WO2014004726A1 - Methods, compositions and kits for the diagnosis, prognosis and monitoring of cancer - Google Patents
Methods, compositions and kits for the diagnosis, prognosis and monitoring of cancer Download PDFInfo
- Publication number
- WO2014004726A1 WO2014004726A1 PCT/US2013/047984 US2013047984W WO2014004726A1 WO 2014004726 A1 WO2014004726 A1 WO 2014004726A1 US 2013047984 W US2013047984 W US 2013047984W WO 2014004726 A1 WO2014004726 A1 WO 2014004726A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- cancer
- dna
- mutations
- panel
- sample
- Prior art date
Links
- 206010028980 Neoplasm Diseases 0.000 title claims abstract description 133
- 238000000034 method Methods 0.000 title claims abstract description 101
- 201000011510 cancer Diseases 0.000 title claims abstract description 77
- 238000003745 diagnosis Methods 0.000 title claims abstract description 25
- 238000004393 prognosis Methods 0.000 title claims abstract description 22
- 238000012544 monitoring process Methods 0.000 title claims abstract description 15
- 239000000203 mixture Substances 0.000 title claims description 29
- 238000001514 detection method Methods 0.000 claims abstract description 53
- 150000007523 nucleic acids Chemical class 0.000 claims abstract description 45
- 108020004707 nucleic acids Proteins 0.000 claims abstract description 44
- 102000039446 nucleic acids Human genes 0.000 claims abstract description 44
- 239000012472 biological sample Substances 0.000 claims abstract description 26
- 206010061309 Neoplasm progression Diseases 0.000 claims abstract description 7
- 230000005751 tumor progression Effects 0.000 claims abstract description 5
- 230000035772 mutation Effects 0.000 claims description 128
- 108020004414 DNA Proteins 0.000 claims description 64
- 108090000623 proteins and genes Proteins 0.000 claims description 46
- 239000000523 sample Substances 0.000 claims description 44
- 239000003153 chemical reaction reagent Substances 0.000 claims description 19
- 238000011282 treatment Methods 0.000 claims description 18
- -1 PBKCA Proteins 0.000 claims description 14
- 210000004027 cell Anatomy 0.000 claims description 14
- 210000002381 plasma Anatomy 0.000 claims description 14
- 210000002966 serum Anatomy 0.000 claims description 13
- 210000004369 blood Anatomy 0.000 claims description 12
- 239000008280 blood Substances 0.000 claims description 12
- 102100030708 GTPase KRas Human genes 0.000 claims description 9
- 210000001519 tissue Anatomy 0.000 claims description 9
- 108010078814 Tumor Suppressor Protein p53 Proteins 0.000 claims description 8
- 102000015098 Tumor Suppressor Protein p53 Human genes 0.000 claims description 8
- 101710182396 Fibroblast growth factor receptor 3 Proteins 0.000 claims description 7
- 101000932478 Homo sapiens Receptor-type tyrosine-protein kinase FLT3 Proteins 0.000 claims description 7
- 101000984753 Homo sapiens Serine/threonine-protein kinase B-raf Proteins 0.000 claims description 7
- 102100020718 Receptor-type tyrosine-protein kinase FLT3 Human genes 0.000 claims description 7
- 102100027103 Serine/threonine-protein kinase B-raf Human genes 0.000 claims description 7
- 231100000588 tumorigenic Toxicity 0.000 claims description 7
- 230000000381 tumorigenic effect Effects 0.000 claims description 7
- 102100028138 F-box/WD repeat-containing protein 7 Human genes 0.000 claims description 6
- 101710105178 F-box/WD repeat-containing protein 7 Proteins 0.000 claims description 6
- 102100027842 Fibroblast growth factor receptor 3 Human genes 0.000 claims description 6
- 101001126417 Homo sapiens Platelet-derived growth factor receptor alpha Proteins 0.000 claims description 6
- 102100025725 Mothers against decapentaplegic homolog 4 Human genes 0.000 claims description 6
- 101710143112 Mothers against decapentaplegic homolog 4 Proteins 0.000 claims description 6
- 108020005187 Oligonucleotide Probes Proteins 0.000 claims description 6
- 108010011536 PTEN Phosphohydrolase Proteins 0.000 claims description 6
- 102000014160 PTEN Phosphohydrolase Human genes 0.000 claims description 6
- 102100030485 Platelet-derived growth factor receptor alpha Human genes 0.000 claims description 6
- 239000012530 fluid Substances 0.000 claims description 6
- 239000002751 oligonucleotide probe Substances 0.000 claims description 6
- 102000004169 proteins and genes Human genes 0.000 claims description 6
- 101000744505 Homo sapiens GTPase NRas Proteins 0.000 claims description 5
- 101001012157 Homo sapiens Receptor tyrosine-protein kinase erbB-2 Proteins 0.000 claims description 5
- 102100030086 Receptor tyrosine-protein kinase erbB-2 Human genes 0.000 claims description 5
- 102000052116 epidermal growth factor receptor activity proteins Human genes 0.000 claims description 5
- 108700015053 epidermal growth factor receptor activity proteins Proteins 0.000 claims description 5
- YOHYSYJDKVYCJI-UHFFFAOYSA-N n-[3-[[6-[3-(trifluoromethyl)anilino]pyrimidin-4-yl]amino]phenyl]cyclopropanecarboxamide Chemical compound FC(F)(F)C1=CC=CC(NC=2N=CN=C(NC=3C=C(NC(=O)C4CC4)C=CC=3)C=2)=C1 YOHYSYJDKVYCJI-UHFFFAOYSA-N 0.000 claims description 5
- 108010009392 Cyclin-Dependent Kinase Inhibitor p16 Proteins 0.000 claims description 4
- WSFSSNUMVMOOMR-UHFFFAOYSA-N Formaldehyde Chemical compound O=C WSFSSNUMVMOOMR-UHFFFAOYSA-N 0.000 claims description 4
- 102100039788 GTPase NRas Human genes 0.000 claims description 4
- 101000916173 Homo sapiens Catenin beta-1 Proteins 0.000 claims description 4
- 101000584633 Homo sapiens GTPase HRas Proteins 0.000 claims description 4
- 101001045751 Homo sapiens Hepatocyte nuclear factor 1-alpha Proteins 0.000 claims description 4
- 101000954986 Homo sapiens Merlin Proteins 0.000 claims description 4
- 102100037106 Merlin Human genes 0.000 claims description 4
- 102100029981 Receptor tyrosine-protein kinase erbB-4 Human genes 0.000 claims description 4
- 101710100963 Receptor tyrosine-protein kinase erbB-4 Proteins 0.000 claims description 4
- 102100033254 Tumor suppressor ARF Human genes 0.000 claims description 4
- 108700020467 WT1 Proteins 0.000 claims description 4
- 102000016914 ras Proteins Human genes 0.000 claims description 4
- 102100028914 Catenin beta-1 Human genes 0.000 claims description 3
- 102100029974 GTPase HRas Human genes 0.000 claims description 3
- 102100022057 Hepatocyte nuclear factor 1-alpha Human genes 0.000 claims description 3
- 238000001574 biopsy Methods 0.000 claims description 3
- 210000001124 body fluid Anatomy 0.000 claims description 3
- 210000001175 cerebrospinal fluid Anatomy 0.000 claims description 3
- 238000000605 extraction Methods 0.000 claims description 3
- 239000012634 fragment Substances 0.000 claims description 3
- 206010003445 Ascites Diseases 0.000 claims description 2
- LFQSCWFLJHTTHZ-UHFFFAOYSA-N Ethanol Chemical compound CCO LFQSCWFLJHTTHZ-UHFFFAOYSA-N 0.000 claims description 2
- 102000035195 Peptidases Human genes 0.000 claims description 2
- 108091005804 Peptidases Proteins 0.000 claims description 2
- 208000002151 Pleural effusion Diseases 0.000 claims description 2
- 101150040459 RAS gene Proteins 0.000 claims description 2
- 101150076031 RAS1 gene Proteins 0.000 claims description 2
- 101150084041 WT1 gene Proteins 0.000 claims description 2
- YTRQFSDWAXHJCC-UHFFFAOYSA-N chloroform;phenol Chemical compound ClC(Cl)Cl.OC1=CC=CC=C1 YTRQFSDWAXHJCC-UHFFFAOYSA-N 0.000 claims description 2
- 230000015271 coagulation Effects 0.000 claims description 2
- 238000005345 coagulation Methods 0.000 claims description 2
- 230000029087 digestion Effects 0.000 claims description 2
- 238000004090 dissolution Methods 0.000 claims description 2
- 238000001914 filtration Methods 0.000 claims description 2
- 230000001926 lymphatic effect Effects 0.000 claims description 2
- 239000012188 paraffin wax Substances 0.000 claims description 2
- 238000001556 precipitation Methods 0.000 claims description 2
- 235000019833 protease Nutrition 0.000 claims description 2
- 238000000746 purification Methods 0.000 claims description 2
- 210000002700 urine Anatomy 0.000 claims description 2
- 102100032610 Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas Human genes 0.000 claims 3
- 101001014590 Homo sapiens Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas Proteins 0.000 claims 3
- 101001014594 Homo sapiens Guanine nucleotide-binding protein G(s) subunit alpha isoforms short Proteins 0.000 claims 3
- 101001014610 Homo sapiens Neuroendocrine secretory protein 55 Proteins 0.000 claims 3
- 101000797903 Homo sapiens Protein ALEX Proteins 0.000 claims 3
- 101000695187 Homo sapiens Protein patched homolog 1 Proteins 0.000 claims 2
- 102100028680 Protein patched homolog 1 Human genes 0.000 claims 2
- 238000011529 RT qPCR Methods 0.000 claims 2
- 101100215673 Arabidopsis thaliana AGL11 gene Proteins 0.000 claims 1
- 108091011896 CSF1 Proteins 0.000 claims 1
- 102000038594 Cdh1/Fizzy-related Human genes 0.000 claims 1
- 108091007854 Cdh1/Fizzy-related Proteins 0.000 claims 1
- 101000823271 Homo sapiens Tyrosine-protein kinase ABL2 Proteins 0.000 claims 1
- 101000997832 Homo sapiens Tyrosine-protein kinase JAK2 Proteins 0.000 claims 1
- 101710177984 Isocitrate dehydrogenase [NADP] Proteins 0.000 claims 1
- 101710102690 Isocitrate dehydrogenase [NADP] cytoplasmic Proteins 0.000 claims 1
- 102100037845 Isocitrate dehydrogenase [NADP], mitochondrial Human genes 0.000 claims 1
- 101710175291 Isocitrate dehydrogenase [NADP], mitochondrial Proteins 0.000 claims 1
- 101710157228 Isoepoxydon dehydrogenase patN Proteins 0.000 claims 1
- 102100028123 Macrophage colony-stimulating factor 1 Human genes 0.000 claims 1
- 241000124008 Mammalia Species 0.000 claims 1
- 206010073150 Multiple endocrine neoplasia Type 1 Diseases 0.000 claims 1
- 101150073911 STK gene Proteins 0.000 claims 1
- 102100022651 Tyrosine-protein kinase ABL2 Human genes 0.000 claims 1
- 102100033444 Tyrosine-protein kinase JAK2 Human genes 0.000 claims 1
- 102000040856 WT1 Human genes 0.000 claims 1
- 239000007864 aqueous solution Substances 0.000 claims 1
- 229940000425 combination drug Drugs 0.000 claims 1
- 230000002452 interceptive effect Effects 0.000 claims 1
- 206010051747 multiple endocrine neoplasia Diseases 0.000 claims 1
- 238000002560 therapeutic procedure Methods 0.000 claims 1
- 230000001173 tumoral effect Effects 0.000 abstract description 4
- 230000001225 therapeutic effect Effects 0.000 abstract description 2
- 108020004999 messenger RNA Proteins 0.000 description 54
- 238000003556 assay Methods 0.000 description 30
- 238000006243 chemical reaction Methods 0.000 description 25
- 108091032973 (ribonucleotides)n+m Proteins 0.000 description 22
- 239000000090 biomarker Substances 0.000 description 18
- 238000003199 nucleic acid amplification method Methods 0.000 description 17
- 238000003752 polymerase chain reaction Methods 0.000 description 17
- 230000003321 amplification Effects 0.000 description 15
- 238000012360 testing method Methods 0.000 description 11
- 108700028369 Alleles Proteins 0.000 description 10
- 101000584612 Homo sapiens GTPase KRas Proteins 0.000 description 8
- 201000010099 disease Diseases 0.000 description 8
- 208000037265 diseases, disorders, signs and symptoms Diseases 0.000 description 8
- 238000004458 analytical method Methods 0.000 description 7
- 238000001356 surgical procedure Methods 0.000 description 7
- 210000004881 tumor cell Anatomy 0.000 description 7
- 239000011541 reaction mixture Substances 0.000 description 6
- 230000008901 benefit Effects 0.000 description 5
- 238000011002 quantification Methods 0.000 description 5
- 108700020796 Oncogene Proteins 0.000 description 4
- 230000001419 dependent effect Effects 0.000 description 4
- 238000011156 evaluation Methods 0.000 description 4
- XLYOFNOQVPJJNP-UHFFFAOYSA-N water Substances O XLYOFNOQVPJJNP-UHFFFAOYSA-N 0.000 description 4
- 108091092584 GDNA Proteins 0.000 description 3
- 206010027476 Metastases Diseases 0.000 description 3
- 206010039491 Sarcoma Diseases 0.000 description 3
- 102100022748 Wilms tumor protein Human genes 0.000 description 3
- 230000009401 metastasis Effects 0.000 description 3
- 231100000350 mutagenesis Toxicity 0.000 description 3
- 238000006467 substitution reaction Methods 0.000 description 3
- 230000003612 virological effect Effects 0.000 description 3
- 101150028074 2 gene Proteins 0.000 description 2
- 102100034540 Adenomatous polyposis coli protein Human genes 0.000 description 2
- 206010064571 Gene mutation Diseases 0.000 description 2
- 101000582631 Homo sapiens Menin Proteins 0.000 description 2
- 101000823316 Homo sapiens Tyrosine-protein kinase ABL1 Proteins 0.000 description 2
- 101150017040 I gene Proteins 0.000 description 2
- 238000002944 PCR assay Methods 0.000 description 2
- 102000006382 Ribonucleases Human genes 0.000 description 2
- 108010083644 Ribonucleases Proteins 0.000 description 2
- 108700028341 SMARCB1 Proteins 0.000 description 2
- 101150008214 SMARCB1 gene Proteins 0.000 description 2
- 102100025746 SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 Human genes 0.000 description 2
- 102100022596 Tyrosine-protein kinase ABL1 Human genes 0.000 description 2
- 230000005856 abnormality Effects 0.000 description 2
- 101150087698 alpha gene Proteins 0.000 description 2
- 238000000137 annealing Methods 0.000 description 2
- 101150118073 cbi gene Proteins 0.000 description 2
- 238000012217 deletion Methods 0.000 description 2
- 230000037430 deletion Effects 0.000 description 2
- 238000013461 design Methods 0.000 description 2
- 238000006073 displacement reaction Methods 0.000 description 2
- 238000003780 insertion Methods 0.000 description 2
- 230000037431 insertion Effects 0.000 description 2
- 230000001613 neoplastic effect Effects 0.000 description 2
- 239000002773 nucleotide Substances 0.000 description 2
- 125000003729 nucleotide group Chemical group 0.000 description 2
- 108700025694 p53 Genes Proteins 0.000 description 2
- 238000003753 real-time PCR Methods 0.000 description 2
- 230000035945 sensitivity Effects 0.000 description 2
- 238000010200 validation analysis Methods 0.000 description 2
- XUHRVZXFBWDCFB-QRTDKPMLSA-N (3R)-4-[[(3S,6S,9S,12R,15S,18R,21R,24R,27R,28R)-12-(3-amino-3-oxopropyl)-6-[(2S)-butan-2-yl]-3-(2-carboxyethyl)-18-(hydroxymethyl)-28-methyl-9,15,21,24-tetrakis(2-methylpropyl)-2,5,8,11,14,17,20,23,26-nonaoxo-1-oxa-4,7,10,13,16,19,22,25-octazacyclooctacos-27-yl]amino]-3-[[(2R)-2-[[(3S)-3-hydroxydecanoyl]amino]-4-methylpentanoyl]amino]-4-oxobutanoic acid Chemical compound CCCCCCC[C@H](O)CC(=O)N[C@H](CC(C)C)C(=O)N[C@H](CC(O)=O)C(=O)N[C@@H]1[C@@H](C)OC(=O)[C@H](CCC(O)=O)NC(=O)[C@@H](NC(=O)[C@H](CC(C)C)NC(=O)[C@@H](CCC(N)=O)NC(=O)[C@H](CC(C)C)NC(=O)[C@@H](CO)NC(=O)[C@@H](CC(C)C)NC(=O)[C@@H](CC(C)C)NC1=O)[C@@H](C)CC XUHRVZXFBWDCFB-QRTDKPMLSA-N 0.000 description 1
- 101150084750 1 gene Proteins 0.000 description 1
- 108700001666 APC Genes Proteins 0.000 description 1
- 206010069754 Acquired gene mutation Diseases 0.000 description 1
- 102000007469 Actins Human genes 0.000 description 1
- 108010085238 Actins Proteins 0.000 description 1
- 101150001086 COB gene Proteins 0.000 description 1
- 108050007957 Cadherin Proteins 0.000 description 1
- 102100025064 Cellular tumor antigen p53 Human genes 0.000 description 1
- 108010077544 Chromatin Proteins 0.000 description 1
- 101000764817 Chromohalobacter salexigens (strain ATCC BAA-138 / DSM 3043 / CIP 106854 / NCIMB 13768 / 1H11) Oxygen-dependent choline dehydrogenase 1 Proteins 0.000 description 1
- 208000005443 Circulating Neoplastic Cells Diseases 0.000 description 1
- RYGMFSIKBFXOCR-UHFFFAOYSA-N Copper Chemical compound [Cu] RYGMFSIKBFXOCR-UHFFFAOYSA-N 0.000 description 1
- 102100034128 Dual specificity phosphatase 28 Human genes 0.000 description 1
- 101000721661 Homo sapiens Cellular tumor antigen p53 Proteins 0.000 description 1
- 101000911952 Homo sapiens Cyclin-dependent kinase 7 Proteins 0.000 description 1
- 101001017423 Homo sapiens Dual specificity phosphatase 28 Proteins 0.000 description 1
- 101001060231 Homo sapiens F-box/WD repeat-containing protein 7 Proteins 0.000 description 1
- 101000972946 Homo sapiens Hepatocyte growth factor receptor Proteins 0.000 description 1
- 101001059454 Homo sapiens Serine/threonine-protein kinase MARK2 Proteins 0.000 description 1
- 102100034343 Integrase Human genes 0.000 description 1
- 101150105104 Kras gene Proteins 0.000 description 1
- 102100030550 Menin Human genes 0.000 description 1
- 241001465754 Metazoa Species 0.000 description 1
- 241001529936 Murinae Species 0.000 description 1
- 101000753280 Mus musculus Angiopoietin-1 receptor Proteins 0.000 description 1
- 108091061960 Naked DNA Proteins 0.000 description 1
- 201000004404 Neurofibroma Diseases 0.000 description 1
- 108091034117 Oligonucleotide Proteins 0.000 description 1
- 108010058765 Oncogene Protein pp60(v-src) Proteins 0.000 description 1
- 238000012408 PCR amplification Methods 0.000 description 1
- 239000012807 PCR reagent Substances 0.000 description 1
- 108091008606 PDGF receptors Proteins 0.000 description 1
- 108010065129 Patched-1 Receptor Proteins 0.000 description 1
- 241000286209 Phasianidae Species 0.000 description 1
- 102000004160 Phosphoric Monoester Hydrolases Human genes 0.000 description 1
- 108090000608 Phosphoric Monoester Hydrolases Proteins 0.000 description 1
- 102000011653 Platelet-Derived Growth Factor Receptors Human genes 0.000 description 1
- 108010092799 RNA-directed DNA polymerase Proteins 0.000 description 1
- 101710151245 Receptor-type tyrosine-protein kinase FLT3 Proteins 0.000 description 1
- 241000283984 Rodentia Species 0.000 description 1
- 101150054830 S100A6 gene Proteins 0.000 description 1
- 102000001332 SRC Human genes 0.000 description 1
- 108060006706 SRC Proteins 0.000 description 1
- 102100028904 Serine/threonine-protein kinase MARK2 Human genes 0.000 description 1
- 102100024547 Tensin-1 Human genes 0.000 description 1
- 108010088950 Tensins Proteins 0.000 description 1
- 108700031765 Von Hippel-Lindau Tumor Suppressor Proteins 0.000 description 1
- 101710127857 Wilms tumor protein Proteins 0.000 description 1
- 239000002253 acid Substances 0.000 description 1
- 150000007513 acids Chemical class 0.000 description 1
- 208000021841 acute erythroid leukemia Diseases 0.000 description 1
- 235000015107 ale Nutrition 0.000 description 1
- 230000004075 alteration Effects 0.000 description 1
- 101150010487 are gene Proteins 0.000 description 1
- 239000011230 binding agent Substances 0.000 description 1
- 239000013060 biological fluid Substances 0.000 description 1
- 230000017531 blood circulation Effects 0.000 description 1
- 210000000481 breast Anatomy 0.000 description 1
- 239000000872 buffer Substances 0.000 description 1
- 230000003197 catalytic effect Effects 0.000 description 1
- 210000003483 chromatin Anatomy 0.000 description 1
- 210000001072 colon Anatomy 0.000 description 1
- 230000002860 competitive effect Effects 0.000 description 1
- 230000006835 compression Effects 0.000 description 1
- 238000007906 compression Methods 0.000 description 1
- 239000013068 control sample Substances 0.000 description 1
- 238000012864 cross contamination Methods 0.000 description 1
- 230000037029 cross reaction Effects 0.000 description 1
- 238000011498 curative surgery Methods 0.000 description 1
- 238000007405 data analysis Methods 0.000 description 1
- 238000011161 development Methods 0.000 description 1
- 239000003814 drug Substances 0.000 description 1
- 238000013399 early diagnosis Methods 0.000 description 1
- 230000014509 gene expression Effects 0.000 description 1
- 230000004077 genetic alteration Effects 0.000 description 1
- 231100000118 genetic alteration Toxicity 0.000 description 1
- 230000007614 genetic variation Effects 0.000 description 1
- 238000003205 genotyping method Methods 0.000 description 1
- PCHJSUWPFVWCPO-UHFFFAOYSA-N gold Chemical compound [Au] PCHJSUWPFVWCPO-UHFFFAOYSA-N 0.000 description 1
- 230000012010 growth Effects 0.000 description 1
- 229940043355 kinase inhibitor Drugs 0.000 description 1
- 210000004072 lung Anatomy 0.000 description 1
- 230000036210 malignancy Effects 0.000 description 1
- 239000011159 matrix material Substances 0.000 description 1
- 238000002703 mutagenesis Methods 0.000 description 1
- 231100000150 mutagenicity / genotoxicity testing Toxicity 0.000 description 1
- 239000013642 negative control Substances 0.000 description 1
- 230000003287 optical effect Effects 0.000 description 1
- 210000000496 pancreas Anatomy 0.000 description 1
- 239000003757 phosphotransferase inhibitor Substances 0.000 description 1
- 208000015768 polyposis Diseases 0.000 description 1
- 239000013641 positive control Substances 0.000 description 1
- 210000002307 prostate Anatomy 0.000 description 1
- 108700042226 ras Genes Proteins 0.000 description 1
- 108020003175 receptors Proteins 0.000 description 1
- 239000013074 reference sample Substances 0.000 description 1
- 230000004043 responsiveness Effects 0.000 description 1
- 238000003757 reverse transcription PCR Methods 0.000 description 1
- 238000012552 review Methods 0.000 description 1
- 238000012216 screening Methods 0.000 description 1
- 238000011896 sensitive detection Methods 0.000 description 1
- 238000012163 sequencing technique Methods 0.000 description 1
- 239000007787 solid Substances 0.000 description 1
- 230000000392 somatic effect Effects 0.000 description 1
- 230000037439 somatic mutation Effects 0.000 description 1
- 238000011895 specific detection Methods 0.000 description 1
- 238000010561 standard procedure Methods 0.000 description 1
- 210000002784 stomach Anatomy 0.000 description 1
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
- C12Q1/6876—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes
- C12Q1/6883—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for diseases caused by alterations of genetic material
- C12Q1/6886—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for diseases caused by alterations of genetic material for cancer
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q2600/00—Oligonucleotides characterized by their use
- C12Q2600/156—Polymorphic or mutational markers
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q2600/00—Oligonucleotides characterized by their use
- C12Q2600/16—Primer sets for multiplex assays
Definitions
- the present invention relates to methods for detecting and/or quantifying tumor-associated nucleic acids in a biological sample using a panel of cancer- specific UBMs, Independent of cancer type, for the molecular detection of a broad range of cancer types tor diagnostic, prognostic and therapeutic purposes.
- the diagnosis can invol ve the evaluation of circulating biomarkers of cancer or tissues biomarkers.
- circulating biomarkers are proteins secreted by tumor cells which can be detected in serum by antibody-based assays such as I d .
- ISA Today there are only a few biomarkers available and they aren't associated with sufficient specificity and/or sensitivity for cancer detection. Most of the biomarkers are usable mainly for disease fol low-up. A further drawback is that they are cancer-type dependent (for example, PSA, CEA, AEP,
- biomarkers When such biomarkers are positive, they are very useful to monitor efficacy of therapeutics and to detect recurrence of cancer.
- one of the problems with their use is that for each type of cancer, not ail patients are positive, and therefore the overall benefit of each biomarker does not exceed more than 30% of all patients for each cancer type, It is known that tumor cells release nucleic acids.
- nucleic acids are round m blood circulation and are called ''circulating DNA or RNA.” Whole circulating DNA.
- RNA or RNA isolated from blood is, for the most part, DNA or RNA released by normal cells and, to a more limited extent, DNA or RNA released by tumor cells (also referred to as tumorigenic, tumor-related or tumor-associated DNA or RNA),
- tumor cells also referred to as tumorigenic, tumor-related or tumor-associated DNA or RNA
- researchers have tried to detect circulating nucleic acids from tumors, and have confirmed the presence of tumor circulating DNA or RNA, identified by molecular abnormalities associated whh tumor cells such as oncogene mutations, by satellite instability, or by irypermethyl iion of genes.
- a number of publications describe methods for detecting cancers based on the identification of specific genetic alterations in circulating DNA or RNA. For instance, US Patent No.
- 5,496,699 discloses a method for detecting imitations m nucleic acid sequences, in particular the sequence of the KRAS gene, in biological fluids such as blood, serum or plasma.
- US Patent No. 5,068,175 discloses a method for detecting the presence of k AS oneogene-rela ed malignancies in whic the gene is quantified in serum or plasma samples.
- WO 01 /42504 discloses the determination of extracellular nucleic acid, lor example DNA of KRA S and APC genes, in serum or plasma samples, for the evaluation of the nsk factors related with a number of neoplastic diseases.
- WO 02/18652 discloses a method of quail/quantitative detection of human ieiomerase RNA and ieiomerase reverse transcriptase R A in plasma or serum for the diagnosis, monitoring, treatment or evaluation of a number of neoplastic diseases.
- AS-PCR presents several drawbacks.
- the detected signal can be skewed due to some PCR artefacts associated with cross-reactions with the PCR primers (e.g., mismatched amplification and/or primer dirner amplification).
- These artefacts usually appear more frequently for PGR reactions using a high number of amplification cycles necessar to detect very low abundant targets, such as circulating DNA.
- the specificity of conventional AS-PC is usually less than 0,1 % and the efficacy often depends on type of mismatch being detected (e.g., ( ⁇ ;( ;. A:A »G:T>C:T).
- the present disclosure relates to sensitive and specific methods for detecting and/or quantifying a panel of target imitations carried by nucleic acid molecules in a biological sample.
- a panel of at least 20, at least, 40, at least 90, at least 380, or at least 600 target imitations are evaluated using the disclosed methods.
- the panel of target mutations presented in Tables 1 and 2, or any combinations thereof, are evaluated using the disclosed methods.
- the nucleic acid molecule is circulating tumor DNA.
- castPCR comparative ailele-speciiic
- TaqMan PGR assay s are used to detect the panel of target mutations in order to scree the sample for the presence of cancer biom.ark.ers.
- castPCR. assays are carried out according to the methods described in, for example, US Application Nos.
- the present disclosure relates to a. method for providing data for diagnosis, prognosis and/or monitoring of cancer nd or tumour progression in a subject.
- the methods comprise performing castPCR assays to detect and/or quantity different target mutations.
- a panel of at least 20, at least 40, at least 90, at least 380, or at least 600 target mutations are evaluated, using the disclosed methods.
- the panel of target mutations presented in Tables 1 and 2, or any combinations thereof, are evaluated using the disclosed methods.
- the different target mutations are found in DNA associated with various cancers.
- the DMA is circulating tumor-associated. DNA .
- kits for performing the methods of the invention to detect and/or quantify a panel of different target mutations carried by nucleic acid molecules in a biological sample from a subject.
- the panel of different target mutations comprises a set of at least 20, at Ieast40, at least 90, at least 380, or at least 600 target mutations.
- the panel of target mutations presented in Tables 1 and 2, or any combinations thereof, are evaluated using the disclosed kits.
- the kits include compositions comprising reagents needed to carry out castPCR assays on the nucleic acid molecules extracted from the biological sample.
- the compositions of the kits are provided in at least one container, such as a vial, a plate, a tube, or a genecard.
- the present disclosure relates to compositions comprising reagents sufficient to carr out the methods disclosed herein for the detection a panel of target mutations.
- a panel of at least 20, at least 40, at least 90, at least 380, or at least 600 different target mutations are evaluated using the disclosed compositions.
- the panel of target mutations presented in Tables I and 2, or any combinations thereof, are evaluated using the disclosed, compositions.
- the compositions further comprise reagents sufficient to cany out castPCR assays for the detection of the selected panel of target mutations.
- Figure 1 Exemplary schematic of cancer ceils releasing naked DNA. which becomes circulating DNA .
- Figure 2 Exemplary schematic of various mutations (e.g., AS, TP53, BRAE, CT NB I , etc.) frequently found in several cancer cell types.
- various mutations e.g., AS, TP53, BRAE, CT NB I , etc.
- ig re 3 Exemplary list of frequent single nucleotide mutations, having a high recurrence, found in various cancer types.
- FIG. 4 Exemplar schematic of ahele-specific PCR (AS--PCR) used for the detection of mutant al leles.
- Figure 5 Exemplary schematic of cast-PCR used for the detection of mutant alleles.
- castPCR involves an a!eie--specific blocker probe with a blocker moiety (e.g., MGB) at the 5' end.
- components of cast-PCR include the following: one locus-specific TaqMan probe (EST); two MGB blockers: one allele- 1 --specific MGB blocker (MOB! and one ailcle-2-specific MGB blocker (MGB2j; 3 PCR primers: one locus-specific PCR primer (LSP); one allele-] -specific primer (ASP1 ) and one ai!cie-2-specific primer (ASP2).
- Figure 6 Exemplary data showing the detection of the relative copy number of KRAS mutant nucleic acids spiked into a sample of wild type nucleic acids (e.g., i copy of mutant DNA in a background of 2.5 pg wild type genomic DNA) using cast-PCR methods.
- wild type nucleic acids e.g., i copy of mutant DNA in a background of 2.5 pg wild type genomic DNA
- Figure 7 Pooled genomic DNA from multiple cell lines carrying KRAS, EGER, and BRAF mutations run against the Universal BioMarker Test Panel.
- the earliest 4 amplification curves come from reference assays which are designed to detect non-mutated loci in ail samples.
- the remainder of the amplification curves come from assay s which target known mutation hot-spots.
- Figure 8 DNA prepared from serum of three individual cancer patients, PreAmpiified with a custom primer pool, and run against the Universal Biornarker Test Panel, In each sample, the earliest 4 amplification curves come from reference assays which are designed to detect non- mutated loci in all samples. In each sample, there is a. single castPCR mutation assay which gives positive amplification, indicating the mutation present in that patient.
- the present disclosure relates to methods, compositions and kits for the diagnosis, prognosis and/or monitoring of cancer and/or tumor progression in a subject
- the present inventio provides methods tor detecting arid/or quantifying tumor-associated nucleic acids in a biological sample using a panel of universal biomarkers (UBM), wherein the confidence level and/or statistical efficacy of the diagnosis or prognosis is at least 50%, at. least 60%, at. least 70%, at least 80%, at least 90%, or at least 99%.
- the methods involve detection of a panel of the most frequent mutations in cancer, having a high recurrence, to screen biological samples from a subject.
- the methods of the invention may be performed by using detection assays, including, but not limited to, nucleic acid amplification, for the detection and/or quantification of a panel of universal biomarkers m a biological sample.
- Nucleic acid amplification can be performed by a variety of methods known by those of skill in the art, including, for example, polymerase chain reaction (PCR).
- PCR polymerase chain reaction
- castPCR which utilizes a combination of factors to improve discrimination of allelic variants during PCR is used for detection of target mutations.
- casiPCR to detect and/or quantify target imitations in a. sample provides a highly specific and sensitive method for rare mutation detection.
- the methods of the invention are used to detect a target mutation that is present at a frequency less than 1 /10, 1/ 100, 1/1,000, 1/10,000, 1 /100,000, 1/1 ,000,000, 1 / 10,000,000, 1/100,000,000 or 1/1,000,000,000, and any ranges in between.
- the methods disclosed herein are based on the probability of occurrence of at least one positive test per tumor.
- RAS and p53 genes are mutated in 50% of ail tumors.
- hot spots account for 50% of all mutations, as well as a number of other mutations (such as those listed in Figure 3 and '" Fables 1 -3) which also exhibit a high rate of occurrence in a variety of cancer types.
- the probability of frequency for the occurrence of at least 1 mutation, out of a panel of multiple mutations (e.g., at least 40, at least 90, at least 380, or at least 600 target mutations), in one given tumor is equal to or higher than at least 30%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80% or at least 90%.
- Mutations are the specific hallmarks of tumors consisting of somatic acquired sequence variations that distinguish a tumor cell from a normal (e.g., wild type) cell.
- biomarkers there are only a few serum-based biomarkers available, of which, most are used mainly for disease follow-up (e.g,, PSA, CEA, AFP, CA 19-9, CA125, etc.). Moreover, the known and/or available biomarkers are mostly cancer-type dependent, and when used, for cancer detection and/or identification, provide a benefit of less than 30% for all patients.
- Cancer is a disease driven by mutagenesis, which subsequently leads to complex genomic aberrations, ft is well known that cancer cells release nucleic acids, such as DNA and/or RNA, which then becomes circulating DNA/RNA (see, for example. Fig. 1 ).
- nucleic acids such as DNA and/or RNA
- circulating tumor-associated DNA or RNA may carry several mutations.
- tumor-associated DNA or RNA is extremely scarce as compared to the total amount of circulating nucleic acids from, for example, whole blood comprised mainly of normal cells, ' This makes the specific detection of target mutations found on tumor-associated nucleic acids obtained from a mixed population of cells, extremely difficult.
- UBM Universal Biomarkers
- the disclosed methods provides the ability to qualitatively and quantitativel monitor a patient ' s blood for the presence of cancer, which may be positive before surgery/treatment and then may decrease or become negative after surgery/treatment.
- the UBM concept is based on the idea of establishing a panel of known mutations, having the highest frequency of occurrence, and then using this panel to screen tumor- associated nucleic acids for the presence of the selected target mutations. In some embodiments, if any one of the mutations is present then the test is considered positive. In other embodiments, it does not necessarily matter which acquired mutation is detected, what is important is the initial detection of any target mutation from the selected panel, in some preferred embodiments, known markers, whenever positive, can be used as standards for validation and efficacy of the UBM methodology.
- the disclosed methods are based on selecting a UBM panel
- the disclosed methods comprise a means for detecting and/or quantifying a target mutatio carried by tumor-associated DNA. in a biological sample from a sirbject, wherein the confidence level and/or statistical efficacy of the diagnosis or prognosis is at least 50%, at least 60%, at least 70%, at least 80%, at least 90%, or at least 99%.
- the method may comprise, for example:
- methods for the detection and/or quantification of such target mutations can involve molecular detection of cancer-associated or umorigenic nucleic acids.
- the molecular detection can involve nucleic acid amplification methods.
- nucleic acid amplification methods can include, for example, polymerase chain reaction (PGR); hgase chain reaction (LC ); ligation detection reaction (LDR); strand displacement amplification (SDA); multiple displacement
- the detection methods involve the highly sensitive method of castPCR (Compeiitrvejjompeiitrve alieie-speafk TaqMan PGR.
- castPCR uses a combination of allele specific primers having 3 5 -end modified bases. In other embodiments, such primers can also possess tails which enable the use of universal forward primers.
- castPCR methods also use a competitor (allele specific binder; ASB) designed as a minor groove binding allele- specific oligonucleotide which competes with mismatched primers based on thermodynamic differences related to the instability of matched vs. mismatched competitors.
- ASB allele specific binder
- aileie-specific primers are designed so that mismatches are placed central ly to confer instability between mismatched primers.
- detection using castPCR involves the use of a common all ele-speci.fi c probe (e.g., a Taqman PCR probe).
- a pair of oligonucleotide probes is provided for each DNA carrying a mutation.
- each pair of oligonucleotide probes presents the same sequence tails. Irs this case, the sequence rails are called universal sequence tails and the quantitative PCR can be carried out with the same primers in every reaction, !n a second embodiment, each pair of oligonucleotide probes presents different sequence tails from the other pairs of oligonucleotide probes. In this case, specific primers are used in each reaction.
- the detection methods are performed m tubes or on plates, such as .microplat.es.
- plates such as .microplat.es.
- niieorpiates include genecards.
- the microplates can be a 96- el , a 384-wcil, or a 1536-well plates.
- the micropiate in addition to the elements necessary for the detection/quantification of each target mutation carried by tumor-associated the nucleic acid of a particular biological sample, the micropiate also comprises positive and negative controls, in particular, for detection by castPCR.
- the methods further comprise a step of providing a biological sample from a subject, in some embodiments, the subject is preferably a mammalian subject, more preferably a human subject.
- the subject may be suffering from a cancer or bearing a cancer or is suspected to suffer f om a cancer or is at a higher risk for developing cancer.
- the subject has been previously treated for a cancer and/or is being monitored for recurrence of cancer (e.g., in remission).
- the subject may not exhibit clinical evidence of disease.
- the subject can be in the course of treatment and/or have received previous treatment(s) for cancer,
- the biological sample from, the subject is a tissue fragment or a bodily fluid
- the sample can be a mixed sample, for example, comprising both normal (i.e., wild type) and tumorigenic cell types.
- the mixed samples comprise less than about 50%, less than about 25%, less than about 20%, less than about 10%, less than about 5%, less than about ! %, less than about 0.1 %, less than about 0,001% tumorigenic cells.
- the biological sample can be whole blood, plasma, serum, ascites, pleural effusion, urine, lymphatic fluid, stool, broncho ⁇ veolar lavage fluid, ductal lavage fluid, cerebrospinal fluid; fresh, frozen or formalin fixed paraffin embedded tissues; surgical fragments, a tum r biopsy, a fine needle aspiration biospy, and/or circulating tumor cells.
- me blood sample can be, for example, plasma or serum.
- the biological sample, including plasma or serum cars be collected using arsy of a myriad of standard methods known in the art.
- the disclosed methods comprise extraction of nucleic acids from the biological sample
- the biological sample is a frozen, fresh or fixed tissue, or blood sample (including serum or plasma).
- the disclosed methods comprise a step whereby nucleic acids are extracted from the biological sample by any method known by the one skilled in the art. For instance, potentially contaminating cells can be removed from a blood sample, e.g. by coagulation then cenirifugation and/or filtration: and, then proteins that may interfere with the detection of circulating nucleic acids can also be removed, e.g. by proteinase digestion.
- the processed blood sample can then be used directly in an amplification reaction (e.g., castPCR) or can be further purified by second or subsequent removal of additional ceils and proteins.
- isolated circulating DNA can be further extracted by phenol chloroform treatment, precipitation in alcohol arsd dissolution in ars aqneons solution.
- the method can comprise an additional step of purification leading to a circulating nucleic acid level of at least 60%, of at least 65%, of at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% pure.
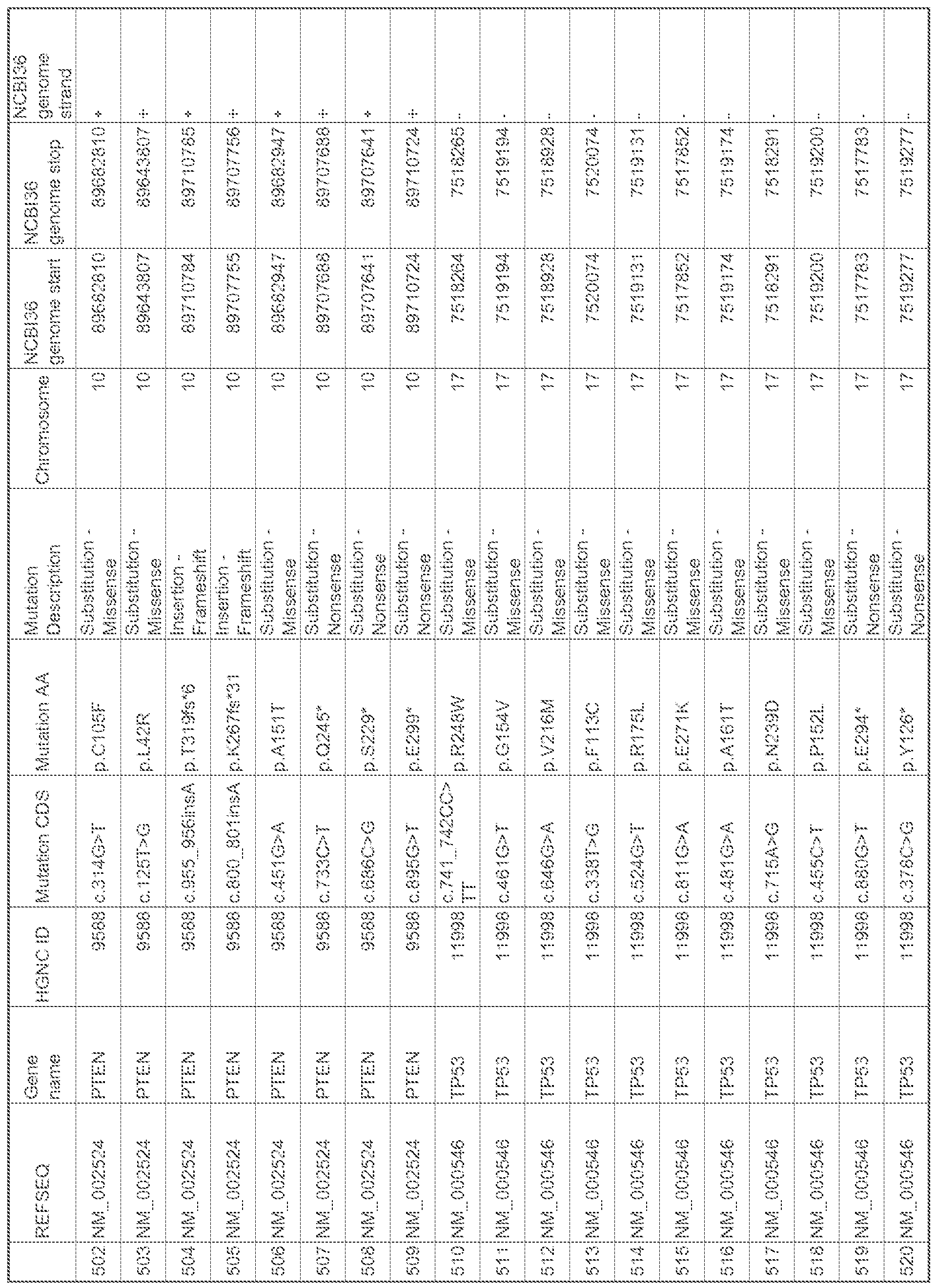
- the UBM panel is selected from a number of target mutations frequently carried by tumor-associated DNA (see, for example, Tables 1-3).
- the target mutations are gene mutations selected from the group consisting of: KRAS, ABLi, SMAD4, NRAS, BRAF, STK1 1 , HRAS, FGFR3, WT1 , CTNNB.1 , MET, TP53, FLT3, VEIL, SRC, APC, RB I , KIT, SMARCB1 , EGFR, CD .N2A, PBKCA, FBXW7, ERBB2, A T1 , i ' .K .
- the set of target mutations earned by circulating tumor-associated DNA. are selected from the group consisting of gene mutations for: KRAS, ABLI , SMAD4, NRAS, BRAF, ST i ! . L .
- TC P i FIR AS, FGFR3, PTCLI, WT1 , CTNNBl, MET, TP53, FLT3, VHP, NF2, SRC, A.PC, RBI, KIT, SMARCB i, EGFR, PTEN, CD .N2A , PI3KCA, FBXW7, CDH.1 , PDGFRA ami ERBB2, Table 3.
- exemplary target mutations Brief description of exemplary target mutations.
- KRAS is Kirsten rat sarcoma viral oncogene homolog. lis NCBI Gene ID is 3845 and the reference mRNA is NM 004985. The mutation position in Table 1 is indicated in this reference mRNA,
- ABL1 s c-abl oncogene 1 , receptor tyrosine kinase. Its NCBI Gene I I ) is 25 and the reference mRNA is XI 416, The mutation position in Table 1 is indicated in this reference mRNA,
- S AD4 is SMAD family member 4. Its NCBI Gene ID is 4089 and the reference mRNA is ' NM 005359. The mntaiion position in Table 1 are indicated in this reference mRNA.
- NRAS neuroblastoma RAS viral (v-ras) oncogene homolog gene. Its NCBI Gene ID is 4893 and the reference mRN A is NM 002524. ' The mutation position m Table 1 are indicated in this reference mRNA .
- BRAF is v-raf murine sarcoma viral oncogene homolog Bl gene. Its NCB Gene ID is 673 and the reference mRNA is NM 004333, The mutation position in Table 1 are indicated i this reference mRNA.
- STKI 1 is serine/threonine kinase I I gene. Its NCBI Gene ID is 6794 and the reference mRNA is NM 000455. The mutation position in Table 1 are indicated in this reference mRNA.
- M EN! is multiple endocrine neoplasia I gene. Its NCBI Gene ID is 4221 and the reference mRNA is ENST00000312049. The mutation position in Table 1 are indicated m this reference mR A,
- TCF1 is HNF1 homeobox A gene. Its CBI Gene I D is 6927 and the reference mRNA is N M 000545, The mutatio position in Table I are indicated in this reference mRNA.
- BRAS is v-Ha-ras Harvey rat sarcoma viral oncogene homolog gene. Its NCBI Gene ID is 3265 and the reference mRN A is M ⁇ 005343. The imitation position in Table 1 are indicated in this reference mRNA.
- FGFR3 is fibroblast growth factor receptor 3 gene. Its NCBI Gene ID is 2261 and the reference mRNA is NM 000142, The mutation position in Table 1 are indicated in this reference mR A,
- MET is met proto-oncogene gene. Its NCBI Gone ID is 4233 and the reference mRNA is NM 000245. The miuation position in Table 1 are indicated in this reference mRNA.
- TP53 is : ⁇ ! ⁇ : ⁇ ⁇ : protein p53 gene.
- NCBI Gene ID is 7157 and the reference mRNA is NM 000546.
- the mutation position in Table 1 are indicated in this reference mRNA .
- FLT3 is fms-related tyrosine kinase 3 gene, its NCBI Gene iD is 2322 and the reference mRNA is Z26652, The mutation position in 1 able I are indicated in this reference mRNA.
- VHL is von Hippel-Lindau tumor suppressor gene.
- lis NCBI Gene ID is 7428 and the reference mRNA Is NM 000551. The mutation position in Table 1 are indicated in this reference mRNA.
- NF2 is neurofibroma 2 gene. Its NCBI Gene ID is 4771 and the reference mRNA is NM 000268. The mutation position in Table 1 are indicated in this reference mRNA . SRC " is v-src sarcoma gene. Its NCBI Gene ID is 6714 and the reference mRNA. is NM 005417. The mutation position n fable I are Indicated in this reference mRNA. APC :.s adenomatosis polyposis cob gene. Its CBI Gene ID Is 324 and the reference mRNA is NM 000038. The mutation position in Table 1 are indicated in this reference mRNA.
- RBI I retoniblastoma I gene. Its NCBI Gene ID is 5925 and the reference mRNA is NM 000321. The mutation position in Table 1 are indicated m this reference mR A, KIT is v-kit Hardy-Znckerrnan 4 felme sarcoma viral oncogene horno!og gene. Its NCBI Gene ID is 3815 and the reference mRNA is NM 000222. The mutation position m Table I are indicated in this reference mRNA.
- SMARCB1 is S W /SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 gene. Its NCBI Gene ID is 6598 and die reference mRNA is NM 003073. The mutation position in Table 1 are indicated in this reference mRNA.
- EGFR epidermal growth actor receptor gene. Its NCBI Gene ID is 1956 and the reference mRNA is N 005228. The mutation position in Table 1 are indicated in this reference mRN A .
- PTEN is phosphatase and tensin homoiog gene.
- NCBI Gene ID is 5728 and the reference rnRNA is NM 000314.
- the mutation position in Table 1 are indicated in this reference mRNA.
- CDKN2A is cyclfn-dependent kinase inhibitor 2A gene. Its NCBI Gene ID is 1 029 and the reference mRNA is NM 000077. The imitation position in Table I are indicated in this reference mRNA.
- PI3KCA is phosphoinosiiide-3-kina.se, catalytic, alpha gene. Its NCBI Gene ID is 5290 and the reference mRNA is NM 0062 1 8. The mutation position m '" Fable 1 are
- FBXW7 is F-box and WD repeat domain containing 7 gene. Its NCBI Gene ID is
- CDH 1 is cadherin 1 gene. Its NCBI Gene 03 is 999 and the reference mRNA is
- NM 004360 The mutation position in Table I are indicated in this reference mRNA.
- PDGFRA platelet-derived growth factor receptor, alpha gene. Its NCBI Gene ID is 5156 and the reference mR A is N M 006206. The mutation position m '" Fable 1 are indicated in this reference mRNA.
- ERBB2 is v-erb ⁇ b2 erythroblastic leukemia viral oncogene homoiog 2 gene. Its NCBI Gene J D is 2064 and the reference mRNA is NM 004448. The mutation position in
- the target mutations of the panel are selected from the group consisting of those disclosed in Table I or 2, or any combinations thereof
- the mutations of the panel are selected in order to obtain a universal blomarker panel allowing the detection of cancer independently of its type.
- the panel is selected based o the most frequent occurrence of mutations in a. variety of cancer types.
- the frequency of occurrence of the target mutations selected for the panel occur in at least 50%, at least 55%, at least 60%, at least 65%>, at least 70%, at least 75% ; , at least 80%, at least 85%, at least 90%, or at least 95% of all tumors and/or cancer types.
- the disclosed methods are performed using a panel of
- the panel of mutations comprises at least 20, at least 40, at least 90, at least 120, at least 140, at least 160, at least 180, at least 200, at least 220, at least 240, at least 260, at least 270, at least 300, at least 320, at least 340, at least 360, at least 380, at least 400, at least 420, at least 460, at least 480, at least 520, at least 560, at least 600, at least 640, at least 660 or at least 700 different mutations.
- the panel may further comprise controls for the detection of wild type sequences and/or additional reference samples as discussed below.
- the target mutations to be detected may comprise a.
- Insertions may typical ly comprise an addition of between 1 and 40, between 5 and 35, between 10 and 30, or between 15 and 25 contiguous base pairs.
- Deletions may typically comprise a removal of between 1 and 40, between 5 and 35, between 10 and 30, or between 15 and 25 contiguous base pairs.
- substitutions may typically comprise a substitution of between 1 and 20, between 5 and 15, or between 8 and 12 contiguous base pairs.
- the target mutations comprise a. single point imitation.
- the nucleic acid sample can be DNA (e.g., gDNA).
- the present invention may be used for the detection of variation in genomic DNA whether human, animal or other.
- the nucleic acid sample can be RNA (e.g., mRNA),
- the DNA or RNA can be circulating DNA or RNA.
- the DNA or RNA can be circulating DNA or RNA, In some embodiments, the circulating DNA or RNA is tumor-related DNA or RNA.
- die tumor-related DNA or RNA is less than about 50%, less than about 25%, less than about 20%, less than about 10%, less than about 5%, less than about 1%, less than about 0.1 %, less than about 0.001 % of the total nucleic acid sample obtained from a biological sample.
- the tumor-related circulating DMA or RNA. compared to wild type DNA or RNA is present at a frequency less than 1/10 copies, 1/100 copies, 1/1,000 copies, 1/10,000 copies, 1/100,000 copies, 1/1,000,000 copies, 1/10,000,000 copies, 1/100,000,000 copies or 1/1,000,000,000 copies.
- the disclosed methods further comprise a step whereby the set of target mutations earned by tumor-associated nucleic acids are compared to a reference or control sample (e.g., wild type sample).
- a reference or control sample e.g., wild type sample.
- the comparison of the set of target imitations earned by tumor-associated nucleic acids to a reference allows the determination of a probability of the subject to be at risk of developing a cancer, to suffer from a cancer, to have a good or bad prognosis (for instance, a complete remittance, or a metastasis), to have a recurrence, or responsiveness to a particular treatment.
- the reference can be a positive reference (e.g., healthy profile, treatment responder profile, no or low risk of metastasis profi le, no or low risk of recurrence profi le) or a negative reference (e.g., cancer profile, treatment non- or low-responder profile, high nsk of metastasis profile, high risk of recurrence profile).
- a positive reference e.g., healthy profile, treatment responder profile, no or low risk of metastasis profi le, no or low risk of recurrence profi le
- a negative reference e.g., cancer profile, treatment non- or low-responder profile, high nsk of metastasis profile, high risk of recurrence profile.
- the present invention provides compositions tor nse in identifying and/or quantitating target imitations in nucleic acid sample based on a panel of UBiVI .
- such compositions can comprise PCR reagents used for the detection and/or quantification of target imitations in a nucleic acid sample based on a panel of UBM.
- these compositions can comprise reagents used for casiPCR.
- the reagents used for castPC can include, tor example: (a) an allele-specific primer; (b) an allele-specific blocker probe; (c) a detector probe; and/or id) a locus-specific primer, wherein the allele-specific primers are targeted to mutation selected from the IJ B panel of target mutations.
- the compositions may further compri.se a. polymerase, dNTPs, reagents and/or buffers suitable for PCR amplification, and/or a template sequence or nucleic acid sample.
- kits for the diagnosis, prognosis and monitoring a cancer and/or tumour progression in a subject In another aspect, the present invention provides kits for providing data for diagnosis, prognosis and monitoring a cancer and/or tumour in a subject is provided. In some embodiments, the kit comprises a microp!ate. In yet other embodiments, the rnicroplate's wel l may comprise:
- reagents used to perform a detection method such as, but not limited to ca&iPC R.
- reagents may include, for example, a pair of primers: reagents for extracting circulating DNA; and/or a combination thereof
- the compositions, methods, and/or kits can be used in detecting circulating ceils in diagnosis.
- the compositions, methods, and/or kits can be used to detect tumor cells in blood for early cancer diagnosis, such for screening, for early detection or early detection of recurrence of cancer and/or tumors.
- the compositions, methods, and/or kits can be used for cancer or disease-associated genetic variation or somatic mutation detection and validation .
- the compositions, methods and/or kits can be used for treatment identification.
- compositions, methods, and/or kits can be used for genotyping tumors and assessing the status of the mutations most used today in clinical management (e.g., EFGR, KRAS) which is typically performed by sequencing and is usually considered extremely long and costly.
- EFGR e.g., EFGR, KRAS
- a panel of 100 patients with frequent solid tumors, treated with curative surgery are initially selected.
- Tumoral DNA is extracted from residual frozen tumors, and blood samples (a minimum of 10 mL) are collected one day before surgery and then after surgery every 2 weeks, and stored as plasma samples.
- the patients' status of current known seric bi.oma.rk.ers is known.
- a test comprising a UB panel of target mutations is first applied to explore tumoral DNA and to assess whether tested samples are found to be positive or negative for any of the UBM panel of target mutations.
- a positive test means detection of at least one mutation out of the 384 total tested . Occurrence frequency is estimated as % of tumor in which the UBM test is positive. If the test is positive for tumoral DNA, then the given mutation is further tested in plasma. Detection is evaluated before treatment and after treatment. Current existent biomarkers, whenever available are used as gold standards to validate the UBM panel.
- UBM test is positive in 50% of tumors. Percentage may be different according to tumor type.
- UBM panel of target mutations Use of cast-PC for detection of a UBM panel of target mutations in biological samples in this example, casi-PCR (see, for example, US Application Nos. 12/641 ,321 and 12/748,329, the contents of which are herein incorporated by reference in their entireties) is used to detect and determine the percentage of tumor cells in biological samples.
- Various biological samples, comprising both tumorigenic and nonnai cells, are obtained and assayed by cast-PCR. for the presence of a number of target mutations associated with cancer based on a panel of Universal Biomarkers (UBM ).
- UBM panel of target mutations can include, for example, any of those mutations shown in Figure 2 as well as those listed in Table 1 and Table 2, or any combinations thereof
- both allele- 1 and alleie-2 assays are performed using the general cast-PCR schema ami reaction conditions indicated below, using gDNA derived from either normal or tumor samples. Assay primers and probes corresponding to the mutations shown in Table I and Table 2 are designed accordingly.
- the cast-PCR assay reaction mixture for analysis of aliele-2 included a tai led allele-2-specific primer (ASP2), one MGB a! !eie-l blocker probe (MGB i), one common locus-specific TaqMan probe (LST) and one common locus-specific primer (LSP).
- ASP2 tai led allele-2-specific primer
- MGB i MGB a! !eie-l blocker probe
- LST common locus-specific TaqMan probe
- LSP common locus-specific primer
- reaction mixtures are prepared, as follows:
- nuclease-free 1.5mL microcentrifuge tube or PCR plate For each sample (to be run in duplicate), the following reagents are pipetted into a nuclease- free 1.5mL microcentrifuge tube or PCR plate:
- #Replicate volumes include 20% excess to compensate for volume loss from pipetting.
- DNA template is added to the reactions as the last step to eliminate template co tamination.
- € When using 48-weii or 96-well plates, reactions are scaled up to 20 ⁇ volumes by doubling the volume of each reaction component.
- Thermal cyclirig is earned out as follows: 95°C/10 min, (92°C/15 sec, 58°C/1 min) for 5 cycles, then (92°C/15 sec, 60°C/ 1 m n) for 45 cycles.
- ⁇ PC reactions were run for a total of 50 cycles. (The 50 cycles can include a pre-run of five cycles at a lower annealing/extension temperature followed by an additional 45 cycles at a higher annealing/extension temperature).
- the ACt between amplification reactions for Allele- 1 assays and Allele-2 assays are calculated.
- the ACt can be defined as the specificity of castPCR (ACt :::: ⁇ ;; : . l: . ; - !i 3 ⁇ 4 ji i - 2 >-
- the 2 ; " ' value is used to estimate the power of discrimination (or selectivity ⁇ which is equal to 1 ⁇ 2" '' or, calculated as % (1 ⁇ 2 ACt x 100).
- preampiification reactions comprising DNA samples in each well are set-up as follows: Reagent Per reaction Final Concentration
- Thermocycling conditions are sei--up on a GeneAmp PC System 9700 to run pre- amplification reactions as follows: 95°C for 10 min, [95 C C for 15 sec - 60°C for 4 mm] for 10 cycles. 99.9°C for 10 uin and 4 " C on hold for np to 24 rs.
- reaction plates are removed from the theraiocyc!er and placed on ice.
- PreAmp 5X is diluted by adding 80 id . of d 20.
- Wells are mixed by pipetting up and down 5 times (being cautious to avoid any cross-contamination) and spun briefly.
- Prearn lification products are stored at 0 " C or used directly in standard castPCR assays.
- 1-20% of diluted PreAmp products are added into each castPCR reaction according to the number of targets (up to 96) and PCR. replicates are applied.
Abstract
Disclosed herein are methods for the diagnosis, prognosis and/or continuous monitoring of cancer and/or tumor progression in a subject. More specifically, the present invention relates to methods for detecting and/or quantifying tumoral-associated nucleic acids in a biological sample using a panel of cancer-specific markers, independent of cancer type, for the molecular detection of a broad range of cancer types for diagnostic, prognostic and therapeutic purposes.
Description
METHODS, COM POSITIONS AND KITS FOR THE DIAGNOSIS, PROGNOSIS
AND MONITORING OF CANCER
FIELD OF THE DISCLOSURE
Disclosed herein are methods for the diagnosis, prognosis and/or continuous monitoring of ca cer and/or tumor progression in a subject using universal biomarkers (UBMs). More specifically, the present invention relates to methods for detecting and/or quantifying tumor-associated nucleic acids in a biological sample using a panel of cancer- specific UBMs, Independent of cancer type, for the molecular detection of a broad range of cancer types tor diagnostic, prognostic and therapeutic purposes.
BACKGROUND INFORMATIO
Early diagnosis, evaluation of prognosis and/or follow-up of cancer are crucial challenges to enhance the effi cacy of medical care of cancer patients and to ameliorate the prognosis of the disease.
The diagnosis can invol ve the evaluation of circulating biomarkers of cancer or tissues biomarkers. Usually, circulating biomarkers are proteins secreted by tumor cells which can be detected in serum by antibody-based assays such as I d . ISA Today, there are only a few biomarkers available and they aren't associated with sufficient specificity and/or sensitivity for cancer detection. Most of the biomarkers are usable mainly for disease fol low-up. A further drawback is that they are cancer-type dependent (for example, PSA, CEA, AEP,
CA19-9, CA125, etc.). When such biomarkers are positive, they are very useful to monitor efficacy of therapeutics and to detect recurrence of cancer. However one of the problems with their use is that for each type of cancer, not ail patients are positive, and therefore the overall benefit of each biomarker does not exceed more than 30% of all patients for each cancer type, It is known that tumor cells release nucleic acids. Such nucleic acids are round m blood circulation and are called ''circulating DNA or RNA." Whole circulating DNA. or RNA isolated from blood is, for the most part, DNA or RNA released by normal cells and, to a more limited extent, DNA or RNA released by tumor cells (also referred to as tumorigenic,
tumor-related or tumor-associated DNA or RNA), For years, researchers have tried to detect circulating nucleic acids from tumors, and have confirmed the presence of tumor circulating DNA or RNA, identified by molecular abnormalities associated whh tumor cells such as oncogene mutations, by satellite instability, or by irypermethyl iion of genes. A number of publications describe methods for detecting cancers based on the identification of specific genetic alterations in circulating DNA or RNA. For instance, US Patent No. 5,496,699 discloses a method for detecting imitations m nucleic acid sequences, in particular the sequence of the KRAS gene, in biological fluids such as blood, serum or plasma. US Patent No. 5,068,175 discloses a method for detecting the presence of k AS oneogene-rela ed malignancies in whic the gene is quantified in serum or plasma samples. WO 01 /42504 discloses the determination of extracellular nucleic acid, lor example DNA of KRA S and APC genes, in serum or plasma samples, for the evaluation of the nsk factors related with a number of neoplastic diseases. WO 02/18652 discloses a method of quail/quantitative detection of human ieiomerase RNA and ieiomerase reverse transcriptase R A in plasma or serum for the diagnosis, monitoring, treatment or evaluation of a number of neoplastic diseases.
However, the detection of turner-related circulating rmcleic acids are difficult to detect and it appears that none of the work-to-date has led to the development of an efficient circulating biornarker useful for diagnosis and/or continuous monitoring of subjects. Ί ne above-mentioned works only disclose methods for detenmning the presence of tumor- associated nucleic acids in some subjects with a particular type of cancer by analyzing the sequence abnormalities in a single target gene. In addition, the detection sensitivity of these methods is too low and, in most cases, the particular assays that are typically used are based on a single target gene (e.g., KRAS or TP53) and are not reliable for clinical use.
In addition, when employing single gene analysis for identification or diagnosis of a particular type of cancer, the methods used for deiecting/quaniifymg the tumor-associated circulating nucleic acid must be highly sensitive and robust. The proposed method lor these types of assays has typically involved the use of quantitative polymerase chain reaction (qPC ) using alieie-specifk primers, also known as ailele-specific PCR (AS-PCR),
However, AS-PCR presents several drawbacks. In particular, the detected signal can be skewed due to some PCR artefacts associated with cross-reactions with the PCR primers (e.g., mismatched amplification and/or primer dirner amplification). These artefacts usually
appear more frequently for PGR reactions using a high number of amplification cycles necessar to detect very low abundant targets, such as circulating DNA. Moreover, the specificity of conventional AS-PC is usually less than 0,1 % and the efficacy often depends on type of mismatch being detected (e.g., ( ·;( ;. A:A »G:T>C:T).
Thus, there is a need in the art to help solve the issue as to which particular mutations, independent of cancer type, should be tested in order to provide some statistical efficacy, as well as what methods to use for the detection of extremely low amounts (e.g., 1 in >1 ,000) of tumorigenic nucleic acids in a mixed sample comprising mainly norma! nucleic acid molecules. SUM ARY OF THE INVENTION
Disclosed herein are methods for the diagnosis, prognosis and/or monitoring of cancer and/or tumor progression in a subject w id; higher degree of statistical confidence level. In some embodiments, the present disclosure relates to sensitive and specific methods for detecting and/or quantifying a panel of target imitations carried by nucleic acid molecules in a biological sample.
In some embodiments, a panel of at least 20, at least, 40, at least 90, at least 380, or at least 600 target imitations are evaluated using the disclosed methods. In some embodiments, the panel of target mutations presented in Tables 1 and 2, or any combinations thereof, are evaluated using the disclosed methods. In some embodiments, the nucleic acid molecule is circulating tumor DNA. In some embodiments, castPCR. (competitive ailele-speciiic
TaqMan PGR) assay s are used to detect the panel of target mutations in order to scree the sample for the presence of cancer biom.ark.ers. In some embodiments, castPCR. assays are carried out according to the methods described in, for example, US Application Nos.
12/641 ,321 and. 12/748,329, the contents of which are herein incorporated by reference in their entireties.
In yet other embodiments, the present disclosure relates to a. method for providing data for diagnosis, prognosis and/or monitoring of cancer nd or tumour progression in a subject. In some embodiments, the methods comprise performing castPCR assays to detect and/or quantity different target mutations. In some embodiments, a panel of at least 20, at least 40, at least 90, at least 380, or at least 600 target mutations are evaluated, using the disclosed methods. In some embodiments, the panel of target mutations presented in Tables 1 and 2, or any combinations thereof, are evaluated using the disclosed methods. In some
embodiments, the different target mutations are found in DNA associated with various cancers. In some embodiments, the DMA is circulating tumor-associated. DNA .
In other embodiments, the present disclosure relates to kits for performing the methods of the invention to detect and/or quantify a panel of different target mutations carried by nucleic acid molecules in a biological sample from a subject. In some
embodiments, the panel of different target mutations comprises a set of at least 20, at Ieast40, at least 90, at least 380, or at least 600 target mutations. Irs some embodiments, the panel of target mutations presented in Tables 1 and 2, or any combinations thereof, are evaluated using the disclosed kits. In other embodiments, the kits include compositions comprising reagents needed to carry out castPCR assays on the nucleic acid molecules extracted from the biological sample. In other embodiments, the compositions of the kits are provided in at least one container, such as a vial, a plate, a tube, or a genecard.
In other embodiments, the present disclosure relates to compositions comprising reagents sufficient to carr out the methods disclosed herein for the detection a panel of target mutations. In some embodiments, a panel of at least 20, at least 40, at least 90, at least 380, or at least 600 different target mutations are evaluated using the disclosed compositions. In some embodiments, the panel of target mutations presented in Tables I and 2, or any combinations thereof, are evaluated using the disclosed, compositions. In some embodiments, the compositions further comprise reagents sufficient to cany out castPCR assays for the detection of the selected panel of target mutations.
Additional advantages of the disclosed methods, compositions and kits are in the description which follows, and in part are understood from the description, or may be learned by practice of the disclosed methods, compositions and/or kits. The advantages of the disclosed methods, compositions and/or kits are realized and attained by means of the elements and combinations particularly pointed out in the appended claims. It is to be understood that both the foregoing general description and the following detailed description are exemplary and explanatory only and are not restrictive.
BRIEF DISCRETION OF THE DRAWINGS
The skilled artisan will understand that the drawings described below are for il lustration purposes only. The drawings are not intended to limit the scope of the present teachings in any way.
Figure 1 : Exemplary schematic of cancer ceils releasing naked DNA. which becomes circulating DNA .
Figure 2: Exemplary schematic of various mutations (e.g., AS, TP53, BRAE, CT NB I , etc.) frequently found in several cancer cell types.
ig re 3: Exemplary list of frequent single nucleotide mutations, having a high recurrence, found in various cancer types.
Figure 4: Exemplar schematic of ahele-specific PCR (AS--PCR) used for the detection of mutant al leles.
Figure 5: Exemplary schematic of cast-PCR used for the detection of mutant alleles. In some exemplary embodiments, castPCR involves an a!!eie--specific blocker probe with a blocker moiety (e.g., MGB) at the 5' end. In some embodiments, components of cast-PCR include the following: one locus-specific TaqMan probe (EST); two MGB blockers: one allele- 1 --specific MGB blocker (MOB!) and one ailcle-2-specific MGB blocker (MGB2j; 3 PCR primers: one locus-specific PCR primer (LSP); one allele-] -specific primer (ASP1 ) and one ai!cie-2-specific primer (ASP2).
Figure 6: Exemplary data showing the detection of the relative copy number of KRAS mutant nucleic acids spiked into a sample of wild type nucleic acids (e.g., i copy of mutant DNA in a background of 2.5 pg wild type genomic DNA) using cast-PCR methods.
Figure 7: Pooled genomic DNA from multiple cell lines carrying KRAS, EGER, and BRAF mutations run against the Universal BioMarker Test Panel. In each sample, the earliest 4 amplification curves come from reference assays which are designed to detect non-mutated loci in ail samples. The remainder of the amplification curves come from assay s which target known mutation hot-spots.
Figure 8: DNA prepared from serum of three individual cancer patients, PreAmpiified with a custom primer pool, and run against the Universal Biornarker Test Panel, In each sample, the earliest 4 amplification curves come from reference assays which are designed to detect non- mutated loci in all samples. In each sample, there is a. single castPCR mutation assay which gives positive amplification, indicating the mutation present in that patient.
DETAILED DESCRIPTION OF THE INVENTION
As ernbodied and broadly described herein, the present disclosure relates to methods, compositions and kits for the diagnosis, prognosis and/or monitoring of cancer and/or tumor progression in a subject ,
In a first aspect, the present inventio provides methods tor detecting arid/or quantifying tumor-associated nucleic acids in a biological sample using a panel of universal biomarkers (UBM), wherein the confidence level and/or statistical efficacy of the diagnosis or prognosis is at least 50%, at. least 60%, at. least 70%, at least 80%, at least 90%, or at least 99%. In some embodiments, the methods involve detection of a panel of the most frequent mutations in cancer, having a high recurrence, to screen biological samples from a subject. Some non-limiting examples of such mutations are presented in Figure 3 and in Tables 1 and 2, A panel of muta tions selected from those listed in Figure 3 and Tables 1 and 2, or any combinations thereof, may be used in the disclosed methods. Using a UBM panel of mutations to screen a biological sample from a subject provides the advantage of providing universal detection of biomarkers independent of the type of cancer being analyzed.
The methods of the invention may be performed by using detection assays, including, but not limited to, nucleic acid amplification, for the detection and/or quantification of a panel of universal biomarkers m a biological sample. Nucleic acid amplification can be performed by a variety of methods known by those of skill in the art, including, for example, polymerase chain reaction (PCR). in some preferred embodiments, castPCR, which utilizes a combination of factors to improve discrimination of allelic variants during PCR is used for detection of target mutations. Using casiPCR to detect and/or quantify target imitations in a. sample provides a highly specific and sensitive method for rare mutation detection. In certain embodiments, the methods of the invention are used to detect a target mutation that is present at a frequency less than 1 /10, 1/ 100, 1/1,000, 1/10,000, 1 /100,000, 1/1 ,000,000, 1 / 10,000,000, 1/100,000,000 or 1/1,000,000,000, and any ranges in between.
Combining UBM analysis with castPCR methodology provides a novel and robust tool based on the highly sensitive detection of a number of hotspot muta tions found in a variety of solid tumors. In some embodiments, the methods disclosed herein are based on the probability of occurrence of at least one positive test per tumor. For example, RAS and p53 genes are mutated in 50% of ail tumors. Moreover, hot spots account for 50% of all mutations, as well as a number of other mutations (such as those listed in Figure 3 and '"Fables 1 -3) which also exhibit a high rate of occurrence in a variety of cancer types. Thus, in some exemplary embodiments, the probability of frequency for the occurrence of at least 1 mutation, out of a panel of multiple mutations (e.g., at least 40, at least 90, at least 380, or at
least 600 target mutations), in one given tumor is equal to or higher than at least 30%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80% or at least 90%.
Use of Universal Biomarkers (UBM) for Detection of Cancer
Mutations are the specific hallmarks of tumors consisting of somatic acquired sequence variations that distinguish a tumor cell from a normal (e.g., wild type) cell.
However, there are only a few serum-based biomarkers available, of which, most are used mainly for disease follow-up (e.g,, PSA, CEA, AFP, CA 19-9, CA125, etc.). Moreover, the known and/or available biomarkers are mostly cancer-type dependent, and when used, for cancer detection and/or identification, provide a benefit of less than 30% for all patients.
Cancer is a disease driven by mutagenesis, which subsequently leads to complex genomic aberrations, ft is well known that cancer cells release nucleic acids, such as DNA and/or RNA, which then becomes circulating DNA/RNA (see, for example. Fig. 1 ). Such circulating tumor-associated DNA or RNA may carry several mutations. However such tumor-associated DNA or RNA is extremely scarce as compared to the total amount of circulating nucleic acids from, for example, whole blood comprised mainly of normal cells, 'This makes the specific detection of target mutations found on tumor-associated nucleic acids obtained from a mixed population of cells, extremely difficult.
The novel concept provided herein, involves the use of a Universal Biomarkers (UBM) to address the challenging issue of a universal test for cancer, which is independent of cancer-type. This is especially important for the detection of early recurrences, amongst the other uses. Thus, i some particular embodiments, tissues are collected during
surgery/treatment and plasma is obtained prior to surgery/treatment and then on a regular basis (e.g,, monthly) following surgei /treatment for analysis using the disclosed methods. Accordingly, and in some embodiments, the disclosed methods provides the ability to qualitatively and quantitativel monitor a patient' s blood for the presence of cancer, which may be positive before surgery/treatment and then may decrease or become negative after surgery/treatment.
The UBM concept is based on the idea of establishing a panel of known mutations, having the highest frequency of occurrence, and then using this panel to screen tumor- associated nucleic acids for the presence of the selected target mutations. In some embodiments, if any one of the mutations is present then the test is considered positive. In
other embodiments, it does not necessarily matter which acquired mutation is detected, what is important is the initial detection of any target mutation from the selected panel, in some preferred embodiments, known markers, whenever positive, can be used as standards for validation and efficacy of the UBM methodology.
In some embodiments, the disclosed methods are based on selecting a UBM panel
(determined using, for example, cancer profiler tools) based on the most frequent, known hot spot mutations across all types of solid tumors (e.g., RAS, BRAF, p53, etc.), and then performing a sensitive method for the detection and/or quantification of such target mutations.
Thus, in another aspect, the disclosed methods comprise a means for detecting and/or quantifying a target mutatio carried by tumor-associated DNA. in a biological sample from a sirbject, wherein the confidence level and/or statistical efficacy of the diagnosis or prognosis is at least 50%, at least 60%, at least 70%, at least 80%, at least 90%, or at least 99%. The method may comprise, for example:
a) extracting circulating DNA from the biological sample; and
b) performing a detection method based on a UBM panel of target mutations.
n some embodiments of the disclosed methods, methods for the detection and/or quantification of such target mutations can involve molecular detection of cancer-associated or umorigenic nucleic acids. In some embodiments, the molecular detection can involve nucleic acid amplification methods. Such nucleic acid amplification methods can include, for example, polymerase chain reaction (PGR); hgase chain reaction (LC ); ligation detection reaction (LDR); strand displacement amplification (SDA); multiple displacement
amplification (MDA); ol igonucleotide ligation assay (OLA); or any other available detection methods known in the art. In preferred embodiments, the detection methods involve the highly sensitive method of castPCR (Compeiitrvejjompeiitrve alieie-speafk TaqMan PGR.
In some embodiments, castPCR uses a combination of allele specific primers having 35 -end modified bases. In other embodiments, such primers can also possess tails which enable the use of universal forward primers. In some embodiments, castPCR methods also use a competitor (allele specific binder; ASB) designed as a minor groove binding allele- specific oligonucleotide which competes with mismatched primers based on thermodynamic differences related to the instability of matched vs. mismatched competitors. In some embodiments, aileie-specific primers are designed so that mismatches are placed central ly to
confer instability between mismatched primers. In some embodiments, detection using castPCR involves the use of a common all ele-speci.fi c probe (e.g., a Taqman PCR probe).
The following is detailed summar regarding performing a castPCR assay. In some embodiments of the disclosed methods, a pair of oligonucleotide probes is provided for each DNA carrying a mutation. In a first embodiment, each pair of oligonucleotide probes presents the same sequence tails. Irs this case, the sequence rails are called universal sequence tails and the quantitative PCR can be carried out with the same primers in every reaction, !n a second embodiment, each pair of oligonucleotide probes presents different sequence tails from the other pairs of oligonucleotide probes. In this case, specific primers are used in each reaction.
In some embodiments, the detection methods are performed m tubes or on plates, such as .microplat.es. Such examples of niieorpiates include genecards. In some embodiments the microplates can be a 96- el , a 384-wcil, or a 1536-well plates. Preferably, in addition to the elements necessary for the detection/quantification of each target mutation carried by tumor-associated the nucleic acid of a particular biological sample, the micropiate also comprises positive and negative controls, in particular, for detection by castPCR.
In some embodiments, the methods further comprise a step of providing a biological sample from a subject, in some embodiments, the subject is preferably a mammalian subject, more preferably a human subject.
in yet other embodiments, the subject may be suffering from a cancer or bearing a cancer or is suspected to suffer f om a cancer or is at a higher risk for developing cancer. In additional embodiments, the subject has been previously treated for a cancer and/or is being monitored for recurrence of cancer (e.g., in remission). In another embodiment, the subject may not exhibit clinical evidence of disease. Alternatively, the subject can be in the course of treatment and/or have received previous treatment(s) for cancer,
In some embodiments, the biological sample from, the subject is a tissue fragment or a bodily fluid, in other embodiments, the sample can be a mixed sample, for example, comprising both normal (i.e., wild type) and tumorigenic cell types. In some embodiments, the mixed samples comprise less than about 50%, less than about 25%, less than about 20%, less than about 10%, less than about 5%, less than about ! %, less than about 0.1 %, less than about 0,001% tumorigenic cells.
In some embodiments the biological sample can be whole blood, plasma, serum, ascites, pleural effusion, urine, lymphatic fluid, stool, broncho ί veolar lavage fluid, ductal
lavage fluid, cerebrospinal fluid; fresh, frozen or formalin fixed paraffin embedded tissues; surgical fragments, a tum r biopsy, a fine needle aspiration biospy, and/or circulating tumor cells. In some preferred embodiments, me blood sample can be, for example, plasma or serum. Irs some embodiments, the biological sample, including plasma or serum, cars be collected using arsy of a myriad of standard methods known in the art.
In some embodiments, the disclosed methods comprise extraction of nucleic acids from the biological sample, In preferred embodiments, the biological sample is a frozen, fresh or fixed tissue, or blood sample (including serum or plasma). In some embodiments, the disclosed methods comprise a step whereby nucleic acids are extracted from the biological sample by any method known by the one skilled in the art. For instance, potentially contaminating cells can be removed from a blood sample, e.g. by coagulation then cenirifugation and/or filtration: and, then proteins that may interfere with the detection of circulating nucleic acids can also be removed, e.g. by proteinase digestion. The processed blood sample can then be used directly in an amplification reaction (e.g., castPCR) or can be further purified by second or subsequent removal of additional ceils and proteins. For instance, isolated circulating DNA can be further extracted by phenol chloroform treatment, precipitation in alcohol arsd dissolution in ars aqneons solution. Accordingly, and in addition to extraction of circulating nucleic acids, the method can comprise an additional step of purification leading to a circulating nucleic acid level of at least 60%, of at least 65%, of at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% pure.
In some preferred embodiments, the UBM panel is selected from a number of target mutations frequently carried by tumor-associated DNA (see, for example, Tables 1-3). In some embodiments the target mutations are gene mutations selected from the group consisting of: KRAS, ABLi, SMAD4, NRAS, BRAF, STK1 1 , HRAS, FGFR3, WT1 , CTNNB.1 , MET, TP53, FLT3, VEIL, SRC, APC, RB I , KIT, SMARCB1 , EGFR, CD .N2A, PBKCA, FBXW7, ERBB2, A T1 , i' .K . ATM, CSF I R, ERBB4, G AS, !TNPIA, IDEi l, JA .2, KDR, M P! .. NOTCH! , NP I, RET, SMQ, UTX, CDFI I, MEN1 , NF2, PDGFRA, PTCFJ, PTEN and TCFL In more preferred embodiments, the set of target mutations earned by circulating tumor-associated DNA. are selected from the group consisting of gene mutations for: KRAS, ABLI , SMAD4, NRAS, BRAF, ST i ! . L . TC P i , FIR AS, FGFR3, PTCLI, WT1 , CTNNBl, MET, TP53, FLT3, VHP, NF2, SRC, A.PC, RBI, KIT, SMARCB i, EGFR, PTEN, CD .N2A , PI3KCA, FBXW7, CDH.1 , PDGFRA ami ERBB2,
Table 3. Brief description of exemplary target mutations.
KRAS is Kirsten rat sarcoma viral oncogene homolog. lis NCBI Gene ID is 3845 and the reference mRNA is NM 004985. The mutation position in Table 1 is indicated in this reference mRNA,
ABL1 :s c-abl oncogene 1 , receptor tyrosine kinase. Its NCBI Gene I I ) is 25 and the reference mRNA is XI 416, The mutation position in Table 1 is indicated in this reference mRNA,
S AD4 is SMAD family member 4. Its NCBI Gene ID is 4089 and the reference mRNA is 'NM 005359. The mntaiion position in Table 1 are indicated in this reference mRNA.
NRAS is neuroblastoma RAS viral (v-ras) oncogene homolog gene. Its NCBI Gene ID is 4893 and the reference mRN A is NM 002524. 'The mutation position m Table 1 are indicated in this reference mRNA .
BRAF is v-raf murine sarcoma viral oncogene homolog Bl gene. Its NCB Gene ID is 673 and the reference mRNA is NM 004333, The mutation position in Table 1 are indicated i this reference mRNA.
STKI 1 is serine/threonine kinase I I gene. Its NCBI Gene ID is 6794 and the reference mRNA is NM 000455. The mutation position in Table 1 are indicated in this reference mRNA.
M EN! is multiple endocrine neoplasia I gene. Its NCBI Gene ID is 4221 and the reference mRNA is ENST00000312049. The mutation position in Table 1 are indicated m this reference mR A,
TCF1 is HNF1 homeobox A gene. Its CBI Gene I D is 6927 and the reference mRNA is N M 000545, The mutatio position in Table I are indicated in this reference mRNA. BRAS is v-Ha-ras Harvey rat sarcoma viral oncogene homolog gene. Its NCBI Gene ID is 3265 and the reference mRN A is M^ 005343. The imitation position in Table 1 are indicated in this reference mRNA.
FGFR3 is fibroblast growth factor receptor 3 gene. Its NCBI Gene ID is 2261 and the reference mRNA is NM 000142, The mutation position in Table 1 are indicated in this reference mR A,
PTC -I Is patched homolog 1 gene. Its NCBI Gene ID is 5727 and the reference mRNA is N M 000264, The mutation position in Table I are indicated in this reference mRNA.
WT1 is Wilms tumor 1 gene, its NCBI Gene ID is 7490 and the reierence mRNA is NM 024426. The mutation position m '"Fable 1 are indicated in this reference mRNA. CTNNB! is catenin (cadherin-associated protein), beta 1 gene. Its NCBI Gene ID is 1499 and the reference mRNA is NM 001904. The imitation position in Table 1 are indicated m this reference mRNA.
MET is met proto-oncogene gene. Its NCBI Gone ID is 4233 and the reference mRNA is NM 000245. The miuation position in Table 1 are indicated in this reference mRNA. TP53 is :■! ·:· ·: protein p53 gene. Its NCBI Gene ID is 7157 and the reference mRNA is NM 000546. The mutation position in Table 1 are indicated in this reference mRNA . FLT3 is fms-related tyrosine kinase 3 gene, its NCBI Gene iD is 2322 and the reference mRNA is Z26652, The mutation position in 1 able I are indicated in this reference mRNA.
VHL is von Hippel-Lindau tumor suppressor gene. lis NCBI Gene ID is 7428 and the reference mRNA Is NM 000551. The mutation position in Table 1 are indicated in this reference mRNA.
NF2 is neurofibroma 2 gene. Its NCBI Gene ID is 4771 and the reference mRNA is NM 000268. The mutation position in Table 1 are indicated in this reference mRNA . SRC" is v-src sarcoma gene. Its NCBI Gene ID is 6714 and the reference mRNA. is NM 005417. The mutation position n fable I are Indicated in this reference mRNA. APC :.s adenomatosis polyposis cob gene. Its CBI Gene ID Is 324 and the reference mRNA is NM 000038. The mutation position in Table 1 are indicated in this reference mRNA.
RBI Is retoniblastoma I gene. Its NCBI Gene ID is 5925 and the reference mRNA is NM 000321. The mutation position in Table 1 are indicated m this reference mR A, KIT is v-kit Hardy-Znckerrnan 4 felme sarcoma viral oncogene horno!og gene. Its NCBI Gene ID is 3815 and the reference mRNA is NM 000222. The mutation position m Table I are indicated in this reference mRNA.
SMARCB1 is S W /SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 gene. Its NCBI Gene ID is 6598 and die reference mRNA is NM 003073. The mutation position in Table 1 are indicated in this reference mRNA.
EGFR is epidermal growth actor receptor gene. Its NCBI Gene ID is 1956 and the
reference mRNA is N 005228. The mutation position in Table 1 are indicated in this reference mRN A .
PTEN is phosphatase and tensin homoiog gene. lis NCBI Gene ID is 5728 and the reference rnRNA is NM 000314. The mutation position in Table 1 are indicated in this reference mRNA.
CDKN2A is cyclfn-dependent kinase inhibitor 2A gene. Its NCBI Gene ID is 1 029 and the reference mRNA is NM 000077. The imitation position in Table I are indicated in this reference mRNA.
PI3KCA is phosphoinosiiide-3-kina.se, catalytic, alpha gene. Its NCBI Gene ID is 5290 and the reference mRNA is NM 0062 1 8. The mutation position m '"Fable 1 are
indicated in this reference mRNA.
FBXW7 is F-box and WD repeat domain containing 7 gene. Its NCBI Gene ID is
55294 and the reference mRNA is NM 033632. The mutatio position in Table 1 are indicated in this reference rnRNA .
CDH 1 is cadherin 1 gene. Its NCBI Gene 03 is 999 and the reference mRNA is
NM 004360. The mutation position in Table I are indicated in this reference mRNA.
PDGFRA is platelet-derived growth factor receptor, alpha gene. Its NCBI Gene ID is 5156 and the reference mR A is N M 006206. The mutation position m '"Fable 1 are indicated in this reference mRNA.
ERBB2 is v-erb~b2 erythroblastic leukemia viral oncogene homoiog 2 gene. Its NCBI Gene J D is 2064 and the reference mRNA is NM 004448. The mutation position in
Table 1 are indicated in this reference mRNA,
Note: References for most of the different mutations listed in Tables 3, as well as others, can be found at, for example, Y^ ,.. v n; e.O
In other preferred embodiments, the target mutations of the panel are selected from the group consisting of those disclosed in Table I or 2, or any combinations thereof
However, this list is a non-limiting list and other novel mutations or those known in the art can be combined with those indicated above. In yet other preferred embodiments, the mutations of the panel are selected in order to obtain a universal blomarker panel allowing the detection of cancer independently of its type. In another embodiment, the panel is selected based o the most frequent occurrence of mutations in a. variety of cancer types. In some embodiments, the frequency of occurrence of the target mutations selected for the panel
occur in at least 50%, at least 55%, at least 60%, at least 65%>, at least 70%, at least 75%;, at least 80%, at least 85%, at least 90%, or at least 95% of all tumors and/or cancer types.
In some embodiments, the disclosed methods are performed using a panel of
mutations associated with cancer, in some embodiments, the panel of mutations comprises at least 20, at least 40, at least 90, at least 120, at least 140, at least 160, at least 180, at least 200, at least 220, at least 240, at least 260, at least 270, at least 300, at least 320, at least 340, at least 360, at least 380, at least 400, at least 420, at least 460, at least 480, at least 520, at least 560, at least 600, at least 640, at least 660 or at least 700 different mutations. In other embodiments, the panel may further comprise controls for the detection of wild type sequences and/or additional reference samples as discussed below.
In some embodiments, the target mutations to be detected may comprise a.
substitution, deletion, or insertion of one or several nucleotides. Insertions may typical ly comprise an addition of between 1 and 40, between 5 and 35, between 10 and 30, or between 15 and 25 contiguous base pairs. Deletions may typically comprise a removal of between 1 and 40, between 5 and 35, between 10 and 30, or between 15 and 25 contiguous base pairs. Substitutions may typically comprise a substitution of between 1 and 20, between 5 and 15, or between 8 and 12 contiguous base pairs. In a preferred embodiment, the target mutations comprise a. single point imitation.
In some embodiments, the nucleic acid sample can be DNA (e.g., gDNA). The present invention may be used for the detection of variation in genomic DNA whether human, animal or other. In yet other embodiments, as in the case of simultaneous analysis of gene expression by RT-PCR, the nucleic acid sample can be RNA (e.g., mRNA), In some embodiments, the DNA or RNA can be circulating DNA or RNA. !n some embodiments, the DNA or RNA can be circulating DNA or RNA, In some embodiments, the circulating DNA or RNA is tumor-related DNA or RNA. In yet other embodiments, die tumor-related DNA or RNA is less than about 50%, less than about 25%, less than about 20%, less than about 10%, less than about 5%, less than about 1%, less than about 0.1 %, less than about 0.001 % of the total nucleic acid sample obtained from a biological sample. In some other embodiments, the tumor-related circulating DMA or RNA. compared to wild type DNA or RNA is present at a frequency less than 1/10 copies, 1/100 copies, 1/1,000 copies, 1/10,000 copies, 1/100,000 copies, 1/1,000,000 copies, 1/10,000,000 copies, 1/100,000,000 copies or 1/1,000,000,000 copies.
In some embodiments; the disclosed methods further comprise a step whereby the set of target mutations earned by tumor-associated nucleic acids are compared to a reference or control sample (e.g., wild type sample). The comparison of the set of target imitations earned by tumor-associated nucleic acids to a reference allows the determination of a probability of the subject to be at risk of developing a cancer, to suffer from a cancer, to have a good or bad prognosis (for instance, a complete remittance, or a metastasis), to have a recurrence, or responsiveness to a particular treatment. In some embodiments, the reference can be a positive reference (e.g., healthy profile, treatment responder profile, no or low risk of metastasis profi le, no or low risk of recurrence profi le) or a negative reference (e.g., cancer profile, treatment non- or low-responder profile, high nsk of metastasis profile, high risk of recurrence profile).
In another aspect, the present invention provides compositions tor nse in identifying and/or quantitating target imitations in nucleic acid sample based on a panel of UBiVI . In some embodiments, such compositions can comprise PCR reagents used for the detection and/or quantification of target imitations in a nucleic acid sample based on a panel of UBM. In some embodiments, these compositions can comprise reagents used for casiPCR. In some embodiments, the reagents used for castPC can include, tor example: (a) an allele-specific primer; (b) an allele-specific blocker probe; (c) a detector probe; and/or id) a locus-specific primer, wherein the allele-specific primers are targeted to mutation selected from the IJ B panel of target mutations. In some embodiments, the compositions may further compri.se a. polymerase, dNTPs, reagents and/or buffers suitable for PCR amplification, and/or a template sequence or nucleic acid sample.
In another aspect, the present invention provides kits for the diagnosis, prognosis and monitoring a cancer and/or tumour progression in a subject. In another aspect, the present invention provides kits for providing data for diagnosis, prognosis and monitoring a cancer and/or tumour in a subject is provided. In some embodiments, the kit comprises a microp!ate. In yet other embodiments, the rnicroplate's wel l may comprise:
(a) a pair of oligonucleotide probes for detecting one target DNA carrying a mutation from a set of at least 90 target circulating DNA carrying a mutation selected f om the group consisting of those listed m Table I or 2; and
(b) one or several reagents used to perform a detection method, such as, but not limited to ca&iPC R.
In some embodiments, such reagents may include, for example, a pair of primers: reagents for extracting circulating DNA; and/or a combination thereof
In some embodiments, the compositions, methods, and/or kits can be used in detecting circulating ceils in diagnosis. In one embodiment, the compositions, methods, and/or kits can be used to detect tumor cells in blood for early cancer diagnosis, such for screening, for early detection or early detection of recurrence of cancer and/or tumors. In some embodiments, the compositions, methods, and/or kits can be used for cancer or disease-associated genetic variation or somatic mutation detection and validation . In some embodiments, the compositions, methods and/or kits can be used for treatment identification. In some embodiments, the compositions, methods, and/or kits can be used for genotyping tumors and assessing the status of the mutations most used today in clinical management (e.g., EFGR, KRAS) which is typically performed by sequencing and is usually considered extremely long and costly.
EXAMPLES
Ex ample 1
Determination of an exemplary UBM Panel
Pilot study on 140 patients:
A panel of 100 patients with frequent solid tumors, treated with curative surgery are initially selected.
Among the 100 panellists, 20 solid tu ors including lung, colon, pancreas, hepatocarcinoma, breast, prostate, stomach, and any other type of tumors are chosen for further analysis.
Tumoral DNA is extracted from residual frozen tumors, and blood samples (a minimum of 10 mL) are collected one day before surgery and then after surgery every 2 weeks, and stored as plasma samples.
Preferably, the patients' status of current known seric bi.oma.rk.ers is known.
A test comprising a UB panel of target mutations is first applied to explore tumoral DNA and to assess whether tested samples are found to be positive or negative for any of the UBM panel of target mutations. A positive test means detection of at least one mutation out of the 384 total tested . Occurrence frequency is estimated as % of tumor in which the UBM test is positive.
If the test is positive for tumoral DNA, then the given mutation is further tested in plasma. Detection is evaluated before treatment and after treatment. Current existent biomarkers, whenever available are used as gold standards to validate the UBM panel.
Efficacy and performance is evaluated:
Percent of tumors in which I B is positive is detennined.
Criteria of successful method : UBM test is positive in 50% of tumors. Percentage may be different according to tumor type.
Percent of patients in which positive UBM tested for tumors and in which mutant DNA is detected in plasma. Decrease of the UBM test in plasma, progressively after surgery, tested every 2 weeks, in correlation with known seric markers ( AFP, PSA, CEA, etc) whenever possible.
Review statistical efficacy to redefine necessary content of the UBM collection of 384 mutations, or if there is a need to design a second UBM panel and to increase the number of mutation tested from, for example, 384 to 2x 384,
Example 2
Use of cast-PC for detection of a UBM panel of target mutations in biological samples in this example, casi-PCR (see, for example, US Application Nos. 12/641 ,321 and 12/748,329, the contents of which are herein incorporated by reference in their entireties) is used to detect and determine the percentage of tumor cells in biological samples. Various biological samples, comprising both tumorigenic and nonnai cells, are obtained and assayed by cast-PCR. for the presence of a number of target mutations associated with cancer based on a panel of Universal Biomarkers (UBM ). The UBM panel of target mutations can include, for example, any of those mutations shown in Figure 2 as well as those listed in Table 1 and Table 2, or any combinations thereof
For detecting and quantitating each mutation in each sample, both allele- 1 and alleie-2 assays are performed using the general cast-PCR schema ami reaction conditions indicated below, using gDNA derived from either normal or tumor samples. Assay primers and probes corresponding to the mutations shown in Table I and Table 2 are designed accordingly.
I General Assay Design:
The general schema for the cast-PCR. assays used in the following examples are illustrated in Fig. 1. For each SNP that was analyzed, allele-specific primers were designed to target a first allele (i.e. allele- 1 ) and a second allele (i.e. a!lele-2). The cast-PCR assay reaction mixture for allele- i analysis included a tailed al ele-i-speciftc primer (ASP1), one MGB aileie-2 blocker probe (MGB2), one common locus-specific TaqMan probe (LST) and one common locus-specific primer (LSP). The cast-PCR assay reaction mixture for analysis of aliele-2 included a tai led allele-2-specific primer (ASP2), one MGB a! !eie-l blocker probe (MGB i), one common locus-specific TaqMan probe (LST) and one common locus-specific primer (LSP).
II. Reaction conditions:
PCR. reaction mixtures are prepared, as follows:
For each sample (to be run in duplicate), the following reagents are pipetted into a nuclease- free 1.5mL microcentrifuge tube or PCR plate:
For Allele- 1 assay:
PCR reaction mix comDonent Volume per i O-μΙ reactions" ί,μί)
Single Reaction Two Replicates
2x Ta jinan Gen«fyping Master Mix 5 12
lOx casiPCR Allele- ! assav 1 2.4
genomic DNA t'5x) s ) 4.8
RNase free water 4,8
For AlJeie-2 assay:
PCR reaction mix component Volume per ]ί)- ί reactions'' ί,μΙ)
Single Reaction Two Replicates*
2x "Taqnian. Gen.otypi.ng Master Mis 12
10.x castPCR Allele-.! assay i. 2.4
genomic DNA t 5 i * 4,8
RNase free water Ί 4,8
#Replicate volumes include 20% excess to compensate for volume loss from pipetting.
*CastPCIl works with 1 - 1 00 ng gDNA sample input. 20 ng gDMA is used as a starting point.
DNA template is added to the reactions as the last step to eliminate template co tamination. € When using 48-weii or 96-well plates, reactions are scaled up to 20 μΐ volumes by doubling the volume of each reaction component.
Plates are loaded and reactions are carried out as follows:
1. 10 pL of PCR reaction mix is transferred into each well of a 48-, 96-, or 384-weli reaction plate.
2. Plates are sealed with the appropriate cover.
3. P!ales are eentrifuged briefly.
4. Plates are loaded into the instrument.
5. Thermal cyclirig is earned out as follows: 95°C/10 min, (92°C/15 sec, 58°C/1 min) for 5 cycles, then (92°C/15 sec, 60°C/ 1 m n) for 45 cycles. { PC reactions were run for a total of 50 cycles. (The 50 cycles can include a pre-run of five cycles at a lower annealing/extension temperature followed by an additional 45 cycles at a higher annealing/extension temperature).
ΙΠ. Data Analysis:
Data from castPC Assays is analyzed as follows:
1. Amplification plots are viewed for the entire plate,
2. Baseline and . threshold values are set.
3. Cq values for Allele- 1 assays and Allele-2 assays are obtained individually.
4. The ACt between amplification reactions for Allele- 1 assays and Allele-2 assays are calculated. The ACt can be defined as the specificity of castPCR (ACt ::: <\;;:.l:. ; - !i ¾ji i - 2 >- The larger the ACt between two alleles, the better assay specificity. The 2; " ' value is used to estimate the power of discrimination (or selectivity} which is equal to ½" '' or, calculated as % (½ACt x 100).
Example 3
Preampiification of DNA samples for use in castPCR assays for the detection of
target mutations using a UB panel
Reagents:
2X PreAmp Master Mix (P/N: 4391 128)
5X PreAmp Primer Pool ( 8 or 96-piex)
10 ng/uL DNA samples
Nuc I ease- free Water
Instrument; GeneAmp PGR System 9700, 7500, 7900 or ViiA7
Procedure:
In a 96-weii plate, preampiification reactions comprising DNA samples in each well are set-up as follows:
Reagent Per reaction Final Concentration
2X PreAmp Master Mix 10 uL I X
SX PreA np Primer Mix 4 :· ! . IX
10 ng/uL DNA sample 6 uL 3 ng/uL
Water 0 id.
Total 20 i d
Plates are sealed with a 96- well Plate Cover, spun quickly, and placed in an ABI PRISM Optical Cover Compression Pad or* the top L
Thermocycling conditions are sei--up on a GeneAmp PC System 9700 to run pre- amplification reactions as follows: 95°C for 10 min, [95CC for 15 sec - 60°C for 4 mm] for 10 cycles. 99.9°C for 10 uin and 4"C on hold for np to 24 rs.
When reactions are complete, reaction plates are removed from the theraiocyc!er and placed on ice. PreAmp 5X is diluted by adding 80 id . of d 20. Wells are mixed by pipetting up and down 5 times (being cautious to avoid any cross-contamination) and spun briefly.
Prearn lification products are stored at 0"C or used directly in standard castPCR assays. For castPCR assays, 1-20% of diluted PreAmp products are added into each castPCR reaction according to the number of targets (up to 96) and PCR. replicates are applied.
Standard castPCR protocols are employed as described in Example l (see also, US Publication No. US-20IG002217I7-A1; published 9/2/2010, the disclosure of which is herein
incorporated by reference in its entirety).
Table i. Lis of 384 exemplary targe ρο:?1;: mutations used ^or^ lui cancer. The Riutation number corresponds to base position in the open reading rame .
Page 44 of 82
Page 46 of 82
Page 47 of 82
Page 43 of 82
Page 49 of 82
Page 51 of 82
Page 53 of 82
Page 54 of 82
Page 55 of 82
Page 57 of 82
Page 58 of 82
Page 60 of 82
Page 62 of 82
Page 64 of 82
Page 65 of 82
Page 66 of 82
Page 67 of 82
Page 69 of 82
Page 70 of 82
Page 71 of 82
Page 72 of 82
Page 73 of 82
Page 76 of 82
Claims
WHAT IS CLAIMED S:
1 , A method for the diagnosis or prognosis of cancer and/or tumor progression in a. su ject comprising the detection of at least one target mutation in a panel of at least 90 target mutations, wherein the confidence level and/or statistical efficacy of the diagnosis or prognosis is at least 50%, at least 60%, at least 70%, at least 80%, at least 90%, or at least 99%.
2, The method of claim 1, wherein said biological sample is a bodily fluid or tissue.
3, The method of claim 2, wherein said bodily fluid is selected from the group consisting of plasma, serum, ascites, pleural effusion, urine, lymphatic fluid, stool, bronchoalveolar lavage fluid, ductal lavage fluid, or cerebrospinal fluid.
4, The method of claim 2, wherein said tissue is frozen, fresh, formalin fixed or paraffin embedded,
5, The method of claim 2, wherein said tissue is selected from the group consisting of a surgical fragment of a tissue, tumor biopsy, fine needle aspiration biopsy, or circulating tumor ceil .
6, The method of claim 1, wherein said nucleic acid is D A.
7, The method of claim 6, wherein said DN A is circulating DNA.
8, The method of claim 7, wherein said circulating DNA is tumorigenic circulating DNA.
9. The method, of claim 1, wherein said extracting is performed by any method known by the one skilled in the art.
10. The method of claim 1, wherein said biological sample is a blood sample,
1 1. 'The method of claim 1 or 10, wherein said step (a) comprises:
(a) removal of potentially contaminating cells from said sample: ami/or
(b) removal of proteins that m y interfere with the detection of target mutations carried by said, nucleic acid obtained from said, sample.
12. The method of claim 1 1 , wherein said removal of contaminating cells is perionned. by coagulation, centrifiigairon arid/or filtration of said sample.
13. The method of claim 1 I , wherein said removal of interfering proteins is done by proteinase digestion of said sample.
14. The method, of claim 1 1, wherein said nucleic acid extracted, from said sample is used directly after removal of contaminating cells and/or proteins.
15. The method, of claim 11, wherein said, nucleic acid extracted from said sample is further purified prior to detection.
16. The method of claim 1 or 15, wherein said nucleic acid extracted in step (a) is further purified by: extraction with phenol chloroform, precipitation in alcohol or dissolution in an aqueous solution.
17. The method of claim 1 , wherein said subject is a mammal.
18. The method of claim 17, wherein said subject is human.
19. The method of claim 1 , wherein said subject has: (a) been diagnosed with cancer; (b) is suspected of having cancer, or (c) has been determined to be at risk for developing cancer.
20, The method of claim I or 15, further comprising a step of purification leading to a nucleic acid purity level of at least 80%, at least 85%, at least 90%, or at least 95% pure.
21. The method, of any preceding claim, wherein the method employs castPCR as the detection method.
22. The method of clarm 21, wherein castPCR is used to deiect a panel of at least 20, at least 90, at least 384 or at least 61 different target mutations.
23. The method of claim 22, wherein said target mutations are in the genes selected from the group consisting of: KJ AS, ABLi, SMAD4, RAS, BRAF, STK I 1, HRAS, FGFR3,
WT I , CTNNBi, MET, TP53, FLT3, VEIL, S C, APC, RBL KIT, SMARCB L EGPR, CDKN2A, PBKCA, FBXW7, ER.BB2, AKTl , ALK, ATM, CSFIR, ERBB4, GNAS, HNF l A, IDH I, JAK2, KDR, MFL, NOTCHL NPM l , RET, SMO, UTX, CDEFi , MEN U NF2, PDGFRA, PTCH, PTEN, TCF1 , or any of the mutations listed in Table 1 or 2, or a y combinations thereof
24. 'The method of step 23, wherein said target mutations are selected from the group consisting of those listed in Table I or Table 2, or any combinations thereof
25. The method of claim 1 , further comprising determining the particular treatment to use on said subject based on said diagnosis and/or prognosis of cancer.
26. A. method for providing data for diagnosis, prognosis and/or monitoring a cancer or tumour in a subject, comprising detecting or quantifying a panel of at least 40, at least 90, at least 380, or at least 600 different target mutations in circulating tumorigenic DNA.
27. A kit for performing the methods of any of the preceding claims.
28. The kit of claim 27, comprising reagents for the detection of a panel of at least 40, at least 90, at least 300, or at least 600 different target mutations carried by DN A in a biological sample of a subject.
29. The method according io claim 1, wherein the subject is bearing a cancer or is suspected to suffer from a cancer, or has been previously treated for a cancer or is in a follow-up step to detect recurrence of cancer, or is in the course of treatment t r cancer.
30. A method of diagnosis, prognosis and monitoring a cancer or tumour in a subject, comprising: detecting or quantifying a panel of at least 90 target mutations carried by tumor DNA in various genes selected from the group consisting h KRAS, ABLE SMAD4, NRAS, BRAF, STKd l , BRAS, FGFR3, WTI , CTNNBI , MET, TP53, FLT3, VHL, SRC, APC, RB I, KIT, S MARCH 1 , EGFR, CDKN2A, PBKCA, FBXW7, ERBB2, A T1 , ALK, ATM, CSFIR, ERBB4, GNAS, HNFl A, iDHl , 3A 2, KDR, MPL, NOTCH 1 , NPM l, RET, SMO, UTX, ( DI U MEN 1 , NF2, PDGFRA, PTC IT PTEN, TCF! , any of the mutations listed in Table 1 and 2, or any combinations thereof; and optionally, comparing the detected set of target mutations carried by tumor DNA to a. reference or contr l sample.
31. The method of claim 30, wherein said method further comprises:
extracting circulating DNA. from a serum sample;
providing a microplate, wherein each mi crop 1 ale well comprises reagents sufficient for detectio of at least one target mutation from said panel of a t least 90 target mutations carried by circulating tumor DNA;
adding said circulating DNA in each rnicropiates' well; and
detecting and/or quantifying said target mutations carried by circulating tumor DNA. in each well,
32. The method of claim 3 1 , wherein said reagents contained in said microplate comprise reagents sufficient to carry out castPCR.
33, A kit used for the diagnosis, prognosis and monitoring a cancer or tumour in a subject, comprising:
a microplate, wherein each microplate well comprises a pair of oligonucleotide probes for detecting one target mutation from the panel of at least 90 target mutations carried by circulating tumor DNA selected in the group consisting of the mutations of the genes KRAS, ABLL SMAD4, RAS, BRA.F, ST .1 1 , HRAS, FGFR3, WT1 , CTNNB1, MET, TP53, FLT3, VHL, SRC, AFC, RBI , KIT, S ARCBE EGFR, CDKN2A, PBKCA, FBXW7, ERBB2, AKTi , ALK, ATM, CSF1 R, ERBB4, GNAS, HNF1A, IDH l, JA.K2, KDR, MPL, NOTCHl , NPM 1, RET, SMC), IJTX, CDH1 , EN I , NF2, PDGFRA, PTCH, PTEN and TCF'l in particular those listed in Table 1 and 2: and
one or several elements selected in the group consisting of a ligation mix, a real-time quantitative PCR mix: a pair of primers; reagents for extracting circulating DNA; reagents sufficient to carry out castPCR or any combinations thereof
34, A. composition used or the diagnosis, prognosis and monitoring a cancer or tumour in a subject using a panel of at least 90 target mutations for the detection of at least one target mutation of said at least 90 target mutations, comprising:
one or several elements selected in the group consisting of a ligation mix,
a real-time quantitative PCR mix;
a pair of primers; reagents for extracting circulating DNA ;
reageiirs sufficient to carry our castPCR or any combi nation of thereof
35. The method of claim 34, further comprising identification of a particular treatment and/or therapy based on said diagnosis and/or prognosis of cancer.
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US201261664501P | 2012-06-26 | 2012-06-26 | |
| US61/664,501 | 2012-06-26 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2014004726A1 true WO2014004726A1 (en) | 2014-01-03 |
Family
ID=49783844
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/US2013/047984 WO2014004726A1 (en) | 2012-06-26 | 2013-06-26 | Methods, compositions and kits for the diagnosis, prognosis and monitoring of cancer |
Country Status (1)
| Country | Link |
|---|---|
| WO (1) | WO2014004726A1 (en) |
Cited By (44)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2015035415A1 (en) | 2013-09-09 | 2015-03-12 | Nantomics, Llc | Method of detecting suceptibility to breast cancer through mutations in the fgfr3 and tp53 genes |
| US9512473B2 (en) | 2008-12-17 | 2016-12-06 | Life Technologies Corporation | Methods, compositions, and kits for detecting allelic variants |
| US9534255B2 (en) | 2008-12-17 | 2017-01-03 | Life Technologies Corporation | Methods, compositions, and kits for detecting allelic variants |
| US9598731B2 (en) | 2012-09-04 | 2017-03-21 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| CN107385097A (en) * | 2017-09-13 | 2017-11-24 | 山东大学第二医院 | Detect the kit of SMAD4 gene V354L site mutations |
| US9902992B2 (en) | 2012-09-04 | 2018-02-27 | Guardant Helath, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US9920366B2 (en) | 2013-12-28 | 2018-03-20 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| CN107949642A (en) * | 2015-05-27 | 2018-04-20 | 奎斯特诊断投资股份有限公司 | For screening the composition and method of solid tumor |
| WO2018083467A1 (en) * | 2016-11-02 | 2018-05-11 | Ucl Business Plc | Method of detecting tumour recurrence |
| US10011870B2 (en) | 2016-12-07 | 2018-07-03 | Natera, Inc. | Compositions and methods for identifying nucleic acid molecules |
| WO2018137685A1 (en) * | 2017-01-25 | 2018-08-02 | The Chinese University Of Hong Kong | Diagnostic applications using nucleic acid fragments |
| US10061890B2 (en) | 2009-09-30 | 2018-08-28 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US10083273B2 (en) | 2005-07-29 | 2018-09-25 | Natera, Inc. | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US10081839B2 (en) | 2005-07-29 | 2018-09-25 | Natera, Inc | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US10113196B2 (en) | 2010-05-18 | 2018-10-30 | Natera, Inc. | Prenatal paternity testing using maternal blood, free floating fetal DNA and SNP genotyping |
| US10174369B2 (en) | 2010-05-18 | 2019-01-08 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US10179937B2 (en) | 2014-04-21 | 2019-01-15 | Natera, Inc. | Detecting mutations and ploidy in chromosomal segments |
| US10227652B2 (en) | 2005-07-29 | 2019-03-12 | Natera, Inc. | System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals |
| US10262755B2 (en) | 2014-04-21 | 2019-04-16 | Natera, Inc. | Detecting cancer mutations and aneuploidy in chromosomal segments |
| CN109837351A (en) * | 2019-03-29 | 2019-06-04 | 南昌艾迪康医学检验实验室有限公司 | Utilize the kit of real-time fluorescence quantitative PCR detection PDGFR α gene relative expression quantity |
| US10316362B2 (en) | 2010-05-18 | 2019-06-11 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10351906B2 (en) | 2014-04-21 | 2019-07-16 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| CN110320363A (en) * | 2014-01-28 | 2019-10-11 | 奎斯特诊断投资股份有限公司 | For detecting adenoma-gland cancer transition method and composition in cancer |
| US10526658B2 (en) | 2010-05-18 | 2020-01-07 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10577655B2 (en) | 2013-09-27 | 2020-03-03 | Natera, Inc. | Cell free DNA diagnostic testing standards |
| US10704086B2 (en) | 2014-03-05 | 2020-07-07 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10894976B2 (en) | 2017-02-21 | 2021-01-19 | Natera, Inc. | Compositions, methods, and kits for isolating nucleic acids |
| US11111543B2 (en) | 2005-07-29 | 2021-09-07 | Natera, Inc. | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US11111544B2 (en) | 2005-07-29 | 2021-09-07 | Natera, Inc. | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US11242569B2 (en) | 2015-12-17 | 2022-02-08 | Guardant Health, Inc. | Methods to determine tumor gene copy number by analysis of cell-free DNA |
| US11306357B2 (en) | 2010-05-18 | 2022-04-19 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US11322224B2 (en) | 2010-05-18 | 2022-05-03 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US11326208B2 (en) | 2010-05-18 | 2022-05-10 | Natera, Inc. | Methods for nested PCR amplification of cell-free DNA |
| US11332785B2 (en) | 2010-05-18 | 2022-05-17 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US11332793B2 (en) | 2010-05-18 | 2022-05-17 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11339429B2 (en) | 2010-05-18 | 2022-05-24 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US11408031B2 (en) | 2010-05-18 | 2022-08-09 | Natera, Inc. | Methods for non-invasive prenatal paternity testing |
| US11459616B2 (en) | 2016-10-24 | 2022-10-04 | The Chinese University Of Hong Kong | Methods and systems for tumor detection |
| US11479812B2 (en) | 2015-05-11 | 2022-10-25 | Natera, Inc. | Methods and compositions for determining ploidy |
| US11485996B2 (en) | 2016-10-04 | 2022-11-01 | Natera, Inc. | Methods for characterizing copy number variation using proximity-litigation sequencing |
| US11525159B2 (en) | 2018-07-03 | 2022-12-13 | Natera, Inc. | Methods for detection of donor-derived cell-free DNA |
| US11913065B2 (en) | 2012-09-04 | 2024-02-27 | Guardent Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11939634B2 (en) | 2010-05-18 | 2024-03-26 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11959139B2 (en) | 2023-05-12 | 2024-04-16 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2010014920A1 (en) * | 2008-07-31 | 2010-02-04 | The Johns Hopkins University | Circulating mutant dna to assess tumor dynamics |
| WO2011150075A2 (en) * | 2010-05-25 | 2011-12-01 | The Johns Hopkins University | Compositions and methods for detecting a neoplasia |
| WO2012001007A1 (en) * | 2010-06-28 | 2012-01-05 | Technische Universität München | Methods and compositions for diagnosing gastrointestinal stromal tumors |
-
2013
- 2013-06-26 WO PCT/US2013/047984 patent/WO2014004726A1/en active Application Filing
Patent Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2010014920A1 (en) * | 2008-07-31 | 2010-02-04 | The Johns Hopkins University | Circulating mutant dna to assess tumor dynamics |
| WO2011150075A2 (en) * | 2010-05-25 | 2011-12-01 | The Johns Hopkins University | Compositions and methods for detecting a neoplasia |
| WO2012001007A1 (en) * | 2010-06-28 | 2012-01-05 | Technische Universität München | Methods and compositions for diagnosing gastrointestinal stromal tumors |
Non-Patent Citations (2)
| Title |
|---|
| GORMALLY ET AL.: "Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance", MUTATION RESEARCH, vol. 635, no. 2-3, 25 January 2007 (2007-01-25), pages 105 - 117 * |
| ZIEGLER ET AL.: "Circulating DNA: a new diagnostic gold mine?", CANCER TREATMENT REVIEWS, vol. 28, no. 5, October 2002 (2002-10-01), pages 255 - 271 * |
Cited By (139)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US10083273B2 (en) | 2005-07-29 | 2018-09-25 | Natera, Inc. | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US11111543B2 (en) | 2005-07-29 | 2021-09-07 | Natera, Inc. | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US11111544B2 (en) | 2005-07-29 | 2021-09-07 | Natera, Inc. | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US10392664B2 (en) | 2005-07-29 | 2019-08-27 | Natera, Inc. | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US10266893B2 (en) | 2005-07-29 | 2019-04-23 | Natera, Inc. | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US10260096B2 (en) | 2005-07-29 | 2019-04-16 | Natera, Inc. | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US10227652B2 (en) | 2005-07-29 | 2019-03-12 | Natera, Inc. | System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals |
| US10081839B2 (en) | 2005-07-29 | 2018-09-25 | Natera, Inc | System and method for cleaning noisy genetic data and determining chromosome copy number |
| US10240202B2 (en) | 2005-11-26 | 2019-03-26 | Natera, Inc. | System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals |
| US11306359B2 (en) | 2005-11-26 | 2022-04-19 | Natera, Inc. | System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals |
| US10597724B2 (en) | 2005-11-26 | 2020-03-24 | Natera, Inc. | System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals |
| US10711309B2 (en) | 2005-11-26 | 2020-07-14 | Natera, Inc. | System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals |
| US9534255B2 (en) | 2008-12-17 | 2017-01-03 | Life Technologies Corporation | Methods, compositions, and kits for detecting allelic variants |
| US9512473B2 (en) | 2008-12-17 | 2016-12-06 | Life Technologies Corporation | Methods, compositions, and kits for detecting allelic variants |
| US10081833B2 (en) | 2008-12-17 | 2018-09-25 | Life Technologies Corporation | Methods, compositions, and kits for detecting allelic variants |
| US10570459B2 (en) | 2008-12-17 | 2020-02-25 | Life Technologies Corporation | Methods, compositions, and kits for detecting allelic variants |
| US11572585B2 (en) | 2008-12-17 | 2023-02-07 | Life Technologies Corporation | Methods, compositions, and kits for detecting allelic variants |
| US10689694B2 (en) | 2008-12-17 | 2020-06-23 | Life Technologies Corporation | Methods, compositions, and kits for detecting allelic variants |
| US11530450B2 (en) | 2008-12-17 | 2022-12-20 | Life Technologies Corporation | Methods, compositions, and kits for detecting allelic variants |
| US10522242B2 (en) | 2009-09-30 | 2019-12-31 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US10061890B2 (en) | 2009-09-30 | 2018-08-28 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US10061889B2 (en) | 2009-09-30 | 2018-08-28 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US10216896B2 (en) | 2009-09-30 | 2019-02-26 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US11306357B2 (en) | 2010-05-18 | 2022-04-19 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US11519035B2 (en) | 2010-05-18 | 2022-12-06 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10174369B2 (en) | 2010-05-18 | 2019-01-08 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US11312996B2 (en) | 2010-05-18 | 2022-04-26 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11326208B2 (en) | 2010-05-18 | 2022-05-10 | Natera, Inc. | Methods for nested PCR amplification of cell-free DNA |
| US11746376B2 (en) | 2010-05-18 | 2023-09-05 | Natera, Inc. | Methods for amplification of cell-free DNA using ligated adaptors and universal and inner target-specific primers for multiplexed nested PCR |
| US11482300B2 (en) | 2010-05-18 | 2022-10-25 | Natera, Inc. | Methods for preparing a DNA fraction from a biological sample for analyzing genotypes of cell-free DNA |
| US10316362B2 (en) | 2010-05-18 | 2019-06-11 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10113196B2 (en) | 2010-05-18 | 2018-10-30 | Natera, Inc. | Prenatal paternity testing using maternal blood, free floating fetal DNA and SNP genotyping |
| US11939634B2 (en) | 2010-05-18 | 2024-03-26 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11286530B2 (en) | 2010-05-18 | 2022-03-29 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11111545B2 (en) | 2010-05-18 | 2021-09-07 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11525162B2 (en) | 2010-05-18 | 2022-12-13 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11322224B2 (en) | 2010-05-18 | 2022-05-03 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US10793912B2 (en) | 2010-05-18 | 2020-10-06 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11332785B2 (en) | 2010-05-18 | 2022-05-17 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US10526658B2 (en) | 2010-05-18 | 2020-01-07 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10774380B2 (en) | 2010-05-18 | 2020-09-15 | Natera, Inc. | Methods for multiplex PCR amplification of target loci in a nucleic acid sample |
| US10538814B2 (en) | 2010-05-18 | 2020-01-21 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10557172B2 (en) | 2010-05-18 | 2020-02-11 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11332793B2 (en) | 2010-05-18 | 2022-05-17 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10731220B2 (en) | 2010-05-18 | 2020-08-04 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11339429B2 (en) | 2010-05-18 | 2022-05-24 | Natera, Inc. | Methods for non-invasive prenatal ploidy calling |
| US10590482B2 (en) | 2010-05-18 | 2020-03-17 | Natera, Inc. | Amplification of cell-free DNA using nested PCR |
| US11408031B2 (en) | 2010-05-18 | 2022-08-09 | Natera, Inc. | Methods for non-invasive prenatal paternity testing |
| US10655180B2 (en) | 2010-05-18 | 2020-05-19 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10597723B2 (en) | 2010-05-18 | 2020-03-24 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10494678B2 (en) | 2012-09-04 | 2019-12-03 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10995376B1 (en) | 2012-09-04 | 2021-05-04 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US9598731B2 (en) | 2012-09-04 | 2017-03-21 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10683556B2 (en) | 2012-09-04 | 2020-06-16 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11913065B2 (en) | 2012-09-04 | 2024-02-27 | Guardent Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11879158B2 (en) | 2012-09-04 | 2024-01-23 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10041127B2 (en) | 2012-09-04 | 2018-08-07 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US9902992B2 (en) | 2012-09-04 | 2018-02-27 | Guardant Helath, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11319598B2 (en) | 2012-09-04 | 2022-05-03 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11773453B2 (en) | 2012-09-04 | 2023-10-03 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10738364B2 (en) | 2012-09-04 | 2020-08-11 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US9840743B2 (en) | 2012-09-04 | 2017-12-12 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10501810B2 (en) | 2012-09-04 | 2019-12-10 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10793916B2 (en) | 2012-09-04 | 2020-10-06 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11319597B2 (en) | 2012-09-04 | 2022-05-03 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10822663B2 (en) | 2012-09-04 | 2020-11-03 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10837063B2 (en) | 2012-09-04 | 2020-11-17 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US9834822B2 (en) | 2012-09-04 | 2017-12-05 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10876172B2 (en) | 2012-09-04 | 2020-12-29 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10876152B2 (en) | 2012-09-04 | 2020-12-29 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10876171B2 (en) | 2012-09-04 | 2020-12-29 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10457995B2 (en) | 2012-09-04 | 2019-10-29 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10501808B2 (en) | 2012-09-04 | 2019-12-10 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11001899B1 (en) | 2012-09-04 | 2021-05-11 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10894974B2 (en) | 2012-09-04 | 2021-01-19 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10947600B2 (en) | 2012-09-04 | 2021-03-16 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10961592B2 (en) | 2012-09-04 | 2021-03-30 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11434523B2 (en) | 2012-09-04 | 2022-09-06 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| WO2015035415A1 (en) | 2013-09-09 | 2015-03-12 | Nantomics, Llc | Method of detecting suceptibility to breast cancer through mutations in the fgfr3 and tp53 genes |
| US10577655B2 (en) | 2013-09-27 | 2020-03-03 | Natera, Inc. | Cell free DNA diagnostic testing standards |
| US11767555B2 (en) | 2013-12-28 | 2023-09-26 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US11434531B2 (en) | 2013-12-28 | 2022-09-06 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US11649491B2 (en) | 2013-12-28 | 2023-05-16 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US9920366B2 (en) | 2013-12-28 | 2018-03-20 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US10883139B2 (en) | 2013-12-28 | 2021-01-05 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US11118221B2 (en) | 2013-12-28 | 2021-09-14 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US11149306B2 (en) | 2013-12-28 | 2021-10-19 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US11149307B2 (en) | 2013-12-28 | 2021-10-19 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US11667967B2 (en) | 2013-12-28 | 2023-06-06 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US11639525B2 (en) | 2013-12-28 | 2023-05-02 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US10889858B2 (en) | 2013-12-28 | 2021-01-12 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US11767556B2 (en) | 2013-12-28 | 2023-09-26 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US10801063B2 (en) | 2013-12-28 | 2020-10-13 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| US11639526B2 (en) | 2013-12-28 | 2023-05-02 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
| CN110320363A (en) * | 2014-01-28 | 2019-10-11 | 奎斯特诊断投资股份有限公司 | For detecting adenoma-gland cancer transition method and composition in cancer |
| US10870880B2 (en) | 2014-03-05 | 2020-12-22 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10982265B2 (en) | 2014-03-05 | 2021-04-20 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11091797B2 (en) | 2014-03-05 | 2021-08-17 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10704085B2 (en) | 2014-03-05 | 2020-07-07 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11091796B2 (en) | 2014-03-05 | 2021-08-17 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US10704086B2 (en) | 2014-03-05 | 2020-07-07 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11447813B2 (en) | 2014-03-05 | 2022-09-20 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11667959B2 (en) | 2014-03-05 | 2023-06-06 | Guardant Health, Inc. | Systems and methods to detect rare mutations and copy number variation |
| US11319595B2 (en) | 2014-04-21 | 2022-05-03 | Natera, Inc. | Detecting mutations and ploidy in chromosomal segments |
| US11486008B2 (en) | 2014-04-21 | 2022-11-01 | Natera, Inc. | Detecting mutations and ploidy in chromosomal segments |
| US10597708B2 (en) | 2014-04-21 | 2020-03-24 | Natera, Inc. | Methods for simultaneous amplifications of target loci |
| US11408037B2 (en) | 2014-04-21 | 2022-08-09 | Natera, Inc. | Detecting mutations and ploidy in chromosomal segments |
| US11414709B2 (en) | 2014-04-21 | 2022-08-16 | Natera, Inc. | Detecting mutations and ploidy in chromosomal segments |
| US11371100B2 (en) | 2014-04-21 | 2022-06-28 | Natera, Inc. | Detecting mutations and ploidy in chromosomal segments |
| US10597709B2 (en) | 2014-04-21 | 2020-03-24 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10351906B2 (en) | 2014-04-21 | 2019-07-16 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US10262755B2 (en) | 2014-04-21 | 2019-04-16 | Natera, Inc. | Detecting cancer mutations and aneuploidy in chromosomal segments |
| US11390916B2 (en) | 2014-04-21 | 2022-07-19 | Natera, Inc. | Methods for simultaneous amplification of target loci |
| US11530454B2 (en) | 2014-04-21 | 2022-12-20 | Natera, Inc. | Detecting mutations and ploidy in chromosomal segments |
| US10179937B2 (en) | 2014-04-21 | 2019-01-15 | Natera, Inc. | Detecting mutations and ploidy in chromosomal segments |
| US11319596B2 (en) | 2014-04-21 | 2022-05-03 | Natera, Inc. | Detecting mutations and ploidy in chromosomal segments |
| US11479812B2 (en) | 2015-05-11 | 2022-10-25 | Natera, Inc. | Methods and compositions for determining ploidy |
| US11946101B2 (en) | 2015-05-11 | 2024-04-02 | Natera, Inc. | Methods and compositions for determining ploidy |
| CN107949642B (en) * | 2015-05-27 | 2022-04-26 | 奎斯特诊断投资股份有限公司 | Compositions and methods for screening for solid tumors |
| CN107949642A (en) * | 2015-05-27 | 2018-04-20 | 奎斯特诊断投资股份有限公司 | For screening the composition and method of solid tumor |
| EP3303610A4 (en) * | 2015-05-27 | 2019-01-02 | Quest Diagnostics Investments Incorporated | Compositions and methods for screening solid tumors |
| US10689710B2 (en) | 2015-05-27 | 2020-06-23 | Quest Diagnostics Investments Incorporated | Methods for screening solid tumors for mutations |
| US11242569B2 (en) | 2015-12-17 | 2022-02-08 | Guardant Health, Inc. | Methods to determine tumor gene copy number by analysis of cell-free DNA |
| US11485996B2 (en) | 2016-10-04 | 2022-11-01 | Natera, Inc. | Methods for characterizing copy number variation using proximity-litigation sequencing |
| US11459616B2 (en) | 2016-10-24 | 2022-10-04 | The Chinese University Of Hong Kong | Methods and systems for tumor detection |
| WO2018083467A1 (en) * | 2016-11-02 | 2018-05-11 | Ucl Business Plc | Method of detecting tumour recurrence |
| US11519028B2 (en) | 2016-12-07 | 2022-12-06 | Natera, Inc. | Compositions and methods for identifying nucleic acid molecules |
| US11530442B2 (en) | 2016-12-07 | 2022-12-20 | Natera, Inc. | Compositions and methods for identifying nucleic acid molecules |
| US10533219B2 (en) | 2016-12-07 | 2020-01-14 | Natera, Inc. | Compositions and methods for identifying nucleic acid molecules |
| US10011870B2 (en) | 2016-12-07 | 2018-07-03 | Natera, Inc. | Compositions and methods for identifying nucleic acid molecules |
| US10577650B2 (en) | 2016-12-07 | 2020-03-03 | Natera, Inc. | Compositions and methods for identifying nucleic acid molecules |
| US11479825B2 (en) | 2017-01-25 | 2022-10-25 | The Chinese University Of Hong Kong | Diagnostic applications using nucleic acid fragments |
| WO2018137685A1 (en) * | 2017-01-25 | 2018-08-02 | The Chinese University Of Hong Kong | Diagnostic applications using nucleic acid fragments |
| US10633713B2 (en) | 2017-01-25 | 2020-04-28 | The Chinese University Of Hong Kong | Diagnostic applications using nucleic acid fragments |
| US10894976B2 (en) | 2017-02-21 | 2021-01-19 | Natera, Inc. | Compositions, methods, and kits for isolating nucleic acids |
| CN107385097A (en) * | 2017-09-13 | 2017-11-24 | 山东大学第二医院 | Detect the kit of SMAD4 gene V354L site mutations |
| US11525159B2 (en) | 2018-07-03 | 2022-12-13 | Natera, Inc. | Methods for detection of donor-derived cell-free DNA |
| CN109837351A (en) * | 2019-03-29 | 2019-06-04 | 南昌艾迪康医学检验实验室有限公司 | Utilize the kit of real-time fluorescence quantitative PCR detection PDGFR α gene relative expression quantity |
| US11959139B2 (en) | 2023-05-12 | 2024-04-16 | Guardant Health, Inc. | Methods and systems for detecting genetic variants |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| WO2014004726A1 (en) | Methods, compositions and kits for the diagnosis, prognosis and monitoring of cancer | |
| US20200385817A1 (en) | Compositions and methods for screening solid tumors | |
| US10400277B2 (en) | DNA mutation detection employing enrichment of mutant polynucleotide sequences and minimally invasive sampling | |
| JP6269494B2 (en) | Method for obtaining information on endometrial cancer, and marker and kit for obtaining information on endometrial cancer | |
| US11384401B2 (en) | Detecting gastrointestinal neoplasms | |
| EP2514836B1 (en) | Prostate cancer markers | |
| EP2698436A1 (en) | Colorectal cancer markers | |
| US10851408B2 (en) | Methods and kits for detecting gene mutations | |
| WO2014173905A2 (en) | Methods and kits for prognosis of stage i nsclc by determining the methylation pattern of cpg dinucleotides | |
| CN109642228B (en) | Methylation control DNA | |
| WO2016044142A1 (en) | Bladder cancer detection and monitoring | |
| US11535897B2 (en) | Composite epigenetic biomarkers for accurate screening, diagnosis and prognosis of colorectal cancer | |
| US20140242583A1 (en) | Assays, methods and compositions for diagnosing cancer | |
| CN116555423A (en) | Lung cancer methylation marker combination, detection product and application thereof | |
| US11542559B2 (en) | Methylation-based biomarkers in breast cancer screening, diagnosis, or prognosis | |
| KR20210146983A (en) | Detection of Pancreatic Coronary Adenocarcinoma in Plasma | |
| EP3075851B1 (en) | Method for acquiring information on gastric cancer and kit for detection of gastric cancer | |
| JP6418594B2 (en) | Method for obtaining information on endometrial cancer, and marker and kit for obtaining information on endometrial cancer | |
| US20240093302A1 (en) | Non-invasive cancer detection based on dna methylation changes | |
| Wang et al. | Highly Selective, Single-Tube Colorimetric Assay for Detection of Multiple Mutations in the Epidermal Growth Factor Receptor Gene | |
| WO2023135600A1 (en) | Personalized cancer management and monitoring based on dna methylation changes in cell-free dna | |
| CN116555422A (en) | Lung cancer methylation marker, detection kit and application thereof | |
| CN115820846A (en) | Thyroid cancer related gene mutation and methylation detection composition and application thereof | |
| CN115820862A (en) | Digital PCR kit for detecting human EGFR gene multiple mutation sites | |
| Class et al. | Patent application title: Detection of hepatitis B virus (HBV) DNA and methylated HBV DNA in urine of patients with HBV-associated hepatocellular carcinoma Inventors: Wei Song (Audubon, PA, US) Surbhi Jain (Doylestown, PA, US) Batbold Boldbaatar (Coatesville, PA, US) Sitong Chen (Audubon, PA, US) Assignees: JBS Science Inc. |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 13809972 Country of ref document: EP Kind code of ref document: A1 |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| 122 | Ep: pct application non-entry in european phase |
Ref document number: 13809972 Country of ref document: EP Kind code of ref document: A1 |