WO2017046994A1 - Il-8-binding antibodies and uses thereof - Google Patents
Il-8-binding antibodies and uses thereof Download PDFInfo
- Publication number
- WO2017046994A1 WO2017046994A1 PCT/JP2016/003616 JP2016003616W WO2017046994A1 WO 2017046994 A1 WO2017046994 A1 WO 2017046994A1 JP 2016003616 W JP2016003616 W JP 2016003616W WO 2017046994 A1 WO2017046994 A1 WO 2017046994A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- antibody
- amino acid
- seq
- region
- antigen
- Prior art date
Links
- 230000027455 binding Effects 0.000 title claims description 489
- 238000009739 binding Methods 0.000 title claims description 320
- 108090001007 Interleukin-8 Proteins 0.000 claims abstract description 56
- 235000001014 amino acid Nutrition 0.000 claims description 294
- 125000003275 alpha amino acid group Chemical group 0.000 claims description 261
- 230000001965 increasing effect Effects 0.000 claims description 168
- 150000001413 amino acids Chemical group 0.000 claims description 136
- 238000000034 method Methods 0.000 claims description 121
- 230000001419 dependent effect Effects 0.000 claims description 120
- 102100026120 IgG receptor FcRn large subunit p51 Human genes 0.000 claims description 80
- 101710177940 IgG receptor FcRn large subunit p51 Proteins 0.000 claims description 79
- DHMQDGOQFOQNFH-UHFFFAOYSA-N Glycine Natural products NCC(O)=O DHMQDGOQFOQNFH-UHFFFAOYSA-N 0.000 claims description 70
- 125000003295 alanine group Chemical group N[C@@H](C)C(=O)* 0.000 claims description 70
- 238000006467 substitution reaction Methods 0.000 claims description 70
- 102000004890 Interleukin-8 Human genes 0.000 claims description 55
- 230000002378 acidificating effect Effects 0.000 claims description 50
- 210000004027 cell Anatomy 0.000 claims description 49
- 102000005962 receptors Human genes 0.000 claims description 36
- 210000002744 extracellular matrix Anatomy 0.000 claims description 34
- 238000004519 manufacturing process Methods 0.000 claims description 33
- 230000002829 reductive effect Effects 0.000 claims description 33
- 108010037362 Extracellular Matrix Proteins Proteins 0.000 claims description 31
- 102000010834 Extracellular Matrix Proteins Human genes 0.000 claims description 31
- 230000002401 inhibitory effect Effects 0.000 claims description 30
- 102000039446 nucleic acids Human genes 0.000 claims description 30
- 108020004707 nucleic acids Proteins 0.000 claims description 30
- 150000007523 nucleic acids Chemical class 0.000 claims description 30
- 239000008194 pharmaceutical composition Substances 0.000 claims description 29
- 125000001493 tyrosinyl group Chemical group [H]OC1=C([H])C([H])=C(C([H])=C1[H])C([H])([H])C([H])(N([H])[H])C(*)=O 0.000 claims description 19
- 101001055222 Homo sapiens Interleukin-8 Proteins 0.000 claims description 16
- 238000012258 culturing Methods 0.000 claims description 16
- 125000000404 glutamine group Chemical group N[C@@H](CCC(N)=O)C(=O)* 0.000 claims description 16
- 125000003630 glycyl group Chemical group [H]N([H])C([H])([H])C(*)=O 0.000 claims description 15
- 239000012636 effector Substances 0.000 claims description 14
- 239000004475 Arginine Substances 0.000 claims description 12
- ODKSFYDXXFIFQN-UHFFFAOYSA-N arginine Natural products OC(=O)C(N)CCCNC(N)=N ODKSFYDXXFIFQN-UHFFFAOYSA-N 0.000 claims description 12
- 238000004113 cell culture Methods 0.000 claims description 12
- 210000000440 neutrophil Anatomy 0.000 claims description 12
- 239000013598 vector Substances 0.000 claims description 12
- OUYCCCASQSFEME-UHFFFAOYSA-N tyrosine Natural products OC(=O)C(N)CC1=CC=C(O)C=C1 OUYCCCASQSFEME-UHFFFAOYSA-N 0.000 claims description 11
- 125000000637 arginyl group Chemical group N[C@@H](CCCNC(N)=N)C(=O)* 0.000 claims description 8
- 230000033115 angiogenesis Effects 0.000 claims description 7
- 230000005012 migration Effects 0.000 claims description 7
- 238000013508 migration Methods 0.000 claims description 7
- ROHFNLRQFUQHCH-UHFFFAOYSA-N Leucine Natural products CC(C)CC(N)C(O)=O ROHFNLRQFUQHCH-UHFFFAOYSA-N 0.000 claims description 6
- DCXYFEDJOCDNAF-UHFFFAOYSA-N Asparagine Natural products OC(=O)C(N)CC(N)=O DCXYFEDJOCDNAF-UHFFFAOYSA-N 0.000 claims description 5
- 235000004279 alanine Nutrition 0.000 claims description 5
- 235000009582 asparagine Nutrition 0.000 claims description 5
- 229960001230 asparagine Drugs 0.000 claims description 5
- ZDXPYRJPNDTMRX-UHFFFAOYSA-N glutamine Natural products OC(=O)C(N)CCC(N)=O ZDXPYRJPNDTMRX-UHFFFAOYSA-N 0.000 claims description 5
- 125000001909 leucine group Chemical group [H]N(*)C(C(*)=O)C([H])([H])C(C([H])([H])[H])C([H])([H])[H] 0.000 claims description 5
- 239000004471 Glycine Substances 0.000 claims description 4
- 125000000613 asparagine group Chemical group N[C@@H](CC(N)=O)C(=O)* 0.000 claims description 4
- 239000003937 drug carrier Substances 0.000 claims description 3
- 239000003814 drug Substances 0.000 abstract description 19
- 239000000427 antigen Substances 0.000 description 371
- 108091007433 antigens Proteins 0.000 description 371
- 102000036639 antigens Human genes 0.000 description 371
- 229940024606 amino acid Drugs 0.000 description 248
- 230000004048 modification Effects 0.000 description 240
- 238000012986 modification Methods 0.000 description 240
- 125000000539 amino acid group Chemical class 0.000 description 205
- COLNVLDHVKWLRT-QMMMGPOBSA-N L-phenylalanine Chemical compound OC(=O)[C@@H](N)CC1=CC=CC=C1 COLNVLDHVKWLRT-QMMMGPOBSA-N 0.000 description 109
- KDXKERNSBIXSRK-UHFFFAOYSA-N Lysine Chemical group NCCCCC(N)C(O)=O KDXKERNSBIXSRK-UHFFFAOYSA-N 0.000 description 107
- WHUUTDBJXJRKMK-UHFFFAOYSA-N Glutamic acid Natural products OC(=O)C(N)CCC(O)=O WHUUTDBJXJRKMK-UHFFFAOYSA-N 0.000 description 105
- 150000002500 ions Chemical class 0.000 description 105
- 241000282414 Homo sapiens Species 0.000 description 97
- WHUUTDBJXJRKMK-VKHMYHEASA-N L-glutamic acid Chemical group OC(=O)[C@@H](N)CCC(O)=O WHUUTDBJXJRKMK-VKHMYHEASA-N 0.000 description 96
- CKLJMWTZIZZHCS-REOHCLBHSA-N L-aspartic acid Chemical group OC(=O)[C@@H](N)CC(O)=O CKLJMWTZIZZHCS-REOHCLBHSA-N 0.000 description 86
- OUYCCCASQSFEME-QMMMGPOBSA-N L-tyrosine Chemical compound OC(=O)[C@@H](N)CC1=CC=C(O)C=C1 OUYCCCASQSFEME-QMMMGPOBSA-N 0.000 description 79
- 230000007935 neutral effect Effects 0.000 description 75
- 125000001360 methionine group Chemical group N[C@@H](CCSC)C(=O)* 0.000 description 67
- KDXKERNSBIXSRK-YFKPBYRVSA-N L-lysine Chemical compound NCCCC[C@H](N)C(O)=O KDXKERNSBIXSRK-YFKPBYRVSA-N 0.000 description 65
- AYFVYJQAPQTCCC-GBXIJSLDSA-N L-threonine Chemical group C[C@@H](O)[C@H](N)C(O)=O AYFVYJQAPQTCCC-GBXIJSLDSA-N 0.000 description 65
- 102100024952 Protein CBFA2T1 Human genes 0.000 description 61
- MTCFGRXMJLQNBG-REOHCLBHSA-N (2S)-2-Amino-3-hydroxypropansäure Chemical group OC[C@H](N)C(O)=O MTCFGRXMJLQNBG-REOHCLBHSA-N 0.000 description 58
- DCXYFEDJOCDNAF-REOHCLBHSA-N L-asparagine Chemical compound OC(=O)[C@@H](N)CC(N)=O DCXYFEDJOCDNAF-REOHCLBHSA-N 0.000 description 58
- AGPKZVBTJJNPAG-WHFBIAKZSA-N L-isoleucine Chemical group CC[C@H](C)[C@H](N)C(O)=O AGPKZVBTJJNPAG-WHFBIAKZSA-N 0.000 description 58
- KZSNJWFQEVHDMF-UHFFFAOYSA-N Valine Chemical group CC(C)C(N)C(O)=O KZSNJWFQEVHDMF-UHFFFAOYSA-N 0.000 description 55
- BHPQYMZQTOCNFJ-UHFFFAOYSA-N Calcium cation Chemical compound [Ca+2] BHPQYMZQTOCNFJ-UHFFFAOYSA-N 0.000 description 53
- 125000000174 L-prolyl group Chemical group [H]N1C([H])([H])C([H])([H])C([H])([H])[C@@]1([H])C(*)=O 0.000 description 53
- 229910001424 calcium ion Inorganic materials 0.000 description 53
- ZDXPYRJPNDTMRX-VKHMYHEASA-N L-glutamine Chemical compound OC(=O)[C@@H](N)CCC(N)=O ZDXPYRJPNDTMRX-VKHMYHEASA-N 0.000 description 52
- XKTZWUACRZHVAN-VADRZIEHSA-N interleukin-8 Chemical compound C([C@H](NC(=O)[C@H](CC(O)=O)NC(=O)[C@H](CC=1C2=CC=CC=C2NC=1)NC(=O)[C@@H](NC(C)=O)CCSC)C(=O)N[C@@H](CC(O)=O)C(=O)N[C@@H](CC(O)=O)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](CC(N)=O)C(=O)N[C@@H](CC=1C=CC=CC=1)C(=O)N[C@@H]([C@@H](C)O)C(=O)NCC(=O)N[C@@H](CCSC)C(=O)N1[C@H](CCC1)C(=O)N1[C@H](CCC1)C(=O)N[C@@H](C)C(=O)N[C@H](CC(O)=O)C(=O)N[C@H](CCC(O)=O)C(=O)N[C@H](CC(O)=O)C(=O)N[C@H](CC=1C=CC(O)=CC=1)C(=O)N[C@H](CO)C(=O)N1[C@H](CCC1)C(N)=O)C1=CC=CC=C1 XKTZWUACRZHVAN-VADRZIEHSA-N 0.000 description 51
- 229940096397 interleukin-8 Drugs 0.000 description 51
- ROHFNLRQFUQHCH-YFKPBYRVSA-N L-leucine Chemical compound CC(C)C[C@H](N)C(O)=O ROHFNLRQFUQHCH-YFKPBYRVSA-N 0.000 description 49
- 108090000765 processed proteins & peptides Proteins 0.000 description 48
- 125000000510 L-tryptophano group Chemical group [H]C1=C([H])C([H])=C2N([H])C([H])=C(C([H])([H])[C@@]([H])(C(O[H])=O)N([H])[*])C2=C1[H] 0.000 description 46
- 229920001184 polypeptide Polymers 0.000 description 46
- 102000004196 processed proteins & peptides Human genes 0.000 description 46
- ODKSFYDXXFIFQN-BYPYZUCNSA-N L-arginine Chemical compound OC(=O)[C@@H](N)CCCN=C(N)N ODKSFYDXXFIFQN-BYPYZUCNSA-N 0.000 description 42
- 108090000623 proteins and genes Proteins 0.000 description 39
- 108010021472 Fc gamma receptor IIB Proteins 0.000 description 37
- 102100029205 Low affinity immunoglobulin gamma Fc region receptor II-b Human genes 0.000 description 37
- 108020003175 receptors Proteins 0.000 description 34
- 108010021468 Fc gamma receptor IIA Proteins 0.000 description 33
- 102100029204 Low affinity immunoglobulin gamma Fc region receptor II-a Human genes 0.000 description 33
- -1 for example Proteins 0.000 description 33
- 235000018102 proteins Nutrition 0.000 description 33
- 102000004169 proteins and genes Human genes 0.000 description 33
- 102100033400 4F2 cell-surface antigen heavy chain Human genes 0.000 description 32
- 101000800023 Homo sapiens 4F2 cell-surface antigen heavy chain Proteins 0.000 description 32
- 102100029185 Low affinity immunoglobulin gamma Fc region receptor III-B Human genes 0.000 description 31
- 230000000694 effects Effects 0.000 description 30
- 230000008030 elimination Effects 0.000 description 29
- 238000003379 elimination reaction Methods 0.000 description 29
- NFGXHKASABOEEW-UHFFFAOYSA-N 1-methylethyl 11-methoxy-3,7,11-trimethyl-2,4-dodecadienoate Chemical compound COC(C)(C)CCCC(C)CC=CC(C)=CC(=O)OC(C)C NFGXHKASABOEEW-UHFFFAOYSA-N 0.000 description 27
- 230000003213 activating effect Effects 0.000 description 26
- 101000917839 Homo sapiens Low affinity immunoglobulin gamma Fc region receptor III-B Proteins 0.000 description 25
- GPRLSGONYQIRFK-UHFFFAOYSA-N hydron Chemical compound [H+] GPRLSGONYQIRFK-UHFFFAOYSA-N 0.000 description 25
- 238000010494 dissociation reaction Methods 0.000 description 24
- 230000005593 dissociations Effects 0.000 description 24
- 230000014759 maintenance of location Effects 0.000 description 23
- 101710099301 Low affinity immunoglobulin gamma Fc region receptor III-A Proteins 0.000 description 20
- 102100029193 Low affinity immunoglobulin gamma Fc region receptor III-A Human genes 0.000 description 19
- 238000003780 insertion Methods 0.000 description 19
- 230000037431 insertion Effects 0.000 description 19
- 238000001727 in vivo Methods 0.000 description 18
- 241001465754 Metazoa Species 0.000 description 16
- 230000035772 mutation Effects 0.000 description 16
- 101001076408 Homo sapiens Interleukin-6 Proteins 0.000 description 15
- 230000008859 change Effects 0.000 description 15
- 230000027948 extracellular matrix binding Effects 0.000 description 15
- 102000052611 human IL6 Human genes 0.000 description 15
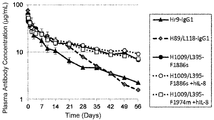
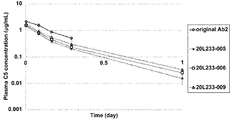
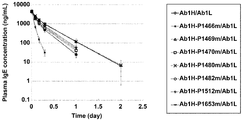
- 230000036470 plasma concentration Effects 0.000 description 14
- 241000699666 Mus <mouse, genus> Species 0.000 description 13
- 230000001225 therapeutic effect Effects 0.000 description 13
- 239000011575 calcium Substances 0.000 description 12
- 208000037265 diseases, disorders, signs and symptoms Diseases 0.000 description 12
- 229940079593 drug Drugs 0.000 description 12
- 206010039073 rheumatoid arthritis Diseases 0.000 description 12
- 210000002966 serum Anatomy 0.000 description 12
- XUJNEKJLAYXESH-REOHCLBHSA-N L-Cysteine Chemical compound SC[C@H](N)C(O)=O XUJNEKJLAYXESH-REOHCLBHSA-N 0.000 description 11
- 238000012217 deletion Methods 0.000 description 11
- 230000037430 deletion Effects 0.000 description 11
- 210000001163 endosome Anatomy 0.000 description 11
- 230000001737 promoting effect Effects 0.000 description 11
- 108010073807 IgG Receptors Proteins 0.000 description 10
- 230000004075 alteration Effects 0.000 description 10
- 230000003247 decreasing effect Effects 0.000 description 10
- 208000035475 disorder Diseases 0.000 description 10
- 230000006870 function Effects 0.000 description 10
- HNDVDQJCIGZPNO-UHFFFAOYSA-N histidine Natural products OC(=O)C(N)CC1=CN=CN1 HNDVDQJCIGZPNO-UHFFFAOYSA-N 0.000 description 10
- 239000000203 mixture Substances 0.000 description 10
- 101100112922 Candida albicans CDR3 gene Proteins 0.000 description 9
- 108060003951 Immunoglobulin Proteins 0.000 description 9
- 125000000291 glutamic acid group Chemical group N[C@@H](CCC(O)=O)C(=O)* 0.000 description 9
- 102000018358 immunoglobulin Human genes 0.000 description 9
- 108010068617 neonatal Fc receptor Proteins 0.000 description 9
- 235000000346 sugar Nutrition 0.000 description 9
- 102000005701 Calcium-Binding Proteins Human genes 0.000 description 8
- 108010045403 Calcium-Binding Proteins Proteins 0.000 description 8
- 102000010781 Interleukin-6 Receptors Human genes 0.000 description 8
- 108010038501 Interleukin-6 Receptors Proteins 0.000 description 8
- 241000699670 Mus sp. Species 0.000 description 8
- 108010029485 Protein Isoforms Proteins 0.000 description 8
- 102000001708 Protein Isoforms Human genes 0.000 description 8
- 238000010276 construction Methods 0.000 description 8
- XUJNEKJLAYXESH-UHFFFAOYSA-N cysteine Natural products SCC(N)C(O)=O XUJNEKJLAYXESH-UHFFFAOYSA-N 0.000 description 8
- 235000018417 cysteine Nutrition 0.000 description 8
- 230000002757 inflammatory effect Effects 0.000 description 8
- 102100035360 Cerebellar degeneration-related antigen 1 Human genes 0.000 description 7
- 102000019034 Chemokines Human genes 0.000 description 7
- 108010012236 Chemokines Proteins 0.000 description 7
- 101000913074 Homo sapiens High affinity immunoglobulin gamma Fc receptor I Proteins 0.000 description 7
- 101000917858 Homo sapiens Low affinity immunoglobulin gamma Fc region receptor III-A Proteins 0.000 description 7
- HNDVDQJCIGZPNO-YFKPBYRVSA-N L-histidine Chemical compound OC(=O)[C@@H](N)CC1=CN=CN1 HNDVDQJCIGZPNO-YFKPBYRVSA-N 0.000 description 7
- 238000007792 addition Methods 0.000 description 7
- 230000004071 biological effect Effects 0.000 description 7
- 239000000243 solution Substances 0.000 description 7
- 210000001519 tissue Anatomy 0.000 description 7
- 238000011830 transgenic mouse model Methods 0.000 description 7
- 125000002987 valine group Chemical group [H]N([H])C([H])(C(*)=O)C([H])(C([H])([H])[H])C([H])([H])[H] 0.000 description 7
- 102000009109 Fc receptors Human genes 0.000 description 6
- 108010087819 Fc receptors Proteins 0.000 description 6
- 102100026122 High affinity immunoglobulin gamma Fc receptor I Human genes 0.000 description 6
- 241000699660 Mus musculus Species 0.000 description 6
- 210000004369 blood Anatomy 0.000 description 6
- 239000008280 blood Substances 0.000 description 6
- 230000014509 gene expression Effects 0.000 description 6
- 230000005847 immunogenicity Effects 0.000 description 6
- 210000002540 macrophage Anatomy 0.000 description 6
- 102000040430 polynucleotide Human genes 0.000 description 6
- 108091033319 polynucleotide Proteins 0.000 description 6
- 239000002157 polynucleotide Substances 0.000 description 6
- 238000004321 preservation Methods 0.000 description 6
- 241000282693 Cercopithecidae Species 0.000 description 5
- 108010021470 Fc gamma receptor IIC Proteins 0.000 description 5
- 241000282412 Homo Species 0.000 description 5
- 125000003290 L-leucino group Chemical group [H]OC(=O)[C@@]([H])(N([H])[*])C([H])([H])C(C([H])([H])[H])([H])C([H])([H])[H] 0.000 description 5
- 102100029206 Low affinity immunoglobulin gamma Fc region receptor II-c Human genes 0.000 description 5
- 239000004472 Lysine Substances 0.000 description 5
- 238000009825 accumulation Methods 0.000 description 5
- 235000003704 aspartic acid Nutrition 0.000 description 5
- 238000003556 assay Methods 0.000 description 5
- OQFSQFPPLPISGP-UHFFFAOYSA-N beta-carboxyaspartic acid Natural products OC(=O)C(N)C(C(O)=O)C(O)=O OQFSQFPPLPISGP-UHFFFAOYSA-N 0.000 description 5
- 239000013060 biological fluid Substances 0.000 description 5
- 238000006243 chemical reaction Methods 0.000 description 5
- 210000002889 endothelial cell Anatomy 0.000 description 5
- 238000012216 screening Methods 0.000 description 5
- 239000007858 starting material Substances 0.000 description 5
- 108091008875 B cell receptors Proteins 0.000 description 4
- OYPRJOBELJOOCE-UHFFFAOYSA-N Calcium Chemical compound [Ca] OYPRJOBELJOOCE-UHFFFAOYSA-N 0.000 description 4
- 102000004127 Cytokines Human genes 0.000 description 4
- 108090000695 Cytokines Proteins 0.000 description 4
- 206010071602 Genetic polymorphism Diseases 0.000 description 4
- 102000010956 Glypican Human genes 0.000 description 4
- 108050001154 Glypican Proteins 0.000 description 4
- 108050007237 Glypican-3 Proteins 0.000 description 4
- 102000009490 IgG Receptors Human genes 0.000 description 4
- ODKSFYDXXFIFQN-BYPYZUCNSA-P L-argininium(2+) Chemical compound NC(=[NH2+])NCCC[C@H]([NH3+])C(O)=O ODKSFYDXXFIFQN-BYPYZUCNSA-P 0.000 description 4
- 241000283973 Oryctolagus cuniculus Species 0.000 description 4
- 241000700159 Rattus Species 0.000 description 4
- 230000004913 activation Effects 0.000 description 4
- 108010081667 aflibercept Proteins 0.000 description 4
- 230000010056 antibody-dependent cellular cytotoxicity Effects 0.000 description 4
- 239000007864 aqueous solution Substances 0.000 description 4
- 210000003719 b-lymphocyte Anatomy 0.000 description 4
- 229910052791 calcium Inorganic materials 0.000 description 4
- 201000011510 cancer Diseases 0.000 description 4
- 235000013922 glutamic acid Nutrition 0.000 description 4
- 239000004220 glutamic acid Substances 0.000 description 4
- 125000000487 histidyl group Chemical group [H]N([H])C(C(=O)O*)C([H])([H])C1=C([H])N([H])C([H])=N1 0.000 description 4
- 210000004408 hybridoma Anatomy 0.000 description 4
- 230000001900 immune effect Effects 0.000 description 4
- 230000000670 limiting effect Effects 0.000 description 4
- 238000005259 measurement Methods 0.000 description 4
- 229910052751 metal Inorganic materials 0.000 description 4
- 229910021645 metal ion Inorganic materials 0.000 description 4
- 238000004064 recycling Methods 0.000 description 4
- 230000009467 reduction Effects 0.000 description 4
- 239000002904 solvent Substances 0.000 description 4
- 238000011282 treatment Methods 0.000 description 4
- 238000002424 x-ray crystallography Methods 0.000 description 4
- 238000011746 C57BL/6J (JAX™ mouse strain) Methods 0.000 description 3
- 241000282472 Canis lupus familiaris Species 0.000 description 3
- 238000002965 ELISA Methods 0.000 description 3
- 101710181478 Envelope glycoprotein GP350 Proteins 0.000 description 3
- 108010067306 Fibronectins Proteins 0.000 description 3
- 102000016359 Fibronectins Human genes 0.000 description 3
- 206010061218 Inflammation Diseases 0.000 description 3
- ONIBWKKTOPOVIA-BYPYZUCNSA-N L-Proline Chemical compound OC(=O)[C@@H]1CCCN1 ONIBWKKTOPOVIA-BYPYZUCNSA-N 0.000 description 3
- 125000002059 L-arginyl group Chemical group O=C([*])[C@](N([H])[H])([H])C([H])([H])C([H])([H])C([H])([H])N([H])C(=N[H])N([H])[H] 0.000 description 3
- 125000002068 L-phenylalanino group Chemical group [H]OC(=O)[C@@]([H])(N([H])[*])C([H])([H])C1=C([H])C([H])=C([H])C([H])=C1[H] 0.000 description 3
- 108700018351 Major Histocompatibility Complex Proteins 0.000 description 3
- 230000010637 Metal Chelating Activity Effects 0.000 description 3
- 206010028980 Neoplasm Diseases 0.000 description 3
- ONIBWKKTOPOVIA-UHFFFAOYSA-N Proline Natural products OC(=O)C1CCCN1 ONIBWKKTOPOVIA-UHFFFAOYSA-N 0.000 description 3
- 101100094105 Saccharomyces cerevisiae (strain ATCC 204508 / S288c) NPL6 gene Proteins 0.000 description 3
- 230000009824 affinity maturation Effects 0.000 description 3
- 230000006229 amino acid addition Effects 0.000 description 3
- 230000005888 antibody-dependent cellular phagocytosis Effects 0.000 description 3
- 230000009286 beneficial effect Effects 0.000 description 3
- 230000008901 benefit Effects 0.000 description 3
- 230000001413 cellular effect Effects 0.000 description 3
- 230000004700 cellular uptake Effects 0.000 description 3
- 230000000875 corresponding effect Effects 0.000 description 3
- 230000001086 cytosolic effect Effects 0.000 description 3
- 230000007423 decrease Effects 0.000 description 3
- 238000011161 development Methods 0.000 description 3
- 230000002349 favourable effect Effects 0.000 description 3
- 210000004602 germ cell Anatomy 0.000 description 3
- 229910052739 hydrogen Inorganic materials 0.000 description 3
- 239000001257 hydrogen Substances 0.000 description 3
- 230000003053 immunization Effects 0.000 description 3
- 230000001976 improved effect Effects 0.000 description 3
- 230000004054 inflammatory process Effects 0.000 description 3
- 210000004698 lymphocyte Anatomy 0.000 description 3
- 230000037353 metabolic pathway Effects 0.000 description 3
- 239000002184 metal Substances 0.000 description 3
- 125000002496 methyl group Chemical group [H]C([H])([H])* 0.000 description 3
- 239000000178 monomer Substances 0.000 description 3
- 230000035790 physiological processes and functions Effects 0.000 description 3
- 125000001500 prolyl group Chemical compound [H]N1C([H])(C(=O)[*])C([H])([H])C([H])([H])C1([H])[H] 0.000 description 3
- 238000004904 shortening Methods 0.000 description 3
- 230000020382 suppression by virus of host antigen processing and presentation of peptide antigen via MHC class I Effects 0.000 description 3
- 210000003556 vascular endothelial cell Anatomy 0.000 description 3
- XLYOFNOQVPJJNP-UHFFFAOYSA-N water Substances O XLYOFNOQVPJJNP-UHFFFAOYSA-N 0.000 description 3
- JKMHFZQWWAIEOD-UHFFFAOYSA-N 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid Chemical compound OCC[NH+]1CCN(CCS([O-])(=O)=O)CC1 JKMHFZQWWAIEOD-UHFFFAOYSA-N 0.000 description 2
- 102100027715 4-hydroxy-2-oxoglutarate aldolase, mitochondrial Human genes 0.000 description 2
- 208000023275 Autoimmune disease Diseases 0.000 description 2
- 108090000342 C-Type Lectins Proteins 0.000 description 2
- 102000003930 C-Type Lectins Human genes 0.000 description 2
- 108050006947 CXC Chemokine Proteins 0.000 description 2
- 102000019388 CXC chemokine Human genes 0.000 description 2
- 102000000584 Calmodulin Human genes 0.000 description 2
- 108010041952 Calmodulin Proteins 0.000 description 2
- 241000282836 Camelus dromedarius Species 0.000 description 2
- 108020004705 Codon Proteins 0.000 description 2
- 108010035532 Collagen Proteins 0.000 description 2
- 102000008186 Collagen Human genes 0.000 description 2
- 108010047041 Complementarity Determining Regions Proteins 0.000 description 2
- SHZGCJCMOBCMKK-UHFFFAOYSA-N D-mannomethylose Natural products CC1OC(O)C(O)C(O)C1O SHZGCJCMOBCMKK-UHFFFAOYSA-N 0.000 description 2
- 108700022150 Designed Ankyrin Repeat Proteins Proteins 0.000 description 2
- 108091006020 Fc-tagged proteins Proteins 0.000 description 2
- PNNNRSAQSRJVSB-SLPGGIOYSA-N Fucose Natural products C[C@H](O)[C@@H](O)[C@H](O)[C@H](O)C=O PNNNRSAQSRJVSB-SLPGGIOYSA-N 0.000 description 2
- 102000003886 Glycoproteins Human genes 0.000 description 2
- 108090000288 Glycoproteins Proteins 0.000 description 2
- 239000007995 HEPES buffer Substances 0.000 description 2
- 101001081225 Homo sapiens 4-hydroxy-2-oxoglutarate aldolase, mitochondrial Proteins 0.000 description 2
- 101001109518 Homo sapiens N-acetylneuraminate lyase Proteins 0.000 description 2
- 101000604027 Homo sapiens Nuclear protein localization protein 4 homolog Proteins 0.000 description 2
- 101000974007 Homo sapiens Nucleosome assembly protein 1-like 3 Proteins 0.000 description 2
- 101001099181 Homo sapiens TATA-binding protein-associated factor 2N Proteins 0.000 description 2
- UFHFLCQGNIYNRP-UHFFFAOYSA-N Hydrogen Chemical compound [H][H] UFHFLCQGNIYNRP-UHFFFAOYSA-N 0.000 description 2
- QNAYBMKLOCPYGJ-REOHCLBHSA-N L-alanine Chemical compound C[C@H](N)C(O)=O QNAYBMKLOCPYGJ-REOHCLBHSA-N 0.000 description 2
- SHZGCJCMOBCMKK-DHVFOXMCSA-N L-fucopyranose Chemical compound C[C@@H]1OC(O)[C@@H](O)[C@H](O)[C@@H]1O SHZGCJCMOBCMKK-DHVFOXMCSA-N 0.000 description 2
- 125000000205 L-threonino group Chemical group [H]OC(=O)[C@@]([H])(N([H])[*])[C@](C([H])([H])[H])([H])O[H] 0.000 description 2
- 108010085895 Laminin Proteins 0.000 description 2
- 102000007547 Laminin Human genes 0.000 description 2
- 108010006444 Leucine-Rich Repeat Proteins Proteins 0.000 description 2
- 102000013519 Lipocalin-2 Human genes 0.000 description 2
- 108010051335 Lipocalin-2 Proteins 0.000 description 2
- 241000282567 Macaca fascicularis Species 0.000 description 2
- 241000282560 Macaca mulatta Species 0.000 description 2
- 241000124008 Mammalia Species 0.000 description 2
- 102100022686 N-acetylneuraminate lyase Human genes 0.000 description 2
- 102100038438 Nuclear protein localization protein 4 homolog Human genes 0.000 description 2
- 239000004365 Protease Substances 0.000 description 2
- 102000016611 Proteoglycans Human genes 0.000 description 2
- 108010067787 Proteoglycans Proteins 0.000 description 2
- 201000004681 Psoriasis Diseases 0.000 description 2
- 241000219061 Rheum Species 0.000 description 2
- KJTLSVCANCCWHF-UHFFFAOYSA-N Ruthenium Chemical compound [Ru] KJTLSVCANCCWHF-UHFFFAOYSA-N 0.000 description 2
- 108010003723 Single-Domain Antibodies Proteins 0.000 description 2
- 102100038917 TATA-binding protein-associated factor 2N Human genes 0.000 description 2
- 108020005038 Terminator Codon Proteins 0.000 description 2
- 101150117115 V gene Proteins 0.000 description 2
- 210000000612 antigen-presenting cell Anatomy 0.000 description 2
- 206010003246 arthritis Diseases 0.000 description 2
- 229910001423 beryllium ion Inorganic materials 0.000 description 2
- 230000004791 biological behavior Effects 0.000 description 2
- 238000004422 calculation algorithm Methods 0.000 description 2
- 238000003776 cleavage reaction Methods 0.000 description 2
- 229920001436 collagen Polymers 0.000 description 2
- 102000006834 complement receptors Human genes 0.000 description 2
- 108010047295 complement receptors Proteins 0.000 description 2
- 238000004132 cross linking Methods 0.000 description 2
- 230000003013 cytotoxicity Effects 0.000 description 2
- 231100000135 cytotoxicity Toxicity 0.000 description 2
- 230000029087 digestion Effects 0.000 description 2
- 201000010099 disease Diseases 0.000 description 2
- 238000005516 engineering process Methods 0.000 description 2
- 230000002708 enhancing effect Effects 0.000 description 2
- 230000029142 excretion Effects 0.000 description 2
- 238000001943 fluorescence-activated cell sorting Methods 0.000 description 2
- 239000000833 heterodimer Substances 0.000 description 2
- 210000003630 histaminocyte Anatomy 0.000 description 2
- 230000036039 immunity Effects 0.000 description 2
- 238000003018 immunoassay Methods 0.000 description 2
- 229940072221 immunoglobulins Drugs 0.000 description 2
- 208000015181 infectious disease Diseases 0.000 description 2
- 230000008595 infiltration Effects 0.000 description 2
- 238000001764 infiltration Methods 0.000 description 2
- 208000027866 inflammatory disease Diseases 0.000 description 2
- 230000003993 interaction Effects 0.000 description 2
- 238000001155 isoelectric focusing Methods 0.000 description 2
- 210000004901 leucine-rich repeat Anatomy 0.000 description 2
- 210000000265 leukocyte Anatomy 0.000 description 2
- 230000008774 maternal effect Effects 0.000 description 2
- 230000007246 mechanism Effects 0.000 description 2
- 230000001404 mediated effect Effects 0.000 description 2
- 210000004379 membrane Anatomy 0.000 description 2
- 239000012528 membrane Substances 0.000 description 2
- 150000002739 metals Chemical class 0.000 description 2
- 229930182817 methionine Natural products 0.000 description 2
- 210000000822 natural killer cell Anatomy 0.000 description 2
- 210000000056 organ Anatomy 0.000 description 2
- 230000003285 pharmacodynamic effect Effects 0.000 description 2
- 230000004962 physiological condition Effects 0.000 description 2
- 238000003127 radioimmunoassay Methods 0.000 description 2
- 238000011160 research Methods 0.000 description 2
- 229910052707 ruthenium Inorganic materials 0.000 description 2
- 230000007017 scission Effects 0.000 description 2
- 230000035945 sensitivity Effects 0.000 description 2
- 238000007920 subcutaneous administration Methods 0.000 description 2
- 239000000126 substance Substances 0.000 description 2
- 238000002198 surface plasmon resonance spectroscopy Methods 0.000 description 2
- 201000000596 systemic lupus erythematosus Diseases 0.000 description 2
- 238000012546 transfer Methods 0.000 description 2
- 206010071198 Allergic hepatitis Diseases 0.000 description 1
- 102000008102 Ankyrins Human genes 0.000 description 1
- 108010049777 Ankyrins Proteins 0.000 description 1
- 102000000412 Annexin Human genes 0.000 description 1
- 108050008874 Annexin Proteins 0.000 description 1
- 108010032595 Antibody Binding Sites Proteins 0.000 description 1
- 101150075175 Asgr1 gene Proteins 0.000 description 1
- 102000005427 Asialoglycoprotein Receptor Human genes 0.000 description 1
- 201000001320 Atherosclerosis Diseases 0.000 description 1
- 102100036597 Basement membrane-specific heparan sulfate proteoglycan core protein Human genes 0.000 description 1
- 208000009137 Behcet syndrome Diseases 0.000 description 1
- ZOXJGFHDIHLPTG-UHFFFAOYSA-N Boron Chemical compound [B] ZOXJGFHDIHLPTG-UHFFFAOYSA-N 0.000 description 1
- 241000283690 Bos taurus Species 0.000 description 1
- 206010006187 Breast cancer Diseases 0.000 description 1
- 208000026310 Breast neoplasm Diseases 0.000 description 1
- 108050005711 C Chemokine Proteins 0.000 description 1
- 102000017483 C chemokine Human genes 0.000 description 1
- 125000001433 C-terminal amino-acid group Chemical group 0.000 description 1
- 102000002110 C2 domains Human genes 0.000 description 1
- 108050009459 C2 domains Proteins 0.000 description 1
- 102000001902 CC Chemokines Human genes 0.000 description 1
- 108010040471 CC Chemokines Proteins 0.000 description 1
- 101150107439 CDR3 gene Proteins 0.000 description 1
- 102000000905 Cadherin Human genes 0.000 description 1
- 108050007957 Cadherin Proteins 0.000 description 1
- 241000283707 Capra Species 0.000 description 1
- 101710167800 Capsid assembly scaffolding protein Proteins 0.000 description 1
- OKTJSMMVPCPJKN-UHFFFAOYSA-N Carbon Chemical compound [C] OKTJSMMVPCPJKN-UHFFFAOYSA-N 0.000 description 1
- 102000016289 Cell Adhesion Molecules Human genes 0.000 description 1
- 108010067225 Cell Adhesion Molecules Proteins 0.000 description 1
- 102100022641 Coagulation factor IX Human genes 0.000 description 1
- 206010009900 Colitis ulcerative Diseases 0.000 description 1
- RYGMFSIKBFXOCR-UHFFFAOYSA-N Copper Chemical group [Cu] RYGMFSIKBFXOCR-UHFFFAOYSA-N 0.000 description 1
- 241000699800 Cricetinae Species 0.000 description 1
- 208000011231 Crohn disease Diseases 0.000 description 1
- 102000001189 Cyclic Peptides Human genes 0.000 description 1
- 108010069514 Cyclic Peptides Proteins 0.000 description 1
- 241001273590 Cyclostomata Species 0.000 description 1
- 201000003883 Cystic fibrosis Diseases 0.000 description 1
- WQZGKKKJIJFFOK-QTVWNMPRSA-N D-mannopyranose Chemical compound OC[C@H]1OC(O)[C@@H](O)[C@@H](O)[C@@H]1O WQZGKKKJIJFFOK-QTVWNMPRSA-N 0.000 description 1
- 108010037897 DC-specific ICAM-3 grabbing nonintegrin Proteins 0.000 description 1
- 108020004414 DNA Proteins 0.000 description 1
- 206010012438 Dermatitis atopic Diseases 0.000 description 1
- 206010012442 Dermatitis contact Diseases 0.000 description 1
- BWGNESOTFCXPMA-UHFFFAOYSA-N Dihydrogen disulfide Chemical compound SS BWGNESOTFCXPMA-UHFFFAOYSA-N 0.000 description 1
- 102000012545 EGF-like domains Human genes 0.000 description 1
- 108050002150 EGF-like domains Proteins 0.000 description 1
- 238000004435 EPR spectroscopy Methods 0.000 description 1
- 241000196324 Embryophyta Species 0.000 description 1
- 102000004190 Enzymes Human genes 0.000 description 1
- 108090000790 Enzymes Proteins 0.000 description 1
- 108010076282 Factor IX Proteins 0.000 description 1
- 108010048049 Factor IXa Proteins 0.000 description 1
- 108010014173 Factor X Proteins 0.000 description 1
- 102000009123 Fibrin Human genes 0.000 description 1
- 108010073385 Fibrin Proteins 0.000 description 1
- BWGVNKXGVNDBDI-UHFFFAOYSA-N Fibrin monomer Chemical compound CNC(=O)CNC(=O)CN BWGVNKXGVNDBDI-UHFFFAOYSA-N 0.000 description 1
- 206010018364 Glomerulonephritis Diseases 0.000 description 1
- 208000005176 Hepatitis C Diseases 0.000 description 1
- 206010019728 Hepatitis alcoholic Diseases 0.000 description 1
- 102000008949 Histocompatibility Antigens Class I Human genes 0.000 description 1
- 101000878605 Homo sapiens Low affinity immunoglobulin epsilon Fc receptor Proteins 0.000 description 1
- 101000845206 Homo sapiens Putative peripheral benzodiazepine receptor-related protein Proteins 0.000 description 1
- 101000845233 Homo sapiens Translocator protein Proteins 0.000 description 1
- 206010020649 Hyperkeratosis Diseases 0.000 description 1
- 241000235789 Hyperoartia Species 0.000 description 1
- 108010054477 Immunoglobulin Fab Fragments Proteins 0.000 description 1
- 102000001706 Immunoglobulin Fab Fragments Human genes 0.000 description 1
- 208000022559 Inflammatory bowel disease Diseases 0.000 description 1
- 102000018682 Interleukin Receptor Common gamma Subunit Human genes 0.000 description 1
- 108010066719 Interleukin Receptor Common gamma Subunit Proteins 0.000 description 1
- 102100026236 Interleukin-8 Human genes 0.000 description 1
- 102000015696 Interleukins Human genes 0.000 description 1
- 108010063738 Interleukins Proteins 0.000 description 1
- 101100507312 Invertebrate iridescent virus 6 EF1 gene Proteins 0.000 description 1
- XEEYBQQBJWHFJM-UHFFFAOYSA-N Iron Chemical group [Fe] XEEYBQQBJWHFJM-UHFFFAOYSA-N 0.000 description 1
- 208000001126 Keratosis Diseases 0.000 description 1
- 208000008839 Kidney Neoplasms Diseases 0.000 description 1
- FFEARJCKVFRZRR-BYPYZUCNSA-N L-methionine Chemical compound CSCC[C@H](N)C(O)=O FFEARJCKVFRZRR-BYPYZUCNSA-N 0.000 description 1
- LRQKBLKVPFOOQJ-YFKPBYRVSA-N L-norleucine Chemical compound CCCC[C@H]([NH3+])C([O-])=O LRQKBLKVPFOOQJ-YFKPBYRVSA-N 0.000 description 1
- QIVBCDIJIAJPQS-VIFPVBQESA-N L-tryptophane Chemical compound C1=CC=C2C(C[C@H](N)C(O)=O)=CNC2=C1 QIVBCDIJIAJPQS-VIFPVBQESA-N 0.000 description 1
- KZSNJWFQEVHDMF-BYPYZUCNSA-N L-valine Chemical compound CC(C)[C@H](N)C(O)=O KZSNJWFQEVHDMF-BYPYZUCNSA-N 0.000 description 1
- 108010001831 LDL receptors Proteins 0.000 description 1
- 102000019298 Lipocalin Human genes 0.000 description 1
- 108050006654 Lipocalin Proteins 0.000 description 1
- 102100038007 Low affinity immunoglobulin epsilon Fc receptor Human genes 0.000 description 1
- 102100024640 Low-density lipoprotein receptor Human genes 0.000 description 1
- 206010058467 Lung neoplasm malignant Diseases 0.000 description 1
- 108090000362 Lymphotoxin-beta Proteins 0.000 description 1
- 108091054437 MHC class I family Proteins 0.000 description 1
- 108010052285 Membrane Proteins Proteins 0.000 description 1
- 102000018697 Membrane Proteins Human genes 0.000 description 1
- 241001529936 Murinae Species 0.000 description 1
- 102000016349 Myosin Light Chains Human genes 0.000 description 1
- 108010067385 Myosin Light Chains Proteins 0.000 description 1
- 241000251752 Myxine glutinosa Species 0.000 description 1
- 125000000729 N-terminal amino-acid group Chemical group 0.000 description 1
- 238000005481 NMR spectroscopy Methods 0.000 description 1
- 102100037369 Nidogen-1 Human genes 0.000 description 1
- 108091060545 Nonsense suppressor Proteins 0.000 description 1
- 108091034117 Oligonucleotide Proteins 0.000 description 1
- 208000007117 Oral Ulcer Diseases 0.000 description 1
- 206010033128 Ovarian cancer Diseases 0.000 description 1
- 206010061535 Ovarian neoplasm Diseases 0.000 description 1
- 241000282579 Pan Species 0.000 description 1
- 241000282577 Pan troglodytes Species 0.000 description 1
- 108090000526 Papain Proteins 0.000 description 1
- 241000282515 Papio hamadryas Species 0.000 description 1
- 102000001675 Parvalbumin Human genes 0.000 description 1
- 108060005874 Parvalbumin Proteins 0.000 description 1
- 241001494479 Pecora Species 0.000 description 1
- 102000057297 Pepsin A Human genes 0.000 description 1
- 108090000284 Pepsin A Proteins 0.000 description 1
- 102000035195 Peptidases Human genes 0.000 description 1
- 108091005804 Peptidases Proteins 0.000 description 1
- 206010057249 Phagocytosis Diseases 0.000 description 1
- 101710130420 Probable capsid assembly scaffolding protein Proteins 0.000 description 1
- 206010060862 Prostate cancer Diseases 0.000 description 1
- 208000000236 Prostatic Neoplasms Diseases 0.000 description 1
- 102000003923 Protein Kinase C Human genes 0.000 description 1
- 108090000315 Protein Kinase C Proteins 0.000 description 1
- 206010038389 Renal cancer Diseases 0.000 description 1
- 241000283984 Rodentia Species 0.000 description 1
- 101710204410 Scaffold protein Proteins 0.000 description 1
- 206010040047 Sepsis Diseases 0.000 description 1
- MTCFGRXMJLQNBG-UHFFFAOYSA-N Serine Natural products OCC(N)C(O)=O MTCFGRXMJLQNBG-UHFFFAOYSA-N 0.000 description 1
- 208000005718 Stomach Neoplasms Diseases 0.000 description 1
- AYFVYJQAPQTCCC-UHFFFAOYSA-N Threonine Natural products CC(O)C(N)C(O)=O AYFVYJQAPQTCCC-UHFFFAOYSA-N 0.000 description 1
- 239000004473 Threonine Substances 0.000 description 1
- 108060008245 Thrombospondin Proteins 0.000 description 1
- 102000002938 Thrombospondin Human genes 0.000 description 1
- 101710120037 Toxin CcdB Proteins 0.000 description 1
- 241000219793 Trifolium Species 0.000 description 1
- 102000013534 Troponin C Human genes 0.000 description 1
- QIVBCDIJIAJPQS-UHFFFAOYSA-N Tryptophan Natural products C1=CC=C2C(CC(N)C(O)=O)=CNC2=C1 QIVBCDIJIAJPQS-UHFFFAOYSA-N 0.000 description 1
- 201000006704 Ulcerative Colitis Diseases 0.000 description 1
- 108091008605 VEGF receptors Proteins 0.000 description 1
- 102000009484 Vascular Endothelial Growth Factor Receptors Human genes 0.000 description 1
- HCHKCACWOHOZIP-UHFFFAOYSA-N Zinc Chemical group [Zn] HCHKCACWOHOZIP-UHFFFAOYSA-N 0.000 description 1
- JLCPHMBAVCMARE-UHFFFAOYSA-N [3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-[[3-[[3-[[3-[[3-[[3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-hydroxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methyl [5-(6-aminopurin-9-yl)-2-(hydroxymethyl)oxolan-3-yl] hydrogen phosphate Polymers Cc1cn(C2CC(OP(O)(=O)OCC3OC(CC3OP(O)(=O)OCC3OC(CC3O)n3cnc4c3nc(N)[nH]c4=O)n3cnc4c3nc(N)[nH]c4=O)C(COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3CO)n3cnc4c(N)ncnc34)n3ccc(N)nc3=O)n3cnc4c(N)ncnc34)n3ccc(N)nc3=O)n3ccc(N)nc3=O)n3ccc(N)nc3=O)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cc(C)c(=O)[nH]c3=O)n3cc(C)c(=O)[nH]c3=O)n3ccc(N)nc3=O)n3cc(C)c(=O)[nH]c3=O)n3cnc4c3nc(N)[nH]c4=O)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)O2)c(=O)[nH]c1=O JLCPHMBAVCMARE-UHFFFAOYSA-N 0.000 description 1
- 239000000556 agonist Substances 0.000 description 1
- 238000012867 alanine scanning Methods 0.000 description 1
- 208000002353 alcoholic hepatitis Diseases 0.000 description 1
- 229910052784 alkaline earth metal Inorganic materials 0.000 description 1
- 150000001342 alkaline earth metals Chemical class 0.000 description 1
- 238000003016 alphascreen Methods 0.000 description 1
- 125000003277 amino group Chemical group 0.000 description 1
- 238000012440 amplified luminescent proximity homogeneous assay Methods 0.000 description 1
- 238000004458 analytical method Methods 0.000 description 1
- 239000002870 angiogenesis inducing agent Substances 0.000 description 1
- 230000009830 antibody antigen interaction Effects 0.000 description 1
- 230000000890 antigenic effect Effects 0.000 description 1
- 229910052787 antimony Inorganic materials 0.000 description 1
- WATWJIUSRGPENY-UHFFFAOYSA-N antimony atom Chemical compound [Sb] WATWJIUSRGPENY-UHFFFAOYSA-N 0.000 description 1
- 208000002399 aphthous stomatitis Diseases 0.000 description 1
- 239000002473 artificial blood Substances 0.000 description 1
- 125000003118 aryl group Chemical group 0.000 description 1
- 210000003567 ascitic fluid Anatomy 0.000 description 1
- 108010006523 asialoglycoprotein receptor Proteins 0.000 description 1
- 208000006673 asthma Diseases 0.000 description 1
- 239000012298 atmosphere Substances 0.000 description 1
- 125000004429 atom Chemical group 0.000 description 1
- 201000008937 atopic dermatitis Diseases 0.000 description 1
- 102000015736 beta 2-Microglobulin Human genes 0.000 description 1
- 108010081355 beta 2-Microglobulin Proteins 0.000 description 1
- 230000033228 biological regulation Effects 0.000 description 1
- 229920001222 biopolymer Polymers 0.000 description 1
- 230000015572 biosynthetic process Effects 0.000 description 1
- 229910052797 bismuth Inorganic materials 0.000 description 1
- JCXGWMGPZLAOME-UHFFFAOYSA-N bismuth atom Chemical compound [Bi] JCXGWMGPZLAOME-UHFFFAOYSA-N 0.000 description 1
- 230000023555 blood coagulation Effects 0.000 description 1
- 210000004204 blood vessel Anatomy 0.000 description 1
- 229910052796 boron Inorganic materials 0.000 description 1
- 206010006451 bronchitis Diseases 0.000 description 1
- 210000004899 c-terminal region Anatomy 0.000 description 1
- 238000004364 calculation method Methods 0.000 description 1
- 229910052799 carbon Inorganic materials 0.000 description 1
- 125000002915 carbonyl group Chemical group [*:2]C([*:1])=O 0.000 description 1
- 150000001768 cations Chemical class 0.000 description 1
- 230000010261 cell growth Effects 0.000 description 1
- 210000000170 cell membrane Anatomy 0.000 description 1
- 230000004663 cell proliferation Effects 0.000 description 1
- 238000001516 cell proliferation assay Methods 0.000 description 1
- 230000017455 cell-cell adhesion Effects 0.000 description 1
- 230000003399 chemotactic effect Effects 0.000 description 1
- 230000035605 chemotaxis Effects 0.000 description 1
- 230000001684 chronic effect Effects 0.000 description 1
- 208000037976 chronic inflammation Diseases 0.000 description 1
- 208000037893 chronic inflammatory disorder Diseases 0.000 description 1
- 235000021277 colostrum Nutrition 0.000 description 1
- 210000003022 colostrum Anatomy 0.000 description 1
- 230000000295 complement effect Effects 0.000 description 1
- 230000004540 complement-dependent cytotoxicity Effects 0.000 description 1
- 238000004590 computer program Methods 0.000 description 1
- 208000010247 contact dermatitis Diseases 0.000 description 1
- 230000002596 correlated effect Effects 0.000 description 1
- 239000012228 culture supernatant Substances 0.000 description 1
- 125000000151 cysteine group Chemical group N[C@@H](CS)C(=O)* 0.000 description 1
- 238000001514 detection method Methods 0.000 description 1
- 238000001962 electrophoresis Methods 0.000 description 1
- 238000010828 elution Methods 0.000 description 1
- 229940088598 enzyme Drugs 0.000 description 1
- 210000000981 epithelium Anatomy 0.000 description 1
- 238000002474 experimental method Methods 0.000 description 1
- 239000013604 expression vector Substances 0.000 description 1
- 210000003722 extracellular fluid Anatomy 0.000 description 1
- 229950003499 fibrin Drugs 0.000 description 1
- 210000002950 fibroblast Anatomy 0.000 description 1
- 239000012530 fluid Substances 0.000 description 1
- 238000009472 formulation Methods 0.000 description 1
- 239000012634 fragment Substances 0.000 description 1
- 125000000524 functional group Chemical group 0.000 description 1
- 230000004927 fusion Effects 0.000 description 1
- 108020001507 fusion proteins Proteins 0.000 description 1
- 102000037865 fusion proteins Human genes 0.000 description 1
- 101150034785 gamma gene Proteins 0.000 description 1
- 206010017758 gastric cancer Diseases 0.000 description 1
- 238000010353 genetic engineering Methods 0.000 description 1
- 230000013595 glycosylation Effects 0.000 description 1
- 238000006206 glycosylation reaction Methods 0.000 description 1
- 229910021480 group 4 element Inorganic materials 0.000 description 1
- 229910021472 group 8 element Inorganic materials 0.000 description 1
- 239000003102 growth factor Substances 0.000 description 1
- 201000010536 head and neck cancer Diseases 0.000 description 1
- 208000014829 head and neck neoplasm Diseases 0.000 description 1
- 230000036541 health Effects 0.000 description 1
- 208000002672 hepatitis B Diseases 0.000 description 1
- 150000002431 hydrogen Chemical class 0.000 description 1
- 125000004435 hydrogen atom Chemical group [H]* 0.000 description 1
- 230000002209 hydrophobic effect Effects 0.000 description 1
- 210000002865 immune cell Anatomy 0.000 description 1
- 230000008105 immune reaction Effects 0.000 description 1
- 230000016784 immunoglobulin production Effects 0.000 description 1
- 230000001506 immunosuppresive effect Effects 0.000 description 1
- 230000006872 improvement Effects 0.000 description 1
- 238000000338 in vitro Methods 0.000 description 1
- 230000001939 inductive effect Effects 0.000 description 1
- 108091008042 inhibitory receptors Proteins 0.000 description 1
- 229940047122 interleukins Drugs 0.000 description 1
- 230000003834 intracellular effect Effects 0.000 description 1
- AGPKZVBTJJNPAG-UHFFFAOYSA-N isoleucine Natural products CCC(C)C(N)C(O)=O AGPKZVBTJJNPAG-UHFFFAOYSA-N 0.000 description 1
- 229960000310 isoleucine Drugs 0.000 description 1
- 201000010982 kidney cancer Diseases 0.000 description 1
- 208000017169 kidney disease Diseases 0.000 description 1
- 238000002372 labelling Methods 0.000 description 1
- 239000003446 ligand Substances 0.000 description 1
- 208000019423 liver disease Diseases 0.000 description 1
- 201000005202 lung cancer Diseases 0.000 description 1
- 208000020816 lung neoplasm Diseases 0.000 description 1
- 230000001926 lymphatic effect Effects 0.000 description 1
- 125000003588 lysine group Chemical group [H]N([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])(N([H])[H])C(*)=O 0.000 description 1
- 238000013507 mapping Methods 0.000 description 1
- 241001515942 marmosets Species 0.000 description 1
- 201000001441 melanoma Diseases 0.000 description 1
- 210000000110 microvilli Anatomy 0.000 description 1
- 235000013336 milk Nutrition 0.000 description 1
- 210000004080 milk Anatomy 0.000 description 1
- 239000008267 milk Substances 0.000 description 1
- 210000001616 monocyte Anatomy 0.000 description 1
- 201000006417 multiple sclerosis Diseases 0.000 description 1
- 238000007857 nested PCR Methods 0.000 description 1
- 108010008217 nidogen Proteins 0.000 description 1
- 230000009871 nonspecific binding Effects 0.000 description 1
- 210000004940 nucleus Anatomy 0.000 description 1
- 230000014207 opsonization Effects 0.000 description 1
- 230000000242 pagocytic effect Effects 0.000 description 1
- 238000004091 panning Methods 0.000 description 1
- 229940055729 papain Drugs 0.000 description 1
- 235000019834 papain Nutrition 0.000 description 1
- 230000036961 partial effect Effects 0.000 description 1
- 230000001717 pathogenic effect Effects 0.000 description 1
- 230000037361 pathway Effects 0.000 description 1
- 229940111202 pepsin Drugs 0.000 description 1
- 210000005259 peripheral blood Anatomy 0.000 description 1
- 239000011886 peripheral blood Substances 0.000 description 1
- 108010049224 perlecan Proteins 0.000 description 1
- 230000035699 permeability Effects 0.000 description 1
- 238000002823 phage display Methods 0.000 description 1
- 210000001539 phagocyte Anatomy 0.000 description 1
- 230000008782 phagocytosis Effects 0.000 description 1
- 230000002399 phagocytotic effect Effects 0.000 description 1
- 230000000144 pharmacologic effect Effects 0.000 description 1
- COLNVLDHVKWLRT-UHFFFAOYSA-N phenylalanine Natural products OC(=O)C(N)CC1=CC=CC=C1 COLNVLDHVKWLRT-UHFFFAOYSA-N 0.000 description 1
- 235000008729 phenylalanine Nutrition 0.000 description 1
- 210000002826 placenta Anatomy 0.000 description 1
- BASFCYQUMIYNBI-UHFFFAOYSA-N platinum Chemical group [Pt] BASFCYQUMIYNBI-UHFFFAOYSA-N 0.000 description 1
- 210000004910 pleural fluid Anatomy 0.000 description 1
- 229910052699 polonium Inorganic materials 0.000 description 1
- HZEBHPIOVYHPMT-UHFFFAOYSA-N polonium atom Chemical compound [Po] HZEBHPIOVYHPMT-UHFFFAOYSA-N 0.000 description 1
- 230000003389 potentiating effect Effects 0.000 description 1
- 125000002924 primary amino group Chemical group [H]N([H])* 0.000 description 1
- 239000000047 product Substances 0.000 description 1
- 230000000770 proinflammatory effect Effects 0.000 description 1
- 230000002035 prolonged effect Effects 0.000 description 1
- 235000019419 proteases Nutrition 0.000 description 1
- 208000005069 pulmonary fibrosis Diseases 0.000 description 1
- 238000005215 recombination Methods 0.000 description 1
- 208000023504 respiratory system disease Diseases 0.000 description 1
- 230000004044 response Effects 0.000 description 1
- 239000000523 sample Substances 0.000 description 1
- 229910052710 silicon Inorganic materials 0.000 description 1
- 239000010703 silicon Substances 0.000 description 1
- 238000002741 site-directed mutagenesis Methods 0.000 description 1
- 210000003491 skin Anatomy 0.000 description 1
- 210000000813 small intestine Anatomy 0.000 description 1
- 241000894007 species Species 0.000 description 1
- 230000002269 spontaneous effect Effects 0.000 description 1
- 230000000638 stimulation Effects 0.000 description 1
- 201000011549 stomach cancer Diseases 0.000 description 1
- 230000008093 supporting effect Effects 0.000 description 1
- 230000001629 suppression Effects 0.000 description 1
- 208000024891 symptom Diseases 0.000 description 1
- 238000012360 testing method Methods 0.000 description 1
- 230000001988 toxicity Effects 0.000 description 1
- 231100000419 toxicity Toxicity 0.000 description 1
- 230000014616 translation Effects 0.000 description 1
- 238000013519 translation Methods 0.000 description 1
- 230000005747 tumor angiogenesis Effects 0.000 description 1
- 238000002255 vaccination Methods 0.000 description 1
- 238000010200 validation analysis Methods 0.000 description 1
- 239000004474 valine Substances 0.000 description 1
- 208000019553 vascular disease Diseases 0.000 description 1
- 210000001325 yolk sac Anatomy 0.000 description 1
Images
Classifications
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K14/00—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- C07K14/195—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from bacteria
- C07K14/235—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from bacteria from Bordetella (G)
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K16/00—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies
- C07K16/18—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans
- C07K16/24—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans against cytokines, lymphokines or interferons
- C07K16/244—Interleukins [IL]
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P1/00—Drugs for disorders of the alimentary tract or the digestive system
- A61P1/02—Stomatological preparations, e.g. drugs for caries, aphtae, periodontitis
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P1/00—Drugs for disorders of the alimentary tract or the digestive system
- A61P1/04—Drugs for disorders of the alimentary tract or the digestive system for ulcers, gastritis or reflux esophagitis, e.g. antacids, inhibitors of acid secretion, mucosal protectants
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P1/00—Drugs for disorders of the alimentary tract or the digestive system
- A61P1/16—Drugs for disorders of the alimentary tract or the digestive system for liver or gallbladder disorders, e.g. hepatoprotective agents, cholagogues, litholytics
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P11/00—Drugs for disorders of the respiratory system
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P11/00—Drugs for disorders of the respiratory system
- A61P11/06—Antiasthmatics
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P13/00—Drugs for disorders of the urinary system
- A61P13/08—Drugs for disorders of the urinary system of the prostate
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P13/00—Drugs for disorders of the urinary system
- A61P13/12—Drugs for disorders of the urinary system of the kidneys
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P15/00—Drugs for genital or sexual disorders; Contraceptives
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P17/00—Drugs for dermatological disorders
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P17/00—Drugs for dermatological disorders
- A61P17/06—Antipsoriatics
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P25/00—Drugs for disorders of the nervous system
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P29/00—Non-central analgesic, antipyretic or antiinflammatory agents, e.g. antirheumatic agents; Non-steroidal antiinflammatory drugs [NSAID]
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
- A61P31/04—Antibacterial agents
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P35/00—Antineoplastic agents
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P37/00—Drugs for immunological or allergic disorders
- A61P37/02—Immunomodulators
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P37/00—Drugs for immunological or allergic disorders
- A61P37/08—Antiallergic agents
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P43/00—Drugs for specific purposes, not provided for in groups A61P1/00-A61P41/00
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P9/00—Drugs for disorders of the cardiovascular system
- A61P9/10—Drugs for disorders of the cardiovascular system for treating ischaemic or atherosclerotic diseases, e.g. antianginal drugs, coronary vasodilators, drugs for myocardial infarction, retinopathy, cerebrovascula insufficiency, renal arteriosclerosis
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K16/00—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K16/00—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies
- C07K16/18—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans
- C07K16/28—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans against receptors, cell surface antigens or cell surface determinants
- C07K16/2866—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans against receptors, cell surface antigens or cell surface determinants against receptors for cytokines, lymphokines, interferons
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K16/00—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies
- C07K16/18—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans
- C07K16/36—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans against blood coagulation factors
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K16/00—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies
- C07K16/40—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against enzymes
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K16/00—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies
- C07K16/42—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against immunoglobulins
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/62—DNA sequences coding for fusion proteins
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/20—Immunoglobulins specific features characterized by taxonomic origin
- C07K2317/24—Immunoglobulins specific features characterized by taxonomic origin containing regions, domains or residues from different species, e.g. chimeric, humanized or veneered
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/30—Immunoglobulins specific features characterized by aspects of specificity or valency
- C07K2317/31—Immunoglobulins specific features characterized by aspects of specificity or valency multispecific
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/50—Immunoglobulins specific features characterized by immunoglobulin fragments
- C07K2317/52—Constant or Fc region; Isotype
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/50—Immunoglobulins specific features characterized by immunoglobulin fragments
- C07K2317/52—Constant or Fc region; Isotype
- C07K2317/526—CH3 domain
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/50—Immunoglobulins specific features characterized by immunoglobulin fragments
- C07K2317/56—Immunoglobulins specific features characterized by immunoglobulin fragments variable (Fv) region, i.e. VH and/or VL
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/50—Immunoglobulins specific features characterized by immunoglobulin fragments
- C07K2317/56—Immunoglobulins specific features characterized by immunoglobulin fragments variable (Fv) region, i.e. VH and/or VL
- C07K2317/565—Complementarity determining region [CDR]
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/50—Immunoglobulins specific features characterized by immunoglobulin fragments
- C07K2317/56—Immunoglobulins specific features characterized by immunoglobulin fragments variable (Fv) region, i.e. VH and/or VL
- C07K2317/567—Framework region [FR]
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/70—Immunoglobulins specific features characterized by effect upon binding to a cell or to an antigen
- C07K2317/72—Increased effector function due to an Fc-modification
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/70—Immunoglobulins specific features characterized by effect upon binding to a cell or to an antigen
- C07K2317/76—Antagonist effect on antigen, e.g. neutralization or inhibition of binding
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/90—Immunoglobulins specific features characterized by (pharmaco)kinetic aspects or by stability of the immunoglobulin
- C07K2317/92—Affinity (KD), association rate (Ka), dissociation rate (Kd) or EC50 value
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2317/00—Immunoglobulins specific features
- C07K2317/90—Immunoglobulins specific features characterized by (pharmaco)kinetic aspects or by stability of the immunoglobulin
- C07K2317/94—Stability, e.g. half-life, pH, temperature or enzyme-resistance
Definitions
- the disclosure provides anti-IL-8 antibodies, pharmaceutical compositions containing the antibodies, nucleic acids encoding the antibodies, and host cells containing the nucleic acids. Production methods and uses of the IL-8 antibodies and pharmaceutical composition in the treatment of for example, IL-8-associated disorders, are also provided.
- Antibodies attract attention as pharmaceuticals because they are highly stable in plasma and have few side effects.
- a number of IgG-type therapeutic antibodies are on the market, and even now many therapeutic antibodies are under development (Reichert et al., Nat. Biotechnol. 23:1073-1078 (2005)(NPL1); Pavlou et al., Eur. J. Pharm. Biopharm. 59(3):389-396 (2005)(NPL2)).
- various techniques are being developed for second-generation therapeutic antibodies; including technologies for improving effector function, antigen-binding ability, pharmacokinetics or stability, and reducing the risk of immunogenicity (Kim et al., Mol. Cells. 20 (1):17-29 (2005)(NPL3)).
- the dosage for therapeutic antibodies is generally very high, and consequently the development of therapeutic antibodies confronts issues such as difficulty in producing subcutaneous formulations and high production costs.
- Methods for improving therapeutic antibody pharmacokinetics, pharmacodynamics, and antigen binding properties provide ways to reduce the dosage and production costs associated with therapeutic antibodies.
- the substitution of amino acid residues in the constant region provides one method for improving antibody pharmacokinetics (Hinton et al., J. Immunol. 176 (1):346-356 (2006)(NPL4); Ghetie et al., Nat. Biotechnol. 15(7):637-640 (1997))(NPL5).
- the technique of affinity maturation provides a method for enhancing antigen-neutralizing ability of an antibody (Rajpal et al., Proc. Natl. Acad. Sci. USA 102(24):8466-8471 (2005)(NPL6); Wu et al., J. Mol. Biol.
- NPL7 may increase the antigen-binding activity by introducing mutation(s) into amino acid residue(s) in the CDRs and/or framework regions of an antibody variable domain. Improving the antigen-binding properties of an antibody may improve the biological activity of the antibody in vitro or reduce the dosage, and may further improve the efficacy in vivo (in the body) (Wu et al., J. Mol. Biol. 368:652-665 (2007)(NPL8)).
- the amount of antigen that can be neutralized by one antibody molecule depends on the affinity of the antibody for the antigen; and thus, it is possible to neutralize an antigen with a small amount of antibody by increasing affinity.
- Antibody affinity for an antigen may routinely be increased using various known methods (see, e.g., Rajpal et al., Proc. Natl. Acad. Sci. USA 102(24):8466-8471 (2005)(NPL6)). Further, it is theoretically possible to neutralize one antigen molecule (2 antigens when an antibody is bivalent) with one antibody molecule, if it can bind covalently to the antigen to make the affinity infinite.
- one limitation for therapeutic antibody development thus far is that one antibody molecule typically only binds to and neutralizes one antigen molecule (2 antigens when an antibody is bivalent).
- an antibody that binds to an antigen in a pH-dependent manner herein below also referred to as “pH-dependent antibody” or “pH-dependent-binding antibody”
- pH-dependent antibody a pH-dependent manner
- pH-dependent-binding antibody enables one antibody molecule to bind to and neutralize multiple antigen molecules
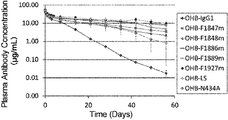
- a pH-dependent antibody binds to an antigen strongly under the neutral pH conditions in the plasma, and dissociates from the antigen under the acidic pH condition within the endosome of a cell. After dissociation from the antigen, the antibody is recycled to the plasma by FcRn and is then free to bind to and neutralize another antigen molecule; and thus one pH-dependent antibody may repeatedly bind to and neutralize multiple antigen molecules.
- IgG antibodies By binding to FcRn, IgG antibodies have long retention in plasma.
- the binding between an IgG antibody and FcRn is strong under an acidic pH conditions (for example, pH 5.8), but there is almost no binding under a neutral pH condition (for example, pH 7.4).
- An IgG antibody is taken up into cells non-specifically, and returned to cell surface by binding to FcRn in the endosome under the acidic pH conditions in the endosome. The IgG then dissociates from the FcRn under the neutral pH conditions in the plasma.
- a pH-dependent antibody that has been modified to increase its FcRn binding under neutral pH conditions has the ability to repeatedly bind to and eliminate antigen molecules from plasma; and thus administration of such an antibody allows antigen elimination from plasma (WO2011/122011(PTL3)).
- a pH-dependent antibody that has been modified to increase its FcRn binding under neutral pH conditions can further accelerate the elimination of the antigen compared to a pH-dependent antibody that comprises the Fc region of a native IgG antibody (WO2011/122011(PTL3)).
- an increase in the binding against an anti-drug antibody (herein below also referred to as "Pre-existing ADA") (for example, rheumatoid factor) present in a patient before administration of a therapeutic antibody has been further reported (WO2013/046722(PTL4), WO2013/046704(PTL5)).
- WO2013/046704(PTL5) reports that an Fc region variant containing specific mutations (represented by two residue modifications of Q438R/S440E according to EU numbering) increase the binding to FcRn under acidic pH conditions and also showed a significant reduction in binding to rheumatoid factor compared to unmodified native Fc.
- WO2013/046704(PTL5) does not specifically demonstrate that this Fc region variant has superior plasma retention to an antibody with native Fc region.
- ADCC antibody-dependent cellular cytotoxicity
- CDC complement-dependent cytotoxicity
- ADCP antibody-dependent cellular phagocytosis
- Fc ⁇ RIa, Fc ⁇ RIIa, Fc ⁇ RIIb, Fc ⁇ RIIIa and Fc ⁇ RIIIb isoforms are reported as Fc ⁇ R family proteins, and their respective allotypes have also been reported (Jefferis et al., Immunol. Lett. 82:57-65 (2002)(NPL13)).
- the balance of the respective affinity of an antibody for an activating receptor comprising Fc ⁇ RIa, Fc ⁇ RIIa, Fc ⁇ RIIIa or Fc ⁇ RIIIb, and an inhibitory receptor comprising Fc ⁇ RIIb is an important element in optimizing the antibody effector functions.
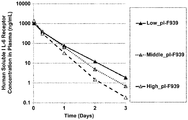
- a soluble antigen may display increased plasma retention and plasma concentration by binding to an antibody that has such a recycling mechanism (for example, an antibody that does not have the characteristics of a pH/Ca concentration-dependent antibody). Accordingly, for example, when a soluble antigen in plasma has multiple types of physiological functions, even if one type of physiological functions is blocked as a result of antibody binding, the plasma concentration of the antigen may worsen the pathogenic symptoms caused by the other physiological functions as a result of the increased plasma retention and/or plasma concentration of the antigen resulting from the antibody binding.
- the extracellular matrix is a structure that covers cells in vivo, and is mainly constituted by glycoproteins such as collagen, proteoglycan, fibronectin, and laminin.
- the role of the ECM in vivo is to create a microenvironment for cells to survive, and the ECM is important in various functions carried out by cells such as, cell proliferation and cell adhesion.
- the ECM has been reported to be involved in the in vivo kinetics of proteins administered to a living body.
- Blood concentration of the VEGF-Trap molecule which is a fusion protein between the VEGF receptor and Fc, when subcutaneously administered was examined (Holash et al., Proc. Natl. Acad. Sci., 99(17):11393-11398 (2002)(NPL21)).
- a modified VEGF-Trap molecule whose pI was reduced by amino acid substitutions has a higher plasma concentration, and its bioavailability could be improved.
- WO2012/093704(PTL18) reports that there is an inverse correlation between antibody binding to the ECM and plasma retention, and consequently, antibody molecules that do not bind to the ECM have better plasma retention when compared to antibodies that bind to the ECM.
- Human IL-8 (Interleukin 8) is a chemokine family member that is 72 or 77 amino acid residues in length.
- the term "chemokine” is a collective term for a family of proteins with a molecular weight of 8-12 kDa and contain 4 cysteine residues that form intermolecular disulfide bonds.
- Chemokines are categorized into CC chemokine, CXC chemokine, C chemokine, CA3C chemokine according to the characteristics of the cysteine arrangement.
- IL-8 is classified as a CXC chemokine, and is also referred to as CXCL8.
- IL-8 exists in solution in monomeric and homodimeric form.
- the IL-8 monomer contains antiparallel ⁇ sheets, and has a structure in which a C-terminal ⁇ helix traverses and covers the ⁇ sheets.
- An IL-8 monomer in the case of the 72 amino acid form of IL-8, comprises two disulfide crosslinks between cysteine 7 and cysteine 34, and between cysteine 9 and cysteine 50.
- IL-8 homodimers are stabilized by noncovalent interactions between the ⁇ sheets of the two monomers, as there is no covalent binding between molecules in homodimers.
- IL-8 expression is induced in various cells such as peripheral blood monocytes, tissue macrophages, NK cells, fibroblasts, and vascular endothelial cells in response to stimulation by inflammatory cytokines (Russo et al., Exp. Rev. Clin. Immunol. 10(5):593-619 (2014)(NPL22)).
- Chemokines are generally not detectable, or only weakly detectable, in normal tissue, but are strongly detected at inflamed sites, and are involved in eliciting inflammation by facilitating infiltration of leukocyte into inflamed tissue sites.
- IL-8 is a proinflammatory chemokine that is known to activate neutrophils, promote expression of cell adhesion molecules, and enhance neutrophil adhesion to vascular endothelial cells.
- IL-8 also has neutrophil chemotactic capacity and IL-8 produced at a damaged tissue facilitates chemotaxis of neutrophils adhered to vascular endothelial cells into the tissue, and induces inflammation along with neutrophil infiltration.
- IL-8 is also known to be a potent angiogenic factor for endothelial cells and is involved in promoting tumor angiogenesis.
- Inflammatory diseases associated with elevated (e.g., excess) IL-8 levels include, inflammatory diseases of the skin such as inflammatory keratosis (e.g., psoriasis), atopic dermatitis, contact dermatitis; chronic inflammatory disorders which are autoimmune diseases, such as rheumatoid arthritis, systemic lupus erythematosus (SLE), and Behcet's disease; inflammatory bowel diseases such as Crohn's disease and ulcerative colitis; inflammatory liver diseases such as hepatitis B, hepatitis C, alcoholic hepatitis, drug-induced allergic hepatitis; inflammatory renal diseases such as glomerulonephritis; inflammatory respiratory diseases such as bronchitis and asthma; inflammatory chronic vascular diseases such as atherosclerosis; multiple sclerosis, oral ulcer, chorditis, and inflammation associated with using artificial organs and/or artificial blood vessels.
- inflammatory keratosis e.g.,
- Elevated (e.g., excess) IL-8 levels are also associated with malignant tumors such as ovarian cancer, lung cancer, prostate cancer, stomach cancer, breast cancer, melanoma, head and neck cancers, and kidney cancer; sepsis due to infection; cystic fibrosis; and pulmonary fibrosis.
- malignant tumors such as ovarian cancer, lung cancer, prostate cancer, stomach cancer, breast cancer, melanoma, head and neck cancers, and kidney cancer
- sepsis due to infection cystic fibrosis
- pulmonary fibrosis See, e.g., Russo et al., Exp. Rev. Clin. Immunol. 10(5):593-619 (2014)(NPL22), which is herein incorporated by reference in its entirety).
- a non-limited objective of embodiments of Disclosure A is to provide molecules with improved pharmacokinetic properties over antibodies, such as ion concentration-dependent antigen binding properties that improve antibody half-life and/or antigen clearance from the plasma.
- a non-limited objective of embodiments of Disclosure B is to provide, safe and more favorable Fc region variants that have increased half-life and decreased binding to pre-existing anti-drug antibodies (ADAs).
- ADAs anti-drug antibodies
- a non-limited objective of embodiments of Disclosure C is to provide anti-IL-8 antibodies that have pH-dependent binding affinity towards IL-8.
- An additional embodiment relates to anti-IL-8 antibodies that have an effect of rapidly eliminating IL-8 compared to a reference antibody when administered to an individual.
- Disclosure C relates to anti-IL-8 antibodies that can stably maintain their IL-8-neutralizing activity when administered to an individual.
- the anti-IL-8 antibodies display reduced immunogenicity.
- Disclosure C relates to a method of producing and using the above-mentioned anti-IL-8 antibodies.
- Another alternative non-limited objective of Disclosure C is, to provide novel anti-IL-8 antibodies that can be included in a pharmaceutical composition.
- an ion concentration-dependent antibody which is an antibody comprising an ion concentration-dependent antigen-binding domain ("an antigen-binding domain whose antigen-binding activity changes according to ion concentration conditions")
- an ion concentration-dependent antibody which is an antibody comprising an ion concentration-dependent antigen-binding domain ("an antigen-binding domain whose antigen-binding activity changes according to ion concentration conditions")
- an ion concentration-dependent antibody with increased pI can further increase the extracellular matrix-binding of the antibody.
- antigen elimination from plasma can be increased, by increasing the binding of the antibody towards extracellular matrix.
- the inventors conducted dedicated research on safe and more favorable Fc region variants that do not show binding to anti-drug antibodies (pre-existing ADA) and that can further improve plasma retention.
- Fc region variants comprising a substitution of position 434 amino acid according to EU numbering with Ala (A) and two specific residue mutations (represented by Q438R/S440E according to EU numbering) as a combination of amino acid residue mutations, are preferred for maintaining significant reduction in the binding to rheumatoid factor, along with achieving a plasma retention of an antibody.
- the inventors generated a number of pH-dependent anti-IL-8 antibodies (anti-IL-8 antibodies that bind to IL-8 in a pH-dependent manner). From the results of various validations, the inventors identified pH-dependent anti-IL-8 antibodies that have an effect of rapidly eliminating IL-8 compared to a reference antibody when administered to an individual. In some embodiments the Disclosure C relates to pH-dependent anti-IL-8 antibodies that can stably maintain their IL-8-neutralizing activity. In additional non-limiting embodiments, the pH-dependent anti-IL-8 antibodies have reduced immunogenicity and excellent expression levels.
- the inventors successfully obtained anti-IL-8 antibodies that comprise an Fc region whose FcRn-binding affinity at acidic pH is increased relative to the FcRn-binding affinity of a native Fc region.
- the inventors successfully obtained anti-IL-8 antibodies that comprise an Fc region whose binding affinity towards pre-existing ADA is reduced relative to the binding affinity of a native Fc region for the pre-existing ADA.
- the inventors successfully obtained anti-IL-8 antibodies comprising an Fc region whose plasma half-life is increased relative to the plasma half-life of a native Fc region.
- the inventors successfully obtained pH-dependent anti-IL-8 antibodies that comprise an Fc region whose binding affinity towards effector receptors is reduced relative to the binding affinity of a native Fc region for the effector receptors.
- the inventors identified nucleic acids encoding the above-mentioned anti-IL-8 antibodies.
- the inventors also obtained hosts comprising the above-mentioned nucleic acids.
- the inventors developed a method for producing the above-mentioned anti-IL-8 antibodies, which comprises culturing the above-mentioned host.
- the inventors developed a method for facilitating the elimination of IL-8 from an individual relative to a reference antibody, which comprises administering the above-mentioned anti-IL-8 antibodies to the individual.
- Disclosure A relates without limitation to, [1] an antibody comprising an antigen-binding domain whose antigen-binding activity changes according to ion concentration conditions, wherein its isoelectric point (pI) is increased by the modification of at least one amino acid residue that may be exposed on the surface of the antibody; [2] the antibody of [1], wherein the antigen is a soluble antigen; [3] the antibody of [1] or [2], wherein the antigen-binding domain is a domain whose antigen-binding activity under a high ion concentration condition is higher than that under a low ion concentration condition; [4] the antibody of any one of [1] to [3], wherein the ion concentration is a hydrogen ion concentration (pH) or a calcium ion concentration; [5] the antibody of [4], wherein the ratio of its KD in an acidic pH range to that in a neutral pH range, KD (acidic pH range) / KD (neutral pH range), for the antigen
- the method of any one of [30], or [30A] to [30C] which further optionally comprises any one or more of: (a) selecting an antibody which can promote elimination of an antigen from plasma; (b) selecting an antibody with enhanced binding activity to an extracellular matrix; (c) selecting an antibody with enhanced Fc ⁇ R-binding activity under a neutral pH condition (e.g., pH 7.4); (d) selecting an antibody with enhanced Fc ⁇ RIIb-binding activity under a neutral pH condition (e.g., pH 7.4); (e) selecting an antibody with maintained or enhanced Fc ⁇ RIIb-binding activity and decreased binding activity to one or more activating Fc ⁇ R, preferably selected from the group consisting of Fc ⁇ RIa, Fc ⁇ RIb, Fc ⁇ RIc, Fc ⁇ RIIIa, Fc ⁇ RIIIb and Fc ⁇ RIIa; (f) selecting an antibody with enhanced FcRn-binding activity under a neutral pH condition (e.g., pH 7.4); (g) selecting an antibody with an antibody with
- Disclosure A relates without limitation to: [A1] an antibody having a constant region, wherein at least one amino acid residue selected from the group of modification sites identical to the modification sites in the group defined in [15] or [16] is modified in the constant region; [A2] the antibody of [A1], which further has a heavy-chain variable region and/or a light-chain variable region, wherein the variable region has CDR(s) and/or FR(s), and wherein at least one amino acid residue selected from the group of modification sites identical to the modification sites in the group defined in [13] or [14] is modified in a CDR and/or a FR; [A3] an antibody having a constant region, wherein at least one amino acid residue selected from the group of modification sites identical to the modification sites in the group defined in [15] or [16] is modified in the constant region so as to increase its pI; [A4] the antibody of [A3], which further has a heavy-chain variable region and/or a light-chain variable region, where
- Disclosure A encompasses combinations of one or multiple elements described in any of [1] to [30], [30A], [30B], [31], [32] and [A1] to [A11] mentioned above, in part or as a whole, as long as such a combination is not technically inconsistent with the common technical knowledge in the art.
- Disclosure A encompasses a method for producing a modified antibody comprising an antigen-binding domain which promotes elimination of an antigen from plasma as compared to that before the antibody modification, wherein the method comprises: (a) modifying at least one amino acid residue that may be exposed on the surface of an antibody, which is: (I) in a position in a CDR or FR selected from the group consisting of: (a) position 1, 3, 5, 8, 10, 12, 13, 15, 16, 18, 19, 23, 25, 26, 39, 41, 42, 43, 44, 46, 68, 71, 72, 73, 75, 76, 77, 81, 82, 82a, 82b, 83, 84, 85, 86, 105, 108, 110, and 112 in a FR of the heavy chain variable region; (b) position 31, 61, 62, 63, 64, 65, and 97 in a CDR of the heavy chain variable region; (c) position 1, 3, 7, 8, 9, 11, 12, 16, 17, 18, 20, 22, 37, 38,
- the method optionally further comprises any one or more of: (e) selecting an antibody which can promote elimination of an antigen from plasma; (f) selecting an antibody with enhanced binding activity to an extracellular matrix; (g) selecting an antibody with enhanced Fc ⁇ R-binding activity under a neutral pH condition (e.g., pH 7.4); (h) selecting an antibody with enhanced Fc ⁇ RIIb-binding activity under a neutral pH condition (e.g., pH 7.4); (i) selecting an antibody with maintained or enhanced Fc ⁇ RIIb-binding activity and decreased binding activity to one or more activating Fc ⁇ R, preferably selected from the group consisting of Fc ⁇ RIa, Fc ⁇ RIb, Fc ⁇ RIc, Fc ⁇ RIIIa, Fc ⁇ RIIIb and Fc ⁇ RIIa; (j) selecting an antibody with enhanced FcRn-binding activity under a neutral pH condition (e.g., pH 7.4); (k) selecting an antibody with an increased isoelectric point (pI); (l) confirming the is
- Another embodiment of Disclosure A relates to, for example, without limitation: [D1] a method for producing a modified antibody, whose half-life in plasma is prolonged or reduced, as compared to that before the modification of the antibody, wherein the method comprises: (a) modifying a nucleic acid encoding the antibody before the modification to change the charge of at least one amino acid residue at a position selected from the group consisting of position 196, 253, 254, 256, 258, 278, 280, 281, 282, 285, 286, 307, 309, 311, 315, 327, 330, 342, 343, 345, 356, 358, 359, 361, 362, 373, 382, 384, 385, 386, 387, 389, 399, 400, 401, 402, 413, 415, 418, 419, 421, 424, 430, 433, 434, and 443, according to EU numbering; (b) culturing a host cell to express the nucleic acid; and (c) collecting the antibody from the
- Disclosure B relates to, for example, without limitation: [33] an Fc region variant comprising an FcRn-binding domain, wherein the FcRn-binding domain comprises Ala at position 434; Glu, Arg, Ser, or Lys at position 438; and Glu, Asp, or Gln at position 440, according to EU numbering; [34] the Fc region variant of [33], wherein the FcRn-binding domain comprises Ala at position 434; Arg or Lys at position 438; and Glu or Asp at position 440, according to EU numbering; [35] the Fc region variant of [33] or [34], wherein the FcRn-binding domain further comprises Ile or Leu at position 428; and/or Ile, Leu, Val, Thr, or Phe at position 436, according to EU numbering; [36] the Fc region variant of [35], wherein the FcRn-binding domain comprises Leu at position 428; and/or Val or
- Disclosure B relates to, for example, without limitation: [B1] use of the Fc region variant of any one of [33] to [43] or the antibody of [44] or [45] in the manufacture of a medicament for increasing retention in plasma; [B2] use of the Fc region variant of any one of [33] to [43] or the antibody of [44] or [45] in the manufacture of a medicament for not significantly increasing the binding activity for an anti-drug antibody (ADA) under a neutral pH condition compared to the Fc region of a native IgG; [B3] use of the Fc region variant of any one of [33] to [43] or the antibody of [44] or [45] for increasing retention in plasma; [B4] use of the Fc region variant of any one of [33] to [43] or the antibody of [44] or [45] for not significantly increasing the binding activity for an anti-drug antibody (ADA) under a neutral pH condition compared to the Fc region of a native IgG; and [B5]
- Disclosure B encompasses combinations of one or multiple elements described in any of [33] to [53] and [B1] to [B5] mentioned above, in part or as a whole, as long as such a combination is not technically inconsistent with the common technical knowledge in the art.
- Disclosure B encompasses an Fc region variant comprising an FcRn-binding domain, wherein the FcRn-binding domain can comprise: (a) Ala at position 434; Glu, Arg, Ser, or Lys at position 438; and Glu, Asp, or Gln at position 440, according to EU numbering; (b) Ala at position 434; Arg or Lys at position 438; and Glu or Asp at position 440, according to EU numbering; (c) Ile or Leu at position 428; Ala at position 434; Ile, Leu, Val, Thr, or Phe at position 436; Glu, Arg, Ser, or Lys at position 438; and Glu, Asp, or Gln at position 440, according to EU numbering; (d) Ile or Leu at position 428; Ala at position 434; Ile, Leu, Val, Thr, or Phe at position 436; Arg or Lys at position 438;
- Disclosure C relates to, for example, without limitation: [54] an isolated anti-IL-8 antibody that binds to human IL-8, which comprises at least one amino acid substitution(s) in at least one of (a) to (f) below, and binds to IL-8 in a pH-dependent manner: (a) HVR-H1 comprising the amino acid sequence of SEQ ID NO:67; (b) HVR-H2 comprising the amino acid sequence of SEQ ID NO:68; (c) HVR-H3 comprising the amino acid sequence of SEQ ID NO:69; (d) HVR-L1 comprising the amino acid sequence of SEQ ID NO:70; (e) HVR-L2 comprising the amino acid sequence of SEQ ID NO:71; and (f) HVR-L3 comprising the amino acid sequence of SEQ ID NO:72; [55] the anti-IL-8 antibody of [54], which comprises an amino acid substitutions of tyrosine at position 9 of the amino acid sequence of SEQ ID NO:68, argin
- the anti-IL-8 antibody of [61] wherein the Fc region comprises amino acid substitution(s) at one or more positions selected from the group consisting of position 235, 236, 239, 327, 330, 331, 428, 434, 436, 438 and 440, according to EU numbering;
- the anti-IL-8 antibody of [62] which comprises an Fc region comprising one or more amino acid substitutions selected from the group consisting of L235R, G236R, S239K, A327G, A330S, P331S, M428L, N434A, Y436T, Q438R and S440E;
- the anti-IL-8 antibody of [63] wherein the Fc region comprises the amino acid substitutions of L235R, G236R, S239K, M428L, N434A, Y436T, Q438R and S440E;
- the anti-IL-8 antibody of [63] wherein the Fc region comprises the amino acid substitution of L235R, G2
- Disclosure C relates to: [C1] use of the anti-IL-8 antibody of any one of [54] to [67] and [C26] to [C31] in the manufacture of a pharmaceutical composition for suppressing accumulation of IL-8 which has a biological activity; [C2] use of the anti-IL-8 antibody of any one of [54] to [67] and [C26] to [C31] for suppressing accumulation of IL-8 which has a biological activity; [C3] use of the anti-IL-8 antibody of any one of [54] to [67] and [C26] to [C31] in the manufacture of a pharmaceutical composition for inhibiting angiogenesis; [C4] use of the anti-IL-8 antibody of any one of [54] to [67] and [C26] to [C31] for inhibiting angiogenesis; [C5] use of the anti-IL-8 antibody of any one of [54] to [67] and [C26] to [C31] in the manufacture of a pharmaceutical composition for inhibiting facilitation of neutrophil migration; [C1] use
- An anti-IL-8 antibody which comprises an Fc region comprising amino acid substitution(s) at one or more positions selected from the group consisting of position 235, 236, 239, 327, 330, 331, 428, 434, 436, 438 and 440, according to EU numbering.
- the anti-IL-8 antibody of [C26] which comprises an Fc region having at least one property selected from the properties of (a) to (f) below: (a) increased binding affinity for FcRn of the Fc region relative to the binding affinity for FcRn of a native Fc region at acidic pH; (b) reduced binding affinity of the Fc region for pre-existing ADA relative to the binding affinity of a native Fc region for the pre-existing ADA; (c) increased plasma half-life of the Fc region relative to the plasma half-life of a native Fc region; (d) reduced plasma clearance of the Fc region relative to the plasma clearance of a native Fc region; (e) reduced binding affinity of the Fc region for an effector receptor relative to the binding affinity of a native Fc region for the effector receptor; and (f) increased binding to extracellular matrix.
- [C28] The anti-IL-8 antibody of [C26] or [C27], which comprises an Fc region comprising one or more amino acid substitution(s) selected from the group consisting of L235R, G236R, S239K, A327G, A330S, P331S, M428L, N434A, Y436T, Q438R and S440E, according to EU numbering.
- the anti-IL-8 antibody of [C28] which comprises an Fc region comprising one or more amino acid substitutions selected from the group consisting of (a) L235R, G236R, S239K, M428L, N434A, Y436T, Q438R and S440E; or (b) L235R, G236R, A327G, A330S, P331S, M428L, N434A, Y436T, Q438R and S440E, according to EU numbering.
- the anti-IL-8 antibody of [C26] that comprises a heavy chain comprising the amino acid sequence of SEQ ID NO:81 and a light chain comprising the amino acid sequence of SEQ ID NO:82.
- [C37] A pharmaceutical composition comprising the anti-IL-8 antibody of any one of [C26] to [C31] and a pharmaceutically acceptable carrier.
- [C38] A method for treating a patient that has a disorder with the presence of excess IL-8, which comprises administering the anti-IL-8 antibody of any one of [C26] to [C31] to the individual.
- [C39] A method for promoting elimination of IL-8 from an individual, which comprises administering the anti-IL-8 antibody of any one of [C26] to [C31] to the individual.
- [C40] A method for inhibiting IL-8, wherein the method comprises contacting the anti-IL 8 antibody of any one of [54] to [67] and [C26] to [C31] with IL-8.
- [C41] The method of [C40], wherein the method inhibits a biological activity of IL-8.
- Disclosure C encompasses combinations of one or multiple elements described in any of [54] to [80] and [C1] to [C41] mentioned above, in part or as a whole, as long as such a combination is not technically inconsistent with the common technical knowledge in the art.
- Fig. 1 shows changes in the plasma concentration of human IL-6 receptor in human FcRn transgenic mice administered with a human IL-6 receptor-binding antibody that binds to human IL-6 receptor in a pH-dependent manner and whose constant region is that of a native IgG1 (Low_pI-IgG1), or an antibody that has increased the pI of the variable region in the antibody (High_pI-IgG1).
- Fig. 2 shows changes in the plasma concentration of human IL-6 receptor in human FcRn transgenic mice administered individually with a human IL-6 receptor-binding antibody that binds to human IL-6 receptor in a pH-dependent manner and has been conferred with binding to FcRn under a neutral pH condition (Low_pI-F939), and antibodies that have increased the pI of the variable region in the antibody (Middle_pI-F939, High_pI-F939).
- Fig. 3 shows changes in the plasma concentration of human IL-6 receptor in human FcRn transgenic mice administered individually with a human IL-6 receptor-binding antibody that binds to human IL-6 receptor in a pH-dependent manner and whose Fc ⁇ R binding under a neutral pH condition is increased (Low_pI-F1180), and antibodies that have increased the pI of the variable region in the antibody (Middle_pI-F1180, High_pI-F1180).
- Fig. 4 shows changes in the plasma concentration of human IL-6 receptor in human FcRn transgenic mice whose soluble human IL-6 receptor concentration in plasma is maintained at a steady state, which have been administered individually with a human IL-6 receptor-binding antibody that binds to human IL-6 receptor in a pH-dependent manner and whose constant region is that of a native IgG1 (Low_pI-IgG1), an antibody that comprises an Fc region variant in which the Fc region in the antibody has increased FcRn binding under a neutral pH condition (Low_pI-F11), and antibodies that have increased the pI of the variable region in these antibodies (High_pI-IgG1, High_pI-F11).
- Fig. 5 shows the extent of extracellular matrix binding of each of the three types of antibodies with different pIs that bind to human IL-6 receptor in a pH-dependent manner (Low_pI-IgG1, Middle_pI-IgG1 and High_pI-IgG1) and the two types of antibodies with different pIs that do not bind to human IL-6 receptor in a pH-dependent manner (Low_pI(NPH)-IgG1 and High_pI(NPH)-IgG1).
- NPH means pH independent within the scope of Disclosure A described herein.
- Fig. 6 shows relative values of the extent of soluble human Fc ⁇ RIIb binding (measured by BIACORE(registered trademark)) of antibodies that comprise an Fc region variant each of whose pI has been increased by modifying one amino acid residue in the constant region of the Ab1H-P600 antibody which binds to IgE in a pH-dependent manner, by setting the value of Ab1H-P600 to 1.00.
- Fig. 7 shows relative values of the rate at which antibodies that comprise an Fc region variant each of whose pI has been increased by modifying one amino acid residue in the constant region of Ab1H-P600 are taken up into cells of an hFc ⁇ RIIb-expressing cell line, respectively, evaluated with the value of Ab1H-P600 set to 1.00.
- Fig. 8 shows the extent of binding of Fv4-IgG1, which has the Fc region of a native human IgG1, to rheumatoid factor in the serum of each RA patient.
- Fig. 9 shows the extent of binding of Fv4-YTE, which has an Fc region variant with increased FcRn binding, to rheumatoid factor in the serum of each RA patient.
- Fig. 10 shows the extent of binding of Fv4-LS, which has an Fc region variant with increased FcRn binding, to rheumatoid factor in the serum of each RA patient.
- Fig. 11 shows the extent of binding of Fv4-N434H, which has an Fc region variant with increased FcRn binding, to rheumatoid factor in the serum of each RA patient.
- Fig. 12 shows the extent of binding of Fv4-F1847m, which has an Fc region variant with increased FcRn binding, to rheumatoid factor in the serum of each RA patient.
- Fig. 13 shows the extent of binding of Fv4-F1848m, which has an Fc region variant with increased FcRn binding, to rheumatoid factor in the serum of each RA patient.
- Fig. 14 shows the extent of binding of Fv4-F1886m, which has an Fc region variant with increased FcRn binding, to rheumatoid factor in the serum of each RA patient.
- Fig. 15 shows the extent of binding of Fv4-F1889m, which has an Fc region variant with increased FcRn binding, to rheumatoid factor in the serum of each RA patient.
- Fig. 16 shows the extent of binding of Fv4-F1927m, which has an Fc region variant with increased FcRn binding, to rheumatoid factor in the serum of each RA patient.
- Fig. 17 shows the extent of binding of Fv4-F1168m, which has an Fc region variant with increased FcRn binding, to rheumatoid factor in the serum of each RA patient.
- Fig. 18 shows average values of the binding of Fv4-IgG1, which has the Fc region of a native human IgG1, and each of the antibodies comprising a novel Fc region variant in which the Fc region has an Fc region variant with increased binding to each FcRn, to rheumatoid factor in the serum of RA patients.
- Fig. 19 shows changes in the plasma concentration of each anti-human IgE antibody in cynomolgus when administered with OHB-IgG1 which is an anti-human IgE antibody and has the Fc region of a native human IgG1, and each of the antibodies comprising a novel Fc region variant in which each the Fc region has an Fc region variant with increased binding to FcRn (OHB-LS, OHB-N434A, OHB-F1847m, OHB-F1848m, OHB-F1886m, OHB-F1889m and OHB-F1927m).
- Fig. 20 shows changes in the plasma concentration of an anti-human IL-6 receptor antibody in human FcRn transgenic mouse when administered with Fv4-IgG1 which is an anti-human IL-6 receptor antibody and has the Fc region of a native human IgG1, or Fv4-F1718 which has increased FcRn binding of the antibody at the acidic pH condition.
- Fig. 21 shows sensorgrams obtained for IL-8 binding of H998/L63 and Hr9 at pH 7.4 and pH 5.8 measured with Biacore.
- Fig. 22 shows changes of the human IL-8 concentration in mouse plasma when H998/L63 or H89/L118 was administered to mice at 2 mg/kg in a mixture with human IL-8.
- Fig. 23 shows changes of the human IL-8 concentration in mouse plasma when H89/L118 was administered to mice at 2 mg/kg or 8 mg/kg in a mixture with human IL-8.
- Fig. 24 shows changes of the human IL-8 concentration in mouse plasma when H89/L118 or H553/L118 was administered to mice at 2 mg/kg or 8 mg/kg in a mixture with human IL-8.
- Fig. 25A shows changes in the relative values of antibody concentration-dependent chemiluminescence with antibody Hr9, H89/L118 or H553/L118 before preservation in plasma.
- Fig. 25B shows changes in the relative values of antibody concentration-dependent chemiluminescence with antibody Hr9, H89/L118 or H553/L118 after one week of preservation in plasma.
- Fig. 25C shows changes in the relative values of antibody concentration-dependent chemiluminescence with antibody Hr9, H89/L118 or H553/L118 after two weeks of preservation in plasma.
- Fig. 26 shows the predicted frequency of ADA occurrence for each anti-IL-8 antibody (hWS4, Hr9, H89/L118, H496/L118 or H553/L118) and the predicted frequency of ADA occurrence for other pre-existing therapeutic antibodies predicted by the EpiMatrix.
- Fig. 27 shows the predicted frequency of ADA occurrence for each anti-IL-8 antibody (H496/L118, H496v1/L118, H496v2/L118, H496v3/L118, H1004/L118 or H1004/L395) and the predicted frequency of ADA occurrence for other pre-existing therapeutic antibodies predicted by EpiMatrix.
- Fig. 28A shows changes in the relative values of antibody concentration-dependent chemiluminescence with antibody Hr9, H89/L118 or H1009/L395-F1886s before preservation in plasma.
- Fig. 28B shows changes in the relative values of antibody concentration-dependent chemiluminescence with antibody Hr9, H89/L118 or H1009/L395-F1886s after one week of preservation in plasma.
- Fig. 28C shows changes in the relative values of antibody concentration-dependent chemiluminescence with antibody Hr9, H89/L118 or H1009/L395-F1886s after two weeks of preservation in plasma.
- Fig. 29 shows changes of the human IL-8 concentration in mouse plasma when mice were administered with each of H1009/L395, H553/L118 and H998/L63 in a mixture with human IL-8.
- Fig. 30 shows the extent of extracellular matrix binding when Hr9, H89/L118 or H1009/L395 was added alone to extracellular matrix, and when they were added in a mixture with human IL-8.
- Fig. 31 shows changes of antibody concentration in mouse plasma when an antibody that has the variable region of H1009/L395 and the Fc region that does not bind to FcRn (F1942m) was administered alone or in a mixture with human IL-8 to human FcRn transgenic mice.
- Fig. 32 shows the predicted frequency of ADA occurrence for H1009/L395 and H1004/L395 and the predicted frequency of ADA occurrence for other pre-existing therapeutic antibodies predicted by EpiMatrix.
- Fig. 33 shows changes in the concentration of the respective anti-human IL-8 antibody in the plasma of cynomolgus when administered with H89/L118-IgG1, which has the variable region of H89/L118 and the Fc region of a native human IgG1, and each antibody that has an Fc region variant with increased binding to FcRn (H89/L118-F1168m, H89/L118-F1847m, H89/L118-F1848m, H89/L118-F1886m, H89/L118-F1889m and H89/L118-F1927m).
- Fig. 34 shows the binding of antibodies that have the variable region of H1009/L395 and whose Fc region is a variant (F1886m, F1886s, or F1974m) to each Fc ⁇ R.
- Fig. 35 shows changes of the human IL-8 concentration in mouse plasma when an anti-IL-8 antibody was administered to human FcRn transgenic mice in a mixture with human IL-8.
- the anti-IL-8 antibody was H1009/L395-IgG1 (2 mg/kg) which comprises the variable region of H1009/L395 and the Fc region of a native human IgG1, or H1009/L395-F1886s (2, 5 or 10 mg/kg) which comprises the variable region of H1009/L395 and the modified Fc region.
- Fig. 36 shows changes in the antibody concentration in the plasma of cynomolgus when administered with Hr9-IgG1 or H89/L118-IgG1, both of which comprise the Fc region of a native human IgG1, or H1009/L395-F1886s or H1009/L395-F1974m, both of which comprise a modified Fc region.
- Fig. 37 shows the IgE plasma concentration time profile of some anti-IgE antibodies in C57BL6J mice in terms of the antibody variable region modification.
- Fig. 38 shows Octet sensorgrams of selected 25 [twenty five] pH-dependent and/or calcium-dependent antigen binding clones.
- Fig. 38B is continuation of Fig. 38A.
- Fig. 38C is continuation of Fig. 38B.
- Fig. 38D is continuation of Fig. 38C.
- Fig. 39 shows the C5 plasma concentration time profile of some anti-C5 bispecific antibodies in C57BL6J mice in terms of the antibody variable region modification.
- Fig. 40 shows the IgE plasma concentration time profile of some anti-IgE antibodies in C57BL6J mice in terms of the antibody constant region modification.
- Disclosure A relates to antibodies comprising an antigen-binding domain whose antigen-binding activity changes according to ion concentration conditions, in which the isoelectric point (pI) is increased by modification of at least one amino acid residue that may be exposed on the antibody surface (herein, also referred to as "ion concentration-dependent antibodies with increased pI" within the scope of Disclosure A; and the antigen-binding domains of the antibodies are also referred to as "ion concentration-dependent antigen-binding domains with increased pI").
- the invention is partly based on the surprising discovery of the inventors that antigen elimination from plasma can be facilitated with an ion concentration-dependent antibody whose isoelectric point (pI) has been increased by the modification of at least one amino acid residue that can be exposed on the antibody surface (for example, when the antibody is administered in vivo); and that binding of an antibody to the extracellular matrix can be increased with an ion concentration-dependent antibody with increased (elevated) pI.
- pI isoelectric point
- the invention is also partly based on the surprising discovery of the inventors that this beneficial effect is brought about by combining two entirely different concepts of: an ion concentration-dependent antigen-binding domain or ion concentration-dependent antibody; and an antibody whose pI is increased by modification of at least one amino acid residue that can be exposed on the surface (herein, also referred to as an "antibody with increased pI" within the scope of Disclosure A; and an antibody whose pI is decreased (reduced) by modification of at least one amino acid residue that can be exposed on the surface is referred to as an "antibody with decreased pI" within the scope of Disclosure A).
- the invention is thus categorized as a type of pioneer invention which can lead to remarkable technological innovation in the field (e.g., medical field) to which Disclosure A belongs.
- an antibody comprising an antigen-binding domain and whose pI is increased by modification of at least one amino acid residue that can be exposed on the antibody surface, which has been further modified so that the antigen-binding activity of the antigen-binding domain changes according to ion concentration conditions, are also included within the scope of Disclosure A described herein (herein, such antibody is also referred to as an "ion concentration-dependent antibody with increased pI" within the scope of the Disclosure A).
- an antibody containing an ion concentration-dependent antigen-binding domain in which at least one amino acid residue that can be exposed on the antibody surface has a charge different from that of the at least one amino acid residue at the corresponding position(s) in an antibody before modification (native antibody (for example, native Ig antibody, preferably native IgG antibody), or reference or parent antibody (e.g., antibody before modification, or antibody prior to or during library construction, or the like)), and whose net antibody pI is increased is also included in Disclosure A described herein (such antibody is also referred to as an "ion concentration-dependent antibody with increased pI" within the scope of Disclosure A described herein).
- an antibody containing an ion concentration-dependent antigen-binding domain, whose pI is increased by modification of at least one amino acid residue that can be exposed on the antibody surface in an antibody before the modification is also included in Disclosure A described herein (such antibody is also referred to as an "ion concentration-dependent antibody with increased pI" within the scope of Disclosure A described herein).
- an antibody containing an ion concentration-dependent antigen-binding domain in which at least one amino acid residue that can be exposed on the antibody surface is modified for the purpose of increasing the pI of the antibody is also included in Disclosure A described herein (such antibody is also referred to as an "ion concentration-dependent antibody with increased pI" within the scope of Disclosure A described herein).
- amino acids include not only natural amino acids but also unnatural amino acids.
- amino acids or amino acid residues may be represented by either one-letter (for example, A) or three-letter codes (for example, Ala), or both (for example, Ala(A)).
- modification of an amino acid may be understood as, without being limited thereto, chemically modifying one or more (for example, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more than 10) specific amino acids (residues) in an antibody amino acid sequence with a molecule or adding, deleting, substituting or inserting one or more (for example, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more than 10) amino acids in an antibody amino acid sequence.
- Amino acid addition, deletion, substitution, or insertion can be carried out to a nucleic acid encoding an amino acid sequence, for example, by site-directed mutagenesis (Kunkel et al., Proc.
- amino acid modification is carried out preferably, without limitation, by substituting one or more amino acid residue in an antibody amino acid sequence with a different amino acid (individually).
- Amino acid addition, deletion, substitution, or insertion, and modification of an amino acid sequence by humanization or chimerization can be carried out by methods known in the art.
- Alteration or modification of an amino acid may also be performed on an antibody variable region or an antibody constant region to be used in preparing recombinant antibodies for the antibodies of Disclosure A or B.
- substitution of amino acids refers to substitution with different amino acids (residues), and can be designed to modify, for example, matters such as in (a) to (c): (a) the polypeptide backbone structure in a region of sheet or helical conformation; (b) charge or hydrophobicity at a target site; or (c) size of a side chain.
- Amino acid residues are classified, based on properties of the side chains in the structure, for example, into the groups of: (1) hydrophobic: norleucine, Met, Ala, Val, Leu, and Ile; (2) neutral, hydrophilic: Cys, Ser, Thr, Asn, and Gln; (3) acidic: Asp and Glu; (4) basic: His, Lys, and Arg; (5) residues that affect the chain orientation: Gly and Pro; and (6) aromatic: Trp, Tyr, and Phe.
- substitution of amino acid residues within each group is referred to as conservative substitution, while substitution of amino acid residues between different groups is referred to as non-conservative substitution.
- substitution of amino acid residues may be conservative substitution, non-conservative substitution, or a combination thereof.
- Several known appropriate methods may be used for substituting amino acids with those other than natural amino acids (Wang et al., Annu. Rev. Biophys. Biomol. Struct. 35:225-249 (2006); Forster et al., Proc. Natl. Acad. Sci. USA 100(11):6353-6357 (2003)).
- antigens may be any ligands, including various cytokines, for example, interleukins, chemokines, and cell growth factors.
- receptors that are present as in a soluble form or have been modified to be a soluble form in biological fluids such as plasma can also be used as antigens.
- soluble receptors include the soluble IL-6 receptor described in Mullberg et al., J. Immunol. 152(10):4958-4968 (1994).
- antigens may be monovalent (for example, soluble IL-6 receptor) or multivalent (for example, IgE).
- antigens that can be bound by an antibody of Disclosures A and B are preferably soluble antigens present in biological fluids (for example, biological fluids illustrated in WO2013/125667, preferably plasma, interstitial fluid, lymphatic fluid, ascitic fluid, or pleural fluid) of subjects (within the scope of Disclosures A and B described herein, subjects to be administered (applied) with the antibody, which can be virtually any animal, for example, a human, mouse, etc.,); however, the antigens may also be membrane antigens.
- biological fluids for example, biological fluids illustrated in WO2013/125667, preferably plasma, interstitial fluid, lymphatic fluid, ascitic fluid, or pleural fluid
- the antigens may also be membrane antigens.
- prolongation of the half-life in plasma or “shortening of the half-life in plasma” of a target molecule (which may be an antigen or antibody), or an equivalent phrase thereof can also be represented more specifically using in addition to the parameter of half-life in plasma (t1/2), any other parameter such as mean retention time in plasma, clearance (CL) in plasma, and area under the concentration curve (AUC) (Pharmacokinetics: Enshuniyoru Rikai (Understanding through practice) Nanzando).
- t1/2 parameter of half-life in plasma
- any other parameter such as mean retention time in plasma, clearance (CL) in plasma, and area under the concentration curve (AUC) (Pharmacokinetics: Enshuniyoru Rikai (Understanding through practice) Nanzando).
- AUC area under the concentration curve
- an “epitope” refers to an antigenic determinant in an antigen and means a site on an antigen at which the antigen-binding domain of an antibody binds.
- an epitope can be defined, for example, based on its structure.
- the epitope may be defined by the antigen-binding activity of an antibody that recognizes the epitope.
- an antigen is a peptide or polypeptide
- the epitope can be specified by the amino acid residues that constitute the epitope.
- an epitope is a sugar chain
- the epitope can be specified based on its specific sugar chain structure.
- An antigen-binding domain of Disclosures A and B may bind to a single epitope or different epitopes on an antigen.
- a linear epitope may be a primary amino acid sequence.
- Such a linear epitope typically contains at least three and commonly at least five, for example, 8 to 10 amino acids or 6 to 20 amino acids as a unique sequence.
- a conformational epitope typically the amino acids that constitute the epitope are not present consecutively as a primary sequence.
- An antibody can recognize a conformational epitope in the three-dimensional structure of a peptide or protein.
- Methods for determining the conformation of an epitope include, but are not limited to, X ray crystallography, two-dimensional nuclear magnetic resonance, site-specific spin labeling, and electron paramagnetic resonance (Epitope Mapping Protocols in Methods in Molecular Biology (1996), Vol. 66, Morris (ed.)).
- an "antibody” is not particularly limited and used in the broadest sense, as long as it can bind to an antigen of target.
- Non-limiting examples of antibodies include widely known common antibodies (for example, native immunoglobulins (abbreviated as "Ig")), and molecules and variants derived therefrom, for example, Fab, Fab', F(ab') 2 , diabodies, ScFv (Holliger et al., Proc. Natl. Acad. Sci.
- bispecific antibodies are not limited but may be prepared, for example, as antibody molecules having the common L chain described in WO2005/035756, or by the method described in WO2008/119353 where two general types of antibodies having an IgG4-like constant regions are mixed to cause an exchange reaction between the two types of such antibodies (known as the "Fab-arm exchange” method to those of ordinary skill in the art).
- they may be antibodies having a structure where the heavy-chain variable region and the light-chain variable region are linked together as a single chain (for example, sc(Fv) 2 ).
- they may be antibody-like molecules (for example, scFv-Fc) that result from linking the Fc region (a constant region that lacks the CH1 domain) to scFv (or sc(Fv) 2 ) where the heavy-chain variable region (VH) is linked to the light-chain variable region (VL).
- Multispecific antibodies consisting of scFv-Fc have an (scFv) 2 -Fc structure where the first and second polypeptides are VH1-linker-VL1-Fc and VH2-linker-VL2-Fc, respectively.
- they may be antibody-like molecules where a single-domain antibody is linked to an Fc region (Marvin et al., Curr. Opin.
- native IgG refers to polypeptides that contain the same amino acid sequence as that of naturally occurring IgG (e.g. native IgG1) and belongs to the class of antibodies encoded substantially by the immunoglobulin gamma gene. Native IgG may be spontaneous mutants thereof and the like.
- the Y-shaped structure of the four chains can be the basic structure.
- the heavy chain and the light chain can be linked together via a disulfide bond (SS bond) and form a heterodimer.
- SS bond disulfide bond
- Such heterodimers may be linked together via a disulfide bond and form a Y-shaped heterotetramer.
- the two heavy chains or light chains may be identical or different from each other.
- an IgG antibody may be cleaved into two units of Fab (region) and a single unit of Fc (region) by papain digestion, which cleaves the hinge region (also referred to as the "hinge" within the scope of Disclosures A and B described herein) where the heavy-chain Fab region is linked to the Fc region.
- the Fab region contains an antigen-binding domain. Since phagocytic cells such as leukocytes and macrophages have receptors that are capable of binding to the Fc region (Fc receptors), and can recognize via the Fc receptors antibodies that are bound to an antigen and phagocytize the antigen (opsonization).
- the Fc region is involved in the mediation of immune reactions such as ADCC or CDC, and has an effector function of inducing a reaction upon an antibody binding to antigens.
- the antibody effector function is known to vary according to the type of immunoglobulin (isotype).
- the Fc region of the IgG class would indicate a region that spans, for example, from cysteine of position 226 or from proline of position 230 (EU numbering) to the C terminus; however, the Fc region is not limited thereto.
- the Fc region can be appropriately obtained by partial digestion of a monoclonal IgG1, IgG2, IgG3, or IgG4 antibody, or others, with a protease such as pepsin, followed by elution of adsorbed fractions from a protein A or protein G column.
- a protease such as pepsin
- library may refer to molecules (populations) such as multiple antibodies that have sequence variability, in which their respective sequences may be the same or different from one another; multiple fusion polypeptides containing the antibodies; or nucleic acids or polynucleotides encoding these amino acid sequences, as described in detail in WO2013/125667 (for example, paragraphs 0121-0125).
- the library may, for example, contain at least 10 4 antibody molecules, more preferably, at least 10 5 antibody molecules, even more preferably, at least 10 6 antibody molecules, particularly preferably, at least 10 7 antibody molecules or more.
- the library may be phage libraries.
- immune libraries that are constructed based on antibody genes derived from lymphocytes of animals immunized with a specific antigen, patients with infection, humans with elevated antibody titer in blood due to vaccination, or patients with cancer or autoimmune disease can be appropriately used as randomized variable region libraries.
- naive libraries containing naive sequences (antibody sequences without bias in the repertoire), which are constructed from antibody genes derived from lymphocytes of healthy persons, can also be appropriately used as randomized variable region libraries (Gejima et al., Human Antibodies 11:121-129 (2002)); Cardoso et al., Scand. J. Immunol. 51:337-344 (2000)).
- Amino acid sequences containing naive sequences can refer to those obtained from such naive libraries.
- synthetic libraries in which the CDR sequence from a V gene of genomic DNA or a reconstructed functional V gene is substituted with a set of synthetic oligonucleotides containing a sequence encoding a codon set of appropriate length can also be appropriately used as randomized variable region libraries.
- synthetic libraries in which the CDR sequence from a V gene of genomic DNA or a reconstructed functional V gene is substituted with a set of synthetic oligonucleotides containing a sequence encoding a codon set of appropriate length can also be appropriately used as randomized variable region libraries.
- a standard way to produce amino acid diversity in the antibody variable region may be to increase variations of amino acid residues at positions that can be exposed on the antibody surface.
- V regions [heavy chain variable region ("VH region”) and light chain variable region (“VL region”)] and constant regions ("C regions")["heavy chain constant region ("CH region”) and light chain constant region (“CL region”)].
- VH region variable chain variable region
- VL region variable chain variable region
- C regions constant regions
- CH region constant chain constant region
- CH region is further divided into three: CH1 to CH3.
- the Fab region of the heavy chain contains VH region and CH1
- Fc region of the heavy chain contains CH2 and CH3.
- the hinge region is located between CH1 and CH2.
- the variable region typically has complementarity determining regions ("CDRs") and framework regions ("FRs").
- the VH region and VL region each have three CDRs (CDR1, CDR2, and CDR3) and four FRs (FR1, FR2, FR3, and FR4).
- CDR1, CDR2, and CDR3 CDR1, CDR2, and CDR3
- FR1, FR2, FR3, and FR4 CDR1, FR2, FR3, and FR4
- the six CDRs in the variable regions of the heavy chain and light chain interact and form the antigen-binding domain of the antibody.
- the antigen-binding affinity is known to be lower as compared to where six CDRs are present, it has still the ability to recognize and bind to the antigen.
- Ig antibodies are classified into several classes (isotypes) based on structural differences in their constant regions. In many mammals, they are categorized into five immunoglobulin classes based on structural differences in the constant region: IgG, IgA, IgM, IgD, and IgE. Furthermore, in the case of human, IgG has four subclasses: IgG1, IgG2, IgG3, and IgG4; and IgA has two subclasses: IgA1 and IgA2.
- the heavy chain is classified into ⁇ chain, ⁇ chain, ⁇ chain, ⁇ chain, and ⁇ chain according to differences in the constant region, and based on these differences, there are five immunoglobulin classes (isotypes): IgG, IgM, IgA, IgD, and IgE.
- immunoglobulin classes are five immunoglobulin classes (isotypes): IgG, IgM, IgA, IgD, and IgE.
- isotypes immunoglobulin classes
- light chains ⁇ chain and ⁇ chain, and all immunoglobulins have either of these two.
- an antibody of Disclosure A or B has a heavy chain
- the heavy chain may be any one of ⁇ chain, ⁇ chain, ⁇ chain, ⁇ chain, and ⁇ chain, or may be derived from any one of them
- an antibody of Disclosure A or B has a light chain
- the light chain may be either ⁇ chain or ⁇ chain, or may be derived from either.
- the antibody may be of any isotype (for example, IgG, IgM, IgA, IgD, or IgE) and of any subclass (for example, human IgG1, IgG2, IgG3, IgG4, IgA1, and IgA2; mouse IgG1, IgG2a, IgG2b, and IgG3), or may be derived from any one of them, but is not limited thereto.
- an "antigen-binding domain” may have any structure as long as it binds to an antigen of interest.
- Such domains may include, for example, the variable regions of antibody heavy chains and light chains (for example, 1 to 6 CDRs); a module of about 35 amino acids referred to as A domain, which is contained in Avimer, a cell membrane protein present in the body (WO2004/044011 and WO2005/040229); Adnectin containing the 10Fn3 domain which binds to the protein in the glycoprotein fibronectin expressed on cell membrane (WO2002/032925); Affibody, having as scaffold the IgG-binding domain constituting a three-helix bundle of 58 amino acids of Protein A (WO1995/001937); Designed Ankyrin Repeat Proteins (DARPins) which are a region exposed on the molecular surface of an Ankyrin repeat (AR) having a structure in which a subunit with a turn containing 33 amino acid residues,
- DARPins Designed An
- Preferred antigen-binding domains of Disclosure A or B may include those having IgG antibody heavy-chain and light-chain variable regions, and more specifically, ScFv, single chain antibodies, Fv, scFv 2 (single chain Fv 2 ), Fab, and F(ab') 2 .
- ion concentration is not particularly limited and refers to hydrogen ion concentration (pH) or metal ion concentration.
- metal ions can be any one of ions of group I elements except hydrogen, such as alkaline metals and copper group elements, group II elements such as alkaline earth metals and zinc group elements, group III elements except boron, group IV elements except carbon and silicon, group VIII elements such as iron group and platinum group elements, elements belonging to subgroup A of groups V, VI, and VII, and metal elements such as antimony, bismuth, and polonium.
- Metal atoms have the property of releasing valence electrons to become cations. This is referred to as ionization tendency. Metals with strong ionization tendency are assumed to be chemically active.
- preferred metal ions may be calcium ion, as described in detail in WO2012/073992 and WO2013/125667.
- ion concentration condition(s) may be a condition that focuses on differences in the biological behavior of an ion concentration-dependent antibody between a low ion concentration and a high ion concentration.
- the antigen-binding activity changes according to the ion concentration condition can mean that the antigen-binding activity of an ion concentration-dependent antigen-binding domain or an ion concentration-dependent antibody of Disclosure A or B changes between a low ion concentration and a high ion concentration.
- Such cases include, for example, those with higher (stronger) or lower (weaker) antigen-binding activity at a high ion concentration than at a low ion concentration, without being limited thereto.
- the ion concentration can be hydrogen ion concentration (pH) or calcium ion concentration.
- the ion concentration-dependent antigen-binding domain may also be referred to as a "pH-dependent antigen-binding domain"; and where the ion concentration is calcium ion concentration, it may also be referred to as a "calcium ion concentration-dependent antigen-binding domain”.
- the ion concentration-dependent antigen-binding domains, ion concentration-dependent antibodies, ion concentration-dependent antigen-binding domains with increased pI, and ion concentration-dependent antibodies with increased pI can be obtained from libraries primarily consisting of antibodies that differ in sequence (have variability) and whose antigen-binding domains contain at least one amino acid residue that causes a change in the antigen-binding activity of the antigen-binding domain or antibody according to the ion concentration condition.
- the antigen-binding domains may be preferably located within the light chain variable region (which may be modified) and/or the heavy chain variable region (which may be modified).
- such light-chain or heavy-chain variable regions may be combined with heavy-chain or light-chain variable regions constructed as a randomized variable region sequence library.
- the ion concentration is hydrogen or calcium ion concentration
- non-limiting examples of the library include, for example, libraries in which heavy chain variable regions constructed as a randomized variable region sequence library are combined with light chain variable region sequences in which amino acid residue(s) in a germ line sequence such as SEQ ID NO:1 (Vk1), SEQ ID NO:2 (Vk2), SEQ ID NO:3 (Vk3), or SEQ ID NO:4 (Vk4) has been substituted with at least one amino acid residue that can alter the antigen-binding activity depending on ion concentrations.
- the library includes, for example, those in which the heavy chain variable region sequence of SEQ ID NO:5 (6RL#9-IgG1) or SEQ ID NO:6 (6KC4-1#85-IgG1) is combined with light chain variable regions constructed as a randomized variable region sequence library or light chain variable regions having a germ line sequence.
- the high calcium ion concentration is not particularly limited to a specific value; however, the concentration may be selected between 100 ⁇ M and 10 mM, between 200 ⁇ M and 5 mM, between 400 ⁇ M and 3 mM, between 200 ⁇ M and 2 mM, or between 400 ⁇ M and 1 mM.
- a concentration selected between 500 ⁇ M and 2.5 mM, which is close to the plasma (blood) concentration of calcium ion in vivo, may be also preferred.
- the low calcium ion concentration is not particularly limited to a specific value; however, the concentration may be selected between 0.1 ⁇ M and 30 ⁇ M, between 0.2 ⁇ M and 20 ⁇ M, between 0.5 ⁇ M and 10 ⁇ M, or between 1 ⁇ M and 5 ⁇ M, or between 2 ⁇ M and 4 ⁇ M.
- a concentration selected between 1 ⁇ M and 5 ⁇ M, which is close to the concentration of calcium ion in early endosomes in vivo, may be also preferred.
- the antigen-binding activity of an antigen-binding domain or antibody containing the domain changes according to the metal ion concentration (for example, calcium ion concentration) condition can be readily determined by known methods, for example, by the methods described herein in the context of Disclosure A, or described in WO2012/073992.
- the antigen-binding activity of an antigen-binding domain or antibody containing the domain can be measured at low and high calcium ion concentrations and compared.
- conditions other than the calcium ion concentration may be preferably the same.
- conditions other than the calcium ion concentration in determining the antigen-binding activity can be appropriately selected by those of ordinary skill in the art.
- the antigen-binding activity can be determined, for example, under the conditions of HEPES buffer at 37°C, or using the BIACORE (GE Healthcare) or others.
- the antigen-binding activity of the ion concentration-dependent antigen-binding domain, ion concentration-dependent antibody, ion concentration-dependent antigen-binding domain with increased pI, or ion concentration-dependent antibody with increased pI is higher under a high calcium ion concentration condition than under a low calcium ion concentration condition.
- the ratio between the antigen-binding activity under a low calcium ion concentration condition and the antigen-binding activity under a high calcium ion concentration condition is not limited; however, the value of the ratio of the KD (dissociation constant) for an antigen under a low calcium ion concentration condition to the KD under a high calcium ion concentration condition, i.e., KD (3 ⁇ M Ca)/KD (2 mM Ca), may be preferably 2 or more, more preferably 10 or more, and still more preferably 40 or more.
- the upper limit of the KD (3 ⁇ M Ca)/KD (2 mM Ca) value is not limited, and may be any value such as 400, 1000, or 10000.
- the dissociation constant (KD) can be used as the value for antigen-binding activity.
- the antigen is a membrane antigen
- the apparent dissociation constant (KD) can be used.
- the dissociation constant (KD) and apparent dissociation constant (KD) can be determined by known methods, for example, by BIACORE (GE healthcare), Scatchard plot, or flow cytometer.
- the dissociation rate constant (kd) can also be used as another indicator to represent the binding activity ratio.
- the value of the ratio of the low-calcium-ion-concentration-condition dissociation rate constant (kd) to the high-calcium-ion-concentration-condition dissociation rate constant (kd), i.e., kd (low calcium ion concentration condition)/kd (high calcium ion concentration condition) may be preferably 2 or more, more preferably 5 or more, still more preferably 10 or more, and yet more preferably 30 or more.
- the upper limit of the kd (low calcium ion concentration condition)/kd (high calcium ion concentration condition) value is not limited, and may be any value such as 50, 100, or 200.
- the dissociation rate constant (kd) can be used as the value for antigen-binding activity.
- the antigen is a membrane antigen, the apparent dissociation rate constant (kd) can be used.
- the dissociation rate constant (kd) and the apparent dissociation rate constant (kd) can be determined by known methods, for example, by BIACORE (GE healthcare) or flow cytometer.
- methods for producing or screening for calcium ion concentration-dependent antigen-binding domains or calcium ion concentration-dependent antibodies whose antigen-binding activity is higher at a high calcium ion concentration condition than at a low calcium ion concentration condition, or libraries thereof are not limited.
- the methods include, for example, those described in WO2012/073992 (for example, paragraphs 0200-0213).
- Such a method may comprise, for example: (a) determining the antigen-binding activity of an antigen-binding domain or antibody at a low calcium ion concentration condition; (b) determining the antigen-binding activity of an antigen-binding domain or antibody at a high calcium ion concentration condition; and (c) selecting an antigen-binding domain or antibody whose antigen-binding activity at a low calcium ion concentration condition is lower than the antigen-binding activity at a high calcium ion concentration condition.
- the method may comprise, for example: (a) contacting an antigen with an antigen-binding domain or antibody, or a library thereof, at a high calcium ion concentration condition; (b) incubating an antigen-binding domain or antibody that bound to the antigen in step (a) at a low calcium ion concentration condition; and (c) isolating an antigen-binding domain or antibody that dissociated in step (b).
- the method may comprise, for example: (a) contacting an antigen with an antigen-binding domain or antibody, or a library thereof at a low calcium ion concentration condition; (b) selecting an antigen-binding domain or antibody that does not bind to the antigen or has a low antigen-binding ability in step (a); (c) allowing the antigen-binding domain or antibody selected in step (b) to bind to the antigen at a high calcium ion concentration condition; and (d) isolating the antigen-binding domain or antibody that bound to the antigen in step (c).
- the method may comprise, for example: (a) contacting an antigen-binding domain or antibody, or a library thereof with an antigen-immobilized column at a high calcium ion concentration condition; (b) eluting an antigen-binding domain or antibody bound to the column in step (a) from the column at a low calcium ion concentration condition; and (c) isolating an antigen-binding domain or antibody eluted in step (b).
- the method may comprise, for example: (a) allowing an antigen-binding domain or antibody, or a library thereof to pass through an antigen-immobilized column at a low calcium ion concentration condition to collect an antigen-binding domain or antibody eluted without binding to the column; (b) allowing an antigen-binding domain or antibody collected in step (a) to bind to the antigen at a high calcium ion concentration condition; and (c) isolating an antigen-binding domain or antibody bound to the antigen in step (b).
- the method may comprise, for example: (a) contacting an antigen with an antigen-binding domain or antibody, or a library thereof at a high calcium ion concentration condition; (b) obtaining an antigen-binding domain or antibody bound to the antigen in step (a); (c) incubating an antigen-binding domain or antibody obtained in step (b) at a low calcium ion concentration; and (d) isolating an antigen-binding domain or antibody whose antigen-binding activity in step (c) is weaker than the criterion selected in step (b).
- Each step of these various screening methods may be repeated several times, or the steps may be combined appropriately to obtain the most suitable molecules.
- the aforementioned conditions may be suitably selected for the low and high calcium ion concentration conditions. Desired calcium ion concentration-dependent antigen-binding domains or calcium ion concentration-dependent antibodies can be obtained thereby.
- the antigen-binding domains or antibodies as a starting material may be, for example, modified antigen-binding domains or antibodies that have an increased pI as a result of modifying the charge of at least one amino acid residue that can be exposed on their surface.
- amino acids that change the binding activity of an ion concentration-dependent antigen-binding domain are introduced into the sequence, they may be introduced in conjunction with a charge modification of at least one amino acid residue that can be exposed on the surface of the antigen-binding domain or antibody so as to increase the pI.
- pre-existing antigen-binding domains or antibodies preexisting libraries (phage library, etc.); antibodies prepared from hybridomas obtained by immunizing animals or from B cells of immunized animals, or libraries thereof; or antigen-binding domains, antibodies, or libraries obtained by introducing natural or unnatural amino acid mutations capable of chelating calcium (described below) thereinto (for example, libraries with an increased content of calcium-chelatable amino acids, or libraries introduced with calcium-chelatable amino acids at specific sites).
- the ion concentration is calcium ion concentration
- the type of amino acids that change the binding activity of ion concentration-dependent antigen-binding domains or ion concentration-dependent antigen-binding domains with increased pI as long as they can form a calcium-binding motif.
- calcium-binding motifs are known to those of ordinary skill in the art (for example, Springer et al. (Cell 102:275-277 (2000)); Kawasaki et al. (Protein Prof. 2:305-490 (1995)); Moncrief et al. (J. Mol. Evol. 30:522-562 (1990)); Chauvaux et al. (Biochem.
- an antigen-binding domain has an arbitrary calcium-binding motif such as of a C-type lectin, for example, ASGPR, CD23, MBR, or DC-SIGN
- the antigen-binding activity of the domain can be changed according to the calcium ion concentration condition.
- Such calcium-binding motifs may include, for example, in addition to those described above, the calcium-binding motif included in the antigen-binding domain shown in SEQ ID NO:7 (which corresponds to "Vk5-2").
- amino acids having a metal-chelating activity may be used as amino acids that change the binding activity of ion concentration-dependent antigen-binding domains or ion concentration-dependent antigen-binding domains of with increased pI.
- any amino acids can be appropriately used as amino acids having a metal-chelating activity, as long as they can form a calcium-binding motif.
- such amino acids include those having an electron-donating property.
- the amino acids preferably include, but are not limited to, Ser (S), Thr (T), Asn (N), Gln (Q), Asp (D), and Glu (E).
- amino acids having a metal-chelating activity in an antigen-binding domain are not limited to specific positions.
- the amino acids may be located at any positions in the heavy chain variable region and/or light chain variable region that may form an antigen-binding domain.
- At least one amino acid residue that causes calcium ion concentration-dependent changes in the antigen-binding activity of an antibody may be contained, for example, in CDR (one or more of CDR1, CDR2, and CDR3) and/or FR (one or more of FR1, FR2, FR3, and FR4) of the heavy chain and/or light chain.
- amino acid residue(s) may be placed, for example, at one or more of positions 95, 96, 100a, and 101 according to Kabat numbering in heavy-chain CDR3; at one or more of positions 30, 31, and 32 according to Kabat numbering in light-chain CDR1; at position 50 according to Kabat numbering in light-chain CDR2; and/or at position 92 according to Kabat numbering in light-chain CDR3.
- Those amino acid residues may be placed alone or in combination.
- Troponin C calmodulin, parvalbumin, myosin light chain, and others are known to have multiple calcium-binding sites and assumed to be derived from a common origin in molecular evolution, and in one embodiment, one or more of light chain CDR1, CDR2, and CDR3 can be designed to contain binding motifs thereof.
- the cadherin domain for example, the cadherin domain; the EF hand contained in calmodulin; the C2 domain contained in Protein kinase C; the Gla domain contained in blood-clotting protein Factor IX; C-type lectin of the asialoglycoprotein receptor or mannose-binding receptor; the A domain contained in the LDL receptor; Annexin; thrombospondin type-3 domain; and EGF-like domain may be suitably used.
- the concentration condition of proton i.e., nucleus of a hydrogen atom
- the concentration condition of proton i.e., nucleus of a hydrogen atom
- pH is defined as -log10aH +
- aH + is nearly equal to the hydrogen ion strength.
- the hydrogen ion concentration condition may be conditions that focus on differences in the biological behavior of a pH-dependent antibody at a high hydrogen ion concentration (acidic pH range) and at a low hydrogen ion concentration (neutral pH range) for the hydrogen ion concentration condition or pH condition.
- the antigen-binding activity at a high hydrogen ion concentration (acidic pH range) condition is lower than the antigen-binding activity at a low hydrogen ion concentration (neutral pH range) condition
- the antigen-binding activity of an ion concentration-dependent antigen-binding domain, an ion concentration-dependent antibody, an ion concentration-dependent antigen-binding domain with increased pI, or an ion concentration-dependent antibody with increased pI is weaker at a pH selected from pH 4.0 to pH 6.5, preferably from pH 4.5 to pH 6.5, more preferably from pH 5.0 to pH 6.5, and still more preferably from pH 5.5 to pH 6.5, than at a pH selected from pH 6.7 to pH 10.0, preferably from pH 6.7 to pH 9.5, more preferably from pH 7.0 to pH 9.0, and still more preferably from pH 7.0 to pH 8.0.
- the above expression can mean that the antigen-binding activity at the pH within early endosomes in vivo is weaker than that at the plasma pH in vivo; and specifically can mean that the antigen-binding activity of an antibody, for example, at pH 5.8 is weaker than that, for example, at pH 7.4.
- the antigen-binding activity of an antigen-binding domain or an antibody containing the domain changes according to the hydrogen ion concentration condition can be readily assessed by known methods, for example, by the assay methods described herein in the context of Disclosure A, or described in WO2009/125825.
- the antigen-binding activity of an antigen-binding domain or an antibody containing the domain toward an antigen of interest may be measured at low and high hydrogen ion concentrations and compared. In this case, it is preferable that conditions other than the hydrogen ion concentration are the same.
- neutral pH range (also referred to as “low hydrogen ion concentration”, “high pH”, “neutral pH condition”, or “neutral pH”) is not particularly limited to a specific value; however, it may be preferably selected from pH 6.7 to pH 10.0, from pH 6.7 to pH 9.5, from pH 7.0 to pH 9.0, or from pH 7.0 to pH 8.0.
- the neutral pH range may be preferably pH 7.4 which is close to the in vivo pH in plasma (blood), but for the convenience of measurement, for example, pH 7.0 may be used.
- “acidic pH range” (also referred to as “high hydrogen ion concentration”, “low pH”, “acidic pH condition”, or “acidic pH”) is not particularly limited to a specific value; however, it may be preferably selected from pH 4.0 to pH 6.5, from pH 4.5 to pH 6.5, pH 5.0 to pH 6.5, or pH 5.5 to pH 6.5.
- the acidic pH range may be preferably pH 5.8 which is close to the in vivo hydrogen ion concentration in the early endosome, but for the convenience of measurement, for example, pH 6.0 may be used.
- the antigen-binding activity of the ion concentration-dependent antigen-binding domain, ion concentration-dependent antibody, ion concentration-dependent antigen-binding domain with increased pI, or ion concentration-dependent antibody with increased pI is higher under a neutral pH condition than under an acidic pH condition.
- the ratio of the antigen-binding activity under a neutral pH condition to the antigen-binding activity under an acidic pH condition is not limited; however, the value of the ratio of the dissociation constant (KD) for an antigen at an acidic pH condition to the KD at a neutral pH condition, i.e., KD (acidic pH range)/KD (neutral pH range), (for example, KD (pH 5.8)/KD (pH 7.4)) may be 2 or more; 10 or more; or 40 or more.
- the upper limit of KD (acidic pH range)/KD (neutral pH range) value is not limited, and may be any value such as 400, 1000, or 10000.
- the dissociation rate constant (kd) as an indicator to represent the above binding activity ratio.
- the value of the ratio of the dissociation rate constant (kd) for an antigen at a high hydrogen ion concentration condition to that at a low hydrogen ion concentration condition i.e., kd (acidic pH range)/kd (neutral pH range) may be 2 or more, 5 or more, 10 or more, or 30 or more.
- the upper limit of the kd (acidic pH range)/kd (neutral pH range) value is not limited, and may be any value such as 50, 100, or 200.
- the value of the antigen-binding activity can be represented by the dissociation rate constant (kd), whereas where the antigen is a membrane antigen, such value can be represented by the apparent dissociation rate constant (apparent kd).
- the dissociation rate constant (kd) and apparent dissociation rate constant (apparent kd) can be determined by known methods, for example, by using the BIACORE (GE healthcare) or a flow cytometer.
- methods for producing or screening for pH-dependent antigen-binding domains or pH-dependent antibodies whose antigen-binding activity is higher under a neutral pH condition than under an acidic pH condition, or libraries thereof are not limited. Such methods include, for example, those described in WO2009/125825 (for example, paragraphs 0158-0190).
- Such a method may comprise, for example: (a) determining the antigen-binding activity of an antigen-binding domain or antibody in an acidic pH condition; (b) determining the antigen-binding activity of an antigen-binding domain or antibody in a neutral pH condition; and (c) selecting an antigen-binding domain or antibody whose antigen-binding activity is lower in the acidic pH condition than in the neutral pH condition.
- the method may comprise, for example: (a) contacting an antigen with an antigen-binding domain or antibody, or a library thereof, in a neutral pH condition; (b) incubating an antigen-binding domain or antibody bound to the antigen in step (a) in an acidic pH condition; and (c) isolating an antigen-binding domain or antibody that dissociated in step (b).
- the method may comprise, for example: (a) contacting an antigen with an antigen-binding domain or antibody, or a library thereof in an acidic pH condition; (b) selecting an antigen-binding domain or antibody that does not bind to the antigen or has a low antigen-binding ability in step (a); (c) allowing the antigen to bind to the antigen-binding domain or antibody selected in step (b) in a neutral pH condition; and (d) isolating an antigen-binding domain or antibody that bound to the antigen in step (c).
- the method may comprise, for example: (a) contacting an antigen-binding domain or antibody, or a library thereof with an antigen-immobilized column in a neutral pH condition; (b) eluting an antigen-binding domain or antibody bound to the column in step (a) from the column in an acidic pH condition; and (c) isolating an antigen-binding domain or antibody eluted in step (b).
- the method may comprise, for example: (a) allowing an antigen-binding domain or antibody, or a library thereof to pass through an antigen-immobilized column in an acidic pH condition to collect an antigen-binding domain or antibody eluted without binding to the column; (b) allowing an antigen-binding domain or antibody collected in step (a) to bind to the antigen in a neutral pH condition; and (c) isolating an antigen-binding domain or antibody bound to the antigen in step (b).
- the method may comprise, for example: (a) contacting an antigen with an antigen-binding domain or antibody, or a library thereof in a neutral pH condition; (b) obtaining an antigen-binding domain or antibody bound to the antigen in step (a); (c) incubating an antigen-binding domain or antibody obtained in step (b) in an acidic pH condition; and (d) isolating an antigen-binding domain or antibody whose antigen-binding activity in step (c) is weaker than the criterion selected in step (b).
- Each step in these various screening methods may be repeated several times, or the steps may be combined.
- the aforementioned conditions may be suitably selected for the acidic and neutral pH conditions. Desired pH-dependent antigen-binding domains or pH-dependent antibodies can be obtained thereby.
- the antigen-binding domains or antibodies as a starting material may be, for example, modified antigen-binding domains or antibodies that have an increased pI as a result of modifying the charge of at least one amino acid residue that can be exposed on their surface.
- amino acids that change the binding activity of an ion concentration-dependent antigen-binding domain are introduced into the sequence, they may be introduced in conjunction with a charge modification of at least one amino acid residue that can be exposed on the surface of the antigen-binding domain or antibody so as to increase the pI.
- pre-existing antigen-binding domains or antibodies pre-existing libraries (phage library, etc.); antibodies prepared from hybridomas obtained by immunizing animals or from B cells of immunized animals, or libraries thereof; or antigen-binding domains, antibodies, or libraries obtained by introducing natural or unnatural amino acid mutations having a side-chain with a pKa of 4.0-8.0 (described below) thereinto (for example, libraries with an increased content of natural or unnatural amino acid mutations with a side-chain pKa of 4.0-8.0, or libraries introduced at specific sites with natural or unnatural amino acid mutations with a side-chain pKa of 4.0-8.0).
- Such a preferred antigen-binding domain can have, for example, an amino acid sequence in which at least one amino acid residue has been substituted with an amino acid(s) with a side-chain pKa of 4.0-8.0 and/or which has been inserted with amino acid(s) with a side-chain pKa of 4.0-8.0, as described in WO2009/125825.
- the site at which the mutation of amino acids with a side-chain pKa of 4.0-8.0 is introduced is not limited, and the mutation may be introduced at any site as long as the antigen-binding activity becomes weaker in an acidic pH range than in a neutral pH range (the KD (acidic pH range)/KD (neutral pH range) value is increased or the kd (acidic pH range)/kd (neutral pH range) value is increased) as compared to before substitution or insertion.
- the site may be within the variable region or CDR(s).
- the number of amino acids that are substituted or inserted can be appropriately determined by those of ordinary skill in the art; and the number may be one or more. Furthermore, it is possible to delete, add, insert, and/or substitute, or modify other amino acids in addition to the substitution or insertion described above.
- Substitution with or insertion of amino acids with a side-chain pKa of 4.0-8.0 may be carried out in a random fashion by scanning methods such as histidine scanning, in which histidine is used instead of alanine in alanine scanning known to those of ordinary skill in the art, and/or antibodies whose KD (acidic pH range)/KD (neutral pH range) value or kd (acidic pH range)/kd (neutral pH range) value has increased as compared to before mutation may be selected from among the antigen-binding domains or antibodies that result from random substitution with or insertion mutations of these amino acids, or libraries thereof.
- scanning methods such as histidine scanning, in which histidine is used instead of alanine in alanine scanning known to those of ordinary skill in the art, and/or antibodies whose KD (acidic pH range)/KD (neutral pH range) value or kd (acidic pH range)/kd (neutral pH range) value has increased as compared to before mutation may be selected
- the antigen-binding domains or antibodies may be preferably those whose antigen-binding activity in a neutral pH range before and after these mutations is not significantly reduced, is not substantially reduced, is substantial identical, or is increased; and in other words, those whose activity may be maintained at at least 10% or higher, preferably 50% or higher, still more preferably 80% or higher, and yet more preferably 90% or higher, or even higher.
- the binding activity of an antigen-binding domain or antibody is decreased due to substitution with or insertion of amino acids with a pKa of 4.0-8.0
- the binding activity may be recovered or increased by e.g., substituting, deleting, adding, or inserting one or more amino acids at sites other than the substitution or insertion sites described above.
- amino acids with a side chain pKa of 4.0-8.0 may be placed at any location within the heavy-chain and/or light-chain variable regions that may form an antigen-binding domain. At least one amino acid residue with a side-chain pKa of 4.0-8.0 may be located, for example, in the CDR (one or more of CDR1, CDR2, and CDR3) and/or FR (one or more of FR1, FR2, FR3, and FR4) of the heavy chain and/or light chain.
- amino acid residues include, but are not limited to, amino acid residues at one or more of positions 24, 27, 28, 31, 32, and 34 according to Kabat numbering in the light-chain variable region CDR1; amino acid residues at one or more of positions 50, 51, 52, 53, 54, 55, and 56 according to Kabat numbering in the light-chain variable region CDR2; and/or amino acid residues at one or more of positions 89, 90, 91, 92, 93, 94, and 95A according to Kabat numbering in the light-chain variable region CDR3.
- Those amino acid residues may be included alone or in combination, as long as the antigen-binding activity of the antibody changes according to the hydrogen ion concentration condition.
- an arbitrary amino acid residue can be suitably used as the amino acid residue that changes the antigen-binding activity of the antigen-binding domain or antibody according to the hydrogen ion concentration condition.
- amino acid residues can include those with a side-chain pKa of 4.0-8.0.
- amino acids having an electron-donating property may include, for example, natural amino acids such as His (H) and Glu (E), and unnatural amino acids such as histidine analogs (US2009/0035836), m-NO2-Tyr (pKa 7.45), 3,5-Br2-Tyr (pKa 7.21), and 3,5-I2-Tyr (pKa 7.38) (Heyl et al., Bioorg. Med. Chem. 11(17):3761-3768 (2003)).
- the amino acid residues may preferably include, for example, amino acids with a side-chain pKa of 6.0-7.0, and in particular His (H).
- isoelectric point may be either a theoretical or an experimentally determined isoelectric point, and it is also referred to as "pI".
- the pI value can be determined experimentally, for example, by isoelectric focusing electrophoresis. Meanwhile, the theoretical pI value can be calculated using gene and amino acid sequence analysis software (Genetyx, etc.).
- whether the pI of an antibody with increased pI or an antibody of Disclosure A has been increased as compared to the antibody before modification can be determined by carrying out, in addition to or instead of the above-described methods, antibody pharmacokinetics test using plasma, for example, from mice, rats, rabbits, dogs, monkeys, or humans, in combination with methods such as BIACORE, cell proliferation assay, ELISA, enzyme immunoassay (EIA), radioimmunoassay (RIA), or fluorescent immunoassay.
- an “amino acid residue that can be exposed on the surface” generally can refer to an amino acid residue located on the surface of a polypeptide constituting an antibody.
- An “amino acid residue located on the surface of a polypeptide” can refer to an amino acid residue whose side chain can be in contact with solvent molecules (which in general may be mostly water molecules). However, the side chain does not necessarily have to be wholly in contact with solvent molecules, and when even a portion of the side chain is in contact with the solvent molecules, the amino acid residue is defined as an "amino acid located on the surface”.
- the amino acid residues located on the surface of a polypeptide can also include amino acid residues located close to the antibody surface and thereby can have a mutual electric charge influence from other amino acid residue(s) whose side chain, even partly, is in contact with the solvent molecules.
- Those of ordinary skill in the art can prepare a homology model of a polypeptide or antibody by for example homology modeling using commercially available softwares. Alternatively, it is possible to use methods such as X-ray crystallography.
- the amino acid residues that may be exposed on the surface can be determined, for example, using coordinates from a three-dimensional model of an antibody using a computer program such as InsightII program (Accelrys).
- Surface-exposed sites may be determined using algorithms known in the technical field (for example, Lee and Richards (J. Mol. Biol. 55:379-400 (1971)); Connolly (J. Appl. Cryst. 16:548-558(1983)). Surface-exposable sites can be determined using software suitable for protein modeling and three-dimensional structure information obtained from the antibody. Software available for such purposes includes, for example, the SYBYL Biopolymer Module software (Tripos Associates). When an algorithm requires a user input size parameter, the "size" of a probe used in the calculation may be set to about 1.4 Angstrom or less in radius. Furthermore, methods for determining surface-exposed regions and areas using software for personal computers have been described by Pacios (Pacios, Comput. Chem18(4):377-386 (1994); J. Mol. Model. 1:46-53 (1995)). Based on such information as described above, appropriate amino acid residues located on the surface of a polypeptide that constitutes an antibody can be selected.
- a method for increasing the pI of a protein is, for example, to reduce the number of amino acids with a negatively charged side chain at a neutral pH condition (for example, aspartic acid and glutamic acid) and/or to increase the number of amino acids with a positively charged side chain (for example, arginine, lysine and histidine).
- Amino acid residues with a negatively charged side chain have a negative charge represented as -1 at a pH condition that is sufficiently higher than their side chain pKa, which is a theory well known to those of ordinary skill in the art.
- the theoretical pKa for the side chain of aspartic acid is 3.9, and the side chain has a negative charge represented as -1 at a neutral pH condition (for example, in a solution of pH 7.0).
- amino acid residues with a positively charged side chain have a positive charge represented as +1 at a pH condition that is sufficiently lower than their side chain pKa.
- the theoretical pKa for the side chain of arginine is 12.5, and the side chain has a positive charge represented as +1 at a neutral pH condition (for example, in a solution of pH 7.0).
- Amino acid residues whose side chain has no charge at a neutral pH condition are known to include 15 types of natural amino acids, i.e., alanine, cysteine, phenylalanine, glycine, isoleucine, leucine, methionine, asparagine, proline, glutamine, serine, threonine, valine, tryptophan, and tyrosine.
- amino acids for changing the pI may be unnatural amino acids.
- a charge alteration of +1 can be conferred to a protein of interest, for example, by substituting amino acids (residues) with non-charged side chains for aspartic acid (residue) or glutamic acid (residue) (whose side chain has a negative charge of -1) in the amino acid sequence of the protein.
- a charge alteration of +1 can be conferred to the protein, for example, by substituting arginine or lysine (whose side chain has a positive charge of +1) for amino acid (residue) whose side chain has no charge.
- a charge alteration of +2 can be conferred at a time to the protein by substituting arginine or lysine (whose side chain has a positive charge of +1) for aspartic acid or glutamic acid (whose side chain has a negative charge of -1).
- amino acids with a side chain having no charge and/or amino acids having a positively charged side chain can be added or inserted into the amino acid sequence of the protein, or amino acids with a side chain having no charge and/or amino acids with a negatively charged side chain present in the amino acid sequence of the protein can be deleted.
- the N-terminal and C-terminal amino acid residues of a protein have a main chain-derived charge (NH 3+ of the amino group at the N-terminus and COO - of the carbonyl group at the C-terminus) in addition to their side chain-derived charges.
- the pI of a protein can also be increased by performing to the main chain-derived functional groups some addition, deletion, substitution, or insertion.
- the effect of changing the net charge or pI of a protein which is obtained by modifying one or more amino acids (residues) in the amino acid sequence with a focus on the presence or magnitude of electrical charges of the amino acids (residues), does not exclusively (or substantially) depend on the antibody-constituting amino acid sequences per se or the type of target antigen, but rather depends on the type and number of amino acid residues that are added, deleted, substituted, or inserted.
- Antibodies which have been modified to have an increased pI by modification on at least one amino acid residue that can be exposed on the antibody surface (“antibodies with increased pI” or "pI-increased antibodies”) can be taken up more rapidly into cells or can promote antigen elimination from the plasma, as described or suggested in, for example, WO2007/114319, WO2009/041643, WO2014/145159, or WO2012/016227.
- the IgG antibody has a sufficiently large molecular weight, and thus its major metabolic pathway is not through renal excretion.
- the IgG antibody which has an Fc region as a part of the molecule, is known to be recycled through a salvage pathway via FcRn, and thus has a long in vivo half-life.
- the IgG antibody is assumed to be mainly metabolized via a metabolic pathway in endothelial cells (He et al., J. Immunol. 160(2):1029-1035 (1998)). Specifically, it is believed that when taken up into endothelial cells nonspecifically, IgG antibodies are recycled by binding to FcRn, while IgG antibodies that could not bind are metabolized.
- the plasma half-life of an IgG antibody may be shortened when its Fc region is modified such that its FcRn-binding activity is reduced.
- the plasma half-life of an antibody with an increased pI has been demonstrated to depend on the pI in a highly correlated manner, as described in e.g., WO2007/114319 and WO2009/041643.
- the plasma half-life of the pI-increased antibodies described in the above documents was reduced without modifying the amino acid sequence constituting Fc which could potentially lead to acquisition of immunogenicity, and this result suggests that the pI-increasing technology is widely applicable even to any types of antibody molecules whose main metabolic pathway is renal excretion, such as scFv, Fab, or Fc fusion proteins.
- the pH concentration in biological fluids is in a neutral pH range.
- biological fluids for example, plasma
- the net positive charge of a pI-increased antibody is increased due to the increased pI, and as a result the antibody is more strongly attracted by physicochemical Coulomb interaction to the endothelial cell surface whose net charge is negative, when compared to antibodies whose pI has not been increased; via non-specific binding, the antibody binds thereto and is taken up into cells, which results in shortening of the antibody half-life in plasma or enhancement of antigen elimination from plasma.
- an antibody enhances uptake into cells of the antibody (or antigen/antibody complex) and/or intracellular permeability, which is considered to result in reducing the antibody concentration in plasma, reducing the antibody bioavailability, and/or shortening the antibody half-life in plasma; and these phenomena are expected to occur commonly in vivo, regardless of cell type, tissue type, organ type, etc.
- an antibody forms a complex with an antigen and is taken up into cells, not only the antibody's pI but also the antigen's pI can have an influence on the decrease or increase of the uptake into cells.
- methods for producing or screening for antibodies with an increased pI may include, for example, those described in WO2007/114319 (for example, paragraphs 0060-0087), WO2009/041643 (for example, paragraphs 0115-), WO2014/145159, and WO2012/016227.
- Such a method may comprise, for example: (a) modifying a nucleic acid that encodes an antibody comprising at least one amino acid residue that can be exposed on the antibody surface such that the charge of the amino acid residue(s) is modified so as to increase the pI of the antibody; (b) culturing a host cell such that the nucleic acid is expressed; and (c) collecting an antibody from the host cell culture.
- the method may comprise, for example: (a') modifying a nucleic acid that encodes an antibody comprising at least one amino acid residue that can be exposed on the antibody surface such that the charge of the amino acid residue(s) is modified; (b') culturing a host cell such that the nucleic acid is expressed; (c') collecting an antibody from the host cell culture; and (d') (optionally confirming or measuring and,) selecting an antibody with a pI increased as compared to an antibody before the modification.
- the antibody as a starting material or the antibody before the modification or the reference antibody may be, for example, an ion concentration-dependent antibody.
- amino acid(s) that change the binding activity of the ion concentration-dependent antigen-binding domain may also be included in the sequence.
- the method may simply be a method that comprises culturing the host cells obtained in step (b) or (b') and collecting an antibody from the cell culture.
- the method may be, for example, a method for producing a multispecific antibody that comprises a first polypeptide and a second polypeptide, and optionally a third polypeptide and a fourth polypeptide, which comprises: (A) modifying nucleic acid(s) that encodes the first polypeptide and/or the second polypeptide, and optionally the third polypeptide and/or the fourth polypeptide, any one or more of which comprises at least one amino acid residue that can be exposed on the polypeptide surface such that the charge of the amino acid residue(s) is modified so as to increase the antibody's pI; (B) culturing a host cell such that the nucleic acid is expressed; and (C) collecting a multispecific antibody from the host cell culture.
- a method for producing a multispecific antibody that comprises a first polypeptide and a second polypeptide, and optionally a third polypeptide and a fourth polypeptide, which comprises: (A) modifying nucleic acid(s) that encodes the first polypeptide
- the method may comprise, for example: (A') modifying nucleic acid(s) that encodes the first polypeptide and/or the second polypeptide, and optionally the third polypeptide and/or the fourth polypeptide, any one or more of which comprises at least one amino acid residue that can be exposed on the polypeptide surface such that the charge of the amino acid residue(s) is altered; (B') culturing a host cell such that the nucleic acid is expressed; (C') collecting a multispecific antibody from the host cell culture; and (D') (optionally confirming and) selecting an antibody whose pI is increased as compared to an antibody before the modification.
- the antibody as a starting material or the antibody before the modification or the reference antibody may be, for example, an ion concentration-dependent antibody.
- amino acid(s) that change the binding activity of the ion concentration-dependent antigen-binding domain may also be included in the sequence.
- the method may simply be a method that comprises culturing the host cells obtained in step (B) or (B') and collecting an antibody from the cell culture.
- the polypeptides whose nucleic acid(s) is to be modified may be preferably a homomultimer of the first polypeptide, a homomultimer of the second polypeptide, or a heteromultimer of the first and second polypeptides (and optionally, a homomultimer of the third polypeptide, a homomultimer of the fourth polypeptide, or a heteromultimer of the third and fourth polypeptides).
- the method may be, for example, a method for producing a humanized or human antibody with shortened half-life in plasma, which comprises: in an antibody which comprises CDR(s) selected from the group consisting of human-derived CDR(s), CDR(s) derived from an animal other than human, and synthetic CDR(s); human-derived FR(s); and a human constant region, (I) modifying at least one amino acid residue that can be exposed on the surface of at least one region selected from the group consisting of the CDR(s), FR(s), and constant region into amino acid residue(s) that has a different charge from the amino acid residue(s) present at the corresponding position(s) before the modification such that the pI of the antibody is increased.
- CDR(s) selected from the group consisting of human-derived CDR(s), CDR(s) derived from an animal other than human, and synthetic CDR(s); human-derived FR(s); and a human constant region
- the method may comprise, for example, in an antibody which comprises CDR(s) selected from the group consisting of human-derived CDR(s), CDR(s) derived from an animal other than human, and synthetic CDR(s); human-derived FR(s); and a human constant region, (I') modifying at least one amino acid residue that can be exposed on the surface of at least one region selected from the group consisting of the CDR(s), FR(s), and constant region into amino acid residue(s) that has a different charge from the amino acid residue(s) present at the corresponding position(s) before the modification; and (II') (optionally confirming and) selecting an antibody whose pI is increased as compared to an antibody before the modification.
- CDR(s) selected from the group consisting of human-derived CDR(s), CDR(s) derived from an animal other than human, and synthetic CDR(s); human-derived FR(s); and a human constant region
- (I') modifying at least one amino acid residue that can
- the antibody as a starting material or the antibody before the modification or the reference antibody may be, for example, an ion concentration-dependent antibody.
- amino acid(s) that change the binding activity of the ion concentration-dependent antigen-binding domain may also be included in the sequence.
- pre-existing antigen-binding domains or antibodies pre-existing libraries (phage library, etc.); antibodies prepared from hybridomas obtained by immunizing animals or from B cells of immunized animals, or libraries thereof; or antigen-binding domains or antibodies or libraries thereof with increased pI, prepared by modifying, in the above-described antigen-binding domains, antibodies, or libraries thereof, at least one amino acid residue that can be exposed on the surface according to for example any one of the above-described embodiments.
- the pI value may be preferably increased, for example, at least by 0.01, 0.03, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, or more, or at least by 0.6, 0.7, 0.8, 0.9, or more, and to significantly shorten the antibody half-life in plasma, the pI value may be increased, for example, by at least by 1.0, 1.1, 1.2, 1.3, 1.4, 1.5, or more, or at least by 1.6, 1.7, 1.8, 1.9, 2.0, 2.1, 2.2, 2.3, 2.4, 2.5, or more, or by 3.0 or more, as compared to the antibodies before modification or alteration (native antibodies (for example, native Ig antibodies, preferably native IgG antibodies), or reference or parent antibodies (e.g., antibodies before antibody modification, or prior to or during library construction)).
- native antibodies for example, native Ig antibodies, preferably native IgG antibodies
- reference or parent antibodies e.g., antibodies before antibody modification, or prior to or during library construction
- antibodies of Disclosure A are beneficial because, in addition to the characteristic of being shuttled between plasma and cellular endosomes and repeated binding to multiple antigens with one single antibody molecule due to the presence of an ion concentration-dependent antigen-binding domain, the antibody's net positive charge is increased as a result of increase in pI and this allows rapid cellular uptake of the antibody. These characteristics would shorten the antibody half-life in plasma, increase the extracellular matrix-binding activity of the antibodies, or enhance antigen elimination from plasma. One may decide on the optimal pI value to take advantage of these characteristics.
- the ion concentration-dependent antibodies of Disclosure A with increased pI may preferably enhance antigen elimination from plasma, for example, by at least 1.1-fold, 1.25-fold, 1.5-fold, 1.75-fold, 2-fold, 2.25-fold, 2.5-fold, 2.75-fold, 3-fold, 3.25-fold, 3.5-fold, 3.75-fold, 4-fold, 4.25-fold, 4.5-fold, 4.75-fold, 5-fold, 5.5-fold, 6-fold, 6.5-fold, 7-fold, 7.5-fold, 8-fold, 8.5-fold, 9-fold, 9.5-fold, or 10-fold or more (when the antibodies are administered in vivo), or their
- the ion concentration-dependent antibodies of Disclosure A with increased pI may preferably enhance antigen elimination from plasma, for example, by at least 1.1-fold, 1.25-fold, 1.5-fold, 1.75-fold, 2-fold, 2.25-fold, 2.5-fold, 2.75-fold, 3-fold, 3.25-fold, 3.5-fold, 3.75-fold, 4-fold, 4.25-fold, 4.5-fold, 4.75-fold, 5-fold, 5.5-fold, 6-fold, 6.5-fold, 7-fold, 7.5-fold, 8-fold, 8.5-fold, 9-fold, 9.5-fold, or 10-fold or more (when the antibodies are administered in vivo), or their extracellular
- assay methods for assessing whether the extracellular matrix-binding activity of antibodies of Disclosure A has been increased as compared to the antibodies before modification or alteration are not limited.
- the assay can be carried out using an ELISA system which detects the binding between an antibody and an extracellular matrix, where an antibody is added to an extracellular matrix-immobilized plate, and a labeled antibody against the antibody is added thereto.
- ECL electrochemiluminescence
- This method can be performed, for example, using an ECL system in which a mixture of an antibody and a ruthenium antibody is added to an extracellular matrix-immobilized plate and the binding between the antibodies and the extracellular matrix is measured based on the electrochemiluminescence of ruthenium.
- concentration of the antibody to be added can be set appropriately; the added concentration can be high in order to increase the sensitivity for detecting extracellular matrix binding.
- extracellular matrices may be derived from animals or plants, as long as they contain glycoproteins such as collagen, proteoglycan, fibronectin, laminin, entactin, fibrin, and perlecan; and animal-derived extracellular matrices may be preferred.
- animal-derived extracellular matrices may be preferred.
- extracellular matrices derived from animals such as humans, mice, rats, monkeys, rabbits, or dogs.
- a human-derived native extracellular matrix may be used as an indicator of antibody pharmacodynamics in human plasma.
- the condition for assessing extracellular matrix-binding of an antibody may be preferably a neutral pH range around pH 7.4, which is the physiological condition; however, the condition does not necessarily have to be a neutral range, and the binding may also be assessed in an acidic pH range (for example, around pH 6.0).
- the assay can be performed in the co-presence of an antigen molecule to which the antibody binds and by assessing the binding activity of the antigen-antibody complex toward the extracellular matrix.
- antibodies of Disclosure A can retain the antigen-binding activity when compared to the antibodies before modification or alteration of at least one amino acid residue to increase pI (native antibodies (for example, native Ig antibodies, preferably native IgG antibodies) or reference antibodies (e.g., antibodies before antibody modification, or prior to or during library construction)).
- “to (substantially) retain the antigen-binding activity” can mean to have an activity of at least 50% or more, preferably 60% or more, more preferably 70% or 75% or more, and still more preferably 80%, 85%, 90%, or 95% or more as compared to the binding activity of the antibodies before modification or alteration.
- antibodies of Disclosure A only need to retain binding activity to a degree that allows them to retain their functions when they bind to antigens; thus, the affinity determined at 37°C under the physiological conditions may be, for example, 100 nM or less, preferably 50 nM or less, more preferably 10 nM or less, and still more preferably 1 nM or less.
- the expression of "modification of at least one amino acid residue that can be exposed on the antibody surface” or an equivalent expression can mean that one or more of addition, deletion, substitution and insertion are performed on at least one amino acid residue that can be exposed on the surface of an antibody. Such modification may preferably include substitution of at least one amino acid residue.
- substitution of amino acid residues can include, for example, substitution of amino acid residues whose side chain has no charge for amino acid residues having a negatively charged side chain, substitution of amino acid residues having a positively charged side chain for amino acid residues whose side chain has no charge, and substitution of amino acid residues having a positively charged side chain for amino acid residues having a negatively charged side chain in the amino acid sequence of an antibody of interest, which can be performed alone or in appropriate combinations.
- the insertion or addition of amino acid residues can include, for example, insertion or addition of amino acids whose side chain has no charge and/or insertion or addition of amino acids having a positively charged side chain in the amino acid sequence of an antibody of interest, which can be performed alone or in appropriate combinations.
- the deletion of amino acid residues can include, for example, deletion of amino acid residues whose side chain has no charge and/or deletion of amino acid residues having a negatively charged side chain in the amino acid sequence of an antibody of interest, which can be performed alone or in appropriate combinations.
- natural amino acids are as follows: an amino acid with a negatively charged side chain can be Glu (E) or Asp (D); an amino acid whose side chain has no charge can be Ala (A), Asn (N), Cys (C), Gln (Q), Gly (G), His (H), Ile (I), Leu (L), Met (M), Phe (F), Pro (P), Ser (S), Thr (T), Trp (W), Tyr (Y), or Val (V); and an amino acid with a positively charged side chain can be His (H), Lys (K), or Arg (R).
- Lys (K) or Arg(R) is preferably selected as an amino acid with a positively charged side chain.
- antibodies of Disclosure A preferably have a variable region and/or a constant region.
- the variable region may preferably have a heavy chain variable region and/or a light chain variable region, and/or may preferably have CDR(s) (for example, one or more of CDR1, CDR2, and CDR3) and/or FR(s) (for example, one or more of FR1, FR2, FR3, and FR4).
- the constant region may preferably have a heavy chain constant region and/or a light chain constant region, and in terms of the sequence and type, it may be, for example, an IgG-type constant region (preferably, human IgG1, human IgG2, human IgG3, or human IgG4-type constant region, human ⁇ chain constant region, and human ⁇ chain constant region). It is also possible to use modified variants of these constant regions.
- IgG-type constant region preferably, human IgG1, human IgG2, human IgG3, or human IgG4-type constant region, human ⁇ chain constant region, and human ⁇ chain constant region. It is also possible to use modified variants of these constant regions.
- the modification of at least one amino acid residue that can be exposed on the antibody surface may be either a modification of a single amino acid or a combination of modifications of multiple amino acids.
- a preferred method can be to introduce a combination of multiple amino acid substitutions at sites where amino acids can be exposed on the antibody surface.
- multiple amino acid substitutions are preferably introduced at positions that are three-dimensionally close to one another.
- amino acids with a positively charged side chain for example, Lys (K) or Arg (R)
- amino acids with a negatively charged side chain for example, Glu (E) or Asp (D)
- amino acids with a negatively charged side chain for example, Glu (E) or Asp (D)
- pre-existing amino acids having a positively charge for example, Lys (K) or Arg (R)
- one or more amino acids which may include amino acids embedded inside the antibody molecule depending on the situation
- amino acids having a positively charge may also be substituted with amino acids having a positively charge to consequently create a dense state of local positive charge in a three-dimensionally proximal location.
- a three-dimensionally proximal location is not particularly limited; but it can mean a state where one or more amino acid substitution is introduced, for example, within 20 Angstroms, preferably within 15 Angstroms, and more preferably within 10 Angstroms. Whether an amino acid substitution site of interest is exposed on the surface of an antibody molecule or whether an amino acid substitution site is close to other amino acid substitution site(s) or the above pre-existing amino acids can be assessed by known methods such as X-ray crystallography.
- methods for giving multiple positive charges at sites three-dimensionally close to one another can include those that use amino acids that originally have a positive charge in the native IgG constant region.
- amino acids include, for example: arginine at positions 255, 292, 301, 344, 355, and 416, according to EU numbering; and lysine at positions 121, 133, 147, 205, 210, 213, 214, 218, 222, 246, 248, 274, 288, 290, 317, 320, 322, 326, 334, 338, 340, 360, 370, 392, 409, 414, and 439, according to EU numbering.
- Multiple positive charges can be given into a three-dimensionally proximal location by substituting with positively charged amino acid(s) at sites three-dimensionally close to these positively charged amino acids.
- antibodies of Disclosure A have a variable region (that may be modified), amino acid residues that are not masked by antigen binding (i.e., that still can be exposed on the surface) may be modified, and/or amino acid modification may not be introduced at sites that are masked by antigen binding or amino acid modification that does not (substantially) inhibit antigen binding may be carried out.
- amino acids of the antigen-binding domain may be modified in such a way that the modification does not (substantially) reduce the binding activity of amino acid residues that can change the antigen-binding activity of the antibody according to the ion concentration condition (for example, those in a calcium-binding motif, or a histidine insertion site and/or a histidine substituted site), or amino acid residues may be modified at sites other than of the amino acid residues that can change the antigen-binding activity of the antibody according to the ion concentration condition.
- the ion concentration condition for example, those in a calcium-binding motif, or a histidine insertion site and/or a histidine substituted site
- the type or the position of amino acid residues that can change the antigen-binding activity of the antibody according to the ion concentration condition may be selected such that the pI of the antibody is not reduced below an acceptable level.
- the pI of an antibody is reduced below an acceptable level, the pI of the overall antibody can be increased by modifying at least one amino acid residue that can be exposed on the surface of the antibody molecule.
- FR sequences with a high pI may be preferably selected from human germline FR sequences or sequences of regions that are equivalent thereto, whose amino acid may be modified in some cases.
- antibodies of Disclosure A have a constant region (that may be modified) having an Fc ⁇ R-binding domain (which may be a binding domain to any of the Fc ⁇ R isoforms and allotypes described below) and/or an FcRn-binding domain
- sites for modification of at least one amino acid residue that can be exposed on the surface of the constant region can be amino acid residues other than those in the Fc ⁇ R-binding domain and/or those in the FcRn-binding domain, if desired.
- modification sites are selected from amino acid residues in the Fc ⁇ R-binding domain and/or in the FcRn-binding domain, it may be preferable to select sites that do not (substantially) affect the binding activity or binding affinity for Fc ⁇ R and/or FcRn, or if they would affect, sites which is biologically or pharmacologically acceptable.
- the site of the at least one amino acid residue that is modified to produce an antibody of Disclosure A whose pI is increased by modification of at least one amino acid residue that can be exposed on the surface of the variable region (that may be modified) is not limited; however, such a site can be selected from the group consisting of, according to Kabat numbering: (a) position 1, 3, 5, 8, 10, 12, 13, 15, 16, 18, 19, 23, 25, 26, 39, 41, 42, 43, 44, 46, 68, 71, 72, 73, 75, 76, 77, 81, 82, 82a, 82b, 83, 84, 85, 86, 105, 108, 110, and 112 in a FR of the heavy chain variable region; (b) position 31, 61, 62, 63, 64, 65, and 97 in a CDR of the heavy chain variable region; (c) position 1, 3, 7, 8, 9, 11, 12, 16, 17, 18, 20, 22, 37, 38, 39, 41, 42, 43, 45, 46, 49, 57, 60, 63
- the following position(s) can be used for aiding in pI increase of an antibody of Disclosure A, in combination with other position(s) which themselves can have sufficient effect of increasing pI of an antibody.
- Such position(s) for aiding in the pI increase can be, for example, as for a light chain variable region, selected from a group consisting of positions 27, 52, 56, 65, and 69, according to Kabat numbering.
- the site of at least one amino acid residue that is modified in the CDR and/or FR is not limited; however, such a site can be selected from the group consisting of: (a) position 8, 10, 12, 13, 15, 16, 18, 23, 39, 41, 43, 44, 77, 82, 82a, 82b, 83, 84, 85, and 105 in the FR of the heavy chain variable region; (b) position 31, 61, 62, 63, 64, 65, and 97 in the CDR of the heavy chain variable region; (c) position 16, 18, 37, 41, 42, 45, 65, 69, 74, 76, 77, 79, and 107 in the FR of the light chain variable region; and(d) position 24, 25, 26, 27, 52, 53, 54, 55, and 56 in the CDR of the light chain variable region. In some embodiments, at least 2, 3, 4, 5, 6, 7, 8, 9, 10 or more than 10 of the above amino acid positions are modified. In some embodiments, 1-20, 1-15, 1-10, or 1-5 of the above
- the type of amino acid after modification in the heavy-chain variable region is, for example: (a) 8K, 8R, 8Q, 8G, 8S, or 8N for position 8; (b) 13K, 13R, 13Q, 13G, 13S, or 13N for position 13; (c) 15K, 15R, 15Q, 15G, 15S, or 15N for position 15; (d) 16K, 16R, 16Q, 16G, 16S, or 16N for position 16; (e) 18K, 18R, 18Q, 18G, 18S, or 18N for position 18; (f) 39K, 39R, 39Q, 39G, 39S, or 39N for position 39; (g) 41K, 41R, 41Q, 41G, 41S, or 41N for position 41; (h) 43K, 43R, 43Q, 43G, 43S, or 43N for position 43; (i) 44K,
- Non-limiting examples of a combination of modified amino acids positions in the heavy-chain variable region is, for example: any two or more of positions selected from the group consisting of positions 16, 43, 64, and 105; any two or more of positions selected from the group consisting of positions 77, 82a, and 82b; positions 77 and 85; positions 41 and 44; positions 82a and 82b; positions 82 and 82b; positions 82b and 83; or positions 63 and 64, according to Kabat numbering, wherein an amino acid at each position after modification can be selected from any of the amino acids described above in terms of the side-chain charge such as Lys (K), Arg (R), Gln (Q), Gly (G), Ser (S), or Asn (N), but is not limited thereto.
- a specific combination can be, for example, 16Q/43R/64K/105Q; 77R/82aN/82bR; 77R/82aG/82bR; 77R/82aS/82bR; 77R/85G; 41R/44R; 82aN/82bR; 82aG/82bR; 82aS/82bR; 82K/82bR; 82bR/83R; 77R/85R; or 63R/64K.
- the type of amino acid after modification in the light-chain variable region is, for example: (a) 16K, 16R, 16Q, 16G, 16S, or 16N for position 16; (b) 18K, 18R, 18Q, 18G, 18S, or 18N for position 18; (c) 24K, 24R, 24Q, 24G, 24S, or 24N for position 24; (d) 25K, 25R, 25Q, 25G, 25S, or 25N for position 25; (e) 26K, 26R, 26Q, 26G, 26S, or 26N for position 26; (f) 27K, 27R, 27Q, 27G, 27S, or 27N for position 27; (g) 37K, 37R, 37Q, 37G, 37S, or 37N for position 37; (h) 41K, 41R, 41Q, 41G, 41S, or 41N for position 41; (i) 42K, 42R, 42Q, 42G, 42S, or 42N for position 42; (j) 45K, 45R, 45
- Non-limiting examples of a combination of modified amino acids positions in the light-chain variable region is, for example: positions 24 and 27; positions 25 and 26; positions 41 and 42; positions 42 and 76; positions 52 and 56; positions 65 and 79; positions 74 and 77; positions 76 and 79; any two or more of positions selected from the group consisting of positions 16, 24, and 27; any two or more of positions selected from the group consisting of positions 24, 27, and 37; any two or more of positions selected from the group consisting of positions 25, 26, and 37; any two or more of positions selected from the group consisting of positions 27, 76, and 79; any two or more of positions selected from the group consisting of positions 41, 74, and 77; any two or more of positions selected from the group consisting of positions 41, 76, and 79; any two or more of positions selected from the group consisting of positions 24, 27, 41, and 42; any two or more of positions selected from the group consisting of positions 24, 27, 52, and 56; any two or more of positions selected from the group consisting of positions 24, 27,
- a specific combination can be, for example, 24R/27Q; 24R/27R; 24K/27K; 25R/26R; 25K/26K; 41R/42K; 42K/76R; 52R/56R; 65R/79K; 74K/77R; 76R/79K; 16K/24R/27R; 24R/27R/37R; 25R/26R/37R; 27R/76R/79K; 41R/74K/77R; 41R/76R/79K; 24R/27R/41R/42K; 24R/27R/52R/56R; 24R/27R/52K/56K; 24R/27R/65R/69R; 24R/27R/74K/77R; 24R/27R/76R/79K; 25R/26R/52R/56R; 25R/26R/52K/56K; 25R/26R/65R/69R; 25R/26R/76R/79K; 27R/
- WO2009/041643 specifically shows that in the heavy-chain FR of a humanized glypican 3 antibody as shown in SEQ ID NO:8, preferred modification sites of amino acid residues that can be exposed on the surface are positions 1, 3, 5, 8, 10, 12, 13, 15, 16, 19, 23, 25, 26, 39, 42, 43, 44, 46, 69, 72, 73, 74, 76, 77, 82, 85, 87, 89, 90, 107, 110, 112, and 114 according to Kabat numbering. It also reports that the amino acid residue at position 97 according to Kabat numbering is preferred because it is exposed on the surface of almost all antibodies.
- WO2009/041643 also shows that the amino acid residues of positions 52, 54, 62, 63, 65, and 66 in the heavy-chain CDR of the antibody are preferred. It also shows that the amino acid residues of positions 1, 3, 7, 8, 9, 11, 12, 16, 17, 18, 20, 22, 43, 44, 45, 46, 48, 49, 50, 54, 62, 65, 68, 70, 71, 73, 74, 75, 79, 81, 82, 84, 85, 86, 90, 105, 108, 110, 111, and 112 according to Kabat numbering in the light-chain FR of a humanized glypican 3 antibody as shown in SEQ ID NO:9 are preferred.
- amino acid residues of positions 24, 27, 33, 55, 59 in the light-chain CDR of this antibody are preferred.
- WO2009/041643 specifically shows that the amino acid residues of positions 31, 64, and 65 according to Kabat numbering in the heavy-chain CDR of an anti-human IL-6 receptor antibody as shown in SEQ ID NO:10 are preferred sites that allow modification of amino acid residues that can be exposed on the surface while maintaining the antigen-binding activity. It also shows that the amino acid residues of positions 24, 27, 53, and 55 according to Kabat numbering in the light chain CDR of an anti-human IL-6 receptor antibody as shown in SEQ ID NO:11 are preferred.
- amino acid residue of position 31 according to Kabat numbering in the heavy-chain CDR of an anti-human IL-6 receptor antibody as shown in SEQ ID NO:12 is a preferred site that allows modification of amino acid residue that can be exposed on the surface while maintaining the antigen-binding activity. It also shows that the amino acid residues of positions 24, 53, 54, and 55 according to Kabat numbering in the light-chain CDR of an anti-human IL-6 receptor antibody as shown in SEQ ID NO:13 are preferred.
- WO2009/041643 also shows that the amino acid residues of positions 61, 62, 64, and 65 according to Kabat numbering in the heavy-chain CDR of an anti-human glypican 3 antibody as shown in SEQ ID NO:14 are preferred sites that allow modification of amino acid residues that can be exposed on the surface while maintaining the antigen-binding activity. It also shows that the amino acid residues of positions 24 and 27 according to Kabat numbering in the light-chain CDR of an anti-human glypican 3 antibody as shown in SEQ ID NO:15 are preferred.
- amino acid residues of positions 61, 62, 64, and 65 according to Kabat numbering in the heavy-chain CDR of an anti-human IL-31 receptor antibody as shown in SEQ ID NO:16 are preferred sites that allow modification of amino acid residues that can be exposed on the surface while maintaining the antigen-binding activity.
- WO2009/041643 also shows that the amino acid residues of positions 24 and 54 according to Kabat numbering in the light-chain CDR of an anti-human IL-31 receptor antibody as shown in SEQ ID NO:17 are preferred.
- WO2007/114319 reports that antibodies hA69-PF, hA69-p18, hA69-N97R, hB26-F123e4, hB26-p15, and hB26-PF, which were produced by modifying the charge of one or more amino acid residues that can be exposed on the surface, showed changes in pI as demonstrated by isoelectric focusing, and had an equivalent binding activity to Factor IXa or Factor X, which are their antigens, compared with that of the antibodies before modification or alteration. It also reports that when these antibodies were administered to mice, the pI of each antibody showed high correlation with their clearance (CL) in plasma, retention time in plasma, and half-life in plasma (T1/2). WO2007/114319 also demonstrates that amino acid residues of positions 10, 12, 23, 39, 43, 97, and 105 in the variable region are preferred as sites for modification of amino acid residues that can be exposed on the surface.
- amino acid residues that can be exposed on the surface of an antibody constant region may be identified to determine the modification sites of at least one amino acid residue for producing an antibody of Disclosure A whose pI has been increased.
- the modification site of at least one amino acid residue that can be exposed on the surface of the constant region is not limited; however, the site can be preferably selected from the group consisting of: position 196, 253, 254, 256, 257, 258, 278, 280, 281, 282, 285, 286, 306, 307, 308, 309, 311, 315, 327, 330, 342, 343, 345, 356, 358, 359, 361, 362, 373, 382, 384, 385, 386, 387, 388, 389, 399, 400, 401, 402, 413, 415, 418, 419, 421, 424, 430, 433, 434, and position 443, according to EU numbering, and may be preferably selected from the group consisting of: position 254, 258, 281, 282, 285, 309, 311, 315, 327, 330, 342, 343, 345, 356, 358, 359, 361, 362, 384, 385, 386, 387, 389, 399, 400
- the type of amino acid after modification at each site can be as follows: 254K, 254R, 254Q, or 254N at position 254; 258K, 258R, 258Q, or 258N at position 258; 281K, 281R, 281Q, or 281N at position 281; 282K, 282R, 282Q, or 282N at position 282; 285K, 285R, 285Q, or 285N at position 285; 309K, 309R, 309Q, or 309N at position 309; 311K, 311R, 311Q, or 311N at position 311; 315K, 315R, 315Q, or 315N at position 315; 327K, 327R, 327Q, or 327N at position 327; 330K, 330R, 330Q, or 330N at position
- the modification site of at least one amino acid residue and the type of amino acid after modification may include 345R or 345K, and/or 430R, 430K, 430G, or 435T, according to EU numbering.
- the antibody's net pI may be increased by modifying at least one amino acid residue that can be exposed on the surface of the variable region (which may be modified) as described above and at least one amino acid residue that can be exposed on the surface of the constant region (which may be modified) as described above.
- the antibody heavy-chain constant region may contain a constant region of the IgG1 type, IgG2 type, IgG3 type, or IgG4 type.
- the heavy-chain constant region may be a human heavy-chain constant region, but is not limited thereto.
- Several allotypes are known to exist for human IgG. Specifically, it has been reported that there are some differences in the amino acid sequence of the human IgG constant region among individuals (Methods Mol. Biol. 882:635-80 (2012); Sequences of proteins of immunological interest, NIH Publication No.91-3242). Examples include human IgG1 constant region (SEQ ID NO:18), human IgG2 constant region (SEQ ID NO:19), human IgG3 constant region (SEQ ID NO:20), and human IgG4 constant region (SEQ ID NO:21).
- G1m1,17 and G1m3 are known for human IgG1.
- the allotypes differ in their amino acid sequences: G1m1,17 has aspartic acid at position 356 and leucine at position 358 according to EU numbering, while G1m3 has glutamic acid at position 356 and methionine at position 358 according to EU numbering.
- G1m1,17 has aspartic acid at position 356 and leucine at position 358 according to EU numbering
- G1m3 has glutamic acid at position 356 and methionine at position 358 according to EU numbering.
- the light-chain constant region of an antibody can include any constant region of the ⁇ chain (IgK) type or ⁇ chain (IgL1, IgL2, IgL3, IgL6, or IgL7) type.
- a light-chain constant region may be preferably a human light-chain constant region, but is not limited thereto.
- Such allotypes include, for example, human ⁇ chain constant region (SEQ ID NO:22) and human ⁇ chain constant region (SEQ ID NO:23).
- the Fc region of a native IgG antibody constitutes a part of the constant region of the native IgG antibody
- the antibodies may have an Fc region contained in the constant region of a native IgG (IgG1, IgG2, IgG3, or IgG4 type) (hereinafter, also collectively referred to as a native (human) IgG (type) Fc region).
- the Fc region of a native IgG can refer to an Fc region consisting of the same amino acid sequence as an Fc region originating from a naturally occurring IgG.
- Fc region of a native human IgG can include the Fc regions contained in the human IgG1 constant region (SEQ ID NO:18), human IgG2 constant region (SEQ ID NO:19), human IgG3 constant region (SEQ ID NO:20), or human IgG4 constant region (SEQ ID NO:21) described above (an Fc region of the IgG class can refer to, for example, from cysteine of position 226 according to EU numbering to the C terminus, or from proline of position 230 according to EU numbering to the C terminus.).
- antibodies of Disclosures A and B may include variants in which one or more modifications selected from amino acid substitution, addition, deletion, or insertion have been made to the constant region of a native (preferably human) IgG (the heavy-chain constant region and/or the light-chain constant region) or in the Fc region of a native (preferably human) IgG.
- WO2013/081143 reports that for example, ion concentration-dependent antibodies capable of forming multivalent immune complexes with a multimeric antigen (multivalent antigen-antibody complexes) and multispecific ion concentration-dependent antibodies or multiparatopic ion concentration-dependent antibodies that can form multivalent immune complexes (multivalent antigen-antibody complexes) by recognizing two or more epitopes on monomeric antigens can bind more strongly to Fc ⁇ R, FcRn, complement receptor, due to the avidity (sum of the strength of binding between multiple epitopes and multiple paratopes) via at least two or more multivalent constant regions (that may be modified) or Fc regions (that may be modified) contained in the antibody molecules, and as a result the antibodies are more rapidly taken up into cells.
- the ion concentration-dependent antibodies described above which are capable of forming multivalent immune complexes with a multimeric antigen or monomeric antigens, can also be used as antibodies of Disclosure A (ion concentration-dependent antibodies with increased pI).
- antibodies of Disclosure A ion concentration-dependent antibodies with increased pI.
- the ion concentration-dependent antibodies with increased pI that can form multivalent immune complexes with a multimeric antigen or monomeric antigens can be more rapidly taken up into cells, as compared to ion concentration-dependent antibodies with increased pI that are incapable of forming multivalent immune complexes.
- the activity of antibodies of Disclosure A to bind to FcRn and/or Fc ⁇ R may be increased under a neutral pH condition and in this case, the ion concentration-dependent antibodies with increased pI that can form multivalent immune complexes with a multimeric antigen or monomeric antigens may be even more rapidly taken up into cells.
- antibodies of Disclosure A may be one-armed antibodies (including all embodiments of the one-armed antibodies described in WO2005/063816).
- one-armed antibodies are antibodies that lack one of the two Fab regions an ordinary IgG antibody has, and can be produced, without limitations, for example, by the methods described in WO2005/063816.
- an IgG-type antibody that has a heavy chain whose structure is, for example, VH-CH1-Hinge-CH2-CH3, when one of the Fab regions is cleaved at a site more to the N terminus than the Hinge (for example, VH or CH1), the antibody will be expressed in a form containing an extra sequence, and when one of the Fab regions is cleaved at a site more to the C terminus than the Hinge (for example, CH2), the Fc region will have an incomplete form.
- the heavy chain after cleavage is linked to the uncleaved heavy chain via intramolecular disulfide bond.
- WO2005/063816 has reported that such one-armed antibodies have an increased stability as compared to Fab molecules.
- Antibodies with an increased or decreased pI can also be generated by preparing such one-armed antibodies.
- the antibody half-life in plasma can be further shortened, cellular uptake of the antibody can be further enhanced, antigen elimination from plasma can be further enhanced, or the antibody's affinity for the extracellular matrix can be further increased, as compared to antibodies with increased pI that do not have an ion concentration-dependent antigen-binding domain.
- an embodiment where the cellular uptake-accelerating effect of one-armed antibodies is expected can be envisaged to be, but is not limited to, a case in which the pI of a soluble antigen is lower than that of the antibodies.
- the net pI of a complex consisting of antibodies and antigens can be calculated by known methods by considering that the complex is a single molecule. In this case, the lower the pI of the soluble antigen is, the lower the net pI of the complex is; and the greater the pI of the soluble antigen is, the greater the net pI of the complex is.
- the Fab of an ordinary IgG-type antibody molecule has a lower pI than that of the Fc
- conversion into a one-armed antibody increases the net pI of the complex consisting of the one-armed antibody and antigen.
- the pI can be expected to be effectively increased by selecting a site which would increase the pI of the one-armed antibody to the desired extent.
- the pI of an antibody can be increased and the accompanying cellular uptake of the antigen may be accelerated by converting the antibody into a one-armed antibody by calculating the theoretical pI of the antibody (theoretical pI of Fc and theoretical pI of Fab) and the theoretical pI of the soluble antigen and predicting the relationship on the difference of their theoretical pI values.
- antibodies of Disclosure A or B may be multispecific antibodies, and the multispecific antibody may be, but is not limited to, a bispecific antibody.
- the multispecific antibody may be a multispecific antibody that contains a first polypeptide and a second polypeptide.
- a multispecific antibody that contains a first polypeptide and a second polypeptide refers to an antibody that binds to at least two or more types of different antigens or at least two or more types of epitopes in a same antigen.
- the first polypeptide and second polypeptide preferably may contain a heavy-chain variable region, and more preferably the variable region contains CDR(s) and/or FR(s).
- the first polypeptide and second polypeptide may preferably each contain a heavy-chain constant region.
- the multispecific antibody may contain a third polypeptide and a fourth polypeptide, each containing a light-chain variable region and preferably also a light-chain constant region. In this case, the first to the fourth polypeptides may assemble together to form a multispecific antibody.
- antibodies of Disclosure A are multispecific antibodies and the multispecific antibodies contain a heavy-chain constant region, to reduce their pI, for example, the following sequences may be used: IgG2 or IgG4 sequence at position 137; IgG1, IgG2, or IgG4 sequence at position 196; IgG2 or IgG4 sequence at position 203; IgG2 sequence at position 214; IgG1, IgG3, or IgG4 sequence at position 217; IgG1, IgG3, or IgG4 sequence at position 233; IgG4 sequence at position 268; IgG2, IgG3, or IgG4 sequence at position 274; IgG1, IgG2, or IgG4 sequence at position 276; IgG4 sequence at position 355; IgG3 sequence at position 392; IgG4 sequence at position 419; or IgG1, IgG2, or IgG4 sequence at position 435.
- the pIs of the two heavy chain constant regions may be the same or different from each other.
- Such heavy-chain constant regions may be IgG1, IgG2, IgG3 and IgG4 heavy-chain constant regions which originally have different pIs.
- Modification sites of at least one amino acid residue for introducing such a pI difference in the constant region may be the position(s) described above or position(s) selected, for example, from the group consisting of position 137, position 196, position 203, position 214, position 217, position 233, position 268, position 274, position 276, position 297, position 355, position 392, position 419, and position 435, according to EU numbering in the heavy-chain constant region as described in WO2009/041643.
- the amino acid residue of position 297 which is a glycosylation site may be modified to remove the sugar chain, since the removal of a sugar chain from the heavy-chain constant region results in a pI difference.
- antibodies of Disclosure A or B may be polyclonal antibodies or monoclonal antibodies, and mammalian-derived monoclonal antibodies are preferred.
- Monoclonal antibodies include those produced by hybridomas or those produced by host cells transformed by genetic engineering techniques with expression vectors carrying antibody genes.
- the antibodies of Disclosure A or B may be, for example, antibodies such as chimeric antibodies, humanized antibodies, or antibodies generated by affinity maturation, or molecules derived therefrom.
- antibodies of Disclosure A or B may be derived, without limitations, from any animal species (for example, human; or nonhuman animals such as mouse, rat, hamster, rabbit, monkey, cynomolgus monkey, Rhesus monkey, hamadryas baboon, chimpanzee, goat, sheep, dog, bovine, or camel), or any birds; and the antibodies are preferably derived from human, monkey, or mouse.
- animal species for example, human; or nonhuman animals such as mouse, rat, hamster, rabbit, monkey, cynomolgus monkey, Rhesus monkey, hamadryas baboon, chimpanzee, goat, sheep, dog, bovine, or camel
- the antibodies are preferably derived from human, monkey, or mouse.
- antibodies of Disclosure A or B may be Ig-type antibodies, and may be preferably IgG-type antibodies.
- the Fc receptor refers to a receptor protein that can bind to the Fc region of an immunoglobulin (antibody) or a molecule derived therefrom, or an Fc region variant.
- Fc receptors for IgG, IgA, IgE, and IgM are known as Fc ⁇ R, Fc ⁇ R, Fc ⁇ R, and Fc ⁇ R, respectively, within the scope of Disclosure A described herein.
- Fc receptors may also be, for example, FcRn (also referred to as "neonatal Fc receptor”), within the scope of Disclosures A and B described herein.
- Fc ⁇ R may refer to a receptor protein that can bind to the Fc region of an IgG1, IgG2, IgG3, or IgG4 antibody or a molecule derived therefrom, or an Fc region variant, and may include any one or more of, or all members of the family of proteins substantially encoded by the Fc ⁇ R gene.
- the family includes, but is not limited to, Fc ⁇ RI (CD64) including isoforms Fc ⁇ RIa, Fc ⁇ RIb, and Fc ⁇ RIc; Fc ⁇ RII (CD32) including isoforms Fc ⁇ RIIa (including allotypes H131 (type H) and R131 (type R)), Fc ⁇ RIIb (including Fc ⁇ RIIb-1 and Fc ⁇ RIIb-2), and Fc ⁇ RIIc; and Fc ⁇ RIII (CD16) including isoforms Fc ⁇ RIIIa (including allotypes V158 and F158) and Fc ⁇ RIIIb (including allotypes Fc ⁇ RIIIb-NA1 and Fc ⁇ RIIIb-NA2), as well as all unidentified human Fc ⁇ Rs and Fc ⁇ R isoforms and allotypes.
- Fc ⁇ RI CD64
- Fc ⁇ RII CD32
- Fc ⁇ RIIa including allotypes H131 (type H) and R131 (type R)
- Fc ⁇ RIIb1 and Fc ⁇ RIIb2 have been reported as splicing variants of human Fc ⁇ RIIb (hFc ⁇ RIIb).
- Fc ⁇ RIIb3 a splicing variant called Fc ⁇ RIIb3 (Brooks et al., J. Exp. Med, 170: 1369-1385 (1989)).
- hFc ⁇ RIIb includes all splicing variants such as those registered in NCBI under NP_001002273.1, NP_001002274.1, NP_001002275.1, NP_001177757.1, and NP_003992.3.
- hFc ⁇ RIIb also includes all genetic polymorphisms already reported, for example, Fc ⁇ RIIb (Li et al., Arthritis Rheum. 48:3242-3252 (2003), Kono et al., Hum. Mol. Genet. 14:2881-2892 (2005); Kyogoku et al., Arthritis Rheum. 46(5):1242-1254 (2002)), as well as all genetic polymorphisms that will be reported in future.
- Fc ⁇ R may be derived from any organism, and may include those derived from humans, mice, rats, rabbits, or monkeys, without being limited thereto.
- Mouse Fc ⁇ Rs include, but are not limited to, Fc ⁇ RI (CD64), Fc ⁇ RII (CD32), Fc ⁇ RIII (CD16) and Fc ⁇ RIII-2 (CD16-2), as well as all unidentified mouse Fc ⁇ Rs, and Fc ⁇ R isoforms and allotypes.
- Such preferred Fc ⁇ R includes, for example, human Fc ⁇ RI (CD64), Fc ⁇ RIIA (CD32), Fc ⁇ RIIB (CD32), Fc ⁇ RIIIA (CD16), or Fc ⁇ RIIIB (CD16). Since Fc ⁇ R is present as a membrane form in vivo, it may be used in experimental systems after being artificially converted into an appropriate soluble form.
- the polynucleotide sequence and amino acid sequence of Fc ⁇ RI may be the sequences shown in NM_000566.3 and NP_000557.1, respectively; the polynucleotide sequence and amino acid sequence of Fc ⁇ RIIA may be the sequences shown in BC020823.1 and AAH20823.1, respectively; the polynucleotide sequence and amino acid sequence of Fc ⁇ RIIB may be the sequences shown in BC146678.1 and AAI46679.1, respectively; the polynucleotide sequence and amino acid sequence of Fc ⁇ RIIIA may be the sequences shown in BC033678.1 and AAH33678.1, respectively; the polynucleotide sequence and amino acid sequence of Fc ⁇ RIIIB may be the sequences shown in BC128562.1 and AAI28563.1, respectively (RefSeq accession numbers are shown).
- Fc ⁇ RIIa has two genetic polymorphisms, in which the amino acid at position 131 of Fc ⁇ RIIa is replaced with histidine (type H) or arginine (type R) (J. Exp. Med. 172:19-25, 1990).
- Fc ⁇ RI which includes Fc ⁇ RIa, Fc ⁇ RIb, and Fc ⁇ RIc
- Fc ⁇ RIII CD16 which includes Fc ⁇ RIIIa (including allotypes V158 and F158)
- the ⁇ chain that binds to the Fc region of IgG is associated with a common ⁇ chain having ITAM which transmits activation signals inside cells.
- Fc ⁇ RIIIb (including allotypes Fc ⁇ RIIIb-NA1 and Fc ⁇ RIIIb-NA2) is a GPI anchor protein.
- the cytoplasmic domain of Fc ⁇ RII CD32 which includes the Fc ⁇ RIIa (including allotypes H131 and R131) and Fc ⁇ RIIc isoforms contains ITAM.
- Fc ⁇ R that has the ability to transduce activation signals as described above is also referred to as an activating Fc ⁇ R within the scope of Disclosures A and B described here.
- the cytoplasmic domain of Fc ⁇ RIIb contains ITIM which transmits inhibitory signals.
- the crosslinking between Fc ⁇ RIIb and B cell receptor (BCR) suppresses the activation signals from BCR, which results in suppression of antibody production by BCR.
- BCR B cell receptor
- the crosslinking of Fc ⁇ RIII and Fc ⁇ RIIb suppresses the phagocytic ability and the ability to produce inflammatory cytokines.
- An Fc ⁇ R that has the ability to transduce inhibitory signals as described above is also referred to as an inhibitory Fc ⁇ Receptor within the scope of Disclosures A and B described herein.
- ELISA and fluorescence activated cell sorting FACS
- ALPHA screen Amplified Luminescent Proximity Homogeneous Assay
- the extracellular domain of human Fc ⁇ R may be used as a soluble antigen (for example, WO2013/047752).
- an acidic or neutral pH condition may suitably be used.
- the binding activity (binding affinity) between an Fc ⁇ R-binding domain and Fc ⁇ R may be assessed, for example, at any temperature between 10°C to 50°C.
- a preferred temperature for determining the binding activity (binding affinity) of a human Fc ⁇ R-binding domain to Fc ⁇ R is, for example, 15°C to 40°C.
- any temperature from 20°C to 35°C for example, such as any one of 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, and 35°C may be used.
- a non-limiting example of such temperature is 25°C.
- the constant region may have an Fc region or an Fc region variant (preferably, a human Fc region or a human Fc region variant), and preferably has an Fc ⁇ R-binding domain within the scope of Disclosure A and an FcRn-binding domain within the scope of Disclosures A and B described herein.
- an antibody of Disclosure A may have an Fc ⁇ R-binding domain, preferably a human Fc ⁇ R-binding domain.
- the Fc ⁇ R-binding domain is not particularly limited as long as the antibody has binding activity to or affinity for Fc ⁇ R at acidic pH and/or neutral pH, and it may be a domain that has an activity to directly or indirectly bind Fc ⁇ R.
- an antibody of Disclosure A has Fc ⁇ R-binding activity
- the Fc ⁇ R-binding activity of the antibody under a neutral pH condition is increased as compared to that of a reference antibody which contains a native IgG constant region.
- the antibody of Disclosure A and the reference antibody which contains a native IgG constant region have identical amino acid sequences in regions (for example, the variable region) other than, preferably, the constant region of the antibody of Disclosure A which has been modified at one or more amino acid residues.
- an antibody of Disclosure A has an Fc ⁇ R-binding activity or an increased Fc ⁇ R-binding activity under a neutral pH condition (e.g., pH 7.4)
- a neutral pH condition e.g., pH 7.4
- the antibody is thought to possess the following properties in combination: the property of being shuttled between plasma and cellular endosome and repeatedly binding to multiple antigens as a single antibody molecule by having an ion concentration-dependent antigen-binding domain; the property of being rapidly taken up into cells by having an increased pI and increased positive charge in the overall antibody; and the property of being rapidly taken up into cells by having an increased Fc ⁇ R-binding activity under a neutral pH condition.
- the antibody half-life in plasma can be further shortened, or the binding activity of the antibody toward the extracellular matrix can be further increased, or antigen elimination from plasma can be further promoted; thus the antibody of Disclosure A is beneficial.
- Those of ordinary skill in the art can routinely determine an optimal pI value for the antibody to take advantage of these properties.
- an Fc ⁇ R-binding domain whose Fc ⁇ R-binding activity is higher than that of the Fc region or constant region of a native human IgG in which the sugar chain linked at position 297 according to EU numbering is a fucose-containing sugar chain can be produced by modifying amino acid residues in the Fc region or constant region of a native human IgG (see WO2013/047752).
- a domain of any structure that binds to Fc ⁇ R can be used as an Fc ⁇ R-binding domain.
- the Fc ⁇ R-binding domain can be produced without the need to introduce an amino acid modification, and alternatively, its affinity for Fc ⁇ R may be increased by introducing an additional modification.
- Such Fc ⁇ R-binding domains can include Fab fragment antibodies that bind to Fc ⁇ RIIIa, camel-derived single domain antibodies, and single chain Fv antibodies described in Schlapschly et al.(Protein Eng. Des. Sel. 22 (3):175-188 (2009), Behar et al. (Protein Eng. Des. Sel. 21(1):1-10 (2008)), and Kipriyanov et al., J Immunol. 169(1):137-144 (2002), and the Fc ⁇ RI-binding cyclic peptide described in Bonetto et al., FASEB J. 23(2):575-585 (2009).
- Fc ⁇ R-binding activity of an Fc ⁇ R-binding domain is higher than that of the Fc region or constant region of a native human IgG in which the sugar chain linked at position 297 according to EU numbering is a fucose-containing sugar chain can be appropriately assessed using the methods described above.
- the starting Fc ⁇ R-binding domain preferably includes, for example, (human) IgG Fc region or (human) IgG constant region.
- any Fc region or constant region can be used as the starting Fc region or starting constant region.
- An Fc region or constant region obtained by further modifying a starting Fc region or starting constant region whose amino acid residue(s) has been already modified from an Fc region or constant region can also be appropriately used as the Fc region or constant region of Disclosure A.
- a starting Fc region or starting constant region may refer to the polypeptide itself, a composition containing the starting Fc region or starting constant region, or an amino acid sequence encoding the starting Fc region or starting constant region.
- the starting Fc region or starting constant region may include known Fc regions or known constant regions produced by recombination technologies.
- the origin of the starting Fc region or starting constant region is not limited, and it can be obtained from any organism of nonhuman animals or from a human.
- the starting Fc ⁇ R-binding domain can be obtained from cynomolgus monkeys, marmosets, Rhesus monkeys, chimpanzees, or humans.
- the starting Fc region or starting constant region can be preferably obtained from human IgG1; however, it is not limited to a particular IgG class. This means that the Fc region of human IgG1, IgG2, IgG3, or IgG4 can be used as an appropriate starting Fc ⁇ R-binding domain, and it also means that within the scope of Disclosure A described herein, an Fc region or constant region of an IgG class or subclass derived from any organism can be preferably used as the starting Fc region or starting constant region. Examples of a native IgG variant or modified form are described in publicly known literature such as Strohl, Curr. Opin. Biotechnol. 20(6):685-691 (2009); Presta, Curr. Opin. Immunol.
- amino acid residues of the starting Fc ⁇ R-binding domain, starting Fc region, or starting constant region may contain, for example, one or more mutations: for example, substitutions with amino acid residues that are different from those in the starting Fc region or starting constant region; insertions of one or more amino acid residues into the amino acid residues in the starting Fc region or starting constant region; or deletions of one or more amino acid residues from those of the starting Fc region or starting constant region.
- the amino acid sequences of Fc regions or constant regions after modifications are preferably amino acid sequences containing at least a portion of an Fc region or constant region that may not occur naturally. Such variants necessarily have a sequence identity or similarity of less than 100% to the starting Fc regions or starting constant regions.
- the variants have an amino acid sequence identity or similarity of about 75% to less than 100%, more preferably about 80% to less than 100%, even more preferably about 85% to less than 100%, still more preferably about 90% to less than 100%, and yet more preferably about 95% to less than 100% to the amino acid sequence of the starting Fc region or starting constant region.
- at least one amino acid is different between a modified Fc region or constant region of Disclosure A and the starting Fc region or starting constant region.
- an Fc region or constant region that has Fc ⁇ R-binding activity in an acidic pH range and/or in a neutral pH range which may be contained in an antibody of Disclosure A, may be obtained by any method.
- a variant of Fc region or constant region that has Fc ⁇ R-binding activity in a neutral pH range may be obtained by modifying amino acids of a human IgG antibody which can be used as the starting Fc region or starting constant region.
- IgG antibody Fc regions or IgG antibody constant regions suitable for modification can include, for example, the Fc regions or constant regions of human IgG (IgG1, IgG2, IgG3, or IgG4, or variants thereof), and mutants spontaneously generated therefrom.
- human IgG1 For the Fc regions or constant regions of human IgG1, human IgG2, human IgG3, or human IgG4 antibodies, a number of allotype sequences due to genetic polymorphism are described in "Sequences of proteins of immunological interest", NIH Publication No.91-3242, and any of them may be used in Disclosure A.
- the amino acid sequence of positions 356 to 358 according to EU numbering may be DEL or EEM.
- the modification into other amino acids is not limited as long as the variants have an Fc ⁇ R-binding activity in a neutral pH range.
- Amino acid position(s) of such modification are reported, for example, in: WO2007/024249, WO2007/021841, WO2006/031370, WO2000/042072, WO2004/029207, WO2004/099249, WO2006/105338, WO2007/041635, WO2008/092117, WO2005/070963, WO2006/020114, WO2006/116260, WO2006/023403, WO2013/047752, WO2006/019447, WO2012/115241, WO2013/125667, WO2014/030728, WO2014/163101, WO2013/118858, and WO2014/030750.
- Sites of amino acid modification in the constant region or Fc region to increase the Fc ⁇ R-binding activity in a neutral pH range can include, for example, one or more positions selected from the group consisting of position: 221, 222, 223, 224, 225, 227, 228, 230, 231, 232, 233, 234, 235, 236, 237, 238, 239, 240, 241, 243, 244, 245, 246, 247, 249, 250, 251, 254, 255, 256, 258, 260, 262, 263, 264, 265, 266, 267, 268, 269, 270, 271, 272, 273, 274, 275, 276, 278, 279, 280, 281, 282, 283, 284, 285, 286, 288, 290, 291, 292, 293, 294, 295, 296, 297, 298, 299, 300, 301, 302, 303, 304, 305, 311, 313, 315, 317, 318, 320, 322, 323, 324, 325,
- Modification of such amino acid residue may increase the Fc ⁇ R binding of the Fc region or constant region of an IgG antibody under a neutral pH condition.
- WO2013/047752 describes, as preferred modifications in an IgG-type constant region or Fc region, for example, modification of one or more amino acid residues selected from the group consisting of: the amino acid at position 221 to either Lys or Tyr; the amino acid at position 222 to any one of Phe, Trp, Glu, and Tyr; the amino acid at position 223 to any one of Phe, Trp, Glu, and Lys; the amino acid at position 224 to any one of Phe, Trp, Glu, and Tyr; the amino acid at position 225 to any one of Glu, Lys, and Trp ; the amino acid at position 227 to any one of Glu, Gly, Lys, and Tyr; the amino acid at position 228 to any one of Glu, Gly, Lys, and Tyr; the amino acid at position 230 to any one of Ala, Glu, Gly
- the number of amino acids to be modified is not particularly limited, and it is possible to modify an amino acid at only one position or amino acids at two or more positions. Combinations of amino acid modifications at two or more positions are shown in Table 5 of WO2013/047752. Modification of these amino acid residues can also be appropriately introduced into the antibodies of Disclosure A.
- the binding activity of (the Fc ⁇ R-binding domain of) the antibody of Disclosure A toward (human) Fc ⁇ R(s), such as any one or more of Fc ⁇ RI, Fc ⁇ RIIa, Fc ⁇ RIIb, Fc ⁇ RIIIa, and Fc ⁇ RIIIb, may be higher than that of (the Fc region or constant region of) a native IgG or that of a reference antibody containing the starting Fc region or starting constant region.
- the Fc ⁇ R-binding activity of (the Fc ⁇ R-binding domain of) an antibody of Disclosure A may be 55% or more, 60% or more, 65% or more, 70% or more, 75% or more, 80% or more, 85% or more, 90% or more, 95% or more, 100% or more, 105% or more, preferably 110% or more, 115% or more, 120% or more, 125% or more, particularly preferably 130% or more, 135% or more, 140% or more, 145% or more, 150% or more, 155% or more, 160% or more, 165% or more, 170% or more, 175% or more, 180% or more, 185% or more, 190% or more, or 195% or more as compared to the Fc ⁇ R-binding activity of the reference antibody, or 2-fold or more, 2.5-fold or more, 3-fold or more, 3.5-fold or more, 4-fold or more, 4.5-fold or more, 5-fold or more, 7.5-fold or more, 10-fold or more, 20-fold or more,
- the level of increase in the binding activity to an inhibitory Fc ⁇ R (Fc ⁇ RIIb-1 and/or Fc ⁇ RIIb-2) (in a neutral pH range) may be greater than the level of increase in the binding activity to an activating Fc ⁇ R (Fc ⁇ RIa: Fc ⁇ RIb; Fc ⁇ RIc; Fc ⁇ RIIIa including allotype V158; Fc ⁇ RIIIa including allotype F158; Fc ⁇ RIIIb including allotype Fc ⁇ RIIIb-NA1; Fc ⁇ RIIIb including allotype Fc ⁇ RIIIb-NA2; Fc ⁇ RIIa including allotype H131; or Fc ⁇ RIIa including allotype R131).
- an antibody of Disclosure A may have binding activity to Fc ⁇ RIIb (including Fc ⁇ RIIb-1 and Fc ⁇ RIIb-2).
- preferred Fc ⁇ R-binding domains of Disclosure A also include, for example, Fc ⁇ R-binding domains whose binding activity to a specific Fc ⁇ R is greater than the binding activity to other Fc ⁇ R (Fc ⁇ R-binding domains having a selective Fc ⁇ R-binding activity).
- Fc ⁇ R-binding domains having a selective Fc ⁇ R-binding activity are used, a single antibody molecule can bind only to a single Fc ⁇ R molecule.
- a single antibody molecule in a state bound to an inhibitory Fc ⁇ R cannot bind to other activating Fc ⁇ Rs, and a single antibody molecule in a state bound to an activating Fc ⁇ R cannot bind to other activating Fc ⁇ Rs or inhibitory Fc ⁇ Rs.
- an activating Fc ⁇ R preferably includes, for example, Fc ⁇ RI (CD64) such as Fc ⁇ RIa, Fc ⁇ RIb, or Fc ⁇ RIc; and Fc ⁇ RIII (CD16) such as Fc ⁇ RIIIa (such as allotype V158 or F158) or Fc ⁇ RIIIb (such as allotype Fc ⁇ RIIIb-NA1 or Fc ⁇ RIIIb-NA2).
- an inhibitory Fc ⁇ R preferably includes, for example, Fc ⁇ RIIb (such as Fc ⁇ RIIb-1 or Fc ⁇ RIIb-2).
- Fc ⁇ R-binding domains that have a greater binding activity to inhibitory Fc ⁇ R than to activating Fc ⁇ R can be used as the selective Fc ⁇ R-binding domain contained in an antibody of Disclosure A.
- Such selective Fc ⁇ R-binding domains can include, for example, Fc ⁇ R-binding domains that have a greater binding activity to Fc ⁇ RIIb (such as Fc ⁇ RIIb-1 and/or Fc ⁇ RIIb-2) than to any one or more of activating Fc ⁇ R selected from the group consisting of: Fc ⁇ RI (CD64) such as Fc ⁇ RIa, Fc ⁇ RIb, or Fc ⁇ RIc; Fc ⁇ RIII (CD16) such as Fc ⁇ RIIIa (such as allotype V158 or F158) or Fc ⁇ RIIIb (such as Fc ⁇ RIIIb-NA1 or Fc ⁇ RIIIb-NA2); Fc ⁇ RII (CD32) such as Fc ⁇ RIIa (including allotype H131 or R131);
- Fc ⁇ R-binding domain has a selective binding activity
- the KD value for activating Fc ⁇ R refers to the KD value for one or more of: Fc ⁇ RIa; Fc ⁇ RIb; Fc ⁇ RIc; Fc ⁇ RIIIa including allotype V158 and/or F158; Fc ⁇ RIIIb including Fc ⁇ RIIIb-NA1 and/or Fc ⁇ RIIIb-NA2; Fc ⁇ RIIa including allotype H131 and/or R131; and Fc ⁇ RIIc; and the KD value for inhibitory Fc ⁇ R refers to the KD value for Fc ⁇ RIIb-1 and/or Fc ⁇ RIIb-2.
- the activating Fc ⁇ R and inhibitory Fc ⁇ R for use in determining the KD values may be selected in any combination.
- the Fc ⁇ R selectivity index can be, for example: 1.2 or greater, 1.3 or greater, 1.4 or greater, 1.5 or greater, 1.6 or greater, 1.7 or greater, 1.8 or greater, 1.9 or greater, 2 or greater, 3 or greater, 5 or greater, 6 or greater, 7 or greater, 8 or greater, 9 or greater, 10 or greater, 15 or greater, 20 or greater, 25 or greater, 30 or greater, 35 or greater, 40 or greater, 45 or greater, 50 or greater, 55 or greater, 60 or greater, 65 or greater, 70 or greater, 75 or greater, 80 or greater, 85 or greater, 90 or greater, 95 or greater, 100 or greater, 110 or greater, 120 or greater, 130 or greater, 140 or greater, 150 or greater, 160 or greater, 170 or greater, 180 or greater, 190 or greater, 200 or greater, 210 or greater, 220 or greater, 230 or greater, 240 or greater, 250 or greater, 260 or greater, 270 or greater, 280 or greater, 290 or greater, 300 or greater, 310 or greater, 320
- an antibody containing) an Fc region variant or constant region variant in which the amino acid at position 238 or 328, according to EU numbering of human IgG (IgG1, IgG2, IgG3, or IgG4) is Asp or Glu, respectively can be preferably used as antibodies of Disclosure A containing an Fc region variant or constant region variant, since as specifically described in WO2013/125667, WO2012/115241, and WO2013/047752, it has a greater binding activity to Fc ⁇ RIIb-1 and/or Fc ⁇ RIIb-2 than to Fc ⁇ RIa, Fc ⁇ RIb, Fc ⁇ RIc, Fc ⁇ RIIIa including allotype V158, Fc ⁇ RIIIa including allotype F158, Fc ⁇ RIIIb including allotype Fc ⁇ RIIIb-NA1, Fc ⁇ RIIIb including allotype Fc ⁇ RIIIb-NA2, Fc ⁇ RIIa including allotype H131, Fc ⁇ RIIa including allotype R131, and/
- the antibodies of Disclosure A have binding activity to all activating Fc ⁇ Rs (herein, which are selected from the group consisting of Fc ⁇ RIa, Fc ⁇ RIb, Fc ⁇ RIc, Fc ⁇ RIIIa, Fc ⁇ RIIIb, Fc ⁇ RIIa) and Fc ⁇ RIIb, and their Fc ⁇ RIIb-binding activity is maintained or increased, and/or their binding activity to all activating Fc ⁇ Rs is reduced, as compared to the reference antibody that contains a native IgG constant region or a native IgG Fc region.
- the antibodies of Disclosure A containing an Fc region variant or constant region variant may be maintained or increased and their binding activity to Fc ⁇ RIIa (type H) and Fc ⁇ RIIa (type R) may be reduced as compared to those of a reference antibody having the constant region or Fc region of a native IgG.
- Such antibodies may have increased binding selectivity to Fc ⁇ RIIb over Fc ⁇ RIIa.
- the extent that the "binding activity to all activating Fc ⁇ Rs is reduced" can be, but is not limited to, 99% or less, 98% or less, 97% or less, 96% or less, 95% or less, 94% or less, 93% or less, 92% or less, 91%or less, 90% or less, 88% or less, 86% or less, 84% or less, 82% or less, 80% or less, 78% or less, 76% or less, 74% or less, 72% or less, 70% or less, 68% or less, 66% or less, 64% or less, 62% or less, 60% or less, 58% or less, 56% or less, 54% or less, 52% or less, 50% or less, 45% or less, 40% or less, 35% or less, 30% or less, 25% or less, 20% or less, 15% or less, 10% or less, 5% or less, 4% or less, 3% or less, 2% or less, 1% or less, 0.5% or less, 0.4% or less, 0.
- the extent that the "Fc ⁇ RIIb-binding activity is maintained or increased”, the “binding activity to Fc ⁇ RIIb is maintained or increased", or the “maintained or increased binding activity to Fc ⁇ RIIb” can be, but is not limited to, 55% or greater, 60% or greater, 65% or greater, 70% or greater, 75% or greater, 80% or greater, 85% or greater, 87% or greater, 88% or greater, 89% or greater, 90% or greater, 91% or greater, 92% or greater, 93% or greater, 94% or greater, 95% or greater, 96% or greater, 97% or greater, 98% or greater, 99% or greater, 99.5% or greater, 100% or greater, 101% or greater, 102% or greater, 103% or greater, 104% or greater, 105% or greater, 106% or greater, 107% or greater, 108% or greater, 109% or greater, 110% or greater, 112% or greater, 114% or greater, 116% or greater, 118% or greater, 120% or greater,
- the extent that the "binding activity to Fc ⁇ RIIa (type H) and Fc ⁇ RIIa (type R) is reduced” or the “reduced binding activity to Fc ⁇ RIIa (type H) and Fc ⁇ RIIa (type R)” can be, but is not limited to, 99% or less, 98% or less, 97% or less, 96% or less, 95% or less, 94% or less, 93% or less, 92% or less, 91% or less, 90% or less, 88% or less, 86% or less, 84% or less, 82% or less, 80% or less, 78% or less, 76% or less, 74% or less, 72% or less, 70% or less, 68% or less, 66% or less, 64% or less, 62% or less, 60% or less, 58% or less, 56% or less, 54% or less, 52% or less, 50% or less, 45% or less, 40% or less, 35% or less, 30% or less, 25% or less, 20% or less,
- modifications that increase binding selectivity to Fc ⁇ RIIb over Fc ⁇ RIIa may be preferred, and modifications that increase binding selectivity to Fc ⁇ RIIb over Fc ⁇ RIIa (type H) may be more preferred, and as reported in WO2013/047752, preferred amino acid substitutions for such modifications may include, for example, according to EU numbering: (a) modification by substituting Gly at position 237 with Trp; (b) modification by substituting Gly at position 237 with Phe; (c) modification by substituting Pro at position 238 with Phe; (d) modification by substituting Asn at position 325 with Met; (e) modification by substituting Ser at position 267 with Ile; (f) modification by substituting Leu at position 328 with Asp; (g) modification by substituting Ser at position 267 with Val; (h) modification by substituting Leu at position 328 with Trp; (i) modification by substituting Ser at position 267 with Gln; (j) modification by substituting Ser
- modifications described above may be at a single position alone or at two or more positions in combination.
- such preferred modifications may include, for example, those shown in Tables 14 to 15, 17 to 24, and 26 to 28 of WO2013/047752, for example, variants of human constant region or human Fc region, in which the amino acid at position 238 according to EU numbering is Asp and the amino acid at position 271 according to EU numbering is Gly in human IgG (IgG1, IgG2, IgG3, or IgG4); in addition, one or more of position(s) 233, 234, 237, 264, 265, 266, 267, 268, 269, 272, 296, 326, 327, 330, 331, 332, 333, and 396, according to EU numbering may be substituted.
- the variants may include, but are not limited to, variants of human constant region or human Fc region that contain one or more of: Asp at position 233, Tyr at position 234, Asp at position 237, Ile at position 264, Glu at position 265, any one of Phe, Met, and Leu at position 266, any one of Ala, Glu, Gly, and Gln at position 267, either Asp or Glu at position 268, Asp at position 269, any one of Asp, Phe, Ile, Met, Asn, and Gln at position 272, Asp at position 296, either Ala or Asp at position 326, Gly at position 327, either Lys or Arg at position 330, Ser at position 331, Thr at position 332, any one of Thr, Lys, and Arg at position 333, and any one of Asp, Glu, Phe, Ile, Lys, Leu, Met, Gln, Arg, and Tyr at position 396, according to EU numbering.
- antibodies of Disclosure A containing an Fc region variant or constant region variant may have maintained or increased binding activity to Fc ⁇ RIIb and reduced binding activity to Fc ⁇ RIIa (type H) and Fc ⁇ RIIa (type R) as compared to a reference antibody containing the constant region or Fc region of a native IgG.
- Preferred sites of amino acid substitution for such variants may be, as reported in WO2014/030728, for example, the amino acid at position 238 according to EU numbering and at least one amino acid position selected from the group consisting of position 233, 234, 235, 237, 264, 265, 266, 267, 268, 269, 271, 272, 274, 296, 326, 327, 330, 331, 332, 333, 334, 355, 356, 358, 396, 409, and 419, according to EU numbering.
- the variants may have Asp at position 238 according to EU numbering, and at least one amino acid selected from the amino acid group of: Asp at position 233, Tyr at position 234, Phe at position 235, Asp at position 237, Ile at position 264, Glu at position 265, Phe, Leu, or Met at position 266, Ala, Glu, Gly, or Gln at position 267, Asp, Gln, or Glu at position 268, Asp at position 269, Gly at position 271, Asp, Phe, Ile, Met, Asn, Pro, or Gln at position 272, Gln at position 274, Asp or Phe at position 296, Ala or Asp at position 326, Gly at position 327, Lys, Arg, or Ser at position 330, Ser at position 331, Lys, Arg, Ser, or Thr at position 332, Lys, Arg, Ser, or Thr at position 333, Arg, Ser, or Thr at position 334, Ala or Gl
- antibodies of Disclosure A containing an Fc region variant or constant region variant may have maintained binding activity to Fc ⁇ RIIb and reduced binding activity to all activating Fc ⁇ Rs, Fc ⁇ RIIa (type R) in particular, as compared to a reference antibody containing the constant region or Fc region of a native IgG.
- Preferred sites of amino acid substitution for such variants may be, as reported in WO2014/163101, for example, in addition to the amino acid at position 238 according to EU numbering), at least one amino acid position selected from positions 235, 237, 241, 268, 295, 296, 298, 323, 324, and 330, according to EU numbering.
- the variants may have Asp at position 238 according to EU numbering, and at least one amino acid selected from the amino acid group of: Phe at position 235; Gln or Asp at position 237; Met or Leu at position 241; Pro at position 268; Met or Val at position 295; Glu, His, Asn, or Asp at position 296; Ala or Met at position 298; Ile at position 323; Asn or His at position 324; and His or Tyr at position 330, according to EU numbering.
- the level of the "maintained binding activity to Fc ⁇ RIIb" can be, but is not limited to, 55% or greater, 60% or greater, 65% or greater, 70% or greater, 75% or greater, 80% or greater, 81% or greater, 82% or greater, 83% or greater, 84% or greater, 85% or greater, 86% or greater, 87% or greater, 88% or greater, 89% or greater, 90% or greater, 91% or greater, 92% or greater, 93% or greater, 94% or greater, 95% or greater, 96% or greater, 97% or greater, 98% or greater, 99% or greater, 99.5% or greater, 100% or greater, 101% or greater, 102% or greater, 103% or greater, 104% or greater, 105% or greater, 106% or greater, 107% or greater, 108% or greater, 109% or greater, 110% or greater, 120% or greater, 130% or greater, 140% or greater, 150% or greater, 175% or greater, or 2-fold or greater.
- the level of the aforementioned "reduced binding activity to all activating Fc ⁇ Rs, Fc ⁇ RIIa (type R) in particular” can be, but is not limited to, 74% or less, 72% or less, 70% or less, 68% or less, 66% or less, 64% or less, 62% or less, 60% or less, 58% or less, 56% or less, 54% or less, 52% or less, 50% or less, 45% or less, 40% or less, 35% or less, 30% or less, 25% or less, 20% or less, 15% or less, 10% or less, 5% or less, 4% or less, 3% or less, 2% or less, 1% or less, 0.5% or less, 0.4% or less, 0.3% or less, 0.2% or less, 0.1% or less, 0.05% or less, 0.01% or less, or 0.005% or less.
- WO2014/030750 also reports variants of the mouse constant region and Fc region.
- antibodies of Disclosure A or B may comprise such a variant.
- FcRn in particular human FcRn, is structurally similar to polypeptides of major histocompatibility complex (MHC) class I, and exhibits 22% to 29% sequence identity with MHC class I molecules (Ghetie et al., Immunol. Today 18(12), 592-598 (1997)).
- FcRn is expressed as a heterodimer consisting of a soluble ⁇ or light chain ( ⁇ 2 microglobulin) complexed with a transmembrane ⁇ or heavy chain.
- the ⁇ chain of FcRn contains three extracellular domains ( ⁇ 1, ⁇ 2, and ⁇ 3), and its short cytoplasmic domain tethers proteins to the cell surface. ⁇ 1 and ⁇ 2 domains interact with the FcRn-binding domain of the antibody Fc region (Raghavan et al., Immunity 1:303-315 (1994)).
- FcRn is expressed in the maternal placenta and yolk sac of mammals, and is involved in mother-to-fetus IgG transfer. In addition, in the small intestines of neonatal rodents where FcRn is expressed, FcRn is involved in transfer of maternal IgG across brush border epithelium from ingested colostrum or milk. FcRn is expressed in a variety of other tissues and endothelial cell systems of various species. FcRn is also expressed in adult human vascular endothelia, muscle vascular system, and liver sinusoidal capillaries. FcRn is believed to play a role in maintaining the plasma IgG concentration by binding to IgG and recycling the IgG to serum. Typically, binding of FcRn to IgG molecules is strictly pH dependent. The optimal binding is observed in an acidic pH range below 7.0.
- polynucleotide and amino acid sequences of human FcRn may be derived, for example, from the precursors shown in NM_004107.4 and NP_004098.1 (containing the signal sequence), respectively (RefSeq accession numbers are shown in parentheses).
- soluble human FcRn capable of forming a complex with human ⁇ 2-microglobulin may be produced for appropriate use in various experimental systems.
- Such soluble human FcRn may be used to assess antibodies or Fc region variants for their FcRn-binding activity.
- FcRn is not particularly limited as long as it is in a form which can bind to the FcRn-binding domain; however, preferred FcRn may be human FcRn.
- an antibody or Fc region variant may have an "FcRn-binding domain", preferably a human FcRn-binding domain.
- the FcRn-binding domain is not particularly limited as long as the antibody has binding activity to or affinity for FcRn at an acidic pH and/or at a neutral pH; or it may be a domain that has the activity to directly or indirectly bind to FcRn.
- Such domains include, but are not limited to, the Fc regions of IgG-type immunoglobulins, albumin, albumin domain 3, anti-FcRn antibodies, anti-FcRn peptides, and anti-FcRn Scaffold molecules, which have the activity of directly binding to FcRn, and molecules that bind to IgG or albumin, which have the activity of binding to FcRn indirectly.
- IgG or albumin which have the activity of binding to FcRn indirectly.
- amino acid residues in the FcRn-binding domain of the antibody or Fc region variant may be modified to have FcRn-binding activity in an acidic pH range and/or in a neutral pH range.
- amino acids of domains that originally have FcRn-binding activity in an acidic pH range and/or in a neutral pH range may be modified to further increase their FcRn-binding activity.
- the FcRn-binding activity in an acidic pH range and/or in a neutral pH range can be compared before and after amino acid modification to find amino acid modifications of interest for the FcRn-binding domains.
- FcRn-binding domains may be preferably regions that directly bind to FcRn.
- Such preferred FcRn-binding domains include, for example, constant regions and Fc regions of antibodies.
- regions capable of binding to a polypeptide having FcRn-binding activity such as albumin and IgG, can indirectly bind to FcRn via albumin, IgG.
- the FcRn-binding regions may be regions that bind to a polypeptide that has binding activity to albumin or IgG.
- FcRn-binding domains whose FcRn-binding activity is greater at a neutral pH are preferred, while to improve antibody retention in plasma, FcRn-binding domains whose FcRn-binding activity is greater at an acidic pH are preferred.
- FcRn-binding domains whose FcRn-binding activity is originally greater at a neutral pH or acidic pH are preferred.
- amino acids of an antibody or Fc region variant may be modified to confer FcRn-binding activity at a neutral pH or acidic pH.
- the assay can be carried out, for example, under the conditions of MES buffer and 37°C, as described in WO2009/125825.
- the FcRn-binding activity of an antibody or Fc region (variant) can be assessed, for example, by loading FcRn as an analyte on an antibody-immobilized chip.
- the FcRn-binding activity of an antibody or Fc region (variant) can be assessed based on the dissociation constant (KD), apparent dissociation constant (apparent KD), dissociation rate (kd), apparent dissociation (apparent kd).
- acidic pH condition or neutral pH condition may be suitably used.
- temperature conditions for measuring the binding activity (binding affinity) between FcRn and the FcRn-binding domain any temperature of 10°C to 50°C may be used.
- a temperature of 15°C to 40°C may be used. More preferably, any temperature from 20°C to 35°C such as any one of 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, and 35°C may be used. A non-limiting example of such temperature can be 25°C.
- antibodies of Disclosure A or B may have an FcRn-binding domain, preferably a human FcRn-binding domain.
- the FcRn-binding domain is not particularly limited as long as the antibodies have binding activity to or affinity for FcRn at an acidic pH and/or a neutral pH, and it may be a domain that has an activity of directly or indirectly binding to FcRn.
- it may be preferable that the antibody of Disclosure A or B has, for example, an increased FcRn-binding activity under a neutral pH condition as compared to a reference antibody containing the constant region of a native IgG (see WO2013/046722).
- the antibody of Disclosure A or B and the reference antibody containing the constant region of a native IgG have identical amino acid sequences in the regions (for example, the variable region) other than, preferably, the constant region of the antibody of Disclosure A or B which has been modified at one or more amino acid residues.
- the antibody of Disclosure A may possess any two or more of the following properties in combination: the property of being shuttled between plasma and cellular endosome and repeatedly binding to multiple antigens as a single antibody molecule by having an ion concentration-dependent antigen-binding domain; the property of being rapidly taken up into cells by having increased pI and increased positive charge in the overall antibody; and the property of being rapidly taken up into cells by having an increased FcRn-binding activity under a neutral pH condition.
- the antibody half-life in plasma can be further shortened, or the binding activity of the antibody toward the extracellular matrix can be further increased, or antigen elimination from plasma can be further promoted.
- Those of ordinary skill in the art can determine an optimal pI value for the antibody of Disclosure A to take advantage of these properties.
- the KD values are
- a domain of any structure that binds to FcRn can be used as an FcRn-binding domain.
- the FcRn-binding domain can be produced without the need to introduce an amino acid modification, or the affinity for FcRn may be increased by introducing an additional modification.
- the starting FcRn-binding domain can include for example, the Fc region or constant region of (human) IgG.
- any Fc region or constant region can be used as the starting Fc region or starting constant region.
- an Fc region or constant region obtained by further modifying a starting Fc region or starting constant region whose amino acid residues have been already modified from an Fc region or constant region can also be appropriately used as the Fc region or constant region.
- the starting Fc region or starting constant region may include known Fc regions produced by recombination.
- a starting Fc region or starting constant region may refer to the polypeptide itself, a composition containing the starting Fc region or starting constant region, or an amino acid sequence encoding the starting Fc region or starting constant region, depending on the context.
- the origin of the starting Fc region or starting constant region is not limited, and it can be obtained from any organism of nonhuman animals or from a human.
- the starting FcRn-binding domain can be obtained from cynomolgus monkeys, marmosets, Rhesus monkeys, chimpanzees, and humans.
- Starting Fc regions or starting constant regions may be obtained from human IgG1, but are not limited to any particular IgG class.
- an Fc region of human IgG1, IgG2, IgG3, or IgG4 can be used as an appropriate starting FcRn-binding domain, and an Fc region or constant region of an IgG class or subclass derived from any organism can be used as a starting Fc region or as a starting constant region.
- native IgG variants or modified forms are described in, for example, Strohl, Curr. Opin. Biotechnol. 20(6):685-691 (2009); Presta, Curr. Opin. Immunol. 20(4):460-470 (2008); Davis et al., Protein Eng. Des. Sel. 23(4):195-202 (2010), WO2009/086320, WO2008/092117; WO2007/041635; and WO2006/105338).
- amino acid residues of the starting FcRn-binding domain, starting Fc region, or starting constant region may contain, for example, one or more mutations: for example, substitution mutations with amino acid residues that are different from the amino acid residues in the starting Fc region or starting constant region; insertions of one or more amino acid residues into the amino acid residues in the starting Fc region or starting constant region; or deletions of one or more amino acid residues from the amino acid residues of the starting Fc region or starting constant region.
- the amino acid sequences of Fc regions or constant regions after modifications may be preferably amino acid sequences containing at least a portion of an Fc region or constant region that does not occur naturally.
- variants necessarily have a sequence identity or similarity of less than 100% to the starting Fc regions or starting constant regions.
- the variants have an amino acid sequence identity or similarity of about 75% to less than 100%, more preferably about 80% to less than 100%, even more preferably about 85% to less than 100%, still more preferably about 90% to less than 100%, and yet more preferably about 95% to less than 100% to the amino acid sequence of the starting Fc region or starting constant region.
- at least one amino acid is different between a modified Fc region or constant region of Disclosure A or B and the starting Fc region or starting constant region.
- an Fc region or constant region that has FcRn-binding activity in an acidic pH range and/or in a neutral pH range may be obtained by any method.
- a variant of Fc region or constant region that has FcRn-binding activity in an acidic pH range and/or in a neutral pH range may be obtained by modifying amino acids of a human IgG-type antibody which can be used as the starting Fc region or starting constant region.
- IgG-type antibody Fc regions or constant regions suitable for modification include, for example, the Fc regions or constant regions of human IgG (IgG1, IgG2, IgG3, and IgG4, and variants thereof), and mutants spontaneously generated therefrom are also included in the IgG Fc regions or constant regions.
- Fc regions or constant regions of human IgG1, human IgG2, human IgG3, and human IgG4 antibodies a number of allotype sequences due to genetic polymorphism are described in "Sequences of proteins of immunological interest", NIH Publication No.91-3242, and any of them may be used in Disclosure A or B.
- the amino acid sequence of positions 356 to 358 according to EU numbering may be DEL or EEM.
- the modification into other amino acids is not particularly limited, as long as the resulting variants have FcRn-binding activity in an acidic pH range and/or in a neutral pH range, and preferably in a neutral pH range.
- Sites of amino acid modification to increase the FcRn-binding activity under a neutral pH condition are described, for example, in WO2013/046722.
- Such modification sites include, for example, one or more positions selected from the group consisting of: position 221 to 225, 227, 228, 230, 232, 233 to 241, 243 to 252, 254 to 260, 262 to 272, 274, 276, 278 to 289, 291 to 312, 315 to 320, 324, 325, 327 to 339, 341, 343, 345, 360, 362, 370, 375 to 378, 380, 382, 385 to 387, 389, 396, 414, 416, 423, 424, 426 to 438, 440, and 442, according to EU numbering in the Fc region or constant region of a human IgG antibody, as described in WO2013/046722.
- WO2013/046722 also describes, as a part of the preferred modifications in the Fc region or constant region, for example, modification of one or more amino acids selected from the group consisting of: the amino acid at position 256 to Pro, the amino acid at position 280 to Lys, the amino acid at position 339 to Thr, the amino acid at position 385 to His, the amino acid at position 428 to Leu, and the amino acid at position 434 to Trp, Tyr, Phe, Ala, or His, according to EU numbering.
- the number of amino acids to be modified is not particularly limited, and modification may be performed at a single position alone or at two or more positions. Modification of these amino acid residues can enhance the FcRn binding of the Fc region or constant region of an IgG-type antibody under a neutral pH condition. Modification of these amino acid residues may also be introduced appropriately into antibodies of Disclosure A or B.
- modification sites for increasing the FcRn-binding activity under an acidic pH condition.
- one or more modification sites that allow an increase in the FcRn binding also in a neutral pH range can be appropriately used in Disclosure A or B.
- modification sites include, for example, those reported in WO2011/122011, WO2013/046722, WO2013/046704, and WO2013/046722.
- the sites of amino acids that allow such modification of the constant region or Fc region of a human IgG-type antibody and the types of amino acids after modification are reported in Table 1 of WO2013/046722.
- WO2013/046722 also describes, as particularly preferred, modification sites in the constant region or Fc region, for example, the location of one or more amino acid positions selected from the group consisting of position 237, 238, 239, 248, 250, 252, 254, 255, 256, 257, 258, 265, 270, 286, 289, 297, 298, 303, 305, 307, 308, 309, 311, 312, 314, 315, 317, 325, 332, 334, 360, 376, 380, 382, 384, 385, 386, 387, 389, 424, 428, 433, 434, and 436, according to EU numbering.
- WO2013/046722 also describes, as a part of the preferred modification in the IgG-type constant region or Fc region, for example, modification of one or more amino acid residues selected from the group consisting of: (a) the amino acid at position 237 to Met; (b) the amino acid at position 238 to Ala; (c) the amino acid at position 239 to Lys; (d) the amino acid at position 248 to Ile; (e) the amino acid at position 250 to any one of Ala, Phe, Ile, Met, Gln, Ser, Val, Trp, and Tyr; (f) the amino acid at position 252 to any one of Phe, Trp, and Tyr; (g) the amino acid at position 254 to Thr; (h) the amino acid at position 255 to Glu; (i) the amino acid at position 256 to any one of Asp, Glu, and Gln;
- the number of amino acids to be modified is not particularly limited, and modification may be performed at a single position alone or at two or more positions. Combinations of amino acid modifications at two or more positions include, for example, those shown in Table 2 of WO2013/046722. Modification of these amino acid residues may also be appropriately introduced into antibodies of Disclosures A and B.
- the FcRn-binding activity of the FcRn-binding domain of an antibody of Disclosure A or B has been increased when compared to that of a reference antibody containing an Fc region or constant region of a native IgG or that of a reference antibody containing a starting Fc region or starting constant region. Namely, the FcRn-binding activity of an Fc region variant or constant region variant of Disclosure A or B, or an antibody containing such variant is greater than that of the reference antibody).
- an antibody of Disclosure A or B can be, for example: 55% or greater, 60% or greater, 65% or greater, 70% or greater, 75% or greater, 80% or greater, 85% or greater, 90% or greater, 95% or greater, 100% or greater, 105% or greater, preferably 110% or greater, 115% or greater, 120% or greater, 125% or greater, more preferably 130% or greater, 135% or greater, 140% or greater, 145% or greater, 150% or greater, 155% or greater, 160% or greater, 165% or greater, 170% or greater, 175% or greater, 180% or greater, 185% or greater, 190% or greater, 195% or greater, 2-fold or greater, 2.5-fold or greater, 3-fold or greater, 3.5-fold or greater, 4-fold or greater, 4.5-fold or greater, or 5-fold or greater.
- amino acid sequences to be modified in an antibody of Disclosure A or B can preferably contain human sequences (sequences found in native human-derived antibodies) in order to not increase the immunogenicity of the antibody when the antibody is administered in vivo (preferably, into a human body).
- mutations may be introduced at positions other than the sites of amino acid modification in such a way that one or more of the FRs (FR1, FR2, FR3, and FR4) is substituted with a human sequence.
- Methods for substituting FR(s) with a human sequence are known in the art and include, but are not limited to that reported in Ono et al., Mol. Immunol. 36(6):387-395 (1999). Humanization methods are known in the art and include, but are not limited to that reported in, Methods 36(1):43-60 (2005).
- the framework region sequences (also referred to as "FR sequences") of the heavy chain and/or light chain variable region of an antibody of Disclosure A or B may contain human germ-line framework sequences.
- the framework sequences are completely human germ-line sequences, the antibody is expected to induce little or no immunogenic reaction when administered to humans (for example, to treat or prevent a certain disease).
- FR sequences preferably can include, for example, fully human FR sequences such as those shown in V-Base (vbase.mrc-cpe.cam.ac.uk/). These FR sequences can be appropriately used for Disclosure A or B.
- the germ-line sequences may be categorized based on their similarity (Tomlinson et al. (J. Mol. Biol. 227:776-798 (1992); Williams et al. (Eur. J. Immunol. 23:1456-1461 (1993); and Cox et al. (Nat. Genetics 7:162-168 (1994)).
- Preferred germ-line sequences can be appropriately selected from V ⁇ , which is categorized into seven subgroups; V ⁇ , which is categorized into ten subgroups; and VH, which is categorized into seven subgroups.
- Fully human VH sequences can preferably include, for example, VH sequences of: subgroup VH1 (for example, VH1-2, VH1-3, VH1-8, VH1-18, VH1-24, VH1-45, VH1-46, VH1-58, and VH1-69); subgroup VH2 (for example, VH2-5, VH2-26, and VH2-70); subgroup VH3 (VH3-7, VH3-9, VH3-11, VH3-13, VH3-15, VH3-16, VH3-20, VH3-21, VH3-23, VH3-30, VH3-33, VH3-35, VH3-38, VH3-43, VH3-48, VH3-49, VH3-53, VH3-64, VH3-66, VH3-72, VH3-73, and VH3-74); subgroup VH4 (VH4-4, VH4-28, VH4-31, VH4-34, VH4-39, VH4-5
- Fully human V ⁇ sequences can preferably include, for example: A20, A30, L1, L4, L5, L8, L9, L11, L12, L14, L15, L18, L19, L22, L23, L24, O2, O4, O8, O12, O14, or O18, which are classified as subgroup Vk1; A1, A2, A3, A5, A7, A17, A18, A19, A23, O1, and O11, which are classified as subgroup Vk2; A11, A27, L2, L6, L10, L16, L20, and L25, which are classified as subgroup Vk3; B3, classified as subgroup Vk4; B2 (also referred to as "Vk5-2"), classified as subgroup Vk5; or A10, A14, and A26, which are classified as subgroup Vk6 (Kawasaki et al.
- Fully human V ⁇ sequences can preferably include, for example: V1-2, V1-3, V1-4, V1-5, V1-7, V1-9, V1-11, V1-13, V1-16, V1-17, V1-18, V1-19, V1-20, and V1-22, which are classified as subgroup VL1; V2-1, V2-6, V2-7, V2-8, V2-11, V2-13, V2-14, V2-15, V2-17, and V2-19, which are classified as subgroup VL2; V3-2, V3-3, and V3-4, which are classified as subgroup VL3; V4-1, V4-2, V4-3, V4-4, and V4-6, which are classified as subgroup VL4; or V5-1, V5-2, V5-4, and V5-6, which are classified as subgroup VL5 (Kawasaki et al. Genome Res. 7:250-261 (1997)).
- these FR sequences are different from one another at one or more amino acid residues.
- These FR sequences can be used in the modification of antibody amino acid residues.
- Fully human FR sequences that may be used in the modification also include, for example, KOL, NEWM, REI, EU, TUR, TEI, LAY, and POM (see, for example, aforementioned Kabat et al. (1991); Wu et al. (J. Exp. Med. 132:211-250 (1970)).
- flexible residues can refer to amino acid residue variations that are present at positions showing high amino acid diversity at which the light chain or heavy chain variable regions have several different amino acids when the amino acid sequences of known and/or native antibodies or antigen-binding domains are compared. Positions showing high diversity are generally located in the CDRs.
- an antibody of Disclosure A or B contains the whole or a portion of the light chain variable region and/or heavy chain variable region
- the antibody may contain one or more appropriate flexible residues, if needed.
- a heavy chain and/or light chain variable region sequence selected to have an FR sequence which originally contains amino acid residues that change the antigen-binding activity of an antibody according to the ion concentration (hydrogen ion concentration or calcium ion concentration) conditions can be designed to contain, other amino acid residues in addition to these amino acid residues.
- the number and locations of the flexible residues can also be determined without being limited to a specific embodiment, as long as the antigen-binding activity of the antibody of Disclosure A or B changes according to the ion concentration condition.
- the CDR sequence and/or FR sequence of a heavy chain and/or light chain may contain at least one flexible residue.
- flexible residues that can be introduced into the light-chain variable region sequence include, but are not limited to, one or more amino acid residue positions shown in Table 1 or Table 2.
- appropriate flexible residues can be introduced, for example, into an ion concentration-dependent antibody or antibody without such ion concentration dependency, containing the whole or a portion of the light chain variable region and/or heavy chain variable region, in which at least one amino acid residue that may be exposed on the antibody surface has been modified such that the pI is increased.
- the pI of the chimeric antibody is increased by modifying one or more amino acid residues that can be exposed on the antibody surface as to produce a humanized antibody of Disclosure A or B with a shortened plasma half-life as compared to the chimeric antibody absent such modification.
- the modification of amino acid residues that can be exposed on the surface of the humanized antibody can be carried out before or concurrently with humanization of the antibody.
- amino acid residues that can be exposed on the surface may be modified to further alter the pI of the humanized antibody.
- the humanized antibodies trastuzumab (antigen: HER2), bevacizumab (antigen: VEGF), and pertuzumab (antigen: HER2), which were humanized using the same human antibody FR sequences, were almost comparable in plasma pharmacokinetics. Specifically, it can be understood that the plasma pharmacokinetics is almost comparable when humanization is performed using the same FR sequences.
- the antigen concentration in plasma is reduced by increasing the antibody's pI by modifying amino acid residues that can be exposed on the antibody surface, in addition to the humanization step.
- human antibodies can be used. By modifying amino acid residues that can be exposed on the surface of a human antibody produced from a human antibody library, a human antibody-producing mouse, a recombinant cell, etc., and increasing the pI of the human antibody, the ability of the originally-produced human antibody to eliminate antigen from plasma can be increased.
- antibodies of Disclosure A may contain modified sugar chains.
- Antibodies with modified sugar chains include, for example, antibodies with modified glycosylation (WO99/54342), antibodies that lack fucose (WO00/61739; WO02/31140, WO2006/067847; WO2006/067913), and antibodies having sugar chains with bisecting GlcNAc (WO02/79255).
- antibodies of Disclosure A or B can be used, for example, in techniques for exhibiting increased antitumor activities against cancer cells or in techniques for promoting elimination of antigens that are harmful to the organism from the plasma.
- Disclosure A or B relate to libraries of the ion concentration-dependent antigen-binding domains with an increased pI or ion concentration-dependent antibodies with an increased pI, as described above.
- Disclosure A or B relates to nucleic acids (polynucleotides) encoding the above-described ion concentration-dependent antigen-binding domains with an increased pI or ion concentration-dependent antibodies with an increased pI.
- the nucleic acids can be obtained using appropriate known methods.
- WO2009/125825, WO2012/073992, WO2011/122011, WO2013/046722, WO2013/046704, WO2000/042072, WO2006/019447, WO2012/115241, WO2013/047752, WO2013/125667, WO2014/030728, WO2014/163101, WO2013/081143, WO2007/114319, WO2009/041643, WO2014/145159, WO2012/016227, and WO2012/093704 can be referred to, each of these are incorporated herein by reference in their entirety.
- nucleic acids of Disclosure A or B can be isolated or purified nucleic acids.
- Nucleic acids encoding the antibodies of Disclosure A or B may be any genes, and may be DNA or RNA, or other nucleic acid analogs.
- the amino acid sequence of the antibody before modification may be a known sequence or the amino acid sequence of an antibody newly obtained.
- antibodies can be obtained from antibody libraries, or by cloning nucleic acids encoding the antibody from hybridomas or B cells that produce monoclonal antibodies.
- the methods for obtaining nucleic acids encoding an antibody from hybridomas may use the techniques of: performing immunization by conventional immunization methods using an antigen of interest or cells expressing the antigen of interest as a sensitizing antigen; fusing the resulting immune cells with known parental cells by conventional cell fusion methods; screening for monoclonal antibody-producing cells (hybridomas) by conventional screening methods; synthesizing cDNAs of the variable region (V region) of the antibody using reverse transcriptase from mRNAs of the obtained hybridomas; and linking the cDNA to a DNA encoding an antibody constant region (C region) of interest.
- Sensitizing antigens which are used to obtain nucleic acids encoding the above-described heavy chain and light chain include, but are not limited to, both complete antigens with immunogenicity and incomplete antigens including haptens which exhibit no immunogenicity. For example, it is possible to use whole proteins of interest or partial peptides of the proteins. In addition, substances that are composed of polysaccharides, nucleic acids, lipids, and other compositions are known to be potential antigens. Thus, in some embodiments, antigens for the antibodies of Disclosure A or B are not particularly limited. The antigens can be prepared by, for example, baculovirus-based methods (see, e.g., WO98/46777).
- Hybridomas can be produced, for example, according to the method of G. Kohler and C. Milstein, Methods Enzymol. 73:3-46 (1981)).
- immunization may be performed by linking the antigen with a macromolecule having immunogenicity, such as albumin.
- soluble antigens can be prepared by linking the antigen with other molecules.
- a transmembrane molecule such as membrane antigens (for example, receptors) is used as an antigen
- a portion of the extracellular region of the membrane antigen can be used as a fragment, or cells expressing the transmembrane molecule on their surface may be used as an immunogen.
- antibody-producing cells can be obtained by immunizing an animal with an appropriate sensitizing antigen described above.
- antibody-producing cells can be prepared by in vitro immunization of lymphocytes that are capable of producing antibodies.
- Various mammals can be used for immunization and other routine antibody producing procedures.
- Commonly used animals include rodents, lagomorphs, and primates.
- the animals may include, for example, rodents such as mice, rats, and hamsters; lagomorphs such as rabbits; and primates including monkeys such as cynomolgus monkeys, rhesus monkeys, baboons, and chimpanzees.
- transgenic animals carrying a human antibody gene repertoire are also known, and these animals can be used to obtain human antibodies (see, e.g., WO96/34096; Mendez et al., Nat. Genet. 15:146-156 (1997); WO93/12227, WO92/03918, WO94/02602, WO96/34096, and WO96/33735).
- desired human antibodies having antigen-binding activity by, for example, sensitizing human lymphocytes in vitro with desired antigens or cells expressing the desired antigens and then fusing the sensitized lymphocytes with human myeloma cells such as U266 (JP Pat. Publ. No. H01-59878).
- Animal immunization can be carried out, for example, by appropriately diluting and suspending a sensitizing antigen in phosphate buffered saline (PBS), physiological saline, or others, and mixing it with an adjuvant to emulsify, if needed; and then injecting it intraperitoneally or subcutaneously into animals. Then, the sensitizing antigen mixed with Freund's incomplete adjuvant can be preferably administered several times every four to 21 days. Antibody production can be confirmed, for example, by measuring the titer of the antibody of interest in animal sera.
- PBS phosphate buffered saline
- an adjuvant to emulsify
- the sensitizing antigen mixed with Freund's incomplete adjuvant can be preferably administered several times every four to 21 days.
- Antibody production can be confirmed, for example, by measuring the titer of the antibody of interest in animal sera.
- Antibody-producing cells obtained from lymphocytes or animals immunized with a desired antigen can be fused with myeloma cells to generate hybridomas using conventional fusing agents (for example, polyethylene glycol) (Goding, Monoclonal Antibodies: Principles and Practice, Academic Press, 1986, 59-103). If needed, hybridomas are cultured and expanded, and the binding specificity of antibodies produced by the hybridomas is assessed by, for instance, immunoprecipitation, radioimmunoassay (RIA), or enzyme-linked immunosorbent assay (ELISA). Then, if needed, antibody-producing hybridomas whose specificity, affinity, or activity of interest has been determined may also be subcloned by methods such as limiting dilution.
- conventional fusing agents for example, polyethylene glycol
- RIA radioimmunoassay
- ELISA enzyme-linked immunosorbent assay
- Nucleic acids encoding the selected antibody can be cloned from hybridomas or antibody-producing cells (sensitized lymphocytes, etc.) using probes that can specifically bind to the antibody (for example, oligonucleotides complementary to sequences encoding the antibody constant regions). Alternatively, the nucleic acids can be cloned from mRNA using RT-PCR. Heavy chains and light chains for use in producing antibodies of Disclosure A or B may be derived from antibodies that, for example, belong to any of Ig antibody classes and subclasses, and IgG may be preferred.
- nucleic acids encoding amino acid sequences that constitute the heavy chain (the whole or a portion thereof) and/or light chain (the whole or a portion thereof) of an antibody of Disclosure A or B are modified by genetic engineering techniques.
- Recombinant antibodies with artificial sequence modification to, for example, reduce heterologous antigenicity against humans, such as chimeric antibodies or humanized antibodies may be appropriately generated by, for example, modifying nucleotide residues encoding amino acid sequences associated with components of antibodies such as mouse antibodies, rat antibodies, rabbit antibodies, hamster antibodies, sheep antibodies, or camel antibodies.
- Chimeric antibodies can be obtained, for example, by ligating a DNA encoding a mouse-derived antibody variable region with a DNA encoding a human antibody constant region and incorporating the ligated DNA coding sequence into an expression vector, then introducing the resulting recombinant vector into a host to express the genes.
- Humanized antibodies which are also referred to as reshaped human antibodies, are antibodies in which human antibody FR(s) are linked in frame with antibody CDR(s) isolated from non-human mammals, such as mice, to form a coding sequence.
- a DNA sequence encoding such a humanized antibody can be synthesized by overlap extension PCR using a number of oligonucleotides as templates.
- a DNA encoding the amino acid sequence of, for example, an antibody variable region of Disclosure A or B may be obtained by overlap extension PCR using a number of oligonucleotides designed to have overlapping nucleotide sequences.
- the overlapping DNA is then linked in frame to a DNA encoding a constant region to form a coding sequence.
- the DNA linked as described above may be then inserted into an expression vector so that the DNA can be expressed, and the resulting vector may be introduced into a host or host cell.
- the antibody encoded by the DNA can be expressed by raising the host or culturing the host cells.
- the expressed antibody can be appropriately purified from culture media of the host or others (EP239400; WO96/02576).
- the FR(s) of a humanized antibody which are linked via CDR(s) may be selected, for example, to allow the CDRs to form an antigen-binding site suitable for the antigen.
- amino acid residues that constitute FR(s) of a variable region of the selected antibody for example, can be modified with appropriate substitution.
- nucleic acid cassettes may be cloned into appropriate vectors.
- vectors such as phagemid vectors are available.
- phagemid vectors can contain various elements including regulatory sequences such as promoters or signal sequences, phenotype selection genes, replication origins, and other necessary elements.
- libraries can be constructed by introducing at least one modified amino acid residue that can be exposed on the surface of antibodies of Disclosure A or B and/or at least one amino acid that can change the antigen-binding activity of antibodies according to the ion concentration condition.
- flexible residues can be added using the method of Kunkel et al. (Methods Enzymol. 154:367-382 (1987)).
- Disclosure A relates to vectors containing nucleic acids encoding an above-described ion concentration-dependent antigen-binding domain with increased pI or an above-described ion concentration-dependent antibody with increased pI.
- the vectors can be obtained by, for example, vectors as described in WO2009/125825, WO2012/073992, WO2011/122011, WO2013/046722, WO2013/046704, WO2000/042072, WO2006/019447, WO2012/115241, WO2013/047752, WO2013/125667, WO2014/030728, WO2014/163101, WO2013/081143, WO2007/114319, WO2009/041643, WO2014/145159, WO2012/016227, or WO2012/093704, each of which is incorporated herein by reference in their entirety.
- nucleic acids encoding embodiments of Disclosure A or B may be operably cloned (inserted) into appropriate vectors and introduced into host cells.
- vectors include the cloning vector, pBluescript vector (Stratagene) or any of various other commercially available vectors.
- expression vectors are useful as vectors containing a nucleic acid for Disclosure A or B.
- Expression vectors can be used to allow polypeptide expression in vitro, in E. coli, in culture cells, or in vivo.
- pBEST vector Promega
- pET vector Invitrogen
- pME18S-FL3 vector GenBank Accession No. AB009864
- pME18S vector Takebe et al., Mol. Cell Biol. 8:466-472 (1988) for in vivo expression.
- DNAs can be inserted into vectors by conventional methods, for example, by ligase reaction using restriction enzyme sites (see, Current protocols in Molecular Biology edit. Ausubel et al. (1987) Publish. John Wiley & Sons. Section 11.4-11.11).
- Disclosure A relates to a host or host cells that comprise a vector containing a nucleic acid encoding an above-described ion concentration-dependent antigen-binding domain with increased pI or an above-described ion concentration-dependent antibody with increased pI.
- the host or host cells can be prepared by, for example, methods described in WO2009/125825, WO2012/073992, WO2011/122011, WO2013/046722, WO2013/046704, WO2000/042072, WO2006/019447, WO2012/115241, WO2013/047752, WO2013/125667, WO2014/030728, WO2014/163101, WO2013/081143, WO2007/114319, WO2009/041643, WO2014/145159, WO2012/016227, or WO2012/093704, each of which is incorporated herein by reference in their entirety.
- the type of host cell of Disclosure A or B is not particularly limited, and the host cells include, for example, bacterial cells such as E. coli, as well as various animal cells.
- the host cells can be appropriately used as production systems for producing and expressing the antibodies. Both eukaryotic and prokaryotic cells can be used.
- Eukaryotic cells for use as host cells include, for example, animal cells, plant cells, and fungal cells.
- animal cells include mammalian cells, for example, CHO (Puck et al., J. Exp. Med. 108:945-956 (1995)), COS, HEK293, 3T3, myeloma, BHK (baby hamster kidney), HeLa, and Vero; amphibian cells, for example, Xenopus oocyte (Valle et al., Nature 291:338-340 (1981)); and insect cells, for example, Sf9, Sf21, and Tn5.
- mammalian cells for example, CHO (Puck et al., J. Exp. Med. 108:945-956 (1995)), COS, HEK293, 3T3, myeloma, BHK (baby hamster kidney), HeLa, and Vero
- amphibian cells for example, Xenopus oocyte (Valle
- Recombinant vectors or others can be introduced into host cells, for example, using calcium phosphate methods, DEAE-dextran methods, methods using cationic liposome DOTAP (Boehringer-Mannheim), electroporation, and lipofection.
- Plant cells that are known to serve as a protein production system include, for example, Nicotiana tabacum-derived cells and duckweed (Lemna minor)-derived cells. Calluses can be cultured from these cells to produce antibodies of Disclosure A or B.
- Fungal cell-based protein production systems include those using yeast cells, for example, cells of genus Saccharomyces such as Saccharomyces cerevisiae and Schizosaccharomyces pombe; and cells of filamentous fungi, for example, genus Aspergillus such as Aspergillus niger. When prokaryotic cells are used, bacterial cell-based production systems can be used.
- Bacterial cell-based production systems include, for example, those using Bacillus subtilis as well as E. coli.
- the host cells are transformed with an expression vector containing a nucleic acid encoding an antibody of Disclosure A or B and cultured to express the nucleic acid.
- culture media may include, for example, DMEM, MEM, RPMI1640, and IMDM, which may be appropriately used in combination with serum supplements such as FBS or fetal calf serum (FCS).
- serum supplements such as FBS or fetal calf serum (FCS).
- the cells may be cultured serum free.
- animals or plants can be used for in vivo production systems for producing antibodies of Disclosure A or B,
- a nucleic acid(s) encoding an antibody of Disclosure A or B can be introduced into such animals or plants to produce the antibody in vivo, and the antibody can then be collected from the animals or plants.
- mammals When animals are used as a host, production systems using mammals or insects are available. Preferred mammals include, but are not limited to, goats, pigs, sheep, mice, and bovines (Vicki Glaser, SPECTRUM Biotechnology Applications (1993)). Transgenic animals can also be used.
- a nucleic acid encoding an antibody of Disclosure A or B is prepared as a fusion gene with a gene encoding a polypeptide that is specifically included in milk, such as goat ⁇ -casein. Then, goat embryos are injected with a polynucleotide fragment containing the fusion gene and transplanted into a female goat.
- the antibody of interest can be obtained from milk produced by the transgenic goats, which are born from goats that received the embryos, or from their offspring. Hormones can be appropriately administered to the transgenic goats to increase the volume of milk containing the antibody produced by the goats (Ebert et al., Bio/Technology 12:699-702 (1994)).
- Insects for use in producing antibodies of Disclosure A or B include, for example, silkworms.
- silkworms When silkworms are used, baculoviruses whose viral genome is inserted with a polynucleotide encoding an antibody of interest is used to infect the silkworm.
- the antibody of interest can be obtained from the body fluids of the infected silkworms (Susumu et al., Nature 315:592-594 (1985)).
- tobacco When plants are used for producing antibodies of Disclosure A or B, tobacco may be used.
- a recombinant vector resulting from insertion of a polynucleotide encoding an antibody of interest into a plant expression vector for example, pMON 530 may be introduced into bacteria such as Agrobacterium tumefaciens.
- the resulting bacteria can be used to infect tobacco, for example, Nicotiana tabacum (Ma et al., Eur. J. Immunol. 24:131-138 (1994)) and the desired antibody is obtained from the leaves of the infected tobacco.
- Such modified bacteria can be also used to infect duckweed (Lemna minor), and the desired antibody is obtained from cloned cells of the infected duckweed (Cox et al., Nat. Biotechnol. 24(12):1591-1597 (2006)).
- suitable secretion signals may be incorporated into the polypeptide of interest.
- signals may be endogenous to the antibody of interest or may be a heterogeneous signal known in the art.
- the antibody of Disclosure A or B produced as described above may be isolated from the inside or outside (such as media and milk) of host cells or a host, and purified to a substantially pure and homogenous antibody.
- the antibodies can be suitably isolated and purified, for example, by appropriately selecting and combining chromatographic columns, filtration, ultrafiltration, salting out, solvent precipitation, solvent extraction, distillation, immunoprecipitation, SDS-polyacrylamide gel electrophoresis, isoelectric focusing, dialysis, recrystallization, and others.
- Chromatography includes, for example, affinity chromatography, ion exchange chromatography, hydrophobic chromatography, gel filtration chromatography, reverse phase chromatography, and adsorption chromatography.
- Such chromatography can be performed, for example, by using liquid chromatography such as HPLC and FPLC.
- Columns for use in affinity chromatography may be Protein A column or Protein G column.
- Protein A column include, for example, Hyper D, POROS, Sepharose F.F. (Pharmacia).
- the antibody can be modified or the peptide can be partially deleted by treating the antibody with appropriate protein modifying enzymes before or after antibody purification, as necessary.
- protein modifying enzymes for example, trypsin, chymotrypsin, lysyl endopeptidase, protein kinases, and glucosidases can be used.
- Disclosure A relates to methods for producing antibodies containing an antigen-binding domain whose antigen-binding activity changes according to the ion concentration condition, which may comprise culturing the host cells or raising the hosts and collecting antibodies from cultures of these cells, materials secreted from the hosts, or by other means known in the art.
- Disclosure A relates to a production method which comprises any one or more steps selected from the group consisting of: (a) selecting an antibody which can promote elimination of an antigen from plasma; (b) selecting an antibody with enhanced binding activity to an extracellular matrix; (c) selecting an antibody with enhanced Fc ⁇ R-binding activity under a neutral pH condition; (d) selecting an antibody with enhanced Fc ⁇ RIIb-binding activity under a neutral pH condition; (e) selecting an antibody with maintained or enhanced Fc ⁇ RIIb-binding activity and decreased binding activity to one or more activating Fc ⁇ R selected from the group consisting of Fc ⁇ RIa, Fc ⁇ RIb, Fc ⁇ RIc, Fc ⁇ RIIIa, Fc ⁇ RIIIb, and Fc ⁇ RIIa; (f) selecting an antibody with enhanced FcRn-binding activity under a neutral pH condition; (g) selecting an antibody with an increased pI; (h) confirming the pI of the collected antibody, and then selecting an antibody with an increased pI; and (i
- the reference antibody includes, but is not limited to, a native antibody (for example, a native Ig antibody, preferably a native IgG antibody) and an antibody before modification (an antibody prior to or during library construction, for example, an ion concentration-dependent antibody prior to increasing its pI, or an antibody with increased pI prior to conferring an ion concentration-dependent antigen-binding domain).
- a native antibody for example, a native Ig antibody, preferably a native IgG antibody
- an antibody before modification an antibody prior to or during library construction, for example, an ion concentration-dependent antibody prior to increasing its pI, or an antibody with increased pI prior to conferring an ion concentration-dependent antigen-binding domain.
- the resulting antibodies may be assessed by antibody pharmacokinetic assay using plasma such as of mice, rats, rabbits, dogs, monkeys, humans, to select antibodies with enhanced antigen elimination from plasma as compared to the reference antibody.
- the resulting antibodies may be compared with a reference antibody in terms of the extracellular matrix-binding ability by electrochemiluminescence or others to select antibodies with increased binding to extracellular matrix.
- the resulting antibodies may be compared with a reference antibody in terms of the binding activity to various Fc ⁇ Rs under a neutral pH condition using BIACORE(registered trademark) or others to select antibodies with increased binding activity to various Fc ⁇ Rs under the neutral pH condition.
- the various Fc ⁇ Rs may be a type of Fc ⁇ R of interest, for example, Fc ⁇ RIIb.
- Fc ⁇ R can be human Fc ⁇ R.
- the resulting antibodies may be compared with a reference antibody in terms of the FcRn-binding activity under a neutral pH condition using BIACORE or other known techniques to select antibodies with increased FcRn-binding activity under the neutral pH condition.
- the FcRn can be human FcRn.
- the resulting antibodies may be evaluated for their pI by isoelectric focusing or others to select antibodies with increased pI as compared to the reference antibody.
- it is possible to select antibodies whose pI value has been increased for example, by at least 0.01, 0.03, 0.05, 0.1, 0.2, 0.3, 0.4, or 0.5 or more, or at least 0.6, 0.7, 0.8, or 0.9 or more; or antibodies whose pI value has been increased by at least 1.0, 1.1, 1.2, 1.3, 1.4, or 1.5 or more, or at least 1.6, 1.7, 1.8, 1.9, 2.0, 2.1, 2.2, 2.3, 2.4, or 2.5 or more, or 3.0 or more.
- the resulting antibodies may be compared with a reference antibody in terms of binding activity to a desired antigen under low and high ion concentration conditions using BIACORE or others to select antibodies whose antigen-binding activity have been changed or increased according to the ion concentration condition.
- the ion concentration may be, for example, hydrogen ion concentration or metal ion concentration.
- the ion concentration is a metal ion concentration, it can be, for example, calcium ion concentration.
- Whether the binding activity has been changed or increased may be assessed based on the presence of, for example: (a) an altered or enhanced antigen uptake by cells; (b) an altered or increased ability to bind to different antigen molecules multiple times; (c) an altered or enhanced reduction of antigen concentration in plasma; or (d) an altered plasma retention of the antibody. Alternatively, any two or more of these selection methods may be appropriately combined, if needed.
- Disclosure A relates to methods for producing or screening for antibodies that contain an antigen-binding domain whose antigen-binding activity changes according to the ion concentration condition and whose pI has been increased by modifying at least one amino acid residue that can be exposed on the antibody surface ("ion concentration-dependent antibodies with increased pI").
- the production methods can be performed, for example, by appropriately combining as needed, the related embodiments described within the scope of Disclosure A herein, for example, the embodiment of methods for producing or screening for antibodies with increased pI described above, as well as the embodiment of methods for producing or screening for calcium ion concentration-dependent antigen-binding domains or calcium ion concentration-dependent antibodies whose antigen-binding activity is higher under a high calcium ion concentration condition than under a low calcium ion concentration condition, or libraries thereof described above and/or the embodiment of methods for producing or screening for pH-dependent antigen-binding domains or pH-dependent antibodies whose antigen-binding activity is higher in a neutral pH condition than in an acidic pH condition, or libraries thereof described above.
- Disclosure A provides a method for producing or screening for an antibody containing an antigen-binding domain whose extracellular matrix-binding activity has been increased, wherein its antigen-binding activity changes according to the ion concentration condition and its pI has been increased by modifying at least one amino acid residue that can be exposed on the antibody surface ("an ion concentration-dependent antibody with increased pI"). Increase in pI of an ion concentration-dependent antibody may be contemplated in the method.
- Such method can be performed, for example, by appropriately combining as needed, related embodiments described within the scope of Disclosure A herein, for example, the embodiment of method for producing or screening for antibodies with an increased pI described above, as well as the embodiment of methods for producing or screening for calcium ion concentration-dependent antigen-binding domains or calcium ion concentration-dependent antibodies whose antigen-binding activity is higher under a high calcium ion concentration condition than under a low calcium ion concentration condition, or libraries thereof described above and/or the embodiment of methods for producing or screening for pH-dependent antigen-binding domains or pH-dependent antibodies whose antigen-binding activity is higher under a neutral pH condition than under an acidic pH condition, or libraries thereof described above.
- the resulting antibodies may be compared with a reference antibody in terms of the extracellular matrix-binding ability by electrochemiluminescence or other known techniques to select antibodies with increased extracellular matrix binding.
- the reference antibody may include, but is not limited to, a native antibody (for example, a native Ig antibody, preferably a native IgG antibody) and an antibody before modification (an antibody prior to or during library construction, for example, an ion concentration-dependent antibody before its pI is increased or an antibody with increased pI before it is conferred with an ion concentration-dependent antigen-binding domain).
- a native antibody for example, a native Ig antibody, preferably a native IgG antibody
- an antibody before modification an antibody prior to or during library construction, for example, an ion concentration-dependent antibody before its pI is increased or an antibody with increased pI before it is conferred with an ion concentration-dependent antigen-binding domain.
- Disclosure A relates to a method for producing an antibody comprising an antigen-binding domain whose antigen-binding activity changes according to ion concentration conditions, wherein the method comprises modifying at least one amino acid residue that may be exposed on the surface of the antibody so as to increase the isoelectric point (pI).
- the amino acid residue modification comprises a modification selected from the group consisting of: (a) substitution of a negatively charged amino acid residue with an uncharged amino acid residue; (b) substitution of a negatively charged amino acid residue with a positively charged amino acid residue; and (c) substitution of an uncharged amino acid residue with a positively charged amino acid residue.
- at least one modified amino acid residue is substituted with histidine.
- the antibody comprises a variable region and/or a constant region, and an amino acid residue is modified in the variable region and/or the constant region.
- at least one amino acid residue modified according to the method is in a position in a CDR or FR selected from the group consisting of: (a) position 1, 3, 5, 8, 10, 12, 13, 15, 16, 18, 19, 23, 25, 26, 39, 41, 42, 43, 44, 46, 68, 71, 72, 73, 75, 76, 77, 81, 82, 82a, 82b, 83, 84, 85, 86, 105, 108, 110, and 112 in a FR of the heavy chain variable region; (b) position 31, 61, 62, 63, 64, 65, and 97 in a CDR of the heavy chain variable region; (c) position 1, 3, 7, 8, 9, 11, 12, 16, 17, 18, 20, 22, 37, 38, 39, 41, 42, 43, 45, 46, 49, 57, 60, 63, 65, 66, 68,
- At least one amino acid residue modified according to the method is in a position in a CDR or FR selected from the group consisting of (a) position 8, 10, 12, 13, 15, 16, 18, 23, 39, 41, 43, 44, 77, 82, 82a, 82b, 83, 84, 85, and 105 in a FR of the heavy chain variable region; (b) position 31, 61, 62, 63, 64, 65, and 97 in a CDR of the heavy chain variable region; (c) position 16, 18, 37, 41, 42, 45, 65, 69, 74, 76, 77, 79, and 107 in a FR of the light chain variable region; and (d) position 24, 25, 26, 27, 52, 53, 54, 55, and 56 in a CDR of the light chain variable region.
- the antigen is a soluble antigen.
- the method further comprises comparing the KD of an antibody produced according to the method for its corresponding antigen in an acidic pH (e.g., pH 5.8) and a neutral pH (e.g., pH 7.4).
- the method comprises selecting an antibody that has a KD (acidic pH range (e.g., pH 5.8)) / KD (neutral pH range (e.g., pH 7.4)), for the antigen of 2 or higher.
- the method further comprises comparing the antigen binding activity of an antibody produced according to the method under a high ion concentration (e.g., a hydrogen ion or calcium ion concentration) and a low ion concentration condition.
- the method further comprises selecting an antibody that has a higher antigen binding activity under a high ion concentration (e.g., 2-fold) than under a low ion concentration.
- the high calcium ion concentration may be selected between 100 ⁇ M and 10 mM, between 200 ⁇ M and 5 mM, between 400 ⁇ M and 3 mM, between 200 ⁇ M and 2 mM, or between 400 ⁇ M and 1 mM.
- a concentration selected between 500 ⁇ M and 2.5 mM, which is close to the plasma (blood) concentration of calcium ion in vivo, may be also preferred.
- the low calcium ion concentration may be selected between 0.1 ⁇ M and 30 ⁇ M, between 0.2 ⁇ M and 20 ⁇ M, between 0.5 ⁇ M and 10 ⁇ M, or between 1 ⁇ M and 5 ⁇ M, or between 2 ⁇ M and 4 ⁇ M.
- a concentration selected between 1 ⁇ M and 5 ⁇ M, which is close to the concentration of calcium ion in early endosomes in vivo, may be also preferred.
- the lower limit of the KD (low calcium ion concentration condition)/KD (high calcium ion concentration condition) is 2 or more, 10 or more, or 40 or more, and the upper limit thereof is 400 or less, 1000 or less, or 10000 or less.
- the lower limit of the kd (low calcium ion concentration condition)/kd (high calcium ion concentration condition) is 2 or more, 5 or more, 10 or more, or 30 or more, and the upper limit thereof is 50 or less, 100 or less, or 200 or less.
- low hydrogen ion concentration may be selected from pH 6.7 to pH 10.0, from pH 6.7 to pH 9.5, from pH 7.0 to pH 9.0, or from pH 7.0 to pH 8.0.
- the low hydrogen ion concentration may be preferably pH 7.4 which is close to the in vivo pH in plasma (blood), but for the convenience of measurement, for example, pH 7.0 may be used.
- high hydrogen ion concentration may be selected from pH 4.0 to pH 6.5, from pH 4.5 to pH 6.5, pH 5.0 to pH 6.5, or pH 5.5 to pH 6.5.
- the acidic pH range may be preferably pH 5.8 which is close to the in vivo hydrogen ion concentration in the early endosome, but for the convenience of measurement, for example, pH 6.0 may be used.
- the lower limit of KD (acidic pH range)/KD (neutral pH range) e.g., KD (pH 5.8)/KD (pH 7.4)
- KD (pH 5.8)/KD (pH 7.4) is 2 or more, 10 or more, or 40 or more
- the upper limit thereof is 400 or less, 1000 or less, or 10000 or less.
- the method further comprises comparing the elimination of antigen from plasma after the administration of an antibody produced according to the method as compared to that when a reference antibody which differs only in that it does not include the modification(s) introduced according to the method, is administered.
- the method further comprises selecting an antibody produced according to the method that promotes elimination of the antigen from plasma (e.g., 2-fold) as compared to an antibody that does not contain the modifications introduced according to the method.
- the method further comprises comparing extracellular matrix-binding of the antibody produced according to the method as compared to the antibody which differs only in that it does not include the modification(s) introduced according to the method.
- the method further comprises selecting an antibody produced according to the method that has increased extracellular matrix-binding (e.g., 2-fold when bound to an antigen) as compared to an antibody which differs only in that it does not include the modification(s) introduced according to the method.
- the antibodies produced according to the method (substantially) retain the antigen-binding activity when compared to the antibodies before modification or alteration of at least one amino acid residue to increase pI (native antibodies (for example, native Ig antibodies, preferably native IgG antibodies) or reference antibodies (e.g., antibodies before antibody modification, or prior to or during library construction)).
- to (substantially) retain the antigen-binding activity can mean to have an activity of at least 50% or more, preferably 60% or more, more preferably 70% or 75% or more, and still more preferably 80%, 85%, 90%, or 95% or more as compared to the binding activity of the antibodies before modification or alteration.
- Disclosure A relates to a method for producing an antibody comprising an antigen-binding domain whose antigen-binding activity changes according to ion concentration conditions, wherein the method comprises modifying at least one amino acid residue that may be exposed on the surface of a constant region of an antibody so as to increase the isoelectric point (pI).
- the amino acid residue modification comprises a modification selected from the group consisting of: (a) substitution of a negatively charged amino acid residue with an uncharged amino acid residue; (b) substitution of a negatively charged amino acid residue with a positively charged amino acid residue; and (c) substitution of an uncharged amino acid residue with a positively charged amino acid residue.
- at least one modified amino acid residue is substituted with histidine.
- the antibody comprises a variable region and/or a constant region, and an amino acid residue is modified in the variable region and/or the constant region.
- at least one amino acid residue modified according to the method is in a position in a constant region selected from the group consisting of position 196, 253, 254, 256, 258, 278, 280, 281, 282, 285, 286, 307, 309, 311, 315, 327, 330, 342, 343, 345, 356, 358, 359, 361, 362, 373, 382, 384, 385, 386, 387, 389, 399, 400, 401, 402, 413, 415, 418, 419, 421, 424, 430, 433, 434, and 443, according to EU numbering.
- At least one amino acid residue modified according to the method is in a position in a constant region selected from the group consisting of position 254, 258, 281, 282, 285, 309, 311, 315, 327, 330, 342, 343, 345, 356, 358, 359, 361, 362, 384, 385, 386, 387, 389, 399, 400, 401, 402, 413, 418, 419, 421, 433, 434, and 443, according to EU numbering.
- At least one amino acid residue modified according to the method is in a position in a constant region selected from the group consisting of position 282, 309, 311, 315, 342, 343, 384, 399, 401, 402, and 413, according to EU numbering.
- the method further comprises comparing the antigen binding activity of an antibody produced according to the method under a high ion concentration (e.g., a hydrogen ion or calcium ion concentration) and a low ion concentration condition.
- the method further comprises selecting an antibody that has a higher antigen binding activity under a high ion concentration than under a low ion concentration.
- the method comprises comparing the elimination of antigen from plasma after the administration of an antibody produced according to the method as compared to that when a reference antibody which differs only in that it does not include the modification(s) introduced according to the method, is administered.
- the method further comprises selecting an antibody produced according to the method that promotes elimination of the antigen from plasma (e.g., 2-fold) as compared to an antibody that does not contain the modifications introduced according to the method.
- the method comprises comparing extracellular matrix-binding of the antibody produced according to the method as compared to the antibody which differs only in that it does not include the modification(s) introduced according to the method.
- the method further comprises selecting an antibody produced according to the method that has increased extracellular matrix-binding binding (e.g., 2-fold when bound to antigen) as compared to an antibody which differs only in that it does not include the modification(s) introduced according to the method.
- the method comprises comparing the Fc gamma receptor (Fc ⁇ R)-binding activity under neutral pH (e.g., pH 7.4) of an antibody produced according to the method with that of a reference antibody comprising a constant region of a native IgG.
- the method comprises selecting an antibody produced according to the method that has enhanced Fc ⁇ R-binding activity under a neutral pH (e.g., pH 7.4) as compared to that of the reference antibody comprising a constant region of a native IgG.
- a neutral pH e.g., pH 7.4
- the selected antibody produced according to the method has enhanced Fc ⁇ RIIb binding activity under neutral pH.
- the selected antibody produced according to the method has binding activity towards one or more activating Fc ⁇ R, preferably selected from the group consisting of Fc ⁇ RIa, Fc ⁇ RIb, Fc ⁇ RIc, Fc ⁇ RIIIa, Fc ⁇ RIIIb and Fc ⁇ RIIa, and towards Fc ⁇ RIIb, and optionally the Fc ⁇ RIIb-binding activity is maintained or enhanced and the binding activity to the activating Fc ⁇ Rs is decreased, as compared to those of a reference antibody which differs only in that its constant region is that of a native IgG.
- the method further comprises comparing FcRn-binding activity under a neutral pH condition of the antibody produced according to the method as compared to a reference antibody which differs only in that its constant region is that of a native IgG. In further embodiments, the method further comprises selecting an antibody produced according to the method that has increased FcRn-binding activity under a neutral pH condition as compared to that of a reference antibody which differs only in that its constant region is that of a native IgG (e.g., 2-fold).
- the antibodies produced according to the method retain the antigen-binding activity when compared to the antibodies before modification or alteration of at least one amino acid residue to increase pI (native antibodies (for example, native Ig antibodies, preferably native IgG antibodies) or reference antibodies (e.g., antibodies before antibody modification, or prior to or during library construction)).
- “to (substantially) retain the antigen-binding activity” can mean to have an activity of at least 50% or more, preferably 60% or more, more preferably 70% or 75% or more, and still more preferably 80%, 85%, 90%, or 95% or more as compared to the binding activity of the antibodies before modification or alteration.
- the method comprises modifying at least one amino acid residue that may be exposed on the surface of a variable region and constant region of an antibody so as to increase the isoelectric point (pI).
- at least one amino acid residue modified according to the method is in a position in a constant region disclosed above.
- at least one amino acid residue modified according to the method is in a position in a variable region disclosed above.
- at least one amino acid residue modified according to the method is in a position in a constant region disclosed above and at least one amino acid residue modified according to the method is in a position in a variable region disclosed above.
- the antigen is a soluble antigen.
- the method further comprises comparing the KD of an antibody produced according to the method for its corresponding antigen in an acidic pH (e.g., pH 5.8) and a neutral pH (e.g., pH 7.4).
- the method comprises selecting an antibody that has a KD (acidic pH range) / KD (neutral pH range), for the antigen of 2 or higher.
- the method further comprises comparing the antigen binding activity of an antibody produced according to the method under a high ion concentration (e.g., a hydrogen ion or calcium ion concentration) condition and a low ion concentration condition.
- the method further comprises selecting an antibody that has a higher antigen binding activity under a high ion concentration than under a low ion concentration.
- the ion concentration is calcium ion concentration
- the high calcium ion concentration may be selected between 100 ⁇ M and 10 mM, between 200 ⁇ M and 5 mM, between 400 ⁇ M and 3 mM, between 200 ⁇ M and 2 mM, or between 400 ⁇ M and 1 mM.
- a concentration selected between 500 ⁇ M and 2.5 mM may be also preferred.
- the low calcium ion concentration may be selected between 0.1 ⁇ M and 30 ⁇ M, between 0.2 ⁇ M and 20 ⁇ M, between 0.5 ⁇ M and 10 ⁇ M, or between 1 ⁇ M and 5 ⁇ M, or between 2 ⁇ M and 4 ⁇ M. A concentration selected between 1 ⁇ M and 5 ⁇ M may be also preferred.
- the lower limit of the KD (low calcium ion concentration condition)/KD (high calcium ion concentration condition) e.g., KD (3 ⁇ M Ca)/KD (2 mM Ca)
- the upper limit thereof is 400 or less, 1000 or less, or 10000 or less.
- the lower limit of the kd (low calcium ion concentration condition)/kd (high calcium ion concentration condition) is 2 or more, 5 or more, 10 or more, or 30 or more, and the upper limit thereof is 50 or less, 100 or less, or 200 or less.
- low hydrogen ion concentration neutral pH range
- the low hydrogen ion concentration may be preferably pH 7.4 which is close to the in vivo pH in plasma (blood), but for the convenience of measurement, for example, pH 7.0 may be used.
- high hydrogen ion concentration (acidic pH range) may be selected from pH 4.0 to pH 6.5, from pH 4.5 to pH 6.5, pH 5.0 to pH 6.5, or pH 5.5 to pH 6.5.
- the acidic pH range may be pH 5.8 or pH 6.0, for example.
- the lower limit of KD (acidic pH range)/KD (neutral pH range) is 2 or more, 10 or more, or 40 or more, and the upper limit thereof is 400 or less, 1000 or less, or 10000 or less.
- the method further comprises comparing the elimination of antigen from plasma after the administration of an antibody produced according to the method as compared to that when a reference antibody which differs only in that it does not include the modification(s) introduced according to the method is administered.
- the method further comprises selecting an antibody produced according to the method that promotes elimination of the antigen from plasma (e.g., 2-fold) as compared to an antibody that does not contain the modifications introduced according to the method.
- the method further comprises comparing extracellular matrix-binding of the antibody produced according to the method as compared to the antibody which differs only in that it does not include the modification(s) introduced according to the method.
- the method further comprises selecting an antibody produced according to the method that has increased extracellular matrix-binding binding (e.g., 5-fold when complexed with antigen) as compared to an antibody which differs only in that it does not include the modification(s) introduced according to the method.
- the method further comprises comparing the Fc gamma receptor (Fc ⁇ R)-binding activity under neutral pH (e.g., pH 7.4) of an antibody produced according to the method with that of a reference antibody comprising a constant region of a native IgG.
- the method comprises selecting an antibody produced according to the method that has enhanced Fc ⁇ R-binding activity under a neutral pH (e.g., pH 7.4) as compared to that of the reference antibody comprising a constant region of a native IgG.
- a neutral pH e.g., pH 7.4
- the selected antibody produced according to the method has enhanced Fc ⁇ RIIb binding activity under neutral pH.
- the selected antibody produced according to the method has binding activity towards one or more activating Fc ⁇ R, preferably selected from the group consisting of Fc ⁇ RIa, Fc ⁇ RIb, Fc ⁇ RIc, Fc ⁇ RIIIa, Fc ⁇ RIIIb and Fc ⁇ RIIa, and towards Fc ⁇ RIIb, and optionally the Fc ⁇ RIIb-binding activity is maintained or enhanced and the binding activity to the activating Fc ⁇ Rs is decreased, as compared to those of a reference antibody which differs only in that its constant region is that of a native IgG.
- the method further comprises comparing FcRn-binding activity under a neutral pH condition of the antibody produced according to the method as compared to a reference antibody which differs only in that its constant region is that of a native IgG.
- the method further comprises selecting an antibody produced according to the method that has increased FcRn-binding activity under a neutral pH condition (e.g., 2-fold) as compared to that of a reference antibody which differs only in that its constant region is that of a native IgG.
- the antibodies produced according to the method retain the antigen-binding activity when compared to the antibodies before modification or alteration of at least one amino acid residue to increase pI (native antibodies (for example, native Ig antibodies, preferably native IgG antibodies) or reference antibodies (e.g., antibodies before antibody modification, or prior to or during library construction)).
- “to (substantially) retain the antigen-binding activity” can mean to have an activity of at least 50% or more, preferably 60% or more, more preferably 70% or 75% or more, and still more preferably 80%, 85%, 90%, or 95% or more as compared to the binding activity of the antibodies before modification or alteration.
- Disclosure A relates to an antibody obtained by the above-described method of Disclosure A for producing or screening antibodies.
- Disclosure A relates to a composition or pharmaceutical composition comprising an antibody of Disclosure A described above.
- the pharmaceutical composition of Disclosure A may be a pharmaceutical composition for accelerating antigen elimination from a biological fluid (preferably, plasma, etc.) of subjects and/or for increasing the extracellular matrix binding (when an antibody of Disclosure A is administered to (applied to) the subject (preferably, in vivo)).
- the pharmaceutical composition of Disclosure A may optionally contain a pharmaceutically acceptable carrier.
- pharmaceutical compositions may typically refer to agents for use in treatment, prevention, diagnosis, or examination of diseases.
- compositions or pharmaceutical compositions of Disclosure A can be suitably formulated. In some embodiments, they can be used parenterally, for example, in a form of a sterile solution or suspension for injection in water or any other pharmaceutically acceptable liquid.
- the compositions can be suitably formulated at a unit dose required for generally accepted pharmaceutical practice, by appropriately combining with pharmaceutically acceptable carriers or media.
- pharmaceutically acceptable carriers or media include, but are not limited to, sterile water, physiological saline, vegetable oils, emulsifiers, suspending agents, surfactants, stabilizers, flavoring agents, excipients, vehicles, preservatives, and binding agents.
- the amount of active ingredient in the compositions may be adjusted in such a way that the dose falls within an appropriate pre-determined range.
- compositions or pharmaceutical compositions of Disclosure A can be administered parenterally.
- the compositions or pharmaceutical compositions may be appropriately prepared as, for example, an injectable, transnasal, transpulmonary, or transdermal composition.
- the compositions or pharmaceutical compositions may be administered systemically or locally, for example, by intravenous injection, intramuscular injection, intraperitoneal injection, or subcutaneous injection.
- the disclosure provides antibodies whose pI is increased by modifying at least one amino acid residue that can be exposed on the surface (antibodies with increased pI); methods for producing these antibodies; or use of these antibodies to enhance antigen elimination from plasma (when the antibodies are administered to the subjects in vivo). It can be understood that the scope of Disclosure A described herein and the contents described in the counterpart Examples herein can be appropriately applied to such embodiments. In other embodiments, the disclosure provides antibodies whose pI is decreased by modifying at least one amino acid residue that can be exposed on the surface ("antibodies with decreased pI"); methods for producing these antibodies; or use of these antibodies to improve plasma retention (when the antibodies are administered to the subjects in vivo).
- the inventors have revealed that cellular internalization of an antibody can be enhanced by increasing its pI by introducing specific amino acid mutations into specific sites in the amino acid sequence of the constant region.
- Those of ordinary skill in the art can understand that antibody plasma retention can be prolonged as the pI has been reduced to suppress cellular internalization of the antibody by introducing amino acids with a different side-chain charge property into the sites described above. It can be understood that the scope of Disclosure A described herein and the contents described in the counterpart Examples herein can be appropriately applied to such embodiments.
- the disclosure provides a method for producing a modified antibody, whose half life in plasma is prolonged or reduced, as compared to that before the modification of the antibody, wherein the method comprises: (a) modifying a nucleic acid encoding the antibody before the modification to change the charge of at least one amino acid residue located at a position selected from the group consisting of position 196, 253, 254, 256, 257, 258, 278, 280, 281, 282, 285, 286, 306, 307, 308, 309, 311, 315, 327, 330, 342, 343, 345, 356, 358, 359, 361, 362, 373, 382, 384, 385, 386, 387, 388, 389, 399, 400, 401, 402, 413, 415, 418, 419, 421, 424, 430, 433, 434, and 443, according to EU numbering; (b) culturing a host cell to express the modified nucleic acid and to produce the antibody; and (c) collecting the
- An additional embodiment provides a method for prolonging or reducing the half-life of an antibody in plasma wherein the method comprises modifying at least one amino acid residue located at a position selected from the group consisting of position 196, 253, 254, 256, 257, 258, 278, 280, 281, 282, 285, 286, 306, 307, 308, 309, 311, 315, 327, 330, 342, 343, 345, 356, 358, 359, 361, 362, 373, 382, 384, 385, 386, 387, 388, 389, 399, 400, 401, 402, 413, 415, 418, 419, 421, 424, 430, 433, 434, and 443, according to EU numbering.
- These methods may further comprise determining that the half life in plasma of the collected and/or modified antibody is prolonged or reduced, as compared to that before the modification of the antibody.
- the change of charge may be achieved by amino acid substitution(s).
- the substituted amino acid residue(s) may be selected from the group consisting of the amino acid residues of group (a) and (b) below, but is not limited thereto: (a) Glu (E) and Asp (D); and (b) Lys (K), Arg (R) and His (H).
- the antibody may be an Ig-type antibody such as an IgG antibody. In some embodiments, the antibody may be a chimeric antibody, humanized antibody, or human antibody. In some embodiments, the antibody may be a multispecific antibody such as a bispecific antibody.
- Disclosure B In non-limited embodiments, Disclosure B relates to Fc region variants, uses thereof, and production methods thereof.
- an "Fc region variant” may refer, for example, to an Fc region modified from the Fc region of a native IgG antibody by modifying at least one amino acid with another amino acid, or may refer to an Fc region modified from such an Fc region variant by additionally modifying at least one amino acid with another amino acid.
- Fc region variants include not only Fc regions that have been introduced with the amino acid modification but also Fc regions containing the same amino acid sequence as an aforementioned Fc region.
- Disclosure B relates to Fc region variants containing an FcRn-binding domain which contains Ala at position 434; any one of Glu, Arg, Ser, and Lys at position 438; and any one of Glu, Asp, and Gln at position 440, according to EU numbering (within the scope of Disclosure B described herein, such an Fc region variant is also referred to as a "novel Fc region variant" for descriptive purposes).
- Fc region variants of Disclosure B can be incorporated into virtually any antibody (e.g., multispecific antibodies such as bispecific antibodies) regardless of the type of the target antigen.
- Anti-factor IXa/factor X bispecific antibodies can be produced using such Fc region variants as shown in Example 20 (e.g., F8M-F1847mv [F8M-F1847mv1 (SEQ ID NO:323) and F8M-F1847mv2 (SEQ ID NO:324) as the heavy chains and F8ML (SEQ ID NO:325) as the light chain]; F8M-F1868mv [ F8M-F1868mv1 (SEQ ID NO:326) and F8M-F1868mv2 (SEQ ID NO:327) as the heavy chains and F8ML (SEQ ID NO:325) as the light chain]; and F8M-F1927mv [ F8M-F1927mv1 (SEQ ID NO:328) and F8M-F19
- WO2013/046704 reports that Fc region variants that have been introduced with a mutation to increase their FcRn binding under acidic conditions in combination with a specific mutation (a representative example is dual-residue mutation Q438R/S440E according to EU numbering) exhibit significantly reduced binding to rheumatoid factor.
- WO2013/046704 does not describe that the Fc region variants whose rheumatoid factor binding has been reduced due to the Q438R/S440E modification are superior in plasma retention as compared to antibodies with a native Fc region.
- Fc region variants that allow improved plasma retention, but do not bind to anti-drug antibodies (pre-existing ADA, etc.).
- Fc region variants that contain combined mutations of amino acid residues, which are a substitution of Ala (A) for the amino acid at position 434 according to EU numbering and a specific dual-residue mutation (a representative example is Q438R/S440E), are preferable for prolonging antibody retention in plasma while maintaining a significantly reduced binding to rheumatoid factor.
- novel Fc region variants of Disclosure B disclosed herein provides an advantageous and surprising improvement over the Fc region variants described in WO2013/046704, which is incorporated herein by reference in their entirety.
- Disclosure B provides novel combinations of amino acid substitutions in the FcRn-binding domain, which increase the FcRn-binding activity of antibodies in an acidic pH range and in a neutral pH range, in particular, in an acidic pH range.
- an Fc region variant of Disclosure B contains Ala at position 434; any one of Glu, Arg, Ser, and Lys at position 438; and any one of Glu, Asp, and Gln at position 440, according to EU numbering; and more preferably contain Ala at position 434; either Arg or Lys at position 438; and either Glu or Asp at position 440, according to EU numbering.
- the Fc region variant of Disclosure B additionally contains either Ile or Leu at position 428, and/or any one of Ile, Leu, Val, Thr, and Phe at position 436, according to EU numbering. More preferably the Fc region variant contains Leu at position 428, and/or either Val or Thr at position 436, according to EU numbering.
- the Fc region variant of Disclosure B can be an Fc region variant of a native Ig antibody, and more preferably the Fc region variant of a native IgG (IgG1, IgG2, IgG3, or IgG4 type) antibody.
- the native Fc region is partly described within the scope of Disclosures A and B, herein. More specifically, in Disclosure B, the native Fc region can refer to an unmodified or naturally-occurring Fc region, and preferably, an unmodified or naturally-occurring Fc region of a native Ig antibody whose Fc region amino acid residues remain unmodified.
- the antibody origin of the Fc region can be an Ig such as IgM or IgG, for example, human IgG1, IgG2, IgG3, or IgG4. In one embodiment, it may be human IgG1.
- a (reference) antibody comprising a native Fc region can refer to an antibody comprising an unmodified or naturally-occurring Fc region.
- Positions 428, 434, 438, and 440 are common to Fc regions of all native human IgG1, IgG2, IgG3, and IgG4 antibodies. However, at position 436 in the Fc region, native human IgG1, IgG2, and IgG4 antibodies share Tyr (Y) whereas native human IgG3 antibody has Phe (F).
- Stapleton et al. (Nature Comm. 599 (2011) reported that human IgG3 allotypes containing the amino acid substitution of R435H according to EU numbering have a plasma half-life in human comparable to that of IgG1.
- the inventors also conceived that plasma retention could be improved by increasing FcRn binding under an acidic condition by introducing the R435H amino acid substitution in combination with the amino acid substitution at position 436.
- WO2013/046704 also specifically reported dual amino acid residue substitutions of Q438R/S440E, Q438R/S440D, Q438K/S440E, and Q438K/S440D according to EU numbering, which result in a significant reduction of the rheumatoid factor binding when combined with an amino acid substitution that can increase the FcRn binding under an acidic condition.
- the FcRn-binding domain of an Fc region variant of Disclosure B may contain a combination of substituted amino acid positions selected from the group consisting of: (a) N434A/Q438R/S440E; (b) N434A/Q438R/ S440D; (c) N434A/Q438K/S440E; (d) N434A/Q438K/S440D; (e) N434A/Y436T/ Q438R/S440E; (f) N434A/Y436T/Q438R/S440D; (g) N434A/Y436T/Q438K/S440E; (h) N434A/Y436T/ Q438K/S440D; (i) N434A/Y436V/Q438R/S440E; (j) N434A/Y436V/ Q438R/S440D; (k) N434A/Y4
- the FcRn-binding domain of an Fc region variant of Disclosure B may contain a combination of substituted amino acids selected from the group consisting of: (a) N434A/Q438R/S440E; (b) N434A/Y436T/Q438R/ S440E; (c) N434A/Y436V/Q438R/S440E; (d) M428L/N434A/Q438R/S440E; (e) M428L/N434A/Y436T/Q438R/S440E; (f) M428L/N434A/ Y436V/Q438R/S440E; (g) L235R/G236R/S239K/M428L/N434A/Y436T/Q438R/S440E; and (h) L235R/G236R/ A327G/A330S/P331S/M428L/N
- the FcRn-binding activity of an Fc region variant of Disclosure B has been increased under an acidic pH condition as compared to the Fc region of a native IgG.
- An increase in the FcRn-binding activity (binding affinity) of an FcRn-binding domain in a pH range may correspond to an increase of the measured FcRn-binding activity (binding affinity) when compared to the measured FcRn-binding activity (binding affinity) of a native FcRn-binding domain.
- KD (native Fc region)/KD (an Fc region variant of Disclosure B) which represents a difference in the binding activity (binding affinity)
- KD (native Fc region)/KD an Fc region variant of Disclosure B
- Such an increase may occur in an acidic pH range and/or in a neutral pH range; however, the increase in an acidic pH range can be preferred from the viewpoint of the action mechanism for Disclosure B.
- the FcRn-binding activity (for example, at pH 6.0 and 25°C) of an Fc region variant of Disclosure B whose FcRn-binding activity has been increased in an acidic pH range is greater than that of the Fc region of a native IgG, for example, by 1.5-fold, 2-fold, 3-fold, 4-fold, 5-fold, 6-fold, 7-fold, 8-fold, 9-fold, 10-fold, 20-fold, 30-fold, 50-fold, 75-fold, 100-fold, 200-fold, 500-fold, 1000-fold or more.
- the increased FcRn-binding activity of an Fc region variant in an acidic pH range may be greater than the FcRn-binding activity of the Fc region of a native IgG by at least 5-fold or at least 10-fold.
- the purity of pharmaceutical proteins in terms of monomer and high-molecular-weight species is also important in developing pharmaceutical agents.
- wild-type IgG1 does not contain a significant amount of high-molecular-weight species, whereas manipulation of the FcRn-binding domain by introducing substitutions can produce a large amount of high-molecular-weight species. In this case, such high-molecular-weight species may have to be removed from the drug substance by purification steps.
- Amino acid substitutions in antibodies can result in negative consequences, such as an increase in the immunogenicity of therapeutic antibodies which in turn can cause a cytokine storm and/or production of anti-drug antibodies (ADAs).
- ADAs anti-drug antibodies
- the clinical utility and efficacy of therapeutic antibodies can be limited by ADAs, since they affect the efficacy and pharmacokinetics of therapeutic antibodies and sometimes lead to serious side effects.
- Many factors influence the immunogenicity of therapeutic antibodies, and the presence of effector T-cell epitopes is one of the factors. Likewise, the presence of pre-existing antibodies against a therapeutic antibody can also be problematic.
- rheumatoid factor an auto-antibody (an antibody directed against a self-protein) against the Fc portion of an antibody (i.e., IgG).
- Rheumatoid factor is found in particular in patients with systemic lupus erythematosus (SLE) or rheumatoid arthritis. In arthritis patients, RF and IgG join to form immune complexes that contribute to the disease process.
- SLE systemic lupus erythematosus
- IgG join to form immune complexes that contribute to the disease process.
- a humanized anti-CD4 IgG1 antibody with a Asn434His mutation has been reported to elicit significant rheumatoid factor binding (Zheng et al., Clin. Pharmacol. Ther. 89(2):283-290 (2011)). Detailed studies have confirmed that the Asn434His mutation in human IgG1 increases the binding of rheumatoid factor to the F
- RF is a polyclonal auto-antibody against human IgG.
- the RF epitope in the human IgG sequence varies among clones; however, the RF epitope seems to be located in the CH2/CH3 interface region as well as in the CH3 domain which may overlap with the FcRn-binding epitope.
- mutations to increase the FcRn-binding activity at a neutral pH may possibly increase the binding activity to specific RF clones as well.
- anti-drug antibody or "ADA” can refer to an endogenous antibody that has binding activity to an epitope located on a therapeutic antibody and thus can bind to the therapeutic antibody.
- pre-existing anti-drug antibody or “pre-existing ADA” can refer to an anti-drug antibody that is present and detectable in the blood of a patient prior to administration of the therapeutic antibody to the patient.
- the pre-existing ADA is a human antibody.
- the pre-existing ADA is rheumatoid factor.
- the binding activity of an antibody Fc region (variant) against a pre-existing ADA can be, for example, represented by electrochemiluminescence (ECL) response at an acidic pH and/or at a neutral pH.
- ECL electrochemiluminescence
- the ECL assay is described, for example, in Moxness et al. (Clin Chem. 51:1983-1985 (2005)) and in Example 6. Assays can be performed, for example, under the conditions of MES buffer and 37°C.
- the antigen-binding activity of antibodies can be determined by, for example, BIACORE(registered trademark) analysis.
- the binding activity to a pre-existing ADA can be assessed at any temperature from 10°C to 50°C.
- the binding activity (binding affinity) of a human Fc region to a human pre-existing ADA is determined at a temperature of 15°C to 40°C, for example, such as between 20°C to 25°C, or 25°C.
- the interaction between a human pre-existing ADA and a human Fc region is measured at pH 7.4 (or pH 7.0) and 25°C.
- the binding activity to (pre-existing) ADA has been significantly increased or an equivalent expression may mean that the measured (pre-existing) ADA-binding activity (binding affinity) (i.e., KD) of an Fc region variant of Disclosure B or an antibody containing it has been increased, for example, by 0.55-fold, 0.6-fold, 0.7-fold, 0.8-fold, 0.9-fold, 1-fold, 1.1-fold, 1.2-fold, 1.3-fold, 1.4-fold, 1.5-fold, 1.6-fold, 1.7-fold, 1.8-fold, 1.9-fold, 2-fold, 2.1-fold, 2.2-fold, or 2.3-fold or more, as compared to the measured (pre-existing) ADA-binding activity (binding affinity) of a reference Fc region variant or a reference antibody containing the reference Fc region variant.
- binding affinity i.e., KD
- the term "patients” or "a patient” is not limited, and can include all humans with a disease who are under treatment with a therapeutic antibody.
- the patients may be humans affected with auto-immune disease, such as an arthritic disease or systemic erythematosus (SLE).
- the arthritic disease can include rheumatoid arthritis.
- the binding activity to a pre-existing ADA is significantly increased in an individual patient may mean that the binding activity of an antibody comprising an Fc region variant (e.g., therapeutic antibody) to a pre-existing ADA measured in a patient has been increased by at least 10%, at least 20%, at least 30%, at least 40%, at least 50%, or at least 60% or more when compared to the binding activity of a reference antibody to the pre-existing ADA.
- this may mean that the ECL reaction for the antibody is preferably above 250, or at least 500, or at least 1000, or even at least 2000.
- this increase may be an increase relative to a reference antibody whose ECL reaction is less than 500 or 250.
- the ECL reaction preferably ranges from less than 250 to at least 250, less than 250 to at least 500, less than 500 to 500 or more, less than 500 to 1000 or more, or less than 500 to at least 2000, without being limited thereto.
- the binding activity to a pre-existing ADA is increased can mean that in a group of patients, the measured proportion of patients who have an ECL reaction of at least 500 (preferably, at least 250) for an antibody comprising an Fc region variant with (a) increased binding activity to FcRn at an acidic pH and (b) increased binding activity to a pre-existing ADA at a neutral pH is elevated as compared to the proportion of patients who have an ECL reaction of at least 500 (preferably, at least 250 or more) for a reference antibody, for example, by at least 10%, at least 20%, at least 30%, at least 40%, at least 50% when compared to the proportion of patients who have an ECL reaction for a reference antibody.
- the binding activity to a pre-existing ADA decreases can mean that the measured binding activity (i.e., KD or ECL reaction) of an antibody comprising an Fc region variant decreases as compared to that of a reference antibody. Such decrease can be observed in an individual patient or in a group of patients.
- the affinity of an antibody comprising an Fc region variant for a pre-existing ADA at a neutral pH significantly decreases in each patient can mean that the measured binding activity to a pre-existing ADA at a neutral pH measured in the patient is decreased as compared to the binding activity of a reference antibody to the pre-existing ADA measured at the neutral pH, for example, by at least 10%, at least 20%, at least 30%, at least 40%, or at least 50%.
- the binding activity of an antibody containing an Fc region variant to a pre-existing ADA significantly decreases in an individual patient can mean that the ECL reaction for the antibody that used to be 500 or more (preferably, 1000 or more, or 2000 or more) is measured to be less than 500, preferably less than 250 as compared to the ECL reaction for a reference antibody, for example.
- the Fc region variants of Disclosure B and antibodies comprising them have low binding activity to a pre-existing ADA at a neutral pH.
- the binding activity of antibodies containing the Fc region variants of Disclosure B to a pre-existing ADA at a neutral pH is lower than or has not significantly been increased, as compared to the binding activity of a reference antibody containing the Fc region of a native IgG to the pre-existing ADA at a neutral pH (e.g., pH 7.4).
- the binding activity (binding affinity) to a pre-existing ADA is low or the affinity is at the baseline level can mean an ECL reaction of less than 500, or less than 250 in an individual patient, but is not limited thereto.
- the binding activity to a pre-existing ADA is low in a group of patients can mean that the ECL reaction is less than 500 in 90%, preferably 95%, and more preferably 98% of the patients in the group, for example.
- Fc region variants of Disclosure B or antibodies containing them whose binding activity to a (pre-existing) ADA in plasma at a neutral pH is not significantly increased, and whose FcRn-binding activity at a neutral pH and/or at an acidic pH is increased.
- the FcRn-binding activity at an acidic pH e.g., pH 5.8 is increased.
- the Fc region variants preferably do not have a significantly increased binding activity to ADA under a neutral pH condition (e.g., pH 7.4) as compared to the Fc region of a native IgG, and the ADA may be a pre-existing ADA, preferably rheumatoid factor (RF).
- the Fc region variants of Disclosure B have an increased FcRn-binding activity under an acidic pH condition as compared to the Fc region of a native IgG, and as a result they exhibit reduced clearance (CL) in plasma, prolonged retention time in plasma, or prolonged half-life in plasma (t1/2). Their correlation is known in the art.
- the Fc region variants of Disclosure B have an increased FcRn-binding activity under an acidic pH condition but do not have a significantly increased ADA-binding activity under a neutral pH condition as compared to the Fc region of a native IgG, and they exhibit reduced clearance (CL) in plasma, prolonged retention time in plasma, or prolonged half-life in plasma (t1/2).
- the ADA may be a pre-existing ADA, preferably rheumatoid factor (RF).
- the Fc region variants of Disclosure B are advantageous, since their plasma retention is improved as compared to a reference Fc region variant comprising a combination of amino acid substitutions N434Y/Y436V/Q438R/S440E according to EU numbering.
- Examples 5 to 7 compare the plasma retention of two Fc region variants: Fc region variant F1718 (Fc region with mutations introduced at four sites: N434Y/Y436V/Q438R/S440E) described in WO2013/046704 and novel Fc region variant F1848m (introduced with mutations at four sites: N434A/Y436V/Q438R/S440E). Difference in amino acid mutation between the two Fc region variants is only at position 434 according to EU numbering, where the introduced amino acid mutation is Y (tyrosine) for F1718 and A (alanine) for F1848m.
- the Fc region variants of Disclosure B can preferably have improved plasma retention as compared to reference Fc region variants containing the combination of amino acid substitutions N434Y/Y436V/Q438R/S440E.
- the experimental results described in Examples (5-2) and (7-3) herein demonstrate that among various Fc region variants, F1847m, F1886m, F1889m, and F1927m are further improved in plasma retention time than F1848m.
- Fc region variants of Disclosure B comprising F1847m, F1886m, F1889m, or F1927m, as well as F1848m have improved plasma retention as compared to reference Fc region variants containing the substitutions N434Y/Y436V/Q438R/S440E.
- Fc region variants that do not bind to effector receptors can be safer and/or more advantageous.
- Fc region variants of Disclosure B have only a weak effector receptor-binding activity or do not bind to effector receptors. Examples of effector receptors include, activating Fc ⁇ R, particularly Fc ⁇ RI, Fc ⁇ RII, and Fc ⁇ RIII.
- Fc ⁇ RI includes Fc ⁇ RIa, Fc ⁇ RIb, and Fc ⁇ RIc, and subtypes thereof.
- Fc ⁇ RII includes Fc ⁇ RIIa (having two allotypes: R131 and H131) and Fc ⁇ RIIb.
- Fc ⁇ RIII includes Fc ⁇ RIIIa (which has two allotypes: V158 and F158) and Fc ⁇ RIIIb (which has two allotypes: Fc ⁇ RIIIb-NA1 and Fc ⁇ RIIIb-NA2).
- Antibodies that have only a weak effector receptor-binding activity or do not bind to the receptors include, for example, antibodies containing a silent Fc region and antibodies that do not have an Fc region (for example, Fab, F(ab)' 2 , scFv, sc(Fv) 2 , and diabodies).
- Fc regions which have only a weak or no effector receptor-binding activity are described, for example, in Strohl et al. (Curr. Op. Biotech. 20(6):685-691 (2009)), and specifically include, for example, deglycosylated Fc regions (N297A and N297Q), and silent Fc regions resulting from manipulation of Fc regions to silence their effector functions (or to suppress immunity) (IgG1-L234A/L235A, IgG1-H268Q/A330S/P331S, IgG1-C226S/C229S, IgG1-C226S/C229S/E233P/L234V/L235A, IgG1-L234F/L235E/P331S, IgG2-V234A/G237A, IgG2-H268Q/V309L/A330S/A331S, IgG4-L235A/G237A/
- WO2008/092117 describes antibodies comprising a silent Fc region that contains a substitution of G236R/L328R, L235G/G236R, N325A/L328R, or N325L/L328R, according to EU numbering.
- WO2000/042072 describes antibodies comprising a silent Fc region that contains substitutions at one or more of positions EU233 (position 233 according to EU numbering), EU234, EU235, and EU237.
- WO2009/011941 describes antibodies comprising a silent Fc region that lacks the residues of EU231 to EU238. Davis et al. (J. Rheum.
- the expression "weak binding to effector receptors” can mean that the effector receptor-binding activity is, for example, 95% or less, preferably 90% or less, 85% or less, 80% or less, 75% or less, more preferably 70% or less, 65% or less, 60% or less, 55% or less, 50% or less, 45% or less, 40% or less, 35% or less, 30% or less, 25% or less, 20% or less, 15% or less, 10% or less, 9% or less, 8% or less, 7% or less, 6% or less, 5% or less, 4% or less, 3% or less, 2% or less, or 1% or less than that of a native IgG or an antibody containing a native IgG Fc region.
- the "silent Fc region” is an Fc region variant containing one or more amino acid substitutions, insertions, additions, deletions, and others that reduce binding to effector receptors as compared to a native Fc region. Since the effector receptor-binding activity can be reduced considerably, such silent Fc regions may no longer bind to the effector receptors.
- the silent Fc regions may include, for example, Fc regions containing amino acid substitutions at one or more positions selected from the group consisting of: EU234, EU235, EU236, EU237, EU238, EU239, EU265, EU266, EU267, EU269, EU270, EU271, EU295, EU296, EU297, EU298, EU300, EU324, EU325, EU327, EU328, EU329, EU331, and EU332. Modification of these amino acid positions may also be appropriately introduced into the Fc region variants of Disclosure B.
- the silent Fc region has a substitution at one or more positions selected from the group consisting of: EU234, EU235, EU236, EU237, EU238, EU239, EU265, EU266, EU267, EU269, EU270, EU271, EU295, EU296, EU297, EU298, EU300, EU324, EU325, EU327, EU328, EU329, EU331, and EU332, and preferably the group consisting of: EU235, EU237, EU238, EU239, EU270, EU298, EU325, and EU329, wherein the substitution is with an amino acid residue selected from the listing below:
- the amino acid at position EU234 is preferably substituted with an amino acid selected from the group consisting of Ala, Arg, Asn, Asp, Gln, Glu, Gly, His, Lys, Met, Phe, Pro, Ser, and Thr.
- the amino acid at position EU235 is preferably substituted with an amino acid selected from the group consisting of Ala, Asn, Asp, Gln, Glu, Gly, His, Ile, Lys, Met, Pro, Ser, Thr, Val, and Arg.
- the amino acid at position EU236 is preferably substituted with an amino acid selected from the group consisting of Arg, Asn, Gln, His, Leu, Lys, Met, Phe, Pro, and Tyr.
- the amino acid at position EU237 is preferably substituted with an amino acid selected from the group consisting of Ala, Asn, Asp, Gln, Glu, His, Ile, Leu, Lys, Met, Pro, Ser, Thr, Val, Tyr, and Arg.
- the amino acid at position EU238 is preferably substituted with an amino acid selected from the group consisting of Ala, Asn, Gln, Glu, Gly, His, Ile, Lys, Thr, Trp, and Arg.
- the amino acid at position EU239 is preferably substituted with an amino acid selected from the group consisting of Gln, His, Lys, Phe, Pro, Trp, Tyr, and Arg.
- the amino acid at position EU265 is preferably substituted with an amino acid selected from the group consisting of Ala, Arg, Asn, Gln, Gly, His, Ile, Leu, Lys, Met, Phe, Ser, Thr, Trp, Tyr, and Val.
- the amino acid at position EU266 is preferably substituted with an amino acid selected from the group consisting of Ala, Arg, Asn, Asp, Gln, Glu, Gly, His, Lys, Phe, Pro, Ser, Thr, Trp, and Tyr.
- the amino acid at position EU267 is preferably substituted with an amino acid selected from the group consisting of Arg, His, Lys, Phe, Pro, Trp, and Tyr.
- the amino acid at position EU269 is preferably substituted with an amino acid selected from the group consisting of Ala, Arg, Asn, Gln, Gly, His, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, and Val.
- the amino acid at position EU270 is preferably substituted with an amino acid selected from the group consisting of Ala, Arg, Asn, Gln, Gly, His, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, and Val.
- the amino acid at position EU271 is preferably substituted with an amino acid selected from the group consisting of Arg, His, Phe, Ser, Thr, Trp, and Tyr.
- the amino acid at position EU295 is preferably substituted with an amino acid selected from the group consisting of Arg, Asn, Asp, Gly, His, Phe, Ser, Trp, and Tyr.
- the amino acid at position EU296 is preferably substituted with an amino acid selected from the group consisting of Arg, Gly, Lys, and Pro.
- the amino acid at position EU297 is preferably substituted with Ala.
- the amino acid at position EU298 is preferably substituted with an amino acid selected from the group consisting of Arg, Gly, Lys, Pro, Trp, and Tyr.
- the amino acid at position EU300 is preferably substituted with an amino acid selected from the group consisting of Arg, Lys, and Pro.
- the amino acid at position EU324 is preferably substituted with either Lys or Pro.
- the amino acid at position EU325 is preferably substituted with an amino acid selected from the group consisting of Ala, Arg, Gly, His, Ile, Lys, Phe, Pro, Thr, Trp, Tyr, and Val.
- the amino acid at position EU327 is preferably substituted with an amino acid selected from the group consisting of Arg, Gln, His, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, and Val.
- the amino acid at position EU328 is preferably substituted with an amino acid selected from the group consisting of Arg, Asn, Gly, His, Lys, and Pro.
- the amino acid at position EU329 is preferably substituted with an amino acid selected from the group consisting of Asn, Asp, Gln, Glu, Gly, His, Ile, Leu, Lys, Met, Phe, Ser, Thr, Trp, Tyr, Val, and Arg.
- amino acid at position EU330 is preferably substituted with either Pro or Ser.
- the amino acid at position EU331 is preferably substituted with an amino acid selected from the group consisting of Arg, Gly, and Lys.
- the amino acid at position EU332 is preferably substituted with an amino acid selected from the group consisting of Arg, Lys, and Pro.
- the silent Fc region preferably may contain a substitution with either Lys or Arg at EU235, a substitution with either Lys or Arg at EU237, a substitution with either Lys or Arg at EU238, a substitution with either Lys or Arg at EU239, a substitution with Phe at EU270, a substitution with Gly at EU298, a substitution with Gly at EU325, or a substitution with either Lys or Arg at EU329. More preferably, the silent Fc region may contain a substitution with arginine at EU235 or a substitution with lysine at EU239. Even more preferably, the silent Fc region may contain L235R/S239K substitutions. Modification of these amino acid residues may also be appropriately introduced into the Fc region variants of Disclosure B.
- antibodies comprising an Fc region variant of Disclosure B have only a weak complement protein-binding activity or do not bind to complement proteins.
- the complement protein is C1q.
- the weak complement protein-binding activity refers to a complement protein-binding activity reduced by 10-fold or more, 50-fold or more, or 100-fold or more when compared to the complement protein-binding activity of a native IgG or an antibody containing a native IgG Fc region.
- the complement protein-binding activity of an Fc region can be reduced by amino acid modifications such as amino acid substitution, insertion, addition, or deletion.
- the Fc region variants of Disclosure B or antibodies comprising the Fc region variants can be assessed for their (human) FcRn-binding activity in a neutral pH range and/or in an acidic pH range in the same manner as described above.
- a method for modifying antibody constant regions to produce the Fc region variants of Disclosure B may be based, for example, on assessment of several constant region isotypes (IgG1, IgG2, IgG3, and IgG4) to select isotypes that have a reduced antigen-binding activity in an acidic pH range and/or have an increased dissociation rate in an acidic pH range.
- An alternative method may be based on introduction of amino acid substitutions into the amino acid sequence of a native IgG isotype to reduce the antigen-binding activity in an acidic pH range (e.g., pH 5.8) and/or to increase the dissociation rate in an acidic pH range.
- the hinge region sequence of an antibody constant region varies greatly across isotypes (IgG1, IgG2, IgG3, and IgG4), and differences in the hinge-region amino acid sequence can have a significant impact on the antigen-binding activity. Therefore, isotypes with reduced antigen-binding activity in an acidic pH range and/or increased dissociation rate in an acidic pH range can be selected by selecting suitable isotypes depending on the type of antigen or epitope. Furthermore, since differences in the hinge-region amino acid sequence can have a significant impact on the antigen-binding activity, amino acid substitutions in the amino acid sequences of native isotypes can be located in the hinge region.
- Disclosure B provides a use of an antibody containing the above-described Fc region variant of Disclosure B to accelerate the release of the antibody that has been internalized into cells in an antigen-bound form to the outside of the cells in an antigen-free form.
- release of an antibody that has been internalized into cells in an antigen-bound form to the outside of the cells in an antigen-free form does not necessarily mean that the antibody that has been internalized into cells in an antigen-bound form is completely released to the outside of the cells in an antigen-free form.
- the proportion of the antibody released in an antigen-free form to the outside of the cells is increased as compared to that before modification of its FcRn-binding domain (for example, before increasing the FcRn-binding activity of the antibody in an acidic pH range). It is preferable that the antibody released to the outside of the cells maintains its antigen-binding activity.
- the “ability to eliminate antigen from plasma” or an equivalent term can refer to the ability to eliminate antigen from plasma when an antibody is administered or secreted in vivo.
- the antibody's ability to eliminate antigen from plasma is increased can mean that when an antibody is administered, for example, the rate of antigen elimination from plasma is increased as compared to that before modification of its FcRn-binding domain.
- the increase in the antibody's activity of antigen elimination from plasma can be assessed, for example, by administering a soluble antigen and the antibody in vivo, and measuring the concentration of the soluble antigen in plasma after administration.
- the soluble antigen may be an antibody-bound or antibody-free antigen, and their concentrations can be determined as "antibody-bound antigen concentration in plasma” and “antibody-free antigen concentration in plasma”, respectively. The latter is synonymous with “free antigen concentration in plasma”.
- the “total antigen concentration in plasma” can refer to the sum of antibody-bound antigen concentration and antibody-free antigen concentration.
- Disclosure B provides a method for prolonging the plasma retention time of an antibody containing the Fc region variant of Disclosure B.
- Native human IgG can bind to FcRn derived from nonhuman animals. For example, since native human IgG can bind to mouse FcRn more strongly than to human FcRn (Ober et al., Intl. Immunol. 13(12):1551-1559 (2001)), the antibodies can be administered to mice for assessing the properties of the antibodies. Alternatively, for example, mice with their own FcRn gene has been disrupted but instead have and express the human FcRn gene as a transgene (Roopenian et al., Meth.Mol. Biol. 602:93-104 (2010)) are also suitable for assessing the antibodies.
- the plasma concentration of free antigen not bound to the antibody or the ratio of free antigen concentration to the total antigen concentration can be determined (e.g., Ng et al., Pharm. Res. 23(1):95-103 (2006)).
- an antigen exhibits a particular function in vivo
- whether the antigen is bound to an antibody that neutralizes the antigen function (antagonistic molecule) can be assessed by testing whether the antigen function is neutralized. Whether the antigen function is neutralized can be evaluated by measuring a particular in vivo marker reflective of the antigen function.
- Whether an antigen is bound to an antibody that activates the antigen function (agonistic molecule) can be assessed by measuring a particular in vivo marker reflective of the antigen function.
- the measurements can be preferably carried out after a certain period of time following antibody administration.
- "after a certain period of time following antibody administration” is not particularly limited, and the period can be appropriately determined by those of ordinary skill in the art depending on the properties of the administered antibody and others, and includes, for example, one day, three days, seven days, 14 days, or 28 days after antibody administration.
- the term "plasma antigen concentration” can refer to either "total antigen concentration in plasma” which is the sum of antibody-bound antigen concentration and antibody-free antigen concentration, or "free antigen concentration in plasma” which is antibody-free antigen concentration.
- a smaller C value indicates a higher efficiency of antigen elimination per antibody, and a larger C value indicates a lower efficiency of antigen elimination per antibody.
- the molar ratio of antigen/antibody is reduced by 2-fold, 5-fold, 10-fold, 20-fold, 50-fold, 100-fold, 200-fold, 500-fold, or 1,000-fold or more, as compared to when a reference antibody containing a native human IgG Fc region as a human FcRn-binding domain is administered.
- the reduction of total antigen concentration in plasma or molar ratio of antigen/antibody can be assessed using methods know in the art, such as that described in Examples 6, 8, and 13 of WO2011/122011. More specifically, they can be assessed based on either an antigen-antibody co-injection model or a steady-state antigen infusion model using the human FcRn transgenic mouse line 32 or 276 (Jackson Laboratories, Methods Mol. Biol. 602:93-104 (2010)), when an antibody of interest in Disclosure B does not cross-react with the mouse counterpart antigen. When the antibody cross-reacts with the mouse counterpart, it can be assessed by simply injecting the antibody into the human FcRn transgenic mouse line 32 or 276 (Jackson Laboratories).
- mice In the co-injection model, a mixture of the antibody and antigen is administered to mice.
- an infusion pump filled with an antigen solution is implanted into mice to achieve a constant antigen concentration in plasma, and then the antibody is injected into the mice. All test antibodies are administered at the same dosage.
- the total antigen concentration in plasma, free antigen concentration in plasma, and antibody concentration in plasma can be measured at appropriate time points.
- the total or free antigen concentration in plasma, or the molar ratio of antigen/antibody can be measured two days, four days, seven days, 14 days, 28 days, 56 days, or 84 days after administration.
- an antigen concentration in plasma in a long period of time can be determined by measuring the total or free antigen concentration in plasma, or the molar ratio of antigen/antibody two days, four days, seven days, 14 days, 28 days, 56 days, or 84 days after antibody administration.
- Whether the antigen concentration in plasma or the molar ratio of antigen/antibody is reduced with the antibody can be determined by assessing such reductions at one or more time points as described above.
- the total or free antigen concentrations in plasma, or the molar ratio of antigen/antibody can be measured 15 minutes, one hour, two hours, four hours, eight hours, 12 hours, or 24 hours after administration.
- an antigen concentration in plasma in a short period of time can be determined by measuring the total or free antigen concentrations in plasma, or the molar ratio of antigen/antibody 15 minutes, one hour, two hours, four hours, eight hours, 12 hours, or 24 hours after administration.
- the plasma retention in human When the plasma retention in human is difficult to determine, it may be predicted based on the plasma retention in mice (for example, normal mice, human antigen-expressing transgenic mice, or human FcRn-expressing transgenic mice) or in monkeys (for example, cynomolgus monkeys).
- Disclosure B relates to an antibody comprising the Fc region variant of Disclosure B described above.
- the various embodiments of the antibodies described within the scope of Disclosures A and B described herein can be applicable without opposing the common technical knowledge in the art and unless there are inconsistencies in the context.
- antibodies comprising an Fc region variant of Disclosure B are useful as therapeutic antibodies for treating human patients with auto-immune diseases, transplantation rejection (graft versus host disease), other inflammatory diseases, or allergy diseases, as described in WO2013/046704.
- antibodies comprising an Fc region variant of Disclosure B may have modified sugar chains.
- Antibodies with modified sugar chains can include, for example, antibodies with modified glycosylation (WO99/54342), antibodies that are deficient in fucose (WO00/61739, WO02/31140, WO2006/067847, WO2006/067913), and antibodies having sugar chains with bisecting GlcNAc (WO02/79255).
- the antibodies may be deglycosylated.
- the antibodies comprise, for example, mutations at the heavy-chain glycosylation site to inhibit glycosylation at such location as described in WO2005/03175.
- Such non-glycosylated antibodies can be prepared by modifying the heavy-chain glycosylation site, i.e., by introducing the N297Q or N297A substitution according to EU numbering, and expressing the proteins in appropriate host cells.
- Disclosure B relates to a composition or a pharmaceutical composition comprising an antibody containing such an Fc region variant.
- the various embodiments of the compositions or pharmaceutical compositions described within the scope of Disclosures A and B herein can be applicable without opposing the common technological knowledge in the art and unless there are inconsistencies in the context.
- Such compositions can be used for enhancing the plasma retention (in subjects, when an antibody of the Disclosure B is administered (applied) to the subjects).
- Disclosure B relates to nucleic acids encoding an Fc region variant or antibodies containing the Fc region variant.
- the various embodiments of the nucleic acids described within the scope of Disclosures A and B described herein can be applicable without opposing the common technical knowledge in the art and unless there are inconsistencies in the context.
- Disclosure B relates to vectors comprising the nucleic acids.
- the various embodiments thereof within the scope of Disclosures A and B described herein can be applicable without opposing the common technical knowledge in the art and unless there are inconsistencies in the context.
- Disclosure B relates to hosts or host cells comprising the vectors.
- the various embodiments thereof within the scope of Disclosures A and B described herein can be applicable without opposing the common technical knowledge in the art and unless there are inconsistencies in the context.
- Disclosure B relates to methods for producing an Fc region variant comprising an FcRn-binding domain or an antibody comprising the Fc region variant, which comprise culturing the host cells described above, or growing the hosts described above and collecting the Fc region variant or antibody comprising the Fc region variant from the cell culture, materials secreted from the hosts.
- Disclosure B may include production methods optionally further comprising any one or more of: (a) selecting an Fc region variant with enhanced FcRn-binding activity under an acidic pH condition as compared to that of an Fc region of a native IgG; (b) selecting an Fc region variant whose binding activity to a (pre-existing) ADA is not significantly enhanced under a neutral pH condition as compared to that of an Fc region of a native IgG; (c) selecting an Fc region variant with increased plasma retention as compared to that of an Fc region of a native IgG; and (d) selecting an antibody comprising an Fc region variant that can promote elimination of an antigen from plasma as compared to a reference antibody comprising an Fc region of a native IgG.
- an antibody comprising the Fc region variant produced in Disclosure B and the "reference antibody comprising the Fc region of a native IgG" are identical to each other except for the Fc region to be compared.
- the FcRn can be human FcRn.
- the antibody may be compared with the reference antibody comprising a native IgG Fc region in terms of the FcRn-binding activity under an acidic pH condition (e.g., pH 5.8) using BIACORE(registered trademark) or other known techniques, to select an Fc region variant or an antibody comprising the Fc region variant whose FcRn-binding activity has been increased under the acidic pH condition.
- an acidic pH condition e.g., pH 5.8
- BIACORE registered trademark
- the antibody may be compared with a reference antibody comprising a native IgG Fc region in terms of the ADA-binding activity under a neutral pH condition by electrochemiluminescence (ECL) or known techniques, to select an Fc region variant or antibodies comprising the Fc region variant whose ADA-binding activity has not been significantly increased under the neutral pH condition.
- ECL electrochemiluminescence
- the antibody may be compared with a reference antibody comprising a native IgG Fc region by conducting antibody pharmacokinetic tests using plasma, for example, from mice, rats, rabbits, dogs, monkeys, or humans, to select Fc region variants or antibodies comprising the Fc region variant which are demonstrated to have improved plasma retention in the subjects.
- the antibody may be compared with a reference antibody comprising a native IgG Fc region by conducting antibody pharmacokinetic tests using plasma, for example, from mice, rats, rabbits, dogs, monkeys, or humans, to select Fc region variants or antibodies comprising the Fc region variant which have enhanced antigen elimination from plasma.
- the selection methods described above may be appropriately combined, if needed.
- Disclosure B relates to a method for producing an Fc region variant comprising an FcRn-binding domain or an antibody comprising the variant, wherein the method comprises substituting amino acids in a way such that the resulting Fc region variant or the antibody comprising the variant comprises Ala at position 434; Glu, Arg, Ser, or Lys at position 438; and Glu, Asp, or Gln at position 440, according to EU numbering.
- such method comprises substituting the amino acids in a way such that the resulting Fc region variant or the antibody comprising the variant further comprises, Ile or Leu at position 428 and/or Ile, Leu, Val, Thr, or Phe at position 436, according to EU numbering.
- the amino acids are substituted in a way such that the resulting Fc region variant or the antibody comprising the variant produced according to the method further comprises Leu at position 428 and/or Val or Thr at position 436, according to EU numbering.
- Disclosure B relates to a method for producing an Fc region variant comprising an FcRn-binding domain or an antibody comprising the variant, wherein the method comprises substituting amino acids in a way such that the resulting Fc region variant or the antibody comprising the variant comprises Ala at position 434; Arg or Lys at position 438; and Glu or Asp at position 440, according to EU numbering.
- such method comprises substituting the amino acids in a way such that the resulting Fc region variant or the antibody comprising the variant further comprises, Ile or Leu at position 428 and/or Ile, Leu, Val, Thr, or Phe at position 436, according to EU numbering.
- the amino acids are substituted in a way such that the resulting Fc region variant or the antibody comprising the variant produced according to the method further comprises Leu at position 428 and/or Val or Thr at position 436, according to EU numbering.
- such method comprises substituting all amino acids at positions 434, 438, and 440 with Ala; Glu, Arg, Ser, or Lys; and Glu, Asp, or Gln, respectively.
- such method comprises substituting the amino acids in a way such that the resulting Fc region variant or the antibody comprising the variant further comprises, Ile or Leu at position 428 and/or Ile, Leu, Val, Thr, or Phe at position 436, according to EU numbering.
- the amino acids are substituted in a way such that the resulting Fc region variant or the antibody comprising the variant produced according to the method further comprises Leu at position 428 and/or Val or Thr at position 436, according to EU numbering.
- Disclosure B relates to an Fc region variant or an antibody comprising the Fc region variant obtained by any of the production methods of Disclosure B described above.
- Disclosure B provides methods for reducing the (pre-existing) ADA-binding activity of antibodies comprising an Fc region variant with increased FcRn-binding activity at an acidic pH; and methods for producing Fc region variants with increased FcRn-binding activity at an acidic pH (e.g., pH 5.8) and reduced pre-existing ADA-binding activity, which comprise: (a) providing an antibody comprising an Fc region (variant) whose FcRn-binding activity at an acidic pH has been increased as compared to a reference antibody; and (b) introducing into the Fc region, according to EU numbering, (i) an amino acid substitution with Ala at position 434; (ii) an amino acid substitution with any one of Glu, Arg, Ser, and Lys at position 438; and (iii) an amino acid substitution with any one of Glu, Asp, and Gln at position 440, (iv) optionally, an amino acid substitution with Ile or Leu at position 428; and
- the Fc domain (variant) in step (a) is preferably a human IgG Fc domain (variant).
- the Fc region (variant) is to contain a combination of substituted amino acids selected from the group consisting of: (a) N434A/Q438R/S440E; (b) N434A/Q438R/S440D; (c) N434A/Q438K/S440E; (d) N434A/ Q438K/S440D; (e) N434A/Y436T/Q438R/S440E; (f) N434A/Y436T/Q438R/S440D; (g) N434A/Y436T/Q438K/S440E; (h) N434A/Y4
- the methods may optionally further comprise: (c) assessing whether the (pre-existing) ADA-binding activity of an antibody comprising a produced Fc region variant is reduced as compared to the binding activity of the reference antibody.
- the methods may be used as a method for enhancing the release of an antibody that has been internalized into cells in an antigen-bound form to the outside of the cells in an antigen-free form, without significantly increasing the (pre-existing) ADA-binding activity of the antibody at a neutral pH.
- Disclosure C also relates to anti-IL-8 antibodies, nucleic acids encoding the antibodies, pharmaceutical compositions comprising the antibodies, methods for producing the antibodies, and uses of the antibodies in treating diseases related to IL-8, as described in detail hereinbelow.
- the meanings of the terms given hereinbelow apply throughout the description of Disclosure C herein, without being contrary to the common technical knowledge of those of ordinary skill in the art as well as embodiments known to those of ordinary skill in the art.
- acidic pH refers to pH that may be selected, for example, from pH 4.0 to pH 6.5.
- acidic pH refers to, but is not limited to, pH 4.0, pH 4.1, pH 4.2, pH 4.3, pH 4.4, pH 4.5, pH 4.6, pH 4.7, pH 4.8, pH 4.9, pH 5.0, pH 5.1, pH 5.2, pH 5.3, pH 5.4, pH 5.5, pH 5.6, pH 5.7, pH 5.8, pH 5.9, pH 6.0, pH 6.1, pH 6.2, pH 6.3, pH 6.4, or pH 6.5.
- the term acidic pH refers to the pH 5.8.
- neutral pH refers to pH that may be selected, for example, from pH 6.7 to pH 10.0.
- neutral pH refers to, but is not limited to, pH 6.7, pH 6.8, pH 6.9, pH 7.0, pH 7.1, pH 7.2, pH 7.3, pH 7.4, pH 7.5, pH 7.6, pH 7.7, pH 7.8, pH 7.9, pH 8.0, pH 8.1, pH 8.2, pH 8.3, pH 8.4, pH 8.5, pH 8.6, pH 8.7, pH 8.8, pH 8.9, pH 9.0, pH 9.5, or pH 10.0.
- the term neutral pH refers to the pH 7.4.
- IL-8 refers to any native IL-8 derived from any vertebrates, primates (e.g., humans, cynomolgus monkeys, rhesus monkeys) and other mammals (e.g., dogs and rabbits), unless otherwise indicated.
- the term “IL-8” encompasses full-length IL-8, unprocessed IL-8 as well as any form of IL-8 that results from processing in the cell.
- the term “IL-8” also encompasses derivatives of native IL-8, for example, splice variants or allelic variants.
- the amino acid sequence of an exemplary human IL-8 is shown in SEQ ID NO:66.
- anti-IL-8 antibody and "an antibody that binds to IL-8” refer to an antibody that is capable of binding to IL-8 with sufficient affinity such that the antibody is useful as a diagnostic and/or therapeutic agent in targeting IL-8.
- the extent of binding of an anti-IL-8 antibody to an unrelated, non-IL-8 protein is, for example, less than about 10% of the binding of the antibody to IL-8.
- Binding affinity refers to intrinsic binding affinity which reflects a 1:1 interaction between members of a binding pair (e.g., antibody and antigen).
- the affinity of a molecule X for its partner Y can generally be represented by the dissociation constant (KD). Binding affinity can be measured using methods known in the art, including those described within the scope of the description of Disclosure C herein.
- an antibody that binds to IL-8 may have a dissociation constant (KD) of, for example, (e.g., 10 -8 M or less, from 10 -8 M to 10 -13 M, from 10 -9 M to 10 -13 M).
- KD dissociation constant
- antibody within the scope of the description of Disclosure C herein is used in the broadest sense and includes, but is not limited to, monoclonal antibodies, polyclonal antibodies, multispecific antibodies (e.g., bispecific antibodies), and antibody fragments so long as they exhibit the desired antigen-binding activity.
- an "antibody that binds to the same epitope” as a reference antibody refers to an antibody that blocks binding of the reference antibody to its antigen by, for example, 50%, 60%, 70%, or 80% or more; and conversely, the reference antibody blocks binding of the antibody to its antigen by, for example, 50%, 60%, 70%, or 80% or more.
- an exemplary competition assay can be used without being limited thereto.
- a “chimeric antibody” refers to an antibody in which a portion of the heavy and/or light chain is derived from a particular source or species, while the remaining portion is derived from a different source or species.
- a “humanized” antibody refers to a chimeric antibody comprising amino acid residues from non-human HVRs and amino acid residues from human FRs.
- a humanized antibody may comprise substantially at least one, and typically two, variable regions, in which all (or substantially all) of the HVRs (e.g., CDRs) correspond to those of a non-human antibody, and all (or substantially all) of the FRs correspond to those of a human antibody.
- a humanized antibody optionally may comprise at least a portion of an antibody constant region derived from a human antibody.
- the term "monoclonal antibody” as used within the scope of the description of Disclosure C herein refers to an antibody obtained from a population of substantially homogeneous antibodies, i.e., the individual antibodies that constitute the population are identical and/or bind the same epitope, except for possible variant antibodies, for example, those containing naturally occurring mutations or arising during production of a monoclonal antibody preparation, which are generally present in minor amounts.
- polyclonal antibody preparations which typically include different antibodies directed against different determinants (epitopes)
- each monoclonal antibody in a monoclonal antibody preparation is directed against a single determinant on an antigen.
- the modifier "monoclonal” indicates the characteristics of an antibody as being obtained from a substantially homogeneous population of antibodies, and is not to be construed as requiring production of the antibody by any specific method.
- the monoclonal antibodies to be used in accordance with Disclosure C may be made by various techniques, including but not limited to the hybridoma method, recombinant DNA methods, phage-display methods, and methods utilizing transgenic animals comprising all or part of the human immunoglobulin loci, such methods and other exemplary methods for making monoclonal antibodies being described herein.
- a native antibody refers to immunoglobulin molecules with various naturally occurring structures.
- a native IgG antibody for example, is a heterotetrameric glycoprotein of about 150,000 daltons composed of two identical light chains and two identical heavy chains that are disulfide-bonded. In the order from N- to C- terminus, each heavy chain has a variable region (VH), which is also referred to as a variable heavy-chain domain or heavy-chain variable domain, followed by three constant domains (CH1, CH2, and CH3).
- VH variable region
- each light chain has a variable region (VL), which is also referred to as a variable light-chain domain or light-chain variable domain, followed by a constant light-chain (CL) domain.
- VL variable region
- An antibody light chain may be assigned to one of the two types, called kappa ( ⁇ ) and lambda ( ⁇ ), based on the amino acid sequence of its constant domain.
- ⁇ kappa
- ⁇ lambda
- Such constant domains for use in the Disclosure C include those of any reported allotype (allele) or any subclass/isotype.
- the heavy-chain constant region includes, but is not limited to, the constant region of a native IgG antibody (IgG1, IgG2, IgG3, and IgG4).
- IgG1 alleles include, for example, IGHG1*01, IGHG1*02, IGHG1*03, IGHG1*04, and IGHG1*05 (see at imgt.org), and any of these can be used as a native human IgG1 sequence.
- the constant domain sequence may be derived from a single allele or subclass/isotype, or from multiple alleles or subclasses/isotypes.
- such antibodies include, but are not limited to, an antibody whose CH1 is derived from IGHG1*01 and CH2 and CH3 are derived from IGHG1*02 and IGHG1*01, respectively.
- Antibody effector functions within the scope of the description of Disclosure C herein refers to biological activities attributable to the Fc region of an antibody, which may vary with the antibody isotype.
- Examples of antibody effector functions include: C1q binding and complement-dependent cytotoxicity; Fc receptor binding; antibody-dependent cell-mediated cytotoxicity (ADCC); phagocytosis; down regulation of cell surface receptors (e.g., B cell receptor); and B cell activation, but are not limited thereto.
- Fc region within the scope of the description of Disclosure C herein is used to define a C-terminal region of an immunoglobulin heavy chain that contains at least a portion of the constant region.
- the term includes native Fc regions and variant Fc regions.
- the native Fc region indicates the Fc region of a native antibody.
- a human IgG heavy-chain Fc region extends from the amino acid residue of Cys226 or Pro230 to the carboxyl terminus of the heavy chain.
- the C-terminal lysine (Lys447) or glycine-lysine (residues 446-447) of the Fc region may or may not be present.
- the numbering of amino acid residues in the Fc region or constant region is according to the EU numbering system, also called the EU index, as described in Kabat et al., Sequences of Proteins of Immunological Interest, 5th Ed. Public Health Service, National Institutes of Health, Bethesda, MD, 1991.
- FR Framework or "FR” within the scope of the description of Disclosure C herein refers to variable domain residues other than hypervariable region (HVR) residues.
- the FR of a variable domain generally consists of four FR domains: FR1, FR2, FR3, and FR4. Accordingly, the HVR and FR sequences generally appear in the following sequence: FR1-H1(L1)-FR2-H2(L2)-FR3-H3(L3)-FR4 in VH (or VL).
- a "human consensus framework" within the scope of the description of Disclosure C herein is a framework that represents the most commonly occurring amino acid residues in a selection of human immunoglobulin VL or VH framework sequences.
- the selection of human immunoglobulin VL or VH sequences is from a subgroup of variable domain sequences.
- the subgroup of sequences is a subgroup according to Kabat et al., Sequences of proteins of Immunological Interest, 5th Ed. Public Health Service, National Institutes of Health, Bethesda, MD (1991).
- the subgroup for the VL is subgroup ⁇ I as in Kabat et al., supra. In one embodiment, the subgroup for the VH is subgroup III as in Kabat et al., supra.
- an "acceptor human framework” for purposes within the scope of the description of Disclosure C herein is a framework comprising the amino acid sequence of a VL or VH framework derived from a human immunoglobulin framework or a human consensus framework.
- An acceptor human framework "derived from” a human immunoglobulin framework or a human consensus framework may comprise the same amino acid sequence thereof, or it may contain existing amino acid sequence substitutions. In some embodiments, the number of existing amino acid substitutions are 10 or less, 9 or less, 8 or less, 7 or less, 6 or less, 5 or less, 4 or less, 3 or less, or 2 or less.
- the VL acceptor human framework is identical to the VL human immunoglobulin framework sequence or human consensus framework sequence.
- variable region or “variable domain” within the scope of the description of Disclosure C herein refers to the domain of an antibody heavy or light chain involved in binding of the antibody to an antigen.
- the variable regions of the heavy chain and light chain (VH and VL, respectively) of a native antibody generally have similar structures, with each domain comprising four conserved framework regions (FRs) and three hypervariable regions (HVRs).
- FRs conserved framework regions
- HVRs hypervariable regions
- antibodies that bind to a particular antigen may be isolated using a VH or VL domain from an antibody that binds the antigen to screen a library of complementary VL or VH domains, respectively. See, e.g., Portolano et al., J. Immunol. 150:880-887 (1993); Clarkson et al., Nature 352:624-628 (1991).
- hypervariable region refers to each of the regions of an antibody variable domain which are hypervariable in sequence ("complementarity determining regions” or “CDRs") and/or form structurally defined loops ("hypervariable loops") and/or contain the antigen-contacting residues ("antigen contacts").
- CDRs complementarity determining regions
- hypervariable loops form structurally defined loops
- antigen contacts Generally, antibodies comprise six hypervariable regions: three in the VH (H1, H2, H3), and three in the VL (L1, L2, L3).
- exemplary HVRs herein include: (a) hypervariable loops in which amino acid residues are 26-32 (L1), 50-52 (L2), 91-96 (L3), 26-32 (H1), 53-55 (H2), and 96-101 (H3) (Chothia and Lesk, J. Mol. Biol. 196:901-917 (1987)); (b) CDRs in which amino acid residues are 24-34 (L1), 50-56 (L2), 89-97 (L3), 31-35b (H1), 50-65 (H2), and 95-102 (H3) (Kabat et al., Sequences of Proteins of Immunological Interest, 5th Ed.
- HVRs and other residues in variable regions are numbered as in Kabat et al., supra.
- mammals include, but are not limited to, domesticated animals (e.g., cows, sheep, cats, dogs, and horses), primates (e.g., humans and non-human primates such as monkeys), rabbits, and rodents (e.g., mice and rats).
- domesticated animals e.g., cows, sheep, cats, dogs, and horses
- primates e.g., humans and non-human primates such as monkeys
- rabbits e.g., mice and rats
- rodents e.g., mice and rats
- an “isolated” antibody within the scope of the description of Disclosure C herein is one which has been separated from a component of its natural environment.
- an antibody is purified to be greater than 95% or 99% in purity as determined, for example, electrophoretically (e.g., SDS-PAGE, isoelectric focusing electrophoresis (IEF), capillary electrophoresis) or chromatographically (e.g., ion exchange or reverse phase HPLC).
- electrophoretically e.g., SDS-PAGE, isoelectric focusing electrophoresis (IEF), capillary electrophoresis
- chromatographically e.g., ion exchange or reverse phase HPLC
- An "isolated" nucleic acid within the scope of the description of Disclosure C herein refers to a nucleic acid molecule that has been separated from a component of its natural environment.
- An isolated nucleic acid includes a nucleic acid molecule contained in cells that ordinarily contain the nucleic acid molecule, but the nucleic acid molecule is present extrachromosomally or at a chromosomal location that is different from its natural chromosomal location.
- isolated nucleic acid encoding an anti-IL-8 antibody within the scope of the description of Disclosure C herein refers to one or more nucleic acid molecules encoding anti-IL-8 antibody heavy and light chains (or fragments thereof), including such nucleic acid(s) in a single vector or separate vectors, nucleic acid(s) present at one or more locations in a host cell.
- host cell refers to cells into which an exogenous nucleic acid has been introduced, including the progeny of such cells.
- Host cells include “transformants” and “transformed cells”, which include the primary transformed cell and progeny derived therefrom without regard to the number of passages.
- a progeny may not be completely identical in its nucleic acid content to a parent cell, but may contain mutations.
- a mutant progeny that has the same function or biological activity as that screened or selected in the originally transformed cell are included herein.
- vector refers to a nucleic acid molecule capable of propagating another nucleic acid to which it is linked.
- the term includes vectors in a self-replicating nucleic acid structure as well as vectors introduced into a host cell and become incorporated into its genome.
- Certain vectors are capable of directing the expression of nucleic acids to which they are operatively linked. Such vectors are referred to herein as "expression vectors.”
- package insert is used to refer to instructions customarily included in commercial packages of therapeutic products, that contain information about the indications, usage, dosage, administration, combination therapy, contraindications and/or warnings concerning the use of such therapeutic products.
- percent (%) amino acid sequence identity with respect to a reference polypeptide sequence is defined as the percentage of amino acid residues in a candidate sequence that are identical to the amino acid residues in the reference polypeptide sequence after sequence alignment, by introducing gaps if necessary and not considering any conservative substitutions as part of the sequence identity, in order to achieve the maximum percent sequence identity. Alignment for purposes of determining percent amino acid sequence identity can be achieved in various ways within the scope of the ability of those of ordinary skill in the art, for instance, by using publicly available computer software such as BLAST, BLAST-2, ALIGN, Megalign (DNASTAR) software, or GENETYX(registered trademark) (Genetyx Co., Ltd.).
- ALIGN-2 sequence comparison computer program ALIGN-2.
- the ALIGN-2 was authored by Genentech, Inc., and the source code has been filed with user documentation in the U.S. Copyright Office, Washington D.C., 20559, where it is registered under U.S. Copyright Registration No. TXU510087.
- the ALIGN-2 program is publicly available from Genentech, Inc., South San Francisco, California, or may be compiled from the source code.
- the ALIGN-2 program should be compiled for use on a UNIX (registered trademark) operating system, including digital UNIX (registered trademark) V4.0D. All sequence comparison parameters are set by the ALIGN-2 program and do not vary.
- the % amino acid sequence identity of a given amino acid sequence A to a given amino acid sequence B is calculated as follows: 100 times the fraction X/Y, where X is the number of amino acid residues scored as identical matches in the program alignment of A and B by the sequence alignment program ALIGN-2, and where Y is the total number of amino acid residues in B. It will be appreciated that where the length of amino acid sequence A is not equal to the length of amino acid sequence B, the % amino acid sequence identity of A to B will not equal the % amino acid sequence identity of B to A. Unless specifically stated otherwise, all % values of amino acid sequence identity are obtained using the ALIGN-2 computer program as demonstrated under the scope of the description of Disclosure C herein.
- a “pharmaceutical composition” generally refers to an agent for treating, preventing, examining, or diagnosing diseases.
- a “pharmaceutically acceptable carrier” refers to an ingredient in a pharmaceutical composition, other than an active ingredient, which is nontoxic to a subject.
- Such pharmaceutically acceptable carriers include, but are not limited to, buffers, excipients, stabilizers, and preservatives.
- treatment refers to a clinical intervention in an attempt to alter the natural course of the individual being treated. Such a clinical intervention can be performed either for prophylaxis or during the course of clinical pathology. Desirable effects of treatment include, but are not limited to, prevention of the occurrence or recurrence of a disease, alleviation of symptoms, diminishment of any direct or indirect pathological consequences of the disease, prevention of metastasis, decrease of the rate of disease progression, amelioration or palliation of the disease state, and remission or improvement of prognosis.
- an antibody of Disclosure C can be used to slow down the progression of a disease or disorder.
- an "effective amount” of an antibody or pharmaceutical composition refers to an amount that is effective when used at doses and for periods of time necessary to achieve the desired therapeutic or prophylactic result.
- Disclosure C is based on the applicability of anti-IL-8 antibodies that have pH-dependent affinity for IL-8 as pharmaceutical compositions.
- the antibodies of Disclosure C are useful, for example, in diagnosing or treating diseases where IL-8 is present in an excessive amount.
- Disclosure C provides an anti-IL-8 antibody having pH-dependent affinity for IL-8.
- Disclosure C provides an anti-IL-8 antibody having pH-dependent affinity for IL-8, which comprises a sequence with at least one, two, three, four, five, six, seven, or eight amino acid substitution(s) within the amino acid sequences of: (a) HVR-H1 which comprises the amino acid sequence of SEQ ID NO:67; (b) HVR-H2 which comprises the amino acid sequence of SEQ ID NO:68; (c) HVR-H3 which comprises the amino acid sequence of SEQ ID NO:69; (d) HVR-L1 which comprises the amino acid sequence of SEQ ID NO:70; (e) HVR-L2 which comprises the amino acid sequence of SEQ ID NO:71; and (f) HVR-L3 which comprises the amino acid sequence of SEQ ID NO:72.
- HVR-H1 which comprises the amino acid sequence of SEQ ID NO:67
- HVR-H2 which comprises the amino acid sequence of SEQ ID NO:68
- HVR-H3 which comprises the amino acid sequence of SEQ ID NO:69
- Disclosure C provides an anti-IL-8 antibody having pH-dependent affinity for IL-8, which comprises at least one amino acid substitution(s) in at least one of the amino acid sequences of: (a) HVR-H1 which comprises the amino acid sequence of SEQ ID NO:67; (b) HVR-H2 which comprises the amino acid sequence of SEQ ID NO:68; (c) HVR-H3 which comprises the amino acid sequence of SEQ ID NO:69; (d) HVR-L1 which comprises the amino acid sequence of SEQ ID NO:70; (e) HVR-L2 which comprises the amino acid sequence of SEQ ID NO:71; and (f) HVR-L3 which comprises the amino acid sequence of SEQ ID NO:72.
- an anti-IL-8 antibody of Disclosure C comprises one or more amino acid substitution(s) at position(s) selected from the group consisting of: (a) aspartic acid at position 1 in the sequence of SEQ ID NO:67; (b) tyrosine at position 2 in the sequence of SEQ ID NO:67; (c) tyrosine at position 3 in the sequence of SEQ ID NO:67; (d) leucine at position 4 in the sequence of SEQ ID NO:67; (e) serine at position 5 in the sequence of SEQ ID NO:67; (f) leucine at position 1 in the sequence of SEQ ID NO:68; (g) isoleucine at position 2 in the sequence of SEQ ID NO:68; (h) arginine at position 3 in the sequence of SEQ ID NO:68; (i) asparagine at position 4 in the sequence of SEQ ID NO:68; (j) lysine at position 5 in the sequence
- an anti-IL-8 antibody of Disclosure C comprises one or more amino acid substitution(s) at position(s) selected from the group consisting of: (a) alanine at position 6 in the sequence of SEQ ID NO:68; (b) glycine at position 8 in the sequence of SEQ ID NO:68; (c) tyrosine at position 9 in the sequence of SEQ ID NO:68; (d) arginine at position 11 in the sequence of SEQ ID NO:68; and (e) tyrosine at position 3 in the sequence of SEQ ID NO:69.
- an anti-IL-8 antibody of Disclosure C comprises combination(s) of amino acid substitutions at positions selected from the group consisting of: (a) alanine at position 6 in the sequence of SEQ ID NO:68; (b) glycine at position 8 in the sequence of SEQ ID NO:68; (c) tyrosine at position 9 in the sequence of SEQ ID NO:68; (d) arginine at position 11 in the sequence of SEQ ID NO:68; and (e) tyrosine at position 3 in the sequence of SEQ ID NO:69.
- an anti-IL-8 antibody of Disclosure C comprises amino acid substitutions at the following positions: (a) tyrosine at position 9 in the sequence of SEQ ID NO:68; (b) arginine at position 11 in the sequence of SEQ ID NO:68; and (c) tyrosine at position 3 in the sequence of SEQ ID NO:69.
- an anti-IL-8 antibody of Disclosure C comprises amino acid substitutions at the following positions: (a) alanine at position 6 in the sequence of SEQ ID NO:68; (b) glycine at position 8 in the sequence of SEQ ID NO:68; (c) tyrosine at position 9 in the sequence of SEQ ID NO:68; (d) arginine at position 11 in the sequence of SEQ ID NO:68; and (e) tyrosine at position 3 in the sequence of SEQ ID NO:69.
- an anti-IL-8 antibody of Disclosure C comprises: (a) substitution of alanine with aspartic acid at position 6 in the sequence of SEQ ID NO:68; (b) substitution of arginine with proline at position 11 in the sequence of SEQ ID NO:68; and (c) substitution of tyrosine with histidine at position 3 in the sequence of SEQ ID NO:69.
- an anti-IL-8 antibody of Disclosure C comprises: (a) substitution of glycine with tyrosine at position 8 in the sequence of SEQ ID NO:68; and (b) substitution of tyrosine with histidine at position 9 in the sequence of SEQ ID NO:68.
- an anti-IL-8 antibody of Disclosure C comprises: (a) substitution of alanine with aspartic acid at position 6 in the sequence of SEQ ID NO:68; (b) substitution of glycine with tyrosine at position 8 in the sequence of SEQ ID NO:68; (c) substitution of tyrosine with histidine at position 9 in the sequence of SEQ ID NO:68; (d) substitution of arginine with proline at position 11 in the sequence of SEQ ID NO:68; and (e) substitution of tyrosine with histidine at position 3 in the sequence of SEQ ID NO:69.
- an anti-IL-8 antibody of Disclosure C comprises HVR-H2 which comprises the amino acid sequence of SEQ ID NO:73.
- an anti-IL-8 antibody of Disclosure C comprises HVR-H3 which comprises the amino acid sequence of SEQ ID NO:74.
- an anti-IL-8 antibody of Disclosure C comprises HVR-H1 comprising the amino acid sequence of SEQ ID NO:67, HVR-H2 comprising the amino acid sequence of SEQ ID NO:73, and HVR-H3 comprising the amino acid sequence of SEQ ID NO:74.
- an anti-IL-8 antibody of Disclosure C comprises one or more amino acid substitution(s) at position(s) selected from the group consisting of: (a) serine at position 8 in the sequence of SEQ ID NO:70; (b) asparagine at position 1 in the sequence of SEQ ID NO:71; (c) leucine at position 5 in the sequence of SEQ ID NO:71; and (d) glutamine at position 1 in the sequence of SEQ ID NO:72.
- the anti-IL-8 antibody comprises a combination of any 2, 3, or all 4 of these substitutions.
- an anti-IL-8 antibody of Disclosure C comprises combination(s) of amino acid substitutions at positions selected from the group consisting of: (a) serine at position 8 in the sequence of SEQ ID NO:70; (b) asparagine at position 1 in the sequence of SEQ ID NO:71; (c) leucine at position 5 in the sequence of SEQ ID NO:71; and (d) glutamine at position 1 in the sequence of SEQ ID NO:72.
- the anti-IL-8 antibody comprises a combination of any 2, 3, or all 4 of these substitutions.
- an anti-IL-8 antibody of Disclosure C comprises amino acid substitutions at the following positions: (a) asparagine at position 1 in the sequence of SEQ ID NO:71; (b) leucine at position 5 in the sequence of SEQ ID NO:71; and (c) glutamine at position 1 in the sequence of SEQ ID NO:72.
- an anti-IL-8 antibody of Disclosure C comprises amino acid substitutions at the following positions: (a) serine at position 8 in the sequence of SEQ ID NO:70; (b) asparagine at position 1 in the sequence of SEQ ID NO:71; (c) leucine at position 5 in the sequence of SEQ ID NO:71; and (d) glutamine at position 1 in the sequence of SEQ ID NO:72.
- an anti-IL-8 antibody of Disclosure C comprises: (a) substitution of asparagine with lysine at position 1 in the sequence of SEQ ID NO:71; (b) substitution of leucine with histidine at position 5 in the sequence of SEQ ID NO:71; and (c) substitution of glutamine with lysine at position 1 in the sequence of SEQ ID NO:72.
- an anti-IL-8 antibody of Disclosure C comprises: (a) substitution of serine with glutamic acid at position 8 in the sequence of SEQ ID NO:70; (b) substitution of asparagine with lysine at position 1 in the sequence of SEQ ID NO:71; (c) substitution of leucine with histidine at position 5 in the sequence of SEQ ID NO:71; and (d) substitution of glutamine with lysine at position 1 in the sequence of SEQ ID NO:72.
- an anti-IL-8 antibody of Disclosure C comprises HVR-L2 comprising the amino acid sequence of SEQ ID NO:75.
- an anti-IL-8 antibody of Disclosure C comprises HVR-L3 comprising the amino acid sequence of SEQ ID NO:76.
- an anti-IL-8 antibody of Disclosure C comprises HVR-L1 comprising the amino acid sequence of SEQ ID NO:70, HVR-L2 comprising the amino acid sequence of SEQ ID NO:75, and HVR-L3 comprising the amino acid sequence of SEQ ID NO:76.
- an anti-IL-8 antibody of Disclosure C comprises amino acid substitutions at the following positions: (a) alanine at position 55 in the sequence of SEQ ID NO:77; (b) glycine at position 57 in the sequence of SEQ ID NO:77; (c) tyrosine at position 58 in the sequence of SEQ ID NO:77; (d) arginine at position 60 in the sequence of SEQ ID NO:77; (e) glutamine at position 84 in the sequence of SEQ ID NO:77; (f) serine at position 87 in the sequence of SEQ ID NO:77; and (g) tyrosine at position 103 in the sequence of SEQ ID NO:77.
- an anti-IL-8 antibody of Disclosure C comprises: (a) substitution of alanine with aspartic acid at position 55 in the sequence of SEQ ID NO:77; (b) substitution of glycine with tyrosine at position 57 in the sequence of SEQ ID NO:77; (c) substitution of tyrosine with histidine at position 58 in the sequence of SEQ ID NO:77; (d) substitution of arginine with proline at position 60 in the sequence of SEQ ID NO:77; (e) substitution of glutamine with threonine at position 84 in the sequence of SEQ ID NO:77; (f) substitution of serine with aspartic acid at position 87 in the sequence of SEQ ID NO:77; (g) and substitution of tyrosine with histidine at position 103 in the sequence of SEQ ID NO:77.
- an anti-IL-8 antibody of Disclosure C comprises a heavy chain variable region comprising the amino acid sequence of SEQ ID NO:78.
- an anti-IL-8 antibody of Disclosure C comprises a light chain variable region comprising the amino acid sequence of SEQ ID NO:79.
- an anti-IL-8 antibody of Disclosure C comprises a heavy chain variable region comprising the amino acid sequence of SEQ ID NO:78 and a light chain variable region comprising the amino acid sequence of SEQ ID NO:79.
- the anti-IL-8 antibody that comprises a heavy chain variable region comprising the amino acid sequence of SEQ ID NO:78 and a light chain variable region comprising the amino acid sequence of SEQ ID NO:79 may be an anti-IL-8 antibody that binds to IL-8 in a pH-dependent manner.
- the anti-IL-8 antibody that comprises a heavy chain variable region comprising the amino acid sequence of SEQ ID NO:78 and a light chain variable region comprising the amino acid sequence of SEQ ID NO:79 may be an anti-IL-8 antibody that maintains the IL-8-neutralizing activity stably in vivo (for example, in plasma).
- the anti-IL-8 antibody that comprises a heavy chain variable region comprising the amino acid sequence of SEQ ID NO:78 and a light chain variable region comprising the amino acid sequence of SEQ ID NO:79 may be an antibody with low immunogenicity.
- anti-IL-8 antibodies of Disclosure C also include those that have pH-dependent affinity for IL-8 and contain at least one amino acid substitution in at least any one amino acid sequence of: (a) HVR-H1 comprising the amino acid sequence of SEQ ID NO:102; (b) HVR-H2 comprising the amino acid sequence of SEQ ID NO:103; (c) HVR-H3 comprising the amino acid sequence of SEQ ID NO:104; (d) HVR-L1 comprising the amino acid sequence of SEQ ID NO:105; (e) HVR-L2 comprising the amino acid sequence of SEQ ID NO:106; and (f) HVR-L3 comprising the amino acid sequence of SEQ ID NO:107.
- anti-IL-8 antibodies of Disclosure C also include those that have pH-dependent affinity for IL-8 and contain at least one amino acid substitution in an amino acid sequence of: (a) HVR-H1 comprising the amino acid sequence of SEQ ID NO:108; (b) HVR-H2 comprising the amino acid sequence of SEQ ID NO:109; (c) HVR-H3 comprising the amino acid sequence of SEQ ID NO:110; (d) HVR-L1 comprising the amino acid sequence of SEQ ID NO:111; (e) HVR-L2 comprising the amino acid sequence of SEQ ID NO:112; and (f) HVR-L3 comprising the amino acid sequence of SEQ ID NO:113.
- anti-IL-8 antibodies of Disclosure C also include those that have pH-dependent affinity for IL-8 and contain at least one amino acid substitution in at least any one amino acid sequence of: (a) HVR-H1 comprising the amino acid sequence of SEQ ID NO:114; (b) HVR-H2 comprising the amino acid sequence of SEQ ID NO:115; (c) HVR-H3 comprising the amino acid sequence of SEQ ID NO:116; (d) HVR-L1 comprising the amino acid sequence of SEQ ID NO:117; (e) HVR-L2 comprising the amino acid sequence of SEQ ID NO:118; and (f) HVR-L3 comprising the amino acid sequence of SEQ ID NO:119.
- anti-IL-8 antibodies of Disclosure C also include those that have pH-dependent affinity for IL-8 and contain at least one amino acid substitution in at least any one amino acid sequence of: (a) HVR-H1 comprising the amino acid sequence of SEQ ID NO:120; (b) HVR-H2 comprising the amino acid sequence of SEQ ID NO:121; (c) HVR-H3 comprising the amino acid sequence of SEQ ID NO:122; (d) HVR-L1 comprising the amino acid sequence of SEQ ID NO:123; (e) HVR-L2 comprising the amino acid sequence of SEQ ID NO:124; and (f) HVR-L3 comprising the amino acid sequence of SEQ ID NO:125.
- anti-IL-8 antibodies of Disclosure C also include those that have pH-dependent affinity for IL-8 and contain at least one amino acid substitution in at least any one amino acid sequence of: (a) HVR-H1 comprising the amino acid sequence of SEQ ID NO:126; (b) HVR-H2 comprising the amino acid sequence of SEQ ID NO:127; (c) HVR-H3 comprising the amino acid sequence of SEQ ID NO:128; (d) HVR-L1 comprising the amino acid sequence of SEQ ID NO:129; (e) HVR-L2 comprising the amino acid sequence of SEQ ID NO:130; and (f) HVR-L3 comprising the amino acid sequence of SEQ ID NO:131.
- anti-IL-8 antibodies of Disclosure C also include those that have pH-dependent affinity for IL-8 and contain at least one amino acid substitution in at least any one amino acid sequence of: (a) HVR-H1 comprising the amino acid sequence of SEQ ID NO:132; (b) HVR-H2 comprising the amino acid sequence of SEQ ID NO:133; (c) HVR-H3 comprising the amino acid sequence of SEQ ID NO:134; (d) HVR-L1 comprising the amino acid sequence of SEQ ID NO:135; (e) HVR-L2 comprising the amino acid sequence of SEQ ID NO:136; and (f) HVR-L3 comprising the amino acid sequence of SEQ ID NO:137.
- anti-IL-8 antibodies of Disclosure C also include those that have pH-dependent affinity for IL-8 and contain at least one amino acid substitution in at least any one amino acid sequence of: (a) HVR-H1 comprising the amino acid sequence of SEQ ID NO:138; (b) HVR-H2 comprising the amino acid sequence of SEQ ID NO:139; (c) HVR-H3 comprising the amino acid sequence of SEQ ID NO:140; (d) HVR-L1 comprising the amino acid sequence of SEQ ID NO:141; (e) HVR-L2 comprising the amino acid sequence of SEQ ID NO:142; and (f) HVR-L3 comprising the amino acid sequence of SEQ ID NO:143.
- an anti-IL-8 antibody of Disclosure C has IL-8-neutralizing activity.
- the IL-8-neutralizing activity refers to an activity of inhibiting the biological activity of IL-8, or may refer to an activity of inhibiting the receptor binding of IL-8.
- an anti-IL-8 antibody of Disclosure C is an anti-IL-8 antibody that binds to IL-8 in a pH-dependent manner.
- an anti-IL-8 antibody that binds to IL-8 in a pH-dependent manner refers to an antibody whose binding affinity for IL-8 at an acidic pH has been reduced as compared to the binding affinity for IL-8 at a neutral pH.
- pH-dependent anti-IL-8 antibodies include antibodies that have a higher affinity for IL-8 at a neutral pH than at an acidic pH.
- an anti-IL-8 antibody of Disclosure C has an IL-8 affinity at a neutral pH that is at least 2 times, 3 times, 5 times, 10 times, 15 times, 20 times, 25 times, 30 times, 35 times, 40 times, 45 times, 50 times, 55 times, 60 times, 65 times, 70 times, 75 times, 80 times, 85 times, 90 times, 95 times, 100 times, 200 times, 400 times, 1000 times, 10000 times or more greater than the affinity at an acidic pH.
- the binding affinity can be measured using, without particular limitations, surface plasmon resonance methods (such as BIACORE(registered trademark)).
- the association rate constant (kon) and dissociation rate constant (koff) can be calculated using the BIACORE(registered trademark) T200 Evaluation Software (GE Healthcare) based on a simple one-to-one Langmuir binding model by fitting the association and dissociation sensorgrams simultaneously.
- the equilibrium dissociation constant (KD) is calculated as a ratio of koff/kon.
- ELISA kinetic exclusion assay
- KinExA TM kinetic exclusion assay
- surface plasmon resonance methods such as BIACORE(registered trademark)
- the pH-dependent IL-8-binding ability refers to the property to bind to IL-8 in a pH-dependent manner. Meanwhile, whether an antibody is capable of binding to IL-8 multiple times can be assessed by the methods described in WO2009/125825.
- an anti-IL-8 antibody of Disclosure C has a small dissociation constant (KD) for IL-8 at a neutral pH.
- KD dissociation constant
- the dissociation constant of an antibody of Disclosure C for IL-8 at a neutral pH is, for example, 0.3 nM or less, but is not limited thereto.
- the dissociation constant of an antibody of Disclosure C for IL-8 at a neutral pH is, for example, 0.1 nM or less, but is not limited thereto.
- the dissociation constant of an antibody of Disclosure C for IL-8 at a neutral pH is, for example, 0.03 nM or less, but is not limited thereto.
- an anti-IL-8 antibody of Disclosure C has a small dissociation constant (KD) for IL-8 at pH 7.4.
- the dissociation constant of an antibody of Disclosure C for IL-8 at pH 7.4 is, for example, 0.3 nM or less, but is not limited thereto.
- the dissociation constant of an antibody of Disclosure C for IL-8 at pH 7.4 is, for example, 0.1 nM or less, but is not limited thereto.
- the dissociation constant of an antibody of Disclosure C for IL-8 at pH 7.4 is, for example, 0.03 nM or less, but is not limited thereto.
- an anti-IL-8 antibody of Disclosure C has a large dissociation constant (KD) for IL-8 at an acidic pH.
- the dissociation constant of an antibody of Disclosure C for IL-8 at an acidic pH is, for example, 3 nM or more, but is not limited thereto.
- the dissociation constant of an antibody of Disclosure C for IL-8 at an acidic pH is, for example, 10 nM or more, but is not limited thereto.
- the dissociation constant of an antibody of Disclosure C for IL-8 at an acidic pH is, for example, 30 nM or more, but is not limited thereto.
- an anti-IL-8 antibody of Disclosure C has a large dissociation constant (KD) for IL-8 at pH 5.8.
- the dissociation constant of an antibody of Disclosure C for IL-8 at pH 5.8 is, for example, 3 nM or more, but is not limited thereto.
- the dissociation constant of an antibody of Disclosure C for IL-8 at pH 5.8 is, for example, 10 nM or more, but is not limited thereto.
- the dissociation constant of an antibody of Disclosure C for IL-8 at pH 5.8 is, for example, 30 nM or more, but is not limited thereto.
- the binding affinity of an anti-IL-8 antibody of Disclosure C for IL-8 is greater at a neutral pH than at an acidic pH.
- the dissociation constant ratio between acidic pH and neutral pH, [KD (acidic pH)/KD (neutral pH)], of an anti-IL-8 antibody of Disclosure C is, for example, 30 or more, but is not limited thereto.
- the dissociation constant ratio between acidic pH and neutral pH, [KD (acidic pH)/KD (neutral pH)], of an anti-IL-8 antibody of Disclosure C is, for example, 100 or more, for example, 200, 300, 400, 500, 600, 700, 800, 900, 1000, 1500, 2000, 2500, 3000, 3500, 4000, 4500, 5000, 5500, 6000, 6500, 7000, 7500, 8000, 8500, 9000, or 9500, but is not limited thereto.
- the dissociation constant ratio between pH 5.8 and pH 7.4, [KD (pH 5.8)/KD (pH 7.4)], of an anti-IL-8 antibody of Disclosure C is 30 or more, but is not limited thereto.
- the dissociation constant ratio between pH 5.8 and pH 7.4, [KD (pH 5.8)/KD (pH 7.4)], of an antibody of Disclosure C is, for example, 100 or more, for example, 200, 300, 400, 500, 600, 700, 800, 900, 1000, 1500, 2000, 2500, 3000, 3500, 4000, 4500, 5000, 5500, 6000, 6500, 7000, 7500, 8000, 8500, 9000, or 9500, but is not limited thereto.
- an anti-IL-8 antibody of Disclosure C has a large dissociation rate constant (koff) at an acidic pH.
- the dissociation rate constant of an antibody of Disclosure C at an acidic pH is, for example, 0.003 (1/s) or more, but is not limited thereto.
- the dissociation rate constant of an antibody of Disclosure C at an acidic pH is, for example, 0.005 (1/s) or more, but is not limited thereto.
- the dissociation rate constant of an antibody of Disclosure C at an acidic pH is, for example, 0.01 (1/s) or more, but is not limited thereto.
- an anti-IL-8 antibody of Disclosure C has a large dissociation rate constant (koff) at pH 5.8.
- the dissociation rate constant of an antibody of Disclosure C at pH 5.8 is, for example, 0.003 (1/s) or more, but is not limited thereto.
- the dissociation rate constant of an antibody of Disclosure C at pH 5.8 is, for example, 0.005 (1/s) or more, but is not limited thereto.
- the dissociation rate constant of an antibody of Disclosure C at pH 5.8 is, for example, 0.01 (1/s) or more, but is not limited thereto.
- the anti-IL-8 antibody of Disclosure C maintains the IL-8-neutralizing activity stably in a solution (for example, in PBS). Whether the activity is maintained stably in a solution can be assessed by measuring whether the IL-8-neutralizing activity of the antibody of Disclosure C added to the solution changes before and after storage for a certain period of time at a certain temperature.
- the storage period is, for example, one, two, three, or four weeks, but is not limited thereto.
- the storage temperature is, for example, 25°C, 30°C, 35°C, 40°C, or 50°C, but is not limited thereto.
- the storage temperature is, for example, 40°C, but is not limited thereto; and the storage period is, for example, two weeks, but is not limited thereto. In one embodiment, the storage temperature is, for example, 50°C, but is not limited thereto; and the storage period is, for example, one week, but is not limited thereto.
- the anti-IL-8 antibody of Disclosure C maintains the IL-8-neutralizing activity stably in vivo (for example, in plasma). Whether the activity is maintained stably in vivo can be assessed by measuring whether the IL-8-neutralizing activity of the antibody of Disclosure C added to plasma of an animal (for example, mouse) or human changes before and after storage for a certain period of time at a certain temperature.
- the storage period is, for example, one, two, three, or four weeks, but is not limited thereto.
- the storage temperature is, for example, 25°C, 30°C, 35°C, or 40°C, but is not limited thereto.
- the storage temperature is, for example, 40°C, but is not limited thereto; and the storage period is, for example, two weeks, but is not limited thereto.
- the rate of cellular uptake of an anti-IL-8 antibody of Disclosure C is greater when the antibody forms a complex with IL-8 than the antibody alone.
- the IL-8 antibody of Disclosure C is more easily taken up into cells when it is complexed with IL-8 outside of cells (for example, in plasma) than when not complexed with IL-8.
- the predicted immunogenicity of an anti-IL-8 antibody of Disclosure C is reduced.
- “Low immunogenicity” may mean, without being limited thereto, for example, that the administered anti-IL-8 antibody does not induce immune response of a living body in at least half or more of the individuals administered with a sufficient amount of the antibody for a sufficient period of time to achieve therapeutic efficacy.
- the induction of immune response may include production of anti-drug antibodies.
- “Low anti-drug antibody production” is interchangeable with "low immunogenicity”.
- the immunogenicity level in human can be estimated with a T cell epitope prediction program.
- T cell epitope prediction programs include Epibase (Lonza), iTope/TCED (Antitope), EpiMatrix (EpiVax), and so on.
- EpiMatrix is a system for predicting the immunogenicity of a protein of interest where sequences of peptide fragments are automatically designed by partitioning the amino acid sequence of a protein being analyzed for its immunogenicity into nine amino acids each to predict their ability to bind to eight major MHC Class II alleles (DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*0801, DRB1*1101, DRB1*1301, and DRB1*1501) (De Groot et al., Clin. Immunol.
- Sequences in which amino acids of the amino acid sequence of an anti-IL-8 antibody have been modified can be analyzed using the above-described T cell epitope prediction programs to design sequences with reduced immunogenicity.
- Preferred sites of amino acid modification to reduce the immunogenicity of the anti-IL-8 antibody of Disclosure C include, but are not limited to, the amino acids at position 81 and/or position 82b according to Kabat numbering in the heavy-chain sequence of the anti-IL-8 antibody shown in SEQ ID NO:78.
- Disclosure C provides methods for enhancing elimination of IL-8 from an individual as compared to when using a reference antibody, comprising administering an anti-IL-8 antibody of Disclosure C to the individual.
- Disclosure C relates to the use of an anti-IL-8 antibody of Disclosure C in the enhancement of the elimination of IL-8 from an individual as compared to when using a reference antibody.
- Disclosure C relates to an anti-IL-8 antibody of Disclosure C for use in the enhancement of the elimination of IL-8 from an individual as compared to when using a reference antibody.
- Disclosure C relates to the use of an anti-IL-8 antibody of Disclosure C in the production of pharmaceutical compositions for enhancing the elimination of IL-8 in vivo as compared to when using a reference antibody.
- Disclosure C relates to pharmaceutical compositions comprising an anti-IL-8 antibody of Disclosure C for enhancing the elimination of IL-8 as compared to when using a reference antibody.
- Disclosure C relates to methods for enhancing the elimination of IL-8 as compared to when using a reference antibody, comprising administering an anti-IL-8 antibody of Disclosure C to a subject.
- the reference antibody refers to an anti-IL-8 antibody before modification to obtain the antibody of Disclosure C, or an antibody whose IL-8 binding affinity is strong at both acidic and neutral pHs.
- the reference antibody may be an antibody comprising the amino acid sequence of SEQ ID NOs:83 and 84, or SEQ ID NOs:89 and 87.
- Disclosure C provides pharmaceutical compositions comprising an anti-IL-8 antibody of Disclosure C, characterized that the anti-IL-8 antibody of Disclosure C binds to IL-8 and then to extracellular matrix. In one embodiment, Disclosure C relates to the use of an anti-IL-8 antibody of Disclosure C in producing pharmaceutical compositions characterized that the anti-IL-8 antibody of Disclosure C binds to IL-8 and then to extracellular matrix.
- the anti-IL-8 antibody may be a humanized antibody.
- the antibody of Disclosure C comprises the heavy chain variable region of any one of the embodiments described above and the light chain variable region of any one of the embodiments described above. In one embodiment, the antibody of Disclosure C comprises each of the heavy-chain variable region of SEQ ID NO:78 and the light-chain variable region of SEQ ID NO:79, and also may comprise post-translational modifications in their sequences.
- an anti-IL-8 antibody according to any one of the embodiments described above may incorporate, singly or in combination, any of the features described in Sections 1 to 7 below.
- an antibody provided in Disclosure C may be a chimeric antibody.
- Certain chimeric antibodies are described, for example, in US Patent No. 4,816,567; and Morrison et al., Proc. Natl. Acad. Sci. USA 81:6851-6855 (1984).
- a chimeric antibody may comprise a non-human variable region (e.g., a variable region derived from a mouse, a rat, a hamster, a rabbit, or a non-human primate such as a monkey) and a human constant region.
- a chimeric antibody is a humanized antibody.
- a non-human antibody is humanized to reduce immunogenicity in humans, while retaining the specificity and affinity of the parental non-human antibody.
- a humanized antibody comprises one or more variable regions in which HVRs, e.g., CDRs (or portions thereof) are derived from a non-human antibody, and FRs (or portions thereof) are derived from human antibody sequences.
- HVRs e.g., CDRs (or portions thereof) are derived from a non-human antibody
- FRs or portions thereof
- a humanized antibody optionally will also comprise at least a portion of a human constant region.
- some FR residues in a humanized antibody may be substituted with corresponding residues from a non-human antibody (e.g., the antibody from which the HVR residues are derived), for example, to retain or improve antibody specificity or affinity.
- a non-human antibody e.g., the antibody from which the HVR residues are derived
- Human framework regions that may be used for humanization include but are not limited to framework regions selected using the "best-fit" method (see, e.g., Sims et al., J. Immunol. 151:2296 (1993)); framework regions derived from the consensus sequence of human antibodies of a particular subgroup of light or heavy chain variable regions (see, e.g., Carter et al., Proc. Natl. Acad. Sci. USA, 89:4285 (1992); and Presta et al., J. Immunol., 151:2623 (1993)); and framework regions derived from screening FR libraries (see, e.g., Baca et al., J. Biol. Chem. 272:10678-10684 (1997) and Rosok et al., J. Biol. Chem. 271:22611-22618 (1996)).
- an antibody provided in Disclosure C may be an antibody fragment.
- Antibody fragments include, but are not limited to, Fab, Fab', Fab'-SH, F(ab') 2 , Fv, and scFv fragments, and other fragments described below.
- Fab fragment antigen
- Fab' fragment antigen binding domain
- Diabodies are antibody fragments with two antigen-binding sites that may be bivalent or bispecific. See, for example, EP 404,097; WO 1993/01161; Hudson et al., Nat. Med. 9:129-134 (2003); and Hollinger et al., Proc. Natl. Acad. Sci. USA 90:6444-6448 (1993). Triabodies and tetrabodies are also described in Hudson et al., Nat. Med. 9:129-134 (2003).
- Single-domain antibodies are antibody fragments comprising all or a portion of the heavy chain variable domain or all or a portion of the light chain variable domain of an antibody.
- a single-domain antibody may be a human single-domain antibody (Domantis, Inc., Waltham, MA; see, e.g., US Patent No. 6,248,516 B1).
- Antibody fragments can be made by various techniques, including but not limited to proteolytic digestion of an intact antibody as well as production by recombinant host cells (e.g., E. coli or phage), as described within the scope of the description of Disclosure C herein.
- an antibody provided in Disclosure C may be a human antibody.
- Human antibodies can be prepared by various techniques known in the art. Human antibodies are described in general terms in van Dijk and van de Winkel, Curr. Opin. Pharmacol. 5:368-74 (2001) and Lonberg, Curr. Opin. Immunol. 20:450-459 (2008). Human antibodies may be prepared by administering an immunogen to a transgenic animal that has been modified to produce intact human antibodies or intact antibodies with human variable regions in response to antigenic challenge. Such animals typically contain all or a portion of the human immunoglobulin loci, which replace the immunoglobulin loci of the animal (non-human), or are present extrachromosomally or integrated randomly into the animal's chromosomes.
- Human variable regions from intact antibodies produced by such animals may be further modified, for example, by combining with a different human constant region.
- Human antibodies can also be prepared by hybridoma-based methods. Human myeloma and mouse-human heteromyeloma cell lines for the production of human monoclonal antibodies are described, for example, in Kozbor, J. Immunol. 133:3001 (1984); Brodeur et al., Monoclonal Antibody Production Techniques and Applications, pp. 51-63 (Marcel Dekker, Inc., New York, 1987); and Boerner et al., J. Immunol. 147:86 (1991). Human antibodies generated via human B-cell hybridoma are described in Li et al., Proc. Natl.
- Additional methods include, for example, the method described in US Patent No. 7,189,826, for the production of monoclonal human IgM antibodies from hybridoma cell lines, as well as, for example, the method described in Ni, Xiandai Mianyixue, 26 (4):265-268 (2006), for human-human hybridomas.
- Human hybridoma technology (trioma technology) is also described in Vollmers et al., Histol. and Histopath. 20(3):927-937 (2005) and Vollmers et al., Methods and Findings in Experimental and Clinical Pharmacology 27(3):185-91 (2005).
- Human antibodies may also be generated by isolating Fv clone variable domain sequences selected from human-derived phage display libraries. Such variable domain sequences can be combined with a desired human constant domain. Techniques for selecting human antibodies from antibody libraries are described below.
- Antibodies of Disclosure C can be isolated by screening combinatorial libraries for antibodies with the desired activity or activities. For example, various methods are known in the art for generating phage display libraries and screening such libraries for antibodies possessing desired binding characteristics. Such methods are reviewed in Hoogenboom et al., in Meth.Mol. Biol. 178:1-37 (O'Brien et al., ed., Human Press, Totowa, NJ, 2001), as wells as, for example, in McCafferty et al., Nature 348:552-554 (1990); Clackson et al., Nature 352:624-628 (1991); Marks et al., J. Mol. Biol.
- repertoires of VH and VL coding sequences may be separately cloned by polymerase chain reaction (PCR) and recombined randomly in phage libraries.
- PCR polymerase chain reaction
- the resulting phage libraries are screened for antigen-binding phage as described in Winter et al., Ann. Rev. Immunol. 12:433-455 (1994).
- Phage typically display antibody fragments, either as single-chain Fv (scFv) fragments or as Fab fragments.
- the naive repertoire can be cloned (for example, from human) to provide a single source of antibodies to a wide range of non-self and also self antigens without any immunization as described by Griffiths et al., EMBO J. 12:725-734 (1993).
- naive libraries can also be constructed synthetically by cloning unrearranged V-gene segments from stem cells, and using PCR primers containing random sequences to encode the highly variable CDR3 regions and to accomplish rearrangement in vitro (see below and Hoogenboom and Winter, J. Mol. Biol., 227:381-388 (1992); patent publications that describe human antibody phage libraries include, for example: US Patent No. 5,750,373; and US Appl. Publ. Nos. 2005/0079574, 2005/0119455, 2005/0266000, 2007/0117126, 2007/0160598, 2007/0237764, 2007/0292936, and 2009/0002360.
- antibodies or antibody fragments isolated from human antibody libraries are considered to be human antibodies or human antibody fragments.
- an antibody provided according to Disclosure C may be, for example, a multispecific antibody such as a bispecific antibody.
- Multispecific antibodies are monoclonal antibodies that have binding specificities for at least two different sites.
- one of the binding specificities is for IL-8, and the others are for any other antigens.
- bispecific antibodies may bind to two different epitopes on IL-8.
- Bispecific antibodies may also be used to localize cytotoxic agents to cells that express IL-8.
- Bispecific antibodies may be prepared as full length antibodies or as antibody fragments.
- Multispecific antibodies include, but are not limited to, recombinant co-expression of two immunoglobulin heavy chain-light chain pairs having different specificities (see Milstein and Cuello, Nature 305:537 (1983)); WO 93/08829; and Traunecker et al., EMBO J. 10:3655 (1991)) and the "knob-in-hole” method (see US Patent No. 5,731,168).
- Multispecific antibodies can be made by using electrostatic steering effects to prepare Fc-heterodimeric molecules (WO 2009/089004A1), by cross-linking two or more antibodies or fragments (US Patent No.
- the antibody or antibody fragment also includes a "Dual Acting Fab” or “DAF” comprising an antigen binding site that binds to IL-8 as well as another, different antigen (see US 2008/0069820, for example).
- Antibody variants Amino acid sequence variants of an antibody can be prepared by introducing appropriate modifications into a nucleotide sequence encoding the antibody, or by peptide synthesis. Such modifications include, for example, deletions, and/or insertions, and/or substitutions of residues in the amino acid sequence of the antibody. A final construct can be attained with any combination of deletion, insertion, and substitution, as long as the final construct is an antibody that has the desired properties described in the context of Disclosure C.
- Disclosure C provides antibody variants having one or more amino acid substitutions. Such substitution sites may be any positions in an antibody. Amino acids for conservative substitutions are shown in Table 10 under the heading of "conservative substitutions”. Amino acids for typical substitutions that result in more substantial changes are shown in Table 10 under the heading of "typical substitutions", and as further described in reference of amino acid side chain classes.
- Amino acids may be grouped according to common side-chain properties: (1) hydrophobic: Norleucine, Met, Ala, Val, Leu, Ile; (2) neutral hydrophilic: Cys, Ser, Thr, Asn, Gln; (3) acidic: Asp, Glu; (4) basic: His, Lys, Arg; (5) residues that influence chain orientation: Gly, Pro; and (6) aromatic: Trp, Tyr, Phe.
- Non-conservative substitutions will entail exchanging a member of one of these classes for another class.
- Amino acid insertions include fusion of a polypeptide comprising one, two, or three to one hundred or more residues at the N terminus and/or C terminus, as well as insertion of one or more amino acid residues into a sequence.
- Antibodies with such terminal insertion include, for example, antibodies with an N-terminal methionyl residue.
- Other insertional variants of the antibody molecule include those that result from N- or C-terminal fusion of the antibody to an enzyme (for example, ADEPT) or a polypeptide that increases plasma half-life of antibody.
- antibodies provided according to Disclosure C may be glycosylated antibodies. Glycosylation sites can be added to or deleted from an antibody by altering amino acid sequences in such a way as to create or remove glycosylation sites.
- Naive antibodies produced by animal cell typically contain a branched, biantennary oligosaccharide, which is attached by an N-linkage to Asn297 of the CH2 domain of the Fc region (see Wright et al. TIBTECH 15:26-32 (1997)).
- the oligosaccharide includes, for example, mannose, N-acetylglucosamine (GlcNAc), galactose, and sialic acid, as well as fucose attached to GlcNAc in the "stem" of the biantennary oligosaccharide structure.
- the oligosaccharide in the antibody of Disclosure C is modified to create antibody variants having certain improved properties.
- Fc region variants In one embodiment, one or more amino acid modifications are introduced into the Fc region of an antibody provided according to Disclosure C, thereby generating an Fc region variant.
- Fc region variants include those that have a modification (for example, a substitution) of one, two, three, or more amino acids in a native human Fc region sequence (for example, the Fc region of human IgG1, IgG2, IgG3, or IgG4).
- An anti-IL-8 antibody of Disclosure C may contain an Fc region having at least one of the following five properties, without being limited thereto: (a) increased binding affinity for FcRn of the Fc region relative to the binding affinity for FcRn of a native Fc region at acidic pH; (b) reduced binding affinity of the Fc region for pre-existing ADA relative to the binding affinity of a native Fc region for the pre-existing ADA; (c) increased plasma half-life of the Fc region relative to the plasma half-life of a native Fc region; (d) reduced plasma clearance of the Fc region relative to the plasma clearance of a native Fc region; and (e) reduced binding affinity of the Fc region for an effector receptor relative to the binding affinity of a native Fc region for the effector receptor.
- Fc region has 2, 3 or 4 of the above-listed properties.
- Fc region variants include those having an increased FcRn-binding affinity at an acidic pH.
- Fc region variants with increased FcRn-binding affinity include, but are not limited to, Fc region variants whose FcRn-binding affinity is increased up to 2-fold, 3-fold, 4-fold, 5-fold, 10-fold, 15-fold, 20-fold, 30-fold, 50-fold, or 100-fold as compared to the FcRn-binding affinity of an antibody comprising the native IgG Fc region.
- an Fc region variant includes a safe and advantageous Fc region variant that does not bind to pre-existing ADA, and at the same time has improved plasma retention.
- ADA refers to an endogenous antibody having binding affinity for an epitope on a therapeutic antibody.
- pre-existing ADA refers to a detectable anti-drug antibody present in a patient's blood prior to administration of a therapeutic antibody to the patient. Pre-existing ADA includes the rheumatoid factor.
- Fc region variants with low binding affinity for pre-existing ADA include, but are not limited to, Fc region variants whose ADA-binding affinity is reduced to 1/10 or less, 1/50 or less, or 1/100 or less as compared to the ADA-binding affinity of an antibody comprising the native IgG Fc region.
- an Fc region variant includes an Fc region variant whose binding affinity for complement proteins is low or that do not bind to complement proteins.
- Complement proteins include C1q.
- Fc region variants with low binding affinity for complement proteins include, but are not limited to, Fc region variants whose binding affinity for complement proteins is reduced to 1/10 or less, 1/50 or less, or 1/100 or less as compared to the complement protein-binding affinity of an antibody comprising a native IgG Fc region.
- an Fc region variant includes an Fc region variant whose binding affinity for effector receptors is low or that does not have the binding affinity for an effector receptor.
- the effector receptors include, but are not limited to, Fc ⁇ RI, Fc ⁇ RII, and Fc ⁇ RIII.
- Fc ⁇ RI includes, but is not limited to, Fc ⁇ RIa, Fc ⁇ RIb, and Fc ⁇ RIc, as well as subtypes thereof.
- Fc ⁇ RII includes, but is not limited to, Fc ⁇ RIIa (which has two allotypes: R131 and H131) and Fc ⁇ RIIb.
- Fc ⁇ RIII includes, but is not limited to, Fc ⁇ RIIIa (which has two allotypes: V158 and F158) and Fc ⁇ RIIIb (which has two allotypes: Fc ⁇ RIIIb-NA1 and Fc ⁇ RIIIb-NA2).
- Fc region variants with low binding affinity for effector receptors include, but are not limited to, Fc region variants whose binding affinity for effector receptors is reduced to at least 1/10 or less, 1/50 or less, or 1/100 or less as compared to the binding affinity of an antibody comprising a native IgG Fc region.
- an Fc region variant includes an Fc region comprising one or more amino acid substitutions at any of the positions of the group consisting of positions 235, 236, 239, 327, 330, 331, 428, 434, 436, 438, and 440, according to EU numbering as compared to the native Fc region.
- an Fc region variant includes an Fc region comprising amino acid substitutions at positions 235, 236, 239, 428, 434, 436, 438, and 440, according to EU numbering as compared to the native Fc region.
- an Fc region variant includes an Fc region comprising amino acid substitutions at positions 235, 236, 327, 330, 331, 428, 434, 436, 438, and 440, according to EU numbering as compared to the native Fc region.
- an Fc region variant includes an Fc region comprising one or more amino acid substitutions selected from the group consisting of: L235R, G236R, S239K, A327G, A330S, P331S, M428L, N434A, Y436T, Q438R, and S440E.
- an Fc region variant includes an Fc region comprising the amino acid substitutions of M428L, N434A, Y436T, Q438R, and S440E.
- an Fc region variant includes an Fc region comprising the amino acid substitutions of L235R, G236R, S239K, M428L, N434A, Y436T, Q438R, and S440E.
- an Fc region variant includes an Fc region comprising the amino acid substitutions of L235R, G236R, A327G, A330S, P331S, M428L, N434A, Y436T, Q438R, and S440E.
- an anti-IL-8 antibody of Disclosure C comprises the amino acid sequence of SEQ ID NO:80 and/or the amino acid sequence of SEQ ID NO:82.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:80 and/or the amino acid sequence of SEQ ID NO:82 may be an anti-IL-8 antibody that binds to IL-8 in a pH-dependent manner.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:80 and/or the amino acid sequence of SEQ ID NO:82 may maintain IL-8-neutralizing activity stably in vivo (for example, in plasma).
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:80 and/or the amino acid sequence of SEQ ID NO:82 may be an antibody with low immunogenicity.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:80 and/or the amino acid sequence of SEQ ID NO:82 may contain an Fc region whose FcRn-binding affinity at an acidic pH (e.g., pH 5.8) is increased as compared to the FcRn-binding affinity of a native Fc region at the acidic pH.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:80 and/or the amino acid sequence of SEQ ID NO:82 may contain an Fc region whose binding affinity for pre-existing ADA is reduced as compared to the binding affinity of a native Fc region for pre-existing ADA.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:80 and/or the amino acid sequence of SEQ ID NO:82 may contain an Fc region whose half-life in plasma is prolonged as compared to that of a native Fc region.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:80 and/or the amino acid sequence of SEQ ID NO:82 may contain an Fc region whose binding affinity for effector receptors is reduced as compared to that of a native Fc region.
- the anti-IL-8 antibody comprises a combination of any 2, 3, 4, 5, 6, or all 7 of above-listed properties.
- an anti-IL-8 antibody of Disclosure C comprises the amino acid sequence of SEQ ID NO:81 and/or the amino acid sequence of SEQ ID NO:82.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:81 and/or the amino acid sequence of SEQ ID NO:82 may be an anti-IL-8 antibody that binds to IL-8 in a pH-dependent manner.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:81 and/or the amino acid sequence of SEQ ID NO:82 may maintain IL-8-neutralizing activity stably in vivo (for example, in plasma).
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:81 and/or the amino acid sequence of SEQ ID NO:82 may be an antibody with low immunogenicity.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:81 and/or the amino acid sequence of SEQ ID NO:82 may contain an Fc region whose FcRn-binding affinity at an acidic pH is increased as compared to the FcRn-binding affinity of a native Fc region.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:81 and/or the amino acid sequence of SEQ ID NO:82 may contain an Fc region whose binding affinity for pre-existing ADA is reduced as compared to the binding affinity of a native Fc region for pre-existing ADA.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:81 and/or the amino acid sequence of SEQ ID NO:82 may contain an Fc region whose half-life in plasma is prolonged as compared to that of a native Fc region.
- the anti-IL-8 antibody comprising the amino acid sequence of SEQ ID NO:81 and/or the amino acid sequence of SEQ ID NO:82 may contain an Fc region whose binding affinity for effector receptors is reduced as compared to that of a native Fc region.
- the anti-IL-8 antibody comprises a combination of any 2, 3, 4, 5, 6, or all 7 of above-listed properties.
- Disclosure C encompasses an antibody variant that possesses some but not all effector functions.
- the antibody variant can be a desirable candidate for cases in which certain effector functions (such as complement and ADCC) are unnecessary or deleterious.
- In vitro and/or in vivo cytotoxicity assays known in the art can routinely be conducted to confirm the reduction/complete loss of CDC and/or ADCC activities.
- Fc receptor (FcR) binding assays can be conducted to confirm that an antibody lacks Fc ⁇ R binding (hence lacking ADCC activity), but retains FcRn binding ability.
- the primary cultured cells for mediating ADCC and NK cells express Fc ⁇ RIII only, whereas monocytes express Fc ⁇ RI, Fc ⁇ RII and Fc ⁇ RIII.
- FcR expression on hematopoietic cells is summarized in Table 3 on page 464 of Ravetch et al., Annu. Rev. Immunol. 9:457-492 (1991).
- Non-limiting examples of in vitro assays for assessing ADCC activity of a molecule of interest are described in US Patent No. 5,500,362 (see, e.g. Hellstrom. et al., Proc. Natl Acad. Sci. USA 83:7059-7063 (1986), Hellstrom et al., Proc. Natl Acad. Sci.
- non-radioactive isotope assays are available for assessing effector cell function (see, for example, ACTI(TM) non-radioactive cytotoxicity assay for flow cytometry (CellTechnology, Inc. Mountain View, CA; and CytoTox 96 TM non-radioactive cytotoxicity assay (Promega, Madison, WI)).
- Effector cells useful for such assays include peripheral blood mononuclear cells (PBMC) and Natural Killer (NK) cells.
- ADCC activity of an antibody variant of interest may be assessed in vivo, for example, in an animal model as disclosed in Clynes et al., Proc. Natl. Acad. Sci. USA 95:652-656 (1998).
- C1q binding assays may also be carried out to confirm that the antibody is unable to bind C1q and lacks CDC activity. See, e.g., C1q and C3c binding ELISA in WO 2006/029879 and WO 2005/100402.
- a CDC assay may be performed (see, e.g., Gazzano-Santoro et al., J. Immunol. Meth.
- FcRn binding and in vivo clearance/half life determinations can be performed using methods known in the art (see, e.g., Petkova et al., Intl. Immunol. 18(12):1759-1769 (2006)).
- Antibodies with reduced effector functions include those with substitution of one or more of Fc region residues at position 238, 265, 269, 270, 297, 327 or 329 (US Patent No. 6,737,056).
- Such Fc region variants include Fc region variants with substitutions at two or more of residues at position 265, 269, 270, 297 or 327, including the so-called "DANA" Fc region variants with substitution of residues 265 and 297 to alanine (US Patent No. 7,332,581).
- Antibody variants with improved or diminished binding to FcR groups are described below. (See, e.g., US Patent No. 6,737,056; WO 2004/056312, and Shields et al., J. Biol. Chem. 9(2):6591-6604 (2001).)
- Antibodies with increased blood half lives and improved FcRn binding at an acidic pH are described in US2005/0014934.
- the described antibodies comprise an Fc region with one or more substitutions that improve binding of the Fc region to FcRn.
- Fc region variants include those with substitutions at one or more of positions selected from 238, 256, 265, 272, 286, 303, 305, 307, 311, 312, 317, 340, 356, 360, 362, 376, 378, 380, 382, 413, 424 or 434 in an Fc region, for example, substitution of position 434 in an Fc region (US Patent No. 7,371,826).
- an antibody provided in Disclosure C may be further modified to contain additional nonproteinaceous moieties that are known in the art and readily available.
- the moieties suitable for derivatization of the antibody include, but are not limited to, water soluble polymers.
- water soluble polymers include, but are not limited to, polyethylene glycol (PEG), copolymers of ethylene glycol or propylene glycol, carboxymethylcellulose, dextran, polyvinyl alcohol, polyvinyl pyrrolidone, poly-1,3-dioxolane, poly-1,3,6-trioxane, ethylene/maleic anhydride copolymer, polyamino acids (either homopolymers or random copolymers), poly(n-vinyl pyrrolidone)polyethylene glycol, propropylene glycol homopolymers, prolylpropylene oxide/ethylene oxide co-polymers, polyoxyethylated polyols (e.g., glycerol), polyvinyl alcohol, and mixtures thereof.
- PEG polyethylene glycol
- copolymers of ethylene glycol or propylene glycol carboxymethylcellulose
- dextran polyvinyl alcohol
- polyvinyl pyrrolidone poly-1,
- the polymer may be of any molecular weight, and may be branched or unbranched.
- the number of polymers attached to the antibody may vary, and if more than one polymers are attached, they can be the same or different molecules. In general, the number and/or type of polymers used for derivertization can be determined based on considerations including, but not limited to, the particular properties or functions of the antibody to be improved, when the antibody derivative is used in a defined therapy, etc.
- conjugates of an anti-IL-8 antibody of Disclosure C and nonproteinaceous moiety that may be selectively heated by exposure to radiation may be provided.
- the nonproteinaceous moiety is, for example, a carbon nanotube (see, e.g., Kam et al., Proc. Natl. Acad. Sci. USA 102:11600-11605 (2005)).
- the radiation may be of any wavelength and includes, without being limited thereto, wavelengths that are harmless to humans but can heat the nonproteinaceous moiety to a temperature so as to kill cells proximal to the antibody-nonproteinaceous moiety.
- Anti-IL-8 antibodies of Disclosure C may be produced using recombinant methods and compositions, for example, as described in US Patent No. 4,816,567.
- One embodiment provides isolated nucleic acid(s) encoding an anti-IL-8 antibody which are presented as Disclosure C.
- Such nucleic acid(s) may encode an amino acid sequence comprising the VL of the antibody and/or an amino acid sequence comprising the VH of the antibody (e.g., the light and/or heavy chains of the antibody).
- one or more vectors e.g., expression vectors
- a host cell comprising such nucleic acid(s) is provided.
- a host cell comprises (e.g., has been transformed with): (1) a vector comprising a nucleic acid that encodes the VL of the antibody and the VH of an antibody, or (2) a first vector comprising a nucleic acid that encodes the VL of an antibody and a second vector comprising a nucleic acid that encodes the VH of the antibody.
- the host is eukaryotic (e.g., a Chinese Hamster Ovary (CHO) cell or lymphoid cell (e.g., Y0, NS0, SP20 cell)).
- eukaryotic e.g., a Chinese Hamster Ovary (CHO) cell or lymphoid cell (e.g., Y0, NS0, SP20 cell)
- lymphoid cell e.g., Y0, NS0, SP20 cell
- a method of producing an anti-IL-8 antibody of Disclosure C comprises culturing a host cell comprising a nucleic acid encoding the anti-IL-8 antibody as provided above, under conditions suitable for expressing the antibody, and optionally recovering the antibody (e.g., from the host cell or host cell culture medium).
- nucleic acid(s) encoding an antibody for example, as described above, is isolated and inserted into one or more vectors for further cloning and/or expression in a host cell.
- nucleic acid(s) may be readily isolated and sequenced using conventional procedures (e.g., by using oligonucleotide probes that specifically bind to nucleic acids encoding the heavy and light chains of the antibody).
- Suitable host cells for cloning or expression of antibody-encoding vectors include prokaryotic or eukaryotic cells described within the scope of the description of Disclosure C herein.
- antibodies may be produced in bacteria, in particular, when glycosylation and Fc effector function are not needed.
- For expression of antibody fragments and polypeptides in bacteria see for example, US Patent Nos. 5,648,237, 5,789,199, and 5,840,523 (See also Charlton, Methods in Molecular Biology, Vol. 248 (B.K.C. Lo, ed., Humana Press, Totowa, NJ, 2003), pp. 245-254, for expression of antibody fragments in E. coli).
- the antibody may be isolated from the bacterial cell paste in a soluble fraction and can be further purified.
- eukaryotic microbes such as filamentous fungi or yeast are suitable hosts for cloning or expression of antibody-encoding vectors, including fungi and yeast strains whose glycosylation pathways have been "humanized", which enable production of antibodies with a partially or fully human glycosylation pattern. See Gerngross, Nat. Biotech. 22:1409-1414 (2004), and Li et al., Nat. Biotech. 24:210-215 (2006).
- Suitable host cells for the expression of a glycosylated antibody are also derived from multicellular organisms (invertebrates and vertebrates). Examples of invertebrate cells include plant and insect cells. Without particular limitations, baculovirus is used in conjunction with insect cells for transfection of Spodoptera frugiperda cells and numerous baculoviral strains have been identified.
- Plant cell cultures can also be utilized as hosts. See US Patent Nos. 5,959,177, 6,040,498, 6,420,548, 7,125,978, and 6,417,429 (describing PLANTIBODIESTM technology for producing antibodies in transgenic plants).
- Vertebrate cells may also be used as hosts.
- mammalian cell lines that are adapted to grow in suspension are useful.
- Other examples of useful mammalian host cells are monkey kidney CV1 line transformed by SV40 (COS-7); human embryonic kidney line (293 cells as described in Graham et al., J. Gen Virol. 36:59 (1977)); baby hamster kidney cells (BHK); mouse sertoli cells (TM4 cells as described in Mather, Biol. Reprod.
- monkey kidney cells (CV1); African green monkey kidney cells (VERO-76); human cervical carcinoma cells (HELA); canine kidney cells (MDCK); buffalo rat liver cells (BRL 3A); human lung cells (W138); human liver cells (Hep G2); mouse mammary tumor (MMT 060562); TRI cells, as described in Mather et al., Annals N.Y. Acad. Sci. 383:44-68 (1982); MRC5 cells; and FS4 cells.
- CV1 monkey kidney cells
- VERO-76 African green monkey kidney cells
- HELA human cervical carcinoma cells
- MDCK canine kidney cells
- BBL 3A buffalo rat liver cells
- W138 human liver cells
- Hep G2 human liver cells
- MMT 060562 mouse mammary tumor
- TRI cells as described in Mather et al., Annals N.Y. Acad. Sci. 383:44-68 (1982); MRC5 cells; and FS4 cells.
- CHO Chinese hamster ovary
- DHFR-CHO cells Urlaub et al., Proc. Natl. Acad. Sci. USA 77:4216 (1980)
- myeloma cell lines such as Y0, NS0 and Sp20, but are not limited thereto.
- CHO Chinese hamster ovary
- myeloma cell lines such as Y0, NS0 and Sp20, but are not limited thereto.
- An antibody of Disclosure C produced by culturing such host cells as described above to carry nucleic acids that encode the antibody under conditions that are suitable for antibody expression may be isolated from inside or outside of the host cells (media, milk, etc.), and purified as a substantially pure homogeneous antibody. Isolation/purification methods that are generally used to purify polypeptides can be appropriately used to isolate and purify the antibody; however, the methods are not limited to the above example.
- the antibody can be appropriately separated and purified, for example, by appropriately selecting and combining column chromatography, filters, ultrafiltration, salting out, solvent precipitation, solvent extraction, distillation, immunoprecipitation, SDS-polyacrylamide gel electrophoresis, isoelectric focusing, dialysis, and recrystallization, without being limited thereto.
- Chromatography includes, but is not limited to, affinity chromatography, ion exchange chromatography, hydrophobic chromatography, gel filtration chromatography, reverse phase chromatography, and adsorption chromatography. Such chromatography can be performed using liquid chromatography, for example, HPLC and FPLC.
- Columns for use in affinity chromatography include, but are not limited to, Protein A column and Protein G column. Protein A columns include, but are not limited to, Hyper D, POROS, Sepharose F. F. (Pharmacia) and so on.
- Disclosure C provides methods for selecting antibodies with increased extracellular matrix-binding and antibodies with enhanced cellular uptake.
- Disclosure C provides methods for producing an anti-IL-8 antibody comprising variable region whose binding activity to IL-8 is in an pH-dependent manner, which comprise the steps of: (a) assessing the binding between anti-IL-8 antibody and extracellular matrix; (b) selecting an anti-IL-8 antibody that strongly binds to extracellular matrix; (c) culturing a host that comprises a vector carrying a nucleic acid encoding the antibody; and (d) isolating the antibody from the culture medium.
- Binding with extracellular matrix can be assessed by any methods without particular limitations, as long as they are known to those of ordinary skill in the art.
- assays can be carried out using an ELISA system for detecting the binding between an antibody and extracellular matrix, where the antibody is added to an extracellular matrix-immobilized plate and a labeled antibody against the antibody is added thereto.
- assays can be performed, for example, using an electrochemiluminescence (ECL) method in which a mixture of the antibody and a ruthenium antibody is added to an extracellular matrix-immobilized plate and the binding between the antibody and extracellular matrix is assessed based on the electrochemiluminescence of ruthenium.
- ECL electrochemiluminescence
- the anti-IL-8 antibody being assessed for extracellular matrix binding in step (a) above may be the antibody by itself or in contact with IL-8.
- "Selecting an anti-IL-8 antibody that strongly binds to extracellular matrix" in step (b) means that an anti-IL-8 antibody is selected based on the criterion that a value representing the binding between extracellular matrix and the anti-IL-8 antibody is higher than a value representing the binding between extracellular matrix and the control antibody in the assessment of extracellular matrix binding, and may be, for example, 2-fold, 3-fold, 4-fold, 5-fold, 6-fold, 7-fold, 8-fold, 9-fold, 10-fold, 20-fold, 50-fold, 100-fold, or more; however, the ratio is not particularly limited to the examples above.
- Disclosure C includes selecting an antibody with a higher value representing extracellular matrix binding from several anti-IL-8 antibodies that are not in contact with IL-8. In another embodiment, Disclosure C includes selecting an antibody with a higher value representing extracellular matrix binding from several anti-IL-8 antibodies that are in contact with IL-8.
- "selecting an anti-IL-8 antibody that strongly binds to extracellular matrix” in step (b) means that an antibody may be selected based on the criterion that the binding between an antibody and extracellular matrix varies depending on the presence of IL-8, when assessing extracellular matrix binding.
- the ratio of a value representing the extracellular matrix binding of an anti-IL-8 antibody in contact with IL-8 to a value representing the extracellular matrix binding of an anti-IL-8 antibody not in contact with IL-8 may be, for example, 2 to 1000. Furthermore, the ratio between the values may be 2, 3, 4, 5, 6, 7, 8, 9, 10, 20, 50, 100, 200, 300, 400, 500, 600, 700, 800, 900, or 1000.
- Anti-IL-8 antibodies provided within the scope of Disclosure C described herein can be identified, screened, or characterized in terms of their physical/chemical properties and/or biological activities by various methods known in the art.
- the antibodies of Disclosure C can be assessed for their antigen-binding activity by known methods, for example, ELISA, Western blotting, kinetic exclusion assay (KinExA TM ), and surface plasmon resonance using a device such as BIACORE (GE Healthcare).
- the binding affinity can be measured using BIACORE T200 (GE Healthcare) in the following manner.
- An appropriate amount of a trapping protein for example, Protein A/G (PIERCE)
- a sensor chip CM4 GE Healthcare
- an antibody of interest is allowed to be captured.
- a diluted antigen solution and running buffer as a reference solution: for example, 0.05% tween20, 20 mM ACES, 150 mM NaCl, pH 7.4
- the sensor chip is regenerated using 10 mM glycine HCl solution (pH 1.5).
- Measurements are performed at a pre-determined temperature (for example, 37°C, 25°C, or 20°C).
- the association rate constant kon (1/Ms) and dissociation rate constant koff (1/s), both of which are kinetic parameters, are calculated from sensorgrams obtained by measurement.
- the KD (M) of each antibody for the antigen is calculated based on these constants.
- Each parameter is calculated using the BIACORE T200 Evaluation Software (GE Healthcare).
- IL-8 can be quantitated as described below.
- An anti-human IL-8 antibody comprising the mouse IgG constant region is immobilized onto a plate.
- a solution comprising IL-8 bound to a humanized anti-IL-8 antibody, which does not compete with the above-described anti-human IL-8 antibody, is aliquoted to the immobilized plate.
- a biotinylated anti-human Ig ⁇ light chain antibody is added and allowed to react for a certain period of time.
- SULFO-Tag-labeled streptavidin is further added and allowed to react for a certain period of time.
- assay buffer is added and immediately measurement is performed with SECTOR Imager 2400 (Meso Scale Discovery).
- assays are provided to identify an anti-IL-8 antibody having a biological activity.
- the biological activity includes, for example, IL-8-neutralizing activity and the activity of blocking IL-8 signals.
- the Disclosure C also provides antibodies with such biological activity in vivo and/or ex vivo.
- the method for determining the level of IL-8 neutralization is not particularly limited and it can also be determined by the methods described below.
- PathHunter TM CHO-K1 CXCR2 ⁇ -Arrestin Cell Line (DiscoveRx, Cat.# 93-0202C2) is an artificial cell line created to express human CXCR2 known as a human IL-8 receptor and emit chemiluminescence when receiving signals by human IL-8.
- human IL-8 is added to a culture medium of the cells, chemiluminescence is emitted from the cells in a manner that depends on the concentration of added human IL-8.
- the chemiluminescence of the cells is reduced or undetectable as compared to when the antibody is not added, since the anti-human IL-8 antibody can block the IL-8 signal transduction.
- the human IL-8-neutralizing activity of the anti-human IL-8 antibody can be assessed by examining the difference described above.
- compositions comprising an anti-IL-8 antibody as described within the scope of the description Disclosure C herein may be prepared by mixing an anti-IL-8 antibody having the desired degree of purity with one or more optional pharmaceutically acceptable carriers (see, e.g., Remington's Pharmaceutical Sciences 16th edition, Osol, A. Ed. (1980)) in the form of lyophilized formulations or aqueous solution formulations.
- pharmaceutically acceptable carriers see, e.g., Remington's Pharmaceutical Sciences 16th edition, Osol, A. Ed. (1980) in the form of lyophilized formulations or aqueous solution formulations.
- Pharmaceutically acceptable carriers are generally nontoxic to recipients at the dosages and concentrations employed, and include without being limited to buffers such as phosphate, citrate, histidine, and other organic acids; antioxidants including ascorbic acid and methionine; preservatives (such as octadecyldimethylbenzyl ammonium chloride; hexamethonium chloride; benzalkonium chloride; benzethonium chloride; phenol, butyl or benzyl alcohol; alkyl parabens such as methyl or propyl paraben; catechol; resorcinol; cyclohexanol; 3-pentanol; and m-cresol); low molecular-weight (less than about 10 residues) polypeptides; proteins such as serum albumin, gelatin, or immunoglobulins; hydrophilic polymers such as polyvinylpyrrolidone; amino acids such as glycine, glutamine, asparagine, histidine,
- Exemplary pharmaceutically acceptable carriers herein further include interstitial drug dispersion agents such as soluble hyaluronidase glycoproteins (sHASEGP), for example, human soluble PH-20 hyaluronidase glycoproteins, such as rHuPH20 (HYLENEX TM , Baxter International, Inc.).
- sHASEGP soluble hyaluronidase glycoproteins
- rHuPH20 HYLENEX TM , Baxter International, Inc.
- Certain exemplary sHASEGPs and methods of use, including rHuPH20 are described in US Appl. Publ. Nos. 2005/0260186 and 2006/0104968.
- a sHASEGP is combined with one or more glycosaminoglycanases such as chondroitinases.
- Exemplary lyophilized antibody formulations are described in US Patent No. 6,267,958.
- Aqueous antibody formulations include those described in US Patent No. 6,171,586 and WO2006/044908; and the WO2006/044908 formulations include a histidine-acetate buffer.
- composition within the scope of Disclosure C herein may also contain more than one active ingredients as necessary for the particular indication being treated, preferably those with complementary activities that do not adversely affect each other.
- active ingredients are suitably present in combination in amounts that are effective for the purpose intended.
- Active ingredients may be entrapped in microcapsules prepared, for example, by coacervation techniques or by interfacial polymerization, for example, hydroxymethylcellulose or gelatin-microcapsules and poly-(methylmethacylate) microcapsules, respectively, in colloidal drug delivery systems (for example, liposomes, albumin microspheres, microemulsions, nano-particles and nanocapsules) or in macroemulsions.
- colloidal drug delivery systems for example, liposomes, albumin microspheres, microemulsions, nano-particles and nanocapsules
- Sustained-release preparations may be prepared. Suitable examples of sustained-release preparations include semipermeable matrices of solid hydrophobic polymers containing the anti-IL8 antibody of the Disclosure C, in which the matrices are in the form of shaped articles, for example, films or microcapsules.
- the formulations to be used for in vivo administration are generally sterile. Sterility may be readily accomplished, for example, by filtration through sterile filtration membranes.
- the anti-IL-8 antibodies provided according to Disclosure C are used in therapeutic methods.
- an anti-IL-8 antibody for use as a pharmaceutical composition is provided.
- an anti-IL-8 antibody for use in treating a disease where IL-8 is present in an excessive amount is provided.
- an anti-IL-8 antibody for use in methods for treating a disease where IL-8 is present in an excessive amount is provided.
- Disclosure C provides methods for treating an individual with a disease where IL-8 is present in an excessive amount (for example, a disease caused by the presence of excessive IL-8), which comprises administering an effective amount of an anti-IL-8 antibody to the individual.
- Disclosure C provides anti-IL-8 antibodies for use in such methods.
- Disclosure C relates to a pharmaceutical composition comprising an effective amount of an anti-IL-8 antibody, which is used to treat a disease where IL-8 is present in an excessive amount. In one embodiment, Disclosure C relates to the use of an anti-IL-8 antibody in producing a pharmaceutical composition for a disease where IL-8 is present in an excessive amount. In one embodiment, Disclosure C relates to the use of an effective amount of an anti-IL-8 antibody in treating a disease where IL-8 is present in an excessive amount.
- Disclosure C provides an anti-IL-8 antibody for use in suppressing the accumulation of IL-8 with biological activity. "Suppressing the accumulation of IL-8” may be achieved by preventing the amount of pre-existing IL-8 in vivo from increasing or by reducing the amount of pre-existing IL-8 in vivo. In one embodiment, the Disclosure C provides an anti-IL-8 antibody for suppressing the accumulation of IL-8 in an individual to suppress the accumulation of IL-8 with biological activity.
- IL-8 present in vivo may refer to IL-8 complexed with anti-IL-8 antibody or free IL-8; alternatively, it may refer to total IL-8 as its sum.
- Disclosure C provides a method for suppressing the accumulation of IL-8 with biological activity, which comprises the step of administering an effective amount of an anti-IL-8 antibody to an individual.
- Disclosure C relates to a pharmaceutical composition for suppressing the accumulation of IL-8 with biological activity, which comprises an effective amount of an anti-IL-8 antibody.
- Disclosure C relates to the use of an anti-IL-8 antibody in producing a pharmaceutical composition for suppressing the accumulation of IL-8 with biological activity.
- Disclosure C relates to the use of an effective amount of an anti-IL-8 antibody in suppressing the accumulation of IL-8 with biological activity.
- an anti-IL-8 antibody of Disclosure C suppresses the accumulation of IL-8 as compared to an anti-IL-8 antibody that does not have pH-dependent binding activity.
- the "individual" is preferably a human.
- Disclosure C provides an anti-IL-8 antibody for use in inhibiting angiogenesis (e.g., neoangiogenesis).
- Disclosure C provides a method for inhibiting neoangiogenesis in an individual which comprises administering an effective amount of an anti-IL-8 antibody to the individual, and also provides an anti-IL-8 antibody for use in the method.
- Disclosure C relates to a pharmaceutical composition for inhibiting neoangiogenesis which comprises an effective amount of an anti-IL-8 antibody.
- Disclosure C relates to the use of an anti-IL-8 antibody in producing a pharmaceutical composition for inhibiting neoangiogenesis.
- Disclosure C relates to the use of an effective amount of an anti-IL-8 antibody in inhibiting neoangiogenesis.
- the "individual" is preferably a human.
- Disclosure C provides an anti-IL-8 antibody for use in inhibiting the facilitation of neutrophil migration.
- Disclosure C provides a method for inhibiting the facilitation of neutrophil migration in an individual, which comprises administering an effective amount of an anti-IL-8 antibody to the individual; and also provides an anti-IL-8 antibody for use in the method.
- Disclosure C relates to pharmaceutical compositions for inhibiting facilitation of neutrophil migration in an individual, which comprise an effective amount of an anti-IL-8 antibody.
- Disclosure C relates to the use of an anti-IL-8 antibody in producing a pharmaceutical composition for inhibiting facilitation of neutrophil migration in an individual.
- Disclosure C relates to the use of an effective amount of an anti-IL-8 antibody in inhibiting facilitation of neutrophil migration in an individual.
- the "individual" is preferably a human.
- Disclosure C provides a pharmaceutical composition comprising an anti-IL-8 antibody provided herein for example, for use in any of the above therapeutic methods.
- a pharmaceutical composition comprises an anti-IL-8 antibodies provided in Disclosure C and a pharmaceutically acceptable carrier.
- An antibody of Disclosure C can be used either alone or in combination with other agents in a therapy.
- an antibody of Disclosure C may be co-administered with at least one additional therapeutic agent.
- An antibody of Disclosure C can be administered by any suitable means, including parenteral, intrapulmonary, and intranasal, and if desired for local treatment, intralesional administration.
- Parenteral infusions include intramuscular, intravenous, intraarterial, intraperitoneal, or subcutaneous administration. Dosing can be by any suitable route, for example, by injections such as intravenous or subcutaneous injections, depending in part on whether the administration is brief or chronic.
- Various dosing schedules include, without being limited to, single or multiple administrations over various time-points, bolus administration, and pulse infusion may be contemplated herein.
- an antibody of Disclosure C is formulated, dosed, and administered in a fashion consistent with good medical practice.
- Factors for consideration in this context include the particular disorder being treated, the particular mammal being treated, the clinical condition of the individual patient, the cause of the disorder, the site of delivery of the pharmaceutical composition, the method of administration, the scheduling of administration, and other factors known to medical practitioners.
- the antibody need not be, but is optionally formulated with one or more agents currently used to prevent or treat the disorder in question.
- the effective amount of such other agents depends on the amount of antibody present in the formulation, the type of disorder or treatment, and other factors discussed above. These may be generally used at the same dosages and via the same administration routes described within the scope of the description of Disclosure C herein, or from 1 to 99% of the dosages described within the scope of the description of Disclosure C herein, or in any dosage and by any route that is empirically/clinically determined to be appropriate.
- an antibody of Disclosure C (when used alone or in combination with one or more other additional therapeutic agents) depends on the type of disease to be treated, the type of antibody, the severity and course of the disease, whether the antibody variant is administered for preventive or therapeutic purposes, previous therapy, the patient's clinical history and response to the antibody, and discretion of the attending physician.
- the antibody is suitably administered to the patient at one time or over a series of treatments.
- about 1 ⁇ g/kg to 15 mg/kg (for example, 0.1 mg/kg to 10 mg/kg) of an antibody can be an initial candidate dose for administration to the patient, for example, by a single administration or several separate administrations, or by continuous infusion.
- One typical daily dose may range from about 1 mg/kg to 100 mg/kg or more, depending on the factors described above.
- the treatment may be generally sustained until a desired suppressive effect of disease symptoms is seen.
- a typical dose of an antibody may fall, for example, in the range from about 0.05 mg/kg to about 10 mg/kg.
- one or more doses of, for example, about 0.5 mg/kg, for example, 2.0 mg/kg, for example, 4.0 mg/kg, or for example, 10 mg/kg (or any combination thereof) may be administered to the patient.
- Such doses may be administered intermittently, for example, every week or every three weeks (for example, in such a manner that the patient receives from about two to about twenty doses, or about six doses of the antibody). It is possible to administer an initial higher loading dose, followed by one or more lower doses; however, other dosage regimens may be useful. The progress of this therapy can be easily monitored by conventional techniques and assays.
- the disclosure provides articles of manufacture comprising materials useful for the treatment, prevention and/or diagnosis of a disorder described above.
- Such an article of manufacture includes a container and a label or package insert on or associated with the container.
- Suitable containers include, for example, bottles, vials, and intravenous solution bags.
- the containers may be formed from various materials such as glass or plastic.
- Such a container holds a composition which is by itself or combined with another composition effective for treating, preventing and/or diagnosing the condition and may have a sterile access port (for example, the container may be an intravenous solution bag or a vial having a stopper pierceable by a hypodermic injection needle).
- At least one active ingredient in the composition is an antibody of Disclosure C.
- the label or package insert indicates that the composition is used for treating the condition of choice.
- the article of manufacture may include: (a) a first container that comprises a composition comprising an antibody of Disclosure C; and (b) a second container that comprises a composition comprising an additional cytotoxic agent or a different therapeutic agent.
- the article of manufacture in the embodiments of Disclosure C may further include a package insert indicating that the compositions can be used to treat a particular condition.
- the article of manufacture may further include, for example, a second (or third) container that comprises a pharmaceutically acceptable buffer such as bacteriostatic water for injection (BWFI), phosphate-buffered saline, Ringer's solution and dextrose solution.
- BWFI bacteriostatic water for injection
- phosphate-buffered saline such as bacteriostatic water for injection (BWFI), phosphate-buffered saline, Ringer's solution and dextrose solution.
- BWFI bacteriostatic water for injection
- the article of manufacture may further include other materials desirable from a commercial perspective or a user
- Disclosure C also includes all combinations of the whole or part of one or more of the entire embodiments described herein, except where there is a technological inconsistency.
- Embodiments of the Disclosures A, B, and C are described with reference to schematic illustrations, which may be exaggerated for clarity.
- Production of pH-dependent human IL-6 receptor-binding human antibodies with increased pI Fv4-IgG1 disclosed in WO2009/125825 is an antibody that binds to the human IL-6 receptor in a pH-dependent manner, and comprises VH3-IgG1 (SEQ ID NO:24) as the heavy chain and VL3-CK (SEQ ID NO:32) as the light chain.
- VH3-IgG1 SEQ ID NO:24
- VL3-CK SEQ ID NO:32
- the variable region of Fv4-IgG1 was introduced with amino acid substitutions that decrease the number of negatively charged amino acids (such as aspartic acid and glutamic acid), while increasing the positively charged amino acids (such as arginine and lysine).
- VH3(High_pI)-IgG1 (SEQ ID NO:25) was produced as a heavy chain with increased pI by substituting glutamic acid at position 16 with glutamine, glutamic acid at position 43 with arginine, glutamine at position 64 with lysine, and glutamic acid at position 105 with glutamine, according to Kabat numbering, in the heavy chain VH3-IgG1.
- VL3(High_pI)-CK (SEQ ID NO:33) was produced as a light chain with increased pI by substituting serine at position 18 with arginine, glutamine at position 24 with arginine, glutamic acid at position 45 with lysine, glutamic acid at position 79 with glutamine, and glutamic acid at position 107 with lysine, according to Kabat numbering, in the light chain VL3-CK.
- modifications that involve substituting alanine at position 80 with proline and alanine at position 83 with isoleucine were simultaneously introduced, although not with the aim to increase the pI.
- the following antibodies were produced by the method of Reference Example 2: (a) Low_pI-IgG1 comprising VH3-IgG1 as the heavy chain and VL3-CK as the light chain; (b) Middle_pI-IgG1 comprising VH3-IgG1 as the heavy chain and VL3(High_pI)-CK as the light chain; and (c) High_pI-IgG1 comprising VH3(High_pI)-IgG1 as the heavy chain and VL3(High_pI)-CK as the light chain.
- WO2011/122011 discloses Fv4-IgG1-F11 (hereinafter, referred to as Low_pI-F11) and Fv4-IgG1-F939 (hereinafter, referred to as Low_pI-F939) whose FcRn-mediated uptake into cells has been enhanced by introducing amino acid substitutions into the Fc region of Fv4-IgG1 and conferring FcRn-binding ability under neutral pH conditions.
- WO2013/125667 discloses Fv4-IgG1-F1180 (hereinafter, referred to as Low_pI-F1180) whose Fc ⁇ R-mediated uptake into cells has been enhanced by introducing amino acid substitutions into the Fc region of Fv4-IgG1 to increase its Fc ⁇ R-binding ability under neutral pH conditions. Simultaneously, amino acid modification for enhancing the plasma retention of the antibody by increasing its FcRn binding under the acidic pH condition in the endosomes was introduced into Fv4-IgG1-F1180. The antibodies shown below were produced by increasing the pI of antibodies containing these novel Fc region variants.
- VH3-IgG1-F11 (SEQ ID NO:30) and VH3-IgG1-F939 (SEQ ID NO:26) in WO2011/122011
- VH3-IgG1-F1180 (SEQ ID NO:28) in WO2013/125667 were each subjected to substitutions of glutamic acid at position 16 with glutamine, glutamic acid at position 43 with arginine, glutamine at position 64 with lysine, and glutamic acid at position 105 with glutamine, according to Kabat numbering, to produce VH3(High_pI)-F11 (SEQ ID NO:31), VH3(High_pI)-F939 (SEQ ID NO:27), and VH3(High_pI)-F1180 (SEQ ID NO:29), respectively, as heavy chains with increased pI.
- the following antibodies were produced by the method of Reference Example 2 using these heavy chains: (1) Low_pI-F939 comprising VH3-IgG1-F939 as the heavy chain and VL3-CK as the light chain; (2) Middle_pI-F939 comprising VH3(High_pI)-F939 as the heavy chain and VL3-CK as the light chain; (3) High_pI-F939 comprising VH3(High_pI)-F939 as the heavy chain and VL3(High_pI)-CK as the light chain; (4) Low_pI-F1180 comprising VH3-IgG1-F1180 as the heavy chain and VL3-CK as the light chain; (5) Middle_pI-F1180 comprising VH3-IgG1-F1180 as the heavy chain and VL3(High_pI)-CK as the light chain; (6) High_pI-F1180 comprising VH3(High_pI)-F1180 as the heavy chain and V
- the theoretical pI for each of the produced antibodies was calculated using GENETYX-SV/RC Ver 9.1.0 (GENETYX CORPORATION) by a method similar to that described previously.
- the calculated theoretical pI values are shown in Table 3.
- the theoretical pI values increased in a stepwise manner in the order of Low_pI, Middle_pI, and High_pI.
- Soluble human IL-6 receptor (also called “hsIL-6R") prepared by the method of Reference Example 3, the anti-human IL-6 receptor antibody, and human immunoglobulin preparation Sanglopor were administered simultaneously to human FcRn transgenic mice (B6.mFcRn-/-.hFcRn Tg line 32 +/+ mouse, Jackson Laboratories; Methods Mol. Biol. 602: 93-104 (2010)), and the subsequent in vivo kinetics of the soluble human IL-6 receptor were evaluated.
- human FcRn transgenic mice B6.mFcRn-/-.hFcRn Tg line 32 +/+ mouse, Jackson Laboratories; Methods Mol. Biol. 602: 93-104 (2010)
- a mixed solution containing the soluble human IL-6 receptor, the anti-human IL-6 receptor antibody, and Sanglopor (at concentrations of 5 ⁇ g/mL, 0.1 mg/mL, and 100 mg/mL, respectively) was administered once at 10 mL/kg through the tail vein. Since the anti-human IL-6 receptor antibody was present in sufficient excess of the soluble human IL-6 receptor, almost all of the soluble human IL-6 receptor was assumed to be bound by the antibody. Blood was collected 15 minutes, seven hours, one day, two days, three days, and seven days after the administration. The collected blood was immediately subjected to centrifugation at 4°C and 15,000 rpm for 15 minutes to obtain the plasma. The separated plasma was stored in a freezer set to -20°C or below until measurements were taken.
- the samples were mixed with a monoclonal anti-human IL-6R antibody (R&D) ruthenium-labeled with SULFO-TAG NHS Ester (Meso Scale Discovery), a biotinylated anti-human IL-6R Antibody (R&D), and Tocilizumab (CAS number: 375823-41-9) which is a human IL-6 receptor-binding antibody, and then they were allowed to react overnight at 37°C. The final concentration of Tocilizumab was adjusted to 333 ⁇ g/mL. Then, the reaction solutions were dispensed into a Streptavidin Gold Multi-ARRAY Plate (Meso Scale Discovery). After another hour of reaction at room temperature, the reaction solution was washed.
- R&D monoclonal anti-human IL-6R antibody
- SULFO-TAG NHS Ester SULFO-TAG NHS Ester
- R&D biotinylated anti-human IL-6R Antibody
- Tocilizumab
- Figs. 1, 2, and 3 show the observed changes in the concentration of the soluble human IL-6 receptor in the plasma of human FcRn transgenic mice after the intravenous administration.
- Fig. 1 shows the effect of enhancing antigen elimination where the pI of the variable region was increased in the case of a native IgG1 constant region.
- Fig. 2 shows the effect of enhancing antigen elimination where the pI of the variable region was increased in an antibody that has been conferred with the ability to bind to FcRn under a neutral pH condition (F939).
- Fig. 3 shows the effect of enhancing antigen elimination where the pI of the variable region was increased in an antibody whose Fc ⁇ R-binding ability under a neutral pH condition has been enhanced (F1180).
- An infusion pump (MINI-OSMOTIC PUMP MODEL 2004; alzet) containing a soluble human IL-6 receptor was implanted subcutaneously on the back of human FcRn transgenic mice (B6.mFcRn-/-.hFcRn Tg line 32 +/+ mouse, Jackson Laboratories; Methods Mol. Biol. 602:93-104 (2010)) to produce model animals whose plasma concentration of the soluble human IL-6 receptor was kept constant.
- Anti-human IL-6 receptor antibodies were administered to the model animals, and the in vivo kinetics of the antibodies after the administration were assessed.
- a monoclonal anti-mouse CD4 antibody obtained by a method known in the art was administered once at 20 mg/kg into the tail vein to suppress the production of neutralizing antibodies potentially producible by the mouse itself against the soluble human IL-6 receptor.
- an infusion pump containing 92.8 ⁇ g/ml of the soluble human IL-6 receptor was implanted subcutaneously on the back of the mice.
- anti-human IL-6 receptor antibodies were administered once at 1 mg/kg into the tail vein.
- Blood was collected from the mice 15 minutes, seven hours, one day, two days, three or four days, six or seven days, 13 or 14 days, 20 or 21 days, and 27 or 28 days after the administration of the anti-human IL-6 receptor antibodies.
- the collected blood was immediately centrifuged at 15,000 rpm and 4°C for 15 minutes to obtain plasma.
- the separated plasma was stored in a freezer at -20°C or below until measurements were taken.
- the final concentration of Tocilizumab was adjusted to 333 ⁇ g/mL. Then, the reaction solutions were dispensed into a Streptavidin Gold Multi-ARRAY Plate (Meso Scale Discovery). After another hour of reaction at room temperature, the reaction solution was washed. Then, immediately after Read Buffer T(x4) (Meso Scale Discovery) was dispensed into the plate, measurement was carried out using the SECTOR Imager 2400 (Meso Scale Discovery). The hsIL-6R concentration was calculated based on the response in the calibration curve using the analytical software SOFT max PRO (Molecular Devices).
- results obtained from these experiments can also be explained as follows: when the Fc region of the administered antibody is that of a native IgG antibody, uptake into the cell is thought to take place mainly by non-specific uptake (pinocytosis).
- the cell membrane is negatively charged, the higher the pI of the administered antibody-antigen complex is (i.e., the charge of the molecule as a whole is inclined toward positive charge), the more readily the complex may approach the cell membrane, and the easier the nonspecific uptake may take place.
- an antibody with increased pI forms a complex with an antigen
- that complex as a whole also has an increased pI in comparison to a complex formed between the original antibody and the antigen; therefore, uptake into cells may be increased. Therefore, by increasing the pI of an antibody that shows pH-dependent antigen binding, the speed or rate of antigen elimination from the plasma can be further accelerated, and the antigen concentration in the plasma can be maintained at a lower level.
- increase of the pI of the antibody was accomplished by introducing amino acid substitutions that decrease the number of negatively charged amino acids and/or increase the number of positively charged amino acids that may be exposed on the surface of the antibody molecule in the antibody variable region.
- effects obtained by such pI increase do not depend primarily (or substantially) on the type of the target antigen or the amino acid sequence that constitutes the antibody, but can be expected to depend on the pI.
- WO2007/114319 and WO2009/041643 describe the following matters in general terms.
- IgG antibodies that have Fc are known to have long half-lives since they are recycled by the salvage pathway of FcRn expressed in cells including the endothelial cells of blood vessels, and IgG is considered to be mainly metabolized in endothelial cells. More specifically, it is thought that IgGs that are non-specifically taken up into endothelial cells are recycled by binding to FcRn, and the molecules that cannot bind FcRn are metabolized.
- IgGs whose FcRn-binding ability has been reduced have shorter blood half-lives, and conversely, the blood half-life can be prolonged by increasing their binding ability toward FcRn.
- Previous methods for controlling the kinetics of IgG in blood involve modifying Fc to change the binding ability toward FcRn; however, the Working Examples of WO2007/114319 (mainly, techniques for substituting amino acids in the FR region) and WO2009/041643 (mainly techniques for substituting amino acids in the CDR region) showed that regardless of the target antigen type, by modifying the pI of the variable region of an antibody, its blood half-life can be controlled without modifying the Fc.
- the rate of non-specific uptake of an IgG antibody into endothelial cells is thought to depend on the physicochemical Coulombic interaction between the negatively charged cell surface and the IgG antibody. Therefore, it is considered that lowering (increasing) the pI of the IgG antibody and thus reducing (increasing) Coulombic interactions decreases (increases) its non-specific uptake into endothelial cells, and consequently decreases (increases) its metabolism in endothelial cells, thereby enabling the control of plasma pharmacokinetics.
- a reduction (an increase) of Coulombic interactions means a decrease (an increase) of the Coulombic force represented as an attractive force and/or an increase (a decrease) of the Columbic force represented as a repulsive force.
- the amino acid substitutions for accomplishing the above may be a single amino acid substitution or a combination of multiple amino acid substitutions.
- a method is provided for introducing a single amino acid substitution or a combination of multiple amino acid substitutions into a position(s) exposed on the antibody molecule surface.
- the multiple amino acid substitutions introduced may be positioned conformationally close to each other.
- the inventors arrived at the idea that, for example, when substituting amino acids that may be exposed on the antibody molecule surface with positively charged amino acids (preferably arginine or lysine) or when using pre-existing positively charged amino acids (preferably arginine or lysine), it may be preferable to further substitute one or more amino acids that are conformationally proximal to those amino acids (in certain cases, even one or more amino acids buried within the antibody molecule) with positively charged amino acids to produce, as a result, a state of locally clustered positive charges at conformationally proximal positions.
- positively charged amino acids preferably arginine or lysine
- pre-existing positively charged amino acids preferably arginine or lysine
- the definition of "conformationally proximal position(s)" is not particularly limited, but for example, it may mean a state where a single amino acid substitution or multiple amino acid substitutions are introduced within 20 Angstroms, preferably within 15 Angstroms, or more preferably within 10 Angstroms of one another. Whether the amino acid substitution of interest is at a position exposed on the antibody molecule surface, or whether the amino acid substitution is proximally positioned can be determined by known methods such as X-ray crystallography.
- the inventors also conceived that by producing an antibody molecule with broad and comprehensive consideration of the effects from charges, which include not only the pI but also the surface charges and local clustering of charges on antibody molecules, the speed of antigen elimination from the plasma can be further accelerated and the antigen concentration in the plasma can be maintained at even lower levels.
- Receptors such as FcRn or Fc ⁇ R are expressed on the cell membrane, and antibodies that have an enhanced affinity toward FcRn or Fc ⁇ R under neutral pH conditions are thought to be taken up into cells mainly through these Fc receptors. Since the cell membrane is negatively charged, the administered antibody-antigen complex approaches the cell membrane more readily when its pI is high (the charge of the molecule as a whole is shifted toward positive charge), and uptake through the Fc receptor may take place more easily. Therefore, antibodies that have an enhanced affinity towards FcRn or Fc ⁇ R under neutral pH conditions as well as an increased pI also show increased uptake into cells through Fc receptors when they form a complex with antigens.
- the speed of antigen elimination from the plasma by antibodies that bind to antigens in a pH-dependent manner and have an enhanced affinity toward FcRn or Fc ⁇ R under neutral pH conditions can be hastened by increasing their pIs, and the plasma antigen concentration can be maintained at lower levels.
- the antibodies to be evaluated were diluted to 30, 10, and 3 ⁇ g/mL using 20 mM ACES buffer at pH 7.4 (ACES-T buffer) containing 150 mM NaCl, 0.05% Tween 20, and 0.01% NaN 3 , and then were further diluted using 20 mM ACES buffer at pH 7.4 containing 150 mM NaCl, 0.01% Tween 20, 0.1% BSA, and 0.01% NaN 3 (Dilution Buffer) to produce a final concentration of 10, 3.3, and 1 ⁇ g/mL, respectively.
- the diluted antibody solutions were added to the plate from which the blocking solution was removed, and this was shaken at room temperature for one hour.
- the antibody solutions were removed, ACES-T buffer containing 0.25% glutaraldehyde was added, and after letting this stand for 10 minutes, the plate was washed with DPBS (manufactured by Wako Pure Chemical Industries) containing 0.05% Tween 20.
- the antibodies for ECL detection were prepared by sulfo-tagging goat anti-human IgG (gamma) (manufactured by Zymed Laboratories) using Sulfo-Tag NHS Ester (manufactured by MSD).
- Antibodies for detection were diluted in a dilution buffer to be 1 ⁇ g/mL, added to the plate, and then shaken in the dark at room temperature for one hour.
- the antibodies for detection were removed, and a 2-fold diluted solution prepared by diluting MSD Read Buffer T (4x) (manufactured by MSD) with ultrapure water was added; and then the amount of luminescence was measured on SECTOR Imager 2400 (manufactured by MSD).
- results obtained from these experiments can also be explained as follows.
- Histidine modifications into the antibody variable region is known to be one method for conferring pH-dependent antigen-binding property to an antibody (see e.g., WO2009/125825).
- Histidine has an imidazoyl group on its side chain, and is uncharged under neutral pH to basic pH conditions, but it is known to be positively charged under acidic pH conditions.
- Using this property of histidine by introducing histidine into the antibody variable region, particularly in the CDR(s) positioned close to the site of interaction with the antigen, one can change the charge environment and conformational environment at the site of interaction with the antigen between the neutral and acidic pH conditions.
- Such antibodies can be expected to have an antigen affinity that changes in a pH-dependent manner.
- Those of ordinary skill in the art will understand that the effects obtained by such introduction(s) of histidine do not primarily (or substantially) depend on the type of target antigen or the amino acid sequence that constitutes the antibody, but depend on the site of histidine introduction or the number of histidine residues introduced.
- WO2009/041643 describes in general terms as follows: protein-protein interactions consist of hydrophobic interactions, electrostatic interactions, and hydrogen bonds, and the strength of such binding can usually be represented using a binding constant (affinity) or an apparent binding constant (avidity). pH-dependent binding, where the strength of binding changes between a neutral pH condition (e.g., pH 7.4) and an acidic pH condition (for example, pH 5.5 to pH 6.0), depends on naturally-occurring protein-protein interactions. For example, the aforementioned binding between an IgG molecule and FcRn, which is known to be a salvage receptor for the IgG molecule, shows strong binding under acidic pH conditions and very weak binding under neutral pH conditions.
- affinity binding constant
- an apparent binding constant for example, pH 5.5 to pH 6.0
- Histidine residues are involved in many of the protein-protein interactions that change in a pH-dependent manner. Since the pKa of a histidine residue is close to 6.0 to 6.5, the state of proton dissociation in the histidine residue changes between neutral and acidic pH conditions. More specifically, the histidine residue is uncharged and neutral under neutral pH conditions, and functions as a hydrogen atom acceptor; while under acidic pH conditions, it is positively charged and functions as a hydrogen atom donor. Also in the IgG molecule-FcRn interaction described above, histidine residues present on the IgG molecule have been reported to be involved in pH-dependent binding (Martin et al., Mol. Cell. 7(4):867-877 (2001)).
- an amino acid residue that is introduced to increase the pI is preferably lysine, arginine, or histidine which have positively charged side chains.
- the standard pKa for these amino acid side chains is 10.5 for lysine, 12.5 for arginine, and 6.0 for histidine (Skoog et al., Trends Anal. Chem.5(4):82-83 (1986)). Based on the acid-base equilibrium theory known in the art, these pKa values mean that in a solution of pH 10.5, 50% of the lysine side chains are positively charged and the remaining 50% are uncharged.
- the above-mentioned theory does not negate the pI-increasing effect of introducing histidine and in the actual three-dimensional structure of the protein. It is sufficiently possible that an amino acid modification introducing histidine, as seen in the case with lysine or arginine, may also to exert an effect of pI increase.
- pI increase by one amino acid substitution in the constant region
- Methods for increasing the pI of an antibody that binds to an antigen in a pH-dependent manner by introducing amino acid substitutions into the antibody variable region have been described.
- methods for increasing the pI of an antibody can also be carried out by performing as few as one amino acid substitution in the antibody constant region.
- amino acid substitutions introduced into the constant region are preferably those that decrease the number of negatively charged amino acids (such as, aspartic acid or glutamic acid) while increasing the positively charged amino acids (such, as arginine or lysine).
- the positions for introducing amino acid substitutions in the constant regions are preferably positions where amino acid side chains may be exposed on the antibody molecule surface.
- Preferable examples include the method of introducing a combination of multiple amino acid substitutions at such positions that may be exposed on the antibody molecule surface.
- the multiple amino acid substitutions introduced are preferably positioned so that they are conformationally close to each other.
- the multiple amino acid substitutions introduced are preferably substitutions to positively charged amino acids, so that in certain cases they result in a state where multiple positive charges are present at conformationally proximal positions.
- a "conformationally proximal position” is not particularly limited, but for example, it may mean a state where a single amino acid substitution or multiple amino acid substitutions are introduced within 20 Angstroms, preferably within 15 Angstroms, or more preferably within 10 Angstroms of one another. Whether the amino acid substitution of interest is at a position exposed on the antibody molecule surface, or whether the multiple positions of amino acid substitutions are proximally positioned can be determined by known methods such as X-ray crystallography.
- the method for conferring multiple positive charges at conformationally proximal positions includes, in addition to the above-mentioned methods, a method of using amino acids that are originally positively charged in an IgG constant region.
- positively charged amino acid positions include (a) arginine at position 255, 292, 301, 344, 355, or 416, according to EU numbering; and (b) lysine at position 121, 133, 147, 205, 210, 213, 214, 218, 222, 246, 248, 274, 288, 290, 317, 320, 322, 326, 334, 338, 340, 360, 370, 392, 409, 414, or 439, according to EU numbering.
- KD (M) for human IgE was calculated for each antibody based on the association rate constant ka (1/Ms) and dissociation rate constant kd (1/s), which are kinetic parameters calculated from the sensorgrams obtained by the measurements.
- the BIACORE T100 Evaluation Software (GE Healthcare) was used to calculate each parameter.
- Affinities of Ab2 and Ab3 for human IgE at pH 7.4 and pH 5.8 were evaluated as follows.
- the binding activity (dissociation constant KD (M)) of anti-hIgE antibodies toward hIgE were evaluated using BIACORE T200 (GE Healthcare). Measurements were carried out using the following two buffers as the running buffers: (1) 1.2 mM CaCl 2 /0.05% tween 20, 20 mM ACES, 150 mM NaCl, pH 7.4; and (2) 1.2 mM CaCl 2 /0.05% tween 20, 20 mM ACES, 150 mM NaCl, pH 5.8.
- a peptide produced by adding biotin to Lys present at the C terminus of a chemically synthesized human glypican 3 (a.k.a., GPC3) protein-derived peptide (having the amino acid sequence of (VDDAPGNSQQATPKDNEISTFHNLGNVHSPLK (SEQ ID NO:44))("biotinylated GPC3 peptide") was added to Sensor chip SA (GE Healthcare) and immobilized onto the chip by utilizing the affinity between streptavidin and biotin.
- An appropriate concentration of hIgE was injected and immobilized onto the chip by capturing of the biotinylated GPC3 peptide.
- Ab1 produced in Example (4-1) is an antibody having native human IgG1 as the constant region.
- Ab1H-P600 was produced by modifying the Fc region of Ab1H, which is the heavy chain of Ab1, through substituting the proline at position 238 according to EU numbering with aspartic acid and substituting the serine at position 298 according to EU numbering with alanine.
- various Fc variants were produced by the method of Reference Example 2 by introducing the various single amino acid substitutions indicated in Tables 7-1 and 7-2 into the Fc region of Ab1H-P600, respectively.
- Ab1L SEQ ID NO:39
- the affinity of these antibodies for hFc ⁇ RII2b was comparable to the P600 variant (data not shown).
- an antibody-antigen complex and a running buffer (as a reference solution) was injected, and interaction was allowed to take place with the hFc ⁇ RIIb captured onto the sensor chip.
- 20 mM N-(2-Acetamido)-2-aminoethanesulfonic acid, 150 mM NaCl, 1.2 mM CaCl 2 , and 0.05% (w/v) Tween 20 at pH 7.4 was used as the running buffer, and the respective buffer was also used to dilute the soluble hFc ⁇ RIIb.
- 10 mM glycine-HCl at pH 1.5 was used. All measurements were carried out at 25°C.
- the BIACORE(registered trademark) sensor chip is known to be negatively charged, and this charged state can be considered to resemble the cell membrane surface. More specifically, the binding of an antigen-antibody complex for hFc ⁇ RIIb fixed onto the negatively charged BIACORE sensor chip is surmised to resemble the manner in which the antigen-antibody complex binds to hFc ⁇ RIIb present on a negatively charged cell membrane surface.
- the antibodies produced by introducing the pI-increasing modification into the Fc region are antibodies in which the charge of the Fc region (constant region) is more positively charged when compared with those before introduction of the modification. Therefore, the Coulombic interaction between the Fc region (positive charge) and the sensor chip surface (negative charge) can be considered to have been strengthened by the pI-increasing amino acid modification. Furthermore, such effects are expected to take place similarly on the negatively charged cell membrane surface; therefore, they are also expected to show an effect of accelerating the speed or rate of uptake into cells in vivo.
- a ratio of above about 1.2 fold or more for the binding to hFc ⁇ RIIb of a variant when compared to the binding to hFc ⁇ RIIb of Ab1H-P600 was considered to have strong charge effect on binding of an antibody to hFc ⁇ RIIb on the sensor chip.
- a modification that is expected to yield a charge effect includes, for example, a modification at position 196, 282, 285, 309, 311, 315, 345, 356, 358, 359, 361, 362, 382, 384, 385, 386, 387, 389, 399, 415, 418, 419, 421, 424, or 443, according to EU numbering.
- the modification is at position 282, 309, 311, 315, 345, 356, 359, 361, 362, 385, 386, 387, 389, 399, 418, 419, or 443.
- the amino acid substitution introduced at such position is preferably arginine or lysine.
- Another example of an amino acid mutation position where such a charge effect can be expected includes the glutamic acid at position 430 according to EU numbering.
- the preferred amino acid substitution to be introduced at position 430 is arginine or lysine which is positively charged, or among uncharged residues, substitution to glycine or threonine is preferred.
- An MDCK (Madin-Darby canine kidney) cell line that constitutively expresses hFc ⁇ RIIb was produced using known methods. Using these cells, intracellular uptake of antigen-antibody complexes was evaluated. Specifically, pHrodoRed (Life Technologies) was used to label human IgE (antigen) according to an established protocol, and antigen-antibody complexes were formed in a culture solution with the antibody concentration being 10.8 mg/mL and the antigen concentration being 12.5 mg/mL.
- the culture solution containing the antigen-antibody complexes was added to culture plates of the above-mentioned MDCK cells which constitutively express hFc ⁇ RIIb and incubated for one hour, and then the fluorescence intensity of the antigen taken up into the cells was quantified using InCell Analyzer 6000 (GE healthcare). The amount of antigen taken up was presented as relative values to the P600 value which is taken as 1.00.
- the antigen and antibodies added to the cell culture solution form antigen-antibody complexes in the culture solution.
- the antigen-antibody complexes bind to hFc ⁇ RIIb expressed on the cell membrane via the antibody Fc region, and are taken up into the cells in a receptor-dependent manner.
- Ab1 used in this experiment is an antibody that binds to the antigen in a pH-dependent manner; therefore, the antibody can dissociate from the antigen. Since the dissociated antigen is labeled with pHrodoRed as described earlier, it fluoresces in the endosomes. Thus, a stronger fluorescence intensity inside the cell compared to the control is thought to indicate that the uptake of the antigen-antibody complexes into the cells is taking place more quickly or more frequently.
- a ratio of above about 1.05 fold or more of the fluorescence intensity of the antigen taken up into the cells of the variants compared to the fluorescence intensity of Ab1H-P600 was considered to have charge effect on an antigen taken up into the cells.
- a ratio of above about 1.5 fold or more of the fluorescence intensity of the antigen taken up into the cells of the variants compared to the fluorescence intensity of Ab1H-P600 was considered to have a strong charge effect on an antigen taken up into the cells.
- An amino acid position modification that shows such effect is, for example, position 253, 254, 256, 258, 281, 282, 285, 286, 307, 309, 311, 315, 327, 330, 358, 384, 385, 387, 399, 400, 421, 433, or 434, according to EU numbering.
- modification is at position 254, 258, 281, 282, 285, 309, 311, 315, 327, 330, 358, 384, 399, 400, 421, 433, or 434, according to EU numbering.
- An amino acid substitution introduced at such a position is preferably arginine or lysine.
- the position where an amino acid substitution is introduced in the constant region with the objective of increasing the pI of the antibody may be, for example, the amino acid residue at position 285 according to EU numbering.
- other examples may include an amino acid substitution of the amino acid residue at position 399 according to EU numbering.
- IgG antibodies taken up into cells are known to be returned to the plasma by binding to FcRn. Therefore, IgG antibodies generally have long plasma half-life compared to proteins that do not bind to FcRn. Methods that utilize this property to enhance plasma retention of antibodies by increasing their FcRn affinity under acidic pH conditions through the introduction of amino acid modifications in the antibody Fc region are known.
- Fc variants with increased FcRn affinity under acidic pH conditions are also known to show undesired affinity towards the rheumatoid factor (RF) (WO2013/046704). Therefore, the following examinations were carried out with an objective of producing Fc variants that can improve plasma retention with decreased or substantially no binding to rheumatoid factor.
- Fv4-IgG1 comprising VH3-IgG1m (SEQ ID NO:46) as the heavy chain and VL3-CK as the light chain
- Fv4-YTE comprising VH3-YTE (SEQ ID NO:47) as the heavy chain and VL3-CK as the light chain
- Fv4-LS comprising VH3-LS (SEQ ID NO:48) as the heavy chain and VL3-CK as the light chain
- Fv4-N434H comprising VH3-N434H (SEQ ID NO:49) as the heavy chain and VL3-CK as the light chain
- Fv4-F1847m comprising VH3-F1847m (SEQ ID NO:50) as the heavy chain and VL3-CK as the light chain
- Fv4-F1848m comprising VH3-F1848m (SEQ ID NO:
- KD (M) for human FcRn was calculated for each antibody based on the association rate constant ka (1/Ms) and dissociation rate constant kd (1/s), which are kinetic parameters calculated from sensorgrams obtained by the measurements.
- the BIACORE T100 Evaluation Software was used to calculate each parameter.
- Anti-drug antibodies affect the efficacy and pharmacokinetics of therapeutic antibodies, and lead to serious side-effects at times; therefore, clinical utility and efficacy of therapeutic antibodies may be limited by production of ADAs.
- Many factors influence the immunogenicity of therapeutic antibodies, and the presence of effector T cell epitopes is one factor.
- the presence of ADA in a patient before administration of the therapeutic antibody also called "Pre-existing ADA" may have similar problems.
- rheumatoid factor which is an autoantibody against human IgG may cause a "pre-existing ADA" problem.
- RA rheumatoid arthritis
- RF rheumatoid factor
- N434H N434H
- the rheumatoid factor is a polyclonal autoantibody against human IgG, and its epitopes in human IgG differ depending on the clone and seem to be positioned in the CH2/CH3 interface region, and in the CH3 domain that may overlap with the FcRn-binding epitope. Therefore, mutations that increase the binding activity (binding affinity) towards FcRn may increase the binding activity (binding affinity) towards specific clones of the rheumatoid factor.
Landscapes
- Health & Medical Sciences (AREA)
- Chemical & Material Sciences (AREA)
- Organic Chemistry (AREA)
- Life Sciences & Earth Sciences (AREA)
- General Health & Medical Sciences (AREA)
- Medicinal Chemistry (AREA)
- Immunology (AREA)
- Genetics & Genomics (AREA)
- Molecular Biology (AREA)
- Engineering & Computer Science (AREA)
- Biochemistry (AREA)
- Biophysics (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Chemical Kinetics & Catalysis (AREA)
- General Chemical & Material Sciences (AREA)
- Nuclear Medicine, Radiotherapy & Molecular Imaging (AREA)
- Pharmacology & Pharmacy (AREA)
- Animal Behavior & Ethology (AREA)
- Public Health (AREA)
- Veterinary Medicine (AREA)
- Pulmonology (AREA)
- Biomedical Technology (AREA)
- Hematology (AREA)
- Urology & Nephrology (AREA)
- Gastroenterology & Hepatology (AREA)
- Biotechnology (AREA)
- General Engineering & Computer Science (AREA)
- Dermatology (AREA)
- Wood Science & Technology (AREA)
- Zoology (AREA)
- Oncology (AREA)
- Reproductive Health (AREA)
- Physics & Mathematics (AREA)
- Neurology (AREA)
- Vascular Medicine (AREA)
- Plant Pathology (AREA)
- Pain & Pain Management (AREA)
- Heart & Thoracic Surgery (AREA)
- Microbiology (AREA)
Priority Applications (18)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| KR1020187032734A KR102538745B1 (ko) | 2015-09-18 | 2016-08-05 | Il-8에 결합하는 항체 및 그의 사용 |
| IL258088A IL258088B2 (en) | 2015-09-18 | 2016-08-05 | Antibodies that bind to IL-8 and their uses |
| BR112018002432A BR112018002432A2 (pt) | 2015-09-18 | 2016-08-05 | anticorpos de ligação à il-8 e usos dos mesmos |
| RU2018113505A RU2728430C2 (ru) | 2015-09-18 | 2016-08-05 | Il-8-связывающие антитела и их применения |
| AU2016323088A AU2016323088B2 (en) | 2015-09-18 | 2016-08-05 | IL-8-binding antibodies and uses thereof |
| MX2018003005A MX2018003005A (es) | 2015-09-18 | 2016-08-05 | Anticuerpos que se unen a interleucina 8 (il-8) y sus usos. |
| EP16753718.2A EP3350202A1 (en) | 2015-09-18 | 2016-08-05 | Il-8-binding antibodies and uses thereof |
| CN201680054135.3A CN108271372B (zh) | 2015-09-18 | 2016-08-05 | Il-8-结合抗体及其应用 |
| CA2993423A CA2993423C (en) | 2015-09-18 | 2016-08-05 | Il-8-binding antibodies and uses thereof |
| KR1020177037003A KR101920175B1 (ko) | 2015-09-18 | 2016-08-05 | Il-8에 결합하는 항체 및 그의 사용 |
| CR20180217A CR20180217A (es) | 2015-09-18 | 2016-08-05 | Anticuerpos que se unen a interleucina 8 (il-8) y sus usos |
| JP2017508124A JP6266164B2 (ja) | 2015-09-18 | 2016-08-05 | Il−8に結合する抗体およびその使用 |
| KR1020237017768A KR20230079500A (ko) | 2015-09-18 | 2016-08-05 | Il-8에 결합하는 항체 및 그의 사용 |
| UAA201804018A UA120981C2 (uk) | 2015-09-18 | 2016-08-05 | Антитіло, що зв'язується з il-8, та його застосування |
| ZA2018/00536A ZA201800536B (en) | 2015-09-18 | 2018-01-25 | Il-8-binding antibodies and uses thereof |
| PH12018500386A PH12018500386A1 (en) | 2015-09-18 | 2018-02-21 | Il-8-binding antibodies and uses thereof |
| CONC2018/0004056A CO2018004056A2 (es) | 2015-09-18 | 2018-04-16 | Anticuerpos que se unen a interleucina 8 (il-8) y sus usos |
| IL285375A IL285375A (en) | 2015-09-18 | 2021-08-04 | Antibodies that bind to il-8 and their uses |
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| JP2015185254 | 2015-09-18 | ||
| JP2015-185254 | 2015-09-18 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2017046994A1 true WO2017046994A1 (en) | 2017-03-23 |
Family
ID=56738152
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/JP2016/003616 WO2017046994A1 (en) | 2015-09-18 | 2016-08-05 | Il-8-binding antibodies and uses thereof |
Country Status (22)
| Country | Link |
|---|---|
| EP (1) | EP3350202A1 (es) |
| JP (4) | JP6266164B2 (es) |
| KR (3) | KR20230079500A (es) |
| CN (2) | CN108271372B (es) |
| AR (1) | AR105634A1 (es) |
| AU (1) | AU2016323088B2 (es) |
| BR (1) | BR112018002432A2 (es) |
| CA (1) | CA2993423C (es) |
| CL (1) | CL2018000699A1 (es) |
| CO (1) | CO2018004056A2 (es) |
| CR (1) | CR20180217A (es) |
| HK (1) | HK1252615A1 (es) |
| IL (2) | IL258088B2 (es) |
| MA (1) | MA42808A (es) |
| MX (2) | MX2018003005A (es) |
| PE (1) | PE20181336A1 (es) |
| PH (1) | PH12018500386A1 (es) |
| RU (1) | RU2728430C2 (es) |
| TW (4) | TWI814162B (es) |
| UA (1) | UA120981C2 (es) |
| WO (1) | WO2017046994A1 (es) |
| ZA (1) | ZA201800536B (es) |
Cited By (25)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US9828429B2 (en) | 2007-09-26 | 2017-11-28 | Chugai Seiyaku Kabushiki Kaisha | Method of modifying isoelectric point of antibody via amino acid substitution in CDR |
| US9868948B2 (en) | 2008-04-11 | 2018-01-16 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecule capable of binding to two or more antigen molecules repeatedly |
| WO2018025982A1 (ja) * | 2016-08-05 | 2018-02-08 | 中外製薬株式会社 | Il-8関連疾患の治療用又は予防用組成物 |
| US9969800B2 (en) | 2015-02-05 | 2018-05-15 | Chugai Seiyaku Kabushiki Kaisha | IL-8 antibodies |
| US10000560B2 (en) | 2014-12-19 | 2018-06-19 | Chugai Seiyaku Kabushiki Kaisha | Anti-myostatin antibodies, polypeptides containing variant Fc regions, and methods of use |
| KR20200014385A (ko) * | 2017-06-05 | 2020-02-10 | 얀센 바이오테크 인코포레이티드 | 이중특이성 항체 생성을 위한 표면 전하 조작 방법 |
| WO2020032230A1 (ja) | 2018-08-10 | 2020-02-13 | 中外製薬株式会社 | 抗cd137抗原結合分子およびその使用 |
| US10604561B2 (en) | 2016-09-16 | 2020-03-31 | Chugai Seiyaku Kabushiki Kaisha | Anti-dengue virus antibodies, polypeptides containing variant Fc regions, and methods of use |
| US10919953B2 (en) | 2012-08-24 | 2021-02-16 | Chugai Seiyaku Kabushiki Kaisha | FcgammaRIIB-specific Fc region variant |
| US10961530B2 (en) | 2013-12-04 | 2021-03-30 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecules, the antigen-binding activity of which varies according to the concentration of compounds, and libraries of said molecules |
| WO2021122733A1 (en) | 2019-12-18 | 2021-06-24 | F. Hoffmann-La Roche Ag | Bispecific anti-ccl2 antibodies |
| US11046784B2 (en) | 2006-03-31 | 2021-06-29 | Chugai Seiyaku Kabushiki Kaisha | Methods for controlling blood pharmacokinetics of antibodies |
| WO2021153723A1 (ja) | 2020-01-31 | 2021-08-05 | 中外製薬株式会社 | 着色を抑制したポリペプチドを含む組成物の製造方法 |
| WO2021162020A1 (ja) | 2020-02-12 | 2021-08-19 | 中外製薬株式会社 | 癌の治療に用いるための抗cd137抗原結合分子 |
| CN113544157A (zh) * | 2019-04-19 | 2021-10-22 | 中外制药株式会社 | 用于检测或捕获样品中的多肽的抗体和组合物,以及用于检测或捕获样品中的多肽的方法 |
| GB2595299A (en) * | 2020-05-21 | 2021-11-24 | Mabsolve Ltd | Modified immunoglobulin FC regions |
| US11236168B2 (en) | 2012-08-24 | 2022-02-01 | Chugai Seiyaku Kabushiki Kaisha | Mouse FcγammaRII-specific Fc antibody |
| US11267868B2 (en) | 2013-04-02 | 2022-03-08 | Chugai Seiyaku Kabushiki Kaisha | Fc region variant |
| US11359009B2 (en) | 2015-12-25 | 2022-06-14 | Chugai Seiyaku Kabushiki Kaisha | Anti-myostatin antibodies and methods of use |
| WO2022244838A1 (ja) | 2021-05-19 | 2022-11-24 | 中外製薬株式会社 | 分子のin vivo薬物動態を予測する方法 |
| WO2022263501A1 (en) | 2021-06-18 | 2022-12-22 | F. Hoffmann-La Roche Ag | Bispecific anti-ccl2 antibodies |
| US11673947B2 (en) | 2012-05-30 | 2023-06-13 | Chugai Seiyaku Kabushiki Kaisha | Target tissue-specific antigen-binding molecule |
| US11891434B2 (en) | 2010-11-30 | 2024-02-06 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecule capable of binding to plurality of antigen molecules repeatedly |
| US11891432B2 (en) | 2018-03-15 | 2024-02-06 | Chugai Seiyaku Kabushiki Kaisha | Anti-dengue virus antibodies having cross-reactivity to Zika virus and methods of use |
| US12122840B2 (en) | 2007-09-26 | 2024-10-22 | Chugai Seiyaku Kabushiki Kaisha | Method of modifying isoelectric point of antibody via amino acid substitution in CDR |
Families Citing this family (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| PE20181336A1 (es) * | 2015-09-18 | 2018-08-21 | Chugai Pharmaceutical Co Ltd | Anticuerpos que se unen a interleucina 8 (il-8) y sus usos |
| JPWO2022025030A1 (es) * | 2020-07-28 | 2022-02-03 |
Citations (118)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US4676980A (en) | 1985-09-23 | 1987-06-30 | The United States Of America As Represented By The Secretary Of The Department Of Health And Human Services | Target specific cross-linked heteroantibodies |
| EP0239400A2 (en) | 1986-03-27 | 1987-09-30 | Medical Research Council | Recombinant antibodies and methods for their production |
| US4816567A (en) | 1983-04-08 | 1989-03-28 | Genentech, Inc. | Recombinant immunoglobin preparations |
| EP0404097A2 (de) | 1989-06-22 | 1990-12-27 | BEHRINGWERKE Aktiengesellschaft | Bispezifische und oligospezifische, mono- und oligovalente Rezeptoren, ihre Herstellung und Verwendung |
| WO1992003918A1 (en) | 1990-08-29 | 1992-03-19 | Genpharm International, Inc. | Transgenic non-human animals capable of producing heterologous antibodies |
| WO1993001161A1 (en) | 1991-07-11 | 1993-01-21 | Pfizer Limited | Process for preparing sertraline intermediates |
| WO1993008829A1 (en) | 1991-11-04 | 1993-05-13 | The Regents Of The University Of California | Compositions that mediate killing of hiv-infected cells |
| WO1993011161A1 (en) | 1991-11-25 | 1993-06-10 | Enzon, Inc. | Multivalent antigen-binding proteins |
| WO1993012227A1 (en) | 1991-12-17 | 1993-06-24 | Genpharm International, Inc. | Transgenic non-human animals capable of producing heterologous antibodies |
| WO1993016185A2 (en) | 1992-02-06 | 1993-08-19 | Creative Biomolecules, Inc. | Biosynthetic binding protein for cancer marker |
| WO1994002602A1 (en) | 1992-07-24 | 1994-02-03 | Cell Genesys, Inc. | Generation of xenogeneic antibodies |
| WO1994029351A2 (en) | 1993-06-16 | 1994-12-22 | Celltech Limited | Antibodies |
| WO1995001937A1 (fr) | 1993-07-09 | 1995-01-19 | Association Gradient | Procede de traitement de residus de combustion et installation de mise en ×uvre dudit procede |
| WO1996002576A1 (fr) | 1994-07-13 | 1996-02-01 | Chugai Seiyaku Kabushiki Kaisha | Anticorps humain reconstitue contre l'interleukine-8 humaine |
| US5500362A (en) | 1987-01-08 | 1996-03-19 | Xoma Corporation | Chimeric antibody with specificity to human B cell surface antigen |
| WO1996034096A1 (en) | 1995-04-28 | 1996-10-31 | Abgenix, Inc. | Human antibodies derived from immunized xenomice |
| WO1996033735A1 (en) | 1995-04-27 | 1996-10-31 | Abgenix, Inc. | Human antibodies derived from immunized xenomice |
| US5571894A (en) | 1991-02-05 | 1996-11-05 | Ciba-Geigy Corporation | Recombinant antibodies specific for a growth factor receptor |
| US5587458A (en) | 1991-10-07 | 1996-12-24 | Aronex Pharmaceuticals, Inc. | Anti-erbB-2 antibodies, combinations thereof, and therapeutic and diagnostic uses thereof |
| US5624821A (en) | 1987-03-18 | 1997-04-29 | Scotgen Biopharmaceuticals Incorporated | Antibodies with altered effector functions |
| US5648237A (en) | 1991-09-19 | 1997-07-15 | Genentech, Inc. | Expression of functional antibody fragments |
| US5731168A (en) | 1995-03-01 | 1998-03-24 | Genentech, Inc. | Method for making heteromultimeric polypeptides |
| WO1998013388A1 (fr) | 1996-09-26 | 1998-04-02 | Chugai Seiyaku Kabushiki Kaisha | Anticorps contre les peptides lies a la parathormone humaine |
| US5750373A (en) | 1990-12-03 | 1998-05-12 | Genentech, Inc. | Enrichment method for variant proteins having altered binding properties, M13 phagemids, and growth hormone variants |
| US5770429A (en) | 1990-08-29 | 1998-06-23 | Genpharm International, Inc. | Transgenic non-human animals capable of producing heterologous antibodies |
| US5789199A (en) | 1994-11-03 | 1998-08-04 | Genentech, Inc. | Process for bacterial production of polypeptides |
| US5821337A (en) | 1991-06-14 | 1998-10-13 | Genentech, Inc. | Immunoglobulin variants |
| WO1998046777A1 (fr) | 1997-04-11 | 1998-10-22 | Centre National De La Recherche Scientifique (Cnrs) | Preparation de recepteurs membranaires a partir de baculovirus extracellulaires |
| US5840523A (en) | 1995-03-01 | 1998-11-24 | Genetech, Inc. | Methods and compositions for secretion of heterologous polypeptides |
| US5959177A (en) | 1989-10-27 | 1999-09-28 | The Scripps Research Institute | Transgenic plants expressing assembled secretory antibodies |
| WO1999054342A1 (en) | 1998-04-20 | 1999-10-28 | Pablo Umana | Glycosylation engineering of antibodies for improving antibody-dependent cellular cytotoxicity |
| US6024956A (en) * | 1994-07-13 | 2000-02-15 | Chugai Pharmaceutical Co., Ltd. | Method to treat inflammation with humanized anti-IL-8-antibodies |
| US6040498A (en) | 1998-08-11 | 2000-03-21 | North Caroline State University | Genetically engineered duckweed |
| US6075181A (en) | 1990-01-12 | 2000-06-13 | Abgenix, Inc. | Human antibodies derived from immunized xenomice |
| WO2000042072A2 (en) | 1999-01-15 | 2000-07-20 | Genentech, Inc. | Polypeptide variants with altered effector function |
| WO2000061739A1 (en) | 1999-04-09 | 2000-10-19 | Kyowa Hakko Kogyo Co., Ltd. | Method for controlling the activity of immunologically functional molecule |
| US6150584A (en) | 1990-01-12 | 2000-11-21 | Abgenix, Inc. | Human antibodies derived from immunized xenomice |
| US6171586B1 (en) | 1997-06-13 | 2001-01-09 | Genentech, Inc. | Antibody formulation |
| US6248516B1 (en) | 1988-11-11 | 2001-06-19 | Medical Research Council | Single domain ligands, receptors comprising said ligands methods for their production, and use of said ligands and receptors |
| US6267958B1 (en) | 1995-07-27 | 2001-07-31 | Genentech, Inc. | Protein formulation |
| WO2002020565A2 (en) | 2000-09-08 | 2002-03-14 | Universität Zürich | Collections of repeat proteins comprising repeat modules |
| WO2002031140A1 (fr) | 2000-10-06 | 2002-04-18 | Kyowa Hakko Kogyo Co., Ltd. | Cellules produisant des compositions d'anticorps |
| WO2002032925A2 (en) | 2000-10-16 | 2002-04-25 | Phylos, Inc. | Protein scaffolds for antibody mimics and other binding proteins |
| US6420548B1 (en) | 1999-10-04 | 2002-07-16 | Medicago Inc. | Method for regulating transcription of foreign genes |
| WO2002079255A1 (en) | 2001-04-02 | 2002-10-10 | Idec Pharmaceuticals Corporation | RECOMBINANT ANTIBODIES COEXPRESSED WITH GnTIII |
| WO2003029462A1 (en) | 2001-09-27 | 2003-04-10 | Pieris Proteolab Ag | Muteins of human neutrophil gelatinase-associated lipocalin and related proteins |
| WO2003105757A2 (en) | 2002-06-12 | 2003-12-24 | Genencor International, Inc. | Methods and compositions for milieu-dependent binding of a targeted agent to a target |
| WO2004029207A2 (en) | 2002-09-27 | 2004-04-08 | Xencor Inc. | Optimized fc variants and methods for their generation |
| US6737056B1 (en) | 1999-01-15 | 2004-05-18 | Genentech, Inc. | Polypeptide variants with altered effector function |
| WO2004044011A2 (en) | 2002-11-06 | 2004-05-27 | Avidia Research Institute | Combinatorial libraries of monomer domains |
| WO2004056312A2 (en) | 2002-12-16 | 2004-07-08 | Genentech, Inc. | Immunoglobulin variants and uses thereof |
| WO2004099249A2 (en) | 2003-05-02 | 2004-11-18 | Xencor, Inc. | Optimized fc variants and methods for their generation |
| WO2005003175A2 (en) | 2003-06-13 | 2005-01-13 | Biogen Idec Ma Inc. | Aglycosyl anti-cd154 (cd40 ligand) antibodies and uses thereof |
| US20050014934A1 (en) | 2002-10-15 | 2005-01-20 | Hinton Paul R. | Alteration of FcRn binding affinities or serum half-lives of antibodies by mutagenesis |
| US20050079574A1 (en) | 2003-01-16 | 2005-04-14 | Genentech, Inc. | Synthetic antibody phage libraries |
| WO2005035756A1 (ja) | 2003-10-10 | 2005-04-21 | Chugai Seiyaku Kabushiki Kaisha | 機能蛋白質を代替する二種特異性抗体 |
| WO2005040229A2 (en) | 2003-10-24 | 2005-05-06 | Avidia, Inc. | Ldl receptor class a and egf domain monomers and multimers |
| US20050119455A1 (en) | 2002-06-03 | 2005-06-02 | Genentech, Inc. | Synthetic antibody phage libraries |
| WO2005063816A2 (en) | 2003-12-19 | 2005-07-14 | Genentech, Inc. | Monovalent antibody fragments useful as therapeutics |
| WO2005070963A1 (en) | 2004-01-12 | 2005-08-04 | Applied Molecular Evolution, Inc | Fc region variants |
| WO2005100402A1 (en) | 2004-04-13 | 2005-10-27 | F.Hoffmann-La Roche Ag | Anti-p-selectin antibodies |
| US20050260186A1 (en) | 2003-03-05 | 2005-11-24 | Halozyme, Inc. | Soluble glycosaminoglycanases and methods of preparing and using soluble glycosaminoglycanases |
| US20050266000A1 (en) | 2004-04-09 | 2005-12-01 | Genentech, Inc. | Variable domain library and uses |
| US6982321B2 (en) | 1986-03-27 | 2006-01-03 | Medical Research Council | Altered antibodies |
| US20060025576A1 (en) | 2000-04-11 | 2006-02-02 | Genentech, Inc. | Multivalent antibodies and uses therefor |
| WO2006020114A2 (en) | 2004-08-04 | 2006-02-23 | Applied Molecular Evolution, Inc. | Variant fc regions |
| WO2006019447A1 (en) | 2004-07-15 | 2006-02-23 | Xencor, Inc. | Optimized fc variants |
| WO2006023403A2 (en) | 2004-08-16 | 2006-03-02 | Medimmune, Inc. | Eph receptor fc variants with enhanced antibody dependent cell-mediated cytotoxicity activity |
| WO2006031370A2 (en) | 2004-08-19 | 2006-03-23 | Genentech, Inc. | Polypeptide variants with altered effector function |
| WO2006029879A2 (en) | 2004-09-17 | 2006-03-23 | F.Hoffmann-La Roche Ag | Anti-ox40l antibodies |
| WO2006044908A2 (en) | 2004-10-20 | 2006-04-27 | Genentech, Inc. | Antibody formulation in histidine-acetate buffer |
| US7041870B2 (en) | 2000-11-30 | 2006-05-09 | Medarex, Inc. | Transgenic transchromosomal rodents for making human antibodies |
| US20060104968A1 (en) | 2003-03-05 | 2006-05-18 | Halozyme, Inc. | Soluble glycosaminoglycanases and methods of preparing and using soluble glycosaminogly ycanases |
| WO2006067913A1 (ja) | 2004-12-22 | 2006-06-29 | Chugai Seiyaku Kabushiki Kaisha | フコーストランスポーターの機能が阻害された細胞を用いた抗体の作製方法 |
| US7087409B2 (en) | 1997-12-05 | 2006-08-08 | The Scripps Research Institute | Humanization of murine antibody |
| WO2006105338A2 (en) | 2005-03-31 | 2006-10-05 | Xencor, Inc. | Fc VARIANTS WITH OPTIMIZED PROPERTIES |
| US7125978B1 (en) | 1999-10-04 | 2006-10-24 | Medicago Inc. | Promoter for regulating expression of foreign genes |
| WO2006116260A2 (en) | 2005-04-26 | 2006-11-02 | Medimmune, Inc. | Modulation of antibody effector function by hinge domain engineering |
| WO2007021841A2 (en) | 2005-08-10 | 2007-02-22 | Macrogenics, Inc. | Identification and engineering of antibodies with variant fc regions and methods of using same |
| WO2007024249A2 (en) | 2004-11-10 | 2007-03-01 | Macrogenics, Inc. | Engineering fc antibody regions to confer effector function |
| US7189826B2 (en) | 1997-11-24 | 2007-03-13 | Institute For Human Genetics And Biochemistry | Monoclonal human natural antibodies |
| US20070061900A1 (en) | 2000-10-31 | 2007-03-15 | Murphy Andrew J | Methods of modifying eukaryotic cells |
| WO2007041635A2 (en) | 2005-10-03 | 2007-04-12 | Xencor, Inc. | Fc variants with optimized fc receptor binding properties |
| US20070117126A1 (en) | 1999-12-15 | 2007-05-24 | Genentech, Inc. | Shotgun scanning |
| US20070160598A1 (en) | 2005-11-07 | 2007-07-12 | Dennis Mark S | Binding polypeptides with diversified and consensus vh/vl hypervariable sequences |
| WO2007092772A2 (en) | 2006-02-03 | 2007-08-16 | Medimmune, Inc. | Protein formulations |
| US20070237764A1 (en) | 2005-12-02 | 2007-10-11 | Genentech, Inc. | Binding polypeptides with restricted diversity sequences |
| WO2007114319A1 (ja) | 2006-03-31 | 2007-10-11 | Chugai Seiyaku Kabushiki Kaisha | 抗体の血中動態を制御する方法 |
| US20070292936A1 (en) | 2006-05-09 | 2007-12-20 | Genentech, Inc. | Binding polypeptides with optimized scaffolds |
| WO2008016854A2 (en) | 2006-08-02 | 2008-02-07 | The Uab Research Foundation | Methods and compositions related to soluble monoclonal variable lymphocyte receptors of defined antigen specificity |
| US20080069820A1 (en) | 2006-08-30 | 2008-03-20 | Genentech, Inc. | Multispecific antibodies |
| WO2008092117A2 (en) | 2007-01-25 | 2008-07-31 | Xencor, Inc. | Immunoglobulins with modifications in the fcr binding region |
| WO2008119353A1 (en) | 2007-03-29 | 2008-10-09 | Genmab A/S | Bispecific antibodies and methods for production thereof |
| US20090002360A1 (en) | 2007-05-25 | 2009-01-01 | Innolux Display Corp. | Liquid crystal display device and method for driving same |
| WO2009011941A2 (en) | 2007-04-04 | 2009-01-22 | The Government Of U.S.A., As Represented By The Secretary, Departmetnt Of Health & Human Services | Monoclonal antibodies against dengue and other viruses with deletion in fc region |
| US20090035836A1 (en) | 2004-03-30 | 2009-02-05 | California Institute Of Technology | Modulating ph-sensitive binding using non-natural amino acids |
| WO2009041643A1 (ja) | 2007-09-26 | 2009-04-02 | Chugai Seiyaku Kabushiki Kaisha | Cdrのアミノ酸置換により抗体の等電点を改変する方法 |
| US7527791B2 (en) | 2004-03-31 | 2009-05-05 | Genentech, Inc. | Humanized anti-TGF-beta antibodies |
| WO2009086320A1 (en) | 2007-12-26 | 2009-07-09 | Xencor, Inc | Fc variants with altered binding to fcrn |
| WO2009089004A1 (en) | 2008-01-07 | 2009-07-16 | Amgen Inc. | Method for making antibody fc-heterodimeric molecules using electrostatic steering effects |
| WO2009125825A1 (ja) | 2008-04-11 | 2009-10-15 | 中外製薬株式会社 | 複数分子の抗原に繰り返し結合する抗原結合分子 |
| WO2011122011A2 (en) | 2010-03-30 | 2011-10-06 | Chugai Seiyaku Kabushiki Kaisha | Antibodies with modified affinity to fcrn that promote antigen clearance |
| WO2012016227A2 (en) | 2010-07-29 | 2012-02-02 | Xencor, Inc. | Antibodies with modified isoelectric points |
| WO2012067176A1 (ja) | 2010-11-17 | 2012-05-24 | 中外製薬株式会社 | 血液凝固第viii因子の機能を代替する機能を有する多重特異性抗原結合分子 |
| WO2012073992A1 (ja) | 2010-11-30 | 2012-06-07 | 中外製薬株式会社 | 複数分子の抗原に繰り返し結合する抗原結合分子 |
| WO2012093704A1 (ja) | 2011-01-07 | 2012-07-12 | 中外製薬株式会社 | 抗体の物性を改善させる方法 |
| WO2012115241A1 (ja) | 2011-02-25 | 2012-08-30 | 中外製薬株式会社 | FcγRIIb特異的Fc抗体 |
| WO2013046704A2 (en) | 2011-09-30 | 2013-04-04 | Chugai Seiyaku Kabushiki Kaisha | THERAPEUTIC ANTIGEN-BINDING MOLECULE WITH A FcRn-BINDING DOMAIN THAT PROMOTES ANTIGEN CLEARANCE |
| WO2013046722A1 (ja) | 2011-09-30 | 2013-04-04 | 中外製薬株式会社 | イオン濃度依存性結合分子ライブラリ |
| WO2013047752A1 (ja) | 2011-09-30 | 2013-04-04 | 中外製薬株式会社 | 抗原の消失を促進する抗原結合分子 |
| WO2013081143A1 (ja) | 2011-11-30 | 2013-06-06 | 中外製薬株式会社 | 免疫複合体を形成する細胞内への運搬体(キャリア)を含む医薬 |
| US20130171138A1 (en) | 2003-05-06 | 2013-07-04 | Syntonix Pharmaceuticals, Inc. | Immunoglobulin Chimeric Monomer-Dimer Hybrids |
| WO2013118858A1 (ja) | 2012-02-09 | 2013-08-15 | 中外製薬株式会社 | 抗体のFc領域改変体 |
| WO2013125667A1 (ja) | 2012-02-24 | 2013-08-29 | 中外製薬株式会社 | FcγRIIBを介して抗原の消失を促進する抗原結合分子 |
| WO2014030750A1 (ja) | 2012-08-24 | 2014-02-27 | 中外製薬株式会社 | マウスFcγRII特異的Fc抗体 |
| WO2014030728A1 (ja) | 2012-08-24 | 2014-02-27 | 中外製薬株式会社 | FcγRIIb特異的Fc領域改変体 |
| WO2014145159A2 (en) | 2013-03-15 | 2014-09-18 | Permeon Biologics, Inc. | Charge-engineered antibodies or compositions of penetration-enhanced targeting proteins and methods of use |
| WO2014163101A1 (ja) | 2013-04-02 | 2014-10-09 | 中外製薬株式会社 | Fc領域改変体 |
Family Cites Families (11)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| JP3865418B2 (ja) * | 1994-07-13 | 2007-01-10 | 中外製薬株式会社 | ヒトインターロイキン−8に対する再構成ヒト抗体 |
| TW416960B (en) * | 1994-07-13 | 2001-01-01 | Chugai Pharmaceutical Co Ltd | Reshaped human antibody to human interleukin-8 |
| CN1156460A (zh) * | 1994-07-13 | 1997-08-06 | 中外制药株式会社 | 抗人白细胞介素-8的重构人抗体 |
| US6025158A (en) * | 1997-02-21 | 2000-02-15 | Genentech, Inc. | Nucleic acids encoding humanized anti-IL-8 monoclonal antibodies |
| US6458355B1 (en) * | 1998-01-22 | 2002-10-01 | Genentech, Inc. | Methods of treating inflammatory disease with anti-IL-8 antibody fragment-polymer conjugates |
| US7771951B2 (en) * | 2001-12-03 | 2010-08-10 | Amgen Fremont Inc. | Antibody categorization based on binding characteristics |
| JP4468803B2 (ja) * | 2002-05-31 | 2010-05-26 | ジーイー・ヘルスケア・バイオ−サイエンシーズ・アーベー | 結合剤を基板表面にカップリングさせる方法 |
| EP2066349B1 (en) * | 2006-09-08 | 2012-03-28 | MedImmune, LLC | Humanized anti-cd19 antibodies and their use in treatment of tumors, transplantation and autoimmune diseases |
| US20110105724A1 (en) * | 2007-08-16 | 2011-05-05 | Stephanie Jane Clegg | Novel compounds |
| KR102605798B1 (ko) * | 2015-02-05 | 2023-11-23 | 추가이 세이야쿠 가부시키가이샤 | 이온 농도 의존적 항원 결합 도메인을 포함하는 항체, Fc 영역 개변체, IL-8에 결합하는 항체, 및 그들의 사용 |
| PE20181336A1 (es) * | 2015-09-18 | 2018-08-21 | Chugai Pharmaceutical Co Ltd | Anticuerpos que se unen a interleucina 8 (il-8) y sus usos |
-
2016
- 2016-08-05 PE PE2018000378A patent/PE20181336A1/es unknown
- 2016-08-05 IL IL258088A patent/IL258088B2/en unknown
- 2016-08-05 KR KR1020237017768A patent/KR20230079500A/ko not_active Application Discontinuation
- 2016-08-05 CA CA2993423A patent/CA2993423C/en active Active
- 2016-08-05 KR KR1020177037003A patent/KR101920175B1/ko active IP Right Grant
- 2016-08-05 EP EP16753718.2A patent/EP3350202A1/en active Pending
- 2016-08-05 JP JP2017508124A patent/JP6266164B2/ja active Active
- 2016-08-05 CN CN201680054135.3A patent/CN108271372B/zh active Active
- 2016-08-05 TW TW110145197A patent/TWI814162B/zh active
- 2016-08-05 RU RU2018113505A patent/RU2728430C2/ru active
- 2016-08-05 TW TW112123388A patent/TW202342532A/zh unknown
- 2016-08-05 AU AU2016323088A patent/AU2016323088B2/en active Active
- 2016-08-05 WO PCT/JP2016/003616 patent/WO2017046994A1/en active Application Filing
- 2016-08-05 AR ARP160102414A patent/AR105634A1/es active IP Right Grant
- 2016-08-05 TW TW107108073A patent/TWI751300B/zh active
- 2016-08-05 CN CN202110827885.2A patent/CN113372443A/zh active Pending
- 2016-08-05 MA MA042808A patent/MA42808A/fr unknown
- 2016-08-05 CR CR20180217A patent/CR20180217A/es unknown
- 2016-08-05 KR KR1020187032734A patent/KR102538745B1/ko not_active Application Discontinuation
- 2016-08-05 TW TW105124915A patent/TWI621628B/zh active
- 2016-08-05 UA UAA201804018A patent/UA120981C2/uk unknown
- 2016-08-05 MX MX2018003005A patent/MX2018003005A/es unknown
- 2016-08-05 BR BR112018002432A patent/BR112018002432A2/pt active Search and Examination
-
2017
- 2017-12-19 JP JP2017242615A patent/JP6903561B2/ja active Active
-
2018
- 2018-01-25 ZA ZA2018/00536A patent/ZA201800536B/en unknown
- 2018-02-21 PH PH12018500386A patent/PH12018500386A1/en unknown
- 2018-03-09 MX MX2022008781A patent/MX2022008781A/es unknown
- 2018-03-16 CL CL2018000699A patent/CL2018000699A1/es unknown
- 2018-04-16 CO CONC2018/0004056A patent/CO2018004056A2/es unknown
- 2018-09-17 HK HK18111915.9A patent/HK1252615A1/zh unknown
-
2021
- 2021-06-23 JP JP2021103813A patent/JP2021168658A/ja active Pending
- 2021-08-04 IL IL285375A patent/IL285375A/en unknown
-
2023
- 2023-09-25 JP JP2023159360A patent/JP2023164732A/ja active Pending
Patent Citations (124)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US4816567A (en) | 1983-04-08 | 1989-03-28 | Genentech, Inc. | Recombinant immunoglobin preparations |
| US4676980A (en) | 1985-09-23 | 1987-06-30 | The United States Of America As Represented By The Secretary Of The Department Of Health And Human Services | Target specific cross-linked heteroantibodies |
| EP0239400A2 (en) | 1986-03-27 | 1987-09-30 | Medical Research Council | Recombinant antibodies and methods for their production |
| US6982321B2 (en) | 1986-03-27 | 2006-01-03 | Medical Research Council | Altered antibodies |
| US5500362A (en) | 1987-01-08 | 1996-03-19 | Xoma Corporation | Chimeric antibody with specificity to human B cell surface antigen |
| US5624821A (en) | 1987-03-18 | 1997-04-29 | Scotgen Biopharmaceuticals Incorporated | Antibodies with altered effector functions |
| US5648260A (en) | 1987-03-18 | 1997-07-15 | Scotgen Biopharmaceuticals Incorporated | DNA encoding antibodies with altered effector functions |
| US6248516B1 (en) | 1988-11-11 | 2001-06-19 | Medical Research Council | Single domain ligands, receptors comprising said ligands methods for their production, and use of said ligands and receptors |
| EP0404097A2 (de) | 1989-06-22 | 1990-12-27 | BEHRINGWERKE Aktiengesellschaft | Bispezifische und oligospezifische, mono- und oligovalente Rezeptoren, ihre Herstellung und Verwendung |
| US6417429B1 (en) | 1989-10-27 | 2002-07-09 | The Scripps Research Institute | Transgenic plants expressing assembled secretory antibodies |
| US5959177A (en) | 1989-10-27 | 1999-09-28 | The Scripps Research Institute | Transgenic plants expressing assembled secretory antibodies |
| US6150584A (en) | 1990-01-12 | 2000-11-21 | Abgenix, Inc. | Human antibodies derived from immunized xenomice |
| US6075181A (en) | 1990-01-12 | 2000-06-13 | Abgenix, Inc. | Human antibodies derived from immunized xenomice |
| US5770429A (en) | 1990-08-29 | 1998-06-23 | Genpharm International, Inc. | Transgenic non-human animals capable of producing heterologous antibodies |
| WO1992003918A1 (en) | 1990-08-29 | 1992-03-19 | Genpharm International, Inc. | Transgenic non-human animals capable of producing heterologous antibodies |
| US5750373A (en) | 1990-12-03 | 1998-05-12 | Genentech, Inc. | Enrichment method for variant proteins having altered binding properties, M13 phagemids, and growth hormone variants |
| US5571894A (en) | 1991-02-05 | 1996-11-05 | Ciba-Geigy Corporation | Recombinant antibodies specific for a growth factor receptor |
| US5821337A (en) | 1991-06-14 | 1998-10-13 | Genentech, Inc. | Immunoglobulin variants |
| WO1993001161A1 (en) | 1991-07-11 | 1993-01-21 | Pfizer Limited | Process for preparing sertraline intermediates |
| US5648237A (en) | 1991-09-19 | 1997-07-15 | Genentech, Inc. | Expression of functional antibody fragments |
| US5587458A (en) | 1991-10-07 | 1996-12-24 | Aronex Pharmaceuticals, Inc. | Anti-erbB-2 antibodies, combinations thereof, and therapeutic and diagnostic uses thereof |
| WO1993008829A1 (en) | 1991-11-04 | 1993-05-13 | The Regents Of The University Of California | Compositions that mediate killing of hiv-infected cells |
| WO1993011161A1 (en) | 1991-11-25 | 1993-06-10 | Enzon, Inc. | Multivalent antigen-binding proteins |
| WO1993012227A1 (en) | 1991-12-17 | 1993-06-24 | Genpharm International, Inc. | Transgenic non-human animals capable of producing heterologous antibodies |
| WO1993016185A2 (en) | 1992-02-06 | 1993-08-19 | Creative Biomolecules, Inc. | Biosynthetic binding protein for cancer marker |
| WO1994002602A1 (en) | 1992-07-24 | 1994-02-03 | Cell Genesys, Inc. | Generation of xenogeneic antibodies |
| WO1994029351A2 (en) | 1993-06-16 | 1994-12-22 | Celltech Limited | Antibodies |
| WO1995001937A1 (fr) | 1993-07-09 | 1995-01-19 | Association Gradient | Procede de traitement de residus de combustion et installation de mise en ×uvre dudit procede |
| US6024956A (en) * | 1994-07-13 | 2000-02-15 | Chugai Pharmaceutical Co., Ltd. | Method to treat inflammation with humanized anti-IL-8-antibodies |
| US6245894B1 (en) | 1994-07-13 | 2001-06-12 | Chugai Seiyaku Kabushiki Kaisha | Reshaped human antibody to human interleukin-8 |
| WO1996002576A1 (fr) | 1994-07-13 | 1996-02-01 | Chugai Seiyaku Kabushiki Kaisha | Anticorps humain reconstitue contre l'interleukine-8 humaine |
| US5789199A (en) | 1994-11-03 | 1998-08-04 | Genentech, Inc. | Process for bacterial production of polypeptides |
| US5731168A (en) | 1995-03-01 | 1998-03-24 | Genentech, Inc. | Method for making heteromultimeric polypeptides |
| US5840523A (en) | 1995-03-01 | 1998-11-24 | Genetech, Inc. | Methods and compositions for secretion of heterologous polypeptides |
| WO1996033735A1 (en) | 1995-04-27 | 1996-10-31 | Abgenix, Inc. | Human antibodies derived from immunized xenomice |
| WO1996034096A1 (en) | 1995-04-28 | 1996-10-31 | Abgenix, Inc. | Human antibodies derived from immunized xenomice |
| US6267958B1 (en) | 1995-07-27 | 2001-07-31 | Genentech, Inc. | Protein formulation |
| WO1998013388A1 (fr) | 1996-09-26 | 1998-04-02 | Chugai Seiyaku Kabushiki Kaisha | Anticorps contre les peptides lies a la parathormone humaine |
| WO1998046777A1 (fr) | 1997-04-11 | 1998-10-22 | Centre National De La Recherche Scientifique (Cnrs) | Preparation de recepteurs membranaires a partir de baculovirus extracellulaires |
| US6171586B1 (en) | 1997-06-13 | 2001-01-09 | Genentech, Inc. | Antibody formulation |
| US7189826B2 (en) | 1997-11-24 | 2007-03-13 | Institute For Human Genetics And Biochemistry | Monoclonal human natural antibodies |
| US7087409B2 (en) | 1997-12-05 | 2006-08-08 | The Scripps Research Institute | Humanization of murine antibody |
| WO1999054342A1 (en) | 1998-04-20 | 1999-10-28 | Pablo Umana | Glycosylation engineering of antibodies for improving antibody-dependent cellular cytotoxicity |
| US6040498A (en) | 1998-08-11 | 2000-03-21 | North Caroline State University | Genetically engineered duckweed |
| WO2000042072A2 (en) | 1999-01-15 | 2000-07-20 | Genentech, Inc. | Polypeptide variants with altered effector function |
| US7332581B2 (en) | 1999-01-15 | 2008-02-19 | Genentech, Inc. | Polypeptide variants with altered effector function |
| US6737056B1 (en) | 1999-01-15 | 2004-05-18 | Genentech, Inc. | Polypeptide variants with altered effector function |
| US7371826B2 (en) | 1999-01-15 | 2008-05-13 | Genentech, Inc. | Polypeptide variants with altered effector function |
| WO2000061739A1 (en) | 1999-04-09 | 2000-10-19 | Kyowa Hakko Kogyo Co., Ltd. | Method for controlling the activity of immunologically functional molecule |
| US6420548B1 (en) | 1999-10-04 | 2002-07-16 | Medicago Inc. | Method for regulating transcription of foreign genes |
| US7125978B1 (en) | 1999-10-04 | 2006-10-24 | Medicago Inc. | Promoter for regulating expression of foreign genes |
| US20070117126A1 (en) | 1999-12-15 | 2007-05-24 | Genentech, Inc. | Shotgun scanning |
| US20060025576A1 (en) | 2000-04-11 | 2006-02-02 | Genentech, Inc. | Multivalent antibodies and uses therefor |
| WO2002020565A2 (en) | 2000-09-08 | 2002-03-14 | Universität Zürich | Collections of repeat proteins comprising repeat modules |
| WO2002031140A1 (fr) | 2000-10-06 | 2002-04-18 | Kyowa Hakko Kogyo Co., Ltd. | Cellules produisant des compositions d'anticorps |
| WO2002032925A2 (en) | 2000-10-16 | 2002-04-25 | Phylos, Inc. | Protein scaffolds for antibody mimics and other binding proteins |
| US20070061900A1 (en) | 2000-10-31 | 2007-03-15 | Murphy Andrew J | Methods of modifying eukaryotic cells |
| US7041870B2 (en) | 2000-11-30 | 2006-05-09 | Medarex, Inc. | Transgenic transchromosomal rodents for making human antibodies |
| WO2002079255A1 (en) | 2001-04-02 | 2002-10-10 | Idec Pharmaceuticals Corporation | RECOMBINANT ANTIBODIES COEXPRESSED WITH GnTIII |
| WO2003029462A1 (en) | 2001-09-27 | 2003-04-10 | Pieris Proteolab Ag | Muteins of human neutrophil gelatinase-associated lipocalin and related proteins |
| US20050119455A1 (en) | 2002-06-03 | 2005-06-02 | Genentech, Inc. | Synthetic antibody phage libraries |
| WO2003105757A2 (en) | 2002-06-12 | 2003-12-24 | Genencor International, Inc. | Methods and compositions for milieu-dependent binding of a targeted agent to a target |
| WO2004029207A2 (en) | 2002-09-27 | 2004-04-08 | Xencor Inc. | Optimized fc variants and methods for their generation |
| US20050014934A1 (en) | 2002-10-15 | 2005-01-20 | Hinton Paul R. | Alteration of FcRn binding affinities or serum half-lives of antibodies by mutagenesis |
| WO2004044011A2 (en) | 2002-11-06 | 2004-05-27 | Avidia Research Institute | Combinatorial libraries of monomer domains |
| WO2004056312A2 (en) | 2002-12-16 | 2004-07-08 | Genentech, Inc. | Immunoglobulin variants and uses thereof |
| US20050079574A1 (en) | 2003-01-16 | 2005-04-14 | Genentech, Inc. | Synthetic antibody phage libraries |
| US20050260186A1 (en) | 2003-03-05 | 2005-11-24 | Halozyme, Inc. | Soluble glycosaminoglycanases and methods of preparing and using soluble glycosaminoglycanases |
| US20060104968A1 (en) | 2003-03-05 | 2006-05-18 | Halozyme, Inc. | Soluble glycosaminoglycanases and methods of preparing and using soluble glycosaminogly ycanases |
| WO2004099249A2 (en) | 2003-05-02 | 2004-11-18 | Xencor, Inc. | Optimized fc variants and methods for their generation |
| US20130171138A1 (en) | 2003-05-06 | 2013-07-04 | Syntonix Pharmaceuticals, Inc. | Immunoglobulin Chimeric Monomer-Dimer Hybrids |
| WO2005003175A2 (en) | 2003-06-13 | 2005-01-13 | Biogen Idec Ma Inc. | Aglycosyl anti-cd154 (cd40 ligand) antibodies and uses thereof |
| WO2005035756A1 (ja) | 2003-10-10 | 2005-04-21 | Chugai Seiyaku Kabushiki Kaisha | 機能蛋白質を代替する二種特異性抗体 |
| WO2005040229A2 (en) | 2003-10-24 | 2005-05-06 | Avidia, Inc. | Ldl receptor class a and egf domain monomers and multimers |
| WO2005063816A2 (en) | 2003-12-19 | 2005-07-14 | Genentech, Inc. | Monovalent antibody fragments useful as therapeutics |
| WO2005070963A1 (en) | 2004-01-12 | 2005-08-04 | Applied Molecular Evolution, Inc | Fc region variants |
| US20090035836A1 (en) | 2004-03-30 | 2009-02-05 | California Institute Of Technology | Modulating ph-sensitive binding using non-natural amino acids |
| US7527791B2 (en) | 2004-03-31 | 2009-05-05 | Genentech, Inc. | Humanized anti-TGF-beta antibodies |
| US20050266000A1 (en) | 2004-04-09 | 2005-12-01 | Genentech, Inc. | Variable domain library and uses |
| WO2005100402A1 (en) | 2004-04-13 | 2005-10-27 | F.Hoffmann-La Roche Ag | Anti-p-selectin antibodies |
| WO2006019447A1 (en) | 2004-07-15 | 2006-02-23 | Xencor, Inc. | Optimized fc variants |
| WO2006020114A2 (en) | 2004-08-04 | 2006-02-23 | Applied Molecular Evolution, Inc. | Variant fc regions |
| WO2006023403A2 (en) | 2004-08-16 | 2006-03-02 | Medimmune, Inc. | Eph receptor fc variants with enhanced antibody dependent cell-mediated cytotoxicity activity |
| WO2006031370A2 (en) | 2004-08-19 | 2006-03-23 | Genentech, Inc. | Polypeptide variants with altered effector function |
| WO2006029879A2 (en) | 2004-09-17 | 2006-03-23 | F.Hoffmann-La Roche Ag | Anti-ox40l antibodies |
| WO2006044908A2 (en) | 2004-10-20 | 2006-04-27 | Genentech, Inc. | Antibody formulation in histidine-acetate buffer |
| WO2007024249A2 (en) | 2004-11-10 | 2007-03-01 | Macrogenics, Inc. | Engineering fc antibody regions to confer effector function |
| WO2006067913A1 (ja) | 2004-12-22 | 2006-06-29 | Chugai Seiyaku Kabushiki Kaisha | フコーストランスポーターの機能が阻害された細胞を用いた抗体の作製方法 |
| WO2006067847A1 (ja) | 2004-12-22 | 2006-06-29 | Chugai Seiyaku Kabushiki Kaisha | フコーストランスポーターの機能が阻害された細胞を用いた抗体の作製方法 |
| WO2006105338A2 (en) | 2005-03-31 | 2006-10-05 | Xencor, Inc. | Fc VARIANTS WITH OPTIMIZED PROPERTIES |
| WO2006116260A2 (en) | 2005-04-26 | 2006-11-02 | Medimmune, Inc. | Modulation of antibody effector function by hinge domain engineering |
| WO2007021841A2 (en) | 2005-08-10 | 2007-02-22 | Macrogenics, Inc. | Identification and engineering of antibodies with variant fc regions and methods of using same |
| WO2007041635A2 (en) | 2005-10-03 | 2007-04-12 | Xencor, Inc. | Fc variants with optimized fc receptor binding properties |
| US20070160598A1 (en) | 2005-11-07 | 2007-07-12 | Dennis Mark S | Binding polypeptides with diversified and consensus vh/vl hypervariable sequences |
| US20070237764A1 (en) | 2005-12-02 | 2007-10-11 | Genentech, Inc. | Binding polypeptides with restricted diversity sequences |
| WO2007092772A2 (en) | 2006-02-03 | 2007-08-16 | Medimmune, Inc. | Protein formulations |
| WO2007114319A1 (ja) | 2006-03-31 | 2007-10-11 | Chugai Seiyaku Kabushiki Kaisha | 抗体の血中動態を制御する方法 |
| US20070292936A1 (en) | 2006-05-09 | 2007-12-20 | Genentech, Inc. | Binding polypeptides with optimized scaffolds |
| WO2008016854A2 (en) | 2006-08-02 | 2008-02-07 | The Uab Research Foundation | Methods and compositions related to soluble monoclonal variable lymphocyte receptors of defined antigen specificity |
| US20080069820A1 (en) | 2006-08-30 | 2008-03-20 | Genentech, Inc. | Multispecific antibodies |
| WO2008092117A2 (en) | 2007-01-25 | 2008-07-31 | Xencor, Inc. | Immunoglobulins with modifications in the fcr binding region |
| WO2008119353A1 (en) | 2007-03-29 | 2008-10-09 | Genmab A/S | Bispecific antibodies and methods for production thereof |
| WO2009011941A2 (en) | 2007-04-04 | 2009-01-22 | The Government Of U.S.A., As Represented By The Secretary, Departmetnt Of Health & Human Services | Monoclonal antibodies against dengue and other viruses with deletion in fc region |
| US20090002360A1 (en) | 2007-05-25 | 2009-01-01 | Innolux Display Corp. | Liquid crystal display device and method for driving same |
| WO2009041643A1 (ja) | 2007-09-26 | 2009-04-02 | Chugai Seiyaku Kabushiki Kaisha | Cdrのアミノ酸置換により抗体の等電点を改変する方法 |
| WO2009086320A1 (en) | 2007-12-26 | 2009-07-09 | Xencor, Inc | Fc variants with altered binding to fcrn |
| WO2009089004A1 (en) | 2008-01-07 | 2009-07-16 | Amgen Inc. | Method for making antibody fc-heterodimeric molecules using electrostatic steering effects |
| WO2009125825A1 (ja) | 2008-04-11 | 2009-10-15 | 中外製薬株式会社 | 複数分子の抗原に繰り返し結合する抗原結合分子 |
| WO2011122011A2 (en) | 2010-03-30 | 2011-10-06 | Chugai Seiyaku Kabushiki Kaisha | Antibodies with modified affinity to fcrn that promote antigen clearance |
| WO2012016227A2 (en) | 2010-07-29 | 2012-02-02 | Xencor, Inc. | Antibodies with modified isoelectric points |
| WO2012067176A1 (ja) | 2010-11-17 | 2012-05-24 | 中外製薬株式会社 | 血液凝固第viii因子の機能を代替する機能を有する多重特異性抗原結合分子 |
| WO2012073992A1 (ja) | 2010-11-30 | 2012-06-07 | 中外製薬株式会社 | 複数分子の抗原に繰り返し結合する抗原結合分子 |
| WO2012093704A1 (ja) | 2011-01-07 | 2012-07-12 | 中外製薬株式会社 | 抗体の物性を改善させる方法 |
| WO2012115241A1 (ja) | 2011-02-25 | 2012-08-30 | 中外製薬株式会社 | FcγRIIb特異的Fc抗体 |
| WO2013046704A2 (en) | 2011-09-30 | 2013-04-04 | Chugai Seiyaku Kabushiki Kaisha | THERAPEUTIC ANTIGEN-BINDING MOLECULE WITH A FcRn-BINDING DOMAIN THAT PROMOTES ANTIGEN CLEARANCE |
| WO2013047752A1 (ja) | 2011-09-30 | 2013-04-04 | 中外製薬株式会社 | 抗原の消失を促進する抗原結合分子 |
| WO2013046722A1 (ja) | 2011-09-30 | 2013-04-04 | 中外製薬株式会社 | イオン濃度依存性結合分子ライブラリ |
| WO2013081143A1 (ja) | 2011-11-30 | 2013-06-06 | 中外製薬株式会社 | 免疫複合体を形成する細胞内への運搬体(キャリア)を含む医薬 |
| WO2013118858A1 (ja) | 2012-02-09 | 2013-08-15 | 中外製薬株式会社 | 抗体のFc領域改変体 |
| WO2013125667A1 (ja) | 2012-02-24 | 2013-08-29 | 中外製薬株式会社 | FcγRIIBを介して抗原の消失を促進する抗原結合分子 |
| WO2014030750A1 (ja) | 2012-08-24 | 2014-02-27 | 中外製薬株式会社 | マウスFcγRII特異的Fc抗体 |
| WO2014030728A1 (ja) | 2012-08-24 | 2014-02-27 | 中外製薬株式会社 | FcγRIIb特異的Fc領域改変体 |
| WO2014145159A2 (en) | 2013-03-15 | 2014-09-18 | Permeon Biologics, Inc. | Charge-engineered antibodies or compositions of penetration-enhanced targeting proteins and methods of use |
| WO2014163101A1 (ja) | 2013-04-02 | 2014-10-09 | 中外製薬株式会社 | Fc領域改変体 |
Non-Patent Citations (193)
| Title |
|---|
| "Current protocols in Molecular Biology", 1987, JOHN WILEY & SONS |
| "Epitope Mapping Protocols in Methods in Molecular Biology", vol. 66, 1996 |
| "Remington's Pharmaceutical Sciences", 1980 |
| "Sequences of Proteins of Immunological Interest", 1987, NATIONAL INSTITUTE OF HEALTH |
| "Sequences of proteins of immunological interest", NIH PUBLICATION NO.91-3242 |
| ADAMS ET AL., CANCER IMMUNOL. IMMUNOTHER., vol. 55, no. 6, 2006, pages 717 - 727 |
| ALMAGRO ET AL., FRONT. BIOSC, vol. 13, 2008, pages 1619 - 1633 |
| BACA ET AL., J. BIOL. CHEM., vol. 272, 1997, pages 10678 - 10684 |
| BAIROCH ET AL., FEBS LETT., vol. 269, 1990, pages 454 - 456 |
| BEHAR ET AL., PROTEIN ENG. DES. SEL., vol. 21, no. 1, 2008, pages 1 - 10 |
| BOERNER ET AL., J. IMMUNOL., vol. 147, 1991, pages 86 |
| BONETTO ET AL., FASEB J., vol. 23, no. 2, 2009, pages 575 - 585 |
| BRENNAN ET AL., SCIENCE, vol. 229, 1985, pages 81 |
| BRENSING-KUPPERS ET AL., GENE, vol. 191, 1997, pages 173 - 181 |
| BRODEUR ET AL.: "Monoclonal Antibody Production Techniques and Applications", 1987, MARCEL DEKKER, INC., pages: 51 - 63 |
| BROOKS ET AL., J. EXP. MED, vol. 170, 1989, pages 1369 - 1385 |
| BRUGGEMANN ET AL., J. EXP. MED., vol. 166, 1987, pages 1351 - 1361 |
| CARDOSO ET AL., SCAND. J. IMMUNOL., vol. 51, 2000, pages 337 - 344 |
| CARTER ET AL., PROC. NATL. ACAD. SCI. USA, vol. 89, 1992, pages 4285 |
| CHARLTON: "Methods in Molecular Biology", vol. 248, 2003, HUMANA PRESS, pages: 245 - 254 |
| CHAUVAUX ET AL., BIOCHEM. J., vol. 265, 1990, pages 261 - 265 |
| CHOTHIA; LESK, J. MOL. BIOL., vol. 196, 1987, pages 901 - 917 |
| CLACKSON ET AL., NATURE, vol. 352, 1991, pages 624 - 628 |
| CLARKSON ET AL., NATURE, vol. 352, 1991, pages 624 - 628 |
| CLIN. IMMUNOL., vol. 131, no. 2, 2009, pages 189 - 201 |
| CLYNES ET AL., NAT. MED., vol. 6, 2000, pages 443 - 446 |
| CLYNES ET AL., PROC. NATL. ACAD. SCI. U SA, vol. 95, 1998, pages 652 - 656 |
| CLYNES ET AL., PROC. NATL. ACAD. SCI. USA, vol. 95, 1998, pages 652 - 656 |
| CONNOLLY, J. APPL. CRYST., vol. 16, 1983, pages 548 - 558 |
| COX ET AL., NAT. BIOTECHNOL., vol. 24, no. 12, 2006, pages 1591 - 1597 |
| COX ET AL., NAT. GENETICS, vol. 7, 1994, pages 162 - 168 |
| CRAGG ET AL., BLOOD, vol. 101, 2003, pages 1045 - 1052 |
| CRAGG ET AL., BLOOD, vol. 103, 2004, pages 2738 - 2743 |
| DALL'ACQUA ET AL., J. BIOL. CHEM., vol. 281, 2006, pages 23514 - 23524 |
| DALL'ACQUA ET AL., J. BIOL. CHEM., vol. 281, 2006, pages 23514 - 235249 |
| DALL'ACQUA ET AL., METHODS, vol. 36, 2005, pages 43 - 60 |
| DAVIS ET AL., J. RHEUM., vol. 34, no. 11, 2007, pages 2204 - 2210 |
| DAVIS ET AL., PROTEIN ENG. DES. SEL., vol. 23, no. 4, 2010, pages 195 - 202 |
| DAVIS, NEW BIOL., vol. 2, 1990, pages 410 - 419 |
| DE GROOT ET AL., CLIN. IMMUNOL., vol. 131, no. 2, 2009, pages 189 - 201 |
| DE HEARD ET AL., METH. MOL. BIOL., vol. 178, 2002, pages 87 - 100 |
| DESAI ET AL., J. IMMUNOL., vol. 178, no. 10, 2007, pages 6217 - 6226 |
| D'MELLO ET AL., J. IMMUNOL. METHODS, vol. 247, no. 1-2, 2001, pages 191 - 203 |
| DUNCAN ET AL., NATURE, vol. 322, 1988, pages 738 - 40 |
| EBERT ET AL., BIO/TECHNOLOGY, vol. 12, 1994, pages 699 - 702 |
| ECONOMOU ET AL., EMBO J., vol. 9, 1990, pages 349 - 354 |
| FELLOUSE, PROC. NATL. ACAD. SCI. USA, vol. 101, no. 34, 2004, pages 12467 - 12472 |
| FLATMAN ET AL., J. CHROMATOGR. B, vol. 848, 2007, pages 79 - 87 |
| FORSTER ET AL., PROC. NATL. ACAD. SCI. USA, vol. 100, no. 11, 2003, pages 6353 - 6357 |
| G. KOHLER; C. MILSTEIN, METHODS ENZYMOL., vol. 73, 1981, pages 3 - 46 |
| GAZZANO-SANTORO ET AL., J. IMMUNOL. METH., vol. 202, 1996, pages 163 |
| GEJIMA ET AL., HUMAN ANTIBODIES, vol. 11, 2002, pages 121 - 129 |
| GERNGROSS, NAT. BIOTECH., vol. 22, 2004, pages 1409 - 1414 |
| GHETIE ET AL., IMMUNOL. TODAY, vol. 18, no. 12, 1997, pages 592 - 598 |
| GHETIE ET AL., NAT. BIOTECHNOL., vol. 15, no. 7, 1997, pages 637 - 640 |
| GODING: "Monoclonal Antibodies: Principles and Practice", 1986, ACADEMIC PRESS, pages: 59 - 103 |
| GRAHAM ET AL., J. GEN VIROL., vol. 36, 1977, pages 59 |
| GRIFFITHS ET AL., EMBO J., vol. 12, 1993, pages 725 - 734 |
| GRUBER ET AL., J. IMMUNOL., vol. 152, 1994, pages 5368 |
| HAYES ET AL., J. BIOL. CHEM., vol. 250, no. 18, 1975, pages 7461 - 7472 |
| HE ET AL., J. IMMUNOL., vol. 160, no. 2, 1998, pages 1029 - 1035 |
| HELLSTROM ET AL., PROC. NATL ACAD. SCI. USA, vol. 82, 1985, pages 1499 - 1502 |
| HELLSTROM. ET AL., PROC. NATL ACAD. SCI. USA, vol. 83, 1986, pages 7059 - 7063 |
| HEYL ET AL., BIOORG. MED. CHEM., vol. 11, no. 17, 2003, pages 3761 - 3768 |
| HINTON ET AL., J. IMMUNOL., vol. 176, no. 1, 2006, pages 346 - 356 |
| HOLASH ET AL., PROC. NATL. ACAD. SCI., vol. 99, no. 17, 2002, pages 11393 - 11398 |
| HOLLIGER ET AL., PROC. NATL. ACAD. SCI. USA, vol. 90, 1993, pages 6444 - 6448 |
| HOLLINGER ET AL., PROC. NATL. ACAD. SCI. USA, vol. 90, 1993, pages 6444 - 6448 |
| HOOGENBOOM ET AL.: "Meth.Mol. Biol.", vol. 178, 2001, HUMAN PRESS, pages: 1 - 37 |
| HOOGENBOOM; WINTER, J. MOL. BIOL., vol. 227, 1992, pages 381 - 388 |
| HOPPE, SEYLER BIOL. CHEM., vol. 374, 1993, pages 1001 - 1022 |
| HUDSON ET AL., NAT. MED., vol. 9, 2003, pages 129 - 134 |
| IGAWA ET AL., NAT. BIOTECHNOL., vol. 28, 2010, pages 1203 - 1207 |
| ITO ET AL., FEBS LETT., vol. 309, no. 1, 1992, pages 85 - 88 |
| J. EXP. MED., vol. 172, 1990, pages 19 - 25 |
| J. MOL. MODEL., vol. 1, 1995, pages 46 - 53 |
| JEFFERIS ET AL., IMMUNOL. LETT., vol. 82, 2002, pages 57 - 65 |
| JENSEN ET AL., MOL. CELL PROTEOMICS, vol. 2, no. 2, 2003, pages 61 - 69 |
| JUNUTULA ET AL., J. IMMUNOL. METHODS, vol. 332, no. 1-2, 2008, pages 41 - 52 |
| KABAT ET AL.: "Sequences of Proteins of Immunological Interest", 1991, NATIONAL INSTITUTES OF HEALTH |
| KABAT: "Sequences of Proteins of Immunological Interest", 1987, NATIONAL INSTITUTE OF HEALTH |
| KAM ET AL., PROC. NATL. ACAD. SCI. USA, vol. 102, 2005, pages 11600 - 11605 |
| KASHMIRI ET AL., METHODS, vol. 36, 2005, pages 25 - 34 |
| KAWASAKI ET AL., EUR. J. IMMUNOL., vol. 31, 2001, pages 1017 - 1028 |
| KAWASAKI ET AL., GENOME RES., vol. 7, 1997, pages 250 - 261 |
| KAWASAKI ET AL., PROTEIN PROF., vol. 2, 1995, pages 305 - 490 |
| KIM ET AL., MOL. CELLS, vol. 20, no. 1, 2005, pages 17 - 29 |
| KINDT ET AL.: "Kuby Immunology", 2007, W.H. FREEMAN AND CO., pages: 91 |
| KIPRIYANOV ET AL., J IMMUNOL., vol. 169, no. 1, 2002, pages 137 - 144 |
| KLIMK ET AL., BR. J. CANCER, vol. 83, 2000, pages 252 - 260 |
| KONO ET AL., HUM. MOL. GENET., vol. 14, 2005, pages 2881 - 2892 |
| KOSTELNY ET AL., J. IMMUNOL., vol. 148, no. 5, 1992, pages 1547 - 1553 |
| KOZBOR: "J. Immunol.", vol. 133, 1984, pages: 3001 |
| KUNKEL ET AL., METHODS ENZYMOL., vol. 154, 1987, pages 367 - 382 |
| KUNKEL ET AL., PROC. NATL. ACAD. SCI. USA, vol. 82, 1985, pages 488 - 492 |
| KYOGOKU ET AL., ARTHRITIS RHEUM., vol. 46, no. 5, 2002, pages 1242 - 1254 |
| LAZAR ET AL., PROC. NAT. ACAD. SCI. USA., vol. 103, 2006, pages 4005 - 4010 |
| LEE ET AL., J. IMMUNOL. METH., vol. 284, no. 1-2, 2004, pages 119 - 132 |
| LEE ET AL., J. MOL. BIOL., vol. 340, no. 5, 2004, pages 1073 - 1093 |
| LEE; RICHARDS, J. MOL. BIOL., vol. 55, 1971, pages 379 - 400 |
| LI ET AL., ARTHRITIS RHEUM., vol. 48, 2003, pages 3242 - 3252 |
| LI ET AL., NAT. BIOTECH., vol. 24, 2006, pages 210 - 215 |
| LI ET AL., PROC. NAT. ACAD. SCI. USA., vol. 109, no. 27, 2012, pages 10966 - 10971 |
| LI ET AL., PROC. NATL. ACAD. SCI. USA, vol. 103, 2006, pages 3557 - 3562 |
| LONBERG, CURR. OPIN. IMMUNOL., vol. 20, 2008, pages 450 - 459 |
| LONBERG, NAT. BIOTECH., vol. 23, 2005, pages 1117 - 1125 |
| MA ET AL., EUR. J. IMMUNOL., vol. 24, 1994, pages 131 - 138 |
| MACCALLUM ET AL., J. MOL. BIOL., vol. 262, 1996, pages 732 - 745 |
| MARKS ET AL., J. MOL. BIOL., vol. 222, 1992, pages 581 - 597 |
| MARKS; BRADBURY: "Meth.Mol. Biol.", vol. 248, 2003, HUMAN PRESS, pages: 161 - 175 |
| MARTIN ET AL., MOL. CELL, vol. 7, no. 4, 2001, pages 867 - 877 |
| MARVIN ET AL., CURR. OPIN. DRUG DISCOV. DEVEL., vol. 9, no. 2, 2006, pages 184 - 193 |
| MATHER ET AL., ANNALS N.Y. ACAD. SCI., vol. 383, 1982, pages 44 - 68 |
| MATHER, BIOL. REPROD., vol. 23, 1980, pages 243 - 251 |
| MATSUDA ET AL., J. EXP. MED., vol. 188, 1998, pages 1973 - 1975 |
| MCCAFFERTY ET AL.: "Nature", vol. 348, 1990, pages: 552 - 554 |
| MENDEZ ET AL., NAT. GENET., vol. 15, 1997, pages 146 - 156 |
| METHODS MOL. BIOL., vol. 602, 2010, pages 93 - 104 |
| METHODS MOL. BIOL., vol. 882, 2012, pages 635 - 680 |
| METHODS, vol. 36, no. 1, 2005, pages 43 - 60 |
| MILSTEIN; CUELLO, NATURE, vol. 305, 1983, pages 537 |
| MONCRIEF ET AL., J. MOL. EVOL., vol. 30, 1990, pages 522 - 562 |
| MORRISON ET AL., PROC. NATL. ACAD. SCI. USA, vol. 81, 1984, pages 6851 - 6855 |
| MOXNESS ET AL., CLIN CHEM., vol. 51, 2005, pages 1983 - 1985 |
| MULLBERG ET AL., J. IMMUNOL., vol. 152, 1994, pages 4958 - 4968 |
| MULLBERG ET AL., J. IMMUNOL., vol. 152, no. 10, 1994, pages 4958 - 4968 |
| NG ET AL., PHARM. RES, vol. 23, no. 1, 2006, pages 95 - 103 |
| NI, XIANDAI MIANYIXUE, vol. 26, no. 4, 2006, pages 265 - 268 |
| OBER ET AL., INTL. IMMUNOL., vol. 13, no. 12, 2001, pages 1551 - 1559 |
| ONO ET AL., MOL. IMMUNOL., vol. 36, no. 6, 1999, pages 387 - 395 |
| ORITA ET AL., BLOOD, vol. 105, 2005, pages 562 - 566 |
| OSBOURN ET AL., METHODS, vol. 36, 2005, pages 61 - 68 |
| PACE ET AL., PROTEIN SCIENCE, vol. 4, 1995, pages 2411 - 2423 |
| PACIOS, COMPUT. CHEM, vol. L8, no. 4, 1994, pages 377 - 386 |
| PADLAN, MOL. IMMUNOL., vol. 28, 1991, pages 489 - 498 |
| PAVLOU ET AL., EUR. J. PHARM. BIOPHARM., vol. 59, no. 3, 2005, pages 389 - 396 |
| PEER ET AL., NATURE NANOTECHNOLOGY, vol. 2, 2007, pages 751 - 760 |
| PETKOVA ET AL., INTL. IMMUNOL., vol. 18, no. 12, 2006, pages 1759 - 1769 |
| PLUCKTHUN: "The Pharmacology of Monoclonal Antibodies", vol. 113, 1994, SPRINGER-VERLAG, pages: 269 - 315 |
| PORTOLANO ET AL., J. IMMUNOL., vol. 150, 1993, pages 880 - 887 |
| PRESTA ET AL., J. IMMUNOL., vol. 151, 1993, pages 2623 |
| PRESTA, CURR. OPIN. IMMUNOL., vol. 20, no. 4, 2008, pages 460 - 470 |
| PROC. NATL. ACAD. SCI. USA, vol. 103, no. 11, 2006, pages 4005 - 4010 |
| PUCK ET AL., J. EXP. MED., vol. 108, 1995, pages 945 - 956 |
| QUEEN ET AL., PROC. NATL ACAD. SCI. USA, vol. 86, 1989, pages 10029 - 10033 |
| RAGHAVAN ET AL., IMMUNITY, vol. 1, 1994, pages 303 - 315 |
| RAJPAL ET AL., PROC. NATL. ACAD. SCI. USA, vol. 102, no. 24, 2005, pages 8466 - 8471 |
| RAVETCH ET AL., ANNU. REV. IMMUNOL., vol. 9, 1991, pages 457 - 492 |
| REICHERT ET AL., NAT. BIOTECHNOL., vol. 23, 2005, pages 1073 - 1078 |
| RIECHMANN ET AL., NATURE, vol. 332, 1988, pages 323 - 329 |
| ROOPENIAN ET AL., METH.MOL. BIOL., vol. 602, 2010, pages 93 - 104 |
| ROSOK ET AL., J. BIOL. CHEM., vol. 271, 1996, pages 22611 - 22618 |
| RUSSO ET AL., EXP. REV. CLIN. IMMUNOL., vol. 10, no. 5, 2014, pages 593 - 619 |
| SCAPPATICCI ET AL., J. NATL. CANCER INST., vol. 99, no. 16, 2007, pages 1232 - 1239 |
| SCHAEFER ET AL., GENOMICS, vol. 25, 1995, pages 638 - 643 |
| SCHLAPSCHLY ET AL., PROTEIN ENG. DES. SEL., vol. 22, no. 3, 2009, pages 175 - 188 |
| SHIELDS ET AL., J. BIOL. CHEM., vol. 276, no. 9, 2001, pages 6591 - 6604 |
| SHIELDS ET AL., J. BIOL. CHEM., vol. 9, no. 2, 2001, pages 6591 - 6604 |
| SHINKAWA ET AL., J. BIOL. CHEM., vol. 278, 2003, pages 3466 - 3473 |
| SIDHU ET AL., J. MOL. BIOL., vol. 338, no. 2, 2004, pages 299 - 310 |
| SIMS ET AL., J. IMMUNOL., vol. 151, 1993, pages 2296 |
| SKOOG ET AL., TRENDS ANAL. CHEM., vol. 5, no. 4, 1986, pages 82 - 83 |
| SKOOG ET AL., TRENDS ANALYT. CHEM., vol. 5, no. 4, 1986, pages 82 - 83 |
| SMITH ET AL., METHODS ENZYMOL., vol. 217, 1993, pages 228 - 257 |
| SPRINGER ET AL., CELL, vol. 102, 2000, pages 275 - 277 |
| STROHL ET AL., CURR. OP. BIOTECH., vol. 20, no. 6, 2009, pages 685 - 691 |
| STROHL, CURR. OPIN. BIOTECHNOL., vol. 20, no. 6, 2009, pages 685 - 691 |
| SUSUMU ET AL., NATURE, vol. 315, 1985, pages 592 - 594 |
| TAKEBE ET AL., MOL. CELL BIOL., vol. 8, 1988, pages 466 - 472 |
| TOMLINSON ET AL., J. MOL. BIOL., vol. 227, 1992, pages 776 - 798 |
| TRAUNECKER ET AL., EMBO J., vol. 10, 1991, pages 3655 |
| TUTT ET AL., J. IMMUNOL., vol. 147, 1991, pages 60 |
| URLAUB ET AL., PROC. NATL. ACAD. SCI. USA, vol. 77, 1980, pages 4216 |
| VALLE ET AL., NATURE, vol. 291, 1981, pages 338 - 340 |
| VAN DIJK; VAN DE WINKEL, CURR. OPIN. PHARMACOL., vol. 5, 2001, pages 368 - 74 |
| VOLLMERS ET AL., HISTOL. AND HISTOPATH., vol. 20, no. 3, 2005, pages 927 - 937 |
| VOLLMERS ET AL., METHODS AND FINDINGS IN EXPERIMENTAL AND CLINICAL PHARMACOLOGY, vol. 27, no. 3, 2005, pages 185 - 91 |
| WALLE ET AL., EXPERT OPIN. BIOL. THER., vol. 7, no. 3, 2007, pages 405 - 418 |
| WANG ET AL., ANNU. REV. BIOPHYS. BIOMOL. STRUCT., vol. 35, 2006, pages 225 - 249 |
| WILLIAMS ET AL., EUR. J. IMMUNOL., vol. 23, 1993, pages 1456 - 1461 |
| WINTER ET AL., ANN. REV. IMMUNOL., vol. 12, 1994, pages 433 - 455 |
| WRIGHT ET AL., TIBTECH, vol. 15, 1997, pages 26 - 32 |
| WU ET AL., J. EXP. MED., vol. 132, 1970, pages 211 - 250 |
| WU ET AL., J. MOL. BIOL., vol. 368, 2007, pages 652 |
| WU ET AL., J. MOL. BIOL., vol. 368, 2007, pages 652 - 665 |
| WURZBURG ET AL., STRUCTURE, vol. 14, no. 6, 2006, pages 1049 - 1058 |
| YAZAKI; WU: "Methods in Molecular Biology", vol. 248, 2003, HUMANA PRESS, pages: 255 - 268 |
| YEUNG ET AL., BIOTECHNOL. PROG., vol. 18, no. 2, 2002, pages 212 - 220 |
| YEUNG ET AL., J. IMMUNOL., vol. 182, 2009, pages 7663 - 7671 |
| ZALEVSKY ET AL., NAT. BIOTECHNOL., vol. 28, 2010, pages 157 - 159 |
| ZHENG ET AL., CLIN. PHARM. & THER., vol. 89, no. 2, 2011, pages 283 - 290 |
| ZHENG ET AL., CLIN. PHARMACOL. THER., vol. 89, no. 2, 2011, pages 283 - 290 |
| ZHENG ET AL., CLINICAL PHARMACOLOGY & THERAPEUTICS, vol. 89, no. 2, 2011, pages 283 - 290 |
Cited By (50)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US11046784B2 (en) | 2006-03-31 | 2021-06-29 | Chugai Seiyaku Kabushiki Kaisha | Methods for controlling blood pharmacokinetics of antibodies |
| US11248053B2 (en) | 2007-09-26 | 2022-02-15 | Chugai Seiyaku Kabushiki Kaisha | Method of modifying isoelectric point of antibody via amino acid substitution in CDR |
| US9828429B2 (en) | 2007-09-26 | 2017-11-28 | Chugai Seiyaku Kabushiki Kaisha | Method of modifying isoelectric point of antibody via amino acid substitution in CDR |
| US12122840B2 (en) | 2007-09-26 | 2024-10-22 | Chugai Seiyaku Kabushiki Kaisha | Method of modifying isoelectric point of antibody via amino acid substitution in CDR |
| US12116414B2 (en) | 2007-09-26 | 2024-10-15 | Chugai Seiyaku Kabushiki Kaisha | Method of modifying isoelectric point of antibody via amino acid substitution in CDR |
| US9868948B2 (en) | 2008-04-11 | 2018-01-16 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecule capable of binding to two or more antigen molecules repeatedly |
| US11359194B2 (en) | 2008-04-11 | 2022-06-14 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecule capable of binding two or more antigen molecules repeatedly |
| US9890377B2 (en) | 2008-04-11 | 2018-02-13 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecule capable of binding to two or more antigen molecules repeatedly |
| US10472623B2 (en) | 2008-04-11 | 2019-11-12 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecule capable of binding two or more antigen molecules repeatedly |
| US11371039B2 (en) | 2008-04-11 | 2022-06-28 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecule capable of binding to two or more antigen molecules repeatedly |
| US11891434B2 (en) | 2010-11-30 | 2024-02-06 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecule capable of binding to plurality of antigen molecules repeatedly |
| US11673947B2 (en) | 2012-05-30 | 2023-06-13 | Chugai Seiyaku Kabushiki Kaisha | Target tissue-specific antigen-binding molecule |
| US10919953B2 (en) | 2012-08-24 | 2021-02-16 | Chugai Seiyaku Kabushiki Kaisha | FcgammaRIIB-specific Fc region variant |
| US11236168B2 (en) | 2012-08-24 | 2022-02-01 | Chugai Seiyaku Kabushiki Kaisha | Mouse FcγammaRII-specific Fc antibody |
| US11267868B2 (en) | 2013-04-02 | 2022-03-08 | Chugai Seiyaku Kabushiki Kaisha | Fc region variant |
| US11912989B2 (en) | 2013-12-04 | 2024-02-27 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecules, the antigen-binding activity of which varies according to the concentration of compounds, and libraries of said molecules |
| US10961530B2 (en) | 2013-12-04 | 2021-03-30 | Chugai Seiyaku Kabushiki Kaisha | Antigen-binding molecules, the antigen-binding activity of which varies according to the concentration of compounds, and libraries of said molecules |
| US10000560B2 (en) | 2014-12-19 | 2018-06-19 | Chugai Seiyaku Kabushiki Kaisha | Anti-myostatin antibodies, polypeptides containing variant Fc regions, and methods of use |
| US11454633B2 (en) | 2014-12-19 | 2022-09-27 | Chugai Seiyaku Kabushiki Kaisha | Anti-myostatin antibodies, polypeptides containing variant Fc regions, and methods of use |
| US10738111B2 (en) | 2014-12-19 | 2020-08-11 | Chugai Seiyaku Kabushiki Kaisha | Anti-myostatin antibodies, polypeptides containing variant Fc regions, and methods of use |
| US11180548B2 (en) | 2015-02-05 | 2021-11-23 | Chugai Seiyaku Kabushiki Kaisha | Methods of neutralizing IL-8 biological activity |
| US10519229B2 (en) | 2015-02-05 | 2019-12-31 | Chugai Seiyaku Kabushiki Kaisha | Nucleic acids encoding IL-8 antibodies |
| US9969800B2 (en) | 2015-02-05 | 2018-05-15 | Chugai Seiyaku Kabushiki Kaisha | IL-8 antibodies |
| US11359009B2 (en) | 2015-12-25 | 2022-06-14 | Chugai Seiyaku Kabushiki Kaisha | Anti-myostatin antibodies and methods of use |
| US11053308B2 (en) | 2016-08-05 | 2021-07-06 | Chugai Seiyaku Kabushiki Kaisha | Method for treating IL-8-related diseases |
| US11780912B2 (en) | 2016-08-05 | 2023-10-10 | Chugai Seiyaku Kabushiki Kaisha | Composition for prophylaxis or treatment of IL-8 related diseases |
| WO2018025982A1 (ja) * | 2016-08-05 | 2018-02-08 | 中外製薬株式会社 | Il-8関連疾患の治療用又は予防用組成物 |
| US11780908B2 (en) | 2016-09-16 | 2023-10-10 | Chugai Seiyaku Kabushiki Kaisha | Anti-dengue virus antibodies, polypeptides containing variant FC regions, and methods of use |
| US10604561B2 (en) | 2016-09-16 | 2020-03-31 | Chugai Seiyaku Kabushiki Kaisha | Anti-dengue virus antibodies, polypeptides containing variant Fc regions, and methods of use |
| US10844113B2 (en) | 2016-09-16 | 2020-11-24 | Chugai Seiyaku Kabushiki Kaisha | Anti-dengue virus antibodies, polypeptides containing variant Fc regions, and methods of use |
| US11192951B2 (en) | 2017-06-05 | 2021-12-07 | Janseen Biotech, Inc. | Methods of engineering surface charge for bispecific antibody production |
| EP3634994A4 (en) * | 2017-06-05 | 2021-06-30 | Janssen Biotech, Inc. | SURFACE LOAD ENGINEERING PROCESSES FOR THE PRODUCTION OF A BISPECIFIC ANTIBODY |
| KR20200014385A (ko) * | 2017-06-05 | 2020-02-10 | 얀센 바이오테크 인코포레이티드 | 이중특이성 항체 생성을 위한 표면 전하 조작 방법 |
| KR102661320B1 (ko) * | 2017-06-05 | 2024-05-03 | 얀센 바이오테크 인코포레이티드 | 이중특이성 항체 생성을 위한 표면 전하 조작 방법 |
| JP7268011B2 (ja) | 2017-06-05 | 2023-05-02 | ヤンセン バイオテツク,インコーポレーテツド | 二重特異性抗体製造のための表面電荷を工学的に操作する方法 |
| JP2020522573A (ja) * | 2017-06-05 | 2020-07-30 | ヤンセン バイオテツク,インコーポレーテツド | 二重特異性抗体製造のための表面電荷を工学的に操作する方法 |
| US11891432B2 (en) | 2018-03-15 | 2024-02-06 | Chugai Seiyaku Kabushiki Kaisha | Anti-dengue virus antibodies having cross-reactivity to Zika virus and methods of use |
| WO2020032230A1 (ja) | 2018-08-10 | 2020-02-13 | 中外製薬株式会社 | 抗cd137抗原結合分子およびその使用 |
| CN113544157A (zh) * | 2019-04-19 | 2021-10-22 | 中外制药株式会社 | 用于检测或捕获样品中的多肽的抗体和组合物,以及用于检测或捕获样品中的多肽的方法 |
| EP3956365A4 (en) * | 2019-04-19 | 2023-07-05 | Chugai Seiyaku Kabushiki Kaisha | ANTIBODIES AND COMPOSITIONS FOR USE IN DETECTING OR CAPTURE OF A POLYPEPTIDE IN A SAMPLE AND METHODS OF DETECTING OR CAPTURE OF A POLYPEPTIDE IN A SAMPLE |
| US11739142B2 (en) | 2019-12-18 | 2023-08-29 | Hoffmann-La Roche Inc. | Bispecific anti-CCL2 antibodies |
| WO2021122733A1 (en) | 2019-12-18 | 2021-06-24 | F. Hoffmann-La Roche Ag | Bispecific anti-ccl2 antibodies |
| US12103967B2 (en) | 2019-12-18 | 2024-10-01 | Hoffmann-La Roche Inc. | Bispecific anti-CCL2 antibodies |
| WO2021153723A1 (ja) | 2020-01-31 | 2021-08-05 | 中外製薬株式会社 | 着色を抑制したポリペプチドを含む組成物の製造方法 |
| WO2021162020A1 (ja) | 2020-02-12 | 2021-08-19 | 中外製薬株式会社 | 癌の治療に用いるための抗cd137抗原結合分子 |
| GB2595299A (en) * | 2020-05-21 | 2021-11-24 | Mabsolve Ltd | Modified immunoglobulin FC regions |
| US12037380B2 (en) | 2020-05-21 | 2024-07-16 | Mabsol Ve Limited | Modified immunoglobulin Fc regions |
| GB2595299B (en) * | 2020-05-21 | 2022-08-03 | Mabsolve Ltd | Modified immunoglobulin FC regions |
| WO2022244838A1 (ja) | 2021-05-19 | 2022-11-24 | 中外製薬株式会社 | 分子のin vivo薬物動態を予測する方法 |
| WO2022263501A1 (en) | 2021-06-18 | 2022-12-22 | F. Hoffmann-La Roche Ag | Bispecific anti-ccl2 antibodies |
Also Published As
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| US11180548B2 (en) | Methods of neutralizing IL-8 biological activity | |
| AU2016323088B2 (en) | IL-8-binding antibodies and uses thereof |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| ENP | Entry into the national phase |
Ref document number: 2017508124 Country of ref document: JP Kind code of ref document: A |
|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 16753718 Country of ref document: EP Kind code of ref document: A1 |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 11201710553Y Country of ref document: SG |
|
| ENP | Entry into the national phase |
Ref document number: 20177037003 Country of ref document: KR Kind code of ref document: A |
|
| ENP | Entry into the national phase |
Ref document number: 2993423 Country of ref document: CA |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 122021001656 Country of ref document: BR |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 12018500386 Country of ref document: PH |
|
| ENP | Entry into the national phase |
Ref document number: 2016323088 Country of ref document: AU Date of ref document: 20160805 Kind code of ref document: A |
|
| WWE | Wipo information: entry into national phase |
Ref document number: MX/A/2018/003005 Country of ref document: MX |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 000378-2018 Country of ref document: PE |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 258088 Country of ref document: IL |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| REG | Reference to national code |
Ref country code: BR Ref legal event code: B01A Ref document number: 112018002432 Country of ref document: BR |
|
| WWE | Wipo information: entry into national phase |
Ref document number: CR2018-000217 Country of ref document: CR |
|
| WWE | Wipo information: entry into national phase |
Ref document number: A201804018 Country of ref document: UA |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 2018113505 Country of ref document: RU Ref document number: 2016753718 Country of ref document: EP |
|
| ENP | Entry into the national phase |
Ref document number: 112018002432 Country of ref document: BR Kind code of ref document: A2 Effective date: 20180205 |