp Lys Thr Lys Val
340 345 350
Leu Leu Glu His His His His His His
355 360
<210> 22
<211> 257
<212> PRT
<213> 人工
<220>
<223> EcSkp-p24/CTD(267-350)/HTLV-II融合蛋白
<400> 22
Met Ala Asp Lys Ile Ala Ile Val Asn Met Gly Ser Leu Phe Gln Gln
1 5 10 15
Val Ala Gln Lys Thr Gly Val Ser Asn Thr Leu Glu Asn Glu Phe Arg
20 25 30
Gly Arg Ala Ser Glu Leu Gln Arg Met Glu Thr Asp Leu Gln Ala Lys
35 40 45
Met Lys Lys Leu Gln Ser Met Lys Ala Gly Ser Asp Arg Thr Lys Leu
50 55 60
Glu Lys Asp Val Met Ala Gln Arg Gln Thr Phe Ala Gln Lys Ala Gln
65 70 75 80
Ala Phe Glu Gln Asp Arg Ala Arg Arg Ser Asn Glu Glu Arg Gly Lys
85 90 95
Leu Val Thr Arg Ile Gln Thr Ala Val Lys Ser Val Ala Asn Ser Gln
100 105 110
Asp Ile Asp Leu Val Val Asp Ala Asn Ala Val Ala Tyr Asn Ser Ser
115 120 125
Asp Val Lys Asp Ile Thr Ala Asp Val Leu Lys Gln Val Lys Gly Gly
130 135 140
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
145 150 155 160
Gly Ser Gly Gly Gly Pro Ser Trp Ala Ala Ile Leu Gln Gly Leu Glu
165 170 175
Glu Pro Tyr Ala Ala Phe Val Glu Arg Leu Asn Val Ala Leu Asp Asn
180 185 190
Gly Leu Pro Glu Gly Thr Pro Lys Glu Pro Ile Leu Arg Ser Leu Ala
195 200 205
Tyr Ser Asn Ala Asn Lys Glu Ala Gln Lys Ile Leu Gln Ala Arg Gly
210 215 220
His Thr Asn Ser Pro Leu Gly Glu Met Leu Arg Thr Ala Gln Ala Trp
225 230 235 240
Thr Pro Lys Asp Lys Thr Lys Val Leu Leu Glu His His His His His
245 250 255
His
<210> 23
<211> 392
<212> PRT
<213> 人工
<220>
<223> EcFkpA-p24/NTD(152-266)/HTLV-II融合蛋白
<400> 23
Met Ala Glu Ala Ala Lys Pro Ala Thr Thr Ala Asp Ser Lys Ala Ala
1 5 10 15
Phe Lys Asn Asp Asp Gln Lys Ser Ala Tyr Ala Leu Gly Ala Ser Leu
20 25 30
Gly Arg Tyr Met Glu Asn Ser Leu Lys Glu Gln Glu Lys Leu Gly Ile
35 40 45
Lys Leu Asp Lys Asp Gln Leu Ile Ala Gly Val Gln Asp Ala Phe Ala
50 55 60
Asp Lys Ser Lys Leu Ser Asp Gln Glu Ile Glu Gln Thr Leu Gln Ala
65 70 75 80
Phe Glu Ala Arg Val Lys Ser Ser Ala Gln Ala Lys Met Glu Lys Asp
85 90 95
Ala Ala Asp Asn Glu Ala Lys Gly Lys Glu Tyr Arg Glu Lys Phe Ala
100 105 110
Lys Glu Lys Gly Val Lys Thr Ser Ser Thr Gly Leu Val Tyr Gln Val
115 120 125
Val Glu Ala Gly Lys Gly Glu Ala Pro Lys Asp Ser Asp Thr Val Val
130 135 140
Val Asn Tyr Lys Gly Thr Leu Ile Asp Gly Lys Glu Phe Asp Asn Ser
145 150 155 160
Tyr Thr Arg Gly Glu Pro Leu Ser Phe Arg Leu Asp Gly Val Ile Pro
165 170 175
Gly Trp Thr Glu Gly Leu Lys Asn Ile Lys Lys Gly Gly Lys Ile Lys
180 185 190
Leu Val Ile Pro Pro Glu Leu Ala Tyr Gly Lys Ala Gly Val Pro Gly
195 200 205
Ile Pro Pro Asn Ser Thr Leu Val Phe Asp Val Glu Leu Leu Asp Val
210 215 220
Lys Pro Ala Pro Lys Ala Asp Ala Lys Pro Glu Ala Asp Ala Lys Ala
225 230 235 240
Ala Asp Ser Ala Lys Lys Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
245 250 255
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Gln Met Lys
260 265 270
Asp Leu Gln Ala Ile Lys Gln Glu Val Ser Ser Ser Ala Leu Gly Ser
275 280 285
Pro Gln Phe Met Gln Thr Leu Arg Leu Ala Val Gln Gln Phe Asp Pro
290 295 300
Thr Ala Lys Asp Leu Gln Asp Leu Leu Gln Tyr Leu Ala Ser Ser Leu
305 310 315 320
Val Val Ser Leu His His Gln Gln Leu Asn Thr Leu Ile Thr Glu Ala
325 330 335
Glu Thr Arg Gly Met Thr Gly Tyr Asn Pro Met Ala Gly Pro Leu Arg
340 345 350
Met Gln Ala Asn Asn Pro Ala Gln Gln Gly Leu Arg Arg Glu Tyr Gln
355 360 365
Asn Leu Trp Leu Ala Ala Phe Ser Thr Leu Pro Gly Asn Thr Arg Asp
370 375 380
Leu Glu His His His His His His
385 390
<210> 24
<211> 288
<212> PRT
<213> 人工
<220>
<223> EcSkp-p24/NTD(152-266)/HTLV-II融合蛋白
<400> 24
Met Ala Asp Lys Ile Ala Ile Val Asn Met Gly Ser Leu Phe Gln Gln
1 5 10 15
Val Ala Gln Lys Thr Gly Val Ser Asn Thr Leu Glu Asn Glu Phe Arg
20 25 30
Gly Arg Ala Ser Glu Leu Gln Arg Met Glu Thr Asp Leu Gln Ala Lys
35 40 45
Met Lys Lys Leu Gln Ser Met Lys Ala Gly Ser Asp Arg Thr Lys Leu
50 55 60
Glu Lys Asp Val Met Ala Gln Arg Gln Thr Phe Ala Gln Lys Ala Gln
65 70 75 80
Ala Phe Glu Gln Asp Arg Ala Arg Arg Ser Asn Glu Glu Arg Gly Lys
85 90 95
Leu Val Thr Arg Ile Gln Thr Ala Val Lys Ser Val Ala Asn Ser Gln
100 105 110
Asp Ile Asp Leu Val Val Asp Ala Asn Ala Val Ala Tyr Asn Ser Ser
115 120 125
Asp Val Lys Asp Ile Thr Ala Asp Val Leu Lys Gln Val Lys Gly Gly
130 135 140
Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly
145 150 155 160
Gly Ser Gly Gly Gly Gln Met Lys Asp Leu Gln Ala Ile Lys Gln Glu
165 170 175
Val Ser Ser Ser Ala Leu Gly Ser Pro Gln Phe Met Gln Thr Leu Arg
180 185 190
Leu Ala Val Gln Gln Phe Asp Pro Thr Ala Lys Asp Leu Gln Asp Leu
195 200 205
Leu Gln Tyr Leu Ala Ser Ser Leu Val Val Ser Leu His His Gln Gln
210 215 220
Leu Asn Thr Leu Ile Thr Glu Ala Glu Thr Arg Gly Met Thr Gly Tyr
225 230 235 240
Asn Pro Met Ala Gly Pro Leu Arg Met Gln Ala Asn Asn Pro Ala Gln
245 250 255
Gln Gly Leu Arg Arg Glu Tyr Gln Asn Leu Trp Leu Ala Ala Phe Ser
260 265 270
Thr Leu Pro Gly Asn Thr Arg Asp Leu Glu His His His His His His
275 280 285
<210> 25
<211> 108
<212> PRT
<213> 人T-细胞嗜淋巴细胞病毒1型
<220>
<221> MISC_FEATURE
<223> Xaa = Cys或Ala或Ser
<400> 25
Ser Leu Ala Ser Gly Lys Ser Leu Leu His Glu Val Asp Lys Asp Ile
1 5 10 15
Ser Gln Leu Thr Gln Ala Ile Val Lys Asn His Lys Asn Leu Leu Lys
20 25 30
Ile Ala Gln Tyr Ala Ala Gln Asn Arg Arg Gly Leu Asp Leu Leu Phe
35 40 45
Trp Glu Gln Gly Gly Leu Xaa Lys Ala Leu Gln Glu Gln Xaa Xaa Phe
50 55 60
Leu Asn Ile Thr Asn Ser His Val Ser Ile Leu Gln Glu Arg Pro Pro
65 70 75 80
Leu Glu Asn Arg Val Leu Thr Gly Trp Gly Leu Asn Trp Asp Leu Gly
85 90 95
Leu Ser Gln Trp Ala Arg Glu Ala Leu Gln Thr Gly
100 105
<210> 26
<211> 304
<212> PRT
<213> 人工
<220>
<223> EcSlyD-gp21(339-446)/HTLV-1融合蛋白
<400> 26
Met Lys Val Ala Lys Asp Leu Val Val Ser Leu Ala Tyr Gln Val Arg
1 5 10 15
Thr Glu Asp Gly Val Leu Val Asp Glu Ser Pro Val Ser Ala Pro Leu
20 25 30
Asp Tyr Leu His Gly His Gly Ser Leu Ile Ser Gly Leu Glu Thr Ala
35 40 45
Leu Glu Gly His Glu Val Gly Asp Lys Phe Asp Val Ala Val Gly Ala
50 55 60
Asn Asp Ala Tyr Gly Gln Tyr Asp Glu Asn Leu Val Gln Arg Val Pro
65 70 75 80
Lys Asp Val Phe Met Gly Val Asp Glu Leu Gln Val Gly Met Arg Phe
85 90 95
Leu Ala Glu Thr Asp Gln Gly Pro Val Pro Val Glu Ile Thr Ala Val
100 105 110
Glu Asp Asp His Val Val Val Asp Gly Asn His Met Leu Ala Gly Gln
115 120 125
Asn Leu Lys Phe Asn Val Glu Val Val Ala Ile Arg Glu Ala Thr Glu
130 135 140
Glu Glu Leu Ala His Gly His Val His Gly Ala His Asp His His His
145 150 155 160
Asp His Asp His Asp Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly
165 170 175
Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Leu Ala Ser
180 185 190
Gly Lys Ser Leu Leu His Glu Val Asp Lys Asp Ile Ser Gln Leu Thr
195 200 205
Gln Ala Ile Val Lys Asn His Lys Asn Leu Leu Lys Ile Ala Gln Tyr
210 215 220
Ala Ala Gln Asn Arg Arg Gly Leu Asp Leu Leu Phe Trp Glu Gln Gly
225 230 235 240
Gly Leu Ala Lys Ala Leu Gln Glu Gln Ala Ala Phe Leu Asn Ile Thr
245 250 255
Asn Ser His Val Ser Ile Leu Gln Glu Arg Pro Pro Leu Glu Asn Arg
260 265 270
Val Leu Thr Gly Trp Gly Leu Asn Trp Asp Leu Gly Leu Ser Gln Trp
275 280 285
Ala Arg Glu Ala Leu Gln Thr Gly Leu Glu His His His His His His
290 295 300
<210> 27
<211> 286
<212> PRT
<213> 人工
<220>
<223> EcSlpA-gp21(339-446)/HTLV-1融合蛋白
<400> 27
Ser Glu Ser Val Gln Ser Asn Ser Ala Val Leu Val His Phe Thr Leu
1 5 10 15
Lys Leu Asp Asp Gly Thr Thr Ala Glu Ser Thr Arg Asn Asn Gly Lys
20 25 30
Pro Ala Leu Phe Arg Leu Gly Asp Ala Ser Leu Ser Glu Gly Leu Glu
35 40 45
Gln His Leu Leu Gly Leu Lys Val Gly Asp Lys Thr Thr Phe Ser Leu
50 55 60
Glu Pro Asp Ala Ala Phe Gly Val Pro Ser Pro Asp Leu Ile Gln Tyr
65 70 75 80
Phe Ser Arg Arg Glu Phe Met Asp Ala Gly Glu Pro Glu Ile Gly Ala
85 90 95
Ile Met Leu Phe Thr Ala Met Asp Gly Ser Glu Met Pro Gly Val Ile
100 105 110
Arg Glu Ile Asn Gly Asp Ser Ile Thr Val Asp Phe Asn His Pro Leu
115 120 125
Ala Gly Gln Thr Val His Phe Asp Ile Glu Val Leu Glu Ile Asp Pro
130 135 140
Ala Leu Glu Gly Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Gly
145 150 155 160
Gly Gly Ser Gly Gly Gly Ser Gly Gly Gly Ser Leu Ala Ser Gly Lys
165 170 175
Ser Leu Leu His Glu Val Asp Lys Asp Ile Ser Gln Leu Thr Gln Ala
180 185 190
Ile Val Lys Asn His Lys Asn Leu Leu Lys Ile Ala Gln Tyr Ala Ala
195 200 205
Gln Asn Arg Arg Gly Leu Asp Leu Leu Phe Trp Glu Gln Gly Gly Leu
210 215 220
Ala Lys Ala Leu Gln Glu Gln Ala Ala Phe Leu Asn Ile Thr Asn Ser
225 230 235 240
His Val Ser Ile Leu Gln Glu Arg Pro Pro Leu Glu Asn Arg Val Leu
245 250 255
Thr Gly Trp Gly Leu Asn Trp Asp Leu Gly Leu Ser Gln Trp Ala Arg
260 265 270
Glu Ala Leu Gln Thr Gly Leu Glu His His His His His His
275 280 285
Claims (6)
1.可溶性HTLV p24抗原,其包含如SEQ ID NO.3或SEQ ID NO.7中指定的HTLV p24的C-末端结构域,其中所述HTLV p24抗原缺乏如SEQ ID NO.2和SEQ ID NO.6中指定的N-末端结构域,且其中所述可溶性HTLV p24抗原融合至寡聚分子伴侣。
2.根据权利要求1所述的可溶性HTLV p24抗原,其中所述寡聚分子伴侣是选自Skp和FkpA的分子伴侣。
3.产生可溶性和免疫反应性HTLV p24抗原的方法,所述方法包括以下步骤
- 培养用表达载体转化的宿主细胞,所述表达载体包含可操作连接的重组DNA分子,所述重组DNA分子编码根据权利要求1至2中任一项所述的HTLV p24抗原
- 表达所述HLTV p24抗原,和
- 纯化所述HLTV p24抗原。
4.HTLV抗原的组合物,其包含根据权利要求1至2中任一项所述的HTLV p24抗原和包含根据SEQ ID NO.25所述的氨基酸序列的HTLV gp21抗原,其中所述p24和gp21抗原被表达为分开的多肽,且其中所述HTLV gp21抗原增强溶解性。
5.根据权利要求1至2中任一项所述的HTLV p24抗原或根据权利要求4所述的HTLV抗原的组合物在制备用于检测抗-HTLV抗体的体外诊断测试中的试剂盒中的用途。
6.用于检测抗-HTLV抗体的试剂盒,其包含至少一种根据权利要求1至2中任一项所述的HTLV p24抗原或根据权利要求4所述的HTLV抗原组合物。
Applications Claiming Priority (3)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| EP14157165.3A EP2913338A1 (en) | 2014-02-28 | 2014-02-28 | Soluable and immunoreactive variants of HTLV capsid antigen p24 |
| EP14157165.3 | 2014-02-28 | ||
| PCT/EP2015/053966 WO2015128394A2 (en) | 2014-02-28 | 2015-02-26 | Soluble and immunoreactive variants of htlv capsid antigen p24 |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| CN106029687A CN106029687A (zh) | 2016-10-12 |
| CN106029687B true CN106029687B (zh) | 2020-07-31 |
Family
ID=50184798
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| CN201580010787.2A Active CN106029687B (zh) | 2014-02-28 | 2015-02-26 | HTLV衣壳抗原p24的可溶性和免疫反应性变体 |
Country Status (7)
| Country | Link |
|---|---|
| US (2) | US10466242B2 (zh) |
| EP (2) | EP2913338A1 (zh) |
| JP (1) | JP6491228B2 (zh) |
| CN (1) | CN106029687B (zh) |
| CA (1) | CA2938712C (zh) |
| ES (1) | ES2662611T3 (zh) |
| WO (1) | WO2015128394A2 (zh) |
Families Citing this family (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN108780092A (zh) * | 2016-03-07 | 2018-11-09 | 豪夫迈·罗氏有限公司 | 抗p53抗体的检测 |
| WO2019164402A1 (en) * | 2018-02-26 | 2019-08-29 | Technische Universiteit Eindhoven | Bioluminescent biosensor for detecting and quantifying biomolecules |
| JP2023522278A (ja) * | 2020-04-23 | 2023-05-29 | エフ. ホフマン-ラ ロシュ アーゲー | 抗体-免疫アッセイでの使用のためのコロナヌクレオカプシド抗原 |
Citations (4)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US6110662A (en) * | 1992-02-24 | 2000-08-29 | Genelabs Technologies, Inc. | HTLV-I/HTLV-II assay and method |
| CN1377276A (zh) * | 1999-08-09 | 2002-10-30 | 特里帕普股份公司 | 含有三肽的药用组合物 |
| CN101289513A (zh) * | 2007-04-20 | 2008-10-22 | 霍夫曼-拉罗奇有限公司 | 病原体原发感染的检测 |
| CN101591665A (zh) * | 2008-05-26 | 2009-12-02 | 霍夫曼-拉罗奇有限公司 | 作为重组蛋白和酶技术的工具的SlpA |
Family Cites Families (7)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| JP2564268B2 (ja) * | 1985-08-28 | 1996-12-18 | 協和醗酵工業株式会社 | 融合抗原ポリペプチド |
| NZ238855A (en) * | 1990-07-18 | 1994-03-25 | Iaf Biochem Int | Peptides, mixtures thereof and compositions useful for detecting htlv-i and htlv-ii infections |
| EP0848820A1 (en) * | 1995-06-05 | 1998-06-24 | Abbott Laboratories | Detection of htlv antibodies utilizing recombinant proteins |
| JP2001501093A (ja) | 1996-09-26 | 2001-01-30 | メディカル リサーチ カウンシル | シャペロン断片 |
| JP2000078973A (ja) * | 1998-09-03 | 2000-03-21 | Tonen Corp | Htlv−i組換え抗原 |
| US6258932B1 (en) * | 1999-08-09 | 2001-07-10 | Tripep Ab | Peptides that block viral infectivity and methods of use thereof |
| ES2372031T3 (es) | 2001-06-22 | 2012-01-13 | F. Hoffmann-La Roche Ag | Complejo soluble que contiene una glicoproteína de superficie retroviral y fkpa o slyd. |
-
2014
- 2014-02-28 EP EP14157165.3A patent/EP2913338A1/en not_active Ceased
-
2015
- 2015-02-26 ES ES15706804.0T patent/ES2662611T3/es active Active
- 2015-02-26 CA CA2938712A patent/CA2938712C/en active Active
- 2015-02-26 WO PCT/EP2015/053966 patent/WO2015128394A2/en active Application Filing
- 2015-02-26 JP JP2016553811A patent/JP6491228B2/ja active Active
- 2015-02-26 CN CN201580010787.2A patent/CN106029687B/zh active Active
- 2015-02-26 EP EP15706804.0A patent/EP3110831B1/en active Active
-
2016
- 2016-08-25 US US15/246,607 patent/US10466242B2/en active Active
-
2019
- 2019-09-20 US US16/577,175 patent/US11567079B2/en active Active
Patent Citations (4)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US6110662A (en) * | 1992-02-24 | 2000-08-29 | Genelabs Technologies, Inc. | HTLV-I/HTLV-II assay and method |
| CN1377276A (zh) * | 1999-08-09 | 2002-10-30 | 特里帕普股份公司 | 含有三肽的药用组合物 |
| CN101289513A (zh) * | 2007-04-20 | 2008-10-22 | 霍夫曼-拉罗奇有限公司 | 病原体原发感染的检测 |
| CN101591665A (zh) * | 2008-05-26 | 2009-12-02 | 霍夫曼-拉罗奇有限公司 | 作为重组蛋白和酶技术的工具的SlpA |
Also Published As
| Publication number | Publication date |
|---|---|
| CA2938712A1 (en) | 2015-09-03 |
| EP3110831B1 (en) | 2018-01-10 |
| JP2017512200A (ja) | 2017-05-18 |
| US10466242B2 (en) | 2019-11-05 |
| US20170184591A1 (en) | 2017-06-29 |
| EP3110831A2 (en) | 2017-01-04 |
| WO2015128394A3 (en) | 2015-11-05 |
| CA2938712C (en) | 2018-10-23 |
| US20200033342A1 (en) | 2020-01-30 |
| JP6491228B2 (ja) | 2019-03-27 |
| EP2913338A1 (en) | 2015-09-02 |
| ES2662611T3 (es) | 2018-04-09 |
| WO2015128394A2 (en) | 2015-09-03 |
| US11567079B2 (en) | 2023-01-31 |
| CN106029687A (zh) | 2016-10-12 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| EP2663642B1 (en) | Treponema pallidum triplet antigen | |
| US11567079B2 (en) | Soluble and immunoreactive variants of HTLV capsid antigen P24 | |
| CA2856187C (en) | Soluble immunoreactive treponema pallidum tpn47 antigens | |
| JP5461423B2 (ja) | 新規風疹e1エンベロープタンパク質変異体および抗風疹抗体の検出におけるその使用 | |
| US10197559B2 (en) | Chaperone-chaperone fusion polypeptides for reduction of interference and stabilization of immunoassays | |
| JP2005139204A (ja) | Hivウイルスに対して免疫反応性である抗体の検出用の合成抗原 | |
| JP6374720B2 (ja) | 抗トレポネーマ属(Treponema)抗体の検出のためのTpN17に基づくイムノアッセイにおける、抗干渉添加剤としてのコレラ菌(Vibriocholerae)リポタンパク質15(Lp15)変異体 | |
| AU627738B2 (en) | Htlv-i / hiv-1 fusion proteins | |
| CN107074921B (zh) | 重组克氏锥虫jl7抗原变体及其用于检测查加斯病的用途 | |
| CN106699895A (zh) | 一种新型融合抗原和包含其的检测试剂盒及应用 |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| C06 | Publication | ||
| PB01 | Publication | ||
| C10 | Entry into substantive examination | ||
| SE01 | Entry into force of request for substantive examination | ||
| REG | Reference to a national code |
Ref country code: HK Ref legal event code: DE Ref document number: 1229824 Country of ref document: HK |
|
| GR01 | Patent grant | ||
| GR01 | Patent grant |