KR20130086386A - Biosynthetically generated pyrroline-carboxy-lysine and site specific protein modifications via chemical derivatization of pyrroline-carboxy-lysine and pyrrolysine residues - Google Patents
Biosynthetically generated pyrroline-carboxy-lysine and site specific protein modifications via chemical derivatization of pyrroline-carboxy-lysine and pyrrolysine residues Download PDFInfo
- Publication number
- KR20130086386A KR20130086386A KR1020137017122A KR20137017122A KR20130086386A KR 20130086386 A KR20130086386 A KR 20130086386A KR 1020137017122 A KR1020137017122 A KR 1020137017122A KR 20137017122 A KR20137017122 A KR 20137017122A KR 20130086386 A KR20130086386 A KR 20130086386A
- Authority
- KR
- South Korea
- Prior art keywords
- formula
- cells
- certain embodiments
- pyrrolysine
- amino acid
- Prior art date
Links
Images
Classifications
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K2/00—Peptides of undefined number of amino acids; Derivatives thereof
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12P—FERMENTATION OR ENZYME-USING PROCESSES TO SYNTHESISE A DESIRED CHEMICAL COMPOUND OR COMPOSITION OR TO SEPARATE OPTICAL ISOMERS FROM A RACEMIC MIXTURE
- C12P21/00—Preparation of peptides or proteins
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07D—HETEROCYCLIC COMPOUNDS
- C07D207/00—Heterocyclic compounds containing five-membered rings not condensed with other rings, with one nitrogen atom as the only ring hetero atom
- C07D207/02—Heterocyclic compounds containing five-membered rings not condensed with other rings, with one nitrogen atom as the only ring hetero atom with only hydrogen or carbon atoms directly attached to the ring nitrogen atom
- C07D207/18—Heterocyclic compounds containing five-membered rings not condensed with other rings, with one nitrogen atom as the only ring hetero atom with only hydrogen or carbon atoms directly attached to the ring nitrogen atom having one double bond between ring members or between a ring member and a non-ring member
- C07D207/20—Heterocyclic compounds containing five-membered rings not condensed with other rings, with one nitrogen atom as the only ring hetero atom with only hydrogen or carbon atoms directly attached to the ring nitrogen atom having one double bond between ring members or between a ring member and a non-ring member with only hydrogen atoms, hydrocarbon or substituted hydrocarbon radicals, directly attached to ring carbon atoms
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07D—HETEROCYCLIC COMPOUNDS
- C07D207/00—Heterocyclic compounds containing five-membered rings not condensed with other rings, with one nitrogen atom as the only ring hetero atom
- C07D207/46—Heterocyclic compounds containing five-membered rings not condensed with other rings, with one nitrogen atom as the only ring hetero atom with hetero atoms directly attached to the ring nitrogen atom
- C07D207/48—Sulfur atoms
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K1/00—General methods for the preparation of peptides, i.e. processes for the organic chemical preparation of peptides or proteins of any length
- C07K1/107—General methods for the preparation of peptides, i.e. processes for the organic chemical preparation of peptides or proteins of any length by chemical modification of precursor peptides
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K5/00—Peptides containing up to four amino acids in a fully defined sequence; Derivatives thereof
- C07K5/02—Peptides containing up to four amino acids in a fully defined sequence; Derivatives thereof containing at least one abnormal peptide link
- C07K5/0215—Peptides containing up to four amino acids in a fully defined sequence; Derivatives thereof containing at least one abnormal peptide link containing natural amino acids, forming a peptide bond via their side chain functional group, e.g. epsilon-Lys, gamma-Glu
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12P—FERMENTATION OR ENZYME-USING PROCESSES TO SYNTHESISE A DESIRED CHEMICAL COMPOUND OR COMPOSITION OR TO SEPARATE OPTICAL ISOMERS FROM A RACEMIC MIXTURE
- C12P13/00—Preparation of nitrogen-containing organic compounds
- C12P13/04—Alpha- or beta- amino acids
- C12P13/08—Lysine; Diaminopimelic acid; Threonine; Valine
Landscapes
- Chemical & Material Sciences (AREA)
- Organic Chemistry (AREA)
- Life Sciences & Earth Sciences (AREA)
- Health & Medical Sciences (AREA)
- Biochemistry (AREA)
- General Health & Medical Sciences (AREA)
- Genetics & Genomics (AREA)
- Engineering & Computer Science (AREA)
- Wood Science & Technology (AREA)
- Zoology (AREA)
- Molecular Biology (AREA)
- Chemical Kinetics & Catalysis (AREA)
- General Chemical & Material Sciences (AREA)
- Biophysics (AREA)
- Medicinal Chemistry (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Biotechnology (AREA)
- Microbiology (AREA)
- Bioinformatics & Cheminformatics (AREA)
- General Engineering & Computer Science (AREA)
- Analytical Chemistry (AREA)
- Peptides Or Proteins (AREA)
- Preparation Of Compounds By Using Micro-Organisms (AREA)
- Pyrrole Compounds (AREA)
- Medicines That Contain Protein Lipid Enzymes And Other Medicines (AREA)
Abstract
생합성적으로 생성되는 천연 아미노산이며 피롤리신 유사체인 피롤린-카르복시-리신 (PCL), 및 PCL을 생합성적으로 생성하는 방법이 본원에 개시된다. 또한, 내부에 혼입된 PCL을 갖는 단백질, 폴리펩티드 및 펩티드, 및 이러한 단백질, 폴리펩티드 및 펩티드에 PCL을 혼입시키는 방법이 본원에 개시된다. 또한, 내부에 혼입된 PCL 또는 피롤리신을 갖는 단백질, 폴리펩티드 및 펩티드의 부위 특이적 유도체화가 본원에 개시된다. 또한, 내부에 혼입된 PCL 또는 피롤리신을 갖는 단백질, 폴리펩티드 및 펩티드의 가교가 본원에 개시된다.Disclosed herein are pyrroline-carboxy-lysine (PCL), a biosynthetically produced natural amino acid and a pyrrolysine analog, and a method for biosynthetically producing PCL. Also disclosed herein are proteins, polypeptides and peptides having PCL incorporated therein, and methods of incorporating PCL into such proteins, polypeptides and peptides. Also disclosed herein are site specific derivatization of proteins, polypeptides and peptides having PCL or pyrrolysine incorporated therein. Also disclosed herein are crosslinking of proteins, polypeptides and peptides having PCL or pyrrolysine incorporated therein.
Description
<관련 출원에 대한 상호 참조><Cross reference to related applications>
본 출원은 35 U.S.C.§119(e) 하에서 2008년 10월 24일자로 출원된 미국 가특허 출원 제61/108,434호에 대한 우선권의 이익을 주장한다. 상기 우선권 출원의 개시내용은 그 전체가 모든 목적을 위해 본원에 참조로 포함된다.This application claims the benefit of priority to U.S. Provisional Patent Application No. 61/108,434, filed Oct. 24, 2008 under 35 U.S.C. §119(e). The disclosure of the priority application is incorporated herein by reference in its entirety for all purposes.
<기술 분야><Technical field>
본 발명은 유전적으로 코딩된 아미노산의 단백질로의 선택적 도입에 관한 것이다. 본 발명은 또한 이러한 아미노산의 화학적 유도체화에 관한 것이다.The present invention relates to the selective introduction of genetically encoded amino acids into proteins. The invention also relates to the chemical derivatization of these amino acids.
패밀리 메타노사르시나(Methanosarcina)의 메탄생성 고세균의 메틸아민 메틸트랜스퍼라제는 천연적으로 피롤리신 (PYL)을 함유한다. 피롤리신은 각각의 mRNA 내의 인-프레임 UAG 코돈에서 번역과 동시에 혼입되는 리신 유사체이고, 이는 22번째 천연 아미노산으로 간주된다.The methanogenic archaea methylamine methyltransferase of the family Methanosarcina naturally contains pyrrolysine (PYL). Pyrrolysine is a lysine analog that is incorporated concurrently with translation at the in-frame UAG codon within each mRNA, which is considered the 22nd natural amino acid.
내부에 혼입된 1개 이상의 PCL을 갖는 단백질 및/또는 폴리펩티드가 본원에 제공되며, 여기서 PCL은 생합성적으로 생성되고 단백질 및/또는 폴리펩티드에 혼입된다. 또한, 내부에 혼입된 1개 이상의 피롤리신 (PYL)을 갖는 단백질 및/또는 폴리펩티드가 본원에 제공되며, 여기서 PYL은 생합성적으로 생성되고 단백질 및/또는 폴리펩티드에 혼입된다. 또한, 내부에 혼입된 1개 이상의 PCL 및 내부에 혼입된 1개 이상의 PYL을 갖는 단백질 및/또는 폴리펩티드가 본원에 제공되며, 여기서 PCL과 PYL은 생합성적으로 생성되고 단백질 및/또는 폴리펩티드에 혼입된다. Provided herein are proteins and/or polypeptides having one or more PCLs incorporated therein, wherein the PCL is produced biosynthetically and incorporated into the protein and/or polypeptide. Also provided herein are proteins and/or polypeptides having at least one pyrrolysine (PYL) incorporated therein, wherein PYL is produced biosynthetically and incorporated into the protein and/or polypeptide. Also provided herein are proteins and/or polypeptides having at least one PCL incorporated therein and at least one PYL incorporated therein, wherein PCL and PYL are produced biosynthetically and incorporated into the protein and/or polypeptide. .
또한, 1개 이상의 PCL 잔기를 갖는 단백질 및/또는 폴리펩티드가 본원에 제공되며, 여기서 PCL은 생합성적으로 생성되고 단백질에 및/또는 폴리펩티드에 혼입되고, 1개 이상의 PCL 잔기는 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, 및 -O-(CH2CH2O)pCH2CH2-X2 (여기서, p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기임)로부터 선택된 기를 단백질 및/또는 폴리펩티드에 커플링시킴으로써 그에 의해 유도체화된다. Also provided herein are proteins and/or polypeptides having one or more PCL residues, wherein PCL is produced biosynthetically and incorporated into the protein and/or polypeptide, and wherein the one or more PCL residues is a label, dye, polymer, Water-soluble polymers, polyalkylene glycols, poly(ethylene glycol), derivatives of poly(ethylene glycol), sugars, lipids, photocrosslinkers, cytotoxic compounds, drugs, affinity labels, photoaffinity labels, reactive compounds; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide, Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 efficacy Agent, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, pressure Tamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radio-imaging probes, other spectroscopic probes, prodrugs, toxins for immunotherapy , Solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 , and Proteins, and a group selected from - -O- (CH 2 CH 2 O )
내부에 혼입된 1개 이상의 PYL 잔기 (여기서, PYL은 생합성적으로 생성되고 단백질 및/또는 폴리펩티드에 혼입됨)를 갖는 이러한 단백질 및/또는 폴리펩티드의 특정 실시양태에서, 피롤리신은 내부에 혼입된 1개 이상의 PCL 잔기를 갖는 단백질 및/또는 폴리펩티드에 대해 상기 주어진 상기 언급된 기 중 하나를 단백질 및/또는 폴리펩티드에 커플링시킴으로써 그에 의해 유도체화된다. In certain embodiments of such proteins and/or polypeptides having one or more PYL residues incorporated therein, wherein PYL is produced biosynthetically and incorporated into the protein and/or polypeptide, pyrrolysine is one incorporated therein. For proteins and/or polypeptides having more than two PCL residues, they are derivatized thereby by coupling to the protein and/or polypeptide one of the aforementioned groups given above.
내부에 혼입된 1개 이상의 PCL 및 1개 이상의 PYL 잔기 (여기서, PCL과 PYL은 생합성적으로 생성되고 단백질 및/또는 폴리펩티드에 혼입됨)를 갖는 이러한 단백질 및/또는 폴리펩티드의 특정 실시양태에서, PCL 및 피롤리신은 내부에 혼입된 1개 이상의 PCL 잔기를 갖는 단백질 및/또는 폴리펩티드에 대해 상기 주어진 상기 언급된 기 중 하나를 단백질 및/또는 폴리펩티드에 커플링시킴으로써 그에 의해 유도체화된다.In certain embodiments of such proteins and/or polypeptides having at least one PCL and at least one PYL residue incorporated therein, wherein PCL and PYL are produced biosynthetically and incorporated into the protein and/or polypeptide, PCL And pyrrolysine is derivatized thereby by coupling to the protein and/or polypeptide one of the aforementioned groups given above for proteins and/or polypeptides having one or more PCL residues incorporated therein.
특정 실시양태에서, 이러한 상기 언급된 생합성은 진핵 세포, 포유동물 세포, 효모 세포 또는 곤충 세포에서 일어난다. 특정 실시양태에서, 세포는 에스케리키아 콜라이 세포이고, 한편 다른 실시양태에서, 효모 세포는 사카로미세스 세레비지애 또는 피키아 파스토랄리스 세포이다. 특정 실시양태에서, 세포는 CHO 세포, HeLa 세포, HEK293F 세포 또는 sf9 세포이다. In certain embodiments, such above-mentioned biosynthesis occurs in eukaryotic cells, mammalian cells, yeast cells or insect cells. In certain embodiments, the cells are Escherichia coli cells, while in other embodiments, the yeast cells are Saccharomyces cerevisiae or Pichia pastoralis cells. In certain embodiments, the cells are CHO cells, HeLa cells, HEK293F cells or sf9 cells.
본원에서 제공되는 한 측면은, 하기 화학식 I 또는 화학식 II의 구조를 갖는 화합물이다.One aspect provided herein is a compound having a structure of formula I or II.
<화학식 I><Formula I>
<화학식 II><Formula II>
상기 식에서,In the above formula,
R1은 H 또는 아미노 말단 변형 기이고;R 1 is H or an amino terminal modified group;
R2는 OH 또는 카르복시 말단 변형 기이고;R 2 is OH or a carboxy terminal modified group;
n은 1 내지 5000의 정수이고;n is an integer from 1 to 5000;
각각의 AA는 아미노산 잔기, 하기 화학식 A-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 및 하기 화학식 B-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기로부터 독립적으로 선택되고;Each AA is independently selected from amino acid residues, pyrrolysine analogue amino acid residues having the structure of formula A-2, and pyrrolysine analogue amino acid residues having the structure of formula B-2;
각각의 BB는 아미노산 잔기, 화학식 A-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 화학식 B-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 C-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 D-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 E-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 F-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 G-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 H-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 I-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 J-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 K-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 및 하기 화학식 L-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기로부터 독립적으로 선택되고;Each BB is an amino acid residue, a pyrrolysine analog having the structure of formula A-2, an amino acid residue of a pyrrolysine analog having the structure of formula B-2, an amino acid residue of a pyrrolysine analog having the structure of formula B-2, and a pyrrolysine analog having the structure of formula C-1 Amino acid residue, pyrrolysine analog amino acid residue having the structure of formula D-1, pyrrolysine analog amino acid residue having the structure of formula E-1, and pyrrolysine analog amino acid residue having the structure of formula F-1 , A pyrrolysine analog amino acid residue having a structure of the following formula G-1, a pyrrolysine analog amino acid residue having a structure of the following formula H-1, a pyrrolysine analog amino acid residue having a structure of the following formula I-1, Independently from the pyrrolysine analog amino acid residue having the structure of formula J-1, the pyrrolysine analog amino acid residue having the structure of formula K-1, and the pyrrolysine analog amino acid residue having the structure of formula L-1 Is selected;
상기 식에서,In the above formula,
R3, R5 및 각각의 R4는 H, -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택되고;R 3, R 5 and each R 4 is H, -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, , Aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1 are independently selected;
R6은 H 또는 C1알킬이고;R 6 is H or C 1 alkyl;
A는 C3-C8시클로알킬, C3-C8헤테로시클로알킬, 5 내지 6원 모노시클릭 아릴, 5 내지 6원 모노시클릭 헤테로아릴, 융합된 9 내지 10원 비시클릭 고리 또는 융합된 13 내지 14원 트리시클릭 고리이고, 여기서 A는 -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택된 1 내지 5개의 치환기로 임의로 치환되고;A is C 3 -C 8 cycloalkyl, C 3 -C 8 heterocycloalkyl, 5-6 membered monocyclic aryl, 5-6 membered monocyclic heteroaryl, fused 9-10 membered bicyclic ring or fused 13 a to 14-membered tricyclic ring, where A is -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, Optionally substituted with 1 to 5 substituents independently selected from aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1;
L은 결합, C1 - 8알킬렌, 할로-치환된-C1 - 8알킬렌, 히드록시-치환된-C1 - 8알킬렌, C2-8알케닐렌, 할로-치환된-C2 - 8알케닐렌, 히드록시-치환된-C2 - 8알케닐렌, -O(CR11R12)k-, -S(CR11R12)k-, -S(O)k(CR11R12)k-, -O(CR11R12)k-NR11C(O)-, -O(CR11R12)kC(O)NR11-, -C(O)-, -C(O)(CR11R12)k-, -C(S)-, -C(S)(CR11R12)k-, -C(O)NR11-, -NR11C(O)-, -NR11(CR11R12)k-, -CONR11(CR11R12)k-, -N(R11)CO(CR11R12)k-, -C(O)NR11(CR11R12)k-, -NR11C(O)(CR11R12)k-로부터 선택되고, 여기서 각각의 R11 및 R12는 독립적으로 H, C1 - 8알킬, 할로-치환된-C1 - 8알킬 또는 히드록시-치환된-C1 - 8알킬이고, k는 1 내지 12의 정수이고;L is a bond, C 1 - 8 alkylene, halo-substituted -C 1 - 8 alkylene, hydroxy-substituted -C 1 - 8 alkylene, C 2-8 alkenylene group, a halo-substituted -C 2 - 8 alkenylene, hydroxy-substituted 2 -C 8 alkenylene, -O (CR 11 R 12) k -, -S (CR 11 R 12) k -, -S (O) k (CR 11 R 12 ) k -, -O(CR 11 R 12 ) k -NR 11 C(O)-, -O(CR 11 R 12 ) k C(O)NR 11 -, -C(O)-, -C( O)(CR 11 R 12 ) k -, -C(S)-, -C(S)(CR 11 R 12 ) k -, -C(O)NR 11 -, -NR 11 C(O)-, -NR 11 (CR 11 R 12 ) k -, -CONR 11 (CR 11 R 12 ) k -, -N(R 11 )CO(CR 11 R 12 ) k -, -C(O)NR 11 (CR 11 R 12) k -, -NR 11 C (O) (CR 11 R 12) k - is selected from wherein each R 11 and R 12 are independently H, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl or hydroxy-substituted -C 1 - 8 is alkyl, k is an integer from 1 to 12;
X1은 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2 및 이들의 임의의 조합물로부터 선택되고, 여기서 p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기이고;X 1 is a label, dye, polymer, water-soluble polymer, polyalkylene glycol, poly(ethylene glycol), derivative of poly(ethylene glycol), sugar, lipid, photocrosslinker, cytotoxic compound, drug, affinity label, photo affinity Label, reactive compound; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide , Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 Agonist, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, Aptamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radiation-imaging probes, other spectroscopic probes, prodrugs, for immunotherapy Toxin, solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 ,- O- (CH 2 CH 2 O) p CH 2 CH 2 -
1개 이상의 AA는 화학식 A-2 또는 화학식 B-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기이거나, 또는 1개 이상의 BB는 화학식 C-1 또는 화학식 D-1 또는 화학식 E-1 또는 화학식 F-1 또는 화학식 G-1 또는 화학식 H-1 또는 화학식 I-1 또는 화학식 J-1 또는 화학식 K-1 또는 화학식 L-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기이다.At least one AA is a pyrrolysine analog amino acid residue having the structure of Formula A-2 or Formula B-2, or at least one BB is Formula C-1 or Formula D-1 or Formula E-1 or Formula F- 1 or a pyrrolysine analog amino acid residue having the structure of formula G-1 or formula H-1 or formula I-1 or formula J-1 or formula K-1 or formula L-1.
상기 화합물의 특정 실시양태에서, 고리 A는 푸란, 티오펜, 피롤, 피롤린, 피롤리딘, 디옥솔란, 옥사졸, 티아졸, 이미다졸, 이미다졸린, 이미다졸리딘, 피라졸, 피라졸린, 피라졸리딘, 이속사졸, 이소티아졸, 옥사디아졸, 트리아졸, 티아디아졸, 피란, 피리딘, 피페리딘, 디옥산, 모르폴린, 디티안, 티오모르폴린, 피리다진, 피리미딘, 피라진, 피페라진, 트리아진, 트리티안, 인돌리진, 인돌, 이소인돌, 인돌린, 벤조푸란, 벤조티오펜, 인다졸, 벤즈이미다졸, 벤즈티아졸, 퓨린, 퀴놀리진, 퀴놀린, 이소퀴놀린, 신놀린, 프탈라진, 퀴나졸린, 퀴녹살린, 나프티리딘, 프테리딘, 퀴누클리딘, 카르바졸, 아크리딘, 페나진, 펜티아진, 페녹사진, 페닐, 인덴, 나프탈렌, 아줄렌, 플루오렌, 안트라센, 페난트라센, 노르보란 및 아다만틴으로부터 선택된다. In certain embodiments of the above compounds, Ring A is furan, thiophene, pyrrole, pyrroline, pyrrolidine, dioxolane, oxazole, thiazole, imidazole, imidazoline, imidazolidine, pyrazole, pyrazole Zoline, pyrazolidine, isoxazole, isothiazole, oxadiazole, triazole, thiadiazole, pyran, pyridine, piperidine, dioxane, morpholine, dithian, thiomorpholine, pyridazine, pyrimidine , Pyrazine, piperazine, triazine, tritian, indolizine, indole, isoindole, indoline, benzofuran, benzothiophene, indazole, benzimidazole, benzthiazole, purine, quinolizine, quinoline, iso Quinoline, cinnoline, phthalazine, quinazoline, quinoxaline, naphthyridine, pteridine, quinuclidine, carbazole, acridine, phenazine, phenthiazin, phenoxazine, phenyl, indene, naphthalene, azulene , Fluorene, anthracene, phenanthracene, norborane and adamantine.
상기 화합물의 다른 실시양태에서, 고리 A는 페닐, 푸란, 티오펜, 피롤, 피롤린, 피롤리딘, 디옥솔란, 옥사졸, 티아졸, 이미다졸, 이미다졸린, 이미다졸리딘, 피라졸, 피라졸린, 피라졸리딘, 이속사졸, 이소티아졸, 옥사디아졸, 트리아졸, 티아디아졸, 피란, 피리딘, 피페리딘, 디옥산, 모르폴린, 디티안, 티오모르폴린, 피리다진, 피리미딘, 피라진, 피페라진, 트리아진 및 트리티안으로부터 선택된다. In other embodiments of the above compounds, Ring A is phenyl, furan, thiophene, pyrrole, pyrroline, pyrrolidine, dioxolane, oxazole, thiazole, imidazole, imidazoline, imidazolidine, pyrazole , Pyrazoline, pyrazolidine, isoxazole, isothiazole, oxadiazole, triazole, thiadiazole, pyran, pyridine, piperidine, dioxane, morpholine, ditian, thiomorpholine, pyridazine, It is selected from pyrimidine, pyrazine, piperazine, triazine and tritian.
상기 화합물의 또 다른 실시양태에서, 고리 A는 인돌리진, 인돌, 이소인돌, 인돌린, 벤조푸란, 벤조티오펜, 인다졸, 벤즈이미다졸, 벤즈티아졸, 퓨린, 퀴놀리진, 퀴놀린, 이소퀴놀린, 신놀린, 프탈라진, 퀴나졸린, 퀴녹살린, 나프티리딘, 프테리딘, 퀴누클리딘, 카르바졸, 아크리딘, 페나진, 펜티아진, 페녹사진, 인덴, 나프탈렌, 아줄렌, 플루오렌, 안트라센, 페난트라센, 노르보란 및 아다만틴으로부터 선택된다. In another embodiment of the above compound, Ring A is indolizine, indole, isoindole, indoline, benzofuran, benzothiophene, indazole, benzimidazole, benzthiazole, purine, quinolizine, quinoline, iso Quinoline, cinnoline, phthalazine, quinazoline, quinoxaline, naphthyridine, pteridine, quinuclidine, carbazole, acridine, phenazine, phenthiazine, phenoxazine, indene, naphthalene, azulene, flu Orene, anthracene, phenanthracene, norborane and adamantine.
상기 화합물의 특정 실시양태에서, 고리 A는 페닐, 나프탈렌 및 피리딘으로부터 선택된다. In certain embodiments of these compounds, Ring A is selected from phenyl, naphthalene and pyridine.
상기 화합물의 특정 실시양태에서, 각각의 BB는 아미노산 잔기, 하기 화학식 A-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 B-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 C-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 D-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 E-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 F-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 G-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 H-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 I-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 J-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 K-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기 및 하기 화학식 L-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기로부터 독립적으로 선택된다.In certain embodiments of the above compounds, each BB is an amino acid residue, a pyrrolysine analog amino acid residue having the structure of formula A-2, a pyrrolysine analog amino acid residue having a structure of formula B-2, formula C A pyrrolysine analog amino acid residue having a structure of -1, a pyrrolysine analog amino acid residue having a structure of the following formula D-2, a pyrrolysine analog amino acid residue having a structure of the following formula E-1, the following formula F-2 A pyrrolysine analog amino acid residue having a structure of, a pyrrolysine analog amino acid residue having a structure of the following formula G-1, a pyrrolysine analog amino acid residue having a structure of the following formula H-2, the structure of the following formula I-1 Having a pyrrolysine analog amino acid residue, a pyrrolysine analog amino acid residue having the structure of the following formula J-2, a pyrrolysine analog amino acid residue having the structure of the following formula K-1, and having the structure of the following formula L-2 It is independently selected from pyrrolysine analogue amino acid residues.
상기 식에서,In the above formula,
R3, R5 및 각각의 R4는 H, -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택되고;R 3, R 5 and each R 4 is H, -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, , Aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1 are independently selected;
R6은 H 또는 C1알킬이고;R 6 is H or C 1 alkyl;
존재할 경우, 각각의 R7은 -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택되고;When present, each R 7 is -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, aryl, heteroaryl , Heterocycloalkyl or cycloalkyl, and -LX 1 are independently selected;
L은 결합, C1 - 8알킬렌, 할로-치환된-C1 - 8알킬렌, 히드록시-치환된-C1 - 8알킬렌, C2-8알케닐렌, 할로-치환된-C2 - 8알케닐렌, 히드록시-치환된-C2 - 8알케닐렌, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), -O(CR11R12)k-, -S(CR11R12)k-, -S(O)k(CR11R12)k-, -O(CR11R12)k-NR11C(O)-, -O(CR11R12)kC(O)NR11-, -C(O)-, -C(O)(CR11R12)k-, -C(S)-, -C(S)(CR11R12)k-, -C(O)NR11-, -NR11C(O)-, -NR11(CR11R12)k-, -CONR11(CR11R12)k-, -N(R11)CO(CR11R12)k-, -C(O)NR11(CR11R12)k-, -NR11C(O)(CR11R12)k-로부터 선택되고, 여기서 각각의 R11 및 R12는 독립적으로 H, C1 - 8알킬, 할로-치환된-C1 - 8알킬 또는 히드록시-치환된-C1 - 8알킬이고, k는 1 내지 12의 정수이고;L is a bond, C 1 - 8 alkylene, halo-substituted -C 1 - 8 alkylene, hydroxy-substituted -C 1 - 8 alkylene, C 2-8 alkenylene group, a halo-substituted -C 2 - 8 alkenylene, hydroxy-substituted 2 -C 8 alkenylene, polyalkylene glycols, poly (ethylene glycol), -O (CR 11 R 12 ) k -, -S (CR 11 R 12) k - , -S(O) k (CR 11 R 12 ) k -, -O(CR 11 R 12 ) k -NR 11 C(O)-, -O(CR 11 R 12 ) k C(O)NR 11- , -C(O)-, -C(O)(CR 11 R 12 ) k -, -C(S)-, -C(S)(CR 11 R 12 ) k -, -C(O)NR 11 -, -NR 11 C(O)-, -NR 11 (CR 11 R 12 ) k -, -CONR 11 (CR 11 R 12 ) k -, -N(R 11 )CO(CR 11 R 12 ) k- , -C(O)NR 11 (CR 11 R 12 ) k -, -NR 11 C(O)(CR 11 R 12 ) k -, wherein each R 11 and R 12 is independently H, C 1 - 8 alkyl, halo-substituted -C 1-8 alkyl, or hydroxy-substituted -C 1 - 8 is alkyl, k is an integer from 1 to 12;
X1은 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2 및 이들의 임의의 조합물로부터 선택되고, 여기서 p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기이다.X 1 is a label, dye, polymer, water-soluble polymer, polyalkylene glycol, poly(ethylene glycol), derivative of poly(ethylene glycol), sugar, lipid, photocrosslinker, cytotoxic compound, drug, affinity label, photo affinity Label, reactive compound; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide , Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 Agonist, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, Aptamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radiation-imaging probes, other spectroscopic probes, prodrugs, for immunotherapy Toxin, solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 ,- O- (CH 2 CH 2 O) p CH 2 CH 2 -
상기 화합물의 특정 실시양태에서, R6은 H이고, 한편 상기 화합물의 다른 실시양태에서, R6은 C1알킬이다. In certain embodiments of these compounds, R 6 is H, while in other embodiments of these compounds, R 6 is C 1 alkyl.
상기 화합물의 특정 실시양태에서, R5는 -LX1이다. 상기 화합물의 특정 실시양태에서, R7은 -LX1이다. 상기 화합물의 특정 실시양태에서, X1은 당, 폴리에틸렌 글리콜, 형광 잔기, 면역조절제, 리보핵산, 데옥시리보핵산, 단백질, 펩티드, 비오틴, 인지질, TLR7 효능제, 면역원성 합텐 또는 고체 지지체이다. 상기 화합물의 특정 실시양태에서, L은 폴리(알킬렌 글리콜), 폴리(에틸렌 글리콜), C1 - 8알킬렌, 할로-치환된-C1 - 8알킬렌 또는 히드록시-치환된-C1 - 8알킬렌이다.In certain embodiments of the above compounds, R 5 is -LX 1 . In certain embodiments of the above compounds, R 7 is -LX 1 . In certain embodiments of the above compounds, X 1 is a sugar, polyethylene glycol, fluorescent moiety, immunomodulator, ribonucleic acid, deoxyribonucleic acid, protein, peptide, biotin, phospholipid, TLR7 agonist, immunogenic hapten or solid support. In certain embodiments of the above compounds, L is a poly (alkylene glycol), poly (ethylene glycol), C 1 - 8 alkylene, halo-substituted -C 1 - 8 alkylene or hydroxy-substituted -C 1 8 is an alkylene group.
본원에서 제공되는 또 다른 측면은, 하기 화학식 I에 따른 구조를 갖는 단백질을 하기 화학식 III 또는 화학식 IV의 시약과 접촉시키는 것을 포함하는, 상기 단백질을 유도체화시키는 방법이다.Another aspect provided herein is a method of derivatizing a protein comprising contacting a protein having a structure according to formula (I) with a reagent of formula (III) or (IV).
<화학식 I><Formula I>
상기 식에서,In the above formula,
R1은 H 또는 아미노 말단 변형 기이고;R 1 is H or an amino terminal modified group;
R2는 OH 또는 카르복시 말단 변형 기이고;R 2 is OH or a carboxy terminal modified group;
n은 1 내지 5000의 정수이고;n is an integer from 1 to 5000;
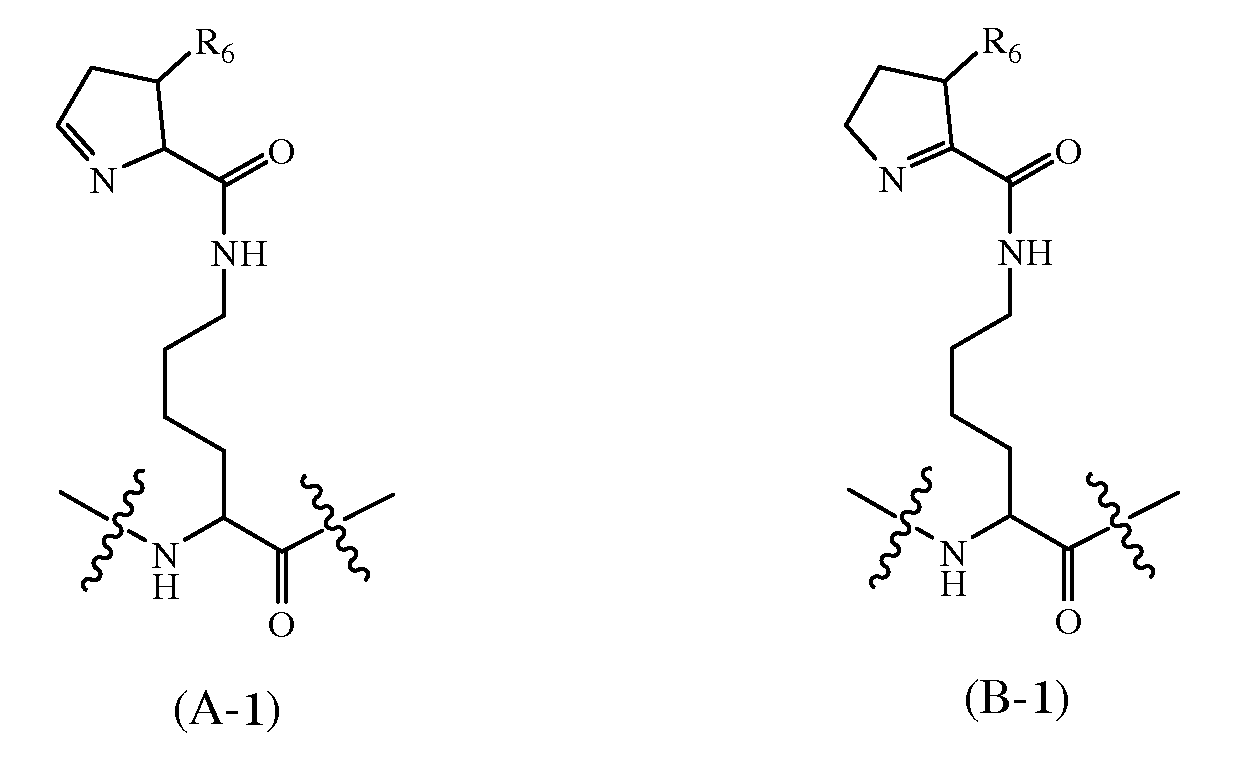
각각의 AA는 아미노산 잔기, 피롤리신 아미노산 잔기, 하기 화학식 A-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 및 하기 화학식 B-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기로부터 독립적으로 선택되고;Each AA is independently selected from an amino acid residue, a pyrrolysine amino acid residue, a pyrrolysine analog amino acid residue having the structure of formula A-1, and a pyrrolysine analog amino acid residue having the structure of formula B-1, ;
R6은 H 또는 C1알킬이고;R 6 is H or C 1 alkyl;
1개 이상의 AA는 피롤리신 아미노산 잔기, 또는 화학식 A-1 또는 화학식 B-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기이다.At least one AA is a pyrrolysine amino acid residue, or a pyrrolysine analog amino acid residue having the structure of Formula A-1 or Formula B-1.
<화학식 III><Formula III>
<화학식 IV><Formula IV>
상기 식에서,In the above formula,
R3, R5 및 각각의 R4는 H, -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택되고;R 3, R 5 and each R 4 is H, -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, , Aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1 are independently selected;
A는 C3-C8시클로알킬, C3-C8헤테로시클로알킬, 5 내지 6원 모노시클릭 아릴, 5 내지 6원 모노시클릭 헤테로아릴, 융합된 9 내지 10원 비시클릭 고리 또는 융합된 13 내지 14원 트리시클릭 고리이고, 여기서 A는 -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택된 1 내지 5개의 치환기로 임의로 치환되고;A is C 3 -C 8 cycloalkyl, C 3 -C 8 heterocycloalkyl, 5-6 membered monocyclic aryl, 5-6 membered monocyclic heteroaryl, fused 9-10 membered bicyclic ring or fused 13 a to 14-membered tricyclic ring, where A is -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, Optionally substituted with 1 to 5 substituents independently selected from aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1;
L은 결합, C1 - 8알킬렌, 할로-치환된-C1 - 8알킬렌, 히드록시-치환된-C1 - 8알킬렌, C2-8알케닐렌, 할로-치환된-C2 - 8알케닐렌, 히드록시-치환된-C2 - 8알케닐렌, 폴리(알킬렌 글리콜), 폴리(에틸렌 글리콜), -O(CR11R12)k-, -S(CR11R12)k-, -S(O)k(CR11R12)k-, -O(CR11R12)k-NR11C(O)-, -O(CR11R12)kC(O)NR11-, -C(O)-, -C(O)(CR11R12)k-, -C(S)-, -C(S)(CR11R12)k-, -C(O)NR11-, -NR11C(O)-, -NR11(CR11R12)k-, -CONR11(CR11R12)k-, -N(R11)CO(CR11R12)k-, -C(O)NR11(CR11R12)k-, -NR11C(O)(CR11R12)k-로부터 선택되고, 여기서 각각의 R11 및 R12는 독립적으로 H, C1 - 8알킬, 할로-치환된-C1 - 8알킬 또는 히드록시-치환된-C1 - 8알킬이고, k는 1 내지 12의 정수이고;L is a bond, C 1 - 8 alkylene, halo-substituted -C 1 - 8 alkylene, hydroxy-substituted -C 1 - 8 alkylene, C 2-8 alkenylene group, a halo-substituted -C 2 - 8 alkenylene, hydroxy-substituted 2 -C 8 alkenylene group, a poly (alkylene glycol), poly (ethylene glycol), -O (CR 11 R 12 ) k -, -S (CR 11 R 12) k -, -S(O) k (CR 11 R 12 ) k -, -O(CR 11 R 12 ) k -NR 11 C(O)-, -O(CR 11 R 12 ) k C(O)NR 11 -, -C(O)-, -C(O)(CR 11 R 12 ) k -, -C(S)-, -C(S)(CR 11 R 12 ) k -, -C(O) NR 11 -, -NR 11 C(O)-, -NR 11 (CR 11 R 12 ) k -, -CONR 11 (CR 11 R 12 ) k -, -N(R 11 )CO(CR 11 R 12 ) k -, -C(O)NR 11 (CR 11 R 12 ) k -, -NR 11 C(O)(CR 11 R 12 ) k -, wherein each of R 11 and R 12 is independently H , C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl or hydroxy-substituted -C 1 - 8 is alkyl, k is an integer from 1 to 12;
X1은 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2 및 이들의 임의의 조합물로부터 선택되고, 여기서 p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기이다.X 1 is a label, dye, polymer, water-soluble polymer, polyalkylene glycol, poly(ethylene glycol), derivative of poly(ethylene glycol), sugar, lipid, photocrosslinker, cytotoxic compound, drug, affinity label, photo affinity Label, reactive compound; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide , Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 Agonist, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, Aptamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radiation-imaging probes, other spectroscopic probes, prodrugs, for immunotherapy Toxin, solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 ,- O- (CH 2 CH 2 O) p CH 2 CH 2 -X 2 , and are selected from any combination thereof, wherein p is 1 to 10,000, X 2 is H, C 1 - 8 alkyl, protecting group or terminal It is a functional group.
상기 언급된 방법의 특정 실시양태에서, 화학식 A-1의 아미노산 잔기는 하기 화학식 A-2 또는 화학식 A-3의 구조를 갖는 아미노산 잔기이다.In certain embodiments of the aforementioned methods, the amino acid residues of formula A-1 are amino acid residues having the structure of formula A-2 or formula A-3 below.
상기 언급된 방법의 특정 실시양태에서, 화학식 B-1의 아미노산 잔기는 하기 화학식 B-2 또는 화학식 B-3의 구조를 갖는 아미노산 잔기이다.In certain embodiments of the aforementioned methods, the amino acid residues of formula B-1 are amino acid residues having the structure of formula B-2 or formula B-3 below.
상기 언급된 방법의 특정 실시양태에서, 화학식 A-1의 아미노산 잔기는 하기 화학식 V의 아미노산의 잔기이고, 화학식 B-1의 아미노산 잔기는 하기 화학식 VI의 아미노산의 잔기이다.In certain embodiments of the above-mentioned methods, the amino acid residues of formula A-1 are those of the amino acids of formula V, and the amino acid residues of formula B-1 are the residues of the amino acid formula VI:
(상기 식에서, R6은 H 또는 C1알킬임)(Wherein, R 6 is H or C 1 alkyl)
특정 실시양태에서, 화학식 V 또는 화학식 VI의 아미노산은 pylB 유전자, pylC 유전자 및 pylD 유전자를 포함하는 세포 내에서 생합성적으로 생성되고, 세포는 전구체를 포함하는 성장 배지와 접촉하고 있다. 다른 실시양태에서, 화학식 V 또는 화학식 VI의 아미노산은 pylC 유전자 및 pylD 유전자를 포함하는 세포 내에서 생합성적으로 생성되고, 세포는 전구체를 포함하는 성장 배지와 접촉하고 있다. In certain embodiments, an amino acid of Formula V or Formula VI is produced biosynthetically within a cell comprising a pylB gene, a pylC gene, and a pylD gene, and the cell is in contact with a growth medium comprising a precursor. In other embodiments, the amino acids of Formula V or Formula VI are produced biosynthetically in cells comprising the pylC gene and the pylD gene, and the cell is in contact with a growth medium comprising the precursor.
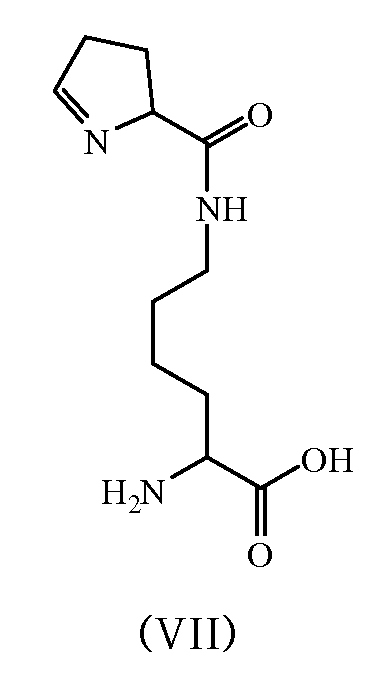
특정 실시양태에서, 화학식 V의 아미노산은 하기 화학식 VII의 구조를 갖는 아미노산이고, 전구체는 오르니틴, 아르기닌, D-오르니틴, D-아르기닌, (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산 또는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다.In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula VII, and the precursor is ornithine, arginine, D-ornithine, D-arginine, (2S)-2-amino-6-(2,5 -Diaminopentanamido)hexanoic acid or (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid.
특정 실시양태에서, 화학식 VI의 아미노산은 하기 화학식 VIII의 구조를 갖는 아미노산이고, 전구체는 오르니틴 또는 아르기닌이고, 전구체는 오르니틴, 아르기닌, D-오르니틴, D-아르기닌, (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산 또는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다.In certain embodiments, the amino acid of Formula VI is an amino acid having the structure of Formula VIII, the precursor is ornithine or arginine, and the precursor is ornithine, arginine, D-ornithine, D-arginine, (2S)-2- Amino-6-(2,5-diaminopentanamido)hexanoic acid or (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid.
특정 실시양태에서, 화학식 V의 아미노산은 화학식 VII의 구조를 갖는 아미노산이고, 전구체는 D-오르니틴 또는 D-아르기닌이다. 특정 실시양태에서, 화학식 VI의 아미노산은 화학식 VIII의 구조를 갖는 아미노산이고, 전구체는 D-오르니틴 또는 D-아르기닌이다. 특정 실시양태에서, 화학식 V의 아미노산은 화학식 VII의 구조를 갖는 아미노산이고, 전구체는 (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산이다. 특정 실시양태에서, 화학식 V의 아미노산은 화학식 VII의 구조를 갖는 아미노산이고, 전구체는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. 특정 실시양태에서, 화학식 VI의 아미노산은 화학식 VIII의 구조를 갖는 아미노산이고, 전구체는 (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산이다. 특정 실시양태에서, 화학식 VI의 아미노산은 화학식 VIII의 구조를 갖는 아미노산이고, 전구체는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. In certain embodiments, the amino acid of formula V is an amino acid having the structure of formula VII and the precursor is D-ornithine or D-arginine. In certain embodiments, the amino acid of Formula VI is an amino acid having the structure of Formula VIII and the precursor is D-ornithine or D-arginine. In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula VII and the precursor is (2S)-2-amino-6-(2,5-diaminopentanamido)hexanoic acid. In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula VII and the precursor is (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid. In certain embodiments, the amino acid of Formula VI is an amino acid having the structure of Formula VIII and the precursor is (2S)-2-amino-6-(2,5-diaminopentanamido)hexanoic acid. In certain embodiments, the amino acid of Formula VI is an amino acid having the structure of Formula VIII, and the precursor is (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid.
특정 실시양태에서, 화학식 V의 아미노산은 화학식 IX의 구조를 갖는 아미노산이고, 전구체는 오르니틴, 아르기닌, D-오르니틴, D-아르기닌 또는 2,5-디아미노-3-메틸펜탄산이다.In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula IX and the precursor is ornithine, arginine, D-ornithine, D-arginine, or 2,5-diamino-3-methylpentanoic acid.
특정 실시양태에서, 화학식 V의 아미노산은 화학식 X의 구조를 갖는 아미노산이고, 전구체는 오르니틴, 아르기닌, D-오르니틴, D-아르기닌 또는 2,5-디아미노-3-메틸펜탄산이다.In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula X and the precursor is ornithine, arginine, D-ornithine, D-arginine, or 2,5-diamino-3-methylpentanoic acid.
특정 실시양태에서, 화학식 V의 아미노산은 화학식 IX의 구조를 갖는 아미노산이고, 전구체는 D-2,5-디아미노-3-메틸펜탄산이다. 특정 실시양태에서, 화학식 V의 아미노산은 화학식 X의 구조를 갖는 아미노산이고, 전구체는 D-2,5-디아미노-3-메틸펜탄산이다. In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula IX and the precursor is D-2,5-diamino-3-methylpentanoic acid. In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula X and the precursor is D-2,5-diamino-3-methylpentanoic acid.
특정 실시양태에서, 화학식 V의 아미노산은 화학식 IX의 구조를 갖는 아미노산이고, 전구체는 (2R,3S)-2,5-디아미노-3-메틸펜탄산이다. 특정 실시양태에서, 화학식 V의 아미노산은 화학식 X의 구조를 갖는 아미노산이고, 전구체는 (2R,3S)-2,5-디아미노-3-메틸펜탄산이다.In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula IX and the precursor is (2R,3S)-2,5-diamino-3-methylpentanoic acid. In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula X and the precursor is (2R,3S)-2,5-diamino-3-methylpentanoic acid.
특정 실시양태에서, 화학식 V의 아미노산은 화학식 IX의 구조를 갖는 아미노산이고 전구체는 (2R,3R)-2,5-디아미노-3-메틸펜탄산이다. 특정 실시양태에서, 화학식 V의 아미노산은 화학식 X의 구조를 갖는 아미노산이고, 전구체는 (2R,3R)-2,5-디아미노-3-메틸펜탄산이다. 특정 실시양태에서, 화학식 V의 아미노산은 화학식 IX의 구조를 갖는 아미노산이고, 전구체는 D-오르니틴 또는 D-아르기닌 또는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. 특정 실시양태에서, 화학식 V의 아미노산은 화학식 X의 구조를 갖는 아미노산이고, 전구체는 D-오르니틴 또는 D-아르기닌 또는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula IX and the precursor is (2R,3R)-2,5-diamino-3-methylpentanoic acid. In certain embodiments, the amino acid of Formula V is an amino acid having the structure of Formula X and the precursor is (2R,3R)-2,5-diamino-3-methylpentanoic acid. In certain embodiments, the amino acid of formula V is an amino acid having the structure of formula IX, and the precursor is D-ornithine or D-arginine or (2S)-2-amino-6-((R)-2,5-di Aminopentanamido)hexanoic acid. In certain embodiments, the amino acid of formula V is an amino acid having the structure of formula X, and the precursor is D-ornithine or D-arginine or (2S)-2-amino-6-((R)-2,5-di Aminopentanamido)hexanoic acid.
상기 언급된 방법의 특정 실시양태에서, 화학식 V, 화학식 VI, 화학식 VII, 화학식 VII, 화학식 IX 또는 화학식 X의 아미노산은 직교 tRNA (O-tRNA) 및 직교 아미노아실 tRNA 신테타제 (O-RS)에 의해 세포 내 단백질에 혼입되고, 여기서 O-RS는 O-tRNA를 화학식 V 또는 화학식 VI의 아미노산으로 아미노아실화시키고, O-tRNA는 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인지한다. In certain embodiments of the aforementioned methods, the amino acids of Formula V, Formula VI, Formula VII, Formula VII, Formula IX or Formula X are directed to orthogonal tRNA (O-tRNA) and orthogonal aminoacyl tRNA synthetase (O-RS). Is incorporated into an intracellular protein, wherein O-RS aminoacylates the O-tRNA to an amino acid of Formula V or Formula VI, and the O-tRNA recognizes one or more selector codons of the mRNA in the cell.
상기 언급된 방법의 특정 실시양태에서, 세포는 추가로 pylS 유전자 및 pylT 유전자를 포함하고, 화학식 V, 화학식 VI, 화학식 VII, 화학식 VII, 화학식 IX 또는 화학식 X의 아미노산은 아미노아실 tRNA 신테타제, 및 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하는 tRNA에 의해 세포 내 단백질에 혼입되고, 여기서 아미노아실 tRNA 신테타제는 pylS 유전자의 유전자 생성물이고 tRNA는 pylT 유전자의 유전자 생성물이다. In certain embodiments of the above-mentioned method, the cell further comprises a pylS gene and a pylT gene, and the amino acid of Formula V, Formula VI, Formula VII, Formula VII, Formula IX or Formula X is an aminoacyl tRNA synthetase, and In the cell, it is incorporated into an intracellular protein by a tRNA that recognizes one or more selector codons of the mRNA, wherein the aminoacyl tRNA synthetase is a gene product of the pylS gene and the tRNA is a gene product of the pylT gene.
상기 언급된 방법의 특정 실시양태에서, 셀렉터 코돈은 앰버 코돈 (TAG)이다. In certain embodiments of the aforementioned method, the selector codon is an amber codon (TAG).
상기 언급된 방법의 특정 실시양태에서, 세포는 원핵 세포이고, 한편 다른 실시양태에서 세포는 진핵 세포이다. 특정 실시양태에서, 세포는 에스케리키아 콜라이 세포이고, 한편 다른 실시양태에서 세포는 포유동물 세포, 효모 세포 또는 곤충 세포이다. 특정 실시양태에서, 효모 세포는 사카로미세스 세레비지애 또는 피키아 파스토랄리스 세포이다. 특정 실시양태에서, 포유동물 세포는 CHO 세포, HeLa 세포 또는 HEK293F 세포이다. 특정 실시양태에서, 곤충 세포는 sf9 세포이다. In certain embodiments of the aforementioned methods, the cells are prokaryotic cells, while in other embodiments the cells are eukaryotic cells. In certain embodiments, the cells are Escherichia coli cells, while in other embodiments the cells are mammalian cells, yeast cells or insect cells. In certain embodiments, the yeast cells are Saccharomyces cerevisiae or Pichia pastoralis cells. In certain embodiments, the mammalian cells are CHO cells, HeLa cells, or HEK293F cells. In certain embodiments, the insect cells are sf9 cells.
본원에서 제공되는 또 다른 측면은 상기 언급된 방법을 이용하여 수득되는 유도체화 단백질이며, 여기서 이러한 유도체화 단백질은 하기 화학식 II에 따른 구조를 갖는다.Another aspect provided herein is a derivatized protein obtained using the above-mentioned method, wherein this derivatized protein has a structure according to formula II below.
<화학식 II><Formula II>
상기 식에서,In the above formula,
R1은 H 또는 아미노 말단 변형 기이고;R 1 is H or an amino terminal modified group;
R2는 OH 또는 카르복시 말단 변형 기이고;R 2 is OH or a carboxy terminal modified group;
n은 1 내지 5000의 정수이고;n is an integer from 1 to 5000;
각각의 BB는 아미노산 잔기, 하기 화학식 A-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 B-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 C-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 D-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 E-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 F-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 G-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 H-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 I-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 J-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 K-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기 및 하기 화학식 L-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기로부터 독립적으로 선택되고;Each BB is an amino acid residue, a pyrrolysine analog amino acid residue having the structure of the following formula A-1, a pyrrolysine analog amino acid residue having the structure of the formula B-1, A new analog amino acid residue, a pyrrolysine analog amino acid residue having a structure of the following formula D-1, a pyrrolysine analog amino acid residue having a structure of the following formula E-1, a pyrrolysine analog having a structure of the following formula F-1 Amino acid residue, a pyrrolysine analog amino acid residue having the structure of the following formula G-1, a pyrrolysine analog amino acid residue having the structure of the following formula H-1, a pyrrolysine analog amino acid residue having the structure of the following formula I-1 , Independent from a pyrrolysine analog amino acid residue having a structure of the following formula (J-1), a pyrrolysine analog amino acid residue having a structure of the following formula (K-1), and a pyrrolysine analog amino acid residue having a structure of formula L-1 Is selected as;
1개 이상의 BB는 화학식 C-1 또는 화학식 D-1 또는 화학식 E-1 또는 화학식 F-1 또는 화학식 G-1 또는 화학식 H-1 또는 화학식 I-1 또는 화학식 J-1 또는 화학식 K-1 또는 화학식 L-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기이다.At least one BB is a formula C-1 or a formula D-1 or a formula E-1 or a formula F-1 or a formula G-1 or a formula H-1 or a formula I-1 or a formula J-1 or a formula K-1 or It is a pyrrolysine analog amino acid residue having the structure of formula L-1.
상기 언급된 방법 및 상기 유도체화 단백질의 특정 실시양태에서, 고리 A는 푸란, 티오펜, 피롤, 피롤린, 피롤리딘, 디옥솔란, 옥사졸, 티아졸, 이미다졸, 이미다졸린, 이미다졸리딘, 피라졸, 피라졸린, 피라졸리딘, 이속사졸, 이소티아졸, 옥사디아졸, 트리아졸, 티아디아졸, 피란, 피리딘, 피페리딘, 디옥산, 모르폴린, 디티안, 티오모르폴린, 피리다진, 피리미딘, 피라진, 피페라진, 트리아진, 트리티안, 인돌리진, 인돌, 이소인돌, 인돌린, 벤조푸란, 벤조티오펜, 인다졸, 벤즈이미다졸, 벤즈티아졸, 퓨린, 퀴놀리진, 퀴놀린, 이소퀴놀린, 신놀린, 프탈라진, 퀴나졸린, 퀴녹살린, 나프티리딘, 프테리딘, 퀴누클리딘, 카르바졸, 아크리딘, 페나진, 펜티아진, 페녹사진, 페닐, 인덴, 나프탈렌, 아줄렌, 플루오렌, 안트라센, 페난트라센, 노르보란 및 아다만틴으로부터 선택된다. In the above-mentioned method and certain embodiments of said derivatized protein, Ring A is furan, thiophene, pyrrole, pyrroline, pyrrolidine, dioxolane, oxazole, thiazole, imidazole, imidazoline, imida Zolidine, pyrazole, pyrazoline, pyrazolidine, isoxazole, isothiazole, oxadiazole, triazole, thiadiazole, pyran, pyridine, piperidine, dioxane, morpholine, ditian, thiomor Foline, pyridazine, pyrimidine, pyrazine, piperazine, triazine, tritian, indolizine, indole, isoindole, indoline, benzofuran, benzothiophene, indazole, benzimidazole, benzthiazole, purine, Quinolizine, quinoline, isoquinoline, cinoline, phthalazine, quinazoline, quinoxaline, naphthyridine, pteridine, quinuclidine, carbazole, acridine, phenazine, phenthiazin, phenoxazine, phenyl , Indene, naphthalene, azulene, fluorene, anthracene, phenanthracene, norborane and adamantine.
상기 언급된 방법 및 상기 유도체화 단백질의 특정 실시양태에서, 고리 A는 페닐, 푸란, 티오펜, 피롤, 피롤린, 피롤리딘, 디옥솔란, 옥사졸, 티아졸, 이미다졸, 이미다졸린, 이미다졸리딘, 피라졸, 피라졸린, 피라졸리딘, 이속사졸, 이소티아졸, 옥사디아졸, 트리아졸, 티아디아졸, 피란, 피리딘, 피페리딘, 디옥산, 모르폴린, 디티안, 티오모르폴린, 피리다진, 피리미딘, 피라진, 피페라진, 트리아진 및 트리티안으로부터 선택된다. In certain embodiments of the aforementioned methods and of the derivatized proteins, Ring A is phenyl, furan, thiophene, pyrrole, pyrroline, pyrrolidine, dioxolane, oxazole, thiazole, imidazole, imidazoline, Imidazolidine, pyrazole, pyrazoline, pyrazolidine, isoxazole, isothiazole, oxadiazole, triazole, thiadiazole, pyran, pyridine, piperidine, dioxane, morpholine, ditian, Selected from thiomorpholine, pyridazine, pyrimidine, pyrazine, piperazine, triazine and tritian.
상기 언급된 방법 및 상기 유도체화 단백질의 특정 실시양태에서, 고리 A는 인돌리진, 인돌, 이소인돌, 인돌린, 벤조푸란, 벤조티오펜, 인다졸, 벤즈이미다졸, 벤즈티아졸, 퓨린, 퀴놀리진, 퀴놀린, 이소퀴놀린, 신놀린, 프탈라진, 퀴나졸린, 퀴녹살린, 나프티리딘, 프테리딘, 퀴누클리딘, 카르바졸, 아크리딘, 페나진, 펜티아진, 페녹사진, 인덴, 나프탈렌, 아줄렌, 플루오렌, 안트라센, 페난트라센, 노르보란 및 아다만틴으로부터 선택된다. In certain embodiments of the aforementioned methods and of the derivatized proteins, Ring A is indolizine, indole, isoindole, indoline, benzofuran, benzothiophene, indazole, benzimidazole, benzthiazole, purine, quie Nolizine, quinoline, isoquinoline, cinnoline, phthalazine, quinazoline, quinoxaline, naphthyridine, pteridine, quinuclidine, carbazole, acridine, phenazine, pentazine, phenoxazine, indene, Naphthalene, azulene, fluorene, anthracene, phenanthracene, norborane and adamantine.
상기 언급된 방법 및 상기 유도체화 단백질의 특정 실시양태에서, 고리 A는 페닐, 나프탈릴 및 피리딜로부터 선택된다. In the aforementioned method and certain embodiments of the derivatized protein, Ring A is selected from phenyl, naphthalyl and pyridyl.
상기 언급된 방법의 특정 실시양태에서 각각의 BB는 아미노산 잔기, 하기 화학식 A-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 B-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 C-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 D-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 E-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 F-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 G-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 H-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 I-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 J-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 K-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기 및 하기 화학식 L-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기로부터 독립적으로 선택된다.In certain embodiments of the above-mentioned method, each BB is an amino acid residue, a pyrrolysine analog amino acid residue having the structure of formula A-2, a pyrrolysine analog amino acid residue having the structure of formula B-2, A pyrrolysine analog amino acid residue having a structure of C-1, a pyrrolysine analog amino acid residue having a structure of formula D-2, a pyrrolysine analog amino acid residue having a structure of formula E-1, the following formula F- A pyrrolysine analog amino acid residue having a structure of 2, a pyrrolysine analog amino acid residue having a structure of the following formula G-1, a pyrrolysine analog amino acid residue having a structure of the following formula H-2, A pyrrolysine analog amino acid residue having a structure, a pyrrolysine analog amino acid residue having a structure of the following formula J-2, a pyrrolysine analog amino acid residue having a structure of the following formula K-1, and the structure of the following formula L-2 It is independently selected from amino acid residues having pyrrolysine analogs.
상기 식에서,In the above formula,
R3, R5 및 각각의 R4는 H, -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택되고;R 3, R 5 and each R 4 is H, -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, , Aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1 are independently selected;
R6은 H 또는 C1알킬이고;R 6 is H or C 1 alkyl;
존재할 경우, 각각의 R7은 -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택되고;When present, each R 7 is -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, aryl, heteroaryl , Heterocycloalkyl or cycloalkyl, and -LX 1 are independently selected;
L은 결합, C1 - 8알킬렌, 할로-치환된-C1 - 8알킬렌, 히드록시-치환된-C1 - 8알킬렌, C2-8알케닐렌, 할로-치환된-C2 - 8알케닐렌, 히드록시-치환된-C2 - 8알케닐렌, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), -O(CR11R12)k-, -S(CR11R12)k-, -S(O)k(CR11R12)k-, -O(CR11R12)k-NR11C(O)-, -O(CR11R12)kC(O)NR11-, -C(O)-, -C(O)(CR11R12)k-, -C(S)-, -C(S)(CR11R12)k-, -C(O)NR11-, -NR11C(O)-, -NR11(CR11R12)k-, -CONR11(CR11R12)k-, -N(R11)CO(CR11R12)k-, -C(O)NR11(CR11R12)k-, -NR11C(O)(CR11R12)k-로부터 선택되고, 여기서 각각의 R11 및 R12는 독립적으로 H, C1 - 8알킬, 할로-치환된-C1 - 8알킬 또는 히드록시-치환된-C1 - 8알킬이고, k는 1 내지 12의 정수이고,L is a bond, C 1 - 8 alkylene, halo-substituted -C 1 - 8 alkylene, hydroxy-substituted -C 1 - 8 alkylene, C 2-8 alkenylene group, a halo-substituted -C 2 - 8 alkenylene, hydroxy-substituted 2 -C 8 alkenylene, polyalkylene glycols, poly (ethylene glycol), -O (CR 11 R 12 ) k -, -S (CR 11 R 12) k - , -S(O) k (CR 11 R 12 ) k -, -O(CR 11 R 12 ) k -NR 11 C(O)-, -O(CR 11 R 12 ) k C(O)NR 11- , -C(O)-, -C(O)(CR 11 R 12 ) k -, -C(S)-, -C(S)(CR 11 R 12 ) k -, -C(O)NR 11 -, -NR 11 C(O)-, -NR 11 (CR 11 R 12 ) k -, -CONR 11 (CR 11 R 12 ) k -, -N(R 11 )CO(CR 11 R 12 ) k- , -C(O)NR 11 (CR 11 R 12 ) k -, -NR 11 C(O)(CR 11 R 12 ) k -, wherein each R 11 and R 12 is independently H, C 1 and 8, and the alkyl, k is an integer from 1 to 12, - 8 alkyl, halo-substituted -C 1 - 8 alkyl or hydroxy-substituted -C 1
X1은 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2 및 이들의 임의의 조합물로부터 선택되고, 여기서 p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기이다.X 1 is a label, dye, polymer, water-soluble polymer, polyalkylene glycol, poly(ethylene glycol), derivative of poly(ethylene glycol), sugar, lipid, photocrosslinker, cytotoxic compound, drug, affinity label, photo affinity Label, reactive compound; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide , Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 Agonist, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, Aptamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radiation-imaging probes, other spectroscopic probes, prodrugs, for immunotherapy Toxin, solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 ,- O- (CH 2 CH 2 O) p CH 2 CH 2 -X 2 , and are selected from any combination thereof, wherein p is 1 to 10,000, X 2 is H, C 1 - 8 alkyl, protecting group or terminal It is a functional group.
상기 언급된 방법 및 상기 유도체화 단백질의 특정 실시양태에서, R6은 H이고, 한편 다른 실시양태에서 R6은 C1알킬이다. In certain embodiments of the aforementioned methods and of such derivatized proteins, R 6 is H, while in other embodiments R 6 is C 1 alkyl.
상기 언급된 방법 및 상기 유도체화 단백질의 특정 실시양태에서, R5는 -LX1이다. 상기 언급된 방법 및 상기 유도체화 단백질의 특정 실시양태에서, X1은 당, 폴리에틸렌 글리콜, 형광 잔기, 면역조절제, 리보핵산, 데옥시리보핵산, 단백질, 펩티드, 비오틴, 인지질, TLR7 효능제, 면역원성 합텐 또는 고체 지지체이다. 특정 실시양태에서, L은 폴리(알킬렌 글리콜), 폴리(에틸렌 글리콜), C1 - 8알킬렌, 할로-치환된-C1 - 8알킬렌 또는 히드록시-치환된-C1 - 8알킬렌이다. In the aforementioned method and certain embodiments of the derivatized protein, R 5 is -LX 1 . In the above-mentioned method and certain embodiments of the derivatized protein, X 1 is a sugar, polyethylene glycol, fluorescent moiety, immunomodulatory agent, ribonucleic acid, deoxyribonucleic acid, protein, peptide, biotin, phospholipid, TLR7 agonist, immunity. It is an original hapten or a solid support. In certain embodiments, L is a poly (alkylene glycol), poly (ethylene glycol), C 1 - 8 alkylene, halo-substituted -C 1 - 8 alkylene or hydroxy-substituted -C 1 - 8 alkyl, It's Len.
상기 언급된 방법의 특정 실시양태에서, 화학식 IV의 시약은In certain embodiments of the aforementioned methods, the reagent of formula IV is
이고, 여기서 L 및 X1은 상기 기재된 바와 같다. Where L and X 1 are as described above.
상기 시약의 특정 실시양태에서, L은 결합이고 X1은 폴리에틸렌 글리콜이다. In certain embodiments of the above reagents, L is a bond and X 1 is polyethylene glycol.
특정 실시양태에서, 화학식 IV의 시약은In certain embodiments, the reagent of formula IV is
이고, 여기서 1개 이상의 폴리에틸렌 글리콜 (PEG) 잔기를 갖는 화합물은 1000 Da 내지 50 kDa 범위의 평균 분자량을 갖고, n은 20 내지 1200이고, exPADRE는 이고, PADRE는 이고, BG1은 이고, BG2는 이고, 여기서 *는 포스포티오에이트 연결부를 표시한다.Wherein the compound having at least one polyethylene glycol (PEG) moiety has an average molecular weight in the range of 1000 Da to 50 kDa, n is 20 to 1200, and exPADRE is And PADRE is And BG1 is And BG2 is And, where * represents a phosphorothioate linkage.
특정 실시양태에서, 화학식 IV의 시약은 하기 구조:In certain embodiments, the reagent of Formula IV has the structure:
를 갖는 화합물이고, 여기서 화합물은 1000 Da 내지 30 kDa 범위의 평균 분자량을 갖고, n은 20 내지 679이다. Wherein the compound has an average molecular weight ranging from 1000 Da to 30 kDa, and n is from 20 to 679.
또 다른 실시양태에서, 화학식 IV의 시약은 하기 구조:In another embodiment, the reagent of Formula IV has the structure:
를 갖는 화합물이고, 여기서 화합물은 1000 Da 내지 45 kDa 범위의 평균 분자량을 갖고, n은 20 내지 1018이다. Wherein the compound has an average molecular weight ranging from 1000 Da to 45 kDa, and n is from 20 to 1018.
본원에서 제공되는 또 다른 측면은 하기 화학식 VII 또는 화학식 VIII의 구조를 갖는 화합물이고, 여기서 화학식 VII 또는 화학식 VIII의 화합물은 pylC 유전자 및 pylD 유전자를 포함하는 세포 내에서 생합성적으로 생성되고, 세포는 전구체를 포함하는 성장 배지와 접촉하고 있다.Another aspect provided herein is a compound having a structure of Formula VII or Formula VIII, wherein the compound of Formula VII or Formula VIII is biosynthetically produced in a cell comprising a pylC gene and a pylD gene, and the cell is a precursor It is in contact with the growth medium containing.
<화학식 VII><Formula VII>
<화학식 VIII><Formula VIII>
상기 화합물의 특정 실시양태에서, 세포는 pylB 유전자, pylC 유전자 및 pylD 유전자를 포함한다. In certain embodiments of the above compounds, the cell comprises a pylB gene, a pylC gene and a pylD gene.
상기 화합물의 특정 실시양태에서, 전구체는 오르니틴 또는 아르기닌이고, 한편 다른 실시양태에서 전구체는 D-오르니틴 또는 D-아르기닌 또는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. In certain embodiments of the above compounds, the precursor is ornithine or arginine, while in other embodiments the precursor is D-ornithine or D-arginine or (2S)-2-amino-6-((R)-2,5 -Diaminopentanamido)hexanoic acid.
상기 화합물의 특정 실시양태에서, 화학식 VII 또는 화학식 VIII의 화합물은 직교 tRNA (O-tRNA) 및 직교 아미노아실 tRNA 신테타제 (O-RS)에 의해 세포 내 단백질에 혼입되고, 여기서 O-RS는 O-tRNA를 화학식 V 또는 화학식 VI의 화합물로 아미노아실화시키고, O-tRNA는 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인지한다.In certain embodiments of the above compounds, the compound of Formula VII or Formula VIII is incorporated into an intracellular protein by an orthogonal tRNA (O-tRNA) and an orthogonal aminoacyl tRNA synthetase (O-RS), wherein O-RS is O -tRNA is aminoacylated with a compound of Formula V or Formula VI, and the O-tRNA recognizes one or more selector codons of the mRNA in the cell.
상기 화합물의 특정 실시양태에서, 세포는 추가로 pylS 유전자 및 pylT 유전자를 포함하고, 화학식 V 또는 화학식 VI의 화합물은 아미노아실 tRNA 신테타제, 및 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하는 tRNA에 의해 세포 내 단백질에 혼입되고, 여기서 아미노아실 tRNA 신테타제는 pylS 유전자의 유전자 생성물이고 tRNA는 pylT 유전자의 유전자 생성물이다. In certain embodiments of the above compounds, the cell further comprises a pylS gene and a pylT gene, and the compound of Formula V or Formula VI is by an aminoacyl tRNA synthetase, and a tRNA that recognizes one or more selector codons of the mRNA in the cell. It is incorporated into an intracellular protein, wherein the aminoacyl tRNA synthetase is the gene product of the pylS gene and the tRNA is the gene product of the pylT gene.
특정 실시양태에서 셀렉터 코돈은 앰버 코돈 (TAG)이다. 특정 실시양태에서, 세포는 원핵 세포이고, 한편 다른 실시양태에서 세포는 진핵 세포이다. 특정 실시양태에서, 세포는 에스케리키아 콜라이 세포이고, 한편 다른 실시양태에서 세포는 포유동물 세포, 효모 세포 또는 곤충 세포이다. 특정 실시양태에서, 효모 세포는 사카로미세스 세레비지애 또는 피키아 파스토랄리스 세포이다. 특정 실시양태에서, 포유동물 세포는 CHO 세포, HeLa 세포 또는 HEK293F 세포이다. 특정 실시양태에서, 곤충 세포는 sf9 세포이다. In certain embodiments the selector codon is an amber codon (TAG). In certain embodiments, the cell is a prokaryotic cell, while in other embodiments the cell is a eukaryotic cell. In certain embodiments, the cells are Escherichia coli cells, while in other embodiments the cells are mammalian cells, yeast cells or insect cells. In certain embodiments, the yeast cells are Saccharomyces cerevisiae or Pichia pastoralis cells. In certain embodiments, the mammalian cells are CHO cells, HeLa cells, or HEK293F cells. In certain embodiments, the insect cells are sf9 cells.
본원에서 제공되는 또 다른 측면은 하기 화학식 VII 또는 화학식 VIII의 구조를 갖는 화합물이고, 여기서 화학식 VII 또는 화학식 VIII의 화합물은 전구체 및 제2 세포를 포함하는 성장 배지와 접촉하고 있는 제1 세포에 의해 생합성적으로 생성 및 분비되고, 여기서 제1 세포는 pylC 유전자 및 pylD 유전자를 포함하는 피더 세포이다.Another aspect provided herein is a compound having the structure of Formula VII or Formula VIII, wherein the compound of Formula VII or Formula VIII is biosynthesized by a first cell in contact with a growth medium comprising a precursor and a second cell. It is produced and secreted by the antagonist, wherein the first cell is a feeder cell containing the pylC gene and the pylD gene.
<화학식 VII><Formula VII>
<화학식 VIII><Formula VIII>
상기 화합물의 특정 실시양태에서, 제1 세포는 pylB 유전자, pylC 유전자 및 pylD 유전자를 포함한다. In certain embodiments of the above compounds, the first cell comprises a pylB gene, a pylC gene and a pylD gene.
상기 화합물의 특정 실시양태에서, 전구체는 오르니틴 또는 아르기닌이다. 상기 화합물의 다른 실시양태에서, 전구체는 D-오르니틴 또는 D-아르기닌이다. 상기 화합물의 다른 실시양태에서, 전구체는 (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산이다. 상기 화합물의 다른 실시양태에서, 전구체는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. In certain embodiments of the above compounds, the precursor is ornithine or arginine. In other embodiments of the above compounds, the precursor is D-ornithine or D-arginine. In another embodiment of the above compound, the precursor is (2S)-2-amino-6-(2,5-diaminopentanamido)hexanoic acid. In another embodiment of the above compound, the precursor is (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid.
상기 화합물의 특정 실시양태에서, 화학식 VII 또는 화학식 VIII의 화합물은 직교 tRNA (O-tRNA) 및 직교 아미노아실 tRNA 신테타제 (O-RS)에 의해 제2 세포 내 단백질에 혼입되고, 여기서 O-RS는 O-tRNA를 화학식 VII 또는 화학식 VIII의 화합물로 아미노아실화시키고, O-tRNA는 제2 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인지한다.In certain embodiments of the above compounds, the compound of Formula VII or Formula VIII is incorporated into a second intracellular protein by an orthogonal tRNA (O-tRNA) and an orthogonal aminoacyl tRNA synthetase (O-RS), wherein the O-RS Aminoacylates the O-tRNA with a compound of Formula VII or Formula VIII, and the O-tRNA recognizes one or more selector codons of the mRNA in the second cell.
상기 화합물의 특정 실시양태에서, 제2 세포는 pylS 유전자 및 pylT 유전자를 포함하고, 화학식 VII 또는 화학식 VIII의 화합물은 제2 세포 내 단백질에 혼입된다. In certain embodiments of the above compounds, the second cell comprises a pylS gene and a pylT gene, and the compound of Formula VII or Formula VIII is incorporated into the protein in the second cell.
상기 화합물의 특정 실시양태에서, 화학식 VII 또는 화학식 VIII의 화합물은 아미노아실 tRNA 신테타제, 및 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하는 tRNA에 의해 제2 세포 내 단백질에 혼입되며, 여기서 아미노아실 tRNA 신테타제는 pylS 유전자의 유전자 생성물이고 tRNA는 pylT 유전자의 유전자 생성물이다. In certain embodiments of the above compounds, the compound of Formula VII or Formula VIII is incorporated into a second intracellular protein by an aminoacyl tRNA synthetase and a tRNA that recognizes one or more selector codons of the mRNA in the cell, wherein the aminoacyl tRNA Synthetase is the gene product of the pylS gene and tRNA is the gene product of the pylT gene.
상기 화합물의 특정 실시양태에서, 셀렉터 코돈은 앰버 코돈 (TAG)이다. In certain embodiments of the above compounds, the selector codon is an amber codon (TAG).
상기 화합물의 특정 실시양태에서, 제1 세포 또는 제2 세포는 원핵 세포이다. 상기 화합물의 특정 실시양태에서, 제1 세포 및 제2 세포는 원핵 세포이다. 상기 화합물의 특정 실시양태에서, 제1 세포 또는 제2 세포는 진핵 세포이다. 상기 화합물의 특정 실시양태에서, 제1 세포 및 제2 세포는 진핵 세포이다. In certain embodiments of the above compounds, the first cell or the second cell is a prokaryotic cell. In certain embodiments of the above compounds, the first and second cells are prokaryotic cells. In certain embodiments of the above compounds, the first cell or the second cell is a eukaryotic cell. In certain embodiments of the above compounds, the first and second cells are eukaryotic cells.
특정 실시양태에서, 원핵 세포는 에스케리키아 콜라이 세포이다. 특정 실시양태에서, 진핵 세포는 포유동물 세포, 효모 세포 또는 곤충 세포이다. 특정 실시양태에서, 효모 세포는 사카로미세스 세레비지애 또는 피키아 파스토랄리스 세포이다. 특정 실시양태에서, 포유동물 세포는 CHO 세포, HeLa 세포 또는 HEK293F 세포이다. 특정 실시양태에서, 곤충 세포는 sf9 세포이다. In certain embodiments, the prokaryotic cell is an Escherichia coli cell. In certain embodiments, the eukaryotic cell is a mammalian cell, a yeast cell, or an insect cell. In certain embodiments, the yeast cells are Saccharomyces cerevisiae or Pichia pastoralis cells. In certain embodiments, the mammalian cells are CHO cells, HeLa cells, or HEK293F cells. In certain embodiments, the insect cells are sf9 cells.
본원에서 제공되는 또 다른 측면은 하기 화학식 IX의 구조를 갖는 화합물이고, 여기서 화학식 IX의 화합물은 pylC 유전자 및 pylD 유전자를 포함하는 세포 내에서 생합성적으로 생성되고, 세포는 2,5-디아미노-3-메틸펜탄산 또는 D-2,5-디아미노-3-메틸펜탄산을 포함하는 성장 배지와 접촉하고 있다.Another aspect provided herein is a compound having the structure of Formula IX, wherein the compound of Formula IX is biosynthetically produced in cells containing the pylC gene and the pylD gene, and the cell is 2,5-diamino- It is in contact with a growth medium containing 3-methylpentanoic acid or D-2,5-diamino-3-methylpentanoic acid.
<화학식 IX><Formula IX>
상기 화합물의 특정 실시양태에서, 2,5-디아미노-3-메틸펜탄산은 (2R,3S)-2,5-디아미노-3-메틸펜탄산이다. In certain embodiments of the above compounds, 2,5-diamino-3-methylpentanoic acid is (2R,3S)-2,5-diamino-3-methylpentanoic acid.
상기 화합물의 특정 실시양태에서, 2,5-디아미노-3-메틸펜탄산은 (2R,3R)-2,5-디아미노-3-메틸펜탄산이다. In certain embodiments of the above compounds, 2,5-diamino-3-methylpentanoic acid is (2R,3R)-2,5-diamino-3-methylpentanoic acid.
상기 화합물의 특정 실시양태에서, 세포는 pylB 유전자, pylC 유전자 및 pylD 유전자를 포함하고, 세포는 오르니틴, 아르기닌, D-오르니틴, D-아르기닌, (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산 2,5-디아미노-3-메틸펜탄산 또는 D-2,5-디아미노-3-메틸펜탄산을 포함하는 성장 배지와 접촉하고 있다.In certain embodiments of the above compounds, the cell comprises a pylB gene, a pylC gene and a pylD gene, and the cell is ornithine, arginine, D-ornithine, D-arginine, (2S)-2-amino-6-(2 ,5-diaminopentanamido)
상기 화합물의 특정 실시양태에서, 화학식 IX의 화합물은 직교 tRNA (O-tRNA) 및 직교 아미노아실 tRNA 신테타제 (O-RS)에 의해 세포 내 단백질에 혼입되고, 여기서 O-RS는 O-tRNA를 화학식 IX의 화합물로 아미노아실화시키고, O-tRNA는 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인지한다. In certain embodiments of the above compounds, the compound of formula IX is incorporated into an intracellular protein by an orthogonal tRNA (O-tRNA) and an orthogonal aminoacyl tRNA synthetase (O-RS), wherein O-RS is an O-tRNA. Aminoacylated with a compound of formula IX, and the O-tRNA recognizes one or more selector codons of the mRNA in the cell.
상기 화합물의 특정 실시양태에서, 세포는 추가로 pylS 유전자 및 pylT 유전자를 포함하고, 화학식 IX의 화합물은 아미노아실 tRNA 신테타제, 및 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하는 tRNA에 의해 세포 내 단백질에 혼입되며, 여기서 아미노아실 tRNA 신테타제는 pylS 유전자의 유전자 생성물이고 tRNA는 pylT 유전자의 유전자 생성물이다. In certain embodiments of the above compounds, the cell further comprises a pylS gene and a pylT gene, and the compound of formula IX is an intracellular protein by an aminoacyl tRNA synthetase, and a tRNA that recognizes one or more selector codons of the mRNA in the cell. Wherein the aminoacyl tRNA synthetase is the gene product of the pylS gene and the tRNA is the gene product of the pylT gene.
특정 실시양태에서, 셀렉터 코돈은 앰버 코돈 (TAG)이다. 특정 실시양태에서, 세포는 원핵 세포이고, 한편 다른 실시양태에서, 세포는 진핵 세포이다. 특정 실시양태에서, 원핵 세포는 에스케리키아 콜라이 세포이다. 특정 실시양태에서, 진핵 세포는 포유동물 세포, 효모 세포 또는 곤충 세포이다. 특정 실시양태에서, 효모 세포는 사카로미세스 세레비지애 또는 피키아 파스토랄리스 세포이다. 특정 실시양태에서, 포유동물 세포는 CHO 세포, HeLa 세포 또는 HEK293F 세포이다. 특정 실시양태에서, 곤충 세포는 sf9 세포이다. In certain embodiments, the selector codon is an amber codon (TAG). In certain embodiments, the cell is a prokaryotic cell, while in other embodiments, the cell is a eukaryotic cell. In certain embodiments, the prokaryotic cell is an Escherichia coli cell. In certain embodiments, the eukaryotic cell is a mammalian cell, a yeast cell, or an insect cell. In certain embodiments, the yeast cells are Saccharomyces cerevisiae or Pichia pastoralis cells. In certain embodiments, the mammalian cells are CHO cells, HeLa cells, or HEK293F cells. In certain embodiments, the insect cells are sf9 cells.
본원에서 제공되는 또 다른 측면은 하기 화학식 IX의 구조를 갖는 화합물이고, 여기서 화학식 IX의 화합물은 2,5-디아미노-3-메틸펜탄산 또는 D-2,5-디아미노-3-메틸펜탄산을 포함하는 성장 배지 및 제2 세포와 접촉하고 있는 제1 세포에 의해 생합성적으로 생성 및 분비되고, 여기서 제1 세포는 pylC 유전자 및 pylD 유전자를 포함하는 피더 세포이다.Another aspect provided herein is a compound having the structure of formula IX, wherein the compound of formula IX is 2,5-diamino-3-methylpentanoic acid or D-2,5-diamino-3-methylphene It is biosynthetically produced and secreted by first cells in contact with the growth medium containing carbonic acid and the second cells, wherein the first cells are feeder cells containing the pylC gene and the pylD gene.
<화학식 IX><Formula IX>
상기 화합물의 특정 실시양태에서, 2,5-디아미노-3-메틸펜탄산은 (2R,3S)-2,5-디아미노-3-메틸펜탄산이다. In certain embodiments of the above compounds, 2,5-diamino-3-methylpentanoic acid is (2R,3S)-2,5-diamino-3-methylpentanoic acid.
상기 화합물의 특정 실시양태에서, 2,5-디아미노-3-메틸펜탄산은 (2R,3R)-2,5-디아미노-3-메틸펜탄산이다. In certain embodiments of the above compounds, 2,5-diamino-3-methylpentanoic acid is (2R,3R)-2,5-diamino-3-methylpentanoic acid.
상기 화합물의 특정 실시양태에서, 세포는 pylB 유전자, pylC 유전자 및 pylD 유전자를 포함하고, 세포는 오르니틴, 아르기닌, D-오르니틴, D-아르기닌, (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산 2,5-디아미노-3-메틸펜탄산 또는 D-2,5-디아미노-3-메틸펜탄산을 포함하는 성장 배지와 접촉하고 있다.In certain embodiments of the above compounds, the cell comprises a pylB gene, a pylC gene and a pylD gene, and the cell is ornithine, arginine, D-ornithine, D-arginine, (2S)-2-amino-6-(2 ,5-diaminopentanamido)
상기 화합물의 특정 실시양태에서, 화학식 IX의 화합물은 직교 tRNA (O-tRNA) 및 직교 아미노아실 tRNA 신테타제 (O-RS)에 의해 제2 세포 내 단백질에 혼입되고, 여기서 O-RS는 O-tRNA를 화학식 IX의 화합물로 아미노아실화시키고, O-tRNA는 제2 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인지한다. In certain embodiments of the above compounds, the compound of formula IX is incorporated into a second intracellular protein by an orthogonal tRNA (O-tRNA) and an orthogonal aminoacyl tRNA synthetase (O-RS), wherein O-RS is O- The tRNA is aminoacylated with the compound of formula IX, and the O-tRNA recognizes one or more selector codons of the mRNA in the second cell.
상기 화합물의 특정 실시양태에서, 제2 세포는 pylS 유전자 및 pylT 유전자를 포함하고, 화학식 IX의 화합물은 제2 세포 내 단백질에 혼입된다. In certain embodiments of the above compounds, the second cell comprises a pylS gene and a pylT gene, and the compound of formula IX is incorporated into the protein in the second cell.
상기 화합물의 특정 실시양태에서, 화학식 IX의 화합물은 아미노아실 tRNA 신테타제, 및 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하는 tRNA에 의해 제2 세포 내 단백질에 혼입되고, 여기서 아미노아실 tRNA 신테타제는 pylS 유전자의 유전자 생성물이고 tRNA는 pylT 유전자의 유전자 생성물이다. In certain embodiments of the above compounds, the compound of formula IX is incorporated into a second intracellular protein by an aminoacyl tRNA synthetase, and a tRNA that recognizes one or more selector codons of the mRNA in the cell, wherein the aminoacyl tRNA synthetase is The gene product of the pylS gene and tRNA is the gene product of the pylT gene.
특정 실시양태에서, 셀렉터 코돈은 앰버 코돈 (TAG)이다. In certain embodiments, the selector codon is an amber codon (TAG).
특정 실시양태에서, 제1 세포 또는 제2 세포는 원핵 세포이다. 특정 실시양태에서, 제1 세포 및 제2 세포는 원핵 세포이다. 특정 실시양태에서, 제1 세포 또는 제2 세포는 진핵 세포이다. 특정 실시양태에서, 제1 세포 및 제2 세포는 진핵 세포이다. 특정 실시양태에서, 원핵 세포는 에스케리키아 콜라이 세포이다. 특정 실시양태에서 진핵 세포는 포유동물 세포, 효모 세포 또는 곤충 세포이다. 특정 실시양태에서, 효모 세포는 사카로미세스 세레비지애 또는 피키아 파스토랄리스 세포이다. 특정 실시양태에서, 포유동물 세포는 CHO 세포, HeLa 세포 또는 HEK293F 세포이다. 특정 실시양태에서, 곤충 세포는 sf9 세포이다. In certain embodiments, the first cell or the second cell is a prokaryotic cell. In certain embodiments, the first and second cells are prokaryotic cells. In certain embodiments, the first cell or the second cell is a eukaryotic cell. In certain embodiments, the first and second cells are eukaryotic cells. In certain embodiments, the prokaryotic cell is an Escherichia coli cell. In certain embodiments the eukaryotic cell is a mammalian cell, a yeast cell or an insect cell. In certain embodiments, the yeast cells are Saccharomyces cerevisiae or Pichia pastoralis cells. In certain embodiments, the mammalian cells are CHO cells, HeLa cells, or HEK293F cells. In certain embodiments, the insect cells are sf9 cells.
본원에서 제공되는 또 다른 측면은 하기 화학식 IV의 화합물이고,Another aspect provided herein is a compound of formula IV,
여기서 1개 이상의 폴리에틸렌 글리콜 (PEG) 잔기를 갖는 화합물은 1000 Da 내지 50 kDa 범위의 평균 분자량을 갖고, n은 20 내지 1200이고, exPADRE는 이고, PADRE는 이고, BG1은 이고, BG2는 이고, 여기서 *는 포스포티오에이트 연결부를 표시한다.Wherein the compound having at least one polyethylene glycol (PEG) moiety has an average molecular weight in the range of 1000 Da to 50 kDa, n is 20 to 1200, and exPADRE is And PADRE is And BG1 is And BG2 is And, where * represents a phosphorothioate linkage.
도 1. 피롤리신 (PYL) 및 피롤리신 유사체 피롤린-카르복시-리신 (PCL: PCL-A 또는 PCL-B)의 구조.
도 2. 피롤리신 (A) 및 PCL (B)에 대한 가능한 생합성 경로의 비교.
도 3. 피롤리신 (A) 및 PCL (B)에 의한 pylT tRNA의 아미노아실화.
도 4A. 포유동물 세포에서의 생합성적으로 유도된 PCL 또는 피롤리신의 혼입을 위한 pylT, pylS, pylB, pylC 및 pylD를 보유하고, 모델 단백질 hRBP4에 대한 단일 부위 TAG 돌연변이 유전자 구축물을 보유하는 플라스미드.
도 4B. hRBP4, mEPO, hEPO 및 mIgG1에 대한 단일 부위 TAG 돌연변이 유전자 구축물. 단일 부위는 화살표에 의해 나타내었다.
도 5. 에스케리키아 콜라이(Escherichia coli) 세포에서의 생합성적으로 유도된 PCL 또는 피롤리신의 혼입을 위한 pylT, pylS, pylB, pylC 및 pylD를 보유하는 플라스미드.
도 6A. HEK293F 세포에서의 hRBP4의 발현. hRBP4 (#1-9)의 TAG 돌연변이 구축물. SDS-PAGE에 이은, 항-His 항체에 의한 웨스턴 블롯. 26 kDa에서의 화살표는 전장 hRBP4를 나타낸다.
도 6B 및 6C. D-오르니틴의 존재 하에 HEK293F 세포에서 생성된 정제된 hRBP4 Phe62PCL의 SDS-PAGE (A) 및 질량 스펙트럼 (B).
도 7. hRBP4 Phe122PCL의 트립신 소화물의 질량 분광측정 분석은 표적 Phe122TAG 부위에의 PCL의 혼입을 나타낸다. YWGVASF*LQK (F* = PCL) (서열 17)의 지정된 MS/MS 스펙트럼.
도 8. hRBP4 Phe122PCL의 트립신 소화물의 질량 스펙트럼 분광측정 분석 (YWGVASF*LQK의 2+ 이온의 TIC 및 EIC) (서열 17)은 표적 Phe122TAG 부위에의 PCL의 혼입을 나타낸다.
도 9. hRBP4 Phe122PCL의 트립신 소화물의 질량 분광측정 분석. 질량 스펙트럼은 표적 Phe122TAG 부위에의 PCL의 혼입을 나타내는 YWGVASF*LQK (서열 17)의 3+ 및 2+ 전구체를 보여준다.
도 10. HEK293F 세포로부터의 용해물에서의, D-오르니틴으로부터 생합성된 PCL의 검출.
도 11. HEK293F 세포에서의 다양한 hRBP4 TAG 돌연변이 단백질로의 N-ε-시클로펜틸옥시카르보닐-L-리신 (CYC)의 혼입. 도 11A는 SDS-PAGE, 및 항-His-태그 및 항-RBP4 항체에 의한 웨스턴 블롯팅에 의해 검출된 바와 같은, hRBP4에서의 다양한 부위에의 CYC 혼입을 보여준다. 도 11B는 정제된 hRBP4 Phe62CYC (돌연변이체 #2)의 SDS-PAGE 결과를 보여준다. 도 11C는 hRBP4 Phe62CYC의 질량 스펙트럼을 보여준다.
도 12. hRBP4 돌연변이체 Phe62CYC의 TAG 부위에의 CYC 혼입의 질량 분광측정 검증.
도 13. 다양한 전구체의 기능으로서의 PCL 혼입 및 HEK293F 세포에 의한 hRBP4 TAG 돌연변이 단백질로의 다양한 피롤리신 유사체 (CYC 포함)의 직접 혼입 (도 13A 및 도 13B), 및 생합성 유전자 pylB, pylC 및 pylD의 다양한 조합을 이용한 PCL 혼입 (도 13C). 도 13A는 항-His-태그 항체에 의한 정제되지 않은 샘플의 웨스턴 블롯이고, 도 13B는 Ni-NTA 정제된 단백질의 SDS-PAGE 겔이다.
도 14. PCL 생합성 (A) 및 다양한 피롤리신 유사체 (B)를 위한 잠재적 전구체.
도 15. HK100 세포에서의 FASTE로의 PCL의 혼입을 위한 다양한 전구체 및 유전자 조합의 평가.
도 16. PCL (PCL-A) 형성에 대한 잠재적 생합성 반응식.
도 17A. 마우스 IgG1의 Fc 도메인의 단일 TAG 코딩 부위 (4 부위)에의 생합성적으로 생성된 PCL의 부위 특이적 혼입.
도 17B. SDS-PAGE에 의해 검출된 바와 같은 에리트로포이에틴 (EPO)의 단일 TAG 코딩 부위 (11 부위)에의 생합성적으로 생성된 PCL의 부위 특이적 혼입.
도 18. 인간 지방산 신테타제 (FAS-TE)의 티오에스테라제 도메인의 단일 TAG 코딩 부위 (2 부위)에의 생합성적으로 생성된 PCL의 부위 특이적 혼입. 도 18은 SDS-PAGE (A) 및 PCL 혼입에 대한 질량 스펙트럼 (B)을 보여준다.
도 19. FKBP-12의 단일 TAG 코딩 부위 (1 부위)에의 생합성적으로 생성된 PCL의 부위 특이적 혼입. 도 19는 SDS-PAGE (A) 및 PCL 혼입에 대한 질량 스펙트럼 (B) 및 FKBP12-I90PCL의 결정 (C)을 보여준다.
도 20. 섬유모세포 성장 인자 21 (FGF21)의 단일 TAG 코딩 부위 (20 부위)에의 생합성적으로 생성된 PCL의 부위 특이적 혼입. SDS-PAGE는 다중 부위에서의 FGF21 내로의 PCL 혼입을 보여준다.
도 21. 도 21A는 pylB의 존재 및 부재 하에 글루타민 Gln21 (CAA)이 TAG 종결 코돈으로 돌연변이된 mTNF-α 내로의 PYL- 또는 PCL-혼입의 SDS-PAGE 분석을 보여준다. 도 21B 단백질의 순도를 평가하는 SDS-PAGE. 도 21C는 단백질의 혼합물에 PYL 및 PCL이 혼입되었고 (도 21C, 하단), PCL이 우세하게 혼입되었음 (도 21C, 상단)을 시사하는, 에스케리키아 콜라이에서 발현된 mEGF Tyr10TAG의 원형 질량 스펙트럼이다.
도 22. 2-아미노-벤즈알데히드에 의한 PCL의 화학적 유도체화에 대한 가능한 반응식.
도 23. 2-아미노 벤즈알데히드, 2-아미노아세토페논 및 2-아미노-5-니트로-벤조페논에 의한 PCL의 유도체화 후에 가정된 단백질 접합체 및 질량 변화.
도 24. 2-아미노-벤즈알데히드 (2-ABA)로 유도체화된 hRBP4 Phe122PCL의 질량 분광측정 분석.
도 25. 2-ABA-유도체화 hRBP4 Phe122PCL 단백질의 트립신 소화물의 질량 분광측정 분석은 TAG 부위에 혼입된 PCL 잔기의 유도체화를 입증한다. YWGVASF*LQK 펩티드 (서열 17).
도 26. 2-ABA에 의한 hRBP4 Phe122PCL의 유도체화의 pH 의존성 평가.
도 27. 단백질 농도에 대한 반응물의 비율의 함수로서의 반응 효율, 및 2-ABA, 2-ANBP와 2-AAP와의 반응성의 평가.
도 28. 4700배를 초과하는 몰비에서 (A : 단백질 대비 4700배 과량의 2-ABA) 및 OMePhe 혼입된 hRBP4 (B: 15400배 과량)에 대한 유도체화.
도 29. 2-아미노-아세토페논 (2-AAP)에 의한 FAS-TE Tyr2454PCL의 유도체화. 미반응 샘플 (A와 C) 및 pH 5.0 (B) 및 pH 7.4 (D)에서의 2-AAP로 유도체화된 샘플의 질량 스펙트럼.
도 30. 2-아미노-벤즈알데히드 또는 2-아미노-벤즈알데히드 유사체에 의한 피롤리신 및/또는 PCL의 화학적 유도체화를 통한 단백질의 부위 특이적 변형에 대한 일반적 반응식.
도 31. 단백질에 혼입된 PCL 잔기를 통해 단백질에 커플링된 관능화된 PEG 중합체, 2-아미노-아세토페논-PEG8 (2-AAP-PEG8; TU3205-044)의 실시양태의 예시.
도 32. 2-AAP-PEG8에 의한 hRBP4 Phe122PCL의 유도체화. 미반응 hRBP4 Phe122PCL 단백질 (C), 및 2-AAP-PEG8이 첨가된 야생형 hRBP4 (D와 E) 및 2-AAP-PEG8이 첨가되지 않은 야생형 hRBP4의 질량 스펙트럼과 비교한, 위치 122에 혼입된 PCL을 갖는 hRBP4의 2-AAP-PEG8에 의한 유도체화 후 pH 7.5 (A) 및 ph 5.0 (B)에서의 질량 스펙트럼.
도 33. 2-AAP-PEG8에 의한 FAS-TE Tyr2454PCL의 유도체화. 미반응 단백질 (A), 및 2-AAP-PEG8 (TU3205-044)과 반응시킨 FAS-TE Tyr2454PCL 단백질 (B)에 대한 질량 스펙트럼. 도 33C 및 33D는 2.4 kDa 2-AAP-PEG (TU3205-048)에 의한 FAS-TE Tyr2454PCL의 유도체화를 보여준다 (도 33C는 실온에서, 도 33D는 4℃에서 유도체화함).
도 34. 나타낸 몰비에서의 0.5 kDa 2-AAP-PEG (2-AAP-PEG8), 2.4 kDa 2-AAP-PEG 및 23 kDa 2-AAP-PEG에 의한 FAS-TE Tyr2454PCL의 PEG화.
도 35. 2-AAP-PEG8에 의한 FGF21 Lys81PCL의 유도체화. 미반응 FGF21 Lys84PCL (A) 및 2-AAP-PEG8에 의한 유도체화 후의 FGF21 Lys84PCL (B)의 질량 스펙트럼.
도 36. FGF21 단백질의 PEG화. PEG-FGF21, 전장 (FL) FGF21-PCL 및 말단절단 (TR) FGF21-PCL을 보여주는, 23 kDa 2-AAP-PEG에 의한 7가지 FGF21 PCL 돌연변이체의 유도체화 후, 부분적 정제 전 (A) 및 후 (B)에 수득된 SDS-PAGE 결과.
도 37. EPO 단백질의 PEG화. 23 kDa 2-AAP-PEG에 의한 마우스 EPO PCL 돌연변이체의 유도체화 후의 SDS-PAGE.
도 38. 아미노 당에 의한 PCL의 유도체화. 내부에 혼입된 PCL을 갖는 단백질 (R1로서 나타냄)이 D-만노사민과 커플링되는 일반화된 반응식.
도 39. D-만노사민에 의한 hRBP4 Phe122PCL의 유도체화. 만노사민과의 반응 후에 위치 122에 혼입된 PCL을 갖는 hRBP4의 질량 스펙트럼.
도 40. D-만노사민에 의한 FAS-TE Leu2222PCL의 유도체화. 위치 2222에 혼입된 PCL을 갖는 미반응 인간 지방산 신테타제 (FAS-TE) (FAS-TE Leu2222PCL/Leu2223Ile) (A) 및 만노사민과 반응시킨 단백질 (B)의 질량 스펙트럼.
도 41. 단백질에 혼입된 PCL을 갖는 올리고사카라이드에 연결된 2-ABA 잔기를 반응시키는 것을 통한 단백질에의 올리고사카라이드의 부위 특이적 부착을 위한 실시양태의 예시.
도 42. 내부에 혼입된 PCL을 갖는 가교 단백질에 의해 형성된 단백질-단백질 접합체 (이종이량체, 이종삼량체, 동종삼량체)의 특정 실시양태의 예시.
도 43. FGF21 Lys84PCL 단백질에 대해 예시된 바와 같은 PCL 특이적, 이관능성 가교제에 의한 동종이량체 형성. 동종이량체를 형성하는 데 사용되는 이관능성 가교제의 비제한적인 예를 A에 나타내고, 상기 이관능성 링커를 사용하여 가교된 FGF-21 Lys84PCL의 반응 혼합물의 질량 스펙트럼을 B에 나타내었다.
도 44. 이관능성 가교제에 의한 FGF-21 PCL 돌연변이 단백질의 동종이량체 형성. 도 44A는 가교된 FGF-21의 질량 스펙트럼을 보여주며, 여기서 반응 조건은 도 43에서 사용된 조건으로부터 변경된다.
도 45. 삼량체를 형성하는 데 사용되는 가교제에 대한 실시양태.
도 46. 부위 특이적 표지 및 본원에서 제공되는 방법을 이용한 표지화의 다양한 실시양태의 예시.
도 47. 도 47A는 비오틴과 접합된 mEGF-Y10PCL의 ESI 질량 분광측정 분석을 보여준다. 도 47B는 양고추냉이 퍼옥시다제 (HRP) 접합된 염소 항-비오틴 항체를 사용한 mEGF-Y10PCL-ABA-비오틴 접합체의 웨스턴 블롯을 보여준다. 도 47C는 플루오레세인과 접합된 mEGF-Y10PCL의 ESI 질량 분광측정 분석을 보여준다. 도 47D는 디사카라이드와 접합된 mEGF-Y10PCL의 ESI 질량 분광측정 분석을 보여준다.
도 48. 도 48A는 모노-니트로페닐 합텐과 접합된 mTNF-Q21PCL의 ESI 질량 분광측정 분석을 보여준다. 도 48B는 모노-니트로페닐 합텐과 접합된 mEGF-Y10PCL의 ESI 질량 분광측정 분석을 보여준다. 도 48C는 디-니트로페닐 합텐과 접합된 mTNF-Q21PCL의 ESI 질량 분광측정 분석을 보여준다. 도 48D는 디-니트로페닐 합텐과 접합된 mEGF-Y10PCL의 ESI 질량 분광측정 분석을 보여준다.
도 49. 도 49A는 TLR7 효능제와 접합된 mEGF-Y10PCL의 ESI 질량 분광측정 분석을 보여주고, 도 49B는 인지질과 접합된 mEGF-Y10PCL의 ESI 질량 분광측정 분석을 보여준다.
도 50. 도 50A 및 도 50B는 2가지 상이한 pH 값 (도 50A: pH 5.0; 도 50B: pH 7.5)에서 PX2-PADRE와 접합된 mTNF-Q21PCL의 MALDI-TOF 질량 분광측정 분석을 보여준다. 도 50C는 BHA-exPADRE와 접합된 mTNF-Q21PCL의 ESI 질량 분광측정 분석을 보여준다.
도 51. 도 51은 BHA-exPADRE의 mEGF-Y10PCL에 대한 커플링을 보여주는 ESI 질량 스펙트럼이다.
도 52. 도 52A는 BHA-BG1 (7.4 kDa) 및 BHA-BG2 (7.4 kDa)의 mTNF-Q21PCL (19.3 kDa)에 대한 커플링의 겔 이동 검정이다. 도 52B는 BHA-BG2 (7.4 kDa)의 mEGF-Y10PCL (7.2 kDa)에 대한 커플링의 겔 이동 검정이다.
도 53. 도 53은 이러한 부위 특이적 배향 부착의 실시양태를 예시한다.
도 54. 도 54A는 2-ABA에 커플링시키고, 이어서 20 mM NaCNBH3으로 1시간 동안 환원시킨 hFGF21-K150PCL의 ESI 질량 분광측정 분석을 보여준다. 도 54B는 10 mM 인산염 완충액 (pH 7.5)으로 투석하고, 50℃에서 1일 동안 인큐베이션한 후의 환원된 hFGF21-K150PCL 2-ABA 접합체의 ESI 질량 분광측정 분석을 보여준다.
도 55. 도 55는 NaCNBH3을 사용한 환원의 존재 및 부재 하에서 PEG화 FGF21에 대한 PCL 연결부의 안정성을 입증한다. 환원된 샘플과 비환원된 샘플에 대한 SDS-PAGE 겔을 도 55A에 나타내었다. 추가로, 도 55B는 4℃, 실온, 37℃ 및 50℃, 및 95℃에서 60시간 동안 인큐베이션한 비환원된 샘플에 대한 SDS-PAGE 겔을 보여준다.
도 56. 2-ABA와의 PCL-A 반응의 NMR 분석.
도 57. 도 57A는 PCL-A 및 2-ABA 사이의 반응으로부터 생성된 생성물의 제안된 구조이다. 도 57B는 PCL-A 및 2-ABA 사이의 반응으로부터 생성된 생성물의 제안된 평형 구조를 보여준다. 도 57C는 환원된 생성물의 제안된 구조이다.
도 58. 2-ABA와의 PCL-B 반응의 NMR 분석.
도 59. mEGF에 혼입된 피롤리신 (Pyl) 및 PCL의 유도체화.Fig. 1. Structure of pyrrolysine (PYL) and pyrrolysine analog pyrroline-carboxy-lysine (PCL: PCL-A or PCL-B).
Figure 2. Comparison of possible biosynthetic pathways for pyrrolysine (A) and PCL (B).
Fig. 3. Aminoacylation of pylT tRNA by pyrrolysine (A) and PCL (B).
Fig. 4A. Plasmids containing pylT, pylS, pylB, pylC and pylD for incorporation of biosynthetically induced PCL or pyrrolysine in mammalian cells, and a single site TAG mutant gene construct for the model protein hRBP4.
Fig. 4B. Single site TAG mutant gene constructs for hRBP4, mEPO, hEPO and mIgG1. Single sites are indicated by arrows.
Fig. 5. Escherichia coli ) Plasmids containing pylT, pylS, pylB, pylC and pylD for incorporation of biosynthetically induced PCL or pyrrolysine in cells.
Fig. 6A. Expression of hRBP4 in HEK293F cells. TAG mutant construct of hRBP4 (#1-9). SDS-PAGE followed by Western blot by anti-His antibody. Arrows at 26 kDa indicate full length hRBP4.
Figures 6B and 6C. SDS-PAGE (A) and mass spectrum (B) of purified hRBP4 Phe62PCL produced in HEK293F cells in the presence of D-ornithine.
Figure 7. Mass spectrometric analysis of the trypsin digest of hRBP4 Phe122PCL shows the incorporation of PCL into the target Phe122TAG site. Designated MS/MS spectrum of YWGVASF*LQK (F* = PCL) (SEQ ID NO: 17).
Figure 8. Mass spectrum spectrometric analysis of trypsin digest of hRBP4 Phe122PCL (TIC and EIC of 2+ ions of YWGVASF*LQK) (SEQ ID NO: 17) shows the incorporation of PCL into the target Phe122TAG site.
Figure 9. Mass spectrometric analysis of trypsin digest of hRBP4 Phe122PCL. The mass spectrum shows the 3+ and 2+ precursors of YWGVASF*LQK (SEQ ID NO: 17) indicating incorporation of PCL into the target Phe122TAG site.
Figure 10. Detection of PCL biosynthesized from D-ornithine in lysates from HEK293F cells.
Figure 11. Incorporation of N-ε-cyclopentyloxycarbonyl-L-lysine (CYC) into various hRBP4 TAG mutant proteins in HEK293F cells. 11A shows CYC incorporation at various sites in hRBP4, as detected by SDS-PAGE, and Western blotting with anti-His-tag and anti-RBP4 antibodies. 11B shows the SDS-PAGE result of purified hRBP4 Phe62CYC (mutant #2). 11C shows the mass spectrum of hRBP4 Phe62CYC.
Figure 12. Mass spectrometric verification of CYC incorporation into the TAG site of the hRBP4 mutant Phe62CYC.
Figure 13. PCL incorporation as a function of various precursors and direct incorporation of various pyrrolysine analogs (including CYC) into hRBP4 TAG mutant proteins by HEK293F cells (Figs. 13A and 13B), and of biosynthetic genes pylB, pylC and pylD PCL incorporation using various combinations (Figure 13C). 13A is a Western blot of a sample that is not purified by an anti-His-tag antibody, and FIG. 13B is an SDS-PAGE gel of a Ni-NTA purified protein.
Figure 14. Potential precursors for PCL biosynthesis (A) and various pyrrolysine analogs (B).
Figure 15. Evaluation of various precursor and gene combinations for incorporation of PCL into FASTE in HK100 cells.
Figure 16. Potential biosynthetic scheme for PCL (PCL-A) formation.
Fig. 17A. Site-specific incorporation of biosynthetically generated PCL into a single TAG coding site (4 sites) of the Fc domain of mouse IgG1.
Fig. 17B. Site-specific incorporation of biosynthetically generated PCL into a single TAG coding site (11 sites) of erythropoietin (EPO) as detected by SDS-PAGE.
Figure 18. Site-specific incorporation of biosynthetically generated PCL into a single TAG coding site (2 sites) of the thioesterase domain of human fatty acid synthetase (FAS-TE). Figure 18 shows the mass spectrum (B) for SDS-PAGE (A) and PCL incorporation.
Fig. 19. Site-specific incorporation of biosynthetically generated PCL into a single TAG coding site (1 site) of FKBP-12. Figure 19 shows the mass spectrum for SDS-PAGE (A) and PCL incorporation (B) and the determination of FKBP12-I90PCL (C).
Fig. 20. Site-specific incorporation of biosynthetically generated PCL into a single TAG coding site (20 sites) of fibroblast growth factor 21 (FGF21). SDS-PAGE shows PCL incorporation into FGF21 at multiple sites.
Figure 21. Figure 21A shows the SDS-PAGE analysis of PYL- or PCL-incorporation into mTNF-α where glutamine Gln21 (CAA) has been mutated with the TAG stop codon in the presence and absence of pylB. Figure 21B SDS-PAGE evaluating the purity of the protein. Figure 21C is a circular mass spectrum of mEGF Tyr10TAG expressed in Escherichia coli, indicating that PYL and PCL were incorporated into a mixture of proteins (Figure 21C, bottom), and PCL was predominantly incorporated (Figure 21C, top). .
Figure 22. Possible reaction scheme for chemical derivatization of PCL with 2-amino-benzaldehyde.
Figure 23. Assumed protein conjugate and mass change after derivatization of PCL with 2-amino benzaldehyde, 2-aminoacetophenone and 2-amino-5-nitro-benzophenone.
Figure 24. Mass spectrometric analysis of hRBP4 Phe122PCL derivatized with 2-amino-benzaldehyde (2-ABA).
Figure 25. Mass spectrometric analysis of the trypsin digest of the 2-ABA-derivatized hRBP4 Phe122PCL protein demonstrates derivatization of PCL residues incorporated in the TAG site. YWGVASF*LQK peptide (SEQ ID NO: 17).
Fig. 26. Evaluation of pH dependence of derivatization of hRBP4 Phe122PCL by 2-ABA.
Figure 27. Evaluation of reaction efficiency as a function of ratio of reactants to protein concentration, and reactivity with 2-ABA, 2-ANBP and 2-AAP.
Fig. 28. Derivatization for hRBP4 (B: 15400-fold excess) with OMePhe and OMePhe at a molar ratio exceeding 4700 times (A: 4700 times excess of 2-ABA compared to protein).
Figure 29. Derivatization of FAS-TE Tyr2454PCL with 2-amino-acetophenone (2-AAP). Mass spectra of unreacted samples (A and C) and samples derivatized with 2-AAP at pH 5.0 (B) and pH 7.4 (D).
Figure 30. General scheme for site-specific modification of proteins through chemical derivatization of pyrrolysine and/or PCL by 2-amino-benzaldehyde or 2-amino-benzaldehyde analogs.
Figure 31. Illustrative of an embodiment of a functionalized PEG polymer, 2-amino-acetophenone-PEG8 (2-AAP-PEG8; TU3205-044), coupled to a protein via a PCL residue incorporated into the protein.
Fig. 32. Derivatization of hRBP4 Phe122PCL by 2-AAP-PEG8. PCL incorporated at position 122 compared to the mass spectra of unreacted hRBP4 Phe122PCL protein (C), and wild-type hRBP4 (D and E) with 2-AAP-PEG8 added and wild-type hRBP4 without 2-AAP-PEG8 added. Mass spectra at pH 7.5 (A) and pH 5.0 (B) after derivatization of hRBP4 with 2-AAP-PEG8 having.
Figure 33. Derivatization of FAS-TE Tyr2454PCL by 2-AAP-PEG8. Mass spectra for unreacted protein (A), and FAS-TE Tyr2454PCL protein (B) reacted with 2-AAP-PEG8 (TU3205-044). Figures 33C and 33D show the derivatization of FAS-TE Tyr2454PCL by 2.4 kDa 2-AAP-PEG (TU3205-048) (Figure 33C at room temperature, Figure 33D at 4°C).
Fig. 34. PEGylation of FAS-TE Tyr2454PCL with 0.5 kDa 2-AAP-PEG (2-AAP-PEG8), 2.4 kDa 2-AAP-PEG and 23 kDa 2-AAP-PEG at the molar ratios shown.
Figure 35. Derivatization of FGF21 Lys81PCL by 2-AAP-PEG8. Mass spectrum of unreacted FGF21 Lys84PCL (A) and FGF21 Lys84PCL (B) after derivatization with 2-AAP-PEG8.
Figure 36. PEGylation of FGF21 protein. After derivatization of 7 FGF21 PCL mutants with 23 kDa 2-AAP-PEG showing PEG-FGF21, full length (FL) FGF21-PCL and truncated (TR) FGF21-PCL, before partial purification (A) and The SDS-PAGE result obtained after (B).
Figure 37. PEGylation of EPO protein. SDS-PAGE after derivatization of mouse EPO PCL mutants by 23 kDa 2-AAP-PEG.
Figure 38. Derivatization of PCL with amino sugars. Generalized reaction scheme in which a protein with PCL incorporated therein ( represented as R 1 ) is coupled with D-mannosamine.
Fig. 39. Derivatization of hRBP4 Phe122PCL by D-mannosamine. Mass spectrum of hRBP4 with PCL incorporated at position 122 after reaction with mannosamine.
Fig. 40. Derivatization of FAS-TE Leu2222PCL by D-mannosamine. Mass spectrum of unreacted human fatty acid synthetase (FAS-TE) (FAS-TE Leu2222PCL/Leu2223Ile) with PCL incorporated at position 2222 (A) and protein reacted with mannosamine (B).
Figure 41. Illustration of an embodiment for site specific attachment of an oligosaccharide to a protein via reacting a 2-ABA moiety linked to an oligosaccharide with PCL incorporated into the protein.
Figure 42. Illustration of certain embodiments of protein-protein conjugates (heterodimers, heterotrimers, homotrimers) formed by crosslinked proteins with PCL incorporated therein.
Figure 43. Homodimer formation by PCL-specific, bifunctional crosslinkers as exemplified for the FGF21 Lys84PCL protein. A non-limiting example of the bifunctional crosslinking agent used to form the homodimer is shown in A, and the mass spectrum of the reaction mixture of FGF-21 Lys84PCL crosslinked using the bifunctional linker is shown in B.
Fig. 44. Homodimer formation of FGF-21 PCL mutant protein by bifunctional crosslinking agent. Figure 44A shows the mass spectrum of crosslinked FGF-21, where the reaction conditions are changed from the conditions used in Figure 43.
Figure 45. Embodiments for crosslinking agents used to form trimers.
Figure 46. Illustration of various embodiments of site specific labeling and labeling using the methods provided herein.
Figure 47. Figure 47A shows the ESI mass spectrometric analysis of mEGF-Y10PCL conjugated with biotin. Figure 47B shows a Western blot of mEGF-Y10PCL-ABA-biotin conjugate using horseradish peroxidase (HRP) conjugated goat anti-biotin antibody. Figure 47C shows the ESI mass spectrometric analysis of mEGF-Y10PCL conjugated with fluorescein. Figure 47D shows the ESI mass spectrometric analysis of mEGF-Y10PCL conjugated with disaccharide.
Figure 48. Figure 48A shows the ESI mass spectrometric analysis of mTNF-Q21PCL conjugated with mono-nitrophenyl hapten. 48B shows the ESI mass spectrometric analysis of mEGF-Y10PCL conjugated with mono-nitrophenyl hapten. 48C shows the ESI mass spectrometric analysis of mTNF-Q21PCL conjugated with di-nitrophenyl hapten. 48D shows the ESI mass spectrometric analysis of mEGF-Y10PCL conjugated with di-nitrophenyl hapten.
Figure 49. Figure 49A shows the ESI mass spectrometric analysis of mEGF-Y10PCL conjugated with a TLR7 agonist, and Figure 49B shows the ESI mass spectrometric analysis of mEGF-Y10PCL conjugated with a phospholipid.
Figure 50. Figure 50A and Figure 50B show MALDI-TOF mass spectrometric analysis of mTNF-Q21PCL conjugated with PX2-PADRE at two different pH values (Figure 50A: pH 5.0; Figure 50B: pH 7.5). Figure 50C shows the ESI mass spectrometric analysis of mTNF-Q21PCL conjugated with BHA-exPADRE.
Fig. 51. Fig. 51 is an ESI mass spectrum showing the coupling of BHA-exPADRE to mEGF-Y10PCL.
Figure 52. Figure 52A is a gel shift assay of the coupling of BHA-BG1 (7.4 kDa) and BHA-BG2 (7.4 kDa) to mTNF-Q21PCL (19.3 kDa). Figure 52B is a gel shift assay of the coupling of BHA-BG2 (7.4 kDa) to mEGF-Y10PCL (7.2 kDa).
Figure 53. Figure 53 illustrates an embodiment of this site specific orientation attachment.
Figure 54. Figure 54A shows the ESI mass spectrometric analysis of hFGF21-K150PCL coupled to 2-ABA and then reduced with 20 mM NaCNBH 3 for 1 hour. 54B shows the ESI mass spectrometric analysis of the reduced hFGF21-K150PCL 2-ABA conjugate after dialysis with 10 mM phosphate buffer (pH 7.5) and incubation at 50° C. for 1 day.
Figure 55. Figure 55 demonstrates the stability of the PCL linkage to PEGylated FGF21 in the presence and absence of reduction with NaCNBH 3. SDS-PAGE gels for the reduced and non-reduced samples are shown in Figure 55A. Additionally, Figure 55B shows SDS-PAGE gels for non-reduced samples incubated at 4°C, room temperature, 37°C and 50°C, and 95°C for 60 hours.
Figure 56. NMR analysis of PCL-A reaction with 2-ABA.
Figure 57. Figure 57A is the proposed structure of the product resulting from the reaction between PCL-A and 2-ABA. 57B shows the proposed equilibrium structure of the product resulting from the reaction between PCL-A and 2-ABA. 57C is the proposed structure of the reduced product.
Fig. 58. NMR analysis of PCL-B reaction with 2-ABA.
Figure 59. Derivatization of pyrrolysine (Pyl) and PCL incorporated in mEGF.
단백질, 폴리펩티드 및/또는 펩티드를 부위 특이적으로 변형시키는 데 사용되는 방법 및 조성물이 본원에 제공되며, 여기서 이러한 방법은 유전적으로 코딩된 피롤리신 또는 피롤린-카르복시-리신 (PCL) 잔기 (여기서, 피롤리신 및 PCL 아미노산은 생합성적으로 생성됨)의 부위 특이적 변형을 수반한다. 생합성적으로 내부에 혼입된 1개 이상의 PCL 잔기 또는 피롤리신을 갖는 단백질, 폴리펩티드 및/또는 펩티드에 부위 특이적으로 커플링되는 다양한 유형의 분자가 본원에서 제공된다. 특정 실시양태에서, 이러한 부위 특이적 변형은 단백질, 폴리펩티드 및/또는 펩티드를 부위 특이적으로 표지화하는 데 이용된다. 특정 실시양태에서, 표지는 형광 잔기, 인광 잔기, 화학발광 잔기, 킬레이트화 잔기, 삽입성 잔기, 방사성 잔기, 발색단 잔기, 방사성 잔기, 회전 표지 잔기, NMR-활성 잔기, PET 또는 MRI 영상화 시약이다. 특정 실시양태에서, 이러한 부위 특이적 변형은 단백질, 폴리펩티드 및/또는 펩티드에 면역 조절제를 부착시키는 데 이용된다. 다른 실시양태에서, 이러한 부위 특이적 변형은 단백질, 폴리펩티드 및/또는 펩티드에 폴리(에틸렌 글리콜) (PEG)을 부착시키는 데 이용된다. 다른 실시양태에서, 이러한 부위 특이적 변형은 단백질, 폴리펩티드 및/또는 펩티드에 당을 부착 (글리코실화)시키는 데 이용된다. Provided herein are methods and compositions used to site-specifically modify proteins, polypeptides and/or peptides, wherein such methods are genetically encoded pyrrolysine or pyrroline-carboxy-lysine (PCL) residues, wherein , Pyrrolysine and PCL amino acids are produced biosynthetically). Various types of molecules are provided herein that are site-specifically coupled to proteins, polypeptides, and/or peptides having one or more PCL residues or pyrrolysine biosynthetically incorporated therein. In certain embodiments, such site-specific modifications are used to site-specifically label proteins, polypeptides and/or peptides. In certain embodiments, the label is a fluorescent moiety, a phosphorescent moiety, a chemiluminescent moiety, a chelating moiety, an intercalating moiety, a radioactive moiety, a chromophore moiety, a radioactive moiety, a rotational label moiety, an NMR-active moiety, a PET or MRI imaging reagent. In certain embodiments, such site-specific modifications are used to attach immune modulators to proteins, polypeptides and/or peptides. In other embodiments, such site specific modifications are used to attach poly(ethylene glycol) (PEG) to proteins, polypeptides and/or peptides. In other embodiments, such site specific modifications are used to attach (glycosylate) sugars to proteins, polypeptides and/or peptides.
다른 실시양태에서, 이러한 부위 특이적 변형은 단백질, 폴리펩티드 및/또는 펩티드를 가교시켜, 그로 인해 이종이량체 및 이종삼량체를 비롯한 (이에 제한되지는 않음) 이종올리고머를 형성하는 데 이용된다. 특정 실시양태에서, 이러한 부위 특이적 변형은 항체를 단백질, 폴리펩티드 및/또는 펩티드에 부위 특이적으로 가교시키는 데 이용된다. 다른 실시양태에서, 이러한 부위 특이적 변형은 단백질, 폴리펩티드 및/또는 펩티드를 가교시켜, 그로 인해 단백질-단백질 접합체, 단백질-폴리펩티드 접합체, 단백질-펩티드 접합체, 폴리펩티드-폴리펩티드 접합체, 폴리펩티드-펩티드 접합체 또는 펩티드-펩티드 접합체를 형성하는 데 이용된다. In other embodiments, such site-specific modifications are used to crosslink proteins, polypeptides and/or peptides, thereby forming heterooligomers, including, but not limited to, heterodimers and heterotrimers. In certain embodiments, such site-specific modifications are used to site-specifically cross-link the antibody to proteins, polypeptides and/or peptides. In other embodiments, such site-specific modifications crosslink proteins, polypeptides and/or peptides, whereby protein-protein conjugates, protein-polypeptide conjugates, protein-peptide conjugates, polypeptide-polypeptide conjugates, polypeptide-peptide conjugates or peptides. -Used to form peptide conjugates.
다른 실시양태에서, 이러한 부위 특이적 변형은 항체를 본원에서 제공되는 독성 단백질에 부위 특이적으로 연결시켜, 그로 인해 항체-약물 접합체를 형성하는 데 이용된다. 다른 실시양태에서, 이러한 부위 특이적 변형은 저분자량 약물에 커플링되는 항체를 단백질에 부위 특이적으로 연결시켜, 그로 인해 항체-약물 접합체를 형성하는 데 이용된다. In other embodiments, such site-specific modifications are used to site-specifically link the antibody to a toxic protein provided herein, thereby forming an antibody-drug conjugate. In other embodiments, such site-specific modifications are used to site-specifically link an antibody coupled to a low molecular weight drug to a protein, thereby forming an antibody-drug conjugate.
다른 실시양태에서, 이러한 부위 특이적 변형은 수용체-리간드를 본원에서 제공되는 독성 단백질에 부위 특이적으로 연결시켜, 그로 인해 수용체-리간드-약물 접합체를 형성하는 데 이용된다. 다른 실시양태에서, 이러한 부위 특이적 변형은 저분자량 약물에 커플링되는 수용체-리간드를 단백질에 부위 특이적으로 연결시켜, 그로 인해 수용체-리간드-약물 접합체를 형성하는 데 이용된다. In other embodiments, such site specific modifications are used to site-specifically link a receptor-ligand to a toxic protein provided herein, thereby forming a receptor-ligand-drug conjugate. In other embodiments, such site specific modifications are used to site-specifically link a receptor-ligand coupled to a low molecular weight drug to a protein, thereby forming a receptor-ligand-drug conjugate.
제공된 방법을 이용하여 내부에 혼입된 피롤리신 및/또는 PCL을 갖는 단백질, 폴리펩티드 및/또는 펩티드가 본원에서 제공된다. 이러한 단백질에는 에리트로포이에틴 (EPO), 섬유모세포 성장 인자 21 (FGF21), 인터페론 알파 (INF-α), 인터류킨 2 (IL-2), 인터류킨 4 (IL-4), 인터류킨 6 (IL-6), 인터류킨 10 (IL-10), 인터류킨 17 (IL-17), 인슐린-유사 성장 인자 1 (IGF-1) 및 인터페론 베타 (INF-β)가 포함되나, 이에 제한되지는 않는다. Proteins, polypeptides and/or peptides having pyrrolysine and/or PCL incorporated therein using the methods provided are provided herein. These proteins include erythropoietin (EPO), fibroblast growth factor 21 (FGF21), interferon alpha (INF-α), interleukin 2 (IL-2), interleukin 4 (IL-4), and interleukin 6 (IL-6). , Interleukin 10 (IL-10), interleukin 17 (IL-17), insulin-like growth factor 1 (IGF-1) and interferon beta (INF-β).
본원에서 제공되는 방법을 이용하여 내부에 혼입되고 본원에서 제공되는 방법을 이용하여 추가로 유도체화된 피롤리신 및/또는 PCL을 갖는 단백질, 폴리펩티드 및/또는 펩티드가 본원에서 제공된다. 이러한 유도체화에는 PEG화가 포함되나, 이에 제한되지는 않는다. 이러한 단백질에는 에리트로포이에틴 (EPO), 섬유모세포 성장 인자 21 (FGF21), 인터페론 알파 (INF-α), 인터류킨 2 (IL-2), 인터류킨 4 (IL-4), 인터류킨 6 (IL-6), 인터류킨 10 (IL-10), 인터류킨 17 (IL-17), 인슐린-유사 성장 인자 1 (IGF-1) 및 인터페론 베타 (INF-β)가 포함되나, 이에 제한되지는 않는다. Provided herein are proteins, polypeptides and/or peptides having pyrrolysine and/or PCL incorporated therein using the methods provided herein and further derivatized using the methods provided herein. Such derivatization includes, but is not limited to, PEGylation. These proteins include erythropoietin (EPO), fibroblast growth factor 21 (FGF21), interferon alpha (INF-α), interleukin 2 (IL-2), interleukin 4 (IL-4), and interleukin 6 (IL-6). , Interleukin 10 (IL-10), interleukin 17 (IL-17), insulin-like growth factor 1 (IGF-1) and interferon beta (INF-β).
내부에 혼입된 피롤리신 및/또는 PCL을 갖는, 본원에서 제공되는 방법을 이용하여 가교된 단백질, 폴리펩티드 및/또는 펩티드가 본원에 추가로 제공된다. 이러한 단백질에는 에리트로포이에틴 (EPO), 섬유모세포 성장 인자 21 (FGF21), 인터페론 알파 (INF-α) 및 인터페론 베타 (INF-β)가 포함되나, 이에 제한되지는 않는다. Further provided herein are proteins, polypeptides and/or peptides crosslinked using the methods provided herein, having pyrrolysine and/or PCL incorporated therein. Such proteins include, but are not limited to, erythropoietin (EPO), fibroblast growth factor 21 (FGF21), interferon alpha (INF-α) and interferon beta (INF-β).
다른 실시양태에서, 이러한 부위 특이적 변형은, 부위 특이적으로 혼입된 피롤리신 또는 PCL의 위치가 단백질, 폴리펩티드 및/또는 펩티드의 고체 지지체 표면 상에의 제어된 배향 및 부착을 가능하게 하는 단백질, 폴리펩티드 및/또는 펩티드를 생성하는 데 이용된다. 특정 실시양태에서, 이러한 고체 지지체는 코팅되거나 코팅되지 않은 플라스틱 마이크로타이터 플레이트, 유리 슬라이드, 실리카 표면, 중합체 비드, 금 입자 또는 나노-입자이다. 특정 실시양태에서, 이러한 제어된 배향 및 부착은 ELISA 또는 다른 항체 검정에 의한 단백질, 폴리펩티드 및 펩티드의 분석에 이용된다. 특정 실시양태에서, 이러한 제어된 배향 및 부착은 고정된 단백질, 폴리펩티드 및/또는 펩티드의 리간드를 정제 및/또는 확인하는 데 이용된다. 특정 실시양태에서, 이러한 제어된 배향 및 부착은, 표면 플라스몬 공명 분석을 이용한 표지를 사용하지 않는 분석을 비롯한 (이에 제한되지는 않음) 소실파(evanescent wave) 분석에 의해 단백질, 폴리펩티드 및 펩티드, 및 이들의 상호작용을 분석하는 데 이용된다. 특정 실시양태에서, 표면 상에의 이러한 제어된 배향 및 부착은, 미세저울 (전기적, 광학적 및/또는 기계적), 적외선 분광분석법, 라만 분광분석법 (표면 증강 라만 분광분석법, 소실 공명, 형광, 간섭측정법, 질량 분광측정법 및 다른 분광분석법 포함)을 사용하여 단백질, 폴리펩티드 및 펩티드, 및 이들의 상호작용을 분석하는 데 이용된다. 이러한 분석 방법은 고정된 단백질, 폴리펩티드 및/또는 펩티드의, 다른 단백질, 폴리펩티드, 펩티드, 핵산, DNA, RNA, 소분자, 약물, 대사물, 당, 탄수화물, 올리고사카라이드, 폴리사카라이드 및/또는 다른 분자와의 상호작용 (이러한 상호작용에 의해 유발된 배좌 변화 포함)을 조사하는 데 이용된다. 이러한 분석 방법은 또한 고정된 단백질, 폴리펩티드 및/또는 펩티드의, 다중 서브-유닛 단백질 복합체와의 상호작용을 조사하고, 천연, 재조합, 합성 또는 태그가 부착된 재조합 분자를 연구하고, 체액, 세포 배양물의 상청액 또는 가공하지 않은 추출물에서 새로운 상호작용 파트너를 발견하고, 소분자, 예컨대 약물 후보의 그의 표적과의 상호작용을 연구하고, 천연 막, 인공 막 또는 소포를 사용하여 막 생화학 또는 막-경계 수용체 상호작용을 연구하고, 복제, 전사 및 번역을 조사하고, 단백질 복합체의 형성 및 그의 DNA와의 상호작용 동안의 분자 관계를 결정하고, DNA 및 RNA의 혼성화를 연구하고, 전체 세포 또는 바이러스를 포함하는 상호작용을 연구하고, 분자 상호작용에 대한 글리코실화의 효과를 연구하고, 세포 표면 탄수화물의 특이적 인식 특성을 결정하는 데 이용된다. In other embodiments, such site-specific modification is a protein in which the position of the site-specifically incorporated pyrrolysine or PCL allows for controlled orientation and attachment of the protein, polypeptide and/or peptide onto the solid support surface. , Polypeptides and/or peptides. In certain embodiments, such solid supports are coated or uncoated plastic microtiter plates, glass slides, silica surfaces, polymer beads, gold particles or nano-particles. In certain embodiments, such controlled orientation and attachment are used for analysis of proteins, polypeptides and peptides by ELISA or other antibody assays. In certain embodiments, such controlled orientation and attachment are used to purify and/or identify ligands of immobilized proteins, polypeptides and/or peptides. In certain embodiments, such controlled orientation and attachment can be achieved by means of evanescent wave analysis, including but not limited to, assays that do not employ labeling using surface plasmon resonance analysis. And to analyze their interactions. In certain embodiments, such controlled orientation and adhesion on a surface is characterized by microbalance (electrical, optical and/or mechanical), infrared spectroscopy, Raman spectroscopy (surface enhanced Raman spectroscopy, vanishing resonance, fluorescence, interferometry). , Mass spectrometry and other spectroscopy) are used to analyze proteins, polypeptides and peptides, and their interactions. Such assay methods include immobilized proteins, polypeptides and/or peptides of other proteins, polypeptides, peptides, nucleic acids, DNA, RNA, small molecules, drugs, metabolites, sugars, carbohydrates, oligosaccharides, polysaccharides and/or other It is used to investigate interactions with molecules (including changes in conformation caused by these interactions). These assay methods also investigate the interaction of immobilized proteins, polypeptides and/or peptides with multiple sub-unit protein complexes, study natural, recombinant, synthetic or tagged recombinant molecules, and in body fluids, cell culture. Discover new interaction partners in supernatant or unprocessed extracts of water, study the interaction of small molecules, such as drug candidates, with their targets, and use natural membranes, artificial membranes or vesicles to interact with membrane biochemistry or membrane-boundary receptors. Study the action, investigate replication, transcription and translation, determine the molecular relationship during the formation of protein complexes and their interactions with DNA, study the hybridization of DNA and RNA, and interactions involving whole cells or viruses , Study the effect of glycosylation on molecular interactions, and determine the specific recognition properties of cell surface carbohydrates.
다른 실시양태에서, 이러한 부위 특이적 변형은 핵산을 단백질에 부위 특이적으로 부착시키는 데 이용된다. 특정 실시양태에서, 이러한 부위 특이적 변형은 핵산을 항체 또는 항체 단편에 부위 특이적으로 부착시키는 데 이용된다. 특정 실시양태에서, 단백질 또는 항체에 부착된 핵산은 혼성화를 통해 DNA 어레이 상의 정의된 위치에 단백질 또는 항체를 고정하는 데 사용된다. 특정 실시양태에서, 단백질 또는 항체에 부착된 핵산은 PCR, 가닥 변위 증폭 (SDA), 리가제 연쇄 반응 (LCR), 인접성 라이게이션 면역-PCR, 롤링 써클 증폭, 전사 매개 증폭, NEN의 티라마이드 신호 증폭 또는 다른 신호 증폭 방법을 통해 단백질 또는 항체 결합을 검출하는 데 사용된다. 특정 실시양태에서, PCR 프로브의 항체에 대한 이러한 부위 특이적 부착은 면역-PCR 반응을 위한 시약을 생성하는 데 이용된다 (문헌 [M. Adler, R. Wacker, Ch.M. Niemeyer, Sensitivity by combination: Immuno-PCR and related technologies, Analyst, 2008, 133, 702-718] 참조). 특정 실시양태에서, 단백질 또는 항체에 부착된 핵산은 다수의 분석물의 동시 면역-PCR 또는 다른 면역-검정 (멀티플렉스화 면역-PCR 또는 멀티플렉스화 면역-검정)을 가능하게 한다.In other embodiments, such site-specific modifications are used to site-specifically attach nucleic acids to proteins. In certain embodiments, such site-specific modifications are used to site-specifically attach nucleic acids to antibodies or antibody fragments. In certain embodiments, the nucleic acid attached to the protein or antibody is used to immobilize the protein or antibody at defined locations on the DNA array through hybridization. In certain embodiments, the nucleic acid attached to the protein or antibody is PCR, strand displacement amplification (SDA), ligase chain reaction (LCR), contiguous ligation immunity-PCR, rolling circle amplification, transcription-mediated amplification, NEN's thyramide signal It is used to detect protein or antibody binding through amplification or other signal amplification methods. In certain embodiments, this site-specific attachment of a PCR probe to an antibody is used to generate reagents for an immuno-PCR reaction (M. Adler, R. Wacker, Ch.M. Niemeyer, Sensitivity by combination). : Immuno-PCR and related technologies, Analyst, 2008, 133, 702-718). In certain embodiments, the nucleic acid attached to the protein or antibody enables simultaneous immuno-PCR or other immuno-assay (multiplexed immuno-PCR or multiplexed immuno-assay) of multiple analytes.
특정 실시양태에서, 단백질 또는 항체에 부착된 핵산은 동종- 및 이종이량체의 형성을 조정한다.In certain embodiments, the nucleic acid attached to the protein or antibody modulates the formation of homo- and heterodimers.
정의Justice
본원에 사용된 용어 "알킬"은 포화 분지형 또는 직쇄 탄화수소를 지칭한다. 본원에 사용된 용어 "C1-C3알킬", "C1-C4알킬", "C1-C5알킬", "C1-C6알킬", "C1-C7알킬" 및 "C1-C8알킬"은 1개 이상, 및 각각 3, 4, 5, 6, 7 또는 8개 이하의 탄소 원자를 함유하는 알킬기를 지칭한다. 본원에 사용된 알킬기의 비제한적인 예에는 메틸, 에틸, n-프로필, 이소프로필, n-부틸, 이소부틸, sec-부틸, t-부틸, n-펜틸, 이소펜틸, 헥실, 헵틸, 옥틸, 노닐, 데실 등이 포함된다.As used herein, the term “alkyl” refers to a saturated branched or straight chain hydrocarbon. As used herein, the terms "C 1 -C 3 alkyl", "C 1 -C 4 alkyl", "C 1 -C 5 alkyl", "C 1 -C 6 alkyl", "C 1 -C 7 alkyl" and “C 1 -C 8 alkyl” refers to an alkyl group containing at least 1 and up to 3, 4, 5, 6, 7 or 8 carbon atoms, respectively. Non-limiting examples of alkyl groups as used herein include methyl, ethyl, n-propyl, isopropyl, n-butyl, isobutyl, sec-butyl, t-butyl, n-pentyl, isopentyl, hexyl, heptyl, octyl, Nonyl, decyl, etc. are included.
본원에 사용된 용어 "알킬렌"은 포화 분지형 또는 직쇄 2가 탄화수소 라디칼을 지칭하며, 여기서 라디칼은 각각의 2개의 탄소 원자로부터 1개의 수소 원자를 제거함으로써 유도된다. 본원에 사용된 용어 "C1-C3알킬렌", "C1-C4알킬렌", "C1-C5알킬렌" 및 "C1-C6알킬렌"은 1개 이상, 및 각각 3, 4, 5 또는 6개의 이하의 탄소 원자를 함유하는 알킬렌기를 지칭한다. 본원에 사용된 알킬렌기의 비제한적인 예에는 메틸렌, 에틸렌, n-프로필렌, 이소프로필렌, n-부틸렌, 이소부틸렌, sec-부틸렌, t-부틸렌, n-펜틸렌, 아이소펜틸렌, 헥실렌 등이 포함된다.The term “alkylene” as used herein refers to a saturated branched or straight chain divalent hydrocarbon radical, wherein the radical is derived by removing one hydrogen atom from each of the two carbon atoms. As used herein, the terms "C 1 -C 3 alkylene", "C 1 -C 4 alkylene", "C 1 -C 5 alkylene" and "C 1 -C 6 alkylene" are at least one, and It refers to an alkylene group containing up to 3, 4, 5 or 6 carbon atoms, respectively. Non-limiting examples of alkylene groups used herein include methylene, ethylene, n-propylene, isopropylene, n-butylene, isobutylene, sec-butylene, t-butylene, n-pentylene, isopentylene , Hexylene, and the like are included.
본원에 사용된 용어 "알콕시"는 -ORa 기를 지칭한다 (여기서, Ra는 본원에 정의된 바와 같은 알킬기임). 본원에 사용된 용어 "C1-C3알콕시", "C1-C4알콕시", "C1-C5알콕시", "C1-C6알콕시", "C1-C7알콕시" 및 "C1-C8알콕시"는, 알킬 잔기가 1개 이상, 및 각각 3, 4, 5, 6, 7 또는 8개 이하의 탄소 원자를 함유하는 알콕시기를 지칭한다. 본원에 사용된 알콕시기의 비제한적인 예에는 메톡시, 에톡시, n-프로폭시, 이소프로폭시, n-부톡시, t-부톡시, 펜톡시, 헥속시, 헵톡시 등이 포함된다.As used herein, the term “alkoxy” refers to the group —OR a , wherein R a is an alkyl group as defined herein. As used herein, the terms "C 1 -C 3 alkoxy", "C 1 -C 4 alkoxy", "C 1 -C 5 alkoxy", "C 1 -C 6 alkoxy", "C 1 -C 7 alkoxy" and “C 1 -C 8 alkoxy” refers to an alkoxy group containing at least one alkyl moiety and up to 3, 4, 5, 6, 7 or 8 carbon atoms, respectively. Non-limiting examples of alkoxy groups as used herein include methoxy, ethoxy, n-propoxy, isopropoxy, n-butoxy, t-butoxy, pentoxy, hexoxy, heptoxy, and the like.
본원에 사용된 용어 "아미노 말단 변형 기"는 말단 아민기와 연결을 형성하는 임의의 분자를 지칭한다. 예로서, 이러한 말단 아민기에는 아민 보호기, 중합체 분자 (여기서, 이러한 중합체 분자에는 폴리펩티드, 폴리뉴클레오티드 및 폴리사카라이드 등이 포함됨)의 말단이 포함되나, 이에 제한되지는 않는다. 아미노 말단 변형 기에는 또한 다양한 수용성 중합체, 펩티드 또는 단백질이 포함되나, 이에 제한되지는 않는다. 단지 예로서 언급하자면, 말단 변형 기에는 폴리에틸렌 글리콜 또는 혈청 알부민이 포함된다. 특정 아미노 말단 변형 기는 혈청 반감기를 증가시키는 것을 비롯하여 (이에 제한되지는 않음) 단백질의 치료적 특성을 변형시키는 데 사용된다. As used herein, the term “amino terminal modifying group” refers to any molecule that forms a linkage with a terminal amine group. By way of example, such terminal amine groups include, but are not limited to, amine protecting groups, the ends of polymer molecules (wherein such polymer molecules include polypeptides, polynucleotides and polysaccharides, etc.). The amino terminal modification group also includes, but is not limited to, a variety of water soluble polymers, peptides or proteins. By way of example only, terminal modifying groups include polyethylene glycol or serum albumin. Certain amino terminal modifying groups are used to modify the therapeutic properties of proteins, including but not limited to increasing serum half-life.
본원에 사용된 용어 "아릴"은 총 5 내지 14개의 고리원을 갖고, 계 내의 1개 이상의 고리가 방향족이고, 계 내의 각각의 고리가 3 내지 7개의 고리원을 함유하는 모노시클릭, 비시클릭 및 트리시클릭 고리계를 지칭한다. 아릴기는 "임의로 치환되며", 여기서 이러한 아릴기는 1개 이상의 치환기를 함유한다. 본원에 달리 정의되지 않는 한, 적합한 치환기는 일반적으로 할로겐, -R, -OR, -SR, -NO2, -CN, -N(R)2, -NRC(O)R, -NRC(S)R, -NRC(O)N(R)2, -NRC(S)N(R)2, -NRCO2R, -NRNRC(O)R, -NRNRC(O)N(R)2, -NRNRCO2R, -C(O)C(O)R, -C(O)CH2C(O)R, -CO2R, -C(O)R, -C(S)R, -C(O)N(R)2, -C(S)N(R)2, -OC(O)N(R)2, -OC(O)R, -C(O)N(OR)R, -C(NOR)R, -S(O)2R, -S(O)3R, -SO2N(R)2, -S(O)R, -NRSO2N(R)2, -NRSO2R, -N(OR)R, -C(=NH)-N(R)2, -P(O)2R, -PO(R)2, -OPO(R)2, -(CH2)0-2NHC(O)R, R로 임의로 치환된 페닐 (Ph), R로 임의로 치환된 -O(Ph), R로 임의로 치환된 -(CH2)1-2(Ph), 또는 R로 임의로 치환된 -CH=CH(Ph)로부터 선택되며, 여기서 각각 독립적으로 출현한 R은 수소, 임의로 치환된 C1-C6알킬, 임의로 치환된 C1-C6알콕시, 치환되지 않은 5 내지 6원 헤테로아릴, 페닐, -O(Ph) 또는 -CH2(Ph)로부터 선택되거나, 또는 동일 치환기 또는 상이한 치환기 상에 독립적으로 출현한 2개의 R은 각각의 R이 결합된 원자(들)와 합쳐져, 질소, 산소, 또는 황으로부터 독립적으로 선택된 0 내지 4개의 헤테로원자를 갖는 임의로 치환된 3 내지 12원 포화, 부분 불포화, 또는 완전 불포화 모노시클릭 또는 비시클릭 고리를 형성한다. 본원에 사용된 아릴기의 비제한적인 예에는 페닐, 나프틸, 플루오레닐, 인데닐, 아줄레닐, 안트라세닐 등이 포함된다. As used herein, the term "aryl" has a total of 5 to 14 ring members, at least one ring in the system is aromatic, and each ring in the system contains 3 to 7 ring members, monocyclic, bicyclic And tricyclic ring systems. Aryl groups are “optionally substituted”, wherein such aryl groups contain one or more substituents. Unless otherwise defined herein, suitable substituents are generally halogen, -R, -OR, -SR, -NO 2 , -CN, -N(R) 2 , -NRC(O)R, -NRC(S) R, -NRC(O)N(R) 2 , -NRC(S)N(R) 2 , -NRCO 2 R, -NRNRC(O)R, -NRNRC(O)N(R) 2 , -NRNRCO 2 R, -C(O)C(O)R, -C(O)CH 2 C(O)R, -CO 2 R, -C(O)R, -C(S)R, -C(O) N(R) 2 , -C(S)N(R) 2 , -OC(O)N(R) 2 , -OC(O)R, -C(O)N(OR)R, -C(NOR )R, -S(O) 2 R, -S(O) 3 R, -SO 2 N(R) 2 , -S(O)R, -NRSO 2 N(R) 2 , -NRSO 2 R,- N(OR)R, -C(=NH)-N(R) 2 , -P(O) 2 R, -PO(R) 2 , -OPO(R) 2 , -(CH 2 ) 0-2 NHC (O)R, phenyl (Ph) optionally substituted with R, -O(Ph) optionally substituted with R, -(CH 2 ) 1-2 (Ph) optionally substituted with R, or- CH=CH(Ph), wherein each independently appearing R is hydrogen, optionally substituted C 1 -C 6 alkyl, optionally substituted C 1 -C 6 alkoxy, unsubstituted 5-6 membered heteroaryl, Two Rs selected from phenyl, -O(Ph) or -CH 2 (Ph), or independently appearing on the same or different substituents, are combined with the atom(s) to which each R is attached, and nitrogen, oxygen , Or an optionally substituted 3-12 membered saturated, partially unsaturated, or fully unsaturated monocyclic or bicyclic ring having 0-4 heteroatoms independently selected from sulfur. Non-limiting examples of aryl groups as used herein include phenyl, naphthyl, fluorenyl, indenyl, azulenyl, anthracenyl, and the like.
사용된 용어 "아릴렌"은 아릴기로부터 유도된 2가 라디칼을 의미한다. The term "arylene" as used refers to a divalent radical derived from an aryl group.
본원에 사용된 "이관능성 링커" (또한 "이관능성 중합체"로도 지칭됨)는 다른 잔기와 특이적으로 반응하여 공유 또는 비공유 결합을 형성할 수 있는 2개의 관능기를 포함하는 링커를 지칭한다. 이러한 잔기에는 아미노산 측쇄 기가 포함되나, 이에 제한되지는 않는다. 단지 예로서 언급하자면, 이관능성 링커는 제1 펩티드 상의 기와 반응성인 관능기, 및 제2 펩티드 상의 기와 반응성인 또 다른 관능기를 가지며, 이에 따라 제1 펩티드, 이관능성 링커 및 제2 펩티드를 포함하는 접합체를 형성한다. 이관능성 링커는 임의의 원하는 길이 또는 분자량을 가지며, 원하는 특정 간격 또는 배좌를 제공하도록 선택된다. As used herein, “bifunctional linker” (also referred to as “bifunctional polymer”) refers to a linker comprising two functional groups capable of reacting specifically with other moieties to form a covalent or non-covalent bond. Such residues include, but are not limited to, amino acid side chain groups. By way of example only, the bifunctional linker has a functional group reactive with the group on the first peptide, and another functional group reactive with the group on the second peptide, and thus a conjugate comprising a first peptide, a bifunctional linker and a second peptide To form. The bifunctional linker has any desired length or molecular weight and is selected to provide the specific spacing or configuration desired.
본원에 사용된 "다관능성 링커" (또한 "다관능성 중합체"로도 지칭됨)는 다른 잔기와 반응하여 공유 또는 비공유 결합을 형성할 수 있는 2개 이상의 관능기를 포함하는 링커를 지칭한다. 이러한 잔기에는 아미노산 측쇄 기가 포함되나, 이에 제한되지는 않는다. 다관능성 링커는 임의의 원하는 길이 또는 분자량을 가지며, 원하는 특정 간격 또는 배좌를 제공하도록 선택된다. As used herein, “multifunctional linker” (also referred to as “multifunctional polymer”) refers to a linker comprising two or more functional groups capable of reacting with other moieties to form covalent or non-covalent bonds. Such residues include, but are not limited to, amino acid side chain groups. The multifunctional linker has any desired length or molecular weight and is selected to provide the specific spacing or configuration desired.
본원에 사용된 용어 "시아노"는 -CN 기를 지칭한다. As used herein, the term "cyano" refers to the group -CN.
본원에 사용된 용어 "시클로알킬"은 포화 또는 부분 불포화, 모노시클릭, 융합된 비시클릭, 융합된 트리시클릭 또는 가교된 폴리시클릭 고리 어셈블리를 지칭한다. 본원에 사용된 용어 "C3-C5시클로알킬", "C3-C6시클로알킬", "C3-C7시클로알킬", "C3-C8시클로알킬", "C3-C9시클로알킬" 및 "C3-C10시클로알킬"은, 포화 또는 부분 불포화, 모노시클릭, 융합된 비시클릭 또는 가교된 폴리시클릭 고리 어셈블리가 3개 이상, 및 5, 6, 7, 8, 9 또는 10개 이하의 탄소 원자를 함유하는 시클로알킬기를 지칭한다. 본원에 사용된 시클로알킬기의 비제한적인 예에는 시클로프로필, 시클로부틸, 시클로펜틸, 시클로헥실, 시클로헵틸, 시클로옥틸, 시클로노닐, 시클로데실, 시클로펜테닐, 시클로헥세닐, 데카히드로나프탈레닐, 2,3,4,5,6,7-헥사히드로-1H-인데닐 등이 포함된다.The term “cycloalkyl,” as used herein, refers to a saturated or partially unsaturated, monocyclic, fused bicyclic, fused tricyclic or bridged polycyclic ring assembly. As used herein, the terms "C 3 -C 5 cycloalkyl", "C 3 -C 6 cycloalkyl", "C 3 -C 7 cycloalkyl", "C 3 -C 8 cycloalkyl", "C 3 -C 9 Cycloalkyl" and "C 3 -C 10 Cycloalkyl" means 3 or more saturated or partially unsaturated, monocyclic, fused bicyclic or bridged polycyclic ring assemblies, and 5, 6, 7, 8, It refers to a cycloalkyl group containing up to 9 or 10 carbon atoms. Non-limiting examples of cycloalkyl groups as used herein include cyclopropyl, cyclobutyl, cyclopentyl, cyclohexyl, cycloheptyl, cyclooctyl, cyclononyl, cyclodecyl, cyclopentenyl, cyclohexenyl, decahydronaphthalenyl, 2,3,4,5,6,7-hexahydro-1H-indenyl, and the like.
본원에 사용된 용어 "시클로덱스트린"은 고리 형성에 있어서 6 내지 8개 이상의 글루코스 분자로 이루어진 시클릭 탄수화물을 지칭한다. 고리의 외부는 수용성 기를 함유하고, 고리의 중심에는 소분자를 수용할 수 있는 상대적으로 비극성인 공동이 있다.As used herein, the term “cyclodextrin” refers to a cyclic carbohydrate consisting of 6 to 8 or more glucose molecules in ring formation. The outside of the ring contains water-soluble groups, and at the center of the ring is a relatively non-polar cavity capable of accommodating small molecules.
본원에 사용된 용어 "할로겐"은 불소 (F), 염소 (Cl), 브롬 (Br) 또는 요오드 (I)를 지칭한다. The term “halogen” as used herein refers to fluorine (F), chlorine (Cl), bromine (Br) or iodine (I).
본원에 사용된 용어 "할로"는 할로겐 라디칼 (플루오로 (-F), 클로로 (-Cl), 브로모 (-Br) 및 요오도 (-I))를 지칭한다. The term “halo” as used herein refers to halogen radicals (fluoro (-F), chloro (-Cl), bromo (-Br) and iodo (-I)).
본원에 사용된 용어 "할로아실"은 -C(O)CH3, -C(O)CF3, -C(O)CH2OCH3 등을 비롯한 (이에 제한되지는 않음) 할로겐 잔기를 함유하는 아실기를 지칭한다. As used herein, the term "haloacyl" contains halogen moieties including, but not limited to , -C(O)CH 3 , -C(O)CF 3 , -C(O)CH 2 OCH 3, etc. Refers to an acyl group.
본원에 사용된 용어 "할로알킬" 또는 "할로-치환된 알킬"은, 1개 이상의 할로기 또는 이들의 조합으로 치환된 본원에 정의된 바와 같은 알킬기를 지칭한다. 본원에 사용된 이러한 분지형 또는 직쇄형 할로알킬기의 비제한적인 예에는 트리플루오로메틸, 펜타플루오로에틸 등을 비롯한 (이에 제한되지는 않음), 1개 이상의 할로기 또는 이들의 조합으로 치환된 메틸, 에틸, 프로필, 이소프로필, 이소부틸 및 n-부틸이 포함된다. The term “haloalkyl” or “halo-substituted alkyl” as used herein refers to an alkyl group as defined herein substituted with one or more halo groups or combinations thereof. As used herein, non-limiting examples of such branched or straight-chain haloalkyl groups include, but are not limited to, trifluoromethyl, pentafluoroethyl, and the like, substituted with one or more halo groups or combinations thereof. Methyl, ethyl, propyl, isopropyl, isobutyl and n-butyl.
본원에 사용된 용어 "할로알콕시"는, 1개 이상의 할로기 또는 이들의 조합으로 치환된 본원에 정의된 바와 같은 알콕시기를 지칭한다. 본원에 사용된 이러한 분지형 또는 직쇄형 할로알콕시기의 비제한적인 예에는 1개 이상의 기 또는 이들의 조합으로 치환된 메톡시, 에톡시, n-프로폭시, 이소프로폭시, n-부톡시, t-부톡시, 펜톡시, 헥속시, 헵톡시 등이 포함된다. The term “haloalkoxy,” as used herein, refers to an alkoxy group, as defined herein, substituted with one or more halo groups or combinations thereof. As used herein, non-limiting examples of such branched or straight-chain haloalkoxy groups include methoxy, ethoxy, n-propoxy, isopropoxy, n-butoxy, substituted with one or more groups or combinations thereof. t-butoxy, pentoxy, hexoxy, heptoxy, and the like.
본원에 사용된 용어 "헤테로알킬"은 1개 이상의 탄소 원자가 1개 이상의 산소, 황, 질소 또는 이들의 조합에 의해 독립적으로 대체된 본원에 정의된 바와 같은 알킬기를 지칭한다.The term “heteroalkyl,” as used herein, refers to an alkyl group as defined herein in which one or more carbon atoms have been independently replaced by one or more oxygen, sulfur, nitrogen, or combinations thereof.
본원에 사용된 용어 "헤테로알킬렌"은 헤테로알킬로부터 유도된 2가 라디칼을 지칭한다.The term “heteroalkylene” as used herein refers to a divalent radical derived from heteroalkyl.
본원에 사용된 용어 "헤테로아릴"은 총 5 내지 14개의 고리원을 갖고, 계 내의 1개 이상의 고리가 방향족이고, 계 내의 하나 이상의 고리가 질소, 산소 및 황으로부터 선택된 1개 이상의 헤테로원자를 함유하고, 계 내의 각각의 고리가 3 내지 7개의 고리원을 함유하는 모노시클릭, 비시클릭 및 트리시클릭 고리계를 지칭한다. 상기 및 본원에 달리 정의되지 않는 한, 헤테로아릴기의 불포화 탄소 원자 상의 적합한 치환기는 일반적으로 할로겐, -R, -OR, -SR, -NO2, -CN, -N(R)2, -NRC(O)R, -NRC(S)R, -NRC(O)N(R)2, -NRC(S)N(R)2, -NRCO2R, -NRNRC(O)R, -NRNRC(O)N(R)2, -NRNRCO2R, -C(O)C(O)R, -C(O)CH2C(O)R, -CO2R, -C(O)R, -C(S)R, -C(O)N(R)2, -C(S)N(R)2, -OC(O)N(R)2, -OC(O)R, -C(O)N(OR)R, -C(NOR)R, -S(O)2R, -S(O)3R, -SO2N(R)2, -S(O)R, -NRSO2N(R)2, -NRSO2R, -N(OR)R, -C(=NH)-N(R)2, -P(O)2R, -PO(R)2, -OPO(R)2, -(CH2)0-2NHC(O)R, R로 임의로 치환된 페닐 (Ph), R로 임의로 치환된 -O(Ph), R로 임의로 치환된 -(CH2)1-2(Ph), 또는 R로 임의로 치환된 -CH=CH(Ph)로부터 선택되고, 여기서 각각 독립적으로 출현한 R은 수소, 임의로 치환된 C1-C6알킬, 임의로 치환된 C1-C6알콕시, 치환되지 않은 5 내지 6원 헤테로아릴, 페닐, -O(Ph) 또는 -CH2(Ph)로부터 선택되거나, 또는 동일 치환기 또는 상이한 치환기 상에 독립적으로 출현한 2개의 R은 각각의 R이 결합된 원자(들)와 합쳐져, 질소, 산소 또는 황으로부터 독립적으로 선택된 0 내지 4개의 헤테로원자를 갖는 임의로 치환된 3 내지 12원 포화, 부분 불포화 또는 완전 불포화 모노시클릭 또는 비시클릭 고리를 형성한다. 본원에 사용된 헤테로아릴기의 비제한적인 예에는 벤조푸라닐, 벤조푸라자닐, 벤족사졸릴, 벤조피라닐, 벤즈티아졸릴, 벤조티에닐, 벤즈아제피닐, 벤즈이미다졸릴, 벤조티오피라닐, 벤조[1,3]디옥솔, 벤조[b]푸릴, 벤조[b]티에닐, 신놀리닐, 푸라지닐, 푸릴, 푸로피리디닐, 이미다졸릴, 인돌릴, 인돌리지닐, 인돌린-2-온, 인다졸릴, 이소인돌릴, 이소퀴놀리닐, 이속사졸릴, 이소티아졸릴, 1,8-나프티리디닐, 옥사졸릴, 옥사인돌릴, 옥사디아졸릴, 피라졸릴, 피롤릴, 프탈라지닐, 프테리디닐, 퓨리닐, 피리딜, 피리다지닐, 피라지닐, 피리미딜, 피리미디닐, 퀴녹살리닐, 퀴놀리닐, 퀴나졸리닐, 4H-퀴놀리지닐, 티아졸릴, 티아디아졸릴, 티에닐, 트리아지닐, 트리아졸릴 및 테트라졸릴이 포함된다. The term “heteroaryl” as used herein has a total of 5 to 14 ring members, at least one ring in the system is aromatic, and at least one ring in the system contains at least one heteroatom selected from nitrogen, oxygen and sulfur. And refers to a monocyclic, bicyclic and tricyclic ring system in which each ring in the system contains 3 to 7 ring members. Unless defined otherwise above and herein, suitable substituents on the unsaturated carbon atom of the heteroaryl group are generally halogen, -R, -OR, -SR, -NO 2 , -CN, -N(R) 2 , -NRC (O)R, -NRC(S)R, -NRC(O)N(R) 2 , -NRC(S)N(R) 2 , -NRCO 2 R, -NRNRC(O)R, -NRNRC(O )N(R) 2 , -NRNRCO 2 R, -C(O)C(O)R, -C(O)CH 2 C(O)R, -CO 2 R, -C(O)R, -C (S)R, -C(O)N(R) 2 , -C(S)N(R) 2 , -OC(O)N(R) 2 , -OC(O)R, -C(O) N(OR)R, -C(NOR)R, -S(O) 2 R, -S(O) 3 R, -SO 2 N(R) 2 , -S(O)R, -NRSO 2 N( R) 2 , -NRSO 2 R, -N(OR)R, -C(=NH)-N(R) 2 , -P(O) 2 R, -PO(R) 2 , -OPO(R) 2 , -(CH 2 ) 0-2 NHC(O)R, phenyl (Ph) optionally substituted with R, -O(Ph) optionally substituted with R, -(CH 2 ) 1-2 ( Ph), or -CH=CH(Ph) optionally substituted with R, wherein each independently appearing R is hydrogen, optionally substituted C 1 -C 6 alkyl, optionally substituted C 1 -C 6 alkoxy, Unsubstituted 5 to 6 membered heteroaryl, phenyl, -O (Ph) or -CH 2 (Ph), or two R independently appearing on the same or different substituents, each R is bonded Combined with the atom(s) to form an optionally substituted 3-12 membered saturated, partially unsaturated or fully unsaturated monocyclic or bicyclic ring having 0-4 heteroatoms independently selected from nitrogen, oxygen or sulfur. Non-limiting examples of heteroaryl groups as used herein include benzofuranyl, benzofurazanyl, benzoxazolyl, benzopyranyl, benzthiazolyl, benzothienyl, benzazepinyl, benzimidazolyl, benzothiopyra Neil, benzo[1,3]dioxole, benzo[b]furyl, benzo[b]thienyl, cinnolinyl, furazinyl, furyl, furopyridinyl, imidazolyl, indolyl, indozinyl, indoline 2-one, indazolyl, isoindolyl, isoquinolinyl, isoxazolyl, isothiazolyl, 1,8-naphthyridinyl, oxazolyl, oxaindolyl, oxadiazolyl, pyrazolyl, pyrrolyl, Phthalasinyl, pteridinil, purinyl, pyridyl, pyridazinyl, pyrazinyl, pyrimidyl, pyrimidinyl, quinoxalinyl, quinolinyl, quinazolinyl, 4H-quinolinyl, thiazolyl, thia Diazolyl, thienyl, triazinyl, triazolyl and tetrazolyl.
본원에 사용된 용어 "헤테로시클로알킬"은 고리 탄소 중 1개 이상이 -O-, -N=, -NR-, -C(O)-, -S-, -S(O)- 또는 -S(O)2- (여기서, R은 수소, C1-C4알킬 또는 질소 보호기임)로부터 선택된 잔기에 의해 대체되되, 단 상기 기의 고리가 2개의 인접한 O 또는 S 원자를 함유하지 않는, 본원에 정의된 바와 같은 시클로알킬을 지칭한다. 본원에 사용된 헤테로시클로알킬기의 비제한적인 예에는 모르폴리노, 피롤리디닐, 피롤리디닐-2-온, 피페라지닐, 피페리디닐, 피페리디닐론, 1,4-디옥사-8-아자-스피로[4.5]데스-8-일, 2H-피롤릴, 2-피롤리닐, 3-피롤리닐, 1,3-디옥솔라닐, 2-이미다졸리닐, 이미다졸리디닐, 2-피라졸리닐, 피라졸리디닐, 1,4-디옥사닐, 1,4-디티아닐, 티오모르폴리닐, 아제파닐, 헥사히드로-1,4-디아제피닐, 테트라히드로푸라닐, 디히드로푸라닐, 테트라히드로티에닐, 테트라히드로피라닐, 디히드로피라닐, 테트라히드로티오피라닐, 티옥사닐, 아제티디닐, 옥세타닐, 티에타닐, 옥세파닐, 티에파닐, 1,2,3,6-테트라히드로피리디닐, 2H-피라닐, 4H-피라닐, 디옥사닐, 1,3-디옥솔라닐, 디티아닐, 디티올라닐, 디히드로피라닐, 디히드로티에닐, 디히드로푸라닐, 이미다졸리닐, 이미다졸리디닐, 3-아자비시클로[3.1.0]헥사닐 및 3-아자비시클로[4.1.0]헵타닐이 포함된다. As used herein, the term "heterocycloalkyl" refers to one or more of the ring carbons being -O-, -N=, -NR-, -C(O)-, -S-, -S(O)-, or -S (O) 2- (wherein R is hydrogen, C 1 -C 4 alkyl or nitrogen protecting group), provided that the ring of the group does not contain two adjacent O or S atoms Refers to cycloalkyl as defined in. Non-limiting examples of heterocycloalkyl groups as used herein include morpholino, pyrrolidinyl, pyrrolidinyl-2-one, piperazinyl, piperidinyl, piperidinylone, 1,4-dioxa-8- Aza-spiro[4.5]des-8-yl, 2H-pyrrolyl, 2-pyrrolinyl, 3-pyrrolinyl, 1,3-dioxolanyl, 2-imidazolinyl, imidazolidinyl, 2 -Pyrazolinyl, pyrazolidinyl, 1,4-dioxanyl, 1,4-ditianyl, thiomorpholinyl, azepanyl, hexahydro-1,4-diazepinyl, tetrahydrofuranyl, dihydro Furanyl, tetrahydrothienyl, tetrahydropyranyl, dihydropyranyl, tetrahydrothiopyranyl, thioxanyl, azetidinyl, oxetanyl, thietanyl, oxepanyl, thiepanyl, 1,2, 3,6-tetrahydropyridinyl, 2H-pyranyl, 4H-pyranyl, dioxanyl, 1,3-dioxolanyl, ditianyl, dithiolanyl, dihydropyranyl, dihydrothienyl, dihydro Furanyl, imidazolinyl, imidazolidinyl, 3-azabicyclo[3.1.0]hexanyl and 3-azabicyclo[4.1.0]heptanyl.
본원에 사용된 용어 "헤테로원자"는 산소, 황, 질소, 인 또는 규소 중 하나 이상을 지칭한다. As used herein, the term “heteroatom” refers to one or more of oxygen, sulfur, nitrogen, phosphorus or silicon.
본원에 사용된 용어 "히드록실"은 -OH 기를 지칭한다. The term "hydroxyl" as used herein refers to the group -OH.
본원에 사용된 용어 "히드록시알킬"은 1개 이상의 히드록실 (히드록실은 본원에 정의된 바와 같음)로 치환된, 본원에 정의된 바와 같은 알킬기를 지칭한다. 분지형 또는 직쇄형 "C1-C6히드록시알킬기의 비제한적인 예에는, 1개 이상의 히드록실기로 독립적으로 치환된 메틸, 에틸, 프로필, 이소프로필, 이소부틸 및 n-부틸이 포함된다. The term “hydroxyalkyl,” as used herein, refers to an alkyl group, as defined herein, substituted with one or more hydroxyls (hydroxyl is as defined herein). Non-limiting examples of branched or straight chain "C 1 -C 6 hydroxyalkyl groups include methyl, ethyl, propyl, isopropyl, isobutyl and n-butyl independently substituted with one or more hydroxyl groups. .
본원에 사용된 용어 "임의로 치환된"은, 언급된 기가 알킬, 알케닐, 알키닐, 시클로알킬, 아릴, 헤테로아릴, 헤테로시클로알킬, 히드록실, 알콕시, 메르캅틸, 시아노, 할로, 카르보닐, 티오카르보닐, 이소시아네이토, 티오시아네이토, 이소티오시아네이토, 니트로, 퍼할로알킬, 퍼플루오로알킬, 및 일치환 및 이치환된 아미노기를 비롯한 아미노, 및 이들의 보호된 유도체로부터 개별적으로 그리고 독립적으로 선택된 1개 이상의 추가적인 기(들)로 치환될 수 있거나 치환되지 않을 수 있음을 의미한다. 임의의 치환기의 비제한적인 예에는 할로, -CN, -OR, -C(O)R, -OC(O)R, -C(O)OR, -OC(O)NHR, -C(O)N(R)2, -SR-, -S(=O)R, -S(=O)2R, -NHR, -N(R)2, -NHC(O)-, NHC(O)O-, -C(O)NH-, S(=O)2NHR, -S(O)2N(R)2, -NHS(=O)2, -NHS(O)2R, C1-C6알킬, C1-C6알콕시, 아릴, 헤테로아릴, 시클로알킬, 헤테로시클로알킬, 할로-치환된 C1-C6알킬, 할로-치환된 C1-C6알콕시 (여기서, 각각의 R은 H, 할로, C1-C6알킬, C1-C6알콕시, 아릴, 헤테로아릴, 시클로알킬, 헤테로시클로알킬, 할로-치환된 C1-C6알킬, 할로-치환된 C1-C6알콕시로부터 독립적으로 선택됨)가 포함된다. As used herein, the term “optionally substituted” means that the group referred to is alkyl, alkenyl, alkynyl, cycloalkyl, aryl, heteroaryl, heterocycloalkyl, hydroxyl, alkoxy, mercaptyl, cyano, halo, carbonyl , Thiocarbonyl, isocyanato, thiocyanato, isothiocyanato, nitro, perhaloalkyl, perfluoroalkyl, and from amino, including mono and disubstituted amino groups, and protected derivatives thereof It means that it may or may not be substituted with one or more additional group(s) selected individually and independently. Non-limiting examples of optional substituents include halo, -CN, -OR, -C(O)R, -OC(O)R, -C(O)OR, -OC(O)NHR, -C(O) N(R) 2 , -SR-, -S(=O)R, -S(=O) 2 R, -NHR, -N(R) 2 , -NHC(O)-, NHC(O)O- , -C(O)NH-, S(=O) 2 NHR, -S(O) 2 N(R) 2 , -NHS(=O) 2 , -NHS(O) 2 R, C 1 -C 6 Alkyl, C 1 -C 6 alkoxy, aryl, heteroaryl, cycloalkyl, heterocycloalkyl, halo-substituted C 1 -C 6 alkyl, halo-substituted C 1 -C 6 alkoxy, wherein each R is H , Halo, C 1 -C 6 alkyl, C 1 -C 6 alkoxy, aryl, heteroaryl, cycloalkyl, heterocycloalkyl, halo-substituted C 1 -C 6 alkyl, halo-substituted C 1 -C 6 alkoxy Independently selected from).
본원에 사용된 용어 "친화성 표지"는 가역적으로 또는 비가역적으로 또 다른 분자에 결합하는 표지를 지칭한다. As used herein, the term “affinity label” refers to a label that reversibly or irreversibly binds to another molecule.
본원에 사용된 용어 "앰버 코돈"은 피롤리신, PCL 및 다른 피롤리신 유사체의 혼입 부위를 지칭하며, 메신저 RNA 내의 뉴클레오티드 트리플렛(triplet)인 UAG에 상응한다. 뉴클레오티드 서열 TAG는 DNA에 코딩되고, 단백질로 번역되는 RNA에 UAG로 전사된다. TAG 및 UAG 코돈은 피롤리신, PCL 및 다른 피롤리신 유사체의 혼입 부위를 지칭하기 위해 본원에서 교환가능하게 사용된다.The term “amber codon” as used herein refers to the site of incorporation of pyrrolysine, PCL and other pyrrolysine analogs, and corresponds to UAG, a nucleotide triplet in messenger RNA. The nucleotide sequence TAG is encoded in DNA and transcribed as UAG into RNA that is translated into protein. The TAG and UAG codons are used interchangeably herein to refer to the site of incorporation of pyrrolysine, PCL and other pyrrolysine analogs.
본원에 사용된 용어 "아미노산"은 자연 발생 아미노산, 비천연 아미노산, 아미노산 유사체, 및 자연 발생 아미노산과 유사한 방식으로 기능하는 아미노산 모방체 (이들의 구조가 D 및 L 입체이성질체 형태를 허용하는 경우, 이러한 모든 입체이성질체 형태로)를 지칭한다. 아미노산은 본원에서 그의 명칭, 그의 통상적으로 공지된 3문자 기호, 또는 IUPAC-IUB 생화학 명명 위원회(Biochemical Nomenclature Commission)에 의해 권고된 1문자 기호에 의해 지칭된다. 자연 발생 아미노산은 유전 암호에 의해 코딩되는 아미노산, 뿐만 아니라 이후 변형되는 코딩된 아미노산이다. 자연 발생 아미노산에는 알라닌 (Ala), 아르기닌 (Arg), 아스파라긴 (Asn), 아스파르트산 (Asp), 시스테인 (Cys), 글루타민 (Gln), 글루탐산 (Glu), 글리신 (Gly), 히스티딘 (His), 이소류신 (Ile), 류신 (Leu), 리신 (Lys), 메티오닌 (Met), 페닐알라닌 (Phe), 프롤린 (Pro), 세린 (Ser), 트레오닌 (Thr), 트립토판 (Trp), 티로신 (Tyr), 발린 (Val), 피롤리신 (Pyl), 셀레노시스테인 (Sec) 및 피롤린-카르복시-리신 (PCL)이 포함되나, 이에 제한되지는 않는다. 변형된 코딩된 아미노산에는 히드록시프롤린, γ-카르복시글루타메이트, O-포스포세린, 아제티딘카르복실산, 2-아미노아디프산, 3-아미노아디프산, 베타-알라닌, 아미노프로피온산, 2-아미노부티르산, 4-아미노부티르산, 6-아미노카프로산, 2-아미노헵탄산, 2-아미노부티르산, 3-아미노부티르산, 2-아미노피멜산, 3급-부틸글리신, 2,4-디아미노이소부티르산, 데스모신, 2,2'-디아미노피멜산, 2,3-디아미노프로피온산, N-에틸글리신, N-메틸글리신, N-에틸아스파라긴, 호모프롤린, 히드록시리신, 알로-히드록시리신, 3-히드록시프롤린, 4-히드록시프롤린, 이소데스모신, 알로-이소류신, N-메틸알라닌, N-메틸글리신, N-메틸이소류신, N-메틸펜틸글리신, N-메틸발린, 나프트알라닌, 노르발린, 노르류신, 오르니틴, 펜틸글리신, 피페콜산과 티오프롤린이 포함되나, 이에 제한되지는 않는다. 용어 아미노산은 또한 특정 유기체에서, 대사물이지만 단백질로의 혼입을 위해 유전 암호에 의해 코딩되지는 않는 자연 발생 아미노산을 포함한다. 이러한 아미노산에는 오르니틴, D-오르니틴 및 D-아르기닌이 포함되나, 이에 제한되지는 않는다. As used herein, the term “amino acid” refers to naturally occurring amino acids, non-natural amino acids, amino acid analogs, and amino acid mimetics that function in a similar manner to naturally occurring amino acids (where their structure allows for D and L stereoisomeric forms, such In all stereoisomeric forms). Amino acids are referred to herein by their name, their commonly known three letter symbols, or single letter symbols recommended by the IUPAC-IUB Biochemical Nomenclature Commission. Naturally occurring amino acids are amino acids that are encoded by the genetic code, as well as encoded amino acids that are subsequently modified. Naturally occurring amino acids include alanine (Ala), arginine (Arg), asparagine (Asn), aspartic acid (Asp), cysteine (Cys), glutamine (Gln), glutamic acid (Glu), glycine (Gly), histidine (His), Isoleucine (Ile), Leucine (Leu), Lysine (Lys), Methionine (Met), Phenylalanine (Phe), Proline (Pro), Serine (Ser), Threonine (Thr), Tryptophan (Trp), Tyrosine (Tyr), Valine (Val), pyrrolysine (Pyl), selenocysteine (Sec) and pyrroline-carboxy-lysine (PCL) are included, but are not limited thereto. Modified encoded amino acids include hydroxyproline, γ-carboxyglutamate, O-phosphoserine, azetidinecarboxylic acid, 2-aminoadipic acid, 3-aminoadipic acid, beta-alanine, aminopropionic acid, 2-amino Butyric acid, 4-aminobutyric acid, 6-aminocaproic acid, 2-aminoheptanoic acid, 2-aminobutyric acid, 3-aminobutyric acid, 2-aminopimelic acid, tert-butylglycine, 2,4-diaminoisobutyric acid, Desmosine, 2,2'-diaminopimelic acid, 2,3-diaminopropionic acid, N-ethylglycine, N-methylglycine, N-ethylasparagine, homoproline, hydroxylysine, allo-hydroxylysine, 3 -Hydroxyproline, 4-hydroxyproline, isodesmosine, allo-isoleucine, N-methylalanine, N-methylglycine, N-methylisoleucine, N-methylpentylglycine, N-methylvaline, naphthalanine, nor Valine, norleucine, ornithine, pentylglycine, pipecolic acid and thioproline are included, but are not limited thereto. The term amino acid also includes naturally occurring amino acids that, in certain organisms, are metabolites but are not encoded by the genetic code for incorporation into proteins. Such amino acids include, but are not limited to, ornithine, D-ornithine and D-arginine.
본원에 사용된 용어 "아미노산 유사체"는, 자연 발생 아미노산과 같은 기본 화학 구조, 단지 예로서 언급하자면 수소, 카르복실기, 아미노기 및 R 기에 결합된 α-탄소를 갖는 화합물을 지칭한다. 아미노산 유사체에는, 가역적으로 또는 비가역적으로 화학적으로 차단되거나, 또는 그의 C-말단 카르복시기, 그의 N-말단 아미노기 및/또는 그의 측쇄 관능기가 화학적으로 변형된 천연 및 비천연 아미노산이 포함된다. 이러한 유사체에는 메티오닌 술폭시드, 메티오닌 술폰, S-(카르복시메틸)-시스테인, S-(카르복시메틸)-시스테인 술폭시드, S-(카르복시메틸)-시스테인 술폰, 아스파르트산-(베타-메틸 에스테르), N-에틸글리신, 알라닌 카르복스아미드, 호모세린, 노르류신, 및 메티오닌 메틸 술포늄이 포함되나, 이에 제한되지는 않는다. 특정 차단제에는 t-부틸옥시카르보닐 (Boc) 및 9-플루오레닐메틸옥시카르보닐 (Fmoc)이 포함되나, 이에 제한되지는 않는다. As used herein, the term “amino acid analog” refers to a compound having a basic chemical structure such as a naturally occurring amino acid, by way of example only hydrogen, a carboxyl group, an amino group and an α-carbon bonded to the R group. Amino acid analogs include natural and non-natural amino acids that are chemically blocked reversibly or irreversibly, or whose C-terminal carboxy group, their N-terminal amino group and/or their side chain functional groups are chemically modified. Such analogs include methionine sulfoxide, methionine sulfone, S-(carboxymethyl)-cysteine, S-(carboxymethyl)-cysteine sulfoxide, S-(carboxymethyl)-cysteine sulfone, aspartic acid-(beta-methyl ester), N-ethylglycine, alanine carboxamide, homoserine, norleucine, and methionine methyl sulfonium are included, but are not limited thereto. Certain blocking agents include, but are not limited to, t-butyloxycarbonyl (Boc) and 9-fluorenylmethyloxycarbonyl (Fmoc).
본원에 사용된 용어 "아미노산 모방체"는 아미노산의 일반적 화학 구조와 상이한 구조를 갖지만, 자연 발생 아미노산과 유사한 방식으로 기능하는 화합물을 지칭한다.As used herein, the term “amino acid mimetic” refers to a compound that has a structure different from the general chemical structure of an amino acid, but functions in a manner similar to a naturally occurring amino acid.
본원에 사용된 용어 "비천연 아미노산"은, 임의의 유기체로부터의 변형되지 않거나 변형된 유전자를 이용하여 임의의 유기체 (동일하든지 상이하든지간에)에서 생합성적으로 생성될 수 없는 아미노산 구조를 나타내도록 의도된다. 추가로, 이러한 "비천연 아미노산"은 단백질로의 혼입을 위한 변형된 tRNA 및 변형된 tRNA 신테타제 (RS)를 필요로 한다는 것이 이해된다. 이러한 "선택된" 직교 tRNA/RS 쌍은 비천연 아미노산에 특이적이고, 슐츠(Schultz) 등에 의해 개발된 선택 절차 또는 유사한 절차에 의해 생성된다. 예로서, 피롤린-카르복시-리신은 한 유기체로부터 숙주 세포로 이동된 유전자에 의해 생합성적으로 생성된다는 점에서, 그리고 천연 tRNA 및 tRNA 신테타제 유전자를 사용하여 단백질에 혼입된다는 점에서 "천연 아미노산"이고, 반면에 p-아미노페닐알라닌 (문헌 [Generation of a bacterium with a 21 amino acid genetic code, Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, Schultz PG. J Am Chem Soc. 2003 Jan 29;125(4):935-9] 참조)은 생합성적으로 생성될지라도, "선택된" 직교 tRNA/tRNA 신테타제 쌍에 의해 단백질에 혼입되기 때문에 "비천연 아미노산"이다.As used herein, the term “non-natural amino acid” is intended to refer to an amino acid structure that cannot be biosynthetically produced in any organism (whether identical or different) using unmodified or modified genes from any organism. do. Additionally, it is understood that such “non-natural amino acids” require modified tRNA and modified tRNA synthetase (RS) for incorporation into proteins. These “selected” orthogonal tRNA/RS pairs are specific for non-natural amino acids and are generated by selection procedures or similar procedures developed by Schultz et al. As an example, pyrroline-carboxy-lysine is a "natural amino acid" in that it is produced biosynthetically by genes transferred from an organism to a host cell, and in that it is incorporated into proteins using natural tRNA and tRNA synthetase genes. And p-aminophenylalanine (Generation of a bacterium with a 21 amino acid genetic code, Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, Schultz PG. J Am Chem. Soc. 2003
본원에 사용된 용어 "아미노산 잔기"는 의 구조를 갖는 잔기를 지칭하며, 여기서 이러한 잔기는 아미노산으로부터 유래되고, R 기는 본원에 기재된 임의의 아미노산의 측쇄이다. 이러한 아미노산 잔기에는 알라니닐, 아르기니닐, 아스파라기닐, 아스파르틸, 시스테이닐, 글루타미닐, 글루타밀, 글리시닐, 히스티디닐, 이소류시닐, 류시닐, 리시닐, 메티오니닐, 페닐알라니닐, 프롤리닐, 세리닐, 트레오니닐, 트립토파닐, 티로시닐, 발리닐, 피로글루타메이트, 포르밀메티오닌, 피로글리시닐 및 셀레노시스테이닐이 포함되나, 이에 제한되지는 않는다. As used herein, the term "amino acid moiety" refers to Refers to a residue having the structure of, wherein such residues are derived from amino acids, and the R group is the side chain of any amino acid described herein. These amino acid residues include alaninyl, arginyl, asparaginyl, aspartyl, cysteinyl, glutaminyl, glutamyl, glycinyl, histidinyl, isoleucinyl, leucinyl, lisinyl, These include methioninyl, phenylalaninyl, prolinyl, serinyl, threoninyl, tryptophanil, tyrosinyl, ballinyl, pyroglutamate, formylmethionine, pyroglycinyl and selenocysteinyl. , But is not limited thereto.
용어 "아미노 말단 변형 기"는 말단 아미노기에 부착될 수 있는 임의의 분자를 지칭한다. 이러한 아미노 말단 변형 기에는 중합체 분자의 말단에 있는 아민 보호기가 포함되나 이에 제한되지는 않으며, 상기 중합체 분자에는 폴리펩티드, 폴리뉴클레오티드 및 폴리사카라이드가 포함되나, 이에 제한되지는 않는다. 말단 변형 기에는 또한 다양한 수용성 중합체, 펩티드 또는 단백질이 포함되나, 이에 제한되지는 않는다. 단지 예로서 언급하자면, 아미노 말단 변형 기에는 폴리에틸렌 글리콜 또는 혈청 알부민이 포함된다. 특정 아미노 말단 변형 기는 단백질, 폴리펩티드 또는 펩티드의 치료 특성을 변형시키는데, 예를 들어 이로 제한되는 것은 아니지만 이러한 단백질, 폴리펩티드 또는 펩티드의 혈청 반감기를 증가시키는데 사용된다. The term “amino terminal modifying group” refers to any molecule that can be attached to a terminal amino group. Such amino terminal modification groups include, but are not limited to, an amine protecting group at the end of the polymer molecule, and the polymer molecule includes, but is not limited to, polypeptides, polynucleotides and polysaccharides. Terminal modification groups also include, but are not limited to, various water soluble polymers, peptides or proteins. By way of example only, amino terminal modifying groups include polyethylene glycol or serum albumin. Certain amino terminal modifying groups are used to modify the therapeutic properties of a protein, polypeptide or peptide, for example, but not limited to, to increase the serum half-life of such a protein, polypeptide or peptide.
본원에 사용된 용어 "항체 단편"은 전장 형태 이외의 임의의 형태의 항체를 지칭한다. 본원에서 항체 단편에는 전장 항체 내에 존재하는 더 작은 성분인 항체, 및 조작된 항체가 포함된다. 항체 단편에는 Fv, Fc, Fab 및 (Fab')2, 단일 쇄 Fv (scFv), 디아바디, 트리아바디, 테트라바디, 이관능성 하이브리드 항체, CDR1, CDR2, CDR3, CDR들의 조합, 가변 영역, 프레임워크 영역, 불변 영역, 중쇄, 경쇄, 및 가변 영역, 및 대안적 스캐폴드 비-항체 분자, 이중특이적 항체 등이 포함되나, 이에 제한되지는 않는다. 또 다른 기능적 하위구조는 펩티드 링커에 의해 공유 결합된 이뮤노글로불린 중쇄 및 경쇄의 가변 영역으로 이루어진 단일 쇄 Fv (scFv)이다. 이러한 작은 (MW < 25,000 Da) 단백질은 일반적으로 단일 폴리펩티드에서 항원에 대한 특이성 및 친화성을 유지하고, 큰 항원-특이적 분자를 위한 편리한 빌딩 블록을 제공할 수 있다. 달리 구체적으로 언급되지 않는다면, 용어 "항체" 또는 "항체들"을 사용하는 내용 및 청구범위는 또한 "항체 단편" 및 "항체 단편들"을 포함한다. The term “antibody fragment” as used herein refers to any form of antibody other than the full-length form. Antibody fragments herein include antibodies, which are smaller components present in full-length antibodies, and engineered antibodies. Antibody fragments include Fv, Fc, Fab and (Fab') 2 , single chain Fv (scFv), diabodies, triabodies, tetrabodies, bifunctional hybrid antibodies, CDR1, CDR2, CDR3, combinations of CDRs, variable regions, frames Work regions, constant regions, heavy chain, light chain, and variable regions, and alternative scaffold non-antibody molecules, bispecific antibodies, and the like. Another functional substructure is the single chain Fv (scFv) consisting of the variable regions of the immunoglobulin heavy and light chains covalently linked by a peptide linker. These small (MW <25,000 Da) proteins generally retain specificity and affinity for the antigen in a single polypeptide and can provide convenient building blocks for large antigen-specific molecules. Unless specifically stated otherwise, content and claims using the term “antibody” or “antibodies” also include “antibody fragments” and “antibody fragments”.
본원에 사용된 용어 "생체이용률"은 물질 또는 그의 활성 잔기가 제약 투여 형태로부터 전달되어 작용 부위에서 또는 일반적 순환으로 이용 가능하게 되는 비율 및 정도를 지칭한다. 생체이용률의 증가는 물질 또는 그의 활성 잔기가 제약 투여 형태로부터 전달되어 작용 부위에서 또는 일반적 순환으로 이용 가능하게 되는 비율 및 정도의 증가를 지칭한다. 예를 들어, 생체이용률의 증가는 혈액 중에서 물질 또는 그의 활성 잔기의 농도가 다른 물질 또는 활성 잔기에 비해 증가되는 것으로 나타내어질 수 있다. As used herein, the term “bioavailability” refers to the rate and extent to which a substance or active moiety thereof is delivered from a pharmaceutical dosage form and is made available at the site of action or in general circulation. An increase in bioavailability refers to an increase in the rate and extent to which a substance or an active moiety thereof is delivered from a pharmaceutical dosage form and becomes available at the site of action or in general circulation. For example, an increase in bioavailability may be indicated by an increase in the concentration of a substance or active moiety thereof in blood compared to other substances or active moieties.
본원에 사용된 용어 "생물학적 활성 분자", "생물학적 활성 잔기" 또는 "생물학적 활성제"는 유기체, 예컨대 이로 제한되는 것은 아니지만 바이러스, 박테리아, 박테리오파지, 트랜스포손, 프리온, 곤충, 진균, 식물, 동물 및 인간과 관련이 있는 생물학적 시스템, 경로, 분자 또는 상호작용의 임의의 물리적 또는 생화학적 성질에 영향을 미칠 수 있는 임의의 물질을 지칭한다. 특히, 본원에 사용된 생물학적 활성 분자에는 인간 또는 다른 동물에서 질환의 진단, 치유, 완화, 치료 또는 예방을 위해, 또는 달리 인간 또는 동물의 신체적 또는 정신적 행복의 개선을 위해 의도된 임의의 물질이 포함되나, 이에 제한되지는 않는다. 생물학적 활성 분자의 예에는 펩티드, 단백질, 효소, DNA, RNA, 소분자 약물, 경질 약물, 연질 약물, 폴리사카라이드, 올리고사카라이드, 디사카라이드, 탄수화물, 무기 원자 또는 분자, 염료, 지질, 뉴클레오시드, 방사성핵종, 올리고뉴클레오티드, 독소, 세포, 바이러스, 리포좀, 마이크로입자 및 미셀이 포함되나, 이에 제한되지는 않는다. 본원에 기재된 방법 및 조성물에 사용하기에 적합한 생물학적 활성제의 부류에는 약물, 전구약물, 방사성핵종, 영상화제, 중합체, 항생제, 살진균제, 항-바이러스성 작용제, 항-염증성 작용제, 항종양제, 심혈관 작용제, 항-불안 작용제, 호르몬, 성장 인자, 스테로이드성 작용제, 마이크로바이알로 유도된 독소 등이 포함되나, 이에 제한되지는 않는다. As used herein, the terms "biologically active molecule", "biologically active moiety" or "biologically active" are organisms such as, but not limited to, viruses, bacteria, bacteriophages, transposons, prions, insects, fungi, plants, animals and humans. Refers to any substance capable of affecting any physical or biochemical property of a biological system, pathway, molecule or interaction with which it is associated. In particular, biologically active molecules as used herein include any substance intended for the diagnosis, cure, alleviation, treatment or prevention of a disease in humans or other animals, or otherwise improving the physical or mental well-being of a human or animal. However, it is not limited thereto. Examples of biologically active molecules include peptides, proteins, enzymes, DNA, RNA, small molecule drugs, hard drugs, soft drugs, polysaccharides, oligosaccharides, disaccharides, carbohydrates, inorganic atoms or molecules, dyes, lipids, nucleos. Seeds, radionuclides, oligonucleotides, toxins, cells, viruses, liposomes, microparticles and micelles are included, but are not limited thereto. Classes of biologically active agents suitable for use in the methods and compositions described herein include drugs, prodrugs, radionuclides, imaging agents, polymers, antibiotics, fungicides, anti-viral agents, anti-inflammatory agents, anti-tumor agents, cardiovascular agents. Agents, anti-anxiety agents, hormones, growth factors, steroidal agents, microvial-induced toxins, and the like.
본원에 사용된 용어 "생물학적 활성의 조절"은 단백질, 폴리펩티드, 펩티드, DNA, RNA, 사카라이드, 당, 대사물, 전구체, 보조인자 또는 다른 생물학적 활성 화학물질 또는 실체의 농도 또는 반응성을 증가 또는 감소시키거나, 단백질, 폴리펩티드, 펩티드, DNA, RNA, 사카라이드, 당, 대사물, 전구체, 보조인자 또는 다른 생물학적 활성 화학물질 또는 실체의 선택성을 변경시키거나, 또는 단백질, 폴리펩티드, 펩티드, DNA, RNA, 사카라이드, 당, 대사물, 전구체, 보조인자 또는 다른 생물학적 활성 화학물질 또는 실체의 기질 선택성을 개선 또는 감소시키는 것을 지칭한다. As used herein, the term "modulation of biological activity" increases or decreases the concentration or reactivity of a protein, polypeptide, peptide, DNA, RNA, saccharide, sugar, metabolite, precursor, cofactor, or other biologically active chemical or entity. Or alter the selectivity of proteins, polypeptides, peptides, DNA, RNA, saccharides, sugars, metabolites, precursors, cofactors or other biologically active chemicals or entities, or proteins, polypeptides, peptides, DNA, RNA , A saccharide, sugar, metabolite, precursor, cofactor, or other biologically active chemical or entity.
본원에 사용된 용어 "생체물질"은 생물학적으로 유래된 물질, 예컨대 이로 제한되는 것은 아니지만, 생물반응기로부터 및/또는 재조합 방법 및 기술로부터 수득된 물질을 지칭한다. As used herein, the term “biomaterial” refers to a biologically derived material, such as, but not limited to, a material obtained from a bioreactor and/or from recombinant methods and techniques.
본원에 사용된 용어 "생물물리학적 프로브"는 물리적 검출 방법에 의해 분자에서의 구조적인 변화를 검출하거나 모니터링하는 프로브를 지칭한다. 이러한 분자에는 단백질, 폴리펩티드, 펩티드, DNA 또는 RNA가 포함되나, 이에 제한되지는 않는다. 이러한 "생물물리학적 프로브"는 또한 단백질, 폴리펩티드, 펩티드, DNA 또는 RNA와 다른 분자, 예컨대 이로 제한되는 것은 아니지만 거대분자의 상호작용을 검출하거나 모니터링하는데 사용된다. 생물물리학적 프로브의 예에는 분자 질량, 핵 스핀, UV 흡광도, 형광, 원편광 이색성, 열 용량, 용융 온도 또는 다른 내인성 분자 특성이 포함되나, 이에 제한되지는 않는다. 생물물리학적 프로브의 예에는 또한 분자에 첨가된 표지가 포함된다. 이러한 프로브에는 스핀 표지, 형광단, 동위원소 표지 및 광활성 기가 포함되나, 이에 제한되지는 않는다. The term “biophysical probe” as used herein refers to a probe that detects or monitors structural changes in a molecule by physical detection methods. Such molecules include, but are not limited to, proteins, polypeptides, peptides, DNA or RNA. Such “biophysical probes” are also used to detect or monitor the interaction of proteins, polypeptides, peptides, DNA or RNA with other molecules, such as, but not limited to, macromolecules. Examples of biophysical probes include, but are not limited to, molecular mass, nuclear spin, UV absorbance, fluorescence, circular dichroism, heat capacity, melting temperature, or other endogenous molecular properties. Examples of biophysical probes also include labels added to the molecule. Such probes include, but are not limited to, spin labels, fluorophores, isotopic labels, and photoactive groups.
본원에 사용된 용어 "생합성적으로 생성된"은 아미노산을 생성하기 위해 세포 또는 효소를 이용하는 임의의 방법을 지칭한다. 이러한 방법은 전구체 및 효소 중 적어도 하나의 사용을 포함한다. 특정 실시양태에서, 이러한 아미노산은 단백질에 혼입된다. 특정 실시양태에서는 아미노산의 생합성 및 혼입이 동일한 세포에서 일어나는 반면, 다른 실시양태에서는 아미노산이 별도의 세포 ("피더 세포")에서 또는 별도의 세포 배양물에서 생합성적으로 생성되어 이마노산이 또 다른 세포에서 단백질에 혼입된다. 후자의 경우, 아미노산은 별도의 세포 배양물로부터 임의로 정제된 후, 정제된 아미노산은 아미노산을 단백질에 혼입시키는 세포 배양물의 배지에 첨가된다.As used herein, the term “biosynthetically produced” refers to any method using cells or enzymes to produce amino acids. This method involves the use of at least one of a precursor and an enzyme. In certain embodiments, such amino acids are incorporated into proteins. In certain embodiments, the biosynthesis and incorporation of amino acids occurs in the same cell, whereas in other embodiments the amino acids are produced biosynthetically in separate cells ("feeder cells") or in separate cell cultures, resulting in imanoic acid being produced in another cell. Is incorporated into the protein. In the latter case, amino acids are optionally purified from a separate cell culture, and then the purified amino acids are added to the medium of the cell culture to incorporate the amino acids into the protein.
본원에 사용된 용어 "비오틴 유사체" (또는 "비오틴 모방체"로도 지칭됨)는 아비딘 및/또는 스트렙타비딘과 높은 친화도로 결합하는, 비오틴 이외의 임의의 분자를 지칭한다. The term “biotin analog” (also referred to as “biotin mimetic”) as used herein refers to any molecule other than biotin that binds avidin and/or streptavidin with high affinity.
용어 "카르복시 말단 변형 기"는 말단 카르복시기에 부착될 수 있는 임의의 분자를 지칭한다. 이러한 카르복시 말단 기에는 중합체 분자의 말단에 있는 카르복실레이트 보호기가 포함되나 이에 제한되지는 않고, 상기 중합체 분자에는 폴리펩티드, 폴리뉴클레오티드 및 폴리사카라이드가 포함되나, 이에 제한되지는 않는다. 말단 변형 기에는 또한 다양한 수용성 중합체, 펩티드 또는 단백질이 포함되나, 이에 제한되지는 않는다. 단지 예로서 언급하자면, 말단 변형 기에는 폴리에틸렌 글리콜 또는 혈청 알부민이 포함된다. 특정 카르복시 말단 변형 기는 단백질, 폴리펩티드 또는 펩티드의 치료 특성을 변형시키는데, 예를 들어 이로 제한되는 것은 아니지만 혈청 반감기를 증가시키는데 사용된다. The term “carboxy terminal modifying group” refers to any molecule that can be attached to a terminal carboxy group. Such carboxy terminal groups include, but are not limited to, a carboxylate protecting group at the end of the polymer molecule, and the polymer molecule includes, but is not limited to, polypeptides, polynucleotides and polysaccharides. Terminal modification groups also include, but are not limited to, various water soluble polymers, peptides or proteins. By way of example only, terminal modifying groups include polyethylene glycol or serum albumin. Certain carboxy terminal modifying groups are used to modify the therapeutic properties of proteins, polypeptides or peptides, for example, but not limited to, to increase serum half-life.
본원에 사용된 용어 "화학 절단성 기" ("화학적으로 불안정한"으로도 지칭됨)는 산, 염기, 산화제, 환원제, 화학적 개시제, 또는 라디칼 개시제에 대한 노출시 파괴 또는 절단되는 기를 지칭한다. As used herein, the term “chemically cleavable group” (also referred to as “chemically labile”) refers to a group that breaks or cleaves upon exposure to an acid, base, oxidizing agent, reducing agent, chemical initiator, or radical initiator.
본원에 사용된 용어 "화학발광 기"는 열을 가하지 않고도 화학 반응의 결과로서 빛을 내는 기를 지칭한다. 단지 예로서 언급하자면, 루미놀 (5-아미노-2,3-디히드로-1,4-프탈라진디온)을 염기 및 금속 촉매의 존재 하에 산화제, 예컨대 과산화수소 (H2O2)와 반응시켜 여기 상태 생성물 (3-아미노프탈레이트, 3-APA)을 생성하고, 후속적으로 검출가능한 광을 방출시킨다. The term “chemiluminescent group” as used herein refers to a group that emits light as a result of a chemical reaction without applying heat. By way of example only, luminol (5-amino-2,3-dihydro-1,4-phthalazinedione) is reacted with an oxidizing agent such as hydrogen peroxide (H 2 O 2) in the presence of a base and a metal catalyst to A state product (3-aminophthalate, 3-APA) is produced, which subsequently emits detectable light.
본원에 사용된 용어 "발색단"은 가시 파장, UV 파장 또는 IR 파장의 광을 흡수하는 분자를 지칭한다. As used herein, the term “chromophore” refers to a molecule that absorbs light in visible, UV or IR wavelengths.
본원에 사용된 용어 "보조인자"는 큰 분자의 작용에 필수적인 원자 또는 분자를 지칭한다. 보조인자에는 무기 이온, 조효소, 단백질, 또는 효소의 활성을 위해 필요한 일부 다른 인자가 포함되나, 이에 제한되지는 않는다. As used herein, the term “cofactor” refers to an atom or molecule essential for the functioning of a large molecule. Cofactors include, but are not limited to, inorganic ions, coenzymes, proteins, or some other factor necessary for the activity of the enzyme.
본원에 사용된 용어 "코폴딩"은 서로 상호작용하는 2개 이상의 분자를 사용하여 언폴딩되거나 부적절하게 폴딩된 분자를 적절하게 폴딩된 분자로 변형시키는 리폴딩 과정, 반응 또는 방법을 지칭한다. 단지 예로서 언급하자면, "코폴딩"은 서로 상호작용하는 2개 이상의 폴리펩티드를 사용하여 언폴딩되거나 부적절하게 폴딩된 폴리펩티드를 원래의 적절하게 폴딩된 폴리펩티드로 변형시킨다. As used herein, the term “cofolding” refers to a refolding process, reaction, or method of transforming an unfolded or improperly folded molecule into an appropriately folded molecule using two or more molecules that interact with each other. By way of example only, "cofolding" uses two or more polypeptides that interact with each other to transform an unfolded or improperly folded polypeptide into an original properly folded polypeptide.
본원에 사용된 용어 "세포독성"은 세포에 해를 입히는 화합물을 지칭한다. As used herein, the term “cytotoxic” refers to a compound that harms cells.
본원에 사용된 용어 "변성 작용제" 또는 "변성제"는 단백질의 가역적 언폴딩을 초래하는 임의의 화합물 또는 물질을 지칭한다. 변성 작용제 또는 변성제의 강도는 특별한 변성 작용제 또는 변성제의 특성 및 농도 둘 다에 의해 결정될 것이다. 예로서, 변성 작용제 또는 변성제에는 카오트로프제, 세정제, 유기, 수혼화성 용매, 인지질 또는 이들의 조합물이 포함되나, 이에 제한되지는 않는다. 카오트로프제의 비제한적인 예에는 우레아, 구아니딘 및 나트륨 티오시아네이트가 포함되나, 이에 제한되지는 않는다. 세정제의 비제한적인 예에는 강성 세정제, 예컨대 나트륨 도데실 술페이트 또는 폴리옥시에틸렌 에테르 (예를 들어, 트윈 또는 트리톤 세정제), 사르코실, 온화한 비-이온성 세정제 (예를 들어, 디기토닌), 온화한 양이온성 세정제, 예컨대 N-2,3-(디올레일옥시)-프로필-N,N,N-트리메틸암모늄, 온화한 이온성 세정제 (예를 들어, 나트륨 콜레이트 또는 나트륨 데옥시콜레이트) 또는 양쪽성 세정제, 예컨대 이로 제한되는 것은 아니지만 술포베타인 (쯔비터젠트(Zwittergent)), 3-(3-클로라미도프로필)디메틸암모니오-1-프로판 술페이트 (CHAPS) 및 3-(3-클로라미도프로필)디메틸암모니오-2-히드록시-1-프로판 술포네이트 (CHAPSO)가 포함되지만 이에 제한되지는 않을 수 있다. 유기, 수혼화성 용매의 비제한적인 예에는 아세토니트릴, 저급 알칸올 (특히, C2-C4 알칸올, 예컨대 에탄올 또는 이소프로판올)이 포함되지만 이에 제한되지는 않거나, 저급 알칸디올 (C2-C4 알칸디올, 예컨대 에틸렌-글리콜)을 변성제로서 사용할 수 있다. 인지질의 비제한적인 예에는 자연 발생 인지질, 예컨대 포스파티딜에탄올아민, 포스파티딜콜린, 포스파티딜세린 및 포스파티딜이노시톨 또는 합성 인지질 유도체 또는 변이체, 예컨대 디헥사노일포스파티딜콜린 또는 디펩타노일포스파티딜콜린이 포함되나, 이에 제한되지는 않는다. As used herein, the term “denaturing agent” or “denaturing agent” refers to any compound or substance that results in reversible unfolding of a protein. The strength of the denaturing agent or denaturing agent will be determined by both the nature and concentration of the particular denaturing agent or denaturing agent. By way of example, denaturing agents or denaturing agents include, but are not limited to, chaotropes, detergents, organic, water-miscible solvents, phospholipids, or combinations thereof. Non-limiting examples of chaotropes include, but are not limited to, urea, guanidine and sodium thiocyanate. Non-limiting examples of detergents include hard detergents such as sodium dodecyl sulfate or polyoxyethylene ether (e.g. Tween or Triton detergents), sarcosyl, mild non-ionic detergents (e.g. digitonin), Mild cationic detergents such as N-2,3-(dioleyyloxy)-propyl-N,N,N-trimethylammonium, mild ionic detergents (e.g. sodium cholate or sodium deoxycholate) or amphoteric detergents , Such as but not limited to sulfobetaine (Zwittergent), 3-(3-chloramidopropyl)dimethylammonio-1-propane sulfate (CHAPS) and 3-(3-chloramido Propyl) dimethylammonio-2-hydroxy-1-propane sulfonate (CHAPSO). Non-limiting examples of organic, water-miscible solvents include, but are not limited to, acetonitrile, lower alkanols (especially C 2 -C 4 alkanols such as ethanol or isopropanol), or lower alkanediols (C 2 -C 4 Alkanedols such as ethylene-glycol) can be used as denaturants. Non-limiting examples of phospholipids include, but are not limited to, naturally occurring phospholipids such as phosphatidylethanolamine, phosphatidylcholine, phosphatidylserine and phosphatidylinositol or synthetic phospholipid derivatives or variants such as dihexanoylphosphatidylcholine or dipeptanoylphosphatidylcholine.
본원에 사용된 용어 "검출가능한 표지"는 분석 기술, 예컨대 이로 제한되는 것은 아니지만 형광, 화학발광, 전자-회전 공명, 자외선/가시선 흡광도 분광분석법, 적외선 분광분석법, 질량 분광측정법, 핵 자기 공명, 자기 공명, 방사측정 방법 및 전기화학적 방법을 이용하여 관찰할 수 있는 표지를 지칭한다. As used herein, the term "detectable label" refers to analytical techniques such as, but not limited to, fluorescence, chemiluminescence, electron-rotating resonance, ultraviolet/visible absorbance spectroscopy, infrared spectroscopy, mass spectrometry, nuclear magnetic resonance, magnetic Refers to a label that can be observed using resonance, radiometric and electrochemical methods.
본원에 사용된 용어 "약물"은 질환 또는 상태의 예방, 진단, 경감, 치료 또는 치유에 사용되는 임의의 물질을 지칭한다. As used herein, the term “drug” refers to any substance used in the prevention, diagnosis, alleviation, treatment or cure of a disease or condition.
본원에 사용된 용어 "염료"는 발색단을 함유하는 가용성 착색 물질을 지칭한다. As used herein, the term “dye” refers to a soluble colored material containing a chromophore.
본원에 사용된 용어 "전자 고밀도 기"는 전자 빔으로 조사될 때 전자를 산재시키는 기를 지칭한다. 이러한 군에는 몰리브덴산암모늄, 비스무트 서브니트레이트 카드뮴 요오다이드, 99%, 카르보히드라지드, 염화제2철 6수화물, 헥사메틸렌 테트라민, 98.5%, 무수 삼염화인듐, 질산란탄, 아세트산납 3수화물, 시트르산납 3수화물, 질산납, 과요오드산, 포스포몰리브덴산, 포스포텅스텐산, 페리시안화칼륨, 페로시안화칼륨, 루테늄 레드, 질산은, 은 단백질화합물 (Ag 검정: 8.0-8.5%) "강성", 은 테트라페닐포르핀 (S-TPPS), 클로로금산나트륨, 텅스텐산나트륨, 질산탈륨, 티오세미카르바지드 (TSC), 우라닐 아세테이트, 우라닐 니트레이트 및 바나딜 술페이트가 포함되나, 이에 제한되지는 않는다. The term “electron high density group” as used herein refers to a group that scatters electrons when irradiated with an electron beam. These groups include ammonium molybdate, bismuth subnitrate cadmium iodide, 99%, carbohydrazide, ferric chloride hexahydrate, hexamethylene tetramine, 98.5%, anhydrous indium trichloride, lanthanum nitrate, lead acetate trihydrate. , Lead citrate trihydrate, lead nitrate, periodic acid, phosphomolybdic acid, phosphotungstic acid, potassium ferricyanide, potassium ferrocyanide, ruthenium red, silver nitrate, silver protein compound (Ag black: 8.0-8.5%) "Stiffness ", silver tetraphenylporphine (S-TPPS), sodium chloroamate, sodium tungstate, thallium nitrate, thiosemicarbazide (TSC), uranyl acetate, uranyl nitrate and vanadyl sulfate are included, It is not limited thereto.
본원에 사용된 용어 "에너지 전달 작용제"는 또 다른 분자로부터 에너지를 공여 또는 수용할 수 있는 분자를 지칭한다. 단지 예로서 언급하자면, 형광 공명 에너지 전달 (FRET)은, 형광 공여 분자의 여기 상태 에너지가 비-방사성에 의해 여기되지 않은 수용 분자에 전달된 다음, 더 긴 파장에서 공여된 에너지를 형광에 의해 방출하는 이중극자-이중극자 커플링 과정이다. As used herein, the term “energy transfer agent” refers to a molecule capable of donating or accepting energy from another molecule. By way of example only, fluorescence resonance energy transfer (FRET) means that the excited state energy of a fluorescent donor molecule is transferred to a recipient molecule that is not excited by non-radioactivity, and then the energy donated at a longer wavelength is emitted by fluorescence. This is a dipole-dipole coupling process.
용어 "개선하다" 또는 "개선하는"은 원하는 효과를 효능 또는 지속시간의 측면에서 증가 또한 연장시키는 것을 의미한다. 예로서, 치료제의 효과를 "개선하는" 것은 질환, 장애 또는 상태의 치료 동안에 치료제의 효과를 효능 또는 지속시간의 측면에서 증가 또는 연장시키는 능력을 지칭한다. 본원에 사용된 "개선 유효량"은 질환, 장애 또는 상태의 치료에서 치료제의 효과를 개선하기에 적절한 양을 지칭한다. 환자에서 사용될 때, 이러한 사용에 효과적인 양은 질환, 장애 또는 상태의 중증도 및 과정, 기존 요법, 환자의 건강 상태 및 약물에 대한 반응, 및 치료하는 의사의 판단에 따라 좌우될 것이다.The terms “improving” or “improving” mean increasing or prolonging the desired effect in terms of efficacy or duration. By way of example, “improving” the effect of a therapeutic agent refers to the ability to increase or prolong the effect of a therapeutic agent in terms of efficacy or duration during treatment of a disease, disorder or condition. As used herein, an “improving effective amount” refers to an amount suitable to improve the effectiveness of a therapeutic agent in the treatment of a disease, disorder or condition. When used in a patient, the amount effective for such use will depend on the severity and course of the disease, disorder or condition, the existing therapy, the patient's health status and response to the drug, and the judgment of the treating physician.
용어 "진핵성"은 계통발생학적 영역 진핵생물(Eucarya)에 속하는 유기체, 예컨대 이로 제한되는 것은 아니지만 동물 (예컨대 이로 제한되는 것은 아니지만, 포유동물, 곤충, 파충류 및 조류(bird)), 섬모충, 식물 (예컨대 이로 제한되는 것은 아니지만, 외자엽식물, 쌍떡잎류 및 조류(algae)), 진균, 효모, 편모충류, 미포자충류, 및 원생생물로부터 기인하는 물질을 지칭한다. The term "eukaryotic" refers to organisms belonging to the phylogenetic domain of Eucarya, such as, but not limited to, animals (such as, but not limited to, mammals, insects, reptiles and birds), ciliated worms, plants. (Such as, but not limited to, exocotyledons, dicotyledons and algae), fungi, yeast, flagella, microspores, and substances resulting from protozoa.
본원에 사용된 용어 "지방산"은 약 C6 이상의 탄화수소 측쇄를 갖는 카르복실산을 지칭한다. The term “fatty acid” as used herein refers to a carboxylic acid having a hydrocarbon side chain of about C 6 or greater.
본원에 사용된 용어 "형광단"은 여기시 광자를 방출하는, 따라서 형광인 분자를 지칭한다. As used herein, the term “fluorophore” refers to a molecule that emits photons upon excitation, and thus is fluorescent.
용어 "관능기", "활성 잔기", "활성화 기", "이탈기", "반응성 부위", "화학 반응성 기" 및 "화학 반응성 잔기"는 화학업계에서 다소 동의어이며, 일부 기능 또는 활성을 수행하고 다른 분자와 반응성인 분자의 일부분을 나타내기 위해 본원에서 사용된다.The terms "functional group", "active moiety", "activating group", "leaving group", "reactive moiety", "chemically reactive group" and "chemically reactive moiety" are somewhat synonymous in the chemical industry and perform some function or activity. And is used herein to refer to a portion of a molecule that is reactive with another molecule.
본원에 사용된 용어 "동일성"은 동일한 2개 이상 서열 또는 하위 서열을 지칭한다. 게다가, 본원에 사용된 용어 "실질적으로 동일한"은 비교 알고리즘을 이용하여 또는 수동 정렬 및 육안 검사에 의해 측정된 바와 같이 비교 범위 또는 지정된 영역에 걸쳐 최대 대응에 대해 비교하고 정렬하였을 때, 동일한 서열 단위를 소정 백분율로 갖는 2개 이상 서열을 지칭한다. 단지 예로서 언급하자면, 2개 이상 서열은 서열 단위가 특정 부위에 걸쳐 약 60% 동일성, 약 65% 동일성, 약 70% 동일성, 약 75% 동일성, 약 80% 동일성, 약 85% 동일성, 약 90% 동일성 또는 약 95% 동일성인 경우 "실질적으로 동일할" 수 있다. 이러한 백분율은 2개 이상 서열의 "% 동일성"을 기재한다. 서열의 동일성은 적어도 약 75-100 서열 단위의 길이인 영역에 걸쳐, 약 50 서열 단위의 길이인 영역에 걸쳐, 또는 달리 명시되지 않는다면 전체 서열에 걸쳐 존재할 수 있다. 이 정의는 또한 시험 서열의 보체를 지칭한다. 단지 예로서 언급하자면, 2개 이상 폴리펩티드 서열은 아미노산 잔기가 동일할 때 동일성이고, 반면에 2개 이상 폴리펩티드 서열은 아미노산 잔기가 특정 부위에 걸쳐 약 60% 동일성, 약 65% 동일성, 약 70% 동일성, 약 75% 동일성, 약 80% 동일성, 약 85% 동일성, 약 90% 동일성 또는 약 95% 동일성인 경우 "실질적으로 동일하다". 동일성은 적어도 약 75-100 아미노산의 길이인 영역에 걸쳐, 약 50 아미노산의 길이인 영역에 걸쳐, 또는 달리 명시되지 않는다면 폴리펩티드 서열의 전체 서열에 걸쳐 존재할 수 있다. 게다가, 단지 예로서 언급하자면, 2개 이상 폴리뉴클레오티드 서열은 핵산 잔기가 동일할 때 동일성이고, 반면에 2개 이상 폴리뉴클레오티드 서열은 핵산 잔기가 특정 부위에 걸쳐 약 60% 동일성, 약 65% 동일성, 약 70% 동일성, 약 75% 동일성, 약 80% 동일성, 약 85% 동일성, 약 90% 동일성 또는 약 95% 동일성인 경우 "실질적으로 동일하다". 동일성은 적어도 약 75-100 핵산의 길이인 영역에 걸쳐, 약 50 핵산의 길이인 영역에 걸쳐, 또는 달리 명시되지 않는다면 폴리뉴클레오티드 서열의 전체 서열에 걸쳐 존재할 수 있다As used herein, the term “identity” refers to two or more identical sequences or subsequences. In addition, the term “substantially identical” as used herein refers to the same sequence unit when compared and aligned for maximum correspondence over a range of comparison or a designated area as measured using a comparison algorithm or by manual alignment and visual inspection. Refers to two or more sequences having a predetermined percentage. By way of example only, two or more sequences have sequence units of about 60% identity, about 65% identity, about 70% identity, about 75% identity, about 80% identity, about 85% identity, about 90 It can be "substantially identical" if it is% identity or about 95% identity. These percentages describe the "% identity" of two or more sequences. The identity of the sequence may exist over a region that is at least about 75-100 sequence units in length, over a region that is about 50 sequence units in length, or across the entire sequence unless otherwise specified. This definition also refers to the complement of the test sequence. By way of example only, two or more polypeptide sequences are identical when the amino acid residues are identical, whereas two or more polypeptide sequences have amino acid residues of about 60% identity, about 65% identity, and about 70% identity over a specific site. , "Substantially identical" when about 75% identity, about 80% identity, about 85% identity, about 90% identity, or about 95% identity. Identity can exist over a region that is at least about 75-100 amino acids in length, over a region that is about 50 amino acids in length, or, unless otherwise specified, over the entire sequence of the polypeptide sequence. Moreover, by way of example only, two or more polynucleotide sequences are homogeneous when the nucleic acid residues are identical, whereas two or more polynucleotide sequences are about 60% identity, about 65% identity over a specific site of the nucleic acid residues, "Substantially identical" when about 70% identity, about 75% identity, about 80% identity, about 85% identity, about 90% identity, or about 95% identity. Identity can exist over a region that is at least about 75-100 nucleic acids in length, over a region that is about 50 nucleic acids in length, or, unless otherwise specified, over the entire sequence of the polynucleotide sequence.
본원에 사용된 용어 "면역원성"은 치료용 약물의 투여에 대한 항체 반응을 지칭한다. 본원에 제공된 치료용 단백질, 폴리펩티드 및 펩티드에 대한 면역원성은 생물학적 유체에서 상기 치료용 단백질, 폴리펩티드 및 펩티드에 대한 항체를 검출하기 위한 정량적 및 정성적 검정을 이용하여 수득된다. 상기 검정에는 방사선면역검정 (RIA), 효소-연결된 면역흡착 검정 (ELISA), 발광 면역검정 (LIA) 및 형광 면역검정 (FIA)이 포함되나, 이에 제한되지는 않는다. 이러한 면역원성의 분석은 본원에 제공된 치료용 단백질, 폴리펩티드 및 펩티드의 투여시의 항체 반응을 대조군 치료용 단백질, 폴리펩티드 또는 펩티드의 투여시의 또는 전달 비히클 또는 전달 완충액의 투여시의 항체 반응과 비교하는 것을 포함한다.As used herein, the term "immunogenicity" refers to an antibody response to administration of a therapeutic drug. Immunogenicity to therapeutic proteins, polypeptides and peptides provided herein is obtained using quantitative and qualitative assays to detect antibodies to the therapeutic proteins, polypeptides and peptides in biological fluids. Such assays include, but are not limited to, radioimmunoassay (RIA), enzyme-linked immunosorbent assay (ELISA), luminescent immunoassay (LIA), and fluorescence immunoassay (FIA). Such immunogenicity assays compare the antibody response upon administration of the therapeutic proteins, polypeptides and peptides provided herein with the antibody responses upon administration of a control therapeutic protein, polypeptide or peptide or upon administration of a delivery vehicle or delivery buffer. Includes that.
본원에 사용된 용어 "삽입제" ("삽입성 기"로도 지칭됨)는 또 다른 분자의 분자내 공간 또는 분자 사이에 분자간 공간에 삽입되는 분자 또는 기를 지칭한다. 단지 예로서 언급하자면, 삽입제 또는 삽입성 기는 DNA 이중 나선의 적층 염기에 삽입되는 분자일 수 있다. As used herein, the term “intercalating agent” (also referred to as “insertable group”) refers to a molecule or group that is inserted into the intramolecular space of another molecule or the intermolecular space between molecules. By way of example only, the intercalating agent or intercalating group may be a molecule that is inserted into the lamination base of a DNA double helix.
본원에 사용된 용어 "표지"는 화합물에 혼입되어 용이하게 검출되는 물질을 지칭하며, 따라서 그의 물리적 분포가 검출 및/또는 모니터링될 수 있다.As used herein, the term “label” refers to a substance that is incorporated into a compound and is easily detected, so that its physical distribution can be detected and/or monitored.
본원에 사용된 용어 "연결부"는 제1 분자의 관능기와 제2 분자의 관능기 사이의 화학 반응으로부터 형성된 결합 또는 화학적 잔기를 지칭한다. 이러한 결합에는 공유 결합 및 비공유 결합이 포함되나 이에 제한되지는 않고, 이러한 화학적 잔기에는 에스테르, 카르보네이트, 이민 포스페이트 에스테르, 히드라존, 아세탈, 오르토에스테르, 펩티드 연결, 올리고뉴클레오티드 연결 및 본원의 표 1에 제시된 것들이 포함되나, 이에 제한되지는 않는다. "가수분해적으로 안정한 연결부"는, 상기 연결부가 물 중에서 실질적으로 안정하고, 사용되는 pH 값에서, 예컨대 이로 제한되는 것은 아니지만 연장된 시간 동안 생리적 조건 하에서 물과 반응하지 않는 것을 의미한다. "가수분해적으로 불안정한 또는 분해성인 연결부"는, 상기 연결부가 물 또는 수용액, 예를 들어 혈액 중에서 분해성인 것을 의미한다. "효소적으로 불안정한 또는 분해성인 연결부"는, 상기 연결부가 하나 이상의 효소에 의해 분해되는 것을 의미한다. 단지 예로서 언급하자면, 특정 PEG 및 관련된 중합체는 중합체 주쇄에서, 또는 PEG 중합체 주쇄와 본원에 제공된 단백질, 폴리펩티드 또는 펩티드의 하나 이상의 말단 관능기 사이의 연결 기에서 분해성인 연결부를 포함한다. 이러한 분해성 연결부에는 PEG 카르복실산 또는 활성화된 PEG 카르복실산과 생물학적 활성제 상의 알콜기의 반응에 의해 형성된 에스테르 연결부가 포함되나 이에 제한되지는 않으며, 이러한 에스테르기는 일반적으로 생리적 조건 하에서 가수분해되어 생물학적 활성제를 방출시킨다. 다른 가수분해적으로 분해성인 연결부에는 카르보네이트 연결부; 아민과 알데히드의 반응으로부터 생성되는 이민 연결부; 알콜과 포스페이트기의 반응에 의해 형성되는 포스페이트 에스테르 연결부; 히드라지드와 알데히드의 반응 생성물인 히드라존 연결부; 알데히드와 알콜의 반응 생성물인 아세탈 연결부; 포르메이트와 알콜의 반응 생성물인 오르토에스테르 연결부; 중합체, 예컨대 PEG의 말단에 있는 아민기를 비롯한 (이에 제한되지는 않음) 아민기, 및 펩티드의 카르복실기에 의해 형성되는 펩티드 연결부; 및 중합체의 말단에 있는 포스포르아미다이트기를 비롯한 (이에 제한되지는 않음) 포스포르아미다이트기, 및 올리고뉴클레오티드의 5' 히드록실기에 의해 형성되는 올리고뉴클레오티드 연결부가 포함되나, 이에 제한되지는 않는다.As used herein, the term “linkage” refers to a bond or chemical moiety formed from a chemical reaction between a functional group of a first molecule and a functional group of a second molecule. Such bonds include, but are not limited to, covalent bonds and non-covalent bonds, and such chemical moieties include esters, carbonates, imine phosphate esters, hydrazones, acetals, orthoesters, peptide linkages, oligonucleotide linkages, and Table 1 herein. Including, but not limited to, those presented in "Hydrolytically stable connection" means that the connection is substantially stable in water and does not react with water under physiological conditions for an extended period of time, such as but not limited to, at the pH value used. "Hydrolytically unstable or degradable connection" means that the connection is degradable in water or an aqueous solution, for example blood. "Enzymatically labile or degradable linkage" means that the linkage is degraded by one or more enzymes. By way of example only, certain PEGs and related polymers contain degradable linkages in the polymer backbone, or at the linking groups between the PEG polymer backbone and one or more terminal functional groups of the proteins, polypeptides or peptides provided herein. Such degradable linkages include, but are not limited to, ester linkages formed by the reaction of PEG carboxylic acid or activated PEG carboxylic acid with an alcohol group on a biologically active agent, and such ester groups are generally hydrolyzed under physiological conditions to obtain a biologically active agent. Release. Other hydrolytically degradable connections include carbonate connections; An imine linkage generated from the reaction of an amine and an aldehyde; A phosphate ester linkage formed by the reaction of an alcohol and a phosphate group; Hydrazone connection, which is a reaction product of hydrazide and aldehyde; An acetal connection part, which is a reaction product of an aldehyde and an alcohol; An orthoester linkage that is a reaction product of formate and alcohol; A peptide linkage formed by an amine group including, but not limited to, an amine group at the end of a polymer such as PEG, and a carboxyl group of the peptide; And a phosphoramidite group, including, but not limited to, a phosphoramidite group at the end of the polymer, and an oligonucleotide linkage formed by the 5′ hydroxyl group of the oligonucleotide, but is not limited thereto. Does not.
본원에 사용된 용어 "배지" 또는 "배지들"은 세포, 및/또는 이러한 세포에 의해 발현 및/또는 분비된 생성물을 성장시키고 수확하는데 사용되는 임의의 배양 배지를 지칭한다. 이러한 "배지" 또는 "배지들"에는 임의의 숙주 세포, 단지 예로서 언급하자면 박테리아 숙주 세포, 효모 숙주 세포, 곤충 숙주 세포, 식물 숙주 세포, 진핵 숙주 세포, 포유동물 숙주 세포, CHO 세포, 원핵 숙주 세포, 에스케리키아 콜라이, 또는 슈도모나스 숙주 세포, 및 세포 내용물을 보충하거나 함유할 수 있는 용액, 고체, 반고체 또는 강직성 지지체가 포함되나, 이에 제한되지는 않는다. 이러한 "배지" 또는 "배지들"에는 증식 단계 전 또는 후의 배지를 비롯한 (이에 제한되지는 않음) 숙주 세포가 성장하여 폴리펩티드가 분비된 배지 또는 배지들이 포함되나, 이에 제한되지는 않는다. 이러한 "배지" 또는 "배지들"에는 또한 숙주 세포 용해물을 함유하는 완충액 또는 시약, 예를 들어 세포내에서 생산된 폴리펩티드가 포함되나 이에 제한되지는 않으며, 숙주 세포는 용해 또는 파괴되어 폴리펩티드를 방출시킨다.As used herein, the term “medium” or “medias” refers to any culture medium used to grow and harvest cells and/or products expressed and/or secreted by such cells. Such "medium" or "medium" includes any host cell, by way of example only a bacterial host cell, a yeast host cell, an insect host cell, a plant host cell, a eukaryotic host cell, a mammalian host cell, a CHO cell, a prokaryotic host. Cells, Escherichia coli, or Pseudomonas host cells, and solutions, solids, semi-solids, or rigid supports that may supplement or contain cell contents. Such “medium” or “medium” includes, but is not limited to, media or media from which the polypeptide is secreted by the growth of host cells, including, but not limited to, media before or after the proliferation step. Such "medium" or "medium" also includes, but is not limited to, a buffer or reagent containing the host cell lysate, for example, a polypeptide produced intracellularly, wherein the host cell is lysed or disrupted to release the polypeptide. Let it.
본원에 사용된 용어 "금속 킬레이트화제"는 금속 이온의 착체를 형성하는 분자를 지칭한다. 예로서, 이러한 분자는 중심 금속 이온과 2개 이상 배위 결합을 형성하고, 임의로 고리 구조체를 형성한다. As used herein, the term “metal chelating agent” refers to a molecule that forms a complex of metal ions. By way of example, these molecules form two or more coordination bonds with the central metal ion, and optionally form a ring structure.
본원에 사용된 용어 "금속-함유 잔기"는 금속 이온, 원자 또는 입자를 함유하는 기를 지칭한다. 이러한 잔기에는 시스플라틴, 킬레이트화된 금속 이온 (예컨대, 니켈, 철 및 백금) 및 금속 나노입자 (예컨대, 니켈, 철 및 백금)가 포함되나, 이에 제한되지는 않는다. As used herein, the term “metal-containing moiety” refers to a group containing a metal ion, atom or particle. Such moieties include, but are not limited to, cisplatin, chelated metal ions (eg nickel, iron and platinum) and metal nanoparticles (eg nickel, iron and platinum).
본원에 사용된 용어 "중원자가 혼입된 잔기"는 통상적으로 탄소보다 무거운 원자의 이온이 혼입된 기를 지칭한다. 이러한 이온 또는 원자에는 규소, 텅스텐, 금, 납 및 우라늄이 포함되나, 이에 제한되지는 않는다. As used herein, the term "moiety incorporating a heavy atom" refers to a group incorporating an ion of an atom heavier than carbon. Such ions or atoms include, but are not limited to, silicon, tungsten, gold, lead, and uranium.
본원에 사용된 용어 "변형된"은 아미노산 또는 아미노산 잔기에 대한 변화의 존재를 지칭하고, 이러한 변화 또는 변형은 화학적 또는 생화학적 과정에 의해 수득된다.As used herein, the term “modified” refers to the presence of a change to an amino acid or amino acid residue, and such change or modification is obtained by chemical or biochemical processes.
본원에 사용된 용어 "조절된 혈청 반감기"는 그의 변형되지 않은 형태에 비해서 변형된 생물학적 활성 분자의 순환 반감기에서의 양성 또는 음성 변화를 지칭한다. 예로서, 변형된 생물학적 활성 분자에는 본원에 제공된 화합물이 포함되나, 이에 제한되지는 않는다. 예로서, 혈청 반감기는 생물학적 활성 분자 또는 변형된 생물학적 활성 분자의 투여 후에 다양한 시점에서 혈액 샘플을 취하고, 각각의 샘플에서 그 분자의 농도를 결정함으로써 측정된다. 혈청 농도와 시간의 상관 관계를 이용하여 혈청 반감기를 계산할 수 있다. 예로서, 조절된 혈청 반감기는 혈청 반감기의 상승일 수 있으며, 이는 개선된 투약 요법을 가능하게 하거나 독성 효과를 피할 수 있게 한다. 이러한 혈청의 증가는 적어도 약 2배, 적어도 약 3배, 적어도 약 5배, 또는 적어도 약 10배일 수 있다.As used herein, the term "regulated serum half-life" refers to a positive or negative change in the circulating half-life of a modified biologically active molecule compared to its unmodified form. By way of example, modified biologically active molecules include, but are not limited to, compounds provided herein. As an example, serum half-life is measured by taking a blood sample at various time points after administration of a biologically active molecule or a modified biologically active molecule, and determining the concentration of that molecule in each sample. The correlation between serum concentration and time can be used to calculate the serum half-life. As an example, a controlled serum half-life can be an increase in serum half-life, which enables improved dosing regimens or avoids toxic effects. This increase in serum may be at least about 2 times, at least about 3 times, at least about 5 times, or at least about 10 times.
본원에 사용된 용어 "나노입자"는 약 500 nm 내지 약 1 nm의 입자 크기를 갖는 입자를 지칭한다. As used herein, the term “nanoparticle” refers to a particle having a particle size of about 500 nm to about 1 nm.
본원에 사용된 용어 "근사-화학량론적"은 화학 반응에 참여하는 화합물의 몰비가 약 0.75 내지 약 1.5인 것을 지칭한다. As used herein, the term “approximate-stoichiometric” refers to a molar ratio of compounds participating in a chemical reaction between about 0.75 and about 1.5.
본원에 사용된 용어 "비-진핵세포"는 비-진핵 유기체를 지칭한다. 예로서, 비-진핵 유기체는 유박테리아 계통발생학적 영역에 속하며, 여기에는 에스케리키아 콜라이, 써무스 테르모필루스(Thermus thermophilus), 또는 바실러스 스테아로써모필루스(Bacillus stearothermophilus), 슈도모나스 플루오레센스(Pseudomonas fluorescens), 슈도모나스 아에루기노사(Pseudomonas aeruginosa), 슈도모나스 푸티다(Pseudomonas putida)가 포함되나 이에 제한되지는 않고, 또는 고세균 계통발생학적 영역에 속하며, 여기에는 메타노코쿠스 잔나쉬이(Methanococcus jannaschii), 메타노박테리움 써모아우토트로피쿰(Methanobacterium thermoautotrophicum), 아르카에오글로부스 풀기두스(Archaeoglobus fulgidus), 피로코쿠스 푸리오수스(Pyrococcus furiosus), 피로코쿠스 호르코쉬이(Pyrococcus horikoshii), 아에로피룸 페르닉스(Aeuropyrum pernix), 또는 할로박테리움, 예컨대 할로페락스 볼카니이(Haloferax volcanii) 및 할로박테리움 종 NRC-1이 포함되나, 이에 제한되지는 않는다.As used herein, the term “non-eukaryotic cell” refers to a non-eukaryotic organism. For example, non-eukaryotic organisms belong to the eubacterial phylogenetic domain, including Escherichia coli, Thermus thermophilus (Thermus thermophilus ), or Bacillus stearomophilus stearothermophilus ), Pseudomonas fluorescens , Pseudomonas aeruginosa ( Pseudomonas) aeruginosa ), Pseudomonas putida ) is included, but not limited thereto, or belongs to the archaeal phylogenetic domain, including metanococcus jannaschii , metanobacterium thermoautotrophicum (Methanobacterium thermoautotrophicum), arcaeoglobus Archaeoglobus fulgidus ), Pyrococcus furiosus ), Pyrococcus horikoshii , Aeuropyrum pernix ), or halobacterium, such as Haloferax volcanii (Haloferax volcanii ) and Halobacterium species NRC-1 are included, but are not limited thereto.
본원에 사용된 용어 "핵산"은 단일- 또는 이중-가닥 형태의 데옥시리보뉴클레오티드, 데옥시리보뉴클레오시드, 리보뉴클레오시드 또는 리보뉴클레오티드 및 이들의 중합체를 지칭한다. 단지 예로서 언급하자면, 이러한 핵산 및 핵산 중합체에는 (i) 참고 핵산과 유사한 특성을 갖는 천연 뉴클레오티드의 유사체; (ii) 올리고뉴클레오티드 유사체, 예컨대 이로 제한되지는 않지만 PNA (펩티도핵산), 안티센스 기술에 사용되는 DNA의 유사체 (포스포로티오에이트, 포스포로아미데이트 등); (iii) 이들의 보존적으로 변형된 변이체 (예컨대 이들로 제한되지는 않지만, 축퇴성 코돈 치환) 및 상보성 서열 및 명백히 나타낸 서열이 포함되나, 이에 제한되지는 않는다. 예로서, 축퇴성 코돈 치환은 하나 이상의 선택된 (또는 모든) 코돈의 제3 위치가 혼합된 염기 및/또는 데옥시이노신 잔기로 치환된 서열을 생성함으로써 달성될 수 있다.The term “nucleic acid” as used herein refers to deoxyribonucleotides, deoxyribonucleosides, ribonucleosides or ribonucleotides and polymers thereof in single- or double-stranded form. By way of example only, such nucleic acids and nucleic acid polymers include (i) analogs of natural nucleotides having properties similar to those of a reference nucleic acid; (ii) oligonucleotide analogs such as, but not limited to, PNA (peptidonucleic acid), analogs of DNA used in antisense technology (phosphorothioate, phosphoroamidate, etc.); (iii) Conservatively modified variants thereof (such as, but not limited to, degenerate codon substitutions) and complementary sequences and clearly indicated sequences. By way of example, degenerate codon substitution can be achieved by generating a sequence in which the third position of one or more selected (or all) codons is substituted with a mixed base and/or deoxyinosine residue.
본원에 사용된 용어 "산화제"는 산화될 화합물로부터 전자를 제거할 수 있는 화합물 또는 물질을 지칭한다. 예로서, 산화제에는 산화된 글루타티온, 시스틴, 시스타민, 산화된 디티오트레이톨, 산화된 에리트레이톨 및 산소가 포함되나, 이에 제한되지는 않는다. 다양한 산화제는 본원에 기재된 방법 및 조성물에 사용하는데 적합하다. As used herein, the term “oxidizing agent” refers to a compound or substance capable of removing electrons from the compound to be oxidized. By way of example, oxidizing agents include, but are not limited to, oxidized glutathione, cystine, cystamine, oxidized dithiothreitol, oxidized erythritol, and oxygen. A variety of oxidizing agents are suitable for use in the methods and compositions described herein.
본원에 사용된 용어 "광친화성 표지"는 빛에 노출시 상기 표지가 친화도를 갖는 분자와의 연결을 형성하는 기를 갖는 표지를 지칭한다. 단지 예로서 언급하자면, 이러한 연결은 공유 또는 비공유일 수 있다. As used herein, the term “photoaffinity label” refers to a label having a group that upon exposure to light, the label forms a linkage with an affinity molecule. By way of example only, these connections can be shared or non-shared.
본원에 사용된 용어 "광케이징 잔기"는 특정 파장에서 발광시 이온 또는 다른 분자와 공유 또는 비공유적으로 결합하는 기를 지칭한다. As used herein, the term “photocausing moiety” refers to a group that binds covalently or non-covalently with an ion or other molecule upon light emission at a specific wavelength.
본원에 사용된 용어 "광분해성 기"는 빛에 노출시 파괴되는 기를 지칭한다. As used herein, the term “photodegradable group” refers to a group that is destroyed upon exposure to light.
본원에 사용된 용어 "광가교제"는 빛에 노출시 반응성이고, 2개 이상의 단량체 또는 중합체 분자와 공유 또는 비공유 결합을 형성하는 2개 이상의 관능기를 포함하는 화합물을 지칭한다. As used herein, the term “photocrosslinking agent” refers to a compound that is reactive upon exposure to light and comprises two or more functional groups that form covalent or non-covalent bonds with two or more monomeric or polymeric molecules.
본원에 사용된 용어 "광이성질체화 잔기"는 빛에 의한 발광시 한 이성질체 형태에서 또 다른 형태로 변화하는 기를 지칭한다. The term “photoisomerizing moiety” as used herein refers to a group that changes from one isomeric form to another upon luminescence by light.
본원에 사용된 용어 "폴리알킬렌 글리콜"은 선형 또는 분지형 중합체 폴리에테르 폴리올을 지칭한다. 이러한 폴리알킬렌 글리콜에는 폴리에틸렌 글리콜, 폴리프로필렌 글리콜, 폴리부틸렌 글리콜 및 이들의 유도체가 포함되나, 이에 제한되지는 않는다.As used herein, the term “polyalkylene glycol” refers to a linear or branched polymeric polyether polyol. Such polyalkylene glycols include, but are not limited to, polyethylene glycol, polypropylene glycol, polybutylene glycol, and derivatives thereof.
본원에 사용된 용어 "중합체"는 반복된 서브유닛으로 이루어진 분자를 지칭한다. 이러한 분자에는 단백질, 폴리펩티드, 펩티드, 폴리뉴클레오티드 또는 폴리사카라이드 또는 폴리알킬렌 글리콜이 포함되나, 이에 제한되지는 않는다. As used herein, the term “polymer” refers to a molecule made up of repeated subunits. Such molecules include, but are not limited to, proteins, polypeptides, peptides, polynucleotides or polysaccharides or polyalkylene glycols.
용어 "폴리펩티드", "펩티드" 및 "단백질"은 아미노산 잔기의 중합체를 지칭하기 위해 본원에서 교환 가능하게 사용된다. 즉, 폴리펩티드에 관한 기재는 펩티드의 기재 및 단백질의 기재에 동일하게 적용되고, 그 반대로도 적용된다. 아미노산 잔기에는 천연 및 비천연 아미노산으로부터 유래된 잔기가 포함된다. 상기 용어는 자연 발생 아미노산 중합체 뿐만 아니라, 1개 이상의 아미노산 잔기가 본원에 제공된 화합물인 아미노산 중합체에 적용된다. 추가로, 이러한 "폴리펩티드", "펩티드" 및 "단백질"은 전장 단백질을 비롯한 임의의 길이의 아미노산 쇄를 포함하며, 여기서 아미노산 잔기는 공유 펩티드 결합 또는 다른 연결에 의해 연결된다. The terms “polypeptide”, “peptide” and “protein” are used interchangeably herein to refer to a polymer of amino acid residues. That is, the description of the polypeptide applies equally to the description of the peptide and the description of the protein, and vice versa. Amino acid residues include those derived from natural and unnatural amino acids. The term applies to naturally occurring amino acid polymers as well as amino acid polymers in which one or more amino acid residues are compounds provided herein. Additionally, such “polypeptide”, “peptide” and “protein” include amino acid chains of any length, including full-length proteins, wherein the amino acid residues are linked by covalent peptide bonds or other linkages.
용어 "번역후 변형된"은 아미노산이 번역에 의해 폴리펩티드 쇄에 혼입된 후에 일어나는 아미노산의 임의의 변형을 지칭한다. 이러한 변형에는 번역후 생체내 변형 및 번역후 시험관내 변형이 포함되나, 이에 제한되지는 않는다. The term “post-translational modified” refers to any modification of an amino acid that occurs after the amino acid has been incorporated into a polypeptide chain by translation. Such modifications include, but are not limited to, post-translational in vivo modifications and post-translational in vitro modifications.
본원에 사용된 용어 "보호된"은 특정 반응 조건 하에서 화학 반응성 관능기의 반응을 방지하는 "보호기" 또는 잔기의 존재를 지칭한다. 보호기는 보호될 화학 반응성 기의 유형에 따라 달라질 것이다. 단지 예로서 언급하자면, (i) 화학 반응성 기가 아민 또는 히드라지드인 경우, 보호기는 tert-부틸옥시카르보닐 (t-Boc) 및 9-플루오레닐메톡시카르보닐 (Fmoc)로부터 선택될 수 있고; (ii) 화학 반응성 기가 티올인 경우, 보호기는 오르토피리딜디술피드일 수 있고; (iii) 화학 반응성 기가 카르복실산, 예컨대 부탄산 또는 프로피온산, 또는 히드록실기인 경우, 보호기는 벤질 또는 알킬기, 예컨대 메틸, 에틸 또는 tert-부틸일 수 있다. 단지 예로서 언급하자면, 차단기/보호기는 또한 아세트아미드, 알릴옥시카르보닐, 알릴 에테르, 벤질, 벤질아민, 벤질리덴아민, 벤질 카르바메이트, 벤질 에스테르, 메틸 에스테르, t-부틸 에스테르, S-t-부틸 에스테르, 2-알킬-1,3-옥사졸린, 디메틸 아세탈, 1,3-디옥산, 1,3-디티안, N,N-디메틸히드라존, 프탈이미드, 트리틸, 파라메톡시벤질 에테르, 카르보벤질옥시, p-톨루엔술폰아미드, 트리플루오로아세트아미드, 트리페닐메틸아민, t-부틸 에테르, 벤질 에테르, t-부틸디메틸실릴 에테르, t-부틸디페닐실릴 에테르, 2-(트리메틸실릴) 에톡시카르보닐, 아세트산 에스테르, 피발산 에스테르, 벤조산 에스테르, 아세토니드, 벤질리덴 아세탈, 및 광분해성 기, 예컨대 Nvoc 및 MeNvoc로부터 선택된다. As used herein, the term “protected” refers to the presence of a “protecting group” or moiety that prevents reaction of a chemically reactive functional group under certain reaction conditions. The protecting group will depend on the type of chemically reactive group to be protected. By way of example only, (i) when the chemically reactive group is an amine or hydrazide, the protecting group may be selected from tert-butyloxycarbonyl (t-Boc) and 9-fluorenylmethoxycarbonyl (Fmoc); (ii) when the chemically reactive group is thiol, the protecting group may be orthopyridyldisulfide; (iii) When the chemically reactive group is a carboxylic acid such as butanoic acid or propionic acid, or a hydroxyl group, the protecting group may be benzyl or an alkyl group such as methyl, ethyl or tert-butyl. By way of example only, the blocker/protecting group is also acetamide, allyloxycarbonyl, allyl ether, benzyl, benzylamine, benzylideneamine, benzyl carbamate, benzyl ester, methyl ester, t-butyl ester, St-butyl Ester, 2-alkyl-1,3-oxazoline, dimethyl acetal, 1,3-dioxane, 1,3-ditian, N,N-dimethylhydrazone, phthalimide, trityl, paramethoxybenzyl ether , Carbobenzyloxy, p-toluenesulfonamide, trifluoroacetamide, triphenylmethylamine, t-butyl ether, benzyl ether, t-butyldimethylsilyl ether, t-butyldiphenylsilyl ether, 2-(trimethyl Silyl) ethoxycarbonyl, acetic acid ester, pivalic acid ester, benzoic acid ester, acetonide, benzylidene acetal, and photodegradable groups such as Nvoc and MeNvoc.
본원에 사용된 용어 "방사성 잔기"는 핵이 핵 방사선, 예컨대 알파 또는 베타 입자, 또는 감마 방사선을 자발적으로 방출시키는 기를 지칭한다. As used herein, the term “radioactive moiety” refers to a group in which the nucleus spontaneously emits nuclear radiation, such as alpha or beta particles, or gamma radiation.
본원에 사용된 용어 "반응성 화합물"은 적절한 조건 하에서 또 다른 원자, 분자 또는 화합물에 대해 반응성인 화합물을 지칭한다. As used herein, the term “reactive compound” refers to a compound that is reactive toward another atom, molecule or compound under appropriate conditions.
용어 "재조합 숙주 세포" ("숙주 세포"로도 지칭됨)는 외인성 폴리뉴클레오티드를 포함하는 세포를 지칭하며, 여기서 외인성 폴리뉴클레오티드를 세포에 삽입하는데 이용되는 방법에는 직접 흡수, 변환, f-교배, 또는 재조합 숙주 세포의 생성을 위해 당업계에 공지된 다른 방법이 포함되나, 이에 제한되지는 않는다. 단지 예로서 언급하자면, 이러한 외인성 폴리뉴클레오티드는 혼입되지 않은 벡터, 예컨대 이들로 제한되는 것은 아니지만 플라스미드일 수 있거나, 숙주 게놈에 혼입될 수 있다. The term “recombinant host cell” (also referred to as “host cell”) refers to a cell comprising an exogenous polynucleotide, wherein the method used to insert the exogenous polynucleotide into the cell includes direct uptake, transformation, f-crossing, or Other methods known in the art for the generation of recombinant host cells include, but are not limited to. By way of example only, such exogenous polynucleotides may be unincorporated vectors, such as, but not limited to, plasmids, or may be incorporated into the host genome.
본원에 사용된 용어 "산화환원-활성제"는 또 다른 분자를 산화 또는 환원시켜 산화환원 활성제가 환원 또는 산화되는 분자를 지칭한다. 산화환원 활성제의 예로는 페로센, 퀴논, Ru2 +/3+ 착체, Co2 +/3+ 착체 및 Os2 +/3+ 착체가 포함되나, 이에 제한되지는 않는다. The term "redox-active agent" as used herein refers to a molecule by which the redox active agent is reduced or oxidized by oxidizing or reducing another molecule. Examples of redox activators include, but are not limited to, ferrocene, quinone, Ru 2 +/3+ complex, Co 2 +/3+ complex, and Os 2 +/3+ complex.
본원에 사용된 용어 "환원제"는 환원시키고자 하는 화합물에 전자를 부가할 수 있는 화합물 또는 물질을 지칭한다. 예로서, 환원제에는 디티오트레이톨 (DTT), 2-메르캅토에탄올, 디티오에리트리톨, 시스테인, 시스테아민 (2-아미노에탄티올) 및 환원된 글루타티온이 포함되나, 이에 제한되지는 않는다. 단지 예로서 언급하자면, 이러한 환원제는 환원된 상태에서 술프히드릴기를 유지하고 분자내 또는 분자간 디술피드 결합을 환원시키는데 사용될 수 있다. As used herein, the term “reducing agent” refers to a compound or substance capable of adding electrons to the compound to be reduced. By way of example, reducing agents include, but are not limited to, dithiothreitol (DTT), 2-mercaptoethanol, dithioerythritol, cysteine, cysteamine (2-aminoethanethiol) and reduced glutathione. By way of example only, such reducing agents can be used to maintain sulfhydryl groups in the reduced state and to reduce intramolecular or intermolecular disulfide bonds.
본원에 사용된 용어 "리폴딩"은 부적절하게 폴딩되거나 언폴딩된 상태를 원래의 또는 적절하게 폴딩된 형태로 변형시키는 임의의 과정, 반응 또는 방법을 기재한다. 단지 예로서 언급하자면, 디술피드 결합과 관련하여 리폴딩은 단백질 또는 폴리펩티드를 함유하는 디술피드 결합을 부적절하게 폴딩되거나 언폴딩된 상태에서 원래의 또는 적절하게 폴딩된 형태로 변형시키는 것이다. 단백질 또는 폴리펩티드를 함유하는 이러한 디술피드 결합에는 본원에 제공된 화합물이 내부에 혼입된 단백질 또는 폴리펩티드가 포함된다. 리폴딩은 종종 단백질의 가용화 및 언폴딩을 위해 단백질 용액에 사전에 첨가된 카오트로픽제, 예컨대 우레아 또는 구아니디늄 히드로클로라이드의 제거에 의해 개시된다. As used herein, the term “refolding” describes any process, reaction or method that transforms an improperly folded or unfolded state into its original or properly folded form. By way of example only, with respect to disulfide bonds, refolding is to modify the disulfide bond containing a protein or polypeptide from an improperly folded or unfolded state to its original or properly folded form. Such disulfide bonds containing proteins or polypeptides include proteins or polypeptides into which the compounds provided herein have been incorporated. Refolding is often initiated by the removal of chaotropic agents, such as urea or guanidinium hydrochloride, previously added to the protein solution for solubilization and unfolding of the protein.
본원에 사용된 용어 "수지"는 고분자량, 불용성 중합체 비드를 지칭한다. 단지 예로서 언급하자면, 이러한 비드는 고체상 펩티드 합성을 위한 지지체로서 또는 정제 이전에 분자의 부착을 위한 부위로서 사용될 수 있다.The term “resin” as used herein refers to high molecular weight, insoluble polymer beads. By way of example only, these beads can be used as a support for solid-phase peptide synthesis or as a site for attachment of molecules prior to purification.
본원에 사용된 용어 "사카라이드"는 일련의 탄수화물, 예컨대 이들로 제한되는 것은 아니지만 당, 모노사카라이드, 올리고사카라이드 및 폴리사카라이드를 지칭한다. As used herein, the term “saccharide” refers to a series of carbohydrates, such as, but not limited to, sugars, monosaccharides, oligosaccharides and polysaccharides.
본원에 사용된 용어 "스핀 표지"는 전자 스핀 공명 분광분석법에 의해 검출될 수 있으며 또 다른 분자에 부착될 수 있는, 쌍을 이루지 않은 전자 스핀을 나타내는 원자 또는 원자의 기 (즉, 안정한 상자성 기)를 함유하는 분자를 지칭한다. 이러한 스핀-표지 분자에는 니트릴 라디칼 및 니트록시드가 포함되나 이에 제한되지는 않고, 단일 스핀 표지 또는 이중 스핀 표지일 수 있다. As used herein, the term “spin label” is an atom or group of atoms that exhibits unpaired electron spin (ie, a stable paramagnetic group) that can be detected by electron spin resonance spectroscopy and attached to another molecule. Refers to a molecule containing. Such spin-labeled molecules include, but are not limited to, nitrile radicals and nitroxides, and may be single spin labels or double spin labels.
본원에 사용된 용어 "화학량론적"은 화학 반응에 참여하는 화합물의 몰비가 약 0.9 내지 약 1.1인 것을 지칭한다. As used herein, the term “stoichiometric” refers to the molar ratio of the compounds participating in the chemical reaction being about 0.9 to about 1.1.
본원에 사용된 용어 "실질적으로 정제된"은 정제 이전에 관심 성분을 정상적으로 동반하거나 그와 상호작용하는 다른 성분들이 실질적으로 또는 본질적으로 없는 관심 성분을 지칭한다. 단지 예로서 언급하자면, 관심 성분의 제조물이 약 30% 미만, 약 25% 미만, 약 20% 미만, 약 15% 미만, 약 10% 미만, 약 5% 미만, 약 4% 미만, 약 3% 미만, 약 2% 미만 또는 약 1% 미만 (건조 중량)의 오염 성분을 함유하는 경우, 관심 성분은 "실질적으로 정제된" 것일 수 있다. 따라서, "실질적으로 정제된" 관심 성분은 약 70%, 약 75%, 약 80%, 약 85%, 약 90%, 약 95%, 약 96%, 약 97%, 약 98%, 약 99% 또는 그 초과의 순도 수준을 가질 수 있다. 단지 예로서 언급하자면, 본원에 제공된 화합물을 함유하는 단백질, 폴리펩티드 또는 펩티드는 원래의 세포 또는 숙주 세포로부터 정제된다. 단지 예로서 언급하자면, 본원에 제공된 화합물을 함유하는 단백질, 폴리펩티드 또는 펩티드의 제조물이 약 30% 미만, 약 25% 미만, 약 20% 미만, 약 15% 미만, 약 10% 미만, 약 5% 미만, 약 4% 미만, 약 3% 미만, 약 2% 미만 또는 약 1% 미만 (건조 중량)의 오염 물질을 함유하는 경우, 상기 제조물은 "실질적으로 정제된" 것이다. 단지 예로서 언급하자면, 본원에 제공된 화합물을 함유하는 단백질, 폴리펩티드 또는 펩티드가 숙주 세포에 의해 재조합적으로 생산될 때, 본원에 제공된 화합물을 함유하는 단백질, 폴리펩티드 또는 펩티드는 세포의 건조 중량의 약 30%, 약 25%, 약 20%, 약 15%, 약 10%, 약 5%, 약 4%, 약 3%, 약 2%, 또는 약 1% 또는 그 미만으로 존재한다. 단지 예로서 언급하자면, 본원에 제공된 화합물을 함유하는 단백질, 폴리펩티드 또는 펩티드가 숙주 세포에 의해 재조합적으로 생산될 때, 본원에 제공된 화합물을 함유하는 단백질, 폴리펩티드 또는 펩티드는 배양 배지에 약 5 g/L, 약 4 g/L, 약 3 g/L, 약 2 g/L, 약 1 g/L, 약 750 mg/L, 약 500 mg/L, 약 250 mg/L, 약 100 mg/L, 약 50 mg/L, 약 10 mg/L 또는 약 1 mg/L 또는 그 미만으로 존재한다. 단지 예로서 언급하자면, 본원에 제공된 화합물을 함유하는 "실질적으로 정제된" 단백질, 폴리펩티드 또는 펩티드는 SDS/PAGE 분석, RP-HPLC, SEC 및 모세관 전기영동을 비롯한 (이에 제한되지는 않음) 적절한 방법에 의해 측정시 약 30%, 약 35%, 약 40%, 약 45%, 약 50%, 약 55%, 약 60%, 약 65%, 약 70%, 약 75%, 약 80%, 약 85%, 약 90, 약 95%, 약 99% 또는 그 초과의 순도 수준을 갖는다. As used herein, the term “substantially purified” refers to an ingredient of interest that is substantially or essentially free of other ingredients that normally accompany or interact with the ingredient of interest prior to purification. By way of example only, the formulation of the component of interest is less than about 30%, less than about 25%, less than about 20%, less than about 15%, less than about 10%, less than about 5%, less than about 4%, less than about 3%. , If it contains less than about 2% or less than about 1% (dry weight) of the contaminating component, the component of interest may be “substantially purified”. Thus, the "substantially purified" component of interest is about 70%, about 75%, about 80%, about 85%, about 90%, about 95%, about 96%, about 97%, about 98%, about 99%. Or it may have a purity level greater than that. By way of example only, a protein, polypeptide or peptide containing a compound provided herein is purified from the original cell or host cell. By way of example only, preparations of proteins, polypeptides or peptides containing the compounds provided herein are less than about 30%, less than about 25%, less than about 20%, less than about 15%, less than about 10%, less than about 5%. , Less than about 4%, less than about 3%, less than about 2% or less than about 1% (by dry weight) of contaminants, the preparation is "substantially purified". By way of example only, when a protein, polypeptide or peptide containing a compound provided herein is recombinantly produced by a host cell, the protein, polypeptide or peptide containing the compound provided herein will be about 30% of the dry weight of the cell. %, about 25%, about 20%, about 15%, about 10%, about 5%, about 4%, about 3%, about 2%, or about 1% or less. By way of example only, when a protein, polypeptide or peptide containing a compound provided herein is recombinantly produced by a host cell, the protein, polypeptide or peptide containing the compound provided herein is about 5 g/g/peptide in the culture medium. L, about 4 g/L, about 3 g/L, about 2 g/L, about 1 g/L, about 750 mg/L, about 500 mg/L, about 250 mg/L, about 100 mg/L, About 50 mg/L, about 10 mg/L or about 1 mg/L or less. By way of example only, "substantially purified" proteins, polypeptides or peptides containing the compounds provided herein are suitable methods including, but not limited to, SDS/PAGE analysis, RP-HPLC, SEC and capillary electrophoresis. As measured by about 30%, about 35%, about 40%, about 45%, about 50%, about 55%, about 60%, about 65%, about 70%, about 75%, about 80%, about 85 %, about 90, about 95%, about 99% or more.
본원에 사용된 용어 "독성 잔기"는 해를 입히거나 사망을 초래할 수 있는 화합물을 지칭한다. As used herein, the term “toxic moiety” refers to a compound that can cause harm or death.
본원에 사용된 용어 "치료하다", "치료하는" 또는 "치료"는 질환 또는 상태 증상을 완화, 감소 또는 개선시키거나, 추가의 증상을 예방하거나, 증상의 기본 대사적 원인을 개선 또는 예방하거나, 질환 또는 상태를 억제하거나, 예를 들어, 질환 또는 상태의 진행을 저지하거나, 질환 또는 상태를 경감시키거나, 질환 또는 상태를 퇴행시키거나, 질환 또는 상태에 의해 초래된 상태를 경감시키거나, 질환 또는 상태의 증상을 중지시키는 것을 포함한다. 용어 "치료하다", "치료하는" 또는 "치료"에는 예방적 및/또는 치유적 치료가 포함되나, 이에 제한되지는 않는다. The terms “treat”, “treating” or “treatment” as used herein alleviate, reduce or ameliorate symptoms of a disease or condition, prevent further symptoms, or improve or prevent underlying metabolic causes of symptoms, or , Inhibiting a disease or condition, for example, arresting the progression of the disease or condition, alleviating the disease or condition, regressing the disease or condition, alleviating the condition caused by the disease or condition, or It includes stopping the symptoms of the disease or condition. The terms “treat”, “treating” or “treatment” include, but are not limited to, prophylactic and/or curative treatment.
본원에 사용된 용어 "수용성 중합체"는 수성 용매에 가용성인 임의의 중합체를 지칭한다. 이러한 수용성 중합체에는 폴리에틸렌 글리콜, 폴리에틸렌 글리콜 프로피온알데히드, 모노 C1-C10알콕시 또는 그의 아릴옥시 유도체, 모노메톡시-폴리에틸렌 글리콜, 폴리비닐 피롤리돈, 폴리비닐 알콜, 폴리아미노산, 디비닐에테르 말레산 무수물, N-(2-히드록시프로필)-메타크릴아미드, 덱스트란, 덱스트란 유도체, 예컨대 덱스트란 술페이트, 폴리프로필렌 글리콜, 폴리프로필렌 옥시드/에틸렌 옥시드 공중합체, 폴리옥시에틸화 폴리올, 헤파린, 헤파린 단편, 폴리사카라이드, 올리고사카라이드, 글리칸, 셀룰로스 및 셀룰로스 유도체, 예컨대 메틸셀룰로스 및 카르복시메틸 셀룰로스 등, 혈청 알부민, 전분 및 전분 유도체, 폴리펩티드, 폴리알킬렌 글리콜 및 그의 유도체, 폴리알킬렌 글리콜 및 그의 유도체의 공중합체, 폴리비닐 에틸 에테르 및 알파-베타-폴리[(2-히드록시에틸)-DL-아스파르트아미드 등 또는 이들의 혼합물이 포함되나, 이에 제한되지는 않는다. 단지 예로서 언급하자면, 이러한 수용성 중합체와 본원에 제공된 화합물을 함유하는 단백질, 폴리펩티드 또는 펩티드의 커플링에 의해, 증가된 수용성, 증가 또는 조절된 혈청 반감기, 비변형 형태에 비해 증가 또는 조절된 치료 반감기, 증가된 생체이용률, 조절된 생물학적 활성, 연장된 순환 시간, 조절된 면역원성, 조절된 물리적 회합 특성, 예컨대 응집 및 다량체 형성, 변경된 수용체 결합, 하나 이상의 결합 파트너와의 변경된 결합, 및 변경된 수용체 이량체화 또는 다량체화 등을 비롯한 (이에 제한되지는 않음) 변화가 일어난다. As used herein, the term “water-soluble polymer” refers to any polymer that is soluble in an aqueous solvent. Such water-soluble polymers include polyethylene glycol, polyethylene glycol propionaldehyde, mono C 1 -C 10 alkoxy or aryloxy derivatives thereof, monomethoxy-polyethylene glycol, polyvinyl pyrrolidone, polyvinyl alcohol, polyamino acids, divinyl ether maleic acid. Anhydride, N-(2-hydroxypropyl)-methacrylamide, dextran, dextran derivatives such as dextran sulfate, polypropylene glycol, polypropylene oxide/ethylene oxide copolymer, polyoxyethylated polyol, Heparin, heparin fragments, polysaccharides, oligosaccharides, glycans, cellulose and cellulose derivatives such as methylcellulose and carboxymethyl cellulose, etc., serum albumin, starch and starch derivatives, polypeptides, polyalkylene glycols and derivatives thereof, polyalkyl Copolymers of ene glycol and derivatives thereof, polyvinyl ethyl ether and alpha-beta-poly[(2-hydroxyethyl)-DL-aspartamide, etc., or mixtures thereof. By way of example only, by coupling of such a water soluble polymer with a protein, polypeptide or peptide containing a compound provided herein, increased water solubility, increased or regulated serum half-life, increased or controlled therapeutic half-life compared to the unmodified form. , Increased bioavailability, regulated biological activity, prolonged circulation time, regulated immunogenicity, regulated physical association properties, such as aggregation and multimer formation, altered receptor binding, altered binding with one or more binding partners, and altered receptors. Changes occur, including, but not limited to, dimerization or multimerization and the like.
본원에 기재된 방법, 조성물 및 조합물의 여타의 목적, 특징 및 장점은 하기 상세한 설명으로부터 명백해질 것이다. 그러나, 본 상세한 설명 및 특정 실시예가 특정의 실시양태를 나타내지만 이는 오로지 예시로서 제공되는 것임을 이해해야 한다. Other objects, features, and advantages of the methods, compositions and combinations described herein will become apparent from the detailed description below. However, it should be understood that although this detailed description and specific examples represent specific embodiments, they are provided by way of example only.
생합성적으로Biosynthetically 생성된 피롤리신 및 Produced pyrrolysine and PCLPCL 의 부위 특이적 혼입Site-specific incorporation of
피롤리신 (PYL)은 메타노사르시나세아에(Methanosarcinaceae) 패밀리의 특정 메탄생성 고세균 및 관련이 없는 두 박테리아 종에서 발견되는, 22번째의 천연의 유전적으로 코딩된 아미노산이다. 구체적으로, 피롤리신은 이러한 고세균 박테리아의 메탄 형성을 개시하는 모노메틸아민 (MMA) 메틸트랜스퍼라제인 MtmB1에서 발견된다 (문헌 [Srinivasan, G., James, C. M., and Krzycki, J. A. (2002), "Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA," Science, 296, 1459-62]; [Soares, J. A., Zhang, L., Pitsch, R. L., Kleinholz, N. M., Jones, R. B., Wolff, J. J., Amster, J., Green-Church, K. B., and Krzycki, J. A. (2005), "The residue mass of L-pyrrolysine in three distinct methylamine methyltransferases," Journal of Biological Chemistry, 280, 36962-9]; [Hao, B., Gong, W., Ferguson, T. K., James, C. M., Krzycki, J. A., and Chan, M. K., (2002), "A new UAG-encoded residue in the structure of a methanogen methyltransferase", Science, 296, 1462-6]; [Krzycki, J. A., (2005), "The direct genetic encoding of pyrrolysine," Current Opinion in Microbiology, 8, 706-12]; [Krzycki, J. A., (2004), "Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases," Current Opinion in Chemical Biology, 8, 484-91], 및 [Ambrogelly, A., Palioura, S., and Soll, D., (2007), "Natural expansion of the genetic code," Nature Chemical Biology 3, 29-35] 참조). 피롤리신은 리신의 ε-아민이 아미드 결합을 통해 4-메틸-피롤린-5-카르복실레이트의 D-이성질체에 연결된 디펩티드인 것으로 여겨진다 (문헌 [Polycarpo, C. R., Herring, S., Berube, A., Wood, J. L., Soll, D., and Ambrogelly, A., (2006), "Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase," FEBS Letters, 580, 6695-700] 참조). 피롤리신의 구조 (도 1)는 MtmB1의 결정 구조 및 잔기의 질량으로부터 추론되었다 (문헌 [J. Biol. Chem. 2005, 44, 36962-36969]; [PNAS, 2007, 104, 1021-1026] 참조). Pyrrolysine (PYL) is the 22nd natural genetically encoded amino acid found in certain methanogenic archaea and two unrelated bacterial species of the Methanosarcinaceae family. Specifically, pyrrolysine is found in MtmB1, a monomethylamine (MMA) methyltransferase that initiates methane formation in these archaea bacteria (Srinivasan, G., James, CM, and Krzycki, JA (2002), " Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA," Science , 296 , 1459-62]; [Soares, JA, Zhang, L., Pitsch, RL, Kleinholz, NM, Jones, RB, Wolff, JJ, Amster, J., Green-Church, KB, and Krzycki, JA (2005), "The residue mass of L-pyrrolysine in three distinct methylamine methyltransferases," Journal of Biological Chemistry , 280 , 36962-9]; [Hao, B., Gong, W., Ferguson, TK, James, CM, Krzycki, JA, and Chan, MK, (2002), "A new UAG-encoded residue in the structure of a methanogen methyltransferase", Science , 296 , 1462-6]; [Krzycki, JA, (2005), "The direct genetic encoding of pyrrolysine," Current Opinion in Microbiology , 8 , 706-12]; [Krzycki, JA, (2004), "Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases," Current Opinion in Chemical Biology , 8 , 484-91], and [Ambrogelly, A., Palioura, S., and Soll, D., (2007), "Natural expansion of the genetic code," Nature Chemical Biology 3 , 29-35). Pyrrolysine is believed to be a dipeptide in which the ε-amine of lysine is linked to the D-isomer of 4-methyl-pyrroline-5-carboxylate via an amide bond (Polycarpo, CR, Herring, S., Berube, A., Wood, JL, Soll, D., and Ambrogelly, A., (2006), "Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase," FEBS Letters , 580 , 6695-700). The structure of pyrrolysine (Fig. 1) was deduced from the crystal structure of MtmB1 and the mass of the moiety (J. Biol. Chem. 2005, 44, 36962-36969]; see [PNAS, 2007, 104, 1021-1026]. ).
MtmB1을 코딩하는 mtmB1 유전자는 정상적으로 기준 종결 코돈인 인-프레임 앰버 (TAG) 코돈을 갖는다. 그러나, mtmB1 mRNA에서, DNA 수준으로 TAG로서 코딩된 UAG 코돈은 MtmB1 단백질의 생산 동안 번역을 종결시키지 않지만, 대신에 UAG 코돈은 단백질에 혼입된 피롤리신을 코딩한다. 피롤리신은 내생적으로 합성된 다음, 동시 번역에 의해 유리 아미노산으로서 이러한 인-프레임 UAG 코돈에 혼입된다. The mtmB1 gene encoding MtmB1 normally has an in-frame amber (TAG) codon, which is a reference stop codon. However, in mtmB1 mRNA, the UAG codon encoded as TAG at the DNA level does not terminate translation during production of the MtmB1 protein, but instead the UAG codon encodes for pyrrolysine incorporated into the protein. Pyrrolysine is synthesized endogenously and then incorporated into this in-frame UAG codon as a free amino acid by simultaneous translation.
피롤리신은 천연 유전자 pylT, pylS, pylB, pylC 및 pylD에 의해 용이하게 생합성 및 혼입된다. pylT는 피롤리실-tRNA를 코딩하고, pylS는 피롤리실-tRNA 신테타제를 코딩하는 반면에, pylB, pylC 및 pylD는 피롤리신의 생합성에 필요한 단백질을 코딩한다. 이들 유전자는 메타노사르시나 마제이(Methanosarcina mazei)로부터 유래된다 (문헌 [Longstaff, D. G., Larue, R. C., Faust, J. E., Mahapatra, A., Zhang, L., Green-Church, K. B., and Krzycki, J. A., (2007), "A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine," Proceedings of the National Academy of Sciences of the United States of America, 104, 1021-6], 및 [Namy, O., Zhou, Y., Gundllapalli, S., Polycarpo, C. R., Denise, A., Rousset, J. P., Soll, D., and Ambrogelly, A., (2007), "Adding pyrrolysine to the Escherichia coli genetic code," FEBS Letters, 581, 5282-8] 참조). pylT 및 pylS 유전자는 pylB, pylC 및 pylD 유전자와 함께 pylTSBCD 유전자 클러스터를 형성하며, 이는 천연 유전 암호 확장 카세트이며, 그의 전달은 UAG 코돈이 UAG 부위에서 단백질에 혼입된 피롤리신 (내생적으로 합성된 유리 아미노산)으로서 번역되도록 한다. Pyrrolysine is readily biosynthesized and incorporated by the natural genes pylT, pylS, pylB, pylC and pylD. pylT encodes pyrrolysyl-tRNA, pylS encodes pyrrolysyl-tRNA synthetase, while pylB, pylC and pylD encode proteins required for the biosynthesis of pyrrolysine. These genes are derived from Methanosarcina mazei (Longstaff, DG, Larue, RC, Faust, JE, Mahapatra, A., Zhang, L., Green-Church, KB, and Krzycki, JA , (2007), "A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine," Proceedings of the National Academy of Sciences of the United States of America , 104 , 1021-6], and [Namy, O., Zhou, Y., Gundllapalli, S., Polycarpo, CR, Denise, A., Rousset, JP, Soll, D., and Ambrogelly, A., (2007), "Adding pyrrolysine to the Escherichia coli genetic code," FEBS Letters , 581 , 5282-8). The pylT and pylS genes together with the pylB, pylC and pylD genes form the pylTSBCD gene cluster, which is a natural genetic coding expansion cassette, and its delivery is pyrrolysine in which the UAG codon is incorporated into the protein at the UAG site (endogenously synthesized Free amino acids).
피롤리신의 생합성은 전구체로서 제안된 D-오르니틴과 함께 천연 유전자 pylB, pylC 및 pylD의 유전자 생성물에 의해 용이해지는 것으로서 제안되었다 (문헌 [Namy, O., Zhou, Y., Gundllapalli, S., Polycarpo, C. R., Denise, A., Rousset, J. P., Soll, D., and Ambrogelly, A., (2007), "Adding pyrrolysine to the Escherichia coli genetic code," FEBS Letters, 581, 5282-8] 참조). 피롤리신에 대한 다양한 전구체, 예컨대 D-글루타메이트, D-이소류신, D-프롤린 및 D-오르니틴이 제안되었지만, D-오르니틴이 천연 유전자 pylT, pylS, pylB, pylC 및 pylD를 가진 플라스미드에 의해 형질전환된 에스케리키아 콜라이에서 피롤리신 생합성을 위한 가장 효과적인 전구체인 것으로 제안되었다. 이 연구는 TAG 혼입 또는 단절 신호에서의 번역초과, 즉, 전장 단백질의 생성을 이용하였고, 피롤리신이 천연 유전자 pylT, pylS, pylB, pylC 및 pylD를 가진 플라스미드에 의해 형질전환된 에스케리키아 콜라이 내에서 전구체로서 D-오르니틴을 사용하여 생성된 단백질에 생합성 및 혼입되었음을 입증하였다. 피롤리신의 혼입은 질량 분광측정법에 의해 직접적으로 검증되지 않았지만, 상기 결론은, 첨가된 D-오르니틴의 부재하에서 피롤리신이 비록 낮은 수준이지만 천연 유전자 pylT, pylS, pylB, pylC 및 pylD를 가진 플라스미드에 의해 형질전환된 에스케리키아 콜라이 내에서 생성된 단백질에 생합성 및 혼입되었다는 점에서, 이전 질량 분광측정법 데이터에 의해 뒷받침되었다 (문헌 [Longstaff, D. G., Larue, R. C., Faust, J. E., Mahapatra, A., Zhang, L., Green-Church, K. B., and Krzycki, J. A., (2007), "A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine," Proceedings of the National Academy of Sciences of the United States of America, 104, 1021-6] 참조).The biosynthesis of pyrrolysine has been suggested as being facilitated by the gene products of the natural genes pylB, pylC and pylD along with D-ornithine proposed as a precursor (Namy, O., Zhou, Y., Gundllapalli, S., Polycarpo, CR, Denise, A., Rousset, JP, Soll, D., and Ambrogelly, A., (2007), "Adding pyrrolysine to the Escherichia coli genetic code," FEBS Letters , 581 , 5282-8). Various precursors for pyrrolysine, such as D-glutamate, D-isoleucine, D-proline and D-ornithine, have been proposed, but D-ornithine is by a plasmid with the natural genes pylT, pylS, pylB, pylC and pylD. It has been proposed to be the most effective precursor for pyrrolysine biosynthesis in transformed Escherichia coli. This study used translating in TAG incorporation or break signal, i.e., the production of full-length proteins, and pyrrolysine in Escherichia coli transformed by a plasmid with the natural genes pylT, pylS, pylB, pylC and pylD. It was demonstrated that it was biosynthesized and incorporated into the produced protein using D-ornithine as a precursor. The incorporation of pyrrolysine was not directly verified by mass spectrometry, but the conclusion is that plasmids with the natural genes pylT, pylS, pylB, pylC and pylD although low levels of pyrrolysine in the absence of added D-ornithine. It was supported by previous mass spectrometry data in that it was biosynthesized and incorporated into proteins produced in Escherichia coli transformed by (Longstaff, DG, Larue, RC, Faust, JE, Mahapatra, A. , Zhang, L., Green-Church, KB, and Krzycki, JA, (2007), "A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine," Proceedings of the National Academy of Sciences of the United States of America , 104 , 1021-6).
그러나, 본원에 제공된 바와 같이, 질량 분광측정법을 이용하여 확인되는 바아 같이, 에스케리키아 콜라이 또는 포유동물 세포로의 pylT, pylS, pylB, pylC 및 pylD 유전자의 혼입, 및 성장 배지로의 D-오르니틴의 첨가에 의해, "탈메틸화 피롤리신"이 생합성 및 혼입된다는 것이 발견되었다 (도 1, PCL-A 및 PCL-B). 이 관측은 Longstaff 등에 의해 제안된 결과에 비추어 볼 때 놀라우며 예측불가능한 것이다. 따라서, 천연 유전자 pylT, pylS, pylB, pylC 및 pylD 및 전구체로서 D-오르니틴을 사용하여, 천연적으로 코딩되고, 생합성적으로 생성되어, 단백질에 혼입된 피롤리신 유사체, 피롤린-카르복시-리신 (PCL)이 본원에 제공된다. 다른 실시양태에서, D-아르기닌이 D-오르니틴에 대한 전구체이기 때문에, 천연 유전자 pylT, pylS, pylB, pylC 및 pylD 및 전구체로서 D-아르기닌을 사용하여, 천연적으로 코딩되고, 생합성적으로 생성되어, 단백질에 혼입된 피롤리신 유사체 또한 본원에 제공된다.However, as provided herein, the incorporation of the pylT, pylS, pylB, pylC and pylD genes into Escherichia coli or mammalian cells, as identified using mass spectrometry, and D-ornis into the growth medium. It was found that by the addition of tin, “demethylated pyrrolysine” was biosynthesized and incorporated (Fig. 1, PCL-A and PCL-B). This observation is surprising and unpredictable in light of the results proposed by Longstaff et al. Thus, using the natural genes pylT, pylS, pylB, pylC and pylD and D-ornithine as a precursor, a pyrrolysine analog, pyrroline-carboxy-, which is naturally encoded, produced biosynthetically, and incorporated into a protein. Lysine (PCL) is provided herein. In another embodiment, since D-arginine is a precursor to D-ornithine, it is naturally encoded and produced biosynthetically, using the natural genes pylT, pylS, pylB, pylC and pylD and D-arginine as a precursor. Thus, pyrrolysine analogs incorporated into proteins are also provided herein.
초기에는 D-오르니틴으로부터의 피롤리신의 피롤린 고리의 형성이 L-오르니틴으로부터 프롤린의 형성과 유사한 것으로 생각되었다 (문헌 [Longstaff, D. G., Larue, R. C., Faust, J. E., Mahapatra, A., Zhang, L., Green-Church, K. B., and Krzycki, J. A., (2007), "A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine," Proceedings of the National Academy of Sciences of the United States of America, 104, 1021-6] 참조). 수많은 박테리아가 L-오르니틴을 1-피롤린-5-카르복실레이트 중간체를 거쳐 L-프롤린으로 전환시킨다. 오르니틴으로부터 1-피롤린-5-카르복실레이트의 형성은 L-프롤린의 생합성에서 2 단계 공정 중 제1 단계이다. L-오르니틴으로부터 1-피롤린-5-카르복실레이트의 형성은 두 가지 경로를 통해 일어날 수 있으며, 제1 경로는 L-오르니틴의 시클로탈아미노화에 의한 1-피롤린-5-카르복실레이트의 형성이고, 제2 경로는 L-오르니틴 아미노 트랜스퍼라제에 의한 L-글루타메이트 세미알데히드의 형성이다. 이어서, 글루타메이트 세미알데히드가 1-피롤린-5-카르복실레이트를 형성한다. 피롤린-5-카르복실레이트 리덕타제에 의한 1-피롤린-5-카르복실레이트의 환원에 의해 L-프롤린이 형성된다. Initially, the formation of the pyrroline ring of pyrrolysine from D-ornithine was thought to be similar to that of proline from L-ornithine (Longstaff, DG, Larue, RC, Faust, JE, Mahapatra, A., Zhang, L., Green-Church, KB, and Krzycki, JA, (2007), "A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine," Proceedings of the National Academy of Sciences of the United States of America , 104 , 1021-6). Numerous bacteria convert L-ornithine to L-proline via the 1-pyrroline-5-carboxylate intermediate. The formation of 1-pyrroline-5-carboxylate from ornithine is the first step in the two-step process in the biosynthesis of L-proline. The formation of 1-pyrroline-5-carboxylate from L-ornithine can occur via two pathways, the first pathway being 1-pyrroline-5-car by cyclodeamination of L-ornithine. It is the formation of boxylate, and the second pathway is the formation of L-glutamate semialdehyde by L-ornithine amino transferase. Subsequently, glutamate semialdehyde forms 1-pyrroline-5-carboxylate. L-proline is formed by reduction of 1-pyrroline-5-carboxylate with pyrroline-5-carboxylate reductase.
최근, 메탄생성 고세균에서 피롤리신의 형성에서 L-오르니틴 아미노 트랜스퍼라제와의 상동성이 확인되지 않았다. 그러나, 이는 유사한 효소와 관련있는 것으로 예상된다. 피롤리신의 생합성은 세포 전구체를 사용하는 일관된 과정 및 pylB, pylC 및 pylD 유전자의 생성물의 일관된 작용인 것으로 생각되며, D-프롤린으로부터 1-피롤린-5-카르복실레이트를 거쳐 피롤리신을 생합성하는 것에 대해 제안된 반응식이 도 2A에 도시되어 있다 (문헌 [Longstaff, D. G., Larue, R. C., Faust, J. E., Mahapatra, A., Zhang, L., Green-Church, K. B., and Krzycki, J. A., (2007), "A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine," Proceedings of the National Academy of Sciences of the United States of America, 104, 1021-6] 참조). pylB, pylD 및 pylD 유전자 생성물은 몇몇 단백질 패밀리의 구성원이다. PylB는 라디칼-촉매된 반응을 매개하는 것으로 공지된 Fe-S 라디칼 SAM 효소 패밀리의 서명 잔기를 갖는다. 또한, PylB는 비오틴 신타제와 일부 서열 상동성을 공유하고, 따라서 PylB가 피롤리신 피롤린 고리의 형성 또는 메틸화에 관련있는 것으로 제안되었다. pylD 유전자 생성물인 PylD는 데히드로게나제의 몇몇 패밀리의 NADH-결합 도메인을 가지며, 이는 PylD가 1-피롤린 이민 결합의 형성에 관련있음을 제안한다. PylC는 카르바모일포스페이트 신테타제 및 D-알라닐-D-알라닌 리가제와 유사하며, 이는 리신과 4-메틸-1-피롤린-5-카르복실레이트의 D-이성질체 사이의 피롤리신 아미드 결합의 형성에서의 역할을 제안한다. 따라서, 추정적 pylB, pylC 및 pylD 유전자 생성물은 라디칼 SAM 단백질, 단백질 형성 아미노산 및 아미노산 데히드로게나제와 각각의 유사성을 가지며, 이로써 피롤리신의 생합성을 위한 유사한 경로에 참여하는 것으로 생각된다.Recently, no homology with L-ornithine amino transferase was confirmed in the formation of pyrrolysine in methanogenic archaea. However, it is expected to be related to similar enzymes. The biosynthesis of pyrrolysine is thought to be a consistent process using cell precursors and a consistent action of the products of the pylB, pylC and pylD genes, and biosynthesis of pyrrolysine from D-proline via 1-pyrroline-5-carboxylate. The proposed scheme for this is shown in Figure 2A (Longstaff, DG, Larue, RC, Faust, JE, Mahapatra, A., Zhang, L., Green-Church, KB, and Krzycki, JA, (2007). ), "A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine," Proceedings of the National Academy of Sciences of the United States of America , 104 , 1021-6). The pylB, pylD and pylD gene products are members of several protein families. PylB has a signature residue of the family of Fe-S radical SAM enzymes known to mediate radical-catalyzed reactions. In addition, PylB shares some sequence homology with biotin synthase, so it has been suggested that PylB is involved in the formation or methylation of the pyrrolysine pyrroline ring. The pylD gene product, PylD, has a NADH-binding domain of several families of dehydrogenases, suggesting that PylD is involved in the formation of 1-pyrroline imine bonds. PylC is similar to carbamoylphosphate synthetase and D-alanyl-D-alanine ligase, which is a pyrrolysine amide between lysine and the D-isomer of 4-methyl-1-pyrroline-5-carboxylate. Suggests a role in the formation of bonds Thus, the putative pylB, pylC and pylD gene products have respective similarities to the radical SAM protein, protein forming amino acid and amino acid dehydrogenase, and are therefore thought to participate in similar pathways for the biosynthesis of pyrrolysine.
그러나, 본원에 제공된 바와 같이, 전구체로서 D-오르니틴을 사용하여 에스케리키아 콜라이 및 HEK293F 세포에서 피롤리신을 생합성하려는 시도는, 피롤리신을 형성하지 않고, 오히려 "탈메틸화 피롤리신" (본원에서 피롤린-카르복시-리신 (PCL) (PCL-A 또는 PCL-B: 도 1 참조)으로도 지칭됨)을 형성한다. 본원에 기재된 바와 같이, PCL-A 또는 PCL-B의 생합성은 pylB 유전자의 존재를 필요로 하지 않고, 따라서 PCL-A 또는 PCL-B의 생합성을 위한 가능한 한 반응식이 도 2B에 도시된다. 임의의 특별한 이론에 구애되는 것은 아니지만, 이 가능한 경로가 내인성 D-아미노산 옥시다제 (E.C. 1.4.3.3)를 통한 D-오르니틴의 5-아미노-2-옥소펜탄산으로의 전환, 및 1-피롤린-2-카르복실레이트 및 물로의 5-아미노-2-옥소펜탄산의 자발적 고리화를 포함한다. 대부분의 유기체에서 이용가능한 이 전구체는 가능하게는 효소 PylD의 보조하에 이중 결합의 재배열에 의해 D-1-피롤린-5-카르복실레이트로 전환될 수 있다. PylC 및 ATP에 의한 D-1-피롤린-5-카르복실레이트의 L-리신의 엡실론 아민으로의 라이게이션은 피롤린-카르복시-리신 (PCL: PCL-A)을 생성할 것이다. 대안적으로, 1-피롤린-2-카르복실레이트의 라이게이션은 PCL-A와 분자량이 동일한 PCL-B를 생성할 수 있다. PylD 및 PylC 둘 다 PCL의 혼입을 위해 필요하다는 관찰, 및 PylS가 PCL의 1-피롤린 고리의 C-5 위치와 등가인 위치에서 sp2 탄소를 갖는 기질 피롤리신 유사체로서 허용하지 못하는 것은 (본원에 제시된 바와 같음), 초기에는 PCL이 주로 PCL-A 형태로 단백질에 혼입될 수 있음을 제안한다. 임의의 특정 이론에 구애되는 것은 아니지만, 탈메틸화 PCL-A 또는 PCL-B는 PylB의 불활성화의 결과로서 첨가된 D-오르니틴의 존재하에, 메틸 공여 PylB 기질의 부재하에, 필요한 보조인자(들)의 부재하에 또는 이들의 조합하에 수득된다. 그러나, 이는 대안적인 메카니즘의 존재를 배제하지 않다. 실제로, 몇몇 중간체를 이용한 혼입 시험 (본원에 제시된 바와 같음)은, 도 2B에서 제안된 것과는 상이한 PCL-A 또는 PCL-B의 생합성 경로를 제안한다. 이 대안적인 경로는 본원에 기재된다. However, as provided herein, an attempt to biosynthesize pyrrolysine in Escherichia coli and HEK293F cells using D-ornithine as a precursor does not form pyrrolysine, but rather “demethylated pyrrolysine” (herein Pyrroline-carboxy-lysine (PCL) (also referred to as PCL-A or PCL-B: see FIG. 1) is formed. As described herein, the biosynthesis of PCL-A or PCL-B does not require the presence of the pylB gene, so a possible scheme for biosynthesis of PCL-A or PCL-B is shown in Figure 2B. While not wishing to be bound by any particular theory, this possible pathway is the conversion of D-ornithine to 5-amino-2-oxopentanoic acid via endogenous D-amino acid oxidase (EC 1.4.3.3), and 1-pi. And spontaneous cyclization of 5-amino-2-oxopentanoic acid with roline-2-carboxylate and water. This precursor, available in most organisms, can be converted to D-1-pyrroline-5-carboxylate, possibly by rearrangement of the double bond with the aid of the enzyme PylD. L-lysine to epsilon amine ligation of D-1-pyrroline-5-carboxylate by PylC and ATP will yield pyrroline-carboxy-lysine (PCL: PCL-A). Alternatively, ligation of 1-pyrroline-2-carboxylate can result in PCL-B having the same molecular weight as PCL-A. The observation that both PylD and PylC are required for the incorporation of PCL, and that PylS is not acceptable as a substrate pyrrolysine analog with a sp2 carbon at a position equivalent to the C-5 position of the 1-pyrroline ring of PCL (here As shown in), it is suggested that in the beginning, PCL can be incorporated into proteins mainly in the form of PCL-A. While not wishing to be bound by any particular theory, the demethylated PCL-A or PCL-B is necessary cofactor(s) in the presence of D-ornithine added as a result of inactivation of PylB, in the absence of a methyl donating PylB substrate, ) In the absence of or in a combination thereof. However, this does not preclude the existence of alternative mechanisms. Indeed, incorporation tests with several intermediates (as presented herein) suggest a different biosynthetic pathway for PCL-A or PCL-B than that suggested in Figure 2B. This alternative route is described herein.
UAG 코돈에 반응하여 피롤리신의 단백질로의 혼입은 천연 유전자 pylT 및 pylS에 의해 용이해지며, 여기서 피롤리신으로서의 UAG 번역은 피롤리실-tRNA 신테타제 (PylS)에 의해 피롤리신으로의 앰버-탈코딩 tRNApyl의 아미노아실화를 필요로 한다는 것이 확인되었다 (도 3A). pylT 유전자는 특정한 천연 저해자 tRNApyl (PylT)을 코딩하며, 그의 CUA 안티코돈은 피롤리신에 상응하는 UAG 센스 코돈과 상보성이다. pylS 유전자는 특정 클래스 II tRNA 신테타제인 피롤리실-tRNA 신테타제 (PylS)를 코딩하며, 이는 tRNApyl에 피롤리신 (화학적으로 또는 생합적적으로 합성됨)을 부여하고, 또한 피롤리신-의존성 ATP:피로포스페이트 교환 반응을 수행한다. 유사하게, UAG 코돈에 반응하여 피롤리신 유사체 PCL-A 또는 PCL-B의 단백질로의 혼입은 천연 유전자 pylT 및 pylS에 의해 용이해지는 것으로 생각된다 (도 3B). Incorporation of pyrrolysine into the protein in response to the UAG codon is facilitated by the natural genes pylT and pylS, where UAG translation as pyrrolysine is amber into pyrrolysine by pyrrolysyl-tRNA synthetase (PylS). It was confirmed that the decoding tRNA pyl requires aminoacylation (Fig. 3A). The pylT gene encodes a specific natural inhibitor tRNA pyl (PylT), whose CUA anticodon is complementary to the UAG sense codon corresponding to pyrrolysine. The pylS gene encodes a specific class II tRNA synthetase, pyrrolysyl-tRNA synthetase (PylS), which confers pyrrolysine (chemically or biochemically synthesized) to the tRNA pyl, and also pyrrolysine- A dependent ATP: pyrophosphate exchange reaction is carried out. Similarly, incorporation of the pyrrolysine analogue PCL-A or PCL-B into the protein in response to the UAG codon is thought to be facilitated by the natural genes pylT and pylS. (Figure 3B).
원핵 및 진핵 세포에 의해 발현된 단백질로의 피롤리신 및 PCL 의 생합성 및 부위-특이적 혼입Biosynthesis and site-specific incorporation of pyrrolysine and PCL into proteins expressed by prokaryotic and eukaryotic cells
생합성적으로 생성된 피롤리신 및/또는 피롤린-카르복시-리신 ((S)-2-아미노-6-(3,4-디히드로-2H-피롤-2-카르복스아미도)헥산산 (PCL-A) 또는 (S)-2-아미노-6-(3,4-디히드로-2H-피롤-5-카르복스아미도)헥산산 (PCL-B))의 부위 특이적 혼입을 위한 방법이 본원에 제공된다. 피롤리신 유사체 PCL-A 및 PCL-B (둘 다 본원에서 PCL로 지칭됨)에는 피롤리신 (PYL)의 메틸기가 결여되어 있다 (도 1). 이러한 방법의 특정 실시양태에서, 진핵 세포는 포유동물 세포, 효모 세포, 곤충 세포, 진균 세포 또는 식물 세포이다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 포유동물 세포에는 인간 배아 신장 (HEK293F) 세포, 인간 상피양 암종 (HeLa 및 GH3) 세포, 원숭이 신장 (COS) 세포, 래트 C6 신경교종 세포, 새끼 햄스터 신장 (BHK-21) 세포 및 차이니즈 햄스터 난소 (CHO) 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 본원에 제공된 방법에 사용된 효모 세포에는 사카로미세스 세레비지애 및 피키아 파스토리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 곤충 세포에는 스포도프테라 프루기페르다(Spodoptera frugiperda) (sf9 및 sf21) 세포, 트리코플루시아 니(Trichoplusia ni) (BTI TN-5B1-4 또는 하이-파이브(High-Five™)) 세포 및 맘메스트라 브라시카에(Mammestra brassicae) 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 원핵 세포는 박테리아이고, 한편 다른 실시양태에서, 본원에 제공된 방법에 사용된 박테리아에는 에스케리키아 콜라이, 미코박테리움 스메그마티스(Mycobacterium smegmatis), 락토코쿠스 락티스(Lactococcus lactis) 및 바실러스 서브틸리스(Bacillus subtilis)가 포함되나, 이에 제한되지는 않는다. Biosynthetically produced pyrrolysine and/or pyrroline-carboxy-lysine ((S)-2-amino-6-(3,4-dihydro-2H-pyrrole-2-carboxamido)hexanoic acid ( Method for site specific incorporation of PCL-A) or (S)-2-amino-6-(3,4-dihydro-2H-pyrrole-5-carboxamido)hexanoic acid (PCL-B)) Is provided herein. The pyrrolysine analogs PCL-A and PCL-B (both referred to herein as PCL) lack the methyl group of pyrrolysine (PYL) (FIG. 1 ). In certain embodiments of this method, the eukaryotic cell is a mammalian cell, a yeast cell, an insect cell, a fungal cell or a plant cell. In other embodiments, the mammalian cells used in the methods provided herein include human embryonic kidney (HEK293F) cells, human epithelial carcinoma (HeLa and GH3) cells, monkey kidney (COS) cells, rat C6 glioma cells, baby hamsters. Kidney (BHK-21) cells and Chinese hamster ovary (CHO) cells. In certain embodiments, yeast cells used in the methods provided herein include, but are not limited to, Saccharomyces cerevisiae and Pichia pastoris cells. In other embodiments, the insect cells used in the methods provided herein include Spodoptera frugiperda (Spodoptera frugiperda ) (sf9 and sf21) cells, Trichoplusia ni (BTI TN-5B1-4 or High-Five™) cells and Mammestra brassicae brassicae ) cells are included, but are not limited thereto. In certain embodiments, the prokaryotic cells are bacteria, while in other embodiments, the bacteria used in the methods provided herein include Escherichia coli, Mycobacterium smegmatis. smegmatis ), Lactococcus lactis ) and Bacillus subtilis .
특정 실시양태에서, 생합성적으로 생성된 피롤리신 및 PCL의 부위 특이적 혼입을 위한 이러한 방법은, 유전자 pylT, pylS, pylB, pylC 및 pylD, 및 원하는 단백질을 위한 유전자를 원핵 세포 및/또는 진핵 세포에 혼입시키고, 임의로 형질감염된 세포의 성장 배지에 피롤리신 또는 PCL을 위한 전구체를 첨가하는 것을 포함한다. 특정 실시양태에서, 전구체는 D-오르니틴이고, 한편 다른 실시양태에서 전구체는 L-오르니틴이다. 특정 실시양태에서, 전구체는 D,L-오르니틴이다. 특정 실시양태에서, 전구체는 D-아르기닌이고, 한편 다른 실시양태에서 전구체는 L-아르기닌이다. 특정 실시양태에서, 전구체는 D,L-아르기닌이다. 특정 실시양태에서, 전구체는 : (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산이다. 특정 실시양태에서, 전구체는 : (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. 특정 실시양태에서, 전구체는 : 2,5-디아미노-3-메틸펜탄산이다. 특정 실시양태에서, 전구체는 (2R,3R)-2,5-디아미노-3-메틸펜탄산이다. 특정 실시양태에서, 진핵 세포는 포유동물 세포, 효모 세포, 곤충 세포, 진균 세포 또는 식물 세포이다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 포유동물 세포에는 인간 배아 신장 HEK293F 세포, 인간 상피양 암종 HeLa 및 GH3 세포, 원숭이 신장 COS 세포, 래트 C6 신경교종 세포, 새끼 햄스터 신장 BHK-21 세포 및 차이니즈 햄스터 난소 CHO 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 본원에 제공된 방법에 사용된 효모 세포에는 사카로미세스 세레비지애 및 피키아 파스토리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 곤충 세포에는 스포도프테라 프루기페르다 sf9 및 sf21 세포, 트리코플루시아 니 (BTI TN-5B1-4 또는 하이-파이브™) 세포 및 맘메스트라 브라시카에 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 원핵 세포는 박테리아이고, 한편 다른 실시양태에서, 본원에 제공된 방법에 사용된 박테리아에는 에스케리키아 콜라이, 미코박테리움 스메그마티스, 락토코쿠스 락티스 및 바실러스 서브틸리스가 포함되나, 이에 제한되지는 않는다. In certain embodiments, such methods for site-specific incorporation of biosynthetically generated pyrrolysine and PCL include the genes pylT, pylS, pylB, pylC and pylD, and the genes for the desired protein in prokaryotic and/or eukaryotic cells. And optionally adding pyrrolysine or a precursor for PCL to the growth medium of the transfected cells. In certain embodiments, the precursor is D-ornithine, while in other embodiments the precursor is L-ornithine. In certain embodiments, the precursor is D,L-ornithine. In certain embodiments, the precursor is D-arginine, while in other embodiments the precursor is L-arginine. In certain embodiments, the precursor is D,L-arginine. In certain embodiments, the precursor is : (2S)-2-Amino-6-(2,5-diaminopentanamido)hexanoic acid. In certain embodiments, the precursor is : (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid. In certain embodiments, the precursor is : It is 2,5-diamino-3-methylpentanoic acid. In certain embodiments, the precursor is (2R,3R)-2,5-diamino-3-methylpentanoic acid. In certain embodiments, the eukaryotic cell is a mammalian cell, yeast cell, insect cell, fungal cell or plant cell. In other embodiments, the mammalian cells used in the methods provided herein include human embryonic kidney HEK293F cells, human epithelial carcinoma HeLa and GH3 cells, monkey kidney COS cells, rat C6 glioma cells, baby hamster kidney BHK-21 cells, and Chinese hamster ovary CHO cells are included, but are not limited thereto. In certain embodiments, yeast cells used in the methods provided herein include, but are not limited to, Saccharomyces cerevisiae and Pichia pastoris cells. In other embodiments, the insect cells used in the methods provided herein include Spodoptera frugiperda sf9 and sf21 cells, Trichoflusia ni (BTI TN-5B1-4 or High-Five™) cells, and Mammestra brassica. Including, but not limited to, cells. In certain embodiments, the prokaryotic cells are bacteria, while in other embodiments, bacteria used in the methods provided herein include Escherichia coli, Mycobacterium smegmatis, Lactococcus lactis, and Bacillus subtilis. , But is not limited thereto.
특정 실시양태에서, 생합성적으로 생성된 피롤리신 및 PCL의 부위 특이적 혼입을 위한 이러한 방법은, 유전자 pylT, pylS, pylB, pylC 및 pylD, 및 원하는 단백질을 위한 유전자를 원핵 세포 및/또는 진핵 세포에 혼입시키고, 형질감염된 세포의 성장 배지에 피롤리신 또는 PCL을 위한 전구체를 첨가하는 것을 포함한다. 특정 실시양태에서, 전구체는 D-오르니틴이고, 한편 다른 실시양태에서 전구체는 L-오르니틴이다. 특정 실시양태에서, 전구체는 D,L-오르니틴이다. 특정 실시양태에서, 전구체는 D-아르기닌이고, 한편 다른 실시양태에서 전구체는 L-아르기닌이다. 특정 실시양태에서, 전구체는 D,L-아르기닌이다. 특정 실시양태에서, 전구체는 (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산이다. 특정 실시양태에서, 전구체는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. 특정 실시양태에서, 전구체는 2,5-디아미노-3-메틸펜탄산이다. 특정 실시양태에서, 전구체는 (2R,3R)-2,5-디아미노-3-메틸펜탄산이다. 특정 실시양태에서, 진핵 세포는 포유동물 세포, 효모 세포, 곤충 세포, 진균 세포 또는 식물 세포이다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 포유동물 세포에는 인간 배아 신장 HEK293F 세포, 인간 상피양 암종 HeLa 및 GH3 세포, 원숭이 신장 COS 세포, 래트 C6 신경교종 세포, 새끼 햄스터 신장 BHK-21 세포 및 차이니즈 햄스터 난소 CHO 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 본원에 제공된 방법에 사용된 효모 세포에는 사카로미세스 세레비지애 및 피키아 파스토리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 곤충 세포에는 스포도프테라 프루기페르다 sf9 및 sf21 세포, 트리코플루시아 니 (BTI TN-5B1-4 또는 하이-파이브™) 세포 및 맘메스트라 브라시카에 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 원핵 세포는 박테리아이고, 한편 다른 실시양태에서, 본원에 제공된 방법에 사용된 박테리아에는 에스케리키아 콜라이, 미코박테리움 스메그마티스, 락토코쿠스 락티스 및 바실러스 서브틸리스가 포함되나, 이에 제한되지는 않는다. In certain embodiments, such methods for site-specific incorporation of biosynthetically generated pyrrolysine and PCL include the genes pylT, pylS, pylB, pylC and pylD, and the genes for the desired protein in prokaryotic and/or eukaryotic cells. Incorporating into cells and adding pyrrolysine or a precursor for PCL to the growth medium of the transfected cells. In certain embodiments, the precursor is D-ornithine, while in other embodiments the precursor is L-ornithine. In certain embodiments, the precursor is D,L-ornithine. In certain embodiments, the precursor is D-arginine, while in other embodiments the precursor is L-arginine. In certain embodiments, the precursor is D,L-arginine. In certain embodiments, the precursor is (2S)-2-amino-6-(2,5-diaminopentanamido)hexanoic acid. In certain embodiments, the precursor is (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid. In certain embodiments, the precursor is 2,5-diamino-3-methylpentanoic acid. In certain embodiments, the precursor is (2R,3R)-2,5-diamino-3-methylpentanoic acid. In certain embodiments, the eukaryotic cell is a mammalian cell, yeast cell, insect cell, fungal cell or plant cell. In other embodiments, the mammalian cells used in the methods provided herein include human embryonic kidney HEK293F cells, human epithelial carcinoma HeLa and GH3 cells, monkey kidney COS cells, rat C6 glioma cells, baby hamster kidney BHK-21 cells, and Chinese hamster ovary CHO cells are included, but are not limited thereto. In certain embodiments, yeast cells used in the methods provided herein include, but are not limited to, Saccharomyces cerevisiae and Pichia pastoris cells. In other embodiments, insect cells used in the methods provided herein include Spodoptera frugiperda sf9 and sf21 cells, Trichoflusia ni (BTI TN-5B1-4 or High-Five™) cells, and Mammestra brassica. Including, but not limited to, cells. In certain embodiments, the prokaryotic cells are bacteria, while in other embodiments, bacteria used in the methods provided herein include Escherichia coli, Mycobacterium smegmatis, Lactococcus lactis, and Bacillus subtilis. , But is not limited thereto.
특정 실시양태에서, 생합성적으로 생성된 PCL의 부위 특이적 혼입을 위한 이러한 방법은, 유전자 pylT, pylS, pylC 및 pylD, 및 원하는 단백질을 위한 유전자를 원핵 세포 및/또는 진핵 세포에 혼입시키고, 성장 배지에 PCL을 위한 전구체로서 D-오르니틴을 첨가하는 것을 포함한다. 특정 실시양태에서, 진핵 세포는 포유동물 세포, 효모 세포, 곤충 세포, 진균 세포 또는 식물 세포이다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 포유동물 세포에는 인간 배아 신장 HEK293F 세포, 인간 상피양 암종 HeLa 및 GH3 세포, 원숭이 신장 COS 세포, 래트 C6 신경교종 세포, 새끼 햄스터 신장 BHK-21 세포 및 차이니즈 햄스터 난소 CHO 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 본원에 제공된 방법에 사용된 효모 세포에는 사카로미세스 세레비지애 및 피키아 파스토리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 곤충 세포에는 스포도프테라 프루기페르다 sf9 및 sf21 세포, 트리코플루시아 니 (BTI TN-5B1-4 또는 하이-파이브™) 세포 및 맘메스트라 브라시카에 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 원핵 세포는 박테리아이고, 한편 다른 실시양태에서, 본원에 제공된 방법에 사용된 박테리아에는 에스케리키아 콜라이, 미코박테리움 스메그마티스, 락토코쿠스 락티스 및 바실러스 서브틸리스가 포함되나, 이에 제한되지는 않는다. 천연 유전자 pylT, pylS, pylC 및 pylD가 사용되는 이러한 실시양태에서, 피롤리신이 생합성적으로 생성되고 혼입되기 보다는, 그 대신에 탈메틸화 피롤리신 유사체 PCL이 생합성적으로 생성되고 혼입된다. In certain embodiments, such a method for site-specific incorporation of biosynthetically generated PCL comprises incorporating the genes pylT, pylS, pylC and pylD, and genes for the desired protein into prokaryotic and/or eukaryotic cells and growing It involves adding D-ornithine as a precursor for PCL to the medium. In certain embodiments, the eukaryotic cell is a mammalian cell, yeast cell, insect cell, fungal cell or plant cell. In other embodiments, the mammalian cells used in the methods provided herein include human embryonic kidney HEK293F cells, human epithelial carcinoma HeLa and GH3 cells, monkey kidney COS cells, rat C6 glioma cells, baby hamster kidney BHK-21 cells, and Chinese hamster ovary CHO cells are included, but are not limited thereto. In certain embodiments, yeast cells used in the methods provided herein include, but are not limited to, Saccharomyces cerevisiae and Pichia pastoris cells. In other embodiments, insect cells used in the methods provided herein include Spodoptera frugiperda sf9 and sf21 cells, Trichoflusia ni (BTI TN-5B1-4 or High-Five™) cells, and Mammestra brassica. Including, but not limited to, cells. In certain embodiments, the prokaryotic cells are bacteria, while in other embodiments, bacteria used in the methods provided herein include Escherichia coli, Mycobacterium smegmatis, Lactococcus lactis, and Bacillus subtilis. , But is not limited thereto. In this embodiment in which the natural genes pylT, pylS, pylC and pylD are used, rather than pyrrolysine produced and incorporated biosynthetically, instead the demethylated pyrrolysine analogue PCL is produced and incorporated biosynthetically.
특정 실시양태에서, 생합성적으로 생성된 피롤리신 및 PCL의 부위 특이적 혼입을 위한 이러한 방법은, 유전자 pylT, pylS, pylC 및 pylD, 및 원하는 단백질을 위한 유전자를 원핵 세포 및/또는 진핵 세포에 혼입시키고, 성장 배지에 피롤리신 및/또는 PCL을 위한 전구체로서 D-오르니틴, L-오르니틴, D,L-오르니틴, D-아르기닌, L-아르기닌, D,L-아르기닌, (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산, (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산 또는 2,5-디아미노-3-메틸펜탄산 또는 (2R,3R)-2,5-디아미노-3-메틸펜탄산을 첨가하는 것을 포함한다. 특정 실시양태에서, 진핵 세포는 포유동물 세포, 효모 세포, 곤충 세포, 진균 세포 또는 식물 세포이다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 포유동물 세포에는 인간 배아 신장 HEK293F 세포, 인간 상피양 암종 HeLa 및 GH3 세포, 원숭이 신장 COS 세포, 래트 C6 신경교종 세포, 새끼 햄스터 신장 BHK-21 세포 및 차이니즈 햄스터 난소 CHO 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 본원에 제공된 방법에 사용된 효모 세포에는 사카로미세스 세레비지애 및 피키아 파스토리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 곤충 세포에는 스포도프테라 프루기페르다 sf9 및 sf21 세포, 트리코플루시아 니 (BTI TN-5B1-4 또는 하이-파이브™) 세포 및 맘메스트라 브라시카에 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 원핵 세포는 박테리아이고, 한편 다른 실시양태에서, 본원에 제공된 방법에 사용된 박테리아에는 에스케리키아 콜라이, 미코박테리움 스메그마티스, 락토코쿠스 락티스 및 바실러스 서브틸리스가 포함되나, 이에 제한되지는 않는다. 천연 유전자 pylT, pylS, pylC 및 pylD가 사용되는 이러한 실시양태에서, 피롤리신이 생합성적으로 생성되고 혼입되기 보다는, 그 대신에 탈메틸화 피롤리신 유사체 PCL이 생합성적으로 생성되고 혼입된다. In certain embodiments, such methods for site-specific incorporation of biosynthetically generated pyrrolysine and PCL include the genes pylT, pylS, pylC and pylD, and the genes for the desired protein into prokaryotic and/or eukaryotic cells. D-ornithine, L-ornithine, D,L-ornithine, D-arginine, L-arginine, D,L-arginine, (2S )-2-amino-6-(2,5-diaminopentanamido)hexanoic acid, (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid or And adding 2,5-diamino-3-methylpentanoic acid or (2R,3R)-2,5-diamino-3-methylpentanoic acid. In certain embodiments, the eukaryotic cell is a mammalian cell, yeast cell, insect cell, fungal cell or plant cell. In other embodiments, the mammalian cells used in the methods provided herein include human embryonic kidney HEK293F cells, human epithelial carcinoma HeLa and GH3 cells, monkey kidney COS cells, rat C6 glioma cells, baby hamster kidney BHK-21 cells, and Chinese hamster ovary CHO cells are included, but are not limited thereto. In certain embodiments, yeast cells used in the methods provided herein include, but are not limited to, Saccharomyces cerevisiae and Pichia pastoris cells. In other embodiments, insect cells used in the methods provided herein include Spodoptera frugiperda sf9 and sf21 cells, Trichoflusia ni (BTI TN-5B1-4 or High-Five™) cells, and Mammestra brassica. Including, but not limited to, cells. In certain embodiments, the prokaryotic cells are bacteria, while in other embodiments, bacteria used in the methods provided herein include Escherichia coli, Mycobacterium smegmatis, Lactococcus lactis, and Bacillus subtilis. , But is not limited thereto. In this embodiment in which the natural genes pylT, pylS, pylC and pylD are used, rather than pyrrolysine produced and incorporated biosynthetically, instead the demethylated pyrrolysine analogue PCL is produced and incorporated biosynthetically.
특정 실시양태에서, 생합성적으로 생성된 피롤리신의 부위 특이적 혼입을 위한 이러한 방법은, 유전자 pylT, pylS, pylC 및 pylD, 및 원하는 단백질을 위한 유전자를 원핵 세포 및/또는 진핵 세포에 혼입시키고, 성장 배지에 피롤리신을 위한 전구체로서 2,5-디아미노-3-메틸펜탄산 또는 (2R,3R)-2,5-디아미노-3-메틸펜탄산을 첨가하는 것을 포함한다. 특정 실시양태에서, 진핵 세포는 포유동물 세포, 효모 세포, 곤충 세포, 진균 세포 또는 식물 세포이다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 포유동물 세포에는 인간 배아 신장 HEK293F 세포, 인간 상피양 암종 HeLa 및 GH3 세포, 원숭이 신장 COS 세포, 래트 C6 신경교종 세포, 새끼 햄스터 신장 BHK-21 세포 및 차이니즈 햄스터 난소 CHO 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 본원에 제공된 방법에 사용된 효모 세포에는 사카로미세스 세레비지애 및 피키아 파스토리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 본원에 제공된 방법에 사용된 곤충 세포에는 스포도프테라 프루기페르다 sf9 및 sf21 세포, 트리코플루시아 니 (BTI TN-5B1-4 또는 하이-파이브™) 세포 및 맘메스트라 브라시카에 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 원핵 세포는 박테리아이고, 한편 다른 실시양태에서, 본원에 제공된 방법에 사용된 박테리아에는 에스케리키아 콜라이, 미코박테리움 스메그마티스, 락토코쿠스 락티스 및 바실러스 서브틸리스가 포함되나, 이에 제한되지는 않는다. In certain embodiments, such a method for site-specific incorporation of biosynthetically generated pyrrolysine incorporates the genes pylT, pylS, pylC and pylD, and genes for the desired protein into prokaryotic and/or eukaryotic cells, It involves adding 2,5-diamino-3-methylpentanoic acid or (2R,3R)-2,5-diamino-3-methylpentanoic acid as a precursor for pyrrolysine to the growth medium. In certain embodiments, the eukaryotic cell is a mammalian cell, yeast cell, insect cell, fungal cell or plant cell. In other embodiments, the mammalian cells used in the methods provided herein include human embryonic kidney HEK293F cells, human epithelial carcinoma HeLa and GH3 cells, monkey kidney COS cells, rat C6 glioma cells, baby hamster kidney BHK-21 cells, and Chinese hamster ovary CHO cells are included, but are not limited thereto. In certain embodiments, yeast cells used in the methods provided herein include, but are not limited to, Saccharomyces cerevisiae and Pichia pastoris cells. In other embodiments, insect cells used in the methods provided herein include Spodoptera frugiperda sf9 and sf21 cells, Trichoflusia ni (BTI TN-5B1-4 or High-Five™) cells, and Mammestra brassica. Including, but not limited to, cells. In certain embodiments, the prokaryotic cells are bacteria, while in other embodiments, bacteria used in the methods provided herein include Escherichia coli, Mycobacterium smegmatis, Lactococcus lactis, and Bacillus subtilis. , But is not limited thereto.
모델 단백질 (hRBP4)의 TAG 코딩된 부위에서 생합성적으로 생성된 PCL의 부위 특이적 혼입은, 천연 유전자 pylT, pylS, pylB, pylC 및 pylD, 및 전구체로서 성장 배지에 첨가된 D-오르니틴을 이용하여 포유동물 세포에서 수행되었다 (실시예 2 참조). 한 실시양태에서, 모델 단백질, hRBP4로의 PCL의 부위 특이적 혼입은 HEK293F 세포를 UAG-인식 tRNA PylT, 그의 특정한 아미노아실-tRNA 신테타제 PylS, 생합성 유전자 pylB, pylC 및 pylD를 코딩하는 DNA 뿐만 아니라, TAG에 의해 코딩된 혼입 부위를 갖는 표적 단백질을 코딩하는 DNA로 동시 형질감염시킴으로써 달성되었다. TAG 코돈에 의해 구체화된 단일 부위에서 모델 단백질 hRBP4로의 포유동물 세포에서의 이러한 생체내 생합성 및 혼입을 위해 사용되는 다른 유전자 구축물이 도 4A에 도시되어 있다. hRBP4, mEPO, hEPO, 및 mIgG1의 Fc 도메인의 다양한 단일 부위 TAG 돌연변이체의 발현을 위해 포유동물에 사용하기 위한 다른 유전자 구축물은 도 4B에 도시되어 있다. 에스케리키아 콜라이 세포에서 생합성적으로 유도된 PCL 또는 피롤리신의 혼입을 위해 pylT, pylS, pylB, pylC 및 pylD를 갖는 플라스미드는 도 5에 도시되어 있다. The site-specific incorporation of PCL biosynthetically generated at the TAG-encoded site of the model protein (hRBP4) uses the natural genes pylT, pylS, pylB, pylC and pylD, and D-ornithine added to the growth medium as a precursor. And carried out in mammalian cells (see Example 2). In one embodiment, the site-specific incorporation of PCL into the model protein, hRBP4, transforms HEK293F cells into UAG-recognized tRNA PylT, its specific aminoacyl-tRNA synthetase PylS, biosynthetic genes pylB, pylC and DNA encoding pylD, as well as This was achieved by co-transfection with DNA encoding the target protein with the incorporation site encoded by the TAG. Another gene construct used for this in vivo biosynthesis and incorporation in mammalian cells into the model protein hRBP4 at a single site specified by the TAG codon is shown in FIG. 4A. Other gene constructs for use in mammals for the expression of various single site TAG mutants of the Fc domain of hRBP4, mEPO, hEPO, and mIgG1 are shown in Figure 4B. Plasmids having pylT, pylS, pylB, pylC and pylD for the incorporation of PCL or pyrrolysine biosynthetically induced in Escherichia coli cells are shown in FIG. 5.
피롤리신의 생합성을 위한 추정적 전구체인 D-오르니틴을 pylT, pylS, pylB, pylC 및 pylD 뿐만 아니라 표적 단백질을 코딩하는 DNA로 형질감염된 HEK293F 세포의 배양 배지에 첨가할 때, 피롤리신이 아니라 PCL이 TAG 코돈의 부위에서 hRBP4에 혼입된다 (실시예 2, 도 6A 참조). 혼입 효율은 도 6A에서 도시된 바와 같이 상이한 혼입 부위에 따라 달라진다.When D-ornithine, a putative precursor for the biosynthesis of pyrrolysine, is added to the culture medium of HEK293F cells transfected with DNA encoding the target protein as well as pylT, pylS, pylB, pylC and pylD, not pyrrolysine but PCL It is incorporated into hRBP4 at the site of this TAG codon (see Example 2, Fig. 6A). The incorporation efficiency depends on the different incorporation sites as shown in Fig. 6A.
모델 단백질 (hRBP4)의 TAG 코딩된 부위에서 생합성적으로 생성된 PCL의 부위 특이적 혼입은 천연 유전자 pylT, pylS, pylC 및 pylD, 및 전구체로서 성장 배지에 첨가된 D-오르니틴을 이용하여 포유동물 세포에서 수행되었다 (실시예 2 참조). 한 실시양태에서, 모델 단백질, hRBP4로의 PCL의 부위 특이적 혼입은 HEK293F 세포를 UAG-인식 tRNA PylT, 그의 특정한 아미노아실-tRNA 신테타제 PylS, 생합성 유전자 pylC 및 pylD를 코딩하는 DNA 뿐만 아니라, TAG에 의해 코딩된 혼입 부위를 갖는 표적 단백질을 코딩하는 DNA로 동시 형질감염시킴으로써 달성되었다. 피롤리신의 생합성을 위한 추정적 전구체인 D-오르니틴을 형질감염된 HEK293F 세포의 배양 배지에 첨가할 때, 피롤리신이 아니라 PCL이 TAG 코돈의 부위에서 hRBP4에 혼입된다. 도 6B는 SDS-PAGE를 도시하고, 도 6C는 D-오르니틴의 부재 (B, 레인 1) 또는 존재 (B, 레인 2) 하에서의 HEK293F 세포에서 생성된 정제된 hRBP4 Phe62PCL (돌연변이체 #2)의 질량 스펙트럼을 도시한다. 화살표는 전장 hRBP4를 나타내고, 수득된 질량은 23166.0 Da이었으며, 이는 예상된 질량 23168 Da과 근사하다. 추가로 도 7-9에 도시된 질량 분광측정법 데이터는 피롤리신이 아니라 PCL이 hRBP4의 잔기 62에서 TAG 부위에 혼입되는 것을 도시한다 (실시예 2 참조). The site-specific incorporation of PCL biosynthetically generated at the TAG-encoded site of the model protein (hRBP4) was performed using the natural genes pylT, pylS, pylC and pylD, and D-ornithine added to the growth medium as a precursor. It was carried out in cells (see Example 2). In one embodiment, the site-specific incorporation of PCL into the model protein, hRBP4, transforms HEK293F cells into UAG-recognized tRNA PylT, its specific aminoacyl-tRNA synthetase PylS, DNA encoding the biosynthetic genes pylC and pylD, as well as TAG. It was achieved by co-transfection with DNA encoding the target protein with the incorporation site encoded by. When D-ornithine, a putative precursor for the biosynthesis of pyrrolysine, is added to the culture medium of transfected HEK293F cells, PCL, not pyrrolysine, is incorporated into hRBP4 at the site of the TAG codon. 6B shows SDS-PAGE, and FIG. 6C shows the purified hRBP4 Phe62PCL (mutant #2) produced in HEK293F cells in the absence (B, lane 1) or presence (B, lane 2) of D-ornithine. Show the mass spectrum. The arrows indicate the full length hRBP4, and the mass obtained was 23166.0 Da, which is close to the expected mass of 23168 Da. Additionally, the mass spectrometry data shown in FIGS. 7-9 show that PCL, but not pyrrolysine, is incorporated into the TAG site at residue 62 of hRBP4 (see Example 2).
도 1은 피롤리신 (PYL) 및 탈메틸화 피롤리신 유사체, 피롤린-카르복시-리신 (PCL)의 구조 (구조 PCL-A 또는 대안적인 구조 PCL-B)를 도시한다. PCL-A의 구조 (또는 대안적인 구조 PCL-B)는 혼입 부위에서 펩티드 단편의 고정밀 질량 분광측정 분석을 이용하여 피롤리신과 구별되었다 (도 7-9). 또한, PCL의 구조는 세포 용해물 중에서 유리 아미노산으로서 질량 분광측정법에 의한 PCL의 검출에 의해 피롤리신과 구별되었다 (도 10, 실시예 3 참조). 또한, 세포 용해물 중에서 PCL의 관찰은, PCL이 번역후 변형에 의해 형성되기 보다는, 유리 아미노산으로서 생합성적으로 생성됨을 입증한다. 도 10A 및 10C는 D-오르니틴으로부터 생합성된 HEK293F 세포로부터의 용해물 중에서 PCL의 검출을 도시한다. 도 10A는 생합성 유전자 pylB , pylC 및 pylD로 형질감염되고 D-오르니틴의 존재 하에서 (하단 트레이스) 및 D-오르니틴의 부재 하에서 (상단 트레이스) 성장한 세포로부터 용해물의 HPLC 트레이스이다. D-오르니틴의 존재 하에서 성장한 세포로부터 수득된 용해물 크로마토그램은 4.13 분 용리 시간에서의 피크 (별표로 표시됨)를 특징으로 하나, 이는 D-오르니틴의 부재 하에서 배양된 동일한 세포의 용해물에서는 존재하지 않는다. 도 10C는 4.13 분에서의 HPLC 피크의 전체 스캔 질량 스펙트럼이고, 여기서 수득된 질량 (m+1)은 PCL에 대한 이론적 질량과 일치한다. 도 10B는 리신이 두 샘플에서 동등하게 풍부함을 도시하는 HPLC 크로마토그램이며, 1.44 분에서의 리신 HPLC의 전체 스캐닝 질량 (m+1) 스펙트럼은 상기 방법의 정확도를 설명한다 (도 10D). 1 shows the structure of pyrrolysine (PYL) and a demethylated pyrrolysine analog, pyrroline-carboxy-lysine (PCL) (structure PCL-A or alternative structure PCL-B). The structure of PCL-A (or alternative structure PCL-B) was distinguished from pyrrolysine using high-precision mass spectrometric analysis of the peptide fragment at the site of incorporation (FIGS. 7-9 ). In addition, the structure of PCL was distinguished from pyrrolysine by detection of PCL by mass spectrometry as a free amino acid in the cell lysate (see Fig. 10 and Example 3). In addition, observation of PCL in cell lysates demonstrates that PCL is produced biosynthetically as a free amino acid rather than formed by post-translational modification. 10A and 10C depict the detection of PCL in lysates from HEK293F cells biosynthesized from D-ornithine. 10A is an HPLC trace of lysates from cells transfected with the biosynthetic genes pylB , pylC and pylD and grown in the presence of D-ornithine (bottom trace) and in the absence of D-ornithine (top trace). The lysate chromatogram obtained from cells grown in the presence of D-ornithine is characterized by a peak at 4.13 min elution time (indicated by an asterisk), which in the lysate of the same cells cultured in the absence of D-ornithine does not exist. 10C is the full scan mass spectrum of the HPLC peak at 4.13 min, where the mass obtained (m+1) is consistent with the theoretical mass for PCL. Figure 10B is an HPLC chromatogram showing that lysine is equally abundant in both samples, and the total scanning mass (m+1) spectrum of lysine HPLC at 1.44 min explains the accuracy of the method (Figure 10D).
도 11은 HEK293F 세포에서 다양한 hRBP4 TAG 돌연변이 단백질로의 N-ε-시클로펜틸옥시카르보닐-L-리신 (CYC)의 혼입을 도시한다. 도 12는 hRBP4 돌연변이체 Phe62TAG의 TAG 부위에서 CYC 혼입의 질량 분광측정 검증을 도시한다. 도 13A 및 도 13B (실시예 4 참조)는 표에 열거되고 도 14A에 도시된 다양한 전구체의 기능으로서의 PCL 혼입을 도시한다. 도 13A 및 도 13B는 또한 HEK293F 세포를 이용하는 hRBP4 TAG 돌연변이 단백질로의 다양한 피롤리신 유사체 (CYC 포함)의 직접 혼입을 도시한다. D-오르니틴이 PCL의 생합성을 위한 전구체임을 입증하기 위해, PCL을 위한 잠재적인 전구체 (도 14A)를 피롤리신 생합성 pylB, pylC 및 pylD 유전자, pylT/pylS tRNA/피롤리실-tRNA 신테타제 쌍 및 hRBP4 TAG 돌연변이체 DNA에 의해 형질감염된 HEK293F 세포의 성장 배지에 첨가하였다. 게다가, pylT/pylS tRNA/aa-tRNA 신테타제 쌍 및 hRBP4 TAG 돌연변이체 DNA로만 형질감염된 HEK293F 세포의 성장 배지에 첨가된 다양한 피롤리신 유사체가 도 14B에 도시되어 있다. 도 13A는 항-His 태그 항체를 갖는 전장 hRBP4 단백질의 웨스턴 블롯이고, 반면에 도 13B는 Ni-NTA 정제 이후 동일한 샘플의 SDS-PAGE이다. D-오르니틴으로 대사될 수 있는 D-아르기닌 (레인 4)이 백그라운드 단백질 생성을 초과하더라도, 도시된 바와 같이 D-오르니틴 (레인 2)이 PCL 생합성을 위한 더 양호한 전구체이다. 피롤리신 유사체 중에서, CYC만이 D-오르니틴과 유사한 수준으로 단백질을 생성하고, 반면에 3-옥소부탄산 유사체 TU3000-016은 측정가능하긴 하지만 매우 낮은 혼입을 나타낸다. 다양한 유사체의 합성 기술은 실시예 33에 제공된다. 모든 레인은 가능하게는 D-오르니틴 및 대사물의 낮은 내인성 수준 또는 PylS 기질로서 허용가능한 배지 성분 때문에 전장 단백질의 낮은 수준의 생성을 나타낸다. 11 depicts the incorporation of N-ε-cyclopentyloxycarbonyl-L-lysine (CYC) into various hRBP4 TAG mutant proteins in HEK293F cells. 12 shows mass spectrometric verification of CYC incorporation at the TAG site of the hRBP4 mutant Phe62TAG. 13A and 13B (see Example 4) show PCL incorporation as a function of the various precursors listed in the table and shown in FIG. 14A. 13A and 13B also depict direct incorporation of various pyrrolysine analogs (including CYC) into hRBP4 TAG mutant proteins using HEK293F cells. To demonstrate that D-ornithine is a precursor for the biosynthesis of PCL, a potential precursor for PCL (Figure 14A) was identified as pyrrolysine biosynthesis pylB, pylC and pylD genes, pylT/pylS tRNA/pyrrolysyl-tRNA synthetase. It was added to the growth medium of HEK293F cells transfected with paired and hRBP4 TAG mutant DNA. In addition, various pyrrolysine analogs added to the growth medium of HEK293F cells transfected only with the pylT/pylS tRNA/aa-tRNA synthetase pair and hRBP4 TAG mutant DNA are shown in FIG. 14B. Figure 13A is a Western blot of full length hRBP4 protein with anti-His tag antibody, whereas Figure 13B is SDS-PAGE of the same sample after Ni-NTA purification. Although D-arginine (lane 4), which can be metabolized to D-ornithine, exceeds background protein production, as shown, D-ornithine (lane 2) is a better precursor for PCL biosynthesis. Of the pyrrolysine analogs, only CYC produces proteins at levels similar to D-ornithine, while the 3-oxobutanoic acid analog TU3000-016, although measurable, shows very low incorporation. Techniques for the synthesis of various analogs are provided in Example 33. All lanes show low levels of production of full-length proteins, possibly due to low endogenous levels of D-ornithine and metabolites or media components acceptable as PylS substrate.
PCL의 혼입은 생합성 유전자 pylB, pylC 및 pylD의 상이한 조합을 이용하여 평가하였다 (도 13C). 데이터는 유전자 pylC 및 pylD만이 PCL 생합성 및 후속적 단백질 혼입에 필수적임을 보여준다. 가장 두드러지게는 D-프롤린, 다른 D-프롤린 유사체 및 D-글루탐산 (도 13 및 14)은 백그라운드 초과의 전장 단백질 생성을 나타내지 않았다. 이는 PCL-A 및 PCL-B의 생합성이 도 2A에서 제안된 경로를 따르지 않음을 제안한다. 그러나, 3,4-디히드로-2H-피롤-5-카르복실레이트 (P2C) 및 1-피롤린-5-카르복실레이트 (P5C)를 이용한 혼입 시험 또한 에스케리키아 콜라이에서 전장 단백질을 생성하는데 실패한 반면에, 합성 PCL-A 및 PCL-B 둘 다 고효율로 혼입되었다 (도 15A, 실시예 15). 따라서, 도 2B에서 제안된 생합성 경로 또한 유망한 경로가 아닐 수 있다. Incorporation of PCL was evaluated using different combinations of biosynthetic genes pylB, pylC and pylD (Fig. 13C). The data show that only genes pylC and pylD are essential for PCL biosynthesis and subsequent protein incorporation. Most notably, D-proline, other D-proline analogs and D-glutamic acid (Figures 13 and 14) did not show full-length protein production above background. This suggests that the biosynthesis of PCL-A and PCL-B does not follow the pathway suggested in Fig. 2A. However, the incorporation test using 3,4-dihydro-2H-pyrrole-5-carboxylate (P2C) and 1-pyrroline-5-carboxylate (P5C) also produced full-length proteins in Escherichia coli. While failing, both synthetic PCL-A and PCL-B were incorporated with high efficiency (Figure 15A, Example 15). Therefore, the biosynthetic pathway proposed in FIG. 2B may not be a promising pathway.
전구체 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산 (Lys-Nε-D-Orn으로도 지칭됨)의 혼입에 의해 상당량의 전장 단백질이 수득되었다 (도 15A, 실시예 15). 게다가, (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산의 혼입이 pylS, pylT 및 PylD 유전자의 존재만을 필요로 하는 것으로 확인되었다 (도 15B, 실시예 15). 이와 함께, 이러한 관측은 도 16A 및 16B에 도시된 대안적인 경로를 제안한다. 이러한 경로는, D-오르니틴이 PylC, 추정적 D-알라닐-D-알라닌 리가제의 도움에 의해 먼저 L-리신의 엡실론 아미노기에 커플링되어 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산을 형성한다는 개념을 기반으로 한다. 도 16A에서, (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산 중간체는 D-오르니틴:산소 옥시도리덕타제 (EC 1.4.3.3) 또는 D-아미노-트랜스아미나제 (EC 2.6.1.21)에 의해 활성화되어, 자발적 고리화에 의해 PCL-B를 형성한다. 대안적으로, 도 16B에서 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산 중간체는 D-오르니틴:2-옥시도글루타레이트 5-트랜스아미나제 (EC 2.6.1.13)에 의해 활성화되어, 자발적 고리화에 의해 PCL-A를 형성한다. 두 고리화 반응 모두 D-글루타메이트 5-세미알데히드의 반응과 유사하다. 고리화 이전의 활성화 단계 중 하나는 추정적으로 PylD에 의해 촉매될 수 있고, 두번째 단계는 PCL의 생합성을 위해 효소를 필요로 한다. 도 16에 도시된 바와 같이, PylB에 의한 PCL-A의 메틸화가 PYL의 생합성을 완성하는 것으로 제안된다. 도 16A의 경로의 경우에는, 이것이 PCL-B로부터 PCL-A를 형성하는 또 다른 발견되어야 하는 Pyl 효소의 존재를 필요로 할 가능성이 있음을 주목한다. 그러나, 이 추가의 효소는 16B에 도시된 대안적인 경로를 필요로 하지 않을 것이다. PylD에 의한 활성화 및 자발적 고리화 이후에 형성된 PCL-A는 직접적으로 PylB에 의해 메틸화되어 피롤리신을 형성할 수 있다. 대안적으로, 임의의 중간체 또는 D-오르니틴은 후속적인 효소가 기질의 이러한 변형을 용인한다면 PylB에 의한 메틸화를 위한 기질일 수 있다. The incorporation of the precursor (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid (also referred to as Lys-Nε-D-Orn) results in a significant amount of full-length protein. Was obtained (Fig. 15A, Example 15). Furthermore, it was confirmed that the incorporation of (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid only required the presence of the pylS, pylT and PylD genes (Fig. 15B). , Example 15). Together, these observations suggest an alternative route shown in Figs. 16A and 16B. This pathway is followed by coupling of D-ornithine to the epsilon amino group of L-lysine first with the help of PylC, putative D-alanyl-D-alanine ligase, and thus (2S)-2-amino-6-(( It is based on the concept of forming R)-2,5-diaminopentanamido)hexanoic acid. In Fig. 16A, (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid intermediate is D-ornithine: oxygen oxidoreductase (EC 1.4.3.3) or Activated by D-amino-transaminase (EC 2.6.1.21), forming PCL-B by spontaneous cyclization. Alternatively, (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid intermediate in FIG. 16B is D-ornithine:2-oxidoglutarate 5- It is activated by transaminase (EC 2.6.1.13), forming PCL-A by spontaneous cyclization. Both cyclization reactions are similar to those of D-glutamate 5-semialdehyde. One of the activation steps prior to cyclization can putatively be catalyzed by PylD, and the second step requires enzymes for the biosynthesis of PCL. As shown in Fig. 16, it is proposed that methylation of PCL-A by PylB completes the biosynthesis of PYL. Note that in the case of the pathway of Figure 16A, it is likely that this will require the presence of another to-be-discovered Pyl enzyme that forms PCL-A from PCL-B. However, this additional enzyme would not require the alternative pathway shown in 16B. PCL-A formed after activation by PylD and spontaneous cyclization can be directly methylated by PylB to form pyrrolysine. Alternatively, any intermediate or D-ornithine can be a substrate for methylation by PylB if the subsequent enzyme tolerates this modification of the substrate.
피롤리실-tRNA 신테타제 PylS는 tRNAPyl을 부여하기 위한 여러 피롤리신 유사체를 허용하는 것으로 확인되었다. 그러나, 이러한 연구는 또한 신테타제 PylS의 인식을 위한 피롤리신 고리 C-5 입체중심의 중요성을 지적하였으며, 여기서 D-형태가 필요로 된다 (문헌 [Polycarpo, C. R., Herring, S., Berube, A., Wood, J. L., Soll, D., and Ambrogelly, A., (2006), "Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase," FEBS Letters, 580, 6695-700] 참조). 본원에 제공된 바와 같이, 이 해석과 일치하게, 방향족 5 및 6-원 고리 피롤리신 유사체를 비롯하여 다양한 피롤리신 유사체를 혼입하고자 하는 시도 (도 13)는, 이러한 방향족 5 및 6-원 고리 유사체가 PylS를 위한 허용가능한 기질이 아님을 입증하였다 (도 14). 이것은 가능하게는 C-5 키랄 중심의 결여 때문이거나, 또는 특정 경우에는 더 큰 크기의 유사체 때문이다. 지적된 바와 같이, PylS를 위한 공지된 기질인 N-ε-시클로펜틸옥시카르보닐-L-리신 (CYC), 및 3-옥소부탄산 유사체인 TU3000-016은 PylT/PylS tRNA/피롤리실-tRNA 신테타제 쌍을 이용하여 혼입시켰다 (도 14). 그러나, N-ε-시클로펜틸옥시카르보닐-L-리신 (CYC)과 크기 및 벌크성이 유사한 유사체는 가능하게는 C-5 입체중심이 결여되었기 때문에 혼입되지 않은 것으로 평가되었다. 따라서, PylS는 피롤린 고리의 부착 지점에서 키랄성에 따라 선택적인 것으로 보인다. 그러나, 합성 PCL-B는 또한 고효율로 혼입되었으며, 이는 C-5 위치에서 sp2 비키랄 탄소가 모든 경우에 혼입을 선호하지 않는 것은 아님을 제안한다. 현재, 합성 PCL-A와 비교할 때 2-ABA를 갖는 합성 PCL-B의 낮은 반응성 (실시예 44, 도 56 및 58), 및 도 16A에서 PCL-B에서 PCL-A로의 전환를 위한 공지된 효소의 부재는 PCL-A로서 혼입된 피롤리신 유사체의 배정을 선호한다. It has been found that the pyrrolysyl-tRNA synthetase PylS allows several pyrrolysine analogs to confer tRNA Pyl. However, these studies also pointed out the importance of the pyrrolysine ring C-5 stereocenter for recognition of the synthetase PylS, where the D-form is required (Polycarpo, CR, Herring, S., Berube, et al. A., Wood, JL, Soll, D., and Ambrogelly, A., (2006), "Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase," FEBS Letters , 580 , 6695-700). As provided herein, consistent with this interpretation, attempts to incorporate a variety of pyrrolysine analogs, including aromatic 5 and 6-membered ring pyrrolysine analogs (Figure 13), show that these aromatic 5 and 6-membered ring analogs Was not an acceptable substrate for PylS (FIG. 14 ). This is possibly due to the lack of a C-5 chiral center, or in certain cases due to a larger sized analog. As noted, N-ε-cyclopentyloxycarbonyl-L-lysine (CYC), a known substrate for PylS, and TU3000-016, a 3-oxobutanoic acid analog, are PylT/PylS tRNA/pyrrolysyl- It was incorporated using a pair of tRNA synthetase (FIG. 14 ). However, analogs similar in size and bulk to N-ε-cyclopentyloxycarbonyl-L-lysine (CYC) were evaluated as not incorporated because possibly lacking the C-5 stereocenter. Thus, PylS appears to be selective depending on chirality at the point of attachment of the pyrroline ring. However, synthetic PCL-B was also incorporated with high efficiency, suggesting that the sp2 achiral carbon at the C-5 position does not favor incorporation in all cases. Currently, the low reactivity of synthetic PCL-B with 2-ABA compared to synthetic PCL-A (Example 44, Figures 56 and 58), and of known enzymes for the conversion of PCL-B to PCL-A in Figure 16A. Absence favors the assignment of pyrrolysine analogs incorporated as PCL-A.
본원에 제공된 바와 같이, 특정 실시양태에서, D-오르니틴이 전구체로서 성장 배지에 첨가될 때, 포유동물 세포 (실시예 2) 및 박테리아 세포 (실시예 8, 9 및 10)에서 피롤리신의 생합성을 위한 천연 유전자 pylB, pylC 및 pylD의 사용이 피롤리신 보다는 PCL을 생성하는 것으로 확인되었다. 또한, 천연 유전자 pylB 및 pylC, 천연 유전자 pylB 및 pylD, 또는 천연 유전자 pylC 및 pylD를 이용하는 PCL의 생합성의 평가는, pylC 및 pylD 유전자 생성물이 존재하는 경우에만 PCL을 생성하였다. 이는, D-오르니틴이 전구체로서 성장 배지에 첨가될 때, pylB의 유전자 생성물이 PCL의 생합성에 필요하지 않음을 제안한다 (도 13C, 실시예 4 참조). 이는 유전자 pylB를 포유동물 세포에 동시 형질감염시키지 않고 수행된, 세 가지 모델 단백질, hRBP4 (도 6, 실시예 2 참조), mIgG1 Fc 도메인 (도 17A, 실시예 5 참조) 및 mEPO (도 17B, 실시예 6 참조)로의 PCL의 혼입에 의해 뒷받침된다. 에스케리키아 콜라이에서, FAS-TE, FGF21 및 FKBP는 천연 유전자 pylB, pylC 및 pylD를 사용하여 PCL을 독점적으로 생성하는 예이다 (도 18B, 19 및 20, 실시예 8, 9 및 10). As provided herein, in certain embodiments, when D-ornithine is added to the growth medium as a precursor, biosynthesis of pyrrolysine in mammalian cells (Example 2) and bacterial cells (Examples 8, 9 and 10). It was found that the use of natural genes pylB, pylC and pylD for pyrrolysine produced PCL. In addition, the evaluation of the biosynthesis of PCL using the natural genes pylB and pylC, the natural genes pylB and pylD, or the natural genes pylC and pylD produced PCL only when the pylC and pylD gene products were present. This suggests that when D-ornithine is added to the growth medium as a precursor, the gene product of pylB is not required for the biosynthesis of PCL (see Fig. 13C, Example 4). This was carried out without co-transfection of the gene pylB into mammalian cells, three model proteins, hRBP4 (see Figure 6, Example 2), mIgG1 Fc domain (see Figure 17A, Example 5) and mEPO (Figure 17B, (See Example 6) is supported by the incorporation of PCL. In Escherichia coli, FAS-TE, FGF21 and FKBP are examples of exclusively generating PCL using the natural genes pylB, pylC and pylD (Figs. 18B, 19 and 20, Examples 8, 9 and 10).
PCL의 생합성은 숙주 세포에 pylB가 아니라 생합성 유전자 pylC 및 pylD의 존재를 필요로 한다. 메타노사르시나 내에서의 피롤리신의 생합성에서, PylD가 데히드로게나제의 NADH-결합 도메인을 함유하고, 따라서 D-프롤린으로부터의 D-1-피롤린-5-카르복실레이트를 생성한다는 것이 제안되었다. 그러나, 성장 배지에 D-프롤린을 첨가하는 것은 유의한 PCL 혼입을 나타내지 않는다 (도 13). 또한, 성장 배지에 3,4-디히드로-2H-피롤-5-카르복실레이트 (본원에서 1-피롤린-2-카르복실레이트; P2C로도 지칭됨) 및 D-1-피롤린-5-카르복실레이트 (P5C)의 첨가는 전장 단백질을 생성하는데 실패한 반면에, (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산은 PCL 함유 단백질을 생성한다. PylC는 D-알라닐-D-알라닌 리가제와 서열 상동성을 가지며, PCL 또는 피롤리신의 생합성에서 리신의 엡실론-아미노기에 D-오르니틴이 부착하는 것을 촉매하여 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산을 제공할 수 있다. 따라서, 이는 본원에서 임의의 이론에 구애되지 않고, 포유동물 또는 에스케리키아 콜라이 세포 내에서 D-오르니틴으로부터 PCL의 생합성이 PylC에 의한 D-오르니틴의 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산으로의 전환과 관련있을 수 있음을 가정한다 (도 16B). 도 16B에 도시된 바와 같이 PylD는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이 PCL-A로 자발적으로 고리화하는 세미알데히드 ((S)-2-아미노-6-((R)-2-아미노-5-옥소펜탄아미도)헥산산)으로 활성화되는 것에 관련있을 수 있다. 대안적으로, PylD가 D-아미노산 트랜스아미나제 또는 D-오르니틴:산소 옥시도리덕타제 유사 활성을 갖는다면 PCL-B는 자발적으로 형성될 것이다 (도 16A). The biosynthesis of PCL requires the presence of the biosynthetic genes pylC and pylD, not pylB, in the host cell. In the biosynthesis of pyrrolysine in metanosarcina, it is found that PylD contains the NADH-binding domain of dehydrogenase, thus producing D-1-pyrroline-5-carboxylate from D-proline. Was proposed. However, adding D-proline to the growth medium did not indicate significant PCL incorporation (FIG. 13 ). In addition, 3,4-dihydro-2H-pyrrole-5-carboxylate (herein 1-pyrroline-2-carboxylate; also referred to as P2C) and D-1-pyrroline-5- The addition of carboxylate (P5C) failed to produce the full-length protein, while (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid produced the PCL-containing protein. do. PylC has sequence homology with D-alanyl-D-alanine ligase, and catalyzes the attachment of D-ornithine to the epsilon-amino group of lysine in the biosynthesis of PCL or pyrrolysine (2S)-2-amino- 6-((R)-2,5-diaminopentanamido)hexanoic acid can be provided. Thus, this is not bound to any theory herein, and the biosynthesis of PCL from D-ornithine in mammalian or Escherichia coli cells is the (2S)-2-amino-6- of D-ornithine by PylC. It is assumed that it may be related to the conversion to ((R)-2,5-diaminopentanamido)hexanoic acid (Figure 16B). As shown in Figure 16B, PylD is (2S)-2-amino-6-((R)-2,5-diaminopentanamido) hexanoic acid spontaneously cyclizes to PCL-A ((S )-2-amino-6-((R)-2-amino-5-oxopentanamido)hexanoic acid). Alternatively, PCL-B will form spontaneously if PylD has a D-amino acid transaminase or D-ornithine:oxygen oxidoreductase-like activity (Figure 16A).
다른 실시양태에서, 에스케리키아 콜라이에서 마우스 EGF 및 마우스 TNF-α의 발현 (도 21 및 실시예 13 및 14)은 TAG 부위에서 혼입된 PCL 또는 피롤리신과의 단백질 혼합물을 생성한다. 피롤리신의 혼입은 pylB 유전자의 존재에 따라 좌우되었지만, PCL 함유 단백질의 균질 제조물은 pylB의 부재시에 관찰되었다 (도 21 및 실시예 11 및 12). 따라서, 이는 피롤리신의 생합성에서 메틸트랜스퍼라제로서의 PylB를 실험적으로 입증한다. 심지어 pylB 유전자의 존재시에, 상대적인 양의 PCL 및 피롤리신을 함유하는 단백질은 전형적으로 더욱 유망한 PCL 단백질에 의한 발효시마다 달라졌다. 이러한 관찰은 메틸트랜스퍼라제 활성 또는 메틸-공여 기질 또는 필요한 보조인자가 에스케리키아 콜라이 및 포유동물 세포에서 효율적 피롤리신 생합성을 위해 제한됨을 제안한다. In another embodiment, expression of mouse EGF and mouse TNF-α in Escherichia coli (Figure 21 and Examples 13 and 14) results in a protein mixture with PCL or pyrrolysine incorporated at the TAG site. The incorporation of pyrrolysine depended on the presence of the pylB gene, but a homogeneous preparation of PCL-containing protein was observed in the absence of pylB (Fig. 21 and Examples 11 and 12). Thus, it experimentally demonstrates PylB as a methyltransferase in the biosynthesis of pyrrolysine. Even in the presence of the pylB gene, proteins containing relative amounts of PCL and pyrrolysine typically differ from fermentation with the more promising PCL protein. These observations suggest that methyltransferase activity or methyl-donating substrates or necessary cofactors are limited for efficient pyrrolysine biosynthesis in Escherichia coli and mammalian cells.
게다가, pylB의 유전자 생성물이 효율적으로 발현되지 않을 수 있다. 따라서, 특정 실시양태에서, 변형된 pylB 유전자는 피롤리신 또는 다른 피롤리신 유사체의 생합성에 사용된다. 따라서, pylB, pylC 및 pylD 유전자 중 하나 이상이 변형된, 에스케리키아 콜라이, 포유동물 및 다른 숙주 세포에서 피롤리신, PCL 및 다른 피롤리신 유사체의 생합성을 위한 방법이 본원에서 제공된다. 이러한 변형은, 다른 유기체, 예컨대 이로 제한되는 것은 아니지만 메타노사르시나에의 다른 종 또는 돌연변이된 유전자로부터의 상동성 유전자를 이용하는 것을 포함할 수 있다. 특정 실시양태에서는 부위 지정 돌연변이유발이 이용되는 반면에, 다른 실시양태에서는 선별과 조합된 랜덤 돌연변이유발을 이용된다. 이러한 방법은 또한 피롤리신, PCL 또는 피롤리신 유사체를 단백질에 혼입시키기 위해 원하는 단백질의 DNA를 첨가하고, pylT 및 pylS 유전자를 포함시키는 것을 포함한다. 특정 실시양태에서, 변형된 pylB 유전자, 천연 pylC, pylD, pylT 및 pylS 유전자를 사용하여, 피롤리신을 생합성적으로 생성하고 혼입시킨다. 다른 실시양태에서, 변형된 pylB 및 pylC 유전자, 천연 pylD, pylT 및 pylS 유전자를 사용하여, PCL 이외의 피롤리신 유사체를 생합성적으로 생성하고 혼입시킨다. 다른 실시양태에서, 변형된 pylB 및 pylD 유전자, 천연 pylC, pylT 및 pylS 유전자를 사용하여, PCL 이외의 피롤리신 유사체를 생합성적으로 생성하고 혼입시킨다. 다른 실시양태에서, pylT, pylS, pylB, pylC 및 pylD 유전자를 임의로 변형시켜, 피롤리신, PCL 또는 다른 피롤리신 유사체의 단백질로의 혼입을 개선시킨다.In addition, the gene product of pylB may not be expressed efficiently. Thus, in certain embodiments, the modified pylB gene is used for biosynthesis of pyrrolysine or other pyrrolysine analogs. Accordingly, provided herein are methods for the biosynthesis of pyrrolysine, PCL and other pyrrolysine analogs in Escherichia coli, mammals and other host cells, wherein one or more of the pylB, pylC and pylD genes have been modified. Such modifications may include the use of homologous genes from other organisms, such as, but not limited to, other species of metanosarcinae or mutated genes. In certain embodiments, site-directed mutagenesis is used, while in other embodiments random mutagenesis in combination with selection is used. This method also includes adding the DNA of the desired protein to incorporate pyrrolysine, PCL or a pyrrolysine analog into the protein, and including the pylT and pylS genes. In certain embodiments, the modified pylB gene, the native pylC, pylD, pylT and pylS genes are used to biosynthetize and incorporate pyrrolysine. In other embodiments, pyrrolysine analogs other than PCL are biosynthetically generated and incorporated using the modified pylB and pylC genes, the native pylD, pylT and pylS genes. In other embodiments, pyrrolysine analogs other than PCL are biosynthetically generated and incorporated using the modified pylB and pylD genes, the native pylC, pylT and pylS genes. In other embodiments, the pylT, pylS, pylB, pylC and pylD genes are optionally modified to improve incorporation of pyrrolysine, PCL or other pyrrolysine analogs into the protein.
게다가, 특정 실시양태의 경우, D-오르니틴으로부터 피롤리신, PCL 및/또는 다른 피롤리신 유사체의 생합성 또는 피롤리신의 생합성에서 중간체의 형성은 숙주 효소 및 단백질의 기능에 의해 제한될 수 있다. 특정 실시양태에서, 하나 이상의 숙주 효소의 낮은 활성 또는 농도는 피롤리신, PCL 또는 다른 피롤리신 유사체의 생합성에 필요한 중간체의 형성을 제한할 수 있다. 특정 실시양태에서, 숙주 효소의 활성은 피롤리신, PCL 또는 다른 피롤리신 유사체를 유도하는 경로로부터 다른 대사 경로로 중간체를 전환시킬 수 있거나, 상기 중간체의 형성을 제한할 수 있다. 따라서, 하나 이상의 숙주 효소가 변형된, 에스케리키아 콜라이, 포유동물 또는 다른 숙주 세포에서 피롤리신, PCL 및 다른 피롤리신 유사체의 생합성을 위한 방법이 본원에 제공된다. 이러한 방법에는 유전적 또는 화학적 수단에 의한 이러한 숙주 효소의 과다발현, 활성화, 저해 또는 억제, 이러한 숙주 효소를 코딩하는 DNA의 첨가, mRNA 번역을 저해하기 위해 침묵 RNA (siRNA)의 첨가, 및 D-오르니틴으로부터 상기 중간체의 형성에 필요한 보조인자의 첨가가 포함되나, 이에 제한되지는 않는다. Furthermore, for certain embodiments, the biosynthesis of pyrrolysine, PCL and/or other pyrrolysine analogs from D-ornithine or the formation of intermediates in the biosynthesis of pyrrolysine may be limited by the function of host enzymes and proteins. . In certain embodiments, the low activity or concentration of one or more host enzymes may limit the formation of intermediates required for the biosynthesis of pyrrolysine, PCL or other pyrrolysine analogs. In certain embodiments, the activity of the host enzyme can convert intermediates from pathways leading to pyrrolysine, PCL or other pyrrolysine analogs to other metabolic pathways, or can limit the formation of such intermediates. Accordingly, provided herein are methods for the biosynthesis of pyrrolysine, PCL and other pyrrolysine analogs in Escherichia coli, mammals or other host cells, in which one or more host enzymes have been modified. These methods include overexpression, activation, inhibition or inhibition of such host enzymes by genetic or chemical means, addition of DNA encoding these host enzymes, addition of silencing RNA (siRNA) to inhibit mRNA translation, and D- The addition of cofactors necessary for the formation of the intermediate from ornithine is included, but is not limited thereto.
TAG 코딩된 부위에서 생합성적으로 생성된 PCL의 부위 특이적 혼입은 또한 마우스 IgG1의 Fc 도메인의 4개의 부위에서 (실시예 5 및 도 17A 참조) 및 에리트로포이에틴 (EPO)의 11개의 부위에서 (실시예 6 및 도 17B 참조) 달성되었다. 두 집합의 단백질에 대한 PCL 혼입은 천연 유전자 pylT, pylS, pylC 및 pylD로 감염된 HEK293F 포유동물 세포에서 전구체로서 배지에 첨가된 D-오르니틴을 사용하여 달성되었다. 도 17A는 4개의 전장 (FL) mFc 단백질을 도시하는 SDS-PAGE이며, 여기서 PCL은 mFc, pCMVpylT, pCMVpylS, pCMVpylC 및 pCMVpylD (도 4A)의 각각의 TAG 돌연변이체 구축물로 동시 형질감염된 HEK293F 세포 및 D-오르니틴의 존재 하에서 성장된 세포를 이용하여 마우스 IgG1 Fc 도메인의 4개의 부위에 혼입되었다. 도 17B는 11개의 전장 (FL) mEPO 단백질을 도시하는 SDS-PAGE이며, 여기서 PCL은 mEPO, pCMVpylT, pCMVpylS, pCMVpylC 및 pCMVpylD의 TAG 돌연변이체 구축물로 동시 형질감염된 HEK293F 세포 및 D-오르니틴의 존재 하에서 성장된 세포를 이용하여 마우스 EPO의 11개의 부위에 혼입되었다. Site-specific incorporation of biosynthetically generated PCL at the TAG-encoded site was also carried out at four sites of the Fc domain of mouse IgG1 (see Example 5 and Figure 17A) and at 11 sites of erythropoietin (EPO) ( See Example 6 and Fig. 17B) was achieved. PCL incorporation for both sets of proteins was achieved using D-ornithine added to the medium as a precursor in HEK293F mammalian cells infected with the native genes pylT, pylS, pylC and pylD. Figure 17A is an SDS-PAGE showing four full-length (FL) mFc proteins, where PCL is HEK293F cells and D co-transfected with respective TAG mutant constructs of mFc, pCMVpylT, pCMVpylS, pCMVpylC and pCMVpylD (Figure 4A). -Cells grown in the presence of ornithine were incorporated into four sites of the mouse IgG1 Fc domain. Figure 17B is an SDS-PAGE showing 11 full-length (FL) mEPO proteins, where PCL is in the presence of HEK293F cells and D-ornithine co-transfected with TAG mutant constructs of mEPO, pCMVpylT, pCMVpylS, pCMVpylC and pCMVpylD. The grown cells were incorporated into 11 sites of mouse EPO.
TAG 코딩된 부위에서 생합성적으로 생성된 PCL의 부위 특이적 혼입은 또한 에스케리키아 콜라이 세포를 이용하여 달성되었다 (실시예 7 참조). pylB, pylC, pylD, pylS 및 pylT를 코딩하는 플라스미드, pARA-pylSTBCD를 구축하였다 (도 6A). 인간 지방산 신테타제 (FAS-TE)의 티오에스테라제 도메인의 2개의 부위 (실시예 8 및 도 18 참조), FKBP-12의 1개의 부위 (실시예 9 및 도 19 참조) 및 섬유모세포 성장 인자 21 (FGF21)의 20개의 부위 (실시예 10 및 도 20 참조)에서의 PCL-A (또는 PCL-B) 혼입은 에스케리키아 콜라이 세포를 pARA-pylSTBCD 및 관심 단백질을 위한 유전자를 가진 제2 발현 플라스미드로 형질전환시키고, 단백질 발현 동안 성장 배지에 D-오르니틴을 첨가함으로써 달성되었다. Site-specific incorporation of biosynthetically generated PCL at the TAG-encoded site was also achieved using Escherichia coli cells (see Example 7). A plasmid encoding pylB, pylC, pylD, pylS and pylT, pARA-pylSTBCD, was constructed (Fig. 6A). Two sites of the thioesterase domain of human fatty acid synthetase (FAS-TE) (see Examples 8 and 18), one site of FKBP-12 (see Examples 9 and 19) and fibroblast growth factor Incorporation of PCL-A (or PCL-B) at 20 sites (see Example 10 and FIG. 20) of 21 (FGF21) results in Escherichia coli cells with a second expression of pARA-pylSTBCD and a gene for the protein of interest. Transformed with plasmid and achieved by adding D-ornithine to the growth medium during protein expression.
도 18은 인간 지방산 신테타제 (FAS-TE)의 티오에스테라제의 2개의 부위에서 PCL 혼입을 위한 SDS-PAGE 및 질량 스펙트럼을 도시한다. FAS-TE Tyr2454PCL이 발현되었고, 가용성 및 불용성 단백질 분획 둘 다 Ni-NTA에 의해 정제하였다. FAS-TE Leu2222PCL/Leu2223Ile는 복제 배양물에서 D-오르니틴의 존재 또는 부재하에 발현되었다. Ni-NTA 용리가 겔 상에 도시된다. 각각에 대해 수득된 질량은 예상된 것과 일치한다. 도 19는 FKBP-12의 1개의 부위에서 PCL 혼입을 위한 SDS-PAGE 및 질량 스펙트럼을 도시한다. 수득된 질량 (12085.6 Da)은 PCL의 단일 부위 혼입을 위해 예상된 것 (12084 Da)과 일치한다. 또한, FKBP12-Ile90PCL의 결정이 도 19에 도시된다. 도 20은 다중 부위에서 FGF21 내로 PCL의 혼입을 보여주는 SDS-PAGE를 도시한다. 18 shows SDS-PAGE and mass spectra for PCL incorporation at two sites of thioesterase of human fatty acid synthetase (FAS-TE). FAS-TE Tyr2454PCL was expressed and both soluble and insoluble protein fractions were purified by Ni-NTA. FAS-TE Leu2222PCL/Leu2223Ile was expressed in clone culture with or without D-ornithine. Ni-NTA elution is shown on the gel. The mass obtained for each is consistent with what is expected. Figure 19 shows SDS-PAGE and mass spectrum for PCL incorporation at one site of FKBP-12. The mass obtained (12085.6 Da) is consistent with that expected for single site incorporation of PCL (12084 Da). Further, the determination of FKBP12-Ile90PCL is shown in Fig. 19. 20 depicts SDS-PAGE showing the incorporation of PCL into FGF21 at multiple sites.
피롤리신 (PYL) 및 PCL의 혼입은 발현계에서 pylB 유전자에 존재에 의존하는 것으로 확인되었다. 도 21A는 TAG 종결 코돈으로 돌연변이된 Gln21의 코돈 (CAA)에 의한 mTNF-α로의 Pyl- 또는 PCL-혼입의 SDS-PAGE 분석을 도시한다 (실시예 13). D-오르니틴이 존재할 때, 레인 2 및 4는 각각 pylB의 존재 및 부재 하에 유사한 단백질 발현 수준을 나타낸다. 레인 3 (pylB 존재) 및 5 (pylB 부재)는 단백질이 D-오르니틴의 부재시에는 발현되지 않음을 보여준다.The incorporation of pyrrolysine (PYL) and PCL was found to depend on the presence of the pylB gene in the expression system. 21A shows SDS-PAGE analysis of Pyl- or PCL-incorporation into mTNF-α by the codon (CAA) of Gln21 mutated to the TAG stop codon (Example 13). When D-ornithine is present,
게다가, mEGF Tyr10TAG는 전구체로서 D-오르니틴을 사용하여 에스케리키아 콜라이에서 pylB, pylC, pylD, pylT 및 pylS 유전자의 존재 하에 발현되었다 (실시예 14). 원형 MS 스펙트럼 (도 21C, 하단)은 PYL 및 PCL이 혼입된 단백질의 혼합물을 제안하였다. 7309 Da에서의 Pyl-함유 mEGF의 MS 피크는 7295 Da에서의 PCL-함유 mEGF의 피크의 대략 6 배만큼 크다. TAG 위치에서의 PYL 및 PCL의 혼입은 mEGF Tyr10TAG 돌연변이 단백질의 N-말단 펩티드 MNSYPGCPSS(PCL)DGYCLNGGVCM (서열 32) 및 MNSYPGCPSS(Pyl)DGYCLNGGVCM (서열 33)의 탠덤 MS 스펙트럼에 의해 검증되었다. 상이한 펩티드의 상대적 전구체 질량 존재도로부터의 정량화에 의해 PYL이 PCL보다 5 내지 10 배 더 풍부한 것으로 나타났고, 이는 원형 질량 측정과 대략 일치한다 (도 21C, 하단). mEGF Tyr10TAG가 pylB의 존재 하에 발현되었을 때, 원형 MS 스펙트럼은 PCL 혼입만을 제안하였다 (도 21C, 상단). In addition, mEGF Tyr10TAG was expressed in the presence of pylB, pylC, pylD, pylT and pylS genes in Escherichia coli using D-ornithine as a precursor (Example 14). The circular MS spectrum (Fig. 21C, bottom) suggested a mixture of proteins in which PYL and PCL were incorporated. The MS peak of the Pyl-containing mEGF at 7309 Da is approximately 6 times as large as the peak of the PCL-containing mEGF at 7295 Da. The incorporation of PYL and PCL at the TAG position was verified by tandem MS spectra of the N-terminal peptides MNSYPGCPSS(PCL )DGYCLNGGVCM (SEQ ID NO:32) and MNSYPGCPSS( Pyl )DGYCLNGGVCM (SEQ ID NO:33) of the mEGF Tyr10TAG mutant protein. Quantification from the relative precursor mass abundance of the different peptides showed that PYL was 5-10 times more abundant than PCL, which is roughly consistent with the circular mass measurements (Figure 21C, bottom). When mEGF Tyr10TAG was expressed in the presence of pylB, the circular MS spectrum suggested only PCL incorporation (Fig. 21C, top).
유사하게, mTNF-α Gln21TAG가 에스케리키아 콜라이에서 pylB, pylC, pylD, pylT 및 pylS 유전자의 존재 하에서 D-오르니틴을 이용하여 발현되었을 때, 탠덤 MS는 mTNF Gln21PCL의 펩티드 NH(Pyl)VEEQLEWLSQR (서열 34) 및 NH(PCL)VEEQLEWLSQR (서열 35)에서 PYL 및 PCL 혼입을 검증하였다. mEGF Tyr10TAG와는 대조적으로, 탠덤 MS에 의해 확인된 상이한 펩티드의 상대적 전구체 질량 존재도로부터의 정량화는, mTNF-α Gln21TAG 단백질에서 PCL이 PYL보다 7 배 더 풍부함을 나타내었다. mTNF-α Gln21TAG는 pylB 유전자의 부재시에 발현되었고, 탠덤 MS 측정에서 상이한 펩티드의 상대적 전구체 질량 존재도로부터의 정량화는 실험의 역동적 범위내에서 PCL 단백질만을 나타낸다. 이러한 실험은 PYL 혼입이 Pyl의 생합성에서 추정적 메틸트랜스퍼라제인 pylB의 존재에 따라 엄격하게 좌우됨을 나타낸다. 그러나, pylB 유전자의 존재 하에 단백질에 혼입된 PCL과 PYL의 상대비는 단백질마다 및 발효 실험마다 달라지는 것으로 보이며, 따라서 발현 벡터의 유형, 성장 조건 및/또는 숙주 세포 배양의 다른 특성에 의존할 수 있다. Similarly, when mTNF-α Gln21TAG was expressed using D-ornithine in the presence of pylB, pylC, pylD, pylT and pylS genes in Escherichia coli, tandem MS showed the peptide NH( Pyl )VEEQLEWLSQR ( SEQ ID NO: 34) and NH ( PCL ) VEEQLEWLSQR (SEQ ID NO: 35) incorporation of PYL and PCL was verified. In contrast to mEGF Tyr10TAG, quantification from the relative precursor mass abundance of the different peptides identified by tandem MS showed that in the mTNF-α Gln21TAG protein, PCL was 7 times more abundant than PYL. mTNF-α Gln21TAG was expressed in the absence of the pylB gene, and quantification from the relative precursor mass abundance of different peptides in tandem MS measurements reveals only the PCL protein within the dynamic range of the experiment. These experiments indicate that PYL incorporation is strictly dependent on the presence of pylB, a putative methyltransferase, in the biosynthesis of Pyl. However, the relative ratio of PCL and PYL incorporated into a protein in the presence of the pylB gene appears to vary from protein to protein and from fermentation experiments, and thus may depend on the type of expression vector, growth conditions and/or other characteristics of the host cell culture. .
하기 화학식 V 및 화학식 VI의 구조를 갖는 아미노산이 본원에 제공되며,Provided herein are amino acids having the structures of Formula V and Formula VI,
<화학식 V><Formula V>
<화학식 VI><Formula VI>
여기서, 상기 화학식 V 또는 화학식 VI의 화합물은 pylB 유전자, pylC 유전자 및 pylD 유전자를 포함하는 세포 내에서 생합성적으로 생성되고, 세포는 전구체를 포함하는 성장 배지와 접촉하고 있다. 한편 다른 실시양태에서, 상기 화학식 V 또는 화학식 VI의 화합물은 pylC 유전자 및 pylD 유전자를 포함하는 세포 내에서 생합성적으로 생성되고, 세포는 전구체를 포함하는 성장 배지와 접촉하고 있다. 이러한 생합성의 특정 실시양태에서, 전구체는 오르니틴 또는 아르기닌이다. 이러한 생합성의 특정 실시양태에서, 전구체는 D-오르니틴 또는 D-아르기닌이다. 이러한 생합성의 특정 실시양태에서, 전구체는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. 이러한 생합성의 다른 실시양태에서, 전구체는 (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산이다. Here, the compound of Formula V or Formula VI is biosynthetically produced in cells containing the pylB gene, the pylC gene, and the pylD gene, and the cells are in contact with the growth medium containing the precursor. Meanwhile, in another embodiment, the compound of Formula V or Formula VI is biosynthetically produced in cells containing the pylC gene and the pylD gene, and the cells are in contact with the growth medium containing the precursor. In certain embodiments of this biosynthesis, the precursor is ornithine or arginine. In certain embodiments of this biosynthesis, the precursor is D-ornithine or D-arginine. In certain embodiments of this biosynthesis, the precursor is (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid. In another embodiment of this biosynthesis, the precursor is (2S)-2-amino-6-(2,5-diaminopentanamido)hexanoic acid.
본원에 또한 제공된 바와 같이, 화학식 V 및 VI의 화합물이 생합성되는 세포는 추가로 pylS 유전자 및 pylT 유전자를 포함하고, 화학식 V 또는 화학식 VI의 화합물은 아미노아실 tRNA 신테타제 및 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하는 tRNA에 의해 세포 내에서 단백질에 혼입된다. 이러한 아미노아실 tRNA 신테타제는 pylS 유전자의 유전자 생성물이고, tRNA는 pylT 유전자의 유전자 생성물이다. 특정 실시양태에서, 상기 화학식 V 또는 화학식 VI의 화합물은 직교 tRNA (O-tRNA) 및 직교 아미노아실 tRNA 신테타제 (O-RS)에 의해 세포 내 단백질에 혼입되며, 여기서 O-RS는 화학식 V 또는 화학식 VI의 화합물에 의해 O-tRNA를 아미노아실화시키고, O-tRNA는 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하였다.As also provided herein, the cells in which the compounds of formulas V and VI are biosynthesized further comprise a pylS gene and a pylT gene, and the compounds of formula V or VI are aminoacyl tRNA synthetase and one or more selectors of mRNA in cells. It is incorporated into the protein within the cell by the tRNA that recognizes the codon. This aminoacyl tRNA synthetase is a gene product of the pylS gene, and tRNA is a gene product of the pylT gene. In certain embodiments, the compound of Formula V or Formula VI is incorporated into an intracellular protein by an orthogonal tRNA (O-tRNA) and an orthogonal aminoacyl tRNA synthetase (O-RS), wherein O-RS is Formula V or The O-tRNA was aminoacylated by the compound of Formula VI, and the O-tRNA recognized one or more selector codons of the mRNA in the cell.
피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물의 혼입을 위한 셀렉터 코돈은 앰버 코돈 (TAG)이다. The selector codon for the incorporation of pyrrolysine, PCL and/or other pyrrolysine analogs such as compounds of formula V and formula VI is the amber codon (TAG).
피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물의 생합성 및/또는 혼입을 위해 사용된 세포는 원핵 세포 또는 진핵 세포이다. 특정 실시양태에서, 원핵 세포에는 에스케리키아 콜라이, 미코박테리움 스메그마티스, 락토코쿠스 락티스 및 바실러스 서브틸리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서 진핵 세포에는 포유동물 세포, 효모 세포, 진균 세포, 식물 세포 또는 곤충 세포가 포함되나, 이에 제한되지는 않는다. 이러한 포유동물 세포에는 인간 배아 신장 (HEK293F) 세포, 인간 상피양 암종 (HeLa 및 GH3) 세포, 원숭이 신장 (COS) 세포, 래트 C6 신경교종 세포, 새끼 햄스터 신장 (BHK-21) 세포 및 차이니즈 햄스터 난소 (CHO) 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 효모 세포에는 사카로미세스 세레비지애 및 피키아 파스토리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 곤충 세포에는 스포도프테라 프루기페르다 sf9 및 sf21 세포, 트리코플루시아 니 (BTI TN-5B1-4 또는 하이-파이브™) 세포 및 맘메스트라 브라시카에 세포가 포함되나, 이에 제한되지는 않는다. Cells used for biosynthesis and/or incorporation of pyrrolysine, PCL and/or other pyrrolysine analogs, such as compounds of formula V and formula VI, are prokaryotic or eukaryotic cells. In certain embodiments, prokaryotic cells include, but are not limited to, Escherichia coli, Mycobacterium smegmatis, Lactococcus lactis, and Bacillus subtilis cells. In other embodiments eukaryotic cells include, but are not limited to, mammalian cells, yeast cells, fungal cells, plant cells, or insect cells. These mammalian cells include human embryonic kidney (HEK293F) cells, human epithelial carcinoma (HeLa and GH3) cells, monkey kidney (COS) cells, rat C6 glioma cells, baby hamster kidney (BHK-21) cells, and Chinese hamster ovary. (CHO) cells are included, but are not limited thereto. In certain embodiments, yeast cells include, but are not limited to, Saccharomyces cerevisiae and Pichia pastoris cells. In other embodiments, the insect cells include Spodoptera frugiperda sf9 and sf21 cells, Trichoflucia ni (BTI TN-5B1-4 or High-Five™) cells, and Mammestra brassicae cells, It is not limited.
본원에 제공된 또 다른 측면에서, 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물은 전구체를 포함하는 성장 배지와 접촉하고 있는 피더 세포에서 생합성되고, 피더 세포는 pylB 유전자, pylC 유전자 및 pylD 유전자를 함유한다. 다른 실시양태에서, 피롤리신, PCL 및/또는 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물은 전구체를 포함하는 성장 배지와 접촉하고 있는 피더 세포에서 생합성되고, 피더 세포는 pylC 유전자 및 pylD 유전자를 함유한다. 이러한 생합성된 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물은 피더 세포로부터 성장 배지로 분비되고, 이로써 이들은 제2 세포에 의해 취해지고, 후속적으로 제2 세포 내에서 합성된 단백질에 혼입된다. 이러한 제2 세포는 pylS 유전자 및 pylT 유전자를 함유한다. 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물은 아미노아실 tRNA 신테타제 및 제2 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하는 tRNA에 의해 제2 세포 내에서 단백질에 혼입된다. 이러한 아미노아실 tRNA 신테타제는 pylS 유전자의 유전자 생성물이고, tRNA는 pylT 유전자의 유전자 생성물이다. 특정 실시양태에서, 화학식 V 또는 화학식 VI의 상기 화합물은 직교 tRNA (O-tRNA) 및 직교 아미노아실 tRNA 신테타제 (O-RS)에 의해 제2 세포 내 단백질에 혼입되며, 여기서 O-RS는 화학식 V 또는 화학식 VI의 화합물에 의해 O-tRNA를 아미노아실화시키고, O-tRNA는 제2 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하였다. In another aspect provided herein, pyrrolysine, PCL and/or other pyrrolysine analogs, such as compounds of Formula V and Formula VI, are biosynthesized in a feeder cell in contact with a growth medium comprising a precursor, and the feeder cell is It contains the pylB gene, the pylC gene and the pylD gene. In other embodiments, pyrrolysine, PCL and/or pyrrolysine analogs, such as compounds of Formula V and Formula VI, are biosynthesized in a feeder cell in contact with a growth medium comprising a precursor, and the feeder cell is the pylC gene and pylD Contains genes. These biosynthesized pyrrolysine, PCL and/or other pyrrolysine analogs, such as compounds of formula V and formula VI, are secreted from feeder cells into the growth medium, whereby they are taken up by the second cell and subsequently second It is incorporated into the protein synthesized in the cell. This second cell contains the pylS gene and the pylT gene. Pyrrolysine, PCL and/or other pyrrolysine analogs, such as compounds of Formula V and Formula VI, are in a second cell by means of an aminoacyl tRNA synthetase and a tRNA that recognizes one or more selector codons of the mRNA in the second cell. It is incorporated into the protein. This aminoacyl tRNA synthetase is a gene product of the pylS gene, and tRNA is a gene product of the pylT gene. In certain embodiments, the compound of Formula V or Formula VI is incorporated into a second intracellular protein by an orthogonal tRNA (O-tRNA) and an orthogonal aminoacyl tRNA synthetase (O-RS), wherein O-RS is the formula O-tRNA was aminoacylated by a compound of V or Formula VI, and the O-tRNA recognized one or more selector codons of the mRNA in the second cell.
피더 세포를 이용하는 이러한 생합성의 특정 실시양태에서, 전구체는 오르니틴 또는 아르기닌이다. 이러한 생합성의 특정 실시양태에서, 전구체는 D-오르니틴 또는 D-아르기닌이다. 특정 실시양태에서, 전구체는 (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산이다. 특정 실시양태에서, 전구체는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. In certain embodiments of such biosynthesis using feeder cells, the precursor is ornithine or arginine. In certain embodiments of this biosynthesis, the precursor is D-ornithine or D-arginine. In certain embodiments, the precursor is (2S)-2-amino-6-(2,5-diaminopentanamido)hexanoic acid. In certain embodiments, the precursor is (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid.
피더 세포를 이용하여 생합성된 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물의 혼입을 위한 셀렉터 코돈은 앰버 코돈 (TAG)이다. The selector codon for the incorporation of pyrrolysine, PCL and/or other pyrrolysine analogs, such as compounds of formula V and formula VI biosynthesized using feeder cells, is the amber codon (TAG).
특정 실시양태에서, 제2 세포는 피더 세포와 동일한 유형의 세포이고, 한편 다른 실시양태에서, 제2 세포는 피더 세포와는 상이한 세포 유형이다. 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물의 이러한 생합성 및/또는 혼입을 위해 사용된 세포는 원핵 세포 또는 진핵 세포이다. 특정 실시양태에서, 원핵 세포에는 에스케리키아 콜라이, 미코박테리움 스메그마티스, 락토코쿠스 락티스 및 바실러스 서브틸리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서 진핵 세포에는 포유동물 세포, 효모 세포, 진균 세포, 식물 세포 또는 곤충 세포가 포함되나, 이에 제한되지는 않는다. 이러한 포유동물 세포에는 인간 배아 신장 (HEK293F) 세포, 인간 상피양 암종 (HeLa 및 GH3) 세포, 원숭이 신장 (COS) 세포, 래트 C6 신경교종 세포, 새끼 햄스터 신장 (BHK-21) 세포 및 차이니즈 햄스터 난소 (CHO) 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 효모 세포에는 사카로미세스 세레비지애 및 피키아 파스토리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 곤충 세포에는 스포도프테라 프루기페르다 sf9 및 sf21 세포, 트리코플루시아 니 (BTI TN-5B1-4 또는 하이-파이브™) 세포 및 맘메스트라 브라시카에 세포가 포함되나, 이에 제한되지는 않는다. In certain embodiments, the second cells are of the same type as the feeder cells, while in other embodiments, the second cells are of a different cell type than the feeder cells. The cells used for this biosynthesis and/or incorporation of pyrrolysine, PCL and/or other pyrrolysine analogs such as compounds of formula V and formula VI are prokaryotic or eukaryotic cells. In certain embodiments, prokaryotic cells include, but are not limited to, Escherichia coli, Mycobacterium smegmatis, Lactococcus lactis, and Bacillus subtilis cells. In other embodiments eukaryotic cells include, but are not limited to, mammalian cells, yeast cells, fungal cells, plant cells, or insect cells. These mammalian cells include human embryonic kidney (HEK293F) cells, human epithelial carcinoma (HeLa and GH3) cells, monkey kidney (COS) cells, rat C6 glioma cells, baby hamster kidney (BHK-21) cells, and Chinese hamster ovary. (CHO) cells are included, but are not limited thereto. In certain embodiments, yeast cells include, but are not limited to, Saccharomyces cerevisiae and Pichia pastoris cells. In other embodiments, the insect cells include Spodoptera frugiperda sf9 and sf21 cells, Trichoflucia ni (BTI TN-5B1-4 or High-Five™) cells, and Mammestra brassicae cells, It is not limited.
본원에 제공된 또 다른 측면에서, 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물은 피더 세포에서 생합성된 다음, 이러한 생합성된 피롤리신 및/또는 PCL, 예컨대 화학식 V 및 화학식 VI의 화합물은 피더 세포 배양물로부터 정제되고, 제2 세포를 함유하는 제2 배양물의 성장 배지에 첨가된다. 이어서, 이러한 정제된 피롤리신, PCL 및/또는 다른 피롤리신 유사체는 제2 세포 내에서 합성된 단백질에 혼입된다. 특정 실시양태에서, 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물은 전구체를 포함하는 성장 배지와 접촉하고 있는 피더 세포에서 생합성되고, 피더 세포는 pylB 유전자, pylC 유전자 및 pylD 유전자를 함유한다. 다른 실시양태에서, 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물은 전구체를 포함하는 성장 배지와 접촉하고 있는 피더 세포에서 생합성되고, 피더 세포는 pylC 유전자 및 pylD 유전자를 함유한다. 이 측면에서 사용된 제2 세포는 pylS 유전자 및 pylT 유전자를 함유한다. 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물은 아미노아실 tRNA 신테타제 및 제2 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하는 tRNA에 의해 제2 세포 내에서 단백질에 혼입된다. 이러한 아미노아실 tRNA 신테타제는 pylS 유전자의 유전자 생성물이고, tRNA는 pylT 유전자의 유전자 생성물이다. 특정 실시양태에서, 상기 화학식 V 또는 화학식 VI의 화합물은 직교 tRNA (O-tRNA) 및 직교 아미노아실 tRNA 신테타제 (O-RS)에 의해 제2 세포 내 단백질에 혼입되며, 여기서 O-RS는 화학식 V 또는 화학식 VI의 화합물에 의해 O-tRNA를 아미노아실화시키고, O-tRNA는 제2 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하였다. In another aspect provided herein, pyrrolysine, PCL and/or other pyrrolysine analogs, such as compounds of Formula V and Formula VI, are biosynthesized in feeder cells, and then such biosynthesized pyrrolysine and/or PCL, such as The compounds of formula (V) and (VI) are purified from the feeder cell culture and added to the growth medium of the second culture containing the second cells. These purified pyrrolysine, PCL and/or other pyrrolysine analogs are then incorporated into the synthesized protein in the second cell. In certain embodiments, pyrrolysine, PCL and/or other pyrrolysine analogs, such as compounds of Formula V and Formula VI, are biosynthesized in a feeder cell in contact with a growth medium comprising a precursor, and the feeder cell is a pylB gene, It contains the pylC gene and the pylD gene. In other embodiments, pyrrolysine, PCL and/or other pyrrolysine analogs, such as compounds of Formula V and Formula VI, are biosynthesized in a feeder cell in contact with a growth medium comprising a precursor, and the feeder cell comprises a pylC gene and It contains the pylD gene. The second cell used in this aspect contains the pylS gene and the pylT gene. Pyrrolysine, PCL and/or other pyrrolysine analogs, such as compounds of Formula V and Formula VI, are in a second cell by means of an aminoacyl tRNA synthetase and a tRNA that recognizes one or more selector codons of the mRNA in the second cell. It is incorporated into the protein. This aminoacyl tRNA synthetase is a gene product of the pylS gene, and tRNA is a gene product of the pylT gene. In certain embodiments, the compound of Formula V or Formula VI is incorporated into a second intracellular protein by an orthogonal tRNA (O-tRNA) and an orthogonal aminoacyl tRNA synthetase (O-RS), wherein O-RS is the formula O-tRNA was aminoacylated by a compound of V or Formula VI, and the O-tRNA recognized one or more selector codons of the mRNA in the second cell.
피더 세포를 이용하는 이러한 생합성의 특정 실시양태에서, 전구체는 오르니틴 또는 아르기닌이다. 이러한 생합성의 특정 실시양태에서, 전구체는 D-오르니틴 또는 D-아르기닌이다. 특정 실시양태에서, 전구체는 (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산이다. 특정 실시양태에서, 전구체는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산이다. In certain embodiments of such biosynthesis using feeder cells, the precursor is ornithine or arginine. In certain embodiments of this biosynthesis, the precursor is D-ornithine or D-arginine. In certain embodiments, the precursor is (2S)-2-amino-6-(2,5-diaminopentanamido)hexanoic acid. In certain embodiments, the precursor is (2S)-2-amino-6-((R)-2,5-diaminopentanamido)hexanoic acid.
피더 세포를 이용하여 생합성된 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물의 혼입을 위한 셀렉터 코돈은 앰버 코돈 (TAG)이다. The selector codon for the incorporation of pyrrolysine, PCL and/or other pyrrolysine analogs, such as compounds of formula V and formula VI biosynthesized using feeder cells, is the amber codon (TAG).
특정 실시양태에서, 제2 세포는 피더 세포와 동일한 유형의 세포이고, 한편 다른 실시양태에서, 제2 세포는 피더 세포와는 상이한 세포 유형이다. 피롤리신, PCL 및/또는 다른 피롤리신 유사체, 예컨대 화학식 V 및 화학식 VI의 화합물의 이러한 생합성 및/또는 혼입에 사용되는 세포는 원핵 세포 또는 진핵 세포이다. 특정 실시양태에서, 원핵 세포에는 에스케리키아 콜라이, 미코박테리움 스메그마티스, 락토코쿠스 락티스 및 바실러스 서브틸리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서 진핵 세포에는 포유동물 세포, 효모 세포, 진균 세포, 식물 세포 또는 곤충 세포가 포함되나, 이에 제한되지는 않는다. 이러한 포유동물 세포에는 인간 배아 신장 (HEK293F) 세포, 인간 상피양 암종 (HeLa 및 GH3) 세포, 원숭이 신장 (COS) 세포, 래트 C6 신경교종 세포, 새끼 햄스터 신장 (BHK-21) 세포 및 차이니즈 햄스터 난소 (CHO) 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 효모 세포에는 사카로미세스 세레비지애 및 피키아 파스토리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 곤충 세포에는 스포도프테라 프루기페르다 sf9 및 sf21 세포, 트리코플루시아 니 (BTI TN-5B1-4 또는 하이-파이브™) 세포 및 맘메스트라 브라시카에 세포가 포함되나, 이에 제한되지는 않는다. In certain embodiments, the second cells are of the same type as the feeder cells, while in other embodiments, the second cells are of a different cell type than the feeder cells. The cells used for this biosynthesis and/or incorporation of pyrrolysine, PCL and/or other pyrrolysine analogs such as compounds of formula V and formula VI are prokaryotic or eukaryotic cells. In certain embodiments, prokaryotic cells include, but are not limited to, Escherichia coli, Mycobacterium smegmatis, Lactococcus lactis, and Bacillus subtilis cells. In other embodiments eukaryotic cells include, but are not limited to, mammalian cells, yeast cells, fungal cells, plant cells, or insect cells. These mammalian cells include human embryonic kidney (HEK293F) cells, human epithelial carcinoma (HeLa and GH3) cells, monkey kidney (COS) cells, rat C6 glioma cells, baby hamster kidney (BHK-21) cells, and Chinese hamster ovary. (CHO) cells are included, but are not limited thereto. In certain embodiments, yeast cells include, but are not limited to, Saccharomyces cerevisiae and Pichia pastoris cells. In other embodiments, the insect cells include Spodoptera frugiperda sf9 and sf21 cells, Trichoflucia ni (BTI TN-5B1-4 or High-Five™) cells, and Mammestra brassicae cells, It is not limited.
또한, 하기 화학식 VII의 구조를 갖는 아미노산이 본원에 제공되며,In addition, an amino acid having the structure of the following formula (VII) is provided herein,
<화학식 VII><Formula VII>
여기서, 화학식 VII의 화합물, 또는 그의 이성질체 또는 호변이성질체는 pylB 유전자, pylC 유전자 및 pylD 유전자를 함유하는 세포 내에서 생합성적으로 생성되고, 세포는 D-오르니틴 또는 D-아르기닌 또는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산 또는 2,5-디아미노-3-메틸펜탄산 또는 (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산을 함유하는 성장 배지와 접촉하고 있다. 한편 다른 실시양태에서, 이러한 화학식 VII의 화합물은 pylC 유전자 및 pylD 유전자를 함유하는 세포 내에서 생합성적으로 생성되고, 세포는 2,5-디아미노-3-메틸펜탄산을 함유하는 성장 배지와 접촉하고 있다. 특정 실시양태에서, 세포는 D-2,5-디아미노-3-메틸펜탄산을 함유하는 성장 배지와 접촉하고 있다. 특정 실시양태에서, 2,5-디아미노-3-메틸펜탄산은 (2R,3S)-2,5-디아미노-3-메틸펜탄산이다. 특정 실시양태에서, 2,5-디아미노-3-메틸펜탄산은 (2R,3R)-2,5-디아미노-3-메틸펜탄산이다.Here, the compound of Formula VII, or an isomer or tautomer thereof, is biosynthetically produced in cells containing the pylB gene, the pylC gene and the pylD gene, and the cell is D-ornithine or D-arginine or (2S)-2 -Amino-6-((R)-2,5-diaminopentanamido)hexanoic acid or 2,5-diamino-3-methylpentanoic acid or (2S)-2-amino-6-(2,5 It is in contact with a growth medium containing -diaminopentanamido)hexanoic acid. Meanwhile, in another embodiment, such a compound of Formula VII is produced biosynthetically in cells containing the pylC gene and the pylD gene, and the cells are contacted with a growth medium containing 2,5-diamino-3-methylpentanoic acid. I'm doing it. In certain embodiments, the cells are contacted with a growth medium containing D-2,5-diamino-3-methylpentanoic acid. In certain embodiments, the 2,5-diamino-3-methylpentanoic acid is (2R,3S)-2,5-diamino-3-methylpentanoic acid. In certain embodiments, the 2,5-diamino-3-methylpentanoic acid is (2R,3R)-2,5-diamino-3-methylpentanoic acid.
화학식 VII의 화합물이 생합성적으로 생성되는 세포는 추가로 pylS 유전자 및 pylT 유전자를 포함하고, 화학식 VII의 화합물은 아미노아실 tRNA 신테타제 및 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하는 tRNA에 의해 세포 내에서 단백질에 혼입된다. 이러한 아미노아실 tRNA 신테타제는 pylS 유전자의 유전자 생성물이고, tRNA는 pylT 유전자의 유전자 생성물이다. 특정 실시양태에서, 상기 화학식 VII의 화합물은 직교 tRNA (O-tRNA) 및 직교 아미노아실 tRNA 신테타제 (O-RS)에 의해 세포 내 단백질에 혼입되며, 여기서 O-RS는 화학식 VII의 화합물에 의해 O-tRNA를 아미노아실화시키고, O-tRNA는 세포에서 mRNA의 하나 이상의 셀렉터 코돈을 인식하였다. Cells in which the compound of Formula VII is biosynthetically produced further contains the pylS gene and the pylT gene, and the compound of Formula VII is intracellular by aminoacyl tRNA synthetase and tRNA that recognizes one or more selector codons of mRNA in the cell. Is incorporated into the protein. This aminoacyl tRNA synthetase is a gene product of the pylS gene, and tRNA is a gene product of the pylT gene. In certain embodiments, the compound of formula VII is incorporated into an intracellular protein by an orthogonal tRNA (O-tRNA) and an orthogonal aminoacyl tRNA synthetase (O-RS), wherein the O-RS is by the compound of formula VII. The O-tRNA was aminoacylated, and the O-tRNA recognized one or more selector codons of the mRNA in the cell.
화학식 VII의 화합물의 혼입을 위한 셀렉터 코돈은 앰버 코돈 (TAG)이다. The selector codon for incorporation of the compound of formula VII is the amber codon (TAG).
화학식 VII의 화합물의 생합성 및/또는 혼입을 위해 사용된 세포는 원핵 세포 또는 진핵 세포이다. 특정 실시양태에서, 원핵 세포에는 에스케리키아 콜라이, 미코박테리움 스메그마티스, 락토코쿠스 락티스 및 바실러스 서브틸리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서 진핵 세포에는 포유동물 세포, 효모 세포, 진균 세포, 식물 세포 또는 곤충 세포가 포함되나, 이에 제한되지는 않는다. 이러한 포유동물 세포에는 인간 배아 신장 (HEK293F) 세포, 인간 상피양 암종 (HeLa 및 GH3) 세포, 원숭이 신장 (COS) 세포, 래트 C6 신경교종 세포, 새끼 햄스터 신장 (BHK-21) 세포 및 차이니즈 햄스터 난소 (CHO) 세포가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 효모 세포에는 사카로미세스 세레비지애 및 피키아 파스토리스 세포가 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 곤충 세포에는 스포도프테라 프루기페르다 sf9 및 sf21 세포, 트리코플루시아 니 (BTI TN-5B1-4 또는 하이-파이브™) 세포 및 맘메스트라 브라시카에 세포가 포함되나, 이에 제한되지는 않는다. Cells used for biosynthesis and/or incorporation of the compounds of formula VII are prokaryotic or eukaryotic cells. In certain embodiments, prokaryotic cells include, but are not limited to, Escherichia coli, Mycobacterium smegmatis, Lactococcus lactis, and Bacillus subtilis cells. In other embodiments eukaryotic cells include, but are not limited to, mammalian cells, yeast cells, fungal cells, plant cells, or insect cells. These mammalian cells include human embryonic kidney (HEK293F) cells, human epithelial carcinoma (HeLa and GH3) cells, monkey kidney (COS) cells, rat C6 glioma cells, baby hamster kidney (BHK-21) cells, and Chinese hamster ovary. (CHO) cells are included, but are not limited thereto. In certain embodiments, yeast cells include, but are not limited to, Saccharomyces cerevisiae and Pichia pastoris cells. In other embodiments, the insect cells include Spodoptera frugiperda sf9 and sf21 cells, Trichoflucia ni (BTI TN-5B1-4 or High-Five™) cells, and Mammestra brassicae cells, It is not limited.
특정 실시양태에서, 1개 이상의 피롤리신, PCL 및/또는 다른 피롤리신 유사체가 본원에 제공된 방법을 이용하여 단백질, 폴리펩티드 및/또는 펩티드에 혼입되고, 이러한 피롤리신 및/또는 PCL은 본원에 제공된 방법을 이용하여 유도체화된다. In certain embodiments, one or more pyrrolysine, PCL and/or other pyrrolysine analogs are incorporated into proteins, polypeptides and/or peptides using the methods provided herein, and such pyrrolysines and/or PCLs are incorporated herein. It is derivatized using the method provided in.
PCLPCL 및 피롤리신의 And of pyrrolysine 유도체화Derivatization
하기 화학식 I에 따른 구조를 갖는 단백질, 폴리펩티드 또는 펩티드가 본원에 제공된다.Provided herein are proteins, polypeptides or peptides having a structure according to Formula I below.
<화학식 I><Formula I>
상기 식에서,In the above formula,
R1은 H 또는 아미노 말단 변형 기이고;R 1 is H or an amino terminal modified group;
R2는 OH 또는 카르복시 말단 변형 기이고;R 2 is OH or a carboxy terminal modified group;
n은 1 내지 5000의 정수이고;n is an integer from 1 to 5000;
각각의 AA는 아미노산 잔기, 하기 화학식 A-1의 구조를 갖는 잔기, 및 하기 화학식 B-1의 구조를 갖는 잔기로부터 독립적으로 선택되고;Each AA is independently selected from an amino acid residue, a residue having the structure of formula A-1, and a residue having the structure of formula B-1;
, 여기서 , here
R6은 H 또는 C1알킬이고, 1개 이상의 AA는 화학식 A-1 또는 화학식 B-1의 구조를 갖는 피롤리신 또는 피롤리신 유사체, 또는 그의 이성질체이다. R 6 is H or C 1 alkyl, and at least one AA is a pyrrolysine or pyrrolysine analog having the structure of formula A-1 or B-1, or an isomer thereof.
이러한 화학식 I의 화합물의 특정 실시양태에서, R6은 H이고, 화학식 A-1의 구조를 갖는 잔기 및 화학식 B-1의 구조를 갖는 잔기는 각각 하기 화학식 A-2 및 화학식 B-2의 구조를 갖는 잔기, 또는 그의 이성질체이다.In certain embodiments of these compounds of formula I, R 6 is H, and the moiety having the structure of formula A-1 and the moiety having the structure of formula B-1 are each of the following structures A-2 and B-2: Is a moiety having, or an isomer thereof.
따라서, 하기 화학식 I에 따른 구조를 갖는 단백질, 폴리펩티드 또는 펩티드가 또한 본원에 제공된다.Accordingly, proteins, polypeptides or peptides having a structure according to formula (I) below are also provided herein.
<화학식 I><Formula I>
상기 식에서,In the above formula,
R1은 H 또는 아미노 말단 변형 기이고;R 1 is H or an amino terminal modified group;
R2는 OH 또는 카르복시 말단 변형 기이고;R 2 is OH or a carboxy terminal modified group;
n은 1 내지 5000의 정수이고;n is an integer from 1 to 5000;
각각의 AA는 아미노산 잔기, 화학식 A-2의 구조를 갖는 잔기 및 화학식 B-2의 구조를 갖는 잔기로부터 독립적으로 선택되고;Each AA is independently selected from an amino acid residue, a residue having the structure of formula A-2, and a residue having the structure of formula B-2;
1개 이상의 AA는 화학식 A-2 또는 화학식 B-2의 구조를 갖는 피롤리신 또는 피롤리신 유사체, 또는 그의 이성질체이다. At least one AA is a pyrrolysine or pyrrolysine analog having the structure of Formula A-2 or Formula B-2, or an isomer thereof.
특정 실시양태에서, n은 1 내지 4000의 정수이다. 특정 실시양태에서 n은 1 내지 3000의 정수이다. 특정 실시양태에서 n은 1 내지 2000의 정수이다. 특정 실시양태에서 n은 1 내지 1000의 정수이다. 특정 실시양태에서 n은 1 내지 700의 정수이다. 특정 실시양태에서 n은 1 내지 800의 정수이다. 특정 실시양태에서 n은 1 내지 600의 정수이다. 특정 실시양태에서 n은 1 내지 500의 정수이다. 특정 실시양태에서 n은 1 내지 400의 정수이다. 특정 실시양태에서 n은 1 내지 300의 정수이다. 특정 실시양태에서 n은 1 내지 200의 정수이다. 특정 실시양태에서 n은 1 내지 100의 정수이다. 특정 실시양태에서 n은 1 내지 90의 정수이다. 특정 실시양태에서 n은 1 내지 80의 정수이다. 특정 실시양태에서 n은 1 내지 70의 정수이다. 특정 실시양태에서 n은 1 내지 60의 정수이다. 특정 실시양태에서 n은 1 내지 50의 정수이다. 특정 실시양태에서 n은 1 내지 40의 정수이다. 특정 실시양태에서 n은 1 내지 30의 정수이다. 특정 실시양태에서 n은 1 내지 20의 정수이다. 특정 실시양태에서 n은 1 내지 10의 정수이다. 특정 실시양태에서 n은 1 내지 5의 정수이다. In certain embodiments, n is an integer from 1 to 4000. In certain embodiments n is an integer from 1 to 3000. In certain embodiments n is an integer from 1 to 2000. In certain embodiments n is an integer from 1 to 1000. In certain embodiments n is an integer from 1 to 700. In certain embodiments n is an integer from 1 to 800. In certain embodiments n is an integer from 1 to 600. In certain embodiments n is an integer from 1 to 500. In certain embodiments n is an integer from 1 to 400. In certain embodiments n is an integer from 1 to 300. In certain embodiments n is an integer from 1 to 200. In certain embodiments n is an integer from 1 to 100. In certain embodiments n is an integer from 1 to 90. In certain embodiments n is an integer from 1 to 80. In certain embodiments n is an integer from 1 to 70. In certain embodiments n is an integer from 1 to 60. In certain embodiments n is an integer from 1 to 50. In certain embodiments n is an integer from 1 to 40. In certain embodiments n is an integer from 1 to 30. In certain embodiments n is an integer from 1 to 20. In certain embodiments n is an integer from 1 to 10. In certain embodiments n is an integer from 1 to 5.
또한, 1개 이상의 피롤리신 및/또는 PCL 잔기를 함유하는 화학식 I의 단백질, 폴리펩티드 및/또는 펩티드를 하기 화학식 III의 구조를 갖는 시약과 혼합하는 것을 포함하는, 화학식 I의 단백질, 폴리펩티드 및/또는 펩티드의 부위 특이적 표지화를 위한 방법이 본원에 제공된다.In addition, proteins, polypeptides and/or proteins of formula I comprising mixing a protein, polypeptide and/or peptide of formula I containing at least one pyrrolysine and/or PCL moiety with a reagent having the structure of formula III Alternatively, methods for site-specific labeling of peptides are provided herein.
<화학식 III><Formula III>
상기 식에서,In the above formula,
R3, R5 및 각각의 R4는 H, -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택되고;R 3, R 5 and each R 4 is H, -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, , Aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1 are independently selected;
L은 결합, C1 - 8알킬렌, 할로-치환된-C1 - 8알킬렌, 히드록시-치환된-C1 - 8알킬렌, C2-8알케닐렌, 할로-치환된-C2 - 8알케닐렌, 히드록시-치환된-C2 - 8알케닐렌, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), -O(CR11R12)k-, -S(CR11R12)k-, -S(O)k(CR11R12)k-, -O(CR11R12)k-NR11C(O)-, -O(CR11R12)kC(O)NR11-, -C(O)-, -C(O)(CR11R12)k-, -C(S)-, -C(S)(CR11R12)k-, -C(O)NR11-, -NR11C(O)-, -NR11(CR11R12)k-, -CONR11(CR11R12)k-, -N(R11)CO(CR11R12)k-, -C(O)NR11(CR11R12)k-, -NR11C(O)(CR11R12)k-로부터 선택되고, 여기서 각각의 R11 및 R12는 독립적으로 H, C1 - 8알킬, 할로-치환된-C1 - 8알킬 또는 히드록시-치환된-C1 - 8알킬이고, k는 1 내지 12의 정수이고;L is a bond, C 1 - 8 alkylene, halo-substituted -C 1 - 8 alkylene, hydroxy-substituted -C 1 - 8 alkylene, C 2-8 alkenylene group, a halo-substituted -C 2 - 8 alkenylene, hydroxy-substituted 2 -C 8 alkenylene, polyalkylene glycols, poly (ethylene glycol), -O (CR 11 R 12 ) k -, -S (CR 11 R 12) k - , -S(O) k (CR 11 R 12 ) k -, -O(CR 11 R 12 ) k -NR 11 C(O)-, -O(CR 11 R 12 ) k C(O)NR 11- , -C(O)-, -C(O)(CR 11 R 12 ) k -, -C(S)-, -C(S)(CR 11 R 12 ) k -, -C(O)NR 11 -, -NR 11 C(O)-, -NR 11 (CR 11 R 12 ) k -, -CONR 11 (CR 11 R 12 ) k -, -N(R 11 )CO(CR 11 R 12 ) k- , -C(O)NR 11 (CR 11 R 12 ) k -, -NR 11 C(O)(CR 11 R 12 ) k -, wherein each R 11 and R 12 is independently H, C 1 - 8 alkyl, halo-substituted -C 1-8 alkyl, or hydroxy-substituted -C 1 - 8 is alkyl, k is an integer from 1 to 12;
X1은 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2 및 이것들의 임의의 조합물 (여기서, p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기임)로부터 선택된다. X 1 is a label, dye, polymer, water-soluble polymer, polyalkylene glycol, poly(ethylene glycol), derivative of poly(ethylene glycol), sugar, lipid, photocrosslinker, cytotoxic compound, drug, affinity label, photo affinity Label, reactive compound; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide , Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 Agonist, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, Aptamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radiation-imaging probes, other spectroscopic probes, prodrugs, for immunotherapy Toxin, solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 ,- O- (CH 2 CH 2 O) p CH 2 CH 2 -X 2 , and any combination of these (where p is 1 to 10,000, X 2 is H, C 1 - 8 alkyl, protecting group or a terminal functional group being ) Is selected from.
추가로, 1개 이상의 피롤리신 및/또는 PCL 잔기를 함유하는 화학식 I의 단백질, 폴리펩티드 및/또는 펩티드를 하기 화학식 IV의 구조를 갖는 시약과 혼합하고 적절한 조건 하에서 반응시키는 것을 포함하는, 화학식 I의 단백질, 폴리펩티드 및/또는 펩티드의 부위 특이적 표지화를 위한 방법이 본원에 제공된다.In addition, formula I, comprising mixing and reacting under appropriate conditions a protein, polypeptide and/or peptide of formula I containing at least one pyrrolysine and/or PCL moiety with a reagent having the structure of formula IV Provided herein are methods for site-specific labeling of proteins, polypeptides and/or peptides of.
<화학식 IV><Formula IV>
상기 식에서,In the above formula,
R5는 -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 선택되고;R 5 is -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, aryl, heteroaryl, heterocycloalkyl or Selected from cycloalkyl, and -LX 1;
A는 C3-C8시클로알킬, C3-C8 헤테로시클로알킬, 5-6원 모노시클릭 아릴, 5-6원 모노시클릭 헤테로아릴, 9-10원 융합된 비시클릭 고리 또는 13-14원 융합된 트리시클릭 고리이고, 여기서 A는 -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택된 1 내지 5개의 치환기로 임의로 치환되고;A is C 3 -C 8 cycloalkyl, C 3 -C 8 heterocycloalkyl, 5-6 membered monocyclic aryl, 5-6 membered monocyclic heteroaryl, 9-10 membered fused bicyclic ring or 13- a 14-membered fused tricyclic ring, where A is -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, Optionally substituted with 1 to 5 substituents independently selected from aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1;
L은 결합, C1 - 8알킬렌, 할로-치환된-C1 - 8알킬렌, 히드록시-치환된-C1 - 8알킬렌, C2-8알케닐렌, 할로-치환된-C2 - 8알케닐렌, 히드록시-치환된-C2 - 8알케닐렌, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), -O(CR11R12)k-, -S(CR11R12)k-, -S(O)k(CR11R12)k-, -O(CR11R12)k-NR11C(O)-, -O(CR11R12)kC(O)NR11-, -C(O)-, -C(O)(CR11R12)k-, -C(S)-, -C(S)(CR11R12)k-, -C(O)NR11-, -NR11C(O)-, -NR11(CR11R12)k-, -CONR11(CR11R12)k-, -N(R11)CO(CR11R12)k-, -C(O)NR11(CR11R12)k-, -NR11C(O)(CR11R12)k-로부터 선택되고, 여기서 각각의 R11 및 R12는 독립적으로 H, C1 - 8알킬, 할로-치환된-C1 - 8알킬 또는 히드록시-치환된-C1 - 8알킬이고, k는 1 내지 12의 정수이고;L is a bond, C 1 - 8 alkylene, halo-substituted -C 1 - 8 alkylene, hydroxy-substituted -C 1 - 8 alkylene, C 2-8 alkenylene group, a halo-substituted -C 2 - 8 alkenylene, hydroxy-substituted 2 -C 8 alkenylene, polyalkylene glycols, poly (ethylene glycol), -O (CR 11 R 12 ) k -, -S (CR 11 R 12) k - , -S(O) k (CR 11 R 12 ) k -, -O(CR 11 R 12 ) k -NR 11 C(O)-, -O(CR 11 R 12 ) k C(O)NR 11- , -C(O)-, -C(O)(CR 11 R 12 ) k -, -C(S)-, -C(S)(CR 11 R 12 ) k -, -C(O)NR 11 -, -NR 11 C(O)-, -NR 11 (CR 11 R 12 ) k -, -CONR 11 (CR 11 R 12 ) k -, -N(R 11 )CO(CR 11 R 12 ) k- , -C(O)NR 11 (CR 11 R 12 ) k -, -NR 11 C(O)(CR 11 R 12 ) k -, wherein each R 11 and R 12 is independently H, C 1 - 8 alkyl, halo-substituted -C 1-8 alkyl, or hydroxy-substituted -C 1 - 8 is alkyl, k is an integer from 1 to 12;
X1은 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2 및 이것들의 임의의 조합물 (여기서, p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기임)로부터 선택된다. X 1 is a label, dye, polymer, water-soluble polymer, polyalkylene glycol, poly(ethylene glycol), derivative of poly(ethylene glycol), sugar, lipid, photocrosslinker, cytotoxic compound, drug, affinity label, photo affinity Label, reactive compound; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide , Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 Agonist, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, Aptamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radiation-imaging probes, other spectroscopic probes, prodrugs, for immunotherapy Toxin, solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 ,- O- (CH 2 CH 2 O) p CH 2 CH 2 -X 2 , and any combination of these (where p is 1 to 10,000, X 2 is H, C 1 - 8 alkyl, protecting group or a terminal functional group being ) Is selected from.
특정 실시양태에서 k는 1 내지 11의 정수이다. 특정 실시양태에서 k는 1 내지 10의 정수이다. 특정 실시양태에서 k는 1 내지 9의 정수이다. 특정 실시양태에서 k는 1 내지 8의 정수이다. 특정 실시양태에서 k는 1 내지 7의 정수이다. 특정 실시양태에서 k는 1 내지 6의 정수이다. 특정 실시양태에서 k는 1 내지 5의 정수이다. 특정 실시양태에서 k는 1 내지 4의 정수이다. 특정 실시양태에서 k는 1 내지 3의 정수이다. 특정 실시양태에서 k는 1 내지 2의 정수이다. In certain embodiments k is an integer from 1 to 11. In certain embodiments k is an integer from 1 to 10. In certain embodiments k is an integer from 1 to 9. In certain embodiments k is an integer from 1 to 8. In certain embodiments k is an integer from 1 to 7. In certain embodiments k is an integer from 1 to 6. In certain embodiments k is an integer from 1 to 5. In certain embodiments k is an integer from 1 to 4. In certain embodiments k is an integer from 1 to 3. In certain embodiments k is an integer from 1 to 2.
특정 실시양태에서 p는 1 내지 8000의 정수이다. 특정 실시양태에서 p는 1 내지 7000의 정수이다. 특정 실시양태에서 p는 1 내지 6000의 정수이다. 특정 실시양태에서 p는 1 내지 5000의 정수이다. 특정 실시양태에서 p는 1 내지 4000의 정수이다. 특정 실시양태에서 p는 1 내지 3000의 정수이다. 특정 실시양태에서 p는 1 내지 2000의 정수이다. 특정 실시양태에서 p는 1 내지 1000의 정수이다. 특정 실시양태에서 p는 1 내지 500의 정수이다. 특정 실시양태에서 p는 1 내지 400의 정수이다. 특정 실시양태에서 p는 1 내지 300의 정수이다. 특정 실시양태에서 p는 1 내지 200의 정수이다. 특정 실시양태에서 p는 1 내지 100의 정수이다. 특정 실시양태에서 p는 1 내지 90의 정수이다. 특정 실시양태에서 p는 1 내지 80의 정수이다. 특정 실시양태에서 p는 1 내지 70의 정수이다. 특정 실시양태에서 p는 1 내지 60의 정수이다. 특정 실시양태에서 p는 1 내지 50의 정수이다. 특정 실시양태에서 p는 1 내지 40의 정수이다. 특정 실시양태에서 p는 1 내지 30의 정수이다. 특정 실시양태에서 p는 1 내지 20의 정수이다. 특정 실시양태에서 p는 1 내지 10의 정수이다. 특정 실시양태에서 p는 1 내지 5의 정수이다. In certain embodiments p is an integer from 1 to 8000. In certain embodiments p is an integer from 1 to 7000. In certain embodiments p is an integer from 1 to 6000. In certain embodiments p is an integer from 1 to 5000. In certain embodiments p is an integer from 1 to 4000. In certain embodiments p is an integer from 1 to 3000. In certain embodiments p is an integer from 1 to 2000. In certain embodiments p is an integer from 1 to 1000. In certain embodiments p is an integer from 1 to 500. In certain embodiments p is an integer from 1 to 400. In certain embodiments p is an integer from 1 to 300. In certain embodiments p is an integer from 1 to 200. In certain embodiments p is an integer from 1 to 100. In certain embodiments p is an integer from 1 to 90. In certain embodiments p is an integer from 1 to 80. In certain embodiments p is an integer from 1 to 70. In certain embodiments p is an integer from 1 to 60. In certain embodiments p is an integer from 1 to 50. In certain embodiments p is an integer from 1 to 40. In certain embodiments p is an integer from 1 to 30. In certain embodiments p is an integer from 1 to 20. In certain embodiments p is an integer from 1 to 10. In certain embodiments p is an integer from 1 to 5.
화학식 IV의 화합물의 비제한적인 예에는 다음이 포함된다:Non-limiting examples of compounds of formula IV include:
상기 식에서, 1개 이상의 폴리에틸렌글리콜 (PEG) 잔기를 갖는 화합물은 1000 Da 내지 50 kDa 범위의 평균 분자량을 갖고, 여기서 n은 20 내지 1200이고, exPADRE는 이고, PADRE는 이고, BG1은 이고, BG2는 이고, 여기서 *는 포스포티오에이트 연결부를 나타낸다.In the above formula, the compound having at least one polyethylene glycol (PEG) moiety has an average molecular weight in the range of 1000 Da to 50 kDa, where n is 20 to 1200, and exPADRE is And PADRE is And BG1 is And BG2 is And, where * represents a phosphorothioate linking moiety.
특정 실시양태에서 본원에 제공된 방법을 이용하여 유도체화된 1개 이상의 피롤리신 및/또는 PCL을 함유하는 단백질, 폴리펩티드 및/또는 펩티드는 하기 화학식 II의 구조를 갖는다.In certain embodiments, proteins, polypeptides and/or peptides containing at least one pyrrolysine and/or PCL derivatized using the methods provided herein have the structure of Formula II:
<화학식 II><Formula II>
상기 식에서,In the above formula,
R1은 H 또는 아미노 말단 변형 기이고;R 1 is H or an amino terminal modified group;
R2는 OH 또는 카르복시 말단 변형 기이고;R 2 is OH or a carboxy terminal modified group;
n은 1 내지 5000의 정수이고;n is an integer from 1 to 5000;
각각의 BB는 아미노산 잔기, 하기 화학식 A-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 B-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 C-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 D-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 E-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 F-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 G-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 H-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 I-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 J-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 하기 화학식 K-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기 및 하기 화학식 L-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 또는 그의 이성질체로부터 독립적으로 선택되고;Each BB is an amino acid residue, a pyrrolysine analog amino acid residue having a structure of the following formula A-2, a pyrrolysine analog amino acid residue having a structure of the following formula B-2, and a pyrrolyse having a structure of the following formula C-1. A new analog amino acid residue, a pyrrolysine analog amino acid residue having a structure of the following formula D-1, a pyrrolysine analog amino acid residue having a structure of the following formula E-1, a pyrrolysine analog having a structure of the following formula F-1 Amino acid residue, a pyrrolysine analog amino acid residue having the structure of the following formula G-1, a pyrrolysine analog amino acid residue having the structure of the following formula H-1, a pyrrolysine analog amino acid residue having the structure of the following formula I-1 , A pyrrolysine analog amino acid residue having a structure of Formula J-1, a pyrrolysine analog amino acid residue having a structure of Formula K-1, and a pyrrolysine analog amino acid residue having a structure of Formula L-1, or Independently selected from its isomers;
, 여기서 , here
R3, R5 및 각각의 R4는 H, -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택되고;R 3, R 5 and each R 4 is H, -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, , Aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1 are independently selected;
R6은 H 또는 C1알킬이고;R 6 is H or C 1 alkyl;
A는 C3-C8시클로알킬, C3-C8 헤테로시클로알킬, 5-6원 모노시클릭 아릴, 5-6원 모노시클릭 헤테로아릴, 9-10원 융합된 비시클릭 고리 또는 13-14원 융합된 트리시클릭 고리이고, 여기서 A는 -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택된 1 내지 5개의 치환기로 임의로 치환되고;A is C 3 -C 8 cycloalkyl, C 3 -C 8 heterocycloalkyl, 5-6 membered monocyclic aryl, 5-6 membered monocyclic heteroaryl, 9-10 membered fused bicyclic ring or 13- a 14-membered fused tricyclic ring, where A is -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, Optionally substituted with 1 to 5 substituents independently selected from aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1;
L은 결합, C1 - 8알킬렌, 할로-치환된-C1 - 8알킬렌, 히드록시-치환된-C1 - 8알킬렌, C2-8알케닐렌, 할로-치환된-C2 - 8알케닐렌, 히드록시-치환된-C2 - 8알케닐렌, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), -O(CR11R12)k-, -S(CR11R12)k-, -S(O)k(CR11R12)k-, -O(CR11R12)k-NR11C(O)-, -O(CR11R12)kC(O)NR11-, -C(O)-, -C(O)(CR11R12)k-, -C(S)-, -C(S)(CR11R12)k-, -C(O)NR11-, -NR11C(O)-, -NR11(CR11R12)k-, -CONR11(CR11R12)k-, -N(R11)CO(CR11R12)k-, -C(O)NR11(CR11R12)k-, -NR11C(O)(CR11R12)k-로부터 선택되고, 여기서 각각의 R11 및 R12는 독립적으로 H, C1 - 8알킬, 할로-치환된-C1 - 8알킬 또는 히드록시-치환된-C1 - 8알킬이고, k는 1 내지 12의 정수이고,L is a bond, C 1 - 8 alkylene, halo-substituted -C 1 - 8 alkylene, hydroxy-substituted -C 1 - 8 alkylene, C 2-8 alkenylene group, a halo-substituted -C 2 - 8 alkenylene, hydroxy-substituted 2 -C 8 alkenylene, polyalkylene glycols, poly (ethylene glycol), -O (CR 11 R 12 ) k -, -S (CR 11 R 12) k - , -S(O) k (CR 11 R 12 ) k -, -O(CR 11 R 12 ) k -NR 11 C(O)-, -O(CR 11 R 12 ) k C(O)NR 11- , -C(O)-, -C(O)(CR 11 R 12 ) k -, -C(S)-, -C(S)(CR 11 R 12 ) k -, -C(O)NR 11 -, -NR 11 C(O)-, -NR 11 (CR 11 R 12 ) k -, -CONR 11 (CR 11 R 12 ) k -, -N(R 11 )CO(CR 11 R 12 ) k- , -C(O)NR 11 (CR 11 R 12 ) k -, -NR 11 C(O)(CR 11 R 12 ) k -, wherein each R 11 and R 12 is independently H, C 1 and 8, and the alkyl, k is an integer from 1 to 12, - 8 alkyl, halo-substituted -C 1 - 8 alkyl or hydroxy-substituted -C 1
X1은 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2 및 이것들의 임의의 조합물 (여기서, p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기임)로부터 선택되고,X 1 is a label, dye, polymer, water-soluble polymer, polyalkylene glycol, poly(ethylene glycol), derivative of poly(ethylene glycol), sugar, lipid, photocrosslinker, cytotoxic compound, drug, affinity label, photo affinity Label, reactive compound; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide , Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 Agonist, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, Aptamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radiation-imaging probes, other spectroscopic probes, prodrugs, for immunotherapy Toxin, solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 ,- O- (CH 2 CH 2 O) p CH 2 CH 2 -X 2 , and any combination of these (where p is 1 to 10,000, X 2 is H, C 1 - 8 alkyl, protecting group or a terminal functional group being ) Is selected from,
1개 이상의 AA는 화학식 A-2 또는 화학식 B-2의 구조를 갖는 피롤리신 유사체 아미노산 잔기이거나, 1개 이상의 BB는 화학식 C-1 또는 화학식 D-1 또는 화학식 E-1 또는 화학식 F-1 또는 화학식 G-1 또는 화학식 H-1 또는 화학식 I-1 또는 화학식 J-1 또는 화학식 K-1 또는 화학식 L-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 또는 그의 이성질체이다.At least one AA is a pyrrolysine analog amino acid residue having the structure of Formula A-2 or Formula B-2, or at least one BB is Formula C-1 or Formula D-1 or Formula E-1 or Formula F-1 Or a pyrrolysine analog amino acid residue having the structure of Formula G-1 or Formula H-1 or Formula I-1 or Formula J-1 or Formula K-1 or Formula L-1, or an isomer thereof.
특정 실시양태에서, k는 1 내지 11의 정수이다. 특정 실시양태에서, k는 1 내지 10의 정수이다. 특정 실시양태에서, k는 1 내지 9의 정수이다. 특정 실시양태에서, k는 1 내지 8의 정수이다. 특정 실시양태에서, k는 1 내지 7의 정수이다. 특정 실시양태에서, k는 1 내지 6의 정수이다. 특정 실시양태에서, k는 1 내지 5의 정수이다. 특정 실시양태에서, k는 1 내지 4의 정수이다. 특정 실시양태에서, k는 1 내지 3의 정수이다. 특정 실시양태에서, k는 1 내지 2의 정수이다.In certain embodiments, k is an integer from 1 to 11. In certain embodiments, k is an integer from 1 to 10. In certain embodiments, k is an integer from 1 to 9. In certain embodiments, k is an integer from 1 to 8. In certain embodiments, k is an integer from 1 to 7. In certain embodiments, k is an integer from 1 to 6. In certain embodiments, k is an integer from 1 to 5. In certain embodiments, k is an integer from 1 to 4. In certain embodiments, k is an integer from 1 to 3. In certain embodiments, k is an integer from 1 to 2.
특정 실시양태에서, p는 1 내지 8000의 정수이다. 특정 실시양태에서, p는 1 내지 7000의 정수이다. 특정 실시양태에서, p는 1 내지 6000의 정수이다. 특정 실시양태에서, p는 1 내지 5000의 정수이다. 특정 실시양태에서, p는 1 내지 4000의 정수이다. 특정 실시양태에서, p는 1 내지 3000의 정수이다. 특정 실시양태에서, p는 1 내지 2000의 정수이다. 특정 실시양태에서, p는 1 내지 1000의 정수이다. 특정 실시양태에서, p는 1 내지 500의 정수이다. 특정 실시양태에서, p는 1 내지 400의 정수이다. 특정 실시양태에서, p는 1 내지 300의 정수이다. 특정 실시양태에서, p는 1 내지 200의 정수이다. 특정 실시양태에서, p는 1 내지 100의 정수이다. 특정 실시양태에서, p는 1 내지 90의 정수이다. 특정 실시양태에서, p는 1 내지 80의 정수이다. 특정 실시양태에서, p는 1 내지 70의 정수이다. 특정 실시양태에서, p는 1 내지 60의 정수이다. 특정 실시양태에서, p는 1 내지 50의 정수이다. 특정 실시양태에서, p는 1 내지 40의 정수이다. 특정 실시양태에서, p는 1 내지 30의 정수이다. 특정 실시양태에서, p는 1 내지 20의 정수이다. 특정 실시양태에서, p는 1 내지 10의 정수이다. 특정 실시양태에서, p는 1 내지 5의 정수이다. In certain embodiments, p is an integer from 1 to 8000. In certain embodiments, p is an integer from 1 to 7000. In certain embodiments, p is an integer from 1 to 6000. In certain embodiments, p is an integer from 1 to 5000. In certain embodiments, p is an integer from 1 to 4000. In certain embodiments, p is an integer from 1 to 3000. In certain embodiments, p is an integer from 1 to 2000. In certain embodiments, p is an integer from 1 to 1000. In certain embodiments, p is an integer from 1 to 500. In certain embodiments, p is an integer from 1 to 400. In certain embodiments, p is an integer from 1 to 300. In certain embodiments, p is an integer from 1 to 200. In certain embodiments, p is an integer from 1 to 100. In certain embodiments, p is an integer from 1 to 90. In certain embodiments, p is an integer from 1 to 80. In certain embodiments, p is an integer from 1 to 70. In certain embodiments, p is an integer from 1 to 60. In certain embodiments, p is an integer from 1 to 50. In certain embodiments, p is an integer from 1 to 40. In certain embodiments, p is an integer from 1 to 30. In certain embodiments, p is an integer from 1 to 20. In certain embodiments, p is an integer from 1 to 10. In certain embodiments, p is an integer from 1 to 5.
특정 실시양태에서, n은 1 내지 4000의 정수이다. 특정 실시양태에서, n은 1 내지 3000의 정수이다. 특정 실시양태에서, n은 1 내지 2000의 정수이다. 특정 실시양태에서, n은 1 내지 1000의 정수이다. 특정 실시양태에서, n은 1 내지 700의 정수이다. 특정 실시양태에서, n은 1 내지 800의 정수이다. 특정 실시양태에서, n은 1 내지 600의 정수이다. 특정 실시양태에서, n은 1 내지 500의 정수이다. 특정 실시양태에서, n은 1 내지 400의 정수이다. 특정 실시양태에서, n은 1 내지 300의 정수이다. 특정 실시양태에서, n은 1 내지 200의 정수이다. 특정 실시양태에서, n은 1 내지 100의 정수이다. 특정 실시양태에서, n은 1 내지 90의 정수이다. 특정 실시양태에서, n은 1 내지 80의 정수이다. 특정 실시양태에서, n은 1 내지 70의 정수이다. 특정 실시양태에서, n은 1 내지 60의 정수이다. 특정 실시양태에서, n은 1 내지 50의 정수이다. 특정 실시양태에서, n은 1 내지 40의 정수이다. 특정 실시양태에서, n은 1 내지 30의 정수이다. 특정 실시양태에서, n은 1 내지 20의 정수이다. 특정 실시양태에서, n은 1 내지 10의 정수이다. 특정 실시양태에서, n은 1 내지 5의 정수이다. In certain embodiments, n is an integer from 1 to 4000. In certain embodiments, n is an integer from 1 to 3000. In certain embodiments, n is an integer from 1 to 2000. In certain embodiments, n is an integer from 1 to 1000. In certain embodiments, n is an integer from 1 to 700. In certain embodiments, n is an integer from 1 to 800. In certain embodiments, n is an integer from 1 to 600. In certain embodiments, n is an integer from 1 to 500. In certain embodiments, n is an integer from 1 to 400. In certain embodiments, n is an integer from 1 to 300. In certain embodiments, n is an integer from 1 to 200. In certain embodiments, n is an integer from 1 to 100. In certain embodiments, n is an integer from 1 to 90. In certain embodiments, n is an integer from 1 to 80. In certain embodiments, n is an integer from 1 to 70. In certain embodiments, n is an integer from 1 to 60. In certain embodiments, n is an integer from 1 to 50. In certain embodiments, n is an integer from 1 to 40. In certain embodiments, n is an integer from 1 to 30. In certain embodiments, n is an integer from 1 to 20. In certain embodiments, n is an integer from 1 to 10. In certain embodiments, n is an integer from 1 to 5.
특정 실시양태에서, 본원에서 제공되는 유도체화 방법에 사용되는 화학식 III의 화합물에는 아미노 당이 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 이러한 아미노 당에는 D-만노사민 및 D-갈락토사민이 포함되나, 이에 제한되지는 않는다. In certain embodiments, compounds of Formula III used in the methods of derivatization provided herein include, but are not limited to, amino sugars. In certain embodiments, such amino sugars include, but are not limited to, D-mannosamine and D-galactosamine.
특정 실시양태에서, 본원에서 제공되는 유도체화 방법에 사용되는 화학식 IV의 화합물에는 2-아미노-벤즈알데히드 (2-ABA), 2-아미노-벤즈알데히드 (2-ABA)의 유도체, 본원에서 제공되는 2-아미노-벤즈알데히드 (2-ABA)의 유도체, 2-아미노-아세토페논 (2-AAP), 2-아미노-아세토페논 (2-AAP)의 유도체, 본원에서 제공되는 2-아미노-아세토페논 (2-AAP)의 유도체, 2-아미노-5-니트로-벤조페논 (2-ANBP), 2-아미노-5-니트로-벤조페논 (2-ANBP)의 유도체 및 본원에서 제공되는 2-아미노-5-니트로-벤조페논 (2-ANBP)의 유도체가 포함되나, 이에 제한되지는 않는다.In certain embodiments, compounds of Formula IV used in the derivatization methods provided herein include 2-amino-benzaldehyde (2-ABA), derivatives of 2-amino-benzaldehyde (2-ABA), 2- Derivatives of amino-benzaldehyde (2-ABA), 2-amino-acetophenone (2-AAP), derivatives of 2-amino-acetophenone (2-AAP), 2-amino-acetophenone provided herein (2- AAP), 2-amino-5-nitro-benzophenone (2-ANBP), 2-amino-5-nitro-benzophenone (2-ANBP) and 2-amino-5-nitro provided herein -Derivatives of benzophenone (2-ANBP) are included, but are not limited thereto.
도 22는 2-아미노-벤즈알데히드 (2-ABA)를 사용한 PCL-A의 화학적 유도체화에 대한 반응식을 보여주며, 여기서 1개 이상의 PCL-A가 단백질에 부위 특이적으로 혼입되었다. 세미알데히드와 2-ABA의 고리화 반응은 프리들랜더(Friedlaender)-유형 반응으로서 퀴나졸린-유형 잔기 (22-A 또는 22-B)를 형성하고, 이것은 적절한 환원제 사용시에 추가로 환원되어 테트라히드로퀴나졸린 유형 잔기 (22-D) 또는 치환된 아닐린 (22-E)을 형성할 수 있다. 대안적으로, 퀴나졸린-유형 잔기 22-B의 추가의 반응으로 융합된 고리 잔기 (22-C)가 형성된다. 퀴나졸린-유형 잔기를 환원시키는데 적합한 환원제에는 시아노수소화붕소나트륨 및 수소화붕소나트륨이 포함되나, 이에 제한되지는 않는다.Figure 22 shows the reaction scheme for the chemical derivatization of PCL-A with 2-amino-benzaldehyde (2-ABA), wherein at least one PCL-A has been site-specifically incorporated into the protein. The cyclization reaction of semialdehyde with 2-ABA forms a quinazoline-type moiety (22-A or 22-B) as a Friedlaender-type reaction, which is further reduced when using an appropriate reducing agent to form tetrahydrogen. Quinazoline type residues (22-D) or substituted anilines (22-E) can be formed. Alternatively, a fused ring moiety (22-C) is formed by further reaction of the quinazoline-type moiety 22-B. Suitable reducing agents for reducing the quinazoline-type moiety include, but are not limited to, sodium cyanoborohydride and sodium borohydride.
추가로, 도 23은 PCL-A와 2-ABA, 2-AAP 또는 2-ANBP의 반응 후에 형성된 다양한 구조의 단백질 접합체를 보여준다. 이러한 기의 부착으로 인한 예상된 질량 증가도 나타냈다. 이러한 기를 사용하여 수득된 구조를 NMR 분광분석법으로 특징규명하였다 (실시예 45 참조). 또한, NaCNBH3 환원은 PCL-기재의 단백질 접합체를 안정화시키고 고온에서일지라도 PCL-ABA 및 PCL-AAP 연결의 해리를 방지하는 것으로 밝혀졌다 (실시예 44 참조).In addition, Figure 23 shows the protein conjugates of various structures formed after the reaction of PCL-A and 2-ABA, 2-AAP or 2-ANBP. The expected mass increase due to the attachment of these groups was also shown. The structure obtained using this group was characterized by NMR spectroscopy (see Example 45). In addition, NaCNBH 3 reduction was found to stabilize PCL-based protein conjugates and prevent dissociation of PCL-ABA and PCL-AAP linkages even at high temperatures (see Example 44).
2-ABA와 Δ1-피롤린-5-카르복실산 (L-1-피롤린-5-카르복실산) (문헌 [Strecker, H. J., (1971), Methods in Enzymology 27B, 254-257], [Vogel, H. J., and Davis, B. D. (1953), "Glutamic gamma-semialdehyde and delta1-pyrroline-5-carboxylic acid, intermediates in the biosynthesis of proline," J. Am. Chem. Soc. 74, 109-112] 및 [Schoepf, C., and Oechler, F., (1936), Ann. Chem. 523, 1]) 및 기타 피롤린 (문헌 [Schoepf, C., and Oechler, F., (1936), Ann. Chem. 523, 1] 및 [Schoepf, C., and Steuer, H., (1947), Ann. Chem. 558, 124] 참조)의 반응은 프롤린 대사 및 생합성에서 Δ1-피롤린-5-카르복실산의 역할을 연구하는데 있어서의 비색 검정으로 이용되어 왔다 (문헌 [Vogel, H. J., and Davis, B. D. (1953), "Glutamic gamma-semialdehyde and Δ1-pyrroline-5-carboxylic acid, intermediates in the biosynthesis of proline," J. Am. Chem. Soc. 74, 109-112], [Strecker, H. J., (1960), "The interconversion of glutamic acid and proline," J. Biol. Chem. 235, 2045-2050], [Mezl, V. A., and Knox, W. E., (1976), "Properties and analysis of a stable derivative of pyrroline-5-carboxylic acid for use in metabolic studies," Analytical Biochemistry 74, 430-40], [Wu, G. Y., and Seifter, S., (1975), "A new method for the preparation of delta 1-pyrroline 5-carboxylic acid and proline," Analytical Biochemistry 67, 413-21], [Williams, I., and Frank, L., (1975), "Improved chemical synthesis and enzymatic assay of Δ1-pyrroline-5-carboxylic acid," Anal. Biochem. 64, 85-97] 및 [Strecker, H. J., (1957), "The interconversion of glutamic acid and proline," J. Biol. Chem. 225, 825] 참조). 반응식은 글루탐산-감마-세미알데히드 중간체의 형성을 보여주지만, 반응은 이러한 중간체의 형성 없이 피롤린-카르복시-리신으로부터 직접적으로 진행될 수 있다. 2-ABA and Δ 1 -pyrroline-5-carboxylic acid (L-1-pyrroline-5-carboxylic acid) (Strecker, HJ, (1971), Methods in Enzymology 27B, 254-257), [Vogel, HJ, and Davis, BD (1953), "Glutamic gamma-semialdehyde and delta1-pyrroline-5-carboxylic acid, intermediates in the biosynthesis of proline," J. Am. Chem. Soc. 74, 109-112] And [Schoepf, C., and Oechler, F., (1936), Ann. Chem. 523, 1]) and other pyrrolines (Schoepf, C., and Oechler, F., (1936), Ann. ... Chem 523, 1] and [Schoepf, C., Steuer and, H., (1947), Ann Chem 558, 124] view) of the reaction is Δ 1 in proline metabolism and biosynthesis-pyrroline-5-carboxylic It has been used as a colorimetric assay in studying the role of acids (Vogel, HJ, and Davis, BD (1953), "Glutamic gamma-semialdehyde and Δ 1 -pyrroline-5-carboxylic acid, intermediates in the biosynthesis. of proline," J. Am. Chem. Soc. 74, 109-112], [Strecker, HJ, (1960), "The interconversion of glutamic acid and proline," J. Biol. Chem. 235, 2045-2050] , [Mezl, VA, and Knox, WE, (1976), "Properties and analysis of a stable derivative of pyrroline-5-carboxylic acid for use in metabolic studies," Analytical Bi ochemistry 74, 430-40], [Wu, GY, and Seifter, S., (1975), "A new method for the preparation of delta 1-pyrroline 5-carboxylic acid and proline," Analytical Biochemistry 67, 413-21 ], [Williams, I., and Frank, L., (1975), "Improved chemical synthesis and enzymatic assay of Δ 1 -pyrroline-5-carboxylic acid," Anal. Biochem. 64, 85-97] and [Strecker, HJ, (1957), "The interconversion of glutamic acid and proline," J. Biol. Chem. 225, 825). The reaction scheme shows the formation of a glutamic acid-gamma-semialdehyde intermediate, but the reaction can proceed directly from pyrroline-carboxy-lysine without the formation of this intermediate.
모델 단백질 hRBP4의 고해상도 질량 분광측정 연구는 2-ABA에 의한 PCL 잔기의 화학적 유도체화를 입증하였고, 여기서 PCL은 단백질 hRBP4에 부위 특이적으로 혼입된 것이었다 (도 25 내지 26, 실시예 11 참조). 도 24는 2-ABA로 유도체화된 hRBP4 Phe122PCL의 질량 분광측정 분석을 보여주며, 여기서 2-ABA로 유도체화된 상기 단백질은 23269.2 Da에서 주요 피크를 생성하였고, 최소량의 미변형 단백질이 23166.8 Da에서 검출되었다. 유도체화된 단백질에 대한 102.4 Da의 질량 증가는 도 23에 나타난 바와 같이 2-ABA 잔기의 부착에 대한 103 Da의 예상 증가와 일치하여, PCL의 부위 특이적 혼입을 갖는 단백질이 PCL 부위에서 부위 특이적으로 변형된다는 것을 입증한다. 2-ABA-유도체화된 hRBP4 Phe122PCL 단백질의 트립신 소화물을 LC-MS 분석한 후 얻은 MS/MS 분석결과 (도 25)는 예상된 YWGVASF*LQK 펩티드를 확인시켜 주었고, 여기서 F*는 2-ABA-변형 PCL과 일치하는 질량을 갖는다. 도 25A는 YWGVASF*LQK 펩티드의 단편화 패턴이며, 여기서 F*는 2-ABA-변형 PCL과 일치하는 질량을 갖는다. 도 25B는 YWGVASF*LQK (F* = PCL 및 PCL-2-ABA 부가물)의 2+ 이온의 TIC (전체 이온 크로마토그램) 및 EIC (추출 이온 크로마토그램)이고, 여기서 유도체화된 종 및 유도체화되지 않은 (검출가능하지 않음) 종에 대한 EIC의 비교는 반응의 완료를 나타낸다. 도 25C는 2-ABA로 유도체화된 hRBP4 Phe122PCL의 질량 분광측정 분석이며, YWGVASF*LQK (F* = PCL-2-ABA 부가물)의 3+ 및 2+ 전구체를 m/z 459.92 (3+) 및 689.37 (2+) 각각에서 보여줌으로써 2-ABA 사용시에 관찰된 반응이 잔기 122의 원하는 TAG 부위에서 혼입된 PCL 잔기와 부위 특이적으로 일어난다는 것을 입증한다.A high-resolution mass spectrometric study of the model protein hRBP4 demonstrated chemical derivatization of PCL residues by 2-ABA, where PCL was site-specific incorporated into the protein hRBP4 (see FIGS. 25-26, Example 11). Figure 24 shows the mass spectrometric analysis of hRBP4 Phe122PCL derivatized with 2-ABA, wherein the protein derivatized with 2-ABA produced a major peak at 23269.2 Da, and the minimum amount of unmodified protein was at 23166.8 Da. Was detected. The increase in mass of 102.4 Da for the derivatized protein is consistent with the expected increase of 103 Da for the attachment of the 2-ABA residue, as shown in FIG. 23, so that the protein with site-specific incorporation of PCL is site-specific at the PCL site. It proves that it is transformed into an enemy. The MS/MS analysis result obtained after LC-MS analysis of the trypsin digest of 2-ABA-derivated hRBP4 Phe122PCL protein (FIG. 25) confirmed the expected YWGVASF*LQK peptide, where F* is 2-ABA- It has a mass consistent with the modified PCL. Figure 25A is the fragmentation pattern of the YWGVASF*LQK peptide, where F* has a mass consistent with the 2-ABA-modified PCL. Figure 25B is the TIC (total ion chromatogram) and EIC (extracted ion chromatogram) of the 2+ ion of YWGVASF*LQK (F* = PCL and PCL-2-ABA adduct), where derivatized species and derivatization Comparison of EIC to non-detectable (not detectable) species indicates completion of the reaction. Figure 25C is a mass spectrometric analysis of hRBP4 Phe122PCL derivatized with 2-ABA, with 3+ and 2+ precursors of YWGVASF*LQK (F* = PCL-2-ABA adduct) m/z 459.92 (3+) And 689.37 (2+), respectively, demonstrating that the reaction observed with 2-ABA occurs site-specific with the incorporated PCL residue at the desired TAG site at residue 122.
유도체화의 pH 의존성의 평가는 도 26에 나타냈다. 위치 122에서 혼입된 PCL을 갖는 hRBP4 (hRBP4 Phe122PCL)의 질량 스펙트럼을 도 26A에 나타냈고, 여기서의 hRBP4 Phe122PCL은 2-ABA와 반응하지 않은 것이며, 도 26B 및 도 26C는 hRBP4 Phe122PCL을 pH 5.0의 아세테이트 완충제 중의 10 mM 2-ABA 및 pH 7.4의 포스페이트 완충제 중의 10 mM 2-ABA 각각과 반응시킨 후의 질량 스펙트럼이다. hRBP4 단백질의 PCL 잔기와 2-ABA의 반응은 pH 5로 조정된 수성 완충제 중에서는 실온에서 최대 95% 완료시까지 신속하게 진행되고, pH 7.4에서는 약간 더 낮은 정도로 대략 87%까지 진행된다. 도 27D 내지 F에 나타난 바와 같이 2-ABA와의 반응은 PCL 잔기에 선택적이며, 여기서 위치 62에서 불활성 비-천연 아미노산인 O-메틸-페닐알라닌 (OMePhe)으로 표지된 hRBP4 및 위치 122에 야생형 페닐알라닌 (Phe) 잔기를 갖는 hRBP4의 경우에는 2-ABA와의 반응이 관찰되지 않았다. The evaluation of the pH dependence of derivatization is shown in FIG. The mass spectrum of hRBP4 (hRBP4 Phe122PCL) with PCL incorporated at position 122 is shown in Figure 26A, where hRBP4 Phe122PCL did not react with 2-ABA, and Figures 26B and 26C show hRBP4 Phe122PCL in acetate at pH 5.0. It is a mass spectrum after reaction with each of 10 mM 2-ABA in buffer and 10 mM 2-ABA in phosphate buffer at pH 7.4. The reaction of 2-ABA with the PCL residue of the hRBP4 protein proceeds rapidly at room temperature to a maximum of 95% completion in an aqueous buffer adjusted to
반응물:단백질 농도 비율의 함수로서의 반응 효율 및 2-ABA-유사 반응물과의 반응성에 대한 평가를 도 27에 나타냈다. 위치 122에서 혼입된 PCL을 갖는 hRBP4 (hRBP4 Phe122PCL)의 0.1 mM 2-아미노-벤즈알데히드 (2-ABA)와의 반응 후 질량 스펙트럼을 도 27A에 나타냈고, hRBP4 Phe122PCL의 0.1 mM 2-아미노-아세토페논 (2-AAP) 및 0.1 mM 2-아미노-5-니트로-벤조페논 (2-ANBP)과의 반응 후 질량 스펙트럼을 도 27B 및 도 27C 각각에 나타냈다. 모든 반응은 pH 5.0의 200 mM 아세트산나트륨에서 수행되었다. 도 23에서와 같이, 단백질 접합체는 정확한 질량에서 예상된 질량 증가분을 갖는 것으로 검출되었다. 또한, 2-ABA 및 2-AAP의 경우에는 88% 초과 전환율의 수율이 달성되었지만, 2-ANBP의 경우에는 낮은 용해도로 인해 약 5%의 수율이 달성되었다. 그러나, 이것은 2-ABA 유사체가 반응물:단백질 농도 비율이 6:1일 때 PCL 혼입 부위에서 효율적으로 반응한다는 것을 입증한다. 반응의 정도는 반응물:단백질 비율이 600:1로 수행된 경우보다 단지 약간만 더 낮았다 (도 26).The evaluation of the reaction efficiency as a function of the reactant:protein concentration ratio and the reactivity with the 2-ABA-like reactant is shown in FIG. 27. The mass spectrum after reaction of hRBP4 (hRBP4 Phe122PCL) with PCL incorporated at position 122 with 0.1 mM 2-amino-benzaldehyde (2-ABA) is shown in Fig.27A, and 0.1 mM 2-amino-acetophenone of hRBP4 Phe122PCL ( 2-AAP) and 0.1 mM 2-amino-5-nitro-benzophenone (2-ANBP) are shown in Figs. 27B and 27C respectively. All reactions were carried out in 200 mM sodium acetate, pH 5.0. As in Figure 23, the protein conjugate was detected to have the expected mass increase at the correct mass. In addition, yields of greater than 88% conversion were achieved for 2-ABA and 2-AAP, but about 5% yield was achieved for 2-ANBP due to low solubility. However, this demonstrates that the 2-ABA analog reacts efficiently at the site of PCL incorporation when the reactant:protein concentration ratio is 6:1. The degree of reaction was only slightly lower than when the reactant:protein ratio was carried out at 600:1 (FIG. 26).
유도체화제를 단백질 대비 6배 내지 600배 몰 과량에 상응하는 0.1 내지 10 mM의 최종 농도 범위로 첨가한 경우에 유도체화의 정도에서의 유의한 개선은 없었다. 또한, 4700 초과의 몰비에서는 리신으로 예상되는 추가의 단백질 잔기가 2-ABA에 의해 유도체화되었다 (도 28). 도 28에서, 10× PBS 중 단백질 대비 상당한 과량의 2-ABA는 PCL 혼입된 hRBP4 (A, 약 17 μM, 단백질 대비 4700배 과량의 2-ABA) 및 OMePhe 혼입된 hRBP4 (B, 약 6.5 μM, 15400배 과량)의 경우에 추가의 부위에서의 접합을 유도했으며, 이는 이러한 추가의 부위에서의 접합이 PCL 이외의 잔기에 의해 매개됨을 예시한다.There was no significant improvement in the degree of derivatization when the derivatization agent was added in a final concentration range of 0.1 to 10 mM corresponding to a molar excess of 6 to 600 times compared to the protein. In addition, at molar ratios above 4700, additional protein residues expected to be lysine were derivatized by 2-ABA (FIG. 28). In Figure 28, a significant excess of 2-ABA compared to protein in 10×PBS is hRBP4 (A, about 17 μM, 4700-fold excess of 2-ABA compared to protein) and OMePhe incorporated hRBP4 (B, about 6.5 μM, compared to protein). 15400 fold excess) induced conjugation at additional sites, exemplifying that conjugation at these additional sites is mediated by residues other than PCL.
본원에서 제공되는 방법의 유용성을 추가로 예시하기 위해서, 도 29는 FAS-TE에 혼입된 PCL을 pH 5.0 및 pH 7.4에서 2-아미노-아세토페논 (2-AAP)으로 유도체화시키기 이전 및 이후의 질량 스펙트럼을 보여준다 (실시예 12 참조). 도 29A는 미반응 FAS-TE Tyr2454PCL의 질량 스펙트럼을 보여주고, 도 29B는 pH 5.0에서의 반응 혼합물의 질량 스펙트럼을 보여준다. 여기서, 관찰가능한 피크 강도의 100%가 33318.8 Da에서 발생되며, 이것은 2-AAP 변형된 FAS-TE Tyr2454PCL에 대해 117 Da의 질량 증가가 예상되는 바와 같이 미반응 물질의 경우보다 116.8 Da 더 크다. 유사하게, pH 7.4에서는 반응이 95% 완료시까지 진행되었다 (도 29C (미반응) 및 도 29D (반응됨)).To further illustrate the utility of the methods provided herein, FIG. 29 shows before and after derivatization of PCL incorporated in FAS-TE with 2-amino-acetophenone (2-AAP) at pH 5.0 and pH 7.4. The mass spectrum is shown (see Example 12). Figure 29A shows the mass spectrum of the unreacted FAS-TE Tyr2454PCL, and Figure 29B shows the mass spectrum of the reaction mixture at pH 5.0. Here, 100% of the observable peak intensity occurs at 33318.8 Da, which is 116.8 Da greater than for the unreacted material, as expected a mass increase of 117 Da for the 2-AAP modified FAS-TE Tyr2454PCL. Similarly, at pH 7.4 the reaction proceeded to 95% completion (Fig. 29C (unreacted) and Fig. 29D (reacted)).
도 30은 2-아미노-벤즈알데히드 또는 2-아미노-벤즈알데히드 유사체를 사용한 피롤리신 및/또는 PCL의 화학적 유도체화를 통해 단백질을 부위 특이적 변형시키기 위한 일반적인 반응식이다. 이러한 유사체의 특정 실시양태가 본원에서 제공된다. 피롤리신 및 PCL의 피롤린 고리와 2-ABA의 반응은 Δ1-피롤린-5-카르복실산과 2-ABA의 반응과 유사하다. 유사하게, 피롤리신 및 PCL과 2-AAP 및 2-ANBP의 반응으로 도 23의 각각의 치환된 잔기로 변형된 단백질이 생성된다.30 is a general reaction scheme for site-specific modification of a protein through chemical derivatization of pyrrolysine and/or PCL using 2-amino-benzaldehyde or 2-amino-benzaldehyde analogue. Certain embodiments of such analogs are provided herein. The reaction of 2-ABA with pyrrolysine and the pyrroline ring of PCL is similar to that of Δ 1 -pyrroline-5-carboxylic acid and 2-ABA. Similarly, the reaction of pyrrolysine and PCL with 2-AAP and 2-ANBP results in a protein modified with each of the substituted residues in FIG. 23.
특정 실시양태에서, 2-ABA 관능성을 이용하여, 내부에 혼입된 피롤리신 또는 PCL을 갖는 단백질, 폴리펩티드 및/또는 펩티드에 다양한 기를 부위 특이적으로 부착한다. 이러한 기에는 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2, 및 이들의 임의의 조합물 (여기서, p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기임)이 포함되나, 이에 제한되지는 않는다.In certain embodiments, the 2-ABA functionality is used to site-specifically attach various groups to proteins, polypeptides and/or peptides with pyrrolysine or PCL incorporated therein. These groups include labels, dyes, polymers, water-soluble polymers, polyalkylene glycols, poly(ethylene glycol), derivatives of poly(ethylene glycol), sugars, lipids, photocrosslinkers, cytotoxic compounds, drugs, affinity labels, photoaffinities. Label, reactive compound; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide , Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 Agonist, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, Aptamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radiation-imaging probes, other spectroscopic probes, prodrugs, for immunotherapy Toxin, solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 ,- O- (CH 2 CH 2 O) p CH 2 CH 2 -X 2, and any combination thereof (wherein, p is 1 to 10,000, X 2 is H, C 1 - 8 alkyl, protecting group or a terminal functional group Im) is included, but is not limited thereto.
특정 실시양태에서, 2-AAP 관능성을 이용하여, 내부에 혼입된 피롤리신 또는 PCL을 갖는 단백질, 폴리펩티드 및/또는 펩티드에 다양한 기를 부위 특이적으로 부착한다. 이러한 기에는 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2, 및 이들의 임의의 조합물 (여기서, p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기임)이 포함되나, 이에 제한되지는 않는다.In certain embodiments, the 2-AAP functionality is used to site-specifically attach various groups to proteins, polypeptides and/or peptides with pyrrolysine or PCL incorporated therein. These groups include labels, dyes, polymers, water-soluble polymers, polyalkylene glycols, poly(ethylene glycol), derivatives of poly(ethylene glycol), sugars, lipids, photocrosslinkers, cytotoxic compounds, drugs, affinity labels, photoaffinities. Label, reactive compound; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide , Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 Agonist, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, Aptamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radiation-imaging probes, other spectroscopic probes, prodrugs, for immunotherapy Toxin, solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 ,- O- (CH 2 CH 2 O) p CH 2 CH 2 -X 2, and any combination thereof (wherein, p is 1 to 10,000, X 2 is H, C 1 - 8 alkyl, protecting group or a terminal functional group Im) is included, but is not limited thereto.
특정 실시양태에서, 2-ANPA 관능성을 이용하여, 내부에 혼입된 피롤리신 또는 PCL을 갖는 단백질, 폴리펩티드 및/또는 펩티드에 다양한 기를 부위 특이적으로 부착한다. 이러한 기에는 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2, 및 이들의 임의의 조합물 (여기서, p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기임)이 포함되나, 이에 제한되지는 않는다.In certain embodiments, the 2-ANPA functionality is used to site-specifically attach various groups to proteins, polypeptides and/or peptides with pyrrolysine or PCL incorporated therein. These groups include labels, dyes, polymers, water-soluble polymers, polyalkylene glycols, poly(ethylene glycol), derivatives of poly(ethylene glycol), sugars, lipids, photocrosslinkers, cytotoxic compounds, drugs, affinity labels, photoaffinities. Label, reactive compound; Resin, peptide, second protein or polypeptide or polypeptide analog, antibody or antibody fragment, metal chelating agent, cofactor, fatty acid, carbohydrate, polynucleotide, DNA, RNA, PCR probe, antisense polynucleotide, ribo-oligonucleotide, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labeling , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or non-covalently interact with other molecules, photocausing moieties, actinic radiation excitable moieties, ligands, photoisomerizing moieties, biotin, biotin analogs , Heavy atom-incorporated moieties, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox-activators, amino thio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups , Chromophore group, chemiluminescent group, fluorescent moiety, electron high density group, magnetic group, intercalating group, chelating group, chromophore, energy transfer agent, biologically active agent, detectable label, small molecule, inhibitory ribonucleic acid, siRNA, radionucleotide , Neutron-capturing agent, biotin derivative, quantum dot(s), nanotransmitter, radiotransmitter, abzyme, enzyme, activated complex activator, virus, toxin, adjuvant, TLR2 agonist, TLR4 agonist, TLR7 Agonist, TLR9 agonist, TLR8 agonist, T-cell epitope, phospholipid, LPS-like molecule, keyhole limpet hemocyanin (KLH), immunogenic hapten, aglycan, allergen, angiostatin, antihormonal, antioxidant, Aptamers, guide RNAs, saponins, shuttle vectors, macromolecules, mimotopes, receptors, reverse micelles, detergents, immune response enhancers, fluorescent dyes, FRET reagents, radiation-imaging probes, other spectroscopic probes, prodrugs, for immunotherapy Toxin, solid support, -CH 2 CH 2 -(OCH 2 CH 2 O) p -OX 2 ,- O- (CH 2 CH 2 O) p CH 2 CH 2 -X 2, and any combination thereof (wherein, p is 1 to 10,000, X 2 is H, C 1 - 8 alkyl, protecting group or a terminal functional group Im) is included, but is not limited thereto.
도 30에 나타낸 임의의 부착 지점 R7 내지 R11을 이용하여 임의의 이들 상기 언급된 기를 2-ABA, 2-AAP 및 2-ANPA에 부착한다. 다른 실시양태에서, 벤젠 고리는 나프탈렌을 비롯한 (이에 제한되지는 않음) 본원에서 제공되는 임의의 다른 고리 구조로 대체된다. 다른 실시양태에서, 벤젠 고리는 당으로 대체된다.Attach any of these above mentioned groups to 2-ABA, 2-AAP and 2-ANPA using any of the attachment points R 7 to R 11 shown in FIG. 30. In other embodiments, the benzene ring is replaced with any other ring structure provided herein, including but not limited to naphthalene. In other embodiments, the benzene ring is replaced with a sugar.
단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 및 PCL의 유도체화를 위해서 본원에서 제공되는 방법에 사용되는 유도체화제의 농도는 약 0.005 mM 내지 약 50 mM의 범위이다. 특정 실시양태에서, 피롤리신 또는 PCL의 유도체화를 위해서 본원에서 제공되는 방법에 사용되는 유도체화제의 농도는 약 0.005 mM 내지 약 25 mM의 범위이다. 특정 실시양태에서, 피롤리신 또는 PCL의 유도체화를 위해서 본원에서 제공되는 방법에 사용되는 유도체화제의 농도는 약 0.005 mM 내지 약 10 mM의 범위이다. 특정 실시양태에서, 피롤리신 또는 PCL의 유도체화를 위해서 본원에서 제공되는 방법에 사용되는 유도체화제의 농도는 약 0.005 mM 내지 약 5 mM의 범위이다. 특정 실시양태에서, 피롤리신 또는 PCL의 유도체화를 위해서 본원에서 제공되는 방법에 사용되는 유도체화제의 농도는 약 0.005 mM 내지 약 2 mM의 범위이다. 특정 실시양태에서, 피롤리신 또는 PCL의 유도체화를 위해서 본원에서 제공되는 방법에 사용되는 유도체화제의 농도는 약 0.005 mM 내지 약 1 mM의 범위이다.The concentration of the derivatizing agent used in the methods provided herein for the derivatization of pyrrolysine and PCL incorporated into proteins, polypeptides and/or peptides ranges from about 0.005 mM to about 50 mM. In certain embodiments, the concentration of the derivatizing agent used in the methods provided herein for derivatization of pyrrolysine or PCL ranges from about 0.005 mM to about 25 mM. In certain embodiments, the concentration of the derivatizing agent used in the methods provided herein for derivatization of pyrrolysine or PCL ranges from about 0.005 mM to about 10 mM. In certain embodiments, the concentration of the derivatizing agent used in the methods provided herein for derivatization of pyrrolysine or PCL ranges from about 0.005 mM to about 5 mM. In certain embodiments, the concentration of the derivatizing agent used in the methods provided herein for the derivatization of pyrrolysine or PCL ranges from about 0.005 mM to about 2 mM. In certain embodiments, the concentration of the derivatizing agent used in the methods provided herein for derivatization of pyrrolysine or PCL ranges from about 0.005 mM to about 1 mM.
본원에서 제공되는 유도체화 방법에서, 내부에 혼입된 1개 이상의 피롤리신 또는 PCL을 갖는 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 10000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 8000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 6000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 4000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 2000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 1000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 800의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 600의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 400의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 200의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 100의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 50의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 25의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 10의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 0.05 내지 약 1의 범위이다.In the derivatization methods provided herein, the molar ratio of the derivatizing agent to the protein, polypeptide or peptide having at least one pyrrolysine or PCL incorporated therein ranges from about 0.05 to about 10000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 8000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 6000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 4000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 2000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 1000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 800. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 600. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 400. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 200. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 100. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 50. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 25. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 10. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 0.05 to about 1.
다른 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 10000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 8000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 6000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 4000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 2000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 1000의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 800의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 600의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 400의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 200의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 100의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 50의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 25의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 1.5 내지 약 10의 범위이다. 특정 실시양태에서, 이러한 단백질, 폴리펩티드 또는 펩티드에 대한 유도체화제의 몰비는 약 6 내지 약 600의 범위이다.In other embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 10000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 8000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 6000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 4000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 2000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 1000. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 800. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 600. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 400. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 200. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 100. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 50. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 25. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 1.5 to about 10. In certain embodiments, the molar ratio of derivatizing agent to such protein, polypeptide or peptide ranges from about 6 to about 600.
본원에서 제공되는 방법 및 조성물을 사용한 단백질, 폴리펩티드 및/또는 펩티드의 유도체화에 사용되는 완충제 용액의 pH는 약 pH 2 내지 약 pH 10이다. 특정 실시양태에서, pH는 약 pH 4 내지 약 pH 8이다. 특정 실시양태에서, pH는 약 pH 4 내지 약 pH 7.5이다. 특정 실시양태에서, pH는 약 pH 4 내지 약 pH 7이다. 특정 실시양태에서, pH는 약 pH 4 내지 약 pH 6.5이다. 특정 실시양태에서, pH는 약 pH 4 내지 약 pH 6이다. 특정 실시양태에서, pH는 약 pH 4 내지 약 pH 5.5이다. 특정 실시양태에서, pH는 약 pH 4 내지 약 pH 5이다. 특정 실시양태에서, pH는 약 pH 4 내지 약 pH 4.5이다. 특정 실시양태에서, pH는 약 pH 6 내지 약 pH 8이다. 특정 실시양태에서, pH는 약 pH 6 내지 약 pH 7.5이다. 특정 실시양태에서, pH는 약 pH 6 내지 약 pH 7이다. 특정 실시양태에서, pH는 약 pH 6 내지 약 pH 6.5이다. 특정 실시양태에서, pH는 약 pH 7 내지 약 pH 8이다. 특정 실시양태에서, pH는 약 pH 7 내지 약 pH 7.5이다.The pH of the buffer solution used for the derivatization of proteins, polypeptides and/or peptides using the methods and compositions provided herein is from about
본원에서 제공되는 유도체화 방법은 단백질에 부위 특이적으로 혼입된 피롤리신 또는 PCL의 유도체화에 유용하다. 특정 실시양태에서, 이러한 유도체화 방법을 이용하여 단백질, 폴리펩티드 및/또는 펩티드에 단일 부위에서 혼입된 PCL 잔기를 유도체화한다. 다른 실시양태에서, 이러한 유도체화 방법을 이용하여 단백질, 폴리펩티드 및/또는 펩티드에 다중 부위에서 혼입된 PCL 잔기를 유도체화한다. 특정 실시양태에서, 본원에서 제공되는 방법 및 조성물을 사용하여 유도체화된 단백질, 폴리펩티드 또는 펩티드는 1개, 2개, 3개, 4개, 5개, 6개, 7개, 8개, 9개, 10개, 11개, 12개, 13개, 14개, 15개 이상의 피롤리신 또는 피롤리신 유사체를 함유한다.The derivatization method provided herein is useful for the derivatization of pyrrolysine or PCL site-specifically incorporated into a protein. In certain embodiments, such methods of derivatization are used to derivatize a PCL residue incorporated at a single site into a protein, polypeptide and/or peptide. In other embodiments, such methods of derivatization are used to derivatize PCL residues incorporated at multiple sites into proteins, polypeptides and/or peptides. In certain embodiments, the protein, polypeptide or peptide derivatized using the methods and compositions provided herein is 1, 2, 3, 4, 5, 6, 7, 8, 9 , 10, 11, 12, 13, 14, 15 or more pyrrolysines or pyrrolysine analogs.
본원에서 제공되는 또 다른 측면에서, 내부에 혼입된 1개 이상의 피롤리신 또는 PCL을 갖는 단백질, 폴리펩티드 및 펩티드는 적어도 1종의 번역 동시 또는 번역후 변형에 의해 유도체화된다. 이러한 번역 동시 또는 번역후 변형의 비제한적인 예에는 글리코실화, 아세틸화, 아실화, 메틸화, 니트로화, 황산화, 지질-변형, 팔미토일화, 팔미테이트 부가, 인산화, 당지질-연결부 변형 등이 포함되나, 이에 제한되지는 않는다. 또한, 이러한 측면에서, 적어도 1종의 이러한 번역 동시 또는 번역후 변형을 함유하는 이러한 단백질, 폴리펩티드 및 펩티드를 생성, 정제, 특징규명 및 사용하는 방법도 포함된다.In another aspect provided herein, proteins, polypeptides and peptides having at least one pyrrolysine or PCL incorporated therein are derivatized by at least one simultaneous or post-translational modification. Non-limiting examples of such simultaneous or post-translational modifications include glycosylation, acetylation, acylation, methylation, nitration, sulfation, lipid-modification, palmitoylation, palmitate addition, phosphorylation, glycolipid-linkage modification, and the like. Included, but not limited to. Also included in this aspect are methods of producing, purifying, characterizing and using such proteins, polypeptides and peptides containing at least one such simultaneous or post-translational modification.
부위 특이적 변형을 갖는 생물치료제Biotherapeutics with site-specific modification
단백질, 폴리펩티드 및/또는 펩티드에 혼입된 PCL 및 피롤리신에 커플링된 거대분자 중합체 PCL incorporated into protein, polypeptide and/or peptide and coupled to pyrrolysine Macromolecular polymer
특정 실시양태에서, 본원에 기재된 조성물, 방법, 기술 및 전략을 이용하여 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 또는 PCL 잔기에 거대분자 중합체를 부가한다. 광범위하게 다양한 거대분자 중합체를 본원에 기재된 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 및 PCL 잔기에 커플링시킬 수 있다. 이러한 변형을 이용하여 이러한 단백질, 폴리펩티드 및/또는 펩티드의 생물학적 특성을 조정하고/하거나 이러한 단백질, 폴리펩티드 및/또는 펩티드에 새로운 생물학적 특성을 제공한다. 특정 실시양태에서, 거대분자 중합체는 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 또는 PCL 잔기(들)로의 직접적인 커플링을 통해 이러한 단백질, 폴리펩티드 및/또는 펩티드에 커플링된다. 다른 실시양태에서, 거대분자 중합체는 피롤리신 또는 PCL 잔기(들)에 커플링된 이관능성, 삼관능성, 사관능성 및 다관능성 링커를 통해 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드에 커플링된다. 다른 실시양태에서, 거대분자 중합체는 피롤리신 또는 PCL 잔기(들)에 커플링된 이관능성 링커를 통해 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드에 커플링된다. 특정 실시양태에서, 이러한 이관능성, 삼관능성, 사관능성 및 다관능성 링커는 모든 말단이 피롤리신 또는 PCL 잔기와의 반응에 특이적인 치환기인 단일관능성 링커이다. 특정 실시양태에서, 이러한 이관능성, 삼관능성, 사관능성 및 다관능성 링커는 1개 이상의 말단이 피롤리신 또는 PCL 잔기(들)과의 반응에 특이적인 치환기이고, 다른 말단은 피롤리신 또는 PCL 잔기(들)과 반응하지 않는 또 다른 기능적 치환기인 이종이관능성 링커이다. 피롤리신 또는 PCL 특이적 반응성 기인 특정 치환기가 본원에서 제공되고, 이러한 이관능성, 삼관능성, 사관능성 및 다관능성 링커에서 사용되는 다른 기능적 치환기 및 생성된 연결부에는 표 1에 기재된 것들이 포함되나, 이에 제한되지는 않는다.In certain embodiments, macromolecular polymers are added to pyrrolysine or PCL residues incorporated into proteins, polypeptides and/or peptides using the compositions, methods, techniques and strategies described herein. A wide variety of macromolecular polymers can be coupled to pyrrolysine and PCL residues incorporated into the proteins, polypeptides and/or peptides described herein. Such modifications are used to modulate the biological properties of such proteins, polypeptides and/or peptides and/or provide new biological properties to such proteins, polypeptides and/or peptides. In certain embodiments, the macromolecular polymer is coupled to such proteins, polypeptides and/or peptides via direct coupling to the pyrrolysine or PCL residue(s) incorporated in the proteins, polypeptides and/or peptides provided herein. . In other embodiments, the macromolecular polymer is coupled to proteins, polypeptides and/or peptides provided herein via bifunctional, trifunctional, tetrafunctional and multifunctional linkers coupled to pyrrolysine or PCL residue(s). do. In other embodiments, the macromolecular polymer is coupled to the proteins, polypeptides and/or peptides provided herein via a bifunctional linker coupled to a pyrrolysine or PCL residue(s). In certain embodiments, such bifunctional, trifunctional, tetrafunctional and polyfunctional linkers are monofunctional linkers whose all ends are specific substituents for reaction with pyrrolysine or PCL moieties. In certain embodiments, such bifunctional, trifunctional, tetrafunctional and multifunctional linkers have at least one end being a substituent specific for reaction with pyrrolysine or PCL residue(s), and the other end is pyrrolysine or PCL. Another functional substituent that does not react with the moiety(s) is a heterobifunctional linker. Certain substituents of pyrrolysine or PCL-specific reactive groups are provided herein, and other functional substituents and resulting linkages used in such bifunctional, trifunctional, tetrafunctional and multifunctional linkers include those described in Table 1, It is not limited.
생물학적 활성 분자, 예컨대 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드에 대한 거대분자 중합체의 공유 부착은 수용해도 (예를 들어, 생리적 환경에서의 수용해도)의 증가, 생체이용률의 증가, 혈청 반감기의 증가, 치료 반감기의 증가, 면역원성의 조정, 생물학적 활성의 조정, 또는 이러한 생물학적 활성 분자의 순환 시간의 연장을 위한 접근법을 나타낸다. 이러한 거대분자 중합체의 중요 특징은 생체적합성, 무독성, 및 면역원성의 부재를 포함하고, 피롤리신 또는 PCL 잔기를 통해 거대분자 중합체에 커플링된 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드의 치료 용도를 위해서는 이러한 거대분자 중합체가 제약상 허용된다.Covalent attachment of macromolecular polymers to biologically active molecules, such as proteins, polypeptides and/or peptides provided herein, increases water solubility (e.g., water solubility in a physiological environment), increases bioavailability, increases serum half-life. An approach for increasing, increasing the therapeutic half-life, modulating immunogenicity, modulating biological activity, or prolonging the circulation time of such biologically active molecules. Important features of such macromolecular polymers include biocompatibility, non-toxicity, and absence of immunogenicity, and treatment of proteins, polypeptides and/or peptides provided herein coupled to the macromolecular polymer via a pyrrolysine or PCL moiety. For use these macromolecular polymers are pharmaceutically acceptable.
본원에 기재된 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 또는 PCL 잔기(들)에 커플링되는 특정 거대분자 중합체는 수용성 중합체이다. 특정 실시양태에서는 이러한 수용성 중합체가 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 또는 PCL 잔기(들)을 통해 커플링되고, 다른 실시양태에서는 이러한 수용성 중합체가 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 또는 PCL 잔기(들)에 커플링된 임의의 관능기 또는 치환기를 통해 이러한 단백질, 폴리펩티드 및/또는 펩티드에 커플링된다. 일부 실시양태에서, 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드는 수용성 중합체에 커플링된 1개 이상의 PCL 잔기 및 수용성 중합체에 연결된 1개 이상의 천연 발생 아미노산을 포함한다.Certain macromolecular polymers coupled to pyrrolysine or PCL residue(s) incorporated into the proteins, polypeptides and/or peptides described herein are water soluble polymers. In certain embodiments, such water soluble polymers are coupled via pyrrolysine or PCL residue(s) incorporated into proteins, polypeptides and/or peptides provided herein, and in other embodiments such water soluble polymers are provided herein. , Pyrrolysine incorporated into the polypeptide and/or peptide, or any functional group or substituent coupled to the PCL residue(s). In some embodiments, proteins, polypeptides and/or peptides provided herein comprise one or more PCL residues coupled to a water soluble polymer and one or more naturally occurring amino acids linked to a water soluble polymer.
단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 또는 PCL 잔기에 커플링된 거대분자 중합체의 구조적 형태에는 선형, 포크형 또는 분지형 중합체가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 이러한 분지형 또는 포크형 수용성 중합체의 주쇄는 2개 내지 약 300개의 말단을 갖는다. 특정 실시양태에서, 이러한 다관능성 중합체 유도체, 예를 들어 (이에 제한되지는 않음) 2개의 말단을 갖는 선형 중합체의 각 말단은 관능기를 포함한다. 특정 실시양태에서는 이러한 관능기가 동일하고, 다른 실시양태에서는 이러한 관능기가 상이하다. 이러한 말단 관능기의 비제한적인 예에는 N-숙신이미딜 카르보네이트, 아민, 히드라지드, 숙신이미딜 프로피오네이트 및 숙신이미딜 부타노에이트, 숙신이미딜 숙시네이트, 숙신이미딜 에스테르, 벤조트리아졸 카르보네이트, 글리시딜 에테르, 옥시카르보닐이미다졸, p-니트로페닐 카르보네이트, 알데히드, 말레이미드, 오르토피리딜-디술피드, 아크릴롤 및 비닐술폰이 포함되나, 이에 제한되지는 않는다. 다른 실시양태에서, 관능기에는 표 1에 기재된 것들이 포함된다.Structural forms of macromolecular polymers coupled to pyrrolysine or PCL residues incorporated into proteins, polypeptides and/or peptides include, but are not limited to, linear, forked or branched polymers. In certain embodiments, the backbone of such a branched or forked water soluble polymer has from 2 to about 300 ends. In certain embodiments, each end of such a polyfunctional polymer derivative, such as, but not limited to, a linear polymer having two ends comprises a functional group. In certain embodiments, these functional groups are the same, and in other embodiments, these functional groups are different. Non-limiting examples of such terminal functional groups include N-succinimidyl carbonate, amine, hydrazide, succinimidyl propionate and succinimidyl butanoate, succinimidyl succinate, succinimidyl ester, benzotria Sol carbonate, glycidyl ether, oxycarbonylimidazole, p-nitrophenyl carbonate, aldehyde, maleimide, orthopyridyl-disulfide, acrylol, and vinylsulfone. Does not. In other embodiments, functional groups include those listed in Table 1.
생물학적 활성 분자, 예컨대 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드에 대한 수용성 중합체 (또한, 본원에서 친수성 중합체라 지칭되기도 함)의 공유 부착은 수용해도 (예를 들어, 생리적 환경에서의 수용해도)의 증가, 생체이용률의 증가, 혈청 반감기의 증가, 치료 반감기의 증가, 면역원성의 조정, 생물학적 활성의 조정, 또는 이러한 생물학적 활성 분자의 순환 시간의 연장을 위한 접근법을 나타낸다. 이러한 수용성 중합체의 중요 특징은 생체적합성, 무독성, 및 면역원성의 부재를 포함하고, 피롤리신 또는 PCL 잔기를 통해 수용성 중합체에 커플링된 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드의 치료 용도를 위해서는 이러한 거대분자 중합체가 제약상 허용된다.The covalent attachment of a water-soluble polymer (also referred to herein as a hydrophilic polymer) to a biologically active molecule, such as a protein, polypeptide and/or peptide provided herein, is a water solubility (e.g., water solubility in a physiological environment). Of, increase in bioavailability, increase in serum half-life, increase in therapeutic half-life, modulate immunogenicity, modulate biological activity, or prolong the circulation time of such biologically active molecules. Important features of such water-soluble polymers include biocompatibility, non-toxicity, and absence of immunogenicity, and for therapeutic use of proteins, polypeptides and/or peptides provided herein coupled to water-soluble polymers via pyrrolysine or PCL moieties. For this, these macromolecular polymers are pharmaceutically acceptable.
피롤리신 또는 PCL 잔기를 통해 단백질, 폴리펩티드 및/또는 펩티드에 커플링된 친수성 중합체에는 폴리알킬 에테르 및 그의 알콕시-캡핑된 유사체, 폴리비닐피롤리돈, 폴리비닐알킬 에테르, 폴리옥사졸린, 폴리알킬 옥사졸린, 폴리히드록시알킬 옥사졸린, 폴리아크릴아미드, 폴리알킬 아크릴아미드, 폴리히드록시알킬 아크릴아미드, 폴리히드록시알킬 아크릴레이트, 폴리시알산 및 그의 유사체, 친수성 펩티드 서열; 폴리사카라이드 및 이들의 유도체, 셀룰로스 및 그의 유도체, 키틴 및 그의 유도체, 하이알루론산 및 그의 유도체, 전분, 알기네이트, 콘드로이틴 술페이트, 알부민, 풀룰란, 카르복시메틸 풀룰란, 폴리아미노산 및 그의 유도체, 말레산 무수물 공중합체, 폴리비닐 알콜 및 그의 공중합체, 폴리비닐 알콜 및 그의 삼원공중합체, 및 이들의 조합물이 포함되나, 이에 제한되지는 않는다.Hydrophilic polymers coupled to proteins, polypeptides and/or peptides via pyrrolysine or PCL moieties include polyalkyl ethers and alkoxy-capped analogs thereof, polyvinylpyrrolidone, polyvinylalkyl ether, polyoxazoline, polyalkyl. Oxazoline, polyhydroxyalkyl oxazoline, polyacrylamide, polyalkyl acrylamide, polyhydroxyalkyl acrylamide, polyhydroxyalkyl acrylate, polysialic acid and its analogs, hydrophilic peptide sequences; Polysaccharides and derivatives thereof, cellulose and derivatives thereof, chitin and derivatives thereof, hyaluronic acid and derivatives thereof, starch, alginate, chondroitin sulfate, albumin, pullulan, carboxymethyl pullulan, polyamino acids and derivatives thereof, maleic acid Anhydride copolymers, polyvinyl alcohol and copolymers thereof, polyvinyl alcohol and terpolymers thereof, and combinations thereof.
이러한 폴리알킬 에테르 및 그의 알콕시-캡핑된 유사체에는 폴리옥시에틸렌 글리콜 (또한, 폴리(에틸렌 글리콜) 또는 PEG라고 공지되어 있음), 폴리옥시에틸렌/프로필렌 글리콜, 및 메톡시 또는 그의 에톡시-캡핑된 유사체가 포함되나, 이에 제한되지는 않는다. 이러한 폴리히드록시알킬 아크릴아미드에는 폴리히드록시프로필메타크릴아미드 및 그의 유도체가 포함되나, 이에 제한되지는 않는다. 이러한 폴리사카라이드 및 그의 유도체에는 덱스트란 및 덱스트란 유도체 (예를 들어 (단지 예시로서), 카르복시메틸덱스트란, 덱스트란 술페이트 및 아미노덱스트란)가 포함된, 이에 제한되지는 않는다. 이러한 셀룰로스 및 그의 유도체에는 카르복시메틸 셀룰로스 및 히드록시알킬 셀룰로스가 포함되나, 이에 제한되지는 않는다. 이러한 키틴 및 그의 유도체에는 키토산, 숙시닐 키토산, 카르복시메틸키틴 및 카르복시메틸키토산이 포함되나, 이에 제한되지는 않는다. 이러한 폴리아미노산 및 그의 유도체에는 폴리글루탐산, 폴리리신, 폴리아스파르트산 및 폴리아스파르트아미드가 포함되나, 이에 제한되지는 않는다. 이러한 말레산 무수물 공중합체에는 스티렌 말레산 무수물 공중합체 및 디비닐에틸 에테르 말레산 무수물 공중합체가 포함되나, 이에 제한되지는 않는다.Such polyalkyl ethers and alkoxy-capped analogs thereof include polyoxyethylene glycol (also known as poly(ethylene glycol) or PEG), polyoxyethylene/propylene glycol, and methoxy or ethoxy-capped analogs thereof. Includes, but is not limited to. Such polyhydroxyalkyl acrylamides include, but are not limited to, polyhydroxypropylmethacrylamide and derivatives thereof. Such polysaccharides and derivatives thereof include, but are not limited to, dextran and dextran derivatives (eg (by way of example only), carboxymethyldextran, dextran sulfate and aminodextran). Such cellulose and its derivatives include, but are not limited to, carboxymethyl cellulose and hydroxyalkyl cellulose. Such chitin and its derivatives include, but are not limited to, chitosan, succinyl chitosan, carboxymethylchitin, and carboxymethylchitosan. Such polyamino acids and derivatives thereof include, but are not limited to, polyglutamic acid, polylysine, polyaspartic acid and polyaspartamide. Such maleic anhydride copolymers include, but are not limited to, styrene maleic anhydride copolymers and divinylethyl ether maleic anhydride copolymers.
일부 실시양태에서, 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 또는 PCL 잔기(들)을 통해 이러한 단백질, 폴리펩티드 및/또는 펩티드에 직접 또는 간접적으로 커플링된 수용성 중합체는 폴리(에틸렌 글리콜) (PEG)이다. 폴리(에틸렌 글리콜)은 생체적합성이라 여겨지며, PEG는 살아있는 조직 또는 유기체에 해를 야기하지 않으면서 이것과 공존이 가능하다. PEG는 또한 실질적으로 비-면역원성이고, 따라서 체내에서 면역 반응을 일으키는 경향이 없다. PEG는 의약품, 인공 이식물, 및 생체적합성, 무독성, 및 면역원성의 부재가 중요한 기타 용도에서 널리 사용되어 온 친수성 중합체이다. PEG 접합체는 실질적인 면역 반응을 일으키지 않거나 혈액 응고 또는 기타 바람직하지 않은 효과를 유발하지 않는 경향이 있다. 따라서, 체내에서 일부 바람직한 기능을 갖는 분자, 예컨대 생물학적 활성제에 부착되는 경우, PEG는 그 작용제를 차폐하는 경향이 있고, 임의의 면역 반응을 감소시키거나 없앨 수 있어서 유기체가 그 작용제의 존재를 허용할 수 있게 한다. In some embodiments, a water-soluble polymer coupled directly or indirectly to such protein, polypeptide and/or peptide via pyrrolysine or PCL residue(s) incorporated into the proteins, polypeptides and/or peptides provided herein is a poly (Ethylene glycol) (PEG). Poly(ethylene glycol) is considered biocompatible, and PEG can coexist with it without causing harm to living tissues or organisms. PEG is also substantially non-immunogenic and therefore does not tend to elicit an immune response in the body. PEG is a hydrophilic polymer that has been widely used in pharmaceuticals, artificial implants, and other applications where biocompatibility, non-toxicity, and absence of immunogenicity are important. PEG conjugates tend not to elicit a substantial immune response or cause blood clotting or other undesirable effects. Thus, when attached to a molecule that has some desirable function in the body, such as a biologically active agent, PEG tends to mask the agent and can reduce or eliminate any immune response so that the organism will allow the presence of the agent. Make it possible.
일부 실시양태에서는 본원에서 제공되는 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 또는 PCL 잔기(들)을 통해 이러한 단백질, 폴리펩티드 및/또는 펩티드에 직접 또는 간접적으로 커플링된 폴리(에틸렌 글리콜)이 직쇄 중합체이고, 다른 실시양태에서는 이러한 PEG 중합체가 분지형 중합체이다. 이러한 직쇄 및 분지형 PEG 중합체의 분자량 또는 분자량 분포는 약 100 Da 내지 약 100,000 Da 또는 그보다 높은 값이다. 일부 실시양태에서, 이러한 PEG 중합체의 분자량 또는 분자량 분포는 약 100 Da 내지 50,000 Da이다. 일부 실시양태에서, 이러한 PEG 중합체의 분자량 또는 분자량 분포는 약 100 Da 내지 40,000 Da이다. 일부 실시양태에서, 이러한 PEG 중합체의 분자량 또는 분자량 분포는 약 1,000 Da 내지 40,000 Da이다. 일부 실시양태에서, 이러한 PEG 중합체의 분자량 또는 분자량 분포는 약 5,000 Da 내지 40,000 Da이다. 일부 실시양태에서, 이러한 PEG 중합체의 분자량 또는 분자량 분포는 약 10,000 Da 내지 40,000 Da이다. 특정 실시양태에서, 이러한 PEG 중합체의 분자량은 100,000 Da, 95,000 Da, 90,000 Da, 85,000 Da, 80,000 Da, 75,000 Da, 70,000 Da, 65,000 Da, 60,000 Da, 55,000 Da, 50,000 Da, 45,000 Da, 40,000 Da, 35,000 Da, 30,000 Da, 25,000 Da, 20,000 Da, 15,000 Da, 10,000 Da, 9,000 Da, 8,000 Da, 7,000 Da, 6,000 Da, 5,000 Da, 4,000 Da, 3,000 Da, 2,000 Da, 1,000 Da, 900 Da, 800 Da, 700 Da, 600 Da, 500 Da, 400 Da, 300 Da, 200 Da 또는 100 Da이다.In some embodiments, poly(ethylene glycol) coupled directly or indirectly to such protein, polypeptide and/or peptide via pyrrolysine or PCL residue(s) incorporated into the proteins, polypeptides and/or peptides provided herein. This is a straight chain polymer, and in other embodiments this PEG polymer is a branched polymer. The molecular weight or molecular weight distribution of these straight chain and branched PEG polymers ranges from about 100 Da to about 100,000 Da or higher. In some embodiments, the molecular weight or molecular weight distribution of such PEG polymers is between about 100 Da and 50,000 Da. In some embodiments, the molecular weight or molecular weight distribution of such PEG polymers is between about 100 Da and 40,000 Da. In some embodiments, the molecular weight or molecular weight distribution of such PEG polymers is between about 1,000 Da and 40,000 Da. In some embodiments, the molecular weight or molecular weight distribution of such PEG polymers is between about 5,000 Da and 40,000 Da. In some embodiments, the molecular weight or molecular weight distribution of such PEG polymers is between about 10,000 Da and 40,000 Da. In certain embodiments, the molecular weight of such PEG polymers is 100,000 Da, 95,000 Da, 90,000 Da, 85,000 Da, 80,000 Da, 75,000 Da, 70,000 Da, 65,000 Da, 60,000 Da, 55,000 Da, 50,000 Da, 45,000 Da, 40,000 Da, 35,000 Da, 30,000 Da, 25,000 Da, 20,000 Da, 15,000 Da, 10,000 Da, 9,000 Da, 8,000 Da, 7,000 Da, 6,000 Da, 5,000 Da, 4,000 Da, 3,000 Da, 2,000 Da, 1,000 Da, 900 Da, 800 Da , 700 Da, 600 Da, 500 Da, 400 Da, 300 Da, 200 Da or 100 Da.
단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 또는 PCL 잔기(들)에 커플링된 PEG 중합체의 구조적 형태에는 선형, 포크형 또는 분지형이 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 이러한 분지형 또는 포크형 PEG 중합체의 주쇄는 2개 내지 약 300개의 말단을 갖는다. 특정 실시양태에서, 이러한 다관능성 중합체 유도체, 예를 들어 (이에 제한되지는 않음) 2개의 말단을 갖는 선형 중합체의 각 말단은 관능기를 포함한다. 특정 실시양태에서는 이러한 관능기가 동일하고, 다른 실시양태에서는 이러한 관능기 중 적어도 하나가 상이하다. 다른 실시양태에서, 이러한 관능기는 상이하다.Structural forms of PEG polymers coupled to pyrrolysine or PCL residue(s) incorporated into proteins, polypeptides and/or peptides include, but are not limited to, linear, forked or branched. In certain embodiments, the backbone of such branched or forked PEG polymers has from 2 to about 300 ends. In certain embodiments, each end of such a polyfunctional polymer derivative, such as, but not limited to, a linear polymer having two ends comprises a functional group. In certain embodiments, these functional groups are the same, and in other embodiments, at least one of these functional groups is different. In other embodiments, these functional groups are different.
특정 실시양태에서, 단백질, 폴리펩티드 및/또는 펩티드와의 커플링 전에 PEG 중합체의 1개 이상의 말단은 관능기를 포함한다. 특정 실시양태에서는 PEG가 1개의 말단에 관능기를 갖는 선형 중합체이고, 다른 실시양태에서는 PEG가 각각의 말단에 관능기를 갖는 선형 중합체여서 이관능성 PEG 중합체를 형성한다. 다른 실시양태에서는 PEG가 1개의 말단에 관능기를 갖는 포크형 중합체이고, 다른 실시양태에서는 PEG가 2개 이상의 말단에 관능기를 갖는 포크형 중합체이다. 다른 실시양태에서, PEG는 각각의 말단에 관능기를 갖는 포크형 중합체여서 다관능성 PEG 중합체를 형성한다. 다른 실시양태에서는 PEG가 1개의 말단에 관능기를 갖는 분지형 중합체이고, 다른 실시양태에서는 PEG가 2개 이상의 말단에 관능기를 갖는 분지형 중합체이다. 다른 실시양태에서, PEG는 각각의 말단에 관능기를 갖는 분지형 중합체여서 다관능성 PEG 중합체를 형성한다. 이러한 상기 언급된 PEG 중합체의 특정 실시양태에서는 관능기가 동일하고, 이러한 관능기의 다른 실시양태에서는 적어도 1개의 관능기가 상이하다. 이러한 상기 언급된 PEG 중합체의 다른 실시양태에서, 관능기는 상이하다. 그러나, PEG 중합체의 적어도 1개의 말단은 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 적어도 1개의 피롤리신 또는 PCL 잔기와의 반응에 이용가능하다.In certain embodiments, at least one end of the PEG polymer comprises a functional group prior to coupling with the protein, polypeptide and/or peptide. In certain embodiments, PEG is a linear polymer having a functional group at one end, and in other embodiments, PEG is a linear polymer having a functional group at each end to form a bifunctional PEG polymer. In other embodiments, the PEG is a forked polymer having a functional group at one end, and in other embodiments, the PEG is a forked polymer having a functional group at two or more ends. In other embodiments, the PEG is a forked polymer having a functional group at each end to form a multifunctional PEG polymer. In other embodiments, the PEG is a branched polymer having a functional group at one end, and in other embodiments, the PEG is a branched polymer having a functional group at two or more ends. In other embodiments, the PEG is a branched polymer having a functional group at each end to form a multifunctional PEG polymer. In certain embodiments of these aforementioned PEG polymers the functional groups are the same, and in other embodiments of these functional groups at least one functional group is different. In other embodiments of these aforementioned PEG polymers, the functional groups are different. However, at least one end of the PEG polymer is available for reaction with at least one pyrrolysine or PCL moiety incorporated in the protein, polypeptide and/or peptide.
말단 관능기의 비제한적인 예에는 N-숙신이미딜 카르보네이트, 아민, 히드라지드, 숙신이미딜 프로피오네이트, 숙신이미딜 부타노에이트, 숙신이미딜 숙시네이트, 숙신이미딜 에스테르, 벤조트리아졸 카르보네이트, 글리시딜 에테르, 옥시카르보닐이미다졸, p-니트로페닐 카르보네이트, 알데히드, 말레이미드, 오르토피리딜-디술피드, 아크릴롤, 비닐술폰, 활성화된 카르보네이트 (예를 들어 (이에 제한되지는 않음), p-니트로페닐 에스테르), 활성화된 에스테르 (예를 들어 (이에 제한되지는 않음), N-히드록시숙신이미드, p-니트로페닐 에스테르), 옥심, 카르보닐, 디카르보닐, 히드록실아민, 히드록실, 메톡시, 벤즈알데히드, 아세토페논, 2-아미노-벤즈알데히드, 2-아미노-아세토페논 및 2-아미노-5-니트로-벤조페논이 포함되나, 이에 제한되지는 않는다.Non-limiting examples of terminal functional groups include N-succinimidyl carbonate, amine, hydrazide, succinimidyl propionate, succinimidyl butanoate, succinimidyl succinate, succinimidyl ester, benzotriazole Carbonate, glycidyl ether, oxycarbonylimidazole, p-nitrophenyl carbonate, aldehyde, maleimide, orthopyridyl-disulfide, acrylol, vinylsulfone, activated carbonate (e.g. For example (but not limited to), p-nitrophenyl ester), activated ester (e.g., but not limited to), N-hydroxysuccinimide, p-nitrophenyl ester), oxime, carbonyl , Dicarbonyl, hydroxylamine, hydroxyl, methoxy, benzaldehyde, acetophenone, 2-amino-benzaldehyde, 2-amino-acetophenone and 2-amino-5-nitro-benzophenone. Does not.
도 31은 단백질에 혼입된 적어도 1개의 PCL 잔기를 통해 상기 단백질에 커플링된 관능화된 PEG 중합체의 한 실시양태, 2-아미노-아세토페논-PEG8 (2-AAP-PEG8; TU3205-044)을 예시한다. 나타낸 바와 같이, 2-AAP-PEG8의 2-아미노-아세토페논 잔기는 PEG8과 단백질 사이의 스페이서로서 퀴나졸린-유형 잔기를 형성한다. 이러한 퀴나졸린-유형 잔기는 추가로 반응하여 융합된 고리 구조를 형성할 수 있다. 2-AAP-PEG8의 커플링의 경우, 이러한 유도체화는 상기 단백질의 질량에 556 Da을 추가한다. 본원에서 제공되는 방법에서 사용되는 다른 관능화된 PEG에는 실시예 20에 기재된 것이 포함되나, 이에 제한되지는 않는다.FIG. 31 is an embodiment of a functionalized PEG polymer coupled to the protein via at least one PCL residue incorporated into the protein, 2-amino-acetophenone-PEG 8 (2-AAP-PEG8; TU3205-044). Illustratively. As shown, the 2-amino-acetophenone residue of 2-AAP-PEG8 forms a quinazoline-type residue as a spacer between PEG 8 and the protein. These quinazoline-type moieties can further react to form a fused ring structure. In the case of the coupling of 2-AAP-PEG8, this derivatization adds 556 Da to the mass of the protein. Other functionalized PEGs used in the methods provided herein include, but are not limited to, those described in Example 20.
도 32는 위치 122에서 혼입된 PCL을 갖는 hRBP4 (hRBP4 Phe122PCL)의 질량 스펙트럼을 pH 7.5 (도 32A) 및 pH 5.0 (도 32B)에서의 2-AAP-PEG8에 의한 유도체화 이전 및 이후에 나타낸 것이다. 이러한 커플링 반응은 pH 7.5 및 pH 5에서 수행되었고, 여기서 89% 내지 100% 전환이 일어나서 미반응 단백질 대비 556 Da의 예상된 질량 증가가 있었다 (도 32C). 야생형 hRBP4는 450배 또는 2300배 과량의 2-AAP-PEG8과 반응하지 않았다 (도 32D 내지 F). PCL 부재시에는 커플링이 일어나지 않기 때문에, 이것은 PCL과 2AAP-PEG8 사이의 커플링 반응의 특이성을 추가로 예시한다.Figure 32 shows the mass spectrum of hRBP4 (hRBP4 Phe122PCL) with PCL incorporated at position 122 before and after derivatization with 2-AAP-PEG8 at pH 7.5 (Figure 32A) and pH 5.0 (Figure 32B). . This coupling reaction was carried out at pH 7.5 and
이러한 표지화 방법의 유용성을 추가로 예시하기 위해서, 에스케리키아 콜라이에서 생성된, 위치 2454에서 혼입된 PCL을 갖는 인간 지방산 신테타제 (FAS-TE) (FAS-TE Tyr2454PCL)의 티오에스테라제 도메인을 2-AAP-PEG8로 유도체화시켰다 (도 33, 실시예 14 참조). 도 33A는 미반응 단백질의 질량 스펙트럼 (질량 33202 Da)을 보여주고, 도 33B는 2-AAP-PEG8 (TU3205-044)와 완료시까지 반응시켜 예상 질량 33756 Da을 갖는 FAS-TE Tyr2454PCL의 질량 스펙트럼을 보여준다. 추가의 예시는 도 33C 및 도 33D에서 제공되며, 이것은 에스케리키아 콜라이에서 생성된, 위치 2454에서 혼입된 PCL을 갖는 FAS-TE (FAS-TE Tyr2454PCL)를 실온에서 (도 33C) 및 4℃에서 (도 33D) 2.4 kDa 2-AAP-PEG (TU3205-048)로 유도체화한 후의 질량 스펙트럼을 보여준다. 이용된 조건하에서, 겨우 대략 25%의 전환율이 달성되어 예상 질량 35585 Da의 단백질을 수득하였다. To further illustrate the utility of this labeling method, the thioesterase domain of human fatty acid synthetase (FAS-TE) (FAS-TE Tyr2454PCL) with PCL incorporated at
또한, 도 34는 2.4 kDa 2-AAP-PEG 및 23 kDa 2-AAP-PEG를 도면에 나타낸 몰비로 사용한 FAS-TE Tyr2454PCL의 PEG화를 보여준다. 0.5 kDa 2-AAP-PEG (2-AAP-PEG8) PEG화 생성물은 SDS-PAGE에서 분리되지 않았지만, 질량 분광분석법으로 입증되었다.In addition, Figure 34 shows the PEGylation of FAS-TE Tyr2454PCL using 2.4 kDa 2-AAP-PEG and 23 kDa 2-AAP-PEG in the molar ratio shown in the figure. The 0.5 kDa 2-AAP-PEG (2-AAP-PEG8) PEGylated product was not separated on SDS-PAGE, but was verified by mass spectrometry.
본원에서 제공되는 방법을 이용하여 섬유모세포 성장 인자 21 (FGF-21)에 20개 위치에서 PCL을 혼입한 후에 상응하는 PCL 잔기를 PEG화하였다 (실시예 15 참조). 도 35는 FGF21 Lys84PCL을 2-AAP-PEG8로 유도체화시키기 이전 및 이후에 얻은 질량 스펙트럼을 보여준다. 도 35A는 미반응 FGF21 Lys84PCL (21235.6 Da)의 질량 스펙트럼이고, 도 35B는 2-AAP-PEG8와 반응시킨 FGF21 Lys84PCL의 질량 스펙트럼이다. PEG화 반응은 완료시까지 진행되어 21792.4 Da의 단백질을 생성했다. 556.8 Da의 증가분은 상기 유도체화에 대해 예상된 556 Da 증가와 일치한다. 도 36은 FGF21 PCL 돌연변이체 7가지를 23 kDa 2-AAP-PEG로 유도체화한 후에 수득된 SDS-PAGE 결과를 보여준다. 도 36A는 23 kDa 2-AAP-PEG와 반응시킨 (pH 7.4, 4℃, 60시간) FGF21 PCL 돌연변이체 (0.1 내지 0.4 mM) 반응 혼합물의 SDS 겔이며, PEG-FGF21, 전장 (FL) FGF21-PCL 및 말단절단 (TR) FGF21-PCL을 보여준다. 도 36B는 PEG-FGF21, 전장 (FL) 및 말단절단 (TR) FGF21의 부분적 정제 후 8가지 FGF21 PCL 돌연변이체의 SDS-PAGE 결과를 보여준다.After incorporation of PCL at 20 positions into fibroblast growth factor 21 (FGF-21) using the method provided herein, the corresponding PCL residues were PEGylated (see Example 15). Figure 35 shows mass spectra obtained before and after derivatization of FGF21 Lys84PCL with 2-AAP-PEG8. Fig. 35A is a mass spectrum of unreacted FGF21 Lys84PCL (21235.6 Da), and Fig. 35B is a mass spectrum of FGF21 Lys84PCL reacted with 2-AAP-PEG8. The PEGylation reaction proceeded to completion, resulting in a protein of 21792.4 Da. The increase of 556.8 Da is consistent with the expected increase of 556 Da for this derivatization. Figure 36 shows the SDS-PAGE results obtained after derivatization of 7 kinds of FGF21 PCL mutants with 23 kDa 2-AAP-PEG. Figure 36A is an SDS gel of a reaction mixture of FGF21 PCL mutant (0.1 to 0.4 mM) reacted with 23 kDa 2-AAP-PEG (pH 7.4, 4° C., 60 hours), PEG-FGF21, full length (FL) FGF21- PCL and truncated (TR) FGF21-PCL are shown. Figure 36B shows the SDS-PAGE results of eight FGF21 PCL mutants after partial purification of PEG-FGF21, full length (FL) and truncated (TR) FGF21.
본원에서 제공되는 방법을 이용하여 마우스 에리트로포이에틴 (EPO)에 11개 위치에서 PCL을 혼입한 후에 3가지 EPO PCL 돌연변이체에 대해 상응하는 PCL 잔기를 PEG화하였다 (실시예 6 참조). 도 37은 마우스 EPO PCL 돌연변이체의 PEG화 후에 수득된 SDS 겔이다. HEK293F 세포에서 3개의 상이한 위치에서 혼입된 PCL을 갖는 마우스 EPO를 2-AAP-mPEG23k (TU3205-052)와 반응시켰다. PEG화된 EPO 및 PEG화되지 않은 EPO를 화살표로 표시했다.After incorporation of PCL at 11 positions into mouse erythropoietin (EPO) using the method provided herein, the corresponding PCL residues were PEGylated for the three EPO PCL mutants (see Example 6). 37 is an SDS gel obtained after PEGylation of mouse EPO PCL mutants. Mouse EPO with PCL incorporated at three different positions in HEK293F cells was reacted with 2-AAP-mPEG23k (TU3205-052). PEGylated EPO and non-PEGylated EPO are indicated by arrows.
특정 실시양태에서, 단백질, 폴리펩티드 및/또는 펩티드에 PCL 잔기(들)을 통해 연결된 이관능성 또는 다관능성 PEG 중합체를 미반응 말단상의 나머지 관능기를 이용하여 추가로 유도체화 또는 치환한다. 특정 실시양태에서, 벤즈알데히드, 벤조페논 또는 아세토페논 잔기와 PCL 잔기(들)의 반응을 통해 단백질, 폴리펩티드 및/또는 펩티드에 연결된 이관능성 또는 다관능성 PEG 중합체를 미반응 말단상의 나머지 관능기를 이용하여 추가로 유도체화 또는 치환한다. In certain embodiments, the bifunctional or polyfunctional PEG polymer linked to the protein, polypeptide and/or peptide via PCL residue(s) is further derivatized or substituted with the remaining functional groups on the unreacted end. In certain embodiments, a bifunctional or polyfunctional PEG polymer linked to a protein, polypeptide and/or peptide via reaction of a benzaldehyde, benzophenone or acetophenone moiety with a PCL moiety(s) is added using the remaining functional groups on the unreacted end. Derivatized or substituted with.
본원에 기재된 방법 및 조성물은 PEG 유도체를 사용한 단백질, 폴리펩티드 및 펩티드의 선택적인 변형에 매우 효율적인 방법을 제공하며, 이것은 아미노산 PCL 또는 피롤리신을 셀렉터 코돈에 대한 반응으로 단백질에 선택적으로 혼입한 후에 적합하게 관능화된 PEG 유도체를 사용하여 상응하는 아미노산 잔기를 변형시키는 것을 포함한다. 따라서, 본원에서는 내부에 혼입된 PCL 또는 피롤리신 잔기(들)을 갖는 단백질, 폴리펩티드 및/또는 펩티드가 제공되고, 또한 본원에서는 이러한 PCL 또는 피롤리신 잔기(들)을 통해 수용성 중합체, 예컨대 폴리(에틸렌 글리콜) (PEG)에 연결된 이러한 단백질, 폴리펩티드 및/또는 펩티드가 제공된다.The methods and compositions described herein provide a highly efficient method for the selective modification of proteins, polypeptides and peptides using PEG derivatives, which are suitably incorporated after selectively incorporating the amino acid PCL or pyrrolysine into the protein in response to a selector codon. It involves modifying the corresponding amino acid residues using functionalized PEG derivatives. Accordingly, provided herein are proteins, polypeptides and/or peptides having PCL or pyrrolysine residue(s) incorporated therein, and also herein is a water-soluble polymer such as poly(s) via such PCL or pyrrolysine residue(s). Such proteins, polypeptides and/or peptides linked to (ethylene glycol) (PEG) are provided.
본원에 기재된 단백질, 폴리펩티드 및/또는 펩티드의 PEG화 정도 (즉, 단백질, 폴리펩티드 및/또는 펩티드에 연결된 PEG 중합체의 수)는 변경된 약리적, 약동학적 또는 약력학적 특징이 제공되도록 조정가능하다. 특정 실시양태에서는 이러한 변경이 이러한 특징에서의 증가이고, 다른 실시양태에서는 이러한 변경이 이러한 특징에서의 감소이다. 특정 실시양태에서, 이러한 특징은 생체내 반감기이다. 일부 실시양태에서, 본원에서 제공되는 방법 및 조성물을 이용하여 PEG화된 단백질, 폴리펩티드 및/또는 펩티드의 반감기는 미변형 단백질, 폴리펩티드 및/또는 펩티드에 비해 적어도 약 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 2배, 5배, 10배, 50배, 100배 또는 적어도 약 200배 증가된다.The degree of PEGylation of the proteins, polypeptides and/or peptides described herein (i.e., the number of PEG polymers linked to the protein, polypeptide and/or peptide) is adjustable to provide altered pharmacological, pharmacokinetic or pharmacodynamic characteristics. In certain embodiments this change is an increase in this characteristic, and in other embodiments this change is a decrease in this characteristic. In certain embodiments, this characteristic is an in vivo half-life. In some embodiments, the half-life of a protein, polypeptide and/or peptide PEGylated using the methods and compositions provided herein is at least about 10%, 20%, 30%, 40 compared to the unmodified protein, polypeptide and/or peptide. %, 50%, 60%, 70%, 80%, 90%, 2 times, 5 times, 10 times, 50 times, 100 times or at least about 200 times.
특정 실시양태에서, 본원에서 제공되는 방법 및 조성물을 이용하여 PEG화된 단백질, 폴리펩티드 및/또는 펩티드는 소수성 크로마토그래피, 친화도 크로마토그래피; 음이온 교환 크로마토그래피, 양이온 교환 크로마토그래피, Q 4급 암모늄 수지, DEAE 세파로스(SEPHAROSE), 실리카에서의 크로마토그래피; 역상 HPLC, 겔 여과 (예를 들어 (이에 제한되지는 않음), 세파덱스(SEPHADEX) G-75, 세파크릴(Sephacryl) S-100, 수퍼덱스(SUPERDEX) 75, 100 및 200 사용), 소수성 상호작용 크로마토그래피, 크기-배제 크로마토그래피, 금속-킬레이트 크로마토그래피; 한외여과/투석여과, 에탄올 침전, 크로마토포커싱, 대체형 크로마토그래피, 전기영동 절차 (예를 들어 (이에 제한되지는 않음), 정제용 등전위 포커싱), 차별적 용해도 (예를 들어 (이에 제한되지는 않음), 황산암모늄 침전) 및 추출을 비롯한 (이에 제한되지는 않음) 방법을 이용하여 정제된다.In certain embodiments, proteins, polypeptides and/or peptides PEGylated using the methods and compositions provided herein may be selected from hydrophobic chromatography, affinity chromatography; Anion exchange chromatography, cation exchange chromatography, Q quaternary ammonium resin, DEAE SEPHAROSE, chromatography on silica; Reverse phase HPLC, gel filtration (e.g., but not limited to), SEPHADEX G-75, Sephacryl S-100,
다른 측면에서, 본원에서 제공되는 방법에 사용되는 PEG 중합체는 주쇄 내에 약하거나 분해가능한 연결부를 갖는다. 단지 예로서 언급하자면, 이러한 PEG 중합체는 중합체 주쇄에 가수분해의 대상인 에스테르 연결부를 가지며, 이러한 가수분해로 상기 연결부가 절단되어 PEG가 부착된 단백질, 폴리펩티드 또는 펩티드가 방출된다.In another aspect, the PEG polymers used in the methods provided herein have weak or degradable linkages within the backbone. By way of example only, such a PEG polymer has an ester linkage to be hydrolyzed in the polymer backbone, and this hydrolysis cleaves the linkage to release the protein, polypeptide or peptide to which the PEG is attached.
다른 측면에서, 본원에 기재된 방법 및 조성물을 사용하여 중합체-단백질 접합체의 실질적으로 균질한 제제를 생성한다. 본원에서 사용된 바와 같이, "실질적으로 균질한"은 중합체:단백질 접합체 분자가 전체 단백질의 절반보다 더 많은 것으로 관찰되는 것을 의미한다. 중합체:단백질 접합체는 생물학적 활성을 갖고, 본원에서 제공되는 본 발명의 "실질적으로 균질한" PEG화 폴리펩티드 제제는 균질한 제제의 이점, 예를 들어 단지 예로서만 언급하자면, 제품번호 마다의(lot to lot) 약동학 예측에 임상적으로 사용하는데 있어서의 용이성을 나타내기에 충분하게 균질한 제제이다.In another aspect, the methods and compositions described herein are used to produce a substantially homogeneous formulation of polymer-protein conjugates. As used herein, “substantially homogeneous” means that polymer:protein conjugate molecules are observed to be more than half of the total protein. Polymer:protein conjugates have biological activity, and the "substantially homogeneous" PEGylated polypeptide formulations of the invention provided herein have the advantage of a homogeneous formulation, for example, lot by lot to lot) It is a sufficiently homogeneous formulation to show ease of clinical use in predicting pharmacokinetics.
다른 측면에서, 본원에 기재된 방법 및 조성물을 사용하여 중합체:단백질 접합체의 실질적으로 균질한 제제를 생성한다. 본원에서 사용된 바와 같이, "실질적으로 균질한"은 중합체:단백질 접합체 분자가 전체 단백질의 절반보다 더 많은 것으로 관찰되는 것을 의미한다. 중합체:단백질 접합체는 생물학적 활성을 갖고, 본원에서 제공되는 본 발명의 "실질적으로 균질한" PEG화 폴리펩티드 제제는 균질한 제제의 이점, 예를 들어 단지 예로서만 언급하자면, 제품번호 마다의 약동학 예측에 임상적으로 사용하는데 있어서의 용이성을 나타내기에 충분하게 균질한 제제이다.In another aspect, the methods and compositions described herein are used to produce a substantially homogeneous formulation of polymer:protein conjugates. As used herein, “substantially homogeneous” means that polymer:protein conjugate molecules are observed to be more than half of the total protein. Polymer:protein conjugates have biological activity, and the "substantially homogeneous" PEGylated polypeptide formulations of the invention provided herein have the advantages of a homogeneous formulation, such as, by way of example only, prediction of pharmacokinetics per product number. It is a sufficiently homogeneous formulation to show ease in clinical use.
또 다른 측면에서, 본원에 기재된 방법 및 조성물을 사용하여 중합체-단백질 접합체 분자의 혼합물을 제조하고, 본원에서 제공되는 이점은 혼합물에 포함시킬 모노-중합체:단백질 접합체의 비율이 선택가능하다는 점이다. 따라서, 특정 실시양태에서, 다양한 수의 중합체 잔기 (즉, 비-, 트리-, 테트라- 등)가 부착된 다양한 단백질의 혼합물을 제조하고, 이들 접합체를 본원에 기재된 방법으로 제조된 모노-중합체-단백질 접합체와 임의로 조합하여 소정 비율의 모노-중합체:단백질 접합체를 갖는 혼합물을 생성한다.In another aspect, the methods and compositions described herein are used to prepare a mixture of polymer-protein conjugate molecules, and an advantage provided herein is that the ratio of mono-polymer:protein conjugates to be included in the mixture is selectable. Thus, in certain embodiments, mixtures of various proteins to which various numbers of polymeric moieties (i.e., non-, tri-, tetra-, etc.) are attached are prepared, and these conjugates are mono-polymers prepared by the methods described herein. It is optionally combined with a protein conjugate to produce a mixture having a proportion of the mono-polymer:protein conjugate.
단백질 분자에 대한 폴리(에틸렌 글리콜) 분자의 비율은 반응 혼합물 중 이들의 농도에 따라 달라질 것이다. 특정 실시양태에서, 최적 (잉여 미반응 단백질 또는 중합체가 최소라는 반응 효율의 측면에서 최적)의 비율은 선택된 폴리(에틸렌 글리콜)의 분자량 및 이용가능한 반응성 기의 수에 따라 결정된다. 중합체의 분자량이 높을 수록 단백질에 부착되는 중합체 분자의 수가 더 적다. 유사하게, 다른 실시양태에서, 이들 파라미터의 최적화시에 PEG 중합체의 분지상태가 고려된다. 이러한 예에서, 분자량이 높을 수록 (또는 분지의 수가 많을 수록) 중합체:단백질 비율이 더 높다.The ratio of poly(ethylene glycol) molecules to protein molecules will depend on their concentration in the reaction mixture. In certain embodiments, the ratio of optimal (optimal in terms of reaction efficiency with minimal excess unreacted protein or polymer) is determined by the molecular weight of the selected poly(ethylene glycol) and the number of reactive groups available. The higher the molecular weight of the polymer, the fewer polymer molecules attached to the protein. Similarly, in other embodiments, the branching state of the PEG polymer is taken into account when optimizing these parameters. In this example, the higher the molecular weight (or the higher the number of branches), the higher the polymer:protein ratio.
단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 및/또는 PCL 과의 아 미노 당의 직접적인 커플링Direct coupling of amino sugars with pyrrolysine and/or PCL incorporated into proteins, polypeptides and/or peptides
본원에서 제공되는 또 다른 측면에서, 내부에 혼입된 1개 이상의 피롤리신 및/또는 PCL 잔기(들)을 갖는 단백질, 폴리펩티드 및/또는 펩티드는 아미노 당으로 유도체화된다. 도 38은 내부에 혼입된 PCL을 갖는 단백질 (R1으로 예시됨)에 D-만노사민이 커플링되는 일반적인 반응식을 보여준다. 이러한 실시양태에서, D-만노사민의 아미노알데히드 잔기가 PCL 잔기와 반응하여 단백질의 질량을 161 Da만큼 증가시킨다. 이러한 실시양태에 사용되는 다른 아미노당에는 만노사민, 갈락토사민 및 글루코사민이 포함되나, 이에 제한되지는 않는다. 도 39는 위치 122에서 혼입된 PCL을 갖는 hRBP4 (hRBP4 Phe122PCL)의 만노사민과의 반응 후 질량 스펙트럼을 보여주고, 도 40은 위치 2222에서 혼입된 PCL을 갖는 인간 지방산 신테타제 (FAS-TE) (FAS-TE Leu2222PCL/Leu2223Ile)의 만노사민과의 반응 이전 (도 40A) 및 이후 (도 40B) 질량 스펙트럼을 보여준다.In another aspect provided herein, proteins, polypeptides and/or peptides having one or more pyrrolysine and/or PCL residue(s) incorporated therein are derivatized with amino sugars. 38 shows a general reaction scheme in which D-mannosamine is coupled to a protein with PCL incorporated therein ( illustrated as R 1 ). In this embodiment, the aminoaldehyde residue of D-mannosamine reacts with the PCL residue to increase the mass of the protein by 161 Da. Other amino sugars used in this embodiment include, but are not limited to, mannosamine, galactosamine and glucosamine. Figure 39 shows the mass spectrum after the reaction of hRBP4 (hRBP4 Phe122PCL) with PCL incorporated at position 122 with mannosamine, and Figure 40 is a human fatty acid synthetase (FAS-TE) with PCL incorporated at
단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 및 PCL 과의 커플링을 통한, 이러한 단백질, 폴리펩티드 및/또는 펩티드의 글리코실화Glycosylation of such proteins, polypeptides and/or peptides via coupling with pyrrolysine and PCL incorporated into proteins, polypeptides and/or peptides
본원에 기재된 방법 및 조성물을 사용하여 사카라이드 잔기를 보유하는 1개 이상의 피롤리신 및/또는 PCL 잔기를 갖는 단백질, 폴리펩티드 및 펩티드를 수득한다. 사카라이드 잔기는 천연 (예를 들어 (이에 제한되지는 않음), N-아세틸글루코사민 및 D-만노사민) 또는 비-천연 (예를 들어 (이에 제한되지는 않음), 3-플루오로갈락토스)일 수 있다. 특정 실시양태에서, 사카라이드는 퀴나졸린-유형 잔기의 형성을 비롯한 (이에 제한되지는 않음) 비-천연 연결에 의해 피롤리신 및/또는 PCL 잔기에 연결된다. 특정 실시양태에서, 사카라이드는 환원제를 사용하여 추가로 환원시킨 퀴나졸린-유형 잔기의 형성을 비롯한 (이에 제한되지는 않음) 비-천연 연결에 의해 피롤리신 및/또는 PCL 잔기에 연결된다. 특정 실시양태에서, 사카라이드는 본원에서 제공되는 바와 같이 추가로 반응하여 융합된 고리계를 형성한 퀴나졸린-유형 잔기의 형성을 비롯한 (이에 제한되지는 않음) 비-천연 연결에 의해 피롤리신 및/또는 PCL 잔기에 연결된다 (도 22, 23 및 30 참조). 특정 실시양태에서, 내부에 혼입된 1개 이상의 피롤리신 및/또는 PCL을 갖는 단백질, 폴리펩티드 및 펩티드로의 글리코실 잔기의 부가를 비롯한 (이에 제한되지는 않음) 사카라이드(들)의 부가가 생체내 일어난다. 다른 실시양태에서, 내부에 혼입된 1개 이상의 피롤리신 및/또는 PCL을 갖는 단백질, 폴리펩티드 및 펩티드로의 글리코실 잔기의 부가를 비롯한 (이에 제한되지는 않음) 사카라이드(들)의 부가가 시험관내 일어난다. 다른 실시양태에서, 피롤리신 및/또는 PCL 잔기에 일단 부착된 사카라이드는 글리코실트랜스퍼라제 및/또는 다른 효소 처리에 의해 추가로 변형되어 내부에 혼입된 1개 이상의 피롤리신 및/또는 PCL을 갖는 단백질, 폴리펩티드 또는 펩티드에 결합된 올리고사카라이드를 생성한다.The methods and compositions described herein are used to obtain proteins, polypeptides and peptides having one or more pyrrolysine and/or PCL residues bearing saccharide residues. The saccharide moiety is natural (e.g., but not limited to), N-acetylglucosamine and D-mannosamine) or non-natural (e.g., but not limited to), 3-fluorogalactose) Can be In certain embodiments, the saccharide is linked to pyrrolysine and/or PCL residues by non-natural linkages, including but not limited to the formation of quinazoline-type residues. In certain embodiments, the saccharide is linked to the pyrrolysine and/or PCL moieties by non-natural linkages, including but not limited to the formation of quinazoline-type moieties that have been further reduced with a reducing agent. In certain embodiments, the saccharide is pyrrolysine by non-natural linkage, including, but not limited to, the formation of quinazoline-type moieties that further react as provided herein to form a fused ring system. And/or to a PCL residue (see FIGS. 22, 23 and 30). In certain embodiments, the addition of saccharide(s), including but not limited to, the addition of glycosyl residues to proteins, polypeptides and peptides having one or more pyrrolysines and/or PCL incorporated therein It occurs in vivo. In other embodiments, the addition of saccharide(s), including but not limited to, the addition of glycosyl residues to proteins, polypeptides and peptides having one or more pyrrolysines and/or PCL incorporated therein Happens in vitro. In other embodiments, the saccharide once attached to the pyrrolysine and/or PCL moiety is further modified by glycosyltransferase and/or other enzymatic treatment to incorporate at least one pyrrolysine and/or PCL It produces an oligosaccharide bound to a protein, polypeptide or peptide having
도 41은 번역 동시 또는 번역후 변형이 퀴나졸린-유형 잔기의 형성을 통해 올리고사카라이드가 단백질, 폴리펩티드 또는 펩티드에 부착되는 것을 포함하는 실시양태를 예시한다. 특정 실시양태에서, 사카라이드는 환원제를 사용하여 추가로 환원시킨 퀴나졸린-유형 잔기의 형성을 비롯한 (이에 제한되지는 않음) 비-천연 연결에 의해 피롤리신 및/또는 PCL 잔기에 연결된다. 특정 실시양태에서, 사카라이드는 본원에서 제공되는 바와 같이 추가로 반응하여 융합된 고리계를 형성한 퀴나졸린-유형 잔기의 형성을 비롯한 (이에 제한되지는 않음) 비-천연 연결에 의해 피롤리신 및/또는 PCL 잔기에 연결된다 (도 22, 23 및 30). 이러한 실시양태에서, 단지 예로서 언급하자면, 올리고사카라이드는 단백질, 폴리펩티드 또는 펩티드에 혼입된 피롤리신 및/또는 PCL 잔기에 접합된 2-아미노-아세토페논 (2-AAP)에 연결된 (GlcNAc-Man)2-Man-GlcNAc-GlcNAc 코어를 포함한다. 다른 실시양태에서, 올리고사카라이드는 2-아미노-벤즈알데히드 (2-ABA)에 연결된 (GlcNAc-Man)2-Man-GlcNAc-GlcNAc 코어를 포함하고, 2-ABA와 단백질, 폴리펩티드 또는 펩티드에 혼입된 피롤리신 및/또는 PCL 잔기의 반응에 의해 단백질 접합체가 형성된다. 다른 실시양태에서, 올리고사카라이드는 2-아미노-벤조페논 (2-ABP)에 연결된 (GlcNAc-Man)2-Man-GlcNAc-GlcNAc 코어를 포함하고, 2-ABP와 단백질, 폴리펩티드 또는 펩티드에 혼입된 피롤리신 및/또는 PCL 잔기의 반응에 의해 단백질 접합체가 형성된다. 이러한 실시양태에서 사용되는 다른 올리고사카라이드는 2-ABA, 2-AAP 또는 2-ANBP 잔기를 보유한다.FIG. 41 illustrates an embodiment in which simultaneous or post-translational modifications include attachment of an oligosaccharide to a protein, polypeptide or peptide through the formation of a quinazoline-type moiety. In certain embodiments, the saccharide is linked to the pyrrolysine and/or PCL moieties by non-natural linkages, including but not limited to the formation of quinazoline-type moieties that have been further reduced with a reducing agent. In certain embodiments, the saccharide is pyrrolysine by non-natural linkage, including, but not limited to, the formation of quinazoline-type moieties that further react as provided herein to form a fused ring system. And/or to a PCL residue (FIGS. 22, 23 and 30 ). In this embodiment, by way of example only, the oligosaccharide is linked to a 2-amino-acetophenone (2-AAP) conjugated to a pyrrolysine and/or PCL residue incorporated into a protein, polypeptide or peptide (GlcNAc- Man) 2 -Man-GlcNAc-GlcNAc core. In another embodiment, the oligosaccharide comprises a (GlcNAc-Man) 2 -Man-GlcNAc-GlcNAc core linked to 2-amino-benzaldehyde (2-ABA), and incorporated into a protein, polypeptide or peptide with 2-ABA. Protein conjugates are formed by reaction of pyrrolysine and/or PCL residues. In other embodiments, the oligosaccharide comprises a (GlcNAc-Man) 2 -Man-GlcNAc-GlcNAc core linked to 2-amino-benzophenone (2-ABP), and is incorporated into a protein, polypeptide or peptide with 2-ABP. Protein conjugates are formed by reaction of pyrrolysine and/or PCL residues. Other oligosaccharides used in this embodiment have 2-ABA, 2-AAP or 2-ANBP residues.
단백질의 배향 공유 부착에 의한, 이종이량체 , 이종삼량체 , 이종다량체 , 동 종이량체, 동종삼량체, 동종다량체, 비대칭 동종이량체, 비대칭 동종삼량체 및 비대칭 동종다량체의 형성 By the covalent attachment of the protein alignment, heterodimer dimer, trimer yijongsam, two kinds of oligomers, such paper-mer, homologous trimer, oligomer homogeneous, asymmetric homodimer, forming a homogeneous asymmetric trimer and asymmetric homogeneous oligomer
본원에서 제공되는 방법을 이용한 또 다른 측면에서, 단백질, 폴리펩티드 및/또는 펩티드로의 1개 이상의 피롤리신 및/또는 PCL의 부위 특이적 혼입을 이용하여 이종이량체, 이종삼량체, 이종다량체, 동종이량체, 동종삼량체, 동종다량체, 비대칭 동종이량체, 비대칭 동종삼량체 및 비대칭 동종다량체를 비롯한 (이에 제한되지는 않음) 단백질-단백질 접합체를 형성한다. 이러한 단백질-단백질 접합체 형성은 내부에 혼입된 피롤리신 및 PCL 잔기(들)을 갖는 단백질들 사이의 부위 특이적 가교를 이용하여 수행된다. 도 42는 내부에 혼입된 PCL 잔기(들)을 갖는 단백질이 가교된, 이러한 단백질-단백질 접합체 형성의 특정 실시양태 (이종이량체, 이종삼량체, 동종삼량체)를 예시한다. 도 42에서는 이러한 가교에 의해 형성된 단백질 접합체 연결부가 단백질들을 함께 연결하는 퀴나졸린-유형 잔기이지만, 다른 실시양태에서는 상기 연결부가 퀴나졸린-유형 잔기의 환원된 형태이다 (도 22, 23 및 30). 다른 실시양태에서, 상기 연결부는 융합된 고리 잔기이다 (도 22, 23 및 30).In another aspect of using the methods provided herein, heterodimers, heterotrimers, heteromultimers, using site-specific incorporation of one or more pyrrolysines and/or PCLs into proteins, polypeptides and/or peptides, It forms protein-protein conjugates, including, but not limited to, homodimers, homotrimers, homomultimers, asymmetric homodimers, asymmetric homotrimers and asymmetric homomultimers. This protein-protein conjugate formation is carried out using site-specific crosslinking between proteins with pyrrolysine and PCL residue(s) incorporated therein. Figure 42 illustrates a specific embodiment (heterodimer, heterotrimer, homotrimer) of such protein-protein conjugate formation, in which proteins with PCL residue(s) incorporated therein are crosslinked. In Figure 42, the protein conjugate linkage formed by such crosslinking is a quinazoline-type residue that connects proteins together, but in other embodiments, the linkage is a reduced form of a quinazoline-type residue (Figs. 22, 23 and 30). In other embodiments, the linkage is a fused ring moiety (FIGS. 22, 23 and 30 ).
이러한 단백질-단백질 접합체의 비제한적인 예에는 다음이 포함되나, 이에 제한되지는 않는다: 단백질-단백질 접합체 시토카인, 성장 인자, 성장 인자 수용체, 인터페론, 인터류킨, 염증성 분자, 종양유전자 생성물, 펩티드 호르몬, 신호 도입 분자, 스테로이드 호르몬 수용체, 전사 활성자, 전사 저해자, 에리트로포이에틴 (EPO), 섬유모세포 성장 인자, 섬유모세포 성장 인자 21 (FGF21), 렙틴, 인슐린, 인간 성장 호르몬, 상피 호중구 활성화 펩티드-78, GROα/MGSA, GROβ, GROγ, MIP-1α, MIP-16, MCP-1, 간세포 성장 인자, 인슐린-유사 성장 인자, 백혈병 억제 인자, 온코스타틴 M, PD-ECSF, PDGF, 플레이오트로핀, SCF, c-kit 리간드, VEGF, G-CSF, IL-1, IL-2, IL-8, IGF-I, IGF-II, FGF (섬유모세포 성장 인자), PDGF, TNF, TGF-α, TGF-ß, EGF (표피 성장 인자), KGF (각질세포 성장 인자), CD40L/CD40, VLA-4/VCAM-1, ICAM-1/LFA-1, 히알루린/CD44, Mos, Ras, Raf, Met; p53, Tat, Fos, Myc, Jun, Myb, Rel, 에스트로겐 수용체, 프로게스테론 수용체, 테스토스테론 수용체, 알도스테론 수용체, LDL 수용체 및/또는 코르티코스테론. 또 다른 세트의 실시양태에서, 단백질은 하기 치료용 단백질 또는 기타 단백질에 상동성이 있다: 알파-1 항-트립신, 안지오스타틴, 항-용혈성 인자, 항체, 아포지단백질, 아포단백질, 심방 나트륨이뇨 인자, 심방 나트륨이뇨 폴리펩티드, 심방 펩티드, C-X-C 케모카인, T39765, NAP-2, ENA-78, Gro-a, Gro-b, Gro-c, IP-10, GCP-2, NAP-4, SDF-1, PF4, MIG, 칼시토닌, c-kit 리간드, 시토카인, CC 케모카인, 단핵구 화학유인물질 단백질-1, 단핵구 화학유인물질 단백질-2, 단핵구 화학유인물질 단백질-3, 단핵구 염증성 단백질-1 알파, 단핵구 염증성 단백질-1 베타, RANTES, 1309, R83915, R91733, HCC1, T58847, D31065, T64262, CD40, CD40 리간드, C-kit 리간드, 콜라겐, 콜로니 자극 인자 (CSF), 보체 인자 5a, 보체 억제제, 보체 수용체 1, 시토카인, 상피 호중구 활성화 펩티드-78, GROα/MGSA, GROβ, GROγ, MIP-1α, MIP-16, MCP-1, 표피 성장 인자 (EGF), 상피 호중구 활성화 펩티드, 박리 독소, 인자 IX, 인자 VII, 인자 VIII, 인자 X, 섬유모세포 성장 인자 (FGF), 피브리노겐, 피브로넥틴, G-CSF, GM-CSF, 글루코세레브로시다제, 고나도트로핀, 성장 인자, 성장 인자 수용체, 헷지호그 단백질, 헤모글로빈, 간세포 성장 인자 (HGF), 히루딘, 인간 혈청 알부민, ICAM-1, ICAM-1 수용체, LFA-1, LFA-1 수용체, 인슐린, 인슐린-유사 성장 인자 (IGF), IGF-I, IGF-II, 인터페론, IFN-α, IFN-ß, IFN-γ, 인터류킨, IL-1, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-11, IL-12, 각질세포 성장 인자 (KGF), 락토페린, 백혈병 억제 인자, 루시퍼라제, 뉴르투린, 호중구 억제 인자 (NIF), 온코스타틴 M, 골형성 단백질, 종양유전자 생성물, 부갑상선 호르몬, PD-ECSF, PDGF, 펩티드 호르몬, 인간 성장 호르몬, 플레이오트로핀, 단백질 A, 단백질 G, 발열성 외독소 A, B 또는 C, 릴랙신, 레닌, SCF, 가용성 보체 수용체 I, 가용성 I-CAM 1, 가용성 인터류킨 수용체, 가용성 TNF 수용체, 소마토메딘, 소마토스타틴, 소마토트로핀, 스트렙토키나제, 초항원, 스타필로코쿠스(Staphylococcal) 장독소, SEA, SEB, SEC1, SEC2, SEC3, SED, SEE, 스테로이드 호르몬 수용체, 수퍼옥시드 디스뮤타제, 독성 쇼크 증후군 독소, 티모신 알파 1, 조직 플라스미노겐 활성자, 종양 성장 인자 (TGF), TGF-α, TGF-ß, 종양 괴사 인자, 종양 괴사 인자 알파, 종양 괴사 인자 베타, 종양 괴사 인자 수용체 (TNFR), VLA-4 단백질, VCAM-1 단백질, 혈관 내피 성장 인자 (VEGF), 유로키나제, Mos, Ras, Raf, Met; p53, Tat, Fos, Myc, Jun, Myb, Rel, 에스트로겐 수용체, 프로게스테론 수용체, 테스토스테론 수용체, 알도스테론 수용체, LDL 수용체 및/또는 코르티코스테론.Non-limiting examples of such protein-protein conjugates include, but are not limited to: protein-protein conjugate cytokines, growth factors, growth factor receptors, interferons, interleukins, inflammatory molecules, oncogene products, peptide hormones, signals. Introducing molecule, steroid hormone receptor, transcription activator, transcription inhibitor, erythropoietin (EPO), fibroblast growth factor, fibroblast growth factor 21 (FGF21), leptin, insulin, human growth hormone, epithelial neutrophil activating peptide-78 , GROα/MGSA, GROβ, GROγ, MIP-1α, MIP-16, MCP-1, hepatocyte growth factor, insulin-like growth factor, leukemia inhibitory factor, oncostatin M, PD-ECSF, PDGF, pleiotropin, SCF, c-kit ligand, VEGF, G-CSF, IL-1, IL-2, IL-8, IGF-I, IGF-II, FGF (fibroblast growth factor), PDGF, TNF, TGF-α, TGF -ß, EGF (epidermal growth factor), KGF (keratinocyte growth factor), CD40L/CD40, VLA-4/VCAM-1, ICAM-1/LFA-1, hyalurin/CD44, Mos, Ras, Raf, Met ; p53, Tat, Fos, Myc, Jun, Myb, Rel, estrogen receptor, progesterone receptor, testosterone receptor, aldosterone receptor, LDL receptor and/or corticosterone. In another set of embodiments, the protein is homologous to the following therapeutic protein or other protein: alpha-1 anti-trypsin, angiostatin, anti-hemolytic factor, antibody, apolipoprotein, apoprotein, atrial natriuretic factor, Atrial natriuretic polypeptide, atrial peptide, CXC chemokine, T39765, NAP-2, ENA-78, Gro-a, Gro-b, Gro-c, IP-10, GCP-2, NAP-4, SDF-1, PF4 , MIG, calcitonin, c-kit ligand, cytokine, CC chemokine, monocyte chemoattractant protein-1, monocyte chemoattractant protein-2, monocyte chemoattractant protein-3, monocyte inflammatory protein-1 alpha, monocyte inflammatory protein- 1 beta, RANTES, 1309, R83915, R91733, HCC1, T58847, D31065, T64262, CD40, CD40 ligand, C-kit ligand, collagen, colony stimulating factor (CSF), complement factor 5a, complement inhibitor, complement receptor 1, cytokine , Epithelial neutrophil activating peptide-78, GROα/MGSA, GROβ, GROγ, MIP-1α, MIP-16, MCP-1, epidermal growth factor (EGF), epithelial neutrophil activating peptide, exfoliation toxin, factor IX, factor VII, factor VIII, factor X, fibroblast growth factor (FGF), fibrinogen, fibronectin, G-CSF, GM-CSF, glucocerebrosidase, gonadotropin, growth factor, growth factor receptor, hedgehog protein, hemoglobin, hepatocyte Growth factor (HGF), hirudin, human serum albumin, ICAM-1, ICAM-1 receptor, LFA-1, LFA-1 receptor, insulin, insulin-like growth factor (IGF), IGF-I, IGF-II, Interferon, IFN-α, IFN-ß, IFN-γ, interleukin, IL-1, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL- 9, IL-10, IL-11, IL-12, keratinocyte growth factor (KGF), lactoferrin, leukemia inhibitory factor, luciferase, neuroturin, Neutrophil inhibitor (NIF), oncostatin M, bone morphogenetic protein, oncogene product, parathyroid hormone, PD-ECSF, PDGF, peptide hormone, human growth hormone, pleiotropin, protein A, protein G, pyrogenic exotoxin A , B or C, relaxin, renin, SCF, soluble complement receptor I, soluble I-CAM 1, soluble interleukin receptor, soluble TNF receptor, somatomedin, somatostatin, somatotropin, streptokinase, superantigen, staphylo Staphylococcal enterotoxin, SEA, SEB, SEC1, SEC2, SEC3, SED, SEE, steroid hormone receptor, superoxide dismutase, toxic shock syndrome toxin, thymosin alpha 1, tissue plasminogen activator, Tumor growth factor (TGF), TGF-α, TGF-ß, tumor necrosis factor, tumor necrosis factor alpha, tumor necrosis factor beta, tumor necrosis factor receptor (TNFR), VLA-4 protein, VCAM-1 protein, vascular endothelial growth Factor (VEGF), urokinase, Mos, Ras, Raf, Met; p53, Tat, Fos, Myc, Jun, Myb, Rel, estrogen receptor, progesterone receptor, testosterone receptor, aldosterone receptor, LDL receptor and/or corticosterone.
이종이량체, 이종삼량체, 이종다량체, 동종이량체, 동종삼량체, 동종다량체, 비대칭 동종이량체, 비대칭 동종삼량체 및 비대칭 동종다량체를 비롯한 (이에 제한되지는 않음) 단백질-단백질 접합체의 형성에 사용되는 가교제는 각각의 말단에 벤즈알데히드, 아세토페논 및/또는 벤조페논 잔기를 보유한다. 이러한 잔기는 본원에서 제공되는 방법을 이용하여 단백질에 혼입된 피롤리신 및/또는 PCL 잔기(들)과 반응하고, 이러한 단백질은 본원에서 제공되는 것들을 포함한다. 동종이량체의 형성에 사용되는 이관능성 가교제의 비제한적인 예를 도 43A에 나타냈다. 이러한 이관능성 링커를 사용하여 위치 84에서 혼입된 PCL 잔기를 갖는 섬유모세포 성장 인자 21 (FGF-21) (FGF21 Lys84PCL)을 가교시켰다. 도 43B는 반응 혼합물의 질량 스펙트럼이며, 여기서는 예상 질량 43037.2 Da의 공유 FGF21 Lys48PCL 이량체에 대한 피크가 우세한 종이다. 미반응 FGF21 Lys84PCL은 21235.2 Da에서 검출되지만, 부착된 가교제의 한쪽 말단으로 변형된 일부 단량체성 FGF21은 21820.0 Da에서 검출된다. 반응 조건을 변형시켜서 FGF21 Lys84PCL을 완전히 반응시켰다. 도 43에서는 이러한 가교에 의해 형성된 단백질 접합체 연결부가 퀴나졸린-유형 잔기이지만, 다른 실시양태에서는 상기 연결부가 퀴나졸린-유형 잔기의 환원된 형태이다 (도 22, 23 및 30). 다른 실시양태에서, 상기 연결부는 융합된 고리 잔기이다 (도 22, 23 및 30).Protein-protein conjugates, including but not limited to heterodimers, heterotrimers, heteromultimers, homodimers, homotrimers, homomultimers, asymmetric homodimers, asymmetric homotrimers and asymmetric homomultimers The crosslinking agent used for the formation of has a benzaldehyde, acetophenone and/or benzophenone residue at each end. These residues react with the pyrrolysine and/or PCL residue(s) incorporated into the protein using the methods provided herein, and such proteins include those provided herein. A non-limiting example of a bifunctional crosslinking agent used in the formation of a homodimer is shown in Fig. 43A. Fibroblast growth factor 21 (FGF-21) (FGF21 Lys84PCL) with a PCL residue incorporated at
도 44A는 반응 혼합물의 질량 스펙트럼을 보여주며, 여기서는 예상 질량 43034 Da의 공유 FGF21 이량체의 피크가 우세한 종이다. 미반응 FGF21 Lys84PCL은 21234 Da에서 검출되지 않지만, 부착된 가교제의 한쪽 말단으로 변형된 일부 단량체성 FGF21은 21818 Da에서 검출된다. 도 43B는 pH 5.0 및 pH 7.5에서의 반응에 대한 SDS-PAGE이며, pH 5.0에서 더 양호하게 전환됨을 나타낸다. PCL-특이적 가교를 이용하여 FGF21의 이량체를 생성하였다. 그러나, 상기 방법은 상기 기재된 임의의 단백질에 적용가능하며, 이량체, 삼량체 및 다량체의 형성에 이용될 수 있다. 이러한 가교를 이용하여 생물학적 활성 단백질의 활성을 강화시킨다. 또한, 이 접근법을 이용하여 구조적 연구를 위해 복합체를 안정화하고/하거나 수용체 유인(decoy)을 안정화한다. 도 45는 가교제를 삼량체 형성에 사용하는 실시양태를 보여준다.44A shows the mass spectrum of the reaction mixture, where the peak of the covalent FGF21 dimer with expected mass of 43034 Da is the dominant species. Unreacted FGF21 Lys84PCL is not detected at 21234 Da, but some monomeric FGF21 modified to one end of the attached crosslinker is detected at 21818 Da. 43B is SDS-PAGE for the reaction at pH 5.0 and pH 7.5, showing better conversion at pH 5.0. A dimer of FGF21 was generated using PCL-specific crosslinking. However, the method is applicable to any of the proteins described above and can be used to form dimers, trimers and multimers. This cross-linking is used to enhance the activity of biologically active proteins. In addition, this approach is used to stabilize complexes and/or to stabilize receptor decoys for structural studies. 45 shows an embodiment in which a crosslinking agent is used to form a trimer.
부위 특이적 표지화Site specific labeling
본원에서 제공되는 방법을 이용한 또 다른 측면에서, 단백질, 폴리펩티드 및/또는 펩티드로의 1개 이상의 피롤리신 및/또는 PCL의 부위 특이적 혼입을 이용하여 이러한 단백질, 폴리펩티드 및/또는 펩티드를 부위 특이적으로 표지화한다. 이러한 표지화는 피롤리신 및/또는 PCL 잔기(들)과 피롤리신 및/또는 PCL 잔기(들)과 선택적으로 반응하는 관능기로 유도체화된 표지의 반응으로 인한 것이다. 사용되는 표지에는 염료, 항체 또는 항체 단편, 금속 킬레이트화제, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 화학 방사선 여기성 잔기, 리간드, 비오틴, 비오틴 유사체, 산화환원-활성제, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 화학발광 기, 전자 고밀도 기, 자성 기, 삽입성 기, 발색단; 에너지 전달 작용제, 양자점(들), 형광 염료, FRET 시약, 및 이들의 임의의 조합물이 포함되나, 이에 제한되지는 않는다. 표지는 또한 본원에서 정의된 바와 같은 임의의 X1이다.In another aspect of using the methods provided herein, site-specific incorporation of one or more pyrrolysines and/or PCLs into proteins, polypeptides and/or peptides is used to site-specificize such proteins, polypeptides and/or peptides. Labeled as a target. This labeling is due to the reaction of a label derivatized with a functional group that selectively reacts with pyrrolysine and/or PCL residue(s) with pyrrolysine and/or PCL residue(s). Labels used include dyes, antibodies or antibody fragments, metal chelating agents, spin labels, fluorophores, metal-containing moieties, radioactive moieties, actinic radiation excitable moieties, ligands, biotin, biotin analogs, redox-activators, isotopic labels. Moieties, biophysical probes, phosphorescent groups, chemiluminescent groups, electron high density groups, magnetic groups, intercalating groups, chromophores; Energy transfer agents, quantum dot(s), fluorescent dyes, FRET reagents, and any combination thereof. The label is also any X 1 as defined herein.
한 실시양태에서, 1개 이상의 피롤리신 및/또는 PCL 잔기를 함유하는 이러한 단백질, 폴리펩티드 및/또는 펩티드의 이러한 부위 특이적 표지화는 피롤리신 및/또는 PCL 잔기(들)과 벤즈알데히드, 아세토페논 또는 벤조페논 잔기로 유도체화된 표지의 반응이다. 사용되는 표지에는 염료, 항체 또는 항체 단편, 금속 킬레이트화제, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 화학 방사선 여기성 잔기, 리간드, 비오틴, 비오틴 유사체, 산화환원-활성제, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 화학발광 기, 전자 고밀도 기, 자성 기, 삽입성 기, 발색단; 에너지 전달 작용제, 양자점(들), 형광 염료, FRET 시약, 및 이들의 임의의 조합물이 포함되나, 이에 제한되지는 않는다. 표지는 또한 본원에서 정의된 바와 같은 임의의 X1이다.In one embodiment, such site-specific labeling of such proteins, polypeptides and/or peptides containing one or more pyrrolysine and/or PCL residues is pyrrolysine and/or PCL residue(s) with benzaldehyde, acetophenone. Or a reaction of a label derivatized with a benzophenone moiety. Labels used include dyes, antibodies or antibody fragments, metal chelating agents, spin labels, fluorophores, metal-containing moieties, radioactive moieties, actinic radiation excitable moieties, ligands, biotin, biotin analogs, redox-activators, isotopic labels. Moieties, biophysical probes, phosphorescent groups, chemiluminescent groups, electron high density groups, magnetic groups, intercalating groups, chromophores; Energy transfer agents, quantum dot(s), fluorescent dyes, FRET reagents, and any combination thereof. The label is also any X 1 as defined herein.
본원에서 제공되는 이러한 부위 특이적 표지화의 추가의 측면에서, 피롤리신 및/또는 PCL 잔기(들)을 피롤리신 및/또는 PCL 잔기(들)과 선택적으로 반응하는 잔기로 유도체화된 표지와 반응시켜 검출 감도를 증가시킨다. 구체적으로, 이러한 반응은 반응 전에 존재하는 반응물의 것과는 상이한 스펙트럼 특성을 갖는 검출가능한 잔기를 형성시키고, 이로 인해 검출가능한 잔기 신호와 관련이 있는 백그라운드 신호를 최소화함으로써 노이즈에 대한 신호의 비율을 증진시킨다. 본원에서 제공되는 이러한 부위 특이적 표지화의 특정 실시양태에서, 피롤리신 또는 PCL 잔기(들)과 벤즈알데히드, 아세토페논 또는 벤조페논 잔기로 유도체화된 표지의 반응은 벤즈알데히드, 아세토페논 또는 벤조페논 잔기의 것과는 상이한 스펙트럼 특성을 갖는 퀴나졸린-유형 잔기를 형성시킨다 (도 22, 23 및 30). 본원에서 제공되는 이러한 부위 특이적 표지화의 특정 실시양태에서, 피롤리신 또는 PCL 잔기(들)과 벤즈알데히드, 아세토페논 또는 벤조페논 잔기로 유도체화된 표지의 반응은 벤즈알데히드, 아세토페논 또는 벤조페논 잔기의 것과는 상이한 스펙트럼 특성을 갖는 환원된 퀴나졸린-유형 잔기를 형성시킨다 (도 22, 23 및 30). 본원에서 제공되는 이러한 부위 특이적 표지화의 특정 실시양태에서, 피롤리신 또는 PCL 잔기(들)과 벤즈알데히드, 아세토페논 또는 벤조페논 잔기로 유도체화된 표지의 반응은 벤즈알데히드, 아세토페논 또는 벤조페논 잔기의 것과는 상이한 스펙트럼 특성을 갖는 융합된 고리 잔기를 형성시킨다 (도 22, 23 및 30).In a further aspect of such site-specific labeling provided herein, with a label derivatized with a residue that selectively reacts with pyrrolysine and/or PCL residue(s) with pyrrolysine and/or PCL residue(s). React to increase detection sensitivity. Specifically, these reactions form detectable moieties with different spectral properties than the reactants present before the reaction, thereby enhancing the ratio of signal to noise by minimizing the background signal associated with the detectable moiety signal. In certain embodiments of such site-specific labeling provided herein, the reaction of a pyrrolysine or PCL moiety(s) with a label derivatized with a benzaldehyde, acetophenone or benzophenone moiety is a reaction of a benzaldehyde, acetophenone or benzophenone moiety. Forms quinazoline-type residues with different spectral properties than those (Figures 22, 23 and 30). In certain embodiments of such site-specific labeling provided herein, the reaction of a pyrrolysine or PCL moiety(s) with a label derivatized with a benzaldehyde, acetophenone or benzophenone moiety is a reaction of a benzaldehyde, acetophenone or benzophenone moiety. Formed reduced quinazoline-type residues with different spectral properties than those (FIGS. 22, 23 and 30 ). In certain embodiments of such site-specific labeling provided herein, the reaction of a pyrrolysine or PCL moiety(s) with a label derivatized with a benzaldehyde, acetophenone or benzophenone moiety is a reaction of a benzaldehyde, acetophenone or benzophenone moiety. Fused ring moieties with different spectral properties than those (FIGS. 22, 23 and 30 ).
특정 실시양태에서는 이러한 표지된 단백질, 폴리펩티드 및/또는 펩티드가 진단 및 영상화에 사용되고, 다른 실시양태에서는 이러한 표지된 단백질, 폴리펩티드 및/또는 펩티드가 고처리량 스크리닝에서 사용된다. 다른 실시양태에서는 이러한 표지된 단백질, 폴리펩티드 및/또는 펩티드가 수송 특성을 평가하는데 사용되고, 다른 실시양태에서는 이러한 표지된 단백질, 폴리펩티드 및/또는 펩티드가 국소화 연구에 사용된다.In certain embodiments, such labeled proteins, polypeptides and/or peptides are used for diagnosis and imaging, and in other embodiments, such labeled proteins, polypeptides and/or peptides are used in high-throughput screening. In other embodiments, such labeled proteins, polypeptides and/or peptides are used to assess transport properties, and in other embodiments, such labeled proteins, polypeptides and/or peptides are used in localization studies.
도 46은 이러한 부위 특이적 표지화의 특정 실시양태를 예시한다. 도 46A는 형광 잔기를 사용한 표지화를 예시한다. 도 46B는 PCL 잔기를 반응시에 형광이 되는 비-형광 잔기와 반응시키는 것을 예시한다. 도 46C는 단백질의 비오티닐화를 예시한다. 도 46D는 방사성 잔기를 사용한 표지화를 예시한다. 도 46E는 1-(2-아미노-5-요오도페닐)에타논을 사용한 표지화를 예시하고, 도 46F는 1-(2-아미노-5-브로모페닐)에타논을 사용한 표지화를 예시하며, 이들 둘다 단백질의 X선 결정 구조를 결정하는 방법을 위한 위상 정보 수득에 사용될 수 있다. 1-(2-아미노-5-요오도페닐)에타논은 또한 방사성 잔기를 사용한 단백질 표지화에 사용될 수도 있다. 도 46에서는 이러한 표지화로 형성된 단백질 접합체 연결부가 퀴나졸린-유형 잔기이지만, 다른 실시양태에서는 상기 연결부가 퀴나졸린-유형 잔기의 환원된 형태이다 (도 22, 23 및 30). 다른 실시양태에서, 상기 연결부는 융합된 고리 잔기이다 (도 22, 23 및 30).46 illustrates certain embodiments of such site specific labeling. 46A illustrates labeling with fluorescent moieties. 46B illustrates the reaction of PCL moieties with non-fluorescent moieties that become fluorescent upon reaction. Figure 46C illustrates the biotinylation of proteins. 46D illustrates labeling with radioactive moieties. Figure 46E illustrates labeling with 1-(2-amino-5-iodophenyl)ethanone, and Figure 46F illustrates labeling with 1-(2-amino-5-bromophenyl)ethanone, Both of these can be used to obtain phase information for a method of determining the X-ray crystal structure of a protein. 1-(2-amino-5-iodophenyl)ethanone can also be used for protein labeling with radioactive moieties. In Figure 46, the protein conjugate linkage formed by this labeling is a quinazoline-type residue, but in other embodiments the linkage is a reduced form of a quinazoline-type residue (Figures 22, 23 and 30). In other embodiments, the linkage is a fused ring moiety (FIGS. 22, 23 and 30 ).
1개 이상의 피롤리신 및/또는 PCL 잔기(들)을 함유하는 이러한 단백질, 폴리펩티드 및/또는 펩티드의 이러한 부위 특이적 표지화의 실시양태에서, 피롤리신 또는 PCL 잔기(들) 자체를 표지된 전구체로 표지화할 수 있다. 도 46G 및 도 46H는 상이하게 표지된 전구체로부터의 방사성 또는 안정적인 동위원소에 의한 표지화를 단지 예로서 보여준다.In embodiments of such site-specific labeling of such proteins, polypeptides and/or peptides containing one or more pyrrolysine and/or PCL residue(s), precursors labeled with pyrrolysine or PCL residue(s) themselves It can be labeled with. 46G and 46H show, by way of example only, labeling by radioactive or stable isotopes from differently labeled precursors.
본 발명의 또 다른 측면은 임의의 이용가능한 단백질에 상동성이지만 1개 이상의 피롤리신 또는 PCL 잔기(들)을 포함하는 단백질의 생성을 제공한다. 예를 들어, 특정 실시양태에서, 1개 이상의 피롤리신 또는 PCL을 포함하고 1종 이상의 치료용 단백질에 상동성인 치료용 단백질이 제조된다. 예를 들어, 한 측면에서, 상기 단백질은 예를 들어 하기하는 치료용 또는 기타 단백질에 상동성이다: 시토카인, 성장 인자, 성장 인자 수용체, 인터페론, 인터류킨, 염증성 분자, 종양유전자 생성물, 펩티드 호르몬, 신호 도입 분자, 스테로이드 호르몬 수용체, 전사 활성자, 전사 저해자, 에리트로포이에틴 (EPO), 섬유모세포 성장 인자, 섬유모세포 성장 인자 21 (FGF21), 렙틴, 인슐린, 인간 성장 호르몬, 상피 호중구 활성화 펩티드-78, GROα/MGSA, GROβ, GROγ, MIP-1α, MIP-16, MCP-1, 간세포 성장 인자, 인슐린-유사 성장 인자, 백혈병 억제 인자, 온코스타틴 M, PD-ECSF, PDGF, 플레이오트로핀, SCF, c-kit 리간드, VEGF, G-CSF, IL-1, IL-2, IL-8, IGF-I, IGF-II, FGF (섬유모세포 성장 인자), PDGF, TNF, TGF-α, TGF-ß, EGF (표피 성장 인자), KGF (각질세포 성장 인자), CD40L/CD40, VLA-4/VCAM-1, ICAM-1/LFA-1, 히알루린/CD44, Mos, Ras, Raf, Met; p53, Tat, Fos, Myc, Jun, Myb, Rel, 에스트로겐 수용체, 프로게스테론 수용체, 테스토스테론 수용체, 알도스테론 수용체, LDL 수용체 및/또는 코르티코스테론. 또 다른 세트의 실시양태에서, 상기 단백질은 예를 들어 하기하는 치료용 또는 기타 단백질에 상동성이다: 알파-1 항-트립신, 안지오스타틴, 항-용혈성 인자, 항체, 아포지단백질, 아포단백질, 심방 나트륨이뇨 인자, 심방 나트륨이뇨 폴리펩티드, 심방 펩티드, C-X-C 케모카인, T39765, NAP-2, ENA-78, Gro-α, Gro-β, Gro-γ, IP-10, GCP-2, NAP-4, SDF-1, PF4, MIG, 칼시토닌, c-kit 리간드, 시토카인, CC 케모카인, 단핵구 화학유인물질 단백질-1, 단핵구 화학유인물질 단백질-2, 단핵구 화학유인물질 단백질-3, 단핵구 염증성 단백질-1 알파, 단핵구 염증성 단백질-1 베타, RANTES, 1309, R83915, R91733, HCC1, T58847, D31065, T64262, CD40, CD40 리간드, C-kit 리간드, 콜라겐, 콜로니 자극 인자 (CSF), 보체 인자 5a, 보체 억제제, 보체 수용체 1, 시토카인, 상피 호중구 활성화 펩티드-78, GROα/MGSA, GROβ, GROγ, MIP-1α, MIP-16, MCP-1, 표피 성장 인자 (EGF), 상피 호중구 활성화 펩티드, 인터페론 알파 (INF-α) 또는 인터페론 베타 (INF-β), 박리 독소, 인자 IX, 인자 VII, 인자 VIII, 인자 X, 섬유모세포 성장 인자 (FGF), 피브리노겐, 피브로넥틴, G-CSF, GM-CSF, 글루코세레브로시다제, 고나도트로핀, 성장 인자, 성장 인자 수용체, 헷지호그 단백질, 헤모글로빈, 간세포 성장 인자 (HGF), 히루딘, 인간 혈청 알부민, ICAM-1, ICAM-1 수용체, LFA-1, LFA-1 수용체, 인슐린, 인슐린-유사 성장 인자 (IGF), IGF-I, IGF-II, 인터페론, IFN-α, IFN-ß, IFN-γ, 인터류킨, IL-1, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-11, IL-12, 각질세포 성장 인자 (KGF), 락토페린, 백혈병 억제 인자, 루시퍼라제, 뉴르투린, 호중구 억제 인자 (NIF), 온코스타틴 M, 골형성 단백질, 종양유전자 생성물, 부갑상선 호르몬, PD-ECSF, PDGF, 펩티드 호르몬, 인간 성장 호르몬, 플레이오트로핀, 단백질 A, 단백질 G, 발열성 외독소 A, B 또는 C, 릴랙신, 레닌, SCF, 가용성 보체 수용체 I, 가용성 I-CAM 1, 가용성 인터류킨 수용체, 가용성 TNF 수용체, 소마토메딘, 소마토스타틴, 소마토트로핀, 스트렙토키나제, 초항원, 스타필로코쿠스 장독소, SEA, SEB, SEC1, SEC2, SEC3, SED, SEE, 스테로이드 호르몬 수용체, 수퍼옥시드 디스뮤타제, 독성 쇼크 증후군 독소, 티모신 알파 1, 조직 플라스미노겐 활성자, 종양 성장 인자 (TGF), TGF-α, TGF-ß, 종양 괴사 인자, 종양 괴사 인자 알파, 종양 괴사 인자 베타, 종양 괴사 인자 수용체 (TNFR), VLA-4 단백질, VCAM-1 단백질, 혈관 내피 성장 인자 (VEGF), 유로키나제, Mos, Ras, Raf, Met; p53, Tat, Fos, Myc, Jun, Myb, Rel, 에스트로겐 수용체, 프로게스테론 수용체, 테스토스테론 수용체, 알도스테론 수용체, LDL 수용체 및/또는 코르티코스테론.Another aspect of the invention provides the production of a protein that is homologous to any available protein but comprises one or more pyrrolysine or PCL residue(s). For example, in certain embodiments, a therapeutic protein comprising at least one pyrrolysine or PCL and homologous to at least one therapeutic protein is prepared. For example, in one aspect, the protein is homologous to a therapeutic or other protein, e.g., a cytokine, a growth factor, a growth factor receptor, an interferon, an interleukin, an inflammatory molecule, an oncogene product, a peptide hormone, a signal. Introducing molecule, steroid hormone receptor, transcription activator, transcription inhibitor, erythropoietin (EPO), fibroblast growth factor, fibroblast growth factor 21 (FGF21), leptin, insulin, human growth hormone, epithelial neutrophil activating peptide-78 , GROα/MGSA, GROβ, GROγ, MIP-1α, MIP-16, MCP-1, hepatocyte growth factor, insulin-like growth factor, leukemia inhibitory factor, oncostatin M, PD-ECSF, PDGF, pleiotropin, SCF, c-kit ligand, VEGF, G-CSF, IL-1, IL-2, IL-8, IGF-I, IGF-II, FGF (fibroblast growth factor), PDGF, TNF, TGF-α, TGF -ß, EGF (epidermal growth factor), KGF (keratinocyte growth factor), CD40L/CD40, VLA-4/VCAM-1, ICAM-1/LFA-1, hyalurin/CD44, Mos, Ras, Raf, Met ; p53, Tat, Fos, Myc, Jun, Myb, Rel, estrogen receptor, progesterone receptor, testosterone receptor, aldosterone receptor, LDL receptor and/or corticosterone. In another set of embodiments, the protein is homologous to a therapeutic or other protein, for example: alpha-1 anti-trypsin, angiostatin, anti-hemolytic factor, antibody, apolipoprotein, apoprotein, atrial sodium Diuretic factor, atrial natriuretic polypeptide, atrial peptide, CXC chemokine, T39765, NAP-2, ENA-78, Gro-α, Gro-β, Gro-γ, IP-10, GCP-2, NAP-4, SDF- 1, PF4, MIG, calcitonin, c-kit ligand, cytokine, CC chemokine, monocyte chemoattractant protein-1, monocyte chemoattractant protein-2, monocyte chemoattractant protein-3, monocyte inflammatory protein-1 alpha, monocyte Inflammatory protein-1 beta, RANTES, 1309, R83915, R91733, HCC1, T58847, D31065, T64262, CD40, CD40 ligand, C-kit ligand, collagen, colony stimulating factor (CSF), complement factor 5a, complement inhibitor, complement receptor 1, cytokine, epidermal neutrophil activation peptide-78, GROα/MGSA, GROβ, GROγ, MIP-1α, MIP-16, MCP-1, epidermal growth factor (EGF), epidermal neutrophil activation peptide, interferon alpha (INF-α) Or interferon beta (INF-β), exfoliating toxin, factor IX, factor VII, factor VIII, factor X, fibroblast growth factor (FGF), fibrinogen, fibronectin, G-CSF, GM-CSF, glucocerebrosidase, Gonadotropin, growth factor, growth factor receptor, hedgehog protein, hemoglobin, hepatocyte growth factor (HGF), hirudin, human serum albumin, ICAM-1, ICAM-1 receptor, LFA-1, LFA-1 receptor, Insulin, insulin-like growth factor (IGF), IGF-I, IGF-II, interferon, IFN-α, IFN-ß, IFN-γ, interleukin, IL-1, IL-2, IL-3, IL-4 , IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-11, IL-12, keratinocytes Growth factor (KGF), lactoferrin, leukemia inhibitory factor, luciferase, neuroturin, neutrophil inhibitor (NIF), oncostatin M, bone morphogenetic protein, oncogene product, parathyroid hormone, PD-ECSF, PDGF, peptide hormone, human Growth hormone, pleiotropin, protein A, protein G, pyrogenic exotoxin A, B or C, relaxin, renin, SCF, soluble complement receptor I, soluble I-CAM 1, soluble interleukin receptor, soluble TNF receptor, soma Tomedin, somatostatin, somatotropin, streptokinase, superantigen, staphylococcal enterotoxin, SEA, SEB, SEC1, SEC2, SEC3, SED, SEE, steroid hormone receptor, superoxide dismutase, toxic shock Syndrome toxin, thymosin alpha 1, tissue plasminogen activator, tumor growth factor (TGF), TGF-α, TGF-ß, tumor necrosis factor, tumor necrosis factor alpha, tumor necrosis factor beta, tumor necrosis factor receptor (TNFR) ), VLA-4 protein, VCAM-1 protein, vascular endothelial growth factor (VEGF), urokinase, Mos, Ras, Raf, Met; p53, Tat, Fos, Myc, Jun, Myb, Rel, estrogen receptor, progesterone receptor, testosterone receptor, aldosterone receptor, LDL receptor and/or corticosterone.
한 측면에서, 본원의 조성물은 1개 이상의 피롤리신 또는 PCL 잔기(들)을 포함하는 단백질, 예를 들어 상기한 임의의 단백질, 및 제약상 허용되는 부형제를 포함한다.In one aspect, a composition herein comprises a protein comprising one or more pyrrolysine or PCL residue(s), such as any of the proteins described above, and a pharmaceutically acceptable excipient.
특정 실시양태에서는 단백질이 치료용 단백질이고, 다른 실시양태에서는 치료용 단백질이 에리트로포이에틴 (EPO), 섬유모세포 성장 인자 21 (FGF21), 인터페론 알파 (INF-α) 또는 인터페론 베타 (INF-β)이다. 특정 실시양태에서, 치료용 단백질은 항체 또는 Fab를 비롯한 (이에 제한되지는 않음) 항체 단편이다. 특정 실시양태에서, 치료용 단백질은 융합 파트너가 변형된 융합 단백질로서 생성된다. 특정 실시양태에서, 치료용 단백질은 Fc 도메인과의 융합물이다.In certain embodiments, the protein is a therapeutic protein, in other embodiments, the therapeutic protein is erythropoietin (EPO), fibroblast growth factor 21 (FGF21), interferon alpha (INF-α) or interferon beta (INF-β). to be. In certain embodiments, the therapeutic protein is an antibody or antibody fragment including, but not limited to, a Fab. In certain embodiments, the therapeutic protein is produced as a fusion protein with a modified fusion partner. In certain embodiments, the therapeutic protein is a fusion with an Fc domain.
단백질 또는 폴리펩티드에 대한 상동성은 예를 들어 디폴트 파라미터를 설정하여 서열 정렬, 예를 들어 BLASTN 또는 BLASTP를 수행함으로써 추론될 수 있다. 예를 들어, 한 실시양태에서, 단백질은 공지의 치료용 단백질 (예를 들어, 진뱅크(Genebank) 또는 기타 이용가능한 데이터베이스에 존재하는 단백질)과 적어도 약 50%, 적어도 약 75%, 적어도 약 80%, 적어도 약 90% 또는 적어도 약 95% 동일하다.Homology to a protein or polypeptide can be inferred, for example, by setting default parameters to perform sequence alignment, eg BLASTN or BLASTP. For example, in one embodiment, the protein is at least about 50%, at least about 75%, at least about 80 with a known therapeutic protein (e.g., a protein present in Genebank or other available databases). %, at least about 90% or at least about 95% identical.
부위 특이적 표지화를 입증하기 위해서, 본원에서 제공되는 방법을 이용하여 PCL을 mEGF에 혼입하였고 (실시예 12 참조), PCL 잔기를 ABA 잔기로 관능화된 폴리에테르를 통해 비오틴에 커플링시켰다 (X3626-140 참조, 실시예 40). 도 47A는 비오틴과 접합된 mEGF-Tyr10PCL의 ESI 질량 분광측정 분석을 보여준다 (실시예 24 참조). 도 47B는 양고추냉이 퍼옥시다제 (HRP) 접합된 염소 항-비오틴 항체를 사용한, mEGF-Tyr10PCL-ABA-비오틴 접합체의 웨스턴 블럿팅을 보여준다. 커플링되지 않은 mEGF-Tyr10PCL 및 플루오레세인-접합된 mEGF-Tyr10PCL은 음성 대조군으로 기능했다.To demonstrate site specific labeling, PCL was incorporated into mEGF using the method provided herein (see Example 12), and the PCL residue was coupled to biotin via a polyether functionalized with an ABA residue (X3626 See -140, Example 40). Figure 47A shows the ESI mass spectrometric analysis of mEGF-Tyr10PCL conjugated with biotin (see Example 24). 47B shows Western blotting of mEGF-Tyr10PCL-ABA-biotin conjugates using horseradish peroxidase (HRP) conjugated goat anti-biotin antibody. Uncoupled mEGF-Tyr10PCL and fluorescein-conjugated mEGF-Tyr10PCL served as negative controls.
부위 특이적 표지화를 추가로 입증하기 위해서, 본원에서 제공되는 방법을 이용하여 PCL을 mEGF에 혼입하였고 (실시예 12 참조), PCL 잔기를 ABA 잔기로 관능화된 폴리에테르를 통해 플루오레세인에 커플링시켰다 (X3757-48 참조, 실시예 41). 도 47C는 플루오레세인과 접합된 mEGF-Tyr10PCL의 ESI 질량 분광측정 분석을 보여준다 (실시예 25 참조). 또한, 본원에서 제공되는 방법을 이용하여 PCL을 mEGF에 혼입하였고 (실시예 12 참조), PCL 잔기를 ABA 잔기로 관능화된 디사카라이드에 커플링시켰다 (3793-050 참조, 실시예 42). 도 47D는 디사카라이드에 접합된 mEGF-Tyr10PCL의 ESI 질량 분광측정 분석을 보여준다 (실시예 26 참조). 디사카라이드의 부착에 사용된 커플링 화학은 다른 당 및 복합 올리고 및 폴리사카라이드의 커플링에 쉽게 적용될 수 있음이 이해된다.To further demonstrate site specific labeling, PCL was incorporated into mEGF using the method provided herein (see Example 12), and the PCL residue was coupled to fluorescein via a polyether functionalized with an ABA residue. Ringed (see X3757-48, Example 41). Figure 47C shows the ESI mass spectrometric analysis of mEGF-Tyr10PCL conjugated with fluorescein (see Example 25). In addition, PCL was incorporated into mEGF using the method provided herein (see Example 12), and the PCL residue was coupled to a disaccharide functionalized with an ABA residue (see 3793-050, Example 42). Figure 47D shows the ESI mass spectrometric analysis of mEGF-Tyr10PCL conjugated to a disaccharide (see Example 26). It is understood that the coupling chemistry used for attachment of the disaccharides can be readily applied to the coupling of other sugars and complex oligos and polysaccharides.
단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 및/또는 피롤리신 유 사체 단백질로의 면역 조절제의 커플링 및 이러한 커플링에 의한 면역원성의 증진Coupling of an immunomodulatory agent to a pyrrolysine and/or pyrrolysine analogue protein incorporated in a protein, polypeptide and/or peptide and enhancement of immunogenicity by such coupling
본원에서 제공되는 또 다른 측면에서, 내부에 혼입된 1개 이상의 피롤리신 및/또는 PCL을 갖는 단백질, 폴리펩티드 및/또는 펩티드를 1개 이상의 면역 조절제로 유도체화한다. 본원에서 제공되는 방법을 이용하여, 면역 자극 잔기를 피롤리신 및/또는 PCL 잔기(들)을 통해 단백질, 폴리펩티드 및/또는 펩티드에 쉽게 커플링시킬 수 있다. 이러한 면역 자극 잔기에는 1개 이상의 니트로기, 지질, 인지질, LPS-유사 분자, 아주반트, 아주반트-유사 분자, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 할로겐, 아릴기, 헤테로아릴기, 시클로알킬기 또는 헤테로시클로알킬기가 포함되나, 이에 제한되지는 않는다. 특정 실시양태에서, 피롤리신 및/또는 PCL에 부착된 면역 조절제를 사용하여, 단백질에 혼입된 p-니트로-페닐알라닌과 유사한 방식으로 자가-항원에 대한 면역 반응을 자극한다 ([Gruenewald J, Tsao ML, Perera R, Dong L, Niessen F, Wen BG, Kubitz DM, Smider VV, Ruf W, Nasoff M, Lerner RA, Schultz PG, Immunochemical termination of self-tolerance, Proc Natl Acad Sci U S A. 2008 Aug 12;105(32):11276-80]). 특정 실시양태에서, 피롤리신 및/또는 PCL에 부착된 면역 조절제를 갖는 자가-항원은 암 백신으로 사용된다. 다른 실시양태에서, 피롤리신 및/또는 PCL에 부착된 면역 조절제를 갖는 항원은 감염성 질환용 백신으로 사용된다. 다른 실시양태에서, 피롤리신 및/또는 PCL에 부착된 면역 조절제를 갖는 항원은 아밀로이드의 형성을 수반하는 질환, 예를 들어 (이에 제한되지는 않음) 알츠하이머병에 대한 백신으로서 사용된다.In another aspect provided herein, proteins, polypeptides and/or peptides having at least one pyrrolysine and/or PCL incorporated therein are derivatized with at least one immunomodulatory agent. Using the methods provided herein, immune stimulating moieties can be readily coupled to proteins, polypeptides and/or peptides via pyrrolysine and/or PCL moiety(s). These immune stimulating moieties include one or more nitro groups, lipids, phospholipids, LPS-like molecules, adjuvants, adjuvant-like molecules, TLR2 agonists, TLR4 agonists, TLR7 agonists, TLR9 agonists, TLR8 agonists, T -Cellular epitope, keyhole limpet hemocyanin (KLH), immunogenic hapten, halogen, aryl group, heteroaryl group, cycloalkyl group or heterocycloalkyl group are included, but are not limited thereto. In certain embodiments, an immune modulator attached to pyrrolysine and/or PCL is used to stimulate an immune response to self-antigens in a manner similar to p-nitro-phenylalanine incorporated into the protein (Gruenewald J, Tsao ML, Perera R, Dong L, Niessen F, Wen BG, Kubitz DM, Smider VV, Ruf W, Nasoff M, Lerner RA, Schultz PG, Immunochemical termination of self-tolerance, Proc Natl Acad Sci US A. 2008
한 실시양태에서, 2-아미노벤즈알데히드 화합물의 라이브러리를 생성하고, 이러한 라이브러리를 관심 항원에 혼입된 피롤리신 또는 PCL에 커플링한 후에 커플링된 생성물을 변형된 항원 및 미변형 항원에 대한 고역가 항체의 생성에 대해 스크리닝하여 면역 자극 잔기의 라이브러리를 생성한다. 이러한 실시양태에서, 2-아미노벤즈알데히드 잔기는 1개 이상의 니트로기, 지질, 인지질, LPS-유사 분자, 아주반트, 아주반트-유사 분자, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 할로겐, 아릴기, 헤테로아릴기, 시클로알킬기 또는 헤테로시클로알킬기를 비롯한 (이에 제한되지는 않음) 다양한 치환기로 치환된다. In one embodiment, a library of 2-aminobenzaldehyde compounds is generated, and after coupling such library to pyrrolysine or PCL incorporated into the antigen of interest, the coupled product is converted to a high titer antibody against the modified and unmodified antigens. Screening for the generation of immunostimulating moieties is generated. In such embodiments, the 2-aminobenzaldehyde moiety is one or more nitro groups, lipids, phospholipids, LPS-like molecules, adjuvants, adjuvant-like molecules, TLR2 agonists, TLR4 agonists, TLR7 agonists, TLR9 agonists. , TLR8 agonist, T-cell epitope, keyhole limpet hemocyanin (KLH), immunogenic hapten, halogen, aryl group, heteroaryl group, cycloalkyl group or heterocycloalkyl group, including, but not limited to, various substituents. Is substituted.
또 다른 실시양태에서, 2-아미노-아세토페논 화합물의 라이브러리를 생성하고, 이러한 라이브러리를 관심 항원에 혼입된 피롤리신 또는 PCL에 커플링한 후에 커플링된 생성물을 변형된 항원 및 미변형 항원에 대한 고역가 항체의 생성에 대해 스크리닝하여 면역 자극 잔기의 라이브러리를 생성한다. 이러한 실시양태에서, 2-아미노-아세토페논 잔기는 1개 이상의 니트로기, 지질, 인지질, LPS-유사 분자, 아주반트, 아주반트-유사 분자, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 할로겐, 아릴기, 헤테로아릴기, 시클로알킬기 또는 헤테로시클로알킬기를 비롯한 (이에 제한되지는 않음) 다양한 치환기로 치환된다.In another embodiment, a library of 2-amino-acetophenone compounds is generated, the library is coupled to the pyrrolysine or PCL incorporated into the antigen of interest, and then the coupled product is transferred to the modified and unmodified antigens. A library of immune stimulating residues is generated by screening for the production of high titer antibodies against. In such embodiments, the 2-amino-acetophenone residue is one or more nitro groups, lipids, phospholipids, LPS-like molecules, adjuvants, adjuvant-like molecules, TLR2 agonists, TLR4 agonists, TLR7 agonists, TLR9. Agonist, TLR8 agonist, T-cell epitope, keyhole limpet hemocyanin (KLH), immunogenic hapten, halogen, aryl group, heteroaryl group, cycloalkyl group or heterocycloalkyl group including, but not limited to, a variety of Substituted with a substituent.
또 다른 실시양태에서, 2-아미노-5-니트로-벤조페논 화합물의 라이브러리를 생성하고, 이러한 라이브러리를 관심 항원에 혼입된 피롤리신 또는 PCL에 커플링한 후에 커플링된 생성물을 변형된 항원 및 미변형 항원에 대한 고역가 항체의 생성에 대해 스크리닝하여 면역 자극 잔기의 라이브러리를 생성한다. 이러한 실시양태에서, 2-아미노-5-니트로-벤조페논 잔기는 1개 이상의 니트로기, 지질, 인지질, LPS-유사 분자, 아주반트, 아주반트-유사 분자, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 할로겐, 아릴기, 헤테로아릴기, 시클로알킬기 또는 헤테로시클로알킬기를 비롯한 (이에 제한되지는 않음) 다양한 치환기로 치환된다.In another embodiment, a library of 2-amino-5-nitro-benzophenone compounds is generated, and after coupling such library to pyrrolysine or PCL incorporated into the antigen of interest, the coupled product is modified antigen and A library of immune stimulating residues is generated by screening for the production of high titer antibodies against unmodified antigens. In this embodiment, the 2-amino-5-nitro-benzophenone moiety is one or more nitro groups, lipids, phospholipids, LPS-like molecules, adjuvants, adjuvant-like molecules, TLR2 agonists, TLR4 agonists, TLR7. Agonists, TLR9 agonists, TLR8 agonists, T-cell epitopes, keyhole limpet hemocyanin (KLH), immunogenic haptens, halogens, aryl groups, heteroaryl groups, cycloalkyl groups, or heterocycloalkyl groups. Not) is substituted with various substituents.
이러한 측면을 입증하기 위해서, 본원에서 제공되는 방법을 이용하여 mTNF-α 및 mEGF에 PCL을 혼입하였고 (실시예 11 및 12 참조), 상기 PCL 잔기를 1개 이상의 니트로페닐기를 함유하는 합텐에 커플링시켰다 (실시예 27 및 28 참조). 도 48A 및 도 48B는 mTNF-Gln21PCL (도 48A) 및 mEGF-Tyr10PCL (도 48B)의 PCL 혼입 부위에서의 모노-니트로페닐 합텐 (3793-001 참조, 실시예 38-8)의 부착을 입증한다. 도 48C 및 도 48D는 mTNF-α (도 48C) 및 mEGF (도 48D)의 PCL 혼입 부위에서의 디-니트로페닐 합텐 (TU3627-088, 실시예 38-7)의 부착을 보여준다. To demonstrate this aspect, PCL was incorporated into mTNF-α and mEGF using the method provided herein (see Examples 11 and 12), and the PCL residue was coupled to a hapten containing at least one nitrophenyl group. (See Examples 27 and 28). Figures 48A and 48B demonstrate the attachment of mono-nitrophenyl hapten (see 3793-001, Examples 38-8) at the site of PCL incorporation of mTNF-Gln21PCL (Figure 48A) and mEGF-Tyr10PCL (Figure 48B). Figures 48C and 48D show the attachment of di-nitrophenyl hapten (TU3627-088, Example 38-7) at the site of PCL incorporation of mTNF-α (Figure 48C) and mEGF (Figure 48D).
도 49A는 mEGF-Tyr10PCL의 PCL 혼입 부위에서의 TLR7 효능제 (X3678-114 참조, 실시예 38-3)의 부착을 입증한다 (실시예 29 참조). 도 49B는 mEGF-Tyr10PCL의 PCL 혼입 부위에서의 인지질 (TU3627-092 참조, 실시예 43-1)의 부착을 입증한다 (실시예 31 참조).Figure 49A demonstrates the attachment of a TLR7 agonist (see X3678-114, Example 38-3) at the site of PCL incorporation of mEGF-Tyr10PCL (see Example 29). Figure 49B demonstrates the adhesion of phospholipids (see TU3627-092, Example 43-1) at the site of PCL incorporation of mEGF-Tyr10PCL (see Example 31).
도 50-51은 mTNF-Gln21PCL 또는 mEGF-Tyr10PCL에 대한 PADRE 펩티드: PX2-PADRE (MW = 1585) (3465-143 참조; 실시예 38-11) 및 BHA-exPADRE (MW = 2060) (3647-104 참조; 실시예 38-10)의 접합을 입증한다. 도 50A 및 도 50B는 2가지 상이한 pH 값에서 PX2-PADRE와의 mTNF-Gln21PCL 접합 반응의 MALDI-TOF 질량 분광측정 분석을 보여주고 (실시예 30 참조), 도 50C는 BHA-exPADRE와의 mTNF-Gln21PCL 접합 반응의 ESI 질량 분광측정 분석을 보여준다 (실시예 30 참조). 도 51은 mEGF-Tyr10PCL에 대한 BHA-exPADRE의 커플링을 보여준다 (실시예 30 참조). PADRE 펩티드의 서열 (서열 28)은 (여기서, X는 시클로헥실-알라닌임)이다. exPADRE 펩티드의 서열 (서열 29)은 (여기서, X는 시클로헥실-알라닌임)이다. 특정 실시양태에서, 공지된 면역원성 펩티드 에피토프는 항원과 커플링되어 상기 항원의 면역원성을 증진시킬 것이다. 특정 실시양태에서, 항원 유래의 펩티드는 면역원성 운반 단백질과 커플링될 것이다. 특정 실시양태에서, 면역원성 운반 단백질은 KLH이다. PADRE 펩티드의 부착에 이용되는 커플링 화학은 PCL 및 피롤리신에 대한 다른 펩티드의 커플링에도 쉽게 적용될 수 있음이 이해된다.Figures 50-51 show PADRE peptides against mTNF-Gln21PCL or mEGF-Tyr10PCL: PX2-PADRE (MW = 1585) (see 3465-143; Example 38-11) and BHA-exPADRE (MW = 2060) (3647-104 See; Examples 38-10) demonstrate conjugation. 50A and 50B show MALDI-TOF mass spectrometric analysis of mTNF-Gln21PCL conjugation reaction with PX2-PADRE at two different pH values (see Example 30), and FIG.50C is mTNF-Gln21PCL conjugation with BHA-exPADRE. The ESI mass spectrometric analysis of the reaction is shown (see Example 30). Figure 51 shows the coupling of BHA-exPADRE to mEGF-Tyr10PCL (see Example 30). The sequence of the PADRE peptide (SEQ ID NO: 28) is (Wherein X is cyclohexyl-alanine). The sequence of the exPADRE peptide (SEQ ID NO: 29) is (Wherein X is cyclohexyl-alanine). In certain embodiments, a known immunogenic peptide epitope will be coupled with an antigen to enhance the immunogenicity of the antigen. In certain embodiments, the antigen-derived peptide will be coupled with an immunogenic carrier protein. In certain embodiments, the immunogenic carrier protein is KLH. It is understood that the coupling chemistry used for attachment of the PADRE peptide can be readily applied to the coupling of PCL and other peptides to pyrrolysine.
면역- PCR : 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 및/또는 피롤리신 유사체에 대한 DNA 및 기타 올리고뉴클레오티드의 커플링 Immuno- PCR : Coupling of DNA and other oligonucleotides to pyrrolysine and/or pyrrolysine analogs incorporated into proteins, polypeptides and/or peptides
본원에서 제공되는 또 다른 측면에서, 내부에 혼입된 1개 이상의 피롤리신 및/또는 PCL을 갖는 단백질, 폴리펩티드 및/또는 펩티드를 본원에서 제공되는 방법을 이용하여 DNA로 유도체화한다. DNA는 단백질, 폴리펩티드 및/또는 펩티드에 혼입된 피롤리신 및/또는 PCL과 반응하는 말단 2-아미노-벤즈알데히드, 2-아미노-아세토페논 또는 2-아미노-5-니트로-벤조페논 잔기를 갖도록 변형되고, 이 DNA는 단백질, 폴리펩티드 및/또는 펩티드에 퀴나졸린-유형 연결부, 환원된 퀴나졸린 연결부 또는 융합된 고리 연결부를 통해 커플링된다 (도 22, 23 및 30 참조).In another aspect provided herein, proteins, polypeptides and/or peptides having at least one pyrrolysine and/or PCL incorporated therein are derivatized to DNA using the methods provided herein. DNA is modified to have a terminal 2-amino-benzaldehyde, 2-amino-acetophenone or 2-amino-5-nitro-benzophenone residue that reacts with pyrrolysine and/or PCL incorporated in the protein, polypeptide and/or peptide. And the DNA is coupled to the protein, polypeptide and/or peptide via a quinazoline-type linkage, a reduced quinazoline linkage, or a fused ring linkage (see FIGS. 22, 23 and 30).
PCL에 대한 DNA의 부착을 입증하기 위해서, 본원에서 제공되는 방법을 이용하여 PCL을 mTNF-α 및 mEGF에 혼입하였고 (실시예 11 및 12 참조), 상기 PCL 잔기를 역시 면역 자극 잔기인 CpG 올리고뉴클레오티드에 커플링시켰다 (실시예 32 참조). BHA-BG1 (3647-057 참조, 실시예 38-12) 및 BHA-BG2 (3597-167 참조, 실시예 38-14)는 PCL 혼입 부위에서 mTNF-Gln21PCL 및 mEGF-Tyr10PCL에 CpG를 부착시키는데 사용된 CpG 시약이었다. mTNF-Gln21PCL (19.3 kDa)에 대한 BHA-BG1 (7.4 kDa) 및 BHA-BG2 (7.4 kDa)의 커플링을 겔 이동 검정 (도 52A)으로 확인하였고, mEGF-Tyr10PCL (7.2 kDa)에 대한 BHA-BG2 (7.4 kDa)의 커플링 (도 52B)도 마찬가지로 확인하였다. BG1의 서열 (서열 30)은 (여기서, *는 포스포티오에이트 연결부를 나타냄)이다. BG2의 서열 (서열 31)은 (여기서, *는 포스포티오에이트 연결부를 나타냄)이다. 특정 실시양태에서, 커플링될 올리고뉴클레오티드는 데옥시리보핵산, 리보핵산, 펩티드 핵산 또는 기타 변형된 올리고뉴클레오티드일 것이다. 처음에는 피롤리신 또는 PCL에 단일 가닥 올리고뉴클레오티드로서 커플링시키고, 이중 가닥 DNA, RNA, PNA 또는 하이브리드 중합체는 상보적 가닥과의 혼성화로 쉽게 제조될 수 있다. CpG 올리고뉴클레오티드의 부착에 사용된 커플링 화학은 다른 올리고뉴클레오티드의 커플링에 쉽게 적용될 수 있다는 것이 이해된다.In order to demonstrate the attachment of DNA to PCL, PCL was incorporated into mTNF-α and mEGF using the method provided herein (see Examples 11 and 12), and the PCL residue was CpG oligonucleotide, which is also an immune stimulating residue. Coupled to (see Example 32). BHA-BG1 (see 3647-057, Example 38-12) and BHA-BG2 (see 3597-167, Example 38-14) were used to attach CpG to mTNF-Gln21PCL and mEGF-Tyr10PCL at the site of PCL incorporation. It was a CpG reagent. The coupling of BHA-BG1 (7.4 kDa) and BHA-BG2 (7.4 kDa) to mTNF-Gln21PCL (19.3 kDa) was confirmed by a gel shift assay (Figure 52A), and BHA- to mEGF-Tyr10PCL (7.2 kDa). The coupling of BG2 (7.4 kDa) (Fig. 52B) was similarly confirmed. The sequence of BG1 (SEQ ID NO: 30) is (Here, * indicates a phosphorothioate linkage). The sequence of BG2 (SEQ ID NO: 31) is (Here, * indicates a phosphorothioate linkage). In certain embodiments, the oligonucleotide to be coupled will be a deoxyribonucleic acid, ribonucleic acid, peptide nucleic acid, or other modified oligonucleotide. Initially coupled to pyrrolysine or PCL as single-stranded oligonucleotides, double-stranded DNA, RNA, PNA or hybrid polymers can be readily prepared by hybridization with complementary strands. It is understood that the coupling chemistry used for the attachment of CpG oligonucleotides can be readily applied to the coupling of other oligonucleotides.
특정 실시양태에서, 단백질, 폴리펩티드 및/또는 펩티드는 항체이고, DNA가 연결된 이러한 항체를 사용하여 면역-PCR 분석을 수행한다. 면역-PCR은 본원에 기재된 방법을 이용하여 DNA를 항체에 연결시킨 후에 DNA 연결된 항체를 표적 항원과 결합시켜서 수행된다. 결합 후, DNA 증폭 기술로 DNA를 증폭시키고, 생성된 증폭 생성물을 전기영동 기술, 마이크로플레이트 방법 또는 실시간 PCR로 분석한다.In certain embodiments, the protein, polypeptide and/or peptide is an antibody, and immuno-PCR analysis is performed using such an antibody to which DNA is linked. Immuno-PCR is performed by ligating DNA to an antibody using the methods described herein, followed by binding of the DNA-linked antibody to a target antigen. After binding, DNA is amplified by DNA amplification technology, and the resulting amplified product is analyzed by electrophoresis technology, microplate method, or real-time PCR.
부위 특이적 및 배향 부착Site specific and orientation attachment
본원에서 제공되는 또 다른 측면에서, 단백질, 폴리펩티드 및/또는 펩티드로의 1개 이상의 피롤리신 및/또는 PCL의 부위 특이적 혼입을 이용하여 지지체 표면과의 부착시에 이러한 단백질, 폴리펩티드 및/또는 펩티드의 방향을 제어한다. 이러한 부착은 피롤리신 또는 PCL 잔기와 벤즈알데히드 잔기, 아세토페논 잔기, 벤조페논 잔기 또는 이들의 조합물로 유도체화된 표면의 반응으로 인한 것이며, 이로써 퀴나졸린 잔기 연결부가 형성되고 단백질이 표면상에서 배향된다. 단백질 내 특정 위치에 피롤리신 및/또는 PCL을 혼입함으로써, 표면에서의 단백질의 방향을 제어한다. 단백질 부착 방향에 대한 이러한 제어를, 단백질 특성을 평가하기 위한 단백질 조작 도구 키트의 성분으로 이용한다. 도 53은 이러한 부위 특이적 배향 부착의 실시양태를 예시하며, 여기서 표면은 단백질에 혼입된 PCL 잔기와 반응하는 20개의 아미노-아세토페논 잔기로 유도체화되어 상기 단백질을 퀴나졸린-유형 연결부를 통해 상기 표면에 부착시킨다. 도 53에서는 형성된 단백질 접합체 연결부가 퀴나졸린-유형 잔기이지만, 다른 실시양태에서는 상기 연결부가 퀴나졸린-유형 잔기의 환원된 형태이다 (도 22, 23 및 30 참조). 다른 실시양태에서, 상기 연결부는 융합된 고리 잔기이다 (도 22, 23 및 30 참조).In another aspect provided herein, site specific incorporation of one or more pyrrolysines and/or PCLs into proteins, polypeptides and/or peptides upon attachment to a support surface, such proteins, polypeptides and/or Controls the orientation of the peptide. This attachment is due to the reaction of a surface derivatized with a pyrrolysine or PCL moiety with a benzaldehyde moiety, acetophenone moiety, benzophenone moiety or a combination thereof, whereby a quinazoline moiety linkage is formed and the protein is oriented on the surface. . By incorporating pyrrolysine and/or PCL at specific positions in the protein, the orientation of the protein on the surface is controlled. This control over the direction of protein attachment is used as a component of a protein engineering tool kit for evaluating protein properties. Figure 53 illustrates an embodiment of such a site-specific orientation attachment, wherein the surface is derivatized with 20 amino-acetophenone residues that react with PCL residues incorporated into the protein, allowing the protein to be described above through a quinazoline-type linkage. Attach it to the surface. In Figure 53 the protein conjugate linkage formed is a quinazoline-type moiety, but in other embodiments the linkage is a reduced form of a quinazoline-type moiety (see FIGS. 22, 23 and 30). In other embodiments, the linkage is a fused ring moiety (see FIGS. 22, 23 and 30).
지지체의 비제한적인 예에는 고체 및 반-고체 매트릭스, 예컨대 에어로젤 및 히드로겔, 수지, 비드, 바이오칩 (예를 들어, 바이오칩으로 코팅된 박막), 미세유체 칩, 규소 칩, 다중 웰 플레이트 (또한, 마이크로타이터 플레이트 또는 마이크로플레이트라고 지칭되기도 함), 막, 세포, 전도성 및 비-전도성 금속, 유리 (예를 들어, 현미경 슬라이드) 및 자성 지지체가 포함되나, 이에 제한되지는 않는다. 본원에 기재된 방법 및 조성물에 사용되는 고체 지지체의 다른 비제한적인 예에는 실리카 겔, 중합체 막, 입자, 유도체화된 플라스틱 필름, 유도체화된 유리, 유도체화된 실리카, 유리 비드, 목화, 플라스틱 비드, 알루미나 겔, 폴리사카라이드, 예컨대 세파로스, 폴리(아크릴레이트), 폴리스티렌, 폴리(아크릴아미드), 폴리올, 아가로스, 한천, 셀룰로스, 덱스트란, 전분, 피콜(FICOLL), 헤파린, 글리코겐, 아밀로펙틴, 만난, 이눌린, 니트로셀룰로스, 디아조셀룰로스, 폴리비닐클로라이드, 폴리프로필렌, 폴리에틸렌 (예를 들어, 폴리(에틸렌 글리콜)), 나일론, 라텍스 비드, 자성 비드, 상자성 비드, 초상자성 비드, 전분 등이 포함된다. 특정 실시양태에서, 본원에 기재된 방법 및 조성물에 사용되는 지지체는 표면 분석에 사용되는 지지체, 예컨대 표면 음파 장치 또는 소실파 분석, 예컨대 표면 플라스몬 공명 분석을 이용하는 장치이다.Non-limiting examples of supports include solid and semi-solid matrices such as aerogels and hydrogels, resins, beads, biochips (e.g., thin films coated with biochips), microfluidic chips, silicon chips, multi-well plates (also (Also referred to as microtiter plates or microplates), membranes, cells, conductive and non-conductive metals, glass (eg, microscope slides), and magnetic supports. Other non-limiting examples of solid supports used in the methods and compositions described herein include silica gel, polymer membranes, particles, derivatized plastic films, derivatized glass, derivatized silica, glass beads, cotton, plastic beads, Alumina gels, polysaccharides such as sepharose, poly(acrylate), polystyrene, poly(acrylamide), polyol, agarose, agar, cellulose, dextran, starch, FICOLL, heparin, glycogen, amylopectin, Mannan, inulin, nitrocellulose, diazocellulose, polyvinyl chloride, polypropylene, polyethylene (e.g., poly(ethylene glycol)), nylon, latex beads, magnetic beads, paramagnetic beads, superparamagnetic beads, starch, etc. do. In certain embodiments, the support used in the methods and compositions described herein is a support used for surface analysis, such as a surface acoustic wave device or an apparatus using vanishing wave analysis, such as surface plasmon resonance analysis.
내부에 혼입된 피롤리신 및/또는 피롤리신 유사체를 갖는 단백질, 폴리펩티드 및/또는 펩티드의 부착에 사용되는 고체 지지체의 표면은 히드록실, 카르복실, 아미노, 티올, 알데히드, 할로겐, 니트로, 시아노, 아미도, 우레아, 카르보네이트, 카르바메이트, 이소시아네이트, 술폰, 술포네이트, 술폰아미드 및 술폭시드를 비롯한 (이에 제한되지는 않음) 반응성 관능기를 갖는다. 이러한 관능기는, 피롤리신 또는 피롤리신 유사체와 반응하여 퀴나졸린 잔기를 형성하는 2-아미노-벤즈알데히드 잔기, 2-아미노-아세토페논 잔기 및/또는 2-아미노-5-니트로-벤조페논 잔기를 공유 부착시키는데 사용된다. 특정 실시양태에서, 이러한 2-아미노-벤즈알데히드 잔기, 2-아미노-아세토페논 잔기 및 2-아미노-5-니트로-벤조페논 잔기는 고체 지지체 반응성 관능기, 예컨대 (이에 제한되지는 않음) 히드록실, 카르복실, 아미노, 티올, 알데히드, 할로겐, 니트로, 시아노, 아미도, 우레아, 카르보네이트, 카르바메이트, 이소시아네이트, 술폰, 술포네이트, 술폰아미드 및 술폭시드와의 반응으로 고체 지지체에 커플링된 중합체 링커의 일부이다.The surface of the solid support used for attachment of proteins, polypeptides and/or peptides having pyrrolysine and/or pyrrolysine analogs incorporated therein is hydroxyl, carboxyl, amino, thiol, aldehyde, halogen, nitro, cyanide. It has reactive functional groups including, but not limited to, furnace, amido, urea, carbonate, carbamate, isocyanate, sulfone, sulfonate, sulfonamide and sulfoxide. Such a functional group comprises a 2-amino-benzaldehyde residue, a 2-amino-acetophenone residue, and/or a 2-amino-5-nitro-benzophenone residue that reacts with pyrrolysine or a pyrrolysine analog to form a quinazoline residue. Used for covalent attachment. In certain embodiments, such 2-amino-benzaldehyde moieties, 2-amino-acetophenone moieties and 2-amino-5-nitro-benzophenone moieties are solid support reactive functional groups such as but not limited to hydroxyl, car Coupled to a solid support by reaction with boxyl, amino, thiol, aldehyde, halogen, nitro, cyano, amido, urea, carbonate, carbamate, isocyanate, sulfone, sulfonate, sulfonamide and sulfoxide. It is part of the polymer linker.
다른 실시양태에서, 고체 지지체의 표면은 여기에 부착된 스트렙타비딘 또는 아비딘을 갖고, 내부에 혼입된 피롤리신 및/또는 피롤리신 유사체를 갖는 단백질, 폴리펩티드 및/또는 펩티드의 부착에 사용되며, 여기서 피롤리신 및/또는 피롤리신 유사체는 비오틴을 상기 단백질, 폴리펩티드 및/또는 펩티드에 부위 특이적으로 부착시키는데 이용된다.In other embodiments, the surface of the solid support has streptavidin or avidin attached thereto, and is used for attachment of proteins, polypeptides and/or peptides having pyrrolysine and/or pyrrolysine analogs incorporated therein, and , Wherein the pyrrolysine and/or pyrrolysine analog is used to site-specifically attach biotin to the protein, polypeptide and/or peptide.
본원에 기재된 방법 및 조성물에 사용되는 다른 지지체는 펩티드 합성에 사용되는 수지, 예를 들어 단지 예로서만 언급하자면, 폴리스티렌, PAM-수지, 폴리하이프(POLYHIPE)™ 수지, 폴리아미드 수지, 폴리(에틸렌 글리콜)이 이식된 폴리스티렌 수지, 폴리디메틸-아크릴아미드 수지 및 PEGA 비드를 포함한다.Other supports used in the methods and compositions described herein include resins used for peptide synthesis, such as polystyrene, PAM-resin, POLYHIPE™ resins, polyamide resins, poly(ethylene, by way of example only). Glycol) implanted polystyrene resin, polydimethyl-acrylamide resin, and PEGA beads.
특정 실시양태에서, 내부에 혼입된 피롤리신 및/또는 PCL을 갖는 단백질, 폴리펩티드 및/또는 펩티드를 고체 지지체에 어레이 포맷으로 침착시킨다. 특정 실시양태에서, 이러한 침착은 지지체 표면과 전달 메카니즘, 예컨대 핀 또는 모세관 사이의 직접적인 표면 접촉, 또는 소형 노즐로부터 고체 표면으로 액체를 이동시키기 위해 압전 및 다른 형태의 추진력을 이용하는 잉크 젯 기술로 수행된다. 접촉 프린팅의 경우, 로봇식 제어 시스템 및 멀티플렉스화 프린트헤드는 자동화된 마이크로어레이 제작을 허용한다. 압전성 추진 기술에 의한 비-접촉 침착의 경우, 로봇식 시스템은 또한 연속식 또는 드랍-온-디맨드(drop-on-demand) 장치를 사용한 자동 마이크로어레이 제작을 허용한다.In certain embodiments, proteins, polypeptides and/or peptides with pyrrolysine and/or PCL incorporated therein are deposited on a solid support in an array format. In certain embodiments, such deposition is carried out by direct surface contact between the support surface and a delivery mechanism, such as a pin or capillary, or by ink jet technology using piezoelectric and other forms of propulsion to move liquid from a small nozzle to a solid surface. . For contact printing, robotic control systems and multiplexed printheads allow automated microarray fabrication. In the case of non-contact deposition by piezoelectric propulsion technology, robotic systems also allow automatic microarray fabrication using continuous or drop-on-demand devices.
실시예Example
본원에 기재된 실시양태 및 하기 실시예는 단지 예시 목적을 위한 것이고, 이에 대한 다양한 변형 또는 변화가 당업자에게 명확할 것이고 본 출원의 사상 및 범위 및 첨부하는 특허청구범위 내에 포함된다는 것을 이해해야 한다. 본원에서 인용된 모든 간행물, 특허 및 특허 출원은 나타낸 바와 같은 목적으로 본원에 참고로 포함된다.It should be understood that the embodiments described herein and the following examples are for illustrative purposes only, and that various modifications or changes thereto will be apparent to those skilled in the art and are included within the spirit and scope of the present application and the appended claims. All publications, patents and patent applications cited herein are incorporated herein by reference for the purposes as indicated.
실시예 1: 포유동물 세포를 사용한, 생합성으로 생성된 PCL 의 단백질로의 부위 특이적 혼입 Example 1 : Site-specific incorporation of biosynthetically generated PCL into protein using mammalian cells
본 실시예는 포유동물 세포로 형질감염된 경우에 표적 단백질 내 TAG 코딩 부위로의 PCL 혼입을 가능하게 하는 유전자 구축물에 관한 기재를 제공한다. 여러 부위에서의 동시 혼입이 가능하지만, 본원에서의 모든 실시예는 단백질 분자 1개 당 단일 부위에서의 PCL 혼입만을 예시한다. 본 실시예는 또한 포유동물 세포를 사용하여 PCL을 단백질에 혼입시키는 일반적인 절차를 기재한다.This example provides a description of a gene construct that enables incorporation of PCL into a TAG coding site in a target protein when transfected with mammalian cells. While simultaneous incorporation at multiple sites is possible, all examples herein illustrate only PCL incorporation at a single site per protein molecule. This example also describes the general procedure for incorporating PCL into proteins using mammalian cells.
구축물:Construct:
메타노사르시나 마제이 Go1 (NC_003901, 하기 참조)의 pylS, pylT, pylB, pylC 및 pylD 뉴클레오티드 서열을 기초로 디자인된 프라이머를 사용한 PCR을 이용하여, 메타노사르시나 마제이 Go1의 게놈 DNA로부터 전장 pylT, 및 pylS, pylT, pylB, pylC 및 pylD의 코딩 영역을 증폭시켰다. 상기 유전자들의 개시 코돈은 ATG로 변화시켰다. pylT를 pCMV 벡터에 CMV 프로모터의 상류에서 클로닝하였다. 전사는 U6 프로모터의 제어를 받았고, 벡터 pCMVU6을 형성하였다. pylS, pylB, pylC 및 pylD를 유사하게 pCMVU6으로 클로닝하였다. pylS, pylB, pylC 및 pylD의 전사는 CMV 프로모터의 제어를 받았다 (도 4A).Using PCR using primers designed based on the nucleotide sequences of pylS, pylT, pylB, pylC and pylD of metanosarcina majei Go1 (NC_003901, see below), full-length pylT from genomic DNA of metanosarcina majei Go1, And coding regions of pylS, pylT, pylB, pylC and pylD were amplified. The start codon of these genes was changed to ATG. pylT was cloned upstream of the CMV promoter into the pCMV vector. Transcription was under the control of the U6 promoter and formed the vector pCMVU6. pylS, pylB, pylC and pylD were similarly cloned into pCMVU6. The transcription of pylS, pylB, pylC and pylD was under the control of the CMV promoter (Fig. 4A).
표적 유전자인 인간 레티놀 결합 단백질 (hRBP4), 인간 및 마우스 에리트로포이에틴 (EPO) 및 mIgG1 Fc (mFc)의 코딩 영역을 C-말단에 His 태그를 갖도록 하여 CMV 프로모터 제어하의 pRS 벡터에 클로닝하였다 (도 4B). TAG 코돈으로 돌연변이시킬 잔기를 표 2에 나타낸 바와 같이 기존의 구조적 모델에서의 용매-노출을 기초로 선택하였다. TAG 코돈을 PCR 방법으로 표적 유전자에 도입하였다.The coding regions of target genes human retinol binding protein (hRBP4), human and mouse erythropoietin (EPO) and mIgG1 Fc (mFc) were cloned into a pRS vector under CMV promoter control by having a His tag at the C-terminus (Fig. 4B). Residues to be mutated to the TAG codon were selected based on solvent-exposure in the existing structural model as shown in Table 2. The TAG codon was introduced into the target gene by PCR.
도 4B는 hRBP4, mEPO, hEPO 및 mIgG1의 TAG 돌연변이 구축물을 보여준다. 돌연변이 부위는 표 2에 기재되어 있고, 도 4B에서 화살표로 표시된다. 각 구축물마다 His 태그를 정제 및 검출 도구로서 C-말단에 부착하였다. 전장 단백질만이 His 태그를 가질 수 있다. pylT, pylS, pylB, pylC 및 pylD를 CMV 제어하의 pCMVU6에 클로닝하였다 (도 4A). 이들을 동시 형질감염 연구에 사용하였다.Figure 4B shows the TAG mutant constructs of hRBP4, mEPO, hEPO and mIgG1. Mutation sites are listed in Table 2 and indicated by arrows in Figure 4B. For each construct, a His tag was attached to the C-terminus as a purification and detection tool. Only full-length proteins can carry His tags. pylT, pylS, pylB, pylC and pylD were cloned into pCMVU6 under CMV control (Fig. 4A). These were used in co-transfection studies.
세포 배양 및 형질감염:Cell culture and transfection:
HEK293F 세포를 293 프리스타일(Freestyle) 발현 배지 중의 현탁액으로서 37℃에서 5% CO2하에 성장시켰다. 형질감염 하루 전에, 세포를 0.7×106개 세포/mL로 분할하였다. 플라스미드 DNA를 퀴아젠(Qiagen) 맥시(Maxi) 플라스미드 제조 키트로 제조하였다. 형질감염을 PEI 방법으로 수행하였다. PEI를 Opti-MEM 중에서 플라스미드 DNA와 2:1의 비율로 혼합하고, DNA 복합체를 세포 배양물 1 mL 당 1 ㎍ 플라스미드 DNA로 HEK293F 세포에 첨가하고, 상기 세포를 4일 동안 5 mM Cyc 또는 D-오르니틴의 존재하에 배양하였다. 세포를 여러가지 플라스미드와 동시 형질감염시킬 때, PEI:플라스미드 DNA의 총량의 비율은 언제나 2:1이었다.HEK293F cells were grown as a suspension in 293 Freestyle expression medium at 37° C. under 5% CO 2. One day before transfection, cells were split at 0.7×10 6 cells/mL. Plasmid DNA was prepared with a Qiagen Maxi plasmid preparation kit. Transfection was carried out by the PEI method. PEI was mixed with plasmid DNA in Opti-MEM at a ratio of 2:1, DNA complex was added to HEK293F cells with 1 μg plasmid DNA per 1 mL of cell culture, and the cells were added to 5 mM Cyc or D- Incubated in the presence of ornithine. When cells were co-transfected with several plasmids, the ratio of the total amount of PEI:plasmid DNA was always 2:1.
단백질 정제 및 분석:Protein purification and analysis:
형질감염 4일 후, 세포 배양물을 2000 g에서 20분 동안 원심분리하고, His 태그가 부착된 단백질의 정제를 위해 배지를 수집하였다. 상기 배지를 10 mM 이미다졸을 함유하는 20 mM Tris-HCl (7.5), 150 mM NaCl로 미리 평형화시킨 Ni-NTA 컬럼에 로딩하였다. 상기 컬럼을 동일 완충제로 세척하고, 용출 완충제 (20 mM Tris-HCl 7.5, 150 mM NaCl, 300 mM 이미다졸)로 용출시켰다. 용출된 단백질을 브래드포드(Bradford) 방법 및 SDS-PAGE로 검정하였다. 일부 경우, 배지 및 정제된 단백질을 His 태그 또는 단백질에 대한 항체를 사용한 웨스턴 블럿팅으로 분석하였다.After 4 days of transfection, the cell culture was centrifuged at 2000 g for 20 minutes, and a medium was collected for purification of the His-tagged protein. The medium was loaded onto a Ni-NTA column previously equilibrated with 20 mM Tris-HCl (7.5) and 150 mM NaCl containing 10 mM imidazole. The column was washed with the same buffer and eluted with an elution buffer (20 mM Tris-HCl 7.5, 150 mM NaCl, 300 mM imidazole). The eluted protein was assayed by Bradford method and SDS-PAGE. In some cases, media and purified proteins were analyzed by Western blotting using His tags or antibodies against the protein.
메타노사르시나 마제이의 게놈 DNA로부터 클로닝된 pyl 유전자의 서열을 아래에 나타냈다:The sequence of the pyl gene cloned from the genomic DNA of metanosarcina majei is shown below:
실시예 2: 포유동물 세포를 사용한, 생합성으로 생성된 PCL 의 hRBP4 로의 부위 특이적 혼입 Example 2: The above portion to the mammal of an animal using the cells generated by biosynthetic PCL hRBP4 specific incorporation
본 실시예는 모델 단백질 인간 RBP4 (인간 레티놀 결합 단백질 4) 내 TAG 코딩 부위에서의 PCL의 부위 특이적 혼입을 입증한다. hRBP4는 분비형 단백질이고, 이것의 구조는 특징규명된 바 있으며, hRBP4에서 용매 노출되는 잔기는 규정되어 있다.This example demonstrates the site-specific incorporation of PCL at the TAG coding site in the model protein human RBP4 (human retinol binding protein 4). hRBP4 is a secreted protein, its structure has been characterized, and the solvent exposed residues in hRBP4 are defined.
hRBP4를 C-말단에 Flag-His 태그를 갖도록 하여 pRS 벡터로 클로닝하였다 (도 4A). 일시적인 형질감염을 이용하여 hRBP4를 HEK293F 세포에서 충분히 발현시켰다. TAG 코돈을 표 2 및 하기 서열에 나타낸 바와 같이 별개의 hRBP4 구축물에서 9개 위치에 도입하였다. 개개의 TAG 코돈은 밑줄을 그은 아미노산 잔기의 코돈에서 도입되었음에 주목한다.hRBP4 was cloned into a pRS vector to have a Flag-His tag at the C-terminus (Fig. 4A). HRBP4 was sufficiently expressed in HEK293F cells using transient transfection. TAG codons were introduced at 9 positions in separate hRBP4 constructs as shown in Table 2 and the sequence below. Note that individual TAG codons were introduced at the codons of the underlined amino acid residues.
Tyr108 및 Phe54는 hRBP4의 레티놀 결합 부위이지만, 모든 다른 TAG 돌연변이는 용매에 노출되는 단백질 잔기에 존재한다. 포유동물 세포에서 발현되는 경우, 이들 TAG 부위에서의 번역 종료는 C-말단 His 태그가 없는 말단절단 단백질을 생성한다. 따라서, 이러한 말단절단 단백질은 항-His 항체에 의해 인식되지 않을 것이다. 전사 진행(read-through) 번역이 His 태그를 갖는 전장 생성물을 생성할 것이다. 따라서, 항-His 항체를 사용한 검출은 전사 진행 활성의 지시자로서 기능한다.Tyr108 and Phe54 are the retinol binding sites of hRBP4, but all other TAG mutations are present at the protein residues exposed to the solvent. When expressed in mammalian cells, termination of translation at these TAG sites results in a truncated protein without a C-terminal His tag. Thus, these truncated proteins will not be recognized by anti-His antibodies. Read-through translation will produce a full-length product with a His tag. Thus, detection with an anti-His antibody serves as an indicator of transcriptional progression activity.
hRBP4hRBP4 의 발현:Manifestations of:
pRSRBP를 HEK293F 세포에 pCMVpyS, pCMVpyB, pCMVpyC 및 pCMVpyD와 함께 동시 형질감염시고, 세포를 실시예 1에 기재된 바와 같이 5 mM D-오르니틴의 존재하에 성장시켰다. 이어서, 웨스턴 블럿팅 및 항-His 항체를 사용하여 His 태그가 부착된 hRBP4를 분석하였고, 말단절단 hRBP4는 His 태그를 함유하지 않기 때문에 TAG 코돈에 혼입된 PCL을 갖는 전장 hRBP4만이 웨스턴 블럿팅으로 검출되었다. 따라서, 전장 hRBP4의 존재는 PCL 혼입을 입증한다. 도 6A에 나타난 바와 같이, 항-His 항체는 전장 hRBP4에 대한 예상 크기인 26 kDa에서의 밴드를 검출하였고, 이는 돌연변이된 TAG 코돈에 PCL이 혼입되었음을 나타낸다. PCL은 9종의 hRBP4 구축물 (표 2)에서 상이한 비율로 혼입되었고, #2, #6 및 #9 돌연변이 구축물에는 더 높은 비율로, #7 및 #8 돌연변이 구축물에는 더 낮은 비율로 혼입되었다. #2, #6 및 #9에 대한 단백질 수율은 4 내지 8 mg/L였고, 야생형 hRBP4의 경우에는 상기 수율의 약 20%였다. 이들 단백질을 정제하고, 질량 분광측정 (MS) 분석을 실시하였다. 단백질에 대해 얻어진 질량은 PCL의 혼입에 대해 예상된 것과 일치하였고, 병렬식 MS 분석으로 예상 부위에서의 PCL 혼입을 확인하였다 (도 6A 내지 11).pRSRBP was co-transfected with pCMVpyS, pCMVpyB, pCMVpyC and pCMVpyD into HEK293F cells, and cells were grown in the presence of 5 mM D-ornithine as described in Example 1. Subsequently, using Western blotting and anti-His antibody, hRBP4 with His tag was analyzed, and since truncated hRBP4 does not contain His tag, only full-length hRBP4 having PCL incorporated in the TAG codon was detected by Western blotting. Became. Thus, the presence of full-length hRBP4 demonstrates PCL incorporation. As shown in Fig. 6A, the anti-His antibody detected a band at 26 kDa, which is the expected size for full-length hRBP4, indicating that PCL was incorporated into the mutated TAG codon. PCL was incorporated at different rates in the 9 hRBP4 constructs (Table 2), higher rates in #2, #6 and #9 mutant constructs and lower in #7 and #8 mutant constructs. Protein yields for #2, #6 and #9 were 4 to 8 mg/L, and for wild-type hRBP4 it was about 20% of the yield. These proteins were purified and subjected to mass spectrometry (MS) analysis. The mass obtained for the protein was consistent with that expected for the incorporation of PCL, and parallel MS analysis confirmed the incorporation of PCL at the expected site (FIGS. 6A to 11 ).
도 6B는 PCL가 hRBP4에 혼입된 HEK293F 세포에서 생성된 hRBP4의 SDS-PAGE를 보여주고, 도 6C는 이것의 질량 스펙트럼을 보여준다. hRBP4를 D-오르니틴의 부재하에 (A, 레인 1) 또는 D-오르니틴의 존재하에 (A, 레인 2) 동시 형질감염된 HEK293F 세포의 배지로부터 정제하고 SDS-PAGE로 분석하였다. 화살표는 전장 hRBP4를 나타낸다. 정제된 단백질을 질량 분광측정법으로 분석하였다 (B): 관찰된 질량은 23166.0 Da으로, 예상 질량 23168 Da과 유사하였다.6B shows the SDS-PAGE of hRBP4 generated in HEK293F cells in which PCL was incorporated into hRBP4, and FIG. 6C shows its mass spectrum. hRBP4 was purified from the medium of co-transfected HEK293F cells in the absence of D-ornithine (A, lane 1) or in the presence of D-ornithine (A, lane 2) and analyzed by SDS-PAGE. Arrows indicate full length hRBP4. The purified protein was analyzed by mass spectrometry (B): The observed mass was 23166.0 Da, which was similar to the expected mass of 23168 Da.
hRBP4hRBP4 에 부위 특이적으로 혼입된 Specifically incorporated in the PCLPCL 의 질량 분광측정 검출:Mass spectrometry detection of:
pylS, pylT, pylC 및 pylD 유전자, 및 hRBP4 돌연변이체 #6에 대한 DNA (표 2: hRBP4 Phe122PCL)로 형질감염된 HEK293F 세포의 500 mL 배양물을 5 mM D-오르니틴이 보충된 배지 중에서 성장시켰다. 상기 배지를 Ni-NTA 컬럼에 2회 운행시켜서 모든 전장 hRBP4 단백질을 포획하였다. 합한 용출 분획물은 전체 단백질을 4.3 mg 함유하였다. 상기 단백질의 관찰된 질량은 23166.8 Da (예상치 23168 Da)이었고, 이는 단일 PCL 잔기의 혼입과 일치하였다 (데이타는 나타내지 않음). 샘플 분취액을 트립신 소화시키고 LC-MS 분석을 실시하였다. 도 7의 MS/MS 스펙트럼은 펩티드 (여기서, 잔기 F*는 PCL의 질량과 일치하는 질량을 가짐)에 배정될 수 있고, 이는 잔기 122의 원하는 TAG 부위에서의 PCL의 부위 특이적 혼입을 확인시켜 준다. 펩티드 에 대한 배정은 하기 기재한다:500 mL cultures of HEK293F cells transfected with DNA for the pylS, pylT, pylC and pylD genes, and hRBP4 mutant #6 (Table 2: hRBP4 Phe122PCL) were grown in medium supplemented with 5 mM D-ornithine. The medium was run twice on a Ni-NTA column to capture all full-length hRBP4 proteins. The combined elution fractions contained 4.3 mg of total protein. The observed mass of the protein was 23166.8 Da (estimated 23168 Da), which was consistent with the incorporation of a single PCL residue (data not shown). Sample aliquots were trypsin digested and LC-MS analysis was performed. The MS/MS spectrum of Figure 7 shows the peptide (Where residue F* has a mass that matches the mass of PCL), which confirms the site-specific incorporation of PCL at the desired TAG site of residue 122. Peptide Assignments to are listed below:
m/z 647.86에서의 에 대한 배정 (F* = PCL)m/z at 647.86 For (F* = PCL)
중성 펩티드 Mr의 모노아이소토픽(monoisotopic) 질량 (계산치): 1273.6819Monoisotopic mass of neutral peptide Mr (calculated value): 1273.6819
가변적 변형:Variable transformation:
F7: F에서의 PCL (F)F7: PCL at F (F)
이온 스코어: 60 예상치: 0.0013Ion Score: 60 Expected: 0.0013
매치 (굵은 글씨): 22/74 단편 이온 (32개의 가장 강력한 피크 사용)Match (bold): 22/74 fragment ions (using 32 strongest peaks)
도 8은 인간 RBP4 Phe122PCL의 트립신 소화물의 질량 분광측정 분석 (의 2+ 이온의 TIC 및 EIC)을 보여주며, 표적 Phe122TAG 부위에서의 PCL의 혼입을 나타낸다. 도 9는 인간 RBP4 Phe122PCL의 트립신 소화물의 질량 분광측정 분석을 보여준다. 질량 스펙트럼은 m/z 425.57 (3+) 및 637.85 (2+) 각각에서 의 3+ 및 2+ 전구체를 보여주고 (F* = PCL), 이것은 표적 Phe122TAG 부위에서의 PCL의 혼입을 추가로 나타낸다.Figure 8 is a mass spectrometric analysis of the trypsin digest of human RBP4 Phe122PCL ( TIC and EIC of the 2+ ion of Phe122TAG), indicating incorporation of PCL at the target Phe122TAG site. Figure 9 shows the mass spectrometric analysis of the trypsin digest of human RBP4 Phe122PCL. Mass spectra at m/z 425.57 (3+) and 637.85 (2+) respectively The 3+ and 2+ precursors of are shown (F* = PCL), which further indicates incorporation of PCL at the target Phe122TAG site.
실시예 3: 포유동물 세포로부터의 용해물 중 생합성으로 생성된 PCL 의 검출 Example 3 : Detection of biosynthetically generated PCL in lysates from mammalian cells
본 실시예는 PCL이 포유동물 세포에서 생합성으로 생성되고 이러한 세포의 용해물 중에서 유리 아미노산으로 검출가능하다는 것을 입증한다. 따라서, 본 실시예는 PCL이 피롤리신 및 다른 20종의 천연 발생 아미노산과 마찬가지로 혼입되고 PCL 혼입이 번역후 단백질 변형의 결과가 아님을 시사한다.This example demonstrates that PCL is produced biosynthetically in mammalian cells and is detectable as a free amino acid in the lysate of such cells. Thus, this example suggests that PCL is incorporated like pyrrolysine and other 20 naturally occurring amino acids, and that PCL incorporation is not the result of post-translational protein modification.
세포 cell 용해물Lysate 중 medium PCLPCL 의 검출Detection of
HEK293F 세포의 60 mL 배양물을 5 mM D-오르니틴의 존재하에 또는 D-오르니틴 없이 pylT, pylS, pylB, pylC 및 pylD 유전자와 함께 성장시켰다. 상기 세포를 수확하고, 이중-탈이온수 250 ㎕ 중에서의 초음파 처리로 용해시켰다. 세포 잔해를 원심분리로 펠렛화하였다. 차가운 메탄올을 최종 농도 80%로 첨가하여 단백질을 침전시키고 원심분리로 제거하였다. 이어서, 가용성 용해물을 스피드백으로 농축시키고, 질량 분광측정법과 조합된 고압 액체 크로마토그래피로 분석하여 PCL의 존재를 확인하였다.60 mL cultures of HEK293F cells were grown with the pylT, pylS, pylB, pylC and pylD genes in the presence of 5 mM D-ornithine or without D-ornithine. The cells were harvested and lysed by sonication in 250 μl of double-deionized water. Cell debris was pelleted by centrifugation. The protein was precipitated by adding cold methanol to a final concentration of 80% and removed by centrifugation. The soluble lysate was then concentrated with a speedbag and analyzed by high pressure liquid chromatography in combination with mass spectrometry to confirm the presence of PCL.
건조된 샘플을 우선 100 ㎕의 98/2 이동상 A/이동상 B (이동상 A: 0.1% 포름산을 함유하는 물; 이동상 B: 0.1% 포름산을 함유하는 아세토니트릴) 중에 재구성하였다. 이어서, 상기 샘플을 20배 희석하고, 2 ㎕를 HPLC에 주입하였다. 분석물의 분리는 다음과 같은 용액 구배를 사용하는 아질런트(Agilent) 조르박스(zorbax) 300SB_C18 역상 HPLC 컬럼에서 수행하였다: 0분 2% B; 5분 2% B; 60분 100% B; 유속: 0.25 mL/분. HPLC 추적도의 비교 (도 10A)는 생합성 유전자 pylB, pylC 및 pylD로 형질감염시키고 D-오르니틴의 존재하에 성장시킨 세포의 용해물 (아래쪽 EIC (추출된 이온 크로마토그램) 추적도)에는 존재하지만 D-오르니틴 없이 성장시킨 세포의 용해물 (위쪽 EIC 추적도)에는 존재하지 않는 제4.13분에서의 피크 (별표로 표시함)를 보여준다. 제4.13분 HPLC 피크의 전체 스캐닝 질량 스펙트럼 (도 10C)은 PYL의 탈메틸화 버전인 PCL에 대한 이론적 질량 (242.14992 Da)과 일치하는 242.14943 Da의 질량을 나타낸다. 리신 (제1.44분에서의 HPLC 피크)은 2가지 샘플 모두에 동일하게 풍부하며 (도 10B), 리신의 전체 질량 스펙트럼 (도 10D)은 내부 보정물로 기능하고 상기 방법의 정확성을 예시한다. 이러한 분석은 PCL이 세포 용해물 중에서 D-오르니틴으로부터 검출가능한 유리 아미노산으로 생성됨을 나타낸다.The dried samples were first reconstituted in 100 μl of 98/2 mobile phase A/mobile phase B (mobile phase A: water containing 0.1% formic acid; mobile phase B: acetonitrile containing 0.1% formic acid). Then, the sample was diluted 20 times, and 2 μl was injected into the HPLC. Separation of analytes was performed on an Agilent zorbax 300SB_C18 reverse phase HPLC column using the following solution gradient: 0
실시예 4: 추정적 PCL 및 피롤리신 전구체, 합성 피롤리신 유사체, 및 생합성 유전자들의 상이한 조합을 사용한 혼입 시험 Example 4 : Incorporation test using putative PCL and pyrrolysine precursors, synthetic pyrrolysine analogs, and different combinations of biosynthetic genes
본 실시예는 D-오르니틴이 인간 RBP4로의 부위 특이적 혼입으로 측정되는 바와 같이 PCL 생합성의 바람직한 전구체임을 입증한다. 또한, 본 실시예에서는 포유동물 세포를 사용한, 모델 단백질 인간 RBP4, 인간 레티놀 결합 단백질 4 내 TAG 코딩 부위에서의 특정 피롤리신 유사체, 예컨대 CYC의 부위 특이적 혼입이 예시된다. 본 실시예는 또한 pylS tRNA 신테타제의 기질 특이성에 대한 통찰을 제공한다.This example demonstrates that D-ornithine is a preferred precursor of PCL biosynthesis as measured by site specific incorporation into human RBP4. In addition, in this Example, the site-specific incorporation of a specific pyrrolysine analog, such as CYC, at the TAG coding site in the model protein human RBP4 and human
N-ε-N-ε- 시클로펜틸옥시카르보닐Cyclopentyloxycarbonyl -L-리신 (-L-lysine ( CYCCYC ) 혼입) Mixed
9종의 hRBP4 TAG 돌연변이체 (표 3)에 대한 DNA 구축물을 개별적으로 실시예 2에 기재된 바와 같이 pylT/pylS DNA와 함께 HEK293F 세포에 동시 형질감염시키고, 피롤리신 유사체인 4 mM N-ε-시클로펜틸옥시카르보닐-L-리신 (CYC) 없이 또는 이것의 존재하에 배양하였다. 배양 배지를 수확하고, 항-His 및 항-hRBP4 항체를 사용한 웨스턴 블럿팅으로 분석하였다 (도 11A). 항-His 항체를 사용한 웨스턴 블럿팅은 9종의 hRBP4 TAG 돌연변이 구축물 중 6종에 대해서 24 kDa의 단백질을 밝혀냈다. 상기 단백질의 크기는 야생형 hRBP4의 크기에 해당되며, 이는 전사 진행 번역을 통한 전장 단백질의 생성을 나타낸다. 전사 진행 활성은 hRBP4 돌연변이 구축물마다 달랐고 (표 3), CYC 및 pylS에 의존적이었다. 특히, 전장 hRBP4 단백질은 돌연변이체 #1, 5 및 7에서는 검출되지 못했다. 고수율의 전장 단백질은 돌연변이체 #2, 6 및 9에서 관찰되었고, 이는 PCL 혼입의 경우에 나타난 결과 (실시예 2, 도 6A)와 유사한 것이었다.DNA constructs for 9 hRBP4 TAG mutants (Table 3) were individually co-transfected into HEK293F cells with pylT/pylS DNA as described in Example 2, and 4 mM N-ε-, a pyrrolysine analogue. Incubated without or in the presence of cyclopentyloxycarbonyl-L-lysine (CYC). Culture medium was harvested and analyzed by Western blotting using anti-His and anti-hRBP4 antibodies (Fig. 11A). Western blotting with anti-His antibody revealed a protein of 24 kDa for 6 out of 9 hRBP4 TAG mutant constructs. The size of the protein corresponds to the size of wild-type hRBP4, indicating the production of a full-length protein through transcriptional progression translation. Transcription progression activity was different for each hRBP4 mutant construct (Table 3) and was CYC and pylS dependent. In particular, the full-length hRBP4 protein was not detected in
CYC를 함유하는 발현된 단백질을 Ni-NTA 크로마토그래피를 이용하여 배지로부터 정제하고 SDS-PAGE 및 이후 쿠마시 블루(Coomassie blue) 염색으로 분석하였다. 도 11B는 정제된 hRBP4 TAG 돌연변이체 #2의 SDS-PAGE 분석을 보여준다. 정제된 제제는 24 kDa 크기의 단일 단백질 밴드를 보여주었다. 정제된 단백질의 질량 스펙트럼은 CYC의 단일 부위 혼입과 일치하였다 (도 11C). Phe62를 대신한 단일 부위 CYC 혼입의 예상 분자량은 (23114.6 Da (야생형) - 165.2 Da (Phe) + 258.3 Da (CYC)) 23208.7 Da이었고, 관찰치는 23182.0 Da이었다. 관찰된 질량은 N-말단 Q 잔기가 피롤리돈 카르복실산으로 고리화되어 18 Da이 빠지고 3개의 무손상 디술피드 결합의 존재로 인해 6 Da이 빠진다는 것을 시사한다 (23208.7 Da - 6 Da - 18 Da = 23184.7 Da). 정제된 단백질의 병렬식 MS 분석은 CYC가 hRBP4 구축물 내 지정된 TAG 부위에 혼입되었음을 입증한다 (도 12). 일시적인 형질감염으로부터의 단백질 수율은 대략 5 mg/L인 것으로 추정되었다. 의 MS/MS 단편화는 다음과 같다:The expressed protein containing CYC was purified from the medium using Ni-NTA chromatography and analyzed by SDS-PAGE followed by Coomassie blue staining. 11B shows SDS-PAGE analysis of purified hRBP4
중성 펩티드 Mr의 모노아이소토픽 질량 (계산치): 3248.5646 Monoisotopic mass of neutral peptide Mr (calculated value): 3248.5646
가변적 변형:Variable transformation:
F7: F에서의 Cyc (F)F7: Cyc at F (F)
Q9 : 탈아미드화 (NQ)Q9: Deamidation (NQ)
N11 : 탈아미드화 (NQ)N11: deamidation (NQ)
M24 : 산화 (M) (중성 손실 0.0000) (표에 나타냄), 63.9983M24: oxidation (M) (neutral loss 0.0000) (shown in table), 63.9983
이온 스코어: 113 예상치: 4.6e-009Ion Score: 113 Expected: 4.6e-009
매치 (굵은 글씨): 110/498 단편 이온 (156개의 가장 강력한 피크 사용)Match (bold): 110/498 fragment ions (using 156 strongest peaks)
추정적 Putative PCLPCL 및 피롤리신 전구체를 사용한 혼입 시험 And incorporation test using a pyrrolysine precursor
전장 hRBP4 Phe122PCL 단백질 (돌연변이체 #6)의 생성을 추정적 PCL 전구체인 D-오르니틴, D-프롤린, D-아르기닌, D-글루탐산, 4-히드록실-D-프롤린 및 2-피롤리돈-5-카르복실산의 존재하에 결정하였다 (도 13A 및 B, 도 14A). pRSRBP를 pCMVpyS, pCMVpyB, pCMVpyC 및 pCMVpyD와 함께 HEK293F 세포로 동시 형질감염시키고, 상기 세포를 실시예 2에 기재된 바와 같이 5 mM의 상기 추정적 전구체의 존재하에 성장시켰다 (도 14A). 이어서, His 태그가 부착되고 PCL이 혼입된 전장 hRBP4를 웨스턴 블럿팅 및 항-His 항체 및 SDS-PAGE로 분석하였다. 도 13B에 나타난 바와 같이, 오직 D-오르니틴만이 Ni-NTA 정제된 샘플의 SDS-PAGE에서 측정가능한 단백질 밴드를 생성하였다. 정제되지 않은 샘플의 웨스턴 블럿팅에 의한 검출 (도 13A)은 모든 다른 전구체의 존재하에서는 전장 hRBP4 단백질이 낮은 수준으로만 형성되었음을 나타냈고, 이는 D-오르니틴이 PCL의 생합성 및 혼입에 가장 효율적인 전구체임을 명확하게 나타낸다. 웨스턴 블럿팅 및 SDS-PAGE 분석으로부터, 5 mM D-오르니틴으로부터의 PCL의 생합성 생성 (레인 2) 및 이후의 단백질 혼입은, pCMVpyS로 형질감염된 세포의 배지에 5 mM로 첨가된 공지의 PylS 기질 CYC의 혼입 (레인 9)보다 더 효율적이라 여겨진다. 본 실시예는 D-오르니틴이 PCL의 생합성에 바람직한 전구체라는 것을 입증한다.The production of the full-length hRBP4 Phe122PCL protein (mutant #6) was determined by the putative PCL precursors D-ornithine, D-proline, D-arginine, D-glutamic acid, 4-hydroxyl-D-proline and 2-pyrrolidone-. It was determined in the presence of 5-carboxylic acid (Figs. 13A and B, Fig. 14A). pRSRBP was co-transfected with HEK293F cells with pCMVpyS, pCMVpyB, pCMVpyC and pCMVpyD, and the cells were grown in the presence of 5 mM of the putative precursor as described in Example 2 (Fig. 14A). Subsequently, full-length hRBP4 with His tag attached and PCL incorporated was analyzed by Western blotting and anti-His antibody and SDS-PAGE. As shown in Fig. 13B, only D-ornithine produced a measurable protein band on the SDS-PAGE of the Ni-NTA purified sample. Detection by Western blotting of the unpurified sample (FIG. 13A) showed that the full-length hRBP4 protein was formed only at low levels in the presence of all other precursors, indicating that D-ornithine is the most efficient precursor for biosynthesis and incorporation of PCL. Is clearly indicated. From Western blotting and SDS-PAGE analysis, biosynthetic production of PCL from 5 mM D-ornithine (lane 2) and subsequent protein incorporation was a known PylS substrate added at 5 mM to the medium of cells transfected with pCMVpyS. It is believed to be more efficient than the incorporation of CYC (lane 9). This example demonstrates that D-ornithine is a preferred precursor for the biosynthesis of PCL.
합성 피롤리신 유사체를 사용한 혼입 시험Incorporation test using synthetic pyrrolysine analogs
상기 실험과 대등하게, 전장 hRBP4 Phe122PCL 단백질 (돌연변이체 #6)의 생성을 또한 단백질 혼입 후의 화학적 유도체화를 위해 아세틸 잔기를 갖도록 디자인된 일련의 합성 피롤리신 유사체의 존재하에 결정하였다 (도 14B). 합성 유사체는 실시예 19에 기재된 바와 같이 제조하였다. pRSRBP를 1개 유전자 카피의 pylT tRNA 및 pylS tRNA 신테타제를 제공하는 pCMVpyS와 함께 HEK293F 세포에 동시 형질감염시켰다. 용해도에 따라, 합성 유사체를 2 또는 5 mM의 최종 농도로 배지에 첨가하였다. 이어서, His 태그가 부착되고 PCL이 혼입된 전장 hRBP4의 검출을 웨스턴 블럿팅 및 항-His 항체 및 SDS-PAGE로 분석하였다. 도 13B에 나타난 바와 같이, 오직 CYC (레인 9)만이 Ni-NTA 정제된 샘플의 분석에서 측정가능한 단백질 밴드를 생성하였다. 정제되지 않은 샘플의 웨스턴 블럿팅 분석 (도 13A)은 TU3000-016 (레인 15) 역시 pylS tRNA 신테타제의 이용가능한 기질임을 시사한다. 그러나, 혼입 효율이 낮고, 핵 자기 공명 분광분석법으로 측정시에 상이한 배치 (TU2982-150)가 분해되어 hRBP4로 훨씬 덜 혼입된 것으로 볼 때 (레인 10) 상기 화합물은 불안정하다. 이전의 공개된 보고와 일치하게, 본 실시예는 고리 잔기 부착 지점의 sp2 탄소를 특징으로 하는 피롤리신 유사체는 pylS tRNA 신테타제의 기질로서 허용되지 않는다는 것을 입증한다.Contrary to the above experiment, the production of full-length hRBP4 Phe122PCL protein (mutant #6) was also determined in the presence of a series of synthetic pyrrolysine analogs designed to have acetyl residues for chemical derivatization after protein incorporation (Fig. 14B). . Synthetic analogs were prepared as described in Example 19. pRSRBP was co-transfected into HEK293F cells with pCMVpyS providing one gene copy of pylT tRNA and pylS tRNA synthetase. Depending on solubility, synthetic analogues were added to the medium at a final concentration of 2 or 5 mM. Subsequently, detection of full-length hRBP4 with His tag attached and PCL incorporated was analyzed by Western blotting and anti-His antibody and SDS-PAGE. 13B, only CYC (lane 9) produced a measurable protein band in the analysis of the Ni-NTA purified sample. Western blotting analysis of the crude sample (FIG. 13A) suggests that TU3000-016 (lane 15) is also a usable substrate for pylS tRNA synthetase. However, the incorporation efficiency is low, and the compound is unstable when it is determined by nuclear magnetic resonance spectroscopy that the different batches (TU2982-150) are decomposed and much less incorporated into hRBP4 (lane 10). Consistent with previous published reports, this example demonstrates that pyrrolysine analogs characterized by the sp2 carbon at the point of attachment of the ring moiety are not acceptable as substrates for pylS tRNA synthetase.
생합성 유전자의 다양한 조합을 사용한 혼입 시험Incorporation test using various combinations of biosynthetic genes
전장 hRBP4 Phe122PCL의 생성을 또한 5 mM D-오르니틴을 함유하는 세포 배양물 및 생합성 유전자 pylB, pylC 및 pylD의 여러 조합을 이용한 동시 형질감염으로 시험하였다 (도 13C). 모든 배양물을 1개 유전자 카피의 pylT tRNA 및 pylS tRNA 신테타제를 제공하는 pCMVpyS와 함께 동시 형질감염시켰다. 전장 hRBP4 단백질은 pylC 및 pylD가 둘다 동시 형질감염된 경우에만 항-His 태그 항체를 사용한 웨스틴 블럿팅으로 검출된다. pylB는 PYL 생합성에 필요하다고 여겨지기는 하지만, PCL 생합성 및 이후의 혼입에 필수적인 것은 아니다. 이러한 관찰결과는 mIgG1 Fc 도메인 단백질 (실시예 5) 및 마우스 및 인간 EPO (실시예 6)의 전장 PCL 돌연변이체의 생성으로 추가로 확인된다. 3가지 예 모두가 오직 유전자 pylC 및 pylD만이 PCL 생합성 및 단백질 혼입에 필수적임을 예시한다.The production of full-length hRBP4 Phe122PCL was also tested by co-transfection using cell cultures containing 5 mM D-ornithine and several combinations of biosynthetic genes pylB, pylC and pylD (Fig. 13C). All cultures were co-transfected with pCMVpyS providing one gene copy of pylT tRNA and pylS tRNA synthetase. Full-length hRBP4 protein is detected by Westin blotting with anti-His tag antibodies only when both pylC and pylD are co-transfected. Although pylB is believed to be necessary for PYL biosynthesis, it is not essential for PCL biosynthesis and subsequent incorporation. These observations are further confirmed by the generation of full-length PCL mutants of mIgG1 Fc domain protein (Example 5) and mouse and human EPO (Example 6). All three examples illustrate that only genes pylC and pylD are essential for PCL biosynthesis and protein incorporation.
실시예 5: 포유동물 세포를 사용한, 생합성으로 생성된 PCL 의 마우스 IgG1 의 Fc 도메인으로의 부위 특이적 혼입 Example 5 : Site-specific incorporation of biosynthetically generated PCL into the Fc domain of mouse IgG1 using mammalian cells
본 실시예는 본원에서 제공되는 방법을 이용하여 포유동물 세포에서 생합성으로 생성된 PCL을 부위 특이적 혼입시키는 것이 일반적인 절차이고, 단백질 hRBP4로 제한되는 것이 아님을 입증한다.This example demonstrates that site-specific incorporation of biosynthetically generated PCL in mammalian cells using the methods provided herein is a general procedure and is not limited to the protein hRBP4.
mIgG1mIgG1 FcFc 의 발현Manifestation of
마우스 IgG1의 Fc 도메인의 K333 (돌연변이체 #1), K336 (돌연변이체 #2), T394 (돌연변이체 #3) 및 L426 (돌연변이체 #4)에서의 TAG 돌연변이체 4종을 생성하였다 (표 2 및 도 17A). pRSFc#1-4 (표 2)를 pCMVpyT, pCMVpyS, pCMVpyC 및 pCMVpyD와 함께 HEK293F 세포에 동시 형질감염시키고, 세포를 5 mM D-오르니틴의 존재하에 성장시켰다 (pCMVpyB는 첨가하지 않음). His 태그가 부착된 Fc 도메인 단백질을 Ni-NTA 크로마토그래피를 이용하여 배지로부터 정제하고 SDS-PAGE로 분석하였다. 각 구축물마다 겔에서의 단백질 밴드의 크기는 전장 단백질에 대한 이들의 예상 크기와 일치하였다 (도 17A). 발현 구축물은 정제를 위한 C-말단 His6 태그를 특징으로 하기 때문에 오직 전장 단백질만이 회수되었다. 도 17A가 나타내는 바와 같이, 돌연변이체 #1의 발현은 성장 배지에 D-오르니틴을 첨가하였는지의 여부에 따라 달라진다. 모든 4종의 돌연변이체의 발현 수준은 유사하였다. Fc 도메인이 HEK293F 세포에서 생성되는 경우에는 이것이 글리코실화되기 때문에 질량 분광측정 분석은 수행하지 않았다.Four types of TAG mutants were generated in the Fc domain of mouse IgG1 K333 (mutant #1), K336 (mutant #2), T394 (mutant #3) and L426 (mutant #4) (Table 2) And Figure 17A). pRSFc#1-4 (Table 2) was co-transfected into HEK293F cells with pCMVpyT, pCMVpyS, pCMVpyC and pCMVpyD, and the cells were grown in the presence of 5 mM D-ornithine (pCMVpyB was not added). The His-tagged Fc domain protein was purified from the medium using Ni-NTA chromatography and analyzed by SDS-PAGE. For each construct, the size of the protein bands in the gel was consistent with their expected size for the full-length protein (Fig. 17A). Only the full length protein was recovered because the expression construct was characterized by a C-terminal His6 tag for purification. As shown in Fig. 17A, the expression of
실시예 6: 포유동물 세포를 사용한, 생합성으로 생성된 PCL 의 에리트로포이에틴 (EPO)으로의 부위 특이적 혼입 Example 6 : Site-specific incorporation of biosynthetically generated PCL into erythropoietin (EPO) using mammalian cells
본 실시예에서는 PCL이 에리트로포이에틴에 부위 특이적으로 혼입되었고, 이것은 본원에서 제공되는 방법이 생합성으로 생성된 PCL을 포유동물 세포 내의 단백질에 부위 특이적으로 혼입시키는 일반적인 절차임을 추가로 입증한다.In this example, PCL was site-specifically incorporated into erythropoietin, which further demonstrates that the method provided herein is a general procedure for site-specific incorporation of biosynthetically generated PCL into proteins in mammalian cells.
마우스 에리트로포이에틴 (Mouse erythropoietin ( EPOEPO )의 발현) Expression
EPO 돌연변이체 단백질로의 PCL 혼입을 HEK293F 세포에서 수행하였다. EPO 수용체 결합 계면에서 떨어진 11개의 표면-노출된 Lys 또는 Arg 잔기에 TAG 돌연변이를 도입하였다. 혼입 부위는 하기 나타냈고 표 2에 기재했다. pylB 유전자를 사용하지 않았다는 점을 제외하고는 실시예 1에 기재된 바와 같이 하여 마우스 EPO 단백질을 발현시켰다: pRSEPO#1-11 (표 2)을 pCMVpyT, pCMVpyS, pCMVpyC 및 pCMVpyD와 함께 HEK293F 세포에 동시 형질감염시키고, 세포를 5 mM D-오르니틴의 존재하에 성장시켰다. His 태그가 부착된 EPO를 Ni-NTA 크로마토그래피를 이용하여 배지로부터 정제하고 SDS-PAGE로 분석하였다. EPO 구축물은 C-말단 His-태그를 함유한다. 따라서, PCL이 성공적으로 혼입된 단백질만이 전장 단백질을 생성할 것이고 Ni-NTA 크로마토그래피로 정제될 것이며, 따라서 SDS-PAGE에서 검출가능한 밴드를 생성할 것이다. 전장 마우스 EPO의 SDS-PAGE는 모든 11종의 돌연변이체에서 PCL이 성공적으로 혼입되었음을 나타낸다 (도 17B). 각 구축물마다 겔에서의 단백질 밴드의 크기는 전장 단백질에 대한 이들의 예상 크기와 일치하였다. 돌연변이체 #1의 경우, 전장 단백질의 형성이 D-오르니틴의 존재 여부에 따라 달라진다는 것이 예시된다. 모든 11종의 돌연변이체 단백질의 발현 수준은 유사하였고, 약 40 mg/L로 발현된 야생형 단백질의 10% 내지 20%였다 (도 17B). EPO는 HEK293F 세포에서 글리코실화되기 때문에 질량 분광측정 분석은 수행하지 않았다. 성숙 인간 (서열 19) 및 마우스 (서열 20) EPO 단백질의 서열을 하기 나타냈고, PCL 혼입 부위는 굵은 글씨로 표시했으며 글리코실화 부위에는 밑줄을 그었다. 돌연변이체의 번호매김은 N-말단에서 시작된다 (또한, 표 2 참조).PCL incorporation into the EPO mutant protein was performed in HEK293F cells. TAG mutations were introduced into 11 surface-exposed Lys or Arg residues away from the EPO receptor binding interface. The incorporation sites are shown below and are shown in Table 2. Mouse EPO protein was expressed as described in Example 1, except that the pylB gene was not used: pRSEPO#1-11 (Table 2) was co-transfected into HEK293F cells with pCMVpyT, pCMVpyS, pCMVpyC and pCMVpyD. Infected and cells were grown in the presence of 5 mM D-ornithine. EPO with His tag was purified from the medium using Ni-NTA chromatography and analyzed by SDS-PAGE. The EPO construct contains a C-terminal His-tag. Thus, only proteins in which PCL has been successfully incorporated will produce full-length proteins and will be purified by Ni-NTA chromatography, thus producing a detectable band on SDS-PAGE. SDS-PAGE of full-length mouse EPO showed successful incorporation of PCL in all 11 mutants (Fig. 17B). For each construct, the size of the protein bands in the gel was consistent with their expected size for the full-length protein. In the case of
실시예 7: 에스케리키아 콜라이 세포를 사용하는 경우, 생합성으로 생성된 PCL을 단백질에 혼입시키기 위한 플라스미드 Example 7 : When using Escherichia coli cells, plasmid for incorporating biosynthetically generated PCL into protein
본 실시예는, 에스케리키아 콜라이 세포로 형질전환된 경우에 동일한 에스케리키아 콜라이 세포로 동시 형질전환된 제2 플라스미드로부터 발현되는 표적 단백질 내 TAG 코딩 부위로 PCL의 혼입이 가능하게 되는 플라스미드에 관한 기재를 제공한다.This Example relates to a plasmid capable of incorporating PCL into a TAG coding site in a target protein expressed from a second plasmid co-transformed with the same Escherichia coli cell when transformed with Escherichia coli cells. Provide a description.
pArapAra -- pylSTBCDpylSTBCD 구축 build
이. 콜라이 세포에서 PCL을 혼입시키기 위한 플라스미드를 도 5에 나타냈고, 다음과 같이 구축하였다: proK 프로모터하에 pylT를 코딩하는 카세트를 프로모터, tRNA, 및 pSUP의 3' 서열의 증폭 (중첩 프라이머 사용)으로 합성하였다. 이어서, 상기 3종의 조각을 ApaLI 및 XhoI에 대한 제한 효소 부위를 코딩하는 말단 프라이머와 혼합하여 전체 삽입물을 합성하였다. ApaLI 및 XhoI로 소화시킨 후, 상기 카세트를 동일 효소로 제조된 pSUP 주쇄에 라이게이션하여 pSUP-pylT를 생성하였다. 엠. 마제이로부터의 pylS, pylB, pylC 및 pylD에 대한 코딩 서열을 미리 제조된 적절한 pCMVU6 플라스미드 (실시예 1)로부터 증폭시키고, 태그 없이 pMH4에 삽입하였다 (문헌 [Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, and Stevens RC, "Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline", Proc Natl Acad Sci USA 2002; 99: 11664-11669] 참조). 이어서, 각각의 전체 프로모터-CDS-종결자를 여러가지 제한 효소 부위를 갖는 프라이머로 증폭시켰다 pylS PCR 생성물을 KpnI 및 SbfI로 소화시키고 KpnI 및 PstI 부위 사이에서 pSUP-pylT로 라이게이션하여 pAra-pylST를 생성하였다. pylB, pylC 및 pylD 생성물을 각각의 효소로 절단하고, NdeI 및 KpnI로 제조된 플라스미드 주쇄로 라이게이션하였다. 이러한 4가지 라이게이션의 플라스미드 생성물을 서열분석 및 진단 PCR로 확인한 후, 전체 카세트를 프라이머로 증폭시켜서 pylD-pylB-pylC 카세트의 양 말단에 KpnI 부위를 부가하였다. 이것을 KpnI로 소화시키고 KpnI 부위에서 pAra-pylST에 라이게이션하여 최종 pAra-pylSTBCD를 생성하였다 (도 5). 상기 최종 플라스미드는 pylD, pylB, pylC 및 pylS를 함유하고, 이것 각각은 별개의 아라비노스-유도성 및 T7 하이브리드 프로모터의 제어를 받고 각각마다 rrnB 종결자 하류 (pMH4 내용물과 동일함) 및 proK 프로모터하의 pylT 유전자 (단일 tRNA 카피를 갖는 pSUP/pSUPAR 내용물과 유사함)를 갖는다 ([Cellitti et al., "In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy", J Am Chem Soc. 2008 Jul 23; 130(29):9268-81]).this. A plasmid for incorporating PCL in E. coli cells is shown in Fig. 5, and was constructed as follows: a cassette encoding pylT under the proK promoter was synthesized by amplification of the 3'sequence of the promoter, tRNA, and pSUP (using overlapping primers). I did. Subsequently, the three fragments were mixed with terminal primers encoding restriction enzyme sites for ApaLI and XhoI to synthesize the entire insert. After digestion with ApaLI and XhoI, the cassette was ligated to the pSUP backbone prepared with the same enzyme to produce pSUP-pylT. M. Coding sequences for pylS, pylB, pylC and pylD from MaJ were amplified from the appropriate pCMVU6 plasmid prepared in advance (Example 1) and inserted into pMH4 without tags (Lesley SA, Kuhn P, Godzik A, Deacon AM , Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, and Stevens RC, "Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline", Proc Natl Acad Sci USA 2002; 99: 11664-11669). Then, each entire promoter-CDS-terminator was amplified with primers having various restriction enzyme sites. The pylS PCR product was digested with KpnI and SbfI and ligated with pSUP-pylT between the KpnI and PstI sites to generate pAra-pylST. The pylB, pylC, and pylD products were digested with respective enzymes and ligated into the plasmid backbone prepared with NdeI and KpnI. After confirming the plasmid products of these four ligations by sequencing and diagnostic PCR, the entire cassette was amplified with primers to add KpnI sites to both ends of the pylD-pylB-pylC cassette. This was digested with KpnI and ligated to pAra-pylST at the KpnI site to produce the final pAra-pylSTBCD (Fig. 5). The final plasmid contains pylD, pylB, pylC and pylS, each of which is under the control of a separate arabinose-inducible and T7 hybrid promoter, each downstream of the rrnB terminator (same as the pMH4 contents) and under the proK promoter. It has a pylT gene (similar to the pSUP/pSUPAR content with a single tRNA copy) ([Cellitti et al., "In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance). spectroscopy", J Am Chem Soc. 2008
실시예 8: 에스케리키아 콜라이 세포를 이용한, 생합성적으로 생성된 PCL 의 FAS-TE에의 부위 특이적 혼입. Example 8 : Site-specific incorporation of biosynthetically generated PCL into FAS-TE using Escherichia coli cells.
본 실시예는 동일한 에스케리키아 콜라이 세포를 공동 형질전환시킨 제2 플라스미드로부터 발현된 인간 지방산 신테타제 (FAS-TE)의 TAG 코딩된 부위에의 PCL의 혼입에 대한 설명을 제공한다.This example provides a description of the incorporation of PCL into the TAG-encoded site of human fatty acid synthetase (FAS-TE) expressed from a second plasmid co-transformed with the same Escherichia coli cells.
인간 지방산 신테타제 (FAS-TE)의 티오에스테라제 도메인 코딩 잔기 2221 내지 2502를 N-말단 태그 (MGDSKIHHHHHHENLYFQG) (서열 21)를 갖는 pMH4 벡터로부터 발현시켰다 (문헌 [Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, and Stevens RC, "Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline", Proc Natl Acad Sci USA 2002; 99: 11664-11669] 참조). TAG 코돈을 PIPE 클로닝을 이용하여 돌연변이화하였고 (문헌 [Klock, HE, Koesema EJ, Knuth MW, and Lesley, SA, "Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts", Proteins. 2008 May 1;71(2):982-94)] 참조), 이는 문헌 [Cellitti et al., "In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy", J Am Chem Soc. 2008 Jul 23;130(29):9268-81]에 보다 상세하게 기재되어 있다. PCL 혼입에 대해 시험한 돌연변이체는 Leu2222TAG/Leu2223Ile 및 Y2454TAG (도 18A)였다. HK100 세포를 pMH4-FAS-TE 플라스미드 및 pAra-pylSTBCD로 공동 형질전환시키고, LB+Kan+Cm 플레이트 상에서 선별하였다. 액체 배양물을 5 mM D-오르니틴 (시그마(Sigma) 또는 노바 바이오켐(Nova Biochem))이 보충된 TB (시그마)+Kan+Cm에서 37℃에서 성장시킨 후 유도하였다. OD595 = 0.8일 때, 세포를 30℃로 옮기고, 15-30 분 뒤에 0.2% 아라비노스로 유도하였다. 유도한 후 원심분리에 의해 수거하기 전에 세포를 대략 20 시간 동안 성장시켰다. 세포를 TBS+5% 글리세롤 (pH 8) 중에서 초음파처리에 의해 용해시켰다. 가용성 단백질 분획을 Ni-NTA (퀴아겐(Qiagen)) 크로마토그래피 상에서 제조자의 프로토콜에 따라 정제하였다. 수율은 FAS-TE Leu2222PCL/Leu2223Ile의 경우에 46-80 mg/L, FAS-TE Tyr2454PCL의 경우에는 155-186 mg/L로, 야생형 단백질의 수율의 50 내지 80%였다. SDS-PAGE에서의 분자 크기 및 질량 분광측정법에 의해 측정된 단백질의 분자량은 목적하는 위치에 PCL이 단독 부위 혼입된 것과 일치하였다 (도 18B).The thioesterase domain coding residues 2221 to 2502 of human fatty acid synthetase (FAS-TE) were expressed from a pMH4 vector with an N-terminal tag (MGDSKIHHHHHHENLYFQG) (SEQ ID NO: 21) (Lesley SA, Kuhn P, Godzik A , Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, and Stevens RC, "Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline", Proc Natl Acad Sci USA 2002; 99:11664-11669). The TAG codon was mutated using PIPE cloning (Klock, HE, Koesema EJ, Knuth MW, and Lesley, SA, "Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts", Proteins. 2008 May 1;71(2):982-94)), which is described in [Cellitti et al., "In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear. magnetic resonance spectroscopy", J Am Chem Soc. 2008
2개의 PCL 혼입 부위 (볼드체 및 밑줄)를 갖는 FAS-TE (서열 22)의 서열을 이하에 제시한다 (Leu2222PCL의 경우, 잔기 Leu2223이 Ile2223로 돌연변이됨): The sequence of FAS-TE (SEQ ID NO: 22) with two PCL incorporation sites (bold and underlined) is shown below (for Leu2222PCL, residue Leu2223 is mutated to Ile2223):
실시예 9: 에스케리키아 콜라이 세포를 이용한, 생합성적으로 생성된 PCL 의 FKBP12에의 부위 특이적 혼입. Example 9 : Site-specific incorporation of biosynthetically generated PCL into FKBP12 using Escherichia coli cells.
본 실시예는 동일한 에스케리키아 콜라이 세포를 공동 형질전환시킨 제2 플라스미드로부터 발현된 FKBP12의 TAG 코딩된 부위에의 PCL의 혼입을 기재한다.This example describes the incorporation of PCL into the TAG-encoded site of FKBP12 expressed from a second plasmid co-transformed with the same Escherichia coli cells.
FKBP12를 N-말단 태그 (MGSSHHHHHHLEVLFQGP) (서열 23)를 갖는 pET 벡터 (노바겐(Novagen))로부터 발현시켰다. TAG 코돈을 PIPE 클로닝을 이용하여 Ile90 위치에 도입하였다 (문헌 [Klock, HE, Koesema EJ, Knuth MW, Lesley, SA, "Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts", Proteins. 2008 May 1;71(2):982-94] 참조). BL21(DE3) 세포 (인비트로겐(Invitrogen))를 pET-FKBP12 및 pAra-pylSTBCD로 공동 형질전환시키고, LB+Kan+Cm 플레이트 상에서 선별하였다. 액체 배양물을 5 mM D-오르니틴이 보충된 TB+Kan+Cm에서 37℃에서 성장시켰다. OD595 = 0.4일 때, 세포를 30℃로 옮기고, 30 분 뒤에 1 mM IPTG로 유도하였다. 세포를 수거하기 전에 20 시간 더 성장시켰다. 세포를 용해시키고, FAS-TE에 대해 정제하였다. 이어서, 정제된 단백질을 TBS로 투석하고, HRV-3C 프로테아제 (1 mg FKBP12 당 2U)로 48 시간 동안 4℃에서 절단하여 His 태그를 제거하였다. 절단된 물질을 Ni-NTA 칼럼으로 흘려보내고, 수집하고, 농축시키고, 추가의 정제를 위해 S75 크기 배제 칼럼 (GE 헬스케어(GE Healthcare))을 통과시켰다. 최종 단백질을 추가로 15-20 mg/ml로 농축시킨 후 결정화하였다. FKBP Ile90PCL의 수율은 120 mg/L (조물질)이었다. 결정화된 FKBP Ile90PCL을 PCL-함유 단백질의 X선 구조를 수득하는데 사용하였다. 도 19A-C는 FKBP12에 생합성적으로 혼입된 PCL에 대한 SDS-PAGE 및 질량 분광측정 데이터 및 결정화 결과를 보여준다. 수득된 질량은 12085.6 Da이었고, 이는 PCL의 단일 부위 혼입에 대해 예상되는 값인 12084 Da과 일치한다.FKBP12 was expressed from a pET vector (Novagen) with an N-terminal tag (MGSSHHHHHHLEVLFQGP) (SEQ ID NO: 23). The TAG codon was introduced at the Ile90 position using PIPE cloning (Reference [Klock, HE, Koesema EJ, Knuth MW, Lesley, SA, "Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts" , Proteins. 2008 May 1;71(2):982-94). BL21(DE3) cells (Invitrogen) were co-transformed with pET-FKBP12 and pAra-pylSTBCD and selected on LB+Kan+Cm plates. Liquid cultures were grown at 37° C. in TB+Kan+Cm supplemented with 5 mM D-ornithine. When OD 595 = 0.4, cells were transferred to 30° C. and induced 30 minutes later with 1 mM IPTG. Cells were grown for an additional 20 hours before harvesting. Cells were lysed and purified for FAS-TE. The purified protein was then dialyzed against TBS and digested with HRV-3C protease (2U per 1 mg FKBP12) at 4° C. for 48 hours to remove the His tag. The cut material was run on a Ni-NTA column, collected, concentrated, and passed through an S75 size exclusion column (GE Healthcare) for further purification. The final protein was further concentrated to 15-20 mg/ml and crystallized. The yield of FKBP Ile90PCL was 120 mg/L (crude). Crystallized FKBP Ile90PCL was used to obtain the X-ray structure of the PCL-containing protein. 19A-C show SDS-PAGE and mass spectrometry data and crystallization results for PCL biosynthetically incorporated in FKBP12. The mass obtained was 12085.6 Da, which is consistent with 12084 Da, the expected value for single site incorporation of PCL.
PCL 혼입 부위 (볼드체 및 밑줄)를 갖는 FKBP12의 서열 (서열 24)을 이하에 제시한다:The sequence of FKBP12 (SEQ ID NO: 24) with the PCL incorporation site (bold and underlined) is shown below:
실시예 10: 에스케리키아 콜라이 세포를 이용한, 생합성적으로 생성된 PCL 의 섬유모세포 성장 인자 21(FGF21)에의 부위 특이적 혼입. Example 10 : Site-specific incorporation of biosynthetically produced PCL into fibroblast growth factor 21 (FGF21) using Escherichia coli cells.
본 실시예에서는 동일한 에스케리키아 콜라이 세포를 공동 형질전환시킨 제2 플라스미드로부터 발현된 인간 섬유모세포 성장 인자, FGF21의 20개의 별개의 TAG 코딩된 부위에의 PCL의 혼입을 제공한다.In this example, the incorporation of PCL into 20 distinct TAG-encoded sites of FGF21, a human fibroblast growth factor, expressed from a second plasmid co-transformed with the same Escherichia coli cells is provided.
섬유모세포 성장 인자 21, FGF21을 N-말단 태그 (MGDSKIHHHHHHENLYFQG) (서열 21) 및 번역된 인간 단백질의 코딩 잔기 33-209를 갖는 pSpeedET 벡터로부터 발현시켰다 (문헌 [Klock, HE, Koesema EJ, Knuth MW, Lesley, SA, "Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts", Proteins. 2008 May 1;71(2):982-94] 참조). PYL 유사체 혼입 및 후속 PEG 부착을 위한 TAG 코돈을 FGF21 (서열 25) aa33-209 구축물의 다음과 같은 20개의 개별 위치에 도입하였다: Ser35, Gln39, Arg47, Gln56, Arg64, Asp74, Lys84, Lys87, Lys97, Arg100, Arg105, His115, Arg124, Glu129, Lys150, Arg154, Leu167, Leu170, Leu181 및 Gln184. FGF21 (33-209 aa) 구축물의 서열을 20개의 별개 구축물에서의 혼입 부위 (볼드체 및 밑줄로 강조함)와 함께 이하에 나타낸다.
HK100 또는 BL21(DE3) 세포를 pSpeedet-FGF21 플라스미드 및 pAra-pylSTBCD로 공동 형질전환시키고, LB+Kan+Cm 상에서 선별하였다. 액체 배양물을 실시예 8에서와 같이 성장시키고, 유도 이전에 5 mM D-오르니틴을 첨가하였다. 30℃로 전환시키기 위한 OD595가 0.2 내지 1.0으로 시험되었고, 15-30 분 뒤에 OD595 = 0.4-2.0에서 0.2% 아라비노스 또는 1 mM IPTG로 후속 유도하였다. 유도 후 세포를 대략 20 시간 동안 성장시킨 후, 수거하였다. 세포를 TBS 중에서 5% 글리세롤, 1% 트리톤 X-114, 또는 2.5% 데옥시콜레이트로 용해시켰다. 이어서, 불용성 펠렛을 TBS+6M 구아니딘-HCl (pH 8)에 재현탁시켰다. 단백질을 Ni-NTA 수지 상에서 정제하고, 칼럼 상에서 또는 칼럼으로부터 용리된 후에 재폴딩시켰다. 후속하여 태그를 TEV 프로테아제로 제거하고, 생성물을 Ni-NTA, 이온 교환, 및 크기 배제 크로마토그래피에 의해 정제하였다.HK100 or BL21(DE3) cells were co-transformed with pSpeedet-FGF21 plasmid and pAra-pylSTBCD and selected on LB+Kan+Cm. Liquid cultures were grown as in Example 8 and 5 mM D-ornithine was added prior to induction. The OD 595 for conversion to 30° C. was tested from 0.2 to 1.0, followed by induction with 0.2% arabinose or 1 mM IPTG at OD595 = 0.4-2.0 after 15-30 minutes. After induction, the cells were grown for approximately 20 hours and then harvested. Cells were lysed with 5% glycerol, 1% Triton X-114, or 2.5% deoxycholate in TBS. The insoluble pellet was then resuspended in TBS+6M guanidine-HCl (pH 8). Proteins were purified on Ni-NTA resin and refolded on or after eluting from the column. The tag was subsequently removed with TEV protease and the product was purified by Ni-NTA, ion exchange, and size exclusion chromatography.
PCL의 FGF21에의 혼입을 보여주는 대표적인 데이터를 도 20에 나타낸다. 예비적 발현 수율을 표 4에 기록하였다. 구축물이 N-말단 His-태그를 포함하는 것을 특징으로 하므로 전장 및 말단절단 FGF21 단백질을 둘 모두 정제하였다. TAG 코돈이 종결 코돈으로서 이용되는 (말단절단 단백질을 생성함) 범위 및 PCL을 혼입하는데 이용되는 (전장 단백질을 생성함) 범위는 상이한 혼입 부위에 따라 의존적이었다 (도 20). 목적하는 전장 단백질의 예상 수율은 4 내지 56 mg/L이지만, 돌연변이체 중 3 종 (표 4)에서는 어떤 FGF21 단백질도 검출할 수 없었고, 나머지 17 종의 돌연변이체에서 수득된 단백질의 총 수율은 5.7 내지 143 mg/L로 다양하였다. 모든 돌연변이체에 대해 PCL 혼입을 질량 분광측정법으로 확인하였다.Representative data showing the incorporation of PCL into FGF21 is shown in FIG. 20. Preliminary expression yields are reported in Table 4. Both full-length and truncated FGF21 proteins were purified as the construct was characterized as containing an N-terminal His-tag. The range in which the TAG codon is used as a stop codon (which produces a truncation protein) and the range used to incorporate PCL (which produces a full-length protein) was dependent on the different incorporation sites (FIG. 20 ). The expected yield of the desired full-length protein was 4 to 56 mg/L, but no FGF21 protein could be detected in three of the mutants (Table 4), and the total yield of the protein obtained from the remaining 17 mutants was 5.7. To 143 mg/L. PCL incorporation was confirmed by mass spectrometry for all mutants.
실시예 11: mTNF -α 에의 PCL 혼입. Example 11: mTNF -α by PCL mixing .
mTNF-α Gln21PCL 돌연변이체를 발현시키기 위해, 이. 콜라이 BL21(DE3) 세포를 pAra-pylSTBCD 및 pET22b 플라스미드 벡터 상의 개별 돌연변이체 mTNF-α 유전자로 공동 형질전환시켰다. 형질전환된 세포를 5 mM D-오르니틴의 존재하에 TB 배지에서 37℃에서 성장시키고, OD600이 0.5에 이르면 1 mM IPTG 및 0.2% (w/v) 아라비노스로 유도하였다. 이어서, 세포를 30℃에서 12-16 시간 동안 계속 진탕시키고 수거하였다. 세포 펠렛을 사용시까지 -20℃에서 저장하였다. mTNF-α의 X선 결정 구조는 N-말단 His6 태그에 이어 인자 Xa 절단 부위 (GIEGR)를 함유하는 재조합 mTNF-α의 단백질 서열 (서열 26)에서 하기 나타낸 바와 같은 PCL 혼입 부위 Lys11 및 Gln21을 나타내었다:To express the mTNF-α Gln21PCL mutant, E. E. coli BL21(DE3) cells were co-transformed with the pAra-pylSTBCD and individual mutant mTNF-α genes on the pET22b plasmid vector. Transformed cells were grown at 37° C. in TB medium in the presence of 5 mM D-ornithine, and when OD 600 reached 0.5, they were induced with 1 mM IPTG and 0.2% (w/v) arabinose. Subsequently, the cells were continuously shaken at 30° C. for 12-16 hours and harvested. Cell pellets were stored at -20°C until use. The X-ray crystal structure of mTNF-α is the N-terminal His 6 tag followed by the PCL incorporation sites Lys11 and Gln21 as shown below in the protein sequence (SEQ ID NO: 26) of the recombinant mTNF-α containing the factor Xa cleavage site (GIEGR). Indicated:
실시예 12: mEGF 에의 PCL 혼입. Example 12 : Incorporation of PCL into mEGF.
mEGF-Tyr10PCL 돌연변이체를 발현시키기 위해, 이. 콜라이 BL21(DE3) 세포를 pAra-pylSTBCD 및 pET22b 플라스미드 벡터 상의 개별 돌연변이체 mEGF 유전자로 공동 형질전환시켰다. 형질전환된 세포를 5 mM D-오르니틴의 존재하에 TB 배지에서 37℃에서 성장시키고, OD600이 0.5에 이르면 1 mM IPTG 및 0.2% (w/v) 아라비노스로 유도하였다. 이어서, 세포를 30℃에서 12-16 시간 동안 계속 진탕하고 수거하였다. 세포 펠렛을 사용시까지 -20℃에서 저장하였다. mEGF의 X선 결정 구조는 C-말단 His6 태그를 갖는 재조합 mEGF의 단백질 서열 (서열 27)에서 하기 나타낸 바와 같은 PCL 혼입 부위 Tyr10 및 Tyr29를 나타내었다:To express the mEGF-Tyr10PCL mutant, E. E. coli BL21(DE3) cells were co-transformed with the pAra-pylSTBCD and individual mutant mEGF genes on the pET22b plasmid vector. Transformed cells were grown at 37° C. in TB medium in the presence of 5 mM D-ornithine, and when OD 600 reached 0.5, they were induced with 1 mM IPTG and 0.2% (w/v) arabinose. Subsequently, the cells were continuously shaken at 30° C. for 12-16 hours and harvested. Cell pellets were stored at -20°C until use. The X-ray crystal structure of mEGF showed PCL incorporation sites Tyr10 and Tyr29 as shown below in the protein sequence (SEQ ID NO: 27) of the recombinant mEGF with a C-terminal His 6 tag:
실시예 13: mTNF -α 에의 피롤리신 ( Pyl ) 또는 PCL 의 혼입. Example 13: mTNF -α to the new pyrrolidine (Pyl), or incorporation of the PCL.
mTNF-α에 PYL을 삽입하기 위해, 이. 콜라이 BL21(DE3) 세포를 Gln21에 대한 코돈 (CAA)이 종결 코돈 (TAG)으로 돌연변이된 pET22b 플라스미드 벡터 상의 돌연변이체 mTNF-α 유전자 뿐만 아니라, M. 마제이(M. mazei) pylS, pylT, pylB, pylC, 및 pylD를 함유하는 pAra-pylSTBCD로 공동 형질전환시켰다. 돌연변이체 mTNF-α에 PCL 만을 혼입시키기 위해, pAra-pylSTBCD를 추정 메틸 트랜스퍼라제 PylB에 대한 유전자가 결여된 pAra-pylSTCD로 대체하였다. 형질전환된 세포를 5 mM D-오르니틴의 존재하에 TB 배지에서 37℃에서 성장시키고, OD600이 0.5에 이르면 1 mM IPTG 및 0.2% (w/v) 아라비노스로 유도하였다. 이어서, 세포를 30℃에서 12-16 시간 동안 계속 진탕하고 수거하였다. 세포 펠렛을 -20℃에서 저장하였다. 빙상에서 세포 펠렛을 15 분 동안 해동시킨 후, 세포를 용해 완충액 (20 mM 트리스/HCl, 50 mM NaCl, pH 8.0)에 습중량 그램 당 3 ml로 재현탁시켰다. 리소자임을 1 mg/ml로 첨가하고, 빙상에서 세포를 2 분 동안 초음파처리하였다. 용해물을 30,000 x g에서 20 분 동안 4℃에서 원심분리하여, 세포 파편을 펠렛화하였다. 50% Ni-NTA 슬러리 (퀴아겐) 1 ml를 맑아진 용해물에 첨가하고, 4℃에서 60 분 동안 진탕하여 부드럽게 혼합하였다. 용해물-Ni-NTA 혼합물을 칼럼에 로딩하고, 통과물을 수집하였다. 수지를 PBS (pH 8.0) 중의 25 mM 이미다졸 20 mL로 세척한 후, 단백질을 PBS (pH 8.0) 중의 250 mM 이미다졸 2.5 ml로 용리시키고, PD-10 칼럼 (GE 헬스케어)을 이용하는 것에 의해 PBS, pH 7.4로 완충액 교환하였다. pylB의 존재 또는 부재하의, 뿐만 아니라 D-오르니틴의 존재 또는 부재하의 mTNF-α Gln21TAG의 발현을 SDS-폴리아크릴아미드 겔 전기영동 (SDS-PAGE)에 의해 분석하였다 (도 21A). 도 21A에서, 레인 1은 표준 씨블루(SeeBlue) 플러스2 예비염색된 표준물질의 분자량이고; 레인 2는 pylB 및 D-오르니틴 둘 모두의 존재하에서의 발현이고; 레인 3은 D-오르니틴이 첨가되지 않은 pylB의 존재하에서의 발현이고; 레인 4는 5 mM의 D-오르니틴이 첨가된 pylB의 부재하에서의 발현이고; 레인 5는 pylB 및 D-오르니틴 둘 모두의 부재하에서의 발현이다. 데이터는 전장 mTNF-α의 발현이 D-오르니틴의 존재에 의존적이며, pylB 유전자의 부재 또는 존재하에서는 유사함을 보여준다.To insert PYL into mTNF-α, E. E. coli BL21(DE3) cells were converted to the mutant mTNF-α gene on the pET22b plasmid vector in which the codon for Gln21 (CAA) was mutated to the stop codon (TAG), as well as M. mazei pylS, pylT, pylB, It was co-transformed with pAra-pylSTBCD containing pylC, and pylD. To incorporate only PCL into the mutant mTNF-α, pAra-pylSTBCD was replaced with pAra-pylSTCD lacking the gene for the putative methyl transferase PylB. Transformed cells were grown at 37° C. in TB medium in the presence of 5 mM D-ornithine, and when OD 600 reached 0.5, they were induced with 1 mM IPTG and 0.2% (w/v) arabinose. Subsequently, the cells were continuously shaken at 30° C. for 12-16 hours and harvested. The cell pellet was stored at -20°C. After thawing the cell pellet on ice for 15 minutes, the cells were resuspended in lysis buffer (20 mM Tris/HCl, 50 mM NaCl, pH 8.0) at 3 ml per gram wet weight. Lysozyme was added at 1 mg/ml, and the cells were sonicated for 2 minutes on ice. The lysate was centrifuged at 30,000 xg for 20 minutes at 4° C. to pellet the cell debris. 1 ml of a 50% Ni-NTA slurry (Qiagen) was added to the cleared lysate and mixed gently by shaking at 4° C. for 60 minutes. The lysate-Ni-NTA mixture was loaded onto the column and the flow-through was collected. After washing the resin with 20 mL of 25 mM imidazole in PBS (pH 8.0), the protein was eluted with 2.5 ml of 250 mM imidazole in PBS (pH 8.0), and by using a PD-10 column (GE Healthcare). The buffer solution was exchanged with PBS, pH 7.4. Expression of mTNF-α Gln21TAG with or without pylB, as well as with or without D-ornithine was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (FIG. 21A ). In Figure 21A,
실시예 14: mEGF 에의 피롤리신 ( Pyl ) 또는 PCL 의 혼입 Example 14 : Incorporation of pyrrolysine (Pyl) or PCL into mEGF
이. 콜라이 BL21(DE3) 세포를 pAra-pylSTBCD 및 pET22b 플라스미드 벡터 상의 개별 돌연변이체 mEGF Tyr10TAG 및 Tyr29TAG 유전자로 공동 형질전환시켰다. 구축물은 둘 모두 5 mM D-오르니틴의 존재하에 TB 배지에서 37℃에서 발현시켰고, OD600이 0.5에 이르면 1 mM IPTG 및 0.2% (w/v) 아라비노스를 첨가하였다. 이어서, 온도를 30℃로 낮추고, 유도 후 16 시간 째에 세포를 수거하였다. 세포 펠렛을 20 mL의 20 mM 트리스/HCl (pH 8.5)에 재현탁시키고, 5 분 동안 초음파처리하였다. 30,000 x g에서 20 분 동안 원심분리한 후, 상청액은 따라내었다. 펠렛을 초음파처리에 의해 2% (v/v) 트리톤-X100을 함유하는 20 mL의 20 mM 트리스/HCl (pH 8.5)에 재현탁시켰다. 30,000 x g에서 20 분 동안 또 다른 라운드의 원심분리를 행한 후, 펠렛을 초음파처리에 의해 10 mL의 8 M 우레아, 20 mM 트리스/HCl, 10 mM β-메르캅토에탄올, pH 8.5에 용해시켰다. 불용성 세포 파편은 원심분리 (30,000 x g, 20 분)에 의해 제거하고, 상청액을 재폴딩 완충액 (100 mM 트리스/HCl, 4 mM 환원 글루타티온, 0.4 mM 산화 글루타티온, 20% (v/v) 에탄올, pH 8.5)으로 2배 희석시켰다. 이어서, 슬라이드-a-라이저 투석 카세트 (3500 Da 분자량 컷오프, 피어스(Pierce))를 이용하여 희석된 샘플을 재폴딩 완충액에 대해 밤새 4℃에서 투석하였다. 불용성 단백질을 30,000 x g에서 20 분 동안 원심분리에 의해 제거하고, 상청액을 β-메르캅토에탄올로 최종 농도 2 mM 까지 보충하였다. 50% Ni-NTA 슬러리 (퀴아겐) 1 ml를 재폴딩된 단백질에 첨가하고, 4℃에서 60 분 동안 진탕하여 부드럽게 혼합하였다. 단백질-Ni-NTA 혼합물을 칼럼에 로딩하고, 통과물을 수집하였다. 수지를 PBS (pH 8.0) 중의 25 mM 이미다졸 20 mL로 세척한 후, 단백질을 PBS (pH 8.0) 중의 250 mM 이미다졸 2.5 ml로 용리시켰다. 마지막으로, 단백질을 PD-10 칼럼 (GE 헬스케어)을 이용하는 것에 의해 PBS (pH 7.4)로 완충액 교환하였다. 단백질 표본의 순도는 SDS-PAGE로 조사하였다 (도 21B). 도 21B에서, 레인 1은 표준 씨블루 플러스2 예비염색된 표준물질의 분자량이고, 레인 2는 Ni-NTA 정제 이후의 mEGF Tyr10TAG 돌연변이체 단백질이다. this. E. coli BL21(DE3) cells were co-transformed with the individual mutant mEGF Tyr10TAG and Tyr29TAG genes on the pAra-pylSTBCD and pET22b plasmid vectors. Both constructs were expressed at 37° C. in TB medium in the presence of 5 mM D-ornithine, and 1 mM IPTG and 0.2% (w/v) arabinose were added when OD 600 reached 0.5. Then, the temperature was lowered to 30° C., and cells were harvested 16 hours after induction. The cell pellet was resuspended in 20 mL of 20 mM Tris/HCl (pH 8.5) and sonicated for 5 minutes. After centrifugation at 30,000 xg for 20 minutes, the supernatant was decanted. The pellet was resuspended in 20 mL of 20 mM Tris/HCl (pH 8.5) containing 2% (v/v) Triton-X100 by sonication. After another round of centrifugation at 30,000 xg for 20 minutes, the pellet was dissolved in 10 mL of 8 M urea, 20 mM Tris/HCl, 10 mM β-mercaptoethanol, pH 8.5 by sonication. Insoluble cell debris was removed by centrifugation (30,000 xg, 20 min), and the supernatant was refolded in buffer (100 mM Tris/HCl, 4 mM reduced glutathione, 0.4 mM glutathione oxide, 20% (v/v) ethanol, pH. 8.5) was diluted 2 times. The diluted samples were then dialyzed against refolding buffer at 4° C. overnight using a slide-a-riser dialysis cassette (3500 Da molecular weight cutoff, Pierce). Insoluble protein was removed by centrifugation at 30,000 xg for 20 minutes, and the supernatant was supplemented with β-mercaptoethanol to a final concentration of 2 mM. 1 ml of 50% Ni-NTA slurry (Qiagen) was added to the refolded protein and mixed gently by shaking at 4° C. for 60 minutes. The protein-Ni-NTA mixture was loaded onto the column and the flow through was collected. After washing the resin with 20 mL of 25 mM imidazole in PBS (pH 8.0), the protein was eluted with 2.5 mL of 250 mM imidazole in PBS (pH 8.0). Finally, the protein was buffer exchanged with PBS (pH 7.4) by using a PD-10 column (GE Healthcare). The purity of the protein sample was investigated by SDS-PAGE (Fig. 21B). In FIG. 21B,
추가로, 상기 mTNFα에 대한 방법을 이용하여 수득한 바와 같이, PylB 발현의 존재 또는 부재하에서 수득한 단백질에 대해 mEGF Tyr10TAG, ESI-MS 스펙트럼을 수득하였다 (도 21C). 도 21C에서 하단 질량 스펙트럼은 PYL 혼입이 pylB 유전자의 존재하에서 우세하게 일어남을 보여주며 (mEGF Tyr10Pyl의 예상 질량 = 7310 Da), 도 21C에서 상단 질량 스펙트럼은 유일한 PCL 혼입은 pylB의 부재하에 일어남을 보여준다 (mEGF Tyr10Pyl의 예상 질량 = 7296 Da). 이와 같이 도 21B에서 관찰된 단백질 중, 레인 2는 PYL이 혼입된 mEGF였다 (추가의 LC-MS/MS 분석에 의해 확인함). 유사하게, 도 21A에서 관찰된 단백질 중, 레인 2는 아마도 PYL이 혼입된 mTNFα일 것이고, 레인 4는 아마도 PCL이 혼입된 mTNFα일 것이다.In addition, as obtained using the method for mTNFα, mEGF Tyr10TAG, ESI-MS spectra were obtained for the protein obtained in the presence or absence of PylB expression (FIG. 21C). In Figure 21C, the lower mass spectrum shows that PYL incorporation occurs predominantly in the presence of the pylB gene (expected mass of mEGF Tyr10Pyl = 7310 Da), and the upper mass spectrum in Figure 21C shows that the only PCL incorporation occurs in the absence of pylB. (Expected mass of mEGF Tyr10Pyl = 7296 Da). As such, of the proteins observed in FIG. 21B,
추가로, 모든 유전자 (M. 마제이 pylS, pylT, pylB, pylC, 및 pylD)를 발현하는 mEGF Tyr10TAG 샘플에의 PCL 및 PYL 혼입의 LC-MS/MS 추출된 이온 크로마토그램 질량 분석으로부터의 PCL/PYL 비율의 정량화는 PYL이 PCL 보다 5 내지 10배 더 풍부한 것으로 나타난 반면, 모든 유전자 (M. 마제이 pylS, pylT, pylB, pylC, 및 pylD)를 발현하는 mTNF Gln21TAG 샘플에의 PCL 및 PYL 혼입의 LC-MS/MS 추출된 이온 크로마토그램 질량 분석으로부터의 PCL/PYL 비율의 정량화는 PCL이 Pyl 보다 약 7 배 더 풍부한 것으로 나타났다. PylB 유전자의 부재하에서의 정량화는 PCL 단백질만을 나타내었는데, 이는 PYL 혼입이 pylB에 절대적으로 의존적임을 나타내며, 이는 추가로 PylB가 실제로 Pyl의 생합성을 위해 요구되는 메틸트랜스퍼라제임을 시사한다.Additionally, LC-MS/MS extracted ion chromatogram PCL/PYL from mass spectrometry of incorporation of PCL and PYL into mEGF Tyr10TAG samples expressing all genes (M. majay pylS, pylT, pylB, pylC, and pylD). Quantification of the ratio showed that PYL was 5 to 10 times more abundant than PCL, while LC- of PCL and PYL incorporation into mTNF Gln21TAG samples expressing all genes (M. majei pylS, pylT, pylB, pylC, and pylD). Quantification of the PCL/PYL ratio from MS/MS extracted ion chromatogram mass spectrometry showed that PCL was about 7 times more abundant than Pyl. Quantification in the absence of the PylB gene revealed only PCL protein, indicating that PYL incorporation was absolutely dependent on pylB, further suggesting that PylB is actually a methyltransferase required for the biosynthesis of Pyl.
실시예 15: 다른 유사체 및 전구체의 혼입: Example 15 : Incorporation of other analogs and precursors :
HK100 세포 (게네혹스(Genehogs)로부터 유래된 것; 인비트로겐)를 pAra-pylSTBCD, pAra-pylSTCD, pAra-pylSTC, pAra-pylSTD, 또는 pSUPAR-pylST 및 pMH4-FASTE-L2222TAG-L2223I로 공동 형질전환시켰다. 세포를 테리픽 브로스(Terrific Broth) (TB) (시그마)의 25 ml 배양물에서 37℃에서 OD595 약 0.6 까지 성장시켰다. 세포를 30℃로 옮기고, 유사체 또는 전구체 화합물을 각각의 개별 배양물에 도 15에 나타낸 농도로 첨가하였다. 평가된 화합물은 N-ε-시클로펜틸옥시카르보닐-L-리신 (CYC; 시그마), D-오르니틴 (켐-임펙스), PCL-A (실시예 36-1, 화합물 3647-125 참조), PCL-B (실시예 36-2, 화합물 3793-011 참조), P2C (실시예 36-2, 화합물 3647-164 참조), P5C (실시예 35-1, 화합물 3793-007 참조) 및 Lys-Nε-D-오르니틴 (실시예 35-2, 화합물 3793-031 참조)이었다. 이어서, 세포를 20 분 뒤에 0.2% 아라비노스로 유도하였다. 18-20 시간 후에, 세포를 원심분리에 의해 수거하였다. 세포를 초음파처리에 의해 용해하고, 자연 조건하에서 Ni-NTA (퀴아겐) 상에서 정제하고, SDS-PAGE 겔에 이어 쿠마시 염색 (0.25 ml Ni-NTA, 0.75 ml 용리, 겔 상 20 ㎕)으로 평가하였다. 도 15A는 나타낸 pyl 유전자 (pylS/pylT or pylB/pylC/pylD/pylS/pylT)의 존재하에 성장시키고 나타낸 화합물을 공급한 세포로부터 정제된 단백질 (FAS-TE)을 나타낸다. Cyc 및 D-오르니틴 (D-Orn)을 양성 대조군으로서 사용하였다. 어떤 화합물도 첨가하지 않은 것을 음성 대조군으로서 나타내었다. 레인 1-11 및 12-18에서의 겔 샘플 용량은 각각 내부적으로 일관되었다. HK100 cells (derived from Genehogs; Invitrogen) were co-transformed with pAra-pylSTBCD, pAra-pylSTCD, pAra-pylSTC, pAra-pylSTD, or pSUPAR-pylST and pMH4-FASTE-L2222TAG-L2223I. . Cells were grown in 25 ml cultures of Terrific Broth (TB) (Sigma) at 37° C. to an OD595 of about 0.6. Cells were transferred to 30° C. and analog or precursor compounds were added to each individual culture at the concentrations shown in FIG. 15. The evaluated compounds were N-ε-cyclopentyloxycarbonyl-L-lysine (CYC; Sigma), D-ornithine (Chem-Impex), PCL-A (see Example 36-1, Compound 3647-125). , PCL-B (see Example 36-2, compound 3793-011), P2C (see Example 36-2, compound 3647-164), P5C (see Example 35-1, compound 3793-007) and Lys- It was Nε-D-ornithine (see Example 35-2, compound 3793-031). The cells were then induced with 0.2% arabinose after 20 minutes. After 18-20 hours, cells were harvested by centrifugation. Cells were lysed by sonication, purified on Ni-NTA (Qiagen) under natural conditions and evaluated by SDS-PAGE gel followed by Coomassie staining (0.25 ml Ni-NTA, 0.75 ml elution, 20 μl on gel) I did. 15A shows a protein (FAS-TE) grown in the presence of the indicated pyl gene (pylS/pylT or pylB/pylC/pylD/pylS/pylT) and purified from cells fed the indicated compound. Cyc and D-ornithine (D-Orn) were used as positive controls. No compound was added as a negative control. The gel sample volumes in lanes 1-11 and 12-18 were internally consistent, respectively.
레인 2 및 3은 단지 pylS 및 pylT 유전자 및, 각각 PCL-B (Lys-P2C) 또는 PCL-A (Lys-P5C)와 함께 성장시킨 세포로부터 수득한 단백질을 나타낸다. 화합물은 둘 모두 단백질에 혼입되어, PylS가 두 화합물을 사용할 수 있음을 보여주었다. 레인 5 내지 11은 pyl 생합성 유전자의 전체 세트 (pylB/pylC/pylD/pylS/pylT)를 함유하는 세포에서 전구체 Lys-Nε-D-Orn (레인 10), 또는 피롤린 고리만을 갖는 전구체: P2C (레인 6) 및 P5C (레인 8)의 존재하에서의 단백질 생합성을 평가한 것을 보여준다. 피롤린 고리만을 갖는 전구체: P2C (레인 6) 및 P5C (레인 8)는 PCL 생합성을 지지하는데 충분하지 않았고, 이는 이들이 경로의 중간체가 아님을 시사한다. 그러나, Lys-Nε-D-Orn (레인 10)은 앰버 코돈에 PCL이 혼입된 단백질을 발현시켰고, 이는 이것이 PCL 생합성 경로의 중간체임을 나타낸다. 표 5는 수득한 단백질의 양 (브래드포드), 관측 질량 및 수득한 PCL:PYL:CYC의 비율을 제시한다.
Lys-Nε-D-Orn이 요구되는 유전자 (pylC 및 pylD) 중 임의의 것의 생합성 경로 상류의 중간체인지를 판단하기 위해, HK100 세포 (게네혹스로부터 유래된 것; 인비트로겐)를 pSUPAR-pylST, pMH4-FASTE-L2222TAG-L2223I, 및 pAra-pylSTB, pAra-pylSTC, pAra-pylSTD, pAra-pylSTCD 또는 pAra-pylSTBCD로 공동 형질전환시켰다. 도 15B는 Lys-Ne-D-Orn로부터 PCL을 형성하는데 pylD (레인 15) 만이 요구됨을 보여주었고, 이는 이것이 생합성 경로에서 pylC의 상류 중간체임을 시사한다. 또한, NMR 평가로 Lys-Ne-D-Orn이 PylD의 기질임을 확인하였다.To determine whether Lys-Nε-D-Orn is an intermediate upstream of the biosynthetic pathway of any of the required genes (pylC and pylD), HK100 cells (derived from Genehox; Invitrogen) were converted to pSUPAR-pylST, pMH4. Cotransformed with -FASTE-L2222TAG-L2223I, and pAra-pylSTB, pAra-pylSTC, pAra-pylSTD, pAra-pylSTCD or pAra-pylSTBCD. Figure 15B showed that only pylD (lane 15) is required to form PCL from Lys-Ne-D-Orn, suggesting that it is an upstream intermediate of pylC in the biosynthetic pathway. In addition, it was confirmed that Lys-Ne-D-Orn is a substrate of PylD by NMR evaluation.
실시예 16: hRBP4 에 혼입된 PCL 의 2-아미노 벤즈알데히드 , 2-아미노-아세토페논 및 2-아미노-5-니트로-벤조페논에 의한 유도체화. Example 16 : Derivatization of PCL incorporated in hRBP4 with 2-amino benzaldehyde , 2-amino- acetophenone and 2-amino-5-nitro-benzophenone.
본 실시예는 PCL을 2-아미노-벤즈알데히드 (2-ABA), 2-아미노-아세토페논 및 2-아미노-5-니트로-벤조페논으로 표지하는 것을 제공한다. 반응은 도 22에 나타낸 일반적인 반응식을 따르는 것으로 보인다. 질량 분광측정법 및 NMR을 이용하여 형성된 구조를 평가하였다. 도 23은 3개의 상이한 2-아미노-벤즈알데히드 잔기로부터 형성된 단백질 접합체를 나타내며, 예상되는 질량 변화 및 관측된 질량 변화를 제공한다. This example provides for labeling PCL with 2-amino-benzaldehyde (2-ABA), 2-amino-acetophenone and 2-amino-5-nitro-benzophenone. The reaction appears to follow the general scheme shown in Figure 22. The structure formed was evaluated using mass spectrometry and NMR. 23 shows protein conjugates formed from three different 2-amino-benzaldehyde residues and provides the expected and observed mass changes.
hRBP4hRBP4 에서 2-2- in ABAABA 에 의해 부위 특이적으로 변형된 Site-specific modified by PCLPCL 의 질량 분광측정 검출Mass spectrometry detection of
레티놀 결합 단백질 (hRBP4)을 실시예 2에 기재한 바와 같이 HEK293F 세포에서 발현시켰다. Phe122 (hRBP4 돌연변이체 #6) 대신 PCL을 혼입시켰고, 이를 질량 분광측정법으로 확인하였다 (도 6A-11). 10 ㎕의 hRBP4 Phe122PCL 저장 용액을 89 ㎕의 200 mM 나트륨 아세테이트 완충액 (pH 5.0) 및 1 ㎕의 1 M 2-ABA 용액과 혼합하고, 실온에서 16 시간 동안 인큐베이션하였다. 반응 혼합물 중 최종 농도는 17 μM hRBP4 Phe122PCL 단백질 및 10 mM 2-아미노-벤즈알데히드 (2-ABA)였다. 유도체화된 hRBP4의 질량 스펙트럼을 도 24에 나타내었고, 이때 수득된 질량은 PCL의 2-ABA 부가물 (23269.2 Da)에 상응하였다. 변형되지 않은 hRBP4는 질량이 23166.8 Da이었고, 따라서 관찰된 102.4 Da의 질량 증가 (예상치 + 103 Da)는 hRBP4가 2-ABA에 의해 변형되었음을 나타낸다. 질량 스펙트럼에서 적어도 96%의 피크 강도는 PCL의 2-ABA 부가물로 인한 것이고, 이는 반응이 거의 완료되었음을 나타낸다. Retinol binding protein (hRBP4) was expressed in HEK293F cells as described in Example 2. PCL was incorporated instead of Phe122 (hRBP4 mutant #6), which was confirmed by mass spectrometry (FIGS. 6A-11). 10 μl of hRBP4 Phe122PCL stock solution was mixed with 89 μl of 200 mM sodium acetate buffer (pH 5.0) and 1 μl of 1 M 2-ABA solution and incubated at room temperature for 16 hours. The final concentration in the reaction mixture was 17 μM hRBP4 Phe122PCL protein and 10 mM 2-amino-benzaldehyde (2-ABA). The mass spectrum of the derivatized hRBP4 is shown in FIG. 24, and the mass obtained at this time corresponds to the 2-ABA adduct of PCL (23269.2 Da). Unmodified hRBP4 had a mass of 23166.8 Da, so the observed 102.4 Da mass increase (expected + 103 Da) indicates that hRBP4 was modified by 2-ABA. The peak intensity of at least 96% in the mass spectrum is due to the 2-ABA adduct of PCL, indicating that the reaction is almost complete.
2-ABA-유도체화된 hRBP4 Phe122PCL 단백질의 트립신 소화의 LC-MS 분석을 행하였고, MS/MS 분석 (도 25A)으로 예상되는 YWGVASF*LQK 펩티드 (서열 17)를 확인하였다 (여기서 F*는 도 23에 나타낸 바와 같은 2-ABA-변형된 PCL의 질량과 일치하는 질량을 가짐). 유도체화되지 않은 PCL 잔기가 검출되지 않는 것으로 반응이 완료되었다. YWGVASF*LQK (F* = PCL-2-ABA 부가물)의 할당된 MS/MS 스펙트럼을 이하에 제공한다:LC-MS analysis of trypsin digestion of 2-ABA-derivated hRBP4 Phe122PCL protein was performed, and YWGVASF * LQK peptide (SEQ ID NO: 17) expected by MS/MS analysis (Fig. 25A) was confirmed (where F * is Fig. It has a mass that matches the mass of the 2-ABA-modified PCL as shown in 23). The reaction was complete with no detection of non-derivatized PCL residues. The assigned MS/MS spectrum of YWGVASF * LQK (F * = PCL-2-ABA adduct) is provided below:
도 25B는 YWGVASF*LQK (F* = PCL 및 PCL-2-ABA 부가물)의 2+ 이온의 TIC 및 EIC로, EIC (추출된 이온 크로마토그램)을 유도체화 및 비유도체화된 (검출되지 않음) 종과 비교한 것은 반응이 완료되었음을 나타내었다. 도 25C는 2-ABA에 의해 유도체화된 hRBP4 Phe122PCL의 질량 분광측정 분석으로, YWGVASF*LQK의 3+ 및 2+ 전구체는 각각 m/z 459.92 (3+) 및 689.37 (2+)에서 나타났다. (F* = PCL-2-ABA 부가물). 본 실시예는 관찰된 2-ABA에 의한 반응이 부위 특이적으로 일어나 PCL 잔기가 잔기 122의 목적하는 TAG 부위에 혼입되었음을 증명하였다. Figure 25B is the TIC and EIC of the 2+ ions of YWGVASF * LQK (F * = PCL and PCL-2-ABA adduct), EIC (extracted ion chromatogram) derivatized and derivatized (not detected). ) Comparison with the species indicated that the reaction was complete. Figure 25C is a mass spectrometric analysis of hRBP4 Phe122PCL derivatized by 2-ABA, where the 3+ and 2+ precursors of YWGVASF * LQK were shown at m/z 459.92 (3+) and 689.37 (2+), respectively. (F * = PCL-2-ABA adduct). This example demonstrated that the observed reaction by 2-ABA occurred site-specifically, and that the PCL residue was incorporated into the desired TAG site of residue 122.
pHpH 의 함수로서의 반응 효율:Reaction efficiency as a function of:
pH의 함수로서의 반응 효율, (도 26)은 17 μM hRBP4 PCL 돌연변이체 단백질을 200 mM 나트륨 아세테이트 완충액 (pH 5.0)에서 10 mM 2-아미노-벤즈알데히드 (2-ABA)와 반응시키거나 반응시키지 않고, 또는 10 mM 2-아미노-벤즈알데히드 (2-ABA) 10x PBS 완충액 (pH 7.4)과 12 시간 동안 실온에서 반응시켜 평가하였다. 반응 혼합물의 질량 스펙트럼은 적어도 87%의 총 피크 강도가 하나의 2-ABA 잔기에 의해 변형된 단백질의 피크 강도 (예상치와 같이 + 102 Da)와 상응함을 나타내었다 (도 26A, B 및 C). pH 7.4 반응 혼합물은 대략 13%의 미반응 단백질을 함유하는 반면 (도 26C), pH 5.0에서의 반응에서는 단지 4.2%의 미반응 단백질이 검출되었는데 (도 26B), 이는 pH 7.4에 비해 pH 5.0에서 PCL의 반응성이 약간 더 높음을 시사한다. Phe62 대신 OMePhe가 혼입된 hRBP4는 10 mM 2-ABA의 존재에 의해서는 변형되지 않았기 때문에, 반응은 PCL의 존재에 특이적이다 (도 26D, E 및 F).The reaction efficiency as a function of pH, (FIG. 26 ), shows that 17 μM hRBP4 PCL mutant protein was reacted with or without 10 mM 2-amino-benzaldehyde (2-ABA) in 200 mM sodium acetate buffer (pH 5.0), Alternatively, it was evaluated by reacting with 10 mM 2-amino-benzaldehyde (2-ABA) 10x PBS buffer (pH 7.4) for 12 hours at room temperature. The mass spectrum of the reaction mixture indicated that a total peak intensity of at least 87% corresponds to the peak intensity of the protein modified by one 2-ABA moiety (+102 Da as expected) (Figures 26A, B and C). . The pH 7.4 reaction mixture contained approximately 13% unreacted protein (Figure 26C), whereas only 4.2% unreacted protein was detected in the reaction at pH 5.0 (Figure 26B), which at pH 5.0 compared to pH 7.4. This suggests that the reactivity of PCL is slightly higher. Since hRBP4 with OMePhe incorporated instead of Phe62 was not modified by the presence of 10 mM 2-ABA, the reaction was specific for the presence of PCL (FIGS. 26D, E and F).
반응물의 단백질 농도에 대한 함수로서의 반응 효율 및 다른 2- ABA -유사 잔 기와의 반응 효율: The reaction efficiency as a function of the protein concentration of the reactants and the reaction efficiency with other 2- ABA -like residues :
17 μM hRBP4 PCL 돌연변이체 단백질을 200 mM 나트륨 아세테이트 완충액 (pH 5.0) 중에서 0.1 mM 2-아미노-벤즈알데히드 (2-ABA), 0.1 mM 2-아미노-아세토페논 (2-AAP) 또는 0.1 mM 2-아미노-5-니트로-벤조페논 (2-ANBP)과 반응시켜, 반응물의 단백질 농도 비율에 대한 함수로서의 반응 효율 및 2-ABA-유사 반응물과의 반응성을 평가하였다. 실온에서 12 시간 후에, 반응 혼합물의 질량 스펙트럼은 접합된 단백질의 예상되는 질량을 나타내었다 (도 27). 2-ABA의 경우, 올바르게 접합된 피크의 상대 강도는 전체 강도의 88%였고; 단지 단백질의 4.2% 만이 미반응된 채로 남았다 (도 27A). 2-AAP의 경우, 93%가 반응된 것으로 보였고; 4.5%는 반응하지 않았다 (도 27B). 2-ANBP의 경우, 단지 5.4% 만이 반응하였고 (도 27C), 이는 반응성 시약의 낮은 용해도, 즉 hRBP4 PCL 돌연변이체 단백질을 함유하는 용액에 첨가하자 마자 2-ANBP가 즉시 침전하기 때문인 것으로 보인다.17 μM hRBP4 PCL mutant protein was added to 0.1 mM 2-amino-benzaldehyde (2-ABA), 0.1 mM 2-amino-acetophenone (2-AAP) or 0.1 mM 2-amino in 200 mM sodium acetate buffer (pH 5.0). By reacting with -5-nitro-benzophenone (2-ANBP), the reaction efficiency as a function of the protein concentration ratio of the reactants and the reactivity with 2-ABA-like reactants were evaluated. After 12 hours at room temperature, the mass spectrum of the reaction mixture showed the expected mass of the conjugated protein (FIG. 27 ). For 2-ABA, the relative intensity of the correctly bonded peak was 88% of the total intensity; Only 4.2% of the protein remained unreacted (FIG. 27A ). For 2-AAP, 93% appeared to be reacted; 4.5% did not react (FIG. 27B). In the case of 2-ANBP, only 5.4% reacted (FIG. 27C ), which appears to be due to the low solubility of the reactive reagent, i.e. 2-ANBP precipitates immediately upon addition to the solution containing the hRBP4 PCL mutant protein.
본 실시예는 PCL 변형된 단백질이 다른 2-아미노-벤즈알데히드 유사체와 반응하고, PCL이 혼입된 부위에서 유도체화됨을 보여준다. 모든 경우에서, 변형된 단백질을 측정한 질량은 도 23에 도시된 구조에서 예상되는 질량과 일치하였다. 데이터는 또한 접합 반응이 높은 효율로 진행되며, 단지 6대 1의 낮은 반응물 대 단백질 비율에서 거의 완료됨을 보여주었다. 데이터는 또한 각각의 단백질 샘플이 단지 한번만 유도체화됨을 보여주었다. 그러나, 매우 높은 반응물 대 단백질 비율 (4700 배) 및 pH 7.5에서 다중 반응물 분자가 부착되는 것으로 관찰되었다 (도 28A). pH 5.0에서 동일한 샘플이 침전되는 것은 역시 다중 반응이 있음을 시사한다. 유사하게 Phe62 대신 OMePhe로 변형된 hRBP4 (6.5 μM)는 pH 7.5에서 15400 배 몰 과량의 2-ABA와 반응시킨 경우에 도 28A에서 관찰되는 것과 유사한 접합 패턴을 나타내었다 (도 28B). 이는 이러한 조건 (단백질보다 많은 몰 과량의 반응물) 하에서의 부착은 PCL 측쇄의 존재에 의존적이지 않지만, 아마도 리신 측쇄를 수반할 것임을 나타낸다. 그러나, 실시예 반응은 단백질에 혼입된 PCL 잔기가 작은 몰 과량으로 첨가된 2-아미노-벤즈알데히드 함유 분자와의 반응에 의해 특이적이고 거의 정량적으로 유도체화될 수 있음을 보여준다.This example shows that the PCL modified protein reacts with other 2-amino-benzaldehyde analogues and is derivatized at the site where PCL is incorporated. In all cases, the measured mass of the modified protein coincided with the mass expected in the structure shown in FIG. 23. The data also showed that the conjugation reaction proceeded with high efficiencies and was nearly complete at only a low reactant to protein ratio of 6 to 1. The data also showed that each protein sample was derivatized only once. However, it was observed that at a very high reactant to protein ratio (4700 fold) and pH 7.5, multiple reactant molecules were attached (FIG. 28A ). Precipitation of the same sample at pH 5.0 suggests that there are also multiple reactions. Similarly, hRBP4 (6.5 μM) modified with OMePhe instead of Phe62 showed a conjugation pattern similar to that observed in FIG. 28A when reacted with a 15400-fold molar excess of 2-ABA at pH 7.5 (FIG. 28B ). This indicates that adhesion under these conditions (molar excess of reactants greater than protein) is not dependent on the presence of PCL side chains, but will probably involve lysine side chains. However, the example reactions show that the PCL moiety incorporated in the protein can be specifically and almost quantitatively derivatized by reaction with a 2-amino-benzaldehyde-containing molecule added in a small molar excess.
실시예 17: FAS - TE 에 혼입된 PCL 의 2-아미노-아세토페논에 의한 유도체화 Example 17 : Derivatization of PCL incorporated in FAS - TE with 2-amino-acetophenone
실시예 8에서와 같이 에스케리키아 콜라이에서 생성된 FAS-TE Tyr2454PCL 16 μM을 1 mM 2-AAP와 16 시간 동안 실온에서 반응시키고, 200 mM 나트륨 아세테이트 완충액 중에서 24 시간 동안 4℃에서 pH 5.0에서 반응시켰다. 도 29A는 미반응된 FAS-TE Tyr2454PCL의 질량 스펙트럼을 나타내고, 도 29B는 반응 혼합물의 질량 스펙트럼을 나타낸다: 관찰가능한 피크 강도의 100%는 미반응 물질의 질량보다 116.8 Da 크고, 2-AAP 변형된 FAS-TE Tyr2454PCL에서 예상되는 116 Da 질량 증가의 오차 이내인 33318.8 Da에서 발생하였다. 유사하게, pH 7.4에서 반응은 95% 완료되었다 (도 29C (미반응) 및 도 29D (반응)).As in Example 8, 16 μM of FAS-TE Tyr2454PCL produced in Escherichia coli was reacted with 1 mM 2-AAP for 16 hours at room temperature, and reacted in 200 mM sodium acetate buffer at 4° C. for 24 hours at pH 5.0. Made it. Figure 29A shows the mass spectrum of unreacted FAS-TE Tyr2454PCL, and Figure 29B shows the mass spectrum of the reaction mixture: 100% of the observable peak intensity is 116.8 Da greater than the mass of the unreacted material, 2-AAP modified It occurred at 33318.8 Da, which is within the error of the 116 Da mass increase expected in FAS-TE Tyr2454PCL. Similarly, at pH 7.4 the reaction was 95% complete (Fig. 29C (unreacted) and Fig. 29D (reacted)).
실시예 18: hRBP4 에 혼입된 PCL 의 2-아미노-아세토페논- PEG8 에 의한 유도체화 Example 18: in the PCL incorporated in hRBP4 2- amino-acetophenone-derivative by PEG8 Chemistry
본 실시예는 hRBP4에 혼입된 PCL이 2-아미노-아세토페논의 폴리에틸렌 글리콜 (PEG) 유도체에 의해 단일 부위에서 완전하게 유도체화될 수 있음을 보여준다. 본 실시예에서, PEG는 8 에틸렌 글리콜 단위를 함유하고, 그의 구조를 도 31에 나타내었다. 본 실시예는 또한, pH 5.0 및 pH 7.5에서 야생형 hRBP4는 단백질 비율에 대한 최대 2300의 시약에 의해 2-AAP-PEG8과 반응하지 않음을 보여준다. This example shows that PCL incorporated in hRBP4 can be completely derivatized at a single site by a polyethylene glycol (PEG) derivative of 2-amino-acetophenone. In this example, PEG contains 8 ethylene glycol units, and its structure is shown in FIG. 31. This example also shows that at pH 5.0 and pH 7.5, wild-type hRBP4 does not react with 2-AAP-PEG8 with reagents of up to 2300 per protein ratio.
hRBP4 돌연변이체 #6을 실시예 2에서와 같이 제조하였다. 2-아미노-아세토페논 PEG8 (TU3205-044)은 실시예 20에 기재한 바와 같이 제조하였다. 실온에서 14 시간 동안 및 4℃에서 72 시간 동안 반응이 진행되도록 한 후, 반응 혼합물에 대한 질량 스펙트럼을 수득하였다.
pH 7.5에서 반응을 수행하는 경우, 10 ㎕의 hRBP4 Phe122PCL (0.22 mg/mL, PBS 중, pH 7.5)을 10 ㎕의 10x PBS로 희석하였다. 0.2 ㎕ 또는 2 ㎕의 2-AAP-PEG8 (물 중)의 100 mM 저장 용액을 각각 1 및 9.1 mM의 최종 농도로 첨가하였다. 단백질 농도는 각각 4.7 μM 및 4.3 μM였고, 반응물은 단백질에 대해 210 또는 2100 몰 과량이 되었다. 도 32A는 210 대 1 비율에서 반응 혼합물의 질량 스펙트럼을 나타낸 것으로 반응이 대략 95% 완료되었음을 나타내었고, 556 Da의 관찰된 질량 증가는 예상치 (도 31)와 동일하였다. 100% 완료는 2100 몰 과량에서의 반응에서 수득되었다 (데이터는 나타내지 않음). When performing the reaction at pH 7.5, 10 µl of hRBP4 Phe122PCL (0.22 mg/mL, in PBS, pH 7.5) was diluted with 10 µl of 10x PBS. 0.2 μl or 2 μl of a 100 mM stock solution of 2-AAP-PEG8 (in water) was added at a final concentration of 1 and 9.1 mM, respectively. Protein concentrations were 4.7 μM and 4.3 μM, respectively, and the reactants were 210 or 2100 molar excesses for the protein. FIG. 32A shows the mass spectrum of the reaction mixture at a 210 to 1 ratio indicating that the reaction was approximately 95% complete, and the observed mass increase of 556 Da was the same as expected (FIG. 31 ). 100% completion was obtained in the reaction at 2100 molar excess (data not shown).
유사하게, 반응을 pH 5.0에서 수행하였다. 이 경우, 10 ㎕의 hRBP4 Phe122PCL (0.22 mg/mL, PBS 중, pH 7.5)을 90 ㎕의 200 mM 나트륨 아세테이트 완충액 (pH 5.0)으로 희석시켰다. 1 ㎕ 또는 10 ㎕의 2-AAP-PEG8 (물 중)의 100 mM 저장 용액을 각각 1 및 9.1 mM의 최종 농도로 첨가하였다. 단백질 농도는 각각 0.94 및 0.86 μM였고, 반응물은 단백질에 대해 1050 또는 10500 몰 과량이 되었다. 도 32B는 1050 대 1 비율에서 반응 혼합물의 질량 스펙트럼을 나타낸다. 100% 완료는 양 반응 모두에서 수득되었고, 관찰된 556 Da의 질량 증가는 예상치 (도 31)와 동일하였다.Similarly, the reaction was carried out at pH 5.0. In this case, 10 μl of hRBP4 Phe122PCL (0.22 mg/mL, pH 7.5 in PBS) was diluted with 90 μl of 200 mM sodium acetate buffer (pH 5.0). 1 μl or 10 μl of a 100 mM stock solution of 2-AAP-PEG8 (in water) was added to a final concentration of 1 and 9.1 mM, respectively. Protein concentrations were 0.94 and 0.86 μM, respectively, and the reactants were 1050 or 10500 molar excesses for the protein. 32B shows the mass spectrum of the reaction mixture at 1050 to 1 ratio. 100% completion was obtained for both reactions and the observed mass increase of 556 Da was the same as expected (Figure 31).
2-AAP-PEG8의 야생형 hRBP4 단백질과의 반응성을 시험하기 위해, 시험 반응을 상기와 유사하게 설정하였다. pH 7.5 반응을 위해, 최종 야생형 단백질 농도를 20 μM로 하고, 2-AAP-PEG8 농도는 1 또는 9.1 mM로 하여, 46 및 460 대 1의 몰 비가 되게 하였다. pH 5 반응은 4 μM 단백질 농도 및 1 및 9.1 mM 2-AAP-PEG (230 및 2300 대 1 반응물 대 단백질)에서 수행하였다. 모든 4 가지의 반응에서, 단지 변형되지 않은 야생형 hRBP4 단백질 만이 예상 질량에서 관찰되었다 (도 32D-F). 본 실시예는 2-AAP-PEG의 커플링 반응이 표적 단백질 내 PCL 잔기의 존재에 매우 특이적임을 보여준다.To test the reactivity of 2-AAP-PEG8 with the wild-type hRBP4 protein, the test reaction was set up similar to the above. For the pH 7.5 reaction, the final wild-type protein concentration was 20 μM, and the 2-AAP-PEG8 concentration was 1 or 9.1 mM, resulting in a molar ratio of 46 and 460 to 1. The
실시예 19: FAS - TE 에 혼입된 PCL 의 다양한 분자량의 2-아미노-아세토페논-PEG에 의한 유도체화 Example 19 : Derivatization of PCL incorporated in FAS - TE with 2-amino-acetophenone-PEG of various molecular weights
본 실시예는 PCL 측쇄의 2-아미노-아세토페논 PEG와의 반응성의 보편성을 보여준다. 본 실시예는 추가로, 생체치료 단백질을 변형시키는데 유용한 충분한 길이의 2-AAP-PEG가 PCL 변형된 단백질에 접합될 수 있음을 보여준다.This example shows the universality of the reactivity of the PCL side chain with 2-amino-acetophenone PEG. This example further shows that 2-AAP-PEG of sufficient length, useful for modifying biotherapeutic proteins, can be conjugated to PCL modified proteins.
실시예 8에 기재한 바와 같이 에스케리키아 콜라이에서 생산한 16 μM의 FAS-TE Tyr2454PCL을 1 mM TU3205-044 (2-AAP-PEG8)와 200 mM 나트륨 아세테이트 완충액 중에서 16 시간 동안 실온에서 pH 5.0에서 반응시켰다. 도 33A는 미반응 FAS-TE Tyr2454PCL의 질량 스펙트럼을 나타내고, 도 33B는 반응 혼합물의 질량 스펙트럼을 나타낸다: 도 33B에서, 관찰가능한 피크 강도의 100%는 미반응 물질의 질량보다 556 Da 큰 33758.4 Da에서 발생하였다. 이러한 질량 차이는 2-AAP-PEG8 변형된 FAS-TE Tyr2454PCL에서 예상되는 것 (도 31)과 같다.As described in Example 8, 16 μM of FAS-TE Tyr2454PCL produced in Escherichia coli was prepared in 1 mM TU3205-044 (2-AAP-PEG8) and 200 mM sodium acetate buffer for 16 hours at pH 5.0 at room temperature. Reacted. Figure 33A shows the mass spectrum of unreacted FAS-TE Tyr2454PCL, and Figure 33B shows the mass spectrum of the reaction mixture: In Figure 33B, 100% of the observable peak intensity is at 33758.4 Da, which is 556 Da greater than the mass of the unreacted material. Occurred. This mass difference is as expected for the 2-AAP-PEG8 modified FAS-TE Tyr2454PCL (Fig. 31).
추가의 실시예에서, FAS-TE의 Tyr2454에 혼입된 PCL을 MW이 0.5 kDa, 2.4 kDa 및 23 kDa인 3개의 상이한 2-AAP-PEG로 유도체화시켰다. FAS-TE 단일 부위 PCL 돌연변이체를 실시예 8에 기재한 바와 같이 에스케리키아 콜라이에서 대략 160 mg/L의 수율 (약 80 중량%의 수율에 상응함)로 생성하였다. 분취량의 FAS-TE Tyr2454PCL (PBS 중 0.16 mM, pH 7.5)을 TU3205-044 (0.5 kDa 2-AAP-PEG), TU3205-048 (2.4 kDa 2-AAP-PEG) 및 TU3205-052 (23 kDa 2-AAP-PEG)와 10:1 또는 100:1의 몰비로 6 일 동안 4℃ 또는 실온에서 반응시켰다. PEG의 구조 및 그의 합성을 실시예 37에 기재한다.In a further example, PCL incorporated into Tyr2454 of FAS-TE was derivatized with three different 2-AAP-PEGs with MW of 0.5 kDa, 2.4 kDa and 23 kDa. FAS-TE single site PCL mutants were generated in Escherichia coli in a yield of approximately 160 mg/L (corresponding to a yield of approximately 80% by weight) as described in Example 8. Aliquots of FAS-TE Tyr2454PCL (0.16 mM in PBS, pH 7.5) were added to TU3205-044 (0.5 kDa 2-AAP-PEG), TU3205-048 (2.4 kDa 2-AAP-PEG) and TU3205-052 (23 kDa 2 -AAP-PEG) and 10:1 or 100:1 were reacted for 6 days at 4°C or room temperature. The structure of PEG and its synthesis are described in Example 37.
SDS-PAGE 분석 (도 34) 이전에, His-태그된 FAS-TE 단백질을 Ni-NTA 비드에 결합시켜 과량의 PEG 시약을 제거하고, PBS 완충액으로 비드를 반복 세척하였다. 겔 및 질량 분광측정 분석을 위해, Ni-NTA 비드에서 과량의 완충액을 제거하고, 이미다졸 완충액으로 단백질을 용리시켰다. 구체적으로, 50 ㎕의 Ni-NTA 비드를 50 ㎕의 반응 혼합물에 첨가하고, 2 시간 동안 인큐베이션하고, 반응 혼합물 및 완충액을 원심분리에 의해 분리하였다. 이어서, 비드를 1 mL PBS로 3 회 세척하고; 70 ㎕의 250 mM 이미다졸 완충액, 20 mM 트리스, pH 8을 비드에 첨가하여 겔 및 질량 분광측정 분석용 단백질을 용리시켰다. 0.5 kDa 2-AAP-PEG에 의한 PEG화는 SDS-PAGE로 분석할 수 없었지만, 질량 분광측정법으로 확인하였다. 반응의 정도는 100:1 실온 반응의 경우 대략 57%였고, 100:1 4℃ 반응의 경우 43%였다 (데이터는 나타내지 않음). 더 큰 2-AAP-PEG의 경우에는 PEG화된 생성물을 SDS-PAGE로 분석할 수 있었다 (도 34). 모든 반응은 불완전했고, 2.4 kDa 및 23 kDa 2-AAP-PEG의 경우 대략 25-30% 진행되었다. PCL 부위에서의 단일 부위 접합 및 반응의 정도는 2.4 kDa 2-AAP-PEG의 경우 질량 분광측정법으로 확인하였다 (도 33). 23 kDa 2-AAP-PEG 유도체화된 단백질에 대한 질량 분광측정 데이터는 PEG가 균질하지 않아 수득할 수 없었다.Prior to SDS-PAGE analysis (FIG. 34 ), His-tagged FAS-TE protein was bound to Ni-NTA beads to remove excess PEG reagent, and the beads were washed repeatedly with PBS buffer. For gel and mass spectrometric analysis, excess buffer was removed from the Ni-NTA beads and the protein was eluted with imidazole buffer. Specifically, 50 μl of Ni-NTA beads were added to 50 μl of the reaction mixture, incubated for 2 hours, and the reaction mixture and buffer were separated by centrifugation. Then, the beads were washed 3 times with 1 mL PBS; 70 μl of 250 mM imidazole buffer, 20 mM Tris,
Ni-NTA 비드 추출의 엄격성이 미반응 FAS-TE의 추출을 도울 수 있고, PEG화가 His-태그의 친화성을 낮출 수 있기 때문에 보고된 반응 수율은 더 낮은 추정치이다 (FAS-TE Leu2222PCL의 PEG화의 경우임; 데이터는 나타내지 않음). 그러나, 추출 이후의 PEG화된 물질의 검출은 PCL-2-AAP-PEG 연결이 안정적임을 보여주었다.The reported reaction yield is a lower estimate because the stringency of the Ni-NTA bead extraction can aid in the extraction of unreacted FAS-TE, and PEGylation can lower the affinity of the His-tag (PEG of FAS-TE Leu2222PCL). In case of anger; data not shown). However, detection of the PEGylated material after extraction showed that the PCL-2-AAP-PEG linkage was stable.
실시예 20: FGF21 에 혼입된 PCL 의 상이한 분자량의 2-아미노-아세토페논- PEG에 의한 유도체화 Example 20 : Derivatization of PCL incorporated in FGF21 with 2-amino-acetophenone- PEG of different molecular weight
본 실시예는 다양한 위치에서 PCL이 혼입된 FGF21이 2-AAP-PEG 시약에 의해 PEG화 될 수 있음을 보여준다. 본 실시예는 또한, PEG화된 FGF21 돌연변이체를 이온 교환 및 크기 배제 크로마토그래피의 조합에 의해 미반응 전장 FGF21 및 말단절단 FGF21로부터 분리할 수 있음을 보여준다.This example shows that FGF21, in which PCL is incorporated at various positions, can be PEGylated by 2-AAP-PEG reagent. This example also shows that PEGylated FGF21 mutants can be separated from unreacted full-length FGF21 and truncated FGF21 by a combination of ion exchange and size exclusion chromatography.
리신 84 위치에 PCL이 혼입된 FGF21을 에스케리키아 콜라이에서 발현시키고, 재폴딩시키고, 단백질을 TEV 프로테아제 절단하지 않는 것을 제외하고 실시예 10에 기재한 바와 같이 정제하였다. 단백질 저장액은 PBS 중 대략 6.6 mg/mL였고, 잔기 84에서 말단절단된 대략 20% FGF21 단백질을 함유하였다. 5 ㎕의 FGF21 저장액을 45 ㎕ 200 mM 나트륨 아세테이트 완충액 (pH 5.0) 및 0.5 ㎕의 100 mM TU3205-044 (2-AAP-PEG8, 실시예 20 참조) 저장 용액과 혼합하였다. 최종 몰비는 1 mM 2-AAP-PEG8 대 대략 30 μM FGF21이었다. 반응을 16 시간 동안 실온에서 진행시키고, 완료되게 하였다.FGF21 with PCL incorporated at
도 35A는 미반응 FGF21의 질량 스펙트럼을 나타내고, 도 35B는 반응 혼합물의 질량 스펙트럼을 나타낸다: PEG화 반응이 진행되어 완료되었고, 미반응 물질의 질량 보다 556 Da 더 큰 21792.4 Da의 단백질이 생성되었다. 이러한 질량 차이는 2-AAP-PEG8 변형된 FGF21에 대한 것으로 예상된다.Figure 35A shows the mass spectrum of unreacted FGF21, and Figure 35B shows the mass spectrum of the reaction mixture: the PEGylation reaction proceeded to completion and produced a protein of 21792.4 Da, which was 556 Da larger than the mass of the unreacted material. This mass difference is expected for the 2-AAP-PEG8 modified FGF21.
다양한 FGF21 PCL 돌연변이체 (실시예 10에서와 같이 발현되고 정제된 것)를 23 kDa 2-AAP-PEG (TU3205-052, 실시예 37-3 참조)로 PEG화하였다. 전형적으로 단백질 농도는 100 및 400 μM 사이였고, 23 kDa 2-AAP-PEG를 PBS 완충액 (pH 7.4) 중에 1 mM의 최종 농도로 첨가하였다. 반응물을 4℃에서 3 일 동안 인큐베이션하였다. 7 종의 대표적인 FGF21 PCL 돌연변이체와의 PEG화 반응의 SDS-PAGE를 도 36A에 나타내었고, 전장 (FL) 미반응 FGF21 및 말단절단 (TR) FGF21로부터 분리한 8 종의 정제된 PEG화된 FGF21 단백질을 도 36B에 나타내었다.Various FGF21 PCL mutants (expressed and purified as in Example 10) were PEGylated with 23 kDa 2-AAP-PEG (see TU3205-052, Example 37-3). Typically the protein concentration was between 100 and 400 μM, and 23 kDa 2-AAP-PEG was added in PBS buffer (pH 7.4) to a final concentration of 1 mM. The reaction was incubated at 4° C. for 3 days. SDS-PAGE of the PEGylation reaction with 7 representative FGF21 PCL mutants is shown in Fig.36A, and 8 types of purified PEGylated FGF21 proteins isolated from full-length (FL) unreacted FGF21 and truncated (TR) FGF21. Is shown in Figure 36B.
실시예 21: EPO 에 혼입된 PCL 의 2-아미노-아세토페논- PEG 에 의한 유도체화 Example 21: in the PCL incorporated into the EPO 2- amino-acetophenone-derivative by PEG Chemistry
PCL을 실시예 6에 기재한 바와 같이 마우스 EPO의 다양한 위치에 혼입시켰다. Ni-NTA 정제한 후, mEPO 구축물 (#6, #9 및 #10 돌연변이체 구축물)을 추가로 PBS 중에서 S-300 겔 여과 칼럼에 의해 정제하였다. 정제된 mEPO 단백질을 대략 1 mg/ml로 농축시켰다. 활성화된 23 kDa 2-AAP-PEG (TU3205-052, 실시예 37-3 참조)를 정제된 단백질에 1 mM로 첨가하고, 표 6에 나타낸 조건에서 인큐베이션하였다.PCL was incorporated into various locations of mouse EPO as described in Example 6. After Ni-NTA purification, mEPO constructs (#6, #9 and #10 mutant constructs) were further purified in PBS by S-300 gel filtration column. The purified mEPO protein was concentrated to approximately 1 mg/ml. Activated 23 kDa 2-AAP-PEG (TU3205-052, see Example 37-3) was added to the purified protein at 1 mM, and incubated under the conditions shown in Table 6.
모든 mEPO 구축물을 칼럼에 단량체로서 구동시켰다. 23 kDa 2-AAP-PEG를 정제된 단백질과 함께 인큐베이션한 경우, SDS-PAGE의 65 kDa 밴드로 이동하였으므로 mEPO는 단일 부위에서 PEG화되었다 (도 37). 반응의 효율은 조건에 따라 10% 내지 15%로 다양하였고, pH 7 및 pH 8.5 보다 pH 5.5에서 더 높은 정도로 PEG화 되었다. 또한, 4℃ 보다 높은 온도는 PEG화 정도를 상당히 증가시키지 않았다.All mEPO constructs were run as monomers on the column. When 23 kDa 2-AAP-PEG was incubated with the purified protein, mEPO was PEGylated at a single site because it moved to the 65 kDa band of SDS-PAGE (FIG. 37). The efficiency of the reaction varied from 10% to 15% depending on the conditions, and was PEGylated to a higher degree at pH 5.5 than at
실시예 22: hRBP 및 FAS - TE 에 혼입된 PCL 의 D- 만노사민에 의한 유도체화. Derivatization of the PCL only D- mixed in TE by industrial Min - hRBP and FAS: Example 22.
본 실시예는 아미노 당, D-만노사민이 2개의 표적 단백질에 혼입된 PCL에 직접 커플링됨을 보여준다. 본 실시예는 PCL 함유 단백질의 글리코실화에 대한 일반적인 반응식을 제시한다 (도 38).This example shows that the amino sugar, D-mannosamine, is directly coupled to PCL incorporated into two target proteins. This example presents a general reaction scheme for the glycosylation of PCL-containing proteins (Figure 38).
인간 RBP4 Phe122PCL (돌연변이체 #6)을 실시예 2에서와 같이 HEK293F 세포에서 제조하였다. 10 ㎕의 170 μM 단백질 저장액을 89 ㎕ 10x PBS 완충액 (pH 7.5)에 첨가하고, 1 ㎕의 1 M D-만노사민과 혼합하였다. hRBP4 Phe122PCL 단백질 (17 μM)을 10 mM D-만노사민과 14 동안 실온에서 인큐베이션한 후 (어떤 반응도 관찰되지 않음), 48 시간 동안 37℃에서 인큐베이션하였다. 도 39는 37℃ 인큐베이션 기간 이후의 반응 혼합물의 질량 스펙트럼을 나타낸다. 23165.6 Da에서의 미반응 hRBP4의 예상 피크 (예상치 23166 Da)에 더하여, D-만노사민 부가물에 대한 추정상 할당 피크가 23300.0 Da에서 관찰되었다. 164.4 Da의 질량 증가는 근접하기는 해도 도 38에 도시된 추정 반응 생성물에 대해 예상되는 증가치 161.1 Da과 동일하지는 않았다. 또한, 종으로 분해된 단백질의 일부가 21007.8 Da에서 검출되었다.Human RBP4 Phe122PCL (mutant #6) was prepared in HEK293F cells as in Example 2. 10 μl of 170 μM protein stock solution was added to 89
제2 실시예에서는, 에스케리키아 콜라이에서 발현시키고 실시예 8에서와 같이 정제한 위치 2222에서 PCL에 의해 변형된 FAS-TE (11 μM)를 10 mM D-만노사민과 함께 72 시간 동안 실온에서 인큐베이션하였다. 미반응 샘플의 질량 스펙트럼을 도 40A에 도시하였고, 이는 미반응 단백질에 상응하는 33250.4 Da에서 예상 신호를 함유하였으며, 33360.4 Da에서의 피크는 샘플 내의 단백질 오염에서 기인한다. 반응 혼합물의 질량 분광측정법은 미반응 단백질에 상응하는 33250.4 Da에서 예상 신호를 나타내었다 (도 40B). 추가의 피크가 출발 물질보다 158.4 Da 더 큰 33408.8 Da에서 발견되었는데, 이는 도 37에 나타낸 반응 생성물의 피크인 것으로 짐작된다. 2개의 샘플을 비교한 결과는, FAS-TE Leu2222PCL이 상기 반응 조건하에서 D-만노사민 부가물로 대략 50%의 수율로 전환되었음을 시사한다.In the second example, FAS-TE (11 μM) modified by PCL at
실시예 23: FGF21 의 PCL 매개 공유 가교 Example 23: PCL-mediated cross-linking of FGF21 shares
본 실시예는 단백질이 2 관능성 PCL-특이적 크로스 링커를 통해 공유적으로 이량체화될 수 있음을 보여준다. 리신 84 위치에 PCL이 혼입된 FGF21을 에스케리키아 콜라이에서 발현시키고, 재폴딩시키고, 단백질을 TEV 프로테아제 절단하지 않는 것을 제외하고 실시예 10에 기재한 바와 같이 정제하였다. FGF21 Lys84PCL을 크로스 링커 TU633-010로 유도체화시켰다 (도 43A; 합성에 대해서는 실시예 37-4 참조). 단백질 저장액은 PBS 중 대략 6.6 mg/mL였고, 잔기 84에서 절단된 대략 20% FGF21 단백질을 함유하였다. 5 ㎕의 FGF21 저장액을 45 ㎕ 200 mM 나트륨 아세테이트 완충액 (pH 5.0)과 혼합하였다. 0.1 ㎕의 크로스 링커 TU633-010 (DMSO 중)의 5 mM 저장 용액을 10 μM 크로스 링커 및 대략 30 μM FGF21의 최종 농도로 첨가하였다. 유사하게, 50 ㎕의 PBS 중의 FGF21 저장 용액 (pH 7.4)을 100 μM의 크로스 링커 및 대략 300 μM의 FGF21 농도에서 1 ㎕의 5 mM TU633-010와 반응시켰다. 200 mM 나트륨 아세테이트 완충액으로 희석한 FGF21 샘플을 대조군으로서 제조하고, 동일하게 처리하였다. 실온에서 16 시간 후에, 분취량의 반응물 및 대조군 샘플을 정제하지 않고 질량 분광측정법 및 SDS-PAGE 분석에 의해 분석하였다. pH 5.0 샘플에 대해 얻은 질량 스펙트럼 (도 43B)은 공유 이량체의 예상 질량 (43037.2 Da; 모든 FGF21 피크의 상대 강도 53%), 한쪽 말단에 크로스 링커가 부착된 FGF21의 예상 질량 (21820.0 Da; 33%) 및 미반응 FGF21 (21235.2 Da; 14%)에서 분명하게 피크를 나타내었다. pH 7.4 샘플의 경우, 공유 이량체는 검출되지 않았고; 28%의 FGF21은 크로스 링커와 단일 반응한 반면, 대부분의 단백질은 반응하지 않았다 (데이터는 나타내지 않음).This example shows that proteins can be covalently dimerized through a bifunctional PCL-specific cross linker. FGF21 with PCL incorporated at
단백질 농도에 대한 반응물의 조정은 공유 이량체의 수율을 더욱 증가시켰다. 구체적으로, 10 ㎕의 FGF21 저장액을 90 ㎕의 200 mM 나트륨 아세테이트 완충액 (pH 5.0)과 혼합시켰다. 0.3 ㎕의 크로스 링커 TU633-010 (DMSO 중)의 5 mM 저장 용액을 15 μM 크로스 링커 및 대략 30 μM FGF21의 최종 농도로 첨가하였다. 하나의 샘플을 4 일 동안 실온에서 인큐베이션하고, 제2 샘플은 4℃에서 인큐베이션하였다. 또한 10x PBS (pH 7.5) 중의 샘플을 제조하고, 동일하게 인큐베이션하였다. 실온에서 인큐베이션한 pH 5.0 샘플의 경우, 43034.4 Da의 예상 질량에서의 공유 FGF21 이량체의 피크가 우세한 종이었고, 미반응 FGF21은 21233.6 Da에서 검출되지 않았다. 한쪽 말단에 크로스 링커가 부착되어 변형된 일부 FGF21이 21818.4 Da에서 검출되었다 (도 44A). 반응은 대략 19%의 FGF21이 미반응인채로 남아 pH 5.0 및 4℃에서와 동일한 정도로 진전되지 않았고; 40%는 한쪽 말단에 크로스 링커가 부착되어 변형된 것인 한편, 대략 41%의 질량 스펙트럼 피크 강도는 공유 이량체의 그것이었다. pH 7.5에서의 반응은 SDS-PAGE에 의해 표시되는 어떤 공유 이량체도 생성하지 않았다 (도 44B).Adjustment of the reactants to protein concentration further increased the yield of covalent dimers. Specifically, 10 μl of FGF21 stock solution was mixed with 90 μl of 200 mM sodium acetate buffer (pH 5.0). A 5 mM stock solution of 0.3 μl of cross linker TU633-010 (in DMSO) was added to a final concentration of 15 μM cross linker and approximately 30 μM FGF21. One sample was incubated at room temperature for 4 days, and the second sample was incubated at 4°C. Further, a sample in 10x PBS (pH 7.5) was prepared and incubated in the same manner. For the pH 5.0 sample incubated at room temperature, the peak of the covalent FGF21 dimer at the expected mass of 43034.4 Da was the dominant species, and unreacted FGF21 was not detected at 21233.6 Da. Some FGF21 modified by attaching a cross linker to one end was detected at 21818.4 Da (FIG. 44A). The reaction did not progress to the same extent as at pH 5.0 and 4° C., leaving approximately 19% of FGF21 unreacted; 40% was modified by attaching a cross linker at one end, while the mass spectrum peak intensity of approximately 41% was that of the covalent dimer. The reaction at pH 7.5 did not produce any covalent dimers as indicated by SDS-PAGE (FIG. 44B ).
다른 다양한 분자에 의한 피롤린 - 카르복시 -리신 ( PCL ) 함유 단백질의 부위 특이적 변형.Site-specific modification of pyrroline - carboxy -lysine ( PCL ) containing proteins by various other molecules.
다른 실시양태에서, PCL-함유 단백질에 대한 본원에서 제공하는 2-아미노벤즈알데히드 (ABA) 접합체 및 2-아미노아세토페논 (AAP) 접합체의 커플링은 10 x 인산염 완충 염수 (PBS) (pH 7.0) 중에서 25℃에서 행하였다. 접합 반응은 10 μM PCL-함유 단백질 및 100 μM ABA/AAP 접합체의 첨가에 의해 개시되었다. 단백질 접합체의 완전한 형성은 전기분무 이온화-질량 분광측정법 (ESI-MS) 또는 매트릭스 보조 레이저 탈착/이온화 (MALDI)에 의해 확인하였다. ABA/AAP DNA 접합체의 커플링은 NuPAGE 4-12% 비스-트리스 겔 (인비트로겐, 캘리포니아주 칼스배드 소재)을 이용한 겔 이동 검정에 의해 분석하였다. 정량적 커플링 이후에, 단백질 접합체를 10 mM 인산나트륨 완충액 (pH 7.5)으로 투석하고, 컷오프가 10 kDa인 아미콘 울트라-4(Amicon Ultra-4) 원심분리 필터 유닛 (밀리포어 코퍼레이션(Millipore Corporation), 메사추세츠주 베드포드 소재)을 이용하여 100 μM로 농축시켰다. 이어서 새롭게 제조한 200 mM NaCNBH3 용액 (10 mM 인산염 완충액에 용해시킨 것, pH 7.5)을 최종 농도 20 mM로 첨가하였다. 환원 반응이 2 - 4 시간 동안 25℃에서 진행되도록 한 후, 6 부피의 10 mM 인산나트륨 완충액 (pH 7.5)을 첨가하여 반응을 켄칭시켰다. NAP-5 칼럼 또는 PD10 칼럼 (GE 헬스케어, 뉴저지주 피스카타웨이 소재)을 이용하여, 환원된 단백질 접합체를 목적하는 완충액으로 마지막 완충액 교환하였다. 그러한 2-아미노벤즈알데히드 (ABA) 접합체 및 2-아미노아세토페논 (AAP) 접합체의 다양한 단백질에의 비제한적인 커플링의 예를 이하에서 제공한다.In another embodiment, the coupling of a 2-aminobenzaldehyde (ABA) conjugate provided herein and a 2-aminoacetophenone (AAP) conjugate to a PCL-containing protein is in 10 x phosphate buffered saline (PBS) (pH 7.0). It was carried out at 25 degreeC. The conjugation reaction was initiated by the addition of 10 μM PCL-containing protein and 100 μM ABA/AAP conjugate. The complete formation of the protein conjugate was confirmed by electrospray ionization-mass spectrometry (ESI-MS) or matrix assisted laser desorption/ionization (MALDI). Coupling of ABA/AAP DNA conjugates was analyzed by gel transfer assay using a NuPAGE 4-12% Bis-Tris gel (Invitrogen, Carlsbad, CA). After quantitative coupling, the protein conjugate is dialyzed with 10 mM sodium phosphate buffer (pH 7.5) and an Amicon Ultra-4 centrifugal filter unit with a cutoff of 10 kDa (Millipore Corporation) , And concentrated to 100 μM using Bedford, MA). Subsequently, a newly prepared 200 mM NaCNBH 3 solution (dissolved in 10 mM phosphate buffer, pH 7.5) was added to a final concentration of 20 mM. After allowing the reduction reaction to proceed at 25° C. for 2 to 4 hours, 6 volumes of 10 mM sodium phosphate buffer (pH 7.5) were added to quench the reaction. Using a NAP-5 column or a PD10 column (GE Healthcare, Piscataway, NJ), the reduced protein conjugate was finally buffer exchanged with the desired buffer. Examples of non-limiting coupling of such 2-aminobenzaldehyde (ABA) conjugates and 2-aminoacetophenone (AAP) conjugates to various proteins are provided below.
실시예 24: 비오틴 시약의 커플링 Example 24 : Coupling of biotin reagent
비오틴이 1개 이상의 PCL 잔기가 내부에 혼입된 단백질에 커플링될 수 있는지 확인하기 위해, ABA-비오틴 시약 (X3626-140, 실시예 40)을 이용하여 mEGF-Tyr10PCL (실시예 12 참조)을 비오틴과 접합시켰다. 비오틴의 커플링은 다음과 같이, 500 μM ABA-비오틴을 인산염 완충 염수 (PBS, pH 7.0) 및 0.5% (v/v) DMSO 중의 10 μM mEGF-Tyr10PCL에 첨가하고, 25℃에서 16 시간 동안 반응시켜 수행하였다. 비오틴 접합체의 완전한 형성은 ESI-MS에 의해 확인하였다 (도 47A; 미반응 단백질에 대해 예상되는 질량 = 7296; 커플링된 단백질에 대해 예상되는 질량 = 7902).In order to confirm whether biotin can be coupled to a protein in which one or more PCL residues are incorporated therein, mEGF-Tyr10PCL (see Example 12) was biotinylated using an ABA-biotin reagent (X3626-140, Example 40). And conjugated. Coupling of biotin is as follows, 500 μM ABA-biotin is added to 10 μM mEGF-Tyr10PCL in phosphate buffered saline (PBS, pH 7.0) and 0.5% (v/v) DMSO and reacted at 25° C. for 16 hours. Was carried out. Complete formation of the biotin conjugate was confirmed by ESI-MS (Figure 47A; expected mass for unreacted protein = 7296; expected mass for coupled protein = 7902).
정량적 커플링 이후에, 새롭게 제조한 200 mM NaCNBH3 용액 (PBS에 용해시킨 것, pH 7.0)을 최종 농도 20 mM로 첨가하였다. 환원 반응이 3 시간 동안 25℃에서 진행되도록 한 후, 6 부피의 PBS (pH 7.0)를 첨가하여 반응을 켄칭시켰다. 과량의 NaCNBH3은 슬라이드-A-라이저 투석 카세트 (3,500 Da 분자량 컷오프, 피어스)를 이용하여 4℃에서 PBS (pH 7.0)에 대해 투석하여 제거하였다. 환원된 비오틴 접합체를 컷오프가 3.5 kDa인 아미콘 울트라-4 원심분리 필터 유닛 (밀리포어 코퍼레이션)을 이용하여 농축시켰다. NuPAGE 4-12% 비스-트리스 겔 (인비트로겐)을 통해 전기영동한 후, 아이블롯(iBlot) 겔 이송 시스템 (인비트로겐)을 이용하여 비오티닐화 단백질을 폴리비닐리덴 디플루오리드 (PVDF) 막으로 옮겼다. 이어서, 비오틴 접합체를 염소 항-비오틴 항체에 접합된 양고추냉이 퍼옥시다제 (HRP) (1:100 희석, 셀 시그날링 테크놀로지스(Cell Signaling Technologies))로 검출하고, 하이퍼필름(Hyperfilm) ECL (GE 헬스케어)을 이용하여 시각화하였다 (도 47B). 커플링되지 않은 mEGF-Tyr10PCL 및 플루오레세인-접합된 mEGF-Tyr10PCL은 음성 대조군으로 제공하였다. 레인 1, 20 pmol mEGF-Tyr10PCL-ABA-비오틴 접합체; 레인 2, 8 pmol mEGF-Tyr10PCL-ABA-비오틴 접합체; 레인 3, 2 nmol mEGF-Y10PCL; 레인 4, 20 pmol mEGF-Tyr10PCL-ABA-플루오레세인 접합체.After quantitative coupling, a freshly prepared 200 mM NaCNBH 3 solution (dissolved in PBS, pH 7.0) was added to a final concentration of 20 mM. After allowing the reduction reaction to proceed at 25° C. for 3 hours, 6 volumes of PBS (pH 7.0) were added to quench the reaction. Excess NaCNBH 3 was removed by dialysis against PBS (pH 7.0) at 4° C. using a Slide-A-Riser dialysis cassette (3,500 Da molecular weight cutoff, Pierce). The reduced biotin conjugate was concentrated using an Amicon Ultra-4 centrifugal filter unit (Millipore Corporation) with a cutoff of 3.5 kDa. After electrophoresis through NuPAGE 4-12% Bis-Tris gel (Invitrogen), biotinylated protein was transferred to a polyvinylidene difluoride (PVDF) membrane using an iBlot gel transfer system (Invitrogen). Moved to. Subsequently, the biotin conjugate was detected with horseradish peroxidase (HRP) conjugated to goat anti-biotin antibody (1:100 dilution, Cell Signaling Technologies), and Hyperfilm ECL (GE Health Care) was used (Fig. 47B). Uncoupled mEGF-Tyr10PCL and fluorescein-conjugated mEGF-Tyr10PCL served as negative controls.
실시예 25: 형광 분자 커플링 Example 25 : Fluorescent molecule coupling
형광 분자가 1개 이상의 PCL 잔기가 내부에 혼입된 단백질에 커플링될 수 있는지 확인하기 위해, ABA-플루오레세인 시약 (3793-050, 실시예 42 참조)을 이용하여 mEGF-Tyr10PCL (실시예 12 참조)을 플루오레세인과 접합시켰다. 플루오레세인의 커플링은 다음과 같이, 1 mM의 ABA-플루오레세인을 인산염 완충 염수 (PBS, pH 7.0) 및 0.5% (v/v) DMSO 중의 10 μM mEGF-Tyr10PCL에 첨가하고, 25℃에서 16 시간 동안 반응시켜 수행하였다. 플루오레세인 접합체의 형성은 ESI-MS에 의해 확인하였다 (도 47C; 커플링되지 않은 단백질에 대해 예상되는 질량 = 7296; 커플링된 단백질에 대해 예상되는 질량 = 8062).In order to confirm whether a fluorescent molecule can be coupled to a protein in which one or more PCL residues are incorporated therein, mEGF-Tyr10PCL (Example 12) using an ABA-fluorescein reagent (3793-050, see Example 42). Reference) was conjugated with fluorescein. Coupling of fluorescein is as follows, 1 mM of ABA-fluorescein is added to 10 μM mEGF-Tyr10PCL in phosphate buffered saline (PBS, pH 7.0) and 0.5% (v/v) DMSO, and at 25° C. It was carried out by reacting at for 16 hours. The formation of the fluorescein conjugate was confirmed by ESI-MS (Figure 47C; expected mass for uncoupled protein = 7296; expected mass for coupled protein = 8062).
플루오레세인 접합체를 20 mM NaCNBH3으로 3 시간 동안 25℃에서 환원시켰다. 6 부피의 PBS (pH 7.0)로 환원 반응을 켄칭시킨 후, 잔여 NaCNBH3은 슬라이드-A-라이저 투석 카세트 (3,500 Da 분자량 컷오프)를 이용하여 4℃에서 PBS (pH 7.0)에 대해 투석하여 제거하였다. 이어서, 컷오프가 3.5 kDa인 아미콘 울트라-4 원심분리 필터 유닛을 이용하여 접합체를 1 μM로 농축시켰다. 스펙트라맥스 플러스(SpectraMax Plus) (몰레큘라 디바이시즈(Molecular Devices))를 이용하여 1 μM mEGF-Tyr10PCL-ABA-플루오레세인 및 10 μM ABA-플루오레세인의 350 - 700 nm 범위에서의 흡수 스펙트럼을 수득하였다. 이들은 둘 모두 500 nm에서 최대 흡수를 나타내었다. The fluorescein conjugate was reduced with 20 mM NaCNBH 3 for 3 hours at 25°C. After quenching the reduction reaction with 6 volumes of PBS (pH 7.0), the remaining NaCNBH 3 was removed by dialysis against PBS (pH 7.0) at 4°C using a slide-A-riser dialysis cassette (3,500 Da molecular weight cutoff). . The conjugate was then concentrated to 1 μM using an Amicon Ultra-4 centrifugal filter unit with a cutoff of 3.5 kDa. Absorption spectra in the 350-700 nm range of 1 μM mEGF-Tyr10PCL-ABA-fluorescein and 10 μM ABA-fluorescein were obtained using SpectraMax Plus (Molecular Devices). Obtained. They both showed maximum absorption at 500 nm.
이어서, 스펙트라맥스 제미니(GEMINI) 형광계 (몰레큘라 디바이시즈) 상에서 형광 스펙트럼을 판독하였다. 1 μM mEGF-Tyr10PCL-ABA-플루오레세인 및 10 μM ABA-플루오레세인에 대한 방출 스펙트럼은, 여기 파장을 490 nm으로 유지하는 한편 방출 파장은 510 nm에서 750 nm으로 2 nm의 단계 크기로 스캐닝하여 수득하였다. 이들은 둘 모두 522 nm에서 최대 방출을 나타내었다. 또한, 1 μM mEGF-Tyr10PCL-ABA-플루오레세인 및 10 μM ABA-플루오레세인에 대한 여기 스펙트럼은, 방출 파장을 522 nm으로 유지하는 한편 여기 파장은 300 nm에서 510 nm으로 2 nm의 단계 크기로 스캐닝하여 수득하였다. Subsequently, the fluorescence spectrum was read on a Spectramax Gemini (GEMINI) fluorescence meter (Molecular Devices). Emission spectra for 1 μM mEGF-Tyr10PCL-ABA-fluorescein and 10 μM ABA-fluorescein, while keeping the excitation wavelength at 490 nm while scanning the emission wavelength from 510 nm to 750 nm with a step size of 2 nm. Obtained by. They both showed maximum emission at 522 nm. In addition, the excitation spectra for 1 μM mEGF-Tyr10PCL-ABA-fluorescein and 10 μM ABA-fluorescein keep the emission wavelength at 522 nm while the excitation wavelength is from 300 nm to 510 nm with a step size of 2 nm. It was obtained by scanning with.
실시예 26: 폴리사카라이드 커플링 Example 26 : Polysaccharide coupling
폴리사카라이드가 1개 이상의 PCL 잔기가 내부에 혼입된 단백질에 커플링될 수 있는지 확인하기 위해, ABA-디사카라이드 (3793-050; 실시예 42, MW 546.52)를 이용하여 mEGF-Tyr10PCL (실시예 12 참조)을 디사카라이드와 접합시켰다. 커플링 반응은 1 mM ABA-디사카라이드를 PBS 및 1% (v/v) DMSO (pH 7.0) 중의 10 μM mEGF-Tyr10PCL 돌연변이체 단백질에 첨가하여 수행하였다. 반응이 실온에서 16 시간 동안 진행되도록 하고, ESI-MS에 의해 분석하였다 (도 47D). 질량 스펙트럼은 접합된 단백질에 대해 예상되는 질량 (7825 Da)에서 주요 피크를 나타내었다. 커플링되지 않은 단백질의 질량은 7296 Da였다.In order to confirm that the polysaccharide can be coupled to the protein in which one or more PCL residues are incorporated therein, mEGF-Tyr10PCL (implemented) using ABA-disaccharide (3793-050; Example 42, MW 546.52). Example 12) was conjugated with a disaccharide. The coupling reaction was performed by adding 1 mM ABA-disaccharide to 10 μM mEGF-Tyr10PCL mutant protein in PBS and 1% (v/v) DMSO (pH 7.0). The reaction was allowed to proceed at room temperature for 16 hours and analyzed by ESI-MS (Figure 47D). The mass spectrum showed a major peak at the expected mass (7825 Da) for the conjugated protein. The mass of the uncoupled protein was 7296 Da.
실시예 27: 면역 조절제: 모노- 니트로페닐 합텐 접합체의 커플링 Example 27 : Immune modulator: mono- nitrophenyl Hapten conjugate coupling
면역 조절제가 1개 이상의 PCL 잔기가 내부에 혼입된 단백질에 커플링될 수 있는지 확인하기 위해, ABA-모노-니트로페닐 합텐 시약 (3793-001, 실시예 38-8)을 이용하여 mTNF-Gln21PCL 및 mEGF-Tyr10PCL (실시예 11 및 12 참조)을 모노-니트로페닐 합텐과 접합시켰다. 모노-니트로페닐 합텐 접합체의 커플링은 상기한 바와 같이 수행하였다. 도 48A는 mTNF-Gln21PCL에 접합된 3793-001의 ESI 질량 스펙트럼이고 (커플링되지 않은 단백질에 대해 예상되는 질량 = 19275; 커플링된 단백질에 대해 예상되는 질량 = 19614), 도 48B는 mEGF-Tyr10PCL에 접합된 3793-001의 ESI 질량 스펙트럼이다 (커플링되지 않은 단백질에 대해 예상되는 질량 = 7296; 커플링된 단백질에 대해 예상되는 질량 = 7635).In order to confirm whether the immunomodulatory agent can be coupled to the protein incorporated therein at least one PCL residue, mTNF-Gln21PCL and mTNF-Gln21PCL using ABA-mono-nitrophenyl hapten reagent (3793-001, Example 38-8) and mEGF-Tyr10PCL (see Examples 11 and 12) was conjugated with mono-nitrophenyl hapten. Coupling of the mono-nitrophenyl hapten conjugate was performed as described above. Figure 48A is the ESI mass spectrum of 3793-001 conjugated to mTNF-Gln21PCL (expected mass for uncoupled protein = 19275; expected mass for coupled protein = 19614), Figure 48B is mEGF-Tyr10PCL Is the ESI mass spectrum of 3793-001 conjugated to (expected mass for uncoupled protein = 7296; expected mass for coupled protein = 7635).
실시예 28: 면역 조절제: 디- 니트로페닐 합텐 접합체의 커플링 Example 28 : Immune modulator: di- nitrophenyl Hapten conjugate coupling
면역 조절제가 1개 이상의 PCL 잔기가 내부에 혼입된 단백질에 커플링될 수 있는지 확인하기 위해, 디-니트로페닐 합텐 (TU3627-088, 실시예 38-7)을 이용하여 mTNF-Gln21PCL 및 mEGF-Tyr10PCL (실시예 11 및 12 참조)을 디-니트로페닐 합텐과 접합시켰다. 디-니트로페닐 합텐 접합체의 커플링은 상기한 바와 같이 수행하였다. 도 48C는 mTNF-Gln21PCL에 접합된 TU3627-088의 ESI 질량 스펙트럼이고 (커플링되지 않은 단백질에 대해 예상되는 질량 = 19275; 커플링된 단백질에 대해 예상되는 질량 = 19688), 도 48D는 mEGF-Tyr10PCL에 접합된 TU3627-088의 ESI 질량 스펙트럼이다 (커플링되지 않은 단백질에 대해 예상되는 질량 = 7296; 커플링된 단백질에 대해 예상되는 질량 = 7709).In order to confirm whether the immunomodulatory agent can be coupled to the protein incorporated therein at least one PCL residue, mTNF-Gln21PCL and mEGF-Tyr10PCL using di-nitrophenyl hapten (TU3627-088, Example 38-7). (See Examples 11 and 12) was conjugated with di-nitrophenyl hapten. Coupling of the di-nitrophenyl hapten conjugate was performed as described above. Figure 48C is the ESI mass spectrum of TU3627-088 conjugated to mTNF-Gln21PCL (expected mass for uncoupled protein = 19275; expected mass for coupled protein = 19688), Figure 48D is mEGF-Tyr10PCL Is the ESI mass spectrum of TU3627-088 conjugated to (expected mass for uncoupled protein = 7296; expected mass for coupled protein = 7709).
실시예 29: 면역 조절제: TLR7 효능제의 커플링 Example 29 : Immune Modulator: Coupling of TLR7 agonists
면역 조절제가 1개 이상의 PCL 잔기가 내부에 혼입된 단백질에 커플링될 수 있는지 확인하기 위해, ABATLR7 효능제 시약 (X3678-114; 실시예 38-3 참조)을 이용하여 mEGF-Y10PCL (실시예 12 참조)을 TLR7 효능제와 접합시켰다. 커플링 반응은 100 μM ABA-TLR7 효능제를 200 mM 나트륨 아세테이트 완충액 및 1% (v/v) DMSO (pH 4.5) 중의 10 μM mEGF-Y10PCL 돌연변이체 단백질에 첨가하여 수행하였다. 반응이 실온에서 16 시간 동안 진행되도록 하고, ESI-MS에 의해 분석하였다 (도 49A). 질량 스펙트럼은 접합된 단백질에 대해 예상되는 질량 (8763 Da)에서 주요 피크를 나타내었고, 커플링되지 않은 단백질의 질량은 7296 Da였다.In order to confirm that the immunomodulatory agent can be coupled to a protein in which one or more PCL residues are incorporated therein, the ABATLR7 agonist reagent (X3678-114; see Example 38-3) was used to use mEGF-Y10PCL (Example 12). Reference) was conjugated with a TLR7 agonist. Coupling reactions were performed by adding 100 μM ABA-TLR7 agonist to 10 μM mEGF-Y10PCL mutant protein in 200 mM sodium acetate buffer and 1% (v/v) DMSO (pH 4.5). The reaction was allowed to proceed at room temperature for 16 hours and analyzed by ESI-MS (Figure 49A). The mass spectrum showed a major peak at the expected mass (8763 Da) for the conjugated protein, and the mass of the uncoupled protein was 7296 Da.
실시예 30: 면역 조절제: PADRE 펩티드의 커플링 Example 30 : Immune Modulator: Coupling of PADRE Peptide
면역 조절제 및 펩티드가 1개 이상의 PCL 잔기가 내부에 혼입된 단백질에 커플링될 수 있는지 확인하기 위해, mTNF-Gln21PCL 및 mEGF-Tyr10PCL (실시예 11 및 12 참조)을 PADRE 펩티드와 접합시켰다. PADRE 펩티드의 커플링은 "다른 다양한 분자에 의한 피롤린-카르복시-리신 (PCL) 함유 단백질의 부위 특이적 변형"이라는 부제 직후의 단락에서 기재한 바와 같이 수행하였다. 도 50A는 pH 5.0에서 PX2-PADRE (3465-143; 실시예 38-11 참조)와 접합된 mTNF-Gln21PCL의 MALDI-TOF 질량 분광측정 분석이고, 도 50B는 pH 7.5에서 PX2-PADRE와 접합된 mTNF-Q21PCL의 MALDI-TOF 질량 분광측정 분석이다. 커플링되지 않은 단백질에 대해 예상되는 질량은 19275 Da이고, 커플링된 단백질에 대해 예상되는 질량은 20842 Da이다. 도 50C는 BHA-exPADRE (3647-104; 실시예 38-10 참조)와 접합된 mTNF-Gln21PCL의 ESI 질량 분광측정 분석이다. 이때 커플링되지 않은 단백질에 대해 예상되는 질량은 19275 Da이고, 커플링된 단백질에 대해 예상되는 질량은 21317 Da이다. 또한, 도 51은 BHA-exPADRE의 mEGF-Tyr10PCL에의 커플링을 보여주는 ESI 질량 스펙트럼이다 (커플링되지 않은 단백질에 대해 예상되는 질량 7296 Da; 커플링된 단백질에 대해 예상되는 질량 9338 Da).In order to confirm that the immunomodulator and peptide can be coupled to the protein with one or more PCL residues incorporated therein, mTNF-Gln21PCL and mEGF-Tyr10PCL (see Examples 11 and 12) were conjugated with the PADRE peptide. Coupling of the PADRE peptide was carried out as described in the paragraph immediately following the subheading "Site-specific modification of pyrroline-carboxy-lysine (PCL) containing proteins by various other molecules". Figure 50A is a MALDI-TOF mass spectrometric analysis of mTNF-Gln21PCL conjugated with PX2-PADRE (3465-143; see Example 38-11) at pH 5.0, and Figure 50B is mTNF conjugated with PX2-PADRE at pH 7.5. MALDI-TOF mass spectrometric analysis of -Q21PCL. The expected mass for the uncoupled protein is 19275 Da, and the expected mass for the coupled protein is 20842 Da. 50C is an ESI mass spectrometric analysis of mTNF-Gln21PCL conjugated with BHA-exPADRE (3647-104; see Example 38-10). At this time, the expected mass for the uncoupled protein is 19275 Da, and the expected mass for the coupled protein is 21317 Da. In addition, Figure 51 is an ESI mass spectrum showing the coupling of BHA-exPADRE to mEGF-Tyr10PCL (expected mass 7296 Da for uncoupled protein; expected mass 9338 Da for coupled protein).
실시예 31: 면역 조절제: 인지질의 커플링 Example 31 : Immunomodulator: Coupling of phospholipids
면역 조절제 및 인지질이 1개 이상의 PCL 잔기가 내부에 혼입된 단백질에 커플링될 수 있는지 확인하기 위해, ABA 인지질 시약 (TU3627-092; 실시예 43-1)을 이용하여 mEGF-Tyr10PCL (실시예 12 참조)을 인지질 (DOPE)과 접합시켰다. mEGF-Tyr10PCL (MW = 7296 Da)에 대한 ABA-DOPE의 커플링은 20 mM HEPES (pH 7.0) 및 1% (v/v) DMSO 중에서 25℃에서 16 시간 동안 수행하였다. 접합 반응은 10 μM mEGF-Tyr10PCL 및 100 μM ABA-DOPE를 첨가하는 것에 의해 개시되었다. 단백질 접합체의 형성은 mEGF-Y10PCL에 대한 DOPE의 접합을 전기분무 이온화-질량 분광측정 (ESI-MS) 분석하는 것에 의해 확인하였다 (도 49B; 커플링되지 않은 단백질에 대해 예상되는 질량 = 7296; 접합된 단백질에 대해 예상되는 질량 = 8227 Da). In order to confirm whether the immunomodulator and phospholipid can be coupled to a protein in which one or more PCL residues are incorporated therein, mEGF-Tyr10PCL (Example 12) was used using ABA phospholipid reagent (TU3627-092; Example 43-1). Reference) was conjugated with phospholipid (DOPE). Coupling of ABA-DOPE to mEGF-Tyr10PCL (MW = 7296 Da) was performed in 20 mM HEPES (pH 7.0) and 1% (v/v) DMSO at 25° C. for 16 hours. The conjugation reaction was initiated by adding 10 μM mEGF-Tyr10PCL and 100 μM ABA-DOPE. The formation of the protein conjugate was confirmed by electrospray ionization-mass spectrometry (ESI-MS) analysis of the conjugation of DOPE to mEGF-Y10PCL (FIG. 49B; expected mass for uncoupled protein = 7296; conjugation) Mass = 8227 Da).
실시예 32: 올리고뉴클레오티드의 단백질: CpG 펩티드에의 커플링 Example 32 : Protein of Oligonucleotide: Coupling to CpG Peptide
올리고뉴클레오티드 및 CpG 면역 조절제가 1개 이상의 PCL 잔기가 내부에 혼입된 단백질에 커플링될 수 있는지 확인하기 위해, CpG 시약 BHA-BG1 (3647-057; 실시예 38-12 참조) 또는 CpG 시약 BHA-BG2 (3597-167; 실시예 38-14 참조)를 사용하여 mTNF-Gln21PCL 및 mEGF-Tyr10PCL (실시예 11 및 12 참조)을 CpG 올리고뉴클레오티드와 접합시켰다. CpG 올리고뉴클레오티드의 커플링은 "다른 다양한 분자에 의한 피롤린-카르복시-리신 (PCL) 함유 단백질의 부위 특이적 변형"이라는 부제 직후의 단락에서 기재한 바와 같이 수행하였다. mTNF-Gln21PCL (19.3 kDa)에 대한 BHA-BG1 (7.4 kDa) 및 BHA-BG2 (7.4 kDa)의 커플링은 겔 이동 검정에 의해 확인하였고 (도 52A), mEGF-Tyr10PCL (7.2 kDa)에 대한 BHA-BG2 (7.4 kDa)의 커플링도 또한 겔 이동 검정에 의해 확인하였다 (도 52B).To ensure that oligonucleotides and CpG immunomodulatory agents can be coupled to proteins in which one or more PCL residues are incorporated, CpG reagent BHA-BG1 (3647-057; see Examples 38-12) or CpG reagent BHA- BG2 (3597-167; see Examples 38-14) was used to conjugate mTNF-Gln21PCL and mEGF-Tyr10PCL (see Examples 11 and 12) with CpG oligonucleotides. Coupling of CpG oligonucleotides was carried out as described in the paragraph immediately following the subheading “Site-specific modification of pyrroline-carboxy-lysine (PCL) containing proteins by various other molecules”. The coupling of BHA-BG1 (7.4 kDa) and BHA-BG2 (7.4 kDa) to mTNF-Gln21PCL (19.3 kDa) was confirmed by gel shift assay (Figure 52A), and BHA to mEGF-Tyr10PCL (7.2 kDa). The coupling of -BG2 (7.4 kDa) was also confirmed by gel shift assay (Figure 52B).
실시예Example 33: 반응성 피롤리신 유사체의 합성 33: Synthesis of reactive pyrrolysine analogs
실시예 33-1: (S)-2-아미노-6-(3- 옥소부탄아미도 ) 헥산산 히드로클로라이드 (TU3000-016)의 합성 Example 33-1: (S)-2-amino-6-(3- oxobutanamido ) hexanoic acid Hydrochloride Synthesis of (TU3000-016)
문헌 [Bing Hao et al., ChemBio 2004, 11, 1317-24] & 그의 자료에 기재된 절차에 따르되, Cbz 기의 제거는 HCl 염이 수득되도록 HCl의 존재하에 행하여 (S)-메틸 6-아미노-2-(2,2,2-트리플루오로아세트아미도)헥사노에이트 히드로클로라이드 (TU2982-126)를 제조하였다.According to the procedure described in the literature [Bing Hao et al.,
40 mL 유리 바이알 내의 (S)-메틸 6-아미노-2-(2,2,2-트리플루오로아세트아미도)헥사노에이트 히드로클로라이드 (0.943 g, 3.22 mmol), N,N-디이소프로필 에틸아민 (DIEA, 1.39 mL) 및 디클로로메탄 (DCM, 10 mL)에 N2 분위기 하에서 디케톤 (0.37 mL)을 첨가하고, 반응물을 주위 온도에서 16 시간 동안 교반하였다. 반응 혼합물을 에틸 아세테이트 (EtOA)로 희석하고, H2O, 1 N HCl, H2O, 포화 aq Na2CO3, 및 포화 aq NaCl로 연속 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 잔류물을 SiO2 플래쉬 크로마토그래피에 의해 정제하여, (S)-메틸-6-(3-옥소부탄아미도)-2-(2,2,2-트리플루오로아세트아미도)헥사노에이트 (TU3000-012)를 황색 오일로서 수득하였다. (S)-methyl 6-amino-2-(2,2,2-trifluoroacetamido)hexanoate hydrochloride (0.943 g, 3.22 mmol), N,N-diisopropyl in a 40 mL glass vial Diketone (0.37 mL) was added to ethylamine (DIEA, 1.39 mL) and dichloromethane (DCM, 10 mL) under N 2 atmosphere, and the reaction was stirred at ambient temperature for 16 hours. The reaction mixture was diluted with ethyl acetate (EtOA), washed successively with H 2 O, 1 N HCl, H 2 O, saturated aq Na 2 CO 3 , and saturated aq NaCl, dried over Na 2 SO 4 , filtered and , Concentrated under reduced pressure. The residue was purified by SiO 2 flash chromatography, (S)-methyl-6-(3-oxobutanamido)-2-(2,2,2-trifluoroacetamido)hexanoate ( TU3000-012) was obtained as a yellow oil.
(S)-메틸 6-(3-옥소부탄아미도)-2-(2,2,2-트리플루오로아세트아미도)헥사노에이트 (0.736 g, 2.16 mmol)를 1 N aq NaOH (6.5 mL) 및 10 mL H2O로 주위 온도에서 18 시간 동안 처리하였다. LC-MS 분석을 이용하여 반응이 완료되었음을 확인하였다. 반응 혼합물을 감압하에 농축시켰다. 잔류물을 과량의 1 N aq HCl로 처리하고, 감압하에 건고상태로 농축시켜, (S)-2-아미노-6-(3-옥소부탄아미도)헥산산 히드로클로라이드 (TU3000-016)를 수득하였다.(S)-methyl 6-(3-oxobutanamido)-2-(2,2,2-trifluoroacetamido)hexanoate (0.736 g, 2.16 mmol) in 1 N aq NaOH (6.5 mL ) And 10 mL H 2 O at ambient temperature for 18 hours. It was confirmed that the reaction was complete using LC-MS analysis. The reaction mixture was concentrated under reduced pressure. The residue was treated with an excess of 1 N aq HCl and concentrated to dryness under reduced pressure to give (S)-2-amino-6-(3-oxobutanamido)hexanoic acid hydrochloride (TU3000-016). I did.
실시예 33-2: (S)-5-(4- 아세틸벤즈아미도 )-1- 카르복시펜탄 -1- 아미늄 클로라이드 (TU3000-004)의 합성Synthesis of (S) -5- (4- acetyl-benz amido) - 1-carboxy-pentane-1-aminium chloride fried (TU3000-004): Example 33-2
(S)-메틸 6-아미노-2-(2,2,2-트리플루오로아세트아미도)헥사노에이트 히드로클로라이드 (TU2982-126) (303 mg, 1.03 mmol), 4-아세틸벤조산 (190 mg), HATU (418 mg), DIEA (523 ㎕) 및 DMF (8 mL)를 합하고, 주위 온도에서 18 시간 동안 교반하였다. 반응 혼합물을 EtOA로 희석하고, H2O, 1 N HCl, H2O, 포화 aq Na2CO3, 및 포화 aq NaCl로 연속 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 잔류물을 SiO2 플래쉬 크로마토그래피에 의해 정제하여, (S)-메틸-6-아미노-2-(2,2,2-트리플루오로아세트아미도)헥사노에이트 히드로클로라이드 (TU2982-136)를 수득하였다.(S)-Methyl 6-amino-2-(2,2,2-trifluoroacetamido)hexanoate hydrochloride (TU2982-126) (303 mg, 1.03 mmol), 4-acetylbenzoic acid (190 mg ), HATU (418 mg), DIEA (523 μL) and DMF (8 mL) were combined and stirred at ambient temperature for 18 hours. The reaction mixture was diluted with EtOA, washed successively with H 2 O, 1 N HCl, H 2 O, saturated aq Na 2 CO 3 , and saturated aq NaCl, dried over Na 2 SO 4 , filtered, and concentrated under reduced pressure. Made it. The residue was purified by SiO 2 flash chromatography to give (S)-methyl-6-amino-2-(2,2,2-trifluoroacetamido)hexanoate hydrochloride (TU2982-136). Obtained.
(S)-메틸 6-(4-아세틸벤즈아미도)-2-(2,2,2-트리플루오로아세트아미도)헥사노에이트 (TU2982-136) (0.473 g)를 10 mL MeOH 중의 1 N aq NaOH (2.36 mL)로 주위 온도에서 18 시간 동안 처리하였다. LC-MS 분석을 이용하여 메틸 에스테르의 가수분해가 완료되었고, 트리플루오로아미드 잔기는 그대로 남아있음을 확인하였다. 반응물을 60℃에서 5 시간 동안 가열하였고, 이 시점에 아미드 가수분해가 거의 완료되었음이 LC-MS 분석에 의해 확인되었다. 반응 혼합물을 냉각시키고 감압하에 농축시켰다. 잔류물을 과량의 1 N aq HCl로 처리하고, 감압하에 건고상태로 농축시켜, ((S)-5-(4-아세틸벤즈아미도)-1-카르복시펜탄-1-아미늄 클로라이드 (TU3000-004)를 연황색 고체로서 수득하였다. (S)-Methyl 6-(4-acetylbenzamido)-2-(2,2,2-trifluoroacetamido)hexanoate (TU2982-136) (0.473 g) was added to 1 in 10 mL MeOH Treated with Naq NaOH (2.36 mL) at ambient temperature for 18 hours. It was confirmed that the hydrolysis of the methyl ester was completed using LC-MS analysis, and that the trifluoroamide residue remained as it was. The reaction was heated at 60° C. for 5 hours, at which point it was confirmed by LC-MS analysis that the amide hydrolysis was almost complete. The reaction mixture was cooled and concentrated under reduced pressure. The residue was treated with an excess of 1 N aq HCl and concentrated to dryness under reduced pressure, ((S)-5-(4-acetylbenzamido)-1-carboxypentane-1-aminium chloride (TU3000- 004) was obtained as a pale yellow solid.
실시예 33-3: ((S)-5-(5- 아세틸티오펜 -2- 카르복스아미도 )-1- 카르복시펜탄 -1-아미늄 클로라이드 (TU3000-006), (S)-5-(3-아세틸벤즈아미도)-1-카르복시펜탄-1-아미늄 클로라이드 (TU3000-008), 및 (S)-5-(4-아세틸-1-메틸-1H-피롤-2-카르복스아미도)-1-카르복시펜탄-1-아미늄 클로라이드 (TU3000-010)의 합성 Example 33-3: ((S)-5-(5- acetylthiophene- 2 -carboxamido )-1- carboxypentane - 1-aminium chloride (TU3000-006), (S)-5- (3-acetylbenzamido)-1-carboxypentane-1-aminium chloride (TU3000-008), and (S)-5-(4-acetyl-1-methyl-1H-pyrrole-2-carboxami Figure) Synthesis of -1-carboxypentane-1-aminium chloride (TU3000-010)
4-아세틸벤조산 대신 상응하는 산을 사용하는 것을 제외하고 TU3000-004와 동일한 방식으로 (S)-5-(5-아세틸티오펜-2-카르복스아미도)-1-카르복시펜탄-1-아미늄 클로라이드 (TU3000-006), (S)-5-(3-아세틸벤즈아미도)-1-카르복시펜탄-1-아미늄 클로라이드 (TU3000-008), 및 (S)-5-(4-아세틸-1-메틸-1H-피롤-2-카르복스아미도)-1-카르복시펜탄-1-아미늄 클로라이드 (TU3000-010)를 제조하였다. 사용한 산은 TU3000-006, TU3000-008, 및 TU3000-010의 경우에 각각 4-아세틸-1-메틸-1H-피롤-2-카르복실산, 5-아세틸티오펜-2-카르복실산 및 3-아세틸벤조산이었다.(S)-5-(5-acetylthiophene-2-carboxamido)-1-carboxypentane-1-amido in the same manner as TU3000-004, except that the corresponding acid is used instead of 4-acetylbenzoic acid. Nium chloride (TU3000-006), (S)-5-(3-acetylbenzamido)-1-carboxypentane-1-aminium chloride (TU3000-008), and (S)-5-(4-acetyl -1-Methyl-1H-pyrrole-2-carboxamido)-1-carboxypentane-1-aminium chloride (TU3000-010) was prepared. The acids used were in the case of TU3000-006, TU3000-008, and TU3000-010, respectively, 4-acetyl-1-methyl-1H-pyrrole-2-carboxylic acid, 5-acetylthiophene-2-carboxylic acid and 3- It was acetylbenzoic acid.
실시예 33-4: (S)-2-아미노-6-(3- 옥소시클로부탄카르복스아미도 ) 헥산산 (TU3205-030)의 합성. Example 33-4: (S)-2-amino-6-(3- oxocyclobutanecarboxamido ) hexanoic acid (TU3205-030).
요오도메탄 (2.0 mL), K2CO3 (5.60 g), (S)-6-(벤질옥시카르보닐아미노)-2-(tert-부톡시카르보닐아미노)헥산산 (1) (노바바이오켐(NovaBiochem), A29340), 및 무수 DMF (20 mL)를 합하고, 주위 온도에서 2 시간 동안 교반하였다. 반응 혼합물을 수성 후처리하여, (S)-메틸 6-(벤질옥시카르보닐아미노)-2-(tert-부톡시카르보닐아미노)헥사노에이트 (TU3000-090)를 맑은 오일로서 수득하였다. Iodomethane (2.0 mL), K 2 CO 3 (5.60 g), (S)-6-(benzyloxycarbonylamino)-2-(tert-butoxycarbonylamino)hexanoic acid (1) (Novabio Chem (NovaBiochem, A29340), and anhydrous DMF (20 mL) were combined and stirred at ambient temperature for 2 hours. The reaction mixture was subjected to aqueous workup to give (S)-methyl 6-(benzyloxycarbonylamino)-2-(tert-butoxycarbonylamino)hexanoate (TU3000-090) as a clear oil.
(S)-메틸 6-(벤질옥시카르보닐아미노)-2-(tert-부톡시카르보닐아미노)헥사노에이트 (8.60 g)를 5% 활성탄상 팔라듐 (1.08 g) 상의 150 mL MeOH 중에서 수소 1 atm 하에 주위 온도에서 3 시간 동안 수소화하였다. 폐 촉매를 진공 여과에 의해 셀라이트 패드를 통과시켜 제거하고, MeOH로 세척하였다. 합한 여과물 및 세척액을 감압하에 농축시켜, (S)-메틸 6-아미노-2-(tert-부톡시카르보닐아미노)헥사노에이트 (TU3000-128)를 맑은 점성 오일로서 수득하였다.(S)-methyl 6-(benzyloxycarbonylamino)-2-(tert-butoxycarbonylamino)hexanoate (8.60 g) was added
20 mL DMF 중의 (S)-메틸 6-아미노-2-(tert-부톡시카르보닐아미노)헥사노에이트의 1 M 저장 용액을 제조하여 사용하였다. (S)-메틸 6-아미노-2-(tert-부톡시카르보닐아미노)헥사노에이트 저장 용액의 2 mL 분취량, 3-옥소시클로부탄카르복실산 (2) (파크웨이(Parkway) *BX-102, 283 mg), HATU (800 mg), DIEA (1.0 mL) 및 8 mL DMF를 40 mL 유리 바이알에서 합하고, 주위 온도에서 밤새 교반하였다. LC-MS 분석에 의해 반응이 완료되었음을 확인하였다. 반응 혼합물을 EtOAc로 희석하고, 물, 포화 aq. NaCl로 연속 세척하고, MgSO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 조 물질을 실리카겔 플래쉬 크로마토그래피 (헥산/EtOAc)에 의해 정제하여, (S)-메틸 2-(tert-부톡시카르보닐아미노)-6-(3-옥소시클로부탄카르복스아미도)헥사노에이트 (TU3000-140)를 맑은 점성 오일로서 수득하였다.A 1 M stock solution of (S)-methyl 6-amino-2-(tert-butoxycarbonylamino)hexanoate in 20 mL DMF was prepared and used. 2 mL aliquots of (S)-methyl 6-amino-2-(tert-butoxycarbonylamino)hexanoate stock solution, 3-oxocyclobutanecarboxylic acid (2) (Parkway * BX -102, 283 mg), HATU (800 mg), DIEA (1.0 mL) and 8 mL DMF were combined in a 40 mL glass vial and stirred at ambient temperature overnight. The reaction was confirmed to be complete by LC-MS analysis. The reaction mixture was diluted with EtOAc, and water, saturated aq. Washed successively with NaCl, dried over MgSO 4 , filtered and concentrated under reduced pressure. The crude material was purified by silica gel flash chromatography (hexane/EtOAc) to obtain (S)-methyl 2-(tert-butoxycarbonylamino)-6-(3-oxocyclobutanecarboxamido)hexanoate. (TU3000-140) was obtained as a clear viscous oil.
(S)-메틸 2-(tert-부톡시카르보닐아미노)-6-(3-옥소시클로부탄카르복스아미도)헥사노에이트 (0.501 g)를 1,4-디옥산 중의 20 mL 4 M HCl로 주위 온도에서 20 분 동안 처리하고, 용매를 감압하에 제거하였다. 생성된 점성 오일을 10 mL CH3CN에 녹이고, 소규모로 제조된 소량의 표제 화합물 결정을 시딩하였다. 생성된 결정을 진공 여과에 의해 수집하고, CH3CN으로 세척하고, 감압하에 건조시켜, (S)-메틸 2-아미노-6-(3-옥소시클로부탄카르복스아미도)헥사노에이트 (TU3205-016)를 무색 고체로서 수득하였다.(S)-Methyl 2-(tert-butoxycarbonylamino)-6-(3-oxocyclobutanecarboxamido)hexanoate (0.501 g) in 20 mL 4 M HCl in 1,4-dioxane The furnace was treated at ambient temperature for 20 minutes, and the solvent was removed under reduced pressure. The resulting viscous oil was dissolved in 10 mL CH 3 CN, and a small amount of crystals of the title compound prepared on a small scale were seeded. The resulting crystals were collected by vacuum filtration, washed with CH 3 CN and dried under reduced pressure, (S)-methyl 2-amino-6-(3-oxocyclobutanecarboxamido)hexanoate (TU3205 -016) was obtained as a colorless solid.
(S)-메틸 2-아미노-6-(3-옥소시클로부탄카르복스아미도)헥사노에이트 (0.297 g) 및 18 mL H2O를 40 mL 유리 바이알에 넣었다. 생성된 맑은 용액에 2.2 mL 1 N NH4OH를 첨가하고, 반응물을 주위 온도에서 22 시간 동안 진탕하였고, 이 시점에LCMS 분석에 의해 반응이 완료되었음을 확인하였다. 반응 혼합물을 동결 및 동결건조시켜, (S)-2-아미노-6-(3-옥소시클로부탄카르복스아미도)헥산산 (TU3205-030)을 무색 결정으로서 수득하였다.(S)-Methyl 2-amino-6-(3-oxocyclobutanecarboxamido)hexanoate (0.297 g) and 18 mL H 2 O were placed in a 40 mL glass vial. 2.2 mL 1 N NH 4 OH was added to the resulting clear solution, and the reaction was shaken at ambient temperature for 22 hours, at which point the reaction was confirmed to be complete by LCMS analysis. The reaction mixture was frozen and lyophilized to give (S)-2-amino-6-(3-oxocyclobutanecarboxamido)hexanoic acid (TU3205-030) as colorless crystals.
실시예Example 34: 반응성 피롤리신 중간체의 합성 34: Synthesis of reactive pyrrolysine intermediate
실시예 34-1: 리튬 2-(4-아세틸-3- 아미노페녹시 )아세테이트 ( TU3205 -042)의 합성 Example 34-1: Synthesis of lithium 2- (4-acetyl-3-aminophenoxy) acetate (TU3205 -042)
1-(4-히드록시-2-니트로페닐)에타논 (카르보코어(Carbocore), 181 mg, 1.00 mmol), 에틸 2-브로모아세테이트 (183 mg, 1.10 mmol), 탄산칼륨 (138 mg. 1.00 mmol) 및 DMF (5 mL)를 20 mL 유리 바이알 내에서 합하고, 60℃에서 2 시간 동안 교반하였고, 이 시점에서 LC-MS 분석은 반응이 완전하게 완료되었음을 나타내었다. 반응 혼합물을 물로 희석하고, EtOAc로 추출하고, 포화 NaCl aq로 세척하였다. 1-(4-히드록시-2-니트로페닐)에타논 (0.802 g, 4.43 mmol), 에틸 2-브로모아세테이트 (0.813 g, 4.87 mmol), 탄산칼륨 (0.612 g. 4.43 mmol) 및 DMF (25 mL)를 이용하여 반응을 반복하고, 동일한 방식으로 후처리하였다. 합한 EtOAc 추출물을 무수 MgSO4 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 에틸 2-(4-아세틸-3-니트로페녹시)아세테이트 (TU3205-034)를 짙은 황색 오일로서 수득하였다. 1-(4-hydroxy-2-nitrophenyl)ethanone (Carbocore, 181 mg, 1.00 mmol), ethyl 2-bromoacetate (183 mg, 1.10 mmol), potassium carbonate (138 mg. 1.00 mmol) and DMF (5 mL) were combined in a 20 mL glass vial and stirred at 60° C. for 2 hours, at which point LC-MS analysis showed the reaction to be complete. The reaction mixture was diluted with water, extracted with EtOAc, and washed with saturated NaCl aq. 1-(4-hydroxy-2-nitrophenyl)ethanone (0.802 g, 4.43 mmol), ethyl 2-bromoacetate (0.813 g, 4.87 mmol), potassium carbonate (0.612 g. 4.43 mmol) and DMF (25 mL) was used to repeat the reaction, followed by work-up in the same manner. The combined EtOAc extracts were dried over anhydrous MgSO 4 , filtered and concentrated under reduced pressure to give ethyl 2-(4-acetyl-3-nitrophenoxy)acetate (TU3205-034) as a dark yellow oil.
MeOH (5 mL) 중의 에틸 2-(4-아세틸-3-니트로페녹시)아세테이트 (TU3205-034) (111 mg)를 10% 목탄상 팔라듐 (11 mg)을 이용하여 H2 기압하에 주위 온도에서 수소화하였다. 30 분 후에, LC-MS 분석에 의해 반응이 완전하게 완료되었음을 확인하였다. TU3205-034 (1.45 g), 10% 목탄상 팔라듐 (140 mg), 및 MeOH (80 mL)를 이용하여 반응을 반복하였다. 2개의 반응 혼합물을 합하고, 여과에 의해 셀라이트 패드를 통해 폐 촉매를 제거하였다. 여과물을 감압하에 농축시켜, 에틸 2-(4-아세틸-3-아미노페녹시)아세테이트 (TU3205-036)를 짙은 황색 오일로서 수득하였다.Ethyl 2-(4-acetyl-3-nitrophenoxy)acetate (TU3205-034) (111 mg) in MeOH (5 mL) was added to 10% palladium on charcoal (11 mg) at ambient temperature under H 2 atm. Hydrogenated. After 30 minutes, the reaction was confirmed to be complete by LC-MS analysis. The reaction was repeated with TU3205-034 (1.45 g), 10% palladium on charcoal (140 mg), and MeOH (80 mL). The two reaction mixtures were combined and the spent catalyst was removed through a pad of celite by filtration. The filtrate was concentrated under reduced pressure to give ethyl 2-(4-acetyl-3-aminophenoxy)acetate (TU3205-036) as a dark yellow oil.
에틸 2-(4-아세틸-3-아미노페녹시)아세테이트 (TU3205-036) (1.02 g)를 THF (17 mL)에 용해시키고, 수성 LiOH (1 M, 4.3 mL)로 주위 온도에서 1 시간 동안 처리하였다. LC-MS 분석은 반응이 완전하게 완료되었음을 나타내었다. 반응 혼합물을 감압하에 농축시켜, 리튬 2-(4-아세틸-3-아미노페녹시)아세테이트 (TU3205-042)를 황색빛 고체로서 수득하였다. Ethyl 2-(4-acetyl-3-aminophenoxy)acetate (TU3205-036) (1.02 g) was dissolved in THF (17 mL) and aqueous LiOH (1 M, 4.3 mL) at ambient temperature for 1 hour. Processed. LC-MS analysis indicated that the reaction was complete. The reaction mixture was concentrated under reduced pressure to give lithium 2-(4-acetyl-3-aminophenoxy)acetate (TU3205-042) as a yellowish solid.
실시예 34-2: 리튬 2-(3-아미노-4- 포르밀페녹시 )아세테이트 ( TU3627 -018)의 합성. Example 34-2: Synthesis of lithium 2- (3-amino-4-formyl-phenoxy) acetate (TU3627 -018).
빙조에서 냉각시키면서 500 mL 둥근바닥 플라스크 내의 4-메톡시-2-니트로벤즈알데히드 (카르보코어, CO-0119, 5.5 g) 및 200 mL DCM에 보론트리브로마이드 (24 g)를 적가하였다. 반응물을 동일한 온도에서 30 분 동안 교반한 다음, 주위 온도에서 3 시간 동안 교반하였다. 반응 혼합물을 조심스럽게 빙수에 붓고, 주위 온도에서 3 일 동안 정치시켰다. 수성 혼합물을 EtOAc로 추출하고, 포화 aq. NaCl로 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 조 물질을 20 → 60% 헥산 중의 EtOAc (Rf. 0.32, 50% 헥산 중의 EtOAc)의 선형 구배를 이용한 SiO2 겔 플래쉬 크로마토그래피에 의해 정제하여, TU3627-002를 오렌지색 결정으로서 수득하였다. While cooling in an ice bath, borontribromide (24 g) was added dropwise to 4-methoxy-2-nitrobenzaldehyde (Carbocore, CO-0119, 5.5 g) and 200 mL DCM in a 500 mL round bottom flask. The reaction was stirred at the same temperature for 30 minutes and then at ambient temperature for 3 hours. The reaction mixture was carefully poured into ice water and allowed to stand at ambient temperature for 3 days. The aqueous mixture was extracted with EtOAc and saturated aq. Washed with NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure. The crude was purified by SiO 2 gel flash chromatography using a linear gradient of 20 to 60% EtOAc in hexane (Rf. 0.32, 50% EtOAc in hexane) to give TU3627-002 as orange crystals.
TU3627-002 (1.87 g), 에틸 브로모아세테이트 (알드리치(Aldrich), 1.5 mL), 탄산칼륨 (2.32 g) 및 DMF를 합하고, 주위 온도에서 18 시간 동안 교반하였다. 반응 혼합물을 EtOAc 및 물 사이에 분배시켰다. 유기층을 분리하고, 포화 aq. NaCl로 세척하고, MgSO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 조 물질을 5 → 35% 헥산 중의 EtOAc (Rf. 0.32, 35% 헥산 중의 EtOAc)의 선형 구배를 이용한 SiO2 겔 플래쉬 크로마토그래피에 의해 정제하여, TU3627-008을 짙은 황색 오일로서 수득하였다. TU3627-002 (1.87 g), ethyl bromoacetate (Aldrich, 1.5 mL), potassium carbonate (2.32 g) and DMF were combined and stirred at ambient temperature for 18 hours. The reaction mixture was partitioned between EtOAc and water. The organic layer was separated, and saturated aq. Washed with NaCl, dried over MgSO 4 , filtered and concentrated under reduced pressure. The crude was purified by SiO 2 gel flash chromatography using a linear gradient from 5 to 35% EtOAc in hexanes (Rf. 0.32, 35% EtOAc in hexanes) to give TU3627-008 as a dark yellow oil.
머릭(Merlic)의 문헌 [C.A.Merlic et al., J.Org.Chem., 1995, 60, 3365-69]에 기재되어 있는 방법을 이용하여 TU3627-008을 환원시켰다. 구체적으로, TU3627-008 (1.717 g), 철 분말 (3.79 g), EtOH (45 mL), H2O (11 mL) 및 진한 HCl (180 ㎕)을 합하고, 2 시간 동안 환류가열하였다. 반응 혼합물을 여과하고, 여과물을 농축시켰다. 잔류물을 5 → 60% 용매 A 중의 용매 B (용매 A: 5% 헥산 중의 NEt3; 용매 B: 5% EtOAc 중의 NEt3) (Rf. 0.34, 35% 헥산 중의 EtOAc)의 선형 구배를 이용한 실리카겔 플래쉬 크로마토그래피에 의해 정제하여, TU3627-014를 밝은 황색 결정으로서 수득하였다. TU3627-008 was reduced using the method described in Merlic et al., J. Org. Chem., 1995, 60, 3365-69. Specifically, TU3627-008 (1.717 g), iron powder (3.79 g), EtOH (45 mL), H 2 O (11 mL) and concentrated HCl (180 μL) were combined and heated to reflux for 2 hours. The reaction mixture was filtered and the filtrate was concentrated. The residue was taken on silica gel using a linear gradient from 5 to 60% solvent B in solvent A (solvent A: NEt 3 in 5% hexane; solvent B: NEt 3 in 5% EtOAc) (Rf. 0.34, 35% EtOAc in hexane). Purification by flash chromatography gave TU3627-014 as light yellow crystals.
5 mL THF 중의 TU3627-014 (0.608 g)를 1 M aq. LiOH의 2.72 mL 분취량으로 주위 온도에서 처리하였다. 20 분 후에, LC-MS 분석에 의해 반응이 완료되었음을 확인하였다. 반응 혼합물을 감압하에 건고상태로 농축시켜, 리튬 2-(3-아미노-4-포르밀페녹시)아세테이트 (TU3627-018)를 황색 고체로서 수득하였다.TU3627-014 (0.608 g) in 5 mL THF was added to 1 M aq. Treated at ambient temperature with 2.72 mL aliquots of LiOH. After 20 minutes, it was confirmed that the reaction was complete by LC-MS analysis. The reaction mixture was concentrated to dryness under reduced pressure to give lithium 2-(3-amino-4-formylphenoxy)acetate (TU3627-018) as a yellow solid.
실시예 34-3: 리튬 4-(3-아세틸-4- 아미노페녹시 ) 부타노에이트 ( TU3627 -064)의 합성. Example 34-3: Synthesis of lithium 4-(3-acetyl-4- aminophenoxy ) butanoate ( TU3627 -064) .
4'-클로로-2'-니트로아세토페논 (바이오넷(Bionet) cat# 3W0333, 1.00 g), 아세트산칼륨 (4.91 g) 및 DMSO의 혼합물을 170℃에서 20 분 동안 마이크로파 조사하에 가열하였다. 반응 혼합물을 EtOAc 및 물 사이에 분배시켰다. 유기층을 분리하고, 포화 aq. NaCl로 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 또 다른 반응을 같은 방식으로 수행하고, 두 반응으로부터의 조 생성물을 합하여, 실리카겔 플래쉬 크로마토그래피 (EtOAc)에 의해 정제하였다. 표제 화합물을 오렌지색 고체로서 수득하였다.A mixture of 4'-chloro-2'-nitroacetophenone (Bionet cat# 3W0333, 1.00 g), potassium acetate (4.91 g) and DMSO was heated at 170° C. for 20 minutes under microwave irradiation. The reaction mixture was partitioned between EtOAc and water. The organic layer was separated, and saturated aq. Washed with NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure. Another reaction was carried out in the same way, and the crude products from the two reactions were combined and purified by silica gel flash chromatography (EtOAc). The title compound was obtained as an orange solid.
4'-히드록시-2'-니트로아세토페논 (1.12 g), 에틸 4-브로모부타노에이트 (1.33 g), 탄산칼륨 (0.94 g) 및 4 mL DMF의 혼합물을 60℃에서 6.5 시간 동안 교반하였다. 반응 혼합물을 EtOAc 및 물 사이에 분배시켰다. 유기층을 분리하고, 포화 aq. NaCl로 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 조 생성물을 실리카겔 플래쉬 크로마토그래피 (헥산/EtOAc)에 의해 정제하여, 표제 화합물 1을 황색 결정으로서 수득하였다.A mixture of 4'-hydroxy-2'-nitroacetophenone (1.12 g), ethyl 4-bromobutanoate (1.33 g), potassium carbonate (0.94 g) and 4 mL DMF was stirred at 60° C. for 6.5 hours I did. The reaction mixture was partitioned between EtOAc and water. The organic layer was separated, and saturated aq. Washed with NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure. The crude product was purified by silica gel flash chromatography (hexane/EtOAc) to give the
TU633-148 (1.48 g)을 본원에서 기재한 바와 같이, 머릭의 문헌 [C.A.Merlic et al., J.Org.Chem., 1995, 60, 3365-69]에 기재된 방법으로 환원시켰다. 5 → 60% 용매 A 중의 용매 B (용매 A: 5% 헥산 중의 NEt3; 용매 B: 5% EtOAc 중의 NEt3)의 선형 구배를 이용한 실리카겔 플래쉬 크로마토그래피 이후에 TU3627-056을 밝은 황색 오일로서 수득하였다.TU633-148 (1.48 g) was reduced by the method described by Merrick, Camerlic et al., J. Org. Chem., 1995, 60, 3365-69, as described herein. TU3627-056 was obtained after flash chromatography on silica gel using a linear gradient of 5→60% solvent B in solvent A (solvent A: NEt 3 in 5% hexane; solvent B: NEt 3 in 5% EtOAc). I did.
3 mL THF 중의 TU3627-056 (0.50 g)을 1 M aq. LiOH의 1.9 mL 분취량으로 주위 온도에서 18 시간 동안 처리하였다. LC-MS 분석에 의해 반응이 완전하게 완료되었음을 확인하였다. 반응 혼합물을 감압하에 건고상태로 농축시켜, 리튬 4-(3-아세틸-4-아미노페녹시)부타노에이트 (TU3627-064)를 황색 고체로서 수득하였다.TU3627-056 (0.50 g) in 3 mL THF was added to 1 M aq. A 1.9 mL aliquot of LiOH was treated at ambient temperature for 18 hours. The reaction was confirmed to be complete by LC-MS analysis. The reaction mixture was concentrated to dryness under reduced pressure to give lithium 4-(3-acetyl-4-aminophenoxy)butanoate (TU3627-064) as a yellow solid.
실시예 34-4: 리튬 4-(3-아미노-4- 포르밀페녹시 ) 부타노에이트 ( TU3627 -074)의 합성. Example 34-4: lithium 4- (3-amino-4-formyl-phenoxy) butanoate: Synthesis of (TU3627 -074).
2-니트로-4-히드록시벤즈알데히드 (2.85 g), 에틸 4-브로모부타노에이트 (3.66 g), 탄산칼륨 (2.84 g) 및 DMF (20 mL)의 혼합물을 주위 온도에서 50 시간 동안 교반하였다. 반응 혼합물을 H2O 및 EtOAc 사이에 분배시켰다. 유기층을 분리하고, 포화 NaHCO3 aq로 세척하였다. 합한 수성층을 EtOAc로 추출하였다. 합한 유기층을 연한 시트르산, 포화 NaCl aq로 세척하고, MgSO4 상에서 건조시키고, 여과하고, 농축시켰다. 잔류물을 20 → 40% 헥산 중의 EtOAc (Rf: 0.35, 35% 헥산 중의 EtOAc)의 선형 구배를 이용한 실리카겔 플래쉬 크로마토그래피에 의해 정제하여, 표제 화합물을 황색 오일로서 수득하였다.A mixture of 2-nitro-4-hydroxybenzaldehyde (2.85 g), ethyl 4-bromobutanoate (3.66 g), potassium carbonate (2.84 g) and DMF (20 mL) was stirred at ambient temperature for 50 hours. . The reaction mixture was partitioned between H 2 O and EtOAc. The organic layer was separated and washed with saturated NaHCO 3 aq. The combined aqueous layer was extracted with EtOAc. The combined organic layers were washed with light citric acid, saturated NaCl aq, dried over MgSO 4 , filtered and concentrated. The residue was purified by silica gel flash chromatography using a linear gradient of 20 to 40% EtOAc in hexanes (Rf: 0.35, 35% EtOAc in hexanes) to give the title compound as a yellow oil.
TU3627-062 (3.89 g)를 본원에서 기재한 바와 같이, 머릭의 문헌 [C.A.Merlic et al., J.Org.Chem., 1995, 60, 3365-69]에 기재된 방법으로 환원시켰다. 20 → 60% 용매 A 중의 용매 B (용매 A: 5% 헥산 중의 NEt3; 용매 B: 5% EtOAc 중의 NEt3)의 선형 구배를 이용한 실리카겔 플래쉬 크로마토그래피 이후에 TU3627-066을 황색 고체로서 수득하였다. Rf: 0.51, 50% 헥산 중 EtOAc.TU3627-062 (3.89 g) was reduced by the method described in Merrick's document Camerlic et al., J. Org. Chem., 1995, 60, 3365-69, as described herein. TU3627-066 was obtained as a yellow solid after silica gel flash chromatography using a linear gradient of solvent B in 20 to 60% solvent A (solvent A: NEt 3 in 5% hexane; solvent B: NEt 3 in 5% EtOAc). . Rf: 0.51, 50% EtOAc in hexane.
6 mL THF 중의 TU3627-066 (1.00 g)을 3.98 mL의 1 M 수성 LiOH로 주위 온도에서 4 시간 동안 처리하였다. 감압하에 대부분의 용매를 제거하고, 생성된 탁한 혼합물을 이중으로 탈이온화된 물로 희석하고, 동결 및 동결건조시켜, 리튬 4-(3-아미노-4-포르밀페녹시)부타노에이트 (TU3627-074)를 흐린 황색 고체로서 수득하였다.TU3627-066 (1.00 g) in 6 mL THF was treated with 3.98 mL of 1 M aqueous LiOH at ambient temperature for 4 hours. Most of the solvent was removed under reduced pressure, and the resulting cloudy mixture was diluted with double deionized water, freeze and lyophilized to obtain lithium 4-(3-amino-4-formylphenoxy)butanoate (TU3627- 074) was obtained as a pale yellow solid.
실시예 34-5: 리튬 3-(3-아세틸-4- 아미노페닐 ) 프로파노에이트 ( X3547 -1)의 합성. Example 34-5: Synthesis of lithium 3-(3-acetyl-4 -aminophenyl ) propanoate ( X3547-1 ) .
압력 튜브 내의 무수 DMF (10 mL) 중의 1-(2-아미노-5-브로모페닐)에타논 (642 mg), Pd(OAc)2 (33.7 mg), 및 P(o톨릴)3 (137 mg) 혼합물에 메틸 크릴레이트 (351 ㎕) 및 TEA (1.4 mL)를 첨가하였다. 혼합물을 N2로 3 분 동안 플러싱한 다음, 밀봉하고, 110℃에서 4 시간 동안 가열하였다. 반응 혼합물을 주위 온도로 냉각시킨 다음, 에틸 아세테이트 및 물 사이에 분배시켰다. 수성층을 에틸 아세테이트로 1회 추출하고, 합한 유기층을 염수로 세척하고, 건조시키고 (Na2SO4), 여과하고, 진공하에 용매를 제거하였다. 조 잔류물을 실리카겔 플래쉬 크로마토그래피 (EtOAc/헥산)에 의해 정제하여, 생성물을 수득하였다. 1-(2-amino-5-bromophenyl)ethanone (642 mg), Pd(OAc) 2 (33.7 mg), and P( o Tolyl) 3 (137 mg) in anhydrous DMF (10 mL) in a pressure tube ) Methyl acrylate (351 μl) and TEA (1.4 mL) were added to the mixture. The mixture was flushed with N 2 for 3 minutes, then sealed and heated at 110° C. for 4 hours. The reaction mixture was cooled to ambient temperature and then partitioned between ethyl acetate and water. The aqueous layer was extracted once with ethyl acetate, and the combined organic layers were washed with brine, dried (Na 2 SO 4 ), filtered and the solvent was removed under vacuum. The crude residue was purified by silica gel flash chromatography (EtOAc/hexane) to give the product.
(E)-메틸 3-(3-아세틸-4-아미노페닐)아크릴레이트 (219 mg)를 수소 풍선과 함께 MeOH 중의 5% Pd/C (21.9 mg)에 의해 실온에서 환원시켜, 여과 및 농축 이후에 생성물 (정량적)을 수득하였다. 생성물을 추가의 정제없이 다음 단계에서 사용하였다.(E)-Methyl 3-(3-acetyl-4-aminophenyl)acrylate (219 mg) was reduced at room temperature with 5% Pd/C (21.9 mg) in MeOH with a hydrogen balloon, after filtration and concentration The product (quantitative) was obtained. The product was used in the next step without further purification.
메틸 3-(3-아세틸-4-아미노페닐)프로파노에이트 (221 mg) 및 4 M LiOH (0.275 mL)를 3 mL THF/물 (v/v = 3/1)에 첨가하였다. 반응이 완료된 후 용매를 감압하에 제거하여, 리튬 3-(3-아세틸-4-아미노페닐)프로파노에이트 (X3547-1)를 수득하였다. Methyl 3-(3-acetyl-4-aminophenyl)propanoate (221 mg) and 4 M LiOH (0.275 mL) were added to 3 mL THF/water (v/v = 3/1). After the reaction was completed, the solvent was removed under reduced pressure to obtain lithium 3-(3-acetyl-4-aminophenyl)propanoate (X3547-1).
실시예 34-6: 리튬 3-(4-아세틸-3- 아미노페닐 ) 프로파노에이트 ( X3547 -8)의 합성. Example 34-6: Synthesis of lithium 3-(4-acetyl-3 -aminophenyl ) propanoate ( X3547-8 ) .
CH3CN (15 mL)에 용해된 에틸 트리페닐포스포라닐리딘 아세테이트 (1.742 g) 용액을 교반하면서 4-브로모-3-니트로벤즈알데히드 (1.150 g)를 함유하는 CH3CN 용액 (10 mL)에 첨가하였다. 반응 혼합물을 밤새 환류시켰다. 혼합물을 냉각시킨 후, 용매를 감압하에 제거하여, 조 고체를 수득하였다. 실리카겔 플래쉬 칼럼 크로마토그래피 이후에 순수한 생성물 X3471-146을 백색 고체로서 수득하였다 (헥산/EtOAc, 9:1).CH 3 CN solution (10 mL) containing 4-bromo-3-nitrobenzaldehyde (1.150 g) while stirring a solution of ethyl triphenylphosphoranilidine acetate (1.742 g) dissolved in CH 3 CN (15 mL) Was added to. The reaction mixture was refluxed overnight. After cooling the mixture, the solvent was removed under reduced pressure to give a crude solid. The pure product X3471-146 was obtained as a white solid after silica gel flash column chromatography (hexane/EtOAc, 9:1).
상기 단계로부터의 생성물 X3471-146 (632 mg) 및 PdCl2(PPh3)2 (148 mg)를 슐렝크(Schlenk) 튜브 내에서 N2 하에 DMF (5.0 mL)에 용해시켰다. 트리부틸(1-에톡시비닐)스탄난 (711 ㎕)을 교반하면서 첨가하였다. 혼합물을 100℃에서 밤새 가열하였다. 혼합물을 냉각시킨 후, 용매를 감압하에 제거하고, DCM으로 희석하고, 물 및 염수로 세척하였다. DCM을 제거한 후, 잔류물을 실리카겔 플래쉬 칼럼 크로마토그래피 (15%-25% 헥산 중의 EtOAc)에 의해 정제하여, 화합물 X3471-154를 수득하였다.The product X3471-146 (632 mg) and PdCl 2 (PPh 3 ) 2 (148 mg) from the above step were dissolved in DMF (5.0 mL) under N 2 in a Schlenk tube. Tributyl(1-ethoxyvinyl)stannan (711 μl) was added with stirring. The mixture was heated at 100° C. overnight. After cooling the mixture, the solvent was removed under reduced pressure, diluted with DCM and washed with water and brine. After removal of DCM, the residue was purified by silica gel flash column chromatography (15%-25% EtOAc in hexane) to give compound X3471-154.
화합물 X3471-154A를 20 mL 1 N HCl로 실온에서 4 시간 동안 처리하였다. 용매를 제거하여 (E)-에틸 3-(4-아세틸-3-니트로페닐)아크릴레이트 X3471-154B를 수득하였다.Compound X3471-154A was treated with 20 mL 1 N HCl at room temperature for 4 hours. Removal of the solvent gave (E)-ethyl 3-(4-acetyl-3-nitrophenyl)acrylate X3471-154B.
화합물 X3471-154B (263 mg)를 5.0 mL MeOH 중의 5% Pd/C (26 mg)를 이용하여 수소하에 1 atm에서 에틸 3-(4-아세틸-3-아미노페닐)프로파노에이트 X3547-2로 환원시켰다. Compound X3471-154B (263 mg) to ethyl 3-(4-acetyl-3-aminophenyl)propanoate X3547-2 at 1 atm under hydrogen using 5% Pd/C (26 mg) in 5.0 mL MeOH. Reduced.
4 M LiOH (0.248 mL), THF (3.0 mL) 및 물 (1.0 mL)의 혼합물을 이용하여 화합물 X3547-2를 실온에서 처리하였다. 반응이 완료된 후, 반응 혼합물을 동결건조시켜, 리튬 3-(4-아세틸-3-아미노페닐)프로파노에이트 (X3547-8)를 수득하였다.Compound X3547-2 was treated at room temperature using a mixture of 4 M LiOH (0.248 mL), THF (3.0 mL) and water (1.0 mL). After the reaction was completed, the reaction mixture was lyophilized to obtain lithium 3-(4-acetyl-3-aminophenyl)propanoate (X3547-8).
실시예 34-7: 리튬 5-(3-아미노-4- 포르밀페닐 ) 펜타노에이트 ( X3547 -30)의 합성. Example 34-7: Lithium 5- (3-amino-4-formylphenyl) pentanoate synthesis of (X3547 -30).
TEA (5.0 mL) 중의 4-브로모-2-니트로벤즈알데히드 (460 mg), Pd(PPh3)2Cl2 (70 mg), 및 CuI (38 mg), 메틸 펜트-4-이노에이트 (269 mg)의 혼합물을 반응이 완료될 때까지 실온에서 교반하였다. 조 잔류물을 실리카겔 플래쉬 크로마토그래피 (15% 헥산 중의 EtOAc)에 의해 정제하여, 메틸 5-(4-포르밀-3-니트로페닐)펜트-4-이노에이트를 수득하였다. 4-bromo-2-nitrobenzaldehyde (460 mg), Pd(PPh 3 ) 2 Cl 2 (70 mg), and CuI (38 mg), methyl pent-4-inoate (269 mg) in TEA (5.0 mL) ) Was stirred at room temperature until the reaction was complete. The crude residue was purified by silica gel flash chromatography (15% EtOAc in hexane) to give methyl 5-(4-formyl-3-nitrophenyl)pent-4-inoate.
메틸 5-(4-포르밀-3-니트로페닐)펜트-4-이노에이트 (522 mg)를 수소 풍선과 함께 MeOH 중의 5% Pd/C (53 mg)에 의해 실온에서 환원시켰다. 여과한 후 감압하에 농축시켜 메틸 5-(3-아미노-4-포르밀페닐)펜타노에이트를 수득하였다.Methyl 5-(4-formyl-3-nitrophenyl)pent-4-inoate (522 mg) was reduced at room temperature with 5% Pd/C in MeOH (53 mg) with a hydrogen balloon. After filtration, it was concentrated under reduced pressure to obtain methyl 5-(3-amino-4-formylphenyl)pentanoate.
메틸 5-(3-아미노-4-포르밀페닐)펜타노에이트 (447 mg)를 8.0 mL THF/물 (v/v = 3/1) 중의 LiOH (88 mg)로 처리하였다. 반응이 완료된 후 용매를 제거하여, 리튬 5-(3-아미노-4-포르밀페닐)펜타노에이트 (X3547-30)를 수득하였다.Methyl 5-(3-amino-4-formylphenyl)pentanoate (447 mg) was treated with LiOH (88 mg) in 8.0 mL THF/water (v/v = 3/1). After the reaction was completed, the solvent was removed to obtain lithium 5-(3-amino-4-formylphenyl)pentanoate (X3547-30).
실시예 34-8: 3-(4-아세틸-3- 아미노페닐 )-2- 아미노프로판산 ( X3179 -96)의 합성. Example 34-8: Synthesis of 3-(4-acetyl-3 -aminophenyl )-2 -aminopropanoic acid ( X3179-96 ) .
수소화나트륨 (광유 중 60%, 1.74 g)을 헥산으로 세척하고, DMF (12 ml) 중에 분산시켰다. DMF (30 ml) 중의 디에틸 아세트아미도말로네이트 (10.4 g) 및 DMF (10 ml) 중의 4-(브로모메틸)-1-플루오로-2-니트로벤젠 (10.2 g)을 연속 첨가하고, 반응 혼합물을 4 시간 동안 실온에서 교반한 후, 감압하에 용매를 제거하였다. 잔류물을 실리카겔 플래쉬 크로마토그래피 (10-20% DCM 중의 EtOAc)에 의해 정제하여, 디에틸 2-아세트아미도-2-(4-플루오로-3-니트로벤질)말로네이트를 수득하였다.Sodium hydride (60% in mineral oil, 1.74 g) was washed with hexane and dispersed in DMF (12 ml). Diethyl acetamidomalonate (10.4 g) in DMF (30 ml) and 4-(bromomethyl)-1-fluoro-2-nitrobenzene (10.2 g) in DMF (10 ml) are added successively, After the reaction mixture was stirred at room temperature for 4 hours, the solvent was removed under reduced pressure. The residue was purified by silica gel flash chromatography (10-20% EtOAc in DCM) to give diethyl 2-acetamido-2-(4-fluoro-3-nitrobenzyl)malonate.
EtOAc 중의 디에틸 2-아세트아미도-2-(4-플루오로-3-니트로벤질)말로네이트 (7.41 g) 및 니트로에탄 (6.0 mL)에 DBU (9.0 mL)를 첨가하고, 혼합물을 실온에서 16 시간 동안 교반하였다. 반응 혼합물을 농축시키고, 잔류물을 메탄올 (35 mL)에 용해시켰다. 수성 H2O2 (30%, 10.2 mL) 및 10% 수성 NaHCO3 (10.2 mL)을 첨가하고, 혼합물을 실온에서 16 시간 동안 교반하였다. 반응 혼합물을 감압하에 농축시켰다. 잔류물을 EtOAc에 용해시키고, 1 N HCl, 염수로 세척하고, MgSO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 잔류물을 실리카겔 플래쉬 크로마토그래피 (10-20% DCM 중의 EtOAc)에 의해 정제하여, 디에틸 2-아세트아미도-2-(4-아세틸-3-니트로벤질)말로네이트를 수득하였다.To diethyl 2-acetamido-2-(4-fluoro-3-nitrobenzyl)malonate (7.41 g) and nitroethane (6.0 mL) in EtOAc was added DBU (9.0 mL) and the mixture was at room temperature Stir for 16 hours. The reaction mixture was concentrated and the residue was dissolved in methanol (35 mL). Aqueous H 2 O 2 (30%, 10.2 mL) and 10% aqueous NaHCO 3 (10.2 mL) were added and the mixture was stirred at room temperature for 16 hours. The reaction mixture was concentrated under reduced pressure. The residue was dissolved in EtOAc, washed with 1 N HCl, brine, dried over MgSO 4 , filtered and concentrated under reduced pressure. The residue was purified by silica gel flash chromatography (10-20% EtOAc in DCM) to give diethyl 2-acetamido-2-(4-acetyl-3-nitrobenzyl)malonate.
디에틸 2-아세트아미도-2-(4-아세틸-3-니트로벤질)말로네이트 (2.0 g)를 10 mL 37% HCl에 용해시키고, 100℃에서 밤새 가열하고, 냉각시켰다. 생성된 고체를 여과에 의해 수집하여, 3-(4-아세틸-3-니트로페닐)-2-아미노프로판산을 수득하였다.Diethyl 2-acetamido-2-(4-acetyl-3-nitrobenzyl)malonate (2.0 g) was dissolved in 10
3-(4-아세틸-3-니트로페닐)-2-아미노프로판산을 MeOH 중의 5% 탄소상 Pd를 이용하여 1 atm H2 하에 환원시켰다. 여과한 후 감압하에 농축시켜 3-(4-아세틸-3-아미노페닐)-2-아미노프로판산 (X3179-96)을 수득하였다.3-(4-acetyl-3-nitrophenyl)-2-aminopropanoic acid was reduced under 1 atm H 2 with 5% Pd on carbon in MeOH. After filtration, it was concentrated under reduced pressure to give 3-(4-acetyl-3-aminophenyl)-2-aminopropanoic acid (X3179-96).
실시예Example 34-9: 1-(2-아미노-5- 34-9: 1-(2-amino-5- 브로모페닐Bromophenyl )) 에타논Ethanone ( ( X1436X1436 -132)의 합성. -132).
1-(2-아미노페닐)에타논 (135 mg) 및 DCM (2.5 mL)이 채워진 플라스크를 -10℃로 냉각시키고, NBS (178 mg)를 수회로 나누어 30 분에 걸쳐 첨가하였다. 반응 혼합물을 10 mL DCM으로 희석하고, 포화 수성 NaHCO3으로 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 표제 화합물을 수득하였다.A flask filled with 1-(2-aminophenyl)ethanone (135 mg) and DCM (2.5 mL) was cooled to -10° C. and NBS (178 mg) was added in several portions over 30 minutes. The reaction mixture was diluted with 10 mL DCM, washed with saturated aqueous NaHCO 3 , dried over Na 2 SO 4 , filtered and concentrated under reduced pressure to give the title compound.
실시예Example 34-10: 1-(2-아미노-5- 34-10: 1-(2-amino-5- 요오도페닐Iodophenyl )) 에타논Ethanone ( ( X1436X1436 -134)의 합성.-134).
1-(2-아미노페닐)에타논 (405 mg) 및 DCM (20 mL)이 채워진 플라스크를 얼음/물 욕조에서 냉각시키고, NIS (675 mg)를 3회로 나누어 첨가하였다. 반응물을 0℃에서 교반하였다. 반응 혼합물을 30 mL DCM으로 희석하고, 포화 수성 NaHCO3으로 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 조 물질을 정제용 RP-HPLC에 의해 정제하여, 표제 화합물을 수득하였다. A flask filled with 1-(2-aminophenyl)ethanone (405 mg) and DCM (20 mL) was cooled in an ice/water bath and NIS (675 mg) was added in 3 portions. The reaction was stirred at 0 °C. The reaction mixture was diluted with 30 mL DCM, washed with saturated aqueous NaHCO 3 , dried over Na 2 SO 4 , filtered and concentrated under reduced pressure. The crude material was purified by preparative RP-HPLC to give the title compound.
실시예Example 34-11: 2-아미노-4- 34-11: 2-amino-4- 브로모벤즈알데히드Bromobenzaldehyde ( ( X3547X3547 -158)의 합성.-158).
철 분말 (1.20 g), 물 (2.0 mL), 및 HCl (12 N, 40 ㎕)을 에탄올 (8.0 mL) 중의 2-니트로벤즈알데히드 (230 mg)의 용액에 연속 첨가하였다. 95℃에서 90 분 동안 교반한 후, 반응 혼합물을 고온상태로 여과하였다. 에탄올로 세척한 후, 여과물을 합하고, 용매를 진공하에 제거하였다. 조 물질을 실리카겔 플래쉬 크로마토그래피 (40:55:5 헥산데틸 아세테이트/트리에틸아민)에 의해 정제하여, 2-아미노-4-브로모벤즈알데히드를 황색 결정으로서 수득하였다. Iron powder (1.20 g), water (2.0 mL), and HCl (12 N, 40 μl) were added successively to a solution of 2-nitrobenzaldehyde (230 mg) in ethanol (8.0 mL). After stirring at 95° C. for 90 minutes, the reaction mixture was filtered at high temperature. After washing with ethanol, the filtrates were combined and the solvent was removed under vacuum. The crude was purified by silica gel flash chromatography (40:55:5 hexanedecyl acetate/triethylamine) to give 2-amino-4-bromobenzaldehyde as yellow crystals.
실시예Example 35: 35: PCLPCL 및 피롤리신 생합성 전구체의 합성 And synthesis of pyrrolysine biosynthetic precursors
실시예Example 35-1: 암모늄 35-1: ammonium DLDL -1--One- 피롤린Pyrroline -5--5- 카르복실레이트Carboxylate (3793-007)의 합성. Synthesis of (3793-007).
H-DL-δ-DL-히드록시Lys-OH (118 mg, 0.5 mmol)의 HCl 염을 양이온 교환 SPE 카트리지에 적용하고, 수산화암모늄을 이용하여 아미노산을 용리시켰다. NaIO4 (107 mg)를 암모늄 용리액에 첨가하고, 반응 혼합물을 실온에서 30 분 동안 교반하였다. 반응 혼합물을 동결건조시키고, 조 생성물을 HCl을 이용하여 pH 2.6으로 산성화하고, 양이온 교환 SPE에 의해 정제하였다. 처음 10 mL의 H2O 용리액을 따라낸 후, 다음 30 ml의 H2O 용리액을 수집하고, 1 N NH4OH( aq ) (0.5 mL)로 즉시 중화시켰다. 동결건조한 후, 목적하는 생성물을 밝은 황색 분말로서 수득하였다.The HCl salt of H-DL-δ-DL-hydroxyLys-OH (118 mg, 0.5 mmol) was applied to a cation exchange SPE cartridge and the amino acids were eluted with ammonium hydroxide. NaIO 4 (107 mg) was added to the ammonium eluent and the reaction mixture was stirred at room temperature for 30 minutes. The reaction mixture was lyophilized and the crude product was acidified to pH 2.6 with HCl and purified by cation exchange SPE. After decanting the first 10 mL of H 2 O eluent, the next 30 ml of H 2 O eluent was collected and neutralized immediately with 1 N NH 4 OH ( aq ) (0.5 mL). After lyophilization, the desired product was obtained as a light yellow powder.
실시예Example 35-2: H-L- 35-2: H-L- LysLys -N-N εε -(D--(D- OrnOrn )-)- OHOH (3793-031)의 합성 Synthesis of (3793-031)
Boc-D-Orn(Boc)-OH (1.097 g)를 DMF (5 mL) 중의 HATU (1.255 g) 및 DIEA (1.53 mL)로 1 시간 동안 처리하였다. 이어서, Boc-Lys-OMe (아세테이트 염, 1.057 g)의 DMF 용액 (5 mL) 및 DIEA (627 ㎕)를 반응 혼합물에 첨가하고, 반응물을 실온에서 2 시간 동안 교반하였다. 반응 혼합물을 10% NaCl( aq ) (30 mL) 및 EtOAc (60 mL) 사이에 분배시켰다. EtOAc 층을 5% 시트르산 (30 mL), 10% NaHCO3(aq) (30 mL), 및 염수 (30 mL)로 세척하였다. EtOAc 층을 Na2SO4 (s) 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 잔류물을 80 g 실리카 칼럼 및 0-15% MeOH/DCM의 구배 용리를 이용한 실리카겔 플래쉬 칼럼 크로마토그래피에 의해 정제하여, Boc-Lys(Boc-D-Orn(Boc))-OMe (3793-026)를 무색 오일로서 수득하였다. Boc-D-Orn(Boc)-OH (1.097 g) was treated with HATU (1.255 g) and DIEA (1.53 mL) in DMF (5 mL) for 1 hour. Then, a DMF solution (5 mL) and DIEA (627 μL) of Boc-Lys-OMe (acetate salt, 1.057 g) were added to the reaction mixture, and the reaction was stirred at room temperature for 2 hours. The reaction mixture was partitioned between 10% NaCl ( aq ) (30 mL) and EtOAc (60 mL). The EtOAc layer was washed with 5% citric acid (30 mL), 10% NaHCO 3(aq) (30 mL), and brine (30 mL). The EtOAc layer was dried over Na 2 SO 4 (s) , filtered and concentrated under reduced pressure. The residue was purified by silica gel flash column chromatography using an 80 g silica column and gradient elution of 0-15% MeOH/DCM, Boc-Lys(Boc-D-Orn(Boc))-OMe (3793-026) Was obtained as a colorless oil.
Boc-Lys(Boc-D-Orn(Boc))-OMe (214 mg)의 65% 아세토니트릴 수용액 (5 mL)에 진한 HCl (1 mL)을 첨가하고, 반응 혼합물을 2 시간 동안 강하게 교반하였다. 반응 혼합물을 동결건조시키고, SCX SPE 카트리지를 이용한 양이온 교환 크로마토그래피에 의해 20% MeCN( aq ) 중의 1 N NH4OH로 정제하였다. 메틸 에스테르를 NH4OH 용리액 중에서 밤새 가수분해하였다. 동결건조시킨 후, H-L-Lys-Nε-(D-Orn)-OH (3793-031)를 백색 분말로서 수득하였다. To 65% aqueous acetonitrile solution (5 mL) of Boc-Lys(Boc-D-Orn(Boc))-OMe (214 mg) was added concentrated HCl (1 mL), and the reaction mixture was stirred vigorously for 2 hours. The reaction mixture was lyophilized and purified with 1 N NH 4 OH in 20% MeCN ( aq ) by cation exchange chromatography using an SCX SPE cartridge. The methyl ester was hydrolyzed in NH 4 OH eluent overnight. After lyophilization, HL-Lys-N ε -(D-Orn)-OH (3793-031) was obtained as a white powder.
실시예 35-3: N-((6- 클로로피리딘 -3-일) 메틸 )-1- 피롤린 -5- 카르복스아미드 (3647-061)의 합성. Example 35-3: N-((6 -chloropyridin- 3-yl) methyl )-1 -pyrroline -5 -carboxamide Synthesis of (3647-061).
디옥산 (10 mL) 중의 t-부틸피로카르보네이트 (6.55 g)를 1 N NaOH( aq ) (50 mL) 중의 DL-δ-DL-히드록시Lys (1.99 g)에 적가하고, 반응물을 실온에서 밤새 교반하였다. 1 N HCl( aq )을 이용하여 반응 혼합물을 pH 3.0으로 산성화하고, EtOAc (200 mL)로 2 회 추출하였다. EtOAc 층을 합하고, Na2SO4 (s) 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 조 생성물을 수득하였다. 조 생성물을 실리카겔 플래쉬 칼럼 크로마토그래피에 의해 0-10% MeOH/DCM의 구배 용리로 정제하여, Boc-DL-δ-DL-히드록시Lys(Boc) (3597-109)을 백색 고체로서 수득하였다.T-butyl pyrocarbonate (6.55 g) in dioxane (10 mL) was added dropwise to DL-δ-DL-hydroxyLys (1.99 g) in 1 N NaOH ( aq ) (50 mL) and the reaction was brought to room temperature. Stir overnight at. The reaction mixture was acidified to pH 3.0 with 1 N HCl ( aq ) and extracted twice with EtOAc (200 mL). The EtOAc layers were combined, dried over Na 2 SO 4 (s) , filtered and concentrated under reduced pressure to give the crude product. The crude product was purified by silica gel flash column chromatography with gradient elution of 0-10% MeOH/DCM to give Boc-DL-δ-DL-hydroxyLys(Boc) (3597-109) as a white solid.
Boc-DL-δ-DL-히드록시Lys(Boc) (435 mg)를 3.5 mL DMF 중의 HATU (456 mg) 및 DIEA (523 ㎕)로 1 시간 동안 처리하였다. 생성된 용액을 1.5 mL DMF 용액 중의 6-클로로피리딘-3-일메틸아민 (143 mg)에 실온에서 첨가하고, 반응물을 2 일 동안 교반하였다. 반응 혼합물을 10% NaCl( aq ) (15 mL) 및 EtOAc (30 mL)로 추출하였다. EtOAc 층을 5% 시트르산 (15 mL), 10% NaHCO3 (15 mL), 및 염수 (15 mL)로 세척하였다. 이어서, EtOAc 층을 Na2SO4 (s) 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 조 생성물을 수득하였다. 조 생성물을 실리카겔 플래쉬 칼럼 크로마토그래피에 의해 0-15% MeOH/DCM의 구배 용리로 정제하여, 목적하는 생성물 (3647-016)을 백색 고체로서 수득하였다.Boc-DL-δ-DL-hydroxyLys(Boc) (435 mg) was treated with HATU (456 mg) and DIEA (523 μL) in 3.5 mL DMF for 1 hour. The resulting solution was added to 6-chloropyridin-3-ylmethylamine (143 mg) in 1.5 mL DMF solution at room temperature and the reaction was stirred for 2 days. The reaction mixture was extracted with 10% NaCl ( aq ) (15 mL) and EtOAc (30 mL). The EtOAc layer was washed with 5% citric acid (15 mL), 10% NaHCO 3 (15 mL), and brine (15 mL). The EtOAc layer was then dried over Na 2 SO 4 (s) , filtered and concentrated under reduced pressure to give the crude product. The crude product was purified by silica gel flash column chromatography with gradient elution of 0-15% MeOH/DCM to give the desired product (3647-016) as a white solid.
상기의 생성물 (3647-016) (263 mg)을 Et2O (7 mL) 및 1 N HCl( aq ) (7 mL) 용액 중에서 실온에서 밤새 교반하였다. 반응 혼합물을 증발시키고 동결건조시킨 후, 잔류물을 양이온 교환 SPE 카트리지에 의해 정제하여, 목적하는 생성물 (3647-037)을 백색 고체로서 수득하였다.The above product (3647-016) (263 mg) was stirred overnight at room temperature in a solution of Et 2 O (7 mL) and 1 N HCl ( aq ) (7 mL). After the reaction mixture was evaporated and lyophilized, the residue was purified by a cation exchange SPE cartridge to give the desired product (3647-037) as a white solid.
상기의 생성물 (3647-037) (69.6 mg)을 2 mL H2O (2 mL)에 용해시키고, 1 N NaOH(aq)를 이용하여 pH를 12로 조정하였다. PL-IO4 수지 (베리언(Varian), 381 mg, 0.48 mmol)를 첨가하고, 반응물을 실온에서 2 시간 동안 교반하였다. 수지를 여과에 의해 제거하고, 여과물의 절반을 정제용 HPLC에 의해 정제하여, 목적하는 생성물 (3647-061)을 백색 고체로서 수득하였다.The above product (3647-037) (69.6 mg) was dissolved in 2 mL H 2 O (2 mL), and the pH was adjusted to 12 using 1 N NaOH (aq). PL-IO 4 resin (Varian, 381 mg, 0.48 mmol) was added and the reaction was stirred at room temperature for 2 hours. The resin was removed by filtration and half of the filtrate was purified by preparative HPLC to give the desired product (3647-061) as a white solid.
실시예Example 36: 36: PCLPCL 모델 화합물의 합성 Synthesis of model compounds
실시예Example 36-1: H-L- 36-1: H-L- LysLys -Nε-(-Nε-( DLDL -1--One- 피롤린Pyrroline -5-카르보닐)--5-carbonyl)- OHOH (3647-125)의 합성. Synthesis of (3647-125).
Boc-DL-δ-DL-히드록시Lys(Boc) (3597-109) (886 mg)를 DIEA (1068 ㎕)의 존재하에서 3.0 mL DMF 중의 HATU (932 mg)에 의해 1 시간 동안 활성화시키고, 3.0 mL DMF (3.0 mL) 중의 Boc-Lys-OMe (549 mg)에 첨가하였다. 반응물을 실온에서 36 시간 동안 교반하였다. 반응 혼합물을 10% NaCl( aq ) (30 mL) 및 EtOAc (60 mL) 사이에 분배시켰다. EtOAc 층을 5% 시트르산 (15 mL), 10% NaHCO3 (15 mL), 및 염수 (15 mL)로 세척하였다. EtOAc 층을 Na2SO4 (s) 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 조 생성물을 수득하였다. 조 생성물을 0-15% MeOH/DCM을 이용한 실리카겔 플래쉬 칼럼 크로마토그래피에 의해 정제한 후, 25% 및 65% MeCN( aq )의 용리를 이용한 RP-C18 SPE에 의해 정제하여, 목적하는 생성물 (3647-099)을 백색 고체로서 수득하였다.Boc-DL-δ-DL-hydroxyLys(Boc) (3597-109) (886 mg) was activated by HATU (932 mg) in 3.0 mL DMF in the presence of DIEA (1068 μl) for 1 hour and 3.0 Added to Boc-Lys-OMe (549 mg) in mL DMF (3.0 mL). The reaction was stirred at room temperature for 36 hours. The reaction mixture was partitioned between 10% NaCl ( aq ) (30 mL) and EtOAc (60 mL). The EtOAc layer was washed with 5% citric acid (15 mL), 10% NaHCO 3 (15 mL), and brine (15 mL). The EtOAc layer was dried over Na 2 SO 4 (s) , filtered and concentrated under reduced pressure to give the crude product. The crude product was purified by silica gel flash column chromatography using 0-15% MeOH/DCM, and then purified by RP-C18 SPE using elution of 25% and 65% MeCN ( aq ), and the desired product (3647 -099) was obtained as a white solid.
1 mL Et2O 중의 Boc-Lys(Boc-DL-δ-DL-히드록시Lys(Boc))-OMe (3647-099) (84.1 mg)에 4 N HCl( aq ) (3 mL)를 첨가하고, 반응물을 실온에서 3 시간 동안 교반하였다. 용매를 제거하여, 61.1 mg의 조 생성물 (3647-120)을 백색 고체로서 수득하였다.To Boc-Lys(Boc-DL-δ-DL-hydroxyLys(Boc))-OMe (3647-099) (84.1 mg) in 1 mL Et 2 O was added 4 N HCl ( aq ) (3 mL) and , The reaction was stirred at room temperature for 3 hours. Removal of the solvent gave 61.1 mg of crude product (3647-120) as a white solid.
상기의 조 생성물 (3647-120) (61.1 mg)을 1 mL H2O에 용해시키고, 1 N NaOH(aq)를 이용하여 pH를 12로 조정하였다. 혼합물을 실온에서 1 시간 동안 교반하여, 메틸 에스테르를 가수분해시켰다. 이어서 NaIO4 (29.9 mg)를 반응 혼합물에 첨가하고, 반응물을 실온에서 추가로 15 분 동안 교반하였다. 반응 혼합물을 1 N HCl(aq)으로 중화시키고, 정제를 위해 양이온 교환 SPE에 로딩하여, H-L-Lys-Nε-(DL-1-피롤린-5-카르보닐)-OH (3647-125)를 수득하였다.The above crude product (3647-120) (61.1 mg) was dissolved in 1 mL H 2 O, and the pH was adjusted to 12 using 1 N NaOH (aq). The mixture was stirred at room temperature for 1 hour to hydrolyze the methyl ester. Then NaIO 4 (29.9 mg) was added to the reaction mixture, and the reaction was stirred at room temperature for an additional 15 minutes. The reaction mixture was neutralized with 1 N HCl (aq) and loaded into cation exchange SPE for purification to obtain HL-Lys-Nε-(DL-1-pyrroline-5-carbonyl)-OH (3647-125). Obtained.
실시예Example 36-2: H-L- 36-2: H-L- LysLys -N-N εε -(-( DLDL -1--One- 피롤린Pyrroline -2-카르보닐)--2-carbonyl)- OHOH (3793-011)의 합성. Synthesis of (3793-011).
문헌 [Gyoergy Szoelloesi et al., Chirality, 2001, 13(10), 619-624]에 기재되어 있는 이하의 방법을 이용하여 나트륨 1-피롤린-2-카르복실레이트 (3647-164)를 제조하였다.Sodium 1-pyrroline-2-carboxylate (3647-164) was prepared using the following method described in the literature [Gyoergy Szoelloesi et al., Chirality, 2001, 13(10), 619-624]. .
트리에틸아민 (8.4 mL)을 에테르 (27 mL) 중의 H-Pro-OMe HCl (7.512 g)에 첨가하고, 반응물을 2 시간 동안 교반하였다. 반응 혼합물을 여과하고, Et2O를 증발에 의해 제거하였다. 잔류물을 진공 증류에 의해 정제하여, 4.119 g의 H-Pro-OMe를 무색 오일로서 수득하였다. t-BuOCl (3.59 mL)를 H-Pro-OMe (4.089 g)의 Et2O (100 mL) 용액에 적가하고, 반응물을 -50℃에서 1 시간 동안 교반하였다. 반응 혼합물을 실온으로 가온하고, 트리에틸아민 (4.64 mL)을 반응 혼합물에 첨가한 후, 3 일 동안 교반하였다. 반응 혼합물을 여과하고, 감압하에 농축시켰다. 잔류물을 진공 증류에 의해 정제하여, 메틸 피롤린-2-카르복실레이트 (3647-154)를 무색 오일로서 수득하였다. Triethylamine (8.4 mL) was added to H-Pro-OMe HCl (7.512 g) in ether (27 mL) and the reaction was stirred for 2 hours. The reaction mixture was filtered and Et 2 O was removed by evaporation. The residue was purified by vacuum distillation to give 4.119 g of H-Pro-OMe as a colorless oil. t-BuOCl (3.59 mL) was added dropwise to a solution of Et 2 O (100 mL) in H-Pro-OMe (4.089 g), and the reaction was stirred at -50°C for 1 hour. The reaction mixture was warmed to room temperature and triethylamine (4.64 mL) was added to the reaction mixture, followed by stirring for 3 days. The reaction mixture was filtered and concentrated under reduced pressure. The residue was purified by vacuum distillation to give methyl pyrroline-2-carboxylate (3647-154) as a colorless oil.
메틸 피롤린-2-카르복실레이트 (2.265 g)를 1 N NaOH( aq ) (17.8 mL)에 용해시키고, 실온에서 1 시간 동안 교반하였다. 반응 혼합물을 동결건조시켜, 조질의 나트륨 피롤린-2-카르복실레이트 (3647-164)를 백색 분말로서 수득하였다.Methyl pyrroline-2-carboxylate (2.265 g ) was dissolved in 1 N NaOH ( aq ) (17.8 mL) and stirred at room temperature for 1 hour. The reaction mixture was lyophilized to give crude sodium pyrroline-2-carboxylate (3647-164) as a white powder.
나트륨 1-피롤린-2-카르복실레이트 (459 mg) 및 DPPA (886 ㎕)를 20 mL DMF 중의 Boc-Lys-OMe (아세테이트 염, 897 mg) 및 DIEA (1184 ㎕)에 첨가하고, 반응물을 N2 하에 24 시간 동안 실온에서 교반하였다. 반응 혼합물을 10% NaCl( aq ) (50 mL) 및 EtOAc (100 mL) 사이에 분배시켰다. EtOAc 층을 Na2SO4 (s) 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 조 생성물을 밝은 오렌지색 오일로서 수득하였다. 조 생성물을 실리카겔 플래쉬 칼럼 크로마토그래피에 의해 0-15% MeOH/DCM의 구배 용리로 정제한 후, RP-C18 SPE에 의해 정제하여, 목적하는 생성물 (3647-167)을 무색 오일로서 수득하였다. Sodium 1-pyrroline-2-carboxylate (459 mg) and DPPA (886 μL) were added to Boc-Lys-OMe (acetate salt, 897 mg) and DIEA (1184 μL) in 20 mL DMF and the reaction was Stirred at room temperature for 24 hours under N 2. The reaction mixture was partitioned between 10% NaCl ( aq ) (50 mL) and EtOAc (100 mL). The EtOAc layer was dried over Na 2 SO 4 (s) , filtered and concentrated under reduced pressure to give the crude product as a light orange oil. The crude product was purified by silica gel flash column chromatography with gradient elution of 0-15% MeOH/DCM and then purified by RP-C18 SPE to give the desired product (3647-167) as a colorless oil.
HCl (진함, 1 mL)을 상기 생성물 (3647-167) (49.3 mg)의 50% MeCN(aq) 용액 (5 mL)에 첨가하고, 반응물을 실온에서 2 시간 동안 교반하였다. 동결건조시킨 후, 잔류물을 양이온 교환 SPE 카트리지에 의해 정제하였다. 메틸 에스테르를 용리액 중에서 NH4OH( aq )로 밤새 가수분해하였다. 동결건조시킨 후, 목적하는 생성물 (3793-011)을 밝은 황색 분말로서 수득하였다.HCl (conc., 1 mL) was added to 50% MeCN(aq) of the product (3647-167) (49.3 mg) It was added to the solution (5 mL) and the reaction was stirred at room temperature for 2 hours. After lyophilization, the residue was purified by a cation exchange SPE cartridge. The methyl ester was hydrolyzed overnight with NH 4 OH ( aq ) in the eluent. After lyophilization, the desired product (3793-011) was obtained as a light yellow powder.
실시예Example 37: 2- 37: 2- AAPAAP -- PEGPEG 시약의 합성 Synthesis of reagents
본원에서 제공하는 실시예에 사용되는 다른 2-AAP-PEG 유도체의 합성을 이하에서 기재한다. 분자량이 대략 500, 2400, 23000 Da 뿐만 아니라 5000 및 10000 Da인 PEG의 2-아미노-아세토페논 유도체를 다음과 같이 합성하였다:Synthesis of other 2-AAP-PEG derivatives used in the examples provided herein is described below. The 2-amino-acetophenone derivatives of PEG with molecular weights of approximately 500, 2400, 23000 Da as well as 5000 and 10000 Da were synthesized as follows:
실시예Example 37-1: 37-1: TU3205TU3205 -044 (0.5 -044 (0.5 kDakDa 2- 2- AAPAAP -- mPEGmPEG )의 합성) Of the synthesis
리튬 2-(4-아세틸-3-아미노페녹시)아세테이트 (TU3205-042) (55.9 mg)를 10 mL 둥근 바닥 플라스크에 채우고, DMF (3 mL) 및 HATU (98.9 mg)를 첨가하였다. 생성된 슬러리를 주위 온도에서 40 분 동안 교반하였다. 이 기간 동안 반응물이 황색 용액으로 바뀌었다. 이어서 반응물에 2 mL DMF 중의 mPEG-아민 (쿠안타 바이오디자인(Quanta Biodesign), MW 383.5, 100 mg)을 첨가하였다. 5 분 후의 LC-MS 분석은 반응이 완료되었음을 나타내었다. 반응 혼합물을 추가로 40 분 동안 교반하고, 감압하에 농축시켰다. 잔류물을 SiO2 플래쉬 크로마토그래피 (3% DCM 중의 MeOH)에 의해 정제하여, TU3205-044 (0.5 kDa 2-AAP-mPEG)를 황색 점성 오일로서 수득하였다.Lithium 2-(4-acetyl-3-aminophenoxy)acetate (TU3205-042) (55.9 mg) was charged to a 10 mL round bottom flask, and DMF (3 mL) and HATU (98.9 mg) were added. The resulting slurry was stirred at ambient temperature for 40 minutes. During this period the reaction turned into a yellow solution. Then to the reaction was added mPEG-amine (Quanta Biodesign, MW 383.5, 100 mg) in 2 mL DMF. LC-MS analysis after 5 minutes indicated the reaction was complete. The reaction mixture was stirred for an additional 40 minutes and concentrated under reduced pressure. The residue was purified by SiO 2 flash chromatography (MeOH in 3% DCM) to give TU3205-044 (0.5 kDa 2-AAP-mPEG) as a yellow viscous oil.
실시예Example 37-2: 37-2: TU3205TU3205 -048 (2.4 -048 (2.4 kDakDa 2- 2- AAPAAP -- PEGPEG )의 합성) Of the synthesis
리튬 2-(4-아세틸-3-아미노페녹시)아세테이트 (TU3205-042) (12.2 mg)를 10 mL 둥근 바닥 플라스크에 채우고, DMF (1 mL) 및 HATU (21.5 mg)를 첨가하였다. 생성된 슬러리를 주위 온도에서 35 분 동안 교반하였다. 이 기간 동안 반응물이 황색 용액으로 바뀌었다. 이어서 반응물에 2 mL DMF 중의 mPEG-아민 (쿠안타 바이오디자인, MW 2209, 100 mg)을 첨가하였다. 반응 혼합물을 18 시간 동안 교반하고, 감압하에 농축시켰다. 잔류물을 SiO2 플래쉬 크로마토그래피 (DCM 중의 MeOH)에 의해 정제하여, TU3205-048 (2.4 kDa 2-AAP-mPEG)을 황색 점성 오일로서 수득하였다. Lithium 2-(4-acetyl-3-aminophenoxy)acetate (TU3205-042) (12.2 mg) was charged to a 10 mL round bottom flask, and DMF (1 mL) and HATU (21.5 mg) were added. The resulting slurry was stirred at ambient temperature for 35 minutes. During this period the reaction turned into a yellow solution. Then to the reaction was added mPEG-amine (Quanta Biodesign,
실시예Example 37-3: 37-3: TU3205TU3205 -052 (23 -052 (23 kDakDa 2- 2- AAPAAP -- PEGPEG )의 합성 ) Of the synthesis
리튬 2-(4-아세틸-3-아미노페녹시)아세테이트 (TU3205-042) (21.5 mg) 및 HATU (38.0 mg)를 10 mL 둥근 바닥 플라스크에 채우고, DMF (0.5 mL)를 첨가하였다. 생성된 슬러리를 주위 온도에서 50 분 동안 교반하였다. 생성된 황색 용액을 20 mL 유리 바이알 내의 5 mL DMF 중의 mPEG-NH2 (레이산 바이오(Laysan Bio), 평균 MW 23k, 평균 n = 520, 0.50 g)에 첨가하였다. 반응물을 주위 온도에서 2.5 시간 동안 진탕하였다. 이어서, 반응 혼합물을 10 mL 물로 희석하고, 각 용액의 2.5 mL 분취량을 PD-10 칼럼 (GE 헬스테크(GE Healthtech))에 적용하고, 목적하는 생성물을 공급자의 지침에 따라 물로 용리하였다. 풀링된 수용액을 풀링하고, 동결시키고, 동결건조시켜, TU3205-052 (23 kDa 2-AAP-mPEG)를 백색 고체로서 수득하였다. (주석; 높은 MW를 특징으로 하는 PEG 시약은 그것이 PCL 함유 단백질에 접합된 이후에만 수득되었다.)Lithium 2-(4-acetyl-3-aminophenoxy)acetate (TU3205-042) (21.5 mg) and HATU (38.0 mg) were charged to a 10 mL round bottom flask, and DMF (0.5 mL) was added. The resulting slurry was stirred at ambient temperature for 50 minutes. The resulting yellow solution was added to mPEG-NH 2 (Laysan Bio, average MW 23k, average n = 520, 0.50 g) in 5 mL DMF in a 20 mL glass vial. The reaction was shaken at ambient temperature for 2.5 hours. The reaction mixture was then diluted with 10 mL water, a 2.5 mL aliquot of each solution was applied to a PD-10 column (GE Healthtech), and the desired product was eluted with water according to the supplier's instructions. The pooled aqueous solution was pooled, frozen and lyophilized to give TU3205-052 (23 kDa 2-AAP-mPEG) as a white solid. (Note; PEG reagent characterized by high MW was obtained only after it was conjugated to PCL containing protein.)
및 And
는 상응하는 10,000 (평균 n = 225) 및 5,000 (평균 n = 111) MW mPEG-NH2를 이용하여 TU3205-052와 유사한 방식으로 제조하였다. (주석; 높은 MW를 특징으로 하는 PEG 시약은 그것이 PCL 함유 단백질에 접합된 이후에만 수득되었다.)Were prepared in a similar manner to TU3205-052 using the corresponding 10,000 (average n = 225) and 5,000 (average n = 111) MW mPEG-NH 2. (Note; PEG reagent characterized by high MW was obtained only after it was conjugated to PCL containing protein.)
실시예Example 37-4: 37-4: TU633TU633 -010: 2--010: 2- 관능성Organoleptic 링커의 합성 Linker synthesis
리튬 2-(4-아세틸-3-아미노페녹시)아세테이트 (TU3205-042) (94.6 mg) 및 HATU (167 mg)를 10 mL 둥근 바닥 플라스크에 채우고, DMF (2 mL)를 첨가하였다. 생성된 슬러리를 주위 온도에서 45 분 동안 교반하였다. 생성된 황색 용액에 1 mL DMF 중의 4,7,10-트리옥사-1,13-트리데칸디아민 (플루카(Fluka), 44 mg)을 첨가하였다. 반응 혼합물을 주위 온도에서 18 시간 동안 교반하고, 감압하에 농축시켰다. 잔류물을 SiO2 플래쉬 크로마토그래피 (DCM 중의 MeOH)에 의해 정제한 후, 정제용 역상 LC 정제하였다. LC 정제된 물질을 EtOAc에 용해시키고, 포화 수성 NaHCO3으로 처리하여 트리플루오로아세트산을 제거하였다. 용매를 증발시켜 2 관능성 링커 TU633-010을 맑은 오일로서 수득하였다.Lithium 2-(4-acetyl-3-aminophenoxy)acetate (TU3205-042) (94.6 mg) and HATU (167 mg) were charged to a 10 mL round bottom flask, and DMF (2 mL) was added. The resulting slurry was stirred at ambient temperature for 45 minutes. To the resulting yellow solution was added 4,7,10-trioxa-1,13-tridecanediamine (Fluka, 44 mg) in 1 mL DMF. The reaction mixture was stirred at ambient temperature for 18 hours and concentrated under reduced pressure. The residue was purified by SiO 2 flash chromatography (MeOH in DCM) followed by preparative reverse phase LC purification. The LC purified material was dissolved in EtOAc and treated with saturated aqueous NaHCO 3 to remove trifluoroacetic acid. Evaporation of the solvent gave the bifunctional linker TU633-010 as a clear oil.
실시예Example 37-5: m- 37-5: m- PEGPEG -- AAPAAP 29k ( 29k ( TU633TU633 -084)의 합성.-084).
리튬 2-(4-아세틸-3-아미노페녹시)아세테이트 (187 mg) 및 HBTU (330 mg)를 20 mL 유리 바이알에 넣고, 10 mL 무수 DMF를 첨가하였다. 생성된 슬러리를 주위 온도에서 교반하였다. 20 분 이내에 반응물이 황색 용액으로 바뀌었고, 80 분 후에 반응 혼합물의 9.5 mL 분취량을 100 mL 둥근 바닥 플라스크 내의 40 mL의 무수 DMF에 용해된 mPEG-NH2 (레이산 바이오, MW 28,700)에 첨가하였다. 반응물을 주위 온도에서 19 시간 동안 진탕하였다. 이어서 반응 혼합물을 24 조각의 PD-10 칼럼 (GE 헬스테크)에 적용하고, 목적하는 생성물을 공급자의 지침에 따라 물로 용리시켰다. 풀링된 용리액을 동결 및 동결건조하여, 백색 고체를 수득하였다. 고체를 이중으로 탈이온화된 물에 용해시키고, MWCO 3500의 투석막을 이용하여 이중으로 탈이온화된 물에 대해 철저히 투석하였다. 투석된 용액을 동결 및 동결건조시켜, TU633-084를 백색의 솜털같은 고체로서 수득하였다. (주석; 높은 MW를 특징으로 하는 PEG 시약은 그것이 PCL 함유 단백질에 접합된 이후에만 수득되었다.)Lithium 2-(4-acetyl-3-aminophenoxy)acetate (187 mg) and HBTU (330 mg) were placed in a 20 mL glass vial, and 10 mL anhydrous DMF was added. The resulting slurry was stirred at ambient temperature. The reaction turned into a yellow solution within 20 minutes, and after 80 minutes a 9.5 mL aliquot of the reaction mixture was added to mPEG-NH 2 (Raysan Bio, MW 28,700) dissolved in 40 mL of anhydrous DMF in a 100 mL round bottom flask. I did. The reaction was shaken at ambient temperature for 19 hours. The reaction mixture was then applied to 24 pieces of PD-10 column (GE Healthtech) and the desired product was eluted with water according to the supplier's instructions. The pooled eluent was frozen and lyophilized to obtain a white solid. The solid was dissolved in double deionized water and thoroughly dialyzed against double deionized water using a dialysis membrane of MWCO 3500. The dialyzed solution was frozen and lyophilized to give TU633-084 as a white fluffy solid. (Note; PEG reagent characterized by high MW was obtained only after it was conjugated to PCL containing protein.)
실시예Example 37-6: 37-6: mPEGmPEG -- AAPAAP -30k (-30k ( TU633TU633 -120)의 합성-120)
리튬 3-(3-아세틸-4-아미노페닐)프로파노에이트 (139 mg) 및 HBTU (247 mg)를 20 mL 유리 바이알에 넣고, 13 mL 무수 DMF를 첨가하였다. 생성된 슬러리를 주위 온도에서 30 분 동안 교반하고, 생성된 용액을 TU633-120, TU633-122, TU633-124 및 TU633-126을 제조하는데 사용하였다. 45 mL 유리 바이알에 mPEG아민 (NOF Corp., SUNBRIGHT MEPA-30T MW 30,298, 3.0 g)을 넣고, 20 mL 무수 DMF를 첨가한 후, 가볍게 가열하여 mPEG아민을 DMF에 용해시켰다. 생성된 mPEG아민 용액에 활성화된 에스테르 용액의 2.2 mL 분취량을 첨가하고, 바이알을 주위 온도에서 18 시간 동안 진탕한 다음, 37℃에서 24 시간 동안 진탕하였다. 반응 혼합물을 투석막 튜빙 (피셔 사이언티픽(Fisher Scientific), cat # 21-152-9, MWCO 3500)으로 옮기고, 이중으로 탈이온화된 물에 대해 2 일에 걸쳐 철저하게 투석하였다. 투석된 용액을 동결 및 동결건조시켜, TU633-120을 백색의 면-유사 고체로서 수득하였다. (주석; 높은 MW를 특징으로 하는 PEG 시약은 그것이 PCL 함유 단백질에 접합된 이후에만 수득되었다.)Lithium 3-(3-acetyl-4-aminophenyl)propanoate (139 mg) and HBTU (247 mg) were placed in a 20 mL glass vial, and 13 mL anhydrous DMF was added. The resulting slurry was stirred at ambient temperature for 30 minutes, and the resulting solution was used to prepare TU633-120, TU633-122, TU633-124 and TU633-126. In a 45 mL glass vial, mPEGamine (NOF Corp., SUNBRIGHT MEPA-30T MW 30,298, 3.0 g) was added, 20 mL of anhydrous DMF was added, and mPEGamine was dissolved in DMF by heating lightly. A 2.2 mL aliquot of the activated ester solution was added to the resulting mPEGamine solution, and the vial was shaken at ambient temperature for 18 hours and then at 37° C. for 24 hours. The reaction mixture was transferred to dialysis membrane tubing (Fisher Scientific, cat # 21-152-9, MWCO 3500) and thoroughly dialyzed against double deionized water over 2 days. The dialyzed solution was frozen and lyophilized to give TU633-120 as a white cotton-like solid. (Note; PEG reagent characterized by high MW was obtained only after it was conjugated to PCL containing protein.)
SUNBRIGHT MEPA-30T 대신, NOF Corp. 사의 상응하는 활성화된 PEG, SUNBRIGHTMEPA-40T (MW 42036), SUNBRIGHT GL2-400PA (MW 42348), SUNBRIGHT GL4-400PA를 각각 이용하는 것을 제외하고 유사한 방식으로 Instead of SUNBRIGHT MEPA-30T, NOF Corp. In a similar manner, except for using the corresponding activated PEG from the company, SUNBRIGHTMEPA-40T (MW 42036), SUNBRIGHT GL2-400PA (MW 42348), SUNBRIGHT GL4-400PA respectively.
및 And
을 제조하였다. Was prepared.
(주석; 높은 MW를 특징으로 하는 PEG 시약은 그것이 PCL 함유 단백질에 접합된 이후에만 수득되었다.)(Note; PEG reagent characterized by high MW was obtained only after it was conjugated to PCL containing protein.)
실시예Example 37-7: 37-7: mPEGmPEG -- ABAABA -30-30 kDkD ( ( TU3627TU3627 -024)의 합성.-024).
표제 화합물은 리튬 3-(3-아세틸-4-아미노페닐)프로파노에이트 대신 리튬 2-(3-아미노-4-포르밀페녹시)아세테이트를 사용하는 것을 제외하고, TU633-120과 동일한 방식으로 제조하였다. (Note; 높은 MW를 특징으로 하는 PEG 시약은 그것이 PCL 함유 단백질에 접합된 이후에만 수득되었다.)The title compound was in the same manner as TU633-120, except that lithium 2-(3-amino-4-formylphenoxy)acetate was used instead of lithium 3-(3-acetyl-4-aminophenyl)propanoate. Was prepared. (Note: The PEG reagent, characterized by high MW, was obtained only after it was conjugated to the PCL containing protein.)
실시예Example 37-8: 37-8: mPEGmPEG -- ABAABA -30k (-30k ( TU3627TU3627 -084)의 합성.-084).
표제 화합물은 리튬 3-(3-아세틸-4-아미노페닐)프로파노에이트 대신 리튬 4-(3-아미노-4-포르밀페녹시)부타노에이트를 사용하는 것을 제외하고, TU633-120과 동일한 방식으로 제조하였다. H NMR 분석에 의해 생성물 내에는 어떤 비변형된 출발 PEG도 존재하지 않음을 확인하였다. 종점 활성: H-NMR에 의해 95% 초과.The title compound is the same as TU633-120, except that lithium 4-(3-amino-4-formylphenoxy)butanoate is used instead of lithium 3-(3-acetyl-4-aminophenyl)propanoate. Prepared in the same way. H NMR analysis confirmed that no unmodified starting PEG was present in the product. End point activity: greater than 95% by H-NMR.
실시예Example 37-9: 37-9: mPEGmPEG -- ABAABA -40k (-40k ( TU3627TU3627 -086)의 합성. -086).
표제 화합물은 SUNBRIGHTMEPA-30T 대신 SUNBRIGHTMEPA-40T를 사용하는 것을 제외하고, TU3627-084와 동일한 방식으로 제조하였다. H NMR 분석에 의해 생성물 내에는 어떤 비변형된 출발 PEG도 존재하지 않음을 확인하였다. 종점 활성: H-NMR에 의해 95% 초과.The title compound was prepared in the same manner as TU3627-084, except that SUNBRIGHTMEPA-40T was used instead of SUNBRIGHTMEPA-30T. H NMR analysis confirmed that no unmodified starting PEG was present in the product. End point activity: greater than 95% by H-NMR.
실시예 37-10: N 1 -(18-(3-아미노-4- 포르밀페녹시 )-15-옥소-4,7,10- 트리옥사 -14-아자옥타데실)-N19-(18-(3-아미노-4-포르밀페녹시)-15-옥소-4,7,10-트리옥사-9,14-디아자옥타데실)-4,7,10,13,16-펜타옥사노나데칸-1,19-디아미드 (X3678-40)의 합성 Example 37-10: N 1 - (18- ( 3- amino-4-formyl-phenoxy) -15- oxo -4,7,10- tree-oxa-14-aza-octadecyl) -N 19 - (18 -(3-amino-4-formylphenoxy)-15-oxo-4,7,10-trioxa-9,14-diazaoctadecyl)-4,7,10,13,16-pentaoxanona Synthesis of decane-1,19-diamide (X3678-40)
리튬 4-(3-아미노-4-포르밀페녹시)부타노에이트 (94.5 mg)를 4 mL DMF 중의 HBTU (151.7 mg)로 활성화시켰다. 디아미도-dPEG™11-디아민 (쿠안타바이오디자인, Cat# 10361, 74.3 mg)을 반응 혼합물에 첨가하고, 실온에서 밤새 교반하였다. DIEA (17.5 ㎕, 0.1 mmol)를 첨가하고, 혼합물을 40℃에서 수 시간 동안 가열하였다. 정제용 RP-HPLC에 의해 표제 화합물을 단리하고, PL-HCO3 MP SPE 카트리지 (배리안 인크.)를 통과시켜 TFA를 제거하였다.Lithium 4-(3-amino-4-formylphenoxy)butanoate (94.5 mg) was activated with HBTU (151.7 mg) in 4 mL DMF. Diamido-dPEG™ 11 -diamine (Quanta Biodesign, Cat# 10361, 74.3 mg) was added to the reaction mixture and stirred at room temperature overnight. DIEA (17.5 μl, 0.1 mmol) was added and the mixture was heated at 40° C. for several hours. The title compound was isolated by preparative RP-HPLC and TFA was removed by passing it through a PL-HCO 3 MP SPE cartridge (Varian Inc.).
실시예Example 38: 면역 조절자를 단백질에 38: immune modulators to proteins 커플링시키기Coupling 위한 시약의 합성 Synthesis of reagents for
실시예 38-1: N-(1-(3-아미노-4- 포르밀페녹시 )-2-옥소-7,10,13- 트리옥사 -3-아자헥사데칸-16-일)-4-((4-(2-(5-아미노-8-메틸벤조[f][1,7]나프티리딘-2-일)에틸)-3-메틸페녹시)메틸)벤즈아미드 (TU3627-042)의 합성. Example 38-1: N- (1- (3- amino-4-formyl-phenoxy) -2-oxo -7,10,13- tree oxa-3-aza-hexadecane -16- yl) -4- ((4-(2-(5-amino-8-methylbenzo[f][1,7]naphthyridin-2-yl)ethyl)-3-methylphenoxy)methyl)benzamide (TU3627-042) synthesis.
10 mL DCM 중의 t-부틸 3-(2-(2-(3-아미노프로폭시)에톡시)에톡시)프로필카르바메이트 (쿠안타바이오디자인, cat # 10225, 500 mg) 및 DIEA (348 ㎕)에 빙조에서 냉각시키면서 2 mL DCM 중의 4-클로로메틸벤조일 클로라이드를 첨가하였다. 반응물을 동일한 온도에서 1 시간 동안 교반하였다. 반응 혼합물을 EtOAc로 희석하고, H2O, 연한 수성 시트르산, 수성 NaHCO3 및 포화 수성 NaCl로 연속 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 생성물 (A)를 맑은 점성 오일로서 수득하였다. T-Butyl 3-(2-(2-(3-aminopropoxy)ethoxy)ethoxy)propylcarbamate (Quantabiodesign,
상기의 생성물 A (303 mg), 화합물 B (200 mg), 탄산세슘 (208 mg) 및 무수 DMSO를 합하고, 반응물을 주위 온도에서 교반하였다. 18 시간 후에, 8 mL DMSO에 용해시킨 추가의 60 mg의 화합물 A 및 추가의 30 mg의 탄산세슘을 반응물에 첨가하고, 반응물을 40 시간 동안 주위 온도에서 교반하였고, 이 즈음에 화합물 B는 단지 흔적량 만이 남았다 (LCMS 분석에 의해 확인됨). 반응 혼합물을 EtOAc로 희석하고, H2O, 포화 수성 NaCl로 연속 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 잔류물을 1 → 5% 용매 A 중의 B (A: DMC; B: 2% MeOH 중의 NH3)의 선형 구배를 이용한 실리카겔 플래쉬 크로마토그래피에 의해 정제하여, 부분적으로 정제된 목적하는 화합물을 수득하였다. MTBE로부터 재결정화한 후, 생성물을 연황색 결정으로서 수득하였다. The above product A (303 mg), compound B (200 mg), cesium carbonate (208 mg) and anhydrous DMSO were combined and the reaction was stirred at ambient temperature. After 18 hours, an additional 60 mg of compound A dissolved in 8 mL DMSO and an additional 30 mg of cesium carbonate were added to the reaction, and the reaction was stirred for 40 hours at ambient temperature, at which time compound B was only traced. Only the amount remained (confirmed by LCMS analysis). The reaction mixture was diluted with EtOAc, washed successively with H 2 O, saturated aqueous NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure. The residue was purified by silica gel flash chromatography using a linear gradient of B in 1→5% solvent A (A: DMC; B: NH 3 in 2% MeOH) to give the desired compound partially purified. After recrystallization from MTBE, the product was obtained as light yellow crystals.
화합물 C의 Boc 기는 주위 온도에서 3 M 메탄올성 HCl로 처리하여 제거하였다. 감압하에 반응 혼합물을 농축시켜 화합물 D를 디히드로클로라이드 염으로서 수득하였다. The Boc group of compound C was removed by treatment with 3 M methanolic HCl at ambient temperature. The reaction mixture was concentrated under reduced pressure to give compound D as a dihydrochloride salt.
리튬 2-(3-아미노-4-포르밀페녹시)아세테이트 (20 mg) 및 HBTU (38 mg)를 20 mL 유리 바이알에 넣고, 2 mL 무수 DMF를 첨가하였다. 생성된 슬러리를 주위 온도에서 30 분 동안 교반한 다음, 화합물 D (38 mg) 및 DIEA (52 ㎕)를 첨가하였다. 반응물을 주위 온도에서 17 시간 동안 교반하였다. LCMS 분석에 의해 출발 물질은 남아있지 않지만, 목적하는 생성물과 함께 비스-아실화된 부산물이 형성되었음을 확인하였다. 반응 혼합물을 EtOAc로 희석하고, H2O 및 포화 수성 NaCl로 연속 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 잔류물을 5 mL THF에 녹이고, 1 M 수성 LiOH의 1 mL 분취량으로 주위 온도에서 2 시간 동안 처리하였고, 이 시점에서의 LC-MS 분석은 비스-아실화된 생성물이 목적하는 생성물로 가수분해되었음을 나타내었다. 반응 혼합물을 EtOAc 및 물 사이에 분배시키고, 유기층을 분리하고, 포화 수성 NaCl로 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 조 물질을 정제를 위해 정제용 RP-HPLC에 적용하였다. 목적하는 생성물을 함유하는 HPLC 용리액을 EtOA로 희석하고, 포화 수성 NaHCO3 및 포화 수성 NaCl로 세척하여 트리플루오로아세트산을 제거하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 표제 화합물 N-(1-(3-아미노-4-포르밀페녹시)-2-옥소-7,10,13-트리옥사-3-아자헥사데칸-16-일)-4-((4-(2-(5-아미노-8-메틸벤조[f][1,7]나프티리딘-2-일)에틸)-3-메틸페녹시)메틸)벤즈아미드 (TU3627-042)를 수득하였다.Lithium 2-(3-amino-4-formylphenoxy)acetate (20 mg) and HBTU (38 mg) were placed in a 20 mL glass vial, and 2 mL anhydrous DMF was added. The resulting slurry was stirred at ambient temperature for 30 minutes, then compound D (38 mg) and DIEA (52 μl) were added. The reaction was stirred at ambient temperature for 17 hours. LCMS analysis confirmed that no starting material remained, but a bis-acylated by-product was formed with the desired product. The reaction mixture was diluted with EtOAc, washed successively with H 2 O and saturated aqueous NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure. The residue was taken up in 5 mL THF and treated with 1 mL aliquots of 1 M aqueous LiOH for 2 hours at ambient temperature, at which point LC-MS analysis showed that the bis-acylated product hydrolyzed to the desired product. Indicated that it has become. The reaction mixture was partitioned between EtOAc and water, the organic layer was separated, washed with saturated aqueous NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure. The crude material was subjected to preparative RP-HPLC for purification. The HPLC eluent containing the desired product was diluted with EtOA, washed with saturated aqueous NaHCO 3 and saturated aqueous NaCl to remove trifluoroacetic acid, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure, the title Compound N-(1-(3-amino-4-formylphenoxy)-2-oxo-7,10,13-trioxa-3-azahexadecane-16-yl)-4-((4-( 2-(5-Amino-8-methylbenzo[f][1,7]naphthyridin-2-yl)ethyl)-3-methylphenoxy)methyl)benzamide (TU3627-042) was obtained.
4-(2-(5-아미노-8- 메틸벤조[f][1,7]나프티리딘 -2-일)에틸)-3- 메틸페놀 (화합 물 B)의 합성 Synthesis of 4-(2-(5-amino-8- methylbenzo[f][1,7]naphthyridin- 2-yl)ethyl)-3 -methylphenol (compound B)
격막으로 캡핑된 둥근 바닥 플라스크에 1-에티닐-4-메톡시-2-메틸벤젠 (시판) (1.1 당량), 3,5-디클로로피콜리노니트릴 (1 당량), 트리에틸아민 (5 당량), 및 무수 DMF (0.2 M)를 첨가하였다. 진공처리 및 질소 플러싱을 3회 행하였다. CuI (0.05 당량) 및 비스(트리페닐포스핀)디클로로-팔라듐(II) (0.05 당량)을 첨가하였다. 격막을 환류 응축기로 교체하고, 플라스크를 60℃에서 밤새 질소 분위기 하에 가열하였다. TLC로 모니터링하여 반응이 완료되면, 플라스크의 내용물을 헥산으로 전처리된 대형 실리카겔 칼럼에 로딩하였다. 플래쉬 크로마토그래피 (실리카겔, 헥산:EtOAc (1:4%))에 의해 생성물 3-클로로-5-((4-메톡시-2-메틸페닐)에티닐)피콜리노니트릴을 수득하였다.1-ethynyl-4-methoxy-2-methylbenzene (commercially available) (1.1 eq), 3,5-dichloropicolinonitrile (1 eq), triethylamine (5 eq) in a round bottom flask capped with a septum , And anhydrous DMF (0.2 M) were added. Vacuum treatment and nitrogen flushing were performed 3 times. CuI (0.05 eq) and bis(triphenylphosphine)dichloro-palladium(II) (0.05 eq) were added. The diaphragm was replaced with a reflux condenser and the flask was heated at 60° C. overnight under a nitrogen atmosphere. When the reaction was completed by monitoring by TLC, the contents of the flask were loaded onto a large silica gel column pretreated with hexane. The product 3-chloro-5-((4-methoxy-2-methylphenyl)ethynyl)picolinonitrile was obtained by flash chromatography (silica gel, hexane:EtOAc (1:4%)).
환류 응축기가 구비된 둥근 바닥 플라스크에 3-클로로-5-((4-메톡시-2-메틸페닐)에티닐)피콜리노니트릴 (이전 단계의 것) (1 당량), tert-부틸 2-(4,4,5,5-테트라메틸-1,3,2-디옥사보롤란-2-일)페닐카르바메이트 (1.25 당량), K3PO4 (2 당량), 트리스(디벤질리덴아세톤)디팔라듐(0) (0.05 당량), 및 2-디시클로헥실포스피노-2',6'-디메톡시비페닐 (0.1 당량)을 첨가하였다. n-부탄올 및 물 (5:2, 0.2 M)을 첨가하고, 내용물을 3회 탈기시켰다 (진공시킨 후 질소 플러싱). 반응 혼합물을 오일조 내에서 질소하에 100℃에서 밤새 강하게 교반하였다. 내용물을 냉각시키고, 물 200 mL에 녹인 후, 메틸렌 클로라이드로 추출하였다. 합한 유기층을 건조시키고 (Na2SO4) 농축시켰다. 플래쉬 크로마토그래피 (실리카겔, 0 - 50% CH2Cl2 중의 EtOAc)에 의해 생성물 2-((4-메톡시-2-메틸페닐)에티닐)-8-메틸벤조[f][1,7]나프티리딘-5-아민을 수득하였다.To a round bottom flask equipped with a reflux condenser, 3-chloro-5-((4-methoxy-2-methylphenyl)ethynyl)picolinonitrile (from the previous step) (1 equivalent), tert-butyl 2-(4) ,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenylcarbamate (1.25 equivalents), K 3 PO 4 (2 equivalents), tris(dibenzylideneacetone) Dipalladium(0) (0.05 eq), and 2-dicyclohexylphosphino-2',6'-dimethoxybiphenyl (0.1 eq) were added. n-butanol and water (5:2, 0.2 M) were added, and the contents were degassed 3 times (vacuum followed by nitrogen flushing). The reaction mixture was stirred vigorously overnight at 100° C. under nitrogen in an oil bath. The contents were cooled, dissolved in 200 mL of water, and extracted with methylene chloride. The combined organic layers were dried (Na 2 SO 4 ) and concentrated. Product 2-((4-methoxy-2-methylphenyl)ethynyl)-8-methylbenzo[f][1,7]naphthy by flash chromatography (silica gel, 0-50% EtOAc in CH 2 Cl 2) Lidin-5-amine was obtained.
둥근 바닥 플라스크에 바(bar)로 교반하면서 2-((4-메톡시-2-메틸페닐)에티닐)-8-메틸벤조[f][1,7]나프티리딘-5-아민 (상기 단계로부터의 것) (1 당량)을 첨가하였다. 에탄올 및 메틸렌 클로라이드 (1:2, 0.2 M)를 첨가한 후, 탄소중 팔라듐 (활성화된 분말, 습식, 탄소상 10%, 0.1 당량)을 첨가하였다. 내용물을 진공처리한 후, 3회 수소 플러싱하였다. 반응 혼합물을 수소 풍선하에 실온에서 밤새 강하게 교반하였다. 이후, 셀라이트 패드를 통해 반응 혼합물을 여과하고, 후속하여 여과물에서 UV 흡수가 일어나지 않을 때까지 셀라이트 패드를 메틸렌 클로라이드 및 EtOAc로 세척하였다. 합한 유기 세척물을 농축시켰다. 플래쉬 크로마토그래피 (실리카겔, 0 - 50% CH2Cl2 중의 EtOAc)에 의해 생성물 2-(4-메톡시-2-메틸페네틸)-8-메틸벤조[f][1,7]나프티리딘-5-아민을 수득하였다.In a round bottom flask with bar stirring, 2-((4-methoxy-2-methylphenyl)ethynyl)-8-methylbenzo[f][1,7]naphthyridin-5-amine (from above step Of) (1 equivalent) was added. After ethanol and methylene chloride (1:2, 0.2 M) were added, palladium in carbon (activated powder, wet, 10% on carbon, 0.1 eq) was added. After vacuuming the contents, hydrogen flushing was performed 3 times. The reaction mixture was stirred vigorously overnight at room temperature under a hydrogen balloon. Thereafter, the reaction mixture was filtered through a pad of celite, and subsequently the pad of celite was washed with methylene chloride and EtOAc until no UV absorption occurred in the filtrate. The combined organic wash was concentrated. Product 2-(4-methoxy-2-methylphenethyl)-8-methylbenzo[f][1,7]naphthyridine- by flash chromatography (silica gel, 0-50% EtOAc in CH 2 Cl 2) 5-amine was obtained.
빙수조 내의 메틸렌 클로라이드 중의 2-(4-메톡시-2-메틸페네틸)-8-메틸벤조[f][1,7]나프티리딘-5-아민의 교반 용액 (0.2 M)에 CH2Cl2 중의 BBr3 (2 당량)의 1 N 용액을 적가 방식으로 첨가하였다. 30 분 이내에 반응물을 메탄올로 켄칭시키고, 진공하에 농축시켜 조 잔류물을 수득하였다. 조 물질을 디클로로메탄 중의 0-20% 메탄올을 이용하여 콤비플래쉬(COMBIFLASH)® 시스템 (ISCO) 상에서 플래쉬 크로마토그래피에 의해 정제하여, 4-(2-(5-아미노-8-메틸벤조[f][1,7]나프티리딘-2-일)에틸)-3-메틸페놀을 백색 고체로서 수득하였다. CH 2 Cl in a stirred solution (0.2 M) of 2-(4-methoxy-2-methylphenethyl)-8-methylbenzo[f][1,7]naphthyridin-5-amine in methylene chloride in an ice water bath a 1 N solution of BBr 3 (2 eq) of 2 was added drop wise. The reaction was quenched with methanol within 30 minutes and concentrated in vacuo to give a crude residue. The crude was purified by flash chromatography on a COMBIFLASH® system (ISCO) using 0-20% methanol in dichloromethane, resulting in 4-(2-(5-amino-8-methylbenzo[f]). [1,7]naphthyridin-2-yl)ethyl)-3-methylphenol was obtained as a white solid.
실시예 38-2: 4-((4-(2-(5-아미노-8- 메틸벤조[f][1,7]나프티리딘 -2-일)에틸)-3-메틸페녹시)메틸)-N-(1-(아미노옥시)-2-옥소-7,10,13-트리옥사-3-아자헥사데칸-16-일)벤즈아미드 히드로클로라이드 (TU3627-044)의 합성. Example 38-2: 4-((4-(2-(5-amino-8- methylbenzo[f][1,7]naphthyridin- 2-yl) ethyl)-3-methylphenoxy)methyl Synthesis of )-N-(1-(aminooxy)-2-oxo-7,10,13-trioxa-3-azahexadecane-16-yl)benzamide hydrochloride (TU3627-044).
HBTU (38 mg), 2-(tert-부톡시카르보닐아미노옥시)아세트산 (플루카, 25 mg), DIEA (44 ㎕) 및 5 mL DMF를 20 mL 유리 바이알 내에서 합하였다. 반응물을 주위 온도에서 30 분 동안 교반한 다음, 화합물 D (38 mg)를 첨가하였다. 반응물을 주위 온도에서 17 시간 동안 교반하였다. LC-MS 분석에 의해 출발 물질은 남아있지 않지만, 목적하는 생성물과 함께 수회 과다아실화된 부산물이 형성되었음을 확인하였다. 반응 혼합물을 EtOAc로 희석하고, H2O 및 포화 수성 NaCl로 연속 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 잔류물을 5 mL THF에 녹이고, 1 M 수성 LiOH의 1 mL 분취량으로 주위 온도에서 2 시간 동안 처리하였고, 이 시점에서의 LC-MS 분석은 과다아실화된 생성물이 목적하는 생성물로 가수분해되었음을 나타내었다. 반응 혼합물을 EtOAc 및 물 사이에 분배시키고, 유기층을 분리하고, 포화 수성 NaCl로 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 조 물질을 5% 용매 A 중의 B (A: DMC; B: 2% MeOH 중의 NH3)를 이용한 실리카겔 플래쉬 크로마토그래피에 의해 정제하여, 목적하는 화합물, E를 수득하였다.HBTU (38 mg), 2-(tert-butoxycarbonylaminooxy)acetic acid (Fluka, 25 mg), DIEA (44 μL) and 5 mL DMF were combined in a 20 mL glass vial. The reaction was stirred at ambient temperature for 30 minutes, then compound D (38 mg) was added. The reaction was stirred at ambient temperature for 17 hours. LC-MS analysis confirmed that there was no starting material left, but several times overacylated by-products were formed with the desired product. The reaction mixture was diluted with EtOAc, washed successively with H 2 O and saturated aqueous NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure. The residue was taken up in 5 mL THF and treated with 1 mL aliquots of 1 M aqueous LiOH for 2 hours at ambient temperature, at which point LC-MS analysis showed that the hyperacylated product was hydrolyzed to the desired product. Indicated. The reaction mixture was partitioned between EtOAc and water, the organic layer was separated, washed with saturated aqueous NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure. The crude was purified by silica gel flash chromatography using B in 5% solvent A (A: DMC; B: NH 3 in 2% MeOH) to give the desired compound, E.
화합물 E (28 mg)를 3 M 메탄올성 HCl로 주위 온도에서 30 분 동안 처리하고, 감압하에 농축시켰다. 이러한 작업을 반복하였다. 표제 화합물 4-((4-(2-(5-아미노-8-메틸벤조[f][1,7]나프티리딘-2-일)에틸)-3-메틸페녹시)메틸)-N-(1-(아미노옥시)-2-옥소-7,10,13-트리옥사-3-아자헥사데칸-16-일)벤즈아미드 히드로클로라이드 (TU3627-044)를 황색 고체로서 수득하였다.Compound E (28 mg) was treated with 3 M methanolic HCl for 30 min at ambient temperature and concentrated under reduced pressure. This operation was repeated. Title compound 4-((4-(2-(5-amino-8-methylbenzo[f][1,7]naphthyridin-2-yl)ethyl)-3-methylphenoxy)methyl)-N-( 1-(Aminooxy)-2-oxo-7,10,13-trioxa-3-azahexadecane-16-yl)benzamide hydrochloride (TU3627-044) was obtained as a yellow solid.
실시예 38-3: N-(58-(3-아미노-4- 포르밀페녹시 )-15,55- 디옥소 -4,7,10,18,21,24,27,30,33,36,39,42,45,48,51-펜타데카옥사-14,54-디아자옥타펜타콘틸)-4-((4-(2-(5-아미노-8-메틸벤조[f][1,7]나프티리딘-2-일)에틸)-3-메틸페녹시)메틸)벤즈아미드 (X3678-114)의 합성. Example 38-3: N- (58- (3- amino-4-formyl-phenoxy) -15,55- dioxo-4,7,10,18,21,24,27,30,33,36 ,39,42,45,48,51-pentadecaoxa-14,54-diazaoctapentacontyl)-4-((4-(2-(5-amino-8-methylbenzo[f][1, Synthesis of 7]naphthyridin-2-yl)ethyl)-3-methylphenoxy)methyl)benzamide (X3678-114).
2.0 mL DMF 중의 dPEG-산 F (100 mg), HATU (52.8 mg) 및 DIEA (97 ㎕)의 혼합물을 실온에서 30 분 동안 교반하였다. 아민 D (109 mg의 화합물 C로부터 제조한 것)를 첨가하고, HPLC로 모니터링하여 완료될 때까지 반응물을 실온에서 교반하였다. 생성물을 정제용 RP-HPLC로 단리하였다. 이전 단계로부터의 생성물 G (138 mg)의 Boc 기를 메탄올 중의 3 N HCl로 처리하여 제거한 후, 건고상태로 농축시켰다. 2.0 mL DMF 중의 리튬 4-(3-아미노-4-포르밀페녹시)부타노에이트 (25.2 mg), HATU (38.0 mg) 및 DIEA (69.7 ㎕)의 혼합물을 실온에서 30 분 동안 교반하였다. 아민 H를 첨가하고, HPLC에 의해 완료된 것으로 모니터링될 때까지 반응물을 실온에서 교반하였다. 생성물 (X3678-114)을 정제용 HPLC로 단리하였다.A mixture of dPEG-acid F (100 mg), HATU (52.8 mg) and DIEA (97 μL) in 2.0 mL DMF was stirred at room temperature for 30 min. Amine D (prepared from 109 mg of compound C) was added and the reaction was stirred at room temperature until completion, monitored by HPLC. The product was isolated by preparative RP-HPLC. The Boc group of product G (138 mg) from the previous step was removed by treatment with 3 N HCl in methanol and then concentrated to dryness. A mixture of lithium 4-(3-amino-4-formylphenoxy)butanoate (25.2 mg), HATU (38.0 mg) and DIEA (69.7 μL) in 2.0 mL DMF was stirred at room temperature for 30 minutes. Amine H was added and the reaction was stirred at room temperature until monitored to be complete by HPLC. The product (X3678-114) was isolated by preparative HPLC.
실시예 38-4: 2-(4-아세틸-3- 아미노페녹시 )-N-(3- 니트로벤질 ) 아세트아미드 (TU633-068)의 합성. Example 38-4: 2-(4-acetyl-3- aminophenoxy )-N-(3 -nitrobenzyl ) acetamide Synthesis of (TU633-068).
리튬 2-(4-아세틸-3-아미노페녹시)아세테이트 (215 mg), HBTU (379 mg), 및 10 mL DMF를 20 mL 유리 바이알 내에서 합하고, 생성된 슬러리를 주위 온도에서 1 시간 동안 교반하였고, 이 시점에서 반응 혼합물은 거의 균질하게 되었다. 이어서, 바이알에 3'-니트로벤질아민 HCl (192 mg) 및 DIEA (191 ㎕)를 첨가하고, 반응물을 주위 온도에서 30 분 동안 교반하였다. 반응 혼합물을 EtOAc로 희석하고, 물, 포화 aq. NaCl로 연속 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켰다. 조 물질을 실리카겔 플래쉬 크로마토그래피 (헥산/EtOAc)에 의해 정제하여, 표제 화합물을 황색 고체로서 수득하였다.Lithium 2-(4-acetyl-3-aminophenoxy)acetate (215 mg), HBTU (379 mg), and 10 mL DMF are combined in a 20 mL glass vial and the resulting slurry is stirred at ambient temperature for 1 hour. And at this point the reaction mixture became almost homogeneous. Then 3'-nitrobenzylamine HCl (192 mg) and DIEA (191 μL) were added to the vial and the reaction was stirred at ambient temperature for 30 minutes. The reaction mixture was diluted with EtOAc, and water, saturated aq. Washed successively with NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure. The crude was purified by silica gel flash chromatography (hexane/EtOAc) to give the title compound as a yellow solid.
실시예 38-5: 2-(3-아미노-4- 포르밀페녹시 )-N-(4- 니트로벤질 ) 아세트아미드 (TU3627-020)의 합성. Example 38-5: Preparation of 2- (3-amino-4-formyl-phenoxy) -N- (4- nitrobenzyl) acetamide (TU3627-020).
리튬 2-(3-아미노-4-포르밀페녹시)아세테이트 (44.2 mg) 및 HBTU (79.6 mg)를 20 mL 유리 바이알에 넣고, 무수 DMF (5 mL)를 첨가하였다. 생성된 슬러리를 주위 온도에서 교반하였고, 10 분 이내에 균질하게 되었다. 15 분 후에, DIEA (50 ㎕) 및 4'-니트로벤질아민 HCl (37.7 mg)을 반응 혼합물에 첨가하고, 반응물을 주위 온도에서 교반하였다. 30 분 후에, LC-MS 분석에 의해 반응이 완료되었음을 확인하였다. 반응 혼합물을 EtOAc 및 물 사이에 분배시켰다. 유기층을 분리하고, 연한 aq. 시트르산, 포화 aq. NaHCO3, 및 포화 aq. NaCl로 연속 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 표제 화합물을 황갈색 고체로서 수득하였다.Lithium 2-(3-amino-4-formylphenoxy)acetate (44.2 mg) and HBTU (79.6 mg) were placed in a 20 mL glass vial, and anhydrous DMF (5 mL) was added. The resulting slurry was stirred at ambient temperature and became homogeneous within 10 minutes. After 15 minutes, DIEA (50 μl) and 4′-nitrobenzylamine HCl (37.7 mg) were added to the reaction mixture and the reaction was stirred at ambient temperature. After 30 minutes, the reaction was confirmed to be complete by LC-MS analysis. The reaction mixture was partitioned between EtOAc and water. The organic layer was separated, and the light aq. Citric acid, saturated aq. NaHCO 3 , and saturated aq. Washed successively with NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure to give the title compound as a tan solid.
실시예 38-6: 2-(3-아미노-4- 포르밀페녹시 )-N-(2-(2,4- 디니트로페닐아미노 )에틸)아세트아미드 (TU3627-022)의 합성. Example 38-6: Preparation of 2- (3-amino-4-formyl-phenoxy) -N- (2- (2,4- dinitrophenyl) ethyl) acetamide By a similar method as in (TU3627-022).
DIEA의 부재하에 4'-니트로벤질아민 HCl 대신 N1-(2,4-디니트로페닐)에탄-1,2-디아민 (오크우드(Oakwood), cat # 015083, 45.2 mg)을 사용하는 것을 제외하고, TU3627-020와 동일한 방식으로 표제 화합물을 제조하였다. TU3627-022를 황색 고체로서 수득하였다. Rf. 0.15 (SiO2, 5% DCM 중 MeOH). Except for the use of N 1 -(2,4-dinitrophenyl)ethane-1,2-diamine (Oakwood, cat # 015083, 45.2 mg) instead of 4'-nitrobenzylamine HCl in the absence of DIEA Then, the title compound was prepared in the same manner as in TU3627-020. TU3627-022 was obtained as a yellow solid. Rf. 0.15 (SiO 2 , MeOH in 5% DCM).
실시예 38-7: 4-(3-아미노-4- 포르밀페녹시 )-N-(2-(2,4- 디니트로페닐아미노 )에틸)부탄아미드 (TU3627-088)의 합성. Example 38-7: 4- (3-amino-4-formyl-phenoxy) -N- (2- (2,4- dinitrophenyl) ethyl) Synthesis of butane amide (TU3627-088).
리튬 4-(3-아미노-4-포르밀페녹시)부타노에이트 (50 mg), HBTU (80 mg) 및 DMF (2 mL)를 20 mL 유리 바이알 내에서 합하고, 주위 온도에서 교반하였다. 30 분 후에, N1-(2,4-디니트로페닐)에탄-1,2-디아민 (오크우드, 45 mg)을 한번에 첨가하고, 반응물을 주위 온도에서 밤새 교반하였다. 반응 혼합물을 EtOAc로 희석하고, 연한 aq. 시트르산, 물, 포화 aq. NaHCO3, 물, 및 포화 aq. NaCl로 연속 세척하고, Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 표제 화합물을 황색 고체로서 수득하였다. Rf: 0.18 (SiO2, EtOAc).Lithium 4-(3-amino-4-formylphenoxy)butanoate (50 mg), HBTU (80 mg) and DMF (2 mL) were combined in a 20 mL glass vial and stirred at ambient temperature. After 30 minutes, N 1 -(2,4-dinitrophenyl)ethane-1,2-diamine (oakwood, 45 mg) was added in one portion and the reaction was stirred at ambient temperature overnight. The reaction mixture was diluted with EtOAc and light aq. Citric acid, water, saturated aq. NaHCO 3 , water, and saturated aq. Washed successively with NaCl, dried over Na 2 SO 4 , filtered and concentrated under reduced pressure to give the title compound as a yellow solid. Rf: 0.18 (SiO 2 , EtOAc).
실시예 38-8: 4-(3-아미노-4- 포르밀페녹시 )-N-(4- 니트로벤질 ) 부탄아미드 (3793-001)의 합성. Example 38-8: 4- (3-amino-4-formyl-phenoxy) -N- (4-nitrobenzyl) butane amide Synthesis of (3793-001).
HATU (114.1 mg)를 리튬 4-(3-아미노-4-포르밀페녹시)-부타노에이트 (68.7 mg)의 1 mL DMF 용액에 첨가하고, 반응물을 실온에서 1 시간 동안 진탕하였다. 이어서, 생성된 용액을 4-니트로벤질아민 HCl 염 (56.6 mg) 및 트리에틸아민 (84 ㎕)의 1 mL DMF 용액에 첨가하였고, 또 다른 1 mL DMF는 전송을 돕도록 하였다. 반응물을 실온에서 2 시간 동안 교반하였다. 완료되면, 반응 혼합물을 4 mL의 10% NaCl(aq) 및 8 mL의 EtOAc 사이에 분배시켰다. 상을 분리시키고, 수성층을 8 mL EtOAc로 다시 추출하였다. 합한 유기층을 Na2SO4 상에서 건조시키고, 여과하고, 감압하에 농축시켜, 조질의 오렌지색 오일을 수득하였다. 조질의 오일을 실리카겔 플래쉬 칼럼 크로마토그래피에 의해 0-10% MeOH/DCM의 구배 용리로 정제하여, 목적하는 생성물을 밝은 황색 고체로서 수득하였다. HATU (114.1 mg) was added to a 1 mL DMF solution of lithium 4-(3-amino-4-formylphenoxy)-butanoate (68.7 mg) and the reaction was shaken at room temperature for 1 hour. The resulting solution was then added to a 1 mL DMF solution of 4-nitrobenzylamine HCl salt (56.6 mg) and triethylamine (84 μL), and another 1 mL DMF was allowed to aid the transfer. The reaction was stirred at room temperature for 2 hours. Upon completion, the reaction mixture was partitioned between 4 mL of 10% NaCl (aq) and 8 mL of EtOAc. The phases were separated and the aqueous layer was extracted again with 8 mL EtOAc. The combined organic layers were dried over Na 2 SO 4 , filtered and concentrated under reduced pressure to give a crude orange oil. The crude oil was purified by silica gel flash column chromatography with gradient elution of 0-10% MeOH/DCM to give the desired product as a light yellow solid.
실시예 38-9: 3-(4-아세틸-3- 아미노페닐 )-2-(4- 니트로벤즈아미도 )프로판산 (X3471-116)의 합성. Example 38-9: Synthesis of 3-(4-acetyl-3 -aminophenyl )-2-(4- nitrobenzamido )propanoic acid (X3471-116).
4-니트로벤조산 (50.1 mg), HATU (114 mg) 및 DIEA (105 ㎕)에 1 mL DMF를 첨가하고, 혼합물을 30 분 동안 교반하였다. 이어서, 생성된 용액을 1.0 mL DMF 중의 3-(4-아세틸-3-아미노페닐)-2-아미노프로판산 (66.7 mg, 1.0 당량)에 첨가하고, LC-MS로 모니터링하면서 반응물을 실온에서 교반하였다. 이어서, 표제 생성물을 정제용 HPLC에 의해 단리하였다.To 4-nitrobenzoic acid (50.1 mg), HATU (114 mg) and DIEA (105 μL) was added 1 mL DMF, and the mixture was stirred for 30 minutes. The resulting solution was then added to 3-(4-acetyl-3-aminophenyl)-2-aminopropanoic acid (66.7 mg, 1.0 eq) in 1.0 mL DMF and the reaction stirred at room temperature while monitoring by LC-MS. I did. The title product was then isolated by preparative HPLC.
실시예 38-10: 4-(3-아미노-4- 포르밀페녹시 ) 부타노일 -AlaGlySerArgSerGly(D-Ala)LysChaValAlaAlaTrpThrLeuLysAla(D-Ala)Gly-OH (3647-104)의 합성. Example 38-10: 4- (3-amino-4-formyl-phenoxy) butanoyl Synthesis of -AlaGlySerArgSerGly (D- Ala) LysChaValAlaAlaTrpThrLeuLysAla (D -Ala) Gly-OH (3647-104).
Fmoc-exPADRE CLEAR 수지 (549.3 mg, 0.1 mmol, 펩티드 인터내셔널 인크.(Peptide International Inc.) 사에서 구입)의 Fmoc 기를 20% 피페리딘/DMF (8 mL x 3)로 제거하였다. 수지를 DMF (1.5 mL x 5)로 세척하였다. 이어서, 수지를 리튬 4-(3-아미노-4-포르밀페녹시)-부타노에이트 (34.4 mg), HBTU (45.5 mg), HOBt (16.2 mg), 및 DIEA (52 ㎕)의 3 mL DMF 용액으로 6 시간 동안 실온에서 처리하였다. 수지를 DMF로 세척하고, H2O/Me2S/TFA (10/10/80 v/v%, 10 mL)를 이용하여 2 시간 동안 실온에서 수지로부터 펩티드를 절단해내었다. 절단 슬러리를 유리 울을 통해 여과하고, 증발에 의해 대부분의 TFA를 여과물로부터 제거하였다. 잔류물을 1 N NaOH( aq )로 중화시키고, 아세토니트릴로 희석한 후, 50% MeCN( aq ) 중의 스펙트라프로(SpectraPro)™ 7 투석막 (MWCO 1000)을 이용하여 투석하였다 (4L x 10). 투석막 내에 남아있는 용액을 동결건조시켜, 목적하는 생성물을 백색 고체로서 수득하였다.The Fmoc group of Fmoc-exPADRE CLEAR resin (549.3 mg, 0.1 mmol, purchased from Peptide International Inc.) was removed with 20% piperidine/DMF (8 mL x 3). The resin was washed with DMF (1.5 mL x 5). The resin was then lithium 4-(3-amino-4-formylphenoxy)-butanoate (34.4 mg), HBTU (45.5 mg), HOBt (16.2 mg), and 3 mL DMF of DIEA (52 μL). The solution was treated for 6 hours at room temperature. The resin was washed with DMF, and peptides were cleaved from the resin at room temperature for 2 hours using H 2 O/Me 2 S/TFA (10/10/80 v/v%, 10 mL). The cut slurry was filtered through glass wool and most of the TFA was removed from the filtrate by evaporation. The residue was neutralized with 1 N NaOH ( aq ) , diluted with acetonitrile, and dialyzed using a
실시예 38-11: 3-(4-아세틸-3- 아미노페닐 ) 프로파노일 -Gly(D-Ala)LysChaValAlaAlaTrpThrLeuLysAla(D-Ala)Gly-OH (3465-143)의 합성. Example 38-11: Synthesis of 3-(4-acetyl-3 -aminophenyl ) propanoyl- Gly(D- Ala)LysChaValAlaAlaTrpThrLeuLysAla(D-Ala)Gly-OH (3465-143).
Fmoc-PADRE CLEAR 수지 (105.3 mg, 0.02 mmol, 펩티드 인터내셔널 인크. 사에서 구입)의 Fmoc 기를 20% 피페리딘/DMF (8 mL x 3)로 제거하였다. 수지를 DMF (1.5 mL x 5)로 세척하였다. 이어서, 수지를 리튬 3-(4-아세틸-3-아미노페닐)프로파노에이트 (5.1 mg), HBTU (9.1 mg), 및 HOBt (3.3 mg)의 0.5 mL DMF 용액과 7 시간 동안 실온에서 커플링시켰다. 수지를 DMF로 세척하고, TIPS/H2O/TFA (5/55/90 v/v%, 3 mL)를 이용하여 2 시간 동안 수지로부터 펩티드를 절단해내었다. 절단 슬러리를 유리 울을 통해 여과하고, 증발에 의해 대부분의 TFA를 여과물로부터 제거하였다. 잔류물을 헥산 (3 mL x 3)으로 세척하고, 50% MeCN( aq )에 용해시켰다. 동결건조시킨 후, 조 생성물을 수득한 다음, 이것을 정제용 HPLC에 의해 정제하여, 목적하는 생성물을 밝은 황색 분말로서 수득하였다.The Fmoc group of the Fmoc-PADRE CLEAR resin (105.3 mg, 0.02 mmol, purchased from Peptide International Inc.) was removed with 20% piperidine/DMF (8 mL x 3). The resin was washed with DMF (1.5 mL x 5). The resin was then coupled with a 0.5 mL DMF solution of lithium 3-(4-acetyl-3-aminophenyl)propanoate (5.1 mg), HBTU (9.1 mg), and HOBt (3.3 mg) at room temperature for 7 hours. Made it. The resin was washed with DMF and the peptide was cleaved from the resin for 2 hours using TIPS/H 2 O/TFA (5/55/90 v/v%, 3 mL). The cut slurry was filtered through glass wool and most of the TFA was removed from the filtrate by evaporation. The residue was washed with hexane (3 mL x 3 ) and dissolved in 50% MeCN ( aq). After lyophilization, a crude product was obtained, which was then purified by preparative HPLC to give the desired product as a light yellow powder.
실시예 38-12: 6-(4-(3-아미노-4- 포르밀페녹시 ) 부탄아미도 ) 헥실 -5'- *T*C*C*A*T*G*A*C*G*T*T*C*C*T*G*A*C*G*T*T-3' (*: 포스포티오에이트) (3647-057)의 합성. Example 38-12: Preparation of 6- (4- (3-amino-4-formyl-phenoxy) butane amido) hexyl -5'- * T * C * C * A * T * G * A * C * G * T * T * C * C * T * G * A * C * G * T * T-3' (*: Phosphothioate) (3647-057) synthesis.
HBTU (6.1 mg), HOBt (2.2 mg), 및 트리에틸아민 (7 ㎕)을 리튬 4-(3-아미노-4-포르밀페녹시)-부타노에이트 (4.6 mg)의 1 mL DMSO 용액에 첨가하고, 반응물을 실온에서 1 시간 동안 진탕하였다. 이어서, 생성된 용액의 276 ㎕ 분취량을 아미노-변형된 BG1 올리고 (12.1 mg, 1.8 μmol, 인테그레이티드 DNA 테크놀로지스, 인크.(Integrated DNA Technologies, Inc.) 사에서 구입) 및 트리에틸아민 (15 ㎕)의 1.2 mL DMSO 용액에 첨가하고, 반응물을 실온에서 2 일 동안 진탕하였다. 반응 혼합물을 물로 희석하고, 슬라이드-A-라이저™ (MWCO 3500)를 이용하여 물 (4 L x 10)에 대해 투석하였다. 투석된 용액을 동결건조시켜, 목적하는 생성물을 백색 고체로서 수득하였다.HBTU (6.1 mg), HOBt (2.2 mg), and triethylamine (7 μl) were added to a 1 mL DMSO solution of lithium 4-(3-amino-4-formylphenoxy)-butanoate (4.6 mg). Was added and the reaction was shaken at room temperature for 1 hour. Then, a 276 μl aliquot of the resulting solution was added to the amino-modified BG1 oligo (12.1 mg, 1.8 μmol, purchased from Integrated DNA Technologies, Inc.) and triethylamine (15). Μl) of 1.2 mL DMSO solution, and the reaction was shaken at room temperature for 2 days. The reaction mixture was diluted with water and dialyzed against water (4 L x 10) using Slide-A-Riser™ (MWCO 3500). The dialyzed solution was lyophilized to give the desired product as a white solid.
실시예 38-13: 6-(3-(4-아세틸-3- 아미노페닐 ) 프로판아미도 ) 헥실 -5'- *T*C*G*T*C*G*T*T*T*T*C*G*G*C*G*C*G*C*G*C*C*G-3' (*:포스포티오에이트) (3597-033)의 합성. Example 38-13: 6-(3-(4-acetyl-3 -aminophenyl ) propanamido ) hexyl- 5'- * T * C * G * T * C * G * T * T * T * T Synthesis of * C * G * G * C * G * C * G * C * G * C * C * G-3' (*:phosphothioate) (3597-033).
HBTU (5.9 mg) 및 HOBt (2.1 mg)를 리튬 3-(4-아세틸-3-아미노페닐)프로파노에이트 (3.3 mg)의 1 mL DMSO 용액에 첨가하고, 반응물을 실온에서 1 시간 동안 진탕하였다. 이어서, 생성된 용액의 20 ㎕ 분취량을 아미노-변형된 BG2 올리고 (1.88 mg, 0.26 μmol, 인테그레이티드 DNA 테크놀로지스, 인크. 사에서 구입) 및 트리에틸아민 (2 ㎕)의 0.2 mL DMSO 용액에 첨가하고, 반응물을 실온에서 20 시간 동안 진탕하였다. 생성된 혼합물을 H2O로 평형화된 NAP-25™ 칼럼에 적용하고, H2O로 용리시켰다. 매 1 mL 분획마다 LC-MS로 모니터링하고, 목적하는 생성물을 함유하는 분획을 합하고 동결건조시켜, 목적하는 생성물을 백색 고체로서 수득하였다.HBTU (5.9 mg) and HOBt (2.1 mg) were added to a 1 mL DMSO solution of lithium 3-(4-acetyl-3-aminophenyl)propanoate (3.3 mg) and the reaction was shaken at room temperature for 1 hour. . Then, a 20 μl aliquot of the resulting solution was added to a 0.2 mL DMSO solution of amino-modified BG2 oligo (1.88 mg, 0.26 μmol, purchased from Integrated DNA Technologies, Inc.) and triethylamine (2 μl). Was added and the reaction was shaken at room temperature for 20 hours. Applying the resulting mixture to a NAP-25 ™ column equilibrated in H 2 O, and eluted with H 2 O. Every 1 mL fraction was monitored by LC-MS, the fractions containing the desired product were combined and lyophilized to give the desired product as a white solid.
실시예 38-14: 6-(4-(3-아미노-4- 포르밀페녹시 ) 부탄아미도 ) 헥실 -5'- *T*C*G*T*C*G*T*T*T*T*C*G*G*C*G*C*G*C*G*C*C*G-3' (*:포스포티오에이트) (3597-167)의 합성. Example 38-14: Preparation of 6- (4- (3-amino-4-upon-formyl phenoxy) butane amido) hexyl -5'- * T * C * G * T * C * G * T * T * T Synthesis of * T * C * G * G * C * G * C * G * C * G * C * C * G-3' (*:phosphothioate) (3597-167).
HATU (7.4 mg)를 리튬 4-(3-아미노-4-포르밀페녹시)-부타노에이트 (5.5 mg)의 1 mL DMSO 용액에 첨가하고, 반응물을 실온에서 1 시간 동안 진탕하였다. 이어서, 생성된 용액의 60 ㎕ 분취량을 아미노-변형된 BG2 올리고 (6.9 mg, 1.0 μmol) 및 트리에틸아민 (7.5 ㎕)의 0.6 mL DMSO 용액에 첨가하고, 반응물을 실온에서 20 시간 동안 진탕하였다. 이어서, HATU 대신 HBTU를 사용하는 것을 제외하고 동일한 방식으로 새롭게 제조한 활성화된 에스테르 용액의 또 다른 60 ㎕ 분취량을 반응 혼합물에 첨가하고, 반응물을 추가로 2 일간 진탕하였다. 반응 혼합물을 3 부분으로 분리시켰다. 각각의 부분을 H2O로 평형화된 NAP-25™ 칼럼에 적용하고, H2O로 용리시켰다. 매 1 mL 분획마다 LC-MS로 모니터링하고, 목적하는 생성물을 함유하는 분획을 합하고 동결건조시켜, 목적하는 생성물을 백색 고체로서 수득하였다.HATU (7.4 mg) was added to a 1 mL DMSO solution of lithium 4-(3-amino-4-formylphenoxy)-butanoate (5.5 mg) and the reaction was shaken at room temperature for 1 hour. Then, a 60 μl aliquot of the resulting solution was added to a 0.6 mL DMSO solution of amino-modified BG2 oligo (6.9 mg, 1.0 μmol) and triethylamine (7.5 μl), and the reaction was shaken at room temperature for 20 hours. . Then, another 60 μl aliquot of the activated ester solution freshly prepared in the same manner except for the use of HBTU instead of HATU was added to the reaction mixture, and the reaction was shaken for an additional 2 days. The reaction mixture was separated into 3 parts. It applied to a NAP-25 ™ column equilibrated in the respective portions of H 2 O and eluted with H 2 O. Every 1 mL fraction was monitored by LC-MS, the fractions containing the desired product were combined and lyophilized to give the desired product as a white solid.
실시예Example 39: 스핀-표지- 39: spin-cover- ABAABA 시약의 합성 Synthesis of reagents
실시예 39-1: N-(3-아미노-4- 포르밀페닐 )-1-히드록시-2,2,5,5- 테트라메틸 -3-피롤린-1-옥실-3-카르복스아미드 (X3626-112b)의 합성. Example 39-1: N- (3- amino-4-formylphenyl) -1-hydroxy-3-pyrroline-1-oxyl -2,2,5,5- tetramethyl-3-carboxamide Synthesis of (X3626-112b).
2,2,5,5-테트라메틸-3-피롤린-1-옥실-3-카르복실산 (55.3 mg), HBTU (102 mg) 및 DIEA (105 ㎕)에 2.0 mL DMF를 첨가하였다. 실온에서 30 분 동안 교반한 후, 2,4-디아미노벤즈알데히드 (40.8 mg)를 첨가하였다. HPLC로 모니터링하여 반응이 완료될 때까지 혼합물을 실온에서 교반하였다. 표제 생성물을 정제용 HPLC로 단리하였다. To 2,2,5,5-tetramethyl-3-pyrroline-1-oxyl-3-carboxylic acid (55.3 mg), HBTU (102 mg) and DIEA (105 μL) were added 2.0 mL DMF. After stirring at room temperature for 30 minutes, 2,4-diaminobenzaldehyde (40.8 mg) was added. The mixture was stirred at room temperature until the reaction was complete, monitored by HPLC. The title product was isolated by preparative HPLC.
실시예Example 40: 비오틴- 40: biotin- ABAABA 시약의 합성 Synthesis of reagents
실시예 40-1: N-(3-아미노-4- 포르밀페닐 )-1-( 비오틴아미도 )-3,6,9,12- 테트라옥사펜타데칸-15-아미드 (X3626-140)의 합성. Example 40-1: N- (3- amino-4-formylphenyl) -1- (biotin amido) -3,6,9,12- tetrahydro-oxa-15-pentadecane amide (X3626-140) of synthesis.
2,4-디아미노벤즈알데히드 (11.5 mg), NHS-dPEG™4 비오틴 (쿠안타바이오디자인, cat # 10200, 50 mg) 및 DMAP (10.4 mg)에 1.0 mL DMF를 첨가하였다. HPLC로 모니터링하여 반응이 완료될 때까지 반응 혼합물을 실온에서 교반하였다. 생성물을 실리카겔 플래쉬 크로마토그래피로 단리하였다. 1.0 mL DMF was added to 2,4-diaminobenzaldehyde (11.5 mg), NHS-
실시예 40-2: N-(1-(3-아미노-4- 포르밀페녹시 )-2-옥소-7,10,13- 트리옥사 -3-아자헥사데칸-16-일)-비오틴아미드 (X3626-142)의 합성. Example 40-2: N- (1- (3- amino-4-formyl-phenoxy) -2-oxo -7,10,13- tree oxa-3-aza-hexadecane -16- yl) - Biotin amide Synthesis of (X3626-142).
리튬 2-(3-아미노-4-포르밀페녹시)아세테이트 (45.3 mg), HBTU (77.8 mg) 및 DIEA (31 ㎕)에 1 mL DMF를 첨가하고, 혼합물을 30 분 동안 교반하였다. 생성된 용액을 1.0 mL DMF 중의 비오틴-dPEG™3-NH3 +TFA (쿠안타바이오디자인, cat# 10193, 100 mg), DIEA (47 ㎕)에 첨가하고, LCMS로 모니터링하면서 반응물을 실온에서 교반하였다. 생성물을 정제용 HPLC로 단리하였다.To lithium 2-(3-amino-4-formylphenoxy)acetate (45.3 mg), HBTU (77.8 mg) and DIEA (31 μL) was added 1 mL DMF, and the mixture was stirred for 30 minutes. The resulting solution was added to Biotin-dPEG™ 3 -NH 3 + TFA (Quanta Biodesign,
실시예Example 41: 형광- 41: fluorescence- PEGPEG -- ABAABA 시약의 합성 Synthesis of reagents
실시예Example 41-1: 41-1: 플루오레세인Fluorescein -- PEGPEG -- ABAABA ( ( X3757X3757 -48)의 합성.-48).
DMF (2.0 mL)를 NHS-플루오레세인 A (23.6 mg), 아민 B (17.6 mg) 및 DIEA (8.7 ㎕)에 첨가하였다. HPLC로 모니터링하여 A가 소모될 때까지 혼합물을 실온에서 교반하였다. 생성물 C를 정제용 HPLC로 단리하였다. ESI-MS (m/z) 679.72 (MH+). 이어서, 생성물 (C, 7.4 mg)를 3 M HCl (1.0 mL)에 용해시키고, 실온에서 5 분간 교반한 후 메탄올을 증발에 의해 제거하였다. 이 작업을 반복하여, Boc 기를 제거하여 아민 D를 수득하였다. 아민 D에 리튬 4-(3-아미노-4-포르밀페녹시)부타노에이트 (2.3 mg), HBTU (3.8 mg), DIEA (7.0 ㎕) 및 DMF (2 mL)를 실온에서 첨가하였다. 이어서, 표제 생성물을 정제용 HPLC로 단리하였다.DMF (2.0 mL) was added to NHS-fluorescein A (23.6 mg), amine B (17.6 mg) and DIEA (8.7 μl). The mixture was stirred at room temperature until A was consumed as monitored by HPLC. Product C was isolated by preparative HPLC. ESI-MS (m/z) 679.72 (MH + ). Then, the product (C, 7.4 mg) was dissolved in 3 M HCl (1.0 mL), stirred at room temperature for 5 minutes, and then methanol was removed by evaporation. This operation was repeated to remove the Boc group to obtain amine D. To amine D lithium 4-(3-amino-4-formylphenoxy)butanoate (2.3 mg), HBTU (3.8 mg), DIEA (7.0 μL) and DMF (2 mL) were added at room temperature. The title product was then isolated by preparative HPLC.
실시예Example 42: 42: 올리고사카라이드Oligosaccharides -- ABAABA 시약의 합성 Synthesis of reagents
실시예 42-1: 3-아미노-4- 포르밀페녹시부티레이트의 Gal - Glu -1-아미드 (3793-050)의 합성. Example 42-1: of 3-amino-4- formylphenoxybutyrate Synthesis of Gal - Glu -1-amide (3793-050).
NaBH3CN (94.3 mg)을 NH4OAc (771 mg) 및 락토스 (180 mg)의 H2O (10 mL) 용액 (pH 7)에 첨가하고, 반응물을 35℃에서 7 일 동안 교반하였다. 반응 혼합물을 동결건조시키고, 잔류물을 H2O에 용해시킨 후, 도웩스(Dowex) 1X8-400 음이온 교환 수지 (OH- 형태, 45 g)를 통과시켜, 과량의 BH3CN-, 그의 부산물 및 아세테이트를 제거하였다. 용리액을 동결건조시켜 조 생성물을 수득하였고, 이것을 도웩스 50WX8-400 수지 (H+ 형태, 30 g)를 사용한 양이온 교환 크로마토그래피에 의해 정제하여, 목적하는 생성물 (3793-048)을 밝은 황색 분말로서 수득하였다.NaBH 3 CN (94.3 mg) was added to a H 2 O (10 mL) solution (pH 7) of NH 4 OAc (771 mg) and lactose (180 mg), and the reaction was stirred at 35° C. for 7 days. The reaction mixture was lyophilized, then the residue was dissolved in H 2 O, also Wexford (Dowex) 1X8-400 anion exchange resin was passed through a (OH- form, 45 g), an excess of BH 3 CN -, its by-product And the acetate was removed. The eluent was lyophilized to give a crude product, which was purified by cation exchange chromatography using Dowex 50WX8-400 resin (H+ form, 30 g) to give the desired product (3793-048) as a light yellow powder. I did.
리튬 3-아미노-4-포르밀페녹시부티레이트 (6.0 mg)를 무수 DMSO (200 ㎕) 중의 HBTU (9.9 mg) 및 DIEA (7.7 ㎕)로 1 시간 동안 처리하였다. 이어서 상기로부터의 Gal-Glu-1-아민 (3793-048) (7.7 mg)의 DMSO (250 ㎕) 용액을 실온에서 반응 혼합물에 첨가한 후, 1 일 동안 교반하였다. 반응 혼합물을 동결건조시키고, NH4OAc 용리하면서 정제용 HPLC-MS에 의해 정제하여, 목적하는 생성물 (3793-050)을 황색 분말로서 수득하였다.Lithium 3-amino-4-formylphenoxybutyrate (6.0 mg) was treated with HBTU (9.9 mg) and DIEA (7.7 μL) in anhydrous DMSO (200 μL) for 1 hour. Subsequently, a DMSO (250 μl) solution of Gal-Glu-1-amine (3793-048) (7.7 mg) from above was added to the reaction mixture at room temperature, followed by stirring for 1 day. The reaction mixture was lyophilized and purified by preparative HPLC-MS eluting with NH 4 OAc to give the desired product (3793-050) as a yellow powder.
실시예Example 43: 인지질- 43: phospholipid- ABAABA 시약의 합성 Synthesis of reagents
실시예Example 43-1: 43-1: DOPEDOPE -- ABAABA ( ( TU3627TU3627 -092)의 합성. -092).
리튬 4-(3-아미노-4-포르밀페녹시)부타노에이트 (34 mg) 및 HBTU (57 mg)를 20 mL 유리 바이알에 넣고, 2 mL DMF를 첨가하였다. 활성화되도록, 반응물을 주위 온도에서 30 분 동안 교반하였다. 별개의 20 mL 바이알에 DOPE (76 mg, 1,2,-디올레오일-sn-글리세로-포스포에탄올아민, NOF Corp.)를 넣은 후, DIEA (35 ㎕) 및 3 mL DCM을 넣었다. 첫번째 바이알 내의 활성화된 에스테르의 황색 용액을 두번째 바이알로 옮기고, 반응물을 주위 온도에서 교반하였다. 24 시간 후, 전체 반응 혼합물을 용매 A (용매 A: 5% DMC 중의 NEt3, 용매 B: 5% MeOH 중의 NEt3)로 평형화된 12 g 미리-패킹된 SiO2 칼럼에 적용하고, 칼럼을 0 → 15% A 중의 B의 선형 구배로 15 분에 걸쳐 용리하여, 부분적으로 정제된 생성물을 밝은 황색의 매우 점성인 오일로서 수득하였다. 이 생성물을 12 g SiO2 칼럼 (용매 A: 5% DMC 중의 NEt3, 용매 B: 5% MeOH 중의 NEt3)을 이용한 플래쉬 크로마토그래피에 의해 다시 정제하고, 2 → 10% A 중의 B의 선형 구배로 15 분에 걸쳐 용리하였다. 순수한 생성물을 함유하는 분획을 감압하에 농축시켜, 표제 화합물의 트리에틸암모늄 염을 수득하였다. Rf: 0.43 (SiO2, 10% DCM 중 MeOH).Lithium 4-(3-amino-4-formylphenoxy)butanoate (34 mg) and HBTU (57 mg) were placed in a 20 mL glass vial, and 2 mL DMF was added. To activate, the reaction was stirred at ambient temperature for 30 minutes. DOPE (76 mg, 1,2,-dioleoyl-sn-glycero-phosphoethanolamine, NOF Corp.) was placed in a separate 20 mL vial, followed by DIEA (35 μL) and 3 mL DCM. The yellow solution of the activated ester in the first vial was transferred to the second vial and the reaction was stirred at ambient temperature. After 24 hours, the entire reaction mixture was applied to a 12 g pre-packed SiO 2 column equilibrated with solvent A (solvent A: NEt 3 in 5% DMC, solvent B: NEt 3 in 5% MeOH) and the column was zero → Elution over 15 minutes with a linear gradient of B in 15% A to give the partially purified product as a light yellow very viscous oil. This product was purified again by flash chromatography using a 12 g SiO 2 column (solvent A: NEt 3 in 5% DMC, solvent B: NEt 3 in 5% MeOH), linear gradient of B in 2 → 10% A And eluted over 15 minutes. Fractions containing pure product were concentrated under reduced pressure to give the triethylammonium salt of the title compound. Rf: 0.43 (SiO 2 , MeOH in 10% DCM).
실시예Example 44: 44: PCLPCL 커플링 연결의 환원 Reduction of coupling connection
PCL-기초 단백질 접합체의 해리를 방지하기 위해 PCL 커플링 연결을 환원시키는 것을 알아내었다. 도 54A는 2-ABA에 커플링시킨 후 20 mM NaCNBH3으로 1 시간 동안 환원시킨 hFGF21-Lys150PCL의 ESI 질량 분광측정 분석을 보여준다. 도 54B는 10 mM 인산염 완충액 (pH 7.5)으로 투석하고 50℃에서 1 일 동안 인큐베이션한 이후의, 환원된 hFGF21-Lys150PCL 2-ABA 접합체의 ESI 질량 분광측정 분석을 보여준다.It was found to reduce the PCL coupling linkage to prevent dissociation of the PCL-based protein conjugate. Figure 54A shows the ESI mass spectrometric analysis of hFGF21-Lys150PCL after coupling to 2-ABA and reduced with 20 mM NaCNBH 3 for 1 hour. Figure 54B shows the ESI mass spectrometric analysis of the reduced hFGF21-Lys150PCL 2-ABA conjugate after dialysis with 10 mM phosphate buffer (pH 7.5) and incubation for 1 day at 50°C.
도 55는 NaCNBH3에 의해 환원되거나 환원되지 않은 PEG화된 FGF21에 대한 PCL 연결의 안정성을 보여준다. FGF21 돌연변이체 Arg154PCL을 30.3 kDa-2-ABA-PEG (TU3627-024; 실시예 37-7 참조)와 반응시키고 정제하였다. 정제된 FGF21Arg154PCL-30.3 kDa-2-ABA-PEG를 20 mM NaBH3CN으로 16 시간 동안 환원시켰다 (실온, pH 7.5, 100 mM 단백질). 샘플을 16 시간 동안 4℃, 실온, 37℃ 및 50℃, 및 95℃에서 5 분 동안 인큐베이션하였다. 환원된 샘플 및 비환원된 샘플의 SDS-PAGE 겔을 도 55A에 나타내었다. 추가로, 도 55B는 60 시간 동안 4℃, 실온, 37℃ 및 50℃, 및 95℃에서 인큐베이션한 비환원된 샘플에 대한 SDS-PAGE 겔을 나타낸다. Figure 55 shows the stability of PCL ligation to PEGylated FGF21, reduced or not reduced by NaCNBH 3. The FGF21 mutant Arg154PCL was reacted with 30.3 kDa-2-ABA-PEG (TU3627-024; see Example 37-7) and purified. Purified FGF21Arg154PCL-30.3 kDa-2-ABA-PEG was reduced with 20 mM NaBH 3 CN for 16 hours (room temperature, pH 7.5, 100 mM protein). Samples were incubated for 5 minutes at 4° C., room temperature, 37° C. and 50° C., and 95° C. for 16 hours. SDS-PAGE gels of the reduced and non-reduced samples are shown in Figure 55A. Additionally, Figure 55B shows SDS-PAGE gels for unreduced samples incubated at 4° C., room temperature, 37° C. and 50° C., and 95° C. for 60 hours.
실시예 45: 2- ABA 에 의해 공유적으로 변형된 PCL -A ( Lys - P5C ) 및 PCL -B (Lys-P2C)의 NMR 연구. Example 45: NMR study of PCL- A ( Lys - P5C ) and PCL- B (Lys-P2C) covalently modified by 2-ABA.
PCL-A (3647-125, 실시예 36-1 참조) 및 PCL-B (3793-011, 실시예 36-2 참조)와 2-아미노벤즈알데히드 (2-ABA)의 반응 및 생성된 PCL-ABA 부가물의 구조를 표준 1D 및 2D 핵 자기 공명 (NMR) 분광분석법에 의해 연구하였다.Reaction of PCL-A (3647-125, see Example 36-1) and PCL-B (3793-011, see Example 36-2) with 2-aminobenzaldehyde (2-ABA) and addition of the resulting PCL-ABA The structure of water was studied by standard 1D and 2D nuclear magnetic resonance (NMR) spectroscopy.
PCL-A 및 PCL-B의 2-ABA와의 반응 및 그 생성물의 후속 특징 규명을 위해, 1H/13C/19F/31P-QNP-저온탐침이 장착된 브루커 아반스(Bruker Avance) 400 MHz NMR 기기 (브루커 바이오스핀, 메사추세츠주 빌레리카 소재) 상에서 300 K에서 NMR 데이터를 수득하였다. 1H 1D 스펙트럼을 전형적으로 16 스캔, 이완 지연 5 s, 16384 복합체 데이터 포인트를 12 ppm의 탐색폭으로 기록하였다. 1H-1H COSY 스펙트럼은 전형적으로 4 스캔, 256 t1 실험으로 기록하고, 1H-1H ROESY 스펙트럼은 8 스캔, 512 t1 실험으로 기록하였다. 1H-13C HMBC 스펙트럼은 전형적으로 32 스캔, 256 t1 실험으로 기록하고, 1H-13C HMQC 스펙트럼은 전형적으로 4 스캔, 128 t1 실험으로 탄소 규모에서는 222 ppm 및 양성자 규모에서는 12 ppm 또는 7.5 ppm의 스펙트럼 폭으로 기록하였다.For the reaction of PCL-A and PCL-B with 2-ABA and subsequent characterization of its products, Bruker Avance equipped with a 1 H/ 13 C/ 19 F/ 31 P-QNP-low temperature probe NMR data were obtained at 300 K on a 400 MHz NMR instrument (Bruker Biospin, Billerica, MA). 1 H 1D spectra were typically recorded with 16 scans, relaxation delay of 5 s, and 16384 complex data points with a search width of 12 ppm. The 1 H- 1 H COSY spectrum was typically recorded as a 4 scan, 256 t 1 experiment, and the 1 H- 1 H ROESY spectrum was recorded as an 8 scan, 512 t 1 experiment. The 1 H- 13 C HMBC spectrum is typically recorded as a 32 scan, 256 t 1 experiment, and the 1 H- 13 C HMQC spectrum is typically a 4 scan, 128 t 1 experiment, 222 ppm on the carbon scale and 12 ppm on the proton scale Or 7.5 ppm of spectral width.
환원된 부가물의 특징을 규명하기 위해, 모든 스펙트럼을 1H/13C/15N-TXI-저온탐침이 장착된 브루커 아반스 600 MHz 기기 상에서 300 K에서 기록하였다. 1H 스펙트럼을 전형적으로 64 스캔, 이완 지연 2 s, 16384 복합체 데이터 포인트를 물분자 억제를 위해 여기 조형하면서 14 ppm의 탐색폭으로 기록하였다. 1H-1H COSY 스펙트럼은 전형적으로 16 스캔, 1024 t1 실험으로 10 ppm의 스펙트럼 폭을 이용하여 기록하였다. 1H-13C HMQC 스펙트럼은 전형적으로 8 스캔, 256 t1 실험으로 탄소 규모에서는 160 ppm 및 양성자 규모에서는 10 ppm의 스펙트럼 폭으로 기록하였다. 1H-13C HMBC 스펙트럼은 전형적으로 300K에서 88 스캔, 256 t1 실험으로, 탄소 규모에서는 180 ppm 및 양성자 규모에서는 10 ppm의 스펙트럼 폭을 이용하여 기록하였다.To characterize the reduced adduct, all spectra were recorded at 300 K on a
PCL-A와 2-ABA의 반응은 다음과 같이 모니터링하였다: 실시예 36-1에 기재한 바와 같이 합성한 1.0 mg의 PCL-A를 D2O 중의 1X PBS 0.5 mL에 용해시켰다. D2O 중의 10 ㎕의 10 mM 3-(트리메틸실릴)프로피온산 (TSP)을 내부 표준으로서, 그리고 NMR로 농도를 측정하기 위해 첨가하였다. 3.7 mg의 2-아미노벤즈알데히드 (2-ABA; 시그마 사에서 구입)를 D2O 중의 1X PBS 0.5 mL 및 10 mM TSP 10 ㎕에 용해시켰다. 양 샘플의 농도를 NMR로 측정하였다. 출발 물질의 NMR 신호 (표 7)를 1H 1D, 1H-1H COSY, 1H-1H ROESY, 1H-13C HMBC 및 1H-13C HMQC 실험을 비롯한 표준 NMR 방법을 이용하여 할당하였다. The reaction of PCL-A and 2-ABA was monitored as follows: 1.0 mg of PCL-A synthesized as described in Example 36-1 was dissolved in 0.5 mL of
반응을 개시하기 위해, 325 ㎕의 PCL-A 용액을 175 ㎕의 2-ABA 용액과 혼합하였다. 생성된 반응 혼합물을 NMR 튜브로 옮기고, PCL-A 및 2-ABA를 대략 1:1 몰비로 함유시켰다. 반응을 실온에서 진행시키고, NMR 스펙트럼을 표시한 시점에 주기적으로 수득하였다 (도 56). 출발 PCL-A 물질 (도트) 및 2-ABA 반응물 (별표)에 대한 반응 신호가 신속하게 사라지고 반응이 진행되어 완료되는 동안, 모든 PCL-A가 전환되었다 (샘플에는 약간 과량의 2-ABA가 존재하였다). 혼합 0.5 시간 후에 수득한 제1 시점에서, 2가지의 새로운 종이 대략 2:1의 비율로 검출되었다 (대표적인 공명을 화살표로 표시함). 소수의 종은 수 일이 경과된 뒤에 주요 종으로 완전하게 전환되었다. To initiate the reaction, 325 μl of PCL-A solution was mixed with 175 μl of 2-ABA solution. The resulting reaction mixture was transferred to an NMR tube and contained PCL-A and 2-ABA in an approximate 1:1 molar ratio. The reaction was allowed to proceed at room temperature, and the NMR spectrum was periodically obtained at the time point indicated (FIG. 56). The reaction signal for the starting PCL-A material (dot) and 2-ABA reactant (asterisk) disappeared quickly and while the reaction proceeded to completion, all PCL-A was converted (there was a slight excess of 2-ABA in the sample. I did). At the first time point obtained after 0.5 hours of mixing, two new species were detected in a ratio of approximately 2:1 (representative resonances are indicated by arrows). A few species have been completely converted to major species after several days have elapsed.
PCL-A와 2-ABA의 최종 반응 생성물은 D2O 및 d6-DMSO 중에서 제조된 샘플을 이용하여 표준 1D 1H 및 13C, 2D 1H-1H COSY, 1H-1H ROESY, 1H-13C HMBC 및 1H-13C HMQC NMR 분광분석법으로 그 특징을 규명하였다. d6-DMSO 중의 샘플은 HPLC로 정제하였다. 양성자 및 탄소 공명의 경우의 신호 할당 및 d6-DMSO 중의 1H-13C HMBC 스펙트럼에서 관찰된 상관관계를 표 8에 요약하였다. The final reaction product of PCL-A and 2-ABA was standard 1D 1 H and 13 C, 2D 1 H- 1 H COSY, 1 H- 1 H ROESY, 1 using samples prepared in D 2 O and d6-DMSO. It was characterized by H- 13 C HMBC and 1 H- 13 C HMQC NMR spectroscopy. Samples in d6-DMSO were purified by HPLC. The signal assignment in the case of proton and carbon resonance and the correlations observed in the 1 H- 13 C HMBC spectrum in d6-DMSO are summarized in Table 8.
원자의 번호매김 및 HMBC에 있어서 선택된 결합 통과 상관관계를 도 57A에 나타내었다. PCL-A 출발 물질에서 관찰된 바와 대조적으로, 탄소 17 상의 2개의 메틸렌 양성자가 2개의 상이한 화학적 이동 값에서 관찰되었다. 이들 양성자는 또한 HMBC 스펙트럼에서 탄소 7에 대해 이종핵 결합 통과 상관관계를 나타내었고, 이는 질소 16 및 탄소 7 사이에 공유 결합이 형성되었음을 시사한다. 탄소 7 상의 양성자는 5.6 ppm에서 공명하며 (양성자 7은 도 201에서 강조표시된 PCL-A/2-ABA 부가물의 신호임), 탄소 2, 8, 9, 5 및 13에 대해 상관관계를 나타내었다. 유사하게, 탄소 2 상의 양성자는 탄소 9에 대해 HMBC 상관관계를 나타내었다. D2O 및 D6-DMSO에서 특징이 규명된 2개의 샘플에 대한 이러한 관찰결과 및 그 밖의 모든 NMR 관찰결과는 PCL-A/2-ABA 부가물의 주요 생성물에 대해 도시한 도 57A에서의 구조와 일치한다. 따라서, 전체적으로 NMR 증거는 PCL-A 및 2-ABA 사이의 반응 생성물의 구조가 다음과 같음을 가리킨다:Figure 57A shows the correlation of atom numbering and selected bond passage for HMBC. In contrast to that observed with the PCL-A starting material, two methylene protons on
또한, 반응 혼합물에서 관찰된 소수 형태의 구조적인 특징을 규명하고자 하였다 (도 56). 소수 형태의 농도가 낮고 그것이 천천히 주요 형태로 전환된다는 사실은 분석을 복잡하게 하였다. 분석은 결론에 이르지 못했다. 소수 및 주요 형태는 둘 모두 탄소 7 상에 하나의 양성자를 가지고, 둘 모두의 화학적 이동은 매우 유사하였다 (도 56에서 5.6 ppm에 화살표로 나타냄). 뿐만 아니라, 공간-통과 ROESY 접촉도 두 형태에서 모두 유사하였다. 탄소 17 상의 2개의 메틸렌 양성자는 퇴화되었고, 이는 그들이 PCL-A/2-ABA 부가물의 소수 형태 뿐만 아니라 미반응 PCL-A의 경우에서도 동일한 화학적 이동을 나타내었음을 의미한다. 그러나, 이들 양성자의 화학적 이동은 주요 형태에서 구분된다. 또한, 이들 양성자는 소수 및 주요 형태 둘 모두에서 탄소 7에 대해 HMBC 상관관계를 보이는데, 이는 탄소 7 및 질소 16 사이의 공유 결합을 시사한다. 소수 형태에서 탄소 17 상의 메틸렌 양성자의 화학적 이동이 퇴화된 것으로 관찰되었는데 이는 탄소 7 및 질소 16 사이의 공유 결합이 반-안정적일 수 있으며, 소수 형태에 대한 NMR 관찰은 2 이상의 종 사이에서의 화학적 교환의 결과임을 암시할 수 있다. 소수 형태는 PCL-A/2-ABA 부가물의 양성자화된 형태와의 화학적 교환에 있어서 주요 형태의 대안적인 반-안정적 입체 이성질체일 수 있다 (도 57B). In addition, it was attempted to clarify the structural characteristics of the hydrophobic form observed in the reaction mixture (FIG. 56). The low concentration of the minority form and the fact that it slowly converts to the main form complicates the analysis. The analysis did not come to a conclusion. Both minority and major forms have one proton on
또한, D2O 중의 합성 PCL-A 및 2-ABA의 용액을 상기한 바와 같이 혼합하고, NMR로 모니터링하여 반응이 진행되어 완료되도록 하였다. D2O 중의 시아노수소화붕소나트륨 (NaCNBH3) 용액의 분취량을 NMR 튜브에 첨가하는 것에 의해 PCL-A/2-ABA 부가물이 새로운 종으로 환원되었다. PCL-A/2-ABA 부가물에 대한 특징인 5.6 ppm에서의 공명 (도 56, 화살표)이 완전히 사라질 때까지 추가의 NaCNBH3 분취량을 첨가하였다. 이어서, 완전히 환원된 PCL-A/2-ABA 부가물의 NMR 샘플을 반복적으로 동결건조시키고, 무수 D2O에 재용해시켜, 샘플을 재농축시키고 물을 제거하였다. 최종 샘플을 0.5 mL의 무수 D2O 중에 재구성하였다. 환원된 형태의 특징을 표준 2D 1H-1H COSY, 1H-13C HMBC 및 1H-13C HMQC NMR 분광분석법으로 규명하였다. 1H-13C HMBC 스펙트럼으로부터의 양성자 및 탄소 공명의 신호 할당 및 이종핵 결합 통과 상관관계를 표 9에 요약하였다.In addition, a solution of synthetic PCL-A and 2-ABA in D 2 O was mixed as described above and monitored by NMR to allow the reaction to proceed and complete. The PCL-A/2-ABA adduct was reduced to a new species by adding an aliquot of a solution of sodium cyanoborohydride (NaCNBH 3 ) in D 2 O to the NMR tube. Additional 3 aliquots of NaCNBH were added until the resonance at 5.6 ppm characteristic for the PCL-A/2-ABA adduct (FIG. 56, arrows) disappeared completely. Then, the NMR sample of the fully reduced PCL-A/2-ABA adduct was repeatedly lyophilized and redissolved in anhydrous D 2 O to reconcentrate the sample and remove water. The final sample was reconstituted in 0.5 mL of anhydrous D 2 O. The reduced form was characterized by standard 2D 1 H- 1 H COSY, 1 H- 13 C HMBC and 1 H- 13 C HMQC NMR spectroscopy. The signal assignment of protons and carbon resonances from the 1 H- 13 C HMBC spectrum and the heteronuclear binding pass-through correlations are summarized in Table 9.
원자의 번호매김은 도 57C에 나타내었다. PCL-A/2-ABA 부가물의 주요 형태에서 관찰된 상관관계에 대한 핵심적인 차이점은 탄소 17 및 탄소 7 상의 메틸렌 양성자 사이에 이종핵 결합 통과 상관관계가 부족하고, 탄소 2 및 탄소 9 상의 양성자 사이에 결합 통과 상관관계가 존재하지 않는다는 것이다. 이러한 관찰결과 및 그 밖의 모든 NMR 관찰결과는 환원된 PCL-A/2-ABA 부가물에 대한 다음과 같은 (또한 도 57C에서의) 구조와 일치한다:The numbering of the atoms is shown in Figure 57C. The key differences in the correlations observed in the major forms of PCL-A/2-ABA adducts are the lack of heteronuclear bond pass-through correlations between the methylene protons on
또한 PCL-B와 2-ABA의 반응을 PCL-A와 2-ABA를 반응시키기 위한 조건과 동일한 조건하에서 연구하였다. 실시예 36-2에서 기재한 바와 같이 합성한 PCL-B를 D2O에 (상기한 바와 같이) 용해시키고, PCL-B 및 2-ABA를 대략 1:1의 몰비로 함유하는 반응 혼합물을 NMR 튜브로 옮겼다. 출발 물질의 신호 할당을 표 10에 열거한다. In addition, the reaction of PCL-B and 2-ABA was studied under the same conditions as those for reacting PCL-A and 2-ABA. PCL-B synthesized as described in Example 36-2 was dissolved in D 2 O (as described above), and a reaction mixture containing PCL-B and 2-ABA in a molar ratio of approximately 1:1 was NMR. Transferred to the tube. The signal assignments of the starting materials are listed in Table 10.
반응을 실온에서 진행시키고, NMR 스펙트럼은 나타낸 시점에 주기적으로 수득하였다 (도 58). PCL-A 샘플과 대조적으로, PCL-B는 2-ABA와 쉽게 반응하지 않았다. 심지어 17 일이 지난 후에도, 다량의 출발 물질 (2-ABA에 대해서는 별표; PCL-B에 대해서는 점)이 여전히 잔존하였고, 단지 소량만이 새로운 종 (화살표)으로 전환되었다. 이러한 종은 추가로 특징을 규명할 수 없었다. 그러나, 2 가지의 반응의 NMR 분석은 PCL-A의 2-ABA와의 반응성이 PCL-B의 반응성보다 훨씬 크다는 것을 분명히 보여주었다. The reaction was allowed to proceed at room temperature, and NMR spectra were obtained periodically at the indicated time points (FIG. 58). In contrast to the PCL-A sample, PCL-B did not react easily with 2-ABA. Even after 17 days, a large amount of starting material (asterisk for 2-ABA; dots for PCL-B) still remained, and only a small amount was converted to a new species (arrow). This species could not be further characterized. However, NMR analysis of the two reactions clearly showed that the reactivity of PCL-A with 2-ABA was much greater than that of PCL-B.
실시예Example 46: 46: mEGFmEGF 에 혼입된 피롤리신 (Pyrrolysine incorporated in ( PylPyl ) 및 ) And PCLPCL 의 of 유도체화Derivatization
피롤리신 (Pyl) 및 PCL의 mEGF에의 혼입은 실시예 14에 기재한 바와 같이 달성화되, 이. 콜라이 BL21(DE3) 세포를 pAra-pylSTBCD 및 pET22b 벡터 상의 돌연변이체 mEGF Tyr10TAG 유전자로 공동 형질전환시켰다. 생성된 mEGF 돌연변이체를 이하에서는 mEGF-Tyr10PCL/Pyl이라 지칭한다. ESI-MS 분석 결과 PCL 및 PYL이 둘 모두 mEGF의 위치 10에 혼입된 것으로 밝혀졌다 (도 59A; mEGF Tyr10PCL에 대해 예상되는 질량 = 7296 Da; mEGF Tyr10Pyl에 대해 예상되는 질량 = 7310 Da). 이에 따라 PCL 또는 PYL이 혼입된 mEGF 단백질의 혼합물이 수득되었다. 이러한 특정 샘플에서, 우세한 종은 내부에 PCL이 혼입된 mEGF였다.Incorporation of pyrrolysine (Pyl) and PCL into mEGF was achieved as described in Example 14, E. E. coli BL21(DE3) cells were cotransformed with the mutant mEGF Tyr10TAG gene on pAra-pylSTBCD and pET22b vector. The resulting mEGF mutant is hereinafter referred to as mEGF-Tyr10PCL/Pyl. ESI-MS analysis revealed that both PCL and PYL were incorporated at
이러한 혼합물의 유도체화를 증명하고, 본원에서 제공하는 방법에 의해 PYL이 변형되었음을 증명하기 위해, 10 x PBS (pH 7.0) 및 1% (v/v) DMSO 중에서 25℃에서 16 시간 동안 mEGF Tyr10PCL/PYL에 대해 ABA 시약 TU3627-014 (실시예 34-2 참조)를 커플링시켰다. 접합 반응은 10 μM mEGF Tyr10PCL/PYL 및 1 mM TU3627-014를 첨가하는 것에 의해 개시되었다. 전기분무 이온화-질량 분광측정법 (ESI-MS)에 의해 단백질 접합체의 형성을 확인하였다 (도 59B). PCL 및 PYL 부가물 사이의 비는 미반응 단백질 내의 PCL 및 PYL의 비와 유사하였고 (도 58A), 이는 PCL 및 Pyl에 대한 반응성이 유사함을 나타낸다.To demonstrate the derivatization of this mixture and to demonstrate that PYL was modified by the method provided herein, mEGF Tyr10PCL/ The ABA reagent TU3627-014 (see Example 34-2) was coupled to PYL. The conjugation reaction was initiated by adding 10 μM mEGF Tyr10PCL/PYL and 1 mM TU3627-014. Formation of the protein conjugate was confirmed by electrospray ionization-mass spectrometry (ESI-MS) (Fig. 59B). The ratio between PCL and PYL adducts was similar to the ratio of PCL and PYL in the unreacted protein (FIG. 58A ), indicating that the reactivity to PCL and Pyl is similar.
본원에서 기재하는 실시예 및 실시양태는 단지 설명을 위한 것으로, 그러한 관점에서 다양한 변형 또는 변화가 당업자에게 제안될 것이며, 이는 본 출원의 취지 및 범위와 첨부하는 특허청구범위의 범주 내에 포함됨을 이해한다.It is understood that the examples and embodiments described herein are for illustrative purposes only, and various modifications or changes will be suggested to those skilled in the art in that respect, which are included within the spirit and scope of the present application and the scope of the appended claims. .
SEQUENCE LISTING <110> IRM Geierstanger, Bernhard <120> BIOSYNTHETICALLY GENERATED PYRROLINE-CARBOXY-LYSINE AND SITE SPECIFIC PROTEIN MODIFICATIONS VIA CHEMICAL DERIVATIZATION OF PYRROLINE-CARBOXY-LYSINE AND PYRROLYSINE RESIDUES <130> PAT053160-WO <160> 35 <170> PatentIn version 3.5 <210> 1 <211> 1365 <212> DNA <213> Methanosarcina mazeii <400> 1 atggataaaa aaccactaaa cactctgata tctgcaaccg ggctctggat gtccaggacc 60 ggaacaattc ataaaataaa acaccacgaa gtctctcgaa gcaaaatcta tattgaaatg 120 gcatgcggag accaccttgt tgtaaacaac tccaggagca gcaggactgc aagagcgctc 180 aggcaccaca aatacaggaa gacctgcaaa cgctgcaggg tttcggatga ggatctcaat 240 aagttcctca caaaggcaaa cgaagaccag acaagcgtaa aagtcaaggt cgtttctgcc 300 cctaccagaa cgaaaaaggc aatgccaaaa tccgttgcga gagccccgaa acctcttgag 360 aatacagaag cggcacaggc tcaaccttct ggatctaaat tttcacctgc gataccggtt 420 tccacccaag agtcagtttc tgtcccggca tctgtttcaa catcaatatc aagcatttct 480 acaggagcaa ctgcatccgc actggtaaaa gggaatacga accccattac atccatgtct 540 gcccctgttc aggcaagtgc ccccgcactt acgaagagcc agactgacag gcttgaagtc 600 ctgttaaacc caaaagatga gatttccctg aattccggca agcctttcag ggagcttgag 660 tccgaattgc tctctcgcag aaaaaaagac ctgcagcaga tctacgcgga agaaagggag 720 aattatctgg ggaaactcga gcgtgaaatt accaggttct ttgtggacag gggttttctg 780 gaaataaaat ccccgatcct gatccctctt gagtatatcg aaaggatggg cattgataat 840 gataccgaac tttcaaaaca gatcttcagg gttgacaaga acttctgcct gagacccatg 900 cttgctccaa acctttacaa ctacctgcgc aagcttgaca gggccctgcc tgatccaata 960 aaaatttttg aaataggccc atgctacaga aaagagtccg acggcaaaga acacctcgaa 1020 gagtttacca tgctgaactt ctgccagatg ggatcgggat gcacacggga aaatcttgaa 1080 agcataatta cggacttcct gaaccacctg ggaattgatt tcaagatcgt aggcgattcc 1140 tgcatggtct atggggatac ccttgatgta atgcacggag acctggaact ttcctctgca 1200 gtagtcggac ccataccgct tgaccgggaa tggggtattg ataaaccctg gataggggca 1260 ggtttcgggc tcgaacgcct tctaaaggtt aaacacgact ttaaaaatat caagagagct 1320 gcaaggtccg agtcttacta taacgggatt tctaccaacc tgtaa 1365 <210> 2 <211> 1050 <212> DNA <213> Methanosarcina mazeii <400> 2 atgatccaga aaatggcaac cgaagaactt gacaggttcg gggagaaaat tattgaaggt 60 tttaaattgt ctgatgatga cctcagggct cttctttctc ttgaattcga agaagagctg 120 gaaaagcttt actatgtagc tagaaaggtc agaaactatt atttcggcaa cagggtgttt 180 cttaactgtt ttatttattt ctcaacttat tgtaaaaacc agtgctcttt ttgctactat 240 aactgtaaaa acgaaattaa ccgctaccgc ctgaccggtg aagaggttaa agagatgtgc 300 aaagccctga aaggtgcagg ctttcacatg atcgacctga caatgggaga ggatccctat 360 tactatgatg accctgaccg cttcgttgaa cttgtcagga cagtaaaaga agaactcggg 420 cttccaataa tgatttctcc gggagttatg gatgacagca ccctcctgaa agccagggaa 480 gaaggagcaa atttctttgc cctttatcag gagacttatg accgcgaact ttatggaaag 540 ctaagggtag gtcagtcctt cgaaggaagg tttaatgccc gcaggtttgc aaaagaacag 600 gggtactgta tagaagacgg cattcttacc ggcgtaggaa atgatatcga atcaactctt 660 atatccctga aggggatgaa agcaaacaat cctgatatgg taagggtaat gacttttctg 720 cctcaggaag gaactccgct tgaaggtttc agcgatagtt caaagctttc ggagctgaaa 780 atcatagcga ttctcaggct catgtttcct gaatgcctga taccggcttc tcttgacctt 840 gaaggcatag acggcatggt gcaccgttta aatgccggag caaatattgt aacctccatc 900 ctcccagatt cacgcctgga aggggttgcc aattacgacc gcggcatgga agagagggac 960 agggacgtta caagcgttgt caaaaggctg aaggttatgg gaatggaacc tgcgccgcag 1020 gctgagtttg agagagtcct ggggtgctaa 1050 <210> 3 <211> 1116 <212> DNA <213> Methanosarcina mazeii <400> 3 atgagagagt cctggggtgc tagcctgaaa acaatatgcc ttataggcgg gaagctgcag 60 ggcttcgagg ctgcatacct atctaagaaa gccggaatga aagtgcttgt aatagacaaa 120 aacccgcagg cgcttataag gaattatgcg gatgagttcc agtgttttaa cataacggaa 180 gagccggaaa aactcgtcgc gatatcaaaa aatgttgatg ccatactgcc ggtaaatgaa 240 aaccttgaat gtatagaatt tctgaattct ataaaagaaa aattctcctg cccggtactt 300 ttcgattttg aagcttacag gatcagcagg gataagagaa aatcaaaaga atacttcgca 360 tccataggaa ccccgacccc tcaggacaaa ccgtcggaac caccttattt tgtaaagcct 420 ccctgcgaaa gcagcagtgt gggagcgaga ataatccatg acaggaaaga gcttaaagag 480 cttgagcccg ggatgctcat agaagaatac gttgaagggg aagtggtctc acttgaggtc 540 ataggggatg gaaataattt tgctgtggta aaggaaaccc ttgtacatat cgatgacacc 600 tatgactgcc atatggtgac ccctctccct ctagaccctt ccttcaggga actatcctac 660 tcccttgcag caaacctgcc cttaaaagga attatggacg tggaagcgat ttccggcccc 720 ctggggttaa aagttattga gatagatgcc cgtttcccga gccagactcc gactgcggtc 780 tattattctt ccgggatcaa cctcatagaa ctcctgttcc gggcttttaa tggaggcata 840 gaagagatca aaactctccc tgaagacagg tactgcattt acgaacatct catgcttgca 900 gaaaatggag tacttatccc tgtgggagaa caggtcctgt ccatgggaaa tgattacggc 960 aattattatg aagaacctgg aatagagatt ttcctgtgca aaggagagaa ccctgtattc 1020 accctggttt tctggggcag agacagggaa gaagctgaag ctagaaaaaa caaagggctt 1080 tcgattctaa aaagccgttt cggagctgct gcataa 1116 <210> 4 <211> 795 <212> DNA <213> Methanosarcina mazeii <400> 4 atggcacttt taaccccaga agacctggaa aatattaaca aacagcttca agaagctgat 60 tctactgtcc gcagagttac agggcttgat ataaaaggta tctgtaaaga tttctacggc 120 acaactccat gctgtgaaaa agtaggtatc gtgcctgtga cctcagggaa cgggatcata 180 gggagctttt ccgaatccct gaatgcaatt gccgggtatt tcgggtttga cagttttatt 240 actgatatgc ctgacgtcag cggatattat gaggcagtaa agaacggagc ccggatcata 300 cttatggcag atgataatac cttccttgcc cacaacctga aaaatggaaa aatcgccaat 360 aaccagccgt gtacaggcat aatttatgct gaaatagctt caagatacct gaaagccgat 420 tccaaagaag tgcttgccgt gggtcttggg aaggttggat ttccgggagc agcccatctc 480 gtacagaaag gcttcaaggt ttacggatat gatgctgaca gaacccttct agaaaaaagc 540 gtttccagcc tcggaattat acctttcaat cccgtcagcc ccgaaggcga caggcaaagg 600 aagttttcca ttattttcga agcaaccccc tgtgcagaca cgattccgga atccgtaatt 660 tcggaaaact gtgtgatttc tacccctggg ataccctgtg caatctcaaa ggagctgcaa 720 aaaaagtgtg gagttgaact tgtaatggaa ccactgggga taggtacagc atcaatgctg 780 tattctgtac tctaa 795 <210> 5 <211> 72 <212> DNA <213> Methanosarcina mazeii <400> 5 ggaaacctga tcatgtagat cgaatggact ctaaatccgt tcagccgggt tagattcccg 60 gggtttccgc ca 72 <210> 6 <211> 223 <212> PRT <213> Homo sapiens <400> 6 Met Lys Thr Phe Ile Leu Leu Leu Trp Val Leu Leu Leu Trp Val Ile 1 5 10 15 Phe Leu Leu Pro Gly Ala Thr Ala Gln Pro Glu Arg Asp Cys Arg Val 20 25 30 Ser Ser Phe Arg Val Lys Glu Asn Phe Asp Lys Ala Arg Phe Ser Gly 35 40 45 Thr Trp Tyr Ala Met Ala Lys Lys Asp Pro Glu Gly Leu Phe Leu Gln 50 55 60 Asp Asn Ile Val Ala Glu Phe Ser Val Asp Glu Thr Gly Gln Met Ser 65 70 75 80 Ala Thr Ala Lys Gly Arg Val Arg Leu Leu Asn Asn Trp Asp Val Cys 85 90 95 Ala Asp Met Val Gly Thr Phe Thr Asp Thr Glu Asp Pro Ala Lys Phe 100 105 110 Lys Met Lys Tyr Trp Gly Val Ala Ser Phe Leu Gln Lys Gly Asn Asp 115 120 125 Asp His Trp Ile Val Asp Thr Asp Tyr Asp Thr Tyr Ala Val Gln Tyr 130 135 140 Ser Cys Arg Leu Leu Asn Leu Asp Gly Thr Cys Ala Asp Ser Tyr Ser 145 150 155 160 Phe Val Phe Ser Arg Asp Pro Asn Gly Leu Pro Pro Glu Ala Gln Lys 165 170 175 Ile Val Arg Gln Arg Gln Glu Glu Leu Cys Leu Ala Arg Gln Tyr Arg 180 185 190 Leu Ile Val His Asn Gly Tyr Cys Asp Gly Arg Ser Glu Arg Asn Leu 195 200 205 Leu Asp Tyr Lys Asp Asp Asp Asp Lys His His His His His His 210 215 220 <210> 7 <211> 672 <212> DNA <213> Homo sapiens <400> 7 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 8 <211> 672 <212> DNA <213> Homo sapiens <400> 8 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg taggccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 9 <211> 672 <212> DNA <213> Homo sapiens <400> 9 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctagctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 10 <211> 672 <212> DNA <213> Homo sapiens <400> 10 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactagg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 11 <211> 672 <212> DNA <213> Homo sapiens <400> 11 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtagtg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 12 <211> 672 <212> DNA <213> Homo sapiens <400> 12 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtacta gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 13 <211> 672 <212> DNA <213> Homo sapiens <400> 13 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctagctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 14 <211> 672 <212> DNA <213> Homo sapiens <400> 14 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtag 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 15 <211> 672 <212> DNA <213> Homo sapiens <400> 15 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tagaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 16 <211> 672 <212> DNA <213> Homo sapiens <400> 16 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttagtgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 17 <211> 10 <212> PRT <213> Homo sapiens <220> <221> MOD_RES <222> (7)..(7) <223> Xaa is a PCL residue <400> 17 Tyr Trp Gly Val Ala Ser Xaa Leu Gln Lys 1 5 10 <210> 18 <211> 29 <212> PRT <213> Homo sapiens <220> <221> MOD_RES <222> (7)..(7) <223> Xaa is Cyc residue <400> 18 Lys Asp Pro Glu Gly Leu Xaa Leu Gln Asp Asn Ile Val Ala Glu Phe 1 5 10 15 Ser Val Asp Glu Thr Gly Gln Met Ser Ala Thr Ala Lys 20 25 <210> 19 <211> 166 <212> PRT <213> Homo sapiens <400> 19 Ala Pro Pro Arg Leu Ile Cys Asp Ser Arg Val Leu Glu Arg Tyr Leu 1 5 10 15 Leu Glu Ala Lys Glu Ala Glu Asn Ile Thr Thr Gly Cys Ala Glu His 20 25 30 Cys Ser Leu Asn Glu Asn Ile Thr Val Pro Asp Thr Lys Val Asn Phe 35 40 45 Tyr Ala Trp Lys Arg Met Glu Val Gly Gln Gln Ala Val Glu Val Trp 50 55 60 Gln Gly Leu Ala Leu Leu Ser Glu Ala Val Leu Arg Gly Gln Ala Leu 65 70 75 80 Leu Val Asn Ser Ser Gln Pro Trp Glu Pro Leu Gln Leu His Val Asp 85 90 95 Lys Ala Val Ser Gly Leu Arg Ser Leu Thr Thr Leu Leu Arg Ala Leu 100 105 110 Gly Ala Gln Lys Glu Ala Ile Ser Pro Pro Asp Ala Ala Ser Ala Ala 115 120 125 Pro Leu Arg Thr Ile Thr Ala Asp Thr Phe Arg Lys Leu Phe Arg Val 130 135 140 Tyr Ser Asn Phe Leu Arg Gly Lys Leu Lys Leu Tyr Thr Gly Glu Ala 145 150 155 160 Cys Arg Thr Gly Asp Arg 165 <210> 20 <211> 166 <212> PRT <213> mouse <400> 20 Ala Pro Pro Arg Leu Ile Cys Asp Ser Arg Val Leu Glu Arg Tyr Leu 1 5 10 15 Leu Glu Ala Lys Glu Ala Glu Asn Val Thr Met Gly Cys Ala Glu Gly 20 25 30 Pro Arg Leu Ser Glu Asn Ile Thr Val Pro Asp Thr Lys Val Asn Phe 35 40 45 Tyr Ala Trp Lys Arg Met Glu Val Glu Glu Gln Ala Ile Glu Val Trp 50 55 60 Gln Gly Leu Ser Leu Leu Ser Glu Ala Ile Leu Gln Ala Gln Ala Leu 65 70 75 80 Leu Ala Asn Ser Ser Gln Pro Pro Glu Thr Leu Gln Leu His Ile Asp 85 90 95 Lys Ala Val Ser Gly Leu Arg Ser Leu Thr Ser Leu Leu Arg Val Leu 100 105 110 Gly Ala Gln Lys Glu Leu Met Ser Pro Pro Asp Thr Thr Pro Pro Ala 115 120 125 Pro Leu Arg Thr Leu Thr Val Asp Thr Phe Cys Lys Leu Phe Arg Val 130 135 140 Tyr Ala Asn Phe Leu Arg Gly Lys Leu Lys Leu Tyr Thr Gly Glu Val 145 150 155 160 Cys Arg Arg Gly Asp Arg 165 <210> 21 <211> 19 <212> PRT <213> Artificial Sequence <220> <223> synthetic construct <400> 21 Met Gly Asp Ser Lys Ile His His His His His His Glu Asn Leu Tyr 1 5 10 15 Phe Gln Gly <210> 22 <211> 301 <212> PRT <213> Homo sapiens <400> 22 Met Gly Ser Asp Lys Ile His His His His His His Glu Asn Leu Tyr 1 5 10 15 Phe Gln Gly Ser Leu Leu Val Asn Pro Glu Gly Pro Thr Leu Met Arg 20 25 30 Leu Asn Ser Val Gln Ser Ser Glu Arg Pro Leu Phe Leu Val His Pro 35 40 45 Ile Glu Gly Ser Thr Thr Val Phe His Ser Leu Ala Ser Arg Leu Ser 50 55 60 Ile Pro Thr Tyr Gly Leu Gln Cys Thr Arg Ala Ala Pro Leu Asp Ser 65 70 75 80 Ile His Ser Leu Ala Ala Tyr Tyr Ile Asp Cys Ile Arg Gln Val Gln 85 90 95 Pro Glu Gly Pro Tyr Arg Val Ala Gly Tyr Ser Tyr Gly Ala Cys Val 100 105 110 Ala Phe Glu Met Cys Ser Gln Leu Gln Ala Gln Gln Ser Pro Ala Pro 115 120 125 Thr His Asn Ser Leu Phe Leu Phe Asp Gly Ser Pro Thr Tyr Val Leu 130 135 140 Ala Tyr Thr Gln Ser Tyr Arg Ala Lys Leu Thr Pro Gly Ser Glu Ala 145 150 155 160 Glu Ala Glu Thr Glu Ala Ile Cys Phe Phe Val Gln Gln Phe Thr Asp 165 170 175 Met Glu His Asn Arg Val Leu Glu Ala Leu Leu Pro Leu Lys Gly Leu 180 185 190 Glu Glu Arg Val Ala Ala Ala Val Asp Leu Ile Ile Lys Ser His Gln 195 200 205 Gly Leu Asp Arg Gln Glu Leu Ser Phe Ala Ala Arg Ser Phe Tyr Tyr 210 215 220 Lys Leu Arg Ala Ala Glu Gln Tyr Thr Pro Lys Ala Lys Tyr His Gly 225 230 235 240 Asn Val Met Leu Leu Arg Ala Lys Thr Gly Gly Ala Tyr Gly Glu Asp 245 250 255 Leu Gly Ala Asp Tyr Asn Leu Ser Gln Val Cys Asp Gly Lys Val Ser 260 265 270 Val His Val Ile Glu Gly Asp His Arg Thr Leu Leu Glu Gly Ser Gly 275 280 285 Leu Glu Ser Ile Ile Ser Ile Ile His Ser Ser Leu Ala 290 295 300 <210> 23 <211> 18 <212> PRT <213> Artificial Sequence <220> <223> synthetic construct <400> 23 Met Gly Ser Ser His His His His His His Leu Glu Val Leu Phe Gln 1 5 10 15 Gly Pro <210> 24 <211> 125 <212> PRT <213> Homo sapiens <400> 24 Met Gly Ser Ser His His His His His His Leu Glu Val Leu Phe Gln 1 5 10 15 Gly Pro Gly Val Gln Val Glu Thr Ile Ser Pro Gly Asp Gly Arg Thr 20 25 30 Phe Pro Lys Arg Gly Gln Thr Cys Val Val His Tyr Thr Gly Met Leu 35 40 45 Glu Asp Gly Lys Lys Phe Asp Ser Ser Arg Asp Arg Asn Lys Pro Phe 50 55 60 Lys Phe Met Leu Gly Lys Gln Glu Val Ile Arg Gly Trp Glu Glu Gly 65 70 75 80 Val Ala Gln Met Ser Val Gly Gln Arg Ala Lys Leu Thr Ile Ser Pro 85 90 95 Asp Tyr Ala Tyr Gly Ala Thr Gly His Pro Gly Ile Ile Pro Pro His 100 105 110 Ala Thr Leu Val Phe Asp Val Glu Leu Leu Lys Leu Glu 115 120 125 <210> 25 <211> 196 <212> PRT <213> Homo sapiens <400> 25 Met Gly Ser Ser His His His His His His Ser Ser Gly Glu Asn Leu 1 5 10 15 Tyr Phe Gln Gly Asp Ser Ser Pro Leu Leu Gln Phe Gly Gly Val Arg 20 25 30 Gln Arg Tyr Leu Tyr Thr Asp Asp Ala Gln Gln Thr Glu Ala His Leu 35 40 45 Glu Ile Arg Glu Asp Gly Thr Val Gly Gly Ala Ala Asp Gln Ser Pro 50 55 60 Glu Ser Leu Leu Gln Leu Lys Ala Leu Lys Pro Gly Val Ile Gln Ile 65 70 75 80 Leu Gly Val Lys Thr Ser Arg Phe Leu Cys Gln Arg Pro Asp Gly Ala 85 90 95 Leu Tyr Gly Ser Leu His Phe Asp Pro Glu Ala Cys Ser Phe Arg Glu 100 105 110 Leu Leu Leu Glu Asp Gly Tyr Asn Val Tyr Gln Ser Glu Ala His Gly 115 120 125 Leu Pro Leu His Leu Pro Gly Asn Lys Ser Pro His Arg Asp Pro Ala 130 135 140 Pro Arg Gly Pro Ala Arg Phe Leu Pro Leu Pro Gly Leu Pro Pro Ala 145 150 155 160 Leu Pro Glu Pro Pro Gly Ile Leu Ala Pro Gln Pro Pro Asp Val Gly 165 170 175 Ser Ser Asp Pro Leu Ser Met Val Gly Pro Ser Gln Gly Arg Ser Pro 180 185 190 Ser Tyr Ala Ser 195 <210> 26 <211> 173 <212> PRT <213> mouse <400> 26 Met Arg Gly Ser His His His His His His Gly Ser Gly Ile Glu Gly 1 5 10 15 Arg Leu Arg Ser Ser Ser Gln Asn Ser Ser Asp Lys Pro Val Ala His 20 25 30 Val Val Ala Asn His Gln Val Glu Glu Gln Leu Trp Leu Ser Gln Arg 35 40 45 Ala Asn Ala Leu Leu Ala Asn Gly Met Asp Leu Lys Asp Asn Gln Leu 50 55 60 Val Val Pro Ala Asp Gly Leu Tyr Leu Val Tyr Ser Gln Val Leu Phe 65 70 75 80 Lys Gly Gln Gly Cys Pro Asp Tyr Val Leu Leu Thr His Thr Val Ser 85 90 95 Arg Phe Ala Ile Ser Tyr Gln Glu Lys Val Asn Leu Leu Ser Ala Val 100 105 110 Lys Ser Pro Cys Pro Lys Asp Thr Pro Glu Gly Ala Glu Leu Lys Pro 115 120 125 Trp Tyr Glu Pro Ile Tyr Leu Gly Gly Val Phe Gln Leu Glu Lys Gly 130 135 140 Asp Gln Leu Ser Ala Glu Val Asn Leu Pro Lys Tyr Leu Asp Phe Ala 145 150 155 160 Glu Ala Ser Gly Gln Val Tyr Phe Gly Val Ile Ala Leu 165 170 <210> 27 <211> 62 <212> PRT <213> mouse <400> 27 Met Asn Ser Tyr Pro Gly Cys Pro Ser Ser Tyr Asp Gly Tyr Cys Leu 1 5 10 15 Asn Gly Gly Val Cys Met His Ile Glu Ser Leu Asp Ser Tyr Thr Cys 20 25 30 Asn Cys Val Ile Gly Tyr Ser Gly Asp Arg Cys Gln Thr Arg Asp Leu 35 40 45 Arg Trp Trp Glu Leu Arg Leu Glu His His His His His His 50 55 60 <210> 28 <211> 14 <212> PRT <213> Artificial Sequence <220> <223> synthetic construct <220> <221> MOD_RES <222> (2)..(2) <223> D-alanine <220> <221> MOD_RES <222> (4)..(4) <223> cyclohexyl-alanine <220> <221> MOD_RES <222> (4)..(4) <223> cyclohexyl-alanine <220> <221> MOD_RES <222> (13)..(13) <223> D-alanine <400> 28 Gly Xaa Lys Xaa Val Ala Ala Trp Thr Leu Lys Ala Xaa Gly 1 5 10 <210> 29 <211> 19 <212> PRT <213> Artificial Sequence <220> <223> synthetic construct <220> <221> MOD_RES <222> (7)..(7) <223> D-alanine <220> <221> MOD_RES <222> (9)..(9) <223> cyclohexyl-alanine <220> <221> MOD_RES <222> (9)..(9) <223> cyclohexyl-alanine <220> <221> MOD_RES <222> (18)..(18) <223> D-alanine <400> 29 Ala Gly Ser Arg Ser Gly Xaa Lys Xaa Val Ala Ala Trp Thr Leu Lys 1 5 10 15 Ala Xaa Gly <210> 30 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> synthetic construct <220> <221> misc_feature <222> (1)..(20) <223> phosphothioate backbone <400> 30 tccatgacgt tcctgacgtt 20 <210> 31 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> synthetic construct <220> <221> misc_feature <222> (1)..(22) <223> phosphothioate backbone <400> 31 tcgtcgtttt cggcgcgcgc cg 22 <210> 32 <211> 22 <212> PRT <213> mouse <220> <221> MOD_RES <222> (11)..(11) <223> PCL <400> 32 Met Asn Ser Tyr Pro Gly Cys Pro Ser Ser Xaa Asp Gly Tyr Cys Leu 1 5 10 15 Asn Gly Gly Val Cys Met 20 <210> 33 <211> 22 <212> PRT <213> mouse <220> <221> MISC_FEATURE <222> (11)..(11) <223> pyrrolysine <400> 33 Met Asn Ser Tyr Pro Gly Cys Pro Ser Ser Xaa Asp Gly Tyr Cys Leu 1 5 10 15 Asn Gly Gly Val Cys Met 20 <210> 34 <211> 14 <212> PRT <213> mouse <220> <221> MISC_FEATURE <222> (3)..(3) <223> pyrrolysine <400> 34 Asn His Xaa Val Glu Glu Gln Leu Glu Trp Leu Ser Gln Arg 1 5 10 <210> 35 <211> 14 <212> PRT <213> mouse <220> <221> MOD_RES <222> (3)..(3) <223> PCL <400> 35 Asn His Xaa Val Glu Glu Gln Leu Glu Trp Leu Ser Gln Arg 1 5 10 SEQUENCE LISTING <110> IRM Geierstanger, Bernhard <120> BIOSYNTHETICALLY GENERATED PYRROLINE-CARBOXY-LYSINE AND SITE SPECIFIC PROTEIN MODIFICATIONS VIA CHEMICAL DERIVATIZATION OF PYRROLINE-CARBOXY-LYSINE AND PYRROLYSINE RESIDUES <130> PAT053160-WO <160> 35 <170> PatentIn version 3.5 <210> 1 <211> 1365 <212> DNA <213> Methanosarcina mazeii <400> 1 atggataaaa aaccactaaa cactctgata tctgcaaccg ggctctggat gtccaggacc 60 ggaacaattc ataaaataaa acaccacgaa gtctctcgaa gcaaaatcta tattgaaatg 120 gcatgcggag accaccttgt tgtaaacaac tccaggagca gcaggactgc aagagcgctc 180 aggcaccaca aatacaggaa gacctgcaaa cgctgcaggg tttcggatga ggatctcaat 240 aagttcctca caaaggcaaa cgaagaccag acaagcgtaa aagtcaaggt cgtttctgcc 300 cctaccagaa cgaaaaaggc aatgccaaaa tccgttgcga gagccccgaa acctcttgag 360 aatacagaag cggcacaggc tcaaccttct ggatctaaat tttcacctgc gataccggtt 420 tccacccaag agtcagtttc tgtcccggca tctgtttcaa catcaatatc aagcatttct 480 acaggagcaa ctgcatccgc actggtaaaa gggaatacga accccattac atccatgtct 540 gcccctgttc aggcaagtgc ccccgcactt acgaagagcc agactgacag gcttgaagtc 600 ctgttaaacc caaaagatga gatttccctg aattccggca agcctttcag ggagcttgag 660 tccgaattgc tctctcgcag aaaaaaagac ctgcagcaga tctacgcgga agaaagggag 720 aattatctgg ggaaactcga gcgtgaaatt accaggttct ttgtggacag gggttttctg 780 gaaataaaat ccccgatcct gatccctctt gagtatatcg aaaggatggg cattgataat 840 gataccgaac tttcaaaaca gatcttcagg gttgacaaga acttctgcct gagacccatg 900 cttgctccaa acctttacaa ctacctgcgc aagcttgaca gggccctgcc tgatccaata 960 aaaatttttg aaataggccc atgctacaga aaagagtccg acggcaaaga acacctcgaa 1020 gagtttacca tgctgaactt ctgccagatg ggatcgggat gcacacggga aaatcttgaa 1080 agcataatta cggacttcct gaaccacctg ggaattgatt tcaagatcgt aggcgattcc 1140 tgcatggtct atggggatac ccttgatgta atgcacggag acctggaact ttcctctgca 1200 gtagtcggac ccataccgct tgaccgggaa tggggtattg ataaaccctg gataggggca 1260 ggtttcgggc tcgaacgcct tctaaaggtt aaacacgact ttaaaaatat caagagagct 1320 gcaaggtccg agtcttacta taacgggatt tctaccaacc tgtaa 1365 <210> 2 <211> 1050 <212> DNA <213> Methanosarcina mazeii <400> 2 atgatccaga aaatggcaac cgaagaactt gacaggttcg gggagaaaat tattgaaggt 60 tttaaattgt ctgatgatga cctcagggct cttctttctc ttgaattcga agaagagctg 120 gaaaagcttt actatgtagc tagaaaggtc agaaactatt atttcggcaa cagggtgttt 180 cttaactgtt ttatttattt ctcaacttat tgtaaaaacc agtgctcttt ttgctactat 240 aactgtaaaa acgaaattaa ccgctaccgc ctgaccggtg aagaggttaa agagatgtgc 300 aaagccctga aaggtgcagg ctttcacatg atcgacctga caatgggaga ggatccctat 360 tactatgatg accctgaccg cttcgttgaa cttgtcagga cagtaaaaga agaactcggg 420 cttccaataa tgatttctcc gggagttatg gatgacagca ccctcctgaa agccagggaa 480 gaaggagcaa atttctttgc cctttatcag gagacttatg accgcgaact ttatggaaag 540 ctaagggtag gtcagtcctt cgaaggaagg tttaatgccc gcaggtttgc aaaagaacag 600 gggtactgta tagaagacgg cattcttacc ggcgtaggaa atgatatcga atcaactctt 660 atatccctga aggggatgaa agcaaacaat cctgatatgg taagggtaat gacttttctg 720 cctcaggaag gaactccgct tgaaggtttc agcgatagtt caaagctttc ggagctgaaa 780 atcatagcga ttctcaggct catgtttcct gaatgcctga taccggcttc tcttgacctt 840 gaaggcatag acggcatggt gcaccgttta aatgccggag caaatattgt aacctccatc 900 ctcccagatt cacgcctgga aggggttgcc aattacgacc gcggcatgga agagagggac 960 agggacgtta caagcgttgt caaaaggctg aaggttatgg gaatggaacc tgcgccgcag 1020 gctgagtttg agagagtcct ggggtgctaa 1050 <210> 3 <211> 1116 <212> DNA <213> Methanosarcina mazeii <400> 3 atgagagagt cctggggtgc tagcctgaaa acaatatgcc ttataggcgg gaagctgcag 60 ggcttcgagg ctgcatacct atctaagaaa gccggaatga aagtgcttgt aatagacaaa 120 aacccgcagg cgcttataag gaattatgcg gatgagttcc agtgttttaa cataacggaa 180 gagccggaaa aactcgtcgc gatatcaaaa aatgttgatg ccatactgcc ggtaaatgaa 240 aaccttgaat gtatagaatt tctgaattct ataaaagaaa aattctcctg cccggtactt 300 ttcgattttg aagcttacag gatcagcagg gataagagaa aatcaaaaga atacttcgca 360 tccataggaa ccccgacccc tcaggacaaa ccgtcggaac caccttattt tgtaaagcct 420 ccctgcgaaa gcagcagtgt gggagcgaga ataatccatg acaggaaaga gcttaaagag 480 cttgagcccg ggatgctcat agaagaatac gttgaagggg aagtggtctc acttgaggtc 540 ataggggatg gaaataattt tgctgtggta aaggaaaccc ttgtacatat cgatgacacc 600 tatgactgcc atatggtgac ccctctccct ctagaccctt ccttcaggga actatcctac 660 tcccttgcag caaacctgcc cttaaaagga attatggacg tggaagcgat ttccggcccc 720 ctggggttaa aagttattga gatagatgcc cgtttcccga gccagactcc gactgcggtc 780 tattattctt ccgggatcaa cctcatagaa ctcctgttcc gggcttttaa tggaggcata 840 gaagagatca aaactctccc tgaagacagg tactgcattt acgaacatct catgcttgca 900 gaaaatggag tacttatccc tgtgggagaa caggtcctgt ccatgggaaa tgattacggc 960 aattattatg aagaacctgg aatagagatt ttcctgtgca aaggagagaa ccctgtattc 1020 accctggttt tctggggcag agacagggaa gaagctgaag ctagaaaaaa caaagggctt 1080 tcgattctaa aaagccgttt cggagctgct gcataa 1116 <210> 4 <211> 795 <212> DNA <213> Methanosarcina mazeii <400> 4 atggcacttt taaccccaga agacctggaa aatattaaca aacagcttca agaagctgat 60 tctactgtcc gcagagttac agggcttgat ataaaaggta tctgtaaaga tttctacggc 120 acaactccat gctgtgaaaa agtaggtatc gtgcctgtga cctcagggaa cgggatcata 180 gggagctttt ccgaatccct gaatgcaatt gccgggtatt tcgggtttga cagttttatt 240 actgatatgc ctgacgtcag cggatattat gaggcagtaa agaacggagc ccggatcata 300 cttatggcag atgataatac cttccttgcc cacaacctga aaaatggaaa aatcgccaat 360 aaccagccgt gtacaggcat aatttatgct gaaatagctt caagatacct gaaagccgat 420 tccaaagaag tgcttgccgt gggtcttggg aaggttggat ttccgggagc agcccatctc 480 gtacagaaag gcttcaaggt ttacggatat gatgctgaca gaacccttct agaaaaaagc 540 gtttccagcc tcggaattat acctttcaat cccgtcagcc ccgaaggcga caggcaaagg 600 aagttttcca ttattttcga agcaaccccc tgtgcagaca cgattccgga atccgtaatt 660 tcggaaaact gtgtgatttc tacccctggg ataccctgtg caatctcaaa ggagctgcaa 720 aaaaagtgtg gagttgaact tgtaatggaa ccactgggga taggtacagc atcaatgctg 780 tattctgtac tctaa 795 <210> 5 <211> 72 <212> DNA <213> Methanosarcina mazeii <400> 5 ggaaacctga tcatgtagat cgaatggact ctaaatccgt tcagccgggt tagattcccg 60 gggtttccgc ca 72 <210> 6 <211> 223 <212> PRT <213> Homo sapiens <400> 6 Met Lys Thr Phe Ile Leu Leu Leu Trp Val Leu Leu Leu Trp Val Ile 1 5 10 15 Phe Leu Leu Pro Gly Ala Thr Ala Gln Pro Glu Arg Asp Cys Arg Val 20 25 30 Ser Ser Phe Arg Val Lys Glu Asn Phe Asp Lys Ala Arg Phe Ser Gly 35 40 45 Thr Trp Tyr Ala Met Ala Lys Lys Asp Pro Glu Gly Leu Phe Leu Gln 50 55 60 Asp Asn Ile Val Ala Glu Phe Ser Val Asp Glu Thr Gly Gln Met Ser 65 70 75 80 Ala Thr Ala Lys Gly Arg Val Arg Leu Leu Asn Asn Trp Asp Val Cys 85 90 95 Ala Asp Met Val Gly Thr Phe Thr Asp Thr Glu Asp Pro Ala Lys Phe 100 105 110 Lys Met Lys Tyr Trp Gly Val Ala Ser Phe Leu Gln Lys Gly Asn Asp 115 120 125 Asp His Trp Ile Val Asp Thr Asp Tyr Asp Thr Tyr Ala Val Gln Tyr 130 135 140 Ser Cys Arg Leu Leu Asn Leu Asp Gly Thr Cys Ala Asp Ser Tyr Ser 145 150 155 160 Phe Val Phe Ser Arg Asp Pro Asn Gly Leu Pro Pro Glu Ala Gln Lys 165 170 175 Ile Val Arg Gln Arg Gln Glu Glu Leu Cys Leu Ala Arg Gln Tyr Arg 180 185 190 Leu Ile Val His Asn Gly Tyr Cys Asp Gly Arg Ser Glu Arg Asn Leu 195 200 205 Leu Asp Tyr Lys Asp Asp Asp Asp Lys His His His His His His 210 215 220 <210> 7 <211> 672 <212> DNA <213> Homo sapiens <400> 7 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 8 <211> 672 <212> DNA <213> Homo sapiens <400> 8 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg taggccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 9 <211> 672 <212> DNA <213> Homo sapiens <400> 9 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctagctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 10 <211> 672 <212> DNA <213> Homo sapiens <400> 10 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactagg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 11 <211> 672 <212> DNA <213> Homo sapiens <400> 11 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtagtg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 12 <211> 672 <212> DNA <213> Homo sapiens <400> 12 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtacta gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 13 <211> 672 <212> DNA <213> Homo sapiens <400> 13 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctagctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 14 <211> 672 <212> DNA <213> Homo sapiens <400> 14 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtag 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 15 <211> 672 <212> DNA <213> Homo sapiens <400> 15 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tagaggctga tcgtccacaa cggttactgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 16 <211> 672 <212> DNA <213> Homo sapiens <400> 16 atgaaaacat tcatactcct gctctgggta ctgctgctct gggttatctt cctgcttccc 60 ggtgccactg ctcagcctga gcgcgactgc cgagtgagca gcttccgagt caaggagaac 120 ttcgacaagg ctcgcttctc tgggacctgg tacgccatgg ccaagaagga ccccgagggc 180 ctctttctgc aggacaacat cgtcgcggag ttctccgtgg acgagaccgg ccagatgagc 240 gccacagcca agggccgagt ccgtcttttg aataactggg acgtgtgcgc agacatggtg 300 ggcaccttca cagacaccga ggaccctgcc aagttcaaga tgaagtactg gggcgtagcc 360 tcctttctcc agaaaggaaa tgatgaccac tggatcgtcg acacagacta cgacacgtat 420 gccgtgcagt actcctgccg cctcctgaac ctcgatggca cctgtgctga cagctactcc 480 ttcgtgtttt cccgggaccc caacggcctg cccccagaag cgcagaagat tgtaaggcag 540 cggcaggagg agctgtgcct ggccaggcag tacaggctga tcgtccacaa cggttagtgc 600 gatggcagat cagaaagaaa ccttttggac tataaagacg atgacgataa gcatcaccat 660 caccatcact aa 672 <210> 17 <211> 10 <212> PRT <213> Homo sapiens <220> <221> MOD_RES <222> (7)..(7) <223> Xaa is a PCL residue <400> 17 Tyr Trp Gly Val Ala Ser Xaa Leu Gln Lys 1 5 10 <210> 18 <211> 29 <212> PRT <213> Homo sapiens <220> <221> MOD_RES <222> (7)..(7) <223> Xaa is Cyc residue <400> 18 Lys Asp Pro Glu Gly Leu Xaa Leu Gln Asp Asn Ile Val Ala Glu Phe 1 5 10 15 Ser Val Asp Glu Thr Gly Gln Met Ser Ala Thr Ala Lys 20 25 <210> 19 <211> 166 <212> PRT <213> Homo sapiens <400> 19 Ala Pro Pro Arg Leu Ile Cys Asp Ser Arg Val Leu Glu Arg Tyr Leu 1 5 10 15 Leu Glu Ala Lys Glu Ala Glu Asn Ile Thr Thr Gly Cys Ala Glu His 20 25 30 Cys Ser Leu Asn Glu Asn Ile Thr Val Pro Asp Thr Lys Val Asn Phe 35 40 45 Tyr Ala Trp Lys Arg Met Glu Val Gly Gln Gln Ala Val Glu Val Trp 50 55 60 Gln Gly Leu Ala Leu Leu Ser Glu Ala Val Leu Arg Gly Gln Ala Leu 65 70 75 80 Leu Val Asn Ser Ser Gln Pro Trp Glu Pro Leu Gln Leu His Val Asp 85 90 95 Lys Ala Val Ser Gly Leu Arg Ser Leu Thr Thr Leu Leu Arg Ala Leu 100 105 110 Gly Ala Gln Lys Glu Ala Ile Ser Pro Pro Asp Ala Ala Ser Ala Ala 115 120 125 Pro Leu Arg Thr Ile Thr Ala Asp Thr Phe Arg Lys Leu Phe Arg Val 130 135 140 Tyr Ser Asn Phe Leu Arg Gly Lys Leu Lys Leu Tyr Thr Gly Glu Ala 145 150 155 160 Cys Arg Thr Gly Asp Arg 165 <210> 20 <211> 166 <212> PRT <213> mouse <400> 20 Ala Pro Pro Arg Leu Ile Cys Asp Ser Arg Val Leu Glu Arg Tyr Leu 1 5 10 15 Leu Glu Ala Lys Glu Ala Glu Asn Val Thr Met Gly Cys Ala Glu Gly 20 25 30 Pro Arg Leu Ser Glu Asn Ile Thr Val Pro Asp Thr Lys Val Asn Phe 35 40 45 Tyr Ala Trp Lys Arg Met Glu Val Glu Glu Gln Ala Ile Glu Val Trp 50 55 60 Gln Gly Leu Ser Leu Leu Ser Glu Ala Ile Leu Gln Ala Gln Ala Leu 65 70 75 80 Leu Ala Asn Ser Ser Gln Pro Pro Glu Thr Leu Gln Leu His Ile Asp 85 90 95 Lys Ala Val Ser Gly Leu Arg Ser Leu Thr Ser Leu Leu Arg Val Leu 100 105 110 Gly Ala Gln Lys Glu Leu Met Ser Pro Pro Asp Thr Thr Pro Pro Ala 115 120 125 Pro Leu Arg Thr Leu Thr Val Asp Thr Phe Cys Lys Leu Phe Arg Val 130 135 140 Tyr Ala Asn Phe Leu Arg Gly Lys Leu Lys Leu Tyr Thr Gly Glu Val 145 150 155 160 Cys Arg Arg Gly Asp Arg 165 <210> 21 <211> 19 <212> PRT <213> Artificial Sequence <220> <223> synthetic construct <400> 21 Met Gly Asp Ser Lys Ile His His His His His His Glu Asn Leu Tyr 1 5 10 15 Phe Gln Gly <210> 22 <211> 301 <212> PRT <213> Homo sapiens <400> 22 Met Gly Ser Asp Lys Ile His His His His His His Glu Asn Leu Tyr 1 5 10 15 Phe Gln Gly Ser Leu Leu Val Asn Pro Glu Gly Pro Thr Leu Met Arg 20 25 30 Leu Asn Ser Val Gln Ser Ser Glu Arg Pro Leu Phe Leu Val His Pro 35 40 45 Ile Glu Gly Ser Thr Thr Val Phe His Ser Leu Ala Ser Arg Leu Ser 50 55 60 Ile Pro Thr Tyr Gly Leu Gln Cys Thr Arg Ala Ala Pro Leu Asp Ser 65 70 75 80 Ile His Ser Leu Ala Ala Tyr Tyr Ile Asp Cys Ile Arg Gln Val Gln 85 90 95 Pro Glu Gly Pro Tyr Arg Val Ala Gly Tyr Ser Tyr Gly Ala Cys Val 100 105 110 Ala Phe Glu Met Cys Ser Gln Leu Gln Ala Gln Gln Ser Pro Ala Pro 115 120 125 Thr His Asn Ser Leu Phe Leu Phe Asp Gly Ser Pro Thr Tyr Val Leu 130 135 140 Ala Tyr Thr Gln Ser Tyr Arg Ala Lys Leu Thr Pro Gly Ser Glu Ala 145 150 155 160 Glu Ala Glu Thr Glu Ala Ile Cys Phe Phe Val Gln Gln Phe Thr Asp 165 170 175 Met Glu His Asn Arg Val Leu Glu Ala Leu Leu Pro Leu Lys Gly Leu 180 185 190 Glu Glu Arg Val Ala Ala Ala Val Asp Leu Ile Ile Lys Ser His Gln 195 200 205 Gly Leu Asp Arg Gln Glu Leu Ser Phe Ala Ala Arg Ser Phe Tyr Tyr 210 215 220 Lys Leu Arg Ala Ala Glu Gln Tyr Thr Pro Lys Ala Lys Tyr His Gly 225 230 235 240 Asn Val Met Leu Leu Arg Ala Lys Thr Gly Gly Ala Tyr Gly Glu Asp 245 250 255 Leu Gly Ala Asp Tyr Asn Leu Ser Gln Val Cys Asp Gly Lys Val Ser 260 265 270 Val His Val Ile Glu Gly Asp His Arg Thr Leu Leu Glu Gly Ser Gly 275 280 285 Leu Glu Ser Ile Ile Ser Ile Ile His Ser Ser Leu Ala 290 295 300 <210> 23 <211> 18 <212> PRT <213> Artificial Sequence <220> <223> synthetic construct <400> 23 Met Gly Ser Ser His His His His His His Leu Glu Val Leu Phe Gln 1 5 10 15 Gly Pro <210> 24 <211> 125 <212> PRT <213> Homo sapiens <400> 24 Met Gly Ser Ser His His His His His His Leu Glu Val Leu Phe Gln 1 5 10 15 Gly Pro Gly Val Gln Val Glu Thr Ile Ser Pro Gly Asp Gly Arg Thr 20 25 30 Phe Pro Lys Arg Gly Gln Thr Cys Val Val His Tyr Thr Gly Met Leu 35 40 45 Glu Asp Gly Lys Lys Phe Asp Ser Ser Arg Asp Arg Asn Lys Pro Phe 50 55 60 Lys Phe Met Leu Gly Lys Gln Glu Val Ile Arg Gly Trp Glu Glu Gly 65 70 75 80 Val Ala Gln Met Ser Val Gly Gln Arg Ala Lys Leu Thr Ile Ser Pro 85 90 95 Asp Tyr Ala Tyr Gly Ala Thr Gly His Pro Gly Ile Ile Pro Pro His 100 105 110 Ala Thr Leu Val Phe Asp Val Glu Leu Leu Lys Leu Glu 115 120 125 <210> 25 <211> 196 <212> PRT <213> Homo sapiens <400> 25 Met Gly Ser Ser His His His His His His Ser Ser Gly Glu Asn Leu 1 5 10 15 Tyr Phe Gln Gly Asp Ser Ser Pro Leu Leu Gln Phe Gly Gly Val Arg 20 25 30 Gln Arg Tyr Leu Tyr Thr Asp Asp Ala Gln Gln Thr Glu Ala His Leu 35 40 45 Glu Ile Arg Glu Asp Gly Thr Val Gly Gly Ala Ala Asp Gln Ser Pro 50 55 60 Glu Ser Leu Leu Gln Leu Lys Ala Leu Lys Pro Gly Val Ile Gln Ile 65 70 75 80 Leu Gly Val Lys Thr Ser Arg Phe Leu Cys Gln Arg Pro Asp Gly Ala 85 90 95 Leu Tyr Gly Ser Leu His Phe Asp Pro Glu Ala Cys Ser Phe Arg Glu 100 105 110 Leu Leu Leu Glu Asp Gly Tyr Asn Val Tyr Gln Ser Glu Ala His Gly 115 120 125 Leu Pro Leu His Leu Pro Gly Asn Lys Ser Pro His Arg Asp Pro Ala 130 135 140 Pro Arg Gly Pro Ala Arg Phe Leu Pro Leu Pro Gly Leu Pro Pro Ala 145 150 155 160 Leu Pro Glu Pro Pro Gly Ile Leu Ala Pro Gln Pro Pro Asp Val Gly 165 170 175 Ser Ser Asp Pro Leu Ser Met Val Gly Pro Ser Gln Gly Arg Ser Pro 180 185 190 Ser Tyr Ala Ser 195 <210> 26 <211> 173 <212> PRT <213> mouse <400> 26 Met Arg Gly Ser His His His His His His His Gly Ser Gly Ile Glu Gly 1 5 10 15 Arg Leu Arg Ser Ser Ser Gln Asn Ser Ser Asp Lys Pro Val Ala His 20 25 30 Val Val Ala Asn His Gln Val Glu Glu Gln Leu Trp Leu Ser Gln Arg 35 40 45 Ala Asn Ala Leu Leu Ala Asn Gly Met Asp Leu Lys Asp Asn Gln Leu 50 55 60 Val Val Pro Ala Asp Gly Leu Tyr Leu Val Tyr Ser Gln Val Leu Phe 65 70 75 80 Lys Gly Gln Gly Cys Pro Asp Tyr Val Leu Leu Thr His Thr Val Ser 85 90 95 Arg Phe Ala Ile Ser Tyr Gln Glu Lys Val Asn Leu Leu Ser Ala Val 100 105 110 Lys Ser Pro Cys Pro Lys Asp Thr Pro Glu Gly Ala Glu Leu Lys Pro 115 120 125 Trp Tyr Glu Pro Ile Tyr Leu Gly Gly Val Phe Gln Leu Glu Lys Gly 130 135 140 Asp Gln Leu Ser Ala Glu Val Asn Leu Pro Lys Tyr Leu Asp Phe Ala 145 150 155 160 Glu Ala Ser Gly Gln Val Tyr Phe Gly Val Ile Ala Leu 165 170 <210> 27 <211> 62 <212> PRT <213> mouse <400> 27 Met Asn Ser Tyr Pro Gly Cys Pro Ser Ser Tyr Asp Gly Tyr Cys Leu 1 5 10 15 Asn Gly Gly Val Cys Met His Ile Glu Ser Leu Asp Ser Tyr Thr Cys 20 25 30 Asn Cys Val Ile Gly Tyr Ser Gly Asp Arg Cys Gln Thr Arg Asp Leu 35 40 45 Arg Trp Trp Glu Leu Arg Leu Glu His His His His His His 50 55 60 <210> 28 <211> 14 <212> PRT <213> Artificial Sequence <220> <223> synthetic construct <220> <221> MOD_RES <222> (2)..(2) <223> D-alanine <220> <221> MOD_RES <222> (4)..(4) <223> cyclohexyl-alanine <220> <221> MOD_RES <222> (4)..(4) <223> cyclohexyl-alanine <220> <221> MOD_RES <222> (13)..(13) <223> D-alanine <400> 28 Gly Xaa Lys Xaa Val Ala Ala Trp Thr Leu Lys Ala Xaa Gly 1 5 10 <210> 29 <211> 19 <212> PRT <213> Artificial Sequence <220> <223> synthetic construct <220> <221> MOD_RES <222> (7)..(7) <223> D-alanine <220> <221> MOD_RES <222> (9)..(9) <223> cyclohexyl-alanine <220> <221> MOD_RES <222> (9)..(9) <223> cyclohexyl-alanine <220> <221> MOD_RES <222> (18)..(18) <223> D-alanine <400> 29 Ala Gly Ser Arg Ser Gly Xaa Lys Xaa Val Ala Ala Trp Thr Leu Lys 1 5 10 15 Ala Xaa Gly <210> 30 <211> 20 <212> DNA <213> Artificial Sequence <220> <223> synthetic construct <220> <221> misc_feature <222> (1)..(20) <223> phosphothioate backbone <400> 30 tccatgacgt tcctgacgtt 20 <210> 31 <211> 22 <212> DNA <213> Artificial Sequence <220> <223> synthetic construct <220> <221> misc_feature <222> (1)..(22) <223> phosphothioate backbone <400> 31 tcgtcgtttt cggcgcgcgc cg 22 <210> 32 <211> 22 <212> PRT <213> mouse <220> <221> MOD_RES <222> (11)..(11) <223> PCL <400> 32 Met Asn Ser Tyr Pro Gly Cys Pro Ser Ser Xaa Asp Gly Tyr Cys Leu 1 5 10 15 Asn Gly Gly Val Cys Met 20 <210> 33 <211> 22 <212> PRT <213> mouse <220> <221> MISC_FEATURE <222> (11)..(11) <223> pyrrolysine <400> 33 Met Asn Ser Tyr Pro Gly Cys Pro Ser Ser Xaa Asp Gly Tyr Cys Leu 1 5 10 15 Asn Gly Gly Val Cys Met 20 <210> 34 <211> 14 <212> PRT <213> mouse <220> <221> MISC_FEATURE <222> (3)..(3) <223> pyrrolysine <400> 34 Asn His Xaa Val Glu Glu Gln Leu Glu Trp Leu Ser Gln Arg 1 5 10 <210> 35 <211> 14 <212> PRT <213> mouse <220> <221> MOD_RES <222> (3)..(3) <223> PCL <400> 35 Asn His Xaa Val Glu Glu Gln Leu Glu Trp Leu Ser Gln Arg 1 5 10
Claims (12)
<화학식 I>
(상기 식에서,
R1은 H 또는 아미노 말단 변형 기이고;
R2는 OH 또는 카르복시 말단 변형 기이고;
n은 1 내지 5000의 정수이고;
각각의 AA는 아미노산 잔기, 피롤리신 아미노산 잔기, 하기 화학식 A-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 및 하기 화학식 B-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기로부터 독립적으로 선택되고;
(여기서, R6은 H 또는 C1알킬임)
1개 이상의 AA는 화학식 A-1 또는 화학식 B-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기이고, 화학식 A-1의 아미노산 잔기는 하기 화학식 V의 아미노산의 잔기이고, 화학식 B-1의 아미노산 잔기는 하기 화학식 VI의 아미노산의 잔기이며,
(여기서, R6은 H 또는 C1알킬임)
화학식 V 또는 화학식 VI의 아미노산은, pylC 유전자 및 pylD 유전자를 포함하고 전구체를 포함하는 성장 배지와 접촉하고 있는 세포 내에서 생합성적으로 생성됨)≪ / RTI >
(I)
(Wherein,
R 1 is H or an amino terminal modification group;
R 2 is OH or a carboxy terminus modification group;
n is an integer from 1 to 5000;
Each AA is independently selected from an amino acid residue, a pyrrolysine amino acid residue, a pyrrolysine analog amino acid residue having the structure of Formula A-1, and a pyrrolysine analog amino acid residue having the structure of Formula B-1, ;
Wherein R 6 is H or C 1 alkyl
At least one AA is a pyrrolysine analog amino acid residue having the structure of Formula A-1 or Formula B-1, the amino acid residue of Formula A-1 is a residue of the amino acid of Formula V, the amino acid residue of Formula B-1 Is a residue of an amino acid of formula
Wherein R 6 is H or C 1 alkyl
Amino acids of formula (V) or formula (VI) are biosynthetically produced in cells in contact with a growth medium comprising a pylC gene and a pylD gene and comprising a precursor)
The compound of claim 1, wherein R 1 is H, the amino acid of formula V is an amino acid having the structure of formula VII, the amino acid of formula VI is an amino acid having the structure of formula VIII, and the precursor is D-ornithine, D Arginine, (2S) -2-amino-6- (2,5-diaminopentaneamido) hexanoic acid or (2S) -2-amino-6-((R) -2,5-diaminopentaneami Fig. 8) Hexanoic acid compound.
화학식 VI의 아미노산이 하기 화학식 X의 구조를 갖는 아미노산이고, 전구체가 D-오르니틴, D-아르기닌, 2,5-디아미노-3-메틸펜탄산, (2R,3S)-2,5-디아미노-3-메틸펜탄산, (2R,3R)-2,5-디아미노-3-메틸펜탄산, (2S)-2-아미노-6-(2,5-디아미노펜탄아미도)헥산산 또는 (2S)-2-아미노-6-((R)-2,5-디아미노펜탄아미도)헥산산인 화합물.
The compound of claim 1, wherein R 1 is C 1 alkyl, the amino acid of formula V is an amino acid having the structure of formula IX, and the precursor is D-ornithine, D-arginine, (2S) -2-amino-6- (2,5-diaminopentaneamido) hexanoic acid or (2S) -2-amino-6-((R) -2,5-diaminopentaneamido) hexanoic acid;
Amino acid of formula VI is an amino acid having a structure of formula X, precursor is D-ornithine, D-arginine, 2,5-diamino-3-methylpentanoic acid, (2R, 3S) -2,5-dia Mino-3-methylpentanoic acid, (2R, 3R) -2,5-diamino-3-methylpentanoic acid, (2S) -2-amino-6- (2,5-diaminopentaneamido) hexanoic acid Or (2S) -2-amino-6-((R) -2,5-diaminopentaneamido) hexanoic acid.
<화학식 I>
(상기 식에서,
R1은 H 또는 아미노 말단 변형 기이고;
R2는 OH 또는 카르복시 말단 변형 기이고;
n은 1 내지 5000의 정수이고;
각각의 AA는 아미노산 잔기, 피롤리신 아미노산 잔기, 하기 화학식 A-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기, 및 하기 화학식 B-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기로부터 독립적으로 선택되고;
R6은 H 또는 C1알킬이고;
1개 이상의 AA는 피롤리신 아미노산 잔기, 또는 화학식 A-1 또는 화학식 B-1의 구조를 갖는 피롤리신 유사체 아미노산 잔기임)
<화학식 III>
<화학식 IV>
(상기 식에서,
R3, R5 및 각각의 R4는 H, -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택되고;
A는 C3-C8시클로알킬, C3-C8헤테로시클로알킬, 5 내지 6원 모노시클릭 아릴, 5 내지 6원 모노시클릭 헤테로아릴, 융합된 9 내지 10원 비시클릭 고리 또는 융합된 13 내지 14원 트리시클릭 고리이고, 여기서 A는 -OH, -NO2, 할로, C1 - 8알킬, 할로-치환된-C1 - 8알킬, 히드록시-치환된-C1 - 8알킬, 아릴, 헤테로아릴, 헤테로시클로알킬 또는 시클로알킬, 및 -LX1로부터 독립적으로 선택된 1 내지 5개의 치환기로 임의로 치환되고;
L은 결합, C1 - 8알킬렌, 할로-치환된-C1 - 8알킬렌, 히드록시-치환된-C1 - 8알킬렌, C2-8알케닐렌, 할로-치환된-C2 - 8알케닐렌, 히드록시-치환된-C2 - 8알케닐렌,
-O(CR11R12)k-, -S(CR11R12)k-, -S(O)k(CR11R12)k-, -O(CR11R12)k-NR11C(O)-, -O(CR11R12)kC(O)NR11-, -C(O)-, -C(O)(CR11R12)k-, -C(S)-, -C(S)(CR11R12)k-, -C(O)NR11-, -NR11C(O)-, -NR11(CR11R12)k-, -CONR11(CR11R12)k-, -N(R11)CO(CR11R12)k-, -C(O)NR11(CR11R12)k-, -NR11C(O)(CR11R12)k-로부터 선택되고, 여기서 각각의 R11 및 R12는 독립적으로 H, C1 - 8알킬, 할로-치환된-C1 - 8알킬 또는 히드록시-치환된-C1 - 8알킬이고, k는 1 내지 12의 정수이고;
X1은 표지, 염료, 중합체, 수용성 중합체, 폴리알킬렌 글리콜, 폴리(에틸렌 글리콜), 폴리(에틸렌 글리콜)의 유도체, 당, 지질, 광가교제, 세포독성 화합물, 약물, 친화성 표지, 광친화성 표지, 반응성 화합물; 수지, 펩티드, 제2 단백질 또는 폴리펩티드 또는 폴리펩티드 유사체, 항체 또는 항체 단편, 금속 킬레이트화제, 보조인자, 지방산, 탄수화물, 폴리뉴클레오티드, DNA, RNA, PCR 프로브, 안티센스 폴리뉴클레오티드, 리보-올리고뉴클레오티드, 데옥시리보-올리고뉴클레오티드, 포스포로티오에이트-변형 DNA, 변형 DNA 및 RNA, 펩티드 핵산, 사카라이드, 디사카라이드, 올리고사카라이드, 폴리사카라이드, 수용성 덴드리머, 시클로덱스트린, 생체물질, 나노입자, 스핀 표지, 형광단, 금속-함유 잔기, 방사성 잔기, 신규 관능기, 다른 분자와 공유 또는 비공유적으로 상호작용하는 기, 광케이징 잔기, 화학 방사선 여기성 잔기, 리간드, 광이성질체화 잔기, 비오틴, 비오틴 유사체, 중원자가 혼입된 잔기, 화학 절단성 기, 광절단성 기, 연장 측쇄, 탄소-연결된 당, 산화환원-활성제, 아미노 티오산, 독성 잔기, 동위원소 표지된 잔기, 생물물리학적 프로브, 인광 기, 발색단 기, 화학발광 기, 형광 잔기, 전자 고밀도 기, 자성 기, 삽입성 기, 킬레이트화 기, 발색단, 에너지 전달 작용제, 생물학적 활성제, 검출가능한 표지, 소분자, 억제성 리보핵산, siRNA, 방사성뉴클레오티드, 중성자-포획제, 비오틴의 유도체, 양자점(들), 나노전달물질, 방사선전달물질, 아브자임, 효소, 활성화된 복합 활성자, 바이러스, 독소, 아주반트, TLR2 효능제, TLR4 효능제, TLR7 효능제, TLR9 효능제, TLR8 효능제, T-세포 에피토프, 인지질, LPS-유사 분자, 키홀 림펫 헤모시아닌 (KLH), 면역원성 합텐, 아글리칸, 알레르겐, 안지오스타틴, 항호르몬, 항산화제, 압타머, 가이드 RNA, 사포닌, 셔틀 벡터, 거대분자, 미모토프, 수용체, 역미셀, 세정제, 면역 반응 강화제, 형광 염료, FRET 시약, 방사선-영상화 프로브, 다른 분광분석법 프로브, 전구약물, 면역요법용 독소, 고체 지지체, -CH2CH2-(OCH2CH2O)p-OX2, -O-(CH2CH2O)pCH2CH2-X2 및 이들의 임의의 조합물로부터 선택되고, 여기서 p는 1 내지 10,000이고, X2는 H, C1 - 8알킬, 보호기 또는 말단 관능기임).A method of derivatizing a protein comprising contacting a protein having a structure according to Formula I with a reagent of Formula III or Formula IV below.
(I)
(Wherein,
R 1 is H or an amino terminal modification group;
R 2 is OH or a carboxy terminus modification group;
n is an integer from 1 to 5000;
Each AA is independently selected from an amino acid residue, a pyrrolysine amino acid residue, a pyrrolysine analog amino acid residue having the structure of Formula A-1, and a pyrrolysine analog amino acid residue having the structure of Formula B-1, ;
R 6 is H or C 1 alkyl;
At least one AA is a pyrrolysine amino acid residue or a pyrrolysine analog amino acid residue having a structure of Formula A-1 or Formula B-1)
(III)
(IV)
(Wherein,
R 3, R 5 and each R 4 is H, -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, , Aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1 ;
A is C 3 -C 8 cycloalkyl, C 3 -C 8 heterocycloalkyl, 5 to 6 membered monocyclic aryl, 5 to 6 membered monocyclic heteroaryl, fused 9 to 10 membered bicyclic ring or fused 13 a to 14-membered tricyclic ring, where A is -OH, -NO 2, halo, C 1 - 8 alkyl, halo-substituted -C 1 - 8 alkyl, hydroxy-substituted -C 1 - 8 alkyl, Optionally substituted with 1 to 5 substituents independently selected from aryl, heteroaryl, heterocycloalkyl or cycloalkyl, and -LX 1 ;
L is a bond, C 1 - 8 alkylene, halo-substituted -C 1 - 8 alkylene, hydroxy-substituted -C 1 - 8 alkylene, C 2-8 alkenylene group, a halo-substituted -C 2 - 8 alkenylene, hydroxy-substituted 2 -C 8 alkenylene,
-O (CR 11 R 12 ) k- , -S (CR 11 R 12 ) k- , -S (O) k (CR 11 R 12 ) k- , -O (CR 11 R 12 ) k -NR 11 C (O)-, -O (CR 11 R 12 ) k C (O) NR 11- , -C (O)-, -C (O) (CR 11 R 12 ) k- , -C (S)-, -C (S) (CR 11 R 12 ) k- , -C (O) NR 11- , -NR 11 C (O)-, -NR 11 (CR 11 R 12 ) k- , -CONR 11 (CR 11 R 12 ) k- , -N (R 11 ) CO (CR 11 R 12 ) k- , -C (O) NR 11 (CR 11 R 12 ) k- , -NR 11 C (O) (CR 11 R 12 ) k - is selected from wherein each R 11 and R 12 are independently H, C 1 - 8 alkyl, and, - 8 alkyl, halo-substituted -C 1 - 8 alkyl or hydroxy-substituted -C 1 k is an integer from 1 to 12;
X 1 is a label, dye, polymer, water soluble polymer, polyalkylene glycol, poly (ethylene glycol), derivative of poly (ethylene glycol), sugar, lipid, photocrosslinker, cytotoxic compound, drug, affinity label, photo affinity Labels, reactive compounds; Resins, peptides, second proteins or polypeptides or polypeptide analogs, antibodies or antibody fragments, metal chelating agents, cofactors, fatty acids, carbohydrates, polynucleotides, DNA, RNA, PCR probes, antisense polynucleotides, ribo-oligonucleotides, deoxy Ribo-oligonucleotides, phosphorothioate-modified DNA, modified DNA and RNA, peptide nucleic acids, saccharides, disaccharides, oligosaccharides, polysaccharides, water soluble dendrimers, cyclodextrins, biomaterials, nanoparticles, spin labels , Fluorophores, metal-containing moieties, radioactive moieties, novel functional groups, groups that covalently or noncovalently interact with other molecules, photocatalytic moieties, chemiradioactive excitation moieties, ligands, photoisomerization moieties, biotin, biotin analogues Residues containing heavy atoms, chemically cleavable groups, photocleavable groups, extended side chains, carbon-linked sugars, redox Active agents, aminothio acids, toxic moieties, isotopically labeled moieties, biophysical probes, phosphorescent groups, chromophore groups, chemiluminescent groups, fluorescent moieties, electron dense groups, magnetic groups, intercalating groups, chelating groups, chromophores , Energy transfer agents, biologically active agents, detectable labels, small molecules, inhibitory ribonucleic acids, siRNAs, radionucleotides, neutron-trapping agents, derivatives of biotin, quantum dot (s), nanotransmitters, radiotransmitters, abzymes, enzymes , Activated complex activators, viruses, toxins, adjuvants, TLR2 agonists, TLR4 agonists, TLR7 agonists, TLR9 agonists, TLR8 agonists, T-cell epitopes, phospholipids, LPS-like molecules, keyhole limpet hemoshi Non- (KLH), immunogenic hapten, aglycan, allergen, angiostatin, anti-hormone, antioxidant, aptamer, guide RNA, saponin, shuttle vector, macromolecule, mimotope, receptor, reverse micelle, detergent, immune booster , brother Dye, FRET reagent, radiation-imaging probe, different probe spectroscopy, prodrugs, toxins for immunotherapy, a solid support, -CH 2 CH 2 - (OCH 2 CH 2 O) p -OX 2, -O- (CH 2 CH 2 O) p CH 2 CH 2 -X 2 , and are selected from any combination thereof, wherein p is 1 to 10,000, X 2 is H, C 1 - 8 alkyl, or a protecting group being the terminal functional group).
인 방법.
(상기 식에서, L 및 X1은 제8항에 기재된 바와 동일함)The method of claim 8, wherein the reagent of Formula IV is
/ RTI >
(Wherein L and X 1 are the same as described in claim 8)
(여기서, exPADRE는 서열 28에 따른 펩티드이고; PADRE는 서열 29에 따른 펩티드이고; BG1은 서열 30에 따른 폴리뉴클레오티드이고; BG2는 서열 31에 따른 폴리뉴클레오티드임)로부터 선택되는 것인 방법.The method of claim 8, wherein the reagent of Formula IV is
Wherein exPADRE is a peptide according to SEQ ID NO: 28; PADRE is a peptide according to SEQ ID NO: 29; BG1 is a polynucleotide according to SEQ ID NO: 30; BG2 is a polynucleotide according to SEQ ID NO: 31.
Applications Claiming Priority (3)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US10843408P | 2008-10-24 | 2008-10-24 | |
| US61/108,434 | 2008-10-24 | ||
| PCT/US2009/061954 WO2010048582A1 (en) | 2008-10-24 | 2009-10-23 | Biosynthetically generated pyrroline-carboxy-lysine and site specific protein modifications via chemical derivatization of pyrroline-carboxy-lysine and pyrrolysine residues |
Related Parent Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| KR1020117011641A Division KR101316159B1 (en) | 2008-10-24 | 2009-10-23 | Biosynthetically generated pyrroline-carboxy-lysine and site specific protein modifications via chemical derivatization of pyrroline-carboxy-lysine and pyrrolysine residues |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| KR20130086386A true KR20130086386A (en) | 2013-08-01 |
Family
ID=41479140
Family Applications (2)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| KR1020137017122A KR20130086386A (en) | 2008-10-24 | 2009-10-23 | Biosynthetically generated pyrroline-carboxy-lysine and site specific protein modifications via chemical derivatization of pyrroline-carboxy-lysine and pyrrolysine residues |
| KR1020117011641A KR101316159B1 (en) | 2008-10-24 | 2009-10-23 | Biosynthetically generated pyrroline-carboxy-lysine and site specific protein modifications via chemical derivatization of pyrroline-carboxy-lysine and pyrrolysine residues |
Family Applications After (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| KR1020117011641A KR101316159B1 (en) | 2008-10-24 | 2009-10-23 | Biosynthetically generated pyrroline-carboxy-lysine and site specific protein modifications via chemical derivatization of pyrroline-carboxy-lysine and pyrrolysine residues |
Country Status (12)
| Country | Link |
|---|---|
| US (2) | US8785151B2 (en) |
| EP (1) | EP2356092B1 (en) |
| JP (1) | JP5542832B2 (en) |
| KR (2) | KR20130086386A (en) |
| CN (1) | CN102264697B (en) |
| AU (1) | AU2009308163B2 (en) |
| BR (1) | BRPI0919932A8 (en) |
| CA (2) | CA2871855C (en) |
| EA (1) | EA021001B1 (en) |
| ES (1) | ES2477543T3 (en) |
| MX (1) | MX342677B (en) |
| WO (1) | WO2010048582A1 (en) |
Families Citing this family (36)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US9023791B2 (en) | 2010-11-19 | 2015-05-05 | Novartis Ag | Fibroblast growth factor 21 mutations |
| EP2750681B1 (en) * | 2011-08-30 | 2020-05-27 | Quanta Biodesign, Ltd. | Branched discrete peg constructs |
| UY34347A (en) | 2011-09-26 | 2013-04-30 | Novartis Ag | DUAL FUNCTION PROTEINS TO TREAT METABOLIC DISORDERS |
| TWI593708B (en) | 2011-09-26 | 2017-08-01 | 諾華公司 | Fusion proteins for treating metabolic disorders |
| CN102504022A (en) * | 2011-11-30 | 2012-06-20 | 苏州元基生物技术有限公司 | Proinsulin containing protecting lysine and preparation method for insulin by utilizing proinsulin |
| EP2820033A4 (en) * | 2012-02-29 | 2015-01-07 | Ambrx Inc | Interleukin-10 polypeptide conjugates and their uses |
| UY35144A (en) | 2012-11-20 | 2014-06-30 | Novartis Ag | APELINE SYNTHETIC LINEAR MIMETICS FOR THE CARDIAC INSUFFICIENCY TREATMENT |
| WO2014124258A2 (en) | 2013-02-08 | 2014-08-14 | Irm Llc | Specific sites for modifying antibodies to make immunoconjugates |
| MX2015010146A (en) | 2013-02-08 | 2016-05-31 | Novartis Ag | Specific sites for modifying antibodies to make immunoconjugates. |
| CN103073627B (en) * | 2013-02-22 | 2014-03-19 | 哈尔滨工业大学 | Method for microwave-assisted reverse micelle separation and purification of kidney bean agglutinin |
| CN105247049B (en) | 2013-03-15 | 2019-09-13 | 米迪缪尼有限公司 | Novel nucleic acid molecule |
| WO2015013168A1 (en) | 2013-07-25 | 2015-01-29 | Novartis Ag | Cyclic polypeptides for the treatment of heart failure |
| US9340582B2 (en) | 2013-07-25 | 2016-05-17 | Novartis Ag | Bioconjugates of synthetic apelin polypeptides |
| WO2015138615A2 (en) * | 2014-03-12 | 2015-09-17 | Irm Llc | Specific sites for modifying antibodies to make immunoconjugates |
| EP3851445A1 (en) | 2014-06-23 | 2021-07-21 | Novartis AG | Site specific protein modifications |
| US10588980B2 (en) | 2014-06-23 | 2020-03-17 | Novartis Ag | Fatty acids and their use in conjugation to biomolecules |
| JP6840668B2 (en) * | 2014-11-14 | 2021-03-10 | ディ.イー.ショー リサーチ, エルエルシーD.E.Shaw Research, Llc | Suppression of interactions between bound particles |
| CA2972871A1 (en) | 2015-01-23 | 2016-07-28 | Novartis Ag | Synthetic apelin fatty acid conjugates with improved half-life |
| EP3393494A1 (en) | 2015-12-22 | 2018-10-31 | Novartis Ag | Methods of treating or ameliorating metabolic disorders using growth differentiation factor 15 (gdf-15) |
| CA3018129A1 (en) * | 2016-03-18 | 2017-09-21 | Department Of Biotechnology | Multi-functional chemical agents, and the method for protein modification |
| US10583199B2 (en) | 2016-04-26 | 2020-03-10 | Northwestern University | Nanocarriers having surface conjugated peptides and uses thereof for sustained local release of drugs |
| EP3468581A1 (en) | 2016-06-13 | 2019-04-17 | Torque Therapeutics, Inc. | Methods and compositions for promoting immune cell function |
| JP2020532565A (en) | 2017-09-05 | 2020-11-12 | トルク セラピューティクス, インコーポレイテッド | Reversible linker and its use |
| JP7285828B2 (en) | 2017-09-05 | 2023-06-02 | トルク セラピューティクス, インコーポレイテッド | Therapeutic protein compositions and methods of making and using them |
| CN108395474B (en) * | 2018-01-30 | 2021-02-19 | 武汉大学 | Method for coupling pyrazole with phenylalanine compound through visible light induction |
| EP3887388A1 (en) | 2018-11-27 | 2021-10-06 | Novartis AG | Cyclic peptides as proprotein convertase subtilisin/kexin type 9 (pcsk9) inhibitors for the treatment of metabolic disorders |
| EP3914282A1 (en) | 2019-01-25 | 2021-12-01 | Ospedale San Raffaele S.r.l. | Inhibitor of dux4 and uses thereof |
| CN112978709A (en) * | 2019-12-12 | 2021-06-18 | 中国科学院大连化学物理研究所 | Carbon quantum dot precursor composition, carbon quantum dot and preparation method thereof |
| JP2023513168A (en) | 2020-02-07 | 2023-03-30 | ノバルティス アーゲー | Targeted plasma proteolysis |
| CN112147132B (en) * | 2020-09-23 | 2021-07-09 | 山东大学 | Preparation method of spectral near-infrared electrochemiluminescence immunosensor |
| TWI741843B (en) * | 2020-10-20 | 2021-10-01 | 新加坡商億恩診斷有限公司 | The protein measurement apparatus and operation method thereof |
| CN113582881B (en) * | 2021-07-29 | 2022-10-11 | 浙江新码生物医药有限公司 | Unnatural amino acid, application thereof, recombinant protein containing unnatural amino acid and recombinant protein conjugate |
| KR20240024235A (en) * | 2021-07-29 | 2024-02-23 | 노보코덱스 바이오파마슈티컬즈 컴퍼니 리미티드 | Non-natural amino acids and uses thereof, recombinant proteins containing them, and recombinant protein conjugates |
| CN113698468B (en) * | 2021-09-01 | 2022-10-11 | 浙江新码生物医药有限公司 | Human interleukin 2-polyethylene glycol conjugate and application thereof |
| EP4389762A1 (en) | 2022-12-23 | 2024-06-26 | Ospedale San Raffaele S.r.l. | Inhibitors of dux4 activity and their use in therapy. |
| CN117417292B (en) * | 2023-02-20 | 2024-07-12 | 浙江新码生物医药有限公司 | Heteroaryl-containing unnatural amino acid and application thereof |
Family Cites Families (6)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US7266613B1 (en) | 2000-08-09 | 2007-09-04 | Microsoft Corporation | Fast dynamic measurement of bandwidth in a TCP network environment |
| CA2444098C (en) | 2001-04-19 | 2016-06-21 | The Scripps Research Institute | Methods and composition for the production of orthogonal trna-aminoacyltrna synthetase pairs |
| EP2392923B1 (en) * | 2003-06-18 | 2014-05-21 | The Scripps Research Institute | Method of producing proteins comprising unnatural amino acid |
| US20060183888A1 (en) * | 2004-08-13 | 2006-08-17 | Chan Michael K | Pyrrolysine synthesis and modification |
| US20060166319A1 (en) * | 2004-08-13 | 2006-07-27 | Chan Michael K | Charging tRNA with pyrrolysine |
| WO2007099854A1 (en) | 2006-02-22 | 2007-09-07 | Riken | METHOD FOR SYNTHESIS OF SUPPRESSOR tRNA, DNA CONSTRUCT, AND PRODUCTION OF PROTEIN HAVING NON-NATURAL AMINO ACID INTEGRATED THEREIN BY USING THE DNA CONSTRUCT |
-
2009
- 2009-10-23 AU AU2009308163A patent/AU2009308163B2/en not_active Ceased
- 2009-10-23 BR BRPI0919932A patent/BRPI0919932A8/en not_active IP Right Cessation
- 2009-10-23 MX MX2011004246A patent/MX342677B/en active IP Right Grant
- 2009-10-23 US US13/125,547 patent/US8785151B2/en not_active Expired - Fee Related
- 2009-10-23 KR KR1020137017122A patent/KR20130086386A/en not_active Application Discontinuation
- 2009-10-23 CN CN200980152207.8A patent/CN102264697B/en not_active Expired - Fee Related
- 2009-10-23 CA CA2871855A patent/CA2871855C/en not_active Expired - Fee Related
- 2009-10-23 EA EA201100659A patent/EA021001B1/en not_active IP Right Cessation
- 2009-10-23 KR KR1020117011641A patent/KR101316159B1/en not_active IP Right Cessation
- 2009-10-23 EP EP09744017.6A patent/EP2356092B1/en not_active Not-in-force
- 2009-10-23 CA CA2742043A patent/CA2742043C/en not_active Expired - Fee Related
- 2009-10-23 WO PCT/US2009/061954 patent/WO2010048582A1/en active Application Filing
- 2009-10-23 ES ES09744017.6T patent/ES2477543T3/en active Active
- 2009-10-23 JP JP2011533389A patent/JP5542832B2/en not_active Expired - Fee Related
-
2014
- 2014-05-15 US US14/279,085 patent/US9624520B2/en not_active Expired - Fee Related
Also Published As
| Publication number | Publication date |
|---|---|
| CN102264697A (en) | 2011-11-30 |
| CA2742043C (en) | 2014-11-25 |
| CA2871855C (en) | 2017-08-15 |
| CA2742043A1 (en) | 2010-04-29 |
| WO2010048582A1 (en) | 2010-04-29 |
| MX342677B (en) | 2016-10-07 |
| JP5542832B2 (en) | 2014-07-09 |
| AU2009308163A1 (en) | 2010-04-29 |
| US20110262963A1 (en) | 2011-10-27 |
| BRPI0919932A2 (en) | 2017-10-24 |
| US9624520B2 (en) | 2017-04-18 |
| BRPI0919932A8 (en) | 2018-02-06 |
| EP2356092A1 (en) | 2011-08-17 |
| KR101316159B1 (en) | 2013-10-15 |
| JP2012506869A (en) | 2012-03-22 |
| EP2356092B1 (en) | 2014-04-16 |
| KR20110089145A (en) | 2011-08-04 |
| EA201100659A1 (en) | 2011-12-30 |
| AU2009308163B2 (en) | 2013-01-10 |
| CA2871855A1 (en) | 2010-04-29 |
| MX2011004246A (en) | 2011-07-13 |
| ES2477543T3 (en) | 2014-07-17 |
| EA021001B1 (en) | 2015-03-31 |
| CN102264697B (en) | 2015-09-09 |
| US20140302553A1 (en) | 2014-10-09 |
| US8785151B2 (en) | 2014-07-22 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| KR101316159B1 (en) | Biosynthetically generated pyrroline-carboxy-lysine and site specific protein modifications via chemical derivatization of pyrroline-carboxy-lysine and pyrrolysine residues | |
| US20220088212A1 (en) | C-terminal lysine conjugated immunoglobulins | |
| KR101423769B1 (en) | Compositions containing, methods involving, and uses of non-natural amino acids and polypeptides | |
| US11753669B2 (en) | Lysine conjugated immunoglobulins | |
| US20090148887A1 (en) | Genetically encoded boronate amino acid | |
| CN117024548A (en) | Activation of bioluminescence by structural complementation | |
| JP2022526939A (en) | Modified cleaving enzyme, its use, and related kits | |
| CN102439014A (en) | Biomolecular labelling using multifunctional biotin analogues | |
| US20050136491A1 (en) | Method of site specific labeling of proteins and uses therefor | |
| AU2013202463A1 (en) | Biosynthetically generated pyrroline-carboxy-lysine and site specific protein modifications via chemical derivatization of pyrroline-carboxy-lysine and pyrrolysine residues | |
| WO2023052526A1 (en) | Methods for preparing pyridazine compounds | |
| JP2023526102A (en) | Site-specific conjugation of glycosylated monoclonal antibodies with transglutaminase | |
| WO2023097604A1 (en) | Isolated polypeptide and use thereof | |
| TW202330571A (en) | Compound having photocleavable site, linker, and screening method using same | |
| Braxton | The progress on mapping ubiquitin signaling using photocrosslinking mono and di-ubiquitin probes and other ubiquitin moieties | |
| JP2013139440A (en) | Improved methods for preparing peptide conjugate and linker |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| A107 | Divisional application of patent | ||
| E902 | Notification of reason for refusal | ||
| E902 | Notification of reason for refusal | ||
| E601 | Decision to refuse application |