WO2019038059A1 - Detergent compositions comprising gh9 endoglucanase variants ii - Google Patents
Detergent compositions comprising gh9 endoglucanase variants ii Download PDFInfo
- Publication number
- WO2019038059A1 WO2019038059A1 PCT/EP2018/071105 EP2018071105W WO2019038059A1 WO 2019038059 A1 WO2019038059 A1 WO 2019038059A1 EP 2018071105 W EP2018071105 W EP 2018071105W WO 2019038059 A1 WO2019038059 A1 WO 2019038059A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- seq
- positions
- region
- amino acids
- endoglucanase
- Prior art date
Links
- 239000003599 detergent Substances 0.000 title claims abstract description 220
- 239000000203 mixture Substances 0.000 title claims abstract description 191
- 108010059892 Cellulase Proteins 0.000 title claims abstract description 151
- FWMNVWWHGCHHJJ-SKKKGAJSSA-N 4-amino-1-[(2r)-6-amino-2-[[(2r)-2-[[(2r)-2-[[(2r)-2-amino-3-phenylpropanoyl]amino]-3-phenylpropanoyl]amino]-4-methylpentanoyl]amino]hexanoyl]piperidine-4-carboxylic acid Chemical compound C([C@H](C(=O)N[C@H](CC(C)C)C(=O)N[C@H](CCCCN)C(=O)N1CCC(N)(CC1)C(O)=O)NC(=O)[C@H](N)CC=1C=CC=CC=1)C1=CC=CC=C1 FWMNVWWHGCHHJJ-SKKKGAJSSA-N 0.000 claims description 411
- 150000001413 amino acids Chemical class 0.000 claims description 298
- 230000004075 alteration Effects 0.000 claims description 164
- 238000006467 substitution reaction Methods 0.000 claims description 116
- 230000000694 effects Effects 0.000 claims description 91
- 229920001285 xanthan gum Polymers 0.000 claims description 72
- 239000000230 xanthan gum Substances 0.000 claims description 60
- 235000010493 xanthan gum Nutrition 0.000 claims description 60
- 229940082509 xanthan gum Drugs 0.000 claims description 60
- 108010068608 xanthan lyase Proteins 0.000 claims description 45
- 230000000593 degrading effect Effects 0.000 claims description 22
- 230000006872 improvement Effects 0.000 claims description 16
- 102220492605 Numb-like protein_S17A_mutation Human genes 0.000 claims description 6
- 102220271883 rs1555611550 Human genes 0.000 claims description 6
- 102220470680 Transforming growth factor beta-1-induced transcript 1 protein_Y60F_mutation Human genes 0.000 claims description 4
- 102200016466 rs104894277 Human genes 0.000 claims description 4
- 102220287170 rs1217007370 Human genes 0.000 claims description 4
- 102200081489 rs121913585 Human genes 0.000 claims description 4
- 102220213081 rs876660092 Human genes 0.000 claims description 4
- 102220279287 rs1554888120 Human genes 0.000 claims description 3
- 102220152923 rs553059467 Human genes 0.000 claims 1
- 238000000034 method Methods 0.000 abstract description 83
- 230000000875 corresponding effect Effects 0.000 description 329
- 235000001014 amino acid Nutrition 0.000 description 325
- 229940024606 amino acid Drugs 0.000 description 289
- 108090000765 processed proteins & peptides Proteins 0.000 description 204
- 102000004196 processed proteins & peptides Human genes 0.000 description 202
- 229920001184 polypeptide Polymers 0.000 description 201
- 210000004027 cell Anatomy 0.000 description 114
- 102000040430 polynucleotide Human genes 0.000 description 75
- 108091033319 polynucleotide Proteins 0.000 description 75
- 239000002157 polynucleotide Substances 0.000 description 75
- -1 xanthanases Proteins 0.000 description 74
- 102000004190 Enzymes Human genes 0.000 description 73
- 108090000790 Enzymes Proteins 0.000 description 73
- DHMQDGOQFOQNFH-UHFFFAOYSA-N Glycine Chemical compound NCC(O)=O DHMQDGOQFOQNFH-UHFFFAOYSA-N 0.000 description 71
- WHUUTDBJXJRKMK-UHFFFAOYSA-N Glutamic acid Natural products OC(=O)C(N)CCC(O)=O WHUUTDBJXJRKMK-UHFFFAOYSA-N 0.000 description 66
- WHUUTDBJXJRKMK-VKHMYHEASA-N L-glutamic acid Chemical compound OC(=O)[C@@H](N)CCC(O)=O WHUUTDBJXJRKMK-VKHMYHEASA-N 0.000 description 66
- MTCFGRXMJLQNBG-REOHCLBHSA-N (2S)-2-Amino-3-hydroxypropansäure Chemical compound OC[C@H](N)C(O)=O MTCFGRXMJLQNBG-REOHCLBHSA-N 0.000 description 65
- 229940088598 enzyme Drugs 0.000 description 65
- OUYCCCASQSFEME-QMMMGPOBSA-N L-tyrosine Chemical compound OC(=O)[C@@H](N)CC1=CC=C(O)C=C1 OUYCCCASQSFEME-QMMMGPOBSA-N 0.000 description 63
- DCXYFEDJOCDNAF-REOHCLBHSA-N L-asparagine Chemical compound OC(=O)[C@@H](N)CC(N)=O DCXYFEDJOCDNAF-REOHCLBHSA-N 0.000 description 62
- KDXKERNSBIXSRK-YFKPBYRVSA-N L-lysine Chemical compound NCCCC[C@H](N)C(O)=O KDXKERNSBIXSRK-YFKPBYRVSA-N 0.000 description 62
- COLNVLDHVKWLRT-QMMMGPOBSA-N L-phenylalanine Chemical compound OC(=O)[C@@H](N)CC1=CC=CC=C1 COLNVLDHVKWLRT-QMMMGPOBSA-N 0.000 description 62
- KDXKERNSBIXSRK-UHFFFAOYSA-N Lysine Natural products NCCCCC(N)C(O)=O KDXKERNSBIXSRK-UHFFFAOYSA-N 0.000 description 62
- CKLJMWTZIZZHCS-REOHCLBHSA-N L-aspartic acid Chemical compound OC(=O)[C@@H](N)CC(O)=O CKLJMWTZIZZHCS-REOHCLBHSA-N 0.000 description 61
- AYFVYJQAPQTCCC-GBXIJSLDSA-N L-threonine Chemical compound C[C@@H](O)[C@H](N)C(O)=O AYFVYJQAPQTCCC-GBXIJSLDSA-N 0.000 description 61
- 108090000623 proteins and genes Proteins 0.000 description 61
- 125000001360 methionine group Chemical group N[C@@H](CCSC)C(=O)* 0.000 description 60
- XUJNEKJLAYXESH-REOHCLBHSA-N L-Cysteine Chemical compound SC[C@H](N)C(O)=O XUJNEKJLAYXESH-REOHCLBHSA-N 0.000 description 59
- 125000003275 alpha amino acid group Chemical group 0.000 description 56
- 240000004808 Saccharomyces cerevisiae Species 0.000 description 42
- 235000014680 Saccharomyces cerevisiae Nutrition 0.000 description 42
- 108091026890 Coding region Proteins 0.000 description 38
- 238000012217 deletion Methods 0.000 description 36
- 230000037430 deletion Effects 0.000 description 36
- 108020004414 DNA Proteins 0.000 description 35
- 239000004753 textile Substances 0.000 description 35
- 239000004744 fabric Substances 0.000 description 34
- 102000013142 Amylases Human genes 0.000 description 33
- 108010065511 Amylases Proteins 0.000 description 33
- 235000019418 amylase Nutrition 0.000 description 33
- 238000004140 cleaning Methods 0.000 description 33
- 238000003780 insertion Methods 0.000 description 32
- 230000037431 insertion Effects 0.000 description 32
- 239000013598 vector Substances 0.000 description 31
- 239000003795 chemical substances by application Substances 0.000 description 30
- 229920000642 polymer Polymers 0.000 description 28
- 239000000126 substance Substances 0.000 description 26
- 230000002538 fungal effect Effects 0.000 description 23
- 150000007523 nucleic acids Chemical group 0.000 description 23
- 239000002689 soil Substances 0.000 description 23
- 239000000975 dye Substances 0.000 description 22
- 230000014509 gene expression Effects 0.000 description 22
- 102000039446 nucleic acids Human genes 0.000 description 22
- 108020004707 nucleic acids Proteins 0.000 description 22
- 239000002773 nucleotide Substances 0.000 description 22
- 125000003729 nucleotide group Chemical group 0.000 description 22
- 102000004169 proteins and genes Human genes 0.000 description 22
- 229940025131 amylases Drugs 0.000 description 20
- 108010084185 Cellulases Proteins 0.000 description 19
- 102000005575 Cellulases Human genes 0.000 description 19
- 108010076504 Protein Sorting Signals Proteins 0.000 description 19
- 235000018102 proteins Nutrition 0.000 description 19
- 108090001060 Lipase Proteins 0.000 description 18
- 102000004882 Lipase Human genes 0.000 description 18
- 239000004367 Lipase Substances 0.000 description 18
- 108090000637 alpha-Amylases Proteins 0.000 description 18
- 235000019421 lipase Nutrition 0.000 description 18
- 239000004094 surface-active agent Substances 0.000 description 18
- 102000035195 Peptidases Human genes 0.000 description 17
- 108091005804 Peptidases Proteins 0.000 description 17
- 238000004061 bleaching Methods 0.000 description 17
- 239000000835 fiber Substances 0.000 description 17
- 230000010076 replication Effects 0.000 description 17
- 239000000523 sample Substances 0.000 description 17
- 102000004139 alpha-Amylases Human genes 0.000 description 16
- 230000001580 bacterial effect Effects 0.000 description 16
- 230000008901 benefit Effects 0.000 description 16
- 239000007844 bleaching agent Substances 0.000 description 16
- 229920002678 cellulose Polymers 0.000 description 16
- 239000001913 cellulose Substances 0.000 description 16
- 235000010980 cellulose Nutrition 0.000 description 16
- 239000007788 liquid Substances 0.000 description 16
- 239000004365 Protease Substances 0.000 description 15
- 229940024171 alpha-amylase Drugs 0.000 description 15
- DBMJMQXJHONAFJ-UHFFFAOYSA-M Sodium laurylsulphate Chemical compound [Na+].CCCCCCCCCCCCOS([O-])(=O)=O DBMJMQXJHONAFJ-UHFFFAOYSA-M 0.000 description 14
- 239000000463 material Substances 0.000 description 14
- 229940083575 sodium dodecyl sulfate Drugs 0.000 description 14
- 235000019333 sodium laurylsulphate Nutrition 0.000 description 14
- 239000004382 Amylase Substances 0.000 description 13
- 239000002253 acid Substances 0.000 description 13
- 235000014113 dietary fatty acids Nutrition 0.000 description 13
- 239000000194 fatty acid Substances 0.000 description 13
- 229930195729 fatty acid Natural products 0.000 description 13
- 239000003752 hydrotrope Substances 0.000 description 13
- 239000002609 medium Substances 0.000 description 13
- 239000013612 plasmid Substances 0.000 description 13
- 239000000047 product Substances 0.000 description 13
- GJCOSYZMQJWQCA-UHFFFAOYSA-N 9H-xanthene Chemical compound C1=CC=C2CC3=CC=CC=C3OC2=C1 GJCOSYZMQJWQCA-UHFFFAOYSA-N 0.000 description 12
- 241000193830 Bacillus <bacterium> Species 0.000 description 12
- ZHNUHDYFZUAESO-UHFFFAOYSA-N Formamide Chemical compound NC=O ZHNUHDYFZUAESO-UHFFFAOYSA-N 0.000 description 12
- 239000002738 chelating agent Substances 0.000 description 12
- 230000008569 process Effects 0.000 description 12
- XLYOFNOQVPJJNP-UHFFFAOYSA-N water Substances O XLYOFNOQVPJJNP-UHFFFAOYSA-N 0.000 description 12
- 240000006439 Aspergillus oryzae Species 0.000 description 11
- 235000002247 Aspergillus oryzae Nutrition 0.000 description 11
- 241000499912 Trichoderma reesei Species 0.000 description 11
- 230000002255 enzymatic effect Effects 0.000 description 11
- 238000009472 formulation Methods 0.000 description 11
- 241000223218 Fusarium Species 0.000 description 10
- 102000003992 Peroxidases Human genes 0.000 description 10
- 108010005400 cutinase Proteins 0.000 description 10
- 239000013604 expression vector Substances 0.000 description 10
- 239000006081 fluorescent whitening agent Substances 0.000 description 10
- 239000012634 fragment Substances 0.000 description 10
- 238000009396 hybridization Methods 0.000 description 10
- 238000004519 manufacturing process Methods 0.000 description 10
- 230000035772 mutation Effects 0.000 description 10
- 238000003860 storage Methods 0.000 description 10
- 238000012546 transfer Methods 0.000 description 10
- 244000063299 Bacillus subtilis Species 0.000 description 9
- 235000014469 Bacillus subtilis Nutrition 0.000 description 9
- 108700020962 Peroxidase Proteins 0.000 description 9
- DNIAPMSPPWPWGF-UHFFFAOYSA-N Propylene glycol Chemical compound CC(O)CO DNIAPMSPPWPWGF-UHFFFAOYSA-N 0.000 description 9
- 241001494489 Thielavia Species 0.000 description 9
- 239000000654 additive Substances 0.000 description 9
- 125000000539 amino acid group Chemical group 0.000 description 9
- 238000003556 assay Methods 0.000 description 9
- 230000015556 catabolic process Effects 0.000 description 9
- 150000001875 compounds Chemical class 0.000 description 9
- 150000004665 fatty acids Chemical class 0.000 description 9
- 150000003839 salts Chemical class 0.000 description 9
- 239000003381 stabilizer Substances 0.000 description 9
- UHPMCKVQTMMPCG-UHFFFAOYSA-N 5,8-dihydroxy-2-methoxy-6-methyl-7-(2-oxopropyl)naphthalene-1,4-dione Chemical compound CC1=C(CC(C)=O)C(O)=C2C(=O)C(OC)=CC(=O)C2=C1O UHPMCKVQTMMPCG-UHFFFAOYSA-N 0.000 description 8
- 241000228212 Aspergillus Species 0.000 description 8
- 241000351920 Aspergillus nidulans Species 0.000 description 8
- 241000194108 Bacillus licheniformis Species 0.000 description 8
- 229920002134 Carboxymethyl cellulose Polymers 0.000 description 8
- 241000588724 Escherichia coli Species 0.000 description 8
- MHAJPDPJQMAIIY-UHFFFAOYSA-N Hydrogen peroxide Chemical compound OO MHAJPDPJQMAIIY-UHFFFAOYSA-N 0.000 description 8
- 125000003412 L-alanyl group Chemical group [H]N([H])[C@@](C([H])([H])[H])(C(=O)[*])[H] 0.000 description 8
- 238000002105 Southern blotting Methods 0.000 description 8
- 230000000996 additive effect Effects 0.000 description 8
- 239000012876 carrier material Substances 0.000 description 8
- 239000002299 complementary DNA Substances 0.000 description 8
- 229920001577 copolymer Polymers 0.000 description 8
- 230000004927 fusion Effects 0.000 description 8
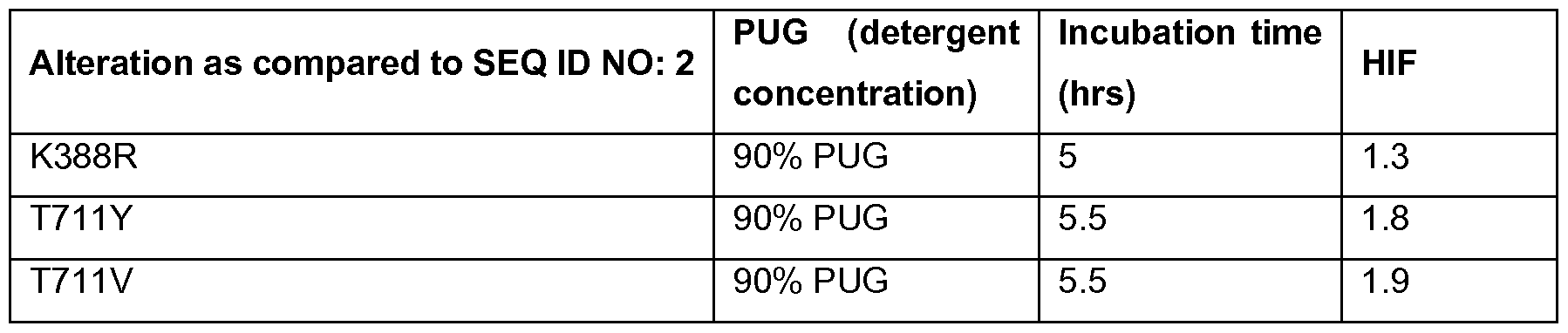
- 238000011534 incubation Methods 0.000 description 8
- 235000019419 proteases Nutrition 0.000 description 8
- 238000002741 site-directed mutagenesis Methods 0.000 description 8
- 239000000758 substrate Substances 0.000 description 8
- 235000000346 sugar Nutrition 0.000 description 8
- 241000228245 Aspergillus niger Species 0.000 description 7
- 241000233866 Fungi Species 0.000 description 7
- 239000004471 Glycine Substances 0.000 description 7
- 108020004711 Nucleic Acid Probes Proteins 0.000 description 7
- 108090000787 Subtilisin Proteins 0.000 description 7
- 230000003197 catalytic effect Effects 0.000 description 7
- 238000010276 construction Methods 0.000 description 7
- 238000006731 degradation reaction Methods 0.000 description 7
- 239000002979 fabric softener Substances 0.000 description 7
- 239000008187 granular material Substances 0.000 description 7
- 238000000338 in vitro Methods 0.000 description 7
- 108020004999 messenger RNA Proteins 0.000 description 7
- 239000002853 nucleic acid probe Substances 0.000 description 7
- 238000003752 polymerase chain reaction Methods 0.000 description 7
- 230000001105 regulatory effect Effects 0.000 description 7
- 238000013518 transcription Methods 0.000 description 7
- 230000035897 transcription Effects 0.000 description 7
- 230000009466 transformation Effects 0.000 description 7
- 241000972773 Aulopiformes Species 0.000 description 6
- LFQSCWFLJHTTHZ-UHFFFAOYSA-N Ethanol Chemical compound CCO LFQSCWFLJHTTHZ-UHFFFAOYSA-N 0.000 description 6
- 241000223221 Fusarium oxysporum Species 0.000 description 6
- 241000221779 Fusarium sambucinum Species 0.000 description 6
- 229920000297 Rayon Polymers 0.000 description 6
- 102000012479 Serine Proteases Human genes 0.000 description 6
- 108010022999 Serine Proteases Proteins 0.000 description 6
- 208000037065 Subacute sclerosing leukoencephalitis Diseases 0.000 description 6
- 206010042297 Subacute sclerosing panencephalitis Diseases 0.000 description 6
- 101710135785 Subtilisin-like protease Proteins 0.000 description 6
- 239000012190 activator Substances 0.000 description 6
- 230000015572 biosynthetic process Effects 0.000 description 6
- 238000004422 calculation algorithm Methods 0.000 description 6
- 239000001768 carboxy methyl cellulose Substances 0.000 description 6
- 235000010948 carboxy methyl cellulose Nutrition 0.000 description 6
- 239000008112 carboxymethyl-cellulose Substances 0.000 description 6
- 229940105329 carboxymethylcellulose Drugs 0.000 description 6
- 230000008859 change Effects 0.000 description 6
- 239000006260 foam Substances 0.000 description 6
- 230000006870 function Effects 0.000 description 6
- 239000011521 glass Substances 0.000 description 6
- 239000004220 glutamic acid Substances 0.000 description 6
- 230000001965 increasing effect Effects 0.000 description 6
- 239000004615 ingredient Substances 0.000 description 6
- 238000004900 laundering Methods 0.000 description 6
- 230000007935 neutral effect Effects 0.000 description 6
- 150000004965 peroxy acids Chemical class 0.000 description 6
- 210000001938 protoplast Anatomy 0.000 description 6
- 235000019515 salmon Nutrition 0.000 description 6
- 239000000243 solution Substances 0.000 description 6
- 241000894007 species Species 0.000 description 6
- 238000005406 washing Methods 0.000 description 6
- 239000004475 Arginine Substances 0.000 description 5
- 102100032487 Beta-mannosidase Human genes 0.000 description 5
- 241000123346 Chrysosporium Species 0.000 description 5
- 241000567178 Fusarium venenatum Species 0.000 description 5
- 241000193385 Geobacillus stearothermophilus Species 0.000 description 5
- 241000223198 Humicola Species 0.000 description 5
- 102000004316 Oxidoreductases Human genes 0.000 description 5
- 108090000854 Oxidoreductases Proteins 0.000 description 5
- 239000002202 Polyethylene glycol Substances 0.000 description 5
- 108010059820 Polygalacturonase Proteins 0.000 description 5
- 239000004372 Polyvinyl alcohol Substances 0.000 description 5
- 241000589516 Pseudomonas Species 0.000 description 5
- 241000235403 Rhizomucor miehei Species 0.000 description 5
- 229920002125 Sokalan® Polymers 0.000 description 5
- 241000194017 Streptococcus Species 0.000 description 5
- 241000187747 Streptomyces Species 0.000 description 5
- 241000223258 Thermomyces lanuginosus Species 0.000 description 5
- 125000000217 alkyl group Chemical group 0.000 description 5
- 230000000844 anti-bacterial effect Effects 0.000 description 5
- ODKSFYDXXFIFQN-UHFFFAOYSA-N arginine Natural products OC(=O)C(N)CCCNC(N)=N ODKSFYDXXFIFQN-UHFFFAOYSA-N 0.000 description 5
- 239000003899 bactericide agent Substances 0.000 description 5
- 108010055059 beta-Mannosidase Proteins 0.000 description 5
- 238000003776 cleavage reaction Methods 0.000 description 5
- 238000004851 dishwashing Methods 0.000 description 5
- 239000002270 dispersing agent Substances 0.000 description 5
- 238000004520 electroporation Methods 0.000 description 5
- 108010093305 exopolygalacturonase Proteins 0.000 description 5
- 239000000499 gel Substances 0.000 description 5
- 150000004676 glycans Chemical class 0.000 description 5
- 125000003630 glycyl group Chemical group [H]N([H])C([H])([H])C(*)=O 0.000 description 5
- 230000002209 hydrophobic effect Effects 0.000 description 5
- 239000003112 inhibitor Substances 0.000 description 5
- 230000010354 integration Effects 0.000 description 5
- 238000002703 mutagenesis Methods 0.000 description 5
- 231100000350 mutagenesis Toxicity 0.000 description 5
- 230000003287 optical effect Effects 0.000 description 5
- 239000002304 perfume Substances 0.000 description 5
- 230000008488 polyadenylation Effects 0.000 description 5
- 229920000728 polyester Polymers 0.000 description 5
- 229920001223 polyethylene glycol Polymers 0.000 description 5
- 229920002451 polyvinyl alcohol Polymers 0.000 description 5
- 229920000036 polyvinylpyrrolidone Polymers 0.000 description 5
- 239000001267 polyvinylpyrrolidone Substances 0.000 description 5
- 235000013855 polyvinylpyrrolidone Nutrition 0.000 description 5
- 230000002265 prevention Effects 0.000 description 5
- 238000011160 research Methods 0.000 description 5
- 102220052102 rs35524245 Human genes 0.000 description 5
- 230000007017 scission Effects 0.000 description 5
- 230000028327 secretion Effects 0.000 description 5
- QUCDWLYKDRVKMI-UHFFFAOYSA-M sodium;3,4-dimethylbenzenesulfonate Chemical compound [Na+].CC1=CC=C(S([O-])(=O)=O)C=C1C QUCDWLYKDRVKMI-UHFFFAOYSA-M 0.000 description 5
- WEAPVABOECTMGR-UHFFFAOYSA-N triethyl 2-acetyloxypropane-1,2,3-tricarboxylate Chemical group CCOC(=O)CC(C(=O)OCC)(OC(C)=O)CC(=O)OCC WEAPVABOECTMGR-UHFFFAOYSA-N 0.000 description 5
- PQHYOGIRXOKOEJ-UHFFFAOYSA-N 2-(1,2-dicarboxyethylamino)butanedioic acid Chemical compound OC(=O)CC(C(O)=O)NC(C(O)=O)CC(O)=O PQHYOGIRXOKOEJ-UHFFFAOYSA-N 0.000 description 4
- 101000757144 Aspergillus niger Glucoamylase Proteins 0.000 description 4
- 241000193744 Bacillus amyloliquefaciens Species 0.000 description 4
- 241000193422 Bacillus lentus Species 0.000 description 4
- 241000193388 Bacillus thuringiensis Species 0.000 description 4
- 241000894006 Bacteria Species 0.000 description 4
- 108091005658 Basic proteases Proteins 0.000 description 4
- 102000016938 Catalase Human genes 0.000 description 4
- 108010053835 Catalase Proteins 0.000 description 4
- 108010008885 Cellulose 1,4-beta-Cellobiosidase Proteins 0.000 description 4
- 241000146399 Ceriporiopsis Species 0.000 description 4
- 241000567163 Fusarium cerealis Species 0.000 description 4
- 241000146406 Fusarium heterosporum Species 0.000 description 4
- 241001480714 Humicola insolens Species 0.000 description 4
- 102100027612 Kallikrein-11 Human genes 0.000 description 4
- QNAYBMKLOCPYGJ-REOHCLBHSA-N L-alanine Chemical compound C[C@H](N)C(O)=O QNAYBMKLOCPYGJ-REOHCLBHSA-N 0.000 description 4
- 108010006035 Metalloproteases Proteins 0.000 description 4
- 102000005741 Metalloproteases Human genes 0.000 description 4
- 108010029182 Pectin lyase Proteins 0.000 description 4
- 229920003171 Poly (ethylene oxide) Polymers 0.000 description 4
- MTCFGRXMJLQNBG-UHFFFAOYSA-N Serine Natural products OCC(N)C(O)=O MTCFGRXMJLQNBG-UHFFFAOYSA-N 0.000 description 4
- 108010056079 Subtilisins Proteins 0.000 description 4
- 102000005158 Subtilisins Human genes 0.000 description 4
- 241001313536 Thermothelomyces thermophila Species 0.000 description 4
- 241000223259 Trichoderma Species 0.000 description 4
- 108700015934 Triose-phosphate isomerases Proteins 0.000 description 4
- 101710152431 Trypsin-like protease Proteins 0.000 description 4
- 238000002835 absorbance Methods 0.000 description 4
- 108010048241 acetamidase Proteins 0.000 description 4
- 150000007513 acids Chemical class 0.000 description 4
- NIXOWILDQLNWCW-UHFFFAOYSA-N acrylic acid group Chemical group C(C=C)(=O)O NIXOWILDQLNWCW-UHFFFAOYSA-N 0.000 description 4
- 235000004279 alanine Nutrition 0.000 description 4
- 229940097012 bacillus thuringiensis Drugs 0.000 description 4
- KGBXLFKZBHKPEV-UHFFFAOYSA-N boric acid Chemical compound OB(O)O KGBXLFKZBHKPEV-UHFFFAOYSA-N 0.000 description 4
- 239000003054 catalyst Substances 0.000 description 4
- 239000000919 ceramic Substances 0.000 description 4
- 238000005352 clarification Methods 0.000 description 4
- 230000021615 conjugation Effects 0.000 description 4
- 238000005260 corrosion Methods 0.000 description 4
- 235000021186 dishes Nutrition 0.000 description 4
- 108010091371 endoglucanase 1 Proteins 0.000 description 4
- 108010091384 endoglucanase 2 Proteins 0.000 description 4
- 238000005516 engineering process Methods 0.000 description 4
- 238000000855 fermentation Methods 0.000 description 4
- 230000004151 fermentation Effects 0.000 description 4
- 235000013922 glutamic acid Nutrition 0.000 description 4
- 238000002744 homologous recombination Methods 0.000 description 4
- 230000006801 homologous recombination Effects 0.000 description 4
- 230000002401 inhibitory effect Effects 0.000 description 4
- JVTAAEKCZFNVCJ-UHFFFAOYSA-N lactic acid Chemical compound CC(O)C(O)=O JVTAAEKCZFNVCJ-UHFFFAOYSA-N 0.000 description 4
- 239000011159 matrix material Substances 0.000 description 4
- 229910052751 metal Inorganic materials 0.000 description 4
- 239000002184 metal Substances 0.000 description 4
- 244000005700 microbiome Species 0.000 description 4
- 235000015097 nutrients Nutrition 0.000 description 4
- 229920001282 polysaccharide Polymers 0.000 description 4
- 239000005017 polysaccharide Substances 0.000 description 4
- 239000000843 powder Substances 0.000 description 4
- 238000003259 recombinant expression Methods 0.000 description 4
- 230000009467 reduction Effects 0.000 description 4
- 238000012216 screening Methods 0.000 description 4
- 239000002904 solvent Substances 0.000 description 4
- 239000000375 suspending agent Substances 0.000 description 4
- 230000002103 transcriptional effect Effects 0.000 description 4
- 230000014616 translation Effects 0.000 description 4
- 238000013519 translation Methods 0.000 description 4
- URAYPUMNDPQOKB-UHFFFAOYSA-N triacetin Chemical compound CC(=O)OCC(OC(C)=O)COC(C)=O URAYPUMNDPQOKB-UHFFFAOYSA-N 0.000 description 4
- OUYCCCASQSFEME-UHFFFAOYSA-N tyrosine Natural products OC(=O)C(N)CC1=CC=C(O)C=C1 OUYCCCASQSFEME-UHFFFAOYSA-N 0.000 description 4
- 108010083879 xyloglucan endo(1-4)-beta-D-glucanase Proteins 0.000 description 4
- 241001019659 Acremonium <Plectosphaerellaceae> Species 0.000 description 3
- 102000057234 Acyl transferases Human genes 0.000 description 3
- 108700016155 Acyl transferases Proteins 0.000 description 3
- 102100034044 All-trans-retinol dehydrogenase [NAD(+)] ADH1B Human genes 0.000 description 3
- 101710193111 All-trans-retinol dehydrogenase [NAD(+)] ADH4 Proteins 0.000 description 3
- 108010037870 Anthranilate Synthase Proteins 0.000 description 3
- 108010017640 Aspartic Acid Proteases Proteins 0.000 description 3
- 102000004580 Aspartic Acid Proteases Human genes 0.000 description 3
- 241001328122 Bacillus clausii Species 0.000 description 3
- 229920002498 Beta-glucan Polymers 0.000 description 3
- 240000008564 Boehmeria nivea Species 0.000 description 3
- OKTJSMMVPCPJKN-UHFFFAOYSA-N Carbon Chemical compound [C] OKTJSMMVPCPJKN-UHFFFAOYSA-N 0.000 description 3
- 229920000742 Cotton Polymers 0.000 description 3
- KCXVZYZYPLLWCC-UHFFFAOYSA-N EDTA Chemical compound OC(=O)CN(CC(O)=O)CCN(CC(O)=O)CC(O)=O KCXVZYZYPLLWCC-UHFFFAOYSA-N 0.000 description 3
- 241000196324 Embryophyta Species 0.000 description 3
- 108010073178 Glucan 1,4-alpha-Glucosidase Proteins 0.000 description 3
- PEDCQBHIVMGVHV-UHFFFAOYSA-N Glycerine Chemical compound OCC(O)CO PEDCQBHIVMGVHV-UHFFFAOYSA-N 0.000 description 3
- 102000005744 Glycoside Hydrolases Human genes 0.000 description 3
- 108010031186 Glycoside Hydrolases Proteins 0.000 description 3
- 102000004157 Hydrolases Human genes 0.000 description 3
- 108090000604 Hydrolases Proteins 0.000 description 3
- 102220468791 Inositol 1,4,5-trisphosphate receptor type 2_Y167A_mutation Human genes 0.000 description 3
- KFZMGEQAYNKOFK-UHFFFAOYSA-N Isopropanol Chemical compound CC(C)O KFZMGEQAYNKOFK-UHFFFAOYSA-N 0.000 description 3
- 125000000570 L-alpha-aspartyl group Chemical group [H]OC(=O)C([H])([H])[C@]([H])(N([H])[H])C(*)=O 0.000 description 3
- HNDVDQJCIGZPNO-YFKPBYRVSA-N L-histidine Chemical compound OC(=O)[C@@H](N)CC1=CN=CN1 HNDVDQJCIGZPNO-YFKPBYRVSA-N 0.000 description 3
- 239000004472 Lysine Substances 0.000 description 3
- 108091028043 Nucleic acid sequence Proteins 0.000 description 3
- 108091034117 Oligonucleotide Proteins 0.000 description 3
- 241000611836 Paenibacillus alginolyticus Species 0.000 description 3
- 102000012288 Phosphopyruvate Hydratase Human genes 0.000 description 3
- 108010022181 Phosphopyruvate Hydratase Proteins 0.000 description 3
- 241000235648 Pichia Species 0.000 description 3
- 239000004952 Polyamide Substances 0.000 description 3
- 102220616113 Sialic acid-binding Ig-like lectin 14_T87R_mutation Human genes 0.000 description 3
- 241000187432 Streptomyces coelicolor Species 0.000 description 3
- 241000187392 Streptomyces griseus Species 0.000 description 3
- 108700005078 Synthetic Genes Proteins 0.000 description 3
- GSEJCLTVZPLZKY-UHFFFAOYSA-N Triethanolamine Chemical compound OCCN(CCO)CCO GSEJCLTVZPLZKY-UHFFFAOYSA-N 0.000 description 3
- 102000005924 Triose-Phosphate Isomerase Human genes 0.000 description 3
- IXKSXJFAGXLQOQ-XISFHERQSA-N WHWLQLKPGQPMY Chemical compound C([C@@H](C(=O)N[C@@H](CC=1C2=CC=CC=C2NC=1)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](CCC(N)=O)C(=O)N[C@@H](CC(C)C)C(=O)N1CCC[C@H]1C(=O)NCC(=O)N[C@@H](CCC(N)=O)C(=O)N[C@@H](CC(O)=O)C(=O)N1CCC[C@H]1C(=O)N[C@@H](CCSC)C(=O)N[C@@H](CC=1C=CC(O)=CC=1)C(O)=O)NC(=O)[C@@H](N)CC=1C2=CC=CC=C2NC=1)C1=CNC=N1 IXKSXJFAGXLQOQ-XISFHERQSA-N 0.000 description 3
- 238000004220 aggregation Methods 0.000 description 3
- 150000001412 amines Chemical class 0.000 description 3
- 239000003945 anionic surfactant Substances 0.000 description 3
- 239000003963 antioxidant agent Substances 0.000 description 3
- 125000000637 arginyl group Chemical group N[C@@H](CCCNC(N)=N)C(=O)* 0.000 description 3
- 239000002585 base Substances 0.000 description 3
- 239000004327 boric acid Substances 0.000 description 3
- 210000004899 c-terminal region Anatomy 0.000 description 3
- 229910052799 carbon Inorganic materials 0.000 description 3
- 229940106157 cellulase Drugs 0.000 description 3
- KRKNYBCHXYNGOX-UHFFFAOYSA-N citric acid Chemical compound OC(=O)CC(O)(C(O)=O)CC(O)=O KRKNYBCHXYNGOX-UHFFFAOYSA-N 0.000 description 3
- 239000003086 colorant Substances 0.000 description 3
- 230000000295 complement effect Effects 0.000 description 3
- 108010092450 endoglucanase Z Proteins 0.000 description 3
- 239000003248 enzyme activator Substances 0.000 description 3
- 239000002532 enzyme inhibitor Substances 0.000 description 3
- 150000002191 fatty alcohols Chemical class 0.000 description 3
- 239000000417 fungicide Substances 0.000 description 3
- 102000006602 glyceraldehyde-3-phosphate dehydrogenase Human genes 0.000 description 3
- 108020004445 glyceraldehyde-3-phosphate dehydrogenase Proteins 0.000 description 3
- 125000003147 glycosyl group Chemical group 0.000 description 3
- 229920000578 graft copolymer Polymers 0.000 description 3
- 230000005764 inhibitory process Effects 0.000 description 3
- 108010076363 licheninase Proteins 0.000 description 3
- 230000000670 limiting effect Effects 0.000 description 3
- 239000003550 marker Substances 0.000 description 3
- 230000004048 modification Effects 0.000 description 3
- 238000012986 modification Methods 0.000 description 3
- 239000002736 nonionic surfactant Substances 0.000 description 3
- 229920000847 nonoxynol Polymers 0.000 description 3
- 239000003921 oil Substances 0.000 description 3
- 239000006072 paste Substances 0.000 description 3
- 150000002978 peroxides Chemical class 0.000 description 3
- 239000000049 pigment Substances 0.000 description 3
- 229920002006 poly(N-vinylimidazole) polymer Polymers 0.000 description 3
- 229920002647 polyamide Polymers 0.000 description 3
- 229920005862 polyol Polymers 0.000 description 3
- 150000003077 polyols Chemical class 0.000 description 3
- 238000000746 purification Methods 0.000 description 3
- 239000002964 rayon Substances 0.000 description 3
- 102220087235 rs864622622 Human genes 0.000 description 3
- 150000003384 small molecules Chemical class 0.000 description 3
- 235000019832 sodium triphosphate Nutrition 0.000 description 3
- 239000007787 solid Substances 0.000 description 3
- YBJHBAHKTGYVGT-ZKWXMUAHSA-N (+)-Biotin Chemical compound N1C(=O)N[C@@H]2[C@H](CCCCC(=O)O)SC[C@@H]21 YBJHBAHKTGYVGT-ZKWXMUAHSA-N 0.000 description 2
- LIPJWTMIUOLEJU-UHFFFAOYSA-N 2-(1,2-diamino-2-phenylethenyl)benzenesulfonic acid Chemical class NC(=C(C=1C(=CC=CC1)S(=O)(=O)O)N)C1=CC=CC=C1 LIPJWTMIUOLEJU-UHFFFAOYSA-N 0.000 description 2
- SMZOUWXMTYCWNB-UHFFFAOYSA-N 2-(2-methoxy-5-methylphenyl)ethanamine Chemical compound COC1=CC=C(C)C=C1CCN SMZOUWXMTYCWNB-UHFFFAOYSA-N 0.000 description 2
- CIEZZGWIJBXOTE-UHFFFAOYSA-N 2-[bis(carboxymethyl)amino]propanoic acid Chemical compound OC(=O)C(C)N(CC(O)=O)CC(O)=O CIEZZGWIJBXOTE-UHFFFAOYSA-N 0.000 description 2
- XWSGEVNYFYKXCP-UHFFFAOYSA-N 2-[carboxymethyl(methyl)amino]acetic acid Chemical compound OC(=O)CN(C)CC(O)=O XWSGEVNYFYKXCP-UHFFFAOYSA-N 0.000 description 2
- OSJPPGNTCRNQQC-UWTATZPHSA-N 3-phospho-D-glyceric acid Chemical compound OC(=O)[C@H](O)COP(O)(O)=O OSJPPGNTCRNQQC-UWTATZPHSA-N 0.000 description 2
- 102000007698 Alcohol dehydrogenase Human genes 0.000 description 2
- 108010021809 Alcohol dehydrogenase Proteins 0.000 description 2
- 101100163849 Arabidopsis thaliana ARS1 gene Proteins 0.000 description 2
- 241001513093 Aspergillus awamori Species 0.000 description 2
- 241000892910 Aspergillus foetidus Species 0.000 description 2
- 241001225321 Aspergillus fumigatus Species 0.000 description 2
- 241001480052 Aspergillus japonicus Species 0.000 description 2
- IJGRMHOSHXDMSA-UHFFFAOYSA-N Atomic nitrogen Chemical compound N#N IJGRMHOSHXDMSA-UHFFFAOYSA-N 0.000 description 2
- 241000223651 Aureobasidium Species 0.000 description 2
- 241000193752 Bacillus circulans Species 0.000 description 2
- 241000193749 Bacillus coagulans Species 0.000 description 2
- 241000193747 Bacillus firmus Species 0.000 description 2
- 101000695691 Bacillus licheniformis Beta-lactamase Proteins 0.000 description 2
- 108010029675 Bacillus licheniformis alpha-amylase Proteins 0.000 description 2
- 241000194107 Bacillus megaterium Species 0.000 description 2
- 241000194103 Bacillus pumilus Species 0.000 description 2
- 101000740449 Bacillus subtilis (strain 168) Biotin/lipoyl attachment protein Proteins 0.000 description 2
- 241000193764 Brevibacillus brevis Species 0.000 description 2
- 125000001433 C-terminal amino-acid group Chemical group 0.000 description 2
- 241000589876 Campylobacter Species 0.000 description 2
- 241000222120 Candida <Saccharomycetales> Species 0.000 description 2
- LZZYPRNAOMGNLH-UHFFFAOYSA-M Cetrimonium bromide Chemical compound [Br-].CCCCCCCCCCCCCCCC[N+](C)(C)C LZZYPRNAOMGNLH-UHFFFAOYSA-M 0.000 description 2
- 241000985909 Chrysosporium keratinophilum Species 0.000 description 2
- 241001674013 Chrysosporium lucknowense Species 0.000 description 2
- 241001556045 Chrysosporium merdarium Species 0.000 description 2
- 241000080524 Chrysosporium queenslandicum Species 0.000 description 2
- 241001674001 Chrysosporium tropicum Species 0.000 description 2
- 241000355696 Chrysosporium zonatum Species 0.000 description 2
- KRKNYBCHXYNGOX-UHFFFAOYSA-K Citrate Chemical compound [O-]C(=O)CC(O)(CC([O-])=O)C([O-])=O KRKNYBCHXYNGOX-UHFFFAOYSA-K 0.000 description 2
- 241000193403 Clostridium Species 0.000 description 2
- 241000222511 Coprinus Species 0.000 description 2
- 244000251987 Coprinus macrorhizus Species 0.000 description 2
- 240000000491 Corchorus aestuans Species 0.000 description 2
- 235000011777 Corchorus aestuans Nutrition 0.000 description 2
- 235000010862 Corchorus capsularis Nutrition 0.000 description 2
- 241001337994 Cryptococcus <scale insect> Species 0.000 description 2
- WQZGKKKJIJFFOK-QTVWNMPRSA-N D-mannopyranose Chemical compound OC[C@H]1OC(O)[C@@H](O)[C@@H](O)[C@@H]1O WQZGKKKJIJFFOK-QTVWNMPRSA-N 0.000 description 2
- 102000053602 DNA Human genes 0.000 description 2
- QXNVGIXVLWOKEQ-UHFFFAOYSA-N Disodium Chemical class [Na][Na] QXNVGIXVLWOKEQ-UHFFFAOYSA-N 0.000 description 2
- 101150015836 ENO1 gene Proteins 0.000 description 2
- 241000194033 Enterococcus Species 0.000 description 2
- 102000010911 Enzyme Precursors Human genes 0.000 description 2
- 108010062466 Enzyme Precursors Proteins 0.000 description 2
- IAYPIBMASNFSPL-UHFFFAOYSA-N Ethylene oxide Chemical group C1CO1 IAYPIBMASNFSPL-UHFFFAOYSA-N 0.000 description 2
- DBVJJBKOTRCVKF-UHFFFAOYSA-N Etidronic acid Chemical compound OP(=O)(O)C(O)(C)P(O)(O)=O DBVJJBKOTRCVKF-UHFFFAOYSA-N 0.000 description 2
- 241000206602 Eukaryota Species 0.000 description 2
- 241000589565 Flavobacterium Species 0.000 description 2
- 241000145614 Fusarium bactridioides Species 0.000 description 2
- 241000223194 Fusarium culmorum Species 0.000 description 2
- 241000223195 Fusarium graminearum Species 0.000 description 2
- 241001112697 Fusarium reticulatum Species 0.000 description 2
- 241001014439 Fusarium sarcochroum Species 0.000 description 2
- 241000223192 Fusarium sporotrichioides Species 0.000 description 2
- 241001465753 Fusarium torulosum Species 0.000 description 2
- 241000605909 Fusobacterium Species 0.000 description 2
- 101150094690 GAL1 gene Proteins 0.000 description 2
- 102100028501 Galanin peptides Human genes 0.000 description 2
- 241000626621 Geobacillus Species 0.000 description 2
- 101100369308 Geobacillus stearothermophilus nprS gene Proteins 0.000 description 2
- 101100080316 Geobacillus stearothermophilus nprT gene Proteins 0.000 description 2
- 102100022624 Glucoamylase Human genes 0.000 description 2
- 241000589989 Helicobacter Species 0.000 description 2
- 101100121078 Homo sapiens GAL gene Proteins 0.000 description 2
- 101001091385 Homo sapiens Kallikrein-6 Proteins 0.000 description 2
- 235000003332 Ilex aquifolium Nutrition 0.000 description 2
- 241000209027 Ilex aquifolium Species 0.000 description 2
- DGAQECJNVWCQMB-PUAWFVPOSA-M Ilexoside XXIX Chemical compound C[C@@H]1CC[C@@]2(CC[C@@]3(C(=CC[C@H]4[C@]3(CC[C@@H]5[C@@]4(CC[C@@H](C5(C)C)OS(=O)(=O)[O-])C)C)[C@@H]2[C@]1(C)O)C)C(=O)O[C@H]6[C@@H]([C@H]([C@@H]([C@H](O6)CO)O)O)O.[Na+] DGAQECJNVWCQMB-PUAWFVPOSA-M 0.000 description 2
- 229920001202 Inulin Polymers 0.000 description 2
- 102100034866 Kallikrein-6 Human genes 0.000 description 2
- 241000235649 Kluyveromyces Species 0.000 description 2
- ODKSFYDXXFIFQN-BYPYZUCNSA-P L-argininium(2+) Chemical compound NC(=[NH2+])NCCC[C@H]([NH3+])C(O)=O ODKSFYDXXFIFQN-BYPYZUCNSA-P 0.000 description 2
- 125000002842 L-seryl group Chemical group O=C([*])[C@](N([H])[H])([H])C([H])([H])O[H] 0.000 description 2
- STECJAGHUSJQJN-USLFZFAMSA-N LSM-4015 Chemical compound C1([C@@H](CO)C(=O)OC2C[C@@H]3N([C@H](C2)[C@@H]2[C@H]3O2)C)=CC=CC=C1 STECJAGHUSJQJN-USLFZFAMSA-N 0.000 description 2
- 108010029541 Laccase Proteins 0.000 description 2
- 241000235087 Lachancea kluyveri Species 0.000 description 2
- 241000186660 Lactobacillus Species 0.000 description 2
- 241000194036 Lactococcus Species 0.000 description 2
- 240000006240 Linum usitatissimum Species 0.000 description 2
- 235000004431 Linum usitatissimum Nutrition 0.000 description 2
- 229920000433 Lyocell Polymers 0.000 description 2
- 241001344133 Magnaporthe Species 0.000 description 2
- 108090000157 Metallothionein Proteins 0.000 description 2
- 241000226677 Myceliophthora Species 0.000 description 2
- 241000187480 Mycobacterium smegmatis Species 0.000 description 2
- FSVCELGFZIQNCK-UHFFFAOYSA-N N,N-bis(2-hydroxyethyl)glycine Chemical compound OCCN(CCO)CC(O)=O FSVCELGFZIQNCK-UHFFFAOYSA-N 0.000 description 2
- QPCDCPDFJACHGM-UHFFFAOYSA-N N,N-bis{2-[bis(carboxymethyl)amino]ethyl}glycine Chemical compound OC(=O)CN(CC(O)=O)CCN(CC(=O)O)CCN(CC(O)=O)CC(O)=O QPCDCPDFJACHGM-UHFFFAOYSA-N 0.000 description 2
- 125000001429 N-terminal alpha-amino-acid group Chemical group 0.000 description 2
- 241000588653 Neisseria Species 0.000 description 2
- 241000233892 Neocallimastix Species 0.000 description 2
- 241000221960 Neurospora Species 0.000 description 2
- 241000221961 Neurospora crassa Species 0.000 description 2
- 241001072230 Oceanobacillus Species 0.000 description 2
- 241000233654 Oomycetes Species 0.000 description 2
- 240000007594 Oryza sativa Species 0.000 description 2
- 235000007164 Oryza sativa Nutrition 0.000 description 2
- 241001236817 Paecilomyces <Clavicipitaceae> Species 0.000 description 2
- 241000194109 Paenibacillus lautus Species 0.000 description 2
- 241000592795 Paenibacillus sp. Species 0.000 description 2
- 241000228143 Penicillium Species 0.000 description 2
- 241000222385 Phanerochaete Species 0.000 description 2
- 241000222393 Phanerochaete chrysosporium Species 0.000 description 2
- 241000235379 Piromyces Species 0.000 description 2
- 229920002319 Poly(methyl acrylate) Polymers 0.000 description 2
- 229920002873 Polyethylenimine Polymers 0.000 description 2
- OFOBLEOULBTSOW-UHFFFAOYSA-N Propanedioic acid Natural products OC(=O)CC(O)=O OFOBLEOULBTSOW-UHFFFAOYSA-N 0.000 description 2
- 108020004511 Recombinant DNA Proteins 0.000 description 2
- 241000235070 Saccharomyces Species 0.000 description 2
- 235000003534 Saccharomyces carlsbergensis Nutrition 0.000 description 2
- 235000001006 Saccharomyces cerevisiae var diastaticus Nutrition 0.000 description 2
- 244000206963 Saccharomyces cerevisiae var. diastaticus Species 0.000 description 2
- 241000204893 Saccharomyces douglasii Species 0.000 description 2
- 241001407717 Saccharomyces norbensis Species 0.000 description 2
- 241001123227 Saccharomyces pastorianus Species 0.000 description 2
- 241000607142 Salmonella Species 0.000 description 2
- 241000222480 Schizophyllum Species 0.000 description 2
- 241000235346 Schizosaccharomyces Species 0.000 description 2
- 101100097319 Schizosaccharomyces pombe (strain 972 / ATCC 24843) ala1 gene Proteins 0.000 description 2
- CDBYLPFSWZWCQE-UHFFFAOYSA-L Sodium Carbonate Chemical compound [Na+].[Na+].[O-]C([O-])=O CDBYLPFSWZWCQE-UHFFFAOYSA-L 0.000 description 2
- 229920002334 Spandex Polymers 0.000 description 2
- 241000191940 Staphylococcus Species 0.000 description 2
- 241000264435 Streptococcus dysgalactiae subsp. equisimilis Species 0.000 description 2
- 241000193996 Streptococcus pyogenes Species 0.000 description 2
- 241000194054 Streptococcus uberis Species 0.000 description 2
- 241000958303 Streptomyces achromogenes Species 0.000 description 2
- 241001468227 Streptomyces avermitilis Species 0.000 description 2
- 241000187398 Streptomyces lividans Species 0.000 description 2
- 241000228341 Talaromyces Species 0.000 description 2
- 241001540751 Talaromyces ruber Species 0.000 description 2
- KKEYFWRCBNTPAC-UHFFFAOYSA-N Terephthalic acid Chemical compound OC(=O)C1=CC=C(C(O)=O)C=C1 KKEYFWRCBNTPAC-UHFFFAOYSA-N 0.000 description 2
- BGRWYDHXPHLNKA-UHFFFAOYSA-N Tetraacetylethylenediamine Chemical compound CC(=O)N(C(C)=O)CCN(C(C)=O)C(C)=O BGRWYDHXPHLNKA-UHFFFAOYSA-N 0.000 description 2
- 241000228178 Thermoascus Species 0.000 description 2
- 241001495429 Thielavia terrestris Species 0.000 description 2
- AYFVYJQAPQTCCC-UHFFFAOYSA-N Threonine Natural products CC(O)C(N)C(O)=O AYFVYJQAPQTCCC-UHFFFAOYSA-N 0.000 description 2
- 239000004473 Threonine Substances 0.000 description 2
- 241001149964 Tolypocladium Species 0.000 description 2
- 241000223260 Trichoderma harzianum Species 0.000 description 2
- 241000378866 Trichoderma koningii Species 0.000 description 2
- 241000223262 Trichoderma longibrachiatum Species 0.000 description 2
- 241000223261 Trichoderma viride Species 0.000 description 2
- 108090000631 Trypsin Proteins 0.000 description 2
- 102000004142 Trypsin Human genes 0.000 description 2
- 241000202898 Ureaplasma Species 0.000 description 2
- 241000589636 Xanthomonas campestris Species 0.000 description 2
- 241000235013 Yarrowia Species 0.000 description 2
- YDONNITUKPKTIG-UHFFFAOYSA-N [Nitrilotris(methylene)]trisphosphonic acid Chemical compound OP(O)(=O)CN(CP(O)(O)=O)CP(O)(O)=O YDONNITUKPKTIG-UHFFFAOYSA-N 0.000 description 2
- 229920006397 acrylic thermoplastic Polymers 0.000 description 2
- 108700014220 acyltransferase activity proteins Proteins 0.000 description 2
- 150000008051 alkyl sulfates Chemical class 0.000 description 2
- 101150078331 ama-1 gene Proteins 0.000 description 2
- 150000001408 amides Chemical class 0.000 description 2
- 230000003321 amplification Effects 0.000 description 2
- 125000000129 anionic group Chemical group 0.000 description 2
- 239000004760 aramid Substances 0.000 description 2
- 235000003704 aspartic acid Nutrition 0.000 description 2
- 229940091771 aspergillus fumigatus Drugs 0.000 description 2
- 229940054340 bacillus coagulans Drugs 0.000 description 2
- 229940005348 bacillus firmus Drugs 0.000 description 2
- OQFSQFPPLPISGP-UHFFFAOYSA-N beta-carboxyaspartic acid Natural products OC(=O)C(N)C(C(O)=O)C(O)=O OQFSQFPPLPISGP-UHFFFAOYSA-N 0.000 description 2
- 239000011230 binding agent Substances 0.000 description 2
- 230000003115 biocidal effect Effects 0.000 description 2
- 150000001720 carbohydrates Chemical class 0.000 description 2
- 125000004432 carbon atom Chemical group C* 0.000 description 2
- 239000003093 cationic surfactant Substances 0.000 description 2
- 229920002301 cellulose acetate Polymers 0.000 description 2
- 230000002759 chromosomal effect Effects 0.000 description 2
- 210000000349 chromosome Anatomy 0.000 description 2
- 239000011248 coating agent Substances 0.000 description 2
- 238000000576 coating method Methods 0.000 description 2
- 239000002361 compost Substances 0.000 description 2
- 238000005520 cutting process Methods 0.000 description 2
- 230000036425 denaturation Effects 0.000 description 2
- 238000004925 denaturation Methods 0.000 description 2
- 239000005547 deoxyribonucleotide Substances 0.000 description 2
- 125000002637 deoxyribonucleotide group Chemical group 0.000 description 2
- 238000001514 detection method Methods 0.000 description 2
- ZBCBWPMODOFKDW-UHFFFAOYSA-N diethanolamine Chemical compound OCCNCCO ZBCBWPMODOFKDW-UHFFFAOYSA-N 0.000 description 2
- XBDQKXXYIPTUBI-UHFFFAOYSA-N dimethylselenoniopropionate Natural products CCC(O)=O XBDQKXXYIPTUBI-UHFFFAOYSA-N 0.000 description 2
- 235000011180 diphosphates Nutrition 0.000 description 2
- PMPJQLCPEQFEJW-GNTLFSRWSA-L disodium;2-[(z)-2-[4-[4-[(z)-2-(2-sulfonatophenyl)ethenyl]phenyl]phenyl]ethenyl]benzenesulfonate Chemical compound [Na+].[Na+].[O-]S(=O)(=O)C1=CC=CC=C1\C=C/C1=CC=C(C=2C=CC(\C=C/C=3C(=CC=CC=3)S([O-])(=O)=O)=CC=2)C=C1 PMPJQLCPEQFEJW-GNTLFSRWSA-L 0.000 description 2
- VUJGKADZTYCLIL-YHPRVSEPSA-L disodium;5-[(4-anilino-6-morpholin-4-yl-1,3,5-triazin-2-yl)amino]-2-[(e)-2-[4-[(4-anilino-6-morpholin-4-yl-1,3,5-triazin-2-yl)amino]-2-sulfonatophenyl]ethenyl]benzenesulfonate Chemical compound [Na+].[Na+].C=1C=C(\C=C\C=2C(=CC(NC=3N=C(N=C(NC=4C=CC=CC=4)N=3)N3CCOCC3)=CC=2)S([O-])(=O)=O)C(S(=O)(=O)[O-])=CC=1NC(N=C(N=1)N2CCOCC2)=NC=1NC1=CC=CC=C1 VUJGKADZTYCLIL-YHPRVSEPSA-L 0.000 description 2
- POULHZVOKOAJMA-UHFFFAOYSA-N dodecanoic acid Chemical compound CCCCCCCCCCCC(O)=O POULHZVOKOAJMA-UHFFFAOYSA-N 0.000 description 2
- 238000005553 drilling Methods 0.000 description 2
- DUYCTCQXNHFCSJ-UHFFFAOYSA-N dtpmp Chemical compound OP(=O)(O)CN(CP(O)(O)=O)CCN(CP(O)(=O)O)CCN(CP(O)(O)=O)CP(O)(O)=O DUYCTCQXNHFCSJ-UHFFFAOYSA-N 0.000 description 2
- 238000010410 dusting Methods 0.000 description 2
- NFDRPXJGHKJRLJ-UHFFFAOYSA-N edtmp Chemical compound OP(O)(=O)CN(CP(O)(O)=O)CCN(CP(O)(O)=O)CP(O)(O)=O NFDRPXJGHKJRLJ-UHFFFAOYSA-N 0.000 description 2
- JBKVHLHDHHXQEQ-UHFFFAOYSA-N epsilon-caprolactam Chemical compound O=C1CCCCCN1 JBKVHLHDHHXQEQ-UHFFFAOYSA-N 0.000 description 2
- 239000003797 essential amino acid Substances 0.000 description 2
- 235000020776 essential amino acid Nutrition 0.000 description 2
- 230000001747 exhibiting effect Effects 0.000 description 2
- 238000000605 extraction Methods 0.000 description 2
- 239000000945 filler Substances 0.000 description 2
- 239000012530 fluid Substances 0.000 description 2
- 239000007850 fluorescent dye Substances 0.000 description 2
- 235000013305 food Nutrition 0.000 description 2
- 108010061330 glucan 1,4-alpha-maltohydrolase Proteins 0.000 description 2
- 239000001087 glyceryl triacetate Substances 0.000 description 2
- 235000013773 glyceryl triacetate Nutrition 0.000 description 2
- KWIUHFFTVRNATP-UHFFFAOYSA-N glycine betaine Chemical compound C[N+](C)(C)CC([O-])=O KWIUHFFTVRNATP-UHFFFAOYSA-N 0.000 description 2
- 239000004519 grease Substances 0.000 description 2
- 229910001385 heavy metal Inorganic materials 0.000 description 2
- HNDVDQJCIGZPNO-UHFFFAOYSA-N histidine Natural products OC(=O)C(N)CC1=CN=CN1 HNDVDQJCIGZPNO-UHFFFAOYSA-N 0.000 description 2
- 229920001519 homopolymer Polymers 0.000 description 2
- 230000003301 hydrolyzing effect Effects 0.000 description 2
- 150000003949 imides Chemical class 0.000 description 2
- 238000001727 in vivo Methods 0.000 description 2
- 229940029339 inulin Drugs 0.000 description 2
- 238000002955 isolation Methods 0.000 description 2
- 239000004310 lactic acid Substances 0.000 description 2
- 235000014655 lactic acid Nutrition 0.000 description 2
- 229940039696 lactobacillus Drugs 0.000 description 2
- 238000007834 ligase chain reaction Methods 0.000 description 2
- 239000004337 magnesium citrate Substances 0.000 description 2
- VZCYOOQTPOCHFL-UPHRSURJSA-N maleic acid Chemical compound OC(=O)\C=C/C(O)=O VZCYOOQTPOCHFL-UPHRSURJSA-N 0.000 description 2
- 239000011976 maleic acid Substances 0.000 description 2
- 150000002739 metals Chemical class 0.000 description 2
- 230000000813 microbial effect Effects 0.000 description 2
- 108010020132 microbial serine proteinases Proteins 0.000 description 2
- 229910052757 nitrogen Inorganic materials 0.000 description 2
- 238000003199 nucleic acid amplification method Methods 0.000 description 2
- 230000037361 pathway Effects 0.000 description 2
- HWGNBUXHKFFFIH-UHFFFAOYSA-I pentasodium;[oxido(phosphonatooxy)phosphoryl] phosphate Chemical compound [Na+].[Na+].[Na+].[Na+].[Na+].[O-]P([O-])(=O)OP([O-])(=O)OP([O-])([O-])=O HWGNBUXHKFFFIH-UHFFFAOYSA-I 0.000 description 2
- 229960003330 pentetic acid Drugs 0.000 description 2
- COLNVLDHVKWLRT-UHFFFAOYSA-N phenylalanine Natural products OC(=O)C(N)CC1=CC=CC=C1 COLNVLDHVKWLRT-UHFFFAOYSA-N 0.000 description 2
- 239000006187 pill Substances 0.000 description 2
- 229920003023 plastic Polymers 0.000 description 2
- 239000004033 plastic Substances 0.000 description 2
- 229920003229 poly(methyl methacrylate) Polymers 0.000 description 2
- 229920005646 polycarboxylate Polymers 0.000 description 2
- 230000004481 post-translational protein modification Effects 0.000 description 2
- 101150054232 pyrG gene Proteins 0.000 description 2
- 238000002708 random mutagenesis Methods 0.000 description 2
- 108091008146 restriction endonucleases Proteins 0.000 description 2
- 238000012552 review Methods 0.000 description 2
- 235000009566 rice Nutrition 0.000 description 2
- 102220285717 rs1555461680 Human genes 0.000 description 2
- 102220026086 rs397518426 Human genes 0.000 description 2
- 230000003248 secreting effect Effects 0.000 description 2
- 230000035945 sensitivity Effects 0.000 description 2
- 150000004760 silicates Chemical class 0.000 description 2
- 239000002002 slurry Substances 0.000 description 2
- 239000011734 sodium Substances 0.000 description 2
- 229910052708 sodium Inorganic materials 0.000 description 2
- 229940079842 sodium cumenesulfonate Drugs 0.000 description 2
- 159000000000 sodium salts Chemical class 0.000 description 2
- 229940048842 sodium xylenesulfonate Drugs 0.000 description 2
- KVCGISUBCHHTDD-UHFFFAOYSA-M sodium;4-methylbenzenesulfonate Chemical compound [Na+].CC1=CC=C(S([O-])(=O)=O)C=C1 KVCGISUBCHHTDD-UHFFFAOYSA-M 0.000 description 2
- QEKATQBVVAZOAY-UHFFFAOYSA-M sodium;4-propan-2-ylbenzenesulfonate Chemical compound [Na+].CC(C)C1=CC=C(S([O-])(=O)=O)C=C1 QEKATQBVVAZOAY-UHFFFAOYSA-M 0.000 description 2
- 239000007921 spray Substances 0.000 description 2
- 229940115922 streptococcus uberis Drugs 0.000 description 2
- 150000005846 sugar alcohols Chemical class 0.000 description 2
- 150000008163 sugars Chemical class 0.000 description 2
- 150000003871 sulfonates Chemical class 0.000 description 2
- 150000003467 sulfuric acid derivatives Chemical class 0.000 description 2
- 238000003786 synthesis reaction Methods 0.000 description 2
- 229920002994 synthetic fiber Polymers 0.000 description 2
- KKEYFWRCBNTPAC-UHFFFAOYSA-L terephthalate(2-) Chemical compound [O-]C(=O)C1=CC=C(C([O-])=O)C=C1 KKEYFWRCBNTPAC-UHFFFAOYSA-L 0.000 description 2
- ISXSCDLOGDJUNJ-UHFFFAOYSA-N tert-butyl prop-2-enoate Chemical compound CC(C)(C)OC(=O)C=C ISXSCDLOGDJUNJ-UHFFFAOYSA-N 0.000 description 2
- 239000002562 thickening agent Substances 0.000 description 2
- VZCYOOQTPOCHFL-UHFFFAOYSA-N trans-butenedioic acid Natural products OC(=O)C=CC(O)=O VZCYOOQTPOCHFL-UHFFFAOYSA-N 0.000 description 2
- 238000010361 transduction Methods 0.000 description 2
- 230000026683 transduction Effects 0.000 description 2
- 229960002622 triacetin Drugs 0.000 description 2
- 239000012588 trypsin Substances 0.000 description 2
- 230000009105 vegetative growth Effects 0.000 description 2
- 229920002554 vinyl polymer Polymers 0.000 description 2
- 239000002023 wood Substances 0.000 description 2
- 210000002268 wool Anatomy 0.000 description 2
- 239000010457 zeolite Substances 0.000 description 2
- 239000002888 zwitterionic surfactant Substances 0.000 description 2
- DIGQNXIGRZPYDK-WKSCXVIASA-N (2R)-6-amino-2-[[2-[[(2S)-2-[[2-[[(2R)-2-[[(2S)-2-[[(2R,3S)-2-[[2-[[(2S)-2-[[2-[[(2S)-2-[[(2S)-2-[[(2R)-2-[[(2S,3S)-2-[[(2R)-2-[[(2S)-2-[[(2S)-2-[[(2S)-2-[[2-[[(2S)-2-[[(2R)-2-[[2-[[2-[[2-[(2-amino-1-hydroxyethylidene)amino]-3-carboxy-1-hydroxypropylidene]amino]-1-hydroxy-3-sulfanylpropylidene]amino]-1-hydroxyethylidene]amino]-1-hydroxy-3-sulfanylpropylidene]amino]-1,3-dihydroxypropylidene]amino]-1-hydroxyethylidene]amino]-1-hydroxypropylidene]amino]-1,3-dihydroxypropylidene]amino]-1,3-dihydroxypropylidene]amino]-1-hydroxy-3-sulfanylpropylidene]amino]-1,3-dihydroxybutylidene]amino]-1-hydroxy-3-sulfanylpropylidene]amino]-1-hydroxypropylidene]amino]-1,3-dihydroxypropylidene]amino]-1-hydroxyethylidene]amino]-1,5-dihydroxy-5-iminopentylidene]amino]-1-hydroxy-3-sulfanylpropylidene]amino]-1,3-dihydroxybutylidene]amino]-1-hydroxy-3-sulfanylpropylidene]amino]-1,3-dihydroxypropylidene]amino]-1-hydroxyethylidene]amino]-1-hydroxy-3-sulfanylpropylidene]amino]-1-hydroxyethylidene]amino]hexanoic acid Chemical compound C[C@@H]([C@@H](C(=N[C@@H](CS)C(=N[C@@H](C)C(=N[C@@H](CO)C(=NCC(=N[C@@H](CCC(=N)O)C(=NC(CS)C(=N[C@H]([C@H](C)O)C(=N[C@H](CS)C(=N[C@H](CO)C(=NCC(=N[C@H](CS)C(=NCC(=N[C@H](CCCCN)C(=O)O)O)O)O)O)O)O)O)O)O)O)O)O)O)N=C([C@H](CS)N=C([C@H](CO)N=C([C@H](CO)N=C([C@H](C)N=C(CN=C([C@H](CO)N=C([C@H](CS)N=C(CN=C(C(CS)N=C(C(CC(=O)O)N=C(CN)O)O)O)O)O)O)O)O)O)O)O)O DIGQNXIGRZPYDK-WKSCXVIASA-N 0.000 description 1
- OCUSNPIJIZCRSZ-ZTZWCFDHSA-N (2s)-2-amino-3-methylbutanoic acid;(2s)-2-amino-4-methylpentanoic acid;(2s,3s)-2-amino-3-methylpentanoic acid Chemical compound CC(C)[C@H](N)C(O)=O.CC[C@H](C)[C@H](N)C(O)=O.CC(C)C[C@H](N)C(O)=O OCUSNPIJIZCRSZ-ZTZWCFDHSA-N 0.000 description 1
- FYGDTMLNYKFZSV-WFYNLLPOSA-N (2s,3r,4s,5s,6r)-2-[(2r,4r,5r,6s)-4,5-dihydroxy-2-(hydroxymethyl)-6-[(2r,3s,4r,5r,6s)-4,5,6-trihydroxy-2-(hydroxymethyl)oxan-3-yl]oxyoxan-3-yl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol Chemical compound O[C@@H]1[C@@H](O)[C@H](O)[C@@H](CO)O[C@H]1OC1[C@@H](CO)O[C@@H](O[C@@H]2[C@H](O[C@H](O)[C@H](O)[C@H]2O)CO)[C@H](O)[C@H]1O FYGDTMLNYKFZSV-WFYNLLPOSA-N 0.000 description 1
- VXWBQOJISHAKKM-UHFFFAOYSA-N (4-formylphenyl)boronic acid Chemical compound OB(O)C1=CC=C(C=O)C=C1 VXWBQOJISHAKKM-UHFFFAOYSA-N 0.000 description 1
- FQVLRGLGWNWPSS-BXBUPLCLSA-N (4r,7s,10s,13s,16r)-16-acetamido-13-(1h-imidazol-5-ylmethyl)-10-methyl-6,9,12,15-tetraoxo-7-propan-2-yl-1,2-dithia-5,8,11,14-tetrazacycloheptadecane-4-carboxamide Chemical compound N1C(=O)[C@@H](NC(C)=O)CSSC[C@@H](C(N)=O)NC(=O)[C@H](C(C)C)NC(=O)[C@H](C)NC(=O)[C@@H]1CC1=CN=CN1 FQVLRGLGWNWPSS-BXBUPLCLSA-N 0.000 description 1
- PSBDWGZCVUAZQS-UHFFFAOYSA-N (dimethylsulfonio)acetate Chemical compound C[S+](C)CC([O-])=O PSBDWGZCVUAZQS-UHFFFAOYSA-N 0.000 description 1
- 108091032973 (ribonucleotides)n+m Proteins 0.000 description 1
- OSSNTDFYBPYIEC-UHFFFAOYSA-N 1-ethenylimidazole Chemical compound C=CN1C=CN=C1 OSSNTDFYBPYIEC-UHFFFAOYSA-N 0.000 description 1
- HZAXFHJVJLSVMW-UHFFFAOYSA-N 2-Aminoethan-1-ol Chemical compound NCCO HZAXFHJVJLSVMW-UHFFFAOYSA-N 0.000 description 1
- PFBBCIYIKJWDIN-BUHFOSPRSA-N 2-[(e)-tetradec-1-enyl]butanedioic acid Chemical compound CCCCCCCCCCCC\C=C\C(C(O)=O)CC(O)=O PFBBCIYIKJWDIN-BUHFOSPRSA-N 0.000 description 1
- URDCARMUOSMFFI-UHFFFAOYSA-N 2-[2-[bis(carboxymethyl)amino]ethyl-(2-hydroxyethyl)amino]acetic acid Chemical compound OCCN(CC(O)=O)CCN(CC(O)=O)CC(O)=O URDCARMUOSMFFI-UHFFFAOYSA-N 0.000 description 1
- MHOFGBJTSNWTDT-UHFFFAOYSA-M 2-[n-ethyl-4-[(6-methoxy-3-methyl-1,3-benzothiazol-3-ium-2-yl)diazenyl]anilino]ethanol;methyl sulfate Chemical compound COS([O-])(=O)=O.C1=CC(N(CCO)CC)=CC=C1N=NC1=[N+](C)C2=CC=C(OC)C=C2S1 MHOFGBJTSNWTDT-UHFFFAOYSA-M 0.000 description 1
- PUAQLLVFLMYYJJ-UHFFFAOYSA-N 2-aminopropiophenone Chemical compound CC(N)C(=O)C1=CC=CC=C1 PUAQLLVFLMYYJJ-UHFFFAOYSA-N 0.000 description 1
- MHKLKWCYGIBEQF-UHFFFAOYSA-N 4-(1,3-benzothiazol-2-ylsulfanyl)morpholine Chemical compound C1COCCN1SC1=NC2=CC=CC=C2S1 MHKLKWCYGIBEQF-UHFFFAOYSA-N 0.000 description 1
- GAUWGYGVFJBRRR-UHFFFAOYSA-N 4-decanoyloxybenzenesulfonic acid Chemical compound CCCCCCCCCC(=O)OC1=CC=C(S(O)(=O)=O)C=C1 GAUWGYGVFJBRRR-UHFFFAOYSA-N 0.000 description 1
- QTMHHQFADWIZCP-UHFFFAOYSA-N 4-decanoyloxybenzoic acid Chemical compound CCCCCCCCCC(=O)OC1=CC=C(C(O)=O)C=C1 QTMHHQFADWIZCP-UHFFFAOYSA-N 0.000 description 1
- CAERUOHSFJZTJD-UHFFFAOYSA-N 4-dodecanoyloxybenzenesulfonic acid Chemical compound CCCCCCCCCCCC(=O)OC1=CC=C(S(O)(=O)=O)C=C1 CAERUOHSFJZTJD-UHFFFAOYSA-N 0.000 description 1
- VNEUMNOZRFLRPI-UHFFFAOYSA-N 4-nonanoyloxybenzenesulfonic acid Chemical compound CCCCCCCCC(=O)OC1=CC=C(S(O)(=O)=O)C=C1 VNEUMNOZRFLRPI-UHFFFAOYSA-N 0.000 description 1
- 101710163881 5,6-dihydroxyindole-2-carboxylic acid oxidase Proteins 0.000 description 1
- UXEKKQRJIGKQNP-UHFFFAOYSA-N 5-(4-phenyltriazol-2-yl)-2-[2-[4-(4-phenyltriazol-2-yl)-2-sulfophenyl]ethenyl]benzenesulfonic acid Chemical compound OS(=O)(=O)C1=CC(N2N=C(C=N2)C=2C=CC=CC=2)=CC=C1C=CC(C(=C1)S(O)(=O)=O)=CC=C1N(N=1)N=CC=1C1=CC=CC=C1 UXEKKQRJIGKQNP-UHFFFAOYSA-N 0.000 description 1
- HRHCYUBSBRXMIN-UHFFFAOYSA-N 5-[(4,6-dianilino-1,3,5-triazin-2-yl)amino]-2-[2-[4-[(4,6-dianilino-1,3,5-triazin-2-yl)amino]-2-sulfophenyl]ethenyl]benzenesulfonic acid Chemical compound C=1C=C(C=CC=2C(=CC(NC=3N=C(NC=4C=CC=CC=4)N=C(NC=4C=CC=CC=4)N=3)=CC=2)S(O)(=O)=O)C(S(=O)(=O)O)=CC=1NC(N=C(NC=1C=CC=CC=1)N=1)=NC=1NC1=CC=CC=C1 HRHCYUBSBRXMIN-UHFFFAOYSA-N 0.000 description 1
- WNUUEQSIAHJIGU-UHFFFAOYSA-N 5-[[4-anilino-6-[2-hydroxyethyl(methyl)amino]-1,3,5-triazin-2-yl]amino]-2-[2-[4-[[4-anilino-6-[2-hydroxyethyl(methyl)amino]-1,3,5-triazin-2-yl]amino]-2-sulfophenyl]ethenyl]benzenesulfonic acid Chemical compound N=1C(NC=2C=C(C(C=CC=3C(=CC(NC=4N=C(N=C(NC=5C=CC=CC=5)N=4)N(C)CCO)=CC=3)S(O)(=O)=O)=CC=2)S(O)(=O)=O)=NC(N(CCO)C)=NC=1NC1=CC=CC=C1 WNUUEQSIAHJIGU-UHFFFAOYSA-N 0.000 description 1
- CNGYZEMWVAWWOB-VAWYXSNFSA-N 5-[[4-anilino-6-[bis(2-hydroxyethyl)amino]-1,3,5-triazin-2-yl]amino]-2-[(e)-2-[4-[[4-anilino-6-[bis(2-hydroxyethyl)amino]-1,3,5-triazin-2-yl]amino]-2-sulfophenyl]ethenyl]benzenesulfonic acid Chemical compound N=1C(NC=2C=C(C(\C=C\C=3C(=CC(NC=4N=C(N=C(NC=5C=CC=CC=5)N=4)N(CCO)CCO)=CC=3)S(O)(=O)=O)=CC=2)S(O)(=O)=O)=NC(N(CCO)CCO)=NC=1NC1=CC=CC=C1 CNGYZEMWVAWWOB-VAWYXSNFSA-N 0.000 description 1
- UZJGVXSQDRSSHU-UHFFFAOYSA-N 6-(1,3-dioxoisoindol-2-yl)hexaneperoxoic acid Chemical compound C1=CC=C2C(=O)N(CCCCCC(=O)OO)C(=O)C2=C1 UZJGVXSQDRSSHU-UHFFFAOYSA-N 0.000 description 1
- RZVAJINKPMORJF-UHFFFAOYSA-N Acetaminophen Chemical compound CC(=O)NC1=CC=C(O)C=C1 RZVAJINKPMORJF-UHFFFAOYSA-N 0.000 description 1
- 229920002126 Acrylic acid copolymer Polymers 0.000 description 1
- 229920002972 Acrylic fiber Polymers 0.000 description 1
- 241000222518 Agaricus Species 0.000 description 1
- 229920000936 Agarose Polymers 0.000 description 1
- 244000198134 Agave sisalana Species 0.000 description 1
- 102100034035 Alcohol dehydrogenase 1A Human genes 0.000 description 1
- GUBGYTABKSRVRQ-XLOQQCSPSA-N Alpha-Lactose Chemical compound O[C@@H]1[C@@H](O)[C@@H](O)[C@@H](CO)O[C@H]1O[C@@H]1[C@@H](CO)O[C@H](O)[C@H](O)[C@H]1O GUBGYTABKSRVRQ-XLOQQCSPSA-N 0.000 description 1
- 241000223600 Alternaria Species 0.000 description 1
- 241000534414 Anotopterus nikparini Species 0.000 description 1
- 101710152845 Arabinogalactan endo-beta-1,4-galactanase Proteins 0.000 description 1
- 240000003291 Armoracia rusticana Species 0.000 description 1
- 241000235349 Ascomycota Species 0.000 description 1
- DCXYFEDJOCDNAF-UHFFFAOYSA-N Asparagine Natural products OC(=O)C(N)CC(N)=O DCXYFEDJOCDNAF-UHFFFAOYSA-N 0.000 description 1
- 241000228215 Aspergillus aculeatus Species 0.000 description 1
- 101000961203 Aspergillus awamori Glucoamylase Proteins 0.000 description 1
- 101000690713 Aspergillus niger Alpha-glucosidase Proteins 0.000 description 1
- 101000756530 Aspergillus niger Endo-1,4-beta-xylanase B Proteins 0.000 description 1
- 101900127796 Aspergillus oryzae Glucoamylase Proteins 0.000 description 1
- 101900318521 Aspergillus oryzae Triosephosphate isomerase Proteins 0.000 description 1
- 108090001008 Avidin Proteins 0.000 description 1
- 108090000145 Bacillolysin Proteins 0.000 description 1
- 101000775727 Bacillus amyloliquefaciens Alpha-amylase Proteins 0.000 description 1
- 108010045681 Bacillus stearothermophilus neutral protease Proteins 0.000 description 1
- 101900040182 Bacillus subtilis Levansucrase Proteins 0.000 description 1
- 108010062877 Bacteriocins Proteins 0.000 description 1
- 241000221198 Basidiomycota Species 0.000 description 1
- 102100030981 Beta-alanine-activating enzyme Human genes 0.000 description 1
- 241000222490 Bjerkandera Species 0.000 description 1
- 241000222478 Bjerkandera adusta Species 0.000 description 1
- 241000283690 Bos taurus Species 0.000 description 1
- 241001453380 Burkholderia Species 0.000 description 1
- 241000589513 Burkholderia cepacia Species 0.000 description 1
- FJUIPMBNXJIKLZ-UHFFFAOYSA-N C1COCCN1C2=NC(=NC(=N2)NC3=CC=C(C=C3)C=CC4=C(C(=C(C=C4)NC5=NC(=NC(=N5)NC6=CC=CC=C6)N7CCOCC7)S(=O)(=O)O)S(=O)(=O)O)NC8=CC=CC=C8 Chemical compound C1COCCN1C2=NC(=NC(=N2)NC3=CC=C(C=C3)C=CC4=C(C(=C(C=C4)NC5=NC(=NC(=N5)NC6=CC=CC=C6)N7CCOCC7)S(=O)(=O)O)S(=O)(=O)O)NC8=CC=CC=C8 FJUIPMBNXJIKLZ-UHFFFAOYSA-N 0.000 description 1
- 101100327917 Caenorhabditis elegans chup-1 gene Proteins 0.000 description 1
- 241000282836 Camelus dromedarius Species 0.000 description 1
- 102100037633 Centrin-3 Human genes 0.000 description 1
- 241001466517 Ceriporiopsis aneirina Species 0.000 description 1
- 241001646018 Ceriporiopsis gilvescens Species 0.000 description 1
- 241001277875 Ceriporiopsis rivulosa Species 0.000 description 1
- 241000524302 Ceriporiopsis subrufa Species 0.000 description 1
- 241000259840 Chaetomidium Species 0.000 description 1
- 241001057137 Chaetomium fimeti Species 0.000 description 1
- 229920002101 Chitin Polymers 0.000 description 1
- 229920001661 Chitosan Polymers 0.000 description 1
- RKWGIWYCVPQPMF-UHFFFAOYSA-N Chloropropamide Chemical compound CCCNC(=O)NS(=O)(=O)C1=CC=C(Cl)C=C1 RKWGIWYCVPQPMF-UHFFFAOYSA-N 0.000 description 1
- 241000233652 Chytridiomycota Species 0.000 description 1
- 108020004638 Circular DNA Proteins 0.000 description 1
- 241000221760 Claviceps Species 0.000 description 1
- 241000228437 Cochliobolus Species 0.000 description 1
- 108020004705 Codon Proteins 0.000 description 1
- 108700010070 Codon Usage Proteins 0.000 description 1
- 241001085790 Coprinopsis Species 0.000 description 1
- 235000001673 Coprinus macrorhizus Nutrition 0.000 description 1
- 241001509964 Coptotermes Species 0.000 description 1
- 241000222356 Coriolus Species 0.000 description 1
- 241001252397 Corynascus Species 0.000 description 1
- 241000221755 Cryphonectria Species 0.000 description 1
- 241001559589 Cullen Species 0.000 description 1
- 102000018832 Cytochromes Human genes 0.000 description 1
- 108010052832 Cytochromes Proteins 0.000 description 1
- 230000004544 DNA amplification Effects 0.000 description 1
- 239000003155 DNA primer Substances 0.000 description 1
- 239000003298 DNA probe Substances 0.000 description 1
- 101100342470 Dictyostelium discoideum pkbA gene Proteins 0.000 description 1
- 108090000204 Dipeptidase 1 Proteins 0.000 description 1
- 241000935926 Diplodia Species 0.000 description 1
- 101710121765 Endo-1,4-beta-xylanase Proteins 0.000 description 1
- 101710132690 Endo-1,4-beta-xylanase A Proteins 0.000 description 1
- 101710147028 Endo-beta-1,4-galactanase Proteins 0.000 description 1
- 101710111935 Endo-beta-1,4-glucanase Proteins 0.000 description 1
- 108010067770 Endopeptidase K Proteins 0.000 description 1
- 101100385973 Escherichia coli (strain K12) cycA gene Proteins 0.000 description 1
- 239000001856 Ethyl cellulose Substances 0.000 description 1
- ZZSNKZQZMQGXPY-UHFFFAOYSA-N Ethyl cellulose Chemical compound CCOCC1OC(OC)C(OCC)C(OCC)C1OC1C(O)C(O)C(OC)C(CO)O1 ZZSNKZQZMQGXPY-UHFFFAOYSA-N 0.000 description 1
- 241000221433 Exidia Species 0.000 description 1
- 241000192125 Firmicutes Species 0.000 description 1
- 101150108358 GLAA gene Proteins 0.000 description 1
- 241000146398 Gelatoporia subvermispora Species 0.000 description 1
- 101100001650 Geobacillus stearothermophilus amyM gene Proteins 0.000 description 1
- 101000892220 Geobacillus thermodenitrificans (strain NG80-2) Long-chain-alcohol dehydrogenase 1 Proteins 0.000 description 1
- 229920001503 Glucan Polymers 0.000 description 1
- WQZGKKKJIJFFOK-GASJEMHNSA-N Glucose Natural products OC[C@H]1OC(O)[C@H](O)[C@@H](O)[C@@H]1O WQZGKKKJIJFFOK-GASJEMHNSA-N 0.000 description 1
- 102220487182 Growth/differentiation factor 9_E53Q_mutation Human genes 0.000 description 1
- 101150009006 HIS3 gene Proteins 0.000 description 1
- 101100295959 Halobacterium salinarum (strain ATCC 700922 / JCM 11081 / NRC-1) arcB gene Proteins 0.000 description 1
- 101100246753 Halobacterium salinarum (strain ATCC 700922 / JCM 11081 / NRC-1) pyrF gene Proteins 0.000 description 1
- 241000238631 Hexapoda Species 0.000 description 1
- 241001497663 Holomastigotoides Species 0.000 description 1
- 101000780443 Homo sapiens Alcohol dehydrogenase 1A Proteins 0.000 description 1
- 101000773364 Homo sapiens Beta-alanine-activating enzyme Proteins 0.000 description 1
- 101000880522 Homo sapiens Centrin-3 Proteins 0.000 description 1
- 101000882901 Homo sapiens Claudin-2 Proteins 0.000 description 1
- 241000223199 Humicola grisea Species 0.000 description 1
- 101001035458 Humicola insolens Endoglucanase-5 Proteins 0.000 description 1
- 229920000663 Hydroxyethyl cellulose Polymers 0.000 description 1
- 239000004354 Hydroxyethyl cellulose Substances 0.000 description 1
- 241000222342 Irpex Species 0.000 description 1
- 241000222344 Irpex lacteus Species 0.000 description 1
- 101710172072 Kexin Proteins 0.000 description 1
- 241001138401 Kluyveromyces lactis Species 0.000 description 1
- 241000824268 Kuma Species 0.000 description 1
- ZDXPYRJPNDTMRX-VKHMYHEASA-N L-glutamine Chemical compound OC(=O)[C@@H](N)CCC(N)=O ZDXPYRJPNDTMRX-VKHMYHEASA-N 0.000 description 1
- 125000003440 L-leucyl group Chemical group O=C([*])[C@](N([H])[H])([H])C([H])([H])C(C([H])([H])[H])([H])C([H])([H])[H] 0.000 description 1
- 125000001176 L-lysyl group Chemical group [H]N([H])[C@]([H])(C(=O)[*])C([H])([H])C([H])([H])C([H])([H])C(N([H])[H])([H])[H] 0.000 description 1
- FFEARJCKVFRZRR-BYPYZUCNSA-N L-methionine Chemical compound CSCC[C@H](N)C(O)=O FFEARJCKVFRZRR-BYPYZUCNSA-N 0.000 description 1
- FBOZXECLQNJBKD-ZDUSSCGKSA-N L-methotrexate Chemical compound C=1N=C2N=C(N)N=C(N)C2=NC=1CN(C)C1=CC=C(C(=O)N[C@@H](CCC(O)=O)C(O)=O)C=C1 FBOZXECLQNJBKD-ZDUSSCGKSA-N 0.000 description 1
- 125000000769 L-threonyl group Chemical group [H]N([H])[C@]([H])(C(=O)[*])[C@](O[H])(C([H])([H])[H])[H] 0.000 description 1
- QIVBCDIJIAJPQS-VIFPVBQESA-N L-tryptophane Chemical compound C1=CC=C2C(C[C@H](N)C(O)=O)=CNC2=C1 QIVBCDIJIAJPQS-VIFPVBQESA-N 0.000 description 1
- 125000003798 L-tyrosyl group Chemical group [H]N([H])[C@]([H])(C(=O)[*])C([H])([H])C1=C([H])C([H])=C(O[H])C([H])=C1[H] 0.000 description 1
- 125000003580 L-valyl group Chemical group [H]N([H])[C@]([H])(C(=O)[*])C(C([H])([H])[H])(C([H])([H])[H])[H] 0.000 description 1
- GUBGYTABKSRVRQ-QKKXKWKRSA-N Lactose Natural products OC[C@H]1O[C@@H](O[C@H]2[C@H](O)[C@@H](O)C(O)O[C@@H]2CO)[C@H](O)[C@@H](O)[C@H]1O GUBGYTABKSRVRQ-QKKXKWKRSA-N 0.000 description 1
- 241000222435 Lentinula Species 0.000 description 1
- 229920002097 Lichenin Polymers 0.000 description 1
- 101710098556 Lipase A Proteins 0.000 description 1
- 101710099648 Lysosomal acid lipase/cholesteryl ester hydrolase Proteins 0.000 description 1
- 102100026001 Lysosomal acid lipase/cholesteryl ester hydrolase Human genes 0.000 description 1
- 101150068888 MET3 gene Proteins 0.000 description 1
- 241001344131 Magnaporthe grisea Species 0.000 description 1
- 229920000057 Mannan Polymers 0.000 description 1
- 241000183011 Melanocarpus Species 0.000 description 1
- 241001184659 Melanocarpus albomyces Species 0.000 description 1
- 241000123315 Meripilus Species 0.000 description 1
- 102000003792 Metallothionein Human genes 0.000 description 1
- 241001465754 Metazoa Species 0.000 description 1
- 241001024304 Mino Species 0.000 description 1
- 241001661345 Moesziomyces antarcticus Species 0.000 description 1
- 241000235395 Mucor Species 0.000 description 1
- WHNWPMSKXPGLAX-UHFFFAOYSA-N N-Vinyl-2-pyrrolidone Chemical compound C=CN1CCCC1=O WHNWPMSKXPGLAX-UHFFFAOYSA-N 0.000 description 1
- 150000001204 N-oxides Chemical class 0.000 description 1
- 238000005481 NMR spectroscopy Methods 0.000 description 1
- 229930193140 Neomycin Natural products 0.000 description 1
- 101100022915 Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) cys-11 gene Proteins 0.000 description 1
- 108090000913 Nitrate Reductases Proteins 0.000 description 1
- 239000000020 Nitrocellulose Substances 0.000 description 1
- 239000006057 Non-nutritive feed additive Substances 0.000 description 1
- 239000004677 Nylon Substances 0.000 description 1
- 108700026244 Open Reading Frames Proteins 0.000 description 1
- 102100028200 Ornithine transcarbamylase, mitochondrial Human genes 0.000 description 1
- 101710113020 Ornithine transcarbamylase, mitochondrial Proteins 0.000 description 1
- 102100037214 Orotidine 5'-phosphate decarboxylase Human genes 0.000 description 1
- 108010055012 Orotidine-5'-phosphate decarboxylase Proteins 0.000 description 1
- 241000283973 Oryctolagus cuniculus Species 0.000 description 1
- 238000012408 PCR amplification Methods 0.000 description 1
- 229910019142 PO4 Inorganic materials 0.000 description 1
- 241000179039 Paenibacillus Species 0.000 description 1
- 206010034133 Pathogen resistance Diseases 0.000 description 1
- 241000222395 Phlebia Species 0.000 description 1
- 241000222397 Phlebia radiata Species 0.000 description 1
- ABLZXFCXXLZCGV-UHFFFAOYSA-N Phosphorous acid Chemical class OP(O)=O ABLZXFCXXLZCGV-UHFFFAOYSA-N 0.000 description 1
- 108091000080 Phosphotransferase Proteins 0.000 description 1
- 241000425347 Phyla <beetle> Species 0.000 description 1
- 241000222350 Pleurotus Species 0.000 description 1
- 244000252132 Pleurotus eryngii Species 0.000 description 1
- 235000001681 Pleurotus eryngii Nutrition 0.000 description 1
- 241001451060 Poitrasia Species 0.000 description 1
- 241000276498 Pollachius virens Species 0.000 description 1
- 229920000805 Polyaspartic acid Polymers 0.000 description 1
- 239000004743 Polypropylene Substances 0.000 description 1
- 229920002396 Polyurea Polymers 0.000 description 1
- 102000001253 Protein Kinase Human genes 0.000 description 1
- 101710194948 Protein phosphatase PhpP Proteins 0.000 description 1
- 241000168225 Pseudomonas alcaligenes Species 0.000 description 1
- 241000589755 Pseudomonas mendocina Species 0.000 description 1
- 241000589630 Pseudomonas pseudoalcaligenes Species 0.000 description 1
- 241000577556 Pseudomonas wisconsinensis Species 0.000 description 1
- 241000383860 Pseudoplectania Species 0.000 description 1
- 241001497658 Pseudotrichonympha Species 0.000 description 1
- 229920001131 Pulp (paper) Polymers 0.000 description 1
- 101710081551 Pyrolysin Proteins 0.000 description 1
- 108020004518 RNA Probes Proteins 0.000 description 1
- 239000003391 RNA probe Substances 0.000 description 1
- 241000235402 Rhizomucor Species 0.000 description 1
- 101000968489 Rhizomucor miehei Lipase Proteins 0.000 description 1
- 101100394989 Rhodopseudomonas palustris (strain ATCC BAA-98 / CGA009) hisI gene Proteins 0.000 description 1
- 101900354623 Saccharomyces cerevisiae Galactokinase Proteins 0.000 description 1
- 101900084120 Saccharomyces cerevisiae Triosephosphate isomerase Proteins 0.000 description 1
- 241000235343 Saccharomycetales Species 0.000 description 1
- 101100022918 Schizosaccharomyces pombe (strain 972 / ATCC 24843) sua1 gene Proteins 0.000 description 1
- 241000223255 Scytalidium Species 0.000 description 1
- 102220616233 Sialic acid-binding Ig-like lectin 14_R170K_mutation Human genes 0.000 description 1
- 239000004115 Sodium Silicate Substances 0.000 description 1
- 241000383403 Solen Species 0.000 description 1
- 108091081024 Start codon Proteins 0.000 description 1
- 241000120569 Streptococcus equi subsp. zooepidemicus Species 0.000 description 1
- 101100309436 Streptococcus mutans serotype c (strain ATCC 700610 / UA159) ftf gene Proteins 0.000 description 1
- 101100370749 Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) trpC1 gene Proteins 0.000 description 1
- 101100242848 Streptomyces hygroscopicus bar gene Proteins 0.000 description 1
- 241001518258 Streptomyces pristinaespiralis Species 0.000 description 1
- CZMRCDWAGMRECN-UGDNZRGBSA-N Sucrose Chemical compound O[C@H]1[C@H](O)[C@@H](CO)O[C@@]1(CO)O[C@@H]1[C@H](O)[C@@H](O)[C@H](O)[C@@H](CO)O1 CZMRCDWAGMRECN-UGDNZRGBSA-N 0.000 description 1
- 229930006000 Sucrose Natural products 0.000 description 1
- QAOWNCQODCNURD-UHFFFAOYSA-L Sulfate Chemical compound [O-]S([O-])(=O)=O QAOWNCQODCNURD-UHFFFAOYSA-L 0.000 description 1
- ULUAUXLGCMPNKK-UHFFFAOYSA-N Sulfobutanedioic acid Chemical compound OC(=O)CC(C(O)=O)S(O)(=O)=O ULUAUXLGCMPNKK-UHFFFAOYSA-N 0.000 description 1
- 241001215623 Talaromyces cellulolyticus Species 0.000 description 1
- 241001136494 Talaromyces funiculosus Species 0.000 description 1
- 239000004098 Tetracycline Substances 0.000 description 1
- 101100157012 Thermoanaerobacterium saccharolyticum (strain DSM 8691 / JW/SL-YS485) xynB gene Proteins 0.000 description 1
- 241000203780 Thermobifida fusca Species 0.000 description 1
- 108090001109 Thermolysin Proteins 0.000 description 1
- 241000223257 Thermomyces Species 0.000 description 1
- 241000183057 Thielavia microspora Species 0.000 description 1
- 241000182980 Thielavia ovispora Species 0.000 description 1
- 241000183053 Thielavia subthermophila Species 0.000 description 1
- 108010022394 Threonine synthase Proteins 0.000 description 1
- 241000222354 Trametes Species 0.000 description 1
- 241000222357 Trametes hirsuta Species 0.000 description 1
- 241000222355 Trametes versicolor Species 0.000 description 1
- 241000217816 Trametes villosa Species 0.000 description 1
- 102000004357 Transferases Human genes 0.000 description 1
- 108090000992 Transferases Proteins 0.000 description 1
- 241000215642 Trichophaea Species 0.000 description 1
- 102220470553 Tryptase delta_Q87E_mutation Human genes 0.000 description 1
- QIVBCDIJIAJPQS-UHFFFAOYSA-N Tryptophan Natural products C1=CC=C2C(CC(N)C(O)=O)=CNC2=C1 QIVBCDIJIAJPQS-UHFFFAOYSA-N 0.000 description 1
- 101150050575 URA3 gene Proteins 0.000 description 1
- 241000082085 Verticillium <Phyllachorales> Species 0.000 description 1
- 241000700605 Viruses Species 0.000 description 1
- 241001507667 Volvariella Species 0.000 description 1
- 241000409279 Xerochrysium dermatitidis Species 0.000 description 1
- 241001523965 Xylaria Species 0.000 description 1
- 229920002000 Xyloglucan Polymers 0.000 description 1
- 241000235015 Yarrowia lipolytica Species 0.000 description 1
- 229910021536 Zeolite Inorganic materials 0.000 description 1
- 241000758405 Zoopagomycotina Species 0.000 description 1
- JLCPHMBAVCMARE-UHFFFAOYSA-N [3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-[[3-[[3-[[3-[[3-[[3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-hydroxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methyl [5-(6-aminopurin-9-yl)-2-(hydroxymethyl)oxolan-3-yl] hydrogen phosphate Polymers Cc1cn(C2CC(OP(O)(=O)OCC3OC(CC3OP(O)(=O)OCC3OC(CC3O)n3cnc4c3nc(N)[nH]c4=O)n3cnc4c3nc(N)[nH]c4=O)C(COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3CO)n3cnc4c(N)ncnc34)n3ccc(N)nc3=O)n3cnc4c(N)ncnc34)n3ccc(N)nc3=O)n3ccc(N)nc3=O)n3ccc(N)nc3=O)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cc(C)c(=O)[nH]c3=O)n3cc(C)c(=O)[nH]c3=O)n3ccc(N)nc3=O)n3cc(C)c(=O)[nH]c3=O)n3cnc4c3nc(N)[nH]c4=O)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)O2)c(=O)[nH]c1=O JLCPHMBAVCMARE-UHFFFAOYSA-N 0.000 description 1
- 238000010521 absorption reaction Methods 0.000 description 1
- 230000002378 acidificating effect Effects 0.000 description 1
- 108010045649 agarase Proteins 0.000 description 1
- 238000000246 agarose gel electrophoresis Methods 0.000 description 1
- 230000002776 aggregation Effects 0.000 description 1
- 238000012867 alanine scanning Methods 0.000 description 1
- 150000001298 alcohols Chemical class 0.000 description 1
- 229910052783 alkali metal Inorganic materials 0.000 description 1
- 238000012870 ammonium sulfate precipitation Methods 0.000 description 1
- 229960000723 ampicillin Drugs 0.000 description 1
- AVKUERGKIZMTKX-NJBDSQKTSA-N ampicillin Chemical compound C1([C@@H](N)C(=O)N[C@H]2[C@H]3SC([C@@H](N3C2=O)C(O)=O)(C)C)=CC=CC=C1 AVKUERGKIZMTKX-NJBDSQKTSA-N 0.000 description 1
- 238000004458 analytical method Methods 0.000 description 1
- 150000008064 anhydrides Chemical class 0.000 description 1
- 230000003254 anti-foaming effect Effects 0.000 description 1
- 229940053200 antiepileptics fatty acid derivative Drugs 0.000 description 1
- 230000000890 antigenic effect Effects 0.000 description 1
- 101150009206 aprE gene Proteins 0.000 description 1
- 239000007864 aqueous solution Substances 0.000 description 1
- 229920006231 aramid fiber Polymers 0.000 description 1
- 101150008194 argB gene Proteins 0.000 description 1
- 229920003235 aromatic polyamide Polymers 0.000 description 1
- 210000004507 artificial chromosome Anatomy 0.000 description 1
- 238000013528 artificial neural network Methods 0.000 description 1
- 235000009582 asparagine Nutrition 0.000 description 1
- 229960001230 asparagine Drugs 0.000 description 1
- 108010019077 beta-Amylase Proteins 0.000 description 1
- MSWZFWKMSRAUBD-UHFFFAOYSA-N beta-D-galactosamine Natural products NC1C(O)OC(CO)C(O)C1O MSWZFWKMSRAUBD-UHFFFAOYSA-N 0.000 description 1
- WQZGKKKJIJFFOK-VFUOTHLCSA-N beta-D-glucose Chemical compound OC[C@H]1O[C@@H](O)[C@H](O)[C@@H](O)[C@@H]1O WQZGKKKJIJFFOK-VFUOTHLCSA-N 0.000 description 1
- 108010051210 beta-Fructofuranosidase Proteins 0.000 description 1
- 108010047754 beta-Glucosidase Proteins 0.000 description 1
- 102000006995 beta-Glucosidase Human genes 0.000 description 1
- 229960003237 betaine Drugs 0.000 description 1
- 239000003139 biocide Substances 0.000 description 1
- 230000009141 biological interaction Effects 0.000 description 1
- 229960002685 biotin Drugs 0.000 description 1
- 235000020958 biotin Nutrition 0.000 description 1
- 239000011616 biotin Substances 0.000 description 1
- 150000001642 boronic acid derivatives Chemical class 0.000 description 1
- 239000006227 byproduct Substances 0.000 description 1
- 102220350531 c.80A>G Human genes 0.000 description 1
- 239000011575 calcium Substances 0.000 description 1
- 229910052791 calcium Inorganic materials 0.000 description 1
- 108010089934 carbohydrase Proteins 0.000 description 1
- 150000004649 carbonic acid derivatives Chemical class 0.000 description 1
- 125000003178 carboxy group Chemical group [H]OC(*)=O 0.000 description 1
- 229920003090 carboxymethyl hydroxyethyl cellulose Polymers 0.000 description 1
- 239000000969 carrier Substances 0.000 description 1
- 210000000085 cashmere Anatomy 0.000 description 1
- 238000012219 cassette mutagenesis Methods 0.000 description 1
- 125000002091 cationic group Chemical group 0.000 description 1
- 230000034303 cell budding Effects 0.000 description 1
- 239000013592 cell lysate Substances 0.000 description 1
- 230000010307 cell transformation Effects 0.000 description 1
- 210000002421 cell wall Anatomy 0.000 description 1
- 230000001413 cellular effect Effects 0.000 description 1
- 229940106135 cellulose Drugs 0.000 description 1
- 229920003086 cellulose ether Polymers 0.000 description 1
- 229920003174 cellulose-based polymer Polymers 0.000 description 1
- 238000005119 centrifugation Methods 0.000 description 1
- 235000013339 cereals Nutrition 0.000 description 1
- 238000006243 chemical reaction Methods 0.000 description 1
- WIIZWVCIJKGZOK-RKDXNWHRSA-N chloramphenicol Chemical compound ClC(Cl)C(=O)N[C@H](CO)[C@H](O)C1=CC=C([N+]([O-])=O)C=C1 WIIZWVCIJKGZOK-RKDXNWHRSA-N 0.000 description 1
- 229960005091 chloramphenicol Drugs 0.000 description 1
- 238000011098 chromatofocusing Methods 0.000 description 1
- 238000004587 chromatography analysis Methods 0.000 description 1
- 239000004927 clay Substances 0.000 description 1
- 238000010367 cloning Methods 0.000 description 1
- 239000000084 colloidal system Substances 0.000 description 1
- 239000013065 commercial product Substances 0.000 description 1
- 238000004590 computer program Methods 0.000 description 1
- 239000000470 constituent Substances 0.000 description 1
- 238000011109 contamination Methods 0.000 description 1
- 230000007797 corrosion Effects 0.000 description 1
- 239000002537 cosmetic Substances 0.000 description 1
- 239000006071 cream Substances 0.000 description 1
- 229930007927 cymene Natural products 0.000 description 1
- 101150005799 dagA gene Proteins 0.000 description 1
- 235000013365 dairy product Nutrition 0.000 description 1
- 238000013461 design Methods 0.000 description 1
- 150000005690 diesters Chemical class 0.000 description 1
- GSPKZYJPUDYKPI-UHFFFAOYSA-N diethoxy sulfate Chemical compound CCOOS(=O)(=O)OOCC GSPKZYJPUDYKPI-UHFFFAOYSA-N 0.000 description 1
- 238000002050 diffraction method Methods 0.000 description 1
- 229940079919 digestives enzyme preparation Drugs 0.000 description 1
- REZZEXDLIUJMMS-UHFFFAOYSA-M dimethyldioctadecylammonium chloride Chemical compound [Cl-].CCCCCCCCCCCCCCCCCC[N+](C)(C)CCCCCCCCCCCCCCCCCC REZZEXDLIUJMMS-UHFFFAOYSA-M 0.000 description 1
- HNPSIPDUKPIQMN-UHFFFAOYSA-N dioxosilane;oxo(oxoalumanyloxy)alumane Chemical compound O=[Si]=O.O=[Al]O[Al]=O HNPSIPDUKPIQMN-UHFFFAOYSA-N 0.000 description 1
- 239000001177 diphosphate Substances 0.000 description 1
- XPPKVPWEQAFLFU-UHFFFAOYSA-J diphosphate(4-) Chemical class [O-]P([O-])(=O)OP([O-])([O-])=O XPPKVPWEQAFLFU-UHFFFAOYSA-J 0.000 description 1
- 230000008034 disappearance Effects 0.000 description 1
- 239000007884 disintegrant Substances 0.000 description 1
- VTIIJXUACCWYHX-UHFFFAOYSA-L disodium;carboxylatooxy carbonate Chemical compound [Na+].[Na+].[O-]C(=O)OOC([O-])=O VTIIJXUACCWYHX-UHFFFAOYSA-L 0.000 description 1
- 125000005066 dodecenyl group Chemical group C(=CCCCCCCCCCC)* 0.000 description 1
- GMSCBRSQMRDRCD-UHFFFAOYSA-N dodecyl 2-methylprop-2-enoate Chemical compound CCCCCCCCCCCCOC(=O)C(C)=C GMSCBRSQMRDRCD-UHFFFAOYSA-N 0.000 description 1
- 238000007876 drug discovery Methods 0.000 description 1
- 102220500059 eIF5-mimic protein 2_S54V_mutation Human genes 0.000 description 1
- 238000002003 electron diffraction Methods 0.000 description 1
- 108010092413 endoglucanase V Proteins 0.000 description 1
- 108010002082 endometriosis protein-1 Proteins 0.000 description 1
- 230000007613 environmental effect Effects 0.000 description 1
- 230000007515 enzymatic degradation Effects 0.000 description 1
- 238000001952 enzyme assay Methods 0.000 description 1
- 150000002148 esters Chemical class 0.000 description 1
- 150000002169 ethanolamines Chemical class 0.000 description 1
- RTZKZFJDLAIYFH-UHFFFAOYSA-N ether Substances CCOCC RTZKZFJDLAIYFH-UHFFFAOYSA-N 0.000 description 1
- 229920001249 ethyl cellulose Polymers 0.000 description 1
- 235000019325 ethyl cellulose Nutrition 0.000 description 1
- 210000003527 eukaryotic cell Anatomy 0.000 description 1
- 238000001704 evaporation Methods 0.000 description 1
- 230000008020 evaporation Effects 0.000 description 1
- 230000007717 exclusion Effects 0.000 description 1
- 108010038658 exo-1,4-beta-D-xylosidase Proteins 0.000 description 1
- 235000019387 fatty acid methyl ester Nutrition 0.000 description 1
- 238000001914 filtration Methods 0.000 description 1
- 238000010230 functional analysis Methods 0.000 description 1
- 108020001507 fusion proteins Proteins 0.000 description 1
- 102000037865 fusion proteins Human genes 0.000 description 1
- 239000010437 gem Substances 0.000 description 1
- 229960002442 glucosamine Drugs 0.000 description 1
- 239000008103 glucose Substances 0.000 description 1
- ZDXPYRJPNDTMRX-UHFFFAOYSA-N glutamine Natural products OC(=O)C(N)CCC(N)=O ZDXPYRJPNDTMRX-UHFFFAOYSA-N 0.000 description 1
- 235000011187 glycerol Nutrition 0.000 description 1
- 150000002334 glycols Chemical class 0.000 description 1
- 230000013595 glycosylation Effects 0.000 description 1
- 238000006206 glycosylation reaction Methods 0.000 description 1
- 238000005469 granulation Methods 0.000 description 1
- 230000003179 granulation Effects 0.000 description 1
- 230000012010 growth Effects 0.000 description 1
- 239000001963 growth medium Substances 0.000 description 1
- 230000007062 hydrolysis Effects 0.000 description 1
- 238000006460 hydrolysis reaction Methods 0.000 description 1
- 235000019447 hydroxyethyl cellulose Nutrition 0.000 description 1
- 229920003088 hydroxypropyl methyl cellulose Polymers 0.000 description 1
- 108010002685 hygromycin-B kinase Proteins 0.000 description 1
- 230000001939 inductive effect Effects 0.000 description 1
- 229910052500 inorganic mineral Inorganic materials 0.000 description 1
- 229910017053 inorganic salt Inorganic materials 0.000 description 1
- 230000017730 intein-mediated protein splicing Effects 0.000 description 1
- 239000000543 intermediate Substances 0.000 description 1
- 230000003834 intracellular effect Effects 0.000 description 1
- 239000001573 invertase Substances 0.000 description 1
- 235000011073 invertase Nutrition 0.000 description 1
- 238000005342 ion exchange Methods 0.000 description 1
- 238000001155 isoelectric focusing Methods 0.000 description 1
- SBUJHOSQTJFQJX-NOAMYHISSA-N kanamycin Chemical compound O[C@@H]1[C@@H](O)[C@H](O)[C@@H](CN)O[C@@H]1O[C@H]1[C@H](O)[C@@H](O[C@@H]2[C@@H]([C@@H](N)[C@H](O)[C@@H](CO)O2)O)[C@H](N)C[C@@H]1N SBUJHOSQTJFQJX-NOAMYHISSA-N 0.000 description 1
- 229960000318 kanamycin Drugs 0.000 description 1
- 229930027917 kanamycin Natural products 0.000 description 1
- 229930182823 kanamycin A Natural products 0.000 description 1
- 239000008101 lactose Substances 0.000 description 1
- 150000002632 lipids Chemical class 0.000 description 1
- 239000006210 lotion Substances 0.000 description 1
- 101150039489 lysZ gene Proteins 0.000 description 1
- 230000014759 maintenance of location Effects 0.000 description 1
- 229930182817 methionine Natural products 0.000 description 1
- 229960000485 methotrexate Drugs 0.000 description 1
- 229920000609 methyl cellulose Polymers 0.000 description 1
- 150000004702 methyl esters Chemical class 0.000 description 1
- 239000001923 methylcellulose Substances 0.000 description 1
- 235000010981 methylcellulose Nutrition 0.000 description 1
- 230000002906 microbiologic effect Effects 0.000 description 1
- 239000011707 mineral Substances 0.000 description 1
- 235000010755 mineral Nutrition 0.000 description 1
- 210000000050 mohair Anatomy 0.000 description 1
- 238000010369 molecular cloning Methods 0.000 description 1
- 150000004682 monohydrates Chemical class 0.000 description 1
- 231100000219 mutagenic Toxicity 0.000 description 1
- 230000003505 mutagenic effect Effects 0.000 description 1
- 229960004927 neomycin Drugs 0.000 description 1
- 101150095344 niaD gene Proteins 0.000 description 1
- MGFYIUFZLHCRTH-UHFFFAOYSA-N nitrilotriacetic acid Chemical compound OC(=O)CN(CC(O)=O)CC(O)=O MGFYIUFZLHCRTH-UHFFFAOYSA-N 0.000 description 1
- 229920001220 nitrocellulos Polymers 0.000 description 1
- 239000004745 nonwoven fabric Substances 0.000 description 1
- SNQQPOLDUKLAAF-UHFFFAOYSA-N nonylphenol Chemical class CCCCCCCCCC1=CC=CC=C1O SNQQPOLDUKLAAF-UHFFFAOYSA-N 0.000 description 1
- 101150105920 npr gene Proteins 0.000 description 1
- 101150017837 nprM gene Proteins 0.000 description 1
- 229920001778 nylon Polymers 0.000 description 1
- JRZJOMJEPLMPRA-UHFFFAOYSA-N olefin Natural products CCCCCCCC=C JRZJOMJEPLMPRA-UHFFFAOYSA-N 0.000 description 1
- 239000011368 organic material Substances 0.000 description 1
- 108090000021 oryzin Proteins 0.000 description 1
- 230000003647 oxidation Effects 0.000 description 1
- 238000007254 oxidation reaction Methods 0.000 description 1
- HFPZCAJZSCWRBC-UHFFFAOYSA-N p-cymene Chemical compound CC(C)C1=CC=C(C)C=C1 HFPZCAJZSCWRBC-UHFFFAOYSA-N 0.000 description 1
- 239000012188 paraffin wax Substances 0.000 description 1
- 230000036961 partial effect Effects 0.000 description 1
- 101150019841 penP gene Proteins 0.000 description 1
- 108040007629 peroxidase activity proteins Proteins 0.000 description 1
- JRKICGRDRMAZLK-UHFFFAOYSA-L peroxydisulfate Chemical compound [O-]S(=O)(=O)OOS([O-])(=O)=O JRKICGRDRMAZLK-UHFFFAOYSA-L 0.000 description 1
- 150000004968 peroxymonosulfuric acids Chemical class 0.000 description 1
- 125000005342 perphosphate group Chemical group 0.000 description 1
- 235000020030 perry Nutrition 0.000 description 1
- 238000002823 phage display Methods 0.000 description 1
- 238000005191 phase separation Methods 0.000 description 1
- JTJMJGYZQZDUJJ-UHFFFAOYSA-N phencyclidine Chemical compound C1CCCCN1C1(C=2C=CC=CC=2)CCCCC1 JTJMJGYZQZDUJJ-UHFFFAOYSA-N 0.000 description 1
- HXITXNWTGFUOAU-UHFFFAOYSA-N phenylboronic acid Chemical class OB(O)C1=CC=CC=C1 HXITXNWTGFUOAU-UHFFFAOYSA-N 0.000 description 1
- 239000010452 phosphate Substances 0.000 description 1
- NBIIXXVUZAFLBC-UHFFFAOYSA-K phosphate Chemical compound [O-]P([O-])([O-])=O NBIIXXVUZAFLBC-UHFFFAOYSA-K 0.000 description 1
- 108010082527 phosphinothricin N-acetyltransferase Proteins 0.000 description 1
- 230000026731 phosphorylation Effects 0.000 description 1
- 238000006366 phosphorylation reaction Methods 0.000 description 1
- 102000020233 phosphotransferase Human genes 0.000 description 1
- 238000005222 photoaffinity labeling Methods 0.000 description 1
- IEQIEDJGQAUEQZ-UHFFFAOYSA-N phthalocyanine Chemical compound N1C(N=C2C3=CC=CC=C3C(N=C3C4=CC=CC=C4C(=N4)N3)=N2)=C(C=CC=C2)C2=C1N=C1C2=CC=CC=C2C4=N1 IEQIEDJGQAUEQZ-UHFFFAOYSA-N 0.000 description 1
- 229920000196 poly(lauryl methacrylate) Polymers 0.000 description 1
- 238000002264 polyacrylamide gel electrophoresis Methods 0.000 description 1
- 229920000058 polyacrylate Polymers 0.000 description 1
- 229920000768 polyamine Polymers 0.000 description 1
- 108010064470 polyaspartate Proteins 0.000 description 1
- 229920000139 polyethylene terephthalate Polymers 0.000 description 1
- 239000005020 polyethylene terephthalate Substances 0.000 description 1
- 229920001155 polypropylene Polymers 0.000 description 1
- 229920001451 polypropylene glycol Polymers 0.000 description 1
- 150000004804 polysaccharides Polymers 0.000 description 1
- 229920001296 polysiloxane Polymers 0.000 description 1
- 229920000136 polysorbate Polymers 0.000 description 1
- 229920006306 polyurethane fiber Polymers 0.000 description 1
- 239000004800 polyvinyl chloride Substances 0.000 description 1
- 229920000915 polyvinyl chloride Polymers 0.000 description 1
- 230000001124 posttranscriptional effect Effects 0.000 description 1
- 239000004300 potassium benzoate Substances 0.000 description 1
- 238000001556 precipitation Methods 0.000 description 1
- 239000002243 precursor Substances 0.000 description 1
- 238000002360 preparation method Methods 0.000 description 1
- 125000002924 primary amino group Chemical group [H]N([H])* 0.000 description 1
- 239000013615 primer Substances 0.000 description 1
- 238000012545 processing Methods 0.000 description 1
- 210000001236 prokaryotic cell Anatomy 0.000 description 1
- 235000019260 propionic acid Nutrition 0.000 description 1
- 239000004405 propyl p-hydroxybenzoate Substances 0.000 description 1
- 235000019833 protease Nutrition 0.000 description 1
- 108060006633 protein kinase Proteins 0.000 description 1
- 238000001742 protein purification Methods 0.000 description 1
- 101150108007 prs gene Proteins 0.000 description 1
- 101150086435 prs1 gene Proteins 0.000 description 1
- 101150070305 prsA gene Proteins 0.000 description 1
- 230000006798 recombination Effects 0.000 description 1
- 238000005215 recombination Methods 0.000 description 1
- 230000008929 regeneration Effects 0.000 description 1
- 238000011069 regeneration method Methods 0.000 description 1
- 230000022532 regulation of transcription, DNA-dependent Effects 0.000 description 1
- 230000003362 replicative effect Effects 0.000 description 1
- 230000004044 response Effects 0.000 description 1
- 230000000717 retained effect Effects 0.000 description 1
- 238000010839 reverse transcription Methods 0.000 description 1
- 108020004418 ribosomal RNA Proteins 0.000 description 1
- 102220220139 rs1060500788 Human genes 0.000 description 1
- 102200131574 rs11556620 Human genes 0.000 description 1
- 102200114213 rs137852930 Human genes 0.000 description 1
- 102220036452 rs137882485 Human genes 0.000 description 1
- 102200065573 rs140660066 Human genes 0.000 description 1
- 102200118280 rs33918343 Human genes 0.000 description 1
- 102200004009 rs36096184 Human genes 0.000 description 1
- 102220243297 rs374524755 Human genes 0.000 description 1
- 102200128586 rs397508464 Human genes 0.000 description 1
- 102220289974 rs757282628 Human genes 0.000 description 1
- 102200025035 rs786203989 Human genes 0.000 description 1
- 102220099575 rs878853725 Human genes 0.000 description 1
- 102220160907 rs886062986 Human genes 0.000 description 1
- 101150025220 sacB gene Proteins 0.000 description 1
- 235000014438 salad dressings Nutrition 0.000 description 1
- 238000000926 separation method Methods 0.000 description 1
- 101150091813 shfl gene Proteins 0.000 description 1
- 239000000344 soap Substances 0.000 description 1
- 229940077386 sodium benzenesulfonate Drugs 0.000 description 1
- 229940001593 sodium carbonate Drugs 0.000 description 1
- 229910000029 sodium carbonate Inorganic materials 0.000 description 1
- 235000017550 sodium carbonate Nutrition 0.000 description 1
- 238000002415 sodium dodecyl sulfate polyacrylamide gel electrophoresis Methods 0.000 description 1
- 235000019795 sodium metasilicate Nutrition 0.000 description 1
- 229940045872 sodium percarbonate Drugs 0.000 description 1
- NTHWMYGWWRZVTN-UHFFFAOYSA-N sodium silicate Chemical compound [Na+].[Na+].[O-][Si]([O-])=O NTHWMYGWWRZVTN-UHFFFAOYSA-N 0.000 description 1
- 229910052911 sodium silicate Inorganic materials 0.000 description 1
- DGSDBJMBHCQYGN-UHFFFAOYSA-M sodium;2-ethylhexyl sulfate Chemical compound [Na+].CCCCC(CC)COS([O-])(=O)=O DGSDBJMBHCQYGN-UHFFFAOYSA-M 0.000 description 1
- OMSMEHWLFJLBSH-UHFFFAOYSA-M sodium;2-hydroxynaphthalene-1-carboxylate Chemical compound [Na+].C1=CC=CC2=C(C([O-])=O)C(O)=CC=C21 OMSMEHWLFJLBSH-UHFFFAOYSA-M 0.000 description 1
- LIAJJWHZAFEJEZ-UHFFFAOYSA-M sodium;2-hydroxynaphthalene-1-sulfonate Chemical compound [Na+].C1=CC=CC2=C(S([O-])(=O)=O)C(O)=CC=C21 LIAJJWHZAFEJEZ-UHFFFAOYSA-M 0.000 description 1
- AXMCIYLNKNGNOT-UHFFFAOYSA-N sodium;3-[[4-[(4-dimethylazaniumylidenecyclohexa-2,5-dien-1-ylidene)-[4-[ethyl-[(3-sulfophenyl)methyl]amino]phenyl]methyl]-n-ethylanilino]methyl]benzenesulfonate Chemical compound [Na+].C=1C=C(C(=C2C=CC(C=C2)=[N+](C)C)C=2C=CC(=CC=2)N(CC)CC=2C=C(C=CC=2)S([O-])(=O)=O)C=CC=1N(CC)CC1=CC=CC(S(O)(=O)=O)=C1 AXMCIYLNKNGNOT-UHFFFAOYSA-N 0.000 description 1
- GSYPNDDXWAZDJB-UHFFFAOYSA-M sodium;4-(3,5,5-trimethylhexanoyloxy)benzenesulfonate Chemical compound [Na+].CC(C)(C)CC(C)CC(=O)OC1=CC=C(S([O-])(=O)=O)C=C1 GSYPNDDXWAZDJB-UHFFFAOYSA-M 0.000 description 1
- MZSDGDXXBZSFTG-UHFFFAOYSA-M sodium;benzenesulfonate Chemical compound [Na+].[O-]S(=O)(=O)C1=CC=CC=C1 MZSDGDXXBZSFTG-UHFFFAOYSA-M 0.000 description 1
- MWNQXXOSWHCCOZ-UHFFFAOYSA-L sodium;oxido carbonate Chemical compound [Na+].[O-]OC([O-])=O MWNQXXOSWHCCOZ-UHFFFAOYSA-L 0.000 description 1
- 238000010563 solid-state fermentation Methods 0.000 description 1
- 238000007614 solvation Methods 0.000 description 1
- 239000004759 spandex Substances 0.000 description 1
- UNFWWIHTNXNPBV-WXKVUWSESA-N spectinomycin Chemical compound O([C@@H]1[C@@H](NC)[C@@H](O)[C@H]([C@@H]([C@H]1O1)O)NC)[C@]2(O)[C@H]1O[C@H](C)CC2=O UNFWWIHTNXNPBV-WXKVUWSESA-N 0.000 description 1
- 229960000268 spectinomycin Drugs 0.000 description 1
- 238000001228 spectrum Methods 0.000 description 1
- 230000002269 spontaneous effect Effects 0.000 description 1
- 238000001694 spray drying Methods 0.000 description 1
- 238000010561 standard procedure Methods 0.000 description 1
- 239000005720 sucrose Substances 0.000 description 1
- 229940117986 sulfobetaine Drugs 0.000 description 1
- BDHFUVZGWQCTTF-UHFFFAOYSA-M sulfonate Chemical compound [O-]S(=O)=O BDHFUVZGWQCTTF-UHFFFAOYSA-M 0.000 description 1
- 150000003457 sulfones Chemical class 0.000 description 1
- 230000002194 synthesizing effect Effects 0.000 description 1
- 239000012209 synthetic fiber Substances 0.000 description 1
- 229920001059 synthetic polymer Polymers 0.000 description 1
- 229960002180 tetracycline Drugs 0.000 description 1
- 229930101283 tetracycline Natural products 0.000 description 1
- 235000019364 tetracycline Nutrition 0.000 description 1
- 150000003522 tetracyclines Chemical class 0.000 description 1
- 150000004685 tetrahydrates Chemical class 0.000 description 1
- 108010031354 thermitase Proteins 0.000 description 1
- 150000003588 threonines Chemical group 0.000 description 1
- 239000000606 toothpaste Substances 0.000 description 1
- 229940034610 toothpaste Drugs 0.000 description 1
- 238000001890 transfection Methods 0.000 description 1
- 230000001131 transforming effect Effects 0.000 description 1
- 150000003626 triacylglycerols Chemical class 0.000 description 1
- UFTFJSFQGQCHQW-UHFFFAOYSA-N triformin Chemical compound O=COCC(OC=O)COC=O UFTFJSFQGQCHQW-UHFFFAOYSA-N 0.000 description 1
- 235000011178 triphosphate Nutrition 0.000 description 1
- 239000001226 triphosphate Substances 0.000 description 1
- 125000002264 triphosphate group Chemical class [H]OP(=O)(O[H])OP(=O)(O[H])OP(=O)(O[H])O* 0.000 description 1
- VRVDFJOCCWSFLI-UHFFFAOYSA-K trisodium 3-[[4-[(6-anilino-1-hydroxy-3-sulfonatonaphthalen-2-yl)diazenyl]-5-methoxy-2-methylphenyl]diazenyl]naphthalene-1,5-disulfonate Chemical compound [Na+].[Na+].[Na+].COc1cc(N=Nc2cc(c3cccc(c3c2)S([O-])(=O)=O)S([O-])(=O)=O)c(C)cc1N=Nc1c(O)c2ccc(Nc3ccccc3)cc2cc1S([O-])(=O)=O VRVDFJOCCWSFLI-UHFFFAOYSA-K 0.000 description 1
- 229960004418 trolamine Drugs 0.000 description 1
- 101150016309 trpC gene Proteins 0.000 description 1
- 238000011144 upstream manufacturing Methods 0.000 description 1
- 235000013311 vegetables Nutrition 0.000 description 1
- 125000000391 vinyl group Chemical group [H]C([*])=C([H])[H] 0.000 description 1
- 230000002087 whitening effect Effects 0.000 description 1
- 101150110790 xylB gene Proteins 0.000 description 1
- 210000005253 yeast cell Anatomy 0.000 description 1
- 150000003751 zinc Chemical class 0.000 description 1
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N9/00—Enzymes; Proenzymes; Compositions thereof; Processes for preparing, activating, inhibiting, separating or purifying enzymes
- C12N9/14—Hydrolases (3)
- C12N9/24—Hydrolases (3) acting on glycosyl compounds (3.2)
- C12N9/2402—Hydrolases (3) acting on glycosyl compounds (3.2) hydrolysing O- and S- glycosyl compounds (3.2.1)
- C12N9/2405—Glucanases
- C12N9/2434—Glucanases acting on beta-1,4-glucosidic bonds
- C12N9/2437—Cellulases (3.2.1.4; 3.2.1.74; 3.2.1.91; 3.2.1.150)
-
- C—CHEMISTRY; METALLURGY
- C11—ANIMAL OR VEGETABLE OILS, FATS, FATTY SUBSTANCES OR WAXES; FATTY ACIDS THEREFROM; DETERGENTS; CANDLES
- C11D—DETERGENT COMPOSITIONS; USE OF SINGLE SUBSTANCES AS DETERGENTS; SOAP OR SOAP-MAKING; RESIN SOAPS; RECOVERY OF GLYCEROL
- C11D3/00—Other compounding ingredients of detergent compositions covered in group C11D1/00
- C11D3/16—Organic compounds
- C11D3/38—Products with no well-defined composition, e.g. natural products
- C11D3/386—Preparations containing enzymes, e.g. protease or amylase
- C11D3/38636—Preparations containing enzymes, e.g. protease or amylase containing enzymes other than protease, amylase, lipase, cellulase, oxidase or reductase
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Y—ENZYMES
- C12Y302/00—Hydrolases acting on glycosyl compounds, i.e. glycosylases (3.2)
- C12Y302/01—Glycosidases, i.e. enzymes hydrolysing O- and S-glycosyl compounds (3.2.1)
- C12Y302/01004—Cellulase (3.2.1.4), i.e. endo-1,4-beta-glucanase
-
- C—CHEMISTRY; METALLURGY
- C11—ANIMAL OR VEGETABLE OILS, FATS, FATTY SUBSTANCES OR WAXES; FATTY ACIDS THEREFROM; DETERGENTS; CANDLES
- C11D—DETERGENT COMPOSITIONS; USE OF SINGLE SUBSTANCES AS DETERGENTS; SOAP OR SOAP-MAKING; RESIN SOAPS; RECOVERY OF GLYCEROL
- C11D2111/00—Cleaning compositions characterised by the objects to be cleaned; Cleaning compositions characterised by non-standard cleaning or washing processes
- C11D2111/10—Objects to be cleaned
- C11D2111/12—Soft surfaces, e.g. textile
Definitions
- the present invention relates to detergent compositions comprising novel GH9 endoglucanase variants exhibiting alterations relative to the parent GH9 endoglucanase in one or more properties including: detergent stability (e.g. improved stability in a detergent composition,) and/or storage stability (e.g. improved storage stability in a detergent composition).
- the present invention further relates to detergent compositions comprising novel GH9 endoglucanase variants having activity on xanthan gum pre-treated with xanthan lyase.
- the invention also relates to methods for producing and using the compositions of the invention. Variants as described herein are particularly suitable for use in cleaning processes and detergent compositions, such as laundry compositions and dish wash compositions, including hand wash and automatic dish wash compositions. Description of the Related Art
- Xanthan gum is a polysaccharide derived from the bacterial coat of Xanthomonas campestris. It is produced by the fermentation of glucose, sucrose, or lactose by the Xanthomonas campestris bacterium. After a fermentation period, the polysaccharide is precipitated from a growth medium with isopropyl alcohol, dried, and ground into a fine powder. Later, it is added to a liquid medium to form the gum.
- Xanthan gum is a natural polysaccharide consisting of different sugars which are connected by several different bonds, such as p-D-mannosyl-p-D-1 ,4-glucuronosyl bonds and -D-glucosyl- -D-1 ,4-glucosyl bonds.
- Xanthan gum is at least partly soluble in water and forms highly viscous solutions or gels. Complete enzymatic degradation of xanthan gum requires several enzymatic activities including xanthan lyase activity and endo- -1 ,4-glu- canase activity.
- Xanthan lyases are enzymes that cleave the p-D-mannosyl-p-D-1 ,4-glucuronosyl bond of xanthan and have been described in the literature.
- Xanthan lyases are known in the art, e.g. two xanthan lyases have been isolated from Paenibacillus alginolyticus XL-1 (e.g. Ruijssenaars ef al. (1999) ⁇ pyruvated mannose-specific xanthan lyase involved in xanthan degradation by Paenibacillus alginolyticus XL-1 ', Appl. Environ. Microbiol.
- Glycoside hydrolases are enzymes that catalyse the hydrolysis of the glycosyl bond to release smaller sugars. There are over 100 classes of glycoside hydrolases which have been classified, see Henrissat ei al. (1991 ) ⁇ classification of glycosyl hydrolases based on amino-acid sequence similarities', J. Biochem. 280: 309-316 and the Uniprot website at www.cazy.org.
- the glycoside hydrolase family 9 (GH9) consists of over 70 different enzymes that are mostly endo-glucanases (EC 3.2.1 .4), cellobiohydrolases (EC 3.2.1.91 ), ⁇ -glucosidases (EC 3.2.1 .21 ) and exo ⁇ -glucosaminidase (EC 3.2.1.165).
- xanthan gum has been used as an ingredient in many consumer products including foods (e.g. as thickening agent in salad dressings and dairy products) and cosmetics (e.g. as stabilizer and thickener in toothpaste and make-up, creams and lotions to prevent ingredients from separating and to provide the right texture of the product).
- xanthan gum has found use in the oil industry as an additive to regulate the viscosity of drilling fluids etc.
- the widespread use of xanthan gum has led to a desire to degrade solutions, gels or mixtures containing xanthan gum thereby allowing easier removal of the byproducts.
- Endoglucanases and xanthan lyases for the degradation of xanthan gum and the use of such enzymes for cleaning purposes, such as the removal of xanthan gum containing stains, and in the drilling and oil industries are known in the art, e.g. WO2013167581 A1 .
- the known xanthan endoglucanase having SEQ ID NO: 2 was found to be sensitive to storage in detergent. To improve the applicability and/or cost and/or the performance of such enzymes there is an ongoing search for variants with altered properties, such as increased stability, e.g. improved stability in a detergent composition. However, mutagenesis of large enzymes followed by purification and functional analysis of mutant libraries can be very expensive and laborious.
- the present invention identifies regions in the protein sequence/structure of the known xanthan endoglucanase having SEQ ID NO: 2 that are relevant for e.g. storage stability, and therefore provides an important guidance on where to mutate an endoglucanase in order to stabilize the molecule in a detergent.
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in a region selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ ID
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglu- canase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in a region selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant having at least 61 %, at least 62%, at least 63%, at least 64%, at least 65%, at least 66%, at least 67%, at least 68%, at least 69%, at least 70%, at least 71 %, at least 72%, at least 73%, at least 74%, at least 75%, at least 76%, at least 77%, at least 78%, at least 79%, at least 80%, at least 81 %, at least 82%, at least 83%, at least 84%, at least 85%, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to SEQ ID NO: 2.
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in a region selected from the group consisting of:
- region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 1 , 2, 3, 4, 5, 6, 7, 8, 9, 10, 1 1 , 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 , 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 , 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 , 42,
- region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 107, 108, 109, 110, 1 11 , 112, 1 13, 1 14, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
- region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 139, 140, 141 , 142, 143, 144, 145, 146,
- iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 252, 253, 254, 255, 256, 257, 258, 259, 260, 261 , 262, 263, 264, 265, 266, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
- v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 302, 303, 304, 305, 306, 307, 308, 309,
- region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 362, 363, 364, 365, 366, 367, 368, 369,
- region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 596, 597, 598, 599, 600, 601 , 602, 603, 604, 605, 606, 607, 608, 609, 610, 61 1 , wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
- region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 661 , 662, 663, 664, 665, 666, 667, 668, 669, 670, 671 , 672, 673, 674, 675, 676, 677, 678, 679, 680, 681 , 682, 683, 684, 685, 686, 687, 688, 689, 690, 691 , 692, 693, 694, 695, 696, 697, 698, 699, 700, 701 , 702, 703, 704, 705, 706, 707, 708, 709, 710, 711 , 712, 713, 714, 715, 716, 717, 718, 719, 720, 721 , 722, 723, 724, 725, 726, 727, 728,
- ix) region 18 corresponding to amino acids 829 to 838 to 1042 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 829, 830, 831 , 832, 833, 834, 835, 836, 837, 838, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2)
- x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions 1043, 1044, 1045, 1046, 1047, 1048, 1049, 1050, 1051 , 1052, 1053, 1054, 1055, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2).
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in: a) one or more regions selected from the group consisting of:
- region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 1 , 2, 3, 4, 5, 6, 7, 8, 9, 10, 1 1 , 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 , 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 , 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 , 42, 43, 44, 45, 46, 47, 48, 49, 50, 51 , 52, 53, 54, 55, 56, 57, 58, 59, 60, 61 , 62, 63, 64, 65, 66, 67, 68, 69, 70, 71 , 72, 73, 74, 75, 76, 77, 78, 79, 80, 81 , 82, 83, 84, 85, 86, 87, 88, 89, 90, 91 , 92, 93, 94, wherein said positions correspond to amino acid positions of
- region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 107, 108, 109, 1 10, 1 11 , 112, 1 13, 1 14, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
- region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 139, 140, 141 , 142, 143, 144, 145, 146, 147, 148, 149, 150, 151 , 152, 153, 154, 155, 156, 157, 158, 159, 160, 161 , 162, 163, 164, 165, 166, 167, 168, 169, 170, 171 , 172, 173, 174, 175, 176, 177, 178, 179, 180, 181 , 182, 183, 184, 185, 186, 187, 188, 189, 190, 191 , 192, 193, 194, 195, 196, 197, 198, 199, 200, 201 , 202, 203, 204, 205, 206, 207, 208, 209,
- iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 252, 253, 254, 255, 256, 257, 258, 259, 260, 261 , 262, 263, 264, 265, 266, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
- v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 302, 303, 304, 305, 306, 307, 308, 309, 310, 311 , 312, 313, 314, 315, 316, 317, 318, 319, 320, 321 , 322, 323, 324, 325, 326, 327, 328, 329,
- region 15 corresponding to amino acids 362 to 546of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 362, 363, 364, 365, 366, 367, 368, 369, 370, 371 , 372, 373, 374, 375, 376, 377, 378, 379, 380, 381 , 382, 383, 384, 385, 386, 387, 388, 389, 390,
- region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 661 , 662, 663, 664, 665, 666, 667, 668,
- ix) region 18 corresponding to amino acids 829 to 838 to 1042 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 829, 830, 831 , 832, 833, 834, 835, 836, 837, 838, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2)
- x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions 1043, 1044, 1045, 1046, 1047, 1048, 1049, 1050, 1051 , 1052, 1053, 1054, 1055, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2).
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglu- canase variant having an alteration (e.g., a substitution, deletion or insertion) at one or more positions selected from the group consisting of positions: 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 31 1 , 313, 387, 388, 390, 403, 408, 410, 416, 448, 451 , 471 , 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 71 1 , 719, 754, 756, 760, 781 , 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglu- canase variant having an alteration (e.g., a substitution, deletion or insertion) at one or more positions se- lected from the group consisting of: S17A, F20P, F20N, F20G, F20Y, K51 Q, K51 H, E53P, E53G, Y55M, V56M, Y60F, S63F, T87R, A186P, K192N, I302D, I302H, I302V, I302M, H311 N, S313D, I387T, K388R, K390Q, I403Y, E408D, E408S, E408P, E408A, E408G, E408N, P410G, Q416S, Q416D, A448E, A448W, A448S, K451 S, G471 S, S472Y,
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglu- canase variant comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions selected from the group consisting of alterations in positions: 17, 20, 302, 31 1 , 313, 408, 476, 602, 688, 697, and 719, wherein numbering is according to SEQ ID NO: 2.
- the endoglucanase variant comprised in the detergent compositions of the invention comprises an alteration at one or more positions selected from the group consisting of alterations in positions: S17A, F20P, I302D, H311 N, S313D, E408D, D476R, I602T, A688G, T697G, and W719R, wherein numbering is according to SEQ ID NO: 2.
- the endoglucanase variant comprised in the detergent compositions of the invention further comprises an alteration at one or more positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651 , 824, 848, 869, 880, 881 , 883, 887, 892, 905, 906, 912, 921 , 928, 934, 937, 948, 956, 999, and 1037.
- the endoglucanase variant comprised in the detergent compositions of the invention further comprises an alteration (e.g., substitution) at one or more positions selected from the group consisting of: N216D, N216Q, A283D, A346D, A559P, A559N, A564E, Y579W, S636K, S636N, F638N, A651 P, A824D, N848D, A869V, R880K, V881 T, V881 Q, T883R, T887K, T892P, N905D, F906A, A912V, K921 R, S928D, Y934G, A937E, K948R, Q956Y, T999R, and A1037E.
- an alteration e.g., substitution
- the endoglucanase variant comprised in the detergent compositions of the invention comprises one or more of the following set of substitutions:
- the endoglucanase variant comprised in the detergent compositions of the invention comprises one or more of the following set of substitutions:
- the endoglucanase variant as described herein is one that does not comprise any amino acid alteration at a position outside of regions 10, 1 1 , 12, 13, 14, 15, 16, 17, 18, and 19.
- the endoglucanase variant thus does not comprise any alteration (e.g., a substitution, deletion or insertion) in a region selected from the group consisting of: region 1 corresponding to amino acids 95 to 105 of SEQ ID NO: 2, region 2 corresponding to amino acids 115 to 138 of SEQ ID NO: 2, region 3 corresponding to amino acids 210 to 251 of SEQ ID NO: 2, region 4 corresponding to amino acids 267 to 301 of SEQ ID NO: 2, region 5 corresponding to amino acids 339 to 361 of SEQ ID NO: 2, region 6 corresponding to amino acids 547 to 595 of SEQ ID NO: 2, region 7 corresponding to amino acids 612 to 660 of SEQ ID NO: 2, region 8 corresponding to amino acids 806 to 828 of SEQ ID NO: 2, and region 9 corresponding to
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant having activity on xanthan gum pre-treated with xanthan lyase; preferably said activity comprises endoglucanase EC 3.2.1.4 activity, further preferably said activity is endoglucanase EC 3.2.1 .4 activity.
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant having an improved stability in a detergent composition compared to a parent endoglucanase (e.g., with SEQ ID NO: 2).
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglu- canase variant having a half-life improvement factor (HIF) of >1.0 relative to a parent endoglucanase, e.g. an endoglucanase of SEQ ID NO: 2.
- HIF half-life improvement factor
- the present invention relates to a detergent composition
- a detergent composition comprising an isolated GH9 endoglucanase variant having activity on xanthan gum pre-treated with xanthan lyase.
- the detergent composition further comprises an isolated polypeptide having xanthan lyase activity.
- the present invention relates to a detergent composition
- a detergent composition comprising one or more detergent components for degrading xanthan gum.
- the present invention relates to use of a detergent composition of the present invention, wherein said use is selected from the group consisting of: use for degrading xanthan gum and use in a cleaning process, such as laundry or hard surface cleaning such as dish wash.
- the present invention further relates to the use of a detergent composition of the invention for degrading xanthan gum, for washing or cleaning textiles and/or hard surfaces, such as dish wash, wherein the composition has an enzyme detergency benefit.
- the present invention also relates to methods of degrading xanthan gum using detergent compositions of the present invention, wherein xanthan gum is on the surface of a hard surface or textile.
- SEQ ID NO: 1 is the DNA sequence of the parent mature endoglucanase from a strain of a Paenibacil- lus sp.
- SEQ ID NO: 2 is the amino acid sequence of mature polypeptide encoded by SEQ ID NO: 1 .
- SEQ ID NO: 3 is the DNA sequence of the alpha-amylase secretion signal from Bacillus licheniformis.
- SEQ ID NO: 4 is the amino acid sequence of the alpha-amylase secretion signal from Bacillus licheniformis.
- cDNA means a DNA molecule that can be prepared by reverse transcription from a mature, spliced, mRNA molecule obtained from a eukaryotic or prokaryotic cell. cDNA lacks intron sequences that may be present in the corresponding genomic DNA.
- the initial, primary RNA transcript is a precursor to mRNA that is processed through a series of steps, including splicing, before appearing as mature spliced mRNA.
- Cleaning or Detergent Application means applying the endoglucanase of the application in any composition for the purpose of cleaning or washing, by hand, machine or automated, a hard surface or a textile.
- Cleaning Composition refers to compositions that find use in the removal of undesired compounds from items to be cleaned, such as textiles, dishes, and hard surfaces.
- the terms encompass any materials/compounds selected for the particular type of cleaning composition desired and the form of the product (e.g., liquid, gel, powder, granulate, paste, or spray compositions) and includes, but is not limited to, detergent compositions (e.g., liquid and/or solid laundry detergents and fine fabric detergents; hard surface cleaning formulations, such as for glass, wood, ceramic and metal counter tops and windows; carpet cleaners; oven cleaners; fabric fresheners; fabric softeners; and textile and laundry pre- spotters, as well as dish wash detergents).
- detergent compositions e.g., liquid and/or solid laundry detergents and fine fabric detergents
- hard surface cleaning formulations such as for glass, wood, ceramic and metal counter tops and windows
- carpet cleaners oven cleaners
- fabric fresheners fabric softeners
- textile and laundry pre- spotters as well as dish wash detergents
- the detergent formulation may contain one or more additional enzymes (such as xanthan lyases, proteases, amylases, lipases, cutinases, cellulases, endoglucanases, xyloglucanases, pectinases, pectin lyases, xanthanases, peroxidases, haloperoxygenases, catalases and mannanases, or any mixture thereof), and/or components such as surfactants, builders, chelators or chelating agents, bleach system or bleach components, polymers, fabric conditioners, foam boosters, suds suppressors, dyes, perfume, tannish inhibitors, optical brighteners, bactericides, fungicides, soil suspending agents, anti-corrosion agents, enzyme inhibitors or stabilizers, enzyme activators, transferase(s), hydrolytic enzymes, oxido reductases, bluing agents and fluorescent dyes, antioxidants, and
- additional enzymes such as xanthan
- Coding sequence means a polynucleotide, which directly specifies the amino acid sequence of a polypeptide.
- the boundaries of the coding sequence are generally determined by an open reading frame, which begins with a start codon such as ATG, GTG, or TTG and ends with a stop codon such as TAA, TAG, or TGA.
- the coding sequence may be a genomic DNA, cDNA, synthetic DNA, or a combination thereof.
- Colour clarification During washing and wearing loose or broken fibers can accumulate on the surface of the fabrics. One consequence can be that the colours of the fabric appear less bright or less intense because of the surface contaminations. Removal of the loose or broken fibers from the textile will partly restore the original colours and looks of the textile.
- colour clarification is meant the partial restoration of the initial colours of textile.
- control sequences means nucleic acid sequences necessary for expression of a polynucleotide encoding a mature polypeptide of the present invention.
- Each control sequence may be native (i.e., from the same gene) or foreign (i.e. , from a different gene) to the polynucleotide encod- ing the polypeptide or native or foreign to each other.
- control sequences include, but are not limited to, a leader, polyadenylation sequence, propeptide sequence, promoter, signal peptide sequence, and transcription terminator.
- the control sequences include a promoter, and transcriptional and trans- lational stop signals.
- the control sequences may be provided with linkers for the purpose of introducing specific restriction sites facilitating ligation of the control sequences with the coding region of the polynucle- otide encoding a polypeptide.
- corresponding to refers to a way of determining the specific amino acid of a sequence wherein reference is made to a specific amino acid sequence.
- reference is made to a specific amino acid sequence.
- the skilled person would be able to align another amino acid sequence to said amino acid sequence that reference has been made to, in order to determine which specific amino acid may be of interest in said another amino acid sequence. Alignment of another amino acid sequence with e.g. the sequence as set forth in SEQ ID NO: 2, or any other sequence listed herein, has been described elsewhere herein. Alternative alignment methods may be used, and are well-known for the skilled person.
- Degrading xanthan gum and xanthan gum degrading activity are used interchangeably and are defined as the depolymerization, degradation or breaking down of xanthan gum into smaller components.
- the degradation of xanthan gum can either be the removal of one or more side chain saccharides, the cutting of the backbone of xanthan gum into smaller components or the removal of one or more side chain saccharides and the cutting of the backbone of xanthan gum into smaller components.
- a preferred assay for measuring degradation of xanthan gum is the reducing sugar assay as described in example 3 herein.
- Non-limiting examples of the xan- than gum degrading activity include endoglucanase EC 3.2.1.4 activity.
- Detergent component the term "detergent component” is defined herein to mean the types of chemicals which can be used in detergent compositions.
- detergent components are surfactants, hydrotropes, builders, co-builders, chelators or chelating agents, bleaching system or bleach components, polymers, fabric hueing agents, fabric conditioners, foam boosters, suds suppressors, dispersants, dye transfer inhibitors, fluorescent whitening agents, perfume, optical brighteners, bactericides, fungicides, soil suspending agents, soil release polymers, anti-redeposition agents, enzyme inhibitors or stabilizers, enzyme activators, antioxidants, and solubilizers.
- the detergent composition may comprise of one or more of any type of detergent component.
- Detergent composition refers to compositions that find use in the removal of undesired compounds from items to be cleaned, such as textiles, dishes, and hard surfaces.
- the detergent composition may be used to e.g. clean textiles, dishes and hard surfaces for both household cleaning and industrial cleaning.
- the terms encompass any materials/compounds selected for the particular type of cleaning composition desired and the form of the product (e.g., liquid, gel, powder, granulate, paste, or spray compositions) and includes, but is not limited to, detergent compositions (e.g., liquid and/or solid laundry detergents and fine fabric detergents; hard surface cleaning formulations, such as for glass, wood, ceramic and metal counter tops and windows; carpet cleaners; oven cleaners; fabric fresheners; fabric softeners; and textile and laundry pre-spotters, as well as dish wash detergents).
- detergent compositions e.g., liquid and/or solid laundry detergents and fine fabric detergents
- hard surface cleaning formulations such as for glass, wood, ceramic and metal counter tops and windows
- carpet cleaners oven cleaners
- fabric fresheners fabric softeners
- textile and laundry pre-spotters as well as dish wash detergents
- the detergent formulation may contain one or more additional enzymes (such as xanthan lyases, proteases, amylases, lipases, cutinases, cellulases, endoglucanases, xyloglucanases, pectinases, pectin lyases, xanthanases, peroxidases, haloperoxygen- ases, catalases and mannanases, or any mixture thereof), and/or components such as surfactants, builders, chelators or chelating agents, bleach system or bleach components, polymers, fabric conditioners, foam boosters, suds suppressors, dyes, perfume, tannish inhibitors, optical brighteners, bactericides, fungicides, soil suspending agents, anti-corrosion agents, enzyme inhibitors or stabilizers, enzyme activators, transferase ⁇ ), hydrolytic enzymes (such as xanthan lyases, proteases, amylases, lipases, cutinases, cellul
- Dish wash refers to all forms of washing dishes, e.g. by hand or automatic dish wash. Washing dishes includes, but is not limited to, the cleaning of all forms of crockery such as plates, cups, glasses, bowls, all forms of cutlery such as spoons, knives, forks and serving utensils as well as ceramics, plastics, metals, china, glass and acrylics.
- Dish washing composition refers to all forms of compositions for cleaning hard surfaces.
- the present invention is not restricted to any particular type of dish wash composition or any particular detergent.
- Endoglucanase means an endo-1 ,4-(1 ,3;1 ,4)-beta-D-glucan 4- glucanohydrolase (EC 3.2.1 .4) that catalyzes endohydrolysis of 1 ,4-beta-D-glycosidic linkages in cellulose, cellulose derivatives (such as carboxymethyl cellulose and hydroxyethyl cellulose), lichenin, beta-1 ,4 bonds in mixed beta-1 ,3 glucans such as cereal beta-D-glucans, xyloglucans, xanthans and other plant material containing cellulosic components.
- Endoglucanase activity can be determined by measuring reduction in substrate viscosity or increase in reducing ends determined by a reducing sugar assay (Zhang ef a/., 2006, Biotechnology Advances 24: 452-481 ).
- a preferred assay for measuring endoglucanase activity is the re- ducing sugar assay as described in example 3 herein.
- Non-limiting examples of endoglucanases include the mature parent endoglucanase having SEQ ID NO: 2.
- Enzyme detergency benefit is defined herein as the advantageous effect an enzyme may add to a detergent compared to the same detergent without the enzyme.
- Important detergency benefits which can be provided by enzymes are stain removal with no or very little visible soils after washing and or cleaning, prevention or reduction of redeposition of soils released in the washing process an effect that also is termed anti-redeposition, restoring fully or partly the whiteness of textiles, which originally were white but after repeated use and wash have obtained a greyish or yellowish appearance an effect that also is termed whitening.
- Textile care benefits which are not directly related to catalytic stain removal or prevention of redeposition of soils are also important for enzyme detergency ben- efits.
- Examples of such textile care benefits are prevention or reduction of dye transfer from one fabric to another fabric or another part of the same fabric an effect that is also termed dye transfer inhibition or anti- backstaining, removal of protruding or broken fibers from a fabric surface to decrease pilling tendencies or remove already existing pills or fuzz an effect that also is termed anti-pilling, improvement of the fabric- softness, colour clarification of the fabric and removal of particulate soils which are trapped in the fibers of the fabric or garment.
- Enzymatic bleaching is a further enzyme detergency benefit where the catalytic activity generally is used to catalyze the formation of bleaching component such as hydrogen peroxide or other peroxides.
- expression includes any step involved in the production of a polypeptide including, but not limited to, transcription, post-transcriptional modification, translation, post-translational modification, and secretion.
- Expression vector means a linear or circular DNA molecule that comprises a polynucleotide encoding a polypeptide and is operably linked to control sequences that provide for its expression.
- fragment means a polypeptide having one or more (e.g. , several) amino acids absent from the amino and/or carboxyl terminus of a mature polypeptide; wherein the fragment has en- doglucanase activity.
- a fragment contains at least 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94% or 95% of the number of amino acids of the mature polypeptide.
- Endoglucanase variant having activity on xanthan gum pre-treated with xanthan lyase The term "Endoglucanase variant having activity on xanthan gum pre-treated with xanthan lyase" or an "endoglucanase having activity on xanthan gum pre-treated with xanthan lyase and belonging to the GH9 class of glycosyl hydrolases" is defined as a polypeptide comprising a domain belonging to the GH9 class of glycosyl hydrolases, and having activity (e.g.
- Xanthan lyase variant having activity on xanthan gum is defined as a polypeptide that cleaves the p-D-mannosyl-p-D-1 ,4-glucuronosyl bond of xanthan (e.g., xanthan lyase EC 4.2.2.12 activity).
- xanthan lyase variants having activity on xanthan gum are xanthan lyase polypeptides as such.
- polypeptides that that cleaves the p-D-mannosyl-p-D-1 ,4-glucuronosyl bond of xanthan are examples of the xanthan lyase polypeptides as such.
- Half-life is the time it takes for an enzyme to lose half of its enzymatic activity under a given set of conditions. It is denoted as T1 ⁇ 2 and is measured in hours (hrs). Half-lives can be calculated at a given detergent concentration and storage temperature for parent (e.g. wild-type) and/or variants, as the degradation follows an exponential decay and the incubation time (hours) is known, i.e., according to the following formulas:
- T112 (wild-type) (Ln (0.5)/Ln (RA-wild-type/100))*Time
- 'RA' is the residual activity in percent and 'Time' is the incubation time in hours.
- Half-life improvement factor the term "Half-life improvement factor” or “HIF” is the improvement of half-life of a variant compared to the parent polypeptide, such as the parent endoglucanase.
- a half-life improvement factor (HIF) under a given set of storage conditions can be calculated as : Tl/2, variant
- the wild-type (wt) is incubated under the same storage condition (detergent concentration and incubation temperature) as the variant.
- the incubation time for wild-type and variant is different e.g. 1 h for wild-type and 138 h for the most stable variants.
- Hard surface cleaning is defined herein as cleaning of hard surfaces wherein hard surfaces may include floors, tables, walls, roofs etc. as well as surfaces of hard objects such as cars (car wash) and dishes (dish wash). Dish washing includes but are not limited to cleaning of plates, cups, glasses, bowls, and cutlery such as spoons, knives, forks, serving utensils, ceramics, plastics, metals, china, glass and acrylics.
- host cell means any cell type that is susceptible to transformation, transfection, transduction, or the like with a nucleic acid construct or expression vector comprising a polynucleotide of the present invention.
- host cell encompasses any progeny of a parent cell that is not identical to the parent cell due to mutations that occur during replication.
- Improved property means a characteristic associated with a variant that is improved compared to the parent. Such improved properties include, but are not limited to, catalytic efficiency, catalytic rate, chemical stability, oxidation stability, pH activity, pH stability, specific activity, sta- bility under storage conditions, chelator stability, substrate binding, substrate cleavage, substrate specificity, substrate stability, surface properties, thermal activity, and thermostability.
- Improved wash performance is defined herein as a (variant) enzyme (also a blend of enzymes, not necessarily only variants but also backbones, and in combination with certain cleaning composition etc.) displaying an alteration of the wash performance of a variant relative to the wash performance of the parent variant e.g. by increased stain removal.
- wash performance includes wash performance in laundry but also e.g. in dish wash.
- Isolated means a substance in a form or environment that does not occur in nature.
- isolated substances include (1 ) any non-naturally occurring substance, (2) any substance including, but not limited to, any enzyme, variant, nucleic acid, protein, peptide or cofactor, that is at least partially removed from one or more or all of the naturally occurring constituents with which it is associated in nature; (3) any substance modified by the hand of man relative to that substance found in nature; or (4) any substance modified by increasing the amount of the substance relative to other components with which it is naturally associated (e.g. , multiple copies of a gene encoding the substance; use of a stronger promoter than the promoter naturally associated with the gene encoding the substance).
- Laundering relates to both household laundering and industrial laundering and means the process of treating textiles with a solution containing a cleaning or detergent composition of the present invention.
- the laundering process can for example be carried out using e.g. a household or an industrial washing machine or can be carried out by hand.
- Mature polypeptide means a polypeptide in its final form following translation and any post-translational modifications, such as N-terminal processing, C-terminal truncation, glycosylation, phosphorylation, etc.
- the mature polypeptide is amino acids 1 to 1055 of SEQ ID NO: 2.
- a host cell may produce a mixture of two of more different mature polypeptides (i.e., with a different C-terminal and/or N-terminal amino acid) expressed by the same polynucleotide. It is also known in the art that different host cells process polypeptides differently, and thus, one host cell expressing a polynucleotide may produce a different mature polypeptide (e.g., having a different C-terminal and/or N-terminal amino acid) as compared to another host cell expressing the same polynucleotide.
- Mature polypeptide coding sequence means a polynucleotide that encodes a mature polypeptide having enzymatic activity such as activity on xanthan gum pre-treated with xanthan lyase or xanthan lyase activity.
- the mature polypeptide coding sequence is nucleotides 1 to 3165 of SEQ ID NO: 1 .
- Mutant means a polynucleotide encoding a variant.
- nucleic acid construct means a nucleic acid molecule, either sin- gle- or double-stranded, which is isolated from a naturally occurring gene or is modified to contain segments of nucleic acids in a manner that would not otherwise exist in nature or which is synthetic, which comprises one or more control sequences.
- operably linked means a configuration in which a control sequence is placed at an appropriate position relative to the coding sequence of a polynucleotide such that the control sequence directs expression of the coding sequence.
- parent means any polypeptide with endoglucanase activity to which an alteration is made to produce the enzyme variants of the present invention.
- the parent is an endoglucanase having the identical amino acid sequence of the variant, but not having the alterations at one or more of the specified positions. It will be understood, that the expression “having iden- tical amino acid sequence” relates to 100% sequence identity.
- Non-limiting examples of parent endoglu- canases include the mature parent endoglucanase having SEQ ID NO: 2.
- Sequence identity The relatedness between two amino acid sequences or between two nucleotide sequences is described by the parameter "sequence identity".
- sequence identity is determined using the Needleman-Wunsch algo- rithm (Needleman and Wunsch, 1970, J. Mol. Biol. 48: 443-453) as implemented in the Needle program of the EMBOSS package (EMBOSS: The European Molecular Biology Open Software Suite, Rice ef a/., 2000, Trends Genet. 16: 276-277), preferably version 5.0.0 or later.
- the parameters used are gap open penalty of 10, gap extension penalty of 0.5, and the EBLOSUM62 (EMBOSS version of BLOSUM62) substitution matrix.
- the output of Needle labeled "longest identity" is used as the percent identity and is calculated as follows:
- the sequence identity between two deoxyribonucleotide sequences is determined using the Needleman-Wunsch algorithm (Needleman and Wunsch, 1970, supra) as implemented in the Needle program of the EMBOSS package (EMBOSS: The European Molecular Biology Open Software Suite, Rice ef a/. , 2000, supra), preferably version 5.0.0 or later.
- the parameters used are gap open penalty of 10, gap extension penalty of 0.5, and the EDNAFULL (EMBOSS version of NCBI NUC4.4) substitution matrix.
- the output of Needle labeled "longest identity" (obtained using the -nobrief option) is used as the percent identity and is calculated as follows:
- very low stringency conditions means for probes of at least 100 nucleotides in length, pre- hybridization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 25% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 45°C.
- low stringency conditions means for probes of at least 100 nucleotides in length, prehybrid- ization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 25% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 50°C.
- medium stringency conditions means for probes of at least 100 nucleotides in length, pre- hybridization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 35% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 55°C.
- medium-high stringency conditions means for probes of at least 100 nucleotides in length, prehybridization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 35% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 60°C.
- high stringency conditions means for probes of at least 100 nucleotides in length, prehybridization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 50% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 65°C.
- very high stringency conditions means for probes of at least 100 nucleotides in length, prehybridization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 50% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 70°C.
- Subsequence means a polynucleotide having one or more (e.g., several) nucleotides absent from the 5' and/or 3' end of a mature polypeptide coding sequence; wherein the subsequence encodes a fragment having enzymatic activity, such as activity on xanthan gum pre-treated with xanthan lyase or xanthan lyase activity.
- Textile means any textile material including yarns, yarn intermediates, fibers, non- woven materials, natural materials, synthetic materials, and any other textile material, fabrics made of these materials and products made from fabrics (e.g., garments and other articles).
- the textile or fabric may be in the form of knits, wovens, denims, non-wovens, felts, yarns, and towelling.
- the textile may be cellulose based such as natural cellulosics, including cotton, flax/linen, jute, ramie, sisal or coir or manmade cellulo- sics (e.g.
- the textile or fabric may also be non-cellulose based such as natural polyamides including wool, camel, cashmere, mohair, rabbit and silk or synthetic polymer such as nylon, aramid, polyester, acrylic, polypropylene and spandex elastane, or blends thereof as well as blend of cellulose based and non- cellulose based fibers.
- non-cellulose based such as natural polyamides including wool, camel, cashmere, mohair, rabbit and silk or synthetic polymer such as nylon, aramid, polyester, acrylic, polypropylene and spandex elastane, or blends thereof as well as blend of cellulose based and non- cellulose based fibers.
- blends are blends of cotton and/or rayon/viscose with one or more companion material such as wool, synthetic fibers (e.g.
- Fabric may be conventional washable laundry, for example stained household laundry.
- fabric or garment it is intended to include the broader term textiles as well.
- Textile care benefits which are not directly related to catalytic stain removal or prevention of redeposition of soils, are also important for enzyme detergency benefits.
- textile care benefits are prevention or reduction of dye transfer from one textile to another textile or another part of the same textile an effect that is also termed dye transfer inhibition or anti-backstaining, removal of protruding or broken fibers from a textile surface to decrease pilling tendencies or remove already existing pills or fuzz an effect that also is termed anti-pilling, improvement of the textile-softness, colour clarification of the textile and removal of particulate soils which are trapped in the fibers of the textile.
- Enzymatic bleach- ing is a further enzyme detergency benefit where the catalytic activity generally is used to catalyze the formation of bleaching component such as hydrogen peroxide or other peroxides or other bleaching species.
- Variant means a polypeptide (e.g., a GH9 endoglucanase polypeptide) comprising an alteration i.e. , a substitution, insertion, and/or deletion, at one or more (e.g. , several) positions.
- a substitution means replacement of the amino acid occupying a position with a different amino acid ;
- a deletion means removal of the amino acid occupying a position; and an insertion means adding one or more (e.g.
- Non-limiting examples of endoglucanase variants as described herein include endoglucanase variants having an activity on xanthan gum pre-treated with xanthan lyase.
- Non-limiting examples of variants as described herein further include variants having at least 20%, e.g., at least 40%, at least 50%, at least 60%, at least 70%, at least 80%, at least 90%, at least 95%, or at least 100% endoglucanase activity of the mature parent having SEQ ID NO: 2.
- a preferred assay for measuring activity on xanthan gum pre-treated with xanthan lyase is disclosed in example 3 herein.
- Stability means resistance or the degree of resistance to change, unfolding, disintegration, denaturation or activity loss.
- Non-limiting examples of stability include conformational stability, storage stability and stability during use, e.g. during a wash process and reflects the stability of a polypeptide (e.g. an endoglucanase variant according to the invention) as a function of time, e.g. how much activity is retained when said polypeptide (e.g. said endoglucanase variant) is kept in solution, in particular in a detergent solution.
- the stability is influenced by many factors, e.g. presence of chelator(s), pH, temperature, detergent composition, e.g. amount of builders, surfactants, chelators etc.
- the endoglucanase stability may be measured using a half-life improvement factor (HIF) as described in example 3 herein.
- the endoglu- canase stability may also be measured using a reducing sugar assay as described in example 3 herein.
- Improved stability is defined herein as increased stability in a detergent composition (e.g. , in solutions), relative to the stability of the parent endoglucanase, relative to an endoglucanase having the identical amino acid sequence of the variant, but not having the alterations at one or more of the specified positions, or relative to SEQ ID NO: 2.
- improved stability and “increased stability” includes “improved chemical stability”, “detergent stability” and “improved detergent stability.
- Improved chemical stability is defined herein as a variant enzyme displaying retention of enzymatic activity after a period of incubation in the presence of a chemical or chemicals, either naturally occurring or synthetic, which reduces the enzymatic activity of the parent enzyme. Improved chemical stability may also result in variants being more able (e.g., better that the parent) to catalyze a reaction in the presence of such chemicals.
- the improved chemical stability is an improved stability in a detergent, in particular in a liquid detergent.
- detergent stability or “improved detergent stability is in particular an improved stability of the endoglucanase compared to the parent endoglucanase, when an endoglucanase variant as described herein is mixed into a liquid detergent formulation, especially into a liquid detergent formulation comprising a chelator (e.g. EDTA or citrate).
- Conformational stability means resistance or a degree of resistance to conformational change, unfolding or disintegration. Accordingly, the term “less conformationally stable” means less resistant or having lesser degree of resistance to conformational change, unfolding or disintegration.
- instability means lack of stability.
- instability include conformational instability, unfolding, denaturation, disintegration, activity loss.
- Wash performance is used as an enzyme's ability to remove stains present on the object to be cleaned during e.g. wash or hard surface cleaning.
- the improvement in the wash performance may be quantified by calculating the so-called intensity value (Int) in 'Automatic Mechanical Stress Assay (AMSA) for laundry' or the remission value (Rem) as defined herein.
- Int intensity value
- AMSA Automatic Mechanical Stress Assay
- Rem remission value
- the mature polypeptide disclosed in SEQ ID NO: 2 is used to determine the corresponding amino acid residue in another endoglucanase.
- the amino acid sequence of another endoglucanase is aligned with the mature polypeptide disclosed in SEQ ID NO: 2, and based on the alignment, the amino acid position number corresponding to any amino acid residue in the mature polypeptide disclosed in SEQ ID NO: 2 is determined using the Needleman-Wunsch algorithm (Needleman and Wunsch, 1970, J. Mol. Biol. 48: 443-453) as implemented in the Needle program of the EMBOSS package (EMBOSS: The European Molecular Biology Open Software Suite, Rice et al., 2000, Trends Genet. 16: 276-277), preferably version 5.0.0 or later.
- the parameters used are gap open penalty of 10, gap extension penalty of 0.5, and the EBLOSUM62 (EMBOSS version of BLOSUM62) substitution matrix.
- Identification of the corresponding amino acid residue in another endoglucanase can be determined by an alignment of multiple polypeptide sequences using several computer programs including, but not limited to, MUSCLE (multiple sequence comparison by log-expectation; version 3.5 or later; Edgar, 2004, Nucleic Acids Research 32: 1792-1797), MAFFT (version 6.857 or later; Katoh and Kuma, 2002, Nucleic Acids Research 30: 3059-3066; Katoh et al.
- MUSCLE multiple sequence comparison by log-expectation; version 3.5 or later
- MAFFT version 6.857 or later
- Katoh and Kuma 2002, Nucleic Acids Research 30: 3059-3066
- proteins of known structure For proteins of known structure, several tools and resources are available for retrieving and generating structural alignments. For example, the SCOP superfamilies of proteins have been structurally aligned, and those alignments are accessible and downloadable.
- Two or more protein structures can be aligned using a variety of algorithms such as the distance alignment matrix (Holm and Sander, 1998, Proteins 33: 88-96) or combinatorial extension (Shindyalov and Bourne, 1998, Protein Engineering 1 1 : 739-747), and implemen- tation of these algorithms can additionally be utilized to query structure databases with a structure of interest in order to discover possible structural homologs (e.g., Holm and Park, 2000, Bioinformatics 16: 566-567).
- the distance alignment matrix Holm and Sander, 1998, Proteins 33: 88-96
- combinatorial extension Shindyalov and Bourne, 1998, Protein Engineering 1 1 : 739-747
- implemen- tation of these algorithms can additionally be utilized
- substitutions For an amino acid substitution, the following nomenclature is used: Original amino acid, position, substituted amino acid. Accordingly, the substitution of threonine at position 226 with alanine is designated as "Thr226Ala” or “T226A”. Multiple mutations are separated by addition marks ("+"), e.g., "Gly205Arg+Ser411 Phe” or “G205R+S411 F", representing substitutions at positions 205 and 41 1 of glycine (G) with arginine (R) and serine (S) with phenylalanine (F), respectively.
- + addition marks
- Insertions For an amino acid insertion, the following nomenclature is used: Original amino acid, position, original amino acid, inserted amino acid. Accordingly, the insertion of lysine after glycine at position 195 is designated “Gly195GlyLys" or “G195GK”. An insertion of multiple amino acids is designated [Original amino acid, position, original amino acid, inserted amino acid #1 , inserted amino acid #2; etc.]. For example, the insertion of lysine and alanine after glycine at position 195 is indicated as "Gly195GlyLysAla" or "G195GKA”.
- Variants comprising multiple alterations are separated by addition marks ("+"), e.g., "Arg170Tyr+Gly195Glu” or “R170Y+G195E” representing a substitution of arginine and glycine at positions 170 and 195 with tyrosine and glutamic acid, respectively.
- Multiple alteration variants may also be separated by a comma (",”), e.g., "Arg170Tyr,Gly195Glu” or "R170Y,G195E” representing a substitution of arginine and glycine at positions 170 and 195 with tyrosine and glutamic acid, respectively.
- the known xanthan endoglucanase having SEQ ID NO: 2 is a large enzyme (>1000 residues), it is therefore extremely laborious and expensive to target its properties for improvement of, e.g. stability in a detergent composition.
- the present invention has targeted specific regions of the endoglucanase which have an effect on the stability of the endoglucanase molecules using protein engineering to a region selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 11 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 8
- the present invention dramatically narrows down the number of residues to target when trying to stabilize endoglucanase molecules using protein engineering.
- regions in the protein sequence of the known xanthan endoglucanase having SEQ ID NO: 2 that are affected when the molecule is incubated in detergent are the following: region 10 corre- sponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2.
- This embodiment relates to an important guidance on where
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in a region selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ ID
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in a region selected from the group consisting of:
- region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 1 , 2, 3, 4, 5, 6, 7, 8, 9, 10, 1 1 , 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 , 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 , 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 , 42, 43, 44, 45, 46, 47, 48, 49, 50, 51 , 52, 53, 54, 55, 56, 57, 58, 59, 60, 61 , 62, 63, 64, 65, 66, 67, 68, 69, 70, 71 , 72, 73, 74, 75, 76, 77, 78, 79, 80, 81 , 82, 83, 84, 85, 86, 87, 88, 89, 90, 91 , 92, 93, 94, wherein said positions correspond to amino acid positions of
- region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 107, 108, 109, 1 10, 1 11 , 112, 1 13, 1 14, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
- region 12 corresponding to amino acids 139 to 209of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 139, 140, 141 , 142, 143, 144, 145, 146, 147, 148, 149, 150, 151 , 152, 153, 154, 155, 156, 157, 158, 159, 160, 161 , 162, 163, 164, 165, 166, 167, 168, 169, 170, 171 , 172, 173, 174, 175, 176, 177, 178, 179, 180, 181 , 182, 183, 184, 185, 186, 187, 188, 189, 190, 191 , 192, 193, 194, 195, 196, 197, 198, 199, 200, 201 , 202, 203, 204, 205, 206, 207, 208, 209,
- v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 302, 303, 304, 305, 306, 307, 308, 309, 310, 311 , 312, 313, 314, 315, 316, 317, 318, 319, 320, 321 , 322, 323, 324, 325, 326, 327, 328, 329, 330, 331 , 332, 333, 334, 335, 336, 337, 338, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
- region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 362, 363, 364, 365, 366, 367, 368, 369, 370, 371 , 372, 373, 374, 375, 376, 377, 378, 379, 380, 381 , 382, 383, 384, 385, 386, 387, 388, 389, 390, 391 , 392, 393, 394, 395, 396, 397, 398, 399, 400, 401 , 402, 403, 404, 405, 406, 407, 408, 409, 410, 411 , 412, 413, 414, 415, 416, 417, 418, 419, 420, 421 , 422, 423, 424, 425, 426, 427, 428, 429, 430, 431 , 432, 433, 434, 4
- region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 596, 597, 598, 599, 600, 601 , 602, 603, 604, 605, 606, 607, 608, 609, 610, 61 1 , wherein said positions correspond to amino acid positions of
- SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2)
- region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 661 , 662, 663, 664, 665, 666, 667, 668, 669, 670, 671 , 672, 673, 674, 675, 676, 677, 678, 679, 680, 681 , 682, 683, 684, 685, 686, 687, 688, 689, 690, 691 , 692, 693, 694, 695, 696, 697, 698, 699, 700, 701 , 702, 703, 704, 705, 706, 707, 708,
- ix) region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 829, 830, 831 , 832, 833, 834, 835, 836, 837, 838, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2)
- x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions 1043, 1044, 1045, 1046, 1047, 1048, 1049, 1050, 1051 , 1052, 1053, 1054, 1055, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g. , using the numbering of SEQ ID NO: 2).
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglu- canase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in one or more regions selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of S
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant as described herein having multiple alterations (such as 2, 3, 4, 5, 6, 7, 8, 9 or 10) in one region (e.g., of SEQ ID NO: 2 or another parent endoglucanase) selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant as described herein having multiple alterations (e.g., 2, 3, 4, 5, 6, 7, 8, 9 or 10) in multiple regions (e.g., 2, 3, 4, 5, 6, 7, 8, 9 or 10) (e.g., of SEQ ID NO: 2 or another parent endoglucanase) selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID
- the present invention relates to detergent compositions comprising endoglucanase variants, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more (e.g., several) positions of the mature parent polypeptide (e.g., SEQ ID NO: 2), wherein each alteration is independently a substitution, insertion or deletion, wherein the variant has endoglucanase activity.
- an alteration e.g., a substitution, deletion or insertion
- SEQ ID NO: 2 e.g., SEQ ID NO: 2
- the variant has sequence identity of at least 60%, e.g., at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99%, but less than 100%, to the amino acid sequence of the parent endoglucanase.
- the variant has at least 60%, e.g., at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, such as at least 96%, at least 97%, at least 98%, or at least 99%, but less than 100%, sequence identity to the mature polypeptide of SEQ ID NO: 2.
- the present invention relates to a detergent composition
- a detergent composition comprising an endoglucanase variant of the invention, wherein said variant has at least 61 %, at least 62%, at least 63%, at least 64%, at least 65%, at least 66%, at least 67%, at least 68%, at least 69%, at least 70%, at least 71 %, at least 72%, at least 73%, at least 74%, at least 75%, at least 76%, at least 77%, at least 78%, at least 79%, at least 80%, at least 81 %, at least 82%, at least 83%, at least 84%, at least 85%, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to SEQ ID NO: 2.
- a variant comprises an alteration at one or more (e.g., several) positions corresponding to positions 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 31 1 , 313, 387, 388, 390, 403, 408, 410, 416, 448, 451 , 471 , 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 71 1 , 719, 754, 756, 760, 781 , 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
- the endoglucanase variant comprised in the detergent compositions of the invention further comprises an alteration at one or more positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651, 824, 848, 869, 880, 881, 883, 887, 892, 905, 906, 912, 921, 928, 934, 937, 948, 956, 999 and 1037 of SEQ ID NO: 2.
- a variant comprises an alteration at two positions corresponding to any of positions 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 311, 313, 387, 388, 390, 403, 408, 410, 416, 448, 451, 471, 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 711, 719, 754, 756, 760, 781, 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
- the endoglucanase variant comprised in the detergent compositions of the invention further comprises an alteration at one or more positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651 , 824, 848, 869, 880, 881 , 883, 887, 892, 905, 906, 912, 921 , 928, 934, 937, 948, 956, 999 and 1037 of SEQ ID NO: 2.
- a variant comprises an alteration at three positions corresponding to any of positions 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 311, 313, 387, 388, 390, 403, 408, 410, 416, 448, 451, 471, 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 711 , 719, 754, 756, 760, 781 , 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
- the endoglucanase variant comprised in the detergent compositions of the invention further comprises an alteration at one or more positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651, 824, 848, 869, 880, 881, 883, 887, 892, 905, 906, 912, 921, 928, 934, 937, 948, 956, 999 and 1037 of SEQ ID NO: 2.
- the variant comprises an alteration in the positions corresponding to: 17+20, 17+51, 17+53, 17+55, 17+56, 17+60, 17+63, 17+79, 17+87, 17+192, 17+302, 7+387, 17+388, 17+390, 17+403, 17+408, 17+410, 17+416, 17+448, 17+451, 17+471, 17+472, 17+507, 17+512, 17+515, 17+538, 17+598, 17+602, 17+605, 17+609, 17+676, 17+694, 17+698, 17+699, 17+711, 17+754, 17+760, 17+781, 17+786, 17+797, 17+834, 17+835, 20+51, 20+53, 20+55, 20+56, 20+60, 20+63, 20+79, 20+87, 20+192, 20+302, 20+387, 20+388, 20+390, 20+403, 20+408,
- 60+408+602 60+408+605, 60+408+609, 60+408+676, 60+408+694, 60+408+698, 60+408+699,
- 60+416+602 60+416+605, 60+416+609, 60+416+676, 60+416+694, 60+416+698, 60+416+699,
- 79+416+834 79+416+835, 79+448+451, 79+448+471 , 79+448+472, 79+448+507, 79+448+512,
Landscapes
- Chemical & Material Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Organic Chemistry (AREA)
- Health & Medical Sciences (AREA)
- Engineering & Computer Science (AREA)
- Wood Science & Technology (AREA)
- Genetics & Genomics (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Zoology (AREA)
- General Health & Medical Sciences (AREA)
- Biochemistry (AREA)
- General Engineering & Computer Science (AREA)
- Microbiology (AREA)
- Biotechnology (AREA)
- Biomedical Technology (AREA)
- Molecular Biology (AREA)
- Medicinal Chemistry (AREA)
- Chemical Kinetics & Catalysis (AREA)
- Oil, Petroleum & Natural Gas (AREA)
- Detergent Compositions (AREA)
Abstract
The present invention relates to detergent compositions comprising endoglucanase variants and methods for use of said compositions.
Description
DETERGENT COMPOSITIONS COMPRISING GH9 ENDOGLUCANASE VARIANTS II
REFERENCE TO A SEQUENCE LISTING
This application contains a Sequence Listing in computer readable form, which is incorporated herein by reference.
BACKGROUND OF THE INVENTION
Field of the Invention
The present invention relates to detergent compositions comprising novel GH9 endoglucanase variants exhibiting alterations relative to the parent GH9 endoglucanase in one or more properties including: detergent stability (e.g. improved stability in a detergent composition,) and/or storage stability (e.g. improved storage stability in a detergent composition). The present invention further relates to detergent compositions comprising novel GH9 endoglucanase variants having activity on xanthan gum pre-treated with xanthan lyase. The invention also relates to methods for producing and using the compositions of the invention. Variants as described herein are particularly suitable for use in cleaning processes and detergent compositions, such as laundry compositions and dish wash compositions, including hand wash and automatic dish wash compositions. Description of the Related Art
Xanthan gum is a polysaccharide derived from the bacterial coat of Xanthomonas campestris. It is produced by the fermentation of glucose, sucrose, or lactose by the Xanthomonas campestris bacterium. After a fermentation period, the polysaccharide is precipitated from a growth medium with isopropyl alcohol, dried, and ground into a fine powder. Later, it is added to a liquid medium to form the gum. Xanthan gum is a natural polysaccharide consisting of different sugars which are connected by several different bonds, such as p-D-mannosyl-p-D-1 ,4-glucuronosyl bonds and -D-glucosyl- -D-1 ,4-glucosyl bonds. Xanthan gum is at least partly soluble in water and forms highly viscous solutions or gels. Complete enzymatic degradation of xanthan gum requires several enzymatic activities including xanthan lyase activity and endo- -1 ,4-glu- canase activity. Xanthan lyases are enzymes that cleave the p-D-mannosyl-p-D-1 ,4-glucuronosyl bond of xanthan and have been described in the literature. Xanthan lyases are known in the art, e.g. two xanthan lyases have been isolated from Paenibacillus alginolyticus XL-1 (e.g. Ruijssenaars ef al. (1999) Ά pyruvated mannose-specific xanthan lyase involved in xanthan degradation by Paenibacillus alginolyticus XL-1 ', Appl. Environ. Microbiol. 65(6): 2446-2452, and Ruijssenaars ef al. (2000), Ά novel gene encoding xanthan lyase of Paenibacillus alginolyticus strain XL-1 ', Appl. Environ. Microbiol. 66(9): 3945-3950). Glycoside hydrolases are enzymes that catalyse the hydrolysis of the glycosyl bond to release smaller sugars. There are over 100
classes of glycoside hydrolases which have been classified, see Henrissat ei al. (1991 ) Ά classification of glycosyl hydrolases based on amino-acid sequence similarities', J. Biochem. 280: 309-316 and the Uniprot website at www.cazy.org. The glycoside hydrolase family 9 (GH9) consists of over 70 different enzymes that are mostly endo-glucanases (EC 3.2.1 .4), cellobiohydrolases (EC 3.2.1.91 ), β-glucosidases (EC 3.2.1 .21 ) and exo^-glucosaminidase (EC 3.2.1.165). In recent years xanthan gum has been used as an ingredient in many consumer products including foods (e.g. as thickening agent in salad dressings and dairy products) and cosmetics (e.g. as stabilizer and thickener in toothpaste and make-up, creams and lotions to prevent ingredients from separating and to provide the right texture of the product). Further xanthan gum has found use in the oil industry as an additive to regulate the viscosity of drilling fluids etc. The widespread use of xanthan gum has led to a desire to degrade solutions, gels or mixtures containing xanthan gum thereby allowing easier removal of the byproducts. Endoglucanases and xanthan lyases for the degradation of xanthan gum and the use of such enzymes for cleaning purposes, such as the removal of xanthan gum containing stains, and in the drilling and oil industries are known in the art, e.g. WO2013167581 A1 .
The known xanthan endoglucanase having SEQ ID NO: 2 was found to be sensitive to storage in detergent. To improve the applicability and/or cost and/or the performance of such enzymes there is an ongoing search for variants with altered properties, such as increased stability, e.g. improved stability in a detergent composition. However, mutagenesis of large enzymes followed by purification and functional analysis of mutant libraries can be very expensive and laborious. SUMMARY OF THE INVENTION
Since the known xanthan endoglucanase having SEQ ID NO: 2 is a large enzyme (>1000 residues), it is difficult and expensive to randomly target its properties for improvement of, e.g., stability in a detergent composition.
In some aspects, the present invention identifies regions in the protein sequence/structure of the known xanthan endoglucanase having SEQ ID NO: 2 that are relevant for e.g. storage stability, and therefore provides an important guidance on where to mutate an endoglucanase in order to stabilize the molecule in a detergent.
In some aspects, the present invention relates to a detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in a region selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ
ID NO: 2.
In some aspects, the present invention relates to a detergent composition comprising an endoglu- canase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in a region selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2; preferably said endoglucanase variant has activity on xanthan gum pre-treated with xanthan lyase.
In some aspects, the present invention relates to a detergent composition comprising an endoglucanase variant having at least 61 %, at least 62%, at least 63%, at least 64%, at least 65%, at least 66%, at least 67%, at least 68%, at least 69%, at least 70%, at least 71 %, at least 72%, at least 73%, at least 74%, at least 75%, at least 76%, at least 77%, at least 78%, at least 79%, at least 80%, at least 81 %, at least 82%, at least 83%, at least 84%, at least 85%, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to SEQ ID NO: 2.
In some aspects, the present invention relates to a detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in a region selected from the group consisting of:
i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 1 , 2, 3, 4, 5, 6, 7, 8, 9, 10, 1 1 , 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 , 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 , 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 , 42,
43, 44, 45, 46, 47, 48, 49, 50, 51 , 52, 53, 54, 55, 56, 57, 58, 59, 60, 61 , 62, 63, 64, 65, 66, 67, 68, 69, 70, 71 , 72, 73, 74, 75, 76, 77, 78, 79, 80, 81 , 82, 83, 84, 85, 86, 87, 88, 89, 90, 91 , 92, 93, 94, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
ii) region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 107, 108, 109, 110, 1 11 , 112, 1 13, 1 14, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
iii) region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 139, 140, 141 , 142, 143, 144, 145, 146,
147, 148, 149, 150, 151 , 152, 153, 154, 155, 156, 157, 158, 159, 160, 161 , 162, 163, 164, 165, 166,
167, 168, 169, 170, 171 , 172, 173, 174, 175, 176, 177, 178, 179, 180, 181 , 182, 183, 184, 185, 186, 187, 188, 189, 190, 191 , 192, 193, 194, 195, 196, 197, 198, 199, 200, 201 , 202, 203, 204, 205, 206, 207, 208, 209, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 252, 253, 254, 255, 256, 257, 258, 259, 260, 261 , 262, 263, 264, 265, 266, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 302, 303, 304, 305, 306, 307, 308, 309,
310, 311 , 312, 313, 314, 315, 316, 317, 318, 319, 320, 321 , 322, 323, 324, 325, 326, 327, 328, 329, 330, 331 , 332, 333, 334, 335, 336, 337, 338, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
vi) region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 362, 363, 364, 365, 366, 367, 368, 369,
370, 371 , 372, 373, 374, 375, 376, 377, 378, 379, 380, 381 , 382, 383, 384, 385, 386, 387, 388, 389, 390, 391 , 392, 393, 394, 395, 396, 397, 398, 399, 400, 401 , 402, 403, 404, 405, 406, 407, 408, 409, 410, 411 , 412, 413, 414, 415, 416, 417, 418, 419, 420, 421 , 422, 423, 424, 425, 426, 427, 428, 429, 430, 431 , 432, 433, 434, 435, 436, 437, 438, 439, 440, 441 , 442, 443, 444, 445, 446, 447, 448, 449, 450, 451 , 452, 453, 454, 455, 456, 457, 458, 459, 460, 461 , 462, 463, 464, 465, 466, 467, 468, 469,
470, 471 , 472, 473, 474, 475, 476, 477, 478, 479, 480, 481 , 482, 483, 484, 485, 486, 487, 488, 489, 490, 491 , 492, 493, 494, 495, 496, 497, 498, 499, 500, 501 , 502, 503, 504, 505, 506, 507, 508, 509, 510, 511 , 512, 513, 514, 515, 516, 517, 518, 519, 520, 521 , 522, 523, 524, 525, 526, 527, 528, 529, 530, 531 , 532, 533, 534, 535, 536, 537, 538, 539, 540, 541 , 542, 543, 544, 545, 546, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID
NO: 2),
vii) region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 596, 597, 598, 599, 600, 601 , 602, 603, 604, 605, 606, 607, 608, 609, 610, 61 1 , wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
viii) region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 661 , 662, 663, 664, 665, 666, 667, 668, 669, 670, 671 , 672, 673, 674, 675, 676, 677, 678, 679, 680, 681 , 682, 683, 684, 685, 686, 687, 688, 689, 690, 691 , 692, 693, 694, 695, 696, 697, 698, 699, 700, 701 , 702, 703, 704, 705, 706, 707, 708, 709, 710, 711 , 712, 713, 714, 715, 716, 717, 718, 719, 720, 721 , 722, 723, 724, 725, 726, 727, 728,
729, 730, 731 , 732, 733, 734, 735, 736, 737, 738, 739, 740, 741 , 742, 743, 744, 745, 746, 747, 748,
749, 750, 751 , 752, 753, 754, 755, 756, 757, 758, 759, 760, 761 , 762, 763, 764, 765, 766, 767, 768, 769, 770, 771 , 772, 773, 774, 775, 776, 777, 778, 779, 780, 781 , 782, 783, 784, 785, 786, 787, 788, 789, 790, 791 , 792, 793, 794, 795, 796, 797, 798, 799, 800, 801 , 802, 803, 804, 805, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
ix) region 18 corresponding to amino acids 829 to 838 to 1042 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 829, 830, 831 , 832, 833, 834, 835, 836, 837, 838, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2)
x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions 1043, 1044, 1045, 1046, 1047, 1048, 1049, 1050, 1051 , 1052, 1053, 1054, 1055, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2).
In some aspects, the present invention relates to a detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in: a) one or more regions selected from the group consisting of:
i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 1 , 2, 3, 4, 5, 6, 7, 8, 9, 10, 1 1 , 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 , 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 , 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 , 42, 43, 44, 45, 46, 47, 48, 49, 50, 51 , 52, 53, 54, 55, 56, 57, 58, 59, 60, 61 , 62, 63, 64, 65, 66, 67, 68, 69, 70, 71 , 72, 73, 74, 75, 76, 77, 78, 79, 80, 81 , 82, 83, 84, 85, 86, 87, 88, 89, 90, 91 , 92, 93, 94, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
ii) region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 107, 108, 109, 1 10, 1 11 , 112, 1 13, 1 14, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
iii) region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 139, 140, 141 , 142, 143, 144, 145, 146, 147, 148, 149, 150, 151 , 152, 153, 154, 155, 156, 157, 158, 159, 160, 161 , 162, 163, 164, 165, 166, 167, 168, 169, 170, 171 , 172, 173, 174, 175, 176, 177, 178, 179, 180, 181 , 182, 183, 184, 185, 186, 187, 188, 189, 190, 191 , 192, 193, 194, 195, 196, 197, 198, 199, 200, 201 , 202, 203, 204, 205, 206, 207, 208, 209, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 252, 253, 254, 255, 256, 257, 258, 259,
260, 261 , 262, 263, 264, 265, 266, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 302, 303, 304, 305, 306, 307, 308, 309, 310, 311 , 312, 313, 314, 315, 316, 317, 318, 319, 320, 321 , 322, 323, 324, 325, 326, 327, 328, 329,
330, 331 , 332, 333, 334, 335, 336, 337, 338, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
vi) region 15 corresponding to amino acids 362 to 546of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 362, 363, 364, 365, 366, 367, 368, 369, 370, 371 , 372, 373, 374, 375, 376, 377, 378, 379, 380, 381 , 382, 383, 384, 385, 386, 387, 388, 389, 390,
391 , 392, 393, 394, 395, 396, 397, 398, 399, 400, 401 , 402, 403, 404, 405, 406, 407, 408, 409, 410, 41 1 , 412, 413, 414, 415, 416, 417, 418, 419, 420, 421 , 422, 423, 424, 425, 426, 427, 428, 429, 430, 431 , 432, 433, 434, 435, 436, 437, 438, 439, 440, 441 , 442, 443, 444, 445, 446, 447, 448, 449, 450, 451 , 452, 453, 454, 455, 456, 457, 458, 459, 460, 461 , 462, 463, 464, 465, 466, 467, 468, 469, 470, 471 , 472, 473, 474, 475, 476, 477, 478, 479, 480, 481 , 482, 483, 484, 485, 486, 487, 488, 489, 490,
491 , 492, 493, 494, 495, 496, 497, 498, 499, 500, 501 , 502, 503, 504, 505, 506, 507, 508, 509, 510, 51 1 , 512, 513, 514, 515, 516, 517, 518, 519, 520, 521 , 522, 523, 524, 525, 526, 527, 528, 529, 530, 531 , 532, 533, 534, 535, 536, 537, 538, 539, 540, 541 , 542, 543, 544, 545, 546, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2), vii) region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 596, 597, 598, 599, 600, 601 , 602, 603, 604, 605, 606, 607, 608, 609, 610, 611 , wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
viii) region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 661 , 662, 663, 664, 665, 666, 667, 668,
669, 670, 671 , 672, 673, 674, 675, 676, 677, 678, 679, 680, 681 , 682, 683, 684, 685, 686, 687, 688, 689, 690, 691 , 692, 693, 694, 695, 696, 697, 698, 699, 700, 701 , 702, 703, 704, 705, 706, 707, 708, 709, 710, 711 , 712, 713, 714, 715, 716, 717, 718, 719, 720, 721 , 722, 723, 724, 725, 726, 727, 728, 729, 730, 731 , 732, 733, 734, 735, 736, 737, 738, 739, 740, 741 , 742, 743, 744, 745, 746, 747, 748, 749, 750, 751 , 752, 753, 754, 755, 756, 757, 758, 759, 760, 761 , 762, 763, 764, 765, 766, 767, 768,
769, 770, 771 , 772, 773, 774, 775, 776, 777, 778, 779, 780, 781 , 782, 783, 784, 785, 786, 787, 788, 789, 790, 791 , 792, 793, 794, 795, 796, 797, 798, 799, 800, 801 , 802, 803, 804, 805, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
ix) region 18 corresponding to amino acids 829 to 838 to 1042 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 829, 830, 831 , 832, 833, 834,
835, 836, 837, 838, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2)
x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions 1043, 1044, 1045, 1046, 1047, 1048, 1049, 1050, 1051 , 1052, 1053, 1054, 1055, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2).
In some aspects, the present invention relates to a detergent composition comprising an endoglu- canase variant having an alteration (e.g., a substitution, deletion or insertion) at one or more positions selected from the group consisting of positions: 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 31 1 , 313, 387, 388, 390, 403, 408, 410, 416, 448, 451 , 471 , 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 71 1 , 719, 754, 756, 760, 781 , 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
In some aspects, the present invention relates to a detergent composition comprising an endoglu- canase variant having an alteration (e.g., a substitution, deletion or insertion) at one or more positions se- lected from the group consisting of: S17A, F20P, F20N, F20G, F20Y, K51 Q, K51 H, E53P, E53G, Y55M, V56M, Y60F, S63F, T87R, A186P, K192N, I302D, I302H, I302V, I302M, H311 N, S313D, I387T, K388R, K390Q, I403Y, E408D, E408S, E408P, E408A, E408G, E408N, P410G, Q416S, Q416D, A448E, A448W, A448S, K451 S, G471 S, S472Y, D476R, Q489P, K507R, K512P, S515V, S538C, Y579W, S598Q, A599S, I602T, I602D, V603P, S605T, G609E, D676H, A688G, Y690F, T694A, T697G, R698W, T699A, T71 1V, T711Y, W719R, K754R, V756H, V756Y, S760G, T781 M, N786K, T797S, A824D, N833D, Q834E, S835D, and F1048W wherein numbering is according to SEQ ID NO: 2.
In one embodiment, the present invention relates to a detergent composition comprising an endoglu- canase variant comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions selected from the group consisting of alterations in positions: 17, 20, 302, 31 1 , 313, 408, 476, 602, 688, 697, and 719, wherein numbering is according to SEQ ID NO: 2.
In a preferred embodiment, the endoglucanase variant comprised in the detergent compositions of the invention comprises an alteration at one or more positions selected from the group consisting of alterations in positions: S17A, F20P, I302D, H311 N, S313D, E408D, D476R, I602T, A688G, T697G, and W719R, wherein numbering is according to SEQ ID NO: 2.
In a preferred embodiment, the endoglucanase variant comprised in the detergent compositions of the invention further comprises an alteration at one or more positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651 , 824, 848, 869, 880, 881 , 883, 887, 892, 905, 906, 912, 921 , 928, 934, 937, 948, 956, 999, and 1037.
In a preferred embodiment, the endoglucanase variant comprised in the detergent compositions of the invention further comprises an alteration (e.g., substitution) at one or more positions selected from the group consisting of: N216D, N216Q, A283D, A346D, A559P, A559N, A564E, Y579W, S636K, S636N, F638N,
A651 P, A824D, N848D, A869V, R880K, V881 T, V881 Q, T883R, T887K, T892P, N905D, F906A, A912V, K921 R, S928D, Y934G, A937E, K948R, Q956Y, T999R, and A1037E.
In a preferred embodiment, the endoglucanase variant comprised in the detergent compositions of the invention comprises one or more of the following set of substitutions:
F20Y+A448S+T781 M
E408D+K451 S
E408D+A448E
E408D
Y579W+E408D
In a preferred embodiment, the endoglucanase variant comprised in the detergent compositions of the invention comprises one or more of the following set of substitutions:
F20P,I302D,S313D,E408D,D476R,Q489P,Y579W,I602T,S636N,T697G,W719R,V756Y,A824D, N848D,V881 Q,T887K,N905D,F906A,A937E,T999R,A1037E,F1048W
F20P,K51 Q,I302D,S313D,E408D,D476R,Q489P,Y579W,I602T,S636K,A651 P,T697G,W719R, V756Y,A824D,N848D,T883R,T887K,F906A,S928D,A937E,A1037E
N216D.S313D,A346D,E408D,D476R,Q489P,A559P,Y579W,I602T,F638N,A651 P,A688G,T697G, W719R,V756H,R880K,T887K,K921 R,S928D,Y934G
F20P,I302D,S313D,A346D,E408D,D476R,Q489P,Y579W,I602T,T697G,W719R,V756Y,N848D, V881 Q,T887K,F906A,A937E
N216D.S313D,E408D,D476R,Q489P,A559P,Y579W,I602T,F638N,A651 P,A688G,T697G,W719R, V756H,Q834E,R880K,T887K,T892P,K921 R,S928D,Y934G
F20P,I302D,S313D,E408D,D476R,Q489P,A559N,Y579W,I602T,S636K,A651 P,T697G,W719R, V756Y,N848D,T883R,T887K,F906A,S928D,A937E
F20P,I302D,S313D,E408D,D476R,Q489P,Y579W,I602T,S636K,A651 P,T697G,W719R,V756Y, N848D,T883R,T887K,F906A,A937E
F20P,I302D,S313D,E408D,Q416S,D476R,Q489P,A559N,Y579W,I602T,S636K,A651 P,T697G, W719R,V756Y,N848D,T883R,T887K,F906A,A937E
F20P,I302D,S313D,E408D,D476R,Y579W,I602T,S636N,T697G,W719R,V756Y,A824D,N848D, V881 Q,T887K,F906A,S928D,A937E,T999R,F1048W
In a particular aspect, the endoglucanase variant as described herein is one that does not comprise any amino acid alteration at a position outside of regions 10, 1 1 , 12, 13, 14, 15, 16, 17, 18, and 19. In this aspect, the endoglucanase variant thus does not comprise any alteration (e.g., a substitution, deletion or insertion) in a region selected from the group consisting of: region 1 corresponding to amino acids 95 to 105 of SEQ ID NO: 2, region 2 corresponding to amino acids 115 to 138 of SEQ ID NO: 2, region 3 corresponding to amino acids 210 to 251 of SEQ ID NO: 2, region 4 corresponding to amino acids 267 to 301 of SEQ ID NO: 2, region 5 corresponding to amino acids 339 to 361 of SEQ ID NO: 2, region 6 corresponding to amino acids 547 to 595 of SEQ ID NO: 2, region 7 corresponding to amino acids 612 to 660 of SEQ ID NO: 2, region 8 corresponding to amino acids 806 to 828 of SEQ ID NO: 2, and region 9 corresponding to amino acids 839 to 1045 of SEQ ID NO: 2.
In some aspects, the present invention relates to a detergent composition comprising an endoglucanase variant having activity on xanthan gum pre-treated with xanthan lyase; preferably said activity comprises endoglucanase EC 3.2.1.4 activity, further preferably said activity is endoglucanase EC 3.2.1 .4 activity.
In some aspects, the present invention relates to a detergent composition comprising an endoglucanase variant having an improved stability in a detergent composition compared to a parent endoglucanase
(e.g., with SEQ ID NO: 2).
In some aspects, the present invention relates to a detergent composition comprising an endoglu- canase variant having a half-life improvement factor (HIF) of >1.0 relative to a parent endoglucanase, e.g. an endoglucanase of SEQ ID NO: 2.
In some aspects, the present invention relates to a detergent composition comprising an isolated GH9 endoglucanase variant having activity on xanthan gum pre-treated with xanthan lyase. In a further aspect, the detergent composition further comprises an isolated polypeptide having xanthan lyase activity.
In some aspects, the present invention relates to a detergent composition comprising one or more detergent components for degrading xanthan gum.
In some aspects, the present invention relates to use of a detergent composition of the present invention, wherein said use is selected from the group consisting of: use for degrading xanthan gum and use in a cleaning process, such as laundry or hard surface cleaning such as dish wash.
In some aspects, the present invention further relates to the use of a detergent composition of the invention for degrading xanthan gum, for washing or cleaning textiles and/or hard surfaces, such as dish wash, wherein the composition has an enzyme detergency benefit.
In some aspects, the present invention also relates to methods of degrading xanthan gum using detergent compositions of the present invention, wherein xanthan gum is on the surface of a hard surface or textile.
OVERVIEW OF SEQUENCE LISTING
SEQ ID NO: 1 is the DNA sequence of the parent mature endoglucanase from a strain of a Paenibacil- lus sp.
SEQ ID NO: 2 is the amino acid sequence of mature polypeptide encoded by SEQ ID NO: 1 .
SEQ ID NO: 3 is the DNA sequence of the alpha-amylase secretion signal from Bacillus licheniformis. SEQ ID NO: 4 is the amino acid sequence of the alpha-amylase secretion signal from Bacillus licheniformis.
DEFINITIONS
cDNA: The term "cDNA" means a DNA molecule that can be prepared by reverse transcription from a mature, spliced, mRNA molecule obtained from a eukaryotic or prokaryotic cell. cDNA lacks intron sequences that may be present in the corresponding genomic DNA. The initial, primary RNA transcript is a precursor to mRNA that is processed through a series of steps, including splicing, before appearing as mature spliced mRNA.
Cleaning or Detergent Application: the term "cleaning or detergent application" means applying the endoglucanase of the application in any composition for the purpose of cleaning or washing, by hand, machine or automated, a hard surface or a textile.
Cleaning Composition: the term "cleaning composition" refers to compositions that find use in the removal of undesired compounds from items to be cleaned, such as textiles, dishes, and hard surfaces. The terms encompass any materials/compounds selected for the particular type of cleaning composition desired and the form of the product (e.g., liquid, gel, powder, granulate, paste, or spray compositions) and includes, but is not limited to, detergent compositions (e.g., liquid and/or solid laundry detergents and fine fabric detergents; hard surface cleaning formulations, such as for glass, wood, ceramic and metal counter tops and windows; carpet cleaners; oven cleaners; fabric fresheners; fabric softeners; and textile and laundry pre- spotters, as well as dish wash detergents). In addition to the endoglucanase, the detergent formulation may contain one or more additional enzymes (such as xanthan lyases, proteases, amylases, lipases, cutinases, cellulases, endoglucanases, xyloglucanases, pectinases, pectin lyases, xanthanases, peroxidases, haloperoxygenases, catalases and mannanases, or any mixture thereof), and/or components such as surfactants, builders, chelators or chelating agents, bleach system or bleach components, polymers, fabric conditioners, foam boosters, suds suppressors, dyes, perfume, tannish inhibitors, optical brighteners, bactericides, fungicides, soil suspending agents, anti-corrosion agents, enzyme inhibitors or stabilizers, enzyme activators, transferase(s), hydrolytic enzymes, oxido reductases, bluing agents and fluorescent dyes, antioxidants, and solubilizers.
Coding sequence: The term "coding sequence" means a polynucleotide, which directly specifies the amino acid sequence of a polypeptide. The boundaries of the coding sequence are generally determined by an open reading frame, which begins with a start codon such as ATG, GTG, or TTG and ends with a stop codon such as TAA, TAG, or TGA. The coding sequence may be a genomic DNA, cDNA, synthetic DNA, or a combination thereof.
Colour clarification: During washing and wearing loose or broken fibers can accumulate on the surface of the fabrics. One consequence can be that the colours of the fabric appear less bright or less intense because of the surface contaminations. Removal of the loose or broken fibers from the textile will partly restore the original colours and looks of the textile. By the term "colour clarification", as used herein, is meant the partial restoration of the initial colours of textile.
Control sequences: The term "control sequences" means nucleic acid sequences necessary for expression of a polynucleotide encoding a mature polypeptide of the present invention. Each control sequence may be native (i.e., from the same gene) or foreign (i.e. , from a different gene) to the polynucleotide encod- ing the polypeptide or native or foreign to each other. Such control sequences include, but are not limited to, a leader, polyadenylation sequence, propeptide sequence, promoter, signal peptide sequence, and transcription terminator. At a minimum, the control sequences include a promoter, and transcriptional and trans- lational stop signals. The control sequences may be provided with linkers for the purpose of introducing specific restriction sites facilitating ligation of the control sequences with the coding region of the polynucle- otide encoding a polypeptide.
Corresponding to: The term "corresponding to" as used herein, refers to a way of determining the
specific amino acid of a sequence wherein reference is made to a specific amino acid sequence. E.g. for the purposes of the present invention, when references are made to specific amino acid positions, the skilled person would be able to align another amino acid sequence to said amino acid sequence that reference has been made to, in order to determine which specific amino acid may be of interest in said another amino acid sequence. Alignment of another amino acid sequence with e.g. the sequence as set forth in SEQ ID NO: 2, or any other sequence listed herein, has been described elsewhere herein. Alternative alignment methods may be used, and are well-known for the skilled person.
Degrading xanthan gum and xanthan gum degrading activity: The terms "degrading xanthan gum" and "xanthan gum degrading activity" are used interchangeably and are defined as the depolymerization, degradation or breaking down of xanthan gum into smaller components. The degradation of xanthan gum can either be the removal of one or more side chain saccharides, the cutting of the backbone of xanthan gum into smaller components or the removal of one or more side chain saccharides and the cutting of the backbone of xanthan gum into smaller components. A preferred assay for measuring degradation of xanthan gum is the reducing sugar assay as described in example 3 herein. Non-limiting examples of the xan- than gum degrading activity include endoglucanase EC 3.2.1.4 activity.
Detergent component: the term "detergent component" is defined herein to mean the types of chemicals which can be used in detergent compositions. Examples of detergent components are surfactants, hydrotropes, builders, co-builders, chelators or chelating agents, bleaching system or bleach components, polymers, fabric hueing agents, fabric conditioners, foam boosters, suds suppressors, dispersants, dye transfer inhibitors, fluorescent whitening agents, perfume, optical brighteners, bactericides, fungicides, soil suspending agents, soil release polymers, anti-redeposition agents, enzyme inhibitors or stabilizers, enzyme activators, antioxidants, and solubilizers. The detergent composition may comprise of one or more of any type of detergent component.
Detergent composition: the term "detergent composition" refers to compositions that find use in the removal of undesired compounds from items to be cleaned, such as textiles, dishes, and hard surfaces. The detergent composition may be used to e.g. clean textiles, dishes and hard surfaces for both household cleaning and industrial cleaning. The terms encompass any materials/compounds selected for the particular type of cleaning composition desired and the form of the product (e.g., liquid, gel, powder, granulate, paste, or spray compositions) and includes, but is not limited to, detergent compositions (e.g., liquid and/or solid laundry detergents and fine fabric detergents; hard surface cleaning formulations, such as for glass, wood, ceramic and metal counter tops and windows; carpet cleaners; oven cleaners; fabric fresheners; fabric softeners; and textile and laundry pre-spotters, as well as dish wash detergents). In addition to containing a GH9 endoglucanase as described herein and/or xanthan lyase, the detergent formulation may contain one or more additional enzymes (such as xanthan lyases, proteases, amylases, lipases, cutinases, cellulases, endoglucanases, xyloglucanases, pectinases, pectin lyases, xanthanases, peroxidases, haloperoxygen- ases, catalases and mannanases, or any mixture thereof), and/or components such as surfactants, builders,
chelators or chelating agents, bleach system or bleach components, polymers, fabric conditioners, foam boosters, suds suppressors, dyes, perfume, tannish inhibitors, optical brighteners, bactericides, fungicides, soil suspending agents, anti-corrosion agents, enzyme inhibitors or stabilizers, enzyme activators, transferase^), hydrolytic enzymes, oxido reductases, bluing agents and fluorescent dyes, antioxidants, and solu- bilizers.
Dish wash: The term "dish wash" refers to all forms of washing dishes, e.g. by hand or automatic dish wash. Washing dishes includes, but is not limited to, the cleaning of all forms of crockery such as plates, cups, glasses, bowls, all forms of cutlery such as spoons, knives, forks and serving utensils as well as ceramics, plastics, metals, china, glass and acrylics.
Dish washing composition: The term "dish washing composition" refers to all forms of compositions for cleaning hard surfaces. The present invention is not restricted to any particular type of dish wash composition or any particular detergent.
Endoglucanase: The term "endoglucanase" or "EG" means an endo-1 ,4-(1 ,3;1 ,4)-beta-D-glucan 4- glucanohydrolase (EC 3.2.1 .4) that catalyzes endohydrolysis of 1 ,4-beta-D-glycosidic linkages in cellulose, cellulose derivatives (such as carboxymethyl cellulose and hydroxyethyl cellulose), lichenin, beta-1 ,4 bonds in mixed beta-1 ,3 glucans such as cereal beta-D-glucans, xyloglucans, xanthans and other plant material containing cellulosic components. Endoglucanase activity can be determined by measuring reduction in substrate viscosity or increase in reducing ends determined by a reducing sugar assay (Zhang ef a/., 2006, Biotechnology Advances 24: 452-481 ). A preferred assay for measuring endoglucanase activity is the re- ducing sugar assay as described in example 3 herein. Non-limiting examples of endoglucanases include the mature parent endoglucanase having SEQ ID NO: 2.
Enzyme detergency benefit: The term "enzyme detergency benefit" is defined herein as the advantageous effect an enzyme may add to a detergent compared to the same detergent without the enzyme. Important detergency benefits which can be provided by enzymes are stain removal with no or very little visible soils after washing and or cleaning, prevention or reduction of redeposition of soils released in the washing process an effect that also is termed anti-redeposition, restoring fully or partly the whiteness of textiles, which originally were white but after repeated use and wash have obtained a greyish or yellowish appearance an effect that also is termed whitening. Textile care benefits, which are not directly related to catalytic stain removal or prevention of redeposition of soils are also important for enzyme detergency ben- efits. Examples of such textile care benefits are prevention or reduction of dye transfer from one fabric to another fabric or another part of the same fabric an effect that is also termed dye transfer inhibition or anti- backstaining, removal of protruding or broken fibers from a fabric surface to decrease pilling tendencies or remove already existing pills or fuzz an effect that also is termed anti-pilling, improvement of the fabric- softness, colour clarification of the fabric and removal of particulate soils which are trapped in the fibers of the fabric or garment. Enzymatic bleaching is a further enzyme detergency benefit where the catalytic activity generally is used to catalyze the formation of bleaching component such as hydrogen peroxide or other
peroxides.
Expression: The term "expression" includes any step involved in the production of a polypeptide including, but not limited to, transcription, post-transcriptional modification, translation, post-translational modification, and secretion.
Expression vector: The term "expression vector" means a linear or circular DNA molecule that comprises a polynucleotide encoding a polypeptide and is operably linked to control sequences that provide for its expression.
Fragment: The term "fragment" means a polypeptide having one or more (e.g. , several) amino acids absent from the amino and/or carboxyl terminus of a mature polypeptide; wherein the fragment has en- doglucanase activity. In one aspect, a fragment contains at least 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94% or 95% of the number of amino acids of the mature polypeptide.
Endoglucanase variant having activity on xanthan gum pre-treated with xanthan lyase: The term "Endoglucanase variant having activity on xanthan gum pre-treated with xanthan lyase" or an "endoglucanase having activity on xanthan gum pre-treated with xanthan lyase and belonging to the GH9 class of glycosyl hydrolases" is defined as a polypeptide comprising a domain belonging to the GH9 class of glycosyl hydrolases, and having activity (e.g. enzymatic activity, xanthan degrading activity, endoglucanase EC 3.2.1 .4 activity) on xanthan gum pre-treated with xanthan lyase. A preferred assay for measuring activity on xanthan gum pre-treated with xanthan lyase is disclosed in example 3 herein.
Xanthan lyase variant having activity on xanthan gum: The term "Xanthan lyase variant having activity on xanthan gum" is defined as a polypeptide that cleaves the p-D-mannosyl-p-D-1 ,4-glucuronosyl bond of xanthan (e.g., xanthan lyase EC 4.2.2.12 activity). Examples of the xanthan lyase variants having activity on xanthan gum, are xanthan lyase polypeptides as such. Thus, polypeptides that that cleaves the p-D-mannosyl-p-D-1 ,4-glucuronosyl bond of xanthan.
Half-life: the term "half-life" is the time it takes for an enzyme to lose half of its enzymatic activity under a given set of conditions. It is denoted as T½ and is measured in hours (hrs). Half-lives can be calculated at a given detergent concentration and storage temperature for parent (e.g. wild-type) and/or variants, as the degradation follows an exponential decay and the incubation time (hours) is known, i.e., according to the following formulas:
T1 /2 (variant) = (Ln (0.5)/Ln (RA-variant/100))*Time
T112 (wild-type) = (Ln (0.5)/Ln (RA-wild-type/100))*Time
Where 'RA' is the residual activity in percent and 'Time' is the incubation time in hours.
Half-life improvement factor: the term "Half-life improvement factor" or "HIF" is the improvement of half-life of a variant compared to the parent polypeptide, such as the parent endoglucanase. A half-life improvement factor (HIF) under a given set of storage conditions (detergent concentration and temperature) can be calculated as :
Tl/2, variant
HIF = Tl/2, wt
where the wild-type (wt) is incubated under the same storage condition (detergent concentration and incubation temperature) as the variant. In the cases where the difference in stability between wild-type and variant is too big to accurately assess half-life for both wild-type and variant using the same incubation time, the incubation time for wild-type and variant is different e.g. 1 h for wild-type and 138 h for the most stable variants.
A preferred way of calculating HIF is also described in example 3 herein.
Hard surface cleaning: The term "Hard surface cleaning" is defined herein as cleaning of hard surfaces wherein hard surfaces may include floors, tables, walls, roofs etc. as well as surfaces of hard objects such as cars (car wash) and dishes (dish wash). Dish washing includes but are not limited to cleaning of plates, cups, glasses, bowls, and cutlery such as spoons, knives, forks, serving utensils, ceramics, plastics, metals, china, glass and acrylics.
Host cell: The term "host cell" means any cell type that is susceptible to transformation, transfection, transduction, or the like with a nucleic acid construct or expression vector comprising a polynucleotide of the present invention. The term "host cell" encompasses any progeny of a parent cell that is not identical to the parent cell due to mutations that occur during replication.
Improved property: The term "improved property" means a characteristic associated with a variant that is improved compared to the parent. Such improved properties include, but are not limited to, catalytic efficiency, catalytic rate, chemical stability, oxidation stability, pH activity, pH stability, specific activity, sta- bility under storage conditions, chelator stability, substrate binding, substrate cleavage, substrate specificity, substrate stability, surface properties, thermal activity, and thermostability.
Improved wash performance: The term "improved wash performance" is defined herein as a (variant) enzyme (also a blend of enzymes, not necessarily only variants but also backbones, and in combination with certain cleaning composition etc.) displaying an alteration of the wash performance of a variant relative to the wash performance of the parent variant e.g. by increased stain removal. The term "wash performance" includes wash performance in laundry but also e.g. in dish wash.
Isolated: The term "isolated" means a substance in a form or environment that does not occur in nature. Non-limiting examples of isolated substances include (1 ) any non-naturally occurring substance, (2) any substance including, but not limited to, any enzyme, variant, nucleic acid, protein, peptide or cofactor, that is at least partially removed from one or more or all of the naturally occurring constituents with which it is associated in nature; (3) any substance modified by the hand of man relative to that substance found in nature; or (4) any substance modified by increasing the amount of the substance relative to other components with which it is naturally associated (e.g. , multiple copies of a gene encoding the substance; use of a stronger promoter than the promoter naturally associated with the gene encoding the substance). An iso- lated substance may be present in a fermentation broth sample.
Laundering: The term "laundering" relates to both household laundering and industrial laundering and means the process of treating textiles with a solution containing a cleaning or detergent composition of the present invention. The laundering process can for example be carried out using e.g. a household or an industrial washing machine or can be carried out by hand.
Mature polypeptide: The term "mature polypeptide" means a polypeptide in its final form following translation and any post-translational modifications, such as N-terminal processing, C-terminal truncation, glycosylation, phosphorylation, etc. In one aspect, the mature polypeptide is amino acids 1 to 1055 of SEQ ID NO: 2.
It is known in the art that a host cell may produce a mixture of two of more different mature polypeptides (i.e., with a different C-terminal and/or N-terminal amino acid) expressed by the same polynucleotide. It is also known in the art that different host cells process polypeptides differently, and thus, one host cell expressing a polynucleotide may produce a different mature polypeptide (e.g., having a different C-terminal and/or N-terminal amino acid) as compared to another host cell expressing the same polynucleotide.
Mature polypeptide coding sequence: The term "mature polypeptide coding sequence" means a polynucleotide that encodes a mature polypeptide having enzymatic activity such as activity on xanthan gum pre-treated with xanthan lyase or xanthan lyase activity. In one aspect, the mature polypeptide coding sequence is nucleotides 1 to 3165 of SEQ ID NO: 1 .
Mutant: The term "mutant" means a polynucleotide encoding a variant.
Nucleic acid construct: The term "nucleic acid construct" means a nucleic acid molecule, either sin- gle- or double-stranded, which is isolated from a naturally occurring gene or is modified to contain segments of nucleic acids in a manner that would not otherwise exist in nature or which is synthetic, which comprises one or more control sequences.
Operably linked: The term "operably linked" means a configuration in which a control sequence is placed at an appropriate position relative to the coding sequence of a polynucleotide such that the control sequence directs expression of the coding sequence.
Parent: The term "parent" or "parent endoglucanase" means any polypeptide with endoglucanase activity to which an alteration is made to produce the enzyme variants of the present invention. In one aspect, the parent is an endoglucanase having the identical amino acid sequence of the variant, but not having the alterations at one or more of the specified positions. It will be understood, that the expression "having iden- tical amino acid sequence" relates to 100% sequence identity. Non-limiting examples of parent endoglu- canases include the mature parent endoglucanase having SEQ ID NO: 2.
Sequence identity: The relatedness between two amino acid sequences or between two nucleotide sequences is described by the parameter "sequence identity". For purposes of the present invention, the sequence identity between two amino acid sequences is determined using the Needleman-Wunsch algo- rithm (Needleman and Wunsch, 1970, J. Mol. Biol. 48: 443-453) as implemented in the Needle program of the EMBOSS package (EMBOSS: The European Molecular Biology Open Software Suite, Rice ef a/., 2000,
Trends Genet. 16: 276-277), preferably version 5.0.0 or later. The parameters used are gap open penalty of 10, gap extension penalty of 0.5, and the EBLOSUM62 (EMBOSS version of BLOSUM62) substitution matrix. The output of Needle labeled "longest identity" (obtained using the -nobrief option) is used as the percent identity and is calculated as follows:
(Identical Residues x 100)/(Length of Alignment - Total Number of Gaps in Alignment)
For purposes of the present invention, the sequence identity between two deoxyribonucleotide sequences is determined using the Needleman-Wunsch algorithm (Needleman and Wunsch, 1970, supra) as implemented in the Needle program of the EMBOSS package (EMBOSS: The European Molecular Biology Open Software Suite, Rice ef a/. , 2000, supra), preferably version 5.0.0 or later. The parameters used are gap open penalty of 10, gap extension penalty of 0.5, and the EDNAFULL (EMBOSS version of NCBI NUC4.4) substitution matrix. The output of Needle labeled "longest identity" (obtained using the -nobrief option) is used as the percent identity and is calculated as follows:
(Identical Deoxyribonucleotides x 100)/(Length of Alignment - Total Number of Gaps in Alignment) Stringency conditions: The different stringency conditions are defined as follows.
The term "very low stringency conditions" means for probes of at least 100 nucleotides in length, pre- hybridization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 25% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 45°C.
The term "low stringency conditions" means for probes of at least 100 nucleotides in length, prehybrid- ization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 25% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 50°C.
The term "medium stringency conditions" means for probes of at least 100 nucleotides in length, pre- hybridization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 35% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 55°C.
The term "medium-high stringency conditions" means for probes of at least 100 nucleotides in length, prehybridization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 35% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 60°C.
The term "high stringency conditions" means for probes of at least 100 nucleotides in length, prehybridization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured
salmon sperm DNA, and 50% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 65°C.
The term "very high stringency conditions" means for probes of at least 100 nucleotides in length, prehybridization and hybridization at 42°C in 5X SSPE, 0.3% SDS, 200 micrograms/ml sheared and denatured salmon sperm DNA, and 50% formamide, following standard Southern blotting procedures for 12 to 24 hours. The carrier material is finally washed three times each for 15 minutes using 2X SSC, 0.2% SDS at 70°C.
Subsequence: The term "subsequence" means a polynucleotide having one or more (e.g., several) nucleotides absent from the 5' and/or 3' end of a mature polypeptide coding sequence; wherein the subsequence encodes a fragment having enzymatic activity, such as activity on xanthan gum pre-treated with xanthan lyase or xanthan lyase activity.
Textile: The term "textile" means any textile material including yarns, yarn intermediates, fibers, non- woven materials, natural materials, synthetic materials, and any other textile material, fabrics made of these materials and products made from fabrics (e.g., garments and other articles). The textile or fabric may be in the form of knits, wovens, denims, non-wovens, felts, yarns, and towelling. The textile may be cellulose based such as natural cellulosics, including cotton, flax/linen, jute, ramie, sisal or coir or manmade cellulo- sics (e.g. originating from wood pulp) including viscose/rayon, ramie, cellulose acetate fibers (tricell), lyocell or blends thereof. The textile or fabric may also be non-cellulose based such as natural polyamides including wool, camel, cashmere, mohair, rabbit and silk or synthetic polymer such as nylon, aramid, polyester, acrylic, polypropylene and spandex elastane, or blends thereof as well as blend of cellulose based and non- cellulose based fibers. Examples of blends are blends of cotton and/or rayon/viscose with one or more companion material such as wool, synthetic fibers (e.g. polyamide fibers, acrylic fibers, polyester fibers, polyvinyl alcohol fibers, polyvinyl chloride fibers, polyurethane fibers, polyurea fibers, aramid fibers), and cellulose-containing fibers (e.g. rayon/viscose, ramie, flax/linen, jute, cellulose acetate fibers, lyocell). Fabric may be conventional washable laundry, for example stained household laundry. When the term fabric or garment is used it is intended to include the broader term textiles as well.
Textile care benefit: "Textile care benefits", which are not directly related to catalytic stain removal or prevention of redeposition of soils, are also important for enzyme detergency benefits. Examples of such textile care benefits are prevention or reduction of dye transfer from one textile to another textile or another part of the same textile an effect that is also termed dye transfer inhibition or anti-backstaining, removal of protruding or broken fibers from a textile surface to decrease pilling tendencies or remove already existing pills or fuzz an effect that also is termed anti-pilling, improvement of the textile-softness, colour clarification of the textile and removal of particulate soils which are trapped in the fibers of the textile. Enzymatic bleach- ing is a further enzyme detergency benefit where the catalytic activity generally is used to catalyze the formation of bleaching component such as hydrogen peroxide or other peroxides or other bleaching species.
Variant: The term "variant" means a polypeptide (e.g., a GH9 endoglucanase polypeptide) comprising an alteration i.e. , a substitution, insertion, and/or deletion, at one or more (e.g. , several) positions. A substitution means replacement of the amino acid occupying a position with a different amino acid ; a deletion means removal of the amino acid occupying a position; and an insertion means adding one or more (e.g. several) amino acids e.g., 1 -5 amino acids adjacent to and immediately following the amino acid occupying a position. Non-limiting examples of endoglucanase variants as described herein include endoglucanase variants having an activity on xanthan gum pre-treated with xanthan lyase. Non-limiting examples of variants as described herein further include variants having at least 20%, e.g., at least 40%, at least 50%, at least 60%, at least 70%, at least 80%, at least 90%, at least 95%, or at least 100% endoglucanase activity of the mature parent having SEQ ID NO: 2. A preferred assay for measuring activity on xanthan gum pre-treated with xanthan lyase is disclosed in example 3 herein.
Stability: The term "stability" means resistance or the degree of resistance to change, unfolding, disintegration, denaturation or activity loss. Non-limiting examples of stability include conformational stability, storage stability and stability during use, e.g. during a wash process and reflects the stability of a polypeptide (e.g. an endoglucanase variant according to the invention) as a function of time, e.g. how much activity is retained when said polypeptide (e.g. said endoglucanase variant) is kept in solution, in particular in a detergent solution. The stability is influenced by many factors, e.g. presence of chelator(s), pH, temperature, detergent composition, e.g. amount of builders, surfactants, chelators etc. The endoglucanase stability may be measured using a half-life improvement factor (HIF) as described in example 3 herein. The endoglu- canase stability may also be measured using a reducing sugar assay as described in example 3 herein.
Improved stability: The term "improved stability" or "increased stability" is defined herein as increased stability in a detergent composition (e.g. , in solutions), relative to the stability of the parent endoglucanase, relative to an endoglucanase having the identical amino acid sequence of the variant, but not having the alterations at one or more of the specified positions, or relative to SEQ ID NO: 2. The terms "improved stability" and "increased stability" includes "improved chemical stability", "detergent stability" and "improved detergent stability.
Improved chemical stability: The term "improved chemical stability" is defined herein as a variant enzyme displaying retention of enzymatic activity after a period of incubation in the presence of a chemical or chemicals, either naturally occurring or synthetic, which reduces the enzymatic activity of the parent enzyme. Improved chemical stability may also result in variants being more able (e.g., better that the parent) to catalyze a reaction in the presence of such chemicals. In a particular aspect of the invention the improved chemical stability is an improved stability in a detergent, in particular in a liquid detergent. The term "detergent stability" or "improved detergent stability is in particular an improved stability of the endoglucanase compared to the parent endoglucanase, when an endoglucanase variant as described herein is mixed into a liquid detergent formulation, especially into a liquid detergent formulation comprising a chelator (e.g. EDTA or citrate).
Conformational stability: The term "conformational stability" means resistance or a degree of resistance to conformational change, unfolding or disintegration. Accordingly, the term "less conformationally stable" means less resistant or having lesser degree of resistance to conformational change, unfolding or disintegration.
Instability: The term "instability" means lack of stability. Non-limiting examples of instability include conformational instability, unfolding, denaturation, disintegration, activity loss.
Wash performance: The term "wash performance" is used as an enzyme's ability to remove stains present on the object to be cleaned during e.g. wash or hard surface cleaning. The improvement in the wash performance may be quantified by calculating the so-called intensity value (Int) in 'Automatic Mechanical Stress Assay (AMSA) for laundry' or the remission value (Rem) as defined herein.
Conventions for designation of variants
For purposes of the present invention, the mature polypeptide disclosed in SEQ ID NO: 2 is used to determine the corresponding amino acid residue in another endoglucanase. The amino acid sequence of another endoglucanase is aligned with the mature polypeptide disclosed in SEQ ID NO: 2, and based on the alignment, the amino acid position number corresponding to any amino acid residue in the mature polypeptide disclosed in SEQ ID NO: 2 is determined using the Needleman-Wunsch algorithm (Needleman and Wunsch, 1970, J. Mol. Biol. 48: 443-453) as implemented in the Needle program of the EMBOSS package (EMBOSS: The European Molecular Biology Open Software Suite, Rice et al., 2000, Trends Genet. 16: 276-277), preferably version 5.0.0 or later. The parameters used are gap open penalty of 10, gap extension penalty of 0.5, and the EBLOSUM62 (EMBOSS version of BLOSUM62) substitution matrix.
Identification of the corresponding amino acid residue in another endoglucanase can be determined by an alignment of multiple polypeptide sequences using several computer programs including, but not limited to, MUSCLE (multiple sequence comparison by log-expectation; version 3.5 or later; Edgar, 2004, Nucleic Acids Research 32: 1792-1797), MAFFT (version 6.857 or later; Katoh and Kuma, 2002, Nucleic Acids Research 30: 3059-3066; Katoh et al. , 2005, Nucleic Acids Research 33: 511 -518; Katoh and Toh, 2007, Bioinformatics 23: 372-374; Katoh et al., 2009, Methods in Molecular Biology 537:_39-64; Katoh and Toh, 2010, Bioinformatics 26:_1899-1900), and EMBOSS EMMA employing ClustalW (1 .83 or later; Thompson ef al., 1994, Nucleic Acids Research 22: 4673-4680), using their respective default parameters.
When the other enzyme has diverged from the mature polypeptide of SEQ ID NO: 2 such that traditional sequence-based comparison fails to detect their relationship (Lindahl and Elofsson, 2000, J. Mol. Biol. 295: 613-615), other pairwise sequence comparison algorithms can be used. Greater sensitivity in sequence- based searching can be attained using search programs that utilize probabilistic representations of polypeptide families (profiles) to search databases. For example, the PSI-BLAST program generates profiles through an iterative database search process and is capable of detecting remote homologs (Altschul ef al. ,
1997, Nucleic Acids Res. 25: 3389-3402). Even greater sensitivity can be achieved if the family or super- family for the polypeptide has one or more representatives in the protein structure databases. Programs such as GenTHREADER (Jones, 1999, J. Mol. Biol. 287: 797-815; McGuffin and Jones, 2003, Bioinformat- ics 19: 874-881 ) utilize information from a variety of sources (PSI-BLAST, secondary structure prediction, structural alignment profiles, and solvation potentials) as input to a neural network that predicts the structural fold for a query sequence. Similarly, the method of Gough ei a/. , 2000, J. Mol. Biol. 313: 903-919, can be used to align a sequence of unknown structure with the superfamily models present in the SCOP database. These alignments can in turn be used to generate homology models for the polypeptide, and such models can be assessed for accuracy using a variety of tools developed for that purpose.
For proteins of known structure, several tools and resources are available for retrieving and generating structural alignments. For example, the SCOP superfamilies of proteins have been structurally aligned, and those alignments are accessible and downloadable. Two or more protein structures can be aligned using a variety of algorithms such as the distance alignment matrix (Holm and Sander, 1998, Proteins 33: 88-96) or combinatorial extension (Shindyalov and Bourne, 1998, Protein Engineering 1 1 : 739-747), and implemen- tation of these algorithms can additionally be utilized to query structure databases with a structure of interest in order to discover possible structural homologs (e.g., Holm and Park, 2000, Bioinformatics 16: 566-567).
In describing the variants of the present invention, the nomenclature described below is adapted for ease of reference. The accepted lUPAC single letter or three letter amino acid abbreviation is employed.
Substitutions. For an amino acid substitution, the following nomenclature is used: Original amino acid, position, substituted amino acid. Accordingly, the substitution of threonine at position 226 with alanine is designated as "Thr226Ala" or "T226A". Multiple mutations are separated by addition marks ("+"), e.g., "Gly205Arg+Ser411 Phe" or "G205R+S411 F", representing substitutions at positions 205 and 41 1 of glycine (G) with arginine (R) and serine (S) with phenylalanine (F), respectively.
Deletions. For an amino acid deletion, the following nomenclature is used: Original amino acid, position, *. Accordingly, the deletion of glycine at position 195 is designated as "Gly195*" or "G195*". Multiple deletions are separated by addition marks ("+"), e.g., "Gly195*+Ser411 *" or "G195*+S41 1 *".
Insertions. For an amino acid insertion, the following nomenclature is used: Original amino acid, position, original amino acid, inserted amino acid. Accordingly, the insertion of lysine after glycine at position 195 is designated "Gly195GlyLys" or "G195GK". An insertion of multiple amino acids is designated [Original amino acid, position, original amino acid, inserted amino acid #1 , inserted amino acid #2; etc.]. For example, the insertion of lysine and alanine after glycine at position 195 is indicated as "Gly195GlyLysAla" or "G195GKA".
In such cases the inserted amino acid residue(s) are numbered by the addition of lower case letters to the position number of the amino acid residue preceding the inserted amino acid residue(s). In the above example, the sequence would thus be:
Parent: Variant:
195 195 195a 195b
G G - K - A
Multiple alterations. Variants comprising multiple alterations are separated by addition marks ("+"), e.g., "Arg170Tyr+Gly195Glu" or "R170Y+G195E" representing a substitution of arginine and glycine at positions 170 and 195 with tyrosine and glutamic acid, respectively. Multiple alteration variants may also be separated by a comma (","), e.g., "Arg170Tyr,Gly195Glu" or "R170Y,G195E" representing a substitution of arginine and glycine at positions 170 and 195 with tyrosine and glutamic acid, respectively.
Different alterations. Where different alterations can be introduced at a position, the different alterations are separated by a comma, e.g., "Arg170Tyr,Glu" represents a substitution of arginine at position 170 with tyrosine or glutamic acid. Thus, "Tyr167Gly,Ala+Arg170Gly,Ala" designates the following variants:
"Tyr167Gly+Arg170Gly", "Tyr167Gly+Arg170Ala", "Tyr167Ala+Arg170Gly", and "Tyr167Ala+Arg 170Ala" .
DETAILED DESCRIPTION OF THE INVENTION
The known xanthan endoglucanase having SEQ ID NO: 2 is a large enzyme (>1000 residues), it is therefore extremely laborious and expensive to target its properties for improvement of, e.g. stability in a detergent composition. In some aspects, the present invention has targeted specific regions of the endoglucanase which have an effect on the stability of the endoglucanase molecules using protein engineering to a region selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 11 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2.
In one embodiment the present invention dramatically narrows down the number of residues to target when trying to stabilize endoglucanase molecules using protein engineering.
Variants
In one embodiment, regions in the protein sequence of the known xanthan endoglucanase having SEQ ID NO: 2 that are affected when the molecule is incubated in detergent, are the following: region 10 corre- sponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to
amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2. This embodiment relates to an important guidance on where to mutate an endoglucanase in order to stabilize its molecule in a detergent.
In one embodiment the present invention relates to a detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in a region selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2; preferably said endoglucanase variant has activity on xanthan gum pre-treated with xanthan lyase, further preferably said activity is a xanthan gum degrading activity.
In one embodiment, the present invention relates to a detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in a region selected from the group consisting of:
i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 1 , 2, 3, 4, 5, 6, 7, 8, 9, 10, 1 1 , 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 , 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 , 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 , 42, 43, 44, 45, 46, 47, 48, 49, 50, 51 , 52, 53, 54, 55, 56, 57, 58, 59, 60, 61 , 62, 63, 64, 65, 66, 67, 68, 69, 70, 71 , 72, 73, 74, 75, 76, 77, 78, 79, 80, 81 , 82, 83, 84, 85, 86, 87, 88, 89, 90, 91 , 92, 93, 94, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
ii) region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 107, 108, 109, 1 10, 1 11 , 112, 1 13, 1 14, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
iii) region 12 corresponding to amino acids 139 to 209of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 139, 140, 141 , 142, 143, 144, 145, 146, 147, 148, 149, 150, 151 , 152, 153, 154, 155, 156, 157, 158, 159, 160, 161 , 162, 163, 164, 165, 166, 167, 168, 169, 170, 171 , 172, 173, 174, 175, 176, 177, 178, 179, 180, 181 , 182, 183, 184, 185, 186, 187, 188, 189, 190, 191 , 192, 193, 194, 195, 196, 197, 198, 199, 200, 201 , 202, 203, 204, 205, 206, 207, 208, 209, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 252, 253, 254, 255, 256, 257, 258, 259, 260, 261 , 262, 263, 264, 265, 266, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 302, 303, 304, 305, 306, 307, 308, 309, 310, 311 , 312, 313, 314, 315, 316, 317, 318, 319, 320, 321 , 322, 323, 324, 325, 326, 327, 328, 329, 330, 331 , 332, 333, 334, 335, 336, 337, 338, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
vi) region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 362, 363, 364, 365, 366, 367, 368, 369, 370, 371 , 372, 373, 374, 375, 376, 377, 378, 379, 380, 381 , 382, 383, 384, 385, 386, 387, 388, 389, 390, 391 , 392, 393, 394, 395, 396, 397, 398, 399, 400, 401 , 402, 403, 404, 405, 406, 407, 408, 409, 410, 411 , 412, 413, 414, 415, 416, 417, 418, 419, 420, 421 , 422, 423, 424, 425, 426, 427, 428, 429, 430, 431 , 432, 433, 434, 435, 436, 437, 438, 439, 440, 441 , 442, 443, 444, 445, 446, 447, 448, 449,
450, 451 , 452, 453, 454, 455, 456, 457, 458, 459, 460, 461 , 462, 463, 464, 465, 466, 467, 468, 469, 470, 471 , 472, 473, 474, 475, 476, 477, 478, 479, 480, 481 , 482, 483, 484, 485, 486, 487, 488, 489, 490, 491 , 492, 493, 494, 495, 496, 497, 498, 499, 500, 501 , 502, 503, 504, 505, 506, 507, 508, 509, 510, 511 , 512, 513, 514, 515, 516, 517, 518, 519, 520, 521 , 522, 523, 524, 525, 526, 527, 528, 529, 530, 531 , 532, 533, 534, 535, 536, 537, 538, 539, 540, 541 , 542, 543, 544, 545, 546, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
vii) region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 596, 597, 598, 599, 600, 601 , 602, 603, 604, 605, 606, 607, 608, 609, 610, 61 1 , wherein said positions correspond to amino acid positions of
SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
viii) region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 661 , 662, 663, 664, 665, 666, 667, 668, 669, 670, 671 , 672, 673, 674, 675, 676, 677, 678, 679, 680, 681 , 682, 683, 684, 685, 686, 687, 688, 689, 690, 691 , 692, 693, 694, 695, 696, 697, 698, 699, 700, 701 , 702, 703, 704, 705, 706, 707, 708,
709, 710, 711 , 712, 713, 714, 715, 716, 717, 718, 719, 720, 721 , 722, 723, 724, 725, 726, 727, 728, 729, 730, 731 , 732, 733, 734, 735, 736, 737, 738, 739, 740, 741 , 742, 743, 744, 745, 746, 747, 748, 749, 750, 751 , 752, 753, 754, 755, 756, 757, 758, 759, 760, 761 , 762, 763, 764, 765, 766, 767, 768, 769, 770, 771 , 772, 773, 774, 775, 776, 777, 778, 779, 780, 781 , 782, 783, 784, 785, 786, 787, 788, 789, 790, 791 , 792, 793, 794, 795, 796, 797, 798, 799, 800, 801 , 802, 803, 804, 805, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID
NO: 2),
ix) region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 829, 830, 831 , 832, 833, 834, 835, 836, 837, 838, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2)
x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions 1043, 1044, 1045, 1046, 1047, 1048, 1049, 1050, 1051 , 1052, 1053, 1054, 1055, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g. , using the numbering of SEQ ID NO: 2).
In one embodiment the present invention relates to a detergent composition comprising an endoglu- canase variant, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more positions in one or more regions selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2; preferably said endoglucanase variant has activity on xanthan gum pre-treated with xan- than lyase, further preferably said activity is a xanthan gum degrading activity.
In one embodiment the present invention relates to a detergent composition comprising an endoglucanase variant as described herein having multiple alterations (such as 2, 3, 4, 5, 6, 7, 8, 9 or 10) in one region (e.g., of SEQ ID NO: 2 or another parent endoglucanase) selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2, preferably said endoglucanase variant has activity on xanthan gum pre-treated with xanthan lyase, further preferably said activity is a xanthan gum degrading activity.
In one embodiment the present invention relates to a detergent composition comprising an endoglucanase variant as described herein having multiple alterations (e.g., 2, 3, 4, 5, 6, 7, 8, 9 or 10) in multiple
regions (e.g., 2, 3, 4, 5, 6, 7, 8, 9 or 10) (e.g., of SEQ ID NO: 2 or another parent endoglucanase) selected from the group consisting of: region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, preferably said endoglucanase variant has activity on xanthan gum pre-treated with xanthan lyase, further preferably said activity is a xanthan gum degrading activity.
In one embodiment, the present invention relates to detergent compositions comprising endoglucanase variants, comprising an alteration (e.g., a substitution, deletion or insertion) at one or more (e.g., several) positions of the mature parent polypeptide (e.g., SEQ ID NO: 2), wherein each alteration is independently a substitution, insertion or deletion, wherein the variant has endoglucanase activity.
In an embodiment, the variant has sequence identity of at least 60%, e.g., at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99%, but less than 100%, to the amino acid sequence of the parent endoglucanase.
In one embodiment, the variant has at least 60%, e.g., at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, such as at least 96%, at least 97%, at least 98%, or at least 99%, but less than 100%, sequence identity to the mature polypeptide of SEQ ID NO: 2.
In one embodiment the present invention relates to a detergent composition comprising an endoglucanase variant of the invention, wherein said variant has at least 61 %, at least 62%, at least 63%, at least 64%, at least 65%, at least 66%, at least 67%, at least 68%, at least 69%, at least 70%, at least 71 %, at least 72%, at least 73%, at least 74%, at least 75%, at least 76%, at least 77%, at least 78%, at least 79%, at least 80%, at least 81 %, at least 82%, at least 83%, at least 84%, at least 85%, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to SEQ ID NO: 2.
In another aspect, a variant comprises an alteration at one or more (e.g., several) positions corresponding to positions 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 31 1 , 313, 387, 388, 390, 403, 408, 410, 416, 448, 451 , 471 , 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 71 1 , 719, 754, 756, 760, 781 , 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
In one embodiment, the endoglucanase variant comprised in the detergent compositions of the invention further comprises an alteration at one or more positions selected from the group consisting of alterations
in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651, 824, 848, 869, 880, 881, 883, 887, 892, 905, 906, 912, 921, 928, 934, 937, 948, 956, 999 and 1037 of SEQ ID NO: 2.
In another aspect, a variant comprises an alteration at two positions corresponding to any of positions 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 311, 313, 387, 388, 390, 403, 408, 410, 416, 448, 451, 471, 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 711, 719, 754, 756, 760, 781, 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
In one embodiment, the endoglucanase variant comprised in the detergent compositions of the invention further comprises an alteration at one or more positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651 , 824, 848, 869, 880, 881 , 883, 887, 892, 905, 906, 912, 921 , 928, 934, 937, 948, 956, 999 and 1037 of SEQ ID NO: 2.
In another aspect, a variant comprises an alteration at three positions corresponding to any of positions 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 311, 313, 387, 388, 390, 403, 408, 410, 416, 448, 451, 471, 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 711 , 719, 754, 756, 760, 781 , 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
In one embodiment, the endoglucanase variant comprised in the detergent compositions of the invention further comprises an alteration at one or more positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651, 824, 848, 869, 880, 881, 883, 887, 892, 905, 906, 912, 921, 928, 934, 937, 948, 956, 999 and 1037 of SEQ ID NO: 2.
Thus, in one embodiment, the variant comprises an alteration in the positions corresponding to: 17+20, 17+51, 17+53, 17+55, 17+56, 17+60, 17+63, 17+79, 17+87, 17+192, 17+302, 7+387, 17+388, 17+390, 17+403, 17+408, 17+410, 17+416, 17+448, 17+451, 17+471, 17+472, 17+507, 17+512, 17+515, 17+538, 17+598, 17+602, 17+605, 17+609, 17+676, 17+694, 17+698, 17+699, 17+711, 17+754, 17+760, 17+781, 17+786, 17+797, 17+834, 17+835, 20+51, 20+53, 20+55, 20+56, 20+60, 20+63, 20+79, 20+87, 20+192, 20+302, 20+387, 20+388, 20+390, 20+403, 20+408, 20+410, 20+416, 20+448, 20+451, 20+471 , 20+472, 20+507, 20+512, 20+515, 20+538, 20+598, 20+602, 20+605, 20+609, 20+676, 20+694, 20+698, 20+699, 20+711, 20+754, 20+760, 20+781, 20+786, 20+797, 20+834, 20+835, 51+53, 51+55, 51+56, 51+60, 51+63, 51+79, 51+87, 51+192, 51+302, 51+387, 51+388, 51+390, 51+403, 51+408, 51+410, 51+416, 51+448, 51+451, 51+471, 51+472, 51+507, 51+512, 51+515, 51+538, 51+598, 51+602, 51+605, 51+609, 51+676, 51+694, 51+698, 51+699, 51+711 , 51+754, 51+760, 51+781, 51+786, 51+797, 51+834, 51+835, 53+55, 53+56, 53+60, 53+63, 53+79, 53+87, 53+192, 53+302, 53+387, 53+388, 53+390, 53+403, 53+408, 53+410, 53+416, 53+448, 53+451 , 53+471 , 53+472, 53+507, 53+512, 53+515, 53+538, 53+598, 53+602, 53+605, 53+609, 3+676, 53+694, 53+698, 53+699, 53+711, 53+754, 53+760, 53+781, 53+786, 53+797, 53+834, 53+835, 55+56, 55+60, 55+63, 55+79, 55+87, 55+192, 55+302, 55+387, 55+388, 55+390, 55+403, 55+408, 55+410, 55+416, 55+448, 55+451, 55+471, 55+472, 55+507, 55+512, 55+515, 55+538,
55+598 55+602, 55+605, 55+609, 55+676, 55+694 55+698, 55+699, 55+71 1 , 55+754, 55+760, 55+781 55+786 55+797, 55+834, 55+835, 56+60, 56+63 56+79, 56+87, 56+192, 56+302, 56+387, 56+388; 56+390 56+403, 56+408, 56+410, 56+416, 56+448 56+451 , 56+471 , 56+472, 56+507, 56+512, 56+515 56+538 56+598, 56+602, 56+605, 56+609, 56+676 56+694, 56+698, 56+699, 56+71 1 , 56+754, 56+760 56+781 56+786, 56+797, 56+834, 56+835, 60+63 , 60+79, 60+87, 60+192, 60+302, 60+387, 60+388; 60+390 60+403, 60+408, 60+410, 60+416, 60+448 60+451 , 60+471 , 60+472, 60+507, 60+512, 60+515 60+538 60+598, 60+602, 60+605, 60+609, 60+676 60+694, 60+698, 60+699, 60+71 1 , 60+754, 60+760 60+781 60+786, 60+797, 60+834, 60+835, 63+79 63+87, 63+192, 63+302, 63+387, 63+388, 63+390 63+403 63+408, 63+410, 63+416, 63+448, 63+451 63+471 , 63+472, 63+507, 63+512, 63+515, 63+538 63+598 63+602, 63+605, 63+609, 63+676, 63+694 63+698, 63+699, 63+71 1 , 63+754, 63+760, 63+781 63+786 63+797, 63+834, 63+835, 79+87, 79+192, 79+302, 79+387, 79+388, 79+390, 79+403, 79+408 79+410 79+416, 79+448, 79+451 , 79+471 , 79+472 79+507, 79+512, 79+515, 79+538, 79+598, 79+602 79+605 79+609, 79+676, 79+694, 79+698, 79+699 79+71 1 , 79+754, 79+760, 79+781 , 79+786, 79+797 79+834 79+835, 87+192, 87+302, 87+387, 87+388 87+390, 87+403, 87+408, 87+410, 87+416, 87+448 87+451 87+471 , 87+472, 87+507, 87+512, 87+515 87+538, 87+598, 87+602, 87+605, 87+609, 87+676 87+694 87+698, 87+699, 87+71 1 , 87+754, 87+760, 87+781 , 87+786, 87+797, 87+834, 87+835, 192+302 192+387 192+388, 192+390, 192+403, 192+408, 192+410, 192+416, 192+448, 192+451 , 192+471 192+472 192+507, 192+512, 192+515, 192+538, 192+598, 192+602, 192+605, 192+609, 192+676 192+694 192+698, 192+699, 192+711 , 192+754, 192+760, 192+781 , 192+786, 192+797, 192+834 192+835 302+387, 302+388, 302+390, 302+403, 302+408, 302+410, 302+416, 302+448, 302+451 302+471 302+472, 302+507, 302+512, 302+515, 302+538, 302+598, 302+602, 302+605, 302+609 302+676 302+694, 302+698, 302+699, 302+71 1 , 302+754, 302+760, 302+781 , 302+786, 302+797 302+834 302+835, 387+388, 387+390, 387+403, 387+408, 387+410, 387+416, 387+448, 387+451 387+471 387+472, 387+507, 387+512, 387+515 , 387+538, 387+598, 387+602, 387+605, 87+609 387+676 387+694, 387+698, 387+699, 387+71 1 , 387+754, 387+760, 387+781 , 387+786, 387+797 387+834 387+835, 388+390, 388+403, 388+408, 388+410, 388+416, 388+448, 388+451 , 388+471 388+472 388+507, 388+512, 388+515, 388+538, 388+598, 388+602, 388+605, 388+609, 388+676 388+694 388+698, 388+699, 388+711 , 388+754, 388+760, 388+781 , 388+786, 388+797, 388+834 388+835 390+403, 390+408, 390+410, 390+416, 390+448, 390+451 , 390+471 , 390+472, 390+507; 390+512 390+515, 390+538, 390+598, 390+602, 390+605, 390+609, 390+676, 390+694, 390+698; 390+699 390+71 1 , 390+754, 390+760, 390+781 , 390+786, 390+797, 390+834, 390+835, 403+408; 403+410 403+416, 403+448, 403+451 , 403+471 , 403+472, 403+507, 403+512, 403+515, 403+538; 403+598 403+602, 403+605, 403+609, 403+676, 403+694, 403+698, 403+699, 403+71 1 , 403+754; 403+760 403+781 , 403+786, 403+797, 403+834, 403+835, 408+410, 408+416, 408+448, 408+451 408+471 408+472, 408+507, 408+512, 408+515, 408+538, 408+598, 408+602, 408+605, 408+609; 408+676 408+694, 408+698, 408+699, 408+71 1 , 408+754, 408+760, 408+781 , 408+786, 408+797
408+835, 410+416, 410+448, 410+451, 410+471, 410+472, 410+507, 410+512, 410+515,
410+598, 410+602, 410+605, 410+609, 410+676, 410+694, 410+698, 410+699, 410+711,
410+760, 410+781, 410+786, 410+797, 410+834, 410+835, 416+448, 416+451, 416+471,
416+507, 416+512, 416+515, 416+538, 416+598, 416+602, 416+605, 416+609, 416+676,
416+698, 416+699, 416+711, 416+754, 416+760, 416+781, 416+786, 416+797, 416+834,
448+451, 448+471, 448+472, 448+507, 448+512, 448+515, 448+538, 448+598, 448+602,
448+609, 448+676, 448+694, 448+698, 448+699, 448+711, 448+754, 448+760, 448+781,
448+797, 448+834, 448+835, 451+471, 451+472, 451+507, 451+512, 451+515, 451+538,
451+602, 451+605, 451+609, 451+676, 451+694, 451+698, 451+699, 451+711, 451+754,
451+781, 451+786, 451+797, 451+834, 451+835, 471+472, 471+507, 471+512, 471+515,
471+598, 471+602, 471+605, 471+609, 471+676, 471+694, 471+698, 471+699, 471+711,
471+760, 471+781, 471+786, 471+797, 471+834, 471+835, 472+507, 472+512, 472+515,
472+598, 472+602, 472+605, 472+609, 472+676, 472+694, 472+698, 472+699, 472+711,
472+760, 472+781, 472+786, 472+797, 472+834, 472+835, 507+512, 507+515, 507+538,
507+602, 507+605, 507+609, 507+676, 507+694, 507+698, 507+699, 507+711, 507+754,
507+781, 507+786, 507+797, 507+834, 507+835, 512+515, 512+538, 512+598, 512+602,
512+609, 512+676, 512+694, 512+698, 512+699, 512+711, 512+754, 512+760, 512+781,
512+797, 512+834, 512+835, 515+538, 515+598, 515+602, 515+605, 515+609, 515+676,
515+698, 515+699, 515+711, 515+754, 515+760, 515+781, 515+786, 515+797, 515+834,
538+598, 538+602, 538+605, 538+609, 538+676, 538+694, 538+698, 538+699, 538+711,
538+760, 538+781, 538+786, 538+797, 538+834, 538+835, 598+602, 598+605, 598+609,
598+694, 598+698, 598+699, 598+711, 598+754, 598+760, 598+781, 598+786, 598+797,
598+835, 602+605, 602+609, 602+676, 602+694, 602+698, 602+699, 602+711, 602+754,
602+781, 602+786, 602+797, 602+834, 602+835, 605+609, 605+676, 605+694, 605+698,
605+711, 605+754, 605+760, 605+781, 605+786, 605+797, 605+834, 605+835, 609+676,
609+698, 609+699, 609+711, 609+754, 609+760, 609+781, 609+786, 609+797, 609+834,
676+694, 676+698, 676+699, 676+711, 676+754, 676+760, 676+781, 676+786, 676+797,
676+835, 694+698, 694+699, 694+711, 694+754, 694+760, 694+781, 694+786, 694+797,
694+835, 698+699, 698+711, 698+754, 698+760, 698+781, 698+786, 698+797, 698+834,
699+711, 699+754, 699+760, 699+781, 699+786, 699+797, 699+834, 699+835, 711+754,
711+781, 711+786, 711+797, 711+834, 711+835, 754+760, 754+781, 754+786, 754+797,
754+835 760+781, 760+786, 760+797, 760+834, 760+835, 781+786, 781+797, 781+834,
786+797, 786+834, 786+835, 797+834, 797+835, 834+835, 17+20+51, 17+20+53, 17+20+55, 17+20+56, 17+20+60, 17+20+63, 17+20+79, 17+20+87, 17+20+192, 17+20+302, 17+20+387, 17+20+388, 17+20+390, 17+20+403, 17+20+408, 17+20+410, 17+20+416, 17+20+448, 17+20+451, 17+20+471, 7+20+472, 17+20+507, 17+20+512, 17+20+515, 17+20+538, 17+20+598, 17+20+602, 17+20+605,
+20+609 17+20+676, 17+20+694, 17+20+698, 17+20+699, 17+20+711 , 17+20+754, 17+20+760+20+781 17+20+786, 17+20+797, 17+20+834, 17+20+835, 17+51 +53, 17+51 +55, 17+51 +56+51 +60, 17+51 +63, 17+51 +79, 17+51 +87, 17+51 +192, 17+51 +302, 17+51 +387, 17+51 +388+51 +390 17+51 +403, 17+51 +408, 17+51 +410, 17+51 +416, 17+51 +448, 17+51 +451 , 17+51 +471+51 +472 17+51 +507, 17+51 +512, 17+51 +515, 17+51 +538, 17+51 +598, 17+51 +602, 17+51 +605+51 +609 17+51 +676, 17+51 +694, 17+51 +698, 17+51 +699, 17+51 +71 1 , 17+51 +754, 17+51 +760+51 +781 17+51 +786, 17+51 +797, 17+51 +834, 17+51 +835, 17+53+55, 17+53+56, 17+53+60+53+63, 17+53+79, 17+53+87, 17+53+192, 17+53+302, 17+53+387, 17+53+388, 17+53+390+53+403 17+53+408, 17+53+410, 17+53+416, 17+53+448, 17+53+451 , 17+53+471 , 17+53+472+53+507 17+53+512, 17+53+515, 17+53+538, 17+53+598, 17+53+602, 17+53+605, 17+53+609+53+676 17+53+694, 17+53+698, 17+53+699, 17+53+71 1 , 17+53+754, 17+53+760, 17+53+781+53+786 17+53+797, 17+53+834, 17+53+835, 17+55+56, 17+55+60, 17+55+63, 17+55+79, 17+55+87+55+192 17+55+302, 17+55+387, 17+55+388, 17+55+390, 17+55+403, 17+55+408, 17+55+410+55+416 17+55+448, 17+55+451 , 17+55+471 , 17+55+472, 17+55+507, 17+55+512, 17+55+515+55+538 17+55+598, 17+55+602, 17+55+605, 17+55+609, 17+55+676, 17+55+694, 17+55+698+55+699 17+55+711 , 17+55+754, 17+55+760, 17+55+781 , 17+55+786, 17+55+797, 17+55+834+55+835 17+56+60, 17+56+63, 17+56+79, 17+56+87, 17+56+192, 17+56+302, 17+56+387+56+388 17+56+390, 17+56+403, 17+56+408, 17+56+410, 17+56+416, 17+56+448, 17+56+451+56+471 17+56+472, 17+56+507, 17+56+512, 17+56+515, 17+56+538, 17+56+598, 17+56+602+56+605 17+56+609, 17+56+676, 17+56+694, 17+56+698, 17+56+699, 17+56+711 , 17+56+754+56+760 17+56+781 , 17+56+786, 17+56+797, 17+56+834, 17+56+835, 17+60+63, 17+60+79+60+87, 17+60+192, 17+60+302, 17+60+387, 17+60+388, 17+60+390, 17+60+403, 17+60+408+60+410 17+60+416, 17+60+448, 17+60+451 , 17+60+471 , 17+60+472, 17+60+507, 17+60+512+60+515 17+60+538, 17+60+598, 17+60+602, 17+60+605, 17+60+609, 17+60+676, 17+60+694+60+698 17+60+699, 17+60+711 , 17+60+754, 17+60+760, 17+60+781 , 17+60+786, 17+60+797+60+834 17+60+835, 17+63+79, 17+63+87, 17+63+192, 17+63+302, 17+63+387, 17+63+388+63+390 17+63+403, 17+63+408, 17+63+410, 17+63+416, 17+63+448, 17+63+451 , 17+63+471+63+472 17+63+507, 17+63+512, 17+63+515, 17+63+538, 17+63+598, 17+63+602, 17+63+605+63+609 17+63+676, 17+63+694, 17+63+698, 17+63+699, 17+63+71 1 , 17+63+754, 17+63+760+63+781 17+63+786, 17+63+797, 17+63+834, 17+63+835, 17+79+87, 17+79+192, 17+79+302+79+387 17+79+388, 17+79+390, 17+79+403, 17+79+408, 17+79+410, 17+79+416, 17+79+448+79+451 17+79+471 , 17+79+472, 17+79+507, 17+79+512, 17+79+515, 17+79+538, 17+79+598+79+602 17+79+605, 17+79+609, 17+79+676, 17+79+694, 17+79+698, 17+79+699, 17+79+71 1+79+754 17+79+760, 17+79+781 , 17+79+786, 17+79+797, 17+79+834, 17+79+835, 17+87+192+87+302 17+87+387, 17+87+388, 17+87+390, 17+87+403, 17+87+408, 17+87+410, 17+87+416+87+448 17+87+451 , 17+87+471 , 17+87+472, 17+87+507, 17+87+512, 17+87+515, 17+87+538
17+87+598, 17+87+602, 17+87+605, 17+87+609, 17+87+676, 17+87+694, 17+87+698, 17+87+699; 17+87+71 1 , 17+87+754, 17+87+760, 17+87+781 , 17+87+786, 17+87+797, 17+87+834, 17+87+835;
17+192+302, 17+192+387, 17+192+388, 17+192+390, 17+192+403, 17+192+408, 17+192+410,
17+192+416, 17+192+448, 17+192+451 , 17+192+471 , 17+192+472, 17+192+507, 17+192+512,
17+192+515, 17+192+538, 17+192+598, 17+192+602, 17+192+605, 17+192+609, 17+192+676,
17+192+694, 17+192+698, 17+192+699, 17+192+711 , 17+192+754, 17+192+760, 17+192+781 ,
17+192+786, 17+192+797, 17+192+834, 17+192+835, 17+302+387, 17+302+388, 17+302+390,
17+302+403, 17+302+408, 17+302+410, 17+302+416, 17+302+448, 17+302+451 , 17+302+471 ,
17+302+472, 17+302+507, 17+302+512, 17+302+515, 17+302+538, 17+302+598, 17+302+602,
17+302+605, 17+302+609, 17+302+676, 17+302+694, 17+302+698, 17+302+699, 17+302+711 ,
17+302+754, 17+302+760, 17+302+781 , 17+302+786, 17+302+797, 17+302+834, 17+302+835,
17+387+388, 17+387+390, 17+387+403, 17+387+408, 17+387+410, 17+387+416, 17+387+448,
17+387+451 , 17+387+471 , 17+387+472, 17+387+507, 17+387+512, 17+387+515, 17+387+538,
17+387+598, 17+387+602, 17+387+605, 17+387+609, 17+387+676, 17+387+694, 17+387+698,
17+387+699, 17+387+71 1 , 17+387+754, 17+387+760, 17+387+781 , 17+387+786, 17+387+797,
17+387+834, 17+387+835, 17+388+390, 17+388+403, 17+388+408, 17+388+410, 17+388+416,
17+388+448, 17+388+451 , 17+388+471 , 17+388+472, 17+388+507, 17+388+512, 17+388+515,
17+388+538, 17+388+598, 17+388+602, 17+388+605, 17+388+609, 17+388+676, 17+388+694,
17+388+698, 17+388+699, 17+388+71 1 , 17+388+754, 17+388+760, 17+388+781 , 17+388+786,
17+388+797, 17+388+834, 17+388+835, 17+390+403, 17+390+408, 17+390+410, 17+390+416,
17+390+448, 17+390+451 , 17+390+471 , 17+390+472, 17+390+507, 17+390+512, 17+390+515,
17+390+538, 17+390+598, 17+390+602, 17+390+605, 17+390+609, 17+390+676, 17+390+694,
17+390+698, 17+390+699, 17+390+71 1 , 17+390+754, 17+390+760, 17+390+781 , 17+390+786,
17+390+797, 17+390+834, 17+390+835, 17+403+408, 17+403+410, 17+403+416, 17+403+448,
17+403+451 , 17+403+471 , 17+403+472, 17+403+507, 17+403+512, 17+403+515, 17+403+538,
17+403+598, 17+403+602, 17+403+605, 17+403+609, 17+403+676, 17+403+694, 17+403+698,
17+403+699, 17+403+71 1 , 17+403+754, 17+403+760, 17+403+781 , 17+403+786, 17+403+797,
17+403+834, 17+403+835, 17+408+410, 17+408+416, 17+408+448, 17+408+451 , 17+408+471 ,
17+408+472, 17+408+507, 17+408+512, 17+408+515, 17+408+538, 17+408+598, 17+408+602,
17+408+605, 17+408+609, 17+408+676, 17+408+694, 17+408+698, 17+408+699, 17+408+711 ,
17+408+754, 17+408+760, 17+408+781 , 17+408+786, 17+408+797, 17+408+834, 17+408+835,
17+410+416, 17+410+448, 17+410+451 , 17+410+471 , 17+410+472, 17+410+507, 17+410+512,
17+410+515, 17+410+538, 17+410+598, 17+410+602, 17+410+605, 17+410+609, 17+410+676,
17+410+694, 17+410+698, 17+410+699, 17+410+711 , 17+410+754, 17+410+760, 17+410+781 ,
17+410+786, 17+410+797, 17+410+834, 17+410+835, 17+416+448, 17+416+451 , 17+416+471 ,
17+416+472, 17+416+507, 17+416+512, 17+416+515, 17+416+538, 17+416+598, 17+416+602,
17+416+605, 17+416+609, 17+416+676, 17+416+694, 17+416+698, 17+416+699, 7+416+711,
17+416+754, 17+416+760, 17+416+781, 17+416+786, 17+416+797, 17+416+834, 17+416+835,
17+448+451, 17+448+471, 17+448+472, 17+448+507, 17+448+512, 17+448+515, 17+448+538,
17+448+598, 17+448+602, 17+448+605, 17+448+609, 17+448+676, 17+448+694, 17+448+698,
17+448+699, 17+448+711, 17+448+754, 17+448+760, 17+448+781, 17+448+786, 17+448+797,
17+448+834, 17+448+835, 17+451+471, 17+451+472, 17+451+507, 17+451+512, 17+451+515,
17+451+538, 17+451+598, 17+451+602, 17+451+605, 17+451+609, 17+451+676, 17+451+694,
17+451+698, 17+451+699, 17+451+711, 17+451+754, 17+451+760, 17+451+781, 17+451+786,
17+451+797, 17+451+834, 17+451+835, 17+471+472, 17+471+507, 17+471+512, 17+471+515,
17+471+538, 17+471+598, 17+471+602, 17+471+605, 17+471+609, 17+471+676, 17+471+694,
17+471+698, 17+471+699, 17+471+711, 17+471+754, 17+471+760, 17+471+781, 17+471+786,
17+471+797, 17+471+834, 17+471+835, 17+472+507, 17+472+512, 17+472+515, 17+472+538,
17+472+598, 17+472+602, 17+472+605, 17+472+609, 17+472+676, 17+472+694, 17+472+698,
17+472+699, 17+472+711, 17+472+754, 17+472+760, 17+472+781, 17+472+786, 17+472+797,
17+472+834, 17+472+835, 17+507+512, 17+507+515, 17+507+538, 17+507+598, 17+507+602,
17+507+605, 17+507+609, 17+507+676, 17+507+694, 17+507+698, 17+507+699, 17+507+711,
17+507+754, 17+507+760, 17+507+781, 17+507+786, 17+507+797, 17+507+834, 17+507+835,
17+512+515, 17+512+538, 17+512+598, 17+512+602, 17+512+605, 17+512+609, 17+512+676,
17+512+694, 17+512+698, 17+512+699, 17+512+711, 17+512+754, 17+512+760, 17+512+781,
17+512+786, 17+512+797, 17+512+834, 17+512+835, 17+515+538, 17+515+598, 17+515+602,
17+515+605, 17+515+609, 17+515+676, 17+515+694, 17+515+698, 17+515+699, 17+515+711,
17+515+754, 17+515+760, 17+515+781, 17+515+786, 17+515+797, 17+515+834, 17+515+835,
17+538+598, 17+538+602, 17+538+605, 17+538+609, 17+538+676, 17+538+694, 17+538+698,
17+538+699, 17+538+711, 17+538+754, 17+538+760, 17+538+781, 17+538+786, 17+538+797,
17+538+834, 17+538+835, 17+598+602, 17+598+605, 17+598+609, 17+598+676, 17+598+694,
17+598+698, 17+598+699, 17+598+711, 17+598+754, 17+598+760, 17+598+781, 17+598+786,
17+598+797, 17+598+834, 17+598+835, 17+602+605, 17+602+609, 17+602+676, 17+602+694,
17+602+698, 17+602+699, 17+602+711, 17+602+754, 17+602+760, 17+602+781, 17+602+786,
17+602+797, 17+602+834, 17+602+835, 17+605+609, 17+605+676, 17+605+694, 17+605+698,
17+605+699, 17+605+711, 17+605+754, 17+605+760, 17+605+781, 17+605+786, 17+605+797,
17+605+834, 17+605+835, 17+609+676, 17+609+694, 17+609+698, 17+609+699, 17+609+711,
17+609+754, 17+609+760, 17+609+781, 17+609+786, 17+609+797, 17+609+834, 17+609+835,
17+676+694, 17+676+698, 17+676+699, 17+676+711, 17+676+754, 17+676+760, 17+676+781,
17+676+786, 17+676+797, 17+676+834, 17+676+835, 17+694+698, 17+694+699, 17+694+711,
17+694+754, 17+694+760, 17+694+781, 17+694+786, 17+694+797, 17+694+834, 17+694+835,
17+698+699, 17+698+711, 17+698+754, 17+698+760, 17+698+781, 17+698+786, 17+698+797,
17+698+834, 17+698+835, 17+699+711, 17+699+754, 17+699+760, 17+699+781, 17+699+786, 17+699+797, 17+699+834, 17+699+835, 17+711+754, 17+711+760, 17+711+781, 17+711+786, 17+711+797, 17+711+834, 17+711+835, 17+754+760, 17+754+781, 17+754+786, 17+754+797, 17+754+834, 17+754+835, 17+760+781, 17+760+786, 17+760+797, 17+760+834, 17+760+835, 17+781+786, 17+781+797, 17+781+834, 17+781+835, 17+786+797, 17+786+834, 17+786+835, 17+797+834, 17+797+835, 17+834+835, 20+51+53, 20+51+55, 20+51+56, 20+51+60, 20+51+63, 20+51+79, 20+51+87, 20+51+192, 20+51+302, 20+51+387, 20+51+388, 20+51+390, 20+51+403, 20+51+408, 20+51+410, 20+51+416, 20+51+448, 20+51+451, 20+51+471, 20+51+472, 20+51+507, 20+51+512 20+51+515, 20+51+538, 20+51+598, 20+51+602, 20+51+605, 20+51+609, 20+51+676, 20+51+694, 20+51+698, 20+51+699, 20+51+711, 20+51+754, 20+51+760, 20+51+781, 20+51+786, 20+51+797, 20+51+834, 20+51+835, 20+53+55, 20+53+56, 20+53+60, 20+53+63, 20+53+79, 20+53+87, 20+53+192, 20+53+302, 20+53+387, 20+53+388, 20+53+390, 20+53+403, 20+53+408, 20+53+410, 20+53+416, 20+53+448, 20+53+451, 20+53+471, 20+53+472, 20+53+507, 20+53+512, 20+53+515, 20+53+538, 20+53+598, 20+53+602, 20+53+605, 20+53+609, 20+53+676, 20+53+694, 20+53+698, 20+53+699, 20+53+711, 20+53+754, 20+53+760, 20+53+781, 20+53+786, 20+53+797, 20+53+834, 20+53+835, 20+55+56, 20+55+60, 20+55+63, 20+55+79, 20+55+87, 20+55+192, 20+55+302, 20+55+387, 20+55+388, 20+55+390, 20+55+403, 20+55+408, 20+55+410, 20+55+416, 20+55+448, 20+55+451 20+55+471, 20+55+472, 20+55+507, 20+55+512, 20+55+515, 20+55+538, 20+55+598, 20+55+602, 20+55+605, 20+55+609, 20+55+676, 20+55+694, 20+55+698, 20+55+699, 20+55+711, 20+55+754, 20+55+760, 20+55+781, 20+55+786, 20+55+797, 20+55+834, 20+55+835, 20+56+60, 20+56+63, 20+56+79, 20+56+87, 20+56+192, 20+56+302, 20+56+387, 20+56+388, 20+56+390, 20+56+403, 20+56+408, 20+56+410, 20+56+416, 20+56+448, 20+56+451, 20+56+471, 20+56+472, 20+56+507, 20+56+512, 20+56+515, 20+56+538, 20+56+598, 20+56+602, 20+56+605, 20+56+609, 20+56+676, 20+56+694, 20+56+698, 20+56+699, 20+56+711, 20+56+754, 20+56+760, 20+56+781, +56+786, 20+56+797, 20+56+834, 20+56+835, 20+60+63, 20+60+79, 20+60+87, 20+60+192, 20+60+302, 20+60+387, 20+60+388, 20+60+390, 20+60+403, 20+60+408, 20+60+410, 20+60+416, 20+60+448, 20+60+451, 20+60+471, 20+60+472, 20+60+507, 20+60+512, 20+60+515, 20+60+538, 20+60+598, 20+60+602, 20+60+605, 20+60+609, 20+60+676, 20+60+694, 20+60+698, 20+60+699, 20+60+711, 20+60+754, 20+60+760, 20+60+781, 20+60+786, 20+60+797, 20+60+834, 20+60+835, 20+63+79, 20+63+87, 20+63+192, 20+63+302, 20+63+387, 20+63+388, 20+63+390, 20+63+403, 20+63+408, 20+63+410, 20+63+416, 20+63+448, 20+63+451, 20+63+471, 20+63+472, 20+63+507, 20+63+512, 20+63+515, 20+63+538, 20+63+598, 20+63+602, 20+63+605, 20+63+609, 20+63+676, 20+63+694, 20+63+698, 20+63+699, 20+63+711, 20+63+754, 20+63+760, 20+63+781, 20+63+786, 20+63+797, 20+63+834, 20+63+835, 20+79+87, 20+79+192, 20+79+302, 20+79+387, 20+79+388, 20+79+390, 20+79+403, 20+79+408, 20+79+410, 20+79+416, 20+79+448, 20+79+451, 20+79+471, 20+79+472, 20+79+507, 20+79+512, 20+79+515, 20+79+538, 20+79+598, 20+79+602, 20+79+605, 20+79+609,
20+79+676, 20+79+694, 20+79+698, 20+79+699, 20+79+71 1 , 20+79+754, 20+79+760, 20+79+781 20+79+786, 20+79+797, 20+79+834, 20+79+835, 20+87+192, 20+87+302, 20+87+387, 20+87+388 20+87+390, 20+87+403, 20+87+408, 20+87+410, 20+87+416, 20+87+448, 20+87+451 , 20+87+471 20+87+472, 20+87+507, 20+87+512, 20+87+515, 20+87+538, 20+87+598, 20+87+602, 20+87+605; 20+87+609, 20+87+676, 20+87+694, 20+87+698, 20+87+699, 20+87+71 1 , 20+87+754, 20+87+760; 20+87+781 , 20+87+786, 20+87+797, 20+87+834, 20+87+835, 20+192+302, 20+192+387, 20+192+388
20+192+390, 20+192+403, 20+192+408, 20+192+410, 20+192+416, 20+192+448, 20+192+451 ,
20+192+471 , 20+192+472, 20+192+507, 20+192+512, 20+192+515, 20+192+538, 20+192+598,
20+192+602, 20+192+605, 20+192+609, 20+192+676, 20+192+694, 20+192+698, 20+192+699
20+192+711 , 20+192+754, 20+192+760, 20+192+781 , 20+192+786, 20+192+797, 20+192+834,
20+192+835, 20+302+387, 20+302+388, 20+302+390, 20+302+403, 20+302+408, 20+302+410,
20+302+416, 20+302+448, 20+302+451 , 20+302+471 , 20+302+472, 20+302+507, 20+302+512,
20+302+515, 20+302+538, 20+302+598, 20+302+602, 20+302+605, 20+302+609, 20+302+676,
20+302+694, 20+302+698, 20+302+699, 20+302+711 , 20+302+754, 20+302+760, 20+302+781 ,
20+302+786, 20+302+797, 20+302+834, 20+302+835, 20+387+388, 20+387+390, 20+387+403,
20+387+408, 20+387+410, 20+387+416, 20+387+448, 20+387+451 , 20+387+471 , 20+387+472,
20+387+507, 20+387+512, 20+387+515, 20+387+538, 20+387+598, 20+387+602, 20+387+605,
20+387+609, 20+387+676, 20+387+694, 20+387+698, 20+387+699, 20+387+71 1 , 20+387+754,
20+387+760, 20+387+781 , 20+387+786, 20+387+797, 20+387+834, 20+387+835, 20+388+390,
20+388+403, 20+388+408, 20+388+410, 20+388+416, 20+388+448, 20+388+451 , 20+388+471 ,
20+388+472, 20+388+507, 20+388+512, 20+388+515, 20+388+538, 20+388+598, 20+388+602,
20+388+605, 20+388+609, 20+388+676, 20+388+694, 20+388+698, 20+388+699, 20+388+711 ,
20+388+754, 20+388+760, 20+388+781 , 20+388+786, 20+388+797, 20+388+834, 20+388+835,
20+390+403, 20+390+408, 20+390+410, 20+390+416, 20+390+448, 20+390+451 , 20+390+471 ,
20+390+472, 20+390+507, 20+390+512, 20+390+515, 20+390+538, 20+390+598, 20+390+602,
20+390+605, 20+390+609, 20+390+676, 20+390+694, 20+390+698, 20+390+699, 20+390+711 ,
20+390+754, 20+390+760, 20+390+781 , 20+390+786, 20+390+797, 20+390+834, 20+390+835,
20+403+408, 20+403+410, 20+403+416, 20+403+448, 20+403+451 , 20+403+471 , 20+403+472,
20+403+507, 20+403+512, 20+403+515, 20+403+538, 20+403+598, 20+403+602, 20+403+605,
20+403+609, 20+403+676, 20+403+694, 20+403+698, 20+403+699, 20+403+71 1 , 20+403+754,
20+403+760, 20+403+781 , 20+403+786, 20+403+797, 20+403+834, 20+403+835, 20+408+410,
20+408+416, 20+408+448, 20+408+451 , 20+408+471 , 20+408+472, 20+408+507, 20+408+512,
20+408+515, 20+408+538, 20+408+598, 20+408+602, 20+408+605, 20+408+609, 20+408+676,
20+408+694, 20+408+698, 20+408+699, 20+408+711 , 20+408+754, 20+408+760, 20+408+781 ,
20+408+786, 20+408+797, 20+408+834, 20+408+835, 20+410+416, 20+410+448, 20+410+451 ,
20+410+471 , 20+410+472, 20+410+507, 20+410+512, 20+410+515, 20+410+538, 20+410+598,
20+410+602, 20+410+605, 20+410+609, 20+410+676, 20+410+694, 20+410+698, 20+410+699,
20+410+711, 20+410+754, 20+410+760, 20+410+781, 20+410+786, 20+410+797, 20+410+834,
20+410+835, 20+416+448, 20+416+451, 20+416+471, 20+416+472, 20+416+507, 20+416+512,
20+416+515, 20+416+538, 20+416+598, 20+416+602, 20+416+605, 20+416+609, 20+416+676,
20+416+694, 20+416+698, 20+416+699, 20+416+711, 20+416+754, 20+416+760, 20+416+781,
20+416+786, 20+416+797, 20+416+834, 20+416+835, 20+448+451 , 20+448+471, 20+448+472,
20+448+507, 20+448+512, 20+448+515, 20+448+538, 20+448+598, 20+448+602, 20+448+605,
20+448+609, 20+448+676, 20+448+694, 20+448+698, 20+448+699, 20+448+711, 20+448+754,
20+448+760, 20+448+781, 20+448+786, 20+448+797, 20+448+834, 20+448+835, 20+451+471,
20+451+472, 20+451+507, 20+451+512, 20+451+515, 20+451+538, 20+451+598, 20+451+602,
20+451+605, 20+451+609, 20+451+676, 20+451+694, 20+451+698, 20+451+699, 20+451+711,
20+451+754, 20+451+760, 20+451+781, 20+451+786, 20+451+797, 20+451+834, 20+451+835,
20+471+472, 20+471+507, 20+471+512, 20+471+515, 20+471+538, 20+471+598, 20+471+602,
20+471+605, 20+471+609, 20+471+676, 20+471+694, 20+471+698, 20+471+699, 20+471+711,
20+471+754, 20+471+760, 20+471+781, 20+471+786, 20+471+797, 20+471+834, 20+471+835,
20+472+507, 20+472+512, 20+472+515, 20+472+538, 20+472+598, 20+472+602, 20+472+605,
20+472+609, 20+472+676, 20+472+694, 20+472+698, 20+472+699, 20+472+711, 20+472+754,
20+472+760, 20+472+781, 20+472+786, 20+472+797, 20+472+834, 20+472+835, 20+507+512,
20+507+515, 20+507+538, 20+507+598, 20+507+602, 20+507+605, 20+507+609, 20+507+676,
20+507+694, 20+507+698, 20+507+699, 20+507+711, 20+507+754, 20+507+760, 20+507+781 ,
20+507+786, 20+507+797, 20+507+834, 20+507+835, 20+512+515, 20+512+538, 20+512+598,
20+512+602, 20+512+605, 20+512+609, 20+512+676, 20+512+694, 20+512+698, 20+512+699,
20+512+711, 20+512+754, 20+512+760, 20+512+781, 20+512+786, 20+512+797, 20+512+834,
20+512+835, 20+515+538, 20+515+598, 20+515+602, 20+515+605, 20+515+609, 20+515+676,
20+515+694, 20+515+698, 20+515+699, 20+515+711, 20+515+754, 20+515+760, 20+515+781,
20+515+786, 20+515+797, 20+515+834, 20+515+835, 20+538+598, 20+538+602, 20+538+605,
20+538+609, 20+538+676, 20+538+694, 20+538+698, 20+538+699, 20+538+711, 20+538+754,
20+538+760, 20+538+781, 20+538+786, 20+538+797, 20+538+834, 20+538+835, 20+598+602,
20+598+605, 20+598+609, 20+598+676, 20+598+694, 20+598+698, 20+598+699, 20+598+711,
20+598+754, 20+598+760, 20+598+781, 20+598+786, 20+598+797, 20+598+834, 20+598+835,
20+602+605, 20+602+609, 20+602+676, 20+602+694, 20+602+698, 20+602+699, 20+602+711,
20+602+754, 20+602+760, 20+602+781, 20+602+786, 20+602+797, 20+602+834, 20+602+835,
20+605+609, 20+605+676, 20+605+694, 20+605+698, 20+605+699, 20+605+711, 20+605+754,
20+605+760, 20+605+781, 20+605+786, 20+605+797, 20+605+834, 20+605+835, 20+609+676,
20+609+694, 20+609+698, 20+609+699, 20+609+711, 20+609+754, 20+609+760, 20+609+781 ,
20+609+786, 20+609+797, 20+609+834, 20+609+835, 20+676+694, 20+676+698, 20+676+699,
+676+711, 20+676+754, 20+676+760, 20+676+781 , 20+676+786, 20+676+797, 20+676+834,+676+835, 20+694+698, 20+694+699, 20+694+711, 20+694+754, 20+694+760, 20+694+781 ,+694+786, 20+694+797, 20+694+834, 20+694+835, 20+698+699, 20+698+711, 20+698+754,+698+760, 20+698+781, 20+698+786, 20+698+797, 20+698+834, 20+698+835, 20+699+711,+699+754, 20+699+760, 20+699+781, 20+699+786, 20+699+797, 20+699+834, 20+699+835,+711+754, 20+711+760, 20+711+781, 20+711+786, 20+711+797, 20+711+834, 20+711+835,+754+760, 20+754+781, 20+754+786, 20+754+797, 20+754+834, 20+754+835, 20+760+781 ,+760+786, 20+760+797, 20+760+834, 20+760+835, 20+781+786, 20+781+797, 20+781+834,+781+835, 20+786+797, 20+786+834, 20+786+835, 20+797+834, 20+797+835, 20+834+835,+53+55, 51+53+56, 51+53+60, 51+53+63, 51+53+7951+53+87, 51+53+192, 51+53+302, 51+53+387+53+388, 51+53+390, 51+53+403, 51+53+408, 51+53+410, 51+53+416, 51+53+448, 51+53+451+53+471, 51+53+472, 51+53+507, 51+53+512, 51+53+515, 51+53+538, 51+53+598, 51+53+602+53+605, 51+53+609, 51+53+676, 51+53+694, 51+53+698, 51+53+699, 51+53+711, 51+53+754+53+760, 51+53+781, 51+53+786, 51+53+797, 51+53+834, 51+53+835, 51+55+56, 51+55+60+55+63, 51+55+79, 51+55+87, 51+55+192, 51+55+302, 51+55+387, 51+55+388, 51+55+390+55+403, 51+55+408, 51+55+410, 51+55+416, 51+55+448, 51+55+451, 51+55+471, 51+55+472+55+507, 51+55+512, 51+55+515, 51+55+538, 51+55+598, 51+55+602, 51+55+605 51+55+609+55+676, 51+55+694, 51+55+698, 51+55+699, 51+55+711, 51+55+754, 51+55+760 51+55+781+55+786, 51+55+797, 51+55+834, 51+55+835, 51+56+60, 51+56+63, 51+56+79, 51+56+87+56+192, 51+56+302, 51+56+387, 51+56+388, 51+56+390, 51+56+403, 51+56+408 51+56+410+56+416, 51+56+448, 51+56+451, 51+56+471, 51+56+472, 51+56+507, 51+56+512 51+56+515+56+538, 51+56+598, 51+56+602, 51+56+605, 51+56+609, 51+56+676, 51+56+694 51+56+698+56+699, 51+56+711, 51+56+754, 51+56+760, 51+56+781, 51+56+786, 51+56+797 51+56+834+56+835, 51+60+63, 51+60+79, 51+60+87, 51+60+192, 51+60+302, 51+60+387, 51+60+388+60+390, 51+60+403, 51+60+408, 51+60+410, 51+60+416, 51+60+448, 51+60+451 51+60+471+60+472, 51+60+507, 51+60+512, 51+60+515, 51+60+538, 51+60+598, 51+60+602 51+60+605+60+609, 51+60+676, 51+60+694, 51+60+698, 51+60+699, 51+60+711, 51+60+754 51+60+760+60+781, 51+60+786, 51+60+797, 51+60+834, 51+60+835, 51+63+79, 51+63+87, 51+63+192+63+302, 51+63+387, 51+63+388, 51+63+390, 51+63+403, 51+63+408, 51+63+410 51+63+416+63+448, 51+63+451, 51+63+471, 51+63+472, 51+63+507, 51+63+512, 51+63+515 51+63+538+63+598, 51+63+602, 51+63+605, 51+63+609, 51+63+676, 51+63+694, 51+63+698 51+63+699+63+711, 51+63+754, 51+63+760, 51+63+781, 51+63+786, 51+63+797, 51+63+834 51+63+835+79+87, 51+79+192, 51+79+302, 51+79+387, 51+79+388, 51+79+390, 51+79+403; 51+79+408+79+410, 51+79+416, 51+79+448, 51+79+451, 51+79+471, 51+79+472, 51+79+507 51+79+512+79+515, 51+79+538, 51+79+598, 51+79+602, 51+79+605, 51+79+609, 51+79+676 51+79+694+79+698, 51+79+699, 51+79+711, 51+79+754, 51+79+760, 51+79+781, 51+79+786 51+79+797
51+79+834, 51+79+835, 51+87+192, 51+87+302, 51+87+387, 51+87+388, 51+87+390, 51+87+403 51+87+408, 51+87+410, 51+87+416, 51+87+448, 51+87+451, 51+87+471, 51+87+472, 51+87+507 51+87+512, 51+87+515, 51+87+538, 51+87+598, 51+87+602, 51+87+605, 51+87+609, 51+87+676 51+87+694, 51+87+698, 51+87+699, 51+87+711, 51+87+754, 51+87+760, 51+87+781, 51+87+786 51+87+797, 51+87+834, 51+87+835, 51 + 192+302, 51+192+387, 51 + 192+388, 51 + 192+390, 51+192+403
51+192+408, 51 + 192+410, 51+192+416, 51+192+448, 51+192+451, 51+192+471, 51
51+192+507, 51+192+512, 51+192+515, 51+192+538, 51+192+598, 51+192+602, 51
51+192+609, 51+192+676, 51+192+694, 51+192+698, 51+192+699, 51+192+711, 51
51+192+760, 51+192+781, 51+192+786, 51+192+797, 51+192+834, 51+192+835, 51
51+302+388, 51+302+390, 51+302+403, 51+302+408, 51+302+410, 51+302+416, 51
51+302+451, 51+302+471, 51+302+472, 51+302+507, 51+302+512, 51+302+515, 51
51+302+598, 51+302+602, 51+302+605, 51+302+609, 51+302+676, 51+302+694, 51
51+302+699, 51+302+711, 51+302+754, 51+302+760, 51+302+781, 51+302+786, 51
51+302+834, 51+302+835, 51+387+388, 51+387+390, 51+387+403, 51+387+408, 51
51+387+416, 51+387+448, 51+387+451, 51+387+471, 51+387+472, 51+387+507, 51
51+387+515, 51+387+538, 51+387+598, 51+387+602, 51+387+605, 51+387+609, 51
51+387+694, 51+387+698, 51+387+699, 51+387+711, 51+387+754, 51+387+760, 51
51+387+786, 51+387+797, 51+387+834, 51+387+835, 51+388+390, 51+388+403, 51
51+388+410, 51+388+416, 51+388+448, 51+388+451, 51+388+471, 51+388+472, 51
51+388+512, 51+388+515, 51+388+538, 51+388+598, 51+388+602, 51+388+605, 51
51+388+676, 51+388+694, 51+388+698, 51+388+699, 51+388+711, 51+388+754, 51
51+388+781, 51+388+786, 51+388+797, 51+388+834, 51+388+835, 51+390+403, 51
51+390+410, 51+390+416, 51+390+448, 51+390+451, 51+390+471, 51+390+472, 51
51+390+512, 51+390+515, 51+390+538, 51+390+598, 51+390+602, 51+390+605, 51
51+390+676, 51+390+694, 51+390+698, 51+390+699, 51+390+711, 51+390+754, 51
51+390+781, 51+390+786, 51+390+797, 51+390+834, 51+390+835, 51+403+408, 51
51+403+416, 51+403+448, 51+403+451, 51+403+471, 51+403+472, 51+403+507, 51
51+403+515, 51+403+538, 51+403+598, 51+403+602, 51+403+605, 51+403+609, 51
51+403+694, 51+403+698, 51+403+699, 51+403+711, 51+403+754, 51+403+760, 51
51+403+786, 51+403+797, 51+403+834, 51+403+835, 51+408+410, 51+408+416, 51
51+408+451, 51+408+471, 51+408+472, 51+408+507, 51+408+512, 51+408+515, 51
51+408+598, 51+408+602, 51+408+605, 51+408+609, 51+408+676, 51+408+694, 51
51+408+699, 51+408+711, 51+408+754, 51+408+760, 51+408+781, 51+408+786, 51
51+408+834, 51+408+835, 51+410+416, 51+410+448, 51+410+451, 51+410+471, 51
51+410+507, 51+410+512, 51+410+515, 51+410+538, 51+410+598, 51+410+602, 51
51+410+609, 51+410+676, 51+410+694, 51+410+698, 51+410+699, 51+410+711, 51
51+410+760, 51+410+781, 51+410+786, 51+410+797, 51+410+834, 51+410+835, 51+416+448,
51+416+451, 51+416+471, 51+416+472, 51+416+507, 51+416+512, 51+416+515, 51+416+538,
51+416+598, 51+416+602, 51+416+605, 51+416+609, 51+416+676, 51+416+694, 51+416+698,
51+416+699, 51+416+711, 51+416+754, 51+416+760, 51+416+781, 51+416+786, 51+416+797,
51+416+834, 51+416+835, 51+448+451, 51+448+471, 51+448+472, 51+448+507, 51+448+512,
51+448+515, 51+448+538, 51+448+598, 51+448+602, 51+448+605, 51+448+609, 51+448+676,
51+448+694, 51+448+698, 51+448+699, 51+448+711, 51+448+754, 51+448+760, 51+448+781,
51+448+786, 51+448+797, 51+448+834, 51+448+835, 51+451+471, 51+451+472, 51+451+507,
51+451+512, 51+451+515, 51+451+538, 51+451+598, 51+451+602, 51+451+605, 51+451+609,
51+451+676, 51+451+694, 51+451+698, 51+451+699, 51+451+711, 51+451+754, 51+451+760,
51+451+781, 51+451+786, 51+451+797, 51+451+834, 51+451+835, 51+471+472, 51+471+507,
51+471+512, 51+471+515, 51+471+538, 51+471+598, 51+471+602, 51+471+605, 51+471+609,
51+471+676, 51+471+694, 51+471+698, 51+471+699, 51+471+711, 51+471+754, 51+471+760,
51+471+781, 51+471+786, 51+471+797, 51+471+834, 51+471+835, 51+472+507, 51+472+512,
51+472+515, 51+472+538, 51+472+598, 51+472+602, 51+472+605, 51+472+609, 51+472+676,
51+472+694, 51+472+698, 51+472+699, 51+472+711, 51+472+754, 51+472+760, 51+472+781,
51+472+786, 51+472+797, 51+472+834, 51+472+835, 51+507+512, 51+507+515, 51+507+538,
51+507+598, 51+507+602, 51+507+605, 51+507+609, 51+507+676, 51+507+694, 51+507+698,
51+507+699, 51+507+711, 51+507+754, 51+507+760, 51+507+781, 51+507+786, 51+507+797,
51+507+834, 51+507+835, 51+512+515, 51+512+538, 51+512+598, 51+512+602, 51+512+605,
51+512+609, 51+512+676, 51+512+694, 51+512+698, 51+512+699, 51+512+711, 51+512+754,
51+512+760, 51+512+781, 51+512+786, 51+512+797, 51+512+834, 51+512+835, 51+515+538,
51+515+598, 51+515+602, 51+515+605, 51+515+609, 51+515+676, 51+515+694, 51+515+698,
51+515+699, 51+515+711, 51+515+754, 51+515+760, 51+515+781, 51+515+786, 51+515+797,
51+515+834, 51+515+835, 51+538+598, 51+538+602, 51+538+605, 51+538+609, 51+538+676,
51+538+694, 51+538+698, 51+538+699, 51+538+711, 51+538+754, 51+538+760, 51+538+781,
51+538+786, 51+538+797, 51+538+834, 51+538+835, 51+598+602, 51+598+605 51+598+609,
51+598+676, 51+598+694, 51+598+698, 51+598+699, 51+598+711, 51+598+754, 51+598+760,
51+598+781 51+598+786, 51+598+797, 51+598+834, 51+598+835, 51+602+605, 51+602+609,
51+602+676, 51+602+694, 51+602+698, 51+602+699, 51+602+711, 51+602+754, 51+602+760,
51+602+781, 51+602+786, 51+602+797, 51+602+834, 51+602+835, 51+605+609, 51+605+676,
51+605+694, 51+605+698, 51+605+699, 51+605+711, 51+605+754, 51+605+760, 51+605+781,
51+605+786, 51+605+797, 51+605+834, 51+605+835, 51+609+676, 51+609+694, 51+609+698,
51+609+699, 51+609+711, 51+609+754, 51+609+760, 51+609+781, 51+609+786, 51+609+797,
51+609+834, 51+609+835, 51+676+694, 51+676+698, 51+676+699, 51+676+711, 51+676+754,
51+676+760, 51+676+781, 51+676+786, 51+676+797, 51+676+834, 51+676+835, 51+694+698,
+694+699, 51+694+711, 51+694+754, 51+694+760, 51+694+781, 51+694+786, 51+694+797,+694+834, 51+694+835, 51+698+699, 51+698+711, 51+698+754, 51+698+760, 51+698+781,+698+786, 51+698+797, 51+698+834, 51+698+835, 51+699+711, 51+699+754, 51+699+760,+699+781, 51+699+786, 51+699+797, 51+699+834, 51+699+835, 51+711+754, 51+711+760,+711+781, 51+711+786, 51+711+797, 51+711+834, 51+711+835, 51+754+760, 51+754+781,+754+786, 51+754+797, 51+754+834, 51+754+835, 51+760+781, 51+760+786, 51+760+797,+760+834, 51+760+835, 51+781+786, 51+781+797, 51+781+834, 51+781+835, 51+786+797,+786+834, 51+786+835, 51+797+834, 51+797+835, 51+834+835, 53+55+56, 53+55+60, 53+55+63+55+79, 53+55+87, 53+55+192, 53+55+302, 53+55+387, 53+55+388, 53+55+390, 53+55+403+55+408, 53+55+410, 53+55+416, 53+55+448, 53+55+451, 53+55+471, 53+55+472, 53+55+507+55+512 53+55+515 53+55+538, 53+55+598, 53+55+602, 53+55+605 53+55+609, 53+55+676+55+694 53+55+698 53+55+699, 53+55+711, 53+55+754, 53+55+760 53+55+781, 53+55+786+55+797 53+55+834 53+55+835, 53+56+60, 53+56+63, 53+56+79 53+56+87, 53+56+192+56+302 53+56+387 53+56+388, 53+56+390, 53+56+403, 53+56+408 53+56+410, 53+56+416+56+448 53+56+451 53+56+471, 53+56+472, 53+56+507, 53+56+512 53+56+515, 53+56+538+56+598 53+56+602 53+56+605, 53+56+609, 53+56+676, 53+56+694 53+56+698, 53+56+699+56+711 53+56+754 53+56+760, 53+56+781, 53+56+786, 53+56+797 53+56+834, 53+56+835+60+63, 53+60+79, 53+60+87, 53+60+192, 53+60+302, 53+60+387, 53+60+388, 53+60+390+60+403 53+60+408 53+60+410, 53+60+416, 53+60+448, 53+60+451 53+60+471, 53+60+472+60+507 53+60+512 53+60+515, 53+60+538, 53+60+598, 53+60+602 53+60+605, 53+60+609+60+676 53+60+694 53+60+698, 53+60+699, 53+60+711, 53+60+754 53+60+760, 53+60+781+60+786 53+60+797 53+60+834, 53+60+835, 53+63+79, 53+63+87, 53+63+192, 53+63+302+63+387 53+63+388 53+63+390, 53+63+403, 53+63+408, 53+63+410 53+63+416, 53+63+448+63+451 53+63+471 53+63+472, 53+63+507, 53+63+512, 53+63+515 53+63+538, 53+63+598+63+602 53+63+605 53+63+609, 53+63+676, 53+63+694, 53+63+698 53+63+699, 53+63+711+63+754 53+63+760 53+63+781, 53+63+786, 53+63+797, 53+63+834, 53+63+835, 53+79+87+79+192 53+79+302 53+79+387, 53+79+388, 53+79+390, 53+79+403 53+79+408, 53+79+410+79+416 53+79+448 53+79+451, 53+79+471, 53+79+472, 53+79+507 53+79+512, 53+79+515+79+538 53+79+598 53+79+602, 53+79+605, 53+79+609, 53+79+676 53+79+694, 53+79+698+79+699 53+79+711 53+79+754, 53+79+760, 53+79+781, 53+79+786 53+79+797, 53+79+834+79+835 53+87+192 53+87+302, 53+87+387, 53+87+388, 53+87+390 53+87+403, 53+87+408+87+410 53+87+416 53+87+448, 53+87+451, 53+87+471, 53+87+472 53+87+507, 53+87+512+87+515 53+87+538 53+87+598, 53+87+602, 53+87+605, 53+87+609 53+87+676, 53+87+694+87+698 53+87+699 53+87+711, 53+87+754, 53+87+760, 53+87+781 53+87+786, 53+87+797+87+834 53+87+835, 53+192+302, 53+192+387, 53+192+388, 53+192+390, 53+192+403+192+408 53+192+410, 53+192+416, 53+192+448, 53+192+451 53+192+471, 53+192+472
53+192+507, 53+192+512, 53+192+515, 53+192+538, 53+192+598, 53+192+602, 53+192+605,
53+192+609, 53+192+676, 53+192+694, 53+192+698, 53+192+699, 53+192+71 1 , 53+192+754,
53+192+760, 53+192+781 , 53+192+786, 53+192+797, 53+192+834, 53+192+835, 53+302+387,
53+302+388, 53+302+390, 53+302+403, 53+302+408, 53+302+410, 53+302+416, 53+302+448,
53+302+451 , 53+302+471 , 53+302+472, 53+302+507, 53+302+512, 53+302+515, 53+302+538,
53+302+598, 53+302+602, 53+302+605, 53+302+609, 53+302+676, 53+302+694, 53+302+698,
53+302+699, 53+302+71 1 , 53+302+754, 53+302+760, 53+302+781 , 53+302+786, 53+302+797,
53+302+834, 53+302+835, 53+387+388, 53+387+390, 53+387+403, 53+387+408, 53+387+410,
53+387+416, 53+387+448, 53+387+451 , 53+387+471 , 53+387+472, 53+387+507, 53+387+512,
53+387+515, 53+387+538, 53+387+598, 53+387+602, 53+387+605, 53+387+609, 53+387+676,
53+387+694, 53+387+698, 53+387+699, 53+387+711 , 53+387+754, 53+387+760, 53+387+781 ,
53+387+786, 53+387+797, 53+387+834, 53+387+835, 53+388+390, 53+388+403, 53+388+408,
53+388+410, 53+388+416, 53+388+448, 53+388+451 , 53+388+471 , 53+388+472, 53+388+507,
53+388+512, 53+388+515, 53+388+538, 53+388+598, 53+388+602, 53+388+605, 53+388+609,
53+388+676, 53+388+694, 53+388+698, 53+388+699, 53+388+711 , 53+388+754, 53+388+760,
53+388+781 , 53+388+786, 53+388+797, 53+388+834, 53+388+835, 53+390+403, 53+390+408,
53+390+410, 53+390+416, 53+390+448, 53+390+451 , 53+390+471 , 53+390+472, 53+390+507,
53+390+512, 53+390+515, 53+390+538, 53+390+598, 53+390+602, 53+390+605, 53+390+609,
53+390+676, 53+390+694, 53+390+698, 53+390+699, 53+390+711 , 53+390+754, 53+390+760,
53+390+781 , 53+390+786, 53+390+797, 53+390+834, 53+390+835, 53+403+408, 53+403+410,
53+403+416, 53+403+448, 53+403+451 , 53+403+471 , 53+403+472, 53+403+507, 53+403+512,
53+403+515, 53+403+538, 53+403+598, 53+403+602, 53+403+605, 53+403+609, 53+403+676,
53+403+694, 53+403+698, 53+403+699, 53+403+711 , 53+403+754, 53+403+760, 53+403+781 ,
53+403+786, 53+403+797, 53+403+834, 53+403+835, 53+408+410, 53+408+416, 53+408+448,
53+408+451 , 53+408+471 , 53+408+472, 53+408+507, 53+408+512, 53+408+515, 53+408+538,
53+408+598, 53+408+602, 53+408+605, 53+408+609 53+408+676, 53+408+694, 53+408+698
53+408+699, 53+408+71 1 , 53+408+754, 53+408+760, 53+408+781 , 53+408+786, 53+408+797,
53+408+834, 53+408+835, 53+410+416, 53+410+448, 53+410+451 , 53+410+471 , 53+410+472,
53+410+507, 53+410+512, 53+410+515, 53+410+538, 53+410+598, 53+410+602, 53+410+605,
53+410+609, 53+410+676, 53+410+694, 53+410+698, 53+410+699, 53+410+71 1 , 53+410+754,
53+410+760, 53+410+781 , 53+410+786, 53+410+797, 53+410+834, 53+410+835, 53+416+448,
53+416+451 , 53+416+471 , 53+416+472, 53+416+507, 53+416+512, 53+416+515, 53+416+538,
53+416+598, 53+416+602, 53+416+605, 53+416+609, 53+416+676, 53+416+694, 53+416+698,
53+416+699, 53+416+71 1 , 53+416+754, 53+416+760, 53+416+781 , 53+416+786, 53+416+797,
53+416+834, 53+416+835, 53+448+451 , 53+448+471 , 53+448+472, 53+448+507, 53+448+512,
53+448+515, 53+448+538, 53+448+598, 53+448+602, 53+448+605, 53+448+609, 53+448+676,
53+448+694, 53+448+698, 53+448+699, 53+448+711, 53+448+754, 53+448+760, 53+448+781 ,
53+448+786, 53+448+797, 53+448+834, 53+448+835, 53+451+471, 53+451+472, 53+451+507,
53+451+512, 53+451+515, 53+451+538, 53+451+598, 53+451+602, 53+451+605, 53+451+609,
53+451+676, 53+451+694, 53+451+698, 53+451+699, 53+451+711, 53+451+754, 53+451+760,
53+451+781, 53+451+786, 53+451+797, 53+451+834, 53+451+835, 53+471+472, 53+471+507,
53+471+512, 53+471+515, 53+471+538, 53+471+598, 53+471+602, 53+471+605, 53+471+609,
53+471+676, 53+471+694, 53+471+698, 53+471+699, 53+471+711, 53+471+754, 53+471+760,
53+471+781, 53+471+786, 53+471+797, 53+471+834, 53+471+835, 53+472+507, 53+472+512,
53+472+515, 53+472+538, 53+472+598, 53+472+602, 53+472+605, 53+472+609, 53+472+676,
53+472+694, 53+472+698, 53+472+699, 53+472+711, 53+472+754, 53+472+760, 53+472+781 ,
53+472+786, 53+472+797, 53+472+834, 53+472+835, 53+507+512, 53+507+515, 53+507+538,
53+507+598, 53+507+602, 53+507+605, 53+507+609, 53+507+676, 53+507+694, 53+507+698,
53+507+699, 53+507+711, 53+507+754, 53+507+760, 53+507+781 , 53+507+786, 53+507+797,
53+507+834, 53+507+835, 53+512+515, 53+512+538, 53+512+598, 53+512+602, 53+512+605,
53+512+609, 53+512+676, 53+512+694, 53+512+698, 53+512+699, 53+512+711, 53+512+754,
53+512+760, 53+512+781, 53+512+786, 53+512+797, 53+512+834, 53+512+835, 53+515+538,
53+515+598, 53+515+602, 53+515+605, 53+515+609, 53+515+676, 53+515+694, 53+515+698,
53+515+699, 53+515+711, 53+515+754, 53+515+760, 53+515+781, 53+515+786, 53+515+797,
53+515+834, 53+515+835, 53+538+598, 53+538+602, 53+538+605, 53+538+609, 53+538+676,
53+538+694, 53+538+698, 53+538+699, 53+538+711, 53+538+754, 53+538+760, 53+538+781 ,
53+538+786, 53+538+797, 53+538+834, 53+538+835, 53+598+602, 53+598+605, 53+598+609,
53+598+676, 53+598+694, 53+598+698, 53+598+699, 53+598+711, 53+598+754, 53+598+760,
53+598+781 , 53+598+786, 53+598+797, 53+598+834, 53+598+835, 53+602+605, 53+602+609,
53+602+676, 53+602+694, 53+602+698, 53+602+699, 53+602+711, 53+602+754, 53+602+760,
53+602+781 , 53+602+786, 53+602+797, 53+602+834, 53+602+835, 53+605+609, 53+605+676,
53+605+694, 53+605+698, 53+605+699, 53+605+711, 53+605+754, 53+605+760, 53+605+781 ,
53+605+786, 53+605+797, 53+605+834, 53+605+835, 53+609+676, 53+609+694, 53+609+698,
53+609+699, 53+609+711, 53+609+754, 53+609+760, 53+609+781 , 53+609+786, 53+609+797,
53+609+834, 53+609+835, 53+676+694, 53+676+698, 53+676+699, 53+676+711, 53+676+754,
53+676+760, 53+676+781, 53+676+786, 53+676+797, 53+676+834, 53+676+835, 53+694+698,
53+694+699, 53+694+711, 53+694+754, 53+694+760, 53+694+781 , 53+694+786, 53+694+797,
53+694+834, 53+694+835, 53+698+699, 53+698+711, 53+698+754, 53+698+760, 53+698+781 ,
53+698+786, 53+698+797, 53+698+834, 53+698+835, 53+699+711, 53+699+754, 53+699+760,
53+699+781 , 53+699+786, 53+699+797, 53+699+834, 53+699+835, 53+711+754, 53+711+760,
53+711+781, 53+711+786, 53+711+797, 53+711+834, 53+711+835, 53+754+760, 53+754+781 ,
53+754+786, 53+754+797, 53+754+834, 53+754+835, 53+760+781 , 53+760+786, 53+760+797,
53+760+834, 53+760+835, 53+781 +786, 53+781 +797, 53+781 +834, 53+781 +835, 53+786+797 53+786+834, 53+786+835, 53+797+834, 53+797+835, 53+834+835, 55+56+60, 55+56+63, 55+56+79
55+56+87, 55+56+192, 55+56+302, 55+56+387, 55+56+388, 55+56+390, 55+56+403, 55+56+408
55+56+410, 55+56+416, 55+56+448, 55+56+451 , 55+56+471 , 55+56+472, 55+56+507, 55+56+512
55+56+515, 55+56+538, 55+56+598, 55+56+602, 55+56+605, 55+56+609, 55+56+676, 55+56+694
55+56+698, 55+56+699, 55+56+711 , 55+56+754, 55+56+760, 55+56+781 , 55+56+786, 55+56+797
55+56+834, 55+56+835, 55+60+63, 55+60+79, 55+60+87, 55+60+192, 55+60+302, 55+60+387
55+60+388, 55+60+390, 55+60+403, 55+60+408, 55+60+410, 55+60+416, 55+60+448, 55+60+451
55+60+471 , 55+60+472, 55+60+507, 55+60+512, 55+60+515, 55+60+538, 55+60+598, 55+60+602
55+60+605, 55+60+609, 5+60+676, 55+60+694, 55+60+698, 55+60+699, 55+60+71 1 , 55+60+754
55+60+760, 55+60+781 , 55+60+786 55+60+797, 55+60+834, 55+60+835, 55+63+79 55+63+87
55+63+192, 55+63+302, 55+63+387, 55+63+388, 55+63+390, 55+63+403, 55+63+408, 55+63+410
55+63+416, 55+63+448, 55+63+451 , 55+63+471 , 55+63+472, 55+63+507, 55+63+512, 55+63+515
55+63+538, 55+63+598, 55+63+602, 55+63+605, 55+63+609, 55+63+676, 55+63+694, 55+63+698
55+63+699, 55+63+711 , 55+63+754, 55+63+760, 55+63+781 , 55+63+786, 55+63+797, 55+63+834
55+63+835, 55+79+87, 55+79+192, 55+79+302, 55+79+387, 55+79+388, 55+79+390, 55+79+403
55+79+408, 55+79+410, 55+79+416, 55+79+448, 55+79+451 , 55+79+471 , 55+79+472, 55+79+507
55+79+512, 55+79+515, 55+79+538, 55+79+598, 55+79+602, 55+79+605, 55+79+609, 55+79+676
55+79+694, 55+79+698, 55+79+699, 55+79+71 1 , 55+79+754, 55+79+760, 55+79+781 , 55+79+786
55+79+797, 55+79+834, 55+79+835, 55+87+192, 55+87+302, 55+87+387, 55+87+388, 55+87+390
55+87+403, 55+87+408, 55+87+410, 55+87+416, 55+87+448, 55+87+451 , 55+87+471 , 55+87+472
55+87+507, 55+87+512, 55+87+515, 55+87+538, 55+87+598, 55+87+602, 55+87+605, 55+87+609
55+87+676, 55+87+694, 55+87+698, 55+87+699, 55+87+71 1 , 55+87+754, 55+87+760, 55+87+781
55+87+786, 55+87+797, 55+87+834, 55+87+835, 55+192+302, 55+192+387, 55+192+388, 55+192+390
55+192+403, 55+192+408, 55+192+410, 55+192+416, 55+192+448, 55+192+451 , 55+192+471
55+192+472, 55+192+507, 55+192+512, 55+192+515, 55+192+538, 55+192+598, 55+192+602
55+192+605, 55+192+609, 55+192+676, 55+192+694, 55+192+698, 55+192+699, 55+192+711
55+192+754, 55+192+760, 55+192+781 , 55+192+786, 55+192+797, 55+192+834, 55+192+835
55+302+387, 55+302+388, 55+302+390, 55+302+403, 55+302+408, 55+302+410, 55+302+416
55+302+448, 55+302+451 , 55+302+471 , 55+302+472, 55+302+507, 55+302+512, 55+302+515
55+302+538, 55+302+598, 55+302+602, 55+302+605, 55+302+609, 55+302+676, 55+302+694
55+302+698, 55+302+699, 55+302+71 1 , 55+302+754, 55+302+760, 55+302+781 , 55+302+786
55+302+797, 55+302+834, 55+302+835, 55+387+388, 55+387+390, 55+387+403, 55+387+408
55+387+410, 55+387+416, 55+387+448, 55+387+451 , 55+387+471 , 55+387+472, 55+387+507
55+387+512, 55+387+515, 55+387+538, 55+387+598, 55+387+602, 55+387+605, 55+387+609
55+387+676, 55+387+694, 55+387+698, 55+387+699, 55+387+711 , 55+387+754, 55+387+760
55+387+781 , 55+387+786, 55+387+797, 55+387+834, 55+387+835, 55+388+390, 55+388+403,
55+388+408, 55+388+410, 55+388+416, 55+388+448, 55+388+451 , 55+388+471, 55+388+472,
55+388+507, 55+388+512, 55+388+515, 55+388+538, 55+388+598, 55+388+602, 55+388+605,
55+388+609, 55+388+676, 55+388+694, 55+388+698, 55+388+699, 55+388+711, 55+388+754,
55+388+760, 55+388+781, 55+388+786, 55+388+797, 55+388+834, 55+388+835, 55+390+403,
55+390+408, 55+390+410, 55+390+416, 55+390+448, 55+390+451, 55+390+471, 55+390+472,
55+390+507, 55+390+512, 55+390+515, 55+390+538, 55+390+598, 55+390+602, 55+390+605,
55+390+609, 55+390+676, 55+390+694, 55+390+698, 55+390+699, 55+390+711, 55+390+754,
55+390+760, 55+390+781, 55+390+786, 55+390+797, 55+390+834, 55+390+835, 55+403+408,
55+403+410, 55+403+416, 55+403+448, 55+403+451 , 55+403+471 , 55+403+472, 55+403+507,
55+403+512, 55+403+515, 55+403+538, 55+403+598, 55+403+602, 55+403+605, 55+403+609,
55+403+676, 55+403+694, 55+403+698, 55+403+699, 55+403+711, 55+403+754, 55+403+760,
55+403+781 , 55+403+786, 55+403+797, 55+403+834, 55+403+835, 55+408+410, 55+408+416,
55+408+448, 55+408+451, 55+408+471, 55+408+472, 55+408+507, 55+408+512, 55+408+515,
55+408+538, 55+408+598, 55+408+602, 55+408+605, 55+408+609, 55+408+676, 55+408+694,
55+408+698, 55+408+699, 55+408+711, 55+408+754, 55+408+760, 55+408+781, 55+408+786,
55+408+797, 55+408+834, 55+408+835, 55+410+416, 55+410+448, 55+410+451, 55+410+471,
55+410+472, 55+410+507, 55+410+512, 55+410+515, 55+410+538, 55+410+598, 55+410+602,
55+410+605, 55+410+609, 55+410+676, 55+410+694, 55+410+698, 55+410+699, 55+410+711,
55+410+754, 55+410+760, 55+410+781, 55+410+786, 55+410+797, 55+410+834, 55+410+835,
55+416+448, 55+416+451, 55+416+471, 55+416+472, 55+416+507, 55+416+512, 55+416+515,
55+416+538, 55+416+598, 55+416+602, 55+416+605, 55+416+609, 55+416+676, 55+416+694,
55+416+698, 55+416+699, 55+416+711, 55+416+754, 55+416+760, 55+416+781, 55+416+786,
55+416+797, 55+416+834, 55+416+835, 55+448+451 , 55+448+471 , 55+448+472, 55+448+507,
55+448+512, 55+448+515, 55+448+538, 55+448+598, 55+448+602, 55+448+605, 55+448+609,
55+448+676, 55+448+694, 55+448+698, 55+448+699, 55+448+711, 55+448+754, 55+448+760,
55+448+781 , 55+448+786, 55+448+797, 55+448+834, 55+448+835, 55+451+471, 55+451+472,
55+451+507, 55+451+512, 55+451+515, 55+451+538, 55+451+598, 55+451+602, 55+451+605,
55+451+609, 55+451+676, 55+451+694, 55+451+698, 55+451+699, 55+451+711, 55+451+754,
55+451+760, 55+451+781, 55+451+786, 55+451+797, 55+451+834, 55+451+835, 55+471+472,
55+471+507, 55+471+512, 55+471+515, 55+471+538, 55+471+598, 55+471+602, 55+471+605,
55+471+609, 55+471+676, 55+471+694, 55+471+698, 55+471+699, 55+471+711, 55+471+754,
55+471+760, 55+471+781, 55+471+786, 55+471+797, 55+471+834, 55+471+835, 55+472+507,
55+472+512, 55+472+515, 55+472+538, 55+472+598, 55+472+602, 55+472+605, 55+472+609,
55+472+676, 55+472+694, 55+472+698, 55+472+699, 55+472+711, 55+472+754, 55+472+760,
55+472+781 , 55+472+786, 55+472+797, 55+472+834, 55+472+835, 55+507+512, 55+507+515,
55+507+538, 55+507+598, 55+507+602, 55+507+605, 55+507+609, 55+507+676, 55+507+694,
55+507+698, 55+507+699, 55+507+71 1 , 55+507+754, 55+507+760, 55+507+781 , 55+507+786,
55+507+797, 55+507+834, 55+507+835, 55+512+515, 55+512+538, 55+512+598, 55+512+602,
55+512+605, 55+512+609, 55+512+676, 55+512+694, 55+512+698, 55+512+699, 55+512+711 ,
55+512+754, 55+512+760, 55+512+781 , 55+512+786, 55+512+797, 55+512+834, 55+512+835,
55+515+538, 55+515+598, 55+515+602, 55+515+605, 55+515+609, 55+515+676, 55+515+694,
55+515+698, 55+515+699, 55+515+71 1 , 55+515+754, 55+515+760, 55+515+781 , 55+515+786,
55+515+797, 55+515+834, 55+515+835, 55+538+598, 55+538+602, 55+538+605, 55+538+609,
55+538+676, 55+538+694, 55+538+698, 55+538+699, 55+538+711 , 55+538+754, 55+538+760,
55+538+781 , 55+538+786, 55+538+797, 55+538+834, 55+538+835, 55+598+602, 55+598+605,
55+598+609, 55+598+676, 55+598+694, 55+598+698, 55+598+699, 55+598+71 1 , 55+598+754,
55+598+760, 55+598+781 , 55+598+786, 55+598+797, 55+598+834, 55+598+835, 55+602+605,
55+602+609, 55+602+676, 55+602+694, 55+602+698, 55+602+699, 55+602+71 1 , 55+602+754,
55+602+760, 55+602+781 , 55+602+786, 55+602+797, 55+602+834, 55+602+835, 55+605+609,
55+605+676, 55+605+694, 55+605+698, 55+605+699, 55+605+711 , 55+605+754, 55+605+760,
55+605+781 , 55+605+786, 55+605+797, 55+605+834, 55+605+835, 55+609+676, 55+609+694,
55+609+698, 55+609+699, 55+609+71 1 , 55+609+754, 55+609+760, 55+609+781 , 55+609+786,
55+609+797, 55+609+834, 55+609+835, 55+676+694, 55+676+698, 55+676+699, 55+676+711 ,
55+676+754, 55+676+760, 55+676+781 , 55+676+786, 55+676+797, 55+676+834, 55+676+835,
55+694+698, 55+694+699, 55+694+71 1 , 55+694+754, 55+694+760, 55+694+781 , 55+694+786,
55+694+797, 55+694+834, 55+694+835, 55+698+699, 55+698+711 , 55+698+754, 55+698+760,
55+698+781 , 55+698+786, 55+698+797, 55+698+834, 55+698+835, 55+699+71 1 , 55+699+754,
55+699+760, 55+699+781 , 55+699+786, 55+699+797, 55+699+834, 55+699+835, 55+711 +754,
55+711 +760, 55+711 +781 , 55+71 1 +786, 55+71 1 +797, 55+71 1 +834, 55+71 1 +835, 55+754+760,
55+754+781 , 55+754+786, 55+754+797, 55+754+834, 55+754+835, 55+760+781 , 55+760+786,
55+760+797, 55+760+834, 55+760+835, 55+781 +786, 55+781 +797, 55+781 +834, 55+781 +835,
55+786+797, 55+786+834, 55+786+835, 55+797+834, 55+797+835, 55+834+835, 56+60+63, 56+60+79,
56+60+87, 56+60+192, 56+60+302, 56+60+387, 56+60+388, 56+60+390, 56+60+403, 56+60+408
56+60+410, 56+60+416, 56+60+448, 56+60+451 , 56+60+471 , 56+60+472, 56+60+507, 56+60+512 56+60+515, 56+60+538, 56+60+598, 56+60+602, 56+60+605, 56+60+609, 56+60+676, 56+60+694
56+60+698, 56+60+699, 56+60+711 , 56+60+754, 56+60+760, 56+60+781 , 56+60+786, 56+60+797
56+60+834, 56+60+835, 56+63+79, 56+63+87, 56+63+192, 56+63+302, 56+63+387, 56+63+388
56+63+390, 56+63+403, 56+63+408, 56+63+410, 56+63+416, 56+63+448, 56+63+451 , 56+63+471
56+63+472, 56+63+507, 56+63+512, 56+63+515, 56+63+538, 56+63+598, 56+63+602, 56+63+605 56+63+609, 56+63+676, 56+63+694, 56+63+698, 56+63+699, 56+63+71 1 , 56+63+754, 56+63+760
56+63+781 , 56+63+786, 56+63+797, 56+63+834, 56+63+835, 56+79+87, 56+79+192, 56+79+302
56+79+387, 56+79+388, 56+79+390, 56+79+403, 56+79+408, 56+79+410, 56+79+416, 56+79+448
56+79+451 , 56+79+471 , 56+79+472, 56+79+507, 56+79+512, 56+79+515, 56+79+538, 56+79+598
56+79+602, 56+79+605, 56+79+609, 56+79+676, 56+79+694, 56+79+698, 56+79+699, 56+79+71 1
56+79+754, 56+79+760, 56+79+781 , 56+79+786, 56+79+797, 56+79+834, 56+79+835, 56+87+192
56+87+302, 56+87+387, 56+87+388, 56+87+390, 56+87+403, 56+87+408, 56+87+410, 56+87+416
56+87+448, 56+87+451 , 56+87+471 , 56+87+472, 56+87+507, 56+87+512, 56+87+515, 56+87+538
56+87+598, 56+87+602, 56+87+605, 56+87+609, 56+87+676, 56+87+694, 56+87+698, 56+87+699
56+87+71 1 , 56+87+754, 56+87+760, 56+87+781 , 56+87+786, 56+87+797, 56+87+834, 56+87+835
56+192+302 56+192+387, 56+192+388, 56+192+390, 56+192+403, 56+192+408, 56+192+410
56+192+416 56+192+448, 56+192+451 , 56+192+471 , 56+192+472, 56+192+507, 56+192+512
56+192+515 56+192+538, 56+192+598, 56+192+602, 56+192+605, 56+192+609, 56+192+676
56+192+694 56+192+698, 56+192+699, 56+192+711 , 56+192+754, 56+192+760, 56+192+781
56+192+786 56+192+797, 56+192+834, 56+192+835, 56+302+387, 56+302+388, 56+302+390
56+302+403 56+302+408, 56+302+410, 56+302+416, 56+302+448, 56+302+451 , 56+302+471
56+302+472 56+302+507, 56+302+512, 56+302+515, 56+302+538, 56+302+598, 56+302+602
56+302+605 56+302+609, 56+302+676, 56+302+694, 56+302+698, 56+302+699, 56+302+711
56+302+754 56+302+760, 56+302+781 , 56+302+786, 56+302+797, 56+302+834, 56+302+835
56+387+388 56+387+390, 56+387+403, 56+387+408, 56+387+410, 56+387+416, 56+387+448
56+387+451 56+387+471 , 56+387+472, 56+387+507, 56+387+512, 56+387+515, 56+387+538
56+387+598 56+387+602, 56+387+605, 56+387+609, 56+387+676, 56+387+694, 56+387+698
56+387+699 56+387+71 1 , 56+387+754, 56+387+760, 56+387+781 , 56+387+786, 56+387+797
56+387+834 56+387+835, 56+388+390, 56+388+403, 56+388+408, 56+388+410, 56+388+416
56+388+448 56+388+451 , 56+388+471 , 56+388+472, 56+388+507, 56+388+512, 56+388+515
56+388+538 56+388+598, 56+388+602, 56+388+605, 56+388+609, 56+388+676, 56+388+694
56+388+698 56+388+699, 56+388+71 1 , 56+388+754, 56+388+760, 56+388+781 , 56+388+786
56+388+797 56+388+834, 56+388+835, 56+390+403, 56+390+408, 56+390+410, 56+390+416
56+390+448 56+390+451 , 56+390+471 , 56+390+472, 56+390+507, 56+390+512, 56+390+515
56+390+538 56+390+598, 56+390+602, 56+390+605, 56+390+609, 56+390+676, 56+390+694
56+390+698 56+390+699, 56+390+71 1 , 56+390+754, 56+390+760, 56+390+781 , 56+390+786
56+390+797 56+390+834, 56+390+835, 56+403+408, 56+403+410, 56+403+416, 56+403+448
56+403+451 56+403+471 , 56+403+472, 56+403+507, 56+403+512, 56+403+515, 56+403+538
56+403+598 56+403+602, 56+403+605, 56+403+609, 56+403+676, 56+403+694, 56+403+698
56+403+699 56+403+71 1 , 56+403+754, 56+403+760, 56+403+781 , 56+403+786, 56+403+797
56+403+834 56+403+835, 56+408+410, 56+408+416, 56+408+448, 56+408+451 , 56+408+471
56+408+472 56+408+507, 56+408+512, 56+408+515, 56+408+538, 56+408+598, 56+408+602
56+408+605 56+408+609, 56+408+676, 56+408+694, 56+408+698, 56+408+699, 56+408+711
56+408+754, 56+408+760, 56+408+781, 56+408+786, 56+408+797, 56+408+834, 56+408+835,
56+410+416, 56+410+448, 56+410+451, 56+410+471, 56+410+472, 56+410+507, 56+410+512,
56+410+515, 56+410+538, 56+410+598, 56+410+602, 56+410+605, 56+410+609, 56+410+676,
56+410+694, 56+410+698, 56+410+699, 56+410+711, 56+410+754, 56+410+760, 56+410+781,
56+410+786, 56+410+797, 56+410+834, 56+410+835, 56+416+448, 56+416+451, 56+416+471,
56+416+472, 56+416+507, 56+416+512, 56+416+515, 56+416+538, 56+416+598, 56+416+602,
56+416+605, 56+416+609, 56+416+676, 56+416+694, 56+416+698, 56+416+699, 56+416+711,
56+416+754, 56+416+760, 56+416+781, 56+416+786, 56+416+797, 56+416+834, 56+416+835,
56+448+451 , 56+448+471, 56+448+472, 56+448+507, 56+448+512, 56+448+515, 56+448+538,
56+448+598, 56+448+602, 56+448+605, 56+448+609, 56+448+676, 56+448+694, 56+448+698,
56+448+699, 56+448+711 , 56+448+754, 56+448+760, 56+448+781 , 56+448+786, 56+448+797,
56+448+834, 56+448+835, 56+451+471, 56+451+472, 56+451+507, 56+451+512, 56+451+515,
56+451+538, 56+451+598, 56+451+602, 56+451+605, 56+451+609, 56+451+676, 56+451+694,
56+451+698, 56+451+699, 56+451+711, 56+451+754, 56+451+760, 56+451+781, 56+451+786,
56+451+797, 56+451+834, 56+451+835, 56+471+472, 56+471+507, 56+471+512, 56+471+515,
56+471+538, 56+471+598, 56+471+602, 56+471+605, 56+471+609, 56+471+676, 56+471+694,
56+471+698, 56+471+699, 6+471+711, 56+471+754, 56+471+760, 56+471+781, 56+471+786,
56+471+797, 56+471+834, 56+471+835, 56+472+507, 56+472+512, 56+472+515, 56+472+538,
56+472+598, 56+472+602, 56+472+605, 56+472+609, 56+472+676, 56+472+694, 56+472+698,
56+472+699, 56+472+711, 56+472+754, 56+472+760, 56+472+781 , 56+472+786, 56+472+797,
56+472+834, 56+472+835, 56+507+512, 56+507+515, 56+507+538, 56+507+598, 56+507+602,
56+507+605, 56+507+609, 56+507+676, 56+507+694, 56+507+698, 56+507+699, 56+507+711,
56+507+754, 56+507+760, 56+507+781, 56+507+786, 56+507+797, 56+507+834, 56+507+835,
56+512+515, 56+512+538, 56+512+598, 56+512+602, 56+512+605, 56+512+609, 56+512+676,
56+512+694, 56+512+698, 56+512+699, 56+512+711, 56+512+754, 56+512+760, 56+512+781,
56+512+786, 56+512+797, 56+512+834, 56+512+835, 56+515+538, 56+515+598, 56+515+602,
56+515+605, 56+515+609, 56+515+676, 56+515+694, 56+515+698, 56+515+699, 56+515+711,
56+515+754, 56+515+760, 56+515+781, 56+515+786, 56+515+797, 56+515+834, 56+515+835,
56+538+598, 56+538+602, 56+538+605, 56+538+609, 56+538+676, 56+538+694, 56+538+698,
56+538+699, 56+538+711, 56+538+754, 56+538+760, 56+538+781 , 56+538+786, 56+538+797,
56+538+834, 56+538+835, 56+598+602, 56+598+605, 56+598+609, 56+598+676, 56+598+694,
56+598+698, 56+598+699, 56+598+711, 56+598+754, 56+598+760, 56+598+781, 56+598+786,
56+598+797, 56+598+834, 56+598+835, 56+602+605, 56+602+609, 56+602+676, 56+602+694,
56+602+698, 56+602+699, 56+602+711, 56+602+754, 56+602+760, 56+602+781, 56+602+786,
56+602+797, 56+602+834, 56+602+835, 56+605+609, 56+605+676, 56+605+694, 56+605+698,
56+605+699, 56+605+711, 56+605+754, 56+605+760, 56+605+781 , 56+605+786, 56+605+797,
56+605+834, 56+605+835, 56+609+676, 56+609+694, 56+609+698, 56+609+699, 56+609+711 ,
56+609+754, 56+609+760, 56+609+781 , 56+609+786, 56+609+797, 56+609+834, 56+609+835,
56+676+694, 56+676+698, 56+676+699, 56+676+711 , 56+676+754, 56+676+760, 56+676+781 ,
56+676+786, 56+676+797, 56+676+834, 56+676+835, 56+694+698, 56+694+699, 56+694+711 ,
56+694+754, 56+694+760, 56+694+781 , 56+694+786, 56+694+797, 56+694+834, 56+694+835,
56+698+699, 56+698+71 1 , 56+698+754, 56+698+760, 56+698+781 , 56+698+786, 56+698+797,
56+698+834, 56+698+835, 56+699+71 1 , 56+699+754, 56+699+760, 56+699+781 , 56+699+786,
56+699+797, 56+699+834, 56+699+835, 56+71 1 +754, 56+71 1 +760, 56+71 1 +781 , 56+711 +786,
56+711 +797, 56+711 +834, 56+71 1 +835, 56+754+760, 56+754+781 , 56+754+786, 56+754+797,
56+754+834, 56+754+835, 56+760+781 , 56+760+786, 56+760+797, 56+760+834, 56+760+835,
56+781 +786, 56+781 +797, 56+781 +834, 56+781 +835, 56+786+797, 56+786+834, 56+786+835,
56+797+834, 56+797+835, 56+834+835, 60+63+79, 60+63+87, 60+63+192, 60+63+302, 60+63+387;
60+63+388, 60+63+390, 60+63+403, 60+63+408, 60+63+410, 60+63+416, 60+63+448, 60+63+451
60+63+471 , 60+63+472, 60+63+507, 60+63+512, 60+63+515, 60+63+538, 60+63+598, 60+63+602; 60+63+605, 60+63+609, 60+63+676, 60+63+694, 60+63+698, 60+63+699, 60+63+711 , 60+63+754;
60+63+760, 60+63+781 , 60+63+786, 60+63+797, 60+63+834, 60+63+835, 60+79+87, 60+79+192;
60+79+302, 60+79+387, 60+79+388, 60+79+390, 60+79+403, 60+79+408, 60+79+410, 60+79+416;
60+79+448, 60+79+451 , 60+79+471 , 60+79+472, 60+79+507, 60+79+512, 60+79+515, 60+79+538;
60+79+598, 60+79+602, 60+79+605, 60+79+609, 60+79+676, 60+79+694, 60+79+698, 60+79+699; 60+79+71 1 , 60+79+754, 60+79+760, 60+79+781 , 60+79+786, 60+79+797, 60+79+834, 60+79+835;
60+87+192, 60+87+302, 60+87+387, 60+87+388, 60+87+390, 60+87+403, 60+87+408, 60+87+410;
60+87+416, 60+87+448, 60+87+451 , 60+87+471 , 60+87+472, 60+87+507, 60+87+512, 60+87+515;
60+87+538, 60+87+598, 60+87+602, 60+87+605, 60+87+609, 60+87+676, 60+87+694, 60+87+698;
60+87+699, 60+87+711 , 60+87+754, 60+87+760, 60+87+781 , 60+87+786, 60+87+797, 60+87+834;
60+87+835, 60+192+302, 60+192+387, 60+192+388, 60+192+390, 60+192+403, 60+192+408,
60+192+410, 60+192+416, 60+192+448, 60+192+451 , 60+192+471 , 60+192+472, 60+192+507,
60+192+512, 60+192+515, 60+192+538, 60+192+598, 60+192+602, 60+192+605, 60+192+609,
60+192+676, 60+192+694, 60+192+698, 60+192+699, 60+192+711 , 60+192+754, 60+192+760,
60+192+781 , 60+192+786, 60+192+797, 60+192+834, 60+192+835, 60+302+387, 60+302+388,
60+302+390, 60+302+403, 60+302+408, 60+302+410, 60+302+416, 60+302+448, 60+302+451 ,
60+302+471 , 60+302+472, 60+302+507, 60+302+512, 60+302+515, 60+302+538, 60+302+598,
60+302+602, 60+302+605, 60+302+609, 60+302+676, 60+302+694, 60+302+698, 60+302+699,
60+302+711 , 60+302+754, 60+302+760, 60+302+781 , 60+302+786, 60+302+797, 60+302+834,
60+302+835, 60+387+388, 60+387+390, 60+387+403, 60+387+408, 60+387+410, 60+387+416,
60+387+448, 60+387+451 , 60+387+471 , 60+387+472, 60+387+507, 60+387+512, 60+387+515,
60+387+538, 60+387+598, 60+387+602, 60+387+605, 60+387+609, 60+387+676, 60+387+694,
60+387+698, 60+387+699, 60+387+711, 60+387+754, 60+387+760, 60+387+781, 60+387+786,
60+387+797, 60+387+834, 60+387+835, 60+388+390, 60+388+403, 60+388+408, 60+388+410,
60+388+416, 60+388+448, 60+388+451, 60+388+471 , 60+388+472, 60+388+507, 60+388+512,
60+388+515, 60+388+538, 60+388+598, 60+388+602, 60+388+605, 60+388+609, 60+388+676,
60+388+694, 60+388+698, 60+388+699, 60+388+711, 60+388+754, 60+388+760, 60+388+781 ,
60+388+786, 60+388+797, 60+388+834, 60+388+835, 60+390+403, 60+390+408, 60+390+410,
60+390+416, 60+390+448, 60+390+451, 60+390+471 , 60+390+472, 60+390+507, 60+390+512,
60+390+515, 60+390+538, 60+390+598, 60+390+602, 60+390+605, 60+390+609, 60+390+676,
60+390+694, 60+390+698, 60+390+699, 60+390+711, 60+390+754, 60+390+760, 60+390+781 ,
60+390+786, 60+390+797, 60+390+834, 60+390+835, 60+403+408, 60+403+410, 60+403+416,
60+403+448, 60+403+451, 60+403+471, 60+403+472, 60+403+507, 60+403+512, 60+403+515,
60+403+538, 60+403+598, 60+403+602, 60+403+605, 60+403+609, 60+403+676, 60+403+694,
60+403+698, 60+403+699, 60+403+711, 60+403+754, 60+403+760, 60+403+781, 60+403+786,
60+403+797, 60+403+834, 60+403+835, 60+408+410, 60+408+416, 60+408+448, 60+408+451 ,
60+408+471 , 60+408+472, 60+408+507, 60+408+512, 60+408+515, 60+408+538, 60+408+598,
60+408+602, 60+408+605, 60+408+609, 60+408+676, 60+408+694, 60+408+698, 60+408+699,
60+408+711, 60+408+754, 60+408+760, 60+408+781 , 60+408+786, 60+408+797, 60+408+834,
60+408+835, 60+410+416, 60+410+448, 60+410+451, 60+410+471, 60+410+472, 60+410+507,
60+410+512, 60+410+515, 60+410+538, 60+410+598, 60+410+602, 60+410+605, 60+410+609,
60+410+676, 60+410+694, 60+410+698, 60+410+699, 60+410+711, 60+410+754, 60+410+760,
60+410+781, 60+410+786, 60+410+797, 60+410+834, 60+410+835, 60+416+448, 60+416+451,
60+416+471, 60+416+472, 60+416+507, 60+416+512, 60+416+515, 60+416+538, 60+416+598,
60+416+602, 60+416+605, 60+416+609, 60+416+676, 60+416+694, 60+416+698, 60+416+699,
60+416+711, 60+416+754, 60+416+760, 60+416+781, 60+416+786, 60+416+797, 60+416+834,
60+416+835, 60+448+451, 60+448+471, 60+448+472, 60+448+507, 60+448+512, 60+448+515,
60+448+538, 60+448+598, 60+448+602, 60+448+605, 60+448+609, 60+448+676, 60+448+694,
60+448+698, 60+448+699, 60+448+711 , 60+448+754, 60+448+760, 60+448+781, 60+448+786,
60+448+797, 60+448+834, 60+448+835, 60+451+471, 60+451+472, 60+451+507, 60+451+512,
60+451+515, 60+451+538, 60+451+598, 60+451+602, 60+451+605, 60+451+609, 60+451+676,
60+451+694, 60+451+698, 60+451+699, 60+451+711, 60+451+754, 60+451+760, 60+451+781,
60+451+786, 60+451+797, 60+451+834, 60+451+835, 60+471+472, 60+471+507, 60+471+512,
60+471+515, 60+471+538, 60+471+598, 60+471+602, 60+471+605, 60+471+609, 60+471+676,
60+471+694, 60+471+698, 60+471+699, 60+471+711, 60+471+754, 60+471+760, 60+471+781,
60+471+786, 60+471+797, 60+471+834, 60+471+835, 60+472+507, 60+472+512, 60+472+515,
60+472+538, 60+472+598, 60+472+602, 60+472+605, 60+472+609, 60+472+676, 60+472+694,
60+472+698, 60+472+699, 60+472+711, 60+472+754, 60+472+760, 60+472+781, 60+472+786,
+472+797 60+472+834 60+472+835 60+507+512 60+507+515 60+507+538 60+507+598+507+602 60+507+605 60+507+609 60+507+676 60+507+694 60+507+698 60+507+699+507+711 60+507+754 60+507+760 60+507+781 60+507+786 60+507+797 60+507+834+507+835 60+512+515 60+512+538 60+512+598 60+512+602 60+512+605 60+512+609+512+676 60+512+694 60+512+698 60+512+699 60+512+711 60+512+754 60+512+760+512+781 60+512+786 60+512+797 60+512+834 60+512+835 60+515+538 60+515+598+515+602 60+515+605 60+515+609 60+515+676 60+515+694 60+515+698 60+515+699+515+711 60+515+754 60+515+760 60+515+781 60+515+786 60+515+797 60+515+834+515+835 60+538+598 60+538+602 60+538+605 60+538+609 60+538+676 60+538+694+538+698 60+538+699 60+538+71 1 60+538+754 60+538+760 60+538+781 60+538+786+538+797 60+538+834 60+538+835 60+598+602 60+598+605 60+598+609 60+598+676+598+694 60+598+698 60+598+699 60+598+711 60+598+754 60+598+760 60+598+781+598+786 60+598+797 60+598+834 60+598+835 60+602+605 60+602+609 60+602+676+602+694 60+602+698 60+602+699 60+602+711 60+602+754 60+602+760 60+602+781+602+786 60+602+797 60+602+834 60+602+835 60+605+609 60+605+676 60+605+694+605+698 60+605+699 60+605+71 1 60+605+754 60+605+760 60+605+781 60+605+786+605+797 60+605+834 60+605+835 60+609+676 60+609+694 60+609+698 60+609+699+609+711 60+609+754 60+609+760 60+609+781 60+609+786 60+609+797 60+609+834+609+835 60+676+694 60+676+698 60+676+699 60+676+711 60+676+754 60+676+760+676+781 60+676+786 60+676+797 60+676+834 60+676+835 60+694+698 60+694+699+694+711 60+694+754 60+694+760 60+694+781 60+694+786 60+694+797 60+694+834+694+835 60+698+699 60+698+71 1 60+698+754 60+698+760 60+698+781 60+698+786+698+797 60+698+834 60+698+835 60+699+711 60+699+754 60+699+760 60+699+781+699+786 60+699+797 60+699+834 60+699+835 60+71 1 +754 60+71 1 +760 60+711 +781+711 +786 60+711 +797 60+71 1 +834 60+71 1 +835 60+754+760 60+754+781 60+754+786+754+797 60+754+834 60+754+835 60+760+781 60+760+786 60+760+797 60+760+834+760+835 60+781 +786 60+781 +797 60+781 +834 60+781 +835 60+786+797 60+786+834+786+835 60+797+834, 60+797+835, 60+834+835, 63+79+87, 63+79+192, 63+79+302, 63+79+387+79+388, 63+79+390, 63+79+403, 63+79+408, 63+79+410, 63+79+416, 63+79+448, 63+79+451+79+471 , 63+79+472, 63+79+507, 63+79+512, 63+79+515, 63+79+538, 63+79+598, 63+79+602+79+605, 63+79+609, 63+79+676, 63+79+694, 63+79+698, 63+79+699, 63+79+711 , 63+79+754+79+760, 63+79+781 , 63+79+786, 63+79+797, 63+79+834, 63+79+835, 63+87+192, 63+87+302+87+387, 63+87+388, 63+87+390, 63+87+403, 63+87+408, 63+87+410, 63+87+416, 63+87+448+87+451 , 63+87+471 , 63+87+472, 63+87+507, 63+87+512, 63+87+515, 63+87+538, 63+87+598+87+602, 63+87+605, 63+87+609, 63+87+676, 63+87+694, 63+87+698, 63+87+699, 63+87+71 1+87+754, 63+87+760, 63+87+781 , 63+87+786, 63+87+797, 63+87+834, 63+87+835, 63+192+302
63+192+387, 63+192+388, 63+192+390, 63+192+403, 63+192+408, 63+192+410, 63+192+416,
63+192+448, 63+192+451 , 63+192+471 , 63+192+472, 63+192+507, 63+192+512, 63+192+515,
63+192+538, 63+192+598, 63+192+602, 63+192+605, 63+192+609, 63+192+676, 63+192+694,
63+192+698, 63+192+699, 63+192+71 1 , 63+192+754, 63+192+760, 63+192+781 , 63+192+786,
63+192+797, 63+192+834, 63+192+835, 63+302+387, 63+302+388, 63+302+390, 63+302+403,
63+302+408, 63+302+410, 63+302+416, 63+302+448, 63+302+451 , 63+302+471 , 63+302+472,
63+302+507, 63+302+512, 63+302+515, 63+302+538 63+302+598, 63+302+602, 63+302+605
63+302+609, 63+302+676, 63+302+694, 63+302+698, 63+302+699, 63+302+71 1 , 63+302+754,
63+302+760, 63+302+781 , 63+302+786, 63+302+797, 63+302+834, 63+302+835, 63+387+388,
63+387+390, 63+387+403, 63+387+408, 63+387+410, 63+387+416, 63+387+448, 63+387+451 ,
63+387+471 , 63+387+472, 63+387+507, 63+387+512, 63+387+515, 63+387+538, 63+387+598,
63+387+602, 63+387+605, 63+387+609, 63+387+676, 63+387+694, 63+387+698, 63+387+699,
63+387+711 , 63+387+754, 63+387+760, 63+387+781 , 63+387+786, 63+387+797, 63+387+834,
63+387+835, 63+388+390, 63+388+403, 63+388+408, 63+388+410, 63+388+416, 63+388+448,
63+388+451 , 63+388+471 , 63+388+472, 63+388+507, 63+388+512, 63+388+515, 63+388+538,
63+388+598, 63+388+602, 63+388+605, 63+388+609, 63+388+676, 63+388+694, 63+388+698,
63+388+699, 63+388+71 1 , 63+388+754, 63+388+760, 63+388+781 , 63+388+786, 63+388+797,
63+388+834, 63+388+835, 63+390+403, 63+390+408, 63+390+410, 63+390+416, 63+390+448,
63+390+451 , 63+390+471 , 63+390+472, 63+390+507, 63+390+512, 63+390+515, 63+390+538,
63+390+598, 63+390+602, 63+390+605, 63+390+609, 63+390+676, 63+390+694, 63+390+698,
63+390+699, 63+390+71 1 , 63+390+754, 63+390+760, 63+390+781 , 63+390+786, 63+390+797,
63+390+834, 63+390+835, 63+403+408, 63+403+410, 63+403+416, 63+403+448, 63+403+451 ,
63+403+471 , 63+403+472, 63+403+507, 63+403+512, 63+403+515, 63+403+538, 63+403+598,
63+403+602, 63+403+605, 63+403+609, 63+403+676, 63+403+694, 63+403+698, 63+403+699,
63+403+711 , 63+403+754, 63+403+760, 63+403+781 , 63+403+786, 63+403+797, 63+403+834,
63+403+835, 63+408+410, 63+408+416, 63+408+448, 63+408+451 , 63+408+471 , 63+408+472,
63+408+507, 63+408+512, 63+408+515, 63+408+538, 63+408+598, 63+408+602, 63+408+605,
63+408+609, 63+408+676, 63+408+694, 63+408+698, 63+408+699, 63+408+71 1 , 63+408+754,
63+408+760, 63+408+781 , 63+408+786, 63+408+797, 63+408+834, 63+408+835, 63+410+416,
63+410+448, 63+410+451 , 63+410+471 , 63+410+472, 63+410+507, 63+410+512, 63+410+515,
63+410+538, 63+410+598, 63+410+602, 63+410+605, 63+410+609, 63+410+676, 63+410+694,
63+410+698, 63+410+699, 63+410+71 1 , 63+410+754, 63+410+760, 63+410+781 , 63+410+786,
63+410+797, 63+410+834, 63+410+835, 63+416+448, 63+416+451 , 63+416+471 , 63+416+472,
63+416+507, 63+416+512, 63+416+515, 63+416+538, 63+416+598, 63+416+602, 63+416+605,
63+416+609, 63+416+676, 63+416+694, 63+416+698, 63+416+699, 63+416+71 1 , 63+416+754,
63+416+760, 63+416+781 , 63+416+786, 63+416+797, 63+416+834, 63+416+835, 63+448+451 ,
63+448+471 , 63+448+472, 63+448+507, 63+448+512, 63+448+515, 63+448+538, 63+448+598,
63+448+602, 63+448+605, 63+448+609, 63+448+676, 63+448+694, 63+448+698, 63+448+699,
63+448+711, 63+448+754, 63+448+760, 63+448+781 , 63+448+786, 63+448+797, 63+448+834,
63+448+835, 63+451+471, 63+451+472, 63+451+507, 63+451+512, 63+451+515, 63+451+538,
63+451+598, 63+451+602, 63+451+605, 63+451+609, 63+451+676, 63+451+694, 63+451+698,
63+451+699, 63+451+711, 63+451+754, 63+451+760, 63+451+781, 63+451+786, 63+451+797,
63+451+834, 63+451+835, 63+471+472, 63+471+507, 63+471+512, 63+471+515, 63+471+538,
63+471+598, 63+471+602, 63+471+605, 63+471+609, 63+471+676, 63+471+694, 63+471+698,
63+471+699, 63+471+711, 63+471+754, 63+471+760, 63+471+781, 63+471+786, 63+471+797,
63+471+834, 63+471+835, 63+472+507, 63+472+512, 63+472+515, 63+472+538, 63+472+598,
63+472+602, 63+472+605, 63+472+609, 63+472+676, 63+472+694, 63+472+698, 63+472+699,
63+472+711, 63+472+754, 63+472+760, 63+472+781 , 63+472+786, 63+472+797, 63+472+834,
63+472+835, 63+507+512, 63+507+515, 63+507+538, 63+507+598, 63+507+602, 63+507+605,
63+507+609, 63+507+676, 63+507+694, 63+507+698, 63+507+699, 63+507+711, 63+507+754,
63+507+760, 63+507+781, 63+507+786, 63+507+797, 63+507+834, 63+507+835, 63+512+515,
63+512+538, 63+512+598, 63+512+602, 63+512+605, 63+512+609, 63+512+676, 63+512+694,
63+512+698, 63+512+699, 63+512+711, 63+512+754, 63+512+760, 63+512+781, 63+512+786,
63+512+797, 63+512+834, 63+512+835, 63+515+538, 63+515+598, 63+515+602, 63+515+605,
63+515+609, 63+515+676, 63+515+694, 63+515+698, 63+515+699, 63+515+711, 63+515+754,
63+515+760, 63+515+781, 63+515+786, 63+515+797, 63+515+834, 63+515+835, 63+538+598,
63+538+602, 63+538+605, 63+538+609, 63+538+676, 63+538+694, 63+538+698, 63+538+699,
63+538+711, 63+538+754, 63+538+760, 63+538+781 , 63+538+786, 63+538+797, 63+538+834,
63+538+835, 63+598+602, 63+598+605, 63+598+609, 63+598+676, 63+598+694, 63+598+698,
63+598+699, 63+598+711, 63+598+754, 63+598+760, 63+598+781 , 63+598+786, 63+598+797,
63+598+834, 63+598+835, 63+602+605, 63+602+609, 63+602+676, 63+602+694, 63+602+698,
63+602+699, 63+602+711, 63+602+754, 63+602+760, 63+602+781 , 63+602+786, 63+602+797,
63+602+834, 63+602+835, 63+605+609, 63+605+676, 63+605+694, 63+605+698, 63+605+699,
63+605+711, 63+605+754, 63+605+760, 63+605+781 , 63+605+786, 63+605+797, 63+605+834,
63+605+835, 63+609+676, 63+609+694, 63+609+698, 63+609+699, 63+609+711, 63+609+754,
63+609+760, 63+609+781, 63+609+786, 63+609+797, 63+609+834, 63+609+835, 63+676+694,
63+676+698, 63+676+699, 63+676+711, 63+676+754, 63+676+760, 63+676+781, 63+676+786,
63+676+797, 63+676+834, 63+676+835, 63+694+698, 63+694+699, 63+694+711, 63+694+754,
63+694+760, 63+694+781, 63+694+786, 63+694+797, 63+694+834, 63+694+835, 63+698+699,
63+698+711, 63+698+754, 63+698+760, 63+698+781 , 63+698+786, 63+698+797, 63+698+834,
63+698+835, 63+699+711, 63+699+754, 63+699+760, 63+699+781 , 63+699+786, 63+699+797,
63+699+834, 63+699+835, 63+711+754, 63+711+760, 63+711+781, 63+711+786, 63+711+797,
63+711 +834, 63+711 +835, 63+754+760, 63+754+781 , 63+754+786, 63+754+797, 63+754+834 ; 63+754+835, 63+760+781 , 63+760+786, 63+760+797, 63+760+834, 63+760+835, 63+781 +786 63+781 +797, 63+781 +834, 63+781 +835, 63+786+797, 63+786+834, 63+786+835, 63+797+834 ; 63+797+835, 63+834+835, 79+87+192, 79+87+302, 79+87+387, 79+87+388, 79+87+390, 79+87+403; 79+87+408, 79+87+410, 79+87+416, 79+87+448, 79+87+451 , 79+87+471 , 79+87+472, 79+87+507; 79+87+512, 79+87+515, 79+87+538, 79+87+598, 79+87+602, 79+87+605, 79+87+609, 79+87+676; 79+87+694, 79+87+698, 79+87+699, 79+87+71 1 , 79+87+754, 79+87+760, 79+87+781 , 79+87+786; 79+87+797, 79+87+834, 79+87+835, 79+192+302, 79+192+387, 79+192+388, 79+192+390, 79+192+403
79+192+408, 79+192+410, 79+192+416, 79+192+448, 79+192+451 , 79+192+471 , 79+192+472,
79+192+507, 79+192+512, 79+192+515, 79+192+538, 79+192+598, 79+192+602, 79+192+605,
79+192+609, 79+192+676, 79+192+694, 79+192+698, 79+192+699, 79+192+71 1 , 79+192+754,
79+192+760, 79+192+781 , 79+192+786, 79+192+797, 79+192+834, 79+192+835, 79+302+387,
79+302+388, 79+302+390, 79+302+403, 79+302+408, 79+302+410, 79+302+416, 79+302+448,
79+302+451 , 79+302+471 , 79+302+472, 79+302+507, 79+302+512, 79+302+515, 79+302+538,
79+302+598, 79+302+602, 79+302+605, 79+302+609, 79+302+676, 79+302+694, 79+302+698,
79+302+699, 79+302+71 1 , 79+302+754, 79+302+760, 79+302+781 , 79+302+786, 79+302+797,
79+302+834, 79+302+835, 79+387+388, 79+387+390, 79+387+403, 79+387+408, 79+387+410,
79+387+416, 79+387+448, 79+387+451 , 79+387+471 , 79+387+472, 79+387+507, 79+387+512,
79+387+515, 79+387+538, 79+387+598, 79+387+602, 79+387+605, 79+387+609, 79+387+676,
79+387+694, 79+387+698, 79+387+699, 79+387+711 , 79+387+754, 79+387+760, 79+387+781 ,
79+387+786, 79+387+797, 79+387+834, 79+387+835, 79+388+390, 79+388+403, 79+388+408,
79+388+410, 79+388+416, 79+388+448, 79+388+451 , 79+388+471 , 79+388+472, 79+388+507,
79+388+512, 79+388+515, 79+388+538, 79+388+598, 79+388+602, 79+388+605, 79+388+609,
79+388+676, 79+388+694, 79+388+698, 79+388+699, 79+388+711 , 79+388+754, 79+388+760,
79+388+781 , 79+388+786, 79+388+797, 79+388+834, 79+388+835, 79+390+403, 79+390+408,
79+390+410, 79+390+416, 79+390+448, 79+390+451 , 79+390+471 , 79+390+472, 79+390+507,
79+390+512, 79+390+515, 79+390+538, 79+390+598, 79+390+602, 79+390+605, 79+390+609,
79+390+676, 79+390+694, 79+390+698, 79+390+699, 79+390+711 , 79+390+754, 79+390+760,
79+390+781 , 79+390+786, 79+390+797, 79+390+834, 79+390+835, 79+403+408, 79+403+410,
79+403+416, 79+403+448, 79+403+451 , 79+403+471 , 79+403+472, 79+403+507, 79+403+512,
79+403+515, 79+403+538, 79+403+598, 79+403+602, 79+403+605, 79+403+609, 79+403+676,
79+403+694, 79+403+698, 79+403+699, 79+403+711 , 79+403+754, 79+403+760, 79+403+781 ,
79+403+786, 79+403+797, 79+403+834, 79+403+835, 79+408+410, 79+408+416, 79+408+448,
79+408+451 , 79+408+471 , 79+408+472, 79+408+507, 79+408+512, 79+408+515, 79+408+538,
79+408+598, 79+408+602, 79+408+605, 79+408+609, 79+408+676, 79+408+694, 79+408+698,
79+408+699, 79+408+71 1 , 79+408+754, 79+408+760, 79+408+781 , 79+408+786, 79+408+797,
79+408+834, 79+408+835, 79+410+416, 79+410+448, 79+410+451, 79+410+471, 79+410+472,
79+410+507, 79+410+512, 79+410+515, 79+410+538, 79+410+598, 79+410+602, 79+410+605,
79+410+609, 79+410+676, 79+410+694, 79+410+698, 79+410+699, 79+410+711, 79+410+754,
79+410+760, 79+410+781, 79+410+786, 79+410+797, 79+410+834, 79+410+835, 9+416+448,
79+416+451, 79+416+471, 79+416+472, 79+416+507, 79+416+512, 79+416+515, 79+416+538,
79+416+598, 79+416+602, 79+416+605, 79+416+609, 79+416+676, 79+416+694, 79+416+698,
79+416+699, 79+416+711, 79+416+754, 79+416+760, 79+416+781, 79+416+786, 79+416+797,
79+416+834, 79+416+835, 79+448+451, 79+448+471 , 79+448+472, 79+448+507, 79+448+512,
79+448+515, 79+448+538, 79+448+598, 79+448+602, 79+448+605, 79+448+609, 79+448+676,
79+448+694, 79+448+698, 79+448+699, 79+448+711, 79+448+754, 79+448+760, 79+448+781 ,
79+448+786, 79+448+797, 79+448+834, 79+448+835, 79+451+471, 79+451+472, 79+451+507,
79+451+512, 79+451+515, 79+451+538, 79+451+598, 79+451+602, 79+451+605, 79+451+609,
79+451+676, 79+451+694, 79+451+698, 79+451+699, 79+451+711, 79+451+754, 79+451+760,
79+451+781, 79+451+786, 79+451+797, 79+451+834, 79+451+835, 79+471+472, 79+471+507,
79+471+512, 79+471+515, 79+471+538, 79+471+598, 79+471+602, 79+471+605, 79+471+609,
79+471+676, 79+471+694, 79+471+698, 79+471+699, 79+471+711, 79+471+754, 79+471+760,
79+471+781, 79+471+786, 79+471+797, 79+471+834, 79+471+835, 79+472+507, 79+472+512,
79+472+515, 79+472+538, 79+472+598, 79+472+602, 79+472+605, 79+472+609, 79+472+676,
79+472+694, 79+472+698, 79+472+699, 79+472+711, 79+472+754, 79+472+760, 79+472+781 ,
79+472+786, 79+472+797, 79+472+834, 79+472+835, 79+507+512, 79+507+515, 79+507+538,
79+507+598, 79+507+602, 79+507+605, 79+507+609, 79+507+676, 79+507+694, 79+507+698,
79+507+699, 79+507+711, 79+507+754, 79+507+760, 79+507+781 , 79+507+786, 79+507+797,
79+507+834, 79+507+835, 79+512+515, 79+512+538, 79+512+598, 79+512+602, 79+512+605,
79+512+609, 79+512+676, 79+512+694, 79+512+698, 79+512+699, 79+512+711, 79+512+754,
79+512+760, 79+512+781, 79+512+786, 79+512+797, 79+512+834, 79+512+835, 79+515+538,
79+515+598, 79+515+602, 79+515+605, 79+515+609, 79+515+676, 79+515+694, 79+515+698,
79+515+699, 79+515+711, 79+515+754, 79+515+760, 79+515+781, 79+515+786, 79+515+797,
79+515+834, 79+515+835, 79+538+598, 79+538+602, 79+538+605, 79+538+609, 79+538+676,
79+538+694, 79+538+698, 79+538+699, 79+538+711, 79+538+754, 79+538+760, 79+538+781 ,
79+538+786, 79+538+797, 79+538+834, 79+538+835, 79+598+602, 79+598+605, 79+598+609,
79+598+676, 79+598+694, 79+598+698, 79+598+699, 79+598+711, 79+598+754, 79+598+760,
79+598+781 , 79+598+786, 79+598+797, 79+598+834, 79+598+835, 79+602+605, 79+602+609,
79+602+676, 79+602+694, 79+602+698, 79+602+699, 79+602+711, 79+602+754, 79+602+760,
79+602+781 , 79+602+786, 79+602+797, 79+602+834, 79+602+835, 79+605+609, 79+605+676,
79+605+694, 79+605+698, 79+605+699, 79+605+711, 79+605+754, 79+605+760, 79+605+781 ,
79+605+786, 79+605+797, 79+605+834, 79+605+835, 79+609+676, 79+609+694, 79+609+698,
79+609+699, 9+609+711 , 79+609+754, 79+609+760, 79+609+781 , 79+609+786, 79+609+797,
79+609+834, 79+609+835, 79+676+694, 79+676+698, 79+676+699, 79+676+71 1 , 79+676+754,
79+676+760, 79+676+781 , 79+676+786, 79+676+797, 79+676+834, 79+676+835, 79+694+698,
79+694+699, 79+694+71 1 , 79+694+754, 79+694+760, 79+694+781 , 79+694+786, 79+694+797,
79+694+834, 79+694+835, 79+698+699, 79+698+711 , 79+698+754, 79+698+760, 79+698+781 ,
79+698+786, 79+698+797, 79+698+834, 79+698+835, 79+699+711 , 79+699+754, 79+699+760,
79+699+781 , 79+699+786, 79+699+797, 79+699+834, 79+699+835, 79+71 1 +754, 79+711 +760,
79+711 +781 , 79+711 +786, 79+71 1 +797, 79+71 1 +834, 79+71 1 +835, 79+754+760, 79+754+781 ,
79+754+786, 79+754+797, 79+754+834, 79+754+835, 79+760+781 , 79+760+786, 79+760+797,
79+760+834, 79+760+835, 79+781 +786, 79+781 +797, 79+781 +834, 79+781 +835, 79+786+797,
79+786+834, 79+786+835, 79+797+834, 79+797+835, 79+834+835, 87+192+302, 87+192+387,
87+192+388, 87+192+390, 87+192+403, 87+192+408, 87+192+410, 87+192+416, 87+192+448,
87+192+451 , 87+192+471 , 87+192+472, 87+192+507, 87+192+512, 87+192+515, 87+192+538,
87+192+598, 87+192+602, 87+192+605, 87+192+609, 87+192+676, 87+192+694, 87+192+698,
87+192+699, 87+192+71 1 , 87+192+754, 87+192+760, 87+192+781 , 87+192+786, 87+192+797,
87+192+834, 87+192+835, 87+302+387, 87+302+388, 87+302+390, 87+302+403, 87+302+408,
87+302+410, 87+302+416, 87+302+448, 87+302+451 , 87+302+471 , 87+302+472, 87+302+507,
87+302+512, 87+302+515, 87+302+538, 87+302+598, 87+302+602, 87+302+605, 87+302+609,
87+302+676, 87+302+694, 87+302+698, 87+302+699, 87+302+711 , 87+302+754, 87+302+760,
87+302+781 , 87+302+786, 87+302+797, 87+302+834, 87+302+835, 87+387+388, 87+387+390,
87+387+403, 87+387+408, 87+387+410, 87+387+416, 87+387+448, 87+387+451 , 87+387+471 ,
87+387+472, 87+387+507, 87+387+512, 87+387+515, 87+387+538, 87+387+598, 87+387+602,
87+387+605, 87+387+609, 87+387+676, 87+387+694, 87+387+698, 87+387+699, 87+387+711 ,
87+387+754, 87+387+760, 87+387+781 , 87+387+786, 87+387+797, 87+387+834, 87+387+835,
87+388+390, 87+388+403, 87+388+408, 87+388+410, 87+388+416, 87+388+448, 87+388+451 ,
87+388+471 , 87+388+472, 87+388+507, 87+388+512, 87+388+515, 87+388+538, 87+388+598,
87+388+602, 87+388+605, 87+388+609, 87+388+676, 87+388+694, 87+388+698, 87+388+699,
87+388+711 , 87+388+754, 87+388+760, 87+388+781 , 87+388+786, 87+388+797, 87+388+834,
87+388+835, 87+390+403, 87+390+408, 87+390+410, 87+390+416, 87+390+448, 87+390+451 ,
87+390+471 , 87+390+472, 87+390+507, 87+390+512, 87+390+515, 87+390+538, 87+390+598,
87+390+602, 87+390+605, 87+390+609, 87+390+676, 87+390+694, 87+390+698, 87+390+699,
87+390+711 , 87+390+754, 87+390+760, 87+390+781 , 87+390+786, 87+390+797, 87+390+834,
87+390+835, 87+403+408, 87+403+410, 87+403+416, 87+403+448, 87+403+451 , 87+403+471 ,
87+403+472, 87+403+507, 87+403+512, 87+403+515, 87+403+538, 87+403+598, 87+403+602,
87+403+605, 87+403+609, 87+403+676, 87+403+694, 87+403+698, 87+403+699, 87+403+711 ,
87+403+754, 87+403+760, 87+403+781 , 87+403+786, 87+403+797, 87+403+834, 87+403+835,
87+408+410, 87+408+416, 87+408+448, 87+408+451 , 87+408+471 , 87+408+472, 87+408+507,
87+408+512, 87+408+515, 87+408+538, 87+408+598, 87+408+602, 87+408+605, 87+408+609,
87+408+676, 87+408+694, 87+408+698, 87+408+699, 87+408+711, 87+408+754, 87+408+760,
87+408+781 , 87+408+786, 87+408+797, 87+408+834, 87+408+835, 87+410+416, 87+410+448,
87+410+451, 87+410+471, 87+410+472, 87+410+507, 87+410+512, 87+410+515, 87+410+538,
87+410+598, 87+410+602, 87+410+605, 87+410+609, 87+410+676, 87+410+694, 87+410+698,
87+410+699, 87+410+711, 87+410+754, 87+410+760, 87+410+781, 87+410+786, 87+410+797,
87+410+834, 87+410+835, 87+416+448, 87+416+451, 87+416+471, 87+416+472, 87+416+507,
87+416+512, 87+416+515, 87+416+538, 87+416+598, 87+416+602, 87+416+605, 87+416+609,
87+416+676, 87+416+694, 87+416+698, 87+416+699, 87+416+711, 87+416+754, 87+416+760,
87+416+781, 87+416+786, 87+416+797, 87+416+834, 87+416+835, 87+448+451, 87+448+471 ,
87+448+472, 87+448+507, 87+448+512, 87+448+515, 87+448+538, 87+448+598, 87+448+602,
87+448+605, 87+448+609, 87+448+676, 87+448+694, 87+448+698, 87+448+699, 87+448+711,
87+448+754, 87+448+760, 87+448+781, 87+448+786, 87+448+797, 87+448+834, 87+448+835,
87+451+471, 87+451+472, 87+451+507, 87+451+512, 87+451+515, 87+451+538, 87+451+598,
87+451+602, 87+451+605, 87+451+609, 87+451+676, 87+451+694, 87+451+698, 87+451+699,
87+451+711, 87+451+754, 87+451+760, 87+451+781, 87+451+786, 87+451+797, 87+451+834,
87+451+835, 87+471+472, 87+471+507, 87+471+512, 87+471+515, 87+471+538, 87+471+598,
87+471+602, 87+471+605, 87+471+609, 87+471+676, 87+471+694, 87+471+698, 87+471+699,
87+471+711, 87+471+754, 87+471+760, 87+471+781, 87+471+786, 87+471+797, 87+471+834,
87+471+835, 87+472+507, 87+472+512, 87+472+515, 87+472+538, 87+472+598, 87+472+602,
87+472+605, 87+472+609, 87+472+676, 87+472+694, 87+472+698, 87+472+699, 87+472+711,
87+472+754, 87+472+760, 87+472+781, 87+472+786, 87+472+797, 87+472+834, 87+472+835,
87+507+512, 87+507+515, 87+507+538, 87+507+598, 87+507+602, 87+507+605, 87+507+609,
87+507+676, 87+507+694, 87+507+698, 87+507+699, 87+507+711, 87+507+754, 87+507+760,
87+507+781 , 87+507+786, 87+507+797, 87+507+834, 87+507+835, 87+512+515, 87+512+538,
87+512+598, 87+512+602, 87+512+605, 87+512+609, 87+512+676, 87+512+694, 87+512+698,
87+512+699, 87+512+711, 87+512+754, 87+512+760, 87+512+781, 87+512+786, 87+512+797,
87+512+834, 87+512+835, 87+515+538, 87+515+598, 87+515+602, 87+515+605, 87+515+609,
87+515+676, 87+515+694, 87+515+698, 87+515+699, 87+515+711, 87+515+754, 87+515+760,
87+515+781, 87+515+786, 87+515+797, 87+515+834, 87+515+835, 87+538+598, 87+538+602,
87+538+605, 87+538+609, 87+538+676, 87+538+694, 87+538+698, 87+538+699, 87+538+711,
87+538+754, 87+538+760, 87+538+781, 87+538+786, 87+538+797, 87+538+834, 87+538+835,
87+598+602, 87+598+605, 87+598+609, 87+598+676, 87+598+694, 87+598+698, 87+598+699,
87+598+711, 87+598+754, 87+598+760, 87+598+781 , 87+598+786, 87+598+797, 87+598+834,
87+598+835, 87+602+605, 87+602+609, 87+602+676, 87+602+694, 87+602+698, 87+602+699,
87+602+711 , 87+602+754, 87+602+760, 87+602+781 , 87+602+786, 87+602+797 87+602+834 87+602+835, 87+605+609, 87+605+676, 87+605+694, 87+605+698, 87+605+699 87+605+711 87+605+754, 87+605+760, 87+605+781 , 87+605+786, 87+605+797, 87+605+834 87+605+835 87+609+676, 87+609+694, 87+609+698, 87+609+699, 87+609+711 , 87+609+754 87+609+760 87+609+781 , 87+609+786, 87+609+797, 87+609+834, 87+609+835, 87+676+694 87+676+698 87+676+699, 87+676+71 1 , 87+676+754, 87+676+760, 87+676+781 , 87+676+786 87+676+797 87+676+834, 87+676+835, 87+694+698, 87+694+699, 87+694+711 , 87+694+754 87+694+760 87+694+781 , 87+694+786, 87+694+797, 87+694+834, 87+694+835, 87+698+699 87+698+711 87+698+754, 87+698+760, 87+698+781 , 87+698+786, 87+698+797, 87+698+834 87+698+835 87+699+711 , 87+699+754, 87+699+760, 87+699+781 , 87+699+786, 87+699+797 87+699+834 87+699+835, 87+711 +754, 87+71 1 +760, 87+71 1 +781 , 87+71 1 +786, 87+71 1 +797 87+711 +834 87+711 +835, 87+754+760, 87+754+781 , 87+754+786, 87+754+797, 87+754+834 87+754+835 87+760+781 , 87+760+786, 87+760+797, 87+760+834, 87+760+835, 87+781 +786 87+781 +797 87+781 +834, 87+781 +835, 87+786+797, 87+786+834, 87+786+835, 87+797+834 87+797+835 87+834+835, 192+302+387, 192+302+388, 192+302+390, 192+302+403, 192+302+408 192+302+410 192+302+416, 192+302+448, 192+302+451 , 192+302+471 192+302+472 192+302+507 192+302+512 192+302+515, 192+302+538, 192+302+598, 192+302+602 192+302+605; 192+302+609 192+302+676 192+302+694, 192+302+698, 192+302+699, 192+302+71 1 192+302+754; 192+302+760 192+302+781 192+302+786, 192+302+797, 192+302+834, 192+302+835 192+387+388 192+387+390 192+387+403 192+387+408, 192+387+410, 192+387+416, 192+387+448 192+387+451 192+387+471 192+387+472 192+387+507, 192+387+512, 192+387+515, 192+387+538 192+387+598 192+387+602 192+387+605 192+387+609, 192+387+676, 192+387+694, 192+387+698 192+387+699 192+387+711 192+387+754 192+387+760, 192+387+781 , 192+387+786, 192+387+797 192+387+834 192+387+835 192+388+390 192+388+403, 192+388+408, 192+388+410, 192+388+416 192+388+448; 192+388+451 192+388+471 192+388+472, 192+388+507, 192+388+512, 192+388+515 192+388+538 192+388+598 192+388+602 192+388+605, 192+388+609, 192+388+676, 192+388+694 192+388+698 192+388+699 192+388+71 1 192+388+754, 192+388+760, 192+388+781 , 192+388+786 192+388+797 192+388+834 192+388+835 192+390+403, 192+390+408, 192+390+410, 192+390+416 192+390+448; 192+390+451 192+390+471 192+390+472, 192+390+507, 192+390+512, 192+390+515 192+390+538; 192+390+598 192+390+602 192+390+605, 192+390+609, 192+390+676, 192+390+694 192+390+698 192+390+699 192+390+71 1 192+390+754, 192+390+760, 192+390+781 , 192+390+786 192+390+797; 192+390+834 192+390+835 192+403+408, 192+403+410, 192+403+416, 192+403+448 192+403+451 192+403+471 192+403+472 192+403+507, 192+403+512, 192+403+515, 192+403+538 192+403+598; 192+403+602 192+403+605 192+403+609, 192+403+676, 192+403+694, 192+403+698 192+403+699; 192+403+711 192+403+754 192+403+760, 192+403+781 , 192+403+786, 192+403+797 192+403+834; 192+403+835 192+408+410 192+408+416, 192+408+448, 192+408+451 , 192+408+471 192+408+472 192+408+507 192+408+512
192+408+515; 192+408+538; 192+408+598, 192+408+602; 192+408+605, 192+408+609; 192+408+676;
192+408+694; 192+408+698; 192+408+699, 192+408+71 1 192+408+754, 192+408+760 ; 192+408+781
192+408+786 192+408+797; 192+408+834, 192+408+835; 192+410+416, 192+410+448; 192+410+451
192+410+471 192+410+472; 192+410+507, 192+410+512 192+410+515, 192+410+538; 192+410+598;
192+410+602; 192+410+605; 192+410+609, 192+410+676; 192+410+694, 192+410+698; 192+410+699 ;
192+410+71 1 192+410+754 , 192+410+760, 192+410+781 192+410+786, 192+410+797 ; 192+410+834 ;
192+410+835; 192+416+448; 192+416+451 , 192+416+471 192+416+472, 192+416+507 ; 192+416+512;
192+416+515; 192+416+538; 192+416+598, 192+416+602; 192+416+605, 192+416+609 ; 192+416+676;
192+416+694; 192+416+698; 192+416+699, 192+416+71 1 192+416+754, 192+416+760 ; 192+416+781
192+416+786; 192+416+797; 192+416+834, 192+416+835; 192+448+451 , 192+448+471 192+448+472;
192+448+507; 192+448+512 192+448+515, 192+448+538; 192+448+598, 192+448+602; 192+448+605 ;
192+448+609; 192+448+676, 192+448+694, 192+448+698; 192+448+699, 192+448+711 192+448+754;
192+448+760; 192+448+781 192+448+786, 192+448+797; 192+448+834, 192+448+835 ; 192+451 +471
192+451 +472; 192+451 +507; 192+451 +512, 192+451 +515; 192+451 +538, 192+451 +598; 192+451 +602;
192+451 +605; 192+451 +609, 192+451 +676, 192+451 +694; 192+451 +698, 192+451 +699; 192+451 +71 1
192+451 +754; 192+451 +760; 192+451 +781 , 192+451 +786; 192+451 +797, 192+451 +834 ; 192+451 +835;
192+471 +472; 192+471 +507; 192+471 +512, 192+471 +515; 192+471 +538, 192+471 +598; 192+471 +602;
192+471 +605; 192+471 +609; 192+471 +676, 192+471 +694; 192+471 +698, 192+471 +699; 192+471 +71 1
192+471 +754; 192+471 +760; 192+471 +781 , 192+471 +786; 192+471 +797, 192+471 +834 ; 192+471 +835;
192+472+507; 192+472+512 192+472+515, 192+472+538 192+472+598, 192+472+602 192+472+605;
192+472+609; 192+472+676; 192+472+694, 192+472+698 192+472+699, 192+472+711 192+472+754;
192+472+760 192+472+781 192+472+786, 192+472+797 192+472+834, 192+472+835 192+507+512
192+507+515; 192+507+538; 192+507+598, 192+507+602 192+507+605, 192+507+609 ; 192+507+676
192+507+694; 192+507+698; 192+507+699, 192+507+71 1 192+507+754, 192+507+760 ; 192+507+781
192+507+786 192+507+797; 192+507+834, 192+507+835; 192+512+515, 192+512+538 192+512+598
192+512+602 192+512+605; 192+512+609, 192+512+676 192+512+694, 192+512+698 192+512+699
192+512+71 1 192+512+754 , 192+512+760, 192+512+781 192+512+786, 192+512+797 192+512+834;
192+512+835 192+515+538; 192+515+598 192+515+602, 192+515+605, 192+515+609 , 192+515+676;
192+515+694; 192+515+698; 192+515+699, 192+515+71 1 192+515+754, 192+515+760; 192+515+781
192+515+786; 192+515+797; 192+515+834, 192+515+835; 192+538+598, 192+538+602 192+538+605;
192+538+609 192+538+676; 192+538+694, 192+538+698 192+538+699, 192+538+711 192+538+754;
192+538+760 192+538+781 192+538+786, 192+538+797 192+538+834, 192+538+835 192+598+602
192+598+605; 192+598+609; 192+598+676, 192+598+694; 192+598+698, 192+598+699 192+598+71 1
192+598+754; 192+598+760; 192+598+781 , 192+598+786 192+598+797, 192+598+834 ; 192+598+835
192+602+605 192+602+609, 192+602+676, 192+602+694; 192+602+698, 192+602+699 192+602+71 1
192+602+754 192+602+760. 192+602+781 , 192+602+786 192+602+797, 192+602+834 192+602+835
192+605+609; 192+605+676 192+605+694; 192+605+698 192+605+699; 192+605+711 192+605+754 ;
192+605+760; 192+605+781 192+605+786 192+605+797; 192+605+834; 192+605+835 ; 192+609+676
192+609+694; 192+609+698 192+609+699; 192+609+71 1 192+609+754; 192+609+760 ; 192+609+781
192+609+786 192+609+797 192+609+834; 192+609+835 192+676+694; 192+676+698 192+676+699
192+676+71 1 192+676+754 192+676+760 192+676+781 192+676+786 192+676+797 192+676+834
192+676+835 192+694+698 192+694+699; 192+694+71 1 192+694+754; 192+694+760 ; 192+694+781
192+694+786 192+694+797; 192+694+834; 192+694+835; 192+698+699 192+698+711 192+698+754 ;
192+698+760 192+698+781 192+698+786 192+698+797 192+698+834 192+698+835 192+699+711
192+699+754; 192+699+760 192+699+781 192+699+786 192+699+797 192+699+834 ; 192+699+835
192+71 1 +754; 192+71 1 +760 192+711 +781 192+71 1 +786 192+711 +797; 192+711 +834; 192+711 +835;
192+754+760; 192+754+781 192+754+786; 192+754+797; 192+754+834; 192+754+835 ; 192+760+781
192+760+786 192+760+797 192+760+834; 192+760+835 192+781 +786 192+781 +797 192+781 +834
192+781 +835 192+786+797 192+786+834 192+786+835 192+797+834; 192+797+835 192+834+835;
302+387+388 302+387+390 302+387+403; 302+387+408 302+387+410; 302+387+416; 302+387+448;
302+387+451 302+387+471 302+387+472 302+387+507 302+387+512 302+387+515; 302+387+538
302+387+598 302+387+602 302+387+605 302+387+609 302+387+676 302+387+694 302+387+698
302+387+699 302+387+71 1 302+387+754; 302+387+760 302+387+781 302+387+786 302+387+797
302+387+834 302+387+835 302+388+390 302+388+403 302+388+408 302+388+410; 302+388+416
302+388+448; 302+388+451 302+388+471 302+388+472 302+388+507 302+388+512 302+388+515
302+388+538 302+388+598 302+388+602 302+388+605 302+388+609 302+388+676 302+388+694
302+388+698 302+388+699 302+388+71 1 302+388+754 302+388+760 302+388+781 302+388+786
302+388+797 302+388+834 302+388+835 302+390+403; 302+390+408; 302+390+410; 302+390+416;
302+390+448; 302+390+451 302+390+471 302+390+472; 302+390+507; 302+390+512; 302+390+515;
302+390+538; 302+390+598 302+390+602 302+390+605; 302+390+609; 302+390+676 302+390+694;
302+390+698 302+390+699 302+390+71 1 302+390+754; 302+390+760; 302+390+781 302+390+786
302+390+797 302+390+834 302+390+835; 302+403+408; 302+403+410; 302+403+416; 302+403+448;
302+403+451 302+403+471 302+403+472; 302+403+507; 302+403+512 302+403+515; 302+403+538;
302+403+598; 302+403+602 302+403+605; 302+403+609; 302+403+676; 302+403+694; 302+403+698;
302+403+699; 302+403+71 1 302+403+754; 302+403+760; 302+403+781 302+403+786; 302+403+797;
302+403+834; 302+403+835 302+408+410; 302+408+416; 302+408+448; 302+408+451 302+408+471
302+408+472; 302+408+507 302+408+512 302+408+515; 302+408+538; 302+408+598; 302+408+602
302+408+605; 302+408+609 302+408+676 302+408+694; 302+408+698 302+408+699; 302+408+71 1
302+408+754; 302+408+760 302+408+781 302+408+786 302+408+797; 302+408+834; 302+408+835;
302+410+416; 302+410+448 302+410+451 302+410+471 302+410+472; 302+410+507 ; 302+410+512
302+410+515; 302+410+538 302+410+598; 302+410+602; 302+410+605; 302+410+609; 302+410+676;
302+410+694; 302+410+698 302+410+699; 302+410+71 1 302+410+754; 302+410+760 ; 302+410+781
302+410+786; 302+410+797; 302+410+834, 302+410+835, 302+416+448, 302+416+451 302+416+471
302+416+472; 302+416+507; 302+416+512, 302+416+515, 302+416+538, 302+416+598, 302+416+602,
302+416+605; 302+416+609; 302+416+676, 302+416+694, 302+416+698, 302+416+699, 302+416+711
302+416+754; 302+416+760; 302+416+781 , 302+416+786 302+416+797, 302+416+834, 302+416+835,
302+448+451 302+448+471 , 302+448+472, 302+448+507, 302+448+512, 302+448+515, 302+448+538,
302+448+598; 302+448+602; 302+448+605, 302+448+609, 302+448+676, 302+448+694, 302+448+698,
302+448+699; 302+448+71 1 ; 302+448+754, 302+448+760, 302+448+781 302+448+786, 302+448+797,
302+448+834; 302+448+835, 302+451 +471 , 302+451 +472, 302+451 +507, 302+451 +512, 302+451 +515,
302+451 +538; 302+451 +598; 302+451 +602, 302+451 +605, 302+451 +609, 302+451 +676, 302+451 +694,
302+451 +698; 302+451 +699; 302+451 +71 1 , 302+451 +754, 302+451 +760, 302+451 +781 302+451 +786,
302+451 +797 302+451 +834; 302+451 +835 302+471 +472, 302+471 +507, 302+471 +512, 302+471 +515,
302+471 +538; 302+471 +598; 302+471 +602, 302+471 +605, 302+471 +609, 302+471 +676, 302+471 +694,
302+471 +698; 302+471 +699; 302+471 +71 1 , 302+471 +754, 302+471 +760, 302+471 +781 302+471 +786
302+471 +797; 302+471 +834 , 302+471 +835, 302+472+507, 302+472+512 302+472+515, 302+472+538
302+472+598 302+472+602; 302+472+605, 302+472+609, 302+472+676 302+472+694, 302+472+698
302+472+699 302+472+71 1 ; 302+472+754, 302+472+760 302+472+781 302+472+786 302+472+797
302+472+834; 302+472+835; 302+507+512, 302+507+515, 302+507+538, 302+507+598, 302+507+602
302+507+605; 302+507+609; 302+507+676, 302+507+694, 302+507+698 302+507+699, 302+507+71 1
302+507+754; 302+507+760, 302+507+781 , 302+507+786 302+507+797 302+507+834, 302+507+835,
302+512+515; 302+512+538, 302+512+598, 302+512+602 302+512+605, 302+512+609, 302+512+676
302+512+694; 302+512+698, 302+512+699, 302+512+71 1 302+512+754, 302+512+760 302+512+781
302+512+786 302+512+797, 302+512+834, 302+512+835 302+515+538, 302+515+598, 302+515+602,
302+515+605; 302+515+609, 302+515+676, 302+515+694, 302+515+698, 302+515+699, 302+515+71 1
302+515+754; 302+515+760, 302+515+781 , 302+515+786 302+515+797, 302+515+834, 302+515+835,
302+538+598 302+538+602, 302+538+605, 302+538+609 302+538+676 302+538+694, 302+538+698
302+538+699 302+538+71 1 , 302+538+754, 302+538+760 302+538+781 302+538+786 302+538+797
302+538+834; 302+538+835, 302+598+602, 302+598+605, 302+598+609 302+598+676 302+598+694,
302+598+698 302+598+699, 302+598+71 1 , 302+598+754, 302+598+760 302+598+781 302+598+786
302+598+797 302+598+834, 302+598+835, 302+602+605 302+602+609 302+602+676 302+602+694,
302+602+698 302+602+699, 302+602+71 1 , 302+602+754, 302+602+760 302+602+781 302+602+786
302+602+797 302+602+834, 302+602+835, 302+605+609, 302+605+676 202+605+694, 302+605+698
302+605+699; 302+605+71 1 , 302+605+754, 302+605+760, 302+605+781 302+605+786 302+605+797
302+605+834; 302+605+835, 302+609+676, 302+609+694, 302+609+698 302+609+699 302+609+71 1
302+609+754; 302+609+760, 302+609+781 , 302+609+786 302+609+797 302+609+834, 302+609+835
302+676+694; 302+676+698, 302+676+699, 302+676+71 1 302+676+754, 302+676+760 302+676+781
302+676+786 302+676+797, 302+676+834, 302+676+835 302+694+698 302+694+699, 302+694+71 1
+694+754 302+694+760 302+694+781 302+694+786 302+694+797 302+694+834 302+694+835+698+699 302+698+71 1 302+698+754 302+698+760 302+698+781 302+698+786 302+698+797+698+834 302+698+835 302+699+71 1 302+699+754 302+699+760 302+699+781 302+699+786+699+797 302+699+834 302+699+835 302+71 1 +754 302+711 +760 302+711 +781 302+711 +786+71 1 +797 302+71 1 +834 302+711 +835 302+754+760 302+754+781 302+754+786 302+754+797+754+834 302+754+835 302+760+781 302+760+786 302+760+797 302+760+834 302+760+835+781 +786 302+781 +797 302+781 +834 302+781 +835 302+786+797 302+786+834 302+786+835+797+834 302+797+835 302+834+835 387+388+390 387+388+403 387+388+408 387+388+410+388+416 387+388+448 387+388+451 387+388+471 387+388+472 387+388+507 387+388+512+388+515 387+388+538 387+388+598 387+388+602 387+388+605 387+388+609 387+388+676+388+694 387+388+698 387+388+699 387+388+71 1 387+388+754 387+388+760 387+388+781+388+786 387+388+797 387+388+834 387+388+835 387+390+403 387+390+408 387+390+410+390+416 387+390+448 387+390+451 387+390+471 387+390+472 387+390+507 387+390+512+390+515 387+390+538 387+390+598 387+390+602 387+390+605 387+390+609 387+390+676+390+694 387+390+698 387+390+699 387+390+711 387+390+754 , 87+390+760 387+390+781+390+786 387+390+797 387+390+834 387+390+835 387+403+408 387+403+410 387+403+416+403+448 387+403+451 387+403+471 387+403+472 387+403+507 387+403+512 387+403+515+403+538 387+403+598 387+403+602 387+403+605 387+403+609 387+403+676 387+403+694+403+698 387+403+699 387+403+71 1 387+403+754 387+403+760 387+403+781 387+403+786+403+797 387+403+834 387+403+835 387+408+410 387+408+416 387+408+448 387+408+451+408+471 387+408+472 387+408+507 387+408+512 387+408+515 387+408+538 387+408+598+408+602 387+408+605 387+408+609 387+408+676 387+408+694 387+408+698 387+408+699+408+71 1 387+408+754 387+408+760 387+408+781 387+408+786 387+408+797 387+408+834+408+835 387+410+416 387+410+448 387+410+451 387+410+471 , 87+410+472 387+410+507+410+512 387+410+515 387+410+538 387+410+598 387+410+602 387+410+605 387+410+609+410+676 387+410+694 387+410+698 387+410+699 387+410+71 1 387+410+754 387+410+760+410+781 387+410+786 387+410+797 387+410+834 387+410+835 387+416+448 387+416+451+416+471 387+416+472 387+416+507 387+416+512 387+416+515 387+416+538 387+416+598+416+602 387+416+605 387+416+609 387+416+676 387+416+694 387+416+698 387+416+699+416+71 1 387+416+754 387+416+760 387+416+781 387+416+786 387+416+797 387+416+834+416+835 387+448+451 387+448+471 387+448+472 387+448+507 387+448+512 387+448+515+448+538 387+448+598 387+448+602 387+448+605 387+448+609 387+448+676 387+448+694+448+698 387+448+699 387+448+71 1 387+448+754 387+448+760 387+448+781 387+448+786+448+797 387+448+834 387+448+835 387+451 +471 387+451 +472 387+451 +507 387+451 +512+451 +515 387+451 +538 387+451 +598 387+451 +602 387+451 +605 387+451 +609 387+451 +676+451 +694 387+451 +698 387+451 +699 387+451 +71 1 387+451 +754 387+451 +760 387+451 +781
387+451 +786; 387+451 +797 387+451 +834; 387+451 +835; 387+471 +472; 387+471 +507 ; 387+471 +512;
387+471 +515; 387+471 +538 387+471 +598; 387+471 +602 387+471 +605; 387+471 +609; 387+471 +676
387+471 +694; 387+471 +698 387+471 +699; 387+471 +71 1 387+471 +754; 387+471 +760; 387+471 +781
387+471 +786 387+471 +797 387+471 +834; 387+471 +835; 387+472+507 387+472+512 387+472+515;
387+472+538 387+472+598 387+472+602 387+472+605 387+472+609 387+472+676 387+472+694;
387+472+698 387+472+699 387+472+71 1 387+472+754; 387+472+760 387+472+781 387+472+786
387+472+797 387+472+834 387+472+835 387+507+512 387+507+515; 387+507+538 387+507+598
387+507+602 387+507+605 387+507+609; 387+507+676 387+507+694; 387+507+698 387+507+699
387+507+71 1 387+507+754 387+507+760 387+507+781 387+507+786 387+507+797 387+507+834;
387+507+835 387+512+515 387+512+538 387+512+598 387+512+602 387+512+605 387+512+609
387+512+676 387+512+694 387+512+698 387+512+699 387+512+71 1 387+512+754 ; 387+512+760
387+512+781 387+512+786 387+512+797 387+512+834 387+512+835 387+515+538; 387+515+598;
387+515+602 387+515+605 387+515+609; 387+515+676 387+515+694; 387+515+698 387+515+699;
387+515+71 1 387+515+754 387+515+760; 387+515+781 387+515+786 387+515+797 ; 387+515+834;
387+515+835; 387+538+598 387+538+602 387+538+605 387+538+609 387+538+676 387+538+694;
387+538+698 387+538+699 387+538+71 1 387+538+754; 387+538+760 387+538+781 387+538+786
387+538+797 387+538+834 387+538+835 387+598+602 387+598+605 387+598+609 387+598+676
387+598+694; 387+598+698 387+598+699 387+598+71 1 387+598+754; 387+598+760 387+598+781
387+598+786 387+598+797 387+598+834 387+598+835 387+602+605 387+602+609 387+602+676
387+602+694 387+602+698 387+602+699 387+602+71 1 387+602+754 387+602+760 387+602+781
387+602+786 387+602+797 387+602+834 387+602+835 387+605+609; 387+605+676 387+605+694;
387+605+698 387+605+699 387+605+71 1 387+605+754; 387+605+760 387+605+781 387+605+786
387+605+797 387+605+834 387+605+835 387+609+676 387+609+694; 387+609+698 387+609+699
387+609+71 1 387+609+754 387+609+760 387+609+781 387+609+786 387+609+797 387+609+834;
387+609+835 387+676+694 387+676+698 387+676+699 387+676+71 1 387+676+754 387+676+760
387+676+781 387+676+786 387+676+797 387+676+834 387+676+835 387+694+698 387+694+699;
387+694+71 1 387+694+754 387+694+760; 387+694+781 387+694+786 387+694+797 387+694+834;
387+694+835; 387+698+699 387+698+71 1 387+698+754 387+698+760 387+698+781 387+698+786
387+698+797 387+698+834 387+698+835 387+699+71 1 387+699+754; 387+699+760 387+699+781
387+699+786 387+699+797 387+699+834 387+699+835 387+711 +754; 387+711 +760 387+711 +781
387+71 1 +786 387+71 1 +797 387+711 +834; 387+71 1 +835 387+754+760; 387+754+781 387+754+786
387+754+797; 387+754+834 387+754+835; 387+760+781 387+760+786 387+760+797 387+760+834
387+760+835 387+781 +786 387+781 +797 387+781 +834 387+781 +835 387+786+797 387+786+834
387+786+835 387+797+834 387+797+835 387+834+835 388+390+403; 388+390+408; 388+390+410;
388+390+416; 388+390+448 388+390+451 388+390+471 388+390+472 388+390+507 388+390+512
388+390+515; 388+390+538 388+390+598 388+390+602 388+390+605 388+390+609 388+390+676
388+390+694; 388+390+698 388+390+699 388+390+71 1 388+390+754; 388+390+760 388+390+781
388+390+786 388+390+797 388+390+834 388+390+835 388+403+408; 388+403+410; 388+403+416;
388+403+448; 388+403+451 388+403+471 388+403+472; 388+403+507; 388+403+512 388+403+515;
388+403+538; 388+403+598; 388+403+602 388+403+605; 388+403+609; 388+403+676 388+403+694;
388+403+698 388+403+699 388+403+71 1 388+403+754; 388+403+760; 388+403+781 388+403+786
388+403+797; 388+403+834 388+403+835; 388+408+410; 388+408+416; 388+408+448 ; 388+408+451
388+408+471 388+408+472 388+408+507; 388+408+512 388+408+515; 388+408+538 388+408+598
388+408+602 388+408+605 388+408+609 388+408+676 388+408+694; 388+408+698 388+408+699
388+408+71 1 388+408+754 388+408+760 388+408+781 388+408+786 388+408+797 388+408+834;
388+408+835 388+410+416 388+410+448; 388+410+451 388+410+471 388+410+472; 388+410+507;
388+410+512 388+410+515 388+410+538; 388+410+598; 388+410+602; 388+410+605 ; 388+410+609;
388+410+676; 388+410+694 388+410+698; 388+410+699; 388+410+71 1 388+410+754 ; 388+410+760;
388+410+781 388+410+786 388+410+797; 388+410+834; 388+410+835; 388+416+448 ; 388+416+451
388+416+471 388+416+472 388+416+507; 388+416+512 388+416+515; 388+416+538; 388+416+598;
388+416+602 388+416+605; 388+416+609; 388+416+676 388+416+694; 388+416+698 388+416+699;
388+416+71 1 388+416+754 388+416+760; 388+416+781 388+416+786 388+416+797 388+416+834;
388+416+835; 388+448+451 388+448+471 388+448+472; 388+448+507; 388+448+512 388+448+515;
388+448+538; 388+448+598; 388+448+602 388+448+605; 388+448+609; 388+448+676 388+448+694;
388+448+698 388+448+699 388+448+71 1 388+448+754; 388+448+760; 388+448+781 388+448+786
388+448+797; 388+448+834 388+448+835; 388+451 +471 388+451 +472; 388+451 +507; 388+451 +512
388+451 +515; 388+451 +538 388+451 +598; 388+451 +602; 388+451 +605; 388+451 +609; 388+451 +676;
388+451 +694; 388+451 +698 388+451 +699; 388+451 +71 1 388+451 +754; 388+451 +760; 388+451 +781
388+451 +786 388+451 +797 388+451 +834; 388+451 +835; 388+471 +472; 388+471 +507; 388+471 +512
388+471 +515; 388+471 +538 388+471 +598; 388+471 +602 388+471 +605; 388+471 +609; 388+471 +676
388+471 +694; 388+471 +698 388+471 +699; 388+471 +71 1 388+471 +754; 388+471 +760; 388+471 +781
388+471 +786 388+471 +797 388+471 +834; 388+471 +835; 388+472+507 388+472+512 388+472+515;
388+472+538 388+472+598 388+472+602 388+472+605 388+472+609 388+472+676 388+472+694
388+472+698 388+472+699 388+472+71 1 388+472+754; 388+472+760 388+472+781 388+472+786
388+472+797 388+472+834 388+472+835 388+507+512 388+507+515; 388+507+538 388+507+598
388+507+602 388+507+605 388+507+609 388+507+676 388+507+694; 388+507+698 388+507+699
388+507+71 1 388+507+754 388+507+760 388+507+781 388+507+786 388+507+797 388+507+834
388+507+835 388+512+515; 388+512+538 388+512+598 388+512+602 388+512+605 388+512+609
388+512+676 388+512+694 388+512+698 388+512+699 388+512+71 1 388+512+754; 388+512+760
388+512+781 388+512+786 388+512+797 388+512+834 388+512+835 388+515+538; 388+515+598;
388+515+602 388+515+605; 388+515+609 388+515+676 388+515+694; 388+515+698 388+515+699;
388+515+71 1 388+515+754 388+515+760 388+515+781 388+515+786 388+515+797 388+515+834;
388+515+835; 388+538+598 388+538+602 388+538+605 388+538+609 388+538+676 388+538+694
388+538+698 388+538+699 388+538+71 1 388+538+754; 388+538+760 388+538+781 388+538+786
388+538+797 388+538+834 388+538+835 388+598+602 388+598+605 388+598+609 388+598+676
388+598+694 388+598+698 388+598+699 388+598+71 1 388+598+754; 388+598+760 388+598+781
388+598+786 388+598+797 388+598+834 388+598+835 388+602+605 388+602+609 388+602+676
388+602+694 388+602+698 388+602+699 388+602+71 1 388+602+754 388+602+760 388+602+781
388+602+786 388+602+797 388+602+834 388+602+835 388+605+609 388+605+676 388+605+694;
388+605+698 388+605+699 388+605+71 1 388+605+754; 388+605+760 388+605+781 388+605+786
388+605+797 388+605+834 388+605+835 388+609+676 388+609+694; 388+609+698 388+609+699
388+609+71 1 388+609+754 388+609+760 388+609+781 388+609+786 388+609+797 388+609+834
388+609+835 388+676+694 388+676+698 388+676+699 388+676+71 1 388+676+754 388+676+760
388+676+781 388+676+786 388+676+797 388+676+834 388+676+835 388+694+698 388+694+699
388+694+71 1 388+694+754; 388+694+760 388+694+781 388+694+786 388+694+797 388+694+834;
388+694+835 388+698+699 388+698+71 1 388+698+754 388+698+760 388+698+781 388+698+786
388+698+797 388+698+834 388+698+835 388+699+71 1 388+699+754; 388+699+760 388+699+781
388+699+786 388+699+797 388+699+834 388+699+835 388+711 +754; 388+711 +760 388+711 +781
388+71 1 +786 388+71 1 +797 388+711 +834; 388+71 1 +835 388+754+760; 388+754+781 388+754+786
388+754+797 388+754+834 388+754+835; 388+760+781 388+760+786 388+760+797 388+760+834
388+760+835 388+781 +786 388+781 +797 388+781 +834 388+781 +835 388+786+797 388+786+834
388+786+835 388+797+834 388+797+835 388+834+835 390+403+408; 390+403+410; 390+403+416;
390+403+448; 390+403+451 390+403+471 390+403+472; 390+403+507; 390+403+512; 390+403+515;
390+403+538; 390+403+598 390+403+602; 390+403+605; 390+403+609; 390+403+676; 390+403+694;
390+403+698; 390+403+699 390+403+71 1 390+403+754; 390+403+760; 390+403+781 390+403+786;
390+403+797; 390+403+834; 390+403+835 390+408+410; 390+408+416; 390+408+448; 390+408+451
390+408+471 390+408+472 390+408+507; 390+408+512 390+408+515; 390+408+538; 390+408+598;
390+408+602; 390+408+605 390+408+609; 390+408+676; 390+408+694; 390+408+698; 390+408+699;
390+408+71 1 390+408+754; 390+408+760 390+408+781 390+408+786; 390+408+797 ; 390+408+834;
390+408+835; 390+410+416 390+410+448 390+410+451 390+410+471 390+410+472; 390+410+507;
390+410+512 390+410+515 390+410+538 390+410+598; 390+410+602; 390+410+605; 390+410+609;
390+410+676; 390+410+694; 390+410+698 390+410+699; 390+410+71 1 390+410+754 ; 390+410+760;
390+410+781 390+410+786 390+410+797 390+410+834; 390+410+835; 390+416+448; 390+416+451
390+416+471 390+416+472 390+416+507 390+416+512 390+416+515; 390+416+538; 390+416+598;
390+416+602; 390+416+605 390+416+609 390+416+676; 390+416+694; 390+416+698; 390+416+699;
390+416+71 1 390+416+754 390+416+760 390+416+781 390+416+786; 390+416+797; 390+416+834;
390+416+835; 390+448+451 390+448+471 390+448+472; 390+448+507; 390+448+512 390+448+515;
390+448+538; 390+448+598 390+448+602 390+448+605; 390+448+609; 390+448+676; 390+448+694;
390+448+698; 390+448+699, 390+448+71 1 390+448+754; 390+448+760, 390+448+781 390+448+786;
390+448+797; 390+448+834, 390+448+835; 390+451 +471 390+451 +472, 390+451 +507 ; 390+451 +512
390+451 +515; 390+451 +538, 390+451 +598; 390+451 +602; 390+451 +605, 390+451 +609; 390+451 +676;
390+451 +694; 390+451 +698, 390+451 +699; 390+451 +71 1 390+451 +754, 390+451 +760; 390+451 +781
390+451 +786; 390+451 +797, 390+451 +834; 390+451 +835; 390+471 +472, 390+471 +507 ; 390+471 +512
390+471 +515; 390+471 +538, 390+471 +598; 390+471 +602; 390+471 +605, 390+471 +609; 390+471 +676;
390+471 +694; 390+471 +698, 390+471 +699; 390+471 +71 1 390+471 +754, 390+471 +760; 390+471 +781
390+471 +786; 390+471 +797, 390+471 +834; 390+471 +835; 390+472+507, 390+472+512 390+472+515;
390+472+538; 390+472+598, 390+472+602; 390+472+605; 390+472+609, 390+472+676; 390+472+694;
390+472+698 390+472+699, 390+472+71 1 390+472+754; 390+472+760, 390+472+781 390+472+786
390+472+797; 390+472+834, 390+472+835 390+507+512 390+507+515, 390+507+538; 390+507+598;
390+507+602; 390+507+605, 390+507+609; 390+507+676; 390+507+694, 390+507+698; 390+507+699;
390+507+71 1 390+507+754, 390+507+760; 390+507+781 390+507+786, 390+507+797; 390+507+834;
390+507+835; 390+512+515, 390+512+538; 390+512+598; 390+512+602, 390+512+605; 390+512+609;
390+512+676; 390+512+694, 390+512+698 390+512+699; 390+512+71 1 , 390+512+754; 390+512+760;
390+512+781 390+512+786, 390+512+797; 390+512+834; 390+512+835, 390+515+538; 390+515+598;
390+515+602; 390+515+605, 390+515+609; 390+515+676; 390+515+694, 390+515+698; 390+515+699;
390+515+71 1 390+515+754 90+515+760, 390+515+781 , 390+515+786, 390+515+797, 390+515+834 ;
390+515+835; 390+538+598, 390+538+602 390+538+605; 390+538+609, 390+538+676 390+538+694;
390+538+698 390+538+699, 390+538+71 1 390+538+754; 390+538+760, 390+538+781 390+538+786
390+538+797; 390+538+834, 390+538+835; 390+598+602 390+598+605, 390+598+609; 390+598+676
390+598+694; 390+598+698, 390+598+699; 390+598+71 1 390+598+754, 390+598+760; 390+598+781
390+598+786 390+598+797, 390+598+834; 390+598+835; 390+602+605, 390+602+609; 390+602+676
390+602+694; 390+602+698, 390+602+699 390+602+71 1 390+602+754, 390+602+760 390+602+781
390+602+786 390+602+797, 390+602+834; 390+602+835 390+605+609, 390+605+676; 390+605+694;
390+605+698; 390+605+699, 390+605+71 1 390+605+754; 390+605+760, 390+605+781 390+605+786;
390+605+797; 390+605+834, 390+605+835; 390+609+676; 390+609+694, 390+609+698; 390+609+699;
390+609+71 1 390+609+754, 390+609+760; 390+609+781 390+609+786, 390+609+797; 390+609+834;
390+609+835; 390+676+694, 390+676+698 390+676+699 390+676+71 1 , 390+676+754; 390+676+760
390+676+781 390+676+786, 390+676+797 390+676+834; 390+676+835, 390+694+698; 390+694+699;
390+694+71 1 390+694+754, 390+694+760; 390+694+781 390+694+786, 390+694+797; 390+694+834;
390+694+835; 390+698+699, 390+698+71 1 390+698+754; 390+698+760, 390+698+781 390+698+786
390+698+797 390+698+834, 390+698+835 390+699+71 1 390+699+754, 390+699+760; 390+699+781
390+699+786 390+699+797, 390+699+834; 390+699+835; 390+71 1 +754, 390+711 +760; 390+711 +781
390+71 1 +786; 390+71 1 +797, 390+711 +834; 390+71 1 +835; 390+754+760, 390+754+781 390+754+786;
390+754+797; 390+754+834, 390+754+835; 390+760+781 390+760+786, 390+760+797; 390+760+834;
390+760+835; 390+781 +786 390+781 +797 390+781 +834; 390+781 +835; 390+786+797 390+786+834; 390+786+835 390+797+834 390+797+835; 390+834+835; 403+408+410; 403+408+416; 403+408+448; 403+408+451 403+408+471 403+408+472; 403+408+507; 403+408+512 403+408+515; 403+408+538; 403+408+598; 403+408+602 403+408+605; 403+408+609; 403+408+676; 403+408+694; 403+408+698; 403+408+699; 403+408+71 1 403+408+754; 403+408+760; 403+408+781 403+408+786; 403+408+797 ; 403+408+834; 403+408+835 403+410+416; 403+410+448; 403+410+451 403+410+471 403+410+472; 403+410+507; 403+410+512 403+410+515; 403+410+538; 403+410+598; 403+410+602; 403+410+605 ; 403+410+609; 403+410+676 403+410+694; 403+410+698; 403+410+699; 403+410+711 403+410+754 ; 403+410+760; 403+410+781 403+410+786; 403+410+797; 403+410+834; 403+410+835 ; 403+416+448; 403+416+451 403+416+471 403+416+472; 403+416+507; 403+416+512 403+416+515; 403+416+538; 403+416+598; 403+416+602 403+416+605; 403+416+609; 403+416+676; 403+416+694 ; 403+416+698; 403+416+699; 403+416+71 1 403+416+754; 403+416+760; 403+416+781 403+416+786; 403+416+797 ; 403+416+834; 403+416+835 403+448+451 403+448+471 403+448+472; 403+448+507 ; 403+448+512 403+448+515; 403+448+538 403+448+598; 403+448+602; 403+448+605; 403+448+609; 403+448+676; 403+448+694; 403+448+698 403+448+699; 403+448+71 1 403+448+754; 403+448+760; 403+448+781 403+448+786; 403+448+797 403+448+834; 403+448+835; 403+451 +471 403+451 +472; 403+451 +507 ; 403+451 +512 403+451 +515 403+451 +538; 403+451 +598; 403+451 +602; 403+451 +605; 403+451 +609; 403+451 +676; 403+451 +694 403+451 +698; 403+451 +699; 403+451 +71 1 403+451 +754 ; 403+451 +760; 403+451 +781 403+451 +786 403+451 +797; 403+451 +834; 403+451 +835; 403+471 +472; 403+471 +507 ; 403+471 +512 403+471 +515; 403+471 +538; 403+471 +598; 403+471 +602; 403+471 +605; 403+471 +609; 403+471 +676; 403+471 +694 403+471 +698; 403+471 +699; 403+471 +71 1 403+471 +754 ; 403+471 +760; 403+471 +781 403+471 +786 403+471 +797; 403+471 +834; 403+471 +835; 403+472+507 ; 403+472+512 403+472+515; 403+472+538 403+472+598; 403+472+602; 403+472+605; 403+472+609; 403+472+676; 403+472+694; 403+472+698 403+472+699; 403+472+71 1 403+472+754; 403+472+760; 403+472+781 403+472+786; 403+472+797 403+472+834; 403+472+835; 403+507+512 403+507+515; 403+507+538; 403+507+598; 403+507+602 403+507+605; 403+507+609; 403+507+676; 403+507+694; 403+507+698; 403+507+699; 403+507+71 1 403+507+754; 403+507+760; 403+507+781 403+507+786; 403+507+797; 403+507+834; 403+507+835 403+512+515; 403+512+538; 403+512+598; 403+512+602; 403+512+605; 403+512+609; 403+512+676 403+512+694; 403+512+698; 403+512+699; 403+512+711 403+512+754; 403+512+760; 403+512+781 403+512+786; 403+512+797; 403+512+834; 403+512+835; 403+515+538; 403+515+598; 403+515+602 403+515+605; 403+515+609; 403+515+676; 403+515+694; 403+515+698; 403+515+699; 403+515+71 1 403+515+754; 403+515+760; 403+515+781 403+515+786; 403+515+797; 403+515+834; 403+515+835 403+538+598; 403+538+602; 403+538+605; 403+538+609; 403+538+676; 403+538+694; 403+538+698; 403+538+699; 403+538+71 1 403+538+754; 403+538+760; 403+538+781 403+538+786; 403+538+797 403+538+834; 403+538+835; 403+598+602; 403+598+605; 403+598+609; 403+598+676; 403+598+694 403+598+698; 403+598+699; 403+598+71 1 403+598+754; 403+598+760;
403+598+781 403+598+786 403+598+797; 403+598+834; 403+598+835; 403+602+605; 403+602+609;
403+602+676; 403+602+694 403+602+698; 403+602+699; 403+602+71 1 403+602+754; 403+602+760;
403+602+781 403+602+786 403+602+797; 403+602+834; 403+602+835; 403+605+609; 403+605+676;
403+605+694; 403+605+698 403+605+699; 403+605+71 1 403+605+754; 403+605+760; 403+605+781
403+605+786; 403+605+797 403+605+834; 403+605+835; 403+609+676; 403+609+694; 403+609+698;
403+609+699; 403+609+71 1 403+609+754 403+609+760; 403+609+781 403+609+786; 403+609+797;
403+609+834; 403+609+835 403+676+694; 403+676+698; 403+676+699; 403+676+711 403+676+754;
403+676+760; 403+676+781 403+676+786 403+676+797; 403+676+834; 403+676+835; 403+694+698;
403+694+699; 403+694+71 1 403+694+754; 403+694+760; 403+694+781 403+694+786; 403+694+797;
403+694+834; 403+694+835 403+698+699; 403+698+71 1 403+698+754; 403+698+760; 403+698+781
403+698+786 403+698+797 403+698+834; 403+698+835; 403+699+71 1 403+699+754; 403+699+760;
403+699+781 403+699+786 403+699+797; 403+699+834; 403+699+835; 403+711 +754; 403+711 +760;
403+71 1 +781 403+71 1 +786 403+711 +797; 403+71 1 +834; 403+711 +835; 403+754+760; 403+754+781
403+754+786; 403+754+797 403+754+834; 403+754+835; 403+760+781 403+760+786; 403+760+797 ;
403+760+834; 403+760+835 403+781 +786; 403+781 +797; 403+781 +834; 403+781 +835; 403+786+797 ;
403+786+834; 403+786+835 403+797+834; 403+797+835; 403+834+835; 408+410+416; 408+410+448;
408+410+451 408+410+471 408+410+472; 408+410+507; 408+410+512 408+410+515; 408+410+538;
408+410+598; 408+410+602 408+410+605; 408+410+609; 408+410+676; 408+410+694 ; 408+410+698;
408+410+699; 408+410+71 1 408+410+754; 408+410+760; 408+410+781 408+410+786; 408+410+797 ;
408+410+834; 408+410+835 408+416+448; 408+416+451 408+416+471 408+416+472; 408+416+507;
408+416+512 408+416+515 408+416+538; 408+416+598; 408+416+602; 408+416+605 ; 408+416+609 ;
408+416+676; 408+416+694 408+416+698; 408+416+699; 408+416+71 1 408+416+754 ; 408+416+760 ;
408+416+781 408+416+786 408+416+797; 408+416+834; 408+416+835; 408+448+451 408+448+471
408+448+472; 408+448+507 408+448+512 408+448+515; 408+448+538; 408+448+598; 408+448+602;
408+448+605; 408+448+609 408+448+676; 408+448+694; 408+448+698; 408+448+699; 408+448+711
408+448+754; 408+448+760 408+448+781 408+448+786; 408+448+797; 408+448+834; 408+448+835;
408+451 +471 408+451 +472 408+451 +507; 408+451 +512 408+451 +515; 408+451 +538; 408+451 +598;
408+451 +602; 408+451 +605 408+451 +609; 408+451 +676; 408+451 +694; 408+451 +698; 408+451 +699;
408+451 +71 1 408+451 +754 408+451 +760; 408+451 +781 408+451 +786; 408+451 +797 ; 408+451 +834;
408+451 +835; 408+471 +472 408+471 +507; 408+471 +512 408+471 +515; 408+471 +538; 408+471 +598;
408+471 +602; 408+471 +605 408+471 +609; 408+471 +676; 408+471 +694; 408+471 +698; 408+471 +699;
408+471 +71 1 408+471 +754 408+471 +760; 408+471 +781 408+471 +786; 408+471 +797 ; 408+471 +834;
408+471 +835; 408+472+507 408+472+512 408+472+515; 408+472+538; 408+472+598; 408+472+602;
408+472+605; 408+472+609 408+472+676; 408+472+694; 408+472+698; 408+472+699; 408+472+71 1
408+472+754; 408+472+760 408+472+781 408+472+786 408+472+797; 408+472+834; 408+472+835;
408+507+512 408+507+515 408+507+538; 408+507+598 408+507+602 408+507+605 408+507+609
408+507+676; 408+507+694 408+507+698; 408+507+699; 408+507+71 1 408+507+754; 408+507+760;
408+507+781 408+507+786 408+507+797; 408+507+834; 408+507+835; 408+512+515; 408+512+538;
408+512+598; 408+512+602 408+512+605; 408+512+609; 408+512+676; 408+512+694 ; 408+512+698;
408+512+699; 408+512+71 1 408+512+754; 408+512+760; 408+512+781 408+512+786 408+512+797;
408+512+834; 408+512+835 408+515+538; 408+515+598; 408+515+602; 408+515+605 ; 408+515+609 ;
408+515+676; 408+515+694 408+515+698; 408+515+699; 408+515+71 1 408+515+754 ; 408+515+760 ;
408+515+781 408+515+786; 408+515+797; 408+515+834; 408+515+835; 408+538+598; 408+538+602;
408+538+605; 408+538+609 408+538+676; 408+538+694; 408+538+698; 408+538+699; 408+538+711
408+538+754; 408+538+760 408+538+781 408+538+786 408+538+797; 408+538+834; 408+538+835;
408+598+602; 408+598+605 408+598+609; 408+598+676; 408+598+694; 408+598+698; 408+598+699;
408+598+71 1 408+598+754 408+598+760; 408+598+781 408+598+786 408+598+797 ; 408+598+834;
408+598+835; 408+602+605 408+602+609; 408+602+676 408+602+694; 408+602+698 408+602+699;
408+602+71 1 408+602+754 408+602+760; 408+602+781 408+602+786 408+602+797 408+602+834;
408+602+835; 408+605+609 408+605+676; 408+605+694; 408+605+698; 408+605+699; 408+605+711
408+605+754; 408+605+760; 408+605+781 408+605+786; 408+605+797; 408+605+834; 408+605+835;
408+609+676; 408+609+694 408+609+698; 408+609+699; 408+609+71 1 408+609+754; 408+609+760;
408+609+781 408+609+786 408+609+797; 408+609+834; 408+609+835; 408+676+694; 408+676+698
408+676+699; 408+676+71 1 408+676+754; 408+676+760; 408+676+781 408+676+786 408+676+797
408+676+834; 408+676+835 408+694+698; 408+694+699; 408+694+71 1 408+694+754; 408+694+760;
408+694+781 408+694+786 408+694+797; 408+694+834; 408+694+835; 408+698+699; 408+698+71 1
408+698+754; 408+698+760 408+698+781 408+698+786 408+698+797 408+698+834; 408+698+835;
408+699+71 1 408+699+754 408+699+760; 408+699+781 408+699+786 408+699+797 ; 408+699+834;
408+699+835; 408+71 1 +754 408+711 +760; 408+71 1 +781 408+711 +786; 408+711 +797 ; 408+711 +834;
408+71 1 +835; 408+754+760 408+754+781 408+754+786; 408+754+797; 408+754+834; 408+754+835;
408+760+781 408+760+786 408+760+797; 408+760+834; 408+760+835; 408+781 +786 408+781 +797;
408+781 +834; 408+781 +835 408+786+797 408+786+834; 408+786+835 408+797+834; 408+797+835;
408+834+835; 410+416+448 410+416+451 410+416+471 410+416+472; 410+416+507 ; 410+416+512;
410+416+515; 410+416+538 410+416+598; 410+416+602; 410+416+605; 410+416+609 ; 410+416+676;
410+416+694; 410+416+698 410+416+699; 410+416+71 1 410+416+754; 410+416+760; 410+416+781
410+416+786; 410+416+797 410+416+834; 410+416+835; 410+448+451 410+448+471 410+448+472;
410+448+507; 410+448+512 410+448+515; 410+448+538; 410+448+598; 410+448+602; 410+448+605;
410+448+609; 410+448+676 410+448+694; 410+448+698; 410+448+699; 410+448+71 1 410+448+754;
410+448+760; 410+448+781 410+448+786; 410+448+797; 410+448+834; 410+448+835; 410+451 +471
410+451 +472; 410+451 +507 410+451 +512; 410+451 +515; 410+451 +538; 410+451 +598; 410+451 +602;
410+451 +605; 410+451 +609; 410+451 +676; 410+451 +694; 410+451 +698; 410+451 +699 ; 410+451 +71 1
410+451 +754 410+451 +760 410+451 +781 410+451 +786 410+451 +797 410+451 +834 ; 410+451 +835;
410+471 +472 410+471 +507 410+471 +512; 410+471 +515; 410+471 +538; 410+471 +598; 410+471 +602;
410+471 +605; 410+471 +609; 410+471 +676; 410+471 +694; 410+471 +698; 410+471 +699 ; 410+471 +71 1
410+471 +754; 410+471 +760 410+471 +781 410+471 +786; 410+471 +797; 410+471 +834 ; 410+471 +835;
410+472+507; 410+472+512 410+472+515; 410+472+538; 410+472+598; 410+472+602; 410+472+605;
410+472+609; 410+472+676 410+472+694; 410+472+698; 410+472+699; 410+472+711 410+472+754
410+472+760; 410+472+781 410+472+786; 410+472+797; 410+472+834; 410+472+835; 410+507+512;
410+507+515; 410+507+538 410+507+598; 410+507+602; 410+507+605; 410+507+609 ; 410+507+676;
410+507+694; 410+507+698 410+507+699; 410+507+71 1 410+507+754; 410+507+760; 410+507+781
410+507+786; 410+507+797 410+507+834; 410+507+835; 410+512+515; 410+512+538; 410+512+598;
410+512+602; 410+512+605; 410+512+609; 410+512+676; 410+512+694; 410+512+698; 410+512+699;
410+512+71 1 410+512+754 410+512+760; 410+512+781 410+512+786; 410+512+797 ; 410+512+834;
410+512+835; 410+515+538 410+515+598; 410+515+602; 410+515+605; 410+515+609 ; 410+515+676;
410+515+694; 410+515+698; 410+515+699; 410+515+71 1 410+515+754; 410+515+760; 410+515+781
410+515+786; 410+515+797 410+515+834; 410+515+835; 410+538+598; 410+538+602; 410+538+605;
410+538+609; 410+538+676 410+538+694; 410+538+698; 410+538+699; 410+538+711 410+538+754;
410+538+760; 410+538+781 410+538+786; 410+538+797; 410+538+834; 410+538+835 ; 410+598+602;
410+598+605; 410+598+609 410+598+676; 410+598+694; 410+598+698; 410+598+699; 410+598+71 1
410+598+754; 410+598+760 410+598+781 410+598+786; 410+598+797; 410+598+834 ; 410+598+835;
410+602+605; 410+602+609 410+602+676; 410+602+694; 410+602+698; 410+602+699; 410+602+71 1
410+602+754; 410+602+760 410+602+781 410+602+786; 410+602+797; 410+602+834 ; 410+602+835;
410+605+609; 410+605+676 410+605+694; 410+605+698; 410+605+699; 410+605+711 410+605+754;
410+605+760; 410+605+781 410+605+786; 410+605+797; 410+605+834; 410+605+835; 410+609+676;
410+609+694; 410+609+698 410+609+699; 410+609+71 1 410+609+754; 410+609+760; 410+609+781
410+609+786; 410+609+797; 410+609+834 410+609+835; 410+676+694; 410+676+698; 410+676+699;
410+676+71 1 410+676+754 410+676+760; 410+676+781 410+676+786; 410+676+797; 410+676+834;
410+676+835; 410+694+698 410+694+699; 410+694+71 1 410+694+754; 410+694+760; 410+694+781
410+694+786; 410+694+797; 410+694+834 410+694+835; 410+698+699; 410+698+711 410+698+754;
410+698+760; 410+698+781 410+698+786 410+698+797; 410+698+834; 410+698+835; 410+699+71 1
410+699+754; 410+699+760 410+699+781 410+699+786; 410+699+797; 410+699+834; 410+699+835;
410+71 1 +754; 410+71 1 +760; 410+71 1 +781 410+71 1 +786; 410+711 +797; 410+711 +834 ; 410+711 +835;
410+754+760; 410+754+781 410+754+786 410+754+797; 410+754+834; 410+754+835; 410+760+781
410+760+786; 410+760+797 410+760+834 410+760+835; 410+781 +786; 410+781 +797; 410+781 +834;
410+781 +835; 410+786+797; 410+786+834 410+786+835; 410+797+834; 410+797+835; 410+834+835;
416+448+451 416+448+471 416+448+472 416+448+507; 416+448+512 416+448+515; 416+448+538;
416+448+598; 416+448+602 416+448+605 416+448+609; 416+448+676; 416+448+694; 416+448+698;
416+448+699 416+448+71 1 416+448+754 416+448+760 416+448+781 416+448+786 416+448+797
416+448+834, 416+448+835, 416+451+471 416+451+472 416+451+507 416+451+512, 416+451+515
41 6+451+538, 416+451+598, 416+451+602 416+451+605 416+451+609 416+451+676, 41 6+451+694
41 6+451+698, 416+451+699, 416+451+711 416+451+754 416+451+760 416+451+781, 41 6+451+786
41 6+451+797, 416+451+834, 416+451+835 416+471+472 416+471+507 416+471+512, 41 6+471+515
41 6+471+538, 416+471+598, 416+471+602 416+471+605 416+471+609 416+471+676, 41 6+471+694
41 6+471+698, 416+471+699, 416+471+711 416+471+754 416+471+760 416+471+781, 41 6+471+786
41 6+471+797, 416+471+834, 416+471+835 416+472+507 416+472+512 416+472+515, 41 6+472+538
41 6+472+598, 416+472+602, 416+472+605 416+472+609 416+472+676 416+472+694, 41 6+472+698
41 6+472+699, 416+472+711, 416+472+754 416+472+760 416+472+781 416+472+786, 41 6+472+797
41 6+472+834, 416+472+835, 416+507+512 416+507+515 416+507+538 416+507+598, 41 6+507+602
41 6+507+605, 416+507+609, 416+507+676 416+507+694 416+507+698 416+507+699, 41 6+507+711
41 6+507+754, 416+507+760, 416+507+781 416+507+786 416+507+797 416+507+834, 41 6+507+835
41 6+512+515, 416+512+538, 416+512+598 416+512+602 416+512+605 416+512+609, 41 6+512+676
41 6+512+694, 416+512+698, 416+512+699 416+512+711 416+512+754 416+512+760, 41 6+512+781
41 6+512+786, 416+512+797, 416+512+834 416+512+835 416+515+538 416+515+598, 41 6+515+602
41 6+515+605, 416+515+609, 416+515+676 416+515+694 416+515+698 416+515+699, 41 6+515+711
41 6+515+754, 416+515+760, 416+515+781 416+515+786 416+515+797 416+515+834, 41 6+515+835
41 6+538+598, 416+538+602, 416+538+605 416+538+609 416+538+676 416+538+694, 41 6+538+698
41 6+538+699, 416+538+711, 416+538+754 416+538+760 416+538+781 416+538+786, 41 6+538+797
41 6+538+834, 416+538+835, 416+598+602 416+598+605 416+598+609 416+598+676, 41 6+598+694
41 6+598+698, 416+598+699, 416+598+711 416+598+754 416+598+760 416+598+781, 41 6+598+786
41 6+598+797, 416+598+834, 416+598+835 416+602+605 416+602+609 416+602+676, 41 6+602+694
41 6+602+698, 416+602+699, 416+602+711 416+602+754 416+602+760 416+602+781, 41 6+602+786
41 6+602+797, 416+602+834, 416+602+835 416+605+609 416+605+676 416+605+694, 41 6+605+698
41 6+605+699, 416+605+711, 416+605+754 416+605+760 416+605+781 416+605+786, 41 6+605+797
41 6+605+834, 416+605+835, 416+609+676 416+609+694 416+609+698 416+609+699, 41 6+609+711
41 6+609+754, 416+609+760, 416+609+781 416+609+786 416+609+797 416+609+834, 41 6+609+835
41 6+676+694, 416+676+698, 416+676+699 416+676+711 416+676+754 416+676+760, 41 6+676+781
41 6+676+786, 416+676+797, 416+676+834 416+676+835 416+694+698 416+694+699, 41 6+694+711
41 6+694+754, 416+694+760, 416+694+781 416+694+786 416+694+797 416+694+834, 41 6+694+835
41 6+698+699, 416+698+711, 416+698+754 416+698+760 416+698+781 416+698+786, 41 6+698+797
41 6+698+834, 416+698+835, 416+699+711 416+699+754 416+699+760 416+699+781, 41 6+699+786
41 6+699+797, 416+699+834, 416+699+835 416+711+754 416+711+760 416+711+781, 41 6+711+786
41 6+711+797, 416+711+834, 416+711+835 416+754+760 416+754+781 416+754+786, 41 6+754+797
41 6+754+834, 416+754+835, 416+760+781 416+760+786 416+760+797 416+760+834, 41 6+760+835
41 6+781+786, 416+781+797, 416+781+834 416+781+835 416+786+797 416+786+834, 41 6+786+835
416+797+834; 416+797+835; 416+834+835, 448+451 +471 448+451 +472; 448+451 +507 448+451 +512;
448+451 +515; 448+451 +538; 448+451 +598, 448+451 +602; 448+451 +605; 448+451 +609 448+451 +676;
448+451 +694; 448+451 +698 448+451 +699, 448+451 +71 1 , 448+451 +754, 448+451 +760 ; 448+451 +781
448+451 +786; 448+451 +797; 448+451 +834, 448+451 +835; 448+471 +472; 448+471 +507 448+471 +512;
448+471 +515; 448+471 +538; 448+471 +598, 448+471 +602; 448+471 +605; 448+471 +609 448+471 +676;
448+471 +694; 448+471 +698; 448+471 +699, 448+471 +71 1 448+471 +754; 448+471 +760 448+471 +781
448+471 +786; 448+471 +797; 448+471 +834, 448+471 +835; 448+472+507; 448+472+512 448+472+515;
448+472+538; 448+472+598; 448+472+602, 448+472+605; 448+472+609; 448+472+676 448+472+694;
448+472+698; 448+472+699; 448+472+71 1 , 448+472+754; 448+472+760; 448+472+781 448+472+786;
448+472+797; 448+472+834; 448+472+835, 448+507+512 448+507+515; 448+507+538 448+507+598;
448+507+602; 448+507+605; 448+507+609, 448+507+676; 448+507+694; 448+507+698 448+507+699;
448+507+71 1 448+507+754; 448+507+760, 448+507+781 448+507+786; 448+507+797 448+507+834;
448+507+835; 448+512+515; 448+512+538, 448+512+598; 448+512+602; 448+512+605 448+512+609 ;
448+512+676; 448+512+694 , 448+512+698, 448+512+699; 448+512+71 1 448+512+754 448+512+760;
448+512+781 448+512+786; 448+512+797, 448+512+834; 448+512+835; 448+515+538 448+515+598;
448+515+602; 448+515+605; 448+515+609, 448+515+676; 448+515+694; 448+515+698 448+515+699 ;
448+515+71 1 448+515+754 ; 448+515+760, 448+515+781 448+515+786; 448+515+797 448+515+834;
448+515+835; 448+538+598; 448+538+602, 448+538+605; 448+538+609; 448+538+676 448+538+694;
448+538+698; 448+538+699; 448+538+71 1 , 448+538+754; 448+538+760; 448+538+781 448+538+786;
448+538+797; 448+538+834; 448+538+835, 448+598+602; 448+598+605; 448+598+609 448+598+676;
448+598+694; 448+598+698; 448+598+699, 448+598+71 1 448+598+754; 448+598+760 448+598+781
448+598+786; 448+598+797; 448+598+834, 448+598+835; 448+602+605; 448+602+609 448+602+676;
448+602+694; 448+602+698; 448+602+699, 448+602+71 1 448+602+754; 448+602+760 448+602+781
448+602+786 448+602+797; 448+602+834, 448+602+835; 448+605+609; 448+605+676 448+605+694;
448+605+698; 448+605+699; 448+605+71 1 , 448+605+754; 448+605+760; 448+605+781 448+605+786;
448+605+797; 448+605+834; 448+605+835, 448+609+676; 448+609+694; 448+609+698 448+609+699;
448+609+71 1 448+609+754; 448+609+760, 448+609+781 448+609+786; 448+609+797 448+609+834;
448+609+835; 448+676+694; 448+676+698, 448+676+699; 448+676+71 1 448+676+754 448+676+760;
448+676+781 448+676+786; 448+676+797, 448+676+834; 448+676+835; 448+694+698 448+694+699;
448+694+71 1 448+694+754; 448+694+760, 448+694+781 448+694+786; 448+694+797 448+694+834;
448+694+835; 448+698+699; 448+698+71 1 , 448+698+754; 448+698+760; 448+698+781 448+698+786
448+698+797; 448+698+834; 448+698+835, 448+699+71 1 448+699+754; 448+699+760 448+699+781
448+699+786; 448+699+797; 448+699+834, 448+699+835; 448+711 +754; 448+711 +760 448+711 +781
448+71 1 +786; 448+71 1 +797; 448+711 +834, 448+71 1 +835; 448+754+760; 448+754+781 448+754+786;
448+754+797; 448+754+834; 448+754+835, 448+760+781 448+760+786; 448+760+797 448+760+834;
448+760+835; 448+781 +786. 448+781 +797, 448+781 +834; 448+781 +835; 448+786+797 448+786+834;
448+786+835; 448+797+834 448+797+835; 448+834+835; 451+471+472; 451 +471 +507 ; 451+471+512;
451+471+515; 451+471+538 451+471+598; 451+471+602; 451+471+605; 451 +471 +609 ; 451+471+676;
451+471+694; 451+471+698 451+471+699; 451+471+711 451+471+754; 451 +471 +760 ; 451+471+781
451+471+786; 451+471+797 451+471+834; 451+471+835; 451+472+507; 451+472+512; 451 +472+515;
451+472+538; 451+472+598 451+472+602; 451+472+605; 451+472+609; 451+472+676; 451 +472+694 ;
451+472+698; 451+472+699 451+472+711 451+472+754; 451+472+760; 451+472+781 451+472+786;
451+472+797; 451+472+834 451+472+835; 451 +507+512 451 +507+515; 451+507+538; 451+507+598;
451+507+602; 451+507+605 451+507+609; 451+507+676; 451+507+694; 451+507+698; 451 +507+699 ;
451+507+711 451+507+754 451+507+760; 451+507+781 451+507+786; 451 +507+797 ; 451+507+834;
451+507+835; 451+512+515 451+512+538; 451+512+598; 451+512+602; 451+512+605; 451+512+609;
451+512+676; 451+512+694 451+512+698; 451+512+699; 451+512+711 451+512+754; 451+512+760;
451+512+781 451+512+786; 451+512+797; 451+512+834; 451+512+835; 451+515+538; 451+515+598;
451+515+602; 451+515+605 451+515+609; 451+515+676; 451+515+694; 451+515+698; 451+515+699;
451+515+711 451+515+754 451+515+760; 451+515+781 451+515+786; 451+515+797 ; 451 +515+834 ;
451+515+835; 451+538+598; 451+538+602; 451+538+605; 451+538+609; 451+538+676; 451+538+694;
451+538+698; 451+538+699 451+538+711 451+538+754; 451+538+760; 451+538+781 451+538+786;
451+538+797; 451+538+834 451+538+835; 451+598+602; 451+598+605; 451 +598+609 ; 451+598+676;
451+598+694; 451+598+698 451+598+699; 451+598+711 451+598+754; 451 +598+760 ; 451+598+781
451+598+786; 451+598+797 451+598+834; 451+598+835; 451+602+605; 451 +602+609 ; 451+602+676;
451+602+694; 451+602+698 451+602+699; 451+602+711 451+602+754; 451+602+760; 451+602+781
451+602+786; 451+602+797 451+602+834; 451+602+835; 451+605+609; 451+605+676; 451+605+694;
451+605+698; 451+605+699 451+605+711 451+605+754; 451+605+760; 451+605+781 451+605+786;
451+605+797; 451+605+834 451+605+835; 451+609+676; 451+609+694; 451+609+698; 451+609+699;
451+609+711 451+609+754 451+609+760; 451+609+781 451+609+786; 451 +609+797 ; 451+609+834;
451+609+835; 451+676+694 451+676+698; 451+676+699; 451+676+711 451 +676+754 ; 451+676+760;
451+676+781 451+676+786 451+676+797; 451+676+834; 451+676+835; 451+694+698; 451+694+699;
451+694+711 451+694+754 451+694+760; 451+694+781 451+694+786; 451 +694+797 ; 451+694+834;
451+694+835; 451+698+699 451+698+711 451+698+754; 451+698+760; 451+698+781 451+698+786;
451+698+797; 451+698+834; 451+698+835; 451+699+711 451+699+754; 451+699+760; 451+699+781
451+699+786; 451+699+797 451+699+834; 451+699+835; 451+711+754; 451+711+760; 451+711+781
451+711+786; 451+711+797 451+711+834; 451+711+835; 451+754+760; 451+754+781 451+754+786;
451+754+797; 451+754+834 451+754+835; 451+760+781 451+760+786; 451 +760+797 ; 451+760+834;
451+760+835; 451+781+786 451+781+797; 451+781+834; 451+781+835; 451 +786+797 ; 451+786+834;
451+786+835; 451+797+834 451+797+835; 451+834+835; 471+472+507; 471+472+512; 471 +472+515;
471+472+538; 471+472+598 471+472+602; 471+472+605; 471+472+609; 471+472+676; 471+472+694;
471+472+698 471+472+699 471+472+711 471+472+754 471+472+760 471+472+781 471+472+786
471+472+797; 471+472+834 471+472+835 471 +507+512 471 +507+515; 471+507+538; 471+507+598;
471+507+602; 471+507+605 471+507+609; 471+507+676; 471+507+694; 471+507+698; 471 +507+699 ;
471+507+711 471+507+754 471+507+760; 471+507+781 471+507+786; 471 +507+797 ; 471 +507+834 ;
471+507+835; 471+512+515 471+512+538; 471+512+598; 471+512+602; 471+512+605; 471+512+609;
471+512+676; 471+512+694 471+512+698; 471+512+699; 471+512+711 471+512+754; 471+512+760;
471+512+781 471+512+786 471+512+797; 471+512+834; 471+512+835; 471+515+538; 471+515+598;
471+515+602; 471+515+605 471+515+609; 471+515+676; 471+515+694; 471+515+698; 471+515+699;
471+515+711 471+515+754 471+515+760; 471+515+781 471+515+786; 471+515+797 ; 471 +515+834 ;
471+515+835; 471+538+598 471+538+602; 471+538+605; 471+538+609; 471+538+676; 471 +538+694 ;
471+538+698; 471+538+699 471+538+711 471+538+754; 471+538+760; 471+538+781 471+538+786;
471+538+797; 471+538+834 471+538+835; 471+598+602; 471+598+605; 471 +598+609 ; 471+598+676;
471+598+694; 471+598+698 471+598+699; 471+598+711 471+598+754; 471 +598+760 ; 471+598+781
471+598+786; 471+598+797 471+598+834; 471+598+835; 471+602+605; 471 +602+609 ; 471+602+676;
471+602+694; 471+602+698 471+602+699; 471+602+711 471+602+754; 471 +602+760 ; 471+602+781
471+602+786 471+602+797 471+602+834; 471+602+835; 471+605+609; 471+605+676; 471+605+694;
471+605+698; 471+605+699 471+605+711 471+605+754; 471+605+760; 471+605+781 471+605+786;
471+605+797; 471+605+834 471+605+835; 471+609+676; 471+609+694; 471+609+698; 471 +609+699 ;
471+609+711 471+609+754 471+609+760; 471+609+781 471+609+786; 471+609+797; 471+609+834;
471+609+835; 471+676+694 471+676+698; 471+676+699; 471+676+711 471 +676+754 ; 471 +676+760 ;
471+676+781 471+676+786 471+676+797; 471+676+834; 471+676+835; 471+694+698; 471 +694+699 ;
471+694+711 471+694+754 471+694+760 471+694+781 471+694+786; 471 +694+797 ; 471+694+834;
471+694+835; 471+698+699 471+698+711 471+698+754; 471+698+760; 471+698+781 471+698+786
471+698+797; 471+698+834 471+698+835; 471+699+711 471+699+754; 471 +699+760 ; 471+699+781
471+699+786; 471+699+797 471+699+834; 471+699+835; 471+711+754; 471+711+760; 471+711+781
471+711+786; 471+711+797 471+711+834; 471+711+835; 471+754+760; 471+754+781 471+754+786;
471+754+797; 471+754+834 471+754+835; 471+760+781 471+760+786; 471 +760+797 ; 471+760+834;
471+760+835; 471+781+786 471+781+797; 471+781+834; 471+781+835; 471 +786+797 ; 471+786+834;
471+786+835; 471+797+834 471+797+835; 471+834+835; 472+507+512 472+507+515; 472+507+538;
472+507+598; 472+507+602 472+507+605; 472+507+609; 472+507+676; 472+507+694; 472+507+698;
472+507+699; 472+507+711 472+507+754; 472+507+760; 472+507+781 472+507+786 472+507+797;
472+507+834; 472+507+835 472+512+515; 472+512+538; 472+512+598; 472+512+602 472+512+605;
472+512+609; 472+512+676 472+512+694; 472+512+698 472+512+699; 472+512+711 472+512+754;
472+512+760; 472+512+781 472+512+786 472+512+797 472+512+834; 472+512+835 ; 472+515+538;
472+515+598; 472+515+602 472+515+605; 472+515+609; 472+515+676; 472+515+694 ; 472+515+698;
472+515+699; 472+515+711 472+515+754; 472+515+760; 472+515+781 472+515+786; 472+515+797;
472+515+834; 472+515+835 472+538+598 472+538+602 472+538+605; 472+538+609; 472+538+676
472+538+694; 472+538+698 472+538+699; 472+538+71 1 472+538+754; 472+538+760 472+538+781
472+538+786 472+538+797 472+538+834; 472+538+835 472+598+602 472+598+605; 472+598+609;
472+598+676 472+598+694 472+598+698 472+598+699; 472+598+71 1 472+598+754; 472+598+760;
472+598+781 472+598+786 472+598+797 472+598+834; 472+598+835 472+602+605; 472+602+609
472+602+676 472+602+694 472+602+698 472+602+699 472+602+71 1 472+602+754; 472+602+760
472+602+781 472+602+786 472+602+797 472+602+834; 472+602+835 472+605+609; 472+605+676;
472+605+694; 472+605+698 472+605+699; 472+605+71 1 472+605+754; 472+605+760; 472+605+781
472+605+786 472+605+797 472+605+834; 472+605+835; 472+609+676 472+609+694; 472+609+698
472+609+699; 472+609+71 1 472+609+754; 472+609+760; 472+609+781 472+609+786 472+609+797 ;
472+609+834; 472+609+835; 472+676+694; 472+676+698 472+676+699 472+676+711 472+676+754;
472+676+760 472+676+781 472+676+786 472+676+797 472+676+834; 472+676+835 472+694+698;
472+694+699; 472+694+71 1 472+694+754; 472+694+760; 472+694+781 472+694+786; 472+694+797;
472+694+834; 472+694+835 472+698+699 472+698+71 1 472+698+754; 472+698+760 472+698+781
472+698+786 472+698+797 472+698+834 472+698+835 472+699+71 1 472+699+754; 472+699+760;
472+699+781 472+699+786 472+699+797 472+699+834; 472+699+835; 472+711 +754; 472+711 +760;
472+71 1 +781 472+71 1 +786 472+711 +797; 472+71 1 +834; 472+711 +835; 472+754+760; 472+754+781
472+754+786; 472+754+797 472+754+834; 472+754+835; 472+760+781 472+760+786 472+760+797
472+760+834; 472+760+835 472+781 +786 472+781 +797 472+781 +834; 472+781 +835 472+786+797
472+786+834 472+786+835 472+797+834; 472+797+835 472+834+835; 507+512+515; 507+512+538;
507+512+598; 507+512+602 507+512+605; 507+512+609; 507+512+676; 507+512+694; 507+512+698;
507+512+699; 507+512+71 1 507+512+754; 507+512+760; 507+512+781 507+512+786 507+512+797;
507+512+834; 507+512+835 507+515+538; 507+515+598; 507+515+602; 507+515+605 ; 507+515+609;
507+515+676; 507+515+694 507+515+698; 507+515+699; 507+515+71 1 507+515+754 ; 507+515+760;
507+515+781 507+515+786 507+515+797; 507+515+834; 507+515+835; 507+538+598; 507+538+602
507+538+605; 507+538+609 507+538+676 507+538+694; 507+538+698 507+538+699; 507+538+71 1
507+538+754; 507+538+760 507+538+781 507+538+786 507+538+797; 507+538+834; 507+538+835;
507+598+602 507+598+605 507+598+609; 507+598+676 507+598+694; 507+598+698 507+598+699;
507+598+71 1 507+598+754 507+598+760; 507+598+781 507+598+786 507+598+797 ; 507+598+834;
507+598+835; 507+602+605 507+602+609; 507+602+676 507+602+694; 507+602+698 507+602+699;
507+602+71 1 507+602+754; 507+602+760 507+602+781 507+602+786 507+602+797 507+602+834;
507+602+835 507+605+609 507+605+676; 507+605+694; 507+605+698; 507+605+699; 507+605+71 1
507+605+754; 507+605+760 507+605+781 507+605+786; 507+605+797; 507+605+834; 507+605+835;
507+609+676; 507+609+694; 507+609+698 507+609+699; 507+609+71 1 507+609+754; 507+609+760;
507+609+781 507+609+786 507+609+797; 507+609+834; 507+609+835; 507+676+694; 507+676+698
507+676+699; 507+676+71 1 507+676+754; 507+676+760 507+676+781 507+676+786 507+676+797
507+676+834; 507+676+835 507+694+698 507+694+699 507+694+71 1 507+694+754 507+694+760
507+694+781 507+694+786 507+694+797; 507+694+834; 507+694+835; 507+698+699 507+698+711
507+698+754; 507+698+760 507+698+781 507+698+786 507+698+797 507+698+834; 507+698+835
507+699+71 1 507+699+754 507+699+760; 507+699+781 507+699+786 507+699+797 ; 507+699+834;
507+699+835; 507+71 1 +754 507+711 +760; 507+71 1 +781 507+711 +786; 507+711 +797 ; 507+711 +834 ;
507+71 1 +835; 507+754+760; 507+754+781 507+754+786; 507+754+797; 507+754+834; 507+754+835;
507+760+781 507+760+786 507+760+797; 507+760+834; 507+760+835; 507+781 +786 507+781 +797
507+781 +834; 507+781 +835 507+786+797 507+786+834; 507+786+835 507+797+834; 507+797+835;
507+834+835; 512+515+538; 512+515+598; 512+515+602; 512+515+605; 512+515+609; 512+515+676;
512+515+694; 512+515+698 512+515+699; 512+515+71 1 512+515+754; 512+515+760 ; 512+515+781
512+515+786; 512+515+797 512+515+834; 512+515+835; 512+538+598 512+538+602 512+538+605;
512+538+609; 512+538+676 512+538+694; 512+538+698 512+538+699; 512+538+711 512+538+754;
512+538+760 512+538+781 512+538+786 512+538+797 512+538+834; 512+538+835 512+598+602
512+598+605; 512+598+609 512+598+676 512+598+694; 512+598+698 512+598+699 ; 512+598+71 1
512+598+754; 512+598+760 512+598+781 512+598+786 512+598+797 512+598+834 ; 512+598+835
512+602+605; 512+602+609 512+602+676 512+602+694; 512+602+698 512+602+699 512+602+71 1
512+602+754; 512+602+760 512+602+781 512+602+786 512+602+797 512+602+834 ; 512+602+835
512+605+609; 512+605+676 512+605+694; 512+605+698; 512+605+699; 512+605+711 512+605+754;
512+605+760; 512+605+781 512+605+786 512+605+797; 512+605+834; 512+605+835 ; 512+609+676
512+609+694; 512+609+698 512+609+699; 512+609+71 1 512+609+754; 512+609+760; 512+609+781
512+609+786 512+609+797 512+609+834; 512+609+835; 512+676+694; 512+676+698 512+676+699
512+676+71 1 512+676+754 512+676+760 512+676+781 512+676+786 512+676+797 512+676+834;
512+676+835 512+694+698 512+694+699 512+694+71 1 512+694+754; 512+694+760; 512+694+781
512+694+786; 512+694+797 512+694+834; 512+694+835; 512+698+699 512+698+71 1 512+698+754;
512+698+760 512+698+781 512+698+786 512+698+797 512+698+834 512+698+835 512+699+71 1
512+699+754; 512+699+760; 512+699+781 512+699+786 512+699+797 512+699+834 ; 512+699+835;
512+71 1 +754; 512+71 1 +760 512+711 +781 512+71 1 +786 512+71 1 +797; 512+711 +834; 512+711 +835;
512+754+760; 512+754+781 512+754+786 512+754+797; 512+754+834; 512+754+835; 512+760+781
512+760+786 512+760+797 512+760+834 512+760+835 512+781 +786 512+781 +797 512+781 +834;
512+781 +835 512+786+797 512+786+834 512+786+835 512+797+834; 512+797+835 512+834+835;
515+538+598; 515+538+602 515+538+605 515+538+609; 515+538+676; 515+538+694; 515+538+698;
515+538+699; 515+538+71 1 515+538+754 515+538+760; 515+538+781 515+538+786; 515+538+797;
515+538+834; 515+538+835 515+598+602 515+598+605; 515+598+609; 515+598+676; 515+598+694;
515+598+698; 515+598+699 515+598+71 1 515+598+754; 515+598+760; 515+598+781 515+598+786;
515+598+797; 515+598+834 515+598+835 515+602+605; 515+602+609; 515+602+676; 515+602+694;
515+602+698; 515+602+699 515+602+71 1 515+602+754; 515+602+760; 515+602+781 515+602+786
515+602+797 515+602+834 515+602+835 515+605+609 515+605+676 515+605+694 515+605+698
515+605+699; 515+605+71 1 515+605+754 515+605+760; 515+605+781 515+605+786; 515+605+797;
515+605+834; 515+605+835 515+609+676; 515+609+694; 515+609+698; 515+609+699; 515+609+711
515+609+754; 515+609+760; 515+609+781 515+609+786; 515+609+797; 515+609+834 ; 515+609+835;
515+676+694; 515+676+698 515+676+699; 515+676+71 1 515+676+754; 515+676+760; 515+676+781
515+676+786 515+676+797 515+676+834; 515+676+835; 515+694+698; 515+694+699; 515+694+71 1
515+694+754; 515+694+760 515+694+781 515+694+786; 515+694+797; 515+694+834 ; 515+694+835;
515+698+699; 515+698+71 1 515+698+754; 515+698+760; 515+698+781 515+698+786 515+698+797;
515+698+834; 515+698+835 515+699+71 1 515+699+754; 515+699+760; 515+699+781 515+699+786;
515+699+797; 515+699+834 515+699+835; 515+71 1 +754; 515+711 +760; 515+711 +781 515+711 +786;
515+71 1 +797; 515+71 1 +834 515+711 +835; 515+754+760; 515+754+781 515+754+786; 515+754+797;
515+754+834; 515+754+835 515+760+781 515+760+786; 515+760+797; 515+760+834 ; 515+760+835;
515+781 +786 515+781 +797 515+781 +834; 515+781 +835; 515+786+797; 515+786+834 ; 515+786+835;
515+797+834; 515+797+835 515+834+835; 538+598+602 538+598+605; 538+598+609; 538+598+676
538+598+694; 538+598+698 538+598+699 538+598+71 1 538+598+754; 538+598+760 538+598+781
538+598+786 538+598+797 538+598+834; 538+598+835 538+602+605 538+602+609 538+602+676
538+602+694; 538+602+698 538+602+699 538+602+71 1 538+602+754; 538+602+760 538+602+781
538+602+786 538+602+797 538+602+834 538+602+835 538+605+609; 538+605+676 538+605+694;
538+605+698 538+605+699 538+605+71 1 538+605+754; 538+605+760; 538+605+781 538+605+786
538+605+797; 538+605+834 538+605+835 538+609+676 538+609+694; 538+609+698 538+609+699;
538+609+71 1 538+609+754; 538+609+760 538+609+781 538+609+786 538+609+797 538+609+834;
538+609+835; 538+676+694 538+676+698 538+676+699 538+676+71 1 538+676+754; 538+676+760
538+676+781 538+676+786 538+676+797 538+676+834 538+676+835 538+694+698; 538+694+699;
538+694+71 1 538+694+754 538+694+760 538+694+781 538+694+786 538+694+797; 538+694+834;
538+694+835; 538+698+699 538+698+71 1 538+698+754; 538+698+760 538+698+781 538+698+786
538+698+797 538+698+834 538+698+835 538+699+71 1 538+699+754; 538+699+760 538+699+781
538+699+786 538+699+797 538+699+834 538+699+835 538+71 1 +754; 538+711 +760; 538+711 +781
538+71 1 +786 538+71 1 +797 538+711 +834 538+71 1 +835; 538+754+760; 538+754+781 538+754+786;
538+754+797; 538+754+834; 538+754+835 538+760+781 538+760+786 538+760+797 538+760+834;
538+760+835 538+781 +786 538+781 +797 538+781 +834; 538+781 +835 538+786+797 538+786+834
538+786+835 538+797+834 538+797+835 538+834+835; 598+602+605 598+602+609 598+602+676
598+602+694; 598+602+698 598+602+699 598+602+71 1 598+602+754; 598+602+760 598+602+781
598+602+786 598+602+797 598+602+834 598+602+835 598+605+609; 598+605+676 598+605+694;
598+605+698 598+605+699 598+605+71 1 598+605+754; 598+605+760; 598+605+781 598+605+786
598+605+797; 598+605+834; 598+605+835 598+609+676 598+609+694; 598+609+698 598+609+699;
598+609+71 1 598+609+754 598+609+760 598+609+781 598+609+786 598+609+797 598+609+834;
598+609+835 598+676+694 598+676+698 598+676+699 598+676+71 1 598+676+754 598+676+760
598+676+781 598+676+786 598+676+797 598+676+834 598+676+835 598+694+698; 598+694+699;
598+694+71 1 598+694+754 598+694+760; 598+694+781 598+694+786 598+694+797 ; 598+694+834;
598+694+835; 598+698+699 598+698+71 1 598+698+754; 598+698+760 598+698+781 598+698+786
598+698+797 598+698+834 598+698+835 598+699+71 1 598+699+754; 598+699+760 598+699+781
598+699+786 598+699+797 598+699+834; 598+699+835 598+711 +754; 598+711 +760; 598+711 +781
598+71 1 +786 598+71 1 +797 598+711 +834; 598+71 1 +835; 598+754+760; 598+754+781 598+754+786;
598+754+797; 598+754+834 598+754+835; 598+760+781 598+760+786 598+760+797 598+760+834;
598+760+835 598+781 +786 598+781 +797 598+781 +834; 598+781 +835 598+786+797 598+786+834
598+786+835 598+797+834; 598+797+835 598+834+835; 602+605+609; 602+605+676 602+605+694;
602+605+698 602+605+699 602+605+71 1 602+605+754; 602+605+760 602+605+781 602+605+786
602+605+797 602+605+834 602+605+835 602+609+676 602+609+694; 602+609+698 602+609+699
602+609+71 1 602+609+754 602+609+760 602+609+781 602+609+786 602+609+797 602+609+834;
602+609+835 602+676+694 602+676+698 602+676+699 602+676+71 1 602+676+754; 602+676+760
602+676+781 602+676+786 602+676+797 602+676+834 602+676+835 602+694+698 602+694+699;
602+694+71 1 602+694+754; 602+694+760; 602+694+781 602+694+786 602+694+797 ; 602+694+834;
602+694+835; 602+698+699 602+698+71 1 602+698+754; 602+698+760 602+698+781 602+698+786
602+698+797 602+698+834 602+698+835 602+699+71 1 602+699+754; 602+699+760 602+699+781
602+699+786 602+699+797 602+699+834 602+699+835 602+711 +754; 602+711 +760; 602+711 +781
602+71 1 +786 602+71 1 +797 602+711 +834; 602+71 1 +835 602+754+760; 602+754+781 602+754+786
602+754+797; 602+754+834 602+754+835; 602+760+781 602+760+786 602+760+797 602+760+834
602+760+835 602+781 +786 602+781 +797 602+781 +834 602+781 +835 602+786+797 602+786+834
602+786+835 602+797+834 602+797+835 602+834+835 605+609+676; 605+609+694; 605+609+698;
605+609+699; 605+609+71 1 605+609+754 605+609+760; 605+609+781 605+609+786 605+609+797;
605+609+834; 605+609+835 605+676+694; 605+676+698 605+676+699; 605+676+71 1 605+676+754;
605+676+760 605+676+781 605+676+786 605+676+797 605+676+834; 605+676+835 605+694+698;
605+694+699; 605+694+71 1 605+694+754 605+694+760; 605+694+781 605+694+786; 605+694+797;
605+694+834; 605+694+835 605+698+699 605+698+71 1 605+698+754; 605+698+760 605+698+781
605+698+786 605+698+797 605+698+834 605+698+835 605+699+71 1 605+699+754; 605+699+760;
605+699+781 605+699+786 605+699+797 605+699+834; 605+699+835; 605+711 +754; 605+711 +760;
605+71 1 +781 605+71 1 +786 605+711 +797 605+71 1 +834; 605+71 1 +835; 605+754+760; 605+754+781
605+754+786; 605+754+797 605+754+834 605+754+835; 605+760+781 605+760+786 605+760+797;
605+760+834; 605+760+835 605+781 +786 605+781 +797 605+781 +834; 605+781 +835; 605+786+797
605+786+834; 605+786+835 605+797+834 605+797+835; 605+834+835; 609+676+694; 609+676+698
609+676+699 609+676+71 1 609+676+754 609+676+760 609+676+781 609+676+786 609+676+797
609+676+834; 609+676+835 609+694+698 609+694+699; 609+694+71 1 609+694+754; 609+694+760;
609+694+781 609+694+786 609+694+797 609+694+834; 609+694+835; 609+698+699 609+698+71 1
609+698+754 609+698+760 609+698+781 609+698+786 609+698+797 609+698+834 609+698+835 609+699+711 609+699+754 609+699+760 609+699+781 609+699+786 609+699+797, 609+699+834 609+699+835 609+711+754 609+711+760 609+711+781 609+711+786 609+711+797, 609+711+834 609+711+835 609+754+760 609+754+781 609+754+786 609+754+797 609+754+834, 609+754+835 609+760+781 609+760+786 609+760+797 609+760+834 609+760+835 609+781+786, 609+781+797 609+781+834 609+781+835 609+786+797 609+786+834 609+786+835 609+797+834, 609+797+835 609+834+835 676+694+698 676+694+699 676+694+711 676+694+754 676+694+760, 676+694+781 676+694+786 676+694+797 676+694+834 676+694+835 676+698+699 676+698+711, 676+698+754 676+698+760 676+698+781 676+698+786 676+698+797 676+698+834 676+698+835, 676+699+711 676+699+754 676+699+760 676+699+781 676+699+786 676+699+797 676+699+834, 676+699+835 676+711+754 676+711+760 676+711+781 676+711+786 676+711+797 676+711+834, 676+711+835 676+754+760 676+754+781 676+754+786 676+754+797 676+754+834 676+754+835, 676+760+781 676+760+786 676+760+797 676+760+834 676+760+835 676+781+786 676+781+797, 676+781+834 676+781+835 676+786+797 676+786+834 676+786+835 676+797+834 676+797+835, 676+834+835 694+698+699 694+698+711 694+698+754 694+698+760 694+698+781 694+698+786, 694+698+797 694+698+834 694+698+835 694+699+711 694+699+754 694+699+760 694+699+781 , 694+699+786 694+699+797 694+699+834 694+699+835 694+711+754 694+711+760 694+711+781, 694+711+786 694+711+797 694+711+834 694+711+835 694+754+760 694+754+781 694+754+786, 694+754+797 694+754+834 694+754+835 694+760+781 694+760+786 694+760+797 694+760+834, 694+760+835 694+781+786 694+781+797 694+781+834 694+781+835 694+786+797 694+786+834, 694+786+835 694+797+834 694+797+835 694+834+835 698+699+711 698+699+754 698+699+760, 698+699+781 698+699+786 698+699+797 698+699+834 698+699+835 968+711+754 698+711+760, 698+711+781 698+711+786 698+711+797 698+711+834 698+711+835 698+754+760 698+754+781 , 698+754+786 698+754+797 698+754+834 698+754+835 698+760+781 698+760+786 698+760+797, 698+760+834 698+760+835 698+781+786 698+781+797 698+781+834 698+781+835 698+786+797, 698+786+834 698+786+835 698+797+834 698+797+835 698+834+835 699+711+754 699+711+760, 699+711+781 699+711+786 699+711+797 699+711+834 699+711+835 699+754+760 699+754+781 , 699+754+786 699+754+797 699+754+834 699+754+835 699+760+781 699+760+786 699+760+797, 699+760+834 699+760+835 699+781+786 699+781+797 699+781+834 699+781+835 699+786+797, 699+786+834 699+786+835 699+797+834 699+797+835 699+834+835 711+754+760 711+754+781, 711+754+786 711+754+797 711+754+834 711+754+835 711+760+781 711+760+786 711+760+797, 711+760+834 711+760+835 711+781+786 711+781+797 711+781+834 711+781+835 711+786+797, 711+786+834 711+786+835 711+797+834 711+797+835 711+834+835 754+760+781 754+760+786, 754+760+797 754+760+834 754+760+835 754+781+786 754+781+797 754+781+834 754+781+835, 754+786+797 754+786+834 754+786+835 754+797+834 754+797+835 754+834+835 , 760+781+786 , 60+781+797 760+781+834 760+781+835 760+786+797, 760+786+834, 760+786+835, 760+797+834, 760+797+835
760+834+835, 781 +786+797, 781 +786+834, 781 +786+835, 781 +797+834, 781 +797+835, 781 +834+835, 786+797+834, 786+797+835, 786+834+835, and 797+834+835 of SEQ ID NO: 2.
In another aspect, a variant comprises an alteration at each position (or at least four positions) corresponding to positions 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 31 1 , 313, 387, 388, 390, 403, 408, 410, 416, 448, 451 , 471 , 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 71 1 , 719, 754, 756, 760, 781 , 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 17. In another aspect, the amino acid at a position corresponding to position 17 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S17A of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 20. In another aspect, the amino acid at a position corresponding to position 20 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution F20P, F20N, F20G, or F20Y, preferably F20P, of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 51. In another aspect, the amino acid at a position corresponding to position 51 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution K51 Q or K51 H, preferably K51 Q, of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 53. In another aspect, the amino acid at a position corresponding to position 53 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution E53P or E53Q, preferably E53P, of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 55. In another aspect, the amino acid at a position corresponding to position 55 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution Y55M of the mature polypeptide of SEQ ID NO:
2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 56. In another aspect, the amino acid at a position corresponding to position 56 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution V56M of the mature polypeptide of SEQ ID NO:
2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 60. In another aspect, the amino acid at a position corresponding to position 60 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution Y60F of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 63. In another aspect, the amino acid at a position corresponding to position 63 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S63F of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 79. In another aspect, the amino acid at a position corresponding to position 79 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S79W of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 87. In another aspect, the amino acid at a position corresponding to position 87 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution T87R of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 186. In another aspect, the amino acid at a position corresponding to position 186 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution A186P of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 192. In another aspect, the amino acid at a position corresponding to position 192 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution K192N of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 302. In another aspect, the amino acid at a position corresponding to position 302 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution I302D, I302V, I302M or I302H, preferably I302H, of the mature polypeptide of SEQ ID NO: 2.
ln another aspect, the variant comprises or consists of an alteration at a position corresponding to position 31 1. In another aspect, the amino acid at a position corresponding to position 31 1 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution H31 1 N, of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 313. In another aspect, the amino acid at a position corresponding to position 302 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S313D of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 387. In another aspect, the amino acid at a position corresponding to position 387 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution I387T of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 388. In another aspect, the amino acid at a position corresponding to position 388 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution K388R of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 390. In another aspect, the amino acid at a position corresponding to position 390 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution K390Q of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 403. In another aspect, the amino acid at a position corresponding to position 403 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution I403Y of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 408. In another aspect, the amino acid at a position corresponding to position 408 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the alteration E408D, E408S, E408P, E408A, E408G, E408N, preferably E408D, of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to
position 410. In another aspect, the amino acid at a position corresponding to position 410 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution P410G of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 416. In another aspect, the amino acid at a position corresponding to position 416 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution Q416S or Q416D, preferably Q416S, of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 448. In another aspect, the amino acid at a position corresponding to position 448 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution A448E, A448S or A448W, preferably A448E, of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 451. In another aspect, the amino acid at a position corresponding to position 451 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution K451 Q and K451 S, preferably K451 Q of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 471. In another aspect, the amino acid at a position corresponding to position 471 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution G471 S of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 472. In another aspect, the amino acid at a position corresponding to position 472 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S472Y of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 476. In another aspect, the amino acid at a position corresponding to position 476 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution D476R of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 489. In another aspect, the amino acid at a position corresponding to position 489 is substituted
with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution Q489P of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 507. In another aspect, the amino acid at a position corresponding to position 507 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution K507R of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 512. In another aspect, the amino acid at a position corresponding to position 512 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution K512P of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 515. In another aspect, the amino acid at a position corresponding to position 515 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S515V of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 538. In another aspect, the amino acid at a position corresponding to position 538 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S538C of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 598. In another aspect, the amino acid at a position corresponding to position 598 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S598Q of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 599. In another aspect, the amino acid at a position corresponding to position 599 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution A599S of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 602. In another aspect, the amino acid at a position corresponding to position 602 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In
another aspect, the variant comprises or consists of the substitution I602T or I602D, preferably I602T, of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 603. In another aspect, the amino acid at a position corresponding to position 603 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution V603P of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 605. In another aspect, the amino acid at a position corresponding to position 605 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S605T of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 609. In another aspect, the amino acid at a position corresponding to position 609 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution G609E of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 676. In another aspect, the amino acid at a position corresponding to position 676 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution D676H of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 688. In another aspect, the amino acid at a position corresponding to position 688 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution A688G of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 690. In another aspect, the amino acid at a position corresponding to position 690 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution Y690F of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 694. In another aspect, the amino acid at a position corresponding to position 694 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution T694A of the mature polypeptide of
SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 697. In another aspect, the amino acid at a position corresponding to position 697 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution T697G of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 698. In another aspect, the amino acid at a position corresponding to position 698 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution R698W of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 699. In another aspect, the amino acid at a position corresponding to position 699 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution T699A of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 71 1. In another aspect, the amino acid at a position corresponding to position 71 1 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution T71 1V or T71 1 T, preferably T71 1V, of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 719. In another aspect, the amino acid at a position corresponding to position 719 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution W719R of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 754. In another aspect, the amino acid at a position corresponding to position 754 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution K754R of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 756. In another aspect, the amino acid at a position corresponding to position 756 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution V756H or V756Y of the mature polypeptide of SEQ ID NO: 2.
ln another aspect, the variant comprises or consists of an alteration at a position corresponding to position 760. In another aspect, the amino acid at a position corresponding to position 760 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S760G of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 781. In another aspect, the amino acid at a position corresponding to position 781 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution T781 M of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 786. In another aspect, the amino acid at a position corresponding to position 786 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution N786K of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 797. In another aspect, the amino acid at a position corresponding to position 797 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution T797S of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 824. In another aspect, the amino acid at a position corresponding to position 824 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution A824D of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 833. In another aspect, the amino acid at a position corresponding to position 833 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution N833D of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 834. In another aspect, the amino acid at a position corresponding to position 834 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution Q834E of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to
position 835. In another aspect, the amino acid at a position corresponding to position 835 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution S835D of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position 1048. In another aspect, the amino acid at a position corresponding to position 1048 is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution F1048W of the mature polypeptide of SEQ ID NO: 2.
In one embodiment, the endoglucanase variant comprised in the detergent compositions of the invention may further comprise an alteration at one or more positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651 , 824, 848, 869, 880, 881 , 883, 887, 892, 905, 906, 912, 921 , 928, 934, 937, 948, 956, 999 and 1037, or corresponding to those positions.
More specifically the endoglucanase variant comprised in the detergent compositions of the invention may further comprise one or more alterations at one or more positions selected from the group consisting of: N216D, N216Q, A283D, A346D, A559P, A559N, A564E, Y579W, S636K, S636N, F638N, A651 P, A824D, N848D, A869V, R880K, V881T, V881 Q, T883R, T887K, T892P, N905D, F906A, A912V, K921 R, S928D, Y934G, A937E, K948R, Q956Y, T999R, and A1037E, or corresponding to those positions.
In another aspect, the variant comprises or consists of an alteration at a position corresponding to position selected from the group consisting of alterations in positions: 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 31 1 , 313, 387, 388, 390, 403, 408, 410, 416, 448, 451 , 471 , 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 71 1 , 719, 754, 756, 760, 781 , 786, 797, 834, 833, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
In another aspect, the amino acid at a position corresponding to any of positions as described above is substituted with Ala, Arg, Asn, Asp, Cys, Gin, Glu, Gly, His, lie, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, or Val. In another aspect, the variant comprises or consists of the substitution selected from the group consisting of: S17A, F20P, F20N, F20G, F20Y, K51 Q, K51 H, E53P, E53G, Y55M, V56M, Y60F, S63F, T87R, A186P, K192N, I302D, I302H, I302V, I302M, H31 1 N, S313D, I387T, K388R, K390Q, I403Y, E408D, E408S, E408P, E408A, E408G, E408N, P410G, Q416S, Q416D, A448E, A448W, A448S, K451 S, G471 S, S472Y, D476R, Q489P, K507R, K512P, S515V, S538C, Y579W, S598Q, A599S, I602T, I602D, V603P, S605T, G609E, D676H, A688G, Y690F, T694A, T697G, R698W, T699A, T711V, T71 1Y, W719R, K754R, V756H, V756Y, S760G, T781 M, N786K, T797S, A824D, N833D, Q834E, S835D and F1048W, wherein numbering is according to SEQ ID NO: 2.
In a particular embodiment, the invention relates to a detergent composition comprising an endoglu- canase variant selected from the group consisting of the endoglucananase variants set forth in Table 2 herein.
ln a particular embodiment, the invention relates to a detergent composition comprising an endoglu- cananase variant selected from the group consisting of the endoglucananase variants set forth in Table 3 herein.
In a particular embodiment, the invention relates to a detergent composition comprising an endoglu- cananase variant selected from the group consisting of the endoglucananase variants set forth in Table 4 herein.
In a particular embodiment, the invention relates to a detergent composition comprising an endoglucananase variant selected from the group consisting of the endoglucananase variants set forth in Table 5 herein.
In a particular embodiment, the invention relates to a detergent composition comprising an endoglucananase variant selected from the group consisting of the endoglucananase variants set forth in Table 6 herein.
The variants may further comprise one or more additional alterations at one or more (e.g., several) other positions.
The amino acid changes may be of a minor nature, that is conservative amino acid substitutions or insertions that do not significantly affect the folding and/or activity of the protein; small deletions, typically of 1 -30 amino acids; small amino- or carboxyl-terminal extensions, such as an amino-terminal methionine residue; a small linker peptide of up to 20-25 residues; or a small extension that facilitates purification by changing net charge or another function, such as a poly-histidine tract, an antigenic epitope or a binding domain.
Examples of conservative substitutions are within the groups of basic amino acids (arginine, lysine and histidine), acidic amino acids (glutamic acid and aspartic acid), polar amino acids (glutamine and aspara- gine), hydrophobic amino acids (leucine, isoleucine and valine), aromatic amino acids (phenylalanine, tryptophan and tyrosine), and small amino acids (glycine, alanine, serine, threonine and methionine). Amino acid substitutions that do not generally alter specific activity are known in the art and are described, for example, by H. Neurath and R.L. Hill, 1979, In, The Proteins, Academic Press, New York. Common substitutions are Ala/Ser, Val/lle, Asp/Glu, Thr/Ser, Ala/Gly, Ala/Thr, Ser/Asn, Ala/Val, Ser/Gly, Tyr/Phe, Ala/Pro, Lys/Arg, Asp/Asn, Leu/lle, Leu Val, Ala/Glu, and Asp/Gly.
Alternatively, the amino acid changes are of such a nature that the physico-chemical properties of the polypeptides are altered. For example, amino acid changes may improve the thermal stability of the polypeptide, alter the substrate specificity, change the pH optimum, and the like.
Essential amino acids in a polypeptide can be identified according to procedures known in the art, such as site-directed mutagenesis or alanine-scanning mutagenesis (Cunningham and Wells, 1989, Science 244: 1081 -1085). In the latter technique, single alanine mutations are introduced at every residue in the molecule, and the resultant mutant molecules are tested for xanthan lyase activity to identify amino acid residues that are critical to the activity of the molecule. See also, Hilton et al., 1996, J. Biol. Chem. 271 : 4699-4708. The
active site of the enzyme or other biological interaction can also be determined by physical analysis of structure, as determined by such techniques as nuclear magnetic resonance, crystallography, electron diffraction, or photoaffinity labeling, in conjunction with mutation of putative contact site amino acids. See, for example, de Vos et al. , 1992, Science 255: 306-312; Smith et al., 1992, J. Mol. Biol. 224: 899-904; Wlodaver ef a/., 1992, FEBS Lett. 309: 59-64. The identity of essential amino acids can also be inferred from an alignment with a related polypeptide.
In one embodiment, the present invention relates to a detergent composition comprising an endoglu- canase variant of the invention, having the total number of alterations compared to SEQ ID NO: 2 between 1 and 20, e.g., between 1 and 10 or between 1 and 5, such as 1 , 2, 3, 4, 5, 6, 7, 8, 9 or 10 alterations.
In one embodiment, the present invention relates to a detergent composition comprising an endoglu- canase variant of the invention, wherein an activity on xanthan gum pre-treated with xanthan lyase is a xanthan degrading activity, preferably said xanthan degrading activity is endoglucanase EC 3.2.1 .4 activity.
In an embodiment, the variant has an improved stability in a detergent composition compared to a parent enzyme (e.g., SEQ ID NO: 2).
In one embodiment, the present invention relates to a detergent composition comprising an endoglucanase variant of the invention, wherein said variant has an improved stability in a detergent composition compared to a parent endoglucanase (e.g., with SEQ ID NO: 2).
In one embodiment, the present invention relates to a detergent composition comprising an endoglucanase variant of the invention, wherein said variant has a half-life improvement factor (HIF) >1 .0; preferably said variant has a half-life improvement factor (HIF) >1.0 relative to a parent endoglucanase, e.g. an endoglucanase of SEQ ID NO: 2. A preferred way of calculating said half-life improvement factor (HIF) is described in example 3 herein. Accordingly, residual activity (RA) can be calculated using the following formula:
, „ Abs(Stress)
RA (%) = J x 100%
Abs(Ref)
wherein Abs(Stress) is the absorbance at 405 nm of the sample in the stress microtiter plate (MTP)
(e.g. incubated at 25°C over-night or any other combination of temperature and time) after subtracting relevant background absorbance contributions, Abs(Ref) is the absorbance at 405 nm of the sample in the reference MTP (e.g. incubated at 5°C over-night) after subtracting relevant background absorbance contributions, wherein half-lives for the degradation of each variant and parent endoglucanase (e.g. at 25°C) are calculated using the following formula:
\n(2) x T
Tl/2 = -
(Abs(Stress)\
{ Abs(Ref) )
wherein 7~ is the incubation time for both the stress and reference MTP, wherein half-life-improvement factors (HIFs) are calculated using the following formula:
Tl/2, variant
HIF
Tl/2, wt
e.g. wherein T1/2 wt (or wild type) is the T1/2 of the mature parent endoglucanase with SEQ ID NO: 2.
In one embodiment, the present invention relates to a detergent composition comprising an endoglucanase variant of the invention, wherein a half-life improvement factor (HIF) is determined after incubation of said endoglucanase variant in a detergent composition at 25°C for a time period from about 17 to about 20 hours.
Parent
The parent endoglucanase may be (a) a polypeptide having at least 60% sequence identity to the mature polypeptide of SEQ ID NO: 2; (b) a polypeptide encoded by a polynucleotide that hybridizes under low stringency conditions with (i) the mature polypeptide coding sequence of SEQ ID NO: 1 , or (ii) the full- length complement of (i); or (c) a polypeptide encoded by a polynucleotide having at least 60% sequence identity to the mature polypeptide coding sequence of SEQ ID NO: 1 .
In an aspect, the parent has a sequence identity to the mature polypeptide of SEQ ID NO: 2 of at least 60%, e.g. , at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100%, which have xanthan lyase activity. In one aspect, the amino acid sequence of the parent differs by up to 10 amino acids, e.g., 1 , 2, 3, 4, 5, 6, 7, 8, 9, or 10, from the mature polypeptide of SEQ ID NO: 2.
In another aspect, the parent comprises or consists of the amino acid sequence of SEQ ID NO: 2. In another aspect, the parent comprises or consists of the mature polypeptide of SEQ ID NO: 2. In another aspect, the parent is a fragment of the mature polypeptide of SEQ ID NO: 2 containing at least 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94% or 95% of the number of amino acids of SEQ ID NO: 2. In another embodiment, the parent is an allelic variant of the mature polypeptide of SEQ ID NO: 2.
In another aspect, the parent is encoded by a polynucleotide that hybridizes under very low stringency conditions, low stringency conditions, medium stringency conditions, medium-high stringency conditions, high stringency conditions, or very high stringency conditions with (i) the mature polypeptide coding sequence of SEQ ID NO: 1 , or (ii) the full-length complement of (i) (Sambrook ef a/., 1989, Molecular Cloning, A Laboratory Manual, 2d edition, Cold Spring Harbor, New York).
The polynucleotide of SEQ ID NO: 1 or a subsequence thereof, as well as the polypeptide of SEQ ID
NO: 2 or a fragment thereof, may be used to design nucleic acid probes to identify and clone DNA encoding a parent from strains of different genera or species according to methods well known in the art. In particular, such probes can be used for hybridization with the genomic DNA or cDNA of a cell of interest, following standard Southern blotting procedures, in order to identify and isolate the corresponding gene therein. Such probes can be considerably shorter than the entire sequence, but should be at least 15, e.g., at least 25, at
least 35, or at least 70 nucleotides in length. Preferably, the nucleic acid probe is at least 100 nucleotides in length, e.g. , at least 200 nucleotides, at least 300 nucleotides, at least 400 nucleotides, at least 500 nucleotides, at least 600 nucleotides, at least 700 nucleotides, at least 800 nucleotides, or at least 900 nucleotides in length. Both DNA and RNA probes can be used. The probes are typically labeled for detecting the corresponding gene (for example, with 32P, 3H, 35S, biotin, or avidin). Such probes are encompassed by the present invention.
A genomic DNA or cDNA library prepared from such other strains may be screened for DNA that hybridizes with the probes described above and encodes a parent. Genomic or other DNA from such other strains may be separated by agarose or polyacrylamide gel electrophoresis, or other separation techniques. DNA from the libraries or the separated DNA may be transferred to and immobilized on nitrocellulose or other suitable carrier material. In order to identify a clone or DNA that hybridizes with SEQ ID NO: 1 or a subsequence thereof, the carrier material is used in a Southern blot.
For purposes of the present invention, hybridization indicates that the polynucleotide hybridizes to a labeled nucleic acid probe corresponding to (i) SEQ ID NO: 1 ; (ii) the mature polypeptide coding sequence of SEQ ID NO: 1 ; (iii) the full-length complement thereof; or (iv) a subsequence thereof; under very low to very high stringency conditions. Molecules to which the nucleic acid probe hybridizes under these conditions can be detected using, for example, X-ray film or any other detection means known in the art.
In one aspect, the nucleic acid probe is the mature polypeptide coding sequence of SEQ ID NO: 1. In another aspect, the nucleic acid probe is a polynucleotide that encodes the polypeptide of SEQ ID NO: 2; the mature polypeptide thereof; or a fragment thereof. In another aspect, the nucleic acid probe is SEQ ID NO: 1.
In another embodiment, the parent is encoded by a polynucleotide having a sequence identity to the mature polypeptide coding sequence of SEQ ID NO: 1 of at least 60%, e.g. , at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100%.
The polypeptide may be a hybrid polypeptide in which a region of one polypeptide is fused at the N-terminus or the C-terminus of a region of another polypeptide.
The parent may be a fusion polypeptide or cleavable fusion polypeptide in which another polypeptide is fused at the N-terminus or the C-terminus of the polypeptide of the present invention. A fusion polypeptide is produced by fusing a polynucleotide encoding another polypeptide to a polynucleotide of the present invention. Techniques for producing fusion polypeptides are known in the art, and include ligating the coding sequences encoding the polypeptides so that they are in frame and that expression of the fusion polypeptide is under control of the same promoter(s) and terminator. Fusion polypeptides may also be constructed using intein technology in which fusion polypeptides are created post-translationally (Cooper ef a/. , 1993, EMBO J. 12: 2575-2583; Dawson et al., 1994, Science 266: 776-779).
A fusion polypeptide can further comprise a cleavage site between the two polypeptides. Upon secretion of the fusion protein, the site is cleaved releasing the two polypeptides. Examples of cleavage sites include, but are not limited to, the sites disclosed in Martin ef al., 2003, J. Ind. Microbiol. Biotechnol. 3: 568- 576; Svetina et al., 2000, J. Biotechnol. 76: 245-251 ; Rasmussen-Wilson et al., 1997, Appl. Environ. Micro- biol. 63: 3488-3493; Ward et al., 1995, Biotechnology 13: 498-503; and Contreras et al., 1991 , Biotechnology 9: 378-381 ; Eaton et al. , 1986, Biochemistry 25: 505-512; Collins-Racie et al., 1995, Biotechnology 13: 982-987; Carter et al. , 1989, Proteins: Structure, Function, and Genetics 6: 240-248; and Stevens, 2003, Drug Discovery World 4: 35-48.
The parent may be obtained from microorganisms of any genus. For purposes of the present invention, the term "obtained from" as used herein in connection with a given source shall mean that the parent encoded by a polynucleotide is produced by the source or by a strain in which the polynucleotide from the source has been inserted. In one aspect, the parent is secreted extracellularly.
The parent may be a bacterial enzyme. For example, the parent may be a Gram-positive bacterial polypeptide such as a Bacillus, Clostridium, Enterococcus, Geobacillus, Lactobacillus, Lactococcus, Oceanobacillus, Staphylococcus, Streptococcus, or Streptomyces enzyme, or a Gram-negative bacterial polypeptide such as a Campylobacter, E. coli, Flavobacterium, Fusobacterium, Helicobacter, llyobacter, Neisseria, Pseudomonas, Salmonella, or Ureaplasma enzyme.
In one aspect, the parent is a Bacillus alkalophilus, Bacillus amyloliquefaciens, Bacillus brevis, Bacillus circulans, Bacillus clausii, Bacillus coagulans, Bacillus firmus, Bacillus lautus, Bacillus lentus, Bacillus li- cheniformis, Bacillus megaterium, Bacillus pumilus, Bacillus stearothermophilus, Bacillus subtilis, or Bacillus thuringiensis enzyme.
In another aspect, the parent is a Streptococcus equisimilis, Streptococcus pyogenes, Streptococcus uberis, or Streptococcus equi subsp. zooepidemicus enzyme.
In another aspect, the parent is a Streptomyces achromogenes, Streptomyces avermitilis, Streptomy- ces coelicolor, Streptomyces griseus, or Streptomyces lividans enzyme.
The parent may be a fungal enzyme. For example, the parent may be a yeast enzyme such as a Candida, Kluyveromyces, Pichia, Saccharomyces, Schizosaccharomyces, or Yarrowia enzyme; or a filamentous fungal enzyme such as an Acremonium, Agaricus, Alternaria, Aspergillus, Aureobasidium, Botry- ospaeria, Ceriporiopsis, Chaetomidium, Chrysosporium, Claviceps, Cochliobolus, Coprinopsis, Cop- totermes, Corynascus, Cryphonectria, Cryptococcus, Diplodia, Exidia, Filibasidium, Fusarium, Gibberella, Holomastigotoides, Humicola, Irpex, Lentinula, Leptospaeria, Magnaporthe, Melanocarpus, Meripilus, Mu- cor, Myceliophthora, Neocallimastix, Neurospora, Paecilomyces, Penicillium, Phanerochaete, Piromyces, Poitrasia, Pseudoplectania, Pseudotrichonympha, Rhizomucor, Schizophyllum, Scytalidium, Talaromyces, Thermoascus, Thielavia, Tolypocladium, Trichoderma, Trichophaea, Verticillium, Volvariella, or Xylaria en- zyme.
ln another aspect, the parent is a Saccharomyces carlsbergensis, Saccharomyces cerevisiae, Saccharomyces diastaticus, Saccharomyces douglasii, Saccharomyces kluyveri, Saccharomyces norbensis, or Saccharomyces oviformis enzyme.
In another aspect, the parent is an Acremonium cellulolyticus, Aspergillus aculeatus, Aspergillus awamori, Aspergillus foetidus, Aspergillus fumigatus, Aspergillus japonicus, Aspergillus nidulans, Aspergillus niger, Aspergillus oryzae, Chrysosporium inops, Chrysosporium keratinophilum, Chrysosporium luck- nowense, Chrysosporium merdarium, Chrysosporium pannicola, Chrysosporium queenslandicum, Chrysosporium tropicum, Chrysosporium zonatum, Fusarium bactridioides, Fusarium cerealis, Fusarium crookwel- lense, Fusarium culmorum, Fusarium graminearum, Fusarium graminum, Fusarium heterosporum, Fusarium negundi, Fusarium oxysporum, Fusarium reticulatum, Fusarium roseum, Fusarium sambucinum, Fusarium sarcochroum, Fusarium sporotrichioides, Fusarium sulphureum, Fusarium torulosum, Fusarium trichothecioides, Fusarium venenatum, Humicola grisea, Humicola insolens, Humicola lanuginosa, Irpex lacteus, Mucor miehei, Myceliophthora thermophila, Neurospora crassa, Penicillium funiculosum, Penicil- lium purpurogenum, Phanerochaete chrysosporium, Thielavia achromatica, Thielavia albomyces, Thielavia albopilosa, Thielavia australeinsis, Thielavia fimeti, Thielavia microspora, Thielavia ovispora, Thielavia peruviana, Thielavia setosa, Thielavia spededonium, Thielavia subthermophila, Thielavia terrestris, Tricho- derma harzianum, Trichoderma koningii, Trichoderma longibrachiatum, Trichoderma reesei, or Trichoderma viride enzyme.
In another aspect, the parent is a Paenibacillus sp. xanthan lyase, e.g. , the xanthan lyase of SEQ ID NO: 2.
It will be understood that for the aforementioned species, the invention encompasses both the perfect and imperfect states, and other taxonomic equivalents, e.g., anamorphs, regardless of the species name by which they are known. Those skilled in the art will readily recognize the identity of appropriate equivalents.
Strains of these species are readily accessible to the public in a number of culture collections, such as the American Type Culture Collection (ATCC), Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ), Centraalbureau Voor Schimmelcultures (CBS), and Agricultural Research Service Patent Culture Collection, Northern Regional Research Center (NRRL).
The parent may be identified and obtained from other sources including microorganisms isolated from nature (e.g., soil, composts, water, etc.) or DNA samples obtained directly from natural materials (e.g. , soil, composts, water, etc.) using the above-mentioned probes. Techniques for isolating microorganisms and DNA directly from natural habitats are well known in the art. A polynucleotide encoding a parent may then be obtained by similarly screening a genomic DNA or cDNA library of another microorganism or mixed DNA sample. Once a polynucleotide encoding a parent has been detected with the probe(s), the polynucleotide can be isolated or cloned by utilizing techniques that are known to those of ordinary skill in the art (see, e.g., Sambrook et ai, 1989, supra).
Preparation of variants
Methods for obtaining a variant having endoglucanase activity, may comprise: (a) introducing into a parent endoglucanase an alteration at one or more (e.g., several) positions corresponding to positions 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 311 , 313, 387, 388, 390, 403, 408, 410, 416, 448, 451 , 471 , 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 711 , 719, 754, 756, 760, 781 , 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2 of the mature polypeptide of SEQ ID NO: 2, wherein the variant has endoglucanase activity; and (b) recovering the variant.
In an embodiment, the variant to be obtained may further comprise an alteration at one or more positions corresponding to positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651 , 824, 848, 869, 880, 881 , 883, 887, 892, 905, 906, 912, 921 , 928, 934, 937, 948, 956, 999 and 1037 of SEQ ID NO: 2 of the mature polypeptide of SEQ ID NO: 2, wherein the variant has endoglucanase activity; and (b) recovering the variant.
The variants can be prepared using any mutagenesis procedure known in the art, such as site-directed mutagenesis, synthetic gene construction, semi-synthetic gene construction, random mutagenesis, shuf- fling, etc.
Site-directed mutagenesis is a technique in which one or more (e.g., several) mutations are introduced at one or more defined sites in a polynucleotide encoding the parent.
Site-directed mutagenesis can be accomplished in vitro by PCR involving the use of oligonucleotide primers containing the desired mutation. Site-directed mutagenesis can also be performed in vitro by cas- sette mutagenesis involving the cleavage by a restriction enzyme at a site in the plasmid comprising a polynucleotide encoding the parent and subsequent ligation of an oligonucleotide containing the mutation in the polynucleotide. Usually the restriction enzyme that digests the plasmid and the oligonucleotide is the same, permitting sticky ends of the plasmid and the insert to ligate to one another. See, e.g. , Scherer and Davis, 1979, Proc. Natl. Acad. Sci. USA 76: 4949-4955; and Barton ei al., 1990, Nucleic Acids Res. 18: 7349-4966.
Site-directed mutagenesis can also be accomplished in vivo by methods known in the art. See, e.g., U.S. Patent Application Publication No. 2004/0171154; Storici ei al., 2001 , Nature Biotechnol. 19: 773-776; Kren ei al. , 1998, Nat. Med. 4: 285-290; and Calissano and Macino, 1996, Fungal Genet. Newslett. 43: 15- 16.
Any site-directed mutagenesis procedure can be used in the present invention. There are many commercial kits available that can be used to prepare variants.
Synthetic gene construction entails in vitro synthesis of a designed polynucleotide molecule to encode a polypeptide of interest. Gene synthesis can be performed utilizing a number of techniques, such as the multiplex microchip-based technology described by Tian et al. (2004, Nature 432: 1050-1054) and similar technologies wherein oligonucleotides are synthesized and assembled upon photo-programmable microflu- idic chips.
Single or multiple amino acid substitutions, deletions, and/or insertions can be made and tested using known methods of mutagenesis, recombination, and/or shuffling, followed by a relevant screening procedure, such as those disclosed by Reidhaar-Olson and Sauer, 1988, Science 241 : 53-57; Bowie and Sauer, 1989, Proc. Natl. Acad. Sci. USA 86: 2152-2156; WO 95/17413; or WO 95/22625. Other methods that can be used include error-prone PCR, phage display (e.g., Lowman et al. , 1991 , Biochemistry 30: 10832-10837; U.S. Patent No. 5,223,409; WO 92/06204) and region-directed mutagenesis (Derbyshire et al., 1986, Gene 46: 145; Ner et al. , 1988, DNA 7: 127).
Mutagenesis/shuffling methods can be combined with high-throughput, automated screening methods to detect activity of cloned, mutagenized polypeptides expressed by host cells (Ness ef a/., 1999, Nature Biotechnology 17: 893-896). Mutagenized DNA molecules that encode active polypeptides can be recovered from the host cells and rapidly sequenced using standard methods in the art. These methods allow the rapid determination of the importance of individual amino acid residues in a polypeptide.
Semi-synthetic gene construction is accomplished by combining aspects of synthetic gene construction, and/or site-directed mutagenesis, and/or random mutagenesis, and/or shuffling. Semi-synthetic con- struction is typified by a process utilizing polynucleotide fragments that are synthesized, in combination with PCR techniques. Defined regions of genes may thus be synthesized de novo, while other regions may be amplified using site-specific mutagenic primers, while yet other regions may be subjected to error-prone PCR or non-error prone PCR amplification. Polynucleotide subsequences may then be shuffled. Embodiments
In one embodiment, the present invention relates to a detergent composition comprising at least one (e.g. 1 , 2, 3, 4, 5, 6, 7, 8, 9 or 10) endoglucanase variant of the invention.
In one embodiment, the present invention relates to a detergent composition comprising at least one (e.g. 1 , 2, 3, 4, 5, 6, 7, 8, 9 or 10) endoglucanase variant of the invention, further comprising one or more detergent components.
In one embodiment, the present invention relates to a detergent composition comprising at least one (e.g. 1 , 2, 3, 4, 5, 6, 7, 8, 9 or 10) endoglucanase variant of the invention, further comprising one or more additional enzymes selected from the group consisting of: xanthan lyases, proteases, amylases, lichenases, lipases, cutinases, cellulases, endoglucanases, xyloglucanases, pectinases, pectin lyases, xanthanases, peroxidases, haloperoxygenases, catalases and mannanases, or any mixture thereof.
In one embodiment, the present invention relates to a detergent composition comprising at least one (e.g. 1 , 2, 3, 4, 5, 6, 7, 8, 9 or 10) endoglucanase variant of the invention, further comprising one or more detergent components and one or more additional enzymes selected from the group consisting of: xanthan lyases, proteases, amylases, lichenases, lipases, cutinases, cellulases, endoglucanases, xyloglucanases, pectinases, pectin lyases, xanthanases, peroxidases, haloperoxygenases, catalases and mannanases, or any mixture thereof.
ln one embodiment, the present invention relates to a detergent composition comprising at least one (e.g. 1 , 2, 3, 4, 5, 6, 7, 8, 9 or 10) endoglucanase variant of the invention, further comprising one or more detergent components, wherein said detergent composition is in form of a bar, a homogenous tablet, a tablet having two or more layers, a pouch having one or more compartments, a regular or compact powder, a granule, a paste, a gel, or a regular, compact or concentrated liquid.
In one embodiment, the present invention relates to use of a detergent composition of the invention, wherein said use is selected from the group of: use for degrading xanthan gum and use in a cleaning process, such as laundry or hard surface cleaning such as dish wash.
In one embodiment, the present invention relates to use of a detergent composition of the invention, wherein said composition has an enzyme detergency benefit
In one embodiment, the present invention relates to a method for degrading xanthan gum comprising: applying a detergent composition of the invention to a xanthan gum.
In one embodiment, the present invention relates to a method for degrading xanthan gum comprising: applying a detergent composition of the invention to a xanthan gum, wherein said xanthan gum is on the surface of a textile or hard surface, such as dish wash.
Polynucleotides
Also disclosed herein are isolated polynucleotides encoding a variant of the present invention. Accordingly, in one aspect, the present disclosure relates to isolated polynucleotides encoding a variant comprising an alteration at one or more positions in a region selected from the group consisting of: i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, ii) region 11 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, iii) region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, vi) region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, vii) region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, viii) region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, ix) region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2.
The techniques used to isolate or clone a polynucleotide are known in the art and include isolation from genomic DNA or cDNA, or a combination thereof. The cloning of the polynucleotides from genomic DNA can be effected, e.g., by using the well-known polymerase chain reaction (PCR) or antibody screening of expression libraries to detect cloned DNA fragments with shared structural features. See, e.g., Innis ef a/., 1990, PCR; A Guide to Methods and Application, Academic Press, New York. Other nucleic acid amplification procedures such as ligase chain reaction (LCR), ligation activated transcription (LAT) and polynucleo- tide-based amplification (NASBA) may be used. The polynucleotides may be cloned in a strain of Bacillus subtilis or E. coli, or a related organism and thus, for example, may be an allelic or species variant of the
polypeptide encoding region of the polynucleotide.
Modification of a polynucleotide encoding a polypeptide of the present invention may be necessary for synthesizing polypeptides substantially similar to the polypeptide. The term "substantially similar" to the polypeptide refers to non-naturally occurring forms of the polypeptide. These polypeptides may differ in some engineered way from the polypeptide isolated from its native source, e.g. , variants that differ in specific activity, thermostability, pH optimum, or the like. The variants may be constructed on the basis of the polynucleotide presented as the mature polypeptide coding sequence of SEQ ID NO: 1 , e.g., a subsequence thereof, and/or by introduction of nucleotide substitutions that do not result in a change in the amino acid sequence of the polypeptide, but which correspond to the codon usage of the host organism intended for production of the enzyme, or by introduction of nucleotide substitutions that may give rise to a different amino acid sequence. For a general description of nucleotide substitution, see, Ford ei a/. , (1991 ), 'Protein Expression and Purification', 2: 95-107.
Nucleic acid constructs
Also disclosed herein are nucleic acid constructs comprising a polynucleotide encoding a variant as described herein operably linked to one or more control sequences that direct the expression of the coding sequence in a suitable host cell under conditions compatible with the control sequences. Accordingly, disclosed are nucleic acid constructs comprising a polynucleotide encoding a variant comprising an alteration at one or more positions in a region selected from the group consisting of: i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, ii) region 11 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, iii) region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, vi) region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, vii) region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, viii) region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, ix) region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2, operably linked to one or more control sequences that direct the expression of the coding sequence in a suitable host cell under conditions compatible with the control sequences.
A polynucleotide may be manipulated in a variety of ways to provide for expression of the polypeptide.
Manipulation of the polynucleotide prior to its insertion into a vector may be desirable or necessary depending on the expression vector. The techniques for modifying polynucleotides utilizing recombinant DNA methods are well known in the art.
The control sequence may be a promoter, a polynucleotide that is recognized by a host cell for expres- sion of a polynucleotide encoding a polypeptide of the present invention. The promoter contains transcrip-
tional control sequences that mediate the expression of the polypeptide. The promoter may be any polynucleotide that shows transcriptional activity in the host cell including mutant, truncated, and hybrid promoters, and may be obtained from genes encoding extracellular or intracellular polypeptides either homologous or heterologous to the host cell.
Examples of suitable promoters for directing transcription of the nucleic acid constructs of the present invention in a bacterial host cell are the promoters obtained from the Bacillus amyloliquefaciens alpha- amylase gene (amyQ), Bacillus licheniformis alpha-amylase gene (amyL), Bacillus licheniformis penicillinase gene (penP), Bacillus stearothermophilus maltogenic amylase gene (amyM), Bacillus subtilis levansu- crase gene (sacB), Bacillus subtilis xylA and xylB genes, Bacillus thuringiensis crylllA gene (Agaisse and Lereclus, 1994, Molecular Microbiology 13: 97-107), E. coli lac operon, E. coli trc promoter (Egon ef al. , 1988, Gene 69: 301-315), Streptomyces coelicolor agarase gene (dagA), and prokaryotic beta-lactamase gene (Villa-Kamaroff et al. , 1978, Proc. Natl. Acad. Sci. USA 75: 3727-3731 ), as well as the tac promoter (DeBoer et al., 1983, Proc. Natl. Acad. Sci. USA 80: 21 -25). Further promoters are described in "Useful proteins from recombinant bacteria" in Gilbert ef al. , 1980, Scientific American 242: 74-94; and in Sambrook ef al., 1989, supra. Examples of tandem promoters are disclosed in WO 99/43835.
Examples of suitable promoters for directing transcription of the nucleic acid constructs of the present invention in a filamentous fungal host cell are promoters obtained from the genes for Aspergillus nidulans acetamidase, Aspergillus niger neutral alpha-amylase, Aspergillus niger acid stable alpha-amylase, Aspergillus niger or Aspergillus awamori glucoamylase (glaA), Aspergillus oryzae TAKA amylase, Aspergillus oryzae alkaline protease, Aspergillus oryzae triose phosphate isomerase, Fusarium oxysporum trypsin-like protease (WO 96/00787), Fusarium venenatum amyloglucosidase (WO 00/56900), Fusarium venenatum Daria (WO 00/56900), Fusarium venenatum Quinn (WO 00/56900), Rhizomucor miehei lipase, Rhizomucor miehei aspartic proteinase, Trichoderma reesei beta-glucosidase, Trichoderma reesei cellobiohydrolase I, Tncftocterma reese/'cellobiohydrolase II, Trichoderma reese/' endoglucanase I, Trichoderma reesei endoglu- canase II, Trichoderma reesei endoglucanase III, Trichoderma reesei endoglucanase IV, Trichoderma reesei endoglucanase V, Trichoderma reesei xylanase I, Trichoderma reesei xylanase II, Trichoderma reesei beta-xylosidase, as well as the NA2-tpi promoter (a modified promoter from an Aspergillus neutral alpha-amylase gene in which the untranslated leader has been replaced by an untranslated leader from an Aspergillus triose phosphate isomerase gene; non-limiting examples include modified promoters from an Aspergillus niger neutral alpha-amylase gene in which the untranslated leader has been replaced by an untranslated leader from an Aspergillus nidulans or Aspergillus oryzae triose phosphate isomerase gene); and mutant, truncated, and hybrid promoters thereof.
In a yeast host, useful promoters are obtained from the genes for Saccharomyces cerevisiae enolase (ENO-1 ), Saccharomyces cerevisiae galactokinase (GAL1 ), Saccharomyces cerevisiae alcohol dehydro- genase/glyceraldehyde-3-phosphate dehydrogenase (ADH1 , ADH2/GAP), Saccharomyces cerevisiae triose phosphate isomerase (TPI), Saccharomyces cerevisiae metallothionein (CUP1 ), and Saccharomyces
cerevisiae 3-phosphoglycerate kinase. Other useful promoters for yeast host cells are described by Romanos et al., 1992, Yeast 8: 423-488.
The control sequence may also be a transcription terminator, which is recognized by a host cell to terminate transcription. The terminator is operably linked to the 3'-terminus of the polynucleotide encoding the polypeptide. Any terminator that is functional in the host cell may be used in the present invention.
Preferred terminators for bacterial host cells are obtained from the genes for Bacillus clausii alkaline protease (aprH), Bacillus licheniformis alpha-amylase (amyL), and Escherichia coli ribosomal RNA (rrnB).
Preferred terminators for filamentous fungal host cells are obtained from the genes for Aspergillus nid- ulans anthranilate synthase, Aspergillus niger glucoamylase, Aspergillus n/geralpha-glucosidase, Aspergil- lus oryzae TAKA amylase, and Fusarium oxysporum trypsin-like protease.
Preferred terminators for yeast host cells are obtained from the genes for Saccharomyces cerevisiae enolase, Saccharomyces cerevisiae cytochrome C (CYC1 ), and Saccharomyces cerevisiae glyceralde- hyde-3-phosphate dehydrogenase. Other useful terminators for yeast host cells are described by Romanos et al., 1992, supra.
The control sequence may also be an mRNA stabilizer region downstream of a promoter and upstream of the coding sequence of a gene which increases expression of the gene.
Examples of suitable mRNA stabilizer regions are obtained from a Bacillus thuringiensis crylllA gene (WO 94/25612) and a Bacillus subtilis SP82 gene (Hue et al. , 1995, Journal of Bacteriology 177: 3465- 3471 ).
The control sequence may also be a leader, a nontranslated region of an mRNA that is important for translation by the host cell. The leader is operably linked to the 5'-terminus of the polynucleotide encoding the polypeptide. Any leader that is functional in the host cell may be used.
Preferred leaders for filamentous fungal host cells are obtained from the genes for Aspergillus oryzae TAKA amylase and Aspergillus nidulans triose phosphate isomerase.
Suitable leaders for yeast host cells are obtained from the genes for Saccharomyces cerevisiae enolase (ENO-1 ), Saccharomyces cerevisiae 3-phosphoglycerate kinase, Saccharomyces cerevisiae alpha- factor, and Saccharomyces cerevisiae alcohol dehydrogenase/glyceraldehyde-3-phosphate dehydrogenase (ADH2/GAP).
The control sequence may also be a polyadenylation sequence, a sequence operably linked to the 3'-terminus of the polynucleotide and, when transcribed, is recognized by the host cell as a signal to add polyadenosine residues to transcribed mRNA. Any polyadenylation sequence that is functional in the host cell may be used.
Preferred polyadenylation sequences for filamentous fungal host cells are obtained from the genes for Aspergillus nidulans anthranilate synthase, Aspergillus niger glucoamylase, Aspergillus niger alpha-gluco- sidase Aspergillus oryzae TAKA amylase, and Fusarium oxysporum trypsin-like protease.
Useful polyadenylation sequences for yeast host cells are described by Guo and Sherman, 1995, Mol.
Cellular Biol. 15: 5983-5990.
The control sequence may also be a signal peptide coding region that encodes a signal peptide linked to the N-terminus of a polypeptide and directs the polypeptide into the cell's secretory pathway. The 5'-end of the coding sequence of the polynucleotide may inherently contain a signal peptide coding sequence naturally linked in translation reading frame with the segment of the coding sequence that encodes the polypeptide. Alternatively, the 5'-end of the coding sequence may contain a signal peptide coding sequence that is foreign to the coding sequence. A foreign signal peptide coding sequence may be required where the coding sequence does not naturally contain a signal peptide coding sequence. Alternatively, a foreign signal peptide coding sequence may simply replace the natural signal peptide coding sequence in order to enhance secretion of the polypeptide. However, any signal peptide coding sequence that directs the expressed polypeptide into the secretory pathway of a host cell may be used.
Effective signal peptide coding sequences for bacterial host cells are the signal peptide coding sequences obtained from the genes for Bacillus NCIB 1 1837 maltogenic amylase, Bacillus licheniformis sub- tilisin, Bacillus licheniformis beta-lactamase, Bacillus stearothermophilus alpha-amylase, Bacillus stea- rothermophilus neutral proteases (nprT, nprS, nprM), and Bacillus subtilis prsA. Further signal peptides are described by Simonen and Palva, 1993, Microbiological Reviews 57: 109-137.
Effective signal peptide coding sequences for filamentous fungal host cells are the signal peptide coding sequences obtained from the genes for Aspergillus niger neutral amylase, Aspergillus n/ger glucoamyl- ase, Aspergillus oryzae TAKA amylase, Humicola insolens cellulase, Humicola insolens endoglucanase V, Humicola lanuginosa lipase, and Rhizomucor miehei aspartic proteinase.
Useful signal peptides for yeast host cells are obtained from the genes for Saccharomyces cerevisiae alpha-factor and Saccharomyces cerevisiae invertase. Other useful signal peptide coding sequences are described by Romanos et ai, 1992, supra.
The control sequence may also be a propeptide coding sequence that encodes a propeptide positioned at the N-terminus of a polypeptide. The resultant polypeptide is known as a proenzyme or propolypeptide (or a zymogen in some cases). A propolypeptide is generally inactive and can be converted to an active polypeptide by catalytic or autocatalytic cleavage of the propeptide from the propolypeptide. The propeptide coding sequence may be obtained from the genes for Bacillus subtilis alkaline protease (aprE), Bacillus subtilis neutral protease (nprT), Myceliophthora thermophila laccase (WO 95/33836), Rhizomucor miehei aspartic proteinase, and Saccharomyces cerevisiae alpha-factor.
Where both signal peptide and propeptide sequences are present, the propeptide sequence is positioned next to the N-terminus of a polypeptide and the signal peptide sequence is positioned next to the N-terminus of the propeptide sequence.
It may also be desirable to add regulatory sequences that regulate expression of the polypeptide rela- tive to the growth of the host cell. Examples of regulatory systems are those that cause expression of the gene to be turned on or off in response to a chemical or physical stimulus, including the presence of a
regulatory compound. Regulatory systems in prokaryotic systems include the lac, tac, and trp operator systems. In yeast, the ADH2 system or GAL1 system may be used. In filamentous fungi, the Aspergillus niger glucoamylase promoter, Aspergillus oryzae TAKA alpha-amylase promoter, and Aspergillus oryzae glu- coamylase promoter may be used. Other examples of regulatory sequences are those that allow for gene amplification. In eukaryotic systems, these regulatory sequences include the dihydrofolate reductase gene that is amplified in the presence of methotrexate, and the metallothionein genes that are amplified with heavy metals. In these cases, the polynucleotide encoding the polypeptide would be operably linked with the regulatory sequence. Expression vectors
Disclosed herein are recombinant expression vectors comprising a polynucleotide encoding a variant as described herein, a promoter, and transcriptional and translational stop signals. Accordingly, the present disclosure relates to recombinant expression vectors comprising a polynucleotide encoding a variant comprising an alteration at one or more positions in a region selected from the group consisting of: i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, ii) region 1 1 corresponding to amino acids 106 to 114 of SEQ ID NO: 2, iii) region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, vi) region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, vii) region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, viii) region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, ix) region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2, a promoter, and transcriptional and translational stop signals.
The various nucleotide and control sequences may be joined together to produce a recombinant ex- pression vector that may include one or more convenient restriction sites to allow for insertion or substitution of the polynucleotide encoding the polypeptide at such sites. Alternatively, the polynucleotide may be expressed by inserting the polynucleotide or a nucleic acid construct comprising the polynucleotide into an appropriate vector for expression. In creating the expression vector, the coding sequence is located in the vector so that the coding sequence is operably linked with the appropriate control sequences for expression.
The recombinant expression vector may be any vector (e.g., a plasmid or virus) that can be conveniently subjected to recombinant DNA procedures and can bring about expression of the polynucleotide. The choice of the vector will typically depend on the compatibility of the vector with the host cell into which the vector is to be introduced. The vector may be a linear or closed circular plasmid.
The vector may be an autonomously replicating vector, i.e., a vector that exists as an extrachromoso- mal entity, the replication of which is independent of chromosomal replication, e.g., a plasmid, an extrachro- mosomal element, a minichromosome, or an artificial chromosome. The vector may contain any means for
assuring self-replication. Alternatively, the vector may be one that, when introduced into the host cell, is integrated into the genome and replicated together with the chromosome(s) into which it has been integrated. Furthermore, a single vector or plasmid or two or more vectors or plasmids that together contain the total DNA to be introduced into the genome of the host cell, or a transposon, may be used.
The vector preferably contains one or more selectable markers that permit easy selection of transformed, transfected, transduced, or the like cells. A selectable marker is a gene the product of which provides for biocide or viral resistance, resistance to heavy metals, prototrophy to auxotrophs, and the like.
Examples of bacterial selectable markers are Bacillus licheniformis or Bacillus subtilis dal genes, or markers that confer antibiotic resistance such as ampicillin, chloramphenicol, kanamycin, neomycin, spec- tinomycin, or tetracycline resistance. Suitable markers for yeast host cells include, but are not limited to, ADE2, HIS3, LEU2, LYS2, MET3, TRP1 , and URA3. Selectable markers for use in a filamentous fungal host cell include, but are not limited to, amdS (acetamidase), argB (ornithine carbamoyltransferase), bar (phosphinothricin acetyltransferase), hph (hygromycin phosphotransferase), niaD (nitrate reductase), pyrG (orotidine-5'-phosphate decarboxylase), sC (sulfate adenyltransferase), and trpC (anthranilate synthase), as well as equivalents thereof. Preferred for use in an Aspergillus cell are Aspergillus nidulans or Aspergillus oryzae amdS and pyrG genes and a Streptomyces hygroscopicus bar gene.
The vector preferably contains an element(s) that permits integration of the vector into the host cell's genome or autonomous replication of the vector in the cell independent of the genome.
For integration into the host cell genome, the vector may rely on the polynucleotide's sequence encod- ing the polypeptide or any other element of the vector for integration into the genome by homologous or non-homologous recombination. Alternatively, the vector may contain additional polynucleotides for directing integration by homologous recombination into the genome of the host cell at a precise location(s) in the chromosome(s). To increase the likelihood of integration at a precise location, the integrational elements should contain a sufficient number of nucleic acids, such as 100 to 10,000 base pairs, 400 to 10,000 base pairs, and 800 to 10,000 base pairs, which have a high degree of sequence identity to the corresponding target sequence to enhance the probability of homologous recombination. The integrational elements may be any sequence that is homologous with the target sequence in the genome of the host cell. Furthermore, the integrational elements may be non-encoding or encoding polynucleotides. On the other hand, the vector may be integrated into the genome of the host cell by non-homologous recombination.
For autonomous replication, the vector may further comprise an origin of replication enabling the vector to replicate autonomously in the host cell in question. The origin of replication may be any plasmid replicator mediating autonomous replication that functions in a cell. The term "origin of replication" or "plasmid replicator" means a polynucleotide that enables a plasmid or vector to replicate in vivo.
Examples of bacterial origins of replication are the origins of replication of plasmids pBR322, pUC19, pACYC177, and pACYC184 permitting replication in E. coli, and pUB1 10, pE194, pTA1060, and ρΑΜβΙ permitting replication in Bacillus.
Examples of origins of replication for use in a yeast host cell are the 2 micron origin of replication, ARS1 , ARS4, the combination of ARS1 and CEN3, and the combination of ARS4 and CEN6.
Examples of origins of replication useful in a filamentous fungal cell are AMA1 and ANSI (Gems ef a/., 1991 , Gene 98: 61-67; Cullen ei a/., 1987, Nucleic Acids Res. 15: 9163-9175; WO 00/24883). Isolation of the AMA1 gene and construction of plasmids or vectors comprising the gene can be accomplished according to the methods disclosed in WO 00/24883.
More than one copy of a polynucleotide of the present invention may be inserted into a host cell to increase production of a polypeptide. An increase in the copy number of the polynucleotide can be obtained by integrating at least one additional copy of the sequence into the host cell genome or by including an amplifiable selectable marker gene with the polynucleotide where cells containing amplified copies of the selectable marker gene, and thereby additional copies of the polynucleotide, can be selected for by cultivating the cells in the presence of the appropriate selectable agent.
The procedures used to ligate the elements described above to construct the recombinant expression vectors of the present invention are well known to one skilled in the art (see, e.g., Sambrook ef a/., 1989, supra).
Host cells
Further disclosed are recombinant host cells, comprising a polynucleotide encoding a variant as described herein operably linked to one or more control sequences that direct the production of a variant of the present invention. Accordingly, in one aspect, the present disclosure relates to recombinant host cells, comprising a polynucleotide encoding a variant comprising an alteration at one or more positions in a region selected from the group consisting of: i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, ii) region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, iii) region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, vi) region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, vii) region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, viii) region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, ix) region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2, operably linked to one or more control sequences that direct the production of a variant comprising an alteration at one or more positions in a selected from the group consisting of: i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, ii) region 11 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, iii) region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, vi) region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, vii) region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, viii) region 17 corresponding to amino acids 661 to 805 of
SEQ ID NO: 2, ix) region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2.
A construct or vector comprising a polynucleotide is introduced into a host cell so that the construct or vector is maintained as a chromosomal integrant or as a self-replicating extra-chromosomal vector as described earlier. The term "host cell" encompasses any progeny of a parent cell that is not identical to the parent cell due to mutations that occur during replication. The choice of a host cell will to a large extent depend upon the gene encoding the polypeptide and its source.
The host cell may be any cell useful in the recombinant production of a polypeptide of the present invention, e.g., a prokaryote or a eukaryote.
The prokaryotic host cell may be any Gram-positive or Gram-negative bacterium. Gram-positive bacteria include, but are not limited to, Bacillus, Clostridium, Enterococcus, Geobacillus, Lactobacillus, Lacto- coccus, Oceanobacillus, Staphylococcus, Streptococcus, and Streptomyces. Gram-negative bacteria include, but are not limited to, Campylobacter, E. coli, Flavobacterium, Fusobacterium, Helicobacter, llyobac- ter, Neisseria, Pseudomonas, Salmonella, and Ureaplasma.
The bacterial host cell may be any Bacillus cell including, but not limited to, Bacillus alkalophilus, Bacillus amyloliquefaciens, Bacillus brevis, Bacillus circulans, Bacillus clausii, Bacillus coagulans, Bacillus fir- mus, Bacillus lautus, Bacillus lentus, Bacillus licheniformis, Bacillus megaterium, Bacillus pumilus, Bacillus stearothermophilus, Bacillus subtilis, and Bacillus thuringiensis cells.
The bacterial host cell may also be any Streptococcus cell including, but not limited to, Streptococcus equisimilis, Streptococcus pyogenes, Streptococcus uberis, and Streptococcus equ/ subsp. zooepidemicus cells.
The bacterial host cell may also be any Streptomyces cell including, but not limited to, Streptomyces achromogenes, Streptomyces avermitilis, Streptomyces coelicolor, Streptomyces griseus, and Streptomy- ces lividans cells.
The introduction of DNA into a Bacillus cell may be effected by protoplast transformation (see, e.g., Chang and Cohen, 1979, Mol. Gen. Genet. 168: 11 1-1 15), competent cell transformation (see, e.g., Young and Spizizen, 1961 , J. Bacteriol. 81 : 823-829, or Dubnau and Davidoff-Abelson, 1971 , J. Mol. Biol. 56: 209- 221 ), electroporation (see, e.g., Shigekawa and Dower, 1988, Biotechniques 6: 742-751 ), or conjugation (see, e.g., Koehler and Thorne, 1987, J. Bacteriol. 169: 5271 -5278). The introduction of DNA into an E. coli cell may be effected by protoplast transformation (see, e.g. , Hanahan, 1983, J. Mol. Biol. 166: 557-580) or electroporation (see, e.g., Dower ei ai , 1988, Nucleic Acids Res. 16: 6127-6145). The introduction of DNA into a Streptomyces cell may be effected by protoplast transformation, electroporation (see, e.g., Gong ei a/., 2004, Folia Microbiol. (Praha) 49: 399-405), conjugation (see, e.g. , Mazodier et ai, 1989, J. Bacteriol. 171 : 3583-3585), or transduction (see, e.g., Burke et ai, 2001 , Proc. Natl. Acad. Sci. USA 98: 6289-6294). The introduction of DNA into a Pseudomonas cell may be effected by electroporation (see, e.g., Choi et ai,
2006, J. Microbiol. Methods 64: 391 -397) or conjugation (see, e.g., Pinedo and Smets, 2005, Appl. Environ. Microbiol. 71 : 51 -57). The introduction of DNA into a Streptococcus cell may be effected by natural competence (see, e.g., Perry and Kuramitsu, 1981 , Infect. Immun. 32: 1295-1297), protoplast transformation (see, e.g., Catt and Jollick, 1991 , Microbios 68: 189-207), electroporation (see, e.g., Buckley et al., 1999, Appl. Environ. Microbiol. 65: 3800-3804), or conjugation (see, e.g., Clewell, 1981 , Microbiol. Rev. 45: 409-436). However, any method known in the art for introducing DNA into a host cell can be used.
The host cell may also be a eukaryote, such as a mammalian, insect, plant, or fungal cell.
The host cell may be a fungal cell. "Fungi" as used herein includes the phyla Ascomycota, Basidiomy- cota, Chytridiomycota, and Zygomycota as well as the Oomycota and all mitosporic fungi (as defined by Hawksworth ef a/., In, Ainsworth and Bisby's Dictionary of The Fungi, 8th edition, 1995, CAB International, University Press, Cambridge, UK).
The fungal host cell may be a yeast cell. "Yeast" as used herein includes ascosporogenous yeast (Endomycetales), basidiosporogenous yeast, and yeast belonging to the Fungi Imperfecti (Blastomycetes). Since the classification of yeast may change in the future, for the purposes of this invention, yeast shall be defined as described in Biology and Activities of Yeast (Skinner, Passmore, and Davenport, editors, Soc. App. Bacteriol. Symposium Series No. 9, 1980).
The yeast host cell may be a Candida, Hansenula, Kluyveromyces, Pichia, Saccharomyces, Schiz- osaccharomyces, or Yarrowia cell, such as a Kluyveromyces lactis, Saccharomyces carlsbergensis, Saccharomyces cerevisiae, Saccharomyces diastaticus, Saccharomyces douglasii, Saccharomyces kluyveri, Saccharomyces norbensis, Saccharomyces oviformis, or Yarrowia lipolytica cell.
The fungal host cell may be a filamentous fungal cell. "Filamentous fungi" include all filamentous forms of the subdivision Eumycota and Oomycota (as defined by Hawksworth et al., 1995, supra). The filamentous fungi are generally characterized by a mycelial wall composed of chitin, cellulose, glucan, chitosan, mannan, and other complex polysaccharides. Vegetative growth is by hyphal elongation and carbon catabolism is obligately aerobic. In contrast, vegetative growth by yeasts such as Saccharomyces cerevisiae is by budding of a unicellular thallus and carbon catabolism may be fermentative.
The filamentous fungal host cell may be an Acremonium, Aspergillus, Aureobasidium, Bjerkandera, Ceriporiopsis, Chrysosporium, Coprinus, Coriolus, Cryptococcus, Filibasidium, Fusarium, Humicola, Mag- naporthe, Mucor, Myceliophthora, Neocallimastix, Neurospora, Paecilomyces, Penicillium, Phanerochaete, Phlebia, Piromyces, Pleurotus, Schizophyllum, Talaromyces, Thermoascus, Thielavia, Tolypocladium, Trametes, or Trichoderma cell.
For example, the filamentous fungal host cell may be an Aspergillus awamori, Aspergillus foetidus, Aspergillus fumigatus, Aspergillus japonicus, Aspergillus nidulans, Aspergillus niger, Aspergillus oryzae, Bjerkandera adusta, Ceriporiopsis aneirina, Ceriporiopsis caregiea, Ceriporiopsis gilvescens, Ceriporiopsis pannocinta, Ceriporiopsis rivulosa, Ceriporiopsis subrufa, Ceriporiopsis subvermispora, Chrysosporium in-
ops, Chrysosporium keratinophilum, Chrysosporium lucknowense, Chrysosporium merdarium, Chryso- sporium pannicola, Chrysosporium queenslandicum, Chrysosporium tropicum, Chrysosporium zonatum, Coprinus cinereus, Coriolus hirsutus, Fusarium bactridioides, Fusarium cerealis, Fusarium crookwellense, Fusarium culmorum, Fusarium graminearum, Fusarium graminum, Fusarium heterosporum, Fusarium negundi, Fusarium oxysporum, Fusarium reticulatum, Fusarium roseum, Fusarium sambucinum, Fusarium sarcochroum, Fusarium sporotrichioides, Fusarium sulphureum, Fusarium torulosum, Fusarium trichothe- cioides, Fusarium venenatum, Humicola insolens, Humicola lanuginosa, Mucor miehei, Myceliophthora thermophila, Neurospora crassa, Penicillium purpurogenum, Phanerochaete chrysosporium, Phlebia radi- ata, Pleurotus eryngii, Thielavia terrestris, Trametes villosa, Trametes versicolor, Trichoderma harzianum, Trichoderma koningii, Trichoderma longibrachiatum, Trichoderma reesei, or Trichoderma viride cell.
Fungal cells may be transformed by a process involving protoplast formation, transformation of the protoplasts, and regeneration of the cell wall in a manner known per se. Suitable procedures for transformation of Aspergillus and Trichoderma host cells are described in EP 238023, Yelton ef a/., 1984, Proc. Natl. Acad. Sci. USA 81 : 1470-1474, and Christensen ef a/., 1988, Bio/Technology 6: 1419-1422. Suitable methods for transforming Fusarium species are described by Malardier ei a/., 1989, Gene 78: 147-156, and WO 96/00787. Yeast may be transformed using the procedures described by Becker and Guarente, In Abelson, J.N. and Simon, M.I., editors, Guide to Yeast Genetics and Molecular Biology, Methods in Enzy- mology, Volume 194, pp 182-187, Academic Press, Inc., New York; Ito et al. , 1983, J. Bacteriol. 153: 163; and Hinnen et al. , 1978, Proc. Natl. Acad. Sci. USA 75: 1920.
Methods of production
Also disclosed are methods of producing (e.g., in vitro or ex vivo methods) a variant, comprising: (a) cultivating a host cell of the present invention under conditions suitable for expression of the variant; and (b) recovering the variant. Accordingly, in one aspect, the present disclosure relates to methods of producing (e.g. in vitro or ex vivo methods) a variant comprising an alteration at one or more positions in a region selected from the group consisting of: i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, ii) region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, iii) region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, vi) region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, vii) region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, viii) region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, ix) region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2, the method comprising cultivating a host cell comprising a polynucleotide encoding the variant, under conditions suitable for expression of the variant; and (b) optionally, recovering the variant.
Further disclosed are methods of producing (e.g., in vitro or ex vivo methods) a variant as described
herein, comprising (a) cultivating a cell, which in its wild-type form produces the polypeptide, under conditions conducive for production of the polypeptide; and (b) recovering the polypeptide. In a preferred aspect, the cell is a Paenibacillus cell, or a Microbactehum cell.
Also disclosed are methods of producing (e.g., in vitro or ex vivo methods) a variant as described herein, comprising (a) cultivating a recombinant host cell of the present invention under conditions conducive for production of the polypeptide; and (b) recovering the polypeptide.
The host cells are cultivated in a nutrient medium suitable for production of the polypeptide using methods known in the art. For example, the cell may be cultivated by shake flask cultivation, or small-scale or large-scale fermentation (including continuous, batch, fed-batch, or solid-state fermentations) in laboratory or industrial fermentors performed in a suitable medium and under conditions allowing the polypeptide to be expressed and/or isolated. The cultivation takes place in a suitable nutrient medium comprising carbon and nitrogen sources and inorganic salts, using procedures known in the art. Suitable media are available from commercial suppliers or may be prepared according to published compositions (e.g. , in catalogues of the American Type Culture Collection). If the polypeptide is secreted into the nutrient medium, the polypep- tide can be recovered directly from the medium. If the polypeptide is not secreted, it can be recovered from cell lysates.
The variant polypeptide may be detected using methods known in the art that are specific for the polypeptides such as methods for determining cellulose or xanthan lyase activity. These detection methods include, but are not limited to, use of specific antibodies, formation of an enzyme product, or disappearance of an enzyme substrate. For example, an enzyme assay may be used to determine the activity of the polypeptide.
The variant polypeptide may be recovered using methods known in the art. For example, the polypeptide may be recovered from the nutrient medium by conventional procedures including, but not limited to, collection, centrifugation, filtration, extraction, spray-drying, evaporation, or precipitation.
The variant polypeptide may be purified by a variety of procedures known in the art including, but not limited to, chromatography (e.g., ion exchange, affinity, hydrophobic, chromatofocusing, and size exclusion), electrophoretic procedures (e.g., preparative isoelectric focusing), differential solubility (e.g. , ammonium sulfate precipitation), SDS-PAGE, or extraction (see, e.g., Protein Purification, Janson and Ryden, editors, VCH Publishers, New York, 1989) to obtain substantially pure polypeptides.
In an alternative aspect, the variant polypeptide is not recovered, but rather a host cell expressing the polypeptide is used as a source of the variant polypeptide.
Compositions
In one certain aspect, the variants according to the invention have improved stability in detergents compared to a parent enzyme or compared to an endoglucanase having the identical amino acid sequence of the variant, but not having an alteration (e.g., a substitution, deletion or insertion) at one or more of the
specified positions or compared to the endoglucanase with SEQ ID NO: 2, wherein activity and/or stability in detergent is measured as disclosed in example 3 herein. Thus, in one embodiment, the present invention relates to detergent compositions comprising a variant comprising an alteration at one or more positions in a region selected from the group consisting of: i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2, ii) region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2, iii) region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2, iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2, v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2, vi) region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2, vii) region 16 corresponding to amino acids 596 to 61 1 of SEQ ID NO: 2, viii) region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2, ix) region 18 corre- sponding to amino acids 829 to 838 of SEQ ID NO: 2, and x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2, wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2.
Besides enzymes the detergent compositions may comprise additional components. The choice of additional components is within the skill of the artisan and includes conventional ingredients, including the exemplary non-limiting components set forth below. The choice of components may include, for fabric care, the consideration of the type of fabric to be cleaned, the type and/or degree of soiling, the temperature at which cleaning is to take place, and the formulation of the detergent product. Although components mentioned below are categorized by general header according to a particular functionality, this is not to be construed as a limitation, as a component may comprise additional functionalities as will be appreciated by the skilled artisan.
The detergent composition may be suitable for the laundering of textiles such as e.g. fabrics, cloths or linen, or for cleaning hard surfaces such as e.g. floors, tables, or dish wash.
Detergent compositions
In one embodiment, a variant as described herein may be added to a detergent composition in an amount corresponding to 0.0001-200 mg of enzyme protein, such as 0.0005-100 mg of enzyme protein, preferably 0.001-30 mg of enzyme protein, more preferably 0.005-8 mg of enzyme protein, even more preferably 0.01 -2 mg of enzyme protein per litre of wash liquor.
A composition for use in automatic dishwash (ADW), for example, may comprise 0.0001 %-50%, such as 0.001 %-20%, such as 0.01 %-10%, such as 0.05-5% of enzyme protein by weight of the composition.
A composition for use in laundry granulation or a solid/granular laundry composition in general, for example, may comprise 0.0001 %-50%, such as 0.001 %-20%, such as 0.01 %-10%, such as 0.05%-5% of enzyme protein by weight of the composition.
A composition for use in laundry liquid, for example, may comprise 0.0001 %-10%, such as 0.001 -7%, such as 0.1 %-5% of enzyme protein by weight of the composition.
The enzyme(s) of the detergent composition of the invention may be stabilized using conventional stabilizing agents, e.g. , a polyol such as propylene glycol or glycerol, a sugar or sugar alcohol, lactic acid,
boric acid, or a boric acid derivative, e.g., an aromatic borate ester, or a phenyl boronic acid derivative such as 4-formylphenyl boronic acid, and the composition may be formulated as described in, for example, WO92/19709 and WO92/19708.
In certain markets different wash conditions and, as such, different types of detergents are used. This is disclosed in e.g. EP 1 025 240. For example, In Asia (Japan) a low detergent concentration system is used, while the United States uses a medium detergent concentration system, and Europe uses a high detergent concentration system.
A low detergent concentration system includes detergents where less than about 800 ppm of detergent components are present in the wash water. Japanese detergents are typically considered low detergent concentration system as they have approximately 667 ppm of detergent components present in the wash water.
A medium detergent concentration includes detergents where between about 800 ppm and about 2000ppm of detergent components are present in the wash water. North American detergents are generally considered to be medium detergent concentration systems as they have approximately 975 ppm of deter- gent components present in the wash water.
A high detergent concentration system includes detergents where greater than about 2000 ppm of detergent components are present in the wash water. European detergents are generally considered to be high detergent concentration systems as they have approximately 4500-5000 ppm of detergent components in the wash water.
Latin American detergents are generally high suds phosphate builder detergents and the range of detergents used in Latin America can fall in both the medium and high detergent concentrations as they range from 1500 ppm to 6000 ppm of detergent components in the wash water. Such detergent compositions are all embodiments of the invention.
A polypeptide of the present invention may also be incorporated in the detergent formulations disclosed in WO97/07202, which is hereby incorporated by reference.
Surfactants
The detergent composition may comprise one or more surfactants, which may be anionic and/or cati- onic and/or non-ionic and/or semi-polar and/or zwitterionic, or a mixture thereof. In a particular embodiment, the detergent composition includes a mixture of one or more non-ionic surfactants and one or more anionic surfactants. The surfactant(s) is typically present at a level of from about 0.1 % to 60% by weight, such as about 1 % to about 40%, or about 3% to about 20%, or about 3% to about 10%. The surfactant(s) is chosen based on the desired cleaning application, and includes any conventional surfactant(s) known in the art. Any surfactant known in the art for use in detergents may be utilized.
When included therein the detergent will usually comprise from about 1 % to about 40% by weight, such as from about 5% to about 30%, including from about 5% to about 15%, or from about 20% to about 25%
of an anionic surfactant. Non-limiting examples of anionic surfactants include sulfates and sulfonates, in particular, linear alkylbenzenesulfonates (LAS), isomers of LAS, branched alkylbenzenesulfonates (BABS), phenylalkanesulfonates, alpha-olefinsulfonates (AOS), olefin sulfonates, alkene sulfonates, alkane-2,3- diylbis(sulfates), hydroxyalkanesulfonates and disulfonates, alkyl sulfates (AS) such as sodium dodecyl sul- fate (SDS), fatty alcohol sulfates (FAS), primary alcohol sulfates (PAS), alcohol ethersulfates (AES or AEOS or FES, also known as alcohol ethoxysulfates or fatty alcohol ether sulfates), secondary alkanesulfonates (SAS), paraffin sulfonates (PS), ester sulfonates, sulfonated fatty acid glycerol esters, alpha-sulfo fatty acid methyl esters (alpha-SFMe or SES) including methyl ester sulfonate (MES), alkyl- or alkenylsuccinic acid, dodecenyl/tetradecenyl succinic acid (DTSA), fatty acid derivatives of amino acids, diesters and monoesters of sulfo-succinic acid or soap, and combinations thereof.
When included therein the detergent will usually comprise from about 0% to about 10% by weight of a cationic surfactant. Non-limiting examples of cationic surfactants include alklydimethylethanolamine quat (ADMEAQ), cetyltrimethylammonium bromide (CTAB), dimethyldistearylammonium chloride (DSDMAC), and alkylbenzyldimethylammonium, alkyl quaternary ammonium compounds, alkoxylated quaternary am- monium (AQA) compounds, and combinations thereof.
When included therein the detergent will usually comprise from about 0.2% to about 40% by weight of a non-ionic surfactant, for example from about 0.5% to about 30%, in particular from about 1 % to about 20%, from about 3% to about 10%, such as from about 3% to about 5%, or from about 8% to about 12%. Non-limiting examples of non-ionic surfactants include alcohol ethoxylates (AE or AEO), alcohol propox- ylates, propoxylated fatty alcohols (PFA), alkoxylated fatty acid alkyl esters, such as ethoxylated and/or propoxylated fatty acid alkyl esters, alkylphenol ethoxylates (APE), nonylphenol ethoxylates (NPE), al- kylpolyglycosides (APG), alkoxylated amines, fatty acid monoethanolamides (FAM), fatty acid diethanola- mides (FADA), ethoxylated fatty acid monoethanolamides (EFAM), propoxylated fatty acid monoethanolamides (PFAM), polyhydroxy alkyl fatty acid amides, or A -acyl A -alkyl derivatives of glucosamine (glu- camides, GA, or fatty acid glucamide, FAGA), as well as products available under the trade names SPAN and TWEEN, and combinations thereof.
When included therein the detergent will usually comprise from about 0% to about 10% by weight of a semipolar surfactant. Non-limiting examples of semipolar surfactants include amine oxides (AO) such as alkyldimethylamineoxide, A -(coco alkyl)-A ,A -dimethylamine oxide and A -(tallow-alkyl)-A ,A -bis(2-hydroxy- ethyl)amine oxide, fatty acid alkanolamides and ethoxylated fatty acid alkanolamides, and combinations thereof.
When included therein the detergent will usually comprise from about 0% to about 10% by weight of a zwitterionic surfactant. Non-limiting examples of zwitterionic surfactants include betaine, alkyldimethylbeta- ine, sulfobetaine, and combinations thereof.
Hydro tropes
A hydrotrope is a compound that solubilises hydrophobic compounds in aqueous solutions (or oppositely, polar substances in a non-polar environment). Typically, hydrotropes have both hydrophilic and a hydrophobic character (so-called amphiphilic properties as known from surfactants); however, the molecular structure of hydrotropes generally do not favor spontaneous self-aggregation, see e.g. review by Hodgdon and Kaler (2007), Current Opinion in Colloid & Interface Science 12: 121 -128. Hydrotropes do not display a critical concentration above which self-aggregation occurs as found for surfactants and lipids forming micel- lar, lamellar or other well defined meso-phases. Instead , many hydrotropes show a continuous-type aggregation process where the sizes of aggregates grow as concentration increases. However, many hydrotropes alter the phase behavior, stability, and colloidal properties of systems containing substances of polar and non-polar character, including mixtures of water, oil, surfactants, and polymers. Hydrotropes are classically used across industries from pharma, personal care, food, to technical applications. Use of hydrotropes in detergent compositions allow for example more concentrated formulations of surfactants (as in the process of compacting liquid detergents by removing water) without inducing undesired phenomena such as phase separation or high viscosity.
The detergent may comprise 0-5% by weight, such as about 0.5 to about 5%, or about 3% to about 5%, of a hydrotrope. Any hydrotrope known in the art for use in detergents may be utilized. Non-limiting examples of hydrotropes include sodium benzene sulfonate, sodium p-toluene sulfonate (STS), sodium xylene sulfonate (SXS), sodium cumene sulfonate (SCS), sodium cymene sulfonate, amine oxides, alcohols and polyglycolethers, sodium hydroxynaphthoate, sodium hydroxynaphthalene sulfonate, sodium ethylhexyl sulfate, and combinations thereof.
Builders and Co-Builders
The detergent composition may comprise about 0-65% by weight, such as about 5% to about 45% of a detergent builder or co-builder, or a mixture thereof. In a dish wash detergent, the level of builder is typically 40-65%, particularly 50-65%. The builder and/or co-builder may particularly be a chelating agent that forms water-soluble complexes with Ca and Mg. Any builder and/or co-builder known in the art for use in laundry detergents may be utilized. Non-limiting examples of builders include zeolites, diphosphates (pyrophosphates), triphosphates such as sodium triphosphate (STP or STPP), carbonates such as sodium car- bonate, soluble silicates such as sodium metasilicate, layered silicates (e.g. , SKS-6 from Hoechst), ethan- olamines such as 2-aminoethan-1 -ol (MEA), diethanolamine (DEA, also known as iminodiethanol), trieth- anolamine (TEA, also known as 2,2',2"-nitrilotriethanol), and carboxymethyl inulin (CMI), and combinations thereof.
The detergent composition may also comprise 0-20% by weight, such as about 5% to about 10%, of a detergent co-builder, or a mixture thereof. The detergent composition may include a co-builder alone, or in
combination with a builder, for example a zeolite builder. Non-limiting examples of co-builders include ho- mopolymers of polyacrylates or copolymers thereof, such as poly(acrylic acid) (PAA) or copoly(acrylic acid/maleic acid) (PAA PMA). Further non-limiting examples include citrate, chelators such as aminocar- boxylates, aminopolycarboxylates and phosphonates, and alkyl- or alkenylsuccinic acid. Additional specific examples include 2,2',2"-nitrilotriacetic acid (NTA), ethylenediaminetetraacetic acid (EDTA), diethylenetri- aminepentaacetic acid (DTPA), iminodisuccinic acid (IDS), ethylenediamine-A .N-disuccinic acid (EDDS), methylglycinediacetic acid (MGDA), glutamic acid-N,N-diacetic acid (GLDA), 1-hydroxyethane-1 ,1-diphos- phonic acid (HEDP), ethylenediaminetetra-(methylenephosphonic acid) (EDTMPA), diethylenetriamine- pentakis(methylenephosphonic acid) (DTPMPA or DTMPA), A -(2-hydroxyethyl)iminodiacetic acid (EDG), aspartic acid-A -monoacetic acid (ASMA), aspartic acid-A ,A -diacetic acid (ASDA), aspartic acid-A -mono- propionic acid (ASMP), iminodisuccinic acid (IDA), A -(2-sulfomethyl)-aspartic acid (SMAS), A -(2-sulfoethyl)- aspartic acid (SEAS), A -(2-sulfomethyl)-glutamic acid (SMGL), A -(2-sulfoethyl)-glutamic acid (SEGL), N- methyliminodiacetic acid (MIDA), a-alanine-A ,A -diacetic acid (a-ALDA), serine-A ,A -diacetic acid (SEDA), isoserine-A ,A -diacetic acid (ISDA), phenylalanine-A ,A -diacetic acid (PHDA), anthranilic acid-A ,A -diacetic acid (ANDA), sulfanilic acid-A ,A -diacetic acid (SLDA), taurine-A ,A -diacetic acid (TUDA) and sulfomethyl- A ,A -diacetic acid (SMDA), A -(2-hydroxyethyl)-ethylidenediamine-A ,A ',A -triacetate (HEDTA), diethanolgly- cine (DEG), diethylenetriamine penta(methylenephosphonic acid) (DTPMP), aminotris(meth- ylenephosphonic acid) (ATMP), and combinations and salts thereof. Further exemplary builders and/or co- builders are described in, e.g., WO 09/102854, US 5977053
Bleaching Systems
The detergent may comprise 0-50% by weight, such as about 0.1 % to about 25%, of a bleaching system. Any bleaching system known in the art for use in laundry detergents may be utilized. Suitable bleaching system components include bleaching catalysts, photobleaches, bleach activators, sources of hydrogen peroxide such as sodium percarbonate and sodium perborates, preformed peracids and mixtures thereof. Suitable preformed peracids include, but are not limited to, peroxycarboxylic acids and salts, percarbonic acids and salts, perimidic acids and salts, peroxymonosulfuric acids and salts, for example, Oxone (R), and mixtures thereof. Non-limiting examples of bleaching systems include peroxide-based bleaching systems, which may comprise, for example, an inorganic salt, including alkali metal salts such as sodium salts of perborate (usually mono- or tetra-hydrate), percarbonate, persulfate, perphosphate, persilicate salts, in combination with a peracid-forming bleach activator. The term bleach activator is meant herein as a compound which reacts with peroxygen bleach like hydrogen peroxide to form a peracid. The peracid thus formed constitutes the activated bleach. Suitable bleach activators to be used herein include those belonging to the class of esters amides, imides or anhydrides. Suitable examples are tetracetylethylene diamine (TAED), sodium 4-[(3,5,5-trimethylhexanoyl)oxy]benzene sulfonate (ISONOBS), diperoxy dodecanoic acid, 4-(dodecanoyloxy)benzenesulfonate (LOBS), 4-(decanoyloxy)benzenesulfonate, 4-(decanoyloxy)benzoate
(DOBS), 4-(nonanoyloxy)-benzenesulfonate (NOBS), and/or those disclosed in W098/17767. A particular family of bleach activators of interest was disclosed in EP624154 and particularly preferred in that family is acetyl triethyl citrate (ATC). ATC or a short chain triglyceride like triacetin has the advantage that it is environmental friendly as it eventually degrades into citric acid and alcohol. Furthermore, acetyl triethyl citrate and triacetin has a good hydrolytical stability in the product upon storage and it is an efficient bleach activator. Finally, ATC provides a good building capacity to the laundry additive. Alternatively, the bleaching system may comprise peroxyacids of, for example, the amide, imide, or sulfone type. The bleaching system may also comprise peracids such as 6-(phthalimido)peroxyhexanoic acid (PAP). The bleaching system may also include a bleach catalyst. In some embodiments the bleach component may be an organic catalyst selected from the group consisting of organic catalysts having the following formulae:
each R is independently a branched alkyl group containing from 9 to 24 carbons or linear alkyl group containing from 11 to 24 carbons, preferably each R is independently a branched alkyl group containing from 9 to 18 carbons or linear alkyl group containing from 1 1 to 18 carbons, more preferably each R is independently selected from the group consisting of 2-propylheptyl, 2-butyloctyl, 2-pentylnonyl, 2-hexyldecyl, n-dodecyl, n-tetradecyl, n-hexadecyl, n-octadecyl, iso-nonyl, iso-decyl, iso- tridecyl and iso-pentadecyl. Other exemplary bleaching systems are described, e.g. in WO2007/087258, WO2007/087244, WO2007/087259 and WO2007/087242. Suitable photobleaches may for example be sulfonated zinc phthalocyanine. Polymers
The detergent may comprise 0-10% by weight, such as 0.5-5%, 2-5%, 0.5-2% or 0.2-1 % of a polymer. Any polymer known in the art for use in detergents may be utilized. The polymer may function as a co- builder as mentioned above, or may provide anti-redeposition, fiber protection, soil release, dye transfer inhibition, grease cleaning and/or anti-foaming properties. Some polymers may have more than one of the above-mentioned properties and/or more than one of the below-mentioned motifs. Exemplary polymers include (carboxymethyl)cellulose (CMC), polyvinyl alcohol) (PVA), poly(vinylpyrrolidone) (PVP), poly(eth- yleneglycol) or poly(ethylene oxide) (PEG), ethoxylated poly(ethyleneimine), carboxymethyl inulin (CMI), and polycarboxylates such as PAA, PAA PMA, poly-aspartic acid, and lauryl methacrylate/acrylic acid copolymers, hydrophobically modified CMC (HM-CMC) and silicones, copolymers of terephthalic acid and oligomeric glycols, copolymers of poly(ethylene terephthalate) and poly(oxyethene terephthalate) (PET-
POET), PVP, poly(vinylimidazole) (PVI), poly(vinylpyridine-A -oxide) (PVPO or PVPNO) and polyvinylpyrrol- idone-vinylimidazole (PVPVI). Further exemplary polymers include sulfonated polycarboxylates, polyethylene oxide and polypropylene oxide (PEO-PPO) and diquaternium ethoxy sulfate. Other exemplary polymers are disclosed in, e.g., WO 2006/130575. Salts of the above-mentioned polymers are also contem- plated .
Fabric hueing agents
The detergent compositions of the present invention may also comprise fabric hueing agents such as dyes or pigments, which when formulated in detergent compositions can deposit onto a fabric when said fabric is contacted with a wash liguor comprising said detergent compositions and thus altering the tint of said fabric through absorption/reflection of visible light. Fluorescent whitening agents emit at least some visible light. In contrast, fabric hueing agents alter the tint of a surface as they absorb at least a portion of the visible light spectrum. Suitable fabric hueing agents include dyes and dye-clay conjugates, and may also include pigments. Suitable dyes include small molecule dyes and polymeric dyes. Suitable small mol- ecule dyes include small molecule dyes selected from the group consisting of dyes falling into the Colour Index (C.I.) classifications of Direct Blue, Direct Red, Direct Violet, Acid Blue, Acid Red, Acid Violet, Basic Blue, Basic Violet and Basic Red, or mixtures thereof, for example as described in WO2005/03274, WO2005/03275, WO2005/03276 and EP1876226 (hereby incorporated by reference). The detergent composition preferably comprises from about 0.00003 wt% to about 0.2 wt%, from about 0.00008 wt% to about 0.05 wt%, or even from about 0.0001 wt% to about 0.04 wt% fabric hueing agent. The composition may comprise from 0.0001 wt% to 0.2 wt% fabric hueing agent, this may be especially preferred when the composition is in the form of a unit dose pouch. Suitable hueing agents are also disclosed in, e.g. WO 2007/087257 and WO2007/087243. Additional Enzymes
The detergent additive as well as the detergent composition may comprise one or more [additional] enzymes such as a xanthan lyase, protease, lipase, cutinase, an amylase, lichenase, carbohydrase, cellu- lase, pectinase, mannanase, arabinase, galactanase, xylanase, oxidase, e.g., a laccase, and/or peroxidase.
In general, the properties of the selected enzyme(s) should be compatible with the selected detergent, (i.e., pH-optimum, compatibility with other enzymatic and non-enzymatic ingredients, etc.), and the en- zyme(s) should be present in effective amounts.
Cellulases
Suitable cellulases include those of bacterial or fungal origin. Chemically modified or protein engineered mutants are included. Suitable cellulases include cellulases from the genera Bacillus, Pseudomo-
nas, Humicola, Fusarium, Thielavia, Acremonium, e.g., the fungal cellulases produced from Humicola in- solens, Myceliophthora thermophila and Fusarium oxysporum disclosed in US 4,435,307, US 5,648,263, US 5,691 ,178, US 5,776,757 and WO 89/09259.
Especially suitable cellulases are the alkaline or neutral cellulases having color care benefits. Examples of such cellulases are cellulases described in EP 0 495 257, EP 0 531 372, WO 96/11262, WO 96/29397, WO 98/08940. Other examples are cellulase variants such as those described in WO 94/07998, EP 0 531 315, US 5,457,046, US 5,686,593, US 5,763,254, WO 95/24471 , WO 98/12307 and PCT/DK98/00299.
Example of cellulases exhibiting endo-beta-1 ,4-glucanase activity (EC 3.2.1.4) are those having described in WO02/099091 .
Other examples of cellulases include the family 45 cellulases described in W096/29397, and especially variants thereof having substitution, insertion and/or deletion at one or more of the positions corresponding to the following positions in SEQ ID NO: 8 of WO 02/099091 : 2, 4, 7, 8, 10, 13, 15, 19, 20, 21 , 25, 26, 29, 32, 33, 34, 35, 37, 40, 42, 42a, 43, 44, 48, 53, 54, 55, 58, 59, 63, 64, 65, 66, 67, 70, 72, 76, 79, 80, 82, 84, 86, 88, 90, 91 , 93, 95, 95d, 95h, 95j, 97, 100, 101 , 102, 103, 113, 114, 1 17, 119, 121 , 133, 136, 137, 138, 139, 140a, 141 , 143a, 145, 146, 147, 150e, 150j, 151 , 152, 153, 154, 155, 156, 157, 158, 159, 160c, 160e, 160k, 161 , 162, 164, 165, 168, 170, 171 , 172, 173, 175, 176, 178, 181 , 183, 184, 185, 186, 188, 191 , 192, 195, 196, 200, and/or 20, preferably selected among P19A, G20K, Q44K, N48E, Q1 19H or Q146 R.
Commercially available cellulases include Celluzyme™, and Carezyme™ (Novozymes A/S), Clazi- nase™, and Puradax HA™ (Genencor International Inc.), and KAC-500(B)™ (Kao Corporation).
Proteases
The additional enzyme may be another protease or protease variant. The protease may be of animal, vegetable or microbial origin, including chemically or genetically modified mutants. Microbial origin is preferred. It may be an alkaline protease, such as a serine protease or a metalloprotease. A serine protease may for example be of the S1 family, such as trypsin, or the S8 family such as subtilisin. A metalloproteases protease may for example be a thermolysin from e.g. family M4, M5, M7 or M8.
The term "subtilases" refers to a sub-group of serine protease according to Siezen et al., Protein Engng. 4 (1991 ) 719-737 and Siezen et al. Protein Science 6 (1997) 501 -523. Serine proteases are a subgroup of proteases characterized by having a serine in the active site, which forms a covalent adduct with the sub- strate. The subtilases may be divided into 6 sub-divisions, i.e. the Subtilisin family, the Thermitase family, the Proteinase K family, the Lantibiotic peptidase family, the Kexin family and the Pyrolysin family. In one aspect of the invention the protease may be a subtilase, such as a subtilisin or a variant hereof. Further the subtilases (and the serine proteases) are characterised by having two active site amino acid residues apart from the serine, namely a histidine and an aspartic acid residue.
Examples of subtilisins are those derived from Bacillus such as subtilisin lentus, Bacillus lentus, subtil-
isin Novo, subtilisin Carlsberg, Bacillus licheniformis, subtilisin BPN', subtilisin 309, subtilisin 147 and sub- tilisin 168 described in WO 89/06279 and protease PD138 (WO 93/18140). Additional serine protease examples are described in WO 98/0201 15, WO 01/44452, WO 01/58275, WO 01/58276, WO 03/006602 and WO 04/099401. An example of a subtilase variants may be those having mutations in any of the positions: 3, 4, 9, 15, 27, 36, 68, 76, 87, 95, 96, 97, 98, 99, 100, 101 , 102, 103, 104, 106, 118, 120, 123, 128, 129, 130, 160, 167, 170, 194, 195, 199, 205, 217, 218, 222, 232, 235, 236, 245, 248, 252 and 274 using the BPN' numbering. More preferred the subtilase variants may comprise the mutations: S3T, V4I, S9R, A15T, K27R, *36D, V68A, N76D, N87S.R, *97E, A98S, S99G,D,A, S99AD, S101 G,M,R S103A, V104I,Y,N, S106A, G118V.R, H120D.N, N123S, S128L, P129Q, S130A, G160D, Y167A, R170S, A194P, G195E, V199M, V205I, L217D, N218D, M222S, A232V, K235L, Q236H, Q245R, N252K, T274A (using BPN' numbering). A further preferred protease is the alkaline protease from Bacillus lentus DSM 5483, as described for example in WO 95/23221 , and variants thereof which are described in WO 92/21760, WO 95/23221 , EP 1921147 and EP 1921 148.
Examples of trypsin-like proteases are trypsin (e.g. of porcine or bovine origin) and the Fusarium pro- tease described in WO 89/06270 and WO 94/25583. Examples of useful proteases are the variants described in WO 92/19729, WO 98/201 15, WO 98/201 16, and WO 98/34946, especially the variants with substitutions in one or more of the following positions: 27, 36, 57, 76, 87, 97, 101 , 104, 120, 123, 167, 170, 194, 206, 218, 222, 224, 235, and 274.
Examples of metalloproteases are the neutral metalloprotease as described in WO 07/044993.
Preferred commercially available protease enzymes include Alcalase™, Coronase™, Duralase™, Du- razym™, Esperase™, Everlase™, Kannase™, Liquanase™, Liquanase Ultra™, Ovozyme™, Polarzyme™, Primase™, Relase™, Savinase™ and Savinase Ultra™, (Novozymes A/S), Axapem™ (Gist-Brocases N.V.), BLAP and BLAP X (Henkel AG & Co. KGaA), Excellase™, FN2™, FN3™, FN4™, Maxaca™, Maxapem™, Maxatase™, Properase™, Purafast™, Purafect™, Purafect OxP™, Purafect Prime™ and Puramax™ (Genencor int.).
Lipases and Cutinases
Suitable lipases and cutinases include those of bacterial or fungal origin. Chemically modified or protein engineered mutant enzymes are included. Examples include lipase from Thermomyces, e.g. from T. lanu- ginosus (previously named Humicola lanuginosa) as described in EP258068 and EP305216, cutinase from Humicola, e.g. H. insolens (WO96/13580), lipase from strains of Pseudomonas (some of these now renamed to Burkholderia), e.g. P. alcaligenes or P. pseudoalcaligenes (EP218272), P. cepacia (EP331376), P. sp. strain SD705 (WO95/06720 & WO96/27002), P. wisconsinensis (WO96/12012), GDSL-type Strepto- myces lipases (W010/065455), cutinase from Magnaporthe grisea (W010/107560), cutinase from Pseudo- monas mendocina (US5,389,536), lipase from Thermobifida fusca (W01 1 /084412), Geobacillus stearother-
mophilus lipase (W01 1/084417), lipase from Bacillus subtilis (W011/084599), and lipase from Streptomy- ces griseus (W01 1/150157) and S. pristinaespiralis (W012/137147).
Further examples are lipases sometimes referred to as acyltransferases or perhydrolases, e.g. acyl- transferases with homology to Candida antarctica lipase A (W010/1 11 143), acyltransferase from Mycobac- terium smegmatis (WO05/56782), perhydrolases from the CE 7 family (WO09/67279), and variants of the M. smegmatis perhydrolase in particular the S54V variant used in the commercial product Gentle Power Bleach from Huntsman Textile Effects Pte Ltd (W010/100028).
Other examples are lipase variants such as those described in EP407225, WO92/05249, WO94/01541 , W094/25578, W095/14783, WO95/30744, W095/35381 , W095/22615, WO96/00292, WO97/04079, WO97/07202, WO00/34450, WO00/60063, WO01/92502, WO07/87508 and WO09/109500.
Preferred commercial lipase products include Lipolase™, Lipex™; Lipolex™ and Lipoclean™ (Novo- zymes A/S), Lumafast (originally from Genencor) and Lipomax (originally from Gist-Brocades).
Amylases
The amylase may be an alpha-amylase, a beta-amylase or a glucoamylase and may be of bacterial or fungal origin. Chemically modified or protein engineered mutants are included. Amylases include, for example, alpha-amylases obtained from Bacillus, e.g. , a special strain of Bacillus licheniformis, described in more detail in GB 1 ,296,839.
Examples of amylases are those having SEQ ID NO: 3 in WO 95/10603 or variants having 90% se- quence identity to SEQ ID NO: 3 thereof. Preferred variants are described in WO 94/02597, WO 94/18314, WO 97/43424 and SEQ ID NO: 4 of WO 99/019467, such as variants with substitutions in one or more of the following positions: 15, 23, 105, 106, 124, 128, 133, 154, 156, 178, 179, 181 , 188, 190, 197, 201 , 202, 207, 208, 209, 21 1 , 243, 264, 304, 305, 391 , 408, and 444 of SEQ ID NO: 3 in WO 95/10603.
Other amylases are variants of SEQ ID NO: 1 of WO 2016/203064 having at least 75% sequence identity to SEQ ID NO: 1 thereof. Preferred variants are variants comprising a modification in one or more positions corresponding to positions 1 , 54, 56, 72, 109, 113, 1 16, 134, 140, 159, 167, 169, 172, 173, 174, 181 , 182, 183, 184, 189, 194, 195, 206, 255, 260, 262, 265, 284, 289, 304, 305, 347, 391 , 395, 439, 469, 444, 473, 476, or 477 of SEQ ID NO: 1 , wherein said alpha-amylase variant has a sequence identity of at least 75% but less than 100% to SEQ ID NO: 1 .
Further amylases which can be used are amylases having SEQ ID NO: 6 in WO 02/010355 or variants thereof having 90% sequence identity to SEQ ID NO: 6. Preferred variants of SEQ ID NO: 6 are those having a deletion in positions 181 and 182 and a substitution in position 193.
Other amylase examples are hybrid alpha-amylase comprising residues 1-33 of the alpha-amylase derived from B. amyloliquefaciens shown in SEQ ID NO: 6 of WO 2006/066594 and residues 36-483 of the B. licheniformis alpha-amylase shown in SEQ ID NO: 4 of WO 2006/066594 or variants having 90% sequence identity thereof. Preferred variants of this hybrid alpha-amylase are those having a substitution, a
deletion or an insertion in one of more of the following positions: G48, T49, G107, H156, A181 , N190, M197, 1201 , A209 and Q264. Most preferred variants of the hybrid alpha-amylase comprising residues 1-33 of the alpha-amylase derived from B. amyloliquefaciens shown in SEQ ID NO: 6 of WO 2006/066594 and residues 36-483 of SEQ ID NO: 4 are those having the substitutions:
M197T;
H156Y+A181T+N190F+A209V+Q264S; or
G48+T49+G107+H156+A181 +N190+1201 +A209+Q264.
Further amylase examples are amylases having SEQ ID NO: 6 in WO 99/019467 or variants thereof having 90% sequence identity to SEQ ID NO: 6. Preferred variants of SEQ ID NO: 6 are those having a substitution, a deletion or an insertion in one or more of the following positions: R181 , G182, H183, G184, N195, I206, E212, E216 and K269. Particularly preferred amylases are those having deletion in positions G182 and H183 or positions H183 and G184.
Additional amylases are those having SEQ ID NO: 1 , SEQ ID NO: 2 or SEQ ID NO: 7 of WO 96/023873 or variants thereof having 90% sequence identity to SEQ ID NO: 1 , SEQ ID NO: 2 or SEQ ID NO: 7. Pre- ferred variants of SEQ ID NO: 1 , SEQ ID NO: 2 or SEQ ID NO: 7 are those having a substitution, a deletion or an insertion in one or more of the following positions: 140, 181 , 182, 183, 184, 195, 206, 212, 243, 260, 269, 304 and 476. More preferred variants are those having a deletion in positions 182 and 183 or positions 183 and 184. Most preferred amylase variants of SEQ ID NO: 1 , SEQ ID NO: 2 or SEQ ID NO: 7 are those having a deletion in positions 183 and 184 and a substitution in positions 140, 195, 206, 243, 260, 304 and 476.
Other amylases which can be used are amylases having SEQ ID NO: 2 of WO 08/153815, SEQ ID NO: 10 in WO 01/66712 or variants thereof having 90% sequence identity to SEQ ID NO: 2 of WO 08/153815 or 90% sequence identity to SEQ ID NO: 10 in WO 01/66712. Preferred variants of SEQ ID NO: 10 in WO 01/66712 are those having a substitution, a deletion or an insertion in one of more of the following positions: 176, 177, 178, 179, 190, 201 , 207, 211 and 264.
Further amylases which can be used are amylases having SEQ ID NO: 2 of WO 09/061380 or variants thereof having 90% sequence identity to SEQ ID NO: 2. Preferred variants of SEQ ID NO: 2 are those having a substitution, a deletion or an insertion in one of more of the following positions: Q87, Q98, S125, N128, T131 , T165, K178, R180, S181 , T182, G183, M201 , F202, N225, S243, N272, N282, Y305, R309, D319, Q320, Q359, K444 and G475. More preferred variants of SEQ ID NO: 2 are those having the substitution in one of more of the following positions: Q87E.R, Q98R, S125A, N128C, T131 I, T165I, K178L, T182G, M201 L, F202Y, N225E.R, N272E.R, S243Q,A,E,D, Y305R, R309A, Q320R, Q359E, K444E and G475K and/or deletion in position R180 and/or S181. Most preferred amylase variants of SEQ ID NO: 2 are those having the substitutions: N128C+K178L+T182G+Y305R+G475K; N128C+K178L+T182G+F202Y+Y305R+D319T+G475K; S125A+N 128C+K178L+T182G+Y305R+G475K;
or S125A+N 128C+T1311+T165I+K178L+T182G+Y305R+G475K wherein the variant optionally further comprises a substitution at position 243 and/or a deletion at position 180 and/or position 181.
Other examples of amylases are the alpha-amylase having SEQ ID NO: 12 in WO01/66712 or a variant having at least 90%, such as at least 95%, sequence identity to SEQ ID NO: 12. Preferred amylase variants are those having a substitution, a deletion or an insertion in one of more of the following positions of SEQ ID NO: 12 in WO01/66712: R28, R118, N174; R181 , G182, D183, G184, G186, W189, N195, M202, Y298, N299, K302, S303, N306, R310, N314; R320, H324, E345, Y396, R400, W439, R444, N445, K446, Q449, R458, N471 , N484. Particular preferred amylases include variants having a deletion of D183 and G184 and having the substitutions R1 18K, N195F, R320K and R458K, and a variant additionally having substitutions in one or more position selected from the group: M9, G149, G182, G186, M202, T257, Y295, N299, M323, E345 and A339, most preferred a variant that additionally has substitutions in all these positions.
Commercially available amylases are Duramyl™, Termamyl™, Fungamyl™, Stainzyme™, Stainzyme Plus™, Natalase™ and BAN™ (Novozymes A S), Rapidase™ and Purastar™ (from Genencor International Inc.).
Peroxidases/Oxidases
Suitable peroxidases/oxidases include those of plant, bacterial or fungal origin. Chemically modified or protein engineered mutants are included. Examples of useful peroxidases include peroxidases from Coprinus, e.g. , from C. cinereus, and variants thereof as those described in WO 93/24618, WO 95/10602, and WO 98/15257.
Commercially available peroxidases include Guardzyme™ (Novozymes A/S).
The detergent enzyme(s) may be included in a detergent composition by adding separate additives containing one or more enzymes, or by adding a combined additive comprising all of these enzymes. A detergent additive of the invention, i.e., a separate additive or a combined additive, can be formulated, for example, as a granulate, liquid, slurry, etc. Preferred detergent additive formulations are granulates, in particular non-dusting granulates, liquids, in particular stabilized liquids, or slurries.
Non-dusting granulates may be produced, e.g., as disclosed in US 4,106,991 and 4,661 ,452 and may optionally be coated by methods known in the art. Examples of waxy coating materials are poly(ethylene oxide) products (polyethyleneglycol, PEG) with mean molar weights of 1000 to 20000; ethoxylated nonylphenols having from 16 to 50 ethylene oxide units; ethoxylated fatty alcohols in which the alcohol contains from 12 to 20 carbon atoms and in which there are 15 to 80 ethylene oxide units; fatty alcohols; fatty acids; and mono- and di- and triglycerides of fatty acids. Examples of film-forming coating materials suitable for application by fluid bed techniques are given in GB 1483591 . Liquid enzyme preparations may, for instance, be stabilized by adding a polyol such as propylene glycol, a sugar or sugar alcohol, lactic acid or boric acid according to established methods. Protected enzymes may be prepared according to the method disclosed in EP 238,216.
Adjunct materials
Any detergent components known in the art for use in laundry detergents may also be utilized. Other optional detergent components include anti-corrosion agents, anti-shrink agents, anti-soil redeposition agents, anti-wrinkling agents, bactericides, binders, corrosion inhibitors, disintegrants/disintegration agents, dyes, enzyme stabilizers (including boric acid, borates, CMC, and/or polyols such as propylene glycol), fabric conditioners including clays, fillers/processing aids, fluorescent whitening agents/optical brighteners, foam boosters, foam (suds) regulators, perfumes, soil-suspending agents, softeners, suds suppressors, tarnish inhibitors, and wicking agents, either alone or in combination. Any ingredient known in the art for use in laundry detergents may be utilized. The choice of such ingredients is well within the skill of the artisan.
Dispersants: The detergent compositions of the present invention can also contain dispersants. In particular powdered detergents may comprise dispersants. Suitable water-soluble organic materials include the homo- or co-polymeric acids or their salts, in which the polycarboxylic acid comprises at least two car- boxyl radicals separated from each other by not more than two carbon atoms. Suitable dispersants are for example described in Powdered Detergents, Surfactant science series volume 71 , Marcel Dekker, Inc.
Dye Transfer Inhibiting Agents: The detergent compositions of the present invention may also include one or more dye transfer inhibiting agents. Suitable polymeric dye transfer inhibiting agents include, but are not limited to, polyvinylpyrrolidone polymers, polyamine N-oxide polymers, copolymers of N-vinylpyrrolidone and N-vinylimidazole, polyvinyloxazolidones and polyvinylimidazoles or mixtures thereof. When present in a subject composition, the dye transfer inhibiting agents may be present at levels from about 0.0001 % to about 10%, from about 0.01 % to about 5% or even from about 0.1 % to about 3% by weight of the composition.
Fluorescent whitening agent: The detergent compositions of the present invention will preferably also contain additional components that may tint articles being cleaned, such as fluorescent whitening agent or optical brighteners. Where present the brightener is preferably at a level of about 0,01 % to about 0,5%. Any fluorescent whitening agent suitable for use in a laundry detergent composition may be used in the composition of the present invention. The most commonly used fluorescent whitening agents are those belonging to the classes of diaminostilbene-sulphonic acid derivatives, diarylpyrazoline derivatives and bisphenyl-dis- tyryl derivatives. Examples of the diaminostilbene-sulphonic acid derivative type of fluorescent whitening agents include the sodium salts of: 4,4'-bis-(2-diethanolamino-4-anilino-s-triazin-6-ylamino)-stilbene-2,2'-di- sulphonate; 4,4'-bis-(2,4-dianilino-s-triazin-6-ylamino)-stilbene-2,2'-disulphonate; 4,4'-bis-(2-anilino-4(N- methyl-N-2-hydroxy-ethylamino)-s-triazin-6-ylamino)-stilbene-2,2'-disulphonate, 4,4'-bis-(4-phenyl-2,1 ,3-tri- azol-2-yl)-stilbene-2,2'-disulphonate; 4,4'-bis-(2-anilino-4-(1-methyl-2-hydroxy-ethylamino)-s-triazin-6-yla- mino)-stilbene-2,2'-disulphonate and 2-(stilbyl-4"-naptho-1 ,2':4,5)-1 ,2,3-trizole-2"-sulphonate. Preferred fluorescent whitening agents are Tinopal DMS and Tinopal CBS available from Ciba-Geigy AG, Basel, Swit- zerland. Tinopal DMS is the disodium salt of 4,4'-bis-(2-morpholino-4 anilino-s-triazin-6-ylamino)-stilbene disulphonate. Tinopal CBS is the disodium salt of 2,2'-bis-(phenyl-styryl) disulphonate. Also preferred are
fluorescent whitening agents is the commercially available Parawhite KX, supplied by Paramount Minerals and Chemicals, Mumbai, India. Other fluorescers suitable for use in the invention include the 1 -3-diaryl pyrazolines and the 7-alkylaminocoumarins. Suitable fluorescent brightener levels include lower levels of from about 0.01 , from 0.05, from about 0.1 or even from about 0.2 wt % to upper levels of 0.5 or even 0.75 wt%.
Soil release polymers: The detergent compositions of the present invention may also include one or more soil release polymers which aid the removal of soils from fabrics such as cotton and polyester based fabrics, in particular the removal of hydrophobic soils from polyester based fabrics. The soil release polymers may for example be non-ionic or anionic terephthalate-based polymers, polyvinyl caprolactam and related copolymers, vinyl graft copolymers, polyester polyamides see for example Chapter 7 in Powdered Detergents, Surfactant science series volume 71 , Marcel Dekker, Inc. Another type of soil release polymers are amphiphilic alkoxylated grease cleaning polymers comprising a core structure and a plurality of alkox- ylate groups attached to that core structure. The core structure may comprise a polyalkylenimine structure or a polyalkanolamine structure as described in detail in WO 2009/087523 (hereby incorporated by refer- ence). Furthermore, random graft co-polymers are suitable soil release polymers Suitable graft co-polymers are described in more detail in WO 2007/138054, WO 2006/108856 and WO 2006/1 13314 (hereby incorporated by reference). Other soil release polymers are substituted polysaccharide structures especially substituted cellulosic structures such as modified cellulose derivatives such as those described in EP 1867808 or WO 2003/040279 (both are hereby incorporated by reference). Suitable cellulosic polymers include cel- lulose, cellulose ethers, cellulose esters, cellulose amides and mixtures thereof. Suitable cellulosic polymers include anionically modified cellulose, nonionically modified cellulose, cationically modified cellulose, zwit- terionically modified cellulose, and mixtures thereof. Suitable cellulosic polymers include methyl cellulose, carboxy methyl cellulose, ethyl cellulose, hydroxyl ethyl cellulose, hydroxyl propyl methyl cellulose, ester carboxy methyl cellulose, and mixtures thereof.
Anti-redeposition agents: The detergent compositions of the present invention may also include one or more anti-redeposition agents such as carboxymethylcellulose (CMC), polyvinyl alcohol (PVA), polyvinylpyrrolidone (PVP), polyoxyethylene and/or polyethyleneglycol (PEG), homopolymers of acrylic acid, copolymers of acrylic acid and maleic acid, and ethoxylated polyethyleneimines. The cellulose-based polymers described under soil release polymers above may also function as anti-redeposition agents.
Other suitable adjunct materials include, but are not limited to, anti-shrink agents, anti-wrinkling agents, bactericides, binders, carriers, dyes, enzyme stabilizers, fabric softeners, fillers, foam regulators, hy- drotropes, perfumes, pigments, sod suppressors, solvents, and structurants for liquid detergents and/or structure elasticizing agents. Formulation of detergent products
The detergent composition may be in any convenient form, e.g., a bar, a homogenous tablet, a tablet
having two or more layers, a pouch having one or more compartments, a regular or compact powder, a granule, a paste, a gel, or a regular, compact or concentrated liquid. There are a number of detergent formulation forms such as layers (same or different phases), pouches, as well as forms for machine dosing unit.
Pouches can be configured as single or multicompartments. It can be of any form, shape and material which is suitable for hold the composition, e.g. without allowing the release of the composition from the pouch prior to water contact. The pouch is made from water soluble film which encloses an inner volume. Said inner volume can be divided into compartments of the pouch. Preferred films are polymeric materials preferably polymers which are formed into a film or sheet. Preferred polymers, copolymers or derivates therof are selected polyacrylates, and water-soluble acrylate copolymers, methyl cellulose, carboxy methyl cellulose, sodium dextrin, ethyl cellulose, hydroxyethyl cellulose, hydroxypropyl methyl cellulose, malto dextrin, poly methacrylates, most preferably polyvinyl alcohol copolymers and, hydroxypropyl methyl cellulose (HPMC). Preferably, the level of polymer in the film for example PVA is at least about 60%. Preferred average molecular weight will typically be about 20,000 to about 150,000. Films can also be of blend compositions comprising hydrolytically degradable and water-soluble polymer blends such as polyactide and polyvinyl alcohol (known under the Trade reference M8630 as sold by Chris Craft In. Prod. Of Gary, Ind., US) plus plasticisers like glycerol, ethylene glycerol, Propylene glycol, sorbitol and mixtures thereof. The pouches can comprise a solid laundry cleaning composition or part components and/or a liquid cleaning composition or part components separated by the water-soluble film. The compartment for liquid components can be different in composition than compartments containing solids. Ref: (US2009/0011970 A1 ).
Detergent ingredients can be separated physically from each other by compartments in water dissolvable pouches or in different layers of tablets. Thereby negative storage interaction between components can be avoided. Different dissolution profiles of each of the compartments can also give rise to delayed dissolution of selected components in the wash solution.
A liquid or gel detergent, which is not unit dosed, may be aqueous, typically containing at least 20% by weight and up to 95% water, such as up to about 70% water, up to about 65% water, up to about 55% water, up to about 45% water, up to about 35% water. Other types of liquids, including without limitation, alkanols, amines, diols, ethers and polyols may be included in an aqueous liquid or gel. An aqueous liquid or gel detergent may contain from 0-30% organic solvent. A liquid or gel detergent may be non-aqueous.
Laundry Soap Bars
The enzymes of the invention may be added to laundry soap bars and used for hand washing laundry, fabrics and/or textiles. The term laundry soap bar includes laundry bars, soap bars, combo bars, syndet bars and detergent bars. The types of bar usually differ in the type of surfactant they contain, and the term laundry soap bar includes those containing soaps from fatty acids and/or synthetic soaps. The laundry soap bar has a physical form which is solid and not a liquid, gel or a powder at room temperature. The term solid
is defined as a physical form which does not significantly change over time, i.e. if a solid object (e.g. laundry soap bar) is placed inside a container, the solid object does not change to fill the container it is placed in. The bar is a solid typically in bar form but can be in other solid shapes such as round or oval.
The laundry soap bar may contain one or more additional enzymes, protease inhibitors such as peptide aldehydes (or hydrosulfite adduct or hemiacetal adduct), boric acid, borate, borax and/or phenylboronic acid derivatives such as 4-formylphenylboronic acid, one or more soaps or synthetic surfactants, polyols such as glycerine, pH controlling compounds such as fatty acids, citric acid, acetic acid and/or formic acid, and/or a salt of a monovalent cation and an organic anion wherein the monovalent cation may be for example Na+, K+ or NHV and the organic anion may be for example formate, acetate, citrate or lactate such that the salt of a monovalent cation and an organic anion may be, for example, sodium formate.
The laundry soap bar may also contain complexing agents like EDTA and HEDP, perfumes and/or different type of fillers, surfactants e.g. anionic synthetic surfactants, builders, polymeric soil release agents, detergent chelators, stabilizing agents, fillers, dyes, colorants, dye transfer inhibitors, alkoxylated polycarbonates, suds suppressers, structurants, binders, leaching agents, bleaching activators, clay soil removal agents, anti-redeposition agents, polymeric dispersing agents, brighteners, fabric softeners, perfumes and/or other compounds known in the art.
The laundry soap bar may be processed in conventional laundry soap bar making equipment such as but not limited to: mixers, plodders, e.g. a two-stage vacuum plodder, extruders, cutters, logo-stampers, cooling tunnels and wrappers. The invention is not limited to preparing the laundry soap bars by any single method. The premix of the invention may be added to the soap at different stages of the process. For example, the premix containing a soap, an enzyme, optionally one or more additional enzymes, a protease inhibitor, and a salt of a monovalent cation and an organic anion may be prepared, and the mixture is then plodded. The enzyme and optional additional enzymes may be added at the same time as the protease inhibitor for example in liquid form. Besides the mixing step and the plodding step, the process may further comprise the steps of milling, extruding, cutting, stamping, cooling and/or wrapping.
Method of producing the composition
The present invention also relates to methods of producing the composition. The method may be relevant for the (storage) stability of the detergent composition: e.g. Soap bar premix method WO2009155557.
Uses
The present invention is also directed to methods for using the detergent compositions thereof. The present invention may be used for example in any detergent application which requires the degradation of xanthan gum.
Use to degrade xanthan gum
Xanthan gum has been used as an ingredient in many consumer products including foods and cosmetics and has found use in the oil industry. Therefore, the degradation of xanthan gum can result in improved cleaning processes, such as the easier removal of stains containing gums, such as xanthan gum. Thus, the present invention is directed to the use of detergent compositions comprising GH9 endoglu- canases (e.g. variants described herein) of the invention to degrade xanthan gum. The present invention is also directed to the use of xanthan lyases in the detergent compositions to degrade xanthan gum. An embodiment is the use of GH9 endoglucanases as described herein (e.g. variants) together with xanthan lyases in detergent compositions to degrade xanthan gum. Degradation of xanthan gum can preferably be meas- ured using the viscosity reduction assay (e.g., ViPr assay) or alternatively as described in example 3 herein.
GH9 endoglucanase activity may alternatively be measured by assessment of reducing ends on xanthan gum pre-treated with xanthan lyase using the colorimetric assay developed by Lever (1972), Anal. Biochem. 47: 273-279, 1972. A preferred embodiment is the use of 0.1 % xanthan gum pre-treated with xanthan lyase. Degradation of xanthan gum pre-treated with xanthan lyase may be determined by calculat- ing difference between blank and sample, wherein a difference of more than 0.5 mAU, preferably more than 0.6 mAU, more preferably more than 0.7 mAU or even more preferably more than 0.8 mAU, shows degradation of xanthan gum pre-treated with xanthan lyase.
Xanthan lyase activity may alternatively be measured by assessment of reducing ends liberated from xanthan gum using the colorimetric assay developed by Lever (1972), Anal. Biochem. 47: 273-279, 1972. A preferred embodiment is the use of 0.1 % xanthan gum. Degradation of xanthan gum may be determined by calculating difference between blank and sample wherein a difference of more than 0.1 mAU, preferably more than 0.15 mAU, more preferably more than 0.2 mAU or even more preferably more than 0.25 mAU shows degradation of xanthan gum.
GH9 endoglucanase (e.g. variants of the present invention) and xanthan lyase activity may alternatively be measured by assessment of reducing ends liberated from xanthan gum using the colorimetric assay developed by Lever (1972), Anal. Biochem. 47: 273-279, 1972. A preferred embodiment is the use of 0.1 % xanthan gum. Degradation of xanthan gum may be determined by calculating difference between blank and sample wherein a difference of more than 0.4 mAU, preferably more than 0.5 mAU, more preferably more than 0.6 mAU or even more preferably more than 0.8 mAU shows degradation of xanthan gum.
The invention also relates to methods for degrading xanthan gum comprising applying a detergent composition comprising one or more GH9 endoglucanases as described herein (e.g. variants) to xanthan gum. An embodiment is a method for degrading xanthan gum comprising applying a detergent composition comprising one or more GH9 endoglucanases as described herein (e.g. variants) together with one or more xanthan lyases to xanthan gum.
Use in detergents
The present invention inter alia relates to the use of GH9 endoglucanases as described herein (e.g. variants of the present invention) in detergent compositions for cleaning processes such as the laundering of textiles and fabrics (e.g. household laundry washing and industrial laundry washing), as well as household and industrial hard surface cleaning, such as dish wash. The GH9 endoglucanases as described herein (e.g. variants of the present invention) may be added to a detergent composition comprising of one or more detergent components.
In some aspects, GH9 endoglucanases as described herein (e.g. variants of the present invention) may be used together with a xanthan lyase(s) in detergent compositions for cleaning processes such as the laundering of textiles and fabrics (e.g. household laundry washing and industrial laundry washing), as well as household and industrial hard surface cleaning, such as dish wash. The GH9 endoglucanases as described herein (e.g. variants of the present invention) together with a xanthan lyase(s) may be added to a detergent composition comprising of one or more detergent components.
The polypeptides described herein (e.g. variants) may be added to and thus become a component of a detergent composition. The detergent composition may be formulated , for example, as a hand or machine laundry detergent composition for both household and industrial laundry cleaning, including a laundry additive composition suitable for pre-treatment of stained fabrics and a rinse added fabric softener composition, or be formulated as a detergent composition for use in general household or industrial hard surface cleaning operations, or be formulated for hand or machine (both household and industrial) dishwashing operations. In a specific aspect, the present invention relates to a detergent additive comprising a polypeptide of the present invention as described herein.
The invention also relates to methods for degrading xanthan gum on the surface of a textile or hard surface, such as dish wash, comprising applying a detergent composition comprising one or more GH9 endoglucanases as described herein (e.g. variants) to xanthan gum. In some aspects, the invention relates a method for degrading xanthan gum on the surface of a textile or hard surface, such as dish wash, comprising applying a detergent composition comprising one or more GH9 endoglucanases as described herein (e.g. variants) together with one or more xanthan lyases to xanthan gum. In some aspects, the invention relates to a detergent composition comprising one or more detergent components as described herein. Also encompassed is the use of GH9 endoglucanases (e.g. variants of the present invention) having an enzyme detergency benefit in the detergent compositions of the invention.
It has been contemplated that the use of a GH9 endoglucanase as described herein (e.g., a) alone gives an enzyme detergency benefit, preferably an enzyme detergency benefit on xanthan gum.
In some aspects, the invention relates to the use of a detergent composition comprising one or more detergent components and an isolated GH9 endoglucanase as described herein (e.g. a variant) together with a xanthan lyase. In some aspects, the invention relates to the use of a detergent composition comprising one or more detergent components and an isolated GH9 endoglucanase (e.g. a variant) together with a
xanthan lyase.
The invention is further defined in the following paragraphs:
1. A detergent composition comprising an endoglucanase variant, comprising an alteration (e.g., a sub- stitution, deletion or insertion) at one or more positions in a region selected from the group consisting of:
i) region 1 corresponding to amino acids 95 to 105 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 95, 96, 97, 98, 99, 100, 101 , 102, 103, 104, 105, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the num- bering of SEQ ID NO: 2),
ii) region 2 corresponding to amino acids 115 to 138 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 115, 116, 1 17, 118, 1 19, 120, 121 , 122, 123, 124, 125, 126, 127, 128, 129, 130, 131 , 132, 133, 134, 135, 136, 137, 138, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2), iii) region 3 corresponding to amino acids 210 to 251 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 210, 21 1 , 212, 213, 214, 215, 216, 217, 218, 219, 220, 221 , 222, 223, 224, 225, 226, 227, 228, 229, 230, 231 , 232, 233, 234, 235, 236, 237, 238, 239, 240, 241 , 242, 243, 244, 245, 246, 247, 248, 249, 250, 251 , wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
iv) region 4 corresponding to amino acids 267 to 301 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 267, 268, 269, 270, 271 , 272, 273, 274, 275, 276, 277, 278, 279, 280, 281 , 282, 283, 284, 285, 286, 287, 288, 289, 290, 291 , 292, 293, 294, 295, 296, 297, 298, 299, 300, 301 , wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
v) region 5 corresponding to amino acids 339 to 361 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 339, 340, 341 , 342, 343, 344, 345, 346, 347, 348, 349, 350, 351 , 352, 353, 354, 355, 356, 357, 358, 359, 360, 361 , wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2), vi) region 6 corresponding to amino acids 547 to 595 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 547, 548, 549, 550, 551 , 552, 553, 554, 555,
556, 557, 558, 559, 560, 561 , 562, 563, 564, 565, 566, 567, 568, 569, 570, 571 , 572, 573, 574, 575, 576, 577, 578, 579, 580, 581 , 582, 583, 584, 585, 586, 587, 588, 589, 590, 591 , 592, 593, 594, 595, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
vii) region 7 corresponding to amino acids 612 to 660 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 612, 613, 614, 615, 616, 617, 618, 619, 620,
621 , 622, 623, 624, 625, 626, 627, 628, 629, 630, 631 , 632, 633, 634, 635, 636, 637, 638, 639, 640, 641 , 642, 643, 644, 645, 646, 647, 648, 649, 650, 651 , 652, 653, 654, 655, 656, 657, 658, 659, 660, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
viii) region 8 corresponding to amino acids 806 to 828 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 806, 807, 808, 809, 810, 81 1 , 812, 813, 814, 815, 816, 817, 818, 819, 820, 821 , 822, 823, 824, 825, 826, 827, 828, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2), and ix) region 9 corresponding to amino acids 839 to 1042 of SEQ ID NO: 2, e.g., said alteration at one or more positions selected from the group consisting of positions: 839, 840, 841 , 842, 843, 844, 845, 846, 847, 848, 849, 850, 851 , 852, 853, 854, 855, 856, 857, 858, 859, 860, 861 , 862, 863, 864, 865, 866, 867, 868, 869, 870, 871 , 872, 873, 874, 875, 876, 877, 878, 879, 880, 881 , 882, 883, 884, 885, 886, 887, 888, 889, 890, 891 , 892, 893, 894, 895, 896, 897, 898, 899, 900, 901 , 902, 903, 904, 905, 906, 907, 908, 909, 910, 911 , 912, 913, 914, 915, 916, 917, 918, 919, 920, 921 , 922, 923, 924, 925, 926, 927, 928, 929, 930, 931 , 932, 933, 934, 935, 936, 937, 938, 939, 940, 941 , 942, 943, 944, 945, 946, 947, 948, 949, 950, 951 , 952, 953, 954, 955, 956, 957, 958, 959, 960, 961 , 962, 963, 964, 965, 966, 967, 968, 969, 970, 971 , 972, 973, 974, 975, 976, 977, 978, 979, 980, 981 , 982, 983, 984, 985, 986, 987, 988, 989, 990, 991 , 992, 993, 994, 995, 996, 997, 998, 999, 1000, 1001 , 1002, 1003, 1004, 1005, 1006, 1007, 1008, 1009, 1010, 101 1 , 1012, 1013, 1014, 1015, 1016, 1017, 1018, 1019, 1020, 1021 , 1022, 1023, 1024, 1025, 1026, 1027, 1028, 1029, 1030, 1031 , 1032, 1033, 1034, 1035, 1036, 1037, 1038, 1039, 1040, 1041 , 1042, wherein said positions correspond to amino acid positions of SEQ ID NO: 2 (e.g., using the numbering of SEQ ID NO: 2),
wherein said variant has at least 60%, e.g., at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99%, and less than 100% sequence identity to SEQ ID NO: 2; preferably said endoglucanase variant has activity on xanthan gum pre-treated with xanthan lyase, further preferably said activity is a xanthan gum degrading activity.
2. The detergent composition comprising an endoglucanase variant of paragraph 1 , which is a variant of a parent endoglucanase selected from the group consisting of:
a) a polypeptide having at least 60% sequence identity to the mature polypeptide of SEQ ID NO: 2; b) a polypeptide encoded by a polynucleotide that hybridizes under low stringency conditions with (i) the mature polypeptide coding sequence of SEQ ID NO: 1 , or (ii) the full-length complement of (i); c) a polypeptide encoded by a polynucleotide having at least 60% identity to the mature polypeptide coding sequence of SEQ ID NO: 1 ; and
d) a fragment of the mature polypeptide of SEQ ID NO: 2, which has endoglucanase activity.
3. The detergent composition comprising an endoglucanase variant of paragraph 2, wherein the parent
endoglucanase having at least 60%, e.g., at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99% or 100% sequence identity to the mature polypeptide of SEQ ID NO: 2.
4. The detergent composition comprising an endoglucanase variant of any of paragraphs 2-3, wherein the parent endoglucanase is encoded by a polynucleotide that hybridizes under low stringency conditions, medium stringency conditions, medium-high stringency conditions, high stringency conditions, or very high stringency conditions with (i) the mature polypeptide coding sequence of SEQ ID NO: 1 or (ii) the full-length complement of (i).
5. The detergent composition comprising an endoglucanase variant of any of paragraphs 2-4, wherein the parent endoglucanase is encoded by a polynucleotide having at least 60%, e.g., at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% sequence identity to the mature polypeptide coding sequence of SEQ ID NO: 1 .
6. The detergent composition comprising an endoglucanase variant of any of paragraphs 2-5, wherein the parent endoglucanase comprises or consists of the mature polypeptide of SEQ ID NO: 2.
7. The detergent composition comprising an endoglucanase variant of any of paragraphs 2-6, wherein the parent endoglucanase is a fragment of the mature polypeptide of SEQ ID NO: 2, wherein the fragment has endoglucanase activity.
8. The detergent composition comprising an endoglucanase variant of any of paragraphs 2-7, which has at least 60%, e.g., at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95% identity, at least 96%, at least 97%, at least 98%, or at least 99%, but less than 100%, sequence identity to the amino acid sequence of the parent endoglucanase.
9. The detergent composition comprising an endoglucanase variant of any of paragraphs 1 -8, wherein said variant has at least 61 %, at least 62%, at least 63%, at least 64%, at least 65%, at least 66%, at least 67%, at least 68%, at least 69%, at least 70%, at least 71 %, at least 72%, at least 73%, at least
74%, at least 75%, at least 76%, at least 77%, at least 78%, at least 79%, at least 80%, at least 81 %, at least 82%, at least 83%, at least 84%, at least 85%, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to SEQ ID NO: 2.
10. The detergent composition comprising an endoglucanase variant of any of paragraphs 1 -9, wherein said alteration at one or more positions is selected from the group consisting of alterations in positions: 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 311 , 313, 387, 388, 390, 403, 408, 410, 416, 448, 451 , 471 , 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 603, 605, 609, 676, 688, 690, 694, 697, 698, 699, 711 , 719, 754, 756, 760, 781 , 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
11 . The detergent composition comprising an endoglucanase variant of any of paragraphs 1-10, wherein
said alteration at one or more positions is selected from the group consisting of: S17A, F20P, F20N, F20G, F20Y, K51 Q, K51 H, E53P, E53G, Y55M, V56M, Y60F, S63F, T87R, A186P, K192N, I302D, I302H, I302V, I302M, H311 N, S313D, I387T, K388R, K390Q, I403Y, E408D, E408S, E408P, E408A, E408G, E408N, P410G, Q416S, Q416D, A448E, A448W, A448S, K451 S, G471 S, S472Y, D476R, Q489P, K507R, K512P, S515V, S538C, Y579W, S598Q, A599S, I602T, I602D, V603P, S605T, G609E, D676H, A688G, Y690F, T694A, T697G, R698W, T699A, T71 1V, T71 1Y, W719R, K754R, V756H, V756Y, S760G, T781 M, N786K, T797S, A824D, N833D, Q834E, S835D and F1048W of SEQ ID NO: 2.
The detergent composition comprising an endoglucanase variant according to any one of paragraphs 1-11 , wherein the variant further comprises an alteration at one or more positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651 , 824, 848, 869, 880, 881 , 883, 887, 892, 905, 906, 912, 921 , 928, 934, 937, 948, 956, 999 and 1037.
The detergent composition comprising an endoglucanase variant according to any one of paragraphs 1-12, further wherein said alteration at one or more positions is selected from the group consisting of: N216D, N216Q, A283D, A346D, A559P, A559N, A564E, Y579W, S636K, S636N, F638N, A651 P, A824D, N848D, A869V, R880K, V881T, V881 Q, T883R, T887K, T892P, N905D, F906A, A912V, K921 R, S928D, Y934G, A937E, K948R, Q956Y, T999R, and A1037E.
The detergent composition comprising an endoglucanase variant according to any one of claims 1 -13, wherein the variant comprises one or more of the following set of substitutions:
F20Y+A448S+T781 M
E408D+K451 S
E408D+A448E
E408D
Y579W+E408D
The detergent composition comprising an endoglucanase variant according to any one of claims 1 -14, wherein the variant comprises one or more of the following set of substitutions:
F20P,I302D,S313D,E408D,Q416S,D476R,Q489P,A559N,Y579W,I602T,S636K,A651 P,T697G, W719R,V756Y,N848D,T883R,T887K,F906A,A937E
F20P,I302D,S313D,E408D,D476R,Y579W,I602T,S636N,T697G,W719R,V756Y,A824D,N848D, V881 Q,T887K,F906A,S928D,A937E,T999R,F1048W
16. The detergent composition comprising an endoglucanase variant of any of paragraphs 1-15, wherein the total number of alterations compared to the parent endoglucanase (e.g., SEQ ID NO: 2) is between 1 and 25, e.g., between 1 and 20, e.g., between 1 and 10 or between 1 and 5, such as 1 , 2, 3, 4, 5, 6, 7, 8, 9 or 10 alterations.
17. The detergent composition comprising an endoglucanase variant of any of paragraphs 1-16, wherein said activity on xanthan gum pre-treated with xanthan lyase is a xanthan degrading activity, preferably said xanthan degrading activity is endoglucanase EC 3.2.1.4 activity.
18. The detergent composition comprising an endoglucanase variant of any of paragraphs 1-17, wherein said variant has an improved stability in a detergent composition compared to a parent endoglucanase (e.g., with SEQ ID NO: 2).
19. The detergent composition comprising an endoglucanase variant of any of paragraphs 1-18, wherein said variant has a half-life improvement factor (HIF) of >1 .0; preferably said variant has a half-life improvement factor (HIF) of >1.0 relative to a parent endoglucanase, e.g. an endoglucanase of SEQ ID NO: 2.
20. The detergent composition comprising an endoglucanase variant of paragraph 19, wherein said half- life improvement factor (HIF) is determined after incubation of said endoglucanase variant in a detergent composition at 25°C for a time period from about 5 to about 140 hours.
21 . The detergent composition of any of paragraphs 1 -20, further comprising one or more detergent components.
22. The detergent composition of any of paragraphs 1 -21 further comprising one or more additional enzymes selected from the group consisting of: endoglucanases, proteases, amylases, lichenases, lipases, cutinases, cellulases, xanthan lyases, xyloglucanases, pectinases, pectin lyases, xanthanases, peroxidases, haloperoxygenases, catalases and mannanases, or any mixture thereof.
23. The detergent composition of any of paragraphs 1-22, further comprising an endoglucanase, such as xanthan endoglucanase.
24. The detergent composition of any of paragraphs 1 -23, wherein said composition is in form of a bar, a homogenous tablet, a tablet having two or more layers, a pouch having one or more compartments, a regular or compact powder, a granule, a paste, a gel, or a regular, compact or concentrated liquid.
25. Use of a detergent composition of any of paragraphs 1-24 for degrading xanthan gum.
26. The use of paragraph 25, wherein said endoglucanase variant has an enzyme detergency benefit.
27. A method for degrading xanthan gum comprising: applying a detergent composition of any of paragraphs 1-24 to a xanthan gum.
28. The method of paragraph 27, wherein said xanthan gum is on a surface or hard surface. The present invention is further described by the following examples that should not be construed as limiting the scope of the invention.
EXAMPLES
Example 1 : Construction of GH9 endoglucanase variants of the mature parent endoglucanase having SEQ ID NO: 2
A linear integration vector-system was used for cloning of the mature parent nucleotide sequence having SEQ ID NO: 1 (same also disclosed as mature peptide within SEQ ID NO: 1 of WO 2013/167581 A1 ) coding for the mature parent polypeptide of the GH9 endoglucanase of SEQ ID NO: 2, and its variants. The linear integration construct was a PCR fusion made by fusing the gene between two Bacillus subtilis homol- ogous chromosomal regions along with strong promoters and a chloramphenicol resistance marker. The fusion was made by Splicing by Overlap Extension (SOE) PCR (Horton, R.M, Hunt, H. D., Ho, S.N., Pullen, J.K. and Pease, L.R. (1989). Engineering hybrid genes without the use of restriction enzymes, or gene splicing were produced by overlap extension (Gene 77: 61-68). The SOE PCR method is also described in patent application WO 2003/095658. The gene was expressed under the control of a triple promoter system (as described in WO 99/43835), consisting of the promoters from Bacillus licheniformis alpha-amylase gene (amyL), Bacillus amyloliquefaciens alpha-amylase gene (amyQ), and the Bacillus thuringiensis crylllA promoter including stabilizing sequence. The gene coding for chloramphenicol acetyltransferase was used as marker (described in e.g. Diderichsen, B.; Poulsen, G.B.; Joergensen, ST.; A useful cloning vector for Bacillus subtilis. Plasmid 30:312, 1993). The final gene constructs were integrated on the Bacillus chromosome by homologous recombination into the pectate lyase locus. The gene fragments were amplified from chromosomal DNA of the corresponding strains with gene specific primers containing overhang to the two flanking vector fragments. All genes were expressed with a Bacillus licheniformis alpha-amylase secretion signal having the nucleotide sequence of SEQ ID NO: 3 and the amino acid sequence of SEQ ID NO: 4 replacing the native secretion signal.
Variants of the mature parent GH9 endoglucanase from Paenibacillus sp-62047 having SEQ ID NO: 2 as described in Examples 3-4 below were made by the megaprimer mutagenesis method using specifically designed mutagenic oligonucleotides introducing desired mutations in the resulting sequence. Design and production methods for such mutagenic oligonucleotides introducing desired mutations into target sequences are well known to those skilled in the art. Consequently, mutagenic oligos were designed and synthesized corresponding to the DNA sequence flanking the desired site(s) of mutation, separated by the
DNA base pairs defining the substitutions. The final expression cassette composed the reference GH9 en- docluconase from Paenibacillus sp-62047 as described above (i.e., parent GH9 endocluconase having SEQ ID NO: 1 ). Successful introduction of the desired substitutions was confirmed by DNA sequencing of the GH9 endoglucanase gene. An aliquot of the PCR product was subsequently transformed into Bacillus sub- Wis. Transformants were selected on LB agar plates supplemented with 10 mM K2P04, 0.4% extra glucose and 6 ig of chloramphenicol per ml. The resulting recombinant Bacillus subtilis clone containing the integrated expression construct was grown in liquid culture as described below. The enzyme containing super- natants were harvested and the enzymes (variants) were either stress tested using a reducing sugar assay or purified as described below.
Variants above were produced by fermentation using standard protocols (TB-glycerol media containing a standard trace metal mix as described in F. William Studier, "Protein production by auto-induction in high- density shaking cultures", Protein Expression and Purification, 41 (2005) 207-234) and grown for 4 days at 30°C before harvested). Supernatants of samples used for stress testing were inoculated from an overnight culture grown at 37°C and subsequently fermented in 96-well plate format (TB-glycerol media described above without calcium in the trace metal mix for 4 days 30°C).
Example 2: Purification of GH9 endoglucanase variants
The culture broth was centrifuged at 13Ό00 rpm (45 min, 18 °C, F12S-6 x 500 rotor) using a Sorval RC-6 plus centrifuge (ThermoFisher Scientific). The supernatant was supplemented with (NH4)2S04 to a final concentration of 0.8 M. The mixture was filtered using 0.2 μητι bottle-top rapid flow filters (Nalgene). The mixture was loaded on a 50 mL Phenyl Sepharose High Performance (GE Healthcare, Uppsala, Sweden) pre-equilibrated with 20 mM Tris-HCI, pH 8.0 with 0.8 M (NH4)2S04. Flowrate was set to 3 mL/min. After protein loading, the flow rate was increased to 5 mL/min and unbound or loosely bound protein was washed out by several column volumes of equilibration buffer. Elution was carried out by step-wise increase of elution buffer (20 mM Tris-HCI, pH 8.0). The target protein eluted during the (75-100%) elution step. Fractions of 8 mL were collected during the purification. The fractions were evaluated using SDS-PAGE (NuPAGE, Invitrogen). Fractions eluting with 20 mM Tris-HCI, pH 8.0 were pooled and desalted on a 350 mL G25 desalting column pre-equilibrated with 20 mM Tris-HCI, pH 8.5. The desalted protein solution was applied on a 20 mL Sourcel 5Q column pre-equilibrated with 20 mM Tris-HCI, pH 8.5 at 2 mL/min. Unbound or loosely bound proteins were washed using at least two column volumes of equilibration buffer until a stable UV baseline was obtained. The flow rate was raised to 4 mL/min and elution was done by a linear NaCI gradient using the elution buffer (20 mM Tris-HCI, pH 8.5 + 750 mM NaCI). 3 mL fractions were collected during the purification. SDS-PAGE was used to evaluate the fractions. Pure fractions were pooled and concentrated if necessary using Vivaspin 20 (10 kDa Cut-off, Sartorius). Protein concentration was determined using absorbance measurements at 280 nm.
Example 3: Detergent Stability Assay
GH9 endoglucanase (EG) activity (EC 3.2.1 .4) was determined by reducing ends on xanthan gum pre- treated with xanthan lyase using the colorimetric assay developed by Lever (1972), Anal. Biochem. 47: 273- 279, 1972. Pre-treated xanthan gum is a modified form of the xanthan sugar, where the terminal pyruvated mannose from side chains is removed (prepared according to Nankai et al. (1999) from the source Keltran). The GH9 mature parent endoglucanase and its variants cleave at beta(1 ,4)-glucosyl bonds in the glucan backbone of pre-treated xanthan gum releasing glucans with a reducing end which can be determined by reaction with p-Hydroxybenzoic acid hydrazide (PAHBAH). The increase of colour is proportional to the enzyme activity under the conditions used in the assay (e.g., Table 1 ) and used to estimate the residual activity (RA), half-life (T1/2) and the half-life improvement factor (HIF).
Table 1 . Description of assays
Method steps:
30 [iL enzyme sample (purified variant, 10-150 ppm) was mixed with 270 [iL detergent using magnetic stirring for 15 minutes in a micro titer plate (MTP). This plate was designated as the "stress MTP". 20 [iL of the mixture was transferred to a new MTP and diluted 100-fold using a 2-step dilution (2 x 10-fold dilution). The sample was diluted into assay buffer (AB): 50 mM MOPS, 4 mM CaCI2, 0.01 % Triton X-100, pH 7.0. This diluted MTP is the "reference MTP" and is stored at 4°C for 5-138 h (at a time interval equal to that of the stress MTP below). The stress MTP was incubated at 25, 26, 28 or 30°C for 5-138 h. After incubation, the stress MTP was initially mixed by magnetic stirring for 15 minutes, and the stress MTP was then diluted 100-fold as described for the reference MTP. To assess the enzymatic activity, 50 [iL of diluted enzyme:de- tergent sample (from both reference and stress MTPs) was mixed with 50 μΙ_ 4 mg/mL modified xanthan gum in PCR plates. The samples were then incubated at 50°C for 1 h. Finally, the level of reducing ends
was estimated by adding 75 μΙ_ PAHBAH solution (15 mg/mL PAHBAH, 50 g/L potassium sodium tartrate tetrahydrate, 20 g/L NaOH) to all samples in the PCR plates. The samples were then incubated at 95°C for 10 min. After cooling down to room temperature, the absorbance at 405 nm was measured. The residual activity (RA) was calculated using the following formula:
RA (%) = (Abs(Stress))/(Abs(Ref)) * 100%
Abs(Stress): The absorbance at 405 nm of the sample in the stress MTP (incubated at 25, 26, 28 or 30°C) after subtracting relevant background absorbance contributions.
Abs(Ref): The absorbance at 405 nm of the sample in the reference MTP (incubated at 4°C).
Also, the half-lives for the degradation of each variant and parent endoglucanase were calculated using the following formula (by applying 1 st order kinetics for the degradation of EG):
T: The incubation time.
Abs(Stress) and Abs(Ref): See above.
Half-life improvement factors (HIFs) can then be calculated as:
Tl/2, variant
HIF = Tl/2, wt
Γ7/2, variant: The half-life for a specific variant
Γ7/2, wt (or wild-type): The half-life for EG wt (EG wild type), wherein said T1/2 wt is
T1/2 of the mature parent endoglucanase with SEQ ID NO: 2.
The HIFs of the tested purified variants are shown in Tables 2-5 below. HIFs in Tables 3-5 were calculated in the same way as in Table 2 except for a slightly increased incubation temperature for variants and controls (26-30 degrees Celsius). All half-life values of the purified variants were calculated relative to the half-life of GH9 wild-type (mature parent endoglucanase with SEQ ID No: 2) incubated at the same detergent concentration and temperature measured as purified protein. HIF of GH9 wild-type in all tables is 1 (per definition).
Table 2. Purified variants of the mature parent GH9 endoglucanase (SEQ ID NO: 2) with corresponding half-life improvement factors (HIF) incubated at 25°C relative to a parent endoglucanase, e.g. an endoglucanase of SEQ ID NO: 2.
PUG (detergent Incubation
Alteration as compared to SEQ ID NO: 2 HIF
concentration) time (hrs)
K507R 90% PUG 16 1 .1
S17A 90% PUG 16 1 .1
S63F 90% PUG 16 1 .1
T694A 90% PUG 16 1 .1
F20Y+A448S+T781 M 90% PUG 16 1 .3
Q834E 90% PUG 16 1 .3
V56M 90% PUG 16 1 .3
I602T 90% PUG 18 1 .4
A448E 90% PUG 16 1 .4
A448W 90% PUG 16 1 .5
F20G 90% PUG 16 1 .5
I403Y 90% PUG 16 1 .5
S538C 90% PUG 16 1 .5
E408N 90% PUG 16 2.2
F20N 90% PUG 16 2.4
K512P 90% PUG 16 2.5
E408G 90% PUG 16 2.9
P410G 90% PUG 16 2.9
E408A 90% PUG 16 3.0
F20P 90% PUG 16 3.3
E408P 90% PUG 16 4.2
E408S 90% PUG 16 5.4
Table 3. Purified variants of the mature parent GH9 endoglucanase (SEQ ID NO: 2) with corresponding half-life improvement factors (HIF) incubated at 26°C relative to a parent endoglucanase, e.g. an endoglucanase of SEQ ID NO: 2.
PUG (detergent Incubation
Alteration as compared to SEQ ID NO: 2 HIF
concentration) time (hrs)
I302H 90% PUG 17.25 1 .1
I387T 90% PUG 19.5 1.1
K51 H 90% PUG 19.5 1 .1
K754R 90% PUG 19.5 1.1
K390Q 90% PUG 18 1.2
S605T 90% PUG 18.5 1.3
I602D 90% PUG 18.5 1 .3
K51 Q 90% PUG 18 1.6
Y55M 90% PUG 18 1 .6
K451 S 90% PUG 18 1.9
Q416S 90% PUG 18 2.2
Q416D 90% PUG 18 2.5
Table 4. Purified variants of the mature parent GH9 endoglucanase (SEQ ID NO: 2) with corresponding half-life improvement factors (HIF) incubated at 28°C relative to a parent endoglucanase, e.g. an endoglucanase of SEQ ID NO:2.
Table 5. Purified variants of the mature parent GH9 endoglucanase (SEQ ID NO: 2) with corresponding half-life improvement factors (HIF) incubated at 30°C relative to a parent endoglucanase, e.g. an endoglucanase of SEQ ID NO: 2.
Example 4: Calculating Half-lives and Half-life improvement factors (HIF) for Xanthan endoglucanase variants
Half-life (T½ (in hours)) was calculated at a given detergent concentration and storage temperature (Persil Universal Gel (PUG) 95%, 30°C, 4 wk or more) for the wild-type controls and/or variants, as the residual activity follows an exponential decay and the incubation time (hours) is known, i.e., according to the following formulas:
T1/2 (variants) = (Ln (0.5)/Ln (RA-variants/100))*Time
T1/2 (Wild-type) = (Ln (0.5)/Ln (RA-wild-type/100))*Time
A half-life improvement factor (HIF) is calculated as HIF = T½ (Variant)/T½ (Wild-type), where the wild- type is incubated under the same storage condition as the variant.
In the cases where the difference in stability between Wild-type and variants is too big to accurately assess half-life for both Wild-type and variant using the same incubation time, the incubation time for Wild- type and variant is different e.g. 1 h for Wild-type and 672 h for the most stable variants, Half-life Improvement Factor of variants were calculated by HIF = T½ (Variant)/T½ (Wild-type) where T½ of the WT was 1 .5 hr.
Table 6
Claims
A detergent composition comprising an endoglucanase variant, comprising an alteration at one or more positions in a region selected from the group consisting of:
i) region 10 corresponding to amino acids 1 to 94 of SEQ ID NO: 2,
ii) region 1 1 corresponding to amino acids 106 to 1 14 of SEQ ID NO: 2,
iii) region 12 corresponding to amino acids 139 to 209 of SEQ ID NO: 2,
iv) region 13 corresponding to amino acids 252 to 266 of SEQ ID NO: 2,
v) region 14 corresponding to amino acids 302 to 338 of SEQ ID NO: 2,
vi) region 15 corresponding to amino acids 362 to 546 of SEQ ID NO: 2,
vii) region 16 corresponding to amino acids 596 to 611 of SEQ ID NO: 2,
viii) region 17 corresponding to amino acids 661 to 805 of SEQ ID NO: 2,
ix) region 18 corresponding to amino acids 829 to 838 of SEQ ID NO: 2, and
x) region 19 corresponding to amino acids 1043 to 1055 of SEQ ID NO: 2,
wherein said variant has at least 60% and less than 100% sequence identity to SEQ ID NO: 2.
The detergent composition comprising an endoglucanase variant according to claim 1 , wherein said variant has at least 61 %, at least 62%, at least 63%, at least 64%, at least 65%, at least 66%, at least 67%, at least 68%, at least 69%, at least 70%, at least 71 %, at least 72%, at least 73%, at least 74%, at least 75%, at least 76%, at least 77%, at least 78%, at least 79%, at least 80%, at least 81 %, at least 82%, at least 83%, at least 84%, at least 85%, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least 91 %, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to SEQ ID NO: 2.
The detergent composition comprising an endoglucanase variant according to any one of claims 1 to
2, wherein said alteration at one or more positions is selected from the group consisting of alterations in positions: 17, 20, 51 , 53, 55, 56, 60, 63, 79, 87, 186, 192, 302, 31 1 , 313, 387, 388, 390, 403, 408, 410, 416, 448, 451 , 471 , 472, 476, 489, 507, 512, 515, 538, 598, 599, 602, 605, 609, 676, 688, 690, 694, 697, 698, 699, 71 1 , 719, 754, 756, 760, 781 , 786, 797, 833, 834, 835 and 1048 of SEQ ID NO: 2, wherein numbering is according to SEQ ID NO: 2.
The detergent composition comprising an endoglucanase variant according to any one of claims 1 to
3, wherein said alteration at one or more positions is selected from the group consisting of: S17A, F20P, F20N, F20G, F20Y, K51 Q, K51 H, E53P, E53G, Y55M, V56M, Y60F, S63F, T87R, A186P, K192N, I320D, I302H, I302V, I302M, H311 N, S313D, I387T, K388R, K390Q, I403Y, E408D, E408S, E408P, E408A, E408G, E408N, P410G, Q416S, Q416D, A448E, A448W, A448S, K451 S, G471 S,
S472Y, D476R, Q489P, K507R, K512P, S515V, S538C, Y579W, S598Q, A599S, I602T, I602D, V603P, S605T, G609E, D676H, A688G, Y690F, T694A, T697G, R698W, T699A, T71 1V, T71 1Y, W719R, K754R, V756H, V756Y, S760G, T781 M, N786K, T797S, A824D, N833D, Q834E, S835D and F1048W, wherein numbering is according to SEQ ID NO: 2.
The detergent composition comprising an endoglucanase variant according to any one of claims 1 to 4, wherein said alteration at one or more positions is selected from the group consisting of alterations in positions: 17, 20, 302, 31 1 , 313, 408, 476, 602, 688, 697, and 719, wherein numbering is according to SEQ ID NO: 2.
The detergent composition comprising an endoglucanase variant according to any one of claims 1 5, wherein said alteration at one or more positions is selected from the group consisting of alterations in positions: S17A, F20P, I302D, H31 1 N, S313D, E408D, D476R, I602T, A688G, T697G, and W719R, wherein numbering is according to SEQ ID NO: 2.
The detergent composition comprising an endoglucanase variant according to any one of claims 1 -6, wherein the variant further comprises an alteration at one or more positions selected from the group consisting of alterations in positions: 216, 283, 346, 559, 564, 579, 636, 638, 651 , 824, 848, 869, 880, 881 , 883, 887, 892, 905, 906, 912, 921 , 928, 934, 937, 948, 956, 999, and 1037 wherein numbering is according to SEQ ID NO: 2.
The detergent composition comprising an endoglucanase variant according to any one of claims 1 to 3, wherein said alteration at one or more positions is selected from the group consisting of: N216D, N216Q, A283D, A346D, A559P, A559N, A564E, Y579W, S636K, S636N, F638N, A651 P, A824D, N848D, A869V, R880K, V881 T, V881 Q, T883R, T887K, T892P, N905D, F906A, A912V, K921 R, S928D, Y934G, A937E, K948R, Q956Y, T999R, and A1037E, wherein numbering is according to SEQ ID NO: 2.
The detergent composition comprising an endoglucanase variant according to any one of claims 1 -8, wherein the variant comprises one or more of the following set of substitutions:
F20Y+A448S+T781 M
E408D+K451 S
E408D+A448E
E408D
Y579W+E408D
10. The detergent composition comprising an endoglucanase variant according to any one of claims 1 -9, wherein the variant comprises one or more of the following set of substitutions:
N216D.S313D,E408D,D476R,Q489P,A559P,Y579W,I602T,F638N,A651 P,A688G,T697G,W719R, V756H,Q834E,R880K,T887K,T892P,K921 R,S928D,Y934G
F20P,I302D,S313D,E408D,D476R,Q489P,A559N,Y579W,I602T,S636K,A651 P,T697G,W719R, V756Y,N848D,T883R,T887K,F906A,S928D,A937E
F20P,I302D,S313D,E408D,D476R,Q489P,Y579W,I602T,S636K,A651 P,T697G,W719R,V756Y, N848D,T883R,T887K,F906A,A937E
F20P,I302D,S313D,E408D,Q416S,D476R,Q489P,A559N,Y579W,I602T,S636K,A651 P,T697G, W719R,V756Y,N848D,T883R,T887K,F906A,A937E
F20P,I302D,S313D,E408D,D476R,Y579W,I602T,S636N,T697G,W719R,V756Y,A824D,N848D, V881 Q,T887K,F906A,S928D,A937E,T999R,F1048W
1 . The detergent composition comprising an endoglucanase variant according to any one of claims 1 to
10, wherein said endoglucanase variant has activity on xanthan gum pre-treated with xanthan lyase, preferably said activity is a xanthan gum degrading activity, further preferably said xanthan gum degrading activity is endoglucanase EC 3.2.1 .4 activity.
2. The detergent composition comprising an endoglucanase variant according to any one of claims 1 to
11 , wherein said variant has an improved stability in a detergent composition compared to a parent endoglucanase having SEQ ID NO: 2.
3. The detergent composition comprising an endoglucanase variant according to any one of claims 1 to
12, wherein said variant has a half-life improvement factor (HIF) of >1 .0 relative to a parent endoglucanase, e.g. an endoglucanase of SEQ ID NO: 2.
4. Use of a detergent composition according to any one of claims 1 to 13 for degrading xanthan gum.
Priority Applications (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| EP18755156.9A EP3673056A1 (en) | 2017-08-24 | 2018-08-03 | Detergent compositions comprising gh9 endoglucanase variants ii |
| US16/638,798 US11624059B2 (en) | 2017-08-24 | 2018-08-03 | Detergent compositions comprising GH9 endoglucanase variants II |
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| DE102017214809 | 2017-08-24 | ||
| DE102017214809.8 | 2017-08-24 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2019038059A1 true WO2019038059A1 (en) | 2019-02-28 |
Family
ID=63207728
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/EP2018/071105 WO2019038059A1 (en) | 2017-08-24 | 2018-08-03 | Detergent compositions comprising gh9 endoglucanase variants ii |
Country Status (3)
| Country | Link |
|---|---|
| US (1) | US11624059B2 (en) |
| EP (1) | EP3673056A1 (en) |
| WO (1) | WO2019038059A1 (en) |
Cited By (11)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| EP3715444A1 (en) | 2019-03-29 | 2020-09-30 | The Procter & Gamble Company | Laundry detergent compositions with stain removal |
| WO2022094588A1 (en) | 2020-10-29 | 2022-05-05 | The Procter & Gamble Company | Cleaning compositions containing alginate lyase enzymes |
| WO2022136389A1 (en) | 2020-12-23 | 2022-06-30 | Basf Se | Amphiphilic alkoxylated polyamines and their uses |
| EP4060010A2 (en) | 2021-03-15 | 2022-09-21 | The Procter & Gamble Company | Cleaning compositions containing polypeptide variants |
| WO2022235720A1 (en) | 2021-05-05 | 2022-11-10 | The Procter & Gamble Company | Methods for making cleaning compositions and detecting soils |
| EP4108767A1 (en) | 2021-06-22 | 2022-12-28 | The Procter & Gamble Company | Cleaning or treatment compositions containing nuclease enzymes |
| WO2023064749A1 (en) | 2021-10-14 | 2023-04-20 | The Procter & Gamble Company | A fabric and home care product comprising cationic soil release polymer and lipase enzyme |
| EP4273210A1 (en) | 2022-05-04 | 2023-11-08 | The Procter & Gamble Company | Detergent compositions containing enzymes |
| EP4273209A1 (en) | 2022-05-04 | 2023-11-08 | The Procter & Gamble Company | Machine-cleaning compositions containing enzymes |
| DE102022116726A1 (en) | 2022-07-05 | 2024-01-11 | Basf Se | Detergents and cleaning agents containing amphiphilic alkoxylated poly(ethylene/propylene)imine copolymers as well as xanthanase and/or mannanase |
| EP4410941A1 (en) | 2023-02-01 | 2024-08-07 | The Procter & Gamble Company | Detergent compositions containing enzymes |
Citations (127)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| GB1296839A (en) | 1969-05-29 | 1972-11-22 | ||
| GB1483591A (en) | 1973-07-23 | 1977-08-24 | Novo Industri As | Process for coating water soluble or water dispersible particles by means of the fluid bed technique |
| US4106991A (en) | 1976-07-07 | 1978-08-15 | Novo Industri A/S | Enzyme granulate composition and process for forming enzyme granulates |
| US4435307A (en) | 1980-04-30 | 1984-03-06 | Novo Industri A/S | Detergent cellulase |
| EP0218272A1 (en) | 1985-08-09 | 1987-04-15 | Gist-Brocades N.V. | Novel lipolytic enzymes and their use in detergent compositions |
| US4661452A (en) | 1984-05-29 | 1987-04-28 | Novo Industri A/S | Enzyme containing granulates useful as detergent additives |
| EP0238216A1 (en) | 1986-02-20 | 1987-09-23 | Albright & Wilson Limited | Protected enzyme systems |
| EP0238023A2 (en) | 1986-03-17 | 1987-09-23 | Novo Nordisk A/S | Process for the production of protein products in Aspergillus oryzae and a promoter for use in Aspergillus |
| EP0258068A2 (en) | 1986-08-29 | 1988-03-02 | Novo Nordisk A/S | Enzymatic detergent additive |
| EP0305216A1 (en) | 1987-08-28 | 1989-03-01 | Novo Nordisk A/S | Recombinant Humicola lipase and process for the production of recombinant humicola lipases |
| WO1989006270A1 (en) | 1988-01-07 | 1989-07-13 | Novo-Nordisk A/S | Enzymatic detergent |
| WO1989006279A1 (en) | 1988-01-07 | 1989-07-13 | Novo-Nordisk A/S | Mutated subtilisin genes |
| EP0331376A2 (en) | 1988-02-28 | 1989-09-06 | Amano Pharmaceutical Co., Ltd. | Recombinant DNA, bacterium of the genus pseudomonas containing it, and process for preparing lipase by using it |
| WO1989009259A1 (en) | 1988-03-24 | 1989-10-05 | Novo-Nordisk A/S | A cellulase preparation |
| EP0407225A1 (en) | 1989-07-07 | 1991-01-09 | Unilever Plc | Enzymes and enzymatic detergent compositions |
| WO1992005249A1 (en) | 1990-09-13 | 1992-04-02 | Novo Nordisk A/S | Lipase variants |
| WO1992006204A1 (en) | 1990-09-28 | 1992-04-16 | Ixsys, Inc. | Surface expression libraries of heteromeric receptors |
| EP0495257A1 (en) | 1991-01-16 | 1992-07-22 | The Procter & Gamble Company | Compact detergent compositions with high activity cellulase |
| WO1992019729A1 (en) | 1991-05-01 | 1992-11-12 | Novo Nordisk A/S | Stabilized enzymes and detergent compositions |
| WO1992019708A1 (en) | 1991-04-30 | 1992-11-12 | The Procter & Gamble Company | Liquid detergents with aromatic borate ester to inhibit proteolytic enzyme |
| WO1992019709A1 (en) | 1991-04-30 | 1992-11-12 | The Procter & Gamble Company | Built liquid detergents with boric-polyol complex to inhibit proteolytic enzyme |
| WO1992021760A1 (en) | 1991-05-29 | 1992-12-10 | Cognis, Inc. | Mutant proteolytic enzymes from bacillus |
| EP0531315A1 (en) | 1990-05-09 | 1993-03-17 | Novo Nordisk As | An enzyme capable of degrading cellulose or hemicellulose. |
| EP0531372A1 (en) | 1990-05-09 | 1993-03-17 | Novo Nordisk As | A cellulase preparation comprising an endoglucanase enzyme. |
| US5223409A (en) | 1988-09-02 | 1993-06-29 | Protein Engineering Corp. | Directed evolution of novel binding proteins |
| WO1993018140A1 (en) | 1992-03-04 | 1993-09-16 | Novo Nordisk A/S | Novel proteases |
| WO1993024618A1 (en) | 1992-06-01 | 1993-12-09 | Novo Nordisk A/S | Peroxidase variants with improved hydrogen peroxide stability |
| WO1994001541A1 (en) | 1992-07-06 | 1994-01-20 | Novo Nordisk A/S | C. antarctica lipase and lipase variants |
| WO1994002597A1 (en) | 1992-07-23 | 1994-02-03 | Novo Nordisk A/S | MUTANT α-AMYLASE, DETERGENT, DISH WASHING AGENT, AND LIQUEFACTION AGENT |
| WO1994007998A1 (en) | 1992-10-06 | 1994-04-14 | Novo Nordisk A/S | Cellulase variants |
| WO1994018314A1 (en) | 1993-02-11 | 1994-08-18 | Genencor International, Inc. | Oxidatively stable alpha-amylase |
| WO1994025578A1 (en) | 1993-04-27 | 1994-11-10 | Gist-Brocades N.V. | New lipase variants for use in detergent applications |
| WO1994025612A2 (en) | 1993-05-05 | 1994-11-10 | Institut Pasteur | Nucleotide sequences for the control of the expression of dna sequences in a cellular host |
| WO1994025583A1 (en) | 1993-05-05 | 1994-11-10 | Novo Nordisk A/S | A recombinant trypsin-like protease |
| EP0624154A1 (en) | 1991-12-13 | 1994-11-17 | The Procter & Gamble Company | Acylated citrate esters as peracid precursors |
| US5389536A (en) | 1986-11-19 | 1995-02-14 | Genencor, Inc. | Lipase from Pseudomonas mendocina having cutinase activity |
| WO1995006720A1 (en) | 1993-08-30 | 1995-03-09 | Showa Denko K.K. | Novel lipase, microorganism producing the lipase, process for producing the lipase, and use of the lipase |
| WO1995010603A1 (en) | 1993-10-08 | 1995-04-20 | Novo Nordisk A/S | Amylase variants |
| WO1995010602A1 (en) | 1993-10-13 | 1995-04-20 | Novo Nordisk A/S | H2o2-stable peroxidase variants |
| WO1995014783A1 (en) | 1993-11-24 | 1995-06-01 | Showa Denko K.K. | Lipase gene and variant lipase |
| WO1995017413A1 (en) | 1993-12-21 | 1995-06-29 | Evotec Biosystems Gmbh | Process for the evolutive design and synthesis of functional polymers based on designer elements and codes |
| WO1995022615A1 (en) | 1994-02-22 | 1995-08-24 | Novo Nordisk A/S | A method of preparing a variant of a lipolytic enzyme |
| WO1995022625A1 (en) | 1994-02-17 | 1995-08-24 | Affymax Technologies N.V. | Dna mutagenesis by random fragmentation and reassembly |
| WO1995023221A1 (en) | 1994-02-24 | 1995-08-31 | Cognis, Inc. | Improved enzymes and detergents containing them |
| WO1995024471A1 (en) | 1994-03-08 | 1995-09-14 | Novo Nordisk A/S | Novel alkaline cellulases |
| WO1995030744A2 (en) | 1994-05-04 | 1995-11-16 | Genencor International Inc. | Lipases with improved surfactant resistance |
| WO1995033836A1 (en) | 1994-06-03 | 1995-12-14 | Novo Nordisk Biotech, Inc. | Phosphonyldipeptides useful in the treatment of cardiovascular diseases |
| WO1995035381A1 (en) | 1994-06-20 | 1995-12-28 | Unilever N.V. | Modified pseudomonas lipases and their use |
| WO1996000292A1 (en) | 1994-06-23 | 1996-01-04 | Unilever N.V. | Modified pseudomonas lipases and their use |
| WO1996000787A1 (en) | 1994-06-30 | 1996-01-11 | Novo Nordisk Biotech, Inc. | Non-toxic, non-toxigenic, non-pathogenic fusarium expression system and promoters and terminators for use therein |
| WO1996011262A1 (en) | 1994-10-06 | 1996-04-18 | Novo Nordisk A/S | An enzyme and enzyme preparation with endoglucanase activity |
| WO1996012012A1 (en) | 1994-10-14 | 1996-04-25 | Solvay S.A. | Lipase, microorganism producing same, method for preparing said lipase and uses thereof |
| WO1996013580A1 (en) | 1994-10-26 | 1996-05-09 | Novo Nordisk A/S | An enzyme with lipolytic activity |
| WO1996023873A1 (en) | 1995-02-03 | 1996-08-08 | Novo Nordisk A/S | Amylase variants |
| WO1996027002A1 (en) | 1995-02-27 | 1996-09-06 | Novo Nordisk A/S | Novel lipase gene and process for the production of lipase with the use of the same |
| WO1996029397A1 (en) | 1995-03-17 | 1996-09-26 | Novo Nordisk A/S | Novel endoglucanases |
| WO1997004079A1 (en) | 1995-07-14 | 1997-02-06 | Novo Nordisk A/S | A modified enzyme with lipolytic activity |
| WO1997007202A1 (en) | 1995-08-11 | 1997-02-27 | Novo Nordisk A/S | Novel lipolytic enzymes |
| US5648263A (en) | 1988-03-24 | 1997-07-15 | Novo Nordisk A/S | Methods for reducing the harshness of a cotton-containing fabric |
| WO1997043424A1 (en) | 1996-05-14 | 1997-11-20 | Genencor International, Inc. | MODIFIED α-AMYLASES HAVING ALTERED CALCIUM BINDING PROPERTIES |
| WO1998008940A1 (en) | 1996-08-26 | 1998-03-05 | Novo Nordisk A/S | A novel endoglucanase |
| WO1998012307A1 (en) | 1996-09-17 | 1998-03-26 | Novo Nordisk A/S | Cellulase variants |
| WO1998015257A1 (en) | 1996-10-08 | 1998-04-16 | Novo Nordisk A/S | Diaminobenzoic acid derivatives as dye precursors |
| WO1998017767A1 (en) | 1996-10-18 | 1998-04-30 | The Procter & Gamble Company | Detergent compositions |
| WO1998020116A1 (en) | 1996-11-04 | 1998-05-14 | Novo Nordisk A/S | Subtilase variants and compositions |
| WO1998020115A1 (en) | 1996-11-04 | 1998-05-14 | Novo Nordisk A/S | Subtilase variants and compositions |
| WO1998034946A1 (en) | 1997-02-12 | 1998-08-13 | Massachusetts Institute Of Technology | Daxx, a novel fas-binding protein that activates jnk and apoptosis |
| WO1999019467A1 (en) | 1997-10-13 | 1999-04-22 | Novo Nordisk A/S | α-AMYLASE MUTANTS |
| WO1999043835A2 (en) | 1998-02-26 | 1999-09-02 | Novo Nordisk Biotech, Inc. | Methods for producing a polypeptide in a bacillus cell |
| US5977053A (en) | 1995-07-31 | 1999-11-02 | Bayer Ag | Detergents and cleaners containing iminodisuccinates |
| WO2000024883A1 (en) | 1998-10-26 | 2000-05-04 | Novozymes A/S | Constructing and screening a dna library of interest in filamentous fungal cells |
| WO2000034450A1 (en) | 1998-12-04 | 2000-06-15 | Novozymes A/S | Cutinase variants |
| EP1025240A2 (en) | 1997-10-23 | 2000-08-09 | Genencor International, Inc. | Multiply-substituted protease variants with altered net charge for use in detergents |
| WO2000056900A2 (en) | 1999-03-22 | 2000-09-28 | Novo Nordisk Biotech, Inc. | Promoter sequences derived from fusarium venenatum and uses thereof |
| WO2000060063A1 (en) | 1999-03-31 | 2000-10-12 | Novozymes A/S | Lipase variant |
| WO2001044452A1 (en) | 1999-12-15 | 2001-06-21 | Novozymes A/S | Subtilase variants having an improved wash performance on egg stains |
| WO2001058276A2 (en) | 2000-02-08 | 2001-08-16 | F Hoffmann-La Roche Ag | Use of acid-stable proteases in animal feed |
| WO2001066712A2 (en) | 2000-03-08 | 2001-09-13 | Novozymes A/S | Variants with altered properties |
| WO2001092502A1 (en) | 2000-06-02 | 2001-12-06 | Novozymes A/S | Cutinase variants |
| WO2002010355A2 (en) | 2000-08-01 | 2002-02-07 | Novozymes A/S | Alpha-amylase mutants with altered stability |
| WO2002099091A2 (en) | 2001-06-06 | 2002-12-12 | Novozymes A/S | Endo-beta-1,4-glucanase from bacillus |
| WO2003006602A2 (en) | 2001-07-12 | 2003-01-23 | Novozymes A/S | Subtilase variants |
| WO2003040279A1 (en) | 2001-11-09 | 2003-05-15 | Unilever Plc | Polymers for laundry applications |
| WO2003095658A1 (en) | 2002-05-07 | 2003-11-20 | Novozymes A/S | Homologous recombination into bacterium for the generation of polynucleotide libraries |
| US20040171154A1 (en) | 2001-07-27 | 2004-09-02 | Francesca Storici | Systems for in vivo site-directed mutagenesis using oligonucleotides |
| WO2004099401A1 (en) | 2003-05-07 | 2004-11-18 | Novozymes A/S | Variant subtilisin enzymes (subtilases) |
| WO2005003274A1 (en) | 2003-06-18 | 2005-01-13 | Unilever Plc | Laundry treatment compositions |
| WO2005003275A1 (en) | 2003-06-18 | 2005-01-13 | Unilever Plc | Laundry treatment compositions |
| WO2005003276A1 (en) | 2003-06-18 | 2005-01-13 | Unilever Plc | Laundry treatment compositions |
| WO2005056782A2 (en) | 2003-12-03 | 2005-06-23 | Genencor International, Inc. | Perhydrolase |
| WO2006066594A2 (en) | 2004-12-23 | 2006-06-29 | Novozymes A/S | Alpha-amylase variants |
| WO2006108856A2 (en) | 2005-04-15 | 2006-10-19 | Basf Aktiengesellschaft | Amphiphilic water-soluble alkoxylated polyalkylenimines with an internal polyethylene oxide block and an external polypropylene oxide block |
| WO2006113314A1 (en) | 2005-04-15 | 2006-10-26 | The Procter & Gamble Company | Liquid laundry detergent compositions with modified polyethyleneimine polymers and lipase enzyme |
| WO2006130575A2 (en) | 2005-05-31 | 2006-12-07 | The Procter & Gamble Company | Polymer-containing detergent compositions and their use |
| WO2007044993A2 (en) | 2005-10-12 | 2007-04-19 | Genencor International, Inc. | Use and production of storage-stable neutral metalloprotease |
| WO2007087257A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | Enzyme and fabric hueing agent containing compositions |
| WO2007087242A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | A composition comprising a lipase and a bleach catalyst |
| WO2007087243A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | Detergent compositions |
| WO2007087508A2 (en) | 2006-01-23 | 2007-08-02 | Novozymes A/S | Lipase variants |
| WO2007087244A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | Detergent compositions |
| WO2007087259A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | Enzyme and photobleach containing compositions |
| WO2007087258A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | A composition comprising a lipase and a bleach catalyst |
| WO2007138054A1 (en) | 2006-05-31 | 2007-12-06 | The Procter & Gamble Company | Cleaning compositions with amphiphilic graft polymers based on polyalkylene oxides and vinyl esters |
| EP1867808A1 (en) | 2006-06-06 | 2007-12-19 | Brose Schliesssysteme GmbH & Co. KG | Motor vehicle lock |
| EP1876226A1 (en) | 2006-07-07 | 2008-01-09 | The Procter and Gamble Company | Detergent compositions |
| WO2008153815A2 (en) | 2007-05-30 | 2008-12-18 | Danisco Us, Inc., Genencor Division | Variants of an alpha-amylase with improved production levels in fermentation processes |
| US20090011970A1 (en) | 2007-07-02 | 2009-01-08 | Marc Francois Theophile Evers | Laundry multi-compartment pouch composition |
| WO2009061380A2 (en) | 2007-11-05 | 2009-05-14 | Danisco Us Inc., Genencor Division | VARIANTS OF BACILLUS sp. TS-23 ALPHA-AMYLASE WITH ALTERED PROPERTIES |
| WO2009067279A1 (en) | 2007-11-21 | 2009-05-28 | E.I. Du Pont De Nemours And Company | Production of peracids using an enzyme having perhydrolysis activity |
| WO2009087523A2 (en) | 2008-01-04 | 2009-07-16 | The Procter & Gamble Company | A laundry detergent composition comprising glycosyl hydrolase |
| WO2009102854A1 (en) | 2008-02-15 | 2009-08-20 | The Procter & Gamble Company | Cleaning compositions |
| WO2009109500A1 (en) | 2008-02-29 | 2009-09-11 | Novozymes A/S | Polypeptides having lipase activity and polynucleotides encoding same |
| WO2009155557A2 (en) | 2008-06-20 | 2009-12-23 | Solae, Llc | Protein hydrolysate compositions stable under acidic conditions |
| WO2010065455A2 (en) | 2008-12-01 | 2010-06-10 | Danisco Us Inc. | Enzymes with lipase activity |
| WO2010100028A2 (en) | 2009-03-06 | 2010-09-10 | Huntsman Advanced Materials (Switzerland) Gmbh | Enzymatic textile bleach-whitening methods |
| WO2010107560A2 (en) | 2009-03-18 | 2010-09-23 | Danisco Us Inc. | Fungal cutinase from magnaporthe grisea |
| WO2010111143A2 (en) | 2009-03-23 | 2010-09-30 | Danisco Us Inc. | Cal a-related acyltransferases and methods of use, thereof |
| WO2011084599A1 (en) | 2009-12-21 | 2011-07-14 | Danisco Us Inc. | Detergent compositions containing bacillus subtilis lipase and methods of use thereof |
| WO2011084417A1 (en) | 2009-12-21 | 2011-07-14 | Danisco Us Inc. | Detergent compositions containing geobacillus stearothermophilus lipase and methods of use thereof |
| WO2011084412A1 (en) | 2009-12-21 | 2011-07-14 | Danisco Us Inc. | Detergent compositions containing thermobifida fusca lipase and methods of use thereof |
| WO2011150157A2 (en) | 2010-05-28 | 2011-12-01 | Danisco Us Inc. | Detergent compositions containing streptomyces griseus lipase and methods of use thereof |
| WO2012137147A1 (en) | 2011-04-08 | 2012-10-11 | Danisco Us, Inc. | Compositions |
| WO2013167581A1 (en) | 2012-05-07 | 2013-11-14 | Novozymes A/S | Polypeptides having xanthan degrading activity and polynucleotides encoding same |
| WO2015001017A2 (en) * | 2013-07-04 | 2015-01-08 | Novozymes A/S | Polypeptides having anti-redeposition effect and polynucleotides encoding same |
| WO2016203064A2 (en) | 2015-10-28 | 2016-12-22 | Novozymes A/S | Detergent composition comprising protease and amylase variants |
| WO2018037062A1 (en) * | 2016-08-24 | 2018-03-01 | Novozymes A/S | Gh9 endoglucanase variants and polynucleotides encoding same |
| WO2018037065A1 (en) * | 2016-08-24 | 2018-03-01 | Henkel Ag & Co. Kgaa | Detergent composition comprising gh9 endoglucanase variants i |
Family Cites Families (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN106414730A (en) | 2014-05-28 | 2017-02-15 | 诺维信公司 | Polypeptides having endoglucanase activity |
-
2018
- 2018-08-03 US US16/638,798 patent/US11624059B2/en active Active
- 2018-08-03 WO PCT/EP2018/071105 patent/WO2019038059A1/en unknown
- 2018-08-03 EP EP18755156.9A patent/EP3673056A1/en active Pending
Patent Citations (135)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| GB1296839A (en) | 1969-05-29 | 1972-11-22 | ||
| GB1483591A (en) | 1973-07-23 | 1977-08-24 | Novo Industri As | Process for coating water soluble or water dispersible particles by means of the fluid bed technique |
| US4106991A (en) | 1976-07-07 | 1978-08-15 | Novo Industri A/S | Enzyme granulate composition and process for forming enzyme granulates |
| US4435307A (en) | 1980-04-30 | 1984-03-06 | Novo Industri A/S | Detergent cellulase |
| US4661452A (en) | 1984-05-29 | 1987-04-28 | Novo Industri A/S | Enzyme containing granulates useful as detergent additives |
| EP0218272A1 (en) | 1985-08-09 | 1987-04-15 | Gist-Brocades N.V. | Novel lipolytic enzymes and their use in detergent compositions |
| EP0238216A1 (en) | 1986-02-20 | 1987-09-23 | Albright & Wilson Limited | Protected enzyme systems |
| EP0238023A2 (en) | 1986-03-17 | 1987-09-23 | Novo Nordisk A/S | Process for the production of protein products in Aspergillus oryzae and a promoter for use in Aspergillus |
| EP0258068A2 (en) | 1986-08-29 | 1988-03-02 | Novo Nordisk A/S | Enzymatic detergent additive |
| US5389536A (en) | 1986-11-19 | 1995-02-14 | Genencor, Inc. | Lipase from Pseudomonas mendocina having cutinase activity |
| EP0305216A1 (en) | 1987-08-28 | 1989-03-01 | Novo Nordisk A/S | Recombinant Humicola lipase and process for the production of recombinant humicola lipases |
| WO1989006270A1 (en) | 1988-01-07 | 1989-07-13 | Novo-Nordisk A/S | Enzymatic detergent |
| WO1989006279A1 (en) | 1988-01-07 | 1989-07-13 | Novo-Nordisk A/S | Mutated subtilisin genes |
| EP0331376A2 (en) | 1988-02-28 | 1989-09-06 | Amano Pharmaceutical Co., Ltd. | Recombinant DNA, bacterium of the genus pseudomonas containing it, and process for preparing lipase by using it |
| US5691178A (en) | 1988-03-22 | 1997-11-25 | Novo Nordisk A/S | Fungal cellulase composition containing alkaline CMC-endoglucanase and essentially no cellobiohydrolase |
| WO1989009259A1 (en) | 1988-03-24 | 1989-10-05 | Novo-Nordisk A/S | A cellulase preparation |
| US5648263A (en) | 1988-03-24 | 1997-07-15 | Novo Nordisk A/S | Methods for reducing the harshness of a cotton-containing fabric |
| US5776757A (en) | 1988-03-24 | 1998-07-07 | Novo Nordisk A/S | Fungal cellulase composition containing alkaline CMC-endoglucanase and essentially no cellobiohydrolase and method of making thereof |
| US5223409A (en) | 1988-09-02 | 1993-06-29 | Protein Engineering Corp. | Directed evolution of novel binding proteins |
| EP0407225A1 (en) | 1989-07-07 | 1991-01-09 | Unilever Plc | Enzymes and enzymatic detergent compositions |
| US5763254A (en) | 1990-05-09 | 1998-06-09 | Novo Nordisk A/S | Enzyme capable of degrading cellulose or hemicellulose |
| US5457046A (en) | 1990-05-09 | 1995-10-10 | Novo Nordisk A/S | Enzyme capable of degrading cellullose or hemicellulose |
| EP0531315A1 (en) | 1990-05-09 | 1993-03-17 | Novo Nordisk As | An enzyme capable of degrading cellulose or hemicellulose. |
| EP0531372A1 (en) | 1990-05-09 | 1993-03-17 | Novo Nordisk As | A cellulase preparation comprising an endoglucanase enzyme. |
| US5686593A (en) | 1990-05-09 | 1997-11-11 | Novo Nordisk A/S | Enzyme capable of degrading cellulose or hemicellulose |
| WO1992005249A1 (en) | 1990-09-13 | 1992-04-02 | Novo Nordisk A/S | Lipase variants |
| WO1992006204A1 (en) | 1990-09-28 | 1992-04-16 | Ixsys, Inc. | Surface expression libraries of heteromeric receptors |
| EP0495257A1 (en) | 1991-01-16 | 1992-07-22 | The Procter & Gamble Company | Compact detergent compositions with high activity cellulase |
| WO1992019708A1 (en) | 1991-04-30 | 1992-11-12 | The Procter & Gamble Company | Liquid detergents with aromatic borate ester to inhibit proteolytic enzyme |
| WO1992019709A1 (en) | 1991-04-30 | 1992-11-12 | The Procter & Gamble Company | Built liquid detergents with boric-polyol complex to inhibit proteolytic enzyme |
| WO1992019729A1 (en) | 1991-05-01 | 1992-11-12 | Novo Nordisk A/S | Stabilized enzymes and detergent compositions |
| WO1992021760A1 (en) | 1991-05-29 | 1992-12-10 | Cognis, Inc. | Mutant proteolytic enzymes from bacillus |
| EP0624154A1 (en) | 1991-12-13 | 1994-11-17 | The Procter & Gamble Company | Acylated citrate esters as peracid precursors |
| WO1993018140A1 (en) | 1992-03-04 | 1993-09-16 | Novo Nordisk A/S | Novel proteases |
| WO1993024618A1 (en) | 1992-06-01 | 1993-12-09 | Novo Nordisk A/S | Peroxidase variants with improved hydrogen peroxide stability |
| WO1994001541A1 (en) | 1992-07-06 | 1994-01-20 | Novo Nordisk A/S | C. antarctica lipase and lipase variants |
| WO1994002597A1 (en) | 1992-07-23 | 1994-02-03 | Novo Nordisk A/S | MUTANT α-AMYLASE, DETERGENT, DISH WASHING AGENT, AND LIQUEFACTION AGENT |
| WO1994007998A1 (en) | 1992-10-06 | 1994-04-14 | Novo Nordisk A/S | Cellulase variants |
| WO1994018314A1 (en) | 1993-02-11 | 1994-08-18 | Genencor International, Inc. | Oxidatively stable alpha-amylase |
| WO1994025578A1 (en) | 1993-04-27 | 1994-11-10 | Gist-Brocades N.V. | New lipase variants for use in detergent applications |
| WO1994025583A1 (en) | 1993-05-05 | 1994-11-10 | Novo Nordisk A/S | A recombinant trypsin-like protease |
| WO1994025612A2 (en) | 1993-05-05 | 1994-11-10 | Institut Pasteur | Nucleotide sequences for the control of the expression of dna sequences in a cellular host |
| WO1995006720A1 (en) | 1993-08-30 | 1995-03-09 | Showa Denko K.K. | Novel lipase, microorganism producing the lipase, process for producing the lipase, and use of the lipase |
| WO1995010603A1 (en) | 1993-10-08 | 1995-04-20 | Novo Nordisk A/S | Amylase variants |
| WO1995010602A1 (en) | 1993-10-13 | 1995-04-20 | Novo Nordisk A/S | H2o2-stable peroxidase variants |
| WO1995014783A1 (en) | 1993-11-24 | 1995-06-01 | Showa Denko K.K. | Lipase gene and variant lipase |
| WO1995017413A1 (en) | 1993-12-21 | 1995-06-29 | Evotec Biosystems Gmbh | Process for the evolutive design and synthesis of functional polymers based on designer elements and codes |
| WO1995022625A1 (en) | 1994-02-17 | 1995-08-24 | Affymax Technologies N.V. | Dna mutagenesis by random fragmentation and reassembly |
| WO1995022615A1 (en) | 1994-02-22 | 1995-08-24 | Novo Nordisk A/S | A method of preparing a variant of a lipolytic enzyme |
| WO1995023221A1 (en) | 1994-02-24 | 1995-08-31 | Cognis, Inc. | Improved enzymes and detergents containing them |
| EP1921147A2 (en) | 1994-02-24 | 2008-05-14 | Henkel Kommanditgesellschaft auf Aktien | Improved enzymes and detergents containing them |
| EP1921148A2 (en) | 1994-02-24 | 2008-05-14 | Henkel Kommanditgesellschaft auf Aktien | Improved enzymes and detergents containing them |
| WO1995024471A1 (en) | 1994-03-08 | 1995-09-14 | Novo Nordisk A/S | Novel alkaline cellulases |
| WO1995030744A2 (en) | 1994-05-04 | 1995-11-16 | Genencor International Inc. | Lipases with improved surfactant resistance |
| WO1995033836A1 (en) | 1994-06-03 | 1995-12-14 | Novo Nordisk Biotech, Inc. | Phosphonyldipeptides useful in the treatment of cardiovascular diseases |
| WO1995035381A1 (en) | 1994-06-20 | 1995-12-28 | Unilever N.V. | Modified pseudomonas lipases and their use |
| WO1996000292A1 (en) | 1994-06-23 | 1996-01-04 | Unilever N.V. | Modified pseudomonas lipases and their use |
| WO1996000787A1 (en) | 1994-06-30 | 1996-01-11 | Novo Nordisk Biotech, Inc. | Non-toxic, non-toxigenic, non-pathogenic fusarium expression system and promoters and terminators for use therein |
| WO1996011262A1 (en) | 1994-10-06 | 1996-04-18 | Novo Nordisk A/S | An enzyme and enzyme preparation with endoglucanase activity |
| WO1996012012A1 (en) | 1994-10-14 | 1996-04-25 | Solvay S.A. | Lipase, microorganism producing same, method for preparing said lipase and uses thereof |
| WO1996013580A1 (en) | 1994-10-26 | 1996-05-09 | Novo Nordisk A/S | An enzyme with lipolytic activity |
| WO1996023873A1 (en) | 1995-02-03 | 1996-08-08 | Novo Nordisk A/S | Amylase variants |
| WO1996027002A1 (en) | 1995-02-27 | 1996-09-06 | Novo Nordisk A/S | Novel lipase gene and process for the production of lipase with the use of the same |
| WO1996029397A1 (en) | 1995-03-17 | 1996-09-26 | Novo Nordisk A/S | Novel endoglucanases |
| WO1997004079A1 (en) | 1995-07-14 | 1997-02-06 | Novo Nordisk A/S | A modified enzyme with lipolytic activity |
| US5977053A (en) | 1995-07-31 | 1999-11-02 | Bayer Ag | Detergents and cleaners containing iminodisuccinates |
| WO1997007202A1 (en) | 1995-08-11 | 1997-02-27 | Novo Nordisk A/S | Novel lipolytic enzymes |
| WO1997043424A1 (en) | 1996-05-14 | 1997-11-20 | Genencor International, Inc. | MODIFIED α-AMYLASES HAVING ALTERED CALCIUM BINDING PROPERTIES |
| WO1998008940A1 (en) | 1996-08-26 | 1998-03-05 | Novo Nordisk A/S | A novel endoglucanase |
| WO1998012307A1 (en) | 1996-09-17 | 1998-03-26 | Novo Nordisk A/S | Cellulase variants |
| WO1998015257A1 (en) | 1996-10-08 | 1998-04-16 | Novo Nordisk A/S | Diaminobenzoic acid derivatives as dye precursors |
| WO1998017767A1 (en) | 1996-10-18 | 1998-04-30 | The Procter & Gamble Company | Detergent compositions |
| WO1998020115A1 (en) | 1996-11-04 | 1998-05-14 | Novo Nordisk A/S | Subtilase variants and compositions |
| WO1998020116A1 (en) | 1996-11-04 | 1998-05-14 | Novo Nordisk A/S | Subtilase variants and compositions |
| WO1998034946A1 (en) | 1997-02-12 | 1998-08-13 | Massachusetts Institute Of Technology | Daxx, a novel fas-binding protein that activates jnk and apoptosis |
| WO1999019467A1 (en) | 1997-10-13 | 1999-04-22 | Novo Nordisk A/S | α-AMYLASE MUTANTS |
| EP1025240A2 (en) | 1997-10-23 | 2000-08-09 | Genencor International, Inc. | Multiply-substituted protease variants with altered net charge for use in detergents |
| WO1999043835A2 (en) | 1998-02-26 | 1999-09-02 | Novo Nordisk Biotech, Inc. | Methods for producing a polypeptide in a bacillus cell |
| WO2000024883A1 (en) | 1998-10-26 | 2000-05-04 | Novozymes A/S | Constructing and screening a dna library of interest in filamentous fungal cells |
| WO2000034450A1 (en) | 1998-12-04 | 2000-06-15 | Novozymes A/S | Cutinase variants |
| WO2000056900A2 (en) | 1999-03-22 | 2000-09-28 | Novo Nordisk Biotech, Inc. | Promoter sequences derived from fusarium venenatum and uses thereof |
| WO2000060063A1 (en) | 1999-03-31 | 2000-10-12 | Novozymes A/S | Lipase variant |
| WO2001044452A1 (en) | 1999-12-15 | 2001-06-21 | Novozymes A/S | Subtilase variants having an improved wash performance on egg stains |
| WO2001058276A2 (en) | 2000-02-08 | 2001-08-16 | F Hoffmann-La Roche Ag | Use of acid-stable proteases in animal feed |
| WO2001058275A2 (en) | 2000-02-08 | 2001-08-16 | F Hoffmann-La Roche Ag | Use of acid-stable subtilisin proteases in animal feed |
| WO2001066712A2 (en) | 2000-03-08 | 2001-09-13 | Novozymes A/S | Variants with altered properties |
| WO2001092502A1 (en) | 2000-06-02 | 2001-12-06 | Novozymes A/S | Cutinase variants |
| WO2002010355A2 (en) | 2000-08-01 | 2002-02-07 | Novozymes A/S | Alpha-amylase mutants with altered stability |
| WO2002099091A2 (en) | 2001-06-06 | 2002-12-12 | Novozymes A/S | Endo-beta-1,4-glucanase from bacillus |
| WO2003006602A2 (en) | 2001-07-12 | 2003-01-23 | Novozymes A/S | Subtilase variants |
| US20040171154A1 (en) | 2001-07-27 | 2004-09-02 | Francesca Storici | Systems for in vivo site-directed mutagenesis using oligonucleotides |
| WO2003040279A1 (en) | 2001-11-09 | 2003-05-15 | Unilever Plc | Polymers for laundry applications |
| WO2003095658A1 (en) | 2002-05-07 | 2003-11-20 | Novozymes A/S | Homologous recombination into bacterium for the generation of polynucleotide libraries |
| WO2004099401A1 (en) | 2003-05-07 | 2004-11-18 | Novozymes A/S | Variant subtilisin enzymes (subtilases) |
| WO2005003274A1 (en) | 2003-06-18 | 2005-01-13 | Unilever Plc | Laundry treatment compositions |
| WO2005003275A1 (en) | 2003-06-18 | 2005-01-13 | Unilever Plc | Laundry treatment compositions |
| WO2005003276A1 (en) | 2003-06-18 | 2005-01-13 | Unilever Plc | Laundry treatment compositions |
| WO2005056782A2 (en) | 2003-12-03 | 2005-06-23 | Genencor International, Inc. | Perhydrolase |
| WO2006066594A2 (en) | 2004-12-23 | 2006-06-29 | Novozymes A/S | Alpha-amylase variants |
| WO2006108856A2 (en) | 2005-04-15 | 2006-10-19 | Basf Aktiengesellschaft | Amphiphilic water-soluble alkoxylated polyalkylenimines with an internal polyethylene oxide block and an external polypropylene oxide block |
| WO2006113314A1 (en) | 2005-04-15 | 2006-10-26 | The Procter & Gamble Company | Liquid laundry detergent compositions with modified polyethyleneimine polymers and lipase enzyme |
| WO2006130575A2 (en) | 2005-05-31 | 2006-12-07 | The Procter & Gamble Company | Polymer-containing detergent compositions and their use |
| WO2007044993A2 (en) | 2005-10-12 | 2007-04-19 | Genencor International, Inc. | Use and production of storage-stable neutral metalloprotease |
| WO2007087257A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | Enzyme and fabric hueing agent containing compositions |
| WO2007087242A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | A composition comprising a lipase and a bleach catalyst |
| WO2007087243A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | Detergent compositions |
| WO2007087508A2 (en) | 2006-01-23 | 2007-08-02 | Novozymes A/S | Lipase variants |
| WO2007087244A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | Detergent compositions |
| WO2007087259A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | Enzyme and photobleach containing compositions |
| WO2007087258A2 (en) | 2006-01-23 | 2007-08-02 | The Procter & Gamble Company | A composition comprising a lipase and a bleach catalyst |
| WO2007138054A1 (en) | 2006-05-31 | 2007-12-06 | The Procter & Gamble Company | Cleaning compositions with amphiphilic graft polymers based on polyalkylene oxides and vinyl esters |
| EP1867808A1 (en) | 2006-06-06 | 2007-12-19 | Brose Schliesssysteme GmbH & Co. KG | Motor vehicle lock |
| EP1876226A1 (en) | 2006-07-07 | 2008-01-09 | The Procter and Gamble Company | Detergent compositions |
| WO2008153815A2 (en) | 2007-05-30 | 2008-12-18 | Danisco Us, Inc., Genencor Division | Variants of an alpha-amylase with improved production levels in fermentation processes |
| US20090011970A1 (en) | 2007-07-02 | 2009-01-08 | Marc Francois Theophile Evers | Laundry multi-compartment pouch composition |
| WO2009061380A2 (en) | 2007-11-05 | 2009-05-14 | Danisco Us Inc., Genencor Division | VARIANTS OF BACILLUS sp. TS-23 ALPHA-AMYLASE WITH ALTERED PROPERTIES |
| WO2009067279A1 (en) | 2007-11-21 | 2009-05-28 | E.I. Du Pont De Nemours And Company | Production of peracids using an enzyme having perhydrolysis activity |
| WO2009087523A2 (en) | 2008-01-04 | 2009-07-16 | The Procter & Gamble Company | A laundry detergent composition comprising glycosyl hydrolase |
| WO2009102854A1 (en) | 2008-02-15 | 2009-08-20 | The Procter & Gamble Company | Cleaning compositions |
| WO2009109500A1 (en) | 2008-02-29 | 2009-09-11 | Novozymes A/S | Polypeptides having lipase activity and polynucleotides encoding same |
| WO2009155557A2 (en) | 2008-06-20 | 2009-12-23 | Solae, Llc | Protein hydrolysate compositions stable under acidic conditions |
| WO2010065455A2 (en) | 2008-12-01 | 2010-06-10 | Danisco Us Inc. | Enzymes with lipase activity |
| WO2010100028A2 (en) | 2009-03-06 | 2010-09-10 | Huntsman Advanced Materials (Switzerland) Gmbh | Enzymatic textile bleach-whitening methods |
| WO2010107560A2 (en) | 2009-03-18 | 2010-09-23 | Danisco Us Inc. | Fungal cutinase from magnaporthe grisea |
| WO2010111143A2 (en) | 2009-03-23 | 2010-09-30 | Danisco Us Inc. | Cal a-related acyltransferases and methods of use, thereof |
| WO2011084599A1 (en) | 2009-12-21 | 2011-07-14 | Danisco Us Inc. | Detergent compositions containing bacillus subtilis lipase and methods of use thereof |
| WO2011084417A1 (en) | 2009-12-21 | 2011-07-14 | Danisco Us Inc. | Detergent compositions containing geobacillus stearothermophilus lipase and methods of use thereof |
| WO2011084412A1 (en) | 2009-12-21 | 2011-07-14 | Danisco Us Inc. | Detergent compositions containing thermobifida fusca lipase and methods of use thereof |
| WO2011150157A2 (en) | 2010-05-28 | 2011-12-01 | Danisco Us Inc. | Detergent compositions containing streptomyces griseus lipase and methods of use thereof |
| WO2012137147A1 (en) | 2011-04-08 | 2012-10-11 | Danisco Us, Inc. | Compositions |
| WO2013167581A1 (en) | 2012-05-07 | 2013-11-14 | Novozymes A/S | Polypeptides having xanthan degrading activity and polynucleotides encoding same |
| WO2015001017A2 (en) * | 2013-07-04 | 2015-01-08 | Novozymes A/S | Polypeptides having anti-redeposition effect and polynucleotides encoding same |
| WO2016203064A2 (en) | 2015-10-28 | 2016-12-22 | Novozymes A/S | Detergent composition comprising protease and amylase variants |
| WO2018037062A1 (en) * | 2016-08-24 | 2018-03-01 | Novozymes A/S | Gh9 endoglucanase variants and polynucleotides encoding same |
| WO2018037065A1 (en) * | 2016-08-24 | 2018-03-01 | Henkel Ag & Co. Kgaa | Detergent composition comprising gh9 endoglucanase variants i |
Non-Patent Citations (100)
| Title |
|---|
| "Powdered Detergents, Surfactant science series", vol. 71, MARCEL DEKKER, INC. |
| "Protein Purification", 1989, VCH PUBLISHERS |
| "Soc. App. Bacteriol. Symposium Series", 1980 |
| ALTSCHUL ET AL., NUCLEIC ACIDS RES., vol. 25, 1997, pages 3389 - 3402 |
| BARTON ET AL., NUCLEIC ACIDS RES., vol. 18, 1990, pages 7349 - 4966 |
| BECKER; GUARENTE: "Guide to Yeast Genetics and Molecular Biology, Methods in Enzymology", vol. 194, ACADEMIC PRESS, INC., pages: 182 - 187 |
| BOWIE; SAUER, PROC. NATL. ACAD. SCI. USA, vol. 86, 1989, pages 2152 - 2156 |
| BUCKLEY ET AL., APPL. ENVIRON. MICROBIOL., vol. 65, 1999, pages 3800 - 3804 |
| BURKE ET AL., PROC. NATL. ACAD. SCI. USA, vol. 98, 2001, pages 6289 - 6294 |
| CALISSANO; MACINO, FUNGAL GENET. NEWSLETT., vol. 43, 1996, pages 15 - 16 |
| CARTER ET AL., PROTEINS: STRUCTURE, FUNCTION, AND GENETICS, vol. 6, 1989, pages 240 - 248 |
| CATT; JOLLICK, MICROBIOS, vol. 68, 1991, pages 189 - 207 |
| CHANG; COHEN, MOL. GEN. GENET., vol. 168, 1979, pages 111 - 115 |
| CHOI ET AL., J. MICROBIOL. METHODS, vol. 64, 2006, pages 391 - 397 |
| CHRISTENSEN ET AL., BIOLTECHNOLOGY, vol. 6, 1988, pages 1419 - 1422 |
| CLEWELL, MICROBIOL. REV., vol. 45, 1981, pages 409 - 436 |
| COLLINS-RACIE ET AL., BIOTECHNOLOGY, vol. 13, 1995, pages 982 - 987 |
| CONTRERAS ET AL., BIOTECHNOLOGY, vol. 9, 1991, pages 378 - 381 |
| COOPER ET AL., EMBO J., vol. 12, 1993, pages 2575 - 2583 |
| CULLEN ET AL., NUCLEIC ACIDS RES., vol. 15, 1987, pages 9163 - 9175 |
| CUNNINGHAM; WELLS, SCIENCE, vol. 244, 1989, pages 1081 - 1085 |
| DAWSON ET AL., SCIENCE, vol. 266, 1994, pages 776 - 779 |
| DE VOS ET AL., SCIENCE, vol. 255, 1992, pages 306 - 312 |
| DEBOER ET AL., PROC. NATL. ACAD. SCI. USA, vol. 80, 1983, pages 21 - 25 |
| DERBYSHIRE ET AL., GENE, vol. 46, 1986, pages 145 |
| DIDERICHSEN, B.; POULSEN, G.B.; JOERGENSEN, S.T.: "A useful cloning vector for Bacillus subtilis", PLASMID, vol. 30, 1993, pages 312, XP024799145, DOI: doi:10.1006/plas.1993.1066 |
| DOWER ET AL., NUCLEIC ACIDS RES., vol. 16, 1988, pages 6127 - 6145 |
| DUBNAU; DAVIDOFF-ABELSON, J. MOL. BIOL., vol. 56, 1971, pages 209 - 221 |
| EATON ET AL., BIOCHEMISTRY, vol. 25, 1986, pages 505 - 512 |
| EDGAR, NUCLEIC ACIDS RESEARCH, vol. 32, 2004, pages 1792 - 1797 |
| EGON ET AL., GENE, vol. 69, 1988, pages 301 - 315 |
| F. WILLIAM STUDIER: "Protein production by auto-induction in high-density shaking cultures", PROTEIN EXPRESSION AND PURIFICATION, vol. 41, 2005, pages 207 - 234 |
| FORD ET AL., PROTEIN EXPRESSION AND PURIFICATION, vol. 2, 1991, pages 95 - 107 |
| GEMS ET AL., GENE, vol. 98, 1991, pages 61 - 67 |
| GILBERT ET AL.: "Useful proteins from recombinant bacteria", SCIENTIFIC AMERICAN, vol. 242, 1980, pages 74 - 94 |
| GONG ET AL., FOLIA MICROBIOL. (PRAHA, vol. 49, 2004, pages 399 - 405 |
| GOUGH ET AL., J. MOL. BIOL., vol. 313, 2000, pages 903 - 919 |
| GUO; SHERMAN, MOL. CELLULAR BIOI., vol. 15, 1995, pages 5983 - 5990 |
| H. NEURATH; R.L. HILL: "The Proteins", 1979, ACADEMIC PRESS |
| HANAHAN, J. MOL. BIOL., vol. 166, 1983, pages 557 - 580 |
| HAWKSWORTH ET AL.: "Ainsworth and Bisby's Dictionary of The Fungi", 1995, CAB INTERNATIONAL, UNIVERSITY PRESS |
| HENRISSAT ET AL.: "A classification of glycosyl hydrolases based on amino-acid sequence similarities", J. BIOCHEM., vol. 280, 1991, pages 309 - 316 |
| HILTON ET AL., J. BIOL. CHEM., vol. 271, 1996, pages 4699 - 4708 |
| HINNEN ET AL., PROC. NATL. ACAD. SCI. USA, vol. 75, 1978, pages 1920 |
| HODGDON; KALER, CURRENT OPINION IN COLLOID & INTERFACE SCIENCE, vol. 12, 2007, pages 121 - 128 |
| HOLM; PARK, BIOINFORMATICS, vol. 16, 2000, pages 566 - 567 |
| HOLM; SANDER, PROTEINS, vol. 33, 1998, pages 88 - 96 |
| HORTON, R.M; HUNT, H. D.; HO, S.N.; PULLEN, J.K.; PEASE, L.R.: "Engineering hybrid genes without the use of restriction enzymes, or gene splicing were produced by overlap extension", GENE, vol. 77, 1989, pages 61 - 68 |
| HUE ET AL., JOURNAL OF BACTERIOLOGY, vol. 177, 1995, pages 3465 - 3471 |
| INNIS ET AL.: "PCR: A Guide to Methods and Application", 1990, ACADEMIC PRESS |
| ITO ET AL., J. BACTERIOL., vol. 153, 1983, pages 163 |
| JONES, J. MOL. BIOL., vol. 287, 1999, pages 797 - 815 |
| KATOH ET AL., METHODS IN MOLECULAR BIOLOGY, vol. 537, 2009, pages 39 - 64 |
| KATOH ET AL., NUCLEIC ACIDS RESEARCH, vol. 33, 2005, pages 511 - 518 |
| KATOH; KUMA, NUCLEIC ACIDS RESEARCH, vol. 30, 2002, pages 3059 - 3066 |
| KATOH; TOH, BIOINFORMATICS, vol. 23, 2007, pages 372 - 374 |
| KATOH; TOH, BIOINFORMATICS, vol. 26, 2010, pages 1899 - 1900 |
| KOEHLER; THORNE, J. BACTERIOL., vol. 169, 1987, pages 5271 - 5278 |
| KREN ET AL., NAT. MED., vol. 4, 1998, pages 285 - 290 |
| LERECLUS, MOLECULAR MICROBIOLOGY, vol. 13, 1994, pages 97 - 107 |
| LEVER, ANAL. BIOCHEM., vol. 47, 1972, pages 273 - 279 |
| LINDAHL; ELOFSSON, J. MOL. BIOL., vol. 295, 2000, pages 613 - 615 |
| LOWMAN ET AL., BIOCHEMISTRY, vol. 30, 1991, pages 10832 - 10837 |
| MALARDIER ET AL., GENE, vol. 78, 1989, pages 147 - 156 |
| MARTIN ET AL., J. IND. MICROBIOL. BIOTECHNOL., vol. 3, 2003, pages 568 - 576 |
| MAZODIER ET AL., J. BACTERIOL., vol. 171, 1989, pages 3583 - 3585 |
| MCGUFFIN; JONES, BIOINFORMATICS, vol. 19, 2003, pages 874 - 881 |
| NEEDLEMAN; WUNSCH, J. MOL. BIOL., vol. 48, 1970, pages 443 - 453 |
| NER ET AL., DNA, vol. 7, 1988, pages 127 |
| NESS ET AL., NATURE BIOTECHNOLOGY, vol. 17, 1999, pages 893 - 896 |
| OLGA V. MOROZ ET AL: "Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase", ACS CATALYSIS, vol. 8, no. 7, 6 July 2018 (2018-07-06), US, pages 6021 - 6034, XP055521064, ISSN: 2155-5435, DOI: 10.1021/acscatal.8b00666 * |
| PERRY; KURAMITSU, INFECT. IMMUN., vol. 32, 1981, pages 1295 - 1297 |
| PINEDO; SMETS, APPL. ENVIRON. MICROBIOL., vol. 71, 2005, pages 51 - 57 |
| RASMUSSEN-WILSON ET AL., APPL. ENVIRON. MICROBIOL., vol. 63, 1997, pages 3488 - 3493 |
| REIDHAAR-OLSON; SAUER, SCIENCE, vol. 241, 1988, pages 53 - 57 |
| RICE ET AL., EMBOSS: THE EUROPEAN MOLECULAR BIOLOGY OPEN SOFTWARE SUITE, 2000 |
| RICE ET AL.: "EMBOSS: The European Molecular Biology Open Software Suite", TRENDS GENET, vol. 16, 2000, pages 276 - 277, XP004200114, DOI: doi:10.1016/S0168-9525(00)02024-2 |
| RICE ET AL.: "EMBOSS: The European Molecular Biology Open Software Suite", TRENDS GENET., vol. 16, 2000, pages 276 - 277, XP004200114, DOI: doi:10.1016/S0168-9525(00)02024-2 |
| ROMANOS ET AL., YEAST, vol. 8, 1992, pages 423 - 488 |
| RUIJSSENAARS ET AL.: "A novel gene encoding xanthan lyase of Paenibacillus alginolyticus strain XL-1", APPL. ENVIRON. MICROBIOL., vol. 66, no. 9, 2000, pages 3945 - 3950, XP002704006, DOI: doi:10.1128/AEM.66.9.3945-3950.2000 |
| RUIJSSENAARS ET AL.: "A pyruvated mannose-specific xanthan lyase involved in xanthan degradation by Paenibacillus alginolyticus XL-1", APPL. ENVIRON. MICROBIOL., vol. 65, no. 6, 1999, pages 2446 - 2452, XP002704005 |
| SAMBROOK ET AL.: "Molecular Cloning, A Laboratory Manual", 1989, COLD SPRING HARBOR |
| SCHERER; DAVIS, PROC. NATL. ACAD. SCI. USA, vol. 76, 1979, pages 4949 - 4955 |
| SHIGEKAWA; DOWER, BIOTECHNIQUES, vol. 6, 1988, pages 742 - 751 |
| SHINDYALOV; BOUME, PROTEIN ENGINEERING, vol. 11, 1998, pages 739 - 747 |
| SIEZEN ET AL., PROTEIN ENGNG., vol. 4, 1991, pages 719 - 737 |
| SIEZEN ET AL., PROTEIN SCIENCE, vol. 6, 1997, pages 501 - 523 |
| SIMONEN; PALVA, MICROBIOLOGICAL REVIEWS, vol. 57, 1993, pages 109 - 137 |
| SMITH ET AL., J. MOT. BIOI., vol. 224, 1992, pages 899 - 904 |
| STEVENS, DRUG DISCOVERY WORLD, vol. 4, 2003, pages 35 - 48 |
| STORICI ET AL., NATURE BIOTECHNOL., vol. 19, 2001, pages 773 - 776 |
| SVETINA ET AL., J. BIOTECHNOL., vol. 76, 2000, pages 245 - 251 |
| THOMPSON ET AL., NUCLEIC ACIDS RESEARCH, vol. 22, 1994, pages 4673 - 4680 |
| TIAN ET AL., NATURE, vol. 432, 2004, pages 1050 - 1054 |
| VILLA-KAMAROFF ET AL., PROC. NATL. ACAD. SCI. USA, vol. 75, 1978, pages 3727 - 3731 |
| WARD ET AL., BIOTECHNOLOGY, vol. 13, 1995, pages 498 - 503 |
| WLODAVER ET AL., FEBS LETT., vol. 309, 1992, pages 59 - 64 |
| YELTON ET AL., PROC. NATL, ACAD. SCI. USA, vol. 81, 1984, pages 1470 - 1474 |
| YOUNG; SPIZIZEN, J. BACTERIOL., vol. 81, 1961, pages 823 - 829 |
| ZHANG ET AL., BIOTECHNOLOGY ADVANCES, vol. 24, 2006, pages 452 - 481 |
Cited By (26)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| EP3715444A1 (en) | 2019-03-29 | 2020-09-30 | The Procter & Gamble Company | Laundry detergent compositions with stain removal |
| WO2020205350A1 (en) | 2019-03-29 | 2020-10-08 | The Procter & Gamble Company | Laundry detergent compositions with stain removal |
| US11111459B2 (en) | 2019-03-29 | 2021-09-07 | The Procter & Gamble Company | Laundry detergent compositions with stain removal |
| CN113574158A (en) * | 2019-03-29 | 2021-10-29 | 宝洁公司 | Laundry detergent composition with soil release effect |
| JP7350881B2 (en) | 2019-03-29 | 2023-09-26 | ザ プロクター アンド ギャンブル カンパニー | Laundry detergent composition with soil removal |
| JP2022527263A (en) * | 2019-03-29 | 2022-06-01 | ザ プロクター アンド ギャンブル カンパニー | Laundry detergent composition with stain removal |
| WO2022094589A1 (en) | 2020-10-29 | 2022-05-05 | The Procter & Gamble Company | Cleaning compositions containing alginase enzymes |
| WO2022094588A1 (en) | 2020-10-29 | 2022-05-05 | The Procter & Gamble Company | Cleaning compositions containing alginate lyase enzymes |
| WO2022094164A1 (en) | 2020-10-29 | 2022-05-05 | The Procter & Gamble Company | Cleaning composition comprising alginate lyase enzymes |
| WO2022094590A1 (en) | 2020-10-29 | 2022-05-05 | The Procter & Gamble Company | Cleaning compositions containing alginate lyase enzymes |
| WO2022094163A1 (en) | 2020-10-29 | 2022-05-05 | The Procter & Gamble Company | Cleaning composition comprising alginate lyase enzymes |
| WO2022136389A1 (en) | 2020-12-23 | 2022-06-30 | Basf Se | Amphiphilic alkoxylated polyamines and their uses |
| EP4060010A2 (en) | 2021-03-15 | 2022-09-21 | The Procter & Gamble Company | Cleaning compositions containing polypeptide variants |
| WO2022197512A1 (en) | 2021-03-15 | 2022-09-22 | The Procter & Gamble Company | Cleaning compositions containing polypeptide variants |
| WO2022235720A1 (en) | 2021-05-05 | 2022-11-10 | The Procter & Gamble Company | Methods for making cleaning compositions and detecting soils |
| EP4095223A1 (en) | 2021-05-05 | 2022-11-30 | The Procter & Gamble Company | Methods for making cleaning compositions and for detecting soils |
| EP4108767A1 (en) | 2021-06-22 | 2022-12-28 | The Procter & Gamble Company | Cleaning or treatment compositions containing nuclease enzymes |
| WO2022272255A1 (en) | 2021-06-22 | 2022-12-29 | The Procter & Gamble Company | Cleaning or treatment compositions containing nuclease enzymes |
| WO2023064749A1 (en) | 2021-10-14 | 2023-04-20 | The Procter & Gamble Company | A fabric and home care product comprising cationic soil release polymer and lipase enzyme |
| EP4273210A1 (en) | 2022-05-04 | 2023-11-08 | The Procter & Gamble Company | Detergent compositions containing enzymes |
| EP4273209A1 (en) | 2022-05-04 | 2023-11-08 | The Procter & Gamble Company | Machine-cleaning compositions containing enzymes |
| WO2023215680A1 (en) | 2022-05-04 | 2023-11-09 | The Procter & Gamble Company | Machine-cleaning compositions containing enzymes |
| WO2023215679A1 (en) | 2022-05-04 | 2023-11-09 | The Procter & Gamble Company | Detergent compositions containing enzymes |
| DE102022116726A1 (en) | 2022-07-05 | 2024-01-11 | Basf Se | Detergents and cleaning agents containing amphiphilic alkoxylated poly(ethylene/propylene)imine copolymers as well as xanthanase and/or mannanase |
| EP4410941A1 (en) | 2023-02-01 | 2024-08-07 | The Procter & Gamble Company | Detergent compositions containing enzymes |
| WO2024163695A1 (en) | 2023-02-01 | 2024-08-08 | The Procter & Gamble Company | Detergent compositions containing enzymes |
Also Published As
| Publication number | Publication date |
|---|---|
| EP3673056A1 (en) | 2020-07-01 |
| US20200239815A1 (en) | 2020-07-30 |
| US11624059B2 (en) | 2023-04-11 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| US11795418B2 (en) | GH9 endoglucanase variants and polynucleotides encoding same | |
| AU2017317564B2 (en) | Detergent composition comprising GH9 endoglucanase variants I | |
| US11624059B2 (en) | Detergent compositions comprising GH9 endoglucanase variants II | |
| AU2017317563B8 (en) | Detergent compositions comprising xanthan lyase variants I | |
| US11359188B2 (en) | Xanthan lyase variants and polynucleotides encoding same | |
| US11512300B2 (en) | Xanthan lyase variants and polynucleotides encoding same | |
| WO2019038060A1 (en) | Detergent composition comprising xanthan lyase variants ii | |
| WO2019162000A1 (en) | Detergent composition comprising xanthan lyase and endoglucanase variants | |
| EP3017032A2 (en) | Polypeptides having anti-redeposition effect and polynucleotides encoding same | |
| WO2018206178A1 (en) | Detergent composition comprising polypeptide comprising carbohydrate-binding domain | |
| US11525128B2 (en) | GH9 endoglucanase variants and polynucleotides encoding same | |
| WO2018206535A1 (en) | Carbohydrate-binding domain and polynucleotides encoding the same |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 18755156 Country of ref document: EP Kind code of ref document: A1 |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| ENP | Entry into the national phase |
Ref document number: 2018755156 Country of ref document: EP Effective date: 20200324 |