WO2000018929A2 - Paramyxovirus vaccines - Google Patents
Paramyxovirus vaccines Download PDFInfo
- Publication number
- WO2000018929A2 WO2000018929A2 PCT/EP1999/007004 EP9907004W WO0018929A2 WO 2000018929 A2 WO2000018929 A2 WO 2000018929A2 EP 9907004 W EP9907004 W EP 9907004W WO 0018929 A2 WO0018929 A2 WO 0018929A2
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- protein
- rsv
- piv3
- muv
- heterochimeric
- Prior art date
Links
- 229960005486 vaccine Drugs 0.000 title claims abstract description 30
- 108090000623 proteins and genes Proteins 0.000 claims abstract description 156
- 102000004169 proteins and genes Human genes 0.000 claims abstract description 144
- 239000012634 fragment Substances 0.000 claims abstract description 125
- 230000002163 immunogen Effects 0.000 claims abstract description 58
- 210000004978 chinese hamster ovary cell Anatomy 0.000 claims abstract description 24
- 230000004927 fusion Effects 0.000 claims abstract description 9
- 239000000203 mixture Substances 0.000 claims abstract description 6
- 208000023504 respiratory system disease Diseases 0.000 claims abstract 2
- 150000001413 amino acids Chemical class 0.000 claims description 75
- 239000013598 vector Substances 0.000 claims description 72
- 210000004027 cell Anatomy 0.000 claims description 66
- 108010068327 4-hydroxyphenylpyruvate dioxygenase Proteins 0.000 claims description 51
- 101710133291 Hemagglutinin-neuraminidase Proteins 0.000 claims description 40
- 108020004414 DNA Proteins 0.000 claims description 34
- 239000012528 membrane Substances 0.000 claims description 33
- 108091006027 G proteins Proteins 0.000 claims description 23
- 108091000058 GTP-Binding Proteins 0.000 claims description 23
- 108020004705 Codon Proteins 0.000 claims description 22
- 102000030782 GTP binding Human genes 0.000 claims description 21
- 102000044159 Ubiquitin Human genes 0.000 claims description 18
- 108090000848 Ubiquitin Proteins 0.000 claims description 18
- 238000000034 method Methods 0.000 claims description 18
- 241000238631 Hexapoda Species 0.000 claims description 17
- 108010076504 Protein Sorting Signals Proteins 0.000 claims description 15
- 238000003776 cleavage reaction Methods 0.000 claims description 14
- 230000007017 scission Effects 0.000 claims description 14
- 238000004519 manufacturing process Methods 0.000 claims description 12
- 241001465754 Metazoa Species 0.000 claims description 11
- 210000004899 c-terminal region Anatomy 0.000 claims description 10
- 229920002704 polyhistidine Polymers 0.000 claims description 10
- 108020004511 Recombinant DNA Proteins 0.000 claims description 9
- 230000008569 process Effects 0.000 claims description 9
- 101710169105 Minor spike protein Proteins 0.000 claims description 8
- 101710081079 Minor spike protein H Proteins 0.000 claims description 8
- 241000711504 Paramyxoviridae Species 0.000 claims description 7
- 208000036142 Viral infection Diseases 0.000 claims description 4
- 239000013604 expression vector Substances 0.000 claims description 4
- 229940035032 monophosphoryl lipid a Drugs 0.000 claims description 4
- 230000009385 viral infection Effects 0.000 claims description 4
- 239000003937 drug carrier Substances 0.000 claims description 2
- 239000003814 drug Substances 0.000 claims 2
- 239000007764 o/w emulsion Substances 0.000 claims 1
- 102000003886 Glycoproteins Human genes 0.000 abstract description 7
- 108090000288 Glycoproteins Proteins 0.000 abstract description 7
- 239000000427 antigen Substances 0.000 abstract description 6
- 102000036639 antigens Human genes 0.000 abstract description 6
- 108091007433 antigens Proteins 0.000 abstract description 6
- 230000003612 virological effect Effects 0.000 abstract description 5
- 235000018102 proteins Nutrition 0.000 description 107
- 239000013612 plasmid Substances 0.000 description 75
- 241000711386 Mumps virus Species 0.000 description 71
- 235000001014 amino acid Nutrition 0.000 description 71
- 229940024606 amino acid Drugs 0.000 description 71
- 230000014509 gene expression Effects 0.000 description 62
- 102000037865 fusion proteins Human genes 0.000 description 42
- 108020001507 fusion proteins Proteins 0.000 description 42
- 241000712079 Measles morbillivirus Species 0.000 description 32
- 210000004379 membrane Anatomy 0.000 description 31
- 241000700605 Viruses Species 0.000 description 27
- 239000000047 product Substances 0.000 description 21
- 210000002966 serum Anatomy 0.000 description 21
- 241000701447 unidentified baculovirus Species 0.000 description 21
- 238000003752 polymerase chain reaction Methods 0.000 description 20
- 108091026890 Coding region Proteins 0.000 description 18
- 238000002965 ELISA Methods 0.000 description 18
- 230000029087 digestion Effects 0.000 description 18
- 108091034117 Oligonucleotide Proteins 0.000 description 17
- JLCPHMBAVCMARE-UHFFFAOYSA-N [3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-[[3-[[3-[[3-[[3-[[3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-hydroxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methyl [5-(6-aminopurin-9-yl)-2-(hydroxymethyl)oxolan-3-yl] hydrogen phosphate Polymers Cc1cn(C2CC(OP(O)(=O)OCC3OC(CC3OP(O)(=O)OCC3OC(CC3O)n3cnc4c3nc(N)[nH]c4=O)n3cnc4c3nc(N)[nH]c4=O)C(COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3CO)n3cnc4c(N)ncnc34)n3ccc(N)nc3=O)n3cnc4c(N)ncnc34)n3ccc(N)nc3=O)n3ccc(N)nc3=O)n3ccc(N)nc3=O)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cc(C)c(=O)[nH]c3=O)n3cc(C)c(=O)[nH]c3=O)n3ccc(N)nc3=O)n3cc(C)c(=O)[nH]c3=O)n3cnc4c3nc(N)[nH]c4=O)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)O2)c(=O)[nH]c1=O JLCPHMBAVCMARE-UHFFFAOYSA-N 0.000 description 16
- 239000002609 medium Substances 0.000 description 16
- 238000010276 construction Methods 0.000 description 15
- 240000004808 Saccharomyces cerevisiae Species 0.000 description 14
- 235000014680 Saccharomyces cerevisiae Nutrition 0.000 description 14
- 108700010070 Codon Usage Proteins 0.000 description 13
- 241000283973 Oryctolagus cuniculus Species 0.000 description 12
- 230000000295 complement effect Effects 0.000 description 12
- 241000699670 Mus sp. Species 0.000 description 11
- 102000003978 Tissue Plasminogen Activator Human genes 0.000 description 11
- 108090000373 Tissue Plasminogen Activator Proteins 0.000 description 11
- 208000015181 infectious disease Diseases 0.000 description 11
- 210000004962 mammalian cell Anatomy 0.000 description 11
- 229960000187 tissue plasminogen activator Drugs 0.000 description 11
- 102100029727 Enteropeptidase Human genes 0.000 description 10
- 108010013369 Enteropeptidase Proteins 0.000 description 10
- 108091028043 Nucleic acid sequence Proteins 0.000 description 10
- 239000002671 adjuvant Substances 0.000 description 10
- 108010088716 attachment protein G Proteins 0.000 description 10
- 239000001963 growth medium Substances 0.000 description 10
- 239000002299 complementary DNA Substances 0.000 description 9
- 102000005396 glutamine synthetase Human genes 0.000 description 9
- 108020002326 glutamine synthetase Proteins 0.000 description 9
- 150000007523 nucleic acids Chemical group 0.000 description 9
- 241000283707 Capra Species 0.000 description 8
- 230000000694 effects Effects 0.000 description 8
- 238000005516 engineering process Methods 0.000 description 8
- 230000004224 protection Effects 0.000 description 8
- 230000004044 response Effects 0.000 description 8
- 238000012163 sequencing technique Methods 0.000 description 8
- 108091032973 (ribonucleotides)n+m Proteins 0.000 description 7
- 241000699666 Mus <mouse, genus> Species 0.000 description 7
- 125000000539 amino acid group Chemical group 0.000 description 7
- 238000011161 development Methods 0.000 description 7
- 230000018109 developmental process Effects 0.000 description 7
- 230000028993 immune response Effects 0.000 description 7
- 238000001890 transfection Methods 0.000 description 7
- 241000894006 Bacteria Species 0.000 description 6
- 206010046865 Vaccinia virus infection Diseases 0.000 description 6
- 238000010367 cloning Methods 0.000 description 6
- 238000002347 injection Methods 0.000 description 6
- 239000007924 injection Substances 0.000 description 6
- SXTAYKAGBXMACB-UHFFFAOYSA-N methionine S-imide-S-oxide Natural products CS(=N)(=O)CCC(N)C(O)=O SXTAYKAGBXMACB-UHFFFAOYSA-N 0.000 description 6
- 238000000746 purification Methods 0.000 description 6
- 230000028327 secretion Effects 0.000 description 6
- 208000007089 vaccinia Diseases 0.000 description 6
- 238000001262 western blot Methods 0.000 description 6
- 238000012286 ELISA Assay Methods 0.000 description 5
- 101710141347 Major envelope glycoprotein Proteins 0.000 description 5
- 108010090054 Membrane Glycoproteins Proteins 0.000 description 5
- 102000012750 Membrane Glycoproteins Human genes 0.000 description 5
- 102100025243 Myeloid cell surface antigen CD33 Human genes 0.000 description 5
- 101710182846 Polyhedrin Proteins 0.000 description 5
- 238000004458 analytical method Methods 0.000 description 5
- 230000005875 antibody response Effects 0.000 description 5
- 230000002238 attenuated effect Effects 0.000 description 5
- 230000001580 bacterial effect Effects 0.000 description 5
- 239000013599 cloning vector Substances 0.000 description 5
- 230000013595 glycosylation Effects 0.000 description 5
- 238000006206 glycosylation reaction Methods 0.000 description 5
- 238000002649 immunization Methods 0.000 description 5
- 239000002502 liposome Substances 0.000 description 5
- 230000003472 neutralizing effect Effects 0.000 description 5
- 238000005457 optimization Methods 0.000 description 5
- 238000002360 preparation method Methods 0.000 description 5
- 239000013605 shuttle vector Substances 0.000 description 5
- MFBOGIVSZKQAPD-UHFFFAOYSA-M sodium butyrate Chemical compound [Na+].CCCC([O-])=O MFBOGIVSZKQAPD-UHFFFAOYSA-M 0.000 description 5
- 238000012546 transfer Methods 0.000 description 5
- FERIUCNNQQJTOY-UHFFFAOYSA-M Butyrate Chemical compound CCCC([O-])=O FERIUCNNQQJTOY-UHFFFAOYSA-M 0.000 description 4
- FERIUCNNQQJTOY-UHFFFAOYSA-N Butyric acid Natural products CCCC(O)=O FERIUCNNQQJTOY-UHFFFAOYSA-N 0.000 description 4
- 241000255581 Drosophila <fruit fly, genus> Species 0.000 description 4
- CKLJMWTZIZZHCS-REOHCLBHSA-N L-aspartic acid Chemical compound OC(=O)[C@@H](N)CC(O)=O CKLJMWTZIZZHCS-REOHCLBHSA-N 0.000 description 4
- 208000005647 Mumps Diseases 0.000 description 4
- 241000710961 Semliki Forest virus Species 0.000 description 4
- HVYWMOMLDIMFJA-DPAQBDIFSA-N cholesterol Chemical compound C1C=C2C[C@@H](O)CC[C@]2(C)[C@@H]2[C@@H]1[C@@H]1CC[C@H]([C@H](C)CCCC(C)C)[C@@]1(C)CC2 HVYWMOMLDIMFJA-DPAQBDIFSA-N 0.000 description 4
- 230000003053 immunization Effects 0.000 description 4
- 208000010805 mumps infectious disease Diseases 0.000 description 4
- 230000002797 proteolythic effect Effects 0.000 description 4
- 238000002415 sodium dodecyl sulfate polyacrylamide gel electrophoresis Methods 0.000 description 4
- 101710160621 Fusion glycoprotein F0 Proteins 0.000 description 3
- 101710154606 Hemagglutinin Proteins 0.000 description 3
- QNAYBMKLOCPYGJ-REOHCLBHSA-N L-alanine Chemical compound C[C@H](N)C(O)=O QNAYBMKLOCPYGJ-REOHCLBHSA-N 0.000 description 3
- 101710093908 Outer capsid protein VP4 Proteins 0.000 description 3
- 101710135467 Outer capsid protein sigma-1 Proteins 0.000 description 3
- 101710176177 Protein A56 Proteins 0.000 description 3
- 102000007056 Recombinant Fusion Proteins Human genes 0.000 description 3
- 108010008281 Recombinant Fusion Proteins Proteins 0.000 description 3
- 235000004279 alanine Nutrition 0.000 description 3
- 230000010056 antibody-dependent cellular cytotoxicity Effects 0.000 description 3
- 229960000074 biopharmaceutical Drugs 0.000 description 3
- 238000004587 chromatography analysis Methods 0.000 description 3
- 210000003527 eukaryotic cell Anatomy 0.000 description 3
- 238000002474 experimental method Methods 0.000 description 3
- 230000036541 health Effects 0.000 description 3
- 238000010438 heat treatment Methods 0.000 description 3
- 239000000185 hemagglutinin Substances 0.000 description 3
- 230000008348 humoral response Effects 0.000 description 3
- 230000035800 maturation Effects 0.000 description 3
- 244000052769 pathogen Species 0.000 description 3
- 239000002243 precursor Substances 0.000 description 3
- 108090000765 processed proteins & peptides Proteins 0.000 description 3
- 238000003156 radioimmunoprecipitation Methods 0.000 description 3
- 210000002345 respiratory system Anatomy 0.000 description 3
- 229920006395 saturated elastomer Polymers 0.000 description 3
- 229940031626 subunit vaccine Drugs 0.000 description 3
- 210000003501 vero cell Anatomy 0.000 description 3
- 206010011416 Croup infectious Diseases 0.000 description 2
- 230000004544 DNA amplification Effects 0.000 description 2
- IAZDPXIOMUYVGZ-UHFFFAOYSA-N Dimethylsulphoxide Chemical compound CS(C)=O IAZDPXIOMUYVGZ-UHFFFAOYSA-N 0.000 description 2
- 101100456896 Drosophila melanogaster metl gene Proteins 0.000 description 2
- 241000701024 Human betaherpesvirus 5 Species 0.000 description 2
- FBOZXECLQNJBKD-ZDUSSCGKSA-N L-methotrexate Chemical compound C=1N=C2N=C(N)N=C(N)C2=NC=1CN(C)C1=CC=C(C(=O)N[C@@H](CCC(O)=O)C(O)=O)C=C1 FBOZXECLQNJBKD-ZDUSSCGKSA-N 0.000 description 2
- 201000005505 Measles Diseases 0.000 description 2
- 125000001429 N-terminal alpha-amino-acid group Chemical group 0.000 description 2
- 108010006232 Neuraminidase Proteins 0.000 description 2
- 102000005348 Neuraminidase Human genes 0.000 description 2
- 208000002606 Paramyxoviridae Infections Diseases 0.000 description 2
- NBIIXXVUZAFLBC-UHFFFAOYSA-N Phosphoric acid Chemical compound OP(O)(O)=O NBIIXXVUZAFLBC-UHFFFAOYSA-N 0.000 description 2
- 229920002684 Sepharose Polymers 0.000 description 2
- FAPWRFPIFSIZLT-UHFFFAOYSA-M Sodium chloride Chemical compound [Na+].[Cl-] FAPWRFPIFSIZLT-UHFFFAOYSA-M 0.000 description 2
- UZQJVUCHXGYFLQ-AYDHOLPZSA-N [(2s,3r,4s,5r,6r)-4-[(2s,3r,4s,5r,6r)-4-[(2r,3r,4s,5r,6r)-4-[(2s,3r,4s,5r,6r)-3,5-dihydroxy-6-(hydroxymethyl)-4-[(2s,3r,4s,5s,6r)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxyoxan-2-yl]oxy-3,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-3,5-dihydroxy-6-(hy Chemical compound O([C@H]1[C@H](O)[C@@H](CO)O[C@H]([C@@H]1O)O[C@H]1[C@H](O)[C@@H](CO)O[C@H]([C@@H]1O)O[C@H]1CC[C@]2(C)[C@H]3CC=C4[C@@]([C@@]3(CC[C@H]2[C@@]1(C=O)C)C)(C)CC(O)[C@]1(CCC(CC14)(C)C)C(=O)O[C@H]1[C@@H]([C@@H](O[C@H]2[C@@H]([C@@H](O[C@H]3[C@@H]([C@@H](O[C@H]4[C@@H]([C@@H](O[C@H]5[C@@H]([C@@H](O)[C@H](O)[C@@H](CO)O5)O)[C@H](O)[C@@H](CO)O4)O)[C@H](O)[C@@H](CO)O3)O)[C@H](O)[C@@H](CO)O2)O)[C@H](O)[C@@H](CO)O1)O)[C@@H]1O[C@H](CO)[C@@H](O)[C@H](O)[C@H]1O UZQJVUCHXGYFLQ-AYDHOLPZSA-N 0.000 description 2
- 230000003321 amplification Effects 0.000 description 2
- 230000000890 antigenic effect Effects 0.000 description 2
- 230000015572 biosynthetic process Effects 0.000 description 2
- 239000000872 buffer Substances 0.000 description 2
- 150000001720 carbohydrates Chemical group 0.000 description 2
- 230000007910 cell fusion Effects 0.000 description 2
- 230000001413 cellular effect Effects 0.000 description 2
- 239000003795 chemical substances by application Substances 0.000 description 2
- 235000012000 cholesterol Nutrition 0.000 description 2
- 230000008878 coupling Effects 0.000 description 2
- 238000010168 coupling process Methods 0.000 description 2
- 238000005859 coupling reaction Methods 0.000 description 2
- 201000010549 croup Diseases 0.000 description 2
- 230000003412 degenerative effect Effects 0.000 description 2
- 229960001760 dimethyl sulfoxide Drugs 0.000 description 2
- 208000037265 diseases, disorders, signs and symptoms Diseases 0.000 description 2
- 238000004520 electroporation Methods 0.000 description 2
- 239000000839 emulsion Substances 0.000 description 2
- 238000005538 encapsulation Methods 0.000 description 2
- 230000002349 favourable effect Effects 0.000 description 2
- 239000003228 hemolysin Substances 0.000 description 2
- 238000003119 immunoblot Methods 0.000 description 2
- 238000001114 immunoprecipitation Methods 0.000 description 2
- 230000006872 improvement Effects 0.000 description 2
- 238000001727 in vivo Methods 0.000 description 2
- 238000005304 joining Methods 0.000 description 2
- 238000011068 loading method Methods 0.000 description 2
- 229920002521 macromolecule Polymers 0.000 description 2
- 229960000485 methotrexate Drugs 0.000 description 2
- 201000009240 nasopharyngitis Diseases 0.000 description 2
- 239000013642 negative control Substances 0.000 description 2
- 238000003199 nucleic acid amplification method Methods 0.000 description 2
- 108020004707 nucleic acids Proteins 0.000 description 2
- 102000039446 nucleic acids Human genes 0.000 description 2
- 230000001717 pathogenic effect Effects 0.000 description 2
- 239000008363 phosphate buffer Substances 0.000 description 2
- 102000004196 processed proteins & peptides Human genes 0.000 description 2
- 230000001681 protective effect Effects 0.000 description 2
- 230000000241 respiratory effect Effects 0.000 description 2
- 125000003607 serino group Chemical group [H]N([H])[C@]([H])(C(=O)[*])C(O[H])([H])[H] 0.000 description 2
- 238000002741 site-directed mutagenesis Methods 0.000 description 2
- 239000011734 sodium Substances 0.000 description 2
- 230000010473 stable expression Effects 0.000 description 2
- 238000006467 substitution reaction Methods 0.000 description 2
- 238000003786 synthesis reaction Methods 0.000 description 2
- 229960000814 tetanus toxoid Drugs 0.000 description 2
- 230000014621 translational initiation Effects 0.000 description 2
- XLYOFNOQVPJJNP-UHFFFAOYSA-N water Substances O XLYOFNOQVPJJNP-UHFFFAOYSA-N 0.000 description 2
- 108010042708 Acetylmuramyl-Alanyl-Isoglutamine Proteins 0.000 description 1
- 241000710929 Alphavirus Species 0.000 description 1
- 102100021257 Beta-secretase 1 Human genes 0.000 description 1
- 101710150192 Beta-secretase 1 Proteins 0.000 description 1
- 241000588807 Bordetella Species 0.000 description 1
- 108091003079 Bovine Serum Albumin Proteins 0.000 description 1
- 102000014914 Carrier Proteins Human genes 0.000 description 1
- 108010078791 Carrier Proteins Proteins 0.000 description 1
- 206010010741 Conjunctivitis Diseases 0.000 description 1
- 238000011537 Coomassie blue staining Methods 0.000 description 1
- 206010011224 Cough Diseases 0.000 description 1
- 241000699802 Cricetulus griseus Species 0.000 description 1
- 102000053602 DNA Human genes 0.000 description 1
- 206010011878 Deafness Diseases 0.000 description 1
- 241000702421 Dependoparvovirus Species 0.000 description 1
- 102100024746 Dihydrofolate reductase Human genes 0.000 description 1
- 241000709661 Enterovirus Species 0.000 description 1
- 241000991587 Enterovirus C Species 0.000 description 1
- 101710121417 Envelope glycoprotein Proteins 0.000 description 1
- 208000000832 Equine Encephalomyelitis Diseases 0.000 description 1
- 102000003951 Erythropoietin Human genes 0.000 description 1
- 108090000394 Erythropoietin Proteins 0.000 description 1
- 208000000666 Fowlpox Diseases 0.000 description 1
- 241000287828 Gallus gallus Species 0.000 description 1
- 108700039691 Genetic Promoter Regions Proteins 0.000 description 1
- 108010043121 Green Fluorescent Proteins Proteins 0.000 description 1
- 102000004144 Green Fluorescent Proteins Human genes 0.000 description 1
- 101000987586 Homo sapiens Eosinophil peroxidase Proteins 0.000 description 1
- 101000920686 Homo sapiens Erythropoietin Proteins 0.000 description 1
- 241000701085 Human alphaherpesvirus 3 Species 0.000 description 1
- 102000004856 Lectins Human genes 0.000 description 1
- 108090001090 Lectins Proteins 0.000 description 1
- 241000186781 Listeria Species 0.000 description 1
- 241000124008 Mammalia Species 0.000 description 1
- 201000009906 Meningitis Diseases 0.000 description 1
- 241000712045 Morbillivirus Species 0.000 description 1
- 239000000020 Nitrocellulose Substances 0.000 description 1
- 230000004989 O-glycosylation Effects 0.000 description 1
- 206010034038 Parotitis Diseases 0.000 description 1
- ISWSIDIOOBJBQZ-UHFFFAOYSA-N Phenol Chemical compound OC1=CC=CC=C1 ISWSIDIOOBJBQZ-UHFFFAOYSA-N 0.000 description 1
- 241000709664 Picornaviridae Species 0.000 description 1
- 241000711902 Pneumovirus Species 0.000 description 1
- 206010037660 Pyrexia Diseases 0.000 description 1
- 208000036002 Rash generalised Diseases 0.000 description 1
- 101000702488 Rattus norvegicus High affinity cationic amino acid transporter 1 Proteins 0.000 description 1
- 208000035415 Reinfection Diseases 0.000 description 1
- 206010062106 Respiratory tract infection viral Diseases 0.000 description 1
- 241001533467 Rubulavirus Species 0.000 description 1
- 241000607142 Salmonella Species 0.000 description 1
- 206010039491 Sarcoma Diseases 0.000 description 1
- 241000607768 Shigella Species 0.000 description 1
- 241000144282 Sigmodon Species 0.000 description 1
- 241000710960 Sindbis virus Species 0.000 description 1
- 102100021696 Syncytin-1 Human genes 0.000 description 1
- 241000700618 Vaccinia virus Species 0.000 description 1
- 108010059722 Viral Fusion Proteins Proteins 0.000 description 1
- 230000002411 adverse Effects 0.000 description 1
- 239000000443 aerosol Substances 0.000 description 1
- 229940037003 alum Drugs 0.000 description 1
- XAGFODPZIPBFFR-UHFFFAOYSA-N aluminium Chemical compound [Al] XAGFODPZIPBFFR-UHFFFAOYSA-N 0.000 description 1
- 229910052782 aluminium Inorganic materials 0.000 description 1
- 239000004411 aluminium Substances 0.000 description 1
- WNROFYMDJYEPJX-UHFFFAOYSA-K aluminium hydroxide Chemical compound [OH-].[OH-].[OH-].[Al+3] WNROFYMDJYEPJX-UHFFFAOYSA-K 0.000 description 1
- 229910021502 aluminium hydroxide Inorganic materials 0.000 description 1
- 229910000147 aluminium phosphate Inorganic materials 0.000 description 1
- 238000010171 animal model Methods 0.000 description 1
- 239000007864 aqueous solution Substances 0.000 description 1
- 229940009098 aspartate Drugs 0.000 description 1
- 238000003556 assay Methods 0.000 description 1
- 239000001506 calcium phosphate Substances 0.000 description 1
- 229910000389 calcium phosphate Inorganic materials 0.000 description 1
- 235000011010 calcium phosphates Nutrition 0.000 description 1
- 235000014633 carbohydrates Nutrition 0.000 description 1
- 238000004113 cell culture Methods 0.000 description 1
- 210000000170 cell membrane Anatomy 0.000 description 1
- 238000007385 chemical modification Methods 0.000 description 1
- 238000006243 chemical reaction Methods 0.000 description 1
- 239000003153 chemical reaction reagent Substances 0.000 description 1
- 239000003638 chemical reducing agent Substances 0.000 description 1
- 238000011097 chromatography purification Methods 0.000 description 1
- 238000000975 co-precipitation Methods 0.000 description 1
- 150000001875 compounds Chemical class 0.000 description 1
- 230000021615 conjugation Effects 0.000 description 1
- 210000000795 conjunctiva Anatomy 0.000 description 1
- 238000012258 culturing Methods 0.000 description 1
- 231100000433 cytotoxic Toxicity 0.000 description 1
- 230000001472 cytotoxic effect Effects 0.000 description 1
- 231100000895 deafness Toxicity 0.000 description 1
- 108020001096 dihydrofolate reductase Proteins 0.000 description 1
- 201000010099 disease Diseases 0.000 description 1
- 230000006806 disease prevention Effects 0.000 description 1
- 208000035475 disorder Diseases 0.000 description 1
- 238000001962 electrophoresis Methods 0.000 description 1
- 206010014599 encephalitis Diseases 0.000 description 1
- 230000002708 enhancing effect Effects 0.000 description 1
- 230000002255 enzymatic effect Effects 0.000 description 1
- 210000002919 epithelial cell Anatomy 0.000 description 1
- 229940105423 erythropoietin Drugs 0.000 description 1
- 238000012869 ethanol precipitation Methods 0.000 description 1
- 239000013613 expression plasmid Substances 0.000 description 1
- 238000000605 extraction Methods 0.000 description 1
- 239000012894 fetal calf serum Substances 0.000 description 1
- 238000004108 freeze drying Methods 0.000 description 1
- 230000002538 fungal effect Effects 0.000 description 1
- 238000002523 gelfiltration Methods 0.000 description 1
- 210000004907 gland Anatomy 0.000 description 1
- 150000004676 glycans Chemical class 0.000 description 1
- 102000035122 glycosylated proteins Human genes 0.000 description 1
- 108091005608 glycosylated proteins Proteins 0.000 description 1
- 239000005090 green fluorescent protein Substances 0.000 description 1
- 230000035931 haemagglutination Effects 0.000 description 1
- 238000003306 harvesting Methods 0.000 description 1
- 208000016354 hearing loss disease Diseases 0.000 description 1
- 230000009083 hemolysis by symbiont of host erythrocytes Effects 0.000 description 1
- 208000002672 hepatitis B Diseases 0.000 description 1
- 239000013628 high molecular weight specie Substances 0.000 description 1
- 125000000487 histidyl group Chemical group [H]N([H])C(C(=O)O*)C([H])([H])C1=C([H])N([H])C([H])=N1 0.000 description 1
- 102000044890 human EPO Human genes 0.000 description 1
- 230000005847 immunogenicity Effects 0.000 description 1
- 230000002480 immunoprotective effect Effects 0.000 description 1
- 230000003308 immunostimulating effect Effects 0.000 description 1
- 238000000338 in vitro Methods 0.000 description 1
- 229940031551 inactivated vaccine Drugs 0.000 description 1
- 238000011534 incubation Methods 0.000 description 1
- 230000006698 induction Effects 0.000 description 1
- 238000011081 inoculation Methods 0.000 description 1
- 238000003780 insertion Methods 0.000 description 1
- 230000037431 insertion Effects 0.000 description 1
- 239000000543 intermediate Substances 0.000 description 1
- 238000007918 intramuscular administration Methods 0.000 description 1
- 238000002356 laser light scattering Methods 0.000 description 1
- 239000002523 lectin Substances 0.000 description 1
- 208000030500 lower respiratory tract disease Diseases 0.000 description 1
- 210000004698 lymphocyte Anatomy 0.000 description 1
- 244000005700 microbiome Species 0.000 description 1
- 239000011859 microparticle Substances 0.000 description 1
- 238000013508 migration Methods 0.000 description 1
- 230000005012 migration Effects 0.000 description 1
- 230000004048 modification Effects 0.000 description 1
- 238000012986 modification Methods 0.000 description 1
- 238000010369 molecular cloning Methods 0.000 description 1
- 229940031348 multivalent vaccine Drugs 0.000 description 1
- BSOQXXWZTUDTEL-ZUYCGGNHSA-N muramyl dipeptide Chemical compound OC(=O)CC[C@H](C(N)=O)NC(=O)[C@H](C)NC(=O)[C@@H](C)O[C@H]1[C@H](O)[C@@H](CO)O[C@@H](O)[C@@H]1NC(C)=O BSOQXXWZTUDTEL-ZUYCGGNHSA-N 0.000 description 1
- 238000011587 new zealand white rabbit Methods 0.000 description 1
- 229920001220 nitrocellulos Polymers 0.000 description 1
- 150000002482 oligosaccharides Polymers 0.000 description 1
- 210000003300 oropharynx Anatomy 0.000 description 1
- 210000001672 ovary Anatomy 0.000 description 1
- 210000003681 parotid gland Anatomy 0.000 description 1
- 238000010647 peptide synthesis reaction Methods 0.000 description 1
- 229920001184 polypeptide Polymers 0.000 description 1
- OXCMYAYHXIHQOA-UHFFFAOYSA-N potassium;[2-butyl-5-chloro-3-[[4-[2-(1,2,4-triaza-3-azanidacyclopenta-1,4-dien-5-yl)phenyl]phenyl]methyl]imidazol-4-yl]methanol Chemical compound [K+].CCCCC1=NC(Cl)=C(CO)N1CC1=CC=C(C=2C(=CC=CC=2)C2=N[N-]N=N2)C=C1 OXCMYAYHXIHQOA-UHFFFAOYSA-N 0.000 description 1
- 238000010814 radioimmunoprecipitation assay Methods 0.000 description 1
- 230000008707 rearrangement Effects 0.000 description 1
- 230000010076 replication Effects 0.000 description 1
- 210000001533 respiratory mucosa Anatomy 0.000 description 1
- 201000005404 rubella Diseases 0.000 description 1
- 229930182490 saponin Natural products 0.000 description 1
- 150000007949 saponins Chemical class 0.000 description 1
- 235000017709 saponins Nutrition 0.000 description 1
- 239000012679 serum free medium Substances 0.000 description 1
- 125000005629 sialic acid group Chemical group 0.000 description 1
- 239000011780 sodium chloride Substances 0.000 description 1
- 239000000243 solution Substances 0.000 description 1
- 238000010561 standard procedure Methods 0.000 description 1
- 239000007858 starting material Substances 0.000 description 1
- 239000007929 subcutaneous injection Substances 0.000 description 1
- 238000010254 subcutaneous injection Methods 0.000 description 1
- 239000000126 substance Substances 0.000 description 1
- 235000000346 sugar Nutrition 0.000 description 1
- 150000008163 sugars Chemical class 0.000 description 1
- 239000006228 supernatant Substances 0.000 description 1
- 239000000725 suspension Substances 0.000 description 1
- 238000012360 testing method Methods 0.000 description 1
- 238000013518 transcription Methods 0.000 description 1
- 230000035897 transcription Effects 0.000 description 1
- 230000014599 transmission of virus Effects 0.000 description 1
- QORWJWZARLRLPR-UHFFFAOYSA-H tricalcium bis(phosphate) Chemical compound [Ca+2].[Ca+2].[Ca+2].[O-]P([O-])([O-])=O.[O-]P([O-])([O-])=O QORWJWZARLRLPR-UHFFFAOYSA-H 0.000 description 1
- 241000701161 unidentified adenovirus Species 0.000 description 1
- 241001529453 unidentified herpesvirus Species 0.000 description 1
- 238000011144 upstream manufacturing Methods 0.000 description 1
- 230000007501 viral attachment Effects 0.000 description 1
- 230000008478 viral entry into host cell Effects 0.000 description 1
Classifications
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K14/00—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- C07K14/005—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from viruses
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K39/00—Medicinal preparations containing antigens or antibodies
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2319/00—Fusion polypeptide
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2760/00—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA ssRNA viruses negative-sense
- C12N2760/00011—Details
- C12N2760/18011—Paramyxoviridae
- C12N2760/18511—Pneumovirus, e.g. human respiratory syncytial virus
- C12N2760/18522—New viral proteins or individual genes, new structural or functional aspects of known viral proteins or genes
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2760/00—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA ssRNA viruses negative-sense
- C12N2760/00011—Details
- C12N2760/18011—Paramyxoviridae
- C12N2760/18511—Pneumovirus, e.g. human respiratory syncytial virus
- C12N2760/18534—Use of virus or viral component as vaccine, e.g. live-attenuated or inactivated virus, VLP, viral protein
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2760/00—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA ssRNA viruses negative-sense
- C12N2760/00011—Details
- C12N2760/18011—Paramyxoviridae
- C12N2760/18611—Respirovirus, e.g. Bovine, human parainfluenza 1,3
- C12N2760/18622—New viral proteins or individual genes, new structural or functional aspects of known viral proteins or genes
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2760/00—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA ssRNA viruses negative-sense
- C12N2760/00011—Details
- C12N2760/18011—Paramyxoviridae
- C12N2760/18611—Respirovirus, e.g. Bovine, human parainfluenza 1,3
- C12N2760/18634—Use of virus or viral component as vaccine, e.g. live-attenuated or inactivated virus, VLP, viral protein
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2760/00—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA ssRNA viruses negative-sense
- C12N2760/00011—Details
- C12N2760/18011—Paramyxoviridae
- C12N2760/18711—Rubulavirus, e.g. mumps virus, parainfluenza 2,4
- C12N2760/18722—New viral proteins or individual genes, new structural or functional aspects of known viral proteins or genes
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2760/00—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA ssRNA viruses negative-sense
- C12N2760/00011—Details
- C12N2760/18011—Paramyxoviridae
- C12N2760/18711—Rubulavirus, e.g. mumps virus, parainfluenza 2,4
- C12N2760/18734—Use of virus or viral component as vaccine, e.g. live-attenuated or inactivated virus, VLP, viral protein
Definitions
- the present invention relates to recombinant heterochimeric paramyxoviridae glycoproteins and their expression in eukaryotic cells, particularly in Chinese Hamster Ovary (CHO) cells.
- the invention further relates to methods for constructing and expressing such heterochimeric proteins, intermediates for use therein, methods to optimize the codon usage of the nucleic acid sequences which encode such heterochimeric proteins and the use of the recombinant proteins as vaccines for the prevention of diseases caused by paramyxoviridae pathogens.
- the mumps (MuV), Measles (MV), the parainfluenza type I (PIVl), type II (PIV2) and type III (PIV3) and the respiratory syncytial (RSV) virus belong to the paramyxoviridae family.
- the MuV is classified in the rubulavirus subclass
- the MV is classified in the Morbillivirus subclass
- the parainfluenza viruses (PIVl, PIV2 and PIV3) are classified in the paramyxovirus subclass while the RSV is attached to the pneumovirus subclass.
- RSV is the most important cause of viral lower respiratory tract disease in infants and children.
- the fusion (F) and the attachment (G) protein which are both viral surface glycoproteins appear to be of potential value for the development of a vaccine against RSV.
- the fusion protein F of RSV contains 574 amino acid residues; amino acids 1 to 21 correspond to the signal peptide and residues 525 to 549 to the membrane anchor domain.
- the molecule presents five potential sites for glycosylation.
- the F protein is synthesized as a 70 kDa precursor (F 0 ) which undergoes proteolytic maturation to yield the F, subunit (48 kDa) and F 2 (23 kDa) linked via disulfide bridges.
- F 0 70 kDa precursor
- F 2 23 kDa
- the attachment or G protein of RSV contains 298 amino acid residues and is heavily glycosylated since half of its molecular mass (90 kDa) is contributed by oligosaccharide side chains, chiefly in the form of O-linked sugars. It has been shown that the G protein, when injected into animals, provides protection against homologous but not heterologous subgroup virus challenge. This protein is extremely variable and there is only a stretch of 13 amino acid residues which is conserved in all RSV.
- the PIV3 is second to RSV as a major agent of severe viral respiratory tract infections in infants.
- the fusion protein F of PIV3 contains 539 amino acid residues; amino acids 1 to 18 correspond to the signal peptide and residues 494 to 516 to the membrane anchor domain.
- the molecule presents 4 potential sites for glycosylation.
- the F protein is synthesized as a 70 kDa precursor (F 0 ) which undergoes proteolytic maturation to yield the Fj (56 kDa) and F 2 (14 kDa) subunits linked via disulfide bridges.
- the protein F when injected into animals, leads to the production of neutralizing antibodies.
- the F protein is involved in cell fusion during viral infection and carries an hemolysin activity. Used alone for immunization, the F protein generates an immune response which is insufficient to confer protection against a challenge with the virus. Complete protection is only acquired by concomitant immunization with the attachment protein HN, another glycoprotein of PIV3.
- the protein HN carries hemagglutinin and neuraminidase activities. It is composed of 572 amino acids; its membrane anchor domain occurs in the N-terminal end of the molecule, between amino acid residues 32 and 53. Four potential sites for glycosylation have been identified. Injection of protein HN into animals generates an immune response and neutralizing antibodies. These antibodies however do not protect completely against a challenge with the virus. Full protection is obtained only by concomitant immunization with the F protein of PIV3.
- the PIVl virus was initially isolated from young children suffering from disorders of the lower respiratory tract. Infection with PIVl causes the majority of cases of croup found for all infections caused by paramyxoviruses. Viral transmission of PIVl is by person to person contact or by aerosol, although the virus does not persist in the environment for long.
- the PIVl virus has two surface glycoproteins, the fusion protein (F) and the attachment protein (HN). These two proteins are the priority targets for the development of a subunit vaccine, the properties of which would be to ensure protection of children from the very first months of life and to prevent reinfection, or at least to prevent the serious complications by restricting viral development to the upper respiratory tract where the consequences would be benign (common cold).
- PIV2 also affects very young children and causes the same type of respiratory discorders, essentially croup, but of less severity.
- the PIV2 virus has two surface glycoproteins (F and HN), which are potential targets for the development of a subunit vaccine.
- the measles virus is an extremely contagious agent which establishes itself in the epithelial cells of the respiratory tract, the oropharynx or the conjunctiva.
- the infection causes fever, cough, head-cold, conjunctivitis and a characteristic generalised rash.
- the measles virus has two surface glycoproteins, which are potential targets for the development of a subunit vaccine.
- the fusion protein (F) is a 550 amino acid long glycosylated molecule and, as for the other paramyxovirus, has to undergo proteolitic cleavage to yield F, and F 2 subunits that are linked via disulfide bridges. This molecule, which carries a haemolysin activity, generates an immune protective response when injected into animals.
- the attachment protein (H) is a 617 amino acid long glycosylated protein, which carries a hemagglutinin activity. This protein leads, when injected into animals, to the production of neutralizing antibodies that are able to inhibit hemagglutination. This immune response protects the animal against a viral challenge.
- the mumps virus is a pathogen causing the contagious infantile illness which consists of the inflammation of parotid glands. During the incubation period following infection, the virus replicates in the respiratory epithelium then disseminates into secretary ducts of the parotid glands. Other glands may become infected thereafter and numerous cases of meningitis have been reported. Among complications related to the infection, encephalitis is a serious one, with a mortality rate of about 1 %; deafness cases have also been reported.
- a vaccine against mumps is available: it is made of an attenuated live virus, produced by culturing infected embryonic chicken cells.
- the vaccine leads to the seroconversion in vaccinated individuals and protects against infection in more than 95% of seronegative persons.
- the vaccine thus reduces significantly the frequencies of complications.
- the fusion protein F of mumps virus contains 538 amino acid residues; amino acids 1 to 26 correspond to the signal peptide and residues 483 to 512 to the membrane anchor domain.
- the molecule presents 7 potential sites for glycosylation.
- the F protein is synthesized as a 65-74 kDa precursor (F 0 ) which undergoes proteolytic maturation to yield the Fj (58-61 kDa) and F 2 (10-16 kDa) subunits linked via disulfide bridges.
- the protein F is involved in cell fusion during viral infection, carries an haemolysin activity and plays a role for viral penetration into cells. It does not however carry the antibody dependent cellular cytotoxicity (ADCC) as observed for another mumps virus glycoprotein, HN.
- ADCC antibody dependent cellular cytotoxicity
- the protein HN (molecular weight 74-80 kDa) carries hemagglutinin and neuraminidase activities which are involved in virus attachment to cells and in the disruption of the host cell membranes. Protein HN (attachment protein or hemagglutinin-neuraminidase) generates neutralizing antibodies and appears important for the development of ADCC. Protein HN is composed of 582 amino acids; it carries a N-terminal anchor domain (residues 33 to 52) and 9 potential sites for glycosylation.
- WO9314207 (Connaught) describes heterochimeric proteins comprising RSV and PIV3 proteins including F(RSV)xHN(PIV3) and F(PIV3)xG(RSV) hybrids, and suggests that such proteins can be expressed from a variety of host cells including bacterial, mammalian, insect, yeast and fungal cells.
- the specific examples describe expression in insect Sf9 and High 5 cells and mammalian Vero cells. There is no specific disclosure of the use of CHO cells. The use of Sf9 and High 5 cells is also described by Du et al, BIO/TECHNOLOGY 12,1994, 813-818.
- Homochimeric paramyxoviridae glycoproteins have also been described by several workers :- WO8905823 (Upjohn) describes RSV FxG and GxF hybrids which can be expressed from bacterial, yeast, mammalian and insect cells.
- Example 7 describes the expression of an RSV FxG protein from CHO cells although there are no details of how successful such expression is.
- WO8910405 (Upjohn) describes PIV3 FxHN and HNxF hybrids which can be expressed from bacterial, yeast, mammalian and insect cells.
- Example 6 describes the expression of a PIV3 FxHN protein from CHO cells, however no details are given quantifying the extent of expression and secretion.
- WO9306218 (SmithKline Beecham Biologicals) describes PIV3 FxHN hybrids which can be expressed in eukaryotic cells including vaccinia, CHO or Vero cells.
- Example B)2 describes the expression of a Fs + a " xHNa " hybrid in CHO cells and indicates that the product was almost evenly distributed between cells and medium. No details are however given quantifying the extent of expression and secretion.
- WO9425600 (SmithKline Beecham Biologicals) describes MuV FxHN and HNxF hybrids which can be expressed in vaccinia, a mammalian cell (such as CHO) or a bacterial cell.
- Examples B) 3 and 4 describe the expression of s + FHNa " xFa " and Fs + a " xHNa ' in CHO cells however no details are given describing the extent of expression and secretion.
- heterochimeric hybrids can be successfully expressed and secreted in both CHO and insect cells.
- the present invention provides a process for preparing a heterochimeric protein or an immunogenic derivative thereof comprising an immunogenic fragment of the fusion (F) protein of RSV, PIVl , PIV2, PIV3, MV or MuV and an immunogenic fragment of the attachment (G, HN or H) protein of RSV, PIVl, PIV2, PIV3, MuV or MV which process comprises expressing recombinant DNA encoding the heterochimeric protein or immunogenic derivative thereof in CHO cells and recovering the protein.
- heterochimeric protein is meant one that does not contain a fusion or attachment protein from the same pathogen.
- This invention also provides novel heterochimeric proteins not previously described in WO 9314207 which can be prepared using the process of the present invention.
- the present invention provides a heterochimeric protein or an immunogenic derivative thereof comprising an immunogenic fragment of the fusion (F) protein of RSV, PIVl, PIV2, PIV3, MV or MuV and an immunogenic fragment of the attachment (G, HN or H) protein of RSV, PIVl , PIV2, PIV3, MuV or MV, with the proviso that where one of the immunogenic fragments is derived from RSV F, RSV HN or PIV3 F, PIV3 HN, the other of the immunogenic fragments is derived from MuV F, MuV HN, MV F, MV H, PIVl F,PIV1 HN, PIV2 F or PIV2 HN.
- an immunogenic fragment of the fusion (F) protein of RSV, PIVl, PIV2, PIV3, MV or MuV is meant a part of the protein which contains at least one antigenic determinant capable of raising an immune response specific to the F protein of RSV, PIVl, PIV2, PIV3, MV or MuV respectively. Included within this definition is the full length F protein, preferably however the immunogenic fragment is lacking the membrane anchor domain at its C-terminal end.
- an immunogenic fragment of the attachment protein (G, HN or H) of RSV, PIVl, PIV2, PIV3, MuV or MV is meant a part of the protein which contains at least one antigenic determinant capable of raising an immune response specific to the G protein of RSV, to the HN protein of PIVl, PIV2, PIV3, MuV or the H protein of MV respectively. Included within this definition is the full length G or HN protein, preferably however the immunogenic fragment is lacking the signal/anchor domain at its N-terminal end.
- heterochimeric protein is linked via an amino acid in the C-terminal part of the immunogenic fragment of the F protein of RSV, PIVl , PIV2, PIV3, MV or MuV to an amino acid in the N-terminal part of the immunogenic fragment of the G protein of RSV, the HN protein of PIVl, PIV2, PIV3, MuV or the H protein of MV.
- the heterochimeric protein commences at its N-terminal end with a signal sequence from the F protein of RSV, PIVl, PIV2, PIV3, MV or MuV. Conveniently this will be part of the corresponding immunogenic fragment of the F protein of RSV, PIVl, PIV2, PIV3, MV or MuV when this fragment is linked via its C-terminal end to the N-terminal end of the immunogenic fragment of the G protein of RSV, the HN protein of PIVl , PIV2, PIV3, MuV or the H protein of MV.
- the heterochimeric protein suitably commences at its N-terminal end with a signal sequence of tissue plasminogen activator (TPA).
- TPA tissue plasminogen activator
- the heterochimeric protein may further comprise a ubiquitin leader sequence which is suitably positioned after any signal sequence as hereinbefore described.
- the ubiquitin leader sequence is linked to the C-terminal end of the signal sequence of TPA.
- the ubiquitin leader sequence is derived from yeast, for example as described in Ecker et al, J.Biological Chemistry, 1988, 264(13), 7715-7719.
- a cleavage site is positioned between the C-terminal end of the ubiquitin sequence and the N-terminal end of the immunogenic fragment of the F protein of RSV, PIVl, PIV2, PIV3, MV or MuV.
- the heterochimeric protein suitably comprises a polyhistidine tail, for example as described in Hochuli et al, BIO/TECHNOLOGY, 1988, 1321-1325.
- the polyhistidine tail preferably comprises from 2 to 6 adjacent histidine residues which is suitably attached at the C- terminal end of the heterochimeric protein.
- a cleavage site is positioned between the polyhistidine tail and the C-terminal end of the immunogenic fragment of the G protein of RSV, the HN protein of PIVl, PIV2, PIV3, MuV or the H protein of MV.
- the cleavage site for the ubiquitin sequence and/or the polyhistidine tail may be chemical or enzymatic and preferably is an enterokinase cleavage site, for example as described in LaVallie et al, BIO-TECHNOLOGY, 1993, 187-193.
- heterochimeric proteins of this invention include: the F protein of RSV lacking its membrane domain linked at its C-terminal end to the HN protein of MuV lacking its signal/anchor domain herein referred to as: Fs + a " RSVxHNs a ' MuV, as well as Fs + a PIV3 x HNs ' a MuV; Fs + a MuV x Gs a RSV; and Fs MuV x HNs a PIV3, and immunogenic derivatives thereof.
- the present invention also provides particular heterochimeric proteins which include:
- the present invention also provides heterochimeric proteins comprising RSV and PFV3 proteins not specifically disclosed in WO9314207, which advantageously can be expressed from CHO cells.
- the heterochimeric proteins of the present invention are immunogenic.
- immunogenic derivative as used herein encompasses any molecule which is a heterochimeric polypeptide which is immunologically reactive with antibodies raised to the heterochimeric protein of the present invention or parts thereof or with antibodies recognising the F protein of RSV, PIVl, PIV2, PIV3, MV or MuV, the G protein of RSV, the HN protein of PIVl , PIV2, PIV3, MuV, the H protein of MV, the RSV virus, the PIVl virus, the PIV2 virus, the PIV3 virus, the MV virus or the MuV virus, or which, when administered to a human, elicits antibodies recognising the F protein of RSV, PIVl, PIV2, PIV3, MV or MuV, the G protein of RSV, the HN protein of PIVl, PIV2, PIV3, MuV, the H protein of MV, the RSV virus, the PIVl virus, the PIV2 virus, the PIV3 virus, the
- immunogenic derivatives which are slightly longer or shorter than the heterochimeric proteins of the present invention may be used.
- Such derivatives may, for example, be prepared by substitution, addition, or rearrangement of amino acids or by chemical modifications thereof including the coupling or for enabling the coupling of the heterochimeric proteins to other carrier proteins such as tetanus toxoid or Hepatitis B surface antigen. All such substitutions and modifications are generally well known to those skilled in the art of peptide chemistry.
- Immunogenic fragments of the heterochimeric proteins which may be useful in the preparation of vaccines may be prepared by expression of the appropriate gene fragments or by peptide synthesis, for example using the Merrifield synthesis (The Peptides, Vol 2., Academic Press, New York, p3).
- recombinant DNA encoding the heterochimeric protein of the invention.
- the recombinant DNA of the invention may form part of a vector, for example a plasmid, especially an expression plasmid from which the heterochimeric protein may be expressed.
- a vector for example a plasmid, especially an expression plasmid from which the heterochimeric protein may be expressed.
- Such vectors also form part of the invention, as do host cells into which the vectors have been introduced.
- cDNA containing the coding sequences of the RSV, PIVl, PIV2, PIV3, MV or MuV fusion and attachment proteins and optionally of the ubiquitin, polyhistidine and enterokinase cleavage sites may be manipulated using standard techniques [see for example Maniatis T. et al Molecular Cloning, Cold Spring Harbor Laboratory, Cold Spring Harbor N.Y. (1982)1 as further described hereinbelow.
- a process of enhancing the protein expression in mammalian cells by optimization of the codon usage of the nucleic acids transfected therein.
- Codon usage involves the replacement of at least one non-preferred or less preferred codon in a natural gene encoding a heterochimeric protein by a preferred codon encoding the same amino acid.
- Highly mammalian-expressed genes have C or G at their degenerative position (third base in the codon) whereas the RSV or PI V3 -prevalent codons have A or T.
- At least one codon, and more prefereably all the codons of the RSV or PIV3 protein can be changed to fit at best the human usage, that is, the one (or ones) that is the most prevalent as shown below.
- Each amino acid encoded by one of these codons are then considered humanised.
- the ratio between the number of humanised codons versus the total number of amino acids gives a percentage of humanisation as shown below.
- the invention also provides DNA encoding a heterochimeric protein or immunogenic derivative thereof in which the codon usage of one or more nucleic acids has been substantially optimised and a process for expressing said DNA in a CHO or insect cell.
- Vectors comprising such DNA, hosts transformed thereby and the truncated or hybrid proteins themselves, expressed as described hereinbelow all form part of the invention.
- plasmids may be constructed which are suitable either for transfer into vaccinia virus or transfection into CHO cells, insect cells or Vero cells. Suitable expression vectors are described hereinbelow. Preferably the proteins of the present invention are expressed in CHO or insect cells.
- a vaccinia transfer plasmid such as pULB 5213 which is a derivative of pSCll (Chakrabati et al, Molecular and Cellular Biology 5, 3403 - 3409, 1985) may be used.
- the protein may be expressed under the control of the vaccinia P ⁇ 5 promoter.
- GS glutamine synthetase
- pEE14 glutamine synthetase vector
- pEE14 may suitably be used so that the protein is expressed under the control of the major immediate early promoter of human cytomegalovirus (hCMV-MIE).
- the coding module is under the control of the Rous Sarcoma Long Terminal Repeat (LTR) promoter.
- LTR Rous Sarcoma Long Terminal Repeat
- the plasmid for expression in CHO-K1 cells carries a GS expression cassette suitable for gene amplification using methionine sulphoximine (MSX).
- the plasmid for expression in CHO-K1 cells carries a DHFR expression cassette suitable for gene amplification using methotrexate (MTX).
- MSX methionine sulphoximine
- MTX methotrexate
- heterochimeric protein of the present invention is carried out in the presence of sodium butyrate and/or dimethyl sulphoxide (DMSO) which may enhance gene expression.
- DMSO dimethyl sulphoxide
- a shuttle vector such as pAcUW51 or pAcGP67 may be used.
- the protein may be expressed under the control of the baculovirus plO promoter or the polyhedrin promoter.
- the expression system may also be a recombinant live microorganism, such as a virus or bacterium.
- the gene of interest can be inserted into the genome of a live recombinant virus or bacterium. Inoculation and in vivo infection with this live vector will lead to in vivo expression of the antigen and induction of immune responses.
- Viruses and bacteria used for this purpose are for instance: poxviruses (e.g; vaccinia, fowlpox, canarypox), alphaviruses (Sindbis virus, Semliki Forest Virus, Dialoguelian Equine Encephalitis Virus), adenoviruses, adeno-associated virus, picornaviruses (poliovirus, rhinovirus), herpesviruses (varicella zoster virus, etc), Listeria, Salmonella, Shigella, BCG. These viruses and bacteria can be virulent, or attenuated in various ways in order to obtain live vaccines. Such live vaccines also form part of the invention.
- poxviruses e.g; vaccinia, fowlpox, canarypox
- alphaviruses Semliki Forest Virus, Kunststoffuelian Equine Encephalitis Virus
- adenoviruses adeno-associated virus
- picornaviruses
- a vaccine composition comprising a heterochimeric protein or immunogenic derivative thereof according to the invention in combination with a pharmaceutically acceptable carrier, a protein according to the invention for use in vaccinating a mammal and the use of a protein according to the invention in the preparation of a vaccine.
- the vaccine of the present invention is combined with other immunogens to afford a polyvalent vaccine.
- the heterochimeric protein is combined with other subcomponents of RSV, PIVl, PIV2, PIV3, MuV or MV, e.g. the single proteins F, G, HN or H or homochimeric proteins such as RSV FxG, PIV3 FxHN or MuV FxHN.
- the invention further provides a vaccine composition
- a vaccine composition comprising a protein according to the invention together with a suitable carrier or adjuvant.
- Vaccine preparation is generally described in New Trends and Developments in Vaccines, edited by Voller et al, University Park Press, Baltimore, Maryland, U.S.A., 1978. Encapsulation within liposomes is described, for example by Fullerton, U.S. Patent 4,235,877.
- an aqueous solution of the protein(s) can be used directly.
- the protein, with or without prior lyophilisation can be mixed, absorbed or adsorbed with any of the various known adjuvants.
- adjuvants include, but are not limited to, aluminium hydroxide, muramyl dipeptide and saponins such as Quil A.
- Particularly preferred adjuvants are MPL (monophosphoryl lipid A) and 3D-MPL (3 deacylated monophosphoryl lipid A) [US patent 4,912,094], optionally formulated with aluminium hudroxide (EP 0 689 454) or oil in water emulsions (WO 95/17210).
- a further preferred adjuvant is known as QS21 which can be obtained by the method disclosed in US patent 5,057,540.
- Use of 3D-MPL is described by Ribi et al. in Microbiology (1986) Levie et al. feds) Amer. Soc. Microbiol.Wash. D.C., 9-13.
- Use of Quil A is disclosed by Dalsgaard et al.,(l911), Acta Vet Scand, 18, 349.
- Use of combined 3D-MPL and QS21 is described in WO 94/00153 (SmithKline Beecham Biologicals s.a).
- QS21 may be advantageously formulated with cholesterol containing liposomes, wherein 3D-MPL is present either in solution or incorporated in the membrane, as described in WO 96/33739.
- a heterochimeric protein of the invention or an immunogenic fragment thereof can be encapsulated within microparticles such as liposomes or associated with oil-in- water emulsions. Encapsulation within liposomes is described by Fullerton in US patent 4,235,877.
- a heterochimeric protein according to the invention or an immunogenic fragment thereof can be conjugated to an immunostimulating macromolecule, such as killed Bordetella or a tetanus toxoid. Conjugation of proteins to macromolecules is disclosed, for example by Likhite in patent 4,372,945 and Armor et al. in US patent 4,474,757.
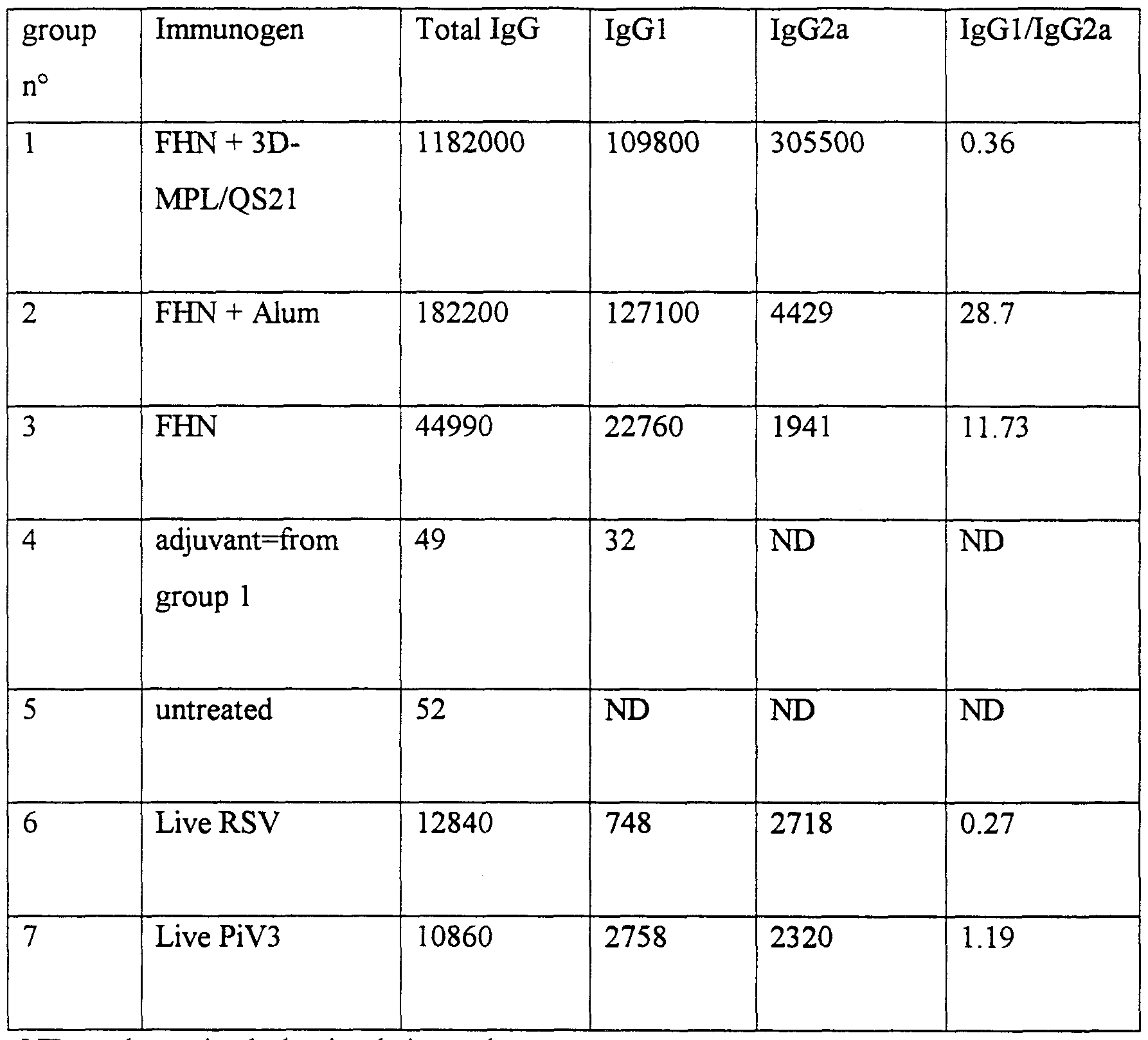
- each vaccine dose is selected as an amount which induces an immunoprotective response without significant, adverse side effects in typical vaccines. Such amount will vary depending upon which specific immunogen is employed and whether or not the vaccine is adjuvanted. Generally, it is expected that each dose will comprise l-1000 ⁇ g of protein, preferably 1-200 ⁇ g. An optimal amount for a particular vaccine can be ascertained by standard studies involving observation of antibody titres and other responses in subjects.
- Fdroso pruified Fa- (drosophila derived); the standard protein in this ELISA assay wherein lul of standard conesponds to lng of product.
- Figure 34B shows humanisation impact on the level of expression of F R svHNp iV3 , where the level of expression was determined by ELISA.
- Fdroso purified Fa-
- lul of standard corresponds to lng of product.
- heterochimeric DNA molecules were constructed combining extracellular domains of the F and the attachment protein for each virus.
- DNA constructs for the PIV3 and MuV have already been described in WO9306218 and WO9425600, respectively.
- the DNA molecule combining the extracellular domains of the RSV F and G proteins were constructed as described below.
- the DNA pieces were first inserted into the mammalian expression vector based on the replicon of the Semliki Forest Virus (pSFVl).
- pSFVl Semliki Forest Virus
- Stable expression in the culture medium of mammalian cell lines is preferred to obtain good quality and quantities of paramyxovirus glycoproteins.
- All the chimeric modules have been inserted in the shuttle vector, the pEE14, which integrates in the genome of mammalian cells such as CHO-K1.
- a quite good expression level was obtained with the RSV FxG homochimeric recombinant protein, however negligible expression was obtained for the FxHN recombinant homochimeric protein of either PIV3 or MuV. Expression of heterochimeric proteins was obtained from CHO cells.
- heterochimeric DNA molecules combining the extracellular domains of the F protein of one virus linked to the extra cellular domain of the HN or G protein of another virus and inserting them into the pEE14 vector for CHO expression it has been possible to raise the expression level of these proteins. These proteins may be used to achieve protection against at least two paramyxoviridae viruses with a single immunogen.
- Some of the chimeric molecules have been inserted into the shuttle vectors, pAcUW51 and pACGP67, which integrate in the genome of bacterial and lepidopteran cells. Surprisingly good expression of heterochimeric proteins was obtained from insect cells.
- a cDNA clone encoding ter alia the F protein of RSV (type RSS-2; received from Dr Pringle, UK) we reconstructed a cDNA module coding for the F protein lacking the membrane anchor sequence.
- Plasmid pNIV2801 was digested with Pstl in order to recover a 1416 bp DNA piece encoding amino acid residues 18 to 489 of the F protein.
- Synthetic oligonucleotides specifying respectively the sequences for amino acids 1 to 17 and 490 to 526, were used to produce the corresponding cDNA fragments by the polymerase chain reaction performed with pNIV2801 DNA as template.
- the primers were designed to generate also unique flanking restriction sites useful for subsequent cloning steps.
- the coding module was assembled, by ligation, from the three DNA pieces described above and introduced into the standard cloning vector pUC19, to create plasmid pNIV2819. This plasmid encodes the RSV F protein carrying its signal sequence but lacking its anchor sequence (figure 1).
- the cDNA module encoding the full length F protein of RSV was constructed as follows. Using two synthetic oligonucleotides, the polymerase chain reaction was performed with pNIV2801 DNA as template to generate a 273 bp DNA fragment encompassing the sequence coding for aa 490 to aa 574 of the F protein, the stop codon and unique restriction sites useful for subsequent cloning steps. This fragment was digested with Nsil and EcoRI and substituted for the Nsil-EcoRl DNA piece present in the coding module of pNIV2819 (figure 2). The resulting plasmid, pNIV2820, thus encodes the RSV F protein carrying both signal and membrane anchor sequences.
- the DNA coding for aa 165 to 176 of the G protein of RSV is fused to the DNA encoding the RSV Fs + a " protein. This part of the G protein is conserved among both subgroups of RSV.
- the starting material, pNIV2819 was digested by Ncol and Smal yielding a 1601 bp fragment. This fragment was subcloned into the Ncol and scl sites of p ⁇ IV103 (a derivative of pULB1221, see European Patent Application No. 186643) leading to pNIV2844. This subcloning allowed to place the translation initiation site of the F protein in a more favourable context according to the model proposed by Kozak (Kozak M, Nature 308, 241-246, 1984).
- a 1605 bp fragment was recovered from pNIV2844 by digestion with Kp and S ⁇ tl and introduced by ligation into pUC19 digested with Kpnl and Sail, creating pNIV2840.
- Plasmid pNIV2875 a derivative of pNIV2820 which carries the DNA coding for the F protein of RSV in which the Spel restriction site has been eliminated by site- directed mutagenesis into the pUC19 vector, has been digested by Hindlll and BspHl, and a 1618 bp fragment has been isolated.
- Plasmid pNIV3229 a derivative of pNIV3215 whose construction has been already described in WO9425600 and which canies the DNA coding for the HN protein of MuV into the pUC19 vector, has been digested with Bbsl and BamHl; a 1580 bp fragment has been isolated.
- Plasmid pIBI-HN a cDNA clone containing the complete coding sequence of protein HN of PIV3 as well as its 3' non coding sequence (received from Dr.K. Dimock, University of Ottawa, Canada), has been digested by Asel and BamHl and a 1468 bp fragment has been isolated.
- Plasmid pNIV2875 (see supra), which carries the DNA encoding the F protein of RSV, in which the unique Spel site has been eliminated by site-directed mutagenesis, inserted into the pUC19 vector, has been digested by BamHl and BspEI, and a 1588 bp fragment has been isolated.
- Both fragments were linked together by two complementary synthetic BspEI-Asel oligonucleotides (Fig5A) and were inserted into the BamHl site of the pUC19 vector leading to pNIV4105 or to pNIV4109 (Fig5B) depending of the orientation of the chimeric module in the vector.
- the chimeric cassette was retrieved by a BamHl digestion from pNIV4109 and inserted into the BamHl site of the pSFVl vector.
- the resulting plasmid, pNIV4110 contains, inserted into the pSFVl vector, the sequence coding for amino acids 1 to 526 of the RSV F protein followed by amino acids 70 to 572 of the PIV3 HN protein. (Fig5C)
- PrV3 fusion protein lacking the membrane anchor domain fused to the RSV attachment protein lacking the signal-anchor domain, F PIV3 (1-492) G ⁇ sv (69-298).
- Plasmid pNIV3310 described in WO9306218 which carries the DNA coding for amino acids 1 to 484 of the PIV3 F protein followed by amino acids 87 to 572 of the PIV3 HN protein into the pIBI vector, was digested by EcoRI and Bgl ⁇ l, and a 1435 bp fragment has been isolated.
- Plasmid pNIV2850 which carries the RSV G protein into the pUC19 vector, has been digested by Mael ⁇ l and Hindl ⁇ l, and a 694 bp fragment has been isolated.
- Both fragments were then linked together by using two complementary BgHl-Maelll synthetic linkers (Fig ⁇ A) and were inserted into the EcoRI-Hbttffll sites of pUC19 vector leading to pNIV4103 (Fig6B).
- the chimeric module was then retrieved from the pUC19 vector by a BamHl-Hindlll digestion. After treating the protruding ends with the Klenow polymerase, the chimeric cassette has been inserted into the Smal site of pSFVl vector.
- Plasmid pNIV3310 (see supra, FHN PIV3 in pIBI) was digested by EcoRI and Bglll and a 1435 bp fragment was isolated. Plasmid pNIV3229 (see supra, HN MuV into pUC19) was digested by Bbsl and Hindl ⁇ l, and a 1610 bp fragment was isolated. Both fragments were linked together by adding two synthetic complementary linkers specifying a Bglll and a Bbsl ends (Fig7A) into the pUC19 vector leading to pNIV4117 (Fig7B).
- the chimeric cassette was retrieved from the pUC19 vector by a BamHl digestion and was inserted into the BamHl site of the pSFVl vector.
- the resulting plasmid pNIV4118 encodes, cloned in the pSFVl vector, the DNA sequence specifying amino acids 1 to 493 of the PIV3 fusion protein linked to amino acids 60 to 582 of the MuV HN protein (Fig7C).
- Plasmid pNIV3221 described in WO9425600 which carries the sequence encoding amino acids 1 to 462 of the MuV fusion protein within the pUC19 vector, has been digested with EcoRI and BsrVl, and a 771 bp fragment has been purified. Plasmid pNIV3221 has been also digested with BsrFl and Pstl, and a 628 bp fragment has been isolated. Plasmid pNIV2850 (see supra, G RSV into the pUC19) has been digested with MaeZ/7 and Hind/77 and a 694 bp fragment has been isolated.
- the three fragments were linked together; the F MUV /G RSV junction was created by adding to the ligation reaction two synthetic complementary oligonucleotide specifying Pstl and Maelll sites (Fig8A), and were inserted into the EcoRI-Ht ⁇ dlll sites of the pBluescript vector leading to pNIV4113(Fig8B).
- the chimeric cassette was recovered from pNIV4113 by a Asp718l digestion and, after treating the protruding ends with the Klenow polymerase, was inserted into the Smal site of the pSFVl vector.
- the resulting plasmid, pNIV4114 contains into the pSFVl vector the sequence specifying amino acids 1 to 482 of the MuV F protein linked to amino acids 69 to 298 of the RSV G protein (Fig8C).
- Plasmid pNIV4113 (see supra, F MuV x G RSV in pBluescript) was digested by Bsal and BamHl, a 1469 bp fragment was isolated.
- the chimeric module was recovered from pNIV4115 by a BamHl digestion and was inserted into BamHl site of pSFVl vector.
- the resulting plasmid, pNIV4116 encodes, in the pSFVl vector, the sequence specifying amino acids 1-482 of the MuV F protein fused to amino acids 54 to 572 of the PIV3 HN protein (Fig9C).
- Plasmid pNIV2857 (Figl ⁇ A), a derivative of pNIV2841 and which contains the DNA sequence coding for amino acids 1 to 526 of the RSV fusion protein linked to amino acids 69 to 298 of the RSV attachment protein, has been digested by Asp718I and Hindlll and a 2180 bp fragment has been isolated. After treating the protruding extremities with Klenow's polymerase, this fragment has been inserted in the Smal site of the pSFVl vector.
- the resulting plasmid pNIV2870 contains in the pSFVl vector, the DNA sequence coding for amino acids 1 to 526 of the RSV fusion protein linked to amino acids 69 to 298 of the RSV attachment protein (Figl ⁇ B).
- Plasmid pNIV4102 (FiglOA, see supra, F RSV x HN MuV into the pUC19 vector) has been digested with BamHl, and after treating the protruding ends with the Klenow polymerase, the chimeric module has been inserted into the Smal site of the glutamine synthetase (GS) vector, pEE14 (Cockett et al, 1990, Bio/Technology 8, 662-667).
- GS glutamine synthetase
- the resulting plasmid pEE14 Fs + a RSV x HN s " a " MuV contains sequences coding for amino acids 1 to 526 of the RSV F protein fused to amino acids 60 to 582 of the MuV HN protein under the control of the major immediate early promoter of the human cytomegalovirus (hCMV-MIE) (Fig 10B).
- Plasmids pNIV4105 and pNIV4109 (FigllA and B, see supra, F RSV X HN PIV3 into the pUC19 vector) were digested by EcoRI and Xhol and a 2032 bp as well as a 1064 bp fragments were isolated. Both fragments were inserted together into the EcoRI site of p ⁇ 14.
- the resulting plasmid pEE14 Fs + a ' RSV x HNs a PIV3 contains sequences coding for amino acids 1 to 526 of the RSV F protein fused to amino acids 70 to 572 of the PIV3 HN protein under the control of the hCMV promoter (Figl lC).
- the PTV3 fusion protein lacking the membrane anchor region linked to the RSV attachment protein lacking the signal-anchor domain, T PIV3 (1-492) G ⁇ (69-298).
- Plasmid pNIV4103 (Figl2A, see supra, F pm x G RSV into the pUC19 vector) was digested by Hindlll and a 2180 bp fragment was isolated. After treating the protruding extremities with the Klenow polymerase, the chimeric module was inserted into the Smal site of the pEE14 vector.
- the resulting plasmid, pEE14 Fs + a ' PIV3 x Gs a RSV contains, under the control of the hCMV promoter, the sequence encoding amino acids 1 to 492 of the PIV3 F protein followed by amino acids 69 to 298 of the RSV G protein (Fig 12B).
- PrV3 fusion protein lacking the membrane anchor domain fused to the MuV hemagglutinin-neuraminidase lacking the signal-anchor domain, F prv3 (1- 493) H MuV (60-582).
- Plasmid pNIV4117 (Figl3A, see supra, Fp IV3 HN MuV into the pUC19 vector) was digested with Hindlll and a 3119 bp fragment was isolated and inserted into the Hindlll site of the pEE14 vector.
- Plasmid pNIV4113 (Figl4A, see supra, F MuV G RSV into the pBluescript vector) has been digested Asp718l, the protruding ends have been treated by the Klenow polymerase. A 2200 bp fragment has been isolated and inserted into the Smal site of pEE14. The resulting plasmid, pEE14 Fs + a " MuV x Gs ' a ' RSV, has, under the control of the hCMV promoter, the sequence encoding amino acids 1 to 482 of the MuV F protein followed by amino acids 69 to 298 of the RSV G protein (Figl4B).
- Plasmid pNIV4115 (Figl5A, see supra, F MuV x HN PIV3 into the pBluescript vector) has been digested with EcoRI and a 3040 bp fragment has been inserted into the EcoRI site of the p ⁇ 14 vector.
- Plasmid pNIV2857 (Figl7A), a derivative of pNIV2841 and which contains the DNA sequence coding for amino acids 1 to 526 of the RSV fusion protein linked to amino acids 69 to 298 of the RSV attachment protein, has been digested by Asp718I and Hindlll and a 2180 bp fragment has been isolated. After treating the protruding extremities with Klenow 's polymerase, this fragment has been inserted the Smal site of the pEE14vector.
- the resulting plasmid, pEE14 Fs + a " RSV x Gs ' a " RSV contains under the control of the hCMV promoter the DNA sequence coding for amino acids 1 to 526 of the RSV fusion protein linked to amino acids 69 to 298 of the RSV attachment protein (Figl7B).
- Plasmid pNIV2852 a derivative of pNIV2820 which carries the DNA encoding the RSV F protein where the translation initiation site is in a more favourable context according to the model proposed by Kozak (Kozak M, Nature 308, 241-246, 1984), has been digested BamHl and BspHI, and a 1588 bp fragment has been isolated.
- Plasmid pIBI-HN a cDNA clone containing the complete coding sequence of the HN protein of PIV3 (received from Dr. K. Dimock, University of Ottawa, Canada) has been digested by Asel and BamHl and a 1468 bp has been isolated.
- Both fragments were linked together by two complementary synthetic BspHI-Asel adaptators (Fig 18 A) and were inserted into the BamHl site of the pUC19 vector leading to pNIV4120 (Figl ⁇ B).
- the chimeric cassette was retrieved by a BamHl digestion from pNIV4120 and inserted into the BamHl compatible Bell site of the pEE14 vector.
- the resulting plasmid pEE14 Fs + a " RSV x HNs a " PIV3 bis contains the sequences coding for amino acids 1 to 526 of the RSV F protein fused to amino acids 70 to 572 of the PIV3 HN protein under the control of the hCMV promoter (Figl8C).
- This construct differs from the earlier pEE14 Fs + a ' RSV x HNs a " PIV3 construct (Il-a) in the F coding region.
- F R syHNpr ⁇ bis the nucleic acid sequence found in F RSV HN PIV3 , ATG GAT CTG (those codons are specifying aa Metl, Asp2 and Leu3) and ACC AGT (specifying aa Thr54 and Ser 55) is replaced by the original sequence of the RSV F protein that is ATG GAG TTG (specifiyng aa Metl, Glu2, Leu3) and ACT AGT (specifying Thr54 and Ser55).
- the resulting plasmid pNIV3340 has been digested by Xhol and BamHl and a 1121 bp fragment has been isolated (Figl9B).
- Plasmid pNIV4120 (see supra) has been digested by Xhol and BamHl and a 2017 bp fragment has been isolated (Figl9C).
- the resulting plasmid pEE14 FRSVs + a " x HNs a " en his contains, under the control of the hCMV promoter, sequences coding for amino acids 1 to 526 of the RSV fusion protein fused to the amino acids 70-572 of the PIV3 HN protein fused to the enterokinase cleavage site, ( ⁇ Asp ⁇ x4 Lys) followed by a polyhistidine tail ( ⁇ his ⁇ x6) and a stop codon (Figl9D).
- a 208 bp fragment conesponding to amino acid 1 to 76 of the ubiquitin protein of Saccharomyces cerevisiae was isolated by a digestion of pNIV3475 ( a derivative of YEPUBSTUALL, a yeast 2 ⁇ vector backbone carrying the yeast ubiquitin) with BamHl and Xbal (Fig 20A).
- Plasmid JW4304 (received from J. Mullins, University of Washington, U.S. A) which encodes the signal domain of the tissue plasminogen activator (sTPA) was digested by Nhel and BamHl and a 5115bp was isolated. Both fragments were linked together using two synthetic complementary Nhel-Xbal adaptators (Fig20B).
- the resulting plasmid pNIV4121 was digested by Hindlll and BamHl. A 330 bp fragment was isolated and inserted into the Hindlll and BamHl sites of the pBluescript vector.
- the resulting plasmid pNIV4122 contains the DNA sequence specifying the signal domain of the tissue plasminogen activator followed by an alanine and a serine residue (those two amino acids are known to produce a good leader cleavage) fused to the yeast ubiquitin (Fig 20C) .
- Plasmid pNIV4122 (Fig 21A, see supra) was digested by Aflll and Spel. A 3212 bp fragment was isolated and linked to synthetic complementary Aflll-Spel adaptators (Fig21B). The entire module was then sequenced. The resulting plasmid pNIV4123 encodes the signal domain of the tissue plasminogen activator linked to the N- terminal 74 aa of the yeast ubiquitin followed by the recognition site of enterokinase ⁇ (Asp)4 Lys ⁇ and amino acid 24 to 55 of the original fusion protein of RSV (Fig21C).
- Plasmid pNIV4123 (Fig 22 A, see supra) was digested by Hindlll, treated by the Klenow polymerase and digested by Spel. A 408 bp fragment has been isolated. Plasmid pNIV4120 (Fig 22B, see supra) has been digested by Xbal, treated by the Klenow polymerase, and digested by Spel. A 5620 bp fragment has been isolated.
- the entire coding module was retrieved from pNIV4124 by a digestion with Xbal and EcoRI and was inserted into the Xbal and EcoRI sites of the pEE14 expression vector.
- the resulting plasmid pEE14 sTPA x UBI x EN x Fs aRSV x HNs " a " PIV3, contains, under the control of the hCMV promoter, the sequence coding for aal-21 of the tissue plasminogen activator followed by an alanine and a serine residue, by the 74 N-terminal amino acids of the yeast ubiquitin, by the recognition cleavage site of the enterokinase ( ⁇ Asp ⁇ 4 Lys), by aa 24-526 of the original RSV fusion protein and by aa 70-572 of the hemagglutin-neuraminidase of PIV3.
- Plasmid pNIV4120 (FIG 23A) was digested by BamHl and a 3114 bp fragment was isolated and inserted into the BamHl site of the baculovirus transfer vector, pAcUW51 (PharMingen).
- the resulting plasmid pNIV4132 (Fig 23B) contains, under the control of the polyhedrin promoter, the sequence coding for amino acids 1-526 of the RSV F protein fused to amino acids 70-572 of the PiV3 HN protein.
- Plasmid pNIV4120 (FIG 24A, see supra) was digested by BamHl and Spel and a 2939 bp fragment was isolated, linked to two complementary synthetic BamHI-Spel adaptators and inserted into the BamHl site of the baculovirus transfer vector, pAcGP67A (PharMingen).
- the resulting plasmid pNIV4136 (Fig 24) contains, under the control of the polyhedrin promoter, the sequence coding for amino acids 1-38 of the Baculovirus gp67 protein, followed by an Alanine and an Aspartate linked to amino acids 25-526 of the RSV F protein fused to amino acids 70-572 of the PiV3 HN protein.
- the pSFVl vector is based on the Semliki Forest Virus (SFV) replicon.
- the DNA of interest is cloned into the pSFVl vector that serves as a template for in vitro synthesis of recombinant RNA.
- the RNA is transfected into mammalian cells such as BHK-21 cells.
- the recombinant RNA in the cells drives its own replication and capping resulting in production of heterologous protein.
- Plasmids pNIV2870 was digested with Pvul; pNIV4106, pNIV4110, pNIV4114, pNIV4116 and pNIV4118 were digested with Spel prior to RNA transcription. After a phenol extraction followed by an ethanol precipitation, 2 ⁇ g of linearized DNA was used as a template for RNA production. About 5 ⁇ g RNA was used to transfect, by electroporation, about 8 10 6 BHK-21 cells. All experimental procedures for RNA production and cell transfection are detailed in Liljestrom and Garoff (Bio/Technology, 1991, 9, 1356).
- pNTV4104, F RSV HN MuV ELISA were done using mAb 2072 anti-HN MuV (Orvell, 1984, J. Immunology 132, 2622-2629) or 20RG45, a goat anti-RSV serum (Fitzgerald, U.S.A.) to coat the microtiter plates and a rabbit polyclonal anti-SBL-1 (MuV) serum or mAb 19 anti-F RSV (G.Taylor, Inst. of Animal Health, Compton Lab. , U.K.) as capture antibody.
- Radioimmunoprecipitation of the 35 S-methionine labelled product was done using mAb2072 (Orvell) and products were resolved onto 7.5% SDS-PAGE.
- ELISA were done using anti-RSV goat serum 20RG45 or mAb anti-HN PIV3 4830 (Rydbeck et al, J. Gen. Virol. 67, 1531-1542, 1986) to coat microtiter plates and mAbl9 anti-F RSV (G.Taylor) or rabbit anti-PIV3 (E.Norrby, Sweden) serum as a capture antibody.
- Radioimmunoprecipitation was done using anti-HN PIV3 mAb4830.
- ELISA were done using mAb anti-F PIV3 4549 (E.Norrby, Sweden) or mAb anti G RSV 858-2 (Chemicon, U.S.A.) to coat microtiter plates and a rabbit anti-PIV3 serum as a capture antibody.
- Radioimmunoprecipitation was done using mAb anti-F PIV3 3283 (Behringwerke).
- ELISA plates were coated with anti-F PIV3 mAb 1031215 (Norrby) or with mAb 2072 anti-HN MuV (Orvel) and rabbit anti-PIV3 sera or rabbit anti-MuV sera were used as capture antibody. Immunoprecipitation of labelled product was done using mAb 2072 anti-HN MuV.
- ELISA plates were coated with anti-F MuV monoclonal 5414 (Orvell) or anti G RSV mAb (Chemicon) and a rabbit anti-SBL-1 serum was used as a capture antibody.
- ELISA plates were coated with anti-F MuV mAb 5414 (Orvell) or mAb anti-HN PIV3 4830 (Norrby) and rabbit anti-SBL-1 serum or a rabbit anti-PIV3 serum as a capture antibody.
- ELISA were done using 20RG45, a goat anti-RSV serum (Fitzgerald, U.S.A.) to coat the microtiter plates and mAbl9 anti-F RSV (G.Taylor, Inst. of Animal Health, Compton Lab. , U.K.) as capture antibody.
- All recombinant plasmids were transfected by calcium phosphate coprecipitation into CHO-KI cells, using 20 ⁇ g DNA per 1.25 10 6 cells.
- the CHO-KI cells were grown in GMEM-S medium.
- the GS transfectants were selected by adding 25 ⁇ M methionine sulfoximine to the culture medium two days after transfection. After ten to fourteen days, resistant colonies were picked and transferred into 96 wells plates. Each transformant was then transferred into 24 wells plates and subsequently to 80 cm 2 flasks. The GS transformants were assayed for the recombinant products when cells reached about 80% confluency. The procedure follows the one described in Cockett et al (Bio/Technology, 1990, 8, 662-667). ELISA and immunoprecipitation of radiolabelled products were done using the same procedures as the ones described above for the pSFVl system.
- the vector pAcUW51 is a shuttle vector for bacteria and lepidopteran cells.
- a heterologous protein coding sequence can be inserted downstream the baculovirus plO promoter or either downstream the polyhedrin promoter.
- the pAcGP67 vector is a shuttle vector for bacteria and lepidopteran cells that contains the gp67 signal sequence upstream a multiple cloning site.
- a heterologous gene can be inserted in one of the cloning site and will be expressed as a gp67 signal peptide fusion protein under the control of the polyhedrin promoter.
- the gp67 signal peptide mediates the secretion of the recombinant protein.
- Either pAcUW51 or pAcGP67 recombinant plasmid can be transfected along with baculovirus linearised DNA into Sf9 cells (Baculogold DNA, PharMingen). This leads to the generation of a recombinant baculovirus stock.
- the expression of the recombinant heterologous protein is obtained by infecting insect cells with the recombinant baculovirus
- Plasmid pNIV4132 or plasmid pNIV4136 were transfected with baculovirus linearised DNA into Sf9 cells.
- Recombinant baculovirus 3546 (derived from cells transfected by pNIV4132) or 5V (derived from cells transfected by pNIV4136) were plaque purified and were used to infect Sf9 or High FiveTM cells (Invitrogen). 24h to 72 h post- infection the cells and the spent culture medium have been collected for ELISA and Western blot analysis.
- ELISA were done using anti-RSV goat serum 20RG45 (Fizgerald) to coat microtiter plates and mAb 19 anti-F RSV (G.Taylor) as a capture antibody.
- Western blots were done using mAbl9 anti-F RSV (G.Taylor) or using anti-RSV goat polyclonal serum 20RG45 (Fizgerald).
- SF9 cells adapted to serum free medium, were infected with the plaque purified recombinant baculovirus V5 or 3546.
- the cells were grown in suspension in 500ml Erlenmeyer flask in SF900II medium (Gibco BRL).
- the medium from virus infected cells were harvested two days post-infection.
- the soluble FRSV-HNPTO product was purified from the medium of infected cells by immunoaffinity chromatography using an anti-F RSV monoclonal antibody, mAb 19.
- the anti-F monoclonal antibody was coupled to Activated CH Sepharose 4B (Pharmacia) following the manufacturer instructions.
- the immunoaffinity gel was washed 3 times with 10 bed volumes of buffer A (20mM phosphate buffer pH 6.4, NaCl 150mM) prior to sample loading. After 16 hours at 4°C, the gel was washed with buffer A and the chimeric product was eluted with lOOmM phosphoric acid. Eluted protein was neutralized immediately with one tenth of volume of 1M phosphate buffer pH 7.
- the baculovirus derived F RSV -HN PJV3 protein purified by immunoaffinity as described above, was used to immunise four BalbC mice and two New Zealand white rabbits. Three sub-cutaneous injections of 20 ⁇ g/ml/dose/rabbit or 6 ⁇ g/100 ⁇ l/dose/mouse were done at three weeks interval. The sera were collected 3 weeks after the second and the third injection and the antibody response was detected using ELISA and Western blots assays. 1) ELISA assays a) Mice response
- the antibody response was followed using a goat anti-RSV serum (2ORG45, Fitzgerald, USA) to coat the microtiter plates and mouse anti-FHN sera as capture antibody.
- the antigens used were either the Fa R s V -Drosophila or CHO derived, the F R sv-HN PiV3 expressed in baculovirus and the medium of CHO cells transfected by the pEE14 was used as a negative control.
- mice sera collected after the second injection showed some but low specific response.
- mice sera collected after the third injection showed a high increase in level of specific antibodies.
- the antibody response was followed using either one of the following ELISA.
- the antigens were the same as the one used to detect the mice antibody response.
- Recombinant Fa-RSV CHO-KI ou Drosophila derived, F RS v-HNpiv 3 baculovirus derived or the CHO-pEE14 spent medium culture were electrophoresed onto a 15% SDS-PAGE and transferred onto a nitrocellulose membrane (Amersham).
- the rabbit anti-HN sera as well as the mouse anti-HN sera detected specifically either the F protein or the F RS V-HNP;V 3 chimera.
- a table showing the comparison of the codon usage found in the F R s V HN PiV3 module with the one found in highly expressed human gene can be found in Fig.26.
- the most prevalent codons found in the F R svHN Pi v 3 module have an A or a T at their third degenerative position, whereas the human prevalent codons have a C or a G.
- the entire coding sequence has been re-engineered to fit at best the human codon usage.
- the re- engineered sequence was obtained using synthetic long oligonucleotides, polymerase chain reaction (PCR) and conventional cloning procedures.
- PCR fragments A, B and C.
- the general strategy to obtain each PCR fragment is schematically represented in Fig 27. It consists of assembling overlapping long oligonucleotides in a first round amplification. The resulting full size fragment is further amplified using two short primers located on each of its extremities.
- the first PCR fragment corresponding to 18 bases encoding restriction sites followed by bases 1 to 1269 of the F R svHN PiV3 followed by 8 bases encoding restriction sites, was obtained by PCR assembly of 18 overlapping oligonucleotides (Fig 28). This fragment has been inserted in the pCRIITOPO cloning vector (Invitrogen). After sequencing the fragment, it was retrieved from the pCRIITOPO vector by a Xbal and BsrGI digestion and inserted into the corresponding sites of pNIV4120.
- the second PCR fragment B corresponding to 13 bases encoding unique restriction sites followed by bases 1264 to 2136 of F R svHN Pi v 3 was obtained by assembling 10 oligonucleotides whose sequences can be found in Fig.30. This fragment has been inserted in the pCRIITOPO vector and sequenced. This fragment has been recovered by a BsrGI and Kpnl digestion.
- the entire F RSY HN P ⁇ codon optimized coding sequence has been obtained by assembling fragment A, B, C as shown in Fig.32.
- pNIV4120 in which the PCR fragment A has replaced the original sequence was digested by BsrGI and EcoRI.
- the original sequence was eliminated and replaced by the BsrGI- Kpnl fragment B and the Kpnl-EcoRI fragment C.
- the codon optimized module was retrieved from the PCRIITOPO vector by a Xbal and an EcoRI and inserted in the corresponding sites of the pEE14 vector.
- the resulting plasmid, PEE14F RSV humHN PiV3 hum encodes for the entire humanized coding sequence.
- the humanized F R svHNp iV3 nucleic acids sequence is shown in Fig. 33.
- the recombinant pEE14 F RSV humHN PiV3 (see construction of fragment A, above, or recombinant pEE14F RSV humHNp iV3 hum see construction of the entire coding sequence, above) was transfected using the FuGene reagent (Boeringer Mannheim), using 5 ⁇ g DNA per 1.25 10° cells.
- the CHO-KI cells were grown in GMEM-S medium.
- the GS transfectants were selected by adding 25 ⁇ M methionine sulfoximine to the culture medium two days after transfection. After ten to fourteen days, resistant colonies were picked and transferred into 96 wells plates. Each transformant was then transferred into 24 wells plates and subsequently to 80 cm 2 flasks.
- the GS transformants were assayed for the recombinant product when cells reached about 80% confluency. The procedure follows the one described in Cockett et al (Bio/Technology, 1990, 8, 662-667). Alternatively, the expression was evaluated three to five days after the addition of sodium butyrate (2mM) in the cell culture.
- ELISA assays were done, using 20RG45, a goat anti-RSV serum (Fizgerald, U.S.A.) to coat the microtiter plates and mAbl9 anti-F RSV (G. Taylor, Inst. of Animal Health, Compton Lab, U.K.) as capture antibody.
- the expression level was estimated using a purified expressed in the Drosophila system.
- PEEMF RSV HN P ⁇ didn't exceed 0.03 mg/L and 0.1 mg/L when sodium butyrate was added to the culture medium.
- the level of expression of the partially humanized product expressed by reached 1 mg/L and up to 3 mg/L when sodium butyrate was added in the culture medium.
- the humanization of the sequence coding for amino acids 1-423 of the 1029 amino acids thus enhanced the level of expression up to 30 fold (see Figure 34a).
- the level of expression of the entirely humanized product expressed by PEE14F RSV humHN PiV3 hum was at least of 2 mg/L and reached up to 50 mg/L when sodium butyrate was added in the culture medium.
- the entire synthetic sequence was recovered by joining four PCR fragments (A, B, C and D).
- the general strategy to obtain each PCR fragment is schematically represented in Fig 36. It consists of assembling overlapping long oligonucleotides in a first round amplification. The resulting full size fragment is further amplified using two short primers located on each of its extremities.
- the first PCR fragment corresponding to 13 bases specifying restriction sites and a Kozak consensus motif followed by bases 1 to 1026 of the F M ⁇ H MV was obtained by PCR assembly of 12 overlapping oligonucleotides (Fig 37). This fragment has been inserted in the pCRIITOPO cloning vector (Invitrogen). After sequencing the fragment, it was retrieved from the pCRIITOPO vector by a Xbal and TspRI digestion and a 963 bp fragment was further purified, leading to fragment A.
- the second PCR fragment B corresponding to bases 965 to 1712 of F M ⁇ H MV was obtained by assembling 9 oligonucleotides whose sequences can be found in Fig.38. After its insertion into the pCRIITOPO vector and its sequencing, this 785 bp fragment has been recovered by a TspRI and Aval digestion. Construction of fragment C
- the third PCR fragment C corresponding to bases 1712 to 2485 has been assembled starting from the 11 oligonucleotides shown in Fig 39. It has been inserted in the pCRIITOPO cloning vector and sequenced. This 774 bp fragment has been retrieved by an Aval and Apal digestion.
- the fourth PCR fragment D corresponding to bases 2485 to 3139 followed by 8 bp specifying a unique restriction site has been assembled starting from the 8 oligonucleotides shown in Fig 40. This fragment has been inserted in the pCRIITOPO vector and sequenced. A 657 bp fragment has been recovered after an Apal and EcoRI digestion.
- the entire F MuV H Mv codon optimised coding sequence has been obtained by assembling fragment A, B, C, D and inserting the module digested by Xbal and EcoR/ into the corresponding sites of the p ⁇ 14 vector (Fig. 41).
- the resulting plasmid, pEE14F MuV humH Mv hum encodes for a humanised sequence coding for aa 1-482 of the mumps virus fusion protein followed by aa 59-617 of the measles virus.
- the humanised and original nucleic and amino acids sequences are shown in Fig. 42.
- CHO cell line expressing secreted recombinant FHN was cultivated in cell factories in G-MEM medium supplemented with 2% FCS, in presence or absence of 1% Butyrate Na.
- FHN was purified by immunoaffinity chromatography by loading spent culture medium onto a Mabl9-sepharose column as described using the same experimental conditions.
- purified FHN migrated on SDS-PAGE, in heating and reducing conditions, mainly as a band of 110 kDa.
- FHN is visualized as a triplet of 110, 120 and 130 kDa when CHO cells are cultivated with butyrate. Heating has a more drastic effect than reducer on the FHN electrophoretic migration.
- FHN aggregates or oligomers were clearly detected in the preparation when electrophoresis proceeded without heating suggesting the presence of FHN aggregates or oligomers. These aggregates did not seem to be contaminated by CHO proteins. Antibodies directed to CHO proteins did not specifically recognize on Western blot any bands. Glycan analysis was performed using several lectins specific for different carbohydrate moieties. Surprisingly, FHN did not carry sialic acids or high-mannose structures but carbohydrates of galactose-acetyl-galactosamine type characteristic of hybrid N- and or O-glycosylations.
- N-terminal microsequence analysis showed mainly the presence of FI subunit in bands of 110-130kDa.
- the F RS vHNpiv 3 protein was purified from the spent medium culture of the CHO-KI cells transfected by the recombinant pEE14 F RS vhumHNpiv 3 hum by immunoaffinity chromatography as described (Purification of the recombinant product expressed in baculovirus recombinant infected SF9 cells).
- the product was injected in 7 groups of Balb Cl mice as descibed in the following table 1. Humoral response directed against the FHN protein
- the humoral response directed against the FHN protein was determined. To this end, ELISA plates were coated with immunoaffinity purified FHN protein.
- ELISA plates were coated with 200ng of immunoaffinity purified FHN protein, plates were then saturated and dilutionsof the mice second bleed sera were then applied. Total IgG were detected using a biotinylated serum directed against mouse IgG.
- ELISA plates were coated with lOOng of immunoaffinity purified FHN protein, plates were then saturated and dilutionsof the mice second bleed sera were then applied. IgGl were detected using a biotinylated serum directed against mouse IgGl .
- ELISA plates were coated with lOOng of immunoaffinity purified FHN protein, plates were then saturated and dilutionsof the mice second bleed sera were then applied. IgG2a were detected using a biotinylated serum directed against mouse IgG2a.
- the IgGl/IgG2a ratio indicates the Thl or Th2 orientation of the immune response
- the total IgG, IgGl and IgG2a was determined for each mouse sera. A mean titer for each group was then calculated and is reported in the table.
Abstract
Description
Claims
Priority Applications (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| AU59808/99A AU5980899A (en) | 1998-09-25 | 1999-09-20 | Novel compounds |
Applications Claiming Priority (4)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| GB9820931.5 | 1998-09-25 | ||
| GBGB9820931.5A GB9820931D0 (en) | 1998-09-25 | 1998-09-25 | Novel compounds |
| GB9906868.6 | 1999-03-24 | ||
| GBGB9906868.6A GB9906868D0 (en) | 1999-03-24 | 1999-03-24 | Novel compounds |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| WO2000018929A2 true WO2000018929A2 (en) | 2000-04-06 |
| WO2000018929A3 WO2000018929A3 (en) | 2000-11-09 |
Family
ID=26314421
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/EP1999/007004 WO2000018929A2 (en) | 1998-09-25 | 1999-09-20 | Paramyxovirus vaccines |
Country Status (3)
| Country | Link |
|---|---|
| AR (1) | AR024213A1 (en) |
| AU (1) | AU5980899A (en) |
| WO (1) | WO2000018929A2 (en) |
Cited By (17)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US6830748B1 (en) | 1997-09-26 | 2004-12-14 | Medimmune Vaccines, Inc. | Recombinant RSV virus expression systems and vaccines |
| WO2008077527A1 (en) * | 2006-12-21 | 2008-07-03 | Pevion Biotech Ltd. | Rsv f-protein and use thereof |
| WO2008133663A2 (en) * | 2006-11-30 | 2008-11-06 | Government Of The United States Of America, As Represented By The Secretary, | Codon modified immunogenic compositions and methods of use |
| US7465574B2 (en) | 1994-09-30 | 2008-12-16 | Medimmune, Llc | Recombinant RSV virus expression systems and vaccines |
| WO2009005917A2 (en) * | 2007-05-29 | 2009-01-08 | Vical Incorporated | Methods of treating measles infectious disease in mammals |
| US7943375B2 (en) | 1998-12-31 | 2011-05-17 | Novartis Vaccines & Diagnostics, Inc | Polynucleotides encoding antigenic HIV type C polypeptides, polypeptides and uses thereof |
| US8133494B2 (en) | 2001-07-05 | 2012-03-13 | Novartis Vaccine & Diagnostics Inc | Expression cassettes endcoding HIV-1 south african subtype C modified ENV proteins with deletions in V1 and V2 |
| US8273361B2 (en) | 2006-09-26 | 2012-09-25 | Infectious Disease Research Institute | Vaccine composition containing synthetic adjuvant |
| US8722064B2 (en) | 2009-06-05 | 2014-05-13 | Infectious Disease Research Institute | Synthetic glucopyranosyl lipid adjuvants |
| EP2811027A1 (en) * | 2004-05-21 | 2014-12-10 | Novartis Vaccines and Diagnostics, Inc. | Alphavirus vectors for RSV and PIV vaccines |
| US8957047B2 (en) | 2013-04-18 | 2015-02-17 | Immune Design Corp. | GLA monotherapy for use in cancer treatment |
| US9044420B2 (en) | 2011-04-08 | 2015-06-02 | Immune Design Corp. | Immunogenic compositions and methods of using the compositions for inducing humoral and cellular immune responses |
| US9463198B2 (en) | 2013-06-04 | 2016-10-11 | Infectious Disease Research Institute | Compositions and methods for reducing or preventing metastasis |
| US9895435B2 (en) | 2012-05-16 | 2018-02-20 | Immune Design Corp. | Vaccines for HSV-2 |
| CN112250768A (en) * | 2020-09-24 | 2021-01-22 | 苏州世诺生物技术有限公司 | Bovine parainfluenza virus recombinant antigen and application thereof |
| WO2021119497A1 (en) * | 2019-12-11 | 2021-06-17 | The United States Of America, As Represented By The Secretary, Department Of Health And Human Services | Mumps and measles virus immunogens and their use |
| WO2022221961A1 (en) * | 2021-04-22 | 2022-10-27 | University Of Saskatchewan | Compositions and methods for preventing rsv and piv3 infections |
Citations (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO1993014207A1 (en) * | 1992-01-06 | 1993-07-22 | Connaught Laboratories Limited | Chimeric immunogens |
| WO1994025600A1 (en) * | 1993-04-30 | 1994-11-10 | Smithkline Beecham Biologicals S.A. | Recombinant antigens from mumps virus and their use in vaccines |
-
1999
- 1999-09-20 AU AU59808/99A patent/AU5980899A/en not_active Abandoned
- 1999-09-20 WO PCT/EP1999/007004 patent/WO2000018929A2/en active Application Filing
- 1999-09-23 AR ARP990104796A patent/AR024213A1/en unknown
Patent Citations (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO1993014207A1 (en) * | 1992-01-06 | 1993-07-22 | Connaught Laboratories Limited | Chimeric immunogens |
| WO1994025600A1 (en) * | 1993-04-30 | 1994-11-10 | Smithkline Beecham Biologicals S.A. | Recombinant antigens from mumps virus and their use in vaccines |
Non-Patent Citations (4)
Cited By (40)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US7465574B2 (en) | 1994-09-30 | 2008-12-16 | Medimmune, Llc | Recombinant RSV virus expression systems and vaccines |
| US7205013B2 (en) | 1997-09-26 | 2007-04-17 | Medimmune Vaccines, Inc. | Recombinant RSV virus expression systems and vaccines |
| US6830748B1 (en) | 1997-09-26 | 2004-12-14 | Medimmune Vaccines, Inc. | Recombinant RSV virus expression systems and vaccines |
| US7943375B2 (en) | 1998-12-31 | 2011-05-17 | Novartis Vaccines & Diagnostics, Inc | Polynucleotides encoding antigenic HIV type C polypeptides, polypeptides and uses thereof |
| US8133494B2 (en) | 2001-07-05 | 2012-03-13 | Novartis Vaccine & Diagnostics Inc | Expression cassettes endcoding HIV-1 south african subtype C modified ENV proteins with deletions in V1 and V2 |
| EP2811027A1 (en) * | 2004-05-21 | 2014-12-10 | Novartis Vaccines and Diagnostics, Inc. | Alphavirus vectors for RSV and PIV vaccines |
| US10765736B2 (en) | 2006-09-26 | 2020-09-08 | Infectious Disease Research Institute | Vaccine composition containing synthetic adjuvant |
| EP3403667A1 (en) | 2006-09-26 | 2018-11-21 | Infectious Disease Research Institute | Vaccine composition containing synthetic adjuvant |
| US9987355B2 (en) | 2006-09-26 | 2018-06-05 | Infectious Disease Research Institute | Vaccine composition containing synthetic adjuvant |
| US10792359B2 (en) | 2006-09-26 | 2020-10-06 | Infectious Disease Research Institute | Methods of using a vaccine composition containing synthetic adjuvant |
| US9950063B2 (en) | 2006-09-26 | 2018-04-24 | Infectious Disease Research Institute | Vaccine composition containing synthetic adjuvant |
| US8840908B2 (en) | 2006-09-26 | 2014-09-23 | Infectious Disease Research Institute | Vaccine composition containing synthetic adjuvant |
| US8273361B2 (en) | 2006-09-26 | 2012-09-25 | Infectious Disease Research Institute | Vaccine composition containing synthetic adjuvant |
| EP3795173A1 (en) | 2006-09-26 | 2021-03-24 | Infectious Disease Research Institute | Vaccine composition containing synthetic adjuvant |
| US9907845B2 (en) | 2006-09-26 | 2018-03-06 | Infectious Disease Research Institute | Methods of using a vaccine composition containing synthetic adjuvant |
| WO2008133663A3 (en) * | 2006-11-30 | 2009-03-19 | Us Gov Health & Human Serv | Codon modified immunogenic compositions and methods of use |
| WO2008133663A2 (en) * | 2006-11-30 | 2008-11-06 | Government Of The United States Of America, As Represented By The Secretary, | Codon modified immunogenic compositions and methods of use |
| EP2368998A1 (en) * | 2006-12-21 | 2011-09-28 | Ruhr-Universität Bochum | RSV F protein and use of same |
| US8372963B2 (en) | 2006-12-21 | 2013-02-12 | Pevion Biotech Ag | RSV F-protein and its use |
| RU2464316C2 (en) * | 2006-12-21 | 2012-10-20 | Певион Биотек Аг | F-protein of respiratory syncytial virus and its use |
| WO2008077527A1 (en) * | 2006-12-21 | 2008-07-03 | Pevion Biotech Ltd. | Rsv f-protein and use thereof |
| WO2009005917A3 (en) * | 2007-05-29 | 2009-05-07 | Vical Inc | Methods of treating measles infectious disease in mammals |
| US20120039935A1 (en) * | 2007-05-29 | 2012-02-16 | Vical Incorporated | Methods of treating measles infectious disease in mammals |
| GB2461832A (en) * | 2007-05-29 | 2010-01-20 | Vical Inc | Methods of treating measles infectious disease in mammals |
| WO2009005917A2 (en) * | 2007-05-29 | 2009-01-08 | Vical Incorporated | Methods of treating measles infectious disease in mammals |
| US9480740B2 (en) | 2009-06-05 | 2016-11-01 | Infectious Disease Research Institute | Synthetic glucopyranosyl lipid adjuvants |
| US9814772B2 (en) | 2009-06-05 | 2017-11-14 | Infectious Disease Research Institute | Synthetic glucopyranosyl lipid adjuvants |
| US8722064B2 (en) | 2009-06-05 | 2014-05-13 | Infectious Disease Research Institute | Synthetic glucopyranosyl lipid adjuvants |
| US10632191B2 (en) | 2009-06-05 | 2020-04-28 | Infectious Disease Research Institute | Synthetic glucopyranosyl lipid adjuvants |
| US9044420B2 (en) | 2011-04-08 | 2015-06-02 | Immune Design Corp. | Immunogenic compositions and methods of using the compositions for inducing humoral and cellular immune responses |
| US9895435B2 (en) | 2012-05-16 | 2018-02-20 | Immune Design Corp. | Vaccines for HSV-2 |
| US8962593B2 (en) | 2013-04-18 | 2015-02-24 | Immune Design Corp. | GLA monotherapy for use in cancer treatment |
| US10342815B2 (en) | 2013-04-18 | 2019-07-09 | Immune Design Corp. | GLA monotherapy for use in cancer treatment |
| US8957047B2 (en) | 2013-04-18 | 2015-02-17 | Immune Design Corp. | GLA monotherapy for use in cancer treatment |
| US10993956B2 (en) | 2013-04-18 | 2021-05-04 | Immune Design Corp. | GLA monotherapy for use in cancer treatment |
| US9463198B2 (en) | 2013-06-04 | 2016-10-11 | Infectious Disease Research Institute | Compositions and methods for reducing or preventing metastasis |
| WO2021119497A1 (en) * | 2019-12-11 | 2021-06-17 | The United States Of America, As Represented By The Secretary, Department Of Health And Human Services | Mumps and measles virus immunogens and their use |
| CN112250768A (en) * | 2020-09-24 | 2021-01-22 | 苏州世诺生物技术有限公司 | Bovine parainfluenza virus recombinant antigen and application thereof |
| CN112250768B (en) * | 2020-09-24 | 2021-05-11 | 苏州世诺生物技术有限公司 | Bovine parainfluenza virus recombinant antigen and application thereof |
| WO2022221961A1 (en) * | 2021-04-22 | 2022-10-27 | University Of Saskatchewan | Compositions and methods for preventing rsv and piv3 infections |
Also Published As
| Publication number | Publication date |
|---|---|
| AU5980899A (en) | 2000-04-17 |
| AR024213A1 (en) | 2002-09-25 |
| WO2000018929A3 (en) | 2000-11-09 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| AU2018226521B2 (en) | Cytomegalovirus antigens and uses thereof | |
| AU2021209185B2 (en) | Stabilized soluble pre-fusion RSV F proteins | |
| JP3290662B2 (en) | Chimeric immunogen | |
| JP6679475B2 (en) | Stabilized soluble pre-fusion RSV F polypeptide | |
| WO2000018929A2 (en) | Paramyxovirus vaccines | |
| EP1766034B1 (en) | Alphavirus vectors for influenza virus vaccines | |
| JP6062245B2 (en) | Recombinant RSV antigen | |
| JPH03501723A (en) | Chimeric glycoproteins containing immunogenic segments of human respiratory syncytial virus glycoproteins | |
| NZ230425A (en) | Production of paramyxovirus fusion (f) protein using recombinant baculovirus expression vector | |
| Ray et al. | Expression of the fusion glycoprotein of human parainfluenza type 3 virus in insect cells by a recombinant baculovirus and analysis of its immunogenic property | |
| CN116034115A (en) | Vaccine compositions, methods and uses thereof | |
| WO1993006218A2 (en) | Chimeras of parainfluenza virus type 3 proteins f and hn as vaccines | |
| JP3940676B2 (en) | Chimeric T helper cell-B cell peptide vaccine against Japanese encephalitis virus | |
| OA18879A (en) | Stabilized soluble pre-fusion RSV F proteins | |
| EA045858B1 (en) | STABILIZED SOLUBLE RSV F-PROTEINS BEFORE FUSION | |
| WO1997028265A9 (en) | Measles immunization by dna transcription unit inoculation | |
| NZ615721A (en) | Immunogenic compositions in particulate form and methods for producing the same |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| AK | Designated states |
Kind code of ref document: A2 Designated state(s): AE AL AM AT AU AZ BA BB BG BR BY CA CH CN CR CU CZ DE DK DM EE ES FI GB GD GE GH GM HR HU ID IL IN IS JP KE KG KP KR KZ LC LK LR LS LT LU LV MD MG MK MN MW MX NO NZ PL PT RO RU SD SE SG SI SK SL TJ TM TR TT TZ UA UG US UZ VN YU ZA ZW |
|
| AL | Designated countries for regional patents |
Kind code of ref document: A2 Designated state(s): GH GM KE LS MW SD SL SZ TZ UG ZW AM AZ BY KG KZ MD RU TJ TM AT BE CH CY DE DK ES FI FR GB GR IE IT LU MC NL PT SE BF BJ CF CG CI CM GA GN GW ML MR NE SN TD TG |
|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application | ||
| DFPE | Request for preliminary examination filed prior to expiration of 19th month from priority date (pct application filed before 20040101) | ||
| AK | Designated states |
Kind code of ref document: A3 Designated state(s): AE AL AM AT AU AZ BA BB BG BR BY CA CH CN CR CU CZ DE DK DM EE ES FI GB GD GE GH GM HR HU ID IL IN IS JP KE KG KP KR KZ LC LK LR LS LT LU LV MD MG MK MN MW MX NO NZ PL PT RO RU SD SE SG SI SK SL TJ TM TR TT TZ UA UG US UZ VN YU ZA ZW |
|
| AL | Designated countries for regional patents |
Kind code of ref document: A3 Designated state(s): GH GM KE LS MW SD SL SZ TZ UG ZW AM AZ BY KG KZ MD RU TJ TM AT BE CH CY DE DK ES FI FR GB GR IE IT LU MC NL PT SE BF BJ CF CG CI CM GA GN GW ML MR NE SN TD TG |
|
| ENP | Entry into the national phase |
Ref country code: JP Ref document number: 2000 572376 Kind code of ref document: A Format of ref document f/p: F |
|
| REG | Reference to national code |
Ref country code: DE Ref legal event code: 8642 |
|
| 122 | Ep: pct application non-entry in european phase |