HLA BINDING PEPTIDES AND THEIR USES
BACKGROUND OF THE INVENTION
The present invention relates to compositions and methods for preventing, treating or diagnosing a number of pathological states such as viral diseases and cancers. In particular, it provides novel peptides capable of binding selected major histocompatibility complex (MHC) molecules and inducing an immune response.
MHC molecules are classified as either Class I or Class II molecules. Class II MHC molecules are expressed primarily on cells involved in initiating and sustaining immune responses, such as T lymphocytes, B lymphocytes, macrophages, etc. Class II MHC molecules are recognized by helper T lymphocytes and induce proliferation of helper T lymphocytes and amplification of the immune response to the particular immunogenic peptide that is displayed. Class I MHC molecules are expressed on almost all nucleated cells and are recognized by cytotoxic T lymphocytes (CTLs), which then destroy the antigen-bearing cells. CTLs are particularly important in tumor rejection and in fighting viral infections.
The CTL recognizes the antigen in the form of a peptide fragment bound to the MHC class I molecules rather than the intact foreign antigen itself. The antigen must normally be endogenously synthesized by the cell, and a portion of the protein antigen is degraded into small peptide fragments in the cytoplasm. Some of these small peptides translocate into a pre-Golgi compartment and interact with class I heavy chains to facilitate proper folding and association with the subunit β2 microglobulin. The peptide-MHC class I complex is then routed to the cell surface for expression and potential recognition by specific CTLs.
Investigations of the crystal structure of the human MHC class I molecule, HLA-A2.1 , indicate that a peptide binding groove is created by the folding of the αl and α2 domains of the class I heavy chain (Bjorkman et al. , Nature 329:506 ( 1987). In these investigations, however, the identity of peptides bound to the groove was not determined.
Buus et al. , Science 242: 1065 (1988) first described a method for acid elution of bound peptides from MHC. Subsequently, Rammensee and his coworkers (Falk
2 et al. , Nature 351 :290 (1991) have developed an approach to characterize naturally processed peptides bound to class I molecules. Other investigators have successfully achieved direct amino acid sequencing of the more abundant peptides in various HPLC fractions by conventional automated sequencing of peptides eluted from class I molecules of the B type (Jardetzky, et al. , Nature 353:326 (1991) and of the A2.1 type by mass spectrometry (Hunt, et al., Science 225: 1261 (1992). A review of the characterization of naturally processed peptides in MHC Class I has been presented by Rδtzschke and Falk (Rόtzschke and Falk, Immunol. Today 12:447 (1991).
Sette et al. , Proc. Natl. Acad. Sci. USA 86:3296 (1989) showed that MHC allele specific motifs could be used to predict MHC binding capacity. Schaeffer et al. ,
Proc. Natl. Acad. Sci. USA 86:4649 (1989) showed that MHC binding was related to immunogenicity . Several authors (De Bruijn et al. , Eur. J. Immunol.. 21:2963-2970 (1991); Pamer et al. , 991 Nature 353:852-955 (1991)) have provided preliminary evidence that class I binding motifs can be applied to the identification of potential immunogenic peptides in animal models. Class I motifs specific for a number of human alleles of a given class I isotype have yet to be described. It is desirable that the combined frequencies of these different alleles should be high enough to cover a large fraction or perhaps the majority of the human outbred population.
Despite the developments in the art, the prior art has yet to provide a useful human peptide-based vaccine or therapeutic agent based on this work. The present invention provides these and other advantages.
SUMMARY OF THE INVENTION The present invention provides compositions comprising immunogenic peptides having binding motifs for HLA molecules. The immunogenic peptides, which bind to the appropriate MHC allele, comprise conserved residues at certain positions which allow the peptides to bind desired HLA molecules.
Epitopes on a number of immunogenic target proteins can be identified using the peptides of the invention. Examples of suitable antigens include prostate cancer specific antigen (PSA), hepatitis B core and surface antigens (HBVc, HBVs) hepatitis C antigens, Epstein-Barr virus antigens, human immunodeficiency type-1 virus (HIV1),
Kaposi's sarcoma herpes virus (KSHV), human papilloma virus (HPV) antigens, Lassa
virus, mycobacterium tuberculosis (MT), p53, CEA, trypanosome surface antigen (TSA) and Her2/neu. The peptides are thus useful in pharmaceutical compositions for both therapeutic and diagnostic applications.
In particular, the invention provides compositions comprising an immunogenic peptide having an HLA binding motif, which immunogenic peptide is a peptide shown in Tables 3-14. Also provided are peptides comprising a conservative substitution of a residue in a peptide shown in Table 3-14. The immunogenic peptide of the invention can be further linked to a second oligopeptide. In some embodiments, the second oligopeptide is a peptide that induces a helper T response. The invention further provides nucleic acid molecules encoding immunogenic peptides as shown in Tables 3-14, or peptides comprising a conservative substitution of a residue of a peptide shown in Table 3-14. The nucleic acid may further comprise a sequence encoding a second immunogenic peptide or peptide that induces a helper T response. The peptides provided here can be used to induce a cytotoxic T cell response either in vivo or in vitro. The methods comprise contacting a cytotoxic T cell with a peptide of the invention.
Definitions The term "peptide" is used interchangeably with "oligopeptide" in the present specification to designate a series of residues, typically L- amino acids, connected one to the other typically by peptide bonds between the alpha-amino and carbonyl groups of adjacent amino acids. The oligopeptides of the invention are less than about 15 residues in length and usually consist of between about 8 and about 11 residues, preferably 9 or 10 residues. An "immunogenic peptide" is a peptide which comprises an allele-specific motif such that the peptide will bind an MHC molecule and induce a CTL response. Immunogenic peptides of the invention are capable of binding to an appropriate HLA molecule and inducing a cytotoxic T cell response against the antigen from which the immunogenic peptide is derived. Immunogenic peptides are conveniently identified using the algorithms of the invention. The algorithms are mathematical procedures that produce a score which
4 enables the selection of immunogenic peptides. Typically one uses the algorithmic score with a "binding threshold" to enable selection of peptides that have a high probability of binding at a certain affinity and will in turn be immunogenic. The algorithm is based upon either the effects on MHC binding of a particular amino acid at a particular position of a peptide or the effects on binding of a particular substitution in a motif containing peptide.
A "conserved residue" is an amino acid which occurs in a significantly higher frequency than would be expected by random distribution at a particular position in a peptide. Typically a conserved residue is one where the MHC structure may provide a contact point with the immunogenic peptide. At least one to three or more, preferably two, conserved residues within a peptide of defined length defines a motif for an immunogenic peptide. These residues are typically in close contact with the peptide binding groove, with their side chains buried in specific pockets of the groove itself. Typically, an immunogenic peptide will comprise up to three conserved residues, more usually two conserved residues. As used herein, "negative binding residues" are amino acids which if present at certain positions will result in a peptide being a nonbinder or poor binder and in turn fail to be immunogenic i.e. induce a CTL response.
The term "motif" refers to the pattern of residues in a peptide of defined length, usually about 8 to about 11 amino acids, which is recognized by a particular MHC allele. The peptide motifs are typically different for each human MHC allele and differ in the pattern of the highly conserved residues and negative residues.
The binding motif for an allele can be defined with increasing degrees of precision. In one case, all of the conserved residues are present in the correct positions in a peptide and there are no negative residues in positions 1,3 and/or 7. The phrases "isolated" or "biologically pure" refer to material which is substantially or essentially free from components which normally accompany it as found in its native state. Thus, the peptides of this invention do not contain materials normally associated with their in situ environment, e.g. , MHC I molecules on antigen presenting cells. Even where a protein has been isolated to a homogenous or dominant band, there are trace contaminants in the range of 5-10% of native protein which co-purify with the desired protein. Isolated peptides of this invention do not contain such endogenous co- purified protein.
5
The term "residue" refers to an amino acid or amino acid mimetic incorporated in an oligopeptide by an amide bond or amide bond mimetic.
DESCRIPTION OF THE PREFERRED EMBODIMENTS The present invention relates to the determination of allele-specific peptide motifs for human Class I MHC (sometimes referred to as HLA) allele subtypes, in particular, peptide motifs recognized by HLA alleles.
For HLA-A2.1 alleles a peptide of 9 amino acids preferrably has the following motif: a first conserved residue at the second position from the N-terminus selected from the group consisting of I, V, A and T and a second conserved residue at the C-terminal position selected from the group consisting of V, L, I, A and M. An alternate motif is one in which the first conserved residue at the second position from the N- terminus selected is from the group consisting of L, M, I, V, A and T and the second conserved residue at the C-terminal position selected from the group consisting of A and M . The amino acid at position 1 is preferrably not an amino acid selected from the group consisting of D, and P. The amino acid at position 3 from the N-terminus is not an amino acid selected from the group consisting of D, E, R, K and H. The amino acid at position 6 from the N-terminus is not an amino acid selected from the group consisting of R, K and H . The amino acid at at position 7 from the N-terminus is not an amino acid selected from the group consisting of R, K, H, D and E. The HLA-A2.1 binding motif for peptide of 10 residues is as follows: a first conserved residue at the second position from the N-terminus selected from the group consisting of L, M, I, V, A, and T, and a second conserved residue at the C-terminal position selected from the group consisting of V, I, L, A and M. The first and second conserved residues are separated by 7 residues. Preferrably, the amino acid at position 1 is not an amino acid selected from the group consisting of D, E and P. The N-terminal residue is not an amino acid selected from the group consisting of D and E. The residue at position 4 from the N-terminus is not an amino acid selected from the group consisting of A, K, R and H. The amino acid at positon 5 from the N-terminus is not P. The amino acid at position 7 from the N-terminus is not an amino acid selected from the group consisting of R, K .and H. The amino acid at position 8 from the N-terminus is not amino acid selected from the group consisting of D, E, R, K and H. The amino acid at position
6
9 from the N-terminus is not an amino acid selected from the group consisting of R, K and
H.
Te motif for HLA-A3.2 comprises from the N-terminus to C-terminus a first conserved residue of L, M, I, V, S, A, T and F at position 2 and a second conserved residue of K, R or Y at the C-terminal end. Other first conserved residues are C, G or D and alternatively E. Other second conserved residues are H or F. The first and second conserved residues are preferably separated by 6 to 7 residues.
The motif for HLA-A1 comprises from the N-terminus to the C-terminus a first conserved residue of T, S or M, a second conserved residue of D or E, and a third conserved residue of Y. Other second conserved residues are A, S or T. The first and second conserved residues are adjacent and are preferably separated from the third conserved residue by 6 to 7 residues. A second motif consists of a first conserved residue of E or D and a second conserved residue of Y where the first and second conserved residues are separated by 5 to 6 residues. The motif for HLA-A11 comprises from the N-terminus to the C-terminus a first conserved residue of T, V, M, L, I, S, A, G, N, C D, or F at position 2 and a C- terminal conserved residue of K, R, Y or H. The first and second conserved residues are preferably separated by 6 or 7 residues.
The motif for HLA-A24.1 comprises from the N-terminus to the C-terminus a first conserved residue of Y, F or W at position 2 and a C terminal conserved residue of
F, I, W, M or L. The first and second conserved residues are preferably separated by 6 to
7 residues.
These motifs are then used to define T cell epitopes from any desired antigen, particularly those associated with human viral diseases, cancers or autoiummune diseases, for which the amino acid sequence of the potential antigen or autoantigen targets is known.
Epitopes on a number of potential target proteins can be identified in this manner. Examples of suitable antigens include prostate specific antigen (PSA), hepatitis B core and surface antigens (HBVc, HBVs) hepatitis C antigens, Epstein-Barr virus antigens, melanoma antigens (e.g., MAGE-1), human immunodeficiency virus (HIV) antigens, human papilloma virus (HPV) antigens, Lassa virus, mycobacterium tuberculosis (MT), p53, CEA, trypanosome surface antigen (TSA) and Her2/neu.
7
Peptides comprising the epitopes from these antigens are synthesized and then tested for their ability to bind to the appropriate MHC molecules in assays using, for example, purified class I molecules and radioiodonated peptides and/or cells expressing empty class I molecules by, for instance, immunofluorescent staining and flow microfluorometry, peptide-dependent class I assembly assays, and inhibition of CTL recognition by peptide competition. Those peptides that bind to the class I molecule are further evaluated for their ability to serve as targets for CTLs derived from infected or immunized individuals, as well as for their capacity to induce primary in vitro or in vivo CTL responses that can give rise to CTL populations capable of reacting with virally infected target cells or tumor cells as potential therapeutic agents.
The MHC class I antigens are encoded by the HLA-A, B, and C loci. HLA-A and B antigens are expressed at the cell surface at approximately equal densities, whereas the expression of HLA-C is significantly lower (perhaps as much as 10-fold lower). Each of these loci have a number of alleles. The peptide binding motifs of the invention are relatively specific for each allelic subtype.
For peptide-based vaccines, the peptides of the present invention preferably comprise a motif recognized by an MHC I molecule having a wide distribution in the human population. Since the MHC alleles occur at different frequencies within different ethnic groups and races, the choice of target MHC allele may depend upon the target population. Table 1 shows the frequency of various alleles at the HLA-A locus products among different races. For instance, the majority of the Caucasoid population can be covered by peptides which bind to four HLA-A allele subtypes, specifically HLA-A2.1 , Al, A3.2, and A24.1. Similarly, the majority of the Asian population is encompassed with the addition of peptides binding to a fifth allele HLA- A 11.2.
TABLE 1
A Allele/Subtvpe N(69)* A(54) C(502)
Al 10.1(7) 1.8(1) 27.4(138)
A2.1 11.5(8) 37.0(20) 39.8(199)
A2.2 10.1(7) 0 3.3(17)
A2.3 1.4(1) 5.5(3) 0.8(4)
A2.4 - - -
A2.5 - - -
A3.1 1.4(1) 0 0.2(0)
A3.2 5.7(4) 5.5(3) 21.5(108)
Al l . l 0 5.5(3) 0
A11.2 5.7(4) 31.4(17) 8.7(44)
Al l .3 0 3.7(2) 0
A23 4.3(3) - 3.9(20)
A24 2.9(2) 27.7(15) 15.3(77)
A24.2 - - -
A24.3 - - -
A25 1.4(1) - 6.9(35)
A26.1 4.3(3) 9.2(5) 5.9(30)
A26.2 7.2(5) - 1.0(5)
A26V - 3.7(2) -
A28.1 10.1(7) - 1.6(8)
A28.2 1.4(1) - 7.5(38)
A29.1 1.4(1) - 1.4(7)
A29.2 10.1(7) 1.8(1) 5.3(27)
A30.1 8.6(6) - 4.9(25)
A30.2 1.4(1) - 0.2(1)
A30.3 7.2(5) - 3.9(20)
A31 4.3(3) 7.4(4) 6.9(35)
A32 2.8(2) - 7.1(36)
Aw33.1 8.6(6) - 2.5(13)
Aw33.2 2.8(2) 16.6(9) 1.2(6)
Aw34.1 1.4(1) - -
Aw34.2 14.5(10) - 0.8(4)
Aw36 5.9(4) - -
Table compiled from B. DuPont, Jmi nunobiologv of HLA. Vol. I, Histocompatibilil
Testing 1987, Springer-Verlag, New York 1989.
N - negroid; A = Asian; C = caucasoid. Numbers in parenthesis represent the number of individuals included in the analysis.
The nomenclature used to describe peptide compounds follows the conventional practice wherein the amino group is presented to the left (the N-terminus)
9 and the carboxyl group to the right (the C-terminus) of each amino acid residue. In the formulae representing selected specific embodiments of the present invention, the amino- and carboxyl-terminal groups, although not specifically shown, are in the form they would assume at physiologic pH values, unless otherwise specified. In the amino acid structure formulae, each residue is generally represented by standard three letter or single letter designations. The L-form of an amino acid residue is represented by a capital single letter or a capital first letter of a three-letter symbol, and the D-form for those amino acids having D-forms is represented by a lower case single letter or a lower case three letter symbol. Gly cine has no asymmetric carbon atom and is simply referred to as "Gly" or G. The procedures used to identify peptides of the present invention generally follow the methods disclosed in Falk et al. , Nature 351:290 (1991), which is incorporated herein by reference. Briefly, the methods involve large-scale isolation of MHC class I molecules, typically by immunoprecipitation or affinity chromatography, from the appropriate cell or cell line. Examples of other methods for isolation of the desired MHC molecule equally well known to the artisan include ion exchange chromatography, lectin chromatography, size exclusion, high performance ligand chromatography, and a combination of all of the above techniques.
In the typical case, immunoprecipitation is used to isolate the desired allele. A number of protocols can be used, depending upon the specificity of the antibodies used. For example, allele-specific mAb reagents can be used for the affinity purification of the
HLA-A, HLA-B,, and HLA-C molecules. Several mAb reagents for the isolation of HLA-A molecules are available. The monoclonal BB7.2 is suitable for isolating HLA-A2 molecules. Affinity columns prepared with these mAbs using standard techniques are successfully used to purify the respective HLA-A allele products. In addition to allele-specific mAbs, broadly reactive anti-HLA-A, B, C mAbs, such as W6/32 and B9.12.1, and one anti-HLA-B, C mAb, B 1.23.2, could be used in alternative affinity purification protocols as described in previous applications.
The peptides bound to the peptide binding groove of the isolated MHC molecules are eluted typically using acid treatment. Peptides can also be dissociated from class I molecules by a variety of standard denaturing means, such as heat, pH, detergents, salts, chaotropic agents, or a combination thereof.
10
Peptide fractions are further separated from the MHC molecules by reversed-phase high performance liquid chromatography (HPLC) and sequenced. Peptides can be separated by a variety of other standard means well known to the artisan, including filtration, ultrafiltration, electrophoresis, size chromatography, precipitation with specific antibodies, ion exchange chromatography, isoelectrofocusing, and the like.
Sequencing of the isolated peptides can be performed according to standard techniques such as Edman degradation (Hunkapiller, M.W. , et al.. Methods Enzymol. 91, 399 [1983]). Other methods suitable for sequencing include mass spectrometry sequencing of individual peptides as previously described (Hunt, et al., Science 225: 1261 (1992), which is incorporated herein by reference). Amino acid sequencing of bulk heterogenous peptides (e.g.. pooled HPLC fractions) from different class I molecules typically reveals a characteristic sequence motif for each class I allele.
Definition of motifs specific for different class I alleles allows the identification of potential peptide epitopes from an antigenic protein whose amino acid sequence is known. Typically, identification of potential peptide epitopes is initially carried out using a computer to scan the amino acid sequence of a desired antigen for the presence of motifs. The epitopic sequences are then synthesized. The capacity to bind MHC Class molecules is measured in a variety of different ways. One means is a Class I molecule binding assay as described in the related applications, noted above. Other alternatives described in the literature include inhibition of antigen presentation (Sette, et al. , J. Immunol. 141:3893 (1991), in vitro assembly assays (Townsend, et al., £eϋ 62:285 (1990), and FACS based assays using mutated ells, such as RMA.S (Melief, et al. , Eur. J. Immunol. 21:2963 (1991)).
Next, peptides that test positive in the MHC class I binding assay are assayed for the ability of the peptides to induce specific CTL responses in vitro. For instance, Antigen-presenting cells that have been incubated with a peptide can be assayed for the ability to induce CTL responses in responder cell populations. Antigen-presenting cells can be normal cells such as peripheral blood mononuclear cells or dendritic cells (Inaba, et al. , J. Exp. Med. 166: 182 (1987); Boog, Eur. J. Immunol. 18:219 [1988]). Alternatively, mutant mammalian cell lines that are deficient in their ability to load class I molecules with internally processed peptides, such as the mouse cell lines RMA-S (Karre, et al.. Nature. 319:675 (1986); Ljunggren, et al., Fur. J. Immunol.
11
21:2963-2970 (1991)), and the human somatic T cell hybrid, T-2 (Cerundolo, et al. , Nature 345:449-452 (1990)) and which have been transfected with the appropriate human class I genes are conveniently used, when peptide is added to them, to test for the capacity of the peptide to induce in vitro primary CTL responses. Other eukaryotic cell lines which could be used include various insect cell lines such as mosquito larvae (ATCC cell lines
CCL 125, 126, 1660, 1591, 6585, 6586), silkworm (ATTC CRL 8851), armyworm (ATCC CRL 1711), moth (ATCC CCL 80) and Drosophila cell lines such as a Schneider cell line (see Schneider J. Embryol. Exp. Morphol. 27:353-365 [1927]).
Peripheral blood lymphocytes are conveniently isolated following simple venipuncture or leukapheresis of normal donors or patients and used as the responder cell sources of CTL precursors. In one embodiment, the appropriate antigen-presenting cells are incubated with 10-100 μM of peptide in serum-free media for 4 hours under appropriate culture conditions. The peptide-loaded antigen-presenting cells are then incubated with the responder cell populations in vitro for 7 to 10 days under optimized culture conditions. Positive CTL activation can be determined by assaying the cultures for the presence of CTLs that kill radiolabeled target cells, both specific peptide-pulsed targets as well as target cells expressing endogenously processed form of the relevant virus or tumor antigen from which the peptide sequence was derived.
Specificity and MHC restriction of the CTL is determined by testing against different peptide target cells expressing appropriate or inappropriate human MHC class I.
The peptides that test positive in the MHC binding assays and give rise to specific CTL responses are referred to herein as immunogenic peptides.
The immunogenic peptides can be prepared synthetically, or by recombinant DNA technology or from natural sources such as whole viruses or tumors. Although the peptide will preferably be substantially free of other naturally occurring host cell proteins and fragments thereof, in some embodiments the peptides can be synthetically conjugated to native fragments or particles.
The polypeptides or peptides can be a variety of lengths, either in their neutral (uncharged) forms or in forms which are salts, and either free of modifications such as glycosylation, side chain oxidation, or phosphorylation or containing these modifications, subject to the condition that the modification not destroy the biological activity of the polypeptides as herein described.
12
Desirably, the peptide will be as small as possible while still maintaining substantially all of the biological activity of the large peptide. When possible, it may be desirable to optimize peptides of the invention to a length of 9 or 10 amino acid residues, commensurate in size with endogenously processed viral peptides or tumor cell peptides that are bound to MHC class I molecules on the cell surface.
Peptides having the desired activity may be modified as necessary to provide certain desired attributes, e.g. , improved pharmacological characteristics, while increasing or at least retaining substantially all of the biological activity of the unmodified peptide to bind the desired MHC molecule and activate the appropriate T cell. For instance, the peptides may be subject to various changes, such as substitutions, either conservative or non-conservative, where such changes might provide for certain advantages in their use, such as improved MHC binding. By conservative substitutions is meant replacing an amino acid residue with another which is biologically and/or chemically similar, e.g., one hydrophobic residue for another, or one polar residue for another. The substitutions include combinations such as Gly, Ala; Val, He, Leu, Met;
Asp, Glu; Asn, Gin; Ser, Thr; Lys, Arg; and Phe, Tyr. The effect of single amino acid substitutions may also be probed using D-amino acids. Such modifications may be made using well known peptide synthesis procedures, as described in e.g. , Merrifield, Science 232:341-347 (1986), Barany and Merrifield, The Peptides. Gross and Meienhofer, eds. (N.Y. , Academic Press), pp. 1-284 (1979); and Stewart and Young, Solid Phase Peptide
Synthesis. (Rockford, 111. , Pierce), 2d Ed. (1984), incorporated by reference herein.
The peptides can also be modified by extending or decreasing the compound's amino acid sequence, e.g. , by the addition or deletion of amino acids. The peptides or analogs of the invention can also be modified by altering the order or composition of certain residues, it being readily appreciated that certain amino acid residues essential for biological activity, e.g., those at critical contact sites or conserved residues, may generally not be altered without an adverse effect on biological activity. The non-critical amino acids need not be limited to those naturally occurring in proteins, such as L-α-amino acids, or their D-isomers, but may include non-natural amino acids as well, such as β-γ-δ-amino acids, as well as many derivatives of L-α-amino acids.
Typically, a series of peptides with single amino acid substitutions are employed to determine the effect of electrostatic charge, hydrophobicity, etc. on binding.
13
For instance, a series of positively charged (e.g. , Lys or Arg) or negatively charged (e.g. , Glu) amino acid substitutions are made along the length of the peptide revealing different patterns of sensitivity towards various MHC molecules and T cell receptors. In addition, multiple substitutions using small, relatively neutral moieties such as Ala, Gly, Pro, or similar residues may be employed. The substitutions may be homo-oligomers or hetero- oligomers. The number and types of residues which are substituted or added depend on the spacing necessary between essential contact points and certain functional attributes which are sought (e.g., hydrophobicity versus hydrophilicity). Increased binding affinity for an MHC molecule or T cell receptor may also be achieved by such substitutions, compared to the affinity of the parent peptide. In any event, such substitutions should employ amino acid residues or other molecular fragments chosen to avoid, for example, steric and charge interference which might disrupt binding.
Amino acid substitutions are typically of single residues. Substitutions, deletions, insertions or any combination thereof may be combined to arrive at a final peptide. Substitutional variants are those in which at least one residue of a peptide has been removed and a different residue inserted in its place. Such substitutions generally are made in accordance with the following Table 2 when it is desired to finely modulate the characteristics of the peptide.
TABLE 2
Original Residue Exemplarv Substitution
Ala Ser
Arg Lys, His
Asn Gin
Asp Glu
Cys Ser
Gin Asn
Glu Asp
Gly Pro
His Lys; Arg
He Leu; Val
Leu He; Val
Lys Arg; His
Met Leu; He
Phe Tyr; Trp
Ser Thr
Thr Ser
Trp Tyr; Phe
Tyr Trp; Phe
Val He; Leu
Substantial changes in function (e.g., affinity for MHC molecules or T cell receptors) are made by selecting substitutions that are less conservative than those in Table 2, i.e. , selecting residues that differ more significantly in their effect on maintaining (a) the structure of the peptide backbone in the area of the substitution, for example as a sheet or helical conformation, (b) the charge or hydrophobicity of the molecule at the target site or (c) the bulk of the side chain. The substimtions which in general are expected to produce the greatest changes in peptide properties will be those in which (a) hydrophilic residue, e.g. seryl, is substituted for (or by) a hydrophobic residue, e.g. leucyl, isoleucyl, phenylalanyl, valyl or alanyl; (b) a residue having an electropositive side chain, e.g., lysl, arginyl, or histidyl, is substituted for (or by) an electronegative residue, e.g. glutamyl or aspartyl; or (c) a residue having a bulky side chain, e.g. phenylalanine, is substituted for (or by) one not having a side chain, e.g., glycine.
The peptides may also comprise isosteres of two or more residues in the immunogenic peptide. An isostere as defined here is a sequence of two or more residues that can be substituted for a second sequence because the steric conformation of the first sequence fits a binding site specific for the second sequence. The term specifically includes peptide backbone modifications well known to those skilled in the art. Such modifications include modifications of the amide nitrogen, the α-carbon, amide carbonyl, complete replacement of the amide bond, extensions, deletions or backbone crosslinks. See, generally. Spatola, Chemistry and Biochemistry of Amino Acids, peptides and Proteins. Vol. VII (Weinstein ed. , 1983).
Modifications of peptides with various amino acid mimetics or unnatural amino acids are particularly useful in increasing the stability of the peptide in vivo. Stability can be assayed in a number of ways. For instance, peptidases and various biological media, such as human plasma and serum, have been used to test stability. See, e.g.. Verhoef et al., Eur. J. Drug Metab. Pharmacokin. 11:291-302 (1986). Half life of the peptides of the present invention is conveniently determined using a 25 % human serum (v/v) assay. The protocol is generally as follows. Pooled human serum (Type AB, non-heat inactivated) is delipidated by centrifugation before use. The serum is then diluted to 25% with RPMI tissue culture media and used to test peptide stability. At predetermined time intervals a small amount of reaction solution is removed and added to either 6% aqueous trichloracetic acid or ethanol. The cloudy reaction sample is cooled
16
(4°C) for 15 minutes and then spun to pellet the precipitated serum proteins. The presence of the peptides is then determined by reversed-phase HPLC using stability-specific chromatography conditions.
The peptides of the present invention or analogs thereof which have CTL stimulating activity may be modified to provide desired attributes other than improved serum half life. For instance, the ability of the peptides to induce CTL activity can be enhanced by linkage to a sequence which contains at least one epitope that is capable of inducing a T helper cell response. Particularly preferred immunogenic peptides/T helper conjugates are linked by a spacer molecule. The spacer is typically comprised of relatively small, neutral molecules, such as amino acids or amino acid mimetics, which are substantially uncharged under physiological conditions. The spacers are typically selected from, e.g. , Ala, Gly, or other neutral spacers of nonpolar amino acids or neutral polar amino acids. It will be understood that the optionally present spacer need not be comprised of the same residues and thus may be a hetero- or homo-oligomer. When present, the spacer will usually be at least one or two residues, more usually three to six residues. Alternatively, the CTL peptide may be linked to the T helper peptide without a spacer.
The immunogenic peptide may be linked to the T helper peptide either directly or via a spacer either at the amino or carboxy terminus of the CTL peptide. The amino terminus of either the immunogenic peptide or the T helper peptide may be acylated.
Exemplary T helper peptides include tetanus toxoid 830-843, influenza 307-319, malaria circumsporozoite 382-398 and 378-389.
In some embodiments it may be desirable to include in the pharmaceutical compositions of the invention at least one component which primes CTL. Lipids have been identified as agents capable of priming CTL in vivo against viral antigens. For example, palmitic acid residues can be attached to the alpha and epsilon amino groups of a Lys residue and then linked, e.g., via one or more linking residues such as Gly, Gly-Gly-, Ser, Ser-Ser, or the like, to an immunogenic peptide. The lipidated peptide can then be injected directly in a micellar form, incorporated into a liposome or emulsified in an adjuvant, e.g. , incomplete Freund's adjuvant. In a preferred embodiment a particularly effective immunogen comprises palmitic acid attached to alpha and epsilon amino groups
17 of Lys, which is attached via linkage, e.g. , Ser-Ser, to the amino terminus of the immunogenic peptide.
As another example of lipid priming of CTL responses, E. coli lipoproteins, such as tripalmitoyl-S-glycerylcysteinlyseryl-serine (P3CSS) can be used to prime virus specific CTL when covalently attached to an appropriate peptide. See, Deres et al. ,
Nature 342:561-564 (1989), incorporated herein by reference. Peptides of the invention can be coupled to P3CSS, for example, and the lipopeptide administered to an individual to specifically prime a CTL response to the target antigen. Further, as the induction of neutralizing antibodies can also be primed with P3CSS conjugated to a peptide which displays an appropriate epitope, the two compositions can be combined to more effectively elicit both humoral and cell-mediated responses to infection.
In addition, additional amino acids can be added to the termini of a peptide to provide for ease of linking peptides one to another, for coupling to a carrier support, or larger peptide, for modifying the physical or chemical properties of the peptide or oligopeptide, or the like. Amino acids such as tyrosine, cysteine, lysine, glutamic or aspartic acid, or the like, can be introduced at the C- or N-terminus of the peptide or oligopeptide. Modification at the C terminus in some cases may alter binding characteristics of the peptide. In addition, the peptide or oligopeptide sequences can differ from the natural sequence by being modified by terminal-NH2 acylation, e.g. , by alkanoyl (C,-C2o) or thioglycolyl acetylation, terminal-carboxyl amidation, e.g. , ammonia, methylamine, etc. In some instances these modifications may provide sites for linking to a support or other molecule.
The peptides of the invention can be prepared in a wide variety of ways. Because of their relatively short size, the peptides can be synthesized in solution or on a solid support in accordance with conventional techniques. Various automatic synthesizers are commercially available and can be used in accordance with known protocols. See, for example, Stewart and Young, Solid Phase Peptide Synthesis. 2d. ed. , Pierce Chemical Co. (1984), supra.
Alternatively, recombinant DNA technology may be employed wherein a nucleotide sequence which encodes an immunogenic peptide of interest is inserted into an expression vector, transformed or transfected into an appropriate host cell and cultivated under conditions suitable for expression. These procedures are generally known in the art,
18 as described generally in Sambrook et al. , Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, New York (1982), which is incorporated herein by reference. Thus, fusion proteins which comprise one or more peptide sequences of the invention can be used to present the appropriate T cell epitope. As the coding sequence for peptides of the length contemplated herein can be synthesized by chemical techniques, for example, the phosphotriester method of Matteucci et al. , J. Am. Chem. Soc. 103:3185 (1981), modification can be made simply by substituting the appropriate base(s) for those encoding the native peptide sequence. The coding sequence can then be provided with appropriate linkers and ligated into expression vectors commonly available in the art, and the vectors used to transform suitable hosts to produce the desired fusion protein. A number of such vectors and suitable host systems are now available. For expression of the fusion proteins, the coding sequence will be provided with operably linked start and stop codons, promoter and terminator regions and usually a replication system to provide an expression vector for expression in the desired cellular host. For example, promoter sequences compatible with bacterial hosts are provided in plasmids containing convenient restriction sites for insertion of the desired coding sequence. The resulting expression vectors are transformed into suitable bacterial hosts. Of course, yeast or mammalian cell hosts may also be used, employing suitable vectors and control sequences. The peptides of the present invention and pharmaceutical and vaccine compositions thereof are useful for administration to mammals, particularly humans, to treat and/or prevent viral infection and cancer. Examples of diseases which can be treated using the immunogenic peptides of the invention include prostate cancer, hepatitis B, hepatitis C, AIDS, renal carcinoma, cervical carcinoma, lymphoma, CMV and condlyloma acuminatum.
For pharmaceutical compositions, the immunogenic peptides of the invention are administered to an individual already suffering from cancer or infected with the virus of interest. Those in the incubation phase or the acute phase of infection can be treated with the immunogenic peptides separately or in conjunction with other treatments, as appropriate. In therapeutic applications, compositions are administered to a patient in an amount sufficient to elicit an effective CTL response to the virus or tumor antigen and to cure or at least partially arrest symptoms and/or complications. An amount adequate to
19 accomplish this is defined as " therapeutical ly effective dose. " Amounts effective for this use will depend on, e.g. , the peptide composition, the manner of administration, the stage and severity of the disease being treated, the weight and general state of health of the patient, and the judgment of the prescribing physician, but generally range for the initial immunization (that is for therapeutic or prophylactic administration) from about 1.0 μg to about 5000 μg of peptide for a 70 kg patient, followed by boosting dosages of from about 1.0 μg to about 1000 μg of peptide pursuant to a boosting regimen over weeks to months depending upon the patient's response and condition by measuring specific CTL activity in the patient's blood. It must be kept in mind that the peptides and compositions of the present invention may generally be employed in serious disease states, that is, life- threatening or potentially life threatening situations. In such cases, in view of the minimization of extraneous substances and the relative nontoxic nature of the peptides, it is possible and may be felt desirable by the treating physician to administer substantial excesses of these peptide compositions. For therapeutic use, administration should begin at the first sign of viral infection or the detection or surgical removal of tumors or shortly after diagnosis in the case of acute infection. This is followed by boosting doses until at least symptoms are substantially abated and for a period thereafter. In chronic infection, loading doses followed by boosting doses may be required. Treatment of an infected individual with the compositions of the invention may hasten resolution of the infection in acutely infected individuals. For those individuals susceptible (or predisposed) to developing chronic infection the compositions are particularly useful in methods for preventing the evolution from acute to chronic infection. Where the susceptible individuals are identified prior to or during infection, for instance, as described herein, the composition can be targeted to them, minimizing need for administration to a larger population.
The peptide compositions can also be used for the treatment of chronic infection and to stimulate the immune system to eliminate virus-infected cells in carriers. It is important to provide an amount of immuno-potentiating peptide in a formulation and mode of administration sufficient to effectively stimulate a cytotoxic T cell response.
Thus, for treatment of chronic infection, a representative dose is in the range of about 1.0 μg to about 5000 μg, preferably about 5 μg to 1000 μg for a 70 kg patient per dose.
20
Immunizing doses followed by boosting doses at established intervals, e.g. , from one to four weeks, may be required, possibly for a prolonged period of time to effectively immunize an individual. In the case of chronic infection, administration should continue until at least clinical symptoms or laboratory tests indicate that the viral infection has been eliminated or substantially abated and for a period thereafter.
The pharmaceutical compositions for therapeutic treatment are intended for parenteral, topical, oral or local administration. Preferably, the pharmaceutical compositions are administered parenterally, e.g. , intravenously, subcutaneously, intradermally, or intramuscularly. Thus, the invention provides compositions for parenteral administration which comprise a solution of the immunogenic peptides dissolved or suspended in an acceptable carrier, preferably an aqueous carrier. A variety of aqueous carriers may be used, e.g., water, buffered water, 0.8% saline, 0.3% glycine, hyaluronic acid and the like. These compositions may be sterilized by conventional, well known sterilization techniques, or may be sterile filtered. The resulting aqueous solutions may be packaged for use as is, or lyophilized, the lyophilized preparation being combined with a sterile solution prior to administration. The compositions may contain pharmaceutically acceptable auxiliary substances as required to approximate physiological conditions, such as pH adjusting and buffering agents, tonicity adjusting agents, wetting agents and the like, for example, sodium acetate, sodium lactate, sodium chloride, potassium chloride, calcium chloride, sorbitan monolaurate, triethanolamine oleate, etc.
The concentration of CTL stimulatory peptides of the invention in the pharmaceutical formulations can vary widely, i.e. , from less than about 0.1 %, usually at or at least about 2% to as much as 20% to 50% or more by weight, and will be selected primarily by fluid volumes, viscosities, etc., in accordance with the particular mode of administration selected.
The peptides of the invention may also be administered via liposomes, which serve to target the peptides to a particular tissue, such as lymphoid tissue, or targeted selectively to infected cells, as well as increase the half-life of the peptide composition. Liposomes include emulsions, foams, micelles, insoluble monolayers, liquid crystals, phospholipid dispersions, lamellar layers and the like. In these preparations the peptide to be delivered is incorporated as part of a liposome, alone or in conjunction with a molecule which binds to, e.g. , a receptor prevalent among lymphoid cells, such as monoclonal
21 antibodies which bind to the CD45 antigen, or with other therapeutic or immunogenic compositions. Thus, liposomes either filled or decorated with a desired peptide of the invention can be directed to the site of lymphoid cells, where the liposomes then deliver the selected therapeutic/immunogenic peptide compositions. Liposomes for use in the invention are formed from standard vesicle-forming lipids, which generally include neutral and negatively charged phospholipids and a sterol, such as cholesterol. The selection of lipids is generally guided by consideration of, e.g. , liposome size, acid lability and stability of the liposomes in the blood stream. A variety of methods are available for preparing liposomes, as described in, e.g. , Szoka et al. , Ann. Rev. Biophys. Bioeng. 9:467 (1980), U.S. Patent Nos. 4,235,871 , 4,501 ,728, 4,837,028, and 5,019,369, incorporated herein by reference.
For targeting to the immune cells, a ligand to be incorporated into the liposome can include, e.g. , antibodies or fragments thereof specific for cell surface determinants of the desired immune system cells. A liposome suspension containing a peptide may be administered intravenously, locally, topically, etc. in a dose which varies according to, inter alia, the manner of administration, the peptide being delivered, and the stage of the disease being treated.
For solid compositions, conventional nontoxic solid carriers may be used which include, for example, pharmaceutical grades of mannitol, lactose, starch, magnesium stearate, sodium saccharin, talcum, cellulose, glucose, sucrose, magnesium carbonate, and the like. For oral administration, a pharmaceutically acceptable nontoxic composition is formed by incorporating any of the normally employed excipients, such as those carriers previously listed, and generally 10-95% of active ingredient, that is, one or more peptides of the invention, and more preferably at a concentration of 25% -75 % . For aerosol administration, the immunogenic peptides are preferably supplied in finely divided form along with a surfactant and propellant. Typical percentages of peptides are 0.01 %-20% by weight, preferably 1 %-10%. The surfactant must, of course, be nontoxic, and preferably soluble in the propellant. Representative of such agents are the esters or partial esters of fatty acids containing from 6 to 22 carbon atoms, such as caproic, octanoic, lauric, palmitic, stearic, linoleic, linolenic, olesteric and oleic acids with an aliphatic polyhydric alcohol or its cyclic anhydride. Mixed esters, such as mixed or natural glycerides may be employed. The surfactant may constitute 0.1 % -20% by weight
22 of the composition, preferably 0.25-5%. The balance of the composition is ordinarily propellant. A carrier can also be included, as desired, as with, e.g. , lecithin for intranasal delivery.
In another aspect the present invention is directed to vaccines which contain as an active ingredient an immunogenically effective amount of an immunogenic peptide as described herein. The peptide(s) may be introduced into a host, including humans, linked to its own carrier or as a homopolymer or heteropolymer of active peptide units. Such a polymer has the advantage of increased immunological reaction and, where different peptides are used to make up the polymer, the additional ability to induce antibodies and/or CTLs that react with different antigenic determinants of the virus or tumor cells. Useful carriers are well known in the art, and include, e.g. , thyroglobulin, albumins such as human serum albumin, tetanus toxoid, polyamino acids such as poly(lysine:glutamic acid), influenza, hepatitis B virus core protein, hepatitis B virus recombinant vaccine and the like. The vaccines can also contain a physiologically tolerable (acceptable) diluent such as water, phosphate buffered saline, or saline, and further typically include an adjuvant. Adjuvants such as incomplete Freund's adjuvant, aluminum phosphate, aluminum hydroxide, or alum are materials well known in the art. And, as mentioned above, CTL responses can be primed by conjugating peptides of the invention to lipids, such as P3CSS. Upon immunization with a peptide composition as described herein, via injection, aerosol, oral, transdermal or other route, the immune system of the host responds to the vaccine by producing large amounts of CTLs specific for the desired antigen, and the host becomes at least partially immune to later infection, or resistant to developing chronic infection.
Vaccine compositions containing the peptides of the invention are administered to a patient susceptible to or otherwise at risk of viral infection or cancer to elicit an immune response against the antigen and thus enhance the patient's own immune response capabilities. Such an amount is defined to be an "immunogenically effective dose. " In this use, the precise amounts again depend on the patient's state of health and weight, the mode of administration, the nature of the formulation, etc. , but generally range from about 1.0 μg to about 5000 μg per 70 kilogram patient, more commonly from about 10 μg to about 500 μg mg per 70 kg of body weight.
23
In some instances it may be desirable to combine the peptide vaccines of the invention with vaccines which induce neutralizing antibody responses to the virus of interest, particularly to viral envelope antigens.
For therapeutic or immunization purposes, nucleic acids encoding one or more of the peptides of the invention can also be admisitered to the patient. A number of methods are conveniently used to deliver the nucleic acids to the patient. For instance, the nulceic acid can be delivered directly, as "naked DNA". This approach is described, for instance, in Wolff et. al. , Science 247: 1465-1468 (1990) as well as U.S. Patent Nos. 5,580,859 and 5,589,466. The nucleic acids can also be administered using ballistic delivery as described, for instance, in U.S. Patent No. 5,204,253. Particles comprised solely of DNA can be administered. Alternatively, DNA can be adhered to particles, such as gold particles. The nucleci acids can also be delivered complexed to cationic compounds, such as cationic lipids. Lipid-mediated gene delivery methods are described, for instance, in WO 96/18372; WO 93/24640; Mannino and Gould-Fogerite (1988) BioTechniques 6(7): 682-691; Rose U.S. Pat No. 5,279,833; WO 91/06309; and Feigner et al. (1987) Proc. Natl. Acad. Sci. USA 84: 7413-7414. The peptides of the invention can also be expressed by attenuated viral hosts, such as vaccinia or fowlpox. This approach involves the use of vaccinia virus as a vector to express nucleotide sequences that encode the peptides of the invention. Upon introduction into an acutely or chronically infected host or into a noninfected host, the recombinant vaccinia virus expresses the immunogenic peptide, and thereby elicits a host CTL response. Vaccinia vectors and methods useful in immunization protocols are described in, e.g. , U.S. Patent No. 4,722,848, incorporated herein by reference. Another vector is BCG (Bacille Calmette Guerin). BCG vectors are described in Stover et al. (Nature 351 :456-460 (1991)) which is incorporated herein by reference. A wide variety of other vectors useful for therapeutic administration or immunization of the peptides of the invention, e.g., Salmonella typhi vectors and the like, will be apparent to those skilled in the art from the description herein. A preferred means of administering nucleic acids encoding the peptides of the invention uses minigene constructs encoding multiple epitopes of the invention. To create a DNA sequence encoding the selected CTL epitopes (minigene) for expression in human cells, the amino acid sequences of the epitopes are reverse translated. A human codon usage table is used to guide the codon choice for each amino acid. These epitope-encoding
24
DNA sequences are directly adjoined, creating a continuous polypeptide sequence. To optimize expression and/or immunogenicity, additional elements can be incorporated into the minigene design. Examples of amino acid sequence that could be reverse translated and included in the minigene sequence include: helper T lymphocyte epitopes, a leader (signal) sequence, and an endoplasmic reticulum retention signal. In addition, MHC presentation of CTL epitopes may be improved by including synthetic (e.g. poly-alanine) or naturally-occurring flanking sequences adjacent to the CTL epitopes.
The minigene sequence is converted to DNA by assembling oligonucleotides that encode the plus and minus strands of the minigene. Overlapping oligonucleotides (30- 100 bases long) are synthesized, phosphorylated, purified and annealed under appropriate conditions using well known techniques, he ends of the oligonucleotides are joined using T4 DNA ligase. This synthetic minigene, encoding the CTL epitope polypeptide, can then cloned into a desired expression vector.
Standard regulatory sequences well known to those of skill in the art are included in the vector to ensure expression in the target cells. Several vector elements are required: a promoter with a down-stream cloning site for minigene insertion; a polyadenylation signal for efficient transcription termination; an E. coli origin of replication; and an E. coli selectable marker (e.g. ampicillin or kanamycin resistance). Numerous promoters can be used for this purpose, e.g., the human cytomegalovirus (hCMV) promoter. See, U.S. Patent Nos. 5,580,859 and 5,589,466 for other suitable promoter sequences.
Additional vector modifications may be desired to optimize minigene expression and immunogenicity. In some cases, introns are required for efficient gene expression, and one or more synthetic or naturally-occurring introns could be incorporated into the transcribed region of the minigene. The inclusion of mRNA stabilization sequences can also be considered for increasing minigene expression. It has recently been proposed that immunostimulatory sequences (ISSs or CpGs) play a role in the immunogenicity of DNA vaccines. These sequences could be included in the vector, outside the minigene coding sequence, if found to enhance immunogenicity. In some embodiments, a bicistronic expression vector, to allow production of the minigene-encoded epitopes and a second protein included to enhance or decrease immunogenicity can be used. Examples of proteins or polypeptides that could beneficially
25 enhance the immune response if co-expressed include cytokines (e.g. , IL2, IL12, GM- CSF), cytokine-inducing molecules (e.g. LeIF) or costimulatory molecules. Helper (HTL) epitopes could be joined to intracellular targeting signals and expressed separately from the CTL epitopes. This would allow direction of the HTL epitopes to a cell compartment different than the CTL epitopes. If required, this could facilitate more efficient entry of HTL epitopes into the MHC class II pathway, thereby improving CTL induction. In contrast to CTL induction, specifically decreasing the immune response by co-expression of immunosuppressive molecules (e.g. TGF-β) may be beneficial in certain diseases. Once an expression vector is selected, the minigene is cloned into the polylinker region downstream of the promoter. This plasmid is transformed into an appropriate E. coli strain, and DNA is prepared using standard techniques. The orientation and DNA sequence of the minigene, as well as all other elements included in the vector, are confirmed using restriction mapping and DNA sequence analysis. Bacterial cells harboring the correct plasmid can be stored as a master cell bank and a working cell bank.
Therapeutic quantities of plasmid DNA are produced by fermentation in E. coli, followed by purification. Aliquots from the working cell bank are used to inoculate fermentation medium (such as Terrific Broth), and grown to saturation in shaker flasks or a bioreactor according to well known techniques. Plasmid DNA can be purified using standard bioseparation technologies such as solid phase anion-exchange resins supplied by Quiagen. If required, supercoiled DNA can be isolated from the open circular and linear forms using gel electrophoresis or other methods.
Purified plasmid DNA can be prepared for injection using a variety of formulations. The simplest of these is reconstitution of lyophilized DNA in sterile phosphate -buffer saline (PBS). A variety of methods have been described, and new techniques may become available. As noted above, nucleic acids are conveniently formulated with cationic lipids. In addition, glycolipids, fusogenic liposomes, peptides and compounds referred to collectively as protective, interactive, non-condensing (PINC) could also be complexed to purified plasmid DNA to influence variables such as stability, intramuscular dispersion, or trafficking to specific organs or cell types.
Target cell sensitization can be used as a functional assay for expression and MHC class I presentation of minigene-encoded CTL epitopes. The plasmid DNA is
26 introduced into a mammalian cell line that is suitable as a target for standard CTL chromium release assays. The transfection method used will be dependent on the final formulation. Electroporation can be used for "naked" DNA, whereas cationic lipids allow direct in vitro transfection. A plasmid expressing green fluorescent protein (GFP) can be co-transfected to allow enrichment of transfected cells using fluorescence activated cell sorting (FACS). These cells are then chromium-51 labeled and used as target cells for epitope-specific CTL lines. Cytolysis, detected by 51Cr release, indicates production of MHC presentation of minigene-encoded CTL epitopes.
In vivo immunogenicity is a second approach for functional testing of minigene DNA formulations. Transgenic mice expressing appropriate human MHC molecules are immunized with the DNA product. The dose and route of administration are formulation dependent (e.g. IM for DNA in PBS, IP for lipid-complexed DNA). Twenty-one days after immunization, splenocytes are harvested and restimulated for 1 week in the presence of peptides encoding each epitope being tested. These effector cells (CTLs) are assayed for cytolysis of peptide-loaded, chromium-51 labeled target cells using standard techniques. Lysis of target cells sensitized by MHC loading of peptides corresponding to minigene-encoded epitopes demonstrates DNA vaccine function for in vivo induction of CTLs.
Antigenic peptides may be used to elicit CTL ex vivo, as well. The resulting CTL, can be used to treat chronic infections (viral or bacterial) or tumors in patients that do not respond to other conventional forms of therapy, or will not respond to a peptide vaccine approach of therapy. Ex vivo CTL responses to a particular pathogen (infectious agent or tumor antigen) are induced by incubating in tissue culture the patient's CTL precursor cells (CTLp) together with a source of antigen-presenting cells (APC) and the appropriate immunogenic peptide. After an appropriate incubation time (typically 1-4 weeks), in which the CTLp are activated and mature and expand into effector CTL, the cells are infused back into the patient, where they will destroy their specific target cell (an infected cell or a tumor cell).
The peptides may also find use as diagnostic reagents. For example, a peptide of the invention may be used to determine the susceptibility of a particular individual to a treatment regimen which employs the peptide or related peptides, and thus may be helpful in modifying an existing treatment protocol or in determining a prognosis for an affected
27 individual. In addition, the peptides may also be used to predict which individuals will be at substantial risk for developing chronic infection.
The following example is offered by way of illustration, not by way of limitation.
Example 1
Class I antigen isolation was carried out as described in the related applications, noted above. Namrally processed peptides were then isolated and sequenced as described there. An allele-specific motif and algorithms were determined and quantitative binding assays were carried out.
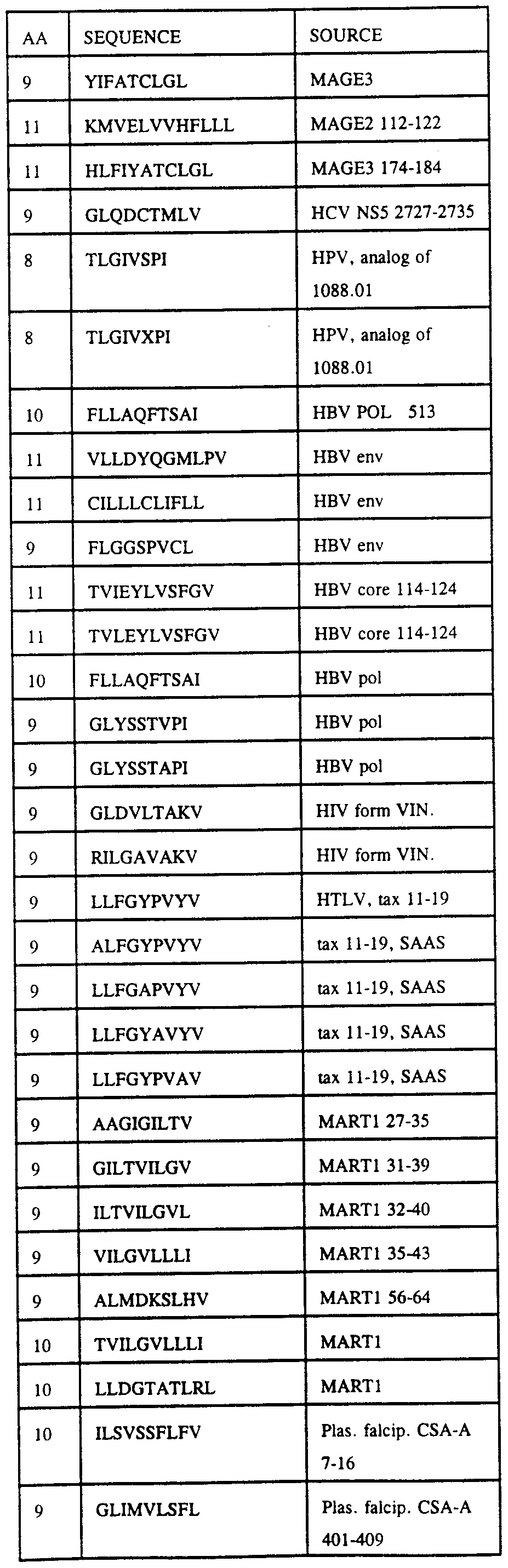
Using the motifs identified above for various HLA alleles, amino acid sequences from a number of antigens were analyzed for the presence of these motifs. Tables 3- ** provide the results of these searches.
The above examples are provided to illustrate the invention but not to limit its scope. Other variants of the invention will be readily apparent to one of ordinary skill in the art and are encompassed by the appended claims. All publications, patents, and patent applications cited herein are hereby incorporated by reference.
Table 3
Sequence Antigen Molecule
FTFSPTYKAFLSK HBV POL
GT PQEHIV KLK HBV POL
FTFSPTYKAFLCK HBV POL
GTLPQEHIV KIK HBV POL
LWSYVNTNMGLK HBV POL
STTD EAYFKDCLFK HBV X
LWSYVNVNMGLK HBV NUC
GTLPQDHIVQKIK HBV POL
STSSC HQSAVRK HBV POL
TTVNAHQILPKVLHK HBV X
RTPARVTGGVFLVDK HBV POL
28
Sequence Antigen Molecule
HTTNFASK HBV ay
FTFSPTYK HBV ayw
PTYKAFLCKQY HBVayw
CTTPAQGTSMY HBVayw
PTSCPPTCPGY HBVayw
FSQFSRGNY HBVayw
LMPLYACIQSK HBVayw
RVTGGVFLVDK HBVayw POL
HTLWKAGILYK HBVayw
10 QTRHYLHTLWK HBVayw
GTDNSWLSRK HBVayw
SYVNTNMGLKF HBVayw
LYSILSPF HBVayw YWGPSLYSIL HBVayw
15 LYSILSPFLPL HBVayw
PYKEFGATVEL HBVayw
CTWMNSTGFTK HCV
MYVGDLCGSVF HCV
VYLLPRRGPRL HCV
20 ITKIQNFRVYY HIV
KVYLA VPAHK HIV
KMIGGIGGFIK HIV
IVASCDKCQLK HIV
KVKQWPLTEEK HIV
25 TVNDIQKLVGK HIV
DVKQLTEAVQK HIV
AWIQDNSDIK HIV TYQIYQEPFK HIV
VTVYYGVPV K HIV
30 LTEDRWNKPQK HIV
ATDIQTKELQK HIV
QTKELQKQITK HIV
29
Sequence Antigen Molecule
WTVQPIVLPEK HIV
QVPLRPMTYK HIV nef 73-82
QVPLYPMTFK HIV nef 73-82
VPLRPMTYK HIV nef 74-82
AVDLYHFLK HIV nef 84-94
AVDLSHFLK HIV nef 84-94
ATLYCVHQR HIV, pl7, 82-90
RLRDLLLIV HIV-1 NL43 768-776
RLRDLLLIVTR HIV-1 NL43 768-778
10 RLRDYLLIVTR HIV-1 NL43 768-778
LRDLLLI TR HIV-1 NL43 769-778
QIYQEPFKNLK HIV-1 RT 507-517
AVFIHNFK HIVcon
RTLNAWVK HIVcon
15 ETAYF1LK HIVcon
RLRPGGKKK HIVgag P17/2
KIRLRPGGKK HIVgag P17/2
KIRLRPGGK HIVgag P17/2
ETTDLYCY HPV16 E7
20
GTLGIVCPICSQK HPV16 E7
30
Sequence Antigen Molecule
LMGTLGIVCPICSQK HPV16 E7
AVCDKCLK HPV16 E6
PYAVCDKCLKF HPV16 E6
HYCYSLYGTTL HPV16 E6
FYSRIREL HPV16 E6
TLEKLTNTGLY HPV18 E6
KTVLELTEVFEFAFK HPV18 E6
TMLCMCCK HPV18 E7
NTSLQDIEITCVYCK HPV18 E6
10 EVFEFAFK HPVI8 E6
KQSSKALQR Leukemia £3A2 CMI
ATGFKQSSK Leukemia t>3A2 CMI
HSATGFKQSSK Leukemia fc>3A2 CMI
FKQSSKALQR Leukemia £3A2 CMI
15 VTCLGLSY MAGE1
ITKKVADLVGFLLLK MAGE1
LVGFLLLK MAGE1
VTKAEMLESVIKNYK MAGE1
TSCILESLFR MAGE1
20 NYKHCFPEI MAGE1
SYVLVTCL MAGE1
ETDPISHTY MAGEl(a)
ETDPTSHLY MAGEl(a)
ETDPTSNTY MAGEl(a)
25 ETDPTSHVY MAGEl(a)
ETDPTSHSY MAGEl(a)
ETDPASHTY MAGEl(a)
EVDPTSHTY MAGEl(a)
ETDPTGHTY MAGEl(a)
30 ETDRTSHTY MAGEl(a)
EADPTSHTY MAGEl(a)
ETVPTSHTY MAGEl(a)
31
Sequence Antigen Molecule
ETDPTSHTY MAGE1 consensus
ETDPTGHSY MAGE1 T(a)
MFPDLESEF MAGE2
TTINYTLWR MAGE2
VIFSKASEY MAGE2
LVHFLLLKY MAGE2
LVHFLLLKY MAGE2
LVHFLLLKYR MAGE2
PVIFSKASEY MAGE2
10 STTINYTLWR MAGE2
WEWPISH MAGE2
EYLQLVFGI MAGE2
IFSKASEYL MAGE2
SFSTTINYTL MAGE2
15 LYILVTCLGL MAGE2
FATCLGLSY MAGE3
WGNWQYFFPVIFSK MAGE3
LIIVLAIIAR MAGE3
YFFPVIFSK MAGE3
20 NWQYFFPVI MAGE3
NWQYFFPVIF MAGE3
IFSKASSSL MAGE3
EVDPTSNTY MAGE41
RYPLTFGWCY nef/182
25 RYPLTFGWC nef/182
ATQIPSYK PAP
LTELYFEK PAP
HSFPHPLY PSA
TQEPALGTTCY PSA
30 VTKFMLCAGRWTGGK PSA
HVISNDVCAQVHPQK PSA
32
Sequence Antigen Molecule
LYDMSLLKNRF PSA
ETDPTGHSY T2 analog of MAGE-3
33
3 8 g S 8 8 g § 8 8
3IE CΛ
Q i
I I ! H
£ - w5 £ = 2 « y ^ ' M
» M Il e*? alt; «•* o *si
*8 5 a 2 s- & S 2 δ'3 3 *l-'sl*ι*. S SιiiϊιϊιN . j J t I w i ω I * I u I ω j ω [ » ' w j 11*» I I > | ω M i .
! I I
I — |Mlθi = lC = I
8 |w ιSι*.3 5 s- -
I I
I ! o lol e lβιβ o — _, o o C'ΣiSIS -l« *5 1*3 » S g o 8 S •fir o = lβ|0, o o — — 1°' SIM o o o I ° o S C s =18
I I
O . o o •^ 00 O . CΛ υ Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24
1 0731 RIL ETELRK 10 C-ERH2 713 3,11 0057 0.11
1 0745 VLV SPNI IVK 10 c EKH2 8.1 3,11 0082~ 0.0072
- 1 1131 SVFQNLQVIR ϊo C-ERB2 423 3,11 0017 0075
1 1133 HTVPWDQLFR 10 c ERB2 478 3.11 oάnT 0072
- - - 1 1127 ILKCGVLIQR 10 c ERB2 14H 3,1 i 0.040 0.0U)5
1.1143 LVSEFSRMAR 10 c ERB2 ~972 3,ϊi 0.0072 003T"
1 1136 GWFCILIKR 10 c ERB2 668 3.11 0018 0.033
1 0726 CVARCPSCVK 10 c ERB2 596 3,11 0.022 0.0O12
1.1137 WPCILIKRR 10 c ERB2 669 3.Ϊ1 0.0030 0.016
1.0728 GILIKRRQQK 10 C-ERB2 672 3,11 0.015 0.0014
1.1129 RTVCAGGCAR 10 < C-ERB2 217 3,11 0.0068 0013
1.1134 GLACHQLCAR 10 C-ERB2 508 3.11 0.011 0
1.1139 KIPVAIKVLR 10 C-ERB2 747 3,11 00009 00099
ro
•»* IΛ Ox IΛ o
f o>
<Λ
©
00 OS
CΛ
H U
0, Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24
10291 VCEΛDYFEY 9 EBNΛ1 409 1 0016
— — 10295 PLRESIVCY 9 EBNΛ1 553 1 "o i "
10681 PVCEADYFEY 10 EBNAl 408 1 0015
10683 CT VΛGVFVY 10 EBNΛ1 50Ϊ" 0014
1.0293 CVFVYGGSK 9 EBNAl 506 3,11 030 0 1
1.1016 KT5LYNLRR 9 EBNΛ1 514 3,11 031 0.12
10297 AIKDLVMTK 9 EBNAl 578 3,11 0048 0034
10687 QTHIFAEVLK 10 EBNAl 567 3,11 0010 021
11124 GTALAIPQCR 10 EBNAl 523 3,11 00028 0056
* IΛ*
* IΛ* o
ON oin
©
00 ON
CΛ υ Peptide Sequence AA Virus Strain Molecule Pos Motif Al A2.1 A3.2 All A24
C
50005 CTELKLSDY 9 FLU Λ NP 44 1 3 6
50006 STLELRSRY 9 FLU Λ NP 377 1 0020
50044 ILRGSVAHK 9 FLU Λ NP 265 3 1 5 00037
50051 RMCNILKGK 9 FLU Λ NP 221 3 027 0062
50046 MQGSTLPR 9 FLU Λ NP 166 3 0031 0 10
50048 MIDCICRFY 9 FLU A NP 32 3 0059 00010
50049 MVLSAFDER 9 FLU A NP 66 3 00016 0041
50054 YIQMCTELK 9 FLU A NP 40 3 00031 0030
50042 CINDRNF R 9 FLU A NP 200 3 00028 0.024
50104 SLMQGSTLPR 10 FLU A NP 165 3 0 12 084
50095 KMIDCtCRFY 10 ' FLU A NP 31 3 050 00079
50096 LILRGSVAHK 10 FLU A NP 264 3 036 0037
50102 RSGAACAAVK 10 FLU A NP 175 3 0019 00046
10
CO 50105 SSTLELRSRY 10 FLU A NP 376 3 00018 0016
50103 RSRYWAIRTR 10 FLU A NP 382 3 0012 0
5.0101 RMVLSAFDER 10 FLU A NP 65 3 00014 0010
50061 FYIQMCΓEL 9 FLU A NP 39 24 2.9
5.0060 AYERMCNIL 9 FLU A NP 218 24 0031
50112 RFYIQMCΓEL 10 FLU A NP 38 24 0 15
IΛ
ON IΛ
-^f
ON. ON o
o
IΛ
©
00
Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24
H υ 1 0155 LLDTASALY 9 HBV adr CORE 420 25 00007 0
1 0186 SLDVSAAFY 9 HBV adr I'OL 10)1 1 172 00037 00006
20125 ΓTTGRTSLY 9 HBV ALL 1.382 1 3 00008 0
20126 MSTTDLEAY 9 HBV adr 1.521 085 <00008 0
— —
1 0208 PTTCRT5LY 9 HBV adr POL 1382 077 0 0
1 0387 LTKQYLNLY 9 HBV adw IΌL 1280 ~0 50 000O3 00075
1 0166 KVCNFTGLY 9 HBV adr POL 629 0068 030 0014
20127 MSPTDLEAY 9 HBV adw 1,550 0067
2-0120 FSQFSRGNY 9 HBV ayw 984 0057
~
20112 PSSWAFAKY 9 HBV adw 316 0054
10119 QSAVRKEAY 9 ' HBV adw 881 ~~0 O25~
1 0174 PLDKGIKPY 9 HBV adr POL 698 0019 <O00O2 <000O2
1 0378 SLMLLYKTY 9 HBV adw POL 1092 0017
10115 ASRDLVYSY 9 HBV ayw 499 0013
2.0124 PSRGRLGLY 9 HBV adr/adw 1,364 0011
2.0121 SSTCRNΪNY 9 HBV adr 1,036 00097
1 0519 DLLDTΛSΛLY 10 HBV adr CORE 419 11 1 0 0
1 0513 LLDPRVRGLY 10 HBV adr ENV 120 63 017 0
2.0239 LSLDVSAAFY 10 HBV ALL 1,000 4 2 <00009 00037
1 0911 FLCQQYLHLY 10 HBV adr POL 1250 1 1 00025 0014 00048 00017
20216 QTFGRKLHLY 10 HBV ayw POL 1087 1 1 00056 0012
10244 KTYCRKLHLY 10 HBV adw 1,098 069 00003 059 022 0
1 0791 KTYCRKLHLY 10 HBV adw POL 1098 0 57 00020 053 035 00001
20242 QTFGRKLHLY 10 HBV ayw 1,087 037 00097 0011
1 0556 KTFGRKLHLY 10 HBV adx POL 1069 ~ 034 00023 0094 0090 0
- -
20241 KTFGRKLHLY 10 HBV adx 1,069 00002 0 15 0095 0
1 0766 QDPRVRALY 10 HBV adw ENV 120 021 0014 0
1 0806 TTPAQGTSMY 10 HBV adw ENV 288 020 0 0
* IΛ*
2.0240 LSδTSRNINY 10 HBV adr 1,035 020 <00009 0 IΛ ** 1 0541 PLDKGIKPYY 10 HBV adr POL 698 — t — ~ 0 16~ 0 0
10238 HSASFCGSIΎ 10 HBV ayw 767 0 15 0 0019 0017 0
O 1 0795 FLTKQYLNLY 10 HBV adw POL 1279 0 12 0 0
20237 RSASPCCSPY 10 HBV adr/adw 718 0 11 0 0033 0020 0
1 0774 WLWGMDIDPY 10 HBV adw CORE 416 0081 <00002 <00002
- _ _
20233 TTPAQCTSMY 10 HBV ayw 288 0066
1 0542 HTLWKAGILY 10 HBV adr
~ θL
~~ 721 0030
20231 T5CPPICPGY 10 HBV adr 226 0018
38
S S
8
CΛ
It A
« e
Q -o < 3
*β|*«!
-- ,03 031 S < < <ι< < < <l<
β I u I > CΛ tt i a i ■>
S.ιo-1? l"<
- i * I 5'
I !
3 3 3 US ;s:ό
-3 ξ" - S S S « ιS,S
S - *J. w ≤, ≤'2
K)ιhjιh i Ml »lκ)|hJ 1 i
' I lolo l I i I i 2i Ξ
i I
Λ I I βlβlo
Si∑ls o j o
S VJ.I|S «
ii 5*
ON roo
IΛ
©
00 ON
CΛ
Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24
H υ 20176 YYPEHLVNHY 10 I IUV ayw 735 24 0040 2 0172 AYRPPNAP1L 10 " ΪJBV ALL 521 24 — — 0022
20171 GYRWMCLRRF 10 HBV AI L 234 24 0011
50115 NFLISLCIHL 10 HBV POL 572 24 — - 00099
1 0377 YVSL LLYK 9 HBV adw POL 10W 3,1 i 031 74
1.0189 LLYKTFCRK 9 HBV adr POL 1066 3~iϊ 5.0 0.30
1.0379 LLYKTYGRK 9 HBV adw POL 1095 3,ii 2.5 0.40
1.0370 VTKYLPLDK 9 HBV adw POL 722 3,1 ϊ 0.014 1.3
1.0176 RHYLHTLWK 9 HBV adr POL 719 3,11 1.2 0.010
1.0367 STVPSFNPK 9 HBV adw POL 668 3,11 0.021 093
1 0215 TTDLEAYFK 9 HBV adr ■X" 1523 3,11 0.0006 0.92
1.0848 YVSLLLLYK 9 HBV adr POL 1061 3,11 0.39 0.92
1.0383 PTYKAFLTK 9 HBV adw POL 1274 3,11 0.17 0.71
1.0987 HLYPVARQR 9 HBV adr POL 1257 3,11 0.54 0.0020
1.0358 STMRQLGRK 9 HBV adw ENV 85 3,11 0.51 0.34
ON 1.0991 ALRFTSARR 9 HBV adr "X" 1488 3,11 0.44 <0.0005
1.0197 PVNRPIDWK 9 HBV adr POL 1197 3,11 0.080 041
1.0369 TVNENRRLK 9 HBV adw POL 703 3,11 0.016 0.40
1.1041 WNHYFQTR 9 HBV adw POL 740 3,11 0030 033
1.0152 STTSTGPCK 9 HBV adr ENV 277 3,11 0.011 0.29
1 0213 QVLPKLLHK 9 HBV adr "X" 1505 3,11 0.10 0.28
1.0172 LTKYLPLDK 9 HBV adr POL 693 3,11 0.0039 0.23
1.0374 CLHQSAVRK 9 HBV adw POL 878 3,11 0.22 0.017
1 0980 WDFSQFSR 9 HBV adr POL 963 3.11 0.011 0.20
1 0382 PLYAC1QAK 9 HBV adw POL 1259 3,11 0.18 0.034
2.0074 YVNTNMGLK 9 HBV ayw CORE 507 3.11 0 16 0048
1 0199 PLYACIQSK 9 HBV adr POL 1230 3.11 0 11 0018
* 1.0972 RLADEGLNR 9 HBV adr POL 601 3,11 0.10 0025 IΛ*
1.0976 AVNHYFKTR ON 9 HBV adr POL 711 3 Ϊ 00071 0098 IΛ 1.0975 RLKUMPAR 9 HBV adr POL 680 3,U 0.095 00002
ON 1.0977 ILYKR.ΩTR ON 9 HBV adr POL 730 3,ii 0095 <00005 o 1.0993 KVFVLGGCR 9 HBV adr "X- 1548 3,ii " 0042 0082
1.0165 NVSIPWTHK 9 HBV adr POL 621 3.11 0072 0076
1.0982 LLLYKTFCR 9 HBV adr POL IOf.5 3.1 1 --- 0072 00045
1 0978 RLVFQTSTR 9 HBV adr POL 757 3,11 0068 0.0032
1.0219 FVLCGCRHK 9 HBV adr "X" 1550 3.11 0065 0019
1 1042 RLVLQTSTR 9 HBV adw POL ~ 78 3,ii 0064 0 (102
©
IΛ
©
00
Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24
H υ 1.1043 MLLYKTYGR 9 HBV adw POL 1094 3.11 0061 0.0032 a. 1.0170 TVNEKRRLK 9 HBV adr POL 674 3,i i 0048 0.037
1 1045 NLYPVARQR 9 HBV adw POL 128β ~ 3~l i 0042 00011
1 1046 LPYRPTTCR 9 HBV adw POL 1407" 3,11 0021 0
1.0845 LVSPGVWIR 9 HBV adr CORE 509 3,ii 00033 0020
1.0981 LVGSSGLPR 9 HBV adr POL 1022 3,11 00008 0015
1.0967 HISCLTFGR 9 HBV adr CORE 494 3,11 0.013 0.011
1.1047 SVPSRLPDR 9 HBV adw POL 1424 3,11 00007 0.010
1.0989 SVPSHLPDR 9 HBV adr POL 1395 3,11 0.0004 0.010
1.0564 TLPQEHIVLK 10 HBV adr POL 1179 3,11 0.092 5.6
2.0205 TVPVFNPHWK 10 ' HBV ayw POL 669 3,11 00067 4 2
1.0543 TLWKΛCILYK 10 HBV adr POL 724 3,11 3 5 1 0
1.0807 SMYPSCCCTK 10 HBV ayw ENV 295 3,11 I S 34 o 1.1153 RLPYRPTTGR 10 HBV adw POL 1406 3,11 2.8 0.030 -a- 1 0584 STTDLEAYFK 10 HBV adr X 1522 3,11 0.0066 2.7
1.0554 LLLYKTPCRK 10 HBV adr POL 1065 3,11 2.5 0.012
1.0799 TVNAHRNLPK 10 HBV adw "X" 1529 3.11 0.82 0.65
1.0586 EAYFKDCLFK 10 HBV adr X 1527 3,11 0.037 0.74
1.1081 LWDFSQFSR 10 HBV adr POL 962 3,11 0.0009 0.63
1.0789 MLLYKTYGRK 10 HBV adw POL 1094 3,11 0.61 0.O20
1 0546 TAYSHLSTSK 10 HBV adr POL 858 3,11 026 0.092
1.0562 SLGIHLNPNK 10 HBV adr POL 1150 3,11 0.20 0.078
1.1152 RLGLYRPLLR 10 HBV adw POL 1397 3.11 0.19 0.0049
1.0547 VTCGVFLVDK 10 HBV adr POL 943 3 I 0.035 0.17
1.1150 R1RTPRTPAR 10 HBV adw POL 962 3,11 0.17 0.0002
1.0581 TVNGHQVLPK 10 HBV adr X 1500 3,11 0.O73 0.092
1.1091 SLPFQFTTGR 10 HBV adr POL 1377 3,11 0.077 0043
1 1072 TLPETTWRR 10 HBV adr CORE 532 3,11 <00003 0075
IΛ 1.1089 GTDNSWLSR 10 HBV adr POL 1320^ 3,Ϊ 1 0.025 0072 IΛ
** 1.1071 STLP.EITWR 10 HBV adr CORE 531 3,11 0.0005 0.068
20210 KVTKYLPLDK 10 HBV ayw POL 721 3,11 0.027 0053
O 1 1148 STRHCDKSFR 10 HBV adw OL 792 3,1 1 0.0057 0038
1.0935 VLSCWWLQFR 10 HBV adw POL 923 3.U 0029 0.0087
1 0781 NVTKYLPLDK 10 HBV adw " i'OL 721 3,1 1 <00004 0023
1.1092 RVCCQLDPΛR 10 HBV adr X 1422 3,1 1 — - 0019 0023
1 0793 SLGIHLNPQK 10 HBV adw poi. M7 » 3.1 1 001 0.114
"
1 0909 YLVSFGVWIR 10 I IBV adr CORE 3.1 1 001 0(XC7
~Ξ
ON roo
IΛ
©
00 ON
CΛ
Hυ Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24 ft.
20207 FVGPLTVNEK 10 HBV ayw OL 698 3,11 00057 0015
1 0535 YVCPLTVNEK 10 HBV adr OL 669 " 3,M 00069 0014 1 1075 RLADECLNRR 10 HBV adr POL 601 3,11 0013 00004
1 1086 IVLKLKQCFR 10 HBV adr POL 118.T 3,11 0013 00024
1 0773 PIPSSWAFAK 10 HBV adw ENV 314 3,11 <00003 0010
1 0778 LTVNENRRLK 10 HBV adw OL 702 3,11 00025 00095
* IΛ*
ON I*Λ*
ON ON o
ON ro o
IΛ
O
00 ON
CΛ
Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24 υ 1 0118 CTCGSSDLY 9 HCV LORF 1123 3 0 0 0010 ft. —
1 0112 NIVDVQYLY 9 HCV NSI /ENV2 47 060 0 0010 20034 VQDCNCSIY 9 HCV 302 0 54 00005 00003
20035 LTPRCMVDY 9 HCV 605 _ 0 7
1 0145 RVCEKMΛLY 9 HCV I ORF 25*88 ~ 0053
1 0140 DWCCSMSY 9 HCV LORF 2416 0039
2.0036 FΠFKIRMY 9 HCV 626 0012
1.0509 CLSAFSLHSY 10 HCV LORF 2888 ~ 041 00002 0.013 00034 00002
1.0489 TLHGPTPLLY 10 HCV LORF 1617 ~~030 0.11 0.0024
2.0037 .EYVLLLFLL 9 HCV 719 24 1.4
20169 MYVGGVEHRL 10 ' HCV 633 24 0026
2.0170 EYVLLLFLLL 10 HCV 719 24 0.010
1 0139 SVPAEILRK 9 HCV LORF 2269 3,11 0.016 0.87
1 0955 QLFTP5PRR 9 HCV ENV1 290 3,11 075 0.033
1.0090 RLGVRATRK 9 HCV CORE 43 3,11 0.74 016
1.0123 LIFCHSKKK 9 HCV LORF 1391 3,11 054 0.19
1.0122 HUPCHSKK 9 HCV LORF 1390 3,11 0.25 0.010
1 0952 KTSERSQPR 9 HCV CORE 51 3,11 0.16 0064
1.0120 AVCTRGVAK 9 HCV LORF 1183 3,11 0.016 0038
1.0143 EVPCVQPEK 9 HCV LORF 2563 3,11 00019 0033
1 0137 ΓΓRVESENK 9 HCV LORF 2241 3,11 0015 0.0079
1 0957 cirrsLTGR 9 HCV LORF 1042 3,ΪI 00095 0011
1.0496 GVAGALVAFK 10 HCV LORF 1858 3,11 087 1.1
1.0480 HLHAPTGSGK 10 HCV LORF 1227 3,11 057 00051
1.1062 RMYVGGVEHR 10 HCV NS1/ENV2 632 3,11 027 0012
1.0485 HUFCHSKKK 10 HCV LORF 1390 3,11 0.27 0.025
1.0484
IΛ TLCFGAYMSK 10 HCV LORF 1261 3,11 0 17 0.13
ON IΛ 1.1067 GVGIYLLPNR 10 HCV LORF 3002 3.11 00029 0032
**
ON 1.1063 LLFLLLADAR 10 HCV NS1/ENV2 723 3,11 0015 0 ON
o
ON ro o
I Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24 oΛ
00 1 0014 FRDYVDRFY 9 HIV GAG 298 0090 ON
CΛ 20129 IYQYMDDLY 9 HIV 875" 0064
1 0028 TVLDVGDAY 9 HIV POL 802 0.018 <0.0002 0.0056 υ 1.0412 VTVLDVCDAY 10 HIV POL 801 028 0 00004 a. 1.0415 VIYQYMDDLY 10 HIV POL 874 i — 025 00007 00090
20252 VTVLDVCDAY 10 HIV 801 0088
1.0431 EVNIVTDSQY 10 HIV POL 1187 0053
1.0441 LVΛVHVΛSCY 10 HIV POL 1329 0039
1.0442 PAETGQEΓAY 10 HIV POL 1345 0013
20251 ISKIGPENPY 10 HIV 742 0.013
20255 QMAVF1HNFK 10 ' HIV 1,432 3 0.61 0.64
20064 RYLKDQQLL 9 HIV 2,778 24 0.76
20134 RYLKDQQLL 9 HIV 2,778 24 032
20065 TYQIYQEPP 9 HIV 1,033 24 030 en 20131 TYQIYQEPF 9 HIV 1,033 24 0.20
20063 IYQEPFKNL 9 HIV 1,036 24 0.052
20132 IYQEPFKNL 9 HIV 1,036 24 0033
20066 IYQYMDDLY 9 HIV 875 24 0013
10247 IYKRWIILCL 10 HIV 266 24 0.017
20190 IYKRWIILGL 10 HIV 266 24 0.014
2.0249 LYPLASLRSL 10 HIV 506 24 0014
1.0069 KLAGRWPVK 9 HIV POL 1358 3,11 2.7 0069
1.0944 ΛVFIHNFKR 9 HIV POL 1434 3,11 0.17 1.8
1.0092 AIFQSSMTK 9 HIV POL 853 3,11 1.1 0.96
1.0046 IVIWGKTPK 9 HIV POL 1075 3,11 0.085 0.37
1.0079 KLTEDRWNK 9 HIV VIF 1712 3,11 0.013 0.27
1.0027 GIPHPAGLK 9 HIV POL 788 3,11 0.23 0.065
1 0059 QIIEQLIKK 9 HIV POL 1215 3,11 00091 0.16
1.0939 KIWPSYKCR 9 HIV CAG 443 3,11 0.12 0.0005
1.0072 IIATDK3TK 9 HIV POL 1458
** 3,11 0.025 0098
IΛ
ON 1.0036 MGYELHPDK 9 HIV POL 925 3,11 0064 0096 IΛ 1.0062 YLAWVPAHK 9 HIV POL 1227 3,11 0077 0057
ON ON 1.0938 KIWPSHKCR 9 HIV GAG 443 3,11 0077 <00005
1 0047 FVNTPPLVK 9 HIV POL 1111 3.11 0012 0066
1.0024 NTPVFAIKK 9 HIV i'OL ~7. 2 3,i l 0033 0060
1.0080 TVQCTHGIK 9 HIV ENV 2420 3.11 00021 0.046
1.0013 ILDIRQCPK 9 HIV GAG 287 3.ΪT 0042 00048
ON ro o
IΛ
©
00 ON
CΛ Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24
1 0015 RDYVDRFYK 9 HIV GAG 2<>Q 3.11 00007 0040 υa. i θQ58 GIIQAQPDK 9 HIV POL Tl99 3,ii <00009 0040
1 0064 VLFLDCIDK 9 HIV POL 1254 3,1 1 0.038 0032
1 0026 LVDFRELNK 9 HIV POL 769 3~ϊl 0011 0.030
1 0078 KWPRRKAK 9 HIV POL 1513 3 1 0029 O.O039
1 0942 MTKILEPFR 9 HIV POL 859 3,11 <0.0008 0016
1.0463 TVYYCVPVWK 10 HIV ENV 2185 3,11 3.8 7.8
1.0418 TVQPIVLPEK 10 HIV POL 935 3,11 0.16 5.6
1.0447 AVFIHNFKRK 10 HIV POL 1434 3,11 066 085
1.0437 KVLFLDCIDK 10 HIV POL 1253 3,11 036 078
1.0408 KLVDFRELNK 10 HIV POL 768 3,11 051 0090
1.0403 KLKPGMDGPK 10 HIV POL 706 3.ΪΪ 0.39 0.076
1.0395 FLGKIWPSYK 10 HIV GAG 440 3,11 0.32 0.024
1 1056 KIQNFRVYYR 10 HIV POL 1474 3,11 0.032 0.21
1.0410 GIPHPAGLKK 10 HIV POL 788 3,11 0011 0.17
1.0426 LVKLWYQLEK 10 HIV POL 1117 3,11 0.056 0.082
1.0398 MIGGICGFIK 10 HIV POL 642 3,11 0.0099 0.055
1.0413 MTKIL.EPFRK 10 HIV POL 859 3,11 0.015 0038
1 0453 WIQDNSDIK 10 HIV POL 1504 3,11 <00005 0.021
1.0394 FLGKIWPSHK 10 HIV GAG 440 3,11 0.020 0.0013
1.1059 IVQQQNNLLR 10 HIV ENV 2741 3,11 00024 0019
1.0417 FTTPDKKHQK 10 HIV POL 909 3,11 <0.0002 0015
1.0405 LVEICTEMEK 10 HIV POL 729 3,11 0.0002 0012
1.0392 LVQNANPDCK 10 HΓV GAG 327 3,11 <0.0002 0011
IΛ
ON IΛ
**
ON ON
O
45
-0
N ft ■*
3io 9 £ 8 2
$ § § 8 |S s «ι o
(117.
3,13. CΛ
7CJ . f o .a 55 C e
8 i m 3
30,30
> >
<!<ι<i< < <
CΛ φvi^ir »*- ac ij. i I '
s SJ 33 S3 13 s. 5! S3 SJ S3 S3 5 5 5 S3 •3
t. g'± O $;s sis a ε -3 z 2 a 3 3 s "l-i 3!K 4'8 o (« o 2 i i
«ιo 1ιβ o o o Il ol o β=rι"sr
O I o o I o i o| o o| o SiSIS
— I wι Si i3 s s o fjo|oιo p| s δ
I I
46
en
,« •I
« g
I !
sis
.lβ !«
OIOIOlo
51=13 »
© IΛ
00
Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24
H 1 0281 GSDCTΠHY 9 P53 226 1 29 5 00010 0029 U a. 1 0667 CTΛKsvTcn. 10 P53 117 1 033 0 0023 0049 0
—
1 0672 RVEGNLRVEY 10 p53 196 1 0022 00014 0.0020
1 0278 RVRAMAΓYK 9 p53 J56 3.11 1 5 073
1 0276 CTYSPALNK 9 P53 124 3,11 046 1 1
1 0285 NTSSSPQPK 9 P53 311 3,11 0.0009 0095
1 0284 RTEEENLRK 9 p53 283 3,11 00015 0.091
1 0287 ELNEALELK 9 P53 343 3,11 0020 00052
1 0678 RTEEENLRKK 10 p53 283 3,11 3.3 0.0080
1 1113 KTYQGSYGFR 10 P53 101 3,11 2 6 088
1 1115 WRRCPHHER 10 P53 172 3,11 0.099 0.0017
1 0679 NTSSSPQPKK 10 p53 311 3,11 00035 0054
1.1121 RVCACPGRDR 10 p53 273 3,11 0.014 0011
1.1116 GLAPPQHLIR 10 p53 187 3,11 0.013 0.0006
* IΛ* I *Λ* o
ON ro o
IΛ o ^» 00 ON CΛ
Peptide Sequence AA Virus Strain Molecule Pos. Motif Al A2.1 A3.2 All A24
U 30175 KGEYFVEMY 9 PAP 322 34 <00002 0.0002 0 ft,
30174 LGEYIRKRY 9 PAP 81 1 078 <00002 0.0002 0
30166 ASCHLTELY 9 PAP 311 1 0.77 <0 002 <00002 0.055 0
30163 ESYKHEQVY 9 PAP 95 1 0098 <00002 0.0002 0
3 237 LSELSLLSLY 10 PAP 238 1 14 00026 00004 0
30235 LSELSLLSLY 10 PAP 238 1 12 00005 0.0004 0
30236 LTQLGMEQHY 10 PAP 70 1 0.62 00005 0.015 0.0024 0.0022
3.0238 KGEYFVEMYY 10 PAP 322 1 0.018 0.0057 0089
30230 LVNEILNHMK 10 PAP 263 3 0.056 0.12
30158 ΛTQIPSYKK 9 PAP 274 11 0.10 1.2
30231 ETLKSEEFQK 10 PAP 170 11 <0.0004 0.014
30161 LYFEKGEYF 9 PAP 318 24 2.5
3.0160 LYCESVHNF 9 PAP 213 24 044
3.0159 FYKDFIΛTL 9 PAP 183 24 0.11
CO 3.0162 VYNCLLPPY 9 PAP 302 24 0032
3.0232 PYASCHLTEL 10 PAP 309 24 0024
T IΛ
ON IΛ
ON ON o
t in
[
NO
8 r
G f § . O
H
CΛ
00
© in
©
.
Table 5
NO
Sequence Sise Antigen Strain Molecule Frβq Pos. Motif A01 A03 All A24 NO
WI
Bind. Bind. Bind. Bind. NO n
EDTPIGHLY 9 MAGE3a 3 analog 161 A01 12.5000
AVDPIGHLY 9 MAGE3a 3 analog 161 A01 8.0000
EVDPIAHLY 9 MAGE3a 3 analog 161 A01 5.5000
FSPAFDNLYY 10 HER-2/neu 1213 A01 5.5000 0.0005 0.0010
EVDAIGHLY 9 MAGE3a 3 analog 161 A01 5.3500
EVDPIGALY 9 - MAGE3a 3 analog 161 A01 5.0000
EVDPIGHAY 9 MAGE3a 3 analog 161 A01 4.6500 o
EADPIGHLY 9 MAGE3a 3 analog 161 A01 3.4500 *
EVDPTGHLY 9 MAGE3a 3 analog 161 A01 2.9500
EVDPIGHSY 9 MAGE3a 3 analog 161 A01 2.6667
EVDPAGHLY 9 MAGE3a 3 analog 161 A01 2.4000
EVDPASNTY 9 MAGE 4 161 A01 1.5000
PLSEDQLLY 9 PAP 147 A01 1.2000 0.0005 0.0001
LSAFSLHSY 9 HCV 2889 A01 0.8100 0.0002 0.0002
IPSYKKLIMY 10 PAP 277 A01 0.5650
YASCHLTELY 10 PAP 310 A01 0.5467 0.0003 0.0002
EVDP1GHLA 9 MAGE3a 3 analog 161 A01 0.3300 o H
CMQIAKGMSY 10 HER-2/neu 826 A01 0.2967 0.0003 0.0001 CΛ NO 00
VGSDCTTIHY 10 P53 225 A01 0.2600 0.0003 0.0003 © in
©
EVAPIGHLY 9 MAGE3a 3 analog 161 A01 0.1800 NO
Table 5
Sequence Sise Antigen Strain Molecule rrβq Pos. Motif A01 A03 All A24
NO
Bind. Biod. Bind. Bind. NO
■ in
ESHPNPECRy 10 HER-2/heu 280 A01 0.180(3 0.0003 0.0003 VO n
*.
ASCVTACPY 9 HER-2/nβu 293 A01 0.0552 0.0074
0.0008
FSPAFDH Y 9 HER-2/nβu 1213 A01 0.0425 0.0002 0.0002
ASPLDSTFY 9 HER-2/nβu 997 A01 0.0290 0.0002 0.0004
RGTQ FENDY 10 HER-2/nβu 103 A01 0.0205 0.0003 0.0015
PASPLDSTFY 10 HER-2/neu 996 A01 0.0148 0.0003 0.0001
PSQKTYQGSY 10 p53 98 A01 0.0140 0.0003 0.0003
KSTKVPAAY 9 HCV 1236 A01 0.0134 0.0009 0.0001
DSSVLCECY 9 HCV 1513 A01 0.0110 0.0002 0.0003
KISEYRHYCY 10 HPV 16 E6 79 A01 0.0090 0.0043 0.0038
NLYVS M LY 10 HBV adw POL 20 1088 A01 0.0090
GTRVRAMAIY 10 p53 154 A01/03 0.0027 0.0365 0.0002
LTCGFADLMGY 11 HCV 126 AOl/11 2.4500 0.0003 0.0120 0.0001
VMAGVGSPY 9 HER-2/neu 773 A01/A03 0.0400 0.0575 0.0079
TLWKAG1LY 9 HBV adr POL 100 724 A03 0.0017 0.2667 0.0016 ►
KLN ASQIY 9 HIV POL 958 A03 0.0070 0.1160 0.0006 o0
LVGFLLLKY 9 MAGE1 1 109 A03 0.0033 0.0563 0.0012 CΛ
NO
00
ILRGTSFVY 9 HBV adr POL 80 1345 A03 0.0017 0.0440 0.0002 o o IΛ
RVLOGLPREY 10 HER-2/neu 545 A03 0.0015 0.0350 0.0050 w
VO
Table 5
Sequence Sise Antigen . Strain Molecule Prβq Pos. Motif A01 A03 All A24
NO NO
Bind. Bind. Bind. Bind. n vo
QLVTQLMPY 9 HER-2/neu 795 A03 0.0024 0.0112 0.0039 in i-
GLNKIVRMY 9 HIV GAG 274 A03 0.0017 0.0103 0.0002
LLGDNQVMPK 10 MAGE2 2 182 A03 0.0093 0.0014
QVRDQAEHLK 10 HIV POL 1419 A03 0.0089 0.0093
LVSAGI K 8 HIV con 1246 A03 0.0091 0.0054
VTDRGRQK 8 HIV con 1153 A03 0.0090 0.0065
TVFDAKRLIGR 11 BLA-A 68 endogenous peptide sequences A03/11 0.1050 1.3000
KTGGPIYKR 9 HLA-Aw68 endogenous peptide sequences A03/11 0.0340 0.8200
SLYTKWHY 9 PSA 237 A03/11 0.0017 0.6750 0.0140 NJ
AVAAVAARR 9 HLA-Aw68 endogenous peptide βequenceβ A03/11 0.1600 0.0825
KIQNFRVYY 9 HIV POL 1474 A03/11 0.0056 0.1190 0.1350
EMLESVIKNYK 11 MAGE1 127 A03/11 0.0087 0.0099
EVAPPEYHRK 10 HLA-Aw68 endogenous peptide βequences All 0.0008 0.0575
ETAYFLLK 8 HIV consensus 1351 All 0.0037 0.0425
RWGLLLALL 9 HER-2 neu 8 A2 1.2567
PYVSRLLGI 9 HER-2/neu 780 A24 0.1650
VYMIMVKCW 9 HER-2/neu 951 A24 0.1640 o
AYSLTLQGL 9 HER-2/neu 440 A24 0.1250
SYGVTVWEL 9 HER-2/neu 907 A24 0.1200 CΛ
NO
0 —0
L ISAWPDSL 10 HER-2/neu 410 A24 0.0835 © uoi
VWSYGVTVW 9 HER-2/neu 905 A24 0.0800 NO
Table 5
Sequence Sise Antigen Strain Molecule rrβq Pos. Motif A01 A03 All A24
Bind. Bind. Bind. Bind. in n
SYGVTVWELM 10 HER-2/neu 907 A24 0.0630
QYLAGLSTL 9 HCV 1777 A24 0.0475
TYLPTNASL 9 HER-2/neu 63 A24 0.0375
EYLVSFGVWI 10 HBV NUC 90 117 A24 0.0335
KFMLCAGRH 9 PSA 190 A24 0.0305
WFHISCLTF 9 HBV NUC 90 102 A24 0.0300
TYSTYGKFL 9 HCV 1296 A24 0.0225
VYMIMVKCWM 10 HER-2/neu 951 A24 0.0218
RFRELVSEF 9 HER-2/neu 968 A24 0.0180
CYGLGMEHL 9 HER-2/neu 342 A24 0.0176
QYSPGQRVEF 10 HCV 2614 A24 0.0175
K HALESIL 9 HER-2/neu 887 A24 0.0149
EYLVPQQGFF 10 HER-2/neu 1022 A24 0.0120
RYSEDPTVPL 10 HER-2/neu 1111 A24 0.0117
RFTHQSDVW 9 HER-2/neu 898 A24 0.0107
-0 o
H
CΛ 00
© o UI t*)
Table 5
Mage vo
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24 NO in
DLVGFLLLK 9 108 3,11 0.0040 0.0014 NO UI
QLVFGIDVK 9 152 3,U 0.0019 0.0051
SLEQRSLHCK 10 2 3,11 0.015 0.015
SLFRAVITKK 10 96 3,11 1.2 0.98
DLVGFLLLKY 10 108 1 0.0068 0.0069 0.0009
MLESVIKNYK 10 128 3,11 0.14 0.027
WEELSVMEVY 10 1 215 1 <0.0009 <0.0002 <0.0002
VYDGREHSAY 10 223 1 <0.0009
LVGFLLLKY 9 109 1 0.0033 0.056 0.0012
LVTCLGLSY 9 171 1 0.0084 0.0014 <0.0002
VLVTCLGLSY 10 170 1 0.0048 0 0.0013 0.0007
FLLLKYRAR 9 1/2/3 112 3,11 0.0007 <0.0005
PTTINFTRQR 10 65 3,11 <0.0002 0.0033
LVGFLLLKYR 10 109 3,11 0.0034 0.0023
EKYLEYGRCR 10 246 3,11 <0.0002 0
ELVHFLLLK 9 2/3 108 3 0.0045 0.0011
AYGEPRKLL 9 231 24 0.0007
SYVLVTCLGL 10 168 24 0.0006 0.0051 n "0
EWPISHLY 9 161 1 0.0028 <0.0002 <0.0002
CΛ NO
EWRIGHLY 9 161 1 0.0002 00
© UI
EVDPASNTY 9 161 1 0.0005 © I
NO
EADPTSNTY 9 5/51 161 1 9.9 0.0006 0.0006 0
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
EVDPIGHVY 9 6 161 1 1.9 <0.0002 <0.0002 0 - 4i». UI
EMLESVIK 8 1 127 3 <0.0003 0 Ui
LVFGIDVK 8 1 153 3 0.0035 0.0037
GVQGPSLK 8 1 266 3 <0.0003 0.0063
VMEVYDGR 8 1 220 3 <0.0003 0.0007
VQEKYLEY 8 1 244 1 0.0018
AYGEPRKL 8 1 231 24 0.0017
VKEADPTGHSY 11 - 1 159 1 <0.0003
IWEELSVMEVY 11 1 214 1 <0.0003 Xjx
EMLESVIKNYK 11 1 127 3 0.0087 0.0099
EADPTSHTY 9 analog 161 1 0.68
EVDPTSNTY 9 analog 161 1 1.8
EALEAQQEA 9 1 14 2.1 0 <0.0002 0
MSLEQRSLH 9 1 1 3 0.0025 0.0003
QSPQGASAF 9 1 56 3 0.0004 0
SAFPTTINF 9 1 62 3 <0.0003 0 0.0003
TSCILESLF 9 1 90 3 <0.0003 0
SCILESLFR 9 1 91 3 <0.0003 0.0026 l-0
LFRAVITKK 9 1 97 3 0.011 0.0005 o
VGFLLLKYR 9 1 110 3 0.0044 0.0051 00
ESVIKNYKH 9 1 130 3 <0.0003 0 o Ui
O
VIKNYIHCF 9 1 132 3 <0.0003 0 w
Table 5
Mag*
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
NO
ASESLQLVF 9 1.2 147 3 <0.0003 0 NO in
LGDNQIMPK 9 183 3 0.0007 0.0048 NO I
VMIAMEGGH 9 200 3 <0.0003 0
YDGREHSAY 9 224 3 <0.0003 0
LTQDLVQEK 9 239 3 <0.0003 0.14
CGVQGPSLK 9 265 3 <0.0003 0.0037
EMLESVIKNY 10 127 1 0.0006 <0.0002 <0.0002 0
KEADPTGHSY 10 *- J 160 1 <0.0005 <0.0002 <0.0002
ASAFPTTINF 10 61 3 <0.0003 <0.0002
AFPTTINFTR 10 63 3 <0.0003 0.0003 ON
PTTINFTRQR 10 65 3 <0.0003 0.0002
STSCILESLF 10 89 3 <0.0003 <0.0002
GFLLLKYRAR 10 111 3 0.0019 0.0008
KAEMLESVIK 10 125 3 <0.0003 0.0097
SVIKNYKHCF 10 131 3 <0.0003 <0.0002
KASESLQLVF 10 146 3 <0.0003 <0.0002 0.0012
DVKEADPTGH 10 158 3 <0.0003 <0.0002
LVMIAMEGGH 10 199 3 0.0008 0.0005
LSVMEVYDGR 10 218 3 <0.0003 0.012 o -0
H
VMEVYDGREH 10 220 3 <0.0003 0.0002 0
CΛ NO
YGRCRTVIPH 10 251 3 <0.0003 <0.0002 00
©"
U
SCGVQGPSLK 10 264 3 0.0005 0.0089 oI
>
NO
Table 5
Mag*
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A 4
VPDSDPARY 9 new 254 1 0.0038
QVPDSDPAR 9 new 254 3 <0.0003 0.0002 Ui
.fe.
VIKVSARVR 9 new 284 3 0.0016 0
PSLREAALR 9 new 296 3 <0.0003 0
EFLWGPRAL 9 new 264 24 0.0006
ETSYVKVLEY 10 new 274 1 0.56
LVQEKYLEYR 10 new 243 3 0.0008 0.0043
QVPDSDPARY 10 new 254 3 0.0014 0.0003
YVKVLEYVIK 10 new 277 3 0.0029 0.0015
YVIKVSARVR 10 new 283 3 0.019 0.0009
RALAETSYVK 10 new 270 11 0.18 0.24
SYVKVLEYVI 10 new 276 24 0.036
FFPSLREAAL 10 new 294 24 0.0044
SVIKNYK 7 1 N POL 131 3,11 0.0006 0.0028
PVTKAEMLESVIK 13 1 n E6 122 3,11 <0.0003 0
ETSYVKVLEYVIK 13 1 n E6 273 3,11 0.0044 0.0003
ITKKVADLVGFLLLK 15 1 n POL 102 3,11 0.40 1.0
VTKAEMLESVIKNYK 15 1 n POL 123 3,11 0.024 0.053
WGNWQYFFPVIFSK 15 3 POL 79 3,11 1.6 0.34 •V
O
PRALAETSY 9 1 new 268 1 <0.0018 <0.0003 <0.0002
CΛ
FATCLGLSY 9 3 171 1 0.038 <0.0003 0.0004 00
©•
UI
LEQRSLHCK 9 1 new 3 3 <0.0002 0 o
Table 5
Maga
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A 4
NO
AEMLESVIK 9 new 126 3 <0.0002 0.0011 NO U *»I.
LESVIKNYK 9 new 129 3 <0.0002 0.0018 NO UI
■
EELSVMEVY 9 new 216 3 <0.0002 0
MEVYDGREH 9 new 221 3 <0.0002 0
DSDPARYEF 9 new 256 3 •θ.0002 0
KVSARVRFF 9 new 285 3 0.0005 0
VSARVRFFF 9 new 286 3 0.0003 0.0026
HSPQGASSF 9 - 2 56 3 <0.0002 0
TTINYTLWR 9 2 66 3 0.089 1.1
00
QEEEGPRMF 9 2 83 3 <0.0002 0
MFPDLESEF 9 2 90 3 <0.0002 0 0.014
SEFQAAISR 9 2 96 3 <0.0002 0.0001
EFQAAISRK 9 2 97 3 <0.0002 0.0002
LVHFLLLKY 9 2,3 109 3 0.043 0.010
AEMLESVLR 9 2 126 3 <0.0002 0
SVLRNCQDF 9 2 131 3 <0.0002 0
VLRNCQDFF 9 2 132 3 <0.0002 0
DFFPVIFSK 9 2 138 3 <0.0002 0.0022
VIFSKASEY 9 •d
2 142 3 0.081 0.033 O H
WE PISH 9 2 159 3 0.0007 0.010
CΛ NO
LGDNQVMPK 9 2 183 3 <0.0002 0.0061 00
©
UI
EGDCAPEEK 9 2,3 205 3 <0.0002 0 ©
UI NO
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
QEEEGPSTF 9 3 83 3 <0.0002 0
UI
TFPDLESEF 9 3 90 3 <0.0002 0 0.0049
SEFQAALSR 9 3 96 3 <0.0002 0
EFQAALSRK 9 3 97 3 <0.0002 0.0001
SWGNWQYF 9 3 131 3 <0.0002 0
WGNWQYFF 9 3 132 3 0.0022 0.0021
YFFPVIFSK 9 3 138 3 0.0020 0.027
ASSSLQLVF 9 , 3 147 3 0.0011 0.0089
LMEVDPIGH 9 3 159 3 <0.0002 0
IIVLAIIAR 9 3 196 3 0.0069 0.0011
VQEKYLEYR 9 1 244 11 <0.0002 0 vo
SNQEEEGPR 9 2 81 11 <0.0002 0
NYKHCFPEI 9 1 new 135 24 4.8
IFGKASESL 9 1 new 143 24 0.0013
GFLIIVLVM 9 1 new 193 24 <0.0002
IFSKASEYL 9 2 143 24 0.023
EYLQLVFGI 9 2 149 24 3.5
NWQYFFPVI 9 3 135 24 0.53
IFSKASSSL 9 3 143 24 0.016 *0 o
LGSWGNHQY 10 3 129 1 <0.0020 <0.0003 0.0012
IFATCLGLSY 10 3 170 1 <0.0002 0.0005 0.0004 00
©
Ui
TSCILESLPR 10 1 new 90 3 <0.0002 0.015 o
UI
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
LESVIKNYKH 10 new 129 3 <0.0002 <0.0002
U ϋl o
REHSAYGEPR 10 new 227 3 <0.0002 <0.0002 Ul
4-.
PDSDPARYEF 10 new 255 3 <0.0002 <0.0002
LEYVIKVSAR 10 new 280 3 <0.0002 <0.0002
VIKVSARVRF 10 new 283 3 <0.0002 <0.0002
KVSARVRFFF 10 new 285 3 0.0013 0.0020
STTINYTLWR 10 2 65 3 0.0014 0.091
SSNQEEEGPR 10 - 2 80 3 <0.0002 <0.0002
RMFPDLESEF 10 2 89 3 <0.0002 <0.0002 0.0016
ESEFQJVAISR 10 2 95 3 <0.0002 <0.0002
SEFQAAISRK 10 2 96 3 0.0012 0.0028 o
ISRKMVELVH 10 2 102 3 <0.0002 <0.0002
VELVHFLLLK 10 2 107 3 0.0009 0.0003
ELVHFLLLKY 10 2,3 108 3 0.0066 0.0003
LVHFLLLKYR 10 2 109 3 0.026 0.0022
HFLLLKYRAR 10 2,3 111 3 0.0014 0.0002
KAEMLESVLR 10 2 125 3 <0.0002 0.0009
ESVLRNCQDF 10 2 130 3 <0.0002 <0.0002
SVLRNCQDFF 10 2 131 3 <0.0002 <0.0002 n
H
NCQDFFPVIF 10 2 135 3 <0.0002 <0.0002 00
QDFFPVIFSK 10 2 137 3 <0.0002 0.0083 © Ui
©
PVIFSKASEY 10 2 141 3 0.016 0.0033 U)
Table 5
Hag*
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A2
KASEYLQLVF 10 2 146 3 <0.0002 <0.0002 0.0030
158 3 <0.0002 <0.0002 i~
E E PISH 10 2
VEWPISHLY 10 2 160 3 <0.0002 <0.0002
ILVTCLGLSY 10 2 170 3 0.0036 0.0002
LLGDNQVMPK 10 2 182 3 0.0093 0.0014
IEGDCAPEEK 10 2 204 3 <0.0002 <0.0002
STFPDLESEF 10 3 89 3 <0.0002 <0.0002
ESEFQAALSR 10 - 3 95 3 <0.0002 <0.0002
SEFQAALSRK 10 3 96 3 0.0010 0.0010
LSRKVAELVH 10 3 102 3 <0.0002 <0.0002 O
AELVHFLLLK 10 3 107 3 0.0008 <0.0002
LVHFLLLKYR 10 3 109 3 0.040 0.0014
GSWGNWQYF 10 3 130 3 0.0020 0.0008
SWGNWQYFF 10 3 131 3 0.0085 0.0067
KASSSLQLVF 10 3 146 3 0.0003 0.0008 0.0021
ELMEVDPIGH 10 3 158 3 <0.0003 <0.0002
MEVDPIGHLY 10 3 160 3 0.0004 0.0004
VDPIGHLYIF 10 3 162 3 <0.0003 <0.0002 o
LIIVLAIIAR 10 3 195 3 0.028 0.0021 H
REGDCAPEEK 10 3 204 3 <0.0003 <0.0002 00
RQPSEGSSSR 10 ©
1 new 74 11 0.0009 0.0009 Ul
LQLVFGIDVK 10 1 new 151 11 0.0050 0.0018
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
NO
NO
RQVPDSDPAR 10 1 new 252 11 <0.0003 <0.0002 USl
NO
MNYPLWSQSY 10 3 new 68 11 <0.0003 <0.0002 uι
GFLIIVLVMI 10 1 new 193 24 0.0008
SFSTTINYTL 10 2 63 24 0.015
EFQAAISRKM 10 2 97 24 <0.0002
LYILVTCLGL 10 2 168 24 0.014
NWQYFFPVIF 10 3 135 24 0.017
AVDPIGHLY 9 , 3 analog 161 1 8.0
EADPIGHLY 9 3 analog 161 1 3.5
EVDPASNTY 9 4 161 1 1.5 3
EDTPIGHLY 9 3 analog 161 1 13
EVDPTGHLY 9 3 analog 161 1 3.0
AADSPSPPH 9 2 55 All
VPISHLYIL 9 2 170 PI
MPKTGLLII 9 2 196 PI
SMLEVFEGR 9 2 226 All
DSVFAHPRK 9 2 236 All
VFAHPRKLL 9 2 238 A24
MQDLVQENY 9 2 247 A01 O H
DPACYEFLW 9 2 265 P2 CΛ
NO 00
FLWGPRALI 9 2 271 A02 © o Ul
ALIETSYVK 9 2 277 A03/A11 Ul
VO
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A NO NO S
TSYVKVLHH 9 2 281 All Ul
VO
Ul
EPHISYPPL 9 2 296 PI
ISYPPLHER 9 2 299 A03/A11
YPPLHERAL 9 2 301 PI
EPVTKAEML 9 2/3 128 PI
VPGSDPACY 9 2/3 261 P2
EGLEARGEA 9 3 14 A03
GLEARGEAL 9 - 3 15 A02
EARGEALGL 9 3 17 A02
ALGLVGAQA 9 3 22 A02/A03 u>
GLVGAQAPλ 9 3 24 A02/A03
LVGAQAPAT 9 3 25 A02
PATEEQEAA 9 3 31 A02/A03
EAASSSSTL 9 3 37 A02
AASSSSTLV 9 3 38 A02
LVEVTLGEV 9 3 45 A02
EVTLGEVPA 9 3 47 A02/A03
VTLGEVPAA 9 3 48 A02/A03 *n0
LPTTMNYPL 9 3 71 PI
PDLESEFQA 9 3 99 A03 CΛ vo 00
©
HFLLLKYRA 9 3 118 A03 Ul
©
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
NO NO
DPIGHLYIF 9 3 170 P2 Ul
NO
GDNQIMPKA 9 3 191 A03 Ul
MPKAGLLII 9 3 1 6 PI
AGLLIIVLA 9 3 199 A03
KIWEELSVL 9 3 220 A02
SVLEVFEGR 9 3 226 A03/A11
EDSILGDPK 9 3 235 A03/A11
SILGDPKKL 9 , 3 237 A02
ILGDPKKLL 9 3 238 A02
FLWGPRALV 9 3 271 A02
O
PRALVETSY 9 3 275 A01
RALVETSYV 9 3 276 A02
ALVETSYVK 9 3 277 A03/A11
LVETSYVKV 9 3 278 A02
YVKVLHHMV 9 3 283 A02
KVLHHMVKI 9 3 285 A02
MVKISGGPH 9 3 290 A03/A11
ISGGPHISY 9 3 293 A01/A03/A11
GPHISYPPL 9 3 296 PI o H
YPPLHEWVL 9 3 301 PI CΛ NO 00
VPISHLYILV 10 2 170 PI © Ui
©
MPKTGLLIIV 10 2 196 PI Ul
NO
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
S;
VFEGREDSVF 10 2 230 A24
HPRKLLMQDL 10 2 241 PI
LMQDLVQENY 10 2 246 A01
EFLWGPRA I 10 2 270 A24
GPRALIETSY 10 2 274 P2
RALIETSYVK 10 2 276 All
SYVKVLHHTL 10 2 282 A24
SYPPLHERAL 10 * 2 300 A24
APEEKIWEEL 10 2/3 216 PI
PLEQRSQHCK 10 3 2 A03/A11
HCKPEEGLBA 10 3 9 A03
EARGEALGLV 10 3 17 A02
RGEALGLVGA 10 3 19 A03
EALGLVGAQA 10 3 21 A02/A03
LGLVGAQAPA 10 3 23 A03
GLVGAQAPAT 10 3 24 A02
QAPATEEQEA 10 3 29 A02/A03
EAASSSSTLV 10 3 37 A02 *3 O
TLVEVTLGEV 10 3 44 A02 H
EVTLGEVPAA 10 3 47 A02/A03 CΛ 00
PDPPQSPQGA 10 3 59 A03 ©
Ul o
LPTTMNYPLW 10 3 71 P2
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24 NO VO
PDLESEFQAA 10 3 99 A03 Ul
VO Ul
YFFPVIFSKA 10 3 145 A03
LGDNQIMPKA 10 3 190 A03
MPKAGLLIIV 10 3 196 PI
EVFEGREDSI 10 3 229 A02
EDSILGDPKK 10 3 235 A03/A11
SILGDPKKLL 10 3 237 A02
ILGDPKKLLT 10 - 3 238 A02
GDPKKLLTQH 10 3 240 A03/A11 ON O
DPKKLLTQHF 10 3 241 P2
LTQHFVQENY 10 3 246 A01/A03/A11
FVQENYLEYR 10 3 250 A03/A11
ACYEFLWGPR 10 3 267 A03/A11
GPRALVETSY 10 3 274 P2
RALVETSYVK 10 3 276 A03/A11
ALVETSYVKV 10 3 277 A02
LVETSYVKVL 10 3 278 A02
YVKVLHHMVK 10 3 283 A03/A11
MVKISGGPHI 10 3 290 A02 O
H
KISGGPHISY 10 3 292 A01 CΛ
NO 00
SPPHSPQGA 9 2 60 P2A ©
Ul
©
Ul
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
VO NO
DPPQSPQGA 9 3 60 P2A i USl
NO
APATEEQQTA 10 2 30 P2A Ul
J-
FPDLESEFQA 10 2/3 98 P2A
APATEEQEAA 10 3 30 P2A
DPIGHLYIFA 10 3 170 P2A
EADPTGHSY 9 1 161 1 0.56 0 0 0.0002 <0.0002
KVADLVGFLL 10 1 105 0.0005 0.041 0.0039 0.0030 0.0070
ASSLPTTMNY 10 , 3 8 1 2.3 0.043
TQDLVQEKY 9 1 240 1 0.57 0.0001 0 0 0
LVQEKYLEY 9 1 243 3 016 0 0.0016 0.0098 0 σv
ILLWQPIPV 9 3 <0.0007 1.4 0.0048 0.0048 0
EVDPIGHLY 9 3 3.7 0.0022
ASSFSTTINY 10 2 8 1 0.016 0 0.0016 0.0054 0
VTCLGLSY 8 1 172 1 0.022 0 0.0001 0.0007 0
SSLPTTMNY 9 3 9 1 0.037 0 0.013 0.12 0
GSWGNWQY 9 3 77 1 0.0059 0 0.0009 0.025 0
DLVQEKYLEY 10 1 new 242 3 0 0 0.0010 0 0
SSFSTTINY 9 2 9 1 0.016 0 0.0095 0.056 0
*
MLESVIKNY 9 1 128 1 0.0016 0.0002 0.0006 0 0 n0
KHVELVHFL 9 2 <0.0007 0.13 0.0007 0 0.0043 CΛ NO 0
KMVELVHFLL 10 2 105 <0.0008 0.071 0.0004 0.0001 0.0008 -—0
©
Ui β
LVFGIBLMBV 10 3 0.0030 0.065 0.0007 0 0 Ul
NO
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al Λ2.1 A3.2 All A24
NO NO
SLFRAVITK 9 96 3,11 <0.0007 0.0001 3.9 2.6 0
Ul
NO
ADLVGFLLLK 10 107 3 0.0012 0.0003 0.0081 0.022 0 Ul
ESLFRAVITK 10 95 3 <0.0008 0 0.0090 0.0052 0
MLESVIKNYK 10 0 0 0.034 0.0045 0
LVGFLLLK 8 109 3 0.0029 0.0002 0.027 0.034 0
TTINFTRQR 9 66 3,11 0 0 0.051 0.40 0
LLGDNQIMPK 10 1/3 182 3,11 <0.0007 0.0001 0.022 0.016 0
SVMEVYDGR 9 219 3,11 <0.0006 0 0.059 0.32 0
HSAYGEPRK 9 229 3 0.0007 0 0.0070 0.0015 0
LLTQDLVQEK 10 238 3,11 <0.0007 0 0.0014 0.011 0 O
00
LTQDLVQEK 9 239 3,11 0.0011 0 0.0002 0.16 0
NYKHCFPEIF 10 135 24 0 0 0 0 0.26
LYIFATCLGL 10 115 24 <0.0007 0 0.0006 0 0.0035
NYPLWSQSY 9 16 24 <0.0006 0 0 0.0001 0.016
SYVLVTCL 8 168 24 0.0029 0.00025 0.0020 0.0002 0.0026
ETSYVKVLEY 10 0.075 0 0.0009 0.0004 0
TSYVKVLEY 9 275 3 0.082 0 0.23 0.013 0
FLWGPRALA 9 <0.0006 0.027 0.0015 0 0
•d
ALAETSYVKV 10 271 <0.0007 0.017 0.0011 0.0029 0 O H
RVRFFFPSLR 10 290 3 <0.0007 0 0.25 0.0035 0 CΛ vo 00
ALAETSYVK 9 <0.0006 0.0002 0.17 0.39 0 © Uol
LTQDLVQEKY 10 239 1 0.041 0 0 0.0002 0 Ul
VO
Table 5
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
NO NO
GFLLLKYRA 9 1 0.0004 0.0002 Ul
NO Ul
CFPEIFGKA 9 1 0 0 £>.
FFFPSLREA 9 1 0 0
FFPSLREAA 9 1 0 0
HCFPEIFGK 9 1 138 3,11 0.0017 0.0022
RSLHCKPEEA 10 1 0.0001 0.0008
EFLWGPRALA 10 1 0 0
RFFFPSLREA 10 , 1 0.0004 0
FPFPSLREAA 10 1 0 0 o
NO
O
H
CΛ
NO 00
©
Ul
©
Ul
NO
Table 5
Sequence Antigen Strain Molecule Position Molif Al A2 A3 All A 24 MJIX. Binding Binding Binding Binding Binding Binding
FSPAFDNLYY c-ErbB2 1213 AOI 5.5000 , 00005 0.0010 5.5000
CMQIAKG SY c ErbB2 826 AOI "0.2967" ~t)(MH)3~ OOlMΪi 02967 NO NO
ESMPNPEGRY c-ErbB2 280 Aθi 0.Ϊ800 00003 00003 "ii J 00 in
NO in
ASCVTACPY c EιbB2 " 293 ΛOI 00552 00(H)8 00074 00552
FSPAFDNLY L ιbB2 12 i 3 AOI 0.0425 ii (MMΪ () (MΪ02 " 00425
ASPLDSTFY t-EιbB2 "997 Alii 00290 (j ()002~ 00004 "00290
RGTOLFEDNY c EιbB2 io A(ii 00205 00003" n (it)i 5 (10205
PASPLDSTΓY c-iϊιbB2 996 λiiP "θ()i48 oiiooi" 00001 00148
LSAFSLHSY iicv 2889 " A«)i )8U)0 001)02 ~ ij iii 102" 08UH)
KSTKVPAAΫ iicv i2Ϊfi A(ii" 7)0J 4 ϊ) 0009 ooooi " IMIM4
DSSVLCECY- "~ iicv Ϊ5M AOI 0.0110 0(KKJ2 O(MM)3 o.oiio
ETDPIGHLY MAGE-3a 3 analog "Ϊ6i A i 12.5000 12.5000
AVDPIGHLY MAGE-3a 3 anaiog "" i6i Aiii 8.0000 " 8 ()()()()
EVDPIAHLY MAGE a " 3 anaiog i6i ΛOI ~5.5θi)0 5.5000
EVDAΪGHLY" MAGE-3a 3 anaiog 161 Aiii 5.35()θ" 53500 σ
EVDPIGALY MAGE-3a 3 analog i6i AOΪ Tθ()OD 50000
EVDPIGUAY MAGE-.la 3 anaiog J6I Aiii 4"65(Hi 46500
EΛDPIGIII.Y AGE-Ja 3 anaiog i6i AOΪ 3.4500 " 4500
EVDPTGHLY MAGE-3a 3~ anaiog 161 Aiii 2.9500 29500
EVDPΪGUS MAGE- a 3" analog i6i Aiii 2.6667" 26667
EVDPAGHLY MAGE-3a 3 anaiog Ϊ6ϊ Aiii 2.4000" 2.4000
EVDPIGHLA MAGE-3a 3 anaiog Ϊ6i Aiii 0.3301) (13101)
EVAPΪGHLY MAGE-Ja 3 anaiog iόi ΛOI O.iWM)" - 01800
EVDPASNTY AGE-4 4 Ϊ6i ΛOI ϊ.500() " I 5000 O
H
VGSDCTTIHY p53 225 AOI 0.2600 0 (KK)3 0.0003 n.2 (itϊ
CΛ
PSQKTYQGSY p53 98 AOI 0.0 ϊ 4(7 0.0003 0.0003 0.0140 NO o^o^
PLSEDQLLΫ PAP 147 AOΪ 1.2000" "0.0005 0.000*1 i.2000 ©
— — n
©
IPSΫKKL1MY~ PAP 277 AOΪ 0.5650 05650 Ol
NO
YASCHLTELY AP
" 310 AOI 0.5467 ό.oδoT 0.0002 0.5467
Table 5
Sequence Antigen Strain Molecnle Position Motif Al A2 A3 All Λ24 Max. Binding Binding Binding Binding Binding Binding
RVLQGLPREY -ER 2 545 A03 0.0015 00350 00050 00350
NO
QLVTQLMPΫ c ERB2 795 A03 0()024 00112 00039 00112 NO in
VMΛGVGSPY c !ΞrbB2~ 773 A03 Ϊ) 04(H) 00575" 00079 00575 NO in
II.WKAGILY iinv adr POL" 724 A0 '" ϋ (iiii 7" 02667 OOOKi 02667
ILRGTSFVY iiiiv adr POL i.345 " A03 ii.iio i 7 00440 ~0(M)02 00440
K UWASQIΫ' HIV POL 958 A03 0.0070 o iifiii" ' (10006 (11 K.O ci.NKIVRMY iiiv GAG 274 A03 0.0017 θi() 00002 00ΪII3
I /GFLLLKY MAGE-I i()9 A(i3 ~ 00033 00563 ~ ϊi.CMl ϊ 2 00561
GTRVRAMAlΫ p53 ~ i54 Λ()3 " U.ΪH>27 (i 0365" (i. 002 01)365
KJQNFRVYY iiiv POL 1474 A03/AII 0.0056 0.U90 "iii 350 il.i350
SLYTKVVHY ~~ PSA "" 237 AO.VAli 0.0017 06750 ~'CMIMU 0.6750
LTCGFADlMGY iicv i26 AU 2.4500 00003 (iiii 20 o.iioiii" 2.45(H)
ETAYFLLK iiiv con 1351 AΪΪ 0(H)37 (λ0425 0.0425
RWGLLLALL c-EibB2 8 A24 i.2567 i.2567
PYVSRLLGI c-EιbB2 780 A24 0.i65() 0.1650
VYMΪMVKCW c-EιbB2 951 " A24 0 K.JO 01 (.10
AYSLTLQGL c-EιbB2 440 A24 01250 i) Ϊ250
SYGVTVWEL c-ErbB2 907 A24 01200 01200
LYISAWPDS c ErbB2 4 iii A24 00835 (10835
VWSYGVTVW c-ErbB2 9115 A24 _ 00800 00800
SYGVTVWELM c-ErbB2 907 A24 00630 " 0(16.10
TYLPT ASL '~ c-ErbB2 63 A24 00375 00375
VYMIMVKCWM c EιbB2 951 A24 00218 (UJ2i8
RFRELVSEF c ErbB2 " 968 A 24 ooiϋo 00IK0 n H
CYGLGMEML c-ErbB2 342 A24 (iiii 76 (iiii 76 c CΛ
NO
KWMALEsiL c ErbB2 ~ ~ 887 A24 00149 00149 00
EYLVPQQGFF -Eι l32 r≡ 1022 "Λ24~ 00121) 0.IH20 en
RYSEDPTVPL . _ c-EιbB2 iiii A24 Oθil7 00117 s NO
RFTUQs v
""
c-ErbB2 898 A24 0.0107 00107
Table 5
m ≥:
NO in
Sequence Antigen Strain Molecule Position Mollf Al A2 A3 Al l A 24 Max.
Binding Binding Binding Binding Binding Binding
EYLVSFGVWI HBV NUC 1 17 A24 00335 0.0335
WFHISCLTF IIBV NUC i()2 A24 0.0300 0.0300
QYLAGLSTL iicv _ _ ____ 1777 A24 0.0475 " 0.0475
TYSTYGKFL iicv J296 ~ A24 00225 0.0225
QYSPGQRVEF iicv 2614 A24 0 (M75 " O.OΪ75
KFMLCAGRW PSA 190 A24 iioo 0.0305 0.0305
n
CΛ NO 00 in
©
73 Table 6
AA SEQUENCE SOURCE
9 GLNKΓVRMY HIV GAG 274
9 KLNWASQIY HIV POL 958
9 KIQNFRVYY HIV POL 1474
9 TLWKAGILY HBV adr POL 724
9 ILRGTSFVY HBV adr POL 1345
9 SLYTKWHY PSA 237
9 NTSSSPQPK p53 311
9 NVKIPVAIK C-ERB2 745
10 TLGFGAYMSK HCV LORF 1261
10 GTRVRAMAIY p53 154
10 EAYSPVSTSK HBV adw POL 887
9 QIT IQNFR HIV POL 1471
9 NITGLILTR HIV ENV 2633
9 FLWEWASVR HBV adr ENV 324
9 RTPSPRRRR HBV adr CORE 549
9 SLARGNQGR HBV adr POL 805
10 VAYQATVCAR HCV LORF 1587
10 KTYQGSYGFR p53 101
9 WMCLRRFII HBV ayw 237
9 WMCLRRFII HBV ayw 237-245
9 KFMLCAGRW PSA 190
10 IMPKTGFLII MAGE 1 188
8 ETAYFLLK HIV con 1351
1 1 LTCGFAD1MGY HCV 126
9 CSPHHTALR HBV NUC;XNUCFUS 48
9 VMPKTGLLI MAGE 2 188
9 VMPKTGLLI MAGE2 188-196
9 VAELVHFLL MAGE 3 106
9 IMPKAGLL1 MAGE 3 188
10 VMPKTGLLII MAGE 2 188
10 VMPKTGLLII MAGE2 188-197
74
AA SEQUENCE SOURCE
9 ASCVTACPY c-ErbB2 293
9 VMAGVGSPY c-ErbB2 773
9 ASPLDSTFY c-ErbB2 997
9 FSPAFDNLY c-ErbB2 1213
9 KSTKVPAAY HCV 1236
9 DSSVLCECY HCV 1513
9 LSAFSLHSY HCV 2889
9 PLSEDQLLY PAP 147
9 YAVCDKCLK HPV 16 E6 67
9 CMSCCRSSR HPV 16 E6 143
9 RWGLLLALL c-ErbB2 8
9 TYLPTNASL c-ErbB2 63
9 CYGLGMEHL c-ErbB2 342
9 AYSLTLQGL c-ErbB2 440
9 PYVSRLLGI c-ErbB2 780
9 KWMALESIL c-ErbB2 887
9 RFTHQSDVW c-ErbB2 898
9 VWSYGVTVW c-ErbB2 905
9 SYGVTVWEL c-ErbB2 907
9 VYMIMVKCW c-ErbB2 951
9 RFRELVSEF c-ErbB2 968
9 WFHISCLTF HBV NUC 102
9 TYSTYGKFL HCV 1296
9 QYLAGLSTL HCV 1777
10 IPSYKKLIMY PAP 277
10 RGTQLFEDNY c-ErbB2 103
10 ESMPNPEGRY c-ErbB2 280
10 CMQIAKGMSY c-ErbB2 826
10 PASPLDSTFY c-ErbB2 996
10 FSPAFDNLYY c-ErbB2 1213
10 PSQKTYQGSY p53 98
10 VGSDCTTIHY p53 225
10 YASCHLTELY PAP 310
10 LYISAWPDSL c-ErbB2 410
75
AA SEQUENCE SOURCE
10 SYGVTVWELM c-ErbB2 907
10 VYMIMVKCWM c-ErbB2 951
10 EYLVPQQGFF c-ErbB2 1022
10 RYSEDPTVPL c-ErbB2 1111
10 EYLVSFGVWI HBV NUC 117
10 QYSPGQRVEF HCV 2614
9 VYNFATCGI LCMV glyco 35
9 GYCLTKWMI LCMV glyco 283
9 MFEALPHII LCMV glyco 7
9 IFALISFLL LCMV glyco 43
9 LFKTTVNSL LCMV glyco 342
9 LYTVKYPNL LCMV nucleo 204
9 PYIACRTSI LCMV nucleo 314
10 GYCLTKWMIL LCMV glyco 283
10 AYLVSIFLHL LCMV glyco 446
9 RWCIPWQRL CEA 10
9 IYPNASLLI CEA 101
9 LWWVNNQSL CEA 177
9 LYGPDAPTI CEA 234
9 VYAEPPKPF CEA 318
9 LWWVNNQSL CEA 355
9 LYGPDDPT1 CEA 412
9 TYYRPGVNL CEA 425
9 LYGPDTPI1 CEA 590
9 QYSWRINGI CEA 624
9 TYACFVSNL CEA 652
9 VWKTWGQYW gplOO 152
9 TWGQYWQFL gplOO 155
9 RYGSFSVTL gplOO 479
9 LMAWLASL gplOO 606
9 HWLRLPRIF gplOO 636
9 SYKHEQVYI PAP 96
9 AMTNLAALF PAP 116
AA SEQUENCE SOURCE
9 TWIG,\APLI PSA 9
9 CYASGWGSI PSA 148
10 YMIMVKCWMI c-ErbB2 952
10 RWCIPWQRLL CEA 10
10 FWNPPTTAKL CEA 27
10 QYSWFVNGTF CEA 268
10 TFQQSTQELF CEA 276
10 VYAEPPKPFI CEA 318
10 YYRPGVNLSL CEA 426
10 QYSWLIDGNI CEA 446
10 SYLSGANLNL CEA 604
10 HFLRNQPLTF gplOO 231
10 LFPPEGVSIW PAP 123
10 TWIGAAPLIL PSA 9
10 HYRKWKDTI PSA 244
9 KLRKPKHKK P. falciparum CSP 104
9 KILSVFFLA P. falciparum EXP-1 2
9 ALFFIIFNK P. falciparum EXP-1 10
9 GTGSGVSSK P. falciparum EXP-1 28
9 VLYNTEKGR P. falciparum EXP-1 99
9 KYKLATSVL P. falciparum EXP-1 73
9 PSENERGYY P. falciparum LSA1 1664
9 FLKENKLNK P. falciparum LSA1 111
9 GVSENIFLK P. falciparum LSA1 105
9 ILVNLLIFH P. falciparum LSA1 12
9 KSLYDEHIK P. falciparum LSA1 1854
77
AA SEQUENCE SOURCE
9 LLIFHINGK P. falciparum LSA1 16
9 QSSLPQDNR P. falciparum LSA1 1676
9 QTNFKSLLR P. falciparum LSA1 94
9 RINEEKHEK P falciparum LSA1 49
9 SLYDEHKK P. falciparum LSA1 1855
9 VLAEDLYGR P. falciparum LSA1 1647
9 VLSHNSYEK P. falciparum LSA1 60
9 FYFILVNLL P. falciparum LSA1 9
9 YYIPHQSSL P. falciparum LSA1 1671
9 PSDGKCNLY P. falciparum TRAP 207
9 LACAGLAYK P. falciparum TRAP 511
9 LLACAGLAY P. falciparum TRAP 510
9 LSTNLPYGR P. falciparum TRAP 122
9 QGINVAFNR P. falciparum TRAP 192
9 RGDNFAVEK P. falciparum TRAP 307
9 RSRKREILH P. falciparum TRAP 262
9 SLLSTNLPY P. falciparum TRAP 120
9 KYLVIVFLI P. falcipamm TRAP 8
9 PYAGEPAPF P. falciparum TRAP 528
78
AA SEQUENCE SOURCE
10 VTCGNGIQVR P. falciparum CSP 375
10 GTGSGVSSKK P. falciparum EXP-1 28
10 LALFFIIFNK P. falciparum EXP-1 9
10 FQDEENIGIY P. falciparum LSA1 1794
10 FILVNLLIFH P. falciparum LSA1 11
10 HVLSHNSYEK P. falciparum LSA1 59
10 KSLYDEHIKK P. falciparum LSA1 1854
10 ALLACAGLAY P. falciparum TRAP 509
10 IIRLHSDASK P. falciparum TRAP 100
10 LLACAGLAYK P. falciparum TRAP 510
10 RLHSDASKNK P. falciparum TRAP 102
9 ILGFVFTLT-NH2 Flu Matrix 59-67
10 KGILGFVFTL- Flu Matrix 57-66 NH2
9 KLQCVPLHV PSA 166-174 P/D
9 KLQCVPLHV PSA 166-174 P/D
9 KLQCVPLHV PSA 166-174 P/D
11 KQVPLRPMTYK 940.03 N-terminal extension
9 LYEIVAKV A2.1 consensus
9 KLAEYVAKV A2.1 consensus
9 KLAEΓVYKV A2.1 consensus
9 KVFEYLINK A3.2 consensus
10 KVFPYALINK A3. consensus
9 AVFAYAAAK A3.2 consensus
9 ALEPAIAKY Al consensus
79
AA SEQUENCE SOURCE
9 YLEPAIAKY Al consensus
9 ALEPYIAKY Al consensus
9 YLEQYIEKY Al consensus
9 GTEKLLAKY Al consensus
9 ATEPAIAKY Al consensus
9 ATNYPAIQK Al l consensus
9 ATNVPAIQK Al l consensus
9 ATNAPYIQK Al l consensus
9 ATNAVYIQK Al l consensus
9 ATNAAYAQK Al l consensus
9 AVNAAYAQK Al l consensus
9 AVNAPYIQK Al l consensus
9 AVNAVYIQK Al l consensus
9 PTDPKLINY Al consensus
9 GTDPKLINY Al consensus
9 YTDPKLINF Al consensus
9 FTDPKLINY Al consensus
9 FTDQAVKY Al consensus
9 YTDQAVIKF Al consensus
9 YTDQKLINF Al consensus
9 STNPKPQKK HCV-core 2-10
11 STNPKPQKKNK HCV-core 2-12
9 SFFPEITYI self pepude of P815 analog, Y2 to F,
9 ATDPNFLLY Al consensus
9 ATDKNFLLY Al consensus
9 ALMEKIYQV A2.1 consensus peptide
9 ALSEKIYQV A2.1 consensus peptide
9 AVYDPIIQK A3.2 consensus peptide
9 AVYDKIIQK A3.2 consensus peptide
9 AVMNPMIQK Al l consensus peptide
80
AA SEQUENCE SOURCE
9 AVMNEMIQK Al l consensus peptide
9 AYMDMVNSF A24 consensus peptide
9 AY1DNVNSF A24 consensus peptide
9 KLAAAAAAK A3 2/A11 poly-A analog
9 DVFRDPALK Aw68 endogenous
9 GYKDGNEYI Lm listenolysin 91- 99
10 MMWYWGPSLY HBV
11 WMMWYWGPSL HBV Y
9 RYLRDQQLL HIV env
8 FLLLKYRA MAGE-1
9 IMPKTGFLI MAGE-1
9 VADLVGFLL MAGE-1
10 IMPKTGFLII MAGE-1
11 FLIIVLVMIAM MAGE-1
11 CILESCFRAVI MAGE-1
9 MYRPDAIQL P Yoeln SSP2 143
10 NYSPNGNTNL P Yoeln SSP2 119
9 KFNPMKTH1 Kd consensus peptide
9 AMKNLDFI Db consensus
9 AMKNLYFI Db consensus analog
11 STLPETYVVRR HCV 141-151 analog
9 QYDDAVYKL Cw4 consensus
10 FQDPQERPRK HPV16 E6
10 VFEFAFKDLF HPV18 E6
9 VVYRDSIPH HPV 18 E6
9 IFEANGNLI Flu HA 240-248
9 IYATVAGSL HA 529-537
81
AA SEQUENCE SOURCE
9 SYIPSAEKI P bergan CS 252- 260
9 KYQAVTTTL Tumour P198 14-22
10 MYPHFMPTNL MCMV pp89 167- 176
9 AYPNVSAKI Lm listenolysm 196- 204
9 AYTGGKINI Lm listenolysm 413- 421
9 SAISSILSK HBV ENV 159
9 QAGFFLLTK HBV ENV 190
9 SALYREALK HBV NUC 64
9 RAKWNNTLK HIV env 370
9 RATQIPSYK PAP 273
9 TAAHCIRNK PSA 58
9 MAVFIHNFK HIV pol 909
9 TAGILELLK HPV 6b El 192
9 RAALLGKFK HPV 6b El 205
9 CATMCRHYK HPV 6b El 406
9 TAACSHEGK Flu HA-1 132
9 NANANSAVK P. fal csp 304
9 GAFKVPGVK LCMV glyco 484
9 RARVHPTTR HBV POL 244
9 CALPFTSAR HBV X 69
9 NMLESILK LCMV nuc 259
9 WMILAAELK LCMV glyco 289
9 EMNLPGRWK HIV pol 107
9 SSLQSKHRK HBV POL 201
9 GSTHVSWPK HBV POL 398
9 TSDLEAYFK HBV X NUC FUS 105
9 ASQIYAGK HIV pol 438
9 ASCDKCQLK HIV pol 769
9 MSLAADLEK LCMV nuc 100
9 VSSKNLMEK Mel tyro 25
82
AA SEQUENCE SOURCE
9 LSTNLPYGK P. fal ssp2 122
9 STDHIPILY Al Nat. Processed
9 STAPPAHGV Breast mucin 9-17
9 LMAVVLASL gplOO
9 WSQKRSFVY gplOO
9 PLDCVLYRY gplOO
10 PSSVGSRSEY gplOO
9 YTAVVPLVY Hu J chain 102-110
83 Table 7
AA SEQUENCE SOURCE
8 LTELYFEK PAP 315
9 TISPSYTYY CEA 419
9 GTGCNGWFY HPV 16/18 El 11
9 LTEMVQWAY HPV6b/ll El 358
9 rrvNNSGSY CEA 289
9 CTGWFMVEA HPV6b/ll El 14
9 ATVQDLKRK HPV6b/ll El 77
10 9 AVESEISPR HPV6b/ll El 101
9 FLNSNMQϋ HPV6b/ll El 393
9 ΓΓRQTVIEH HPV6b/ll El 341
9 ΓVGPPDTGK HPV 6b/ 11 El 476
9 KLIEPLSLY HPV6b/ll El 254
15 9 KLWLHGTPK HPV 6b/ 11 El 462
9 KMSIKQWIK HPV6b/ll El 420
9 WAGFGIHH HPV6b/ll El 238
9 HLFGYSWYK CEA 61
9 ISPSYTYYR CEA 420
20 9 HTQVLFIAK CEA 636
9 ITVYAEPPK CEA 316
9 rrvsAELPK CEA 494
9 RLQLSNGNR CEA 190
9 RLQLSNGNR CEA 546
25 9 RINGIPQQH CEA 628
9 SNMQAKYVK HPV 6b/ 11 El 396
9 EWITRQTVI HPV6b/ll El 339
9 FFERLSSSL HPV6b/ll El 613
9 NWKPIVQFL HPV6b/ll El 439
30 10 PTISPSYTYY CEA 418
10 PTISPLNTSY CEA 240
10 HSASNPSPQY CEA 616
10 KLIEPLSLYA HPV6b/ll El 254
10 AIVGPPDTGK HPV6b/ll El 475
35 10 DCATMCRHYK HPV6b/16El 405
10 KLWLHGTPKK HPV 6b/ 11 El 462
10 WVVAGFGIHH HPV6b/ll El 237
84
AA SEQUENCE SOURCE
10 TΓΓVSAELPK CEA 493
10 TFWNPPTTAK CEA 26
10 TISPSYTYYR CEA 419
10 TISPLNTSYR CEA 241
10 RTLTLFNVTR , CEA 198
10 RTLTLFNVTR CEA 554
10 RTLTLLSVTR CEA 376
10 ATPGPAYSGR CEA 89
10 ASGHSRTTVK CEA 483
10 10 QFLRHQNIEF HPV6b/ll El 445
10 TFTFPNPFPF HPV 6b/ 11 El 586
9 RVDCTPLMY ProstCa PSM 463
9 LLSLYGIHK ProstCa PAP 243
9 SIVLPFDCR ProstCa PSM 590
15 9 KSLYESWTK Prost Ca PSM 491
9 SMKHPQEMK ProstCa PSM 615
9 SLYESWTKK ProstCa PSM 492
9 YSLVHNLTK Prost Ca PSM 471
9 HLTELYFEK ProstCa PAP 314
20 9 RATQIPSYK ProstCa PAP 273
9 ASGRARYTK ProstCa PSM 531
9 SLYGIHKQk ProstCa PAP 245
9 RDYAVVLRK ProstCa PSM 598
9 SSHDLMLLR ProstCa PSA 113
25 9 GAAPLILSR ProstCa PSA 12
9 KIVIARYGK ProstCa PSM 199
9 RAAPLLLAR Prost Ca PAP 2
9 VVLRKYADK ProstCa PSM 602
9 GLPDRPFYR ProstCa PSM 680
30 9 WLDRSVLAK ProstCa PAP 25
9 KVFRGNKVK ProstCa PSM 207
9 IVRSFGTLK ProstCa PSM 398
9 KIYSISMKH ProstCa PSM 610
9 RSVLAKELK ProstCa PAP 28
35 9 STNEVTRIY ProstCa PSM 348
9 GFFLLGFLF ProstCa PSM 31
85
AA SEQUENCE SOURCE
9 LYSDPADYF Prost Ca PSM 227
9 KYADKIYSI Prost Ca PSM 606
9 NYARTEDFF Prost Ca PSM 178
9 AYINADSSI Prost Ca PSM 448
9 SASFCGSPY HBV POL 165
9 AFTFSPTYK HBV POL 655
9 SVVRRAFPH HBV POL 524
9 RWMCLRRFI HBV ENV 236
9 SWLSLLVPF HBV ENV 334
10 9 SWWTSLNFL HBV ENV 197
9 PWTHKVGNF HBV POL 51
9 SFCGSPYSW HBV POL 167
10 NADSSIEGNY Prost Ca PSM 451
10 GLDSVELAHY Prost Ca PSM 104
15 10 RATQIPSYKK Prost Ca PAP 273
10 LGFLFGWFD Prost Ca PSM 35
10 SSIEGNYTLR Prost Ca PSM 454
10 KSLYESWTKK Prost Ca PSM 491
10 SLLSLYGIHK Prost Ca PAP 242
20 10 FLYNFTQIPH Prost Ca PSM 73
10 VIYAPSSHNK Prost Ca PSM 690
10 AVVLRKYADK Prost Ca PSM 601
10 KSPDEGFEGK Prost Ca PSM 482
10 IVRSFGTLKK Prost Ca PSM 398
25 10 RIYNVIGTLR Prost Ca PSM 354
10 LSLYGIHKQK Prost Ca PAP 244
10 MSLLKNRFLR Prost Ca PSA 99
10 ISMKHPQEMK Prost Ca PSM 614
10 RAVCGGVLVH Prost Ca PSA 43
30 10 GSAPPDSSWR Prost Ca PSM 311
10 SIPVHPIGYY Prost Ca PSM 291
10 CSGKIVIARY Prost Ca PSM 196
10 ETYELVEKFY Prost Ca PSM 557
10 RLLQERGVAY Prost Ca PSM 440
35 10 FYDPMFKYHL Prost Ca PSM 565
10 TYSVSFDSLF Prost Ca PSM 624
86
AA SEQUENCE SOURCE
10 LYNFTQIPHL Prost.Ca PSM 74
10 GWRPRRTILF Prost.Ca PSM 409
10 FAAPFTQCGY HBV POL 631
10 RWMCLRRFII HBV ENV 236
10 WFVGLSPTVW HBV ENV 345
10 SWPKFAVPNL HBV POL 392
10 VFADATPTGW HBV POL 686
9 FIFHKFQTK HTLV-I tax 276
9 FLTNVPYKR HTLV-I tax 182
10 9 ΓΓWDPIDGR HTLV-1 tax 54
9 SALQFLIPR HTLV-I tax 66
9 LSFPDPGLR HTLV-I tax 131
9 QSSSFIFHK HTLV-I tax 272
9 GLCSARLHR HTLV-1 tax 34
15 9 RLPSFPTQR HTLV-I tax 74
9 AMRKYSPFR HTLV-I tax 108
9 ISGGLCSAR HTLV-I tax 31
9 ALFTAQEAK HPV 16 El 69
9 ATMCRHYKR HPV 16 El 406
20 9 FMSFLTALK HPV 16 El 453
9 GVSFSELVR HPV 16 El 216
9 KAAMLAKFK HPV 16 El 204
9 LTNILNVLK HPV 16 El 191
9 LVRPFKSNK HPV 16 El 222
25 9 MSFLTALKR HPV 16 El 454
9 NSNASAFLK HPV 16 El 386
9 QMSMSQWIK HPV 16 El 419
9 RLKAICIEK HPV 16 El 109
9 SLFGMSLMK HPV 16 El 484
30 9 SMSQWKYR HPV 16 El 421
9 TAAALYWYK HPV 16 El 315
9 VVLLLVRYK HPV 16 El 274
9 ALLRYKCGK HPV 18 El 284
9 ATMCKHYRR HPV 18 El 413
35 9 CATMCKHYR HPV 18 El 412
9 FITFLGALK HPV 18 El 460
87
AA SEQUENCE SOURCE
9 GVLILALLR HPV 18 El 279
9 KLRAGQNHR HPV 18 El 647
9 LILALLRYK HPV 18 El 281
9 LTTNIHPAK HPV 18 El 571
9 NMSQWIRFR HPV 18 El 428
9 NSNAAAFLK HPV 18 El 393
9 SVAALYWYR HPV 18 El 322
9 WTYFDTYMR HPV 18 El 536
9 YVQAIVDKK HPV 18 El 19
10 9 ID NFDIPK GCDFP-15 36
9 VLAVQTELK GCDFP-15 55
10 IIKNFDIPK GCDFP-15 35
10 TACLCDDNPK GCDFP-15 87
10 AVLAVQTELK GCDFP-15 54
15 10 TFYWDFYTNR GCDFP-15 97
9 ASCHLTELY PAP 311
10 KGEYFVEMYY PAP 322
10 LTAAHCIRNK PSA 57
9 PLYDMSLLK PSA 95
20 9 QVHPQKVTK PSA 182
9 SLLKNRFLR PSA 100
9 YTKVVHYRK PSA 239
9 TLWKAGILY HBV pol 150
9 SLYTKVVHY PSA 237
25 9 PVNRPIDWK HBV POL 612
9 RHYLHTLWK HBV POL 719
11 HTLWKAGILYK HBV POL 149
11 GTDNSVVLSRK HBV POL 735
1 1 RVTGGVFLVDK HBV POL 357
30 8 ATQIPSYK PAP 274
9 WMNSTGFTK HCV consensus
9 RVLEDGVNY HCV consensus
9 RLLAPΓΓAY HCV consensus
9 GVLAALAAY HCV consensus
35 9 RVCEKMALY HCV consensus
88 TABLE 8
PEPTIDE AA SEQUENCE
1235.01 10 AVFDRKSDAK
26.0149 9 CALRFTSAR
26.0153 9 SSAGPCALR
F104.02 9 SLTPPHSAK
F105.01 9 AIFQSSMTK
F105.02 9 GIFQSSMTK
10 F105.03 9 AAFQSSMTK
F105.04 9 AIAQSSMTK
F105.05 9 AIFASSMTK
F105.06 9 AIFQASMTK
F105.07 9 AIFQSAMTK
15 F105.08 9 AIFQSSATK
F105.O9 9 AIFQSSMAK
F105.10 9 AIFQSSMTA
F105.l l 9 FIFQSSMTK
F105.12 9 SIFQSSMTK
20 F105.14 9 ANFQSSMTK
F105.16 9 AIFQCSMTK
F105.17 9 AIFQSSMTR
F105.19 9 AIFQSSMTY
F105.20 9 AILQSSMTR
25 F105.21 9 AIFQRSMTR
F105.24 10 PAIFQSSMTK
F105.25 10 AIFQSSMTKI
27.0103 9 AIILHQQQK
27.0104 9 YGFRLGFLH
30 27.0108 9 SSCMGGMNR
27.0235 10 TCTYSPALNK
27.0239 10 NSSCMGGMNR
27.0240 10 SSCMGGMNRR
27.0250 10 KSKKGQSTSR
35 27.0252 10 TSRHKKLMFK
28.0062 8 FMFSPTYK
28.0063 8 FVFSPTYK
28.0066 8 TMLXMXXK
89
PEPTIDE AA SEQUENCE
28 0322 9 SMICSWRR
28 0323 9 SVICSWRR
28 0324 9 KVGNFTGLK
28 0325 9 KVGNFTGLR
28 0326 9 VVFFSQFSR
28 0327 9 SVNRPIDWK
28 0328 9 TLWKAGILK
28 0329 9 TLWKAGILR
28 0330 9 TMWKAGILY
10 28 0331 9 TVWKAGILY
28 0332 9 RMYLHTLWK
28 0333 9 RVYLHTLWK
28 0334 9 AMTFSPTYK
28 0335 9 AVTFSPTYK
15 28 0336 9 SVVRRAFPR
28 0337 9 SVVRRAFPK
28 0338 9 ISEYRHYXY
28 0339 9 GTGXNGWFY
28 0340 9 ASXHLTELY
20 28 0341 9 ASXDKXQLK
28 0371 9 RVXEKMALY
28 0372 9 XTGWFMVEA
28 0374 9 HISXLTFGR
28 0375 9 AVXTRGVAK
25 28 0377 9 HLIFXHSKK
28 0378 9 HTMLXMXXK
28 0381 9 RLKAD IEK
28 0383 9 TLFXASDAK
28 0384 9 ALLRYKXGK
30 28 0387 9 ATMXRHYKR
28 0388 9 XATMXRHYK
28 0390 9 ATMXKHYRR
28 0391 9 LLAXAGLAY
28 0392 9 LAXAGLAYK
35 28 0393 9 SIVLPFDXR
28 0394 9 AAXWWAGIK
28 0628 10 OMFTFSPTYK
90
PEPTIDE AA SEQUENCE
28.0629 10 QVFTFSPTYK
28.0630 10 TMWKAGILYK
28.0631 10 TVWKAGILYK
28.0632 10 VMGGVFLVDK
28.0633 10 VVGGVFLVDK
28.0635 10 SVLPETTVVR
28.0638 10 HTLWKAGILK
28.0640 10 HMLWKAGILY
28.0395 9 SAD(SVVRR
10 28.0644 10 GTFNSVVLSR
28.0645 10 YMFDVVLGAK
28.0646 10 MMWYWGPSLK
28.0647 10 MMWYWGPSLR
28.0665 10 IVGGWEXEK
15 28.0667 10 IILEXVYXK
28.0668 10 SIPHAAXHK
28.0670 10 ΓVXPIXSQK
28.0671 10 LIRXLRXQK
28.0672 10 XTYSPALNK
20 28.0675 10 TVXAGGXAR
28.0676 10 HISXLTFGR
28.0677 10 XVNXSQFLR
28.0678 10 LIFXHSKKK
28.0679 10 FVLGGXRHK
25 28.0713 10 TSAIXSVVRR
28.0714 10 HLIFXHSKKK
28.0715 10 LLIRXINXQK
28.0716 10 GΓVXPIXSQK
28.0717 10 LLIRXLRXQK
30 28.0718 10 SLEQRSLHXK
28.0720 10 RTVGGWEXEK
28.0721 10 DIILEXVYXK
28.0722 10 XVYXKQQLLR
28.0723 10 RAVXGGVLVH
35 28.0725 10 LTAAHXIRNK
28.0728 10 KAAXWWAGIK
28.0730 10 VVRRXPHHER
91
PEPTIDE AA SEQUENCE
28 0731 10 LLGIWGXSGK
28 0732 10 TTLFXASDAK
28 0734 10 RTVXAGGXAR
28 0736 10 GTQRXEKXSK
28 0737 10 LVQNANPDXK
28 0738 10 VTXGNGIQVR
28 0739 10 DXATMXRHYK
28 0740 10 GLAXHQLXAR
28 0741 10 ALLAXAGLAY
10 28 0742 10 LLAXAGLAYK
28 0743 10 XVARXPSGVK
28 0745 10 LVEIXTEMEK
28 0746 10 LLNWXMQIAK
28 0824 1 1 HMLWKAGILYK
15 28 0825 11 HVLWKAGILYK
28 0826 11 SMLPETTVVRR
28 0827 11 SVLPETTVVRR
28 0828 11 GMDNSVVLSRK
28 0829 1 1 GVDNSVVLSRK
20 28 0830 11 GTFNSVVLSRK
28 0369 9 GLAXHQLXA
1259 02 9 DTVDTVLEK
1259 10 9 PVTIGECPK
1259 14 10 FTAVGKEFNK
25 1259 16 11 RTLDFHDSNVK
1259 21 11 KTRPILSPLTK
1259 26 11 GTHPSSSAGLK
1259 28 11 ILWILDRLFFK
1259 29 9 WILDRLFFK
30 1259 30 11 CIYRRFKYGLK
1259 31 9 KSMREEYRK
1259 33 9 YIQMCTELK
1259 37 10 MVMELVRMK
1259 38 9 VMELVRMIK
35 1259 41 11 LIRPNENPAHK
26 0023 8 VSFGVWIR
26 0024 8 VSIPWTHK
92
PEPTIDE AA SEQUENCE
26 0026 8 ASFCGSPY
26 0035 9 TSPYELSLY
26 0036 9 TSIPFLHEY
26 0041 9 FNDPGPGTY
26 0045 9 YVDLGALRY
26 0051 9 DADRSFIEY
26 0055 9 NMDKAVKLY
26 0056 9 TTDNFYRNY
26 0058 9 HSAEALQKY
10 26 0059 9 LTAGLDFAY
26 0061 9 LTYKYNQFY
26 0062 9 CSNDKSLVY
26 0063 9 RSARASSRY
26 0065 9 ASADKPYSY
15 26 0067 9 STTAGPNEY
26 0069 9 LSGNGHFHY
26 0073 9 NTFVQANLY
26 0074 9 GTATYLPPY
26 0081 9 RLDAFRQTY
20 26 0082 9 KAEVHTFYY
26 0083 9 VAEGDTVIY
26 0084 9 LTEIDIRDY
26 0085 9 HTEFEGQVY
26 0086 9 VSDGGPNLY
25 26 0092 9 ΠEDQYNRY
26 0093 9 FLDQWWTEY
26 0095 9 FVEDPNGKY
26 0096 9 ISDESYRVY
26 0156 9 YLAEADLSY
30 26 0197 9 ALLAVGATK
26 0198 9 ALNFPGSQK
26 0199 9 AVGATKVPR
26 0203 9 FSVSVSQLR
26 0204 9 GTATLRLVK
35 26 0205 9 GVSRQLRTK
26 0207 9 LIYRRRLMK
26 021 1 9 OLVLHOILK
93
PEPTIDE AA SEQUENCE
26 0212 9 SSHWLRLPR
26 0214 9 TMEVTVYHR
26 0216 9 VLASLIYRR
26 0217 9 VSCQGGLPK
26 0218 9 VVLASLIYR
26 0227 9 GTQCALTRR
26 0251 9 FTIPYWDWR
26 0252 9 GTPEGPLRR
26 0253 9 KSYLEQASR
10 26 0255 9 LVSLLCRHK
26 0256 9 MVPFIPLYR
26 0258 9 QTSAGHFPR
26 0259 9 SIFEQWLRR
26 0260 9 SLLCRHKRK
15 26 0261 9 SSWQIVCSR
26 0267 10 NMQIGGVLTY
26 0273 10 RMAQNFAMRY
26 0274 10 FTVQGSLSGY
26 0275 10 QTSPYELSLY
20 26 0276 10 SSNAILSLSY
26 0280 10 TSQPWWPADY
26 0284 10 VSDVSIIIPY
26 0285 10 ASDAQSANKY
26 0286 10 FTETNLAGEY
25 26 0287 10 YVDGFEPNGY
26 0291 10 FNDPGPGTYY
26 0296 10 FLDQWWTEYY
26 0299 10 AAEFATETAY
26 0309 10 NAEVVLNQLY
30 26 0311 10 FVDGDSLFEY
26 0316 10 PSEDAQVAVY
26 0317 10 MSDNIRTGLY
26 0318 10 ESELREILNY
26 0319 10 CMESVRNGTY
35 26 0320 10 KTENGΓΓRLY
26 0321 10 LTEIDIRDYY
26 0397 10 LLVLMAWLA
94
PEPTIDE AA SEQUENCE
26 0424 10 AVVLASLIYR
26 0425 10 GALLAVGATK
26 0426 10 GTATLRLVKR
26 0427 10 HTMEVTVYHR
26 0428 10 IALNFPGSQK
26 0432 10 QLRALDGGNK
26 0433 10 QVPLDCVLYR
26 0434 10 SLIYRRRLMK
26 0435 10 SSSHWLRLPR
10 26 0438 10 TVSCQGGLPK
26 0442 10 VVLASLIYRR
26 0466 10 YVKVLHHTLK
26 0473 10 LIGCWYCRRR
26 0474 10 LLIGCWYCRR
15 26 0485 10 SSMHNALHIY
26 0504 10 CVSSKNLMEK
26 0510 10 FSSWQΓVCSR
26 0511 10 GLVSLLCRHK
26 0518 10 YMVPFIPLYR
20 26 0535 11 GVWIRTPPAYR
26 0539 11 RLVVDFSQFSR
26 0545 1 1 TLPETTVVRRR
26 0549 11 LLPIFFCLWVY
11 STLPETTWRR
25
26 0550 11 RAFPHCLAFSY
95
*
O o o co n o o o o o o p . o vn 00 O o o o o o o o o o l o o o O o O o o o o O
3 o o O l o o o o v v d
ω 01
4→ oo J~ C f CM
H
ON 00 R
£ s s 51 ? SI S 3 5 VO δ ω
«M N M *N M
■8 σ» o» t CM OΛ σ> o» «* CM *
> >
2 > -I u CO •J •J u ω u iJ
I u u
Q u •J §1 (0 Q fa. I O
8 u •J W Bu H § H ϋ
OT to (0 n I ω U ω •J < 3
2 X •J ϋ t >- b.
2 II K O •J co Si s u u X
Mage
Sequence Aλ Strain Mol. POB. Motif λl A2.1 A3.2 All A24
0 NO
AISRKMVELV 10 2 101 2.1 NO n
MVELVHFLLL 10 2 106 2.1 0.0017 NO in
KLPGLLSROL 10 2 135 2.1 0
LLSROLQQSL 10 2 139 2.1 0.0007
SLPTTMNYPL 10 3 63 2.1 0.0035
DLESEFQAAL 10 3 93 2.1 0.0001
ALSRKVAELV 10 3 101 2.1 0.0001
KVAELVHFLL 10 3 105 2.1 0.012
VIFSKASSSL 10 3 142 2.1 0
SLQLVFGIEL 10 3 150 2.1 0.0049
LMEVDPIGHL 10 3 159 2.1 0.0005 NO
ON
FLIIVLVMI 9 194 2.1 0.0005
GLLGDNQIM 9 181 2.1 0.0051
SLHCKPEEA 9 7 2.1 0.013 <0.0002 0
ALGLVCVQA 9 22 2.1 0.015 <0.0002 <0.0002
CKPEEALEA 9 10 Random <0.0002
QQBALGLVC 9 19 Random <0.0002
VQAATSSSS 9 28 Random <0.0002
PLVLGTLEB 9 37 Random <0.0002 "o0
VPTAGSTDP 9 46 Random <0.0002
CΛ
PQSPQGASA 9 55 Random <0.0002 NO 00
©
FPTTINFTR 9 64 Random <0.0002 in
©
1*1 NO
Mage
Sequence AA Strain Mol. Poβ. Motif Al A2.1 A3.2 All A24
NO
QRQPSEGSS 9 73 Random <0 0002 NO in
SREEEGPST 9 82 Random <0 0002 NO in
AVITKKVAD 9 100 Random <0.0002
EMLESVIKN 9 127 Random <0.0002 0
YKHCFPEIF 9 136 Random <0.0002
GKASESLQL 9 145 Random <0.0002
VFGIDVKEA 9 154 Random <0.0002 <0.0002 0
DPTGHSYVL 9 163 Random <0.0002
VTCLGLSYD 9 172 Random <0.0002
PKTGFLIIV 9 190 Random <0.0002 NO
LVMIAMEGG 9 199 Random <0.0002
HAPEEEIHE 9 208 Random <0.0002
ELSVMEVYD 9 217 Random <0.0002
GREHSAYGE 9 226 Random <0.0002
PRKLLTQDL 9 235 Random 0.0002
VQEKYLEYG 9 244 Random <0.0002
RCRTVIPHA 9 253 Random <0.0002
MSSCGVQGP 9 262 Random <0.0002
ILESLFRAVI 10 93 2.1 0.0002 O H
FLIIVLVMIA 10 194 2.1 0.0003 0.0093 0.0030
CΛ
LVFGIDVKEA 10 153 2.1 0.0002 <0.0002 0 NO 00
©
EVYDGRBHSA 10 222 2.1 0 <0.0002 0 ion
NO
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
NO
GVQGPSLKPA 10 266 2.1 0.0001 NO in
QLVFGIDV 8 152 2.1 0 NO n
KLLTQDLV 8 237 2.1 0.0004
GLLGDNQI 8 181 2.1 0
DLVGFLLL 8 108 2.1 0
GLSYDGLL 8 176 2.1 0.0001
DLVQEKYL 8 242 2.1 0
LLGDNQIM 8 182 2.1 0
FLIIVLVM 8 194 2.1 0
ALBAQQEA 8 15 2.1 0 NO 00
TLEEVPTA 8 42 2.1 0
IMPKTGFL 8 188 2.1 0.0001
PVTKAEML 8 122 2.1 0
IVLVMIAM 8 197 2.1 0.0001
AVITKKVA 8 100 2.1 0
EIHBELSV 8 213 2.1 0
LIIVLVMI 8 195 2.1 0.0001
IIVLVMIA 8 196 2.1 0.0002
SLFRAVITKKV 11 96 2.1 0.0001 0
O
H
LLLKYRAREPV 11 113 2.1 0.0001
CΛ
YLEYGRCRTVI 11 248 2.1 NO
0.0006 00 β
Ul
ALBAQQEALGL 11 15 2.1 0.0001 o W
NO
Mage
Sequence AA Strain Mol. Poβ. Motif Al A2.1 A3.2 All A24
FLIIVLVMIAM 194 2.1 0.0041 NO NO
VLGTLEEVPTA 39 2.1 0.0002 in
NO n i-
QLVFGIDVKEA 152 2.1 0.0001
AVITKKVADLV 100 2.1 0
PVTKAEMLESV 122 2.1 0
KVADLVGFLLL 105 2.1 0.020
GVQGPSLKPAM 266 2.1 0
LVGFLLLKYRA 109 2.1 0.0004
LVMIAMEGGHA 199 2.1 0.0005
CILESLFRAVX 92 2.1 0.0030
EALEAQQEA 9 14 2.1 0 <0.0002 0 NO NO
EAQQEALGL 9 17 2.1 0 •θ.0002
AATSSSSPL 9 30 2.1 0 <0.0002
ATSSSSPLV 9 31 2.1 0.0007
GTLEEVPTA 9 41 2.1 0.013 <0.0002 0
GASAFPTTI 9 60 2.1 0 <0.0002
STSCILESL 9 89 2.1 0.0002
RAVITKKVA 9 99 2.1 0 <0.0002 0
ITKKVADLV 9 102 2.1 0 o
RAREPVTKA 9 118 2.1 0
KAEMLESVI 9 125 2.1 0 <0.0002 CΛ
NO 00
KASESLQLV 9 146 2.1 0.0009 © o in l
NO
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
PTGHSYVLV 9 164 2.1 0 NO NO
KTGFLIIVL 9 191 2.1 0.0006 CΛ
NO
LIIVLVMIA 0 0.0022 0.0006 in
9 195 2.1
IIVLVMIAM 9 196 2.1 0.0007
MIAMEGGHA 9 201 2.1 0.0005 <0.0002 0.0002
EIWEELSVM 9 213 2.1 0
SAYGEPRKL 9 230 2.1 0.0002 <0.0002
YLEYGRCRT 9 248 2.1 0
EALGLVCVQA 10 21 2.1 0.0005 <0.0002 0
QAATSSSSPL 10 29 2.1 0 <0.0002 o o
VTKAEMLESV 10 123 2.1 0
EADPTGHSYV 10 161 2.1 0
VLGTLEEVPT 10 39 2.1 0.0004
SAFPTTIN^ 10 62 2.1 0
GIDVKEADPT 10 156 2.1 0
PTGHSYVLVT 10 164 2.1 0
FLWGPRALA 9 new 265 2.1 0.042 0.0017 0
LAETSYVKV 9 new 272 2.1 0
YVKVLEYVI 9 new 277 2.1 0.0002
RVRFFFPSL 9 new 290 2.1 0.0001 n "0
LAETSYVKVL 10 new 272 2.1 0 <0.0002 CΛ NO 00
VLEYVIKVSA 10 new 280 2.1 0.0002 0.0002 0 © ion l
NO
Mage
Sequence AA Strain Mol. Poa. Motif Al A2.1 A3.2 All A24
AALREEEEGV 10 new 301 2.1 0 NO NO J≥
SMHCKPEEV 9 new (a) 7 2.1 0.018 CΛ
NO CΛ
AMGLVCVQV 9 new (a) 22 2.1 0.012
LMLGTLEEV 9 new (a) 38 2.1 0.13
LQLVFGIDV 9 new 151 2.1 0.0004
GLSYDGLLG 9 new 176 2.1 0
GLSYDGLLV 9 new (a) 176 2.1 0.0047
LLGDNQIMP 9 new 182 2.1 0.0001
LLGDNQIMV 9 new (a) 182 2.1 0.043
WEELSVMEV 9 new 215 2.1 0
WMELSVMEV 9 new (a) 215 2.1 0.041
RKLLTQDLV 9 new 236 2.1 0
YEFLWGPRA 9 new 262 2.1 0
YMFLWGPRV 9 new (a) 262 2.1 0.22
AATSSSSPLV 10 new 30 2.1 0
ATSSSSPLVL 10 new 31 2.1 0
KMADLVGFLV 10 new (a) 105 2.1 1.5
VADLVGFLLL 10 new 106 2.1 0.0008 0.0003
SESLQLVFGI 10 new 148 2.1 0
O
VMVTCLGLSV 10 new (a) 170 2.1 0.30 H
QIMPKTGFLI 10 new 187 2.1 0.0009 CΛ NO 0-
^0
QMMPKTGFLV 10 new (a) 187 2.1 0.050 © CΛ s NO
Mage
Sequence AA Strain Mol. Poβ. Motif Al A2.1 A3.2 All A24
NO
KTGFLIIVLV 10 1 new 191 2.1 0.0012 NO
C £Λ:
LIIVLVMIAM 10 1 new 195 2.1 0.0003 NO CΛ
4-.
VMIAMEGGHV 10 1 new (a) 200 2.1 0.053
SAYGEPRKLL 10 1 new 230 2.1 0 0.0008
ALAETSYVKVL 11 1 N 270 2.1 0.012
KMVELVHFLLL 11 2 52 2.1 0.67
ELMEVDPIGHL 11 3 105 2.1 0.026
HLYIFATCLGL 11 3 114 2.1 0.041
LLLKYRAREPV 11 3 60 2.1 0.0001
QLVFGIELMEV 11 3 99 2.1 0.34 o
NJ
IMPKAGLLIIV 11 3 135 2.1 0.013
VLVTCLGLSYDGL 13 1 n B6 170 2.1 0.0017
KLLTQDLVQEKYL 13 1 n E6 237 2.1 0.0060
DLVQEKYLEYRQV 13 1 n E6 242 2.1 0
SLFRAVITKKVADLV 15 1 n POL 96 2.1 0.0004 .
DLESEFQAAISRKMV 15 2 POL 40 2.1 0
MLGSWGNWQYFFPV 15 3 POL 75 2.1 0.012
GASSFSTTI 9 2 60 2.1 0 0.0002
DLESEFQAA 9 2.3 93 2.1 0 n H
QAAISRKMV 9 2 99 2.1 0
CΛ
KAEMLESVL 9 2 125 2.1 0 0 NO go ©
KASEYLQLV 9 2 146 2.1 0.011 CΛ
O Ul
NO
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
NO
QLVFGIEW 9 2 152 2.1 0.0038 NO
≥ CΛ:
WPISHLYI 9 2 162 2.1 0.0002 NO CΛ
PISHLYILV 9 2 164 2.1 0.0005
HLYILVTCL 9 2 167 2.1 0.0034
YILVTCLGL 9 2 169 2.1 0.0014
GLLGDNQVM 9 2 181 2.1 0.0038
QVMPKTGLL 9 2 187 2.1 0
VMPKTGLLI 9 2 188 2.1 0.0010 0.230
KTGLLIIVL 9 2 191 2.1 0.0002
GLLIIVLAI 9 2.3 193 2.1 0.0002 o
LLIIVLAII 9 2,3 194 2.1 0.0001 >
LIIVLAIIA 9 2,3 195 2.1 0.0008
IIVLAIIAI 9 2 196 2.1 0.0009
IIAIEGDCA 9 2 201 2.1 0
GASSLPTTM 9 3 60 2.1 0 0.0010
QAALSRKVA 9 3 99 2.1 0
VAELVHFLL 9 3 106 2.1 0 0.039
KAEMLGS 9 3 125 2.1 0
KASSSLQLV 9 3 146 2.1 0.0005 n
QLVFGIELM 9 3 152 2.1 0.0010
CΛ
PIGHLYIFA 9 3 164 2.1 0 NO 00
©
IMPKAGLLI 9 3 188 2.1 0.0064 CΛ β Ul
NO
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All Λ2
KAGLLIIVL 9 3 191 2.1 0.0002 0 NO
NO
S
IIAREGDCA 9 3 201 2.1 0 CΛ:
NO CΛ
EALEAQQEAL 10 1 new 14 2.1 0 0
EAQQEALGLV 10 1 new 17 2.1 0
DLESEFQAAI 10 2 93 2.1 0
AAISRKMVBL 10 2 100 2.1 0 0
VIFSKASEYL 10 2 142 2.1 0.0014
YLQLVFGIEV 10 2 150 2.1 0.37
LVFGIEWEV 10 2 153 2.1 0.012
GIEWEWPI 10 2 156 2.1 <0.0002 o
WE PISHL 10 2 159 2.1 <0.0002
E PISHLYI 10 2 161 2.1 <0.0002
WPISHLYIL 10 2 162 2.1 0.0002
PISHLYILVT 10 2 164 2.1 0.0003
QVMPKTGLLI 10 2 187 2.1 0.0002
VMPKTGLLII 10 2 188 2.1 0.0009 0.058
KTGLLIIVLA 10 2 191 2.1 <0.0002
GLLIIVLAII 10 2,3 193 2.1 0.0005
LLIIVLAIIA 10 2,3 194 2.1 <0.0002 -a o
LIIVLAIIAI 10 2 195 2.1 0.0013 H
CΛ
AIIAIEGDCA 10 2 200 2.1 0.0023 NO 00
AALSRKVAEL 10 3 100 2.1 0.0007 0 © o CΛ l
NO
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
VAELVHFLLL 10 3 106 2.1 0.0009 0.0018 NO NO
VTKAEMLGSV 10 3 123 2.1 <0.0002 CΛ
NO C*Λ-.
GIELMEVDPI 10 3 156 2.1 <0.0002
EVDPIGHL I 10 3 161 2.1 <0.0002
PIGHLYIFAT 10 3 164 2.1 0.0003
QIMPKAGLLI 10 3 187 2.1 0.0006
IMPKAGLLII 10 3 188 2.1 0.0015
KAGLLIIVLA 10 3 191 2.1 <0.0002
AIIAREGDCA 10 3 200 2.1 <0.0002
FLWGPRALI 9 2 271 A02
GLEARGEAL 9 3 15 A02 o
EARGEALGL 9 3 17 A02
ALGLVGAQA 9 3 22 A02/A03
GLVGAQAPA 9 3 24 A02 A03
LVGAQAPAT 9 3 25 A02
PATEEQEAA 9 3 31 A02/A03
EAASSSSTL 9 3 37 A02
AASSSSTLV 9 3 38 A02
LVEVTLGEV 9 3 45 A02 n
EVTLGEVPA 9 3 47 A02 A03 H
CΛ
VTLGEVPAA 9 3 48 A02 A03 NO go ©
KIWEELSVL 9 3 220 A02 CΛ
O Ul
NO
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
SILGDPKKL 9 3 237 A02 NO NO
ILGDPKKLL 9 3 238 A02 CΛ
NO CΛ
FLWGPRALV 9 3 271 A02 *.
RALVETSYV 9 3 276 A02
LVETSYVKV 9 3 278 A02
YVKVLHHMV 9 3 283 A02
KVLHHMVKI 9 3 285 A02
EARGEALGLV 10 3 17 A02
EALGLVGAQA 10 3 21 A02/A03
GLVGAQAPAT 10 3 24 A02
QAPATEEQEA 10 3 29 A02/A03 o O
EAASSSSTLV 10 3 37 A02
TLVEVTLGEV 10 3 44 A02
EVTLGEVPAA 10 3 47 A02/A03
EVFEGREDSI 10 3 229 A02
SILGDPKKLL 10 3 237 A02
ILGDPKKLLT 10 3 238 A02
ALVETSYVKV 10 3 277 A02
LVETSYVKVL 10 3 278 A02
MVKISGGPHI 10 3 290 A02 n ►T3
LVLGTLEEV 9 1 38 2.1 <0.0006 0.032 0 0 0.0003 CΛ
NO OO
KVADLVGFLL 10 1 105 0.0005 0.041 0.0039 0.0030 0.0070 ©
CΛ
© Ul
NO
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
LVFGIELMEV 10 3 153 2.1 0.17 j£
I LWQPI V 9 3 <0.0007 1.4 0.0048 0.0048 0 *o
EVDPIGHLY 9 3 3.7 0.0022
KMVELVHFL 9 2 <0.0007 0.13 0.0007 0 0.0043
KMVELVHFLL 10 2 105 <0.0008 0.071 0.0004 0.0001 0.0008
LVFGIBLMBV 10 3 0.0030 0.065 0.0007 0 0
KVAELVHFL 9 3 105 2.1 0 0.073 0.011 0.0047 0.0005
CILESLFRA 9 1 92 2.1 0.0001 0.073 0 0.0002 0
VMIAMEGGHA 10 1 200 2.1 <0.00008 0.0023 0 0 0
MLESVIKNYK 10 1 0 0 0.034 0.0045 0
ETSYVKVLBY 10 1 0.075 0 0.0009 0.0004 0
KVLEYVIKV 9 1 new 279 2.1 <0.0005 0.095 0.022 0.015 0
FLHGPRALA 9 1 <0.0006 0.027 0.0015 0 0
ALREBEEGV 9 1 302 2.1 <0.0006 0.0056 0 0 0
ALAETSYVKV 10 1 271 <0.0007 0.017 0.0011 0.0029 0
YVIKVSARV 9 1 283 2.1 0.0005 0.018 0 0 0
RALAETSYV 9 1 270 2.1 <0.0006 0.014 0.0003 0.0005 0
ALAETSYVK 9 1 <0.0006 0.0002 0.17 0.39 0
VLGTLEEV 8 1 39 2.1 <0.0007 0.0088 0 0 0 n
SLQLVFGI 8 1 150 2.1 <0.0007 0.0094 0 0.0001 0
ILESLFRA 8 1 93 2.1 <0.0004 0.0017 0.0003 0 0.0001 00
FLLLKYRA 8 1 112 2.1 0.0036 0.0007 ©-
0.0003 0.0001 0 CΛ s
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
NO
GLVCVQAA 8 24 2.1 0.0016 0.0008 0.0008 0 0 NO
CΛ
VLVTCLGL 8 170 2.1 <0.0007 0.0010 0.0001 0 0 NO CΛ
KVADLVGFL 9 105 2.1 <0.0008 0.0091 0.0013 0.0005 0
YVLVTCLGL 9 169 2.1
IMPKTGFLI 9 188 2.1 <0.0008 0.0035 0 0 3.2
GLLGDNQIM 9 A2.1 <0.0008 0.0054 0 0 0.0002
GLVCVQAAT 9 24 2.1 0.0030 0.0007 0.0026 0 0.0001
VADLVGFLL 9 106 2.1 0.032 0.0011 0.0054 0.0008 0.0007
YLEYGRCRTV 10 248 2.1 0.0008 0.0097 0.0001 0 0
SLQLVFGIDV 10 150 2.1 0.0028 0.0047 0.0013 0.0001 0.0001 o 00
IMPKTGFLII 10 188 2.1 <0.0008 0.0007 0 0 0.050
ALGLVCVQAA 10 22 A2.1 0.0011 0.0002 0.0003 0 0
EIHEBLSVMEV 11 213 A2.1 0.0007 0.013 0.0001 0.0001 0
FLIIVLVMIAM 11 A2.1 0.023 0.0031 0.016 0.0014 0.0011
VIPHAMSSCGV 11 257 2.1 <0.0009 1.4 0 0 0
CILESCFRAVI 11 A2.1 0.079 0.0017 0.058 0.0005 0.0008
QIMPKTGFLII 11 187 2.1 <0.0009 0.0003 0 0 0.0030
GFLLLKYRA 9 0.0004 0.0002
CFPBIFGKA 9 0 0 n
FFFPSLREA 9 0 0 H
CΛ
FFPSLREAA 9 0 0 NO 00
©
RSLHCKPEEA 10 0.0001 0.0008 CΛ
© Ul
NO
Mage
Sequence AA Strain Mol. Pos. Motif Al A2.1 A3.2 All A24
0 0 NO
EFLWGPRALA 10 1 NO
≤ CΛ:
RFFFPSLREA 10 1 0.0004 0 NO CΛ
FFFPSLREAA 10 1 0 0
o NO
O H
CΛ NO 00
© CΛ
O Ul
NO
Sequence Antigen Strain Molecule Position Motif Al A2 Λ3 A l l A24 Mux.
Binding Binding Binding Binding Binding Binding
ALF GF GAA HIV MN gpI6U 518 A02 0 4950 j 0 4950 NO NO
MLQLTVWGI HIV N gplβO 566 A02 0 2450 0 2 150 CΛ
NO IV 0 i%3 0 1961 CΛ RVIEVLQRA H MN gplβO 829 A 2
KLTPLCVTL HIV MN gpl60 12(1 A02 (1 1600 0 1600
LLIAARIVEL HIV MN gpl60 776 A02 0.1550 0 1550
SLLNATDIAV HIV MN gpl60 814 A02 O.Ϊ05U 0 1050
ALFLGFLGA HIV MN gplβO 518 A02 0.0945 0 0945 HMLQLTV GI HIV MN gpl60 565 A02 0.0677 0 0677 NATDIAV HIV MN gplβO 815 A02 0 (1607 00607
ALLYKLDIV IIIV MN g l60 Ϊ 79 A02 " (1.0362 0 0362 WLWYIKIFI HIV MN gpI60 679 ~A02 0.0155 0 0 55
TIIVHLNESV HIV MN gplβO 288 A02 0.0350 00150
LLQYWSQEL HIV MN gpl60 800 A02 0.0265 0 0265 IMIVGGLVGL HIV MN gpl60 687 A02 0.0252 00252
LLYKLDIVSI HIV MN gpl60 180 A02 0.0245 0 0245
FLAI IWVDL HIV MN gpl60 753 A02 0.0233 00231
TLQCKIKQII HIV MN gp!60 415 A02 0.i)20() — 00200
GLVGLRIVFA HIV MN g I60 692 A02 0.0195 0 0195
FLGAAGSTM HIV MN gplβO 523 A02 "o.ϊϊii o 0 0190
I ISLWDQSL HIV MN gplβO 107 A02 00179 00179
TVWGIKQLQA HIV MN g l 0 570 A02 0.0150 0 0150
LLGRRGWEV HIV MN
• gpl60 785 A02 Iiii i 42 00142
AVLSIVNRV HIV MN g i60 701 A02 0.0132 0.0132 o
CΛ NO OO
Co oΛ
Ul
NO
Sequence Antigen Strain Molecule Position Motif Al A2 A3 All A24 Mux.
Binding Binding Binding Binding Binding Binding
FIMIVGGLV HIV MN gplόO 686 A02 0.0131 001 1
LLNATDIAVA HIV MN g lβO 815 A02 0.0117 0(1117 NO NO FLYGALLLA PLP Human 80 A02 " i .906 1.9000
CΛ
SLLTFMIAA PLP Human 253 A02 0.5300 NO
— _ _ 0.5300 CΛ
4^
FMIAATYNFAV PLP Human 257 A02 0.495(1 (14950
- - -
RMYGVLPWI PLP Human 205 A02 (11650 0.1650
IAATYNFAV PLP Human 259 A02 0054(1 00540
GLLECCARCLV PLP Human 2 A02 0.1)515 00 15
YALTWWLL PLP Human 157 A02 0.(14 i 5 00 J5
ALTWWLLV PLP Human 158 A02 0.0.390 00390
FLYGALLL PLP Human 80 A02 0.0345 0(13 5
SLCADARMYGV PLP Human 199 A02 (1.0140 0.0140
LLVFACSAV PLP
Human 164 A02 αoiiπ 00107
"o0
CΛ NO 00
© o CΛ
Ul
NO
112 Table 10
AA SEQUENCE SOURCE
9 YIFATCLGL MAGE 3 109
9 IMPKTGFLI MAGE 1 188
10 IMPKTGFLII MAGE 1 188
15 ' MLGSWGNWQYFFPV MAGE 3 POL 75
9 VMPKTGLLI MAGE 2 188
9 IMPKAGLLI MAGE 3 188
10 10 IMPKAGLLII MAGE 3 188
9 RLWHYPCTV HCV Env2 614
9 RLWHYPCTI HCV Env2 614
9 FLLLADAR1 HCV Env2
9 GVWPLLLLL HCV Env2 792
15 9 GMWPLLLLL HCV Env2 792
9 YLNTPGLPV HCV NS3/NS4 1542
9 YMNTPGLPV HCV NS3/NS4 1542
9 VILDSFDPL HCV NS5 2251
9 ILMTHFFSI HCV NS5 2843
20 9 ILMTHFFSV HCV NS5 2843
9 LMAVVLASL gplOO 606
9 SLSLGFLFL PAP 13
10 YMIMVKCWMI c-ErbB2 952
10 GLHGQDLFGI PAP 196
25 9 AILSVSSFL P. falcipamm CSP 6
9 GLIMVLSFL P. falcipamm CSP 425
9 VLLGGVGLV P. falcipamm EXP-1 91
9 GLLGNVSTV P. falcipamm EXP-1 83
9 LLGNVSTVL P. falcipamm EXP-1 84
30 9 VLAGLLGNV P. falcipamm EXP-1 80
113
AA SEQUENCE SOURCE
9 KILSVFFLA P. falcipamm EXP-1 2
9 FLIFFDLFL P. falcipamm TRAP 14
9 LIFFDLFLV P. falcipamm TRAP 15
9 FMKAVCVEV P. falcipamm TRAP 230
9 LLMDCSGSI P. falcipamm TRAP 51
10 ILSVSSFLFV P. falcipamm CSP 7
10 VLLGGVGLVL P. falcipamm EXP-1 91
10 GLLGNVSTVL P. falcipamm EXP-1 83
10 FLIFFDLFLV P. falcipamm TRAP 14
10 10 GLALLACAGL P. falcipamm TRAP 507
9 KIWEELSML MAGE2 220
9 TLMSAMTNL Prost.Ca PAP 112
9 LLLARAASL Prost.Ca PAP 6
9 ALDVYNGLL Prost.Ca PAP 299
15 9 VTWIGAAPL PSA 8
10 ALIETSYVKV MAGE2 277
10 SLSLGFLFLL Prost.Ca PAP 13
10 RTLMSAMTNL PAP 111
10 FLPSDFFPSV(CONH2) HBc 18-27
20 10 FLPSDFFPSV-NH2 HBc 18-27
9 ILGFVFTLT-NH2 Flu Matrix 59-67
10 KGILGFVFTL-NH2 Flu Matrix 57-66
11 FLPSDFFPSVR HBc 18-28
9 FLPSDFFPS HBc 18-26
25 9 GILGKVFTL Flu Matrix 58-66 analog
9 FLSKQYLNL HBV polymerase
9 KLQCVPLHV PSA 166-174 P/D
114
AA SEQUENCE SOURCE
9 KLQCVPLHV PSA 166-174 P/D
9 KLQCVPLHV PSA 166-174 P/D
9 KLYEIVAKV A2 1 consensus
9 KLAEYVAKV A2 1 consensus
9 KLAEΓVYKV A2 1 consensus
9 TLTSCNTSV HIV gp 120 env RE trans 197
9 ALMEKIYQV A2 1 consensus peptide
9 ALSEKIYQV A2 1 consensus peptide
9 FLMSYFPSV 941 01 9-mer analog
10 9 FLPSYFPSV 941 01 9-mer analog
10 FLMSDYFPSV 941 01 M2 analog
9 FLYCYFALV Chiron consensus
9 FMYCYFALV Chiron consensus
10 SLVGFGILCV Chiron consensus
15 10 SLMGCGLFWV Chiron consensus
8 GLLGPLLV HBVadr-ENV
9 AMAKAAAAI A2 1 poly-A
10 MMWYWGPSLY HBV
9 FLPSYFPSA analog of 994 02 chiron comb
20 9 FAPSYFPSV analog of 994 02 chiron comb
9 FLPSYFPSS analog of 994 02 chiron comb
9 FSPSYFPSV analog of 994 02 chiron comb
9 IMPKTGFLI MAGE-1
9 VADLVGFLL MAGE-1
25 11 EΓWEELSVMEV MAGE-1
11 FLIIVLVMIAM MAGE-1
11 VIPHAMSSCGV MAGE-1
11 CILESCFRAVI MAGE-1
AA SEQUENCE SOURCE
9 YIFATCLGL MAGE3
11 KMVELWHFLLL MAGE2 112-122
11 HLFIYATCLGL MAGE3 174-184
9 GLQDCTMLV HCV NS5 2727-2735
8 TLGIVSPI HPV, analog of 1088.01
8 TLGΓVXPI HPV, analog of 1088.01
10 FLLAQFTSAI HBV POL 513
11 VLLDYQGMLPV HBV env
11 CILLLCLIFLL HBV env
10 9 FLGGSPVCL HBV env
11 TVIEYLVSFGV HBV core 114-124
1 1 TVLEYLVSFGV HBV core 114-124
10 FLLAQFTSAI HBV pol
9 GLYSSTVPI HBV pol
15 9 GLYSSTAPI HBV pol
9 GLDVLTAKV HIV form VIN.
9 RILGAVAKV HIV form VIN.
9 LLFGYPVYV HTLV, tax 11-19
9 ALFGYPVYV tax 11-19, SAAS
20 9 LLFGAPVYV tax 11-19, SAAS
9 LLFGYAVYV tax 11-19, SAAS
9 LLFGYPVAV tax 11-19, SAAS
9 AAGIGILTV MARTI 27-35
9 GILTVILGV MARTI 31-39
25 9 ILTVILGVL MARTI 32-40
9 VILGVLLLI MARTI 35-43
9 ALMDKSLHV MARTI 56-64
10 TVILGVLLL1 MARTI
10 LLDGTATLRL MARTI
30 10 ILSVSSFLFV Plas. falcip. CSA-A 7-16
9 GLIMVLSFL Plas. falcip. CSA-A 401-409
116
AA SEQUENCE SOURCE
9 IMVLSFLFL Plas. falcip. CSA-A 403-411
10 FLIFFDLFLV Plas. falcip. TRAP-A 14-23
9 FMKAVCVEV Plas. falcip. TRAP-A 200-207
9 IMPGQEAGL gplOO
9 GLGQVPLΓV gplOO
9 LMAVVLASL gplOO
9 RLMKQDFSV gplOO
9 HLAVIGALL gplOO
9 LLAVGATKV gplOO
10 9 MLGTHTMEV gplOO
10 LLDGTATLRL gplOO
10 VLYRYGSFSV gpioo
10 VLPSPACQLV gpioo
10 SLADTNSLAV gplOO
15 10 VLMAVVLASL gplOO
10 LMAVVLASLI gplOO
10 RLDCWRGGQV gplOO
10 AMLGTHTMEV gplOO
10 ALDGGNKHFL gplOO
20 9 YLEPGPVTA gplOO
10 LLNATALAVA
11 SLLNATAIAVA
9 KTWGQYWQV gplOO
9 ΓΓDQVPFSV gplOO
25 9 YLEPGPVTA gplOO
10 LLDGTATLRL gplOO
10 VLYRYGSFSV gplOO
10 ALDGGNKHFL gpioo
9 GILTVILGV MARTI 31-39
30 9 YMNGTMSQV Human Tyrosinase
9 MLLAVLYBL Human Tyrosinase
9 LLWSFQTSA Human Tyrosinase
117
AA SEQUENCE SOURCE
9 YLTLAKHTI Human Tyrosinase
9 FLPWHRLFL Human Tyrosinase
9 FLLRWEQEI Human Tyrosinase
9 RIWSWLLGA Human Tyrosinase
9 LLGAAMVGA Human Tyrosinase
9 AMVGAVLTA Human Tyrosinase
9 VLTALLAGL Human Tyrosinase
9 ALLAGLVSL Human Tyrosinase
9 LLAGLVSLL Human Tyrosinase
10 10 BLLWSFQTSA Human Tyrosinase
10 WMHYYVSMDA Human Tyrosinase
10 FLPWHRLFLL Human Tyrosinase
10 WLLGAAMVGA Human Tyrosinase
10 AMVGAVLTAL Human Tyrosinase
15 10 VLTALLAGLV Human Tyrosinase
10 TALLAGLVSL Human Tyrosinase
10 ALLAGLVSLL Human Tyrosinase
9 NLTDALLQV P. falcipamm SSP2 132
9 SAWENVKNV P. falcipamm SSP2 218
20 10 FLIFFDLFLV P. falcipamm SSP2 14
9 NLNDNAIHL P. falcipamm SSP2 80
10 YLLMDCSGSI P. falcipamm SSP2 51
25
118 Table 11
AA SEQUENCE SOURCE
9 ALYWFRTGI HPVόb/11 El 319
LLDGNPMSI HPV6b/ll El 540
9 NAWGMVLLV HPV6b/ll El 270
9 SLYAHIQWL HPV6b/ll El 260
9 TLIKCPPLL HPV6b/ll El 556
10 9 GIYDALFDI PSMAg 707
9 YLSGANLNL CEA 605
9 VLYGPDTPI CEA 589
9 IMIGVLVGV CEA 691
9 LLTFWNPPT CEA 24
15 9 KLTEMVQWA HPV6b/ll El
357
9 YMDTYMRNL HPV6b/ll El
532
10 NLLDGNPMSI HPV6b/ll El 539
10 SLYAHIQWLT HPV6b/llEl 260
10 TLKCPPLLV HPV6b/ll El 556
20 10 MVFELANSrV PSMAg 583
10 YLWWVNNQSL CEA 176
10 YLWWVNNQSL CEA 354
10 YLWWVNGQSL CEA 532
10 GIMIGVLVGV CEA 690
25 10 VLYGPDAPTI CEA 233
10 KLIEPLSLYA HPV6b/ll El 254
10 WLCAGALVLA PSMAg 20
10 IMIGVLVGVA CEA 691
119
AA SEQUENCE SOURCE
9 YLYQLSPPI HTLV-I tax 155
9 LLFEEYTNI HTLV-I tax 307
9 QLGAFLTNV HTLV-I tax 178
9 TLTAWQNGL HTLV-I tax 226
9 ALQFLIPRL HTLV-I tax 67
9 TLGQHLPTL HTLV-1 tax 123
9 FAFKDLFVV HPV 18 E6
47
9 RLLQLLFRA GCDFP-15 2
9 CMWKTYLI GCDFP-15 65
10 9 LLLVLCLQL GCDFP-15 15
9 ILYAHIQCL HPV18 El 266
9 SLACSWGMV HPV16 El 266
9 CLYLHIQSL HPV 16 El 259
9 YLVSPLSDI HPV16 El 90
15 9 VMFLRYQGV HPV16 El 443
9 KLLSKLLCV HPV16 El 292
9 ALDGNPISI HPV18 El 546
9 AVFKDTYGL HPV18 El 216
9 LLTTNIHPA HPV18 El 570
20 9 LLQQYCLYL HPV 16 El
AA SEQUENCE SOURCE
9 AMLAKFKEL HPV16 El 206
9 ALDGNLVSM HPV 16 El 539
9 FLGALKSFL HPV18 El 463
9 FIHFIQGAV HPV18 El 497
10 TLLLVLCLQL GCDFP-15
14
10 LLFRASPATL GCDFP-15 6
10 SLMKFLQGSV HPV16 El 489
10 SLACSWGMVV HPV16 El 266
10 FLQGSVICFV HPV16 El 493
10 10 FIQGAVISFV HPV18 El 500
10 KLLCVSPMCM HPV16 El 296
10 FILYAHIQCL HPV18 El 265
10 FVNSTSHFWL HPV18 El 508
10 ILLTTNIHPA HPV18 El 569
15 10 TLLQQYCLYL HPV16 El 253
9 GLLGWSPQA HBV ENV 62
9 GLACHQLCA HER2/neu
9 ILDEAYVMA HER2/neu
9 SIISAVVGI HER2/neu
20 9 VVLGWFGI HER2/neu
9 YMIMVKCWM HER2/neu
10 ALCRWGLLLA HER2/neu
10 QLFEDNYALA HER2/neu
121
AA SEQUENCE SOURCE
9 HMWNFISG1 HCV consensus
9 VIYQYMDDL HIV POL 358
9 SLYNTVATL HIV GAG 77
10 TVWGKQLQA HIV ENV
735
9 LLLEAGALV MSH 99
9 VLETAVGLL MSH 92
9 CLALSDLLV MSH 79
9 FLSLGLVSL MSH 45
9 SLVENALW MSH 52
10 9 AIIDPLIYA MSH 291
9 FLCWGPFFL MSH 251
9 FLALIICNA MSH 283
9 TILLGIFFL MSH 244
9 RLLGSLNST MSH 9
15 9 SLYNTVATL HIV pl7/5B 77-8
9 VIYQYMDDL HIV RT/50A 346-
9 ILKEPVHGV HIV RT/IV9 476-
122 Table 12
PEPTIDE NO PEPTIDE LENGTH SEQUENCE
1237 01 9 FLWGPQALV
1237 02 9 FLWGPNALV
1237 03 9 FLWGPHALV
1237 04 9 FLWGPKALV
1237 05 9 FLWGPFALV
26 0158 9 AVIGALLAV
26 0172 9 LLHLAVIGA
26 0186 9 SLADTNSLA
26 0192 9 VMGTTLAEM
26 0240 9 LLAVLYCLL
26 0383 10 FLRNQPLTFA
26 0390 10 HLAVIGALLA
26 0395 10 LAVIGALLAV
26 0418 10 TLAEMSTPEA
26 0423 10 YLAEADLSYT
26 0497 10 MLLAVLYCLL
1183 10 10 VLYRYGSFSV
27 0007 9 ILSSLGLPV
27 0012 9 LLFLGWFL
27 0019 9 GLYGAQYDV
27 0022 9 FVVALIPLV
27 0023 9 GLMTAVYLV
27 0027 9 ALVLLMLPV
27 0028 9 ILLSIARVV
27 0029 9 SLYFGGICV
27 0030 9 QLIPCMDVV
27 0031 9 VLQQSTYQL
27 0032 9 AIHNVVHAI
27 0034 9 GLHGVGVSV
27 0035 9 GLVDFVKHI
27 0036 9 LLFRFMRPL
27 0038 9 LMLPGMNGI
27 0043 9 TVLRFVPPL
27 0044 9 MLGNAPSVV
27 0050 9 YLDLALMSV
27 0064 9 RMPEAAPPV
123
PEPTIDE NO PEPTIDE LENGTH SEQUENCE
27 0082 9 FLLPDAQSI
27 0083 9 MTYAAPLFV
27 0088 9 LLPLGYPFV
27 0089 9 GLYYLTTEV
27 0090 9 MALLRLPLV
27 0091 9 RLPLVLPAV
27 0093 9 RMFAANLGV
27 0095 9 RLLDDTPEV
27 0096 9 YLYVHSPAL
27 0100 9 GLYLSQIAV
27 0101 9 YLSQIAVLL
27 0102 9 SLAGFVRML
27 0137 10 ATYDKGILTV
27 0146 10 KIFMLVTAVV
27 0151 10 FLLADERVRV
27 0153 10 MLATDLSLRV
27 0154 10 RLQPQVGWEV
27 0161 10 FLMPVEDVF1
27 0165 10 RMSRVTTFTV
27 0168 10 LALVLLMLPV
27 0169 10 ALVLLMLPVV
27 0170 10 GΓVSGILLSI
27 0171 10 SLYFGGICVI
27 0173 10 QLIPCMDVVL
27 0181 10 LLFRFMRPLI
27 0183 10 VLLEDGGVEV
27 0184 10 AMPAYNWMTV
27 0186 10 GLAGTVLRFV
27 0188 10 VLIAFGRFPI
27 0189 10 FLTCDANLAV
27 0197 10 AIAWGAWGEV
27 0204 10 LLLETSWEA1
27 0217 10 RMPEAAPPVA
27 0223 10 WMAETTLGRV
27 0226 10 AMALLRLPLV
27 0229 10 FMSLAOFVRM
27 0266 11 SLLTEVETYVL
124
PEPTIDE NO PEPTIDE LENGTH SEQUENCE
27 0268 GILGFVFTLTV
27 0269 VLDVGDAYFSV
27 0271 KIWEELSMLEV
27 0272 STLVEVTLGEV
27 0273 GLAPPQHLIRV
27 0274 HLIRVEGNLRV
27 0005 9 YLLALRYLA
27 0013 9 GLYRQWALA
27 0017 9 LLWQDPVPA
27 0040 9 ALLSDWLPA
27 0045 9 WLLIDTSNA
27 0046 9 MLASTLTDA
27 0081 9 YLSEGDMAA
27 0094 9 LLACAVIHA
27 0144 10 LLCCSGVATA
27 0191 10 LLATVFKLTA
27 0192 10 KLTADGVLTA
27 0195 10 GLGGLGLFFA
28 0064 8 TLGIVXPI
28 0065 8 ALGTTXYA
28 0293 9 FLLTRILTV
28 0294 9 ALMPLYACV
28 0295 9 LLAQFTSAV
28 0296 9 LLPFVQWFV
28 0297 9 FLLAQFTSV
28 0298 9 KLHLYSHPV
28 0299 9 KLFLYSHPI
28 0300 9 LLSSNLSWV
28 0301 9 FLLSLGIHV
28 0302 9 MMWYWGPSV
28 0303 9 VLQAGFFLV
28 0304 9 PLLPIFFCV
28 0305 9 FLLPIFFCL
28 0306 9 VLLDYQGMV
28 0307 9 YMDDVVLGV
28 0308 9 YMFDVVLGA
28 0309 9 GLLGWSPOV
125
PEPTIDE NO PEPTIDE LENGTH SEQUENCE
28 0342 9 YMIMVKXWM
28 0343 9 YIFATXLGL
28 0345 9 SLHXKPEEA
28 0346 9 ALGLVXVQA
28 0348 9 LLMDXSGSI
28 0349 9 FAFRDLXIV
28 0352 9 GTLGΓVXPI
28 0353 9 TLGΓVXPIX
28 0354 9 LLWFHISXL
28 0355 9 KLTPLXVTL
28 0356 9 ALVEIXTEM
28 0357 9 LTFGWXFKL
28 0359 9 KLQXVDLHV
28 0360 9 FMKAVXVEV
28 0361 9 LLQQYXLYL
28 0362 9 XLYLHIQSL
28 0363 9 SLAXSWGMV
28 0364 9 ILYAHIQXL
28 0365 9 KLLSKLLXV
28 0366 9 PLLPIFFXL
28 0367 9 TLIKXPPLL
28 0368 9 ALMPLYAXI
28 0370 9 XILESLFRA
28 0609 10 FLLAQFTSAV
28 0610 10 YLHTLWKAGV
28 0611 10 YLFTLWKAGI
28 0612 10 YLLTLWKAGI
28 0613 10 LLFYQGMLPV
28 0614 10 LLLYQGMLPV
28 0615 10 LLVLQAGFFV
28 0616 10 ILLLCLIFLV
28 0650 10 ALXRWGLLL
28 0651 10 KLPDLXTEL
28 0652 10 HLYQGXQVV
28 0653 10 XILESLFRA
28 0654 10 KLQXVDLHV
28 0655 10 YIFATXLGL
126
PEPTIDE NO PEPTIDE LENGTH SEQUENCE
Fill 01 9 SLYNTVATL
Fill 02 9 ALYNTVATL
Fill 04 9 SLANTVATL
Fill 06 9 SLFNAVATL
Fill 07 9 SLFNLLATL
Fill 10 9 SLFNTIAVL
Fill 11 9 SLFNAVAVL
Fill 09 9 SLFNTIWL
Fill 12 9 SLFNAIAVL
Fill 13 9 SLFNTVAVL
Fill 14 9 SLFNTVCVI
Fill 15 9 SLHNTVATL
Fill 17 9 SLHNTVAVL
Fill 18 9 SLYATVATL
Fill 19 9 SLYNAVATL
Fill 21 9 SLYNTAATL
Fill 22 9 SLYNTIAVL
Fill 23 9 SLYNTSATL
Fill 25 9 SLYNTVAVL
Fill 26 9 SLYNTVATA
Fill 27 9 SLYNAIATL
Fill 28 9 SLYNLVAVL
Fill 29 9 SLFNLLAVL
FI 1132 9 SLFNTVVTL
Fill 34 9 SLYNTVAAL
1039031 9 MMWYWGPSL
121140 10 SLLNATAIAV
10 TIHDIILECV
9 FAFRDLCIV
9 GTLGIVCPI
A SEQUENCE SOURCE A
9 IPQSLDSWW HBV ENV 191
9 IPIPSSWAF HBV ENV 313
9 TPARVTGGV HBV POL
365
9 LPIFFCLWV HBV ENV 379
9 HPAAMPHLL HBV POL
440
10 9 FPHCLAFSY HBV POL
541
9 DPSRGRLGL HBV POL
789
9 QPRGRRQPI HCV Core 57
9 SPRGSRPSW HCV Core 99
9 DPRRRSRNL HCV Core 111
15 9 LPGCSFSIF HCV Core 168
9 YPCTVNFTI HCV E2 622
9 LPALSTGLI HCV E2 681
9 HPNIEEVAL HCV NS3 1358
9 SPGALVVGV HCV NS4 1887
128
A SEQUENCE SOURCE
A
9 SPGQRVEFL HCV NS5 2615
9 APTLWARMI HCV NS5 2835
9 FPRIWLHJL HIV VPR 34
9 SPTRRELQV HIV POL 37
9 FPVRPQVPL HIV NEF 84
9 RPQVPLRPM HIV NEF 87
9 KPCVKLTPL HIV ENV 123
9 SPRTLNAWV HIV GAG
153
9 FPISPIETV HIV POL 171
10 9 SPAIFQSSM HIV POL 327
9 NPDIVIYQY HIV POL 346
9 GPGHKARVL HIV GAG
360
9 LPEKDSWTV HIV POL 417
9 YPLASLRSL HIV GAG
507
15 9 VPRRKAKII HIV POL 991
9 TPTLHEYML HPV 16 E7 5
9 KPLNPAEKL HPV 18 E6 110
9 NPAEKLRHL HPV 18 E6
113
9 VPISHLYIL MAGE2 170
20
9 MPKTGLLII MAGE2 196
129
A SEQUENCE SOURCE A
9 DPACYEFLW MAGE2 265
9 EPHISYPPL MAGE2 296
9 YPPLHERAL MAGE2 301
9 LPTTMNYPL MAGE3 71
9 DPIGHLYIF MAGE3 170
9 MPKAGLLII MAGE3 196
9 GPHISYPPL MAGE3 296
9 HPSDGKCNL P. falciparum S
9 RPRGDNFAV P. falciparum S
10 9 QPRPRGDNF P. falciparum S
9 LPNDKSDRY P. falciparum S
10 LPLDKGIKPY HBV POL
123
10 TPARVTGGVF HBV POL 365
10 FPHCLAFSYM HBV POL
541
15 10 LPRRGPRLGV HCV Core 37
10 APLGGAARAL HCV Core 142
10 LPGCSFSIFL HCV Core 168
10 VPASQVCGPV HCV E2 497
10 YPCTVNFTIF HCV E2 622
130
A SEQUENCE SOURCE
A
10 SPLLLSTTEW HCV E2 663
10 RPSGMFDSSV HCV NS3 1506
10 LPVCQDHLEF HCV NS3
1547
10 KPTLHGPTPL HCV NS3 1614
10 TPLLYRLGAV HCV NS3 1621
10 NPAIASLMAF HCV NS4 1783
10 LPAILSPGAL HCV NS4 1882
10 SPGALVVGVV HCV NS4 1887
10 APTLWARMIL HCV NS5 2835
10 10 IPVGEIYKRW HIV GAG
261
10 YPLASLRSLF HIV GAG
507
10 APTKAKRRVV HIV ENV 547
10 VPISHLYILV MAGE2 170
10 MPKTGLLIIV MAGE2 196
15 10 HPRKLLMQDL MAGE2 241
10 LPTTMNYPLW MAGE3 71
10 MPKAGLLIIV MAGE3 196
131
A SEQUENCE SOURCE A
10 IPYSPLSPKV P. falciparum S
10 TPYAGEPAPF P. falciparum S
9 FPDHQLDPA HBV ENV 14
9 YPALMPLYA HBV POL
640
9 LPVCAFSSA HBV X 58
9 APLGGAARA HCV 142
9 DPTTPLARA HCV 2806
9 FPYLVAYQA HCV 1582
9 LPAILSPGA HCV 1882
10 9 NPAIASLMA HCV 1783
9 TPIDTTIMA HCV 2551
9 TPLLYRLGA HCV 1621
9 WPLLLLLLA HCV 793
9 NPYNTPVFA HIV POL 225
15 9 APLLLARAA PAP 4
9 HPQWVLTAA PSA 52
10 IPIPSSWAFA HBV ENV 313
10 TPPAYRPPNA HBV NUC 128
10 APFTQCGYPA HBV POL
633
20 10 LPIHTAELLA HBV POL
712
10 GPCALRFTSA HBV X 67
132
A SEQUENCE SOURCE A
10 DPTTPLARAA HCV 2806
10 IPQAVVDMVA HCV 339
10 LPCSFTTLPA HCV 674
10 QPEKGGRKPA HCV 2567
10 VPHPNIEEVA HCV 1356
10 IPAETGQETA HIV POL 820
10 LPQGWKGSPA HIV POL 320
10 FPDLESEFQA MAGE2/3 98
10 DPIGHLYIFA MAGE3 170
10 9 EPLSLYAHI HPV 6b/ 11 El
2
9 PPLLVTSNI HPV 6b/ 11 El
5
9 SPRLDAIKL HPV 6b/ 11 El 1
9 TPKKNCIAI HPV 6b/ 11 El
4
9 FPFDRNGNA HPV 6b/ 11 El
5
15 10 CPPLLVTSNI HPV 6b/ 11 El
5
10 FPFDRNGNAV HPV 6b/ 11 El 5
8 GPLLVLQA HBV ENV 173
8 IPIPSSWA HBV ENV
313
133
A SEQUENCE SOURCE A
8 VPFVQWFV HBV ENV 340
8 LPIFFCLW HBV ENV 379
8 RPPNAPIL HBV NUC
133
8 MPLSYQHF HBV POL 1
8 HPAAMPHL HBV POL
429
8 SPFLLAQF HBV POL
511
8 YPALMPLY HBV POL
640
8 SPTYKAFL HBV POL
659
8 VPSALNPA HBV POL
769
10 8 HPvhAGPI HIV con. GAG
8 GPGvRyPL HIV con. NEF
8 SPIETVPV HIV con. POL
8 NPYNTPVF HIV con. POL
8 LPIQKETW HIV con.
POL
134
A SEQUENCE SOURCE A
8 VPRRKaKi HIV con. POL
8 VpLQLPPl HIV con. REV
8 VPLAMKLI P. falciparum
8 LPYGRTNL P. falciparum
8 RPRGDNFA P. falciparum
8 IPQQEPNI P. falciparum
8 TPFAGEPA P. falciparum
9 SPINTIAEA HPV 6b El 93
9 SPISNVANA HPV 11 El 93
10 9 SPRLDAIKL HPV 6b/ 11 El 1
9 EPLSLYAHI HPV 6b/ 11 El 2
9 EPPKIQSGV HPV 6b/ 11 El
3
9 IPFLTKFKL HPV 6b El 455
9 TPKKNCIAI HPV 6b/ 11 El 4
15 9 QPLTDAKVA HPV 11 El
512
9 PPLLVTSNI HPV 6b/ 11 El
5
135
A SEQUENCE SOURCE A
9 FPFDRNGNA HPV 6b/ 11 El
5
9 APLILSRIV PSA 14
9 HPEDTGQVF PSA 78
9 HPLYDMSLL PSA 94
9 HPQKVTKFM PSA 184
9 GPLVCNGVL PSA 211
9 RPSLYTKVV PSA 235
9 FPPEGVSIW PAP 124
9 NPILLWQPI PAP 133
10 9 LPFRNCPRF PAP 156
9 IPSYKKLIM PAP 277
9 LPPYASCHL PAP 307
9 SPSCPLERF PAP 348
9 CPLERFAEL PAP 351
15 9 GPTLIGANA gplOO 74
9 LPDGQVIWV gplOO 97
9 VPLAHSSSA gplOO 198
9 QPLTFALQL gplOO 236
9 DPSGYLAEA gplOO 246
20 9 EPGPVTAQV gplOO 282
9 MPTAESTGM gplOO 366
9 TPAEVSIVV gplOO 401
9 LPKEACMEI gplOO 520
9 LPSPACQLV gplOO 545
25 9 VPLIVGILL gplOO 596
9 LPHSSSHWL gplOO 630
136
A SEQUENCE SOURCE A
9 CPIGENSPL gplOO 647
9 SPLLSGQQV gplOO 653
9 MPREDAHFI MARTI 1
9 APLGPQFPF Tyrosinase 6
9 IPIGTYGQM Tyrosinase 1
9 TPMFNDINI Tyrosinase 1
9 LPWHRLFLL Tyrosinase 2
9 IPYWDWRDA Tyrosinase 2
9 SPASFFSSW Tyrosinase 2
10 9 LPSSADVEF Tyrosinase 3
9 SPLTGIADA Tyrosinase 3
9 DPIFLLHHA Tyrosinase 3
9 IPLYRNGDF Tyrosinase 4
9 YPELPKPSI CEA 141
15 9 LPVSPRLQL CEA 185
9 LPVSPRLQL CEA 363
9 NPPAQYSWL CEA 442
9 LPVSPRLQL CEA 541
9 IPQQHTQVL CEA 632
20 9 NPPAQYSWF CEA 264
9 LPSIPVHPI Prost.Ca PSM
9 IPVHPIGYY Prost.Ca PSM
9 RPFYRHVIY Prost.Ca PSM
9 TPKHNMKAF Prost.Ca PSM
25 9 FPGIYDALF Prost.Ca PSM
9 RPRWLCAGA Prost.Ca PSM
9 DPLTPGYPA Prost.Ca PSM
137
A SEQUENCE SOURCE A
9 RPRRTILFA Prost.Ca PSM
9 LPFDCRDYA Prost.Ca PSM
9 LPIHTAELL HBV POL
712
10 GPDAPTISPL CEA 236
10 IPQQHTQVLF CEA 632
10 QPIPVHTVPL Prost.Ca PAP
10 HPYKDFIATL Prost.Ca PAP
10 LPGCSPSCPL Prost.Ca PAP
10 LPSWATEDTM Prost.Ca PAP
10 10 VPLSEDQLLY Prost.Ca PAP
10 FPHPLYDMSL Prost.Ca PSA
10 RPGDDSSHDL Prost.Ca PSA
10 HPQKVTKFML Prost.Ca PSA
10 LPFDCRDYAV Prost.Ca PSM
15 10 YPNKTHPNYI Prost.Ca PSM
10 SPEFSGMPRI Prost.Ca PSM
10 RPRWLCAGAL Prost.Ca PSM
10 TPKHNMKAFL Prost.Ca PSM
10 RPFYRHVIYA Prost.Ca PSM
20 10 HPAAMPHLLV HBV POL
429
9 SPREGPLPA HER2/neu 1151
9 KPDLSYMPI HER2/neu 605
9 HPPPAFSPA HER2/neu 1208
138
A SEQUENCE SOURCE A
9 GPLPAARPA HER2/neu 1155
9 APQPHPPPA HER2/neu 1204
9 EPLTPSGAM HER2/neu 698
9 LPTHDPSPL HER2/neu 1101
9 DPLNNTTPV HER2/neu 121
9 SPLTSIISA HER2/neu 649
9 SPKANKEIL HER2/neu 760
9 LPTNASLSF HER2/neu 65
9 CPSGVKPDL HER2/neu 600
10 9 SPLAPSEGA HER2/neu 1073
9 MPNQAQMRI HER2/neu 706
9 LPAARPAGA HER2/neu 1157
9 LPQPPICTI HER2/neu 941
9 SPAFDNLYY HER2/neu 1214
139
A SEQUENCE SOURCE A
9 TPTAENPEY HER2/neu 1240
9 LPSETDGYV HER2/neu 1120
10 LPTNASLSFL HER2/neu 65
10 CPAEQRASPL HER2/neu 642
10 KPCARVCYGL HER2/neu 336
10 APQPHPPPAF HER2/neu 1204
10 SPGGLRELQL HER2/neu 133
10 SPLTSIISAV HER2/neu 649
10 MPNQAQMRIL HER2/neu 706
10 10 SPYVSRLLGI HER2/neu 779
10 HPPPAFSPAF HER2/neu 1208
10 SPREGPLPAA HER2/neu 1151
10 NPHQALLHTA HER2/neu 488
10 MPYGCLLDHV HER2/neu
801
140
A SEQUENCE SOURCE A
10 GPASPLDSTF HER2/neu 995
9 LPTTLFQPV HTLV-I tax
21
9 IPPSFLQAM HTLV-I tax
10
9 FPGFGQSLL HTLV-I tax
4
9 WPLLPHVIF HTLV-I tax 16
9 SPPITWPLL HTLV-I tax 16
9 VPYKRIEEL HTLV-I tax
18
9 RPQNLYTLW HTLV-I tax
13
9 CPKDGQPSL HTLV-I tax 26
10 9 RPNDEVTAV GCDFP-15
47
9 SPATLLLVL GCDFP-15 11
9 WPYLHNRLV HPV16 El
576
9 QPFILYAHI HPV18 El 263
9 SPRLKAICI HPV16 El 107
141
A SEQUENCE SOURCE A
9 SPLGERLEV HPV 18 El 97
9 SPRLQEISL HPV 18 El 110
9 RPIVQFLRY HPV 18 El
447
10 WPYLHNRLVV HPV 16 El
576
10 WPYLESRITV HPV 18 El
583
10 QPPKLRSSVA HPV18 El 315
10 EPPKLRSTAA HPV 16 El 308
9 DPSRGRLGL HBV POL
778
9 HPAAMPHLL HBV POL
429
10 9 IPIPSSWAF HBV ENV 313
10 TPARVTGGVF HBV POL
354
10 FPHCLAFSYM HBV POL
530
9 LPVCAFSSA HBV X 58
9 YPALMPLYA HBV POL 640
A SEQUENCE SOURCE A
9 HPQWVLTAA PSA 52
9 HPSDGKCNL Pf SSP2 206
9 RPRGDNFAV Pf SSP2 305
9 QPRPRGDNF Pf SSP2 303
10 TPYAGEPAPF Pf SSP2 539
9 GPHISYPPL MAGE3 296
9 YPPLHERAL MAGE2 301
9 VPISHLYIL MAGE2 170
9 EPHISYPPL MAGE2 296
10 9 LPTTMNYPL MAGE3 71
9 MPKAGLLII MAGE3 196
143 Table 14
PEPTIDE AA SEQUENCE
25.0129 9 LPPLERLTL
26.0445 10 EPGPVTAQVV
26.0448 10 LPRIFCSCPI
26.0449 10 LPSPACQLVL
26.0455 10 VPLAHSSSAF
26.0458 10 VPRNQDWLGV
10 26.0476 10 APPAYEKLSA
26.0478 10 MPREDAHFIY
26.0519 10 APAFLPWHRL
26.0522 10 GPNCTERRLL
26.0523 10 IPLYRNGDFF
15 26.0529 10 TPRLPSSADV
19.0101 9 TPAEVSIVV
26.0554 11 APFTQCGYPAL
26.0561 11 NPADDPSRGRL
26.0564 11 RPPNAPILSTL
20 26.0566 11 SPFLLAQFTSA
26.0567 11 SPHHTALRQAI