KR101961653B1 - SNP molecular marker for selecting cultivars of sweet potatoes and uses thereof - Google Patents
SNP molecular marker for selecting cultivars of sweet potatoes and uses thereof Download PDFInfo
- Publication number
- KR101961653B1 KR101961653B1 KR1020170148336A KR20170148336A KR101961653B1 KR 101961653 B1 KR101961653 B1 KR 101961653B1 KR 1020170148336 A KR1020170148336 A KR 1020170148336A KR 20170148336 A KR20170148336 A KR 20170148336A KR 101961653 B1 KR101961653 B1 KR 101961653B1
- Authority
- KR
- South Korea
- Prior art keywords
- snp
- primer set
- seq
- nos
- dna
- Prior art date
Links
Images
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
- C12Q1/6876—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes
- C12Q1/6888—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for detection or identification of organisms
- C12Q1/6895—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for detection or identification of organisms for plants, fungi or algae
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q2600/00—Oligonucleotides characterized by their use
- C12Q2600/13—Plant traits
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q2600/00—Oligonucleotides characterized by their use
- C12Q2600/156—Polymorphic or mutational markers
Landscapes
- Chemical & Material Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Analytical Chemistry (AREA)
- Engineering & Computer Science (AREA)
- Organic Chemistry (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Health & Medical Sciences (AREA)
- Biotechnology (AREA)
- Zoology (AREA)
- Wood Science & Technology (AREA)
- Immunology (AREA)
- Mycology (AREA)
- Microbiology (AREA)
- Molecular Biology (AREA)
- Botany (AREA)
- Biophysics (AREA)
- Physics & Mathematics (AREA)
- Biochemistry (AREA)
- Bioinformatics & Cheminformatics (AREA)
- General Engineering & Computer Science (AREA)
- General Health & Medical Sciences (AREA)
- Genetics & Genomics (AREA)
- Measuring Or Testing Involving Enzymes Or Micro-Organisms (AREA)
Abstract
Description
본 발명은 고구마 품종 판별용 SNP 분자 마커 및 이의 용도에 관한 것이다. 보다 상세하게는 고구마 품종 판별용 SNP 분자 마커 증폭용 프라이머 세트, 상기 프라이머 세트를 포함하는 키트 및 상기 프라이머 세트를 이용한 고구마 품종 판별 방법에 관한 것이다.The present invention relates to SNP molecular markers for the identification of sweet potato varieties and uses thereof. More particularly, the present invention relates to a primer set for SNP molecular marker amplification for the identification of sweet potato varieties, a kit comprising the primer set, and a method for identifying sweet potato varieties using the primer set.
육종에 이용되고 있는 대부분의 작물에서 해마다 많은 양의 유전자원이 수집되고 있으며, 수집된 유전자원의 종내 혹은 아종내 자원구분은 정확한 유전자원의 관리를 위하여 필요하며 출원되는 품종의 신속한 판별은 신품종 보호를 위하여 매우 중요하다. Most of the crops used for breeding are collecting a large amount of genetic resources every year. The classification of the genetic resources within or within the species is necessary for the management of accurate genetic resources. It is very important for.
또한, 국제 식물 신품종 보호동맹(International Union for the Protection of New Varieties of Plants, UPOV)에 우리나라가 가입하고 종자산업법이 발효되면서 육종가의 권리 보호 및 농가소득 증대를 위하여 과학적인 국내외 농산물 품종 구분 필요성이 대두 되었으며, 영양번식작물인 고구마의 경우에도 품종과 자원에 대한 변별력이 있는 식별시스템이 필요한 실정이다.In addition, as Korea joined the International Union for the Protection of New Varieties of Plants (UPOV) and the Seed Industry Act came into effect, the necessity of sorting scientific and domestic agricultural products was needed to protect breeder rights and increase farm income. In addition, even in the case of sweet potatoes, a nutritional breeding crop, an identification system with discriminative power of varieties and resources is needed.
현재 품종 구별은 전통적인 멘델의 법칙을 적용한 표현형 위주의 형태학적 형질 및 효소나 단백질 변이와 같은 생화학적 특성에 의해 이루어지고 있다. 그러나, 이와 같은 방식은 변이의 빈도가 낮고 전 작물에 대한 적용에 한계가 있으며, 생산자의 재배 조건에 따라 다른 결과를 초래할 수 있어서 정확한 품종 판별이 곤란하기 때문에 분자 수준에서의 품종 구별 방법이 활발하게 연구되고 있다. 예를 들면, 네덜란드에서는 장미 및 토마토에 대해 각 품종별 DNA 프로파일을 데이터베이스화하였고, 일본은 벼 등의 작물에 대한 품종 식별용 분자 마커를 개발하여 품종 보호에 활용하고 있다.Current breeds are distinguished by phenotypic traits based on traditional Mendelian laws and biochemical characteristics such as enzymes and protein mutations. However, such a method has a low frequency of mutation, has limited application to whole crops, and may result in different results depending on the cultivation conditions of the producer. Research. For example, in the Netherlands, DNA profiles for each variety of roses and tomatoes are databaseed, and in Japan, molecular markers for breed identification for rice and other crops have been developed and used to protect varieties.
분자표지는 DNA 염기서열의 차이를 대상으로 모든 조직에서 탐지할 수 있으며, 환경적인 영향과 유전자 간의 다면 발현에 의한 영향을 받지 않는다는 장점을 가지고 있다. 현재 주요 작물에 대한 게놈 유전자 지도 작성이 활발하게 진행되고 있고, 분자표지는 육종의 선발 과정에 실제로 활용되고 있다. 식물의 육종 연구에 이용할 수 있는 분자표지가 다양하게 개발되어 왔으며 개발된 분자표지를 대량으로 검증하고 판별할 수 있는 기술과 실험 기자재도 매우 빠르게 개발되고 있다. 시간이 많이 소요되는 기술보다는 PCR을 이용한 RAPD(Random Amplified polymorphic DNA), AFLP(Amplified fragment length polymorphism), SSR(Simple Sequence Repeat) 및 SNP(Single nucleotide polymorphism) 분석 등 다양한 분자표지 기술이 폭넓게 이용되고 있다. 분자표지 중 공우성 표지(codominant marker)를 이용하면 유전자의 동형접합(homozygous) 여부를 판별할 수 있으며, 형질이 발현되기 전에 조사가 가능하여 품종 육성에 있어서 빠른 판별이 가능하다는 장점이 있다.Molecular markers have the advantage that they can be detected in all tissues by differences in DNA sequence and are not affected by environmental influences and multiple expression of genes. Genomic genetic mapping of major crops is currently underway, and molecular markers are being used in the selection process of breeding. Numerous molecular markers have been developed for the study of plant breeding, and techniques and laboratory equipment for massively verifying and identifying the developed molecular markers are being developed very quickly. A variety of molecular labeling techniques such as Random Amplified Polymorphic DNA (RAPD), Amplified Fragment Length Polymorphism (AFLP), Simple Sequence Repeat (SSR) and Single Nucleotide Polymorphism (SNP) . The use of a codominant marker in the molecular markers can discriminate homozygous of the gene, and it is possible to carry out a search before the expression of the trait, thereby enabling rapid discrimination in breeding.
SNP(Single nucleotide polymorphism)는 개체 간의 DNA에 나타나는 하나의 염기 차이로, 매우 다양한 종의 게놈에 걸쳐서 존재한다. 식물체 게놈에 나타나는 빈번한 SNP는 유전자 지도제작(mapping), 분자표지 보조 육종(marker assisted breeding) 및 유전자지도 기반 클로닝(map-based cloning)을 가능하게 한다. SNP는 게놈 유전자 표지 중 가장 많으며, 이러한 SNP를 검출하기 위해서 실험의 용이성 및 비용을 고려한 다양한 SNP 검출 방법과 실험장비가 개발되고 있다.Single nucleotide polymorphism (SNP) is a single nucleotide difference in DNA between individuals that exists across a wide variety of species genomes. Frequent SNPs appearing in plant genomes enable gene mapping, marker assisted breeding, and map-based cloning. SNPs are the most common genomic markers, and various SNP detection methods and experimental equipments have been developed in consideration of the ease and cost of the experiment to detect such SNPs.
한국등록특허 제1403494호에는 '고구마의 품종 판별을 위한 SSR 프라이머 및 이를 이용한 고구마의 품종 판별 방법'이 개시되어 있고, 한국원예학회지(박권우 외, 5:112-113, 1996)에 '고구마 품종 구분 및 분류를 위한 RAPD법 이용'이 개시어 있다. 그러나, 고구마 품종 판별을 위하여 SSR(Simple Sequence Repeat)법 또는 RAPD(Random Amplified Polymorphic DNA)법 등은 다배수성에 따른 품종 특이적 단일 밴드 형성이 어려워 혼합된 자원들로부터 각각의 자원을 식별키가 어렵고 재현성이 낮은 단점이 있다.Korean Patent No. 1403494 discloses 'SSR primer for discriminating sweet potato varieties and a method for discriminating sweet potato varieties using the SSR primer.' The Korean Society of Horticulture (Park, Kwon Woo et al., 5: 112-113, 1996) And the use of the RAPD method for classification. However, SSR (Simple Sequence Repeat) method or Random Amplified Polymorphic DNA (RAPD) method is difficult to discriminate the sweet potato varieties, The reproducibility is low.
한편, 고구마의 경우 특허권 및 로열티 문제로 고구마 품종에 대한 변별력 요구가 급증하고 있으나, 고구마는 동일 품종 내에서도 형태적 구분이 어려워 판별에 어려움을 겪고 있다. 더욱이, 현재 고구마 재배 시 단일 품종화가 이루어지지 않고 있으며 혼재되어 존재하여 상품의 단일화나 규격화가 힘든 상태이다.On the other hand, in the case of sweet potatoes, the demand for discrimination of sweet potato varieties is increasing rapidly due to patent rights and royalties, but sweet potatoes are difficult to distinguish even in the same varieties. Furthermore, there is no single varietalization of sweet potatoes at present, and it is difficult to unify or standardize the products because they exist in a mixed state.
이에, 고구마의 산업적 이용도 증대 및 새로운 시장개척 등을 위한 고구마 품종 판별 마커의 개발이 필요한 실정이다.Therefore, it is necessary to develop a marking system for sweet potato varieties to increase the industrial utilization of sweet potatoes and to develop new markets.
이러한 배경하에서, 본 발명자들은 고구마 품종 판별 분자 마커를 개발하기 위하여, 고구마 유전체 염기서열 비교분석 및 다형성 확인을 통하여 신규 SNP 분자 마커를 도출하고, 이를 토대로 다양한 원산의 고구마 품종간의 SNP 분자 마커 증폭용 프라이머 세트를 개발함으로써, 본 발명을 완성하게 되었다.Under these circumstances, the inventors of the present invention derived novel SNP molecular markers through comparative analysis of sweet potato genome sequence and polymorphism in order to develop a sweet potato varietal discriminating marker, and based on this, a primer for SNP molecular marker amplification The present invention has been completed.
본 발명의 목적은 고구마 품종 판별용 SNP 분자 마커 증폭용 프라이머 세트, 상기 프라이머 세트를 포함하는 키트 및 상기 프라이머 세트를 이용한 고구마 품종 판별 방법을 제공하기 위한 것이다.It is an object of the present invention to provide a primer set for SNP molecular marker amplification for the identification of sweet potato varieties, a kit containing the primer set, and a method for distinguishing sweet potato varieties using the primer set.
상기의 목적을 달성하기 위한 하나의 양태로서, 본 발명은 서열번호 1, 2, 3 및 4으로 표시되는 프라이머 세트; 서열번호 5, 6, 7 및 8로 표시되는 프라이머 세트; 서열번호 9, 10, 11 및 12로 표시되는 프라이머 세트; 서열번호 13, 14, 15 및 16으로 표시되는 프라이머 세트; 서열번호 17, 18, 19 및 20으로 표시되는 프라이머 세트; 서열번호 21, 22, 23 및 24로 표시되는 프라이머 세트; 서열번호 25, 26, 27 및 28로 표시되는 프라이머 세트; 서열번호 29, 30, 31 및 32로 표시되는 프라이머 세트; 서열번호 33, 34, 35 및 36으로 표시되는 프라이머 세트; 서열번호 37, 38, 39 및 40으로 표시되는 프라이머 세트; 서열번호 41, 42, 43 및 44로 표시되는 프라이머 세트; 서열번호 45, 46, 47 및 48로 표시되는 프라이머 세트; 서열번호 49, 50, 51 및 52로 표시되는 프라이머 세트; 서열번호 53, 54, 55 및 56으로 표시되는 프라이머 세트; 서열번호 57, 58, 59 및 60으로 표시되는 프라이머 세트; 서열번호 61, 62, 63 및 64로 표시되는 프라이머 세트; 서열번호 65, 66, 67 및 68로 표시되는 프라이머 세트; 서열번호 69, 70, 71 및 72로 표시되는 프라이머 세트; 서열번호 73, 74, 75 및 76으로 표시되는 프라이머 세트; 서열번호 77, 78, 79 및 80으로 표시되는 프라이머 세트; 서열번호 81, 82, 83 및 84로 표시되는 프라이머 세트; 서열번호 85, 86, 87 및 88로 표시되는 프라이머 세트; 서열번호 89, 90, 91 및 92로 표시되는 프라이머 세트; 서열번호 93, 94, 95 및 96으로 표시되는 프라이머 세트; 서열번호 97, 98, 99 및 100으로 표시되는 프라이머 세트; 서열번호 101, 102, 103 및 104로 표시되는 프라이머 세트; 서열번호 105, 106, 107 및 108로 표시되는 프라이머 세트; 서열번호 109, 110, 111 및 112로 표시되는 프라이머 세트; 서열번호 113, 114, 115 및 116으로 표시되는 프라이머 세트; 서열번호 117, 118, 119 및 120으로 표시되는 프라이머 세트; 서열번호 121, 122, 123 및 124로 표시되는 프라이머 세트; 서열번호 125, 126, 127 및 128로 표시되는 프라이머 세트; 서열번호 129, 130, 131 및 132로 표시되는 프라이머 세트; 서열번호 133, 134, 135 및 136으로 표시되는 프라이머 세트; 서열번호 137, 138, 139 및 140으로 표시되는 프라이머 세트; 서열번호 141, 142, 143 및 144로 표시되는 프라이머 세트; 서열번호 145, 146, 147 및 148로 표시되는 프라이머 세트; 서열번호 149, 150, 151 및 152로 표시되는 프라이머 세트; 서열번호 153, 154, 155 및 156으로 표시되는 프라이머 세트; 서열번호 157, 158, 159 및 160으로 표시되는 프라이머 세트; 서열번호 161, 162, 163 및 164로 표시되는 프라이머 세트; 서열번호 165, 166, 167 및 168로 표시되는 프라이머 세트; 서열번호 169, 170, 171 및 172로 표시되는 프라이머 세트; 서열번호 173, 174, 175 및 176으로 표시되는 프라이머 세트; 서열번호 177, 178, 179 및 180으로 표시되는 프라이머 세트; 서열번호 181, 182, 183 및 184로 표시되는 프라이머 세트; 및 서열번호 185, 186, 187 및 188로 표시되는 프라이머 세트;를 포함하는, 고구마 품종 판별용 프라이머 세트를 제공한다. According to one aspect of the present invention, there is provided a primer set comprising: a primer set represented by SEQ ID NOS: 1, 2, 3 and 4; A primer set represented by SEQ ID NOS: 5, 6, 7, and 8; A primer set represented by SEQ ID NOS: 9, 10, 11, and 12; A primer set represented by SEQ ID NOs: 13, 14, 15, and 16; A primer set represented by SEQ ID NOs: 17, 18, 19, and 20; A primer set represented by SEQ ID NOS: 21, 22, 23 and 24; A primer set represented by SEQ ID NOS: 25, 26, 27, and 28; A primer set represented by SEQ ID NOs: 29, 30, 31, and 32; A primer set represented by SEQ ID NOS: 33, 34, 35, and 36; A primer set represented by SEQ ID NOS: 37, 38, 39, and 40; A primer set represented by SEQ ID NOS: 41, 42, 43, and 44; A primer set represented by SEQ ID NOS: 45, 46, 47, and 48; A primer set represented by SEQ ID NOS: 49, 50, 51, and 52; A primer set represented by SEQ ID NOS: 53, 54, 55, and 56; A primer set represented by SEQ ID NOS: 57, 58, 59, and 60; A primer set represented by SEQ ID NOS: 61, 62, 63 and 64; A primer set represented by SEQ ID NOS: 65, 66, 67, and 68; A primer set represented by SEQ ID NOS: 69, 70, 71 and 72; A primer set represented by SEQ ID NOS: 73, 74, 75, and 76; A primer set represented by SEQ ID NOS: 77, 78, 79 and 80; A primer set represented by SEQ ID NOS: 81, 82, 83 and 84; A primer set represented by SEQ ID NOS: 85, 86, 87 and 88; A primer set represented by SEQ ID NOS: 89, 90, 91, and 92; A primer set represented by SEQ ID NOS: 93, 94, 95, and 96; A primer set represented by SEQ ID NOS: 97, 98, 99 and 100; A primer set represented by SEQ ID NOS: 101, 102, 103, and 104; A primer set represented by SEQ ID NOS: 105, 106, 107, and 108; A primer set represented by SEQ ID NOS: 109, 110, 111, and 112; A primer set represented by SEQ ID NOS: 113, 114, 115, and 116; A primer set represented by SEQ ID NOS: 117, 118, 119, and 120; A primer set represented by SEQ ID NOS: 121, 122, 123, and 124; A primer set represented by SEQ ID NOS: 125, 126, 127, and 128; A primer set represented by SEQ ID NOs: 129, 130, 131, and 132; A primer set represented by SEQ ID NOS: 133, 134, 135, and 136; A primer set represented by SEQ ID NOS: 137, 138, 139, and 140; A primer set represented by SEQ ID NOS: 141, 142, 143 and 144; A primer set represented by SEQ ID NOS: 145, 146, 147, and 148; A primer set represented by SEQ ID NOs: 149, 150, 151, and 152; A primer set represented by SEQ ID NOS: 153, 154, 155, and 156; A primer set represented by SEQ ID NOS: 157, 158, 159, and 160; A primer set represented by SEQ ID NOS: 161, 162, 163, and 164; A primer set represented by SEQ ID NOS: 165, 166, 167, and 168; A primer set represented by SEQ ID NOs: 169, 170, 171, and 172; A primer set represented by SEQ ID NOS: 173, 174, 175, and 176; A primer set represented by SEQ ID NOS: 177, 178, 179, and 180; A primer set represented by SEQ ID NOS: 181, 182, 183, and 184; And a primer set set forth in SEQ ID NOS: 185, 186, 187, and 188.
다른 하나의 양태로서, 본 발명은 상기 프라이머 세트 및 증폭 반응을 수행하기 위한 시약을 포함하는, 고구마 품종 판별용 키트를 제공한다.In another aspect, the present invention provides a kit for discriminating sweet potato varieties comprising the primer set and a reagent for carrying out an amplification reaction.
또 다른 하나의 양태로서, 본 발명은 a) 고구마 시료에서 게놈 DNA를 분리하는 단계; b) 상기 분리된 게놈 DNA를 주형으로 하고, 제1항의 프라이머 세트를 이용하여 PCR(polymerase chain reaction)을 수행하는 단계; 및 c) 상기 b) 단계의 증폭된 PCR 산물을 검출하는 단계;를 포함하는, 고구마 품종 판별 방법을 제공한다.In another aspect, the present invention provides a method for producing a sweet potato comprising: a) separating genomic DNA from a sweet potato sample; b) performing PCR (polymerase chain reaction) using the separated genomic DNA as a template and using the primer set of claim 1; And c) detecting the amplified PCR product of step b).
본 발명에 따른, 고구마 품종 판별용 SNP 분자 마커 증폭용 프라이머 세트를 이용하여 상용되고 있는 고구마의 원산지에 따른 품종 판별 및 순도 검정과 더불어, 품종 보호와 종자 관리체계 확립을 위한 품종의 구별성(distinctness), 균일성(uniformity) 및 안정성(stability) 검정에 유용하게 활용될 수 있다. 또한, 본발명에 따른 고구마 품종 판별용 SNP 분자 마커는 수입 개방화 시대에 우리나라 육성 품종에 대한 지적재산권 확보 및 향후 고구마 품종 등록 시 품종 보증의 표지 인자로 활용 가능하다.Using the primer set for SNP molecular marker amplification for the identification of sweet potato varieties according to the present invention, the identification of varieties and purity according to the origin of sweet potatoes which are commonly used, and the discrimination of varieties for establishing the variety protection and seed management system ), Uniformity, and stability. In addition, SNP molecular markers for the identification of sweet potato varieties according to the present invention can be utilized as a marking factor for the assurance of intellectual property rights for the cultivars cultivated in Korea during the import opening period and the breed guarantee for the registration of the sweet potato variety in the future.
도 1은 본 발명의 일실시예에 따라 Fluidigm 플랫폼을 이용한 각 SNP의 유전자형 분석 방법을 통해 고구마 품종 판별용 SNP 분자 마커 증폭용 프라이머 세트를 이용하여 분석한 유전형 분석(genotyping) 결과를 나타낸 도이다.
도 2는 본 발명의 일실시예에 따라 고구마 품종 판별용 SNP 분자 마커 증폭용 프라이머 세트에 의해 구별되는 다양한 원산의 고구마 유전자원 계통수를 나타낸 도이다.FIG. 1 is a diagram showing genotyping results obtained by analyzing genotypes of SNPs using a Fluidigm platform according to an embodiment of the present invention and using a primer set for SNP molecular marker amplification for identifying sweet potato varieties.
FIG. 2 is a diagram showing a sweet potato genetic circle of various native species distinguished by a primer set for SNP molecular marker amplification for the identification of sweet potato varieties according to an embodiment of the present invention.
본 발명은 고구마 품종 판별용 SNP 분자 마커 및 이의 용도에 관한 것으로, 보다 상세하게는 고구마 품종 판별용 SNP 분자 마커 증폭용 프라이머 세트, 상기 프라이머 세트를 포함하는 키트 및 상기 프라이머 세트를 이용한 고구마 품종 판별 방법에 관한 것이다.More particularly, the present invention relates to a primer set for SNP molecular marker amplification for identifying sweet potato varieties, a kit including the primer set, and a method for discriminating sweet potato varieties using the primer set .
하나의 양태로서, 본 발명은 서열번호 1, 2, 3 및 4으로 표시되는 프라이머 세트; 서열번호 5, 6, 7 및 8로 표시되는 프라이머 세트; 서열번호 9, 10, 11 및 12로 표시되는 프라이머 세트; 서열번호 13, 14, 15 및 16으로 표시되는 프라이머 세트; 서열번호 17, 18, 19 및 20으로 표시되는 프라이머 세트; 서열번호 21, 22, 23 및 24로 표시되는 프라이머 세트; 서열번호 25, 26, 27 및 28로 표시되는 프라이머 세트; 서열번호 29, 30, 31 및 32로 표시되는 프라이머 세트; 서열번호 33, 34, 35 및 36으로 표시되는 프라이머 세트; 서열번호 37, 38, 39 및 40으로 표시되는 프라이머 세트; 서열번호 41, 42, 43 및 44로 표시되는 프라이머 세트; 서열번호 45, 46, 47 및 48로 표시되는 프라이머 세트; 서열번호 49, 50, 51 및 52로 표시되는 프라이머 세트; 서열번호 53, 54, 55 및 56으로 표시되는 프라이머 세트; 서열번호 57, 58, 59 및 60으로 표시되는 프라이머 세트; 서열번호 61, 62, 63 및 64로 표시되는 프라이머 세트; 서열번호 65, 66, 67 및 68로 표시되는 프라이머 세트; 서열번호 69, 70, 71 및 72로 표시되는 프라이머 세트; 서열번호 73, 74, 75 및 76으로 표시되는 프라이머 세트; 서열번호 77, 78, 79 및 80으로 표시되는 프라이머 세트; 서열번호 81, 82, 83 및 84로 표시되는 프라이머 세트; 서열번호 85, 86, 87 및 88로 표시되는 프라이머 세트; 서열번호 89, 90, 91 및 92로 표시되는 프라이머 세트; 서열번호 93, 94, 95 및 96으로 표시되는 프라이머 세트; 서열번호 97, 98, 99 및 100으로 표시되는 프라이머 세트; 서열번호 101, 102, 103 및 104로 표시되는 프라이머 세트; 서열번호 105, 106, 107 및 108로 표시되는 프라이머 세트; 서열번호 109, 110, 111 및 112로 표시되는 프라이머 세트; 서열번호 113, 114, 115 및 116으로 표시되는 프라이머 세트; 서열번호 117, 118, 119 및 120으로 표시되는 프라이머 세트; 서열번호 121, 122, 123 및 124로 표시되는 프라이머 세트; 서열번호 125, 126, 127 및 128로 표시되는 프라이머 세트; 서열번호 129, 130, 131 및 132로 표시되는 프라이머 세트; 서열번호 133, 134, 135 및 136으로 표시되는 프라이머 세트; 서열번호 137, 138, 139 및 140으로 표시되는 프라이머 세트; 서열번호 141, 142, 143 및 144로 표시되는 프라이머 세트; 서열번호 145, 146, 147 및 148로 표시되는 프라이머 세트; 서열번호 149, 150, 151 및 152로 표시되는 프라이머 세트; 서열번호 153, 154, 155 및 156으로 표시되는 프라이머 세트; 서열번호 157, 158, 159 및 160으로 표시되는 프라이머 세트; 서열번호 161, 162, 163 및 164로 표시되는 프라이머 세트; 서열번호 165, 166, 167 및 168로 표시되는 프라이머 세트; 서열번호 169, 170, 171 및 172로 표시되는 프라이머 세트; 서열번호 173, 174, 175 및 176으로 표시되는 프라이머 세트; 서열번호 177, 178, 179 및 180으로 표시되는 프라이머 세트; 서열번호 181, 182, 183 및 184로 표시되는 프라이머 세트; 및 서열번호 185, 186, 187 및 188로 표시되는 프라이머 세트;를 포함하는, 고구마 품종 판별용 프라이머 세트를 제공한다.In one aspect, the present invention provides a primer set comprising a set of primers represented by SEQ ID NOS: 1, 2, 3 and 4; A primer set represented by SEQ ID NOS: 5, 6, 7, and 8; A primer set represented by SEQ ID NOS: 9, 10, 11, and 12; A primer set represented by SEQ ID NOs: 13, 14, 15, and 16; A primer set represented by SEQ ID NOs: 17, 18, 19, and 20; A primer set represented by SEQ ID NOS: 21, 22, 23 and 24; A primer set represented by SEQ ID NOS: 25, 26, 27, and 28; A primer set represented by SEQ ID NOs: 29, 30, 31, and 32; A primer set represented by SEQ ID NOS: 33, 34, 35, and 36; A primer set represented by SEQ ID NOS: 37, 38, 39, and 40; A primer set represented by SEQ ID NOS: 41, 42, 43, and 44; A primer set represented by SEQ ID NOS: 45, 46, 47, and 48; A primer set represented by SEQ ID NOS: 49, 50, 51, and 52; A primer set represented by SEQ ID NOS: 53, 54, 55, and 56; A primer set represented by SEQ ID NOS: 57, 58, 59, and 60; A primer set represented by SEQ ID NOS: 61, 62, 63 and 64; A primer set represented by SEQ ID NOS: 65, 66, 67, and 68; A primer set represented by SEQ ID NOS: 69, 70, 71 and 72; A primer set represented by SEQ ID NOS: 73, 74, 75, and 76; A primer set represented by SEQ ID NOS: 77, 78, 79 and 80; A primer set represented by SEQ ID NOS: 81, 82, 83 and 84; A primer set represented by SEQ ID NOS: 85, 86, 87 and 88; A primer set represented by SEQ ID NOS: 89, 90, 91, and 92; A primer set represented by SEQ ID NOS: 93, 94, 95, and 96; A primer set represented by SEQ ID NOS: 97, 98, 99 and 100; A primer set represented by SEQ ID NOS: 101, 102, 103, and 104; A primer set represented by SEQ ID NOS: 105, 106, 107, and 108; A primer set represented by SEQ ID NOS: 109, 110, 111, and 112; A primer set represented by SEQ ID NOS: 113, 114, 115, and 116; A primer set represented by SEQ ID NOS: 117, 118, 119, and 120; A primer set represented by SEQ ID NOS: 121, 122, 123, and 124; A primer set represented by SEQ ID NOS: 125, 126, 127, and 128; A primer set represented by SEQ ID NOs: 129, 130, 131, and 132; A primer set represented by SEQ ID NOS: 133, 134, 135, and 136; A primer set represented by SEQ ID NOS: 137, 138, 139, and 140; A primer set represented by SEQ ID NOS: 141, 142, 143 and 144; A primer set represented by SEQ ID NOS: 145, 146, 147, and 148; A primer set represented by SEQ ID NOs: 149, 150, 151, and 152; A primer set represented by SEQ ID NOS: 153, 154, 155, and 156; A primer set represented by SEQ ID NOS: 157, 158, 159, and 160; A primer set represented by SEQ ID NOS: 161, 162, 163, and 164; A primer set represented by SEQ ID NOS: 165, 166, 167, and 168; A primer set represented by SEQ ID NOs: 169, 170, 171, and 172; A primer set represented by SEQ ID NOS: 173, 174, 175, and 176; A primer set represented by SEQ ID NOS: 177, 178, 179, and 180; A primer set represented by SEQ ID NOS: 181, 182, 183, and 184; And a primer set set forth in SEQ ID NOS: 185, 186, 187, and 188.
본 발명에서 용어 "판별"은 고구마 품종 시료로부터, 본 발명의 프라이머 세트에 의해 품종의 다양성을 판단하여 구별하는 것을 의미하며, '구별'과 혼용하여 사용할 수 있다.In the present invention, the term " discrimination " means discrimination of varieties from sweet potato cultivar samples by the primer set of the present invention to discriminate them and can be used in combination with 'discrimination'.
본 발명에서 용어 "프라이머(primer)"는 카피하려는 핵산 가닥에 상보적인 단일 가닥 올리고뉴클레오티드 서열을 말하며, 프라이머 연장 산물의 합성을 위한 개시점으로 작용한다. 상기 프라이머의 길이 및 서열은 연장 산물의 합성을 시작하도록 허용해야 하며, 프라이머의 구체적인 길이 및 서열은 요구되는 DNA 또는 RNA 표적의 복합도(complexity) 뿐만 아니라 온도 및 이온 강도와 같은 프라이머 이용 조건에 의존할 것이다.The term " primer " in the present invention refers to a single-stranded oligonucleotide sequence complementary to a nucleic acid strand to be copied and serves as a starting point for the synthesis of a primer extension product. The length and sequence of the primer should allow for the start of synthesis of the extended product and the specific length and sequence of the primer will depend on the complexity of the desired DNA or RNA target as well as the primer usage conditions such as temperature and ionic strength something to do.
본 발명에 있어서, 상기 프라이머는 DNA 삽입(intercalating) 형광, 인광 또는 방사성을 발하는 물질을 더 포함할 수 있으며, 이에 제한되지 않는다. 바람직하게는, 상기 표지 물질은 FAM 또는 HEX이다. 표적 서열의 증폭 시 대립유전자(allele) 특이적인 프라이머의 5'-말단에 FAM 또는 HEX를 표지하여 PCR을 수행하면 표적 서열이 검출 가능한 형광 표지 물질로 표지될 수 있다.In the present invention, the primer may further include, but is not limited to, a substance that emits intercalating fluorescence, phosphorescence, or radioactive. Preferably, the labeling substance is FAM or HEX. When the target sequence is amplified, FAM or HEX is labeled at the 5'-end of the allele-specific primer, and PCR is carried out, whereby the target sequence can be labeled with a detectable fluorescent labeling substance.
본 발명에서 용어 "프라이머 세트"는 복수의 프라이머를 의미한다. 또한, 프라이머 세트는 프라이머를 수용하기 위한 컨테이너를 더 포함할 수 있다. 프라이머는 짧은 자유 3' 말단 수산화기(free 3' hydroxyl group)를 가지는 염기 서열로 상보적인 주형(template)과 염기쌍을 형성할 수 있고 주형 가닥 복사를 위한 시작 지점으로 기능을 하는 짧은 염기 서열을 의미한다. 프라이머는 적절한 완충용액 및 온도에서 중합반응을 위한 시약과 4가지 뉴클레오사이트 트리포스페이트의 존재하에서 DNA 합성을 개시할 수 있다. 프라이머는 올리고뉴클레오티드일 수 있으며, 올리고뉴클레오티드는 뉴클레오티드 유사체(analogue), 예를 들어, 포스포로티오에이트(phosphorothioate), 알킬포스포로티오에이트 또는 펩티드 헥산(peptide nucleic acid)를 포함할 수 있거나 삽입물질(intercalating agent)을 포함할 수 있다. 프라이머는 포스포르아미다이트법, 포스포디 에스테르법, 디에틸포스모르아미다이트법 등을 이용하는 화학 합성법을 통하여 제조될 수 있다. 또한, 프라이머 염기서열은 당해 분야에 공지된 수단에 의해 변형될 수 있다.The term " primer set " in the present invention means a plurality of primers. In addition, the primer set may further include a container for receiving the primer. Primer refers to a short nucleotide sequence that can form a base pair with a complementary template with a short free 3 'hydroxyl group and serves as a starting point for template strand copying . The primer can initiate DNA synthesis in the presence of a reagent for polymerization and four nucleoside triphosphates at the appropriate buffer solution and temperature. The primer may be an oligonucleotide and the oligonucleotide may comprise a nucleotide analogue such as phosphorothioate, alkylphosphorothioate or peptide nucleic acid, intercalating agent. The primer can be produced through a chemical synthesis method using a phosphoramidite method, a phosphodiester method, a diethylphosphoramidite method, or the like. In addition, the primer sequence may be modified by means known in the art.
본 발명의 프라이머 세트는 서열번호 1 내지 188의 총 47개 세트의 프라이머 세트로 이루어진 군으로부터 선택되는 하나 이상의 프라이머 세트를 포함하는 것일 수 있으며, 상기 각 프라이머 세트는 4개의 프라이머가 하나의 세트를 이루는 것일 수 있다(표 3).The primer set of the present invention may comprise at least one set of primers selected from the group consisting of a total of 47 sets of primers set forth in SEQ ID NOS: 1 to 188, wherein each set of primers has a set of four primers (Table 3).
상기 프라이머 세트는 서열번호 189 내지 235의 염기서열로 이루어진 폴리뉴클레오티드를 증폭시킬 수 있으며, 상기 서열번호 189 내지 235의 염기서열로 이루어진 폴리뉴클레오티드는 고구마 품종 간 구별을 위한 SNP(single nuclotide polymorphism) 부위를 포함한 것일 수 있다(표 2).The primer set may amplify a polynucleotide comprising the nucleotide sequence of SEQ ID NOS: 189 to 235, and the polynucleotide comprising the nucleotide sequence of SEQ ID NOS: 189 to 235 may comprise a single nucleotide polymorphism (SNP) region for distinguishing between sweet potato varieties (Table 2).
상기 'SNP'란 유전체 상에서 A, T, C, G로 구성되는 염기서열의 한 개가 다른 염기서열로 변한 것을 의미한다.The 'SNP' means that one of the nucleotide sequences consisting of A, T, C, and G on the genome is changed to another nucleotide sequence.
본 발명에 있어서, SNP(single nucleotide polymorphism) 위치 염기를 포함하는 상기 서열번호 189 내지 235의 염기서열로 이루어진 폴리뉴클레오티드로부터 선택되는 하나 이상의 폴리뉴클레오티드 또는 이의 상보적 폴리뉴클레오티드를 포함하는, 고구마 품종 판별용 SNP 조성물을 제공할 수 있다.In the present invention, there is provided a method for discriminating sweet potato varieties comprising at least one polynucleotide selected from the polynucleotides consisting of the nucleotide sequences of SEQ ID NOS: 189 to 235 including the SNP (single nucleotide polymorphism) position base or a complementary polynucleotide thereof SNP < / RTI >
상기 서열번호 189 내지 235는 상기 각각의 SNP 부위의 염기서열을 포함하는 폴리뉴클레오티드이다. 상기 서열번호 189 내지 235는 다형성 부위를 포함하는 다형성 서열이다. 다형성 서열(polymorphic sequence)이란 폴리뉴클레오티드 서열 중에 SNP를 나타내는 다형성 부위(polymorphic site)를 포함하는 서열을 말한다. 상기 폴리뉴클레오티드 서열은 DNA 또는 RNA가 될 수 있다.SEQ ID NOS: 189 to 235 are polynucleotides comprising the nucleotide sequences of the respective SNP sites. SEQ ID NOS: 189 to 235 are polymorphic sequences including polymorphic sites. A polymorphic sequence refers to a sequence comprising a polymorphic site representing a SNP in a polynucleotide sequence. The polynucleotide sequence may be DNA or RNA.
본 발명의 일실시예에 따른 SNP 조성물에 있어서, 상기 SNP 위치 염기는 서열번호 189부터 서열번호 235까지 모두 21번째로, 다형성 염기 정보는 표 2의 SNP 염기서열 정보에 [/]로 표시하였다.In the SNP composition according to an embodiment of the present invention, the SNP position base is 21st from SEQ ID NO: 189 to SEQ ID NO: 235, and the polymorphic base information is indicated by [/] in the SNP nucleotide sequence information of Table 2.
본 발명에 따른 고구마 품종 판별용 SNP를 구성하는 각 단일 SNP의 폴리뉴클레오티드는 바람직하게는 10개 이상의 연속 염기이며, 보다 바람직하게는 15개 이상의 연속 염기이며, 보다 더 바람직하게는 20개 이상의 연속 염기이며, 가장 바람직하게는 서열번호 189 내지 235의 각 서열이다.The polynucleotide of each SNP constituting SNP for the sweet potato variety according to the present invention is preferably at least 10 continuous bases, more preferably at least 15 continuous bases, even more preferably at least 20 continuous bases And most preferably the respective sequences of SEQ ID NOS: 189 to 235.
본 발명은 또한, 상기 SNP의 폴리뉴클레오티드, 이에 의해 코딩되는 폴리펩티드 또는 이의 cDNA를 포함하는 고구마 품종 판별용 마이크로어레이를 제공할 수 있다. 바람직하게는, 상기 폴리뉴클레오티드는 아미노-실란, 폴리-L-라이신 또는 알데히드의 활성기가 코팅된 기판에 고정될 수 있으나, 이에 제한되지는 않는다. 바람직하게는, 상기 기판은 실리콘 웨이퍼, 유리, 석영, 금속 또는 플라스틱일 수 있으나, 이에 제한되지는 않는다. 상기 폴리뉴클레오티드를 기판에 고정화시키는 방법으로는 피에조일렉트릭(piezoelectric) 방식을 이용한 마이크로피펫팅(micropipetting) 법, 핀(pin) 형태의 스폿터(spotter)를 이용한 방법 등을 사용할 수 있다.The present invention can also provide a microarray for distinguishing sweet potato varieties comprising a polynucleotide of the SNP, a polypeptide encoded thereby, or a cDNA thereof. Preferably, the polynucleotide may be immobilized on a substrate coated with an activator of amino-silane, poly-L-lysine or aldehyde, but is not limited thereto. Preferably, the substrate may be a silicon wafer, glass, quartz, metal or plastic, but is not limited thereto. As a method for immobilizing the polynucleotide on a substrate, a micropipetting method using a piezo electric method, a method using a pin-type spotter can be used.
본 발명에 따른 마이크로어레이는 본 발명에 따른 폴리뉴클레오티드 또는 그의 상보적 폴리뉴클레오티드, 그에 의해 코딩되는 폴리펩티드 또는 그의 cDNA를 이용하여 본 분야의 당업자에게 알려져있는 통상적인 방법에 의해 제조될 수 있다.The microarray according to the present invention can be produced by a conventional method known to those skilled in the art using a polynucleotide according to the present invention or a complementary polynucleotide thereof, a polypeptide encoded thereby, or cDNA thereof.
다른 하나의 양태로서, 본 발명은 상기 프라이머 세트 및 증폭 반응을 수행하기 위한 시약을 포함하는, 고구마 품종 판별용 키트를 제공한다. In another aspect, the present invention provides a kit for discriminating sweet potato varieties comprising the primer set and a reagent for carrying out an amplification reaction.
본 발명의 키트에서 상기 증폭 반응을 수행하기 위한 시약은 DNA 폴리머라제, dNTPs 및 버퍼(buffer) 등을 포함할 수 있다. 상기 dNTP 혼합물은 dATP, dCTP, dGTP, dTTP를 포함하며, 내열성 DNA 중합효소는 Taq DNA 중합효소, Tth DNA 중합효소 등 시판되는 내열성 중합효소를 이용할 수 있다. 또한, 본 발명의 키트는 최적의 반응 수행 조건을 기재한 사용자 안내서를 추가로 포함할 수 있다. 안내서는 키트 사용법, 예를 들면, PCR 완충액 제조 방법, 제시되는 반응 조건 등을 설명하는 인쇄물이다. 안내서는 팜플렛 또는 전단지 형태의 안내 책자, 키트에 부착된 라벨 및 키트를 포함하는 패키지의 표면상에 설명을 포함한다. 또한, 안내서는 인터넷과 같이 전기 매체를 통해 공개되거나 제공되는 정보를 포함한다.In the kit of the present invention, the reagent for carrying out the amplification reaction may include a DNA polymerase, dNTPs and a buffer. The dNTP mixture may include dATP, dCTP, dGTP, dTTP, and the heat-resistant DNA polymerase may be a commercially available heat-resistant DNA polymerase such as Taq DNA polymerase or Tth DNA polymerase. In addition, the kit of the present invention may further include a user guide describing optimal reaction performing conditions. The manual is a printed document that explains how to use the kit, for example, how to prepare PCR buffer, the reaction conditions presented, and so on. The manual includes instructions on the surface of the package including a brochure or leaflet in the form of a brochure, a label attached to the kit, and a kit. In addition, the brochure includes information that is disclosed or provided through an electronic medium such as the Internet.
또 다른 하나의 양태로서, 본 발명은 a) 고구마 시료에서 게놈 DNA를 분리하는 단계; b) 상기 분리된 게놈 DNA를 주형으로 하고, 제1항의 프라이머 세트를 이용하여 PCR(polymerase chain reaction)을 수행하는 단계; 및 c) 상기 b) 단계의 증폭된 PCR 산물을 검출하는 단계;를 포함하는, 고구마 품종 판별 방법을 제공한다.In another aspect, the present invention provides a method for producing a sweet potato comprising: a) separating genomic DNA from a sweet potato sample; b) performing PCR (polymerase chain reaction) using the separated genomic DNA as a template and using the primer set of claim 1; And c) detecting the amplified PCR product of step b).
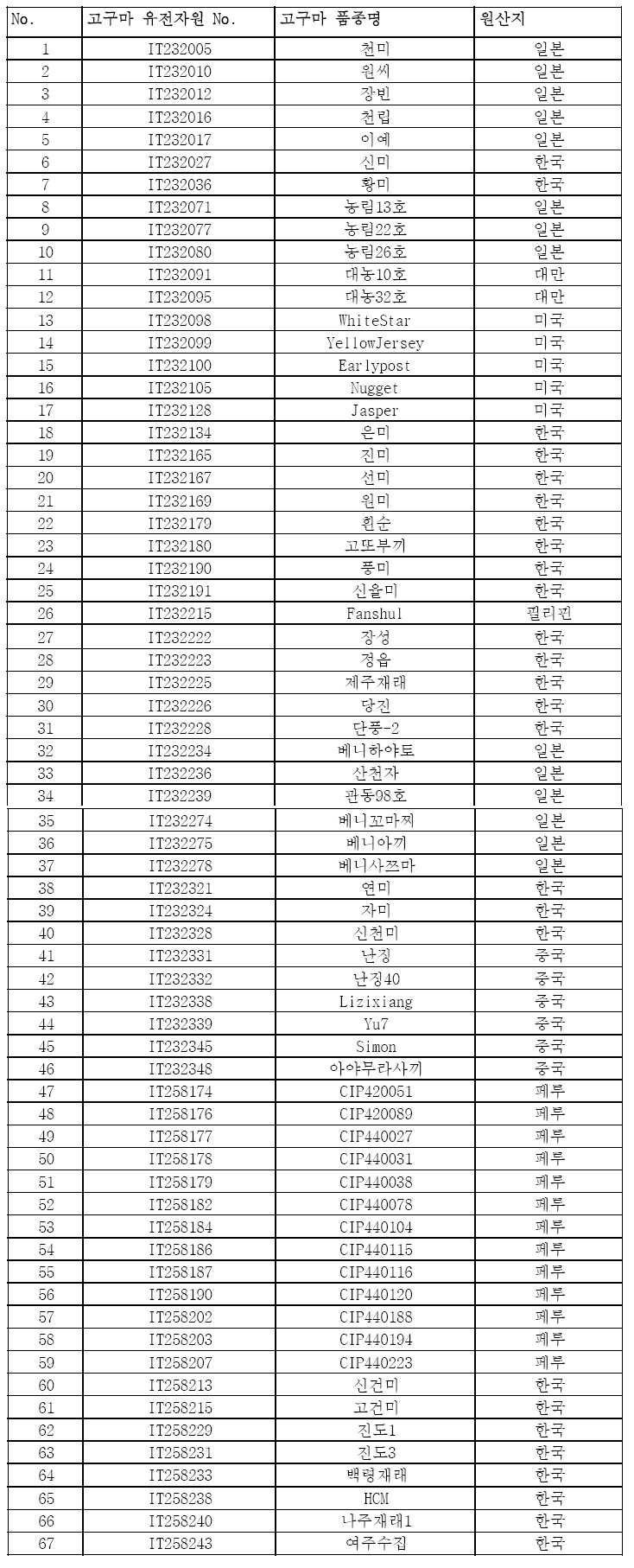
본 발명의 고구마 품종 판별 방법에 있어서, 상기 고구마 시료는 이에 제한되지는 않으나, 하기 표 1의 다양한 원산의 고구마 67자원(품종 56자원 및 재래종 11자원)일 수 있다.In the sweet potato variety identification method of the present invention, the sweet potato sample may be, but not limited to, the sweet potato resource 67 (variety 56 resource and
본 발명의 방법에 있어서, 고구마 시료로부터 게놈 DNA를 분리하는 방법은 당업계에 알려진 통상적인 방법을 통하여 이루어질 수 있으며, 그 방법은 페놀/클로로포름 추출법, SDS 추출법(Tai et al., Plant Mol. Biol. Reporter, 8: 297-303, 1990), CTAB 분리법(Cetyl Trimethyl Ammonium Bromide; Murray et al., Nuc. Res., 4321-4325, 1980), 또는 상업적으로 판매되는 DNA 추출 키트를 이용하여 수행할 수 있다.In the method of the present invention, the method of isolating the genomic DNA from the sweet potato sample can be performed by a conventional method known in the art, and the method can be selected from phenol / chloroform extraction method, SDS extraction method (Tai et al., Plant Mol. Biol Reporter, 8: 297-303, 1990), Cetyl Trimethyl Ammonium Bromide (Murray et al., Nuc. Res., 4321-4325, 1980), or commercially available DNA extraction kits .
상기 분리된 고구마 시료의 게놈 DNA를 주형으로 하고, 본 발명의 일실시예에 따른 하나 이상의 프라이머 세트를 이용하여 증폭 반응을 수행하여 표적 서열을 증폭할 수 있다. 표적 핵산을 증폭하는 방법은 중합효소연쇄반응(PCR), 리가아제 연쇄반응(ligase chain reaction), 핵산 서열 기재 증폭(nucleic acid sequence-based amplification), 전사 기재 증폭 시스템(transcription-based amplification system), 가닥 치환 증폭(strand displacement amplification) 또는 Qβ 복제효소(replicase)를 통한 증폭 또는 당업계에 알려진 핵산 분자를 증폭하기 위한 임의의 기타 적당한 방법이 있다. 이 중에서, PCR이란 중합효소를 이용하여 표적 핵산에 특이적으로 결합하는 프라이머 쌍으로부터 표적 핵산을 증폭하는 방법이다. 이러한 PCR 방법은 당업계에 잘 알려져있으며, 상업적으로 이용가능한 키트를 이용할 수도 있다. PCR은 PCR 반응에 필요한 당업계에 공지된 여러 성분을 포함하는 PCR 반응 혼합액을 이용하여 수행될 수 있다. 상기 PCR 반응 혼합액에는 분석하고자 하는 고구마 시료에서 추출된 게놈 DNA와 본 발명에서 제공되는 프라이머 세트 이외에 적당량의 DNA 중합효소, dNTP, PCR 완충용액 및 물을 포함한다. 상기 PCR 완충용액은 트리스-HCl(Tris-HCl), MgCl2, KCl 등을 포함한다. The target sequence may be amplified by performing amplification reaction using the genomic DNA of the separated sweet potato sample as a template and using at least one primer set according to an embodiment of the present invention. Methods for amplifying a target nucleic acid include polymerase chain reaction (PCR), ligase chain reaction, nucleic acid sequence-based amplification, transcription-based amplification system, Strand displacement amplification or amplification with Q [beta] replicase, or any other suitable method for amplifying nucleic acid molecules known in the art. Among them, PCR is a method of amplifying a target nucleic acid from a pair of primers that specifically bind to a target nucleic acid using a polymerase. Such PCR methods are well known in the art, and commercially available kits may be used. The PCR can be carried out using a PCR reaction mixture containing various components known in the art necessary for the PCR reaction. The PCR reaction mixture includes a suitable amount of DNA polymerase, dNTP, PCR buffer solution and water in addition to the genomic DNA extracted from sweet potato samples to be analyzed and the primer set provided in the present invention. The PCR buffer comprises Tris -HCl (Tris-HCl), MgCl 2, KCl and the like.
본 발명의 방법에 있어서, 상기 증폭된 표적 서열은 검출가능한 표지 물질로 표지될 수 있다. 상기 표지 물질은 형광, 인광 또는 방사성을 발하는 물질일 수 있으나, 이에 제한되지 않는다. 상기 표지 물질은 FAM 또는 HEX일 수 있다. 표적 서열의 증폭시 프라이머의 5'-말단에 FAM 또는 HEX를 표지하여 PCR을 수행하면 표적 서열이 검출가능한 형광 표지 물질로 표지될 수 있다. 또한, 방사성 물질을 이용한 표지는 PCR 수행시 32P 또는 35S 등과 같은 방사성 동위원소를 PCR 반응액에 첨가하면 증폭 산물이 합성되면서 방사성이 증폭 산물에 혼입되어 증폭 산물이 방사성으로 표지될 수 있다. 표적 서열을 증폭하기 위해 이용된 하나 이상의 프라이머 세트는 상기에 기재된 바와 같다.In the method of the present invention, the amplified target sequence may be labeled with a detectable labeling substance. The labeling substance may be a fluorescent, phosphorescent or radioactive substance, but is not limited thereto. The labeling substance may be FAM or HEX. When the target sequence is amplified, PCR is carried out by labeling FAM or HEX at the 5'-end of the primer, and the target sequence may be labeled with a detectable fluorescent labeling substance. When the radioactive isotope such as 32 P or 35 S is added to the PCR reaction solution, the amplification product may be synthesized and the radioactive substance may be incorporated into the amplification product and the amplification product may be labeled as radioactive. One or more sets of primers used to amplify the target sequence are as described above.
본 발명의 방법에 있어서, 고구마 품종 판별을 위한 방법은 상기 증폭 산물을 검출하는 단계를 포함하며, 상기 증폭 산물의 검출은 시퀀싱, DNA 칩, 겔 전기영동, 방사성 측정, 형광 측정 또는 인광 측정을 통해 수행될 수 있으나, 이에 제한되지 않는다. 증폭 산물을 검출하는 방법 중의 하나로서, 겔 전기영동을 수행할 수 있다. 겔 전기영동은 증폭 산물의 크기에 따라 아가로스 겔 전기영동 또는 아크릴아미드 겔 전기영동을 이용할 수 있다. 또한, 형광 측정 방법은 프라이머의 5'-말단에 FAM 또는 HEX를 표지하여 PCR을 수행하면 표적 서열이 검출 가능한 형광 표지 물질로 표지되며, 이렇게 표지된 형광은 형광 측정기를 이용하여 측정할 수 있다. 또한, 방사성 측정 방법은 PCR 수행시 32P 또는 35S 등과 같은 방사성 동위원소를 PCR 반응액에 첨가하여 증폭 산물을 표지한 후, 방사성 측정기구, 예를 들면, 가이거 계수기(Geiger counter) 또는 액체 섬광 계수기(liquid scintillation counter)를 이용하여 방사성을 측정할 수 있다.In the method of the present invention, the method for distinguishing sweet potato varieties comprises detecting the amplification product, wherein the amplification product is detected by sequencing, DNA chip, gel electrophoresis, radioactivity measurement, fluorescence measurement or phosphorescence measurement But is not limited thereto. As one method of detecting the amplification product, gel electrophoresis can be performed. Gel electrophoresis can be performed using agarose gel electrophoresis or acrylamide gel electrophoresis depending on the size of the amplification product. In addition, in the fluorescence measurement method, FAM or HEX is labeled at the 5'-end of the primer and when the PCR is performed, the target sequence is labeled with a fluorescent label capable of detecting the fluorescence, and the fluorescence thus labeled can be measured using a fluorescence analyzer. In addition, in the case of performing the PCR, the radioactive isotope such as 32 P or 35 S is added to the PCR reaction solution to mark the amplification product, and then a radioactive measurement device such as a Geiger counter or liquid scintillation counter The radioactivity can be measured using a liquid scintillation counter.
이하, 실시예를 통하여 본 발명의 구성 및 효과를 더욱 상세히 설명하고자 한다. 이들 실시예는 오로지 본 발명을 예시하기 위한 것일 뿐, 본 발명의 범위가 이들 실시예에 의해 한정되는 것은 아니다.Hereinafter, the constitution and effects of the present invention will be described in more detail through examples. These examples are only for illustrating the present invention, and the scope of the present invention is not limited by these examples.
실시예 1: 재료 준비 및 DNA 추출Example 1: Material preparation and DNA extraction
실시예 1-1: 시험재료 Example 1-1: Test material
고구마 품종 판별용 SNP 분자 마커 개발을 위해, 국립식량과학원 바이오에너지작물연구소에 국가등록 관리중인 다양한 원산의 고구마 67자원(품종 56자원 및 재래종 11자원)을 이용하였다(표 1).In order to develop the SNP molecular markers for sweet potato varieties, 67 varieties of native potatoes (56 varieties and 11 indigenous varieties) under the National Register of the National Institute of Food Science and Technology were used.
실시예 1-2: DNA 추출Example 1-2: DNA extraction
상기 실시예 1-1의 67자원의 고구마 품종에 대하여 식물체의 게놈 DNA(genomic DNA)는 Gentra Puregene Cell kit for plant(QIAGEN)을 사용하여 추출하였다. 추출한 각 고구마의 DNA는 ND-1000 spectrometer(NanoDrop Technologies, Wilmington, DE, USA)를 이용하여 고구마 DNA의 품질 및 양을 확인하였고, PCR 증폭 반응 수행을 위해 DNA 농도를 50 ng/㎕로 정량한 후 사용하였다.Genomic DNA of the plants was extracted using the Gentra Puregene Cell kit for plant (QIAGEN) for the potato varieties of 67 resources of Example 1-1. The DNA of each sweet potato extracted was checked for the quality and quantity of sweet potato DNA using ND-1000 spectrometer (NanoDrop Technologies, Wilmington, DE, USA), and the DNA concentration was determined to be 50 ng / Respectively.
실시예 2: 고구마 품종 판별용 신규 SNP 분자 마커 개발 및 프라이머 디자인Example 2: Development of novel SNP molecular markers for sweet potato variety identification and primer design
고구마 품종 판별용 신규 SNP 분자 마커를 개발하기 위해, 상기 실시예 1-1에서 준비한 다양한 원산의 고구마 67자원(품종 56자원 및 재래종 11자원)을 대상으로 GBS(Genotyping By Sequencing) 방법을 통해 96개의 후보 SNP(Single Nucleotide Polymorphism) 부위 및 주변 염기서열 정보를 확보하였다. 상기 확보한 후보 SNP 부위를 대상으로 도출한 대립유전자좌별 증폭 프라이머 세트를 제작한 후 Fluidigm 기기를 이용하여 고구마 품종 판별 분석에 이용하였다. 상기 96개 후보 SNP 부위 중 대상자원간 유전적 차이가 뚜렷이 관찰되고, 확인된 유전형에 기반하여 계통수를 작성하여 본 결과를 토대로 고구마 품종 판별에 사용이 가능한 47개의 SNP 부위에 대한 대립유전자좌별 증폭 프라이머 세트를 최종 선발하였다(표 2 및 표 3). In order to develop a new SNP molecular marker for the identification of sweet potato varieties, a variety of 96 native potatoes (56 varieties and 11 native varieties) prepared in Example 1-1 were subjected to GIP (Genotyping By Sequencing) The candidate SNP (Single Nucleotide Polymorphism) site and surrounding nucleotide sequence information were obtained. An amplification primer set for allelic loci derived from the obtained candidate SNP region was prepared and used for discrimination of sweet potato varieties using Fluidigm instrument. Genetic differences between the target candidates of the 96 candidate SNP regions were clearly observed. Based on the results, the amplification primers of allelic loci for 47 SNP regions, which can be used for the identification of sweet potato varieties, (Table 2 and Table 3).
(Marker name)(Marker name)
좌측 플랭킹 서열 [A/B] 우측 플랭킹 서열 SNP molecular marker base sequence information (5 '- >3')
Left flanking sequence [A / B] Right flanking sequence
*[A/B]: 염기 A가 염기 B로 점돌연변이된 단일염기 다형성(single nucleotide polymorphism, SNP)을 가짐을 의미한다.* [A / B]: means that nucleotide A has a single nucleotide polymorphism (SNP) that is point mutated to base B.
(Marker name)(Marker name)
번호number
번호number
번호number
번호number
* A1: SNP 부위의 첫 번째 대립유전자(allele).* A1: the first allele in the SNP region.
* A2: SNP 부위의 두 번째 대립유전자(allele).A2: A second allele at the SNP site.
실시예 3: 고구마 품종 내에서 유전적 다양성 평가 및 유전형 분석Example 3: Genetic diversity evaluation and genotyping analysis in sweet potato varieties
상기 실시예 2에서 신규 개발된 고구마 품종 판별용 SNP 분자 마커 증폭용 프라이머 세트의 유용성에 대한 평가 및 다양한 원산의 고구마 67자원(품종 56자원 및 재래종 11자원) 간의 유전적 다양성을 연구하기 위해, PowerMarker(ver 3.25)를 이용하여 주요 대립인자 빈도(major allele frequency, MAF), 유전적 다양성(Genetic diversity, GD), 다형성 정보(Polymorphic information content, PIC) 등을 분석하였다. 또한, PowerMarker에 포함되어 있는 shared allele genetic distance를 이용하여 고구마 67자원에 대한 각각의 유전적 거리를 분석한 후 UPGMA 방법을 이용하여 계통수(phylogenetic tree)를 작성하여 계통분류학적 분석을 수행하였다. 또한, 각 SNP의 유전형 분석 방법은 Fluidigm 기기를 이용하여 실시하였으며, 고구마 67자원을 대상으로 상기 신규 개발된 고구마 품종 판별용 SNP 분자 마커 증폭용 프라이머 세트를 사용하여 유전형 분석(genotyping)을 수행하였다(도 1). 이후, 계통수를 작성한 후 최종적으로 다양한 원산의 고구마 67자원에 대한 품종을 판별 분석하였다.In order to evaluate the usefulness of a primer set for SNP molecular marker amplification for the identification of sweet potato varieties newly developed in Example 2 and to study the genetic diversity among various native sweet potato 67 resources (56 varieties and 11 indigenous varieties), PowerMarker Major allele frequency (MAF), genetic diversity (GD), and polymorphic information content (PIC) were analyzed using the data set (ver 3.25). In addition, phylogenetic analysis was performed by using the shared allele genetic distance included in PowerMarker to analyze the genetic distances of 67 resources of sweet potato, and then creating a phylogenetic tree using the UPGMA method. Genotype analysis of each SNP was performed using a Fluidigm instrument, and genotyping was performed using a primer set for amplifying a SNP molecular marker for the identification of the newly developed sweet potato varieties in the potato 67 resource 1). After that, the tree species were finally identified and the varieties of 67 different potatoes of various native species were identified and analyzed.
신규 개발된 고구마 품종 판별용 SNP 분자 마커 증폭용 프라이머 세트를 이용하여 67자원의 다양한 고구마 품종에 대하여 유전적 다양성을 분석한 결과, 평균 MAF(Major allele frequency)는 0.674 이었으며, SP-SNP_11 부위에서 0.993으로 가장 높게 확인되었다. 분석에 사용한 마커에 대한 유전적 다양성을 나타내는 GD(Genetic diversity) 및 PIC(Polymorphism Informatino Content)는 각각 0.015~0.500, 및 0.015~0.375의 범위를 나타내었으며, SP_SNP_9, SP_SNP_24, SP_SNP_29, SP_SNP_34, SP_SNP_35, SP_SNP_37, SP_SNP_38, SP_SNP_39, 및 SP_SNP_45 부위에서 GD 및 PIC 두 값 모두 최대의 값을 보였다(표 4).Genetic diversity analysis of 67 varieties of sweet potato varieties using primer set for SNP molecular markers for identification of newly developed sweet potato varieties revealed that the mean MAF (major allele frequency) was 0.674 and 0.993 at the SP-SNP_11 site , Respectively. Genetic diversity (GD) and polymorphism informatino content (PIC), which indicate the genetic diversity of the markers used in the analysis, ranged from 0.015 to 0.500 and 0.015 to 0.375, respectively. SP_SNP_9, SP_SNP_24, SP_SNP_29, SP_SNP_34, SP_SNP_35, SP_SNP_37 , SP_SNP_38, SP_SNP_39, and SP_SNP_45 showed the highest values for both GD and PIC (Table 4).
또한, 상기 고구마 품종 판별용 SNP 분자 마커 증폭용 프라이머 세트를 이용하여 Fluidigm 플랫폼을 이용한 각 SNP의 유전자형 분석을 수행하고, 상기 분석한 유전형 분석(genotyping) 결과를 토대로 계통수를 작성하여 분석해 본 결과, 다양한 원산의 총 67자원의 고구마 대상 자원들이 가지고 있는 유전자형에 따라 명확하게 구별되었다(도 2). Also, genotype analysis of each SNP using the Fluidigm platform was performed using the primer set for SNP molecular marker amplification for the sweet potato variety identification, and the phylogenetic tree was created and analyzed based on the genotyping result. As a result, A total of 67 resources in the wild were clearly distinguished according to the genotype of the sweet potato target resources (Fig. 2).
이에 따라, 상기 신규 개발된 고구마 품종 판별용 SNP 분자 마커 및 이의 증폭용 프라이머 세트를 다양한 원산의 고구마 품종을 판별 및 구별하는데 유용하게 사용할 수 있음을 알 수 있었다. Accordingly, it was found that the newly developed SNP molecular markers for distinguishing sweet potato varieties and their primer sets for amplification can be used for distinguishing and distinguishing sweet potato varieties of various native ones.
(Marker name)(Marker name)
a) GN(Genotype Number): 유전자형 수; b) MAF(major allele frequency): 주요 대립인자 빈도; c) GD(Genetic diversity): 유전적 다양성; d) OH(Observed heterozygosity): 관찰된 이형접합 기대치; e) PIC(Polymorphic information content): 다형성 정보. a) GN (Genotype Number): number of genotypes; b) MAF (major allele frequency): Major allele frequency; c) Genetic diversity (GD): genetic diversity; d) OH (Observed heterozygosity): observed heterozygosity expectation; e) Polymorphic information content (PIC): Polymorphic information.
<110> Republic of Korea <120> SNP molecular marker for selecting cultivars of sweet potatoes and uses thereof <130> P17R12C1235 <160> 235 <170> KoPatentIn 3.0 <210> 1 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_1_F <400> 1 acagacctca ggattgggaa aa 22 <210> 2 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_1_R <400> 2 catagagctc gggagggca 19 <210> 3 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_1_A1 <400> 3 ctatgtccct aaccggggc 19 <210> 4 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_1_A2 <400> 4 actatgtccc taaccgggga 20 <210> 5 <211> 16 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_2_F <400> 5 atgcgcgcaa gcagtt 16 <210> 6 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_2_R <400> 6 accacggaag cagctaggat 20 <210> 7 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_2_A1 <400> 7 gccttcacca gagttacttc cttc 24 <210> 8 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_2_A2 <400> 8 gccttcacca gagttacttc cttt 24 <210> 9 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_3_F <400> 9 cgtctcttct ttccccgca 19 <210> 10 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_3_R <400> 10 ctggaactcc ggtagggtac t 21 <210> 11 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_3_A1 <400> 11 agcagccccg acattcg 17 <210> 12 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_3_A2 <400> 12 agcagccccg acattca 17 <210> 13 <211> 16 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_4_F <400> 13 gcgaagttcc agcccc 16 <210> 14 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_4_R <400> 14 gcagcctgtg ttgcatgtgt 20 <210> 15 <211> 28 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_4_A1 <400> 15 gtgtcatgat gaatatgtcc atggaaac 28 <210> 16 <211> 28 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_4_A2 <400> 16 gtgtcatgat gaatatgtcc atggaaaa 28 <210> 17 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_5_F <400> 17 tcgtttcttc tcctgaacgg a 21 <210> 18 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_5_R <400> 18 cacgagtgcc actgaaatcc aa 22 <210> 19 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_5_A1 <400> 19 tggcgtaaac tcagcatggt 20 <210> 20 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_5_A2 <400> 20 ggcgtaaact cagcatggc 19 <210> 21 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_6_F <400> 21 caaacttgtt ttggaaagag cgtc 24 <210> 22 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_6_R <400> 22 ttggtggcct gagttccga 19 <210> 23 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_6_A1 <400> 23 cgtaatctat tgcagcagcc tttttc 26 <210> 24 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_6_A2 <400> 24 cgtaatctat tgcagcagcc ttttta 26 <210> 25 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_7_F <400> 25 cctgcatgca ttctatgtca ttgat 25 <210> 26 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_7_R <400> 26 cagcacacga gacaacaccg 20 <210> 27 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_7_A1 <400> 27 gcaaagaaag cagtggaagc aatc 24 <210> 28 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_7_A2 <400> 28 gcaaagaaag cagtggaagc aata 24 <210> 29 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_8_F <400> 29 ccgcagctat acatggttga a 21 <210> 30 <211> 32 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_8_R <400> 30 caaacaggat ggaatcattt gaaactgatc aa 32 <210> 31 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_8_A1 <400> 31 cttcaggtgt ctcttatgta agttgc 26 <210> 32 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_8_A2 <400> 32 ccttcaggtg tctcttatgt aagttgt 27 <210> 33 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_9_F <400> 33 agcatgggat gagatagatc caa 23 <210> 34 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_9_R <400> 34 cttggaggct gcaatctgct 20 <210> 35 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_9_A1 <400> 35 ccaaaatcat caggagaagg atacaca 27 <210> 36 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_9_A2 <400> 36 ccaaaatcat caggagaagg atacact 27 <210> 37 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_10_F <400> 37 cacaacggat aacaaaagaa agagga 26 <210> 38 <211> 33 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_10_R <400> 38 gatgatctgt acacgcaaat aaataaactc aca 33 <210> 39 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_10_A1 <400> 39 aaccgcaatt cctgagcttc ta 22 <210> 40 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_10_A2 <400> 40 ccgcaattcc tgagcttctg 20 <210> 41 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_11_F <400> 41 tgttagaaca tgttttgacc agca 24 <210> 42 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_11_R <400> 42 ctcgttgcat tgtggacgct 20 <210> 43 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_11_A1 <400> 43 ccgtctcaaa ttatgttgga tgagag 26 <210> 44 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_11_A2 <400> 44 accgtctcaa attatgttgg atgagaa 27 <210> 45 <211> 15 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_12_F <400> 45 cgcagccact gcctc 15 <210> 46 <211> 16 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_12_R <400> 46 gcggcagggt tgcgaa 16 <210> 47 <211> 15 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_12_A1 <400> 47 gcctccgcct ccacg 15 <210> 48 <211> 15 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_12_A2 <400> 48 gcctccgcct ccacc 15 <210> 49 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_13_F <400> 49 gctgaagaaa gcaagcagca 20 <210> 50 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_13_R <400> 50 agtgaatcgg ctccgaagga a 21 <210> 51 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_13_A1 <400> 51 atcagcagtg cccatggat 19 <210> 52 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_13_A2 <400> 52 tcagcagtgc ccatggag 18 <210> 53 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_14_F <400> 53 cctctgtgca ataaaatcag agga 24 <210> 54 <211> 36 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_14_R <400> 54 gcagcactat tttgattacc tctgataatt tttcat 36 <210> 55 <211> 32 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_14_A1 <400> 55 acagttgaaa gaaattgtta tatccaacct tc 32 <210> 56 <211> 33 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_14_A2 <400> 56 gacagttgaa agaaattgtt atatccaacc tta 33 <210> 57 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_15_F <400> 57 tgacttccct tccacggtaa 20 <210> 58 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_15_R <400> 58 cccgaacaga gctacctagc a 21 <210> 59 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_15_A1 <400> 59 aggtcacact gcgctgaa 18 <210> 60 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_15_A2 <400> 60 aggtcacact gcgctgac 18 <210> 61 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_16_F <400> 61 ctggctcagt ttaattcatt gcac 24 <210> 62 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_16_R <400> 62 ccccagggca taatcctgag a 21 <210> 63 <211> 36 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_16_A1 <400> 63 tgattcttta aagctttact gtctcttttg aataaa 36 <210> 64 <211> 36 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_16_A2 <400> 64 tgattcttta aagctttact gtctcttttg aataat 36 <210> 65 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_17_F <400> 65 acaagcattg aagagtcttc ttcag 25 <210> 66 <211> 31 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_17_R <400> 66 ggtcaaataa tagcagctat ggttttagct c 31 <210> 67 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_17_A1 <400> 67 tttgatggcc tttcatgagc ac 22 <210> 68 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_17_A2 <400> 68 gtttgatggc ctttcatgag cag 23 <210> 69 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_18_F <400> 69 cagataatag agcatggtgc aca 23 <210> 70 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_18_R <400> 70 agcagcgggt tacggga 17 <210> 71 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_18_A1 <400> 71 accatatgcc tcaatcgtat ataggac 27 <210> 72 <211> 28 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_18_A2 <400> 72 caccatatgc ctcaatcgta tataggat 28 <210> 73 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_19_F <400> 73 catcaacaag attcaggaat ccca 24 <210> 74 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_19_R <400> 74 cccacactgc agctagctag t 21 <210> 75 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_19_A1 <400> 75 cccactagca ggaatcctaa ttcc 24 <210> 76 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_19_A2 <400> 76 cccactagca ggaatcctaa ttca 24 <210> 77 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_20_F <400> 77 aacttacgcg atcccctct 19 <210> 78 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_20_R <400> 78 cacgggcagc attcaactga 20 <210> 79 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_20_A1 <400> 79 cctctgcaga accaattgca atg 23 <210> 80 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_20_A2 <400> 80 cctctgcaga accaattgca ata 23 <210> 81 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_21_F <400> 81 gttgcaagca cagtgatgaa ag 22 <210> 82 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_21_R <400> 82 cagaagccac cttgttcccc 20 <210> 83 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_21_A1 <400> 83 ggtattgcag cgtttacacc a 21 <210> 84 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_21_A2 <400> 84 ggtattgcag cgtttacacc t 21 <210> 85 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_22_F <400> 85 aaccatcact acaataagca tggg 24 <210> 86 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_22_R <400> 86 cgacgaggca gcagactgta 20 <210> 87 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_22_A1 <400> 87 tgggtataac acacaatggg aataca 26 <210> 88 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_22_A2 <400> 88 tgggtataac acacaatggg aatacg 26 <210> 89 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_23_F <400> 89 gagcatggct gcgtatctg 19 <210> 90 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_23_R <400> 90 gtttgatcac tgcacgtctc catt 24 <210> 91 <211> 31 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_23_A1 <400> 91 gcaaatatta caaatatgtg ggtcatcctt t 31 <210> 92 <211> 31 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_23_A2 <400> 92 gcaaatatta caaatatgtg ggtcatcctt g 31 <210> 93 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_24_F <400> 93 gcgatttgcc tgctgcg 17 <210> 94 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_24_R <400> 94 agccatggag ctgagactga a 21 <210> 95 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_24_A1 <400> 95 ggtgaggacg tttgatccca c 21 <210> 96 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_24_A2 <400> 96 ggtgaggacg tttgatccca g 21 <210> 97 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_25_F <400> 97 agcacatccg agatcagaaa tttt 24 <210> 98 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_25_R <400> 98 tgcagcgtgt aatacttgct tttgt 25 <210> 99 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_25_A1 <400> 99 tggaacgtgc caaggcttat ataa 24 <210> 100 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_25_A2 <400> 100 tggaacgtgc caaggcttat atac 24 <210> 101 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_26_F <400> 101 acacatggca gcatgtttac g 21 <210> 102 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_26_R <400> 102 tggcacattc ccagtactgg t 21 <210> 103 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_26_A1 <400> 103 gatgtggatt gtcactttca attctct 27 <210> 104 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_26_A2 <400> 104 gatgtggatt gtcactttca attctcc 27 <210> 105 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_27_F <400> 105 gaatccggag aattatctgt ggc 23 <210> 106 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_27_R <400> 106 tcccacccga gtttcatacc c 21 <210> 107 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_27_A1 <400> 107 agctttgatt acccgacgga c 21 <210> 108 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_27_A2 <400> 108 agctttgatt acccgacgga t 21 <210> 109 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_28_F <400> 109 tccctcgttc ggccatg 17 <210> 110 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_28_R <400> 110 cagccgtgtc gaacataggg 20 <210> 111 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_28_A1 <400> 111 ccaagaagct gaaacgagtc g 21 <210> 112 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_28_A2 <400> 112 gccaagaagc tgaaacgagt ca 22 <210> 113 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_29_F <400> 113 ttgcaataaa tccagagaat gggaat 26 <210> 114 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_29_R <400> 114 gccaccggct ttgttcaca 19 <210> 115 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_29_A1 <400> 115 ttcaaccaag gatattcagg tagacag 27 <210> 116 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_29_A2 <400> 116 ttcaaccaag gatattcagg tagacaa 27 <210> 117 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_30_F <400> 117 cagcaaatgg caagtgagag a 21 <210> 118 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_30_R <400> 118 tgtctttggc tcccccagt 19 <210> 119 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_30_A1 <400> 119 aggtgtgttc ttcacaagga cc 22 <210> 120 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_30_A2 <400> 120 aggtgtgttc ttcacaagga ca 22 <210> 121 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_31_F <400> 121 cgatgaacgt atgccagaat gc 22 <210> 122 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_31_R <400> 122 gacgtacagc caagcagcc 19 <210> 123 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_31_A1 <400> 123 cagttggact catccacctt gt 22 <210> 124 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_31_A2 <400> 124 agttggactc atccaccttg c 21 <210> 125 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_32_F <400> 125 ccaactcagg taaagctaca tgaa 24 <210> 126 <211> 32 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_32_R <400> 126 agggaatgca atcataaata aagcatgaaa ca 32 <210> 127 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_32_A1 <400> 127 aactatctcc aaccattgta ggtcttc 27 <210> 128 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_32_A2 <400> 128 actatctcca accattgtag gtcttg 26 <210> 129 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_33_F <400> 129 tctgaaggag cagcagga 18 <210> 130 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_33_R <400> 130 agcactgtgt agtggacagc a 21 <210> 131 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_33_A1 <400> 131 agagttggcc gcatttcct 19 <210> 132 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_33_A2 <400> 132 gagttggccg catttccc 18 <210> 133 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_34_F <400> 133 cctctttctg ctgctgct 18 <210> 134 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_34_R <400> 134 tgtccccgtt caataccagc t 21 <210> 135 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_34_A1 <400> 135 ctccaaccat gacaagaaag cg 22 <210> 136 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_34_A2 <400> 136 ctccaaccat gacaagaaag ca 22 <210> 137 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_35_F <400> 137 gacaaatgag tcctgatatg aacca 25 <210> 138 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_35_R <400> 138 ccggtgttgc gagaatgtgt 20 <210> 139 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_35_A1 <400> 139 cagtctttac aatgtgaccg tgaatt 26 <210> 140 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_35_A2 <400> 140 cagtctttac aatgtgaccg tgaatc 26 <210> 141 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_36_F <400> 141 gccaagaagg gtcctgc 17 <210> 142 <211> 37 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_36_R <400> 142 ggaatcacta taactatctc tcatattaat ggaagca 37 <210> 143 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_36_A1 <400> 143 ctcccaaagg aaagaatgca cc 22 <210> 144 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_36_A2 <400> 144 ctcccaaagg aaagaatgca ct 22 <210> 145 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_37_F <400> 145 gcaatttcac catcaagaac aactc 25 <210> 146 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_37_R <400> 146 gctgtcaaag ggtgctgct 19 <210> 147 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_37_A1 <400> 147 ccatgcctct tccaccaaga g 21 <210> 148 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_37_A2 <400> 148 ccatgcctct tccaccaaga c 21 <210> 149 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_38_F <400> 149 gaaatggatc ctttgcagaa acc 23 <210> 150 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_38_R <400> 150 aatctgaggg ccgagcca 18 <210> 151 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_38_A1 <400> 151 ccaaacagga tcgctatgcc 20 <210> 152 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_38_A2 <400> 152 cccaaacagg atcgctatgc t 21 <210> 153 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_39_F <400> 153 gcacatcgtt gcacctgt 18 <210> 154 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_39_R <400> 154 ggaatccatg cccgcgt 17 <210> 155 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_39_A1 <400> 155 ttttctcacc gggcacgat 19 <210> 156 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_39_A2 <400> 156 ttctcaccgg gcacgac 17 <210> 157 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_40_F <400> 157 gtctgttatt tgctttaaag tccgc 25 <210> 158 <211> 37 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_40_R <400> 158 ttcactattg agcagcaaat aagttaatga tttttca 37 <210> 159 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_40_A1 <400> 159 tccgcagttg tgacattata tttgc 25 <210> 160 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_40_A2 <400> 160 gtccgcagtt gtgacattat atttgt 26 <210> 161 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_41_F <400> 161 gttctcgatc ccctcgttg 19 <210> 162 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_41_R <400> 162 ccactcccat cctctggagc 20 <210> 163 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_41_A1 <400> 163 gccagcctaa agaaaacgca tc 22 <210> 164 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_41_A2 <400> 164 gccagcctaa agaaaacgca ta 22 <210> 165 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_42_F <400> 165 tgctgcctat atgatcgact gag 23 <210> 166 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_42_R <400> 166 gcactccaat ctgcaaaagc atatca 26 <210> 167 <211> 30 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_42_A1 <400> 167 gatgatgaaa tgactgtttc ttctctgttt 30 <210> 168 <211> 29 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_42_A2 <400> 168 atgatgaaat gactgtttct tctctgttg 29 <210> 169 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_43_F <400> 169 cgcagatatt gggaagctga taa 23 <210> 170 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_43_R <400> 170 acgtccattg atgaggctgg a 21 <210> 171 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_43_A1 <400> 171 acatggtaag agatgagccc g 21 <210> 172 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_43_A2 <400> 172 ataaacatgg taagagatga gccca 25 <210> 173 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_44_F <400> 173 ggcagagagc atgtgcaata ataa 24 <210> 174 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_44_R <400> 174 aggctgcaat aggacctgaa ca 22 <210> 175 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_44_A1 <400> 175 ggtttaatgt acgccccatg tgta 24 <210> 176 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_44_A2 <400> 176 gtttaatgta cgccccatgt gtt 23 <210> 177 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_45_F <400> 177 tcgatgttcc tgataacctc agtac 25 <210> 178 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_45_R <400> 178 tgcagcaaca tagacatact gaagtga 27 <210> 179 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_45_A1 <400> 179 gactggtcag ttcacctcat tatctat 27 <210> 180 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_45_A2 <400> 180 gactggtcag ttcacctcat tatctac 27 <210> 181 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_46_F <400> 181 ggcagaatgc acgtcgattt 20 <210> 182 <211> 16 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_46_R <400> 182 ggcgatcagg cgaccg 16 <210> 183 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_46_A1 <400> 183 gaagaacgcg ctcgatttcg 20 <210> 184 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_46_A2 <400> 184 tgaagaacgc gctcgatttc a 21 <210> 185 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_47_F <400> 185 ctgtgaagga acgtcgttta gtaat 25 <210> 186 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_47_R <400> 186 acgcccaccc tttctcct 18 <210> 187 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_47_A1 <400> 187 ggtttctaca atgggtactg caac 24 <210> 188 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_47_A2 <400> 188 tggtttctac aatgggtact gcaat 25 <210> 189 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_1 <400> 189 cagtaatcgg gatcaagaag tccccggtta gggacatagt t 41 <210> 190 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_2 <400> 190 ttcaccagag ttacttcctt tgctttctga caactcatta c 41 <210> 191 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_3 <400> 191 ggtgtgcgag ggcgacctga tgaatgtcgg ggctgctgcg g 41 <210> 192 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_4 <400> 192 gatgaatatg tccatggaaa acaggatcaa tatccatgtt t 41 <210> 193 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_5 <400> 193 atggcgtaaa ctcagcatgg ctacaaagag ctttttcttg a 41 <210> 194 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_6 <400> 194 tctattgcag cagccttttt aacattggag aactcggaac t 41 <210> 195 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_7 <400> 195 aagaaagcag tggaagcaat atgccccggt gttgtctcgt g 41 <210> 196 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_8 <400> 196 catttgaaac tgatcaataa acaacttaca taagagacac c 41 <210> 197 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_9 <400> 197 tcatcaggag aaggatacac tatgttattt accaaagtga a 41 <210> 198 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_10 <400> 198 accgcaattc ctgagcttct gtgtgagttt atttatttgc g 41 <210> 199 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_11 <400> 199 tcaaattatg ttggatgaga aatgatgcag cgtccacaat g 41 <210> 200 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_12 <400> 200 gtggctgtgg atgcggtggc ggtggaggcg gaggcagtgg c 41 <210> 201 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_13 <400> 201 tctgtcttaa ctgcagattc ctccatgggc actgctgatt g 41 <210> 202 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_14 <400> 202 taatttttca taagtagttg taaggttgga tataacaatt t 41 <210> 203 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_15 <400> 203 gtaaggtcac actgcgctga caaatctgag aattccccac t 41 <210> 204 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_16 <400> 204 ttactgtctc ttttgaataa tgtttggttt cataagataa a 41 <210> 205 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_17 <400> 205 gcagctatgg ttttagctct ctgctcatga aaggccatca a 41 <210> 206 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_18 <400> 206 tgcctcaatc gtatatagga taaccagaat gagatcatac a 41 <210> 207 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_19 <400> 207 actagcagga atcctaattc atagccttta aacgacttga c 41 <210> 208 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_20 <400> 208 tctgcagaac caattgcaat actcattctc cgcttccaat c 41 <210> 209 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_21 <400> 209 ggtattgcag cgtttacacc ttcgccaagg ggaacaaggt g 41 <210> 210 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_22 <400> 210 ataacacaca atgggaatac gaaaggaatt actccgtaat a 41 <210> 211 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_23 <400> 211 caaatatgtg ggtcatcctt gcatttgtac gacaaagatc a 41 <210> 212 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_24 <400> 212 ggtgaggacg tttgatccca gagagatttc tggagaattt a 41 <210> 213 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_25 <400> 213 aattaatttt gaagaaatat gtatataagc cttggcacgt t 41 <210> 214 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_26 <400> 214 ttcgtcatac catataggat ggagaattga aagtgacaat c 41 <210> 215 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_27 <400> 215 agctttgatt acccgacgga tacattgctg ccgggtatga a 41 <210> 216 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_28 <400> 216 ccaagaagct gaaacgagtc actgctcatc tcggccacga a 41 <210> 217 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_29 <400> 217 caaggatatt caggtagaca aggatcagtc agttgctcag t 41 <210> 218 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_30 <400> 218 tttggctccc ccagtttagc tgtccttgtg aagaacacac c 41 <210> 219 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_31 <400> 219 cctttgggca gtgcatcgga gcaaggtgga tgagtccaac t 41 <210> 220 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_32 <400> 220 ctccaaccat tgtaggtctt gctgtttcat gctttattta t 41 <210> 221 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_33 <400> 221 agcattgatc attaactcgt gggaaatgcg gccaactctt t 41 <210> 222 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_34 <400> 222 atctttagct tgttagatag tgctttcttg tcatggttgg a 41 <210> 223 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_35 <400> 223 tcagtttcag aggttgaagt gattcacggt cacattgtaa a 41 <210> 224 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_36 <400> 224 tcatattaat ggaagcagta agtgcattct ttcctttggg a 41 <210> 225 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_37 <400> 225 gctgctgaga actatcatcg gtcttggtgg aagaggcatg g 41 <210> 226 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_38 <400> 226 cccaaacagg atcgctatgc tctccgcact tctccccaat g 41 <210> 227 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_39 <400> 227 taccgctgca cggtggcgcc gtcgtgcccg gtgagaaaac a 41 <210> 228 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_40 <400> 228 cagttgtgac attatatttg taatgaaaaa tcattaactt a 41 <210> 229 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_41 <400> 229 ccagcctaaa gaaaacgcat agagcacaag gcttcaccga g 41 <210> 230 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_42 <400> 230 atgactgttt cttctctgtt gtgctgtgat tatattcatt c 41 <210> 231 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_43 <400> 231 acatggtaag agatgagccc aacactgcaa ccctcaacga c 41 <210> 232 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_44 <400> 232 ttaatgtacg ccccatgtgt ttgttcaggt cctattgcag c 41 <210> 233 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_45 <400> 233 gaagtgaaaa tcaatacgaa gtagataatg aggtgaactg a 41 <210> 234 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_46 <400> 234 gcgatcaggc gaccgccgcg tgaaatcgag cgcgttcttc a 41 <210> 235 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_47 <400> 235 ttctacaatg ggtactgcaa tggttatgca tatgaagaag a 41 <110> Republic of Korea <120> SNP molecular marker for selecting cultivars of sweet potatoes and uses <130> P17R12C1235 <160> 235 <170> KoPatentin 3.0 <210> 1 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_1_F <400> 1 acagacctca ggattgggaa aa 22 <210> 2 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_1_R <400> 2 catagagctc gggagggca 19 <210> 3 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_1_A1 <400> 3 ctatgtccct aaccggggc 19 <210> 4 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_1_A2 <400> 4 actatgtccc taaccgggga 20 <210> 5 <211> 16 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_2_F <400> 5 atgcgcgcaa gcagtt 16 <210> 6 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_2_R <400> 6 accacggaag cagctaggat 20 <210> 7 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_2_A1 <400> 7 gccttcacca gagttacttc cttc 24 <210> 8 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_2_A2 <400> 8 gccttcacca gagttacttc cttt 24 <210> 9 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_3_F <400> 9 cgtctcttct ttccccgca 19 <210> 10 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_3_R <400> 10 ctggaactcc ggtagggtac t 21 <210> 11 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_3_A1 <400> 11 agcagccccg acattcg 17 <210> 12 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_3_A2 <400> 12 agcagccccg acattca 17 <210> 13 <211> 16 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_4_F <400> 13 gcgaagttcc agcccc 16 <210> 14 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_4_R <400> 14 gcagcctgtg ttgcatgtgt 20 <210> 15 <211> 28 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_4_A1 <400> 15 gtgtcatgat gaatatgtcc atggaaac 28 <210> 16 <211> 28 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_4_A2 <400> 16 gtgtcatgat gaatatgtcc atggaaaa 28 <210> 17 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_5_F <400> 17 tcgtttcttc tcctgaacgg a 21 <210> 18 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_5_R <400> 18 cacgagtgcc actgaaatcc aa 22 <210> 19 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_5_A1 <400> 19 tggcgtaaac tcagcatggt 20 <210> 20 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_5_A2 <400> 20 ggcgtaaact cagcatggc 19 <210> 21 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_6_F <400> 21 caaacttgtt ttggaaagag cgtc 24 <210> 22 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_6_R <400> 22 ttggtggcct gagttccga 19 <210> 23 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_6_A1 <400> 23 cgtaatctat tgcagcagcc tttttc 26 <210> 24 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_6_A2 <400> 24 cgtaatctat tgcagcagcc ttttta 26 <210> 25 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_7_F <400> 25 cctgcatgca ttctatgtca ttgat 25 <210> 26 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_7_R <400> 26 cagcacacga gacaacaccg 20 <210> 27 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_7_A1 <400> 27 gcaaagaaag cagtggaagc aatc 24 <210> 28 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_7_A2 <400> 28 gcaaagaaag cagtggaagc aata 24 <210> 29 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_8_F <400> 29 ccgcagctat acatggttga a 21 <210> 30 <211> 32 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_8_R <400> 30 caaacaggat ggaatcattt gaaactgatc aa 32 <210> 31 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_8_A1 <400> 31 cttcaggtgt ctcttatgta agttgc 26 <210> 32 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_8_A2 <400> 32 ccttcaggtg tctcttatgt aagttgt 27 <210> 33 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_9_F <400> 33 agcatgggat gagatagatc caa 23 <210> 34 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_9_R <400> 34 cttggaggct gcaatctgct 20 <210> 35 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_9_A1 <400> 35 ccaaaatcat caggagaagg atacaca 27 <210> 36 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_9_A2 <400> 36 ccaaaatcat caggagaagg atacact 27 <210> 37 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_10_F <400> 37 cacaacggat aacaaaagaa agagga 26 <210> 38 <211> 33 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_10_R <400> 38 gatgatctgt acacgcaaat aaataaactc aca 33 <210> 39 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_10_A1 <400> 39 aaccgcaatt cctgagcttc ta 22 <210> 40 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_10_A2 <400> 40 ccgcaattcc tgagcttctg 20 <210> 41 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_11_F <400> 41 tgttagaaca tgttttgacc agca 24 <210> 42 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_11_R <400> 42 ctcgttgcat tgtggacgct 20 <210> 43 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_11_A1 <400> 43 ccgtctcaaa ttatgttgga tgagag 26 <210> 44 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_11_A2 <400> 44 accgtctcaa attatgttgg atgagaa 27 <210> 45 <211> 15 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_12_F <400> 45 cgcagccact gcctc 15 <210> 46 <211> 16 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_12_R <400> 46 gcggcagggt tgcgaa 16 <210> 47 <211> 15 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_12_A1 <400> 47 gcctccgcct ccacg 15 <210> 48 <211> 15 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_12_A2 <400> 48 gcctccgcct ccacc 15 <210> 49 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_13_F <400> 49 gctgaagaaa gcaagcagca 20 <210> 50 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_13_R <400> 50 agtgaatcgg ctccgaagga a 21 <210> 51 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_13_A1 <400> 51 atcagcagtg cccatggat 19 <210> 52 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_13_A2 <400> 52 tcagcagtgc ccatggag 18 <210> 53 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_14_F <400> 53 cctctgtgca ataaaatcag agga 24 <210> 54 <211> 36 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_14_R <400> 54 gcagcactat tttgattacc tctgataatt tttcat 36 <210> 55 <211> 32 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_14_A1 <400> 55 acagttgaaa gaaattgtta tatccaacct tc 32 <210> 56 <211> 33 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_14_A2 <400> 56 gacagttgaa agaaattgtt atatccaacc tta 33 <210> 57 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_15_F <400> 57 tgacttccct tccacggtaa 20 <210> 58 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_15_R <400> 58 cccgaacaga gctacctagc a 21 <210> 59 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_15_A1 <400> 59 aggtcacact gcgctgaa 18 <210> 60 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_15_A2 <400> 60 aggtcacact gcgctgac 18 <210> 61 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_16_F <400> 61 ctggctcagt ttaattcatt gcac 24 <210> 62 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_16_R <400> 62 ccccagggca taatcctgag a 21 <210> 63 <211> 36 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_16_A1 <400> 63 tgattcttta aagctttact gtctcttttg aataaa 36 <210> 64 <211> 36 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_16_A2 <400> 64 tgattcttta aagctttact gtctcttttg aataat 36 <210> 65 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_17_F <400> 65 acaagcattg aagagtcttc ttcag 25 <210> 66 <211> 31 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_17_R <400> 66 ggtcaaataa tagcagctat ggttttagct c 31 <210> 67 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_17_A1 <400> 67 tttgatggcc tttcatgagc ac 22 <210> 68 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_17_A2 <400> 68 gtttgatggc ctttcatgag cag 23 <210> 69 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_18_F <400> 69 cagataatag agcatggtgc aca 23 <210> 70 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_18_R <400> 70 agcagcgggt tacggga 17 <210> 71 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_18_A1 <400> 71 accatatgcc tcaatcgtat ataggac 27 <210> 72 <211> 28 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_18_A2 <400> 72 caccatatgc ctcaatcgta tataggat 28 <210> 73 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_19_F <400> 73 catcaacaag attcaggaat ccca 24 <210> 74 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_19_R <400> 74 cccacactgc agctagctag t 21 <210> 75 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_19_A1 <400> 75 cccactagca ggaatcctaa ttcc 24 <210> 76 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_19_A2 <400> 76 cccactagca ggaatcctaa ttca 24 <210> 77 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_20_F <400> 77 aacttacgcg atcccctct 19 <210> 78 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_20_R <400> 78 cacgggcagc attcaactga 20 <210> 79 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_20_A1 <400> 79 cctctgcaga accaattgca atg 23 <210> 80 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_20_A2 <400> 80 cctctgcaga accaattgca ata 23 <210> 81 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_21_F <400> 81 gttgcaagca cagtgatgaa ag 22 <210> 82 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_21_R <400> 82 cagaagccac cttgttcccc 20 <210> 83 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_21_A1 <400> 83 ggtattgcag cgtttacacc a 21 <210> 84 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_21_A2 <400> 84 ggtattgcag cgtttacacc t 21 <210> 85 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_22_F <400> 85 aaccatcact acaataagca tggg 24 <210> 86 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_22_R <400> 86 cgacgaggca gcagactgta 20 <210> 87 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_22_A1 <400> 87 tgggtataac acacaatggg aataca 26 <210> 88 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_22_A2 <400> 88 tgggtataac acacaatggg aatacg 26 <210> 89 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_23_F <400> 89 gagcatggct gcgtatctg 19 <210> 90 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_23_R <400> 90 gtttgatcac tgcacgtctc catt 24 <210> 91 <211> 31 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_23_A1 <400> 91 gcaaatatta caaatatgtg ggtcatcctt t 31 <210> 92 <211> 31 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_23_A2 <400> 92 gcaaatatta caaatatgtg ggtcatcctt g 31 <210> 93 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_24_F <400> 93 gcgatttgcc tgctgcg 17 <210> 94 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_24_R <400> 94 agccatggag ctgagactga a 21 <210> 95 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_24_A1 <400> 95 ggtgaggacg tttgatccca c 21 <210> 96 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_24_A2 <400> 96 ggtgaggacg tttgatccca g 21 <210> 97 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_25_F <400> 97 agcacatccg agatcagaaa tttt 24 <210> 98 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_25_R <400> 98 tgcagcgtgt aatacttgct tttgt 25 <210> 99 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_25_A1 <400> 99 tggaacgtgc caaggcttat ataa 24 <210> 100 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_25_A2 <400> 100 tggaacgtgc caaggcttat atac 24 <210> 101 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_26_F <400> 101 acacatggca gcatgtttac g 21 <210> 102 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_26_R <400> 102 tggcacattc ccagtactgg t 21 <210> 103 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_26_A1 <400> 103 gatgtggatt gtcactttca attctct 27 <210> 104 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_26_A2 <400> 104 gatgtggatt gtcactttca attctcc 27 <210> 105 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_27_F <400> 105 gaatccggag aattatctgt ggc 23 <210> 106 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_27_R <400> 106 tcccacccga gtttcatacc c 21 <210> 107 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_27_A1 <400> 107 agctttgatt acccgacgga c 21 <210> 108 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_27_A2 <400> 108 agctttgatt acccgacgga t 21 <210> 109 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_28_F <400> 109 tccctcgttc ggccatg 17 <210> 110 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_28_R <400> 110 cagccgtgtc gaacataggg 20 <210> 111 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_28_A1 <400> 111 ccaagaagct gaaacgagtc g 21 <210> 112 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_28_A2 <400> 112 gccaagaagc tgaaacgagt ca 22 <210> 113 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_29_F <400> 113 ttgcaataaa tccagagaat gggaat 26 <210> 114 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_29_R <400> 114 gccaccggct ttgttcaca 19 <210> 115 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_29_A1 <400> 115 ttcaaccaag gatattcagg tagacag 27 <210> 116 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_29_A2 <400> 116 ttcaaccaag gatattcagg tagacaa 27 <210> 117 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_30_F <400> 117 cagcaaatgg caagtgagag a 21 <210> 118 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_30_R <400> 118 tgtctttggc tcccccagt 19 <210> 119 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_30_A1 <400> 119 aggtgtgttc ttcacaagga cc 22 <210> 120 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_30_A2 <400> 120 aggtgtgttc ttcacaagga ca 22 <210> 121 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_31_F <400> 121 cgatgaacgt atgccagaat gc 22 <210> 122 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_31_R <400> 122 gacgtacagc caagcagcc 19 <210> 123 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_31_A1 <400> 123 cagttggact catccacctt gt 22 <210> 124 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_31_A2 <400> 124 agttggactc atccaccttg c 21 <210> 125 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_32_F <400> 125 ccaactcagg taaagctaca tgaa 24 <210> 126 <211> 32 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_32_R <400> 126 agggaatgca atcataaata aagcatgaaa ca 32 <210> 127 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_32_A1 <400> 127 aactatctcc aaccattgta ggtcttc 27 <210> 128 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_32_A2 <400> 128 actatctcca accattgtag gtcttg 26 <210> 129 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_33_F <400> 129 tctgaaggag cagcagga 18 <210> 130 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_33_R <400> 130 agcactgtgt agtggacagc a 21 <210> 131 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_33_A1 <400> 131 agagttggcc gcatttcct 19 <210> 132 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_33_A2 <400> 132 gagttggccg catttccc 18 <210> 133 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_34_F <400> 133 cctctttctg ctgctgct 18 <210> 134 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_34_R <400> 134 tgtccccgtt caataccagc t 21 <210> 135 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_34_A1 <400> 135 ctccaaccat gacaagaaag cg 22 <210> 136 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_34_A2 <400> 136 ctccaaccat gacaagaaag ca 22 <210> 137 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_35_F <400> 137 gacaaatgag tcctgatatg aacca 25 <210> 138 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_35_R <400> 138 ccggtgttgc gagaatgtgt 20 <210> 139 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_35_A1 <400> 139 cagtctttac aatgtgaccg tgaatt 26 <210> 140 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_35_A2 <400> 140 cagtctttac aatgtgaccg tgaatc 26 <210> 141 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_36_F <400> 141 gccaagaagg gtcctgc 17 <210> 142 <211> 37 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_36_R <400> 142 ggaatcacta taactatctc tcatattaat ggaagca 37 <210> 143 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_36_A1 <400> 143 ctcccaaagg aaagaatgca cc 22 <210> 144 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_36_A2 <400> 144 ctcccaaagg aaagaatgca ct 22 <210> 145 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_37_F <400> 145 gcaatttcac catcaagaac aactc 25 <210> 146 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_37_R <400> 146 gctgtcaaag ggtgctgct 19 <210> 147 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_37_A1 <400> 147 ccatgcctct tccaccaaga g 21 <210> 148 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_37_A2 <400> 148 ccatgcctct tccaccaaga c 21 <210> 149 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_38_F <400> 149 gaaatggatc ctttgcagaa acc 23 <210> 150 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_38_R <400> 150 aatctgaggg ccgagcca 18 <210> 151 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_38_A1 <400> 151 ccaaacagga tcgctatgcc 20 <210> 152 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_38_A2 <400> 152 cccaaacagg atcgctatgc t 21 <210> 153 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_39_F <400> 153 gcacatcgtt gcacctgt 18 <210> 154 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_39_R <400> 154 ggaatccatg cccgcgt 17 <210> 155 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_39_A1 <400> 155 ttttctcacc gggcacgat 19 <210> 156 <211> 17 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_39_A2 <400> 156 ttctcaccgg gcacgac 17 <210> 157 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_40_F <400> 157 gtctgttatt tgctttaaag tccgc 25 <210> 158 <211> 37 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_40_R <400> 158 ttcactattg agcagcaaat aagttaatga tttttca 37 <210> 159 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_40_A1 <400> 159 tccgcagttg tgacattata tttgc 25 <210> 160 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_40_A2 <400> 160 gtccgcagtt gtgacattat atttgt 26 <210> 161 <211> 19 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_41_F <400> 161 gttctcgatc ccctcgttg 19 <210> 162 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_41_R <400> 162 ccactcccat cctctggagc 20 <210> 163 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_41_A1 <400> 163 gccagcctaa agaaaacgca tc 22 <210> 164 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_41_A2 <400> 164 gccagcctaa agaaaacgca ta 22 <210> 165 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_42_F <400> 165 tgctgcctat atgatcgact gag 23 <210> 166 <211> 26 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_42_R <400> 166 gcactccaat ctgcaaaagc atatca 26 <210> 167 <211> 30 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_42_A1 <400> 167 gatgatgaaa tgactgtttc ttctctgttt 30 <210> 168 <211> 29 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_42_A2 <400> 168 atgatgaaat gactgtttct tctctgttg 29 <210> 169 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_43_F <400> 169 cgcagatatt gggaagctga taa 23 <210> 170 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_43_R <400> 170 acgtccattg atgaggctgg a 21 <210> 171 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_43_A1 <400> 171 acatggtaag agatgagccc g 21 <210> 172 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_43_A2 <400> 172 ataaacatgg taagagatga gccca 25 <210> 173 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_44_F <400> 173 ggcagagagc atgtgcaata ataa 24 <210> 174 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_44_R <400> 174 aggctgcaat aggacctgaa ca 22 <210> 175 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_44_A1 <400> 175 ggtttaatgt acgccccatg tgta 24 <210> 176 <211> 23 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_44_A2 <400> 176 gtttaatgta cgccccatgt gtt 23 <210> 177 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_45_F <400> 177 tcgatgttcc tgataacctc agtac 25 <210> 178 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_45_R <400> 178 tgcagcaaca tagacatact gaagtga 27 <210> 179 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_45_A1 <400> 179 gactggtcag ttcacctcat tatctat 27 <210> 180 <211> 27 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_45_A2 <400> 180 gactggtcag ttcacctcat tatctac 27 <210> 181 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_46_F <400> 181 ggcagaatgc acgtcgattt 20 <210> 182 <211> 16 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_46_R <400> 182 ggcgatcagg cgaccg 16 <210> 183 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_46_A1 <400> 183 gaagaacgcg ctcgatttcg 20 <210> 184 <211> 21 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_46_A2 <400> 184 tgaagaacgc gctcgatttc a 21 <210> 185 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_47_F <400> 185 ctgtgaagga acgtcgttta gtaat 25 <210> 186 <211> 18 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_47_R <400> 186 acgcccaccc tttctcct 18 <210> 187 <211> 24 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_47_A1 <400> 187 ggtttctaca atgggtactg caac 24 <210> 188 <211> 25 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_47_A2 <400> 188 tggtttctac aatgggtact gcaat 25 <210> 189 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_1 <400> 189 cagtaatcgg gatcaagaag tccccggtta gggacatagt t 41 <210> 190 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_2 <400> 190 ttcaccagag ttacttcctt tgctttctga caactcatta c 41 <210> 191 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_3 <400> 191 ggtgtgcgag ggcgacctga tgaatgtcgg ggctgctgcg g 41 <210> 192 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_4 <400> 192 gatgaatatg tccatggaaa acaggatcaa tatccatgtt t 41 <210> 193 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_5 <400> 193 atggcgtaaa ctcagcatgg ctacaaagag ctttttcttg a 41 <210> 194 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_6 <400> 194 tctattgcag cagccttttt aacattggag aactcggaac t 41 <210> 195 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_7 <400> 195 aagaaagcag tggaagcaat atgccccggt gttgtctcgt g 41 <210> 196 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_8 <400> 196 catttgaaac tgatcaataa acaacttaca taagagacac c 41 <210> 197 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_9 <400> 197 tcatcaggag aaggatacac tatgttattt accaaagtga a 41 <210> 198 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_10 <400> 198 accgcaattc ctgagcttct gtgtgagttt atttatttgc g 41 <210> 199 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_11 <400> 199 tcaaattatg ttggatgaga aatgatgcag cgtccacaat g 41 <210> 200 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_12 <400> 200 gtggctgtgg atgcggtggc ggtggaggcg gaggcagtgg c 41 <210> 201 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_13 <400> 201 tctgtcttaa ctgcagattc ctccatgggc actgctgatt g 41 <210> 202 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_14 <400> 202 taatttttca taagtagttg taaggttgga tataacaatt t 41 <210> 203 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_15 <400> 203 gtaaggtcac actgcgctga caaatctgag aattccccac t 41 <210> 204 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_16 <400> 204 ttactgtctc ttttgaataa tgtttggttt cataagataa a 41 <210> 205 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_17 <400> 205 gcagctatgg ttttagctct ctgctcatga aaggccatca a 41 <210> 206 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_18 <400> 206 tgcctcaatc gtatatagga taaccagaat gagatcatac a 41 <210> 207 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_19 <400> 207 actagcagga atcctaattc atagccttta aacgacttga c 41 <210> 208 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_20 <400> 208 tctgcagaac caattgcaat actcattctc cgcttccaat c 41 <210> 209 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_21 <400> 209 ggtattgcag cgtttacacc ttcgccaagg ggaacaaggt g 41 <210> 210 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_22 <400> 210 ataacacaca atgggaatac gaaaggaatt actccgtaat a 41 <210> 211 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_23 <400> 211 caaatatgtg ggtcatcctt gcatttgtac gacaaagatc a 41 <210> 212 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_24 <400> 212 ggtgaggacg tttgatccca gagagatttc tggagaattt a 41 <210> 213 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_25 <400> 213 aattaatttt gaagaaatat gtatataagc cttggcacgt t 41 <210> 214 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_26 <400> 214 ttcgtcatac catataggat ggagaattga aagtgacaat c 41 <210> 215 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_27 <400> 215 agctttgatt acccgacgga tacattgctg ccgggtatga a 41 <210> 216 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_28 <400> 216 ccaagaagct gaaacgagtc actgctcatc tcggccacga a 41 <210> 217 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_29 <400> 217 caaggatatt caggtagaca aggatcagtc agttgctcag t 41 <210> 218 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_30 <400> 218 tttggctccc ccagtttagc tgtccttgtg aagaacacac c 41 <210> 219 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_31 <400> 219 cctttgggca gtgcatcgga gcaaggtgga tgagtccaac t 41 <210> 220 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_32 <400> 220 ctccaaccat tgtaggtctt gctgtttcat gctttattta t 41 <210> 221 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_33 <400> 221 agcattgatc attaactcgt gggaaatgcg gccaactctt t 41 <210> 222 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_34 <400> 222 atctttagct tgttagatag tgctttcttg tcatggttgg a 41 <210> 223 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_35 <400> 223 tcagtttcag aggttgaagt gattcacggt cacattgtaa a 41 <210> 224 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_36 <400> 224 tcatattaat ggaagcagta agtgcattct ttcctttggg a 41 <210> 225 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_37 <400> 225 gctgctgaga actatcatcg gtcttggtgg aagaggcatg g 41 <210> 226 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_38 <400> 226 cccaaacagg atcgctatgc tctccgcact tctccccaat g 41 <210> 227 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_39 <400> 227 taccgctgca cggtggcgcc gtcgtgcccg gtgagaaaac a 41 <210> 228 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_40 <400> 228 cagttgtgac attatatttg taatgaaaaa tcattaactt a 41 <210> 229 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_41 <400> 229 ccagcctaaa gaaaacgcat agagcacaag gcttcaccga g 41 <210> 230 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_42 <400> 230 atgactgttt cttctctgtt gtgctgtgat tatattcatt c 41 <210> 231 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_43 <400> 231 acatggtaag agatgagccc aacactgcaa ccctcaacga c 41 <210> 232 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_44 <400> 232 ttaatgtacg ccccatgtgt ttgttcaggt cctattgcag c 41 <210> 233 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_45 <400> 233 gaagtgaaaa tcaatacgaa gtagataatg aggtgaactg a 41 <210> 234 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_46 <400> 234 gcgatcaggc gaccgccgcg tgaaatcgag cgcgttcttc a 41 <210> 235 <211> 41 <212> DNA <213> Artificial Sequence <220> <223> SP_SNP_47 <400> 235 ttctacaatg ggtactgcaa tggttatgca tatgaagaag a 41
Claims (5)
[표 1]
A primer set represented by SEQ ID NOS: 1, 2, 3, and 4; A primer set represented by SEQ ID NOS: 5, 6, 7, and 8; A primer set represented by SEQ ID NOS: 9, 10, 11, and 12; A primer set represented by SEQ ID NOs: 13, 14, 15, and 16; A primer set represented by SEQ ID NOs: 17, 18, 19, and 20; A primer set represented by SEQ ID NOS: 21, 22, 23 and 24; A primer set represented by SEQ ID NOS: 25, 26, 27, and 28; A primer set represented by SEQ ID NOs: 29, 30, 31, and 32; A primer set represented by SEQ ID NOS: 33, 34, 35, and 36; A primer set represented by SEQ ID NOS: 37, 38, 39, and 40; A primer set represented by SEQ ID NOS: 41, 42, 43, and 44; A primer set represented by SEQ ID NOS: 45, 46, 47, and 48; A primer set represented by SEQ ID NOS: 49, 50, 51, and 52; A primer set represented by SEQ ID NOS: 53, 54, 55, and 56; A primer set represented by SEQ ID NOS: 57, 58, 59, and 60; A primer set represented by SEQ ID NOS: 61, 62, 63 and 64; A primer set represented by SEQ ID NOS: 65, 66, 67, and 68; A primer set represented by SEQ ID NOS: 69, 70, 71 and 72; A primer set represented by SEQ ID NOS: 73, 74, 75, and 76; A primer set represented by SEQ ID NOS: 77, 78, 79 and 80; A primer set represented by SEQ ID NOS: 81, 82, 83 and 84; A primer set represented by SEQ ID NOS: 85, 86, 87 and 88; A primer set represented by SEQ ID NOS: 89, 90, 91, and 92; A primer set represented by SEQ ID NOS: 93, 94, 95, and 96; A primer set represented by SEQ ID NOS: 97, 98, 99 and 100; A primer set represented by SEQ ID NOS: 101, 102, 103, and 104; A primer set represented by SEQ ID NOS: 105, 106, 107, and 108; A primer set represented by SEQ ID NOS: 109, 110, 111, and 112; A primer set represented by SEQ ID NOS: 113, 114, 115, and 116; A primer set represented by SEQ ID NOS: 117, 118, 119, and 120; A primer set represented by SEQ ID NOS: 121, 122, 123, and 124; A primer set represented by SEQ ID NOS: 125, 126, 127, and 128; A primer set represented by SEQ ID NOs: 129, 130, 131, and 132; A primer set represented by SEQ ID NOS: 133, 134, 135, and 136; A primer set represented by SEQ ID NOS: 137, 138, 139, and 140; A primer set represented by SEQ ID NOS: 141, 142, 143 and 144; A primer set represented by SEQ ID NOS: 145, 146, 147, and 148; A primer set represented by SEQ ID NOs: 149, 150, 151, and 152; A primer set represented by SEQ ID NOS: 153, 154, 155, and 156; A primer set represented by SEQ ID NOS: 157, 158, 159, and 160; A primer set represented by SEQ ID NOS: 161, 162, 163, and 164; A primer set represented by SEQ ID NOS: 165, 166, 167, and 168; A primer set represented by SEQ ID NOs: 169, 170, 171, and 172; A primer set represented by SEQ ID NOS: 173, 174, 175, and 176; A primer set represented by SEQ ID NOS: 177, 178, 179, and 180; A primer set represented by SEQ ID NOS: 181, 182, 183, and 184; And a primer set represented by SEQ ID NOS: 185, 186, 187, and 188; and a primer set for discriminating 67 kinds of sweet potato varieties shown in Table 1 below;
[Table 1]
A kit for distinguishing sweet potato varieties comprising the primer set of claim 1 and a reagent for carrying out an amplification reaction.
3. The kit according to claim 2, wherein the reagent for carrying out the amplification reaction comprises a DNA polymerase, dNTPs and a buffer.
b) 상기 분리된 게놈 DNA를 주형으로 하고, 제1항의 프라이머 세트를 이용하여 PCR(polymerase chain reaction)을 수행하는 단계; 및
c) 상기 b) 단계의 증폭된 PCR 산물을 검출하는 단계;를 포함하는, 고구마 품종 판별 방법.
a) separating the genomic DNA from the sweet potato sample;
b) performing PCR (polymerase chain reaction) using the separated genomic DNA as a template and using the primer set of claim 1; And
and c) detecting the amplified PCR product of step b).
Priority Applications (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| KR1020170148336A KR101961653B1 (en) | 2017-11-08 | 2017-11-08 | SNP molecular marker for selecting cultivars of sweet potatoes and uses thereof |
Applications Claiming Priority (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| KR1020170148336A KR101961653B1 (en) | 2017-11-08 | 2017-11-08 | SNP molecular marker for selecting cultivars of sweet potatoes and uses thereof |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| KR101961653B1 true KR101961653B1 (en) | 2019-03-26 |
Family
ID=65949668
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| KR1020170148336A KR101961653B1 (en) | 2017-11-08 | 2017-11-08 | SNP molecular marker for selecting cultivars of sweet potatoes and uses thereof |
Country Status (1)
| Country | Link |
|---|---|
| KR (1) | KR101961653B1 (en) |
Cited By (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| KR102198566B1 (en) * | 2019-09-30 | 2021-01-05 | 서울시립대학교 산학협력단 | Tetra primers ARMS-PCR molecular marker for discriminating cultivars of sweet potato and uses thereof |
| KR20210045755A (en) * | 2019-10-17 | 2021-04-27 | 대한민국(농촌진흥청장) | SNP marker for discriminating cultivars of sweet potatoes and method for discriminating cultivars using the marker |
| CN115976251A (en) * | 2022-09-24 | 2023-04-18 | 中国热带农业科学院热带作物品种资源研究所 | InDel marker for cassava genetic diversity analysis and application thereof |
Citations (4)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| KR20060023378A (en) * | 2004-09-09 | 2006-03-14 | 대한민국(강원대학교 총장) | Method for producing a marker for detecting polymorphism in a plant |
| KR20110108809A (en) * | 2010-03-29 | 2011-10-06 | 고려대학교 산학협력단 | Sweetpotato srd1 cdna and high-numbered transgenic plants using the same |
| KR20130075986A (en) * | 2011-12-28 | 2013-07-08 | 대한민국(관리부서:농촌진흥청장) | Ssr primer for the identification of sweet potatoes and method for identifying sweet potatoes |
| KR101796260B1 (en) * | 2016-06-01 | 2017-11-13 | 순천대학교 산학협력단 | SNP marker set for identifying strawberry cultivars and method for identifying strawberry cultivars using the same |
-
2017
- 2017-11-08 KR KR1020170148336A patent/KR101961653B1/en active IP Right Grant
Patent Citations (5)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| KR20060023378A (en) * | 2004-09-09 | 2006-03-14 | 대한민국(강원대학교 총장) | Method for producing a marker for detecting polymorphism in a plant |
| KR20110108809A (en) * | 2010-03-29 | 2011-10-06 | 고려대학교 산학협력단 | Sweetpotato srd1 cdna and high-numbered transgenic plants using the same |
| KR20130075986A (en) * | 2011-12-28 | 2013-07-08 | 대한민국(관리부서:농촌진흥청장) | Ssr primer for the identification of sweet potatoes and method for identifying sweet potatoes |
| KR101403494B1 (en) | 2011-12-28 | 2014-06-10 | 대한민국 | SSR Primer for the Identification of Sweet Potatoes and Method for Identifying Sweet Potatoes |
| KR101796260B1 (en) * | 2016-06-01 | 2017-11-13 | 순천대학교 산학협력단 | SNP marker set for identifying strawberry cultivars and method for identifying strawberry cultivars using the same |
Non-Patent Citations (1)
| Title |
|---|
| 박권우 외, 한국원예학회지, 5:112-113, 1996 |
Cited By (5)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| KR102198566B1 (en) * | 2019-09-30 | 2021-01-05 | 서울시립대학교 산학협력단 | Tetra primers ARMS-PCR molecular marker for discriminating cultivars of sweet potato and uses thereof |
| KR20210045755A (en) * | 2019-10-17 | 2021-04-27 | 대한민국(농촌진흥청장) | SNP marker for discriminating cultivars of sweet potatoes and method for discriminating cultivars using the marker |
| KR102267332B1 (en) | 2019-10-17 | 2021-06-21 | 대한민국 | SNP marker for discriminating cultivars of sweet potatoes and method for discriminating cultivars using the marker |
| CN115976251A (en) * | 2022-09-24 | 2023-04-18 | 中国热带农业科学院热带作物品种资源研究所 | InDel marker for cassava genetic diversity analysis and application thereof |
| CN115976251B (en) * | 2022-09-24 | 2023-06-30 | 中国热带农业科学院热带作物品种资源研究所 | InDel marker for cassava genetic diversity analysis and application thereof |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| KR101331740B1 (en) | SSR primer derived from Paeonia lactiflora and use thereof | |
| KR101954673B1 (en) | Molecular marker for discriminating Codonopsis lanceolata cultivars and uses thereof | |
| KR101961653B1 (en) | SNP molecular marker for selecting cultivars of sweet potatoes and uses thereof | |
| KR101976974B1 (en) | SSR molecular markers for discriminating grape cultivars and uses thereof | |
| KR101778877B1 (en) | Markers for discrimination of Whangkeumbae and Minibae | |
| KR102009712B1 (en) | Primer set for discriminating Zizyphus jujuba 'Bokjo' cultivar and uses thereof | |
| KR20160082292A (en) | Molecular Markers for Selecting radish genetic resources and use thereof | |
| KR102335806B1 (en) | Molecular marker based on chloroplast genome sequence for discriminating Zizyphus jujuba 'SanJo' cultivar and uses thereof | |
| KR102126809B1 (en) | SSR molecular markers for discriminating of Angelica gigas Nakai and uses thereof | |
| KR102296889B1 (en) | Molecular marker for selecting Gray leaf spot resistant or susceptible tomato cultivar and selection method using the same molecular marker | |
| KR102174274B1 (en) | Molecular marker for discriminating Zizyphus jujuba 'Boeun' and 'Chuseok' cultivar and uses thereof | |
| KR102267332B1 (en) | SNP marker for discriminating cultivars of sweet potatoes and method for discriminating cultivars using the marker | |
| KR101779030B1 (en) | Molecular marker for selecting plant with enhanced zinc content and uses thereof | |
| CN113637790B (en) | KASP molecular marker of stripe rust resistance gene YrAS2388R, primer, kit and application | |
| KR102309663B1 (en) | Fluidigm based SNP chip for genotype-identifying and classifying Panax ginseng cultivar or resource and uses thereof | |
| KR102019966B1 (en) | Specific SSR primer for discrimination of Astragalus membransis and uses thereof | |
| KR102159634B1 (en) | Marker derived from complete sequencing of chloroplast genome of three Sanguisorba species, primer set for discrimination of Sanguisorba species and uses thereof | |
| KR102380784B1 (en) | Molecular marker for discriminating genetic resources of Peucedanum japonicum and uses thereof | |
| KR102313439B1 (en) | Molecular marker for distinguishing Gentina macrophylla, Aconitum pseudolaeve and Aconitum longecassidatum | |
| KR102164175B1 (en) | Marker derived from complete sequencing of chloroplast genome of Cynanchum wilfordii and Cynanchum auriculatum, primer set for discrimination of Cynanchum species and uses thereof | |
| KR102235439B1 (en) | Marker derived from complete sequencing of chloroplast genome of Cynanchum wilfordii and Cynanchum auriculatum, primer set for discrimination of Cynanchum species and uses thereof | |
| KR102030608B1 (en) | Chloroplast genome sequence-derived primer set for discriminating Korean spring orchid and uses thereof | |
| KR102451456B1 (en) | Primer set for discriminating genetic polymorphism of Codonopsis lanceolata and uses thereof | |
| KR102535529B1 (en) | SINGLE NUCLEOTIDE POLYMORPHISM MARKER SET FOR LINE PURITY CHECKING AND EARLY FIXED LINE SELECTING IN WATERMELON (Citrullus lanatus) | |
| KR102665741B1 (en) | Molecular marker for discriminating bacterial wilt-resistant pepper and uses thereof |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| E701 | Decision to grant or registration of patent right | ||
| GRNT | Written decision to grant |