WO2013173638A1 - Compositions and methods for modulating smn gene family expression - Google Patents
Compositions and methods for modulating smn gene family expression Download PDFInfo
- Publication number
- WO2013173638A1 WO2013173638A1 PCT/US2013/041440 US2013041440W WO2013173638A1 WO 2013173638 A1 WO2013173638 A1 WO 2013173638A1 US 2013041440 W US2013041440 W US 2013041440W WO 2013173638 A1 WO2013173638 A1 WO 2013173638A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- single stranded
- stranded oligonucleotide
- nucleotide
- nucleotides
- oligonucleotide
- Prior art date
Links
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/113—Non-coding nucleic acids modulating the expression of genes, e.g. antisense oligonucleotides; Antisense DNA or RNA; Triplex- forming oligonucleotides; Catalytic nucleic acids, e.g. ribozymes; Nucleic acids used in co-suppression or gene silencing
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K31/00—Medicinal preparations containing organic active ingredients
- A61K31/70—Carbohydrates; Sugars; Derivatives thereof
- A61K31/7088—Compounds having three or more nucleosides or nucleotides
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K48/00—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P21/00—Drugs for disorders of the muscular or neuromuscular system
- A61P21/02—Muscle relaxants, e.g. for tetanus or cramps
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07H—SUGARS; DERIVATIVES THEREOF; NUCLEOSIDES; NUCLEOTIDES; NUCLEIC ACIDS
- C07H21/00—Compounds containing two or more mononucleotide units having separate phosphate or polyphosphate groups linked by saccharide radicals of nucleoside groups, e.g. nucleic acids
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/113—Non-coding nucleic acids modulating the expression of genes, e.g. antisense oligonucleotides; Antisense DNA or RNA; Triplex- forming oligonucleotides; Catalytic nucleic acids, e.g. ribozymes; Nucleic acids used in co-suppression or gene silencing
- C12N15/1131—Non-coding nucleic acids modulating the expression of genes, e.g. antisense oligonucleotides; Antisense DNA or RNA; Triplex- forming oligonucleotides; Catalytic nucleic acids, e.g. ribozymes; Nucleic acids used in co-suppression or gene silencing against viruses
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/113—Non-coding nucleic acids modulating the expression of genes, e.g. antisense oligonucleotides; Antisense DNA or RNA; Triplex- forming oligonucleotides; Catalytic nucleic acids, e.g. ribozymes; Nucleic acids used in co-suppression or gene silencing
- C12N15/1131—Non-coding nucleic acids modulating the expression of genes, e.g. antisense oligonucleotides; Antisense DNA or RNA; Triplex- forming oligonucleotides; Catalytic nucleic acids, e.g. ribozymes; Nucleic acids used in co-suppression or gene silencing against viruses
- C12N15/1132—Non-coding nucleic acids modulating the expression of genes, e.g. antisense oligonucleotides; Antisense DNA or RNA; Triplex- forming oligonucleotides; Catalytic nucleic acids, e.g. ribozymes; Nucleic acids used in co-suppression or gene silencing against viruses against retroviridae, e.g. HIV
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/10—Type of nucleic acid
- C12N2310/11—Antisense
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/31—Chemical structure of the backbone
- C12N2310/315—Phosphorothioates
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/32—Chemical structure of the sugar
- C12N2310/323—Chemical structure of the sugar modified ring structure
- C12N2310/3231—Chemical structure of the sugar modified ring structure having an additional ring, e.g. LNA, ENA
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/34—Spatial arrangement of the modifications
- C12N2310/343—Spatial arrangement of the modifications having patterns, e.g. ==--==--==--
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/35—Nature of the modification
- C12N2310/351—Conjugate
- C12N2310/3517—Marker; Tag
Definitions
- the invention relates to oligonucleotide based compositions, as well as methods of using oligonucleotide based compositions for treating disease.
- SMA Spinal muscular atrophy
- SMA type I is the most severe form and is one of the most common causes of infant mortality, with symptoms of muscle weakness and difficulty breathing occurring at birth.
- SMA type II occurs later, with muscle weakness and other symptoms developing from ages 6 month to 2 years. Symptoms appear in SMA type III during childhood and in SMA type IV, the mildest form, during adulthood. All four types of SMA have been found to be associated with mutations in the SMN gene family, particularly SMN1.
- SNS motor neuron
- snRNPs small nuclear ribonucleoproteins
- snRNPs protein-RNA complexes that bind with pre-mRNA to form a spliceosome, which then splices the pre-mRNA, most often resulting in removal of introns.
- SMNl and SMN2 are a result of a gene duplication at 5ql3 in humans.
- a lack of SMN activity results in widespread splicing defects, especially in spinal motor neurons, and degeneration of the spinal cord lower motor neurons.
- aspects of the invention disclosed herein provide methods and compositions that are useful for upregulating the expression of certain genes in cells.
- single stranded oligonucleotides are provided that target a PRC2-associated region of a gene and thereby cause upregulation of the gene.
- methods and related single stranded oligonucleotides that are useful for selectively inducing expression of particular splice variants of genes.
- the methods are useful for controlling the levels in a cell of particular protein isoforms encoded by the splice variants.
- the methods are useful for inducing expression of proteins to levels sufficient to treat disease.
- single stranded oligonucleotides are provided that target a
- PRC2-associated region of a SMN gene e.g., human SMNl, human SMN2
- methods are provided for increasing expression of full-length SMN protein in a cell for purposes of treating SMA.
- aspects of the invention disclosed herein provide methods and compositions that are useful for upregulating SMNl or SMN2 in cells.
- single stranded oligonucleotides are provided that target a PRC2-associated region of the gene encoding SMNl or SMN2.
- these single stranded oligonucleotides activate or enhance expression of SMNl or SMN2 by relieving or preventing PRC2 mediated repression of SMNl or SMN2.
- the methods comprise delivering to the cell a first single stranded oligonucleotide complementary with a PRC2-associated region of an SMN gene, eg.., a PRC2-associated region of SMNl or SMN2, and a second single stranded
- oligonucleotide complementary with a splice control sequence of a precursor mRNA of an SMN gene e.g., a precursor mRNA of SMNl or SMN2
- a precursor mRNA of SMNl or SMN2 in amounts sufficient to increase expression of a mature mRNA of SMNl or SMN2 that comprises (or includes) exon 7 in the cell.
- single stranded oligonucleotides that have a region of complementarity that is complementarty with (e.g., at least 8 consecutive nucleotides of ) a PRC2-associated region of an SMN gene, e.g., a PRC2- associated region of the nucleotide sequence set forth as SEQ ID NO: 1, 2, 4, or 5.
- the oligonucleotide has at least one of the following features: a) a sequence that is 5 'X-Y-Z, in which X is any nucleotide and in which X is at the 5 ' end of the oligonucleotide, Y is a nucleotide sequence of 6 nucleotides in length that is not a human seed sequence of a microRNA, and Z is a nucleotide sequence of 1 to 23 nucleotides in length; b) a sequence that does not comprise three or more consecutive guanosine
- the single stranded oligonucleotide has at least two of features a), b), c), d), and e), each independently selected. In some embodiments, the single stranded
- oligonucleotide has at least three of features a), b), c), d), and e), each independently selected. In some embodiments, the single stranded oligonucleotide has at least four of features a), b), c), d), and e), each independently selected. In some embodiments, the single stranded oligonucleotide has each of features a), b), c), d), and e). In certain embodiments, the oligonucleotide has the sequence 5 'X-Y-Z, in which the oligonucleotide is 8-50 nucleotides in length.
- single stranded oligonucleotides have a sequence X-Y-Z, in which X is any nucleotide, Y is a nucleotide sequence of 6 nucleotides in length that is not a seed sequence of a human microRNA, and Z is a nucleotide sequence of 1 to 23 nucleotides in length, in which the single stranded oligonucleotide is complementary with a PRC2-associated region of an SMN gene, e.g., a PRC2-associated region of the nucleotide sequence set forth as SEQ ID NO: 1, 2, 4, or 5.
- single stranded oligonucleotides have a sequence 5 ' -X-Y-Z, in which X is any nucleotide, Y is a nucleotide sequence of 6 nucleotides in length that is not a seed sequence of a human microRNA, and Z is a nucleotide sequence of 1 to 23 nucleotides in length, in which the single stranded oligonucleotide is complementary with at least 8 consecutive nucleotides of a PRC2-associated region of an SMN gene, e.g., a PRC2-associated region of the nucleotide sequence set forth as SEQ ID NO: 1, 2, 4, or 5.
- Y is a sequence selected from Table 1.
- the PRC2-associated region is a sequence listed in any one of SEQ ID NOS: 9 to 18.
- the single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 13087, or a fragment thereof that is at least 8 nucleotides. In some embodiments, the single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 13087, in which the 5' end of the nucleotide sequence provided is the 5 ' end of the oligonucleotide. In some
- the region of complementarity (e.g., the at least 8 consecutive nucleotides) is also present within the nucleotide sequence set forth as SEQ ID NO: 7 or 8.
- a PRC2-associated region is a sequence listed in any one of SEQ ID NOS: 9 to 14.
- the single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 8329 and 13093 to
- the single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 8329 and 13093 to 13094, wherein the 5' end of the nucleotide sequence provided is the 5' end of the oligonucleotide.
- nucleotide sequence set forth as SEQ ID NO: 7.
- a PRC2-associated region is a sequence listed in any one of SEQ ID NOS: 15 to 18.
- the single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 1158-1159, 1171, 1482-1483, 1485-1486, 2465-2471, 2488-2490, 2542-2546, 2656-2657, 2833-2835, 3439-3440, 3916- 3918, 4469-4472, 4821, 5429, 5537, 6061, 7327, 8330-13061, and 13062-13087 or a fragment thereof that is at least 8 nucleotides.
- the at least 8 consecutive nucleotides are present within the nucleotide sequence set forth as SEQ ID NO: 8.
- the single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 13087.
- the oligonucleotide is up to 50 nucleotides in length.
- the single stranded oligonucleotide comprises a fragment of at least 8 nucleotides of a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 13087.
- a single stranded oligonucleotide comprises a nucleotide sequence as set forth in Table 4. In some embodiments, the single stranded oligonucleotide comprises a fragment of at least 8 nucleotides of a nucleotide sequence as set forth in Table 4. In some embodiments, a single stranded oligonucleotide consists of a nucleotide sequence as set forth in Table 4.
- compounds that comprise the general formula A-B-C, wherein A is a single stranded oligonucleotide complementary with at least 8 consecutive nucleotides of a PRC2-associated region of a gene, B is a linker, and C is a single stranded oligonucleotide complementary to a splice control sequence of a precursor mR A of the gene.
- B comprises an oligonucleotide, peptide, low pH labile bond, or disulfide bond.
- the splice control sequence resides in an exon of the gene.
- the splice control sequence traverses an intron-exon junction of the gene. In some embodiments, the splice control sequence resides in an intron of the gene. In some embodiments, the splice control sequence comprises at least one hnR AP binding sequence. In some embodiments, hybridization of an oligonucleotide having the sequence of C with the splice control sequence of the precursor mRNA in a cell results in inclusion of a particular exon in a mature mRNA that arises from processing of the precursor mRNA in the cell.
- hybridization of an oligonucleotide having the sequence of C with the splice control sequence of the precursor mRNA in a cell results in exclusion of a particular intron or exon in a mature mRNA that arises from processing of the precursor mRNA in the cell.
- the gene is SMNl or SMN2.
- the splice control sequence resides in intron 6, intron 7, exon 7, exon 8 or at the junction of intron 7 and exon 8 of SMNl or SMN2.
- the splice control sequence comprises the sequence: CAGCAUUAUGAAAG (SEQ ID NO: 13100).
- B comprises a sequence selected from: TCACTTTCATAATGCTGG (SEQ ID NO: 13088); TCACTTTCATAATGC (SEQ ID NO: 13089); CACTTTCATAATGCT (SEQ ID NO:
- A has a sequence 5'-X-Y-Z, wherein X is any nucleotide, Y is a nucleotide sequence of 6 nucleotides in length that is not a seed sequence of a human microRNA, and Z is a nucleotide sequence of 1-23 nucleotides in length.
- the PRC2-associated region of an SMN2 gene is a PRC2-associated region within SEQ ID NO: 1, 2, 4 or 5.
- Y is a sequence selected from Table
- the PRC2-associated region is a sequence set forth in any one of SEQ ID NOS: 9 to 23.
- A comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 8329 and 13088 to 13094 or a fragment thereof that is at least 8 nucleotides.
- A comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 8329 and 13088 to 13094, wherein the 5' end of the nucleotide sequence provided is the 5' end of A.
- the at least 8 consecutive nucleotides are also present within the nucleotide sequence set forth as SEQ ID NO: 7.
- the PRC2-associated region is a sequence set forth in any one of SEQ ID NOS: 24 to 29.
- A comprises a nucleotide sequence as set forth in any one of
- the at least 8 consecutive nucleotides are present within the nucleotide sequence set forth as SEQ ID NO: 8.
- A does not comprise three or more consecutive guanosine nucleotides.
- A does not comprise four or more consecutive guanosine nucleotides.
- a or C is 8 to 30 nucleotides in length. In some embodiments, A is 8 to 10 nucleotides in length and all but 1,
- B is an oligonucleotide comprising
- B is more susceptible to cleavage in a mammalian extract than A and C.
- A comprises a nucleotide sequence selected from
- GCTUTGGGAAGUAUG (SEQ ID NO: 11394), CUTUGGGAAGTATG (SEQ ID NO: 11395) and GGTACATGAGTGGCT (SEQ ID NO: 11419);
- B comprises the nucleotide sequence TTTT or UUUU; and
- C comprises the nucleotide sequence
- TCACTTTCATAATGCTGG (SEQ ID NO: 13088); TCACTTTCATAATGC (SEQ ID NO: 13089); CACTTTCATAATGCT (SEQ ID NO: 13090); ACTTTCATAATGCTG (SEQ ID NO: 13091); or CTTTCATAATGCTGG (SEQ ID NO: 13092), and wherein the 3' end of A is linked to the 5' end of B, and the 3' end of B is linked to 5' end of C.
- the single stranded oligonucleotide does not comprise three or more consecutive guanosine nucleotides. In some embodiments, the single stranded oligonucleotide does not comprise four or more consecutive guanosine nucleotides.
- the single stranded oligonucleotide is 8 to 30 nucleotides in length. In some embodiments, the single stranded oligonucleotide is up to 50 nucleotides in length. In some embodiments, the single stranded oligonucleotide is 8 to 10 nucleotides in length and all but 1, 2, or 3 of the nucleotides of the complementary sequence of the PRC2- associated region are cytosine or guanosine nucleotides.
- the single stranded oligonucleotide is complementary with at least 8 consecutive nucleotides of a PRC2-associated region of an SMN gene, e.g., a PRC2- associated region of a nucleotide sequence set forth as SEQ ID NO: 1, 2, 4, or 5, in which the nucleotide sequence of the single stranded oligonucleotide comprises one or more of a nucleotide sequence selected from the group consisting of
- At least one nucleotide of the oligonucleotide is a nucleotide analogue.
- the at least one nucleotide analogue results in an increase in Tm of the oligonucleotide in a range of 1 to 5 °C compared with an oligonucleotide that does not have the at least one nucleotide analogue.
- At least one nucleotide of the oligonucleotide comprises a 2' O-methyl. In some embodiments, each nucleotide of the oligonucleotide comprises a 2' O- methyl. In some embodiments, the oligonucleotide comprises at least one ribonucleotide, at least one deoxyribonucleotide, or at least one bridged nucleotide. In some embodiments, the bridged nucleotide is a LNA nucleotide, a cEt nucleotide or a ENA modified nucleotide. In some embodiments, each nucleotide of the oligonucleotide is a LNA nucleotide.
- the nucleotides of the oligonucleotide comprise alternating deoxyribonucleotides and 2'-fluoro-deoxyribonucleotides. In some embodiments, the nucleotides of the oligonucleotide comprise alternating deoxyribonucleotides and 2'-0- methyl nucleotides. In some embodiments, the nucleotides of the oligonucleotide comprise alternating deoxyribonucleotides and ENA nucleotide analogues. In some embodiments, the nucleotides of the oligonucleotide comprise alternating deoxyribonucleotides and LNA nucleotides. In some embodiments, the 5' nucleotide of the oligonucleotide is a
- the nucleotides of the oligonucleotide comprise alternating LNA nucleotides and 2 '-O-methyl nucleotides.
- the 5' nucleotide of the oligonucleotide is a LNA nucleotide.
- the nucleotides of the oligonucleotide comprise deoxyribonucleotides flanked by at least one LNA nucleotide on each of the 5' and 3' ends of the deoxyribonucleotides.
- the single stranded oligonucleotide comprises modified internucleotide linkages (e.g., phosphorothioate internucleotide linkages or other linkages) between at least two, at least three, at least four, at least five or more nucleotides. In some embodiments, the single stranded oligonucleotide comprises modified internucleotide linkages (e.g., phosphorothioate internucleotide linkages or other linkages) between between all nucleotides.
- modified internucleotide linkages e.g., phosphorothioate internucleotide linkages or other linkages
- the nucleotide at the 3' position of the oligonucleotide has a 3' hydroxyl group. In some embodiments, the nucleotide at the 3' position of the
- the oligonucleotide has a 3' thiophosphate.
- the single stranded oligonucleotide has a biotin moiety or other moiety conjugated to its 5' or 3' nucleotide.
- the single stranded oligonucleotide has cholesterol, Vitamin A, folate, sigma receptor ligands, aptamers, peptides, such as CPP, hydrophobic molecules, such as lipids, ASGPR or dynamic polyconjugates and variants thereof at its 5' or 3' end.
- compositions are provided that comprise any of the oligonucleotides disclosed herein, and a carrier.
- compositions are provided that comprise any of the oligonucleotides in a buffered solution.
- the oligonucleotide is conjugated to the carrier.
- the carrier is a peptide.
- the carrier is a steroid.
- pharmaceutical compositions are provided that comprise any of the oligonucleotides disclosed herein, and a pharmaceutically acceptable carrier.
- kits that comprise a container housing any of the compositions disclosed herein.
- methods of increasing expression of SMNl or SMN2 in a cell involve delivering any one or more of the single stranded oligonucleotides disclosed herein into the cell.
- delivery of the single stranded oligonucleotide into the cell results in a level of expression of SMNl or SMN2 that is greater (e.g., at least 50% greater) than a level of expression of SMNl or SMN2 in a control cell that does not comprise the single stranded oligonucleotide.
- methods of increasing levels of SMNl or SMN2 in a subject are provided.
- methods of treating a condition e.g., Spinal muscular atrophy
- the methods involve
- aspects of the invention relate to methods of increasing expression of SMN protein in a cell.
- the method comprise delivering to the cell a first single stranded oligonucleotide complementary with at least 8 consecutive nucleotides of a PRC2- associated region of SMN2 and a second single stranded oligonucleotide complementary with a splice control sequence of a precursor mRNA of SMN2, in amounts sufficient to increase expression of a mature mRNA of SMN2 that comprises exon 7 in the cell.
- the region of complementarity with at least 8 consecutive nucleotides of a PRC2-associated region of SMN 2 has at least 1, at least 2, at least 3, at least 4, at least 5, at least 6, at least 7, at least 8, or more mismatches with a corresponding region of SMN1.
- splice control sequence refers to a nucleotide sequence that when present in a precursor mR A influences splicing of that precursor mR A in a cell.
- a splice control sequence includes one or more binding sites for a molecule that regulates mRNA splicing, such as a hnRNAP protein.
- a splice control sequence comprises the sequence CAG or AAAG. In some embodiments, a splice control sequence resides in an exon (e.g., an exon of SMN1 or SMN2, such as exon 7 or exon 8). In some embodiments, a splice control sequence traverses an intron-exon junction (e.g., an intron-exon junction of SMN1 or SMN2, such as the intron 6/exon 7 junction or the intron 7/exon 8 junction). In some embodiments, a splice control sequence resides in an intron (e.g., an intron of SMN1 or SMN2, such as intron 6 or intron 7). In some embodiments, a splice control sequence comprises the sequence: CAGCAUUAUGAAAG (SEQ ID NO: 13100) or a portion thereof.
- the second single stranded oligonucleotide is splice switching oligonucleotide that comprises a sequence selected from: TCACTTTCATAATGCTGG (SEQ ID NO: 13088); TCACTTTCATAATGC (SEQ ID NO: 13089);
- the second single stranded oligonucleotide is 8 to 30 nucleotides in length.
- the first single stranded oligonucleotide has a sequence 5'-X-
- the PRC2-associated region of an SMN2 gene is a PRC2-associated region within SEQ ID NO: 1, 2, 4 or 5.
- Y is a sequence selected from Table 1.
- the PRC2-associated region is a sequence set forth in any one of SEQ ID NOS: 9 to 23.
- the first single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 8329 and 13088 to 13094 or a fragment thereof that is at least 8 nucleotides.
- the first single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 8329 and 13088 to 13094, wherein the 5' end of the nucleotide sequence provided is the 5' end of the first single stranded oligonucleotide.
- the at least 8 consecutive nucleotides are also present within the nucleotide sequence set forth as SEQ ID NO: 7.
- the PRC2-associated region is a sequence set forth in any one of SEQ ID NOS: 24 to 29.
- the first single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 1158-1159, 1171, 1482-1483, 1485-1486, 2465-2471, 2488-2490, 2542-2546, 2656-2657, 2833-2835, 3439- 3440, 3916-3918, 4469-4472, 4821, 5429, 5537, 6061, 7327, 8330-13061, and 13062-13087 or a fragment thereof that is at least 8 nucleotides.
- the at least 8 consecutive nucleotides are present within the nucleotide sequence set forth as SEQ ID NO: 8.
- the first single stranded oligonucleotide does not comprise three or more consecutive guanosine nucleotides.
- the first single stranded oligonucleotide does not comprise four or more consecutive guanosine nucleotides.
- the first single stranded oligonucleotide is 8 to 30 nucleotides in length.
- the first single stranded oligonucleotide is 8 to 10 nucleotides in length and all but 1, 2, or 3 of the nucleotides of the complementary sequence of the PRC2- associated region are cytosine or guanosine nucleotides.
- the first single stranded oligonucleotide and the second single stranded oligonucleotide are delivered to the cell simultaneously.
- the cell is in a subject and the step of delivering to the cell comprises administering the first single stranded oligonucleotide and the second single stranded oligonucleotide to the subject as a co-formulation.
- the first single stranded oligonucleotide is covalently linked to the second single stranded oligonucleotide through a linker.
- the linker comprises an oligonucleotide, a peptide, a low pH-labile bond, or a disulfide bond. In some embodiments, the linker comprises an oligonucleotide, optionally wherein the oligonucleotide comprises 1 to 10 thymidines or uridines. In some embodiments, the linker is more susceptible to cleavage in a mammalian extract than the first and second single stranded oligonucleotides. In some embodiments, the first single stranded
- oligonucleotide is not covalently linked to the second single stranded oligonucleotide.
- the first single stranded oligonucleotide and the second single stranded oligonucleotide are delivered to the cell separately.
- methods for treating spinal muscular atrophy in a subject.
- the methods comprise administering to the subject a first single stranded oligonucleotide complementary with at least 8 consecutive nucleotides of a PRC2-associated region of SMN2 and a second single stranded oligonucleotide complementary with a splice control sequence of a precursor mRNA of SMN2, in amounts sufficient to increase expression of SMN protein in the subject.
- methods for treating spinal muscular atrophy in a subject that involve administering to the subject a first single stranded oligonucleotide complementary with a PRC2-associated region of SMN2 and a second single stranded oligonucleotide complementary with a splice control sequence of a precursor mRNA of SMN2, in amounts sufficient to increase expression of SMN protein in the subject.
- compositions are provided that comprise a first single stranded oligonucleotide complementary with at least 8 consecutive nucleotides of a PRC2-associated region of SMN2, and a second single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of SMN2.
- compositions are provided that comprise a single stranded
- kits comprising single stranded oligonucleotides that regulate SMNl or SMN2 expression are also provided.
- compositions are provided that comprise any of the oligonucleotides or compounds disclosed herein, and a carrier.
- compositions are provided that comprise any of the oligonucleotides or compounds in a buffered solution.
- the oligonucleotide is conjugated to the carrier.
- the carrier is a peptide.
- the carrier is a steroid.
- pharmaceutical compositions are provided that comprise any of the oligonucleotides disclosed herein, and a pharmaceutically acceptable carrier.
- compositions that comprise a first single stranded oligonucleotide complementary with at least 8 consecutive nucleotides of a PRC2-associated region of SMN2, and a second single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of SMN2.
- the splice control sequence resides in an exon of SMN2.
- the exon is exon 7 or exon 8.
- the splice control sequence traverses an intron-exon junction of SMN2.
- the intron-exon junction is the intron 6/exon 7 junction or the intron 7/exon 8 junction.
- the splice control sequence resides in an intron of SMN2.
- the intron is intron 6 or intron 7 (SEQ ID NO: 13101).
- the splice control sequence comprises the sequence: CAGCAUUAUGAAAG (SEQ ID NO: 13100) or a portion thereof.
- the splice control sequence comprises at least one hnRNAP binding sequence.
- the second single stranded oligonucleotide comprises a sequence selected from: TCACTTTCATAATGCTGG (SEQ ID NO: 13088); TCACTTTCATAATGC (SEQ ID NO: 13089); CACTTTCATAATGCT (SEQ ID NO: 13090); ACTTTCATAATGCTG (SEQ ID NO: 13091); and
- the first single stranded oligonucleotide has a sequence 5'-X-Y-Z, wherein X is any nucleotide, Y is a nucleotide sequence of 6 nucleotides in length that is not a seed sequence of a human microRNA, and Z is a nucleotide sequence of 1-23 nucleotides in length.
- the PRC2-associated region of SMN2 is a PRC2-associated region within SEQ ID NO: 1,2, 4 or 5.
- Y is a sequence selected from Table 1.
- the PRC2-associated region is a sequence set forth in any one of SEQ ID NOS: 9 to 23.
- the first single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 8329 and 13093 to 13094 or a fragment thereof that is at least 8 nucleotides.
- the first single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 8329 and 13093 to 13094, wherein the 5' end of the nucleotide sequence provided is the 5' end of the first single stranded oligonucleotide.
- the at least 8 consecutive nucleotides are also present within the nucleotide sequence set forth as SEQ ID NO: 7.
- the PRC2-associated region is a sequence set forth in any one of SEQ ID NOS: 24 to 29.
- the first single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 1158-1159, 1171, 1482-1483, 1485-1486, 2465-2471, 2488-2490, 2542-2546, 2656-2657, 2833-2835, 3439-3440, 3916-3918, 4469-4472, 4821, 5429, 5537, 6061, 7327, 8330-13061, and 13062-13087 or a fragment thereof that is at least 8 nucleotides.
- the at least 8 consecutive nucleotides are present within the nucleotide sequence set forth as SEQ ID NO: 8.
- the first single stranded oligonucleotide does not comprise three or more consecutive guanosine nucleotides. In some embodiments, the first single stranded oligonucleotide does not comprise four or more consecutive guanosine nucleotides. In some embodiments, the first and/or second single stranded oligonucleotide is 8 to 30 nucleotides in length. In some embodiments, the first single stranded oligonucleotide is 8 to 10 nucleotides in length and all but 1, 2, or 3 of the nucleotides of the complementary sequence of the PRC2-associated region are cytosine or guanosine nucleotides.
- the first single stranded oligonucleotide is covalently linked to the second single stranded oligonucleotide through a linker.
- the linker comprises an oligonucleotide, a peptide, a low pH-labile bond, or a disulfide bond.
- the linker comprises an oligonucleotide, optionally wherein the
- oligonucleotide comprises 1 to 10 thymidines or uridines.
- the linker is more susceptible to cleavage in a mammalian extract than the first and second single stranded oligonucleotides.
- the first single stranded oligonucleotide is not covalently linked to the second single stranded oligonucleotide.
- the composition further comprises a carrier.
- the carrier is a
- Further aspects of the invention provide methods for selecting oligonucleotides for activating or enhancing expression of SMNl or SMN2.
- methods are provided for selecting a set of oligonucleotides that is enriched in candidates (e.g., compared with a random selection of oligonucleotides) for activating or enhancing expression of SMNl or SMN2.
- the methods may be used to establish sets of clinical candidates that are enriched in oligonucleotides that activate or enhance expression of SMNl or SMN2.
- Such libraries may be utilized, for example, to identify lead oligonucleotides for developing therapeutics to treat SMNl or SMN2.
- oligonucleotide chemistries are provided that are useful for controlling the pharmacokinetics, biodistribution, bioavailability and/or efficacy of the single stranded oligonucleotides for activating expression of SMNl or SMN2.
- kits are provided that comprise a container housing any of the compositions disclosed herein.
- kits are provided that comprise a first container housing first single stranded oligonucleotide complementary with at least 8 consecutive nucleotides of a PRC2-associated region of a gene; and a second container housing a second single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of the gene.
- the splice control sequence resides in an exon of the gene.
- the splice control sequence traverses an intron-exon junction of the gene. In some embodiments, the splice control sequence resides in an intron of the gene. In some embodiments, the splice control sequence comprises at least one hnRNAP binding sequence. In some embodiments, hybridization of an oligonucleotide having the sequence of C with the splice control sequence of the precursor mRNA in a cell results in inclusion of a particular exon in a mature mRNA that arises from processing of the precursor mRNA in the cell.
- hybridization of an oligonucleotide having the sequence of C with the splice control sequence of the precursor mRNA in a cell results in exclusion of a particular intron or exon in a mature mRNA that arises from processing of the precursor mRNA in the cell.
- the gene is SMN1 or SMN2.
- the splice control sequence resides in intron 6, intron 7, exon 7, exon 8 or at the junction of intron 7 and exon 8.
- the splice control sequence comprises the sequence:
- the second single stranded oligonucleotide comprises a sequence selected from: TCACTTTCATAATGCTGG (SEQ ID NO: 13088); TCACTTTCATAATGC (SEQ ID NO: 13089);
- the first single stranded oligonucleotide has a sequence 5'-X-Y-Z, wherein X is any nucleotide, Y is a nucleotide sequence of 6 nucleotides in length that is not a seed sequence of a human microRNA, and Z is a nucleotide sequence of 1-23 nucleotides in length.
- the PRC2-associated region of an SMN2 gene is a PRC2-associated region within SEQ ID NO: 1, 2, 4 or 5.
- Y is a sequence selected from Table 1.
- the PRC2-associated region is a sequence set forth in any one of SEQ ID NOS: 9 to 23.
- the first single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 8329 and 13093 to 13094 or a fragment thereof that is at least 8 nucleotides.
- the first single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 30 to 8329 and 13093 to 13094, wherein the 5 ' end of the nucleotide sequence provided is the 5' end of the first single stranded oligonucleotide.
- the at least 8 consecutive nucleotides are also present within the nucleotide sequence set forth as SEQ ID NO: 7.
- the PRC2-associated region is a sequence set forth in any one of SEQ ID NOS: 24 to 29.
- the first single stranded oligonucleotide comprises a nucleotide sequence as set forth in any one of SEQ ID NOS: 1158-1159, 1171, 1482-1483, 1485-1486, 2465-2471, 2488-2490, 2542-2546, 2656-2657, 2833-2835, 3439-3440, 3916-3918, 4469-4472, 4821, 5429, 5537, 6061, 7327, 8330-13061, and 13062-13087 or a fragment thereof that is at least 8 nucleotides.
- the at least 8 consecutive nucleotides are present within the nucleotide sequence set forth as SEQ ID NO: 8.
- FIG. 1 provides a schematic of SMN 1 and SMN2 mRNA processing
- FIG. 2 provides a table outlining genotypes and patent information, including SMA classification, of cell lines tested in Example 2. Baseline SMN protein levels in the cell lines are also depicted.
- FIG. 3 depicts results of RT-PCR assays showing effects on SMN mRNA expression of oligonucleotides directed against a PRC2-associated region of SMN2 (oligos 1-52 and 59-
- FIG. 4 depicts results of RT-PCR assays showing effects on SMN mRNA expression of oligonucleotides directed against a PRC2-associated region of SMN2 (oligos 1-52 and 59-
- FIG. 5 shows that splice switching oligonucleotides (oligoes 53-58) increase expression of full length SMN2. Results are based on a gel separation analysis of PCR products obtained following a Ddel restriction digest. Two cell lines were tested, 3813 and
- Oligo 84 which targets a PRC2-associated region of SMN2, did not exhibit an increase in full length SMN2 expression when delivered alone to cells.
- FIG. 6 provides results of an SMN ELISA (Enzo) showing that certain
- oligonucleotides directed against a PRC2-associated region of SMN2 alone do not significantly increase SMN2 protein 24h post-transfection in certain SMA patient fibroblasts
- FIG. 7 provides results of an SMN ELISA showing that oligonucleotides directed against a PRC2-associated region of SMN2 in combination with a splice switching oligonucleotide (oligo 53) significantly increase SMN2 protein 24h post-transfection in SMA patient fibroblasts (compared to Lipofectamine treated cells - dashed line).
- FIG. 8 provides results of an SMN ELISA showing that oligonucleotides directed against a PRC2-associated region of SMN2 in combination with a splice switching oligonucleotide (oligo 54) significantly increase SMN2 protein 24h post-transfection in SMA patient fibroblasts (compared to Lipofectamine treated cells - dashed line).
- FIG. 9 provides results of an RT-PCR assay showing that oligonucleotides directed against a PRC2-associated region of SMN2 in combination with a splice switching oligonucleotide (oligo 53 or 54) significantly increase SMN2 protein 24h post-transfection in SMA patient fibroblasts (compared to negative control oligo and Lipofectamine treated cells).
- oligonucleotides directed against a PRC2-associated region of SMN2 in combination with a splice switching oligonucleotide (oligo 53 or 54) significantly increase SMN2 protein 24h post-transfection in SMA patient fibroblasts (compared to negative control oligo and Lipofectamine treated cells).
- oligonucleotides (DL design) were tested.
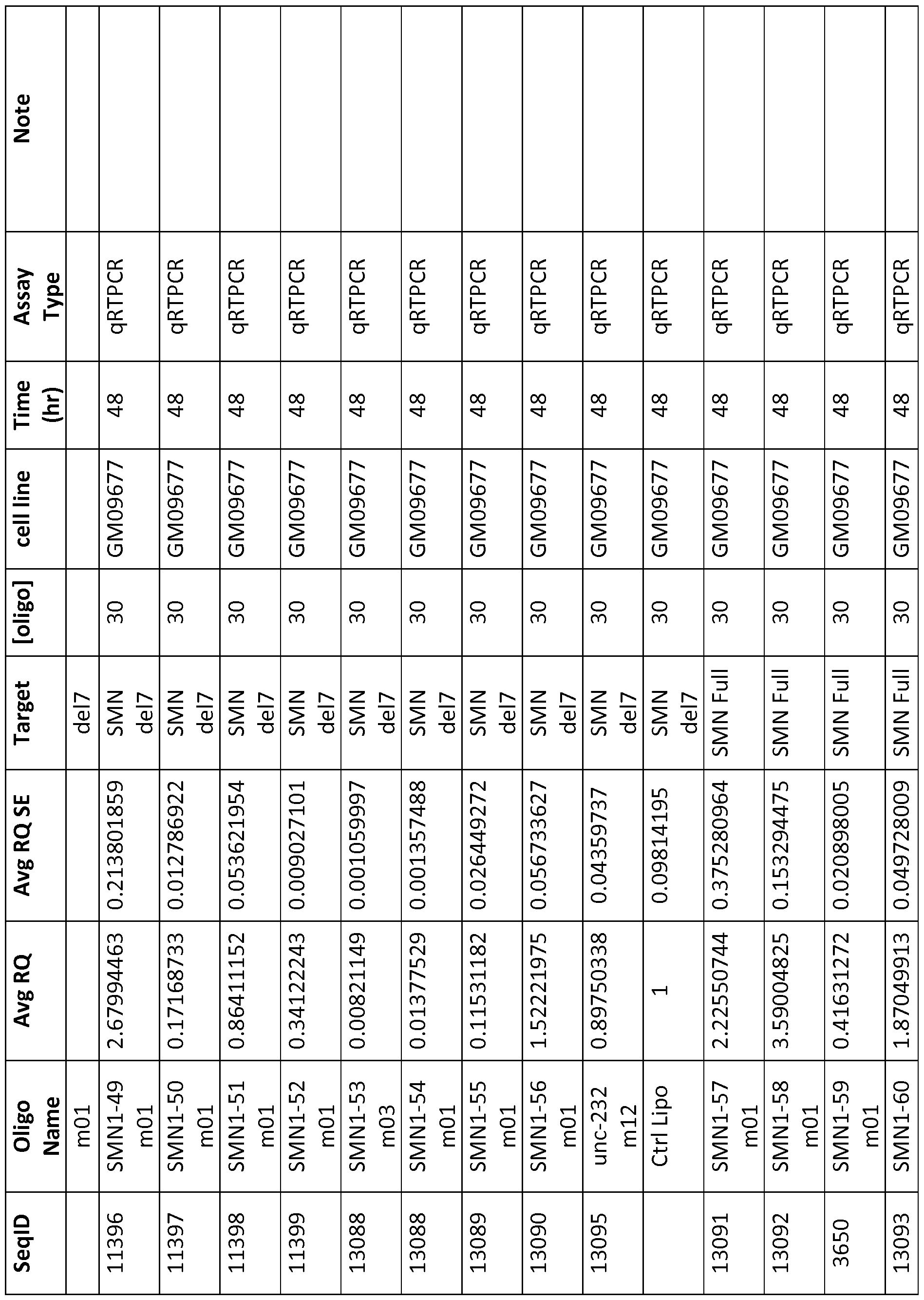
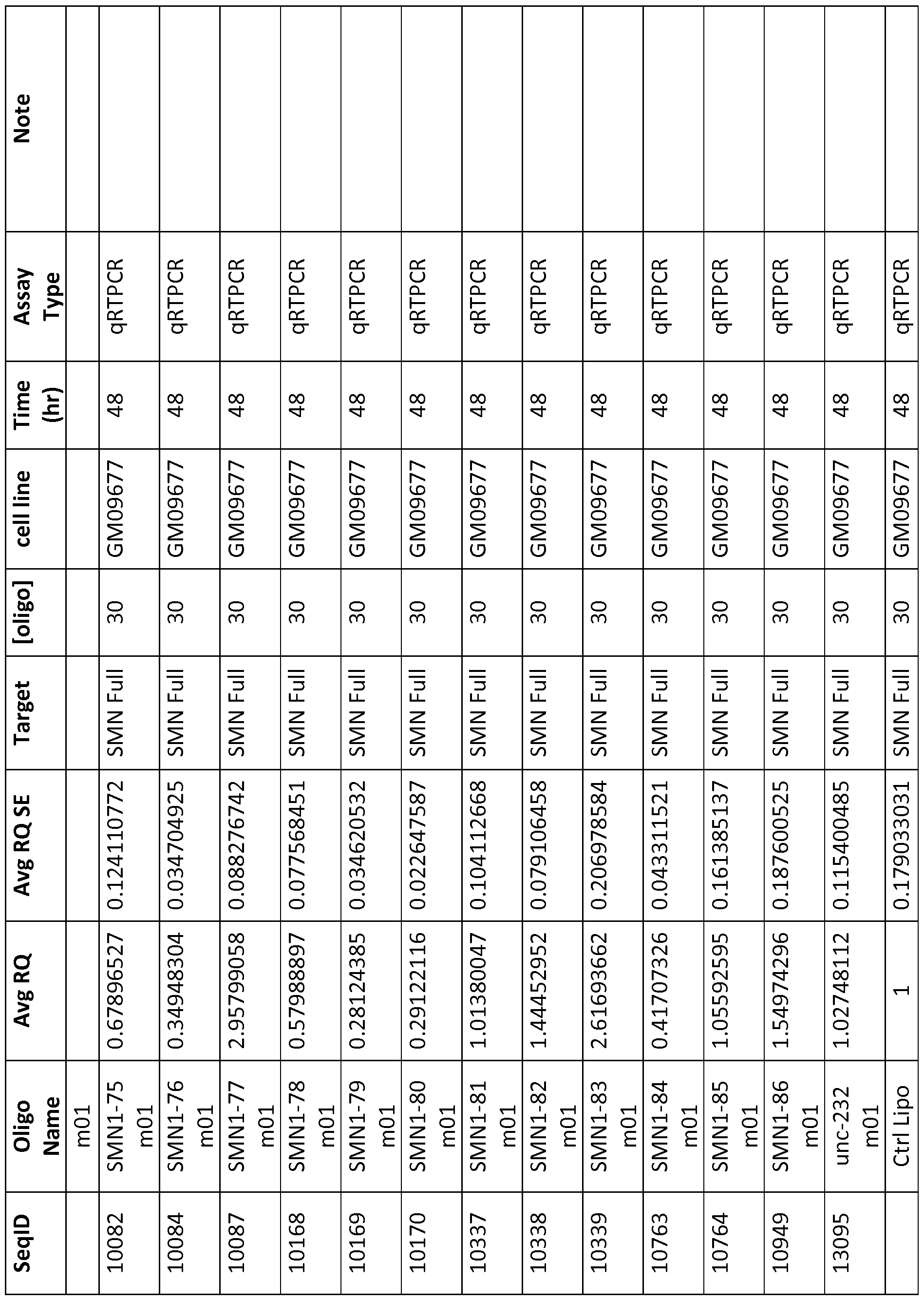
- Table 2 Oligonucleotide sequences made for testing in the lab.
- RQ column 2
- RQ SE column 3
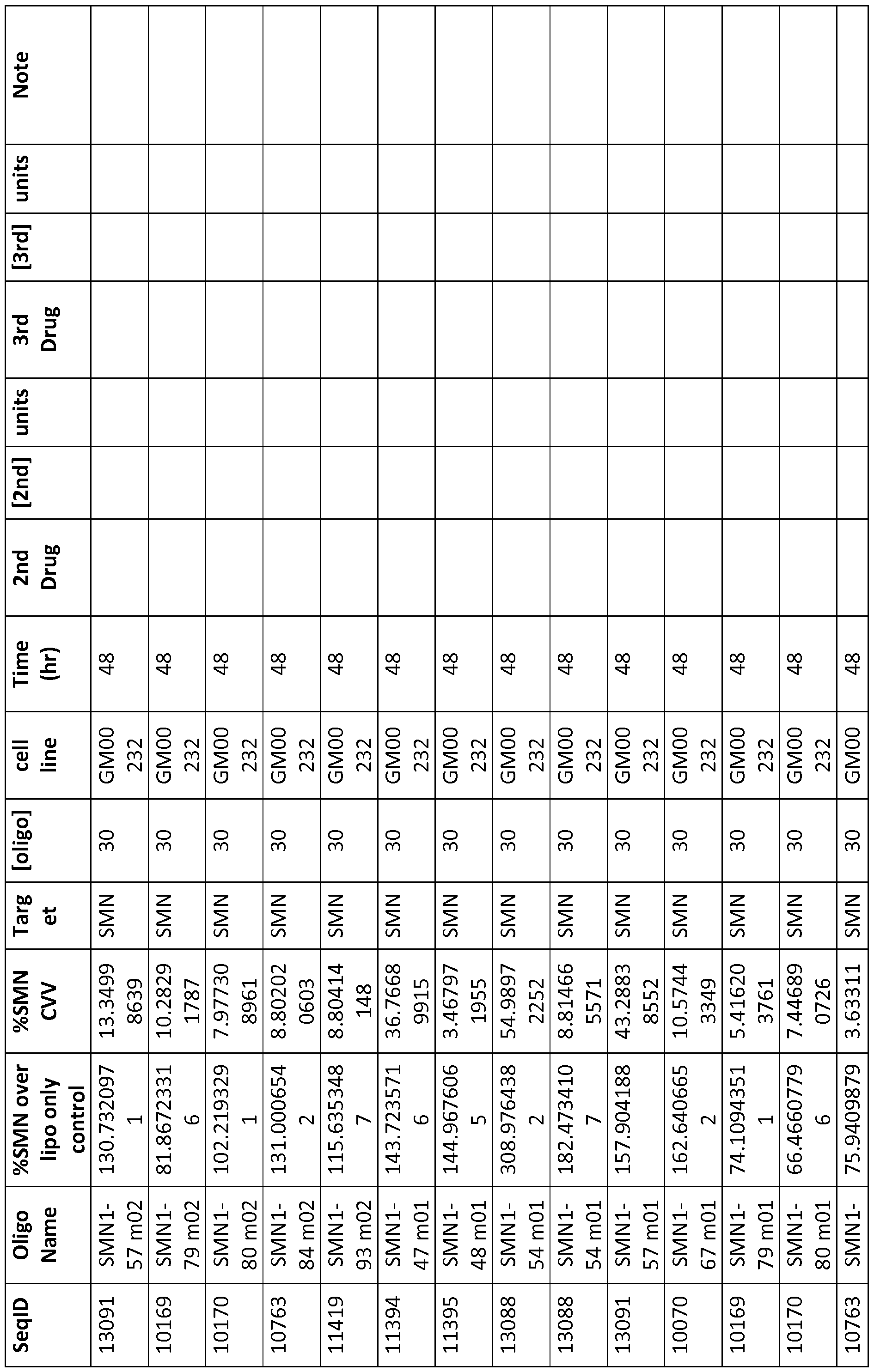
- Table 2 shows the activity of the oligo relative to a control well (usually carrier alone) and the standard error or the triplicate replicates of the experiment, [oligo] is shown in nanomolar for in vitro experiments and in milligrams per kilogram of body weight for in vivo experiments.
- Table 4 Oligonucleotide sequences made for testing human cells obtained from subjects with Spinal Muscular Atrophy.
- the table shows the sequence of the modified nucleotides, where InaX represents an LNA nucleotide with 3' phosphorothioate linkage, omeX is a 2'-0-methyl nucleotide, dX is a deoxy nucleotide.
- An s at the end of a nucleotide code indicates that the nucleotide had a 3' phosphorothioate linkage.
- the "-Sup" at the end of the sequence marks the fact that the 3 ' end lacks either a phosphate or thiophosphate on the 3' linkage.
- the Formatted Sequence column shows the sequence of the oligonucleotide, including modified nucleotides, for the oligonucleotides tested in Table 2, 5, 6 and 7.
- Appendix A contains Table 5, which shows RT-PCR data from
- Appendix B contains Table 6, which shows RT-PCR data from
- combination treatments e.g., two oligonucleotides, an oligonucleotide and a drug.
- Appendix C contains Table 7, which shows ELISA data from
- SEQID sequence identifier of base sequence of oligonucleotide used
- Oligo Name name of oligonucleotide
- Avg RQ average relative quantification of RT-PCR based expression levels of target gene(s);
- Avg RQ SE standard error of mean of relative quantification of RT-PCR based expression level
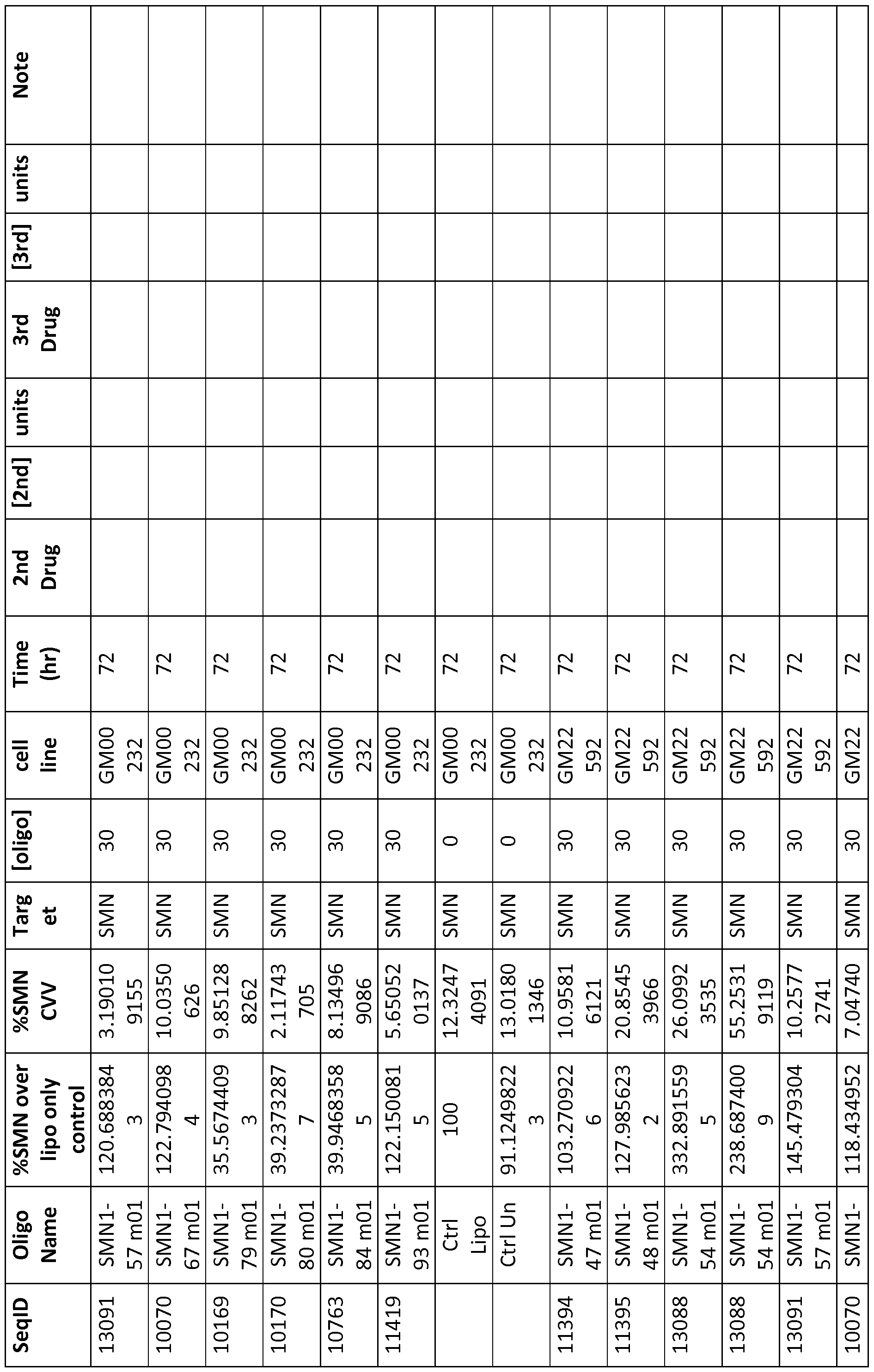
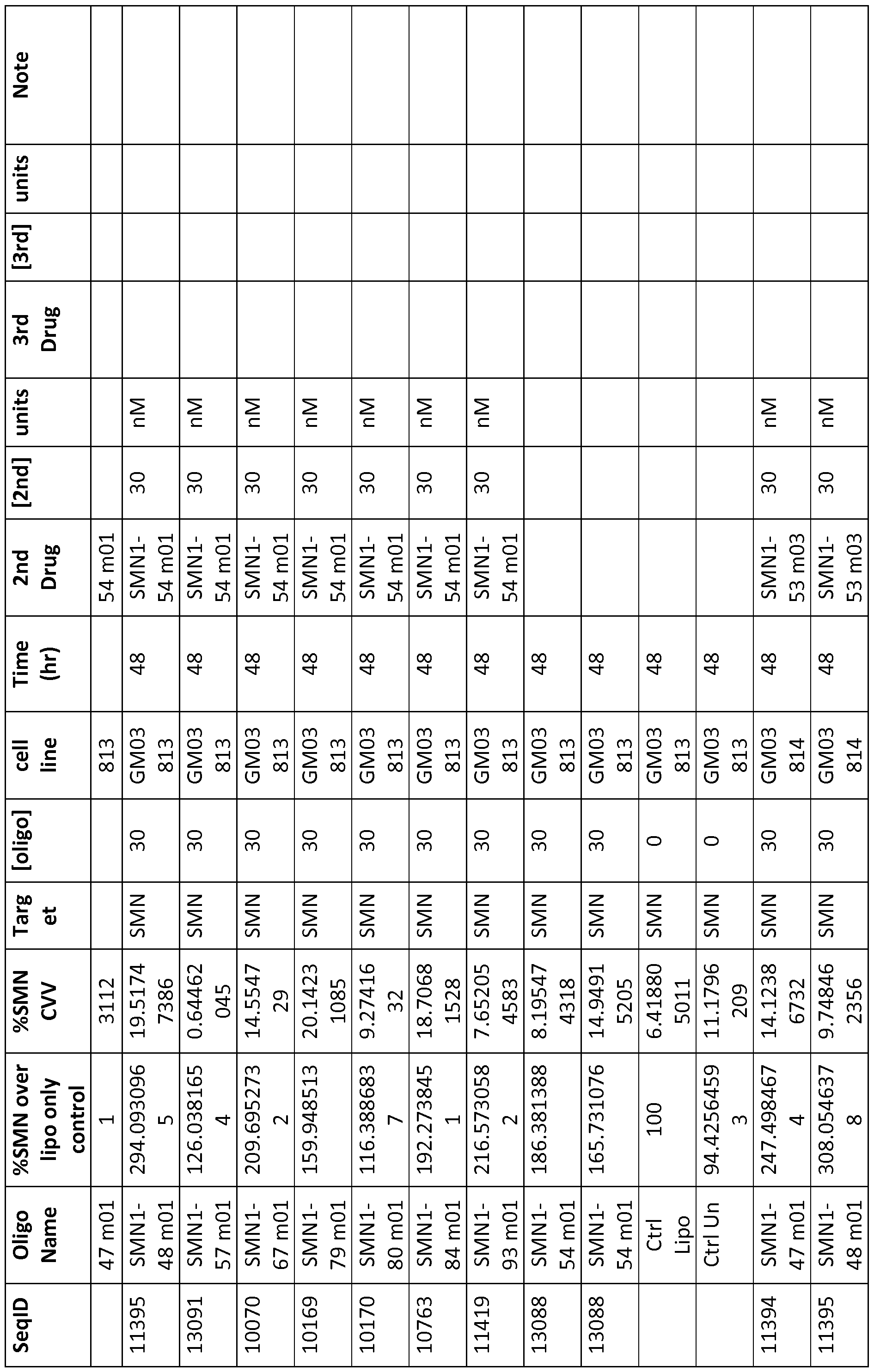
- %SMN over lipo only control refers to the ratio of SMN protein levels (ng/mg total protein) when compared to Lipofectamine2000 (transfection reagent) treated cells converted into %
- %SMN CVV refers to coefficient of variation
- Exp # Experiment reference number
- Target target gene
- [oligo] concentration of oligonucleotide used in nM unless otherwise indicated
- Cell Line cell line used
- Assay Type assay used
- Time(hr) time of assay following treatment
- 2 nd Drug name of second oligonugonu
- SMA Spinal muscular atrophy
- MIMs 253300, 253550, and 253400 types I, II, and III
- type I generally understood as being the most severe.
- Loss of function of the SMNl gene is responsible for SMA. Humans have an extra SMN gene copy, called SMN2. Both SMN genes reside within a segmental duplication on Chromosome 5ql3 as inverted repeats.
- SMNl and SMN2 are almost identical. In some cases, SMNl and SMN2 differ by 11 nucleotide substitutions, including seven in intron 6, two in intron 7, one in coding exon 7, and one in non-coding exon 8.
- the substitution in exon 7 involves a translationally silent C to T transition compared with SMNl, that results in alternative splicing because the substitution disrupts recognition of the upstream 3' splice site, in which exon 7 is frequently skipped during precursor mR A splicing. Consequently, SMN2 encodes primarily the exon 7-skipped protein isoform (SMNA7), which is unstable, mislocalized, and only partially functional.
- SMNA7 exon 7-skipped protein isoform
- Methods and related single stranded oligonucleotides that are useful for selectively inducing expression of particular splice variants of SMNl or SMN2 are provided herein.
- the methods are useful for controlling the levels in a cell of particular SMN protein isoforms encoded by the splice variants.
- the methods are useful for inducing expression of SMN proteins to levels sufficient to treat SMA.
- methods are provided for increasing expression of full-length SMN protein in a cell for purposes of treating SMA.
- the methods comprise delivering to the cell a first single stranded oligonucleotide complementary with a PRC2- associated region of SMNl or SMN2 and a second single stranded oligonucleotide complementary with a splice control sequence of a precursor mRNA of SMNl or SMN2, in amounts sufficient to increase expression of a mature mRNA of SMNl or SMN2 that comprises (or includes) exon 7 in the cell.
- PRC2 Polycomb repressive complex 2
- Polycomb repressive complex 2 (PRC2) is a histone methyltransferase and a known epigenetic regulator involved in silencing of genomic regions through methylation of histone H3.
- PRC2 interacts with long noncoding RNAs (IncRNAs), such as RepA, Xist, and Tsix, to catalyze
- PRC2 contains four subunits, Eed, Suzl2, RbAp48, and Ezh2. Aspects of the invention relate to the recognition that single stranded oligonucleotides that bind to PRC2-associated regions of RNAs (e.g., IncRNAs) that are expressed from within a genomic region that encompasses or that is in functional proximity to the SMNl or SMN2 gene can induce or enhance expression of SMNl or SMN2. In some embodiments, this upregulation is believed to result from inhibition of PRC2 mediated repression of SMNl or SMN2.
- RNAs e.g., IncRNAs
- PRC2-associated region refers to a region of a nucleic acid that comprises or encodes a sequence of nucleotides that interact directly or indirectly with a component of PRC2.
- a PRC2-associated region may be present in a RNA (e.g., a long non- coding RNA (IncRNA)) that interacts with a PRC2.
- a PRC2-associated region may be present in a DNA that encodes an RNA that interacts with PRC2. In some cases, the PRC2- associated region is equivalently referred to as a PRC2-interacting region.
- a PRC2-associated region is a region of an RNA that crosslinks to a component of PRC2 in response to in situ ultraviolet irradiation of a cell that expresses the RNA, or a region of genomic DNA that encodes that RNA region.
- a PRC2-associated region is a region of an RNA that immunoprecipitates with an antibody that targets a component of PRC2, or a region of genomic DNA that encodes that RNA region.
- a PRC2-associated region is a region of an RNA that immunoprecipitates with an antibody that binds specifically to SUZ12, EED, EZH2 or RBBP4 (which as noted above are components of PRC2), or a region of genomic DNA that encodes that RNA region.
- a PRC2-associated region is a region of an RNA that is protected from nucleases (e.g., RNases) in an RNA-immunoprecipitation assay that employs an antibody that targets a component of PRC2, or a region of genomic DNA that encodes that protected RNA region.
- a PRC2-associated region is a region of an RNA that is protected from nucleases (e.g., RNases) in an RNA-immunoprecipitation assay that employs an antibody that targets SUZ12, EED, EZH2 or RBBP4, or a region of genomic DNA that encodes that protected RNA region.
- a PRC2-associated region is a region of an RNA within which occur a relatively high frequency of sequence reads in a sequencing reaction of products of an RNA-immunoprecipitation assay that employs an antibody that targets a component of PRC2, or a region of genomic DNA that encodes that RNA region.
- a PRC2- associated region is a region of an RNA within which occur a relatively high frequency of sequence reads in a sequencing reaction of products of an R A-immunoprecipitation assay that employs an antibody that binds specifically to SUZ12, EED, EZH2 or RBBP4, or a region of genomic DNA that encodes that protected RNA region.
- the PRC2-associated region may be referred to as a "peak.”
- a PRC2-associated region comprises a sequence of 40 to 60 nucleotides that interact with PRC2 complex. In some embodiments, a PRC2-associated region comprises a sequence of 40 to 60 nucleotides that encode an RNA that interacts with PRC2. In some embodiments, a PRC2-associated region comprises a sequence of up to 5kb in length that comprises a sequence (e.g. , of 40 to 60 nucleotides) that interacts with
- a PRC2-associated region comprises a sequence of up to 5kb in length within which an RNA is encoded that has a sequence (e.g., of 40 to 60 nucleotides) that is known to interact with PRC2. In some embodiments, a PRC2-associated region comprises a sequence of about 4kb in length that comprise a sequence (e.g., of 40 to 60 nucleotides) that interacts with PRC2. In some embodiments, a PRC2-associated region comprises a sequence of about 4kb in length within which an RNA is encoded that includes a sequence (e.g., of 40 to 60 nucleotides) that is known to interact with PRC2.
- a PRC2-associated region has a sequence as set forth in any one of SEQ ID NOS: 9 to 29. In some embodiments, a PRC2-associated region has a sequence as set forth in any one of SEQ ID NOS: 24 to 29.
- single stranded oligonucleotides are provided that specifically bind to, or are complementary to, a PRC2-associated region in a genomic region that encompasses or that is in proximity to the SMN1 or SMN2 gene. In some embodiments, single stranded oligonucleotides are provided that specifically bind to, or are complementary to, a PRC2-associated region that has a sequence as set forth in any one of SEQ ID NOS: 9 to 29.
- single stranded oligonucleotides are provided that specifically bind to, or are complementary to, a PRC2-associated region that has a sequence as set forth in any one of SEQ ID NOS: 9 to 29 combined with up to 2kb, up to 5kb, or up to lOkb of flanking sequences from a corresponding genomic region to which these SEQ IDs map (e.g., in a human genome).
- single stranded oligonucleotides have a sequence as set forth in any one of SEQ ID NOS: 30 to 13087.
- single stranded oligonucleotides have a sequence as set forth in Table 2.
- a PRC2 associated region of SMN1 or SMN2 against which a single stranded oligonucleotide is complementary is selected from SEQ ID NOS: 24-29.
- a single stranded oligonucleotide that is complementary with a PRC2 associated region of SMNl or SMN2 comprises a sequence selected from SEQ ID NOS: 1158-1159, 1171, 1482-1483, 1485-1486, 2465-2471, 2488-2490, 2542-2546, 2656-2657, 2833-2835, 3439-3440, 3916- 3918, 4469-4472, 4821, 5429, 5537, 6061, 7327, 8330-13061, and 13062-13087.
- a single stranded oligonucleotide that is complementary with a PRC2 associated region of SMNl or SMN2 comprises a sequence selected from 11395, 11394, 10169,
- these oligonucleotides are able to interfere with the binding of and function of PRC2, by preventing recruitment of PRC2 to a specific chromosomal locus.
- a single administration of single stranded oligonucleotides designed to specifically bind a PRC2-associated region IncRNA can stably displace not only the IncRNA, but also the PRC2 that binds to the IncRNA, from binding chromatin. After displacement, the full complement of PRC2 is not recovered for up to 24 hours.
- IncRNA can recruit PRC2 in a cis fashion, repressing gene expression at or near the specific chromosomal locus from which the IncRNA was transcribed.
- Methods of modulating gene expression are provided, in some embodiments, that may be carried out in vitro, ex vivo, or in vivo. It is understood that any reference to uses of compounds throughout the description contemplates use of the compound in preparation of a pharmaceutical composition or medicament for use in the treatment of condition ⁇ e.g., Spinal muscular atrophy) associated with decreased levels or activity of SMNl or SMN2. Thus, as one nonlimiting example, this aspect of the invention includes use of such single stranded oligonucleotides in the preparation of a medicament for use in the treatment of disease, wherein the treatment involves upregulating expression of SMNl or SMN2.
- methods are provided for selecting a candidate oligonucleotide for activating expression of SMNl or SMN2.
- the methods generally involve selecting as a candidate oligonucleotide, a single stranded oligonucleotide comprising a nucleotide sequence that is complementary to a PRC2-associated region ⁇ e.g., a nucleotide sequence as set forth in any one of SEQ ID NOS: 9 to 29).
- sets of oligonucleotides may be selected that are enriched ⁇ e.g., compared with a random selection of oligonucleotides) in oligonucleotides that activate expression of SMNl or SMN2.
- single Stranded Oligonucleotides for Modulating Expression of SMNl or SMN2 are provided for modulating expression of SMNl or SMN2 in a cell.
- expression of SMNl or SMN2 is upregulated or increased.
- single stranded oligonucleotides complementary to these PRC2- associated regions inhibit the interaction of PRC2 with long RNA transcripts such that gene expression is upregulated or increased.
- oligonucleotides complementary to these PRC2-associated regions inhibit the interaction of PRC2 with long RNA transcripts, resulting in reduced methylation of histone H3 and reduced gene inactivation, such that gene expression is upregulated or increased.
- this interaction may be disrupted or inhibited due to a change in the structure of the long RNA that prevents or reduces binding to PRC2.
- the oligonucleotide may be selected using any of the methods disclosed herein for selecting a candidate oligonucleotide for activating expression of SMNl or SMN2.
- the single stranded oligonucleotide may comprise a region of complementarity that is complementary with a PRC2-associated region of a nucleotide sequence set forth in any one of SEQ ID NOS: 1 to 8.
- oligonucleotide may be complementary with at least 6, e.g., at least 7, at least 8, at least 9, at least 10, at least 15 or more consecutive nucleotides of the PRC2-associated region.
- the PRC2-associated region may map to a position in a chromosome between 50 kilobases upstream of a 5 '-end of the SMNl or SMN2 gene and 50 kilobases downstream of a 3 '-end of the SMNl or SMN2 gene.
- the PRC2-associated region may map to a position in a chromosome between 25 kilobases upstream of a 5 '-end of the SMNl or SMN2 gene and 25 kilobases downstream of a 3 '-end of the SMNl or SMN2 gene.
- the PRC2-associated region may map to a position in a chromosome between 12 kilobases upstream of a 5 '-end of the SMNl or SMN2 gene and 12 kilobases downstream of a 3 '-end of the SMNl or SMN2 gene.
- the PRC2-associated region may map to a position in a chromosome between 5 kilobases upstream of a 5 '-end of the SMNl or SMN2 gene and 5 kilobases downstream of a 3 '-end of the SMNl or SMN2 gene.
- the genomic position of the selected PRC2-associated region relative to the SMNl or SMN2 gene may vary.
- the PRC2-associated region may be upstream of the 5 ' end of the SMNl or SMN2 gene.
- the PRC2-associated region may be downstream of the 3 ' end of the SMNl or SMN2 gene.
- the PRC2-associated region may be within an intron of the SMNl or SMN2 gene.
- the PRC2-associated region may be within an exon of the SMNl or SMN2 gene.
- the PRC2-associated region may traverse an intron-exon junction, a 5 '-UTR- exon junction or a 3 '-UTR-exon junction of the SMNl or SMN2 gene.
- the single stranded oligonucleotide may comprise a sequence having the formula X- Y-Z, in which X is any nucleotide, Y is a nucleotide sequence of 6 nucleotides in length that is not a human seed sequence of a microRNA, and Z is a nucleotide sequence of varying length.
- X is the 5 ' nucleotide of the oligonucleotide.
- the oligonucleotide when X is anchored at the 5 ' end of the oligonucleotide, the oligonucleotide does not have any nucleotides or nucleotide analogs linked 5 ' to X.
- the single stranded oligonucleotide has a sequence 5 'X-Y-Z and is 8-50 nucleotides in length.
- the Y sequence may be a nucleotide sequence of 6 nucleotides in length set forth in Table 1.
- the single stranded oligonucleotide may have a sequence that does not contain guanosine nucleotide stretches (e.g., 3 or more, 4 or more, 5 or more, 6 or more consecutive guanosine nucleotides).
- guanosine nucleotide stretches e.g., 3 or more, 4 or more, 5 or more, 6 or more consecutive guanosine nucleotides.
- oligonucleotides having guanosine nucleotide stretches have increased non-specific binding and/or off-target effects, compared with oligonucleotides that do not have guanosine nucleotide stretches.
- the single stranded oligonucleotide may have a sequence that has less than a threshold level of sequence identity with every sequence of nucleotides, of equivalent length, that map to a genomic position encompassing or in proximity to an off-target gene.
- an oligonucleotide may be designed to ensure that it does not have a sequence that maps to genomic positions encompassing or in proximity with all known genes (e.g. , all known protein coding genes) other than SMNl or SMN2.
- an oligonucleotide may be designed to ensure that it does not have a sequence that maps to any other known PRC2-associated region, particularly PRC2-associated regions that are functionally related to any other known gene (e.g., any other known protein coding gene). In either case, the oligonucleotide is expected to have a reduced likelihood of having off-target effects.
- the threshold level of sequence identity may be 50%, 60%, 70%, 80%, 85%, 90%, 95%, 99% or 100% sequence identity.
- the single stranded oligonucleotide may have a sequence that is complementary to a PRC2-associated region that encodes an RNA that forms a secondary structure comprising at least two single stranded loops.
- oligonucleotides that are complementary to a PRC2-associated region that encodes an RNA that forms a secondary structure comprising one or more single stranded loops e.g., at least two single stranded loops
- have a greater likelihood of being active e.g., of being capable of activating or enhancing expression of a target gene
- the secondary structure may comprise a double stranded stem between the at least two single stranded loops. Accordingly, the region of
- complementarity between the oligonucleotide and the PRC2-associated region may be at a location of the PRC2 associated region that encodes at least a portion of at least one of the loops. In some cases, the region of complementarity between the oligonucleotide and the PRC2-associated region may be at a location of the PRC2-associated region that encodes at least a portion of at least two of the loops. In some cases, the region of complementarity between the oligonucleotide and the PRC2-associated region may be at a location of the PRC2 associated region that encodes at least a portion of the double stranded stem.
- a PRC2-associated region (e.g., of an lncRNA) is identified (e.g., using RIP- Seq methodology or information derived therefrom).
- the predicted secondary structure RNA (e.g., lncRNA) containing the PRC2-associated region is determined using RNA secondary structure prediction algorithms, e.g., RNAfold, mfold.
- oligonucleotides are designed to target a region of the RNA that forms a secondary structure comprising one or more single stranded loop (e.g., at least two single stranded loops) structures which may comprise a double stranded stem between the at least two single stranded loops.
- the single stranded oligonucleotide may have a sequence that is has greater than 30% G-C content, greater than 40% G-C content, greater than 50% G-C content, greater than 60% G-C content, greater than 70% G-C content, or greater than 80% G-C content.
- the single stranded oligonucleotide may have a sequence that has up to 100% G-C content, up to 95% G-C content, up to 90% G-C content, or up to 80% G-C content.
- the oligonucleotide is 8 to 10 nucleotides in length, all but 1, 2, 3, 4, or 5 of the nucleotides of the complementary sequence of the PRC2-associated region are cytosine or guanosine nucleotides.
- the sequence of the PRC2-associated region to which the single stranded oligonucleotide is complementary comprises no more than 3 nucleotides selected from adenine and uracil.
- the single stranded oligonucleotide may be complementary to a chromosome of a different species (e.g., a mouse, rat, rabbit, goat, monkey, etc.) at a position that encompasses or that is in proximity to that species' homo log of SMN1 or SMN2.
- the single stranded oligonucleotide may be complementary to a human genomic region encompassing or in proximity to the SMN1 or SMN2 gene and also be complementary to a mouse genomic region encompassing or in proximity to the mouse homolog of SMN1 or SMN2.
- the single stranded oligonucleotide may be complementary to a sequence as set forth in SEQ ID NO: 1, 2, 4, or 5, which is a human genomic region encompassing or in proximity to the SMN1 or SMN2 gene, and also be complementary to a sequence as set forth in SEQ ID NO:7 or 8, which is a mouse genomic region encompassing or in proximity to the mouse homolog of the SMN1 or SMN2 gene.
- Oligonucleotides having these characteristics may be tested in vivo or in vitro for efficacy in multiple species (e.g., human and mouse). This approach also facilitates development of clinical candidates for treating human disease by selecting a species in which an appropriate animal exists for the disease.
- the region of complementarity of the single stranded oligonucleotide is complementary with at least 8 to 15, 8 to 30, 8 to 40, or 10 to 50, or 5 to 50, or 5 to 40 bases, e.g., 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or 50 consecutive nucleotides of a PRC2-associated region.
- the region of complementarity is complementary with at least 8 consecutive nucleotides of a PRC2-associated region.
- the sequence of the single stranded oligonucleotide is based on an RNA sequence that binds to PRC2, or a portion thereof, said portion having a length of from 5 to 40 contiguous base pairs, or about 8 to 40 bases, or about 5 to 15, or about 5 to 30, or about 5 to 40 bases, or about 5 to 50 bases.
- RNA sequence that binds to PRC2
- portion having a length of from 5 to 40 contiguous base pairs, or about 8 to 40 bases, or about 5 to 15, or about 5 to 30, or about 5 to 40 bases, or about 5 to 50 bases.
- Complementary refers to the capacity for precise pairing between two nucleotides.
- nucleotide at a certain position of an oligonucleotide is capable of hydrogen bonding with a nucleotide at the same position of PRC2-associated region
- the single stranded nucleotide and PRC2-associated region are considered to be complementary to each other at that position.
- the single stranded nucleotide and PRC2-associated region are complementary to each other when a sufficient number of corresponding positions in each molecule are occupied by nucleotides that can hydrogen bond with each other through their bases.
- “complementary" is a term which is used to indicate a sufficient degree of complementarity or precise pairing such that stable and specific binding occurs between the single stranded nucleotide and PRC2-associated region.
- a base at one position of a single stranded nucleotide is capable of hydrogen bonding with a base at the corresponding position of a PRC2-associated region, then the bases are considered to be complementary to each other at that position. 100% complementarity is not required.
- the single stranded oligonucleotide may be at least 80% complementary to
- the single stranded oligonucleotide may contain 1 , 2 or 3 base mismatches compared to the portion of the consecutive nucleotides of a PRC2-associated region. In some embodiments the single stranded oligonucleotide may have up to 3 mismatches over 15 bases, or up to 2 mismatches over 10 bases.
- a complementary nucleotide sequence need not be 100% complementary to that of its target to be specifically hybridizable.
- a complementary nucleic acid sequence for purposes of the present disclosure is specifically hybridizable when binding of the sequence to the target molecule (e.g., IncRNA) interferes with the normal function of the target (e.g., IncRNA) to cause a loss of activity (e.g., inhibiting PRC2-associated repression with consequent up-regulation of gene expression) and there is a sufficient degree of complementarity to avoid non-specific binding of the sequence to non-target sequences under conditions in which avoidance of non-specific binding is desired, e.g., under physiological conditions in the case of in vivo assays or therapeutic treatment, and in the case of in vitro assays, under conditions in which the assays are performed under suitable conditions of stringency.
- the target molecule e.g., IncRNA
- the single stranded oligonucleotide is 7, 8, 9, 10, 1 1 , 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 , 22, 23, 24, 25, 26, 27, 28, 29, 30, 35, 40, 45, 50 or more nucleotides in length. In a preferred embodiment, the oligonucleotide is 8 to 30 nucleotides in length.
- the PRC2-associated region occurs on the same DNA strand as a gene sequence (sense). In some embodiments, the PRC2-associated region occurs on the opposite DNA strand as a gene sequence (anti-sense). Oligonucleotides complementary to a PRC2-associated region can bind either sense or anti-sense sequences.
- Base pairings may include both canonical Watson-Crick base pairing and non-Watson-Crick base pairing (e.g., Wobble base pairing and Hoogsteen base pairing).
- adenosine-type bases are complementary to thymidine -type bases (T) or uracil-type bases (U), that cytosine-type bases (C) are complementary to guanosine-type bases (G), and that universal bases such as 3-nitropyrrole or 5-nitroindole can hybridize to and are considered complementary to any A, C, U, or T.
- Inosine (I) has also been considered in the art to be a universal base and is considered complementary to any A, C, U or T.
- any one or more thymidine (T) nucleotides (or modified nucleotide thereof) or uridine (U) nucleotides (or a modified nucleotide thereof) in a sequence provided herein, including a sequence provided in the sequence listing, may be replaced with any other nucleotide suitable for base pairing (e.g., via a Watson-Crick base pair) with an adenosine nucleotide.
- any one or more thymidine (T) nucleotides (or modified nucleotide thereof) or uridine (U) nucleotides (or a modified nucleotide thereof) in a sequence provided herein, including a sequence provided in the sequence listing, may be suitably replaced with a different pyrimidine nucleotide or vice versa.
- any one or more thymidine (T) nucleotides (or modified nucleotide thereof) in a sequence provided herein, including a sequence provided in the sequence listing may be suitably replaced with a uridine (U) nucleotide (or a modified nucleotide thereof) or vice versa.
- GC content of the single stranded oligonucleotide is preferably between about 30-60 %. Contiguous runs of three or more Gs or Cs may not be preferable in some embodiments. Accordingly, in some embodiments, the oligonucleotide does not comprise a stretch of three or more guanosine nucleotides.
- the single stranded oligonucleotide specifically binds to, or is complementary to an RNA that is encoded in a genome (e.g., a human genome) as a single contiguous transcript (e.g., a non-spliced RNA).
- a genome e.g., a human genome
- a single contiguous transcript e.g., a non-spliced RNA
- the single stranded oligonucleotide specifically binds to, or is complementary to an RNA that is encoded in a genome (e.g., a human genome), in which the distance in the genome between the 5 'end of the coding region of the RNA and the 3 ' end of the coding region of the RNA is less than 1 kb, less than 2 kb, less than 3 kb, less than 4 kb, less than 5 kb, less than 7 kb, less than 8 kb, less than 9 kb, less than 10 kb, or less than 20 kb.
- a genome e.g., a human genome
- single stranded oligonucleotides disclosed herein may increase expression of mRNA corresponding to the gene by at least about 50% (i.e. 150% of normal or 1.5 fold), or by about 2 fold to about 5 fold. In some embodiments, expression may be increased by at least about 15 fold, 20 fold, 30 fold, 40 fold, 50 fold or 100 fold, or any range between any of the foregoing numbers. It has also been found that increased mRNA expression has been shown to correlate to increased protein expression.
- the oligonucleotides will upregulate gene expression and may specifically bind or specifically hybridize or be complementary to the PRC2 binding RNA that is transcribed from the same strand as a protein coding reference gene.
- the oligonucleotide may bind to a region of the PRC2 binding RNA that originates within or overlaps an intron, exon, intron exon junction, 5' UTR, 3' UTR, a translation initiation region, or a translation termination region of a protein coding sense strand of a reference gene (refGene).
- the oligonucleotides will upregulate gene expression and may specifically bind or specifically hybridize or be complementary to a PRC2 binding RNA that transcribed from the opposite strand (the antisense strand) of a protein coding reference gene.
- the oligonucleotide may bind to a region of the PRC2 binding RNA that originates within or overlaps an intron, exon, intron exon junction, 5' UTR, 3' UTR, a translation initiation region, or a translation termination region of a protein coding antisense strand of a reference gene.
- oligonucleotides described herein may be modified, e.g., comprise a modified sugar moiety, a modified internucleoside linkage, a modified nucleotide and/or combinations thereof.
- the oligonucleotides can exhibit one or more of the following properties: do not induce substantial cleavage or degradation of the target RNA; do not cause
- RNAse H pathway do not activate RNAse H pathway; do not activate RISC; do not recruit any Argonaute family protein; are not cleaved by Dicer; do not mediate alternative splicing; are not immune stimulatory; are nuclease resistant; have improved cell uptake compared to unmodified oligonucleotides; are not toxic to cells or mammals; may have improved endosomal exit; do interfere with interaction of IncRNA with PRC2, preferably the Ezh2 subunit but optionally the Suzl2, Eed, RbAp46/48 subunits or accessory factors such as Jarid2; do decrease histone H3 lysine27 methylation and/or do upregulate gene expression.
- PRC2 preferably the Ezh2 subunit but optionally the Suzl2, Eed, RbAp46/48 subunits or accessory factors such as Jarid2; do decrease histone H3 lysine27 methylation and/or do upregulate gene expression.
- Oligonucleotides that are designed to interact with RNA to modulate gene expression are a distinct subset of base sequences from those that are designed to bind a DNA target (e.g. , are complementary to the underlying genomic DNA sequence from which the RNA is transcribed).
- aspects of the invention provide strategies for targeting SMN1 or SMN2 precursor mRNA to affect splicing to minimize exon skipping. Accordingly, aspects of the invention provide therapeutic compounds useful for the treatment of SMA.
- oligonucleotides referred to herein as "splice switching oligonucleotides” are provided that modulate SMN2 splicing.
- Methods and related compositions, compounds, and kits are provided, in some embodiments, that are useful for increasing expression of full-length.
- the methods generally involve delivering to a cell a first single stranded oligonucleotide complementary with at least 8 consecutive nucleotides of a PRC2- associated region of SMN2 and a second single stranded oligonucleotide complementary with a splice control sequence of a precursor mRNA of SMN2, in amounts sufficient to increase expression of a mature mRNA of SMN2 that comprises (or includes) exon 7 in the cell. Any of the single stranded oligonucleotides that are complementary with at least 8 consecutive nucleotides of a PRC2-associated region of SMNl or SMN2 may be used.
- single stranded oligonucleotides that are complementary with a splice control sequence may alternatively be referred herein, as splice switching oligonucleotides.
- Splice switching oligonucleotides typically comprise a sequence complementary to a splice control sequence (e.g., a intronic splicing silencer sequence) of a precursor mRNA, and are capable of binding to and affecting processing of the precursor mRNA.
- Splice switching oligonucleotides may be complementary with a region of an exon, a region of an intron or an intron/exon junction.
- the splice control sequence comprises the sequence: CAGCAUUAUGAAAG (SEQ ID NO: 13100) or a portion thereof. In some embodiments, the splice control sequence comprises at least one hnRNAP binding sequence. In some embodiments, splice switching oligonucleotides that target SMNl or SMN2 function based on the premise that there is a competition between the 3' splice sites of exons 7 and 8 for pairing with the 5' splice site of exon 6, so impairing the recognition of the 3' splice site of exon 8 favors exon 7 inclusion.
- splice switching oligonucleotides are provided that promote SMN2 exon 7 inclusion and full-length SMN protein expression, in which the oligonucleotides are complementary to the intron 7/exon 8 junction.
- splice switching oligonucleotide are composed of a segment complementary to an exon of SMNl or SMN2 (e.g., exon 7).
- splice switching oligonucleotides comprise a tail (e.g., a non-complementary tail) consisting of RNA sequences with binding motifs recognized by a serine/arginine-rich (SR) protein.
- SR serine/arginine-rich
- splice switching oligonucleotides are complementary (at least partially) with an intronic splicing silencer (ISS).
- the ISS is in intron 6 or intron 7 of SMNl or SMN2.
- splice switching oligonucleotides comprise an antisense moiety complementary to a target exon or intron (e.g., of SMNl or SMN2) and a minimal RS domain peptide similar to the splicing activation domain of SR proteins.
- the splice switching oligonucleotide is 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 35, 40, 45, 50 or more nucleotides in length. In one embodiment, the oligonucleotide is 8 to 30 nucleotides in length.
- any of the oligonucleotides disclosed herein may be linked to one or more other oligonucleotides disclosed herein by a linker, e.g. , a cleavable linker.
- a linker e.g. , a cleavable linker.
- compounds are provided that comprise a single stranded oligonucleotide complementary with a PRC2-associated region of a gene that is linked via a linker to a single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of the gene.
- compounds are provided that have the general formula A-B-C, in which A is a single stranded oligonucleotide complementary with a PRC2- associated region of a gene, B is a linker, and C is a single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of the gene.
- linker B comprises an oligonucleotide, peptide, low pH labile bond, or disulfide bond.
- the compounds is orientated as 5'-A-B-C-3'. In some embodiments, the compound is orientated as 3'-A-B-C-5'.
- the 3' end of A is linked to the 5' end of B, and the 3' end of B is linked to 5' end of C.
- the 5' end of A is linked to the 3 ' end of B, and the 5 ' end of B is linked to 3 ' end of C.
- the 5 ' end of A is linked to the 5' end of B, and/or the 3' end of B is linked to the 3' end of C.
- the 3 ' end of A is linked to the 3' end of B, and/or the 5' end of B is linked to the 5' end of C.
- linker generally refers to a chemical moiety that is capable of covalently linking two or more oligonucleotides.
- at least one bond comprised or contained within the linker is capable of being cleaved (e.g., in a biological context, such as in a mammalian extract, such as an endosomal extract), such that at least two
- oligonucleotides are no longer covalently linked to one another after bond cleavage.
- a provided linker may include a region that is non- cleavable, as long as the linker also comprises at least one bond that is cleavable.
- the linker comprises a polypeptide that is more susceptible to cleavage by an endopeptidase in the mammalian extract than the oligonucleotides.
- the endopeptidase may be a trypsin, chymotrypsin, elastase, thermolysin, pepsin, or
- the endopeptidase may be a cathepsin B, cathepsin D, cathepsin L, cathepsin C, papain, cathepsin S or endosomal acidic insulinase.
- the linker comprise a peptide having an amino acid sequence selected from: ALAL, APISFFELG, FL, GFN, R/KXX, GRWHTVGLRWE, YL, GF, and FF, in which X is any amino acid.
- the linker comprises the formula -(CH 2 ) compassionS-S(CH 2 ) m -, wherein n and m are independently integers from 0 to 10.
- the linker may comprise an oligonucleotide that is more susceptible to cleavage by an endonuclease in the mammalian extract than the oligonucleotides.
- the linker may have a nucleotide sequence comprising from 1 to 10 thymidines or uridines.
- the linker may have a nucleotide sequence comprising
- the linker may have a nucleotide sequence comprising from 1 to 10 thymidines or uridines linked through phosphodiester internucleotide linkages.
- the linker may have a nucleotide sequence comprising from 1 to 10 thymidines or uridines linked through phosphorothioate
- At least one linker is 2-fold, 3-fold, 4-fold, 5-fold, 10-fold or more sensitive to enzymatic cleavage in the presence of a mammalian extract than at least two oligonucleotides. It should be appreciated that different linkers can be designed to be cleaved at different rates and/or by different enzymes in compounds comprising two or more linkers. Similarly different linkers can be designed to be sensitive to cleavage in different tissues, cells or subcellular compartments in compounds comprising two or more linkers.
- linkers are stable (e.g., more stable than the oligonucleotides they link together) in plasma, blood or serum which are richer in exonucleases, and less stable in the intracellular environments which are relatively rich in endonucleases.
- a linker is considered “non-cleavable” if the linker's half-life is at least 24, or 28, 32, 36, 48, 72, 96 hours or longer under the conditions described here, such as in liver homogenates.
- a linker is considered “cleavable” if the half-life of the linker is at most 10, or 8, 6, 5 hours or shorter.
- the linker is a nuclease-cleavable oligonucleotide linker.
- the nuclease-cleavable linker contains one or more phosphodiester bonds in the oligonucleotide backbone.
- the linker may contain a single
- phosphodiester bridge or 2, 3, 4, 5, 6, 7 or more phosphodiester linkages for example as a string of 1-10 deoxynucleotides, e.g., dT, or ribonucleotides, e.g., rU, in the case of RNA linkers.
- the cleavable linker contains one or more phosphodiester linkages.
- the cleavable linker may consist of phosphorothioate linkages only.
- phosphorothioate-linked deoxynucleotides which in some embodiments are cleaved relatively slowly by nucleases (thus termed
- noncleavable phosphorothioate-linked rU undergoes relatively rapid cleavage by ribonucleases and therefore is considered cleavable herein in some embodiments. It is also possible to combine dN and rN into the linker region, which are connected by phosphodiester or phosphorothioate linkages. In other embodiments, the linker can also contain chemically modified nucleotides, which are still cleavable by nucleases, such as, e.g., 2'-0-modified analogs. In particular, 2'-0-methyl or 2'-fluoro nucleotides can be combined with each other or with dN or rN nucleotides.
- a linker is a part of the compound that is usually not complementary to a target, although it could be. This is because the linker is generally cleaved prior to action of the oligonucleotides on the target, and therefore, the linker identity with respect to a target is inconsequential. Accordingly, in some embodiments, a linker is an (oligo)nucleotide linker that is not complementary to any of the targets against which the oligonucleotides are designed.
- the cleavable linker is an oligonucleotide linker that contains a continuous stretch of deliberately introduced Rp phosphorothioate stereoisomers (e.g., 4, 5, 6, 7 or longer stretches).
- the Rp stereoisoform unlike Sp isoform, is known to be susceptible to nuclease cleavage (Krieg et al, 2003, Oligonucleotides, 13:491-499).
- Such a linker would not include a racemic mix of PS linkaged oligonucleotides since the mixed linkages are relatively stable and are not likely to contain long stretches of the Rp stereoisomers, and therefore, considered “non-cleavable" herein.
- a linker comprises a stretch of 4, 5, 6, 7 or more phosphorothioated nucleotides, wherein the stretch does not contain a substantial amount or any of the Sp stereoisoform. The amount could be considered substantial if it exceeds 10% on a per-mole basis.
- the linker is a non-nucleotide linker, for example, a single phosphodiester bridge.
- the linker can be designed so as to undergo a chemical or enzymatic cleavage reaction.
- Chemical reactions involve, for example, cleavage in acidic environments (e.g., endosomes), reductive cleavage (e.g., cytosolic cleavage) or oxidative cleavage (e.g., in liver microsomes).
- the cleavage reaction can also be initiated by a rearrangement reaction.
- Enzymatic reactions can include reactions mediated by nucleases, peptidases, proteases, phosphatases, oxidases, reductases, etc.
- a linker can be pH-sensitive, cathepsin- sensitive, or predominantly cleaved in endosomes and/or cytosol.
- the linker comprises a peptide. In certain embodiments, the linker comprises a peptide which includes a sequence that is cleavable by an endopeptidase. In addition to the cleavable peptide sequence, the linker may comprise additional amino acid residues and/or non-peptide chemical moieties, such as an alkyl chain. In certain
- the linker comprises Ala-Leu-Ala-Leu, which is a substrate for cathepsin B. See, for example, the maleimidocaproyl-Arg-Arg-Ala-Leu-Ala-Leu linkers described in
- a cathepsin B- cleavable linker is cleaved in tumor cells.
- the linker comprises Ala- Pro-Ile-Ser-Phe-Phe-Glu-Leu-Gly, which is a substrate for cathepsins D, L, and B (see, for example, Fischer et al, Chembiochem 2006, 7, 1428-1434).
- a cathepsin-cleavable linker is cleaved in HeLA cells.
- the linker comprises Phe-Lys, which is a substrate for cathepsin B.
- the linker comprises Phe-Lys-p-aminobenzoic acid (PABA). See, e.g., the maleimidocaproyl-Phe-Lys-PABA linker described in Walker et al, Bioorg. Med. Chem. Lett. 2002, 12, 217-219.
- the linker comprises Gly-Phe-2- naphthylamide, which is a substrate for cathepsin C (see, for example, Berg et al. Biochem. J. 1994, 300, 229-235).
- a cathepsin C-cleavable linker is cleaved in hepatocytes.
- the linker comprises a cathepsin S cleavage site.
- the linker comprises Gly-Arg-Trp-His-Thr-Val-Gly-Leu- Arg-Trp-Glu, Gly-Arg-Trp-Pro-Pro-Met-Gly-Leu-Pro-Trp-Glu, or Gly-Arg-Trp-His-Pro- Met-Gly-Ala-Pro-Trp-Glu, for example, as described in Lutzner et al, J. Biol. Chem. 2008, 283, 36185-36194.
- a cathepsin S-cleavable linker is cleaved in antigen presenting cells.
- the linker comprises a papain cleavage site.
- Papain typically cleaves a peptide having the sequence -R/K-X-X (see Chapman et al, Annu. Rev. Physiol 1997, 59, 63-88).
- a papain-cleavable linker is cleaved in endosomes.
- the linker comprises an endosomal acidic insulinase cleavage site.
- the linker comprises Tyr-Leu, Gly-Phe, or Phe-Phe (see, e.g., Authier et al, FEBS Lett. 1996, 389, 55-60).
- an endosomal acidic insulinase-cleavable linker is cleaved in hepatic cells.
- the linker is pH sensitive. In certain embodiments, the linker comprises a low pH-labile bond.
- a low-pH labile bond is a bond that is selectively broken under acidic conditions (pH ⁇ 7). Such bonds may also be termed endosomally labile bonds, because cell endosomes and lysosomes have a pH less than 7.
- the linker comprises an amine, an imine, an ester, a benzoic imine, an amino ester, a diortho ester, a polyphosphoester, a polyphosphazene, an acetal, a vinyl ether, a hydrazone, an azidomethyl-methylmaleic anhydride, a thiopropionate, a masked endosomo lytic agent or a citraconyl group.
- the linker comprises a low pH-labile bond selected from the following: ketals that are labile in acidic environments (e.g., pH less than 7, greater than about 4) to form a diol and a ketone; acetals that are labile in acidic environments (e.g., pH less than 7, greater than about 4) to form a diol and an aldehyde; imines or iminiums that are labile in acidic environments (e.g., pH less than 7, greater than about 4) to form an amine and an aldehyde or a ketone; silicon-oxygen-carbon linkages that are labile under acidic condition; silicon-nitrogne (silazane) linkages; silicon-carbon linkages (e.g., arylsilanes, vinylsilanes, and allylsilanes); maleamates (amide bonds synthesized from maleic anhydride derivatives and amines); ortho esters; hydrazones; activate
- the linker comprises a masked endosomolytic agent.
- Endosomolytic polymers are polymers that, in response to a change in pH, are able to cause disruption or lysis of an endosome or provide for escape of a normally membrane- impermeable compound, such as a polynucleotide or protein, from a cellular internal membrane-enclosed vesicle, such as an endosome or lysosome.
- a normally membrane- impermeable compound such as a polynucleotide or protein
- fusogenic compounds including fusogenic peptides. Fusogenic peptides can facilitate endosomal release of agents such as oligomeric compounds to the cytoplasm. See, for example, US Patent Application Publication Nos. 20040198687, 20080281041,
- the linker can also be designed to undergo an organ/ tissue-specific cleavage. For example, for certain targets, which are expressed in multiple tissues, only the knock-down in liver may be desirable, as knock-down in other organs may lead to undesired side effects.
- linkers susceptible to liver-specific enzymes such as pyrrolase (TPO) and glucose-6- phosphatase (G-6-Pase), can be engineered, so as to limit the antisense effect to the liver mainly.
- linkers not susceptible to liver enzymes but susceptible to kidney- specific enzymes can be engineered, so that the antisense effect is limited to the kidneys mainly.
- intestine-specific peptidases cleaving Phe-Ala and Leu- Ala could be considered for orally administered multimeric oligonucleotides.
- an enzyme recognition site into the linker, which is recognized by an enzyme over-expressed in tumors, such as plasmin (e.g., PHEA-D-Val-Leu- Lys recognition site), tumor-specific knock-down should be feasible.
- the linker can also contain a targeting signal, such as N-acetyl galactosamine for liver targeting, or folate, vitamin A or RGD-peptide in the case of tumor or activated macrophage targeting.
- a targeting signal such as N-acetyl galactosamine for liver targeting, or folate, vitamin A or RGD-peptide in the case of tumor or activated macrophage targeting.
- the cleavable linker is organ- or tissue-specific, for example, liver-specific, kidney-specific, intestine-specific, etc.
- the oligonucleotides can be linked through any part of the individual oligonucleotide, e.g., via the phosphate, the sugar (e.g., ribose, deoxyribose), or the nucleobase.
- the linker when linking two oligonucleotides together, can be attached e.g.
- the linker when linking two oligonucleotides together, can attach internal residues of each oligonucleotides, e.g., via a modified nucleobase.
- the linker can attach internal residues of each oligonucleotides, e.g., via a modified nucleobase.
- OLIGONUCLEOTIDE COMPOUNDS the contents of which relating to linkers and related chemistries are incorporated herein by referenced in its entirety.
- the target selection methods may generally involve steps for selecting single stranded oligonucleotides having any of the structural and functional characteristics disclosed herein.
- the methods involve one or more steps aimed at identifying oligonucleotides that target a PRC2-associated region that is functionally related to SMNl or SMN2, for example a PRC2-associated region of a IncRNA that regulates expression of SMNl or SMN2 by facilitating ⁇ e.g., in a czs-regulatory manner) the recruitment of PRC2 to the SMNl or SMN2 gene.
- Such oligonucleotides are expected to be candidates for activating expression of SMNl or SMN2 because of their ability to hybridize with the PRC2-associated region of a nucleic acid ⁇ e.g., a IncRNA).
- this hybridization event is understood to disrupt interaction of PRC2 with the nucleic acid ⁇ e.g., a IncRNA) and as a result disrupt recruitment of PRC2 and its associated co-repressors ⁇ e.g., chromatin remodeling factors) to the SMNl or SMN2 gene locus.
- Methods of selecting a candidate oligonucleotide may involve selecting a PRC2- associated region ⁇ e.g., a nucleotide sequence as set forth in any one of SEQ ID NOS: 9 to 29) that maps to a chromosomal position encompassing or in proximity to the SMNl or SMN2 gene ⁇ e.g., a chromosomal position having a sequence as set forth in any one of SEQ ID NOS: 1 to 8).
- the PRC2-associated region may map to the strand of the chromosome comprising the sense strand of the SMNl or SMN2 gene, in which case the candidate oligonucleotide is complementary to the sense strand of the SMNl or SMN2 gene (i.e., is antisense to the SMNl or SMN2 gene).
- the PRC2-associated region may map to the strand of the first chromosome comprising the antisense strand of the SMNl or SMN2 gene, in which case the oligonucleotide is complementary to the antisense strand (the template strand) of the SMNl or SMN2 gene (i.e., is sense to the SMNl or SMN2 gene).
- Methods for selecting a set of candidate oligonucleotides that is enriched in oligonucleotides that activate expression of SMNl or SMN2 may involve selecting one or more PRC2-associated regions that map to a chromosomal position that encompasses or that is in proximity to the SMNl or SMN2 gene and selecting a set of oligonucleotides, in which each oligonucleotide in the set comprises a nucleotide sequence that is complementary with the one or more PRC2-associated regions.
- a set of oligonucleotides that is enriched in oligonucleotides that activate expression of refers to a set of oligonucleotides that has a greater number of oligonucleotides that activate expression of a target gene (e.g., SMN1 or SMN2) compared with a random selection of a target gene (e.g., SMN1 or SMN2) compared with a random selection of a target gene (e.g., SMN1 or SMN2) compared with a random selection of a target gene (e.g., SMN1 or SMN2) compared with a random selection of a target gene (e.g., SMN1 or SMN2) compared with a random selection of a target gene (e.g., SMN1 or SMN2) compared with a random selection of a target gene (e.g., SMN1 or SMN2) compared with a random selection of
- oligonucleotides of the same physicochemical properties e.g. , the same GC content, T m , length etc.
- design and/or synthesis of a single stranded oligonucleotide involves design and/or synthesis of a sequence that is complementary to a nucleic acid or PRC2- associated region described by such sequence information
- the skilled person is readily able to determine the complementary sequence, e.g., through understanding of Watson Crick base pairing rules which form part of the common general knowledge in the field.
- design and/or synthesis of a single stranded oligonucleotide involves manufacture of an oligonucleotide from starting materials by techniques known to those of skill in the art, where the synthesis may be based on a sequence of a PRC2- associated region, or portion thereof.
- Methods of design and/or synthesis of a single stranded oligonucleotide may involve one or more of the steps of:
- Single stranded oligonucleotides so designed and/or synthesized may be useful in method of modulating gene expression as described herein.
- oligonucleotides of the invention are synthesized chemically.
- Oligonucleotides used to practice this invention can be synthesized in vitro by well-known chemical synthesis techniques.
- Oligonucleotides of the invention can be stabilized against nucleolytic degradation such as by the incorporation of a modification, e.g., a nucleotide modification.
- nucleic acid sequences of the invention include a phosphorothioate at least the first, second, or third internucleotide linkage at the 5 ' or 3' end of the nucleotide sequence.
- the nucleic acid sequence can include a 2'-modified nucleotide, e.g., a 2'-deoxy, 2'- deoxy-2'-fluoro, 2'-0-methyl, 2'-0-methoxyethyl (2'-0-MOE), 2'-0-aminopropyl (2'-0-AP), 2 * -0-dimethylaminoethyl (2 * -0-DMAOE), 2 * -0-dimethylaminopropyl (2 * -0-DMAP), 2 * -0- dimethylaminoethyloxyethyl (2'-0-DMAEOE), or 2'-0 ⁇ N-methylacetamido (2'-0 ⁇ NMA).
- a 2'-modified nucleotide e.g., a 2'-deoxy, 2'- deoxy-2'-fluoro, 2'-0-methyl, 2'-0-methoxyethyl (2'-0-MOE), 2'-0-aminoprop
- the nucleic acid sequence can include at least one 2'-0-methyl-modified nucleotide, and in some embodiments, all of the nucleotides include a 2'-0-methyl modification.
- the nucleic acids are "locked,” i.e., comprise nucleic acid analogues in which the ribose ring is "locked” by a methylene bridge connecting the 2'- O atom and the 4'-C atom.
- any of the modified chemistries or formats of single stranded oligonucleotides described herein can be combined with each other, and that one, two, three, four, five, or more different types of modifications can be included within the same molecule.
- the method may further comprise the steps of amplifying the synthesized single stranded oligonucleotide, and/or purifying the single stranded
- oligonucleotide (or amplified single stranded oligonucleotide), and/or sequencing the single stranded oligonucleotide so obtained.
- the process of preparing a single stranded oligonucleotide may be a process that is for use in the manufacture of a pharmaceutical composition or medicament for use in the treatment of disease, optionally wherein the treatment involves modulating expression of a gene associated with a PRC2-associated region.
- a PRC2-associated region may be, or have been, identified, or obtained, by a method that involves identifying RNA that binds to PRC2.
- Such methods may involve the following steps: providing a sample containing nuclear ribonucleic acids, contacting the sample with an agent that binds specifically to PRC2 or a subunit thereof, allowing complexes to form between the agent and protein in the sample, partitioning the complexes, synthesizing nucleic acid that is complementary to nucleic acid present in the complexes.
- single stranded oligonucleotide is based on a PRC2-associated region, or a portion of such a sequence, it may be based on information about that sequence, e.g., sequence information available in written or electronic form, which may include sequence information contained in publicly available scientific publications or sequence databases.
- Nucleotide Analogues are examples of nucleotide Analogues.
- the oligonucleotide may comprise at least one ribonucleotide, at least one deoxyribonucleotide, and/or at least one bridged nucleotide.
- the oligonucleotide may comprise a bridged nucleotide, such as a locked nucleic acid (LNA) nucleotide, a constrained ethyl (cEt) nucleotide, or an ethylene bridged nucleic acid (ENA) nucleotide.
- LNA locked nucleic acid
- cEt constrained ethyl
- ENA ethylene bridged nucleic acid
- the oligonucleotide comprises a nucleotide analog disclosed in one of the following United States Patent or Patent Application Publications: US 7,399,845, US 7,741,457, US 8,022,193, US 7,569,686, US 7,335,765, US 7,314,923, US 7,335,765, and US 7,816,333, US 20110009471, the entire contents of each of which are incorporated herein by reference for all purposes.
- the oligonucleotide may have one or more 2' O-methyl nucleotides.
- the oligonucleotide may consist entirely of 2' O-methyl nucleotides.
- the single stranded oligonucleotide has one or more nucleotide analogues.
- the single stranded oligonucleotide may have at least one nucleotide analogue that results in an increase in T m of the oligonucleotide in a range of 1°C, 2 °C, 3°C, 4 °C, or 5°C compared with an oligonucleotide that does not have the at least one nucleotide analogue.
- the single stranded oligonucleotide may have a plurality of nucleotide analogues that results in a total increase in T m of the oligonucleotide in a range of 2 °C, 3 °C, 4 °C, 5 °C, 6 °C, 7 °C, 8 °C, 9 °C, 10 °C, 15 °C, 20 °C, 25 °C, 30 °C, 35 °C, 40 °C, 45 °C or more compared with an oligonucleotide that does not have the nucleotide analogue.
- the oligonucleotide may be of up to 50 nucleotides in length in which 2 to 10, 2 to 15, 2 to 16, 2 to 17, 2 to 18, 2 to 19, 2 to 20, 2 to 25, 2 to 30, 2 to 40, 2 to 45, or more nucleotides of the oligonucleotide are nucleotide analogues.
- the oligonucleotide may be of 8 to 30 nucleotides in length in which 2 to 10, 2 to 15 , 2 to 16, 2 to 17, 2 to 18, 2 to 19, 2 to 20, 2 to 25, 2 to 30 nucleotides of the oligonucleotide are nucleotide analogues.
- the oligonucleotide may be of 8 to 15 nucleotides in length in which 2 to 4, 2 to 5, 2 to 6, 2 to 7, 2 to 8, 2 to 9, 2 to 10, 2 to 11, 2 to 12, 2 to 13, 2 to 14 nucleotides of the oligonucleotide are nucleotide analogues.
- the oligonucleotides may have every nucleotide except 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 nucleotides modified.
- the oligonucleotide may consist entirely of bridged nucleotides (e.g., LNA nucleotides, cEt nucleotides, ENA nucleotides).
- the oligonucleotide may comprise alternating deoxyribonucleotides and 2'-fluoro-deoxyribonucleotides.
- the oligonucleotide may comprise alternating deoxyribonucleotides and 2'-0-methyl nucleotides.
- the oligonucleotide may comprise alternating deoxyribonucleotides and ENA nucleotide analogues.
- the oligonucleotide may comprise alternating deoxyribonucleotides and LNA nucleotides.
- the oligonucleotide may comprise alternating LNA nucleotides and 2'-0- methyl nucleotides.
- the oligonucleotide may have a 5 ' nucleotide that is a bridged nucleotide (e.g., a LNA nucleotide, cEt nucleotide, ENA nucleotide).
- the oligonucleotide may have a 5 ' nucleotide that is a deoxyribonucleotide.
- the oligonucleotide may comprise deoxyribonucleotides flanked by at least one bridged nucleotide (e.g., a LNA nucleotide, cEt nucleotide, ENA nucleotide) on each of the 5 ' and 3 ' ends of the deoxyribonucleotides.
- the oligonucleotide may comprise
- deoxyribonucleotides fianked by 1 , 2, 3, 4, 5, 6, 7, 8 or more bridged nucleotides (e.g. , LNA nucleotides, cEt nucleotides, ENA nucleotides) on each of the 5 ' and 3 ' ends of the deoxyribonucleotides.
- the 3 ' position of the oligonucleotide may have a 3 ' hydroxyl group.
- the 3 ' position of the oligonucleotide may have a 3 ' thiophosphate.
- the oligonucleotide may be conjugated with a label.
- the oligonucleotide may be conjugated with a label.
- oligonucleotide may be conjugated with a biotin moiety, cholesterol, Vitamin A, folate, sigma receptor ligands, aptamers, peptides, such as CPP, hydrophobic molecules, such as lipids, ASGPR or dynamic polyconjugates and variants thereof at its 5 ' or 3 ' end.
- the single stranded oligonucleotide comprises one or more modifications comprising: a modified sugar moiety, and/or a modified internucleoside linkage, and/or a modified nucleotide and/or combinations thereof. It is not necessary for all positions in a given oligonucleotide to be uniformly modified, and in fact more than one of the
- modifications described herein may be incorporated in a single oligonucleotide or even at within a single nucleoside within an oligonucleotide.
- the single stranded oligonucleotides are chimeric
- oligonucleotides that contain two or more chemically distinct regions, each made up of at least one nucleotide. These oligonucleotides typically contain at least one region of modified nucleotides that confers one or more beneficial properties (such as, for example, increased nuclease resistance, increased uptake into cells, increased binding affinity for the target) and a region that is a substrate for enzymes capable of cleaving R A:DNA or RNA:R A hybrids. Chimeric single stranded oligonucleotides of the invention may be formed as composite structures of two or more oligonucleotides, modified oligonucleotides,
- oligonucleosides and/or oligonucleotide mimetics as described above.
- Such compounds have also been referred to in the art as hybrids or gapmers.
- Representative United States patents that teach the preparation of such hybrid structures comprise, but are not limited to, US patent nos. 5,013,830; 5,149,797; 5, 220,007; 5,256,775; 5,366,878; 5,403,711; 5,491,133;
- the single stranded oligonucleotide comprises at least one nucleotide modified at the 2' position of the sugar, most preferably a 2'-0-alkyl, 2'-0-alkyl-0- alkyl or 2'-fluoro-modified nucleotide.
- RNA modifications include 2'-fluoro, 2'-amino and 2' O-methyl modifications on the ribose of pyrimidines, abasic residues or an inverted base at the 3' end of the RNA.
- modified oligonucleotides include those comprising modified backbones, for example, phosphorothioates, phosphotriesters, methyl phosphonates, short chain alkyl or cycloalkyl intersugar linkages or short chain heteroatomic or heterocyclic intersugar linkages. Most preferred are oligonucleotides with
- phosphorothioate backbones and those with heteroatom backbones particularly CH 2 -NH-O- CH 2 , CH, ⁇ N(CH 3 ) ⁇ 0 ⁇ CH 2 (known as a methylene(methylimino) or MMI backbone, CH 2 - O-N (CH 3 )-CH 2 , CH 2 -N (CH 3 )-N (CH 3 )-CH 2 and O-N (CH 3 )- CH 2 -CH 2 backbones, wherein the native phosphodiester backbone is represented as O- P— O- CH,); amide backbones (see De Mesmaeker et al. Ace. Chem. Res.
- PNA peptide nucleic acid
- Phosphorus-containing linkages include, but are not limited to, phosphorothioates, chiral phosphorothioates, phosphorodithioates, phosphotriesters, aminoalkylphosphotriesters, methyl and other alkyl phosphonates comprising 3'alkylene phosphonates and chiral phosphonates, phosphinates, phosphoramidates comprising 3 '-amino phosphoramidate and aminoalkylphosphoramidates, thionophosphoramidates, thionoalkylphosphonates, thionoalkylphosphotriesters, and boranophosphates having normal 3'-5' linkages, 2'-5' linked analogs of these, and those having inverted polarity wherein the adjacent pairs of nucleoside units are linked 3*-5* to 5*-3* or 2*-5* to 5*-2*; see US patent nos. 3,687,808; 4,469,863;
- Morpholino-based oligomeric compounds are described in Dwaine A. Braasch and David R. Corey, Biochemistry, 2002, 41(14), 4503-4510); Genesis, volume 30, issue 3, 2001 ; Heasman, J., Dev. Biol, 2002, 243, 209-214; Nasevicius et al, Nat. Genet., 2000, 26, 216- 220; Lacerra et al, Proc. Natl. Acad. Sci., 2000, 97, 9591-9596; and U.S. Pat. No. 5,034,506, issued Jul. 23, 1991.
- the morpholino-based oligomeric compound is a phosphorodiamidate morpholino oligomer (PMO) (e.g. , as described in Iverson, Curr. Opin. Mol. Ther., 3 :235-238, 2001 ; and Wang et al, J. Gene Med., 12:354-364, 2010; the disclosures of which are incorporated herein by reference in their entireties).
- PMO phosphorodiamidate morpholino oligomer
- Cyclohexenyl nucleic acid oligonucleotide mimetics are described in Wang et al, J. Am. Chem. Soc, 2000, 122, 8595-8602.
- Modified oligonucleotide backbones that do not include a phosphorus atom therein have backbones that are formed by short chain alkyl or cycloalkyl internucleoside linkages, mixed heteroatom and alkyl or cycloalkyl internucleoside linkages, or one or more short chain heteroatomic or heterocyclic internucleoside linkages.
- These comprise those having morpholino linkages (formed in part from the sugar portion of a nucleoside); siloxane backbones; sulfide, sulfoxide and sulfone backbones; formacetyl and thioformacetyl backbones; methylene formacetyl and thioformacetyl backbones; alkene containing backbones; sulfamate backbones; methyleneimino and methylenehydrazino backbones;
- Modified oligonucleotides are also known that include oligonucleotides that are based on or constructed from arabinonucleotide or modified arabinonucleotide residues.
- Arabinonucleosides are stereoisomers of ribonucleosides, differing only in the configuration at the 2'-position of the sugar ring.
- a 2'-arabino modification is 2'-F arabino.
- the modified oligonucleotide is 2'-fluoro-D-arabinonucleic acid (FANA) (as described in, for example, Lon et al, Biochem., 41 :3457-3467, 2002 and Min et al., Bioorg. Med. Chem. Lett., 12:2651-2654, 2002; the disclosures of which are incorporated herein by reference in their entireties). Similar modifications can also be made at other positions on the sugar, particularly the 3' position of the sugar on a 3' terminal nucleoside or in 2'-5' linked oligonucleotides and the 5' position of 5' terminal nucleotide.
- WO 99/67378 discloses arabinonucleic acids (ANA) oligomers and their analogues for improved sequence specific inhibition of gene expression via association to complementary messenger RNA.
- ENAs ethylene-bridged nucleic acids
- Preferred ENAs include, but are not limited to, 2'-0,4'-C-ethylene -bridged nucleic acids.
- LNAs examples include compounds of the following general formula.
- -CH 2 -0-, -CH 2 -S-, -CH 2 -N(H)-, -CH 2 -N(R)-, -CH 2 -CH 2 - or -CH 2 -CH- (if part of a double bond), -CH CH-, where R is selected from hydrogen and Ci_4-alkyl; Z and Z* are independently selected among an internucleoside linkage, a terminal group or a protecting group; B constitutes a natural or non-natural nucleotide base moiety; and the asymmetric groups may be found in either orientation.
- the LNA used in the oligonucleotides described herein comprises at least one LNA unit according any of the formulas
- the Locked Nucleic Acid (LNA) used in the oligonucleotides described herein comprises at least one Locked Nucleic Acid (LNA) unit according any of the formulas shown in Scheme 2 of PCT/DK2006/000512.
- the LNA used in the oligomer of the invention comprises internucleoside linkages selected from -0-P(O) 2 -O-, -0-P(0,S)-0-, -0-P(S) 2 -O-, -S-P(0) 2 -0-, -S-P(0,S)-0-, -S-P(S) 2 -0-, -0-P(O) 2 -S-, -0-P(0,S)-S-, -S-P(0) 2 -S-, -0-PO(R H )-0-, o- PO(OCH 3 )-0-, -0-PO(NR H )-0-, -0-PO(OCH 2 CH 2 S-R)-O-, -0-PO(BH 3 )-0-, -0-PO(NHR H )- 0-, -0-P(0) 2 -NR H -, -NR H -P(0) 2 -0-, -NR H -CO
- thio-LNA comprises a locked nucleotide in which at least one of X or Y in the general formula above is selected from S or -CH 2 -S-.
- Thio-LNA can be in both beta-D and alpha-L-configuration.
- amino-LNA comprises a locked nucleotide in which at least one of X or Y in the general formula above is selected from -N(H)-, N(R)-, CH 2 -N(H)-, and -CH 2 -N(R)- where R is selected from hydrogen and Ci_4-alkyl.
- Amino-LNA can be in both beta-D and alpha-L-configuration.
- Oxy-LNA comprises a locked nucleotide in which at least one of X or Y in the general formula above represents -O- or -CH 2 -0-. Oxy-LNA can be in both beta-D and alpha-L-configuration.
- ena-LNA comprises a locked nucleotide in which Y in the general formula above is -CH 2 -0- (where the oxygen atom of -CH 2 -0- is attached to the 2'-position relative to the base B).
- LNAs are described in additional detail herein.
- One or more substituted sugar moieties can also be included, e.g. , one of the following at the 2' position: OH, SH, SCH 3 , F, OCN, OCH 3 OCH 3 , OCH 3 0(CH 2 )n CH 3 , 0(CH 2 )n NH 2 or 0(CH 2 )n CH 3 where n is from 1 to about 10; CI to CIO lower alkyl, alkoxyalkoxy, substituted lower alkyl, alkaryl or aralkyl; CI; Br; CN; CF 3 ; OCF 3 ; 0-, S-, or N-alkyl; 0-, S-, or N-alkenyl; SOCH 3 ; S0 2 CH 3 ; ON0 2 ; N0 2 ; N 3 ; NH2; heterocycloalkyl; heterocycloalkaryl; aminoalkylamino; polyalkylamino; substituted silyl; an RNA cleaving group;
- a preferred modification includes 2'-methoxyethoxy [2'-0-CH 2 CH 2 OCH 3 , also known as 2'-0-(2-methoxyethyl)] (Martin et al, Helv. Chim. Acta, 1995, 78, 486).
- Other preferred modifications include 2'- methoxy (2'-0-CH 3 ), 2'-propoxy (2'-OCH 2 CH 2 CH 3 ) and 2'-fluoro (2'-F). Similar
- Oligonucleotides may also have sugar mimetics such as cyclobutyls in place of the pentofuranosyl group.
- Single stranded oligonucleotides can also include, additionally or alternatively, nucleobase (often referred to in the art simply as “base”) modifications or substitutions.
- nucleobase often referred to in the art simply as “base”
- “unmodified” or “natural” nucleobases include adenine (A), guanine (G), thymine (T), cytosine (C) and uracil (U).
- Modified nucleobases include nucleobases found only infrequently or transiently in natural nucleic acids, e.g., hypoxanthine, 6-methyladenine, 5-Me pyrimidines, particularly 5-methylcytosine (also referred to as 5-methyl-2' deoxycytosine and often referred to in the art as 5-Me-C), 5-hydroxymethylcytosine (HMC), glycosyl HMC and gentobiosyl HMC, isocytosine, pseudoisocytosine, as well as synthetic nucleobases, e.g., 2-aminoadenine, 2- (methylamino)adenine, 2-(imidazolylalkyl)adenine, 2- (aminoalklyamino)adenine or other heterosubstituted alkyladenines, 2-thiouracil, 2- thiothymine, 5-bromouracil, 5-hydroxymethyluracil, 5-propynyluracil, 8-azaguanine
- both a sugar and an internucleoside linkage, i.e., the backbone, of the nucleotide units are replaced with novel groups.
- the base units are maintained for hybridization with an appropriate nucleic acid target compound.
- an oligomeric compound an oligonucleotide mimetic that has been shown to have excellent hybridization properties, is referred to as a peptide nucleic acid (PNA).
- PNA peptide nucleic acid
- the sugar- backbone of an oligonucleotide is replaced with an amide containing backbone, for example, an aminoethylglycine backbone.
- the nucleobases are retained and are bound directly or indirectly to aza nitrogen atoms of the amide portion of the backbone.
- PNA compounds include, but are not limited to, US patent nos. 5,539,082; 5,714,331; and 5,719,262, each of which is herein incorporated by reference. Further teaching of PNA compounds can be found in Nielsen et al, Science, 1991, 254, 1497-1500.
- Single stranded oligonucleotides can also include one or more nucleobase (often referred to in the art simply as “base”) modifications or substitutions.
- nucleobases comprise the purine bases adenine (A) and guanine (G), and the pyrimidine bases thymine (T), cytosine (C) and uracil (U).
- Modified nucleobases comprise other synthetic and natural nucleobases such as 5-methylcytosine (5- me-C), 5-hydroxymethyl cytosine, xanthine, hypoxanthine, 2-aminoadenine, 6-methyl and other alkyl derivatives of adenine and guanine, 2-propyl and other alkyl derivatives of adenine and guanine, 2-thiouracil, 2-thiothymine and 2-thiocytosine, 5-halouracil and cytosine, 5-propynyl uracil and cytosine, 6-azo uracil, cytosine and thymine, 5 -uracil (pseudo-uracil), 4-thiouracil, 8-halo, 8-amino, 8-thiol, 8- thioalkyl, 8-hydroxyl and other 8- substituted adenines and guanines, 5-halo particularly 5- bromo, 5-trifluoromethyl and other 5-substi
- nucleobases comprise those disclosed in United States Patent No. 3,687,808, those disclosed in "The Concise Encyclopedia of Polymer Science And Engineering", pages 858-859, Kroschwitz, ed. John Wiley & Sons, 1990;, those disclosed by Englisch et al, Angewandle Chemie, International Edition, 1991, 30, page 613, and those disclosed by Sanghvi, Chapter 15, Antisense Research and Applications," pages 289- 302, Crooke, and Lebleu, eds., CRC Press, 1993. Certain of these nucleobases are particularly useful for increasing the binding affinity of the oligomeric compounds of the invention.
- 5-substituted pyrimidines 6-azapyrimidines and N-2, N-6 and 0-6 substituted purines, comprising 2-aminopropyladenine, 5-propynyluracil and 5- propynylcytosine.
- 5- methylcytosine substitutions have been shown to increase nucleic acid duplex stability by 0.6-1.2 ⁇ 0>C (Sanghvi, et al, eds, "Antisense Research and Applications," CRC Press, Boca Raton, 1993, pp. 276-278) and are presently preferred base substitutions, even more particularly when combined with 2'-0-methoxyethyl sugar modifications. Modified nucleobases are described in US patent nos.
- the single stranded oligonucleotides are chemically linked to one or more moieties or conjugates that enhance the activity, cellular distribution, or cellular uptake of the oligonucleotide.
- one or more single stranded oligonucleotides, of the same or different types, can be conjugated to each other; or single stranded
- oligonucleotides can be conjugated to targeting moieties with enhanced specificity for a cell type or tissue type.
- moieties include, but are not limited to, lipid moieties such as a cholesterol moiety (Letsinger et al, Proc. Natl. Acad. Sci. USA, 1989, 86, 6553-6556), cholic acid (Manoharan et al, Bioorg. Med. Chem. Let., 1994, 4, 1053-1060), a thioether, e.g., hexyl-S- tritylthiol (Manoharan et al, Ann. N. Y. Acad.
- Acids Res., 1990, 18, 3777-3783 a polyamine or a polyethylene glycol chain (Mancharan et al, Nucleosides & Nucleotides, 1995, 14, 969-973), or adamantane acetic acid (Manoharan et al., Tetrahedron Lett., 1995, 36, 3651-3654), a palmityl moiety (Mishra et al., Biochim. Biophys. Acta, 1995, 1264, 229-237), or an octadecylamine or hexylamino-carbonyl-t oxycholesterol moiety (Crooke et al., J. Pharmacol. Exp.
- conjugate groups of the invention include intercalators, reporter molecules, polyamines, polyamides, polyethylene glycols, polyethers, groups that enhance the pharmacodynamic properties of oligomers, and groups that enhance the pharmacokinetic properties of oligomers.
- Typical conjugate groups include cholesterols, lipids, phospholipids, biotin, phenazine, folate, phenanthridine, anthraquinone, acridine, fluoresceins, rhodamines, coumarins, and dyes.
- Groups that enhance the pharmacodynamic properties include groups that improve uptake, enhance resistance to degradation, and/or strengthen sequence-specific hybridization with the target nucleic acid.
- Groups that enhance the pharmacokinetic properties include groups that improve uptake, distribution, metabolism or excretion of the compounds of the present invention. Representative conjugate groups are disclosed in International Patent Application No. PCT/US92/09196, filed Oct. 23, 1992, and U.S. Pat. No. 6,287,860, which are incorporated herein by reference.
- Conjugate moieties include, but are not limited to, lipid moieties such as a cholesterol moiety, cholic acid, a thioether, e.g., hexyl-5-tritylthiol, a thiocholesterol, an aliphatic chain, e.g., dodecandiol or undecyl residues, a phospholipid, e.g., di-hexadecyl-rac- glycerol or triethylammonium 1,2- di-O-hexadecyl-rac-glycero-3-H-phosphonate, a polyamine or a polyethylene glycol chain, or adamantane acetic acid, a palmityl moiety, or an octadecylamine or hexylamino-carbonyl-oxy cholesterol moiety.
- lipid moieties such as a cholesterol moiety, cholic acid, a thioether,
- single stranded oligonucleotide modification include modification of the 5' or 3' end of the oligonucleotide.
- the 3' end of the oligonucleotide comprises a hydroxyl group or a thiophosphate.
- additional molecules e.g. a biotin moiety or a fluorophor
- the single stranded oligonucleotide comprises a biotin moiety conjugated to the 5' nucleotide.
- the single stranded oligonucleotide comprises locked nucleic acids (LNA), ENA modified nucleotides, 2'-0-methyl nucleotides, or 2'-fluoro- deoxyribonucleotides. In some embodiments, the single stranded oligonucleotide comprises alternating deoxyribonucleotides and 2'-fluoro-deoxyribonucleotides. In some embodiments, the single stranded oligonucleotide comprises alternating deoxyribonucleotides and 2'-0- methyl nucleotides.
- the single stranded oligonucleotide comprises alternating deoxyribonucleotides and ENA modified nucleotides. In some embodiments, the single stranded oligonucleotide comprises alternating deoxyribonucleotides and locked nucleic acid nucleotides. In some embodiments, the single stranded oligonucleotide comprises alternating locked nucleic acid nucleotides and 2'-0-methyl nucleotides.
- the 5' nucleotide of the oligonucleotide is a
- the 5' nucleotide of the oligonucleotide is a locked nucleic acid nucleotide.
- the nucleotides of the oligonucleotide comprise deoxyribonucleotides flanked by at least one locked nucleic acid nucleotide on each of the 5' and 3 ' ends of the deoxyribonucleotides.
- the nucleotide at the 3' position of the oligonucleotide has a 3 ' hydroxyl group or a 3' thiophosphate.
- the single stranded oligonucleotide comprises
- the single stranded oligonucleotide comprises phosphorothioate internucleotide linkages between at least two nucleotides. In some embodiments, the single stranded oligonucleotide comprises phosphorothioate internucleotide linkages between all nucleotides.
- the single stranded oligonucleotide can have any combination of modifications as described herein.
- the oligonucleotide may comprise a nucleotide sequence having one or more of the following modification patterns.
- XXXXXXx in which "X” denotes a nucleotide analogue, (X) denotes an optional nucleotide analogue, and "x" denotes a DNA or R A nucleotide unit.
- X denotes a nucleotide analogue
- X denotes an optional nucleotide analogue
- x denotes a DNA or R A nucleotide unit.
- methods for increasing expression of SMN protein in a cell.
- the methods involve delivering to the cell a first single stranded oligonucleotide complementary with a PRC2-associated region of SMNl or SMN2 and a second single stranded oligonucleotide complementary with a splice control sequence of a precursor mRNA of SMNl or SMN2, in amounts sufficient to increase expression of a mature mRNA of SMNl or SMN2 that comprises (or includes) exon 7 in the cell.
- the first and second single stranded oligonucleotides may be delivered together or separately.
- the first and second single stranded oligonucleotides may be linked together, or unlinked, i.e., separate.
- methods for treating spinal muscular atrophy in a subject.
- the methods involve administering to a subject a first single stranded oligonucleotide complementary with a PRC2-associated region of SMNl or SMN2 and a second single stranded oligonucleotide complementary with a splice control sequence of a precursor mRNA of SMNl or SMN2, in amounts sufficient to increase expression of full length SMN protein in the subject to levels sufficient to improve one or more conditions associated with SMA.
- the first and second single stranded oligonucleotides may be administered together or separately.
- the first and second single stranded oligonucleotides may be linked together, or unlinked, i.e., separate.
- the first single stranded oligonucleotide may be administered within 1 hour, 2 hours, 3 hours, 4 hours, 8 hours, 12 hours, 24 hours, 48 hours or more of administration of the second single stranded oligonucleotide.
- the first single stranded oligonucleotide may be administered before or after the second single stranded oligonucleotide.
- the oligonucleotides may be administered once or on multiple occasions depending on the needs of the subject and/or judgment of the treating physician. In some cases, the oligonucleotides may be administered in cycles.
- the administration cycles may vary; for example, the administration cycle may be 2 nd oligo - 1 st oligo - 2 nd oligo - 1 st oligo and so on; or 1 st oligo-2 nd oligo- 1 st oligo-2 nd oligo, and so on; or 1 st oligo - 2 nd oligo - 2 nd oligo -1 st oligo- 1 st oligo - 2 nd oligo - 2 nd oligo -1 st oligo, and so on.
- the skilled artisan will be capable of selecting administration cycles and intervals between each administration that are appropriate for treating a particular subject.
- the invention relates to methods for modulating gene expression in a cell (e.g., a cell for which SMNl or SMN2 levels are reduced) for research purposes (e.g., to study the function of the gene in the cell).
- the invention relates to methods for modulating gene expression in a cell (e.g., a cell for which SMNl or SMN2 levels are reduced) for gene or epigenetic therapy.
- the cells can be in vitro, ex vivo, or in vivo (e.g., in a subject who has a disease resulting from reduced expression or activity of SMNl or SMN2.
- methods for modulating gene expression in a cell comprise delivering a single stranded oligonucleotide as described herein.
- delivery of the single stranded oligonucleotide to the cell results in a level of expression of gene that is at least 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 100%, 200% or more greater than a level of expression of gene in a control cell to which the single stranded
- oligonucleotide has not been delivered.
- delivery of the single stranded oligonucleotide to the cell results in a level of expression of gene that is at least 50%> greater than a level of expression of gene in a control cell to which the single stranded oligonucleotide has not been delivered.
- methods comprise administering to a subject (e.g. a human) a composition comprising a single stranded oligonucleotide as described herein to increase protein levels in the subject.
- a subject e.g. a human
- the increase in protein levels is at least 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 100%, 200%, or more, higher than the amount of a protein in the subject before administering.
- the methods include introducing into the cell a single stranded oligonucleotide that is sufficiently complementary to a PRC2-associated region (e.g., of a long non-coding R A) that maps to a genomic position encompassing or in proximity to the SMNl or SMN2 gene.
- a PRC2-associated region e.g., of a long non-coding R A
- a condition e.g. , Spinal muscular atrophy
- SMNl or SMN2 a condition associated with decreased levels of expression of SMNl or SMN2 in a subject
- the method comprising administering a single stranded oligonucleotide as described herein.
- a subject can include a non-human mammal, e.g. mouse, rat, guinea pig, rabbit, cat, dog, goat, cow, or horse.
- a subject is a human.
- Single stranded oligonucleotides have been employed as therapeutic moieties in the treatment of disease states in animals, including humans.
- Single stranded oligonucleotides can be useful therapeutic modalities that can be configured to be useful in treatment regimes for the treatment of cells, tissues and animals, especially humans.
- an animal preferably a human, suspected of having Spinal muscular atrophy is treated by administering single stranded oligonucleotide in accordance with this invention.
- the methods comprise the step of administering to the animal in need of treatment, a therapeutically effective amount of a single stranded oligonucleotide as described herein.
- oligonucleotides described herein can be formulated for administration to a subject for treating a condition ⁇ e.g., Spinal muscular atrophy) associated with decreased levels of SMN protein. It should be understood that the formulations, compositions and methods can be practiced with any of the oligonucleotides disclosed herein. In some embodiments, formulations are provided that comprise a first single stranded oligonucleotide complementary with a PRC2-associated region of a gene and a second single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of the gene.
- formulations comprise a first single stranded oligonucleotide complementary with a PRC2-associated region of a gene that is linked via a linker with a second single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of the gene.
- a first single stranded oligonucleotide complementary with a PRC2-associated region of a gene is linked with a second single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of the gene, and in other embodiments, the single stranded oligonucleotides are not linked.
- Single stranded oligonucleotides that are not linked may be administered to a subject or delivered to a cell simultaneously (e.g., within the same composition) or separately.
- the formulations may conveniently be presented in unit dosage form and may be prepared by any methods well known in the art of pharmacy.
- the amount of active ingredient ⁇ e.g. , an oligonucleotide or compound of the invention
- the amount of active ingredient which can be combined with a carrier material to produce a single dosage form will vary depending upon the host being treated, the particular mode of administration, e.g., intradermal or inhalation.
- the amount of active ingredient which can be combined with a carrier material to produce a single dosage form will generally be that amount of the compound which produces a therapeutic effect, e.g. tumor regression.
- compositions of this invention can be prepared according to any method known to the art for the manufacture of pharmaceuticals. Such formulations can contain sweetening agents, flavoring agents, coloring agents and preserving agents. A formulation can be admixtured with nontoxic pharmaceutically acceptable excipients which are suitable for manufacture. Formulations may comprise one or more diluents, emulsifiers, preservatives, buffers, excipients, etc. and may be provided in such forms as liquids, powders, emulsions, lyophilized powders, sprays, creams, lotions, controlled release formulations, tablets, pills, gels, on patches, in implants, etc.
- a formulated single stranded oligonucleotide composition can assume a variety of states.
- the composition is at least partially crystalline, uniformly crystalline, and/or anhydrous (e.g., less than 80, 50, 30, 20, or 10% water).
- the single stranded oligonucleotide is in an aqueous phase, e.g., in a solution that includes water.
- the aqueous phase or the crystalline compositions can, e.g. , be incorporated into a delivery vehicle, e.g., a liposome (particularly for the aqueous phase) or a particle (e.g., a microparticle as can be appropriate for a crystalline composition).
- the single stranded oligonucleotide composition is formulated in a manner that is compatible with the intended method of administration.
- the composition is prepared by at least one of the following methods: spray drying, lyophilization, vacuum drying, evaporation, fluid bed drying, or a combination of these techniques; or sonication with a lipid, freeze-drying, condensation and other self-assembly.
- a single stranded oligonucleotide preparation can be formulated or administered (together or separately) in combination with another agent, e.g., another therapeutic agent or an agent that stabilizes a single stranded oligonucleotide, e.g., a protein that complexes with single stranded oligonucleotide.
- another agent e.g., another therapeutic agent or an agent that stabilizes a single stranded oligonucleotide, e.g., a protein that complexes with single stranded oligonucleotide.
- Still other agents include chelators, e.g., EDTA (e.g., to
- RNAse inhibitors e.g., a broad specificity
- RNAse inhibitor such as RNAsin
- the other agent used in combination with the single stranded oligonucleotide is an agent that also regulates SMN expression.
- the other agent is a growth hormone, a histone deacetylase inhibitor, a hydroxycarbamide (hydroxyurea), a natural polyphenol compound (e.g., resveratrol, curcumin), prolactin, or salbutamol.
- histone deacetylase inhibitors examples include aliphatic compounds (e.g., butyrates (e.g., sodium butyrate and sodium phenylbutyrate) and valproic acid), benzamides (e.g., M344), and hydroxamic acids (e.g., CBHA, SBHA, Entinostat (MS-275)) Panobinostat (LBH-589), Trichostatin A, Vorinostat (SAHA)),
- aliphatic compounds e.g., butyrates (e.g., sodium butyrate and sodium phenylbutyrate) and valproic acid
- benzamides e.g., M344
- hydroxamic acids e.g., CBHA, SBHA, Entinostat (MS-275)
- Panobinostat LH-589
- Trichostatin A Vorinostat (SAHA)
- the single stranded oligonucleotide preparation includes another single stranded oligonucleotide, e.g. , a second single stranded oligonucleotide that modulates expression and/or mR A processing of a second gene or a second single stranded
- the single stranded oligonucleotide preparation includes at least a second therapeutic agent ⁇ e.g., an agent other than an oligonucleotide).
- a composition that includes a single stranded oligonucleotide can be delivered to a subject by a variety of routes.
- routes include: intravenous, intradermal, topical, rectal, parenteral, anal, intravaginal, intranasal, pulmonary, ocular.
- therapeutically effective amount is the amount of oligonucleotide present in the composition that is needed to provide the desired level of SMNl or SMN2 expression in the subject to be treated to give the anticipated physiological response.
- physiologically effective amount is that amount delivered to a subject to give the desired palliative or curative effect.
- pharmaceutically acceptable carrier means that the carrier can be administered to a subject with no significant adverse toxico logical effects to the subject.
- compositions suitable for administration can be incorporated into pharmaceutical compositions suitable for administration.
- Such compositions typically include one or more species of single stranded oligonucleotide and a pharmaceutically acceptable carrier.
- pharmaceutically acceptable carrier is intended to include any and all solvents, dispersion media, coatings, antibacterial and antifungal agents, isotonic and absorption delaying agents, and the like, compatible with pharmaceutical administration.
- the use of such media and agents for pharmaceutically active substances is well known in the art. Except insofar as any conventional media or agent is incompatible with the active compound, use thereof in the compositions is contemplated. Supplementary active compounds can also be incorporated into the compositions.
- compositions of the present invention may be administered in a number of ways depending upon whether local or systemic treatment is desired and upon the area to be treated. Administration may be topical (including ophthalmic, vaginal, rectal, intranasal, transdermal), oral or parenteral. Parenteral administration includes intravenous drip, subcutaneous, intraperitoneal or intramuscular injection, or intrathecal or
- the route and site of administration may be chosen to enhance targeting.
- intramuscular injection into the muscles of interest would be a logical choice.
- Lung cells might be targeted by administering the single stranded oligonucleotide in aerosol form.
- the vascular endothelial cells could be targeted by coating a balloon catheter with the single stranded oligonucleotide and mechanically introducing the oligonucleotide.
- Topical administration refers to the delivery to a subject by contacting the formulation directly to a surface of the subject.
- the most common form of topical delivery is to the skin, but a composition disclosed herein can also be directly applied to other surfaces of the body, e.g., to the eye, a mucous membrane, to surfaces of a body cavity or to an internal surface.
- the most common topical delivery is to the skin.
- the term encompasses several routes of administration including, but not limited to, topical and transdermal. These modes of administration typically include penetration of the skin's permeability barrier and efficient delivery to the target tissue or stratum.
- Topical administration can be used as a means to penetrate the epidermis and dermis and ultimately achieve systemic delivery of the composition.
- Topical administration can also be used as a means to selectively deliver oligonucleotides to the epidermis or dermis of a subject, or to specific strata thereof, or to an underlying tissue.
- Formulations for topical administration may include transdermal patches, ointments, lotions, creams, gels, drops, suppositories, sprays, liquids and powders.
- Conventional pharmaceutical carriers, aqueous, powder or oily bases, thickeners and the like may be necessary or desirable.
- Coated condoms, gloves and the like may also be useful.
- Transdermal delivery is a valuable route for the administration of lipid soluble therapeutics.
- the dermis is more permeable than the epidermis and therefore absorption is much more rapid through abraded, burned or denuded skin.
- Inflammation and other physiologic conditions that increase blood flow to the skin also enhance transdermal adsorption. Absorption via this route may be enhanced by the use of an oily vehicle
- transdermal route provides a potentially effective means to deliver a composition disclosed herein for systemic and/or local therapy.
- iontophoresis transfer of ionic solutes through biological membranes under the influence of an electric field
- phonophoresis or sonophoresis use of ultrasound to enhance the absorption of various therapeutic agents across biological membranes, notably the skin and the cornea
- optimization of vehicle characteristics relative to dose position and retention at the site of administration may be useful methods for enhancing the transport of topically applied compositions across skin and mucosal sites.
- oligonucleotides administered through these membranes may have a rapid onset of action, provide therapeutic plasma levels, avoid first pass effect of hepatic metabolism, and avoid exposure of the oligonucleotides to the hostile gastrointestinal (GI) environment. Additional advantages include easy access to the membrane sites so that the oligonucleotide can be applied, localized and removed easily.
- GI gastrointestinal
- compositions can be targeted to a surface of the oral cavity, e.g. , to sublingual mucosa which includes the membrane of ventral surface of the tongue and the floor of the mouth or the buccal mucosa which constitutes the lining of the cheek.
- the sublingual mucosa is relatively permeable thus giving rapid absorption and acceptable bioavailability of many agents. Further, the sublingual mucosa is convenient, acceptable and easily accessible.
- a pharmaceutical composition of single stranded oligonucleotide may also be administered to the buccal cavity of a human being by spraying into the cavity, without inhalation, from a metered dose spray dispenser, a mixed micellar pharmaceutical formulation as described above and a propellant.
- the dispenser is first shaken prior to spraying the pharmaceutical formulation and propellant into the buccal cavity.
- compositions for oral administration include powders or granules, suspensions or solutions in water, syrups, slurries, emulsions, elixirs or non-aqueous media, tablets, capsules, lozenges, or troches.
- carriers that can be used include lactose, sodium citrate and salts of phosphoric acid.
- Various disintegrants such as starch, and lubricating agents such as magnesium stearate, sodium lauryl sulfate and talc, are commonly used in tablets.
- useful diluents are lactose and high molecular weight polyethylene glycols.
- the nucleic acid compositions can be combined with emulsifying and suspending agents. If desired, certain sweetening and/or flavoring agents can be added.
- Parenteral administration includes intravenous drip, subcutaneous, intraperitoneal or intramuscular injection, intrathecal or intraventricular administration.
- parental administration involves administration directly to the site of disease (e.g. injection into a tumor).
- Formulations for parenteral administration may include sterile aqueous solutions which may also contain buffers, diluents and other suitable additives.
- Intraventricular injection may be facilitated by an intraventricular catheter, for example, attached to a reservoir.
- the total concentration of solutes should be controlled to render the preparation isotonic.
- any of the single stranded oligonucleotides described herein can be administered to ocular tissue.
- the compositions can be applied to the surface of the eye or nearby tissue, e.g., the inside of the eyelid.
- ointments or droppable liquids may be delivered by ocular delivery systems known to the art such as applicators or eye droppers.
- Such compositions can include mucomimetics such as hyaluronic acid, chondroitin sulfate, hydroxypropyl methylcellulose or poly( vinyl alcohol), preservatives such as sorbic acid, EDTA or benzylchronium chloride, and the usual quantities of diluents and/or carriers.
- the single stranded oligonucleotide can also be administered to the interior of the eye, and can be introduced by a needle or other delivery device which can introduce it to a selected area or structure.
- Pulmonary delivery compositions can be delivered by inhalation by the patient of a dispersion so that the composition, preferably single stranded oligonucleotides, within the dispersion can reach the lung where it can be readily absorbed through the alveolar region directly into blood circulation. Pulmonary delivery can be effective both for systemic delivery and for localized delivery to treat diseases of the lungs.
- Pulmonary delivery can be achieved by different approaches, including the use of nebulized, aerosolized, micellular and dry powder-based formulations. Delivery can be achieved with liquid nebulizers, aerosol-based inhalers, and dry powder dispersion devices. Metered-dose devices are preferred. One of the benefits of using an atomizer or inhaler is that the potential for contamination is minimized because the devices are self-contained. Dry powder dispersion devices, for example, deliver agents that may be readily formulated as dry powders. A single stranded oligonucleotide composition may be stably stored as lyophilized or spray-dried powders by itself or in combination with suitable powder carriers.
- the delivery of a composition for inhalation can be mediated by a dosing timing element which can include a timer, a dose counter, time measuring device, or a time indicator which when incorporated into the device enables dose tracking, compliance monitoring, and/or dose triggering to a patient during administration of the aerosol medicament.
- a dosing timing element which can include a timer, a dose counter, time measuring device, or a time indicator which when incorporated into the device enables dose tracking, compliance monitoring, and/or dose triggering to a patient during administration of the aerosol medicament.
- the term “powder” means a composition that consists of finely dispersed solid particles that are free flowing and capable of being readily dispersed in an inhalation device and subsequently inhaled by a subject so that the particles reach the lungs to permit penetration into the alveoli.
- the powder is said to be "respirable.”
- the average particle size is less than about 10 ⁇ in diameter preferably with a relatively uniform spheroidal shape distribution. More preferably the diameter is less than about 7.5 ⁇ m and most preferably less than about 5.0 ⁇ m.
- the particle size distribution is between about 0.1 ⁇ m and about 5 ⁇ m in diameter, particularly about 0.3 ⁇ m to about 5 ⁇ m.
- dry means that the composition has a moisture content below about 10% by weight (% w) water, usually below about 5% w and preferably less it than about 3% w.
- a dry composition can be such that the particles are readily dispersible in an inhalation device to form an aerosol.
- HSA human serum albumin
- Suitable H adjusters or buffers include organic salts prepared from organic acids and bases, such as sodium citrate, sodium ascorbate, and the like; sodium citrate is preferred.
- Pulmonary administration of a micellar single stranded oligonucleotide formulation may be achieved through metered dose spray devices with propellants such as tetrafluoroethane, heptafluoroethane, dimethylfluoropropane, tetrafluoropropane, butane, isobutane, dimethyl ether and other non-CFC and CFC propellants.
- propellants such as tetrafluoroethane, heptafluoroethane, dimethylfluoropropane, tetrafluoropropane, butane, isobutane, dimethyl ether and other non-CFC and CFC propellants.
- Exemplary devices include devices which are introduced into the vasculature, e.g. , devices inserted into the lumen of a vascular tissue, or which devices themselves form a part of the vasculature, including stents, catheters, heart valves, and other vascular devices. These devices, e.g., catheters or stents, can be placed in the vasculature of the lung, heart, or leg.
- Other devices include non-vascular devices, e.g., devices implanted in the
- the device can release a therapeutic substance in addition to a single stranded oligonucleotide, e.g., a device can release insulin.
- unit doses or measured doses of a composition that includes single stranded oligonucleotide are dispensed by an implanted device.
- the device can include a sensor that monitors a parameter within a subject.
- the device can include pump, e.g., and, optionally, associated electronics.
- Tissue e.g., cells or organs can be treated with a single stranded oligonucleotide, ex vivo and then administered or implanted in a subject.
- the tissue can be autologous, allogeneic, or xenogeneic tissue.
- tissue can be treated to reduce graft v. host disease .
- the tissue is allogeneic and the tissue is treated to treat a disorder characterized by unwanted gene expression in that tissue.
- tissue e.g., hematopoietic cells, e.g., bone marrow hematopoietic cells, can be treated to inhibit unwanted cell proliferation.
- the single stranded oligonucleotide treated cells are insulated from other cells, e.g., by a semi-permeable porous barrier that prevents the cells from leaving the implant, but enables molecules from the body to reach the cells and molecules produced by the cells to enter the body.
- the porous barrier is formed from alginate.
- a contraceptive device is coated with or contains a single stranded oligonucleotide. Exemplary devices include condoms, diaphragms, IUD
- the invention features a method of administering a single stranded oligonucleotide ⁇ e.g., as a compound or as a component of a composition) to a subject ⁇ e.g., a human subject).
- the methods involve administering a compound (e.g., a compound of the general formula A-B-C, as disclosed herein, or an single stranded oligonucleotide,) in a unit dose to a subject.
- the unit dose is between about 10 mg and 25 mg per kg of bodyweight. In one embodiment, the unit dose is between about 1 mg and 100 mg per kg of bodyweight.
- the unit dose is between about 0.1 mg and 500 mg per kg of bodyweight. In some embodiments, the unit dose is more than 0.001, 0.005, 0.01, 0.05, 0.1, 0.5, 1, 2, 5, 10, 25, 50 or 100 mg per kg of bodyweight.
- the defined amount can be an amount effective to treat or prevent a disease or disorder, e.g., a disease or disorder associated with the SMN1 or SMN2.
- the unit dose for example, can be administered by injection ⁇ e.g., intravenous or intramuscular), an inhaled dose, or a topical application.
- the unit dose is administered daily. In some embodiments, less frequently than once a day, e.g., less than every 2, 4, 8 or 30 days. In another embodiment, the unit dose is not administered with a frequency ⁇ e.g., not a regular frequency). For example, the unit dose may be administered a single time. In some embodiments, the unit dose is administered more than once a day, e.g. , once an hour, two hours, four hours, eight hours, twelve hours, etc.
- a subject is administered an initial dose and one or more maintenance doses of a single stranded oligonucleotide.
- the maintenance dose or doses are generally lower than the initial dose, e.g., one-half less of the initial dose.
- a maintenance regimen can include treating the subject with a dose or doses ranging from 0.0001 to 100 mg/kg ofbody weight per day, e.g., 100, 10, 1, 0.1, 0.01, 0.001, or 0.0001 mg per kg of bodyweight per day.
- the maintenance doses may be administered no more than once every 1, 5, 10, or 30 days. Further, the treatment regimen may last for a period of time which will vary depending upon the nature of the particular disease, its severity and the overall condition of the patient.
- the dosage may be delivered no more than once per day, e.g., no more than once per 24, 36, 48, or more hours, e.g., no more than once for every 5 or 8 days.
- the patient can be monitored for changes in his condition and for alleviation of the symptoms of the disease state.
- the dosage of the oligonucleotide may either be increased in the event the patient does not respond significantly to current dosage levels, or the dose may be decreased if an alleviation of the symptoms of the disease state is observed, if the disease state has been ablated, or if undesired side-effects are observed.
- the effective dose can be administered in a single dose or in two or more doses, as desired or considered appropriate under the specific circumstances. If desired to facilitate repeated or frequent infusions, implantation of a delivery device, e.g., a pump, semipermanent stent ⁇ e.g., intravenous, intraperitoneal, intracisternal or intracapsular), or reservoir may be advisable.
- a delivery device e.g., a pump, semipermanent stent ⁇ e.g., intravenous, intraperitoneal, intracisternal or intracapsular
- reservoir may be advisable.
- the pharmaceutical composition includes a plurality of single stranded oligonucleotide species.
- the pharmaceutical composition comprises a first single stranded oligonucleotide complementary with a PRC2-associated region of a gene (e.g., SMN1 or SMN2), and a second single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of a gene (e.g., SMN1 or SMN2).
- the pharmaceutical composition includes a compound comprising the general formula A-B-C, in which A is a single stranded oligonucleotide complementary with a PRC2-associated region of a gene, B is a linker, and C is a single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of the gene.
- the single stranded oligonucleotide species has sequences that are non-overlapping and non-adjacent to another species with respect to a naturally occurring target sequence ⁇ e.g., a PRC2-associated region).
- the plurality of single stranded oligonucleotide species is specific for different PRC2-associated regions.
- the single stranded oligonucleotide is allele specific. In some cases, a patient is treated with a single stranded oligonucleotide in conjunction with other therapeutic modalities.
- the patient undergo maintenance therapy to prevent the recurrence of the disease state, wherein the compound of the invention is administered in maintenance doses, ranging from 0.0001 mg to 100 mg per kg of body weight.
- the concentration of the single stranded oligonucleotide composition is an amount sufficient to be effective in treating or preventing a disorder or to regulate a physiological condition in humans.
- concentration or amount of single stranded oligonucleotide administered will depend on the parameters determined for the agent and the method of administration, e.g. nasal, buccal, pulmonary.
- nasal formulations may tend to require much lower concentrations of some ingredients in order to avoid irritation or burning of the nasal passages. It is sometimes desirable to dilute an oral formulation up to 10-100 times in order to provide a suitable nasal formulation.
- treatment of a subject with a therapeutically effective amount of a single stranded oligonucleotide can include a single treatment or, preferably, can include a series of treatments.
- the effective dosage of a single stranded oligonucleotide used for treatment may increase or decrease over the course of a particular treatment.
- the subject can be monitored after administering a single stranded oligonucleotide composition. Based on information from the monitoring, an additional amount of the single stranded
- oligonucleotide composition can be administered.
- Dosing is dependent on severity and responsiveness of the disease condition to be treated, with the course of treatment lasting from several days to several months, or until a cure is effected or a diminution of disease state is achieved.
- Optimal dosing schedules can be calculated from measurements of SMN1 or SMN2 expression levels in the body of the patient. Persons of ordinary skill can easily determine optimum dosages, dosing
- the animal models include transgenic animals that express a human SMN1 or SMN2.
- the composition for testing includes a single stranded oligonucleotide that is complementary, at least in an internal region, to a sequence that is conserved between SMN1 or SMN2 in the animal model and the SMN1 or SMN2 in a human.
- the administration of the single stranded oligonucleotide composition is parenteral, e.g.
- intravenous ⁇ e.g., as a bolus or as a diffusible infusion
- intradermal intraperitoneal
- intramuscular intrathecal
- intraventricular intracranial
- subcutaneous transmucosal
- buccal sublingual
- endoscopic rectal
- oral vaginal
- topical pulmonary, intranasal, urethral or ocular.
- Administration can be provided by the subject or by another person, e.g., a health care provider.
- the composition can be provided in measured doses or in a dispenser which delivers a metered dose. Selected modes of delivery are discussed in more detail below.
- kits comprising a container housing a composition comprising a single stranded oligonucleotide.
- the kits comprise a container housing a single stranded oligonucleotide complementary with of a PRC2-associated region of a gene; and a second container housing a single stranded oligonucleotide complementary to a splice control sequence of a precursor mR A of the gene.
- kits comprise a container housing a single stranded oligonucleotide complementary with of a PRC2-associated region of a gene and a single stranded oligonucleotide complementary to a splice control sequence of a precursor mRNA of the gene.
- the composition is a pharmaceutical composition comprising a single stranded oligonucleotide and a pharmaceutically acceptable carrier.
- the individual components of the pharmaceutical composition may be provided in one container. Alternatively, it may be desirable to provide the components of the pharmaceutical composition separately in two or more containers, e.g., one container for single stranded oligonucleotides, and at least another for a carrier compound.
- the kit may be packaged in a number of different configurations such as one or more containers in a single box.
- the different components can be combined, e.g., according to instructions provided with the kit.
- the components can be combined according to a method described herein, e.g., to prepare and administer a pharmaceutical composition.
- the kit can also include a delivery device.
- Example 1 Oligonucleotides targeting PRC2-associated regions that upregulate SMN1.
- MA TERIALS AND METHODS Real Time PCR
- Human hepatocyte Hep3B, human hepatocyte HepG2 cells, mouse hepatoma Hepal-6 cells, and human renal proximal tubule epithelial cells (RPTEC) were cultured using conditions known in the art (see, e.g. Current Protocols in Cell Biology).
- Other cell lines tested were neuronal cell lines (SK-N-AS, U-87) and SMN patient fibroblasts. Details of the cell lines used in the experiments described herein are provided in Table 8.
- Oligonucleotides were designed within PRC2-interacting regions in order to upregulate SMNl .
- the sequence and structure of each oligonucleotide is shown in Table 2.
- the following table provides a description of the nucleotide analogs, modifications and intranucleotide linkages used for certain oligonucleotides tested and described in Table 2.
- Oligonucleotides were designed as candidates for upregulating SMNl expression. A total of 52 single stranded oligonucleotides were designed to be complementary to a PRC2- interacting region within a sequence as set forth in SEQ ID NO: 1, 2, 4, or 5.
- Oligonucleotides were tested in at least duplicate. The sequence and structural features of the oligonucleotides are set forth in Table 2. Briefly, cells were transfected in vitro with the oligonucleotides as described above. SMNl expression in cells following treatment was evaluated by qRT-PCR. Oligonucleotides that upregulated SMNl expression were identified. Further details are outlined in Table 2.
- Table 5 shows further results from experiments in which oligonucleotides were transfected into cells at a particular concentration [oligo] and 48 or 72h later RNA was prepared and qRTPCR assays carried out to determine mRNA levels of full length (FL) or delta7 SMN.
- oligos were administered gymnotically to cells at ⁇ and R A harvested 9 days post treatment.
- the cell lines tested were neuronal cell lines (SK-N- AS, U-87) and SMN patient fibroblasts.
- Table 6 shows results from experiments in which oligonucleotides were transfected into cells in combination with either one or two more oligos or small molecule compounds at a particular concentration ([oligo], [2nd], [3rd]) and 48 or 72h later RNA was prepared and qRTPCR assays carried out to determine mRNA levels of full length (FL) or delta7 SMN.
- the cell lines tested were SMN patient fibroblasts.
- Table 7 shows results from experiments in which oligonucleotides were transfected into cells in combination with either one or two more oligos or as dimers or by gymnotic treatment at a particular concentration ([oligo], [2nd], [3rd]) and 24, 48, 72 or 216h later cell lysates were prepared and ELISA assays carried out to determine SMN protein levels.
- the cell lines tested were SMN patient fibroblasts.
- GGGGGU GGGGUA, GGGUAC, GGGUAU, GGGUCA, GGGUCC, GGGUCG, GGGUGA, GGGUGC, GGGU UA, GGGU UG, GGUAAA, GGUAAC, GGUAAG, GGUAAU, GGUACA, GGUACC, GGUACG,
- GGUACU GGUAGC, GGUAGG, GGUAGU, GGUAUA, GGUAUC, GGUAUG, GGUCAA, GGUCAC, GGUCAG, GGUCAU, GGUCCA, GGUCCG, GGUCCU, GGUCGA, GGUCGC, GGUCGG, GGUCGU, GGUCUC, GGUCU U, GGUGAA, GGUGAC, GGUGAU, GGUGCA, GGUGCC, GGUGGC, GGUGUA, GGUGUC, GGU UAA, GGU UAG, GGU UAU, GGUUCA, GGU UCC, GGU UCG, GGU UGC, GGU UUC, GGUU UU, GUAAAA, GUAAAG, GUAAAU, GUAACC, GUAACG, GUAACU, GUAAGA, GUAAGC, GUAAGG, GUAAGU, GUAAUA, GUAAUC, GUAAUG, GUAAUU,
- GUAGGU GUAGUA, GUAGUC, GUAUAA, GUAUAC, GUAUAG, GUAUAU, GUAUCA, GUAUCG, GUAUCU, GUAUGA, GUAUGC, GUAUGG, GUAUUA, GUAU UG, G UAU UU, GUCAAA, GUCAAG, GUCAAU, GUCACA, GUCACC, GUCACG, GUCAGA, GUCAGC, GUCAGG, GUCAUA, GUCAUC, GUCAUG, GUCCAA, GUCCAC, GUCCAU, GUCCCC, GUCCCU, GUCCGA, GUCCGC, G UCCGG, GUCCGU, GUCCUA, GUCCUG, GUCCU U, GUCGAA, GUCGAC, GUCGAG, GUCGAU, GUCGCA, GUCGCC, GUCGCG, GUCGCU, GUCGGA, GUCGGC, GUCGGG,
- GUGCAU GUGCCC
- GUGCCG GUGCGA
- GUGCGG GUGCGU
- GUGCUA GUGCUC
- GUGCUG GUGCAU
- GUGGAG GUGGCG, GUGGCU, GUGGGU, GUGGUC, GUGGUG, GUGUAA, GUGUAG, GUGUCG, GUGUGA, GUGUGC, GUGUGU, GUGUUG, GUGU UU, GU UAAA, GU UAAC, GUUAAG, GU UACA, GU UACC, GUUACG, GU UACU, GU UAGA, GUUAGC, GUUAGU, GUUAUA, GUUAUC, GUUAUG,
- GU UAUU GUUCAA, GUUCAC, GUUCAG, GU UCCA, GUUCCG, GUUCGA, GU UCGC, GU UCGG, GU UCGU, GUUCUA, GUUCUG, GUUGAA, GU UGAC, GUUGAG, GUUGAU, GUUGCG, GUUGCU, GUUGGA, GU UGGC, GU UGGU, GU UGUC, GUUGUG, GUUGU U, GUU UAA, GU UAC, GU UUAG, GUU UAU, GU UUCA, GUUUCC, GU UUCU, GU UUGA, GUU UGC, GUU UGG, GUU UGU, GU UUUA, GU UU UC, GU UU UU, UAAAAA, UAAAAC, UAAAAG, UAAAAU, UAAACA, UAAACC, UAAACG, UAAACU,
- Oligo RQ RQ SE Gene Expt Type Cell [Oligo] Assay Coordinates_g Name Name Line/Tissue Type
- SMN1 1.9782541 1.3529511 SMN1 in vitro Hep3B 50 qRTPCR SMN1:21170U20 -14 54 56
- SeqID range 30 to 8329, 13088-13094 SeqIDs w/o G Runs: 30-142, 156-560, 575-780, 794-912, 926-1013, 1027-1078, 1092-1286, 1300-1335, 1349- 1385, 1399-1453, 1460-1527, 1548-1555, 1571-1653, 1675-1691, 1706-1802, 1816-1883, 1897-2009, 2023-2141, 2165-2289, 2303-2320, 2334-2447, 2461-2494, 2508-2526, 2540- 2545, 2571-2635, 2651-2670, 2689-2763, 2772-2814, 2828-2854, 2868-3030, 3044-3256, 3270-3360, 3374-3400, 3414-3722, 3737, 3759-3783, 3797-3970, 3986-4059, 4073-4153, 4175-4240, 4255-4415, 4438-4441, 4456-4472, 4484-
- SeqID range 1158-1159, 1171, 1482-1483, 1485-1486, 2465-2471, 2488-2490, 2542-2546, 2656-2657, 2833-2835, 3439-3440, 3916-3918, 4469-4472, 4821, 5429, 5537, 6061, 7327, 8330-13061, 13062-13087
- Example 2 Selective upregulation of exon 7 containing SMN2 transcripts using oligonucleotides targeting PRC2-interacting regions that upregulate SMN2 and splice- switching oligonucleotides.
- Oligonucleotides targeting PRC2-interacting regions (IncR A peaks) in the SMN1/2 gene loci were designed. These oligos were synthesized with various DNA base
- SSO Splice switching oligos
- SMA fibroblast cell lines and one lymphoblast cell line were obtained from the Coriell Institute (FIG. 2).
- the cells were either transfected with the oligos using Lipofectamine 2000 (Fibroblasts) or by electroporation or unassisted delivery (lymphoblast) to ascertain effects of the oligonucleotides on SMNl/2 mRNA and protein expression. All experiments were carried out as biological triplicates.
- transfections were performed using Lipofectamie2000 per manufacturer's instructions with oligos at either ⁇ or 30nM.
- SMN ELISA to determine SMN protein was carried out per manufacturer's instructions (SMN ELISA kit #ADI-900-209, Enzo Life Sciences). Briefly, SMN fibroblasts were cultures at 40,000 cells/well of a 24-well tissue culture coated plate on day 1. Cells were transfected with the oligos using Lipofectamine2000 on day 2 and cell lysates prepared at 24 and 48 hours post-treatment. ELISA was carried out per manufacturer's instructions.
- Splice switching assay (Ddel assay) - SMN2-derived transcripts contain a unique Ddel restriction element introduced because of a nucleotide polymorphism not present in SMNl and are differentiated from SMNl -derived transcripts because of the faster migration of the SMN2 products.
- Ddel assay Splice switching assay
- SMA fibroblasts were treated with oligonucleotides targeting PRC2-interacting regions with or without SSO at 30nM each as described before.
- RT-PCR was carried out with an SMN exon 5 forward primer and an exon 8 reverse primer to generate cDNAs that were then digested with Ddel.
- the SMNl transcript if present, migrates at a slower rate than the Ddel- digested SMN2 transcript and is seen as the first band from the top of the gel.
- the second band from the top indicates full length SMN2 (accurately spliced form) and the third band indicates the incorrectly spliced SMN2delta7. (FIG. 5)
- the SMN1 gene is often mutated in such a way that it is unable to correctly code the SMN protein - due to either a deletion encompassing at least a portion of exon 7 or to other mutations.
- SMA patients generally retain at least one copy of the SMN2 gene (with many having 2 or more copies) that still expresses small amounts of SMN protein.
- the SMN2 gene has a C to T mutation (compared with SMN1) in exon 7 that alters splicing of its precursor mRNA such that exon 7 is spliced out at a high frequency. Consequently, only about 10% of the normal levels of full length SMN protein are produced from SMN2. (See FIG. 1)
- SMA fibroblast cell lines and one lymphoblast cell line were obtained from the Coriell Institute (FIG. 2).
- Cells were transfected with oligonucleotides (oligos 1-52 and 59- 101) directed against a PRC2-associated region of SMN2 and RT-PCR assays were conducted to evaluate effects on SMN mRNA expression. (See FIGS. 3 and 4 for results in cell lines 3814 and 3813).
- oligonucleotides oligos 1-52 and 59- 101
- RT-PCR assays were conducted to evaluate effects on SMN mRNA expression.
- oligonucleotides (oligos 53-58) directed at a splice control sequence in intron 7 of SMN2 and RT-PCR assays were conducted to evaluate effects on SMN mRNA expression (See FIGS. 3 and 4 for results in cell lines 3814 and 3813).
- Splice switching oligonucleotides (oligos 53- 58) were found to increase expression of full length SMN2 based on a gel separation analysis of PCR products obtained following a Ddel restriction digest; whereas certain
- SMN ELISA (Enzo) assays were conducted and revealed that certain oligonucleotides directed against a PRC2-associated region of SMN2 alone did not significantly increase full length SMN protein 24h post-transfection in certain SMA patient fibroblasts.
- FIG. 6 the same SMN ELISA assays showed that oligonucleotides directed against a PRC2-associated region of SMN2 in combination with a splice switching oligonucleotide (oligo 53 or 54) significantly increase full length SMN protein 24h post-transfection in SMA patient fibroblasts above that observed with splice switching oligonucleotides alone.
- RT-PCR assays were conducted and showed that oligonucleotides directed against a PRC2-associated region of SMN2 in combination with a splice switching oligonucleotide (oligo 53 or 54) significantly increased SMN2 protein 24h post-transfection in SMA patient fibroblasts.
- oligo 53 or 54 a splice switching oligonucleotide
- FIG. 9. These experiments were conducted modified oligonucleotides with either alternating LNA and 2'OMe nucleotides or alternating DNA and LNA nucleotides.
- Example 2 show that certain oligos targeting PRC2 associated regions of SMN2 induce SMN RNA expression (e.g.., of the SMNA7 transcript) SMA patient-derived fibroblasts.
- SMN RNA expression e.g.., of the SMNA7 transcript
- results show that, in some embodiments, splice- switching oligos may not induce SMN RNA expression, but rather shift SMN RNA splicing to the full-length transcript.
- combination of splice switching oligos with PRC2-associated region targeting oligos substantially increases full length SMN protein in cells from SMA patients.
- Table 4 Oligonucleotide sequences made for testing human cells obtained from subjects with Spinal Muscular Atrophy (See Table 3 for structural features of formatted sequence).
Abstract
Description
Claims
Priority Applications (14)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| DK13790819.0T DK2850186T3 (en) | 2012-05-16 | 2013-05-16 | COMPOSITIONS AND PROCEDURES FOR MODULATING SMN GENFAMILY EXPRESSION |
| AU2013262649A AU2013262649A1 (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating smn gene family expression |
| KR1020147035185A KR20150030205A (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating smn gene family expression |
| JP2015512858A JP2015523854A (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating SMN gene family expression |
| EP13790819.0A EP2850186B1 (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating smn gene family expression |
| SG11201407483YA SG11201407483YA (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating smn gene family expression |
| EA201492123A EA201492123A1 (en) | 2012-05-16 | 2013-05-16 | COMPOSITIONS AND METHODS FOR MODULATING THE EXPRESSION OF THE SMN GENES FAMILY |
| US14/401,194 US10059941B2 (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating SMN gene family expression |
| CN201380037632.9A CN104540947A (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating SMN gene family expression |
| CA2873794A CA2873794A1 (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating smn gene family expression |
| IL235676A IL235676A0 (en) | 2012-05-16 | 2014-11-13 | Compositions and methods for modulating smn gene family expression |
| ZA2014/09228A ZA201409228B (en) | 2012-05-16 | 2014-12-15 | Compositions and methods for modulating smn gene family expression |
| HK15109139.6A HK1208700A1 (en) | 2012-05-16 | 2015-09-17 | Compositions and methods for modulating smn gene family expression smn |
| US15/872,684 US10837014B2 (en) | 2012-05-16 | 2018-01-16 | Compositions and methods for modulating SMN gene family expression |
Applications Claiming Priority (6)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US201261647858P | 2012-05-16 | 2012-05-16 | |
| US61/647,858 | 2012-05-16 | ||
| US201261719394P | 2012-10-27 | 2012-10-27 | |
| US61/719,394 | 2012-10-27 | ||
| US201361785529P | 2013-03-14 | 2013-03-14 | |
| US61/785,529 | 2013-03-14 |
Related Parent Applications (2)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/US2013/041452 Continuation-In-Part WO2013173645A1 (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating utrn expression |
| US14/401,196 Continuation-In-Part US10058623B2 (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating UTRN expression |
Related Child Applications (2)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| US14/401,194 A-371-Of-International US10059941B2 (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating SMN gene family expression |
| US15/872,684 Continuation-In-Part US10837014B2 (en) | 2012-05-16 | 2018-01-16 | Compositions and methods for modulating SMN gene family expression |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2013173638A1 true WO2013173638A1 (en) | 2013-11-21 |
Family
ID=49584305
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/US2013/041440 WO2013173638A1 (en) | 2012-05-16 | 2013-05-16 | Compositions and methods for modulating smn gene family expression |
Country Status (13)
| Country | Link |
|---|---|
| US (1) | US10059941B2 (en) |
| EP (1) | EP2850186B1 (en) |
| JP (1) | JP2015523854A (en) |
| KR (1) | KR20150030205A (en) |
| CN (1) | CN104540947A (en) |
| AU (1) | AU2013262649A1 (en) |
| CA (1) | CA2873794A1 (en) |
| DK (1) | DK2850186T3 (en) |
| EA (1) | EA201492123A1 (en) |
| HK (1) | HK1208700A1 (en) |
| SG (1) | SG11201407483YA (en) |
| WO (1) | WO2013173638A1 (en) |
| ZA (1) | ZA201409228B (en) |
Cited By (21)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN103911346A (en) * | 2014-03-27 | 2014-07-09 | 江苏雄鸣医药科技有限公司 | Method of knocking out spinal muscular atrophy SMN genes and cell model |
| US9328346B2 (en) | 2010-11-12 | 2016-05-03 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| WO2017040271A1 (en) * | 2015-08-28 | 2017-03-09 | Sarepta Therapeutics, Inc. | Modified antisense oligomers for exon inclusion in spinal muscular atrophy |
| US9920317B2 (en) | 2010-11-12 | 2018-03-20 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| US10059941B2 (en) | 2012-05-16 | 2018-08-28 | Translate Bio Ma, Inc. | Compositions and methods for modulating SMN gene family expression |
| US10058623B2 (en) | 2012-05-16 | 2018-08-28 | Translate Bio Ma, Inc. | Compositions and methods for modulating UTRN expression |
| US10174328B2 (en) | 2013-10-04 | 2019-01-08 | Translate Bio Ma, Inc. | Compositions and methods for treating amyotrophic lateral sclerosis |
| US10174323B2 (en) | 2012-05-16 | 2019-01-08 | The General Hospital Corporation | Compositions and methods for modulating ATP2A2 expression |
| US10174315B2 (en) | 2012-05-16 | 2019-01-08 | The General Hospital Corporation | Compositions and methods for modulating hemoglobin gene family expression |
| WO2019048645A1 (en) | 2017-09-08 | 2019-03-14 | Mina Therapeutics Limited | Stabilized cebpa sarna compositions and methods of use |
| WO2019048632A1 (en) | 2017-09-08 | 2019-03-14 | Mina Therapeutics Limited | Stabilized hnf4a sarna compositions and methods of use |
| US10357543B2 (en) | 2015-11-16 | 2019-07-23 | Ohio State Innovation Foundation | Methods and compositions for treating disorders and diseases using Survival Motor Neuron (SMN) protein |
| US10655128B2 (en) | 2012-05-16 | 2020-05-19 | Translate Bio Ma, Inc. | Compositions and methods for modulating MECP2 expression |
| EP3471779A4 (en) * | 2016-06-16 | 2020-07-08 | Ionis Pharmaceuticals, Inc. | Combinations for the modulation of smn expression |
| US10837014B2 (en) | 2012-05-16 | 2020-11-17 | Translate Bio Ma, Inc. | Compositions and methods for modulating SMN gene family expression |
| US10851371B2 (en) | 2015-04-10 | 2020-12-01 | Ionis Pharmaceuticals, Inc. | Modulation of SMN expression |
| US10858650B2 (en) | 2014-10-30 | 2020-12-08 | The General Hospital Corporation | Methods for modulating ATRX-dependent gene repression |
| US10900036B2 (en) | 2015-03-17 | 2021-01-26 | The General Hospital Corporation | RNA interactome of polycomb repressive complex 1 (PRC1) |
| WO2021032777A1 (en) | 2019-08-19 | 2021-02-25 | Mina Therapeutics Limited | Oligonucleotide conjugate compositions and methods of use |
| US11299737B1 (en) | 2020-02-28 | 2022-04-12 | Ionis Pharmaceuticals, Inc. | Compounds and methods for modulating SMN2 |
| EP4035659A1 (en) | 2016-11-29 | 2022-08-03 | PureTech LYT, Inc. | Exosomes for delivery of therapeutic agents |
Families Citing this family (15)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| EP2983676B1 (en) * | 2013-04-12 | 2019-03-20 | The Curators of the University of Missouri | Smn2 element 1 antisense compositions and methods and uses thereof |
| US11249071B2 (en) | 2015-04-24 | 2022-02-15 | California Institute Of Technology | Reactivation of x chromosome genes |
| US20180312839A1 (en) * | 2015-10-26 | 2018-11-01 | Translate Bio Ma, Inc. | Methods and compositions for increasing smn expression |
| WO2018081661A1 (en) | 2016-10-27 | 2018-05-03 | California Institute Of Technology | Hdac inhibitor compositions for reactivation of the x chromosome |
| BR112019008810A2 (en) | 2016-11-01 | 2019-07-16 | Univ New York State Res Found | 5-halouracil modified microplates and their use in cancer treatment |
| EP3599844A4 (en) * | 2017-06-07 | 2021-01-13 | University of Massachusetts | Anti-adam33 oligonucleotides and related methods |
| WO2019075357A1 (en) * | 2017-10-12 | 2019-04-18 | Wave Life Sciences Ltd. | Oligonucleotide compositions and methods thereof |
| AU2019350786A1 (en) | 2018-09-26 | 2021-05-13 | Greenlight Biosciences, Inc. | Control of Coleopteran insects |
| CA3118148A1 (en) | 2018-11-08 | 2020-05-14 | Greenlight Biosciences, Inc. | Control of insect infestation |
| EP3908671A4 (en) * | 2019-01-09 | 2022-10-05 | Coyote Bioscience USA Inc. | Methods and systems for identification of spinal muscular atrophy |
| JP2022544538A (en) * | 2019-08-15 | 2022-10-19 | バイオジェン・エムエイ・インコーポレイテッド | Combination therapy for spinal muscular atrophy |
| CN113444722A (en) * | 2020-03-24 | 2021-09-28 | 中国科学院脑科学与智能技术卓越创新中心 | Application of single base editing mediated splicing repair in preparation of drugs for treating spinal muscular atrophy |
| TW202302854A (en) * | 2021-02-19 | 2023-01-16 | 美商佛羅里達大學研究基金會公司 | Methods and compositions to confer regulation to gene therapy cargoes by heterologous use of alternative splicing cassettes |
| WO2023020421A1 (en) * | 2021-08-16 | 2023-02-23 | Hangzhou Jiayin Biotech Ltd. | Regulation of imprinting silenced genes by smn1 and snrpn expression and uses thereof |
| CA3230274A1 (en) * | 2021-09-03 | 2023-03-09 | Alfica Sehgal | Modulation of gene transcription using antisense oligonucleotides targeting regulatory rnas |
Citations (134)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US3687808A (en) | 1969-08-14 | 1972-08-29 | Univ Leland Stanford Junior | Synthetic polynucleotides |
| US4469863A (en) | 1980-11-12 | 1984-09-04 | Ts O Paul O P | Nonionic nucleic acid alkyl and aryl phosphonates and processes for manufacture and use thereof |
| US4476301A (en) | 1982-04-29 | 1984-10-09 | Centre National De La Recherche Scientifique | Oligonucleotides, a process for preparing the same and their application as mediators of the action of interferon |
| US4587044A (en) | 1983-09-01 | 1986-05-06 | The Johns Hopkins University | Linkage of proteins to nucleic acids |
| US4605735A (en) | 1983-02-14 | 1986-08-12 | Wakunaga Seiyaku Kabushiki Kaisha | Oligonucleotide derivatives |
| US4667025A (en) | 1982-08-09 | 1987-05-19 | Wakunaga Seiyaku Kabushiki Kaisha | Oligonucleotide derivatives |
| US4762779A (en) | 1985-06-13 | 1988-08-09 | Amgen Inc. | Compositions and methods for functionalizing nucleic acids |
| US4824941A (en) | 1983-03-10 | 1989-04-25 | Julian Gordon | Specific antibody to the native form of 2'5'-oligonucleotides, the method of preparation and the use as reagents in immunoassays or for binding 2'5'-oligonucleotides in biological systems |
| US4828979A (en) | 1984-11-08 | 1989-05-09 | Life Technologies, Inc. | Nucleotide analogs for nucleic acid labeling and detection |
| US4835263A (en) | 1983-01-27 | 1989-05-30 | Centre National De La Recherche Scientifique | Novel compounds containing an oligonucleotide sequence bonded to an intercalating agent, a process for their synthesis and their use |
| US4845205A (en) | 1985-01-08 | 1989-07-04 | Institut Pasteur | 2,N6 -disubstituted and 2,N6 -trisubstituted adenosine-3'-phosphoramidites |
| US4876335A (en) | 1986-06-30 | 1989-10-24 | Wakunaga Seiyaku Kabushiki Kaisha | Poly-labelled oligonucleotide derivative |
| US4904582A (en) | 1987-06-11 | 1990-02-27 | Synthetic Genetics | Novel amphiphilic nucleic acid conjugates |
| US4948882A (en) | 1983-02-22 | 1990-08-14 | Syngene, Inc. | Single-stranded labelled oligonucleotides, reactive monomers and methods of synthesis |
| US4958013A (en) | 1989-06-06 | 1990-09-18 | Northwestern University | Cholesteryl modified oligonucleotides |
| US5013830A (en) | 1986-09-08 | 1991-05-07 | Ajinomoto Co., Inc. | Compounds for the cleavage at a specific position of RNA, oligomers employed for the formation of said compounds, and starting materials for the synthesis of said oligomers |
| US5023243A (en) | 1981-10-23 | 1991-06-11 | Molecular Biosystems, Inc. | Oligonucleotide therapeutic agent and method of making same |
| US5034506A (en) | 1985-03-15 | 1991-07-23 | Anti-Gene Development Group | Uncharged morpholino-based polymers having achiral intersubunit linkages |
| US5082830A (en) | 1988-02-26 | 1992-01-21 | Enzo Biochem, Inc. | End labeled nucleotide probe |
| US5109124A (en) | 1988-06-01 | 1992-04-28 | Biogen, Inc. | Nucleic acid probe linked to a label having a terminal cysteine |
| US5112963A (en) | 1987-11-12 | 1992-05-12 | Max-Planck-Gesellschaft Zur Foerderung Der Wissenschaften E.V. | Modified oligonucleotides |
| US5118802A (en) | 1983-12-20 | 1992-06-02 | California Institute Of Technology | DNA-reporter conjugates linked via the 2' or 5'-primary amino group of the 5'-terminal nucleoside |
| US5130302A (en) | 1989-12-20 | 1992-07-14 | Boron Bilogicals, Inc. | Boronated nucleoside, nucleotide and oligonucleotide compounds, compositions and methods for using same |
| US5134066A (en) | 1989-08-29 | 1992-07-28 | Monsanto Company | Improved probes using nucleosides containing 3-dezauracil analogs |
| US5138045A (en) | 1990-07-27 | 1992-08-11 | Isis Pharmaceuticals | Polyamine conjugated oligonucleotides |
| US5149797A (en) | 1990-02-15 | 1992-09-22 | The Worcester Foundation For Experimental Biology | Method of site-specific alteration of rna and production of encoded polypeptides |
| US5166315A (en) | 1989-12-20 | 1992-11-24 | Anti-Gene Development Group | Sequence-specific binding polymers for duplex nucleic acids |
| US5175273A (en) | 1988-07-01 | 1992-12-29 | Genentech, Inc. | Nucleic acid intercalating agents |
| US5177196A (en) | 1990-08-16 | 1993-01-05 | Microprobe Corporation | Oligo (α-arabinofuranosyl nucleotides) and α-arabinofuranosyl precursors thereof |
| US5185444A (en) | 1985-03-15 | 1993-02-09 | Anti-Gene Deveopment Group | Uncharged morpolino-based polymers having phosphorous containing chiral intersubunit linkages |
| US5188897A (en) | 1987-10-22 | 1993-02-23 | Temple University Of The Commonwealth System Of Higher Education | Encapsulated 2',5'-phosphorothioate oligoadenylates |
| US5214136A (en) | 1990-02-20 | 1993-05-25 | Gilead Sciences, Inc. | Anthraquinone-derivatives oligonucleotides |
| US5214134A (en) | 1990-09-12 | 1993-05-25 | Sterling Winthrop Inc. | Process of linking nucleosides with a siloxane bridge |
| US5216141A (en) | 1988-06-06 | 1993-06-01 | Benner Steven A | Oligonucleotide analogs containing sulfur linkages |
| US5218105A (en) | 1990-07-27 | 1993-06-08 | Isis Pharmaceuticals | Polyamine conjugated oligonucleotides |
| US5220007A (en) | 1990-02-15 | 1993-06-15 | The Worcester Foundation For Experimental Biology | Method of site-specific alteration of RNA and production of encoded polypeptides |
| US5235033A (en) | 1985-03-15 | 1993-08-10 | Anti-Gene Development Group | Alpha-morpholino ribonucleoside derivatives and polymers thereof |
| US5245022A (en) | 1990-08-03 | 1993-09-14 | Sterling Drug, Inc. | Exonuclease resistant terminally substituted oligonucleotides |
| US5254469A (en) | 1989-09-12 | 1993-10-19 | Eastman Kodak Company | Oligonucleotide-enzyme conjugate that can be used as a probe in hybridization assays and polymerase chain reaction procedures |
| US5256775A (en) | 1989-06-05 | 1993-10-26 | Gilead Sciences, Inc. | Exonuclease-resistant oligonucleotides |
| US5258506A (en) | 1984-10-16 | 1993-11-02 | Chiron Corporation | Photolabile reagents for incorporation into oligonucleotide chains |
| US5262536A (en) | 1988-09-15 | 1993-11-16 | E. I. Du Pont De Nemours And Company | Reagents for the preparation of 5'-tagged oligonucleotides |
| US5264564A (en) | 1989-10-24 | 1993-11-23 | Gilead Sciences | Oligonucleotide analogs with novel linkages |
| US5264562A (en) | 1989-10-24 | 1993-11-23 | Gilead Sciences, Inc. | Oligonucleotide analogs with novel linkages |
| US5264423A (en) | 1987-03-25 | 1993-11-23 | The United States Of America As Represented By The Department Of Health And Human Services | Inhibitors for replication of retroviruses and for the expression of oncogene products |
| US5272250A (en) | 1992-07-10 | 1993-12-21 | Spielvogel Bernard F | Boronated phosphoramidate compounds |
| US5276019A (en) | 1987-03-25 | 1994-01-04 | The United States Of America As Represented By The Department Of Health And Human Services | Inhibitors for replication of retroviruses and for the expression of oncogene products |
| US5278302A (en) | 1988-05-26 | 1994-01-11 | University Patents, Inc. | Polynucleotide phosphorodithioates |
| US5292873A (en) | 1989-11-29 | 1994-03-08 | The Research Foundation Of State University Of New York | Nucleic acids labeled with naphthoquinone probe |
| US5317098A (en) | 1986-03-17 | 1994-05-31 | Hiroaki Shizuya | Non-radioisotope tagging of fragments |
| US5321131A (en) | 1990-03-08 | 1994-06-14 | Hybridon, Inc. | Site-specific functionalization of oligodeoxynucleotides for non-radioactive labelling |
| US5367066A (en) | 1984-10-16 | 1994-11-22 | Chiron Corporation | Oligonucleotides with selectably cleavable and/or abasic sites |
| US5371241A (en) | 1991-07-19 | 1994-12-06 | Pharmacia P-L Biochemicals Inc. | Fluorescein labelled phosphoramidites |
| US5391723A (en) | 1989-05-31 | 1995-02-21 | Neorx Corporation | Oligonucleotide conjugates |
| US5399676A (en) | 1989-10-23 | 1995-03-21 | Gilead Sciences | Oligonucleotides with inverted polarity |
| US5403711A (en) | 1987-11-30 | 1995-04-04 | University Of Iowa Research Foundation | Nucleic acid hybridization and amplification method for detection of specific sequences in which a complementary labeled nucleic acid probe is cleaved |
| US5405939A (en) | 1987-10-22 | 1995-04-11 | Temple University Of The Commonwealth System Of Higher Education | 2',5'-phosphorothioate oligoadenylates and their covalent conjugates with polylysine |
| US5405938A (en) | 1989-12-20 | 1995-04-11 | Anti-Gene Development Group | Sequence-specific binding polymers for duplex nucleic acids |
| US5414077A (en) | 1990-02-20 | 1995-05-09 | Gilead Sciences | Non-nucleoside linkers for convenient attachment of labels to oligonucleotides using standard synthetic methods |
| US5432272A (en) | 1990-10-09 | 1995-07-11 | Benner; Steven A. | Method for incorporating into a DNA or RNA oligonucleotide using nucleotides bearing heterocyclic bases |
| US5434257A (en) | 1992-06-01 | 1995-07-18 | Gilead Sciences, Inc. | Binding compentent oligomers containing unsaturated 3',5' and 2',5' linkages |
| US5451463A (en) | 1989-08-28 | 1995-09-19 | Clontech Laboratories, Inc. | Non-nucleoside 1,3-diol reagents for labeling synthetic oligonucleotides |
| US5455233A (en) | 1989-11-30 | 1995-10-03 | University Of North Carolina | Oligoribonucleoside and oligodeoxyribonucleoside boranophosphates |
| US5457187A (en) | 1993-12-08 | 1995-10-10 | Board Of Regents University Of Nebraska | Oligonucleotides containing 5-fluorouracil |
| US5459255A (en) | 1990-01-11 | 1995-10-17 | Isis Pharmaceuticals, Inc. | N-2 substituted purines |
| US5466677A (en) | 1993-03-06 | 1995-11-14 | Ciba-Geigy Corporation | Dinucleoside phosphinates and their pharmaceutical compositions |
| US5470967A (en) | 1990-04-10 | 1995-11-28 | The Dupont Merck Pharmaceutical Company | Oligonucleotide analogs with sulfamate linkages |
| US5476925A (en) | 1993-02-01 | 1995-12-19 | Northwestern University | Oligodeoxyribonucleotides including 3'-aminonucleoside-phosphoramidate linkages and terminal 3'-amino groups |
| US5484908A (en) | 1991-11-26 | 1996-01-16 | Gilead Sciences, Inc. | Oligonucleotides containing 5-propynyl pyrimidines |
| US5486603A (en) | 1990-01-08 | 1996-01-23 | Gilead Sciences, Inc. | Oligonucleotide having enhanced binding affinity |
| US5489677A (en) | 1990-07-27 | 1996-02-06 | Isis Pharmaceuticals, Inc. | Oligonucleoside linkages containing adjacent oxygen and nitrogen atoms |
| US5491133A (en) | 1987-11-30 | 1996-02-13 | University Of Iowa Research Foundation | Methods for blocking the expression of specifically targeted genes |
| US5502177A (en) | 1993-09-17 | 1996-03-26 | Gilead Sciences, Inc. | Pyrimidine derivatives for labeled binding partners |
| US5510475A (en) | 1990-11-08 | 1996-04-23 | Hybridon, Inc. | Oligonucleotide multiple reporter precursors |
| US5512439A (en) | 1988-11-21 | 1996-04-30 | Dynal As | Oligonucleotide-linked magnetic particles and uses thereof |
| US5512667A (en) | 1990-08-28 | 1996-04-30 | Reed; Michael W. | Trifunctional intermediates for preparing 3'-tailed oligonucleotides |
| US5514785A (en) | 1990-05-11 | 1996-05-07 | Becton Dickinson And Company | Solid supports for nucleic acid hybridization assays |
| US5519126A (en) | 1988-03-25 | 1996-05-21 | University Of Virginia Alumni Patents Foundation | Oligonucleotide N-alkylphosphoramidates |
| US5525711A (en) | 1994-05-18 | 1996-06-11 | The United States Of America As Represented By The Secretary Of The Department Of Health And Human Services | Pteridine nucleotide analogs as fluorescent DNA probes |
| US5525465A (en) | 1987-10-28 | 1996-06-11 | Howard Florey Institute Of Experimental Physiology And Medicine | Oligonucleotide-polyamide conjugates and methods of production and applications of the same |
| US5539082A (en) | 1993-04-26 | 1996-07-23 | Nielsen; Peter E. | Peptide nucleic acids |
| US5541307A (en) | 1990-07-27 | 1996-07-30 | Isis Pharmaceuticals, Inc. | Backbone modified oligonucleotide analogs and solid phase synthesis thereof |
| US5545730A (en) | 1984-10-16 | 1996-08-13 | Chiron Corporation | Multifunctional nucleic acid monomer |
| US5550111A (en) | 1984-07-11 | 1996-08-27 | Temple University-Of The Commonwealth System Of Higher Education | Dual action 2',5'-oligoadenylate antiviral derivatives and uses thereof |
| US5552540A (en) | 1987-06-24 | 1996-09-03 | Howard Florey Institute Of Experimental Physiology And Medicine | Nucleoside derivatives |
| US5561225A (en) | 1990-09-19 | 1996-10-01 | Southern Research Institute | Polynucleotide analogs containing sulfonate and sulfonamide internucleoside linkages |
| US5565350A (en) | 1993-12-09 | 1996-10-15 | Thomas Jefferson University | Compounds and methods for site directed mutations in eukaryotic cells |
| US5565552A (en) | 1992-01-21 | 1996-10-15 | Pharmacyclics, Inc. | Method of expanded porphyrin-oligonucleotide conjugate synthesis |
| US5571799A (en) | 1991-08-12 | 1996-11-05 | Basco, Ltd. | (2'-5') oligoadenylate analogues useful as inhibitors of host-v5.-graft response |
| US5574142A (en) | 1992-12-15 | 1996-11-12 | Microprobe Corporation | Peptide linkers for improved oligonucleotide delivery |
| US5578718A (en) | 1990-01-11 | 1996-11-26 | Isis Pharmaceuticals, Inc. | Thiol-derivatized nucleosides |
| US5580731A (en) | 1994-08-25 | 1996-12-03 | Chiron Corporation | N-4 modified pyrimidine deoxynucleotides and oligonucleotide probes synthesized therewith |
| US5585481A (en) | 1987-09-21 | 1996-12-17 | Gen-Probe Incorporated | Linking reagents for nucleotide probes |
| US5587371A (en) | 1992-01-21 | 1996-12-24 | Pharmacyclics, Inc. | Texaphyrin-oligonucleotide conjugates |
| US5587361A (en) | 1991-10-15 | 1996-12-24 | Isis Pharmaceuticals, Inc. | Oligonucleotides having phosphorothioate linkages of high chiral purity |
| US5596086A (en) | 1990-09-20 | 1997-01-21 | Gilead Sciences, Inc. | Modified internucleoside linkages having one nitrogen and two carbon atoms |
| US5595726A (en) | 1992-01-21 | 1997-01-21 | Pharmacyclics, Inc. | Chromophore probe for detection of nucleic acid |
| US5596091A (en) | 1994-03-18 | 1997-01-21 | The Regents Of The University Of California | Antisense oligonucleotides comprising 5-aminoalkyl pyrimidine nucleotides |
| US5597696A (en) | 1994-07-18 | 1997-01-28 | Becton Dickinson And Company | Covalent cyanine dye oligonucleotide conjugates |
| US5599923A (en) | 1989-03-06 | 1997-02-04 | Board Of Regents, University Of Tx | Texaphyrin metal complexes having improved functionalization |
| US5602240A (en) | 1990-07-27 | 1997-02-11 | Ciba Geigy Ag. | Backbone modified oligonucleotide analogs |
| US5608046A (en) | 1990-07-27 | 1997-03-04 | Isis Pharmaceuticals, Inc. | Conjugated 4'-desmethyl nucleoside analog compounds |
| US5610289A (en) | 1990-07-27 | 1997-03-11 | Isis Pharmaceuticals, Inc. | Backbone modified oligonucleotide analogues |
| US5614617A (en) | 1990-07-27 | 1997-03-25 | Isis Pharmaceuticals, Inc. | Nuclease resistant, pyrimidine modified oligonucleotides that detect and modulate gene expression |
| US5618704A (en) | 1990-07-27 | 1997-04-08 | Isis Pharmacueticals, Inc. | Backbone-modified oligonucleotide analogs and preparation thereof through radical coupling |
| US5623070A (en) | 1990-07-27 | 1997-04-22 | Isis Pharmaceuticals, Inc. | Heteroatomic oligonucleoside linkages |
| US5623065A (en) | 1990-08-13 | 1997-04-22 | Isis Pharmaceuticals, Inc. | Gapped 2' modified oligonucleotides |
| US5625050A (en) | 1994-03-31 | 1997-04-29 | Amgen Inc. | Modified oligonucleotides and intermediates useful in nucleic acid therapeutics |
| US5633360A (en) | 1992-04-14 | 1997-05-27 | Gilead Sciences, Inc. | Oligonucleotide analogs capable of passive cell membrane permeation |
| US5652355A (en) | 1992-07-23 | 1997-07-29 | Worcester Foundation For Experimental Biology | Hybrid oligonucleotide phosphorothioates |
| US5652356A (en) | 1995-08-17 | 1997-07-29 | Hybridon, Inc. | Inverted chimeric and hybrid oligonucleotides |
| US5663312A (en) | 1993-03-31 | 1997-09-02 | Sanofi | Oligonucleotide dimers with amide linkages replacing phosphodiester linkages |
| US5677439A (en) | 1990-08-03 | 1997-10-14 | Sanofi | Oligonucleotide analogues containing phosphate diester linkage substitutes, compositions thereof, and precursor dinucleotide analogues |
| US5677437A (en) | 1990-07-27 | 1997-10-14 | Isis Pharmaceuticals, Inc. | Heteroatomic oligonucleoside linkages |
| US5681941A (en) | 1990-01-11 | 1997-10-28 | Isis Pharmaceuticals, Inc. | Substituted purines and oligonucleotide cross-linking |
| US5688941A (en) | 1990-07-27 | 1997-11-18 | Isis Pharmaceuticals, Inc. | Methods of making conjugated 4' desmethyl nucleoside analog compounds |
| US5700922A (en) | 1991-12-24 | 1997-12-23 | Isis Pharmaceuticals, Inc. | PNA-DNA-PNA chimeric macromolecules |
| US5714331A (en) | 1991-05-24 | 1998-02-03 | Buchardt, Deceased; Ole | Peptide nucleic acids having enhanced binding affinity, sequence specificity and solubility |
| US5719262A (en) | 1993-11-22 | 1998-02-17 | Buchardt, Deceased; Ole | Peptide nucleic acids having amino acid side chains |
| US5750692A (en) | 1990-01-11 | 1998-05-12 | Isis Pharmaceuticals, Inc. | Synthesis of 3-deazapurines |
| WO1999067378A1 (en) | 1998-06-19 | 1999-12-29 | Mcgill University | ANTISENSE OLIGONUCLEOTIDE CONSTRUCTS BASED ON β-ARABINOFURANOSE AND ITS ANALOGUES |
| EP0999270A1 (en) * | 1998-10-09 | 2000-05-10 | Institut National De La Sante Et De La Recherche Medicale (Inserm) | Method of regulating SMN gene expression |
| US6287860B1 (en) | 2000-01-20 | 2001-09-11 | Isis Pharmaceuticals, Inc. | Antisense inhibition of MEKK2 expression |
| US20040198687A1 (en) | 2003-04-04 | 2004-10-07 | Rozema David B. | Endosomolytic polymers |
| WO2005042777A2 (en) | 2003-10-24 | 2005-05-12 | Expresson Biosystems Limited | App/ena antisense |
| US20070292408A1 (en) | 2004-12-03 | 2007-12-20 | University Of Massachusetts | Spinal Muscular Atrophy (SMA) treatment via targeting of SMN2 splice site inhibitory sequences |
| US7314923B2 (en) | 1999-02-12 | 2008-01-01 | Daiichi Sankyo Company, Limited | Nucleoside and oligonucleotide analogues |
| WO2008022309A2 (en) | 2006-08-18 | 2008-02-21 | F. Hoffmann-La Roche Ag | Polyconjugates for in vivo delivery of polynucleotides |
| US7399845B2 (en) | 2006-01-27 | 2008-07-15 | Isis Pharmaceuticals, Inc. | 6-modified bicyclic nucleic acid analogs |
| US20080281041A1 (en) | 1999-06-07 | 2008-11-13 | Rozema David B | Reversibly Masked Polymers |
| US7569686B1 (en) | 2006-01-27 | 2009-08-04 | Isis Pharmaceuticals, Inc. | Compounds and methods for synthesis of bicyclic nucleic acid analogs |
| WO2011032109A1 (en) * | 2009-09-11 | 2011-03-17 | Sma Foundation | Biomarkers for spinal muscular atrophy |
| US20110086833A1 (en) * | 2008-05-27 | 2011-04-14 | Paushkin Sergey V | Methods for treating spinal muscular atrophy |
| US9209196B2 (en) | 2011-11-30 | 2015-12-08 | Sharp Kabushiki Kaisha | Memory circuit, method of driving the same, nonvolatile storage device using the same, and liquid crystal display device |
Family Cites Families (266)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO1983001451A1 (en) | 1981-10-23 | 1983-04-28 | Molecular Biosystems Inc | Oligonucleotide therapeutic agent and methods of making same |
| DE3329892A1 (en) | 1983-08-18 | 1985-03-07 | Köster, Hubert, Prof. Dr., 2000 Hamburg | METHOD FOR PRODUCING OLIGONUCLEOTIDES |
| US6753423B1 (en) | 1990-01-11 | 2004-06-22 | Isis Pharmaceuticals, Inc. | Compositions and methods for enhanced biostability and altered biodistribution of oligonucleotides in mammals |
| US5914396A (en) | 1990-01-11 | 1999-06-22 | Isis Pharmaceuticals, Inc. | 2'-O-modified nucleosides and phosphoramidites |
| US6395492B1 (en) | 1990-01-11 | 2002-05-28 | Isis Pharmaceuticals, Inc. | Derivatized oligonucleotides having improved uptake and other properties |
| US6005087A (en) | 1995-06-06 | 1999-12-21 | Isis Pharmaceuticals, Inc. | 2'-modified oligonucleotides |
| WO1992000386A1 (en) | 1990-06-27 | 1992-01-09 | The Trustees Of Columbia University In The City Of New York | Methods for detecting spinal muscular atrophy type i and types ii/iii and marker therefor |
| JP3514779B2 (en) | 1991-04-17 | 2004-03-31 | 株式会社アズウェル | Method for testing apolipoprotein E genotype and primers and probes suitable for the test |
| US7015315B1 (en) | 1991-12-24 | 2006-03-21 | Isis Pharmaceuticals, Inc. | Gapped oligonucleotides |
| WO1994008003A1 (en) | 1991-06-14 | 1994-04-14 | Isis Pharmaceuticals, Inc. | ANTISENSE OLIGONUCLEOTIDE INHIBITION OF THE ras GENE |
| US5965722A (en) | 1991-05-21 | 1999-10-12 | Isis Pharmaceuticals, Inc. | Antisense inhibition of ras gene with chimeric and alternating oligonucleotides |
| US5661134A (en) | 1991-10-15 | 1997-08-26 | Isis Pharmaceuticals, Inc. | Oligonucleotides for modulating Ha-ras or Ki-ras having phosphorothioate linkages of high chiral purity |
| US6831166B2 (en) | 1992-10-23 | 2004-12-14 | Isis Pharmaceuticals, Inc. | Derivatized oligonucleotides having improved uptake and other properties |
| US8153602B1 (en) | 1991-11-19 | 2012-04-10 | Isis Pharmaceuticals, Inc. | Composition and methods for the pulmonary delivery of nucleic acids |
| US20070032446A1 (en) | 1991-12-24 | 2007-02-08 | Isis Pharmaceuticals, Inc. | Gapped 2' modified oligonucleotides |
| CA2126691C (en) | 1991-12-24 | 2003-05-06 | Phillip Dan Cook | Gapped 2' modified oligonucleotides |
| TW244371B (en) | 1992-07-23 | 1995-04-01 | Tri Clover Inc | |
| US6346614B1 (en) | 1992-07-23 | 2002-02-12 | Hybridon, Inc. | Hybrid oligonucleotide phosphorothioates |
| RU95104940A (en) | 1992-07-27 | 1997-01-10 | Хайбрайдон | Method of incorporation of alkylphosphonothioate or arylphosphonothioate internucleotide linkage in oligonucleotide, method of oligonucleotide synthesis, method of gene expression inhibition, treatment method |
| KR960701077A (en) | 1993-01-25 | 1996-02-24 | 다알렌 반스톤 | Oligonucleotide Alkylphosphonate and Alkylphosphonothioate (ALIGLPHOSPHONATES AND ALKYLPHOSPHONOTHIOATES) |
| PT698092E (en) | 1993-05-11 | 2007-10-29 | Univ North Carolina | Antisense oligonucleotides which combat aberrant splicing and methods of using the same |
| FR2720757B1 (en) | 1994-06-03 | 1996-08-02 | Inst Nat Sante Rech Med | Method and probes for the detection of markers linked to the locus of infantile spinal muscular atrophies. |
| EP0708178A1 (en) | 1994-10-19 | 1996-04-24 | Institut National De La Sante Et De La Recherche Medicale (Inserm) | Survival motor neuron (SMN) gene: a gene for spinal muscular atrophy |
| US6608035B1 (en) | 1994-10-25 | 2003-08-19 | Hybridon, Inc. | Method of down-regulating gene expression |
| JP4293636B2 (en) | 1996-02-14 | 2009-07-08 | アイシス・ファーマシューティカルス・インコーポレーテッド | Oligonucleotide with sugar-modified gap |
| US6015710A (en) | 1996-04-09 | 2000-01-18 | The University Of Texas System | Modulation of mammalian telomerase by peptide nucleic acids |
| US5898031A (en) | 1996-06-06 | 1999-04-27 | Isis Pharmaceuticals, Inc. | Oligoribonucleotides for cleaving RNA |
| US5912332A (en) | 1996-07-26 | 1999-06-15 | Hybridon, Inc. | Affinity-based purification of oligonucleotides using soluble multimeric oligonucleotides |
| CA2294900A1 (en) | 1997-07-02 | 1999-01-14 | Sdg, Inc. | Targeted liposomal constructs for diagnostic and therapeutic uses |
| JPH1133863A (en) | 1997-07-23 | 1999-02-09 | Howa Mach Ltd | Tool replacing device and extruding device for replacing tool |
| US7572582B2 (en) | 1997-09-12 | 2009-08-11 | Exiqon A/S | Oligonucleotide analogues |
| US6794499B2 (en) * | 1997-09-12 | 2004-09-21 | Exiqon A/S | Oligonucleotide analogues |
| US7715989B2 (en) | 1998-04-03 | 2010-05-11 | Elitech Holding B.V. | Systems and methods for predicting oligonucleotide melting temperature (TmS) |
| US6503754B1 (en) | 2000-09-07 | 2003-01-07 | Isis Pharmaceuticals, Inc. | Antisense modulation of BH3 interacting domain death agonist expression |
| US6646113B1 (en) * | 1998-09-17 | 2003-11-11 | The Trustees Of The University Of Pennsylvania | Nucleic acid molecule encoding human survival of motor neuron-interacting protein 1 (SIP1) deletion mutants |
| US6210892B1 (en) | 1998-10-07 | 2001-04-03 | Isis Pharmaceuticals, Inc. | Alteration of cellular behavior by antisense modulation of mRNA processing |
| US6465628B1 (en) | 1999-02-04 | 2002-10-15 | Isis Pharmaceuticals, Inc. | Process for the synthesis of oligomeric compounds |
| US9029523B2 (en) | 2000-04-26 | 2015-05-12 | Ceres, Inc. | Promoter, promoter control elements, and combinations, and uses thereof |
| US20040002153A1 (en) | 1999-07-21 | 2004-01-01 | Monia Brett P. | Modulation of PTEN expression via oligomeric compounds |
| US6284538B1 (en) | 1999-07-21 | 2001-09-04 | Isis Pharmaceuticals, Inc. | Antisense inhibition of PTEN expression |
| WO2001007662A1 (en) | 1999-07-22 | 2001-02-01 | Genaissance Pharmaceuticals, Inc. | Drug target isogenes: polymorphisms in the prostaglandin-endoperoxide synthase 2 gene |
| US6677445B1 (en) | 1999-08-27 | 2004-01-13 | Chiron Corporation | Chimeric antisense oligonucleotides and cell transfecting formulations thereof |
| IL132972A0 (en) | 1999-11-16 | 2001-03-19 | Yissum Res Dev Co | Pharmaceutical compositions comprising acetylcholinesterase antisense deoxynucleotides for the treatment of muscular and neuromuscular disorders |
| US6187545B1 (en) * | 2000-01-19 | 2001-02-13 | Isis Pharmaceuticals Inc. | Antisense modulation of pepck-cytosolic expression |
| EP1133993A1 (en) | 2000-03-10 | 2001-09-19 | Max-Planck-Gesellschaft zur Förderung der Wissenschaften e.V. | Substances for the treatment of spinal muscular atrophy |
| EP1268768A2 (en) | 2000-03-27 | 2003-01-02 | University Of Delaware | Targeted chromosomal genomic alterations with modified single stranded oligonucleotides |
| WO2001072765A1 (en) | 2000-03-28 | 2001-10-04 | Isis Pharmaceuticals, Inc. | ALTERATION OF CELLULAR BEHAVIOR BY ANTISENSE MODULATION OF mRNA PROCESSING |
| EP1268856A2 (en) | 2000-04-07 | 2003-01-02 | Epigenomics AG | Detection of single nucleotide polymorphisms (snp's) and cytosine-methylations |
| WO2001083740A2 (en) | 2000-05-04 | 2001-11-08 | Avi Biopharma, Inc. | Splice-region antisense composition and method |
| CA2410514A1 (en) | 2000-05-30 | 2001-12-06 | Advanced Research & Technology Institute | Compositions and methods for identifying agents which modulate pten function and pi-3 kinase pathways |
| US6727355B2 (en) | 2000-08-25 | 2004-04-27 | Jcr Pharmaceuticals Co., Ltd. | Pharmaceutical composition for treatment of Duchenne muscular dystrophy |
| WO2002038738A2 (en) | 2000-11-09 | 2002-05-16 | Cold Spring Harbor Laboratory | Chimeric molecules to modulate gene expression |
| CA2429473C (en) | 2000-11-20 | 2011-02-15 | The Board Of Trustees Of The University Of Illinois | Membrane scaffold proteins |
| US20040058356A1 (en) | 2001-03-01 | 2004-03-25 | Warren Mary E. | Methods for global profiling gene regulatory element activity |
| US6825338B2 (en) * | 2001-03-30 | 2004-11-30 | Isis Pharmaceuticals, Inc. | Labeled oligonucleotides, methods for making same, and compounds useful therefor |
| EP1446500B1 (en) | 2001-05-08 | 2008-08-20 | Darwin Molecular Corporation | A method for regulating immune function in primates using the foxp3 protein |
| US20050176025A1 (en) | 2001-05-18 | 2005-08-11 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of B-cell CLL/Lymphoma-2 (BCL-2) gene expression using short interfering nucleic acid (siNA) |
| WO2002103015A2 (en) | 2001-06-14 | 2002-12-27 | Active Pass Pharmaceuticals, Inc. | Abca10 transporter |
| US7407943B2 (en) | 2001-08-01 | 2008-08-05 | Isis Pharmaceuticals, Inc. | Antisense modulation of apolipoprotein B expression |
| US7259150B2 (en) | 2001-08-07 | 2007-08-21 | Isis Pharmaceuticals, Inc. | Modulation of apolipoprotein (a) expression |
| MXPA04004056A (en) | 2001-10-29 | 2004-09-10 | Univ Mcgill | Acyclic linker-containing oligonucleotides and uses thereof. |
| WO2003056030A2 (en) * | 2001-11-08 | 2003-07-10 | The Johns Hopkins University | Methods and systems of nucleic acid sequencing |
| US20030198983A1 (en) | 2002-02-01 | 2003-10-23 | Affymetrix, Inc. | Methods of genetic analysis of human genes |
| US20080207542A1 (en) | 2002-03-26 | 2008-08-28 | Sirna Therapeutics, Inc. | RNA inteference mediated inhibition of hepatitis C virus (HVC) gene expression using short interfering nucleic acid (siNA) |
| SI2264172T1 (en) | 2002-04-05 | 2017-12-29 | Roche Innovation Center Copenhagen A/S | Oligomeric compounds for the modulation of hif-1alpha expression |
| US20050130924A1 (en) | 2002-06-26 | 2005-06-16 | Monia Brett P. | Antisense inhibition via RNAse H-independent reduction in mRNA |
| WO2004011613A2 (en) | 2002-07-29 | 2004-02-05 | Epigenesis Pharmaceuticals, Inc. | Composition & methods for treatment and screening |
| US20050003369A1 (en) | 2002-10-10 | 2005-01-06 | Affymetrix, Inc. | Method for depleting specific nucleic acids from a mixture |
| US20040219565A1 (en) | 2002-10-21 | 2004-11-04 | Sakari Kauppinen | Oligonucleotides useful for detecting and analyzing nucleic acids of interest |
| WO2004044136A2 (en) * | 2002-11-05 | 2004-05-27 | Isis Pharmaceuticals, Inc. | Compositions comprising alternating 2’-modified nucleosides for use in gene modulation |
| EP2305812A3 (en) | 2002-11-14 | 2012-06-06 | Dharmacon, Inc. | Fuctional and hyperfunctional sirna |
| US7655785B1 (en) | 2002-11-14 | 2010-02-02 | Rosetta Genomics Ltd. | Bioinformatically detectable group of novel regulatory oligonucleotides and uses thereof |
| US7250496B2 (en) | 2002-11-14 | 2007-07-31 | Rosetta Genomics Ltd. | Bioinformatically detectable group of novel regulatory genes and uses thereof |
| AU2003281969B2 (en) | 2002-11-18 | 2011-01-27 | Roche Innovation Center Copenhagen A/S | Amino-LNA, thio-LNA and alpha-L-oxy-LN |
| JP2006518197A (en) | 2003-02-10 | 2006-08-10 | サンタリス・ファルマ・アクティーゼルスカブ | Oligomeric compounds for modification of lath expression |
| WO2004083430A2 (en) | 2003-03-21 | 2004-09-30 | Santaris Pharma A/S | SHORT INTERFERING RNA (siRNA) ANALOGUES |
| FR2853663B1 (en) | 2003-04-14 | 2007-08-31 | Aventis Pharma Sa | PROCESS FOR OBTAINING MASTOCYTE LINES FROM PORK TISSUES AND PROCESS FOR PRODUCING HEPARIN TYPE MOLECULES |
| US8092992B2 (en) | 2003-05-29 | 2012-01-10 | Salk Institute For Biological Studies | Transcriptional regulation of gene expression by small double-stranded modulatory RNA |
| WO2004113867A2 (en) | 2003-06-16 | 2004-12-29 | University Of Massachusetts | Exon analysis |
| WO2005013901A2 (en) | 2003-07-31 | 2005-02-17 | Isis Pharmaceuticals, Inc. | Oligomeric compounds and compositions for use in modulation of small non-coding rnas |
| US7888497B2 (en) | 2003-08-13 | 2011-02-15 | Rosetta Genomics Ltd. | Bioinformatically detectable group of novel regulatory oligonucleotides and uses thereof |
| WO2005030928A2 (en) * | 2003-09-23 | 2005-04-07 | Chihiro Koike | PORCINE INVARIANT CHAIN PROTEIN, FULL LENGTH cDNA, GENOMIC ORGANIZATION, AND REGULATORY REGION |
| EP1675953A2 (en) | 2003-10-23 | 2006-07-05 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF RAS GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US8188254B2 (en) | 2003-10-30 | 2012-05-29 | Coley Pharmaceutical Gmbh | C-class oligonucleotide analogs with enhanced immunostimulatory potency |
| EP1756137A4 (en) | 2003-11-05 | 2007-10-31 | Univ Texas | Diagnostic and therapeutic methods and compositions involving pten and breast cancer |
| CA2550258A1 (en) | 2003-12-23 | 2005-07-07 | Santaris Pharma A/S | Oligomeric compounds for the modulation of bcl-2 |
| US20050244851A1 (en) | 2004-01-13 | 2005-11-03 | Affymetrix, Inc. | Methods of analysis of alternative splicing in human |
| EP1713332A4 (en) | 2004-01-23 | 2010-08-18 | Avi Biopharma Inc | Antisense oligomers and methods for inducing immune tolerance and immunosuppression |
| US20050164209A1 (en) | 2004-01-23 | 2005-07-28 | Bennett C. F. | Hepatocyte free uptake assays |
| DK1713912T3 (en) | 2004-01-30 | 2013-12-16 | Santaris Pharma As | Modified Short Interfering RNA (Modified siRNA) |
| WO2005089169A2 (en) | 2004-03-12 | 2005-09-29 | Exelixis, Inc. | Mptens as modifiers of the pten pathway and methods of use |
| WO2005094370A2 (en) | 2004-03-29 | 2005-10-13 | The General Hospital Corporation | Oligonucleotide complex compositions and methods of use as gene alteration tools |
| US7374927B2 (en) | 2004-05-03 | 2008-05-20 | Affymetrix, Inc. | Methods of analysis of degraded nucleic acid samples |
| AU2005248147A1 (en) | 2004-05-11 | 2005-12-08 | Alphagen Co., Ltd. | Polynucleotides for causing RNA interference and method for inhibiting gene expression using the same |
| US7687616B1 (en) | 2004-05-14 | 2010-03-30 | Rosetta Genomics Ltd | Small molecules modulating activity of micro RNA oligonucleotides and micro RNA targets and uses thereof |
| US20050265927A1 (en) | 2004-05-17 | 2005-12-01 | Yale University | Intranasal delivery of nucleic acid molecules |
| HUE028632T2 (en) | 2004-06-28 | 2016-12-28 | Univ Western Australia | Antisense oligonucleotides for inducing exon skipping and methods of use thereof |
| US8029985B2 (en) | 2004-09-01 | 2011-10-04 | Vybion, Inc. | Amplified bioassay |
| US9550990B2 (en) | 2004-12-10 | 2017-01-24 | Ionis Pharmaceuticals, Inc. | Regulation of epigenetic control of gene expression |
| US7879992B2 (en) | 2005-01-31 | 2011-02-01 | Isis Pharmaceuticals, Inc. | Modification of MyD88 splicing using modified oligonucleotides |
| CA2596506C (en) | 2005-02-09 | 2021-04-06 | Avi Biopharma, Inc. | Antisense composition and method for treating muscle atrophy |
| US8999943B2 (en) | 2005-03-14 | 2015-04-07 | Board Of Regents, The University Of Texas System | Antigene oligomers inhibit transcription |
| US7449297B2 (en) * | 2005-04-14 | 2008-11-11 | Euclid Diagnostics Llc | Methods of copying the methylation pattern of DNA during isothermal amplification and microarrays |
| US8877721B2 (en) | 2005-04-15 | 2014-11-04 | The Regents Of The University Of California | Small activating RNA molecules and methods of use |
| US8586719B2 (en) | 2005-04-27 | 2013-11-19 | Pacific Arrow Limited | Triterpenes for modulating gene expression and cell membrane, and as antiprotozoal agents |
| DK3308788T3 (en) * | 2005-06-23 | 2019-01-02 | Biogen Ma Inc | COMPOSITIONS AND PROCEDURES FOR MODULATING SMN2 SPLASH |
| US8067571B2 (en) | 2005-07-13 | 2011-11-29 | Avi Biopharma, Inc. | Antibacterial antisense oligonucleotide and method |
| WO2007031091A2 (en) | 2005-09-15 | 2007-03-22 | Santaris Pharma A/S | Rna antagonist compounds for the modulation of p21 ras expression |
| EP2431467A3 (en) | 2005-11-17 | 2012-05-02 | Board Of Regents, The University Of Texas | Modulation of gene expression by oligomers targeted to chromosomal DNA |
| CA2630952A1 (en) | 2005-11-21 | 2007-05-24 | Johnson & Johnson Research Pty Limited | Multitargeting interfering rnas having two active strands and methods for their design and use |
| WO2007087113A2 (en) | 2005-12-28 | 2007-08-02 | The Scripps Research Institute | Natural antisense and non-coding rna transcripts as drug targets |
| US20090011004A1 (en) | 2005-12-30 | 2009-01-08 | Philadelphia Health & Education Corp., D/B/A/ Drexel University Of College Of Medicine | Improved carriers for delivery of nucleic acid agents to cells and tissues |
| CA2640058C (en) | 2006-01-27 | 2018-04-24 | Isis Pharmaceuticals, Inc. | Oligomeric compounds and compositions for the use in modulation of micrornas |
| US8362229B2 (en) | 2006-02-08 | 2013-01-29 | Quark Pharmaceuticals, Inc. | Tandem siRNAS |
| EP2194129A3 (en) | 2006-04-03 | 2012-12-26 | Santaris Pharma A/S | Pharmaceutical composition comprising anti-miRNA antisense oligonucleotides |
| ATE500265T1 (en) | 2006-04-07 | 2011-03-15 | Univ Goettingen Georg August | SYNTHETIC MECP2 SEQUENCE FOR PROTEIN REPLACEMENT THERAPY |
| AU2007258117B2 (en) | 2006-05-05 | 2013-05-30 | Isis Pharmaceuticals, Inc. | Compounds and methods for modulating gene expression |
| WO2008002996A2 (en) | 2006-06-27 | 2008-01-03 | Shanghai Institutes For Biological Sciences | Rheumatoid arthritis t cell vaccine |
| WO2008024499A2 (en) | 2006-08-25 | 2008-02-28 | Duke University | Methods for in vivo identification of endogenous mrna targets of micrornas |
| WO2008025069A1 (en) | 2006-08-28 | 2008-03-06 | The Walter And Eliza Hall Institute Of Medical Research | Methods of modulating cellular activity and compositions therefor |
| WO2008029619A1 (en) | 2006-09-07 | 2008-03-13 | Daiichi Sankyo Company, Limited | Ena antisense oligonucleotide having sequence-specific action |
| WO2008036282A1 (en) | 2006-09-18 | 2008-03-27 | The Regents Of The University Of Californina | Upregulating activity or expression of bdnf to mitigate cognitive impairment in asymptomatic huntington's subjects |
| US9550988B2 (en) | 2006-10-18 | 2017-01-24 | Ionis Pharmaceuticals, Inc. | Antisense compounds |
| JP2010509923A (en) | 2006-11-23 | 2010-04-02 | ミルクス セラピューティクス アンパーツゼルスカブ | Oligonucleotides for altering the activity of target RNA |
| WO2008066784A2 (en) | 2006-11-27 | 2008-06-05 | Ludwig Institute For Cancer Research | Expression of foxp3 by cancer cells |
| AU2008206168B2 (en) | 2007-01-19 | 2012-01-19 | The Regents Of The University Of Michigan | ADRB2 cancer markers |
| ITMI20070127A1 (en) | 2007-01-29 | 2008-07-30 | Fond I R C C S | E-O PEPTIDES PROTEINS FOR THE PREVENTION AND-OR CARE OF NEURODEGENERATIVE DISEASES |
| WO2008103763A2 (en) * | 2007-02-20 | 2008-08-28 | Sequenom, Inc. | Methods and compositions for cancer diagnosis and treatment based on nucleic acid methylation |
| US7858592B2 (en) | 2007-02-26 | 2010-12-28 | The Board Of Regents Of The University Of Texas System | Interfering RNAs against the promoter region of P53 |
| DK2149605T3 (en) | 2007-03-22 | 2013-09-30 | Santaris Pharma As | Short RNA antagonist compounds to modulate the desired mRNA |
| WO2008141282A2 (en) | 2007-05-11 | 2008-11-20 | The Regents Of The University Of Michigan | Materials and methods for foxp3 tumor suppression |
| EP2714904B1 (en) | 2007-06-14 | 2017-04-12 | Mirx Therapeutics ApS | Oligonucleotides for modulation of target rna activity |
| US20090082297A1 (en) | 2007-06-25 | 2009-03-26 | Lioy Daniel T | Compositions and Methods for Regulating Gene Expression |
| EP2170363B1 (en) * | 2007-06-29 | 2018-08-08 | Sarepta Therapeutics, Inc. | Tissue specific peptide conjugates and methods |
| WO2009027349A2 (en) | 2007-08-24 | 2009-03-05 | Oryzon Genomics Sa | Treatment and prevention of neurodegenerative diseases |
| JP2010539990A (en) | 2007-10-04 | 2010-12-24 | ボード オブ リージェンツ ザ ユニバーシティー オブ テキサス システム | Method for regulating gene expression using agRNA and gapmer targeting antisense transcript |
| EP2205737B1 (en) | 2007-10-04 | 2013-02-13 | Santaris Pharma A/S | Micromirs |
| ITRM20070523A1 (en) | 2007-10-05 | 2009-04-06 | Consiglio Nazionale Ricerche | CODIFYING NUCLEIC ACID FOR A REGULATING PROTEIN SPECIFIC TO THE TRANSCRIPTION OF THE UROPHINE PROTEIN BY IT CODIFIED AND ITS APPLICATIONS |
| US9029337B2 (en) | 2007-11-09 | 2015-05-12 | Isis Pharmaceuticals, Inc. | Modulation of factor 7 expression |
| US8637478B2 (en) | 2007-11-13 | 2014-01-28 | Isis Pharmaceuticals, Inc. | Compounds and methods for modulating protein expression |
| US8937217B2 (en) | 2007-12-18 | 2015-01-20 | E. I. Du Pont De Nemours And Company | Down-regulation of gene expression using artificial microRNAs |
| WO2009090182A1 (en) | 2008-01-14 | 2009-07-23 | Santaris Pharma A/S | C4'-substituted - dna nucleotide gapmer oligonucleotides |
| US8361980B2 (en) | 2008-03-07 | 2013-01-29 | Santaris Pharma A/S | Pharmaceutical compositions for treatment of microRNA related diseases |
| EP2265717A4 (en) | 2008-03-07 | 2012-09-12 | Senesco Technologies Inc | Use of sirna to achieve down regulation of an endogenous gene in combination with the use of a sense construct to achieve expression of a desired polynucleotide |
| US8846639B2 (en) | 2008-04-04 | 2014-09-30 | Isis Pharmaceutical, Inc. | Oligomeric compounds comprising bicyclic nucleosides and having reduced toxicity |
| AU2009235941A1 (en) | 2008-04-07 | 2009-10-15 | Riken | RNA molecules and uses thereof |
| US8916532B2 (en) | 2008-04-28 | 2014-12-23 | The Trustees Of The University Of Pennsylvania | Methods for enhancing utrophin production via inhibition of microRNA |
| US8222221B2 (en) | 2008-06-04 | 2012-07-17 | The Board Of Regents Of The University Of Texas System | Modulation of gene expression through endogenous small RNA targeting of gene promoters |
| CA2635187A1 (en) | 2008-06-05 | 2009-12-05 | The Royal Institution For The Advancement Of Learning/Mcgill University | Oligonucleotide duplexes and uses thereof |
| WO2010000665A1 (en) | 2008-06-30 | 2010-01-07 | Santaris Pharma A/S | Antidote oligomers |
| AU2009276763B2 (en) | 2008-07-29 | 2015-07-16 | The Board Of Regents Of The University Of Texas Sytem | Selective inhibition of polyglutamine protein expression |
| CN104557914A (en) | 2008-09-26 | 2015-04-29 | 新加坡科技研究局 | 3-deazaneplanocin derivatives |
| ES2727549T3 (en) | 2008-10-03 | 2019-10-17 | Curna Inc | Treatment of diseases related to apolipoprotein a1 by inhibition of the natural antisense transcript to apolipoprotein a1 |
| KR20110071017A (en) | 2008-10-16 | 2011-06-27 | 마리나 바이오테크, 인크. | Processes and compositions for liposomal and efficient delivery of gene silencing therapeutics |
| PE20120115A1 (en) | 2008-11-17 | 2012-02-20 | Hoffmann La Roche | COMPOSITIONS AND METHODS TO INHIBIT THE EXPRESSION OF FACTOR VII GENES |
| GB0821457D0 (en) | 2008-11-24 | 2008-12-31 | Trillion Genomics Ltd | Oligonucleotides |
| CN102307997B (en) | 2008-12-04 | 2018-03-30 | 库尔纳公司 | By suppressing to treat the related disease of Sirtuin 1 (SIRT1) for the natural antisense transcript of Sirtuin 1 |
| MX2011005910A (en) | 2008-12-04 | 2011-06-17 | Opko Curna Llc | Treatment of erythropoietin (epo) related diseases by inhibition of natural antisense transcript to epo. |
| US8927511B2 (en) | 2008-12-04 | 2015-01-06 | Curna, Inc. | Treatment of vascular endothelial growth factor (VEGF) related diseases by inhibition of natural antisense transcript to VEGF |
| KR101840618B1 (en) | 2008-12-04 | 2018-03-20 | 큐알엔에이, 인크. | Treatment of tumor suppressor gene related diseases by inhibition of natural antisense transcript to the gene |
| CA2750561C (en) | 2009-01-26 | 2017-10-10 | Protiva Biotherapeutics, Inc. | Compositions and methods for silencing apolipoprotein c-iii expression |
| CN102387817B (en) | 2009-02-12 | 2018-01-30 | 库尔纳公司 | By suppressing to treat the related diseases of BDNF for the natural antisense transcript of neurotrophic factor derived from brain (BDNF) |
| WO2010093906A2 (en) | 2009-02-12 | 2010-08-19 | Curna, Inc. | Treatment of glial cell derived neurotrophic factor (gdnf) related diseases by inhibition of natural antisense transcript to gdnf |
| WO2010093860A2 (en) | 2009-02-13 | 2010-08-19 | The General Hospital Corporation | Isolation of factors that associate directly or indirectly with chromatin |
| US20110319317A1 (en) | 2009-03-04 | 2011-12-29 | Opko Curna, Llc | Treatment of sirtuin 1 (sirt1) related diseases by inhibition of natural antisense transcript to sirt1 |
| EP2408919B1 (en) | 2009-03-16 | 2017-10-18 | CuRNA, Inc. | Treatment of nuclear factor (erythroid-derived 2)-like 2 (nrf2) related diseases by inhibition of natural antisense transcript to nrf2 |
| CA2755404C (en) | 2009-03-17 | 2020-03-24 | Joseph Collard | Treatment of delta-like 1 homolog (dlk1) related diseases by inhibition of natural antisense transcript to dlk1 |
| US20120149756A1 (en) | 2009-04-10 | 2012-06-14 | Associatin Institut de Myologie | Tricyclo-dna antisense oligonucleotides, compositions, and methods for the treatment of disease |
| WO2010124231A2 (en) | 2009-04-24 | 2010-10-28 | The Board Of Regents Of The University Of Texas System | Modulation of gene expression using oligomers that target gene regions downstream of 3' untranslated regions |
| NO2424987T3 (en) | 2009-05-01 | 2018-04-14 | ||
| EP2427553A4 (en) | 2009-05-06 | 2012-11-07 | Opko Curna Llc | Treatment of lipid transport and metabolism gene related diseases by inhibition of natural antisense transcript to a lipid transport and metabolism gene |
| CN102803492B (en) | 2009-05-06 | 2016-06-29 | 库尔纳公司 | TTP relevant disease is treated for the natural antisense transcript of triple four proline (TTP) by suppression |
| KR101742334B1 (en) | 2009-05-08 | 2017-06-01 | 큐알엔에이, 인크. | Treatment of dystrophin family related diseases by inhibition of natural antisense transcript to dmd family |
| DK2432881T3 (en) | 2009-05-18 | 2018-02-26 | Curna Inc | TREATMENT OF REPROGRAMMING FACTOR-RELATED DISEASES BY INHIBITING NATURAL ANTISENSE TRANSCRIPTS TO A REPROGRAMMING FACTOR |
| US8895527B2 (en) | 2009-05-22 | 2014-11-25 | Curna, Inc. | Treatment of transcription factor E3 (TFE3) and insulin receptor substrate 2(IRS2) related diseases by inhibition of natural antisense transcript to TFE3 |
| WO2010138806A2 (en) | 2009-05-28 | 2010-12-02 | Curna, Inc. | Treatment of antiviral gene related diseases by inhibition of natural antisense transcript to an antiviral gene |
| US8951981B2 (en) | 2009-06-16 | 2015-02-10 | Curna, Inc. | Treatment of paraoxonase 1 (PON1) related diseases by inhibition of natural antisense transcript to PON1 |
| KR101801404B1 (en) | 2009-06-16 | 2017-12-20 | 큐알엔에이, 인크. | Treatment of collagen gene related diseases by inhibition of natural antisense transcript to a collagen gene |
| KR101807323B1 (en) | 2009-06-24 | 2017-12-08 | 큐알엔에이, 인크. | Ttreatment of tumor necrosis factor receptor 2 (tnfr2) related diseases by inhibition of natural antisense transcript to tnfr2 |
| CA2765815A1 (en) | 2009-06-26 | 2010-12-29 | Opko Curna, Llc | Treatment of down syndrome gene related diseases by inhibition of natural antisense transcript to a down syndrome gene |
| BR112012000421A2 (en) | 2009-07-06 | 2019-09-24 | Alnylam Pharmaceuticals Inc | compositions and methods for enhancing the production of a biological product. |
| CA2768598A1 (en) | 2009-07-22 | 2011-01-27 | Cenix Bioscience Gmbh | Delivery system and conjugates for compound delivery via naturally occurring intracellular transport routes |
| WO2011010706A1 (en) | 2009-07-23 | 2011-01-27 | 武田薬品工業株式会社 | Fgf21 cis-element binding substance |
| US20120252869A1 (en) | 2009-07-24 | 2012-10-04 | Opko Curna, Llc | Treatment of sirtuin (sirt) related diseases by inhibition of natural antisense transcript to a sirtuin (sirt) |
| ES2585360T3 (en) | 2009-08-05 | 2016-10-05 | Curna, Inc. | Treatment of diseases related to an insulin gene (INS) by inhibition of natural antisense transcription in an insulin gene (INS) |
| CN104313027B (en) | 2009-08-11 | 2018-11-20 | 库尔纳公司 | By inhibiting the natural antisense transcript of adiponectin (ADIPOQ) to treat adiponectin (ADIPOQ) related disease |
| KR101805213B1 (en) | 2009-08-21 | 2017-12-06 | 큐알엔에이, 인크. | Treatment of 'c terminus of hsp70-interacting protein' (chip) related diseases by inhibition of natural antisense transcript to chip |
| JP5964232B2 (en) | 2009-08-25 | 2016-08-03 | カッパーアールエヌエー,インコーポレイテッド | Treatment of IQGAP-related diseases by inhibition of natural antisense transcripts against 'IQ motif-containing GTPase-activating protein' (IQGAP) |
| WO2011025862A2 (en) | 2009-08-28 | 2011-03-03 | Curna, Inc. | Treatment of atp-binding cassette sub-family b member 1 (abcb1) related diseases by inhibition of natural antisense transcript to abcb1 |
| CA2775111C (en) | 2009-09-25 | 2019-12-31 | Opko Curna, Llc | Treatment of filaggrin (flg) related diseases by modulation of flg expression and activity |
| WO2011038205A2 (en) | 2009-09-25 | 2011-03-31 | Curna, Inc. | Treatment of growth hormone (gh) related diseases by inhibition of natural antisense transcript to gh |
| WO2011048125A1 (en) | 2009-10-20 | 2011-04-28 | Santaris Pharma A/S | Oral delivery of therapeutically effective lna oligonucleotides |
| KR101138048B1 (en) | 2009-11-06 | 2012-04-23 | 성균관대학교산학협력단 | Novel peptides upregulating BDNF expression and pharmaceutical composition for protection and therapy against Alzheimer's disease and Parkinson's disease comprising the same |
| CA2782366A1 (en) | 2009-12-16 | 2011-07-14 | Opko Curna, Llc | Treatment of membrane bound transcription factor peptidase, site 1 (mbtps1) related diseases by inhibition of natural antisense transcript to mbtps1 |
| CA2782373C (en) | 2009-12-23 | 2019-03-26 | Opko Curna, Llc | Treatment of hepatocyte growth factor (hgf) related diseases by inhibition of natural antisense transcript to hgf |
| EP2515947B1 (en) | 2009-12-23 | 2021-10-06 | CuRNA, Inc. | Treatment of uncoupling protein 2 (ucp2) related diseases by inhibition of natural antisense transcript to ucp2 |
| EP2519634B1 (en) | 2009-12-29 | 2016-06-01 | CuRNA, Inc. | TREATMENT OF TUMOR PROTEIN 63 (p63) RELATED DISEASES BY INHIBITION OF NATURAL ANTISENSE TRANSCRIPT TO p63 |
| EP2519633B1 (en) | 2009-12-29 | 2017-10-25 | CuRNA, Inc. | Treatment of nuclear respiratory factor 1 (nrf1) related diseases by inhibition of natural antisense transcript to nrf1 |
| DK2519632T3 (en) | 2009-12-31 | 2018-07-23 | Curna Inc | TREATMENT OF INSULIN RECEPTOR SUBSTRATE 2- (IRS2) RELATED DISEASES BY INHIBITION OF NATURAL ANTISENSE TRANSCRIPTION TO IRS2 AND TRANSCRIPTION FACTOR E3 (TFE3) |
| JP5886757B2 (en) | 2010-01-04 | 2016-03-16 | カッパーアールエヌエー,インコーポレイテッド | Treatment of interferon regulatory factor 8 (IRF8) related diseases by inhibition of natural antisense transcripts against interferon regulatory factor 8 (IRF8) |
| US8912157B2 (en) | 2010-01-06 | 2014-12-16 | Curna, Inc. | Treatment of pancreatic developmental gene related diseases by inhibition of natural antisense transcript to a pancreatic developmental gene |
| US9200277B2 (en) | 2010-01-11 | 2015-12-01 | Curna, Inc. | Treatment of sex hormone binding globulin (SHBG) related diseases by inhibition of natural antisense transcript to SHBG |
| DK2529015T3 (en) | 2010-01-25 | 2018-02-26 | Curna Inc | TREATMENT OF RNASE H1-RELATED DISEASES BY INHIBITION OF NATURAL ANTISENSE TRANSCRIPT TO RNASE H1 |
| US9574191B2 (en) | 2010-02-03 | 2017-02-21 | The Board Of Regents Of The University Of Texas System | Selective inhibition of polyglutamine protein expression |
| US20130059902A1 (en) | 2010-02-08 | 2013-03-07 | Isis Pharmaceuticals, Inc. | Methods and compositions useful in treatment of diseases or conditions related to repeat expansion |
| WO2011097582A2 (en) | 2010-02-08 | 2011-08-11 | Opko Curna Llc | Treatment of arachidonate 12-lipoxygenase, 12r type (alox12b) related diseases by inhibition of natural antisense transcript to alox12b |
| JP5976548B2 (en) | 2010-02-22 | 2016-08-23 | カッパーアールエヌエー,インコーポレイテッド | Treatment of pyrroline-5-carboxylate reductase 1 (PYCR1) related diseases by inhibition of natural antisense transcripts against PYCR1 |
| EP2550360B1 (en) | 2010-03-24 | 2017-08-30 | Mirrx Therapeutics A/s | Bivalent antisense oligonucleotides |
| CA2795145C (en) | 2010-04-02 | 2019-01-22 | Curna, Inc. | Treatment of colony-stimulating factor 3 (csf3) related diseases by inhibition of natural antisense transcript to csf3 |
| KR101900962B1 (en) | 2010-04-09 | 2018-09-20 | 큐알엔에이, 인크. | Treatment of fibroblast growth factor 21 (fgf21) related diseases by inhibition of natural antisense transcript to fgf21 |
| US9145556B2 (en) | 2010-04-13 | 2015-09-29 | Life Technologies Corporation | Compositions and methods for inhibition of nucleic acids function |
| KR101936011B1 (en) | 2010-05-03 | 2019-01-07 | 큐알엔에이, 인크. | Treatment of sirtuin (sirt) related diseases by inhibition of natural antisense transcript to a sirtuin (sirt) |
| TWI531370B (en) | 2010-05-14 | 2016-05-01 | 可娜公司 | Treatment of par4 related diseases by inhibition of natural antisense transcript to par4 |
| WO2011146675A2 (en) | 2010-05-19 | 2011-11-24 | Opko Curna Llc | Treatment of lim homeobox 2 (lhx2) related diseases by inhibition of natural antisense transcript to lhx2 |
| TW201201819A (en) | 2010-05-19 | 2012-01-16 | Opko Curna Llc | Treatment of BCL2-like 11 (BCL2L11) related diseases by inhibition of natural antisense transcript to BCL2L11 |
| WO2011150005A2 (en) | 2010-05-26 | 2011-12-01 | Opko Curna Llc | Treatment of atonal homolog 1 (atoh1) related diseases by inhibition of natural antisense transcript to atoh1 |
| KR101902197B1 (en) | 2010-05-26 | 2018-10-01 | 큐알엔에이, 인크. | Treatment of methionine sulfoxide reductase a (msra) related diseases by inhibition of natural antisense transcript to msra |
| WO2011159836A2 (en) | 2010-06-15 | 2011-12-22 | Isis Pharmaceuticals, Inc. | Compounds and methods for modulating interaction between proteins and target nucleic acids |
| US20120004278A1 (en) | 2010-06-18 | 2012-01-05 | The Board Of Trustees Of The Leland Stanford Junior University | Linc rnas in cancer diagnosis and treatment |
| WO2011163499A2 (en) | 2010-06-23 | 2011-12-29 | Opko Curna, Llc | Treatment of sodium channel, voltage-gated, alpha subunit (scna) related diseases by inhibition of natural antisense transcript to scna |
| GB201010557D0 (en) | 2010-06-23 | 2010-08-11 | Mina Therapeutics Ltd | RNA molecules and uses thereof |
| CN102309757B (en) | 2010-07-09 | 2014-09-17 | 中国科学院上海巴斯德研究所 | Novel regulatory factor of FOXP3 and regulatory T cells, and use thereof |
| TW201209163A (en) | 2010-07-12 | 2012-03-01 | Opko Curna Llc | Treatment of BCL2 binding component 3 (BBC3) related diseases by inhibition of natural antisense transcript to BBC3 |
| JP5998131B2 (en) | 2010-07-14 | 2016-09-28 | カッパーアールエヌエー,インコーポレイテッド | DISCSLARGEHOMOLOG (DLG) Treatment of DLG-related diseases by inhibition of natural antisense transcripts on DLG1 |
| WO2012027033A1 (en) * | 2010-07-19 | 2012-03-01 | Isis Pharmaceuticals, Inc. | Compounds and methods for modulating target nuclear and sub-nuclear nucleic acid molecules in cells and animals |
| KR20180105730A (en) | 2010-07-19 | 2018-09-28 | 아이오니스 파마수티컬즈, 인코포레이티드 | Modulation of dystrophia myotonica-protein kinase (dmpk) expression |
| WO2012018881A2 (en) | 2010-08-03 | 2012-02-09 | Alnylam Pharmaceuticals, Inc. | Methods and compositions for the regulation of rna |
| WO2012024478A2 (en) | 2010-08-19 | 2012-02-23 | Opko Curna Llc | Treatment of nicotinamide phosphoribosyltransferase (nampt) related diseases by inhibition of natural antisense transcript to nampt |
| EP2614369B1 (en) | 2010-09-10 | 2016-02-03 | Epizyme, Inc. | Method for determining the suitability of inhibitors of human ezh2 in treatment |
| KR101886457B1 (en) | 2010-10-06 | 2018-08-07 | 큐알엔에이, 인크. | Treatment of sialidase 4 (neu4) related diseases by inhibition of natural antisense transcript to neu4 |
| JP6049623B2 (en) | 2010-10-22 | 2016-12-21 | カッパーアールエヌエー,インコーポレイテッド | Treatment of IDUA-related diseases by inhibition of natural antisense transcripts to α-L-iduronidase (IDUA) |
| CN103201387B (en) | 2010-10-27 | 2018-02-02 | 库尔纳公司 | IFRD1 relevant diseases are treated by suppressing the natural antisense transcript of interferon correlative development regulatory factor 1 (IFRD1) |
| WO2012064806A2 (en) | 2010-11-11 | 2012-05-18 | The University Of North Carolina At Chapel Hill | Methods and compositions for unsilencing imprinted genes |
| AU2011325956B2 (en) | 2010-11-12 | 2016-07-14 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| US9920317B2 (en) | 2010-11-12 | 2018-03-20 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| US10000752B2 (en) | 2010-11-18 | 2018-06-19 | Curna, Inc. | Antagonat compositions and methods of use |
| CA2818824A1 (en) | 2010-11-23 | 2012-05-31 | Joseph Collard | Treatment of nanog related diseases by inhibition of natural antisense transcript to nanog |
| CA2822462A1 (en) | 2010-12-20 | 2012-06-28 | The General Hospital Corporation | Polycomb-associated non-coding rnas |
| US9206479B2 (en) | 2011-02-09 | 2015-12-08 | University Of Rochester | Methods and compositions related to Staufen 1 binding sites formed by duplexing Alu elements |
| WO2012144220A1 (en) | 2011-04-22 | 2012-10-26 | Oncotherapy Science, Inc. | Ezh2 as target gene for cancer therapy and diagnosis |
| US8557787B2 (en) | 2011-05-13 | 2013-10-15 | The Board Of Trustees Of The Leland Stanford Junior University | Diagnostic, prognostic and therapeutic uses of long non-coding RNAs for cancer and regenerative medicine |
| RU2620980C2 (en) | 2011-06-09 | 2017-05-30 | Курна, Инк. | Treatment of diseases associated with frataxin (fxn), by inhibiting natural antisense fxn transcript |
| WO2012174610A1 (en) | 2011-06-24 | 2012-12-27 | Murdoch Childrens Research Institute | Treatment and diagnosis of epigenetic disorders and conditions |
| AU2012279114B2 (en) | 2011-07-05 | 2017-07-20 | The General Hospital Corporation | RNA-YY1 interactions |
| EA029151B1 (en) | 2011-09-06 | 2018-02-28 | Курна, Инк. | TREATMENT OF DISEASES RELATED TO ALPHA SUBUNITS OF VOLTAGE-GATED SODIUM CHANNELS (SCNxA) WITH SMALL MOLECULES |
| US9580708B2 (en) | 2011-09-14 | 2017-02-28 | Rana Therapeutics, Inc. | Multimeric oligonucleotides compounds |
| ITMI20120275A1 (en) | 2012-02-24 | 2013-08-25 | Biogenera Societa A Responsabilita Limitata | OLIGONUCLEOTIDS FOR THE MODULATION OF GENE EXPRESSION AND THEIR USES |
| WO2013138374A2 (en) | 2012-03-15 | 2013-09-19 | Curna, Inc. | Treatment of brain derived neurotrophic factor (bdnf) related diseases by inhibition of natural antisense transcript to bdnf |
| JP2015523855A (en) | 2012-05-16 | 2015-08-20 | ラナ セラピューティクス インコーポレイテッド | Compositions and methods for modulating APOA1 and ABCA1 expression |
| US20150232836A1 (en) | 2012-05-16 | 2015-08-20 | Rana Therapeutics, Inc. | Compositions and methods for modulating gene expression |
| EA201492123A1 (en) | 2012-05-16 | 2015-10-30 | Рана Терапьютикс, Инк. | COMPOSITIONS AND METHODS FOR MODULATING THE EXPRESSION OF THE SMN GENES FAMILY |
| EP2850184A4 (en) | 2012-05-16 | 2016-01-27 | Rana Therapeutics Inc | Compositions and methods for modulating gene expression |
| WO2013173608A1 (en) | 2012-05-16 | 2013-11-21 | Rana Therapeutics, Inc. | Compositions and methods for modulating mecp2 expression |
| EP3511416A1 (en) | 2012-05-16 | 2019-07-17 | Translate Bio MA, Inc. | Compositions and methods for modulating gene expression |
| JP2015519057A (en) | 2012-05-16 | 2015-07-09 | ラナ セラピューティクス インコーポレイテッド | Compositions and methods for modulating PTEN expression |
| KR20160074368A (en) | 2012-05-16 | 2016-06-28 | 라나 테라퓨틱스, 인크. | Compositions and methods for modulating utrn expression |
| US20150133529A1 (en) | 2012-05-16 | 2015-05-14 | Rana Therapeutics, Inc. | Compositions and methods for modulating bdnf expression |
| JP2015518710A (en) | 2012-05-16 | 2015-07-06 | ラナ セラピューティクス インコーポレイテッド | Compositions and methods for regulating hemoglobin gene family expression |
| CA2873766A1 (en) | 2012-05-16 | 2013-11-21 | Rana Therapeutics Inc. | Compositions and methods for modulating atp2a2 expression |
| US9567581B2 (en) | 2012-08-07 | 2017-02-14 | The General Hospital Corporation | Selective reactivation of genes on the inactive X chromosome |
| AU2013315225B2 (en) | 2012-09-14 | 2018-11-08 | Translate Bio Ma, Inc. | Multimeric oligonucleotide compounds |
| EP2920304B1 (en) | 2012-11-15 | 2019-03-06 | Roche Innovation Center Copenhagen A/S | Oligonucleotide conjugates |
| US10590412B2 (en) | 2013-04-19 | 2020-03-17 | Ionis Pharmaceuticals, Inc. | Compositions and methods for modulation nucleic acids through nonsense mediated decay |
| AU2014274730A1 (en) | 2013-06-07 | 2016-01-21 | Rana Therapeutics, Inc. | Compositions and methods for modulating FOXP3 expression |
| CN105324119A (en) | 2013-06-16 | 2016-02-10 | 国立大学法人东京医科齿科大学 | Double-stranded antisense nucleic acid with exon-skipping effect |
| WO2015035476A1 (en) | 2013-09-16 | 2015-03-19 | University Of Western Sydney | Modulation of gene expression |
-
2013
- 2013-05-16 EA EA201492123A patent/EA201492123A1/en unknown
- 2013-05-16 KR KR1020147035185A patent/KR20150030205A/en not_active Application Discontinuation
- 2013-05-16 CA CA2873794A patent/CA2873794A1/en not_active Abandoned
- 2013-05-16 JP JP2015512858A patent/JP2015523854A/en active Pending
- 2013-05-16 EP EP13790819.0A patent/EP2850186B1/en not_active Not-in-force
- 2013-05-16 WO PCT/US2013/041440 patent/WO2013173638A1/en active Application Filing
- 2013-05-16 CN CN201380037632.9A patent/CN104540947A/en active Pending
- 2013-05-16 SG SG11201407483YA patent/SG11201407483YA/en unknown
- 2013-05-16 US US14/401,194 patent/US10059941B2/en active Active
- 2013-05-16 AU AU2013262649A patent/AU2013262649A1/en not_active Abandoned
- 2013-05-16 DK DK13790819.0T patent/DK2850186T3/en active
-
2014
- 2014-12-15 ZA ZA2014/09228A patent/ZA201409228B/en unknown
-
2015
- 2015-09-17 HK HK15109139.6A patent/HK1208700A1/en unknown
Patent Citations (156)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US3687808A (en) | 1969-08-14 | 1972-08-29 | Univ Leland Stanford Junior | Synthetic polynucleotides |
| US4469863A (en) | 1980-11-12 | 1984-09-04 | Ts O Paul O P | Nonionic nucleic acid alkyl and aryl phosphonates and processes for manufacture and use thereof |
| US5023243A (en) | 1981-10-23 | 1991-06-11 | Molecular Biosystems, Inc. | Oligonucleotide therapeutic agent and method of making same |
| US4476301A (en) | 1982-04-29 | 1984-10-09 | Centre National De La Recherche Scientifique | Oligonucleotides, a process for preparing the same and their application as mediators of the action of interferon |
| US4667025A (en) | 1982-08-09 | 1987-05-19 | Wakunaga Seiyaku Kabushiki Kaisha | Oligonucleotide derivatives |
| US4789737A (en) | 1982-08-09 | 1988-12-06 | Wakunaga Seiyaku Kabushiki Kaisha | Oligonucleotide derivatives and production thereof |
| US4835263A (en) | 1983-01-27 | 1989-05-30 | Centre National De La Recherche Scientifique | Novel compounds containing an oligonucleotide sequence bonded to an intercalating agent, a process for their synthesis and their use |
| US4605735A (en) | 1983-02-14 | 1986-08-12 | Wakunaga Seiyaku Kabushiki Kaisha | Oligonucleotide derivatives |
| US5541313A (en) | 1983-02-22 | 1996-07-30 | Molecular Biosystems, Inc. | Single-stranded labelled oligonucleotides of preselected sequence |
| US4948882A (en) | 1983-02-22 | 1990-08-14 | Syngene, Inc. | Single-stranded labelled oligonucleotides, reactive monomers and methods of synthesis |
| US4824941A (en) | 1983-03-10 | 1989-04-25 | Julian Gordon | Specific antibody to the native form of 2'5'-oligonucleotides, the method of preparation and the use as reagents in immunoassays or for binding 2'5'-oligonucleotides in biological systems |
| US4587044A (en) | 1983-09-01 | 1986-05-06 | The Johns Hopkins University | Linkage of proteins to nucleic acids |
| US5118802A (en) | 1983-12-20 | 1992-06-02 | California Institute Of Technology | DNA-reporter conjugates linked via the 2' or 5'-primary amino group of the 5'-terminal nucleoside |
| US5550111A (en) | 1984-07-11 | 1996-08-27 | Temple University-Of The Commonwealth System Of Higher Education | Dual action 2',5'-oligoadenylate antiviral derivatives and uses thereof |
| US5578717A (en) | 1984-10-16 | 1996-11-26 | Chiron Corporation | Nucleotides for introducing selectably cleavable and/or abasic sites into oligonucleotides |
| US5552538A (en) | 1984-10-16 | 1996-09-03 | Chiron Corporation | Oligonucleotides with cleavable sites |
| US5367066A (en) | 1984-10-16 | 1994-11-22 | Chiron Corporation | Oligonucleotides with selectably cleavable and/or abasic sites |
| US5545730A (en) | 1984-10-16 | 1996-08-13 | Chiron Corporation | Multifunctional nucleic acid monomer |
| US5258506A (en) | 1984-10-16 | 1993-11-02 | Chiron Corporation | Photolabile reagents for incorporation into oligonucleotide chains |
| US4828979A (en) | 1984-11-08 | 1989-05-09 | Life Technologies, Inc. | Nucleotide analogs for nucleic acid labeling and detection |
| US4845205A (en) | 1985-01-08 | 1989-07-04 | Institut Pasteur | 2,N6 -disubstituted and 2,N6 -trisubstituted adenosine-3'-phosphoramidites |
| US5235033A (en) | 1985-03-15 | 1993-08-10 | Anti-Gene Development Group | Alpha-morpholino ribonucleoside derivatives and polymers thereof |
| US5034506A (en) | 1985-03-15 | 1991-07-23 | Anti-Gene Development Group | Uncharged morpholino-based polymers having achiral intersubunit linkages |
| US5185444A (en) | 1985-03-15 | 1993-02-09 | Anti-Gene Deveopment Group | Uncharged morpolino-based polymers having phosphorous containing chiral intersubunit linkages |
| US4762779A (en) | 1985-06-13 | 1988-08-09 | Amgen Inc. | Compositions and methods for functionalizing nucleic acids |
| US5317098A (en) | 1986-03-17 | 1994-05-31 | Hiroaki Shizuya | Non-radioisotope tagging of fragments |
| US4876335A (en) | 1986-06-30 | 1989-10-24 | Wakunaga Seiyaku Kabushiki Kaisha | Poly-labelled oligonucleotide derivative |
| US5013830A (en) | 1986-09-08 | 1991-05-07 | Ajinomoto Co., Inc. | Compounds for the cleavage at a specific position of RNA, oligomers employed for the formation of said compounds, and starting materials for the synthesis of said oligomers |
| US5286717A (en) | 1987-03-25 | 1994-02-15 | The United States Of America As Represented By The Department Of Health And Human Services | Inhibitors for replication of retroviruses and for the expression of oncogene products |
| US5276019A (en) | 1987-03-25 | 1994-01-04 | The United States Of America As Represented By The Department Of Health And Human Services | Inhibitors for replication of retroviruses and for the expression of oncogene products |
| US5264423A (en) | 1987-03-25 | 1993-11-23 | The United States Of America As Represented By The Department Of Health And Human Services | Inhibitors for replication of retroviruses and for the expression of oncogene products |
| US4904582A (en) | 1987-06-11 | 1990-02-27 | Synthetic Genetics | Novel amphiphilic nucleic acid conjugates |
| US5552540A (en) | 1987-06-24 | 1996-09-03 | Howard Florey Institute Of Experimental Physiology And Medicine | Nucleoside derivatives |
| US5585481A (en) | 1987-09-21 | 1996-12-17 | Gen-Probe Incorporated | Linking reagents for nucleotide probes |
| US5188897A (en) | 1987-10-22 | 1993-02-23 | Temple University Of The Commonwealth System Of Higher Education | Encapsulated 2',5'-phosphorothioate oligoadenylates |
| US5405939A (en) | 1987-10-22 | 1995-04-11 | Temple University Of The Commonwealth System Of Higher Education | 2',5'-phosphorothioate oligoadenylates and their covalent conjugates with polylysine |
| US5525465A (en) | 1987-10-28 | 1996-06-11 | Howard Florey Institute Of Experimental Physiology And Medicine | Oligonucleotide-polyamide conjugates and methods of production and applications of the same |
| US5112963A (en) | 1987-11-12 | 1992-05-12 | Max-Planck-Gesellschaft Zur Foerderung Der Wissenschaften E.V. | Modified oligonucleotides |
| US5491133A (en) | 1987-11-30 | 1996-02-13 | University Of Iowa Research Foundation | Methods for blocking the expression of specifically targeted genes |
| US5403711A (en) | 1987-11-30 | 1995-04-04 | University Of Iowa Research Foundation | Nucleic acid hybridization and amplification method for detection of specific sequences in which a complementary labeled nucleic acid probe is cleaved |
| US5082830A (en) | 1988-02-26 | 1992-01-21 | Enzo Biochem, Inc. | End labeled nucleotide probe |
| US5519126A (en) | 1988-03-25 | 1996-05-21 | University Of Virginia Alumni Patents Foundation | Oligonucleotide N-alkylphosphoramidates |
| US5453496A (en) | 1988-05-26 | 1995-09-26 | University Patents, Inc. | Polynucleotide phosphorodithioate |
| US5278302A (en) | 1988-05-26 | 1994-01-11 | University Patents, Inc. | Polynucleotide phosphorodithioates |
| US5109124A (en) | 1988-06-01 | 1992-04-28 | Biogen, Inc. | Nucleic acid probe linked to a label having a terminal cysteine |
| US5216141A (en) | 1988-06-06 | 1993-06-01 | Benner Steven A | Oligonucleotide analogs containing sulfur linkages |
| US5175273A (en) | 1988-07-01 | 1992-12-29 | Genentech, Inc. | Nucleic acid intercalating agents |
| US5262536A (en) | 1988-09-15 | 1993-11-16 | E. I. Du Pont De Nemours And Company | Reagents for the preparation of 5'-tagged oligonucleotides |
| US5512439A (en) | 1988-11-21 | 1996-04-30 | Dynal As | Oligonucleotide-linked magnetic particles and uses thereof |
| US5599923A (en) | 1989-03-06 | 1997-02-04 | Board Of Regents, University Of Tx | Texaphyrin metal complexes having improved functionalization |
| US5391723A (en) | 1989-05-31 | 1995-02-21 | Neorx Corporation | Oligonucleotide conjugates |
| US5256775A (en) | 1989-06-05 | 1993-10-26 | Gilead Sciences, Inc. | Exonuclease-resistant oligonucleotides |
| US4958013A (en) | 1989-06-06 | 1990-09-18 | Northwestern University | Cholesteryl modified oligonucleotides |
| US5416203A (en) | 1989-06-06 | 1995-05-16 | Northwestern University | Steroid modified oligonucleotides |
| US5451463A (en) | 1989-08-28 | 1995-09-19 | Clontech Laboratories, Inc. | Non-nucleoside 1,3-diol reagents for labeling synthetic oligonucleotides |
| US5134066A (en) | 1989-08-29 | 1992-07-28 | Monsanto Company | Improved probes using nucleosides containing 3-dezauracil analogs |
| US5254469A (en) | 1989-09-12 | 1993-10-19 | Eastman Kodak Company | Oligonucleotide-enzyme conjugate that can be used as a probe in hybridization assays and polymerase chain reaction procedures |
| US5399676A (en) | 1989-10-23 | 1995-03-21 | Gilead Sciences | Oligonucleotides with inverted polarity |
| US5264564A (en) | 1989-10-24 | 1993-11-23 | Gilead Sciences | Oligonucleotide analogs with novel linkages |
| US5264562A (en) | 1989-10-24 | 1993-11-23 | Gilead Sciences, Inc. | Oligonucleotide analogs with novel linkages |
| US5292873A (en) | 1989-11-29 | 1994-03-08 | The Research Foundation Of State University Of New York | Nucleic acids labeled with naphthoquinone probe |
| US5455233A (en) | 1989-11-30 | 1995-10-03 | University Of North Carolina | Oligoribonucleoside and oligodeoxyribonucleoside boranophosphates |
| US5405938A (en) | 1989-12-20 | 1995-04-11 | Anti-Gene Development Group | Sequence-specific binding polymers for duplex nucleic acids |
| US5130302A (en) | 1989-12-20 | 1992-07-14 | Boron Bilogicals, Inc. | Boronated nucleoside, nucleotide and oligonucleotide compounds, compositions and methods for using same |
| US5166315A (en) | 1989-12-20 | 1992-11-24 | Anti-Gene Development Group | Sequence-specific binding polymers for duplex nucleic acids |
| US5486603A (en) | 1990-01-08 | 1996-01-23 | Gilead Sciences, Inc. | Oligonucleotide having enhanced binding affinity |
| US5587469A (en) | 1990-01-11 | 1996-12-24 | Isis Pharmaceuticals, Inc. | Oligonucleotides containing N-2 substituted purines |
| US5578718A (en) | 1990-01-11 | 1996-11-26 | Isis Pharmaceuticals, Inc. | Thiol-derivatized nucleosides |
| US5750692A (en) | 1990-01-11 | 1998-05-12 | Isis Pharmaceuticals, Inc. | Synthesis of 3-deazapurines |
| US5459255A (en) | 1990-01-11 | 1995-10-17 | Isis Pharmaceuticals, Inc. | N-2 substituted purines |
| US5681941A (en) | 1990-01-11 | 1997-10-28 | Isis Pharmaceuticals, Inc. | Substituted purines and oligonucleotide cross-linking |
| US5220007A (en) | 1990-02-15 | 1993-06-15 | The Worcester Foundation For Experimental Biology | Method of site-specific alteration of RNA and production of encoded polypeptides |
| US5149797A (en) | 1990-02-15 | 1992-09-22 | The Worcester Foundation For Experimental Biology | Method of site-specific alteration of rna and production of encoded polypeptides |
| US5366878A (en) | 1990-02-15 | 1994-11-22 | The Worcester Foundation For Experimental Biology | Method of site-specific alteration of RNA and production of encoded polypeptides |
| US5214136A (en) | 1990-02-20 | 1993-05-25 | Gilead Sciences, Inc. | Anthraquinone-derivatives oligonucleotides |
| US5414077A (en) | 1990-02-20 | 1995-05-09 | Gilead Sciences | Non-nucleoside linkers for convenient attachment of labels to oligonucleotides using standard synthetic methods |
| US5321131A (en) | 1990-03-08 | 1994-06-14 | Hybridon, Inc. | Site-specific functionalization of oligodeoxynucleotides for non-radioactive labelling |
| US5563253A (en) | 1990-03-08 | 1996-10-08 | Worcester Foundation For Biomedical Research | Linear aminoalkylphosphoramidate oligonucleotide derivatives |
| US5541306A (en) | 1990-03-08 | 1996-07-30 | Worcester Foundation For Biomedical Research | Aminoalkylphosphotriester oligonucleotide derivatives |
| US5536821A (en) | 1990-03-08 | 1996-07-16 | Worcester Foundation For Biomedical Research | Aminoalkylphosphorothioamidate oligonucleotide deratives |
| US5470967A (en) | 1990-04-10 | 1995-11-28 | The Dupont Merck Pharmaceutical Company | Oligonucleotide analogs with sulfamate linkages |
| US5514785A (en) | 1990-05-11 | 1996-05-07 | Becton Dickinson And Company | Solid supports for nucleic acid hybridization assays |
| US5610289A (en) | 1990-07-27 | 1997-03-11 | Isis Pharmaceuticals, Inc. | Backbone modified oligonucleotide analogues |
| US5623070A (en) | 1990-07-27 | 1997-04-22 | Isis Pharmaceuticals, Inc. | Heteroatomic oligonucleoside linkages |
| US5218105A (en) | 1990-07-27 | 1993-06-08 | Isis Pharmaceuticals | Polyamine conjugated oligonucleotides |
| US5602240A (en) | 1990-07-27 | 1997-02-11 | Ciba Geigy Ag. | Backbone modified oligonucleotide analogs |
| US5138045A (en) | 1990-07-27 | 1992-08-11 | Isis Pharmaceuticals | Polyamine conjugated oligonucleotides |
| US5541307A (en) | 1990-07-27 | 1996-07-30 | Isis Pharmaceuticals, Inc. | Backbone modified oligonucleotide analogs and solid phase synthesis thereof |
| US5608046A (en) | 1990-07-27 | 1997-03-04 | Isis Pharmaceuticals, Inc. | Conjugated 4'-desmethyl nucleoside analog compounds |
| US5489677A (en) | 1990-07-27 | 1996-02-06 | Isis Pharmaceuticals, Inc. | Oligonucleoside linkages containing adjacent oxygen and nitrogen atoms |
| US5688941A (en) | 1990-07-27 | 1997-11-18 | Isis Pharmaceuticals, Inc. | Methods of making conjugated 4' desmethyl nucleoside analog compounds |
| US5614617A (en) | 1990-07-27 | 1997-03-25 | Isis Pharmaceuticals, Inc. | Nuclease resistant, pyrimidine modified oligonucleotides that detect and modulate gene expression |
| US5677437A (en) | 1990-07-27 | 1997-10-14 | Isis Pharmaceuticals, Inc. | Heteroatomic oligonucleoside linkages |
| US5618704A (en) | 1990-07-27 | 1997-04-08 | Isis Pharmacueticals, Inc. | Backbone-modified oligonucleotide analogs and preparation thereof through radical coupling |
| US5567810A (en) | 1990-08-03 | 1996-10-22 | Sterling Drug, Inc. | Nuclease resistant compounds |
| US5677439A (en) | 1990-08-03 | 1997-10-14 | Sanofi | Oligonucleotide analogues containing phosphate diester linkage substitutes, compositions thereof, and precursor dinucleotide analogues |
| US5245022A (en) | 1990-08-03 | 1993-09-14 | Sterling Drug, Inc. | Exonuclease resistant terminally substituted oligonucleotides |
| US5623065A (en) | 1990-08-13 | 1997-04-22 | Isis Pharmaceuticals, Inc. | Gapped 2' modified oligonucleotides |
| US5177196A (en) | 1990-08-16 | 1993-01-05 | Microprobe Corporation | Oligo (α-arabinofuranosyl nucleotides) and α-arabinofuranosyl precursors thereof |
| US5512667A (en) | 1990-08-28 | 1996-04-30 | Reed; Michael W. | Trifunctional intermediates for preparing 3'-tailed oligonucleotides |
| US5214134A (en) | 1990-09-12 | 1993-05-25 | Sterling Winthrop Inc. | Process of linking nucleosides with a siloxane bridge |
| US5561225A (en) | 1990-09-19 | 1996-10-01 | Southern Research Institute | Polynucleotide analogs containing sulfonate and sulfonamide internucleoside linkages |
| US5596086A (en) | 1990-09-20 | 1997-01-21 | Gilead Sciences, Inc. | Modified internucleoside linkages having one nitrogen and two carbon atoms |
| US5432272A (en) | 1990-10-09 | 1995-07-11 | Benner; Steven A. | Method for incorporating into a DNA or RNA oligonucleotide using nucleotides bearing heterocyclic bases |
| US5510475A (en) | 1990-11-08 | 1996-04-23 | Hybridon, Inc. | Oligonucleotide multiple reporter precursors |
| US5714331A (en) | 1991-05-24 | 1998-02-03 | Buchardt, Deceased; Ole | Peptide nucleic acids having enhanced binding affinity, sequence specificity and solubility |
| US5371241A (en) | 1991-07-19 | 1994-12-06 | Pharmacia P-L Biochemicals Inc. | Fluorescein labelled phosphoramidites |
| US5571799A (en) | 1991-08-12 | 1996-11-05 | Basco, Ltd. | (2'-5') oligoadenylate analogues useful as inhibitors of host-v5.-graft response |
| US5587361A (en) | 1991-10-15 | 1996-12-24 | Isis Pharmaceuticals, Inc. | Oligonucleotides having phosphorothioate linkages of high chiral purity |
| US5484908A (en) | 1991-11-26 | 1996-01-16 | Gilead Sciences, Inc. | Oligonucleotides containing 5-propynyl pyrimidines |
| US5700922A (en) | 1991-12-24 | 1997-12-23 | Isis Pharmaceuticals, Inc. | PNA-DNA-PNA chimeric macromolecules |
| US5565552A (en) | 1992-01-21 | 1996-10-15 | Pharmacyclics, Inc. | Method of expanded porphyrin-oligonucleotide conjugate synthesis |
| US5595726A (en) | 1992-01-21 | 1997-01-21 | Pharmacyclics, Inc. | Chromophore probe for detection of nucleic acid |
| US5587371A (en) | 1992-01-21 | 1996-12-24 | Pharmacyclics, Inc. | Texaphyrin-oligonucleotide conjugates |
| US5633360A (en) | 1992-04-14 | 1997-05-27 | Gilead Sciences, Inc. | Oligonucleotide analogs capable of passive cell membrane permeation |
| US5434257A (en) | 1992-06-01 | 1995-07-18 | Gilead Sciences, Inc. | Binding compentent oligomers containing unsaturated 3',5' and 2',5' linkages |
| US5272250A (en) | 1992-07-10 | 1993-12-21 | Spielvogel Bernard F | Boronated phosphoramidate compounds |
| US5652355A (en) | 1992-07-23 | 1997-07-29 | Worcester Foundation For Experimental Biology | Hybrid oligonucleotide phosphorothioates |
| US5574142A (en) | 1992-12-15 | 1996-11-12 | Microprobe Corporation | Peptide linkers for improved oligonucleotide delivery |
| US5476925A (en) | 1993-02-01 | 1995-12-19 | Northwestern University | Oligodeoxyribonucleotides including 3'-aminonucleoside-phosphoramidate linkages and terminal 3'-amino groups |
| US5466677A (en) | 1993-03-06 | 1995-11-14 | Ciba-Geigy Corporation | Dinucleoside phosphinates and their pharmaceutical compositions |
| US5663312A (en) | 1993-03-31 | 1997-09-02 | Sanofi | Oligonucleotide dimers with amide linkages replacing phosphodiester linkages |
| US5539082A (en) | 1993-04-26 | 1996-07-23 | Nielsen; Peter E. | Peptide nucleic acids |
| US5502177A (en) | 1993-09-17 | 1996-03-26 | Gilead Sciences, Inc. | Pyrimidine derivatives for labeled binding partners |
| US5719262A (en) | 1993-11-22 | 1998-02-17 | Buchardt, Deceased; Ole | Peptide nucleic acids having amino acid side chains |
| US5457187A (en) | 1993-12-08 | 1995-10-10 | Board Of Regents University Of Nebraska | Oligonucleotides containing 5-fluorouracil |
| US5565350A (en) | 1993-12-09 | 1996-10-15 | Thomas Jefferson University | Compounds and methods for site directed mutations in eukaryotic cells |
| US5599928A (en) | 1994-02-15 | 1997-02-04 | Pharmacyclics, Inc. | Texaphyrin compounds having improved functionalization |
| US5596091A (en) | 1994-03-18 | 1997-01-21 | The Regents Of The University Of California | Antisense oligonucleotides comprising 5-aminoalkyl pyrimidine nucleotides |
| US5625050A (en) | 1994-03-31 | 1997-04-29 | Amgen Inc. | Modified oligonucleotides and intermediates useful in nucleic acid therapeutics |
| US5525711A (en) | 1994-05-18 | 1996-06-11 | The United States Of America As Represented By The Secretary Of The Department Of Health And Human Services | Pteridine nucleotide analogs as fluorescent DNA probes |
| US5597696A (en) | 1994-07-18 | 1997-01-28 | Becton Dickinson And Company | Covalent cyanine dye oligonucleotide conjugates |
| US5591584A (en) | 1994-08-25 | 1997-01-07 | Chiron Corporation | N-4 modified pyrimidine deoxynucleotides and oligonucleotide probes synthesized therewith |
| US5580731A (en) | 1994-08-25 | 1996-12-03 | Chiron Corporation | N-4 modified pyrimidine deoxynucleotides and oligonucleotide probes synthesized therewith |
| US5652356A (en) | 1995-08-17 | 1997-07-29 | Hybridon, Inc. | Inverted chimeric and hybrid oligonucleotides |
| WO1999067378A1 (en) | 1998-06-19 | 1999-12-29 | Mcgill University | ANTISENSE OLIGONUCLEOTIDE CONSTRUCTS BASED ON β-ARABINOFURANOSE AND ITS ANALOGUES |
| EP0999270A1 (en) * | 1998-10-09 | 2000-05-10 | Institut National De La Sante Et De La Recherche Medicale (Inserm) | Method of regulating SMN gene expression |
| US7314923B2 (en) | 1999-02-12 | 2008-01-01 | Daiichi Sankyo Company, Limited | Nucleoside and oligonucleotide analogues |
| US7335765B2 (en) | 1999-02-12 | 2008-02-26 | Daiichi Sankyo Company, Limited | Nucleoside and oligonucleotide analogues |
| US20110009471A1 (en) | 1999-02-12 | 2011-01-13 | Daiichi Sankyo Company, Limited | Oligonucleotide analogues and methods utilizing the same |
| US7816333B2 (en) | 1999-02-12 | 2010-10-19 | Daiichi Sankyo Company, Limited | Oligonucleotide analogues and methods utilizing the same |
| US20080281041A1 (en) | 1999-06-07 | 2008-11-13 | Rozema David B | Reversibly Masked Polymers |
| US6287860B1 (en) | 2000-01-20 | 2001-09-11 | Isis Pharmaceuticals, Inc. | Antisense inhibition of MEKK2 expression |
| US20040198687A1 (en) | 2003-04-04 | 2004-10-07 | Rozema David B. | Endosomolytic polymers |
| WO2005042777A2 (en) | 2003-10-24 | 2005-05-12 | Expresson Biosystems Limited | App/ena antisense |
| US20070292408A1 (en) | 2004-12-03 | 2007-12-20 | University Of Massachusetts | Spinal Muscular Atrophy (SMA) treatment via targeting of SMN2 splice site inhibitory sequences |
| US8022193B2 (en) | 2006-01-27 | 2011-09-20 | Isis Pharmaceuticals, Inc. | 6-modified bicyclic nucleic acid analogs |
| US7569686B1 (en) | 2006-01-27 | 2009-08-04 | Isis Pharmaceuticals, Inc. | Compounds and methods for synthesis of bicyclic nucleic acid analogs |
| US7741457B2 (en) | 2006-01-27 | 2010-06-22 | Isis Pharmaceuticals, Inc. | 6-modified bicyclic nucleic acid analogs |
| US7399845B2 (en) | 2006-01-27 | 2008-07-15 | Isis Pharmaceuticals, Inc. | 6-modified bicyclic nucleic acid analogs |
| US20090023890A1 (en) | 2006-08-18 | 2009-01-22 | Monahan Sean D | Membrane Active Heteropolymers |
| US20080152661A1 (en) | 2006-08-18 | 2008-06-26 | Rozema David B | Polyconjugates for In Vivo Delivery of Polynucleotides |
| WO2008022309A2 (en) | 2006-08-18 | 2008-02-21 | F. Hoffmann-La Roche Ag | Polyconjugates for in vivo delivery of polynucleotides |
| US20110086833A1 (en) * | 2008-05-27 | 2011-04-14 | Paushkin Sergey V | Methods for treating spinal muscular atrophy |
| WO2011032109A1 (en) * | 2009-09-11 | 2011-03-17 | Sma Foundation | Biomarkers for spinal muscular atrophy |
| US9209196B2 (en) | 2011-11-30 | 2015-12-08 | Sharp Kabushiki Kaisha | Memory circuit, method of driving the same, nonvolatile storage device using the same, and liquid crystal display device |
Non-Patent Citations (42)
| Title |
|---|
| "Antisense Research and Applications", 1993, CRC PRESS, pages: 276 - 278 |
| "The Concise Encyclopedia of Polymer Science And Engineering", 1990, JOHN WILEY & SONS, pages: 858 - 859 |
| AUTHIER ET AL., FEBS LETT., vol. 389, 1996, pages 55 - 60 |
| BURGHES; MCGOVERN, GENES DEV., vol. 24, no. 15, 2010, pages 1574 - 1579 |
| CROOKE ET AL., J. PHARMACOL. EXP. THER., vol. 277, 1996, pages 923 - 937 |
| DE MESMAEKER ET AL., ACE. CHEM. RES., vol. 28, 1995, pages 366 - 374 |
| DWAINE A. BRAASCH; DAVID R. COREY, BIOCHEMISTRY, vol. 41, no. 14, 2002, pages 4503 - 4510 |
| ENGLISCH ET AL.: "Angewandle Chemie, International Edition", vol. 30, 1991, pages: 613 |
| GEBEYEHU, G. ET AL., NUCL. ACIDS RES., vol. 15, 1987, pages 4513 |
| GENESIS, vol. 30, no. 3, 2001 |
| HEASMAN, J., DEV. BIOL., vol. 243, 2002, pages 209 - 214 |
| HORIE ET AL., NUCLEIC ACIDS SYMP. SER (OXF, vol. 49, 2005, pages 171 - 172 |
| IVERSON, CURR. OPIN. MOL. THER., vol. 3, 2001, pages 235 - 238 |
| KABANOV ET AL., FEBS LETT., vol. 259, 1990, pages 327 - 330 |
| KANHERE, ADITI ET AL.: "Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2", CELL PRESS, vol. 38, no. 5, 11 June 2010 (2010-06-11), pages 675 - 688, XP055177784 * |
| KOIZUMI, CURR. OPIN. MOL. THER., vol. 8, 2006, pages 144 - 149 |
| KORNBERG: "DNA Replication", 1980, W. H. FREEMAN & CO., pages: 75 - 77 |
| LACERRA ET AL., PROC. NATL. ACAD. SCI., vol. 97, 2000, pages 9591 - 9596 |
| LETSINGER ET AL., PROC. NATL. ACAD. SCI. USA, vol. 86, 1989, pages 6553 - 6556 |
| LON ET AL., BIOCHEM., vol. 41, 2002, pages 3457 - 3467 |
| MANCHARAN ET AL., NUCLEOSIDES & NUCLEOTIDES, vol. 14, 1995, pages 969 - 973 |
| MANOHARAN ET AL., ANN. N. Y. ACAD. SCI., vol. 660, 1992, pages 306 - 309 |
| MANOHARAN ET AL., BIOORG. MED. CHEM. LET., vol. 3, 1993, pages 2765 - 2770 |
| MANOHARAN ET AL., BIOORG. MED. CHEM. LET., vol. 4, 1994, pages 1053 - 1060 |
| MANOHARAN ET AL., TETRAHEDRON LETT., vol. 36, 1995, pages 3651 - 3654 |
| MIN ET AL., BIOORG. MED. CHEM. LETT., vol. 12, 2002, pages 2651 - 2654 |
| MISHRA ET AL., BIOCHIM. BIOPHYS. ACTA, vol. 1264, 1995, pages 229 - 237 |
| MORITA ET AL., NUCLEIC ACID RES., vol. 1, 2001, pages 241 - 242 |
| NASEVICIUS ET AL., NAT. GENET., vol. 26, 2000, pages 216 - 220 |
| NIELSEN ET AL., SCIENCE, vol. 254, 1991, pages 1497 |
| NIELSEN ET AL., SCIENCE, vol. 254, 1991, pages 1497 - 1500 |
| OBERHAUSER ET AL., NUCL. ACIDS RES., vol. 20, 1992, pages 533 - 538 |
| SANGHVI: "Antisense Research and Applications", 1993, CRC PRESS, pages: 289 - 302 |
| See also references of EP2850186A4 * |
| SHEA ET AL., NUCL. ACIDS RES., vol. 18, 1990, pages 3777 - 3783 |
| SKORDIS ET AL., PNAS, vol. 100, no. 7, 2003, pages 4114 - 4119 |
| SURONO ET AL., HUM. GENE THER., vol. 15, 2004, pages 749 - 757 |
| SVINARCHUK ET AL., BIOCHIMIE, vol. 75, 1993, pages 49 - 54 |
| WANG ET AL., J. AM. CHEM. SOC., vol. 122, 2000, pages 8595 - 8602 |
| WANG ET AL., J. GENE MED., vol. 12, 2010, pages 354 - 364 |
| WANG, KEVIN C. ET AL.: "Molecular mechanisms of long noncoding RNAs", CELL PRESS, vol. 43, no. 6, 16 September 2011 (2011-09-16), pages 904 - 914, XP028294655 * |
| WILLIAMS ET AL., J NEUROSCI., vol. 29, no. 24, 2009, pages 7633 - 7638 |
Cited By (33)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US10119144B2 (en) | 2010-11-12 | 2018-11-06 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| US9328346B2 (en) | 2010-11-12 | 2016-05-03 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| US11066673B2 (en) | 2010-11-12 | 2021-07-20 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| US10358644B2 (en) | 2010-11-12 | 2019-07-23 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| US9816094B2 (en) | 2010-11-12 | 2017-11-14 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| US9856479B2 (en) | 2010-11-12 | 2018-01-02 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| US9920317B2 (en) | 2010-11-12 | 2018-03-20 | The General Hospital Corporation | Polycomb-associated non-coding RNAs |
| US10053694B2 (en) | 2010-11-12 | 2018-08-21 | The General Hospital Corporation | Polycomb-associated non-coding RNAS |
| US10059941B2 (en) | 2012-05-16 | 2018-08-28 | Translate Bio Ma, Inc. | Compositions and methods for modulating SMN gene family expression |
| US10058623B2 (en) | 2012-05-16 | 2018-08-28 | Translate Bio Ma, Inc. | Compositions and methods for modulating UTRN expression |
| US10837014B2 (en) | 2012-05-16 | 2020-11-17 | Translate Bio Ma, Inc. | Compositions and methods for modulating SMN gene family expression |
| US10655128B2 (en) | 2012-05-16 | 2020-05-19 | Translate Bio Ma, Inc. | Compositions and methods for modulating MECP2 expression |
| US10174323B2 (en) | 2012-05-16 | 2019-01-08 | The General Hospital Corporation | Compositions and methods for modulating ATP2A2 expression |
| US10174315B2 (en) | 2012-05-16 | 2019-01-08 | The General Hospital Corporation | Compositions and methods for modulating hemoglobin gene family expression |
| US11788089B2 (en) | 2012-05-16 | 2023-10-17 | The General Hospital Corporation | Compositions and methods for modulating MECP2 expression |
| US10174328B2 (en) | 2013-10-04 | 2019-01-08 | Translate Bio Ma, Inc. | Compositions and methods for treating amyotrophic lateral sclerosis |
| CN103911346B (en) * | 2014-03-27 | 2016-09-07 | 江苏雄鸣医药科技有限公司 | A kind of cell model for treating spinal muscular atrophy drug screening |
| CN103911346A (en) * | 2014-03-27 | 2014-07-09 | 江苏雄鸣医药科技有限公司 | Method of knocking out spinal muscular atrophy SMN genes and cell model |
| US10858650B2 (en) | 2014-10-30 | 2020-12-08 | The General Hospital Corporation | Methods for modulating ATRX-dependent gene repression |
| US10900036B2 (en) | 2015-03-17 | 2021-01-26 | The General Hospital Corporation | RNA interactome of polycomb repressive complex 1 (PRC1) |
| US10851371B2 (en) | 2015-04-10 | 2020-12-01 | Ionis Pharmaceuticals, Inc. | Modulation of SMN expression |
| US10905709B2 (en) | 2015-08-28 | 2021-02-02 | Sarepta Therapeutics, Inc. | Modified antisense oligomers for exon inclusion in spinal muscular atrophy |
| WO2017040271A1 (en) * | 2015-08-28 | 2017-03-09 | Sarepta Therapeutics, Inc. | Modified antisense oligomers for exon inclusion in spinal muscular atrophy |
| US10357543B2 (en) | 2015-11-16 | 2019-07-23 | Ohio State Innovation Foundation | Methods and compositions for treating disorders and diseases using Survival Motor Neuron (SMN) protein |
| US11198867B2 (en) | 2016-06-16 | 2021-12-14 | Ionis Pharmaceuticals, Inc. | Combinations for the modulation of SMN expression |
| EP3471779A4 (en) * | 2016-06-16 | 2020-07-08 | Ionis Pharmaceuticals, Inc. | Combinations for the modulation of smn expression |
| EP4035659A1 (en) | 2016-11-29 | 2022-08-03 | PureTech LYT, Inc. | Exosomes for delivery of therapeutic agents |
| WO2019048632A1 (en) | 2017-09-08 | 2019-03-14 | Mina Therapeutics Limited | Stabilized hnf4a sarna compositions and methods of use |
| EP4183882A1 (en) | 2017-09-08 | 2023-05-24 | MiNA Therapeutics Limited | Stabilized hnf4a sarna compositions and methods of use |
| EP4219715A2 (en) | 2017-09-08 | 2023-08-02 | MiNA Therapeutics Limited | Stabilized cebpa sarna compositions and methods of use |
| WO2019048645A1 (en) | 2017-09-08 | 2019-03-14 | Mina Therapeutics Limited | Stabilized cebpa sarna compositions and methods of use |
| WO2021032777A1 (en) | 2019-08-19 | 2021-02-25 | Mina Therapeutics Limited | Oligonucleotide conjugate compositions and methods of use |
| US11299737B1 (en) | 2020-02-28 | 2022-04-12 | Ionis Pharmaceuticals, Inc. | Compounds and methods for modulating SMN2 |
Also Published As
| Publication number | Publication date |
|---|---|
| EP2850186B1 (en) | 2018-12-19 |
| KR20150030205A (en) | 2015-03-19 |
| EP2850186A1 (en) | 2015-03-25 |
| DK2850186T3 (en) | 2019-04-08 |
| EA201492123A1 (en) | 2015-10-30 |
| EP2850186A4 (en) | 2016-01-27 |
| CA2873794A1 (en) | 2013-11-21 |
| ZA201409228B (en) | 2016-07-27 |
| US20150252364A1 (en) | 2015-09-10 |
| US10059941B2 (en) | 2018-08-28 |
| HK1208700A1 (en) | 2016-03-11 |
| SG11201407483YA (en) | 2014-12-30 |
| CN104540947A (en) | 2015-04-22 |
| JP2015523854A (en) | 2015-08-20 |
| AU2013262649A1 (en) | 2015-01-22 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| US11788089B2 (en) | Compositions and methods for modulating MECP2 expression | |
| US10059941B2 (en) | Compositions and methods for modulating SMN gene family expression | |
| US10174328B2 (en) | Compositions and methods for treating amyotrophic lateral sclerosis | |
| US10058623B2 (en) | Compositions and methods for modulating UTRN expression | |
| US10174323B2 (en) | Compositions and methods for modulating ATP2A2 expression | |
| WO2013173647A1 (en) | Compositions and methods for modulating apoa1 and abca1 expression | |
| US10837014B2 (en) | Compositions and methods for modulating SMN gene family expression | |
| EP2850188A1 (en) | Compositions and methods for modulating hemoglobin gene family expression | |
| EP2850183A1 (en) | Compositions and methods for modulating gene expression | |
| WO2013173605A1 (en) | Compositions and methods for modulating pten expression | |
| EP3004354A1 (en) | Compositions and methods for modulating foxp3 expression | |
| WO2013173601A1 (en) | Compositions and methods for modulating bdnf expression |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 13790819 Country of ref document: EP Kind code of ref document: A1 |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 235676 Country of ref document: IL |
|
| ENP | Entry into the national phase |
Ref document number: 2015512858 Country of ref document: JP Kind code of ref document: A Ref document number: 2873794 Country of ref document: CA |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 14401194 Country of ref document: US |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 2013790819 Country of ref document: EP |
|
| ENP | Entry into the national phase |
Ref document number: 20147035185 Country of ref document: KR Kind code of ref document: A |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 201492123 Country of ref document: EA |
|
| ENP | Entry into the national phase |
Ref document number: 2013262649 Country of ref document: AU Date of ref document: 20130516 Kind code of ref document: A |
|
| REG | Reference to national code |
Ref country code: BR Ref legal event code: B01A Ref document number: 112014028637 Country of ref document: BR |
|
| ENP | Entry into the national phase |
Ref document number: 112014028637 Country of ref document: BR Kind code of ref document: A2 Effective date: 20141117 |