US20030192072A1 - Methods for making plants tolerant to glyphosate and compositions thereof - Google Patents
Methods for making plants tolerant to glyphosate and compositions thereof Download PDFInfo
- Publication number
- US20030192072A1 US20030192072A1 US10/413,909 US41390903A US2003192072A1 US 20030192072 A1 US20030192072 A1 US 20030192072A1 US 41390903 A US41390903 A US 41390903A US 2003192072 A1 US2003192072 A1 US 2003192072A1
- Authority
- US
- United States
- Prior art keywords
- glyphosate
- plant
- sequence
- dna molecule
- dna
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Abandoned
Links
Images
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N9/00—Enzymes; Proenzymes; Compositions thereof; Processes for preparing, activating, inhibiting, separating or purifying enzymes
- C12N9/10—Transferases (2.)
- C12N9/1085—Transferases (2.) transferring alkyl or aryl groups other than methyl groups (2.5)
- C12N9/1092—3-Phosphoshikimate 1-carboxyvinyltransferase (2.5.1.19), i.e. 5-enolpyruvylshikimate-3-phosphate synthase
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/63—Introduction of foreign genetic material using vectors; Vectors; Use of hosts therefor; Regulation of expression
- C12N15/79—Vectors or expression systems specially adapted for eukaryotic hosts
- C12N15/82—Vectors or expression systems specially adapted for eukaryotic hosts for plant cells, e.g. plant artificial chromosomes (PACs)
- C12N15/8241—Phenotypically and genetically modified plants via recombinant DNA technology
- C12N15/8261—Phenotypically and genetically modified plants via recombinant DNA technology with agronomic (input) traits, e.g. crop yield
- C12N15/8271—Phenotypically and genetically modified plants via recombinant DNA technology with agronomic (input) traits, e.g. crop yield for stress resistance, e.g. heavy metal resistance
- C12N15/8274—Phenotypically and genetically modified plants via recombinant DNA technology with agronomic (input) traits, e.g. crop yield for stress resistance, e.g. heavy metal resistance for herbicide resistance
- C12N15/8275—Glyphosate
Definitions
- This invention relates in general to plant molecular biology and plant genetic engineering for herbicide resistance and, more particularly, to a novel glyphosate resistant 5-enolpyruvylshikimate-3-phosphate synthase from Eleusine indica .
- Plant genetic engineering methods can be used to transfer the glyphosate resistant 5-enolpyruvylshikimate-3-phosphate synthase gene isolated and purified from Eleusine indica into crop and ornamental plants of economic importance.
- N-phosphonomethylglycine also known as glyphosate
- Glyphosate is a well known herbicide that has activity on a broad spectrum of plant species.
- Glyphosate is the active ingredient of Roundup® (Monsanto Co.), a safe herbicide having a desirably short half life in the environment. When applied onto a plant surface, glyphosate moves systemically through the plant. Glyphosate is toxic to plants by inhibiting the shikimic acid pathway that provides a precursor for the synthesis of aromatic amino acids.
- glyphosate affects the conversion of phosphoenolpyruvate and 3-phosphoshikimic acid to 5-enolpyruvyl-3-phosphoshikimic acid by inhibiting the enzyme 5-enolpyruvyl-3-phosphoshikimic acid (hereinafter referred to as EPSP synthase or EPSPS).
- EPSP synthase EPSPS
- the term “glyphosate” should be considered to include any herbicidally effective form of N-phosphonomethylglycine (including any salt thereof) and other forms that result in the production of the glyphosate anion in planta.
- glyphosate tolerance has been genetically engineered into corn (U.S. Pat. No. 5,554,798), wheat (Zhou et al. Plant Cell Rep. 15:159-163 (1995), soybean (WO 9200377) and canola (WO 9204449).
- Variants of the wild-type EPSPS enzyme are glyphosate-resistant as a result of alterations in the EPSPS amino acid coding sequence (Kishore et al., Annu. Rev. Biochem. 57:627-663 (1988); Schulz et al., Arch. Microbiol. 137:121-123 (1984); Sost et al., FEBS Lett. 173:238-241 (1984); Kishore et al., In “Biotechnology for Crop Protection” ACS Symposium Series No. 379. Eds. Hedlin et al., 37-48 (1988).
- variants typically have a higher K i for glyphosate than the wild-type EPSPS enzyme that confers the glyphosate-tolerant phenotype, but these variants are also characterized by a high K m for PEP that makes the enzyme kinetically less efficient.
- the apparent K m for PEP and the apparent K i for glyphosate for the native EPSPS from E. coli are 10 ⁇ M and 0.5 ⁇ M while for a glyphosate-resistant isolate having a single amino acid substitution of an alanine for the glycine at position 96 these values are 220 ⁇ M and 4.0 mM, respectively.
- a number of glyphosate-resistant plant variant EPSPS genes have been constructed by mutagenesis.
- Eleusine indica is commonly referred to as “goose grass” and may also be known as “yard grass”. It is a common monocotyledonous plant found world wide. As a member of the Poaceae family, the grass family, it is related to many well known crop plants.
- Eleusine indica is most closely related to the millets, that include Sorghum bicolor (sorghum or great millet), Zea mays (maize), Pennisetum americanum (pearl millet), Eleusine coracana (finger millet), Setaria italica (foxtail millet), Paspalum scrobiculatum (kodo millet), Echinochloa frumentacea (barnyhard millet) and Eragrostis tef (teff) (Chennaveeraiah et al., In “Chromosome engineering in plants: genetics, breeding and evolution”, Cytogenetics of Minor Millets, in Tsuchiya et al., eds Elsevier Sci Pub Amsterdam, 613-627 (1991).
- Eleusine indica has been shown to hybridize with Eleusine coracana (finger millet), an important cultivated millet of India and East Africa (Chennaveeraiah et al., Euphytica 2-3:489-495, (1974).
- Classical plant breeding methods can be used to transfer the genes and traits of interest from Eleusine indica into agronomic crop plants within the family Poaceae.
- the present invention herein provides a method for plant tolerance to glyphosate herbicide by the expression of an isolated DNA molecule encoding a naturally occurring glyphosate resistant EPSPS enzyme.
- the enzyme and the DNA is isolated from Eleusine species, more particularly Eleusine indica ( E. indica ).
- the first aspect of the present invention described herein provides a method to cause plants to be tolerant to glyphosate herbicide by the insertion of a recombinant DNA molecule into the nuclear genome of a plant cell, the recombinant DNA molecule comprising:
- a promoter that functions in plant cells to cause the production of an RNA molecule operably linked to,
- a DNA molecule transcribing an RNA encoding for a chloroplast transit peptide and a E. indica glyphosate resistant EPSPS enzyme; operably linked to, a 3′ non-translated region that functions in plant cells to cause the polyadenylation of the 3′ end of the RNA molecule.
- the promoter used in the DNA molecule is expressed in a constitutive fashion.
- suitable promoters that function effectively in this capacity include cauliflower mosaic virus 19S promoter, cauliflower mosaic virus 35S promoter, figwort mosaic virus 34S promoter, sugarcane bacilliform virus promoter, commelina yellow mottle virus promoter, small subunit of ribulose-1,5-bisphosphate carboxylase promoter, rice cytosolic triosephosphate isomerase promoter, adenine phosphoribosyltransferae promoter, rice actin 1 promoter, maize ubiquitin promoter, mannopine synthase promoter and octopine synthase promoter.
- a Promoter may also comprise leader sequences and intron sequences useful in the invention.
- a DNA molecule that encodes a chloroplast transit peptide sequence can be isolated from EPSPS genes purified from various plant species including E. indica as well as from various plant genes whose protein products have been shown to be transported into the chloroplast.
- a DNA molecule that encodes a glyphosate resistant EPSPS enzyme isolated from Eleusine species, more particularly from E. indica, comprising SEQ ID NO: 7 is an object of the invention and a DNA molecule substantially homologous to the DNA molecule isolated from E. indica or a portion thereof identified as SEQ ID NO: 6.
- the 3′ non-translated region can be obtained from various genes that are expressed in plant cells.
- the nopaline synthase 3′ untranslated, the 3′ untranslated region from pea small subunit Rubisco gene, the wheat heat shock protein 17.9 3′ untranslated region, the 3′ untranslated region from soybean 7S seed storage protein gene are commonly used in this capacity.
- the invention also relates to a glyphosate tolerant transgenic crop plant cell, a glyphosate tolerant crop plant and crop plant parts, crop seeds and progeny thereof comprising the recombinant DNA molecule of the present invention.
- a DNA molecule that encodes a naturally occurring plant derived glyphosate resistant EPSPS enzyme wherein the glyphosate resistant EPSPS enzyme has a K m for phosphoenolpyruvate (PEP) of less than 10 ⁇ M. More preferably, a DNA molecule that encodes a naturally occurring plant derived glyphosate resistant EPSPS enzyme wherein the glyphosate resistant EPSPS enzyme has a K m for PEP of less than 10 ⁇ M and the K m for PEP is not more than about 2 ⁇ of the naturally occurring plant derived glyphosate sensitive EPSPS enzyme.
- PEP phosphoenolpyruvate
- PEP phosphoenolpyruvate
- the invention also relates to the homologous genetic elements regulating expression of the E. indica glyphosate resistant EPSPS gene.
- These elements include but are not limited to the DNA sequences of a promoter, a 5′ untranslated region, a chloroplast transit peptide, an intron, and a 3′ untranslated region of E. indica EPSPS glyphosate resistance gene.
- a DNA molecule that encodes a glyphosate resistant EPSPS enzyme purified from the genome of Eleusine species, more particularly from E. indica glyphosate resistant biotype provided by the ATCC deposit #PTA-2177 is an object of the invention.
- FIG. 1 Eleusine indica (glyphosate tolerant) EPSP synthase DNA sequence (SEQ ID NO: 6).
- FIG. 2 Deduced amino acid sequence for the mature protein-coding region of the Eleusine indica (glyphosate tolerant biotype) EPSP synthase gene (SEQ ID NO: 7).
- FIG. 3 Growth rates on glyphosate containing media of transgenic E. coli on expressing the E. indica glyphosate sensitive EPSPS enzyme and the E. indica glyphosate resistant EPSPS enzyme.
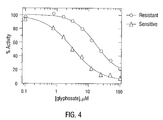
- FIG. 4 Glyphosate inhibition study comparing the EPSP synthase activities detectable in extracts prepared from the glyphosate-sensitive and tolerant E. indica biotypes.
- FIG. 5 Plasmid map of pMON45364
- FIG. 6 Plasmid map of pMON45365
- FIG. 7 Plasmid map of pMON45367
- FIG. 8 Plasmid map of pMON45369
- FIG. 9 The amino acid sequence deduced from the cDNA sequence of the mature EPSP synthase protein sequence derived from the glyphosate-tolerant E. indica biotype (top row) aligned with that of the glyphosate sensitive E. indica biotype (bottom row).
- cDNA library refers to a collection of cDNA fragments, each cloned into a separate vector molecule.
- chimeric refers to a fusion nucleic acid or protein sequence.
- a chimeric nucleic acid coding sequence is comprised of two or more sequences joined in-frame that encode a chimeric protein.
- a chimeric gene refers to the multiple genetic elements derived from heterologous sources comprising a gene.
- coding sequence refers to the region of continuous sequential nucleic acid triplets encoding a protein, polypeptide, or peptide sequence.
- Codon refers to a sequence of three nucleotides that specify a particular amino acid.
- construct refers to the heterologous genetic elements operably linked to each other making up a recombinant DNA molecule.
- C-terminal region refers to the region of a peptide, polypeptide, or protein chain from the middle thereof to the end that carries the amino acid having a free carboxyl group.
- encoding DNA refers to chromosomal DNA, plasmid DNA, cDNA, or synthetic DNA that encodes any of the proteins discussed herein.
- endogenous refers to materials originating from within an organism or cell.
- Endonuclease refers to an enzyme that hydrolyzes double stranded DNA at internal locations.
- Exogenous refers to materials originating from outside of an organism or cell. This typically applies to nucleic acid molecules used in producing transformed or transgenic host cells and plants.
- Example refers to the portion of a gene that is actually translated into protein, i.e. a coding sequence.
- expression refers to the transcription of a gene to produce the corresponding mRNA.
- a fragment of a EPSPS gene is a portion of a full-length EPSPS gene nucleic acid that is of at least a minimum length capable of expressing a protein with EPSPS activity.
- gene refers to chromosomal DNA, plasmid DNA, cDNA, synthetic DNA, or other DNA that encodes a peptide, polypeptide, protein, or RNA molecule, and regions flanking the coding sequence involved in the regulation of expression.
- the term “genome” as it applies to viruses encompasses all of the nucleic acid sequence contained within the capsid of the virus.
- the term “genome” as it applies to bacteria encompasses both the chromosome and plasmids within a bacterial host cell. Encoding nucleic acids of the present invention introduced into bacterial host cells can therefore be either chromosomally-integrated or plasmid-localized.
- the term “genome” as it applies to plant cells encompasses not only chromosomal DNA found within the nucleus, but organelle DNA found within subcellular components of the cell. Nucleic acids of the present invention introduced into plant cells can therefore be either chromosomally-integrated or organelle-localized.
- Glyphosate refers to N-phosphonomethylglycine and its' salts
- Glyphosate is the active ingredient of Roundup® herbicide (Monsanto Co.).
- Plant treatments with “glyphosate” refer to treatments with the Roundup® or Roundup Ultra® herbicide formulation, unless otherwise stated.
- Glyphosate as N-phosphonomethylglycine and its' salts are components of synthetic culture media used for the selection of bacteria and plant tolerance to glyphosate or used to determine enzyme resistance in in vitro biochemical assays.
- Heterologous DNA refers to DNA from a source different than that of the recipient cell.
- “Homologous DNA” refers to DNA from the same source as that of the recipient cell.
- Hybridization refers to the ability of a strand of nucleic acid to join with a complementary strand via base pairing. Hybridization occurs when complementary sequences in the two nucleic acid strands bind to one another.
- Identity refers to the degree of similarity between two nucleic acid or protein sequences.
- An alignment of the two sequences is performed by a suitable computer program.
- a widely used and accepted computer program for performing sequence alignments is CLUSTALW v1.6 (Thompson, et al. Nucl. Acids Res., 22: 4673-4680, 1994).
- the number of matching bases or amino acids is divided by the total number of bases or amino acids, and multiplied by 100 to obtain a percent identity. For example, if two 580 base pair sequences had 145 matched bases, they would be 25 percent identical. If the two compared sequences are of different lengths, the number of matches is divided by the shorter of the two lengths.
- the shorter sequence is less than 150 bases or 50 amino acids in length, the number of matches are divided by 150 (for nucleic acid bases) or 50 (for amino acids), and multiplied by 100 to obtain a percent identity.
- Intron refers to a portion of a gene not translated into protein, even though it is transcribed into RNA.
- isolated nucleic acid is one that has been substantially separated or purified away from other nucleic acid sequences in the cell of the organism that the nucleic acid naturally occurs, i.e., other chromosomal and extrachromosomal DNA and RNA, by conventional nucleic acid-purification methods.
- the term also embraces recombinant nucleic acids and chemically synthesized nucleic acids.
- “Native” refers to a naturally-occurring (“wild-type”) nucleic acid or polypeptide.
- N-terminal region refers to the region of a peptide, polypeptide, or protein chain from the amino acid having a free amino group to the middle of the chain.
- Nucleic acid refers to deoxyribonucleic acid (DNA) and ribonucleic acid (RNA).
- a “nucleic acid segment” or a “nucleic acid molecule segment” is a nucleic acid molecule that has been isolated free of total genomic DNA of a particular species, or that has been synthesized. Included with the term “nucleic acid segment” are DNA segments, recombinant vectors, plasmids, cosmids, phagemids, phage, viruses, et cetera.
- nucleic-Acid Hybridization is functionally defined with regard to the hybridization of a nucleic-acid probe to a target nucleic acid (i.e., to a particular nucleic-acid sequence of interest) by the specific hybridization procedure discussed in Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989), at 9.52-9.55. See also, Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989) at 9.47-9.52, 9.56-9.58; Kanehisa, Nucl. Acids Res. 12:203-213, (1984); and Wetmur and Davidson, J. Mol. Biol. 31:349-370, (1968).
- “Nucleotide Sequence Variants” Using well-known methods, the skilled artisan can readily produce nucleotide and amino acid sequence variants of EPSPS genes and proteins, respectively.
- “Variant” DNA molecules are DNA molecules containing minor changes in a native EPSPS gene sequence, i.e., changes that one or more nucleotides of a native EPSPS gene sequence is deleted, added, and/or substituted, such that the variant EPSPS gene encodes a protein that retains EPSPS activity.
- Variant DNA molecules can be produced, for example, by standard DNA mutagenesis techniques or by chemically synthesizing the variant DNA molecule or a portion thereof.
- nucleic acids are discussed, for example, in Beaucage et al, Tetra. Letts. 22:1859-1862 (1981), and Matteucci et al., J. Am. Chem. Soc. 103:3185-(1981).
- Chemical synthesis of nucleic acids can be performed, for example, on automated oligonucleotide synthesizers.
- Such variants preferably do not change the reading frame of the protein-coding region of the nucleic acid and preferably encode a protein having no amino acid changes.
- Nucleic acid sequence variants are most often created for the purposes of modification of the sequence to add or delete restriction endonuclease sites or to affect transcription or translation of the nucleic acid molecule.
- amino-acid substitutions are preferably substitutions of single amino-acid residue for another amino-acid residue at any position within the protein. Substitutions, deletions, insertions or any combination thereof can be combined to arrive at a final construct.
- Open reading frame refers to a region of DNA or RNA encoding a peptide, polypeptide, or protein.
- a first nucleic-acid sequence is “operably” linked with a second nucleic-acid sequence when the first nucleic-acid sequence is placed in a functional relationship with the second nucleic-acid sequence.
- a promoter is operably linked to a protein-coding sequence if the promoter effects the transcription or expression of the coding sequence.
- operably linked DNA sequences are contiguous and, where necessary to join two protein-coding regions, in reading frame.
- “Overexpression” refers to the expression of a polypeptide or protein encoded by a DNA introduced into a host cell, wherein said polypeptide or protein is either not normally present in the host cell, or wherein said polypeptide or protein is present in said host cell at a higher level than that normally expressed from the endogenous gene encoding said polypeptide or protein.
- Plant expression vector refers to chimeric DNA molecules comprising the regulatory elements that are operably linked to provide the expression of a transgene product in plants.
- “Plasmid” refers to a circular, extrachromosomal, self-replicating piece of DNA.
- Polyadenylation signal or “polyA signal” refers to a nucleic acid sequence located 3′ to a coding region that causes the addition of adenylate nucleotides to the 3′ end of the mRNA transcribed from the coding region.
- PCR Polymerase chain reaction
- promoter refers to a nucleic acid sequence, usually found upstream (5′) to a coding sequence, that controls expression of the coding sequence by controlling production of messenger RNA (mRNA) by providing the recognition site for RNA polymerase and/or other factors necessary for start of transcription at the correct site.
- mRNA messenger RNA
- a promoter or promoter region includes variations of promoters derived by means of ligation to various regulatory sequences, random or controlled mutagenesis, and addition or duplication of enhancer sequences.
- the promoter region disclosed herein, and biologically functional equivalents thereof, are responsible for driving the transcription of coding sequences under their control when introduced into a host as part of a suitable recombinant vector, as demonstrated by its ability to produce mRNA.
- a “recombinant” nucleic acid is made by an artificial combination of two otherwise separated segments of sequence, e.g., by chemical synthesis or by the manipulation of isolated segments of nucleic acids by genetic engineering techniques.
- recombinant DNA construct refers to any agent such as a plasmid, cosmid, virus, autonomously replicating sequence, phage, or linear or circular single-stranded or double-stranded DNA or RNA nucleotide sequence, derived from any source, capable of genomic integration or autonomous replication, comprising a DNA molecule that one or more DNA sequences have been linked in a functionally operative manner.
- recombinant DNA constructs or vectors are capable of introducing a 5′ regulatory sequence or promoter region and a DNA sequence for a selected gene product into a cell in such a manner that the DNA sequence is transcribed into a functional mRNA that is translated and therefore expressed.
- Recombinant DNA constructs or recombinant vectors may be constructed to be capable of expressing antisense RNAs, in order to inhibit translation of a specific RNA of interest.
- Regeneration refers to the process of growing a plant from a plant cell (e.g., plant protoplast or explant).
- Reporter refers to a gene and corresponding gene product that when expressed in transgenic organisms produces a product detectable by chemical or molecular methods or produces an observable phenotype.
- Restriction enzyme refers to an enzyme that recognizes a specific palindromic sequence of nucleotides in double stranded DNA and cleaves both strands; also called a restriction endonuclease. Cleavage typically occurs within the restriction site.
- “Selectable marker” refers to a nucleic acid sequence whose expression confers a phenotype facilitating identification of cells containing the nucleic acid sequence. Selectable markers include those that confer resistance to toxic chemicals (e.g. ampicillin resistance, kanamycin resistance), complement a nutritional deficiency (e.g. uracil, histidine, leucine), or impart a visually distinguishing characteristic (e.g. color changes or fluorescence).
- toxic chemicals e.g. ampicillin resistance, kanamycin resistance
- complement a nutritional deficiency e.g. uracil, histidine, leucine
- impart a visually distinguishing characteristic e.g. color changes or fluorescence
- Useful dominant selectable marker genes include genes encoding antibiotic resistance genes (e.g., resistance to hygromycin, kanamycin, bleomycin, G418, streptomycin or spectinomycin); and herbicide resistance genes (e.g., phosphinothricin acetyltransferase).
- antibiotic resistance genes e.g., resistance to hygromycin, kanamycin, bleomycin, G418, streptomycin or spectinomycin
- herbicide resistance genes e.g., phosphinothricin acetyltransferase
- the term “specific for (a target sequence)” indicates that a probe or primer hybridizes under given hybridization conditions only to the target sequence in a sample comprising the target sequence.
- Tolerant refers to a reduced toxic effect of glyphosate on the growth and development of microorganisms and plants.
- Transcription refers to the process of producing an RNA copy from a DNA template.
- Transformation refers to a process of introducing an exogenous nucleic acid sequence (e.g., a vector, recombinant nucleic acid molecule) into a cell or protoplast that exogenous nucleic acid is incorporated into a chromosome or is capable of autonomous replication.
- exogenous nucleic acid sequence e.g., a vector, recombinant nucleic acid molecule
- Transformed or “transgenic” refers to a cell, tissue, organ, or organism into that has been introduced a foreign nucleic acid, such as a recombinant vector.
- a “transgenic” or “transformed” cell or organism also includes progeny of the cell or organism and progeny produced from a breeding program employing such a “transgenic” plant as a parent in a cross and exhibiting an altered phenotype resulting from the presence of the foreign nucleic acid.
- transgene refers to any nucleic acid sequence nonnative to a cell or organism transformed into said cell or organism. “Transgene” also encompasses the component parts of a native plant gene modified by insertion of a nonnative nucleic acid sequence by directed recombination.
- translation refers to the production the corresponding gene product, i.e., a peptide, polypeptide, or protein from a mRNA.
- Vector refers to a plasmid, cosmid, bacteriophage, or virus that carries foreign DNA into a host organism.
- a polypeptide is “isolated” if it has been separated from the cellular components (nucleic acids, lipids, carbohydrates, and other polypeptides) that naturally accompany it or that is chemically synthesized or recombinant.
- a monomeric polypeptide is isolated when at least 60% by weight of a sample is composed of the polypeptide, preferably 90% or more, more preferably 95% or more, and most preferably more than 99%.
- Protein purity or homogeneity is indicated, for example, by polyacrylamide gel electrophoresis of a protein sample, followed by visualization of a single polypeptide band upon staining the polyacrylamide gel; high pressure liquid chromatography; or other conventional methods.
- Coat proteins can be purified by any of the means known in the art, for example as described in Guide to Protein Purification, ed. Guide to Protein Purification, ed. Manual Repeat Analysis, ed. Deutscher, Meth. Enzymol. 185, Academic Press, San Diego, 1990; and Scopes, Protein Purification: Principles and Practice, Springer Verlag, New York, 1982.
- Labeling There are a variety of conventional methods and reagents for labeling polypeptides and fragments thereof Typical labels include radioactive isotopes, ligands or ligand receptors, fluorophores, chemiluminescent agents, and enzymes. Methods for labeling and guidance in the choice of labels appropriate for various purposes are discussed, e.g., in Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989) and Ausubel et al., Greene Publishing and Wiley-Interscience, New York, (1992).
- Polypeptide fragments The present invention also encompasses fragments of an E indica EPSPS that lacks at least one residue of a native full-length E. indica EPSPS protein, but that specifically maintains EPSPS activity.
- Transit peptide or targeting peptide sequence generally refer to peptide sequences that when linked to a protein of interest directs the protein to a particular tissue, cell, subcellular location, or cell organelle. Examples include, but are not limited to, chloroplast transit peptides, nuclear targeting signals, and vacuolar signals.
- the chloroplast transit peptide is of particular utility in the present invention to direct expression of the EPSPS enzyme to the chloroplast.
- plant encompasses any higher plant and progeny thereof, including monocots (e.g., corn, rice, wheat, barley, etc.), dicots (e.g., soybean, cotton, tomato, potato, Arabidopsis, tobacco, etc.), gymnosperms (pines, firs, cedars, etc) and includes parts of plants, including reproductive units of a plant (e.g., seeds, bulbs, tubers, or other parts or tissues from that the plant can be reproduced), fruits and flowers.
- monocots e.g., corn, rice, wheat, barley, etc.
- dicots e.g., soybean, cotton, tomato, potato, Arabidopsis, tobacco, etc.
- gymnosperms pines, firs, cedars, etc
- Exogenous genetic material may be transferred into a plant by the use of a DNA vector designed for such a purpose by methods that utilize Agrobacterium, particle bombardment or other methods known to those skilled in the art.
- a particularly preferred subgroup of exogenous material comprises a nucleic acid molecule of the present invention. Design of such a vector is generally within the skill of the art (Plant Molecular Biology: A Laboratory Manual, eds. Clark, Springer, New York (1997).
- Examples of such plants in to which exogenous genetic material may be transferred include, without limitation, alfalfa, Arabidopsis, barley, Brassica, broccoli, cabbage, citrus, cotton, garlic, oat, oilseed rape, onion, canola, flax, maize, an ornamental annual and ornamental perennial plant, pea, peanut, pepper, potato, rice, rye, sorghum, soybean, strawberry, sugarcane, sugar beet, tomato, wheat, poplar, pine, fir, eucalyptus, apple, lettuce, lentils, grape, banana, tea, turf grasses, sunflower, oil palm, Phaseolus etc.
- the particular promoters selected for use in embodiments of the present invention should be capable of causing the production of sufficient expression to, in the case of the DNA molecule, generate protein expression in vegetative and reproductive tissues of a transformed plant.
- the DNA molecule will typically contain a constitutive promoter, a structural DNA sequence encoding a herbicide resistant enzyme, and a 3′ non-translated region.
- constitutive promoters that are active in plant cells have been described.
- Suitable promoters for constitutive expression in plants of herbicide tolerance for the DNA molecule include, but are not limited to, the cauliflower mosaic virus (CaMV) 35S promoter (Odell et al.
- Figwort mosaic virus FMV 35S
- sugarcane bacilliform virus promoter Bouhida et al., J. Gen. Virol. 74:15-22 (1993)
- commelina yellow mottle virus promoter Medberry et al., Plant J. 3:619-626 (1993)
- ssRUBISCO light-inducible promoter from the small subunit of the ribulose-1,5-bis-phosphate carboxylase
- sucrose synthase promoter Yang et al., Proc. Natl. Acad. Sci. U.S.A. 87: 4144-4148 (1990)
- the R gene complex promoter Chandler et al., The Plant Cell 1: 1175-1183 (1989)
- chlorophyll ⁇ / ⁇ binding protein gene promoter et cetera
- promoters Comparative analysis of constitutive promoters by the expression of reporter genes such as the uidA ( ⁇ -glucuronidase) gene from E. coli has been performed with many of these and other promoters (Li et al. Mol. Breeding 3:1-14 (1997); Wen et al. Chinese J. of Bot. 5:102-109 (1993).
- Promoters that are known or are found to cause transcription of DNA in plant cells can be used in the present invention. Such promoters may be obtained from a variety of sources such as plants and plant viruses.
- promoters may be identified for use in the current invention by screening a plant cDNA library for genes that are selectively or preferably expressed in the target tissues or cells.
- promoters utilized in the present invention have relatively high expression in these specific tissues.
- promoters for genes with tissue- or cell-specific or -enhanced expression. Examples of such promoters reported in the literature include the chloroplast glutamine synthetase GS2 promoter from pea (Edwards et al., Proc. Natl. Acad. Sci. U.S.A. 87: 3459-3463 (1990), the chloroplast fructose-1,6-biphosphatase (FBPase) promoter from wheat (Lloyd et al., Mol.
- promoters for the chlorophyll ⁇ / ⁇ -binding proteins may also be utilized in the present invention, such as the promoters for LhcB gene and PsbP gene from white mustard ( Sinapis alba ) (Kretsch et al., Plant Mol. Biol. 28: 219-229 (1995).
- organ-specific promoters e.g., Roshal et al., EMBO J. 6:1155-(1987); Schernthaner et al., EMBO J. 7:1249-1255 (1988); Bustos et al., Plant Cell 1:839.-853 (1989).
- the promoters utilized in the present invention have relatively high expression in these specific tissues.
- a number of promoters for genes with tuber-specific or -enhanced expression are known, including the class I patatin promoter (Bevan et al., EMBO J. 8:1899-1906 (1986); Jefferson et al., Plant Mol. Biol.
- the promoter for the potato tuber ADPGPP genes both the large and small subunits, the sucrose synthase promoter (Salanoubat et al.,, Gene 60:47-56 (1987); Salanoubat et al., Gene 84:181-185 (1989), the promoter for the major tuber proteins including the 22 kD protein complexes and proteinase inhibitors (Hannapel, Plant Physiol. 101:703-704 (1993), the promoter for the granule bound starch synthase gene (GBSS) (Visser et al., Plant Mol. Biol.
- Genomic clones for zein genes have been isolated (Pedersen et al., Cell 29:1015-1026 (1982), and the promoters from these clones, including the 15 kD, 16 kD, 19 kD, 22 kD, 27 kD, and gamma genes, could also be used.
- Other promoters known to function, for example, in Zea mays include the promoters for the following genes: waxy, Brittle, Shrunken 2, Branching enzymes I and II, starch synthases, debranching enzymes, oleosins, glutelins, and sucrose synthases.
- a particularly preferred promoter for Zea mays endosperm expression is the promoter for the glutelin gene from rice, more particularly the Osgt-1 promoter (Zheng et al., Mol. Cell Biol. 13:5829-5842 (1993).
- promoters suitable for expression in wheat include those promoters for the ADPglucose pyrosynthase (ADPGPP) subunits, the granule bound and other starch synthase, the branching and debranching enzymes, the embryogenesis-abundant proteins, the gliadins, and the glutenins.
- ADPGPP ADPglucose pyrosynthase
- promoters in rice include those promoters for the ADPGPP subunits, the granule bound and other starch synthase, the branching enzymes, the debranching enzymes, sucrose synthases, and the glutelins.
- a particularly preferred promoter is the promoter for rice glutelin, Osgt-1 gene.
- promoters for barley include those for the ADPGPP subunits, the granule bound and other starch synthase, the branching enzymes, the debranching enzymes, sucrose synthases, the hordeins, the embryo globulins, and the aleurone specific proteins.
- Root specific promoters may also be used.
- An example of such a promoter is the promoter for the acid chitinase gene (Samac et al., Plant Mol. Biol. 25:587-596 (1994). Expression in root tissue could also be accomplished by utilizing the root specific subdomains of the CaMV 35S promoter that have been identified (Lam et al., Proc. Natl. Acad. Sci. U.S.A. 86: 7890-7894 (1989).
- Other root cell specific promoters include those reported by Conkling et al. (Plant Physiol. 93: 1203-1211 (1990).
- the 5′ non-translated leader sequence can be derived from the promoter selected to express the heterologous gene sequence of the DNA molecule of the present invention, and can be specifically modified if desired so as to increase translation of mRNA.
- the 5′ non-translated regions can also be obtained from plant viral RNAs (Tobacco mosaic virus, Tobacco etch virus, Maize dwarf mosaic virus, Alfalfa mosaic virus, among others) from suitable eukaryotic genes, plant genes (wheat and maize chlorophyll a/b binding protein gene leader), or from a synthetic gene sequence.
- the present invention is not limited to constructs wherein the non-translated region is derived from the 5′ non-translated sequence that accompanies the promoter sequence.
- the leader sequence could also be derived from an unrelated promoter or coding sequence.
- Leader sequences useful in context of the present invention comprise the maize Hsp70 leader (U.S. Pat. No. 5,362,865 and U.S. Pat. No. 5,859,347), and the TMV omega element (Gallie et al., The Plant Cell 1:301-311 (1989).
- a vector or construct may also include various regulatory elements.
- Intron sequences are known in the art to aid in the expression of transgenes in monocot plant cells. Examples of such introns include the Adh intron 1 (Callis et al., Genes and Develop. 1:1183-1200 (1987), the sucrose synthase intron (Vasil et al., Plant Physiol. 91:1575-1579 (1989), U.S. Pat. No. 5,955,330), first intron of the rice actin gene (U.S. Pat. No. 5,641,876).
- a vector may also include a transit peptide nucleic acid sequence.
- the glyphosate target in plants the 5-enolpyruvyl-shikimate-3-phosate synthase (EPSPS) enzyme, is located in the chloroplast.
- EPSPS 5-enolpyruvyl-shikimate-3-phosate synthase
- CTP chloroplast transit peptide
- examples of other such chloroplast proteins include the small subunit (SSU) of Ribulose-1,5,-bisphosphate carboxylase, Ferredoxin, Ferredoxin oxidoreductase, the light-harvesting complex protein I and protein II, and Thioredoxin F.
- non-chloroplast proteins may be targeted to the chloroplast by use of protein fusions with a CTP and that a CTP sequence is sufficient to target a protein to the chloroplast.
- a suitable chloroplast transit peptide such as, the Arabidopsis thaliana EPSPS CTP (Klee et al., Mol. Gen. Genet. 210:437-442 (1987), and the Petunia hybrida EPSPS CTP (della-Cioppa et al., Proc. Natl. Acad. Sci.
- the termination of transcription is accomplished by a 3′ non-translated DNA sequence operably linked in the chimeric vector to the gene of interest.
- the 3′ non-translated region of a recombinant DNA molecule contains a polyadenylation signal that functions in plants to cause the addition of adenylate nucleotides to the 3′ end of the RNA.
- the 3′ non-translated region can be obtained from various genes that are expressed in plant cells.
- the nopaline synthase 3′ untranslated region (Fraley et al., Proc. Natl. Acad. Sci.
- DNA constructs for glyphosate tolerance designed for expression in plastids will necessarily contain genetic elements that function in plastids.
- a vector may also include a screenable or scorable marker gene.
- Screenable or scorable markers may be used to monitor expression.
- Exemplary markers include a ⁇ -glucuronidase or uidA gene (GUS) that encodes an enzyme for that various chromogenic substrates are known (Jefferson, Plant Mol. Biol, Rep. 5:387-405 (1987); Jefferson et al., EMBO J.
- tyrosinase gene (Katz et al., J. Gen. Microbiol. 129:2703-2714 (1983) that encodes an enzyme capable of oxidizing tyrosine to DOPA and dopaquinone that in turn condenses to melanin; green flourescence protein (Elliot et al., Plant cell Rep. 18:707-714 (1999) and an ⁇ -galactosidase.
- selectable or screenable marker genes are also genes that encode a secretable marker whose secretion can be detected as a means of identifying or selecting for transformed cells. Examples include markers that encode a secretable antigen that can be identified by antibody interaction, or even secretable enzymes that can be detected catalytically.
- Secretable proteins fall into a number of classes, including small, diffusible proteins that are detectable, (e.g., by ELISA), small active enzymes that are detectable in extracellular solution (e.g., ⁇ -amylase, ⁇ -lactamase, phosphinothricin transferase), or proteins that are inserted or trapped in the cell wall (such as proteins that include a leader sequence such as that found in the expression unit of extension or tobacco PR-S).
- small active enzymes that are detectable in extracellular solution
- proteins that are inserted or trapped in the cell wall such as proteins that include a leader sequence such as that found in the expression unit of extension or tobacco PR-S.
- Other possible selectable and/or screenable marker genes will be apparent to those of skill in the art.
- Suitable methods are believed to include virtually any method shown effective in introducing the nucleic acid molecules into a plant cell, such as by Agrobacterium infection or direct delivery of nucleic acid molecules.
- Acceleration methods include, for example, microprojectile bombardment and the like.
- microprojectile bombardment One example of a method for delivering transforming nucleic acid molecules to plant cells is microprojectile bombardment. This method has been reviewed by Yang et al., Particle Bombardment Technology for Gene Transfer, Oxford Press, Oxford, England (1994).
- Non-biological particles that may be coated with nucleic acids and delivered into cells by a propelling force.
- Exemplary particles include those comprised of tungsten, gold, platinum, and the like.
- a particular advantage of microprojectile bombardment, in addition to it being an effective means of reproducibly transforming monocots, is that neither the isolation of protoplasts (Cristou et al., Plant Physiol.
- An illustrative embodiment of a method for delivering DNA into Zea mays cells by acceleration is a biolistics ⁇ -particle delivery system, that can be used to propel particles coated with DNA through a screen, such as a stainless steel or Nytex screen, onto a filter surface covered with corn cells cultured in suspension.
- a screen such as a stainless steel or Nytex screen
- Gordon-Kamm et al. describes the basic procedure for coating tungsten particles with DNA (Gordon-Kamm et al., Plant Cell 2:603-618 (1990). The screen disperses the tungsten nucleic acid particles so that they are not delivered to the recipient cells in large aggregates.
- a particle delivery system suitable for use with the present invention is the helium acceleration PDS-1000/He gun is available from Bio-Rad Laboratories (Bio-Rad, Hercules, Calif.) (Sanford et al., Technique 3:3-16 (1991).
- cells in suspension may be concentrated on filters.
- Filters containing the cells to be bombarded are positioned at an appropriate distance below the microprojectile stopping plate. If desired, one or more screens are also positioned between the gun and the cells to be bombarded.
- immature embryos or other target cells may be arranged on solid culture medium.
- the cells to be bombarded are positioned at an appropriate distance below the microprojectile stopping plate.
- one or more screens are also positioned between the acceleration device and the cells to be bombarded.
- bombardment transformation one may optimize the pre-bombardment culturing conditions and the bombardment parameters to yield the maximum numbers of stable transformants.
- Both the physical and biological parameters for bombardment are important in this technology. Physical factors are those that involve manipulating the DNA/microprojectile precipitate or those that affect the flight and velocity of either the macro- or microprojectiles.
- Biological factors include all steps involved in manipulation of cells before and immediately after bombardment, the osmotic adjustment of target cells to help alleviate the trauma associated with bombardment, and also the nature of the transforming DNA, such as linearized DNA or intact supercoiled plasmids. It is believed that pre-bombardment manipulations are especially important for successful transformation of immature embryos.
- plastids can be stably transformed.
- Method disclosed for plastid transformation in higher plants include particle gun delivery of DNA containing a selectable marker and targeting of the DNA to the plastid genome through homologous recombination (Svab et al. Proc. Natl. Acad. Sci. (U.S.A.) 87:8526-8530 (1990); Svab et al., Proc. Natl. Acad. Sci. (U.S.A.) 90:913-917 (1993); (Staub et al., EMBO J. 12:601-606 (1993).
- Agrobacterium-mediated transfer is a widely applicable system for introducing genes into plant cells because the DNA can be introduced into whole plant tissues, thereby bypassing the need for regeneration of an intact plant from a protoplast.
- the use of Agrobacterium-mediated plant integrating vectors to introduce DNA into plant cells is well known in the art. See, for example the methods described by Fraley et al., Bio/Technology 3:629-635 (1985) and Rogers et al., Methods Enzymol. 153:253-277 (1987). Further, the integration of the T-DNA is a relatively precise process resulting in few rearrangements.
- the region of DNA to be transferred is defined by the border sequences, and intervening DNA is usually inserted into the plant genome as described (Seemann et al., Mol. Gen. Genet. 205:34 (1986).
- Modern Agrobacterium transformation vectors are capable of replication in E. coli as well as Agrobacterium, allowing for convenient manipulations as described (Klee et al., In: Plant DNA Infectious Agents, Hohn and Schell, eds., Springer-Verlag, New York, pp. 179-203 (1985). Moreover, technological advances in vectors for Agrobacterium-mediated gene transfer have improved the arrangement of genes and restriction sites in the vectors to facilitate construction of vectors capable of expressing various polypeptide coding genes.
- the vectors described have convenient multi-linker regions flanked by a promoter and a polyadenylation site for direct expression of inserted polypeptide coding genes and are suitable for present purposes (Rogers et al., Methods Enzymol. 153:253-277 (1987).
- Agrobacterium containing both armed and disarmed Ti genes can be used for the transformations. In those plant varieties where Agrobacterium-mediated transformation is efficient, it is the method of choice because of the facile and defined nature of the gene transfer.
- a transgenic plant formed using Agrobacterium transformation methods typically contains a single genetic locus on one chromosome. Such transgenic plants can be referred to as being hemizygous for the added gene. More preferred is a transgenic plant that is homozygous for the added structural gene; i.e., a transgenic plant that contains two added genes, one gene at the same locus on each chromosome of a chromosome pair.
- a homozygous transgenic plant can be obtained by sexually mating (selfing) an independent segregant transgenic plant that contains a single added gene, germinating some of the seed produced and analyzing the resulting plants for the gene of interest.
- transgenic plants can also be mated to produce offspring that contain two independently segregating exogenous genes. Selfing of appropriate progeny can produce plants that are homozygous for both exogenous genes.
- Back-crossing to a parental plant and out-crossing with a non-transgenic plant are also contemplated, as is vegetative propagation. Descriptions of other breeding methods that are commonly used for different traits and crops can be found in Fehr, In: Breeding Methods for Cultivar Development, Wilcox J. ed., American Society of Agronomy, Madison Wis. (1987).
- Transformation of plant protoplasts can be achieved using methods based on calcium phosphate precipitation, polyethylene glycol treatment, electroporation, and combinations of these treatments (see, e.g., Potrykus et al., Mol. Gen. Genet. 205:193-200 (1986); Lorz et al., Mol. Gen. Genet. 199:178 (1985); Fromm et al., Nature 319:791 (1986); Uchimiya et al., Mol. Gen. Genet. 204:204 (1986); Marcotte et al., Nature 335:454-457 (1988).
- Application of these systems to different plant varieties depends upon the ability to regenerate that particular plant strain from protoplasts.
- Other methods of cell transformation can also be used and include but are not limited to introduction of DNA into plants by direct DNA transfer into pollen (Hess et al., Intern Rev. Cytol. 107:367 (1987); Luo et al., Plant Mol Biol. Reporter 6:165 (1988), by direct injection of DNA into reproductive organs of a plant (Pena et al., Nature 325:274 (1987), or by direct injection of DNA into the cells of immature embryos followed by the rehydration of desiccated embryos (Neuhaus et al., Theor. Appl. Genet. 75:30 (1987).
- the development or regeneration of plants containing the foreign, exogenous gene is well known in the art.
- the regenerated plants are self-pollinated to provide homozygous transgenic plants. Otherwise, pollen obtained from the regenerated plants is crossed to seed-grown plants of agronomically important lines. Conversely, pollen from plants of these important lines is used to pollinate regenerated plants.
- a transgenic plant of the present invention containing a desired exogenous nucleic acid is cultivated using methods well known to one skilled in the art.
- Transformation of monocotyledons using electroporation, particle bombardment, and Agrobacterium have also been reported. Transformation and plant regeneration have been achieved in asparagus (Bytebier et al., Proc. Natl. Acad. Sci.
- Assays for gene expression based on the transient expression of cloned nucleic acid vectors have been developed by introducing the nucleic acid molecules into plant cells by polyethylene glycol treatment, electroporation, or particle bombardment (Marcotte et al., Nature 335:454-457 (1988); Marcotte et al., Plant Cell 1:523-532 (1989); McCarty et al., Cell 66:895-905 (1991); Hattori et al., Genes Dev. 6:609-618 (1992); Goff et al., EMBO J. 9:2517-2522 (1990).
- Transient expression systems may be used to functionally dissect gene constructs (see generally, Mailga et al., Methods in Plant Molecular Biology, Cold Spring Harbor Press (1995). It is understood that any of the nucleic acid molecules of the present invention can be introduced into a plant cell in a permanent or transient manner in combination with other genetic elements such as promoters, leaders, transit peptide sequences, enhancers, introns, 3′ nontranslated regions and other elements known to those skilled in the art that are useful for control of transgene expression in plants.
- Eleusine indica has been shown to hybridize with Eleusine coracana (finger millet), an important cultivated millet of India and East Africa (Chennaveeraiah et al., Euphytica 2-3:489-495, (1974).
- Classical plant breeding methods can be used to transfer the gene and the glyphosate tolerant phenotype to crop plants within the family Poaceae.
- the DNA molecules of the EPSPS glyphosate resistance gene of E. indica (SEQ ID NO: 6) can be used as a probe to identify other like DNA molecules by standard methods. Oligonucleotide DNA molecules homologous or complementary to the EPSPS glyphosate resistance gene of E.
- indica can be used in a marker assisted breeding method (Simple sequence repeat DNA marker analysis, in “DNA markers: Protocols, applications, and overviews: (1997) 173-185, Cregan, et al., eds., Wiley-Liss NY ) to assist in the breeding of this gene into related and heterologous crop species.
- a marker assisted breeding method Simple sequence repeat DNA marker analysis, in “DNA markers: Protocols, applications, and overviews: (1997) 173-185, Cregan, et al., eds., Wiley-Liss NY .
- Plant species containing a naturally occurring EPSPS enzyme resistant to glyphosate have not been previously reported.
- the subject of this invention is the EPSPS enzyme isolated from Eleusine indica that has been shown to be resistant to glyphosate and the expression of the DNA molecule encoding this EPSPS enzyme in other plants that then confers glyphosate tolerance to those recipient plants.
- the glyphosate resistant EPSPS enzyme isolated from Eleusine indica glyphosate tolerant biotype has a novel Km with respect to binding of PEP as compared to other plant EPSP Synthases that have been modified for glyphosate resistance by a single amino acid substitution of a proline to serine substitution in the active site of the enzyme. The K m for PEP of the E.
- indica glyphosate resistant enzyme is little changed from the E. indica glyphosate sensitive EPSPS enzyme.
- this gene is from a monocot plant and hence may not need nucleic sequence modification to affect expression in transgenic monocot crop plants.
- the E. indica glyphosate tolerant EPSPS enzyme amino acid sequence can be modified by site directed mutation to include other known substitutions.
- the present invention also provides for parts of the plants of the present invention.
- Plant parts include seed, endosperm, ovule and pollen.
- the plant part is a seed.
- Eleusine indica plants tolerant to glyphosate were collected from a site near Johor, Malaysia. Glyphosate tolerant biotypes of E. indica are identified and numbered. Seed is collected from each biotype and planted in pots in the greenhouse. Clones are generated for each plant by excising 10-20 tillers and transplanting these in separate pots. Glyphosate sensitive and tolerant individual plants are then identified by treatment with glyphosate at either 0.5 kg active ingredient (ai)/hectare (ha) or 2.0 kg ai/ha, respectively. A corresponding clone for glyphosate tolerant and glyphosate sensitive E. indica biotype is left untreated. These clones are used as the source of fresh tissue for enzyme analysis and gene isolation.
- RNAs are extracted from frozen crown samples using the RNeasy Plant Mini Kit (cat. #74904, Qiagen Inc., Valencia, Calif.) per manufacturer's instructions. Oligo.dT-primed first-strand cDNAs are prepared from 5 ⁇ g samples of total RNA using the Superscript Pre-Amplification System (cat. #18089-011, Life Technologies, Rockville, Md.) per manufacturer's instructions.
- PCR amplifications are then performed using the Expand High Fidelity PCR System (cat. #1 732 641, Roche Molecular Biochemicals, Indianapolis, Ind.) per manufacturer's instructions.
- a thermal profile of 94° C. for 20 seconds, followed by 60° C. for 1 minute, then 72° C. for 1 minute 30 seconds is used for the initial 30 cycles with a 0.5° C. decrease in annealing temperature per cycle. This is followed by 10 additional cycles of 94° C. for 20 seconds, 45° C. for 1 minute, then 72° C. for 1 minute 30 seconds.
- RT-PCR products are then purified by agarose gel electrophoresis using a QIAquick Gel Extraction Kit (cat. #28704, Qiagen Inc., Valencia, Calif.) then directly cloned into the pCR2.1-TOPO vector (cat. #K4500-40, Invitrogen, Carlsbad, Calif.). The identity of the cloned RT-PCR products is confirmed by DNA sequence analysis (ABI PrismTM 377, Perkin Elmer, Foster City, Calif.).
- the remainder of the 3′ end of the EPSP synthase coding region is generated using the 3′ RACE System for Rapid Amplification of cDNA Ends (cat. #18373-027, Life Technologies, Rockville, Md.), using the gene-specific oligonucleotide of SEQ ID NO: 3.
- the cDNA is prepared according to manufacturer's instructions using 5 ⁇ g of total RNA isolated from crown tissues as previously described.
- PCR amplifications are conducted in 50 ⁇ l reactions including 5 ⁇ l first-strand cDNA reaction, 20 picomoles of each primer, 10 mM Tris.HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl 2 , 200 ⁇ M dNTPs, and 2.5 units Taq polymerase.
- a thermal profile of 94° C. for 20 seconds, followed by 57° C. for 1 minute, then 72° C. for 1 minute 30 seconds is used for 35 cycles.
- the identity of the 3′-RACE products is confirmed by DNA sequence analysis (ABI PrismTM 377, Perkin Elmer, Foster, Calif.).
- the remainder of the 5′ end of the E. indica EPSP synthase mature protein coding region is generated using the SMART RACE cDNA Amplification Kit (cat. #K1811-1, Clontech Laboratories Inc., Palo Alto, Calif.), using the gene-specific oligonucleotides of SEQ ID NO: 4 and SEQ ID NO: 5.
- the cDNA is prepared according to manufacturer's instructions using 150 ng of polyA+ mRNA isolated from crown tissues using an Oligotex mRNA Midi Kit (cat. #28704, Qiagen Inc., Valencia, Calif.). (SEQ ID NO: 4) 5′-GGCTGCTGTCAATGTCGCATTGCAGTTCC-3′ (SEQ ID NO: 5) 5′-CTCTTTCGCATCCTTCTCAACTGGGAACTTGC-3′
- PCR reactions are conducted as recommended by the manufacturer, except that the Expand High Fidelity PCR System (cat. #1 732 641, Roche Molecular Biochemicals, Indianapolis, Ind.) is used and DMSO is included in all reactions at a final concentration of 5.0% to facilitate the amplification of GC-rich sequences.
- the synthetic DNA oligonucleotide described in SEQ ID NO: 4 is used in the primary amplifications, then second round (“nested”) amplifications are performed using the oligonucleotide described in SEQ ID NO: 5, with a 1 ⁇ l aliquot of 1:100 dilution of the primary PCR reactions.
- the identity of the 5′-RACE products is confirmed by DNA sequence analysis (ABI PrismTM 377, Perkin Elmer, Foster City, Calif.).
- EPSP synthase enzyme from the glyphosate tolerant E. indica biotype confers increased glyphosate tolerance in transgenic E. coli.
- E. coli is useful as a heterologous expression system for testing glyphosate resistant enzymes.
- the EPSP synthase mature protein-coding regions isolated from the glyphosate tolerant and glyphosate sensitive E. indica biotypes can be directly compared for their ability to confer tolerance to glyphosate in transgenic hosts.
- E. coli strain SR481 are transformed with the glyphosate resistant EPSPS gene (Ei.EPSPS:glyR) and the glyphosate sensitive EPSPS gene (Ei.EPSPS:glyS) purified from E.
- This nucleotide sequence includes 1) flanking BglII and Nde1 endonuclease sites, 2) a ribosome binding site, and 3) an unstructured region 5′ to the ribosome binding element (Balbas, P. et. al., in “Methods in Enzymology” (D. V. Goeddel, ed.)185: 15-37, 1990). This was inserted by ligation to facilitate expression and cloning at the ATG start codon of an open reading frame. A multiple cloning site is positioned immediately downstream of this Ndel site, followed by the rho-independant transcriptional terminator element of the E.
- This vector when it operbly contains the EPSP synthase coding sequences of the present invention is employed for the inducible expression of glyphosate resistant and glyphosate sensitive EPSP synthase cDNAs in E. coli.
- Other commercially available inducible E. coli expression vectors are suitable for testing the EPSP synthases from E. indica.
- E. coli expression vectors carrying the EPSP synthase mature protein coding sequences from the tolerant and sensitive E. indica biotypes the oligonucleotide primers of SEQ ID NO: 9 and SEQ ID NO: 10. 5′-GCAATTCCATATGGCGGGCGCGGAGGAGGTGGTGCT-3′ (SEQ ID NO: 9) 5′-GACTAGGAATTCTTAGTTCTTTTGACGAAAGTGCTCAGCACGTCGAAG-3′, (SEQ ID NO: 10)
- RT-PCR reactions are employed in RT-PCR reactions to generate expression cassettes suitable for cloning into pMON45337 cut with the restriction enzymes Nde1 and EcoR1.
- RT-PCR reactions are performed with total RNAs extracted from frozen crown samples using the RNeasy Plant Mini Kit (cat. #74904, Qiagen Inc., Valencia, Calif.) per manufacturer's instructions.
- Oligo.dT-primed first-strand cDNAs are prepared from 5 ⁇ g samples of total RNA using the Superscript Pre-Amplification System (cat. #18089-011, Life Technologies, Rockville, Md.) per manufacturer's instructions. Two ⁇ l of first-strand cDNA are then used to generate E. indica EPSP synthase expression cassettes via polymerase chain reaction.
- oligonucleotides are added in 50 ⁇ l RT-PCR reactions at a final concentration of 0.4 ⁇ M.
- PCR amplifications are then performed using the Expand High Fidelity PCR System (cat. #1 732 641, Roche Molecular Biochemicals, Indianapolis, Ind.) per manufacturer's instructions, using a thermal profile of 94° C. for 30 seconds, then 57° C. for 2 minutes, followed by 75° C. for 3 minutes, for a total of 35 cycles.
- the resulting PCR products are digested with Nde I and EcoRI, then ligated into pMON45337, resulting in the E. coli expression vectors pMON45364 (FIG. 5) and pMON45365 (FIG.
- E. indica EPSP synthase isolated from the resistant and sensitive biotype, respectively.
- Expression of the two enzymes in E. coli will thus be directed by the Lac operon and trpA gene genetic elements described above for pMON45337.
- the accuracy of the cloned sequences are confirmed by DNA sequence analysis (ABI PrismTM 377, Perkin Elmer, Foster, Calif.).
- pMON45337, pMON45364, and pMON45365 are all transformed into the E. coli strain SR481, an aroA-strain lacking endogenous EPSP synthase activity (Padgette et al., Arch. Biochem. Biophys. 258:564-573 (1987).
- coli SR481 cells transformed with pMON45337, pMON45364 (Ei.EPSPS:glypR), and pMON45365 (Ei.EPSPS:glypS) are grown in Terrific Broth (Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989), supplemented with 1.0 mM IPTG, 50 ⁇ g/ml ampicillin, and 100 ⁇ g/ml each of L-phenylalanine, L-tyrosine, and L-tryptophan. O.D. 595 measurements are taken on all of the overnight cultures to confirm similar cell densities.
- E.EPSPS glypR
- pMON45365 Ei.EPSPS:glypS
- coli SR48 1-pMON45364 and SR481-pMON45365 cells 14 ml culture tubes (cat.# 60818-725, VWR Scientific, West Chester, Pa.) each containing 3.0 ml of minimal M9 media (Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989) supplemented with 50 ⁇ g/ml ampicillin, 1.0 mM IPTG, and either 0.0, 0.5, 1.5, or 5.0 mM glyphosate (N-phosphonomethyl glycine or a salt thereof) are inoculated with 100 ⁇ l of undiluted overnight culture per tube. Each experimental condition is performed in triplicate to confirm the reproducibility of the experiment.
- Homogenates are prepared from 0.5 g tissue per sample in 25 ml extraction buffer (100 mM TrisCl, 10% glycerol, 1 mM EDTA, 1 mM benzamidine, 1 mM dithiothreitol, 1 mM 4-(2-aminoethyl)-benzenesulfonyl floride HCl, 0.1 mM leupeptin, pH 7.4) at 4° C. using a model PT3000 Polytron homogenizer (Brinkman Instuments Inc., Westbury, N.Y.).
- reaction include 50 mM HEPES, pH 7.0, 5 mM potassium fluoride, 1 mM shikimate-3-phosphate, 0.5 mM [1- 14 C]-phosphoenolpyruvate (29.0 mCi/mmol cyclohexylammonium salt; #CFQ10004, Amersham Life Science, Inc., Arlington Heights, Ill.), and 0.1 mM ammonium molybdate).
- Reactions are quenched with the addition of 50 ⁇ l 9:1 ethanol: 0.1 M acetic acid. Thirty ⁇ l of quenched reaction is then injected onto a Synchropak AX100 anion exchange column (cat. #942804, P.J.
- FIG. 4 shows data generated for a typical glyphosate inhibition study, comparing the EPSP synthase activities detectable in extracts prepared from the glyphosate sensitive and tolerant E. indica biotypes.
- E.EPSPS glyphosate resistant EPSP synthase gene from E. indica
- a monocot vector that utilizes a plant expression cassette that contains a promoter (P) and first intron (I) from the rice (Os) actin gene P-Os.Act1/I-Os.Act1 (U.S. Pat. No.
- TS plastid transit peptide sequence
- Arabidopsis thaliana Arabidopsis thaliana
- TS-At.EPSPS:CTP Arabidopsis thaliana
- T-AGRTU.nos polyadenylation/termination region from the Agrobacterium tumefaciens nopaline synthase gene
- This expression cassette may be combined with a second transgene expression cassette by plant breeding, plant transformation, or by joining in a DNA construct that comprises a plant DNA virus promoter, for example, the cauliflower mosaic virus (CaMV) 35S promoter containing a tandem duplication of the enhancer region, operably connected to a Zea mays Hsp70 intron, operably connected to a nucleic acid sequence encoding an Arabidopsis thaliana EPSPS chloroplast transit peptide sequence, operably connected to a E. indica glyphosate resistant EPSPS coding sequence, operably connected to a nopaline synthase transcriptional terminator.
- CaMV cauliflower mosaic virus
- Other combinations of genetic elements are known and those skilled in the art of plant molecular biology can easily construct plant expression vectors that will express the E. indica EPSPS glyphosate resistant enzyme at sufficient levels to confer glyphosate the transformed plant.
- a plant expression vector that utilizes the promoter and 5′ untranslated region (including intron I) of the plant elongation factor 1 ⁇ gene (Elf ⁇ -A1) as described in U.S. Pat. No. 5,177,011 or more specifically, dicot vector of the present invention which utilizes the Arabidopsis thaliana Elf ⁇ -A1 promoter and intron sequence (P-At.Elf1a/I-At.Elf1a), (Axelos et al., Mol. Gen. Genet.
- This expression cassette may be combined with a second transgene expression cassette by plant breeding, plant transformation, or by joining in a DNA construct that comprises a plant DNA virus promoter, for example, the Figwort mosaic virus (FMV) 34S promoter, operably connected to a nucleic acid sequence encoding an Arabidopsis thaliana EPSPS chloroplast transit peptide sequence, operably connected to a E. indica glyphosate resistant EPSPS coding sequence, operably connected to a nopaline synthase transcriptional terminator.
- FMV Figwort mosaic virus
- Other combinations of genetic elements are known and those skilled in the art of plant molecular biology can easily construct plant expression vectors that will express the E. indica EPSPS glyphosate resistant enzyme at sufficient levels to confer glyphosate tolerance to the transformed plant.
- oligonucleotide primers of SEQ ID NO: 11 and SEQ ID NO: 12 are employed in PCR reactions to generate an expression cassette suitable for direct cloning into a monocot vector and a dicot vector.
- 5′-GCAATTCGCATGCCGGGCGCGGAGGAGGTGGTGCT-3′ SEQ ID NO: 11
- 5′-GACTAGGAATTCTTAGTTCTTTTGACGAAAGTGCTCAGCACGTCGAAG-3′ SEQ ID NO: 12
- PCR reactions are performed using 300-500 ng of pMON45364 plasmid DNA as template to amplify the E. indica EPSP synthase mature protein coding region, flanked by Sph1 and EcoR1 restriction cleavage sites.
- the oligonucleotides are added in 50 ⁇ l PCR reactions at a final concentration of 0.4 ⁇ M.
- PCR amplifications are then performed using the Expand High Fidelity PCR System (cat. #1 732 641, Roche Molecular Biochemicals, Indianapolis, Ind.) per manufacturer's instructions, using a thermal profile of 94° C. for 30 seconds, then 57° C. for 2 minutes, followed by 75° C.
- PCR products are digested with Sph1 and EcoR1, then ligated into pMON45366 and pMON45368 resulting in the plant expression vectors pMON45367 (FIG. 7) and pMON45369 (FIG. 8), respectively.
- the accuracy of the cloned sequences are confirmed by DNA sequence analysis (ABI PrismTM 377, Perkin Elmer, Foster City, Calif.).
- Transgenic corn can be produced by particle bombardment transformation methods as described in U.S. Pat. No. 5,424,412.
- the plant expression vector (pMON45367) contains the glyphosate resistant E. indica EPSPS mature protein coding sequence in an expression cassette suitable for expression in monocot plants.
- the pMON45367 plasmid DNA is digested with Not1 and Pme1 restriction endonucleases to complete digestion.
- the 3.3 kb expression cassette is agarose gel purified, then bombarded into embryogenic corn tissue culture cells using a Biolistic® (Dupont, Wilmington, Del.) particle gun with purified isolated DNA fragment.
- Transformed cells are selected on glyphosate (N-phosphonomethyl glycine and its salts) containing media and whole plants are regenerated then grown under greenhouse conditions. Fertile seed is collected, planted and the glyphosate tolerant phenotype is back crossed into commercially acceptable corn germplasm by methods known in the art of corn breeding (Sprague et al., Corn and Corn Improvement 3 rd Edition, Am. Soc. Agron. Publ (1988).
- Transgenic corn plants can be produced by an Agrobacterium mediated transformation method.
- a disarmed Agrobacterium strain C58 (ABI) harboring a binary vector (pMON45367) is used for all the experiments.
- the pMON45367 is transferred into Agrobacterium by a triparental mating method (Ditta et al., Proc. Natl. Acad. Sci. 77:7347-7351). Liquid cultures of Agrobacterium are initiated from glycerol stocks or from a freshly streaked plate and grown overnight at 26° C.-28° C.
- the embryos are then transferred to delay media (N6 1-100-12/micro/Carb 500/20 ⁇ M AgNO3) and incubated at 28 ° C. for 4 to 5 days. All subsequent cultures are kept at this temperature. Coleoptiles are removed one week after inoculation.
- the embryos are transferred to the first selection medium (N61-0-12/Carb 500/0.5 mM glyphosate).
- Two weeks later, surviving tissue are transferred to the second selection medium (N61-0-12/Carb 500/1.0 mM glyphosate). Subculture surviving callus every 2 weeks until events can be identified. This will take 3 subcultures on 1.0 mM glyphosate. Once events are identified, bulk up the tissue to regenerate.
- callus tissues are transferred to the regeneration medium (MSOD 0.1 ⁇ M ABA) and incubated for two weeks.
- the regenerating calli are transferred to a high sucrose medium and incubated for two weeks.
- the plantlets are transferred to MSOD media in culture vessel and kept for two weeks. Then the plants with roots are transferred into soil.
- R 0 plants Three R 0 plants are regenerated for any given transgenic event. These three plants are expected to be near isogenic because they are thought to be derived from a single transgenic plant cell. Thus, one plant is used as a non-sprayed control and the remaining two plants are treated with glyphosate (as Roundup® herbicide). Plants are most effectively treated with glyphosate at V2-V6 stage. Glyphosate (as Roundup® herbicide) is administered through the use of a linear track sprayer set to deliver a 16, 32 or 64 oz./A rate of glyphosate.
- the R 0 plants produced are allowed to self, then R 1 plants are screened using spray applications of glyphosate and the rating system as described for the R 0 screen.
- An increase in whole-plant tolerance to the herbicide, as compared to non-transgenic control plants, is used to assess the utility of the E. indica EPSP synthase enzyme for the generation of glyphosate tolerance in planta.
- Immature embryos of wheat ( Triticum aestivum L) cultivar Bobwhite are isolated from the immature caryopsis 13-15 days after pollination, and cultured on CM4C (Table 3) for 3-4 days. The embryos showing active cell division, but no apparent callus formation are selected for Agrobacterium infection.
- a disarmed Agrobacterium strain C58 (ABI) harboring a binary vector of interest (pMON45367) is used for all the experiments.
- the pMON45367 is transferred into Agrobacterium by a triparental mating method (Ditta et al., Proc. Natl. Acad. Sci. 77:7347-7351). Liquid cultures of Agrobacterium are initiated from glycerol stocks or from a freshly streaked plate and grown overnight at 26° C.-28° C.
- the immature embryos cultured in CM4C medium are transferred into sterile petri plates (16 ⁇ 20 mm) or wells of a 6-well cell culture plate (Costar Corporation, Cambridge, Mass.) containing 10 ml of inoculation medium per petri plate or 5 ml per cell culture cluster plate.
- An equal amount of the Agrobacterium cell suspension is added such that the final concentration of Agrobacterium cells is an OD 600 of 0.5.
- pluronic F68 is added to the inoculation mixture at a final concentration of 0.01%.
- the ratio between the Agrobacterium and immature embryos is about 10 ml: 20-200 IEs.
- the inoculation is allowed to proceed at 23° C.-26° C. from 5-60 minutes.
- the remaining Agrobacterium cells are removed from the explants by using vacuum aspiration equipment.
- a piece of sterile Whatman No. 1 filter paper (to fit the size of the petri plate) is placed in each of 60 ⁇ 15 or 60 ⁇ 20 mm petri dishes. Two hundred ⁇ l of sterile water is placed in the middle of the filter paper.
- the inoculated immature embryos are placed in the plates.
- 20-50 explants are grouped as one stack (about 1 cm in size and 60-80 mg/stack), with 4-5 stacks on each plate.
- the plates are immediately covered with Parafilm® and then co-cultivated in the dark at 24° C.-26° C. for 2-3 days.
- the co-cultivated PCIEs are transferred CM4C+500 mg/l carbenicillin medium (delay medium) at dark. After 7 days on the delay medium, the immature embryos are transferred to CM4C supplemented with 2 mM glyphosate and 500 mg/l carbenicillin for selection for one week. Then calli are transferred to MMS0.2C+0.1 mM glyphosate +250 mg/l carbenicillin medium for 2 weeks under light for further selection. Embryogenic calli are transferred to a second regeneration medium MMS0C with lower glyphosate concentration (0.02 mM) and 500 mg/L carbenicillin for plant regeneration. Those embryogenic calli are transferred onto fresh medium every two weeks. Regenerated plantlets are transferred to Sundae cups (Sweetheart Cup Company, Chicago, Ill.) containing the second regeneration medium for further growth and selection. When roots are well established from transgenic plants the plants are transferred to soil for further evaluation.
- Novel glyphosate-resistant EPSP synthases can be designed based on the E. indica glyphosate resistant EPSPS.
- the amino acid sequence deduced from the cDNA sequence shows that two amino acid substitutions distinguish the mature EPSP protein sequence derived from the glyphosate-tolerant E. indica biotype (top row, FIG. 9) from that of the glyphosate sensitive E. indica biotype EPSPS protein sequence (bottom row, FIG. 9).
- indica glypR was found to not have a major effect on the K m indicating that this enzyme will continue to function well in the plant chloroplast. It required a double mutation in the Z. mays EPSPS enzyme to achieve a low K m for PEP. TABLE 4 Comparison of the apparent K m for PEP and apparent K i for glyphosate of the E. indica glyphosate resistant EPSPS with other known plant EPSPS modified for glyphosate resistance. EPSPS enzyme K m PEP ( ⁇ M) K i Glyphosate ( ⁇ M) E. indica glypS 5 0.05 E. indica glypR (Pro-Ser) 7 1 Z.
- glypS mays glypS (wt) 5 0.2 Z. mays glypR (Thr-Ile, Pro-Ser) 5 60 P. hybrida glypS (wt) 5 0.4 P. hybrida glypR (Pro-Ser) 44 3 P. hybrida glypR (Gly-Ala) 200 2000 P. hybrida glypR (Gly-Ala, Pro- 340 8500 Ser)
- K m (PEP) determinations for the different enzymes are performed at saturating shikimate-3-phosphate (S3P) concentrations, which is determined according to standard methods (Fersht, Enzyme Structure and Mechanism, W.H. Freeman and Co., Ltd., San Francisco, Calif., 1977). A series of PEP concentrations are tested, such that the final range of concentrations spans one order of magnitude above and below the experimentally determined K m .
- K i (glyphosate) is determined in a similar manner, at saturating S3P concentrations, except that velocity vs.
- PEP is determined for a range of glyphosate concentrations (Orsi in “Methods in Enzymology” (Purich, ed.), vol. 63, pg. 159-183, 1979). Calculations, graphical representation, and statistical analysis of enzyme kinetic data are performed using GraFit version 3.0 software (Erithacus Software Ltd., Staines, U.K.).
- transgenic plants resistant to glyphosate are made by transformation with an E. indica glyphosate resistant EPSPS gene construct that can include additional modification of the naturally occurring amino acid sequence.
- These changes are be made by site-directed mutagenesis of the codons of DNA sequence to incorporate other known amino acid substitutions in glyphosate resistant plant EPSPSs, such as the threonine to isoleucine substitution at 103 (U.S. Pat. No. 6,040,497), and glycine to alanine substitution at 102 (U.S. Pat. No. 5,188,642) in the catalytic domain of the E. indica EPSPS amino acid seqeunce.
- the catalytic domain of the E. indica EPSPS can be modified to the amino acid sequence of the catalytic domain of the Agrobacterium strain CP4 glyphosate resistant EPSPS (U.S. Pat. No. 5,633,435) by the same methods.
- This modification will result in the plant derived EPSPS possessing similar PEP binding and glyphosate resistance as the CP4 glyphosate resistant EPSPS that has been used in cotton, corn, canola, soybeans, potato, wheat, sugarbeet and other agronomically important crop plant to impart plant tolerance to glyphosate.
- Modification of plant EPSP Synthases to the CP4 EPSPS catalytic domain sequence may comprise the deletion of an amino acid and the substitution of other amino acids.
- the deletion of the amino acid at 107 of the E. indica EPSPS sequence (FIG. 2) or the same relative amino acid position in other plant EPSP Synthases that can in addition to the deletion include substitutions of an alanine for a glycine at 102, glycine for alanine at 104, cysteine for methionine at 105, methionine for alanine at 110, or glycine for alanine at 111.
- E. indica EPSPS regulatory sequences can be isolated by any number of methods known to those of skill in the art for genomic library preparation.
- E. indica genomic DNA is isolated by a CsCl purification protocol according to Current Protocols in Molecular Biology, ed. Ausubel et al., Greene Publishing and Wiley-Interscience, New York, 1992 (with periodic updates); or by a CTAB purification method (Rogers et al., Plant Mol. Biol., 5:69, 1988). Reagents are available commercially (see, for example Sigma Chemical Co., St. Louis, Mo.).
- genomic DNA libraries are prepared according to manufacturer instructions (Genome WalkerTM, CloneTech Laboratories, Inc, Palo Alto, Calif.). In separate reactions, genomic DNA is subjected to restriction enzyme digestion overnight at 37° C. with the following blunt-end endonucleases: EcoRV, Sca1, Dra1, PvuII, or Stu1 (CloneTech Laboratories, Inc. Palo Alto, Calif.). The reaction mixtures are extracted with phenol:chloroform, ethanol precipitated, and resuspended in Tris-EDTA buffer (10 mM Tris-.HCI, pH 8.0, 1 mM EDTA).
- the purified blunt-ended genomic DNA fragments are then ligated to the Genome WalkerTM adapters and ligation of the resulting DNA fragments to adapters were done according to the manufacturer's protocol. After ligation, each reaction is heated treated (70° C. for 5 min) to terminate the reaction and then diluted 10-fold in Tris-EDTA buffer. One ⁇ l of each respective ligation is then amplified in a 50 ⁇ l reaction according to manufacturer's recommended protocol using an adaptor-specific oligonucleotide (supplied by manufacturer) and an E. indica EPSP synthase gene-specific oligonucleotide, such as SEQ ID NO 4.
- PCR products representing 5′ regions of the E. indica EPSP synthase gene are then purified by agarose gel electrophoresis using a QIAquick Gel Extraction Kit (cat. #28704, Qiagen Inc., Valencia, Calif.) then directly cloned into the pCR2.1-TOPO vector (cat. #K4500-40, Invitrogen, Carlsbad, Calif.).
- E. indica Genome WalkerTM libraries and methods that are used to isolate the E. indica EPSP synthase 5′ region can be used to isolate the 3′ region of E. indica EPSP synthase gene, by substituting gene-specific primers (first round SEQ ID NO: 13 and second round SEQ ID NO: 14) that anneal to the 3′ end of the gene.
- Amplification products are cloned and verified as for the 5′ end of the E. indica EPSP synthase gene. 5′-TGCAATCCGGACTGAGCTAACAAAGC-3′ (SEQ ID NO: 13) 5′-ACTGCATTATCACACCGCCCGAGAAG-3′ (SEQ ID NO: 14)
- the translation initiation codon is determined for the E. indica EPSP synthase gene by inspection, anticipating an initiation codon approximately 63 codons upstream of the start of the mature protein codon region, based on comparison to the maize EPSP synthase gene. Primers are then designed to amplify approximately 2.5 kb of the 5′ region beginning at the initiation codon. These primers incorporate restriction sites for cloning into expression vectors, for example, placing a EcoR1 site in the 5′ end of promoter region the E. indica EPSP synthase gene and an Nco1 site incorporating the translation start. Such primers are added in a 50 ⁇ l RT-PCR reaction at a final concentration of 25 ⁇ M with 50 ng of E.
- PCR amplifications are then performed using the Expand High Fidelity PCR System (cat. #1 732 641, Roche Molecular Biochemicals, Indianapolis, Ind.) per manufacturer's instructions.
- a thermal profile of 94° C. for 30 seconds, followed by 60° C. for 30seconds, then 72° C. for 3 minutes is used for thirty cycles. This is followed by a cycle of 72° C. for 3 minutes.
- the gel purified amplification product is then digested with Pst1 and Nco1.
- the 3′ end of the E. indica EPSP synthase gene is amplified using two gene specific primers (SEQ ID NO: 15 and SEQ ID NO: 16) which incorporate a BamH1 site immediately downstream of the translation stop codon and a Pst1 approximately 650 bases downstream of the translation stop codon.
- the product is amplified as for the 5′ end of the E. indica EPSP synthase gene except an extension time of 1 minute is used.
- the gel purified product is digested with BamH1 and Pst1.
- SEQ ID NO: 15 5′-CTAAGGATCCTCTGTGCCTGCCTCATGAAGAGAGTT-3′
- SEQ ID NO: 16 5′-TGATCTGCAGGCAAGTGTCTTACCCTTACCCTTCTG-3′
- the 5′ (EcoR1/Nco1 fragment) regulatory and 3′ (BamH1/Pst1 fragment) regulatory regions of the E. indica EPSP synthase gene can be ligated to a compatibly digested vector and a coding region to generate a transgene capable of expressing a transcript under control of E. indica EPSP synthase gene regulatory elements.
- a coding region would be the A. fumefaciens strain CP4 EPSP synthase gene (U.S. Pat. No. 5,633,435) expressed under the control of the E. indica regulatory sequences.
- the basal expression of the E. indica EPSP synthase gene promoter may be modified to enhance its expression.
- Methods known to those of skill in the art can be used to insert enhancing elements (for example, subdomains of the CaMV 35S promoter, Benfey et. al, 1990 EMBO J. 9: 1677-1684) into the E. indica EPSP synthase gene 5′ sequence to generate a promoter which encompasses the temporal and spatial expression of the E. indica EPSP synthase gene but have quantitatively higher levels of expression.
- tissue specific modifications of the E. indica EPSP synthase 5′ region expression can be accomplished with elements determined to specifically activate or repress gene expression (for example, pollen specific elements, Eyal et al., 1995 Plant Cell 7: 373-384).
Landscapes
- Health & Medical Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Genetics & Genomics (AREA)
- Engineering & Computer Science (AREA)
- Chemical & Material Sciences (AREA)
- Zoology (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Wood Science & Technology (AREA)
- Organic Chemistry (AREA)
- General Engineering & Computer Science (AREA)
- Biomedical Technology (AREA)
- Biotechnology (AREA)
- Molecular Biology (AREA)
- Microbiology (AREA)
- Biochemistry (AREA)
- General Health & Medical Sciences (AREA)
- Medicinal Chemistry (AREA)
- Cell Biology (AREA)
- Physics & Mathematics (AREA)
- Biophysics (AREA)
- Plant Pathology (AREA)
- Breeding Of Plants And Reproduction By Means Of Culturing (AREA)
- Micro-Organisms Or Cultivation Processes Thereof (AREA)
- Agricultural Chemicals And Associated Chemicals (AREA)
- Enzymes And Modification Thereof (AREA)
Abstract
The methods and materials disclosed herein are directed to glyphosate herbicide tolerance in plants. In particular, the isolation of a glyphosate resistant EPSP synthase coding sequence and its regulatory elements from Eleusine indica. The coding sequence and regulatory sequences are useful to genetically engineer plants for tolerance to glyphosate herbicide.
Description
- This invention relates in general to plant molecular biology and plant genetic engineering for herbicide resistance and, more particularly, to a novel glyphosate resistant 5-enolpyruvylshikimate-3-phosphate synthase from Eleusine indica. Plant genetic engineering methods can be used to transfer the glyphosate resistant 5-enolpyruvylshikimate-3-phosphate synthase gene isolated and purified from Eleusine indica into crop and ornamental plants of economic importance.
- N-phosphonomethylglycine, also known as glyphosate, is a well known herbicide that has activity on a broad spectrum of plant species. Glyphosate is the active ingredient of Roundup® (Monsanto Co.), a safe herbicide having a desirably short half life in the environment. When applied onto a plant surface, glyphosate moves systemically through the plant. Glyphosate is toxic to plants by inhibiting the shikimic acid pathway that provides a precursor for the synthesis of aromatic amino acids. Specifically, glyphosate affects the conversion of phosphoenolpyruvate and 3-phosphoshikimic acid to 5-enolpyruvyl-3-phosphoshikimic acid by inhibiting the enzyme 5-enolpyruvyl-3-phosphoshikimate synthase (hereinafter referred to as EPSP synthase or EPSPS). For purposes of the present invention, the term “glyphosate” should be considered to include any herbicidally effective form of N-phosphonomethylglycine (including any salt thereof) and other forms that result in the production of the glyphosate anion in planta.
- Through plant genetic engineering methods, it is possible to produce glyphosate tolerant plants by inserting into the plant genome a DNA molecule that causes the production of higher levels of wild-type EPSPS (Shah et al., Science 233:478-481 (1986). Glyphosate tolerance can also be achieved by the expression of EPSPS variants that have lower affinity for glyphosate and therefore retain their catalytic activity in the presence of glyphosate (U.S. Pat. No. 4,940,835, U.S. Pat. No. 5,094,945, U.S. Pat. No. 5,633,435). Enzymes that degrade glyphosate in the plant tissues (U.S. Pat. No. 5,463,175) are also capable of conferring cellular tolerance to glyphosate. Such genes, therefore, allow for the production of transgenic crops that are tolerant to glyphosate, thereby allowing glyphosate to be used for effective weed control with minimal concern of crop damage. For example, glyphosate tolerance has been genetically engineered into corn (U.S. Pat. No. 5,554,798), wheat (Zhou et al. Plant Cell Rep. 15:159-163 (1995), soybean (WO 9200377) and canola (WO 9204449).
- Variants of the wild-type EPSPS enzyme are glyphosate-resistant as a result of alterations in the EPSPS amino acid coding sequence (Kishore et al., Annu. Rev. Biochem. 57:627-663 (1988); Schulz et al., Arch. Microbiol. 137:121-123 (1984); Sost et al., FEBS Lett. 173:238-241 (1984); Kishore et al., In “Biotechnology for Crop Protection” ACS Symposium Series No. 379. Eds. Hedlin et al., 37-48 (1988). These variants typically have a higher K i for glyphosate than the wild-type EPSPS enzyme that confers the glyphosate-tolerant phenotype, but these variants are also characterized by a high Km for PEP that makes the enzyme kinetically less efficient. For example, the apparent Km for PEP and the apparent Ki for glyphosate for the native EPSPS from E. coli are 10 μM and 0.5 μM while for a glyphosate-resistant isolate having a single amino acid substitution of an alanine for the glycine at position 96 these values are 220 μM and 4.0 mM, respectively. A number of glyphosate-resistant plant variant EPSPS genes have been constructed by mutagenesis.
- A variety of native and variant EPSPS enzymes have been expressed in transgenic plants in order to confer glyphosate tolerance (Singh, et al., In “Biosynthesis and Molecular Regulation of Amino Acids in Plants”, Amer Soc Plant Phys. Pubs (1992). Examples of some of these EPSP Synthases and methods for preparing transgenic plants resistant to glyphosate include those described and/or isolated in accordance with U.S. Pat. No. 4,940,835, U.S. Pat. No. 4,971,908, U.S. Pat. No. 5,145,783, U.S. Pat. No. 5,188,642, U.S. Pat. No. 5,310,667, U.S. Pat. No. 5,312,910, and U.S. Pat. No. 6,40,497. They can also be derived from a structurally distinct class of non-homologous EPSPS genes, such as the naturally occurring class II EPSPS genes isolated from Agrobacterium sp. strain CP4 as described in U.S. Pat. No. 5,633,435 and U.S. Pat. No. 5,627,061.
- Eleusine indica is commonly referred to as “goose grass” and may also be known as “yard grass”. It is a common monocotyledonous plant found world wide. As a member of the Poaceae family, the grass family, it is related to many well known crop plants. Eleusine indica is most closely related to the millets, that include Sorghum bicolor (sorghum or great millet), Zea mays (maize), Pennisetum americanum (pearl millet), Eleusine coracana (finger millet), Setaria italica (foxtail millet), Paspalum scrobiculatum (kodo millet), Echinochloa frumentacea (barnyhard millet) and Eragrostis tef (teff) (Chennaveeraiah et al., In “Chromosome engineering in plants: genetics, breeding and evolution”, Cytogenetics of Minor Millets, in Tsuchiya et al., eds Elsevier Sci Pub Amsterdam, 613-627 (1991). Eleusine indica has been shown to hybridize with Eleusine coracana (finger millet), an important cultivated millet of India and East Africa (Chennaveeraiah et al., Euphytica 2-3:489-495, (1974). Classical plant breeding methods can be used to transfer the genes and traits of interest from Eleusine indica into agronomic crop plants within the family Poaceae.
- In its broadest sense, the present invention herein provides a method for plant tolerance to glyphosate herbicide by the expression of an isolated DNA molecule encoding a naturally occurring glyphosate resistant EPSPS enzyme. The enzyme and the DNA is isolated from Eleusine species, more particularly Eleusine indica (E. indica).
- In the first aspect of the present invention described herein provides a method to cause plants to be tolerant to glyphosate herbicide by the insertion of a recombinant DNA molecule into the nuclear genome of a plant cell, the recombinant DNA molecule comprising:
- a promoter that functions in plant cells to cause the production of an RNA molecule; operably linked to,
- a DNA molecule transcribing an RNA encoding for a chloroplast transit peptide and a E. indica glyphosate resistant EPSPS enzyme; operably linked to, a 3′ non-translated region that functions in plant cells to cause the polyadenylation of the 3′ end of the RNA molecule.
- Typically, the promoter used in the DNA molecule is expressed in a constitutive fashion. Examples of suitable promoters that function effectively in this capacity include cauliflower mosaic virus 19S promoter, cauliflower mosaic virus 35S promoter, figwort mosaic virus 34S promoter, sugarcane bacilliform virus promoter, commelina yellow mottle virus promoter, small subunit of ribulose-1,5-bisphosphate carboxylase promoter, rice cytosolic triosephosphate isomerase promoter, adenine phosphoribosyltransferae promoter,
rice actin 1 promoter, maize ubiquitin promoter, mannopine synthase promoter and octopine synthase promoter. A Promoter may also comprise leader sequences and intron sequences useful in the invention. - A DNA molecule that encodes a chloroplast transit peptide sequence can be isolated from EPSPS genes purified from various plant species including E. indica as well as from various plant genes whose protein products have been shown to be transported into the chloroplast.
- A DNA molecule that encodes a glyphosate resistant EPSPS enzyme isolated from Eleusine species, more particularly from E. indica, comprising SEQ ID NO: 7 is an object of the invention and a DNA molecule substantially homologous to the DNA molecule isolated from E. indica or a portion thereof identified as SEQ ID NO: 6.
- The 3′ non-translated region can be obtained from various genes that are expressed in plant cells. The nopaline synthase 3′ untranslated, the 3′ untranslated region from pea small subunit Rubisco gene, the wheat heat shock protein 17.9 3′ untranslated region, the 3′ untranslated region from soybean 7S seed storage protein gene are commonly used in this capacity.
- The invention also relates to a glyphosate tolerant transgenic crop plant cell, a glyphosate tolerant crop plant and crop plant parts, crop seeds and progeny thereof comprising the recombinant DNA molecule of the present invention.
- A DNA molecule that encodes a naturally occurring plant derived glyphosate resistant EPSPS enzyme, wherein the glyphosate resistant EPSPS enzyme has a K m for phosphoenolpyruvate (PEP) of less than 10 μM. More preferably, a DNA molecule that encodes a naturally occurring plant derived glyphosate resistant EPSPS enzyme wherein the glyphosate resistant EPSPS enzyme has a Km for PEP of less than 10 μM and the Km for PEP is not more than about 2× of the naturally occurring plant derived glyphosate sensitive EPSPS enzyme.
- A DNA molecule that encodes a naturally occurring glyphosate resistant EPSPS enzyme derived from Eleusine species, wherein the glyphosate resistant EPSPS enzyme has a K m for phosphoenolpyruvate (PEP) of less than 10 μM. More preferably, a DNA molecule that encodes a naturally occurring glyphosate resistant EPSPS enzyme derived from Eleusine species, wherein the glyphosate resistant EPSPS enzyme has a Km for PEP of less than 10 μM and the Km for PEP is not more than about 2× of the naturally occurring plant derived glyphosate sensitive EPSPS enzyme.
- A DNA molecule that encodes a naturally occurring glyphosate resistant EPSPS enzyme derived from E. indica, wherein the naturally occurring glyphosate resistant EPSPS enzyme amino acid sequence has been modified by amino acid substitutions selected from the group consisting of threonine to isoleucine at amino acid position 103 and glycine to alanine at amino acid position 102.
- The invention also relates to the homologous genetic elements regulating expression of the E. indica glyphosate resistant EPSPS gene. These elements include but are not limited to the DNA sequences of a promoter, a 5′ untranslated region, a chloroplast transit peptide, an intron, and a 3′ untranslated region of E. indica EPSPS glyphosate resistance gene. A DNA molecule that encodes a glyphosate resistant EPSPS enzyme purified from the genome of Eleusine species, more particularly from E. indica glyphosate resistant biotype provided by the ATCC deposit #PTA-2177 is an object of the invention.
- The following drawings form part of the present specification and are included to further demonstrate certain aspects of the present invention. The invention may be better understood by reference to one or more of these drawings in combination with the detailed description of specific embodiments presented herein.
- FIG. 1. Eleusine indica (glyphosate tolerant) EPSP synthase DNA sequence (SEQ ID NO: 6).
- FIG. 2. Deduced amino acid sequence for the mature protein-coding region of the Eleusine indica (glyphosate tolerant biotype) EPSP synthase gene (SEQ ID NO: 7).
- FIG. 3. Growth rates on glyphosate containing media of transgenic E. coli on expressing the E. indica glyphosate sensitive EPSPS enzyme and the E. indica glyphosate resistant EPSPS enzyme.
- FIG. 4. Glyphosate inhibition study comparing the EPSP synthase activities detectable in extracts prepared from the glyphosate-sensitive and tolerant E. indica biotypes.
- FIG. 5. Plasmid map of pMON45364
- FIG. 6. Plasmid map of pMON45365
- FIG. 7. Plasmid map of pMON45367
- FIG. 8. Plasmid map of pMON45369
- FIG. 9. The amino acid sequence deduced from the cDNA sequence of the mature EPSP synthase protein sequence derived from the glyphosate-tolerant E. indica biotype (top row) aligned with that of the glyphosate sensitive E. indica biotype (bottom row).
- This application claims the benefit of U.S. Provisional Application No. 60/188,093, filed Mar. 9, 2000.
- The following definitions and methods are provided to better define the present invention and to guide those of ordinary skill in the art in the practice of the present invention. Unless otherwise noted, terms are to be understood according to conventional usage by those of ordinary skill in the relevant art. Definitions of common terms in molecular biology may also be found in Rieger et al., Glossary of Genetics: Classical and Molecular, 5th edition, Springer-Verlag: New York, (1991); and Lewin, Genes V, Oxford University Press: New York, (1994). The nomenclature for DNA bases as set forth at 37 CFR § 1.822 is used. The standard one- and three-letter nomenclature for amino acid residues is used.
- “cDNA library” refers to a collection of cDNA fragments, each cloned into a separate vector molecule.
- The term “chimeric” refers to a fusion nucleic acid or protein sequence. A chimeric nucleic acid coding sequence is comprised of two or more sequences joined in-frame that encode a chimeric protein. A chimeric gene refers to the multiple genetic elements derived from heterologous sources comprising a gene.
- The phrases “coding sequence”, “open reading frame”, and “structural sequence” refer to the region of continuous sequential nucleic acid triplets encoding a protein, polypeptide, or peptide sequence.
- “Codon” refers to a sequence of three nucleotides that specify a particular amino acid.
- “Complementarity” and “complement” when referring to nucleic acid sequences, refers to the specific binding of adenine to thymine (or uracil in RNA) and cytosine to guanine on opposite strands of DNA or RNA.
- “Construct” refers to the heterologous genetic elements operably linked to each other making up a recombinant DNA molecule.
- “C-terminal region” refers to the region of a peptide, polypeptide, or protein chain from the middle thereof to the end that carries the amino acid having a free carboxyl group.
- The term “encoding DNA” refers to chromosomal DNA, plasmid DNA, cDNA, or synthetic DNA that encodes any of the proteins discussed herein.
- The term “endogenous” refers to materials originating from within an organism or cell.
- “Endonuclease” refers to an enzyme that hydrolyzes double stranded DNA at internal locations.
- “Exogenous” refers to materials originating from outside of an organism or cell. This typically applies to nucleic acid molecules used in producing transformed or transgenic host cells and plants.
- “Exon” refers to the portion of a gene that is actually translated into protein, i.e. a coding sequence.
- The term “expression” refers to the transcription of a gene to produce the corresponding mRNA.
- “Fragments”. A fragment of a EPSPS gene is a portion of a full-length EPSPS gene nucleic acid that is of at least a minimum length capable of expressing a protein with EPSPS activity.
- The term “gene” refers to chromosomal DNA, plasmid DNA, cDNA, synthetic DNA, or other DNA that encodes a peptide, polypeptide, protein, or RNA molecule, and regions flanking the coding sequence involved in the regulation of expression.
- The term “genome” as it applies to viruses encompasses all of the nucleic acid sequence contained within the capsid of the virus. The term “genome” as it applies to bacteria encompasses both the chromosome and plasmids within a bacterial host cell. Encoding nucleic acids of the present invention introduced into bacterial host cells can therefore be either chromosomally-integrated or plasmid-localized. The term “genome” as it applies to plant cells encompasses not only chromosomal DNA found within the nucleus, but organelle DNA found within subcellular components of the cell. Nucleic acids of the present invention introduced into plant cells can therefore be either chromosomally-integrated or organelle-localized.
- “Glyphosate” refers to N-phosphonomethylglycine and its' salts, Glyphosate is the active ingredient of Roundup® herbicide (Monsanto Co.). Plant treatments with “glyphosate” refer to treatments with the Roundup® or Roundup Ultra® herbicide formulation, unless otherwise stated. Glyphosate as N-phosphonomethylglycine and its' salts (not formulated Roundup® herbicide) are components of synthetic culture media used for the selection of bacteria and plant tolerance to glyphosate or used to determine enzyme resistance in in vitro biochemical assays.
- “Heterologous DNA” refers to DNA from a source different than that of the recipient cell.
- “Homologous DNA” refers to DNA from the same source as that of the recipient cell.
- “Hybridization” refers to the ability of a strand of nucleic acid to join with a complementary strand via base pairing. Hybridization occurs when complementary sequences in the two nucleic acid strands bind to one another.
- “Identity” refers to the degree of similarity between two nucleic acid or protein sequences. An alignment of the two sequences is performed by a suitable computer program. A widely used and accepted computer program for performing sequence alignments is CLUSTALW v1.6 (Thompson, et al. Nucl. Acids Res., 22: 4673-4680, 1994). The number of matching bases or amino acids is divided by the total number of bases or amino acids, and multiplied by 100 to obtain a percent identity. For example, if two 580 base pair sequences had 145 matched bases, they would be 25 percent identical. If the two compared sequences are of different lengths, the number of matches is divided by the shorter of the two lengths. For example, if there are 100 matched amino acids between 200 and a 400 amino acid proteins, they are 50 percent identical with respect to the shorter sequence. If the shorter sequence is less than 150 bases or 50 amino acids in length, the number of matches are divided by 150 (for nucleic acid bases) or 50 (for amino acids), and multiplied by 100 to obtain a percent identity.
- “Intron” refers to a portion of a gene not translated into protein, even though it is transcribed into RNA.
- “Isolated” An “isolated” nucleic acid is one that has been substantially separated or purified away from other nucleic acid sequences in the cell of the organism that the nucleic acid naturally occurs, i.e., other chromosomal and extrachromosomal DNA and RNA, by conventional nucleic acid-purification methods. The term also embraces recombinant nucleic acids and chemically synthesized nucleic acids.
- “Native” The term “native” refers to a naturally-occurring (“wild-type”) nucleic acid or polypeptide.
- “N-terminal region” refers to the region of a peptide, polypeptide, or protein chain from the amino acid having a free amino group to the middle of the chain.
- “Nucleic acid” refers to deoxyribonucleic acid (DNA) and ribonucleic acid (RNA).
- Nucleic acid codes: A=adenosine; C=cytosine; G=guanosine; T=thymidine. Codes used for synthesis of oligonucleotides: N=equimolar A, C, G, and T; I=deoxyinosine; K=equimolar G and T; R=equimolar A and G; S=equimolar C and G; W=equimolar A and T; Y=equimolar C and T.
- A “nucleic acid segment” or a “nucleic acid molecule segment” is a nucleic acid molecule that has been isolated free of total genomic DNA of a particular species, or that has been synthesized. Included with the term “nucleic acid segment” are DNA segments, recombinant vectors, plasmids, cosmids, phagemids, phage, viruses, et cetera. “Nucleic-Acid Hybridization”; “Stringent Conditions”; “Specific” The term “stringent conditions” is functionally defined with regard to the hybridization of a nucleic-acid probe to a target nucleic acid (i.e., to a particular nucleic-acid sequence of interest) by the specific hybridization procedure discussed in Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989), at 9.52-9.55. See also, Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989) at 9.47-9.52, 9.56-9.58; Kanehisa, Nucl. Acids Res. 12:203-213, (1984); and Wetmur and Davidson, J. Mol. Biol. 31:349-370, (1968).
- “Nucleotide Sequence Variants” Using well-known methods, the skilled artisan can readily produce nucleotide and amino acid sequence variants of EPSPS genes and proteins, respectively. “Variant” DNA molecules are DNA molecules containing minor changes in a native EPSPS gene sequence, i.e., changes that one or more nucleotides of a native EPSPS gene sequence is deleted, added, and/or substituted, such that the variant EPSPS gene encodes a protein that retains EPSPS activity. Variant DNA molecules can be produced, for example, by standard DNA mutagenesis techniques or by chemically synthesizing the variant DNA molecule or a portion thereof. Methods for chemical synthesis of nucleic acids are discussed, for example, in Beaucage et al, Tetra. Letts. 22:1859-1862 (1981), and Matteucci et al., J. Am. Chem. Soc. 103:3185-(1981). Chemical synthesis of nucleic acids can be performed, for example, on automated oligonucleotide synthesizers. Such variants preferably do not change the reading frame of the protein-coding region of the nucleic acid and preferably encode a protein having no amino acid changes. Nucleic acid sequence variants are most often created for the purposes of modification of the sequence to add or delete restriction endonuclease sites or to affect transcription or translation of the nucleic acid molecule.
- “Amino-acid substitutions”, “Amino-acid variants”, are preferably substitutions of single amino-acid residue for another amino-acid residue at any position within the protein. Substitutions, deletions, insertions or any combination thereof can be combined to arrive at a final construct.
- “Open reading frame (ORF)” refers to a region of DNA or RNA encoding a peptide, polypeptide, or protein.
- “Operably Linked”. A first nucleic-acid sequence is “operably” linked with a second nucleic-acid sequence when the first nucleic-acid sequence is placed in a functional relationship with the second nucleic-acid sequence. For instance, a promoter is operably linked to a protein-coding sequence if the promoter effects the transcription or expression of the coding sequence. Generally, operably linked DNA sequences are contiguous and, where necessary to join two protein-coding regions, in reading frame.
- “Overexpression” refers to the expression of a polypeptide or protein encoded by a DNA introduced into a host cell, wherein said polypeptide or protein is either not normally present in the host cell, or wherein said polypeptide or protein is present in said host cell at a higher level than that normally expressed from the endogenous gene encoding said polypeptide or protein.
- “Plant expression vector” refers to chimeric DNA molecules comprising the regulatory elements that are operably linked to provide the expression of a transgene product in plants.
- “Plasmid” refers to a circular, extrachromosomal, self-replicating piece of DNA.
- “Polyadenylation signal” or “polyA signal” refers to a nucleic acid sequence located 3′ to a coding region that causes the addition of adenylate nucleotides to the 3′ end of the mRNA transcribed from the coding region.
- “Polymerase chain reaction (PCR)” refers to an enzymatic technique to create multiple copies of one sequence of nucleic acid. Copies of DNA sequence are prepared by shuttling a DNA polymerase between two amplimers. The basis of this amplification method is multiple cycles of temperature changes to denature, then re-anneal amplimers, followed by extension to synthesize new DNA strands in the region located between the flanking amplimers.
- The term “promoter” or “promoter region” refers to a nucleic acid sequence, usually found upstream (5′) to a coding sequence, that controls expression of the coding sequence by controlling production of messenger RNA (mRNA) by providing the recognition site for RNA polymerase and/or other factors necessary for start of transcription at the correct site. As contemplated herein, a promoter or promoter region includes variations of promoters derived by means of ligation to various regulatory sequences, random or controlled mutagenesis, and addition or duplication of enhancer sequences. The promoter region disclosed herein, and biologically functional equivalents thereof, are responsible for driving the transcription of coding sequences under their control when introduced into a host as part of a suitable recombinant vector, as demonstrated by its ability to produce mRNA.
- “Recombinant”. A “recombinant” nucleic acid is made by an artificial combination of two otherwise separated segments of sequence, e.g., by chemical synthesis or by the manipulation of isolated segments of nucleic acids by genetic engineering techniques.
- The term “recombinant DNA construct” or “recombinant vector” refers to any agent such as a plasmid, cosmid, virus, autonomously replicating sequence, phage, or linear or circular single-stranded or double-stranded DNA or RNA nucleotide sequence, derived from any source, capable of genomic integration or autonomous replication, comprising a DNA molecule that one or more DNA sequences have been linked in a functionally operative manner. Such recombinant DNA constructs or vectors are capable of introducing a 5′ regulatory sequence or promoter region and a DNA sequence for a selected gene product into a cell in such a manner that the DNA sequence is transcribed into a functional mRNA that is translated and therefore expressed. Recombinant DNA constructs or recombinant vectors may be constructed to be capable of expressing antisense RNAs, in order to inhibit translation of a specific RNA of interest.
- “Regeneration” refers to the process of growing a plant from a plant cell (e.g., plant protoplast or explant).
- “Reporter” refers to a gene and corresponding gene product that when expressed in transgenic organisms produces a product detectable by chemical or molecular methods or produces an observable phenotype.
- “Restriction enzyme” refers to an enzyme that recognizes a specific palindromic sequence of nucleotides in double stranded DNA and cleaves both strands; also called a restriction endonuclease. Cleavage typically occurs within the restriction site.
- “Selectable marker” refers to a nucleic acid sequence whose expression confers a phenotype facilitating identification of cells containing the nucleic acid sequence. Selectable markers include those that confer resistance to toxic chemicals (e.g. ampicillin resistance, kanamycin resistance), complement a nutritional deficiency (e.g. uracil, histidine, leucine), or impart a visually distinguishing characteristic (e.g. color changes or fluorescence). Useful dominant selectable marker genes include genes encoding antibiotic resistance genes (e.g., resistance to hygromycin, kanamycin, bleomycin, G418, streptomycin or spectinomycin); and herbicide resistance genes (e.g., phosphinothricin acetyltransferase). A useful strategy for selection of transformants for herbicide resistance is described, e.g., in Vasil, Cell Culture and Somatic Cell Genetics of Plants, Vols. I-III, Laboratory Procedures and Their Applications Academic Press, New York (1984).
- The term “specific for (a target sequence)” indicates that a probe or primer hybridizes under given hybridization conditions only to the target sequence in a sample comprising the target sequence.
- “Tolerant” refers to a reduced toxic effect of glyphosate on the growth and development of microorganisms and plants.
- “Transcription” refers to the process of producing an RNA copy from a DNA template.
- “Transformation” refers to a process of introducing an exogenous nucleic acid sequence (e.g., a vector, recombinant nucleic acid molecule) into a cell or protoplast that exogenous nucleic acid is incorporated into a chromosome or is capable of autonomous replication.
- “Transformed” or “transgenic” refers to a cell, tissue, organ, or organism into that has been introduced a foreign nucleic acid, such as a recombinant vector. A “transgenic” or “transformed” cell or organism also includes progeny of the cell or organism and progeny produced from a breeding program employing such a “transgenic” plant as a parent in a cross and exhibiting an altered phenotype resulting from the presence of the foreign nucleic acid.
- The term “transgene” refers to any nucleic acid sequence nonnative to a cell or organism transformed into said cell or organism. “Transgene” also encompasses the component parts of a native plant gene modified by insertion of a nonnative nucleic acid sequence by directed recombination.
- The term “translation” refers to the production the corresponding gene product, i.e., a peptide, polypeptide, or protein from a mRNA.
- “Vector” refers to a plasmid, cosmid, bacteriophage, or virus that carries foreign DNA into a host organism.
- “Isolated,” “Purified,” “Homogeneous” Polypeptides. A polypeptide is “isolated” if it has been separated from the cellular components (nucleic acids, lipids, carbohydrates, and other polypeptides) that naturally accompany it or that is chemically synthesized or recombinant. A monomeric polypeptide is isolated when at least 60% by weight of a sample is composed of the polypeptide, preferably 90% or more, more preferably 95% or more, and most preferably more than 99%. Protein purity or homogeneity is indicated, for example, by polyacrylamide gel electrophoresis of a protein sample, followed by visualization of a single polypeptide band upon staining the polyacrylamide gel; high pressure liquid chromatography; or other conventional methods. Coat proteins can be purified by any of the means known in the art, for example as described in Guide to Protein Purification, ed. Deutscher, Meth. Enzymol. 185, Academic Press, San Diego, 1990; and Scopes, Protein Purification: Principles and Practice, Springer Verlag, New York, 1982.
- “Labeling”. There are a variety of conventional methods and reagents for labeling polypeptides and fragments thereof Typical labels include radioactive isotopes, ligands or ligand receptors, fluorophores, chemiluminescent agents, and enzymes. Methods for labeling and guidance in the choice of labels appropriate for various purposes are discussed, e.g., in Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989) and Ausubel et al., Greene Publishing and Wiley-Interscience, New York, (1992).
- “Mature protein coding region”. This term refers to the sequence of the processed protein product of EPSPS remaining after the chloroplast transit peptide sequence has been removed.
- “Polypeptide fragments”. The present invention also encompasses fragments of an E indica EPSPS that lacks at least one residue of a native full-length E. indica EPSPS protein, but that specifically maintains EPSPS activity.
- “Transit peptide or targeting peptide sequence”. These terms generally refer to peptide sequences that when linked to a protein of interest directs the protein to a particular tissue, cell, subcellular location, or cell organelle. Examples include, but are not limited to, chloroplast transit peptides, nuclear targeting signals, and vacuolar signals. The chloroplast transit peptide is of particular utility in the present invention to direct expression of the EPSPS enzyme to the chloroplast.
- The term “plant” encompasses any higher plant and progeny thereof, including monocots (e.g., corn, rice, wheat, barley, etc.), dicots (e.g., soybean, cotton, tomato, potato, Arabidopsis, tobacco, etc.), gymnosperms (pines, firs, cedars, etc) and includes parts of plants, including reproductive units of a plant (e.g., seeds, bulbs, tubers, or other parts or tissues from that the plant can be reproduced), fruits and flowers.
- Exogenous genetic material may be transferred into a plant by the use of a DNA vector designed for such a purpose by methods that utilize Agrobacterium, particle bombardment or other methods known to those skilled in the art. A particularly preferred subgroup of exogenous material comprises a nucleic acid molecule of the present invention. Design of such a vector is generally within the skill of the art (Plant Molecular Biology: A Laboratory Manual, eds. Clark, Springer, New York (1997). Examples of such plants in to which exogenous genetic material may be transferred, include, without limitation, alfalfa, Arabidopsis, barley, Brassica, broccoli, cabbage, citrus, cotton, garlic, oat, oilseed rape, onion, canola, flax, maize, an ornamental annual and ornamental perennial plant, pea, peanut, pepper, potato, rice, rye, sorghum, soybean, strawberry, sugarcane, sugar beet, tomato, wheat, poplar, pine, fir, eucalyptus, apple, lettuce, lentils, grape, banana, tea, turf grasses, sunflower, oil palm, Phaseolus etc.
- The particular promoters selected for use in embodiments of the present invention should be capable of causing the production of sufficient expression to, in the case of the DNA molecule, generate protein expression in vegetative and reproductive tissues of a transformed plant. The DNA molecule will typically contain a constitutive promoter, a structural DNA sequence encoding a herbicide resistant enzyme, and a 3′ non-translated region. A number of constitutive promoters that are active in plant cells have been described. Suitable promoters for constitutive expression in plants of herbicide tolerance for the DNA molecule include, but are not limited to, the cauliflower mosaic virus (CaMV) 35S promoter (Odell et al. Nature 313:801-812 (1985), the Figwort mosaic virus (FMV) 35S (Sanger et al. Plant Mol. Biol. 14:433-443 (1990), the sugarcane bacilliform virus promoter (Bouhida et al., J. Gen. Virol. 74:15-22 (1993), the commelina yellow mottle virus promoter (Medberry et al., Plant J. 3:619-626 (1993), the light-inducible promoter from the small subunit of the ribulose-1,5-bis-phosphate carboxylase (ssRUBISCO) (Coruzzi et al., EMBO J. 3:1671-1679 (1984), the rice cytosolic triosephosphate isomerase (TPI) promoter (Xu et al. Plant Physiol. 106:459-467 (1994), the adenine phosphoribosyltransferase (APRT) promoter of Arabidopsis (Moffatt et al. Gene 143:211-216 (1994), the rice actin 1 gene promoter (Zhong et al. Plant Sci. 116:73-84 (1996), the mannopine synthase and octopine synthase promoters (Ni et al. Plant J. 7:661-676 (1995), the Adh promoter (Walker et al., Proc. Natl. Acad. Sci. U.S.A. 84: 6624-6628 (1987), the sucrose synthase promoter (Yang et al., Proc. Natl. Acad. Sci. U.S.A. 87: 4144-4148 (1990), the R gene complex promoter (Chandler et al., The Plant Cell 1: 1175-1183 (1989), and the chlorophyll α/β binding protein gene promoter, et cetera These promoters have been used to create DNA vectors that have been expressed in plants; see, e.g., PCT publication WO 8402913. All of these promoters have been used to create various types of plant-expressible recombinant DNA vectors. Comparative analysis of constitutive promoters by the expression of reporter genes such as the uidA (β-glucuronidase) gene from E. coli has been performed with many of these and other promoters (Li et al. Mol. Breeding 3:1-14 (1997); Wen et al. Chinese J. of Bot. 5:102-109 (1993). Promoters that are known or are found to cause transcription of DNA in plant cells can be used in the present invention. Such promoters may be obtained from a variety of sources such as plants and plant viruses. In addition to promoters that are known to cause transcription of DNA in plant cells, other promoters may be identified for use in the current invention by screening a plant cDNA library for genes that are selectively or preferably expressed in the target tissues or cells. For the purpose of expression in source tissues of the plant, such as the leaf, seed, root or stem, it is preferred that the promoters utilized in the present invention have relatively high expression in these specific tissues. For this purpose, one may choose from a number of promoters for genes with tissue- or cell-specific or -enhanced expression. Examples of such promoters reported in the literature include the chloroplast glutamine synthetase GS2 promoter from pea (Edwards et al., Proc. Natl. Acad. Sci. U.S.A. 87: 3459-3463 (1990), the chloroplast fructose-1,6-biphosphatase (FBPase) promoter from wheat (Lloyd et al., Mol. Gen. Genet. 225: 209-216 (1991), the nuclear photosynthetic ST-LS1 promoter from potato (Stockhaus et al., EMBO J. 8: 2445-2451, (1989), the serine/threonine kinase (PAL) promoter and the glucoamylase (CHS) promoter from Arabidopsis thaliana. Also reported to be active in photosynthetically active tissues are the ribulose-1,5-bisphosphate carboxylase (RBCS) promoter from eastern larch (Larix laricina), the promoter for the Cab gene, Cab6, from pine (Yamamoto et al., Plant Cell Physiol. 35: 773-778 (1994), the promoter for the Cab-1 gene from wheat (Fejes et al., Plant Mol. Biol. 15: 921-932 (1990), the promoter for the Cab-1 gene from spinach (Lubberstedt et al., Plant Physiol. 104:997-1006 (1994), the promoter for the Cab1R gene from rice (Luan et al., Plant Cell. 4:971-981 (1992), the pyruvate, orthophosphate dikinase (PPDK) promoter from Zea mays (Matsuoka et al., Proc. Natl. Acad. Sci. U.S.A. 90: 9586-9590 (1993), the promoter for the tobacco Lhcb1*2 gene (Cerdan et al., Plant Mol. Biol. 33:245-255 (1997), the Arabidopsis thaliana Suc2 sucrose-H+ symporter promoter (Truernit et al., Planta. 196:564-570 (1995), and the promoter for the thylakoid membrane protein genes from spinach (PsaD, PsaF, PsaE, PC, FNR, AtpC, AtpD, Cab, RbcS).
- Other promoters for the chlorophyll α/β-binding proteins may also be utilized in the present invention, such as the promoters for LhcB gene and PsbP gene from white mustard ( Sinapis alba) (Kretsch et al., Plant Mol. Biol. 28: 219-229 (1995). A variety of plant gene promoters that are regulated in response to environmental, hormonal, chemical, and/or developmental signals, also can be used for expression of RNA-binding protein genes in plant cells, including promoters regulated by (1) heat (Callis et al., Plant Physiol. 88:965-968 (1988), (2) light (e.g., pea RbcS-3A promoter, Kuhlemeier et al., Plant Cell 1:471-478 (1989); maize RbcS promoter, Schaffner et al., Plant Cell 3:997-1012 (1991); (3) hormones, such as abscisic acid (Marcotte et al., Plant Cell 1:969-976 (1989), (4) wounding (e.g., WunI, Siebertz et al., Plant Cell 1:961-968 (1989); or (5) chemicals, such as methyl jasminate, salicylic acid, steroid hormones, alcohol, Safeners (Gatz, Curr. Opin. Biotech 7:168-172 (1996), WO 9706269), or it may also be advantageous to employ (6) organ-specific promoters (e.g., Roshal et al., EMBO J. 6:1155-(1987); Schernthaner et al., EMBO J. 7:1249-1255 (1988); Bustos et al., Plant Cell 1:839.-853 (1989).
- For the purpose of expression in sink tissues of the plant, such as the tuber of the potato plant, the fruit of tomato, or the seed of soybean, canola, cotton, Zea mays, wheat, rice, and barley, it is preferred that the promoters utilized in the present invention have relatively high expression in these specific tissues. A number of promoters for genes with tuber-specific or -enhanced expression are known, including the class I patatin promoter (Bevan et al., EMBO J. 8:1899-1906 (1986); Jefferson et al., Plant Mol. Biol. 14:995-1006 (1990), the promoter for the potato tuber ADPGPP genes, both the large and small subunits, the sucrose synthase promoter (Salanoubat et al.,, Gene 60:47-56 (1987); Salanoubat et al., Gene 84:181-185 (1989), the promoter for the major tuber proteins including the 22 kD protein complexes and proteinase inhibitors (Hannapel, Plant Physiol. 101:703-704 (1993), the promoter for the granule bound starch synthase gene (GBSS) (Visser et al., Plant Mol. Biol. 17:691-699 (1991), and other class I and II patatins promoters (Koster-Topfer et al., Mol. Gen. Genet. 219:390-396 (1989); Mignery et al., Gene 62:27-44 (1988). Other promoters can also be used to express a protein in specific tissues, such as seeds or fruits. The promoter for β-conglycinin (Chen et al., Dev. Genet. 10:112-122 (1989) or other seed-specific promoters such as the napin and phaseolin promoters, can be used. The zeins are a group of storage proteins found in Zea mays endosperm. Genomic clones for zein genes have been isolated (Pedersen et al., Cell 29:1015-1026 (1982), and the promoters from these clones, including the 15 kD, 16 kD, 19 kD, 22 kD, 27 kD, and gamma genes, could also be used. Other promoters known to function, for example, in Zea mays include the promoters for the following genes: waxy, Brittle, Shrunken 2, Branching enzymes I and II, starch synthases, debranching enzymes, oleosins, glutelins, and sucrose synthases. A particularly preferred promoter for Zea mays endosperm expression is the promoter for the glutelin gene from rice, more particularly the Osgt-1 promoter (Zheng et al., Mol. Cell Biol. 13:5829-5842 (1993). Examples of promoters suitable for expression in wheat include those promoters for the ADPglucose pyrosynthase (ADPGPP) subunits, the granule bound and other starch synthase, the branching and debranching enzymes, the embryogenesis-abundant proteins, the gliadins, and the glutenins. Examples of such promoters in rice include those promoters for the ADPGPP subunits, the granule bound and other starch synthase, the branching enzymes, the debranching enzymes, sucrose synthases, and the glutelins. A particularly preferred promoter is the promoter for rice glutelin, Osgt-1 gene. Examples of such promoters for barley include those for the ADPGPP subunits, the granule bound and other starch synthase, the branching enzymes, the debranching enzymes, sucrose synthases, the hordeins, the embryo globulins, and the aleurone specific proteins.
- Root specific promoters may also be used. An example of such a promoter is the promoter for the acid chitinase gene (Samac et al., Plant Mol. Biol. 25:587-596 (1994). Expression in root tissue could also be accomplished by utilizing the root specific subdomains of the CaMV 35S promoter that have been identified (Lam et al., Proc. Natl. Acad. Sci. U.S.A. 86: 7890-7894 (1989). Other root cell specific promoters include those reported by Conkling et al. (Plant Physiol. 93: 1203-1211 (1990).
- The 5′ non-translated leader sequence can be derived from the promoter selected to express the heterologous gene sequence of the DNA molecule of the present invention, and can be specifically modified if desired so as to increase translation of mRNA. For a review of optimizing expression of transgenes, see Koziel et al., (Plant Mol. Biol. 32:393-405 (1996). The 5′ non-translated regions can also be obtained from plant viral RNAs (Tobacco mosaic virus, Tobacco etch virus, Maize dwarf mosaic virus, Alfalfa mosaic virus, among others) from suitable eukaryotic genes, plant genes (wheat and maize chlorophyll a/b binding protein gene leader), or from a synthetic gene sequence. The present invention is not limited to constructs wherein the non-translated region is derived from the 5′ non-translated sequence that accompanies the promoter sequence. The leader sequence could also be derived from an unrelated promoter or coding sequence. Leader sequences useful in context of the present invention comprise the maize Hsp70 leader (U.S. Pat. No. 5,362,865 and U.S. Pat. No. 5,859,347), and the TMV omega element (Gallie et al., The Plant Cell 1:301-311 (1989).
- A vector or construct may also include various regulatory elements. Intron sequences are known in the art to aid in the expression of transgenes in monocot plant cells. Examples of such introns include the Adh intron 1 (Callis et al., Genes and Develop. 1:1183-1200 (1987), the sucrose synthase intron (Vasil et al., Plant Physiol. 91:1575-1579 (1989), U.S. Pat. No. 5,955,330), first intron of the rice actin gene (U.S. Pat. No. 5,641,876).
- A vector may also include a transit peptide nucleic acid sequence. The glyphosate target in plants, the 5-enolpyruvyl-shikimate-3-phosate synthase (EPSPS) enzyme, is located in the chloroplast. Many chloroplast-localized proteins, including EPSPS, are expressed from nuclear genes as precursors and are targeted to the chloroplast by a chloroplast transit peptide (CTP) that is removed during the import steps. Examples of other such chloroplast proteins include the small subunit (SSU) of Ribulose-1,5,-bisphosphate carboxylase, Ferredoxin, Ferredoxin oxidoreductase, the light-harvesting complex protein I and protein II, and Thioredoxin F. It has been demonstrated in vivo and in vitro that non-chloroplast proteins may be targeted to the chloroplast by use of protein fusions with a CTP and that a CTP sequence is sufficient to target a protein to the chloroplast. Incorporation of a suitable chloroplast transit peptide, such as, the Arabidopsis thaliana EPSPS CTP (Klee et al., Mol. Gen. Genet. 210:437-442 (1987), and the Petunia hybrida EPSPS CTP (della-Cioppa et al., Proc. Natl. Acad. Sci. USA 83:6873-6877 (1986) has been show to target heterologous EPSPS protein sequences to chloroplasts in transgenic plants. The production of glyphosate tolerant plants by expression of a fusion protein comprising an amino-terminal CTP with a glyphosate resistant EPSPS enzyme is well known by those skilled in the art, (U.S. Pat. No. 5,627,061, U.S. Pat. No. 5,633,435, U.S. Pat. No. 5,312,910, EP 0218571, EP 189707, EP 508909, and EP 924299). Those skilled in the art will recognize that various chimeric constructs can be made that utilize the functionality of a particular CTP to import glyphosate resistant EPSPS enzymes into the plant cell chloroplast.
- The termination of transcription is accomplished by a 3′ non-translated DNA sequence operably linked in the chimeric vector to the gene of interest. The 3′ non-translated region of a recombinant DNA molecule contains a polyadenylation signal that functions in plants to cause the addition of adenylate nucleotides to the 3′ end of the RNA. The 3′ non-translated region can be obtained from various genes that are expressed in plant cells. The nopaline synthase 3′ untranslated region (Fraley et al., Proc. Natl. Acad. Sci. 80:4803-4807 (1983), the 3′ untranslated region from pea small subunit Rubisco gene (Coruzzi et al., EMBO J. 3:1671-1679 (1994), the 3′ untranslated region from soybean 7S seed storage protein gene (Schuler et al., Nuc Acids Res. 10:8225-8244 (1982) are commonly used in this capacity. The 3′ transcribed, non-translated regions containing the polyadenylate signal of Agrobacterium tumor-inducing (Ti) plasmid genes are also suitable.
- The aforesaid described genetic elements and other regulatory elements of similar function may be substituted when appropriate by those skilled in the art of plant molecular biology to provide necessary function to the plant expression cassette. DNA constructs for glyphosate tolerance designed for expression in plastids will necessarily contain genetic elements that function in plastids.
- A vector may also include a screenable or scorable marker gene. Screenable or scorable markers may be used to monitor expression. Exemplary markers include a β-glucuronidase or uidA gene (GUS) that encodes an enzyme for that various chromogenic substrates are known (Jefferson, Plant Mol. Biol, Rep. 5:387-405 (1987); Jefferson et al., EMBO J. 6:3901-3907 (1987); an R-locus gene, that encodes a product that regulates the production of anthocyanin pigments (red color) in plant tissues (Dellaporta et al., Stadler Symposium 11:263-282 (1988); a β-lactamase gene (Sutcliffe et al., Proc. Natl. Acad. Sci. (U.S.A.) 75:3737-3741 (1978); a gene that encodes an enzyme for that various chromogenic substrates are known (e.g., PADAC, a chromogenic cephalosporin); a luciferase gene (Ow et al., Science 234:856-859 (1986); a xylE gene (Zukowsky et al., Proc. Natl. Acad. Sci. (U.S.A.) 80:1101-1105 (1983) that encodes a catechol dioxygenase that can convert chromogenic catechols; an α-amylase gene (Ikatu et al., Bio/Technol. 8:241-242 (1990); a tyrosinase gene (Katz et al., J. Gen. Microbiol. 129:2703-2714 (1983) that encodes an enzyme capable of oxidizing tyrosine to DOPA and dopaquinone that in turn condenses to melanin; green flourescence protein (Elliot et al., Plant cell Rep. 18:707-714 (1999) and an α-galactosidase.
- Included within the terms “selectable or screenable marker genes” are also genes that encode a secretable marker whose secretion can be detected as a means of identifying or selecting for transformed cells. Examples include markers that encode a secretable antigen that can be identified by antibody interaction, or even secretable enzymes that can be detected catalytically. Secretable proteins fall into a number of classes, including small, diffusible proteins that are detectable, (e.g., by ELISA), small active enzymes that are detectable in extracellular solution (e.g., α-amylase, β-lactamase, phosphinothricin transferase), or proteins that are inserted or trapped in the cell wall (such as proteins that include a leader sequence such as that found in the expression unit of extension or tobacco PR-S). Other possible selectable and/or screenable marker genes will be apparent to those of skill in the art.
- There are many methods for introducing transforming nucleic acid molecules into plant cells. Suitable methods are believed to include virtually any method shown effective in introducing the nucleic acid molecules into a plant cell, such as by Agrobacterium infection or direct delivery of nucleic acid molecules.
- Four general methods for direct delivery of a gene into cells have been described: (1) chemical methods (Graham et al., Virology 54:536-539 (1973); (2) physical methods such as microinjection (Capecchi, Cell 22:479-488 (1980); electroporation (Wong et al., Biochem. Biophys. Res. Commun. 107:584-587 (1982); Fromm et al., Proc. Natl. Acad. Sci. (U.S.A.) 82:5824-5828 (1985); (U.S. Pat. No. 5,384,253); and the gene gun (Johnston et al., Methods Cell Biol. 43:353-365 (1994); (3) viral vectors (Clapp, Clin. Perinatol. 20:155-168 (1993); Lu et al., J. Exp. Med. 178:2089-2096 (1993); Eglitis et al., Biotechniques 6:608-614 (1988); and (4) receptor-mediated mechanisms (Curiel et al., Hum. Gen. Ther. 3:147-154 (1992), Wagner et al., Proc. Natl. Acad. Sci. USA 89:6099-6103 (1992).
- Acceleration methods that may be used include, for example, microprojectile bombardment and the like. One example of a method for delivering transforming nucleic acid molecules to plant cells is microprojectile bombardment. This method has been reviewed by Yang et al., Particle Bombardment Technology for Gene Transfer, Oxford Press, Oxford, England (1994). Non-biological particles (microprojectiles) that may be coated with nucleic acids and delivered into cells by a propelling force. Exemplary particles include those comprised of tungsten, gold, platinum, and the like. A particular advantage of microprojectile bombardment, in addition to it being an effective means of reproducibly transforming monocots, is that neither the isolation of protoplasts (Cristou et al., Plant Physiol. 87:671-674 (1988), nor the susceptibility of Agrobacterium infection are required. An illustrative embodiment of a method for delivering DNA into Zea mays cells by acceleration is a biolistics α-particle delivery system, that can be used to propel particles coated with DNA through a screen, such as a stainless steel or Nytex screen, onto a filter surface covered with corn cells cultured in suspension. Gordon-Kamm et al., describes the basic procedure for coating tungsten particles with DNA (Gordon-Kamm et al., Plant Cell 2:603-618 (1990). The screen disperses the tungsten nucleic acid particles so that they are not delivered to the recipient cells in large aggregates. A particle delivery system suitable for use with the present invention is the helium acceleration PDS-1000/He gun is available from Bio-Rad Laboratories (Bio-Rad, Hercules, Calif.) (Sanford et al., Technique 3:3-16 (1991).
- For the bombardment, cells in suspension may be concentrated on filters. Filters containing the cells to be bombarded are positioned at an appropriate distance below the microprojectile stopping plate. If desired, one or more screens are also positioned between the gun and the cells to be bombarded.
- Alternatively, immature embryos or other target cells may be arranged on solid culture medium. The cells to be bombarded are positioned at an appropriate distance below the microprojectile stopping plate. If desired, one or more screens are also positioned between the acceleration device and the cells to be bombarded. Through the use of techniques set forth herein one may obtain up to 1000 or more foci of cells transiently expressing a marker gene. The number of cells in a focus that express the exogenous gene product 48 hours post-bombardment often range from one to ten and average one to three.
- In bombardment transformation, one may optimize the pre-bombardment culturing conditions and the bombardment parameters to yield the maximum numbers of stable transformants. Both the physical and biological parameters for bombardment are important in this technology. Physical factors are those that involve manipulating the DNA/microprojectile precipitate or those that affect the flight and velocity of either the macro- or microprojectiles. Biological factors include all steps involved in manipulation of cells before and immediately after bombardment, the osmotic adjustment of target cells to help alleviate the trauma associated with bombardment, and also the nature of the transforming DNA, such as linearized DNA or intact supercoiled plasmids. It is believed that pre-bombardment manipulations are especially important for successful transformation of immature embryos.
- In another alternative embodiment, plastids can be stably transformed. Method disclosed for plastid transformation in higher plants include particle gun delivery of DNA containing a selectable marker and targeting of the DNA to the plastid genome through homologous recombination (Svab et al. Proc. Natl. Acad. Sci. (U.S.A.) 87:8526-8530 (1990); Svab et al., Proc. Natl. Acad. Sci. (U.S.A.) 90:913-917 (1993); (Staub et al., EMBO J. 12:601-606 (1993). The methods disclosed in U.S. Pat. No. 5,451,513, U.S. Pat. No. 5,545,818, U.S. Pat. No. 5,877,402, U.S. Pat. No. 5,932479, and WO 99/05265.
- Accordingly, it is contemplated that one may wish to adjust various aspects of the bombardment parameters in small scale studies to fully optimize the conditions. One may particularly wish to adjust physical parameters such as gap distance, flight distance, tissue distance, and helium pressure. One may also minimize the trauma reduction factors by modifying conditions that influence the physiological state of the recipient cells and that may therefore influence transformation and integration efficiencies. For example, the osmotic state, tissue hydration and the subculture stage or cell cycle of the recipient cells may be adjusted for optimum transformation. The execution of other routine adjustments will be known to those of skill in the art in light of the present disclosure.
- Agrobacterium-mediated transfer is a widely applicable system for introducing genes into plant cells because the DNA can be introduced into whole plant tissues, thereby bypassing the need for regeneration of an intact plant from a protoplast. The use of Agrobacterium-mediated plant integrating vectors to introduce DNA into plant cells is well known in the art. See, for example the methods described by Fraley et al., Bio/Technology 3:629-635 (1985) and Rogers et al., Methods Enzymol. 153:253-277 (1987). Further, the integration of the T-DNA is a relatively precise process resulting in few rearrangements. The region of DNA to be transferred is defined by the border sequences, and intervening DNA is usually inserted into the plant genome as described (Spielmann et al., Mol. Gen. Genet. 205:34 (1986).
- Modern Agrobacterium transformation vectors are capable of replication in E. coli as well as Agrobacterium, allowing for convenient manipulations as described (Klee et al., In: Plant DNA Infectious Agents, Hohn and Schell, eds., Springer-Verlag, New York, pp. 179-203 (1985). Moreover, technological advances in vectors for Agrobacterium-mediated gene transfer have improved the arrangement of genes and restriction sites in the vectors to facilitate construction of vectors capable of expressing various polypeptide coding genes. The vectors described have convenient multi-linker regions flanked by a promoter and a polyadenylation site for direct expression of inserted polypeptide coding genes and are suitable for present purposes (Rogers et al., Methods Enzymol. 153:253-277 (1987). In addition, Agrobacterium containing both armed and disarmed Ti genes can be used for the transformations. In those plant varieties where Agrobacterium-mediated transformation is efficient, it is the method of choice because of the facile and defined nature of the gene transfer.
- A transgenic plant formed using Agrobacterium transformation methods typically contains a single genetic locus on one chromosome. Such transgenic plants can be referred to as being hemizygous for the added gene. More preferred is a transgenic plant that is homozygous for the added structural gene; i.e., a transgenic plant that contains two added genes, one gene at the same locus on each chromosome of a chromosome pair. A homozygous transgenic plant can be obtained by sexually mating (selfing) an independent segregant transgenic plant that contains a single added gene, germinating some of the seed produced and analyzing the resulting plants for the gene of interest.
- It is also to be understood that two different transgenic plants can also be mated to produce offspring that contain two independently segregating exogenous genes. Selfing of appropriate progeny can produce plants that are homozygous for both exogenous genes. Back-crossing to a parental plant and out-crossing with a non-transgenic plant are also contemplated, as is vegetative propagation. Descriptions of other breeding methods that are commonly used for different traits and crops can be found in Fehr, In: Breeding Methods for Cultivar Development, Wilcox J. ed., American Society of Agronomy, Madison Wis. (1987).
- Transformation of plant protoplasts can be achieved using methods based on calcium phosphate precipitation, polyethylene glycol treatment, electroporation, and combinations of these treatments (see, e.g., Potrykus et al., Mol. Gen. Genet. 205:193-200 (1986); Lorz et al., Mol. Gen. Genet. 199:178 (1985); Fromm et al., Nature 319:791 (1986); Uchimiya et al., Mol. Gen. Genet. 204:204 (1986); Marcotte et al., Nature 335:454-457 (1988). Application of these systems to different plant varieties depends upon the ability to regenerate that particular plant strain from protoplasts. Illustrative methods for the regeneration of cereals from protoplasts are described (Fujimura et al., Plant Tissue Culture Letters 2:74 (1985); Toriyama et al., Theor Appl. Genet. 205:34 (1986); Yamada et al., Plant Cell Rep. 4:85 (1986); Abdullah et al., Biotechnology 4:1087 (1986).
- Other methods of cell transformation can also be used and include but are not limited to introduction of DNA into plants by direct DNA transfer into pollen (Hess et al., Intern Rev. Cytol. 107:367 (1987); Luo et al., Plant Mol Biol. Reporter 6:165 (1988), by direct injection of DNA into reproductive organs of a plant (Pena et al., Nature 325:274 (1987), or by direct injection of DNA into the cells of immature embryos followed by the rehydration of desiccated embryos (Neuhaus et al., Theor. Appl. Genet. 75:30 (1987).
- The regeneration, development, and cultivation of plants from single plant protoplast transformants or from various transformed explants is well known in the art (Weissbach et al., In: Methods for Plant Molecular Biology, Academic Press, San Diego, Calif., (1988). This regeneration and growth process typically includes the steps of selection of transformed cells, culturing those individualized cells through the usual stages of embryonic development through the rooted plantlet stage. Transgenic embryos and seeds are similarly regenerated. The resulting transgenic rooted shoots are thereafter planted in an appropriate plant growth medium such as soil.
- The development or regeneration of plants containing the foreign, exogenous gene is well known in the art. Preferably, the regenerated plants are self-pollinated to provide homozygous transgenic plants. Otherwise, pollen obtained from the regenerated plants is crossed to seed-grown plants of agronomically important lines. Conversely, pollen from plants of these important lines is used to pollinate regenerated plants. A transgenic plant of the present invention containing a desired exogenous nucleic acid is cultivated using methods well known to one skilled in the art.
- Methods for transforming dicots, primarily by use of Agrobacterium tumefaciens, and obtaining transgenic plants have been published for cotton (U.S. Pat. No. 5,004,863, U.S. Pat. No. 5,159,135, U.S. Pat. No. 5,518,908); soybean (U.S. Pat. No. 5,569,834, U.S. Pat. No. 5,416,011, McCabe et. al., Bio/Technology 6:923 (1988), Christou et al., Plant Physiol. 87:671-674 (1988); Brassica (U.S. Pat. No. 5,463,174); peanut (Cheng et al., Plant Cell Rep. 15:653-657 (1996), McKently et al., Plant Cell Rep. 14:699-703 (1995); and pea (Grant et al., Plant Cell Rep. 15:254-258, (1995).
- Transformation of monocotyledons using electroporation, particle bombardment, and Agrobacterium have also been reported. Transformation and plant regeneration have been achieved in asparagus (Bytebier et al., Proc. Natl. Acad. Sci. (USA) 84:5354-5349 (1987); barley (Wan et al., Plant Physiol 104:37-48 (1994); Zea mays (Rhodes et al., Science 240:204-207 (1988), Gordon-Kamm et al., Plant Cell 2:603-618 (1990), Fromm et al., Bio/Technology 8:833-839 (1990), Koziel et al., Bio/Technology 11: 194-200 (1993), Armstrong et al., Crop Science 35:550-557 (1995); oat (Somers et al., Bio/Technology 10:1589-1594 (1992); orchard grass (Horn et al., Plant Cell Rep. 7:469-472 (1988); rice (Toriyama et al., Theor Appl. Genet. 205:34-(1986), Part et al., Plant Mol. Biol. 32:1135-1148, (1996), Abedinia et al., Aust. J. Plant Physiol. 24:133-141 (1997), Battraw et al., Plant Mol. Biol.. 15:527-538 (1990), Christou et al., Bio/Technology 9:957-962 (1991); rye (De la Pena et al., Nature 325:274-276 (1987); sugarcane (Bower et al., Plant J. 2:409-416 (1992); tall fescue (Wang et al., Bio/Technology 10:691-696 (1992); and wheat (Vasil et al., Bio/Technology 10:667-674 (1992); U.S. Pat. No. 5,631,152).
- Assays for gene expression based on the transient expression of cloned nucleic acid vectors have been developed by introducing the nucleic acid molecules into plant cells by polyethylene glycol treatment, electroporation, or particle bombardment (Marcotte et al., Nature 335:454-457 (1988); Marcotte et al., Plant Cell 1:523-532 (1989); McCarty et al., Cell 66:895-905 (1991); Hattori et al., Genes Dev. 6:609-618 (1992); Goff et al., EMBO J. 9:2517-2522 (1990). Transient expression systems may be used to functionally dissect gene constructs (see generally, Mailga et al., Methods in Plant Molecular Biology, Cold Spring Harbor Press (1995). It is understood that any of the nucleic acid molecules of the present invention can be introduced into a plant cell in a permanent or transient manner in combination with other genetic elements such as promoters, leaders, transit peptide sequences, enhancers, introns, 3′ nontranslated regions and other elements known to those skilled in the art that are useful for control of transgene expression in plants.
- Eleusine indica has been shown to hybridize with Eleusine coracana (finger millet), an important cultivated millet of India and East Africa (Chennaveeraiah et al., Euphytica 2-3:489-495, (1974). Classical plant breeding methods can be used to transfer the gene and the glyphosate tolerant phenotype to crop plants within the family Poaceae. The DNA molecules of the EPSPS glyphosate resistance gene of E. indica (SEQ ID NO: 6) can be used as a probe to identify other like DNA molecules by standard methods. Oligonucleotide DNA molecules homologous or complementary to the EPSPS glyphosate resistance gene of E. indica can be used in a marker assisted breeding method (Simple sequence repeat DNA marker analysis, in “DNA markers: Protocols, applications, and overviews: (1997) 173-185, Cregan, et al., eds., Wiley-Liss NY ) to assist in the breeding of this gene into related and heterologous crop species.
- In addition to the above discussed procedures, practitioners are familiar with the standard resource materials that describe specific conditions and procedures for the construction, manipulation and isolation of macromolecules (e.g., DNA molecules, plasmids, etc.), generation of recombinant organisms and the screening and isolating of clones, (see for example, Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989); Mailga et al., Methods in Plant Molecular Biology, Cold Spring Harbor Press (1995); Birren et al., Genome Analysis: Detecting Genes, 1, Cold Spring Harbor, New York (1998); Birren et al., Genome Analysis: Analyzing DNA, 2, Cold Spring Harbor, New York (1998); Clark et al., Plant Molecular Biology: A Laboratory Manual, Springer, New York (1997); and Innis et al., PCR Protocols: A Guide to Methods and Applications, Academic Press: San Diego, (1990).
- Plant species containing a naturally occurring EPSPS enzyme resistant to glyphosate have not been previously reported. The subject of this invention is the EPSPS enzyme isolated from Eleusine indica that has been shown to be resistant to glyphosate and the expression of the DNA molecule encoding this EPSPS enzyme in other plants that then confers glyphosate tolerance to those recipient plants. The glyphosate resistant EPSPS enzyme isolated from Eleusine indica glyphosate tolerant biotype has a novel Km with respect to binding of PEP as compared to other plant EPSP Synthases that have been modified for glyphosate resistance by a single amino acid substitution of a proline to serine substitution in the active site of the enzyme. The Km for PEP of the E. indica glyphosate resistant enzyme is little changed from the E. indica glyphosate sensitive EPSPS enzyme. In addition, this gene is from a monocot plant and hence may not need nucleic sequence modification to affect expression in transgenic monocot crop plants. The E. indica glyphosate tolerant EPSPS enzyme amino acid sequence can be modified by site directed mutation to include other known substitutions.
- The present invention also provides for parts of the plants of the present invention. Plant parts, without limitation, include seed, endosperm, ovule and pollen. In a particularly preferred embodiment of the present invention, the plant part is a seed.
- The following examples are included to demonstrate examples of certain preferred embodiments of the invention. It should be appreciated by those of skill in the art that the techniques disclosed in the examples that follow represent approaches the inventors have found function well in the practice of the invention, and thus can be considered to constitute examples of preferred modes for its practice. However, those of skill in the art should, in light of the present disclosure, appreciate that many changes can be made in the specific embodiments that are disclosed and still obtain a like or similar result without departing from the spirit and scope of the invention. All references cited herein are hereby expressly incorporated herein by reference.
- Seeds from glyphosate tolerant Eleusine indica plants were deposited with the American Type Culture Collection (ATCC, 10801 University Blvd, Manassas, Va., U.S.A., 20110-2209) and assigned ATCC No. PTA-2177. The deposit will be maintained in the depository for a period of 30 years, or 5 years after the last request, or for the effective life of the patent, whichever is longer, and will be replaced as necessary during that period.
- Eleusine indica plants tolerant to glyphosate were collected from a site near Johor, Malaysia. Glyphosate tolerant biotypes of E. indica are identified and numbered. Seed is collected from each biotype and planted in pots in the greenhouse. Clones are generated for each plant by excising 10-20 tillers and transplanting these in separate pots. Glyphosate sensitive and tolerant individual plants are then identified by treatment with glyphosate at either 0.5 kg active ingredient (ai)/hectare (ha) or 2.0 kg ai/ha, respectively. A corresponding clone for glyphosate tolerant and glyphosate sensitive E. indica biotype is left untreated. These clones are used as the source of fresh tissue for enzyme analysis and gene isolation.
- Construction of a cDNA library from the glyphosate tolerant E. indica biotype and the glyphosate sensitive E. indica biotype is performed by isolating total RNA from the crown tissues. The crown tissues are dissected from the plants, then flash-frozen with liquid nitrogen and maintained at −80° C. until needed. Total RNAs are extracted from frozen crown samples using the RNeasy Plant Mini Kit (cat. #74904, Qiagen Inc., Valencia, Calif.) per manufacturer's instructions. Oligo.dT-primed first-strand cDNAs are prepared from 5 μg samples of total RNA using the Superscript Pre-Amplification System (cat. #18089-011, Life Technologies, Rockville, Md.) per manufacturer's instructions. Two μl of first-strand cDNA are then used to generate partial E. indica EPSP synthase cDNAs via polymerase chain reaction using a modification of the “touchdown PCR” technique (Don et al., Nucl. Acids Res. 19:4008, 1991). Degenerate oligonucleotide pools of SEQ ID NO: 1 and SEQ ID NO: 2 are added in a 50 μl RT-PCR reaction at a final concentration of 25 μM.
5′-TNWSNGTNGARGCNGAYAARGT-3′ (SEQ ID NO: 1) 5′-GCCATNGCCATNCKRTGRTCRTC-3′ (SEQ ID NO: 2) - PCR amplifications are then performed using the Expand High Fidelity PCR System (cat. #1 732 641, Roche Molecular Biochemicals, Indianapolis, Ind.) per manufacturer's instructions. A thermal profile of 94° C. for 20 seconds, followed by 60° C. for 1 minute, then 72° C. for 1
minute 30 seconds is used for the initial 30 cycles with a 0.5° C. decrease in annealing temperature per cycle. This is followed by 10 additional cycles of 94° C. for 20 seconds, 45° C. for 1 minute, then 72° C. for 1minute 30 seconds. - RT-PCR products are then purified by agarose gel electrophoresis using a QIAquick Gel Extraction Kit (cat. #28704, Qiagen Inc., Valencia, Calif.) then directly cloned into the pCR2.1-TOPO vector (cat. #K4500-40, Invitrogen, Carlsbad, Calif.). The identity of the cloned RT-PCR products is confirmed by DNA sequence analysis (ABI Prism™ 377, Perkin Elmer, Foster City, Calif.).
- The remainder of the 3′ end of the EPSP synthase coding region is generated using the 3′ RACE System for Rapid Amplification of cDNA Ends (cat. #18373-027, Life Technologies, Rockville, Md.), using the gene-specific oligonucleotide of SEQ ID NO: 3. The cDNA is prepared according to manufacturer's instructions using 5 μg of total RNA isolated from crown tissues as previously described.
- 5′-GTGAAAGCAGAGCATTCTGATAGC-3′ (SEQ ID NO: 3)
- PCR amplifications are conducted in 50 μl reactions including 5 μl first-strand cDNA reaction, 20 picomoles of each primer, 10 mM Tris.HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl 2, 200 μM dNTPs, and 2.5 units Taq polymerase. A thermal profile of 94° C. for 20 seconds, followed by 57° C. for 1 minute, then 72° C. for 1
minute 30 seconds is used for 35 cycles. The identity of the 3′-RACE products is confirmed by DNA sequence analysis (ABI Prism™ 377, Perkin Elmer, Foster, Calif.). - The remainder of the 5′ end of the E. indica EPSP synthase mature protein coding region is generated using the SMART RACE cDNA Amplification Kit (cat. #K1811-1, Clontech Laboratories Inc., Palo Alto, Calif.), using the gene-specific oligonucleotides of SEQ ID NO: 4 and SEQ ID NO: 5. The cDNA is prepared according to manufacturer's instructions using 150 ng of polyA+ mRNA isolated from crown tissues using an Oligotex mRNA Midi Kit (cat. #28704, Qiagen Inc., Valencia, Calif.).
(SEQ ID NO: 4) 5′-GGCTGCTGTCAATGTCGCATTGCAGTTCC-3′ (SEQ ID NO: 5) 5′-CTCTTTCGCATCCTTCTCAACTGGGAACTTGC-3′ - PCR reactions are conducted as recommended by the manufacturer, except that the Expand High Fidelity PCR System (cat. #1 732 641, Roche Molecular Biochemicals, Indianapolis, Ind.) is used and DMSO is included in all reactions at a final concentration of 5.0% to facilitate the amplification of GC-rich sequences. The synthetic DNA oligonucleotide described in SEQ ID NO: 4 is used in the primary amplifications, then second round (“nested”) amplifications are performed using the oligonucleotide described in SEQ ID NO: 5, with a 1 μl aliquot of 1:100 dilution of the primary PCR reactions. The identity of the 5′-RACE products is confirmed by DNA sequence analysis (ABI Prism™ 377, Perkin Elmer, Foster City, Calif.).
- The significant overlap of sequences generated by RT-PCR, 3′ RACE, and 5′ RACE allows for the unambiguous assembly of the sequences into a single DNA sequence containing the entire open reading frame for the mature protein, using the SEQMan II software package (DNASTAR Inc., Madison, Wis.). The DNA sequence corresponding to the mature protein-coding region of the Eleusine indica (glyphosate tolerant biotype) EPSP synthase gene (SEQ ID NO: 6) is shown in FIG. 1.
- The deduced amino acid sequence for the mature protein-coding region of the Eleusine indica (glyphosate tolerant biotype) EPSP synthase gene (SEQ ID NO: 7) for this protein is shown in FIG. 2.
- EPSP synthase enzyme from the glyphosate tolerant E. indica biotype confers increased glyphosate tolerance in transgenic E. coli. E. coli is useful as a heterologous expression system for testing glyphosate resistant enzymes. The EPSP synthase mature protein-coding regions isolated from the glyphosate tolerant and glyphosate sensitive E. indica biotypes, can be directly compared for their ability to confer tolerance to glyphosate in transgenic hosts. E. coli (strain SR481) are transformed with the glyphosate resistant EPSPS gene (Ei.EPSPS:glyR) and the glyphosate sensitive EPSPS gene (Ei.EPSPS:glyS) purified from E. indica. The growth rate differentials of transformed cell lines grown in the presence of glyphosate contained in the culture medium is used as a measure of the resistance of the EPSPS enzyme to the inhibitory effects of glyphosate (Rogers et al., Appl. Enviro. Microbiol. 46:37-43 (1983). An appropriate E. coli expression vector that carries the native promoter and operator sequence from the E. coli lac operon (Dickson et al., Science 187:27-35 (1975) including the sequence
(SEQ ID NO:8) 5′-AGATCTCCTAGGGCTTAATTAATTAAGCACTAGTCACACAGGAGGTA ATTCATATG-3′ - is contained in pMON45337. This nucleotide sequence includes 1) flanking BglII and Nde1 endonuclease sites, 2) a ribosome binding site, and 3) an unstructured region 5′ to the ribosome binding element (Balbas, P. et. al., in “Methods in Enzymology” (D. V. Goeddel, ed.)185: 15-37, 1990). This was inserted by ligation to facilitate expression and cloning at the ATG start codon of an open reading frame. A multiple cloning site is positioned immediately downstream of this Ndel site, followed by the rho-independant transcriptional terminator element of the E. coli trpA gene (T. Sato et al., J. Biochem. (Tokyo), 101:525-534, (1987). This vector when it operbly contains the EPSP synthase coding sequences of the present invention is employed for the inducible expression of glyphosate resistant and glyphosate sensitive EPSP synthase cDNAs in E. coli. Other commercially available inducible E. coli expression vectors are suitable for testing the EPSP synthases from E. indica.
- DNA manipulations and transformations of E. coli are performed according to standard procedures (Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989). To construct E. coli expression vectors carrying the EPSP synthase mature protein coding sequences from the tolerant and sensitive E. indica biotypes, the oligonucleotide primers of SEQ ID NO: 9 and SEQ ID NO: 10.
5′-GCAATTCCATATGGCGGGCGCGGAGGAGGTGGTGCT-3′ (SEQ ID NO: 9) 5′-GACTAGGAATTCTTAGTTCTTTTGACGAAAGTGCTCAGCACGTCGAAG-3′, (SEQ ID NO: 10) - These sequences are employed in RT-PCR reactions to generate expression cassettes suitable for cloning into pMON45337 cut with the restriction enzymes Nde1 and EcoR1. RT-PCR reactions are performed with total RNAs extracted from frozen crown samples using the RNeasy Plant Mini Kit (cat. #74904, Qiagen Inc., Valencia, Calif.) per manufacturer's instructions. Oligo.dT-primed first-strand cDNAs are prepared from 5 μg samples of total RNA using the Superscript Pre-Amplification System (cat. #18089-011, Life Technologies, Rockville, Md.) per manufacturer's instructions. Two μl of first-strand cDNA are then used to generate E. indica EPSP synthase expression cassettes via polymerase chain reaction. The oligonucleotides are added in 50 μl RT-PCR reactions at a final concentration of 0.4 μM. PCR amplifications are then performed using the Expand High Fidelity PCR System (cat. #1 732 641, Roche Molecular Biochemicals, Indianapolis, Ind.) per manufacturer's instructions, using a thermal profile of 94° C. for 30 seconds, then 57° C. for 2 minutes, followed by 75° C. for 3 minutes, for a total of 35 cycles. The resulting PCR products are digested with Nde I and EcoRI, then ligated into pMON45337, resulting in the E. coli expression vectors pMON45364 (FIG. 5) and pMON45365 (FIG. 6), which contain the mature protein coding region for E. indica EPSP synthase isolated from the resistant and sensitive biotype, respectively. Expression of the two enzymes in E. coli will thus be directed by the Lac operon and trpA gene genetic elements described above for pMON45337. The accuracy of the cloned sequences are confirmed by DNA sequence analysis (ABI Prism™ 377, Perkin Elmer, Foster, Calif.). pMON45337, pMON45364, and pMON45365 are all transformed into the E. coli strain SR481, an aroA-strain lacking endogenous EPSP synthase activity (Padgette et al., Arch. Biochem. Biophys. 258:564-573 (1987).
- To directly compare the glyphosate tolerance of E. coli aroA-cells expressing the EPSP synthase gene isolated from the glyphosate sensitive E. indica biotype with cells expressing the EPSP synthase gene isolated from the glyphosate tolerant biotype, growth rates are compared for the two cell lines in the presence of increasing concentrations of glyphosate (FIG. 3). Growth rates are also monitored for E. coli SR481 cells transformed with pMON45337 (empty vector) in the absence of glyphosate as a negative control. Fresh overnight cultures of E. coli SR481 cells transformed with pMON45337, pMON45364 (Ei.EPSPS:glypR), and pMON45365 (Ei.EPSPS:glypS) are grown in Terrific Broth (Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989), supplemented with 1.0 mM IPTG, 50 μg/ml ampicillin, and 100 μg/ml each of L-phenylalanine, L-tyrosine, and L-tryptophan. O.D.595 measurements are taken on all of the overnight cultures to confirm similar cell densities. For E. coli SR48 1-pMON45364 and SR481-pMON45365 cells, 14 ml culture tubes (cat.# 60818-725, VWR Scientific, West Chester, Pa.) each containing 3.0 ml of minimal M9 media (Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989) supplemented with 50 μg/ml ampicillin, 1.0 mM IPTG, and either 0.0, 0.5, 1.5, or 5.0 mM glyphosate (N-phosphonomethyl glycine or a salt thereof) are inoculated with 100 μl of undiluted overnight culture per tube. Each experimental condition is performed in triplicate to confirm the reproducibility of the experiment. Where t=0, 12, 24, 28, 32, and 48 hours past the onset of growth in minimal media, 100 μl aliquots from each tube are removed and O.D.595 measurements are taken immediately. A typical result from these analyses is shown in FIG. 3, where an approximately three-fold increase in tolerance to glyphosate is observed due to the expression of the EPSP synthase enzyme from the glyphosate tolerant E. indica biotype.
- Kinetic characterization of the E. indica glyphosate-resistant EPSP synthase activity in plant and bacterial extracts. Kinetic characterization of the glyphosate-resistant E. indica enzyme are performed using both partially purified plant extracts as well as bacterial extracts prepared from cells expressing the cloned sequence on a suitable vector such as pMON45365. Parameters that describe the enzyme's resistance to glyphosate-mediated inhibition and the affinity for the substrate phosphoenolpyruvate (PEP) are of particular interest, given that glyphosate is a competitive inhibitor of EPSP synthase with respect to PEP (Boocock, M. et al., FEBS Letts. 154:127-133 (1983).
- Preparation of extracts and radiometric EPSP synthase assays are performed using methods adapted from published procedures (Padgette et al., J. Biol. Chem. 266:22364-22369 (1991). Crown regions are dissected from whole plants, pulverized under liquid nitrogen with a mortar and pestle, then stored at −80° C. prior to extraction. Homogenates are prepared from 0.5 g tissue per sample in 25 ml extraction buffer (100 mM TrisCl, 10% glycerol, 1 mM EDTA, 1 mM benzamidine, 1 mM dithiothreitol, 1 mM 4-(2-aminoethyl)-benzenesulfonyl floride HCl, 0.1 mM leupeptin, pH 7.4) at 4° C. using a model PT3000 Polytron homogenizer (Brinkman Instuments Inc., Westbury, N.Y.). Debris is removed by 0.2 μm filtration, then the resulting supernatant is concentrated and desalted using an Ultrafree-15 centrifugal filtration unit (cat. #UFV2-BGC-10, Millipore Corp., Bedford, Mass.). Final sample volumes are approximately 0.5 ml. Protein concentrations are determined spectrophotometrically using the Bio-Rad protein assay reagent (cat. #500-0006, Bio-Rad Laboratories, Hercules, Calif.). EPSPS specific activities are determined by assaying 10 μl extract at 25° C. for 5-15 min. (50 μl reactions include 50 mM HEPES, pH 7.0, 5 mM potassium fluoride, 1 mM shikimate-3-phosphate, 0.5 mM [1- 14C]-phosphoenolpyruvate (29.0 mCi/mmol cyclohexylammonium salt; #CFQ10004, Amersham Life Science, Inc., Arlington Heights, Ill.), and 0.1 mM ammonium molybdate). Reactions are quenched with the addition of 50 μl 9:1 ethanol: 0.1 M acetic acid. Thirty μ l of quenched reaction is then injected onto a Synchropak AX100 anion exchange column (cat. #942804, P.J. Cobert Associates, Inc., St. Louis, Mo.) equilibrated with 0.235 M potassium phosphate buffer, pH 6.5, and eluted isocratically with the same buffer. A model D525 radioactive flow detector (Packard Instrument Co., Downer's Grove, Ill.) is used to determine production of [14C]-EPSP in the reaction.
- For determination of I 50 (glyphosate) values, the assays as described, are performed in the presence of increasing concentrations of glyphosate and the resulting activities analyzed and plotted using GraFit version 3.0 software (Erithacus Software Ltd., Staines, U.K.). FIG. 4 shows data generated for a typical glyphosate inhibition study, comparing the EPSP synthase activities detectable in extracts prepared from the glyphosate sensitive and tolerant E. indica biotypes. These data demonstrate the difference in sensitivity to glyphosate for the activities present in the two biotypes, with the sensitive biotype EPSPS activity having an I50 (glyphosate) of approximately 3.0 μM and the tolerant biotype EPSPS approximately 16 μM.
- Activities of the enzymes extracted from several different tolerant and sensitive E. indica individuals are compared in the presence and absence of 1.6 μM glyphosate (Table 1), showing similar sensitivity to glyphosate and very low plant-to-plant variation exhibited among individuals from the same biotype.
TABLE 1 Percent-maximal EPSPS activity in extracts from different E. indica individuals assayed in the presence 1.6 uM glyphosate EPSPS Activity, biotype-individual (% maximal @ 1.6 uM glyphosate) Sensitive - #1 51.6 Sensitive - #2 55.6 Sensitive - #3 57.4 Tolerant - #1 76.3 Tolerant - #2 86.0 Tolerant - #3 82.4 - Similar analyses are performed using extracts prepared from E. coli strain SR481 cells expressing the cloned E. indica sensitive and resistant EPSPS enzymes from the expression vectors pMON45365 and pMON45364, respectively. Fresh bacterial overnight cultures are grown at 37° C. in Terrific Broth, (Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press (1989), supplemented with 50 μg/ml ampicillin and 100 μg/ml each of L-phenylalanine, L-tyrosine, and L-tryptophan. Overnight cultures are used to inoculate large-scale cultures containing the same media at a 1:100 dilution, then grown to an O.D.595 of 0.6 at 37° C. with vigorous shaking. The culture is inducted by the addition of IPTG to 1.0 mM final concentration then incubation is continued for an additional 4 hours. Cells are then pelleted by centrifugation at 10,000×g for 5 minutes, then washed twice in ice-cold 0.9% NaCl. Excess wash buffer is removed by aspiration, then pellets are flash-frozen in liquid nitrogen and stored at −80° C. prior to use. Bacterial extracts are prepared using the same extraction buffer used for plant extracts, with 3 ml buffer added per gram of pelleted cells. Cells are lysed using a French press (model # J5-598A, American Instrument Co., Silver Springs, Md.), then extracts are centrifuged at 14,000×g for 10 minutes to remove cell debris. The supernatants are then desalted using a PD-10 column (cat.# 17-0851-01, Amersham Pharmacia Biotech Inc., Piscataway, N.J.). EPSP synthase assays are performed as described above for plant-derived extracts. Table 2 illustrates a typical result for bacterial extracts expressing the sensitive and resistant E. indica EPSP synthase enzymes assayed in the presence and absence of 1.6 μM glyphosate. These data indicate that similar inhibition kinetics are obtained when these results are compared to the respective activities detected in plant extracts.
TABLE 2 EPSPS activity inaroA - bacterial extracts transformed with pMON45364 or pMON45365 assayed in the presence and absence of glyphosate activity % maximal activity sample (nMol/min/mg protein) (1.6 uM glyphosate) SR481(pMON45364) −glyphosate 34.3 +glyphosate 24.9 72.6 SR481(pMON45365) −glyphosate 27.1 +glyphosate 14.4 53.1 - The use of the glyphosate resistant EPSP synthase gene from E. indica (Ei.EPSPS:glypR) to develop glyphosate tolerant crop plants involves the construction of plant transformation vectors that include the appropriate combination of genetic elements necessary to direct adequate expression levels in target tissues. For monocotyledonous crop plants, a monocot vector that utilizes a plant expression cassette that contains a promoter (P) and first intron (I) from the rice (Os) actin gene (P-Os.Act1/I-Os.Act1 (U.S. Pat. No. 5,641,876), plastid transit peptide sequence (TS) from the Arabidopsis thaliana (At) EPSP synthase gene (TS-At.EPSPS:CTP) (Klee et al. Mol. Gen. Genet. 210:437-442), a E. indica glyphosate resistant EPSPS coding sequence, and the polyadenylation/termination (T) region from the Agrobacterium tumefaciens nopaline synthase gene (T-AGRTU.nos), would be an appropriate choice. This expression cassette may be combined with a second transgene expression cassette by plant breeding, plant transformation, or by joining in a DNA construct that comprises a plant DNA virus promoter, for example, the cauliflower mosaic virus (CaMV) 35S promoter containing a tandem duplication of the enhancer region, operably connected to a Zea mays Hsp70 intron, operably connected to a nucleic acid sequence encoding an Arabidopsis thaliana EPSPS chloroplast transit peptide sequence, operably connected to a E. indica glyphosate resistant EPSPS coding sequence, operably connected to a nopaline synthase transcriptional terminator. Other combinations of genetic elements are known and those skilled in the art of plant molecular biology can easily construct plant expression vectors that will express the E. indica EPSPS glyphosate resistant enzyme at sufficient levels to confer glyphosate the transformed plant.
- For specific dicotyledonous species, a plant expression vector that utilizes the promoter and 5′ untranslated region (including intron I) of the plant elongation factor 1α gene (Elfα-A1) as described in U.S. Pat. No. 5,177,011 or more specifically, dicot vector of the present invention which utilizes the Arabidopsis thaliana Elfα-A1 promoter and intron sequence (P-At.Elf1a/I-At.Elf1a), (Axelos et al., Mol. Gen. Genet. 219: 106-112 (1989); Genbank accession #U63815), the chloroplast transit peptide from the Arabidopsis thaliana EPSP synthase gene (TS-At.EPSPS:CTP2), and the polyadenylation/3′ termination region from the Pisum sativum ribulose-1,5-bisphosphate carboxylase gene (T-Ps.RbcS:E9). This expression cassette may be combined with a second transgene expression cassette by plant breeding, plant transformation, or by joining in a DNA construct that comprises a plant DNA virus promoter, for example, the Figwort mosaic virus (FMV) 34S promoter, operably connected to a nucleic acid sequence encoding an Arabidopsis thaliana EPSPS chloroplast transit peptide sequence, operably connected to a E. indica glyphosate resistant EPSPS coding sequence, operably connected to a nopaline synthase transcriptional terminator. Other combinations of genetic elements are known and those skilled in the art of plant molecular biology can easily construct plant expression vectors that will express the E. indica EPSPS glyphosate resistant enzyme at sufficient levels to confer glyphosate tolerance to the transformed plant.
- To construct monocotyledonous and dicotyledonous plant expression vectors carrying the DNA encoding the EPSP synthase mature protein coding sequence isolated from the glyphosate tolerant E. indica biotype (SEQ ID NO. 6, FIG. 1), the oligonucleotide primers of SEQ ID NO: 11 and SEQ ID NO: 12 are employed in PCR reactions to generate an expression cassette suitable for direct cloning into a monocot vector and a dicot vector.
5′-GCAATTCGCATGCCGGGCGCGGAGGAGGTGGTGCT-3′ (SEQ ID NO: 11) 5′-GACTAGGAATTCTTAGTTCTTTTGACGAAAGTGCTCAGCACGTCGAAG-3′ (SEQ ID NO: 12) - The PCR reactions are performed using 300-500 ng of pMON45364 plasmid DNA as template to amplify the E. indica EPSP synthase mature protein coding region, flanked by Sph1 and EcoR1 restriction cleavage sites. The oligonucleotides are added in 50 μl PCR reactions at a final concentration of 0.4 μM. PCR amplifications are then performed using the Expand High Fidelity PCR System (cat. #1 732 641, Roche Molecular Biochemicals, Indianapolis, Ind.) per manufacturer's instructions, using a thermal profile of 94° C. for 30 seconds, then 57° C. for 2 minutes, followed by 75° C. for 3 minutes, for a total of 20-35 cycles. The resulting PCR products are digested with Sph1 and EcoR1, then ligated into pMON45366 and pMON45368 resulting in the plant expression vectors pMON45367 (FIG. 7) and pMON45369 (FIG. 8), respectively. The accuracy of the cloned sequences are confirmed by DNA sequence analysis (ABI Prism™ 377, Perkin Elmer, Foster City, Calif.).
- Transgenic corn can be produced by particle bombardment transformation methods as described in U.S. Pat. No. 5,424,412. The plant expression vector (pMON45367) contains the glyphosate resistant E. indica EPSPS mature protein coding sequence in an expression cassette suitable for expression in monocot plants. The pMON45367 plasmid DNA is digested with Not1 and Pme1 restriction endonucleases to complete digestion. The 3.3 kb expression cassette is agarose gel purified, then bombarded into embryogenic corn tissue culture cells using a Biolistic® (Dupont, Wilmington, Del.) particle gun with purified isolated DNA fragment. Transformed cells are selected on glyphosate (N-phosphonomethyl glycine and its salts) containing media and whole plants are regenerated then grown under greenhouse conditions. Fertile seed is collected, planted and the glyphosate tolerant phenotype is back crossed into commercially acceptable corn germplasm by methods known in the art of corn breeding (Sprague et al., Corn and Corn Improvement 3rd Edition, Am. Soc. Agron. Publ (1988).
- Transgenic corn plants can be produced by an Agrobacterium mediated transformation method. A disarmed Agrobacterium strain C58 (ABI) harboring a binary vector (pMON45367) is used for all the experiments. The pMON45367 is transferred into Agrobacterium by a triparental mating method (Ditta et al., Proc. Natl. Acad. Sci. 77:7347-7351). Liquid cultures of Agrobacterium are initiated from glycerol stocks or from a freshly streaked plate and grown overnight at 26° C.-28° C. with shaking (approximately 150 rpm) to mid-log growth phase in liquid LB medium, pH 7.0 containing 50 mg/l kanamycin, 50 mg/l streptomycin and spectinomycin and 25 mg/l chioramphenicol with 200 μM acetosyringone (AS). The Agrobacterium cells are resuspended in the inoculation medium (liquid CM4C) and the density is adjusted to OD 660 of 1. Freshly isolated Type II immature HiII×LH198 and HiII corn embryos are inoculated with Agrobacterium containing pMON45367 and co-cultured 2-3 days in the dark at 23 ° C. The embryos are then transferred to delay media (N6 1-100-12/micro/Carb 500/20 μM AgNO3) and incubated at 28 ° C. for 4 to 5 days. All subsequent cultures are kept at this temperature. Coleoptiles are removed one week after inoculation. The embryos are transferred to the first selection medium (N61-0-12/Carb 500/0.5 mM glyphosate). Two weeks later, surviving tissue are transferred to the second selection medium (N61-0-12/Carb 500/1.0 mM glyphosate). Subculture surviving callus every 2 weeks until events can be identified. This will take 3 subcultures on 1.0 mM glyphosate. Once events are identified, bulk up the tissue to regenerate. For regeneration, callus tissues are transferred to the regeneration medium (MSOD 0.1 μM ABA) and incubated for two weeks. The regenerating calli are transferred to a high sucrose medium and incubated for two weeks. The plantlets are transferred to MSOD media in culture vessel and kept for two weeks. Then the plants with roots are transferred into soil.
- Three R 0 plants are regenerated for any given transgenic event. These three plants are expected to be near isogenic because they are thought to be derived from a single transgenic plant cell. Thus, one plant is used as a non-sprayed control and the remaining two plants are treated with glyphosate (as Roundup® herbicide). Plants are most effectively treated with glyphosate at V2-V6 stage. Glyphosate (as Roundup® herbicide) is administered through the use of a linear track sprayer set to deliver a 16, 32 or 64 oz./A rate of glyphosate. Vegetative tolerance to the glyphosate is visually evaluated a week after spray based on a scale of 0 to 5 (0=No observable/vegetative effect of glyphosate; 1=Chlorosis observed; 2=Advanced chlorosis, minor necrosis; 3=Advanced chlorosis, moderate necrosis; 4=Advanced chlorosis, severe necrosis; 5=No live tissue remaining). The R0 plants produced are allowed to self, then R1 plants are screened using spray applications of glyphosate and the rating system as described for the R0 screen. An increase in whole-plant tolerance to the herbicide, as compared to non-transgenic control plants, is used to assess the utility of the E. indica EPSP synthase enzyme for the generation of glyphosate tolerance in planta.
- Immature embryos of wheat ( Triticum aestivum L) cultivar Bobwhite are isolated from the immature caryopsis 13-15 days after pollination, and cultured on CM4C (Table 3) for 3-4 days. The embryos showing active cell division, but no apparent callus formation are selected for Agrobacterium infection.
TABLE 3 Supplemental Components in Basal Media1 Components CM4 CM4C MMS.2C MMS0 2,4-D (mg/l) 0.5 0.5 0.2 — Pichloram (mg/l)2 2.2 2.2 Maltose (g/l) 40.0 40.0 40.0 40.0 Glutamine (g/l) 0.5 0.5 Magnesium Chloride (g/l) 0.75 0.7 Casein Hydrolysate (g/l) 0.1 0.1 MES (g/l) 1.95 1.95 1.95 Ascorbic Acid (mg/l)2 100.0 100.0 100.0 Gelling Agent (g/l)3 2(P) 2(P) 2(G) 2(G) - A disarmed Agrobacterium strain C58 (ABI) harboring a binary vector of interest (pMON45367) is used for all the experiments. The pMON45367 is transferred into Agrobacterium by a triparental mating method (Ditta et al., Proc. Natl. Acad. Sci. 77:7347-7351). Liquid cultures of Agrobacterium are initiated from glycerol stocks or from a freshly streaked plate and grown overnight at 26° C.-28° C. with shaking (approximately 150 rpm) to mid-log phase (OD 660=1-1.5) in liquid LB medium, pH 7.0 containing 50 mg/l kanamycin, 50 mg/l streptomycin and spectinomycin and 25 mg/l chloramphenicol with 200 μM acetosyringone (AS). The Agrobacterium cells are resuspended in the inoculation medium (liquid CM4C) and the density is adjusted to OD660 of 1. The immature embryos cultured in CM4C medium are transferred into sterile petri plates (16×20 mm) or wells of a 6-well cell culture plate (Costar Corporation, Cambridge, Mass.) containing 10 ml of inoculation medium per petri plate or 5 ml per cell culture cluster plate. An equal amount of the Agrobacterium cell suspension is added such that the final concentration of Agrobacterium cells is an OD600 of 0.5. In most experiments, pluronic F68 is added to the inoculation mixture at a final concentration of 0.01%. The ratio between the Agrobacterium and immature embryos is about 10 ml: 20-200 IEs. The inoculation is allowed to proceed at 23° C.-26° C. from 5-60 minutes.
- After the inoculation period, the remaining Agrobacterium cells are removed from the explants by using vacuum aspiration equipment. A piece of sterile Whatman No. 1 filter paper (to fit the size of the petri plate) is placed in each of 60×15 or 60×20 mm petri dishes. Two hundred μl of sterile water is placed in the middle of the filter paper. After 2-3 minutes, the inoculated immature embryos are placed in the plates. Usually, 20-50 explants are grouped as one stack (about 1 cm in size and 60-80 mg/stack), with 4-5 stacks on each plate. The plates are immediately covered with Parafilm® and then co-cultivated in the dark at 24° C.-26° C. for 2-3 days.
- The co-cultivated PCIEs are transferred CM4C+500 mg/l carbenicillin medium (delay medium) at dark. After 7 days on the delay medium, the immature embryos are transferred to CM4C supplemented with 2 mM glyphosate and 500 mg/l carbenicillin for selection for one week. Then calli are transferred to MMS0.2C+0.1 mM glyphosate +250 mg/l carbenicillin medium for 2 weeks under light for further selection. Embryogenic calli are transferred to a second regeneration medium MMS0C with lower glyphosate concentration (0.02 mM) and 500 mg/L carbenicillin for plant regeneration. Those embryogenic calli are transferred onto fresh medium every two weeks. Regenerated plantlets are transferred to Sundae cups (Sweetheart Cup Company, Chicago, Ill.) containing the second regeneration medium for further growth and selection. When roots are well established from transgenic plants the plants are transferred to soil for further evaluation.
- Novel glyphosate-resistant EPSP synthases can be designed based on the E. indica glyphosate resistant EPSPS. The amino acid sequence deduced from the cDNA sequence shows that two amino acid substitutions distinguish the mature EPSP protein sequence derived from the glyphosate-tolerant E. indica biotype (top row, FIG. 9) from that of the glyphosate sensitive E. indica biotype EPSPS protein sequence (bottom row, FIG. 9). The substitution of a serine for a proline at position 107 of the E. indica EPSPS amino acid sequence and in the corresponding amino acid position in both higher plant and bacterial EPSP synthase enzymes is known to result in the enzyme having resistance to glyphosate (Padgette et al., J. Biol. Chem. 266:22364-22369 (1991); U.S. Pat. No. 4,535,060). All catalytic domain single amino acid substitution EPSP synthase variants characterized to date that exhibit increased tolerance to glyphosate have a higher K; for glyphosate, but also have an increase in the apparent Km for PEP and reduced Vmax, thereby lowering the catalytic efficiency (Vmax/Km) of the enzyme (Kishore et al., Annu. Rev. Biochem. 57:627-663 (1988); glyphosate (Padgette et al., J. Biol. Chem. 266:22364-22369 (1991). In contrast, the E. indica glyphosate resistant EPSP synthase (E. indica glypR) exhibits a high affinity for PEP, while retaining significant catalytic efficiency in the presence of glyphosate (TABLE 4). The engineered petunia (Petunia hybrida) and corn (Zea mays) glyphosate resistant (glypR) variants that in studies by others (U.S. Pat. No. 5,866,774; U.S. Pat. No. 6,040,497) have shown to confer a high level of glyphosate tolerance in transgenic plants were assayed for affinity for PEP and inhibition by glyphosate. All of the naturally occurring wild type (wt) Z. mays glyphosate sensitive (Z. mays glypS), wild type P. hybrida glyphosate sensitive (P. hybrida glypS) and the E. indica glypS and E. indica glypR wild type EPSPS enzymes have very similar Km values for PEP. The single amino acid substitutions engineered into the catalytic domain of P hybrida EPSPS enzyme drastically increases the Km for PEP. The single amino acid substitution found in this domain in the naturally occurring variant of E. indica glypR was found to not have a major effect on the Km indicating that this enzyme will continue to function well in the plant chloroplast. It required a double mutation in the Z. mays EPSPS enzyme to achieve a low Km for PEP.
TABLE 4 Comparison of the apparent Km for PEP and apparent Ki for glyphosate of the E. indica glyphosate resistant EPSPS with other known plant EPSPS modified for glyphosate resistance. EPSPS enzyme Km PEP (μM) Ki Glyphosate (μM) E. indica glypS 5 0.05 E. indica glypR (Pro-Ser) 7 1 Z. mays glypS (wt) 5 0.2 Z. mays glypR (Thr-Ile, Pro-Ser) 5 60 P. hybrida glypS (wt) 5 0.4 P. hybrida glypR (Pro-Ser) 44 3 P. hybrida glypR (Gly-Ala) 200 2000 P. hybrida glypR (Gly-Ala, Pro- 340 8500 Ser) - K m(PEP) determinations for the different enzymes are performed at saturating shikimate-3-phosphate (S3P) concentrations, which is determined according to standard methods (Fersht, Enzyme Structure and Mechanism, W.H. Freeman and Co., Ltd., San Francisco, Calif., 1977). A series of PEP concentrations are tested, such that the final range of concentrations spans one order of magnitude above and below the experimentally determined Km. Ki (glyphosate), with respect to PEP, is determined in a similar manner, at saturating S3P concentrations, except that velocity vs. [PEP] is determined for a range of glyphosate concentrations (Orsi in “Methods in Enzymology” (Purich, ed.), vol. 63, pg. 159-183, 1979). Calculations, graphical representation, and statistical analysis of enzyme kinetic data are performed using GraFit version 3.0 software (Erithacus Software Ltd., Staines, U.K.).
- It is anticipated by this study that transgenic plants resistant to glyphosate are made by transformation with an E. indica glyphosate resistant EPSPS gene construct that can include additional modification of the naturally occurring amino acid sequence. These changes are be made by site-directed mutagenesis of the codons of DNA sequence to incorporate other known amino acid substitutions in glyphosate resistant plant EPSPSs, such as the threonine to isoleucine substitution at 103 (U.S. Pat. No. 6,040,497), and glycine to alanine substitution at 102 (U.S. Pat. No. 5,188,642) in the catalytic domain of the E. indica EPSPS amino acid seqeunce. Furthermore, it is anticipated that the catalytic domain of the E. indica EPSPS, as well as other plant EPSP Synthases, can be modified to the amino acid sequence of the catalytic domain of the Agrobacterium strain CP4 glyphosate resistant EPSPS (U.S. Pat. No. 5,633,435) by the same methods. This modification will result in the plant derived EPSPS possessing similar PEP binding and glyphosate resistance as the CP4 glyphosate resistant EPSPS that has been used in cotton, corn, canola, soybeans, potato, wheat, sugarbeet and other agronomically important crop plant to impart plant tolerance to glyphosate. Modification of plant EPSP Synthases to the CP4 EPSPS catalytic domain sequence may comprise the deletion of an amino acid and the substitution of other amino acids. In particular the deletion of the amino acid at 107 of the E. indica EPSPS sequence (FIG. 2) or the same relative amino acid position in other plant EPSP Synthases that can in addition to the deletion, include substitutions of an alanine for a glycine at 102, glycine for alanine at 104, cysteine for methionine at 105, methionine for alanine at 110, or glycine for alanine at 111. Previous random and site-directed mutations in the conserved region (catalytic domain) of bacterial and plant EPSPSs have shown that modifications that increase the Ki for glyphosate while keeping the Km for PEP low are important for an enzyme that is useful for genetically modifying plants for glyphosate tolerance (U.S. Pat. No. 5,866,775).
- E. indica EPSPS regulatory sequences can be isolated by any number of methods known to those of skill in the art for genomic library preparation. For genomic libraries of the present invention, E. indica genomic DNA is isolated by a CsCl purification protocol according to Current Protocols in Molecular Biology, ed. Ausubel et al., Greene Publishing and Wiley-Interscience, New York, 1992 (with periodic updates); or by a CTAB purification method (Rogers et al., Plant Mol. Biol., 5:69, 1988). Reagents are available commercially (see, for example Sigma Chemical Co., St. Louis, Mo.). The genomic DNA libraries are prepared according to manufacturer instructions (Genome Walker™, CloneTech Laboratories, Inc, Palo Alto, Calif.). In separate reactions, genomic DNA is subjected to restriction enzyme digestion overnight at 37° C. with the following blunt-end endonucleases: EcoRV, Sca1, Dra1, PvuII, or Stu1 (CloneTech Laboratories, Inc. Palo Alto, Calif.). The reaction mixtures are extracted with phenol:chloroform, ethanol precipitated, and resuspended in Tris-EDTA buffer (10 mM Tris-.HCI, pH 8.0, 1 mM EDTA). The purified blunt-ended genomic DNA fragments are then ligated to the Genome Walker™ adapters and ligation of the resulting DNA fragments to adapters were done according to the manufacturer's protocol. After ligation, each reaction is heated treated (70° C. for 5 min) to terminate the reaction and then diluted 10-fold in Tris-EDTA buffer. One μl of each respective ligation is then amplified in a 50 μl reaction according to manufacturer's recommended protocol using an adaptor-specific oligonucleotide (supplied by manufacturer) and an E. indica EPSP synthase gene-specific oligonucleotide, such as SEQ ID NO 4. One μl of each primary reaction is diluted 50-fold and 1 μl of this dilution is then amplified in a secondary amplification using a “nested” adaptor-specific oligonucleotide (supplied by manufacturer) and a “nested” gene-specific oligonucleotide such as SEQ ID NO 5. PCR products, representing 5′ regions of the E. indica EPSP synthase gene are then purified by agarose gel electrophoresis using a QIAquick Gel Extraction Kit (cat. #28704, Qiagen Inc., Valencia, Calif.) then directly cloned into the pCR2.1-TOPO vector (cat. #K4500-40, Invitrogen, Carlsbad, Calif.). The identity of the cloned PCR products is confirmed by DNA sequence analysis (ABI Prism™ 377, Perkin Elmer, Foster City, Calif.). The same E. indica Genome Walker™ libraries and methods that are used to isolate the E. indica EPSP synthase 5′ region can be used to isolate the 3′ region of E. indica EPSP synthase gene, by substituting gene-specific primers (first round SEQ ID NO: 13 and second round SEQ ID NO: 14) that anneal to the 3′ end of the gene. Amplification products are cloned and verified as for the 5′ end of the E. indica EPSP synthase gene.
5′-TGCAATCCGGACTGAGCTAACAAAGC-3′ (SEQ ID NO: 13) 5′-ACTGCATTATCACACCGCCCGAGAAG-3′ (SEQ ID NO: 14) - The translation initiation codon is determined for the E. indica EPSP synthase gene by inspection, anticipating an initiation codon approximately 63 codons upstream of the start of the mature protein codon region, based on comparison to the maize EPSP synthase gene. Primers are then designed to amplify approximately 2.5 kb of the 5′ region beginning at the initiation codon. These primers incorporate restriction sites for cloning into expression vectors, for example, placing a EcoR1 site in the 5′ end of promoter region the E. indica EPSP synthase gene and an Nco1 site incorporating the translation start. Such primers are added in a 50 μl RT-PCR reaction at a final concentration of 25 μM with 50 ng of E. indica genomic DNA. PCR amplifications are then performed using the Expand High Fidelity PCR System (cat. #1 732 641, Roche Molecular Biochemicals, Indianapolis, Ind.) per manufacturer's instructions. A thermal profile of 94° C. for 30 seconds, followed by 60° C. for 30seconds, then 72° C. for 3 minutes is used for thirty cycles. This is followed by a cycle of 72° C. for 3 minutes. The gel purified amplification product is then digested with Pst1 and Nco1.
- The 3′ end of the E. indica EPSP synthase gene is amplified using two gene specific primers (SEQ ID NO: 15 and SEQ ID NO: 16) which incorporate a BamH1 site immediately downstream of the translation stop codon and a Pst1 approximately 650 bases downstream of the translation stop codon. The product is amplified as for the 5′ end of the E. indica EPSP synthase gene except an extension time of 1 minute is used. The gel purified product is digested with BamH1 and Pst1.
(SEQ ID NO: 15) 5′-CTAAGGATCCTCTGTGCCTGCCTCATGAAGAGAGTT-3′ (SEQ ID NO: 16) 5′-TGATCTGCAGGCAAGTGTCTTACCCTTACCCTTCTG-3′ - The 5′ (EcoR1/Nco1 fragment) regulatory and 3′ (BamH1/Pst1 fragment) regulatory regions of the E. indica EPSP synthase gene can be ligated to a compatibly digested vector and a coding region to generate a transgene capable of expressing a transcript under control of E. indica EPSP synthase gene regulatory elements. An example of such is a coding region would be the A. fumefaciens strain CP4 EPSP synthase gene (U.S. Pat. No. 5,633,435) expressed under the control of the E. indica regulatory sequences.
- The basal expression of the E. indica EPSP synthase gene promoter may be modified to enhance its expression. Methods known to those of skill in the art can be used to insert enhancing elements (for example, subdomains of the CaMV 35S promoter, Benfey et. al, 1990 EMBO J. 9: 1677-1684) into the E. indica EPSP synthase gene 5′ sequence to generate a promoter which encompasses the temporal and spatial expression of the E. indica EPSP synthase gene but have quantitatively higher levels of expression. Similarly, tissue specific modifications of the E. indica EPSP synthase 5′ region expression can be accomplished with elements determined to specifically activate or repress gene expression (for example, pollen specific elements, Eyal et al., 1995 Plant Cell 7: 373-384).
- From the foregoing, it will be seen that this invention is one well adapted to attain all the end and object herein above set forth together with advantages that are obvious and that are inherent to the invention.
- The embodiments described above are provided to better elucidate the practice of the present invention. Many possible embodiments may be made of the invention without departing from the scope thereof, it should be understood that these embodiments are provided for illustrative purposes only, and are not intended to limit the scope of the invention.
- It will be understood that certain features and subcombinations are of utility and may be employed without reference to other features and subcombinations. This is contemplated by and is within the scope of the claims. All publications and published patent documents cited in this specification are incorporated herein by reference to the same extent as if each individual publication or patent application was specifically and individually indicated to be incorporated by reference.
-
1 16 1 22 DNA Artificial sequence fully synthetic DNA primer 1 tnw sng tng arg cng aya arg t 22 Xaa Xaa Xaa Xaa Xaa Xaa Xaa 1 5 2 23 DNA Artificial sequence fully synthetic DNA primer 2 gcc atn gcc atn ckr tgr tcr tc 23 Ala Xaa Ala Xaa Xaa Xaa Xaa 1 5 3 24 DNA Artificial sequence fully synthetic DNA primer 3 gtg aaa gca gag cat tct gat agc 24 Val Lys Ala Glu His Ser Asp Ser 1 5 4 29 DNA Artificial sequence fully synthetic DNA primer 4 ggc tgc tgt caa tgt cgc att gca gtt cc 29 Gly Cys Cys Gln Cys Arg Ile Ala Val 1 5 5 32 DNA Artificial sequence fully synthetic DNA primer 5 ctc ttt cgc atc ctt ctc aac tgg gaa ctt gc 32 Leu Phe Arg Ile Leu Leu Asn Trp Glu Leu 1 5 10 6 1338 DNA Eleusine indica 6 gcgggcgcgg aggaggtggt gctgcagccc atcaaggaga tctccggcgt cgtgaagctg 60 ccggggtcca agtcgctctc caaccggatc ctcctgctct ccgccctcgc cgagggaaca 120 actgtggtgg ataacctttt aaacagtgag gacgtccact acatgctcgg ggccctgaaa 180 accctcggac tctctgtgga agcggacaaa gctgccaaaa gagcggtagt tgttggctgt 240 ggtggcaagt tcccagttga gaaggatgcg aaagaggagg tgcagctctt cttggggaat 300 gctggaactg caatgcgatc attgacagca gccgtaactg ctgctggagg aaatgcaact 360 tatgtgcttg atggagtgcc aagaatgcgg gagagaccca ttggcgactt ggttgtcgga 420 ttgaaacagc ttggtgcgga tgttgattgt ttccttggca ctgactgccc acctgttcgt 480 gtcaagggaa tcggagggct acctggtggc aaggttaagt tatctggttc catcagcagt 540 cagtacttga gtgccttgct gatggctgct cctttagctc ttggggatgt ggagattgaa 600 atcattgata aactgatctc catcccttat gttgaaatga cattgagatt gatggagcgt 660 tttggcgtga aagcagagca ttctgatagc tgggacagat tctacatcaa gggaggtcaa 720 aaatacaagt cccctaaaaa tgcctacgtg gaaggtgatg cctcaagtgc gagctatttc 780 ttggctggtg ctgcaatcac tggagggact gtgactgttg aaggttgtgg caccaccagt 840 ctgcagggtg atgtgaaatt tgccgaggta ctcgagatga tgggagcgaa ggttacatgg 900 actgaaacta gcgtaactgt taccggtcca caacgtgagc catttgggag gaaacaccta 960 aaagctattg atgttaacat gaacaaaatg cccgatgtcg ccatgactct tgccgtggtt 1020 gccctatttg ctgatggccc aactgctatc agagatgtgg cttcctggag agtaaaggag 1080 accgagagga tggttgcaat ccggactgag ctaacaaagc tgggagcgtc ggtcgaggaa 1140 ggactggact actgcattat cacaccgccc gagaagctga acgtaacggc catcgacacc 1200 tacgatgacc acaggatggc catggccttc tccctcgccg cctgcgccga cgtgcctgtg 1260 accatccggg accccggctg cacccgcaag accttcccag actacttcga cgtgctgagc 1320 actttcgtca agaactaa 1338 7 445 PRT Eleusine indica 7 Ala Gly Ala Glu Glu Val Val Leu Gln Pro Ile Lys Glu Ile Ser Gly 1 5 10 15 Val Val Lys Leu Pro Gly Ser Lys Ser Leu Ser Asn Arg Ile Leu Leu 20 25 30 Leu Ser Ala Leu Ala Glu Gly Thr Thr Val Val Asp Asn Leu Leu Asn 35 40 45 Ser Glu Asp Val His Tyr Met Leu Gly Ala Leu Lys Thr Leu Gly Leu 50 55 60 Ser Val Glu Ala Asp Lys Ala Ala Lys Arg Ala Val Val Val Gly Cys 65 70 75 80 Gly Gly Lys Phe Pro Val Glu Lys Asp Ala Lys Glu Glu Val Gln Leu 85 90 95 Phe Leu Gly Asn Ala Gly Thr Ala Met Arg Ser Leu Thr Ala Ala Val 100 105 110 Thr Ala Ala Gly Gly Asn Ala Thr Tyr Val Leu Asp Gly Val Pro Arg 115 120 125 Met Arg Glu Arg Pro Ile Gly Asp Leu Val Val Gly Leu Lys Gln Leu 130 135 140 Gly Ala Asp Val Asp Cys Phe Leu Gly Thr Asp Cys Pro Pro Val Arg 145 150 155 160 Val Lys Gly Ile Gly Gly Leu Pro Gly Gly Lys Val Lys Leu Ser Gly 165 170 175 Ser Ile Ser Ser Gln Tyr Leu Ser Ala Leu Leu Met Ala Ala Pro Leu 180 185 190 Ala Leu Gly Asp Val Glu Ile Glu Ile Ile Asp Lys Leu Ile Ser Ile 195 200 205 Pro Tyr Val Glu Met Thr Leu Arg Leu Met Glu Arg Phe Gly Val Lys 210 215 220 Ala Glu His Ser Asp Ser Trp Asp Arg Phe Tyr Ile Lys Gly Gly Gln 225 230 235 240 Lys Tyr Lys Ser Pro Lys Asn Ala Tyr Val Glu Gly Asp Ala Ser Ser 245 250 255 Ala Ser Tyr Phe Leu Ala Gly Ala Ala Ile Thr Gly Gly Thr Val Thr 260 265 270 Val Glu Gly Cys Gly Thr Thr Ser Leu Gln Gly Asp Val Lys Phe Ala 275 280 285 Glu Val Leu Glu Met Met Gly Ala Lys Val Thr Trp Thr Glu Thr Ser 290 295 300 Val Thr Val Thr Gly Pro Gln Arg Glu Pro Phe Gly Arg Lys His Leu 305 310 315 320 Lys Ala Ile Asp Val Asn Met Asn Lys Met Pro Asp Val Ala Met Thr 325 330 335 Leu Ala Val Val Ala Leu Phe Ala Asp Gly Pro Thr Ala Ile Arg Asp 340 345 350 Val Ala Ser Trp Arg Val Lys Glu Thr Glu Arg Met Val Ala Ile Arg 355 360 365 Thr Glu Leu Thr Lys Leu Gly Ala Ser Val Glu Glu Gly Leu Asp Tyr 370 375 380 Cys Ile Ile Thr Pro Pro Glu Lys Leu Asn Val Thr Ala Ile Asp Thr 385 390 395 400 Tyr Asp Asp His Arg Met Ala Met Ala Phe Ser Leu Ala Ala Cys Ala 405 410 415 Asp Val Pro Val Thr Ile Arg Asp Pro Gly Cys Thr Arg Lys Thr Phe 420 425 430 Pro Asp Tyr Phe Asp Val Leu Ser Thr Phe Val Lys Asn 435 440 445 8 56 DNA Artificial sequence fully synthetic DNA leader sequence 8 agatctccta gggcttaatt aattaagcac tagtcacaca ggaggtaatt catatg 56 9 36 DNA Artificial sequence fully synthetic DNA sequence 9 gca att cca tat ggc ggg cgc gga gga ggt ggt gct 36 Ala Ile Pro Tyr Gly Gly Arg Gly Gly Gly Gly Ala 1 5 10 10 48 DNA Artificial sequence fully synthetic DNA sequence 10 gac tag gaa ttc tta gtt ctt ttg acg aaa gtg ctc agc acg tcg aag 48 Asp Glu Phe Leu Val Leu Leu Thr Lys Val Leu Ser Thr Ser Lys 1 5 10 15 11 35 DNA Artificial sequence Fully synthetic DNA sequence 11 gca att cgc atg ccg ggc gcg gag gag gtg gtg ct 35 Ala Ile Arg Met Pro Gly Ala Glu Glu Val Val 1 5 10 12 48 DNA Artificial sequence Fully synthetic DNA primer 12 gac tag gaa ttc tta gtt ctt ttg acg aaa gtg ctc agc acg tcg aag 48 Asp Glu Phe Leu Val Leu Leu Thr Lys Val Leu Ser Thr Ser Lys 1 5 10 15 13 26 DNA Artificial sequence fully synthetic DNA sequence 13 tgc aat ccg gac tga gct aac aaa gc 26 Cys Asn Pro Asp Ala Asn Lys 1 5 14 26 DNA Artificial sequence fully synthetic DNA sequence 14 act gca tta tca cac cgc ccg aga ag 26 Thr Ala Leu Ser His Arg Pro Arg 1 5 15 36 DNA Artificial sequence fully synthetic DNA sequence 15 cta agg atc ctc tgt gcc tgc ctc atg aag aga gtt 36 Leu Arg Ile Leu Cys Ala Cys Leu Met Lys Arg Val 1 5 10 16 36 DNA Artificial sequence fully synthetic DNA sequence 16 tga tct gca ggc aag tgt ctt acc ctt acc ctt ctg 36 Ser Ala Gly Lys Cys Leu Thr Leu Thr Leu Leu 1 5 10
Claims (26)
1. A DNA molecule that encodes a naturally occurring glyphosate resistant EPSPS enzyme derived from a glyphosate tolerant plant, wherein the glyphosate resistant EPSPS enzyme has a Km for phosphoenolpyruvate (PEP) of less than 10 μM.
2. A DNA molecule of claim 1 that encodes a naturally occurring glyphosate resistant EPSPS enzyme derived from a glyphosate tolerant plant, wherein the glyphosate resistant EPSPS enzyme has a Km for PEP of less than 10 μM and the Km for PEP is not more than about twice of the Km for PEP of a naturally occurring glyphosate sensitive EPSPS enzyme derived from a glyphosate sensitive plant.
3. A DNA molecule of claim 2 , wherein said plant is Eleusine species.
4. A DNA molecule of claim 1 , wherein said naturally occurring glyphosate resistant EPSPS enzyme is modified by a substitution or a deletion of at least one amino acid in a catalytic domain.
5. A DNA molecule of claim 4 , wherein said substitution is selected from the group consisting of glycine to alanine 102 and threonine to isoleucine 103 of SEQ ID NO: 7.
6. A DNA molecule that encodes a naturally occurring glyphosate resistant EPSPS enzyme of SEQ ID NO: 7.
7. A DNA molecule of claim 6 that encodes a naturally occurring glyphosate resistant EPSPS enzyme of SEQ ID NO: 7, wherein the DNA molecule is substantially homologous to SEQ ID NO: 6.
8. A recombinant DNA molecule comprising: a promoter that functions in plant cells to cause the production of an RNA sequence, operably linked to; a structural DNA sequence that causes the production of an RNA sequence that encodes an EPSPS enzyme comprising the sequence of SEQ ID NO: 7, operably linked to; a 3′ non-translated region that functions in plant cells to cause the addition of polyadenyl nucleotides to the 3′end of the RNA sequence, wherein the promoter is heterologous with respect to the structural DNA sequence and selected so as to cause sufficient expression of the polypeptide to enhance the glyphosate tolerance of a transgenic plant cell containing said recombinant DNA molecule.
9. A recombinant DNA molecule of claim 8 , wherein said structural DNA sequence encodes a fusion polypeptide comprising an amino-terminal chloroplast transit peptide and an EPSPS enzyme comprising the sequence of SEQ ID NO: 7.
10. A method of producing glyphosate tolerant plants comprising the steps of:
a) inserting into the genome of a plant cell a recombinant DNA molecule comprising: a promoter that functions in plant cells to cause the production of a RNA sequence, operably linked to; a structural DNA sequence that caused the production of a RNA sequence that encodes an EPSPS enzyme having the sequence of SEQ ID NO: 7, operably linked to; a 3′ non-translated region that functions in plant cells to cause the addition of polyadenyl nucleotides the 3′end of the RNA sequence; where the promoter is heterologous with respect to the structural DNA sequence and adapted to cause sufficient expression of the polypeptide to enhance the glyphosate tolerance of a plant cell transformed with the DNA molecule;
b) obtaining a transformed plant cell; and
c) regenerating from the transformed plant cell a genetically transformed plant which has increased tolerance to glyphosate herbicide.
11. A method of claim 10 , wherein the structural DNA sequence encodes a fusion polypeptide comprising an amino-terminal chloroplast transit peptide and an EPSPS enzyme comprising the sequence of SEQ ID NO: 7.
12. A glyphosate tolerant plant cell comprising a recombinant DNA molecule of claim 8 or 9.
13. A glyphosate tolerant plant cell of claim 12 selected from the group consisting of corn, wheat, rice, millet, sugarcane, barley, oat, rye, turf grasses, asparagus, soybean, cotton, sugar beet, oilseed rape, canola, flax, sunflower, potato, tobacco, tomato, alfalfa, forest trees, fruit trees, ornamental annuals, and ornamental perennials.
14. A glyphosate tolerant plant comprising the plant cells of claim 12 .
15. A glyphosate tolerant plant of claim 14 selected from the group consisting of corn, wheat, rice, millet, sugarcane, barley, oat, rye, turf grasses, asparagus, soybean, cotton, sugar beet, oilseed rape, canola, flax, sunflower, potato, tobacco, tomato, alfalfa, forest trees, fruit trees, ornamental annuals, and ornamental perennials.
16. A recombinant DNA molecule comprising: a promoter that functions in plant cells to cause the production of an RNA sequence, operably linked to; a structural DNA sequence that causes the production of an RNA sequence which encodes an EPSPS enzyme having the sequence of SEQ ID NO: 7, operably linked to; a 3′ non-translated region that functions in plant cells to cause the addition of polyadenyl nucleotides the 3′ end of the RNA sequence, wherein the promoter is homologous with respect to the structural DNA sequence.
17. A DNA molecule of claim 16 wherein the structural DNA sequence encodes a fusion polypeptide comprising an amino-terminal chloroplast transit peptide and the EPSPS enzyme comprising the sequence of SEQ ID NO: 7.
18. A glyphosate tolerant transgenic plant cell comprising a DNA molecule of claim 16 or 17.
19. A glyphosate tolerant transgenic plant cell of claim 18 selected from the group consisting of corn, wheat, rice, millet, sugarcane, barley, oat, rye, turf grasses, asparagus, soybean, cotton, sugar beet, oilseed rape, canola, flax, sunflower, potato, tobacco, tomato, alfalfa, forest trees, fruit trees, ornamental annuals, ornamental perennials.
20. A glyphosate tolerant transgenic plant comprising plant cells of claim 18 .
21. Glyphosate tolerant transgenic plant of claim 20 selected from the group consisting of corn, wheat, rice, millet, sugarcane, barley, oat, rye, turf grasses, asparagus, soybean, cotton, sugar beet, oilseed rape, canola, flax, sunflower, potato, tobacco, tomato, alfalfa, forest trees, fruit trees, ornamental annuals, ornamental perennials.
22. The seed of a glyphosate tolerant transgenic plant of claim 15 .
23. The seed of a glyphosate tolerant transgenic plant of claim 21 .
24. A DNA molecule comprising the promoter region located 5′ to the DNA molecule of claim 3 .
25. A DNA molecule comprising the chloroplast transit peptide coding region located 5′ to the DNA molecule of claim 3 .
26. A DNA molecule comprising the 3′ untranslated region located 3′ to the DNA molecule of claim 3 .
Priority Applications (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US10/413,909 US20030192072A1 (en) | 2000-03-09 | 2003-04-15 | Methods for making plants tolerant to glyphosate and compositions thereof |
Applications Claiming Priority (3)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US18809300P | 2000-03-09 | 2000-03-09 | |
| US09/800,130 US6803501B2 (en) | 2000-03-09 | 2001-03-06 | Methods for making plants tolerant to glyphosate and compositions thereof using a DNA encoding an EPSPS enzyme from Eleusine indica |
| US10/413,909 US20030192072A1 (en) | 2000-03-09 | 2003-04-15 | Methods for making plants tolerant to glyphosate and compositions thereof |
Related Parent Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| US09/800,130 Continuation US6803501B2 (en) | 2000-03-09 | 2001-03-06 | Methods for making plants tolerant to glyphosate and compositions thereof using a DNA encoding an EPSPS enzyme from Eleusine indica |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| US20030192072A1 true US20030192072A1 (en) | 2003-10-09 |
Family
ID=22691747
Family Applications (3)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| US09/800,130 Expired - Fee Related US6803501B2 (en) | 2000-03-09 | 2001-03-06 | Methods for making plants tolerant to glyphosate and compositions thereof using a DNA encoding an EPSPS enzyme from Eleusine indica |
| US10/413,909 Abandoned US20030192072A1 (en) | 2000-03-09 | 2003-04-15 | Methods for making plants tolerant to glyphosate and compositions thereof |
| US10/803,156 Abandoned US20040148650A1 (en) | 2000-03-09 | 2004-03-17 | Methods for making plants tolerant to glyphosate and compositions thereof |
Family Applications Before (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| US09/800,130 Expired - Fee Related US6803501B2 (en) | 2000-03-09 | 2001-03-06 | Methods for making plants tolerant to glyphosate and compositions thereof using a DNA encoding an EPSPS enzyme from Eleusine indica |
Family Applications After (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| US10/803,156 Abandoned US20040148650A1 (en) | 2000-03-09 | 2004-03-17 | Methods for making plants tolerant to glyphosate and compositions thereof |
Country Status (10)
| Country | Link |
|---|---|
| US (3) | US6803501B2 (en) |
| EP (1) | EP1261695B1 (en) |
| AR (2) | AR028243A1 (en) |
| AT (1) | ATE298364T1 (en) |
| AU (2) | AU4200501A (en) |
| BR (1) | BR0109118A (en) |
| CA (1) | CA2401093A1 (en) |
| DE (1) | DE60111613T2 (en) |
| WO (1) | WO2001066704A2 (en) |
| ZA (1) | ZA200206788B (en) |
Cited By (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20070180574A1 (en) * | 2006-01-23 | 2007-08-02 | Donald Penner | Methods for breeding glyphosate resistant plants and compositions thereof |
| US20100199363A1 (en) * | 2006-05-12 | 2010-08-05 | Hartley Carol J | Enzymes for degrading herbicides |
Families Citing this family (396)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US7462481B2 (en) | 2000-10-30 | 2008-12-09 | Verdia, Inc. | Glyphosate N-acetyltransferase (GAT) genes |
| FR2848570B1 (en) | 2002-12-12 | 2005-04-01 | Bayer Cropscience Sa | EXPRESSION CASSETTE ENCODING A 5-ENOL PYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS) AND HERBICIDE TOLERANT PLANTS CONTAINING THE SAME |
| EP1594961B1 (en) * | 2003-02-18 | 2013-12-25 | Monsanto Technology LLC | Glyphosate resistant class i 5-enolpyruvylshikimate-3-phosphate synthase (epsps) |
| CA2521284C (en) | 2003-04-29 | 2014-07-08 | Pioneer Hi-Bred International, Inc. | Novel glyphosate-n-acetyltransferase (gat) genes |
| WO2005068642A2 (en) | 2003-10-01 | 2005-07-28 | Board Of Trustees Operating Michigan State University | Bacterial synthesis of 1,2,4-butanetriol enantiomers |
| EP2868749B1 (en) | 2004-01-20 | 2017-10-04 | Monsanto Technology LLC | Chimeric promoters for use in plants |
| EP2557166B1 (en) * | 2004-04-06 | 2016-03-23 | Fibria Celulose S/A | Cambium/Xylem-preferred promoters and uses thereof |
| US7405074B2 (en) | 2004-04-29 | 2008-07-29 | Pioneer Hi-Bred International, Inc. | Glyphosate-N-acetyltransferase (GAT) genes |
| NZ568867A (en) | 2005-08-24 | 2010-12-24 | Pioneer Hi Bred Int | Compositions providing tolerance to multiple herbicides and methods of use thereof |
| EP2843053A1 (en) * | 2006-02-17 | 2015-03-04 | Monsanto Technology LLC | Chimeric regulatory sequences comprising introns for plant gene expression |
| EP1996009A4 (en) | 2006-03-02 | 2009-09-30 | Athenix Corp | Methods and compositions for improved enzyme activity in transgenic plant |
| US20110165641A1 (en) * | 2006-03-31 | 2011-07-07 | The Board Of Trustees Of Michigan State University | Synthesis of 1,2,4-Butanetriol Enantiomers from Carbohydrates |
| CA2653231C (en) | 2006-05-12 | 2016-04-12 | Monsanto Technology Llc | Methods and compositions for obtaining marker-free transgenic plants |
| US9045765B2 (en) * | 2006-06-09 | 2015-06-02 | Athenix Corporation | EPSP synthase genes conferring herbicide resistance |
| AU2007257704A1 (en) | 2006-06-13 | 2007-12-21 | Athenix Corporation | Improved EPSP synthases: compositions and methods of use |
| US7951995B2 (en) | 2006-06-28 | 2011-05-31 | Pioneer Hi-Bred International, Inc. | Soybean event 3560.4.3.5 and compositions and methods for the identification and detection thereof |
| US20110076730A1 (en) * | 2006-07-19 | 2011-03-31 | Board Of Trustees Of Michigan State University | Microbial synthesis of d-1,2,4-butanetriol |
| WO2008070179A2 (en) | 2006-12-06 | 2008-06-12 | Monsanto Technology, Llc | Genes and uses for plant improvement |
| CL2007003743A1 (en) | 2006-12-22 | 2008-07-11 | Bayer Cropscience Ag | COMPOSITION THAT INCLUDES FENAMIDONA AND AN INSECTICIDE COMPOUND; AND METHOD TO CONTROL FITOPATOGENOS CULTURES AND INSECTS FACING OR PREVENTIVELY. |
| CL2007003744A1 (en) | 2006-12-22 | 2008-07-11 | Bayer Cropscience Ag | COMPOSITION THAT INCLUDES A 2-PYRIDILMETILBENZAMIDE DERIVATIVE AND AN INSECTICIDE COMPOUND; AND METHOD TO CONTROL FITOPATOGENOS CULTURES AND INSECTS FACING OR PREVENTIVELY. |
| US7838729B2 (en) | 2007-02-26 | 2010-11-23 | Monsanto Technology Llc | Chloroplast transit peptides for efficient targeting of DMO and uses thereof |
| EP1969929A1 (en) | 2007-03-12 | 2008-09-17 | Bayer CropScience AG | Substituted phenylamidines and their use as fungicides |
| JP2010520899A (en) | 2007-03-12 | 2010-06-17 | バイエル・クロツプサイエンス・アクチエンゲゼルシヤフト | Dihalophenoxyphenylamidine and its use as a fungicide |
| BRPI0808798A2 (en) | 2007-03-12 | 2014-10-07 | Bayer Cropscience Ag | 3,5-DISSUBSTITUTED PHENOXYPHENYLAMIDINS AND THEIR USE AS FUNGICIDES |
| EP1969934A1 (en) | 2007-03-12 | 2008-09-17 | Bayer CropScience AG | 4-cycloalkyl or 4-aryl substituted phenoxy phenylamidines and their use as fungicides |
| BRPI0810654B1 (en) | 2007-04-19 | 2016-10-04 | Bayer Cropscience Ag | thiadiazolyloxyphenylamidines, their use and their method of preparation, composition and method for combating unwanted microorganisms, seed resistant to unwanted microorganism, as well as method for protecting said seed against microorganisms |
| DE102007045955A1 (en) | 2007-09-26 | 2009-04-09 | Bayer Cropscience Ag | Active agent combination, useful e.g. for combating animal pests and treating seeds of transgenic plants, comprises substituted amino-furan-2-one compound and at least one compound e.g. diazinon, isoxathion, carbofuran or aldicarb |
| DE102007045953B4 (en) | 2007-09-26 | 2018-07-05 | Bayer Intellectual Property Gmbh | Drug combinations with insecticidal and acaricidal properties |
| DE102007045956A1 (en) | 2007-09-26 | 2009-04-09 | Bayer Cropscience Ag | Combination of active ingredients with insecticidal and acaricidal properties |
| DE102007045919B4 (en) | 2007-09-26 | 2018-07-05 | Bayer Intellectual Property Gmbh | Drug combinations with insecticidal and acaricidal properties |
| DE102007045920B4 (en) | 2007-09-26 | 2018-07-05 | Bayer Intellectual Property Gmbh | Synergistic drug combinations |
| DE102007045922A1 (en) | 2007-09-26 | 2009-04-02 | Bayer Cropscience Ag | Drug combinations with insecticidal and acaricidal properties |
| DE102007045957A1 (en) | 2007-09-26 | 2009-04-09 | Bayer Cropscience Ag | Active agent combination, useful e.g. for combating animal pests e.g. insects and treating seeds of transgenic plants, comprises substituted amino-furan-2-one compound and at least one compound e.g. benzoyl urea, buprofezin and cyromazine |
| PT2205749T (en) | 2007-09-27 | 2016-07-20 | Dow Agrosciences Llc | Engineered zinc finger proteins targeting 5-enolpyruvyl shikimate-3-phosphate synthase genes |
| EP2090168A1 (en) | 2008-02-12 | 2009-08-19 | Bayer CropScience AG | Method for improving plant growth |
| EP2072506A1 (en) | 2007-12-21 | 2009-06-24 | Bayer CropScience AG | Thiazolyloxyphenylamidine or thiadiazolyloxyphenylamidine und its use as fungicide |
| CN101932701A (en) * | 2008-02-01 | 2010-12-29 | 阿森尼克斯公司 | Directed evolution of GRG31 and GRG36 EPSP synthases |
| ES2562908T3 (en) | 2008-04-07 | 2016-03-09 | Monsanto Technology Llc | Plant regulatory elements and their uses |
| US7947877B2 (en) | 2008-05-14 | 2011-05-24 | Monosanto Technology LLC | Plants and seeds of spring canola variety SCV328921 |
| US7935870B2 (en) | 2008-05-14 | 2011-05-03 | Monsanto Technology Llc | Plants and seeds of spring canola variety SCV354718 |
| US7964774B2 (en) | 2008-05-14 | 2011-06-21 | Monsanto Technology Llc | Plants and seeds of spring canola variety SCV384196 |
| US8829282B2 (en) | 2008-05-14 | 2014-09-09 | Monsanto Technology, Llc | Plants and seeds of spring canola variety SCV425044 |
| EP2168434A1 (en) | 2008-08-02 | 2010-03-31 | Bayer CropScience AG | Use of azols to increase resistance of plants of parts of plants to abiotic stress |
| US9371564B2 (en) | 2008-08-08 | 2016-06-21 | Bayer Bioscience N.V. | Methods for plant fiber characterization and identification |
| US20110190365A1 (en) | 2008-08-14 | 2011-08-04 | Bayer Crop Science Ag | Insecticidal 4-phenyl-1H-pyrazoles |
| DE102008041695A1 (en) | 2008-08-29 | 2010-03-04 | Bayer Cropscience Ag | Methods for improving plant growth |
| EP2733212B1 (en) | 2008-09-26 | 2018-09-05 | BASF Agrochemical Products, B.V. | Herbicide-resistant ahas-mutants and methods of use |
| EP2201838A1 (en) | 2008-12-05 | 2010-06-30 | Bayer CropScience AG | Active ingredient-beneficial organism combinations with insecticide and acaricide properties |
| EP2198709A1 (en) | 2008-12-19 | 2010-06-23 | Bayer CropScience AG | Method for treating resistant animal pests |
| EP2204094A1 (en) | 2008-12-29 | 2010-07-07 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants Introduction |
| EP2223602A1 (en) | 2009-02-23 | 2010-09-01 | Bayer CropScience AG | Method for improved utilisation of the production potential of genetically modified plants |
| AU2009335333B2 (en) | 2008-12-29 | 2015-04-09 | Bayer Intellectual Property Gmbh | Method for improved use of the production potential of genetically modified plants |
| EP2039772A2 (en) | 2009-01-06 | 2009-03-25 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants introduction |
| EP2039770A2 (en) | 2009-01-06 | 2009-03-25 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants |
| EP2039771A2 (en) | 2009-01-06 | 2009-03-25 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants |
| AR074941A1 (en) | 2009-01-07 | 2011-02-23 | Bayer Cropscience Sa | TRANSPLASTOMIC PLANTS EXEMPTED FROM SELECTOR MARKER |
| CN102355820B (en) | 2009-01-19 | 2013-10-16 | 拜尔农作物科学股份公司 | Cyclic diones and their use as insecticides, acaricides and/or fungicides |
| EP2227951A1 (en) | 2009-01-23 | 2010-09-15 | Bayer CropScience AG | Application of enaminocarbonyl compounds for combating viruses transmitted by insects |
| EP2100506A2 (en) | 2009-01-23 | 2009-09-16 | Bayer CropScience AG | Uses of fluopyram |
| PT2391608E (en) | 2009-01-28 | 2013-05-13 | Bayer Cropscience Ag | Fungicide n-cycloalkyl-n-bicyclicmethylene-carboxamide derivatives |
| AR075126A1 (en) | 2009-01-29 | 2011-03-09 | Bayer Cropscience Ag | METHOD FOR THE BEST USE OF THE TRANSGENIC PLANTS PRODUCTION POTENTIAL |
| EP2218717A1 (en) | 2009-02-17 | 2010-08-18 | Bayer CropScience AG | Fungicidal N-((HET)Arylethyl)thiocarboxamide derivatives |
| BRPI1006006B1 (en) | 2009-02-17 | 2018-05-22 | Bayer Intellectual Property Gmbh | COMPOUNDS, FUNGICIDE COMPOSITION AND METHOD FOR THE CONTROL OF PHYTOPATHOGENIC CROPS FUNGI |
| TW201031331A (en) | 2009-02-19 | 2010-09-01 | Bayer Cropscience Ag | Pesticide composition comprising a tetrazolyloxime derivative and a fungicide or an insecticide active substance |
| US20120277117A1 (en) | 2009-02-27 | 2012-11-01 | Adel Zayed | Hydroponic apparatus and methods of use |
| DE102009001469A1 (en) | 2009-03-11 | 2009-09-24 | Bayer Cropscience Ag | Improving utilization of productive potential of transgenic plant by controlling e.g. animal pest, and/or by improving plant health, comprises treating the transgenic plant with active agent composition comprising prothioconazole |
| DE102009001681A1 (en) | 2009-03-20 | 2010-09-23 | Bayer Cropscience Ag | Improving utilization of production potential of a transgenic plant by controlling animal pests, phytopathogenic fungi, microorganisms and/or improving plant health, comprises treating plant with a drug composition comprising iprovalicarb |
| DE102009001730A1 (en) | 2009-03-23 | 2010-09-30 | Bayer Cropscience Ag | Improving utilization of production potential of a transgenic plant by controlling animal pests, phytopathogenic fungi and/or microorganisms and/or the plant health, comprises treating plant with a drug composition comprising spiroxamine |
| DE102009001732A1 (en) | 2009-03-23 | 2010-09-30 | Bayer Cropscience Ag | Improving the production potential of transgenic plant, by combating e.g. animal pests and/or microorganism, and/or increasing plant health, comprises treating the plants with active agent composition comprising trifloxystrobin |
| DE102009001728A1 (en) | 2009-03-23 | 2010-09-30 | Bayer Cropscience Ag | Improving the production potential of transgenic plant, by combating e.g. animal pests and/or microorganism, and/or increasing plant health, comprises treating the plants with active agent composition comprising fluoxastrobin |
| WO2010108505A1 (en) | 2009-03-25 | 2010-09-30 | Bayer Cropscience Ag | Active ingredient combinations having insecticidal and acaricidal properties |
| WO2010108506A1 (en) | 2009-03-25 | 2010-09-30 | Bayer Cropscience Ag | Active ingredient combinations having insecticidal and acaricidal properties |
| AU2009342807B2 (en) | 2009-03-25 | 2015-04-02 | Bayer Cropscience Aktiengesellschaft | Synergistic combinations of active ingredients |
| BRPI0924451B1 (en) | 2009-03-25 | 2017-12-26 | Bayer Intellectual Property Gmbh | Combinations of active substances and their uses, as well as methods for the control of animal pests and method for the manufacture of insecticides and acaricides |
| AP3073A (en) | 2009-03-25 | 2014-12-31 | Bayer Cropscience Ag | Active ingredient combinations with insecticidal and acaricidal properties |
| EP2232995A1 (en) | 2009-03-25 | 2010-09-29 | Bayer CropScience AG | Method for improved utilisation of the production potential of transgenic plants |
| EP2239331A1 (en) | 2009-04-07 | 2010-10-13 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants |
| EP2245936A1 (en) | 2009-04-27 | 2010-11-03 | Bayer CropScience AG | Use of 4-aza indole derivatives for the reduction of mycotoxin contamination |
| WO2010127797A2 (en) | 2009-05-06 | 2010-11-11 | Bayer Cropscience Ag | Cyclopentanedione compounds and their use as insecticides, acaricides and/or fungicides |
| EP2251331A1 (en) | 2009-05-15 | 2010-11-17 | Bayer CropScience AG | Fungicide pyrazole carboxamides derivatives |
| AR076839A1 (en) | 2009-05-15 | 2011-07-13 | Bayer Cropscience Ag | FUNGICIDE DERIVATIVES OF PIRAZOL CARBOXAMIDAS |
| US8471100B2 (en) | 2009-05-15 | 2013-06-25 | University Of Tennessee Research Foundation | Environmental stress-inducible promoter and its application in crops |
| EP2255626A1 (en) | 2009-05-27 | 2010-12-01 | Bayer CropScience AG | Use of succinate dehydrogenase inhibitors to increase resistance of plants or parts of plants to abiotic stress |
| UA106618C2 (en) | 2009-06-02 | 2014-09-25 | Баєр Кропсаєнс Аг | APPLICATION OF SUCCINDEGYDROGENASE INHIBITORS FOR SCLEROTINIA SUBSCRIPTION CONTROL |
| MX2011013224A (en) | 2009-06-09 | 2012-06-01 | Pioneer Hi Bred Int | Early endosperm promoter and methods of use. |
| US8071848B2 (en) | 2009-06-17 | 2011-12-06 | Monsanto Technology Llc | Plants and seeds of spring canola variety SCV218328 |
| JP2012531216A (en) | 2009-07-01 | 2012-12-10 | バイエル・クロップサイエンス・エヌ・ヴェー | Methods and means for obtaining plants with enhanced tolerance to glyphosate |
| EP2453750A2 (en) | 2009-07-16 | 2012-05-23 | Bayer CropScience AG | Synergistic active substance combinations containing phenyl triazoles |
| WO2011015524A2 (en) | 2009-08-03 | 2011-02-10 | Bayer Cropscience Ag | Fungicide heterocycles derivatives |
| MX374069B (en) | 2009-08-04 | 2020-07-30 | Evogene Ltd | ISOLATED POLYNUCLEOTIDES AND POLYPEPTIDES, AND METHODS OF USING THEM TO IMPROVE ABIOTIC STRESS TOLERANCE, YIELD, GROWTH RATE, VIGOR, BIOMASS, OIL CONTENT AND/OR NITROGEN USE EFFICIENCY OF PLANTS. |
| EP2292094A1 (en) | 2009-09-02 | 2011-03-09 | Bayer CropScience AG | Active compound combinations |
| WO2011050271A1 (en) | 2009-10-23 | 2011-04-28 | Monsanto Technology Llc | Methods and compositions for expression of transgenes in plants |
| MX2012004819A (en) | 2009-10-26 | 2012-06-25 | Pioneer Hi Bred Int | Somatic ovule specific promoter and methods of use. |
| EP2343280A1 (en) | 2009-12-10 | 2011-07-13 | Bayer CropScience AG | Fungicide quinoline derivatives |
| EP2338890A1 (en) | 2009-12-22 | 2011-06-29 | Bayer CropScience AG | 4,7-Diazaindole derivatives and their use as fungicides |
| US20130012546A1 (en) | 2009-12-28 | 2013-01-10 | Christian Beier | Fungicide hydroximoyl-tetrazole derivatives |
| CN105399666A (en) | 2009-12-28 | 2016-03-16 | 拜尔农科股份公司 | Fungicide hydroxyimino-heterocyclic derivatives |
| MX2012007539A (en) | 2009-12-28 | 2012-07-23 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives. |
| CA3017321C (en) | 2010-01-14 | 2021-07-20 | Monsanto Technology Llc | Plant regulatory elements and uses thereof |
| EP2525658B1 (en) | 2010-01-22 | 2017-03-01 | Bayer Intellectual Property GmbH | Acaricides and/or insecticidal agent combinations |
| AR080022A1 (en) | 2010-01-26 | 2012-03-07 | Pioneer Hi Bred Int | SEQUENCES OF POLINUCLEOTIDES AND POLYPEPTIDES ASSOCIATED WITH THE TOLERANCE TO HERBICIDES |
| US8148611B2 (en) | 2010-02-26 | 2012-04-03 | Monsanto Technology Llc | Plants and seeds of spring canola variety SCV453784 |
| US8143488B2 (en) | 2010-02-26 | 2012-03-27 | Monsanto Technoloy LLC | Plants and seeds of spring canola variety SCV470336 |
| US8138394B2 (en) | 2010-02-26 | 2012-03-20 | Monsanto Technology Llc | Plants and seeds of spring canola variety SCV431158 |
| AR080443A1 (en) | 2010-03-04 | 2012-04-11 | Bayer Cropscience Ag | 2-AMIDOBENCIMIDAZOLES REPLACED WITH FLURUOALQUILO |
| US8581048B2 (en) | 2010-03-09 | 2013-11-12 | Monsanto Technology, Llc | Plants and seeds of spring canola variety SCV119103 |
| US8153865B2 (en) | 2010-03-11 | 2012-04-10 | Monsanto Technology Llc | Plants and seeds of spring canola variety SCV152154 |
| BR112012023551A2 (en) | 2010-03-18 | 2015-09-15 | Bayer Ip Gmbh | aryl and hetaryl sulfonamides as active agents against abiotic stress in plants |
| JP2013523795A (en) | 2010-04-06 | 2013-06-17 | バイエル・インテレクチユアル・プロパテイー・ゲー・エム・ベー・ハー | Use of 4-phenylbutyric acid and / or salt thereof to enhance stress tolerance of plants |
| AU2011237909B2 (en) | 2010-04-09 | 2015-08-20 | Bayer Intellectual Property Gmbh | Use of derivatives of the (1-cyanocyclopropyl)phenylphosphinic acid, the esters thereof and/or the salts thereof for enhancing the tolerance of plants to abiotic stress |
| JP2013525401A (en) | 2010-04-28 | 2013-06-20 | バイエル・クロップサイエンス・アーゲー | Fungicide hydroxymoyl-heterocyclic derivative |
| BR112012027558A2 (en) | 2010-04-28 | 2015-09-15 | Bayer Cropscience Ag | '' Compound of formula (I), fungicidal composition and method for the control of crop phytogenic fungi '' |
| WO2011134911A2 (en) | 2010-04-28 | 2011-11-03 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| UA110703C2 (en) | 2010-06-03 | 2016-02-10 | Байєр Кропсайнс Аг | Fungicidal n-[(trisubstitutedsilyl)methyl]carboxamide |
| PL2576517T3 (en) | 2010-06-03 | 2015-06-30 | Bayer Ip Gmbh | N-[(het)arylalkyl)]pyrazole (thio)carboxamides and their heterosubstituted analogues |
| ES2533026T3 (en) | 2010-06-03 | 2015-04-07 | Bayer Intellectual Property Gmbh | N - [(het) arylalkyl)] pyrazole (thio) carboxamides and their hetero substituted analogs |
| US8653114B2 (en) | 2010-06-03 | 2014-02-18 | Bayer Intellectual Property Gmbh | O-cyclopropylcyclohexyl-carboxanilides and their use as fungicides |
| CN103119169B (en) | 2010-06-09 | 2018-11-20 | 拜尔作物科学公司 | Methods and tools for modifying plant genomes in nucleotide sequences commonly used in plant genome engineering |
| WO2011154158A1 (en) | 2010-06-09 | 2011-12-15 | Bayer Bioscience N.V. | Methods and means to modify a plant genome at a nucleotide sequence commonly used in plant genome engineering |
| JP2013532648A (en) | 2010-07-20 | 2013-08-19 | バイエル・クロップサイエンス・アーゲー | Benzocycloalkenes as antibacterial agents |
| US9139842B2 (en) | 2010-08-13 | 2015-09-22 | Pioneer Hi-Bred International, Inc. | Methods and compositions for targeting sequences of interest to the chloroplast |
| PT2611925T (en) | 2010-08-30 | 2018-03-02 | Dow Agrosciences Llc | Sugarcane bacilliform viral (scbv) enhancer and its use in plant functional genomics |
| UA109669C2 (en) | 2010-09-03 | 2015-09-25 | Байєр Інтеллектуал Проперті Гмбх | DITIINETRA (THIO) CARBOXYMIDES FOR THE FIGHT AGAINST PHYTOPATHOGENIC MUSHROOMS |
| CN103228141B (en) | 2010-09-03 | 2016-04-20 | 拜耳知识产权有限责任公司 | Substituted fused pyrimidinones and dihydropyrimidinones |
| JP2012062267A (en) | 2010-09-15 | 2012-03-29 | Bayer Cropscience Ag | Pesticidal pyrroline n-oxide derivative |
| JP2012082186A (en) | 2010-09-15 | 2012-04-26 | Bayer Cropscience Ag | Insecticidal arylpyrrolidines |
| EP2460406A1 (en) | 2010-12-01 | 2012-06-06 | Bayer CropScience AG | Use of fluopyram for controlling nematodes in nematode resistant crops |
| WO2012038480A2 (en) | 2010-09-22 | 2012-03-29 | Bayer Cropscience Ag | Use of biological or chemical control agents for controlling insects and nematodes in resistant crops |
| PL2624699T3 (en) | 2010-10-07 | 2019-05-31 | Bayer Cropscience Ag | Fungicide composition comprising a tetrazolyloxime derivative and a thiazolylpiperidine derivative |
| JP2013541554A (en) | 2010-10-21 | 2013-11-14 | バイエル・インテレクチユアル・プロパテイー・ゲー・エム・ベー・ハー | N-benzyl heterocyclic carboxamides |
| AR083501A1 (en) | 2010-10-21 | 2013-02-27 | Bayer Cropscience Ag | 1- (CARBON HETEROCICLE) PIPERIDINS |
| WO2012059497A1 (en) | 2010-11-02 | 2012-05-10 | Bayer Cropscience Ag | N-hetarylmethyl pyrazolylcarboxamides |
| WO2012062749A1 (en) | 2010-11-12 | 2012-05-18 | Bayer Cropscience Ag | Benzimidazolidinones that can be used as fungicides |
| BR112013011937B1 (en) | 2010-11-15 | 2018-05-15 | Bayer Intellectual Property Gmbh | CYANOENAMINE DERIVATIVES, COMPOSITION AND METHOD FOR CONTROLING PHYTOPATHOGENIC FUNGES, USE OF CYANOENAMINS AND PROCESS FOR PREPARING COMPOSITIONS TO CONTROL PHYTOPATHOGENIC FUNGES |
| BR112013012082A2 (en) | 2010-11-15 | 2016-07-19 | Bayer Ip Gmbh | 5-halopyrazole carboxamides |
| MX2013005258A (en) | 2010-11-15 | 2013-07-05 | Bayer Ip Gmbh | N-ARIL PIRAZOL (UNCLE) CARBOXAMIDS. |
| CN103354807A (en) | 2010-11-15 | 2013-10-16 | 拜耳知识产权有限责任公司 | Cyanoenamines and their use as fungicides |
| WO2012065945A1 (en) | 2010-11-15 | 2012-05-24 | Bayer Cropscience Ag | 5-halogenopyrazole(thio)carboxamides |
| EP2454939A1 (en) | 2010-11-18 | 2012-05-23 | Bayer CropScience AG | Post-harvest treatment |
| WO2012071040A1 (en) | 2010-11-24 | 2012-05-31 | Pioneer Hi-Bred International, Inc. | Brassica gat event dp-073496-4 and compositions and methods for the identification and/or detection thereof |
| CA2818918A1 (en) | 2010-11-24 | 2012-05-31 | Pioneer Hi-Bred International, Inc. | Brassica gat event dp-061061-7 and compositions and methods for the identification and/or detection thereof |
| WO2012072547A1 (en) | 2010-11-30 | 2012-06-07 | Bayer Cropscience Ag | Pyrimidine derivatives and use thereof as pesticides |
| EP2460407A1 (en) | 2010-12-01 | 2012-06-06 | Bayer CropScience AG | Agent combinations comprising pyridylethyl benzamides and other agents |
| KR20180096815A (en) | 2010-12-01 | 2018-08-29 | 바이엘 인텔렉쳐 프로퍼티 게엠베하 | Use of fluopyram for controlling nematodes in crops and for increasing yield |
| AR083987A1 (en) | 2010-12-01 | 2013-04-10 | Bayer Cropscience Ag | USED PIRAZOLCARBOXILIC ACID AMIDAS FOR REDUCTION OF MICOTOXIN POLLUTION IN PLANTS |
| TWI667347B (en) | 2010-12-15 | 2019-08-01 | 瑞士商先正達合夥公司 | Soybean event syht0h2 and compositions and methods for detection thereof |
| EP2655635A1 (en) | 2010-12-22 | 2013-10-30 | Pioneer Hi-Bred International, Inc. | Viral promoter, truncations thereof, and methods of use |
| US8962916B2 (en) | 2010-12-22 | 2015-02-24 | Pioneer Hi Bred International Inc | Viral promoter, truncations thereof, and methods of use |
| EP2474542A1 (en) | 2010-12-29 | 2012-07-11 | Bayer CropScience AG | Fungicide hydroximoyl-tetrazole derivatives |
| WO2012089757A1 (en) | 2010-12-29 | 2012-07-05 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| EP2471363A1 (en) | 2010-12-30 | 2012-07-04 | Bayer CropScience AG | Use of aryl-, heteroaryl- and benzylsulfonamide carboxylic acids, -carboxylic acid esters, -carboxylic acid amides and -carbonitriles and/or its salts for increasing stress tolerance in plants |
| WO2012088645A1 (en) | 2010-12-31 | 2012-07-05 | Bayer Cropscience Ag | Method for improving plant quality |
| CA2826276C (en) | 2011-02-15 | 2020-03-24 | Pioneer Hi-Bred International, Inc. | Root-preferred promoter and methods of use |
| MX336181B (en) | 2011-02-15 | 2016-01-11 | Bayer Ip Gmbh | Synergistic combinations containing a dithiino-tetracarboxamide fungicide and a herbicide, safener or plant growth regulator. |
| EP2494867A1 (en) | 2011-03-01 | 2012-09-05 | Bayer CropScience AG | Halogen-substituted compounds in combination with fungicides |
| EP2683239A1 (en) | 2011-03-10 | 2014-01-15 | Bayer Intellectual Property GmbH | Use of lipochito-oligosaccharide compounds for safeguarding seed safety of treated seeds |
| JP2014509599A (en) | 2011-03-14 | 2014-04-21 | バイエル・インテレクチユアル・プロパテイー・ゲー・エム・ベー・ハー | Fungicide hydroxymoyl-tetrazole derivative |
| EP2502495A1 (en) | 2011-03-16 | 2012-09-26 | Bayer CropScience AG | Use of a dithiino-tetracarboxamide for the protection of harvested products against phytopathogenic fungi |
| KR102003175B1 (en) | 2011-03-25 | 2019-07-24 | 몬산토 테크놀로지 엘엘씨 | Plant regulatory elements and uses thereof |
| MX350825B (en) | 2011-03-31 | 2017-09-22 | Bayer Ip Gmbh | Herbicidally and fungicidally active 3-phenylisoxazoline-5-carbox amides and 3-phenylisoxazoline-5-thioamides. |
| US8513487B2 (en) | 2011-04-07 | 2013-08-20 | Zenon LISIECZKO | Plants and seeds of spring canola variety ND-662c |
| EP2694494A1 (en) | 2011-04-08 | 2014-02-12 | Bayer Intellectual Property GmbH | Fungicide hydroximoyl-tetrazole derivatives |
| US8513494B2 (en) | 2011-04-08 | 2013-08-20 | Chunren Wu | Plants and seeds of spring canola variety SCV695971 |
| AR085585A1 (en) | 2011-04-15 | 2013-10-09 | Bayer Cropscience Ag | VINIL- AND ALQUINILCICLOHEXANOLES SUBSTITUTED AS ACTIVE PRINCIPLES AGAINST STRIPS ABIOTIQUE OF PLANTS |
| EP2511255A1 (en) | 2011-04-15 | 2012-10-17 | Bayer CropScience AG | Substituted prop-2-in-1-ol and prop-2-en-1-ol derivatives |
| AR085568A1 (en) | 2011-04-15 | 2013-10-09 | Bayer Cropscience Ag | 5- (BICYCLE [4.1.0] HEPT-3-EN-2-IL) -PENTA-2,4-DIENOS AND 5- (BICYCLE [4.1.0] HEPT-3-EN-2-IL) -PENT- 2-IN-4-INOS REPLACED AS ACTIVE PRINCIPLES AGAINST ABIOTIC STRESS OF PLANTS |
| AR090010A1 (en) | 2011-04-15 | 2014-10-15 | Bayer Cropscience Ag | 5- (CICLOHEX-2-EN-1-IL) -PENTA-2,4-DIENOS AND 5- (CICLOHEX-2-EN-1-IL) -PENT-2-EN-4-INOS REPLACED AS ACTIVE PRINCIPLES AGAINST THE ABIOTIC STRESS OF PLANTS, USES AND TREATMENT METHODS |
| ES2561296T3 (en) | 2011-04-22 | 2016-02-25 | Bayer Intellectual Property Gmbh | Combinations of an active compound comprising a carboximide derivative and a fungicidal compound |
| US8507761B2 (en) | 2011-05-05 | 2013-08-13 | Teresa Huskowska | Plants and seeds of spring canola variety SCV372145 |
| US8513495B2 (en) | 2011-05-10 | 2013-08-20 | Dale Burns | Plants and seeds of spring canola variety SCV291489 |
| US9845477B2 (en) | 2011-05-13 | 2017-12-19 | Monsanto Technology Llc | Plant regulatory elements and uses thereof |
| WO2012168124A1 (en) | 2011-06-06 | 2012-12-13 | Bayer Cropscience Nv | Methods and means to modify a plant genome at a preselected site |
| CN103957711A (en) | 2011-07-04 | 2014-07-30 | 拜耳知识产权有限责任公司 | Use of substituted isoquinolinones, isoquinolindiones, isoquinolintriones and dihydroisoquinolinones or in each case salts thereof as active agents against abiotic stress in plants |
| UA115128C2 (en) | 2011-07-27 | 2017-09-25 | Байєр Інтеллектуал Проперті Гмбх | Seed dressing for controlling phytopathogenic fungi |
| WO2013020985A1 (en) | 2011-08-10 | 2013-02-14 | Bayer Intellectual Property Gmbh | Active compound combinations comprising specific tetramic acid derivatives |
| CN103717612A (en) | 2011-08-12 | 2014-04-09 | 拜尔作物科学公司 | Guard cell-specific expression of transgenes in cotton |
| WO2013026836A1 (en) | 2011-08-22 | 2013-02-28 | Bayer Intellectual Property Gmbh | Fungicide hydroximoyl-tetrazole derivatives |
| CN103890181A (en) | 2011-08-22 | 2014-06-25 | 拜尔作物科学公司 | Methods and means for modifying plant genomes |
| EP2561759A1 (en) | 2011-08-26 | 2013-02-27 | Bayer Cropscience AG | Fluoroalkyl-substituted 2-amidobenzimidazoles and their effect on plant growth |
| AR087718A1 (en) | 2011-08-31 | 2014-04-09 | Monsanto Technology Llc | MOLECULAR MARKERS ASSOCIATED WITH CANCRO RESISTANCE OF THE SIZE OF SOY |
| EP2753177A1 (en) | 2011-09-09 | 2014-07-16 | Bayer Intellectual Property GmbH | Acyl-homoserine lactone derivatives for improving plant yield |
| JP6002225B2 (en) | 2011-09-12 | 2016-10-05 | バイエル・インテレクチュアル・プロパティ・ゲゼルシャフト・ミット・ベシュレンクテル・ハフツングBayer Intellectual Property GmbH | Bactericidal 4-substituted-3- {phenyl [(heterocyclylmethoxy) imino] methyl} -1,2,4-oxadiazol-5 (4H) -one derivatives |
| IN2014CN01862A (en) | 2011-09-15 | 2015-05-29 | Bayer Ip Gmbh | |
| MX357299B (en) | 2011-09-16 | 2018-07-04 | Bayer Ip Gmbh | Use of acylsulfonamides for improving plant yield. |
| EP2755484A1 (en) | 2011-09-16 | 2014-07-23 | Bayer Intellectual Property GmbH | Use of 5-phenyl- or 5-benzyl-2 isoxazoline-3 carboxylates for improving plant yield |
| EA029005B1 (en) | 2011-09-16 | 2018-01-31 | Байер Интеллектчуал Проперти Гмбх | Use of phenylpyrazolin-3-carboxylates for improving plant yield |
| AR087971A1 (en) | 2011-09-23 | 2014-04-30 | Bayer Ip Gmbh | USE OF ACID DERIVATIVES 1-PHENYL-PIRAZOL-3-CARBOXILIC 4-SUBSTITUTED AS ACTIVE PRINCIPLES AGAINST PLANTS ABIOTIC STRESS |
| AU2012320554B2 (en) | 2011-10-04 | 2017-11-09 | Bayer Intellectual Property Gmbh | RNAi for the control of fungi and oomycetes by inhibiting saccharopine dehydrogenase gene |
| WO2013050324A1 (en) | 2011-10-06 | 2013-04-11 | Bayer Intellectual Property Gmbh | Combination, containing 4-phenylbutyric acid (4-pba) or a salt thereof (component (a)) and one or more selected additional agronomically active compounds (component(s) (b)), that reduces abiotic plant stress |
| WO2013075817A1 (en) | 2011-11-21 | 2013-05-30 | Bayer Intellectual Property Gmbh | Fungicide n-[(trisubstitutedsilyl)methyl]-carboxamide derivatives |
| TW201335140A (en) | 2011-11-25 | 2013-09-01 | 拜耳智慧財產有限公司 | 2-iodoimidazole derivatives |
| MX2014006073A (en) | 2011-11-25 | 2014-08-08 | Bayer Ip Gmbh | Novel heterocyclic alkanol-derivatives. |
| US9725414B2 (en) | 2011-11-30 | 2017-08-08 | Bayer Intellectual Property Gmbh | Fungicidal N-bicycloalkyl and N-tricycloalkyl pyrazole-4-(thio)carboxamide derivatives |
| EP2601839A1 (en) | 2011-12-08 | 2013-06-12 | Bayer CropScience AG | Synergisitic fungicidal combinations containing phosphorous acid derivative and zoxamide |
| EP2606732A1 (en) | 2011-12-19 | 2013-06-26 | Bayer CropScience AG | Use of an anthranilic diamide derivatives with heteroaromatic and heterocyclic substituents in combination with a biological control agent |
| US9414595B2 (en) | 2011-12-19 | 2016-08-16 | Bayer Cropscience Ag | Use of anthranilic acid diamide derivatives for pest control in transgenic crops |
| US20130167262A1 (en) | 2011-12-21 | 2013-06-27 | The Curators Of The University Of Missouri | Soybean variety s05-11268 |
| WO2013096810A1 (en) | 2011-12-21 | 2013-06-27 | The Curators Of The University Of Missouri | Soybean variety s05-11482 |
| EP2797891B1 (en) | 2011-12-29 | 2015-09-30 | Bayer Intellectual Property GmbH | Fungicidal 3-[(pyridin-2-ylmethoxyimino)(phenyl)methyl]-2-substituted-1,2,4-oxadiazol-5(2h)-one derivatives |
| KR102028893B1 (en) | 2011-12-29 | 2019-10-07 | 바이엘 인텔렉쳐 프로퍼티 게엠베하 | Fungicidal 3-[(1,3-thiazol-4-ylmethoxyimino)(phenyl)methyl]-2-substituted-1,2,4-oxadiazol-5(2h)-one derivatives |
| BR112014016791A2 (en) | 2012-01-06 | 2019-09-24 | Pioneer Hi Bred Int | isolated nucleic acid molecule, expression cassette, vector, plant cell, plant, transgenic seed, method for expression of a polynucleotide in a plant or plant cell, method for expression of a polynucleotide, preferably in egg tissues of a plant |
| US9006515B2 (en) | 2012-01-06 | 2015-04-14 | Pioneer Hi Bred International Inc | Pollen preferred promoters and methods of use |
| HUE036328T2 (en) | 2012-02-22 | 2018-06-28 | Bayer Cropscience Ag | Use of fluopyram for controlling wood diseases in grape |
| PE20190345A1 (en) | 2012-02-27 | 2019-03-07 | Bayer Ip Gmbh | ACTIVE COMPOUND COMBINATIONS |
| CA2865977A1 (en) | 2012-02-29 | 2013-09-06 | Dow Agrosciences Llc | Sugarcane bacilliform viral (scbv) enhancer and its use in plant functional genomics |
| WO2013135724A1 (en) | 2012-03-14 | 2013-09-19 | Bayer Intellectual Property Gmbh | Pesticidal arylpyrrolidines |
| WO2013139949A1 (en) | 2012-03-23 | 2013-09-26 | Bayer Intellectual Property Gmbh | Compositions comprising a strigolactame compound for enhanced plant growth and yield |
| US9357778B2 (en) | 2012-04-12 | 2016-06-07 | Bayer Cropscience Ag | N-acyl-2-(cyclo)alkypyrrolidines and piperidines useful as fungicides |
| CN102643804B (en) * | 2012-04-12 | 2014-04-02 | 北京奥瑞金种业股份有限公司 | Method for culturing roundup ready transgene corns |
| US20150080337A1 (en) | 2012-04-20 | 2015-03-19 | Bayer Cropscience | N-cycloalkyl-n-[(trisubstitutedsilylphenyl)methylene]-(thio)carboxamide derivatives |
| US9663793B2 (en) | 2012-04-20 | 2017-05-30 | Monsanto Technology, Llc | Plant regulatory elements and uses thereof |
| WO2013156559A1 (en) | 2012-04-20 | 2013-10-24 | Bayer Cropscience Ag | N-cycloalkyl-n-[(heterocyclylphenyl)methylene]-(thio)carboxamide derivatives |
| US11518997B2 (en) | 2012-04-23 | 2022-12-06 | BASF Agricultural Solutions Seed US LLC | Targeted genome engineering in plants |
| US8859857B2 (en) | 2012-04-26 | 2014-10-14 | Monsanto Technology Llc | Plants and seeds of spring canola variety SCV259778 |
| US8835720B2 (en) | 2012-04-26 | 2014-09-16 | Monsanto Technology Llc | Plants and seeds of spring canola variety SCV967592 |
| US8802935B2 (en) | 2012-04-26 | 2014-08-12 | Monsanto Technology Llc | Plants and seeds of spring canola variety SCV942568 |
| US8878009B2 (en) | 2012-04-26 | 2014-11-04 | Monsanto Technology, LLP | Plants and seeds of spring canola variety SCV318181 |
| EP2662370A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | 5-Halogenopyrazole benzofuranyl carboxamides |
| EP2662361A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | Pyrazol indanyl carboxamides |
| EP2662364A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | Pyrazole tetrahydronaphthyl carboxamides |
| CN104768934B (en) | 2012-05-09 | 2017-11-28 | 拜耳农作物科学股份公司 | Pyrazole indanyl carboxamide |
| EP2662360A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | 5-Halogenopyrazole indanyl carboxamides |
| CN104364236B (en) | 2012-05-09 | 2018-01-16 | 拜尔农作物科学股份公司 | 5 halo-pyrazole dihydro indenyl formamides |
| EP2662363A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | 5-Halogenopyrazole biphenylcarboxamides |
| EP2662362A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | Pyrazole indanyl carboxamides |
| AR091104A1 (en) | 2012-05-22 | 2015-01-14 | Bayer Cropscience Ag | COMBINATIONS OF ACTIVE COMPOUNDS THAT INCLUDE A LIPO-CHYTOOLIGOSACARIDE DERIVATIVE AND A NEMATICIDE, INSECTICIDE OR FUNGICIDE COMPOUND |
| WO2013188291A2 (en) | 2012-06-15 | 2013-12-19 | E. I. Du Pont De Nemours And Company | Methods and compositions involving als variants with native substrate preference |
| EP2871958A1 (en) | 2012-07-11 | 2015-05-20 | Bayer CropScience AG | Use of fungicidal combinations for increasing the tolerance of a plant towards abiotic stress |
| EP2892345A1 (en) | 2012-09-05 | 2015-07-15 | Bayer CropScience AG | Use of substituted 2-amidobenzimidazoles, 2-amidobenzoxazoles and 2-amidobenzothiazoles or salts thereof as active substances against abiotic plant stress |
| CN104797564B (en) | 2012-09-25 | 2018-05-04 | 拜尔农作物科学股份公司 | The 3- phenyl-isoxazole oxazoline -5- thioamides of the 3- phenyl-isoxazoles oxazoline -5- formamides and 5- epoxides of weeding and antifungal 5- epoxides-substituted-substituted |
| US20150259696A1 (en) | 2012-10-11 | 2015-09-17 | Shane E. Abbitt | Guard cell promoters and uses thereof |
| EP2908642B1 (en) | 2012-10-19 | 2022-02-23 | Bayer Cropscience AG | Method for enhancing tolerance to abiotic stress in plants by using carboxamide or thiocarboxamide derivatives |
| WO2014060520A1 (en) | 2012-10-19 | 2014-04-24 | Bayer Cropscience Ag | Method for treating plants against fungi resistant to fungicides using carboxamide or thiocarboxamide derivatives |
| UA114937C2 (en) | 2012-10-19 | 2017-08-28 | Байєр Кропсайнс Аг | Method of plant growth promotion using carboxamide derivatives |
| EP2908639A1 (en) | 2012-10-19 | 2015-08-26 | Bayer Cropscience AG | Active compound combinations comprising carboxamide derivatives |
| PL2908643T3 (en) | 2012-10-19 | 2019-08-30 | Bayer Cropscience Ag | Active compound combinations comprising carboxamide derivatives and a biological control agent |
| EP2735231A1 (en) | 2012-11-23 | 2014-05-28 | Bayer CropScience AG | Active compound combinations |
| WO2014079957A1 (en) | 2012-11-23 | 2014-05-30 | Bayer Cropscience Ag | Selective inhibition of ethylene signal transduction |
| BR112015012054A2 (en) | 2012-11-30 | 2017-07-11 | Bayer Cropscience Ag | fungicide or binary pesticide mixture |
| US9775351B2 (en) | 2012-11-30 | 2017-10-03 | Bayer Cropscience Ag | Ternary fungicidal and pesticidal mixtures |
| WO2014083031A2 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropscience Ag | Binary pesticidal and fungicidal mixtures |
| EA030235B1 (en) | 2012-11-30 | 2018-07-31 | Байер Кропсайенс Акциенгезельшафт | Ternary fungicidal mixtures |
| US9615578B2 (en) | 2012-11-30 | 2017-04-11 | Bayer Cropscience Ag | Binary fungicidal mixtures |
| JP2016500368A (en) | 2012-12-05 | 2016-01-12 | バイエル・クロップサイエンス・アクチェンゲゼルシャフト | As substituted 1- (arylethynyl)-, 1- (heteroarylethynyl)-, 1- (heterocyclic ethynyl)-and 1- (cycloalkenylethynyl) -cyclohexanol as active agents against abiotic plant stress use |
| EP2740356A1 (en) | 2012-12-05 | 2014-06-11 | Bayer CropScience AG | Substituted (2Z)-5(1-Hydroxycyclohexyl)pent-2-en-4-inic acid derivatives |
| EP2740720A1 (en) | 2012-12-05 | 2014-06-11 | Bayer CropScience AG | Substituted bicyclic and tricyclic pent-2-en-4-inic acid derivatives and their use for enhancing the stress tolerance in plants |
| AR093909A1 (en) | 2012-12-12 | 2015-06-24 | Bayer Cropscience Ag | USE OF ACTIVE INGREDIENTS TO CONTROL NEMATODES IN CULTURES RESISTANT TO NEMATODES |
| AR093996A1 (en) | 2012-12-18 | 2015-07-01 | Bayer Cropscience Ag | BACTERICIDAL COMBINATIONS AND BINARY FUNGICIDES |
| CA3062365C (en) | 2012-12-19 | 2022-03-29 | Monsanto Technology Llc | Plant regulatory elements and uses thereof |
| CN104995174A (en) | 2012-12-19 | 2015-10-21 | 拜耳作物科学股份公司 | Difluoromethyl-nicotinic-tetrahydronaphtyl carboxamides |
| AU2013361220A1 (en) | 2012-12-21 | 2015-04-02 | Pioneer Hi-Bred International, Inc. | Compositions and methods for auxin-analog conjugation |
| EP2964614A1 (en) | 2013-03-07 | 2016-01-13 | Bayer Cropscience AG | Fungicidal 3-{phenyl[(heterocyclylmethoxy)imino]methyl}-heterocycle derivatives |
| US9273322B2 (en) | 2013-03-12 | 2016-03-01 | Pioneer Hi Bred International Inc | Root-preferred promoter and methods of use |
| US9243258B2 (en) | 2013-03-12 | 2016-01-26 | Pioneer Hi Bred International Inc | Root-preferred promoter and methods of use |
| WO2014159306A1 (en) | 2013-03-13 | 2014-10-02 | Pioneer Hi-Bred International, Inc. | Glyphosate application for weed control in brassica |
| CN108823203B (en) | 2013-03-14 | 2022-08-12 | 孟山都技术公司 | Plant regulatory elements and their uses |
| MX369750B (en) | 2013-03-14 | 2019-11-20 | Pioneer Hi Bred Int | Compositions and methods to control insect pests. |
| US20160053277A1 (en) | 2013-03-14 | 2016-02-25 | Pioneer Hi-Bred International, Inc. | Compositions Having Dicamba Decarboxylase Activity and Methods of Use |
| US9617553B2 (en) | 2013-03-14 | 2017-04-11 | Monsanto Technology Llc | Plant regulatory elements and uses thereof |
| EP2970935A1 (en) | 2013-03-14 | 2016-01-20 | Pioneer Hi-Bred International, Inc. | Compositions having dicamba decarboxylase activity and methods of use |
| CA2901316A1 (en) | 2013-03-15 | 2014-09-25 | Pioneer Hi-Bred International, Inc. | Phi-4 polypeptides and methods for their use |
| WO2014161821A1 (en) | 2013-04-02 | 2014-10-09 | Bayer Cropscience Nv | Targeted genome engineering in eukaryotes |
| US9550752B2 (en) | 2013-04-12 | 2017-01-24 | Bayer Cropscience Aktiengesellschaft | Triazolinthione derivatives |
| CN105308032B (en) | 2013-04-12 | 2017-05-24 | 拜耳作物科学股份公司 | Novel triazole derivatives |
| AR095867A1 (en) | 2013-04-19 | 2015-11-18 | Bayer Cropscience Ag | METHOD FOR IMPROVED USE OF THE POTENTIAL OF PRODUCTION OF TRANSGENIC PLANTS |
| JP2016519687A (en) | 2013-04-19 | 2016-07-07 | バイエル・クロップサイエンス・アクチェンゲゼルシャフト | Binary insecticide or pesticide mixture |
| TW201507722A (en) | 2013-04-30 | 2015-03-01 | Bayer Cropscience Ag | N-(2-halogen-2-phenethyl)carboxamides as nematicides and endoparasiticides |
| WO2014177514A1 (en) | 2013-04-30 | 2014-11-06 | Bayer Cropscience Ag | Nematicidal n-substituted phenethylcarboxamides |
| US10059999B2 (en) | 2013-06-10 | 2018-08-28 | Monsanto Technology Llc | Molecular markers associated with soybean tolerance to low iron growth conditions |
| EP3013802B1 (en) | 2013-06-26 | 2019-08-14 | Bayer Cropscience AG | N-cycloalkyl-n-[(bicyclylphenyl)methylene]-(thio)carboxamide derivatives |
| EP3019012A1 (en) | 2013-07-09 | 2016-05-18 | Bayer CropScience AG | Use of selected pyridone carboxamides or salts thereof as active substances against abiotic plant stress |
| US20160219812A1 (en) | 2013-07-25 | 2016-08-04 | Pioneer Hi-Bred International, Inc. | Method for producing hybrid brassica seed |
| EP2837287A1 (en) | 2013-08-15 | 2015-02-18 | Bayer CropScience AG | Use of prothioconazole for increasing root growth of Brassicaceae |
| CA2920339C (en) | 2013-08-16 | 2023-10-24 | Pioneer Hi-Bred International, Inc. | Insecticidal proteins and methods for their use |
| US20160201073A1 (en) | 2013-09-11 | 2016-07-14 | Pioneer Hi-Bred International, Inc. | Plant regulatory elements and methods of use thereof |
| CA3223359A1 (en) | 2013-09-13 | 2015-03-19 | Pioneer Hi-Bred International, Inc. | Insecticidal proteins and methods for their use |
| US10093907B2 (en) | 2013-09-24 | 2018-10-09 | Basf Se | Hetero-transglycosylase and uses thereof |
| CN105874062B (en) | 2013-10-18 | 2021-07-20 | 先锋国际良种公司 | Glyphosate-N-acetyltransferase (GLYAT) sequences and methods of use |
| BR112016008036A2 (en) | 2013-10-25 | 2018-01-16 | Pioneer Hi Bred Int | method of identifying a first plant, kit, isolated polynucleotide, plant, germplasm or soybean |
| US10070645B2 (en) | 2013-12-05 | 2018-09-11 | Bayer Cropscience Aktiengesellschaft | N-cycloalkyl-N-{[2-(1-substitutedcycloalkyl)phenyl]methylene}-(thio)carboxamide derivatives |
| US10071967B2 (en) | 2013-12-05 | 2018-09-11 | Bayer Cropscience Aktiengesellschaft | N-cycloalkyl-N-{[2-(1-substitutedcycloalkyl)phenyl]methylene}-(thio)carboxamide derivatives |
| EP3102684B1 (en) | 2014-02-07 | 2020-05-06 | Pioneer Hi-Bred International, Inc. | Insecticidal proteins and methods for their use |
| MX2016010187A (en) | 2014-02-07 | 2017-07-11 | Pioneer Hi Bred Int | INSECTICIDED PROTEINS AND METHODS FOR USE. |
| WO2015164457A1 (en) | 2014-04-22 | 2015-10-29 | E. I. Du Pont De Nemours And Company | Plastidic carbonic anhydrase genes for oil augmentation in seeds with increased dgat expression |
| AR101214A1 (en) | 2014-07-22 | 2016-11-30 | Bayer Cropscience Ag | CIANO-CICLOALQUILPENTA-2,4-DIENOS, CIANO-CICLOALQUILPENT-2-EN-4-INAS, CIANO-HETEROCICLILPENTA-2,4-DIENOS AND CYANO-HETEROCICLILPENT-2-EN-4-INAS REPLACED AS ACTIVE PRINCIPLES PLANTS ABIOTIC |
| WO2016022516A1 (en) | 2014-08-08 | 2016-02-11 | Pioneer Hi-Bred International, Inc. | Ubiquitin promoters and introns and methods of use |
| WO2016044092A1 (en) | 2014-09-17 | 2016-03-24 | Pioneer Hi Bred International Inc | Compositions and methods to control insect pests |
| CN113372421B (en) | 2014-10-16 | 2024-08-06 | 先锋国际良种公司 | Insecticidal proteins and methods of use thereof |
| AR103024A1 (en) | 2014-12-18 | 2017-04-12 | Bayer Cropscience Ag | SELECTED PYRIDONCARBOXAMIDS OR ITS SALTS AS ACTIVE SUBSTANCES AGAINST ABIOTIC PLANTS STRESS |
| WO2016099916A1 (en) | 2014-12-19 | 2016-06-23 | E. I. Du Pont De Nemours And Company | Polylactic acid compositions with accelerated degradation rate and increased heat stability |
| UA126544C2 (en) | 2015-01-15 | 2022-11-02 | Піонір Хай-Бред Інтернешнл, Інк. | RECOMBINANT POLYNUCLEOTIDE ENCODING INSECTICIDE POLYPEPTIDE AND METHOD OF ITS USE |
| EP3274462A4 (en) | 2015-03-26 | 2018-12-26 | The Texas A&M University System | Conversion of lignin into bioplastics and lipid fuels |
| EP3283476B1 (en) | 2015-04-13 | 2019-08-14 | Bayer Cropscience AG | N-cycloalkyl-n-(biheterocyclyethylene)-(thio)carboxamide derivatives |
| BR112017024948A2 (en) | 2015-05-19 | 2018-07-31 | Pioneer Hi Bred Int | insecticide proteins and methods for their use |
| CN107771181A (en) | 2015-06-16 | 2018-03-06 | 先锋国际良种公司 | Compositions and methods for controlling insect pests |
| RU2762832C2 (en) | 2015-08-06 | 2021-12-23 | Пайонир Хай-Бред Интернэшнл, Инк. | Insecticide proteins of plant origin and their application methods |
| WO2017060321A1 (en) | 2015-10-09 | 2017-04-13 | Bayer Cropscience Aktiengesellschaft | Use of pydiflumetofen for the reduction of mycotoxin contamination in plants |
| CA3004056C (en) | 2015-12-22 | 2024-01-23 | Pioneer Hi-Bred International, Inc. | Tissue-preferred promoters and methods of use |
| CA3004914A1 (en) | 2016-02-05 | 2017-08-10 | Pioneer Hi-Bred International, Inc. | Genetic loci associated with brown stem rot resistance in soybean and methods of use |
| ES2900300T3 (en) | 2016-03-11 | 2022-03-16 | Monsanto Technology Llc | Regulatory elements of plants and their uses |
| EP3960863B1 (en) | 2016-05-04 | 2024-11-13 | Pioneer Hi-Bred International, Inc. | Insecticidal proteins and methods for their use |
| NZ748198A (en) | 2016-05-24 | 2023-05-26 | Monsanto Technology Llc | Plant regulatory elements and uses thereof |
| EP3472323A1 (en) | 2016-06-16 | 2019-04-24 | Pioneer Hi-Bred International, Inc. | Compositions and methods to control insect pests |
| CA3026653A1 (en) | 2016-06-24 | 2017-12-28 | Pioneer Hi-Bred International, Inc. | Plant regulatory elements and methods of use thereof |
| US20190359992A1 (en) | 2016-06-28 | 2019-11-28 | Cellectis | Altering expression of gene products in plants through targeted insertion of nucleic acid sequences |
| WO2018005411A1 (en) | 2016-07-01 | 2018-01-04 | Pioneer Hi-Bred International, Inc. | Insecticidal proteins from plants and methods for their use |
| WO2018013333A1 (en) | 2016-07-12 | 2018-01-18 | Pioneer Hi-Bred International, Inc. | Compositions and methods to control insect pests |
| EP3490379A1 (en) | 2016-07-29 | 2019-06-05 | Bayer CropScience Aktiengesellschaft | Active compound combinations and methods to protect the propagation material of plants |
| WO2018054829A1 (en) | 2016-09-22 | 2018-03-29 | Bayer Cropscience Aktiengesellschaft | Novel triazole derivatives and their use as fungicides |
| BR112019005668A2 (en) | 2016-09-22 | 2019-06-04 | Bayer Ag | new triazole derivatives |
| US20190225974A1 (en) | 2016-09-23 | 2019-07-25 | BASF Agricultural Solutions Seed US LLC | Targeted genome optimization in plants |
| CA3041351A1 (en) | 2016-10-26 | 2018-05-03 | Bayer Cropscience Aktiengesellschaft | Use of pyraziflumid for controlling sclerotinia spp in seed treatment applications |
| MX387927B (en) | 2016-11-01 | 2025-03-19 | Pioneer Hi Bred Int | INSECTICIDAL PROTEINS AND METHODS FOR THEIR USE. |
| CA3046145A1 (en) | 2016-12-08 | 2018-06-14 | Bayer Cropscience Aktiengesellschaft | Use of insecticides for controlling wireworms |
| EP3332645A1 (en) | 2016-12-12 | 2018-06-13 | Bayer Cropscience AG | Use of substituted pyrimidine diones or their salts as agents to combat abiotic plant stress |
| WO2018108627A1 (en) | 2016-12-12 | 2018-06-21 | Bayer Cropscience Aktiengesellschaft | Use of substituted indolinylmethyl sulfonamides, or the salts thereof for increasing the stress tolerance of plants |
| ES2986585T3 (en) | 2017-01-19 | 2024-11-12 | Monsanto Technology Llc | Regulatory elements of plants and their uses |
| WO2018183050A2 (en) * | 2017-03-30 | 2018-10-04 | Pioneer Hi-Bred International, Inc. | Improved plant epsp synthases and methods of use |
| CA3066872A1 (en) | 2017-06-13 | 2018-12-20 | Bayer Aktiengesellschaft | Herbicidally active 3-phenylisoxazoline-5-carboxamides of tetrahydro and dihydrofuran carboxamides |
| PT3638665T (en) | 2017-06-13 | 2021-10-19 | Bayer Cropscience Ag | Herbicidally active 3-phenylisoxazoline-5-carboxamides of tetrahydro and dihydrofuran carboxylic acids and esters |
| WO2019025153A1 (en) | 2017-07-31 | 2019-02-07 | Bayer Cropscience Aktiengesellschaft | USE OF SUBSTITUTED N-SULFONYL-N'-ARYLDIAMINOALKANES AND N-SULFONYL-N'-HETEROARYL DIAMINOALKANES OR THEIR SALTS TO INCREASE STRESSTOLERANCE IN PLANTS |
| EP3668845B1 (en) | 2017-08-17 | 2024-06-26 | Bayer Aktiengesellschaft | Herbicidally active 3-phenyl-5-trifluormethylisoxazolin-5-carboxamides of cyclopentyl carboxylic acids and esters |
| CA3076831A1 (en) | 2017-09-25 | 2019-03-28 | Pioneer Hi-Bred International, Inc. | Tissue-preferred promoters and methods of use |
| US11473099B2 (en) | 2017-11-01 | 2022-10-18 | Monsanto Technology, Llc | Methods and compositions for herbicide tolerance in plants |
| EP3360417A1 (en) | 2017-11-02 | 2018-08-15 | Bayer CropScience Aktiengesellschaft | Use of sulfonylindol as herbicide |
| EP3737747A1 (en) | 2018-01-12 | 2020-11-18 | The Texas A&M University System | Increasing plant bioproduct yield |
| ES2937924T3 (en) | 2018-01-25 | 2023-04-03 | Bayer Ag | 3-Phenyl-isoxazole-yl-5-carboxamides of cyclopentenylcarboxylic acid derivatives with herbicidal activity |
| CN111868247B (en) | 2018-03-12 | 2025-01-10 | 先锋国际良种公司 | Methods for plant transformation |
| US11525144B2 (en) | 2018-03-14 | 2022-12-13 | Pioneer Hi-Bred International, Inc. | Insecticidal proteins from plants and methods for their use |
| WO2019178038A1 (en) | 2018-03-14 | 2019-09-19 | Pioneer Hi-Bred International, Inc. | Insecticidal proteins from plants and methods for their use |
| WO2019219584A1 (en) | 2018-05-15 | 2019-11-21 | Bayer Aktiengesellschaft | New spiro cyclohexyl pyrrolin-2-ones and their use as herbicides |
| WO2019219588A1 (en) | 2018-05-15 | 2019-11-21 | Bayer Aktiengesellschaft | Specifically substituted 2-alkyl-6-alkoxyphenyl-3-pyrrolin-2-ones and their use as herbicides |
| WO2019219585A1 (en) | 2018-05-15 | 2019-11-21 | Bayer Aktiengesellschaft | New 3-(4-alkynyl-6-alkoxy-2-chlorophenyl)-3-pyrrolin-2-ones and their use as herbicides |
| AR115086A1 (en) | 2018-05-15 | 2020-11-25 | Bayer Ag | PYRROLIN-2-ONAS SUBSTITUTED WITH 2-BROMO-6-ALCOXIFENIL AND ITS USE AS HERBICIDES |
| BR112020023800A2 (en) | 2018-05-22 | 2021-02-23 | Pioneer Hi-Bred International, Inc. | plant regulatory elements and methods of using them |
| WO2019228787A1 (en) | 2018-05-29 | 2019-12-05 | Bayer Aktiengesellschaft | Specifically substituted 2-alkyl-6-alkoxyphenyl-3-pyrrolin-2-ones and their use as herbicides |
| WO2019228788A1 (en) | 2018-05-29 | 2019-12-05 | Bayer Aktiengesellschaft | 2-bromo-6-alkoxyphenyl-substituted pyrrolin-2-ones and their use as herbicides |
| WO2019233863A1 (en) | 2018-06-04 | 2019-12-12 | Bayer Aktiengesellschaft | Herbicidally active bicyclic benzoylpyrazoles |
| EP3833747A1 (en) | 2018-06-28 | 2021-06-16 | Pioneer Hi-Bred International, Inc. | Methods for selecting transformed plants |
| UA128698C2 (en) | 2018-07-26 | 2024-10-02 | Баєр Акціенгезельшафт | Use of the succinate dehydrogenase inhibitor fluopyram for controlling root rot complex and/or seedling disease complex caused by rhizoctonia solani, fusarium species and pythium species in brassicaceae species |
| US20220039383A1 (en) | 2018-09-17 | 2022-02-10 | Bayer Aktiengesellschaft | Use of the Succinate Dehydrogenase Inhibitor Fluopyram for Controlling Claviceps Purpurea and Reducing Sclerotia in Cereals |
| WO2020057939A1 (en) | 2018-09-17 | 2020-03-26 | Bayer Aktiengesellschaft | Use of the fungicide isoflucypram for controlling claviceps purpurea and reducing sclerotia in cereals |
| BR112021008329A2 (en) | 2018-10-31 | 2021-08-03 | Pioneer Hi-Bred International, Inc. | compositions and methods for ochrobactrum-mediated plant transformation |
| CN119040379A (en) | 2019-01-10 | 2024-11-29 | 孟山都技术公司 | Plant regulatory elements and their uses |
| AU2020209871B2 (en) | 2019-01-14 | 2025-07-17 | Bayer Aktiengesellschaft | Herbicidal substituted n-tetrazolyl aryl carboxamides |
| WO2020169509A1 (en) | 2019-02-20 | 2020-08-27 | Bayer Aktiengesellschaft | Herbicidally active 4-(4-trifluormethyl-6-cycloropylpyrazolyl)pyrimidines |
| US12146147B2 (en) | 2019-03-11 | 2024-11-19 | Pioneer Hi-Bred International, Inc. | Methods for clonal plant production |
| DK3937637T3 (en) | 2019-03-12 | 2023-07-24 | Bayer Ag | HERBICIDELY ACTIVE 3-PHENYLISOXAZOLINE-5-CARBOXAMIDES OF S-CONTAINING CYCLOPENTENYLCARBONY ACID ESTERS |
| BR112021013653A2 (en) | 2019-03-15 | 2021-09-14 | Bayer Aktiengesellschaft | 5-SPIROCYCLO-HEXYL-3-PYROLIN-2-ONES SUBSTITUTED BY 3-(2-BROMO-4-ALKYNYL-6-ALKOXYPHENIL) AND THEIR USE AS HERBICIDES |
| EA202192467A1 (en) | 2019-03-15 | 2022-02-16 | Байер Акциенгезельшафт | SPECIFICLY SUBSTITUTED 3-(2-HALOGEN-6-ALKYL-4-PROPYNYLPHENYL)-3-PYRRROLIN-2-ONES AND THEIR USE AS HERBICIDES |
| AU2020244063A1 (en) | 2019-03-15 | 2021-10-07 | Bayer Aktiengesellschaft | Specifically substituted 3-(2-alkoxy-6-alkyl-4-propinylphenyl)-3-pyrrolin-2-ones and their use as herbicides |
| EP3938347A1 (en) | 2019-03-15 | 2022-01-19 | Bayer Aktiengesellschaft | Specifically substituted 3-phenyl-5-spirocyclopentyl-3-pyrrolin-2-ones and their use as herbicides |
| BR112021012818A2 (en) | 2019-03-15 | 2021-11-03 | Bayer Ag | New 3-(2-bromo-4-alkynyl-6-alkoxyphenyl)-3-pyrrolin-3-ones and their use as herbicides |
| US20220170033A1 (en) | 2019-03-27 | 2022-06-02 | Pioneer Hi-Bred International, Inc. | Plant explant transformation |
| WO2020198496A1 (en) | 2019-03-28 | 2020-10-01 | Pioneer Hi-Bred International, Inc. | Modified agrobacterium strains and use thereof for plant transformation |
| US12180187B2 (en) | 2019-06-03 | 2024-12-31 | Bayer Aktiengesellschaft | 1-phenyl-5-azinylpyrazolyl-3-oxyalkyl acids and their use for controlling unwanted plant growth |
| JP7631346B2 (en) | 2019-12-19 | 2025-02-18 | バイエル・アクチエンゲゼルシヤフト | 1,5-diphenylpyrazolyl-3-oxyalkyl acids and 1-phenyl-5-thienylpyrazolyl-3-oxyalkyl acids and their use for controlling undesirable plant growth - Patents.com |
| UA129254C2 (en) | 2020-04-07 | 2025-02-26 | Байєр Акцієнгезелльшафт | SUBSTITUTED ISOPHTHALIC ACID DIAMIDES |
| ES2976966T3 (en) | 2020-04-07 | 2024-08-13 | Bayer Ag | Substituted isophthalic acid diamides and their use as herbicides |
| WO2021204669A1 (en) | 2020-04-07 | 2021-10-14 | Bayer Aktiengesellschaft | Substituted isophthalic acid diamides |
| WO2021204667A1 (en) | 2020-04-07 | 2021-10-14 | Bayer Aktiengesellschaft | Substituted isophthalic acid diamides |
| WO2021204884A1 (en) | 2020-04-09 | 2021-10-14 | Bayer Aktiengesellschaft | 3-(4-alkenyl-phenyl)-3-pyrrolin-2-ones and their use as herbicides |
| WO2021209486A1 (en) | 2020-04-15 | 2021-10-21 | Bayer Aktiengesellschaft | Specifically substituted pyrroline-2-ones and their use as herbicides |
| AU2021263052A1 (en) | 2020-04-29 | 2022-12-08 | Bayer Aktiengesellschaft | 1-pyrazinylpyrazolyl-3-oxyalkyl acids and their derivatives, and their use for control of undesired plant growth |
| CN111394369B (en) * | 2020-05-07 | 2022-03-22 | 海南波莲水稻基因科技有限公司 | Glyphosate-resistant EPSPS mutant gene, plant genetic transformation screening vector containing glyphosate-resistant EPSPS mutant gene and application of glyphosate-resistant EPSPS mutant gene |
| US20230180758A1 (en) | 2020-05-27 | 2023-06-15 | Bayer Aktiengesellschaft | Substituted pyrroline-2-ones and their use as herbicides |
| CN115867564A (en) | 2020-07-14 | 2023-03-28 | 先锋国际良种公司 | Insecticidal proteins and methods of use thereof |
| US20240002870A1 (en) | 2020-09-30 | 2024-01-04 | Pioneer Hi-Bred International, Inc. | Rapid transformation of monocot leaf explants |
| JP2023549462A (en) | 2020-10-23 | 2023-11-27 | バイエル・アクチエンゲゼルシヤフト | 1-(Pyridyl)-5-azinylpyrazole derivatives and their use for the control of undesirable plant growth |
| EP4026833A1 (en) | 2021-01-12 | 2022-07-13 | Bayer Aktiengesellschaft | Herbicidally active 2-(het)arylmethyl pyrimidines |
| US20240263206A1 (en) | 2021-03-26 | 2024-08-08 | Flagship Pioneering Innovations Vii, Llc | Compositions and methods for producing circular polyribonucleotides |
| US20240181079A1 (en) | 2021-03-26 | 2024-06-06 | Flagship Pioneering Innovations Vii, Llc | Production of circular polyribonucleotides in a eukaryotic system |
| EP4314289A1 (en) | 2021-03-26 | 2024-02-07 | Flagship Pioneering Innovations VII, LLC | Production of circular polyribonucleotides in a prokaryotic system |
| WO2022253700A1 (en) | 2021-06-01 | 2022-12-08 | Bayer Aktiengesellschaft | Specifically substituted pyrroline-2-ones and their use as herbicides |
| EP4358718A1 (en) | 2021-06-25 | 2024-05-01 | Bayer Aktiengesellschaft | (1,4,5-trisubstituted-1h-pyrazole-3-yl)oxy-2-alkoxy alkyl acids and their derivatives, their salts and their use as herbicidal agents |
| WO2023274869A1 (en) | 2021-06-29 | 2023-01-05 | Bayer Aktiengesellschaft | 3-(4-alkenyl-phenyl)-3-pyrrolino-2-ones and their use as herbicides |
| AR126252A1 (en) | 2021-07-08 | 2023-10-04 | Bayer Ag | SUBSTITUTED BENZOIC ACID AMIDES |
| EP4169382A1 (en) | 2021-10-22 | 2023-04-26 | Bayer AG | Use of cyclobutrifluram for the reduction of mycotoxin contamination in plants |
| UY39997A (en) | 2021-11-01 | 2023-05-15 | Flagship Pioneering Innovations Vii Llc | POLYNUCLEOTIDES FOR MODIFYING ORGANISMS |
| AU2022400175A1 (en) | 2021-12-01 | 2024-07-18 | Bayer Aktiengesellschaft | (1,4,5-trisubstituted-1h-pyrazole-3-yl)oxy-2-alkoxythio alkyl acids and derivatives thereof, their salts and their use as herbicidal active agents |
| MX2024009021A (en) | 2022-01-20 | 2024-08-06 | Flagship Pioneering Innovations Vii Llc | POLYNUCLEOTIDES TO MODIFY ORGANISMS. |
| US12004476B2 (en) | 2022-01-26 | 2024-06-11 | Monsanto Technology Llc. | Cotton variety 20R750B3XF |
| CN120265780A (en) | 2022-09-09 | 2025-07-04 | 埃朗根-纽伦堡弗里德里希-亚历山大大学 | Plant regulatory elements and their uses |
| WO2024078871A1 (en) | 2022-10-14 | 2024-04-18 | Bayer Aktiengesellschaft | 1-pyridyl-5-phenylpyrazolyl-3-oxy- and -3-thioalkyl acids and derivatives and their use for controlling undesired plant growth |
| AR132586A1 (en) | 2023-05-03 | 2025-07-16 | Flagship Pioneering Innovations Vii Llc | ARTIFICIAL AMALGAVIRUS SATELLITE RNA |
| WO2024229359A2 (en) | 2023-05-03 | 2024-11-07 | Flagship Pioneering Innovations Vii, Llc | Artificial tymovirales satellite rnas |
| AR132592A1 (en) | 2023-05-03 | 2025-07-16 | Flagship Pioneering Innovations Vii Llc | Artificial Ghabviral Satellite Arn |
| AR132590A1 (en) | 2023-05-03 | 2025-07-16 | Flagship Pioneering Innovations Vii Llc | PARTITIVE SATELLITE RNA AMPLIFICATION SYSTEMS FOR PLANTS |
| AR132587A1 (en) | 2023-05-03 | 2025-07-16 | Flagship Pioneering Innovations Vii Llc | Artificial Martelliviral Satellite Arn |
| WO2024229403A1 (en) | 2023-05-03 | 2024-11-07 | Flagship Pioneering Innovations Vii, Llc | Endornaviral satellite rna amplification systems for plants |
| WO2024229347A1 (en) | 2023-05-03 | 2024-11-07 | Flagship Pioneering Innovations Vii, Llc | Artificial tombusviridae satellite rnas |
| AR132589A1 (en) | 2023-05-03 | 2025-07-16 | Flagship Pioneering Innovations Vii Llc | Satellite RNA of artificial solemoviridae |
| AR132591A1 (en) | 2023-05-03 | 2025-07-16 | Flagship Pioneering Innovations Vii Llc | SATELLITE RNA OF ARTIFICIAL SECOVIRIDAE |
| AU2024283657A1 (en) | 2023-06-04 | 2026-01-22 | Jiangsu Flag Chemical Industry Co., Ltd. | 3-phenylisoxazoline-5-carboxamide compound, and preparation method therefor, herbicidal composition thereof and use thereof |
| US20250075226A1 (en) | 2023-08-29 | 2025-03-06 | University Of Freiburg | Proteins for regulation of symbiotic infection and associated regulatory elements |
| WO2026006779A1 (en) | 2024-06-27 | 2026-01-02 | The Regents Of The University Of California | Transgenic plants expressing fern rapid non-photochemical quenching (npq) proteins |
Citations (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US5627061A (en) * | 1990-08-31 | 1997-05-06 | Monsanto Company | Glyphosate-tolerant 5-enolpyruvylshikimate-3-phosphate synthases |
Family Cites Families (4)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US4940835A (en) * | 1985-10-29 | 1990-07-10 | Monsanto Company | Glyphosate-resistant plants |
| FR2736926B1 (en) * | 1995-07-19 | 1997-08-22 | Rhone Poulenc Agrochimie | 5-ENOL PYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE MUTEE, CODING GENE FOR THIS PROTEIN AND PROCESSED PLANTS CONTAINING THIS GENE |
| AU6882298A (en) * | 1997-04-03 | 1998-10-22 | Dekalb Genetics Corporation | Glyphosate resistant maize lines |
| GB9711015D0 (en) * | 1997-05-28 | 1997-07-23 | Zeneca Ltd | Improvements in or relating to organic compounds |
-
2001
- 2001-03-06 BR BR0109118-2A patent/BR0109118A/en not_active IP Right Cessation
- 2001-03-06 EP EP01913329A patent/EP1261695B1/en not_active Expired - Lifetime
- 2001-03-06 AU AU4200501A patent/AU4200501A/en active Pending
- 2001-03-06 WO PCT/US2001/007135 patent/WO2001066704A2/en not_active Ceased
- 2001-03-06 AT AT01913329T patent/ATE298364T1/en not_active IP Right Cessation
- 2001-03-06 US US09/800,130 patent/US6803501B2/en not_active Expired - Fee Related
- 2001-03-06 AU AU2001242005A patent/AU2001242005B2/en not_active Ceased
- 2001-03-06 DE DE60111613T patent/DE60111613T2/en not_active Expired - Lifetime
- 2001-03-06 CA CA002401093A patent/CA2401093A1/en not_active Abandoned
- 2001-03-09 AR ARP010101130A patent/AR028243A1/en active IP Right Grant
-
2002
- 2002-08-23 ZA ZA200206788A patent/ZA200206788B/en unknown
-
2003
- 2003-04-15 US US10/413,909 patent/US20030192072A1/en not_active Abandoned
-
2004
- 2004-03-17 US US10/803,156 patent/US20040148650A1/en not_active Abandoned
-
2008
- 2008-10-01 AR ARP080104300A patent/AR068711A2/en unknown
Patent Citations (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US5627061A (en) * | 1990-08-31 | 1997-05-06 | Monsanto Company | Glyphosate-tolerant 5-enolpyruvylshikimate-3-phosphate synthases |
Cited By (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20070180574A1 (en) * | 2006-01-23 | 2007-08-02 | Donald Penner | Methods for breeding glyphosate resistant plants and compositions thereof |
| US7906709B2 (en) | 2006-01-23 | 2011-03-15 | Board Of Trustees Of Michigan State University | Methods for breeding glyphosate resistant plants and compositions thereof |
| US20100199363A1 (en) * | 2006-05-12 | 2010-08-05 | Hartley Carol J | Enzymes for degrading herbicides |
Also Published As
| Publication number | Publication date |
|---|---|
| US20030188346A1 (en) | 2003-10-02 |
| US20040148650A1 (en) | 2004-07-29 |
| BR0109118A (en) | 2002-11-26 |
| WO2001066704A2 (en) | 2001-09-13 |
| EP1261695A2 (en) | 2002-12-04 |
| DE60111613D1 (en) | 2005-07-28 |
| AU4200501A (en) | 2001-09-17 |
| AU2001242005B2 (en) | 2006-04-27 |
| AR068711A2 (en) | 2009-12-02 |
| CA2401093A1 (en) | 2001-09-13 |
| DE60111613T2 (en) | 2006-05-18 |
| US6803501B2 (en) | 2004-10-12 |
| ATE298364T1 (en) | 2005-07-15 |
| EP1261695B1 (en) | 2005-06-22 |
| WO2001066704A3 (en) | 2002-02-21 |
| AR028243A1 (en) | 2003-04-30 |
| ZA200206788B (en) | 2003-11-24 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| US6803501B2 (en) | Methods for making plants tolerant to glyphosate and compositions thereof using a DNA encoding an EPSPS enzyme from Eleusine indica | |
| AU2001242005A1 (en) | Methods for making plants tolerant to glyphosate and compositions thereof | |
| AU2001242005A2 (en) | Methods for making plants tolerant to glyphosate and compositions thereof | |
| US5972644A (en) | Cyanobacterial and plant acetyl-CoA carboxylase | |
| CA2671203C (en) | Improved grg23 epsp synthases: compositions and methods of use | |
| CA2603465C (en) | Identification of a new class of epsp synthases | |
| EP1994158B1 (en) | Epsp synthase enzyme domains conferring glyphosate resistance | |
| EP2078754B1 (en) | GRG23 and GRG51 genes conferring herbicide resistance | |
| US20070295251A1 (en) | Epsp synthases: compositions and methods of use | |
| US20090151018A1 (en) | Gdc-1 genes conferring herbicide resistance | |
| US20110223647A1 (en) | GRG36: Novel EPSP Synthase Gene Conferring Herbicide Resistance | |
| US20020062499A1 (en) | Methods for translational repression of gene expression in plants | |
| EP1582583A2 (en) | Methods for making plants tolerant to glyphosate and compositions thereof | |
| US20040205847A1 (en) | GDC-1 genes conferring herbicide resistance | |
| HK1134108B (en) | Improved grg23 epsp synthases: compositions and methods of use | |
| HK1131183A (en) | Epsp synthase enzyme domains conferring glyphosate resistance |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| STCB | Information on status: application discontinuation |
Free format text: ABANDONED -- FAILURE TO RESPOND TO AN OFFICE ACTION |