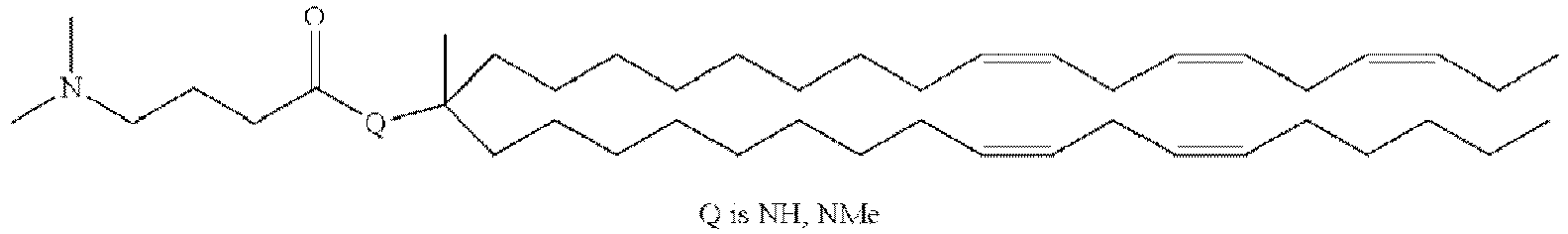
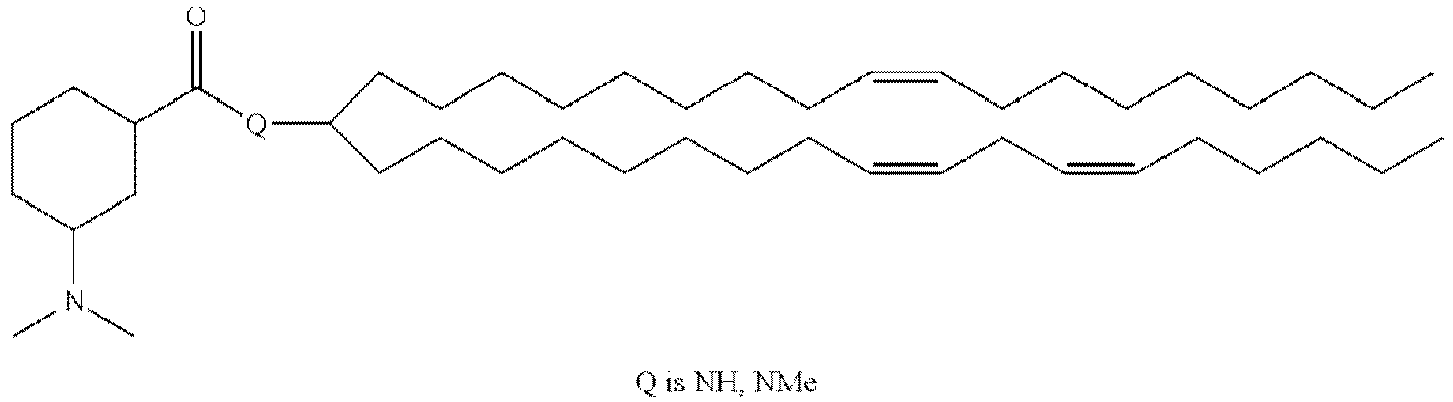
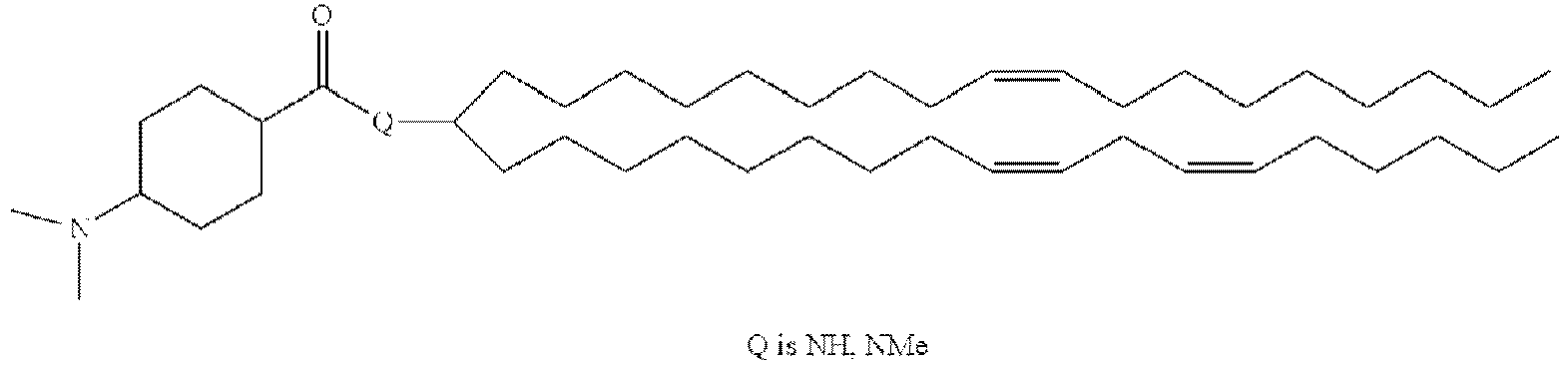
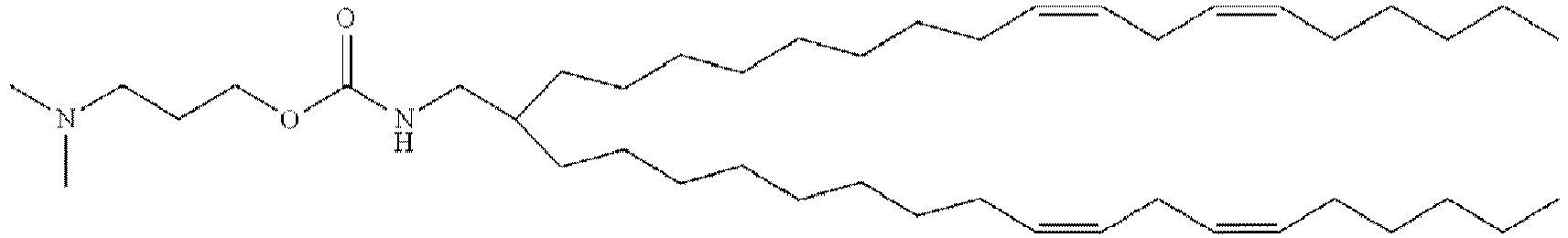
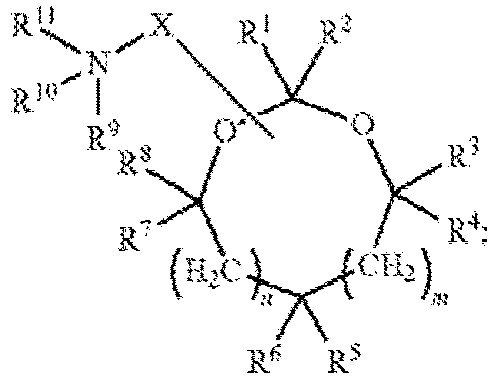
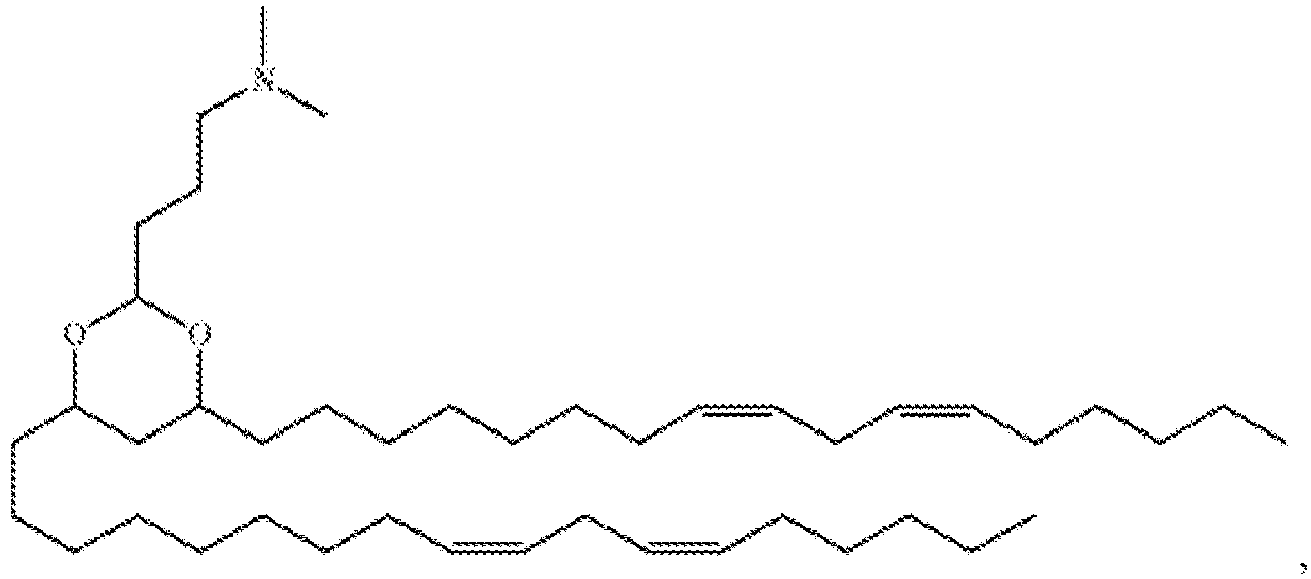
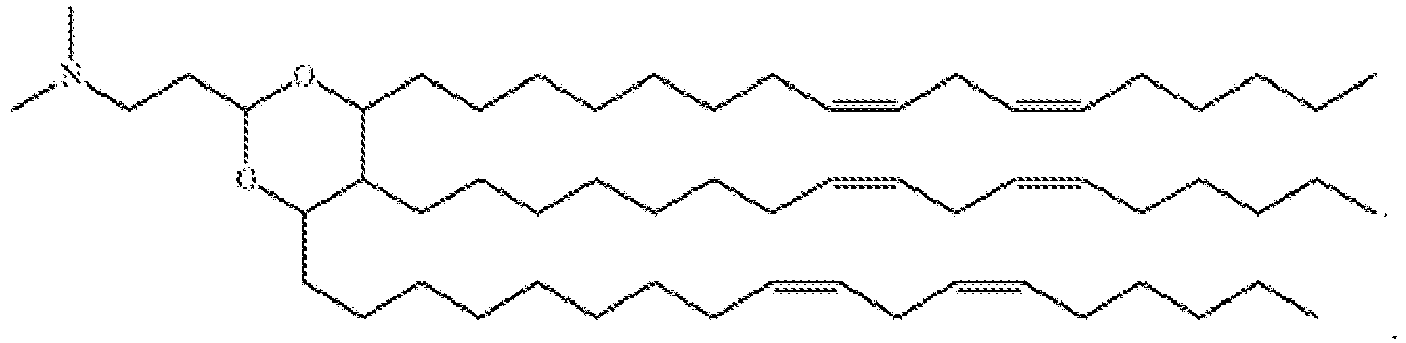
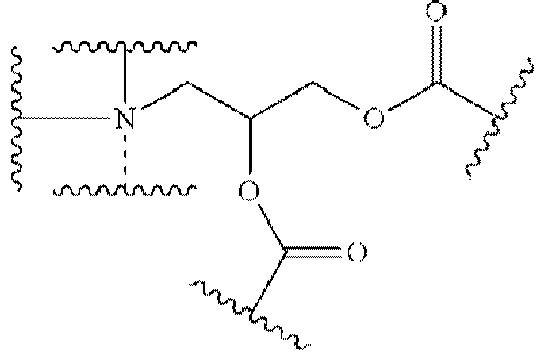
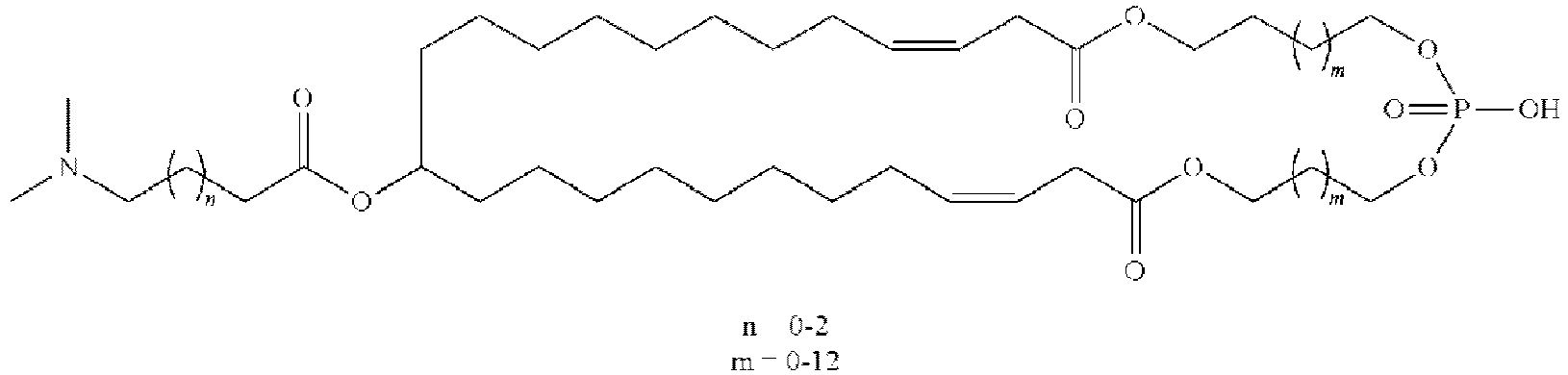
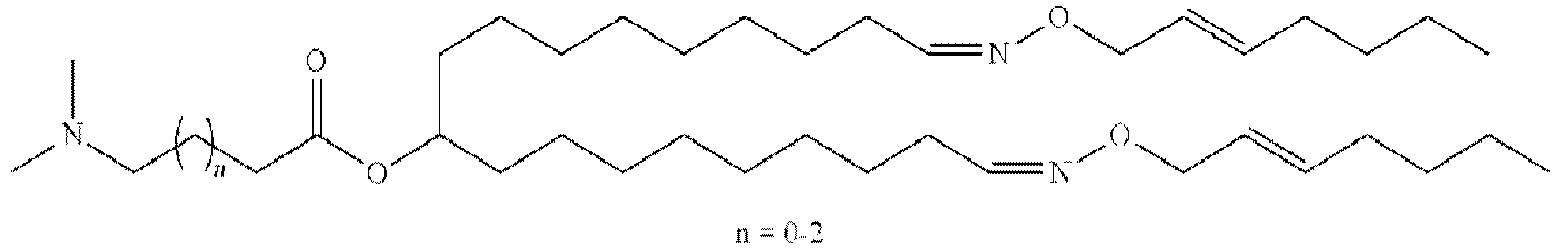
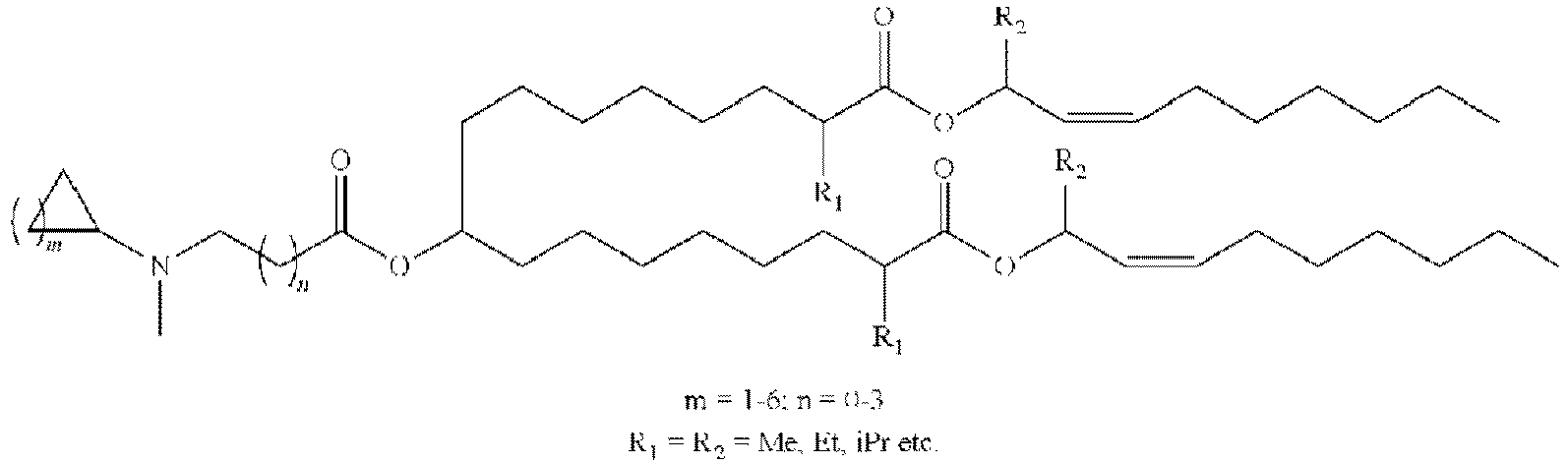
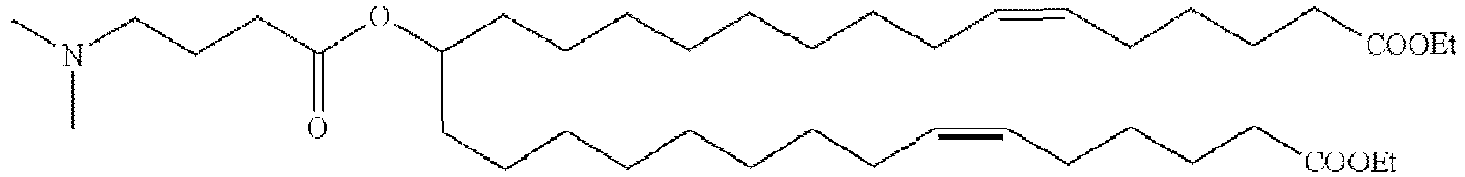
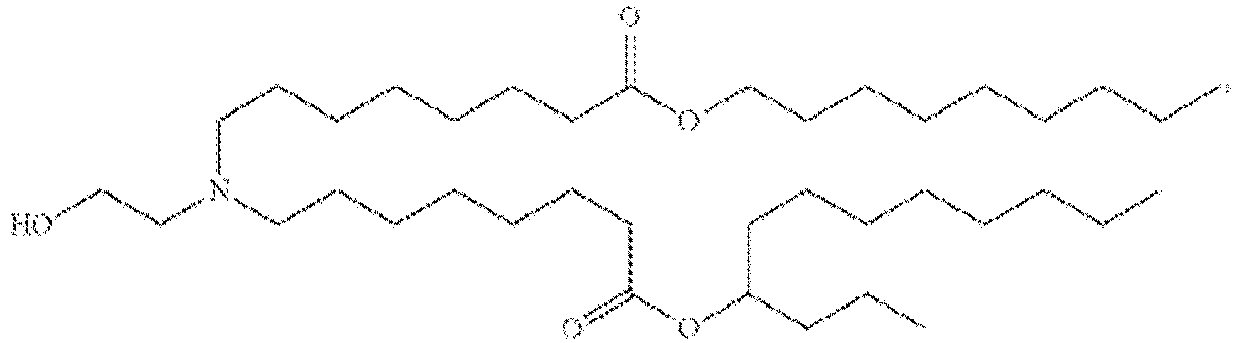
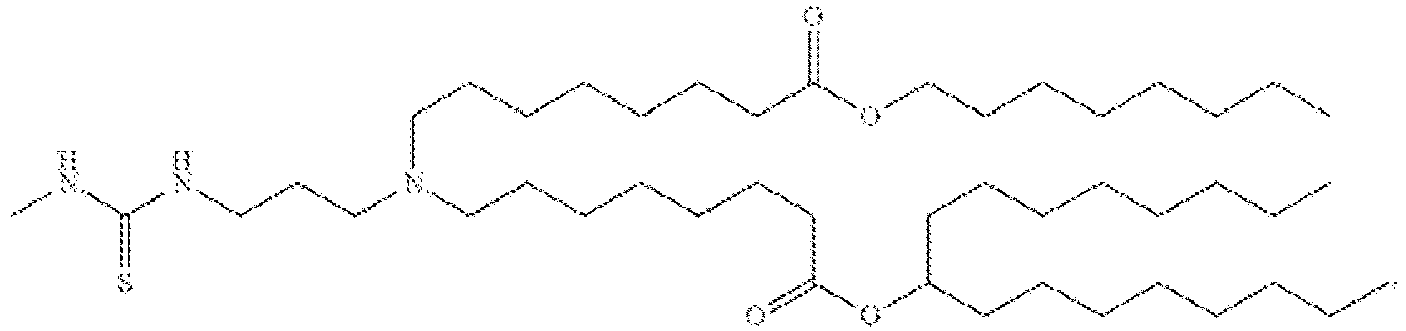
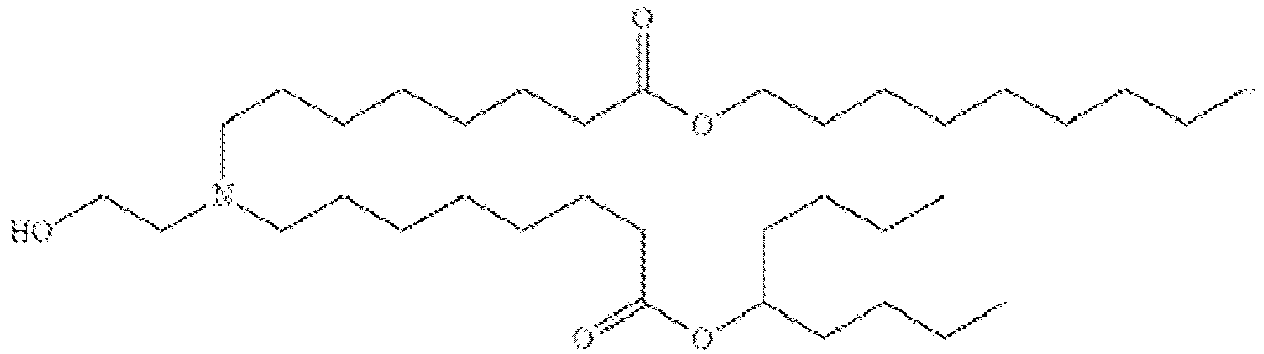
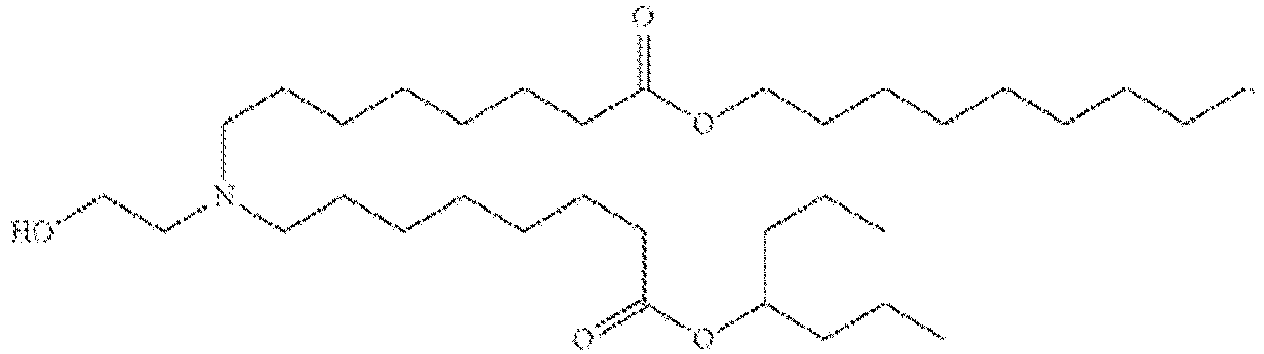
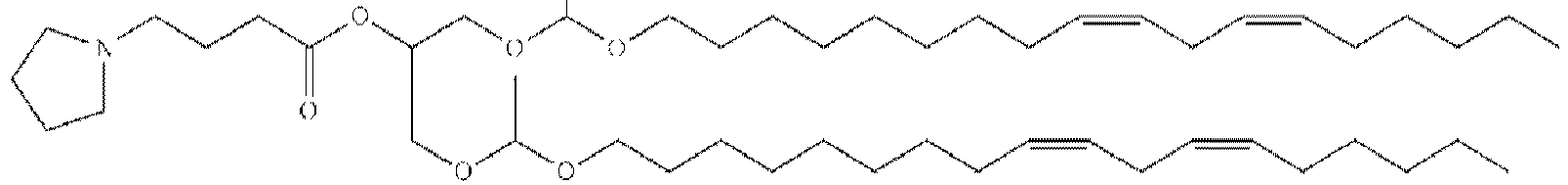
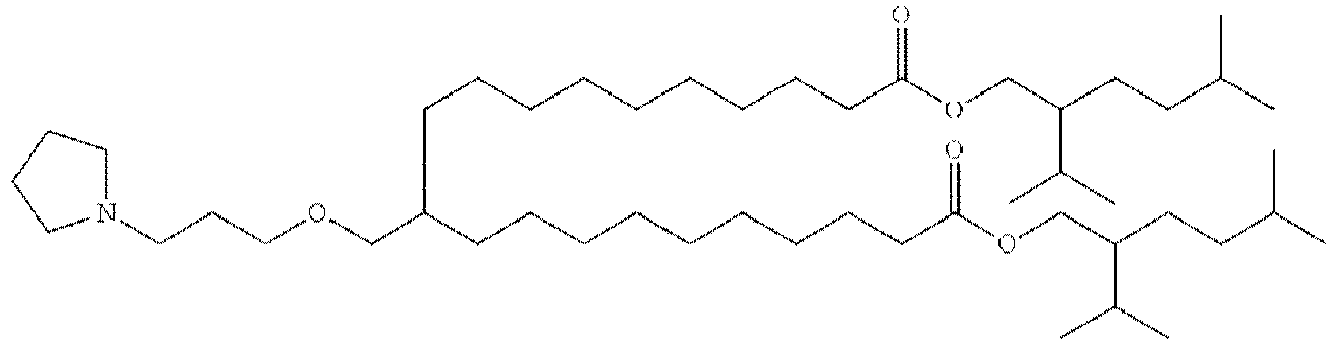
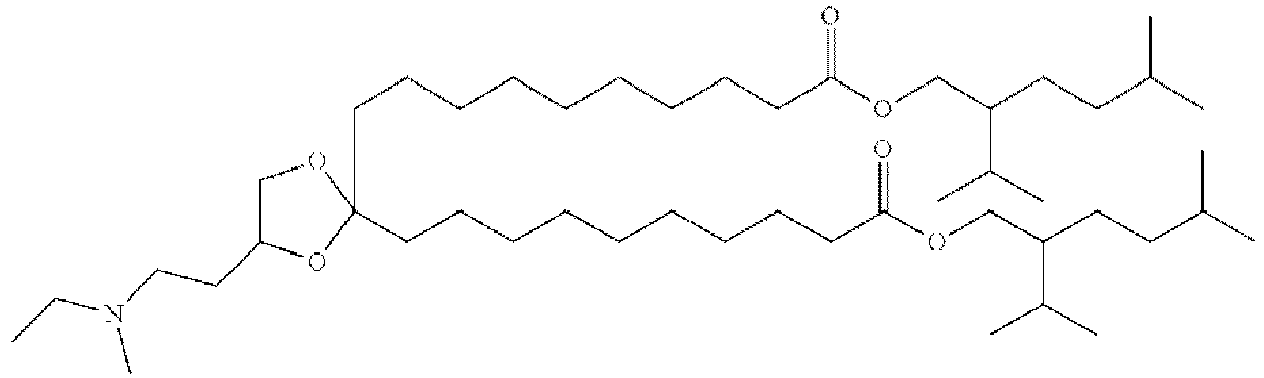
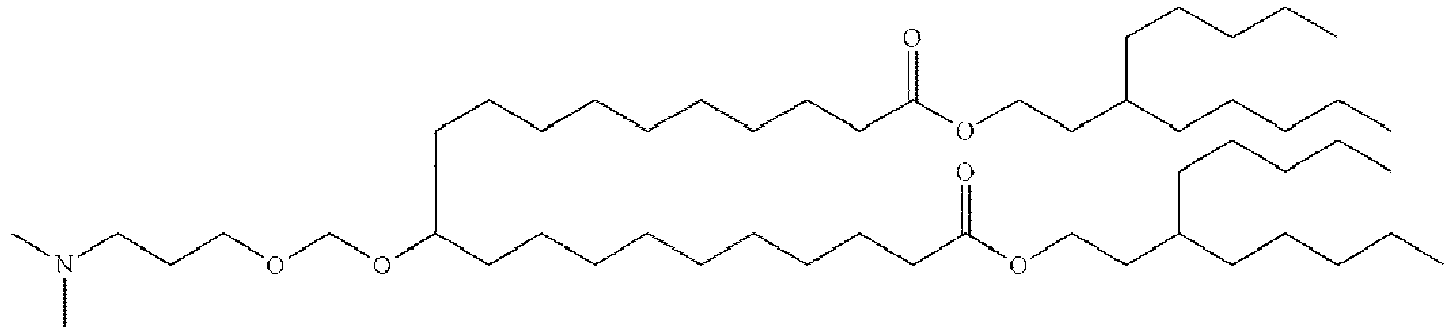
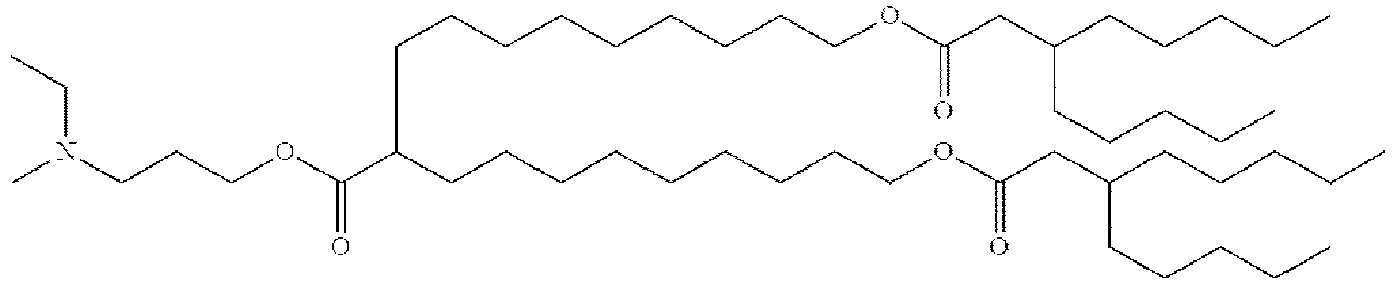
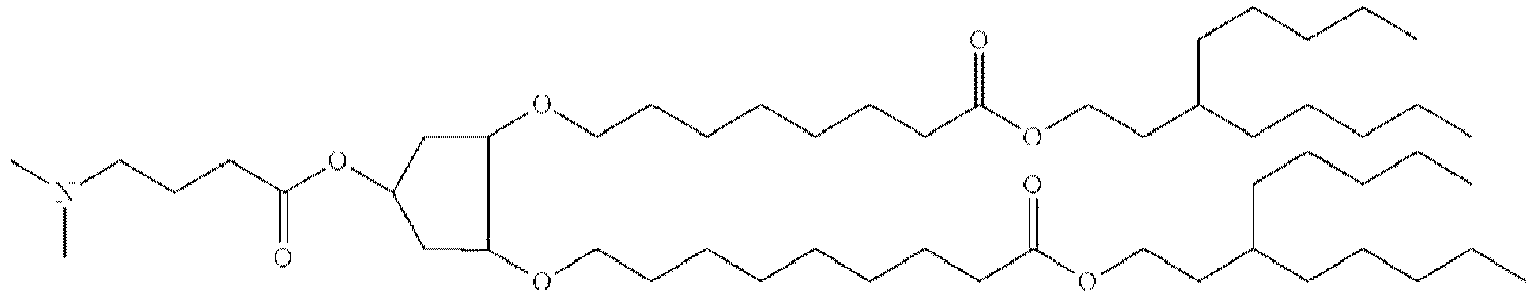
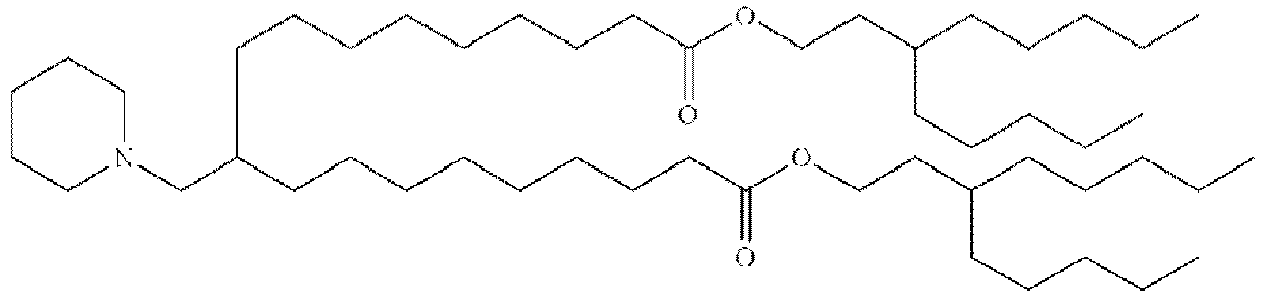
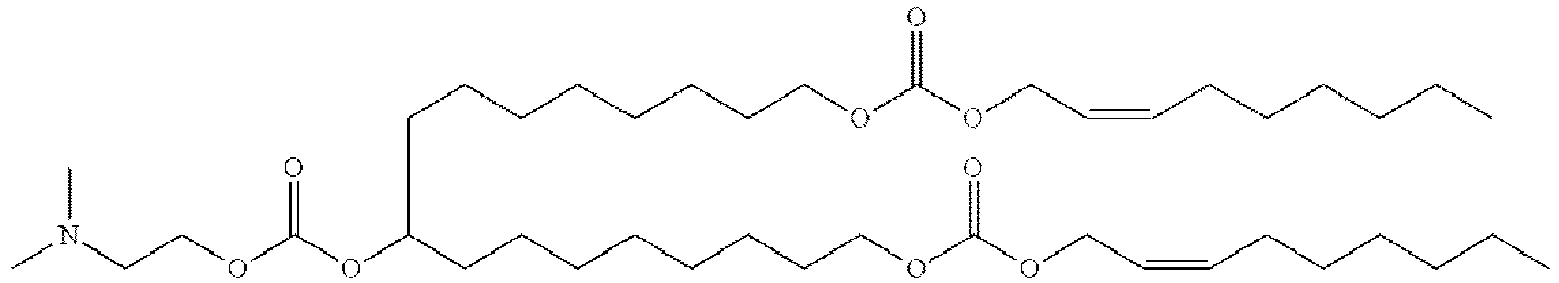
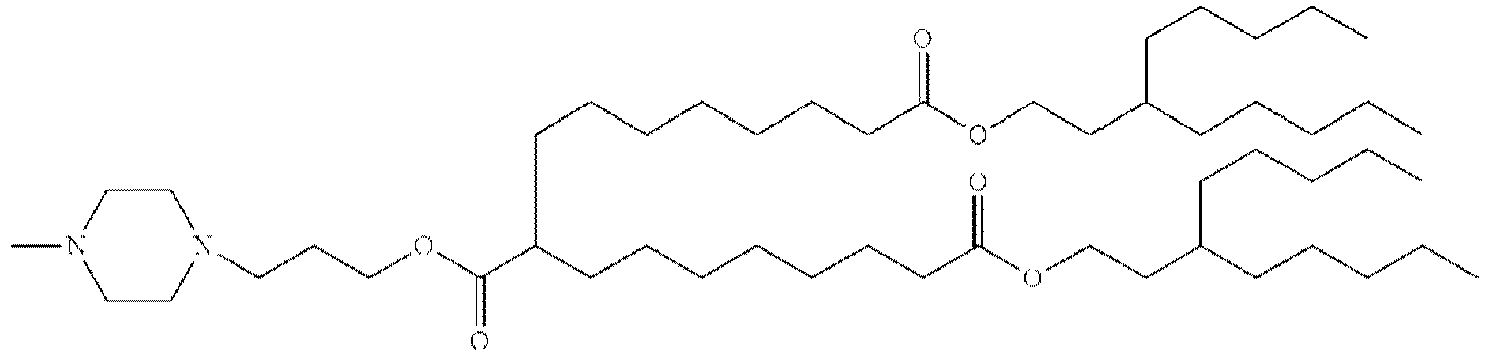
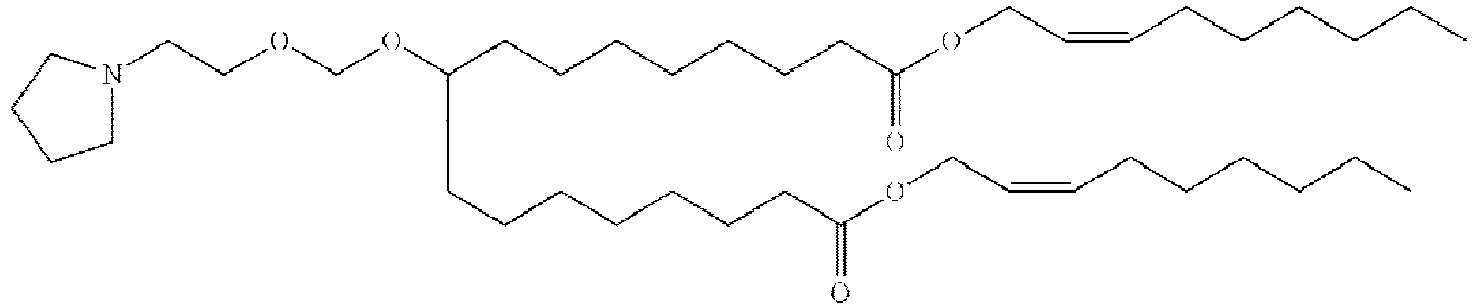
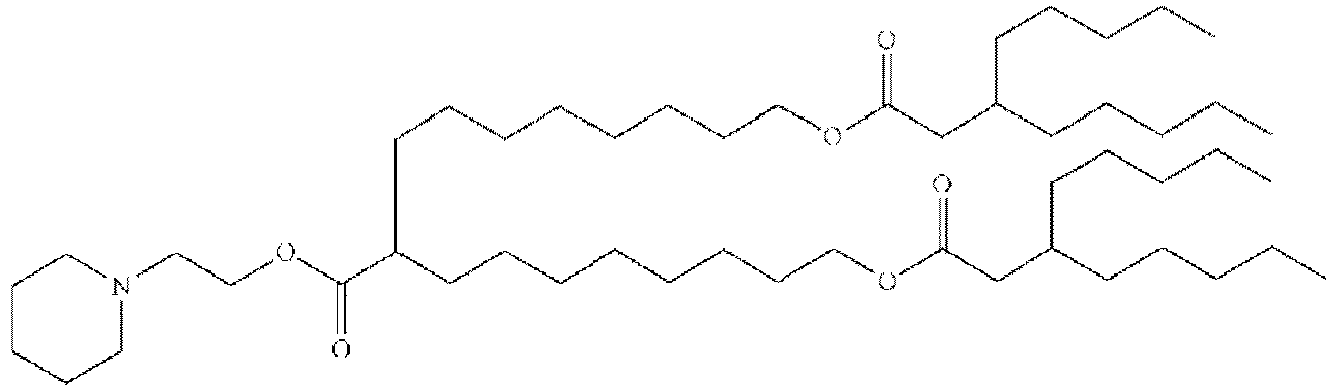
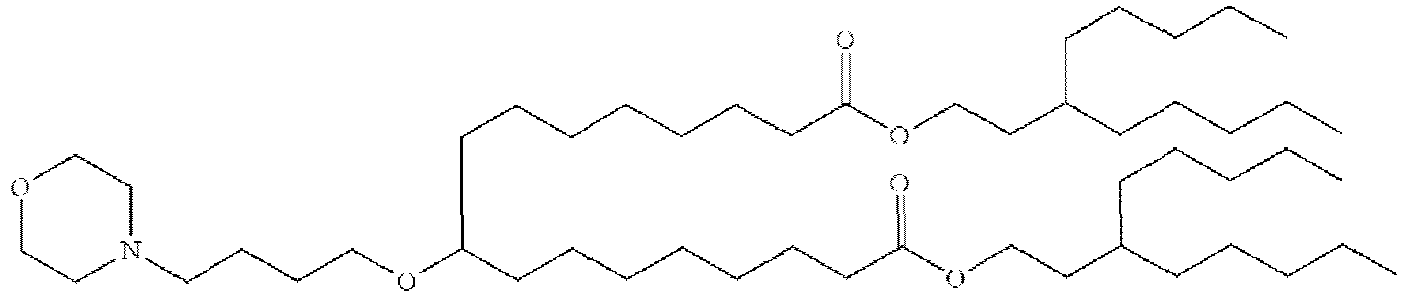
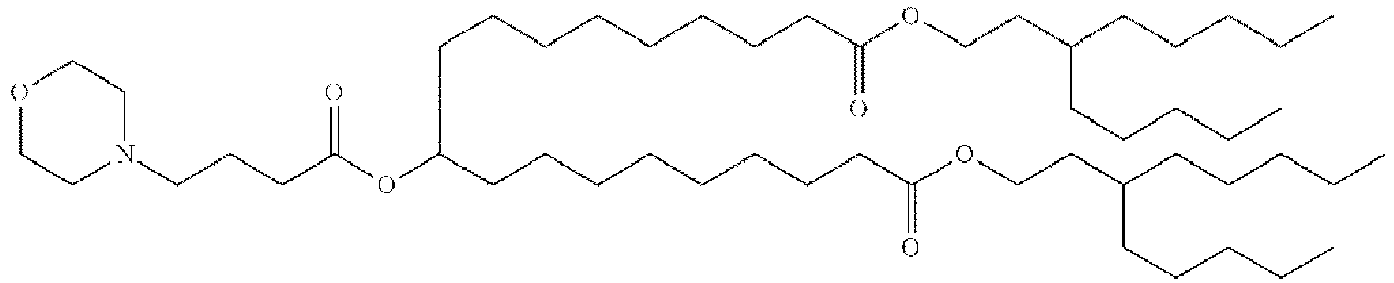
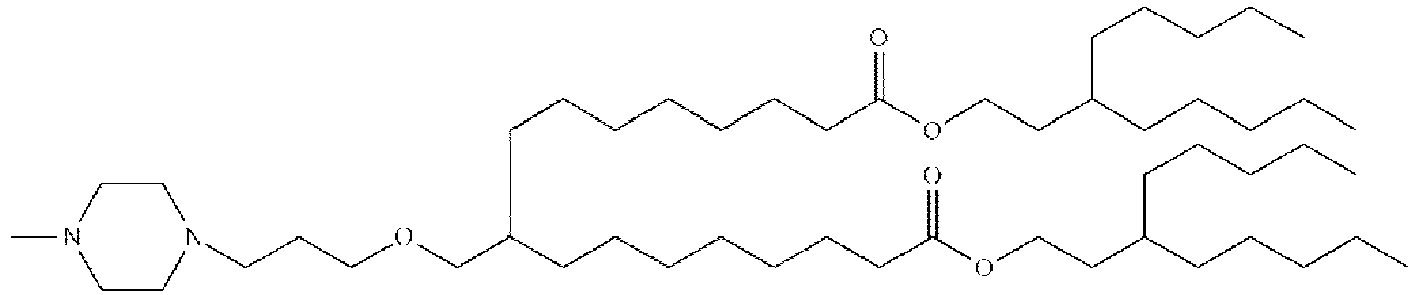
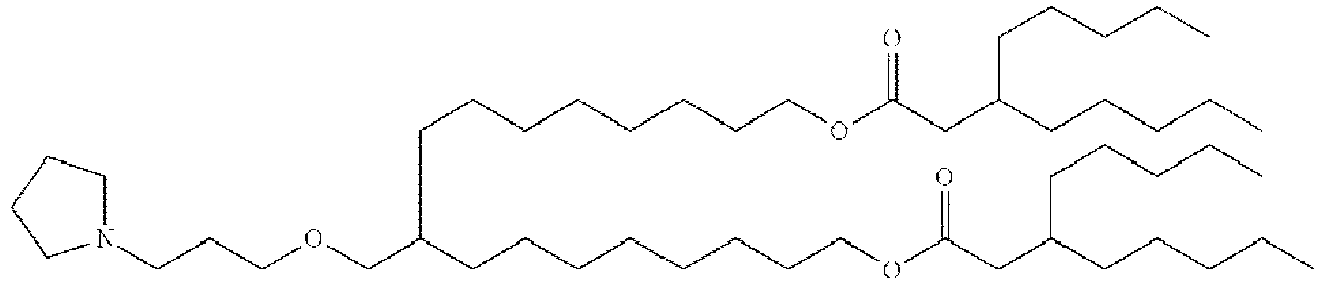
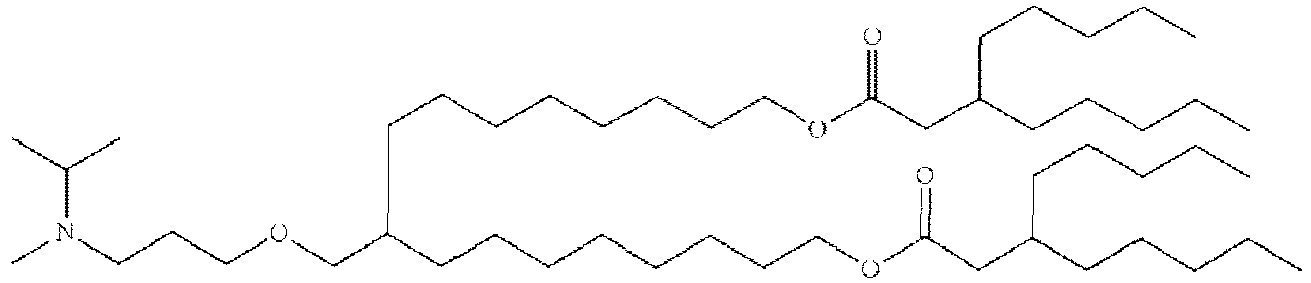
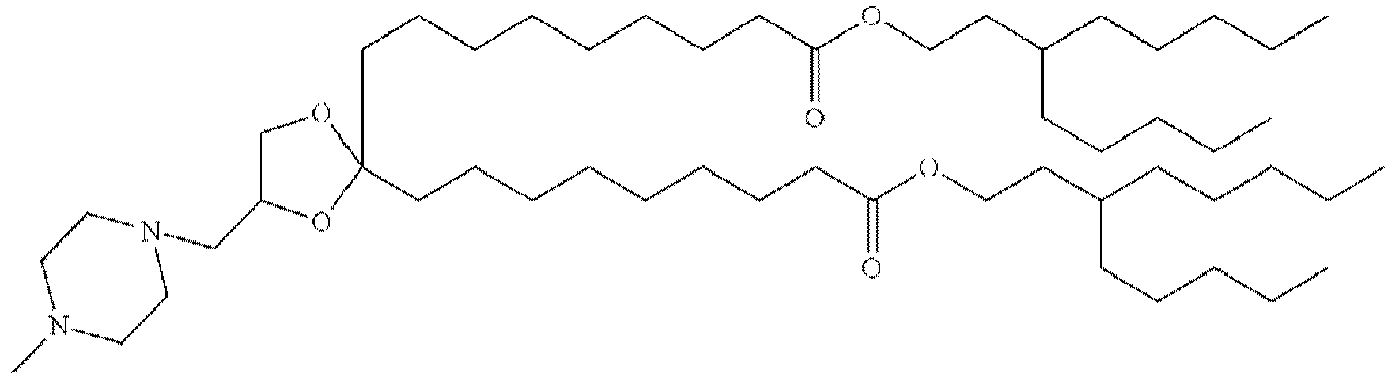
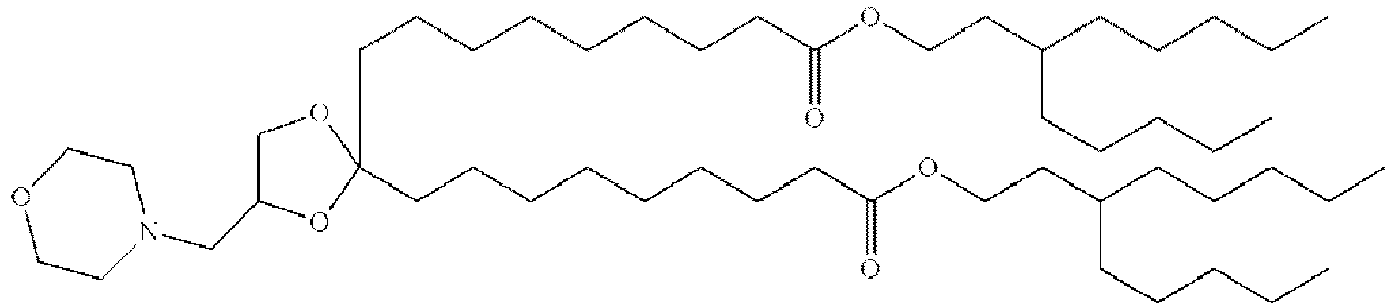
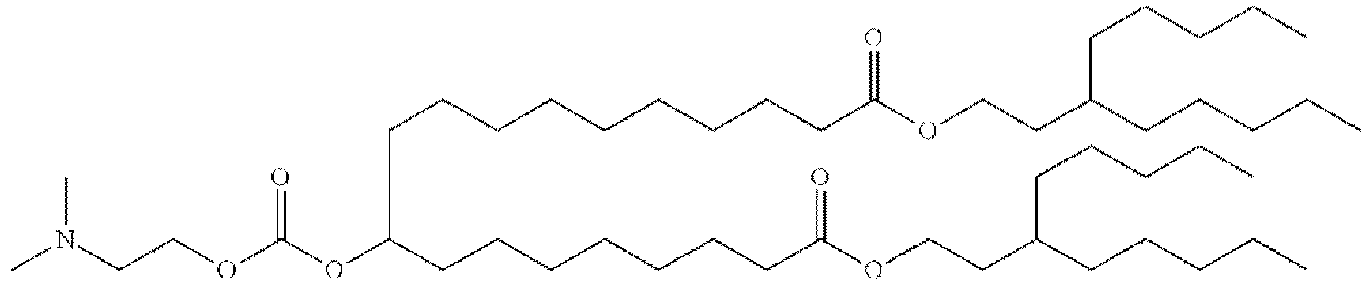
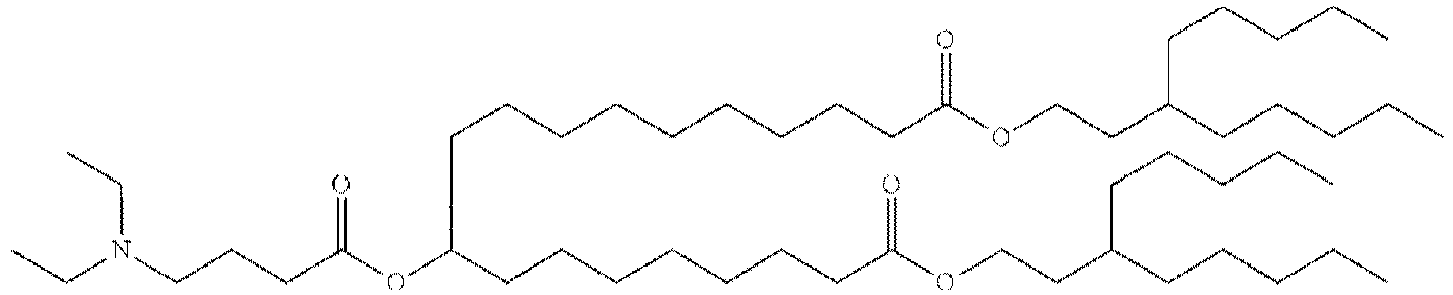
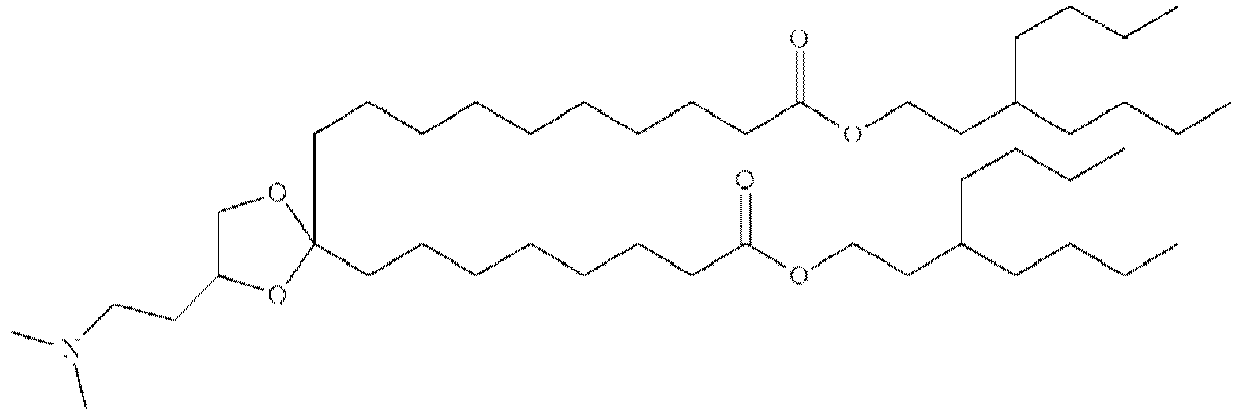
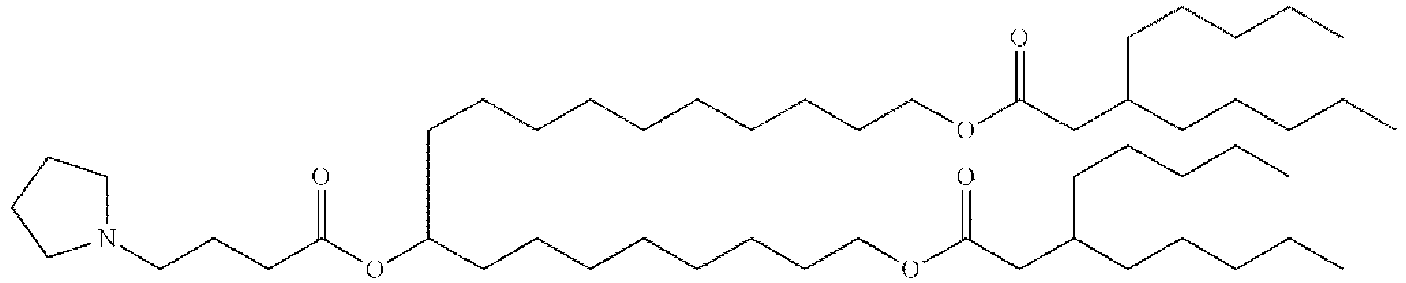
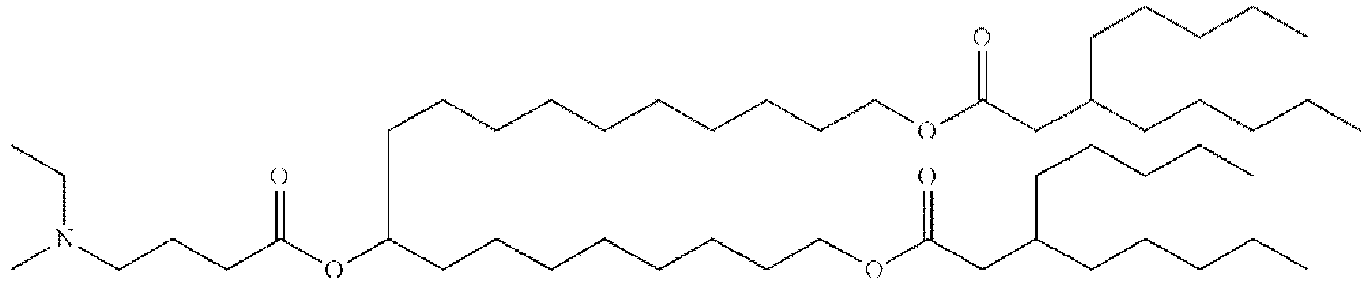
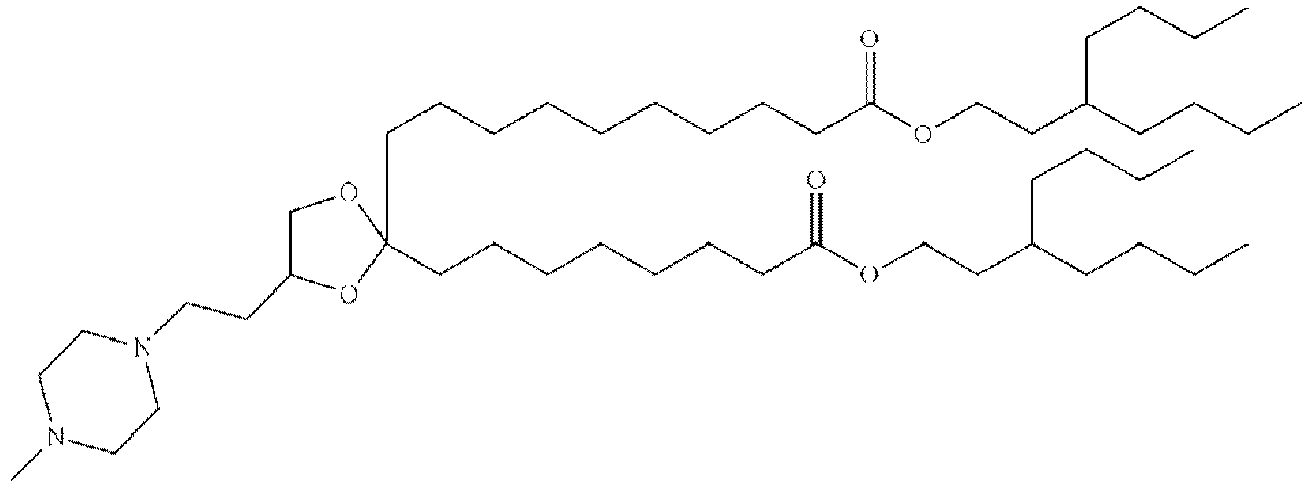
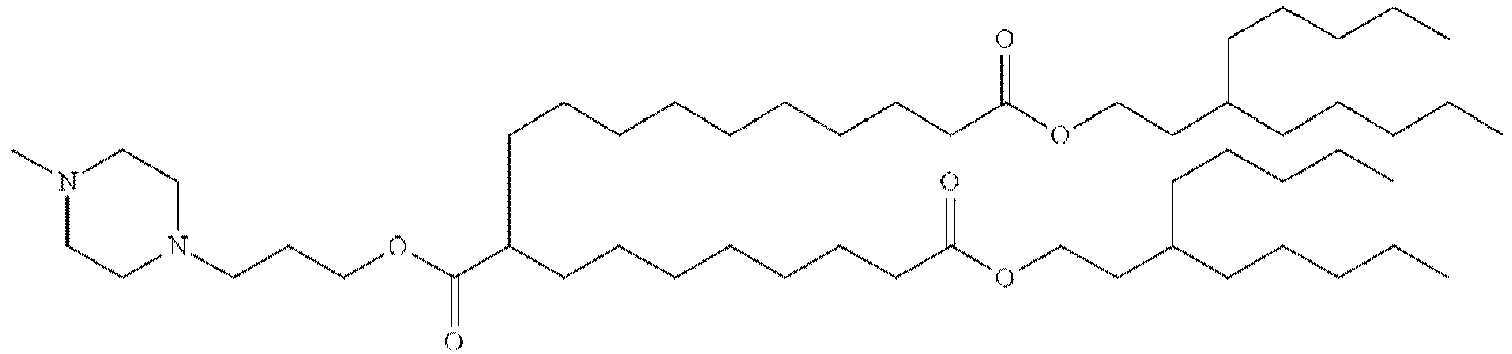
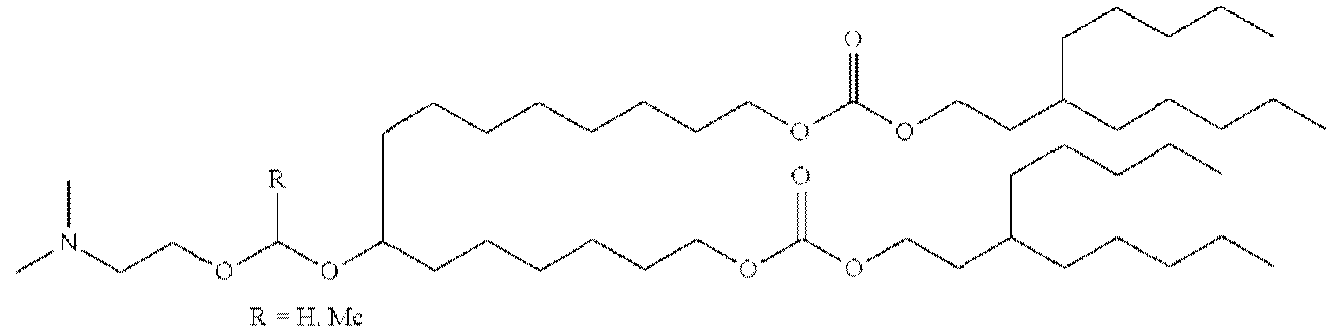
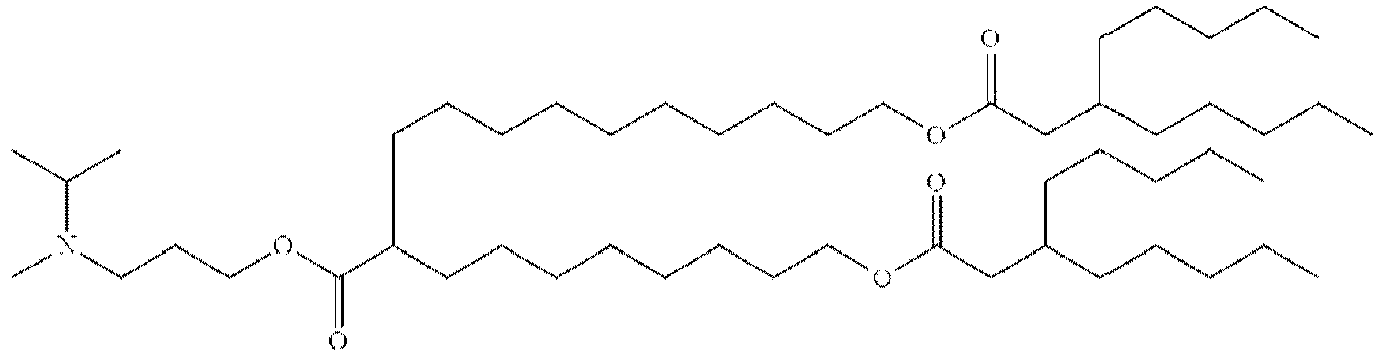
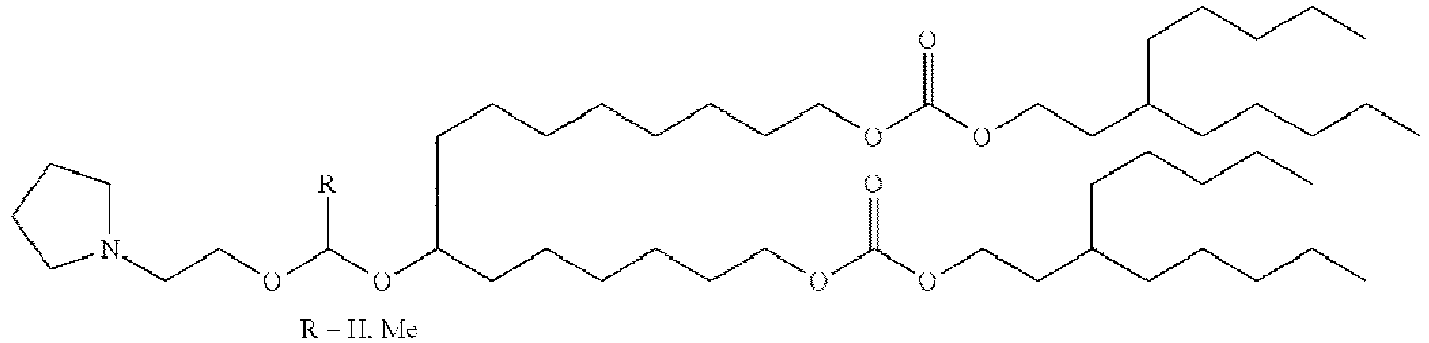
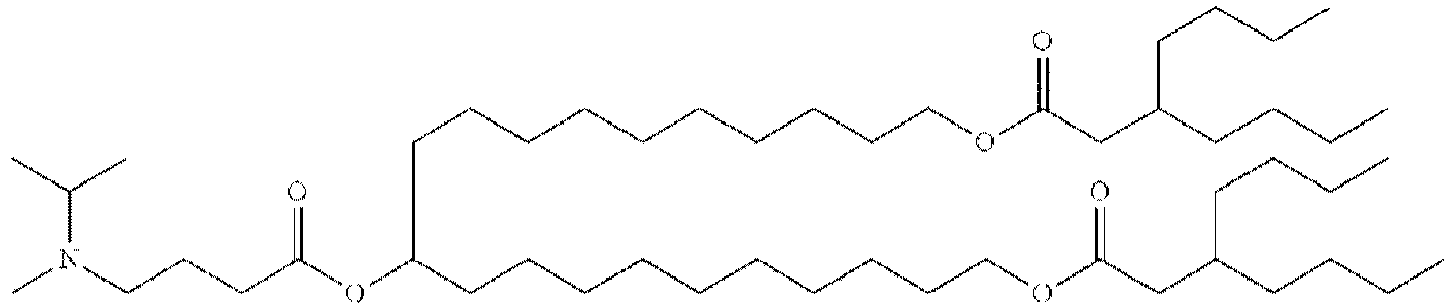
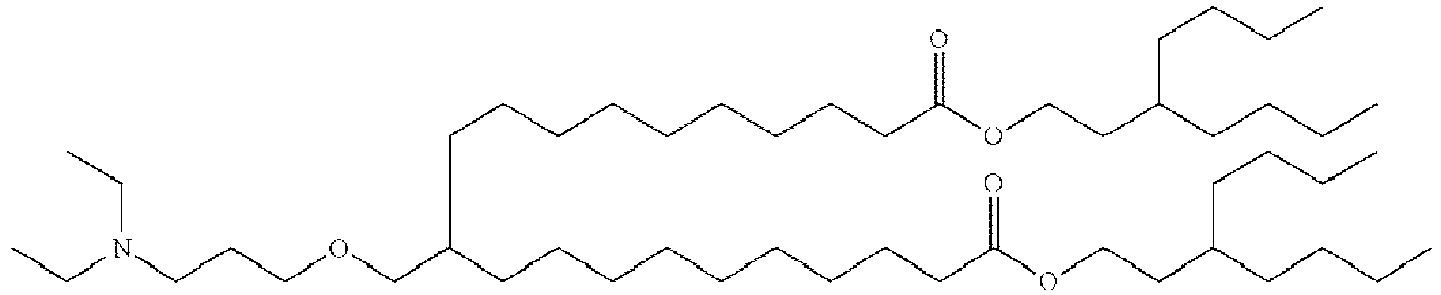
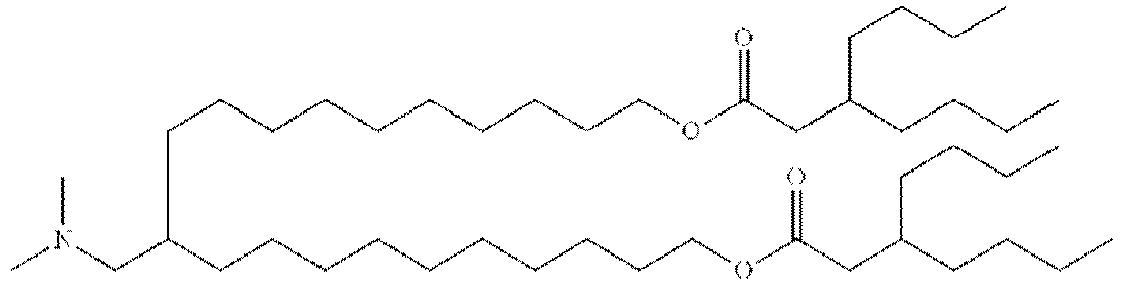
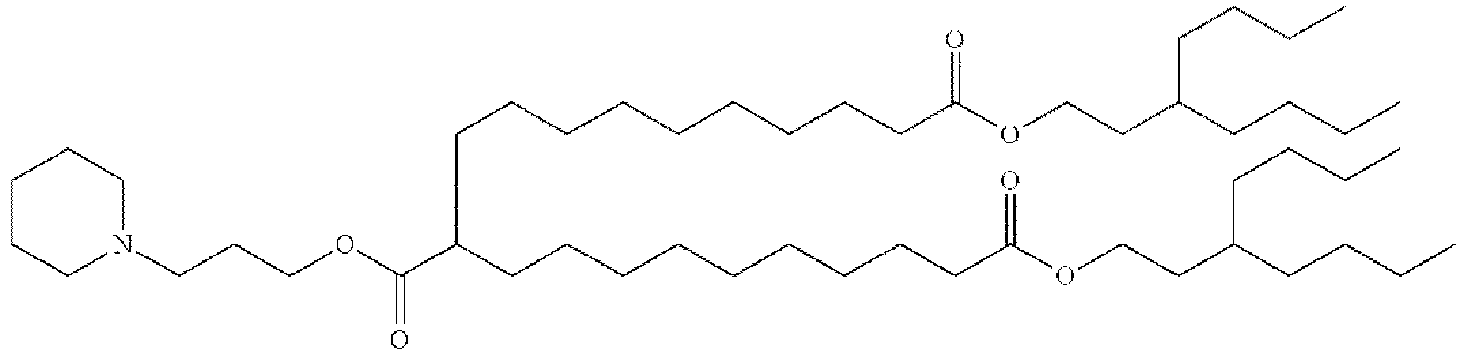
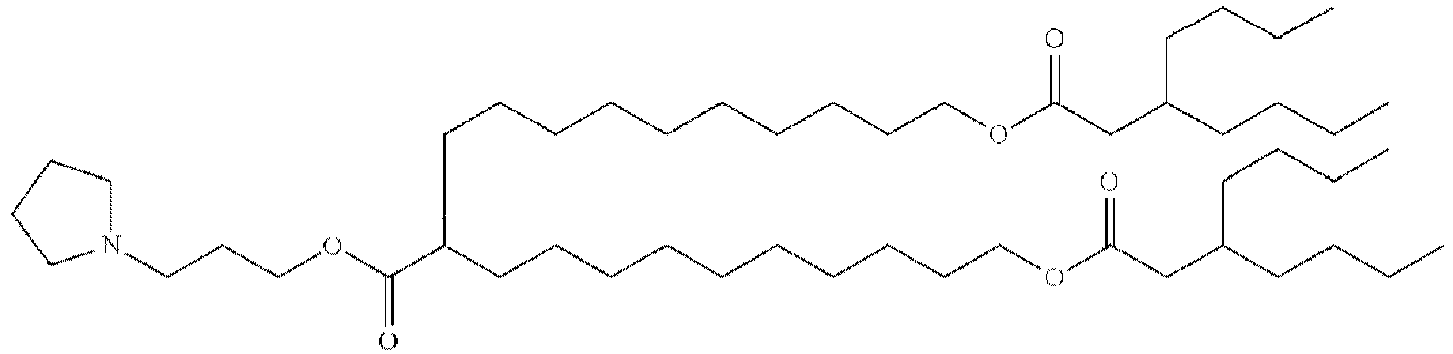
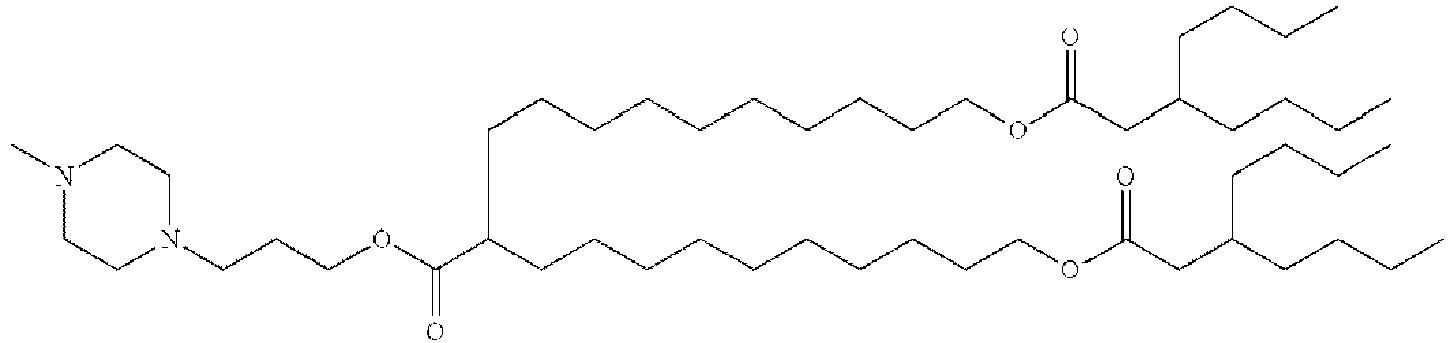
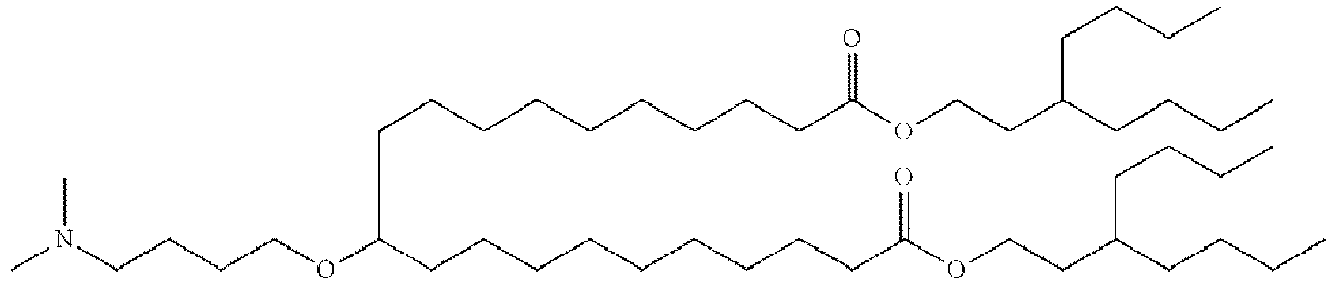
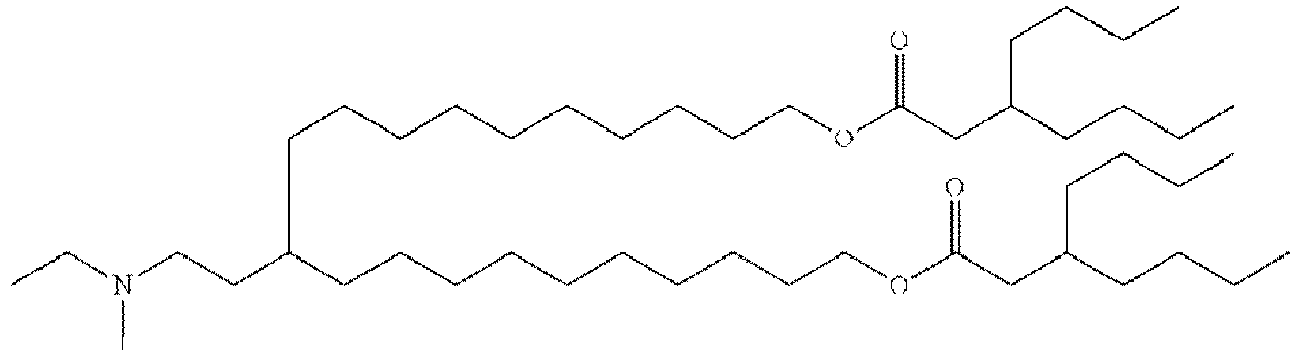
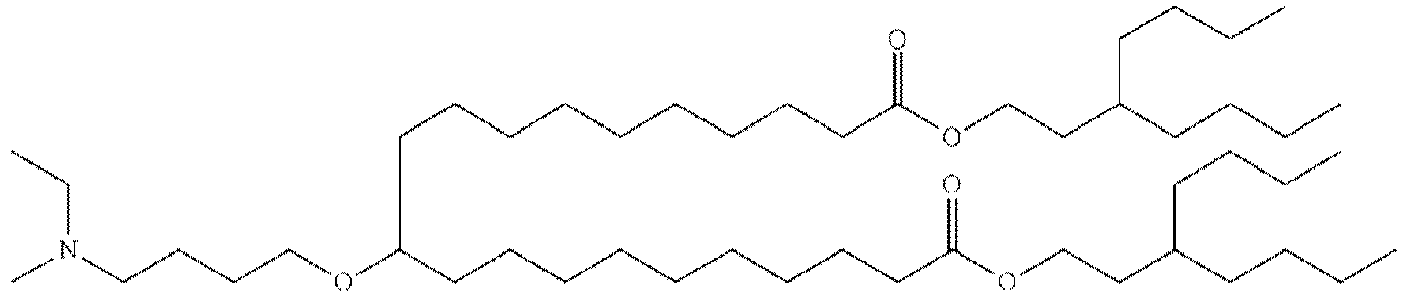
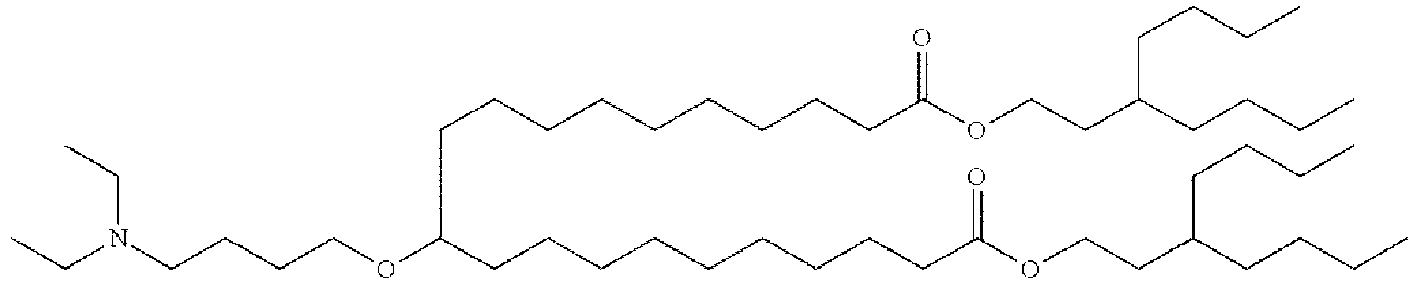
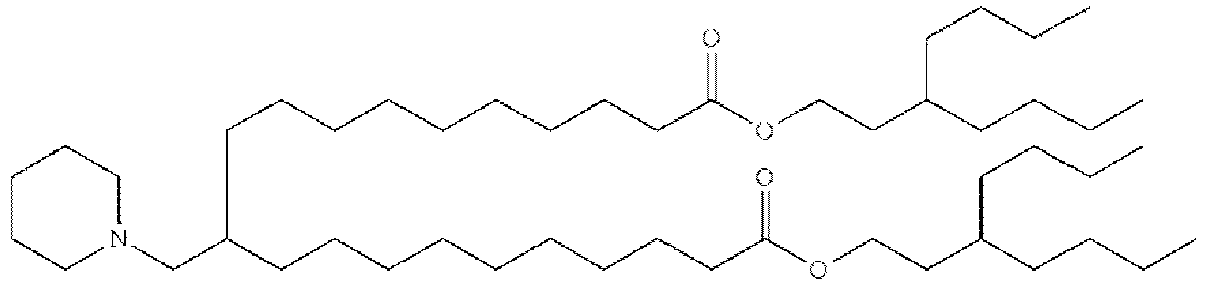
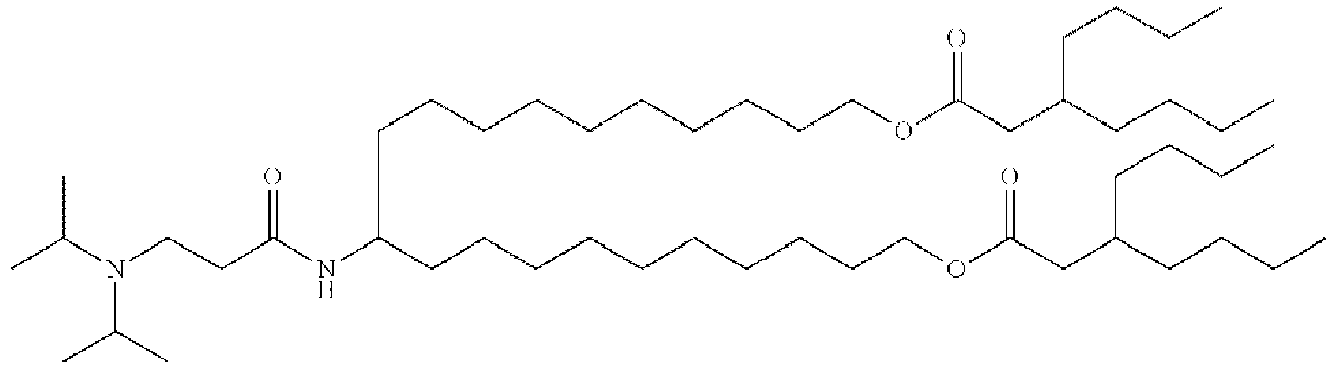
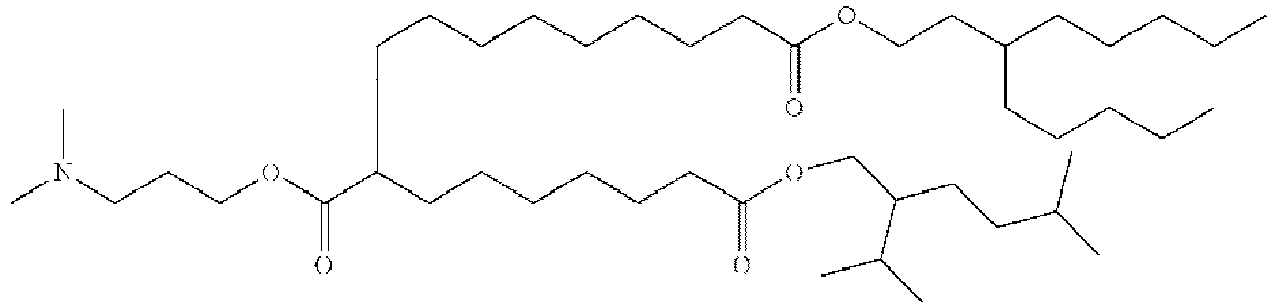
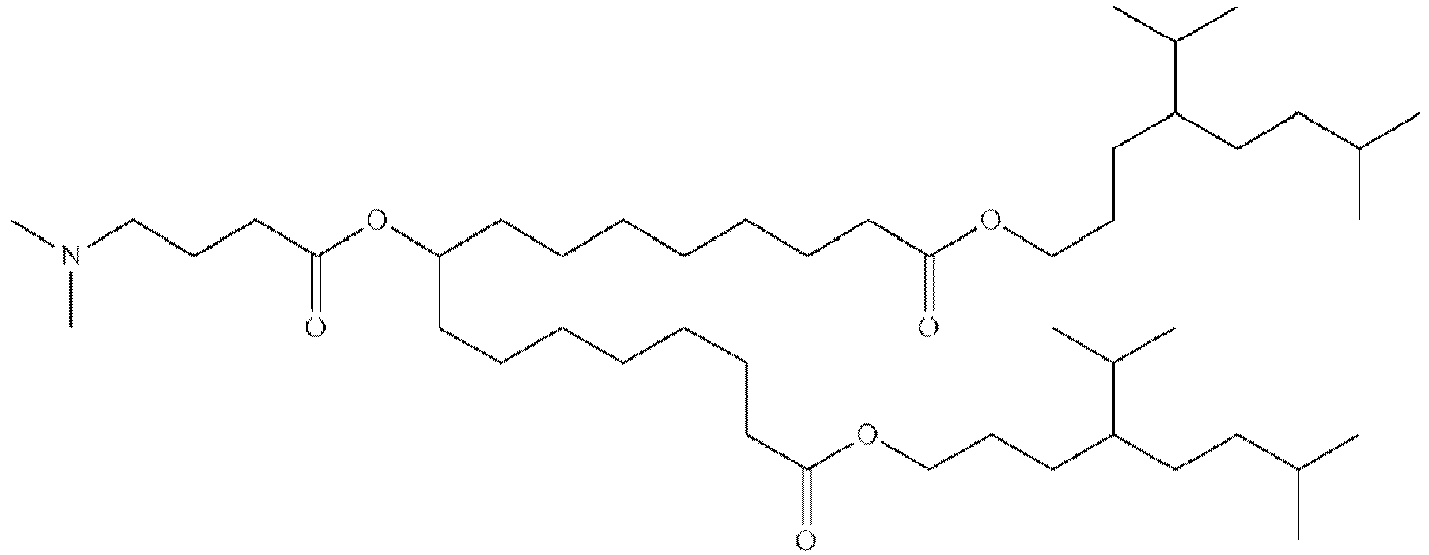
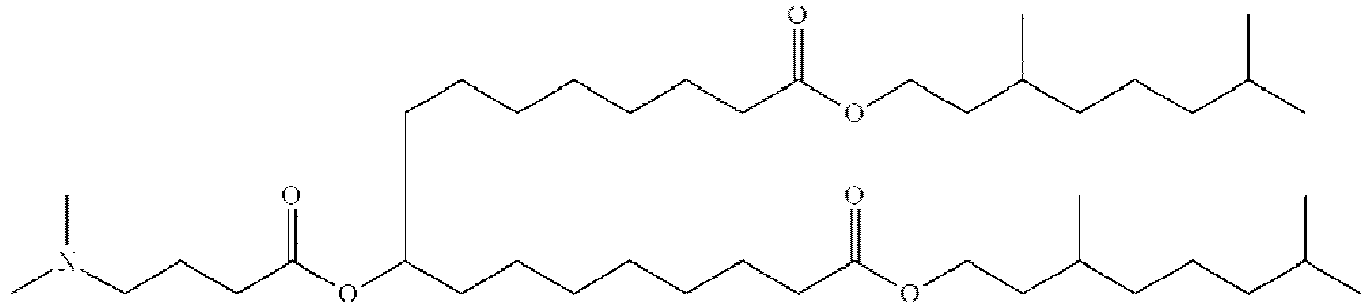
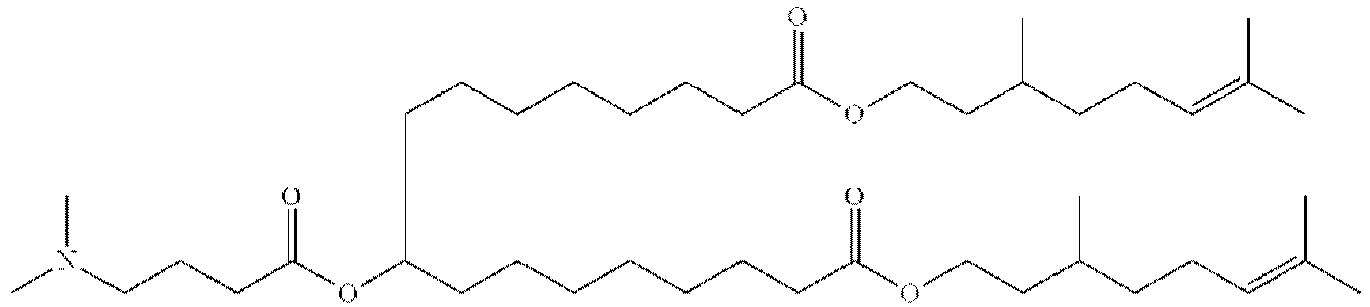
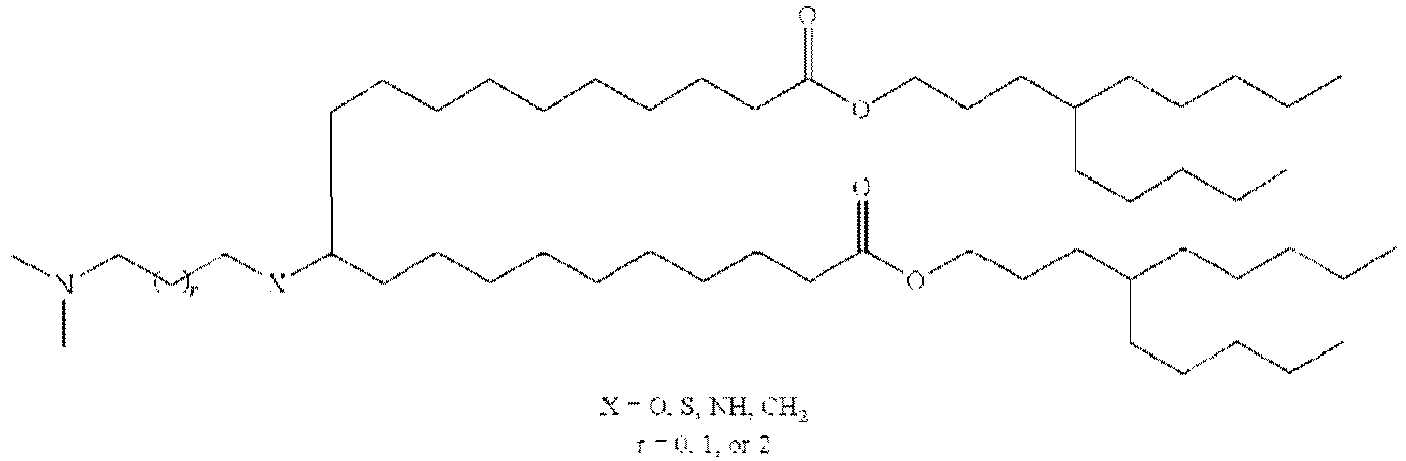
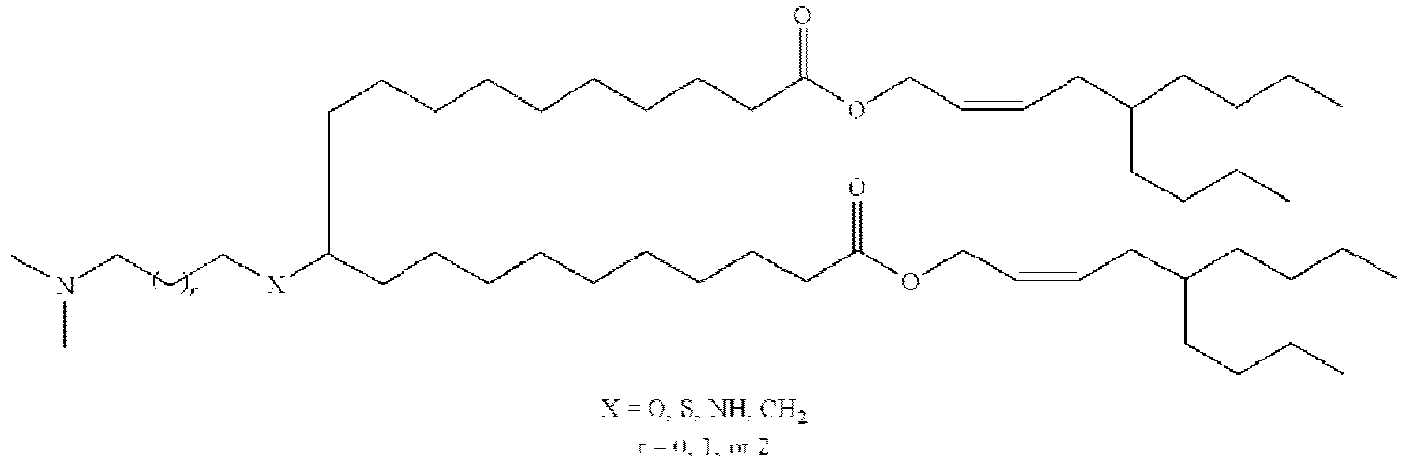
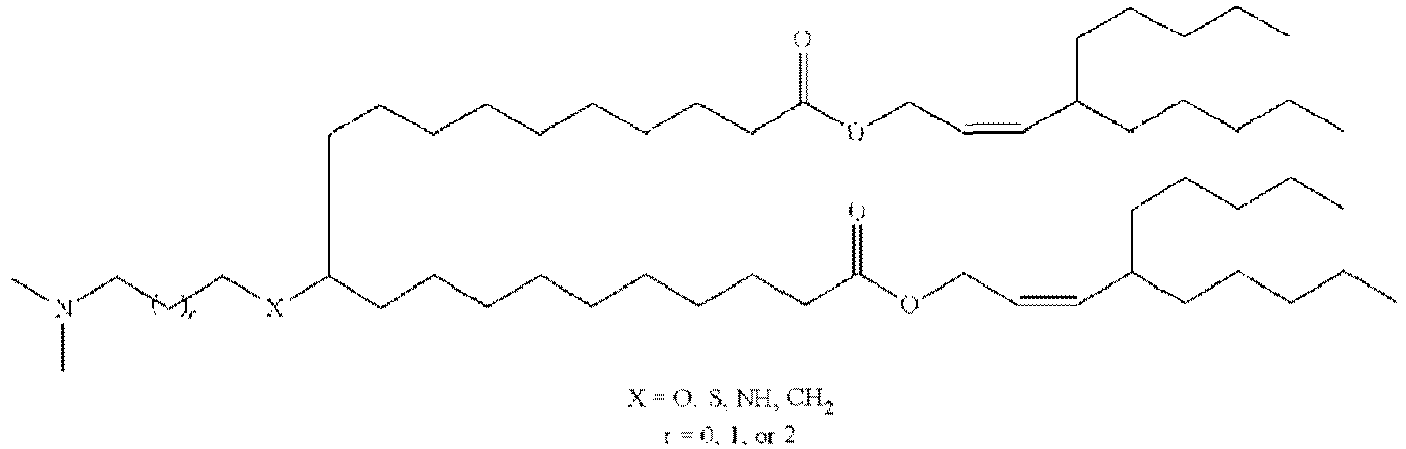
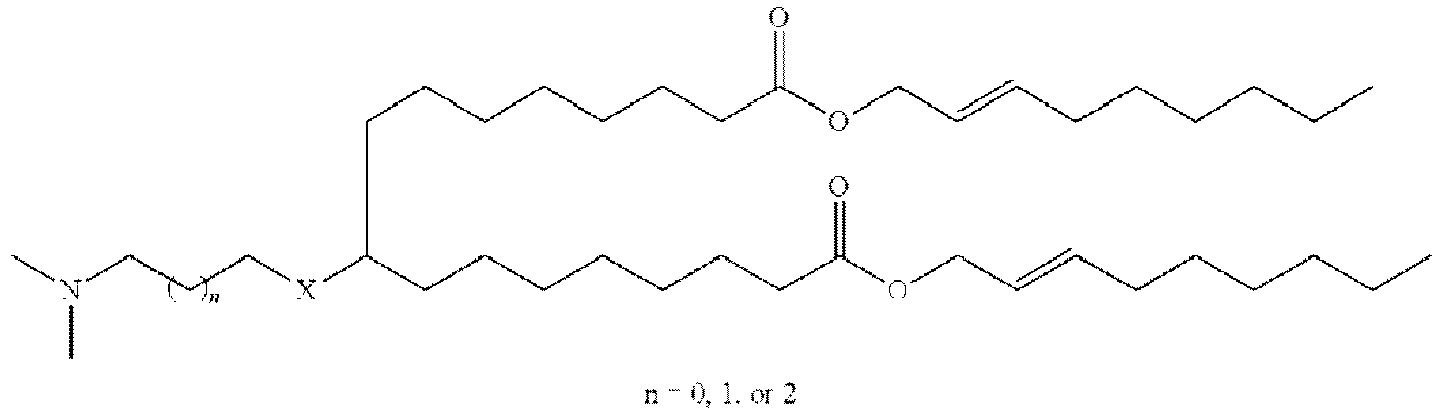
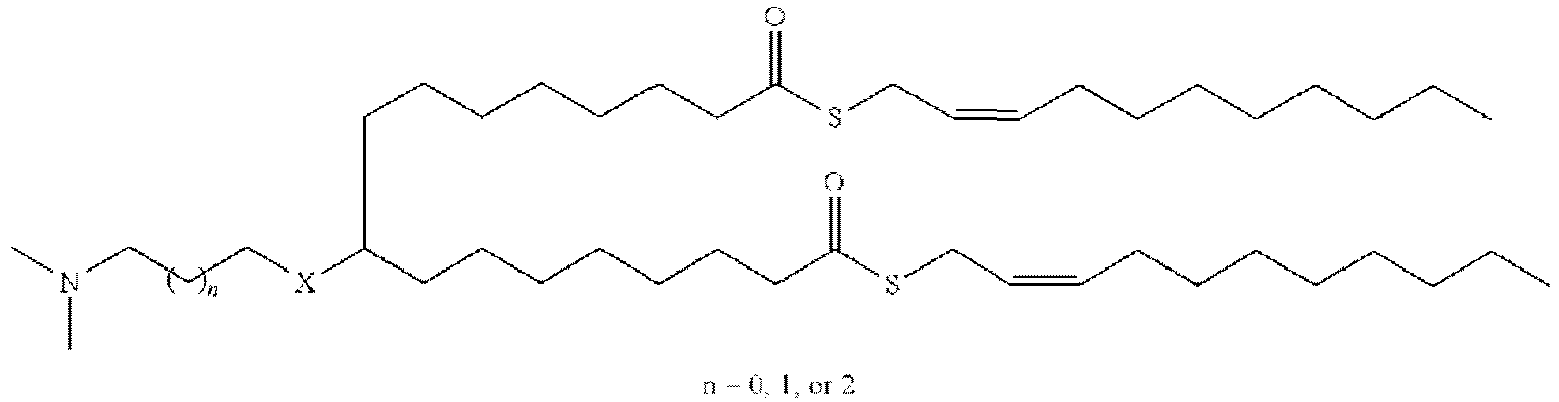
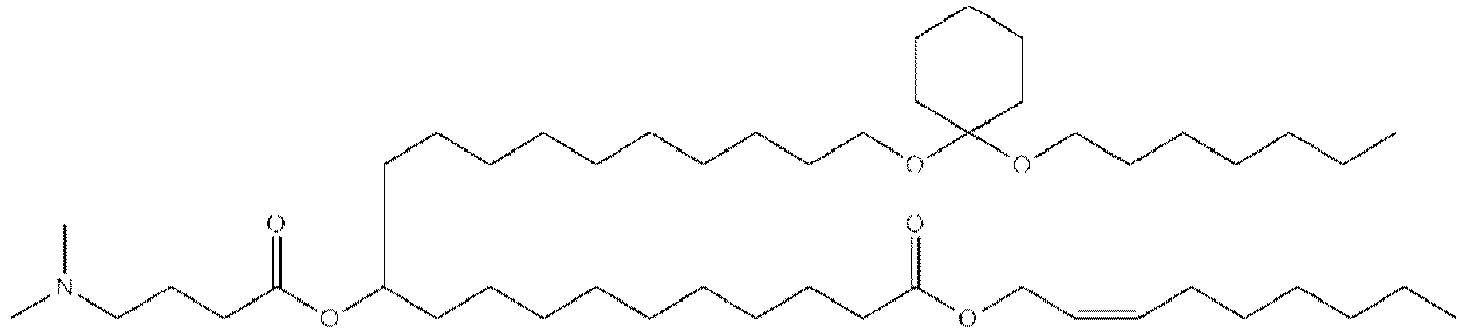
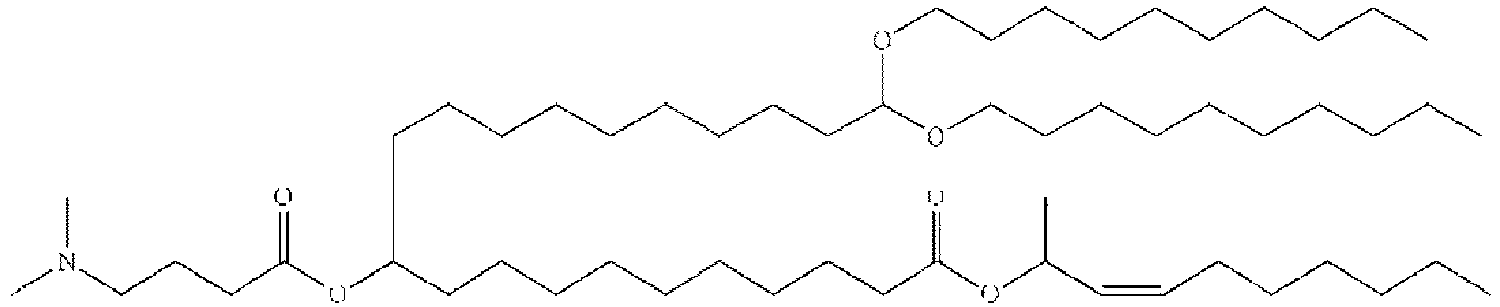
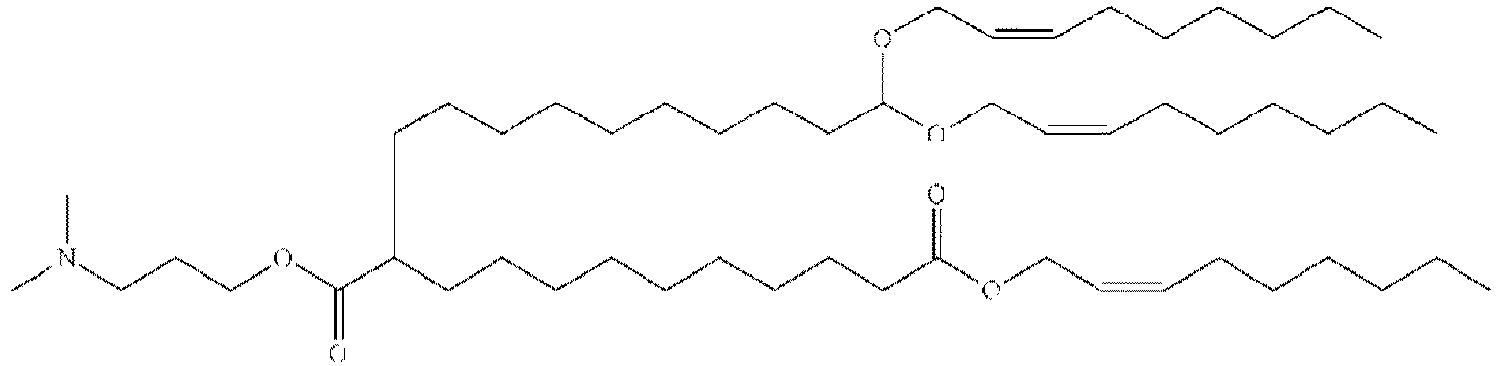
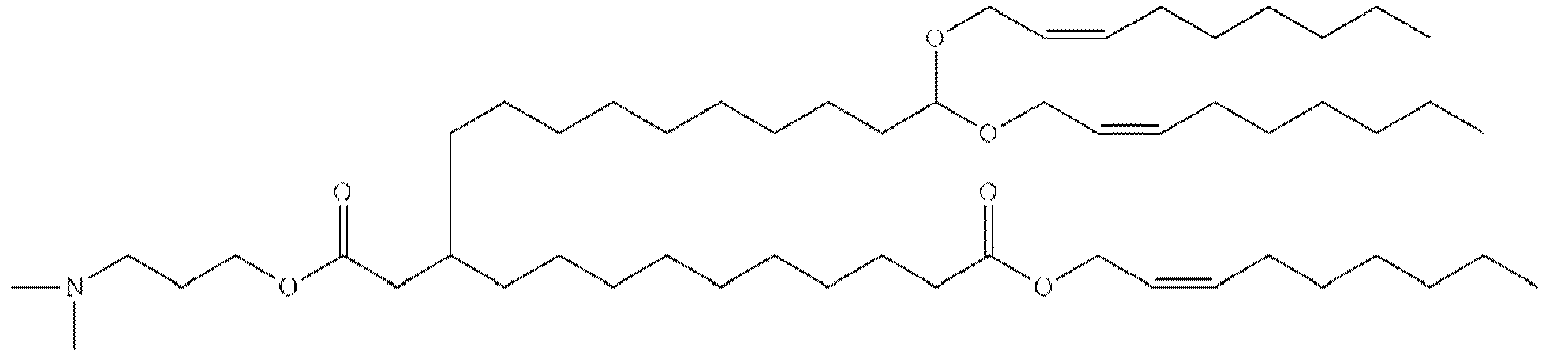
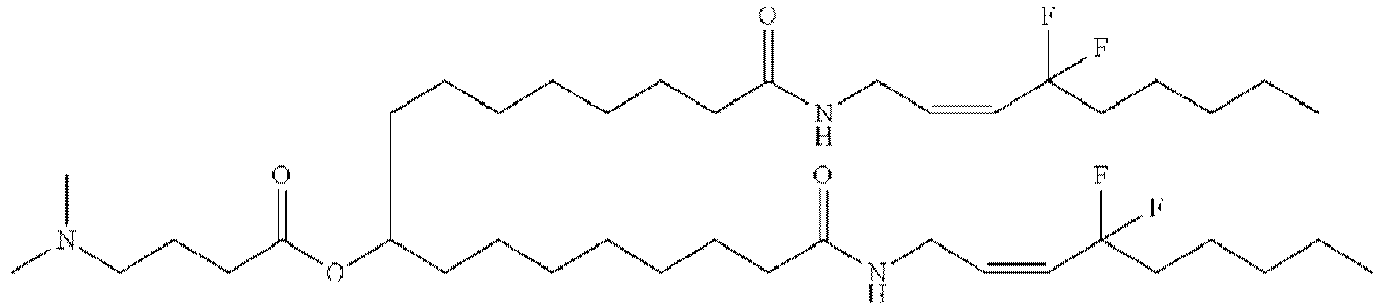
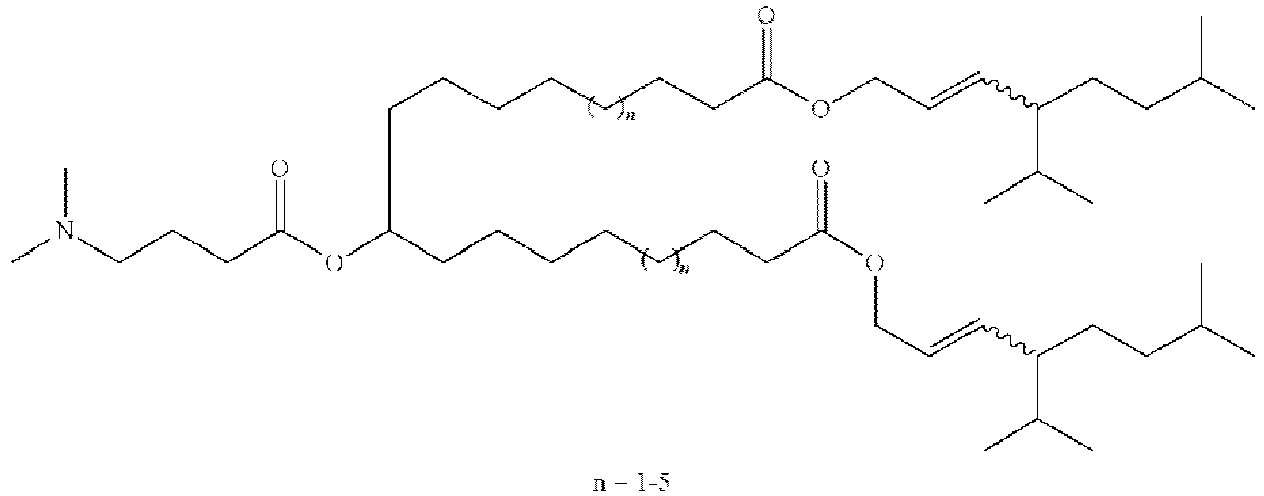
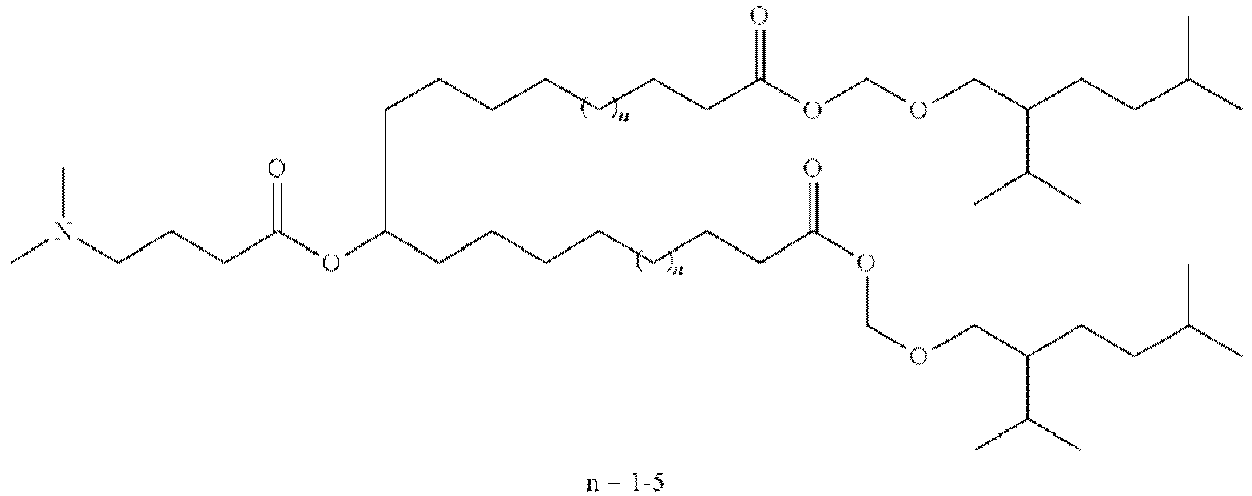
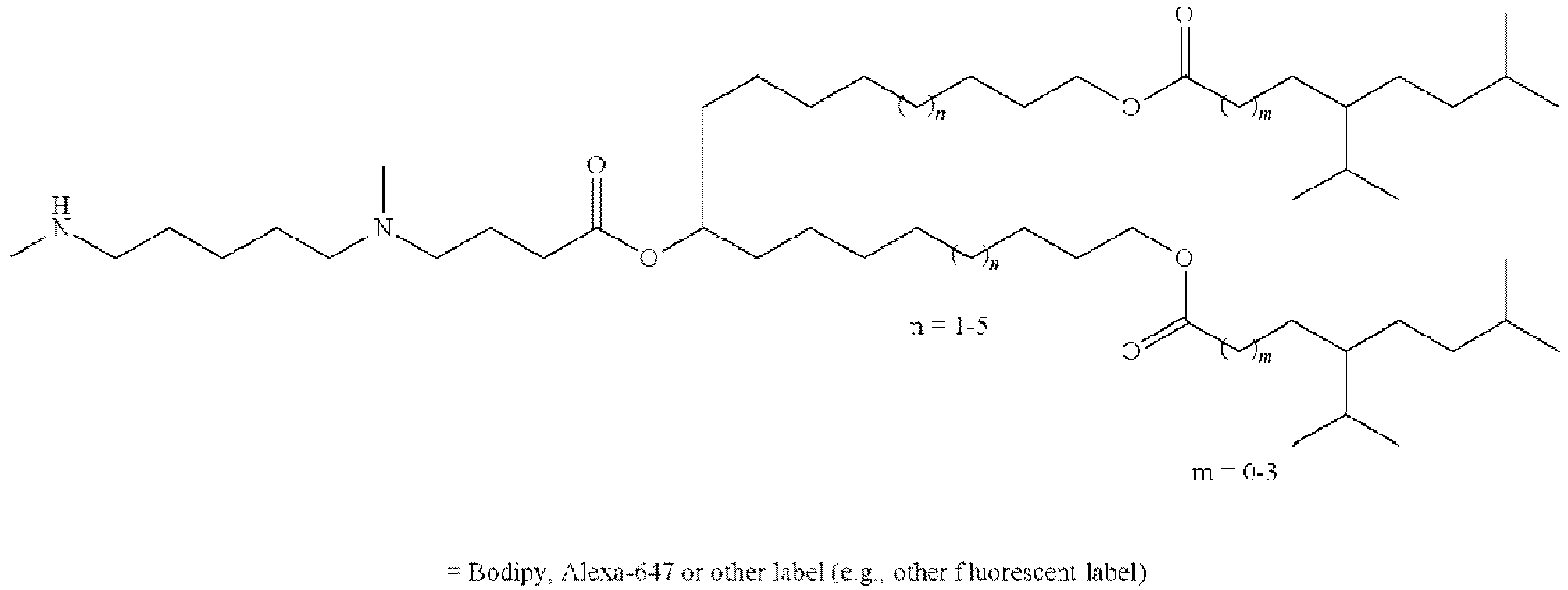
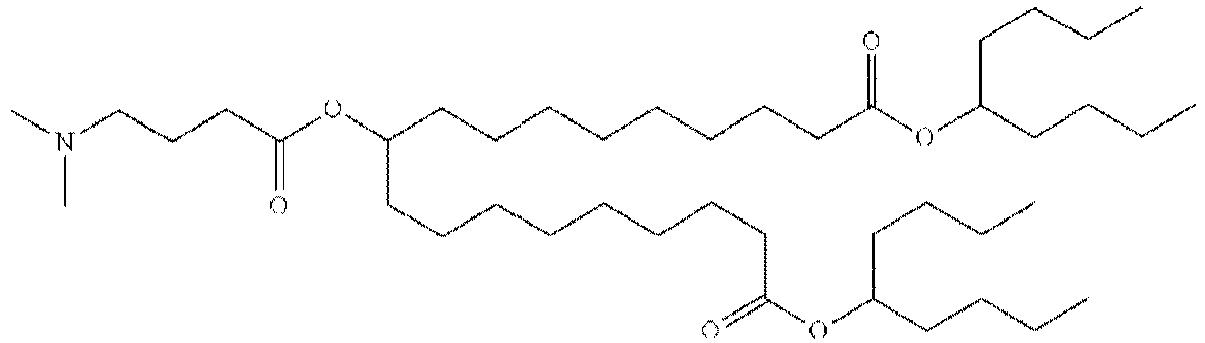
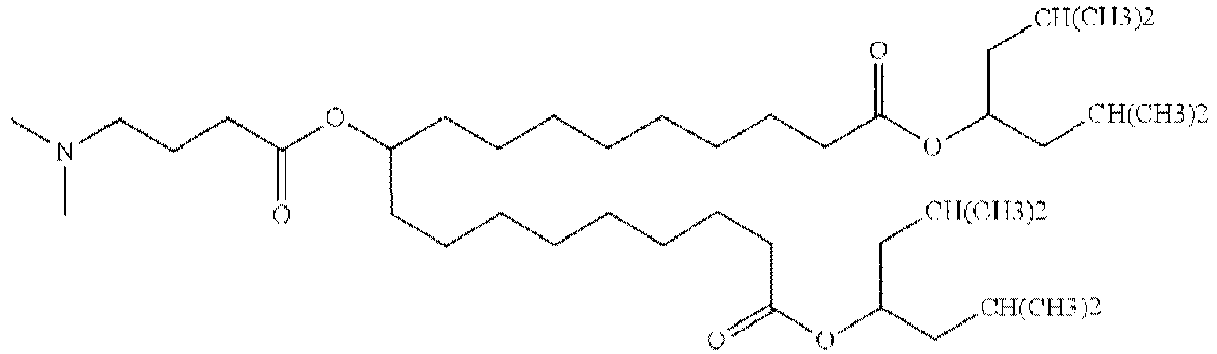
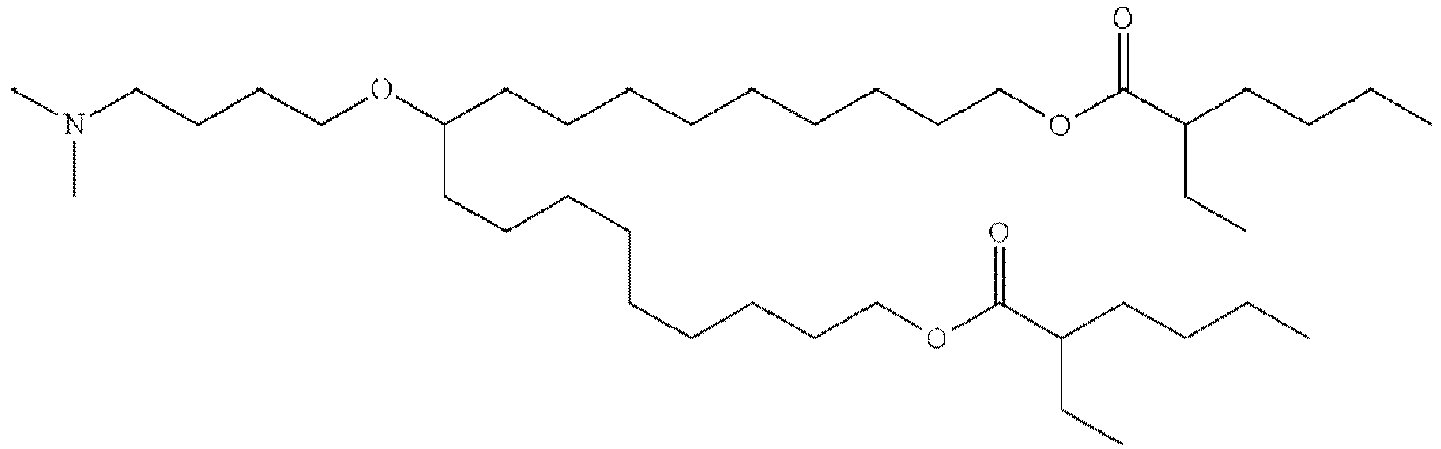
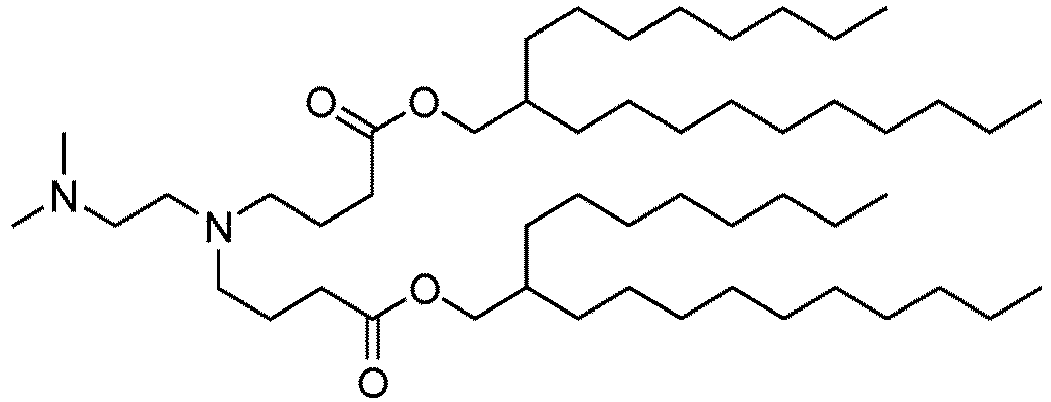
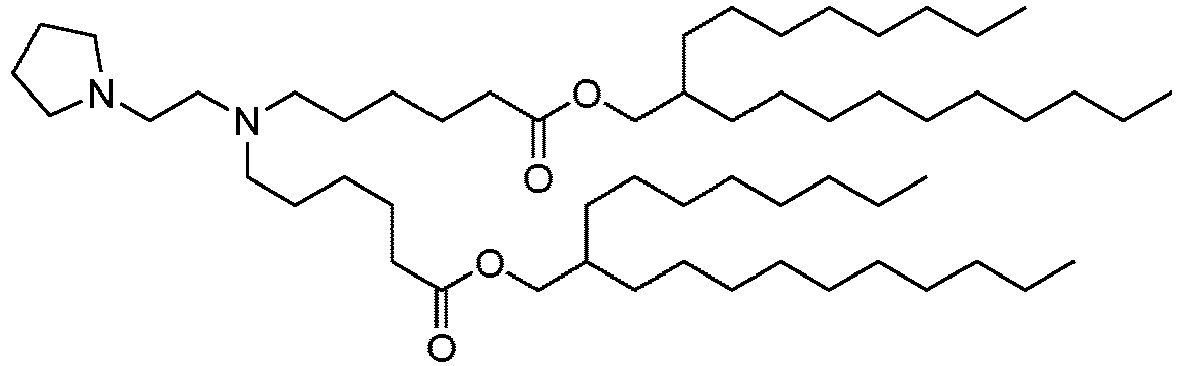
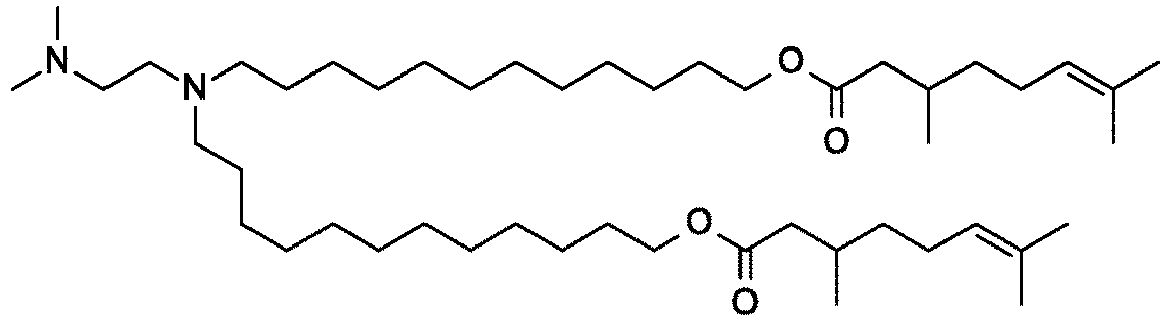
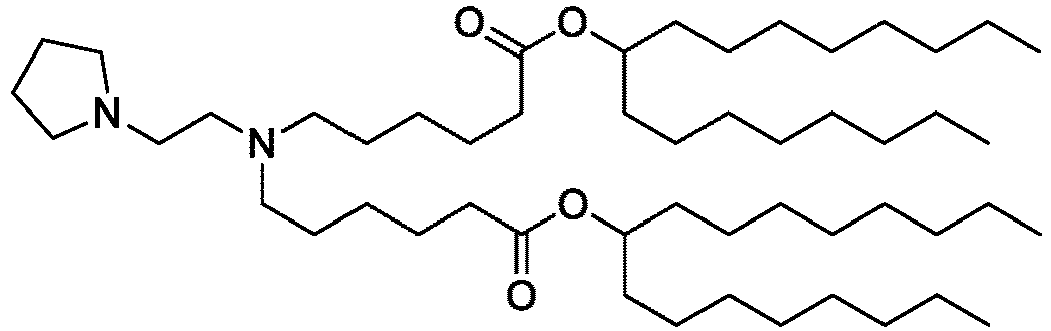
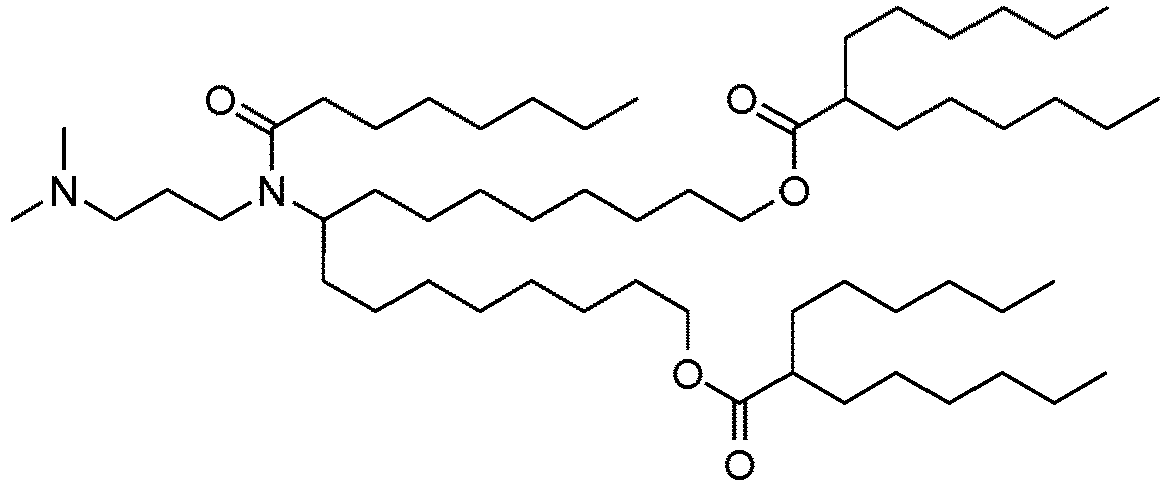
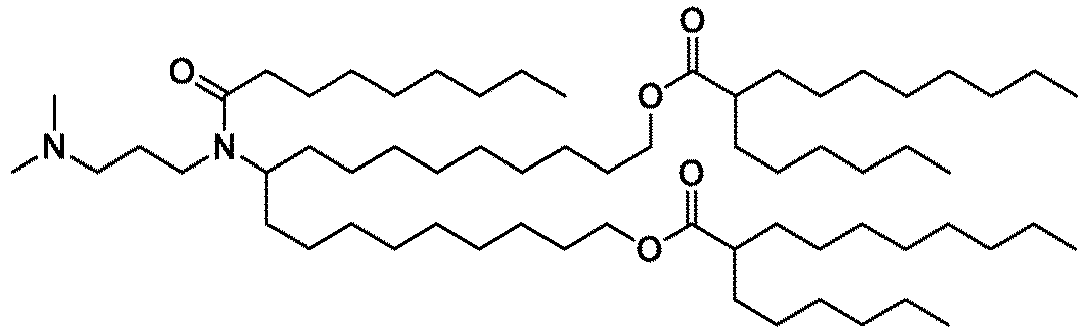
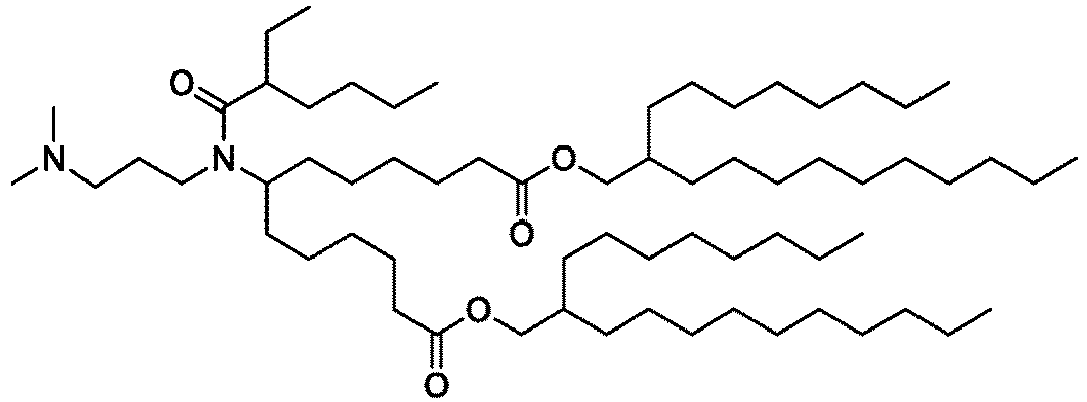
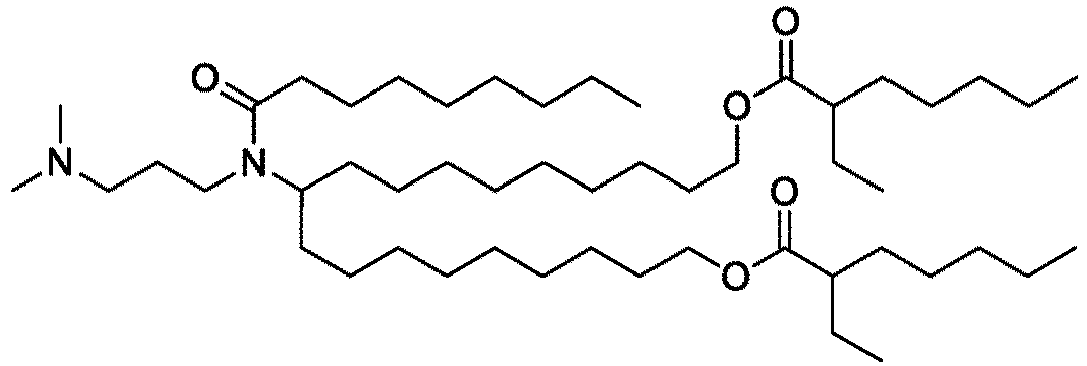
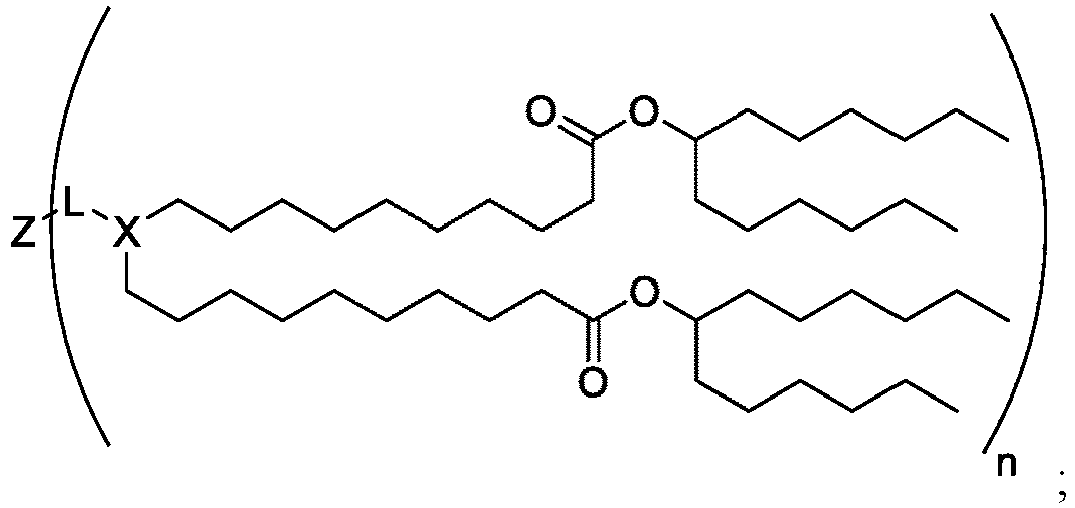
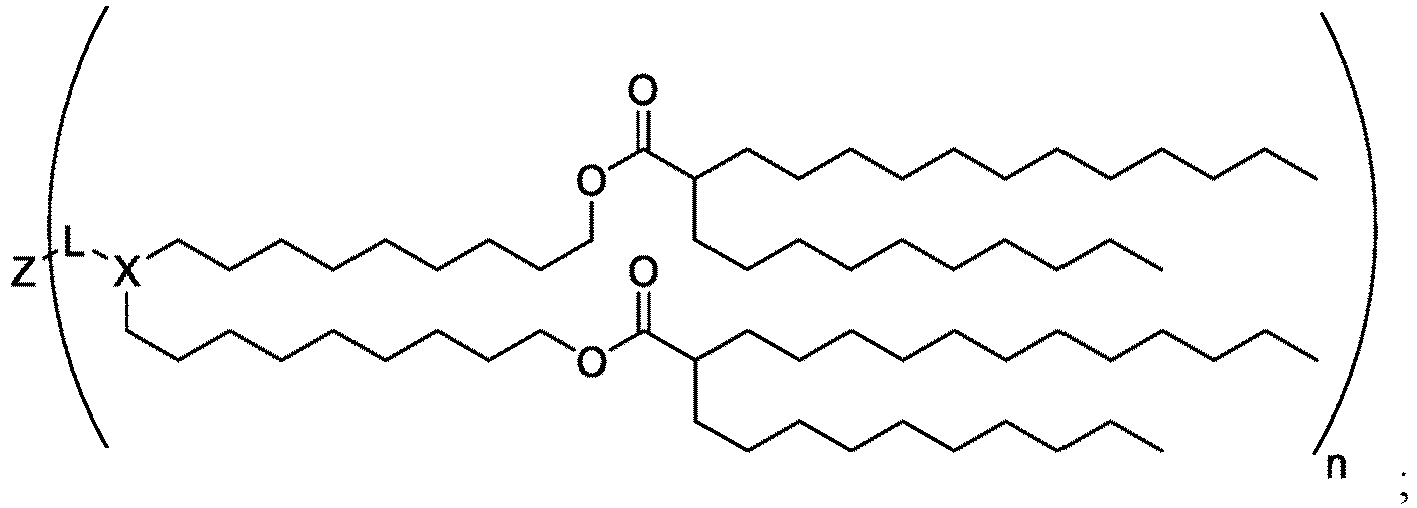
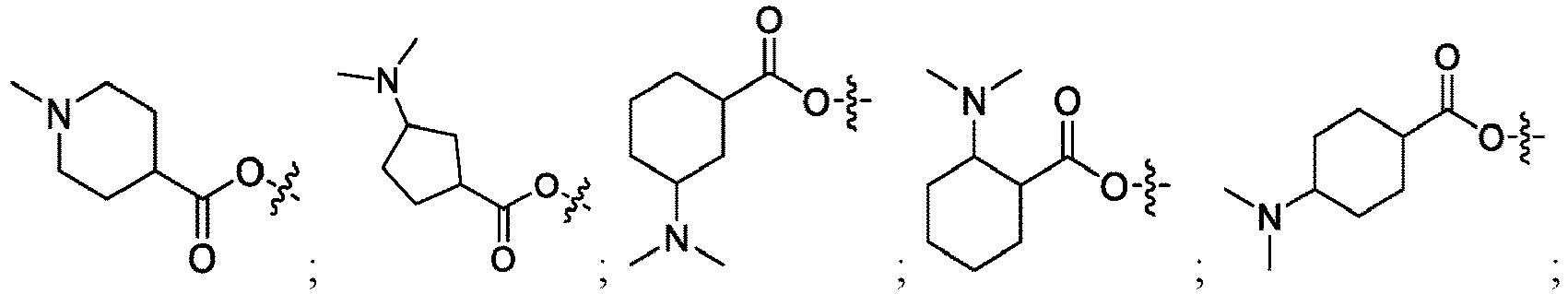
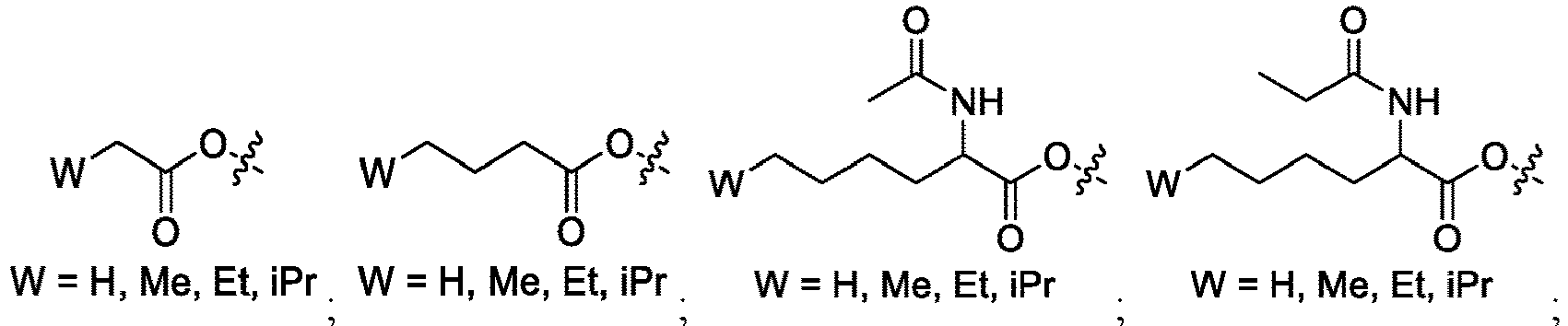
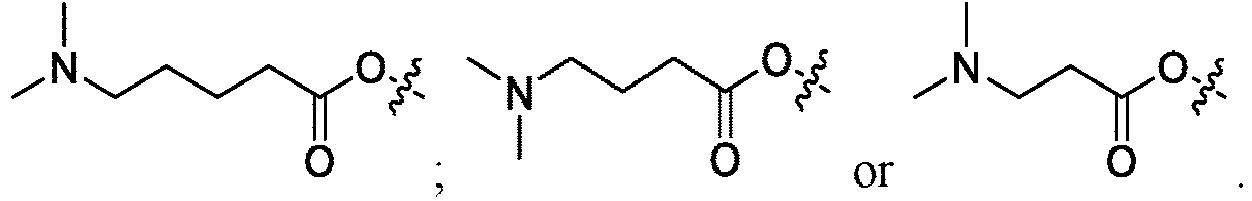
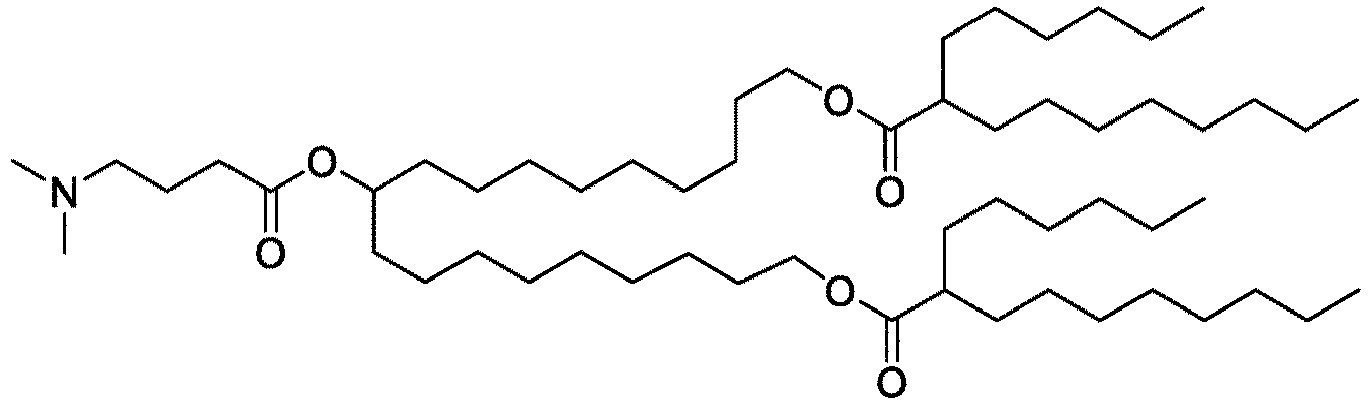
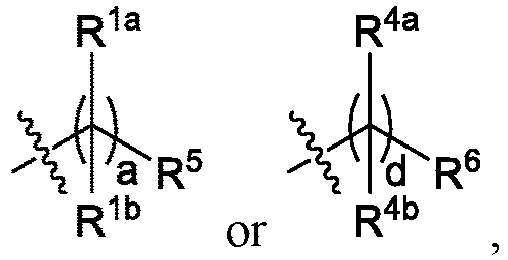
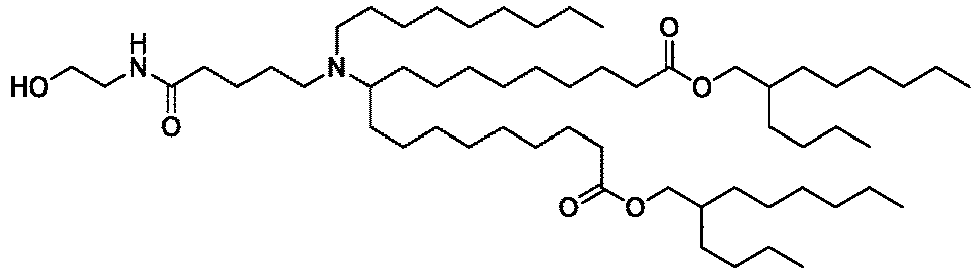
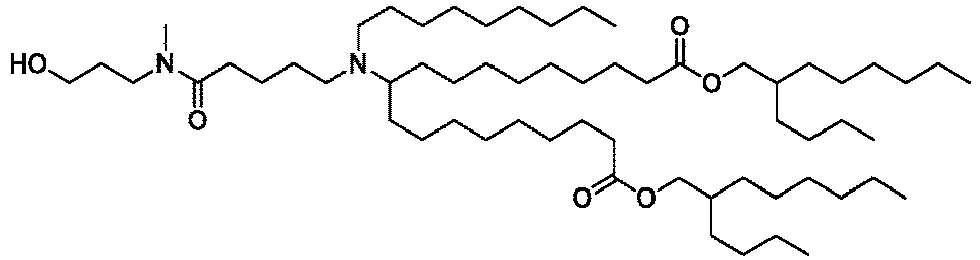
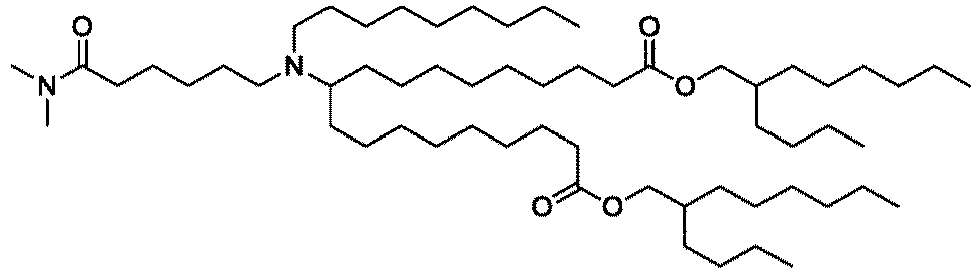
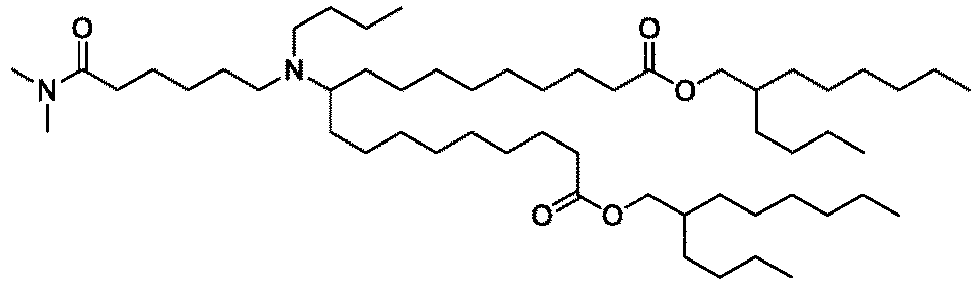
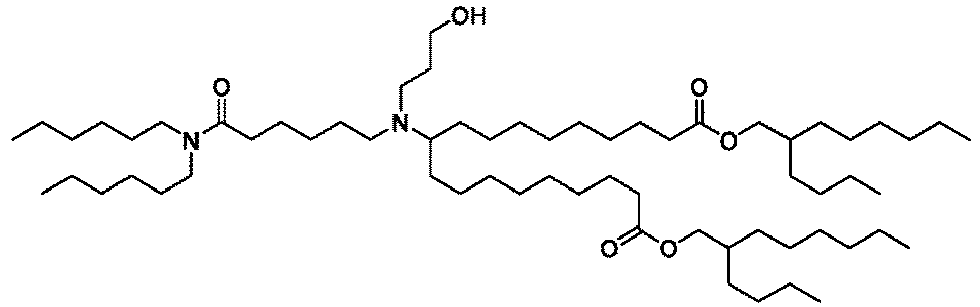
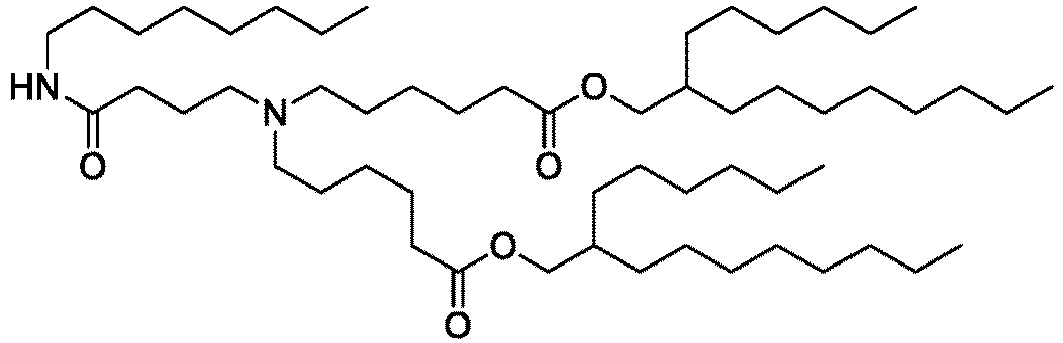
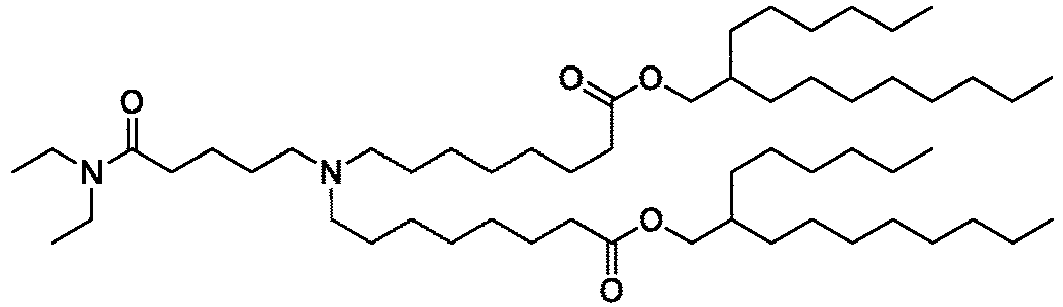
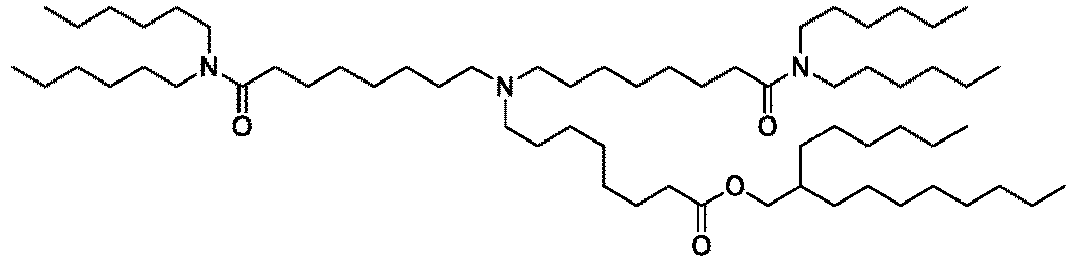
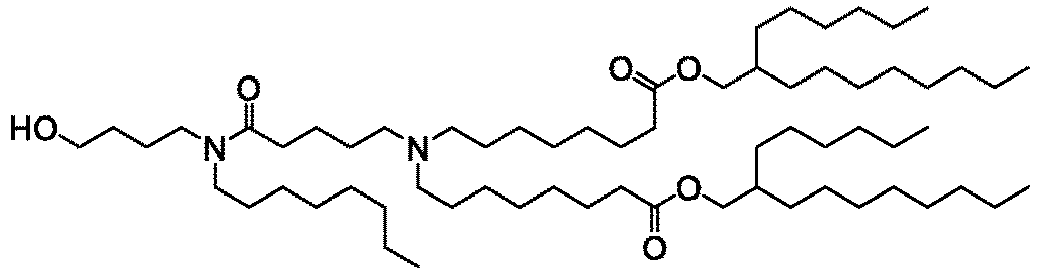
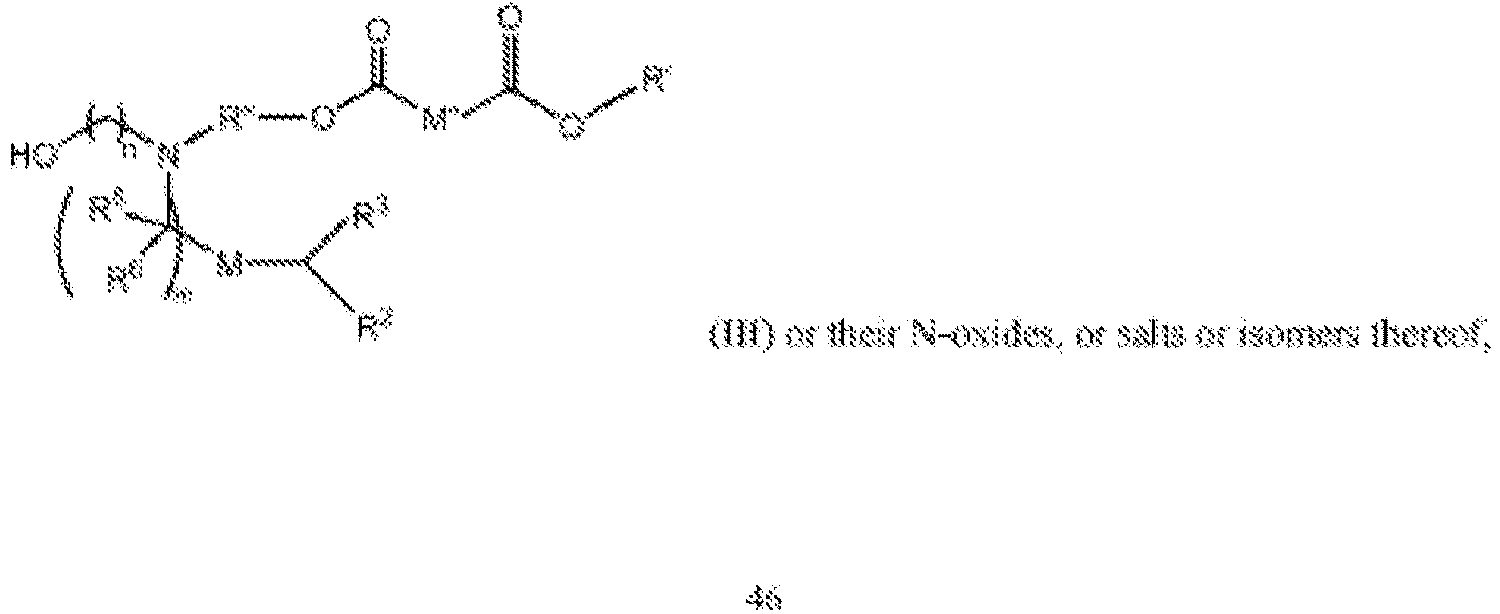
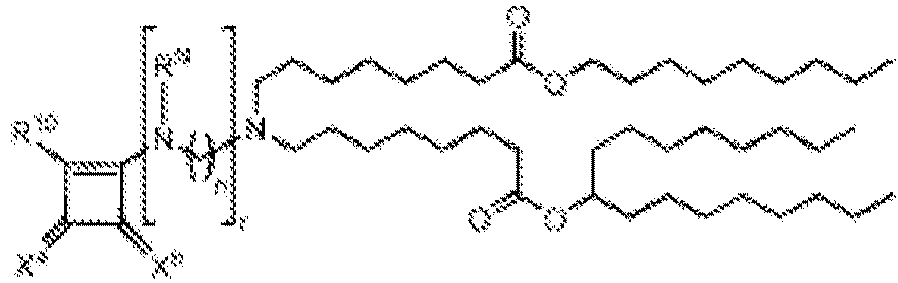
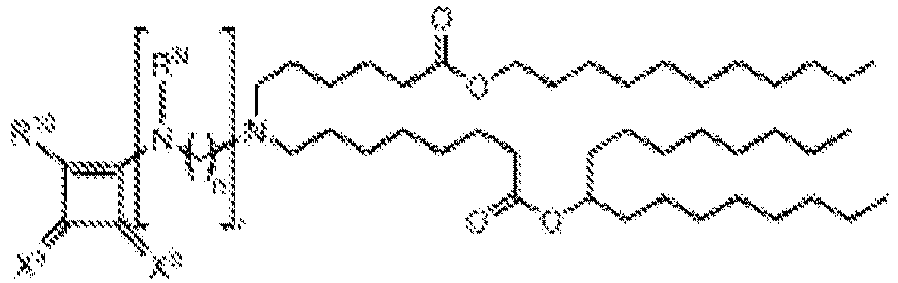
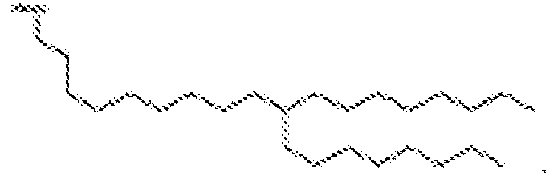
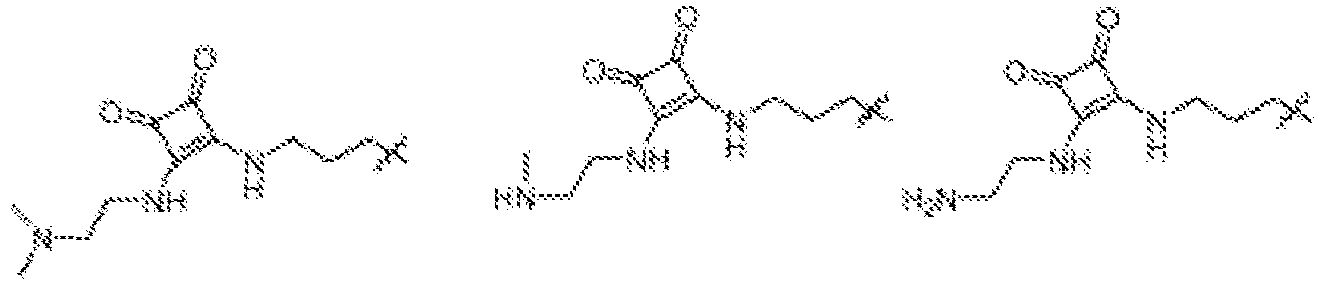
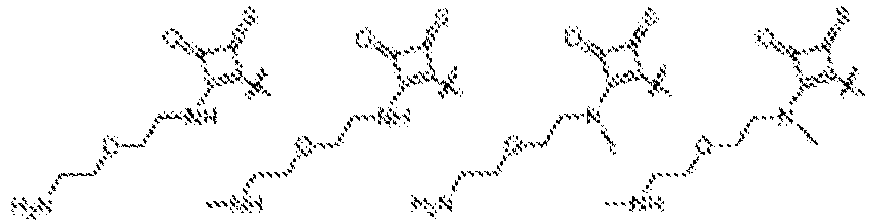
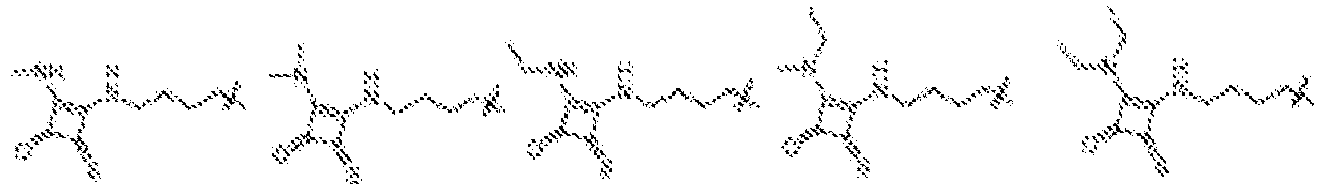
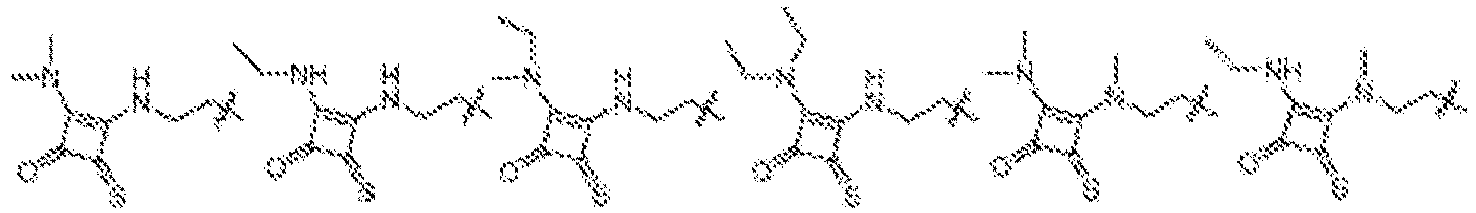
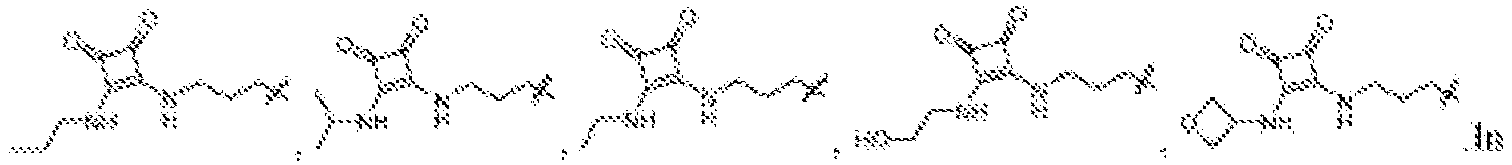
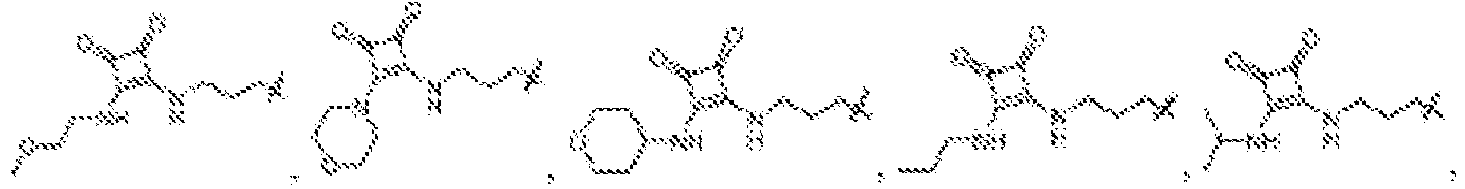
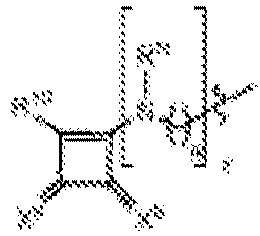
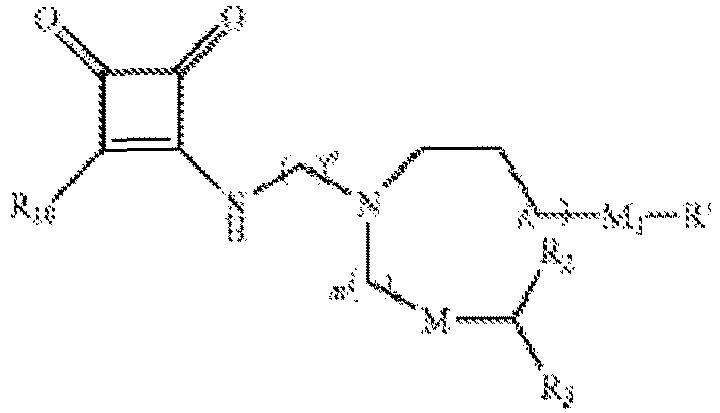
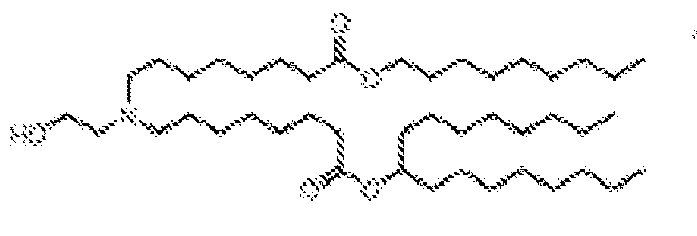
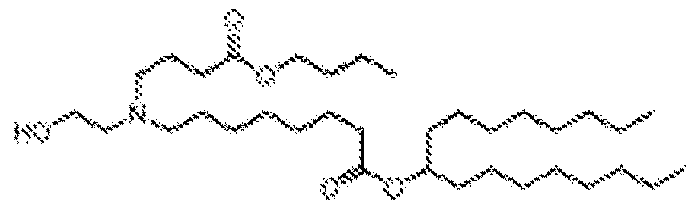
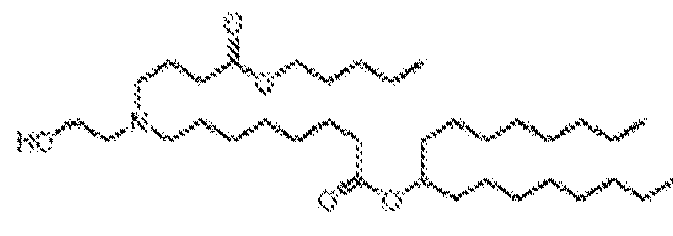
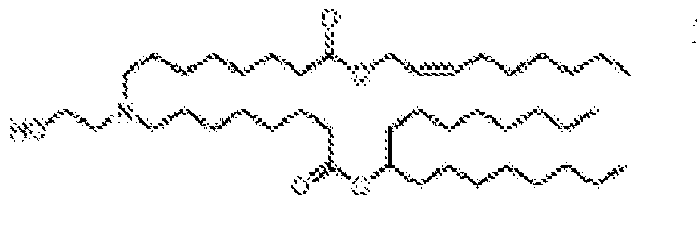
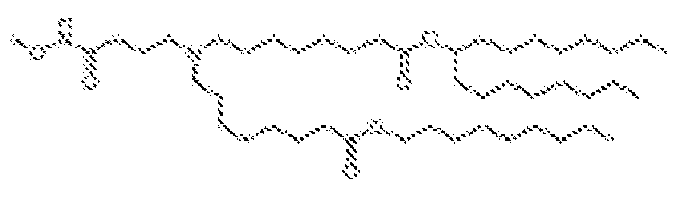
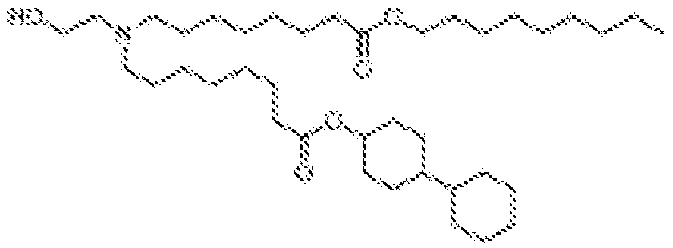
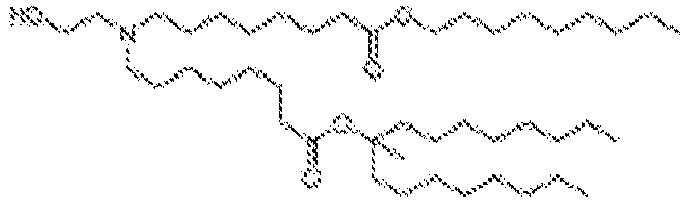
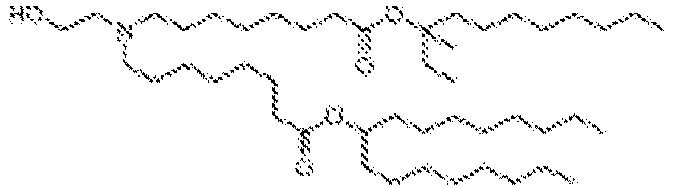
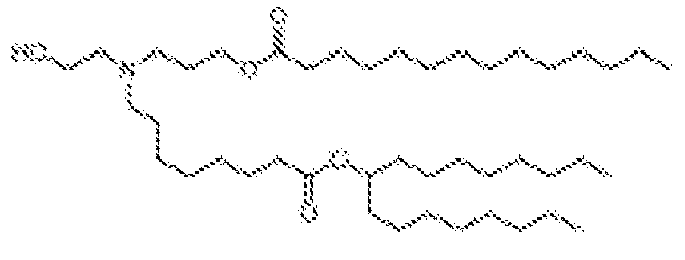
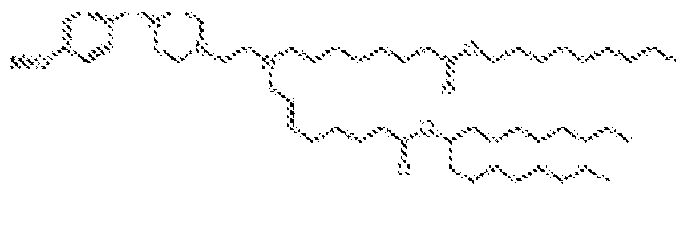
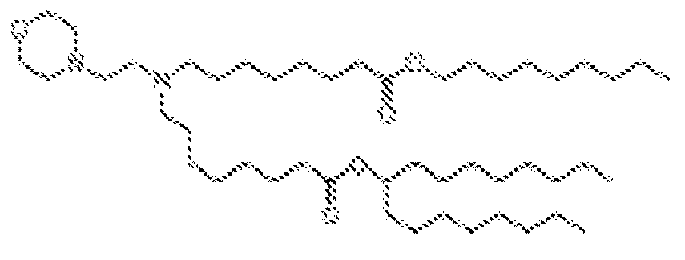
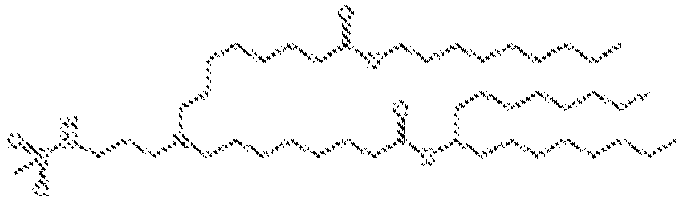
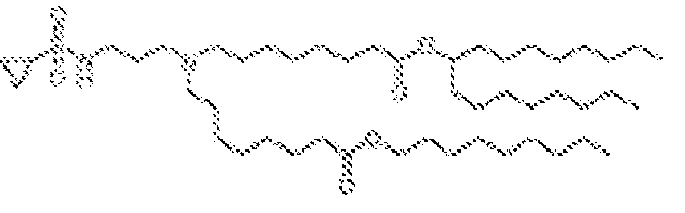
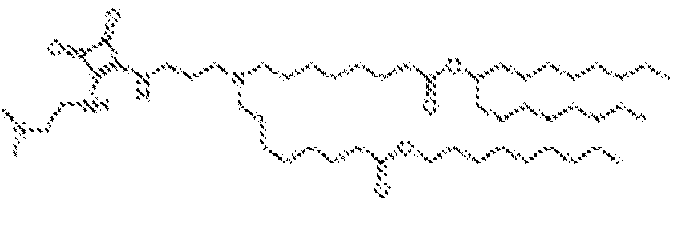
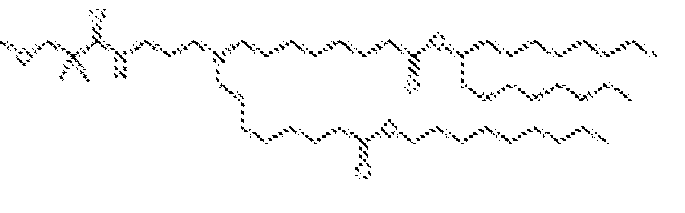
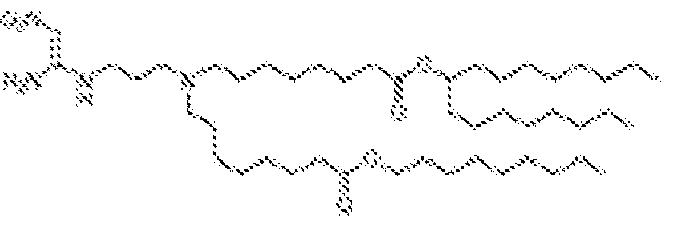
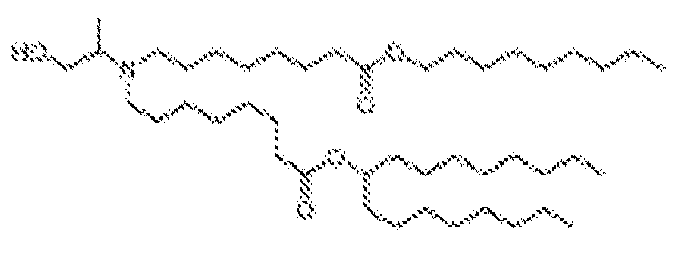
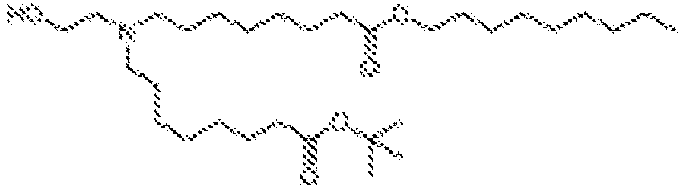
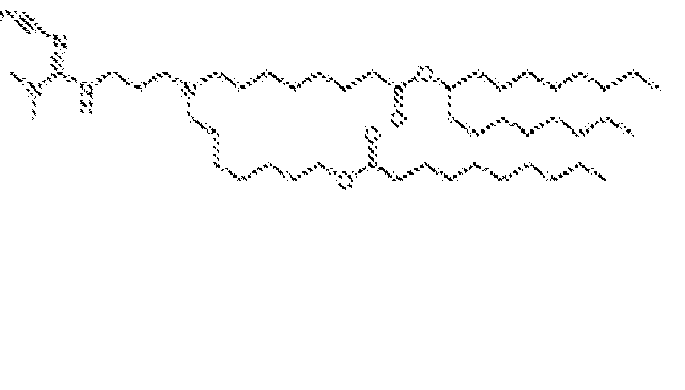
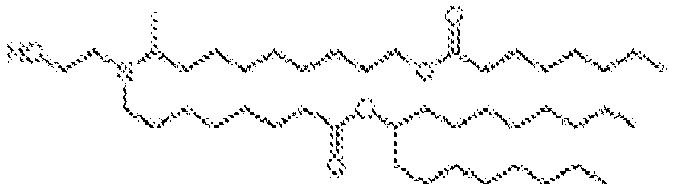
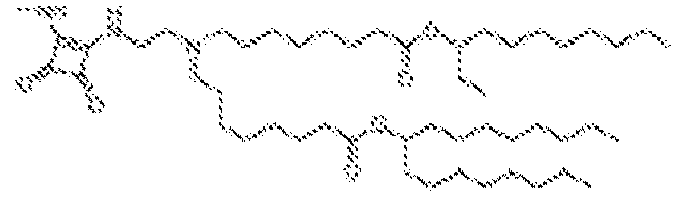
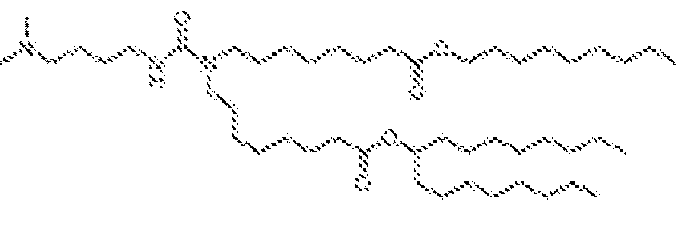
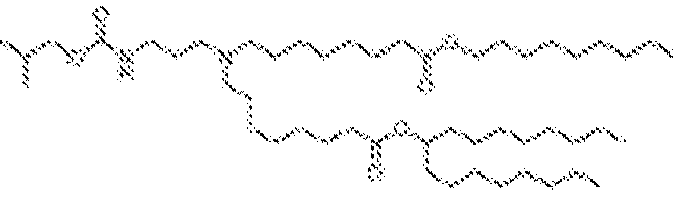
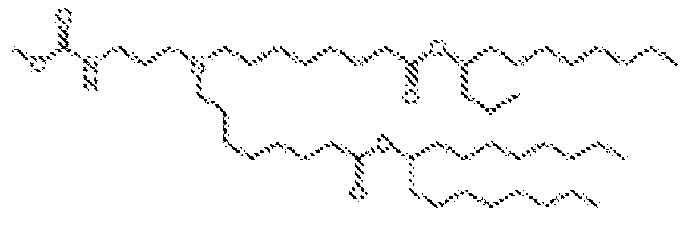
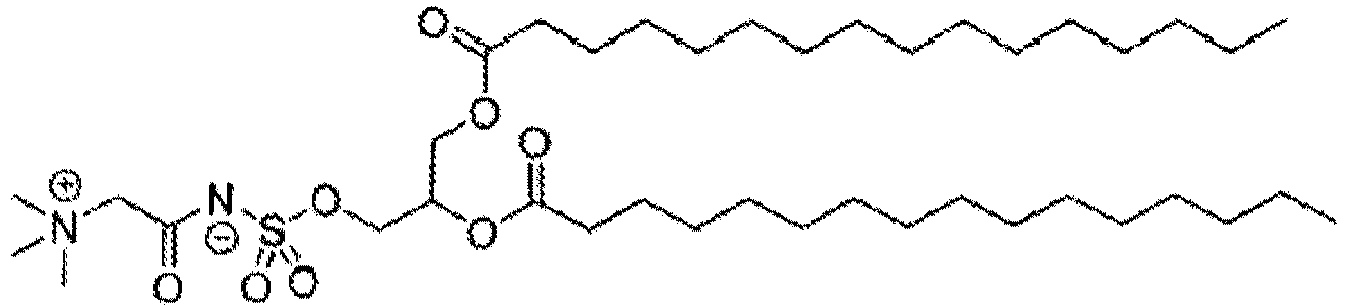
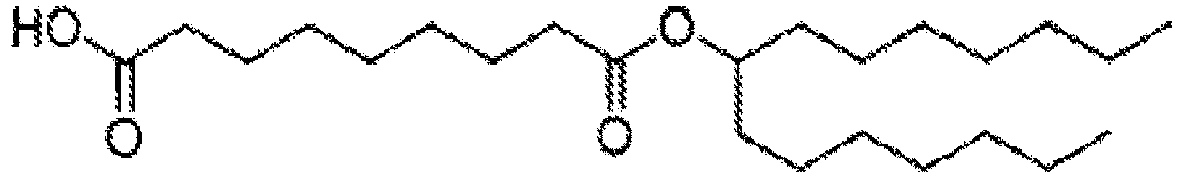
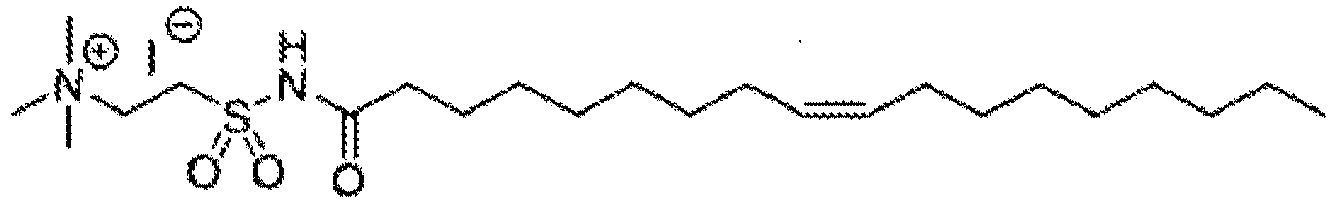
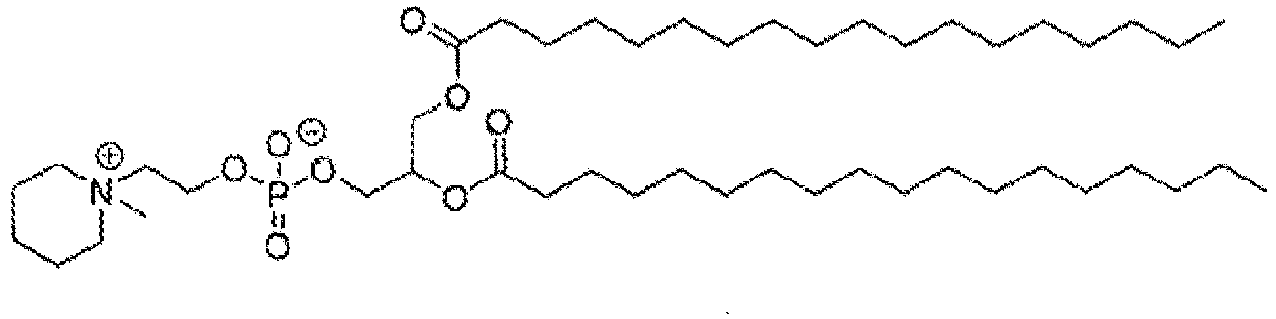
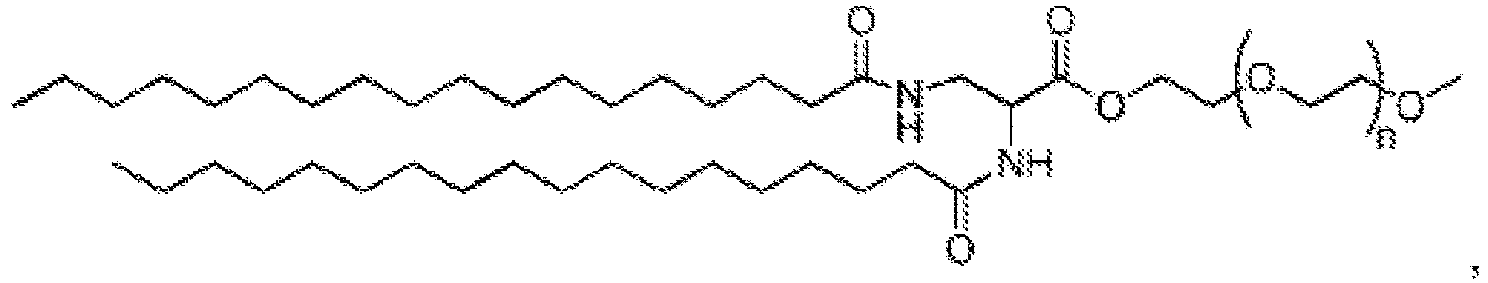
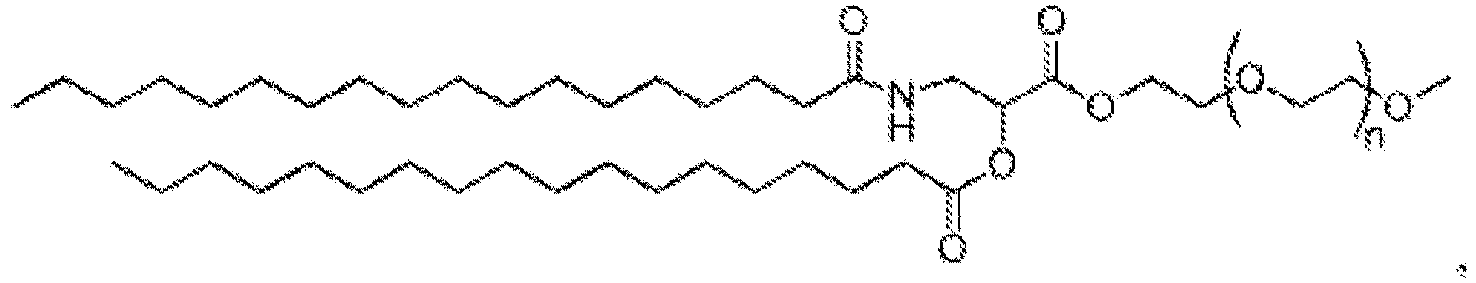
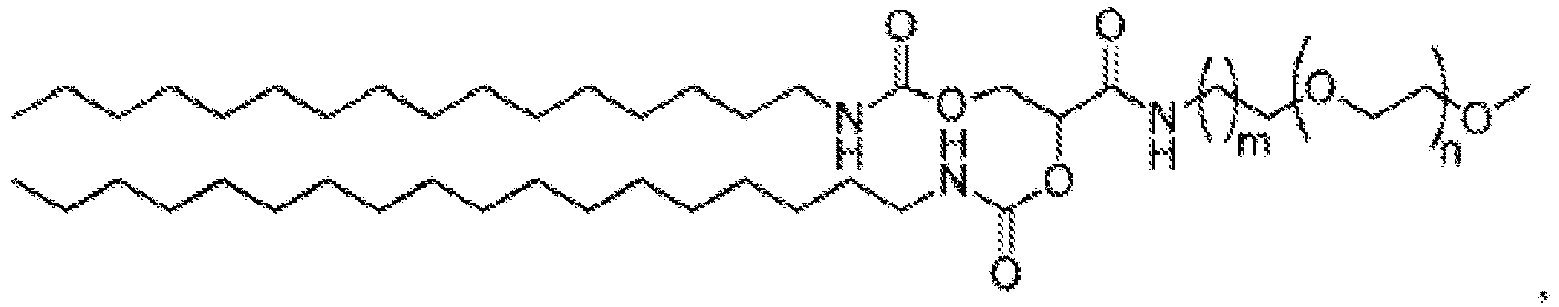
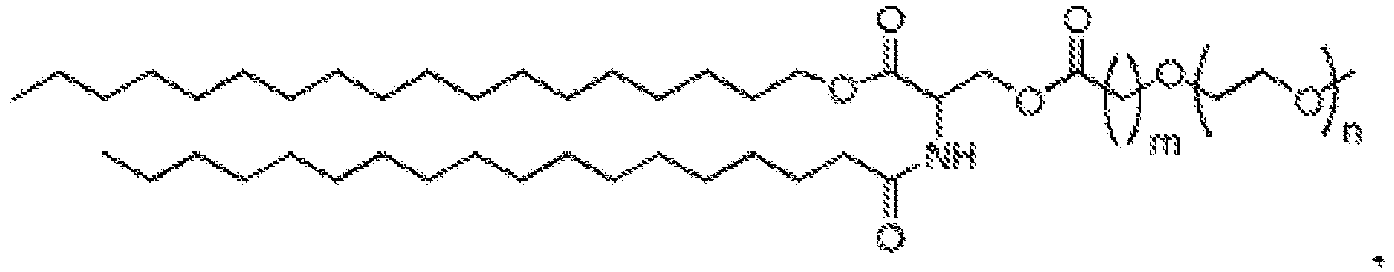
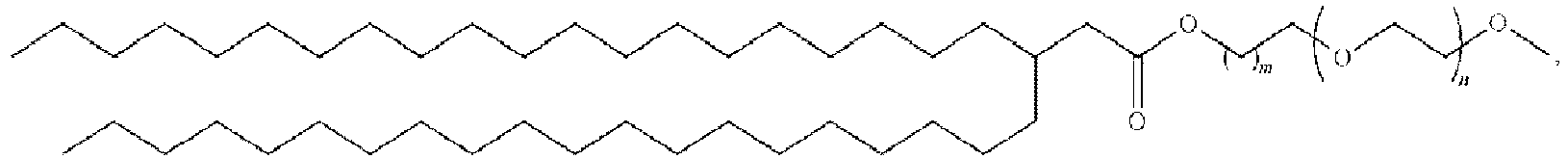
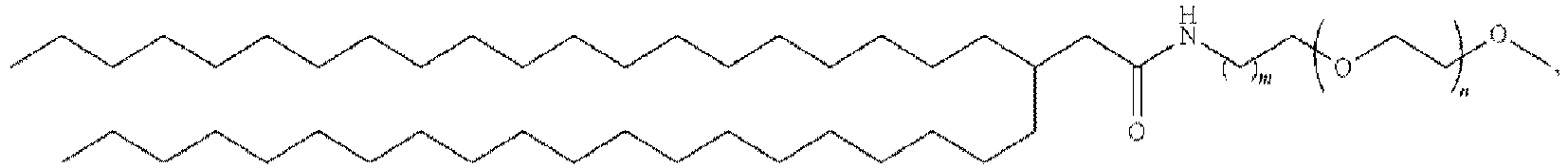
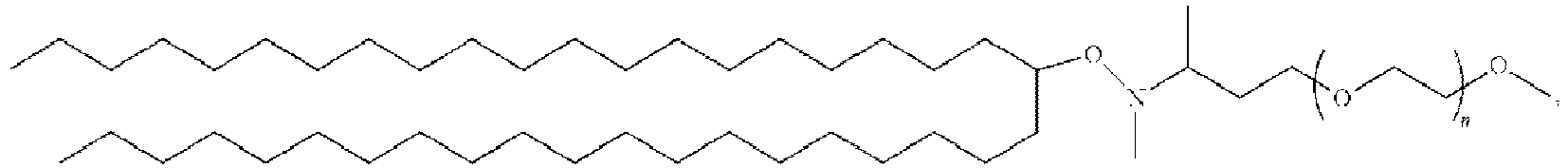
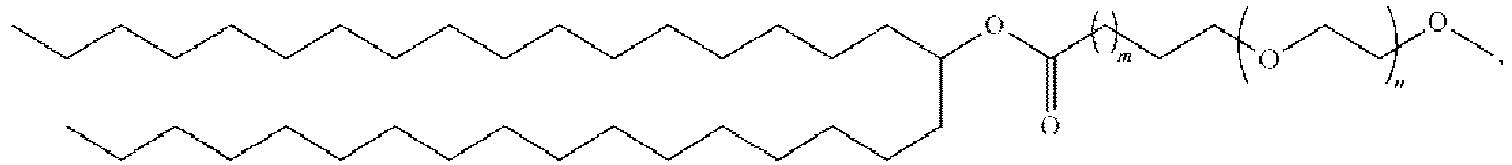
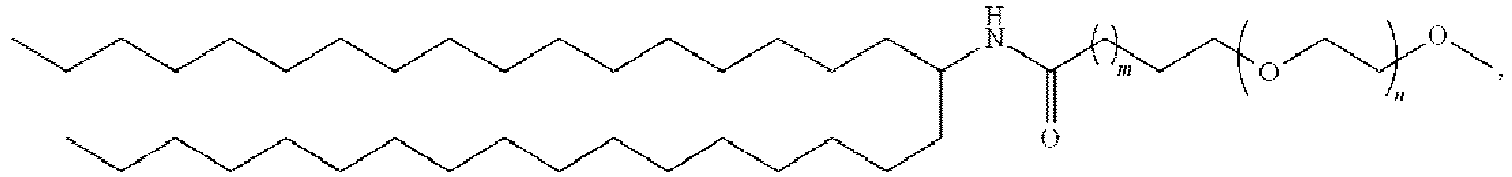
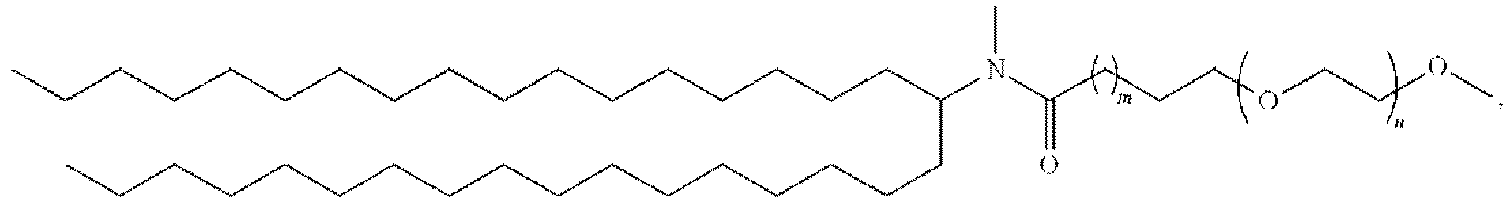
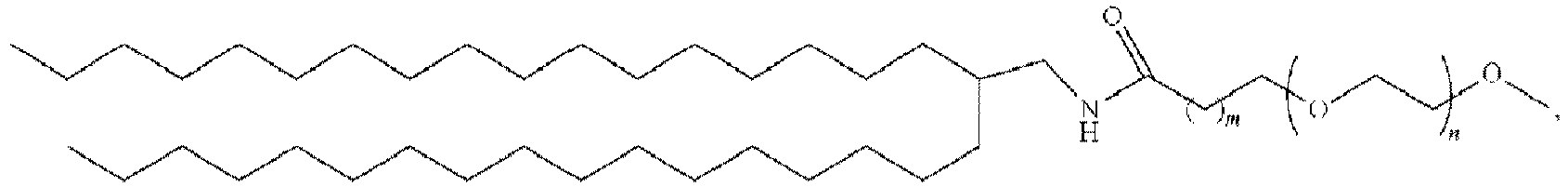
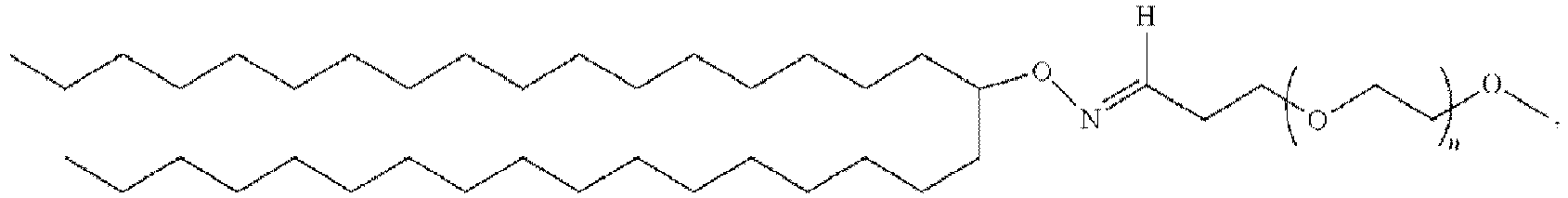
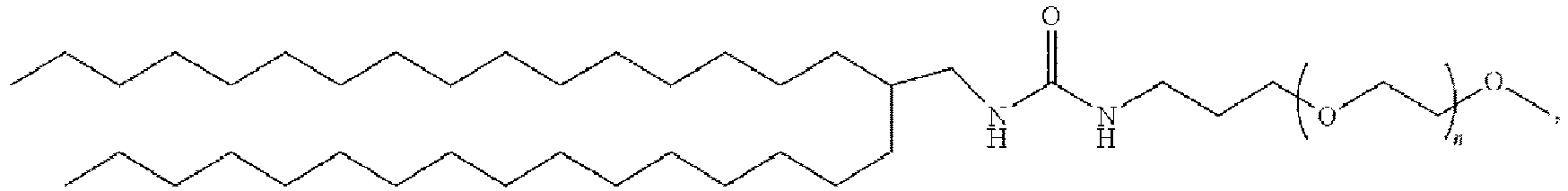
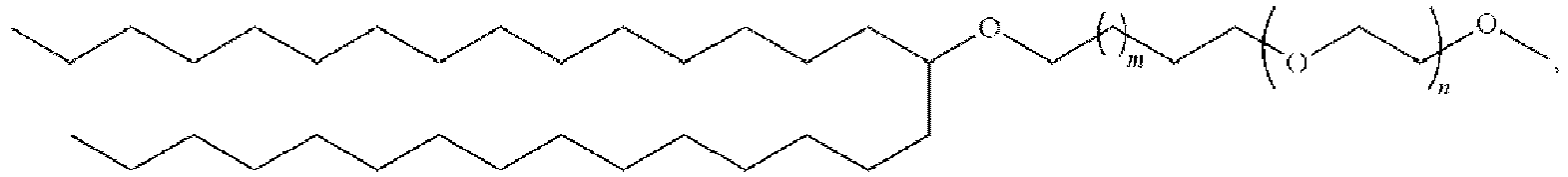
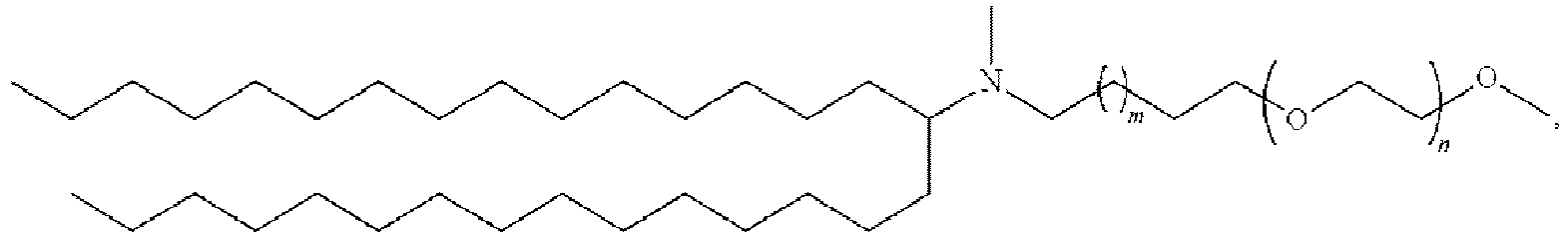
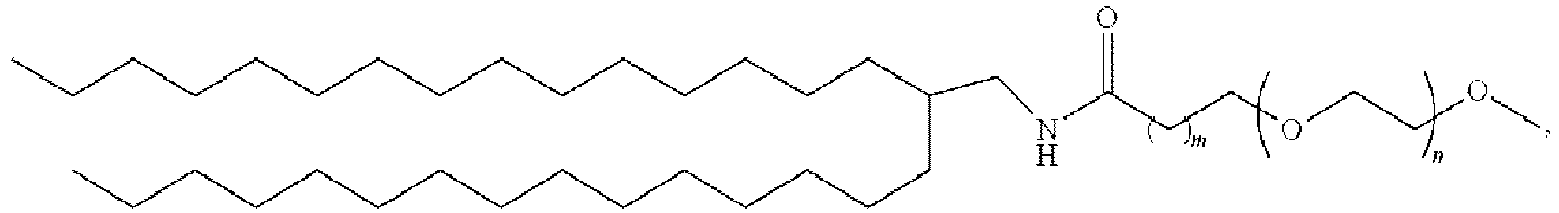
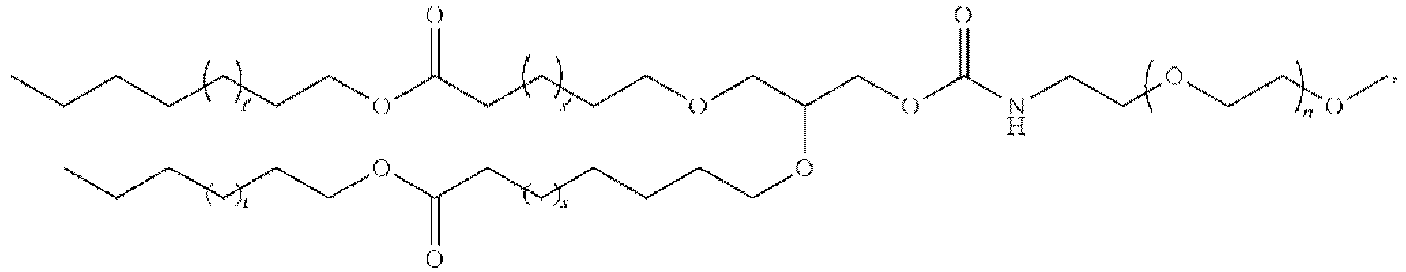
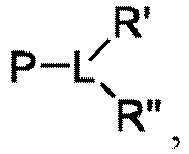
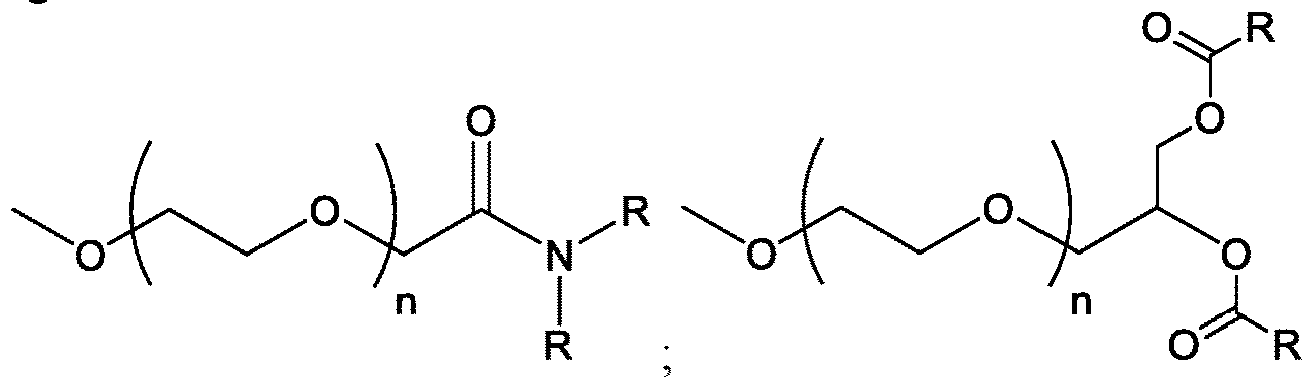
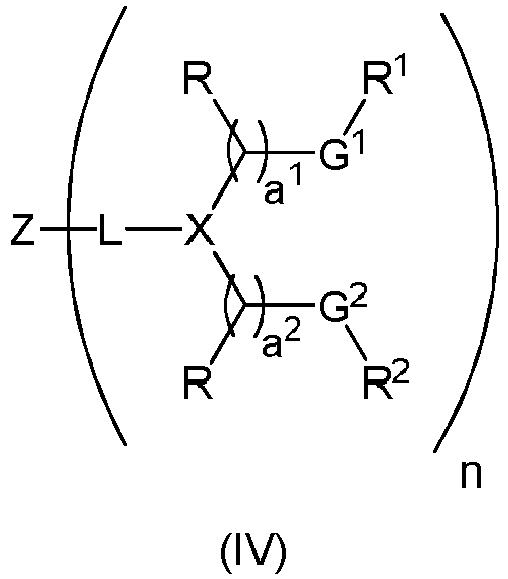
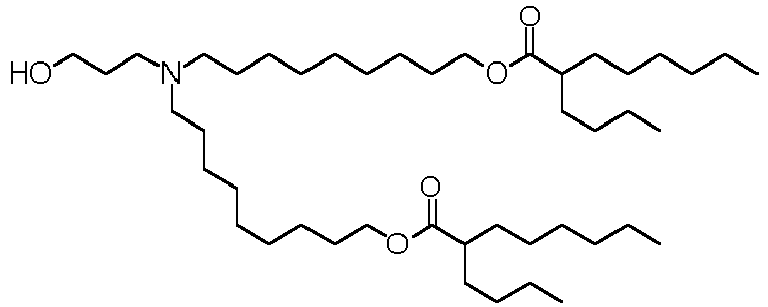
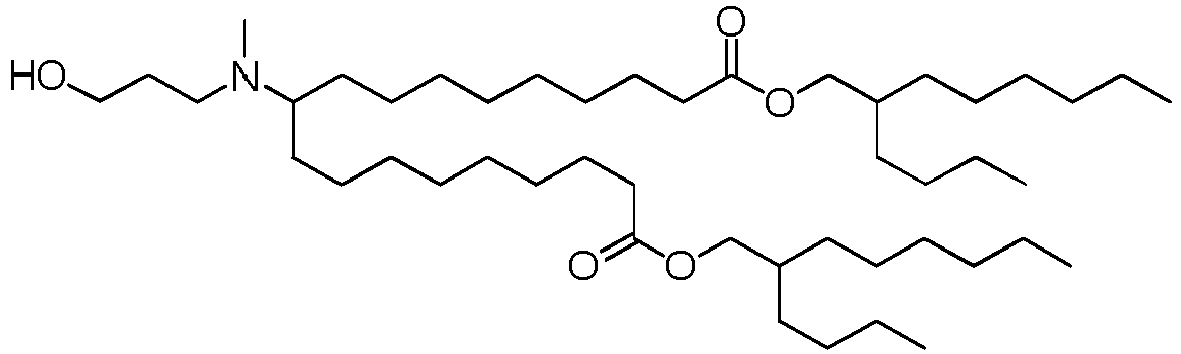
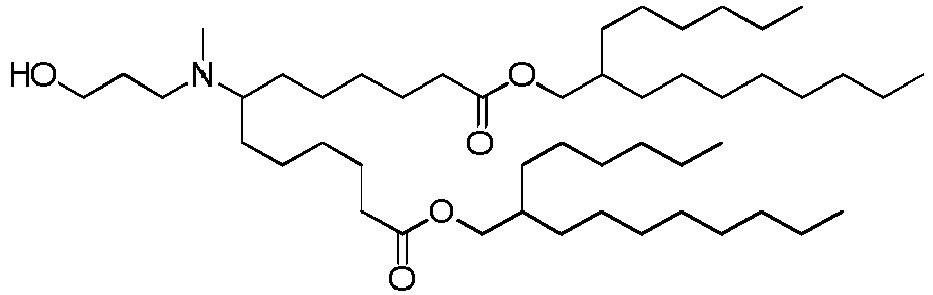
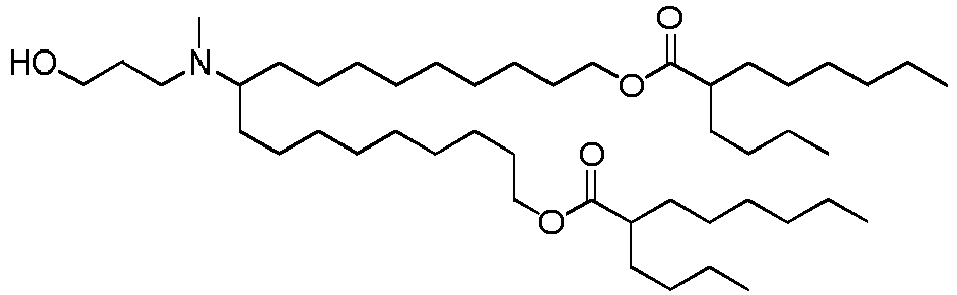
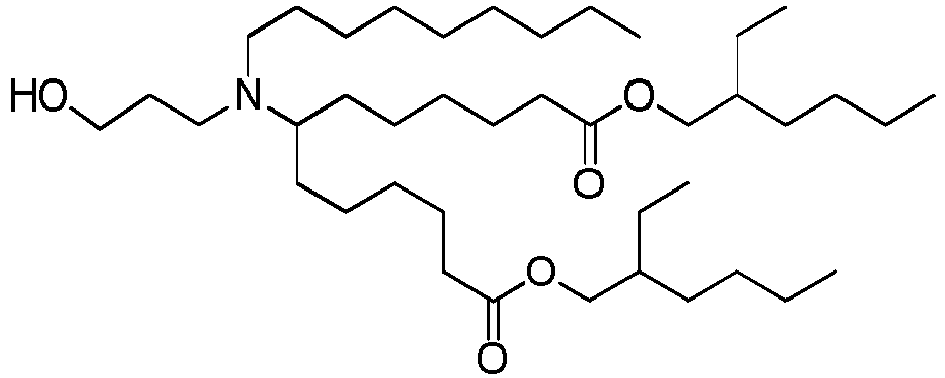
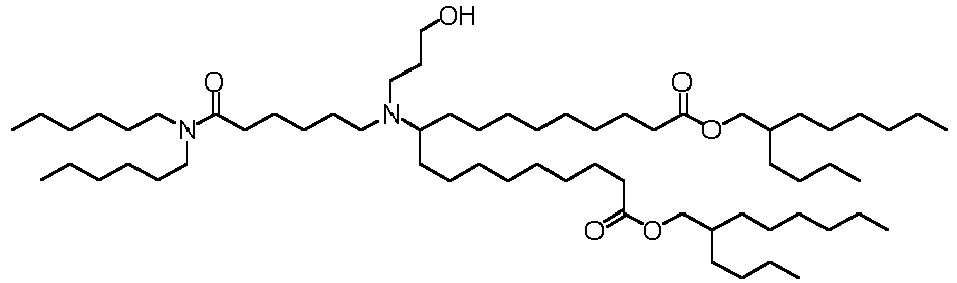
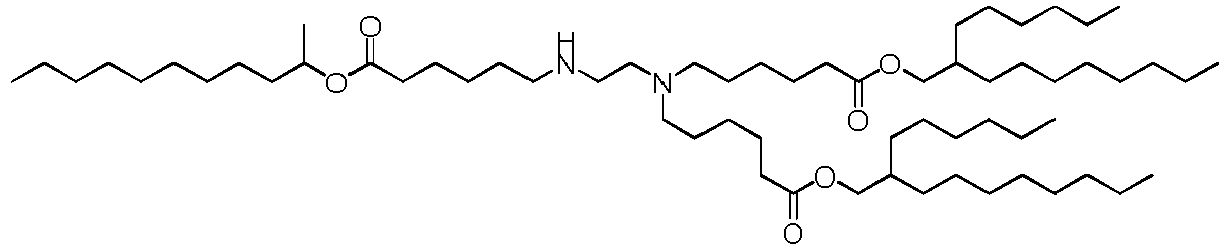
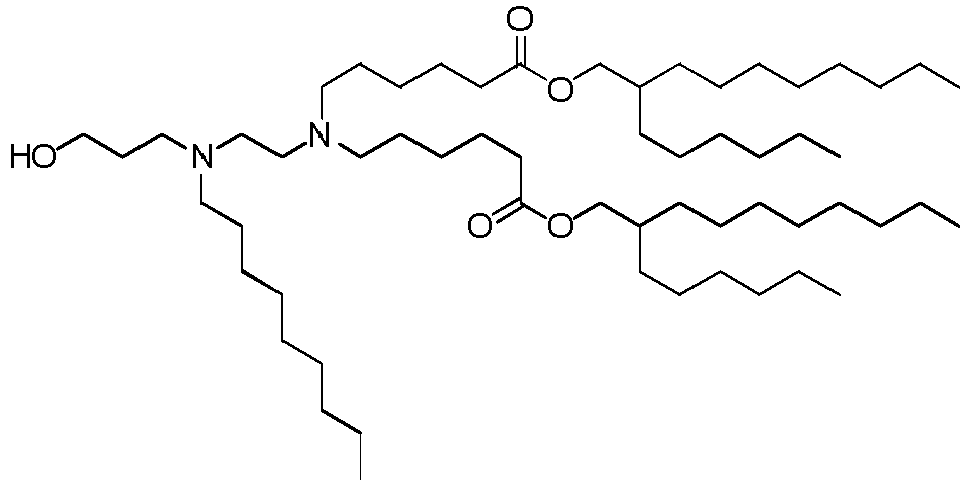
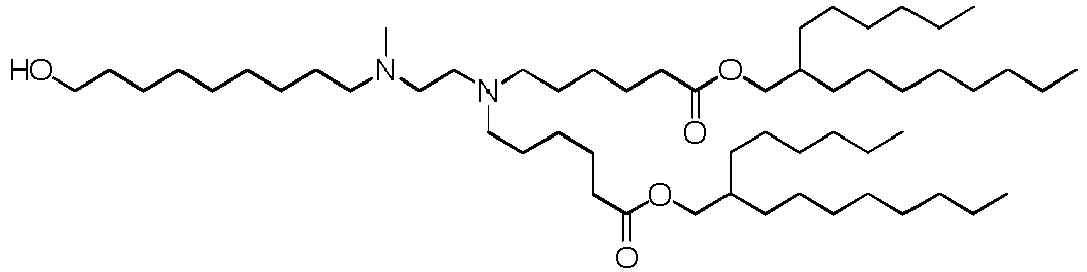
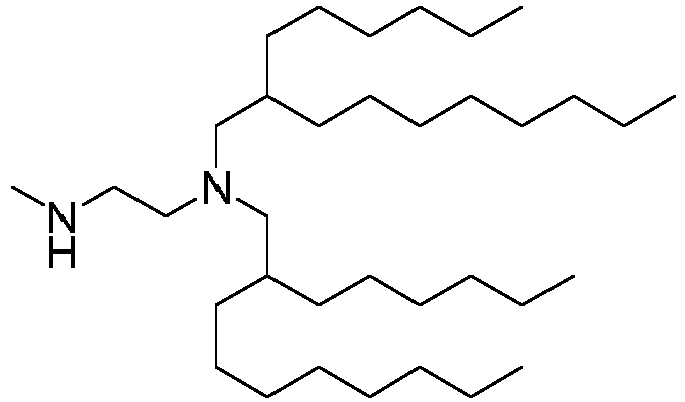
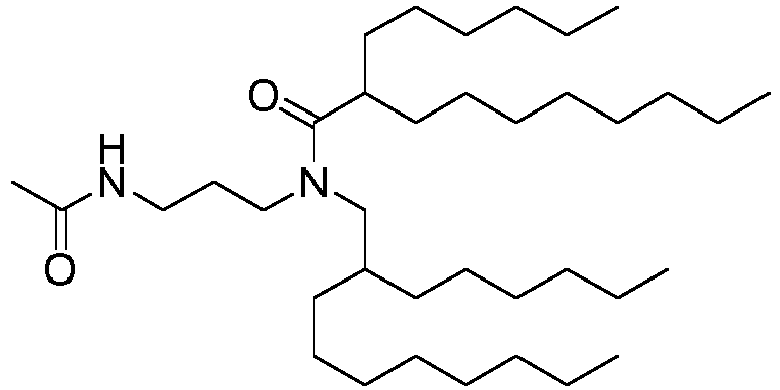
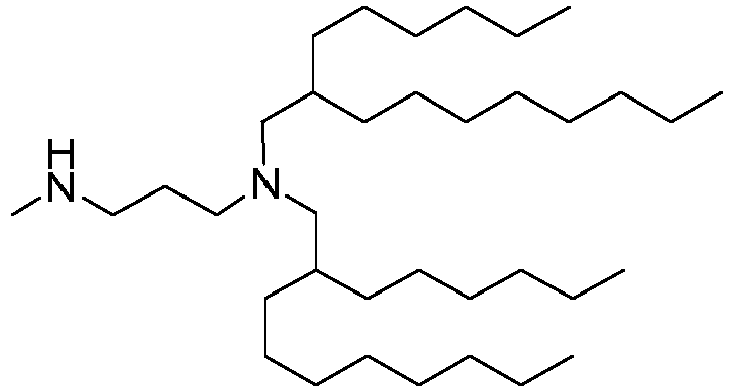
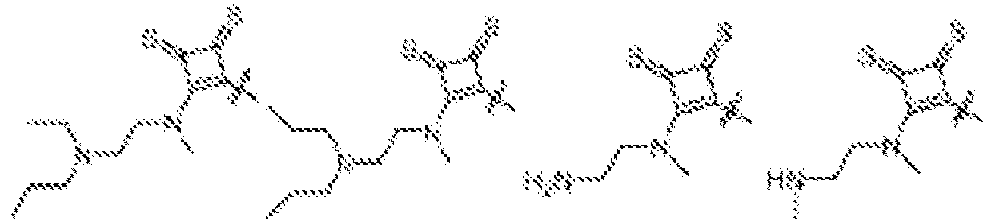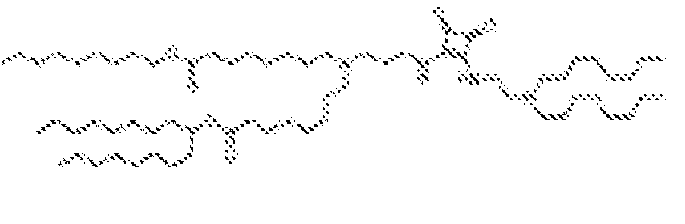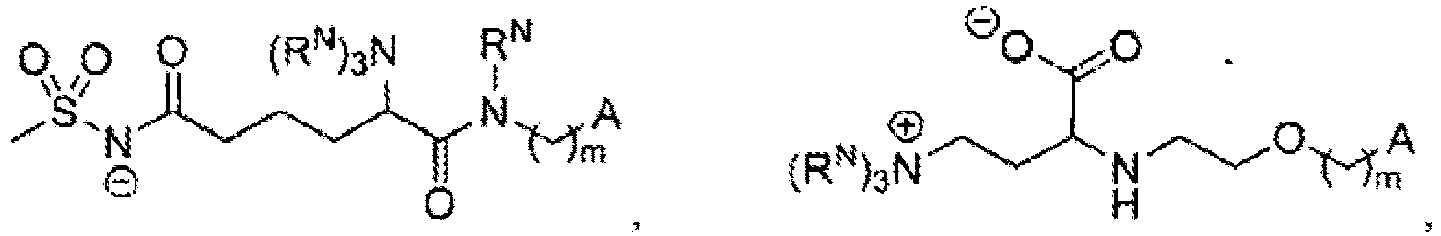EP4013385B1 - Improved lipid nanoparticles for delivery of nucleic acids - Google Patents
Improved lipid nanoparticles for delivery of nucleic acids Download PDFInfo
- Publication number
- EP4013385B1 EP4013385B1 EP20765121.7A EP20765121A EP4013385B1 EP 4013385 B1 EP4013385 B1 EP 4013385B1 EP 20765121 A EP20765121 A EP 20765121A EP 4013385 B1 EP4013385 B1 EP 4013385B1
- Authority
- EP
- European Patent Office
- Prior art keywords
- alkyl
- independently
- lipid
- occurrence
- carbon
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Active
Links
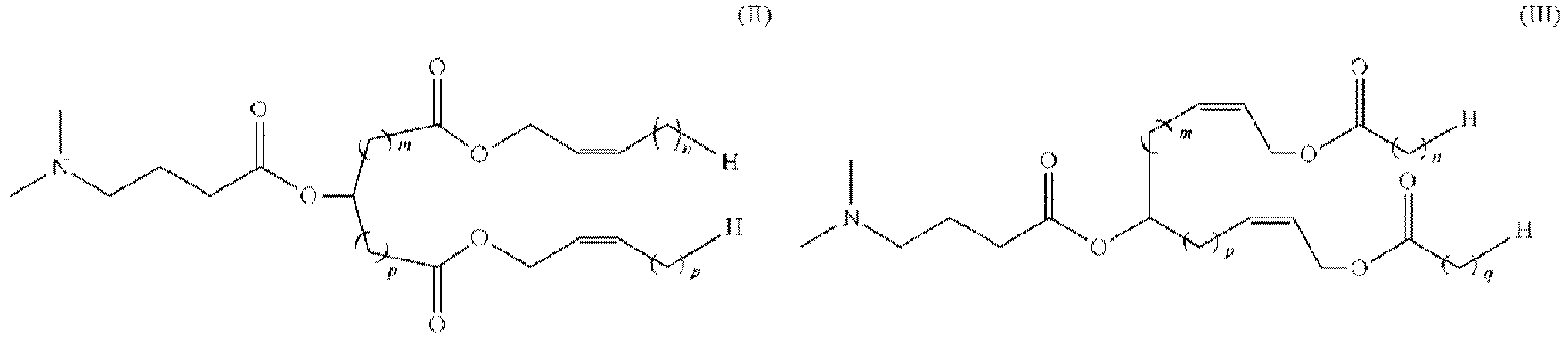
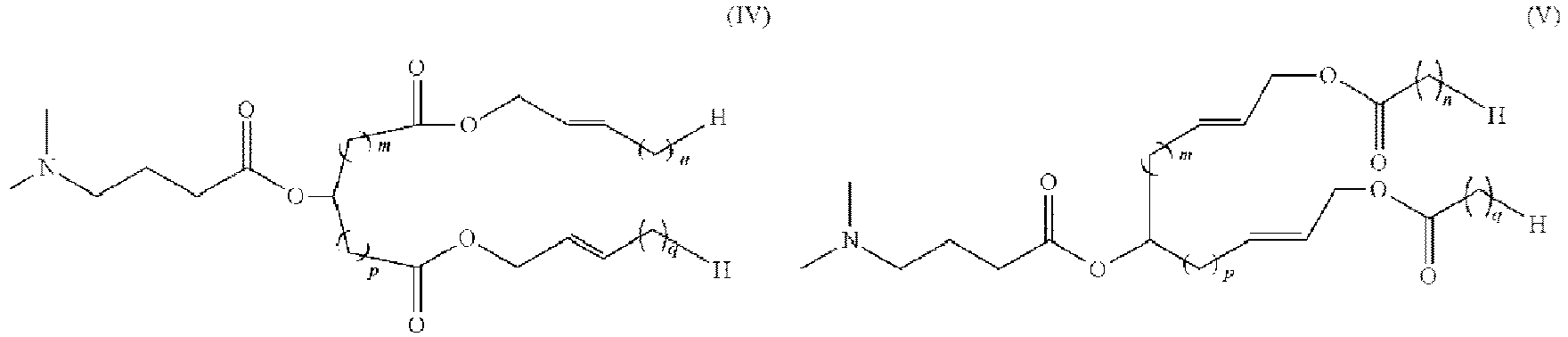
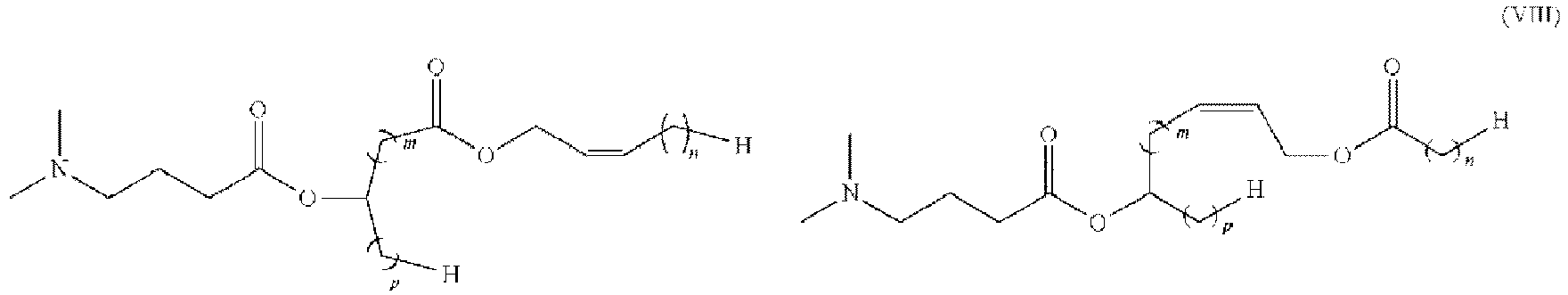
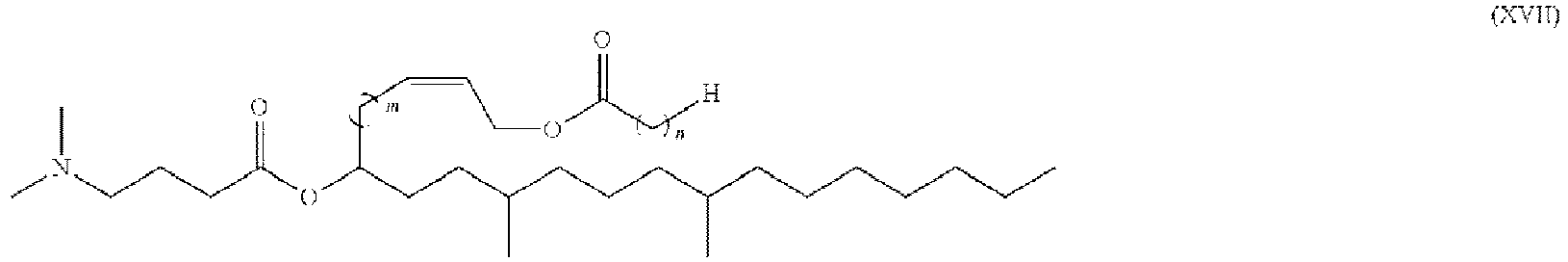
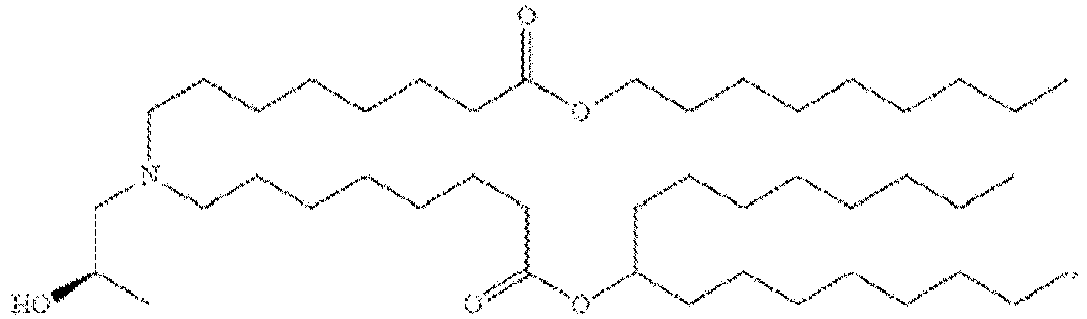
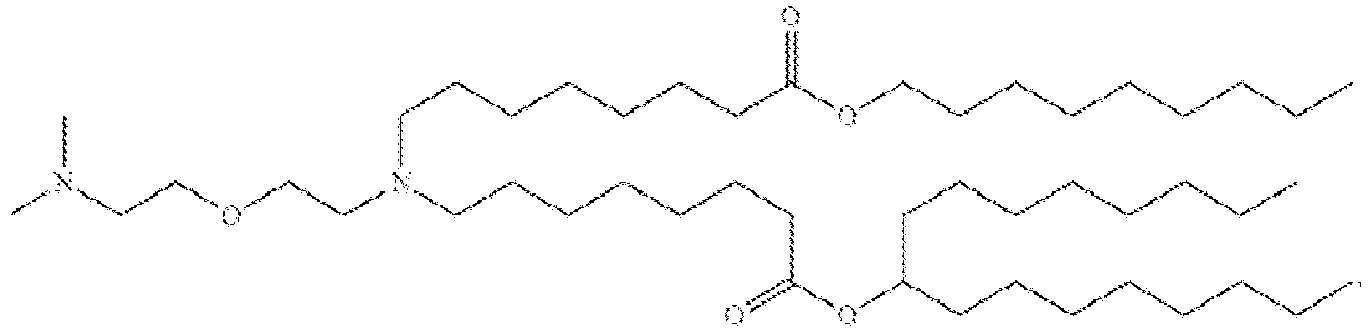
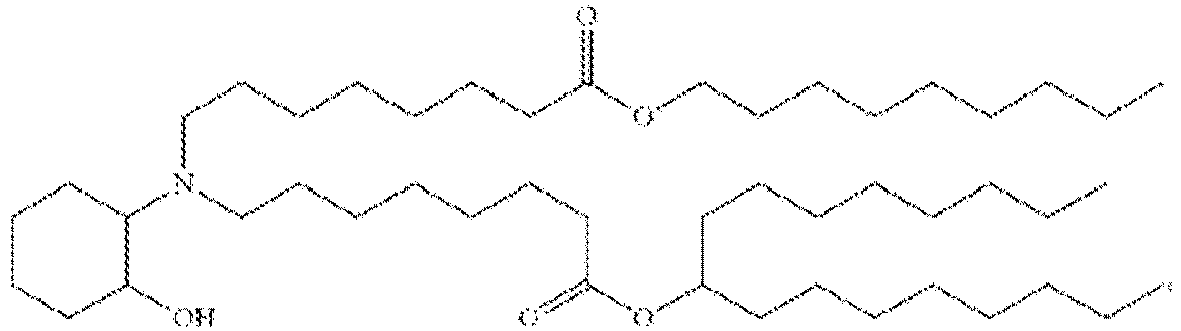
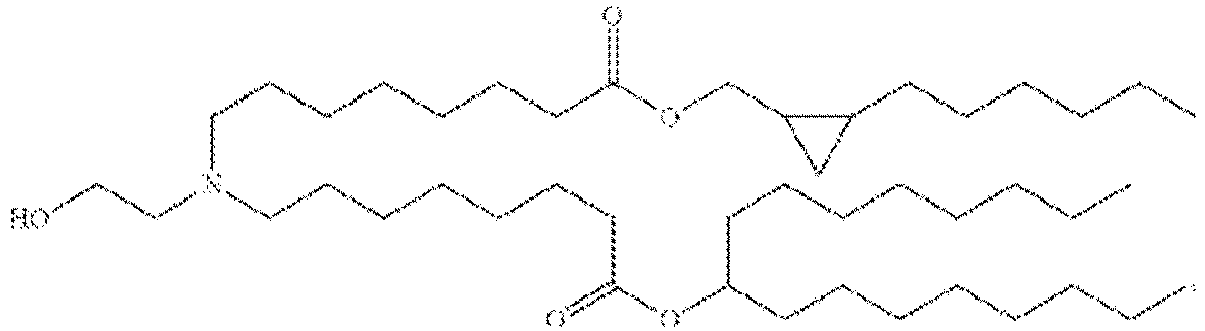
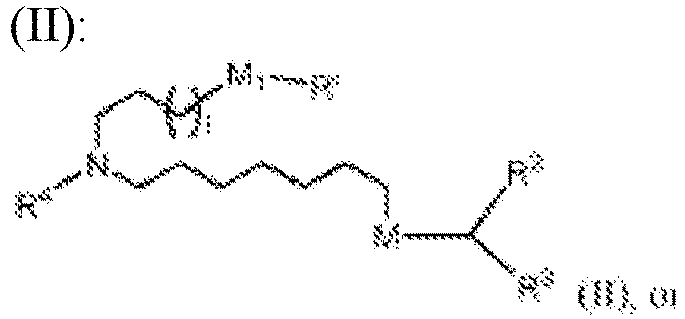
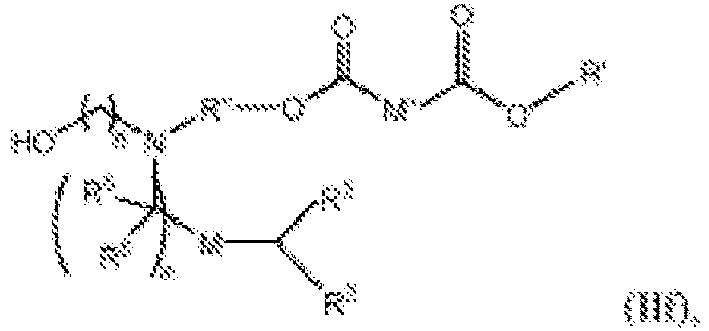
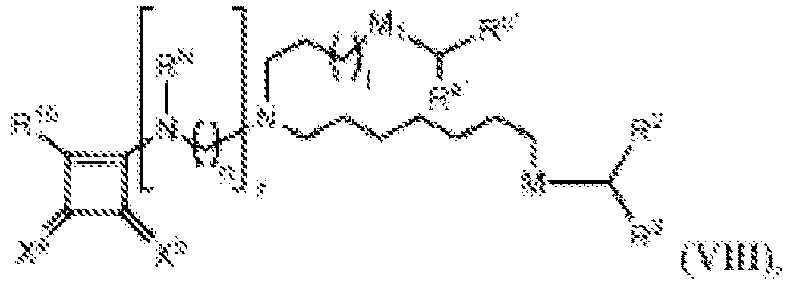
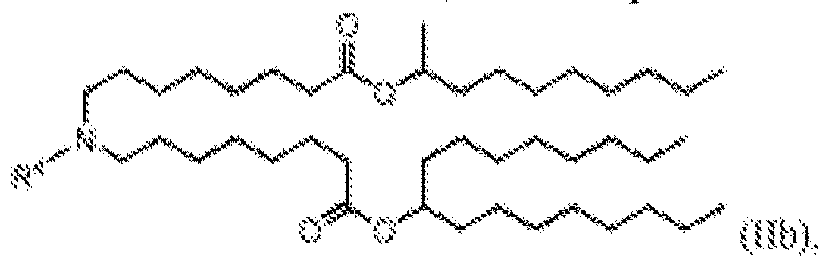
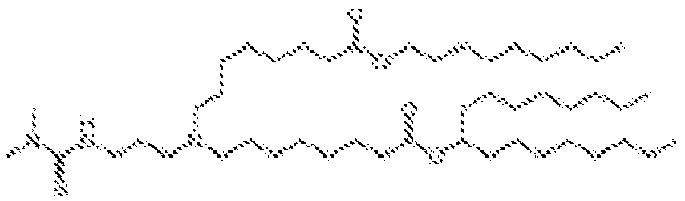
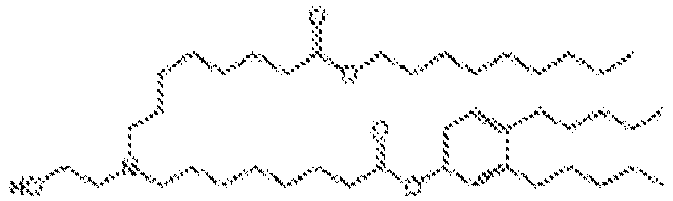
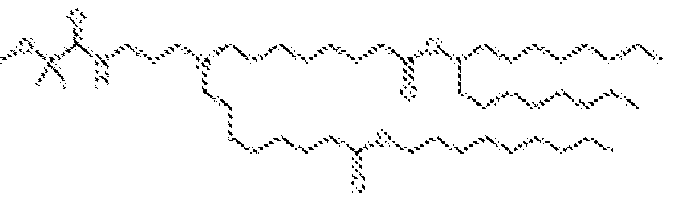
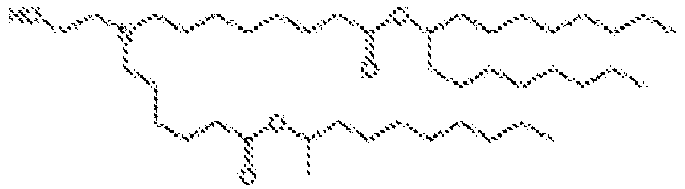
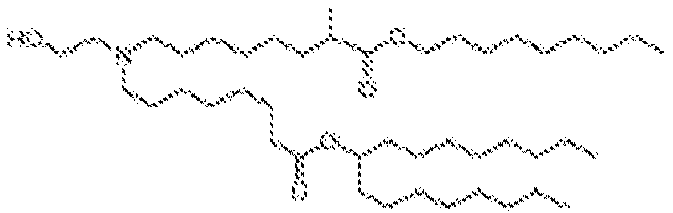
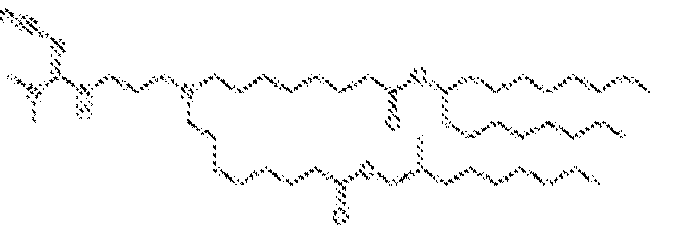
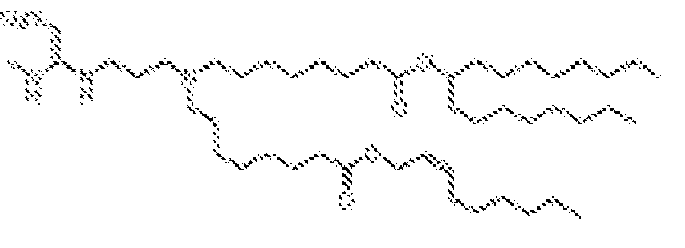
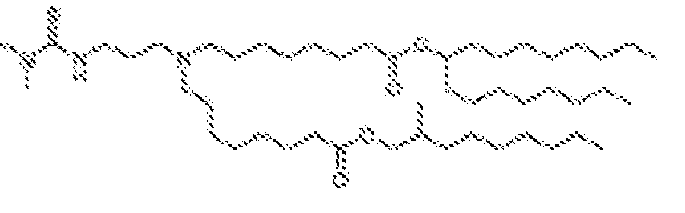
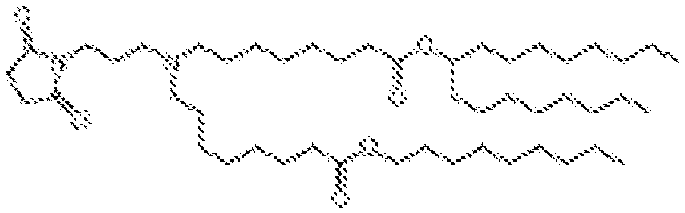
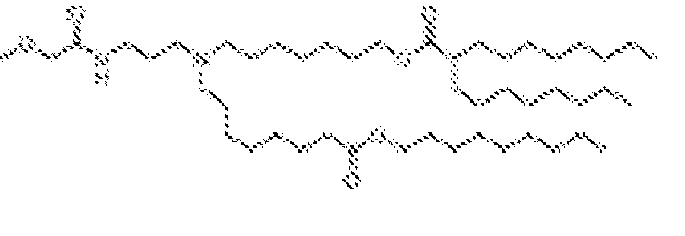
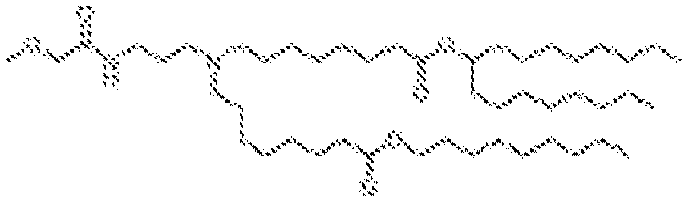
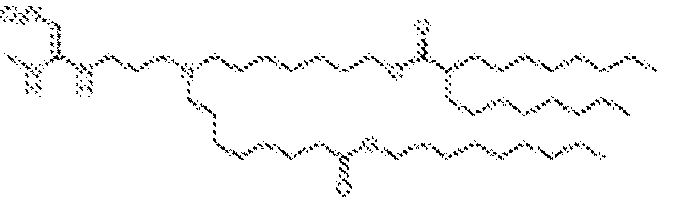
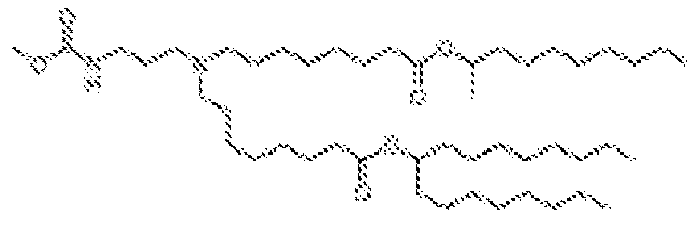
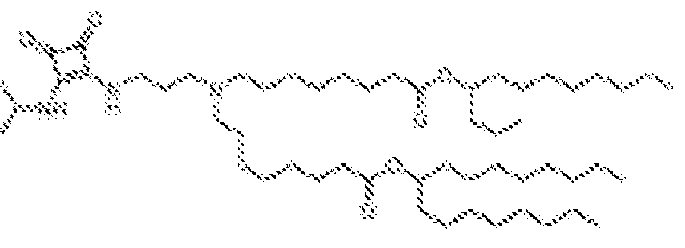
- 150000002632 lipids Chemical class 0.000 title claims description 513
- 239000002105 nanoparticle Substances 0.000 title claims description 129
- 150000007523 nucleic acids Chemical class 0.000 title claims description 95
- 102000039446 nucleic acids Human genes 0.000 title claims description 92
- 108020004707 nucleic acids Proteins 0.000 title claims description 92
- 238000012384 transportation and delivery Methods 0.000 title description 31
- 230000001976 improved effect Effects 0.000 title description 12
- 125000000217 alkyl group Chemical group 0.000 claims description 404
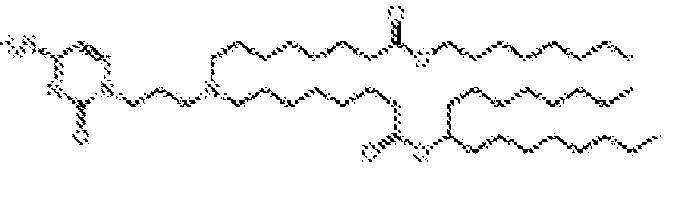
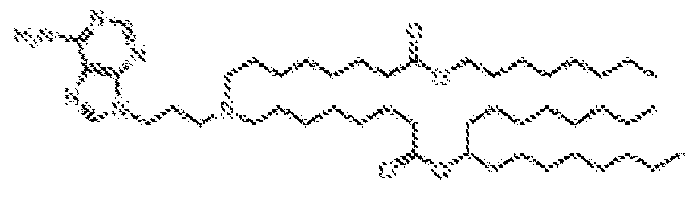
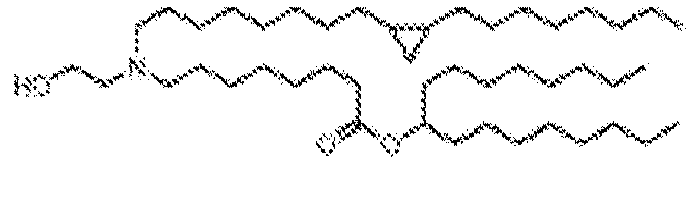
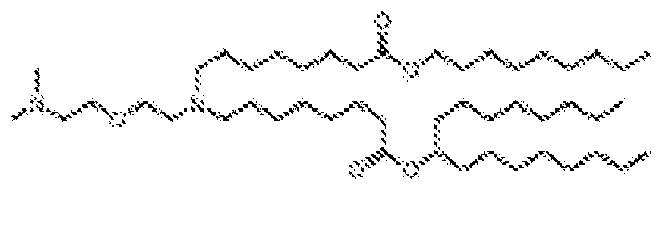
- -1 cationic lipid Chemical class 0.000 claims description 389
- 125000003342 alkenyl group Chemical group 0.000 claims description 200
- 125000004400 (C1-C12) alkyl group Chemical group 0.000 claims description 143
- 150000003839 salts Chemical class 0.000 claims description 121
- 229910052799 carbon Inorganic materials 0.000 claims description 117
- 230000007935 neutral effect Effects 0.000 claims description 105
- 150000001721 carbon Chemical group 0.000 claims description 99
- 108020004999 messenger RNA Proteins 0.000 claims description 92
- 229920000642 polymer Polymers 0.000 claims description 91
- 108090000623 proteins and genes Proteins 0.000 claims description 83
- 238000000034 method Methods 0.000 claims description 78
- 125000004169 (C1-C6) alkyl group Chemical group 0.000 claims description 72
- 125000002947 alkylene group Chemical group 0.000 claims description 65
- 125000002496 methyl group Chemical group [H]C([H])([H])* 0.000 claims description 64
- 239000011203 carbon fibre reinforced carbon Substances 0.000 claims description 55
- 229910052757 nitrogen Inorganic materials 0.000 claims description 52
- 125000003118 aryl group Chemical group 0.000 claims description 48
- 125000000753 cycloalkyl group Chemical group 0.000 claims description 42
- 102000004169 proteins and genes Human genes 0.000 claims description 42
- 125000001424 substituent group Chemical group 0.000 claims description 40
- HVYWMOMLDIMFJA-DPAQBDIFSA-N cholesterol Chemical compound C1C=C2C[C@@H](O)CC[C@]2(C)[C@@H]2[C@@H]1[C@@H]1CC[C@H]([C@H](C)CCCC(C)C)[C@@]1(C)CC2 HVYWMOMLDIMFJA-DPAQBDIFSA-N 0.000 claims description 36
- 150000003431 steroids Chemical class 0.000 claims description 31
- 125000004433 nitrogen atom Chemical group N* 0.000 claims description 29
- 241000288906 Primates Species 0.000 claims description 27
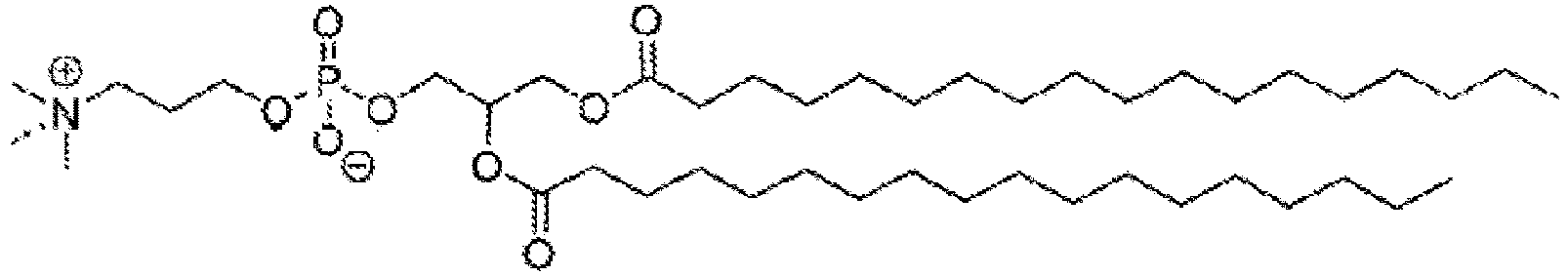
- NRJAVPSFFCBXDT-HUESYALOSA-N 1,2-distearoyl-sn-glycero-3-phosphocholine Chemical compound CCCCCCCCCCCCCCCCCC(=O)OC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CCCCCCCCCCCCCCCCC NRJAVPSFFCBXDT-HUESYALOSA-N 0.000 claims description 24
- 230000000692 anti-sense effect Effects 0.000 claims description 21
- 239000002245 particle Substances 0.000 claims description 21
- 229920006395 saturated elastomer Polymers 0.000 claims description 21
- 125000000524 functional group Chemical group 0.000 claims description 20
- 235000012000 cholesterol Nutrition 0.000 claims description 18
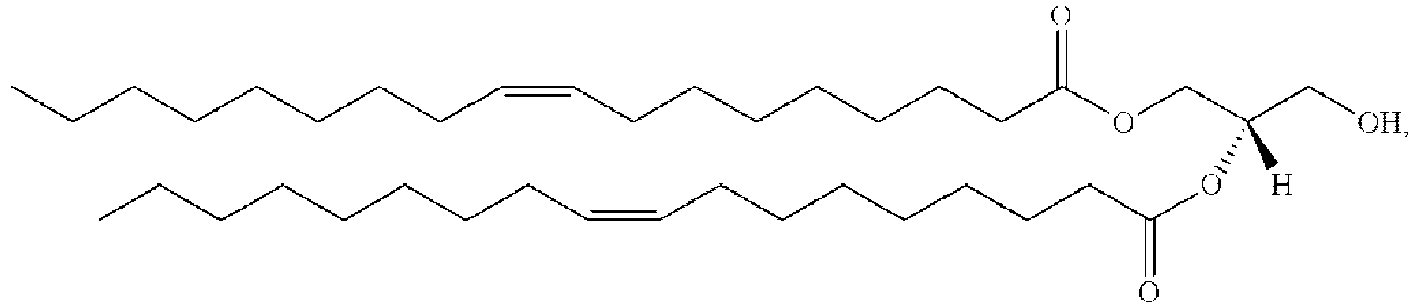
- MWRBNPKJOOWZPW-CLFAGFIQSA-N dioleoyl phosphatidylethanolamine Chemical compound CCCCCCCC\C=C/CCCCCCCC(=O)OCC(COP(O)(=O)OCCN)OC(=O)CCCCCCC\C=C/CCCCCCCC MWRBNPKJOOWZPW-CLFAGFIQSA-N 0.000 claims description 18
- 201000010099 disease Diseases 0.000 claims description 16
- 208000037265 diseases, disorders, signs and symptoms Diseases 0.000 claims description 16
- 125000003161 (C1-C6) alkylene group Chemical group 0.000 claims description 14
- 125000004450 alkenylene group Chemical group 0.000 claims description 14
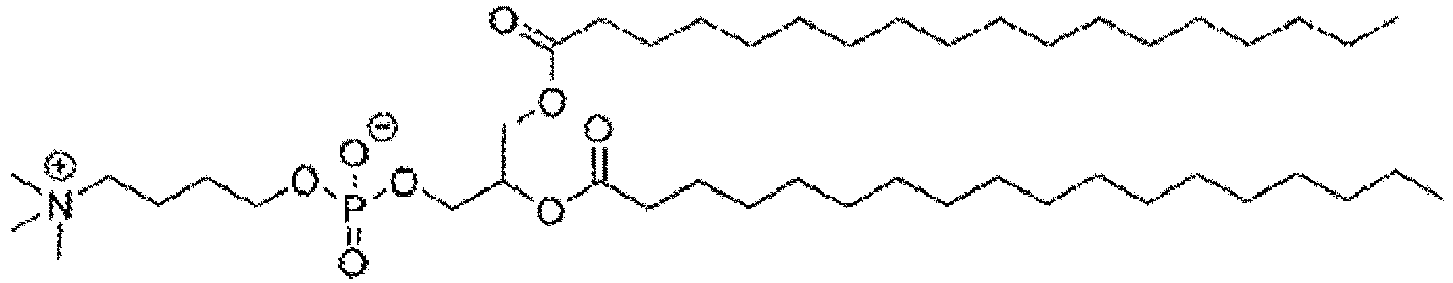
- KILNVBDSWZSGLL-KXQOOQHDSA-N 1,2-dihexadecanoyl-sn-glycero-3-phosphocholine Chemical compound CCCCCCCCCCCCCCCC(=O)OC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CCCCCCCCCCCCCCC KILNVBDSWZSGLL-KXQOOQHDSA-N 0.000 claims description 13
- 125000002993 cycloalkylene group Chemical group 0.000 claims description 13
- SNKAWJBJQDLSFF-NVKMUCNASA-N 1,2-dioleoyl-sn-glycero-3-phosphocholine Chemical compound CCCCCCCC\C=C/CCCCCCCC(=O)OC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CCCCCCC\C=C/CCCCCCCC SNKAWJBJQDLSFF-NVKMUCNASA-N 0.000 claims description 10
- 125000004103 aminoalkyl group Chemical group 0.000 claims description 10
- 125000005724 cycloalkenylene group Chemical group 0.000 claims description 10
- 125000002373 5 membered heterocyclic group Chemical group 0.000 claims description 9
- 125000002768 hydroxyalkyl group Chemical group 0.000 claims description 9
- WTJKGGKOPKCXLL-RRHRGVEJSA-N phosphatidylcholine Chemical compound CCCCCCCCCCCCCCCC(=O)OC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CCCCCCCC=CCCCCCCCC WTJKGGKOPKCXLL-RRHRGVEJSA-N 0.000 claims description 9
- 125000006686 (C1-C24) alkyl group Chemical group 0.000 claims description 8
- 125000006710 (C2-C12) alkenyl group Chemical group 0.000 claims description 8
- 125000004070 6 membered heterocyclic group Chemical group 0.000 claims description 8
- 125000003710 aryl alkyl group Chemical group 0.000 claims description 8
- 125000003341 7 membered heterocyclic group Chemical group 0.000 claims description 7
- 125000004406 C3-C8 cycloalkylene group Chemical group 0.000 claims description 6
- 125000004474 heteroalkylene group Chemical group 0.000 claims description 6
- DSNRWDQKZIEDDB-GCMPNPAFSA-N [(2r)-3-[2,3-dihydroxypropoxy(hydroxy)phosphoryl]oxy-2-[(z)-octadec-9-enoyl]oxypropyl] (z)-octadec-9-enoate Chemical compound CCCCCCCC\C=C/CCCCCCCC(=O)OC[C@H](COP(O)(=O)OCC(O)CO)OC(=O)CCCCCCC\C=C/CCCCCCCC DSNRWDQKZIEDDB-GCMPNPAFSA-N 0.000 claims description 5
- 125000004183 alkoxy alkyl group Chemical group 0.000 claims description 5
- 150000001412 amines Chemical class 0.000 claims description 5
- SLKDGVPOSSLUAI-PGUFJCEWSA-N 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine zwitterion Chemical compound CCCCCCCCCCCCCCCC(=O)OC[C@H](COP(O)(=O)OCCN)OC(=O)CCCCCCCCCCCCCCC SLKDGVPOSSLUAI-PGUFJCEWSA-N 0.000 claims description 4
- LVNGJLRDBYCPGB-UHFFFAOYSA-N 1,2-distearoylphosphatidylethanolamine Chemical compound CCCCCCCCCCCCCCCCCC(=O)OCC(COP([O-])(=O)OCC[NH3+])OC(=O)CCCCCCCCCCCCCCCCC LVNGJLRDBYCPGB-UHFFFAOYSA-N 0.000 claims description 4
- BIABMEZBCHDPBV-MPQUPPDSSA-N 1,2-palmitoyl-sn-glycero-3-phospho-(1'-sn-glycerol) Chemical compound CCCCCCCCCCCCCCCC(=O)OC[C@H](COP(O)(=O)OC[C@@H](O)CO)OC(=O)CCCCCCCCCCCCCCC BIABMEZBCHDPBV-MPQUPPDSSA-N 0.000 claims description 4
- GZDFHIJNHHMENY-UHFFFAOYSA-N Dimethyl dicarbonate Chemical compound COC(=O)OC(=O)OC GZDFHIJNHHMENY-UHFFFAOYSA-N 0.000 claims description 4
- NONFBHXKNNVFMO-UHFFFAOYSA-N [2-aminoethoxy(tetradecanoyloxy)phosphoryl] tetradecanoate Chemical compound CCCCCCCCCCCCCC(=O)OP(=O)(OCCN)OC(=O)CCCCCCCCCCCCC NONFBHXKNNVFMO-UHFFFAOYSA-N 0.000 claims description 4
- 125000004453 alkoxycarbonyl group Chemical group 0.000 claims description 4
- 125000004448 alkyl carbonyl group Chemical group 0.000 claims description 4
- 125000005197 alkyl carbonyloxy alkyl group Chemical group 0.000 claims description 4
- 125000005196 alkyl carbonyloxy group Chemical group 0.000 claims description 4
- 125000003837 (C1-C20) alkyl group Chemical group 0.000 claims description 2
- 108020005544 Antisense RNA Proteins 0.000 claims description 2
- 238000002296 dynamic light scattering Methods 0.000 claims description 2
- 230000002163 immunogen Effects 0.000 claims description 2
- 150000001875 compounds Chemical class 0.000 description 182
- 125000000623 heterocyclic group Chemical group 0.000 description 104
- 229910052739 hydrogen Inorganic materials 0.000 description 90
- 125000006592 (C2-C3) alkenyl group Chemical group 0.000 description 83
- 125000006273 (C1-C3) alkyl group Chemical group 0.000 description 77
- 229920001223 polyethylene glycol Polymers 0.000 description 76
- 239000002202 Polyethylene glycol Substances 0.000 description 74
- 229920002477 rna polymer Polymers 0.000 description 71
- 239000000203 mixture Substances 0.000 description 68
- 125000004432 carbon atom Chemical group C* 0.000 description 65
- 125000001072 heteroaryl group Chemical group 0.000 description 60
- 230000014509 gene expression Effects 0.000 description 53
- 125000001570 methylene group Chemical group [H]C([H])([*:1])[*:2] 0.000 description 48
- 239000001257 hydrogen Substances 0.000 description 44
- 125000004435 hydrogen atom Chemical group [H]* 0.000 description 44
- 239000002773 nucleotide Substances 0.000 description 44
- 125000003729 nucleotide group Chemical group 0.000 description 43
- 108020004459 Small interfering RNA Proteins 0.000 description 40
- 150000004665 fatty acids Chemical class 0.000 description 39
- 238000000338 in vitro Methods 0.000 description 39
- 239000004055 small Interfering RNA Substances 0.000 description 39
- 102000053602 DNA Human genes 0.000 description 38
- 108020004414 DNA Proteins 0.000 description 38
- 150000001204 N-oxides Chemical class 0.000 description 38
- 235000014113 dietary fatty acids Nutrition 0.000 description 38
- 229930195729 fatty acid Natural products 0.000 description 38
- 239000000194 fatty acid Substances 0.000 description 38
- 108091034117 Oligonucleotide Proteins 0.000 description 37
- 125000000304 alkynyl group Chemical group 0.000 description 34
- 125000001495 ethyl group Chemical group [H]C([H])([H])C([H])([H])* 0.000 description 33
- WRIDQFICGBMAFQ-UHFFFAOYSA-N (E)-8-Octadecenoic acid Natural products CCCCCCCCCC=CCCCCCCC(O)=O WRIDQFICGBMAFQ-UHFFFAOYSA-N 0.000 description 32
- LQJBNNIYVWPHFW-UHFFFAOYSA-N 20:1omega9c fatty acid Natural products CCCCCCCCCCC=CCCCCCCCC(O)=O LQJBNNIYVWPHFW-UHFFFAOYSA-N 0.000 description 32
- QSBYPNXLFMSGKH-UHFFFAOYSA-N 9-Heptadecensaeure Natural products CCCCCCCC=CCCCCCCCC(O)=O QSBYPNXLFMSGKH-UHFFFAOYSA-N 0.000 description 32
- 239000005642 Oleic acid Substances 0.000 description 32
- ZQPPMHVWECSIRJ-UHFFFAOYSA-N Oleic acid Natural products CCCCCCCCC=CCCCCCCCC(O)=O ZQPPMHVWECSIRJ-UHFFFAOYSA-N 0.000 description 32
- 125000004429 atom Chemical group 0.000 description 32
- 125000002091 cationic group Chemical group 0.000 description 32
- 125000002887 hydroxy group Chemical group [H]O* 0.000 description 32
- QXJSBBXBKPUZAA-UHFFFAOYSA-N isooleic acid Natural products CCCCCCCC=CCCCCCCCCC(O)=O QXJSBBXBKPUZAA-UHFFFAOYSA-N 0.000 description 32
- ZQPPMHVWECSIRJ-KTKRTIGZSA-N oleic acid Chemical compound CCCCCCCC\C=C/CCCCCCCC(O)=O ZQPPMHVWECSIRJ-KTKRTIGZSA-N 0.000 description 32
- 235000021313 oleic acid Nutrition 0.000 description 32
- 229910052760 oxygen Inorganic materials 0.000 description 32
- 150000003904 phospholipids Chemical class 0.000 description 32
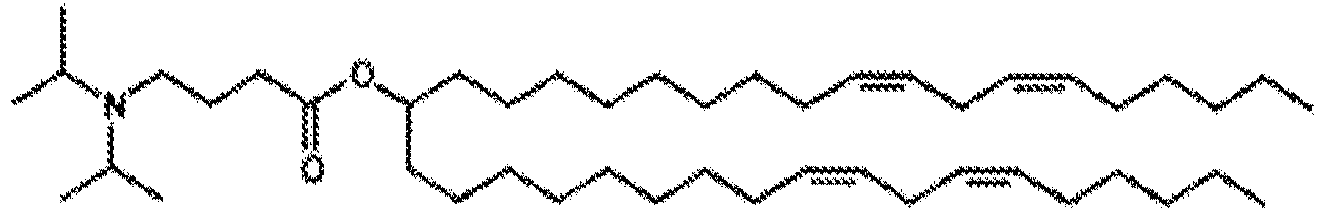
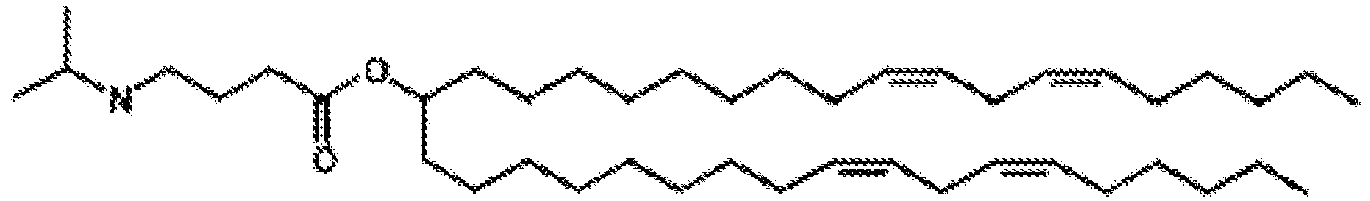
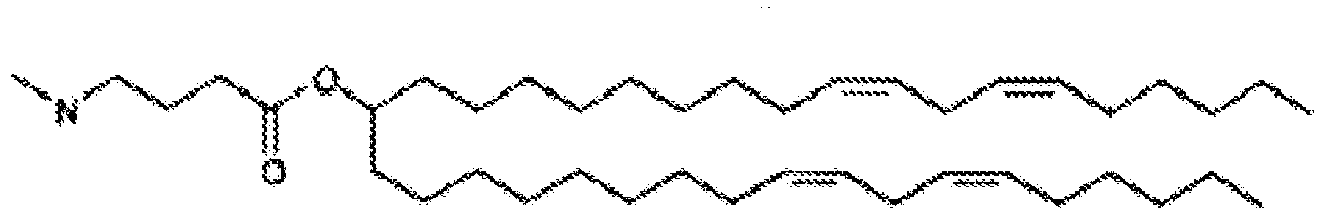
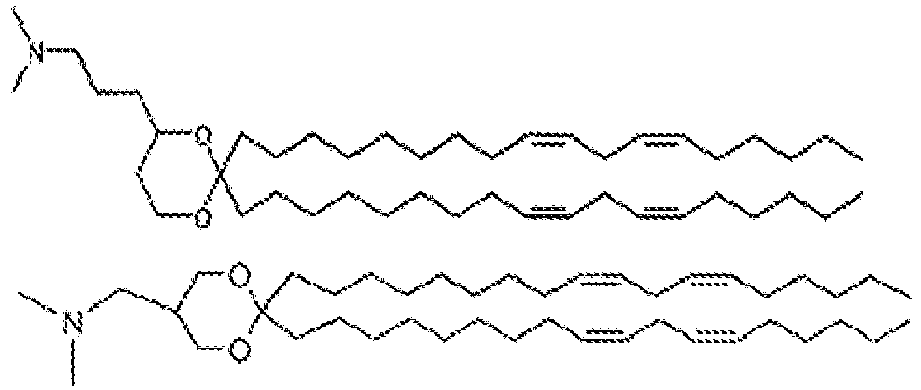
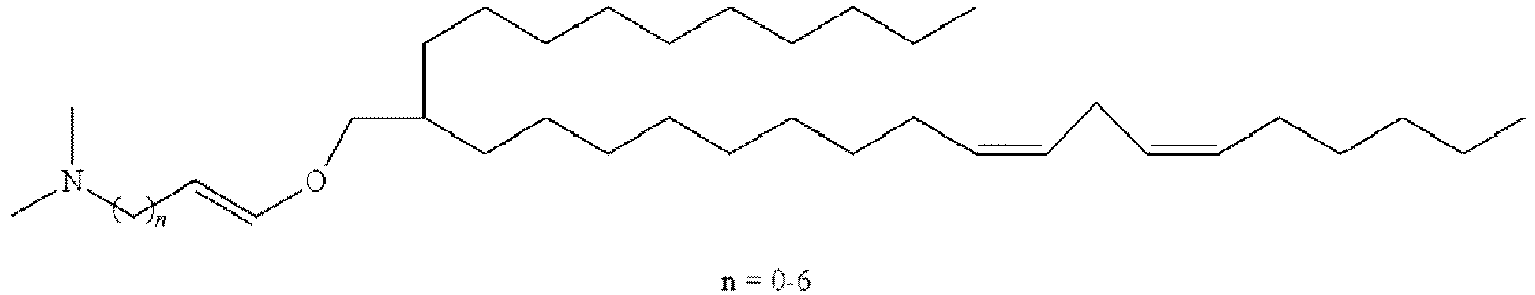
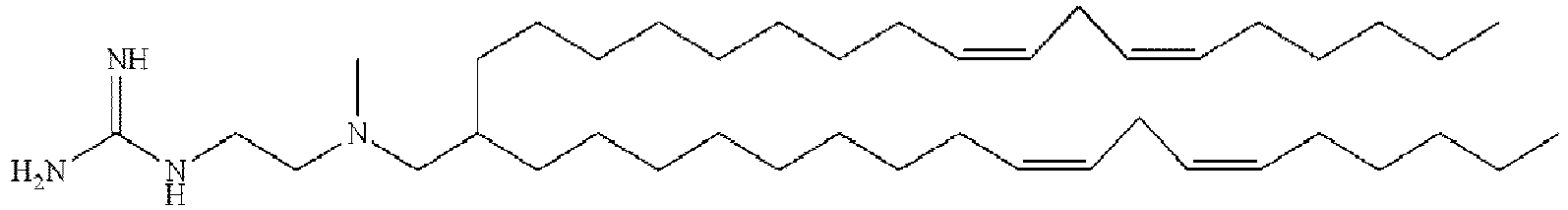
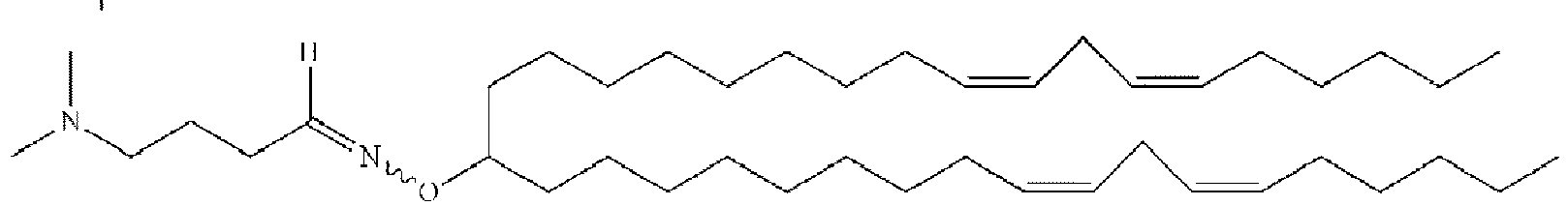
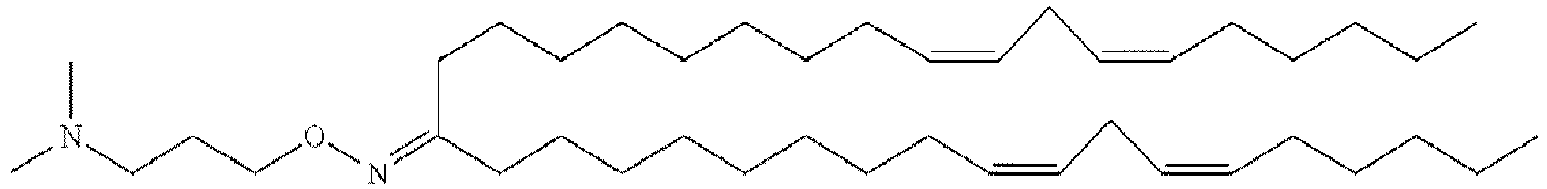
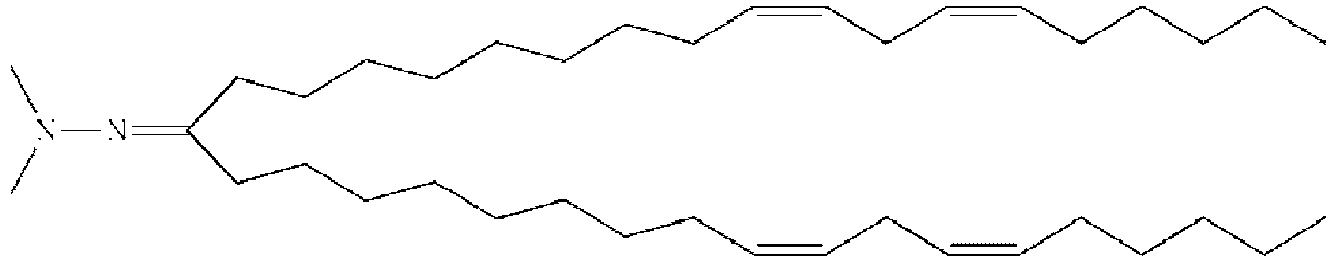
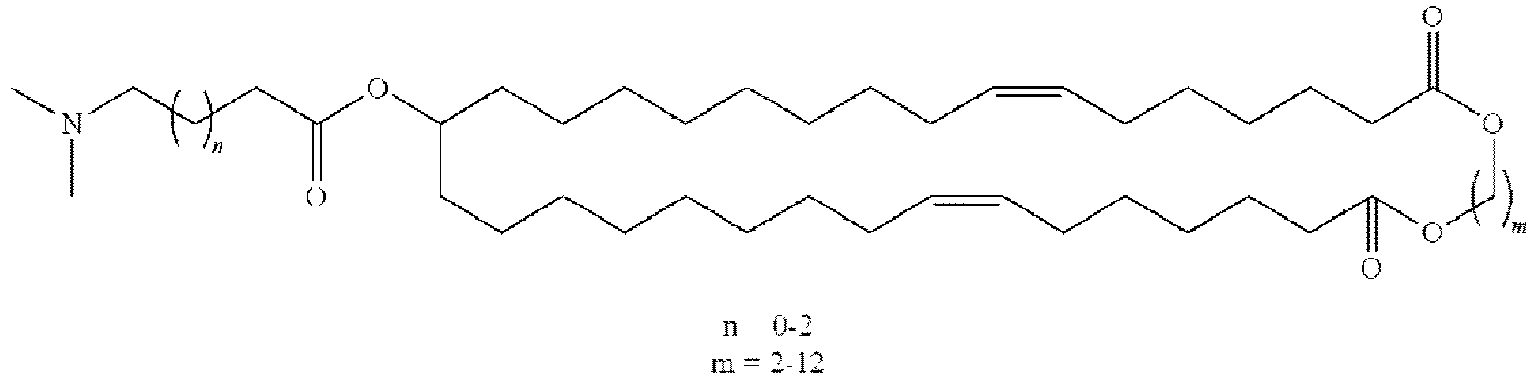
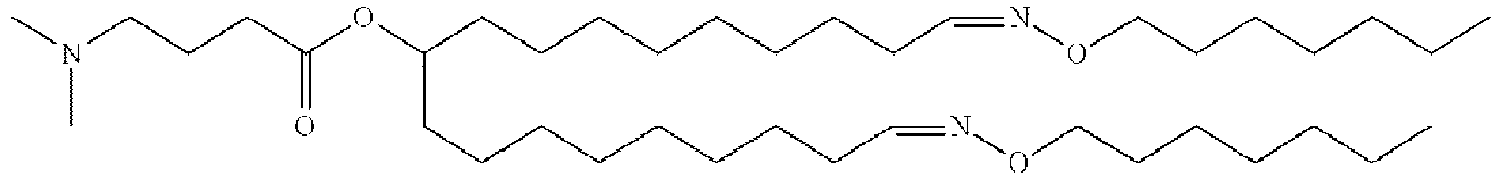
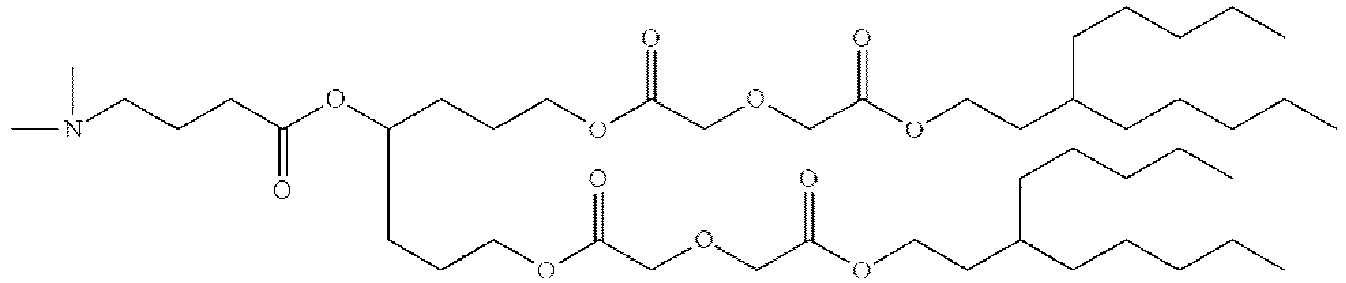
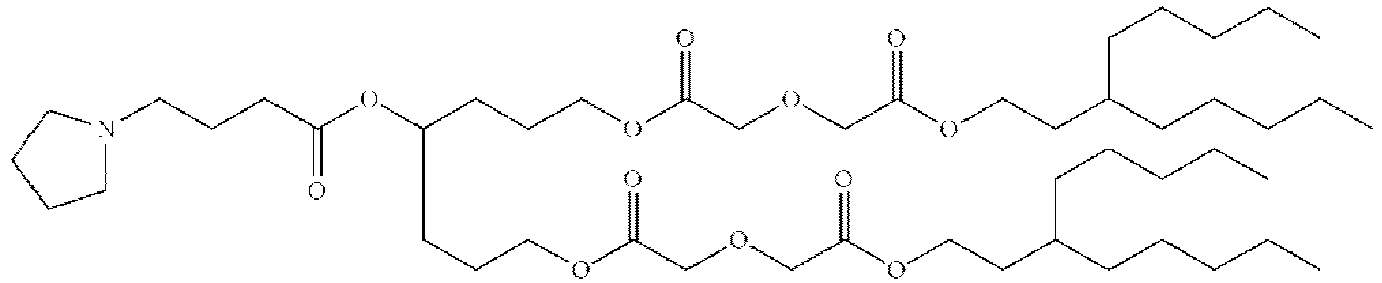
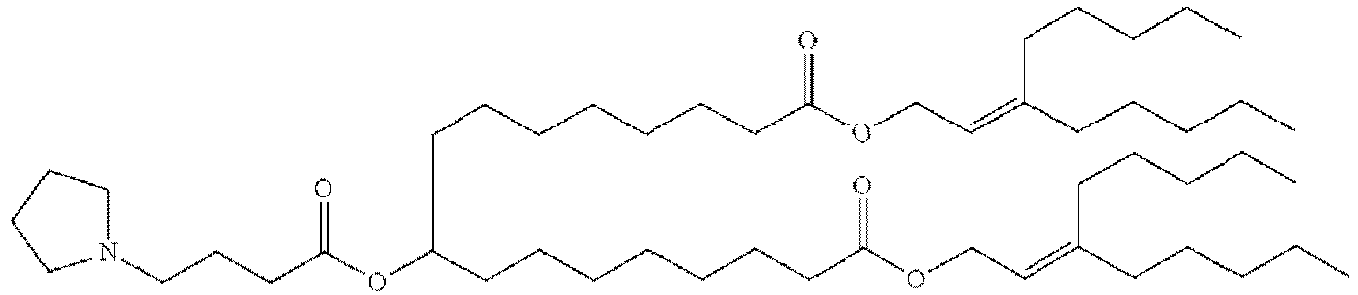
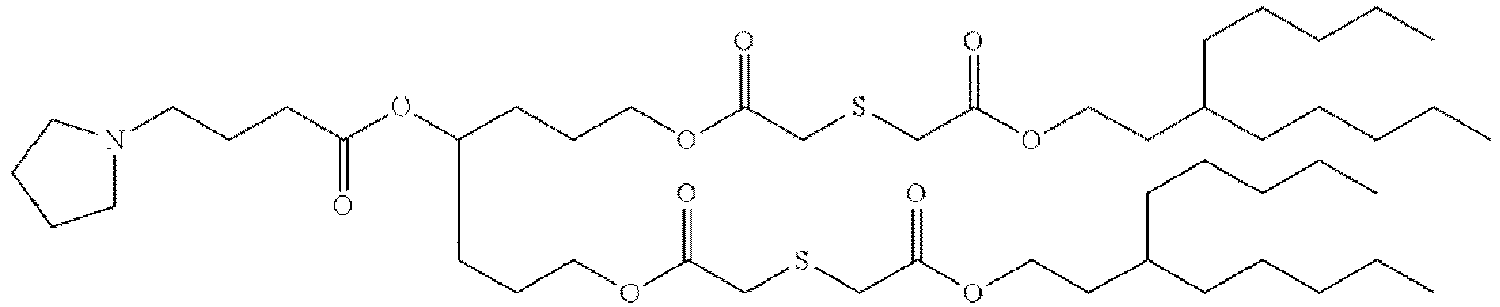
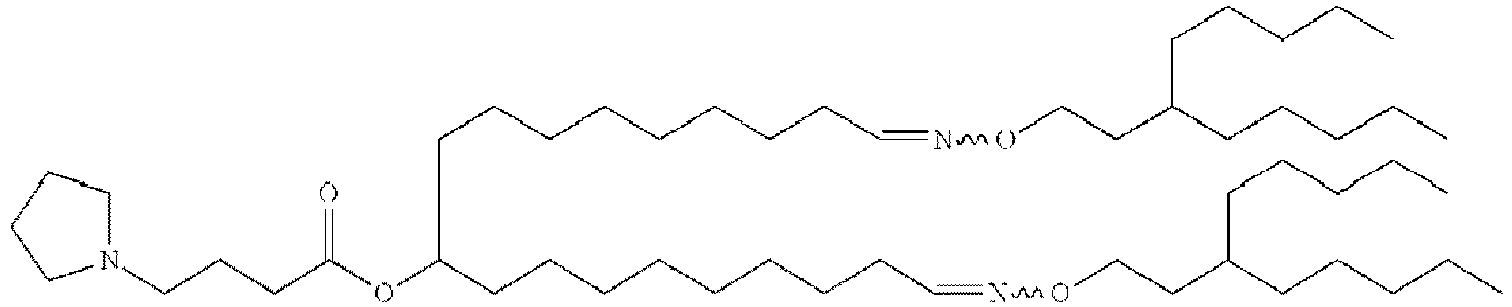
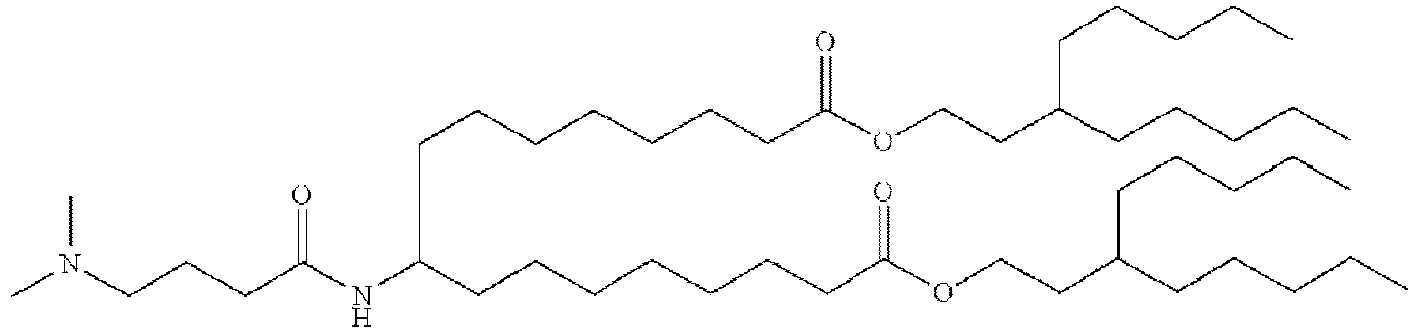
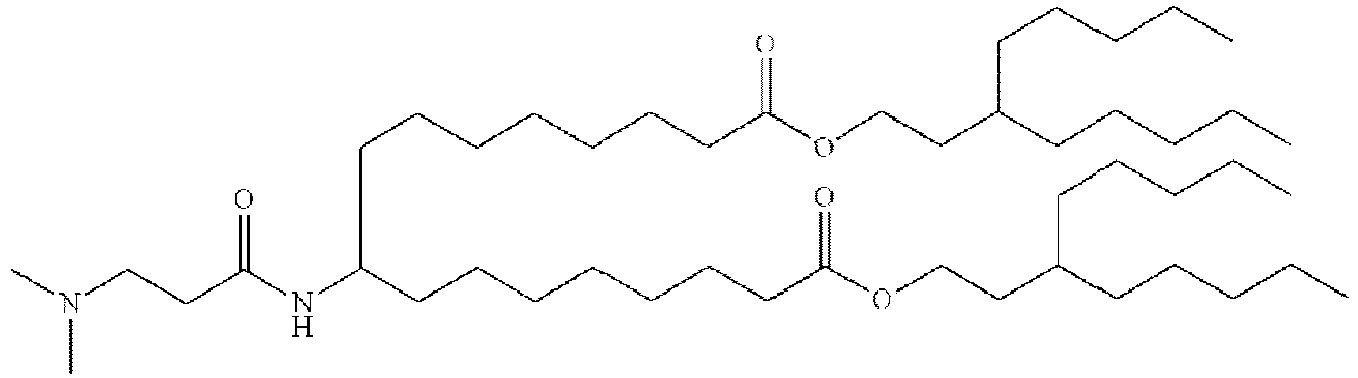
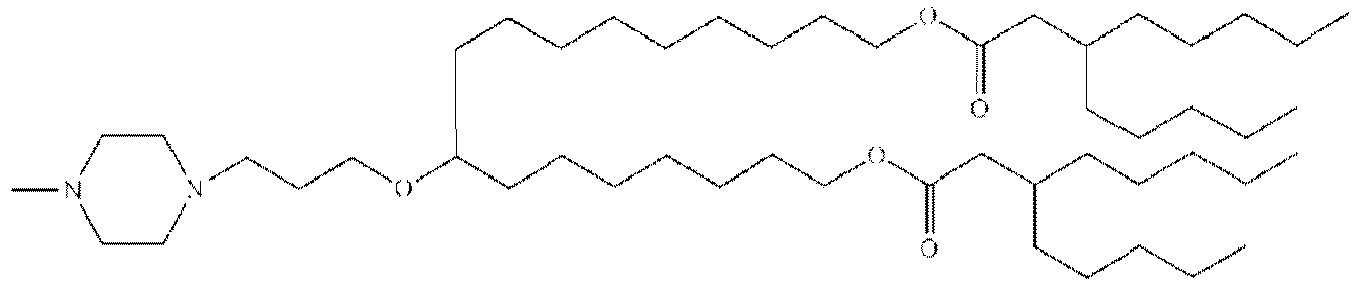
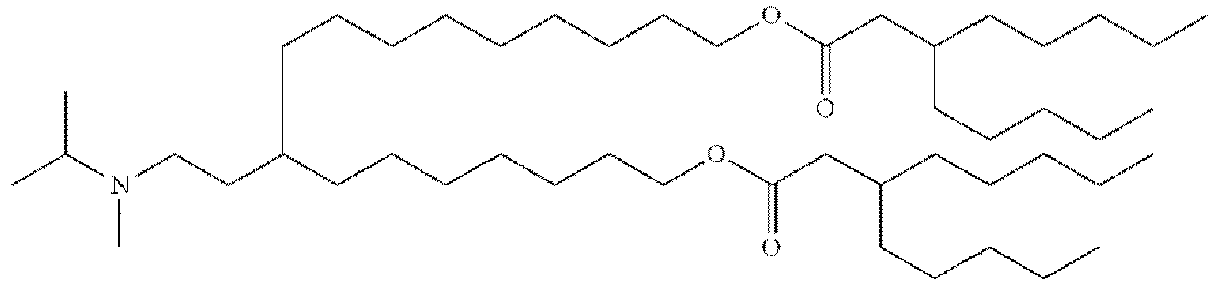
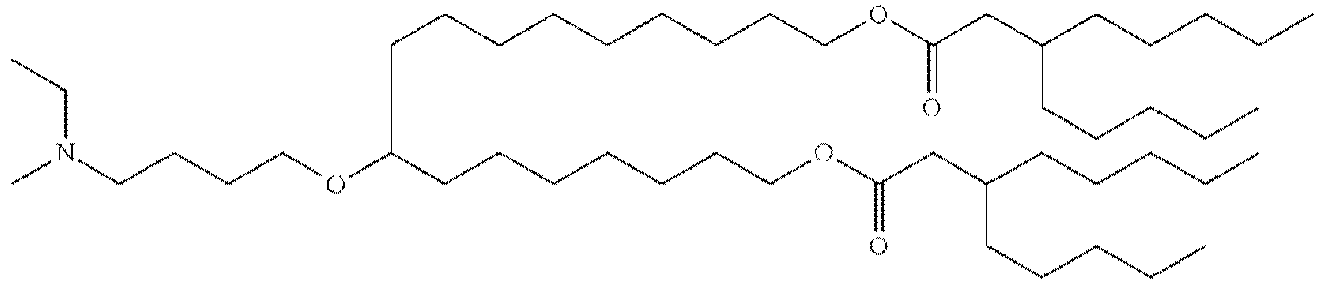
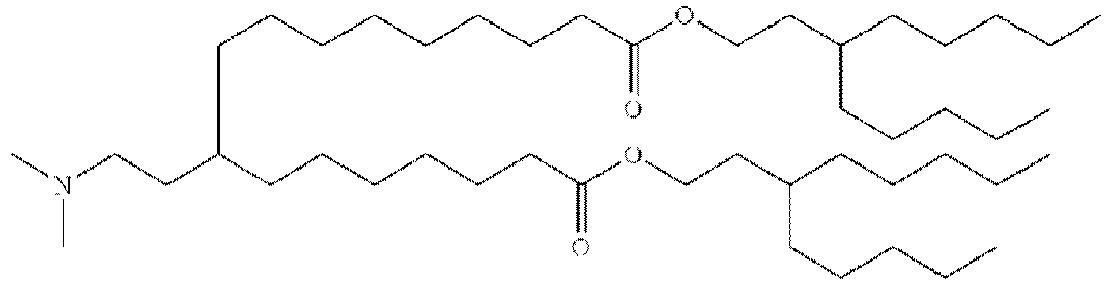
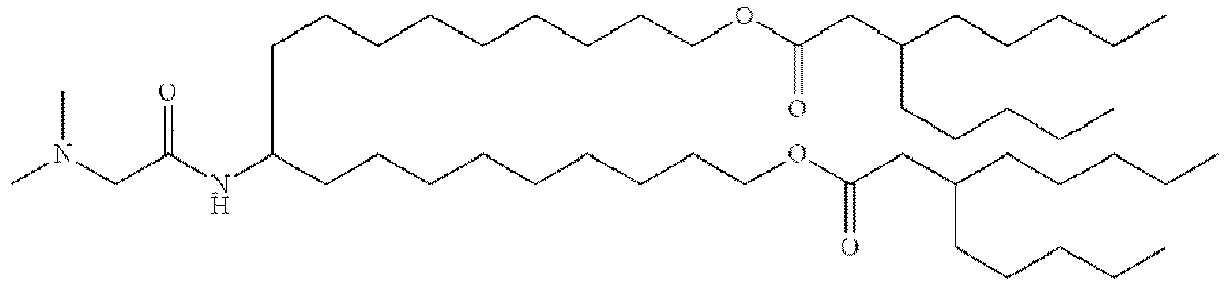
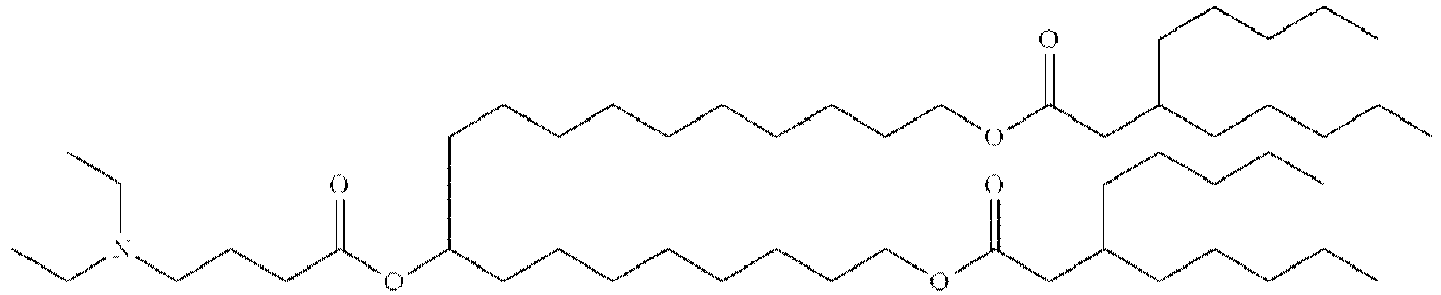
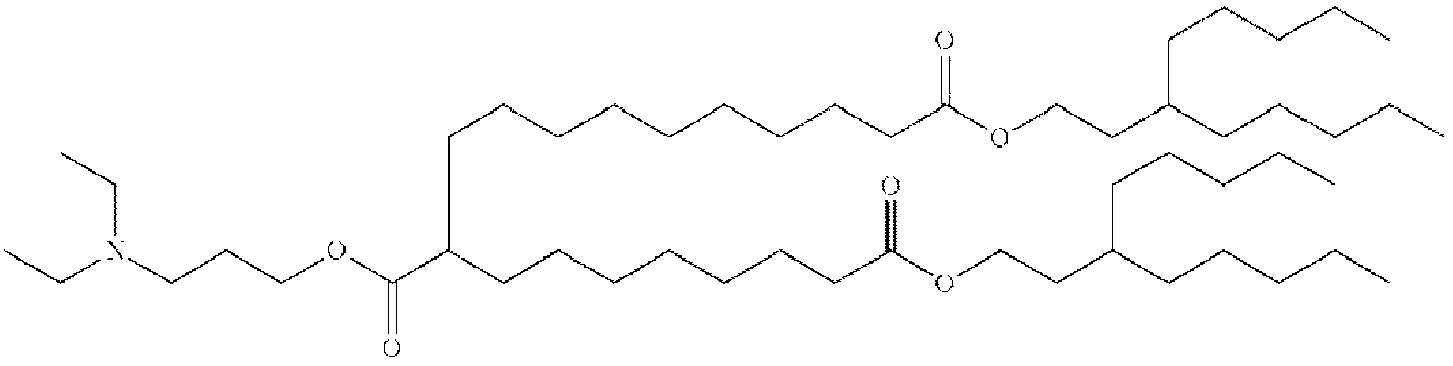
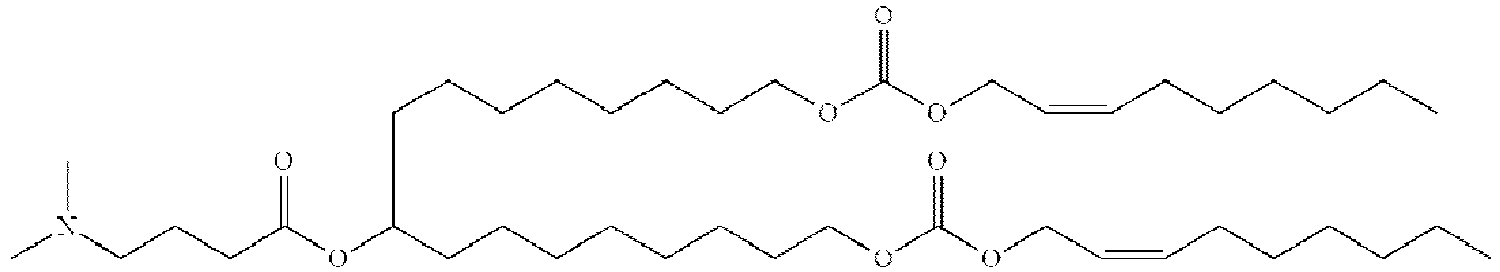
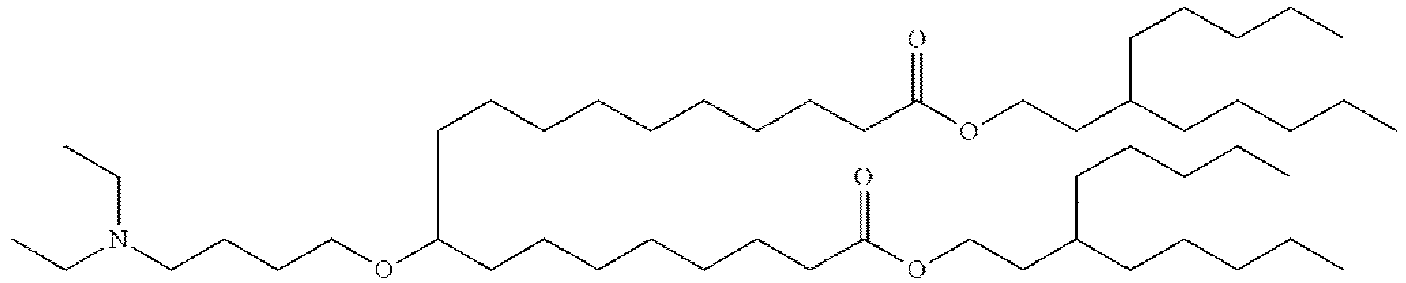
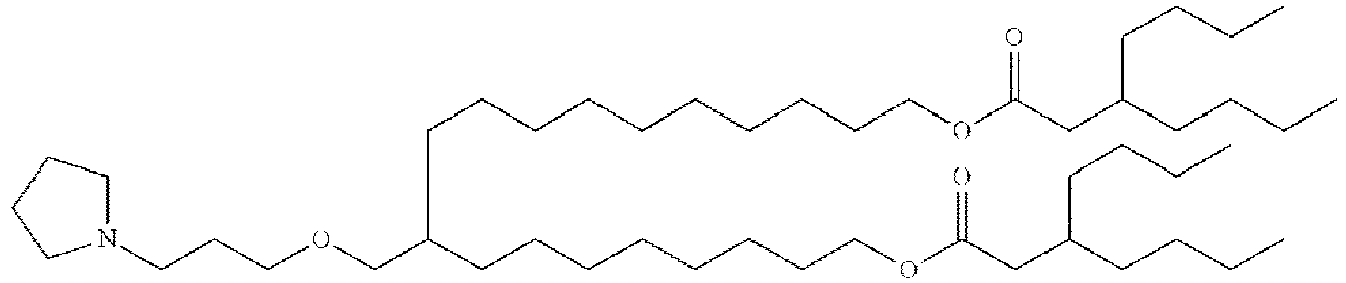
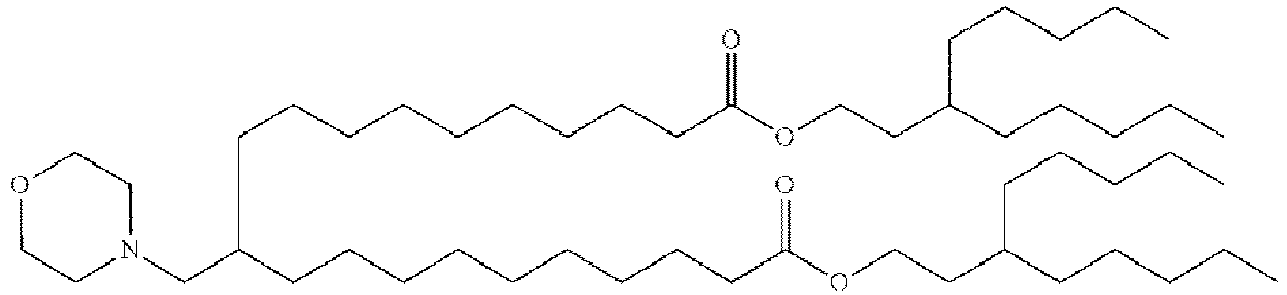
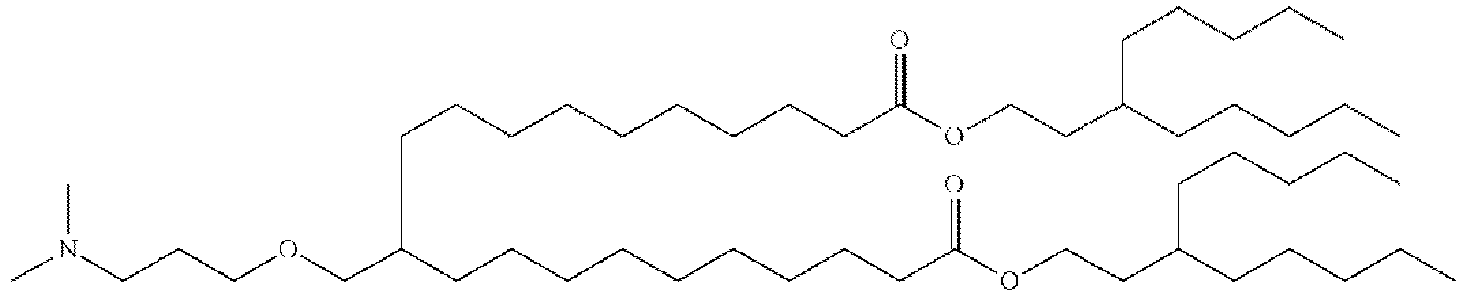
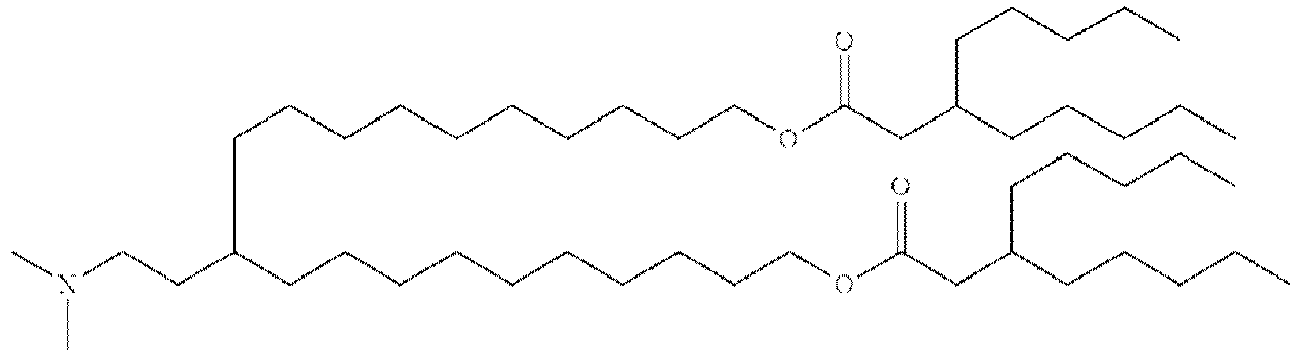
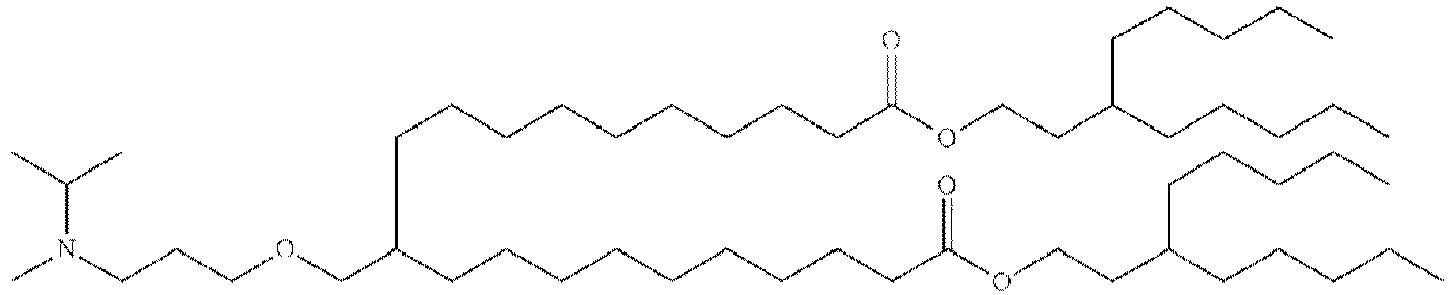
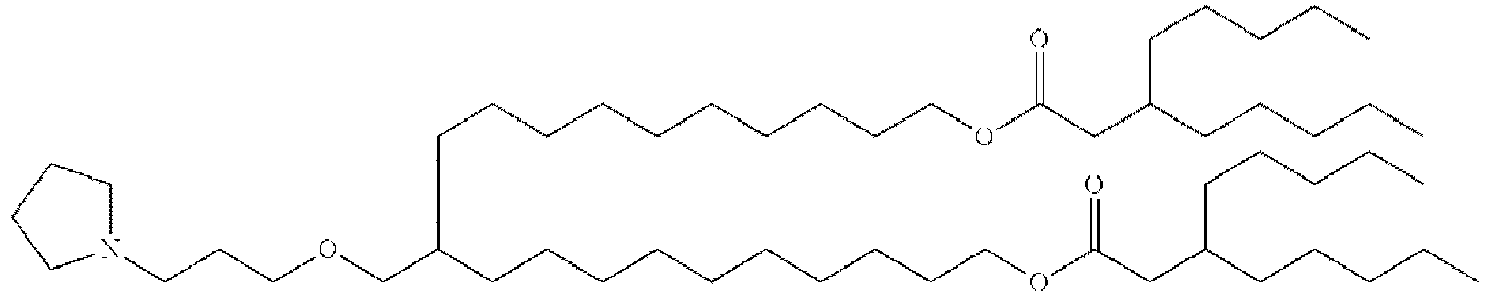
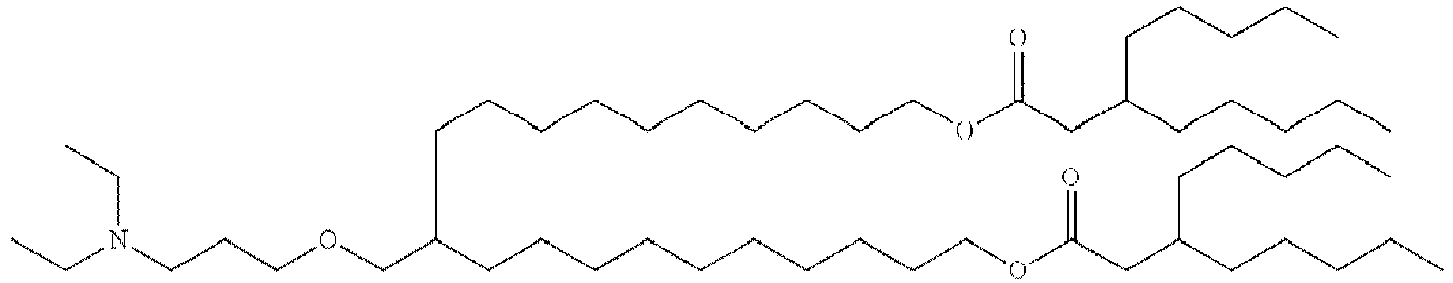
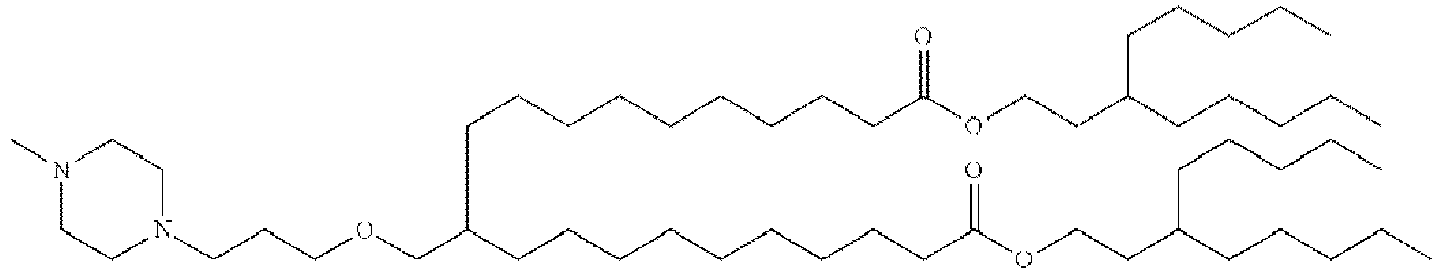
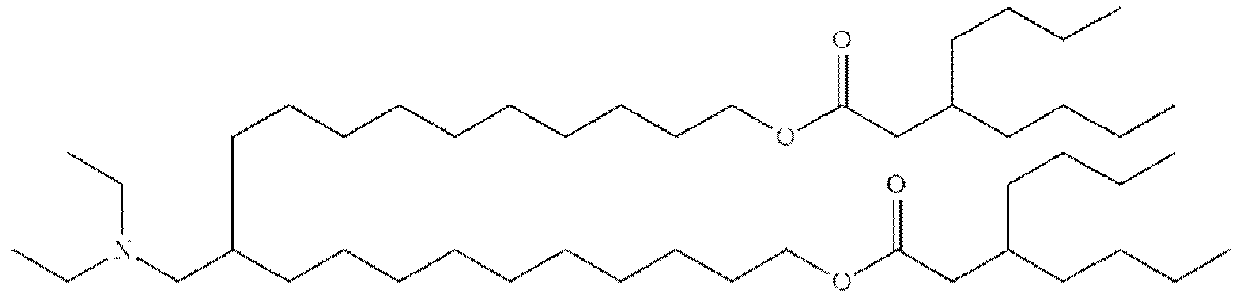
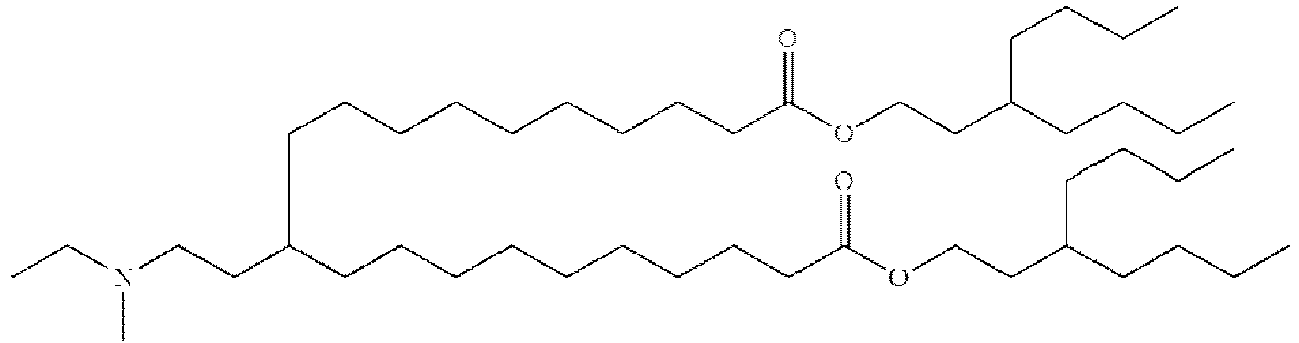
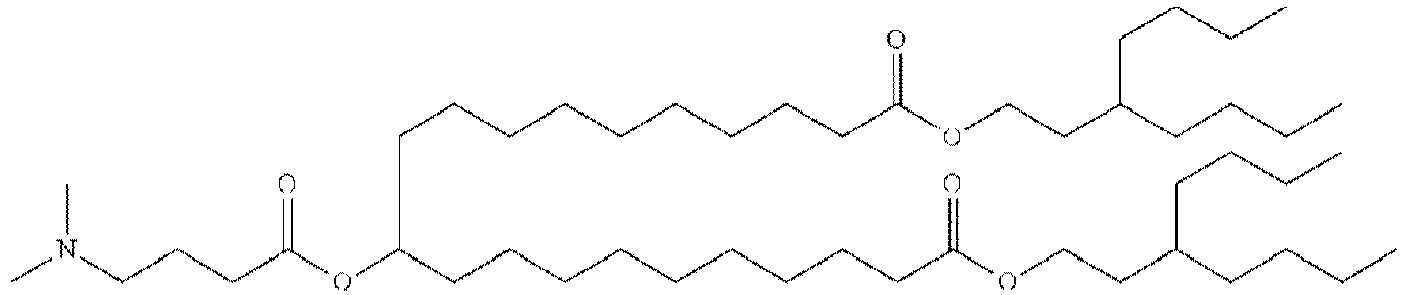
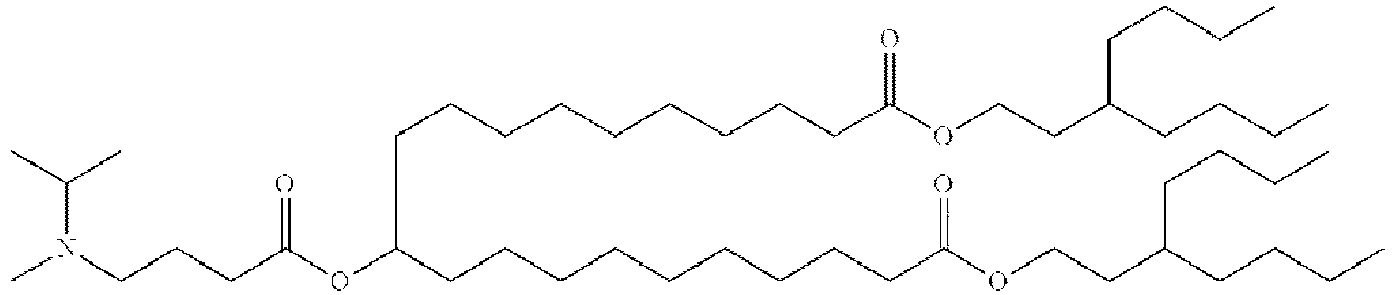
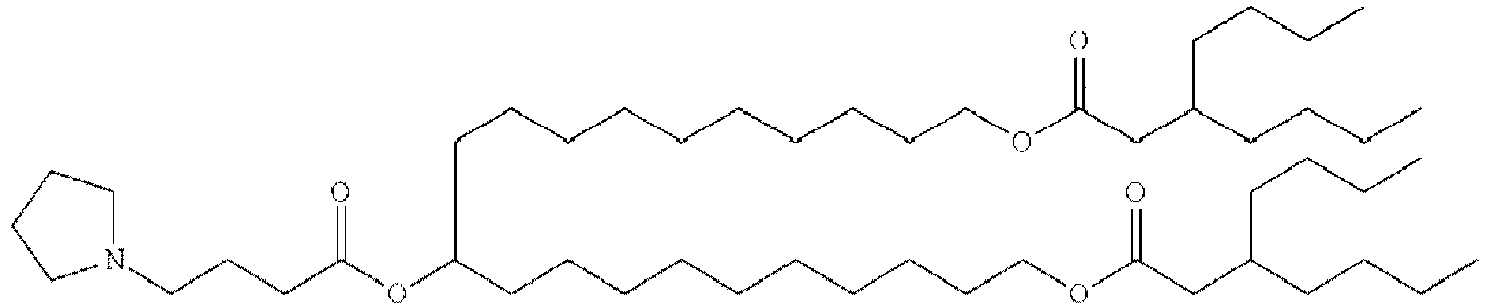
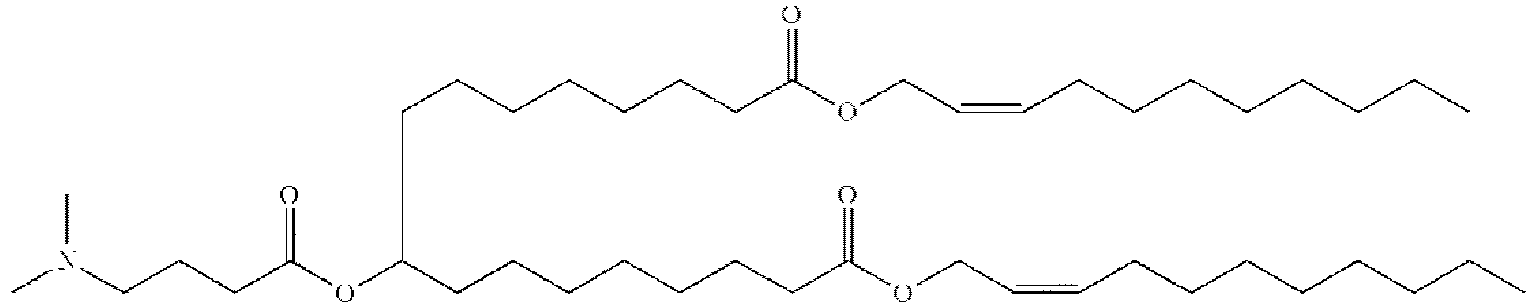
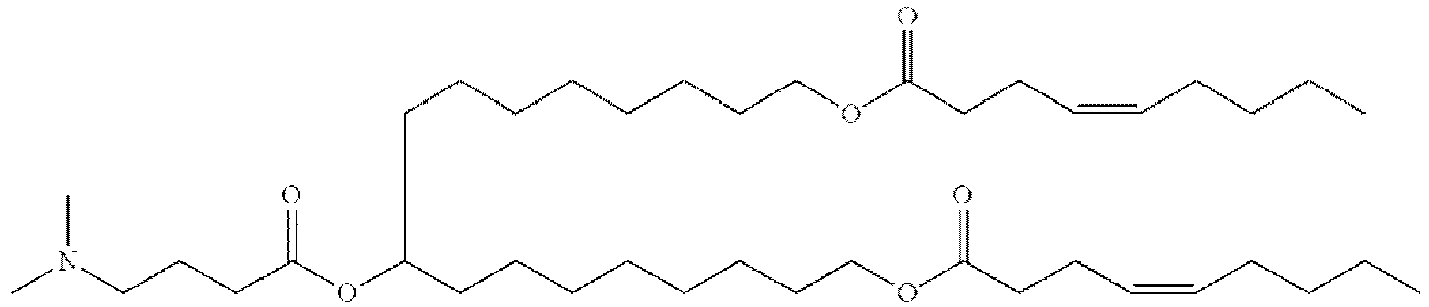
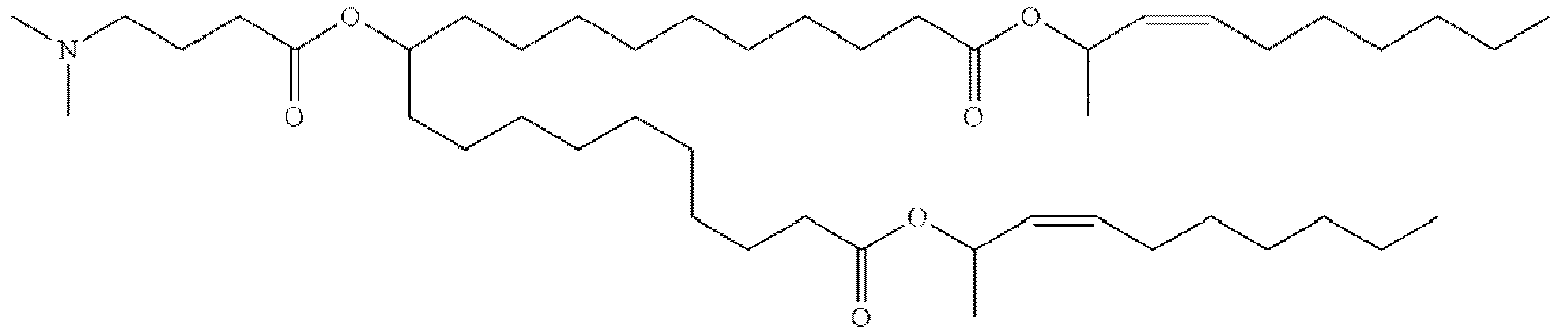
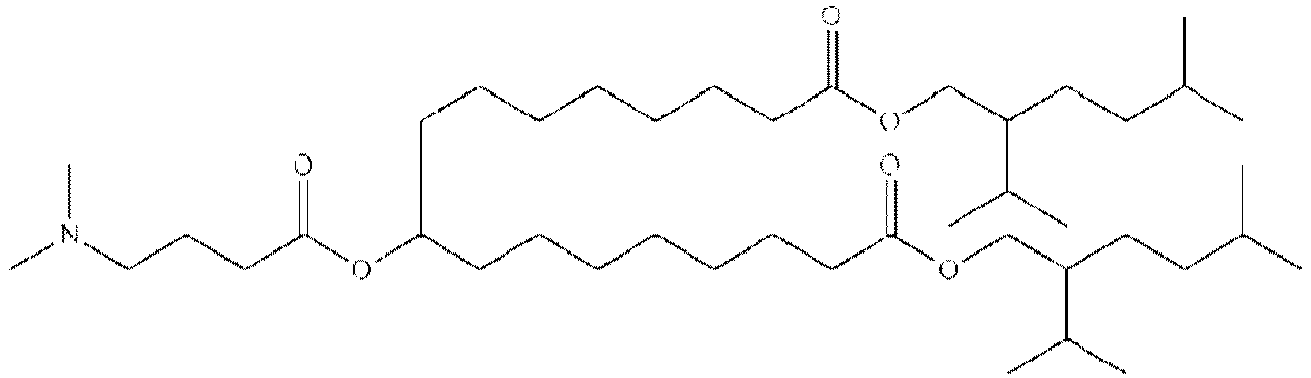
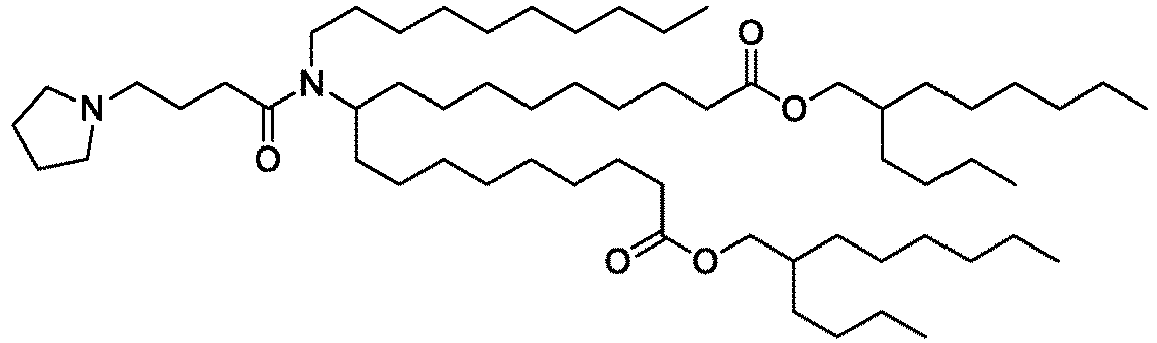
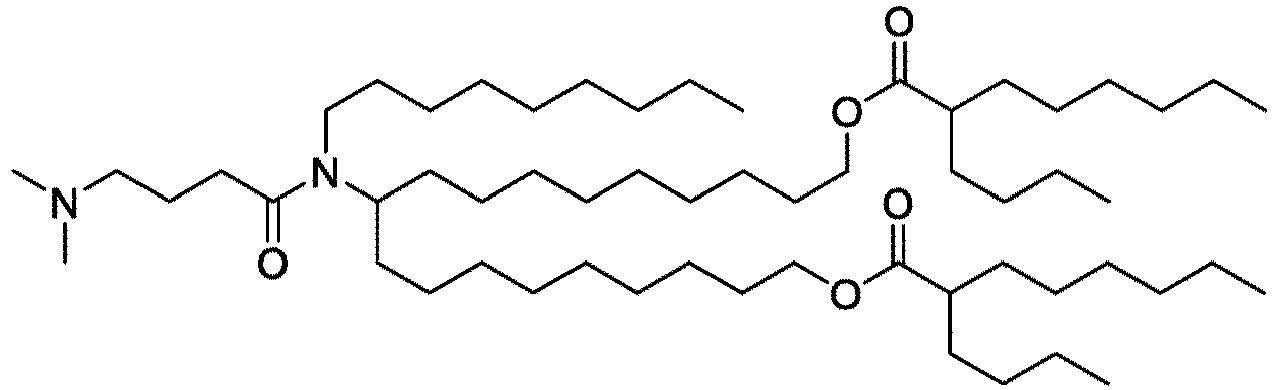
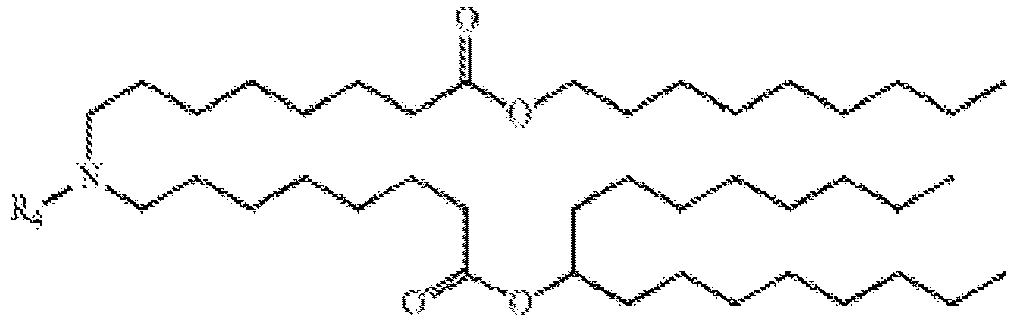
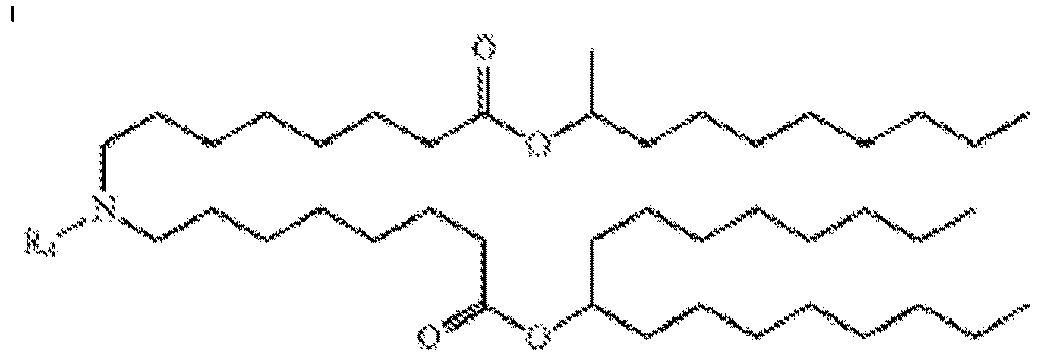
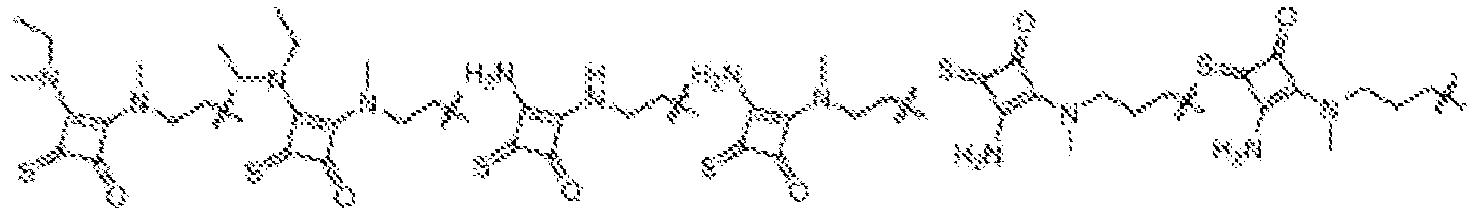
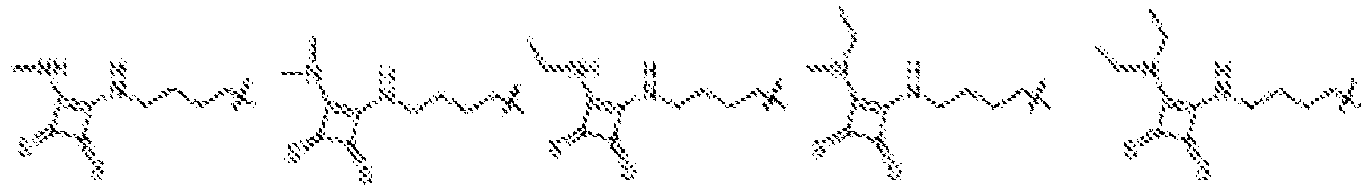
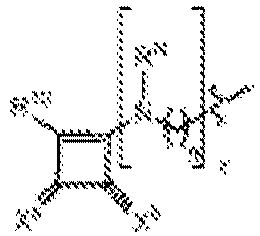
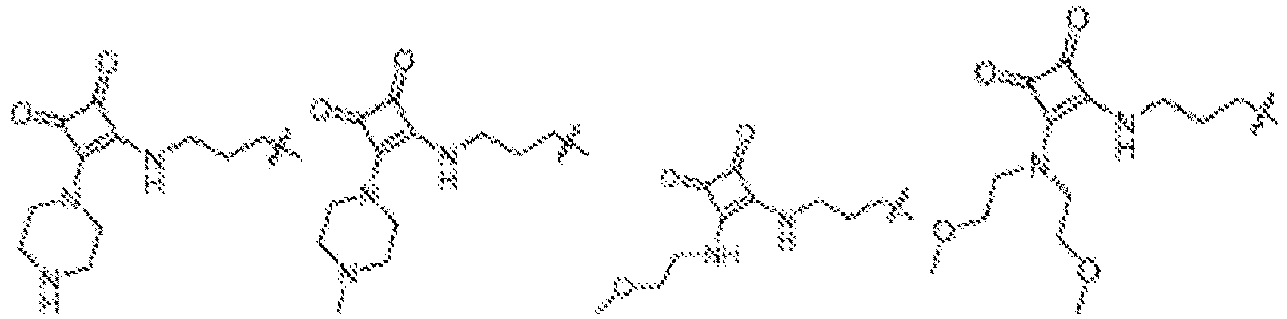
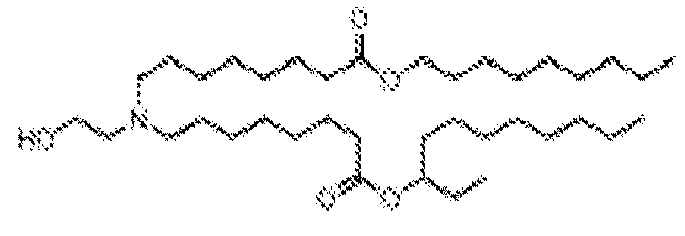
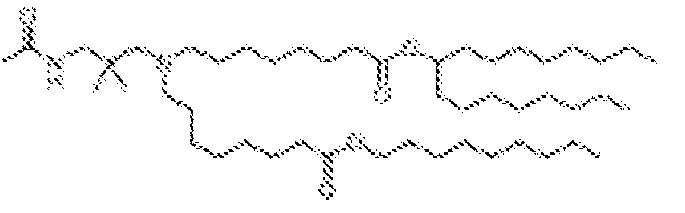
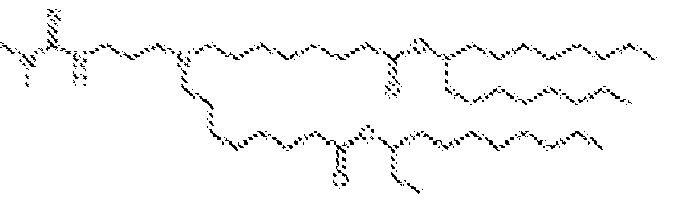
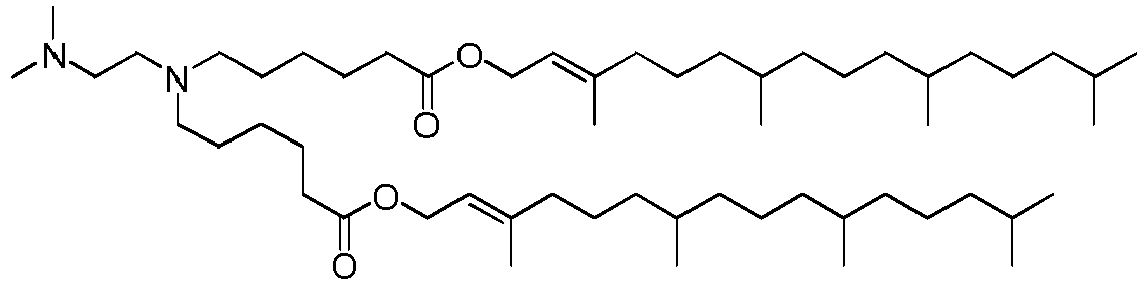
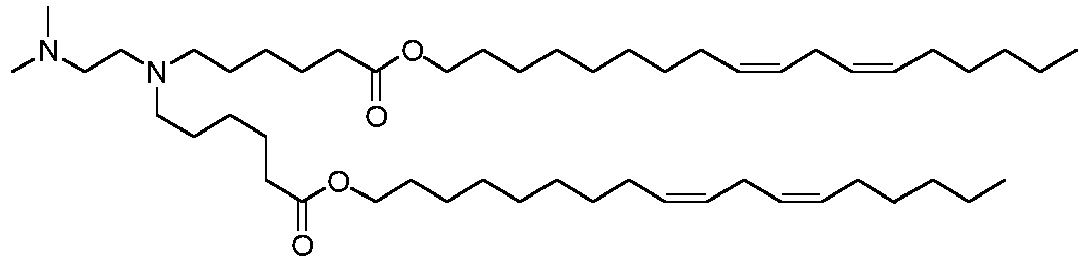
- NRLNQCOGCKAESA-KWXKLSQISA-N [(6z,9z,28z,31z)-heptatriaconta-6,9,28,31-tetraen-19-yl] 4-(dimethylamino)butanoate Chemical compound CCCCC\C=C/C\C=C/CCCCCCCCC(OC(=O)CCCN(C)C)CCCCCCCC\C=C/C\C=C/CCCCC NRLNQCOGCKAESA-KWXKLSQISA-N 0.000 description 30
- 101100439665 Arabidopsis thaliana SWI2 gene Proteins 0.000 description 29
- 239000003814 drug Substances 0.000 description 29
- 235000018102 proteins Nutrition 0.000 description 29
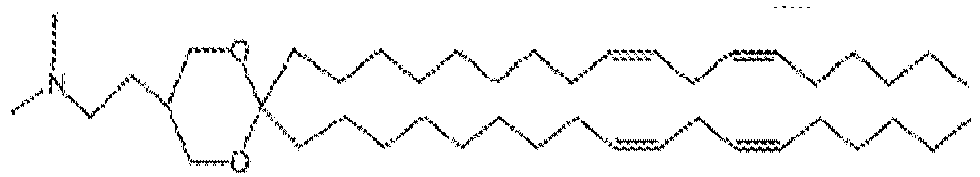
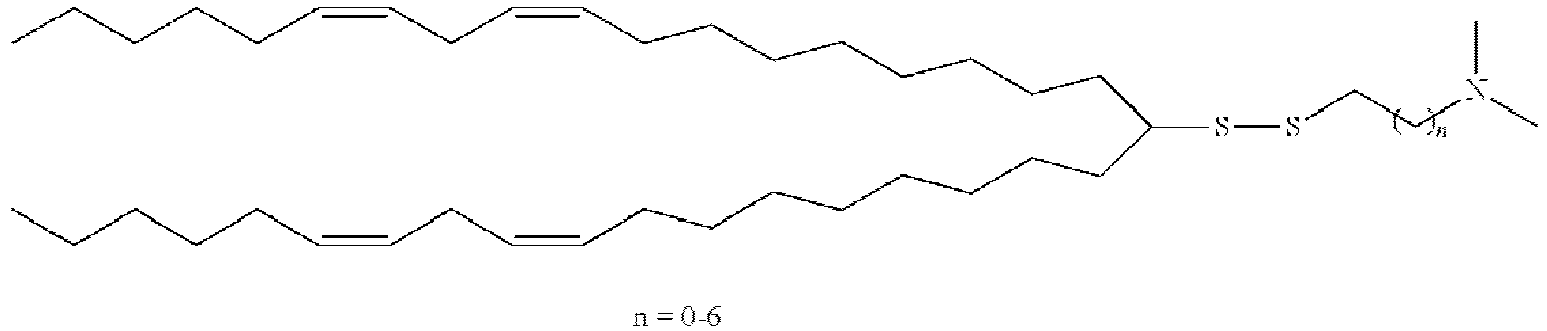
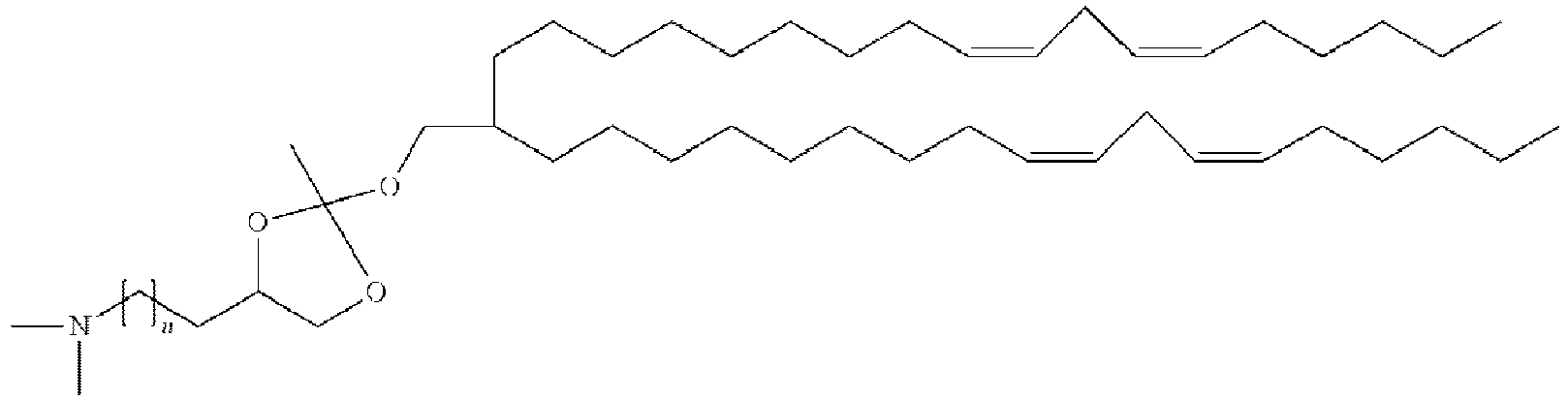
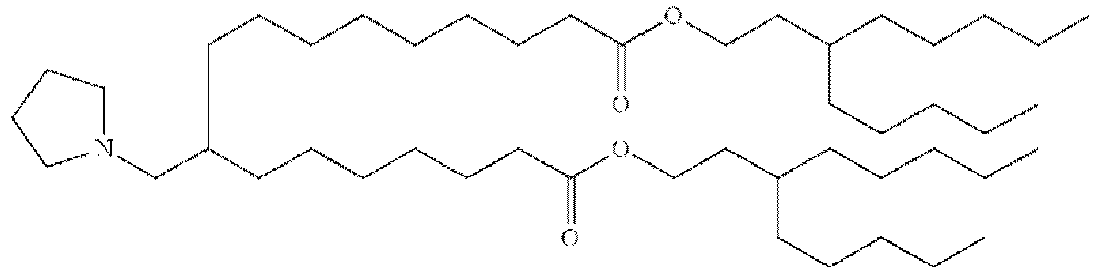
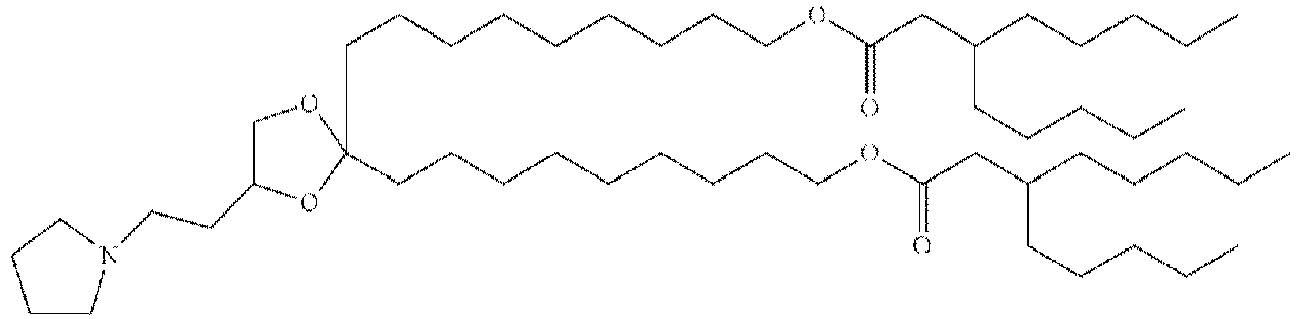
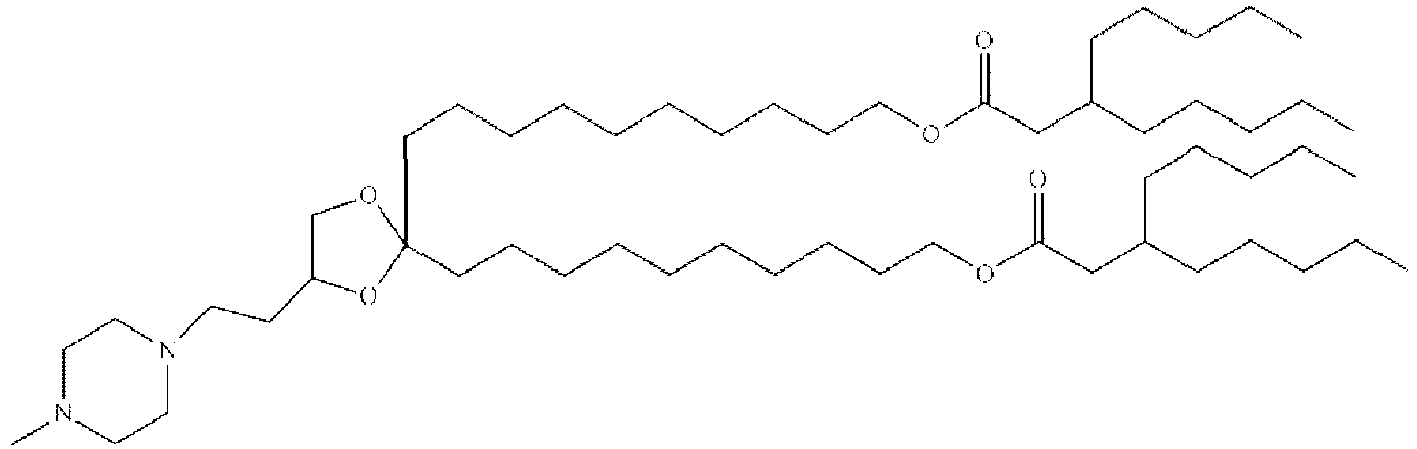
- LRFJOIPOPUJUMI-KWXKLSQISA-N 2-[2,2-bis[(9z,12z)-octadeca-9,12-dienyl]-1,3-dioxolan-4-yl]-n,n-dimethylethanamine Chemical compound CCCCC\C=C/C\C=C/CCCCCCCCC1(CCCCCCCC\C=C/C\C=C/CCCCC)OCC(CCN(C)C)O1 LRFJOIPOPUJUMI-KWXKLSQISA-N 0.000 description 28
- 229930182558 Sterol Natural products 0.000 description 28
- 235000003702 sterols Nutrition 0.000 description 28
- 241000124008 Mammalia Species 0.000 description 27
- 238000009472 formulation Methods 0.000 description 27
- 150000003432 sterols Chemical class 0.000 description 27
- 125000000882 C2-C6 alkenyl group Chemical group 0.000 description 25
- 125000003545 alkoxy group Chemical group 0.000 description 25
- 125000002924 primary amino group Chemical group [H]N([H])* 0.000 description 24
- 229910052717 sulfur Chemical group 0.000 description 23
- 239000000460 chlorine Substances 0.000 description 22
- 239000013612 plasmid Substances 0.000 description 22
- 239000000651 prodrug Substances 0.000 description 22
- 229940002612 prodrug Drugs 0.000 description 22
- 229940124597 therapeutic agent Drugs 0.000 description 22
- 238000013518 transcription Methods 0.000 description 22
- 230000035897 transcription Effects 0.000 description 22
- 125000003282 alkyl amino group Chemical group 0.000 description 21
- 125000001436 propyl group Chemical group [H]C([*])([H])C([H])([H])C([H])([H])[H] 0.000 description 21
- 125000004209 (C1-C8) alkyl group Chemical group 0.000 description 20
- 210000004027 cell Anatomy 0.000 description 20
- 125000000592 heterocycloalkyl group Chemical group 0.000 description 20
- 229910052801 chlorine Inorganic materials 0.000 description 19
- 125000004093 cyano group Chemical group *C#N 0.000 description 19
- 229910052731 fluorine Inorganic materials 0.000 description 19
- 125000001280 n-hexyl group Chemical group C(CCCCC)* 0.000 description 19
- 125000004043 oxo group Chemical group O=* 0.000 description 19
- 239000000074 antisense oligonucleotide Substances 0.000 description 18
- 238000012230 antisense oligonucleotides Methods 0.000 description 18
- 229910052794 bromium Inorganic materials 0.000 description 18
- 230000000295 complement effect Effects 0.000 description 18
- 125000004108 n-butyl group Chemical group [H]C([H])([H])C([H])([H])C([H])([H])C([H])([H])* 0.000 description 18
- OKTJSMMVPCPJKN-UHFFFAOYSA-N Carbon Chemical group [C] OKTJSMMVPCPJKN-UHFFFAOYSA-N 0.000 description 17
- 238000012228 RNA interference-mediated gene silencing Methods 0.000 description 17
- JLCPHMBAVCMARE-UHFFFAOYSA-N [3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-[[3-[[3-[[3-[[3-[[3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-[[5-(2-amino-6-oxo-1H-purin-9-yl)-3-hydroxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(6-aminopurin-9-yl)oxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-5-(4-amino-2-oxopyrimidin-1-yl)oxolan-2-yl]methyl [5-(6-aminopurin-9-yl)-2-(hydroxymethyl)oxolan-3-yl] hydrogen phosphate Polymers Cc1cn(C2CC(OP(O)(=O)OCC3OC(CC3OP(O)(=O)OCC3OC(CC3O)n3cnc4c3nc(N)[nH]c4=O)n3cnc4c3nc(N)[nH]c4=O)C(COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3COP(O)(=O)OC3CC(OC3CO)n3cnc4c(N)ncnc34)n3ccc(N)nc3=O)n3cnc4c(N)ncnc34)n3ccc(N)nc3=O)n3ccc(N)nc3=O)n3ccc(N)nc3=O)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cc(C)c(=O)[nH]c3=O)n3cc(C)c(=O)[nH]c3=O)n3ccc(N)nc3=O)n3cc(C)c(=O)[nH]c3=O)n3cnc4c3nc(N)[nH]c4=O)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)n3cnc4c(N)ncnc34)O2)c(=O)[nH]c1=O JLCPHMBAVCMARE-UHFFFAOYSA-N 0.000 description 17
- 125000004663 dialkyl amino group Chemical group 0.000 description 17
- 230000009368 gene silencing by RNA Effects 0.000 description 17
- 229910052740 iodine Inorganic materials 0.000 description 17
- 125000000547 substituted alkyl group Chemical group 0.000 description 17
- 125000005842 heteroatom Chemical group 0.000 description 16
- 238000012360 testing method Methods 0.000 description 16
- 125000002877 alkyl aryl group Chemical group 0.000 description 15
- 230000000670 limiting effect Effects 0.000 description 15
- 108700011259 MicroRNAs Proteins 0.000 description 14
- 125000002252 acyl group Chemical group 0.000 description 14
- 125000005843 halogen group Chemical group 0.000 description 14
- 239000002679 microRNA Substances 0.000 description 14
- 239000000523 sample Substances 0.000 description 14
- 230000014616 translation Effects 0.000 description 14
- 102000040650 (ribonucleotides)n+m Human genes 0.000 description 13
- 108091032973 (ribonucleotides)n+m Proteins 0.000 description 13
- 229940024606 amino acid Drugs 0.000 description 13
- 235000001014 amino acid Nutrition 0.000 description 13
- 125000001047 cyclobutenyl group Chemical group C1(=CCC1)* 0.000 description 13
- 238000001727 in vivo Methods 0.000 description 13
- 125000001449 isopropyl group Chemical group [H]C([H])([H])C([H])(*)C([H])([H])[H] 0.000 description 13
- 230000004048 modification Effects 0.000 description 13
- 238000012986 modification Methods 0.000 description 13
- 230000001225 therapeutic effect Effects 0.000 description 13
- 108020000948 Antisense Oligonucleotides Proteins 0.000 description 12
- IJGRMHOSHXDMSA-UHFFFAOYSA-N Atomic nitrogen Chemical group N#N IJGRMHOSHXDMSA-UHFFFAOYSA-N 0.000 description 12
- 229910003827 NRaRb Inorganic materials 0.000 description 12
- 150000001413 amino acids Chemical class 0.000 description 12
- 230000027455 binding Effects 0.000 description 12
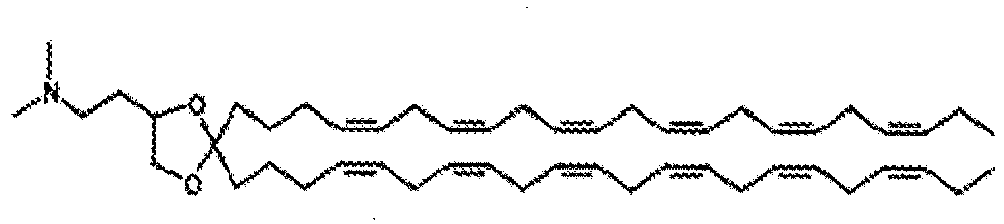
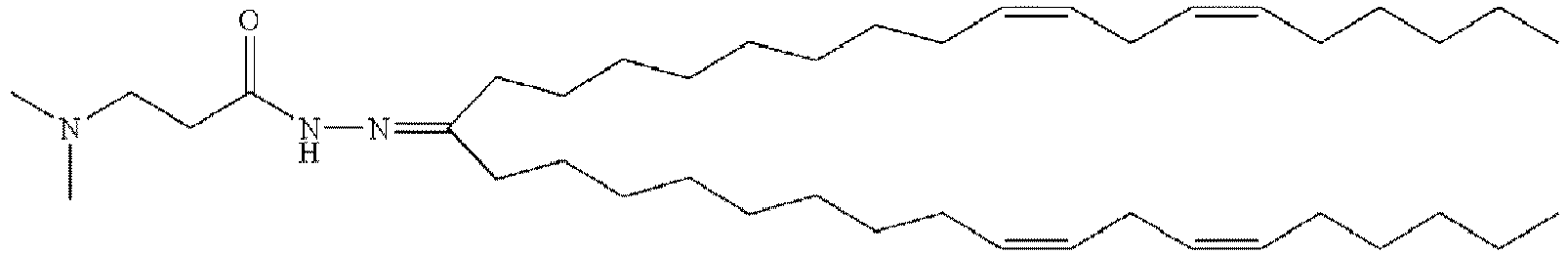
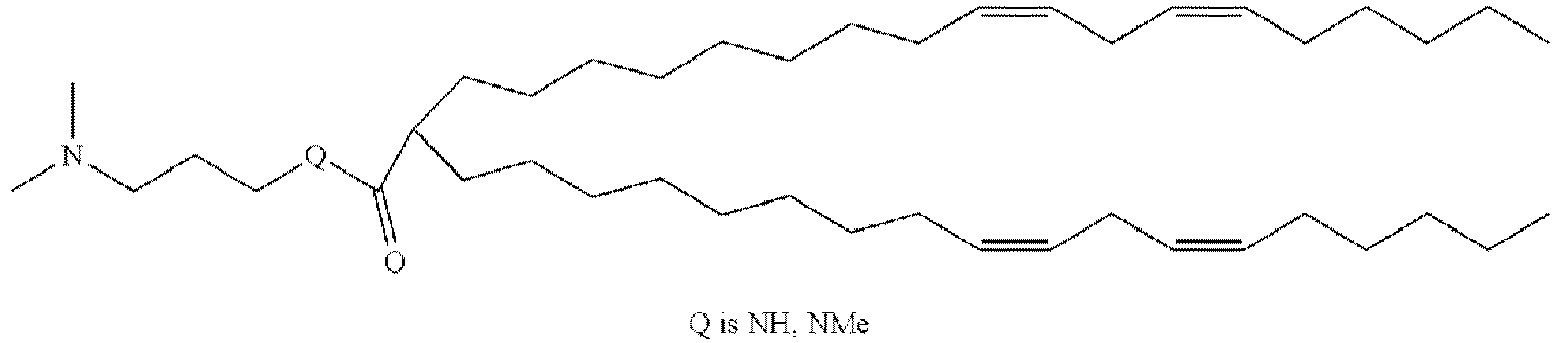
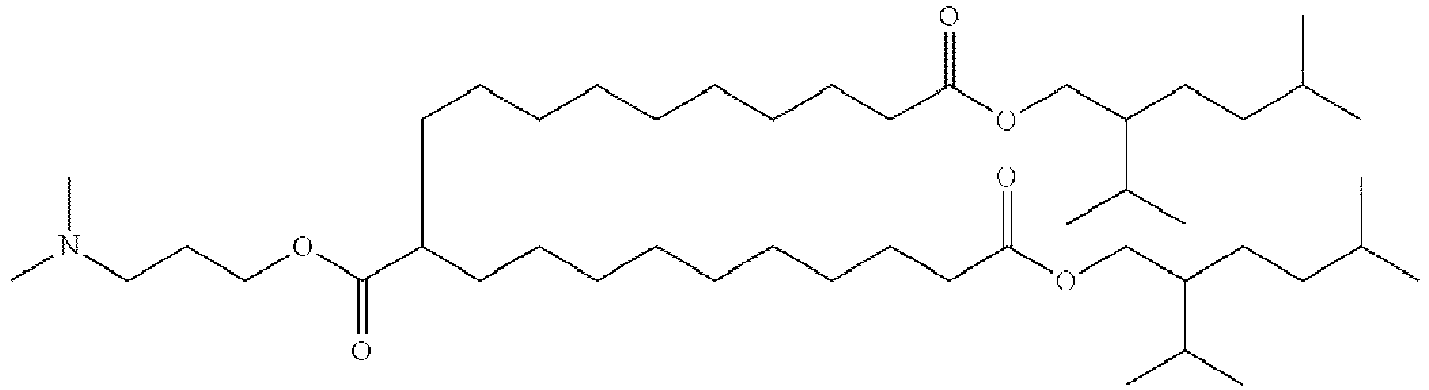
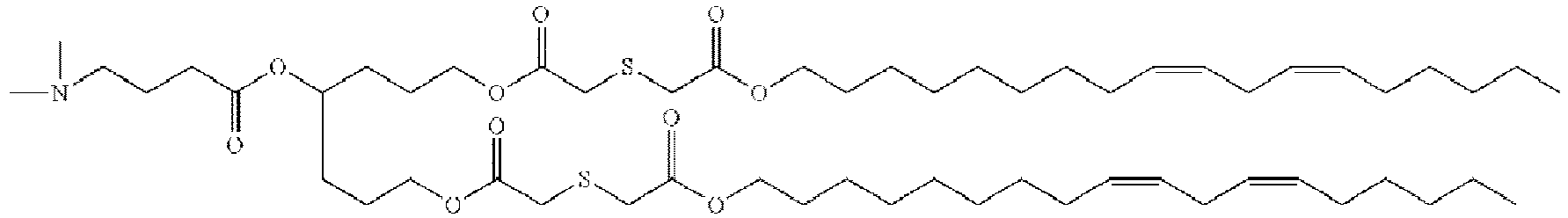
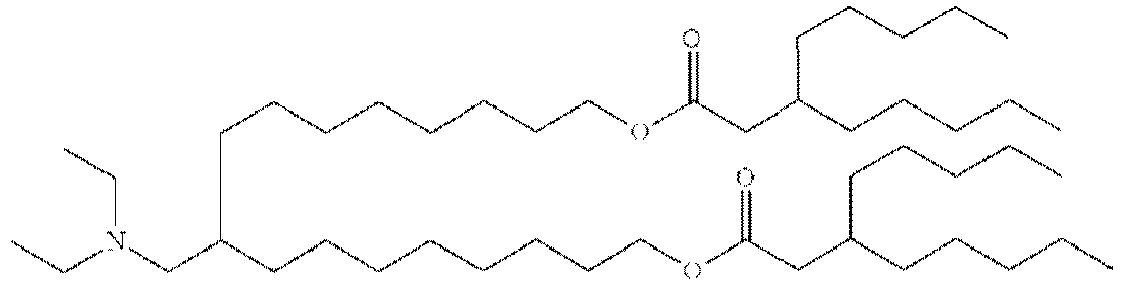
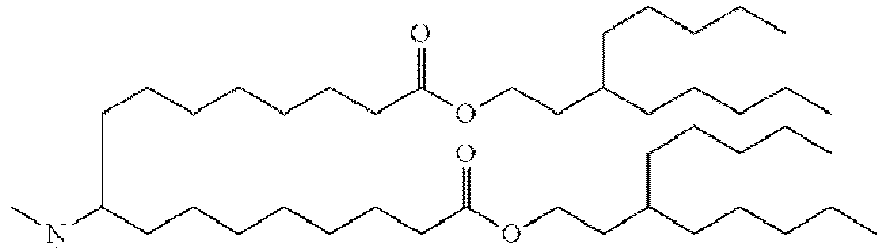
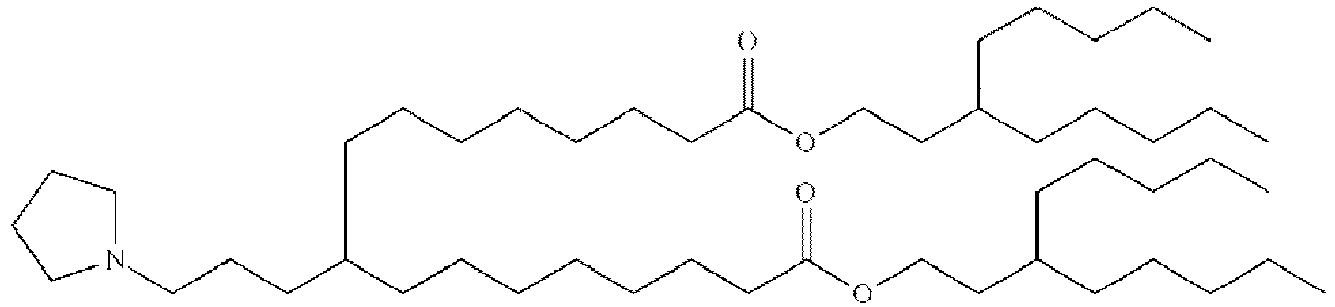
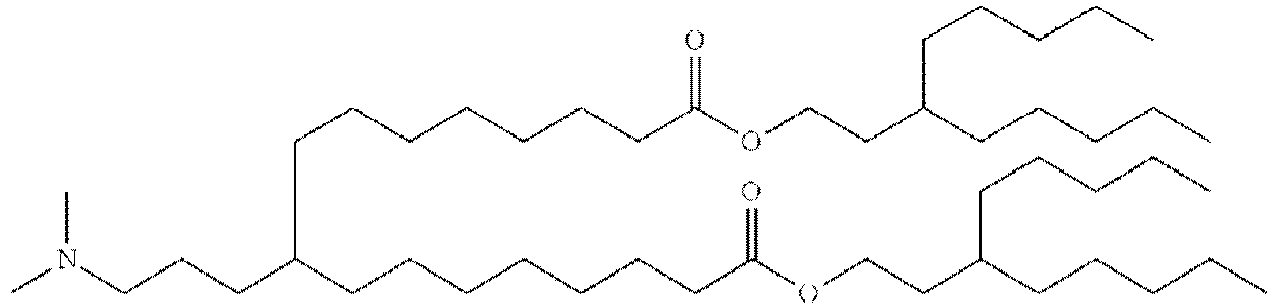
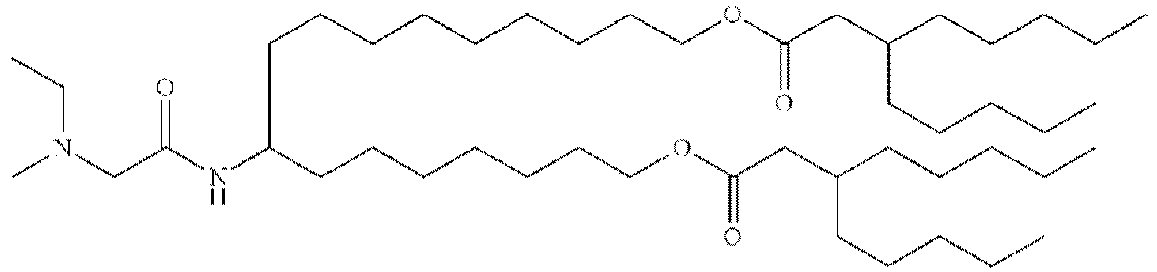
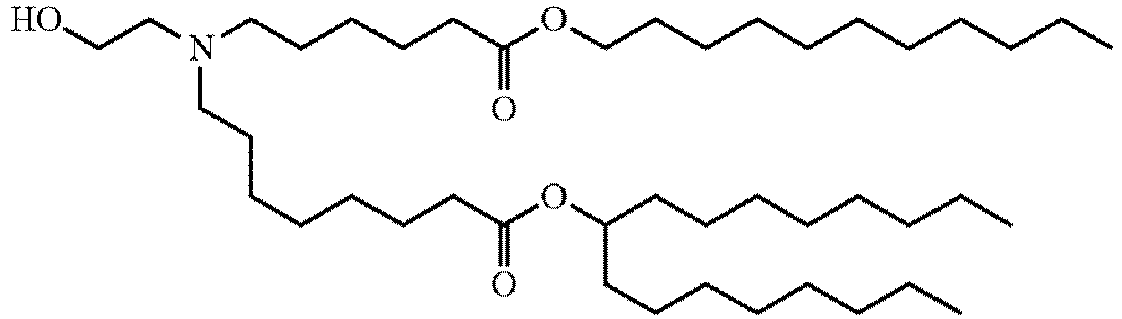
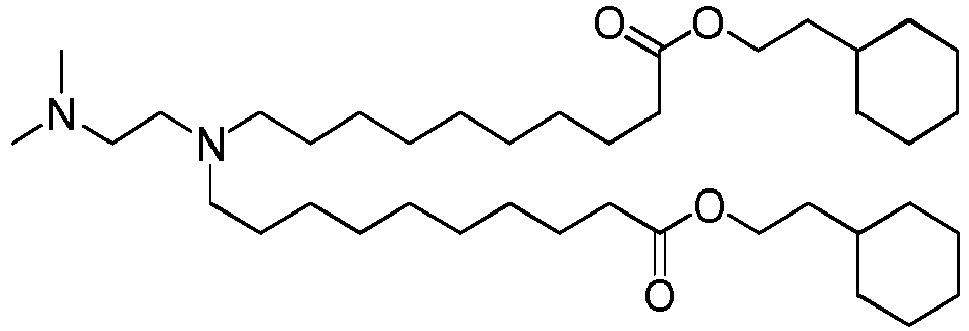
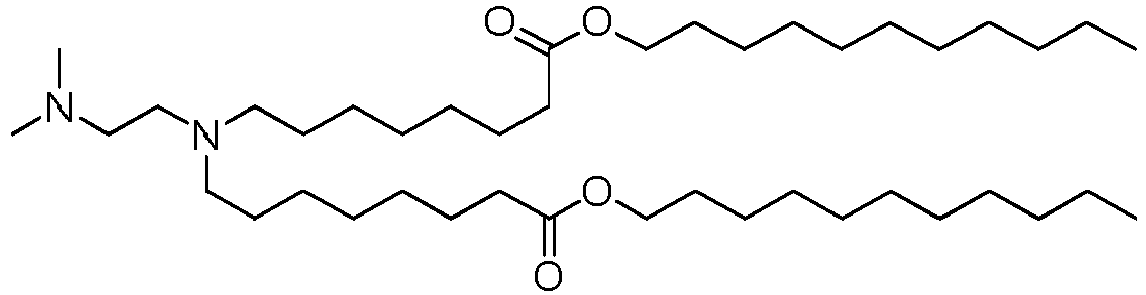
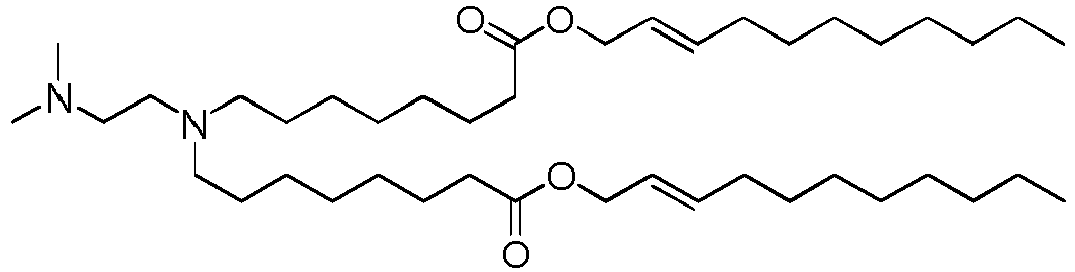
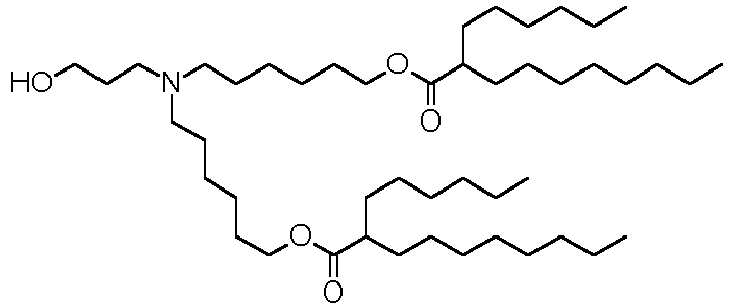
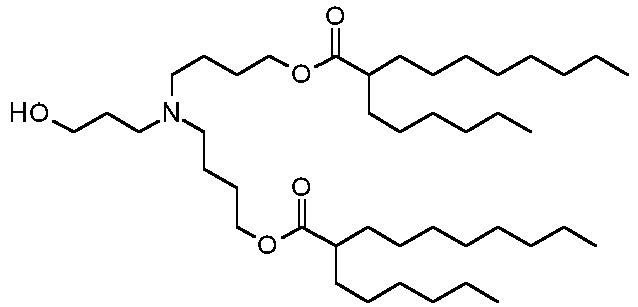
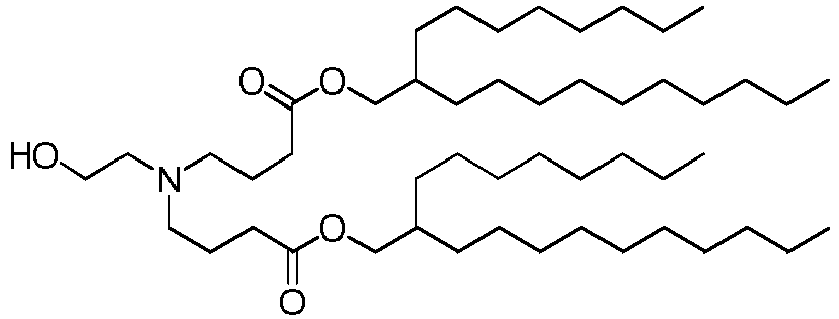
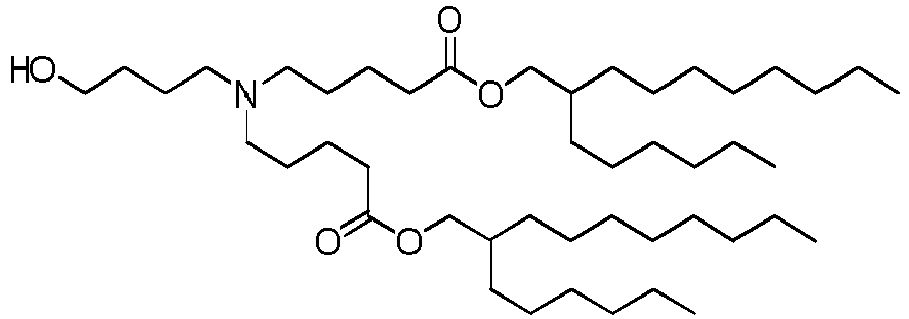
- DGNMJYUPWDTKJB-ZDSKVHJSSA-N bis[(z)-non-2-enyl] 9-[4-(dimethylamino)butanoyloxy]heptadecanedioate Chemical compound CCCCCC\C=C/COC(=O)CCCCCCCC(OC(=O)CCCN(C)C)CCCCCCCC(=O)OC\C=C/CCCCCC DGNMJYUPWDTKJB-ZDSKVHJSSA-N 0.000 description 12
- 102000040430 polynucleotide Human genes 0.000 description 12
- 108091033319 polynucleotide Proteins 0.000 description 12
- 239000002157 polynucleotide Substances 0.000 description 12
- 229910052702 rhenium Inorganic materials 0.000 description 12
- 230000000694 effects Effects 0.000 description 11
- 230000030279 gene silencing Effects 0.000 description 11
- 125000000999 tert-butyl group Chemical group [H]C([H])([H])C(*)(C([H])([H])[H])C([H])([H])[H] 0.000 description 11
- 238000003556 assay Methods 0.000 description 10
- 125000004123 n-propyl group Chemical group [H]C([H])([H])C([H])([H])C([H])([H])* 0.000 description 10
- 238000002360 preparation method Methods 0.000 description 10
- 238000003786 synthesis reaction Methods 0.000 description 10
- 238000013519 translation Methods 0.000 description 10
- QVGXLLKOCUKJST-UHFFFAOYSA-N atomic oxygen Chemical group [O] QVGXLLKOCUKJST-UHFFFAOYSA-N 0.000 description 9
- 239000013068 control sample Substances 0.000 description 9
- 238000005538 encapsulation Methods 0.000 description 9
- 125000003187 heptyl group Chemical group [H]C([*])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H] 0.000 description 9
- 125000000959 isobutyl group Chemical group [H]C([H])([H])C([H])(C([H])([H])[H])C([H])([H])* 0.000 description 9
- 125000001400 nonyl group Chemical group [H]C([*])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H] 0.000 description 9
- 241001465754 Metazoa Species 0.000 description 8
- 229910004749 OS(O)2 Inorganic materials 0.000 description 8
- 230000015572 biosynthetic process Effects 0.000 description 8
- 239000003153 chemical reaction reagent Substances 0.000 description 8
- UYTPUPDQBNUYGX-UHFFFAOYSA-N guanine Chemical class O=C1NC(N)=NC2=C1N=CN2 UYTPUPDQBNUYGX-UHFFFAOYSA-N 0.000 description 8
- 125000005645 linoleyl group Chemical group 0.000 description 8
- 108090000765 processed proteins & peptides Proteins 0.000 description 8
- 238000000746 purification Methods 0.000 description 8
- LFQSCWFLJHTTHZ-UHFFFAOYSA-N Ethanol Chemical compound CCO LFQSCWFLJHTTHZ-UHFFFAOYSA-N 0.000 description 7
- 239000013543 active substance Substances 0.000 description 7
- 125000001316 cycloalkyl alkyl group Chemical group 0.000 description 7
- 150000002148 esters Chemical class 0.000 description 7
- 238000010348 incorporation Methods 0.000 description 7
- 125000000956 methoxy group Chemical group [H]C([H])([H])O* 0.000 description 7
- 230000008685 targeting Effects 0.000 description 7
- 210000001519 tissue Anatomy 0.000 description 7
- 125000004178 (C1-C4) alkyl group Chemical group 0.000 description 6
- 241000699666 Mus <mouse, genus> Species 0.000 description 6
- 125000001931 aliphatic group Chemical group 0.000 description 6
- 238000010171 animal model Methods 0.000 description 6
- 239000003795 chemical substances by application Substances 0.000 description 6
- 238000009826 distribution Methods 0.000 description 6
- 230000006870 function Effects 0.000 description 6
- PEDCQBHIVMGVHV-UHFFFAOYSA-N glycerol group Chemical group OCC(O)CO PEDCQBHIVMGVHV-UHFFFAOYSA-N 0.000 description 6
- RAXXELZNTBOGNW-UHFFFAOYSA-N imidazole Natural products C1=CNC=N1 RAXXELZNTBOGNW-UHFFFAOYSA-N 0.000 description 6
- 125000005647 linker group Chemical group 0.000 description 6
- 238000004519 manufacturing process Methods 0.000 description 6
- 238000002156 mixing Methods 0.000 description 6
- 125000003835 nucleoside group Chemical group 0.000 description 6
- 239000001301 oxygen Substances 0.000 description 6
- 238000006467 substitution reaction Methods 0.000 description 6
- 125000005913 (C3-C6) cycloalkyl group Chemical group 0.000 description 5
- LRYZPFWEZHSTHD-HEFFAWAOSA-O 2-[[(e,2s,3r)-2-formamido-3-hydroxyoctadec-4-enoxy]-hydroxyphosphoryl]oxyethyl-trimethylazanium Chemical class CCCCCCCCCCCCC\C=C\[C@@H](O)[C@@H](NC=O)COP(O)(=O)OCC[N+](C)(C)C LRYZPFWEZHSTHD-HEFFAWAOSA-O 0.000 description 5
- 241000282693 Cercopithecidae Species 0.000 description 5
- UFHFLCQGNIYNRP-UHFFFAOYSA-N Hydrogen Chemical compound [H][H] UFHFLCQGNIYNRP-UHFFFAOYSA-N 0.000 description 5
- 206010028980 Neoplasm Diseases 0.000 description 5
- 108091034057 RNA (poly(A)) Proteins 0.000 description 5
- 241000283984 Rodentia Species 0.000 description 5
- 239000002253 acid Substances 0.000 description 5
- 125000000732 arylene group Chemical group 0.000 description 5
- 230000008901 benefit Effects 0.000 description 5
- 230000001413 cellular effect Effects 0.000 description 5
- 229940079593 drug Drugs 0.000 description 5
- 238000005516 engineering process Methods 0.000 description 5
- 239000012530 fluid Substances 0.000 description 5
- 125000005549 heteroarylene group Chemical group 0.000 description 5
- 238000007901 in situ hybridization Methods 0.000 description 5
- 230000001939 inductive effect Effects 0.000 description 5
- 230000005764 inhibitory process Effects 0.000 description 5
- 238000002955 isolation Methods 0.000 description 5
- 239000003446 ligand Substances 0.000 description 5
- 210000002381 plasma Anatomy 0.000 description 5
- 230000003389 potentiating effect Effects 0.000 description 5
- 230000008569 process Effects 0.000 description 5
- 102000004196 processed proteins & peptides Human genes 0.000 description 5
- 230000002829 reductive effect Effects 0.000 description 5
- 241000894007 species Species 0.000 description 5
- 125000000446 sulfanediyl group Chemical group *S* 0.000 description 5
- 238000012385 systemic delivery Methods 0.000 description 5
- 125000006732 (C1-C15) alkyl group Chemical group 0.000 description 4
- HZAXFHJVJLSVMW-UHFFFAOYSA-N 2-Aminoethan-1-ol Chemical compound NCCO HZAXFHJVJLSVMW-UHFFFAOYSA-N 0.000 description 4
- 125000006538 C11 alkyl group Chemical group [H]C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])* 0.000 description 4
- 125000006539 C12 alkyl group Chemical group [H]C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])* 0.000 description 4
- 108090000994 Catalytic RNA Proteins 0.000 description 4
- 102000053642 Catalytic RNA Human genes 0.000 description 4
- LYCAIKOWRPUZTN-UHFFFAOYSA-N Ethylene glycol Chemical compound OCCO LYCAIKOWRPUZTN-UHFFFAOYSA-N 0.000 description 4
- 108020005004 Guide RNA Proteins 0.000 description 4
- 241000282412 Homo Species 0.000 description 4
- 101710163270 Nuclease Proteins 0.000 description 4
- 108091093037 Peptide nucleic acid Proteins 0.000 description 4
- NQRYJNQNLNOLGT-UHFFFAOYSA-N Piperidine Chemical compound C1CCNCC1 NQRYJNQNLNOLGT-UHFFFAOYSA-N 0.000 description 4
- 101710124239 Poly(A) polymerase Proteins 0.000 description 4
- 239000004698 Polyethylene Substances 0.000 description 4
- 108020000999 Viral RNA Proteins 0.000 description 4
- GFFGJBXGBJISGV-UHFFFAOYSA-N adenyl group Chemical class N1=CN=C2N=CNC2=C1N GFFGJBXGBJISGV-UHFFFAOYSA-N 0.000 description 4
- RYYVLZVUVIJVGH-UHFFFAOYSA-N caffeine Chemical compound CN1C(=O)N(C)C(=O)C2=C1N=CN2C RYYVLZVUVIJVGH-UHFFFAOYSA-N 0.000 description 4
- 230000015556 catabolic process Effects 0.000 description 4
- 230000019522 cellular metabolic process Effects 0.000 description 4
- 230000004700 cellular uptake Effects 0.000 description 4
- 229940106189 ceramide Drugs 0.000 description 4
- 238000006243 chemical reaction Methods 0.000 description 4
- 125000000113 cyclohexyl group Chemical group [H]C1([H])C([H])([H])C([H])([H])C([H])(*)C([H])([H])C1([H])[H] 0.000 description 4
- 125000001559 cyclopropyl group Chemical group [H]C1([H])C([H])([H])C1([H])* 0.000 description 4
- 238000006731 degradation reaction Methods 0.000 description 4
- 238000011161 development Methods 0.000 description 4
- 230000018109 developmental process Effects 0.000 description 4
- POULHZVOKOAJMA-UHFFFAOYSA-N dodecanoic acid Chemical compound CCCCCCCCCCCC(O)=O POULHZVOKOAJMA-UHFFFAOYSA-N 0.000 description 4
- IPCSVZSSVZVIGE-UHFFFAOYSA-N hexadecanoic acid Chemical compound CCCCCCCCCCCCCCCC(O)=O IPCSVZSSVZVIGE-UHFFFAOYSA-N 0.000 description 4
- 230000028993 immune response Effects 0.000 description 4
- 230000003308 immunostimulating effect Effects 0.000 description 4
- 230000002401 inhibitory effect Effects 0.000 description 4
- 230000003834 intracellular effect Effects 0.000 description 4
- 210000004185 liver Anatomy 0.000 description 4
- 108091070501 miRNA Proteins 0.000 description 4
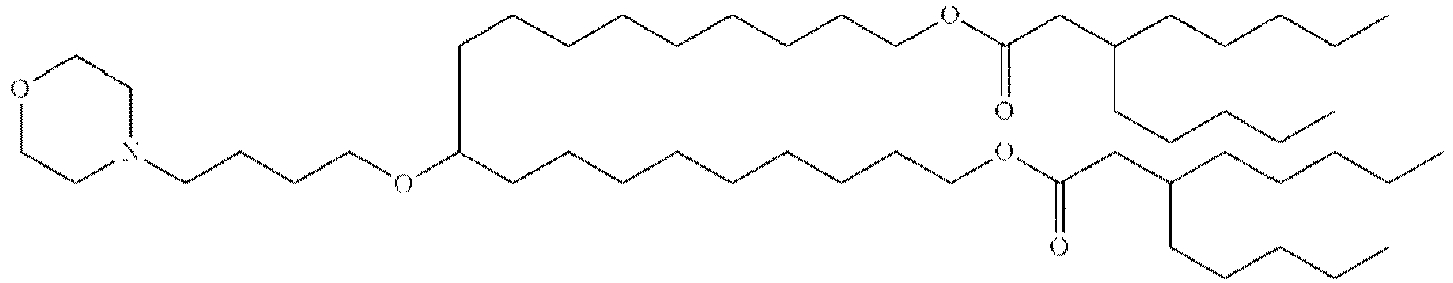
- GLGLUQVVDHRLQK-WRBBJXAJSA-N n,n-dimethyl-2,3-bis[(z)-octadec-9-enoxy]propan-1-amine Chemical compound CCCCCCCC\C=C/CCCCCCCCOCC(CN(C)C)OCCCCCCCC\C=C/CCCCCCCC GLGLUQVVDHRLQK-WRBBJXAJSA-N 0.000 description 4
- 125000000449 nitro group Chemical group [O-][N+](*)=O 0.000 description 4
- QJGQUHMNIGDVPM-UHFFFAOYSA-N nitrogen group Chemical group [N] QJGQUHMNIGDVPM-UHFFFAOYSA-N 0.000 description 4
- 239000002777 nucleoside Substances 0.000 description 4
- 150000003833 nucleoside derivatives Chemical class 0.000 description 4
- 125000002347 octyl group Chemical group [H]C([*])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H] 0.000 description 4
- 150000002888 oleic acid derivatives Chemical class 0.000 description 4
- 239000000546 pharmaceutical excipient Substances 0.000 description 4
- 125000001997 phenyl group Chemical group [H]C1=C([H])C([H])=C(*)C([H])=C1[H] 0.000 description 4
- 125000002467 phosphate group Chemical group [H]OP(=O)(O[H])O[*] 0.000 description 4
- 229920001184 polypeptide Polymers 0.000 description 4
- 238000001243 protein synthesis Methods 0.000 description 4
- 150000003254 radicals Chemical group 0.000 description 4
- 108091092562 ribozyme Proteins 0.000 description 4
- 125000006413 ring segment Chemical group 0.000 description 4
- 239000000758 substrate Substances 0.000 description 4
- GETQZCLCWQTVFV-UHFFFAOYSA-N trimethylamine Chemical compound CN(C)C GETQZCLCWQTVFV-UHFFFAOYSA-N 0.000 description 4
- 125000000008 (C1-C10) alkyl group Chemical group 0.000 description 3
- 125000006619 (C1-C6) dialkylamino group Chemical group 0.000 description 3
- 125000006656 (C2-C4) alkenyl group Chemical group 0.000 description 3
- LDGWQMRUWMSZIU-LQDDAWAPSA-M 2,3-bis[(z)-octadec-9-enoxy]propyl-trimethylazanium;chloride Chemical compound [Cl-].CCCCCCCC\C=C/CCCCCCCCOCC(C[N+](C)(C)C)OCCCCCCCC\C=C/CCCCCCCC LDGWQMRUWMSZIU-LQDDAWAPSA-M 0.000 description 3
- KDCGOANMDULRCW-UHFFFAOYSA-N 7H-purine Chemical compound N1=CNC2=NC=NC2=C1 KDCGOANMDULRCW-UHFFFAOYSA-N 0.000 description 3
- QTBSBXVTEAMEQO-UHFFFAOYSA-N Acetic acid Chemical compound CC(O)=O QTBSBXVTEAMEQO-UHFFFAOYSA-N 0.000 description 3
- 241000894006 Bacteria Species 0.000 description 3
- 108020004705 Codon Proteins 0.000 description 3
- 108020004635 Complementary DNA Proteins 0.000 description 3
- 108090000695 Cytokines Proteins 0.000 description 3
- 102000004127 Cytokines Human genes 0.000 description 3
- 238000002965 ELISA Methods 0.000 description 3
- 102000004190 Enzymes Human genes 0.000 description 3
- 108090000790 Enzymes Proteins 0.000 description 3
- 241000206602 Eukaryota Species 0.000 description 3
- 238000000636 Northern blotting Methods 0.000 description 3
- MUBZPKHOEPUJKR-UHFFFAOYSA-N Oxalic acid Chemical compound OC(=O)C(O)=O MUBZPKHOEPUJKR-UHFFFAOYSA-N 0.000 description 3
- OFOBLEOULBTSOW-UHFFFAOYSA-N Propanedioic acid Natural products OC(=O)CC(O)=O OFOBLEOULBTSOW-UHFFFAOYSA-N 0.000 description 3
- RWRDLPDLKQPQOW-UHFFFAOYSA-N Pyrrolidine Chemical compound C1CCNC1 RWRDLPDLKQPQOW-UHFFFAOYSA-N 0.000 description 3
- NINIDFKCEFEMDL-UHFFFAOYSA-N Sulfur Chemical group [S] NINIDFKCEFEMDL-UHFFFAOYSA-N 0.000 description 3
- FZWLAAWBMGSTSO-UHFFFAOYSA-N Thiazole Chemical compound C1=CSC=N1 FZWLAAWBMGSTSO-UHFFFAOYSA-N 0.000 description 3
- 108020004566 Transfer RNA Proteins 0.000 description 3
- ZMANZCXQSJIPKH-UHFFFAOYSA-N Triethylamine Chemical compound CCN(CC)CC ZMANZCXQSJIPKH-UHFFFAOYSA-N 0.000 description 3
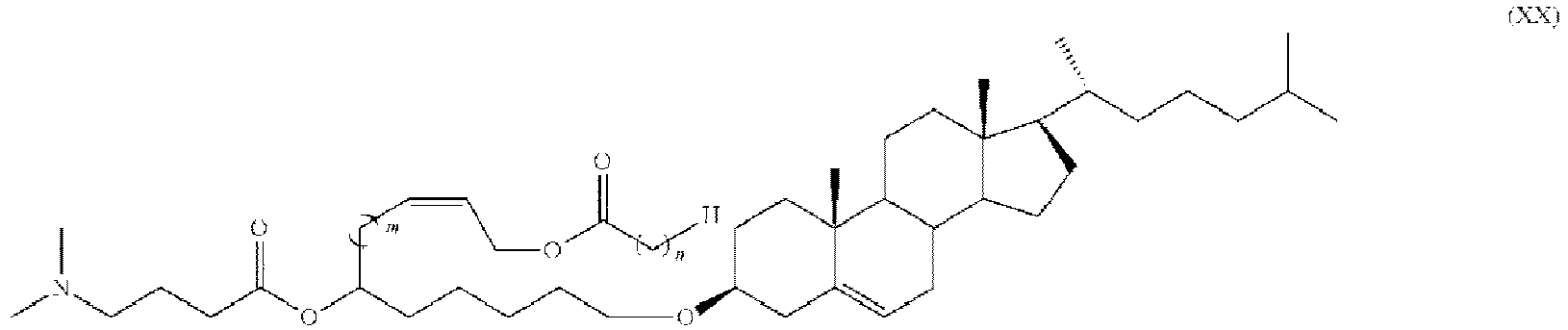
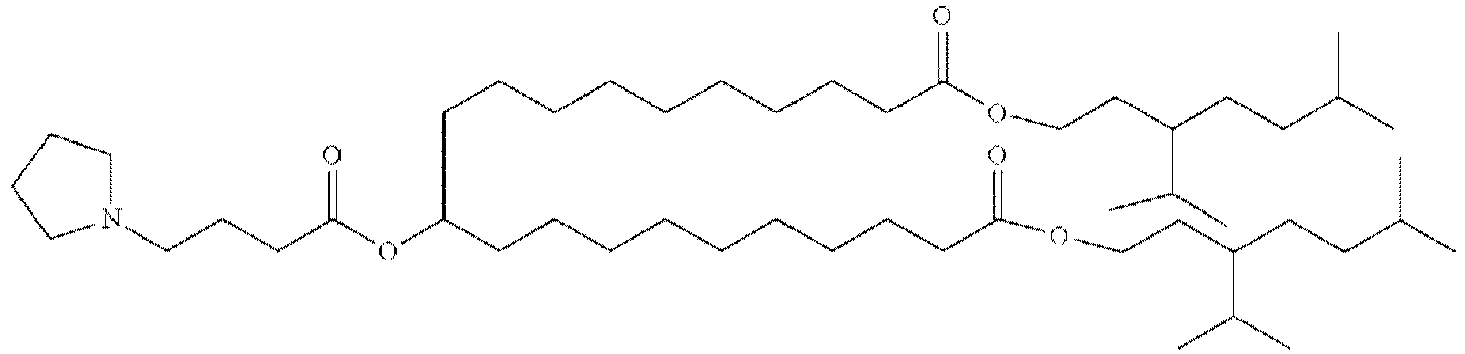
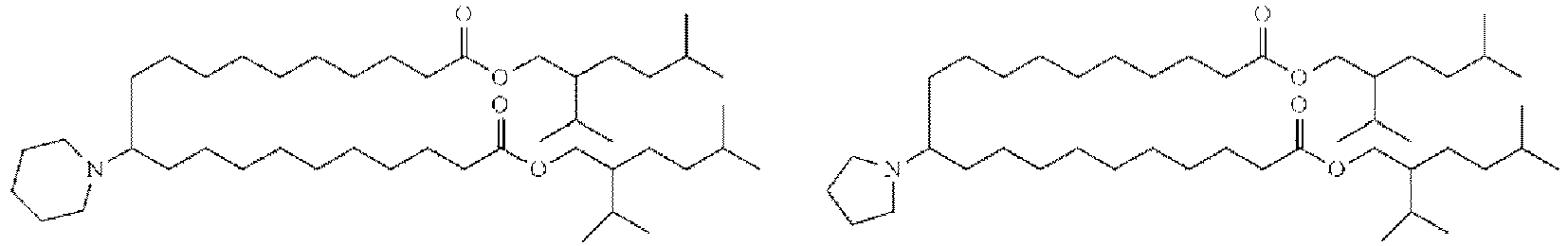
- HIHOWBSBBDRPDW-PTHRTHQKSA-N [(3s,8s,9s,10r,13r,14s,17r)-10,13-dimethyl-17-[(2r)-6-methylheptan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1h-cyclopenta[a]phenanthren-3-yl] n-[2-(dimethylamino)ethyl]carbamate Chemical compound C1C=C2C[C@@H](OC(=O)NCCN(C)C)CC[C@]2(C)[C@@H]2[C@@H]1[C@@H]1CC[C@H]([C@H](C)CCCC(C)C)[C@@]1(C)CC2 HIHOWBSBBDRPDW-PTHRTHQKSA-N 0.000 description 3
- 150000003973 alkyl amines Chemical class 0.000 description 3
- 125000005600 alkyl phosphonate group Chemical group 0.000 description 3
- AWUCVROLDVIAJX-UHFFFAOYSA-N alpha-glycerophosphate Natural products OCC(O)COP(O)(O)=O AWUCVROLDVIAJX-UHFFFAOYSA-N 0.000 description 3
- 238000013459 approach Methods 0.000 description 3
- 125000001797 benzyl group Chemical group [H]C1=C([H])C([H])=C(C([H])=C1[H])C([H])([H])* 0.000 description 3
- 125000002619 bicyclic group Chemical group 0.000 description 3
- 125000000484 butyl group Chemical group [H]C([*])([H])C([H])([H])C([H])([H])C([H])([H])[H] 0.000 description 3
- 238000010804 cDNA synthesis Methods 0.000 description 3
- 201000011510 cancer Diseases 0.000 description 3
- 125000003178 carboxy group Chemical group [H]OC(*)=O 0.000 description 3
- 230000000739 chaotic effect Effects 0.000 description 3
- KRKNYBCHXYNGOX-UHFFFAOYSA-N citric acid Chemical compound OC(=O)CC(O)(C(O)=O)CC(O)=O KRKNYBCHXYNGOX-UHFFFAOYSA-N 0.000 description 3
- 238000003776 cleavage reaction Methods 0.000 description 3
- 239000003184 complementary RNA Substances 0.000 description 3
- 125000004122 cyclic group Chemical group 0.000 description 3
- 125000000000 cycloalkoxy group Chemical group 0.000 description 3
- 125000002704 decyl group Chemical group [H]C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])* 0.000 description 3
- 150000001982 diacylglycerols Chemical class 0.000 description 3
- 239000003085 diluting agent Substances 0.000 description 3
- 125000002147 dimethylamino group Chemical group [H]C([H])([H])N(*)C([H])([H])[H] 0.000 description 3
- 230000002708 enhancing effect Effects 0.000 description 3
- 230000002255 enzymatic effect Effects 0.000 description 3
- 230000009088 enzymatic function Effects 0.000 description 3
- 238000004128 high performance liquid chromatography Methods 0.000 description 3
- CBOIHMRHGLHBPB-UHFFFAOYSA-N hydroxymethyl Chemical compound O[CH2] CBOIHMRHGLHBPB-UHFFFAOYSA-N 0.000 description 3
- 238000001114 immunoprecipitation Methods 0.000 description 3
- 239000003112 inhibitor Substances 0.000 description 3
- 238000002347 injection Methods 0.000 description 3
- 239000007924 injection Substances 0.000 description 3
- 230000007246 mechanism Effects 0.000 description 3
- 238000002515 oligonucleotide synthesis Methods 0.000 description 3
- 150000007530 organic bases Chemical class 0.000 description 3
- 230000001717 pathogenic effect Effects 0.000 description 3
- 230000000144 pharmacologic effect Effects 0.000 description 3
- 238000012247 phenotypical assay Methods 0.000 description 3
- 230000004962 physiological condition Effects 0.000 description 3
- 125000004193 piperazinyl group Chemical group 0.000 description 3
- 238000002264 polyacrylamide gel electrophoresis Methods 0.000 description 3
- 239000002243 precursor Substances 0.000 description 3
- 150000003212 purines Chemical class 0.000 description 3
- 230000001603 reducing effect Effects 0.000 description 3
- 230000007017 scission Effects 0.000 description 3
- 230000009870 specific binding Effects 0.000 description 3
- 235000000346 sugar Nutrition 0.000 description 3
- 239000011593 sulfur Chemical group 0.000 description 3
- 238000002560 therapeutic procedure Methods 0.000 description 3
- 239000001226 triphosphate Substances 0.000 description 3
- YWWVWXASSLXJHU-AATRIKPKSA-N (9E)-tetradecenoic acid Chemical compound CCCC\C=C\CCCCCCCC(O)=O YWWVWXASSLXJHU-AATRIKPKSA-N 0.000 description 2
- CITHEXJVPOWHKC-UUWRZZSWSA-N 1,2-di-O-myristoyl-sn-glycero-3-phosphocholine Chemical compound CCCCCCCCCCCCCC(=O)OC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CCCCCCCCCCCCC CITHEXJVPOWHKC-UUWRZZSWSA-N 0.000 description 2
- FVXDQWZBHIXIEJ-LNDKUQBDSA-N 1,2-di-[(9Z,12Z)-octadecadienoyl]-sn-glycero-3-phosphocholine Chemical compound CCCCC\C=C/C\C=C/CCCCCCCC(=O)OC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CCCCCCC\C=C/C\C=C/CCCCC FVXDQWZBHIXIEJ-LNDKUQBDSA-N 0.000 description 2
- MWRBNPKJOOWZPW-NYVOMTAGSA-N 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine zwitterion Chemical compound CCCCCCCC\C=C/CCCCCCCC(=O)OC[C@H](COP(O)(=O)OCCN)OC(=O)CCCCCCC\C=C/CCCCCCCC MWRBNPKJOOWZPW-NYVOMTAGSA-N 0.000 description 2
- RVHYPUORVDKRTM-UHFFFAOYSA-N 1-[2-[bis(2-hydroxydodecyl)amino]ethyl-[2-[4-[2-[bis(2-hydroxydodecyl)amino]ethyl]piperazin-1-yl]ethyl]amino]dodecan-2-ol Chemical compound CCCCCCCCCCC(O)CN(CC(O)CCCCCCCCCC)CCN(CC(O)CCCCCCCCCC)CCN1CCN(CCN(CC(O)CCCCCCCCCC)CC(O)CCCCCCCCCC)CC1 RVHYPUORVDKRTM-UHFFFAOYSA-N 0.000 description 2
- 125000004973 1-butenyl group Chemical group C(=CCC)* 0.000 description 2
- WTJKGGKOPKCXLL-VYOBOKEXSA-N 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine Chemical compound CCCCCCCCCCCCCCCC(=O)OC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CCCCCCC\C=C/CCCCCCCC WTJKGGKOPKCXLL-VYOBOKEXSA-N 0.000 description 2
- KSXTUUUQYQYKCR-LQDDAWAPSA-M 2,3-bis[[(z)-octadec-9-enoyl]oxy]propyl-trimethylazanium;chloride Chemical compound [Cl-].CCCCCCCC\C=C/CCCCCCCC(=O)OCC(C[N+](C)(C)C)OC(=O)CCCCCCC\C=C/CCCCCCCC KSXTUUUQYQYKCR-LQDDAWAPSA-M 0.000 description 2
- WALUVDCNGPQPOD-UHFFFAOYSA-M 2,3-di(tetradecoxy)propyl-(2-hydroxyethyl)-dimethylazanium;bromide Chemical compound [Br-].CCCCCCCCCCCCCCOCC(C[N+](C)(C)CCO)OCCCCCCCCCCCCCC WALUVDCNGPQPOD-UHFFFAOYSA-M 0.000 description 2
- SXGMVGOVILIERA-UHFFFAOYSA-N 2,3-diaminobutanoic acid Chemical compound CC(N)C(N)C(O)=O SXGMVGOVILIERA-UHFFFAOYSA-N 0.000 description 2
- WXTMDXOMEHJXQO-UHFFFAOYSA-N 2,5-dihydroxybenzoic acid Chemical compound OC(=O)C1=CC(O)=CC=C1O WXTMDXOMEHJXQO-UHFFFAOYSA-N 0.000 description 2
- OYIFNHCXNCRBQI-UHFFFAOYSA-N 2-aminoadipic acid Chemical compound OC(=O)C(N)CCCC(O)=O OYIFNHCXNCRBQI-UHFFFAOYSA-N 0.000 description 2
- RDFMDVXONNIGBC-UHFFFAOYSA-N 2-aminoheptanoic acid Chemical compound CCCCCC(N)C(O)=O RDFMDVXONNIGBC-UHFFFAOYSA-N 0.000 description 2
- ILBCSMHIEBDGJY-UHFFFAOYSA-N 3-[4-(3-aminopropylamino)butylamino]propylcarbamic acid Chemical compound NCCCNCCCCNCCCNC(O)=O ILBCSMHIEBDGJY-UHFFFAOYSA-N 0.000 description 2
- PECYZEOJVXMISF-UHFFFAOYSA-N 3-aminoalanine Chemical compound [NH3+]CC(N)C([O-])=O PECYZEOJVXMISF-UHFFFAOYSA-N 0.000 description 2
- QCXJEYYXVJIFCE-UHFFFAOYSA-N 4-acetamidobenzoic acid Chemical compound CC(=O)NC1=CC=C(C(O)=O)C=C1 QCXJEYYXVJIFCE-UHFFFAOYSA-N 0.000 description 2
- ODHCTXKNWHHXJC-VKHMYHEASA-N 5-oxo-L-proline Chemical compound OC(=O)[C@@H]1CCC(=O)N1 ODHCTXKNWHHXJC-VKHMYHEASA-N 0.000 description 2
- SLXKOJJOQWFEFD-UHFFFAOYSA-N 6-aminohexanoic acid Chemical compound NCCCCCC(O)=O SLXKOJJOQWFEFD-UHFFFAOYSA-N 0.000 description 2
- QGZKDVFQNNGYKY-UHFFFAOYSA-N Ammonia Chemical compound N QGZKDVFQNNGYKY-UHFFFAOYSA-N 0.000 description 2
- QGZKDVFQNNGYKY-UHFFFAOYSA-O Ammonium Chemical compound [NH4+] QGZKDVFQNNGYKY-UHFFFAOYSA-O 0.000 description 2
- CIWBSHSKHKDKBQ-JLAZNSOCSA-N Ascorbic acid Chemical compound OC[C@H](O)[C@H]1OC(=O)C(O)=C1O CIWBSHSKHKDKBQ-JLAZNSOCSA-N 0.000 description 2
- YDNKGFDKKRUKPY-JHOUSYSJSA-N C16 ceramide Natural products CCCCCCCCCCCCCCCC(=O)N[C@@H](CO)[C@H](O)C=CCCCCCCCCCCCCC YDNKGFDKKRUKPY-JHOUSYSJSA-N 0.000 description 2
- 125000006374 C2-C10 alkenyl group Chemical group 0.000 description 2
- 125000005865 C2-C10alkynyl group Chemical group 0.000 description 2
- 125000003601 C2-C6 alkynyl group Chemical group 0.000 description 2
- OYPRJOBELJOOCE-UHFFFAOYSA-N Calcium Chemical compound [Ca] OYPRJOBELJOOCE-UHFFFAOYSA-N 0.000 description 2
- KXDHJXZQYSOELW-UHFFFAOYSA-N Carbamic acid Chemical group NC(O)=O KXDHJXZQYSOELW-UHFFFAOYSA-N 0.000 description 2
- 108091006146 Channels Proteins 0.000 description 2
- HEDRZPFGACZZDS-UHFFFAOYSA-N Chloroform Chemical compound ClC(Cl)Cl HEDRZPFGACZZDS-UHFFFAOYSA-N 0.000 description 2
- RGHNJXZEOKUKBD-SQOUGZDYSA-N D-gluconic acid Chemical compound OC[C@@H](O)[C@@H](O)[C@H](O)[C@@H](O)C(O)=O RGHNJXZEOKUKBD-SQOUGZDYSA-N 0.000 description 2
- 108090000626 DNA-directed RNA polymerases Proteins 0.000 description 2
- 102000004163 DNA-directed RNA polymerases Human genes 0.000 description 2
- XULFJDKZVHTRLG-JDVCJPALSA-N DOSPA trifluoroacetate Chemical compound [O-]C(=O)C(F)(F)F.CCCCCCCC\C=C/CCCCCCCCOCC(C[N+](C)(C)CCNC(=O)C(CCCNCCCN)NCCCN)OCCCCCCCC\C=C/CCCCCCCC XULFJDKZVHTRLG-JDVCJPALSA-N 0.000 description 2
- XBPCUCUWBYBCDP-UHFFFAOYSA-N Dicyclohexylamine Chemical compound C1CCCCC1NC1CCCCC1 XBPCUCUWBYBCDP-UHFFFAOYSA-N 0.000 description 2
- VZCYOOQTPOCHFL-OWOJBTEDSA-N Fumaric acid Chemical compound OC(=O)\C=C\C(O)=O VZCYOOQTPOCHFL-OWOJBTEDSA-N 0.000 description 2
- DSLZVSRJTYRBFB-UHFFFAOYSA-N Galactaric acid Natural products OC(=O)C(O)C(O)C(O)C(O)C(O)=O DSLZVSRJTYRBFB-UHFFFAOYSA-N 0.000 description 2
- WHUUTDBJXJRKMK-UHFFFAOYSA-N Glutamic acid Natural products OC(=O)C(N)CCC(O)=O WHUUTDBJXJRKMK-UHFFFAOYSA-N 0.000 description 2
- AEMRFAOFKBGASW-UHFFFAOYSA-N Glycolic acid Chemical compound OCC(O)=O AEMRFAOFKBGASW-UHFFFAOYSA-N 0.000 description 2
- 229930186217 Glycolipid Natural products 0.000 description 2
- NYHBQMYGNKIUIF-UUOKFMHZSA-N Guanosine Chemical compound C1=NC=2C(=O)NC(N)=NC=2N1[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O NYHBQMYGNKIUIF-UUOKFMHZSA-N 0.000 description 2
- VEXZGXHMUGYJMC-UHFFFAOYSA-N Hydrochloric acid Chemical compound Cl VEXZGXHMUGYJMC-UHFFFAOYSA-N 0.000 description 2
- DGAQECJNVWCQMB-PUAWFVPOSA-M Ilexoside XXIX Chemical compound C[C@@H]1CC[C@@]2(CC[C@@]3(C(=CC[C@H]4[C@]3(CC[C@@H]5[C@@]4(CC[C@@H](C5(C)C)OS(=O)(=O)[O-])C)C)[C@@H]2[C@]1(C)O)C)C(=O)O[C@H]6[C@@H]([C@H]([C@@H]([C@H](O6)CO)O)O)O.[Na+] DGAQECJNVWCQMB-PUAWFVPOSA-M 0.000 description 2
- XEEYBQQBJWHFJM-UHFFFAOYSA-N Iron Chemical compound [Fe] XEEYBQQBJWHFJM-UHFFFAOYSA-N 0.000 description 2
- LPHGQDQBBGAPDZ-UHFFFAOYSA-N Isocaffeine Natural products CN1C(=O)N(C)C(=O)C2=C1N(C)C=N2 LPHGQDQBBGAPDZ-UHFFFAOYSA-N 0.000 description 2
- KFZMGEQAYNKOFK-UHFFFAOYSA-N Isopropanol Chemical compound CC(C)O KFZMGEQAYNKOFK-UHFFFAOYSA-N 0.000 description 2
- CKLJMWTZIZZHCS-REOHCLBHSA-N L-aspartic acid Chemical compound OC(=O)[C@@H](N)CC(O)=O CKLJMWTZIZZHCS-REOHCLBHSA-N 0.000 description 2
- WHUUTDBJXJRKMK-VKHMYHEASA-N L-glutamic acid Chemical compound OC(=O)[C@@H](N)CCC(O)=O WHUUTDBJXJRKMK-VKHMYHEASA-N 0.000 description 2
- KDXKERNSBIXSRK-YFKPBYRVSA-N L-lysine Chemical compound NCCCC[C@H](N)C(O)=O KDXKERNSBIXSRK-YFKPBYRVSA-N 0.000 description 2
- 239000005639 Lauric acid Substances 0.000 description 2
- 239000000232 Lipid Bilayer Substances 0.000 description 2
- 108060001084 Luciferase Proteins 0.000 description 2
- 239000005089 Luciferase Substances 0.000 description 2
- KDXKERNSBIXSRK-UHFFFAOYSA-N Lysine Natural products NCCCCC(N)C(O)=O KDXKERNSBIXSRK-UHFFFAOYSA-N 0.000 description 2
- AFVFQIVMOAPDHO-UHFFFAOYSA-N Methanesulfonic acid Chemical compound CS(O)(=O)=O AFVFQIVMOAPDHO-UHFFFAOYSA-N 0.000 description 2
- DBTDEFJAFBUGPP-UHFFFAOYSA-N Methanethial Chemical compound S=C DBTDEFJAFBUGPP-UHFFFAOYSA-N 0.000 description 2
- 241000699670 Mus sp. Species 0.000 description 2
- CRJGESKKUOMBCT-VQTJNVASSA-N N-acetylsphinganine Chemical compound CCCCCCCCCCCCCCC[C@@H](O)[C@H](CO)NC(C)=O CRJGESKKUOMBCT-VQTJNVASSA-N 0.000 description 2
- QIAFMBKCNZACKA-UHFFFAOYSA-N N-benzoylglycine Chemical compound OC(=O)CNC(=O)C1=CC=CC=C1 QIAFMBKCNZACKA-UHFFFAOYSA-N 0.000 description 2
- UEEJHVSXFDXPFK-UHFFFAOYSA-N N-dimethylaminoethanol Chemical compound CN(C)CCO UEEJHVSXFDXPFK-UHFFFAOYSA-N 0.000 description 2
- KSPIYJQBLVDRRI-UHFFFAOYSA-N N-methylisoleucine Chemical compound CCC(C)C(NC)C(O)=O KSPIYJQBLVDRRI-UHFFFAOYSA-N 0.000 description 2
- PVNIIMVLHYAWGP-UHFFFAOYSA-N Niacin Chemical compound OC(=O)C1=CC=CN=C1 PVNIIMVLHYAWGP-UHFFFAOYSA-N 0.000 description 2
- 108091028043 Nucleic acid sequence Proteins 0.000 description 2
- ZCQWOFVYLHDMMC-UHFFFAOYSA-N Oxazole Chemical compound C1=COC=N1 ZCQWOFVYLHDMMC-UHFFFAOYSA-N 0.000 description 2
- 235000021314 Palmitic acid Nutrition 0.000 description 2
- ISWSIDIOOBJBQZ-UHFFFAOYSA-N Phenol Chemical compound OC1=CC=CC=C1 ISWSIDIOOBJBQZ-UHFFFAOYSA-N 0.000 description 2
- NBIIXXVUZAFLBC-UHFFFAOYSA-N Phosphoric acid Chemical compound OP(O)(O)=O NBIIXXVUZAFLBC-UHFFFAOYSA-N 0.000 description 2
- GLUUGHFHXGJENI-UHFFFAOYSA-N Piperazine Chemical compound C1CNCCN1 GLUUGHFHXGJENI-UHFFFAOYSA-N 0.000 description 2
- 229920003171 Poly (ethylene oxide) Polymers 0.000 description 2
- 108091036407 Polyadenylation Proteins 0.000 description 2
- ZLMJMSJWJFRBEC-UHFFFAOYSA-N Potassium Chemical compound [K] ZLMJMSJWJFRBEC-UHFFFAOYSA-N 0.000 description 2
- JUJWROOIHBZHMG-UHFFFAOYSA-N Pyridine Chemical compound C1=CC=NC=C1 JUJWROOIHBZHMG-UHFFFAOYSA-N 0.000 description 2
- CZPWVGJYEJSRLH-UHFFFAOYSA-N Pyrimidine Chemical compound C1=CN=CN=C1 CZPWVGJYEJSRLH-UHFFFAOYSA-N 0.000 description 2
- ODHCTXKNWHHXJC-GSVOUGTGSA-N Pyroglutamic acid Natural products OC(=O)[C@H]1CCC(=O)N1 ODHCTXKNWHHXJC-GSVOUGTGSA-N 0.000 description 2
- LCTONWCANYUPML-UHFFFAOYSA-N Pyruvic acid Chemical compound CC(=O)C(O)=O LCTONWCANYUPML-UHFFFAOYSA-N 0.000 description 2
- 108091000106 RNA cap binding Proteins 0.000 description 2
- 102000028391 RNA cap binding Human genes 0.000 description 2
- 108091028664 Ribonucleotide Proteins 0.000 description 2
- 108091081021 Sense strand Proteins 0.000 description 2
- VYPSYNLAJGMNEJ-UHFFFAOYSA-N Silicium dioxide Chemical compound O=[Si]=O VYPSYNLAJGMNEJ-UHFFFAOYSA-N 0.000 description 2
- 108091027967 Small hairpin RNA Proteins 0.000 description 2
- 235000021355 Stearic acid Nutrition 0.000 description 2
- QAOWNCQODCNURD-UHFFFAOYSA-N Sulfuric acid Chemical compound OS(O)(=O)=O QAOWNCQODCNURD-UHFFFAOYSA-N 0.000 description 2
- WYURNTSHIVDZCO-UHFFFAOYSA-N Tetrahydrofuran Chemical compound C1CCOC1 WYURNTSHIVDZCO-UHFFFAOYSA-N 0.000 description 2
- RYYWUUFWQRZTIU-UHFFFAOYSA-N Thiophosphoric acid Chemical class OP(O)(S)=O RYYWUUFWQRZTIU-UHFFFAOYSA-N 0.000 description 2
- 108091036066 Three prime untranslated region Proteins 0.000 description 2
- DTQVDTLACAAQTR-UHFFFAOYSA-M Trifluoroacetate Chemical compound [O-]C(=O)C(F)(F)F DTQVDTLACAAQTR-UHFFFAOYSA-M 0.000 description 2
- DTQVDTLACAAQTR-UHFFFAOYSA-N Trifluoroacetic acid Chemical compound OC(=O)C(F)(F)F DTQVDTLACAAQTR-UHFFFAOYSA-N 0.000 description 2
- 108091023045 Untranslated Region Proteins 0.000 description 2
- ISAKRJDGNUQOIC-UHFFFAOYSA-N Uracil Chemical compound O=C1C=CNC(=O)N1 ISAKRJDGNUQOIC-UHFFFAOYSA-N 0.000 description 2
- DRTQHJPVMGBUCF-XVFCMESISA-N Uridine Chemical compound O[C@@H]1[C@H](O)[C@@H](CO)O[C@H]1N1C(=O)NC(=O)C=C1 DRTQHJPVMGBUCF-XVFCMESISA-N 0.000 description 2
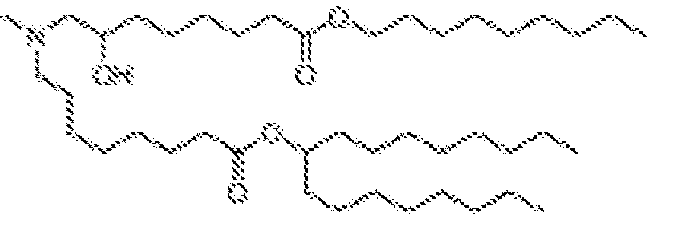
- ATBOMIWRCZXYSZ-XZBBILGWSA-N [1-[2,3-dihydroxypropoxy(hydroxy)phosphoryl]oxy-3-hexadecanoyloxypropan-2-yl] (9e,12e)-octadeca-9,12-dienoate Chemical class CCCCCCCCCCCCCCCC(=O)OCC(COP(O)(=O)OCC(O)CO)OC(=O)CCCCCCC\C=C\C\C=C\CCCCC ATBOMIWRCZXYSZ-XZBBILGWSA-N 0.000 description 2
- NYDLOCKCVISJKK-WRBBJXAJSA-N [3-(dimethylamino)-2-[(z)-octadec-9-enoyl]oxypropyl] (z)-octadec-9-enoate Chemical compound CCCCCCCC\C=C/CCCCCCCC(=O)OCC(CN(C)C)OC(=O)CCCCCCC\C=C/CCCCCCCC NYDLOCKCVISJKK-WRBBJXAJSA-N 0.000 description 2
- ODHCTXKNWHHXJC-UHFFFAOYSA-N acide pyroglutamique Natural products OC(=O)C1CCC(=O)N1 ODHCTXKNWHHXJC-UHFFFAOYSA-N 0.000 description 2
- 230000009471 action Effects 0.000 description 2
- 230000003213 activating effect Effects 0.000 description 2
- 125000004423 acyloxy group Chemical group 0.000 description 2
- OIRDTQYFTABQOQ-KQYNXXCUSA-N adenosine Chemical compound C1=NC=2C(N)=NC=NC=2N1[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O OIRDTQYFTABQOQ-KQYNXXCUSA-N 0.000 description 2
- WNLRTRBMVRJNCN-UHFFFAOYSA-N adipic acid Chemical compound OC(=O)CCCCC(O)=O WNLRTRBMVRJNCN-UHFFFAOYSA-N 0.000 description 2
- 230000002776 aggregation Effects 0.000 description 2
- 238000004220 aggregation Methods 0.000 description 2
- 150000001345 alkine derivatives Chemical class 0.000 description 2
- MBMBGCFOFBJSGT-KUBAVDMBSA-N all-cis-docosa-4,7,10,13,16,19-hexaenoic acid Chemical compound CC\C=C/C\C=C/C\C=C/C\C=C/C\C=C/C\C=C/CCC(O)=O MBMBGCFOFBJSGT-KUBAVDMBSA-N 0.000 description 2
- QWCKQJZIFLGMSD-UHFFFAOYSA-N alpha-aminobutyric acid Chemical compound CCC(N)C(O)=O QWCKQJZIFLGMSD-UHFFFAOYSA-N 0.000 description 2
- DTOSIQBPPRVQHS-PDBXOOCHSA-N alpha-linolenic acid Chemical compound CC\C=C/C\C=C/C\C=C/CCCCCCCC(O)=O DTOSIQBPPRVQHS-PDBXOOCHSA-N 0.000 description 2
- 150000001408 amides Chemical class 0.000 description 2
- 125000000539 amino acid group Chemical group 0.000 description 2
- 125000003277 amino group Chemical group 0.000 description 2
- 229960002684 aminocaproic acid Drugs 0.000 description 2
- 238000004458 analytical method Methods 0.000 description 2
- 125000000129 anionic group Chemical group 0.000 description 2
- 239000007864 aqueous solution Substances 0.000 description 2
- YZXBAPSDXZZRGB-DOFZRALJSA-N arachidonic acid Chemical compound CCCCC\C=C/C\C=C/C\C=C/C\C=C/CCCC(O)=O YZXBAPSDXZZRGB-DOFZRALJSA-N 0.000 description 2
- HONIICLYMWZJFZ-UHFFFAOYSA-N azetidine Chemical compound C1CNC1 HONIICLYMWZJFZ-UHFFFAOYSA-N 0.000 description 2
- WPYMKLBDIGXBTP-UHFFFAOYSA-N benzoic acid Chemical compound OC(=O)C1=CC=CC=C1 WPYMKLBDIGXBTP-UHFFFAOYSA-N 0.000 description 2
- 230000003115 biocidal effect Effects 0.000 description 2
- 210000004369 blood Anatomy 0.000 description 2
- 239000008280 blood Substances 0.000 description 2
- 210000004556 brain Anatomy 0.000 description 2
- 229960001948 caffeine Drugs 0.000 description 2
- VJEONQKOZGKCAK-UHFFFAOYSA-N caffeine Natural products CN1C(=O)N(C)C(=O)C2=C1C=CN2C VJEONQKOZGKCAK-UHFFFAOYSA-N 0.000 description 2
- 239000011575 calcium Substances 0.000 description 2
- 229910052791 calcium Inorganic materials 0.000 description 2
- 238000004364 calculation method Methods 0.000 description 2
- 239000002775 capsule Substances 0.000 description 2
- 125000004452 carbocyclyl group Chemical group 0.000 description 2
- CREMABGTGYGIQB-UHFFFAOYSA-N carbon carbon Chemical group C.C CREMABGTGYGIQB-UHFFFAOYSA-N 0.000 description 2
- 125000002915 carbonyl group Chemical group [*:2]C([*:1])=O 0.000 description 2
- 150000001732 carboxylic acid derivatives Chemical class 0.000 description 2
- 239000000969 carrier Substances 0.000 description 2
- 230000004663 cell proliferation Effects 0.000 description 2
- ZVEQCJWYRWKARO-UHFFFAOYSA-N ceramide Natural products CCCCCCCCCCCCCCC(O)C(=O)NC(CO)C(O)C=CCCC=C(C)CCCCCCCCC ZVEQCJWYRWKARO-UHFFFAOYSA-N 0.000 description 2
- 150000001783 ceramides Chemical class 0.000 description 2
- 150000003841 chloride salts Chemical class 0.000 description 2
- 229960001231 choline Drugs 0.000 description 2
- OEYIOHPDSNJKLS-UHFFFAOYSA-N choline Chemical compound C[N+](C)(C)CCO OEYIOHPDSNJKLS-UHFFFAOYSA-N 0.000 description 2
- 238000000576 coating method Methods 0.000 description 2
- 239000002299 complementary DNA Substances 0.000 description 2
- 239000000356 contaminant Substances 0.000 description 2
- OPTASPLRGRRNAP-UHFFFAOYSA-N cytosine Chemical compound NC=1C=CNC(=O)N=1 OPTASPLRGRRNAP-UHFFFAOYSA-N 0.000 description 2
- GHVNFZFCNZKVNT-UHFFFAOYSA-N decanoic acid Chemical compound CCCCCCCCCC(O)=O GHVNFZFCNZKVNT-UHFFFAOYSA-N 0.000 description 2
- 230000003247 decreasing effect Effects 0.000 description 2
- 230000001419 dependent effect Effects 0.000 description 2
- 238000013461 design Methods 0.000 description 2
- 125000005265 dialkylamine group Chemical group 0.000 description 2
- JXTHNDFMNIQAHM-UHFFFAOYSA-N dichloroacetic acid Chemical compound OC(=O)C(Cl)Cl JXTHNDFMNIQAHM-UHFFFAOYSA-N 0.000 description 2
- HPNMFZURTQLUMO-UHFFFAOYSA-N diethylamine Chemical compound CCNCC HPNMFZURTQLUMO-UHFFFAOYSA-N 0.000 description 2
- XBDQKXXYIPTUBI-UHFFFAOYSA-N dimethylselenoniopropionate Natural products CCC(O)=O XBDQKXXYIPTUBI-UHFFFAOYSA-N 0.000 description 2
- ZUOUZKKEUPVFJK-UHFFFAOYSA-N diphenyl Chemical compound C1=CC=CC=C1C1=CC=CC=C1 ZUOUZKKEUPVFJK-UHFFFAOYSA-N 0.000 description 2
- PMMYEEVYMWASQN-UHFFFAOYSA-N dl-hydroxyproline Natural products OC1C[NH2+]C(C([O-])=O)C1 PMMYEEVYMWASQN-UHFFFAOYSA-N 0.000 description 2
- UKMSUNONTOPOIO-UHFFFAOYSA-N docosanoic acid Chemical compound CCCCCCCCCCCCCCCCCCCCCC(O)=O UKMSUNONTOPOIO-UHFFFAOYSA-N 0.000 description 2
- 239000003937 drug carrier Substances 0.000 description 2
- 239000010408 film Substances 0.000 description 2
- 125000001153 fluoro group Chemical group F* 0.000 description 2
- 239000012634 fragment Substances 0.000 description 2
- DSLZVSRJTYRBFB-DUHBMQHGSA-N galactaric acid Chemical compound OC(=O)[C@H](O)[C@@H](O)[C@@H](O)[C@H](O)C(O)=O DSLZVSRJTYRBFB-DUHBMQHGSA-N 0.000 description 2
- BTCSSZJGUNDROE-UHFFFAOYSA-N gamma-aminobutyric acid Chemical compound NCCCC(O)=O BTCSSZJGUNDROE-UHFFFAOYSA-N 0.000 description 2
- 238000012226 gene silencing method Methods 0.000 description 2
- 238000010362 genome editing Methods 0.000 description 2
- 235000011187 glycerol Nutrition 0.000 description 2
- 229960004956 glycerylphosphorylcholine Drugs 0.000 description 2
- KWIUHFFTVRNATP-UHFFFAOYSA-N glycine betaine Chemical compound C[N+](C)(C)CC([O-])=O KWIUHFFTVRNATP-UHFFFAOYSA-N 0.000 description 2
- 229910052736 halogen Inorganic materials 0.000 description 2
- 150000002367 halogens Chemical class 0.000 description 2
- FUZZWVXGSFPDMH-UHFFFAOYSA-N hexanoic acid Chemical compound CCCCCC(O)=O FUZZWVXGSFPDMH-UHFFFAOYSA-N 0.000 description 2
- 125000004051 hexyl group Chemical group [H]C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])* 0.000 description 2
- WGCNASOHLSPBMP-UHFFFAOYSA-N hydroxyacetaldehyde Natural products OCC=O WGCNASOHLSPBMP-UHFFFAOYSA-N 0.000 description 2
- VKOBVWXKNCXXDE-UHFFFAOYSA-N icosanoic acid Chemical compound CCCCCCCCCCCCCCCCCCCC(O)=O VKOBVWXKNCXXDE-UHFFFAOYSA-N 0.000 description 2
- 150000007529 inorganic bases Chemical class 0.000 description 2
- 238000001990 intravenous administration Methods 0.000 description 2
- SUMDYPCJJOFFON-UHFFFAOYSA-N isethionic acid Chemical compound OCCS(O)(=O)=O SUMDYPCJJOFFON-UHFFFAOYSA-N 0.000 description 2
- KQNPFQTWMSNSAP-UHFFFAOYSA-N isobutyric acid Chemical compound CC(C)C(O)=O KQNPFQTWMSNSAP-UHFFFAOYSA-N 0.000 description 2
- JJWLVOIRVHMVIS-UHFFFAOYSA-N isopropylamine Chemical compound CC(C)N JJWLVOIRVHMVIS-UHFFFAOYSA-N 0.000 description 2
- JVTAAEKCZFNVCJ-UHFFFAOYSA-N lactic acid Chemical compound CC(O)C(O)=O JVTAAEKCZFNVCJ-UHFFFAOYSA-N 0.000 description 2
- 125000005644 linolenyl group Chemical group 0.000 description 2
- 239000002502 liposome Substances 0.000 description 2
- KWGKDLIKAYFUFQ-UHFFFAOYSA-M lithium chloride Chemical compound [Li+].[Cl-] KWGKDLIKAYFUFQ-UHFFFAOYSA-M 0.000 description 2
- 210000005228 liver tissue Anatomy 0.000 description 2
- 238000004020 luminiscence type Methods 0.000 description 2
- 210000004962 mammalian cell Anatomy 0.000 description 2
- 230000001404 mediated effect Effects 0.000 description 2
- 230000002503 metabolic effect Effects 0.000 description 2
- BDAGIHXWWSANSR-UHFFFAOYSA-N methanoic acid Natural products OC=O BDAGIHXWWSANSR-UHFFFAOYSA-N 0.000 description 2
- 238000007069 methylation reaction Methods 0.000 description 2
- 125000002950 monocyclic group Chemical group 0.000 description 2
- 239000000178 monomer Substances 0.000 description 2
- 125000002757 morpholinyl group Chemical group 0.000 description 2
- 125000001419 myristoyl group Chemical group O=C([*])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H] 0.000 description 2
- WQEPLUUGTLDZJY-UHFFFAOYSA-N n-Pentadecanoic acid Natural products CCCCCCCCCCCCCCC(O)=O WQEPLUUGTLDZJY-UHFFFAOYSA-N 0.000 description 2
- XTEGVFVZDVNBPF-UHFFFAOYSA-N naphthalene-1,5-disulfonic acid Chemical compound C1=CC=C2C(S(=O)(=O)O)=CC=CC2=C1S(O)(=O)=O XTEGVFVZDVNBPF-UHFFFAOYSA-N 0.000 description 2
- VVGIYYKRAMHVLU-UHFFFAOYSA-N newbouldiamide Natural products CCCCCCCCCCCCCCCCCCCC(O)C(O)C(O)C(CO)NC(=O)CCCCCCCCCCCCCCCCC VVGIYYKRAMHVLU-UHFFFAOYSA-N 0.000 description 2
- QIQXTHQIDYTFRH-UHFFFAOYSA-N octadecanoic acid Chemical compound CCCCCCCCCCCCCCCCCC(O)=O QIQXTHQIDYTFRH-UHFFFAOYSA-N 0.000 description 2
- OQCDKBAXFALNLD-UHFFFAOYSA-N octadecanoic acid Natural products CCCCCCCC(C)CCCCCCCCC(O)=O OQCDKBAXFALNLD-UHFFFAOYSA-N 0.000 description 2
- WWZKQHOCKIZLMA-UHFFFAOYSA-N octanoic acid Chemical compound CCCCCCCC(O)=O WWZKQHOCKIZLMA-UHFFFAOYSA-N 0.000 description 2
- 210000000056 organ Anatomy 0.000 description 2
- 239000003960 organic solvent Substances 0.000 description 2
- PXQPEWDEAKTCGB-UHFFFAOYSA-N orotic acid Chemical compound OC(=O)C1=CC(=O)NC(=O)N1 PXQPEWDEAKTCGB-UHFFFAOYSA-N 0.000 description 2
- SECPZKHBENQXJG-FPLPWBNLSA-N palmitoleic acid Chemical compound CCCCCC\C=C/CCCCCCCC(O)=O SECPZKHBENQXJG-FPLPWBNLSA-N 0.000 description 2
- 230000036961 partial effect Effects 0.000 description 2
- 239000008194 pharmaceutical composition Substances 0.000 description 2
- 150000008103 phosphatidic acids Chemical class 0.000 description 2
- 150000008104 phosphatidylethanolamines Chemical class 0.000 description 2
- 150000008106 phosphatidylserines Chemical class 0.000 description 2
- 125000002525 phosphocholine group Chemical group OP(=O)(OCC[N+](C)(C)C)O* 0.000 description 2
- 125000003386 piperidinyl group Chemical group 0.000 description 2
- 229920000765 poly(2-oxazolines) Polymers 0.000 description 2
- 229920000191 poly(N-vinyl pyrrolidone) Polymers 0.000 description 2
- 229920001308 poly(aminoacid) Polymers 0.000 description 2
- 229920002187 poly[N-2-(hydroxypropyl) methacrylamide] polymer Polymers 0.000 description 2
- 230000008488 polyadenylation Effects 0.000 description 2
- 125000003367 polycyclic group Chemical group 0.000 description 2
- 229920000223 polyglycerol Polymers 0.000 description 2
- 238000006116 polymerization reaction Methods 0.000 description 2
- 229920002451 polyvinyl alcohol Polymers 0.000 description 2
- 238000012877 positron emission topography Methods 0.000 description 2
- 239000011591 potassium Substances 0.000 description 2
- 229910052700 potassium Inorganic materials 0.000 description 2
- 238000011809 primate model Methods 0.000 description 2
- WGYKZJWCGVVSQN-UHFFFAOYSA-N propylamine Chemical compound CCCN WGYKZJWCGVVSQN-UHFFFAOYSA-N 0.000 description 2
- 125000002568 propynyl group Chemical group [*]C#CC([H])([H])[H] 0.000 description 2
- 150000003230 pyrimidines Chemical class 0.000 description 2
- 125000000719 pyrrolidinyl group Chemical group 0.000 description 2
- 230000002285 radioactive effect Effects 0.000 description 2
- 230000014493 regulation of gene expression Effects 0.000 description 2
- 230000008960 regulation of mRNA stability Effects 0.000 description 2
- 230000001105 regulatory effect Effects 0.000 description 2
- 230000004044 response Effects 0.000 description 2
- 239000002336 ribonucleotide Substances 0.000 description 2
- 108020004418 ribosomal RNA Proteins 0.000 description 2
- YGSDEFSMJLZEOE-UHFFFAOYSA-N salicylic acid Chemical compound OC(=O)C1=CC=CC=C1O YGSDEFSMJLZEOE-UHFFFAOYSA-N 0.000 description 2
- FSYKKLYZXJSNPZ-UHFFFAOYSA-N sarcosine Chemical compound C[NH2+]CC([O-])=O FSYKKLYZXJSNPZ-UHFFFAOYSA-N 0.000 description 2
- CXMXRPHRNRROMY-UHFFFAOYSA-N sebacic acid Chemical compound OC(=O)CCCCCCCCC(O)=O CXMXRPHRNRROMY-UHFFFAOYSA-N 0.000 description 2
- 210000002966 serum Anatomy 0.000 description 2
- AWUCVROLDVIAJX-GSVOUGTGSA-N sn-glycerol 3-phosphate Chemical group OC[C@@H](O)COP(O)(O)=O AWUCVROLDVIAJX-GSVOUGTGSA-N 0.000 description 2
- 239000011734 sodium Substances 0.000 description 2
- 229910052708 sodium Inorganic materials 0.000 description 2
- 239000000243 solution Substances 0.000 description 2
- 239000008117 stearic acid Substances 0.000 description 2
- 125000003696 stearoyl group Chemical group O=C([*])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H] 0.000 description 2
- 238000007920 subcutaneous administration Methods 0.000 description 2
- 239000000126 substance Substances 0.000 description 2
- 125000003107 substituted aryl group Chemical group 0.000 description 2
- 208000024891 symptom Diseases 0.000 description 2
- 239000003826 tablet Substances 0.000 description 2
- RAOIDOHSFRTOEL-UHFFFAOYSA-N tetrahydrothiophene Chemical compound C1CCSC1 RAOIDOHSFRTOEL-UHFFFAOYSA-N 0.000 description 2
- YAPQBXQYLJRXSA-UHFFFAOYSA-N theobromine Chemical compound CN1C(=O)NC(=O)C2=C1N=CN2C YAPQBXQYLJRXSA-UHFFFAOYSA-N 0.000 description 2
- ZMZDMBWJUHKJPS-UHFFFAOYSA-N thiocyanic acid Chemical compound SC#N ZMZDMBWJUHKJPS-UHFFFAOYSA-N 0.000 description 2
- 125000003396 thiol group Chemical group [H]S* 0.000 description 2
- UMGDCJDMYOKAJW-UHFFFAOYSA-N thiourea group Chemical group NC(=S)N UMGDCJDMYOKAJW-UHFFFAOYSA-N 0.000 description 2
- RWQNBRDOKXIBIV-UHFFFAOYSA-N thymine Chemical compound CC1=CNC(=O)NC1=O RWQNBRDOKXIBIV-UHFFFAOYSA-N 0.000 description 2
- JOXIMZWYDAKGHI-UHFFFAOYSA-N toluene-4-sulfonic acid Chemical compound CC1=CC=C(S(O)(=O)=O)C=C1 JOXIMZWYDAKGHI-UHFFFAOYSA-N 0.000 description 2
- VZCYOOQTPOCHFL-UHFFFAOYSA-N trans-butenedioic acid Natural products OC(=O)C=CC(O)=O VZCYOOQTPOCHFL-UHFFFAOYSA-N 0.000 description 2
- 230000003827 upregulation Effects 0.000 description 2
- 239000013598 vector Substances 0.000 description 2
- XLYOFNOQVPJJNP-UHFFFAOYSA-N water Substances O XLYOFNOQVPJJNP-UHFFFAOYSA-N 0.000 description 2
- DRAFVCKNYNQOKR-GFCCVEGCSA-N (1-methoxycarbonylcyclopropyl) 3-[(1r)-1-phenylethyl]imidazole-4-carboxylate Chemical compound C=1N=CN([C@H](C)C=2C=CC=CC=2)C=1C(=O)OC1(C(=O)OC)CC1 DRAFVCKNYNQOKR-GFCCVEGCSA-N 0.000 description 1
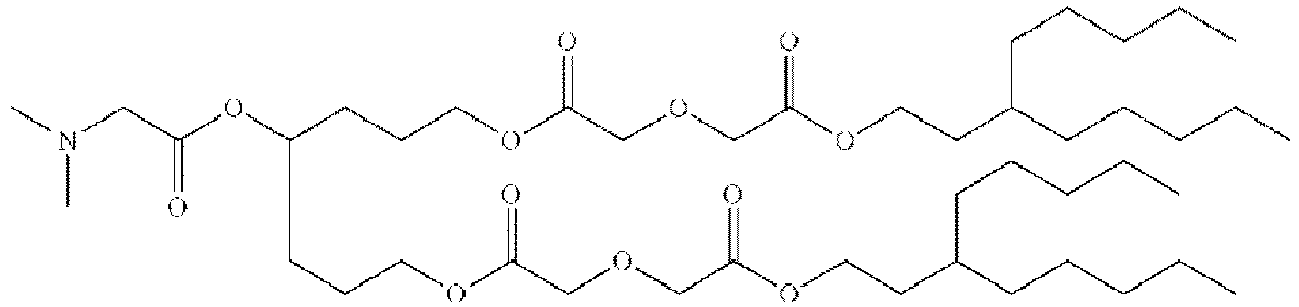
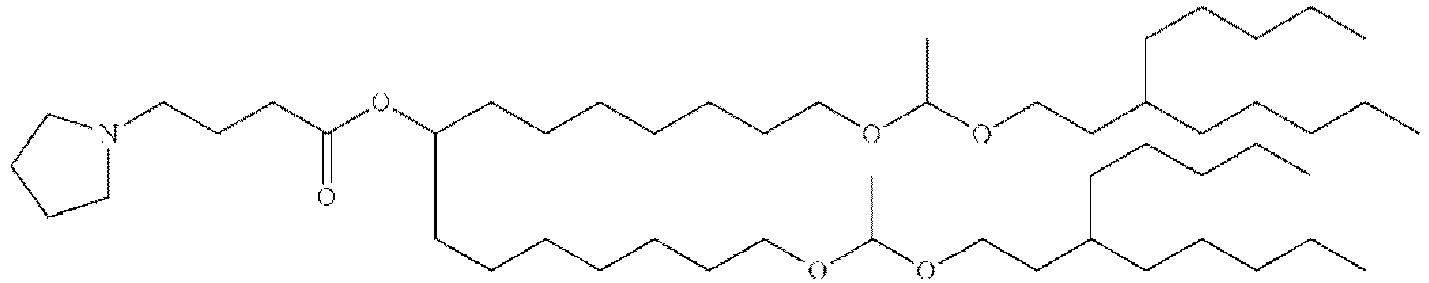
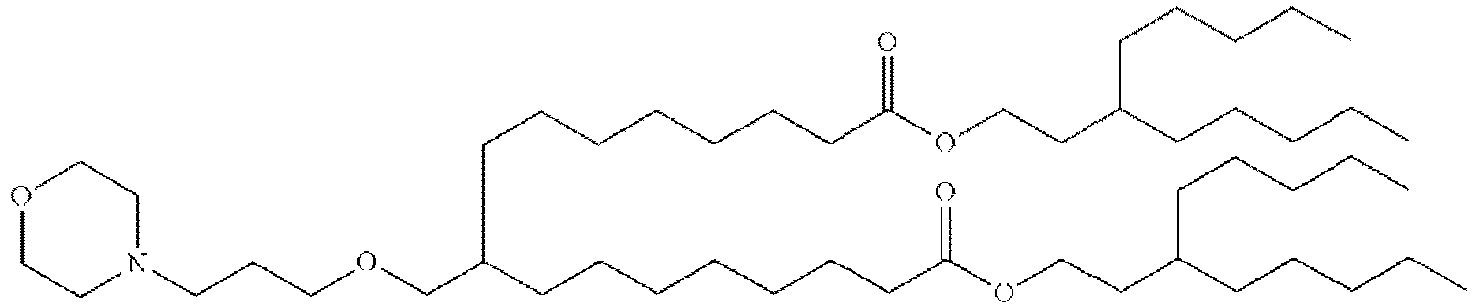
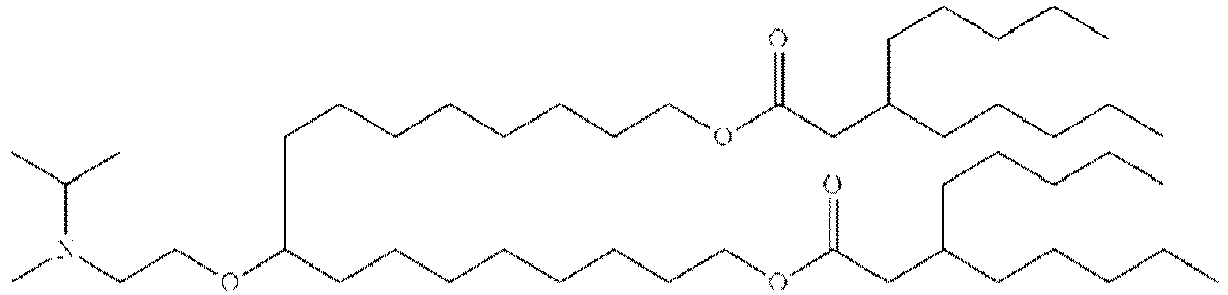
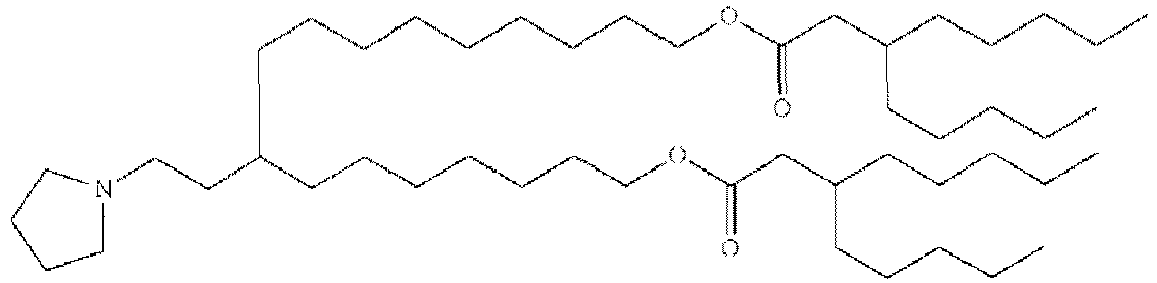
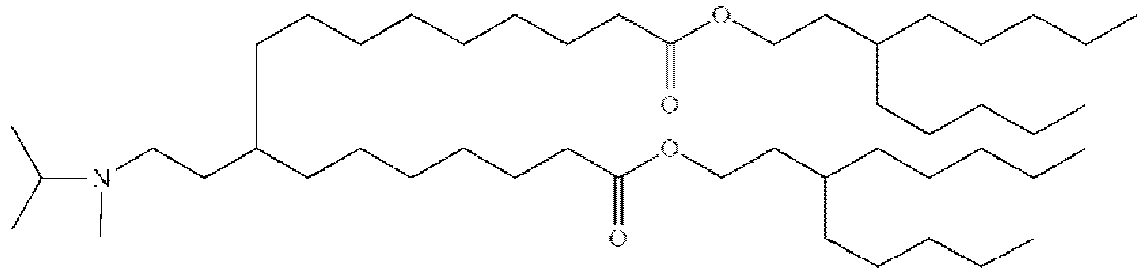
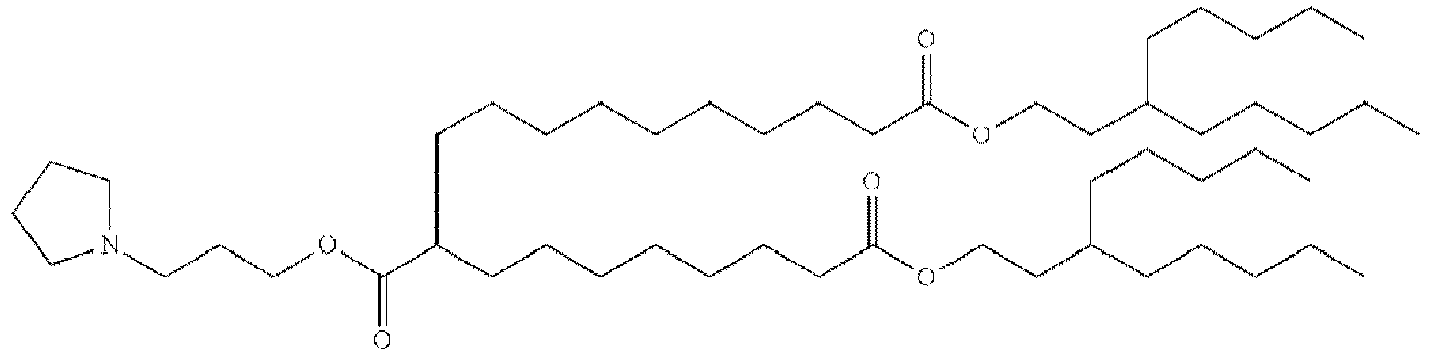
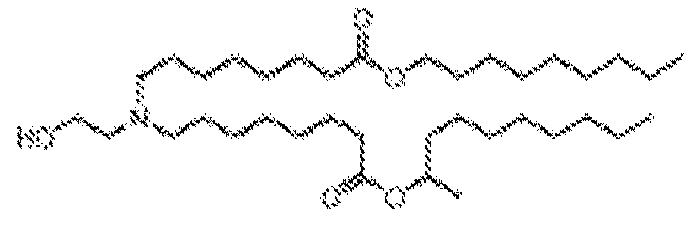
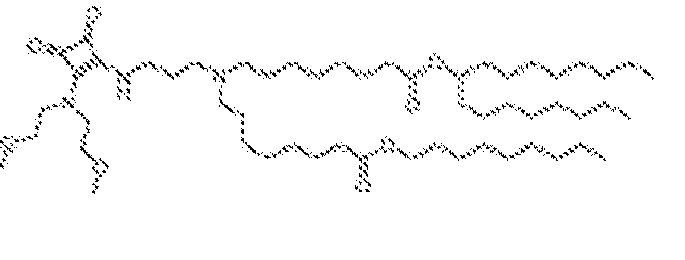
- RVIZTCLKCHZBMR-KWXKLSQISA-N (12z,15z)-1-(dimethylamino)-2-[(9z,12z)-octadeca-9,12-dienoxy]henicosa-12,15-dien-4-one Chemical compound CCCCC\C=C/C\C=C/CCCCCCCCOC(CN(C)C)CC(=O)CCCCCCC\C=C/C\C=C/CCCCC RVIZTCLKCHZBMR-KWXKLSQISA-N 0.000 description 1
- JTERLNYVBOZRHI-PPBJBQABSA-N (2-aminoethoxy)[(2r)-2,3-bis[(5z,8z,11z,14z)-icosa-5,8,11,14-tetraenoyloxy]propoxy]phosphinic acid Chemical compound CCCCC\C=C/C\C=C/C\C=C/C\C=C/CCCC(=O)OC[C@H](COP(O)(=O)OCCN)OC(=O)CCC\C=C/C\C=C/C\C=C/C\C=C/CCCCC JTERLNYVBOZRHI-PPBJBQABSA-N 0.000 description 1
- IHNKQIMGVNPMTC-UHFFFAOYSA-N (2-hydroxy-3-octadecanoyloxypropyl) 2-(trimethylazaniumyl)ethyl phosphate Chemical compound CCCCCCCCCCCCCCCCCC(=O)OCC(O)COP([O-])(=O)OCC[N+](C)(C)C IHNKQIMGVNPMTC-UHFFFAOYSA-N 0.000 description 1
- XLKQWAMTMYIQMG-SVUPRYTISA-N (2-{[(2r)-2,3-bis[(4z,7z,10z,13z,16z,19z)-docosa-4,7,10,13,16,19-hexaenoyloxy]propyl phosphonato]oxy}ethyl)trimethylazanium Chemical compound CC\C=C/C\C=C/C\C=C/C\C=C/C\C=C/C\C=C/CCC(=O)OC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CC\C=C/C\C=C/C\C=C/C\C=C/C\C=C/C\C=C/CC XLKQWAMTMYIQMG-SVUPRYTISA-N 0.000 description 1
- QBYIENPQHBMVBV-HFEGYEGKSA-N (2R)-2-hydroxy-2-phenylacetic acid Chemical compound O[C@@H](C(O)=O)c1ccccc1.O[C@@H](C(O)=O)c1ccccc1 QBYIENPQHBMVBV-HFEGYEGKSA-N 0.000 description 1
- BJBUEDPLEOHJGE-UHFFFAOYSA-N (2R,3S)-3-Hydroxy-2-pyrolidinecarboxylic acid Natural products OC1CCNC1C(O)=O BJBUEDPLEOHJGE-UHFFFAOYSA-N 0.000 description 1
- JSPNNZKWADNWHI-PNANGNLXSA-N (2r)-2-hydroxy-n-[(2s,3r,4e,8e)-3-hydroxy-9-methyl-1-[(2r,3r,4s,5s,6r)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxyoctadeca-4,8-dien-2-yl]heptadecanamide Chemical compound CCCCCCCCCCCCCCC[C@@H](O)C(=O)N[C@H]([C@H](O)\C=C\CC\C=C(/C)CCCCCCCCC)CO[C@@H]1O[C@H](CO)[C@@H](O)[C@H](O)[C@H]1O JSPNNZKWADNWHI-PNANGNLXSA-N 0.000 description 1
- OPCHFPHZPIURNA-MFERNQICSA-N (2s)-2,5-bis(3-aminopropylamino)-n-[2-(dioctadecylamino)acetyl]pentanamide Chemical compound CCCCCCCCCCCCCCCCCCN(CC(=O)NC(=O)[C@H](CCCNCCCN)NCCCN)CCCCCCCCCCCCCCCCCC OPCHFPHZPIURNA-MFERNQICSA-N 0.000 description 1
- YPJJGMCMOHDOFZ-ZETCQYMHSA-N (2s)-2-(1-benzothiophen-3-ylamino)propanoic acid Chemical compound C1=CC=C2C(N[C@@H](C)C(O)=O)=CSC2=C1 YPJJGMCMOHDOFZ-ZETCQYMHSA-N 0.000 description 1
- BVAUMRCGVHUWOZ-ZETCQYMHSA-N (2s)-2-(cyclohexylazaniumyl)propanoate Chemical compound OC(=O)[C@H](C)NC1CCCCC1 BVAUMRCGVHUWOZ-ZETCQYMHSA-N 0.000 description 1
- IYKLZBIWFXPUCS-VIFPVBQESA-N (2s)-2-(naphthalen-1-ylamino)propanoic acid Chemical compound C1=CC=C2C(N[C@@H](C)C(O)=O)=CC=CC2=C1 IYKLZBIWFXPUCS-VIFPVBQESA-N 0.000 description 1
- CNMAQBJBWQQZFZ-LURJTMIESA-N (2s)-2-(pyridin-2-ylamino)propanoic acid Chemical compound OC(=O)[C@H](C)NC1=CC=CC=N1 CNMAQBJBWQQZFZ-LURJTMIESA-N 0.000 description 1
- MRTPISKDZDHEQI-YFKPBYRVSA-N (2s)-2-(tert-butylamino)propanoic acid Chemical compound OC(=O)[C@H](C)NC(C)(C)C MRTPISKDZDHEQI-YFKPBYRVSA-N 0.000 description 1
- NPDBDJFLKKQMCM-SCSAIBSYSA-N (2s)-2-amino-3,3-dimethylbutanoic acid Chemical compound CC(C)(C)[C@H](N)C(O)=O NPDBDJFLKKQMCM-SCSAIBSYSA-N 0.000 description 1
- VEVRNHHLCPGNDU-MUGJNUQGSA-N (2s)-2-amino-5-[1-[(5s)-5-amino-5-carboxypentyl]-3,5-bis[(3s)-3-amino-3-carboxypropyl]pyridin-1-ium-4-yl]pentanoate Chemical compound OC(=O)[C@@H](N)CCCC[N+]1=CC(CC[C@H](N)C(O)=O)=C(CCC[C@H](N)C([O-])=O)C(CC[C@H](N)C(O)=O)=C1 VEVRNHHLCPGNDU-MUGJNUQGSA-N 0.000 description 1
- WAMWSIDTKSNDCU-ZETCQYMHSA-N (2s)-2-azaniumyl-2-cyclohexylacetate Chemical compound OC(=O)[C@@H](N)C1CCCCC1 WAMWSIDTKSNDCU-ZETCQYMHSA-N 0.000 description 1
- LJRDOKAZOAKLDU-UDXJMMFXSA-N (2s,3s,4r,5r,6r)-5-amino-2-(aminomethyl)-6-[(2r,3s,4r,5s)-5-[(1r,2r,3s,5r,6s)-3,5-diamino-2-[(2s,3r,4r,5s,6r)-3-amino-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-6-hydroxycyclohexyl]oxy-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl]oxyoxane-3,4-diol;sulfuric ac Chemical compound OS(O)(=O)=O.N[C@@H]1[C@@H](O)[C@H](O)[C@H](CN)O[C@@H]1O[C@H]1[C@@H](O)[C@H](O[C@H]2[C@@H]([C@@H](N)C[C@@H](N)[C@@H]2O)O[C@@H]2[C@@H]([C@@H](O)[C@H](O)[C@@H](CO)O2)N)O[C@@H]1CO LJRDOKAZOAKLDU-UDXJMMFXSA-N 0.000 description 1
- VDYVTMXBGOIUMS-KWXKLSQISA-N (6z,9z,29z,32z)-19-[(dimethylamino)methyl]octatriaconta-6,9,29,32-tetraene-18,21-dione Chemical compound CCCCC\C=C/C\C=C/CCCCCCCC(=O)CC(CN(C)C)C(=O)CCCCCCC\C=C/C\C=C/CCCCC VDYVTMXBGOIUMS-KWXKLSQISA-N 0.000 description 1
- YUFFSWGQGVEMMI-JLNKQSITSA-N (7Z,10Z,13Z,16Z,19Z)-docosapentaenoic acid Chemical compound CC\C=C/C\C=C/C\C=C/C\C=C/C\C=C/CCCCCC(O)=O YUFFSWGQGVEMMI-JLNKQSITSA-N 0.000 description 1
- OYHQOLUKZRVURQ-NTGFUMLPSA-N (9Z,12Z)-9,10,12,13-tetratritiooctadeca-9,12-dienoic acid Chemical compound C(CCCCCCC\C(=C(/C\C(=C(/CCCCC)\[3H])\[3H])\[3H])\[3H])(=O)O OYHQOLUKZRVURQ-NTGFUMLPSA-N 0.000 description 1
- 125000006698 (C1-C3) dialkylamino group Chemical group 0.000 description 1
- 125000000923 (C1-C30) alkyl group Chemical group 0.000 description 1
- 125000006833 (C1-C5) alkylene group Chemical group 0.000 description 1
- 125000006552 (C3-C8) cycloalkyl group Chemical group 0.000 description 1
- LVNGJLRDBYCPGB-LDLOPFEMSA-N (R)-1,2-distearoylphosphatidylethanolamine Chemical compound CCCCCCCCCCCCCCCCCC(=O)OC[C@H](COP([O-])(=O)OCC[NH3+])OC(=O)CCCCCCCCCCCCCCCCC LVNGJLRDBYCPGB-LDLOPFEMSA-N 0.000 description 1
- MIOPJNTWMNEORI-GMSGAONNSA-N (S)-camphorsulfonic acid Chemical compound C1C[C@@]2(CS(O)(=O)=O)C(=O)C[C@@H]1C2(C)C MIOPJNTWMNEORI-GMSGAONNSA-N 0.000 description 1
- BJEPYKJPYRNKOW-REOHCLBHSA-N (S)-malic acid Chemical compound OC(=O)[C@@H](O)CC(O)=O BJEPYKJPYRNKOW-REOHCLBHSA-N 0.000 description 1
- MAUMSNABMVEOGP-UHFFFAOYSA-N (methyl-$l^{2}-azanyl)methane Chemical group C[N]C MAUMSNABMVEOGP-UHFFFAOYSA-N 0.000 description 1
- UKAUYVFTDYCKQA-UHFFFAOYSA-N -2-Amino-4-hydroxybutanoic acid Natural products OC(=O)C(N)CCO UKAUYVFTDYCKQA-UHFFFAOYSA-N 0.000 description 1
- WBYWAXJHAXSJNI-VOTSOKGWSA-M .beta-Phenylacrylic acid Natural products [O-]C(=O)\C=C\C1=CC=CC=C1 WBYWAXJHAXSJNI-VOTSOKGWSA-M 0.000 description 1
- SSCDRSKJTAQNNB-DWEQTYCFSA-N 1,2-di-(9Z,12Z-octadecadienoyl)-sn-glycero-3-phosphoethanolamine Chemical compound CCCCC\C=C/C\C=C/CCCCCCCC(=O)OC[C@H](COP(O)(=O)OCCN)OC(=O)CCCCCCC\C=C/C\C=C/CCCCC SSCDRSKJTAQNNB-DWEQTYCFSA-N 0.000 description 1
- LZLVZIFMYXDKCN-QJWFYWCHSA-N 1,2-di-O-arachidonoyl-sn-glycero-3-phosphocholine Chemical compound CCCCC\C=C/C\C=C/C\C=C/C\C=C/CCCC(=O)OC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CCC\C=C/C\C=C/C\C=C/C\C=C/CCCCC LZLVZIFMYXDKCN-QJWFYWCHSA-N 0.000 description 1
- XXKFQTJOJZELMD-JICBSJGISA-N 1,2-di-[(9Z,12Z,15Z)-octadecatrienoyl]-sn-glycero-3-phosphocholine Chemical compound CC\C=C/C\C=C/C\C=C/CCCCCCCC(=O)OC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CCCCCCC\C=C/C\C=C/C\C=C/CC XXKFQTJOJZELMD-JICBSJGISA-N 0.000 description 1
- PORPENFLTBBHSG-MGBGTMOVSA-N 1,2-dihexadecanoyl-sn-glycerol-3-phosphate Chemical compound CCCCCCCCCCCCCCCC(=O)OC[C@H](COP(O)(O)=O)OC(=O)CCCCCCCCCCCCCCC PORPENFLTBBHSG-MGBGTMOVSA-N 0.000 description 1
- UHUSDOQQWJGJQS-QNGWXLTQSA-N 1,2-dioctadecanoyl-sn-glycerol Chemical compound CCCCCCCCCCCCCCCCCC(=O)OC[C@H](CO)OC(=O)CCCCCCCCCCCCCCCCC UHUSDOQQWJGJQS-QNGWXLTQSA-N 0.000 description 1
- JEJLGIQLPYYGEE-XIFFEERXSA-N 1,2-dipalmitoyl-sn-glycerol Chemical compound CCCCCCCCCCCCCCCC(=O)OC[C@H](CO)OC(=O)CCCCCCCCCCCCCCC JEJLGIQLPYYGEE-XIFFEERXSA-N 0.000 description 1
- TZCPCKNHXULUIY-RGULYWFUSA-N 1,2-distearoyl-sn-glycero-3-phosphoserine Chemical compound CCCCCCCCCCCCCCCCCC(=O)OC[C@H](COP(O)(=O)OC[C@H](N)C(O)=O)OC(=O)CCCCCCCCCCCCCCCCC TZCPCKNHXULUIY-RGULYWFUSA-N 0.000 description 1
- JFBCSFJKETUREV-LJAQVGFWSA-N 1,2-ditetradecanoyl-sn-glycerol Chemical compound CCCCCCCCCCCCCC(=O)OC[C@H](CO)OC(=O)CCCCCCCCCCCCC JFBCSFJKETUREV-LJAQVGFWSA-N 0.000 description 1
- UPNNXUSUOSTIIM-UHFFFAOYSA-N 1,2-dithietane Chemical compound C1CSS1 UPNNXUSUOSTIIM-UHFFFAOYSA-N 0.000 description 1
- LUNCLNIYHUMRFU-UHFFFAOYSA-N 1-(trimethylazaniumyl)butan-2-yl hydrogen phosphate Chemical class C[N+](C)(C)CC(CC)OP(O)([O-])=O LUNCLNIYHUMRFU-UHFFFAOYSA-N 0.000 description 1
- VXNZUUAINFGPBY-UHFFFAOYSA-N 1-Butene Chemical group CCC=C VXNZUUAINFGPBY-UHFFFAOYSA-N 0.000 description 1
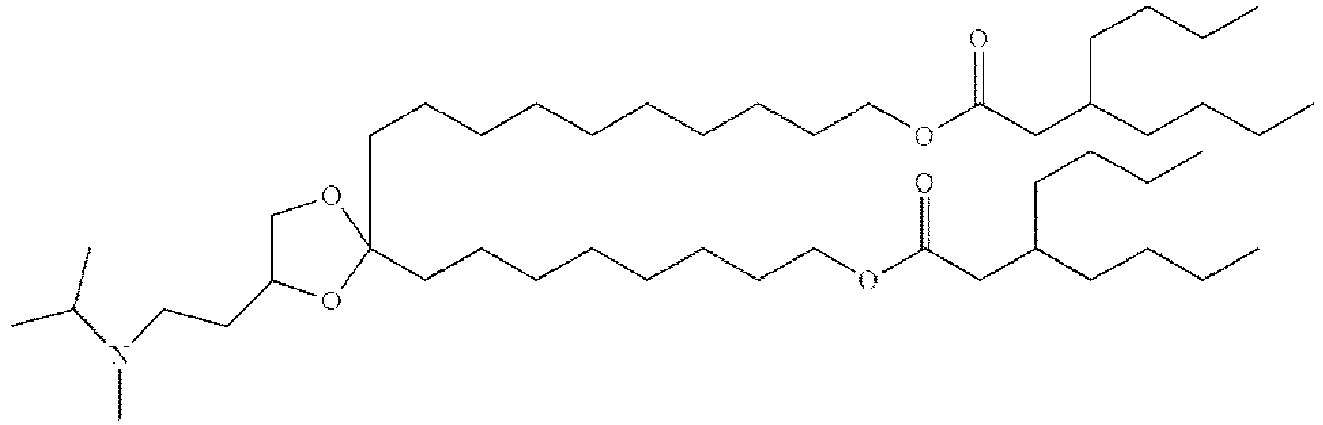
- BUOBCSGIAFXNKP-KWXKLSQISA-N 1-[2,2-bis[(9z,12z)-octadeca-9,12-dienyl]-1,3-dioxolan-4-yl]-n,n-dimethylmethanamine Chemical compound CCCCC\C=C/C\C=C/CCCCCCCCC1(CCCCCCCC\C=C/C\C=C/CCCCC)OCC(CN(C)C)O1 BUOBCSGIAFXNKP-KWXKLSQISA-N 0.000 description 1
- PLKOSISDOAHHCI-QYCRHRGJSA-N 1-[2,3-bis[(9z,12z)-octadeca-9,12-dienoxy]propyl]-4-methylpiperazine Chemical compound CCCCC\C=C/C\C=C/CCCCCCCCOCC(OCCCCCCCC\C=C/C\C=C/CCCCC)CN1CCN(C)CC1 PLKOSISDOAHHCI-QYCRHRGJSA-N 0.000 description 1
- UHDGCWIWMRVCDJ-UHFFFAOYSA-N 1-beta-D-Xylofuranosyl-NH-Cytosine Natural products O=C1N=C(N)C=CN1C1C(O)C(O)C(CO)O1 UHDGCWIWMRVCDJ-UHFFFAOYSA-N 0.000 description 1
- 125000004972 1-butynyl group Chemical group [H]C([H])([H])C([H])([H])C#C* 0.000 description 1
- SJJCQDRGABAVBB-UHFFFAOYSA-N 1-hydroxy-2-naphthoic acid Chemical compound C1=CC=CC2=C(O)C(C(=O)O)=CC=C21 SJJCQDRGABAVBB-UHFFFAOYSA-N 0.000 description 1
- RTBFRGCFXZNCOE-UHFFFAOYSA-N 1-methylsulfonylpiperidin-4-one Chemical compound CS(=O)(=O)N1CCC(=O)CC1 RTBFRGCFXZNCOE-UHFFFAOYSA-N 0.000 description 1
- 125000006023 1-pentenyl group Chemical group 0.000 description 1
- FRPZMMHWLSIFAZ-UHFFFAOYSA-N 10-undecenoic acid Chemical compound OC(=O)CCCCCCCCC=C FRPZMMHWLSIFAZ-UHFFFAOYSA-N 0.000 description 1
- VGONTNSXDCQUGY-RRKCRQDMSA-N 2'-deoxyinosine Chemical group C1[C@H](O)[C@@H](CO)O[C@H]1N1C(N=CNC2=O)=C2N=C1 VGONTNSXDCQUGY-RRKCRQDMSA-N 0.000 description 1
- 125000006069 2,3-dimethyl-2-butenyl group Chemical group 0.000 description 1
- OGNSCSPNOLGXSM-UHFFFAOYSA-N 2,4-diaminobutyric acid Chemical compound NCCC(N)C(O)=O OGNSCSPNOLGXSM-UHFFFAOYSA-N 0.000 description 1
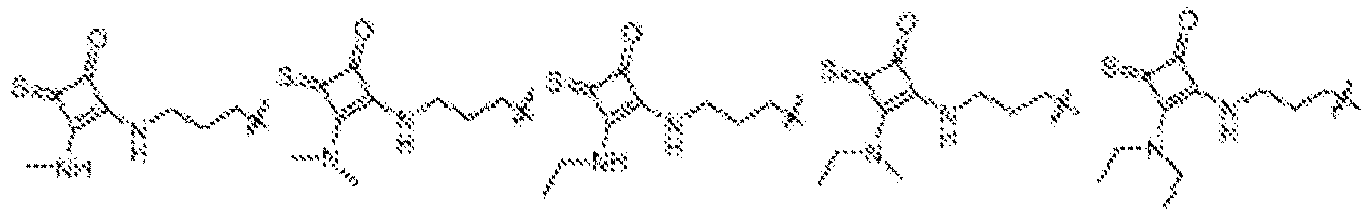
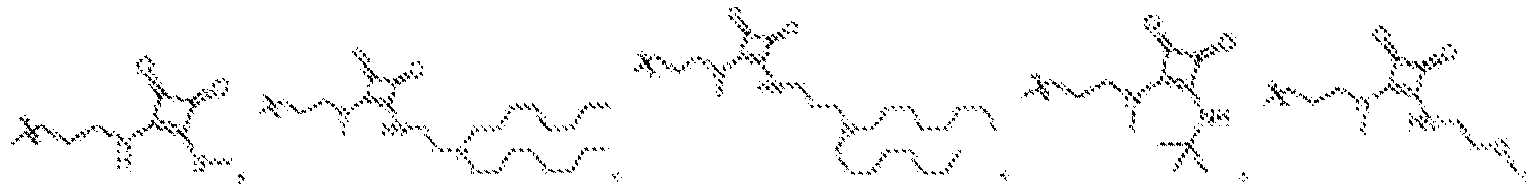
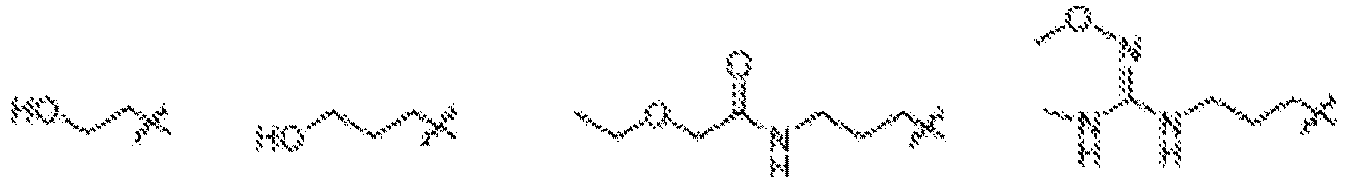
- NDWGZPKSWNQVOQ-UHFFFAOYSA-N 2-(diethylamino)-4-sulfanylidenecyclobut-2-en-1-one Chemical group C(C)N(C=1C(C(C=1)=S)=O)CC NDWGZPKSWNQVOQ-UHFFFAOYSA-N 0.000 description 1
- SBIIXADGZNPZFF-KWXKLSQISA-N 2-(dimethylamino)-3-[(9z,12z)-octadeca-9,12-dienoxy]-2-[[(9z,12z)-octadeca-9,12-dienoxy]methyl]propan-1-ol Chemical compound CCCCC\C=C/C\C=C/CCCCCCCCOCC(CO)(N(C)C)COCCCCCCCC\C=C/C\C=C/CCCCC SBIIXADGZNPZFF-KWXKLSQISA-N 0.000 description 1
- JZAVUVDMHXUDTP-UHFFFAOYSA-N 2-(ethylamino)-4-sulfanylidenecyclobut-2-en-1-one Chemical group C(C)NC=1C(C(C=1)=S)=O JZAVUVDMHXUDTP-UHFFFAOYSA-N 0.000 description 1
- FUOOLUPWFVMBKG-UHFFFAOYSA-N 2-Aminoisobutyric acid Chemical compound CC(C)(N)C(O)=O FUOOLUPWFVMBKG-UHFFFAOYSA-N 0.000 description 1
- NUHBVWKTEJNULY-UHFFFAOYSA-N 2-[(4-oxocyclohexyl)amino]acetic acid Chemical compound OC(=O)CNC1CCC(=O)CC1 NUHBVWKTEJNULY-UHFFFAOYSA-N 0.000 description 1
- COUVCUNDLBYGMZ-HDXUUTQWSA-N 2-amino-2-[[(9z,12z)-octadeca-9,12-dienoxy]methyl]-3-octoxypropan-1-ol Chemical compound CCCCCCCCOCC(N)(CO)COCCCCCCCC\C=C/C\C=C/CCCCC COUVCUNDLBYGMZ-HDXUUTQWSA-N 0.000 description 1
- MSWZFWKMSRAUBD-IVMDWMLBSA-N 2-amino-2-deoxy-D-glucopyranose Chemical compound N[C@H]1C(O)O[C@H](CO)[C@@H](O)[C@@H]1O MSWZFWKMSRAUBD-IVMDWMLBSA-N 0.000 description 1
- LDHYTBAFXANWKM-UHFFFAOYSA-N 2-amino-3,7-dihydropurin-6-one Chemical compound O=C1NC(N)=NC2=C1NC=N2.O=C1NC(N)=NC2=C1N=CN2 LDHYTBAFXANWKM-UHFFFAOYSA-N 0.000 description 1
- HKMQLTCTBJOAQB-CLFAGFIQSA-N 2-amino-3-[(z)-octadec-9-enoxy]-2-[[(z)-octadec-9-enoxy]methyl]propan-1-ol Chemical compound CCCCCCCC\C=C/CCCCCCCCOCC(N)(CO)COCCCCCCCC\C=C/CCCCCCCC HKMQLTCTBJOAQB-CLFAGFIQSA-N 0.000 description 1
- IFPQOXNWLSRZKX-UHFFFAOYSA-N 2-amino-4-(diaminomethylideneamino)butanoic acid Chemical compound OC(=O)C(N)CCN=C(N)N IFPQOXNWLSRZKX-UHFFFAOYSA-N 0.000 description 1
- LRQKBLKVPFOOQJ-UHFFFAOYSA-N 2-aminohexanoic acid Chemical compound CCCCC(N)C(O)=O LRQKBLKVPFOOQJ-UHFFFAOYSA-N 0.000 description 1
- XRKBQVGBWJWJJJ-UHFFFAOYSA-N 2-aminooctadecanoic acid Chemical compound CCCCCCCCCCCCCCCCC(N)C(O)=O XRKBQVGBWJWJJJ-UHFFFAOYSA-N 0.000 description 1
- WTOFYLAWDLQMBZ-UHFFFAOYSA-N 2-azaniumyl-3-thiophen-2-ylpropanoate Chemical compound OC(=O)C(N)CC1=CC=CS1 WTOFYLAWDLQMBZ-UHFFFAOYSA-N 0.000 description 1
- XLPHMKQBBCKEFO-DHYROEPTSA-N 2-azaniumylethyl [(2r)-2,3-bis(3,7,11,15-tetramethylhexadecanoyloxy)propyl] phosphate Chemical compound CC(C)CCCC(C)CCCC(C)CCCC(C)CC(=O)OC[C@H](COP(O)(=O)OCCN)OC(=O)CC(C)CCCC(C)CCCC(C)CCCC(C)C XLPHMKQBBCKEFO-DHYROEPTSA-N 0.000 description 1
- 125000004974 2-butenyl group Chemical group C(C=CC)* 0.000 description 1
- 125000000069 2-butynyl group Chemical group [H]C([H])([H])C#CC([H])([H])* 0.000 description 1
- ASJSAQIRZKANQN-CRCLSJGQSA-N 2-deoxy-D-ribose Chemical compound OC[C@@H](O)[C@@H](O)CC=O ASJSAQIRZKANQN-CRCLSJGQSA-N 0.000 description 1
- BFSVOASYOCHEOV-UHFFFAOYSA-N 2-diethylaminoethanol Chemical compound CCN(CC)CCO BFSVOASYOCHEOV-UHFFFAOYSA-N 0.000 description 1
- 229940013085 2-diethylaminoethanol Drugs 0.000 description 1
- NYCRCTMDYITATC-UHFFFAOYSA-N 2-fluorophenylalanine Chemical compound OC(=O)C(N)CC1=CC=CC=C1F NYCRCTMDYITATC-UHFFFAOYSA-N 0.000 description 1
- 125000006029 2-methyl-2-butenyl group Chemical group 0.000 description 1
- 125000003229 2-methylhexyl group Chemical group [H]C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])(C([H])([H])[H])C([H])([H])* 0.000 description 1
- KPGXRSRHYNQIFN-UHFFFAOYSA-N 2-oxoglutaric acid Chemical compound OC(=O)CCC(=O)C(O)=O KPGXRSRHYNQIFN-UHFFFAOYSA-N 0.000 description 1
- 125000006024 2-pentenyl group Chemical group 0.000 description 1
- MGADZUXDNSDTHW-UHFFFAOYSA-N 2H-pyran Chemical compound C1OC=CC=C1 MGADZUXDNSDTHW-UHFFFAOYSA-N 0.000 description 1
- 108020005345 3' Untranslated Regions Proteins 0.000 description 1
- RLCKHJSFHOZMDR-PWCSWUJKSA-N 3,7R,11R,15-tetramethyl-hexadecanoic acid Chemical compound CC(C)CCC[C@@H](C)CCC[C@@H](C)CCCC(C)CC(O)=O RLCKHJSFHOZMDR-PWCSWUJKSA-N 0.000 description 1
- BBTSKBDLIAXROH-UHFFFAOYSA-N 3-(diethylamino)-4-sulfanylidenecyclobut-2-en-1-one Chemical group C(C)N(C1=CC(C1=S)=O)CC BBTSKBDLIAXROH-UHFFFAOYSA-N 0.000 description 1
- NFWLMJZHLNTLDM-UHFFFAOYSA-N 3-(diethylamino)cyclobut-3-ene-1,2-dione Chemical group CCN(CC)C1=CC(=O)C1=O NFWLMJZHLNTLDM-UHFFFAOYSA-N 0.000 description 1
- NWEFFLUSWNOSBH-UHFFFAOYSA-N 3-(diethylamino)cyclobut-3-ene-1,2-dithione Chemical group CCN(CC)C1=CC(=S)C1=S NWEFFLUSWNOSBH-UHFFFAOYSA-N 0.000 description 1
- OSJONOUUSQIDHR-UHFFFAOYSA-N 3-(ethylamino)-4-sulfanylidenecyclobut-2-en-1-one Chemical group C(C)NC1=CC(C1=S)=O OSJONOUUSQIDHR-UHFFFAOYSA-N 0.000 description 1
- IOYFHTXSNCBMRK-UHFFFAOYSA-N 3-(ethylamino)cyclobut-3-ene-1,2-dione Chemical group CCNC1=CC(=O)C1=O IOYFHTXSNCBMRK-UHFFFAOYSA-N 0.000 description 1
- DOICVEDRHVWXCR-UHFFFAOYSA-N 3-(ethylamino)cyclobut-3-ene-1,2-dithione Chemical group CCNC1=CC(=S)C1=S DOICVEDRHVWXCR-UHFFFAOYSA-N 0.000 description 1
- HXVVOLDXHIMZJZ-UHFFFAOYSA-N 3-[2-[2-[2-[bis[3-(dodecylamino)-3-oxopropyl]amino]ethyl-[3-(dodecylamino)-3-oxopropyl]amino]ethylamino]ethyl-[3-(dodecylamino)-3-oxopropyl]amino]-n-dodecylpropanamide Chemical compound CCCCCCCCCCCCNC(=O)CCN(CCC(=O)NCCCCCCCCCCCC)CCN(CCC(=O)NCCCCCCCCCCCC)CCNCCN(CCC(=O)NCCCCCCCCCCCC)CCC(=O)NCCCCCCCCCCCC HXVVOLDXHIMZJZ-UHFFFAOYSA-N 0.000 description 1
- HVCOBJNICQPDBP-UHFFFAOYSA-N 3-[3-[3,5-dihydroxy-6-methyl-4-(3,4,5-trihydroxy-6-methyloxan-2-yl)oxyoxan-2-yl]oxydecanoyloxy]decanoic acid;hydrate Chemical compound O.OC1C(OC(CC(=O)OC(CCCCCCC)CC(O)=O)CCCCCCC)OC(C)C(O)C1OC1C(O)C(O)C(O)C(C)O1 HVCOBJNICQPDBP-UHFFFAOYSA-N 0.000 description 1
- VZTQZXGJOSFBTJ-UHFFFAOYSA-N 3-[bis(2-methoxyethyl)amino]cyclobut-3-ene-1,2-dione Chemical group COCCN(CCOC)C1=CC(=O)C1=O VZTQZXGJOSFBTJ-UHFFFAOYSA-N 0.000 description 1
- PKXRZLCKEAZQPI-CLFAGFIQSA-N 3-[bis[(z)-octadec-9-enyl]amino]propane-1,2-diol Chemical compound CCCCCCCC\C=C/CCCCCCCCN(CC(O)CO)CCCCCCCC\C=C/CCCCCCCC PKXRZLCKEAZQPI-CLFAGFIQSA-N 0.000 description 1
- XABCFXXGZPWJQP-UHFFFAOYSA-N 3-aminoadipic acid Chemical compound OC(=O)CC(N)CCC(O)=O XABCFXXGZPWJQP-UHFFFAOYSA-N 0.000 description 1
- BMYNFMYTOJXKLE-UHFFFAOYSA-N 3-azaniumyl-2-hydroxypropanoate Chemical compound NCC(O)C(O)=O BMYNFMYTOJXKLE-UHFFFAOYSA-N 0.000 description 1
- UOQHWNPVNXSDDO-UHFFFAOYSA-N 3-bromoimidazo[1,2-a]pyridine-6-carbonitrile Chemical compound C1=CC(C#N)=CN2C(Br)=CN=C21 UOQHWNPVNXSDDO-UHFFFAOYSA-N 0.000 description 1
- ASBJGPTTYPEMLP-UHFFFAOYSA-N 3-chloroalanine Chemical compound ClCC(N)C(O)=O ASBJGPTTYPEMLP-UHFFFAOYSA-N 0.000 description 1
- 125000006027 3-methyl-1-butenyl group Chemical group 0.000 description 1
- 125000003469 3-methylhexyl group Chemical group [H]C([H])([H])C([H])([H])C([H])([H])C([H])(C([H])([H])[H])C([H])([H])C([H])([H])* 0.000 description 1
- OSWFIVFLDKOXQC-UHFFFAOYSA-N 4-(3-methoxyphenyl)aniline Chemical compound COC1=CC=CC(C=2C=CC(N)=CC=2)=C1 OSWFIVFLDKOXQC-UHFFFAOYSA-N 0.000 description 1
- CMUHFUGDYMFHEI-QMMMGPOBSA-N 4-amino-L-phenylalanine Chemical compound OC(=O)[C@@H](N)CC1=CC=C(N)C=C1 CMUHFUGDYMFHEI-QMMMGPOBSA-N 0.000 description 1
- WUBBRNOQWQTFEX-UHFFFAOYSA-N 4-aminosalicylic acid Chemical compound NC1=CC=C(C(O)=O)C(O)=C1 WUBBRNOQWQTFEX-UHFFFAOYSA-N 0.000 description 1
- 229960000549 4-dimethylaminophenol Drugs 0.000 description 1
- VHYFNPMBLIVWCW-UHFFFAOYSA-N 4-dimethylaminopyridine Substances CN(C)C1=CC=NC=C1 VHYFNPMBLIVWCW-UHFFFAOYSA-N 0.000 description 1
- XWHHYOYVRVGJJY-QMMMGPOBSA-N 4-fluoro-L-phenylalanine Chemical compound OC(=O)[C@@H](N)CC1=CC=C(F)C=C1 XWHHYOYVRVGJJY-QMMMGPOBSA-N 0.000 description 1
- 125000004217 4-methoxybenzyl group Chemical group [H]C1=C([H])C(=C([H])C([H])=C1OC([H])([H])[H])C([H])([H])* 0.000 description 1
- 108020003589 5' Untranslated Regions Proteins 0.000 description 1
- YWWVWXASSLXJHU-UHFFFAOYSA-N 9E-tetradecenoic acid Natural products CCCCC=CCCCCCCCC(O)=O YWWVWXASSLXJHU-UHFFFAOYSA-N 0.000 description 1
- QTBSBXVTEAMEQO-UHFFFAOYSA-M Acetate Chemical compound CC([O-])=O QTBSBXVTEAMEQO-UHFFFAOYSA-M 0.000 description 1
- 229930024421 Adenine Natural products 0.000 description 1
- 108700028369 Alleles Proteins 0.000 description 1
- 108020004491 Antisense DNA Proteins 0.000 description 1
- 108091023037 Aptamer Proteins 0.000 description 1
- 101001053401 Arabidopsis thaliana Acid beta-fructofuranosidase 3, vacuolar Proteins 0.000 description 1
- 101001053395 Arabidopsis thaliana Acid beta-fructofuranosidase 4, vacuolar Proteins 0.000 description 1
- 239000004475 Arginine Substances 0.000 description 1
- 108090000749 Aurora kinase B Proteins 0.000 description 1
- 102100032306 Aurora kinase B Human genes 0.000 description 1
- 108091032955 Bacterial small RNA Proteins 0.000 description 1
- 235000021357 Behenic acid Nutrition 0.000 description 1
- 239000005711 Benzoic acid Substances 0.000 description 1
- 102000004506 Blood Proteins Human genes 0.000 description 1
- 108010017384 Blood Proteins Proteins 0.000 description 1
- 241000283690 Bos taurus Species 0.000 description 1
- 101000856746 Bos taurus Cytochrome c oxidase subunit 7A1, mitochondrial Proteins 0.000 description 1
- DPUOLQHDNGRHBS-UHFFFAOYSA-N Brassidinsaeure Natural products CCCCCCCCC=CCCCCCCCCCCCC(O)=O DPUOLQHDNGRHBS-UHFFFAOYSA-N 0.000 description 1
- 239000002126 C01EB10 - Adenosine Substances 0.000 description 1
- 125000003358 C2-C20 alkenyl group Chemical group 0.000 description 1
- 125000004648 C2-C8 alkenyl group Chemical group 0.000 description 1
- 125000001313 C5-C10 heteroaryl group Chemical group 0.000 description 1
- 125000000041 C6-C10 aryl group Chemical group 0.000 description 1
- PCBZRNYXXCIELG-WYFCWLEVSA-N COC1=CC=C(C[C@H](NC(=O)OC2CCCC3(C2)OOC2(O3)C3CC4CC(C3)CC2C4)C(=O)N[C@@H]2[C@@H](CO)O[C@H]([C@@H]2O)N2C=NC3=C2N=CN=C3N(C)C)C=C1 Chemical compound COC1=CC=C(C[C@H](NC(=O)OC2CCCC3(C2)OOC2(O3)C3CC4CC(C3)CC2C4)C(=O)N[C@@H]2[C@@H](CO)O[C@H]([C@@H]2O)N2C=NC3=C2N=CN=C3N(C)C)C=C1 PCBZRNYXXCIELG-WYFCWLEVSA-N 0.000 description 1
- QCMYYKRYFNMIEC-UHFFFAOYSA-N COP(O)=O Chemical class COP(O)=O QCMYYKRYFNMIEC-UHFFFAOYSA-N 0.000 description 1
- 108091033409 CRISPR Proteins 0.000 description 1
- LSPHULWDVZXLIL-UHFFFAOYSA-N Camphoric acid Natural products CC1(C)C(C(O)=O)CCC1(C)C(O)=O LSPHULWDVZXLIL-UHFFFAOYSA-N 0.000 description 1
- 241000282472 Canis lupus familiaris Species 0.000 description 1
- 241000283707 Capra Species 0.000 description 1
- 239000005632 Capric acid (CAS 334-48-5) Substances 0.000 description 1
- 239000005635 Caprylic acid (CAS 124-07-2) Substances 0.000 description 1
- KXDHJXZQYSOELW-UHFFFAOYSA-M Carbamate Chemical compound NC([O-])=O KXDHJXZQYSOELW-UHFFFAOYSA-M 0.000 description 1
- 239000004215 Carbon black (E152) Substances 0.000 description 1
- OKTJSMMVPCPJKN-NJFSPNSNSA-N Carbon-14 Chemical compound [14C] OKTJSMMVPCPJKN-NJFSPNSNSA-N 0.000 description 1
- ZAMOUSCENKQFHK-UHFFFAOYSA-N Chlorine atom Chemical compound [Cl] ZAMOUSCENKQFHK-UHFFFAOYSA-N 0.000 description 1
- 108010009685 Cholinergic Receptors Proteins 0.000 description 1
- WBYWAXJHAXSJNI-SREVYHEPSA-N Cinnamic acid Chemical compound OC(=O)\C=C/C1=CC=CC=C1 WBYWAXJHAXSJNI-SREVYHEPSA-N 0.000 description 1
- 108020004638 Circular DNA Proteins 0.000 description 1
- 108091028075 Circular RNA Proteins 0.000 description 1
- 108020004394 Complementary RNA Proteins 0.000 description 1
- RYGMFSIKBFXOCR-UHFFFAOYSA-N Copper Chemical compound [Cu] RYGMFSIKBFXOCR-UHFFFAOYSA-N 0.000 description 1
- MIKUYHXYGGJMLM-GIMIYPNGSA-N Crotonoside Natural products C1=NC2=C(N)NC(=O)N=C2N1[C@H]1O[C@@H](CO)[C@H](O)[C@@H]1O MIKUYHXYGGJMLM-GIMIYPNGSA-N 0.000 description 1
- XZMCDFZZKTWFGF-UHFFFAOYSA-N Cyanamide Chemical group NC#N XZMCDFZZKTWFGF-UHFFFAOYSA-N 0.000 description 1
- 108050006400 Cyclin Proteins 0.000 description 1
- UHDGCWIWMRVCDJ-PSQAKQOGSA-N Cytidine Natural products O=C1N=C(N)C=CN1[C@@H]1[C@@H](O)[C@@H](O)[C@H](CO)O1 UHDGCWIWMRVCDJ-PSQAKQOGSA-N 0.000 description 1
- SNDPXSYFESPGGJ-SCSAIBSYSA-N D-2-aminopentanoic acid Chemical compound CCC[C@@H](N)C(O)=O SNDPXSYFESPGGJ-SCSAIBSYSA-N 0.000 description 1
- RGHNJXZEOKUKBD-UHFFFAOYSA-N D-gluconic acid Natural products OCC(O)C(O)C(O)C(O)C(O)=O RGHNJXZEOKUKBD-UHFFFAOYSA-N 0.000 description 1
- AEMOLEFTQBMNLQ-AQKNRBDQSA-N D-glucopyranuronic acid Chemical compound OC1O[C@H](C(O)=O)[C@@H](O)[C@H](O)[C@H]1O AEMOLEFTQBMNLQ-AQKNRBDQSA-N 0.000 description 1
- NYHBQMYGNKIUIF-UHFFFAOYSA-N D-guanosine Natural products C1=2NC(N)=NC(=O)C=2N=CN1C1OC(CO)C(O)C1O NYHBQMYGNKIUIF-UHFFFAOYSA-N 0.000 description 1
- HMFHBZSHGGEWLO-SOOFDHNKSA-N D-ribofuranose Chemical compound OC[C@H]1OC(O)[C@H](O)[C@@H]1O HMFHBZSHGGEWLO-SOOFDHNKSA-N 0.000 description 1
- 108091027757 Deoxyribozyme Proteins 0.000 description 1
- YZCKVEUIGOORGS-OUBTZVSYSA-N Deuterium Chemical compound [2H] YZCKVEUIGOORGS-OUBTZVSYSA-N 0.000 description 1
- FEWJPZIEWOKRBE-JCYAYHJZSA-N Dextrotartaric acid Chemical compound OC(=O)[C@H](O)[C@@H](O)C(O)=O FEWJPZIEWOKRBE-JCYAYHJZSA-N 0.000 description 1
- BVTJGGGYKAMDBN-UHFFFAOYSA-N Dioxetane Chemical compound C1COO1 BVTJGGGYKAMDBN-UHFFFAOYSA-N 0.000 description 1
- 235000021294 Docosapentaenoic acid Nutrition 0.000 description 1
- 108010024212 E-Selectin Proteins 0.000 description 1
- 102100023471 E-selectin Human genes 0.000 description 1
- 241000196324 Embryophyta Species 0.000 description 1
- 108010042407 Endonucleases Proteins 0.000 description 1
- 102000004533 Endonucleases Human genes 0.000 description 1
- 241000283086 Equidae Species 0.000 description 1
- URXZXNYJPAJJOQ-UHFFFAOYSA-N Erucic acid Natural products CCCCCCC=CCCCCCCCCCCCC(O)=O URXZXNYJPAJJOQ-UHFFFAOYSA-N 0.000 description 1
- 102000003951 Erythropoietin Human genes 0.000 description 1
- 108090000394 Erythropoietin Proteins 0.000 description 1
- PIICEJLVQHRZGT-UHFFFAOYSA-N Ethylenediamine Chemical compound NCCN PIICEJLVQHRZGT-UHFFFAOYSA-N 0.000 description 1
- 101710091918 Eukaryotic translation initiation factor 4E Proteins 0.000 description 1
- 102100027304 Eukaryotic translation initiation factor 4E Human genes 0.000 description 1
- 101710126428 Eukaryotic translation initiation factor 4E-2 Proteins 0.000 description 1
- 101710126416 Eukaryotic translation initiation factor 4E-3 Proteins 0.000 description 1
- 101710126432 Eukaryotic translation initiation factor 4E1 Proteins 0.000 description 1
- 101710133325 Eukaryotic translation initiation factor NCBP Proteins 0.000 description 1
- 101710190212 Eukaryotic translation initiation factor isoform 4E Proteins 0.000 description 1
- 101710124729 Eukaryotic translation initiation factor isoform 4E-2 Proteins 0.000 description 1
- 241000282326 Felis catus Species 0.000 description 1
- NIGWMJHCCYYCSF-UHFFFAOYSA-N Fenclonine Chemical compound OC(=O)C(N)CC1=CC=C(Cl)C=C1 NIGWMJHCCYYCSF-UHFFFAOYSA-N 0.000 description 1
- PXGOKWXKJXAPGV-UHFFFAOYSA-N Fluorine Chemical compound FF PXGOKWXKJXAPGV-UHFFFAOYSA-N 0.000 description 1
- BDAGIHXWWSANSR-UHFFFAOYSA-M Formate Chemical compound [O-]C=O BDAGIHXWWSANSR-UHFFFAOYSA-M 0.000 description 1
- 102000004300 GABA-A Receptors Human genes 0.000 description 1
- 108090000839 GABA-A Receptors Proteins 0.000 description 1
- IAJILQKETJEXLJ-UHFFFAOYSA-N Galacturonsaeure Natural products O=CC(O)C(O)C(O)C(O)C(O)=O IAJILQKETJEXLJ-UHFFFAOYSA-N 0.000 description 1
- JZNWSCPGTDBMEW-UHFFFAOYSA-N Glycerophosphorylethanolamin Natural products NCCOP(O)(=O)OCC(O)CO JZNWSCPGTDBMEW-UHFFFAOYSA-N 0.000 description 1
- ZWZWYGMENQVNFU-UHFFFAOYSA-N Glycerophosphorylserin Natural products OC(=O)C(N)COP(O)(=O)OCC(O)CO ZWZWYGMENQVNFU-UHFFFAOYSA-N 0.000 description 1
- 101000851176 Homo sapiens Pro-epidermal growth factor Proteins 0.000 description 1
- 101001098868 Homo sapiens Proprotein convertase subtilisin/kexin type 9 Proteins 0.000 description 1
- LCWXJXMHJVIJFK-UHFFFAOYSA-N Hydroxylysine Natural products NCC(O)CC(N)CC(O)=O LCWXJXMHJVIJFK-UHFFFAOYSA-N 0.000 description 1
- PMMYEEVYMWASQN-DMTCNVIQSA-N Hydroxyproline Chemical compound O[C@H]1CN[C@H](C(O)=O)C1 PMMYEEVYMWASQN-DMTCNVIQSA-N 0.000 description 1
- 208000035150 Hypercholesterolemia Diseases 0.000 description 1
- 206010061218 Inflammation Diseases 0.000 description 1
- UGQMRVRMYYASKQ-KQYNXXCUSA-N Inosine Chemical compound O[C@@H]1[C@H](O)[C@@H](CO)O[C@H]1N1C2=NC=NC(O)=C2N=C1 UGQMRVRMYYASKQ-KQYNXXCUSA-N 0.000 description 1
- 229930010555 Inosine Natural products 0.000 description 1
- 102100034343 Integrase Human genes 0.000 description 1
- 108010064593 Intercellular Adhesion Molecule-1 Proteins 0.000 description 1
- 102100037877 Intercellular adhesion molecule 1 Human genes 0.000 description 1
- SNDPXSYFESPGGJ-BYPYZUCNSA-N L-2-aminopentanoic acid Chemical compound CCC[C@H](N)C(O)=O SNDPXSYFESPGGJ-BYPYZUCNSA-N 0.000 description 1
- JUQLUIFNNFIIKC-YFKPBYRVSA-N L-2-aminopimelic acid Chemical compound OC(=O)[C@@H](N)CCCCC(O)=O JUQLUIFNNFIIKC-YFKPBYRVSA-N 0.000 description 1
- QUOGESRFPZDMMT-UHFFFAOYSA-N L-Homoarginine Natural products OC(=O)C(N)CCCCNC(N)=N QUOGESRFPZDMMT-UHFFFAOYSA-N 0.000 description 1
- AGPKZVBTJJNPAG-UHNVWZDZSA-N L-allo-Isoleucine Chemical compound CC[C@@H](C)[C@H](N)C(O)=O AGPKZVBTJJNPAG-UHNVWZDZSA-N 0.000 description 1
- ZGUNAGUHMKGQNY-ZETCQYMHSA-N L-alpha-phenylglycine zwitterion Chemical compound OC(=O)[C@@H](N)C1=CC=CC=C1 ZGUNAGUHMKGQNY-ZETCQYMHSA-N 0.000 description 1
- 150000008575 L-amino acids Chemical group 0.000 description 1
- ODKSFYDXXFIFQN-BYPYZUCNSA-P L-argininium(2+) Chemical compound NC(=[NH2+])NCCC[C@H]([NH3+])C(O)=O ODKSFYDXXFIFQN-BYPYZUCNSA-P 0.000 description 1
- RHGKLRLOHDJJDR-BYPYZUCNSA-N L-citrulline Chemical compound NC(=O)NCCC[C@H]([NH3+])C([O-])=O RHGKLRLOHDJJDR-BYPYZUCNSA-N 0.000 description 1
- HNDVDQJCIGZPNO-YFKPBYRVSA-N L-histidine Chemical compound OC(=O)[C@@H](N)CC1=CN=CN1 HNDVDQJCIGZPNO-YFKPBYRVSA-N 0.000 description 1
- QUOGESRFPZDMMT-YFKPBYRVSA-N L-homoarginine Chemical compound OC(=O)[C@@H](N)CCCCNC(N)=N QUOGESRFPZDMMT-YFKPBYRVSA-N 0.000 description 1
- FFFHZYDWPBMWHY-VKHMYHEASA-N L-homocysteine Chemical compound OC(=O)[C@@H](N)CCS FFFHZYDWPBMWHY-VKHMYHEASA-N 0.000 description 1
- UKAUYVFTDYCKQA-VKHMYHEASA-N L-homoserine Chemical compound OC(=O)[C@@H](N)CCO UKAUYVFTDYCKQA-VKHMYHEASA-N 0.000 description 1
- FFEARJCKVFRZRR-BYPYZUCNSA-N L-methionine Chemical compound CSCC[C@H](N)C(O)=O FFEARJCKVFRZRR-BYPYZUCNSA-N 0.000 description 1
- QEFRNWWLZKMPFJ-ZXPFJRLXSA-N L-methionine (R)-S-oxide Chemical compound C[S@@](=O)CC[C@H]([NH3+])C([O-])=O QEFRNWWLZKMPFJ-ZXPFJRLXSA-N 0.000 description 1
- QEFRNWWLZKMPFJ-UHFFFAOYSA-N L-methionine sulphoxide Natural products CS(=O)CCC(N)C(O)=O QEFRNWWLZKMPFJ-UHFFFAOYSA-N 0.000 description 1
- SNDPXSYFESPGGJ-UHFFFAOYSA-N L-norVal-OH Natural products CCCC(N)C(O)=O SNDPXSYFESPGGJ-UHFFFAOYSA-N 0.000 description 1
- LRQKBLKVPFOOQJ-YFKPBYRVSA-N L-norleucine Chemical compound CCCC[C@H]([NH3+])C([O-])=O LRQKBLKVPFOOQJ-YFKPBYRVSA-N 0.000 description 1
- 108091026898 Leader sequence (mRNA) Proteins 0.000 description 1
- WHXSMMKQMYFTQS-UHFFFAOYSA-N Lithium Chemical compound [Li] WHXSMMKQMYFTQS-UHFFFAOYSA-N 0.000 description 1
- 239000004472 Lysine Substances 0.000 description 1
- FYYHWMGAXLPEAU-UHFFFAOYSA-N Magnesium Chemical compound [Mg] FYYHWMGAXLPEAU-UHFFFAOYSA-N 0.000 description 1
- 108060004795 Methyltransferase Proteins 0.000 description 1
- 108091007780 MiR-122 Proteins 0.000 description 1
- 206010048723 Multiple-drug resistance Diseases 0.000 description 1
- UQOFGTXDASPNLL-XHNCKOQMSA-N Muscarine Chemical compound C[C@@H]1O[C@H](C[N+](C)(C)C)C[C@H]1O UQOFGTXDASPNLL-XHNCKOQMSA-N 0.000 description 1
- VEYYWZRYIYDQJM-ZETCQYMHSA-N N(2)-acetyl-L-lysine Chemical compound CC(=O)N[C@H](C([O-])=O)CCCC[NH3+] VEYYWZRYIYDQJM-ZETCQYMHSA-N 0.000 description 1
- OLNLSTNFRUFTLM-UHFFFAOYSA-N N-ethylasparagine Chemical compound CCNC(C(O)=O)CC(N)=O OLNLSTNFRUFTLM-UHFFFAOYSA-N 0.000 description 1
- YPIGGYHFMKJNKV-UHFFFAOYSA-N N-ethylglycine Chemical compound CC[NH2+]CC([O-])=O YPIGGYHFMKJNKV-UHFFFAOYSA-N 0.000 description 1
- 108010065338 N-ethylglycine Proteins 0.000 description 1
- HTLZVHNRZJPSMI-UHFFFAOYSA-N N-ethylpiperidine Chemical compound CCN1CCCCC1 HTLZVHNRZJPSMI-UHFFFAOYSA-N 0.000 description 1
- AKCRVYNORCOYQT-YFKPBYRVSA-N N-methyl-L-valine Chemical compound CN[C@@H](C(C)C)C(O)=O AKCRVYNORCOYQT-YFKPBYRVSA-N 0.000 description 1
- MBBZMMPHUWSWHV-BDVNFPICSA-N N-methylglucamine Chemical compound CNC[C@H](O)[C@@H](O)[C@H](O)[C@H](O)CO MBBZMMPHUWSWHV-BDVNFPICSA-N 0.000 description 1
- RHGKLRLOHDJJDR-UHFFFAOYSA-N Ndelta-carbamoyl-DL-ornithine Natural products OC(=O)C(N)CCCNC(N)=O RHGKLRLOHDJJDR-UHFFFAOYSA-N 0.000 description 1
- GRYLNZFGIOXLOG-UHFFFAOYSA-N Nitric acid Chemical compound O[N+]([O-])=O GRYLNZFGIOXLOG-UHFFFAOYSA-N 0.000 description 1
- 241000283973 Oryctolagus cuniculus Species 0.000 description 1
- 238000012408 PCR amplification Methods 0.000 description 1
- WIHSZOXPODIZSW-KJIWEYRQSA-N PE(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) Chemical compound CC\C=C/C\C=C/C\C=C/CCCCCCCC(=O)OC[C@H](COP(O)(=O)OCCN)OC(=O)CCCCCCC\C=C/C\C=C/C\C=C/CC WIHSZOXPODIZSW-KJIWEYRQSA-N 0.000 description 1
- 229910019142 PO4 Inorganic materials 0.000 description 1
- 235000021319 Palmitoleic acid Nutrition 0.000 description 1
- 241001494479 Pecora Species 0.000 description 1
- 229930182555 Penicillin Natural products 0.000 description 1
- JGSARLDLIJGVTE-MBNYWOFBSA-N Penicillin G Chemical compound N([C@H]1[C@H]2SC([C@@H](N2C1=O)C(O)=O)(C)C)C(=O)CC1=CC=CC=C1 JGSARLDLIJGVTE-MBNYWOFBSA-N 0.000 description 1
- 241000577979 Peromyscus spicilegus Species 0.000 description 1
- 108010039918 Polylysine Proteins 0.000 description 1
- 102000015623 Polynucleotide Adenylyltransferase Human genes 0.000 description 1
- 108010024055 Polynucleotide adenylyltransferase Proteins 0.000 description 1
- 102100038955 Proprotein convertase subtilisin/kexin type 9 Human genes 0.000 description 1
- 229930185560 Pseudouridine Natural products 0.000 description 1
- PTJWIQPHWPFNBW-UHFFFAOYSA-N Pseudouridine C Natural products OC1C(O)C(CO)OC1C1=CNC(=O)NC1=O PTJWIQPHWPFNBW-UHFFFAOYSA-N 0.000 description 1
- IWYDHOAUDWTVEP-UHFFFAOYSA-N R-2-phenyl-2-hydroxyacetic acid Natural products OC(=O)C(O)C1=CC=CC=C1 IWYDHOAUDWTVEP-UHFFFAOYSA-N 0.000 description 1
- 102000000574 RNA-Induced Silencing Complex Human genes 0.000 description 1
- 108010016790 RNA-Induced Silencing Complex Proteins 0.000 description 1
- PYMYPHUHKUWMLA-LMVFSUKVSA-N Ribose Natural products OC[C@@H](O)[C@@H](O)[C@@H](O)C=O PYMYPHUHKUWMLA-LMVFSUKVSA-N 0.000 description 1
- 108010077895 Sarcosine Proteins 0.000 description 1
- 108020004682 Single-Stranded DNA Proteins 0.000 description 1
- 108091081024 Start codon Proteins 0.000 description 1
- KDYFGRWQOYBRFD-UHFFFAOYSA-N Succinic acid Natural products OC(=O)CCC(O)=O KDYFGRWQOYBRFD-UHFFFAOYSA-N 0.000 description 1
- 241000282898 Sus scrofa Species 0.000 description 1
- 230000024932 T cell mediated immunity Effects 0.000 description 1
- FEWJPZIEWOKRBE-UHFFFAOYSA-N Tartaric acid Natural products [H+].[H+].[O-]C(=O)C(O)C(O)C([O-])=O FEWJPZIEWOKRBE-UHFFFAOYSA-N 0.000 description 1
- DHXVGJBLRPWPCS-UHFFFAOYSA-N Tetrahydropyran Chemical compound C1CCOCC1 DHXVGJBLRPWPCS-UHFFFAOYSA-N 0.000 description 1
- YPWFISCTZQNZAU-UHFFFAOYSA-N Thiane Chemical compound C1CCSCC1 YPWFISCTZQNZAU-UHFFFAOYSA-N 0.000 description 1
- 239000004473 Threonine Substances 0.000 description 1
- 108700019146 Transgenes Proteins 0.000 description 1
- GSEJCLTVZPLZKY-UHFFFAOYSA-N Triethanolamine Chemical compound OCCN(CCO)CCO GSEJCLTVZPLZKY-UHFFFAOYSA-N 0.000 description 1
- YZCKVEUIGOORGS-NJFSPNSNSA-N Tritium Chemical compound [3H] YZCKVEUIGOORGS-NJFSPNSNSA-N 0.000 description 1
- XSQUKJJJFZCRTK-UHFFFAOYSA-N Urea Natural products NC(N)=O XSQUKJJJFZCRTK-UHFFFAOYSA-N 0.000 description 1
- HCHKCACWOHOZIP-UHFFFAOYSA-N Zinc Chemical compound [Zn] HCHKCACWOHOZIP-UHFFFAOYSA-N 0.000 description 1
- NJFCSWSRXWCWHV-USYZEHPZSA-N [(2R)-2,3-bis(octadec-1-enoxy)propyl] 2-(trimethylazaniumyl)ethyl phosphate Chemical compound CCCCCCCCCCCCCCCCC=COC[C@H](COP([O-])(=O)OCC[N+](C)(C)C)OC=CCCCCCCCCCCCCCCCC NJFCSWSRXWCWHV-USYZEHPZSA-N 0.000 description 1
- SUTHKQVOHCMCCF-QZNUWAOFSA-N [(2r)-3-[2-aminoethoxy(hydroxy)phosphoryl]oxy-2-docosa-2,4,6,8,10,12-hexaenoyloxypropyl] docosa-2,4,6,8,10,12-hexaenoate Chemical compound CCCCCCCCCC=CC=CC=CC=CC=CC=CC(=O)OC[C@H](COP(O)(=O)OCCN)OC(=O)C=CC=CC=CC=CC=CC=CCCCCCCCCC SUTHKQVOHCMCCF-QZNUWAOFSA-N 0.000 description 1
- HCAJCMUKLZSPFT-KWXKLSQISA-N [3-(dimethylamino)-2-[(9z,12z)-octadeca-9,12-dienoyl]oxypropyl] (9z,12z)-octadeca-9,12-dienoate Chemical compound CCCCC\C=C/C\C=C/CCCCCCCC(=O)OCC(CN(C)C)OC(=O)CCCCCCC\C=C/C\C=C/CCCCC HCAJCMUKLZSPFT-KWXKLSQISA-N 0.000 description 1
- HMNZFMSWFCAGGW-XPWSMXQVSA-N [3-[hydroxy(2-hydroxyethoxy)phosphoryl]oxy-2-[(e)-octadec-9-enoyl]oxypropyl] (e)-octadec-9-enoate Chemical compound CCCCCCCC\C=C\CCCCCCCC(=O)OCC(COP(O)(=O)OCCO)OC(=O)CCCCCCC\C=C\CCCCCCCC HMNZFMSWFCAGGW-XPWSMXQVSA-N 0.000 description 1
- 230000001594 aberrant effect Effects 0.000 description 1
- 230000002159 abnormal effect Effects 0.000 description 1
- 238000010521 absorption reaction Methods 0.000 description 1
- 125000002777 acetyl group Chemical group [H]C([H])([H])C(*)=O 0.000 description 1
- 102000034337 acetylcholine receptors Human genes 0.000 description 1
- 150000007513 acids Chemical class 0.000 description 1
- 125000000641 acridinyl group Chemical group C1(=CC=CC2=NC3=CC=CC=C3C=C12)* 0.000 description 1
- 230000004913 activation Effects 0.000 description 1
- 125000005073 adamantyl group Chemical group C12(CC3CC(CC(C1)C3)C2)* 0.000 description 1
- 125000000848 adenin-9-yl group Chemical group [H]N([H])C1=C2N=C([H])N(*)C2=NC([H])=N1 0.000 description 1
- 229960000643 adenine Drugs 0.000 description 1
- 229960005305 adenosine Drugs 0.000 description 1
- 235000011037 adipic acid Nutrition 0.000 description 1
- 239000001361 adipic acid Substances 0.000 description 1
- 229960000250 adipic acid Drugs 0.000 description 1
- 239000002671 adjuvant Substances 0.000 description 1
- 230000002411 adverse Effects 0.000 description 1
- 238000001042 affinity chromatography Methods 0.000 description 1
- 150000001298 alcohols Chemical class 0.000 description 1
- 235000010443 alginic acid Nutrition 0.000 description 1
- 239000000783 alginic acid Substances 0.000 description 1
- 229920000615 alginic acid Polymers 0.000 description 1
- 229960001126 alginic acid Drugs 0.000 description 1
- 150000004781 alginic acids Chemical class 0.000 description 1
- 125000002355 alkine group Chemical group 0.000 description 1
- 125000000278 alkyl amino alkyl group Chemical group 0.000 description 1
- 150000001350 alkyl halides Chemical class 0.000 description 1
- 125000004419 alkynylene group Chemical group 0.000 description 1
- JAZBEHYOTPTENJ-JLNKQSITSA-N all-cis-5,8,11,14,17-icosapentaenoic acid Chemical compound CC\C=C/C\C=C/C\C=C/C\C=C/C\C=C/CCCC(O)=O JAZBEHYOTPTENJ-JLNKQSITSA-N 0.000 description 1
- HMFHBZSHGGEWLO-UHFFFAOYSA-N alpha-D-Furanose-Ribose Natural products OCC1OC(O)C(O)C1O HMFHBZSHGGEWLO-UHFFFAOYSA-N 0.000 description 1
- HSFWRNGVRCDJHI-UHFFFAOYSA-N alpha-acetylene Natural products C#C HSFWRNGVRCDJHI-UHFFFAOYSA-N 0.000 description 1
- 150000001371 alpha-amino acids Chemical class 0.000 description 1
- BJEPYKJPYRNKOW-UHFFFAOYSA-N alpha-hydroxysuccinic acid Natural products OC(=O)C(O)CC(O)=O BJEPYKJPYRNKOW-UHFFFAOYSA-N 0.000 description 1
- 235000020661 alpha-linolenic acid Nutrition 0.000 description 1
- 229910000147 aluminium phosphate Inorganic materials 0.000 description 1
- 125000003368 amide group Chemical group 0.000 description 1
- 229960004909 aminosalicylic acid Drugs 0.000 description 1
- 229910021529 ammonia Inorganic materials 0.000 description 1
- JFCQEDHGNNZCLN-UHFFFAOYSA-N anhydrous glutaric acid Natural products OC(=O)CCCC(O)=O JFCQEDHGNNZCLN-UHFFFAOYSA-N 0.000 description 1
- 239000000427 antigen Substances 0.000 description 1
- 210000000612 antigen-presenting cell Anatomy 0.000 description 1
- 108091007433 antigens Proteins 0.000 description 1
- 102000036639 antigens Human genes 0.000 description 1
- 239000003816 antisense DNA Substances 0.000 description 1
- 230000006907 apoptotic process Effects 0.000 description 1
- 235000021342 arachidonic acid Nutrition 0.000 description 1
- 229940114079 arachidonic acid Drugs 0.000 description 1
- ODKSFYDXXFIFQN-UHFFFAOYSA-N arginine Natural products OC(=O)C(N)CCCNC(N)=N ODKSFYDXXFIFQN-UHFFFAOYSA-N 0.000 description 1
- 229960003121 arginine Drugs 0.000 description 1
- 235000010323 ascorbic acid Nutrition 0.000 description 1
- 239000011668 ascorbic acid Substances 0.000 description 1
- 229960005070 ascorbic acid Drugs 0.000 description 1
- 235000003704 aspartic acid Nutrition 0.000 description 1
- 229960005261 aspartic acid Drugs 0.000 description 1
- 150000001540 azides Chemical class 0.000 description 1
- 230000037429 base substitution Effects 0.000 description 1
- 229940116226 behenic acid Drugs 0.000 description 1
- UPABQMWFWCMOFV-UHFFFAOYSA-N benethamine Chemical compound C=1C=CC=CC=1CNCCC1=CC=CC=C1 UPABQMWFWCMOFV-UHFFFAOYSA-N 0.000 description 1
- JUHORIMYRDESRB-UHFFFAOYSA-N benzathine Chemical compound C=1C=CC=CC=1CNCCNCC1=CC=CC=C1 JUHORIMYRDESRB-UHFFFAOYSA-N 0.000 description 1
- SRSXLGNVWSONIS-UHFFFAOYSA-N benzenesulfonic acid Chemical compound OS(=O)(=O)C1=CC=CC=C1 SRSXLGNVWSONIS-UHFFFAOYSA-N 0.000 description 1
- 229940092714 benzenesulfonic acid Drugs 0.000 description 1
- 125000000499 benzofuranyl group Chemical group O1C(=CC2=C1C=CC=C2)* 0.000 description 1
- 235000010233 benzoic acid Nutrition 0.000 description 1
- 229960004365 benzoic acid Drugs 0.000 description 1
- 150000001558 benzoic acid derivatives Chemical class 0.000 description 1
- 125000004196 benzothienyl group Chemical group S1C(=CC2=C1C=CC=C2)* 0.000 description 1
- 125000003354 benzotriazolyl group Chemical group N1N=NC2=C1C=CC=C2* 0.000 description 1
- MSWZFWKMSRAUBD-UHFFFAOYSA-N beta-D-galactosamine Natural products NC1C(O)OC(CO)C(O)C1O MSWZFWKMSRAUBD-UHFFFAOYSA-N 0.000 description 1
- DRTQHJPVMGBUCF-PSQAKQOGSA-N beta-L-uridine Natural products O[C@H]1[C@@H](O)[C@H](CO)O[C@@H]1N1C(=O)NC(=O)C=C1 DRTQHJPVMGBUCF-PSQAKQOGSA-N 0.000 description 1
- WGDUUQDYDIIBKT-UHFFFAOYSA-N beta-Pseudouridine Natural products OC1OC(CN2C=CC(=O)NC2=O)C(O)C1O WGDUUQDYDIIBKT-UHFFFAOYSA-N 0.000 description 1
- 150000001576 beta-amino acids Chemical class 0.000 description 1
- OQFSQFPPLPISGP-UHFFFAOYSA-N beta-carboxyaspartic acid Natural products OC(=O)C(N)C(C(O)=O)C(O)=O OQFSQFPPLPISGP-UHFFFAOYSA-N 0.000 description 1
- 229960003237 betaine Drugs 0.000 description 1
- 230000004071 biological effect Effects 0.000 description 1
- 239000004305 biphenyl Substances 0.000 description 1
- 235000010290 biphenyl Nutrition 0.000 description 1
- 238000004820 blood count Methods 0.000 description 1
- KDYFGRWQOYBRFD-NUQCWPJISA-N butanedioic acid Chemical compound O[14C](=O)CC[14C](O)=O KDYFGRWQOYBRFD-NUQCWPJISA-N 0.000 description 1
- 125000000480 butynyl group Chemical group [*]C#CC([H])([H])C([H])([H])[H] 0.000 description 1
- LSPHULWDVZXLIL-QUBYGPBYSA-N camphoric acid Chemical compound CC1(C)[C@H](C(O)=O)CC[C@]1(C)C(O)=O LSPHULWDVZXLIL-QUBYGPBYSA-N 0.000 description 1
- KHAVLLBUVKBTBG-UHFFFAOYSA-N caproleic acid Natural products OC(=O)CCCCCCCC=C KHAVLLBUVKBTBG-UHFFFAOYSA-N 0.000 description 1
- 125000003917 carbamoyl group Chemical group [H]N([H])C(*)=O 0.000 description 1
- 125000002837 carbocyclic group Chemical group 0.000 description 1
- BVKZGUZCCUSVTD-UHFFFAOYSA-N carbonic acid Chemical compound OC(O)=O BVKZGUZCCUSVTD-UHFFFAOYSA-N 0.000 description 1
- 150000007942 carboxylates Chemical class 0.000 description 1
- 150000001768 cations Chemical class 0.000 description 1
- 238000004113 cell culture Methods 0.000 description 1
- 230000024245 cell differentiation Effects 0.000 description 1
- 210000000170 cell membrane Anatomy 0.000 description 1
- 108091092328 cellular RNA Proteins 0.000 description 1
- 229930183167 cerebroside Natural products 0.000 description 1
- RIZIAUKTHDLMQX-UHFFFAOYSA-N cerebroside D Natural products CCCCCCCCCCCCCCCCC(O)C(=O)NC(C(O)C=CCCC=C(C)CCCCCCCCC)COC1OC(CO)C(O)C(O)C1O RIZIAUKTHDLMQX-UHFFFAOYSA-N 0.000 description 1
- 229930016911 cinnamic acid Natural products 0.000 description 1
- 235000013985 cinnamic acid Nutrition 0.000 description 1
- 230000004087 circulation Effects 0.000 description 1
- BJBUEDPLEOHJGE-IUYQGCFVSA-N cis-3-hydroxy-D-proline zwitterion Chemical compound O[C@H]1CCN[C@H]1C(O)=O BJBUEDPLEOHJGE-IUYQGCFVSA-N 0.000 description 1
- SECPZKHBENQXJG-UHFFFAOYSA-N cis-palmitoleic acid Natural products CCCCCCC=CCCCCCCCC(O)=O SECPZKHBENQXJG-UHFFFAOYSA-N 0.000 description 1
- 229960002173 citrulline Drugs 0.000 description 1
- 235000013477 citrulline Nutrition 0.000 description 1
- 238000010367 cloning Methods 0.000 description 1
- 230000008045 co-localization Effects 0.000 description 1
- 239000011248 coating agent Substances 0.000 description 1
- 239000003086 colorant Substances 0.000 description 1
- 229940125904 compound 1 Drugs 0.000 description 1
- 229940125782 compound 2 Drugs 0.000 description 1
- 229940126214 compound 3 Drugs 0.000 description 1
- 230000001268 conjugating effect Effects 0.000 description 1
- 238000013270 controlled release Methods 0.000 description 1
- 238000007796 conventional method Methods 0.000 description 1
- 229910052802 copper Inorganic materials 0.000 description 1
- 239000010949 copper Substances 0.000 description 1
- 125000004966 cyanoalkyl group Chemical group 0.000 description 1
- 239000000625 cyclamic acid and its Na and Ca salt Substances 0.000 description 1
- 230000001351 cycling effect Effects 0.000 description 1
- 238000006352 cycloaddition reaction Methods 0.000 description 1
- 125000001995 cyclobutyl group Chemical group [H]C1([H])C([H])([H])C([H])(*)C1([H])[H] 0.000 description 1
- 125000000582 cycloheptyl group Chemical group [H]C1([H])C([H])([H])C([H])([H])C([H])([H])C([H])(*)C([H])([H])C1([H])[H] 0.000 description 1
- HCAJEUSONLESMK-UHFFFAOYSA-N cyclohexylsulfamic acid Chemical compound OS(=O)(=O)NC1CCCCC1 HCAJEUSONLESMK-UHFFFAOYSA-N 0.000 description 1
- 125000000640 cyclooctyl group Chemical group [H]C1([H])C([H])([H])C([H])([H])C([H])([H])C([H])(*)C([H])([H])C([H])([H])C1([H])[H] 0.000 description 1
- 125000001511 cyclopentyl group Chemical group [H]C1([H])C([H])([H])C([H])([H])C([H])(*)C1([H])[H] 0.000 description 1
- 229940104302 cytosine Drugs 0.000 description 1
- 230000001086 cytosolic effect Effects 0.000 description 1
- 229960002887 deanol Drugs 0.000 description 1
- 125000004855 decalinyl group Chemical group C1(CCCC2CCCCC12)* 0.000 description 1
- 230000007423 decrease Effects 0.000 description 1
- 230000007812 deficiency Effects 0.000 description 1
- 230000003111 delayed effect Effects 0.000 description 1
- YSMODUONRAFBET-UHFFFAOYSA-N delta-DL-hydroxylysine Natural products NCC(O)CCC(N)C(O)=O YSMODUONRAFBET-UHFFFAOYSA-N 0.000 description 1
- 239000005547 deoxyribonucleotide Substances 0.000 description 1
- 125000002637 deoxyribonucleotide group Chemical group 0.000 description 1
- 238000001514 detection method Methods 0.000 description 1
- 239000003599 detergent Substances 0.000 description 1
- 229910052805 deuterium Inorganic materials 0.000 description 1
- 150000001985 dialkylglycerols Chemical class 0.000 description 1
- YRTMEEURRDTMST-UHFFFAOYSA-N diazetidine Chemical compound C1CNN1 YRTMEEURRDTMST-UHFFFAOYSA-N 0.000 description 1
- UMGXUWVIJIQANV-UHFFFAOYSA-M didecyl(dimethyl)azanium;bromide Chemical compound [Br-].CCCCCCCCCC[N+](C)(C)CCCCCCCCCC UMGXUWVIJIQANV-UHFFFAOYSA-M 0.000 description 1
- ZBCBWPMODOFKDW-UHFFFAOYSA-N diethanolamine Chemical compound OCCNCCO ZBCBWPMODOFKDW-UHFFFAOYSA-N 0.000 description 1
- 230000029087 digestion Effects 0.000 description 1
- 239000000539 dimer Substances 0.000 description 1
- PSLWZOIUBRXAQW-UHFFFAOYSA-M dimethyl(dioctadecyl)azanium;bromide Chemical compound [Br-].CCCCCCCCCCCCCCCCCC[N+](C)(C)CCCCCCCCCCCCCCCCCC PSLWZOIUBRXAQW-UHFFFAOYSA-M 0.000 description 1
- UAKOZKUVZRMOFN-JDVCJPALSA-M dimethyl-bis[(z)-octadec-9-enyl]azanium;chloride Chemical compound [Cl-].CCCCCCCC\C=C/CCCCCCCC[N+](C)(C)CCCCCCCC\C=C/CCCCCCCC UAKOZKUVZRMOFN-JDVCJPALSA-M 0.000 description 1
- 231100000676 disease causative agent Toxicity 0.000 description 1
- 239000002270 dispersing agent Substances 0.000 description 1
- 235000020669 docosahexaenoic acid Nutrition 0.000 description 1
- 229940090949 docosahexaenoic acid Drugs 0.000 description 1
- MOTZDAYCYVMXPC-UHFFFAOYSA-N dodecyl hydrogen sulfate Chemical compound CCCCCCCCCCCCOS(O)(=O)=O MOTZDAYCYVMXPC-UHFFFAOYSA-N 0.000 description 1
- 239000010459 dolomite Substances 0.000 description 1
- 229910000514 dolomite Inorganic materials 0.000 description 1
- 230000002222 downregulating effect Effects 0.000 description 1
- 230000003828 downregulation Effects 0.000 description 1
- 238000009510 drug design Methods 0.000 description 1
- 239000000975 dye Substances 0.000 description 1
- JAZBEHYOTPTENJ-UHFFFAOYSA-N eicosapentaenoic acid Natural products CCC=CCC=CCC=CCC=CCC=CCCCC(O)=O JAZBEHYOTPTENJ-UHFFFAOYSA-N 0.000 description 1
- 235000020673 eicosapentaenoic acid Nutrition 0.000 description 1
- 229960005135 eicosapentaenoic acid Drugs 0.000 description 1
- 239000003995 emulsifying agent Substances 0.000 description 1
- 210000001163 endosome Anatomy 0.000 description 1
- 230000007515 enzymatic degradation Effects 0.000 description 1
- DPUOLQHDNGRHBS-KTKRTIGZSA-N erucic acid Chemical compound CCCCCCCC\C=C/CCCCCCCCCCCC(O)=O DPUOLQHDNGRHBS-KTKRTIGZSA-N 0.000 description 1
- YSMODUONRAFBET-UHNVWZDZSA-N erythro-5-hydroxy-L-lysine Chemical compound NC[C@H](O)CC[C@H](N)C(O)=O YSMODUONRAFBET-UHNVWZDZSA-N 0.000 description 1
- 210000003743 erythrocyte Anatomy 0.000 description 1
- 229940105423 erythropoietin Drugs 0.000 description 1
- AFAXGSQYZLGZPG-UHFFFAOYSA-N ethanedisulfonic acid Chemical compound OS(=O)(=O)CCS(O)(=O)=O AFAXGSQYZLGZPG-UHFFFAOYSA-N 0.000 description 1
- CCIVGXIOQKPBKL-UHFFFAOYSA-M ethanesulfonate Chemical compound CCS([O-])(=O)=O CCIVGXIOQKPBKL-UHFFFAOYSA-M 0.000 description 1
- 125000005678 ethenylene group Chemical group [H]C([*:1])=C([H])[*:2] 0.000 description 1
- 125000005469 ethylenyl group Chemical group 0.000 description 1
- 125000002534 ethynyl group Chemical group [H]C#C* 0.000 description 1
- 210000003527 eukaryotic cell Anatomy 0.000 description 1
- 239000013604 expression vector Substances 0.000 description 1
- 238000000605 extraction Methods 0.000 description 1
- 239000003925 fat Substances 0.000 description 1
- 235000019197 fats Nutrition 0.000 description 1
- 150000002190 fatty acyls Chemical group 0.000 description 1
- 230000002349 favourable effect Effects 0.000 description 1
- 239000011737 fluorine Substances 0.000 description 1
- 235000019264 food flavour enhancer Nutrition 0.000 description 1
- 235000003599 food sweetener Nutrition 0.000 description 1
- 235000019253 formic acid Nutrition 0.000 description 1
- 125000002485 formyl group Chemical group [H]C(*)=O 0.000 description 1
- 239000001530 fumaric acid Substances 0.000 description 1
- 125000002541 furyl group Chemical group 0.000 description 1
- 229960003692 gamma aminobutyric acid Drugs 0.000 description 1
- 150000002270 gangliosides Chemical class 0.000 description 1
- 238000003197 gene knockdown Methods 0.000 description 1
- 238000007429 general method Methods 0.000 description 1
- 229960005219 gentisic acid Drugs 0.000 description 1
- 239000000174 gluconic acid Substances 0.000 description 1
- 235000012208 gluconic acid Nutrition 0.000 description 1
- 229960002442 glucosamine Drugs 0.000 description 1
- 229940097043 glucuronic acid Drugs 0.000 description 1
- 235000013922 glutamic acid Nutrition 0.000 description 1
- 239000004220 glutamic acid Substances 0.000 description 1
- JEJLGIQLPYYGEE-UHFFFAOYSA-N glycerol dipalmitate Natural products CCCCCCCCCCCCCCCC(=O)OCC(CO)OC(=O)CCCCCCCCCCCCCCC JEJLGIQLPYYGEE-UHFFFAOYSA-N 0.000 description 1
- 150000002327 glycerophospholipids Chemical class 0.000 description 1
- 125000003738 guanin-9-yl group Chemical group O=C1N([H])C(N([H])[H])=NC2=C1N=C([H])N2[*] 0.000 description 1
- 229940029575 guanosine Drugs 0.000 description 1
- 229940093915 gynecological organic acid Drugs 0.000 description 1
- 210000002216 heart Anatomy 0.000 description 1
- 125000004415 heterocyclylalkyl group Chemical group 0.000 description 1
- 125000005980 hexynyl group Chemical group 0.000 description 1
- HNDVDQJCIGZPNO-UHFFFAOYSA-N histidine Natural products OC(=O)C(N)CC1=CN=CN1 HNDVDQJCIGZPNO-UHFFFAOYSA-N 0.000 description 1
- 229960002885 histidine Drugs 0.000 description 1
- 238000009396 hybridization Methods 0.000 description 1
- XGIHQYAWBCFNPY-AZOCGYLKSA-N hydrabamine Chemical compound C([C@@H]12)CC3=CC(C(C)C)=CC=C3[C@@]2(C)CCC[C@@]1(C)CNCCNC[C@@]1(C)[C@@H]2CCC3=CC(C(C)C)=CC=C3[C@@]2(C)CCC1 XGIHQYAWBCFNPY-AZOCGYLKSA-N 0.000 description 1
- BHEPBYXIRTUNPN-UHFFFAOYSA-N hydridophosphorus(.) (triplet) Chemical compound [PH] BHEPBYXIRTUNPN-UHFFFAOYSA-N 0.000 description 1
- 229930195733 hydrocarbon Natural products 0.000 description 1
- 150000002430 hydrocarbons Chemical group 0.000 description 1
- 230000007062 hydrolysis Effects 0.000 description 1
- 238000006460 hydrolysis reaction Methods 0.000 description 1
- QJHBJHUKURJDLG-UHFFFAOYSA-N hydroxy-L-lysine Natural products NCCCCC(NO)C(O)=O QJHBJHUKURJDLG-UHFFFAOYSA-N 0.000 description 1
- 229960002591 hydroxyproline Drugs 0.000 description 1
- 238000003384 imaging method Methods 0.000 description 1
- 230000005934 immune activation Effects 0.000 description 1
- 239000012535 impurity Substances 0.000 description 1
- 238000000099 in vitro assay Methods 0.000 description 1
- 238000005462 in vivo assay Methods 0.000 description 1
- 125000003392 indanyl group Chemical group C1(CCC2=CC=CC=C12)* 0.000 description 1
- 125000001041 indolyl group Chemical group 0.000 description 1
- 230000006698 induction Effects 0.000 description 1
- 208000015181 infectious disease Diseases 0.000 description 1
- 230000004054 inflammatory process Effects 0.000 description 1
- 239000003999 initiator Substances 0.000 description 1
- 230000015788 innate immune response Effects 0.000 description 1
- 229960003786 inosine Drugs 0.000 description 1
- 230000003993 interaction Effects 0.000 description 1
- 238000001361 intraarterial administration Methods 0.000 description 1
- 238000007918 intramuscular administration Methods 0.000 description 1
- 238000007912 intraperitoneal administration Methods 0.000 description 1
- PNDPGZBMCMUPRI-UHFFFAOYSA-N iodine Chemical compound II PNDPGZBMCMUPRI-UHFFFAOYSA-N 0.000 description 1
- 239000003456 ion exchange resin Substances 0.000 description 1
- 229920003303 ion-exchange polymer Polymers 0.000 description 1
- 229910052742 iron Inorganic materials 0.000 description 1
- 125000005468 isobutylenyl group Chemical group 0.000 description 1
- RGXCTRIQQODGIZ-UHFFFAOYSA-O isodesmosine Chemical compound OC(=O)C(N)CCCC[N+]1=CC(CCC(N)C(O)=O)=CC(CCC(N)C(O)=O)=C1CCCC(N)C(O)=O RGXCTRIQQODGIZ-UHFFFAOYSA-O 0.000 description 1
- 125000002183 isoquinolinyl group Chemical group C1(=NC=CC2=CC=CC=C12)* 0.000 description 1
- 239000007951 isotonicity adjuster Substances 0.000 description 1
- 125000000842 isoxazolyl group Chemical group 0.000 description 1
- SBUJHOSQTJFQJX-NOAMYHISSA-N kanamycin Chemical compound O[C@@H]1[C@@H](O)[C@H](O)[C@@H](CN)O[C@@H]1O[C@H]1[C@H](O)[C@@H](O[C@@H]2[C@@H]([C@@H](N)[C@H](O)[C@@H](CO)O2)O)[C@H](N)C[C@@H]1N SBUJHOSQTJFQJX-NOAMYHISSA-N 0.000 description 1
- 229930027917 kanamycin Natural products 0.000 description 1
- 229960000318 kanamycin Drugs 0.000 description 1
- 229930182823 kanamycin A Natural products 0.000 description 1
- 210000003734 kidney Anatomy 0.000 description 1
- 239000004310 lactic acid Substances 0.000 description 1
- 235000014655 lactic acid Nutrition 0.000 description 1
- 229940099563 lactobionic acid Drugs 0.000 description 1
- 238000011031 large-scale manufacturing process Methods 0.000 description 1
- 125000000400 lauroyl group Chemical group O=C([*])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H] 0.000 description 1
- 210000000265 leukocyte Anatomy 0.000 description 1
- 229960004488 linolenic acid Drugs 0.000 description 1
- 238000009630 liquid culture Methods 0.000 description 1
- 229910052744 lithium Inorganic materials 0.000 description 1
- 229960003646 lysine Drugs 0.000 description 1
- VLBPIWYTPAXCFJ-XMMPIXPASA-N lysophosphatidylcholine O-16:0/0:0 Chemical compound CCCCCCCCCCCCCCCCOC[C@@H](O)COP([O-])(=O)OCC[N+](C)(C)C VLBPIWYTPAXCFJ-XMMPIXPASA-N 0.000 description 1
- VWHRYODZTDMVSS-QMMMGPOBSA-N m-fluoro-L-phenylalanine Chemical compound OC(=O)[C@@H](N)CC1=CC=CC(F)=C1 VWHRYODZTDMVSS-QMMMGPOBSA-N 0.000 description 1
- 239000011777 magnesium Substances 0.000 description 1
- 229910052749 magnesium Inorganic materials 0.000 description 1
- 159000000003 magnesium salts Chemical class 0.000 description 1
- VZCYOOQTPOCHFL-UPHRSURJSA-N maleic acid Chemical compound OC(=O)\C=C/C(O)=O VZCYOOQTPOCHFL-UPHRSURJSA-N 0.000 description 1
- 239000011976 maleic acid Substances 0.000 description 1
- 239000001630 malic acid Substances 0.000 description 1
- 235000011090 malic acid Nutrition 0.000 description 1
- 229960002510 mandelic acid Drugs 0.000 description 1
- 239000012528 membrane Substances 0.000 description 1
- 229940098779 methanesulfonic acid Drugs 0.000 description 1
- 229930182817 methionine Natural products 0.000 description 1
- WBYWAXJHAXSJNI-UHFFFAOYSA-N methyl p-hydroxycinnamate Natural products OC(=O)C=CC1=CC=CC=C1 WBYWAXJHAXSJNI-UHFFFAOYSA-N 0.000 description 1
- 230000011987 methylation Effects 0.000 description 1
- 125000004092 methylthiomethyl group Chemical group [H]C([H])([H])SC([H])([H])* 0.000 description 1
- 108091051828 miR-122 stem-loop Proteins 0.000 description 1
- 230000003278 mimic effect Effects 0.000 description 1
- 150000007522 mineralic acids Chemical class 0.000 description 1
- 230000000116 mitigating effect Effects 0.000 description 1
- 238000009126 molecular therapy Methods 0.000 description 1
- 235000021281 monounsaturated fatty acids Nutrition 0.000 description 1
- 238000010172 mouse model Methods 0.000 description 1
- XVUQPECVOGMPRU-ZPPAUJSGSA-N n,n-dimethyl-1,2-bis[(9z,12z)-octadeca-9,12-dienoxy]propan-1-amine Chemical compound CCCCC\C=C/C\C=C/CCCCCCCCOC(C)C(N(C)C)OCCCCCCCC\C=C/C\C=C/CCCCC XVUQPECVOGMPRU-ZPPAUJSGSA-N 0.000 description 1
- OZBZDYGIYDRTBV-RSLAUBRISA-N n,n-dimethyl-1,2-bis[(9z,12z,15z)-octadeca-9,12,15-trienoxy]propan-1-amine Chemical compound CC\C=C/C\C=C/C\C=C/CCCCCCCCOC(C)C(N(C)C)OCCCCCCCC\C=C/C\C=C/C\C=C/CC OZBZDYGIYDRTBV-RSLAUBRISA-N 0.000 description 1
- NFQBIAXADRDUGK-KWXKLSQISA-N n,n-dimethyl-2,3-bis[(9z,12z)-octadeca-9,12-dienoxy]propan-1-amine Chemical compound CCCCC\C=C/C\C=C/CCCCCCCCOCC(CN(C)C)OCCCCCCCC\C=C/C\C=C/CCCCC NFQBIAXADRDUGK-KWXKLSQISA-N 0.000 description 1
- UKXOXMLXFQEEQJ-KWXKLSQISA-N n,n-dimethyl-2,3-bis[[(9z,12z)-octadeca-9,12-dienyl]sulfanyl]propan-1-amine Chemical compound CCCCC\C=C/C\C=C/CCCCCCCCSCC(CN(C)C)SCCCCCCCC\C=C/C\C=C/CCCCC UKXOXMLXFQEEQJ-KWXKLSQISA-N 0.000 description 1
- DDBRXOJCLVGHLX-UHFFFAOYSA-N n,n-dimethylmethanamine;propane Chemical class CCC.CN(C)C DDBRXOJCLVGHLX-UHFFFAOYSA-N 0.000 description 1
- ZUHZZVMEUAUWHY-UHFFFAOYSA-N n,n-dimethylpropan-1-amine Chemical class CCCN(C)C ZUHZZVMEUAUWHY-UHFFFAOYSA-N 0.000 description 1
- 125000003136 n-heptyl group Chemical group [H]C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])* 0.000 description 1
- BDERNNFJNOPAEC-UHFFFAOYSA-N n-propyl alcohol Natural products CCCO BDERNNFJNOPAEC-UHFFFAOYSA-N 0.000 description 1
- KVBGVZZKJNLNJU-UHFFFAOYSA-N naphthalene-2-sulfonic acid Chemical compound C1=CC=CC2=CC(S(=O)(=O)O)=CC=C21 KVBGVZZKJNLNJU-UHFFFAOYSA-N 0.000 description 1
- 125000001624 naphthyl group Chemical group 0.000 description 1
- 229930014626 natural product Natural products 0.000 description 1
- GVUGOAYIVIDWIO-UFWWTJHBSA-N nepidermin Chemical compound C([C@@H](C(=O)N[C@@H]([C@@H](C)CC)C(=O)NCC(=O)N[C@@H](CCC(O)=O)C(=O)N[C@@H](CCCNC(N)=N)C(=O)N[C@@H](CS)C(=O)N[C@@H](CCC(N)=O)C(=O)N[C@@H](CC=1C=CC(O)=CC=1)C(=O)N[C@@H](CCCNC(N)=N)C(=O)N[C@@H](CC(O)=O)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](CCCCN)C(=O)N[C@@H](CC=1C2=CC=CC=C2NC=1)C(=O)N[C@@H](CC=1C2=CC=CC=C2NC=1)C(=O)N[C@@H](CCC(O)=O)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](CCCNC(N)=N)C(O)=O)NC(=O)CNC(=O)[C@@H](NC(=O)[C@@H](NC(=O)[C@H](CS)NC(=O)[C@H](CC(N)=O)NC(=O)[C@H](CS)NC(=O)[C@H](C)NC(=O)[C@H](CC=1C=CC(O)=CC=1)NC(=O)[C@H](CCCCN)NC(=O)[C@H](CC(O)=O)NC(=O)[C@H](CC(C)C)NC(=O)[C@H](C)NC(=O)[C@H](CCC(O)=O)NC(=O)[C@@H](NC(=O)[C@H](CC=1C=CC(O)=CC=1)NC(=O)[C@H](CCSC)NC(=O)[C@H](CS)NC(=O)[C@@H](NC(=O)CNC(=O)[C@H](CC(O)=O)NC(=O)[C@H](CC=1NC=NC=1)NC(=O)[C@H](CC(C)C)NC(=O)[C@H](CS)NC(=O)[C@H](CC=1C=CC(O)=CC=1)NC(=O)CNC(=O)[C@H](CC(O)=O)NC(=O)[C@H](CC=1NC=NC=1)NC(=O)[C@H](CO)NC(=O)[C@H](CC(C)C)NC(=O)[C@H]1N(CCC1)C(=O)[C@H](CS)NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](CO)NC(=O)[C@H](CC(O)=O)NC(=O)[C@H](CO)NC(=O)[C@@H](N)CC(N)=O)C(C)C)[C@@H](C)CC)C(C)C)C(C)C)C1=CC=C(O)C=C1 GVUGOAYIVIDWIO-UFWWTJHBSA-N 0.000 description 1
- 229960003512 nicotinic acid Drugs 0.000 description 1
- 235000001968 nicotinic acid Nutrition 0.000 description 1
- 239000011664 nicotinic acid Substances 0.000 description 1
- 229910017604 nitric acid Inorganic materials 0.000 description 1
- 125000006574 non-aromatic ring group Chemical group 0.000 description 1
- 108091027963 non-coding RNA Proteins 0.000 description 1
- 102000042567 non-coding RNA Human genes 0.000 description 1
- 230000009871 nonspecific binding Effects 0.000 description 1
- 231100000252 nontoxic Toxicity 0.000 description 1
- 230000003000 nontoxic effect Effects 0.000 description 1
- 125000002868 norbornyl group Chemical group C12(CCC(CC1)C2)* 0.000 description 1
- 210000004940 nucleus Anatomy 0.000 description 1
- 229960002446 octanoic acid Drugs 0.000 description 1
- 239000003921 oil Substances 0.000 description 1
- 229960002969 oleic acid Drugs 0.000 description 1
- 238000005457 optimization Methods 0.000 description 1
- 150000007524 organic acids Chemical class 0.000 description 1
- 235000005985 organic acids Nutrition 0.000 description 1
- 150000002894 organic compounds Chemical class 0.000 description 1
- 125000000962 organic group Chemical group 0.000 description 1
- 229960005010 orotic acid Drugs 0.000 description 1
- 235000006408 oxalic acid Nutrition 0.000 description 1
- 229940116315 oxalic acid Drugs 0.000 description 1
- 125000002971 oxazolyl group Chemical group 0.000 description 1
- AHHWIHXENZJRFG-UHFFFAOYSA-N oxetane Chemical compound C1COC1 AHHWIHXENZJRFG-UHFFFAOYSA-N 0.000 description 1
- 230000003647 oxidation Effects 0.000 description 1
- 238000007254 oxidation reaction Methods 0.000 description 1
- 125000004430 oxygen atom Chemical group O* 0.000 description 1
- 230000020477 pH reduction Effects 0.000 description 1
- 229940098695 palmitic acid Drugs 0.000 description 1
- 125000001312 palmitoyl group Chemical group O=C([*])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H] 0.000 description 1
- WLJNZVDCPSBLRP-UHFFFAOYSA-N pamoic acid Chemical compound C1=CC=C2C(CC=3C4=CC=CC=C4C=C(C=3O)C(=O)O)=C(O)C(C(O)=O)=CC2=C1 WLJNZVDCPSBLRP-UHFFFAOYSA-N 0.000 description 1
- 210000000496 pancreas Anatomy 0.000 description 1
- FJKROLUGYXJWQN-UHFFFAOYSA-N papa-hydroxy-benzoic acid Natural products OC(=O)C1=CC=C(O)C=C1 FJKROLUGYXJWQN-UHFFFAOYSA-N 0.000 description 1
- 244000052769 pathogen Species 0.000 description 1
- 230000006320 pegylation Effects 0.000 description 1
- 229960001639 penicillamine Drugs 0.000 description 1
- 229940049954 penicillin Drugs 0.000 description 1
- 235000019371 penicillin G benzathine Nutrition 0.000 description 1
- 125000001147 pentyl group Chemical group C(CCCC)* 0.000 description 1
- 125000005981 pentynyl group Chemical group 0.000 description 1
- 238000011170 pharmaceutical development Methods 0.000 description 1
- NBIIXXVUZAFLBC-UHFFFAOYSA-K phosphate Chemical compound [O-]P([O-])([O-])=O NBIIXXVUZAFLBC-UHFFFAOYSA-K 0.000 description 1
- 239000010452 phosphate Substances 0.000 description 1
- 235000021317 phosphate Nutrition 0.000 description 1
- 229940067605 phosphatidylethanolamines Drugs 0.000 description 1
- 229940067626 phosphatidylinositols Drugs 0.000 description 1
- 150000003905 phosphatidylinositols Chemical class 0.000 description 1
- 150000008298 phosphoramidates Chemical class 0.000 description 1
- 229910052698 phosphorus Inorganic materials 0.000 description 1
- 125000000587 piperidin-1-yl group Chemical group [H]C1([H])N(*)C([H])([H])C([H])([H])C([H])([H])C1([H])[H] 0.000 description 1
- 102000028499 poly(A) binding Human genes 0.000 description 1
- 108091023021 poly(A) binding Proteins 0.000 description 1
- 229920000724 poly(L-arginine) polymer Polymers 0.000 description 1
- 229920001606 poly(lactic acid-co-glycolic acid) Polymers 0.000 description 1
- 229920000768 polyamine Polymers 0.000 description 1
- 108010011110 polyarginine Proteins 0.000 description 1
- 229920000656 polylysine Polymers 0.000 description 1
- 102000054765 polymorphisms of proteins Human genes 0.000 description 1
- 108010055896 polyornithine Proteins 0.000 description 1
- 229920002714 polyornithine Polymers 0.000 description 1
- 235000020777 polyunsaturated fatty acids Nutrition 0.000 description 1
- 230000001124 posttranscriptional effect Effects 0.000 description 1
- WSHYKIAQCMIPTB-UHFFFAOYSA-M potassium;2-oxo-3-(3-oxo-1-phenylbutyl)chromen-4-olate Chemical compound [K+].[O-]C=1C2=CC=CC=C2OC(=O)C=1C(CC(=O)C)C1=CC=CC=C1 WSHYKIAQCMIPTB-UHFFFAOYSA-M 0.000 description 1
- OXCMYAYHXIHQOA-UHFFFAOYSA-N potassium;[2-butyl-5-chloro-3-[[4-[2-(1,2,4-triaza-3-azanidacyclopenta-1,4-dien-5-yl)phenyl]phenyl]methyl]imidazol-4-yl]methanol Chemical compound [K+].CCCCC1=NC(Cl)=C(CO)N1CC1=CC=C(C=2C(=CC=CC=2)C2=N[N-]N=N2)C=C1 OXCMYAYHXIHQOA-UHFFFAOYSA-N 0.000 description 1
- 238000001556 precipitation Methods 0.000 description 1
- 239000003755 preservative agent Substances 0.000 description 1
- 230000002335 preservative effect Effects 0.000 description 1
- 150000003141 primary amines Chemical class 0.000 description 1
- MFDFERRIHVXMIY-UHFFFAOYSA-N procaine Chemical compound CCN(CC)CCOC(=O)C1=CC=C(N)C=C1 MFDFERRIHVXMIY-UHFFFAOYSA-N 0.000 description 1
- 229960004919 procaine Drugs 0.000 description 1
- 238000012545 processing Methods 0.000 description 1
- 125000001325 propanoyl group Chemical group O=C([*])C([H])([H])C([H])([H])[H] 0.000 description 1
- 125000006410 propenylene group Chemical group 0.000 description 1
- 235000019260 propionic acid Nutrition 0.000 description 1
- 125000005470 propylenyl group Chemical group 0.000 description 1
- 125000006239 protecting group Chemical group 0.000 description 1
- PTJWIQPHWPFNBW-GBNDHIKLSA-N pseudouridine Chemical compound O[C@@H]1[C@H](O)[C@@H](CO)O[C@H]1C1=CNC(=O)NC1=O PTJWIQPHWPFNBW-GBNDHIKLSA-N 0.000 description 1
- 125000000561 purinyl group Chemical group N1=C(N=C2N=CNC2=C1)* 0.000 description 1
- 125000003373 pyrazinyl group Chemical group 0.000 description 1
- 125000003226 pyrazolyl group Chemical group 0.000 description 1
- 125000002098 pyridazinyl group Chemical group 0.000 description 1
- UMJSCPRVCHMLSP-UHFFFAOYSA-N pyridine Natural products COC1=CC=CN=C1 UMJSCPRVCHMLSP-UHFFFAOYSA-N 0.000 description 1
- 125000004076 pyridyl group Chemical group 0.000 description 1
- 125000000714 pyrimidinyl group Chemical group 0.000 description 1
- 125000000168 pyrrolyl group Chemical group 0.000 description 1
- 229940107700 pyruvic acid Drugs 0.000 description 1
- IUVKMZGDUIUOCP-BTNSXGMBSA-N quinbolone Chemical compound O([C@H]1CC[C@H]2[C@H]3[C@@H]([C@]4(C=CC(=O)C=C4CC3)C)CC[C@@]21C)C1=CCCC1 IUVKMZGDUIUOCP-BTNSXGMBSA-N 0.000 description 1
- 125000002943 quinolinyl group Chemical group N1=C(C=CC2=CC=CC=C12)* 0.000 description 1
- 125000001567 quinoxalinyl group Chemical group N1=C(C=NC2=CC=CC=C12)* 0.000 description 1
- 239000002516 radical scavenger Substances 0.000 description 1
- 239000011541 reaction mixture Substances 0.000 description 1
- 102000005962 receptors Human genes 0.000 description 1
- 108020003175 receptors Proteins 0.000 description 1
- 230000009467 reduction Effects 0.000 description 1
- 238000009877 rendering Methods 0.000 description 1
- 239000011347 resin Substances 0.000 description 1
- 229920005989 resin Polymers 0.000 description 1
- 238000010839 reverse transcription Methods 0.000 description 1
- 238000012552 review Methods 0.000 description 1
- 239000002342 ribonucleoside Substances 0.000 description 1
- 125000002652 ribonucleotide group Chemical group 0.000 description 1
- 238000007363 ring formation reaction Methods 0.000 description 1
- 238000011808 rodent model Methods 0.000 description 1
- 229960004889 salicylic acid Drugs 0.000 description 1
- 235000003441 saturated fatty acids Nutrition 0.000 description 1
- 150000004671 saturated fatty acids Chemical class 0.000 description 1
- 229940116353 sebacic acid Drugs 0.000 description 1
- 150000003335 secondary amines Chemical class 0.000 description 1
- 125000003607 serino group Chemical group [H]N([H])[C@]([H])(C(=O)[*])C(O[H])([H])[H] 0.000 description 1
- 239000000377 silicon dioxide Substances 0.000 description 1
- 238000001542 size-exclusion chromatography Methods 0.000 description 1
- IUVFCFQZFCOKRC-IPKKNMRRSA-M sodium;[(2r)-2,3-bis[[(z)-octadec-9-enoyl]oxy]propyl] 2,3-dihydroxypropyl phosphate Chemical compound [Na+].CCCCCCCC\C=C/CCCCCCCC(=O)OC[C@H](COP([O-])(=O)OCC(O)CO)OC(=O)CCCCCCC\C=C/CCCCCCCC IUVFCFQZFCOKRC-IPKKNMRRSA-M 0.000 description 1
- 239000007790 solid phase Substances 0.000 description 1
- 239000002904 solvent Substances 0.000 description 1
- 238000003797 solvolysis reaction Methods 0.000 description 1
- 150000003408 sphingolipids Chemical class 0.000 description 1
- 239000003381 stabilizer Substances 0.000 description 1
- 229960004274 stearic acid Drugs 0.000 description 1
- 230000004936 stimulating effect Effects 0.000 description 1
- KDYFGRWQOYBRFD-UHFFFAOYSA-L succinate(2-) Chemical compound [O-]C(=O)CCC([O-])=O KDYFGRWQOYBRFD-UHFFFAOYSA-L 0.000 description 1
- 150000008163 sugars Chemical class 0.000 description 1
- 125000004434 sulfur atom Chemical group 0.000 description 1
- 239000004094 surface-active agent Substances 0.000 description 1
- 239000000375 suspending agent Substances 0.000 description 1
- 239000003765 sweetening agent Substances 0.000 description 1
- 208000011580 syndromic disease Diseases 0.000 description 1
- 230000002194 synthesizing effect Effects 0.000 description 1
- 230000009885 systemic effect Effects 0.000 description 1
- 235000002906 tartaric acid Nutrition 0.000 description 1
- 239000011975 tartaric acid Substances 0.000 description 1
- 229960001367 tartaric acid Drugs 0.000 description 1
- 238000001447 template-directed synthesis Methods 0.000 description 1
- NPDBDJFLKKQMCM-UHFFFAOYSA-N tert-butylglycine Chemical compound CC(C)(C)C(N)C(O)=O NPDBDJFLKKQMCM-UHFFFAOYSA-N 0.000 description 1
- 150000003512 tertiary amines Chemical class 0.000 description 1
- TUNFSRHWOTWDNC-HKGQFRNVSA-N tetradecanoic acid Chemical compound CCCCCCCCCCCCC[14C](O)=O TUNFSRHWOTWDNC-HKGQFRNVSA-N 0.000 description 1
- YLQBMQCUIZJEEH-UHFFFAOYSA-N tetrahydrofuran Natural products C=1C=COC=1 YLQBMQCUIZJEEH-UHFFFAOYSA-N 0.000 description 1
- 125000001712 tetrahydronaphthyl group Chemical group C1(CCCC2=CC=CC=C12)* 0.000 description 1
- 125000000147 tetrahydroquinolinyl group Chemical group N1(CCCC2=CC=CC=C12)* 0.000 description 1
- 229960004559 theobromine Drugs 0.000 description 1
- 125000001544 thienyl group Chemical group 0.000 description 1
- XSROQCDVUIHRSI-UHFFFAOYSA-N thietane Chemical compound C1CSC1 XSROQCDVUIHRSI-UHFFFAOYSA-N 0.000 description 1
- 150000003573 thiols Chemical class 0.000 description 1
- YSMODUONRAFBET-WHFBIAKZSA-N threo-5-hydroxy-L-lysine Chemical compound NC[C@@H](O)CC[C@H](N)C(O)=O YSMODUONRAFBET-WHFBIAKZSA-N 0.000 description 1
- 229940113082 thymine Drugs 0.000 description 1
- 230000000699 topical effect Effects 0.000 description 1
- 231100000419 toxicity Toxicity 0.000 description 1
- 230000001988 toxicity Effects 0.000 description 1
- 230000005026 transcription initiation Effects 0.000 description 1
- 230000002103 transcriptional effect Effects 0.000 description 1
- 230000010474 transient expression Effects 0.000 description 1
- 230000009752 translational inhibition Effects 0.000 description 1
- 230000014621 translational initiation Effects 0.000 description 1
- 125000001425 triazolyl group Chemical group 0.000 description 1
- 125000005457 triglyceride group Chemical group 0.000 description 1
- 235000011178 triphosphate Nutrition 0.000 description 1
- YFTHZRPMJXBUME-UHFFFAOYSA-N tripropylamine Chemical compound CCCN(CCC)CCC YFTHZRPMJXBUME-UHFFFAOYSA-N 0.000 description 1
- LENZDBCJOHFCAS-UHFFFAOYSA-N tris Chemical compound OCC(N)(CO)CO LENZDBCJOHFCAS-UHFFFAOYSA-N 0.000 description 1
- DCXXMTOCNZCJGO-UHFFFAOYSA-N tristearoylglycerol Chemical compound CCCCCCCCCCCCCCCCCC(=O)OCC(OC(=O)CCCCCCCCCCCCCCCCC)COC(=O)CCCCCCCCCCCCCCCCC DCXXMTOCNZCJGO-UHFFFAOYSA-N 0.000 description 1
- 229910052722 tritium Inorganic materials 0.000 description 1
- 229960004418 trolamine Drugs 0.000 description 1
- 229960000281 trometamol Drugs 0.000 description 1
- 229960002703 undecylenic acid Drugs 0.000 description 1
- 241001515965 unidentified phage Species 0.000 description 1
- 235000021122 unsaturated fatty acids Nutrition 0.000 description 1
- 150000004670 unsaturated fatty acids Chemical class 0.000 description 1
- 238000011144 upstream manufacturing Methods 0.000 description 1
- 229940035893 uracil Drugs 0.000 description 1
- DRTQHJPVMGBUCF-UHFFFAOYSA-N uracil arabinoside Natural products OC1C(O)C(CO)OC1N1C(=O)NC(=O)C=C1 DRTQHJPVMGBUCF-UHFFFAOYSA-N 0.000 description 1
- 125000000845 uracil-1-yl group Chemical group [*]N1C(=O)N([H])C(=O)C([H])=C1[H] 0.000 description 1
- 229940045145 uridine Drugs 0.000 description 1
- 229960005486 vaccine Drugs 0.000 description 1
- 125000000391 vinyl group Chemical group [H]C([*])=C([H])[H] 0.000 description 1
- 230000003612 virological effect Effects 0.000 description 1
- 239000001993 wax Substances 0.000 description 1
- 239000000080 wetting agent Substances 0.000 description 1
- 239000011701 zinc Substances 0.000 description 1
- 229910052725 zinc Inorganic materials 0.000 description 1
Images
Classifications
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K31/00—Medicinal preparations containing organic active ingredients
- A61K31/70—Carbohydrates; Sugars; Derivatives thereof
- A61K31/7088—Compounds having three or more nucleosides or nucleotides
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K31/00—Medicinal preparations containing organic active ingredients
- A61K31/70—Carbohydrates; Sugars; Derivatives thereof
- A61K31/7088—Compounds having three or more nucleosides or nucleotides
- A61K31/7105—Natural ribonucleic acids, i.e. containing only riboses attached to adenine, guanine, cytosine or uracil and having 3'-5' phosphodiester links
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K47/00—Medicinal preparations characterised by the non-active ingredients used, e.g. carriers or inert additives; Targeting or modifying agents chemically bound to the active ingredient
- A61K47/06—Organic compounds, e.g. natural or synthetic hydrocarbons, polyolefins, mineral oil, petrolatum or ozokerite
- A61K47/16—Organic compounds, e.g. natural or synthetic hydrocarbons, polyolefins, mineral oil, petrolatum or ozokerite containing nitrogen, e.g. nitro-, nitroso-, azo-compounds, nitriles, cyanates
- A61K47/18—Amines; Amides; Ureas; Quaternary ammonium compounds; Amino acids; Oligopeptides having up to five amino acids
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K47/00—Medicinal preparations characterised by the non-active ingredients used, e.g. carriers or inert additives; Targeting or modifying agents chemically bound to the active ingredient
- A61K47/06—Organic compounds, e.g. natural or synthetic hydrocarbons, polyolefins, mineral oil, petrolatum or ozokerite
- A61K47/28—Steroids, e.g. cholesterol, bile acids or glycyrrhetinic acid
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K47/00—Medicinal preparations characterised by the non-active ingredients used, e.g. carriers or inert additives; Targeting or modifying agents chemically bound to the active ingredient
- A61K47/30—Macromolecular organic or inorganic compounds, e.g. inorganic polyphosphates
- A61K47/34—Macromolecular compounds obtained otherwise than by reactions only involving carbon-to-carbon unsaturated bonds, e.g. polyesters, polyamino acids, polysiloxanes, polyphosphazines, copolymers of polyalkylene glycol or poloxamers
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K9/00—Medicinal preparations characterised by special physical form
- A61K9/0012—Galenical forms characterised by the site of application
- A61K9/0019—Injectable compositions; Intramuscular, intravenous, arterial, subcutaneous administration; Compositions to be administered through the skin in an invasive manner
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K9/00—Medicinal preparations characterised by special physical form
- A61K9/10—Dispersions; Emulsions
- A61K9/127—Synthetic bilayered vehicles, e.g. liposomes or liposomes with cholesterol as the only non-phosphatidyl surfactant
- A61K9/1271—Non-conventional liposomes, e.g. PEGylated liposomes or liposomes coated or grafted with polymers
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K9/00—Medicinal preparations characterised by special physical form
- A61K9/48—Preparations in capsules, e.g. of gelatin, of chocolate
- A61K9/50—Microcapsules having a gas, liquid or semi-solid filling; Solid microparticles or pellets surrounded by a distinct coating layer, e.g. coated microspheres, coated drug crystals
- A61K9/51—Nanocapsules; Nanoparticles
- A61K9/5107—Excipients; Inactive ingredients
- A61K9/5123—Organic compounds, e.g. fats, sugars
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K9/00—Medicinal preparations characterised by special physical form
- A61K9/48—Preparations in capsules, e.g. of gelatin, of chocolate
- A61K9/50—Microcapsules having a gas, liquid or semi-solid filling; Solid microparticles or pellets surrounded by a distinct coating layer, e.g. coated microspheres, coated drug crystals
- A61K9/51—Nanocapsules; Nanoparticles
- A61K9/5107—Excipients; Inactive ingredients
- A61K9/513—Organic macromolecular compounds; Dendrimers
- A61K9/5146—Organic macromolecular compounds; Dendrimers obtained otherwise than by reactions only involving carbon-to-carbon unsaturated bonds, e.g. polyethylene glycol, polyamines, polyanhydrides
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07C—ACYCLIC OR CARBOCYCLIC COMPOUNDS
- C07C235/00—Carboxylic acid amides, the carbon skeleton of the acid part being further substituted by oxygen atoms
- C07C235/02—Carboxylic acid amides, the carbon skeleton of the acid part being further substituted by oxygen atoms having carbon atoms of carboxamide groups bound to acyclic carbon atoms and singly-bound oxygen atoms bound to the same carbon skeleton
- C07C235/04—Carboxylic acid amides, the carbon skeleton of the acid part being further substituted by oxygen atoms having carbon atoms of carboxamide groups bound to acyclic carbon atoms and singly-bound oxygen atoms bound to the same carbon skeleton the carbon skeleton being acyclic and saturated
- C07C235/08—Carboxylic acid amides, the carbon skeleton of the acid part being further substituted by oxygen atoms having carbon atoms of carboxamide groups bound to acyclic carbon atoms and singly-bound oxygen atoms bound to the same carbon skeleton the carbon skeleton being acyclic and saturated having the nitrogen atom of at least one of the carboxamide groups bound to an acyclic carbon atom of a hydrocarbon radical substituted by singly-bound oxygen atoms
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07C—ACYCLIC OR CARBOCYCLIC COMPOUNDS
- C07C271/00—Derivatives of carbamic acids, i.e. compounds containing any of the groups, the nitrogen atom not being part of nitro or nitroso groups
- C07C271/06—Esters of carbamic acids
- C07C271/08—Esters of carbamic acids having oxygen atoms of carbamate groups bound to acyclic carbon atoms
- C07C271/10—Esters of carbamic acids having oxygen atoms of carbamate groups bound to acyclic carbon atoms with the nitrogen atoms of the carbamate groups bound to hydrogen atoms or to acyclic carbon atoms
- C07C271/12—Esters of carbamic acids having oxygen atoms of carbamate groups bound to acyclic carbon atoms with the nitrogen atoms of the carbamate groups bound to hydrogen atoms or to acyclic carbon atoms to hydrogen atoms or to carbon atoms of unsubstituted hydrocarbon radicals
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07C—ACYCLIC OR CARBOCYCLIC COMPOUNDS
- C07C275/00—Derivatives of urea, i.e. compounds containing any of the groups, the nitrogen atoms not being part of nitro or nitroso groups
- C07C275/04—Derivatives of urea, i.e. compounds containing any of the groups, the nitrogen atoms not being part of nitro or nitroso groups having nitrogen atoms of urea groups bound to acyclic carbon atoms
- C07C275/06—Derivatives of urea, i.e. compounds containing any of the groups, the nitrogen atoms not being part of nitro or nitroso groups having nitrogen atoms of urea groups bound to acyclic carbon atoms of an acyclic and saturated carbon skeleton
- C07C275/14—Derivatives of urea, i.e. compounds containing any of the groups, the nitrogen atoms not being part of nitro or nitroso groups having nitrogen atoms of urea groups bound to acyclic carbon atoms of an acyclic and saturated carbon skeleton being further substituted by nitrogen atoms not being part of nitro or nitroso groups
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07C—ACYCLIC OR CARBOCYCLIC COMPOUNDS
- C07C323/00—Thiols, sulfides, hydropolysulfides or polysulfides substituted by halogen, oxygen or nitrogen atoms, or by sulfur atoms not being part of thio groups
- C07C323/23—Thiols, sulfides, hydropolysulfides or polysulfides substituted by halogen, oxygen or nitrogen atoms, or by sulfur atoms not being part of thio groups containing thio groups and nitrogen atoms, not being part of nitro or nitroso groups, bound to the same carbon skeleton
- C07C323/39—Thiols, sulfides, hydropolysulfides or polysulfides substituted by halogen, oxygen or nitrogen atoms, or by sulfur atoms not being part of thio groups containing thio groups and nitrogen atoms, not being part of nitro or nitroso groups, bound to the same carbon skeleton at least one of the nitrogen atoms being part of any of the groups, X being a hetero atom, Y being any atom
- C07C323/40—Y being a hydrogen or a carbon atom
- C07C323/41—Y being a hydrogen or an acyclic carbon atom
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07C—ACYCLIC OR CARBOCYCLIC COMPOUNDS
- C07C333/00—Derivatives of thiocarbamic acids, i.e. compounds containing any of the groups, the nitrogen atom not being part of nitro or nitroso groups
- C07C333/02—Monothiocarbamic acids; Derivatives thereof
- C07C333/04—Monothiocarbamic acids; Derivatives thereof having nitrogen atoms of thiocarbamic groups bound to hydrogen atoms or to acyclic carbon atoms
Definitions
- Embodiments of the present invention generally relate to lipid nanoparticles (LNPs) having improved properties.
- LNPs are useful for facilitating the intracellular delivery of therapeutic agents, such as nucleic acids (e.g., oligonucleotides, messenger RNA), to primates, including humans.
- therapeutic agents such as nucleic acids (e.g., oligonucleotides, messenger RNA), to primates, including humans.
- nucleic acid based therapeutics have enormous potential but there remains a need for more effective delivery of nucleic acids to appropriate sites within a cell or organism in order to realize this potential.
- Therapeutic nucleic acids include, e.g., messenger RNA (mRNA), antisense oligonucleotides, ribozymes, DNAzymes, plasmids, immune stimulating nucleic acids, antagomir, antimir, mimic, supermir, and aptamers.
- nucleic acids such as mRNA or plasmids
- mRNA or plasmids can be used to effect expression of specific cellular products as would be useful in the treatment of, for example, diseases related to a deficiency of a protein or enzyme.
- the therapeutic applications of translatable nucleotide delivery are extremely broad as constructs can be synthesized to produce any chosen protein sequence, whether or not indigenous to the system.
- the expression products of the nucleic acid can augment existing levels of protein, replace missing or non-functional versions of a protein, or introduce new protein and associated functionality in a cell or organism.
- RNAs are susceptible to nuclease digestion in plasma.
- free RNAs have limited ability to gain access to the intracellular compartment where the relevant translation machinery resides.
- Lipid nanoparticles formed from cationic lipids with other lipid components, such as neutral lipids, cholesterol, PEG, PEGylated lipids, and oligonucleotides have been used to protect the RNAs in plasma and facilitate the cellular uptake of the oligonucleotides.
- WO2018081480A1 discloses lipid nanoparticle comprising 40-50 mol.% cationic lipid, neutral lipid, steroid, polymer conjugated lipid, and atherapeutic agent or its salt encapsulated within or associated with lipid nanoparticle.
- lipid nanoparticle formulations have shown tremendous promise for enhancing nucleic acid therapies in both in vitro and in vivo animal models, the performance in rodent models vastly exceeds that observed in non-human primate models in nearly every measure, including toxicity and tolerability, pharmacokinetics, tissue targeting and efficacy. Notably, achieving therapeutically relevant outcomes at tolerable dose levels in primate models remains a significant challenge. Thus, there remains a need for improved lipid nanoparticles for the delivery of oligonucleotides in primates such that an efficacious and reproducible therapeutic result can be realized. Embodiments of the present disclosure provide these and related advantages.
- LNPs lipid nanoparticles
- modified nucleosides into in vitro transcribed mRNA can be used to prevent recognition and activation of RNA sensors, thus mitigating this undesired immunostimulatory activity and enhancing translation capacity (see e.g . Kariko, K. And Weissman, D.
- nucleic acid refers to a polymer containing at least two deoxyribonucleotides or ribonucleotides in either single- or double-stranded form and includes DNA, RNA, and hybrids thereof.
- DNA may be in the form of antisense molecules, plasmid DNA, cDNA, PCR products, or vectors.
- RNA may be in the form of small hairpin RNA (shRNA), messenger RNA (mRNA), self amplifying RNA (saRNA), small activating RNA, antisense RNA, miRNA, micRNA, multivalent RNA, dicer substrate RNA or viral RNA (vRNA), and combinations thereof.
- Nucleotides contain a sugar deoxyribose (DNA) or ribose (RNA), a base, and a phosphate group. Nucleotides are linked together through the phosphate groups.
- Bases include purines and pyrimidines, which further include natural compounds adenine, thymine, guanine, cytosine, uracil, inosine, and natural analogs, and synthetic derivatives of purines and pyrimidines, which include, but are not limited to, modifications which place new reactive groups such as, but not limited to, amines, alcohols, thiols, carboxylates, and alkylhalides.
- neutral lipid refers to any of a number of lipid species that exist either in an uncharged or neutral zwitterionic form at a selected pH.
- lipids include, but are not limited to, phosphotidylcholines such as 1,2-Distearoyl- sn -glycero-3-phosphocholine (DSPC), 1,2-Dipalmitoyl- sn -glycero-3-phosphocholine (DPPC), 1,2-Dimyristoyl- sn -glycero-3-phosphocholine (DMPC), 1-Palmitoyl-2-oleoyl- sn -glycero-3-phosphocholine (POPC), 1,2-dioleoyl- sn -glycero-3-phosphocholine (DOPC), phophatidylethanolamines such as 1,2-Dioleoyl- sn -glycero-3-phosphoethanolamine (DOPE), s
- DOPE 1,2-D
- nucleic acids when present in the lipid nanoparticles, are resistant in aqueous solution to degradation with a nuclease.
- Lipids and their method of preparation are disclosed in, e.g., U.S. Patent Nos. 8,569,256 , 5,965,542 and U.S. Patent Publication Nos.
- lipid encapsulated refers to a lipid nanoparticle that provides an active agent or therapeutic agent, such as a nucleic acid (e.g., mRNA), with full encapsulation, partial encapsulation, or both.
- a nucleic acid e.g., mRNA
- the nucleic acid is fully encapsulated in the lipid nanoparticle.
- aqueous solution refers to a composition comprising water.
- “Serum-stable” in relation to nucleic acid-lipid nanoparticles means that the nucleotide is not significantly degraded after exposure to a serum or nuclease assay that would significantly degrade free DNA or RNA.
- Suitable assays include, for example, a standard serum assay, a DNAse assay, or an RNAse assay.
- Local delivery refers to delivery of an active agent directly to a target site within an organism.
- an agent can be locally delivered by direct injection into a disease site such as a tumor, other target site such as a site of inflammation, or a target organ such as the liver, heart, pancreas, kidney, and the like.
- Local delivery can also include topical applications or localized injection techniques such as intramuscular, subcutaneous or intradermal injection. Local delivery does not preclude a systemic pharmacological effect.
- amino acid refers to naturally-occurring and non-naturally occurring amino acids.
- An amino acid lipid can be made from a genetically encoded amino acid, a naturally occurring non-genetically encoded amino acid, or a synthetic amino acid.
- amino acids include Ala, Arg, Asn, Asp, Cys, Gln, Glu, Gly, His, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, and Val.
- amino acids also include azetidine, 2-aminooctadecanoic acid, 2-aminoadipic acid, 3-aminoadipic acid, 2,3-diaminopropionic acid, 2-aminobutyric acid, 4-aminobutyric acid, 2,3-diaminobutyric acid, 2,4-diaminobutyric acid, 2-aminoisobutyric acid, 4-aminoisobutyric acid, 2-aminopimelic acid, 2,2'-diaminopimelic acid, 6-aminohexanoic acid, 6-aminocaproic acid, 2-aminoheptanoic acid, desmosine, omithine, citrulline, N-methylisoleucine, norleucine, tert-leucine, phenylglycine, t-butylglycine, N-methylglycine, sacrosine, N-ethylglycine, cyclohexylglycine
- Alkyl refers to a straight or branched hydrocarbon chain radical consisting solely of carbon and hydrogen atoms, which is saturated or unsaturated (i.e., contains one or more double (alkenyl) and/or triple bonds (alkynyl)), having, for example, from one to twenty-four carbon atoms (C 1 -C 24 alkyl), four to twenty carbon atoms (C 4 -C 20 alkyl), six to sixteen carbon atoms (C 6 -C 16 alkyl), six to nine carbon atoms (C 6 -C 9 alkyl), one to fifteen carbon atoms (C 1 -C 15 alkyl),one to twelve carbon atoms (Ci-C 12 alkyl), one to eight carbon atoms (C 1 -C 8 alkyl) or one to six carbon atoms (C 1 -C 6 alkyl) and which is attached to the rest of the molecule by a single bond, e.g., methyl, ethyl, n
- Alkylene or "alkylene chain” refers to a straight or branched divalent hydrocarbon chain linking the rest of the molecule to a radical group, consisting solely of carbon and hydrogen, which is saturated or unsaturated (i.e., contains one or more double (alkenylene) and/or triple bonds (alkynylene)), and having, for example, from one to twenty-four carbon atoms (C 1 -C 24 alkylene), one to fifteen carbon atoms (C 1 -C 15 alkylene),one to twelve carbon atoms (C 1 -C 12 alkylene), one to eight carbon atoms (Ci-C 8 alkylene), one to six carbon atoms (C 1 -C 6 alkylene), two to four carbon atoms (C 2 -C 4 alkylene), one to two carbon atoms (C 1 -C 2 alkylene), e.g., methylene, ethylene, propylene, n-butylene, ethenylene, propenylene,
- the alkylene chain is attached to the rest of the molecule through a single or double bond and to the radical group through a single or double bond.
- the points of attachment of the alkylene chain to the rest of the molecule and to the radical group can be through one carbon or any two carbons within the chain. Unless stated otherwise specifically in the specification, an alkylene chain may be optionally substituted.
- alkenyl refers to an alkyl, as defined above, containing at least one double bond between adjacent carbon atoms. Alkenyls include both cis and trans isomers. Representative straight chain and branched alkenyls include, but are not limited to, ethylenyl, propylenyl, 1-butenyl, 2-butenyl, isobutylenyl, 1-pentenyl, 2-pentenyl, 3-methyl-1-butenyl, 2-methyl-2-butenyl, 2,3-dimethyl-2-butenyl, and the like.
- Alkoxy refers to an alkyl, cycloalkyl, alkenyl, or alkynyl group covalently bonded to an oxygen atom.
- alkynyl includes any alkyl or alkenyl, as defined above, which additionally contains at least one triple bond between adjacent carbons.
- Representative straight chain and branched alkynyls include, without limitation, acetylenyl, propynyl, 1-butynyl, 2-butynyl, 1-pentynyl, 2-pentynyl, 3-methyl-1 butynyl, and the like.
- Aryl refers to any stable monocyclic, bicyclic, or polycyclic carbon ring system of from 4 to 12 atoms in each ring, wherein at least one ring is aromatic.
- Some examples of an aryl include phenyl, naphthyl, tetrahydro-naphthyl, indanyl, and biphenyl. Where an aryl substituent is bicyclic and one ring is non-aromatic, it is understood that attachment is to the aromatic ring. An aryl may be substituted or unsubstituted.
- Cyano refers to a functional group of the formula -CN.
- Cycloalkyl or “carbocyclic ring” refers to a stable non-aromatic monocyclic or polycyclic hydrocarbon radical consisting solely of carbon and hydrogen atoms, which may include fused or bridged ring systems, having from three to fifteen carbon atoms, preferably having from three to ten carbon atoms, and which is saturated or unsaturated and attached to the rest of the molecule by a single bond.
- Monocyclic radicals include, for example, cyclopropyl, cyclobutyl, cyclopentyl, cyclohexyl, cycloheptyl, and cyclooctyl.
- Polycyclic radicals include, for example, adamantyl, norbornyl, decalinyl, 7,7-dimethyl-bicyclo[2.2.1]heptanyl, and the like. Unless otherwise stated specifically in the specification, a cycloalkyl group may be optionally substituted.
- Cycloalkylene is a divalent cycloalkyl group. Unless otherwise stated specifically in the specification, a cycloalkylene group may be optionally substituted.
- diacylglycerol or “DAG” includes a compound having 2 fatty acyl chains, both of which have independently between 2 and 30 carbons bonded to the 1- and 2-position of glycerol by ester linkages.
- the acyl groups can be saturated or have varying degrees of unsaturation. Suitable acyl groups include, but are not limited to, lauroyl (C12), myristoyl (C14), palmitoyl (C16), stearoyl (C18), and icosoyl (C20).
- the fatty acid acyl chains of one compound are the same, i.e., both myristoyl (i.e., dimyristoyl), both stearoyl (i.e., distearoyl), etc.
- alkylphosphate refers to ---O---P(Q')(Q")-O---R, wherein Q' and Q" are each independently O, S, N(R) 2 , optionally substituted alkyl or alkoxy; and R is optionally substituted alkyl, ⁇ -aminoalkyl or ⁇ -(substituted)aminoalkyl.
- “Pharmaceutically acceptable salt” includes both acid and base addition salts.
- Treating covers the treatment of the disease or condition of interest in a mammal, preferably a human, having the disease or condition of interest, and includes:
- Embodiments disclosed herein are directed to methods of using LNPs for delivery of a therapeutic agent, such as a nucleic acid, to a primate, such as a human, for treatment of various diseases treatable with the nucleic acid.
- a therapeutic agent such as a nucleic acid
- a primate such as a human
- the disclosed methods are surprisingly more effective for delivery of therapeutic agents to primates, compared with delivery of the same therapeutic agent to a non-primate, such as a mouse.
- some methods include use of LNPs having a diameter smaller than typical LNPs, for example a mean particle diameter ranging from about 40-70 nm, or for instance, a mean particle diameter ranging from about 50-70 nm, and such LNPs have unexpectedly improved delivery in primates relative to rodent.
- Another embodiment is directed to a method for delivering a nucleic acid to a primate in need thereof, comprising administering a lipid nanoparticle (LNP) to the primate, the LNP comprising:
- the polymer-conjugated lipid has the following structure: wherein:
- P comprises a polyethylene glycol polymer, such as a hydroxyl or alkoxyl-terminating polyethylene glycol polymer.
- the polymer conjugated lipid has the following structure: wherein n is an integer ranging from 40 to 50.
- Asymmetric polymer conjugated lipids wherein R' and R" are different are also included in various embodiments, such as wherein R' is 12 and R" is 13, or R' is 13 and R" is 14, or R' is 11 and R" is 12, or R' is 10 and R" is 11 and the like
- the cationic lipid is selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319).
- the lipid nanoparticle has a polydispersity value of less than 0.4.
- the lipid nanoparticle has a net neutral charge at a neutral pH.
- the lipid nanoparticle has a mean diameter of 40-200 nm..
- Lipid nanoparticles may comprise one or more lipid species, including, but not limited to, cationic/ionizable lipids, neutral lipids, structural lipids, phospholipids, and helper lipids. Any of these lipids may be conjugated to polyethylene glycol (PEG) and thus may be referred to as PEGylated lipids or PEG-modified lipids.
- PEG polyethylene glycol
- the lipid nanoparticle comprises a cationic lipid and a neutral lipid.
- the LNP comprises a cationic lipid and a DSPC substitute.
- the LNP comprises a cationic lipid and a fatty acid.
- the LNP a cationic lipid and oleic acid.
- the LNP comprises a cationic lipid and an analog of oleic acid.
- the lipid nanoparticle formulation comprises a cationic lipid, a neutral lipid, and a structural lipid.
- the LNP comprises a cationic lipid, a fatty acid, and a structural lipid.
- the LNP comprises a cationic lipid, oleic acid, and a structural lipid.
- the LNP comprises a cationic lipid, an analog of oleic acid, and a structural lipid.
- the LNP comprises a cationic lipid, a fatty acid, and a sterol.
- the LNP comprises a cationic lipid, oleic acid, and a sterol.
- the LNP comprises a cationic lipid, oleic acid, and cholesterol.
- the LNP comprises a cationic lipid, oleic acid, and cholesterol.
- the lipid nanoparticle comprises a cationic lipid, a neutral lipid, and a PEGylated lipid.
- the LNP formulation comprises a cationic lipid, a neutral lipid, and a PEG-OH lipid.
- the lipid nanoparticle comprises a cationic lipid, a fatty acid, and a PEG-OH lipid.
- the lipid nanoparticle comprises a cationic lipid, oleic acid, and a PEG-OH lipid.
- the lipid nanoparticle comprises a cationic lipid, an analog of oleic acid, and a PEG-OH lipid.
- the LNP comprises a cationic lipid, a fatty acid (e.g., oleic acid or an analog thereof), a structural lipid, and a PEG lipid.
- the LNP comprises a cationic lipid, a fatty acid (e.g., oleic acid or an analog thereof), a structural lipid, and a PEG-OH lipid.
- the LNP comprises a cationic lipid, oleic acid, a structural lipid (e.g., a sterol), and a PEG-OH lipid.
- the LNP comprises a cationic lipid, oleic acid, and a structural lipid (e.g., cholesterol). In certain embodiments, the LNP comprises one or more cationic or neutral lipids, a fatty acid (e.g., oleic acid), and a PEG lipid. In certain embodiments, the LNP comprises one or more cationic or neutral lipids, a fatty acid (e.g., oleic acid), and a PEG-OH lipid.
- the ratio of PEG in the LNPs may be increased or decreased and/or the carbon chain length of the alkyl portion of the PEG lipid may be varied from C8 to C18 (eight to eighteen carbons) to alter the pharmacokinetics and/or biodistribution of the LNPs.
- LNPs may contain 0.1% to 3.0%, 1.0% to 3.5%, 1.5% to 4.0%, 2.0% to 4.5%, 2.0% to 3.0%, 2.5% to 5.0%, and/or 3.0% to 6.0% of PEGylated lipid relative to the other components.
- LNPs may contain 0.5% to 3.0%, 1.0% to 3.5%, 1.5% to 4.0%, 2.0% to 4.5%, 2.0% to 3.0%, 2.5% to 5.0%, and/or 3.0% to 6.0% of PEG-c-DOMG (R-3-[( ⁇ -methoxy-poly(ethyleneglycol)2000)carbamoyl)]-1,2-dimyristyloxypropyl-3-amine) (also referred to herein as PEG-DOMG) as compared to the cationic lipid, DSPC, and cholesterol.
- PEG-c-DOMG R-3-[( ⁇ -methoxy-poly(ethyleneglycol)2000)carbamoyl)]-1,2-dimyristyloxypropyl-3-amine
- the PEG-c-DOMG may be replaced with a PEG lipid such as, but not limited to, PEG-DSG (1,2-distearoyl-sn-glycerol, methoxypolyethylene glycol), DMG-PEG (1,2-dimyristoyl-sn-glycerol) and/or PEG-DPG (1,2-dipalmitoyl-sn-glycerol, methoxypolyethylene glycol).
- the cationic lipid may be selected from any lipid known in the art such as, but not limited to, DLin-MC3-DMA, DLin-DMA, C12-200, and DLin-KC2-DMA.
- the lipid nanoparticle does not contain a PEG lipid. In certain embodiments, the lipid nanoparticle contains a PEG lipid such as a PEG-OH lipid. Incorporation of PEG-OH lipids in the nanoparticle formulation can improve the pharmacokinetics and/or biodistribution of the LNPs. For example, incorporation of PEG-OH lipids in the nanoparticle formulation can reduce the ABC effect.
- LNPs may contain 0.5% to 3.0%, 1.0% to 3.5%, 1.5% to 4.0%, 2.0% to 4.5%, 2.0% to 5.0%, 2.5% to 5.0%, and/or 3.0% to 6.0% of the lipid molar ratio of PEG-OH lipid to the other components (e.g., the cationic, neutral, and structural lipids).
- the other components e.g., the cationic, neutral, and structural lipids.
- LNPs include 35-65% of a cationic lipid selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319).
- DLin-KC2-DMA 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane
- DLin-MC3-DMA dilinoleyl-methyl-4-dimethylaminobutyrate
- L319 di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoy
- LNPs include 45-65% of a cationic lipid, 5-10% of the neutral lipid, 25-40% of the structural lipid, and 0.5-10% of the PEG or PEGylated lipid on a molar basis. In some embodiments, LNPs include 45-65% of a cationic lipid, 5-10% of the neutral lipid, 25-40% of the structural lipid, and 0.5-10% of a PEG-OH lipid on a molar basis. In some embodiments, LNPs include 45-65% of a cationic lipid, 5-10% of the neutral lipid, and 25-40% of the structural lipid on a molar basis.
- LNPs include 60% of a cationic lipid, 7.5% of the neutral lipid, 31% of a structural lipid, and 1.5% of the PEG or PEGylated lipid on a molar basis. In some embodiments, LNPs include 60% of a cationic lipid, 7.5% of the neutral lipid, 31% of a structural lipid, and 1.5% of a PEG-OH lipid on a molar basis. In some embodiments, LNPs include 60% of a cationic lipid, 9% of the neutral lipid, and 31% of a structural lipid on a molar basis.
- LNPs include 50% of a cationic lipid, 10% of the neutral lipid, 38.5% of the structural lipid, and 1.5% of the PEG or PEGylated lipid on a molar basis. In some embodiments, LNPs include 50% of a cationic lipid, 10% of the neutral lipid, 38.5% of a structural lipid, and 1.5% of a PEG-OH lipid on a molar basis. In some embodiments, LNPs include 50% of a cationic lipid, 10% of the neutral lipid, and 40% of a structural lipid on a molar basis.
- LNPs include 57.2% of a cationic lipid selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319).
- DLin-KC2-DMA 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane
- DLin-MC3-DMA dilinoleyl-methyl-4-dimethylaminobutyrate
- L319 di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butano
- LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% neutral lipid; 20-55% structural lipid; 0.1-15% PEGylated lipid. In some embodiments, LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% neutral lipid (e.g., phospholipid or fatty acid); 20-55% structural lipid; and 0.1-15% PEG-OH lipid.
- LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% oleic acid; 20-55% structural lipid (e.g., sterols); and 0.1-15% PEG-OH lipid. In some embodiments, LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% oleic acid; and 20-55% structural lipid (e.g., sterols).
- Non-limiting examples of lipid nanoparticle compositions and methods of making them are described, for example, in Semple et al. (2010) Nat. Biotechnol. 28:172-176 ; Jayarama et al. (2012), Angew. Chem. Int. Ed., 51: 8529-8533 ; and Maier et al. (2013) Molecular Therapy 21, 1570-1578 .
- LNPs may comprise a cationic lipid, a PEG lipid (e.g., PEG-OH lipid) and optionally comprise a neutral lipid (e.g., phospholipid or fatty acid).
- LNPs may comprise a cationic lipid, a PEG lipid (e.g., PEG-OH lipid) and a structural lipid (e.g., a sterol) and optionally comprise a neutral lipid (e.g., phospholipid or fatty acid).
- the LNPs described herein may be four component lipid nanoparticles.
- a 4 component LNP may comprise four different lipids selected from any described herein. The four components do not include the payload.
- the lipid nanoparticle may comprise a cationic lipid, a neutral lipid, a PEG lipid, and a structural lipid.
- the lipid nanoparticle comprises a cationic lipid, a fatty acid, a PEG lipid, and a structural lipid.
- the lipid nanoparticle comprises a cationic lipid, a fatty acid, a PEG-OH lipid, and a structural lipid.
- Each possibility represents a separate embodiment of the present invention.
- the LNPs described herein may be three component lipid nanoparticles.
- a three component LNP may comprise three different lipids described herein.
- the lipid nanoparticle may comprise a cationic lipid, a neutral lipid (e.g., phospholipid or fatty acid), and a structural lipid.
- the lipid nanoparticle comprises a cationic lipid, a fatty acid, and a structural lipid.
- the lipid nanoparticle comprises a cationic lipid, a phospholipid, and a structural lipid.
- the LNP formulation may be formulated by the methods described in International Publication Nos. WO2011127255 or WO2008103276 .
- LNP formulations as described in WO2011127255 and/or WO2008103276 are examples of LNP formulations as described in WO2011127255 and/or WO2008103276 .
- the lipid nanoparticle may be formulated by the methods described in US Patent Publication No US2013/0156845 or International Publication No WO2013/093648 or WO2012024526 .
- lipid nanoparticles described herein may be made in a sterile environment by the system and/or methods described in US Patent Publication No. US20130164400 .
- the LNP formulation may be formulated in a nanoparticle such as a nucleic acid-lipid nanoparticle described in U.S. Pat. No. 8,492,359 .
- the lipid nanoparticle may comprise one or more active agents or therapeutic agents (e.g., RNA); one or more cationic lipids comprising from about 50 mol % to about 85 mol % of the total lipid present in the particle; one or more neutral lipid lipids comprising from about 13 mol % to about 49.5 mol % of the total lipid present in the particle; and one or more structural lipids that inhibit aggregation of particles comprising from about 0.5 mol % to about 2 mol % of the total lipid present in the particle.
- active agents or therapeutic agents e.g., RNA
- one or more cationic lipids comprising from about 50 mol % to about 85 mol % of the total lipid present in the particle
- one or more neutral lipid lipids comprising from about 13 mol % to about 49.5 mol % of the total lipid present in the particle
- structural lipids that inhibit aggregation of particles comprising from about 0.5 mol % to about 2
- the LNP formulation may be formulated by the methods described in International Publication Nos. WO2011127255 or WO2008103276 .
- LNP formulations as described in WO2011 127255 and/or WO2008103276 may comprise a polycationic composition.
- the polycationic composition may be selected from formula 1-60 of US Patent Publication No. US20050222064 .
- LNPs comprise the lipid KL52 (an amino-lipid disclosed in U.S. Application Publication No. 2012/0295832 ). Activity and/or safety (as measured by examining one or more of ALT/AST, white blood cell count and cytokine induction) of LNP administration may be improved by incorporation of such lipids.
- LNPs comprising KL52 may be administered intravenously and/or in one or more doses. In some embodiments, administration of LNPs comprising KL52 results in equal or improved mRNA and/or protein expression as compared to LNPs comprising MC3.
- the LNP may include a cationic peptide or a polypeptide such as, but not limited to, polylysine, polyornithine and/or polyarginine and the cationic peptides described in International Pub. No. WO2012013326 or US Patent Pub. No. US20130142818 .
- the lipid nanoparticle includes a neutral lipid such as, but not limited to, cholesterol or dioleoyl phosphatidylethanolamine (DOPE).
- DOPE dioleoyl phosphatidylethanolamine
- a nanoparticle composition may be relatively homogenous.
- a polydispersity index may be used to indicate the homogeneity of a nanoparticle composition, e.g., the particle size distribution of the nanoparticle compositions.
- a small (e.g., less than 0.3) polydispersity index generally indicates a narrow particle size distribution.
- a nanoparticle composition may have a polydispersity index from about 0 to about 0.25, such as 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.10, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.20, 0.21, 0.22, 0.23, 0.24, or 0.25.
- the polydispersity index of a nanoparticle composition may be from about 0.10 to about 0.20, or about 0.05 to about 0.15, or less than about 0.1, or less than about 0.15. Each possibility represents a separate embodiment of the present invention.
- the zeta potential of a nanoparticle composition may be used to indicate the electrokinetic potential of the composition.
- the zeta potential may describe the surface charge of a nanoparticle composition.
- Nanoparticle compositions with relatively low charges at physiological pH, positive or negative, are generally desirable, as more highly charged species may interact undesirably with cells, tissues, and other elements in the body.
- the zeta potential of a nanoparticle composition may be from about -10 mV to about +20 mV, from about -10 mV to about +15 mV, from about -10 mV to about +10 mV, from about -10 mV to about +5 mV, from about -10 mV to about 0 mV, from about -10 mV to about -5 mV, from about -5 mV to about +20 mV, from about -5 mV to about +15 mV, from about -5 mV to about +10 mV, from about -5 mV to about +5 mV, from about -5 mV to about 0 mV, from about 0 mV to about +20 mV, from about 0 mV to about +15 mV, from about 0 mV to about +10 mV, from about 0 mV to about +5 mV, from about 0 mV to about +20 mV
- the efficiency of encapsulation of a therapeutic agent describes the amount of therapeutic agent that is encapsulated or otherwise associated with a nanoparticle composition after preparation, relative to the initial amount provided.
- the encapsulation efficiency is desirably high (e.g., close to 100%).
- the encapsulation efficiency may be measured, for example, by comparing the amount of therapeutic agent in a solution containing the nanoparticle composition before and after breaking up the nanoparticle composition with one or more organic solvents or detergents. Fluorescence may be used to measure the amount of free therapeutic agent (e.g., nucleic acids) in a solution.
- the encapsulation efficiency of a therapeutic agent may be at least 50%, for example 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%.
- the encapsulation efficiency may be at least 80%.
- the encapsulation efficiency may be at least 90%.
- the encapsulation efficiency may be at least 95%.
- microfluidic mixers may include, but are not limited to a slit interdigitial micromixer including, but not limited to those manufactured by Microinnova (Allerheiligen bei Wildon, Austria) and/or a staggered herringbone micromixer (SHM) ( Zhigaltsev, I.V. et al., Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing have been published (Langmuir. 2012. 28:3633-40 ; Belliveau, N. M.
- the lipid nanoparticles may be formulated using a micromixer such as, but not limited to, a Slit Interdigital Microstructured Mixer (SIMM-V2) or a Standard Slit Interdigital Micro Mixer (SSIMM) or Caterpillar (CPMM) or Impinging jet (UMM) from the Institut für Mikrotechnik Mainz GmbH, Mainz Germany).
- a micromixer such as, but not limited to, a Slit Interdigital Microstructured Mixer (SIMM-V2) or a Standard Slit Interdigital Micro Mixer (SSIMM) or Caterpillar (CPMM) or Impinging jet (UMM) from the Institut für Mikrotechnik Mainz GmbH, Mainz Germany).
- the lipid nanoparticles are created using microfluidic technology (see Whitesides, George M. The Origins and the Future of Microfluidics. Nature, 2006 442: 368-373 ; and Abraham et al. Chaotic Mixer for Microchannels. Science, 2002 295: 647-651 ).
- controlled microfluidic formulation includes a passive method for mixing streams of steady pressure-driven flows in micro channels at a low Reynolds number (See e.g., Abraham et al. Chaotic Mixer for Microchannels. Science, 2002 295: 647651 ).
- Cationic lipids useful in embodiments of the present invention are neutral while in circulation but become positively charged upon acidification of the endosome.
- a positive charge on the LNP may promote association with the negatively charged cell membrane to enhance cellular uptake.
- Cationic lipids may also combine with negatively charged lipids to induce nonbilayer structures that facilitate intracellular delivery.
- Suitable cationic lipids for use in making the LNPs disclosed herein can be ionizable cationic lipids, as disclosed herein.
- the cationic lipids may be prepared according to the procedures set forth in the Examples or according to methods known or derivable by one of ordinary skill in the art.
- LNPs may comprise, in molar percentages, 35 to 45% cationic lipid, 40% to 50% cationic lipid, 45% to 55% cationic lipid, 50% to 60% cationic lipid and/or 55% to 65% cationic lipid.
- the ratio of lipid to nucleic acid (e.g., mRNA) in lipid nanoparticles may be 5:1 to 20:1, 10:1 to 25:1, 15:1 to 40:1, 20:1 to 30:1, 25:1 to 50:1, 30:1 to 60:1 and/or at least 40:1.
- Such lipids include, but are not limited to, N,N-dioleyl-N,N-dimethylammonium chloride (DODAC); N-(2,3-dioleyloxy)propyl)-N,N,N-trimethylammonium chloride (DOTMA); N,N-distearyl-N,N-dimethylammonium bromide (DDAB); N-(2,3dioleoyloxy)propyl)-N,N,N-trimethylammonium chloride (DOTAP); 3-(N---(N',N'dimethylaminoethane)-carbamoyl)cholesterol (DC-Chol), N-(1-(2,3-dioleoyloxy)propyl)N-2-(sperminecarboxamido)ethyl)-N,N-dimethylammonium trifluoracetate (DOSPA), dioctadecylamidoglycyl carboxyspermine (DOGS
- the cationic lipid for use in any of the described embodiments is independently an amino lipid.
- Suitable amino lipids include those described in WO 2010/054401 and WO 2012/016184 .
- Representative amino lipids include, but are not limited to, 1,2-dilinoleyoxy-3-(dimethylamino)acetoxypropane (DLin-DAC), 1,2-dilinoleyoxy-3morpholinopropane (DLin-MA), 1,2-dilinoleoyl-3-dimethylaminopropane (DLinDAP), 1,2-dilinoleylthio-3-dimethylaminopropane (DLin-S-DMA), 1-linoleoyl-2-linoleyloxy-3dimethylaminopropane (DLin-2-DMAP), 1,2-dilinoleyloxy-3-trimethylaminopropane chloride salt (DLin-TMA.Cl), 1,2-dilinoleoyl-3-trimethyl
- the cationic lipid has the following structure: or a pharmaceutically acceptable salt, tautomer, prodrug or stereoisomer thereof, wherein: R 1 and R 2 are independently selected and are H or C 1 -C 3 alkyls. R 3 and R 4 are independently selected and are alkyl groups having from about 10 to about 20 carbon atoms, wherein at least one of R 4 and R 4 comprises at least two sites of unsaturation. In one embodiment, R 3 and R 4 are both the same, for example, in some embodiments R 3 and R 4 are both linoleyl (i.e., C18), etc.
- R 3 and R 4 are different, for example, in some embodiments R 3 is tetradectrienyl (C14) and R 4 is linoleyl (C18).
- the cationic lipid(s) of the present invention are symmetrical, i.e., R 3 and R 4 are the same.
- both R 3 and R 4 comprise at least two sites of unsaturation.
- R 3 and R 4 are independently selected from dodecadienyl, tetradecadienyl, hexadecadienyl, linoleyl, and icosadienyl.
- R 3 and R 4 are both linoleyl.
- R 4 and R 4 comprise at least three sites of unsaturation and are independently selected from, e.g., dodecatrienyl, tetradectrienyl, hexadecatrienyl, linolenyl, and icosatrienyl.
- the cationic lipid is DLin-K-DMA. In one embodiment, a cationic lipid is DLin-KC2-DMA (DLin-K-DMA above, wherein n is 2).
- the cationic has the following structure: or a pharmaceutically acceptable salt, tautomer, prodrug or stereoisomer thereof, wherein:
- the cationic lipid has one of the following structures:
- the cationic lipid is DLin-M-C3-DMA, MC3 or M-C3 and has been described in WO 2010/054401 , and WO 2010/144740 A1 .
- the cationic lipid has one of the following structures:
- the cationic lipid has the structure: or
- the cationic lipid is a cyclic lipid having the following structure: or a pharmaceutically acceptable salt, tautomer, prodrug or stereoisomer thereof, wherein:
- the cationic lipid is selected from the compounds:
- the cationic lipid has a structure of one of the following compounds, and salts thereof:
- the cationic lipid has the structure of Formula I: or a pharmaceutically acceptable salt, tautomer or stereoisomer thereof, wherein:
- one of L 1 or L 2 is a carbon-carbon double bond. In other embodiments, both L 1 and L 2 are a carbon-carbon double bond.
- the lipid compounds of Formula (I) have the following Formula (Ib):
- e, f, g and h are each independently an integer from 4 to 10.
- b is 1. In other embodiments, b is 2. In more embodiments, b is 3. In yet other embodiments, b is 4. In some embodiments, b is 5. In other embodiments, b is 6. In more embodiments, b is 7. In yet other embodiments, b is 8. In some embodiments, b is 9. In other embodiments, b is 10. In more embodiments, b is 11. In yet other embodiments, b is 12. In some embodiments, b is 13. In other embodiments, b is 14. In more embodiments, b is 15. In yet other embodiments, b is 16.
- At least one of R 1b , R 2b , R 3b and R 4b is H or R 1b , R 2b , R 3b and R 4b are H at each occurrence.
- G 3 is C 2 -C 4 alkylene, for example C 3 alkylene.
- the lipid compound has one of the structures set forth in Table 2 below Table 2: Representative Lipids of Formula (II) No.
- the cationic lipid has a structure of Formula (III): or a pharmaceutically acceptable salt or stereoisomer thereof, wherein:
- the lipid has one of the following Formulae (IIIA) or (IIIB): wherein:
- the lipid has Formula (IIIA), and in other embodiments, the lipid has Formula (IIIB).
- the lipid has one of the following Formulae (IIIC) or (IIID): wherein y and z are each independently integers ranging from 1 to 12.
- the lipid has one of the following Formulae (IIIE) or (IIIF):
- the lipid has one of the following Formulae (IIIG), (IIIH), (IIII), or (IIIJ):
- y and z are each independently an integer ranging from 2 to 10.
- y and z are each independently an integer ranging from 4 to 9 or from 4 to 6.
- G 3 is unsubstituted. In other embodiments, G3 is substituted. In various different embodiments, G 3 is linear C 1 -C 24 alkylene or linear C 1 -C 24 alkenylene.
- R 1 or R 2 is C 6 -C 24 alkenyl.
- R 1 and R 2 each, independently have the following structure: wherein:
- R 1 or R 2 has one of the following structures:
- a cationic lipid has one of the structures set forth in Table 3 below.
- Table 3 Representative Compounds of Formula (III) No. Structure pKa III-1 5.89 III-2 6.05 III-3 6.09 III-4 5.60 III-5 5.59 III-6 5.42 III-7 6.11 III-8 5.84 III-9 - III-10 - III-11 - III-12 - III-13 - III-14 - III-15 I 6.14 III-16 6.31 III-17 6.28 III-18 - III-19 - III-20 6.36 III-21 - III-22 6.10 III-23 5.98 III-24 - III-25 6.22 III-26 5.84 III-27 5.77 III-28 - III-29 - III-30 6.09 III-31 - III-32 - III-33 - III-34 - III-35 - III-36 - III-37 - III-38 - III-39 - III-40 - III-41 - III-42 - III-43 - III-44 - III-45 - III-46 - III-47 - III
- b 1 and b 2 are 0. In different embodiments, b 1 and b 2 are 1.
- Z is alkyl, cycloalkyl or a monovalent moiety comprising at least one polar functional group when n is 1. In other embodiments, Z is alkyl.
- the sum of a 1 +c 1 +d 1 is an integer from 20 to 30, and the sum of a 2 +c 2 +d 2 is an integer from 18 to 30. In other embodiments, the sum of a 1 +c 1 +d 1 is an integer from 20 to 30, and the sum of a 2 +c 2 +d 2 is an integer from 20 to 30. In more embodiments of Formula (V), the sum of a 1 + b 1 + c 1 or the sum of a 2 + b 2 + c 2 is an integer from 12 to 26.
- b 1 and b 2 are 0. In different embodiments b 1 and b 2 are 1.
- Z is alkyl or a monovalent moiety comprising at least one polar functional group when n is 1; or Z is alkylene or a polyvalent moiety comprising at least one polar functional group when n is greater than 1.
- Z is alkyl, cycloalkyl or a monovalent moiety comprising at least one polar functional group when n is 1. In other embodiments, Z is alkyl.
- R 1 and R 2 independently have one of the following structures: or
- the compound has one of the following structures: or
- n is 1. In other of the foregoing embodiments of Formula (IV) or (V), n is greater than 1.
- Z has the following structure: wherein:
- Z has the following structure: wherein:
- Z-L has one of the following structures:
- X is CH and Z-L has one of the following structures:
- a cationic lipid has one of the structures set forth in Table 4 below.
- Table 4 Representative Compounds of Formula (IV) or (V) No. Structure IV-1 IV-2 IV-3
- the cationic lipid has the following Formula (VI): or a pharmaceutically acceptable salt, tautomer or stereoisomer thereof, wherein:
- the compound has one of the following Formulas (VIA) or (VIB):
- the compound has Formula (VIA). In other embodiments, the compound has Formula (VIB).
- one of L 1 or L 2 is a direct bond.
- a "direct bond” means the group (e.g., L 1 or L 2 ) is absent.
- each of L 1 and L 2 is a direct bond.
- R 3a is H or C 1 -C 12 alkyl
- R 3b together with the carbon atom to which it is bound is taken together with an adjacent R 3b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- the compound has one of the following Formulas (VIC) or (VID): wherein e, f, g and h are each independently an integer from 1 to 12.
- the compound has Formula (VIC). In other embodiments, the compound has Formula (VID).
- a, b, c and d are each independently an integer from 2 to 12 or an integer from 4 to 12. In other embodiments, a, b, c and d are each independently an integer from 8 to 12 or 5 to 9. In some certain embodiments, a is 0. In some embodiments, a is 1. In other embodiments, a is 2. In more embodiments, a is 3. In yet other embodiments, a is 4. In some embodiments, a is 5. In other embodiments, a is 6. In more embodiments, a is 7. In yet other embodiments, a is 8. In some embodiments, a is 9. In other embodiments, a is 10. In more embodiments, a is 11. In yet other embodiments, a is 12. In some embodiments, a is 13. In other embodiments, a is 14. In more embodiments, a is 15. In yet other embodiments, a is 16.
- d is 0. In some embodiments, d is 1. In other embodiments, d is 2. In more embodiments, d is 3. In yet other embodiments, d is 4. In some embodiments, d is 5. In other embodiments, d is 6. In more embodiments, d is 7. In yet other embodiments, d is 8. In some embodiments, d is 9. In other embodiments, d is 10. In more embodiments, d is 11. In yet other embodiments, d is 12. In some embodiments, d is 13. In other embodiments, d is 14. In more embodiments, d is 15. In yet other embodiments, d is 16.
- e is 1. In other embodiments, e is 2. In more embodiments, e is 3. In yet other embodiments, e is 4. In some embodiments, e is 5. In other embodiments, e is 6. In more embodiments, e is 7. In yet other embodiments, e is 8. In some embodiments, e is 9. In other embodiments, e is 10. In more embodiments, e is 11. In yet other embodiments, e is 12.
- a and d are the same. In some other embodiments, b and c are the same. In some other specific embodiments a and d are the same and b and c are the same.
- the sum of a and b and the sum of c and d are factors which may be varied to obtain a lipid having the desired properties.
- a and b are chosen such that their sum is an integer ranging from 14 to 24.
- c and d are chosen such that their sum is an integer ranging from 14 to 24.
- the sum of a and b and the sum of c and d are the same.
- the sum of a and b and the sum of c and d are both the same integer which may range from 14 to 24.
- a. b, c and d are selected such that the sum of a and b and the sum of c and d is 12 or greater.
- At least one of R 1b , R 2b , R 3b and R 4b is H or R 1b , R 2b , R 3b and R 4b are H at each occurrence.
- R 1b together with the carbon atom to which it is bound is taken together with an adjacent R 1b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- R 4b together with the carbon atom to which it is bound is taken together with an adjacent R 4b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- R 5 and R 6 are not particularly limited in the foregoing embodiments. In certain embodiments one of R 5 or R 6 is methyl. In other embodiments each of R 5 or R 6 is methyl.
- R b is branched C 3 -C 15 alkyl.
- R b has one of the following structures: or
- R 8 is OH
- R 11 is benzyl.
- G 3 is C 2 -C 5 alkylene, for example C 2 -C 4 alkylene, C 3 alkylene or C 4 alkylene.
- R 8 is OH.
- G 2 is absent and R 7 is C 1 -C 2 alkylene, such as methyl.
- the compound has one of the structures set forth in Table 5 below.
- Table 5 Representative Compounds of Formula (VI) No. Structure VI-1 VI-2 VI-3 VI-4 VI-5 VI-6 VI-7 VI-8 VI-9 VI-10 VI-11 VI-12 VI-13 VI-14 VI-15 VI-16 VI-17 VI-18 VI-19 VI-20 VI-21 VI-22 VI-23 VI-24 VI-25 VI-26 VI-27 VI-28 VI-29 VI-30 VI-31 VI-32 VI-33 VI-34 VI-35 VI-36 VI-37
- the cationic lipid has the following Formula (VII): or a pharmaceutically acceptable salt or stereoisomer thereof, wherein:
- G 1 , G 1' , G 2 and G 2' are each independently C 2 -C 8 alkylene, for example C 4 -C 8 alkylene.
- R 1 or R 2 are each, at each occurrence, independently branched C 6 -C 24 alkyl.
- R 1 and R 2 at each occurrence independently have the following structure: wherein:
- At least one occurrence of R 7a is H.
- R 7a is H at each occurrence.
- at least one occurrence of R 7b is C 1 -C 8 alkyl.
- C 1 -C 8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
- R 1 or R 2 at each occurrence independently has one of the following structures: or
- R b , R c , R e and R f when present, are each independently C 3 -C 12 alkyl.
- R b , R c , R e and R f when present, are n-hexyl and in other embodiments R b , R c , R e and R f , when present, are n-octyl.
- the cationic lipid has the following Formula (VIII): or a pharmaceutically acceptable salt or stereoisomer thereof, wherein:
- G 3 is C 1 -C 12 heteroalkylene, for example C 1 -C 12 aminylalkylene.
- At least one occurrence of R 7a is H.
- R 7a is H at each occurrence.
- at least one occurrence of R 7b is C 1 -C 8 alkyl.
- C 1 -C 8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
- R 1 , R 2 and R 3 each, independently have one of the following structures: or
- R 1 and R 2 and R 3 are each, independently, branched C 6 -C 24 alkyl and R 3 is C 1 -C 24 alkyl or C 2 -C 24 alkenyl.
- G 3 is unsubstituted.
- G 3 is C 2 -C 12 alkylene, for example, in some embodiments G 3 is C 3 -C 7 alkylene or in other embodiments G 3 is C 3 -C 12 alkylene. In some embodiments, G 3 is C 2 or C 3 alkylene.
- the compound has one of the following Formulas (IXB), (IXC), (IXD) or (IXE):
- R 1 or R 2 has one of the following structures:
- R 4 is substituted or unsubstituted: methyl, ethyl, propyl, n-butyl, n-hexyl, n-octyl or n-nonyl.
- R 4 is unsubstituted.
- R 1 and R 2 are each independently branched, saturated or unsaturated C 12 -C 30 alkyl, C 12 -C 20 alkyl, or C 15 -C 20 alkyl. In some specific embodiments, R 1 and R 2 are each saturated. In certain embodiments, at least one of R 1 and R 2 is unsaturated.
- At least one R is OH. In other embodiments, each R is H.
- the cationic lipid has the following Formula (XI): or a pharmaceutically acceptable salt, tautomer or stereoisomer thereof, wherein:
- the compound has the following structure (IA): wherein y1 and z1 are each independently integers ranging from 2 to 12, for example an integer from 2 to 6, for example 4.
- the compound has one of the following Formulas (IB), (IC), (ID) or (IE):
- the compound has one of the following Formulas (XIF), (XIG), (XIH) or (XU): wherein y1 and z1 are each independently integers ranging from 2 to 12, for example an integer from 2 to 6, for example 4.
- R 1 or R 2 has one of the following structures:
- R b , R c , R e and R f are each independently C 3 -C 12 alkyl.
- R b , R c , R e and R f are n-hexyl and in other embodiments of Formula (XI) R b , R c , R e and R f are n-octyl.
- the compound has one of the structures set forth in Table 10 below.
- Table 10 Representative Compounds of Formula (XI) No. Structure XI-1 XI-2 XI-3 XI-4 XI-5 XI-6 XI-7 XI-8 XI-9 XI-10 XI-11 XI-12 XI-13 XI-14 XI-15 XI-16 XI-17 XI-18 XI-19
- G 3 is unsubstituted.
- G 3 is C 1 -C 12 alkylene, for example, G 3 is C 3 -C 5 alkylene or G 3 is C 3 -C 12 alkylene.
- the cationic lipid has the following Formula (XIIA): or a pharmaceutically acceptable salt, tautomer or stereoisomer thereof, wherein y2 and z2 are each independently integers ranging from 1 to 12.
- the compound has one of the following Formulas (XIIB) or (XIIC):
- the compound has Formula (XIIB), in other embodiments, the compound has Formula (XIIC).
- the compound has one of the following Formulas (XIID) or (XIIE): wherein y2 and z2 are each independently integers ranging from 1 to 12.
- y2 and z2 are each independently an integer ranging from 2 to 12, for example from 2 to 10, from 2 to 8, from 4 to 7 or from 4 to 10.
- y2 is 4, 5, 6, 7, 8, 9, 10, 11 or 12.
- z2 is 4, 5, 6, 7, 8, 9, 10, 11 or 12.
- y2 and z2 are the same, while in other embodiments of Formula (XII), y2 and z2 are different.
- R 1 or R 2 is branched C 6 -C 24 alkyl.
- R 1 and R 2 each, independently have the following structure: wherein:
- R 3b has one of the following structures:
- R 1 is optionally substituted C 6 -C 18 alkyl or C 14 -C 18 alkenyl. In certain embodiments, R 1 is C 8 alkyl, C 9 alkyl, C 10 alkyl, C 12 alkyl, C 14 alkyl, or C 16 alkyl. In some more specific embodiments, R 1 is C 16 alkenyl. In certain more specific embodiments, R 1 is unbranched. In some embodiments, R 1 is branched. In certain embodiments, R 1 is unsubstituted.
- R 4 and R 5 join, along with the N to which they are attached, to form a heterocyclyl.
- the heterocyclyl is a 5-membered heterocyclyl.
- the heterocyclyl has the following structure:
- the compound has one of the structures set forth in Table 12 below. Table 12.
- the lipid compound has the following structure: or salts or isomers thereof, wherein:
- another subset of compounds of Formula (I) includes those in which
- a subset of compounds of Formula (III) includes those in which, when R4 is -(CH2)nQ, -(CH2)nCHQR, -CHQR, or -CQ(R)2, then (i) Q is not -N(R)2 when n is 1, 2, 3, 4 or 5, or (ii) Q is not 5, 6, or 7-membered heterocycloalkyl when n is 1 or 2.
- Another subset of compounds of Formula (III) includes those in which
- another subset of compounds of Formula (III) includes those in which
- a subset of compounds of Formula (I) includes those of Formula (lid): or its N-oxide, or a salt or isomer thereof, wherein n is 2, 3, or 4; and m, R', R", and R2 through R6 are as described herein.
- each of R2 and R3 may be independently selected from the group consisting of C5-14 alkyl and C5-14 alkenyl.
- a subset of compounds of Formula (I) includes those of Formula (Ilg): or its N-oxide, or a salt or isomer thereof, wherein 1, m, M, Mi, R', R2 and R3 are as described herein.
- each of R2 and R3 may be independently selected from the group consisting of C5-14 alkyl and C5-14 alkenyl, 1 is selected from 1, 2, 3, 4, and 5, and m is selected from 5, 6, 7, 8, and 9.
- a subset of compounds of Formula (VI) includes those of Formula (VI-a): r its N-oxide, or a salt or isomer thereof, wherein
- a subset of compounds of Formula (VI) includes those of Formula (VIII):
- the compounds of any one of formula (I), (IA), (VI), (Vl-a), (VII) or (VIII) include one or more of the following features when applicable.
- M and M' are independently -C(0)0- or -OC(O)-.
- At least one of M and M' is -C(0)0- or -OC(O)-.
- At least one of M and M' is -OC(O)-.
- one of M and M' is -0C(0)-M"-C(0)0-, in which M" is a bond, Ci-i3 alkyl or C2-13 alkenyl.
- M" is C1-6 alkyl or C2-6 alkenyl.
- M" is C1-4 alkyl or C2-4 alkenyl.
- M" is Ci alkyl.
- M" is C2 alkyl.
- M" is C3 alkyl.
- M" is C4 alkyl.
- M" is C2 alkenyl.
- M" is C3 alkenyl.
- M" is C4 alkenyl.
- R4 is not hydrogen
- Q is OH
- Q is -0(CH2)nOR.
- Q is -N(R)R8.
- n 3.
- n 4.
- At least one R5 is hydroxyl.
- one R5 is hydroxyl.
- At least one R6 is hydroxyl.
- one R6 is hydroxyl.
- one of R5 and R6 is hydroxyl.
- one R5 is hydroxyl and each R6 is hydrogen.
- one R6 is hydroxyl and each R5 is hydrogen.
- Rx is Ci-6 alkyl. In some embodiments, Rx is Ci-3 alkyl. For example, Rx is methyl. For example, Rx is ethyl. For example, Rx is propyl.
- R' is Ci-ib alkyl, C2-18 alkenyl, -R*YR", or - YR".
- R2 and R3 are independently C3-14 alkyl or C3-14 alkenyl.
- Rib is Ci-14 alkyl. In some embodiments, Rlb is C2-14 alkyl. In some embodiments, Rib is C3-14 alkyl. In some embodiments, Rlb is Ci-8 alkyl. In some embodiments, Rib is C1-5 alkyl. In some embodiments, Rlb is C1-3 alkyl. In some embodiments, Rlb is selected from Ci alkyl, C2 alkyl, C3 alkyl, C4 alkyl, and C5 alkyl. For example, in some embodiments, Rlb is Ci alkyl. For example, in some embodiments, Rlb is C2 alkyl. For example, in some embodiments, Rib is C3 alkyl. For example, in some embodiments, Rlb is C4 alkyl. For example, in some embodiments, Rlb is C5 alkyl.
- R1 is different from -(CHR5R6)m-M-CR2R3R7.
- -CHRlaRIb- is different from -(CHR5R6)m-M-CR2R3R7.
- R7 is H. In some embodiments, R7 is selected from C1-3 alkyl. For example, in some embodiments, R7 is Ci alkyl. For example, in some embodiments, R7 is C2 alkyl. For example, in some embodiments, R7 is C3 alkyl. In some embodiments, R7 is selected from C4 alkyl, C4 alkenyl, C5 alkyl, C5 alkenyl, Ce alkyl, Ce alkenyl, C7 alkyl, C7 alkenyl, C9 alkyl, C9 alkenyl, C11 alkyl, C11 alkenyl, C17 alkyl, C17 alkenyl, Cie alkyl, and Cie alkenyl.
- the compounds of Formula (I) are of Formula (Ila): or their N-oxides, or salts or isomers thereof, wherein R4 is as described herein.
- the compounds of Formula (I) are of Formula (lib): or their N-oxides, or salts or isomers thereof, wherein R4 is as described herein.
- the compounds of Formula (I) are of Formula (lie) or (He): (lie) (He) or their N-oxides, or salts or isomers thereof, wherein R4 is as described herein.
- the compounds of Formula (I) are of Formula (Ilf): wherein M is -C(0)0- or -OC(O)-, M" is C1-6 alkyl or C2-6 alkenyl, R2 and R3 are independently selected from the group consisting of C5-14 alkyl and C5-14 alkenyl, and n is selected from 2, 3, and 4.
- the compounds of Formula (I) are of Formula (lid): (lid), or their N-oxides, or salts or isomers thereof, wherein n is 2, 3, or 4; and m, R', R", and R2 through R6 are as described herein.
- each of R2 and R3 may be independently selected from the group consisting of C5-14 alkyl and C5-14 alkenyl.
- the compounds of Formula (I) are of Formula (Ilg): r their N-oxides, or salts or isomers thereof, wherein 1 is selected from 1, 2, 3, 4, and 5; m is selected from 5, 6, 7, 8, and 9; Mi is a bond or M'; M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -P(0)(0R')0-, -S-S-, an aryl group, and a heteroaryl group; and R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, and C2-14 alkenyl.
- M is Ci-6 alkyl (e.g., C 1-4 alkyl) or C2-6 alkenyl (e.g. C2-4 alkenyl).
- R2 and R3 are independently selected from the group consisting of C5-14 alkyl and C5-14 alkenyl.
- a subset of compounds of Formula (VI) includes those of Formula (Vllb-l): (Vllb-l), or its N-oxide, or a salt or isomer thereof.
- a subset of compounds of Formula (VI) includes those of Formula (Vlld): (Vlld), or its N-oxide, or a salt or isomer thereof.
- a subset of compounds of Formula (VI) includes those of Formula (VUId): r its N-oxide, or a salt or isomer thereof.
- the compounds of any one of formulae (I), (IA), (IB), (II), (Ila), (lib), (lie), (lid), (He), (Ilf), (Ilg), (III), (VI), (Vl-a), (VII), (VIII), (Vila), (Villa), (VUIb), (Vllb-l), (VIIb-2), (VIIb-3), (Vile), (Vlld), (VIIIc), or (VUId) include one or more of the following features when applicable.
- R4 is selected from the group consisting of a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)0C(R12)2(CH2)n-oQ, -CHQR, and - CQ(R)2, where Q is selected from a C3-6 carbocycle, 5- to 14- membered aromatic or non-aromatic heterocycle having one or more heteroatoms selected from N, O, S, and P, -OR, -0(CH2)nN(R)2, -C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -N(R)2, - N(R)S(0)2R8, -C(0)N(R)2, -N(R)C(0)R, -N(R)S(0)2R, - N(R)C(0)N(R)2, - N(R)C(S)N(R)2, and -C(R)2, where Q
- n is independently selected from 1, 2, 3, 4, and 5.
- R4 is -C(0)NQR, where Q is -(CH2)nN(R)2. In a further embodiments, R4 is -C(0)NH(CH2)3N(CH3)2, -C(0)NH(CH2)4N(CH3)2, or - C(0)NH(CH 2 )2N(CH3)2.
- the disclosure provides a compound having the Formula (I), wherein R4 is selected from the group consisting of -(CH2)nQ, - (CH2)nCHQR, -CHQR, and -CQ(R)2, where Q is -N(R)2, and n is selected from 1, 2, 3, 4, and 5.
- the disclosure provides a compound having the Formula (I), wherein R2 and R3 are independently selected from the group consisting of C2-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle, and R4 is - (CH2)nQ or -(CH2)nCHQR, where Q is -N(R)2, and n is selected from 3, 4, and 5.
- R2 and R3 are independently selected from the group consisting of C2-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle, and R4 is - (CH2)nQ or -(CH2)nCHQR, where Q is -N(R)2, and n is selected from 3, 4, and 5.
- R2 and R3 are independently selected from the group consisting of C2-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle.
- R2 and R3 are independently selected from the group consisting of C2-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle.
- R2 and R3 are independently selected from the group consisting of C2-14 alkyl, and C2-14 alkenyl. In some embodiments, R2 and R3 are independently selected from the group consisting of -R*YR", -YR", and -R * OR" . In some embodiments, R2 and R3 together with the atom to which they are attached, form a heterocycle or carbocycle.
- R1 is selected from the group consisting of C5-20 alkyl and C5-20 alkenyl. In some embodiments, R1 is C5-20 alkyl substituted with hydroxyl.
- R1 is selected from the group consisting of -R*YR", -YR", and -R"M'R ⁇
- R1 is selected from -R*YR" and -YR".
- Y is a cyclopropyl group.
- R* is Cx alkyl or Cx alkenyl.
- R" is C3-12 alkyl.
- R" is C3 alkyl.
- R" is C4-8 alkyl (e.g., C4, C5, Ce, C7, or Cs alkyl).
- R is (CH2)qOR*, q is selected from 1, 2, and 3, and R* is C1-12 alkyl substituted with one or more substituents selected from the group consisting of amino, Ci-Ce alkylamino, and C1-C6 dialkylamino.
- R is (CFh)qOR*, q is selected from 1, 2, and 3 and R* is C1-12 alkyl substituted with C1-C6 dialkylamino.
- R is (CH2)qOR*, q is selected from 1, 2, and 3 and R* is C1-3 alkyl substituted with C1-C6 dialkylamino.
- R is (CH2)qOR*, q is selected from 1, 2, and 3 and R* is C1-3 alkyl substituted with dimethylamino (e.g., dimethylaminoethanyl).
- R1 is C5-20 alkyl. In some embodiments, R1 is G, alkyl. In some embodiments, R1 is Cs alkyl. In other embodiments, R1 is C9 alkyl. In certain embodiments, R1 is C 14 alkyl. In other embodiments, R1 is Cie alkyl.
- R1 is C5-20 alkenyl. In certain embodiments, R1 is Cie alkenyl. In some embodiments, R1 is linoleyl.
- R1 is branched (e.g., decan-2 -yl, undecan-3-yl, dodecan-4-yl, tridecan-5-yl, tetradecan-6-yl, 2-methylundecan-3-yl, 2-methyldecan-2-yl, 3-methylundecan-3-yl, 4-methyldodecan-4-yl, or heptadeca-9-yl).
- R1 is branched (e.g., decan-2 -yl, undecan-3-yl, dodecan-4-yl, tridecan-5-yl, tetradecan-6-yl, 2-methylundecan-3-yl, 2-methyldecan-2-yl, 3-methylundecan-3-yl, 4-methyldodecan-4-yl, or heptadeca-9-yl).
- R1 is branched (e.g., decan-2 -yl, undecan-3-yl, dodecan-4-yl, tridecan-5-
- R1 is unsubstituted C5-20 alkyl or C5-20 alkenyl.
- R' is substituted C5-20 alkyl or C5-20 alkenyl (e.g., substituted with a C3-6 carbocycle such as l-cyclopropylnonyl or substituted with OH or alkoxy).
- R1 is
- R1 is -R"M'R ⁇
- M' is - OC(0)-M"-C(0)0-.
- R 1 is , wherein x1 is an integer between 1 and 13 (e.g., selected from 3, 4, 5, and 6), x2 is an integer between 1 and 13 (e.g., selected from 1, 2, and 3), and x3 is an integer between 2 and 14 (e.g., selected from 4, 5, and 6).
- x1 is selected from 3, 4, 5, and 6, x2 is selected from 1, 2, and 3, and x3 is selected from 4, 5, and 6.
- R1 is different from -(CHR5R6)m-M-CR2R3R7.
- R" is selected from the group consisting of C3-12 alkyl and C3- 12 alkenyl. In some embodiments, R" is Cs alkyl. In some embodiments, R" adjacent to Y is Ci alkyl. In some embodiments, R" adjacent to Y is C4-9 alkyl (e.g., C4, C5, Ce, Ci or Cs or C9 alkyl).
- R" is substituted C3-12 alkyl (e.g., C3-12 alkyl substituted with, e.g., an hydroxyl).
- R" is substituted C3-12 alkyl (e.g., C3-12 alkyl substituted with, e.g., an hydroxyl).
- R" is
- R' is selected from C4 alkyl, C4 alkenyl, C5 alkyl, C5 alkenyl, C6 alkyl, Ce alkenyl, C7 alkyl, C7 alkenyl, C9 alkyl, C9 alkenyl, C 11 alkyl, C 11 alkenyl, C 17 alkyl, C17 alkenyl, Cie alkyl, and Cie alkenyl, each of which is either linear or branched.
- R' is C4 alkyl or C4 alkenyl. In some embodiments, R' is C5 alkyl or C5 alkenyl. In some embodiments, R' is G, alkyl or G, alkenyl. In some embodiments, R' is C7 alkyl or C7 alkenyl. In some embodiments, R' is Cs alkyl or Cs alkenyl. In some embodiments, R' is C9 alkylor C9 alkenyl. In some embodiments, R' is C10 alkyl or C 10 alkenyl. In some embodiments, R' is C 11 alkyl or C11 alkenyl.
- R' is linear. In some embodiments, R' is branched.
- R' is and M' is -OC(O-), In other embodiments, R' is and M' is -C(O)O-.
- R' is selected from C11 alkyl and C 11 alkenyl. In other embodiments, R' is selected from C12 alkyl, C12 alkenyl, C13 alkyl, C13 alkenyl, C14 alkyl, C14 alkenyl, C15 alkyl, C15 alkenyl, Ci6 alkyl, Ci6 alkenyl, C17 alkyl, C 17 alkenyl, Cie alkyl, and Cie alkenyl. In certain embodiments, R' is linear C4-18 alkyl or C4-18 alkenyl. In certain embodiments, R' is linear C4-18 alkyl or C4-18 alkenyl. In certain embodiments,
- M is -C(O). In some embodiments, M is -OC(O)- and M' is -C(0)0-. In some embodiments, M is -C(0)0- and M' is -OC(O)-. In some embodiments, M and M' are each -OC(O)-. In some embodiments, M and M' are each - C(0)0-.
- R8 is 3-(bis(2-methoxyethyl)amino)cyclobut-3-ene-1,2-dione.
- R8 is cyclobutenyl substituted with one or more of oxo, and heterocycloalkyl.
- R8 is cyclobutenyl substituted with one or more of oxo, and piperidinyl, piperazinyl, or morpholinyl.
- R8 is cyclobutenyl substituted with one or more of oxo, and heterocycloalkyl, wherein heterocycloalkyl is further substituted, e.g., with one or more C1-3 alkyl.
- R8 is cyclobutenyl substituted with one or more of oxo, and
- heterocycloalkyl wherein heterocycloalkyl (e.g., piperidinyl, piperazinyl, or morpholinyl) is further substituted with methyl.
- heterocycloalkyl e.g., piperidinyl, piperazinyl, or morpholinyl
- Q is -NHR8, in which R8 is a heteroaryl optionally substituted with one or more substituents selected from amino (NH2), mono- or di-alkylamino, C1-3 alkyl and halo.
- R8 is thiazole or imidazole.
- Q is -NHR8 and R8 is purine.
- R is C 1-6 alkyl substituted with one or more substituents selected from the group consisting of C 1-3 alkoxyl, amino, and C1-C3 dialkylamino.
- R4 is -CH2CH(OH)CH3, - CH(CH3)CH20H, or -CH2CH(OH)CH2CH3.
- R4 is selected from any of the following groups:
- R4 is selected from any of the following groups: some embodiments, is selected from any of the following groups
- the lipid compound has the following structure:
- R 10 is selected from the group consisting of hydroxyl, amino, alkylamino, dialkylamino, NH-heterocyclyl and heterocyclyl, wherein the alkyl portion of the alkylamino and dialkylamino are optionally substituted with hydroxyl, alkoxy, amino, alkylamino and/or dialkylamino.
- the cationic lipid compound has the following structure:
- the cationic lipid compound has the following structure:
- the cationic lipid compound has the following structure:
- the cationic lipid compound has the following structure:
- the cationic lipid compound has the following structure:
- the cationic lipid has one of the following structures: Cpd Structure Cpd Structure 1 32 2 33 3 34 4 35 5 36 6 37 7 38 8 39 9 40 10 41 11 42 12 43 13 44 14 45 15 46 16 47 17 48 18 49 19 50 20 51 21 52 22 53 23 54 24 55 25 56 26 57 27 58 28 59 29 60 30 61 31
- the cationic lipid has one of the following structures: Cpd Structure Cpd Structure 62 64 63
- the cationic lipid has one of the following structures: Cpd Structure Cpd Structure 65 M2 66 213 67 214 68 215 69 216 70 217 71 218 72 219 73 220 74 221 75 212 76 223 77 224 78 225 79 226 80 227 81 228 82 229 83 230 84 231 85 232 86 233 87 234 88 235 89 236 90 237 91 238 92 239 93 240 94 241 95 242 96 243 97 244 98 245 99 246 100 247 101 248 102 249 103 250 104 251 105 252 106 253 107 254 108 255 109 256 110 257 111 258 112 259 113 260 114 261 115 262 116 263 117 264 118 265 119 266 120 267 121 26S 122 269 123 270 124 271 125 27
- the cationic lipid has the following structure:
- the LNPs comprise a neutral lipid.
- the molar ratio of the cationic lipid to the neutral lipid ranges from about 2:1 to about 8:1.
- the neutral lipid is present in any of the foregoing LNPs in a concentration ranging from 5 to 10 mol percent, from 5 to 15 mol percent, 7 to 13 mol percent, or 9 to 11 mol percent. In certain specific embodiments, the neutral lipid is present in a concentration of about 9.5, 10 or 10.5 mol percent.
- the molar ratio of cationic lipid to the neutral lipid ranges from about 4.1:1.0 to about 4.9:1.0, from about 4.5:1.0 to about 4.8:1.0, or from about 4.7:1.0 to 4.8:1.0. In some embodiments, the molar ratio of total cationic lipid to the neutral lipid ranges from about 4.1:1.0 to about 4.9: 1.0, from about 4.5:1.0 to about 4.8:1.0, or from about 4.7:1.0 to 4.8:1.0.
- the neutral lipid is 1,2-distearoyl-sn-glycero-3phosphocholine (DSPC). In some embodiments, the neutral lipid is selected from DSPC, DPPC, DMPC, DOPC, POPC, DOPE and SM. In some embodiments, the neutral lipid is DSPC.
- neutral lipids useful in the present invention are DSPC analogs wherein the phosphocholine moiety is replaced by a different zwitterionic group.
- the different zwitterionic group is not a phosphocholine group.
- a neutral lipid useful in the present invention is a compound of formula: or a salts thereof, wherein:
- Z is an amino acid or a derivative thereof.
- Z is of one of the following formulas: wherein R O is hydrogen, optionally substituted alkyl or an oxygen protecting group.
- a compound of said formula is of one of the following: or a salt thereof.
- a compound of formula is of one of the following formulas: or a salt thereof.
- a compound of formula is one of the following: or salts thereof.
- an oleic acid analog can comprise a modified oleic acid tail, a modified carboxylic acid moiety, or both.
- an analog of oleic acid is a compound of formula: or a salt thereof, wherein:
- the compound of said formula is one of the following: or salts thereof.
- an oleic acid analog is a compound wherein the carboxylic acid moiety of oleic acid replaced by a different group.
- an oleic acid analog useful in the present invention is one of the following: or salts thereof.
- Phospholipids are any lipids that comprise a phosphate group. Phospholipids are a subset of neutral lipids.
- the lipid component of a nanoparticle composition may include one or more phospholipids, such as one or more (poly)unsaturated lipids. Phospholipids may assemble into one or more lipid bilayers. In general, phospholipids may include a phospholipid moiety and one or more fatty acid moieties.
- a phospholipid moiety may be selected from the non-limiting group consisting of phosphatidyl choline, phosphatidyl ethanolamine, phosphatidyl glycerol, phosphatidyl serine, phosphatidic acid, 2-lysophosphatidyl choline, and a sphingomyelin.
- a fatty acid moiety may be selected from the non-limiting group consisting of lauric acid, myristic acid, myristoleic acid, palmitic acid, palmitoleic acid, stearic acid, oleic acid, linoleic acid, alpha-linolenic acid, erucic acid, phytanoic acid, arachidic acid, arachidonic acid, eicosapentaenoic acid, behenic acid, docosapentaenoic acid, and docosahexaenoic acid.
- Non-natural species including natural species with modifications and substitutions including branching, oxidation, cyclization, and alkynes are also contemplated.
- a phospholipid may be functionalized with or cross-linked to one or more alkynes (e.g., an alkenyl group in which one or more double bonds is replaced with a triple bond).
- alkynes e.g., an alkenyl group in which one or more double bonds is replaced with a triple bond.
- an alkyne group may undergo a copper-catalyzed cycloaddition upon exposure to an azide.
- Such reactions may be useful in functionalizing a lipid bilayer of a nanoparticle composition to facilitate membrane permeation or cellular recognition or in conjugating a nanoparticle composition to a useful component such as a targeting or imaging moiety (e.g., a dye).
- a targeting or imaging moiety e.g., a dye
- Phospholipids useful in the compositions and methods may be selected from the nonlimiting group consisting of 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC),
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine DOPE
- 1,2-dilinoleoyl-sn-glycero-3-phosphocholine DLPC
- 1,2-dimyristoyl-sn-glycero-phosphocholine DMPC
- 1,2 dioleoyl-sn-glycero-3-phosphocholine DOPC
- 1,2-dipalmitoyl-sn-glycero-3-phosphocholine DPPC
- 1,2-diundecanoyl-sn-glycero-phosphocholine DUPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine POPC
- 1,2-di-O-octadecenyl-sn-glycero-3-phosphocholine (18:0 Diether PC
- a nanoparticle composition includes DSPC. In certain embodiments, a nanoparticle composition includes DOPE. In some embodiments, a nanoparticle composition includes both DSPC and DOPE.
- phospholipids examples include, but are not limited to, the following: or salts thereof.
- neutral/non-cationic lipids include, but are not limited to, the following:
- any of the disclosed lipid nanoparticles comprise a steroid or steroid analogue.
- the steroid or steroid analogue is cholesterol.
- the steroid is present in a concentration ranging from 35 to 49 molar percent, 37 to 46 molar percent, from 38 to 44 molar percent, from 38 to 40 molar percent, from 40 to 42 molar percent, from 42 to 44 molar percent, or from 44 to 46 molar percent.
- the steroid is present in a concentration of 37, 38, 39, 40, 41, 42, 43, 44, 45, or 46 molar percent.
- the molar ratio of cationic lipid to the steroid ranges from 1.0:0.9 to 1.0:1.2, or from 1.0:1.0 to 1.0:1.2. In some of these embodiments, the molar ratio of cationic lipid to cholesterol ranges from about 5:1 to 1:1. In certain embodiments, the steroid is present in a concentration ranging from 35 to 45 mol percent of the steroid.
- the molar ratio of total cationic to the steroid ranges from 1.0:0.9 to 1.0:1.2, or from 1.0:1.0 to 1.0:1.2. In some of these embodiments, the molar ratio of total cationic lipid to cholesterol ranges from about 5:1 to 1:1. In certain embodiments, the steroid is present in a concentration ranging from 35 to 45 mol percent of the steroid.
- polymer-conjugated lipids useful in various methods, such as delivery of a therapeutic nucleic acid to a primate.
- One such polymer-conjugated lipid is a compound having the following structure: or a salt thereof, wherein:
- the R' and R" moieties are collectively referred to as the di-acyl chains of a polymer conjugated lipid.
- a C12 di-acyl chain polymer conjugated lipid refers to a polymer-conjugated lipid, such as the above structure, having two C12 acyl chains (e.g., the R' and R" moieties).
- a C12/14 di-acyl chain polymer-conjugated lipid refers to a polymer-conjugated lipid, such as the above structure, having one C12 acyl chain and one C 14 acyl chain (e.g., the R' and R" moieties).
- Other polymer-conjugated lipids are identified similarly.
- n is an integer from 40 to 50.
- R′′′ is H or CH 3 .
- the total number of carbon atoms collectively in both of R' and R" ranges from 16 to 22, 16 to 21, 16 to 20, 18 to 23, 18 to 22, 18 to 21, 19 to 23, 19 to 22, 19 to 21, 20 to 23, or 20 to 22.
- LNPs comprising the foregoing polymer-conjugated lipid are also provided.
- the LNPs comprise a polymer conjugated lipid.
- the polymer conjugated lipid is a pegylated lipid.
- some embodiments include a pegylated diacylglycerol (PEG-DAG) such as 1-(monomethoxy-polyethyleneglycol)-2,3-dimyristoylglycerol (PEG-DMG), a pegylated phosphatidylethanoloamine (PEG-PE), a PEG succinate diacylglycerol (PEG-S-DAG) such as 4-O-(2',3'-di(tetradecanoyloxy)propyl-]-O-( ⁇ -methoxy(polyethoxy)ethyl)butanedioate (PEG-S-DMG), a pegylated ceramide (PEG-cer), or a PEG dialkoxypropylcarbamate such as ⁇ -methoxy
- a polymer conjugated lipid may be selected from the non-limiting group consisting of PEGylated phosphatidylethanolamines, PEGmodified phosphatidic acids, PEGylated ceramides, PEGylated dialkylamines, PEGylated diacylglycerols, PEGylated dialkylglycerols, and mixtures thereof.
- a PEG lipid may be PEG-c-DOMG, PEG-DLPE, PEG-DMPE, PEG-DPPC, or a PEG-DSPE lipid.
- PEGylated lipids are a modified form of PEG DMG.
- PEG-DMG has the following structure:
- PEG lipids useful in the present invention can be PEGylated lipids described in International Publication No. WO2012/099755 . Any of these exemplary PEG lipids described herein may be modified to comprise a hydroxyl group on the PEG chain.
- the PEG lipid is a PEG-OH lipid.
- a "PEG-OH lipid" (also referred to herein as "hydroxy-PEGylated lipid”) is a PEGylated lipid having one or more hydroxyl (-OH) groups on the lipid.
- the PEG-OH lipid includes one or more hydroxyl groups on the PEG chain.
- a PEG-OH or hydroxy-PEGylated lipid comprises an -OH group at the terminus of the PEG chain.
- a PEG lipid useful in the present invention is a compound of formula: or salts thereof, wherein:
- the PEGylated lipid is of one of the following formulas:
- a PEG lipid useful in embodiments of the present invention is a PEGylated fatty acid. In certain embodiments, a PEG lipid useful in embodiments of the present invention is a compound of formula: or salts thereof, wherein:
- a compound of said formula is of one of the following compounds: or a salt thereof,
- r is an integer between 1 and 100.
- the present invention relates to a compound of formula: or a pharmaceutically acceptable salt thereof, wherein:
- the polymer conjugated lipid is selected from: and a pharmaceutically acceptable salts thereof; wherein
- the LNPs further comprise a polymer conjugated lipid compound of formula: or a pharmaceutically acceptable salt thereof, wherein
- the LNPs comprise a polymer conjugated lipid compound selected from: and wherein
- PEG lipids include, but are not limited to:
- the ratio of polymer conjugated lipid in the LNPs may be increased or decreased to alter the pharmacokinetics and/or biodistribution of the LNPs.
- LNPs may contain from 0.1 to 5.0, from 1.0 to 3.5, from 1.5 to 4.0, from 2.0 to 4.5, from .0 to 3.0, from 2.5 to 5.0, and/or from 3.0 to 6.0 molar percent of the polymer conjugated lipid to the other components.
- the polymer conjugated lipid is present in a concentration ranging from 1.0 to 3.0 molar percent.
- the LNP comprises from 2.2 to 3.3, from 2.3 to 2.8, from 2.1 to 2.5, or from 2.5 to 2.9 molar percent of polymer conjugated lipid.
- the polymer conjugated lipid is present in a concentration of about 2.0 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.3 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.4 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.5 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.6 molar percent.
- the polymer conjugated lipid is present in a concentration of about 2.7 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.8 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 3.0 molar percent.
- the polymer conjugated lipid has one of the following structures: or wherein n is an integer ranging from 30 to 60.
- the polymer conjugated lipid has the following structure: wherein n is an integer ranging from 40 to 50, and each R is a saturated alkyl having from 8 to 14 carbon atoms, or 8 to 12 carbon atoms, or 8 carbon atoms, or 10 carbon atoms, or 12 carbon atoms.
- lipid nanoparticles are associated with a nucleic acid, resulting in a nucleic acid-lipid nanoparticle.
- the nucleic acid is fully encapsulated in the lipid nanoparticle.
- nucleic acid is meant to include any oligonucleotide or polynucleotide. Fragments containing up to 50 nucleotides are generally termed oligonucleotides, and longer fragments are called polynucleotides. In particular embodiments, oligonucletoides are 15-50 nucleotides in length.
- Highly preferred target regions of the niRNA include those regions at or near the AUG translation initiation codon and those sequences that are substantially complementary to 5' regions of the mRNA.
- These secondary structure analyses and target site selection considerations can be performed, for example, using v.4 of the OLIGO primer analysis software (Molecular Biology Insights) and/or the BLASTN 2.0.5 algorithm software ( Altschul et al, Nucleic Acids Res. 1997, 25(17):3389-402 ).
- An antagomir can include ligand-conjugated monomer subunits and monomers for oligonucleotide synthesis. Exemplary monomers are described in U.S. Patent Application Publication No. 2005/0107325 .
- An antagomir can have a ZXY structure, such as is described in WO 2004/080406 .
- An antagomir can be complexed with an amphipathic moiety. Exemplary amphipathic moieties for use with oligonucleotide agents are described in WO 2004/080406 .
- Nucleic acids associated with lipid nanoparticles may be immunostimulatory, including immunostimulatory oligonucleotides (ISS; single-or double- stranded) capable of inducing an immune response when administered to a subject, which may be a mammal or other patient.
- ISS immunostimulatory oligonucleotides
- ISS include, e.g., certain palindromes leading to hairpin secondary structures (see Yamamoto S., et al. (1992) J . Immunol. 148: 4072-4076 ), or CpG motifs, as well as other known ISS features (such as multi-G domains, see WO 96/1 1266 ).
- the immune response may be an innate or an adaptive immune response.
- the immune system is divided into a more innate immune system, and acquired adaptive immune system of vertebrates, the latter of which is further divided into humoral cellular components.
- the immune response may be mucosal.
- an immunostimulatory nucleic acid is only immunostimulatory when administered in combination with a lipid nanoparticle, and is not immunostimulatory when administered in its "free form.”
- Such an oligonucleotide is considered to be immunostimulatory.
- the immunostimulatory nucleic acid or oligonucleotide comprises at least one CpG dinucleotide.
- the oligonucleotide or CpG dinucleotide may be unmethylated or methylated.
- the immunostimulatory nucleic acid comprises at least one CpG dinucleotide having a methylated cytosine.
- the nucleic acid comprises a single CpG dinucleotide, wherein the cytosine in said CpG dinucleotide is methylated.
- the nucleic acid comprises the sequence 5' TAACGTTGAGGGGCAT 3'.
- a supermir refers to a single stranded, double stranded or partially double stranded oligomer or polymer of ribonucleic acid (RNA) or deoxyribonucleic acid (DNA) or both or modifications thereof, which has a nucleotide sequence that is substantially identical to an miRNA and that is antisense with respect to its target.
- This term includes oligonucleotides composed of naturally-occurring nucleobases, sugars and covalent internucleoside (backbone) linkages and which contain at least one non-naturally-occurring portion which functions similarly.
- modified or substituted oligonucleotides are preferred over native forms because of desirable properties such as, for example, enhanced cellular uptake, enhanced affinity for nucleic acid target and increased stability in the presence of nucleases.
- the supermir does not include a sense strand, and in another preferred embodiment, the supermir does not self-hybridize to a significant extent.
- a supermir can have secondary structure, but it is substantially single-stranded under physiological conditions.
- An supermir that is substantially single-stranded is single-stranded to the extent that less than about 50% (e.g., less than about 40%, 30%, 20%, 10%, or 5%) of the supermir is duplexed with itself.
- the supermir can include a hairpin segment, e.g., sequence, preferably at the 3' end can self hybridize and form a duplex region, e.g., a duplex region of at least 1, 2, 3, or 4 and preferably less than 8, 7, 6, or n nucleotides, e.g., 5 nucleotides.
- the duplexed region can be connected by a linker, e.g., a nucleotide linker, e.g., 3, 4, 5, or 6 dTs, e.g., modified dTs.
- miRNA mimics represent a class of molecules that can be used to imitate the gene silencing ability of one or more miRNAs.
- miRNA mimic refers to synthetic non-coding RNAs (i.e. the miRNA is not obtained by purification from a source of the endogenous miRNA) that are capable of entering the RNAi pathway and regulating gene expression.
- miRNA mimics can be designed as mature molecules (e.g. single stranded) or mimic precursors (e.g., pri- or pre-miRNAs).
- miRNA mimics are double stranded molecules (e.g., with a duplex region of between about 16 and about 3 1 nucleotides in length) and contain one or more sequences that have identity with the mature strand of a given miRNA.
- Modifications can comprise 2' modifications (including 2'-O methyl modifications and 2' F modifications) on one or both strands of the molecule and internucleotide modifications (e.g. phosphorothioate modifications) that enhance nucleic acid stability and/or specificity.
- miRNA mimics can include overhangs. The overhangs can consist of 1-6 nucleotides on either the 3' or 5' end of either strand and can be modified to enhance stability or functionality.
- inhibitors are synonymous and refer to oligonucleotides or modified oligonucleotides that interfere with the ability of specific miRNAs.
- the inhibitors are nucleic acid or modified nucleic acids in nature including oligonucleotides comprising RNA, modified RNA, DNA, modified DNA, locked nucleic acids (LNAs), or any combination of the above.
- Modifications include 2' modifications (including 2'-O alkyl modifications and 2' F modifications) and internucleotide modifications (e.g. phosphorothioate modifications) that can affect delivery, stability, specificity, intracellular compartmentalization, or potency.
- miRNA inhibitors can comprise conjugates that can affect delivery, intracellular compartmentalization, stability, and/or potency.
- Inhibitors can adopt a variety of configurations including single stranded, double stranded (RNA/RNA or RNA/DNA duplexes), and hairpin designs, in general, microRNA inhibitors comprise contain one or more sequences or portions of sequences that are complementary or partially complementary with the mature strand (or strands) of the miRNA to be targeted, in addition, the miRNA inhibitor may also comprise additional sequences located 5' and 3' to the sequence that is the reverse complement of the mature miRNA.
- the additional sequences may be the reverse complements of the sequences that are adjacent to the mature miRNA in the pri-miRNA from which the mature miRNA is derived, or the additional sequences may be arbitrary sequences (having a mixture of A, G, C, or U). In some embodiments, one or both of the additional sequences are arbitrary sequences capable of forming hairpins. Thus, in some embodiments, the sequence that is the reverse complement of the miRNA is flanked on the 5' side and on the 3' side by hairpin structures.
- Micro-RNA inhibitors when double stranded, may include mismatches between nucleotides on opposite strands. Furthermore, micro-RNA inhibitors may be linked to conjugate moieties in order to facilitate uptake of the inhibitor into a cell.
- a micro-RNA inhibitor may be linked to cholesteryl 5-(bis(4-methoxyphenyl)(phenyl)methoxy)-3 hydroxypentylcarbamate) which allows passive uptake of a micro-RNA inhibitor into a cell.
- Micro-RNA inhibitors including hairpin miRNA inhibitors, are described in detail in Vermeulen et al., "Double-Stranded Regions Are Essential Design Components Of Potent Inhibitors of RISC Function," RNA 13: 723-730 (2007 ) and in WO2007/095387 and WO 2008/036825 .
- a person of ordinary skill in the art can select a sequence from the database for a desired miRNA and design an inhibitor useful for the methods disclosed herein.
- Ul adaptor inhibit polyA sites and are bifunctional oligonucleotides with a target domain complementary to a site in the target gene's terminal exon and a 'Ul domain' that binds to the Ul smaller nuclear RNA component of the Ul snRNP ( Goraczniak, et al., 2008, Nature Biotechnology, 27(3), 257-263 ).
- Ul snRNP is a ribonucleoprotein complex that functions primarily to direct early steps in spliceosome formation by binding to the pre-mRNA exon- intron boundary ( Brown and Simpson, 1998, Annu Rev Plant Physiol Plant Mol Biol 49:77-95 ).
- oligonucleotides 2-11 of the 5'end of Ul snRNA base pair bind with the 5'ss of the pre mRNA.
- oligonucleotides are Ul adaptors.
- the Ul adaptor can be administered in combination with at least one other iRNA agent.
- the invention is directed to a method for administering a therapeutic agent to a patient in need thereof, the method comprising preparing or providing any of the foregoing LNPs and/or administering a composition comprising the same to the patient.
- the therapeutic agent is effective to treat the disease.
- the lipid nanoparticles of embodiments of the present invention may be administered alone or may be formulated as pharmaceutical compositions.
- Pharmaceutical compositions of certain embodiments comprise a lipid nanoparticle according to any of the foregoing embodiments and one or more pharmaceutically acceptable carrier, diluent or excipient.
- the lipid nanoparticle may be present in an amount which is effective to deliver the therapeutic agent, e.g., for treating a particular disease or condition of interest. Appropriate concentrations and dosages can be readily determined by one skilled in the art.
- lipid nanoparticles of some embodiments can be carried out via any of the accepted modes of administration of agents for serving similar utilities.
- the pharmaceutical compositions of some embodiments may be formulated into preparations in solid, semi-solid, liquid or gaseous forms, such as tablets, capsules, powders, granules, ointments, solutions, suspensions, suppositories, injections, inhalants, gels, microspheres, and aerosols.
- Typical routes of administering such pharmaceutical compositions include, without limitation, oral, topical, transdermal, inhalation, parenteral, sublingual, buccal, rectal, vaginal, and intranasal.
- parenteral includes subcutaneous injections, intravenous, intramuscular, intradermal, intrasternal injection or infusion techniques.
- Pharmaceutical compositions of some embodiments are formulated so as to allow the active ingredients contained therein to be bioavailable upon administration of the composition to a patient.
- Compositions that may be administered to a subject or patient may take the form of one or more dosage units, where for example, a tablet may be a single dosage unit, and a container comprising LNPs in aerosol form may hold a plurality of dosage units.
- Actual methods of preparing such dosage forms are known, or will be apparent, to those skilled in this art; for example, see Remington: The Science and Practice of Pharmacy, 20th Edition (Philadelphia College of Pharmacy and Science, 2000 ).
- the composition to be administered will typically contain a therapeutically effective amount of a lipid nanoparticle of any of the embodiments disclosed herein, comprising a therapeutic agent, or a pharmaceutically acceptable salt thereof, for treatment of a disease or condition of interest.
- a pharmaceutical composition of some embodiments may be in the form of a solid or liquid.
- the carrier(s) are particulate, so that the compositions are, for example, in tablet or powder form.
- the carrier(s) may be liquid, with the compositions being, for example, an oral syrup, injectable liquid or an aerosol, which is useful in, for example, inhalatory administration.
- the pharmaceutical composition may be formulated into a powder, granule, compressed tablet, pill, capsule, chewing gum, wafer or the like form.
- a solid composition will typically contain one or more inert diluents or edible carriers.
- binders such as carboxymethylcellulose, ethyl cellulose, microcrystalline cellulose, gum tragacanth or gelatin; excipients such as starch, lactose or dextrins, disintegrating agents such as alginic acid, sodium alginate, Primogel, corn starch and the like; lubricants such as magnesium stearate or Sterotex; glidants such as colloidal silicon dioxide; sweetening agents such as sucrose or saccharin; a flavoring agent such as peppermint, methyl salicylate or orange flavoring; and a coloring agent.
- excipients such as starch, lactose or dextrins, disintegrating agents such as alginic acid, sodium alginate, Primogel, corn starch and the like
- lubricants such as magnesium stearate or Sterotex
- glidants such as colloidal silicon dioxide
- sweetening agents such as sucrose or saccharin
- a flavoring agent such as peppermint, methyl sal
- the pharmaceutical composition when in the form of a capsule, for example, a gelatin capsule, it may contain, in addition to materials of the above type, a liquid carrier such as polyethylene glycol or oil.
- a liquid carrier such as polyethylene glycol or oil.
- the pharmaceutical composition may be in the form of a liquid, for example, an elixir, syrup, solution, emulsion or suspension.
- the liquid may be for oral administration or for delivery by injection, as two examples.
- preferred composition contain, in addition to the present compounds, one or more of a sweetening agent, preservatives, dye/colorant and flavor enhancer.
- a surfactant, preservative, wetting agent, dispersing agent, suspending agent, buffer, stabilizer and isotonic agent may be included.
- the liquid pharmaceutical compositions of some embodiments may include one or more of the following adjuvants: sterile diluents such as water for injection, saline solution, preferably physiological saline, Ringer's solution, isotonic sodium chloride, fixed oils such as synthetic mono or diglycerides which may serve as the solvent or suspending medium, polyethylene glycols, glycerin, propylene glycol or other solvents; antibacterial agents such as benzyl alcohol or methyl paraben; antioxidants such as ascorbic acid or sodium bisulfite; chelating agents such as ethylenediaminetetraacetic acid; buffers such as acetates, citrates or phosphates and agents for the adjustment of tonicity such as sodium chloride or dextrose; agents to act as cryoprotectants such as sucrose or trehalose.
- the parenteral preparation can be enclosed in ampoules, disposable syringes or multiple dose vials made of
- a liquid pharmaceutical composition of certain embodiments intended for either parenteral or oral administration should contain an amount of a lipid nanoparticle of the invention such that a suitable dosage will be obtained.
- the pharmaceutical composition of embodiments of the invention may be intended for topical administration, in which case the carrier may suitably comprise a solution, emulsion, ointment or gel base.
- the base for example, may comprise one or more of the following: petrolatum, lanolin, polyethylene glycols, bee wax, mineral oil, diluents such as water and alcohol, and emulsifiers and stabilizers.
- Thickening agents may be present in a pharmaceutical composition for topical administration.
- the composition may include a transdermal patch or iontophoresis device.
- compositions of some embodiments may be intended for rectal administration, in the form, for example, of a suppository, which will melt in the rectum and release the drug.
- the composition for rectal administration may contain an oleaginous base as a suitable nonirritating excipient.
- bases include, without limitation, lanolin, cocoa butter and polyethylene glycol.
- the pharmaceutical composition of other embodiments may include various materials, which modify the physical form of a solid or liquid dosage unit.
- the composition may include materials that form a coating shell around the active ingredients.
- the materials that form the coating shell are typically inert, and may be selected from, for example, sugar, shellac, and other enteric coating agents.
- the active ingredients may be encased in a gelatin capsule.
- the pharmaceutical composition of embodiments in solid or liquid form may include an agent that binds to the LNP or therapeutic agent, and thereby assists in the delivery of the LNP or therapeutic agent.
- Suitable agents that may act in this capacity include a monoclonal or polyclonal antibody, or a protein.
- the compounds of the invention and one or more additional active agents can be administered at essentially the same time, i.e., concurrently, or at separately staggered times, i.e., sequentially; combination therapy is understood to include all these regimens.
- the indicated cationic lipid e.g. III-45
- DSPC cationic lipid
- cholesterol lipid-lipid
- PEG-lipid lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid-lipid
- LNP Lipid nanoparticles
- Figures 10 and 11 provide an expanded view of the 12 hour tissue sample, better demonstrating the difference in LNP density in the sinusoidal space.
- LNPs were formulated according to standard methods as described herein in Example 1. Control subjects receive a 5 mL/kg saline injection. Non-control animals are nominally dosed at 1.0 mg/kg RNA with a dose volume of 5 mL/kg.
- LNP formulations contained an mRNA expression vector for human immunoglobulin G, type 1 (IgG1). LNPs were synthesized according to standard methods known to those skilled in the art, or as described herein in Example 1, using cationic lipd III-45 and PEG lipid with C14 di-acyl chains as described above and size of 70 nm (LNP 9-1). Another LNP test group had the same composition but smaller LNP diameter of 54 nm (LNP 9-2). Non-control animals were dosed at 0.5 mg/kg or 2.0 mg/kg RNA with a dose volume of 5 mL/kg.
- IgG1 human immunoglobulin G
Landscapes
- Health & Medical Sciences (AREA)
- Chemical & Material Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Organic Chemistry (AREA)
- Medicinal Chemistry (AREA)
- Pharmacology & Pharmacy (AREA)
- Epidemiology (AREA)
- Animal Behavior & Ethology (AREA)
- General Health & Medical Sciences (AREA)
- Public Health (AREA)
- Veterinary Medicine (AREA)
- Engineering & Computer Science (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Dermatology (AREA)
- Dispersion Chemistry (AREA)
- Optics & Photonics (AREA)
- Physics & Mathematics (AREA)
- Biomedical Technology (AREA)
- Nanotechnology (AREA)
- General Chemical & Material Sciences (AREA)
- Molecular Biology (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Chemical Kinetics & Catalysis (AREA)
- Oil, Petroleum & Natural Gas (AREA)
- Biochemistry (AREA)
- Inorganic Chemistry (AREA)
- Pharmaceuticals Containing Other Organic And Inorganic Compounds (AREA)
- Medicinal Preparation (AREA)
- Medicines That Contain Protein Lipid Enzymes And Other Medicines (AREA)
- Micro-Organisms Or Cultivation Processes Thereof (AREA)
- Organic Low-Molecular-Weight Compounds And Preparation Thereof (AREA)
- Polyethers (AREA)
- Saccharide Compounds (AREA)
Description
- Embodiments of the present invention generally relate to lipid nanoparticles (LNPs) having improved properties. The LNPs are useful for facilitating the intracellular delivery of therapeutic agents, such as nucleic acids (e.g., oligonucleotides, messenger RNA), to primates, including humans.
- There are many challenges associated with the delivery of nucleic acids to affect a desired response in a biological system. Nucleic acid based therapeutics have enormous potential but there remains a need for more effective delivery of nucleic acids to appropriate sites within a cell or organism in order to realize this potential. Therapeutic nucleic acids include, e.g., messenger RNA (mRNA), antisense oligonucleotides, ribozymes, DNAzymes, plasmids, immune stimulating nucleic acids, antagomir, antimir, mimic, supermir, and aptamers. Some nucleic acids, such as mRNA or plasmids, can be used to effect expression of specific cellular products as would be useful in the treatment of, for example, diseases related to a deficiency of a protein or enzyme. The therapeutic applications of translatable nucleotide delivery are extremely broad as constructs can be synthesized to produce any chosen protein sequence, whether or not indigenous to the system. The expression products of the nucleic acid can augment existing levels of protein, replace missing or non-functional versions of a protein, or introduce new protein and associated functionality in a cell or organism.
- However, problems currently face the use of oligonucleotides in therapeutic contexts. First, free RNAs are susceptible to nuclease digestion in plasma. Second, free RNAs have limited ability to gain access to the intracellular compartment where the relevant translation machinery resides. Lipid nanoparticles formed from cationic lipids with other lipid components, such as neutral lipids, cholesterol, PEG, PEGylated lipids, and oligonucleotides have been used to protect the RNAs in plasma and facilitate the cellular uptake of the oligonucleotides.
-
WO2018081480A1 discloses lipid nanoparticle comprising 40-50 mol.% cationic lipid, neutral lipid, steroid, polymer conjugated lipid, and atherapeutic agent or its salt encapsulated within or associated with lipid nanoparticle. - Additionally, while lipid nanoparticle formulations have shown tremendous promise for enhancing nucleic acid therapies in both in vitro and in vivo animal models, the performance in rodent models vastly exceeds that observed in non-human primate models in nearly every measure, including toxicity and tolerability, pharmacokinetics, tissue targeting and efficacy. Notably, achieving therapeutically relevant outcomes at tolerable dose levels in primate models remains a significant challenge. Thus, there remains a need for improved lipid nanoparticles for the delivery of oligonucleotides in primates such that an efficacious and reproducible therapeutic result can be realized. Embodiments of the present disclosure provide these and related advantages.
- The subject matter of the invention is as set out in the appended claims.
- One aspect of the invention relates to lipid nanoparticles (LNPs) for use in a method of treating or preventing a disease in a primate in need thereof, wherein the method comprises administering the LNPs to the primate, each of the LNPs comprising:
- i) a nucleic acid, or a pharmaceutically acceptable salt thereof, encapsulated within the LNP;
- ii) a cationic lipid;
- iii) a neutral lipid;
- iv) a steroid; and
- v) a polymer-conjugated lipid,
- Preferred embodiments are set forth in the dependent claims.
- Any references in the description to methods of treatment refer to the compounds, pharmaceutical compositions and medicaments of the present invention for use in a method for treatment of the human (or animal) body by therapy.
- The following embodiments are not according to the invention unless they are embraced by the claims.
- Embodiments of the present disclosure provide improved lipid nanoparticles (LNPs) and methods of use of the same, for example, for delivery of nucleic acid therapeutic agents to human and/or non-human primates. In an exemplary embodiment, a method for delivering a nucleic acid to a primate in need thereof is disclosed, the method comprising administering a lipid nanoparticle (LNP) to the primate, the LNP comprising:
- i) a nucleic acid, or a pharmaceutically acceptable salt thereof, encapsulated within the LNP;
- ii) a cationic lipid;
- iii) a neutral lipid;
- iv) a steroid; and
- v) from 2.0 to 3.5 mol percent of a polymer-conjugated lipid based on total mol of lipids in the LNP.
- In other embodiments, the present disclosure is directed to a method for delivering a nucleic acid to a primate in need thereof, comprising administering a lipid nanoparticle (LNP) to the primate, the LNP comprising:
- i) a nucleic acid, or a pharmaceutically acceptable salt thereof, encapsulated within the LNP;
- ii) a cationic lipid;
- iii) a neutral lipid;
- iv) a steroid; and
- v) a polymer-conjugated lipid,
- In still more exemplary embodiments, the present disclosure provides a method for delivering a nucleic acid to a primate in need thereof, comprising administering a lipid nanoparticle (LNP) to the primate, the LNP comprising:
- i) a nucleic acid, or a pharmaceutically acceptable salt thereof, encapsulated within the LNP;
- ii) a cationic lipid;
- iii) a neutral lipid;
- iv) a steroid; and
- v) a polymer-conjugated lipid having the following structure:
- P is a polymer;
- L is a trivalent linker of 1 to 15 atoms in length; and
- R' and R" are each independently a saturated alkyl having from 8 to 14 carbon atoms, provided that the total number of carbon atoms collectively in both of R' and R" is no more than 27.
- Further embodiments are directed to improved components for lipid nanoparticles, as well as lipid nanoparticles comprising the same and use of the same. For example, one embodiment is directed to a compound having the following structure:
- These and other aspects of various embodiments will be apparent upon reference to the following detailed description.
- In the figures, identical reference numbers identify similar elements. The sizes and relative positions of elements in the figures are not necessarily drawn to scale and some of these elements are arbitrarily enlarged and positioned to improve figure legibility. Further, the particular shapes of the elements as drawn are not intended to convey any information regarding the actual shape of the particular elements, and have been solely selected for ease of recognition in the figures.
-
Figures 1 and 2 show relative concentrations of expressed luciferase in mouse liver for different embodiments of lipid nanoparticles.. -
Figure 3 and 4 show relative concentrations of expressed luciferase in mouse liver for different embodiments of lipid nanoparticles as a function of the quantity of PEG lipid in the LNP. -
Figure 5 shows levels of IgG1 present in non-human primate blood plasma for different embodiments of lipid nanoparticles. -
Figure 6 plots the concentration of amino lipid in non-human primate blood plasma for different embodiments of lipid nanoparticles. -
Figure 7 plots the concentration of amino lipids in non-human primate liver for different embodiments of lipid nanoparticles as a function of time. -
Figures 8 - 11 show in situ hybridization images demonstrating the distribution of LNPs in certain liver tissue regions for different embodiments of the LNP. -
Figure 12 shows cytokine data for monkeys treated with the LNPs of example 4. -
Figure 13 compares plasma IgG1 levels for two diferent sizes of LNPs. -
Figure 14 presents igG expression in mice for two different sizes of LNPs. -
Figure 15 is cytokine data for two different LNP sizes. -
Figure 16 shows in situ hybridization images demonstrating the distribution of LNPs in certain liver tissue regions for different sizes of LNPs. -
Figure 17 is igG expression in NHPs for two different LNPs. -
Figure 18 is igG expression in mice for two different LNPs. -
Figure 19 presents igG expression data for LNPs 10-1 and 10-2. - In the following description, certain specific details are set forth in order to provide a thorough understanding of various embodiments of the invention. However, one skilled in the art will understand that the invention may be practiced without these details.
- The invention is defined by the claims. Any subject-matter falling outside the scope of the claims is provided for information purposes only.
- In particular embodiments, the present invention provides lipid nanoparticles and methods for the in vitro and in vivo delivery of mRNA and/or other oligonucleotides. In some embodiments, these improved lipid nanoparticle compositions are useful for expression of protein encoded by mRNA. In other embodiments, these improved lipid nanoparticles are useful for upregulation of endogenous protein expression by delivering miRNA inhibitors targeting one specific miRNA or a group of miRNA regulating one target mRNA or several mRNA. In other embodiments, these improved lipid nanoparticles are useful for upregulation of endogenous protein expression by delivering smaRNA targeting a gene promotor or group of gene promotors. In other embodiments, these improved lipid nanoparticles are useful for down-regulating (e.g., silencing) the protein levels and/or mRNA levels of target genes. In some other embodiments, the lipid nanoparticles are also useful for delivery of mRNA, self amplifying RNA (saRNA) and plasmids for expression of transgenes. In yet other embodiments, the lipid nanoparticles are useful for inducing a pharmacological effect resulting from expression of a protein, e.g., increased production of red blood cells through the delivery of a suitable erythropoietin mRNA, or protection against infection through delivery of mRNA encoding for a suitable antigen or antibody. In yet other embodiments, the lipid nanoparticles can be employed in gene editing applications, for example those based on Clustered Regularly Interspaced Short Palindrome Repeats (CRISPR) methods, through the delivery of mRNA capable of expressing Cas9 in combination with an appropriate single guide RNA (sgRNA). Gene editing approaches can be used to treat, for example, hypercholesterolemia by targeting an appropriate gene target, e.g., PCSK9 in a murine model for the disease. The lipid nanoparticles of embodiments of the present invention may be used for a variety of purposes, including the delivery of encapsulated or associated (e.g., complexed) therapeutic agents such as nucleic acids to cells, both in vitro and in vivo. Accordingly, embodiments of the present invention provide a method for administering a therapeutic agent to a patient, for example a primate, in need thereof, the method comprising administering a lipid nanoparticle as described herein to the patient.
- As described herein, embodiments of the lipid nanoparticles of the present invention are particularly useful for the delivery of nucleic acids, including, e.g., mRNA, guide RNA, circular RNA, antisense oligonucleotide, plasmid DNA, closed ended DNA (ceDNA), circular DNA, microRNA (miRNA), miRNA inhibitors (antagomirs/antimirs), messenger-RNA-interfering complementary RNA (micRNA), self amplifying RNA (saRNA), small activating RNA (smaRNA), DNA, multivalent RNA, dicer substrate RNA, complementary DNA (cDNA), peptide nucleic acid (PNA) etc. Therefore, the lipid nanoparticles of embodiments of the present invention may be used to induce expression of a desired protein both in vitro and in vivo by contacting cells with a lipid nanoparticle. The expressed protein may have a biological effect, such as inducing an immune response. Alternatively, the lipid nanoparticles and compositions of embodiments of the present invention may be used to decrease the expression of target genes and proteins both in vitro and in vivo by contacting cells with a lipid nanoparticle. The lipid nanoparticles and compositions of embodiments of the present invention may also be used for co-delivery of different nucleic acids (e.g., mRNA and plasmid DNA) separately or in combination, such as may be useful to provide an effect requiring colocalization of different nucleic acids (e.g. mRNA encoding for a suitable gene modifying enzyme with an associated guide RNA sequence if applicable, and optionally, DNA segment(s) for incorporation into the host genome).
- Nucleic acids for use with embodiments of this invention may be prepared according to the techniques described herein. For mRNA, the primary methodology of preparation is, but not limited to, enzymatic synthesis (also termed in vitro transcription) which currently represents the most efficient method to produce long sequence-specific mRNA. In vitro transcription describes a process of template-directed synthesis of RNA molecules from an engineered DNA template comprised of an upstream bacteriophage promoter sequence (e.g. including but not limited to that from the T7, T3 and SP6 coliphage) linked to a downstream sequence encoding the gene of interest. Template DNA can be prepared for in vitro transcription from a number of sources with appropriate techniques which are well known in the art including, but not limited to, plasmid DNA and polymerase chain reaction amplification (see Linpinsel, J.L and Conn, G.L., General protocols for preparation of plasmid DNA template and Bowman, J.C., Azizi, B., Lenz, T.K., Ray, P., and Williams, L.D. in RNA in vitro transcription and RNA purification by denaturing PAGE in Recombinant and in vitro RNA syntheses Methods v. 941 Conn G.L. (ed), New York, N.Y. Humana Press, 2012).
- Transcription of the RNA occurs in vitro using the linearized DNA template in the presence of the corresponding RNA polymerase and adenosine, guanosine, uridine and cytidine ribonucleoside triphosphates (rNTPs) under conditions that support polymerase activity while minimizing potential degradation of the resultant mRNA transcripts. In vitro transcription can be performed using a variety of commercially available kits including, but not limited to RiboMax Large Scale RNA Production System (Promega), MegaScript Transcription kits (Life Technologies) as well as with commercially available reagents including RNA polymerases and rNTPs. The methodology for in vitro transcription of mRNA is well known in the art. (see, e.g. Losick, R., 1972, In vitro transcription, Ann Rev Biochem v.41 409-46; Kamakaka, R. T. and Kraus, W. L. 2001. In Vitro Transcription. Current Protocols in Cell Biology. 2:11.6:11.6.1-11.6.17; Beckert, B. And Masquida, B.,(2010) Synthesis of RNA by In Vitro Transcription in RNA in Methods in Molecular Biology v. 703 (Neilson, H. Ed), New York, N.Y. Humana Press, 2010; Brunelle, J.L. and Green, R., 2013, Chapter Five - In vitro transcription from plasmid or PCR-amplified DNA, Methods in Enzymology v. 530, 101-114.
- The desired in vitro transcribed mRNA is then purified from the undesired components of the transcription or associated reactions (including unincorporated rNTPs, protein enzyme, salts, short RNA oligos, etc.). Techniques for the isolation of the mRNA transcripts are well known in the art. Well known procedures include phenol/chloroform extraction or precipitation with either alcohol (e.g., ethanol, isopropanol) in the presence of monovalent cations or lithium chloride. Additional, non-limiting examples of purification procedures which can be used include size exclusion chromatography (Lukavsky, P.J. and Puglisi, J.D., 2004, Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides, RNA v.10, 889-893), silica-based affinity chromatography and polyacrylamide gel electrophoresis (Bowman, J.C., Azizi, B., Lenz, T.K., Ray, P., and Williams, L.D. in RNA in vitro transcription and RNA purification by denaturing PAGE in Recombinant and in vitro RNA syntheses Methods v. 941 Conn G.L. (ed), New York, N.Y. Humana Press, 2012 ). Purification can be performed using a variety of commercially available kits including, but not limited to SV Total Isolation System (Promega) and In Vitro Transcription Cleanup and Concentration Kit (Norgen Biotek).
- Furthermore, while reverse transcription can yield large quantities of mRNA, the products can contain a number of aberrant RNA impurities associated with undesired polymerase activity which may need to be removed from the full-length mRNA preparation. These include short RNAs that result from abortive transcription initiation as well as double-stranded RNA (dsRNA) generated by RNA-dependent RNA polymerase activity, RNA-primed transcription from RNA templates and self-complementary 3' extension. It has been demonstrated that these contaminants with dsRNA structures can lead to undesired immunostimulatory activity through interaction with various innate immune sensors in eukaryotic cells that function to recognize specific nucleic acid structures and induce potent immune responses. This in turn, can dramatically reduce mRNA translation when protein synthesis is reduced during the innate cellular immune response. Therefore, additional techniques to remove these dsRNA contaminants have been developed and are known in the art including but not limited to scaleable HPLC purification (see, e.g., Kariko, K., Muramatsu, H., Ludwig, J. and Weissman, D., 2011, Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA, Nucl Acid Res, v. 39 e142; Weissman, D., Pardi, N., Muramatsu, H., and Kariko, K., HPLC Purification of in vitro transcribed long RNA in Synthetic Messenger RNA and Cell Metabolism Modulation in Methods in Molecular Biology v.969 (Rabinovich, P.H. Ed), 2013). Purified mRNA has been reported to be translated at much greater levels, particularly in primary cells and in vivo.
- A significant variety of modifications have been described in the art which are used to alter specific properties of in vitro transcribed mRNA, and improve its utility. These include, but are not limited to modifications to the 5' and 3' termini of the mRNA. Endogenous eukaryotic mRNA typically contain a cap structure on the 5'-end of a mature molecule which plays an important role in mediating binding of the mRNA Cap Binding Protein (CBP), which is in turn responsible for enhancing mRNA stability in the cell and efficiency of mRNA translation. Therefore, highest levels of protein expression are achieved with capped mRNA transcripts. The 5'-cap contains a 5'-5'-triphosphate linkage between the 5'-most nucleotide and guanine nucleotide. The conjugated guanine nucleotide is methylated at the N7 position. Additional modifications include methylation of the ultimate and penultimate most 5'-nucleotides on the 2'-hydroxyl group.
- Multiple distinct cap structures can be used to generate the 5'-cap of in vitro transcribed synthetic mRNA. 5'-capping of synthetic mRNA can be performed co-transcriptionally with chemical cap analogs (i.e., capping during in vitro transcription). For example, CleanCap® technology provides high efficiency capping (90%+) in a co-transcriptional reaction using commercially available reagents with an AG initiator to provide a
natural Cap 1 structure with a 2'-O-methyl group and N7 methyl on separate guanine components. As another example, the Anti-Reverse Cap Analog (ARCA) cap contains a 5'-5'-triphosphate guanine-guanine linkage where one guanine contains an N7 methyl group as well as a 3'-O-methyl group. However, up to 20% of transcripts remain uncapped during this co-transcriptional process and the synthetic cap analog is not identical to the 5'-cap structure of an authentic cellular mRNA, potentially reducing translatability and cellular stability. Alternatively, synthetic mRNA molecules may also be enzymatically capped post-transcriptionally. These may generate a more authentic 5'-cap structure that more closely mimics, either structurally or functionally, the endogenous 5'-cap which have enhanced binding of cap binding proteins, increased half-life, reduced susceptibility to 5' endonucleases and/or reduced 5' decapping. Numerous synthetic 5'-cap analogs have been developed and are known in the art to enhance mRNA stability and translatability (see, e.g., Grudzien-Nogalska, E., Kowalska, J., Su, W., Kuhn, A.N., Slepenkov, S.V., Darynkiewicz, E., Sahin, U., Jemielity, J., and Rhoads, R.E., Synthetic mRNAs with superior translation and stability properties in Synthetic Messenger RNA and Cell Metabolism Modulation in Methods in Molecular Biology v.969 (Rabinovich, P.H. Ed), 2013). - On the 3'-terminus, a long chain of adenine nucleotides (poly-A tail) is normally added to mRNA molecules during RNA processing. Immediately after transcription, the 3' end of the transcript is cleaved to free a 3' hydroxyl to which poly-A polymerase adds a chain of adenine nucleotides to the RNA in a process called polyadenylation. The poly-A tail has been extensively shown to enhance both translational efficiency and stability of mRNA (see Bernstein, P. and Ross, J., 1989, Poly (A), poly (A) binding protein and the regulation of mRNA stability, Trends Bio Sci v. 14 373-377; Guhaniyogi, J. And Brewer, G., 2001, Regulation of mRNA stability in mammalian cells, Gene, v. 265, 11-23; Dreyfus, M. And Regnier, P., 2002, The poly (A) tail of mRNAs: Bodyguard in eukaryotes, scavenger in bacteria, Cell, v.111, 611-613).
- Poly (A) tailing of in vitro transcribed mRNA can be achieved using various approaches including, but not limited to, cloning of a poly (T) tract into the DNA template or by post-transcriptional addition using Poly (A) polymerase. The first case allows in vitro transcription of mRNA with poly (A) tails of defined length, depending on the size of the poly (T) tract, but requires additional manipulation of the template. The latter case involves the enzymatic addition of a poly (A) tail to in vitro transcribed mRNA using poly (A) polymerase which catalyzes the incorporation of adenine residues onto the 3'termini of RNA, requiring no additional manipulation of the DNA template, but results in mRNA with poly(A) tails of heterogeneous length. 5'-capping and 3'-poly (A) tailing can be performed using a variety of commercially available kits including, but not limited to Poly (A) Polymerase Tailing kit (EpiCenter), mMESSAGE mMACHINE T7 Ultra kit and Poly (A) Tailing kit (Life Technologies) as well as with commercially available reagents, various ARCA caps, Poly (A) polymerase, etc.
- In addition to 5' cap and 3' poly adenylation, other modifications of the in vitro transcripts have been reported to provide benefits as related to efficiency of translation and stability. It is well known in the art that pathogenic DNA and RNA can be recognized by a variety of sensors within eukaryotes and trigger potent innate immune responses. The ability to discriminate between pathogenic and self DNA and RNA has been shown to be based, at least in part, on structure and nucleoside modifications since most nucleic acids from natural sources contain modified nucleosides In contrast, in vitro synthesized RNA lacks these modifications, thus rendering it immunostimulatory which in turn can inhibit effective mRNA translation as outlined above. The introduction of modified nucleosides into in vitro transcribed mRNA can be used to prevent recognition and activation of RNA sensors, thus mitigating this undesired immunostimulatory activity and enhancing translation capacity (see e.g. Kariko, K. And Weissman, D. 2007, Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development, Curr Opin Drug Discov Devel, v.10 523-532; Pardi, N., Muramatsu, H., Weissman, D., Kariko, K., In vitro transcription of long RNA containing modified nucleosides in Synthetic Messenger RNA and Cell Metabolism Modulation in Methods in Molecular Biology v.969 (Rabinovich, P.H. Ed), 2013); Kariko, K., Muramatsu, H., Welsh, F.A., Ludwig, J., Kato, H., Akira, S., Weissman, D., 2008, Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability, Mol Ther v.16, 1833-1840. The modified nucleosides and nucleotides used in the synthesis of modified RNAs can be prepared monitored and utilized using general methods and procedures known in the art. A large variety of nucleoside modifications are available that may be incorporated alone or in combination with other modified nucleosides to some extent into the in vitro transcribed mRNA (see, e.g.,
U.S. Pub. No. 2012/0251618 ). In vitro synthesis of nucleoside-modified mRNA have been reported to have reduced ability to activate immune sensors with a concomitant enhanced translational capacity. - Other components of mRNA which can be modified to provide benefit in terms of translatability and stability include the 5' and 3' untranslated regions (UTR). Optimization of the UTRs (favorable 5' and 3' UTRs can be obtained from cellular or viral RNAs), either both or independently, have been shown to increase mRNA stability and translational efficiency of in vitro transcribed mRNA (see, e.g., Pardi, N., Muramatsu, H., Weissman, D., Kariko, K., In vitro transcription of long RNA containing modified nucleosides in Synthetic Messenger RNA and Cell Metabolism Modulation in Methods in Molecular Biology v.969 (Rabinovich, P.H. Ed), 2013).
- In addition to mRNA, other nucleic acid payloads may be used for this invention. For oligonucleotides, methods of preparation include but are not limited to chemical synthesis and enzymatic, chemical cleavage of a longer precursor, in vitro transcription as described above, etc. Methods of synthesizing DNA and RNA nucleotides are widely used and well known in the art (see, e.g., Gait, M. J. (ed.) Oligonucleotide synthesis: a practical approach, Oxford [Oxfordshire], Washington, D.C.: IRL Press, 1984; and Herdewijn, P. (ed.) Oligonucleotide synthesis: methods and applications, Methods in Molecular Biology, v. 288 (Clifton, N.J.) Totowa, N.J.: Humana Press, 2005.
- For plasmid DNA, preparation for use with embodiments of this invention commonly utilizes but is not limited to expansion and isolation of the plasmid DNA in vitro in a liquid culture of bacteria containing the plasmid of interest. The presence of a gene in the plasmid of interest that encodes resistance to a particular antibiotic (penicillin, kanamycin, etc.) allows those bacteria containing the plasmid of interest to selectively grow in antibiotic-containing cultures. Methods of isolating plasmid DNA are widely used and well known in the art (see, e.g., Heilig, J., Elbing, K. L. and Brent, R (2001) Large-Scale Preparation of Plasmid DNA. Current Protocols in Molecular Biology. 41:II:1.7:1.7.1-1.7.16; Rozkov, A., Larsson, B., Gillström, S., Björnestedt, R. and Schmidt, S. R. (2008), Large-scale production of endotoxin-free plasmids for transient expression in mammalian cell culture. Biotechnol. Bioeng., 99: 557-566; and
U.S. Pat. No. 6,197,553 B1 ). Plasmid isolation can be performed using a variety of commercially available kits including, but not limited to Plasmid Plus (Qiagen), GenJET plasmid MaxiPrep (Thermo) and PureYield MaxiPrep (Promega) kits as well as with commercially available reagents. - As used herein, the following terms have the meanings ascribed to them unless specified otherwise.
- Unless the context requires otherwise, throughout the present specification and claims, the word "comprise" and variations thereof, such as, "comprises" and "comprising" are to be construed in an open and inclusive sense, that is, as "including, but not limited to".
- Reference throughout this specification to "one embodiment" or "an embodiment" means that a particular feature, structure or characteristic described in connection with the embodiment is included in at least one embodiment of the present invention. Thus, the appearances of the phrases "in one embodiment" or "in an embodiment" in various places throughout this specification are not necessarily all referring to the same embodiment. Furthermore, the particular features, structures, or characteristics may be combined in any suitable manner in one or more embodiments.
- Unless defined otherwise, all technical and scientific terms used herein have the same meaning as is commonly understood by one of skill in the art to which this invention belongs. As used in the specification and claims, the singular form "a", "an" and "the" include plural references unless the context clearly dictates otherwise.
- The phrase "induce expression of a desired protein" refers to the ability of a nucleic acid to increase expression of the desired protein. To examine the extent of protein expression, a test sample (e.g., a sample of cells in culture expressing the desired protein) or a test mammal (e.g., a mammal such as a human or an animal model such as a rodent (e.g. mouse) or a non-human primate (e.g., monkey) model) is contacted with a nucleic acid (e.g., nucleic acid in combination with a lipid of the present invention). Expression of the desired protein in the test sample or test animal is compared to expression of the desired protein in a control sample (e.g. a sample of cells in culture expressing the desired protein) or a control mammal (e.g., a mammal such as a human or an animal model such as a rodent (e.g., mouse) or non-human primate (e.g., monkey) model) that is not contacted with or administered the nucleic acid. When the desired protein is present in a control sample or a control mammal, the expression of a desired protein in a control sample or a control mammal may be assigned a value of 1.0. In particular embodiments, inducing expression of a desired protein is achieved when the ratio of desired protein expression in the test sample or the test mammal to the level of desired protein expression in the control sample or the control mammal is greater than 1, for example, about 1.1, 1.5, 2.0. 5.0 or 10.0. When a desired protein is not present in a control sample or a control mammal, inducing expression of a desired protein is achieved when any measurable level of the desired protein in the test sample or the test mammal is detected. One of ordinary skill in the art will understand appropriate assays to determine the level of protein expression in a sample, for example dot blots, northern blots, in situ hybridization, ELISA, immunoprecipitation, enzyme function, and phenotypic assays, or assays based on reporter proteins that can produce fluorescence or luminescence under appropriate conditions.
- The phrase "inhibiting expression of a target gene" refers to the ability of a nucleic acid to silence, reduce, or inhibit the expression of a target gene. To examine the extent of gene silencing, a test sample (e.g., a sample of cells in culture expressing the target gene) or a test mammal (e.g., a mammal such as a human or an animal model such as a rodent (e.g., mouse) or a non-human primate (e.g., monkey) model) is contacted with a nucleic acid that silences, reduces, or inhibits expression of the target gene. Expression of the target gene in the test sample or test animal is compared to expression of the target gene in a control sample (e.g., a sample of cells in culture expressing the target gene) or a control mammal (e.g., a mammal such as a human or an animal model such as a rodent (e.g., mouse) or non-human primate (e.g., monkey) model) that is not contacted with or administered the nucleic acid. The expression of the target gene in a control sample or a control mammal may be assigned a value of 100%. In particular embodiments, silencing, inhibition, or reduction of expression of a target gene is achieved when the level of target gene expression in the test sample or the test mammal relative to the level of target gene expression in the control sample or the control mammal is about 95%, 90%, 85%, 80%, 75%, 70%, 65%, 60%, 55%, 50%, 45%, 40%, 35%, 30%, 25%, 20%, 15%, 10%, 5%, or 0%. In other words, the nucleic acids are capable of silencing, reducing, or inhibiting the expression of a target gene by at least about 5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100% in a test sample or a test mammal relative to the level of target gene expression in a control sample or a control mammal not contacted with or administered the nucleic acid. Suitable assays for determining the level of target gene expression include, without limitation, examination of protein or mRNA levels using techniques known to those of skill in the art, such as, e.g., dot blots, northern blots, in situ hybridization, ELISA, immunoprecipitation, enzyme function, as well as phenotypic assays known to those of skill in the art.
- An "effective amount" or "therapeutically effective amount" of an active agent or therapeutic agent such as a therapeutic nucleic acid is an amount sufficient to produce the desired effect, e.g. an increase or inhibition of expression of a target sequence in comparison to the normal expression level detected in the absence of the nucleic acid. An increase in expression of a target sequence is achieved when any measurable level is detected in the case of an expression product that is not present in the absence of the nucleic acid. In the case where the expression product is present at some level prior to contact with the nucleic acid, an in increase in expression is achieved when the fold increase in value obtained with a nucleic acid such as mRNA relative to control is about 1.05, 1.1, 1.2, 1.3, 1.4, 1.5, 1.75, 2, 2.5, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, 30, 40, 50, 75, 100, 250, 500, 750, 1000, 5000, 10000 or greater. Inhibition of expression of a target gene or target sequence is achieved when the value obtained with a nucleic acid such as antisense oligonucleotide relative to the control is about 95%, 90%, 85%, 80%, 75%, 70%, 65%, 60%, 55%, 50%, 45%, 40%, 35%, 30%, 25%, 20%, 15%, 10%, 5%, or 0%. Suitable assays for measuring expression of a target gene or target sequence include, e.g., examination of protein or RNA levels using techniques known to those of skill in the art such as dot blots, northern blots, in situ hybridization, ELISA, immunoprecipitation, enzyme function, fluorescence or luminescence of suitable reporter proteins, as well as phenotypic assays known to those of skill in the art.
- The term "nucleic acid" as used herein refers to a polymer containing at least two deoxyribonucleotides or ribonucleotides in either single- or double-stranded form and includes DNA, RNA, and hybrids thereof. DNA may be in the form of antisense molecules, plasmid DNA, cDNA, PCR products, or vectors. RNA may be in the form of small hairpin RNA (shRNA), messenger RNA (mRNA), self amplifying RNA (saRNA), small activating RNA, antisense RNA, miRNA, micRNA, multivalent RNA, dicer substrate RNA or viral RNA (vRNA), and combinations thereof. Nucleic acids include nucleic acids containing known nucleotide analogs or modified backbone residues or linkages, which are synthetic, naturally occurring, and non-naturally occurring, and which have similar binding properties as the reference nucleic acid. Examples of such analogs include, without limitation, phosphorothioates, phosphoramidates, methyl phosphonates, chiral-methyl phosphonates, 2'-O-methyl ribonucleotides, and peptide-nucleic acids (PNAs). Unless specifically limited, the term encompasses nucleic acids containing known analogues of natural nucleotides that have similar binding properties as the reference nucleic acid. Unless otherwise indicated, a particular nucleic acid sequence also implicitly encompasses conservatively modified variants thereof (e.g., degenerate codon substitutions), alleles, orthologs, single nucleotide polymorphisms, and complementary sequences as well as the sequence explicitly indicated. Specifically, degenerate codon substitutions may be achieved by generating sequences in which the third position of one or more selected (or all) codons is substituted with mixed-base and/or deoxyinosine residues (Batzer et al., Nucleic Acid Res., 19:5081 (1991); Ohtsuka et al., J. Biol. Chem., 260:2605-2608 (1985); Rossolini et al., Mol. Cell. Probes, 8:91-98 (1994)). "Nucleotides" contain a sugar deoxyribose (DNA) or ribose (RNA), a base, and a phosphate group. Nucleotides are linked together through the phosphate groups. "Bases" include purines and pyrimidines, which further include natural compounds adenine, thymine, guanine, cytosine, uracil, inosine, and natural analogs, and synthetic derivatives of purines and pyrimidines, which include, but are not limited to, modifications which place new reactive groups such as, but not limited to, amines, alcohols, thiols, carboxylates, and alkylhalides.
- The term "gene" refers to a nucleic acid (e.g., DNA or RNA) sequence that comprises partial length or entire length coding sequences necessary for the production of a polypeptide or precursor polypeptide, or provides regulation of gene expression. "Gene" can refer to both coding and non-coding (does not encode a protein sequence) sequences of nucleic acids. For example, a non-coding "gene" may be transcribed into functional RNA products, including regulatory RNA, transfer RNA (tRNA), microRNA (miRNA), and ribosomal RNA (rRNA).
- "Gene product," as used herein, refers to a product of a gene such as an RNA transcript, including coding and non-coding variants, or a polypeptide.
- The term "lipid" refers to a group of organic compounds that include, but are not limited to, esters of fatty acids and are generally characterized by being poorly soluble in water, but soluble in many organic solvents. They are usually divided into at least three classes: (1) "simple lipids," which include fats and oils as well as waxes; (2) "compound lipids," which include phospholipids and glycolipids; and (3) "derived lipids" such as steroids.
-
- A "cationic lipid" refers to a lipid capable of being positively charged. Exemplary cationic lipids include one or more amine group(s) which bear the positive charge. Preferred cationic lipids are ionizable such that they can exist in a positively charged or neutral form depending on pH. The ionization of the cationic lipid affects the surface charge of the lipid nanoparticle under different pH conditions. This charge state can influence plasma protein absorption, blood clearance and tissue distribution (Semple, S.C., et al., Adv. Drug Deliv Rev 32:3-17 (1998)) as well as the ability to form nonbilayer structures (Hafez, I.M., et al., Gene Ther 8:1188-1196 (2001)) critical to the intracellular delivery of nucleic acids.
- An "anionic lipid" refers to a lipid capable of being negatively charged. Exemplary anionic lipids include one or more phosphate group(s) which bear a negative charge, for example at physiological pHs. In some embodiments, the anionic lipid does not include a serine moiety, including phosphatidylserine lipids.
- "Phosphatidylglycerol lipid" refers to a lipid with a structure that generally comprises a glycerol 3-phosphate backbone which is attached to saturated or unsaturated fatty acids via and ester linkage. Exemplary phosphatidylglycerol lipids have the following structure:
- The term "polymer conjugated lipid" refers to a molecule comprising both a lipid portion and a polymer portion. An example of a polymer conjugated lipid is a pegylated lipid. The term "pegylated lipid" refers to a molecule comprising both a lipid portion and a polyethylene glycol portion. Pegylated lipids are known in the art and include 1-(monomethoxy-polyethyleneglycol)-2,3-dimyristoylglycerol (PEG-DMG) and the like. The term "pegylated lipid" is used interchangeably with "PEGylated lipid."
- The term "neutral lipid" refers to any of a number of lipid species that exist either in an uncharged or neutral zwitterionic form at a selected pH. At physiological pH, such lipids include, but are not limited to, phosphotidylcholines such as 1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-Dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), phophatidylethanolamines such as 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), sphingomyelins (SM), ceramides, steroids such as sterols and their derivatives. Neutral lipids may be synthetic or naturally derived. Neutral lipids include those lipids sometimes referred to as 'non-cationic' lipids.
- The term "charged lipid" refers to any of a number of lipid species that exist in either a positively charged or negatively charged form independent of the pH within a useful physiological range, e.g., pH ~3 to pH ~9. Charged lipids may be synthetic or naturally derived. Examples of charged lipids include phosphatidylserines, phosphatidic acids, phosphatidylglycerols, phosphatidylinositols, sterol hemisuccinates, dialkyl trimethylammonium-propanes, (e.g., DOTAP, DOTMA), dialkyl dimethylaminopropanes, ethyl phosphocholines, dimethylaminoethane carbamoyl sterols (e.g., DC-Chol).
- The term "lipid nanoparticle" refers to particles having at least one dimension on the order of nanometers (e.g., 1-1,000 nm) which include one or more specified lipids. In some embodiments, lipid nanoparticles are included in a formulation that can be used to deliver an active agent or therapeutic agent, such as a nucleic acid (e.g., mRNA) to a target site of interest (e.g., cell, tissue, organ, tumor, and the like). In some embodiments, the lipid nanoparticles of the invention comprise a nucleic acid. Such lipid nanoparticles typically comprise a cationic lipid and one or more excipient selected from neutral lipids, charged lipids, steroids and polymer conjugated lipids. In some embodiments, the active agent or therapeutic agent, such as a nucleic acid, may be encapsulated in the lipid portion of the lipid nanoparticle or an aqueous space enveloped by some or all of the lipid portion of the lipid nanoparticle, thereby protecting it from enzymatic degradation or other undesirable effects induced by the mechanisms of the host organism or cells, e.g., an adverse immune response.
- In various embodiments, the lipid nanoparticles have a mean diameter of from about 30 nm to about 150 nm, from about 40 nm to about 150 nm, from about 50 nm to about 150 nm, from about 60 nm to about 130 nm, from about 70 nm to about 110 nm, from about 70 nm to about 100 nm, from about 80 nm to about 100 nm, from about 90 nm to about 100 nm, from about 70 to about 90 nm, from about 80 nm to about 90 nm, from about 70 nm to about 80 nm, from about 40 nm to about 50 nm, from about 40 nm to about 60 nm, from about 40 nm to about 70 nm, from about 40 nm to about 80 nm, from about 45 nm to about 50 nm, from about 45 nm to about 55 nm, from about 45 nm to about 60 nm, from about 45 nm to about 65 nm, from about 45 nm to about 70 nm, from about 50 nm to about 70 nm, from about 50 nm to about 60 nm, from about 60 nm to about 70 nm, from about 55 nm to about 65 nm, or about 30 nm, 35 nm, 40 nm, 45 nm, 50 nm, 55 nm, 60 nm, 65 nm, 70 nm, 75 nm, 80 nm, 85 nm, 90 nm, 95 nm, 100 nm, 105 nm, 110 nm, 115 nm, 120 nm, 125 nm, 130 nm, 135 nm, 140 nm, 145 nm, or 150 nm and are substantially non-toxic (not according to the invention unless embraced by the claims). In certain embodiments, nucleic acids, when present in the lipid nanoparticles, are resistant in aqueous solution to degradation with a nuclease. Lipids and their method of preparation are disclosed in, e.g.,
U.S. Patent Nos. 8,569,256 ,5,965,542 andU.S. Patent Publication Nos. 2016/0199485 ,2016/0009637 ,2015/0273068 ,2015/0265708 ,2015/0203446 ,2015/0005363 ,2014/0308304 ,2014/0200257 ,2013/086373, 2013/0338210 ,2013/0323269 ,2013/0245107 ,2013/0195920 ,2013/0123338 ,2013/0022649 ,2013/0017223 ,2012/0295832 ,2012/0183581 ,2012/0172411 ,2012/0027803 ,2012/0058188 ,2011/0311583 ,2011/0311582 ,2011/0262527 ,2011/0216622 ,2011/0117125 ,2011/0091525 ,2011/0076335 ,2011/0060032 ,2010/0130588 ,2007/0042031 ,2006/0240093, 2006/0083780 ,2006/0008910 ,2005/0175682 ,2005/017054 ,2005/0118253 ,2005/0064595 ,2004/0142025 ,2007/0042031 ,1999/009076 andPCT Pub. Nos. WO 99/39741 WO 2017/117528 ,WO 2017/004143 ,WO 2017/075531 ,WO 2015/199952 ,WO 2014/008334 ,WO 2013/086373 ,WO 2013/086322 ,WO 2013/016058 ,WO 2013/086373 ,WO2011/141705 , andWO 2001/07548 - Other exemplary lipids and their manufacture are described in the art, for example in
U.S. Patent Application Publication No. U.S. 2012/0276209 , Semple et al., 2010, Nat Biotechnol., 28(2):172-176; Akinc et al., 2010, Mol Ther., 18(7): 1357-1364; Basha et al., 2011, Mol Ther, 19(12): 2186-2200; Leung et al., 2012, J Phys Chem C Nanomater Interfaces, 116(34): 18440-18450; Lee et al., 2012, Int J Cancer., 131(5): E781-90; Belliveau et al., 2012, Mol Ther nucleic Acids, 1: e37; Jayaraman et al., 2012, Angew Chem Int Ed Engl., 51(34): 8529-8533; Mui et al., 2013, Mol Ther Nucleic Acids. 2, e139; Maier et al., 2013, Mol Ther., 21(8): 1570-1578; and Tam et al., 2013, Nanomedicine, 9(5): 665-74. Lipids and their manufacture can be found, for example, inU.S. Pub. No. 2015/0376115 and2016/0376224 . - As used herein, "lipid encapsulated" refers to a lipid nanoparticle that provides an active agent or therapeutic agent, such as a nucleic acid (e.g., mRNA), with full encapsulation, partial encapsulation, or both. In an embodiment, the nucleic acid (e.g., mRNA) is fully encapsulated in the lipid nanoparticle.
- As used herein, the term "aqueous solution" refers to a composition comprising water.
- "Serum-stable" in relation to nucleic acid-lipid nanoparticles means that the nucleotide is not significantly degraded after exposure to a serum or nuclease assay that would significantly degrade free DNA or RNA. Suitable assays include, for example, a standard serum assay, a DNAse assay, or an RNAse assay.
- "Systemic delivery," as used herein, refers to delivery of a therapeutic product that can result in a broad exposure of an active agent within an organism. Some techniques of administration can lead to the systemic delivery of certain agents, but not others. Systemic delivery means that a useful, preferably therapeutic, amount of an agent is exposed to most parts of the body. Systemic delivery of lipid nanoparticles can be by any means known in the art including, for example, intravenous, intraarterial, subcutaneous, and intraperitoneal delivery. In some embodiments, systemic delivery of lipid nanoparticles is by intravenous delivery.
- "Local delivery," as used herein, refers to delivery of an active agent directly to a target site within an organism. For example, an agent can be locally delivered by direct injection into a disease site such as a tumor, other target site such as a site of inflammation, or a target organ such as the liver, heart, pancreas, kidney, and the like. Local delivery can also include topical applications or localized injection techniques such as intramuscular, subcutaneous or intradermal injection. Local delivery does not preclude a systemic pharmacological effect.
- "Amino acid" refers to naturally-occurring and non-naturally occurring amino acids. An amino acid lipid can be made from a genetically encoded amino acid, a naturally occurring non-genetically encoded amino acid, or a synthetic amino acid. Examples of amino acids include Ala, Arg, Asn, Asp, Cys, Gln, Glu, Gly, His, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, and Val. Examples of amino acids also include azetidine, 2-aminooctadecanoic acid, 2-aminoadipic acid, 3-aminoadipic acid, 2,3-diaminopropionic acid, 2-aminobutyric acid, 4-aminobutyric acid, 2,3-diaminobutyric acid, 2,4-diaminobutyric acid, 2-aminoisobutyric acid, 4-aminoisobutyric acid, 2-aminopimelic acid, 2,2'-diaminopimelic acid, 6-aminohexanoic acid, 6-aminocaproic acid, 2-aminoheptanoic acid, desmosine, omithine, citrulline, N-methylisoleucine, norleucine, tert-leucine, phenylglycine, t-butylglycine, N-methylglycine, sacrosine, N-ethylglycine, cyclohexylglycine, 4-oxo-cyclohexylglycine, N-ethylasparagine, cyclohexylalanine, t-butylalanine, naphthylalanine, pyridylalanine, 3-chloroalanine, 3-benzothienylalanine, 4-halophenylalanine, 4-chlorophenylalanine, 2-fluorophenylalanine, 3-fluorophenylalanine, 4-fluorophenylalanine, penicillamine, 2-thienylalanine, methionine, methionine sulfoxide, homoarginine, norarginine, nor-norarginine, N-acetyllysine, 4-aminophenylalanine, N-methylvaline, homocysteine, homoserine, hydroxylysine, allo-hydroxylysine, 3-hydroxyproline, 4-hydroxyproline, isodesmosine, allo-isoleucine, 6-N-methyllysine, norvaline, 0-allyl-serine, 0-allyl-threonine, alpha-aminohexanoic acid, alpha-aminovaleric acid, pyroglutamic acid, and derivatives thereof. "Amino acid" includes alpha- and beta- amino acids. Examples of amino acid residues can be found in Fasman, CRC Practical Handbook of Biochemistry and Molecular Biology, CRC Press, Inc. (1989).
- "Alkyl" refers to a straight or branched hydrocarbon chain radical consisting solely of carbon and hydrogen atoms, which is saturated or unsaturated (i.e., contains one or more double (alkenyl) and/or triple bonds (alkynyl)), having, for example, from one to twenty-four carbon atoms (C1-C24 alkyl), four to twenty carbon atoms (C4-C20 alkyl), six to sixteen carbon atoms (C6-C16 alkyl), six to nine carbon atoms (C6-C9 alkyl), one to fifteen carbon atoms (C1-C15 alkyl),one to twelve carbon atoms (Ci-C12 alkyl), one to eight carbon atoms (C1-C8 alkyl) or one to six carbon atoms (C1-C6 alkyl) and which is attached to the rest of the molecule by a single bond, e.g., methyl, ethyl, n propyl, 1 methylethyl (iso propyl), n butyl, n pentyl, 1,1-dimethylethyl (t butyl), 3-methylhexyl, 2-methylhexyl, ethenyl, prop-1-enyl, but-1-enyl, pent-1-enyl, penta-1,4-dienyl, ethynyl, propynyl, butynyl, pentynyl, hexynyl, and the like. Unless stated otherwise specifically in the specification, an alkyl group is optionally substituted.
- "Alkylene" or "alkylene chain" refers to a straight or branched divalent hydrocarbon chain linking the rest of the molecule to a radical group, consisting solely of carbon and hydrogen, which is saturated or unsaturated (i.e., contains one or more double (alkenylene) and/or triple bonds (alkynylene)), and having, for example, from one to twenty-four carbon atoms (C1-C24 alkylene), one to fifteen carbon atoms (C1-C15 alkylene),one to twelve carbon atoms (C1-C12 alkylene), one to eight carbon atoms (Ci-C8 alkylene), one to six carbon atoms (C1-C6 alkylene), two to four carbon atoms (C2-C4 alkylene), one to two carbon atoms (C1-C2 alkylene), e.g., methylene, ethylene, propylene, n-butylene, ethenylene, propenylene, n-butenylene, propynylene, n-butynylene, and the like. The alkylene chain is attached to the rest of the molecule through a single or double bond and to the radical group through a single or double bond. The points of attachment of the alkylene chain to the rest of the molecule and to the radical group can be through one carbon or any two carbons within the chain. Unless stated otherwise specifically in the specification, an alkylene chain may be optionally substituted.
- The term "alkenyl" refers to an alkyl, as defined above, containing at least one double bond between adjacent carbon atoms. Alkenyls include both cis and trans isomers. Representative straight chain and branched alkenyls include, but are not limited to, ethylenyl, propylenyl, 1-butenyl, 2-butenyl, isobutylenyl, 1-pentenyl, 2-pentenyl, 3-methyl-1-butenyl, 2-methyl-2-butenyl, 2,3-dimethyl-2-butenyl, and the like.
- "Alkoxy" refers to an alkyl, cycloalkyl, alkenyl, or alkynyl group covalently bonded to an oxygen atom.
- "Alkanoyloxy" refers to -O-C(=O)-alkyl groups.
- "Alkylamino" refers to the group -NRR', where R and R' are each either hydrogen or alkyl, and at least one of R and R' is alkyl. Alkylamino includes groups such as piperidino wherein R and R' form a ring. The term "alkylaminoalkyl" refers to -alkyl-NRR'.
- The term "alkynyl" includes any alkyl or alkenyl, as defined above, which additionally contains at least one triple bond between adjacent carbons. Representative straight chain and branched alkynyls include, without limitation, acetylenyl, propynyl, 1-butynyl, 2-butynyl, 1-pentynyl, 2-pentynyl, 3-methyl-1 butynyl, and the like.
- The terms "acyl," "carbonyl," and "alkanoyl" refer to any alkyl, alkenyl, or alkynyl wherein the carbon at the point of attachment is substituted with an oxo group, as defined below. The following are non-limiting examples of acyl, carbonyl or alkanoyl groups: -C(=O)alkyl, -C(=O)alkenyl, and -C(=O)alkynyl.
- "Aryl" refers to any stable monocyclic, bicyclic, or polycyclic carbon ring system of from 4 to 12 atoms in each ring, wherein at least one ring is aromatic. Some examples of an aryl include phenyl, naphthyl, tetrahydro-naphthyl, indanyl, and biphenyl. Where an aryl substituent is bicyclic and one ring is non-aromatic, it is understood that attachment is to the aromatic ring. An aryl may be substituted or unsubstituted.
- "Carboxyl" refers to a functional group of the formula -C(=O)OH.
- "Cyano" refers to a functional group of the formula -CN.
- "Cycloalkyl" or "carbocyclic ring" refers to a stable non-aromatic monocyclic or polycyclic hydrocarbon radical consisting solely of carbon and hydrogen atoms, which may include fused or bridged ring systems, having from three to fifteen carbon atoms, preferably having from three to ten carbon atoms, and which is saturated or unsaturated and attached to the rest of the molecule by a single bond. Monocyclic radicals include, for example, cyclopropyl, cyclobutyl, cyclopentyl, cyclohexyl, cycloheptyl, and cyclooctyl. Polycyclic radicals include, for example, adamantyl, norbornyl, decalinyl, 7,7-dimethyl-bicyclo[2.2.1]heptanyl, and the like. Unless otherwise stated specifically in the specification, a cycloalkyl group may be optionally substituted.
- "Cycloalkylene" is a divalent cycloalkyl group. Unless otherwise stated specifically in the specification, a cycloalkylene group may be optionally substituted.
- The term "diacylglycerol" or "DAG" includes a compound having 2 fatty acyl chains, both of which have independently between 2 and 30 carbons bonded to the 1- and 2-position of glycerol by ester linkages. The acyl groups can be saturated or have varying degrees of unsaturation. Suitable acyl groups include, but are not limited to, lauroyl (C12), myristoyl (C14), palmitoyl (C16), stearoyl (C18), and icosoyl (C20). In preferred embodiments, the fatty acid acyl chains of one compound are the same, i.e., both myristoyl (i.e., dimyristoyl), both stearoyl (i.e., distearoyl), etc.
- The term "heterocycle" or "heterocyclyl" refers to an aromatic or nonaromatic ring system of from five to twenty-two atoms, wherein from 1 to 4 of the ring atoms are heteroatoms selected from oxygen, nitrogen, and sulfur. Thus, a heterocycle may be a heteroaryl or a dihydro or tetrathydro version thereof. Heterocycles include, but are not limited to, pyrrolidine, tetryhydrofuran, thiolane, azetidine, oxetane, thietane, diazetidine, dioxetane, dithietane, piperidine, tetrahydrofuran, pyran, tetrahydropyran, thiacyclohexane, tetrahydrothiophene, pyridine, pyrimidine and the like.
- "Heteroaryl" refers to any stable monocyclic, bicyclic, or polycyclic carbon ring system of from 4 to 12 atoms in each ring, wherein at least one ring is aromatic and contains from 1 to 4 heteroatoms selected from oxygen, nitrogen and sulfur. Some examples of a heteroaryl include acridinyl, quinoxalinyl, pyrazolyl, indolyl, benzotriazolyl, furanyl, thienyl, benzothienyl, benzofuranyl, quinolinyl, isoquinolinyl, oxazolyl, isoxazolyl, pyrazinyl, pyridazinyl, pyridinyl, pyrimidinyl, pyrrolyl, and tetrahydroquinolinyl. A heteroaryl includes the N-oxide derivative of a nitrogen-containing heteroaryl.
- The terms "alkylamine" and "dialkylamine" refer to ---NH(alkyl) and ---N(alkyl)2 radicals respectively.
- The term "alkylphosphate" refers to ---O---P(Q')(Q")-O---R, wherein Q' and Q" are each independently O, S, N(R)2, optionally substituted alkyl or alkoxy; and R is optionally substituted alkyl, ω-aminoalkyl or ω-(substituted)aminoalkyl.
- The term "alkylphosphorothioate" refers to an alkylphosphate wherein at least one of Q' or Q" is S.
- The term "alkylphosphonate" refers to an alkylphosphate wherein at least one of Q' or Q" is alkyl.
- "Hydroxyalkyl" refers to an ---O-alkyl radical.
- The term "alkylheterocycle" refers to an alkyl where at least one methylene has been replaced by a heterocycle.
- The term "ω-aminoalkyl" refers to -alkyl-NH2 radical. And the term "ω-(substituted)aminoalkyl refers to an ω-aminoalkyl wherein at least one of the H on N has been replaced with alkyl.
- The term "ω-phosphoalkyl" refers to -alkyl-O---P(Q')(Q")-O---R, wherein Q' and Q" are each independently O or S and R optionally substituted alkyl.
- The term "ω-thiophosphoalkyl" refers to ω-phosphoalkyl wherein at least one of Q' or Q" is S.
- The term "substituted" used herein means any of the above groups (e.g., alkyl, alkylene, cycloalkyl or cycloalkylene) wherein at least one hydrogen atom is replaced by a bond to a non-hydrogen atom such as, but not limited to: a halogen atom such as F, Cl, Br, or I; oxo groups (=O); hydroxyl groups (-OH); C1-C12 alkyl groups; cycloalkyl groups; -(C=O)OR'; -O(C=O)R'; -C(=O)R'; -OR'; -S(O)xR'; -S-SR'; -C(=O)SR'; -SC(=O)R'; -NR'R'; -NR'C(=O)R'; -C(=O)NR'R'; -NR'C(=O)NR'R'; -OC(=O)NR'R'; -NR'C(=O)OR'; -NR'S(O)xNR'R'; -NR'S(O)xR'; and -S(O)xNR'R', wherein: R' is, at each occurrence, independently H, C1-C15 alkyl or cycloalkyl, and x is 0, 1 or 2. In some embodiments the substituent is a C1-C12 alkyl group. In other embodiments, the substituent is a cycloalkyl group. In other embodiments, the substituent is a halo group, such as fluoro. In other embodiments, the substituent is an oxo group. In other embodiments, the substituent is a hydroxyl group. In other embodiments, the substituent is an alkoxy group (-OR). In other embodiments, the substituent is a carboxyl group. In other embodiments, the substituent is an amine group(-NR'R').
- "Optional" or "optionally" (e.g., optionally substituted) means that the subsequently described event of circumstances may or may not occur, and that the description includes instances where said event or circumstance occurs and instances in which it does not. For example, "optionally substituted alkyl" means that the alkyl radical may or may not be substituted and that the description includes both substituted alkyl radicals and alkyl radicals having no substitution.
- "Prodrug" is meant to indicate a compound, such as a therapeutic agent, that may be converted under physiological conditions or by solvolysis to a biologically active compound of the invention. Thus, the term "prodrug" refers to a metabolic precursor of a compound of the invention that is pharmaceutically acceptable. A prodrug may be inactive when administered to a subject in need thereof, but is converted in vivo to an active compound of the invention. Prodrugs are typically rapidly transformed in vivo to yield the parent compound of the invention, for example, by hydrolysis in blood. The prodrug compound often offers advantages of solubility, tissue compatibility or delayed release in a mammalian organism (see, Bundgard, H., Design of Prodrugs (1985), pp. 7-9, 21-24 (Elsevier, Amsterdam)). A discussion of prodrugs is provided in Higuchi, T., et al., A.C.S. Symposium Series, Vol. 14, and in Bioreversible Carriers in Drug Design, Ed. Edward B. Roche, American Pharmaceutical Association and Pergamon Press, 1987.
- The term "prodrug" is also meant to include any covalently bonded carriers, which release the active compound of the invention in vivo when such prodrug is administered to a mammalian subject. Prodrugs (e.g., a prodrug of a therapeutic agent) may be prepared by modifying functional groups present in the compound of the invention in such a way that the modifications are cleaved, either in routine manipulation or in vivo, to the parent compound of the invention. Prodrugs include compounds wherein a hydroxy, amino or mercapto group is bonded to any group such that, when the prodrug is administered to a mammalian subject, cleaves to form a free hydroxy, free amino or free mercapto group, respectively. Examples of prodrugs include, but are not limited to, acetate, formate and benzoate derivatives of alcohol or amide derivatives of amine functional groups in the therapeutic agents of the invention and the like.
- Embodiments of the invention disclosed herein are also meant to encompass all pharmaceutically acceptable lipid nanoparticles and components thereof (e.g., cationic lipid, therapeutic agent, etc.) being isotopically-labelled by having one or more atoms replaced by an atom having a different atomic mass or mass number. Examples of isotopes that can be incorporated into the disclosed compounds include isotopes of hydrogen, carbon, nitrogen, oxygen, phosphorous, fluorine, chlorine, and iodine, such as 2H, 3H, 11C, 13C, 14C, 13N, 15N, 15O, 17O, 18O, 31P, 32P, 35S, 18F, 36Cl, 123I, and 125I, respectively. These radiolabeled LNPs could be useful to help determine or measure the effectiveness of the compounds, by characterizing, for example, the site or mode of action, or binding affinity to pharmacologically important site of action. Certain isotopically-labelled LNPs, for example, those incorporating a radioactive isotope, are useful in drug and/or substrate tissue distribution studies. The radioactive isotopes tritium, i.e., 3H, and carbon-14, that is, 14C, are particularly useful for this purpose in view of their ease of incorporation and ready means of detection.
- Substitution with heavier isotopes such as deuterium, that is, 2H, may afford certain therapeutic advantages resulting from greater metabolic stability, for example, increased in vivo half-life or reduced dosage requirements, and hence may be preferred in some circumstances.
- Substitution with positron emitting isotopes, such as 11C, 18F, 15O and 13N, can be useful in Positron Emission Topography (PET) studies for examining substrate receptor occupancy. Isotopically-labeled compounds of used in the present disclosure can generally be prepared by conventional techniques known to those skilled in the art or by processes analogous to those described in the Examples as set out below using an appropriate isotopically-labeled reagent in place of the non-labeled reagent previously employed.
- "Stable compound" and "stable structure" are meant to indicate a compound that is sufficiently robust to survive isolation to a useful degree of purity from a reaction mixture, and formulation into an efficacious therapeutic agent.
- "Mammal" includes humans and both domestic animals such as laboratory animals and household pets (e.g., cats, dogs, swine, cattle, sheep, goats, horses, rabbits), and non-domestic animals such as wildlife and the like. "Primate" includes both human and non-human primates.
- "Pharmaceutically acceptable carrier, diluent or excipient" includes without limitation any adjuvant, carrier, excipient, glidant, sweetening agent, diluent, preservative, dye/colorant, flavor enhancer, surfactant, wetting agent, dispersing agent, suspending agent, stabilizer, isotonic agent, solvent, or emulsifier which has been approved by the United States Food and Drug Administration as being acceptable for use in humans or domestic animals.
- "Pharmaceutically acceptable salt" includes both acid and base addition salts.
- "Pharmaceutically acceptable acid addition salt" refers to those salts which retain the biological effectiveness and properties of the free bases, which are not biologically or otherwise undesirable, and which are formed with inorganic acids such as, but are not limited to, hydrochloric acid, hydrobromic acid, sulfuric acid, nitric acid, phosphoric acid and the like, and organic acids such as, but not limited to, acetic acid, 2,2-dichloroacetic acid, adipic acid, alginic acid, ascorbic acid, aspartic acid, benzenesulfonic acid, benzoic acid, 4-acetamidobenzoic acid, camphoric acid, camphor-10-sulfonic acid, capric acid, caproic acid, caprylic acid, carbonic acid, cinnamic acid, citric acid, cyclamic acid, dodecylsulfuric acid, ethane-1,2-disulfonic acid, ethanesulfonic acid, 2-hydroxyethanesulfonic acid, formic acid, fumaric acid, galactaric acid, gentisic acid, glucoheptonic acid, gluconic acid, glucuronic acid, glutamic acid, glutaric acid, 2-oxo-glutaric acid, glycerophosphoric acid, glycolic acid, hippuric acid, isobutyric acid, lactic acid, lactobionic acid, lauric acid, maleic acid, malic acid, malonic acid, mandelic acid, methanesulfonic acid, mucic acid, naphthalene-1,5-disulfonic acid, naphthalene-2-sulfonic acid, 1-hydroxy-2-naphthoic acid, nicotinic acid, oleic acid, orotic acid, oxalic acid, palmitic acid, pamoic acid, propionic acid, pyroglutamic acid, pyruvic acid, salicylic acid, 4-aminosalicylic acid, sebacic acid, stearic acid, succinic acid, tartaric acid, thiocyanic acid, p-toluenesulfonic acid, trifluoroacetic acid, undecylenic acid, and the like.
- "Pharmaceutically acceptable base addition salt" refers to those salts which retain the biological effectiveness and properties of the free acids, which are not biologically or otherwise undesirable. These salts are prepared from addition of an inorganic base or an organic base to the free acid. Salts derived from inorganic bases include, but are not limited to, the sodium, potassium, lithium, ammonium, calcium, magnesium, iron, zinc, copper, manganese, aluminum salts and the like. Preferred inorganic salts are the ammonium, sodium, potassium, calcium, and magnesium salts. Salts derived from organic bases include, but are not limited to, salts of primary, secondary, and tertiary amines, substituted amines including naturally occurring substituted amines, cyclic amines and basic ion exchange resins, such as ammonia, isopropylamine, trimethylamine, diethylamine, triethylamine, tripropylamine, diethanolamine, ethanolamine, deanol, 2-dimethylaminoethanol, 2-diethylaminoethanol, dicyclohexylamine, lysine, arginine, histidine, caffeine, procaine, hydrabamine, choline, betaine, benethamine, benzathine, ethylenediamine, glucosamine, methylglucamine, theobromine, triethanolamine, tromethamine, purines, piperazine, piperidine, N-ethylpiperidine, polyamine resins and the like. Particularly preferred organic bases are isopropylamine, diethylamine, ethanolamine, trimethylamine, dicyclohexylamine, choline and caffeine.
- A "pharmaceutical composition" refers to a formulation of an LNP of the invention and a medium generally accepted in the art for the delivery of the biologically active compound to mammals, e.g., humans. Such a medium includes all pharmaceutically acceptable carriers, diluents or excipients therefor.
- "Effective amount" or "therapeutically effective amount" refers to that amount of a compound of the invention which, when administered to a mammal, preferably a human, is sufficient to effect treatment in the mammal, preferably a human. The amount of a lipid nanoparticle of the invention which constitutes a "therapeutically effective amount" will vary depending on the compound, the condition and its severity, the manner of administration, and the age of the mammal to be treated, but can be determined routinely by one of ordinary skill in the art having regard to his own knowledge and to this disclosure.
- "Treating" or "treatment" as used herein covers the treatment of the disease or condition of interest in a mammal, preferably a human, having the disease or condition of interest, and includes:
- (i) preventing the disease or condition from occurring in a mammal, in particular, when such mammal is predisposed to the condition but has not yet been diagnosed as having it;
- (ii) inhibiting the disease or condition, i.e., arresting its development;
- (iii) relieving the disease or condition, i.e., causing regression of the disease or condition; or
- (iv) relieving the symptoms resulting from the disease or condition, i.e., relieving pain without addressing the underlying disease or condition. As used herein, the terms "disease" and "condition" may be used interchangeably or may be different in that the particular malady or condition may not have a known causative agent (so that etiology has not yet been worked out) and it is therefore not yet recognized as a disease but only as an undesirable condition or syndrome, wherein a more or less specific set of symptoms have been identified by clinicians.
- Embodiments disclosed herein are directed to methods of using LNPs for delivery of a therapeutic agent, such as a nucleic acid, to a primate, such as a human, for treatment of various diseases treatable with the nucleic acid. The present Applicant has discovered that the disclosed methods are surprisingly more effective for delivery of therapeutic agents to primates, compared with delivery of the same therapeutic agent to a non-primate, such as a mouse. For example, some methods include use of LNPs having a diameter smaller than typical LNPs, for example a mean particle diameter ranging from about 40-70 nm, or for instance, a mean particle diameter ranging from about 50-70 nm, and such LNPs have unexpectedly improved delivery in primates relative to rodent. Other methods comprise use of LNPs with higher concentrations of PEGylated lipid (e.g., from about 2.0 to 3.5%). Othere exemplary methods comprise delivering LNPs to primates, wherein the LNPs include a PEGylated lipid having two acyl chains independently comprising from 8 to 14 carbon atoms, with the sum of the carbon atoms in the acyl chains not exceeding 27. The LNPs can be delivered intraveneously or via other administration routes known in the art. Further details of these exemplary embodiments, and others, will be apparent in view of the details described herein.
- Accordingly, in one embodiment is provided a method for delivering a nucleic acid to a primate in need thereof, comprising administering a lipid nanoparticle (LNP) to the primate, the LNP comprising:
- i) a nucleic acid, or a pharmaceutically acceptable salt thereof, encapsulated within the LNP;
- ii) a cationic lipid;
- iii) a neutral lipid;
- iv) a steroid; and
- v) from 2.0 to 3.5 mol percent of a polymer-conjugated lipid based on total mol of lipids in the LNP.
- The mol percent of polymer-conjugated lipid is determined based on the total mol percent of lipid present in the LNP. For this calculation, all lipid components, including for example, cationic lipid, neutral lipid, steroid and any other lipids, such as anionic or other lipids, are included in the calculation.
- In certain embodiments, the LNP comprises from 2.0 to 3.4 mol of the polymer conjugated lipid. In other embodiments, the LNP comprises from 2.1 to 3.5 mol of the polymer conjugated lipid. In more embodiments, the LNP comprises from 2.2 to 3.3 mol percent of the polymer-conjugated lipid, for example 2.3 to 2.8 mol percent of the polymer-conjugated lipid. In other embodiments, the LNP comprises from 2.1 to 2.5 mol percent of the polymer-conjugated lipid. In other different embodiments, the LNP comprises from 2.5 to 2.9 mol percent of the polymer-conjugated lipid. In other embodiments, the LNP comprises from 2.4 to 2.6 mol percent of the polymer conjugated lipid, from 2.6 to 2.8 mol percent of the polymer conjugated lipid, from 2.4 to 2.5 mol percent of the polymer conjugated lipid or from 2.5 to 2.7 mol percent of the polymer conjugated lipid. In still different embodiments, the LNP comprises about 2.3, about 2.35, about 2.4, about 2.45, about 2.5, about 2.55, about 2.6, about 2.65 about 2.7, about 2.75 or about 2.8 mol percent of the polymer-conjugated lipid.
- Another embodiment is directed to a method for delivering a nucleic acid to a primate in need thereof, comprising administering a lipid nanoparticle (LNP) to the primate, the LNP comprising:
- i) a nucleic acid, or a pharmaceutically acceptable salt thereof, encapsulated within the LNP;
- ii) a cationic lipid;
- iii) a neutral lipid;
- iv) a steroid; and
- v) a polymer-conjugated lipid,
- In certain embodiments, the mean particle diameter ranges from 45 nm to 70 nm, 50 nm to 70 nm, 55 nm to 65 nm, from 50 nm to 60 nm or from 60 nm to 70 nm. In different embodiments, the mean particle diameter ranges from 45 nm to 50 nm, 50 nm to 55 nm, from 55 nm to 60 nm, from 60 nm to 65 nm or from 65 nm to 70 nm. In still more embodiments, the mean particle diameter is about 45 nm, 46 nm, 47 nm, 48 nm, 49 nm, 50 nm, about 51 nm, about 52 nm, about 53 nm, about 54 nm, about 55 nm, about 56 nm, about 57 nm, about 58 nm, about 59 nm, about 60 nm, about 61 nm, about 62 nm, about 63 nm, about 64 nm or about 65 nm, about 66 nm, about 67 nm, about 68 nm, about 69 nm or about 70 nm (not according to the invention unless embraced by the claims).
-
- P is a polymer;
- L is a trivalent linker of 1 to 15 atoms in length; and
- R' and R" are each independently a saturated alkyl having from 8 to 14 carbon atoms.
- In some embodiments, P comprises a polyethylene glycol polymer, for example a hydroxyl or alkoxyl-terminating (PEG-OR) polyethylene glycol polymer. A hydroxyl-terminating polyethylene glycol polymer (PEG-OH) is a polyethylene glycol polymer which terminates with a hydroxyl group, while an alkoxyl-terminating polyethylene glycol polymer (PEG-OR) is a polyethylene glycol polymer which terminates with an alkoxyl group, such as methoxy.
- Any suitable linker can be used for L. In some exemplary embodiments, L comprises amide, ester and/or carbamate functional groups. For example, in some embodiments the polymer conjugated lipid has one of the following structures:
- In other more specific embodiments, the polymer conjugated lipid has the following structure:
-
- R3 is -ORO;
- RO is hydrogen or alkyl;
- r is an integer from 30 to 60, inclusive;
- R5 is C10-20 alkyl.
- For example, in certain embodiments:
- R3 is OH or OCH3;
- R5 is C18, C19 or C20; and
- r is selected such that
- In yet other embodiments is provided a method for delivering a nucleic acid to a primate in need thereof, comprising administering a lipid nanoparticle (LNP) to the primate, the LNP comprising:
- i) a nucleic acid, or a pharmaceutically acceptable salt thereof, encapsulated within the LNP;
- ii) a cationic lipid;
- iii) a neutral lipid;
- iv) a steroid; and
- v) a polymer-conjugated lipid having the following structure:
- P is a polymer;
- L is a trivalent linker of 1 to 15 atoms in length; and
- R' and R" are each independently a saturated alkyl having from 8 to 14 carbon atoms, provided that the total number of carbon atoms collectively in both of R' and R" is no more than 27.
- In certain embodiments of the foregoing, P comprises a polyethylene glycol polymer, such as a hydroxyl or alkoxyl-terminating polyethylene glycol polymer.
-
-
- In certain of the foregoing embodiments, the total number of carbon atoms in R' and R" ranges from 16 to 25, 16 to 24, 17 to 24 or 18 to 24. For example, in some embodiments:
- a) R' and R" are each a saturated alkyl having 8 carbon atoms;
- b) R' and R" are each a saturated alkyl having 9 carbon atoms;
- c) R' and R" are each a saturated alkyl having 10 carbon atoms;
- d) R' and R" are each a saturated alkyl having 11 carbon atoms;
- e) R' and R" are each a saturated alkyl having 12 carbon atoms; or
- f) R' and R" are each a saturated alkyl having 13 carbon atoms.
- Asymmetric polymer conjugated lipids, wherein R' and R" are different are also included in various embodiments, such as wherein R' is 12 and R" is 13, or R' is 13 and R" is 14, or R' is 11 and R" is 12, or R' is 10 and R" is 11 and the like
- In some embodiments, the lipid nanoparticle comprises a cationic lipid, a PEGylated lipid, a sterol and a neutral lipid. In some embodiments, the lipid nanoparticle comprises a molar ratio of about 20-60% cationic lipid: 5-25% neutral lipid: 25-55% sterol; and 0.1-15% PEGylated lipid. In some embodiments, the cationic lipid is an ionizable cationic lipid. In some embodiments, the neutral lipid is a phospholipid. In some embodiments, the sterol is a cholesterol. In some embodiments, the cationic lipid is selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319). In some embodiments, the lipid nanoparticle has a polydispersity value of less than 0.4. In some embodiments, the lipid nanoparticle has a net neutral charge at a neutral pH. In some embodiments, the lipid nanoparticle has a mean diameter of 40-200 nm..
- Lipid nanoparticles may comprise one or more lipid species, including, but not limited to, cationic/ionizable lipids, neutral lipids, structural lipids, phospholipids, and helper lipids. Any of these lipids may be conjugated to polyethylene glycol (PEG) and thus may be referred to as PEGylated lipids or PEG-modified lipids.
- The formation of the lipid nanoparticle (LNP) may be accomplished by methods known in the art and/or as described in
U.S. Pub. No. 2012/0178702 . - A lipid nanoparticle formulation may be influenced by, but not limited to, the selection of the cationic lipid component, the degree of cationic lipid saturation, the selection of the neutral lipid component, the degree of neutral lipid saturation, the selection of the structural lipid component, the nature of the PEGylation, ratio of all components and biophysical parameters such as size. In certain non-limiting examples, a LNP comprises four basic components: (1) a cationic lipid; (2) a neutral lipid (e.g., a phospholipid such as DSPC); (3) a structural lipid (e.g., a sterol such as cholesterol); and (4) a PEGylated lipid. In one example by Semple et al. (Nature Biotech. 2010 28:172-176), the lipid nanoparticle formulation is composed of molar ratios as follows: 57.1% cationic lipid, 7.1% dipalmitoylphosphatidylcholine, 34.3% cholesterol, and 1.4% PEG-c-DMA. As another example, changing the composition of the cationic lipid can more effectively deliver siRNA to various antigen presenting cells (Basha et al., Mol Ther. 2011 19:2186-2200).
- In certain embodiments, the lipid nanoparticle comprises a cationic lipid and a neutral lipid. In certain embodiments, the LNP comprises a cationic lipid and a DSPC substitute. In certain embodiments, the LNP comprises a cationic lipid and a fatty acid. In certain embodiments, the LNP a cationic lipid and oleic acid. In certain embodiments, the LNP comprises a cationic lipid and an analog of oleic acid.
- In certain embodiments, the lipid nanoparticle formulation comprises a cationic lipid, a neutral lipid, and a structural lipid. In certain embodiments, the LNP comprises a cationic lipid, a fatty acid, and a structural lipid. In certain embodiments, the LNP comprises a cationic lipid, oleic acid, and a structural lipid. In certain embodiments, the LNP comprises a cationic lipid, an analog of oleic acid, and a structural lipid. In certain embodiments, the LNP comprises a cationic lipid, a fatty acid, and a sterol. In certain embodiments, the LNP comprises a cationic lipid, oleic acid, and a sterol. In certain embodiments, the LNP comprises a cationic lipid, oleic acid, and cholesterol.
- In certain embodiments, the lipid nanoparticle comprises a cationic lipid, a neutral lipid, and a PEGylated lipid. In certain embodiments, the LNP formulation comprises a cationic lipid, a neutral lipid, and a PEG-OH lipid. In certain embodiments, the lipid nanoparticle comprises a cationic lipid, a fatty acid, and a PEG-OH lipid. In certain embodiments, the lipid nanoparticle comprises a cationic lipid, oleic acid, and a PEG-OH lipid. In certain embodiments, the lipid nanoparticle comprises a cationic lipid, an analog of oleic acid, and a PEG-OH lipid.
- In certain embodiments, the lipid nanoparticle comprises a cationic lipid, a neutral lipid (e.g., a phospholipid or fatty acid), a structural lipid, and a PEG lipid. In certain embodiments, the lipid nanoparticle formulation comprises a cationic lipid, a neutral lipid (e.g., phospholipid or fatty acid), a structural lipid, and a PEG-OH lipid. In certain embodiments, the LNP comprises a cationic lipid, a neutral lipid (e.g., phospholipid or fatty acid), and a structural lipid. In certain embodiments, the LNP comprises a cationic lipid, a fatty acid (e.g., oleic acid or an analog thereof), a structural lipid, and a PEG lipid. In certain embodiments, the LNP comprises a cationic lipid, a fatty acid (e.g., oleic acid or an analog thereof), a structural lipid, and a PEG-OH lipid. In certain embodiments, the LNP comprises a cationic lipid, oleic acid, a structural lipid (e.g., a sterol), and a PEG-OH lipid. In certain embodiments, the LNP comprises a cationic lipid, oleic acid, and a structural lipid (e.g., cholesterol). In certain embodiments, the LNP comprises one or more cationic or neutral lipids, a fatty acid (e.g., oleic acid), and a PEG lipid. In certain embodiments, the LNP comprises one or more cationic or neutral lipids, a fatty acid (e.g., oleic acid), and a PEG-OH lipid.
- In some embodiments, the LNP comprises a fatty acid. In certain embodiments, the fatty acid is a monounsaturated fatty acid. In certain embodiments, the fatty acid is a polyunsaturated fatty acid. In some embodiments, the LNP comprises oleic acid. In certain embodiments, the LNP comprises one or more cationic or neutral lipids, and a fatty acid (e.g., oleic acid). In certain embodiments, the LNP comprises one or more cationic or neutral lipids, and oleic acid. In certain embodiments, when the LNP includes oleic acid, the LNP does not include a phospholipid. In certain embodiments, when the LNP includes oleic acid, the LNP does not include DSPC. In certain embodiments, when the LNP includes a fatty acid, the LNP does not include a phospholipid. In certain embodiments, when the LNP includes a fatty acid, the LNP does not include DSPC.
- In some embodiments, LNPs may comprise, in molar percentages, 35 to 45% cationic lipid, 40% to 50% cationic lipid, 45% to 55% cationic lipid, 50% to 60% cationic lipid and/or 55% to 65% cationic lipid. In some embodiments, the ratio of lipid to nucleic acid (e.g., mRNA) in lipid nanoparticles may be 5:1 to 20:1, 10:1 to 25:1, 15:1 to 40:1, 20:1 to 30:1, 25:1 to 50:1, 30:1 to 60:1 and/or at least 40:1.
- In some embodiments, the ratio of PEG in the LNPs may be increased or decreased and/or the carbon chain length of the alkyl portion of the PEG lipid may be varied from C8 to C18 (eight to eighteen carbons) to alter the pharmacokinetics and/or biodistribution of the LNPs. In certain embodiments, LNPs may contain 0.1% to 3.0%, 1.0% to 3.5%, 1.5% to 4.0%, 2.0% to 4.5%, 2.0% to 3.0%, 2.5% to 5.0%, and/or 3.0% to 6.0% of PEGylated lipid relative to the other components. As a non-limiting example, LNPs may contain 0.5% to 3.0%, 1.0% to 3.5%, 1.5% to 4.0%, 2.0% to 4.5%, 2.0% to 3.0%, 2.5% to 5.0%, and/or 3.0% to 6.0% of PEG-c-DOMG (R-3-[(ω-methoxy-poly(ethyleneglycol)2000)carbamoyl)]-1,2-dimyristyloxypropyl-3-amine) (also referred to herein as PEG-DOMG) as compared to the cationic lipid, DSPC, and cholesterol. In some embodiments, the PEG-c-DOMG may be replaced with a PEG lipid such as, but not limited to, PEG-DSG (1,2-distearoyl-sn-glycerol, methoxypolyethylene glycol), DMG-PEG (1,2-dimyristoyl-sn-glycerol) and/or PEG-DPG (1,2-dipalmitoyl-sn-glycerol, methoxypolyethylene glycol). The cationic lipid may be selected from any lipid known in the art such as, but not limited to, DLin-MC3-DMA, DLin-DMA, C12-200, and DLin-KC2-DMA. In certain embodiments, the lipid nanoparticle does not contain a PEG lipid. In certain embodiments, the lipid nanoparticle contains a PEG lipid such as a PEG-OH lipid. Incorporation of PEG-OH lipids in the nanoparticle formulation can improve the pharmacokinetics and/or biodistribution of the LNPs. For example, incorporation of PEG-OH lipids in the nanoparticle formulation can reduce the ABC effect. In certain embodiments, LNPs may contain 0.5% to 3.0%, 1.0% to 3.5%, 1.5% to 4.0%, 2.0% to 4.5%, 2.0% to 5.0%, 2.5% to 5.0%, and/or 3.0% to 6.0% of the lipid molar ratio of PEG-OH lipid to the other components (e.g., the cationic, neutral, and structural lipids). Each possibility represents a separate embodiment of the present invention.
- In some embodiments, a LNP comprises at least one lipid. In certain embodiments, the lipids is selected from cationic/ionizable lipids, neutral lipids (e.g., fatty acids and phospholipids), PEG lipids (e.g., PEG-OH lipids, methyl PEG (mPEG) lipids, ethyl PEG lipids, and other derivatized PEG lipid conjugates), and structural lipids (e.g., sterols). The lipid may be selected from, but is not limited to, DLin-DMA, DLin-K-DMA, 98N12-5, C12-200, DLin-MC3-DMA, DLin-KC2-DMA, DODMA, PLGA, PEG, PEG-DMG, PEGylated lipids, and amino alcohol lipids. In some embodiments, the lipid may be a cationic lipid, such as, but not limited to, DLin-DMA, DLin-D-DMA, DLin-MC3-DMA, DLin-KC2-DMA, DODMA, and amino alcohol lipids. The amino alcohol cationic lipid may be the lipids described in and/or made by the methods described in US Patent Publication No.
US2013/0150625 . As a non-limiting example, the cationic lipid may be 2-amino-3-[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]-2-{[(9Z,2Z)-octadeca-9,12-dien-1-yloxy]methyl}propan-1-ol (Compound 1 inUS2013/0150625 ); 2-amino-3-[(9Z)-octadec-9-en-1-yloxy]-2-{[(9Z)-octadec-9-en-1-yloxy]methyl}propan-1-ol (Compound 2 inUS20130150625 ); 2-amino-3-[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]-2-[(octyloxy)methyl]propan-1-ol (Compound 3 inUS2013/0150625 ); and 2-(dimethylamino)-3 -[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]-2-{[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]methyl}propan-1-ol (Compound 4 inUS2013/0150625 ); or any pharmaceutically acceptable salt or stereoisomer thereof. Each possibility represents a separate embodiment of the present invention. - Lipid nanoparticle formulations can comprise a lipid, in particular, an ionizable cationic lipid, for example, 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), or di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319), and further comprise a neutral lipid (e.g., phospholipid or fatty acid), a structural lipid (e.g., a sterol such as cholesterol), and a molecule capable of reducing particle aggregation, for example, a PEG or PEGylated lipid (e.g., mPEG lipid or PEG-OH lipid). In certain embodiments, the formulation does not contain the PEG lipid.
- In some embodiments, the LNP formulation consists essentially of a molar ratio of 20-60% cationic lipid; 5-25% neutral lipid; 25-55% sterol; 0.1-15% PEG lipid. In some embodiments, the LNP formulation consists essentially of a molar ratio of 20-60% cationic lipid; 5-25% neutral lipid; 25-55% sterol; 0.1-15% mPEG lipid. In some embodiments, the LNP formulation consists essentially of in a molar ratio of 20-60% cationic lipid; 5-25% neutral lipid; and 25-55% sterol. In certain embodiments, the neutral lipid is a fatty acid. In certain embodiments, the neutral lipid is oleic acid or an analog thereof. In certain embodiments, the PEG lipid is a mPEG lipid or a PEG-OH lipid.
- In some embodiments, a LNP consists essentially of (i) at least one lipid selected from the group consisting of 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319); (ii) a neutral lipid selected from DSPC, DPPC, POPC, DOPE, and SM; (iii) a sterol, e.g., cholesterol; and (iv) a PEG-lipid, e.g., PEG-DMG or PEG-cDMA, in a molar ratio of 20-60% cationic lipid; 5-25% neutral lipid; 25-55% sterol; 0.1-15% PEG-lipid. Each possibility represents a separate embodiment of the present invention.
- In some embodiments, a LNP consists essentially of (i) at least one lipid selected from the group consisting of 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319); (ii) a neutral lipid as a DSPC substitute (e.g., a different phospholipid, or a fatty acid); (iii) a structural lipid (e.g., a sterol such as cholesterol); and (iv) a PEG-lipid or a PEG-OH lipid (e.g., PEG-DMG or PEG-cDMA), in a molar ratio of 20-60% cationic lipid; 5-25% DSPC substitute; 25-55% structural lipid; 0.1-15% PEG-lipid. Each possibility represents a separate embodiment of the present invention.
- In some embodiments, a LNP includes 25% to 75% on a molar basis of a cationic lipid. The cationic lipid may be selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319), e.g., 35 to 65%, 45 to 65%, 60%, 57.5%, 50% or 40% on a molar basis. Each possibility represents a separate embodiment of the present invention.
- In some embodiments, a LNP includes 0.5% to 15% on a molar basis of the neutral lipid, e.g., 3 to 12%, 5 to 10% or 15%, 10%, or 7.5% on a molar basis. In certain embodiments, the neutral lipid is a phospholipid. In certain embodiments, the neutral lipid is a DSPC substitute (e.g., a phospholipid other than DSPC, %or a fatty acid). In certain embodiments, the neutral lipid is a fatty acid (e.g., oleic acid or an analog thereof). Other examples of neutral lipids include, without limitation, POPC, DPPC, DOPE and SM. In some embodiments, a LNP includes 0.5% to 15% on a molar basis of a fatty acid, e.g., 3 to 12%, 5 to 10% or 15%, 10%, or 7.5% on a molar basis. In some embodiments, a LNP includes 0.5% to 15% on a molar basis of oleic acid, e.g., 3 to 12%, 5 to 10% or 15%, 10%, or 7.5% on a molar basis. In some embodiments, a LNP includes 0.5% to 15% on a molar basis of an analog of oleic acid, e.g., 3 to 12%, 5 to 10% or 15%, 10%, or 7.5% on a molar basis.
- In some embodiments, the formulation includes 5% to 50% on a molar basis of the structural lipid, e.g., 15 to 45%, 20 to 40%, 41%, 38.5%, 35%, or 31% on a molar basis. In some embodiments, the formulation includes 5% to 50% on a molar basis of a sterol, e.g., 15 to 45%, 20 to 40%, 41%, 38.5%, 35%, or 31% on a molar basis. In some other embodiments, the formulation includes about 35%, about 36%, about 37%, about 38%, about 39%, about 40%, about 41%, about 42%, about 43%, about 44% or about 45% on a molar basis. A non-limiting example of a sterol is cholesterol.
- In some embodiments, a LNP includes 0.5% to 20% on a molar basis of the PEG or PEGylated lipid, e.g., 0.5 to 10%, 0.5 to 5%, 1.5%, 0.5%, 1.5%, 2.0%, 2.5%, 3.0%3.5%, or 5% on a molar basis. In some embodiments, a PEG or PEGylated lipid comprises a PEG molecule of an average molecular weight of 2,000 Da. In some embodiments, a PEG or PEGylated lipid comprises a PEG molecule of an average molecular weight of less than 2,000, for example, around 1,500 Da, around 1,000 Da, or around 500 Da. Non-limiting examples of PEGylated lipids include PEG-distearoyl glycerol (PEG-DMG) (also referred herein as Cmpd422), PEG-cDMA (further discussed in Reyes et al. J. Controlled Release, 107, 276-287 (2005). As described herein, any PEG lipids or PEGylated lipids may be PEG-OH lipids. In some embodiments, a LNP includes 0.5% to 20% on a molar basis of a PEG-OH lipid, e.g., 0.5 to 10%, 0.5 to 5%, 1.5%, 0.5%, 1.5%, 3.5%, or 5% on a molar basis.
- In some embodiments, LNPs include 25-75% of a cationic lipid, 0.5-15% of the neutral lipid; 5-50% of the structural lipid, and 0.5-20% of the PEG or PEGylated lipid on a molar basis. In some embodiments, LNPs include 25-75% of a cationic lipid, 0.5-15% of the neutral lipid; 5-50% of the structural lipid, and 0.5-20% of a PEG-OH lipid on a molar basis. In some embodiments, LNPs include 25-75% of a cationic lipid, 0.5-15% of the neutral lipid, and 5-50% of the structural lipid on a molar basis. In some embodiments, LNPs include 25-75% of a cationic lipid selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319).
- In some embodiments, LNPs include 35-65% of a cationic lipid, 3-12% of the neutral lipid, 15-45% of the structural lipid, and 0.5-10% of the PEG or PEGylated lipid on a molar basis. In some embodiments, LNPs include 35-65% of a cationic lipid, 3-12% of the neutral lipid, 15-45% of the structural lipid, and 0.5-10% of the PEG-OH lipid on a molar basis. In some embodiments, LNPs include 35-65% of a cationic lipid, 3-12% of the neutral lipid, and 15-45% of the structural lipid on a molar basis. In some embodiments, LNPs include 35-65% of a cationic lipid selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319). Each possibility represents a separate embodiment of the present invention.
- In some embodiments, LNPs include 45-65% of a cationic lipid, 5-10% of the neutral lipid, 25-40% of the structural lipid, and 0.5-10% of the PEG or PEGylated lipid on a molar basis. In some embodiments, LNPs include 45-65% of a cationic lipid, 5-10% of the neutral lipid, 25-40% of the structural lipid, and 0.5-10% of a PEG-OH lipid on a molar basis. In some embodiments, LNPs include 45-65% of a cationic lipid, 5-10% of the neutral lipid, and 25-40% of the structural lipid on a molar basis. In some embodiments, LNPs include 45-65% of a cationic lipid selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319). Each possibility represents a separate embodiment of the present invention.
- In some embodiments, LNPs include 60% of a cationic lipid, 7.5% of the neutral lipid, 31% of a structural lipid, and 1.5% of the PEG or PEGylated lipid on a molar basis. In some embodiments, LNPs include 60% of a cationic lipid, 7.5% of the neutral lipid, 31% of a structural lipid, and 1.5% of a PEG-OH lipid on a molar basis. In some embodiments, LNPs include 60% of a cationic lipid, 9% of the neutral lipid, and 31% of a structural lipid on a molar basis. In some embodiments, LNPs include 60% of a cationic lipid selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319). Each possibility represents a separate embodiment of the present invention.
- In some embodiments, LNPs include 50% of a cationic lipid, 10% of the neutral lipid, 38.5% of the structural lipid, and 1.5% of the PEG or PEGylated lipid on a molar basis. In some embodiments, LNPs include 50% of a cationic lipid, 10% of the neutral lipid, 38.5% of a structural lipid, and 1.5% of a PEG-OH lipid on a molar basis. In some embodiments, LNPs include 50% of a cationic lipid, 10% of the neutral lipid, and 40% of a structural lipid on a molar basis. In some embodiments, LNPs include 50% of a cationic lipid selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319). Each possibility represents a separate embodiment of the present invention.
- In some embodiments, LNPs include 40% of a cationic lipid, 15% of the neutral lipid, 40% of the structural lipid, and 5% of the PEG or PEGylated lipid on a molar basis. In some embodiments, LNPs include 40% of a cationic lipid, 15% of the neutral lipid, 40% of the structural lipid, and 5% of a PEG-OH lipid on a molar basis. In some embodiments, LNPs include 40% of a cationic lipid, 20% of the neutral lipid, 40% of the structural lipid on a molar basis. In some embodiments, LNPs include 40% of a cationic lipid selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319). Each possibility represents a separate embodiment of the present invention.
- In some embodiments, LNPs include 57.2% of a cationic lipid, 7.1% of the neutral lipid 34.3% of the sterol, and 1.4% of the PEG or PEGylated lipid on a molar basis. In some embodiments, LNPs include 57.2% of a cationic lipid, 7.1% of the neutral lipid, 34.3% of the structural lipid, and 1.4% of the PEG-OH lipid on a molar basis. In some embodiments, LNPs include 57.2% of a cationic lipid, 8.5% of the neutral lipid, and 34.3% of the structural lipid on a molar basis. In some embodiments, LNPs include 57.2% of a cationic lipid selected from 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA), dilinoleyl-methyl-4-dimethylaminobutyrate (DLin-MC3-DMA), and di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319). Each possibility represents a separate embodiment of the present invention.
- In some embodiments, LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% neutral lipid; 20-55% structural lipid; 0.1-15% PEGylated lipid. In some embodiments, LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% neutral lipid (e.g., phospholipid or fatty acid); 20-55% structural lipid; and 0.1-15% PEG-OH lipid. In some embodiments, LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% neutral lipid (e.g., phospholipid or fatty acid); 20-55% structural lipid (e.g., sterols); and 0.1-15% PEG-OH lipid. In some embodiments, LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% neutral lipid (e.g., phospholipid or fatty acid); and 20-55% structural lipid (e.g., sterols). In some embodiments, LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% fatty acid (e.g., oleic acid or analog thereof); 20-55% structural lipid (e.g., sterols); and 0.1-15% PEG-OH lipid. In some embodiments, LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% fatty acid (e.g., oleic acid or analog thereof); and 20-55% structural lipid (e.g., sterols). In some embodiments, LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% oleic acid; 20-55% structural lipid (e.g., sterols); and 0.1-15% PEG-OH lipid. In some embodiments, LNPs consists essentially of a lipid mixture in molar ratios of 20-70% cationic lipid; 5-45% oleic acid; and 20-55% structural lipid (e.g., sterols).
- Non-limiting examples of lipid nanoparticle compositions and methods of making them are described, for example, in Semple et al. (2010) Nat. Biotechnol. 28:172-176; Jayarama et al. (2012), Angew. Chem. Int. Ed., 51: 8529-8533; and Maier et al. (2013) Molecular Therapy 21, 1570-1578.
- In some embodiments, LNPs may comprise a cationic lipid, a PEG lipid (e.g., PEG-OH lipid) and optionally comprise a neutral lipid (e.g., phospholipid or fatty acid). In some embodiments, LNPs may comprise a cationic lipid, a PEG lipid (e.g., PEG-OH lipid) and a structural lipid (e.g., a sterol) and optionally comprise a neutral lipid (e.g., phospholipid or fatty acid).
- Lipid nanoparticles described herein may comprise 2 or more components (e.g., lipids), not including the payload. In certain embodiments, the LNP comprises two components (e.g., lipids), not including the payload. In certain embodiments, the lipid nanoparticle comprises 5 components (e.g., lipids), not including the payload. In certain embodiments, the LNP comprises 6 components (e.g., lipids), not including the payload.
- In some embodiments, the LNPs described herein may be four component lipid nanoparticles. A 4 component LNP may comprise four different lipids selected from any described herein. The four components do not include the payload. The lipid nanoparticle may comprise a cationic lipid, a neutral lipid, a PEG lipid, and a structural lipid. In certain embodiments, the lipid nanoparticle comprises a cationic lipid, a fatty acid, a PEG lipid, and a structural lipid. In certain embodiments, the lipid nanoparticle comprises a cationic lipid, a fatty acid, a PEG-OH lipid, and a structural lipid. Each possibility represents a separate embodiment of the present invention.
- In some embodiments, the LNPs described herein may be three component lipid nanoparticles. A three component LNP may comprise three different lipids described herein. The lipid nanoparticle may comprise a cationic lipid, a neutral lipid (e.g., phospholipid or fatty acid), and a structural lipid. In certain embodiments, the lipid nanoparticle comprises a cationic lipid, a fatty acid, and a structural lipid. In certain embodiments, the lipid nanoparticle comprises a cationic lipid, a phospholipid, and a structural lipid.
- In one embodiment, the LNP formulation may be formulated by the methods described in International Publication Nos.
WO2011127255 orWO2008103276 . As a non-limiting example, LNP formulations as described inWO2011127255 and/orWO2008103276 . - In one embodiment, the lipid nanoparticle may be formulated by the methods described in
US Patent Publication No US2013/0156845 or International Publication NoWO2013/093648 orWO2012024526 . - The lipid nanoparticles described herein may be made in a sterile environment by the system and/or methods described in US Patent Publication No.
US20130164400 . - In one embodiment, the LNP formulation may be formulated in a nanoparticle such as a nucleic acid-lipid nanoparticle described in
U.S. Pat. No. 8,492,359 . - As a non-limiting example, the lipid nanoparticle may comprise one or more active agents or therapeutic agents (e.g., RNA); one or more cationic lipids comprising from about 50 mol % to about 85 mol % of the total lipid present in the particle; one or more neutral lipid lipids comprising from about 13 mol % to about 49.5 mol % of the total lipid present in the particle; and one or more structural lipids that inhibit aggregation of particles comprising from about 0.5 mol % to about 2 mol % of the total lipid present in the particle.
- In one embodiment, the LNP formulation may be formulated by the methods described in International Publication Nos.
WO2011127255 orWO2008103276 . - As a non-limiting example, LNP formulations as described in
WO2011 127255 and/orWO2008103276 . In one embodiment, LNP formulations described herein may comprise a polycationic composition. As a non-limiting example, the polycationic composition may be selected from formula 1-60 of US Patent Publication No.US20050222064 . - In some embodiments, LNPs comprise the lipid KL52 (an amino-lipid disclosed in
U.S. Application Publication No. 2012/0295832 ). Activity and/or safety (as measured by examining one or more of ALT/AST, white blood cell count and cytokine induction) of LNP administration may be improved by incorporation of such lipids. LNPs comprising KL52 may be administered intravenously and/or in one or more doses. In some embodiments, administration of LNPs comprising KL52 results in equal or improved mRNA and/or protein expression as compared to LNPs comprising MC3. - As a non-limiting example, the LNP may include a cationic peptide or a polypeptide such as, but not limited to, polylysine, polyornithine and/or polyarginine and the cationic peptides described in International Pub. No.
WO2012013326 or US Patent Pub. No.US20130142818 . In some embodiments, the lipid nanoparticle includes a neutral lipid such as, but not limited to, cholesterol or dioleoyl phosphatidylethanolamine (DOPE). - A nanoparticle composition may be relatively homogenous. A polydispersity index may be used to indicate the homogeneity of a nanoparticle composition, e.g., the particle size distribution of the nanoparticle compositions. A small (e.g., less than 0.3) polydispersity index generally indicates a narrow particle size distribution. A nanoparticle composition may have a polydispersity index from about 0 to about 0.25, such as 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.10, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.20, 0.21, 0.22, 0.23, 0.24, or 0.25. In some embodiments, the polydispersity index of a nanoparticle composition may be from about 0.10 to about 0.20, or about 0.05 to about 0.15, or less than about 0.1, or less than about 0.15. Each possibility represents a separate embodiment of the present invention.
- The zeta potential of a nanoparticle composition may be used to indicate the electrokinetic potential of the composition. For example, the zeta potential may describe the surface charge of a nanoparticle composition. Nanoparticle compositions with relatively low charges at physiological pH, positive or negative, are generally desirable, as more highly charged species may interact undesirably with cells, tissues, and other elements in the body. In some embodiments, the zeta potential of a nanoparticle composition may be from about -10 mV to about +20 mV, from about -10 mV to about +15 mV, from about -10 mV to about +10 mV, from about -10 mV to about +5 mV, from about -10 mV to about 0 mV, from about -10 mV to about -5 mV, from about -5 mV to about +20 mV, from about -5 mV to about +15 mV, from about -5 mV to about +10 mV, from about -5 mV to about +5 mV, from about -5 mV to about 0 mV, from about 0 mV to about +20 mV, from about 0 mV to about +15 mV, from about 0 mV to about +10 mV, from about 0 mV to about +5 mV, from about +5 mV to about +20 mV, from about +5 mV to about +15 mV, or from about +5 mV to about +10 mV. Each possibility represents a separate embodiment of the present invention.
- The efficiency of encapsulation of a therapeutic agent describes the amount of therapeutic agent that is encapsulated or otherwise associated with a nanoparticle composition after preparation, relative to the initial amount provided. The encapsulation efficiency is desirably high (e.g., close to 100%). The encapsulation efficiency may be measured, for example, by comparing the amount of therapeutic agent in a solution containing the nanoparticle composition before and after breaking up the nanoparticle composition with one or more organic solvents or detergents. Fluorescence may be used to measure the amount of free therapeutic agent (e.g., nucleic acids) in a solution. For the nanoparticle compositions described herein, the encapsulation efficiency of a therapeutic agent may be at least 50%, for example 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%. In some embodiments, the encapsulation efficiency may be at least 80%. In certain embodiments, the encapsulation efficiency may be at least 90%. In certain embodiments, the encapsulation efficiency may be at least 95%. Each possibility represents a separate embodiment of the present invention.
- A nanoparticle composition may optionally comprise one or more coatings. For example, a nanoparticle composition may be formulated in a capsule, film, or tablet having a coating. A capsule, film, or tablet including a composition described herein may have any useful size, tensile strength, hardness, or density.
- In some embodiments, such LNPs are synthesized using methods comprising microfluidic mixers. Exemplary microfluidic mixers may include, but are not limited to a slit interdigitial micromixer including, but not limited to those manufactured by Microinnova (Allerheiligen bei Wildon, Austria) and/or a staggered herringbone micromixer (SHM) (Zhigaltsev, I.V. et al., Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing have been published (Langmuir. 2012. 28:3633-40; Belliveau, N. M. et al., Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Molecular Therapy-Nucleic Acids. 2012. 1:e37; Chen, D. et al., Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J Am Chem Soc. 2012. 134(16):6948-51).
- In some embodiments, methods of LNP generation comprising SHM, further comprise the mixing of at least two input streams wherein mixing occurs by microstructure-induced chaotic advection (MICA). According to this method, fluid streams flow through channels present in a herringbone pattern causing rotational flow and folding the fluids around each other. This method may also comprise a surface for fluid mixing wherein the surface changes orientations during fluid cycling. Methods of generating LNPs using SHM include those disclosed in
U.S. Application Publication Nos. 2004/0262223 and2012/0276209 . - In one embodiment, the lipid nanoparticles may be formulated using a micromixer such as, but not limited to, a Slit Interdigital Microstructured Mixer (SIMM-V2) or a Standard Slit Interdigital Micro Mixer (SSIMM) or Caterpillar (CPMM) or Impinging jet (UMM) from the Institut für Mikrotechnik Mainz GmbH, Mainz Germany).
- In one embodiment, the lipid nanoparticles are created using microfluidic technology (see Whitesides, George M. The Origins and the Future of Microfluidics. Nature, 2006 442: 368-373; and Abraham et al. Chaotic Mixer for Microchannels. Science, 2002 295: 647-651). As a non-limiting example, controlled microfluidic formulation includes a passive method for mixing streams of steady pressure-driven flows in micro channels at a low Reynolds number (See e.g., Abraham et al. Chaotic Mixer for Microchannels. Science, 2002 295: 647651).
- In one embodiment, a therapeutic nucleic acid (e.g., mRNA) may be formulated in lipid nanoparticles created using a micromixer chip such as, but not limited to, those from Harvard Apparatus (Holliston, Mass.) or Dolomite Microfluidics (Royston, UK). A micromixer chip can be used for rapid mixing of two or more fluid streams with a split and recombine mechanism.
- Cationic lipids useful in embodiments of the present invention are neutral while in circulation but become positively charged upon acidification of the endosome. A positive charge on the LNP may promote association with the negatively charged cell membrane to enhance cellular uptake. Cationic lipids may also combine with negatively charged lipids to induce nonbilayer structures that facilitate intracellular delivery. Suitable cationic lipids for use in making the LNPs disclosed herein can be ionizable cationic lipids, as disclosed herein. The cationic lipids may be prepared according to the procedures set forth in the Examples or according to methods known or derivable by one of ordinary skill in the art.
- In some embodiments, LNPs may comprise, in molar percentages, 35 to 45% cationic lipid, 40% to 50% cationic lipid, 45% to 55% cationic lipid, 50% to 60% cationic lipid and/or 55% to 65% cationic lipid. In some embodiments, the ratio of lipid to nucleic acid (e.g., mRNA) in lipid nanoparticles may be 5:1 to 20:1, 10:1 to 25:1, 15:1 to 40:1, 20:1 to 30:1, 25:1 to 50:1, 30:1 to 60:1 and/or at least 40:1.
- Such lipids include, but are not limited to, N,N-dioleyl-N,N-dimethylammonium chloride (DODAC); N-(2,3-dioleyloxy)propyl)-N,N,N-trimethylammonium chloride (DOTMA); N,N-distearyl-N,N-dimethylammonium bromide (DDAB); N-(2,3dioleoyloxy)propyl)-N,N,N-trimethylammonium chloride (DOTAP); 3-(N---(N',N'dimethylaminoethane)-carbamoyl)cholesterol (DC-Chol), N-(1-(2,3-dioleoyloxy)propyl)N-2-(sperminecarboxamido)ethyl)-N,N-dimethylammonium trifluoracetate (DOSPA), dioctadecylamidoglycyl carboxyspermine (DOGS), 1,2-dioleoyl-3-dimethylammonium propane (DODAP), N,N-dimethyl-2,3-dioleoyloxy)propylamine (DODMA), and N-(1,2-dimyristyloxyprop-3-yl)-N,N-dimethyl-N-hydroxyethyl ammonium bromide (DMRIE).
- Additionally, a number of commercial preparations of cationic lipids are available which can be used in any of the described embodiments. These include, for example, LIPOFECTIN® (commercially available cationic liposomes comprising DOTMA and 1,2-dioleoyl-sn-3phosphoethanolamine (DOPE), from GIBCO/BRL, Grand Island, N.Y.); LIPOFECTAMINE® (commercially available cationic liposomes comprising N-(1-(2,3dioleyloxy)propyl)-N-(2-(sperminecarboxamido)ethyl)-N,N-dimethylammonium trifluoroacetate (DOSPA) and (DOPE), from GIBCOBRL); and TRANSFECTAM® (commercially available cationic lipids comprising dioctadecylamidoglycyl carboxyspermine (DOGS) in ethanol from Promega Corp., Madison, Wis.). The following lipids are cationic and have a positive charge at below physiological pH: DODAP, DODMA, DMDMA, 1,2-dilinoleyloxy-N,N-dimethylaminopropane (DLinDMA), 1,2-dilinolenyloxy-N,N-dimethylaminopropane (DLenDMA).
- In one specific embodiment, the cationic lipid for use in any of the described embodiments is independently an amino lipid. Suitable amino lipids include those described in
WO 2010/054401 andWO 2012/016184 . Representative amino lipids include, but are not limited to, 1,2-dilinoleyoxy-3-(dimethylamino)acetoxypropane (DLin-DAC), 1,2-dilinoleyoxy-3morpholinopropane (DLin-MA), 1,2-dilinoleoyl-3-dimethylaminopropane (DLinDAP), 1,2-dilinoleylthio-3-dimethylaminopropane (DLin-S-DMA), 1-linoleoyl-2-linoleyloxy-3dimethylaminopropane (DLin-2-DMAP), 1,2-dilinoleyloxy-3-trimethylaminopropane chloride salt (DLin-TMA.Cl), 1,2-dilinoleoyl-3-trimethylaminopropane chloride salt (DLin-TAP.Cl), 1,2-dilinoleyloxy-3-(N-methylpiperazino)propane (DLin-MPZ), 3-(N,Ndilinoleylamino)-1,2-propanediol (DLinAP), 3-(N,N-dioleylamino)-1,2-propanediol (DOAP), 1,2-dilinoleyloxo-3-(2-N,N-dimethylamino)ethoxypropane (DLin-EG-DMA), and 2,2-dilinoleyl-4-dimethylaminomethyl-[1,3]-dioxolane (DLin-K-DMA).In some of the described embodiments, the cationic lipid has the following formula: - R3 and R4 are either the same or different and independently optionally substituted C1-C6 alkyl, optionally substituted C2-C6 alkenyl, or optionally substituted C2-C6 alkynyl or R3 and R4 may join to form an optionally substituted heterocyclic ring of 4 to 6 carbon atoms and 1 or 2 heteroatoms chosen from nitrogen and oxygen;
- R5 is either absent or present and when present is hydrogen or C1-C6 alkyl; m, n, and p are either the same or different and independently either 0 or 1 with the proviso that m, n, and p are not simultaneously 0; q is 0, 1, 2, 3, or 4; and
- Y and Z are either the same or different and independently O, S, or NH. In one embodiment, R1 and R2 are each linoleyl, and the amino lipid is a dilinoleyl amino lipid. In one embodiment, the amino lipid is a dilinoleyl amino lipid. In various other embodiments, the cationic lipid has the following structure:
wherein:- R1 and R2 are independently selected from the group consisting of H, and C1-C3 alkyls;
- R3 and R4 are independently selected from the group consisting of alkyl groups having from about 10 to about 20 carbon atoms, wherein at least one of R3 and R4 comprises at least two sites of unsaturation. (e.g., R3 and R4 may be, for example, dodecadienyl, tetradecadienyl, hexadecadienyl, linoleyl, and icosadienyl. In a preferred embodiment, R3 and R4 are both linoleyl. R3 and R4 may comprise at least three sites of unsaturation (e.g., R3 and R4 may be, for example, dodecatrienyl, tetradectrienyl, hexadecatrienyl, linolenyl, and icosatrienyl).
- In some embodiments, the cationic lipid has the following structure:
wherein:
R1 and R2 are independently selected and are H or C1-C3 alkyls. R3 and R4 are independently selected and are alkyl groups having from about 10 to about 20 carbon atoms, wherein at least one of R4 and R4 comprises at least two sites of unsaturation. In one embodiment, R3 and R4 are both the same, for example, in some embodiments R3 and R4 are both linoleyl (i.e., C18), etc. In another embodiment, R3 and R4 are different, for example, in some embodiments R3 is tetradectrienyl (C14) and R4 is linoleyl (C18). In a preferred embodiment, the cationic lipid(s) of the present invention are symmetrical, i.e., R3 and R4 are the same. In another preferred embodiment, both R3 and R4 comprise at least two sites of unsaturation. In some embodiments, R3 and R4 are independently selected from dodecadienyl, tetradecadienyl, hexadecadienyl, linoleyl, and icosadienyl. In a preferred embodiment, R3 and R4 are both linoleyl. In some embodiments, R4 and R4 comprise at least three sites of unsaturation and are independently selected from, e.g., dodecatrienyl, tetradectrienyl, hexadecatrienyl, linolenyl, and icosatrienyl. -
- Xaa is a D- or L-amino acid residue having the formula -NRN-CR1R2-C(C=O)-, or a peptide or a peptide of amino acid residues having the formula ---(NRN-CR1R2---(C=O)}n---, wherein n is 2 to 20;
- R1 is independently, for each occurrence, a non-hydrogen, substituted or unsubstituted side chain of an amino acid;
- R2 and RN are independently, for each occurrence, hydrogen, an organic group consisting of carbon, oxygen, nitrogen, sulfur, and hydrogen atoms, or any combination of the foregoing, and having from 1 to 20 carbon atoms, C(1-5)alkyl, cycloalkyl, cycloalkylalkyl, C(3-5)alkenyl, C(3-5)alkynyl, C(1-5)alkanoyl, C(1-5)alkanoyloxy, C(1-5)alkoxy, C(1-5)alkoxy-C(1-5)alkyl, C(1-5)alkoxy-C(1-5)alkoxy, C(1-5)alkyl-amino-C(1-5)alkyl-, C(1-5)dialkyl-amino-C(1-5)alkyl-, nitro-C(1-5)alkyl, cyano-C(1-5)alkyl, aryl-C(1-5)alkyl, 4-biphenyl-C(1-5)alkyl, carboxyl, or hydroxyl;
- Z is NH, O, S, -CH2S-, -CH2S(O)-, or an organic linker consisting of 1-40 atoms selected from hydrogen, carbon, oxygen, nitrogen, and sulfur atoms (preferably, 2 is NH or O);
- Rx and Ry are, independently, (i) a lipophilic tail derived from a lipid (which can be naturally-occurring or synthetic), phospholipid, glycolipid, triacylglycerol, glycerophospholipid, sphingolipid, ceramide, sphingomyelin, cerebroside, or ganglioside, wherein the tail optionally includes a steroid; (ii) an amino acid terminal group selected from hydrogen, hydroxyl, amino, and an organic protecting group; or (iii) a substituted or unsubstituted C(3-22)alkyl, C(6-12)cycloalkyl, C(6-12)cycloalkyl-C(3-22)alkyl, C(3-22)alkenyl, C(3-22)alkynyl, C(3-22)alkoxy, or C(6-12)-alkoxy-C(3-22)alkyl;
- one of Rx and Ry is a lipophilic tail as defined above and the other is an amino acid terminal group, or both Rx and Ry are lipophilic tails;
- at least one of Rx and Ry is interrupted by one or more biodegradable groups (e.g., -OC(O)-, -C(O)O-, -SC(O)-, -C(O)S-, -OC(S)-, -C(S)O-, -S-S-, -C(R5)=N-, -N=C(R5)--, -C(R3)=N-O-, -O-N=C(R5)-, -C(O)(NR5)-, -N(R5)C(O)-, -C(S)(NR5)-, - N(R5)C(O)-, -N(R5)C(O)N(R5)-, -OC(O)O-, -OSi(R5)2O-, -C(O)(CR3R4)C(O)O-, - OC(O)(CR3R4)C(O)- or
- wherein R11 is a C2-C8 alkyl or alkenyl and each occurrence of R5 is, independently, H or alkyl; and each occurrence of R3 and R4 are, independently H, halogen, OH, alkyl, alkoxy, --NH2, alkylamino, or dialkylamino; or R3 and R4, together with the carbon atom to which they are directly attached, form a cycloalkyl group (in one preferred embodiment, each occurrence of R3 and R4 are, independently H or C1-C4 alkyl)); and Rx and Ry each, independently, optionally have one or more carbon-carbon double bonds.
-
- R1 and R2 are independently alkyl, alkenyl or alkynyl, and each can be optionally substituted;
- R3 and R4 are independently a C1-C6 alkyl, or R3 and R4 can be taken together to form an optionally substituted heterocyclic ring.
-
- In one embodiment, the cationic lipid is DLin-K-DMA. In one embodiment, a cationic lipid is DLin-KC2-DMA (DLin-K-DMA above, wherein n is 2).
-
- R1 and R2 are each independently for each occurrence optionally substituted C10-C30 alkyl, optionally substituted C10-C30 alkenyl, optionally substituted C10-C30 alkynyl or optionally substituted C10-C30 acyl, or linker-ligand;
- R3 is H, optionally substituted C1-C10 alkyl, optionally substituted C2-C10 alkenyl, optionally substituted C2-C10 alkynyl, alkylhetrocycle, alkylphosphate, alkylphosphorothioate, alkylphosphorodithioate, alkylphosphonate, alkylamine, hydroxyalkyl, ω-aminoalkyl, ω-(substituted)aminoalkyl, ω-phosphoalkyl, ω-thiophosphoalkyl, optionally substituted polyethylene glycol (PEG, mw 100-40K), optionally substituted mPEG (mw 120-40K), heteroaryl, or heterocycle, or linker-ligand, for example in some embodiments R3 is (CH3)2N(CH2)n-, wherein n is 1, 2, 3 or 4;
- E is O, S, N(Q), C(O), OC(O), C(O)O, N(Q)C(O), C(O)N(Q), (Q)N(CO)O, O(CO)N(Q), S(O), NS(O)2N(Q), S(O)2, N(Q)S(O)2, SS, O=N, aryl, heteroaryl, cyclic or heterocycle, for example -C(O)O, wherein - is a point of connection to R3; and
- Q is H, alkyl, ω-aminoalkyl, ω-(substituted)aminoalkyl, ω-phosphoalkyl or ω-thiophosphoalkyl.
-
- E is O, S, N(Q), C(O), N(Q)C(O), C(O)N(Q), (Q)N(CO)O, O(CO)N(Q), S(O), NS(O)2N(Q), S(O)2, N(Q)S(O)2, SS, O=N, aryl, heteroaryl, cyclic or heterocycle;
- Q is H, alkyl, ω-amninoalkyl, ω-(substituted)amninoalky, ω-phosphoalkyl or ω-miophosphoalkyl;
- R1 and R2 and Rx are each independently for each occurrence H, optionally substituted C1-C10 alkyl, optionally substituted C10-C30 alkyl, optionally substituted C10-C30 alkenyl, optionally substituted C10-C30 alkynyl, optionally substituted C10-C30 acyl, or linker-ligand, provided that at least one of R1, R2 and Rx is not H;
- R3 is H, optionally substituted C1-C10 alkyl, optionally substituted C2-C10 alkenyl, optionally substituted C2-C10 alkynyl, alkylhetrocycle, alkylphosphate, alkylphosphorothioate, alkylphosphorodithioate, alkylphosphonate, alkylamine, hydroxyalkyl, ω-aminoalkyl, ω-(substituted)aminoalkyl, ω-phosphoalkyl, ω-thiophosphoalkyl, optionally substituted polyethylene glycol (PEG, mw 100-40K), optionally substituted mPEG (mw 120-40K), heteroaryl, or heterocycle, or linker-ligand; and
- n is 0, 1, 2, or 3.
-
- In some embodiments, the cationic lipid is DLin-M-C3-DMA, MC3 or M-C3 and has been described in
WO 2010/054401 , andWO 2010/144740 A1 . -
-
- R1, R2, R3, R4, R5, R6, R7 and R8 are independently selected from the group consisting of hydrogen, optionally substituted C7-C30 alkyl, optionally substituted C7-C30 alkenyl and optionally substituted C7-C30 alkynyl:
- provided that (a) at least two of R1, R2, R3, R4, R5, R6, R7 and R8 are not hydrogen, and (b) two of the at least two of R1, R2, R3, R4, R5, R6, R7 and R8 that are not hydrogen are present in a 1,3 arrangement, a 1,4 arrangement or a 1,5 arrangement with respect to each other;
- X is selected from the group consisting of Ci-Cs alkyl, C2-C6 alkenyl and C2-C6 alkynyl;
- R9, R10, and R11 are independently selected from the group consisting of hydrogen, optionally substituted C1-C7, alkyl, optionally substituted C2-C7, alkenyl and option ally substituted C2-C7, alkynyl, provided that one of R9, R10, and R11 may be absent; and n and m are each independently 0 or 1.
-
-
- R1 is independently selected from -(CH2)2-N(R)2, -(CH2)2-N(R)-(CH2)2-N(R)2, wherein
- R is independently selected from -H, C6-40 alkyl, C6-40 alkenyl and C6-40 alkynyl, provided that -N(R)2 is not NH2;
- R2 is C6-40 alkyl, C6-40 alkenyl or C6-40 alkynyl; and
- m is 0 or 1.
-
-
- R' is absent, hydrogen, or alkyl;
- with respect to R1 and R2,
- (i) R1 and R2 are each, independently, optionally substituted alkyl, alkenyl, alkynyl, cycloalkyl, cycloalkylalkyl, or heterocycle;
- (ii) R1 and R2, together with the nitrogen atom to which they are attached, form an optionally substituted heterocyclic ring; or
- (iii) one of R1 and R2 is optionally substituted alkyl, alkenyl, alkynyl, cycloalkyl, cycloalkylalkyl, or heterocycle, and the other forms a 4-10 member heterocyclic ring or heteroaryl with (a) the adjacent nitrogen atom and (b) the (R)a group adjacent to the nitrogen atom;
- each occurrence of Ris, independently, -(CR3R4)-;
- each occurrence of R3 and R4 are, independently H, OH, alkyl, alkoxy, - NH2, alkylamino, or dialkylamino;
- or R3 and R4, together with the carbon atom to which they are directly attached, form a cycloalkyl group, wherein no more than three R groups in each chain attached to the carbon C* are cycloalkyl;
- the dashed line to Q is absent or a bond;
- when the dashed line to Q is absent then Q is absent or is -O-, -NH-, -S-, - C(O)O-, -OC(O)-, -C(O)N(R4)-, -N(R5)C(O)-, -S-S-, -OC(O)O-, -O-N=C(R5)-, - C(R5)=N-O-, -OC(O)N(R5)-, -N(R5)C(O)N(R5)-, -N(R5)C(O)O-, -C(O)S-, -C(S)O- or - C(R5)=N-O-C(O)-; or
- when the dashed line to Q is a bond then (i) b is 0 and (ii) Q and the tertiary carbon adjacent to it (C*) form a substituted or unsubstituted, mono- or bi-cyclic heterocyclic group having from 5 to 10 ring atoms;
- Q1 and Q2 are each, independently, absent, -O-, -S-, -OC(O)-, -C(O)O-, - SC(O)-, -C(O)S-, -OC(S)-, -C(S)O-, -S-S-, -C(O)(NR5)-, -N(R5)C(O)-, -C(S)(NR5)-, - N(R5)C(O)-, -N(R5)C(O)N(R5)-, or -OC(O)O-;
- Q3 and Q4 are each, independently, H, -(CR3R4)-, aryl, or a cholesterol moiety;
- each occurrence of A1, A2, A3 and A4 is, independently, -(CR5R5-CR5=CR5)-;
- each occurrence of R5 is, independently, H or alkyl;
- M1 and M2 are each, independently, a biodegradable group; wherein
- the biodegradable group is selected from -OC(O)-, -C(O)O-, -SC(O)-, - C(O)S-, -OC(S)-, -C(S)O-, -S-S-, -C(R5)=N-, -N=C(R5)-, -C(R5)=N-O-, -O-N=C(R5)-, - C(O)(NR5)-, -N(R5)C(O)-, -C(S)(NR5)-, -N(R5)C(O)-, -N(R5)C(O)N(R5)-, -OC(O)O-, - OSi(R5)2O-, -C(O)(CR3R4)C(O)O-, and -OC(O)(CR3R4)C(O)-;
- Z is absent, alkylene or -O-P(O)(OH)-O-;
- each ------ attached to Z is an optional bond, such that when Z is absent, Q3 and Q4 are not directly covalently bound together;
- a is 1, 2, 3, 4, 5 or 6;
- b is 0, 1, 2, or 3;
- c, d, e, f, i, j, m, n, q and r are each, independently, 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10;
- g and h are each, independently, 0, 1 or 2;
- k and 1 are each, independently, 0 or 1, where at least one of k and 1 is 1; and
- o and p are each, independently, 0, 1 or 2,
- wherein
- (i) the compound does not contain the following moiety:
- (i) the compound does not contain the following moiety:
- wherein ---- is an optional bond; and
- Q3 and Q4 are each, independently, separated from the tertiary carbon atom marked with an asterisk (*) by a chain of 8 or more atoms.
-
- (i) in structure (II), (IV), (VI) and (VII), m and p are both 4 or greater;
- (ii) in structure (VIII), (X), (XII), (XIV), (XVI), (XVIII), (XXI) and (XXIII), m is 4 or greater; and
- (iii) in structure (VIII), (IX), (XII) and (XIII), p is 8 or greater (e.g., 12 or 14 or greater).
-
-
- R1 is selected from the group consisting of C5-30 alkyl, C5-20alkenyl, - R*YR", -YR", and -R"M'R';
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of a C3-6 carbocycle, -(CH2)nQ, - (CH2)nCHQR, -CHQR, -CQ(R)2, and unsubstituted C1-6 alkyl, where Q is selected from a carbocycle, heterocycle, -OR, -O(CH2)nN(R)2, -C(O)OR, -OC(O)R, -CX3, -CX2H, - CXH2, -CN, -N(R)2, -C(O)N(R)2, -N(R)C(O)R, -N(R)S(O)2R, -N(R)C(O)N(R)2, - N(R)C(S)N(R)2, -N(R)R8, O(CH2)nOR, -N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, - OC(O)N(R)2, -N(R)C(O)OR, -N(OR)C(O)R, -N(OR)S(O)2R, -N(OR)C(O)OR, - N(OR)C(O)N(R)2, -N(OR)C(S)N(R)2, -N(OR)C(=NR9)N(R)2, -N(OR)C(=CHR9)N(R)2, -C(=NR9)N(R)2, -C(=NR9)R, -C(O)N(R)OR, and -C(R)N(R)2C(O)OR, and each n is independently selected from 1, 2, 3, 4, and 5;
- each R5 is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(O)O-, -OC(O)-, - C(O)N(R')-, -N(R')C(O)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, -P(O)(OR')O-, - S(O)2-, -S-S-, an aryl group, and a heteroaryl group;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, NO2, C1-6 alkyl, -OR, - S(O)2R, -S(O)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- each R is independently selected from the group consisting of C1-3alkyl, C2-3 alkenyl, and H;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-14 alkyl and C3-14 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and
- m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13.
-
-
- R' is absent, hydrogen, or C1-C4 alkyl;
- with respect to R1 and R2,
- (i) R1 and R2 are each, independently, optionally substituted alkyl, alkenyl, alkynyl, cycloalkylalkyl, heterocycle, or R10;
- (ii) R1 and R2, together with the nitrogen atom to which they are attached, form an optionally substituted heterocylic ring; or
- (iii) one of R1 and R2 is optionally substituted alkyl, alkenyl, alkynyl, cycloalkyl, cycloalkylalkyl, or heterocycle, and the other forms a 4-10 member heterocyclic ring or heteroaryl with (a) the adjacent nitrogen atom and (b) the (R)a group adjacent to the nitrogen atom;
- each occurrence of R is, independently, -(CR3R4)-;
- each occurrence of R3 and R4 are, independently H, halogen, OH, alkyl, alkoxy, -NH2, R10, alkylamino, or dialkylamino;
- each occurrence of R10 is independently selected from PEG and polymers based on poly(oxazoline), poly(ethylene oxide), poly(vinyl alcohol), poly(glycerol), poly(N-vinylpyrrolidone), poly[N-(2-hydroxypropyl)methacrylamide] and poly(amino acid)s, wherein (i) the PEG or polymer is linear or branched, (ii) the PEG or polymer is polymerized by n subunits, (iii) n is a number-averaged degree of polymerization between 10 and 200 units, and (iv) the compound of said formula has at most two R10 groups;
- the dashed line to Q is absent or a bond;
- when the dashed line to Q is absent then Q is absent or is -O-, -NH-, -S-, - C(O)-, -C(O)O-, -OC(O)-, -C(O)N(R4)-, -N(R5)C(O)-, -S-S-, -OC(O)O-, -O-N=C(R5)-, - C(R5)=N-O-, -OC(O)N(R5)-, -N(R5)C(O)N(R5)-, -N(R5)C(O)O-, -C(O)S-, -C(S)O- or - C(R5)=N-O-C(O)-; or
- when the dashed line to Q is a bond then (i) b is 0 and (ii) Q and the tertiary carbon adjacent to it (C*) form a substituted or unsubstituted, mono- or bi-cyclic heterocyclic group having from 5 to 10 ring atoms;
- each occurrence of R5 is, independently, H or C1-C4 alkyl;
- M1 and M2 are each, independently, a biodegradable group selected from - OC(O)-, -C(O)O-, -SC(O)-, -C(O)S-, -OC(S)-, -C(S)O-, -S-S-, -C(R5)=N-, -N=C(R5)-, - C(R5)=N-O-, -O-N=C(R5)-, -C(O)(NR5)-, -N(R5)C(O)-, -C(S)(NR5)-, -N(R5)C(O)-, - N(R5)C(O)N(R5)-, -OC(O)O-, -OSi(R5)2O-, -C(O)(CR3R4)C(O)O-, and - OC(O)(CR3R4)C(O)-, or
- each occurrence of Rz is, independently, C1-C8 alkyl;
- a is 1, 2, 3, 4, 5 or 6;
- b is 0, 1, 2, or 3;
- L1 and L2 are each, independently, C1-C5 alkylene or C2-C5 alkenylene;
- X and Y are each, independently, alkylene or alkenylene; and
- Z1 and Z2 are each, independently, C8-C14 alkyl or C8-C14 alkenyl, wherein the alkenyl group may optionally be substituted with one or two fluorine atoms at the alpha position to a double bond which is between the double bond and the terminus of Z1 or Z2, and with the proviso that the terminus of at least one of Z1 and Z2 is separated from the group M1 or M2 by at least 8 carbon atoms.
-
-
-
-
-
-
- R' is absent, hydrogen, or C1-C4 alkyl;
- with respect to R1 and R2,
- R' is absent, hydrogen, or alkyl;
- with respect to R1 and R2,
- (i) R1 and R2 are each, independently, optionally substituted alkyl, alkenyl, alkynyl, cycloalkyl, cycloalkylalkyl, heterocycle, or R10;
- (ii) R1 and R2, together with the nitrogen atom to which they are attached, form an optionally substituted heterocylic ring; or
- (iii) one of R1 and R2 is optionally substituted alkyl, alkenyl, alkynyl, cycloalkyl, cycloalkylalkyl, or heterocycle, and the other forms a 4-10 member heterocyclic ring or heteroaryl with (a) the adjacent nitrogen atom and (b) the (R)a group adjacent to the nitrogen atom;
- each occurrence of R is, independently, -(CR3R4)-;
- each occurrence of R3 and R4 are, independently hydrogen, OH, alkyl, alkoxy, -NH2, R10, alkylamino, or dialkylamino;
- each occurrence of R10 is independently selected from PEG and polymers based on poly(oxazoline), poly(ethylene oxide), poly(vinyl alcohol), poly(glycerol), poly(N-vinylpyrrolidone), poly[N-(2-hydroxypropyl)methacrylamide] and poly(amino acid)s, wherein (i) the PEG or polymer is linear or branched, (ii) the PEG or polymer is polymerized by n subunits, (iii) n is a number-averaged degree of polymerization between 10 and 200 units, and (iv) wherein the compound of said formula has at most two R10 groups;
- the dashed line to Q is absent or a bond;
- when the dashed line to Q is absent then Q is absent or is -O-, -NH-, -S-, - C(O)-, -C(O)O, -OC(O)-, -C(O)N(R4)-, -N(R5)C(O)-, -S-S-, -OC(O)O-, -O-N=C(R5)-, - C(R5)=N-O-, -OC(O)N(R5)-, -N(R5)C(O)N(R5)-, -N(R5)C(O)O-, -C(O)S-, -C(S)O- or - C(R5)=N-O-C(O)-; or
- when the dashed line to Q is a bond then (i) b is 0 and (ii) Q and the tertiary carbon adjacent to it (C*) form a substituted or unsubstituted, mono- or bi-cyclic heterocyclic group having from 5 to 10 ring atoms;
- each occurrence of R5 is, independently, hydrogen or alkyl;
- X and Y are each, independently, -(CR6R7)c-;
- each occurrence of R6 and R7 are, independently hydrogen, OH, alkyl, alkoxy, -NH2, alkylamino, or dialkylamino;
- M1 and M2 are each, independently, a biodegradable group;
- a is 1, 2, 3, 4, 5 or 6;
- b is 0, 1, 2, or 3;
- each occurrence of c is, independently, 2-10; and
- Z1 and Z2 are each, independently (i) C3-C10 cycloalkyl, (ii) C3-C10cycloalkyl(C1-C6 alkyl), or (iii)
- wherein each of R8 and R9 is a C2-C8 alkyl.
-
-
- one of L1 or L2 is -O(C=O)-, -(C=O)O-, -C(=O)-, -O-, -S(O)x-, -S-S-, -C(=O)S-, SC(=O)-, -NRaC(=O)-, -C(=O)NRa-, NRaC(=O)NRa-, -OC(=O)NRa- or -NRaC(=O)O-, and the other of L1 or L2 is -O(C=O)-, -(C=O)O-, -C(=O)-, -0-, -S(O)x-, -S-S-, -C(=O)S-, SC(=O)-, -NRaC(=O)-, -C(=O)NRa-, NRaC(=O)NRa-, -OC(=O)NRa-or
-NRaC(=O)O- or a direct bond; - Ra is H or C1-C12 alkyl;
- R1a and R1b are, at each occurrence, independently either (a) H or C1-C12 alkyl, or (b) R1a is H or C1-C12 alkyl, and R1b together with the carbon atom to which it is bound is taken together with an adjacent R1b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R2a and R2b are, at each occurrence, independently either (a) H or C1-C12 alkyl, or (b) R2a is H or C1-C12 alkyl, and R2b together with the carbon atom to which it is bound is taken together with an adjacent R2b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R3a and R3b are, at each occurrence, independently either (a) H or C1-C12 alkyl, or (b) R3a is H or C1-C12 alkyl, and R3b together with the carbon atom to which it is bound is taken together with an adjacent R3b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R4a and R4b are, at each occurrence, independently either (a) H or C1-C12 alkyl, or (b) R4a is H or C1-C12 alkyl, and R4b together with the carbon atom to which it is bound is taken together with an adjacent R4b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R5 and R6 are each independently methyl or cycloalkyl;
- R7 is, at each occurrence, independently H or C1-C12 alkyl;
- R8 and R9 are each independently unsubstituted C1-C12 alkyl; or R8 and R9, together with the nitrogen atom to which they are attached, form a 5, 6 or 7-membered heterocyclic ring comprising one nitrogen atom;
- a and d are each independently an integer from 0 to 24;
- b and c are each independently an integer from 1 to 24;
- e is 1 or 2; and
- x is 0, 1 or 2.
- In some embodiments of Formula (I), L1 and L2 are independently - O(C=O)- or -(C=O)O-.
- In certain embodiments of Formula (I), at least one of R1a, R2a, R3a or R4a is C1-C12 alkyl, or at least one of L1 or L2 is -O(C=O)- or -(C=O)O-. In other embodiments, R1a and R1b are not isopropyl when a is 6 or n-butyl when a is 8.
- In still further embodiments of Formula (I), at least one of R1a, R2a, R3a or R4a is C1-C12 alkyl, or at least one of L1 or L2 is -O(C=O)- or -(C=O)O-; and
R1a and R1b are not isopropyl when a is 6 or n-butyl when a is 8. - In other embodiments of Formula (1), R8 and R9 are each independently unsubstituted C1-C12 alkyl; or R8 and R9, together with the nitrogen atom to which they are attached, form a 5, 6 or 7-membered heterocyclic ring comprising one nitrogen atom;
In certain embodiments of Formula (I), any one of L1 or L2 may be -O(C=O)- or a carbon-carbon double bond. L1 and L2 may each be -O(C=O)- or may each be a carbon-carbon double bond. - In some embodiments of Formula (I), one of L1 or L2 is -O(C=O)-. In other embodiments, both L1 and L2 are -O(C=O)-.
- In some embodiments of Formula (I), one of L1 or L2 is -(C=O)O-. In other embodiments, both L1 and L2 are -(C=O)O-.
- In some other embodiments of Formula (I), one of L1 or L2 is a carbon-carbon double bond. In other embodiments, both L1 and L2 are a carbon-carbon double bond.
- In still other embodiments of Formula (I), one of L1 or L2 is -O(C=O)- and the other of L1 or L2 is -(C=O)O-. In more embodiments, one of L1 or L2 is -O(C=O)- and the other of L1 or L2 is a carbon-carbon double bond. In yet more embodiments, one of L1 or L2 is -(C=O)O- and the other of L1 or L2 is a carbon-carbon double bond.
- It is understood that "carbon-carbon" double bond, as used throughout the specification, refers to one of the following structures:
-
-
-
- In certain embodiments of the lipid compound of Formula (I), a, b, c and d are each independently an integer from 2 to 12 or an integer from 4 to 12. In other embodiments, a, b, c and d are each independently an integer from 8 to 12 or 5 to 9. In some certain embodiments, a is 0. In some embodiments, a is 1. In other embodiments, a is 2. In more embodiments, a is 3. In yet other embodiments, a is 4. In some embodiments, a is 5. In other embodiments, a is 6. In more embodiments, a is 7. In yet other embodiments, a is 8. In some embodiments, a is 9. In other embodiments, a is 10. In more embodiments, a is 11. In yet other embodiments, a is 12. In some embodiments, a is 13. In other embodiments, a is 14. In more embodiments, a is 15. In yet other embodiments, a is 16.
- In some other embodiments of Formula (I), b is 1. In other embodiments, b is 2. In more embodiments, b is 3. In yet other embodiments, b is 4. In some embodiments, b is 5. In other embodiments, b is 6. In more embodiments, b is 7. In yet other embodiments, b is 8. In some embodiments, b is 9. In other embodiments, b is 10. In more embodiments, b is 11. In yet other embodiments, b is 12. In some embodiments, b is 13. In other embodiments, b is 14. In more embodiments, b is 15. In yet other embodiments, b is 16.
- In some more embodiments of Formula (I), c is 1. In other embodiments, c is 2. In more embodiments, c is 3. In yet other embodiments, c is 4. In some embodiments, c is 5. In other embodiments, c is 6. In more embodiments, c is 7. In yet other embodiments, c is 8. In some embodiments, c is 9. In other embodiments, c is 10. In more embodiments, c is 11. In yet other embodiments, c is 12. In some embodiments, c is 13. In other embodiments, c is 14. In more embodiments, c is 15. In yet other embodiments, c is 16.
- In some certain other embodiments of Formula (I), d is 0. In some embodiments, d is 1. In other embodiments, d is 2. In more embodiments, d is 3. In yet other embodiments, d is 4. In some embodiments, d is 5. In other embodiments, d is 6. In more embodiments, d is 7. In yet other embodiments, d is 8. In some embodiments, d is 9. In other embodiments, d is 10. In more embodiments, d is 11. In yet other embodiments, d is 12. In some embodiments, d is 13. In other embodiments, d is 14. In more embodiments, d is 15. In yet other embodiments, d is 16.
- In some other various embodiments of Formula (I), a and d are the same. In some other embodiments, b and c are the same. In some other specific embodiments, a and d are the same and b and c are the same.
- The sum of a and b and the sum of c and d in Formula (I) are factors which may be varied to obtain a lipid of Formula (I) having the desired properties. In one embodiment, a and b are chosen such that their sum is an integer ranging from 14 to 24. In other embodiments, c and d are chosen such that their sum is an integer ranging from 14 to 24. In further embodiment, the sum of a and b and the sum of c and d are the same. For example, in some embodiments the sum of a and b and the sum of c and d are both the same integer which may range from 14 to 24. In still more embodiments, a. b, c and d are selected such the sum of a and b and the sum of c and d is 12 or greater.
- In some embodiments of Formula (I), e is 1. In other embodiments, e is 2.
- The substituents at R1a, R2a, R3a and R4a of Formula (I) are not particularly limited. In certain embodiments R1a, R2a, R3a and R4a are H at each occurrence. In certain other embodiments at least one of R1a, R2a, R3a and R4a is C1-C12 alkyl. In certain other embodiments at least one of R1a, R2a, R3a and R4a is C1-C8 alkyl. In certain other embodiments at least one of R1a, R2a, R3a and R4a is C1-C6 alkyl. In some of the foregoing embodiments, the C1-C8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
- In certain embodiments of Formula (I), R1a, R1b, R4a and R4b are C1-C12 alkyl at each occurrence.
- In further embodiments of Formula (I), at least one of R1b, R2b, R3b and R4b is H or R1b, R2b, R3b and R4b are H at each occurrence.
- In certain embodiments of Formula (I), R1b together with the carbon atom to which it is bound is taken together with an adjacent R1b and the carbon atom to which it is bound to form a carbon-carbon double bond. In other embodiments of the foregoing R4b together with the carbon atom to which it is bound is taken together with an adjacent R4b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- The substituents at R5 and R6 of Formula (I) are not particularly limited in the foregoing embodiments. In certain embodiments one or both of R5 or R6 is methyl. In certain other embodiments one or both of R5 or R6 is cycloalkyl for example cyclohexyl. In these embodiments the cycloalkyl may be substituted or not substituted. In certain other embodiments the cycloalkyl is substituted with C1-C12 alkyl, for example tert-butyl.
- The substituents at R7 are not particularly limited in the foregoing embodiments of Formula I. In certain embodiments at least one R7 is H. In some other embodiments, R7 is H at each occurrence. In certain other embodiments R7 is C1-C12 alkyl.
- In certain other of the foregoing embodiments of Formula (I), one of R8 or R9 is methyl. In other embodiments, both R8 and R9 are methyl.
- In some different embodiments of Formula (I), R8 and R9, together with the nitrogen atom to which they are attached, form a 5, 6 or 7-membered heterocyclic ring. In some embodiments of the foregoing, R8 and R9, together with the nitrogen atom to which they are attached, form a 5-membered heterocyclic ring, for example a pyrrolidinyl ring.
- In various different embodiments, the lipid of Formula (I) has one of the structures set forth in Table 1 below.
Table 1: Representative Lipids of Formula (I) No. Structure pKa I-1 - I-2 5.64 I-3 7.15 I-4 6.43 I-5 6.28 I-6 6.12 I-7 - I-8 - I-9 - I-10 - I-11 6.36 I-12 - I-13 6.51 I-14 - I-15 6.30 I-16 6.63 I-17 - I-18 - I-13 6.72 I-20 6.44 I-21 6.28 I-22 6.53 I-23 6.24 I-24 6.28 I-25 6.20 I-26 6.89 I-27 6.30 I-28 6.20 I-29 6.22 I-30 - I-31 6.33 I-32 6.47 I-33 6.27 I-34 - I-35 6.21 I-36 - I-37 - I-38 6.24 I-39 5.82 I-40 6.38 I-41 5.91 -
- one of L1 or L2 is -O(C=O)-, -(C=O)O-, -C(=O)-, -O-, -S(O)x-, -S-S-, -C(=O)S-, SC(=O)-, -NRaC(=O)-, -C(=O)NRa-, NRaC(=O)NRa-, -OC(=O)NRa- or -NRaC(=O)O-, and the other of L1 or L2 is -O(C=O)-, -(C=O)O-, -C(=O)-, -O-, -S(O)x-, -S-S-, -C(=O)S-, SC(=O)-, -NRaC(=O)-, -C(=O)NRa-, ,NRaC(=O)NRa-, -OC(=O)NRa- or
-NRaC(=O)O- or a direct bond; - G1 is C1-C2 alkylene, -(C=O)-, -O(C=O)-, -SC(=O)-, -NRaC(=O)- or a direct bond;
- G2 is -C(=O)-, -(C=O)O-, -C(=O)S-, -C(=O)NRa- or a direct bond;
- G3 is C1-C6 alkylene;
- Ra is H or C1-C12 alkyl;
- R1a and R1b are, at each occurrence, independently either: (a) H or C1-C12 alkyl; or (b) R1a is H or C1-C12 alkyl, and R1b together with the carbon atom to which it is bound is taken together with an adjacent R1b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R2a and R2b are, at each occurrence, independently either: (a) H or C1-C12 alkyl; or (b) R2a is H or C1-C12 alkyl, and R2b together with the carbon atom to which it is bound is taken together with an adjacent R2b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R3a and R3b are, at each occurrence, independently either (a): H or C1-C12 alkyl; or (b) R3a is H or C1-C12 alkyl, and R3b together with the carbon atom to which it is bound is taken together with an adjacent R3b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R4a and R4b are, at each occurrence, independently either: (a) H or C1-C12 alkyl; or (b) R4a is H or C1-C12 alkyl, and R4b together with the carbon atom to which it is bound is taken together with an adjacent R4b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R5 and R6 are each independently H or methyl;
- R7 is C4-C20 alkyl;
- R8 and R9 are each independently C1-C12 alkyl; or R8 and R9, together with the nitrogen atom to which they are attached, form a 5, 6 or 7-membered heterocyclic ring;
- a, b, c and d are each independently an integer from 1 to 24; and
- x is 0, 1 or 2.
- In some embodiments of Formula (II), L1 and L2 are each independently -O(C=O)-, -(C=O)O- or a direct bond. In other embodiments, G1 and G2 are each independently -(C=O)- or a direct bond. In some different embodiments, L1 and L2 are each independently -O(C=O)-, -(C=O)O- or a direct bond; and G1 and G2 are each independently -(C=O)- or a direct bond.
- In some different embodiments of Formula (II), L1 and L2 are each independently -C(=O)-, -O-, -S(O)x-, -S-S-, -C(=O)S-, -SC(=O)-, -NRa-, -NRaC(=O)-, -C(=O)NRa-, -NRaC(=O)NRa, -OC(=O)NRa-, -NRaC(=O)O-, -NRaS(O)xNRa-, -NRaS( O)x- or -S(O)xNRa-.
-
- In some embodiments of Formula (II), the lipid compound has Formula (IIA). In other embodiments, the lipid compound has Formula (IIB).
- In any of the foregoing embodiments of Formula (II), one of L1 or L2 is -O(C=O)-. For example, in some embodiments each of L1 and L2 are -O(C=O)-.
- In some different embodiments of Formula (II), one of L1 or L2 is -(C=O)O-. For example, in some embodiments each of L1 and L2 is -(C=O)O-.
- In different embodiments of Formula (II), one of L1 or L2 is a direct bond. As used herein, a "direct bond" means the group (e.g., L1 or L2) is absent. For example, in some embodiments each of L1 and L2 is a direct bond.
- In other different embodiments of Formula (II), for at least one occurrence of R1a and R1b, R1a is H or C1-C12 alkyl, and R1b together with the carbon atom to which it is bound is taken together with an adjacent R1b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- In still other different embodiments of Formula (II), for at least one occurrence of R4a and R4b, R4a is H or C1-C12 alkyl, and R4b together with the carbon atom to which it is bound is taken together with an adjacent R4b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- In more embodiments of Formula (II), for at least one occurrence of R2a and R2b, R2a is H or C1-C12 alkyl, and R2b together with the carbon atom to which it is bound is taken together with an adjacent R2b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- In other different embodiments of Formula (II), for at least one occurrence of R3a and R3b, R3a is H or C1-C12 alkyl, and R3b together with the carbon atom to which it is bound is taken together with an adjacent R3b and the carbon atom to which it is bound to form a carbon-carbon double bond.
-
- In some embodiments of Formula (II), the lipid compound has Formula (IIC). In other embodiments, the lipid compound has Formula (IID).
- In various embodiments of Formulae (IIC) or (IID), e, f, g and h are each independently an integer from 4 to 10.
- In certain embodiments of Formula (II), a, b, c and d are each independently an integer from 2 to 12 or an integer from 4 to 12. In other embodiments, a, b, c and d are each independently an integer from 8 to 12 or 5 to 9. In some certain embodiments, a is 0. In some embodiments, a is 1. In other embodiments, a is 2. In more embodiments, a is 3. In yet other embodiments, a is 4. In some embodiments, a is 5. In other embodiments, a is 6. In more embodiments, a is 7. In yet other embodiments, a is 8. In some embodiments, a is 9. In other embodiments, a is 10. In more embodiments, a is 11. In yet other embodiments, a is 12. In some embodiments, a is 13. In other embodiments, a is 14. In more embodiments, a is 15. In yet other embodiments, a is 16.
- In some embodiments of Formula (II), b is 1. In other embodiments, b is 2. In more embodiments, b is 3. In yet other embodiments, b is 4. In some embodiments, b is 5. In other embodiments, b is 6. In more embodiments, b is 7. In yet other embodiments, b is 8. In some embodiments, b is 9. In other embodiments, b is 10. In more embodiments, b is 11. In yet other embodiments, b is 12. In some embodiments, b is 13. In other embodiments, b is 14. In more embodiments, b is 15. In yet other embodiments, b is 16.
- In some embodiments of Formula (II), c is 1. In other embodiments, c is 2. In more embodiments, c is 3. In yet other embodiments, c is 4. In some embodiments, c is 5. In other embodiments, c is 6. In more embodiments, c is 7. In yet other embodiments, c is 8. In some embodiments, c is 9. In other embodiments, c is 10. In more embodiments, c is 11. In yet other embodiments, c is 12. In some embodiments, c is 13. In other embodiments, c is 14. In more embodiments, c is 15. In yet other embodiments, c is 16.
- In some certain embodiments of Formula (II), d is 0. In some embodiments, d is 1. In other embodiments, d is 2. In more embodiments, d is 3. In yet other embodiments, d is 4. In some embodiments, d is 5. In other embodiments, d is 6. In more embodiments, d is 7. In yet other embodiments, d is 8. In some embodiments, d is 9. In other embodiments, d is 10. In more embodiments, d is 11. In yet other embodiments, d is 12. In some embodiments, d is 13. In other embodiments, d is 14. In more embodiments, d is 15. In yet other embodiments, d is 16.
- In some embodiments of Formula (II), e is 1. In other embodiments, e is 2. In more embodiments, e is 3. In yet other embodiments, e is 4. In some embodiments, e is 5. In other embodiments, e is 6. In more embodiments, e is 7. In yet other embodiments, e is 8. In some embodiments, e is 9. In other embodiments, e is 10. In more embodiments, e is 11. In yet other embodiments, e is 12.
- In some embodiments of Formula (II), f is 1. In other embodiments, f is 2. In more embodiments, f is 3. In yet other embodiments, f is 4. In some embodiments, f is 5. In other embodiments, f is 6. In more embodiments, f is 7. In yet other embodiments, f is 8. In some embodiments, f is 9. In other embodiments, f is 10. In more embodiments, f is 11. In yet other embodiments, f is 12.
- In some embodiments of Formula (II), g is 1. In other embodiments, g is 2. In more embodiments, g is 3. In yet other embodiments, g is 4. In some embodiments, g is 5. In other embodiments, g is 6. In more embodiments, g is 7. In yet other embodiments, g is 8. In some embodiments, g is 9. In other embodiments, g is 10. In more embodiments, g is 11. In yet other embodiments, g is 12.
- In some embodiments of Formula (II), h is 1. In other embodiments, e is 2. In more embodiments, h is 3. In yet other embodiments, h is 4. In some embodiments, e is 5. In other embodiments, h is 6. In more embodiments, h is 7. In yet other embodiments, h is 8. In some embodiments, h is 9. In other embodiments, h is 10. In more embodiments, h is 11. In yet other embodiments, h is 12.
- In some other various embodiments of Formula (II), a and d are the same. In some other embodiments, b and c are the same. In some other specific embodiments and a and d are the same and b and c are the same.
- The sum of a and b and the sum of c and d of Formula (II) are factors which may be varied to obtain a lipid having the desired properties. In one embodiment, a and b are chosen such that their sum is an integer ranging from 14 to 24. In other embodiments, c and d are chosen such that their sum is an integer ranging from 14 to 24. In further embodiment, the sum of a and b and the sum of c and d are the same. For example, in some embodiments the sum of a and b and the sum of c and d are both the same integer which may range from 14 to 24. In still more embodiments, a. b, c and d are selected such that the sum of a and b and the sum of c and d is 12 or greater.
- The substituents at R1a, R2a, R3a and R4a of Formula (II) are not particularly limited. In some embodiments, at least one of R1a, R2a, R3a and R4a is H. In certain embodiments R1a, R2a, R3a and R4a are H at each occurrence. In certain other embodiments at least one of R1a, R2a, R3a and R4a is C1-C12 alkyl. In certain other embodiments at least one of R1a, R2a, R3a and R4a is Ci-Cs alkyl. In certain other embodiments at least one of R1a, R2a, R3a and R4a is C1-C6 alkyl. In some of the foregoing embodiments, the C1-C8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
- In certain embodiments of Formula (II), R1a, R1b, R4a and R4b are C1-C12 alkyl at each occurrence.
- In further embodiments of Formula (II), at least one of R1b, R2b, R3b and R4b is H or R1b, R2b, R3b and R4b are H at each occurrence.
- In certain embodiments of Formula (II), R1b together with the carbon atom to which it is bound is taken together with an adjacent R1b and the carbon atom to which it is bound to form a carbon-carbon double bond. In other embodiments of the foregoing R4b together with the carbon atom to which it is bound is taken together with an adjacent R4b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- The substituents at R5 and R6 of Formula (II) are not particularly limited in the foregoing embodiments. In certain embodiments one of R5 or R6 is methyl. In other embodiments each of R5 or R6 is methyl.
- The substituents at R7 of Formula (II) are not particularly limited in the foregoing embodiments. In certain embodiments R7 is C6-C16 alkyl. In some other embodiments, R7 is C6-C9 alkyl. In some of these embodiments, R7 is substituted with -(C=O)ORb, -O(C=O)Rb, -C(=O)Rb, -ORb, -S(O)xRb, -S-SRb, -C(=O)SRb, -SC(=O)Rb, -NRaRb, -NRaC(=O)Rb, -C(=O)NRaRb, -NRaC(=O)NRaRb, -OC(=O)NRaRb, -NRaC(=O)ORb, -NRaS(O)xNRaRb, =NRaS(O)xRb or -S(O)xNRaRb, wherein: Ra is H or C1-C12 alkyl; Rb is C1-C15 alkyl; and x is 0, 1 or 2. For example, in some embodiments R7 is substituted with -(C=O)ORb or -O(C=O)Rb.
-
- In certain other of the foregoing embodiments of Formula (II), one of R8 or R9 is methyl. In other embodiments, both R8 and R9 are methyl.
- In some different embodiments of Formula (II), R8 and R9, together with the nitrogen atom to which they are attached, form a 5, 6 or 7-membered heterocyclic ring. In some embodiments of the foregoing, R8 and R9, together with the nitrogen atom to which they are attached, form a 5-membered heterocyclic ring, for example a pyrrolidinyl ring. In some different embodiments of the foregoing, R8 and R9, together with the nitrogen atom to which they are attached, form a 6-membered heterocyclic ring, for example a piperazinyl ring.
- In still other embodiments of the foregoing lipids of Formula (II), G3 is C2-C4 alkylene, for example C3 alkylene. In various different embodiments, the lipid compound has one of the structures set forth in Table 2 below
Table 2: Representative Lipids of Formula (II) No. Structure pKa II-1 5.64 II-2 - II-3 - II-4 - II-5 6.27 II-6 6.14 II-7 5.93 II-8 5.35 II-9 6.27 II-10 6.16 II-11 6.13 II-12 6.21 II-13 6.22 II-14 6.33 II-15 6.32 II-16 6.37 II-17 6.27 II-18 - II-19 - II-20 - II-21 - II-22 - II-23 - II-24 6.14 II-25 - II-26 - II-27 - II-28 - II-29 - II-30 - II-31 - II-32 - II-33 - II-34 - II-35 5.97 II-36 6.13 II-37 5.61 II-38 6.45 II-39 6.45 II-40 6.57 II-41 - II-42 - II-43 - II-44 - II-45 - II-46 - -
- one of L1 or L2 is -O(C=O)-, -(C=O)O-, -C(=O)-, -O-, -S(O)x-, -S-S-, -C(=O)S-, SC(=O)-, -NRaC(=O)-, -C(=O)NRa-, NRaC(=O)NRa-, -OC(=O)NRa- or -NRaC(=O)O-, and the other of L1 or L2 is -O(C=O)-, -(C=O)O-, -C(=O)-, -O-, -S(O)x-, -S-S-, -C(=O)S-, SC(=0)-, -NRaC(=O)-, -C(=O)NRa-, ,NRaC(=O)NRa-, -OC(=O)NRa- or
-NRaC(=O)O- or a direct bond; - G1 and G2 are each independently unsubstituted C1-C12 alkylene or C1-C12 alkenylene;
- G3 is C1-C24 alkylene, C1-C24 alkenylene, C3-C8 cycloalkylene, C3-C8 cycloalkenylene;
- Ra is H or C1-C12 alkyl;
- R1 and R2 are each independently C6-C24 alkyl or C6-C24 alkenyl;
- R3 is H, OR5, CN, -C(=O)OR4, -OC(=O)R4 or -NR5C(=O)R4;
- R4 is C1-C12 alkyl;
- R5 is H or C1-C6 alkyl; and
- x is 0, 1 or 2.
-
- A is a 3 to 8-membered cycloalkyl or cycloalkylene ring;
- R6 is, at each occurrence, independently H, OH or C1-C24 alkyl;
- n is an integer ranging from 1 to 15.
- In some of the foregoing embodiments of Formula (III), the lipid has Formula (IIIA), and in other embodiments, the lipid has Formula (IIIB).
-
- In any of the foregoing embodiments of Formula (III), one of L1 or L2 is -O(C=O)-. For example, in some embodiments each of L1 and L2 are -O(C=O)-. In some different embodiments of any of the foregoing, L1 and L2 are each independently -(C=O)O- or -O(C=O)-. For example, in some embodiments each of L1 and L2 is -(C=O)O-.
-
-
- In some of the foregoing embodiments of Formula (III), n is an integer ranging from 2 to 12, for example from 2 to 8 or from 2 to 4. For example, in some embodiments, n is 3, 4, 5 or 6. In some embodiments, n is 3. In some embodiments, n is 4. In some embodiments, n is 5. In some embodiments, n is 6.
- In some other of the foregoing embodiments of Formula (III), y and z are each independently an integer ranging from 2 to 10. For example, in some embodiments, y and z are each independently an integer ranging from 4 to 9 or from 4 to 6.
- In some of the foregoing embodiments of Formula (III), R6 is H. In other of the foregoing embodiments, R6 is C1-C24 alkyl. In other embodiments, R6 is OH.
- In some embodiments of Formula (III), G3 is unsubstituted. In other embodiments, G3 is substituted. In various different embodiments, G3 is linear C1-C24 alkylene or linear C1-C24 alkenylene.
-
- R7a and R7b are, at each occurrence, independently H or C1-C12 alkyl; and
- a is an integer from 2 to 12,
- wherein R7a, R7b and a are each selected such that R1 and R2 each independently comprise from 6 to 20 carbon atoms. For example, in some embodiments a is an integer ranging from 5 to 9 or from 8 to 12.
- In some of the foregoing embodiments of Formula (III), at least one occurrence of R7a is H. For example, in some embodiments, R7a is H at each occurrence. In other different embodiments of the foregoing, at least one occurrence of R7b is C1-C8 alkyl. For example, in some embodiments, C1-C8 alkyl is methyl, ethyl, n-propyl, isopropyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
-
- In some of the foregoing embodiments of Formula (III), R3 is OH, CN, -C(=O)OR4, -OC(=O)R4 or -NHC(=O)R4. In some embodiments, R4 is methyl or ethyl.
- In various different embodiments, a cationic lipid has one of the structures set forth in Table 3 below.
Table 3: Representative Compounds of Formula (III) No. Structure pKa III-1 5.89 III-2 6.05 III-3 6.09 III-4 5.60 III-5 5.59 III-6 5.42 III-7 6.11 III-8 5.84 III-9 - III-10 - III-11 - III-12 - III-13 - III-14 - III-15 6.14 III-16 6.31 III-17 6.28 III-18 - III-19 - III-20 6.36 III-21 - III-22 6.10 III-23 5.98 III-24 - III-25 6.22 III-26 5.84 III-27 5.77 III-28 - III-29 - III-30 6.09 III-31 - III-32 - III-33 - III-34 - III-35 - III-36 - III-37 - III-38 - III-39 - III-40 - III-41 - III-42 - III-43 - III-44 - III-45 - III-46 - III-47 - III-48 - III-49 - -
- one of G1 or G2 is, at each occurrence, -O(C=O)-, -(C=O)O-, -C(=O)-, -O-, -S(O)y-, -S-S-, -C(=O)S-, SC(=O)-, -N(Ra)C(=O)-, -C(=O)N(Ra)-, -N(Ra)C(=O)N(Ra)-, -OC(=O)N(Ra)- or -N(Ra)C(=O)O-, and the other of G1 or G2 is, at each occurrence, -O(C=O)-, -(C=O)O-, -C(=O)-, -O-, -S(O)y-, -S-S-, -C(=O)S-, -SC(=O)-, -N(Ra)C(=O)-, -C(=O)N(Ra)-, -N(Ra)C(=O)N(Ra)-, -OC(=O)N(Ra)- or -N(Ra)C(=O)O- or a direct bond;
- L is, at each occurrence, ~O(C=O)-, wherein ~ represents a covalent bond to X;
- X is CRa;
- Z is alkyl, cycloalkyl or a monovalent moiety comprising at least one polar functional group when n is 1; or Z is alkylene, cycloalkylene or a polyvalent moiety comprising at least one polar functional group when n is greater than 1;
- Ra is, at each occurrence, independently H, C1-C12 alkyl, C1-C12 hydroxylalkyl, C1-C12 aminoalkyl, C1-C12 alkylaminylalkyl, C1-C12 alkoxyalkyl, C1-C12 alkoxycarbonyl, C1-C12 alkylcarbonyloxy, C1-C12 alkylcarbonyloxyalkyl or C1-C12 alkylcarbonyl;
- R is, at each occurrence, independently either: (a) H or C1-C12 alkyl; or (b) R together with the carbon atom to which it is bound is taken together with an adjacent R and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R1 and R2 have, at each occurrence, the following structure, respectively:
- a1 and a2 are, at each occurrence, independently an integer from 3 to 12;
- b1 and b2 are, at each occurrence, independently 0 or 1;
- c1 and c2 are, at each occurrence, independently an integer from 5 to 10;
- d1 and d2 are, at each occurrence, independently an integer from 5 to 10;
- y is, at each occurrence, independently an integer from 0 to 2; and
- n is an integer from 1 to 6,
- wherein each alkyl, alkylene, hydroxylalkyl, aminoalkyl, alkylaminylalkyl, alkoxyalkyl, alkoxycarbonyl, alkylcarbonyloxy, alkylcarbonyloxyalkyl and alkylcarbonyl is optionally substituted with one or more substituent.
- In some embodiments of Formula (IV), G1 and G2 are each independently -O(C=O)- or -(C=O)O-.
- In other embodiments of Formula (IV), X is CH.
- In different embodiments of Formula (IV), the sum of a1 + b1 + c1 or the sum of a2 + b2 + c2 is an integer from 12 to 26.
- In still other embodiments of Formula (IV), a1 and a2 are independently an integer from 3 to 10. For example, in some embodiments a1 and a2 are independently an integer from 4 to 9.
- In various embodiments of Formula (IV), b1 and b2 are 0. In different embodiments, b1 and b2 are 1.
- In more embodiments of Formula (IV), c1, c2, d1 and d2 are independently an integer from 6 to 8.
- In other embodiments of Formula (IV), c1 and c2 are, at each occurrence, independently an integer from 6 to 10, and d1 and d2 are, at each occurrence, independently an integer from 6 to 10.
- In other embodiments of Formula (IV), c1 and c2 are, at each occurrence, independently an integer from 5 to 9, and d1 and d2 are, at each occurrence, independently an integer from 5 to 9.
- In more embodiments of Formula (IV), Z is alkyl, cycloalkyl or a monovalent moiety comprising at least one polar functional group when n is 1. In other embodiments, Z is alkyl.
- In various embodiments of the foregoing Formula (IV), R is, at each occurrence, independently either: (a) H or methyl; or (b) R together with the carbon atom to which it is bound is taken together with an adjacent R and the carbon atom to which it is bound to form a carbon-carbon double bond. In certain embodiments, each R is H. In other embodiments at least one R together with the carbon atom to which it is bound is taken together with an adjacent R and the carbon atom to which it is bound to form a carbon-carbon double bond.
-
-
-
- one of G1 or G2 is, at each occurrence, -O(C=O)-, -(C=O)O-, -C(=O)-, -O-, -S(O)y-, -S-S-, -C(=O)S-, SC(=O)-, -N(Ra)C(=O)-, -C(=O)N(Ra)-, -N(Ra)C(=O)N(Ra)-, -OC(=O)N(Ra)- or -N(Ra)C(=O)O-, and the other of G1 or G2 is, at each occurrence, -O(C=O)-, -(C=O)O-, -C(=O)-, -O-, -S(O)y-, -S-S-, -C(=O)S-, -SC(=O)-, -N(Ra)C(=O)-, -C(=O)N(Ra)-, -N(Ra)C(=O)N(Ra)-, -OC(=O)N(Ra)- or -N(Ra)C(=O)O- or a direct bond;
- L is, at each occurrence, ~O(C=O)-, wherein ~ represents a covalent bond to X;
- X is CRa;
- Z is alkyl, cycloalkyl or a monovalent moiety comprising at least one polar functional group when n is 1; or Z is alkylene, cycloalkylene or a polyvalent moiety comprising at least one polar functional group when n is greater than 1;
- Ra is, at each occurrence, independently H, C1-C12 alkyl, C1-C12 hydroxylalkyl, C1-C12 aminoalkyl, C1-C12 alkylaminylalkyl, C1-C12 alkoxyalkyl, C1-C12 alkoxycarbonyl, C1-C12 alkylcarbonyloxy, C1-C12 alkylcarbonyloxyalkyl or C1-C12 alkylcarbonyl;
- R is, at each occurrence, independently either: (a) H or C1-C12 alkyl; or (b) R together with the carbon atom to which it is bound is taken together with an adjacent R and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R1 and R2 have, at each occurrence, the following structure, respectively:
- R' is, at each occurrence, independently H or C1-C12 alkyl;
- a1 and a2 are, at each occurrence, independently an integer from 3 to 12;
- b1 and b2 are, at each occurrence, independently 0 or 1;
- c1 and c2 are, at each occurrence, independently an integer from 2 to 12;
- d1 and d2 are, at each occurrence, independently an integer from 2 to 12;
- y is, at each occurrence, independently an integer from 0 to 2; and
- n is an integer from 1 to 6,
- wherein a1, a2, c1, c2, d1 and d2 are selected such that the sum of a1+c1+d1 is an integer from 18 to 30, and the sum of a2+c2+d2 is an integer from 18 to 30, and wherein each alkyl, alkylene, hydroxylalkyl, aminoalkyl, alkylaminylalkyl, alkoxyalkyl, alkoxycarbonyl, alkylcarbonyloxy, alkylcarbonyloxyalkyl and alkylcarbonyl is optionally substituted with one or more substituent.
- In certain embodiments of Formula (V), G1 and G2 are each independently -O(C=O)- or -(C=O)O-.
- In other embodiments of Formula (V), X is CH.
- In some embodiments of Formula (V), the sum of a1+c1+d1 is an integer from 20 to 30, and the sum of a2+c2+d2 is an integer from 18 to 30. In other embodiments, the sum of a1+c1+d1 is an integer from 20 to 30, and the sum of a2+c2+d2 is an integer from 20 to 30. In more embodiments of Formula (V), the sum of a1 + b1 + c1 or the sum of a2 + b2 + c2 is an integer from 12 to 26. In other embodiments, a1, a2, c1, c2, d1 and d2 are selected such that the sum of a1+c1+d1 is an integer from 18 to 28, and the sum of a2+c2+d2 is an integer from 18 to 28,
In still other embodiments of Formula (V), a1 and a2 are independently an integer from 3 to 10, for example an integer from 4 to 9. - In yet other embodiments of Formula (V), b1 and b2 are 0. In different embodiments b1 and b2 are 1.
- In certain other embodiments of Formula (V), c1, c2, d1 and d2 are independently an integer from 6 to 8.
- In different other embodiments of Formula (V), Z is alkyl or a monovalent moiety comprising at least one polar functional group when n is 1; or Z is alkylene or a polyvalent moiety comprising at least one polar functional group when n is greater than 1.
- In more embodiments of Formula (V), Z is alkyl, cycloalkyl or a monovalent moiety comprising at least one polar functional group when n is 1. In other embodiments, Z is alkyl.
- In other different embodiments of Formula (V), R is, at each occurrence, independently either: (a) H or methyl; or (b) R together with the carbon atom to which it is bound is taken together with an adjacent R and the carbon atom to which it is bound to form a carbon-carbon double bond. For example in some embodiments each R is H. In other embodiments at least one R together with the carbon atom to which it is bound is taken together with an adjacent R and the carbon atom to which it is bound to form a carbon-carbon double bond.
- In more embodiments, each R' is H.
- In certain embodiments of Formula (V), the sum of a1+c1+d1 is an integer from 20 to 25, and the sum of a2+c2+d2 is an integer from 20 to 25.
-
-
- In any of the foregoing embodiments of Formula (IV) or (V), n is 1. In other of the foregoing embodiments of Formula (IV) or (V), n is greater than 1.
- In more of any of the foregoing embodiments of Formula (IV) or (V), Z is a mono- or polyvalent moiety comprising at least one polar functional group. In some embodiments, Z is a monovalent moiety comprising at least one polar functional group. In other embodiments, Z is a polyvalent moiety comprising at least one polar functional group.
- In more of any of the foregoing embodiments of Formula (IV) or (V), the polar functional group is a hydroxyl, alkoxy, ester, cyano, amide, amino, alkylaminyl, heterocyclyl or heteroaryl functional group.
- In any of the foregoing embodiments of Formula (IV) or (V), Z is hydroxyl, hydroxylalkyl, alkoxyalkyl, amino, aminoalkyl, alkylaminyl, alkylaminylalkyl, heterocyclyl or heterocyclylalkyl.
-
- R5 and R6 are independently H or C1-C6 alkyl;
- R7 and R8 are independently H or C1-C6 alkyl or R7 and R8, together with the nitrogen atom to which they are attached, join to form a 3-7 membered heterocyclic ring; and
- x is an integer from 0 to 6.
-
- R5 and R6 are independently H or C1-C6 alkyl;
- R7 and R8 are independently H or C1-C6 alkyl or R7 and R8, together with the nitrogen atom to which they are attached, join to form a 3-7 membered heterocyclic ring; and
- x is an integer from 0 to 6.
-
- R5 and R6 are independently H or C1-C6 alkyl;
- R7 and R8 are independently H or C1-C6 alkyl or R7 and R8, together with the nitrogen atom to which they are attached, join to form a 3-7 membered heterocyclic ring; and
- x is an integer from 0 to 6.
- In some other embodiments of Formula (IV) or (V), Z is hydroxylalkyl, cyanoalkyl or an alkyl substituted with one or more ester or amide groups.
-
-
-
-
-
-
- L1 and L2 are each independently -O(C=O)-, -(C=O)O-, -C(=O)-, -O-, -S(O)x-, -S-S-, -C(=O)S-, -SC(=O)-, -NRaC(=O)-, -C(=O)NRa-, -NRaC(=O)NRa-, -OC(=O)NRa-, -NRaC(=O)O- or a direct bond;
- G1 is C1-C2 alkylene, -(C=O)-, -O(C=O)-, -SC(=O)-, -NRaC(=O)- or a direct bond;
- G2 is -C(=O)-, -(C=O)O-, -C(=O)S-, -C(=O)NRa- or a direct bond;
- G3 is C1-C6 alkylene;
- Ra is H or C1-C12 alkyl;
- R1a and R1b are, at each occurrence, independently either: (a) H or C1-C12 alkyl; or (b) R1a is H or C1-C12 alkyl, and R1b together with the carbon atom to which it is bound is taken together with an adjacent R1b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R2a and R2b are, at each occurrence, independently either: (a) H or C1-C12 alkyl; or (b) R2a is H or C1-C12 alkyl, and R2b together with the carbon atom to which it is bound is taken together with an adjacent R2b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R3a and R3b are, at each occurrence, independently either (a): H or C1-C12 alkyl; or (b) R3a is H or C1-C12 alkyl, and R3b together with the carbon atom to which it is bound is taken together with an adjacent R3b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R4a and R4b are, at each occurrence, independently either: (a) H or C1-C12 alkyl; or (b) R4a is H or C1-C12 alkyl, and R4b together with the carbon atom to which it is bound is taken together with an adjacent R4b and the carbon atom to which it is bound to form a carbon-carbon double bond;
- R5 and R6 are each independently H or methyl;
- R7 is H or C1-C20 alkyl;
- R8 is OH, -N(R9)(C=O)R10, -(C=O)NR9R10, -NR9R10, -(C=O)OR11 or -O(C=O)R11, provided that G3 is C4-C6 alkylene when R8 is -NR9R10,
- R9 and R10 are each independently H or C1-C12 alkyl;
- R11 is aralkyl;
- a, b, c and d are each independently an integer from 1 to 24; and
- x is 0, 1 or 2,
- In some embodiments, L1 and L2 are each independently -O(C=O)-, -(C=O)O- or a direct bond. In other embodiments, G1 and G2 are each independently -(C=O)- or a direct bond. In some different embodiments, L1 and L2 are each independently -O(C=O)-, -(C=O)O- or a direct bond; and G1 and G2 are each independently - (C=O)- or a direct bond.
- In some different embodiments, L1 and L2 are each independently -C(=O)-, -O-, -S(O)x-, -S-S-, -C(=O)S-, -SC(=O)-, -NRa-, -NRaC(=O)-, -C(=O)NRa-, -NRaC(=O)NRa, -OC(=O)NRa-, -NRaC(=O)O-, -NRaS(O)xNRa-, -NRaS(O)x- or -S(O)xNRa-.
-
- In some embodiments, the compound has Formula (VIA). In other embodiments, the compound has Formula (VIB).
- In any of the foregoing embodiments, one of L1 or L2 is -O(C=O)-. For example, in some embodiments each of L1 and L2 are -O(C=O)-.
- In some different embodiments of any of the foregoing, one of L1 or L2 is -(C=O)O-. For example, in some embodiments each of L1 and L2 is -(C=O)O-.
- In different embodiments, one of L1 or L2 is a direct bond. As used herein, a "direct bond" means the group (e.g., L1 or L2) is absent. For example, in some embodiments each of L1 and L2 is a direct bond.
- In other different embodiments of the foregoing, for at least one occurrence of R1a and R1b, R1a is H or C1-C12 alkyl, and R1b together with the carbon atom to which it is bound is taken together with an adjacent R1b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- In still other different embodiments, for at least one occurrence of R4a and R4b, R4a is H or C1-C12 alkyl, and R4b together with the carbon atom to which it is bound is taken together with an adjacent R4b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- In more embodiments, for at least one occurrence of R2a and R2b, R2a is H or C1-C12 alkyl, and R2b together with the carbon atom to which it is bound is taken together with an adjacent R2b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- In other different embodiments of any of the foregoing, for at least one occurrence of R3a and R3b, R3a is H or C1-C12 alkyl, and R3b together with the carbon atom to which it is bound is taken together with an adjacent R3b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- It is understood that "carbon-carbon" double bond refers to one of the following structures:
-
- In some embodiments, the compound has Formula (VIC). In other embodiments, the compound has Formula (VID).
- In various embodiments of the compounds of Formulas (VIC) or (VID), e, f, g and h are each independently an integer from 4 to 10.
-
- In certain embodiments of the foregoing, a, b, c and d are each independently an integer from 2 to 12 or an integer from 4 to 12. In other embodiments, a, b, c and d are each independently an integer from 8 to 12 or 5 to 9. In some certain embodiments, a is 0. In some embodiments, a is 1. In other embodiments, a is 2. In more embodiments, a is 3. In yet other embodiments, a is 4. In some embodiments, a is 5. In other embodiments, a is 6. In more embodiments, a is 7. In yet other embodiments, a is 8. In some embodiments, a is 9. In other embodiments, a is 10. In more embodiments, a is 11. In yet other embodiments, a is 12. In some embodiments, a is 13. In other embodiments, a is 14. In more embodiments, a is 15. In yet other embodiments, a is 16.
- In some embodiments, b is 1. In other embodiments, b is 2. In more embodiments, b is 3. In yet other embodiments, b is 4. In some embodiments, b is 5. In other embodiments, b is 6. In more embodiments, b is 7. In yet other embodiments, b is 8. In some embodiments, b is 9. In other embodiments, b is 10. In more embodiments, b is 11. In yet other embodiments, b is 12. In some embodiments, b is 13. In other embodiments, b is 14. In more embodiments, b is 15. In yet other embodiments, b is 16.
- In some embodiments, c is 1. In other embodiments, c is 2. In more embodiments, c is 3. In yet other embodiments, c is 4. In some embodiments, c is 5. In other embodiments, c is 6. In more embodiments, c is 7. In yet other embodiments, c is 8. In some embodiments, c is 9. In other embodiments, c is 10. In more embodiments, c is 11. In yet other embodiments, c is 12. In some embodiments, c is 13. In other embodiments, c is 14. In more embodiments, c is 15. In yet other embodiments, c is 16.
- In some certain embodiments, d is 0. In some embodiments, d is 1. In other embodiments, d is 2. In more embodiments, d is 3. In yet other embodiments, d is 4. In some embodiments, d is 5. In other embodiments, d is 6. In more embodiments, d is 7. In yet other embodiments, d is 8. In some embodiments, d is 9. In other embodiments, d is 10. In more embodiments, d is 11. In yet other embodiments, d is 12. In some embodiments, d is 13. In other embodiments, d is 14. In more embodiments, d is 15. In yet other embodiments, d is 16.
- In some embodiments, e is 1. In other embodiments, e is 2. In more embodiments, e is 3. In yet other embodiments, e is 4. In some embodiments, e is 5. In other embodiments, e is 6. In more embodiments, e is 7. In yet other embodiments, e is 8. In some embodiments, e is 9. In other embodiments, e is 10. In more embodiments, e is 11. In yet other embodiments, e is 12.
- In some embodiments, f is 1. In other embodiments, f is 2. In more embodiments, f is 3. In yet other embodiments, f is 4. In some embodiments, f is 5. In other embodiments, f is 6. In more embodiments, f is 7. In yet other embodiments, f is 8. In some embodiments, f is 9. In other embodiments, f is 10. In more embodiments, f is 11. In yet other embodiments, f is 12.
- In some embodiments, g is 1. In other embodiments, g is 2. In more embodiments, g is 3. In yet other embodiments, g is 4. In some embodiments, g is 5. In other embodiments, g is 6. In more embodiments, g is 7. In yet other embodiments, g is 8. In some embodiments, g is 9. In other embodiments, g is 10. In more embodiments, g is 11. In yet other embodiments, g is 12.
- In some embodiments, h is 1. In other embodiments, e is 2. In more embodiments, h is 3. In yet other embodiments, h is 4. In some embodiments, e is 5. In other embodiments, h is 6. In more embodiments, h is 7. In yet other embodiments, h is 8. In some embodiments, h is 9. In other embodiments, h is 10. In more embodiments, h is 11. In yet other embodiments, h is 12.
- In some other various embodiments, a and d are the same. In some other embodiments, b and c are the same. In some other specific embodiments a and d are the same and b and c are the same.
- The sum of a and b and the sum of c and d are factors which may be varied to obtain a lipid having the desired properties. In one embodiment, a and b are chosen such that their sum is an integer ranging from 14 to 24. In other embodiments, c and d are chosen such that their sum is an integer ranging from 14 to 24. In further embodiment, the sum of a and b and the sum of c and d are the same. For example, in some embodiments the sum of a and b and the sum of c and d are both the same integer which may range from 14 to 24. In still more embodiments, a. b, c and d are selected such that the sum of a and b and the sum of c and d is 12 or greater.
- The substituents at R1a, R2a, R3a and R4a are not particularly limited. In some embodiments, at least one of R1a, R2a, R3a and R4a is H. In certain embodiments R1a, R2a, R3a and R4a are H at each occurrence In certain other embodiments at least one of R1a, R2a, R3a and R4a is C1-C12 alkyl. In certain other embodiments at least one of R1a, R2a, R3a and R4a is Ci-Cs alkyl. In certain other embodiments at least one of R1a, R2a, R3a and R4a is C1-C6 alkyl. In some of the foregoing embodiments, the C1-C8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
- In certain embodiments of the foregoing, R1a, R1b, R4a and R4b are C1-C12 alkyl at each occurrence.
- In further embodiments of the foregoing, at least one of R1b, R2b, R3b and R4b is H or R1b, R2b, R3b and R4b are H at each occurrence.
- In certain embodiments of the foregoing, R1b together with the carbon atom to which it is bound is taken together with an adjacent R1b and the carbon atom to which it is bound to form a carbon-carbon double bond. In other embodiments of the foregoing R4b together with the carbon atom to which it is bound is taken together with an adjacent R4b and the carbon atom to which it is bound to form a carbon-carbon double bond.
- The substituents at R5 and R6 are not particularly limited in the foregoing embodiments. In certain embodiments one of R5 or R6 is methyl. In other embodiments each of R5 or R6 is methyl.
- The substituents at R7 are not particularly limited in the foregoing embodiments. In certain embodiments R7 is C6-C16 alkyl. In some other embodiments, R7 is C6-C9 alkyl. In some of these embodiments, R7 is substituted with -(C=O)ORb, -O(C=O)Rb, -C(=O)Rb, -ORb, -S(O)xRb, -S-SRb, -C(=O)SRb, -SC(=O)Rb, -NRaRb, -NRaC(=O)Rb, -C(=O)NRaRb, -NRaC(=O)NRaRb, -OC(=O)NRaRb, -NRaC(=O)ORb, -NRaS(O)xNRaRb, -NRaS(O)xRb or -S(O)xNRaRb, wherein: Ra is H or C1-C12 alkyl; Rb is C1-C15 alkyl; and x is 0, 1 or 2. For example, in some embodiments R7 is substituted with -(C=O)ORb or -O(C=O)Rb.
-
- In certain embodiments, R8 is OH.
- In other embodiments, R8 is -N(R9)(C=O)R10. In some other embodiments, R8 is -(C=O)NR9R10. In still more embodiments, R8 is -NR9R10. In some of the foregoing embodiments, R9 and R10 are each independently H or C1-C8 alkyl, for example H or C1-C3 alkyl. In more specific of these embodiments, the C1-C8 alkyl or C1-C3 alkyl is unsubstituted or substituted with hydroxyl. In other of these embodiments, R9 and R10 are each methyl.
- In yet more embodiments, R8 is -(C=O)OR11. In some of these embodiments R11 is benzyl.
-
- In still other embodiments of the foregoing compounds, G3 is C2-C5 alkylene, for example C2-C4 alkylene, C3 alkylene or C4 alkylene. In some of these embodiments, R8 is OH. In other embodiments, G2 is absent and R7 is C1-C2 alkylene, such as methyl.
- In various different embodiments, the compound has one of the structures set forth in Table 5 below.
Table 5: Representative Compounds of Formula (VI) No. Structure VI-1 VI-2 VI-3 VI-4 VI-5 VI-6 VI-7 VI-8 VI-9 VI-10 VI-11 VI-12 VI-13 VI-14 VI-15 VI-16 VI-17 VI-18 VI-19 VI-20 VI-21 VI-22 VI-23 VI-24 VI-25 VI-26 VI-27 VI-28 VI-29 VI-30 VI-31 VI-32 VI-33 VI-34 VI-35 VI-36 VI-37 -
- X and X' are each independently N or CR;
- Y and Y' are each independently absent, -O(C=O)-, -(C=O)O- or NR, provided that:
- a)Y is absent when X is N;
- b) Y' is absent when X' is N;
- c) Y is -O(C=O)-, -(C=O)O- or NR when X is CR; and
- d) Y' is -O(C=O)-, -(C=O)O- or NR when X' is CR,
- L1 and L1' are each independently -O(C=O)R1, -(C=O)OR1, -C(=O)R1, -OR1, -S(O)zR1, -S-SR1, -C(=O)SR1, -SC(=O)R1, -NRaC(=O)R1, -C(=O)NRbRc, -NRaC(=O)NRbRc, -OC(=O)NRbRc or -NRaC(=O)OR1;
- L2 and L2' are each independently -O(C=O)R2, -(C=O)OR2, -C(=O)R2, -OR2, -S(O)zR2, -S-SR2, -C(=O)SR2, -SC(=O)R2, -NRdC(=O)R2, -C(=O)NReRf, -NRdC(=O)NReRf, -OC(=O)NReRf;-NRdC(=O)OR2 or a direct bond to R2;
- G1, G1', G2 and G2' are each independently C2-C12 alkylene or C2-C12 alkenylene;
- G3 is C2-C24 heteroalkylene or C2-C24 heteroalkenylene;
- Ra, Rb, Rd and Re are, at each occurrence, independently H, C1-C12 alkyl or C2-C12 alkenyl;
- Rc and Rf are, at each occurrence, independently C1-C12 alkyl or C2-C12 alkenyl;
- R is, at each occurrence, independently H or C1-C12 alkyl;
- R1 and R2 are, at each occurrence, independently branched C6-C24 alkyl or branched C6-C24 alkenyl;
- z is 0, 1 or 2, and
- In other different embodiments of Formula (VII):
- X and X' are each independently N or CR;
- Y and Y' are each independently absent or NR, provided that:
- a)Y is absent when X is N;
- b) Y' is absent when X' is N,
- c) Y is NR when X is CR; and
- d) Y' is NR when X' is CR,
- L1 and L1' are each independently -O(C=O)R1, -(C=O)OR1, -C(=O)R1, -OR1, -S(O)zR1, -S-SR1, -C(=O)SR1, -SC(=O)R1, -NRaC(=O)R1, -C(=O)NRbRc, -NRaC(=O)NRbRc, -OC(=O)NRbRc or -NRaC(=O)OR1;
- L2 and L2' are each independently -O(C=O)R2, -(C=O)OR2, -C(=O)R2, -OR2, -S(O)zR2, -S-SR2, -C(=O)SR2, -SC(=O)R2, -NRdC(=O)R2, -C(=O)NReRf, -NRdC(=O)NReRf, -OC(=O)NReRf;-NRdC(=O)OR2 or a direct bond to R2;
- G1, G1', G2 and G2' are each independently C2-C12 alkylene or C2-C12 alkenylene;
- G3 is C2-C24 alkyleneoxide or C2-C24 alkenyleneoxide;
- Ra, Rb, Rd and Re are, at each occurrence, independently H, C1-C12 alkyl or C2-C12 alkenyl;
- Rc and Rf are, at each occurrence, independently C1-C12 alkyl or C2-C12 alkenyl;
- R is, at each occurrence, independently H or C1-C12 alkyl;
- R1 and R2 are, at each occurrence, independently branched C6-C24 alkyl or branched C6-C24 alkenyl;
- z is 0, 1 or 2, and
- In some embodiments, G3 is C2-C24 alkyleneoxide or C2-C24 alkenyleneoxide. In certain embodiments, G3 is unsubstituted. In other embodiments, G3 is substituted, for example substituted with hydroxyl. In more specific embodiments G3 is C2-C12 alkyleneoxide, for example, in some embodiments G3 is C3-C7 alkyleneoxide or in other embodiments G3 is C3-C12 alkyleneoxide.
- In other embodiments, G3 is C2-C24 alkyleneaminyl or C2-C24 alkenyleneaminyl, for example C6-C12 alkyleneaminyl. In some of these embodiments, G3 is unsubstituted. In other of these embodiments, G3 is substituted with C1-C6 alkyl.
- In some embodiments, X and X' are each N, and Y and Y' are each absent. In other embodiments, X and X' are each CR, and Y and Y' are each NR. In some of these embodiments, R is H.
- In certain embodiments, X and X' are each CR, and Y and Y' are each independently -O(C=O)- or -(C=O)O-.
- In some of the foregoing embodiments, the compound has one of the following Formulas (VIIA), (VIIB), (VIIC), (VIID), (VIIE), (VIIF), (VIIG) or (VIIH):
- In some of the foregoing embodiments, L1 and L1' are each independently -O(C=O)R1, -(C=O)OR1 or -C(=O)NRbRc, and L2 and L2' are each independently - O(C=O)R2, -(C=O)OR2 or -C(=O)NReRf For example, in some embodiments L1 and L1' are each -(C=O)OR1, and L2 and L2' are each -(C=O)OR2.. In other embodiments L1 and L1' are each -(C=O)OR1, and L2 and L2' are each -C(=O)NReRf. In other embodiments L1 and L1' are each -C(=O)NRbRc, and L2 and L2' are each -C(=O)NReRf.
- In some embodiments of the foregoing, G1, G1', G2 and G2' are each independently C2-C8 alkylene, for example C4-C8 alkylene.
-
- R7a and R7b are, at each occurrence, independently H or C1-C12 alkyl; and
- a is an integer from 2 to 12,
- In some of the foregoing embodiments, at least one occurrence of R7a is H. For example, in some embodiments, R7a is H at each occurrence. In other different embodiments of the foregoing, at least one occurrence of R7b is C1-C8 alkyl. For example, in some embodiments, C1-C8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
-
- In some of the foregoing embodiments, Rb, Rc, Re and Rf, when present, are each independently C3-C12 alkyl. For example, in some embodiments Rb, Rc, Re and Rf, when present, are n-hexyl and in other embodiments Rb, Rc, Re and Rf, when present, are n-octyl.
-
-
- X is N, and Y is absent; or X is CR, and Y is NR;
- L1 is -O(C=O)R1, -(C=O) OR1, -C(=O)R1, -OR1, -S(O)xR1, -S-SR1, -C(=O)SR1, -SC(=O)R1, -NRaC(=O)R1, -C(=O)NRbRc, -NRaC(=O)NRbRc, -OC(=O)NRbRc or -NRaC(=O)OR1;
- L2 is -O(C=O)R2, -(C=O)OR2, -C(=O)R2, -OR2, -S(O)xR2, -S-SR2, -C(=O)SR2, -SC(=O)R2, -NRdC(=O)R2, -C(=O)NReRf, -NRdC(=O)NReRf, -OC(=O)NReRf; -NRdC(=O)OR2 or a direct bond to R2;
- L3 is -O(C=O)R3 or -(C=O)OR3;
- G1 and G2 are each independently C2-C12 alkylene or C2-C12 alkenylene;
- G3 is C1-C24 alkylene, C2-C24 alkenylene, C1-C24 heteroalkylene or C2-C24 heteroalkenylene;
- Ra, Rb, Rd and Re are each independently H or C1-C12 alkyl or C1-C12 alkenyl;
- Rc and Rf are each independently C1-C12 alkyl or C2-C12 alkenyl;
- each R is independently H or C1-C12 alkyl;
- R1, R2 and R3 are each independently C1-C24 alkyl or C2-C24 alkenyl; and
- x is 0, 1 or 2, and
- wherein each alkyl, alkenyl, alkylene, alkenylene, heteroalkylene and heteroalkenylene is independently substituted or unsubstituted unless otherwise specified.
- In more embodiments of Formula (VIII):
- X is N, and Y is absent; or X is CR, and Y is NR;
- L1 is -O(C=O)R1, -(C=O)OR1, -C(=O)R1, -OR1, -S(O)xR1, -S-SR1, -C(=O)SR1, -SC(=O)R1, -NRaC(=O)R1, -C(=O)NRbRc, -NRaC(=O)NRbRc, -OC(=O)NRbRc or -NRaC(=O)OR1;
- L2 is -O(C=O)R2, -(C=O)OR2, -C(=O)R2, -OR2, -S(O)xR2, -S-SR2, -C(=O)SR2, -SC(=O)R2, -NRdC(=O)R2, -C(=O)NReRf, -NRdC(=O)NReRf, -OC(=O)NReRf; -NRdC(=O)OR2 or a direct bond to R2;
- L3 is -O(C=O)R3 or -(C=O)OR3;
- G1 and G2 are each independently C2-C12 alkylene or C2-C12 alkenylene;
- G3 is C1-C24 alkylene, C2-C24 alkenylene, C1-C24 heteroalkylene or C2-C24 heteroalkenylene when X is CR, and Y is NR; and G3 is C1-C24 heteroalkylene or C2-C24 heteroalkenylene when X is N, and Y is absent;
- Ra, Rb, Rd and Re are each independently H or C1-C12 alkyl or C1-C12 alkenyl;
- Rc and Rf are each independently C1-C12 alkyl or C2-C12 alkenyl;
- each R is independently H or C1-C12 alkyl;
- R1, R2 and R3 are each independently C1-C24 alkyl or C2-C24 alkenyl; and
- x is 0, 1 or 2, and
- wherein each alkyl, alkenyl, alkylene, alkenylene, heteroalkylene and heteroalkenylene is independently substituted or unsubstituted unless otherwise specified.
- In other embodiments of Formula (VIII):
- X is N and Y is absent, or X is CR and Y is NR;
- L1 is -O(C=O)R1, -(C=O)OR1, -C(=O)R1, -OR1, -S(O)xR1, -S-SR1, -C(=O)SR1, -SC(=O)R1, -NRaC(=O)R1, -C(=O)NRbRc, -NRaC(=O)NRbRc, -OC(=O)NRbRc or -NRaC(=O)OR1;
- L2 is -O(C=O)R2, -(C=O)OR2, -C(=O)R2, -OR2, -S(O)xR2, -S-SR2, -C(=O)SR2, -SC(=O)R2, -NRdC(=O)R2, -C(=O)NReRf, -NRdC(=O)NReRf, -OC(=O)NReRf; -NRdC(=O)OR2 or a direct bond to R2;
- L3 is -O(C=O)R3 or -(C=O)OR3;
- G1 and G2 are each independently C2-C12 alkylene or C2-C12 alkenylene;
- G3 is C1-C24 alkylene, C2-C24 alkenylene, C1-C24 heteroalkylene or C2-C24 heteroalkenylene;
- Ra, Rb, Rd and Re are each independently H or C1-C12 alkyl or C1-C12 alkenyl;
- Rc and Rf are each independently C1-C12 alkyl or C2-C12 alkenyl;
- each R is independently H or C1-C12 alkyl;
- R1, R2 and R3 are each independently branched C6-C24 alkyl or branched C6-C24 alkenyl; and
- x is 0, 1 or 2, and
- In certain embodiments, G3 is unsubstituted. In more specific embodiments G3 is C2-C12 alkylene, for example, in some embodiments G3 is C3-C7 alkylene or in other embodiments G3 is C3-C12 alkylene. In some embodiments, G3 is C2 or C3 alkylene.
- In other embodiments, G3 is C1-C12 heteroalkylene, for example C1-C12 aminylalkylene.
- In certain embodiments, X is N and Y is absent. In other embodiments, X is CR and Y is NR, for example in some of these embodiments R is H.
-
- In some of the foregoing embodiments, L1 is -O(C=O)R1, -(C=O)OR1 or -C(=O)NRbRc, and L2 is -O(C=O)R2, -(C=O)OR2 or -C(=O)NReRf. In other specific embodiments, L1 is -(C=O)OR1 and L2 is -(C=O)OR2. In any of the foregoing embodiments, L3 is -(C=O)OR3.
- In some of the foregoing embodiments, G1 and G2 are each independently C2-C12 alkylene, for example C4-C10 alkylene.
-
- R7a and R7b are, at each occurrence, independently H or C1-C12 alkyl; and
- a is an integer from 2 to 12,
- In some of the foregoing embodiments, at least one occurrence of R7a is H. For example, in some embodiments, R7a is H at each occurrence. In other different embodiments of the foregoing, at least one occurrence of R7b is C1-C8 alkyl. For example, in some embodiments, C1-C8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
- In some of the foregoing embodiments, X is CR, Y is NR and R3 is C1-C12 alkyl, such as ethyl, propyl or butyl. In some of these embodiments, R1 and R2 are each independently branched C6-C24 alkyl.
-
- In certain embodiments, R1 and R2 and R3 are each, independently, branched C6-C24 alkyl and R3 is C1-C24 alkyl or C2-C24 alkenyl.
- In some of the foregoing embodiments, Rb, Rc, Re and Rf are each independently C3-C12 alkyl. For example, in some embodiments Rb, Rc, Re and Rf are n-hexyl and in other embodiments Rb, Rc, Re and Rf are n-octyl.
-
-
- L1 is -O(C=O)R1, -(C=O)OR1, -C(=O)R1, -OR1, -S(O)xR1, -S-SR1, -C(=O)SR1, -SC(=O)R1, -NRaC(=O)R1, -C(=O)NRbRc, -NRaC(=O)NRbRc, - OC(=O)NRbRc or -NRaC(=O)OR1;
- L2 is -O(C=O)R2, -(C=O)OR2, -C(=O)R2, -OR2, -S(O)xR2, -S-SR2, -C(=O)SR2, -SC(=O)R2, -NRdC(=O)R2, -C(=O)NReRf, -NRdC(=O)NReRf, - OC(=O)NReRf;
-NRdC(=O)OR2 or a direct bond to R2; - G1 and G2 are each independently C2-C12 alkylene or C2-C12 alkenylene;
- G3 is C1-C24 alkylene, C2-C24 alkenylene, C3-C8 cycloalkylene or C3-C8 cycloalkenylene;
- Ra, Rb, Rd and Re are each independently H or C1-C12 alkyl or C1-C12 alkenyl;
- Rc and Rf are each independently C1-C12 alkyl or C2-C12 alkenyl;
- R1 and R2 are each independently branched C6-C24 alkyl or branched C6-C24 alkenyl;
- R3 is -N(R4)R5;
- R4 is C1-C12 alkyl;
- R5 is substituted C1-C12 alkyl; and
- x is 0, 1 or 2, and
- In certain embodiments, G3 is unsubstituted. In more specific embodiments G3 is C2-C12 alkylene, for example, in some embodiments G3 is C3-C7 alkylene or in other embodiments G3 is C3-C12 alkylene. In some embodiments, G3 is C2 or C3 alkylene.
- In some of the foregoing embodiments, the compound has the following Formula (IXA):
- In some of the foregoing embodiments, L1 is -O(C=O)R1, -(C=O)OR1 or -C(=O)NRbRc, and L2 is -O(C=O)R2, -(C=O)OR2 or -C(=O)NReRf. For example, in some embodiments L1 and L2 are -(C=O)OR1 and -(C=O)OR2, respectively. In other embodiments L1 is -(C=O)OR1 and L2 is -C(=O)NReRf. In other embodiments L1 is -C(=O)NRbRc and L2 is -C(=O)NReRf.
-
- In some of the foregoing embodiments, the compound has Formula (IXB), in other embodiments, the compound has Formula (IXC) and in still other embodiments the compound has the Formula (IXD). In other embodiments, the compound has Formula (IXE).
-
- In some of the foregoing embodiments, y and z are each independently an integer ranging from 2 to 10, 2 to 8, from 4 to 10 or from 4 to 7. For example, in some embodiments, y is 4, 5, 6, 7, 8, 9, 10, 11 or 12. In some embodiments, a is 4, 5, 6, 7, 8, 9, 10, 11 or 12. In some embodiments, y and z are the same, while in other embodiments y and z are different.
-
- R7a and R7b are, at each occurrence, independently H or C1-C12 alkyl; and
- a is an integer from 2 to 12,
- In some of the foregoing embodiments, at least one occurrence of R7a is H. For example, in some embodiments, R7a is H at each occurrence. In other different embodiments of the foregoing, at least one occurrence of R7b is C1-C8 alkyl. For example, in some embodiments, C1-C8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
-
- In some of the foregoing embodiments, Rb, Rc, Re and Rf are each independently C3-C12 alkyl. For example, in some embodiments Rb, Rc, Re and Rf are n-hexyl and in other embodiments Rb, Rc, Re and Rf are n-octyl.
- In any of the foregoing embodiments, R4 is substituted or unsubstituted: methyl, ethyl, propyl, n-butyl, n-hexyl, n-octyl or n-nonyl. For example, in some embodiments R4 is unsubstituted. In other R4 is substituted with one or more substituents selected from the group consisting of -ORg, -NRgC(=O)Rh, -C(=O)NRgRh, - C(=O)Rh, -OC(=O)Rh, -C(=O)ORh and -ORiOH, wherein:
- Rg is, at each occurrence independently H or C1-C6 alkyl;
- Rh is at each occurrence independently C1-C6 alkyl; and
- Ri is, at each occurrence independently C1-C6 alkylene.
- In other of the foregoing embodiments, R5 is substituted: methyl, ethyl, propyl, n-butyl, n-hexyl, n-octyl or n-nonyl. In some embodiments, R5 is substituted ethyl or substituted propyl. In other different embodiments, R5 is substituted with hydroxyl. In still more embodiments, R5 is substituted with one or more substituents selected from the group consisting of -ORg, -NRgC(=O)Rh, -C(=O)NRgRh, -C(=O)Rh, - OC(=O)Rh, -C(=O)ORh and -ORiOH, wherein:
- Rg is, at each occurrence independently H or C1-C6 alkyl;
- Rh is at each occurrence independently C1-C6 alkyl; and
- Ri is, at each occurrence independently C1-C6 alkylene.
- In other embodiments, R4 is unsubstituted methyl, and R5 is substituted: methyl, ethyl, propyl, n-butyl, n-hexyl, n-octyl or n-nonyl. In some of these embodiments, R5 is substituted with hydroxyl.
-
-
-
- G1 is -OH, -NR3R4, -(C=O)NR5 or -NR3(C=O)R5;
- G2 is -CH2- or -(C=O)-;
- R is, at each occurrence, independently H or OH;
- R1 and R2 are each independently branched, saturated or unsaturated C12-C36 alkyl;
- R3 and R4 are each independently H or straight or branched, saturated or unsaturated C1-C6 alkyl;
- R5 is straight or branched, saturated or unsaturated C1-C6 alkyl; and
- n is an integer from 2 to 6.
- In some embodiments, R1 and R2 are each independently branched, saturated or unsaturated C12-C30 alkyl, C12-C20 alkyl, or C15-C20 alkyl. In some specific embodiments, R1 and R2 are each saturated. In certain embodiments, at least one of R1 and R2 is unsaturated.
-
-
- R6 and R7 are, at each occurrence, independently H or straight or branched, saturated or unsaturated C1-C14 alkyl;
- a and b are each independently an integer ranging from 1 to 15,
- provided that R6 and a, and R7 and b, are each independently selected such that R1 and R2, respectively, are each independently branched, saturated or unsaturated C12-C36 alkyl.
- In some of the foregoing embodiments, the compound has the following Formula (XB):
R8, R9, R10 and R11 are each independently straight or branched, saturated or unsaturated C4-C12 alkyl, provided that R8 and R9, and R10 and R11, are each independently selected such that R1 and R2, respectively, are each independently branched, saturated or unsaturated C12-C36 alkyl. In some embodiments of (XB), R8, R9, R10 and R11 are each independently straight or branched, saturated or unsaturated C6-C10 alkyl. In certain embodiments of (XB), at least one of R8, R9, R10 and R11 is unsaturated. In other certain specific embodiments of (XB), each of R8, R9, R10 and R11 is saturated. - In some of the foregoing embodiments, the compound has Formula (XA), and in other embodiments, the compound has Formula (XB).
- In some of the foregoing embodiments, G1 is -OH, and in some embodiments G1 is -NR3R4. For example, in some embodiments, G1 is -NH2, -NHCH3 or -N(CH3)2. In certain embodiments, G1 is -(C=O)NR5. In certain other embodiments, G1 is -NR3(C=O)R5. For example, in some embodiments G1 is -NH(C=O)CH3 or -NH(C=O)CH2CH2CH3.
- In some of the foregoing embodiments, G2 is -CH2-. In some different embodiments, G2 is -(C=O)-.
- In some of the foregoing embodiments, n is an integer ranging from 2 to 6, for example, in some embodiments n is 2, 3, 4, 5 or 6. In some embodiments, n is 2. In some embodiments, n is 3. In some embodiments, n is 4.
- In certain of the foregoing embodiments, at least one of R1, R2, R3, R4 and R5 is unsubstituted. For example, in some embodiments, R1, R2, R3, R4 and R5 are each unsubstituted. In some embodiments, R3 is substituted. In other embodiments R4 is substituted. In still more embodiments, R5 is substituted. In certain specific embodiments, each of R3 and R4 are substituted. In some embodiments, a substituent on R3, R4 or R5 is hydroxyl. In certain embodiments, R3 and R4 are each substituted with hydroxyl.
- In some of the foregoing embodiments, at least one R is OH. In other embodiments, each R is H.
-
-
- L1 is -O(C=O)R1, -(C=O)OR1, -C(=O)R1, -OR1, -S(O)xR1, -S-SR1, -C(=O)SR1, -SC(=O)R1, -NRaC(=O)R1, -C(=O)NRbRc, -NRaC(=O)NRbRc, -OC(=O)NRbRc or -NRaC(=O)OR1;
- L2 is -O(C=O)R2, -(C=O)OR2, -C(=O)R2, -OR2, -S(O)xR2, -S-SR2, -C(=O)SR2, -SC(=O)R2, -NRdC(=O)R2, -C(=O)NReRf, -NRcC(=O)NReRf, -OC(=O)NReRf; -NRdC(=O)OR2 or a direct bond to R2;
- G1a and G2a are each independently C2-C12 alkylene or C2-C12 alkenylene;
- G3 is C1-C24 alkylene, C2-C24 alkenylene, C3-C8 cycloalkylene or C3-C8 cycloalkenylene;
- Ra, Rb, Rd and Re are each independently H or C1-C12 alkyl or C2-C12 alkenyl;
- Rc and Rf are each independently C1-C12 alkyl or C2-C12 alkenyl;
- R1 and R2 are each independently branched C6-C24 alkyl or branched C6-C24 alkenyl;
- R3a is -C(=O)N(R4a)R5a or -C(=O)OR6;
- R4a is C1-C12 alkyl;
- R5a is H or Ci-Cs alkyl or C2-C8 alkenyl;
- R6 is H, aryl or aralkyl; and
- x is 0, 1 or 2, and
- In certain embodiments of Formula (XI), G3 is unsubstituted. In more specific embodiments of Formula (XI), G3 is C3-C12 alkylene. In some embodiments of Formula (XI), G3 is C2 or C3 alkylene.
-
- In some of the foregoing embodiments of Formula (XI), L1 is -O(C=O)R1, -(C=O)OR1 or -C(=O)NRbRc, and L2 is -O(C=O)R2, -(C=O)OR2 or -C(=O)NReRf. For example, in some embodiments of Formula (XI) L1 and L2 are -(C=O)OR1 and - (C=O)OR2, respectively. In other embodiments of Formula (XI) L1 is -(C=O)OR1 and L2 is -C(=O)NReRf. In other embodiments of Formula (XI) L1 is -C(=O)NRbRc and L2 is -C(=O)NReRf.
-
- In some of the foregoing embodiments, the compound has Formula (XIB), in other embodiments, the compound has Formula (XIC) and in still other embodiments the compound has the Formula (XID). In other embodiments, the compound has Formula (XIE).
-
- In some of the foregoing embodiments of Formula (XI), y1 and z1 are each independently an integer ranging from 2 to 10, 2 to 8, from 4 to 10 or from 4 to 7. For example, in some embodiments of Formula (XI), y1 is 4, 5, 6, 7, 8, 9, 10, 11 or 12. In some embodiments of Formula (XI), z1 is 4, 5, 6, 7, 8, 9, 10, 11 or 12. In some embodiments of Formula (XI), y1 and z1 are the same, while in other embodiments of Formula (XI) y1 and z1 are different.
-
- R7a and R7b are, at each occurrence, independently H or C1-C12 alkyl; and
- a is an integer from 2 to 12,
- In some of the foregoing embodiments of Formula (XI), at least one occurrence of R7a is H. For example, in some embodiments of Formula (XI), R7a is H at each occurrence. In other different embodiments of the foregoing, at least one occurrence of R7b is C1-C8 alkyl. For example, in some embodiments, C1-C8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
-
- In some of the foregoing embodiments of Formula (XI), Rb, Rc, Re and Rf are each independently C3-C12 alkyl. For example, in some embodiments of Formula (XI) Rb, Rc, Re and Rf are n-hexyl and in other embodiments of Formula (XI) Rb, Rc, Re and Rf are n-octyl.
- In some of the foregoing embodiments of Formula (XI), R3a is -C(=O)N(R4a)R5a. In more specific embodiments of Formula (XI), R4a is ethyl, propyl, n-butyl, n-hexyl, n-octyl or n-nonyl. In certain embodiments of Formula (XI), R5a is H, methyl, ethyl, propyl, n-butyl, n-hexyl or n-octyl. In some of these embodiments of Formula (XI), R4a and/or R5a is optionally substituted with a substituent, for example hydroxyl.
- In some embodiments of Formula (XI), R3a is -C(=O)OR6. In certain embodiments of Formula (XI), R6 is benzyl and in other embodiments R6 is H.
- In some of the foregoing embodiments of Formula (XI), R4a, R5a and R6 are independently optionally substituted with one or more substituents selected from the group consisting of -ORg, -NRgC(=O)Rh, -C(=O)NRgRh, -C(=O)Rh, -OC(=O)Rh, - C(=O)ORh and -ORiOH, wherein:
- Rg is, at each occurrence independently H or C1-C6 alkyl;
- Rh is at each occurrence independently C1-C6 alkyl; and
- Ri is, at each occurrence independently C1-C6 alkylene.
-
-
-
- L1 is -O(C=O)R1, -(C=O)OR1, -C(=O)R1, -OR1, -S(O)xR1, -S-SR1, -C(=O)SR1, -SC(=O)R1, -NRaC(=O)R1, -C(=O)NRbRc, -NRaC(=O)NRbRc, -OC(=O)NRbRc or -NRaC(=O)OR1;
- L2 is -O(C=O)R2, -(C=O)OR2, -C(=O)R2, -OR2, -S(O)xR2, -S-SR2, -C(=O)SR2, -SC(=O)R2, -NRdC(=O)R2, -C(=O)NReRf, -NRcC(=O)NReRf, -OC(=O)NReRf; -NRdC(=O)OR2 or a direct bond;
- G1b and G2b are each independently C1-C12 alkylene or C2-C12 alkenylene;
- G3 is C1-C24 alkylene, C2-C24 alkenylene, C3-C8 cycloalkylene, C3-C8 cycloalkenylene;
- Ra, Rb, Rd and Re are each independently H or C1-C12 alkyl or C2-C12 alkenyl;
- Rc and Rf are each independently C1-C12 alkyl or C2-C12 alkenyl;
- R1 and R2 are each independently branched C6-C24 alkyl or branched C6-C24 alkenyl;
- R3b is -NR4bC(=O)R5b;
- R4b is H, C1-C12 alkyl or C2-C12 alkenyl;
- R5b is C2-C12 alkyl or C2-C12 alkenyl when R4b is H; or R5 is C1-C12 alkyl or C2-C12 alkenyl when R4b is C1-C12 alkyl or C2-C12 alkenyl; and
- x is 0, 1 or 2, and
- In certain embodiments of Formula (XII), G3 is unsubstituted. In more specific embodiments of Formula (XII) G3 is C1-C12 alkylene, for example, G3 is C3-C5 alkylene or G3 is C3-C12 alkylene.
-
- In some of the foregoing embodiments of Formula (XIIA), L1 and L2 are each independently -O(C=O)R1 or -(C=O)OR1.
-
- In some of the foregoing embodiments, the compound has Formula (XIIB), in other embodiments, the compound has Formula (XIIC).
-
- In some of the foregoing embodiments of Formula (XII), y2 and z2 are each independently an integer ranging from 2 to 12, for example from 2 to 10, from 2 to 8, from 4 to 7 or from 4 to 10. For example, in some embodiments of structure (II), y2 is 4, 5, 6, 7, 8, 9, 10, 11 or 12. In some embodiments of Formula (XII), z2 is 4, 5, 6, 7, 8, 9, 10, 11 or 12. In some embodiments of Formula (XII), y2 and z2 are the same, while in other embodiments of Formula (XII), y2 and z2 are different.
-
- R7a and R7b are, at each occurrence, independently H or C1-C12 alkyl; and
- a is an integer from 2 to 12,
- In some of the foregoing embodiments of Formula (XII), at least one occurrence of R7a is H. For example, in some embodiments of Formula (XII), R7a is H at each occurrence. In other different embodiments of the foregoing, at least one occurrence of R7b is C1-C8 alkyl. For example, in some embodiments of Formula (XII), C1-C8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
-
- In some of the foregoing embodiments of Formula (XII), R4b is H, methyl, ethyl, propyl or octyl. In some embodiments of Formula (XII), R5b is methyl, ethyl, propyl, heptyl or octyl, for example n-heptyl or n-octyl.
- In certain related embodiments of Formula (XII), R4b and R5b are independently optionally substituted with one or more substituents selected from the group consisting of -ORg, -NRgC(=O)Rh, -C(=O)NRgRh, -C(=O)Rh, -OC(=O)Rh, -C(=O)OR h and -ORhOH, wherein:
- Rg is, at each occurrence independently H or C1-C6 alkyl;
- Rh is at each occurrence independently C1-C6 alkyl; and
- R1 is, at each occurrence independently C1-C6 alkylene.
-
- In various different embodiments, the compound of Formula (XII) has one of the structures set forth in Table 11 below.
Table 11: Representative Compounds of Formula (XII) No. Structure XII-1 XII-2 XII-3 XII-4 XII-5 XII-6 XII-7 XII-8 XII-9 XII-10 XII-11 XII-12 XII-13 XII-14 XII-15 XII-16 XII-17 XII-18 XII-19 XII-20 -
- R1 is optionally substituted C1-C24 alkyl or optionally substituted C2-C24 alkenyl;
- R2 and R3 are each independently optionally substituted C1-C36 alkyl;
- R4 and R5 are each independently optionally substituted C1-C6 alkyl, or R4 and R3 join, along with the N to which they are attached, to form a heterocyclyl or heteroaryl;
- L1, L2, and L3 are each independently optionally substituted C1-C18 alkylene;
- G1 is a direct bond, -(CH2)nO(C=O)-, -(CH2)n(C=O)O-, or -(C=O)-;
- G2 and G3 are each independently -(C=O)O- or -O(C=O)-; and
- n is an integer greater than 0.
-
-
- In some embodiments, R1 is optionally substituted C6-C18 alkyl or C14-C18 alkenyl. In certain embodiments, R1 is C8 alkyl, C9 alkyl, C10 alkyl, C12 alkyl, C14 alkyl, or C16 alkyl. In some more specific embodiments, R1 is C16 alkenyl. In certain more specific embodiments, R1 is unbranched. In some embodiments, R1 is branched. In certain embodiments, R1 is unsubstituted.
- In some embodiments, G1 is a direct bond, -(CH2)nO(C=O)-, or - (CH2)n(C=O)O-. In certain embodiments, G1 is a direct bond. In some more specific embodiments, G1 is -(CH2)n(C=O)O- and n is greater than 1. In some embodiments, n is 1-20. In some embodiments n is 1-10. In some embodiments n is 5-11. In some embodiments, n is 6-10. In certain more specific embodiments, n is 5, 6, 7, 8, 9, or 10. In some embodiments, n is 5. In some embodiments, n is 6. In some embodiments, n is 7. In certain embodiments, n is 8. In some embodiments, n is 9. In some embodiments, n is 10.
- In some embodiments, L1 is C1-C6 alkylene. In certain embodiments, L1 is C2 alkylene, C3 alkylene, or C4 alkylene. In some more specific embodiments, L1 is unbranched. In certain more specific embodiments, L1 is unsubstituted.
- In some embodiments, R2 is C8-C24 alkyl. In some embodiments, R3 is C8-C24 alkyl. In some more specific embodiments, R2 and R3 are both C8-C24 alkyl. In some embodiments, R2 and R3 are each independently C11 alkyl, C12 alkyl, C13 alkyl, C14 alkyl, C15 alkyl, C16 alkyl, C18 alkyl, or C20 alkyl. In certain embodiments, R2 is branched. In more specific embodiments, R3 is branched. In some more specific embodiments, R2 and R3 each independently have one of the following structures:
R6 and R7 are each independently C2-C12 alkyl. -
- In some embodiments, L2 and L3 are each independently C4-C10 alkylene. In certain embodiments, L2 and L3 are both C5 alkylene. In some more specific embodiments, L2 and L3 are both C6 alkylene. In certain embodiments, L2 and L3 are both C8 alkylene. In some more specific embodiments, L2 and L3 are both C9 alkylene. In some embodiments, L2 is unbranched. In some embodiments, L3 is unbranched. In more specific embodiments, L2 is unsubstituted. In some embodiments, L2 is unsubstituted.
- In some embodiments, R4 and R5 are each independently C1-C6 alkyl. In more specific embodiments, R4 and R5 are both methyl. In certain embodiments, R4 and R5 are both ethyl. In certain embodiments, R4 is methyl and R5 is n-butyl. In some embodiments, R4 and R5 are both n-butyl. In different embodiments, R4 is methyl and R5 is n-hexyl.
-
- In various different embodiments, the compound has one of the structures set forth in Table 12 below.
Table 12. Representative Lipid Compounds No. Structure pKa XIII-1 - XIII-2 - XIII-3 - XIII-4 - XIII-5 - XIII-6 - XIII-7 6.74 XIII-8 6.68 XIII-9 6.83 XIII-10 - XIII-11 - XIII-12 - XIII-13 - XIII-14 - XIII-15 - XIII-16 6.77 XIII-17 - XIII-18 6.47 XIII-19 - XIII-20 6.84 XIII-21 - XIII-22 - XIII-23 - XIII-24 - XIII-25 6.20 XIII-26 - XIII-27 - XIII-28 - XIII-29 6.81 XIII-30 6.47 XIII-31 5.05 XIII-32 6.41 XIII-33 6.19 XIII-34 - XIII-35 - XIII-36 - XIII-37 - XIII-38 - XIII-39 - XIII-40 - -
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR" and YR";
- R4 is selected from the group consisting of C3-6 carbocycle, -(CH2)nQ, - (CH2)nCHQR, -CHQR, -CQR2, and unsubstituted C1-6 alkyl, where Q is selected from a carbocycle, heterocycle, -OR, -N(R)2, -C(O)NR2, -N(R)C(O)R, -N(R)S(O)2R, - N(R)C(O)N(R)2, -N(R)C(S)N(R)2, -and N(R)R8, and each n is independently selected from 1, 2, 3, 4, and 5;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- Each R is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- Each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", and H;
- Each R" is independently selected from the group consisting of C3-14 alkyl and C3-14 alkenyl;
- Each R* is independently selected from the group consisting of C1-12 alkyl and C2-12 alkenyl;
- Each Y is independently C3-6 carbocycle;
- 1 is selected from 1, 2, 3, 4, and 5
- m is selected from 5, 6, 7. 8, and 9;
- M1 is a bond of M'; and
- M and M' are independently selected from -C(O)O-, -OC(O)-, - C(O)N(R')-, -P(O)(OR')O-, -S-S-, an aryl group, and a heteroaryl group.
-
-
-
-
-
- In a certain embodiment, the lipid compound has formula, wherein R4 is selected from the group consisting of -(CH2)nQ, -(CH2)nCHQR, -CHQR, and -CQ(R)2, where Q is -N(R)R8.
- In some embodiments, M and M' are independently -C(O)O- or -OC(O)-
-
-
-
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R'M'R';
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of hydrogen, a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R10)2(CH2)n-oQ, -CHQR, -CQ(R)2, -C(0)NQR and unsubstituted Ci-e alkyl, where Q is selected from a carbocycle, heterocycle, -OR, - 0(CH2)nN(R)2, -C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -N(R)2, -C(0)N(R)2, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -N(R)R8, -N(R)S(0)2R8, -0(CH2)nOR, -N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, -OC(0)N(R)2, - N(R)C(0)OR, -N(OR)C(0)R, -N(OR)S(0)2R, -N(OR)C(0)OR, -N(OR)C(0)N(R)2, - N(OR)C(S)N(R)2, -N(OR)C(=NR9)N(R)2, -N(OR)C(=CHR9)N(R)2, -C(=NR9)N(R)2, - C(=NR9)R, -C(0)N(R)OR, -(CH2)nN(R)2 and -C(R)N(R)2C(0)OR, each 0 is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5;
- each R5 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R ,
- -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, -P(0)(0R')0-, - S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, NO2, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- R10 is selected from the group consisting of H, OH, C1-3 alkyl, and C2-3 alkenyl;
- each R is independently selected from the group consisting of C1-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, (CH2)qOR*, and H,
- and each q is independently selected from 1, 2, and 3;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and
- C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and
- C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13; and wherein when R4 is - (CH2)nQ, -(CH2)nCHQR, -CHQR, or -CQ(R)2, then (i) Q is not -N(R)2 when n is 1, 2, 3, 4 or 5, or (ii) Q is not 5, 6, or 7-membered heterocycloalkyl when n is 1 or 2.
-
- or a salt or isomer thereof, wherein
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R'M'R';
- R2 and R3 are independently selected from the group consisting of H, Ci-i4 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of hydrogen, a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R10)2(CH2)n-oQ, -CHQR, -CQ(R)2, -C(0)NQR and unsubstituted Ci-e alkyl, where Q is selected from a carbocycle, heterocycle, -OR, - 0(CH2)nN(R)2, -C(0)0R, -0C(0)R, -CX3, -CX2H, -CXH2, -CN, -N(R)2, -C(0)N(R)2, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -N(R)R8, -N(R)S(0)2R8, -0(CH2)n0R, -N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, -0C(0)N(R)2, - N(R)C(0)0R, -N(OR)C(0)R, -N(0R)S(0)2R, -N(0R)C(0)0R, -N(0R)C(0)N(R)2, - N(OR)C(S)N(R)2, -N(OR)C(=NR9)N(R)2, -N(OR)C(=CHR9)N(R)2, -C(=NR9)N(R)2, - C(=NR9)R, -C(O)N(R)OR, -(CH2)nN(R)2 and -C(R)N(R)2C(0)0R, each 0 is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5;
- Rx is selected from the group consisting of C1-6 alkyl, C2-6 alkenyl, - (CH2)vOH, and -(CH2)VN(R)2,
- wherein v is selected from 1, 2, 3, 4, 5, and 6;
- each R5 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C 1-13 alkyl or C2-i3 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, N02, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- R10 is selected from the group consisting of H, OH, C1-3 alkyl, and C2-3 alkenyl;
- each R is independently selected from the group consisting of C1-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, (CH2)qOR*. and H,
- and each q is independently selected from 1, 2, and 3;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-i8 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and
- C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and
- C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13.
- Other aspects the disclosure relate to a compound of Formula (I), wherein R4 is selected from the group consisting -(CH2)nQ, -(CH2)nCHQR, - (CH2)oC(R12)2(CH2)n-oQ, -CHQR, -CQ(R)2, and -C(0)NQR, where Q is - (CH2)nN(R)2.
- Other aspects the disclosure relate to a compound of Formula (III), wherein R4 is selected from the group consisting -(CH2)nQ, -(CH2)nCHQR, - (CH2)oC(R12)2(CH2)n-oQ, -CHQR, -CQ(R)2, and -C(0)NQR, where Q is - (CH2)nN(R)2.
- In some embodiments, a subset of compounds of Formula (I) includes those in which when R4 is -(CH2)nQ, -(CH2)nCHQR, -CHQR, or -CQ(R)2, then (i) Q is not -N(R)2 when n is 1, 2, 3, 4 or 5, or (ii) Q is not 5, 6, or 7-membered heterocycloalkyl when n is 1 or 2.
- For example, when R4 is -(CH2)nQ, -(CH2)nCHQR, - (CH2)oC(R10)2(CH2)n-oQ, -CHQR, or -CQ(R)2, then (i) Q is not -N(R)2 when n is 1, 2, 3, 4 or 5, or (ii) Q is not 5, 6, or 7-membered heterocycloalkyl when n is 1 or 2.
- In another embodiments, another subset of compounds of Formula (I) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R'M'R';
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of hydrogen, a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R10)2(CH2)n-oQ,-CHQR, -CQ(R)2, -C(0)NQR and unsubstituted Ci-e alkyl, where Q is selected from a C3-6 carbocycle, a 5- to 14-membered heteroaryl having one or more heteroatoms selected from N, O, and S, -OR, - 0(CH2)nN(R)2, -C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -CRN(R)2C(0)OR, - N(R)R8, -N(R)S(0)2R8, -0(CH2)nOR, -N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, - OC(0)N(R)2, -N(R)C(0)OR, -N(OR)C(0)R, -N(OR)S(0)2R, -N(OR)C(0)OR, - N(OR)C(0)N(R)2, -N(OR)C(S)N(R)2, -N(OR)C(=NR9)N(R)2, -N(0R)C(=CHR9)N(R)2, -C(=NR9)N(R)2, -C(=NR9)R, -C(0)N(R)OR, -(CH2)nN(R)2, and a 5- to
- 14-membered heterocycloalkyl having one or more heteroatoms selected fromN, O, and S which is substituted with one or more substituents selected from oxo (=0), OH, amino, mono- or di-alkylamino, and Ci-3 alkyl, each o is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5;
- each R5 is independently selected from the group consisting of OH, Ci-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, NO2, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- R10 is selected from the group consisting of H, OH, C 1-3 alkyl, and C2-3 alkenyl;
- each R is independently selected from the group consisting of C1-6 alkyl, C 1-3 alkyl-aryl, C2-3 alkenyl, (CH2)qOR*, and H;
- each R' is independently selected from the group consisting of Ci-ib alkyl, C2-18 alkenyl, -R*YR", -YR", and H,
- and each q is independently selected from 1, 2, and 3;
- each R" is independently selected from the group consisting of C3-15 alkyl and C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13,
- or their N-oxides, or salts or isomers thereof.
- In yet another embodiments, another subset of compounds of Formula (I) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R'M'R';
- R2 and R3 are independently selected from the group consisting of H, Ci-i4 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of hydrogen, a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R10)2(CH2)n-oQ,-CHQR, -CQ(R)2, -C(0)NQR and unsubstituted Ci-e alkyl, where Q is selected from a C3-6 carbocycle, a 5- to 14-membered heterocycle having one or more heteroatoms selected from N, O, and S, -OR, -0(CH2)nN(R)2, -C(0)0R, -0C(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -CRN(R)2C(0)0R, - N(R)R8, -N(R)S(0)2R8, -0(CH2)n0R, -N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, - 0C(0)N(R)2, -N(R)C(0)0R, -N(0R)C(0)R, -N(OR)S(0)2R, -N(0R)C(0)0R, - N(0R)C(0)N(R)2, -N(OR)C(S)N(R)2, -N(OR)C(=NR9)N(R)2, -N(OR)C(=CHR9)N(R)2, -C(=NR9)R, -C(0)N(R)0R, -(CH2)nN(R)2 and -C(=NR9)N(R)2, each 0 is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5; and when Q is a 5- to 14-membered heterocycle and (i) R4 is -(CH2)nQ in which n is 1 or 2, or (ii) R4 is -(CH2)nCHQR in which n is 1, or (iii) R4 is -CHQR, and -CQ(R)2, then Q is either a 5- to 14-membered heteroaryl or 8- to 14-membered heterocycloalkyl;
- each R5 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-i3 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, N02, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- R10 is selected from the group consisting of H, OH, C1-3 alkyl, and C2-3 alkenyl;
- each R is independently selected from the group consisting of C1-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, (CH2)qOR*. and H,
- and each q is independently selected from 1, 2, and 3;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-i8 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13,
- or their N-oxides, or salts or isomers thereof.
- In still another embodiments, another subset of compounds of Formula (I) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R'M'R';
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of hydrogen, a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R10)2(CH2)n-oQ,-CHQR, -CQ(R)2, -C(0)NQR and unsubstituted Ci-e alkyl, where Q is selected from a C3-6 carbocycle, a 5- to 14-membered heteroaryl having one or more heteroatoms selected from N, O, and S, -OR, - 0(CH2)nN(R)2, -C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -CRN(R)2C(0)OR, - N(R)R8, -N(R)S(0)2R8, -0(CH2)nOR, -N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, - OC(0)N(R)2, -N(R)C(0)OR, -N(OR)C(0)R, -N(OR)S(0)2R, -N(OR)C(0)OR, - N(OR)C(0)N(R)2, -N(OR)C(S)N(R)2, -N(OR)C(=NR9)N(R)2, -N(0R)C(=CHR9)N(R)2, -C(=NR9)R, -C(0)N(R)OR, -(CH2)nN(R)2, each 0 is independently selected from 1, 2, 3, and 4, and -C(=NR9)N(R)2, and each n is independently selected from 1, 2, 3, 4, and 5;
- each R5 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, NO2, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- R10 is selected from the group consisting of H, OH, C1-3 alkyl, and C2-3 alkenyl, each R is independently selected from the group consisting of C 1 -6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, (CH2)qOR*, and H,
- and each q is independently selected from 1, 2, and 3;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13,
- or their N-oxides, or salts or isomers thereof.
- In still another embodiments, another subset of compounds of Formula (I) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R"M'R';
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of hydrogen, a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R10)2(CH2)n-oQ, -CHQR, -CQ(R)2, -C(0)NQR and unsubstituted Ci-e alkyl, where Q is selected from a carbocycle, -OR, - 0(CH2)nN(R)2, -C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -N(R)R8, -N(R)S(0)2R8, -0(CH2)nOR, -N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, -OC(0)N(R)2, - N(R)C(0)OR, -N(OR)C(0)R, -N(OR)S(0)2R, -N(OR)C(0)OR, -N(OR)C(0)N(R)2, - N(OR)C(S)N(R)2, -N(OR)C(=NR9)N(R)2, -N(OR)C(=CHR9)N(R)2, -C(=NR9)N(R)2, - C(=NR9)R, -C(0)N(R)OR, -(CH2)nN(R)2 and -C(R)N(R)2C(0)OR, each 0 is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5;
- each R5 is independently selected from the group consisting of OH,CI-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH,CI-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, NO2, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- R10 is selected from the group consisting of H, OH, C1-3 alkyl, and C2-3 alkenyl;
- each R is independently selected from the group consisting of C1-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, and H;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", (CH2)qOR*. and H,
- and each q is independently selected from 1, 2, and 3;
- each R" is independently selected from the group consisting of C3-15 alkyl and
- C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and
- C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13.
- In yet another embodiments, another subset of compounds of Formula (I) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R"M'R';
- R2 and R3 are independently selected from the group consisting of H, C2-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is -(CH2)nQ or -(CH2)nCHQR, where Q is -N(R)2, and n is selected from 3, 4, and 5; each R5 is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- each R is independently selected from the group consisting of C1-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, and H;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and C1-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13,
- or their N-oxides, or salts or isomers thereof.
- In still another embodiment, another subset of compounds of Formula (I) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R"M'R';
- R2 and R3 are independently selected from the group consisting of C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of -(CH2)nQ, -(CH2)nCHQR, - CHQR, and -CQ(R)2, where Q is -N(R)2, and n is selected from 1, 2, 3, 4, and 5;
- each R5 is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- each R is independently selected from the group consisting of C1-6 alkyl, C 1-3 alkyl-aryl, C2-3 alkenyl, and H;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and C1-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13,
- or their N-oxides, or salts or isomers thereof.
- In still another embodiment, another subset of compounds of Formula (I) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R"M'R';
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is -C(0)NQR, where Q is selected from a carbocycle, heterocycle, - C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, -(CH2)nN(R)2, - C(=NR9)N(R)2, -C(=NR9)R, -C(0)N(R)OR, and -C(R)N(R)2C(0)OR, and each n is independently selected from 1, 2, 3, 4, and 5;
- each R5 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R9 is selected from the group consisting of H, CN, NO2, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- each R is independently selected from the group consisting of Ci-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, and H;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", (CH2)qOR*, and H, and each q is independently selected from 1, 2, and 3;
- each R" is independently selected from the group consisting of C3-15 alkyl and
- C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and
- C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13.
- In some embodiments, a subset of compounds of Formula (III) includes those in which, when R4 is -(CH2)nQ, -(CH2)nCHQR, -CHQR, or -CQ(R)2, then (i) Q is not -N(R)2 when n is 1, 2, 3, 4 or 5, or (ii) Q is not 5, 6, or 7-membered heterocycloalkyl when n is 1 or 2.
- In another embodiments, another subset of compounds of Formula (III) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R"M'R';
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of hydrogen, a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R10)2(CH2)n-oQ,-CHQR, -CQ(R)2, -C(0)NQR and unsubstituted Ci-e alkyl, where Q is selected from a C3-6 carbocycle, a 5- to 14-membered heteroaryl having one or more heteroatoms selected from N, O, and S, -OR, - 0(CH2)nN(R)2, -C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -CRN(R)2C(0)OR, - N(R)R8, -N(R)S(0)2R8, -0(CH2)nOR, -N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, - OC(0)N(R)2, -N(R)C(0)OR, -N(OR)C(0)R, -N(OR)S(0)2R, -N(OR)C(0)OR, - N(OR)C(0)N(R)2, -N(OR)C(S)N(R)2, -N(OR)C(=NR9)N(R)2, -N(0R)C(=CHR9)N(R)2, -C(=NR9)N(R)2, -C(=NR9)R, -C(0)N(R)OR, -(CH2)nN(R)2 and a 5- to 14-membered heterocycloalkyl having one or more heteroatoms selected fromN, O, and S which is substituted with one or more substituents selected from oxo (=0), OH, amino, mono- or di-alkylamino, and C1-3 alkyl, each 0 is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5;
- Rx is selected from the group consisting of Ci-6 alkyl, C2-6 alkenyl, - (CfkXOH, and -(CH2)VN(R)2,
- wherein v is selected from 1, 2, 3, 4, 5, and 6;
- each R5 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, NO2, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- R10 is selected from the group consisting of H, OH, C1-3 alkyl, and C2-3 alkenyl;
- each R is independently selected from the group consisting of C1-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, (CH2)qOR*, and H;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", and H,
- and each q is independently selected from 1, 2, and 3;
- each R" is independently selected from the group consisting of C3-15 alkyl and C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13,
- or their N-oxides, or salts or isomers thereof.
- In yet another embodiments, another subset of compounds of Formula (III) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R'M'R';
- R2 and R3 are independently selected from the group consisting of H, Ci-i4 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of hydrogen, a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R12)2(CH2)n-oQ,-CHQR, -CQ(R)2, -C(0)NQR and unsubstituted Ci-e alkyl, where Q is selected from a C3-6 carbocycle, a 5- to 14-membered heterocycle having one or more heteroatoms selected from N, O, and S, -OR, -0(CH2)nN(R)2, -C(0)0R, -0C(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -CRN(R)2C(0)0R, - N(R)R8, -N(R)S(0)2R8, -0(CH2)n0R, -N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, - 0C(0)N(R)2, -N(R)C(0)0R, -N(OR)C(0)R, -N(OR)S(0)2R, -N(0R)C(0)0R, - N(0R)C(0)N(R)2, -N(OR)C(S)N(R)2, -N(OR)C(=NR9)N(R)2, -N(OR)C(=CHR9)N(R)2, -C(=NR9)R, -C(0)N(R)OR, -(CH2)nN(R)2 and -C(=NR9)N(R)2, each 0 is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3,
- 4, and 5; and when Q is a 5- to 14-membered heterocycle and (i) R4 is - (CH2)nQ in which n is 1 or 2, or (ii) R4 is -(CH2)nCHQR in which n is 1, or (iii) R4 is - CHQR, and -CQ(R)2, then Q is either a 5- to 14-membered heteroaryl or 8- to 14-membered heterocycloalkyl;
- Rx is selected from the group consisting of Ci-6 alkyl, C2-6 alkenyl, - (CH2)vOH, and -(CH2)VN(R)2,
- wherein v is selected from 1, 2, 3, 4, 5, and 6;
- each R5 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-i3 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, N02, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- R12 is selected from the group consisting of H, OH, C1-3 alkyl, and C2-3 alkenyl;
- each R is independently selected from the group consisting of C1-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, (CH2)qOR*. and H,
- and each q is independently selected from 1, 2, and 3;
- each R' is independently selected from the group consisting of Ci-ib alkyl, C2-18 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13,
- or their N-oxides, or salts or isomers thereof.
- In still another embodiments, another subset of compounds of Formula (III) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R"M'R';
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of hydrogen, a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R12)2(CH2)n-oQ,-CHQR, -CQ(R)2, -C(0)NQR and unsubstituted Ci-e alkyl, where Q is selected from a C3-6 carbocycle, a 5- to 14-membered heteroaryl having one or more heteroatoms selected from N, O, and S, -OR, - 0(CH2)nN(R)2, -C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -CRN(R)2C(0)OR, - N(R)R8, -N(R)S(0)2R8, -0(CH2)nOR, -N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, - OC(0)N(R)2, -N(R)C(0)OR, -N(OR)C(0)R, -N(OR)S(0)2R, -N(OR)C(0)OR, - N(OR)C(0)N(R)2, -N(OR)C(S)N(R)2, -N(OR)C(=NR9)N(R)2, -N(0R)C(=CHR9)N(R)2, -C(=NR9)R, -C(0)N(R)OR, -(CH2)nN(R)2, each 0 is independently selected from 1, 2, 3, and 4, and -C(=NR9)N(R)2, each 0 is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5;
- Rx is selected from the group consisting of C1-6 alkyl, C2-6 alkenyl, - (CH2)vOH, and -(CH2)VN(R)2,
- wherein v is selected from 1, 2, 3, 4, 5, and 6;
- each R5 is independently selected from the group consisting of OH, C 1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, NO2, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- R12 is selected from the group consisting of H, OH, C1-3 alkyl, and C2-3 alkenyl;
- each R is independently selected from the group consisting of C1-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, (CH2)qOR*, and H,
- and each q is independently selected from 1, 2, and 3;
- each R' is independently selected from the group consisting of Ci-is alkyl, C2-18 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13,
- or their N-oxides, or salts or isomers thereof.
- In still another embodiments, another subset of compounds of Formula (III) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R"M'R';
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of hydrogen, a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R12)2(CH2)n-oQ, -CHQR, -CQ(R)2, -C(0)NQR and unsubstituted Ci-e alkyl, where Q is selected from a carbocycle, -OR, - 0(CH2)nN(R)2, -C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -
- N(R)C(S)N(R)2, -N(R)R8, -N(R)S(0)2R8, -0(CH2)n0R, - N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, -0C(0)N(R)2, -N(R)C(0)0R, - N(0R)C(0)R, -N(0R)S(0)2R, -N(0R)C(0)0R, -N(0R)C(0)N(R)2, -N(OR)C(S)N(R)2, - N(OR)C(=NR9)N(R)2, -N(OR)C(=CHR9)N(R)2, -C(=NR9)N(R)2, -C(=NR9)R, - C(0)N(R)0R, -(CH2)nN(R)2 and -C(R)N(R)2C(0)0R, each o is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5;
- Rx is selected from the group consisting of Ci-6 alkyl, C2-6 alkenyl, - (CH2)vOH, and -(CH2)VN(R)2,
- wherein v is selected from 1, 2, 3, 4, 5, and 6;
- each R5 is independently selected from the group consisting of OH, Ci-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, Ci-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-i3 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- R8 is selected from the group consisting of C3-6 carbocycle and heterocycle;
- R9 is selected from the group consisting of H, CN, N02, C1-6 alkyl, -OR, -S(0)2R, -S(0)2N(R)2, C2-6 alkenyl, C3-6 carbocycle and heterocycle;
- R12 is selected from the group consisting of H, OH, C 1-3 alkyl, and C2-3 alkenyl;
- each R is independently selected from the group consisting of C1-6 alkyl, C 1-3 alkyl-aryl, C2-3 alkenyl, and H;
- each R' is independently selected from the group consisting of Ci-ib alkyl, C2-ie alkenyl, -R*YR", -YR", (CH2)qOR*, and H,
- and each q is independently selected from 1, 2, and 3;
- each R" is independently selected from the group consisting of C3-15 alkyl and
- C3-15 alkenyl;
- each R* is independently selected from the group consisting of Ci-i2 alkyl and
- C2-i2 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13.
- In yet another embodiments, another subset of compounds of Formula (III) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R"M'R';
- R2 and R3 are independently selected from the group consisting of H, C2-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is -(CH2)nQ or -(CH2)nCHQR, where Q is -N(R)2, and n is selected from 3, 4, and 5; Rx is selected from the group consisting of Ci-6 alkyl, C2-6 alkenyl, - (CH2)vOH, and -(CH2)VN(R)2,
- wherein v is selected from 1, 2, 3, 4, 5, and 6;
- each R5 is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-i3 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- each R is independently selected from the group consisting of C1-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, and H;
- each R' is independently selected from the group consisting of Ci-is alkyl, C2-i8 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and C3-lsalkenyl;
- each R* is independently selected from the group consisting of Ci-i2 alkyl and Ci-i2 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13,
- or their N-oxides, or salts or isomers thereof.
- In still another embodiments, another subset of compounds of Formula (III) includes those in which
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R"M'R';
- R2 and R3 are independently selected from the group consisting of C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- R4 is selected from the group consisting of -(CH2)nQ, -(CH2)nCHQR, - CHQR, and -CQ(R)2, where Q is -N(R)2, and n is selected from 1, 2, 3, 4, and 5;
- Rx is selected from the group consisting of Ci-6 alkyl, C2-6 alkenyl, - (CH2)vOH, and -(CH2)VN(R)2,
- wherein v is selected from 1, 2, 3, 4, 5, and 6;
- each R5 is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- each R is independently selected from the group consisting of C1-6 alkyl, C1-3 alkyl-aryl, C2-3 alkenyl, and H;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and C3-15 alkenyl;
- each R* is independently selected from the group consisting of C 1-12 alkyl and C1-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I; and m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13,
- or their N-oxides, or salts or isomers thereof.In certain embodiments, a subset of compounds of Formula (I) includes those of Formula (IA):
- or its N-oxide, or a salt or isomer thereof, wherein 1 is selected from 1, 2, 3, 4, and 5; m is selected from 5, 6, 7, 8, and 9; Mi is a bond or M'; R4 is hydrogen, unsubstituted C1-3 alkyl, -(CH2)oC(R12)2(CH2)n-oQ, -C(0)NQR or -(CH2)nQ, in which Q is OH, -NHC(S)N(R)2, -NHC(0)N(R)2, -N(R)C(0)R, -N(R)S(0)2R, -N(R)R8, - NHC(=NR9)N(R)2, -NHC(=CHR9)N(R)2, -0C(0)N(R)2, -N(R)C(0)0R, -(CH2)nN(R)2, heteroaryl or heterocycloalkyl; M and M' are independently selected from -C(0)0-, - OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R , -P(0)(0R')0-, -S-S-, an aryl group, and a heteroaryl group,; and R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, and C2-i4 alkenyl. For example, m is 5, 7, or 9. For example, Q is OH, -NHC(S)N(R)2, or -NHC(0)N(R)2. For example, Q is -N(R)C(0)R, or - N(R)S(0)2R.
- In certain embodiments, a subset of compounds of Formula (I) includes those of Formula (IB):
- -P(0)(0R')0-, -S-S-, an aryl group, and a heteroaryl group; and R2 and R3 are independently selected from the group consisting of H, Ci-i4 alkyl, and C2-14 alkenyl. For example, m is 5, 7, or 9. In certain embodiments, a subset of compounds of Formula (I) includes those of Formula
- NHC(=CHR9)N(R)2, -0C(0)N(R)2, -N(R)C(0)0R, -(CH2)nN(R)2, heteroaryl or
- heterocycloalkyl; M and M' are independently selected from -C(0)0-, - OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -P(0)(0R')0-, -S-S-, an aryl group, and a heteroaryl group; and R2 and R3 are independently selected from the group consisting of H, CI-M alkyl, and C2-M alkenyl.
-
- In certain embodiments, a subset of compounds of Formula (I) includes those of Formula (lid):
-
- or its N-oxide, or a salt or isomer thereof, wherein n is 2, 3, or 4; and m, M, M", R', R", and R2 through R6 are as described herein. For example, each of R2 and R3 may be independently
- selected from the group consisting of C5-14 alkyl and CS-14 alkenyl, and n is selected from 2, 3, and 4.
- In another embodiment, a subset of compounds of Formula (I) includes those of Formula (Ilg):
-
- R1 is selected from the group consisting of C5-30 alkyl, C5-20 alkenyl, - R*YR", -YR", and -R'M'R';
- R2 and R3 are independently selected from the group consisting of H, C1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle;
- each R5 is independently selected from the group consisting of OH, C 1-3 alkyl, C2-3 alkenyl, and H;
- each R6 is independently selected from the group consisting of OH, C1-3 alkyl, C2-3 alkenyl, and H;
- M and M' are independently selected from -C(0)0-, -OC(O)-, -0C(0)-M"-C(0)0-, -C(0)N(R')-, -N(R')C(0)-, -C(O)-, -C(S)-, -C(S)S-, -SC(S)-, -CH(OH)-, - P(0)(0R')0-, -S(0)2-, -S-S-, an aryl group, and a heteroaryl group, in which M" is a bond, C1-13 alkyl or C2-13 alkenyl;
- R7 is selected from the group consisting of C1-3 alkyl, C2-3 alkenyl, and H;
- each R is independently selected from the group consisting of H, C 1-3 alkyl, and C2-3 alkenyl;
- RN is H, or Ci-3 alkyl;
- each R' is independently selected from the group consisting of C1-18 alkyl, C2-18 alkenyl, -R*YR", -YR", and H;
- each R" is independently selected from the group consisting of C3-15 alkyl and
- C3-15 alkenyl;
- each R* is independently selected from the group consisting of C1-12 alkyl and
- C2-12 alkenyl;
- each Y is independently a C3-6 carbocycle;
- each X is independently selected from the group consisting of F, Cl, Br, and I;
- Xa and Xb are each independently O or S;
- R10 is selected from the group consisting of H, halo, -OH, R, -N(R)2, - CN, -N3, -C(0)0H, -C(0)0R, -0C(0)R, -OR, -SR, -S(0)R, -S(0)0R, -S(0)20R, -N02, - S(0)2N(R)2, -N(R)S(0)2R, -NH(CH2)tiN(R)2, -NH(CH2)PiO(CH2)qiN(R)2, - NH(CH2)SIOR, -N((CH2)SIOR)2, -N(R)-carbocycle, -N(R)-heterocycle, -N(R)-aryl, - N(R)-heteroaryl, -N(R)(CH2)ti-carbocycle, -N(R)(CH2)ti-heterocycle, -N(R)(CH2)ti-aryl, -N(R)(CH2)u-heteroaryl, a carbocycle, a heterocycle, aryl and heteroaryl;
- m is selected from 5, 6, 7, 8, 9, 10, 11, 12, and 13;
- n is selected from 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10;
- r is 0 or 1;
- t1 is selected from 1, 2, 3, 4, and 5;
- p1 is selected from 1, 2, 3, 4, and 5;
- q1 is selected from 1, 2, 3, 4, and 5; and
- s1 is selected from 1, 2, 3, 4, and 5.
-
- Rla and Rib are independently selected from the group consisting of C 1-14 alkyl and C2-14 alkenyl; and
- R2 and R3 are independently selected from the group consisting of C 1-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle.
-
- or its N-oxide, or a salt or isomer thereof, wherein
- 1 is selected from 1, 2, 3, 4, and 5;
- Mi is a bond or M'; and
- R2 and R3 are independently selected from the group consisting of H, Ci- i4 alkyl, and C2-14 alkenyl.
-
- or its N-oxide, or a salt or isomer thereof, wherein
- 1 is selected from 1, 2, 3, 4, and 5;
- Mi is a bond or M'; and
- Ra and Rb are independently selected from the group consisting of C1-14 alkyl and C2-14 alkenyl; and
- R2 and R3 are independently selected from the group consisting of C1-14 alkyl, and C2-14 alkenyl.
- The compounds of any one of formula (I), (IA), (VI), (Vl-a), (VII) or (VIII) include one or more of the following features when applicable.
- In some embodiments, Mi is M'.
- In some embodiments, M and M' are independently -C(0)0- or -OC(O)-.
- In some embodiments, at least one of M and M' is -C(0)0- or -OC(O)-.
- In certain embodiments, at least one of M and M' is -OC(O)-.
- In certain embodiments, M is -OC(O)- and M' is -C(0)0-. In some embodiments, M is -C(0)0- and M' is -OC(O)-. In certain embodiments, M and M' are each -OC(O)-. In some embodiments, M and M' are each -C(0)0-.
- In certain embodiments, at least one of M and M' is -0C(0)-M"-C(0)0-.
- In some embodiments, M and M' are independently -S-S-.
- In some embodiments, at least one of M and M' is -S-S.
- In some embodiments, one of M and M' is -C(0)0- or -OC(O)- and the other is -S-S-. For example, M is -C(0)0- or -OC(O)- and M' is -S-S- or M' is -C(0)0-, or -OC(O)- and M is -S-S-.
- In some embodiments, one of M and M' is -0C(0)-M"-C(0)0-, in which M" is a bond, Ci-i3 alkyl or C2-13 alkenyl. In other embodiments, M" is C1-6 alkyl or C2-6 alkenyl. In certain embodiments, M" is C1-4 alkyl or C2-4 alkenyl. For example, in some embodiments, M" is Ci alkyl. For example, in some embodiments, M" is C2 alkyl. For example, in some embodiments, M" is C3 alkyl. For example, in some embodiments, M" is C4 alkyl. For example, in some embodiments, M" is C2 alkenyl. For example, in some embodiments, M" is C3 alkenyl. For example, in some embodiments, M" is C4 alkenyl.
- In some embodiments, 1 is 1, 3, or 5.
- In some embodiments, R4 is hydrogen.
- In some embodiments, R4 is not hydrogen.
- In some embodiments, R4 is unsubstituted methyl or -(CH2)nQ, in which Q is OH, -NHC(S)N(R)2, -NHC(0)N(R)2, -N(R)C(0)R, or -N(R)S(0)2R.
- In some embodiments, Q is OH.
- In some embodiments, Q is -NHC(S)N(R)2.
- In some embodiments, Q is -NHC(0)N(R)2.
- In some embodiments, Q is -N(R)C(0)R.
- In some embodiments, Q is -N(R)S(0)2R.
- In some embodiments, Q is -0(CH2)nN(R)2.
- In some embodiments, Q is -0(CH2)nOR.
- In some embodiments, Q is -N(R)R8.
- In some embodiments, Q is -NHC(=NR9)N(R)2.
- In some embodiments, Q is -NHC(=CHR9)N(R)2.
- In some embodiments, Q is -OC(0)N(R)2.
- In some embodiments, Q is -N(R)C(0)OR.
- In some embodiments, n is 2.
- In some embodiments, n is 3.
- In some embodiments, n is 4.
- In some embodiments, Mi is absent.
- In some embodiments, at least one R5 is hydroxyl. For example, one R5 is hydroxyl.
- In some embodiments, at least one R6 is hydroxyl. For example, one R6 is hydroxyl.
- In some embodiments one of R5 and R6 is hydroxyl. For example, one R5 is hydroxyl and each R6 is hydrogen. For example, one R6 is hydroxyl and each R5 is hydrogen.
- In some embodiments, Rx is Ci-6 alkyl. In some embodiments, Rx is Ci-3 alkyl. For example, Rx is methyl. For example, Rx is ethyl. For example, Rx is propyl.
- In some embodiments, Rx is -(CFkXOFl and, v is 1, 2 or 3. For example, Rx is methanoyl. For example, Rx is ethanoyl. For example, Rx is propanoyl.
- In some embodiments, Rx is -(CH2)vN(R)2, v is 1, 2 or 3 and each R is H or methyl. For example, Rx is methanamino, methylmethanamino, or dimethylmethanamino. For example, Rx is aminomethanyl, methylaminomethanyl, or dimethylaminomethanyl. For example, Rx is aminoethanyl, methylaminoethanyl, or dimethylaminoethanyl. For example, Rx is
aminopropanyl, methylaminopropanyl, or dimethylaminopropanyl. - In some embodiments, R' is Ci-ib alkyl, C2-18 alkenyl, -R*YR", or - YR".
- In some embodiments, R2 and R3 are independently C3-14 alkyl or C3-14 alkenyl.
- In some embodiments, Rib is Ci-14 alkyl. In some embodiments, Rlb is C2-14 alkyl. In some embodiments, Rib is C3-14 alkyl. In some embodiments, Rlb is Ci-8 alkyl. In some embodiments, Rib is C1-5 alkyl. In some embodiments, Rlb is C1-3 alkyl. In some embodiments, Rlb is selected from Ci alkyl, C2 alkyl, C3 alkyl, C4 alkyl, and C5 alkyl. For example, in some embodiments, Rlb is Ci alkyl. For example, in some embodiments, Rlb is C2 alkyl. For example, in some embodiments, Rib is C3 alkyl. For example, in some embodiments, Rlb is C4 alkyl. For example, in some embodiments, Rlb is C5 alkyl.
- In some embodiments, R1 is different from -(CHR5R6)m-M-CR2R3R7.
- In some embodiments, -CHRlaRIb- is different from -(CHR5R6)m-M-CR2R3R7.
- In some embodiments, R7 is H. In some embodiments, R7 is selected from C1-3 alkyl. For example, in some embodiments, R7 is Ci alkyl. For example, in some embodiments, R7 is C2 alkyl. For example, in some embodiments, R7 is C3 alkyl. In some embodiments, R7 is selected from C4 alkyl, C4 alkenyl, C5 alkyl, C5 alkenyl, Ce alkyl, Ce alkenyl, C7 alkyl, C7 alkenyl, C9 alkyl, C9 alkenyl, C11 alkyl, C11 alkenyl, C17 alkyl, C17 alkenyl, Cie alkyl, and Cie alkenyl.
- In some embodiments, Rb is Ci-i4 alkyl. In some embodiments, Rb is C2-14 alkyl. In some embodiments, Rb is C3-14 alkyl. In some embodiments, Rb is Ci-8 alkyl. In some embodiments, Rb is C1-5 alkyl. In some embodiments, Rb is C1-3 alkyl. In some embodiments, Rb is selected from Ci alkyl, C2 alkyl, C3 alkyl, C4 alkyl and C5 alkyl. For example, in some embodiments, Rb is Ci alkyl. For example, in some embodiments, Rb is C2 alkyl. For example, some embodiments, Rb is C3 alkyl. For example, some embodiments, Rb is C4 alkyl.
-
-
-
-
- In a further embodiment, the compounds of Formula (I) are of Formula (lid):
or their N-oxides, or salts or isomers thereof, wherein n is 2, 3, or 4; and m, R', R", and R2 through R6 are as described herein. For example, each of R2 and R3 may be independently selected from the group consisting of C5-14 alkyl and C5-14 alkenyl. - In a further embodiment, the compounds of Formula (I) are of Formula (Ilg):
-
-
-
-
-
-
-
-
-
-
-
- The compounds of any one of formulae (I), (IA), (IB), (II), (Ila), (lib), (lie), (lid), (He), (Ilf), (Ilg), (III), (VI), (Vl-a), (VII), (VIII), (Vila), (Villa), (VUIb), (Vllb-l), (VIIb-2), (VIIb-3), (Vile), (Vlld), (VIIIc), or (VUId) include one or more of the following features when applicable.
- In some embodiments, R4 is selected from the group consisting of a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)0C(R12)2(CH2)n-oQ, -CHQR, and - CQ(R)2, where Q is selected from a C3-6 carbocycle, 5- to 14- membered aromatic or non-aromatic heterocycle having one or more heteroatoms selected from N, O, S, and P, -OR, -0(CH2)nN(R)2, -C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -N(R)2, - N(R)S(0)2R8, -C(0)N(R)2, -N(R)C(0)R, -N(R)S(0)2R, - N(R)C(0)N(R)2, - N(R)C(S)N(R)2, and -C(R)N(R)2C(0)OR, each 0 is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5.
- In some embodiments, R4 is selected from the group consisting of a C3-6 carbocycle, - (CH2)nQ, -(CHQnCHQR, -(CH2)0C(R12)2(CH2)n-oQ, -CHQR, and - CQ(R)2, where Q is selected from a C3-6 carbocycle, a 5- to 14-membered heteroaryl having one or more heteroatoms selected from N, O, and S, -OR, -0(CH2)nN(R)2, - C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, -N(R)S(0)2R8, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, - N(R)C(S)N(R)2, -C(R)N(R)2C(0)OR, and a 5- to l4-membered heterocycloalkyl having one or more heteroatoms selected from N, O, and S which is substituted with one or more substituents selected from oxo (=0), OH, amino, and C1-3 alkyl, each 0 is independently selected from 1, 2,
- 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5.
- In some embodiments, R4 is selected from the group consisting of a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)0C(R12)2(CH2)n-oQ, -CHQR, and - CQ(R)2, where Q is selected from a C3-6 carbocycle, a 5- to 14-membered heterocycle having one or more heteroatoms selected from N, O, and S, -OR, -0(CH2)nN(R)2, - C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, -N(R)S(0)2R8, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -C(R)N(R)2C(0)OR, each 0 is independently selected from 1, 2, 3, and 4, and
each n is independently selected from 1, 2, 3, 4, and 5; and when Q is a 5-to 14-membered heterocycle and (i) R4 is -(CH2)nQ in which n is 1 or 2, or (ii) R4 is - (CH2)nCHQR in which n is 1, or (iii) R4 is -CHQR, and -CQ(R)2, then Q is either a 5-to 14-membered heteroaryl or 8- to 14-membered heterocycloalkyl. - In some embodiments, R4 is selected from the group consisting of a C3-6 carbocycle, -(CH2)nQ, -(CH2)nCHQR, -(CH2)oC(R12)2(CH2)n-oQ, -CHQR, and - CQ(R)2, where Q is selected from a C3-6 carbocycle, a 5- to 14-membered heteroaryl having one or more heteroatoms selected from N, O, and S, -OR, -0(CH2)nN(R)2, - C(0)OR, -OC(0)R, -CX3, -CX2H, -CXH2, -CN, -C(0)N(R)2, -N(R)S(0)2R8, - N(R)C(0)R, -N(R)S(0)2R, -N(R)C(0)N(R)2, -N(R)C(S)N(R)2, -C(R)N(R)2C(0)OR, each 0 is independently selected from 1, 2, 3, and 4, and each n is independently selected from 1, 2, 3, 4, and 5.
- In some embodiments, R4 is -(CH2)nQ, where Q is -N(R)S(0)2R8 and n is selected from 1, 2, 3, 4, and 5. In a further embodiment, R4 is -(CH2)nQ, where Q is - N(R)S(0)2R8, in whichR8 is a C3-6 carbocycle such as C3-6 cycloalkyl, and n is selected from 1, 2, 3, 4, and 5.
- For example, R4 is -(CH2)3NHS(0)2R8 and R8 is cyclopropyl.
- In some embodiments, R4 is -(CH2)oC(R12)2(CH2)n-oQ, where Q is - N(R)C(0)R, n is selected from 1, 2, 3, 4, and 5, and 0 is selected from 1, 2, 3, and 4. In a further embodiment, R4 is -(CH2)oC(R12)2(CH2)n-oQ, where Q is -N(R)C(0)R, wherein R is C1-C3 alkyl and n is selected from 1, 2, 3, 4, and 5, and 0 is selected from 1, 2, 3, and 4. In a another embodiment, R4 is is -(CH2)oC(R12)2(CH2)n-oQ, where Q is -N(R)C(0)R, wherein R is C1-C3 alkyl, n is 3, and 0 is 1.
- In some embodiments, R12 is H, OH, C1-3 alkyl, or C2-3 alkenyl. For example, R4 is 3-acetamido-2,2-dimethylpropyl.
- In some embodiments, R4 is -C(0)NQR, where Q is -(CH2)nN(R)2. In a further embodiments, R4 is -C(0)NH(CH2)3N(CH3)2, -C(0)NH(CH2)4N(CH3)2, or - C(0)NH(CH2)2N(CH3)2.
- In some embodiments, one R12 is H and one R12 is C1-3 alkyl or C2-3 alkenyl. In some embodiments, each R12 is is C1-3 alkyl or C2-3 alkenyl. In some embodiments, each R12 is is C1-3 alkyl (e.g. methyl, ethyl or propyl). For example, one R12 is methyl and one R12 is ethyl or propyl. For example, one R12 is ethyl and one R12 is methyl or propyl. For example, one R12 is propyl and one R12 is methyl or ethyl. For example, each R12 is methyl. For example, each R12 is ethyl. For example, each R12 is propyl.
- In some embodiments, one R12 is H and one R12 is OH. In some embodiments, each R12 is is OH.
- In some embodiments, R4 is unsubstituted C1-4 alkyl, e.g., unsubstituted methyl.
- In some embodiments, R4 is hydrogen.
- In certain embodiments, the disclosure provides a compound having the Formula (I), wherein R4 is -(CF JnQ or -(CH2)nCHQR, where Q is -N(R)2, and n is selected from 3, 4, and 5.
- In certain embodiments, the disclosure provides a compound having the Formula (I), wherein R4 is selected from the group consisting of -(CH2)nQ, - (CH2)nCHQR, -CHQR, and -CQ(R)2, where Q is -N(R)2, and n is selected from 1, 2, 3, 4, and 5.
- In certain embodiments, the disclosure provides a compound having the Formula (I), wherein R2 and R3 are independently selected from the group consisting of C2-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle, and R4 is - (CH2)nQ or -(CH2)nCHQR, where Q is -N(R)2, and n is selected from 3, 4, and 5.
- In certain embodiments, R2 and R3 are independently selected from the group consisting of C2-14 alkyl, C2-14 alkenyl, -R*YR", -YR", and -R*OR", or R2 and R3, together with the atom to which they are attached, form a heterocycle or carbocycle. In some embodiments,
- R2 and R3 are independently selected from the group consisting of C2-14 alkyl, and C2-14 alkenyl. In some embodiments, R2 and R3 are independently selected from the group consisting of -R*YR", -YR", and -R * OR" . In some embodiments, R2 and R3 together with the atom to which they are attached, form a heterocycle or carbocycle.
- In some embodiments, R1 is selected from the group consisting of C5-20 alkyl and C5-20 alkenyl. In some embodiments, R1 is C5-20 alkyl substituted with hydroxyl.
- In other embodiments, R1 is selected from the group consisting of -R*YR", -YR", and -R"M'R\
- In certain embodiments, R1 is selected from -R*YR" and -YR". In some
embodiments, Y is a cyclopropyl group. In some embodiments, R* is Cx alkyl or Cx alkenyl. In certain embodiments, R" is C3-12 alkyl. For example, in some embodiments, R" is C3 alkyl. For example, in some embodiments, R" is C4-8 alkyl (e.g., C4, C5, Ce, C7, or Cs alkyl). - In some embodiments, R is (CH2)qOR*, q is selected from 1, 2, and 3, and R* is C1-12 alkyl substituted with one or more substituents selected from the group consisting of amino, Ci-Ce alkylamino, and C1-C6 dialkylamino. For example, R is (CFh)qOR*, q is selected from 1, 2, and 3 and R* is C1-12 alkyl substituted with C1-C6 dialkylamino. For example, R is (CH2)qOR*, q is selected from 1, 2, and 3 and R* is C1-3 alkyl substituted with C1-C6 dialkylamino. For example, R is (CH2)qOR*, q is selected from 1, 2, and 3 and R* is C1-3 alkyl substituted with dimethylamino (e.g., dimethylaminoethanyl).
- In some embodiments, R1 is C5-20 alkyl. In some embodiments, R1 is G, alkyl. In some embodiments, R1 is Cs alkyl. In other embodiments, R1 is C9 alkyl. In certain
embodiments, R1 isC 14 alkyl. In other embodiments, R1 is Cie alkyl. -
- In some embodiments, R1 is C5-20 alkenyl. In certain embodiments, R1 is Cie alkenyl. In some embodiments, R1 is linoleyl.
-
-
- In other embodiments, R1 is -R"M'R\ In certain embodiments, M' is - OC(0)-M"-C(0)0-. For example, R1 is
13 (e.g., selected from 3, 4, 5, and 6), x2 is an integer between 1 and 13 (e.g., selected from 1, 2, and 3), and x3 is an integer between 2 and 14 (e.g., selected from 4, 5, and 6). For example, x1 is selected from 3, 4, 5, and 6, x2 is selected from 1, 2, and 3, and x3 is selected from 4, 5, and 6. - In other embodiments, R1 is different from -(CHR5R6)m-M-CR2R3R7.
- In some embodiments, R' is selected from -R*YR" and -YR". In some
- embodiments, Y is C3-8 cycloalkyl. In some embodiments, Y is Ce-io aryl. In some
- embodiments, Y is a cyclopropyl group. In some embodiments, Y is a cyclohexyl group. In certain embodiments, R* is Ci alkyl.
- In some embodiments, R" is selected from the group consisting of C3-12 alkyl and C3- 12 alkenyl. In some embodiments, R" is Cs alkyl. In some embodiments, R" adjacent to Y is Ci
alkyl. In some embodiments, R" adjacent to Y is C4-9 alkyl (e.g., C4, C5, Ce, Ci or Cs or C9 alkyl). -
- In some embodiments, R' is selected from C4 alkyl and C4 alkenyl. In certain embodiments, R' is selected from C5 alkyl and C5 alkenyl. In some embodiments, R' is selected from C6 alkyl and Ce alkenyl. In some embodiments, R' is selected from C7 alkyl and C7 alkenyl. In some embodiments, R' is selected from C9 alkyl and C9 alkenyl.
- In some embodiments, R' is selected from C4 alkyl, C4 alkenyl, C5 alkyl, C5 alkenyl, C6 alkyl, Ce alkenyl, C7 alkyl, C7 alkenyl, C9 alkyl, C9 alkenyl, C 11 alkyl, C 11 alkenyl, C 17 alkyl, C17 alkenyl, Cie alkyl, and Cie alkenyl, each of which is either linear or branched.
- In some embodiments, R' is C4 alkyl or C4 alkenyl. In some embodiments, R' is C5 alkyl or C5 alkenyl. In some embodiments, R' is G, alkyl or G, alkenyl. In some embodiments, R' is C7 alkyl or C7 alkenyl. In some embodiments, R' is Cs alkyl or Cs alkenyl. In some embodiments, R' is C9 alkylor C9 alkenyl. In some embodiments, R' is C10 alkyl or
C 10 alkenyl. In some embodiments, R' is C 11 alkyl or C11 alkenyl. - In some embodiments, R' is linear. In some embodiments, R' is branched.
-
-
- In other embodiments, R' is selected from C11 alkyl and C 11 alkenyl. In other embodiments, R' is selected from C12 alkyl, C12 alkenyl, C13 alkyl, C13 alkenyl, C14 alkyl, C14 alkenyl, C15 alkyl, C15 alkenyl, Ci6 alkyl, Ci6 alkenyl, C17 alkyl, C 17 alkenyl, Cie alkyl, and Cie alkenyl. In certain embodiments, R' is linear C4-18 alkyl or C4-18 alkenyl. In certain
- embodiments, R' is branched (e.g., decan-2-yl, undecan-3-yl, dodecan-4-yl, tridecan-5-yl, tetradecan-6-yl, 2-methylundecan-3-yl, 2-methyldecan-2-yl, 3-methylundecan-3-yl, 4-
- methyldodecan-4-yl or heptadeca-9-yl). In certain embodiments, R' is
- In certain embodiments, R' is unsubstituted Ci-ie alkyl. In certain embodiments, R' is substituted Ci-ie alkyl (e.g., C1-15 alkyl substituted with, e.g., an alkoxy such as methoxy, or a C3-6 carbocycle such as l-cyclopropylnonyl, or C(0)0-alkyl or 0C(0)-alkyl such as C(0)0CH3
or OC(O)CH3). For example, R' is -
- In some embodiments, R" is selected from the group consisting of C3-15 alkyl and C3-15 alkenyl. In some embodiments, R" is C3 alkyl, C4 alkyl, C5 alkyl, Ce alkyl, C7 alkyl, or Cs alkyl. In some embodiments, R" is C9 alkyl, C10 alkyl, C11 alkyl, C12 alkyl, C13 alkyl, C14 alkyl, or C15 alkyl.
- In some embodiments, M' is -C(0)0-. In some embodiments, M' is - OC(O)-. In some embodiments, M' is -0C(0)-M"-C(0)0-. In some embodiments, M' is - S-S-.
- In some embodiments, M' is -C(0)0-, -OC(O)-, or -0C(0)-M"-C(0)0-. In some embodiments wherein M' is -0C(0)-M"-C(0)0-, M" is Ci-4 alkyl or C2-4 alkenyl.
- In other embodiments, M' is an aryl group or heteroaryl group. For example, in some embodiments, M' is selected from the group consisting of phenyl, oxazole, and thiazole.
- In some embodiments, M is -C(0)0-. In some embodiments, M is -OC(O)-. In some embodiments, M is -C(0)N(R')-. In some embodiments, M is -P(0)(0R')0-. In some embodiments, M is -0C(0)-M"-C(0)0-. In some embodiments, M is -S-S-.
- In some embodiments, M is -C(O). In some embodiments, M is -OC(O)- and M' is -C(0)0-. In some embodiments, M is -C(0)0- and M' is -OC(O)-. In some embodiments, M and M' are each -OC(O)-. In some embodiments, M and M' are each - C(0)0-.
- In other embodiments, M is an aryl group or heteroaryl group. For example, in some embodiments, M is selected from the group consisting of phenyl, oxazole, and thiazole.
- In some embodiments, M is the same as M'. In other embodiments, M is different from M'.
- In some embodiments, M" is a bond. In some embodiments, M" is C1-13 alkyl or C2-13 alkenyl. In some embodiments, M" is C1-6 alkyl or C2-6 alkenyl. In certain embodiments, M" is linear alkyl or alkenyl. In certain embodiments, M" is branched, e.g., -CH(CH3)CH2-.
- In some embodiments, each R5 is H. In some embodiments, each R6 is H. In certain such embodiments, each R5 and each R6 is H.
- In some embodiments, R7 is H. In other embodiments, R7 is Ci-3 alkyl (e.g., methyl, ethyl, propyl, or i-propyl).
- In some embodiments, R2 and R3 are independently C5-14 alkyl or C5-14 alkenyl.
- In some embodiments, R2 and R3 are the same. In some embodiments, R2 and R3 are C8 alkyl. In certain embodiments, R2 and R3 are C2 alkyl. In other embodiments, R2 and R3 are C3 alkyl. In some embodiments, R2 and R3 are C4 alkyl. In certain embodiments, R2 and R3 are C5 alkyl. In other embodiments, R2 and R3 are Ce alkyl. In some embodiments, R2 and R3 are C7 alkyl.
- In other embodiments, R2 and R3 are different. In certain embodiments, R2 is G alkyl. In some embodiments, R3 is C1-7 (e.g., Ci, C2, C3, C4, C5, Ce, or C7 alkyl) or C9 alkyl.
- In some embodiments, R3 is Ci alkyl. In some embodiments, R3 is C2 alkyl. In some embodiments, R3 is C3 alkyl. In some embodiments, R3 is C4 alkyl. In some embodiments, R3 is C5 alkyl. In some embodiments, R3 is G, alkyl. In some embodiments, R3 is C7 alkyl. In some embodiments, R3 is C9 alkyl.
- In some embodiments, R7 and R3 are H.
- In certain embodiments, R2 is H.
- In some embodiments, m is 5, 6, 7, 8, or 9. In some embodiments, m is 5, 7, or 9.
- For example, in some embodiments, m is 5. For example, in some embodiments, m is 7. For example, in some embodiments, m is 9.
- In some embodiments, R4 is selected from -(CH2)nQ and -(CH2)nCHQR.
- In some embodiments, Q is selected from the group consisting of -OR, - OH, -0(CH2)nN(R)2, -0C(0)R, -CX3, -CN, -N(R)C(0)R, -N(H)C(0)R, -N(R)S(0)2R, -N(H)S(0)2R, -N(R)C(0)N(R)2, -N(H)C(0)N(R)2, -N(H)C(0)N(H)(R), - N(R)C(S)N(R)2, -N(H)C(S)N(R)2, -N(H)C(S)N(H)(R), -C(R)N(R)2C(0)0R, - N(R)S(0)2R8, a carbocycle, and a heterocycle.
- In certain embodiments, Q is -N(R)R8, -N(R)S(0)2R8, -0(CH2)n0R, - N(R)C(=NR9)N(R)2, -N(R)C(=CHR9)N(R)2, -0C(0)N(R)2, or -N(R)C(0)0R.
-
- In certain embodiments, Q is thiourea or an isostere thereof, e.g., H or - NHC(=NR9)N(R)2.
- In certain embodiments, Q is -C(=NR9)N(R)2. For example, when Q is - C(=NR9)N(R)2, n is 4 or 5. For example, R9 is -S(0)2N(R)2.
- In certain embodiments, Q is -C(=NR9)R or -C(0)N(R)OR, e g., -CH(=N-OCH3), -C(0)NH-OH, -C(0)NH-OCH3, -C(0)N(CH3)-OH, or -C(0)N(CH3)-0CH3.
- In certain embodiments, Q is -OH.
- In certain embodiments, Q is a substituted or unsubstituted 5- to 10-membered heteroaryl, e.g., Q is a triazole, an imidazole, a pyrimidine, a purine, 2-amino-1 9-dihydro-6//-purin-6-one-9-yl (or guanin-9-yl), adenin-9-yl, cytosin-l-yl, or uracil-1-yl, each of which is optionally substituted with one or more substituents selected from alkyl, OH, alkoxy, -alkyl-OH, -alkyl-O-alkyl, and the substituent can be further substituted. In certain embodiments, Q is a substituted 5- to 14-membered heterocycloalkyl, e.g., substituted with one or more substituents selected from oxo (=0), OH, amino, mono- or di-alkylamino, and Ci-3 alkyl. For example, Q is 4-methylpiperazinyl, 4-(4-methoxybenzyl)piperazinyl, isoindolin-2-yl-l,3-dione, pyrrolidin-l-yl-2,5-dione, or imidazolidin-3-yl-2,4-dione.
- In certain embodiments, Q is -NHR8, in which R8 is a C3-6 cycloalkyl optionally substituted with one or more substituents selected from oxo (=0), amino (NH2), mono- or di-alkylamino, Ci-3 alkyl and halo. For example, R8 is cyclobutenyl, e.g., 3-(dimethylamino)-cyclobut-3-ene-4-yl-1,2-dione. In further embodiments, R8 is a C3-6 cycloalkyl optionally substituted with one or more substituents selected from oxo (=0), thio (=S), amino (NH2), mono- or di-alkylamino, Ci-3 alkyl, heterocycloalkyl, and halo, wherein the mono- or di-alkylamino, Ci-3 alkyl, and heterocycloalkyl are further substituted. For example R8 is cyclobutenyl substituted with one or more of oxo, amino, and alkylamino, wherein the alkylamino is further substituted, e.g., with one or more of Ci-3 alkoxy, amino, mono- or di-alkylamino, and halo. For example, R8 is 3-(((dimethylamino)ethyl)amino)cyclobut-3-enyl-l,2-dione. For example R8 is cyclobutenyl substituted with one or more of oxo, and alkylamino.
- For example, R8 is 3-(ethylamino)cyclobut-3-ene-1,2-dione. For example R8 is cyclobutenyl substituted with one or more of oxo, thio, and alkylamino. For example R8 is 3-(ethylamino)-4-thioxocyclobut-2-en-1-one or 2-(ethylamino)-4-thioxocyclobut-2-en-1-one. For example R8 is cyclobutenyl substituted with one or more of thio, and alkylamino. For example R8 is 3-(ethylamino)cyclobut-3-ene-1,2-dithione. For example R8 is cyclobutenyl substituted with one or more of oxo and dialkylamino. For example R8 is 3-(diethylamino)cyclobut-3-ene-1,2-dione. For example, R8 is cyclobutenyl substituted with one or more of oxo, thio, and dialkylamino.
- For example, R8 is 2-(diethylamino)-4-thioxocyclobut-2-en-l-one or 3-(diethylamino)-4-thioxocyclobut-2-en-l-one. For example, R8 is cyclobutenyl substituted with one or more of thio, and dialkylamino. For example, R8 is 3-(diethylamino)cyclobut-3-ene-1,2-dithione. For example, R8 is cyclobutenyl substituted with one or more of oxo and alkylamino or dialkylamino, wherein alkylamino or dialkylamino is further substituted, e.g. with one or more alkoxy. For example, R8 is 3-(bis(2-methoxyethyl)amino)cyclobut-3-ene-1,2-dione. For example, R8 is cyclobutenyl substituted with one or more of oxo, and heterocycloalkyl. For example, R8 is cyclobutenyl substituted with one or more of oxo, and piperidinyl, piperazinyl, or morpholinyl. For example, R8 is cyclobutenyl substituted with one or more of oxo, and heterocycloalkyl, wherein heterocycloalkyl is further substituted, e.g., with one or more C1-3 alkyl. For example, R8 is cyclobutenyl substituted with one or more of oxo, and
- heterocycloalkyl, wherein heterocycloalkyl (e.g., piperidinyl, piperazinyl, or morpholinyl) is further substituted with methyl.
- In certain embodiments, Q is -NHR8, in which R8 is a heteroaryl optionally substituted with one or more substituents selected from amino (NH2), mono- or di-alkylamino, C1-3 alkyl and halo. For example, R8 is thiazole or imidazole.
- In certain embodiments, Q is -NHR8 and R8 is purine.
- In certain embodiments, Q is -NHC(=NR9)N(R)2 in which R9 is CN, Ci-6 alkyl, NO2, -S(0)2N(R)2, -OR, -S(0)2R, or H. For example, Q is - NHC(=NR9)N(CH3)2, -NHC(=NR9)NHCH3, -NHC(=NR9)NH2. In some embodiments, Q is -NHC(=NR9)N(R)2 in which R9 is CN and R is Ci-3 alkyl substituted with mono- or di-alkylamino, e.g., R is
((dimethylamino)ethyl)amino. In some embodiments, Q is - NHC(=NR9)N(R)2 in which R9 is Ci-6 alkyl, NO2, -S(0)2N(R)2, -OR, -S(0)2R, or H and R is Ci-3 alkyl substituted with mono- or di-alkylamino, e.g., R is ((dimethylamino)ethyl)amino. - In certain embodiments, Q is -NHC(=CHR9)N(R)2, in which R9 is NO2, CN, Ci-6 alkyl, -S(0)2N(R)2, -OR, -S(0)2R, or H. For example, Q is - NHC(=CHR9)N(CH3)2, -NHC(=CHR9)NHCH3, or -NHC(=CHR9)NH2.
- In certain embodiments, Q is -OC(0)N(R)2, -N(R)C(0)OR, - N(OR)C(0)OR, such as -OC(0)NHCH3, -N(OH)C(0)OCH3, -N(OH)C(0)CH3, - N(0CH3)C(0)0CH3, -N(0CH3)C(0)CH3, -N(0H)S(0)2CH3, or -NHC(0)OCH3.
- In certain embodiments, Q is -N(R)C(0)R, in which R is alkyl optionally substituted with Ci-3 alkoxyl or S(0)zCi-3 alkyl, in which z is 0, 1, or 2.
- In certain embodiments, Q is an unsubstituted or substituted C6-10 aryl (such as phenyl) or C3-6 cycloalkyl.
- In some embodiments, n is 1. In other embodiments, n is 2. In further embodiments, n is 3. In certain other embodiments, n is 4. In some embodiments, n is 5. For example, in
some embodiments, R4 is -(Cth^OH. For example, in some embodiments, R4 is -(CFh^OFl. - For example, in some embodiments, R4 is -(CFh^OFl. For example, in some embodiments, R4 is -(CH2)5OH. For example, in some embodiments, R4 is benzyl. For example, in some embodiments, R4 may be 4-methoxybenzyl.
- In some embodiments, R4 is a C3-6 carbocycle. In some embodiments, R4 is a C3-6 cycloalkyl. For example, in some embodiments, R4 is cyclohexyl optionally substituted with e.g., OH, halo, C1-6 alkyl, etc. For example, in some embodiments, R4 is 2-hydroxy cyclohexyl.
- In some embodiments, R is H.
- In some embodiments, R is C1-3 alkyl substituted with mono- or dialkylamino, e.g.,
- R is ((dimethylamino)ethyl)amino.
- In some embodiments, R is C 1-6 alkyl substituted with one or more substituents selected from the group consisting of C 1-3 alkoxyl, amino, and C1-C3 dialkylamino.
- In some embodiments, R is unsubstituted C1-3 alkyl or unsubstituted C2-3 alkenyl.
- For example, in some embodiments R4 is -CH2CH(OH)CH3, - CH(CH3)CH20H, or -CH2CH(OH)CH2CH3.
- In some embodiments, R is substituted C1-3 alkyl, e.g., CH2OH. For example, in some embodiments, R4 is -CH2CH(OH)CH2OH, -(CH2)3NHC(0)CH20H, - (CH2)3NHC(0)CH20Bn, -(CH2)20(CH2)20H, -(CTH^NHCTBOCTB, - (Ca^NHCTBOCTBCTB, CH2SCH3, CH2S(0)CH3, CH2S(0)2CH3, or -CH(CH2OH)2.
-
-
-
-
-
-
-
- In one embodiment, R10 is selected from the group consisting of hydroxyl, amino, alkylamino, dialkylamino, NH-heterocyclyl and heterocyclyl, wherein the alkyl portion of the alkylamino and dialkylamino are optionally substituted with hydroxyl, alkoxy, amino, alkylamino and/or dialkylamino.In one embodiment, the cationic lipid compound has the following structure:
-
-
-
-
-
-
-
- In some embodiments, the cationic lipid has one of the following structures:
Cpd Structure Cpd Structure 65 M2 66 213 67 214 68 215 69 216 70 217 71 218 72 219 73 220 74 221 75 212 76 223 77 224 78 225 79 226 80 227 81 228 82 229 83 230 84 231 85 232 86 233 87 234 88 235 89 236 90 237 91 238 92 239 93 240 94 241 95 242 96 243 97 244 98 245 99 246 100 247 101 248 102 249 103 250 104 251 105 252 106 253 107 254 108 255 109 256 110 257 111 258 112 259 113 260 114 261 115 262 116 263 117 264 118 265 119 266 120 267 121 26S 122 269 123 270 124 271 125 272 126 273 127 274 128 275 129 276 130 277 131 278 132 279 133 280 134 281 135 282 136 283 137 284 138 285 139 286 140 287 141 288 142 289 143 290 144 291 145 292 146 293 147 294 148 295 i49 296 150 297 151 298 152 299 153 300 154 301 155 302 156 303 157 304 158 305 159 306 160 307 161 308 162 309 163 310 164 311 165 312 166 313 167 314 168 315 168 316 170 317 171 318 172 319 173 320 174 321 175 322 176 323 177 324 178 325 179 326 180 327 181 328 182 329 183 330 184 331 185 332 186 333 187 334 189 335 189 336 190 337 191 338 192 339 193 340 194 341 195 342 196 343 197 344 198 345 199 344 200 347 201 348 202 349 203 350 204 351 205 352 206 353 207 354 208 355 209 356 210 357 211 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 -
- In various embodiments, the LNPs comprise a neutral lipid. In various embodiments, the molar ratio of the cationic lipid to the neutral lipid ranges from about 2:1 to about 8:1. In certain embodiments, the neutral lipid is present in any of the foregoing LNPs in a concentration ranging from 5 to 10 mol percent, from 5 to 15 mol percent, 7 to 13 mol percent, or 9 to 11 mol percent. In certain specific embodiments, the neutral lipid is present in a concentration of about 9.5, 10 or 10.5 mol percent. In some embodiments, the molar ratio of cationic lipid to the neutral lipid ranges from about 4.1:1.0 to about 4.9:1.0, from about 4.5:1.0 to about 4.8:1.0, or from about 4.7:1.0 to 4.8:1.0. In some embodiments, the molar ratio of total cationic lipid to the neutral lipid ranges from about 4.1:1.0 to about 4.9: 1.0, from about 4.5:1.0 to about 4.8:1.0, or from about 4.7:1.0 to 4.8:1.0.
- Exemplary neutral lipids include, for example, distearoylphosphatidylcholine (DSPC), dioleoylphosphatidylcholine (DOPC), dipalmitoylphosphatidylcholine (DPPC), dioleoylphosphatidylglycerol (DOPG), dipalmitoylphosphatidylglycerol (DPPG), dioleoyl-phosphatidylethanolamine (DOPE), palmitoyloleoylphosphatidylcholine (POPC), palmitoyloleoyl-phosphatidylethanolamine (POPE) and dioleoyl-phosphatidylethanolamine 4-(N-maleimidomethyl)-cyclohexane-1carboxylate (DOPE-mal), dipalmitoyl phosphatidyl ethanolamine (DPPE), dimyristoylphosphoethanolamine (DMPE), distearoyl-phosphatidylethanolamine (DSPE), 16-O-monomethyl PE, 16-O-dimethyl PE, 18-1-trans PE, 1-stearioyl-2-oleoylphosphatidyethanol amine (SOPE), and 1,2-dielaidoyl-sn-glycero-3-phophoethanolamine (transDOPE). In one embodiment, the neutral lipid is 1,2-distearoyl-sn-glycero-3phosphocholine (DSPC). In some embodiments, the neutral lipid is selected from DSPC, DPPC, DMPC, DOPC, POPC, DOPE and SM. In some embodiments, the neutral lipid is DSPC.
- In certain embodiments, neutral lipids useful in the present invention are DSPC analogs wherein the phosphocholine moiety is replaced by a different zwitterionic group. In certain embodiments, the different zwitterionic group is not a phosphocholine group. In certain embodiments, a neutral lipid useful in the present invention is a compound of formula:
- Z is a zwitterionic moiety,
- m is 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10;
- A is of the formula:
- each instance of L2 is independently a bond or optionally substituted C1-6 alkylene, wherein one methylene unit of the optionally substituted C1-6 alkylene is optionally replaced with -O-, -N(RN )-, -S-, -C(O)-, -C(O)N(RN)-, -NRNC(O)-, -C(O)O-, -OC(O)-, -OC(O)O-, -OC(O)N(RN)-, -NRC(O)O-, or -NRNC(O)N(R N)-;
- each instance of R2 is independently optionally substituted C1-30 alkyl, optionally substituted C1-30 alkenyl, or optionally substituted C1-30 alkynyl; optionally wherein one or more methylene units of R2 are independently replaced with optionally substituted carbocyclylene, optionally substituted heterocyclylene, optionally substituted arylene, optionally substituted heteroarylene, -N(RN)-, -O-, -S-, -C(O)-, -C(O)N(RN)-, - NRNC(O)-, -NRNC(O)N(RN)-, -C(O)O-, -OC(O)-, -OC(O)O-, -OC(O)N(RN)-, - NRNC(O)O-, -C(O)S- -SC(O)-, -C(=NRN)-, -C(=NRN)N(RN)-, -NRNC(=N RN)-, - NRNC(=NRN)N(RN)-, - C(S)-, C(S)N(RN)-, -NRNC(S)-, -NRNC(S)N(RN)-, -S(O)-, - OS(O)-, -S(O)O-, -OS(O)O-, -OS(O)2-, -S(O)2O-, -OS(O)2O -, -N(RN)S(O)-, - S(O)N(RN)-, -N(RN)S(O)N(RN)-, -OS(O)N(RN)-, -N(RN)S(O)O-, -S(O)2-, -N(RN)S(O)2-,-S(O)2N(RN)-, -N(RN)S(O)2N(RN)-, -OS(O)2N(RN)-, or -N(RN)S(O)2O-;
- each instance of RN is independently hydrogen, optionally substituted alkyl, or a nitrogen protecting group;
- Ring B is optionally substituted carbocyclyl, optionally substituted heterocyclyl, optionally substituted aryl, or optionally substituted hctcroaryl; and
- p is 1 or 2.
- In certain embodiments, Z is an amino acid or a derivative thereof. In certain embodiments, Z is of one of the following formulas:
-
-
- Other neutral lipids useful in the present invention include analogs of oleic acid. As described herein, an oleic acid analog can comprise a modified oleic acid tail, a modified carboxylic acid moiety, or both. In certain embodiments, an analog of oleic acid is a compound of formula:
- R4 is optionally substituted, C1-40 alkyl; optionally substituted, C2-20 alkenyl;
- optionally substituted, C2-40 alkynyl; wherein at least one methylene group of R4 is independently replaced with optionally substituted carbocyclylene, optionally substituted heterocyclylene, optionally substituted arylene, optionally substituted heteroarylene, - N(RN)-, -O-, -S-, -C(O)-, -C(O)N(RN)-, -NRNC(O)-, -NRNC(O)N(RN)-, -C(O)O-, - OC(O)-, -OC(O)O-, -OC(O)N(RN)-, -NRNC(O)O-, -C(O)S-, -SC(O)-, -C(=NRN)-, - C(=NRN)N(RN)-, -NRNC(=N RN)-, -NRNC(=NRN)N(RN)-, - C(S)-, C(S)N(RN)-, - NRNC(S)-, -NRNC(S)N(RN)-, -S(O)-, OS(O)-, -S(O)O-, -OS(O)O-, -OS(O)2-, -S(O)2O-, - OS(O)2O -, -N(RN)S(O)-, -S(O)N(RN)-, -N(RN)S(O)N(RN)-, -OS(O)N(RN)-, - N(RN)S(O)O-, -S(O)2-, -N(RN)S(O)2-,-S(O) 2N(RN)-, -N(RN)S(O)2N(RN)-, - OS(O)2N(RN)-, or -N(RN)S(O)2O-; and
each instance of R is independently hydrogen, optionally substituted alkyl, or a nitrogen protecting group. -
-
- Phospholipids, as defined herein, are any lipids that comprise a phosphate group. Phospholipids are a subset of neutral lipids. The lipid component of a nanoparticle composition may include one or more phospholipids, such as one or more (poly)unsaturated lipids. Phospholipids may assemble into one or more lipid bilayers. In general, phospholipids may include a phospholipid moiety and one or more fatty acid moieties. A phospholipid moiety may be selected from the non-limiting group consisting of phosphatidyl choline, phosphatidyl ethanolamine, phosphatidyl glycerol, phosphatidyl serine, phosphatidic acid, 2-lysophosphatidyl choline, and a sphingomyelin. A fatty acid moiety may be selected from the non-limiting group consisting of lauric acid, myristic acid, myristoleic acid, palmitic acid, palmitoleic acid, stearic acid, oleic acid, linoleic acid, alpha-linolenic acid, erucic acid, phytanoic acid, arachidic acid, arachidonic acid, eicosapentaenoic acid, behenic acid, docosapentaenoic acid, and docosahexaenoic acid. Non-natural species including natural species with modifications and substitutions including branching, oxidation, cyclization, and alkynes are also contemplated. For example, a phospholipid may be functionalized with or cross-linked to one or more alkynes (e.g., an alkenyl group in which one or more double bonds is replaced with a triple bond). Under appropriate reaction conditions, an alkyne group may undergo a copper-catalyzed cycloaddition upon exposure to an azide. Such reactions may be useful in functionalizing a lipid bilayer of a nanoparticle composition to facilitate membrane permeation or cellular recognition or in conjugating a nanoparticle composition to a useful component such as a targeting or imaging moiety (e.g., a dye). Each possibility represents a separate embodiment of the present invention.
- Phospholipids useful in the compositions and methods may be selected from the nonlimiting group consisting of 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC),
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE);1,2-dilinoleoyl-sn-glycero-3-phosphocholine (DLPC); 1,2-dimyristoyl-sn-glycero-phosphocholine (DMPC); 1,2 dioleoyl-sn-glycero-3-phosphocholine (DOPC); 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC); 1,2-diundecanoyl-sn-glycero-phosphocholine (DUPC); 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC); 1,2-di-O-octadecenyl-sn-glycero-3-phosphocholine (18:0 Diether PC); l-oleoyl-2-cholesterylhemisuccinoyl-sn-glycero-3-phosphocholine (OChemsPC); 1-hexadecyl-sn-glycero-3-phosphocholine (CI 6 Lyso PC); 1,2-dilinolenoyl-sn-glycero-3-phosphocholine; 1,2-diarachidonoyl-sn-glycero-3-phosphocholine; 1,2-didocosahexaenoyl-sn-glycero-3-phosphocholine; 1,2-diphytanoyl-sn-glycero-3-phosphoethanolamine (ME 16.0 PE); 1,2-distearoyl-sn-glycero-3-phosphoethanolamine; 1,2-dilinoleoyl-sn-glycero-3-phosphoethanolamine; 1,2-dilinolenoyl-sn-glycero-3-phosphoethanolamine; 1,2-diarachidonoyl-sn-glycero-3-phosphoethanolamine; 1,2-didocosahexaenoyl-sn-glycero-3-phosphoethanolamine; or 1,2-dioleoyl-sn-glycero-3-phospho-rac-(1 -glycerol) sodium salt (DOPG), and sphingomyelin.
- In some embodiments, a nanoparticle composition includes DSPC. In certain embodiments, a nanoparticle composition includes DOPE. In some embodiments, a nanoparticle composition includes both DSPC and DOPE.
-
-
- In various embodiments any of the disclosed lipid nanoparticles comprise a steroid or steroid analogue. In certain embodiments, the steroid or steroid analogue is cholesterol. In some embodiments, the steroid is present in a concentration ranging from 35 to 49 molar percent, 37 to 46 molar percent, from 38 to 44 molar percent, from 38 to 40 molar percent, from 40 to 42 molar percent, from 42 to 44 molar percent, or from 44 to 46 molar percent. In certain specific embodiments, the steroid is present in a concentration of 37, 38, 39, 40, 41, 42, 43, 44, 45, or 46 molar percent.
- In certain embodiments, the molar ratio of cationic lipid to the steroid ranges from 1.0:0.9 to 1.0:1.2, or from 1.0:1.0 to 1.0:1.2. In some of these embodiments, the molar ratio of cationic lipid to cholesterol ranges from about 5:1 to 1:1. In certain embodiments, the steroid is present in a concentration ranging from 35 to 45 mol percent of the steroid.
- In certain embodiments, the molar ratio of total cationic to the steroid ranges from 1.0:0.9 to 1.0:1.2, or from 1.0:1.0 to 1.0:1.2. In some of these embodiments, the molar ratio of total cationic lipid to cholesterol ranges from about 5:1 to 1:1. In certain embodiments, the steroid is present in a concentration ranging from 35 to 45 mol percent of the steroid.
-
- R' and R" are each independently a saturated alkyl having from 8 to 12 carbon atoms, provided that the total number of carbon atoms collectively in both of R' and R" is no more than 23;
- R‴ is H or C1-C6 alkyl; and
- n is an integer ranging from 30 to 60.
- As used herein, the R' and R" moieties are collectively referred to as the di-acyl chains of a polymer conjugated lipid. For example, a C12 di-acyl chain polymer conjugated lipid refers to a polymer-conjugated lipid, such as the above structure, having two C12 acyl chains (e.g., the R' and R" moieties). Similarly, a C12/14 di-acyl chain polymer-conjugated lipid refers to a polymer-conjugated lipid, such as the above structure, having one C12 acyl chain and one
C 14 acyl chain (e.g., the R' and R" moieties). Other polymer-conjugated lipids are identified similarly. - In some embodiments, n is an integer from 40 to 50.
- In other embodiments, R‴ is H or CH3.
- In various different embodiments, the total number of carbon atoms collectively in both of R' and R" ranges from 16 to 22, 16 to 21, 16 to 20, 18 to 23, 18 to 22, 18 to 21, 19 to 23, 19 to 22, 19 to 21, 20 to 23, or 20 to 22.
- In still more embodiments:
- a) R' and R" are each a saturated alkyl having 8 carbon atoms;
- b) R' and R" are each a saturated alkyl having 9 carbon atoms;
- c) R' and R" are each a saturated alkyl having 10 carbon atoms; or
- d) R' and R" are each a saturated alkyl having 11 carbon atoms.
- LNPs comprising the foregoing polymer-conjugated lipid are also provided.
- In some embodiments, the LNPs comprise a polymer conjugated lipid. In various other embodiments the polymer conjugated lipid is a pegylated lipid. For example, some embodiments include a pegylated diacylglycerol (PEG-DAG) such as 1-(monomethoxy-polyethyleneglycol)-2,3-dimyristoylglycerol (PEG-DMG), a pegylated phosphatidylethanoloamine (PEG-PE), a PEG succinate diacylglycerol (PEG-S-DAG) such as 4-O-(2',3'-di(tetradecanoyloxy)propyl-]-O-(ω-methoxy(polyethoxy)ethyl)butanedioate (PEG-S-DMG), a pegylated ceramide (PEG-cer), or a PEG dialkoxypropylcarbamate such as ω-methoxy(polyethoxy)ethyl-N-(2,3-di(tetradecanoxy)propyl)carbamate or 2,3-di(tetradecanoxy)propyl-N-(ω-methoxy(polyethoxy)ethyl)carbamate.
- In yet more embodiments, a polymer conjugated lipid may be selected from the non-limiting group consisting of PEGylated phosphatidylethanolamines, PEGmodified phosphatidic acids, PEGylated ceramides, PEGylated dialkylamines, PEGylated diacylglycerols, PEGylated dialkylglycerols, and mixtures thereof. For example, a PEG lipid may be PEG-c-DOMG, PEG-DLPE, PEG-DMPE, PEG-DPPC, or a PEG-DSPE lipid.
-
- In one embodiment, PEG lipids useful in the present invention can be PEGylated lipids described in International Publication No.
WO2012/099755 . Any of these exemplary PEG lipids described herein may be modified to comprise a hydroxyl group on the PEG chain. In certain embodiments, the PEG lipid is a PEG-OH lipid. As generally defined herein, a "PEG-OH lipid" (also referred to herein as "hydroxy-PEGylated lipid") is a PEGylated lipid having one or more hydroxyl (-OH) groups on the lipid. In certain embodiments, the PEG-OH lipid includes one or more hydroxyl groups on the PEG chain. In certain embodiments, a PEG-OH or hydroxy-PEGylated lipid comprises an -OH group at the terminus of the PEG chain. Each possibility represents a separate embodiment of the present invention. -
- R3 is -ORO;
- RO is hydrogen, optionally substituted alkyl, or an oxygen protecting group;
- r is an integer between 1 and 150, inclusive;
- L1 is optionally substituted C1-10alkylene, wherein at least one methylene of the optionally substituted C1-10alkylene is independently replaced with optionally substituted carbocyclyclene, optionally substituted heterocyclylene, optionally substituted arylene, optionally substituted heteroarylene, -O-, -N(RN)-, -S-, -C(O)-, - C(O)N(RN)-, -NRNC(O )-, - C(O)O-, -OC(O)-, -OC (O)O-, -OC(O)N(RN) -, -NRNC(O)O -, or -NRNC(O)N(RN )-;
- D is a moiety obtained by click chemistry or a moiety cleavable under physiological conditions;
- m is 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10;
- A is of the formula:
- each instance of L2 is independently a bond or optionally substituted C 1-6 alkylene, wherein one methylene unit of the optionally substituted C1-6 alkylene is optionally replaced with -O-, -N(RN )-, -S-, -C(O)-, -C(O)N(RN)-, -NRNC(O)-, -C(O)O-, -OC(O)-, -OC(O)O-, -OC(O)N(RN)-, -NRC(O)O-, or -NRNC(O)N(R N)-;
- each instance of R2 is independently optionally substituted C 1-30 alkyl, optionally substituted C1-30 alkenyl, or optionally substituted C1-30 alkynyl; optionally wherein one or more methylene units of R2 are independently replaced with optionally substituted carbocyclylene, optionally substituted heterocyclylene, optionally substituted arylene, optionally substituted heteroarylene, -N(RN)-, -O-, -S-, -C(O)-, -C(O)N(RN)-, - NRNC(O)-, -NRNC(O)N(RN)-, -C(O)O-, -OC(O)-, -OC(O)O-, -OC(O)N(RN)-, - NRNC(O)O-, -C(O)S- -SC(O)-, -C(=NRN)-, -C(=NRN)N(RN)-, -NRNC(=N RN)-, - NRNC(=NRN)N(RN)-, - C(S)-, C(S)N(RN)-, -NRNC(S)-, -NRNC(S)N(RN)-, -S(O)-, - OS(O)-, -S(O)O-, -OS(O)O-, -OS(O)2-, -S(O)2O-, -OS(O)2O -, -N(RN)S(O)-, - S(O)N(RN)-, -N(RN)S(O)N(RN)-, -OS(O)N(RN)-, -N(RN)S(O)O-, -S(O)2-, -N(RN)S(O)2-,-S(O)2N(RN)-, -N(RN)S(O) 2N(RN)-, -OS(O)2N(RN)-, or -N(RN)S(O)2O-;
- each instance of RN is independently hydrogen, optionally substituted alkyl, or a nitrogen protecting group;
- Ring B is optionally substituted carbocyclyl, optionally substituted heterocyclyl, optionally substituted aryl, or optionally substituted hctcroaryl; and
- p is
1or 2. -
-
- R3 is -ORO;
- RO is hydrogen, optionally substituted alkyl or an oxygen protecting group;
- r is an integer between 1 and 100, inclusive;
- R5 is optionally substituted C10-40 alkyl, optionally substituted C10-40 alkenyl, or optionally substituted C10-40 alkynyl; and optionally one or more methylene groups of R5 are replaced with optionally substituted carbocyclylene, optionally substituted heterocyclylene, optionally substituted arylene, optionally substituted heteroarylene, -N(RN)-, -O-, -S-, -C(O)-, -C(O)N(RN)-, -NRNC(O)-, -NRNC(O)N(RN)-, - C(O)O-, -OC(O)-, -OC(O)O-, -OC(O)N(RN)-, -NRNC(O)O-, -C(O)S- -SC(O)-, - C(=NRN)-, -C(=NRN)N(RN)-, -NRNC(=N RN)-, -NRNC(=NRN)N(RN)-, - C(S)-, C(S)N(RN)-, -NRNC(S)-, -NRNC(S)N(RN)-, -S(O)-, -OS(O)-, -S(O)O-, -OS(O)O-, - OS(O)2-, -S(O)2O-, -OS(O)2O -, -N(RN)S(O)-, -S(O)N(RN)-, -N(RN)S(O)N(RN)-, - OS(O)N(RN)-, -N(RN)S(O)O-, -S(O)2-, -N(RN)S(O)2-,-S(O) 2N(RN)-, -N(RN)S(O) 2N(RN)-, -OS(O)2N(RN)-, or -N(RN)S(O)2O-; and
- each instance of RN is independently hydrogen, optionally substituted alkyl, or a nitrogen protecting group.
-
- Wherein r is an integer between 1 and 100.
-
- each of R1 and R2, independently, is a C10 to C30 aliphatic group, where the aliphatic group is optionally substituted by one or more groups each independently selected from Ra; and where the aliphatic group is optionally interrupted by cycloalkylene, -O-, -S-, -C(O)-, -OC(O)-,-C(O)O-, -N(Rc)-, -C(O)N(Rc)-, or -N(Rc)C(O)-
- X is -(CRaRb )i-, -O-, -S-, -C(O)-, -N(Rc )-, -OC(O)-, -C(O)O-, -OC(O)O-, -C(O)N(Rc)-,-N(Rc)C(O)-, -OC(O)N(Rc)-, -N(Rc)C(O)O-, -N(Rc)C(O)N(Rc)-, - SC(O)N(Rc)-, or -N(Rc)C(O)S-;
- Y is -(CRaRb )i-, -O-, -S-, -C(O)-, -N(Rc )-, -OC(O)-, -C(O)O-, -OC(O)O-, -C(O)N(Rc)-,-N(Rc)C(O)-, -OC(O)N(Rc)-, -N(Rc)C(O)O-, -N(Rc)C(O)N(Rc)-, - SC(O)N(Rc)-, or -N(Rc)C(O)S-;
- L is -L'-Z'-(L2-Z2) -L3- ;
- L1 is a bond, -(CR'R5')i-, or -(CR5R5')i-(C(Ra)=C(Rb))k-(C≡C)k, -(CRaRb )j -;
- Z1 is -O-, -S-, -N(Rc)-, -OC(O)-, -C(O)O-, -OC(O)O-, -OC(O)N(Rc)-, - N(Rc)C(O)O-, -N(Rc)C(O)-, -C(O)N(Rc)-, -N=C(Ra)-, -C(Ra)=N-, -O-N=C(Ra)-, or -ON(Rc)-;
- L2 is -(CRaRb)p- or -(CRaRb)j-(C(Ra)=C(Rb))k-(C≡C)k-(CRaR )j;
- Z2 is -O-, -S-, -N(Rc)-, -OC(O)-, -C(O)O-, -OC(O)O-, -OC(O)N(Rc)-, - N(Rc)C(O)O-, -N(Rc)C(O)-, -C(O)N(Rc)-, -N=C(Ra)-, -C(Ra)=N-, -O-N=C(Ra)-, or -ON(Rc)-;
- L3 is -(CRaRb)i-;
- each A, independently, is -L4-, -NH-(L4)q -(CRaRb )r-C(O)- or -C(O)-(CRaRb )r -(L4)q -NH-; where each q, independently, is 0, 1, 2, 3, or 4; and each r, independently, is 0, 1, 2, 3, or 4;
- each L4, independently, is -(CRaRb)sO- or -O(CRaRb)s-; where each s, independently, is 0, 1, 2, 3, or 4;
- R3 is -H, -R , or -OR ;
- each of R4 and R4' , independently, is -H, halo, cyano, hydroxy, nitro, alkyl, alkenyl, alkynyl, cycloalkyl, alkoxy, or cycloalkoxy;
- each R5 and each R5 , independently, is -H, halo, cyano, hydroxy, nitro, alkyl, alkenyl, alkynyl, or cycloalkyl;
- or R4 and one R5, taken together, can form a 5- to 8-membered cycloalkyl or heterocyclic ring;
- each Ra , independently, is -H, halo, cyano, hydroxy, nitro, amino, alkylamino, dialkylamino, alkyl, alkenyl, alkynyl, cycloalkyl, alkoxy, cycloalkoxy, aryl, heteroaryl, or heterocyclyl;
- each Rb , independently, is -H, halo, cyano, hydroxy, nitro, amino, alkylamino, dialkylamino, alkyl, alkenyl, alkynyl, cycloalkyl, alkoxy, cycloalkoxy, aryl, heteroaryl, or heterocyclyl;
- each Rc is -H, alkyl, acyl, cycloalkyl, alkenyl, alkynyl, aryl, heteroaryl, or heterocyclyl;
- a is 0 or 1;
- b is an integer from 1 to 1,000;
- c is 0 or 1;
- each occurrence of i, independently, is 1, 2, 3, 4, 5, or 6;
- each occurrence of j , independently, is 0, 1, 2, or 3;
- each occurrence of k, independently, is 0, 1, 2, or 3; and
- p is 1 to 10; with the proviso that
- (i) X and Y are not simultaneously -CH2-; and
- (ii) when a is 1 and L1 is -CH2-, then
- (a) X and Y are not simultaneously -O-; and
- (b) X and Y are not simultaneously -C(O)O-.
-
- n is an integer from 1 to 1,000; and
- m is 1, 2, 3, 4, 5, or 6.
-
- R1 and R2 are each, independently, a C10 to C30 aliphatic group, wherein each aliphatic group is optionally substituted by one or more groups each independently selected from Ra;
- L is -L1-Z1-(L2-Z2)c-L3-;
- L1 is a bond, -(CR5R5')i-, or -(CR5R5')i-(C(Ra)=C(Rb))k-(C≡C)k-(CRaRb)j-;
- Z1 is -O-, -S-, -N(Rc)-, -OC(O)-, -C(O)O-, -OC(O)O-, -N(Rc)C(O)O-, - N(Rc)C(O)N(Rc)-, -N(Rc)C(O)-, -C(O)N(Rc)-, -N=C(Ra)-, -C(Ra)=N-, -O-N=C(Ra)-, - O-N(Rc)-; heteroaryl, or heterocyclyl;
- L2 is -(CRaRb)p- or -(CRaRb)j-(C(Ra)=C(Rb))k-(C≡C)k-(CRaRb)j-;
- Z2 is -O-, -S-, -N(Rc)-, -OC(O)-, -C(O)O-, -OC(O)O-, -OC(O)N(Rc)-, - N(Rc)C(O)O-, -N(Rc)C(O)-, -C(O)N(Rc)-, -N=C(Ra)-, -C(Ra)=N-, -O-N=C(Ra)-, -ON(Rc)-, heteroaryl, or heterocyclyl;
- L3 is -(CRaRb)i-;
- each A, independently, is -L4-, -NH-(L4)q-(CRaRb)r-C(O)-, or -C(O)-(CRaRb)r-(L4)q-NH-; wherein each q, independently, is 0, 1, 2, 3, or 4; and each r, independently, is 0, 1, 2, 3, or 4;
- each L4, independently, is -(CRaRb)sO- or -O(CRaRb)s-, wherein each s, independently, is 0, 1, 2, 3, or 4;
- R3 is H, -Rc, or -ORc;
- each occurrence of R5 and R5' is, independently, H, halo, cyano, hydroxy, nitro, alkyl, alkenyl, alkynyl, or cycloalkyl;
- each occurrence of Ra and Rb is, independently, H, halo, cyano, hydroxy, nitro, amino, alkylamino, dialkylamino, alkyl, alkenyl, alkynyl, cycloalkyl, alkoxy, aryl, heteroaryl, or heterocyclyl;
- each Rc is, independently, H, alkyl, acyl, cycloalkyl, alkenyl, alkynyl, aryl, heteroaryl, or heterocyclyl;
- b ranges from 5 to about 500;
- c is 0 or 1;
- each i is, independently, 1, 2, 3, 4, 5, 6, 7, 8, or 9;
- each occurrence of j and k, independently, is 0, 1, 2, or 3; and
- p is an integer from 1 to 10;
or - R1 and R2 are each, independently C10 to C30 aliphatic group;
- L is -L1-Z1-L3-;
- L1 is a bond or -(CR5R5')i-;
- Z1 is -N(Rc)-, -N(Rc)C(O)O-, -N(Rc)C(O)N(Rc)-, -N(Rc)C(O)-, or - N=C(Ra)-, wherein the leftmost nitrogen atom in Z1 is bound to L1 or if L1 is a bond, then to the central tertiary carbon atom of formula (II)), or
- Z1 is a nitrogen-containing heteroaryl or heterocyclyl, wherein the nitrogen atom of the heteroaryl or heterocyclyl is bound to L1 or if L1 is a bond, then to the central tertiary carbon atom of formula (II));
- L3 is -(CRaRb)i-;
- each A is, independently, -L4-;
- b ranges from about 5 to about 500;
- each L4, independently, is -OCH2CH2-, -CH2CH2O-, -OCH(CH3)CH2- or -OCH2CH(CH3)-;
- R3 is -ORc;
- each occurrence of Ra, Rc, R5 and R5' is, independently, H or alkyl; and i is 2, 3, 4 or 5;
or - R1 and R2 are each, independently C12 to C20 alkyl or C12 to C20alkenyl;
- L is -L1-Z1-L2-Z2-L3-;
- L1 is a bond or -(CR5R5')i-;
- Z1 is -N(Rc)-, -N(Rc)C(O)O-, -N(Rc)C(O)N(Rc)-, -N(Rc)C(O)-, or - N=C(Ra)-, wherein the leftmost nitrogen atom in Z1 is bound to L1 or if L1 is a bond, then to the central tertiary carbon atom of formula (II)), or
- Z1 is a nitrogen-containing heteroaryl or heterocyclyl, wherein the nitrogen atom of the heteroaryl or heterocyclyl is bound to L1 or if L1 is a bond, then to the central tertiary carbon atom of formula (II);
- L2 is -(CRaRb)p;
- Z2 is -O-, -C(O)O-, -C(O)N(Rc)-, or heteroaryl;
- L3 is -(CRaRb)i-;
- each A is, independently, -L4-;
- b ranges from about 5 to about 500;
- each L4, independently, is -OCH2CH2-, -CH2CH2O-, -OCH(CH3)CH2- or -OCH2CH(CH3)-;
- R3 is -ORc;
- each occurrence of Ra, Rb, Rc, R5 and R5' is, independently, H or alkyl;
- i is 2, 3, 4 or 5; and
- p is 1 to 10.
-
- n is an integer from 1 to 1,000;
- m is 0, 1, 2, 3, 4, 5, or 6;
- and pharmaceutically acceptable salts thereof.
-
- n is an integer from 10 to 100 (e.g. 20-50 or 40-50);
- s, s', t and t' are independently 0, 1, 2, 3, 4, 5, 6 or 7; and m is 1, 2, 3, 4, 5, or 6.
-
- In some embodiments, the ratio of polymer conjugated lipid in the LNPs may be increased or decreased to alter the pharmacokinetics and/or biodistribution of the LNPs. In certain embodiments, LNPs may contain from 0.1 to 5.0, from 1.0 to 3.5, from 1.5 to 4.0, from 2.0 to 4.5, from .0 to 3.0, from 2.5 to 5.0, and/or from 3.0 to 6.0 molar percent of the polymer conjugated lipid to the other components. In various embodiments, the polymer conjugated lipid is present in a concentration ranging from 1.0 to 3.0 molar percent. In certain specific embodiments, the LNP comprises from 2.2 to 3.3, from 2.3 to 2.8, from 2.1 to 2.5, or from 2.5 to 2.9 molar percent of polymer conjugated lipid. In yet more specific embodiments the polymer conjugated lipid is present in a concentration of about 2.0 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.3 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.4 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.5 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.6 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.7 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 2.8 molar percent. In some embodiments, the polymer conjugated lipid is present in a concentration of about 3.0 molar percent.
- In certain embodiments, the molar ratio of cationic lipid to the polymer conjugated lipid ranges from about 35:1 to about 15:1. In some embodiments, the molar ratio of cationic lipid to polymer conjugated lipid ranges from about 100:1 to about 10:1.
-
- P is a polymer;
- L is a trivalent linker of 1 to 15 atoms in length; and
- R' and R" are each independently a saturated alkyl having from 8 to 14 carbon atoms.
-
-
- R8 and R9 are each independently a straight or branched, saturated or unsaturated alkyl chain containing from 8 to 30 carbon atoms, wherein the alkyl chain is optionally interrupted by one or more ester bonds; and
- n has a mean value ranging from 30 to 60, or 15 to 25, or 100 to 125.
- In some embodiments, R8 and R9 are each independently straight, saturated alkyl chains containing from 8 to 16 carbon atoms. In other embodiments, the average n ranges from 42 to 55, for example, the average w is 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54 or 55. In some specific embodiments, the average w is about 49.
-
- R' and R" are each independently a saturated alkyl having from 8 to 12 carbon atoms;
- R‴ is H or C1-C6 alkyl; and
- n is an integer ranging from 30 to 60.
-
-
- In certain embodiments, lipid nanoparticles are associated with a nucleic acid, resulting in a nucleic acid-lipid nanoparticle. In particular embodiments, the nucleic acid is fully encapsulated in the lipid nanoparticle. As used herein, the term "nucleic acid" is meant to include any oligonucleotide or polynucleotide. Fragments containing up to 50 nucleotides are generally termed oligonucleotides, and longer fragments are called polynucleotides. In particular embodiments, oligonucletoides are 15-50 nucleotides in length.
- The terms "polynucleotide" and "oligonucleotide" refer to a polymer or oligomer of nucleotide or nucleoside monomers consisting of naturally occurring bases, sugars and intersugar (backbone) linkages. The terms "polynucleotide" and "oligonucleotide" also includes polymers or oligomers comprising non-naturally occurring monomers, or portions thereof, which function similarly. Such modified or substituted oligonucleotides are often preferred over native forms because of properties such as, for example, enhanced cellular uptake and increased stability in the presence of nucleases.
- In some embodiments the nucleic acid is selected from antisense, self amplifying RNA and messenger RNA. For example, messenger RNA may be used to induce an immune response (e.g., as a vaccine), for example by translation of immunogenic proteins.
- In other embodiments, the nucleic acid is mRNA, and the mRNA to lipid ratio in the LNP (i.e., N/P, were N represents the moles of cationic lipid and P represents the moles of phosphate present as part of the nucleic acid backbone) range from 2:1 to 30:1, for example 3:1 to 22:1. In other embodiments, N/P ranges from 6:1 to 20:1 or 2: 1 to 12:1. Exemplary N/P ranges include about 3:1. About 6:1, about 9:1, about 12:1 and about 22:1.
- The nucleic acid that is present in a lipid-nucleic acid particle includes any form of nucleic acid that is known. The nucleic acids used herein can be single-stranded DNA or RNA, or double- stranded DNA or RNA, or DNA-RNA hybrids. Examples of double- stranded DNA include structural genes, genes including control and termination regions, and self-replicating systems such as viral or plasmid DNA. Examples of double- stranded RNA include siRNA and other RNA interference reagents. Single- stranded nucleic acids include, e.g., messenger RNA, antisense oligonucleotides, ribozymes, microRNA, and triplex-forming oligonucleotides. The nucleic acid that is present in a lipid-nucleic acid particle may include one or more of the oligonucleotide modifications described below.
- Nucleic acids may be of various lengths, generally dependent upon the particular form of nucleic acid. For example, in particular embodiments, plasmids or genes may be from about 1,000 to 100,000 nucleotide residues in length. In particular embodiments, oligonucleotides may range from about 10 to 100 nucleotides in length. In various related embodiments, oligonucleotides, single- stranded, double- stranded, and triple- stranded, may range in length from about 10 to about 50 nucleotides, from about 20 o about 50 nucleotides, from about 15 to about 30 nucleotides, from about 20 to about 30 nucleotides in length.
- In particular embodiments, the oligonucleotide (or a strand thereof) specifically hybridizes to or is complementary to a target polynucleotide. "Specifically hybridizable" and "complementary" are terms which are used to indicate a sufficient degree of complementarity such that stable and specific binding occurs between the DNA or RNA target and the oligonucleotide. It is understood that an oligonucleotide need not be 100% complementary to its target nucleic acid sequence to be specifically hybridizable. An oligonucleotide is specifically hybridizable when binding of the oligonucleotide to the target interferes with the normal function of the target molecule to cause a loss of utility or expression therefrom, and there is a sufficient degree of complementarity to avoid non-specific binding of the oligonucleotide to non-target sequences under conditions in which specific binding is desired, i.e., under physiological conditions in the case of in vivo assays or therapeutic treatment, or, in the case of in vitro assays, under conditions in which the assays are conducted. Thus, in other embodiments, this oligonucleotide includes 1, 2, or 3 base substitutions, e.g. mismatches, as compared to the region of a gene or mRNA sequence that it is targeting or to which it specifically hybridizes.
- In particular embodiments, nucleic acid-lipid nanoparticles are associated with RNA interference (RNAi) molecules. RNA interference methods using RNAi molecules may be used to disrupt the expression of a gene or polynucleotide of interest. Small interfering RNA (siRNA) has essentially replaced antisense ODN and ribozymes as the next generation of targeted oligonucleotide drugs under development.
- SiRNAs are RNA duplexes normally 16-30 nucleotides long that can associate with a cytoplasmic multi-protein complex known as RNAi-induced silencing complex (RISC). RISC loaded with siRNA mediates the degradation of homologous mRNA transcripts, therefore siRNA can be designed to knock down protein expression with high specificity. Unlike other antisense technologies, siRNA function through a natural mechanism evolved to control gene expression through non-coding RNA. This is generally considered to be the reason why their activity is more potent in vitro and in vivo than either antisense ODN or ribozymes. A variety of RNAi reagents, including siRNAs targeting clinically relevant targets, are currently under pharmaceutical development, as described, e.g., in de Fougerolles, A. et al., Nature Reviews 6:443-453 (2007).
- While the first described RNAi molecules were RNA:RNA hybrids comprising both an RNA sense and an RNA antisense strand, it has now been demonstrated that DNA sense:RNA antisense hybrids, RNA sense:DNA antisense hybrids, and DNA:DNA hybrids are capable of mediating RNAi (Lamberton, J.S. and Christian, A.T., (2003) Molecular Biotechnology24: 111-119). Thus, the use of RNAi molecules comprising any of these different types of double-stranded molecules is contemplated. In addition, it is understood that RNAi molecules may be used and introduced to cells in a variety of forms. Accordingly, as used herein, RNAi molecules encompasses any and all molecules capable of inducing an RNAi response in cells, including, but not limited to, double-stranded oligonucleotides comprising two separate strands, i.e. a sense strand and an antisense strand, e.g., small interfering RNA (siRNA); double-stranded oligonucleotide comprising two separate strands that are linked together by non-nucleotidyl linker; oligonucleotides comprising a hairpin loop of complementary sequences, which forms a double-stranded region, e.g., shRNAi molecules, and expression vectors that express one or more polynucleotides capable of forming a double-stranded polynucleotide alone or in combination with another polynucleotide.
- A "single strand siRNA compound" as used herein, is an siRNA compound which is made up of a single molecule. It may include a duplexed region, formed by intra-strand pairing, e.g., it may be, or include, a hairpin or pan-handle structure. Single strand siRNA compounds may be antisense with regard to the target molecule
- A single strand siRNA compound may be sufficiently long that it can enter the RISC and participate in RISC mediated cleavage of a target mRNA. A single strand siRNA compound is at least 14, and in other embodiments at least 15, 20, 25, 29, 35, 40, or 50 nucleotides in length. In certain embodiments, it is less than 200, 100, or 60 nucleotides in length.
- Hairpin siRNA compounds will have a duplex region equal to or at least 17, 18, 19, 29, 21, 22, 23, 24, or 25 nucleotide pairs. The duplex region will may be equal to or less than 200, 100, or 50, in length. In certain embodiments, ranges for the duplex region are 15-30, 17 to 23, 19 to 23, and 19 to 2 1 nucleotides pairs in length. The hairpin may have a single strand overhang or terminal unpaired region. In certain embodiments, the overhangs are 2-3 nucleotides in length. In some embodiments, the overhang is at the sense side of the hairpin and in some embodiments on the antisense side of the hairpin.
- A "double stranded siRNA compound" as used herein, is an siRNA compound which includes more than one, and in some cases two, strands in which interchain hybridization can form a region of duplex structure.
- The antisense strand of a double stranded siRNA compound may be equal to or at least, 14, 15, 16 17, 18, 19, 25, 29, 40, or 60 nucleotides in length. It may be equal to or less than 200, 100, or 50, nucleotides in length. Ranges may be 17 to 25, 19 to 23, and 19 to21 nucleotides in length. As used herein, term "antisense strand" means the strand of an siRNA compound that is sufficiently complementary to a target molecule, e.g. a target RNA.
- The sense strand of a double stranded siRNA compound may be equal to or at least 14, 15, 16 17, 18, 19, 25, 29, 40, or 60 nucleotides in length. It may be equal to or less than 200, 100, or 50, nucleotides in length. Ranges may be 17 to 25, 19 to 23, and 19 to 2 1 nucleotides in length.
- The double strand portion of a double stranded siRNA compound may be equal to or at least, 14, 15, 16 17, 18, 19, 20, 21, 22, 23, 24, 25, 29, 40, or 60 nucleotide pairs in length. It may be equal to or less than 200, 100, or 50, nucleotides pairs in length. Ranges may be 15-30, 17 to 23, 19 to 23, and 19 to 2 1 nucleotides pairs in length.
- In many embodiments, the siRNA compound is sufficiently large that it can be cleaved by an endogenous molecule, e.g., by Dicer, to produce smaller siRNA compounds, e.g., siRNAs agents.
- The sense and antisense strands may be chosen such that the double-stranded siRNA compound includes a single strand or unpaired region at one or both ends of the molecule. Thus, a double-stranded siRNA compound may contain sense and antisense strands, paired to contain an overhang, e.g., one or two 5' or 3' overhangs, or a 3' overhang of 1 - 3 nucleotides. The overhangs can be the result of one strand being longer than the other, or the result of two strands of the same length being staggered. Some embodiments will have at least one 3' overhang. In one embodiment, both ends of an siRNA molecule will have a 3' overhang. In some embodiments, the overhang is 2 nucleotides.
- In certain embodiments, the length for the duplexed region is between 15 and 30, or 18, 19, 20, 21, 22, and 23 nucleotides in length, e.g., in the ssiRNA compound range discussed above. ssiRNA compounds can resemble in length and structure the natural Dicer processed products from long dsiRNAs. Embodiments in which the two strands of the ssiRNA compound are linked, e.g., covalently linked are also included. Hairpin, or other single strand structures which provide the required double stranded region, and a 3' overhang are also contemplated.
- The siRNA compounds described herein, including double-stranded siRNA compounds and single-stranded siRNA compounds can mediate silencing of a target RNA, e.g., mRNA, e.g., a transcript of a gene that encodes a protein. For convenience, such mRNA is also referred to herein as mRNA to be silenced. Such a gene is also referred to as a target gene. In general, the RNA to be silenced is an endogenous gene or a pathogen gene. In addition, RNAs other than mRNA, e.g., tRNAs, and viral RNAs, can also be targeted.
- As used herein, the phrase "mediates RNAi" refers to the ability to silence, in a sequence specific manner, a target RNA. While not wishing to be bound by theory, it is believed that silencing uses the RNAi machinery or process and a guide RNA, e.g., an ssiRNA compound of 2 1 to 23 nucleotides.
- In one embodiment, an siRNA compound is "sufficiently complementary" to a target RNA, e.g., a target mRNA, such that the siRNA compound silences production of protein encoded by the target mRNA. In another embodiment, the siRNA compound is "exactly complementary" to a target RNA, e.g., the target RNA and the siRNA compound anneal, for example to form a hybrid made exclusively of Watson-Crick base pairs in the region of exact complementarity. A "sufficiently complementary" target RNA can include an internal region (e.g., of at least 10 nucleotides) that is exactly complementary to a target RNA. Moreover, in certain embodiments, the siRNA compound specifically discriminates a single-nucleotide difference. In this case, the siRNA compound only mediates RNAi if exact complementary is found in the region (e.g., within 7 nucleotides of) the single-nucleotide difference.
- Micro RNAs (miRNAs) are a highly conserved class of small RNA molecules that are transcribed from DNA in the genomes of plants and animals, but are not translated into protein. Processed miRNAs are single stranded -17-25 nucleotide (nt) RNA molecules that become incorporated into the RNA-induced silencing complex (RISC) and have been identified as key regulators of development, cell proliferation, apoptosis and differentiation. They are believed to play a role in regulation of gene expression by binding to the 3'-untranslated region of specific mRNAs. RISC mediates down-regulation of gene expression through translational inhibition, transcript cleavage, or both. RISC is also implicated in transcriptional silencing in the nucleus of a wide range of eukaryotes.
- In one embodiment, a nucleic acid is an antisense oligonucleotide directed to a target polynucleotide. The term "antisense oligonucleotide" or simply "antisense" is meant to include oligonucleotides that are complementary to a targeted polynucleotide sequence. Antisense oligonucleotides are single strands of DNA or RNA that are complementary to a chosen sequence, e.g. a target gene mRNA. Antisense oligonucleotides are thought to inhibit gene expression by binding to a complementary mRNA. Binding to the target mRNA can lead to inhibition of gene expression either by preventing translation of complementary mRNA strands by binding to it, or by leading to degradation of the target mRNA. Antisense DNA can be used to target a specific, complementary (coding or non-coding) RNA. If binding takes places this DNA/RNA hybrid can be degraded by the enzyme RNase H. In particular embodiments, antisense oligonucleotides contain from about 10 to about 50 nucleotides, more preferably about 15 to about 30 nucleotides. The term also encompasses antisense oligonucleotides that may not be exactly complementary to the desired target gene. Thus, instances where non-target specific-activities are found with antisense, or where an antisense sequence containing one or more mismatches with the target sequence is the most preferred for a particular use, are contemplated.
- Antisense oligonucleotides have been demonstrated to be effective and targeted inhibitors of protein synthesis, and, consequently, can be used to specifically inhibit protein synthesis by a targeted gene. The efficacy of antisense oligonucleotides for inhibiting protein synthesis is well established. For example, the synthesis of polygalactauronase and the
muscarine type 2 acetylcholine receptor are inhibited by antisense oligonucleotides directed to their respective mRNA sequences (U. S. Patent 5,739,119 andU. S. Patent 5,759,829 ). Further, examples of antisense inhibition have been demonstrated with the nuclear protein cyclin, the multiple drug resistance gene (MDG1), ICAM-1, E-selectin, STK-1, striatal GABA A receptor and human EGF (Jaskulski et al., Science. 1988 ; Vasanthakumar and Ahmed, Cancer Commun. 1989;1(4):225-32; Peris et al, Brain Res Mol Brain Res. 1998 ;U. S. Patent 5,801,154 ;U.S. Patent 5,789,573 ;U. S. Patent 5,718,709 andU.S. Patent 5,610,288 ). Furthermore, antisense constructs have also been described that inhibit and can be used to treat a variety of abnormal cellular proliferations, e.g. cancer (U. S. Patent 5,747,470 ;U. S. Patent 5,591,317 andU. S. Patent 5,783,683 ). - Methods of producing antisense oligonucleotides are known in the art and can be readily adapted to produce an antisense oligonucleotide that targets any polynucleotide sequence. Selection of antisense oligonucleotide sequences specific for a given target sequence is based upon analysis of the chosen target sequence and determination of secondary structure, Tm, binding energy, and relative stability. Antisense oligonucleotides may be selected based upon their relative inability to form dimers, hairpins, or other secondary structures that would reduce or prohibit specific binding to the target mRNA in a host cell. Highly preferred target regions of the niRNA include those regions at or near the AUG translation initiation codon and those sequences that are substantially complementary to 5' regions of the mRNA. These secondary structure analyses and target site selection considerations can be performed, for example, using v.4 of the OLIGO primer analysis software (Molecular Biology Insights) and/or the BLASTN 2.0.5 algorithm software (Altschul et al, Nucleic Acids Res. 1997, 25(17):3389-402).
- Antagomirs are RNA-like oligonucleotides that harbor various modifications for RNAse protection and pharmacologic properties, such as enhanced tissue and cellular uptake. They differ from normal RNA by, for example, complete 2'-O-methylation of sugar, phosphorothioate backbone and, for example, a cholesterol-moiety at 3'-end. Antagomirs may be used to efficiently silence endogenous miRNAs by forming duplexes comprising the antagomir and endogenous miRNA, thereby preventing miRNA-induced gene silencing. An example of antagomir-mediated miRNA silencing is the silencing of miR-122, described in Krutzfeldt et al, Nature, 2005, 438: 685-689. Antagomir RNAs may be synthesized using standard solid phase oligonucleotide synthesis protocols. See
U.S. Patent Application Publication Nos. 2007/0123482 and2007/0213292 . - An antagomir can include ligand-conjugated monomer subunits and monomers for oligonucleotide synthesis. Exemplary monomers are described in
U.S. Patent Application Publication No. 2005/0107325 . An antagomir can have a ZXY structure, such as is described inWO 2004/080406 . An antagomir can be complexed with an amphipathic moiety. Exemplary amphipathic moieties for use with oligonucleotide agents are described inWO 2004/080406 . - Aptamers are nucleic acid or peptide molecules that bind to a particular molecule of interest with high affinity and specificity (Tuerk and Gold, Science 249:505 (1990); Ellington and Szostak, Nature 346:818 (1990). DNA or RNA aptamers have been successfully produced which bind many different entities from large proteins to small organic molecules. See Eaton, Curr. Opin. Chem. Biol. 1:10-16 (1997), Famulok, Curr. Opin. Struct. Biol. 9:324-9(1999), and Hermann and Patel, Science 287:820-5 (2000). Aptamers may be RNA or DNA based, and may include a riboswitch. A riboswitch is a part of an mRNA molecule that can directly bind a small target molecule, and whose binding of the target affects the gene's activity. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, depending on the presence or absence of its target molecule. Generally, aptamers are engineered through repeated rounds of in vitro selection or equivalently, SELEX (systematic evolution of ligands by exponential enrichment) to bind to various molecular targets such as small molecules, proteins, nucleic acids, and even cells, tissues and organisms. The aptamer may be prepared by any known method, including synthetic, recombinant, and purification methods, and may be used alone or in combination with other aptamers specific for the same target.
- Further, as described more fully herein, the term "aptamer" specifically includes "secondary aptamers" containing a consensus sequence derived from comparing two or more known aptamers to a given target.
- According to another embodiment, nucleic acid-lipid nanoparticles are associated with ribozymes. Ribozymes are RNA molecules complexes having specific catalytic domains that possess endonuclease activity (Kim and Cech, Proc Natl Acad Sci USA. 1987 Dec;84(24):8788-92; Forster and Symons, Cell. 1987 ). For example, a large number of ribozymes accelerate phosphoester transfer reactions with a high degree of specificity, often cleaving only one of several phosphoesters in an oligonucleotide substrate (Cech et al, Cell. 1981 Dec;27(3 Pt 2):487-96; Michel and Westhof, J Mol Biol. 1990 ; Reinhold-Hurek and Shub, Nature. 1992 May 14;357(6374): 173-6). This specificity has been attributed to the requirement that the substrate bind via specific base-pairing interactions to the internal guide sequence ("IGS") of the ribozyme prior to chemical reaction.
- At least six basic varieties of naturally-occurring enzymatic RNAs are known presently. Each can catalyze the hydrolysis of RNA phosphodiester bonds in trans (and thus can cleave other RNA molecules) under physiological conditions. In general, enzymatic nucleic acids act by first binding to a target RNA. Such binding occurs through the target binding portion of a enzymatic nucleic acid which is held in close proximity to an enzymatic portion of the molecule that acts to cleave the target RNA. Thus, the enzymatic nucleic acid first recognizes and then binds a target RNA through complementary base-pairing, and once bound to the correct site, acts enzymatically to cut the target RNA. Strategic cleavage of such a target RNA will destroy its ability to direct synthesis of an encoded protein. After an enzymatic nucleic acid has bound and cleaved its RNA target, it is released from that RNA to search for another target and can repeatedly bind and cleave new targets.
- The enzymatic nucleic acid molecule may be formed in a hammerhead, hairpin, a hepatitis d virus, group I intron or RNaseP RNA (in association with an RNA guide sequence) or Neurospora VS RNA motif, for example. Specific examples of hammerhead motifs are described by Rossi et al. Nucleic Acids Res. 1992 Sep 11;20(17):4559-65. Examples of hairpin motifs are described by
Hampel et al. (Eur. Pat. Appl. Publ. No. EP 0360257 ), Hampel and Tritz, Biochemistry 1989 ; Hampel et al, Nucleic Acids Res. 1990 Jan 25;18(2):299-304 and U. S. Patent5,631,359 . An example of the hepatitis d virus motif is described by Perrotta and Been, Biochemistry. 1992 ; an example of the RNaseP motif is described by Guerrier-Takada et al, Cell. 1983 Dec;35(3 Pt 2):849-57; Neurospora VS RNA ribozyme motif is described by Collins (Saville and Collins, Cell. 1990 May 18;61(4):685-96; Saville and Collins, Proc Natl Acad Sci USA. 1991 ; Collins and Olive, Biochemistry. 1993 Mar 23;32(ll):2795-9); and an example of the Group I intron is described inU. S. Patent 4,987,071 . Important characteristics of enzymatic nucleic acid molecules used are that they have a specific substrate binding site which is complementary to one or more of the target gene DNA or RNA regions, and that they have nucleotide sequences within or surrounding that substrate binding site which impart an RNA cleaving activity to the molecule. Thus the ribozyme constructs need not be limited to specific motifs mentioned herein. - Methods of producing a ribozyme targeted to any polynucleotide sequence are known in the art. Ribozymes may be designed as described in Int. Pat. Appl. Publ. Nos.
WO 93/23569 WO 94/02595 - Ribozyme activity can be optimized by altering the length of the ribozyme binding arms or chemically synthesizing ribozymes with modifications that prevent their degradation by serum ribonucleases (see e.g., Int. Pat. Appl. Publ. Nos.
WO 92/07065 WO 93/15187 WO 91/03162 92110298.4 U.S. Patent 5,334,711 ; and Int. Pat. Appl. Publ. No.WO 94/13688 - Nucleic acids associated with lipid nanoparticles may be immunostimulatory, including immunostimulatory oligonucleotides (ISS; single-or double- stranded) capable of inducing an immune response when administered to a subject, which may be a mammal or other patient. ISS include, e.g., certain palindromes leading to hairpin secondary structures (see Yamamoto S., et al. (1992) J . Immunol. 148: 4072-4076), or CpG motifs, as well as other known ISS features (such as multi-G domains, see
WO 96/1 1266 - The immune response may be an innate or an adaptive immune response. The immune system is divided into a more innate immune system, and acquired adaptive immune system of vertebrates, the latter of which is further divided into humoral cellular components. In particular embodiments, the immune response may be mucosal.
- In particular embodiments, an immunostimulatory nucleic acid is only immunostimulatory when administered in combination with a lipid nanoparticle, and is not immunostimulatory when administered in its "free form." Such an oligonucleotide is considered to be immunostimulatory.
- Immunostimulatory nucleic acids are considered to be non-sequence specific when it is not required that they specifically bind to and reduce the expression of a target polynucleotide in order to provoke an immune response. Thus, certain immunostimulatory nucleic acids may comprise a sequence corresponding to a region of a naturally occurring gene or mRNA, but they may still be considered non-sequence specific immunostimulatory nucleic acids.
- In one embodiment, the immunostimulatory nucleic acid or oligonucleotide comprises at least one CpG dinucleotide. The oligonucleotide or CpG dinucleotide may be unmethylated or methylated. In another embodiment, the immunostimulatory nucleic acid comprises at least one CpG dinucleotide having a methylated cytosine. In one embodiment, the nucleic acid comprises a single CpG dinucleotide, wherein the cytosine in said CpG dinucleotide is methylated. In a specific embodiment, the nucleic acid comprises the sequence 5' TAACGTTGAGGGGCAT 3'. In an alternative embodiment, the nucleic acid comprises at least two CpG dinucleotides, wherein at least one cytosine in the CpG dinucleotides is methylated. In a further embodiment, each cytosine in the CpG dinucleotides present in the sequence is methylated. In another embodiment, the nucleic acid comprises a plurality of CpG dinucleotides, wherein at least one of said CpG dinucleotides comprises a methylated cytosine.
- Because transcription factors recognize their relatively short binding sequences, even in the absence of surrounding genomic DNA, short oligonucleotides bearing the consensus binding sequence of a specific transcription factor can be used as tools for manipulating gene expression in living cells. This strategy involves the intracellular delivery of such "decoy oligonucleotides", which are then recognized and bound by the target factor. Occupation of the transcription factor's DNA-binding site by the decoy renders the transcription factor incapable of subsequently binding to the promoter regions of target genes. Decoys can be used as therapeutic agents, either to inhibit the expression of genes that are activated by a transcription factor, or to upregulate genes that are suppressed by the binding of a transcription factor. Examples of the utilization of decoy oligonucleotides may be found in Mann et al., J . Clin. Invest., 2000, 106: 1071-1075.
- A supermir refers to a single stranded, double stranded or partially double stranded oligomer or polymer of ribonucleic acid (RNA) or deoxyribonucleic acid (DNA) or both or modifications thereof, which has a nucleotide sequence that is substantially identical to an miRNA and that is antisense with respect to its target. This term includes oligonucleotides composed of naturally-occurring nucleobases, sugars and covalent internucleoside (backbone) linkages and which contain at least one non-naturally-occurring portion which functions similarly. Such modified or substituted oligonucleotides are preferred over native forms because of desirable properties such as, for example, enhanced cellular uptake, enhanced affinity for nucleic acid target and increased stability in the presence of nucleases. In a preferred embodiment, the supermir does not include a sense strand, and in another preferred embodiment, the supermir does not self-hybridize to a significant extent. A supermir can have secondary structure, but it is substantially single-stranded under physiological conditions. An supermir that is substantially single-stranded is single-stranded to the extent that less than about 50% (e.g., less than about 40%, 30%, 20%, 10%, or 5%) of the supermir is duplexed with itself. The supermir can include a hairpin segment, e.g., sequence, preferably at the 3' end can self hybridize and form a duplex region, e.g., a duplex region of at least 1, 2, 3, or 4 and preferably less than 8, 7, 6, or n nucleotides, e.g., 5 nucleotides. The duplexed region can be connected by a linker, e.g., a nucleotide linker, e.g., 3, 4, 5, or 6 dTs, e.g., modified dTs. In another embodiment the supermir is duplexed with a shorter oligo, e.g., of 5, 6, 7, 8, 9, or 10 nucleotides in length, e.g., at one or both of the 3' and 5' end or at one end and in the non-terminal or middle of the supermir.
- miRNA mimics represent a class of molecules that can be used to imitate the gene silencing ability of one or more miRNAs. Thus, the term "microRNA mimic" refers to synthetic non-coding RNAs (i.e. the miRNA is not obtained by purification from a source of the endogenous miRNA) that are capable of entering the RNAi pathway and regulating gene expression. miRNA mimics can be designed as mature molecules (e.g. single stranded) or mimic precursors (e.g., pri- or pre-miRNAs). miRNA mimics can be comprised of nucleic acid (modified or modified nucleic acids) including oligonucleotides comprising, without limitation, RNA, modified RNA, DNA, modified DNA, locked nucleic acids, or 2'-O,4'-C-ethylene-bridged nucleic acids (ENA), or any combination of the above (including DNA-RNA hybrids). In addition, miRNA mimics can comprise conjugates that can affect delivery, intracellular compartmentalization, stability, specificity, functionality, strand usage, and/or potency. In one design, miRNA mimics are double stranded molecules (e.g., with a duplex region of between about 16 and about 3 1 nucleotides in length) and contain one or more sequences that have identity with the mature strand of a given miRNA. Modifications can comprise 2' modifications (including 2'-O methyl modifications and 2' F modifications) on one or both strands of the molecule and internucleotide modifications (e.g. phosphorothioate modifications) that enhance nucleic acid stability and/or specificity. In addition, miRNA mimics can include overhangs. The overhangs can consist of 1-6 nucleotides on either the 3' or 5' end of either strand and can be modified to enhance stability or functionality. In one embodiment, a miRNA mimic comprises a duplex region of between 16 and 3 1 nucleotides and one or more of the following chemical modification patterns: the sense strand contains 2'-O-methyl modifications of
nucleotides 1 and 2 (counting from the 5' end of the sense oligonucleotide), and all of the Cs and Us; the antisense strand modifications can comprise 2' F modification of all of the Cs and Us, phosphorylation of the 5' end of the oligonucleotide, and stabilized internucleotide linkages associated with a 2 nucleotide 3' overhang. - The terms "antimir," "microRNA inhibitor," "miR inhibitor," or "inhibitor," are synonymous and refer to oligonucleotides or modified oligonucleotides that interfere with the ability of specific miRNAs. In general, the inhibitors are nucleic acid or modified nucleic acids in nature including oligonucleotides comprising RNA, modified RNA, DNA, modified DNA, locked nucleic acids (LNAs), or any combination of the above. Modifications include 2' modifications (including 2'-O alkyl modifications and 2' F modifications) and internucleotide modifications (e.g. phosphorothioate modifications) that can affect delivery, stability, specificity, intracellular compartmentalization, or potency. In addition, miRNA inhibitors can comprise conjugates that can affect delivery, intracellular compartmentalization, stability, and/or potency. Inhibitors can adopt a variety of configurations including single stranded, double stranded (RNA/RNA or RNA/DNA duplexes), and hairpin designs, in general, microRNA inhibitors comprise contain one or more sequences or portions of sequences that are complementary or partially complementary with the mature strand (or strands) of the miRNA to be targeted, in addition, the miRNA inhibitor may also comprise additional sequences located 5' and 3' to the sequence that is the reverse complement of the mature miRNA. The additional sequences may be the reverse complements of the sequences that are adjacent to the mature miRNA in the pri-miRNA from which the mature miRNA is derived, or the additional sequences may be arbitrary sequences (having a mixture of A, G, C, or U). In some embodiments, one or both of the additional sequences are arbitrary sequences capable of forming hairpins. Thus, in some embodiments, the sequence that is the reverse complement of the miRNA is flanked on the 5' side and on the 3' side by hairpin structures. Micro-RNA inhibitors, when double stranded, may include mismatches between nucleotides on opposite strands. Furthermore, micro-RNA inhibitors may be linked to conjugate moieties in order to facilitate uptake of the inhibitor into a cell. For example, a micro-RNA inhibitor may be linked to cholesteryl 5-(bis(4-methoxyphenyl)(phenyl)methoxy)-3 hydroxypentylcarbamate) which allows passive uptake of a micro-RNA inhibitor into a cell. Micro-RNA inhibitors, including hairpin miRNA inhibitors, are described in detail in Vermeulen et al., "Double-Stranded Regions Are Essential Design Components Of Potent Inhibitors of RISC Function," RNA 13: 723-730 (2007) and in
WO2007/095387 andWO 2008/036825 . A person of ordinary skill in the art can select a sequence from the database for a desired miRNA and design an inhibitor useful for the methods disclosed herein. - Ul adaptor inhibit polyA sites and are bifunctional oligonucleotides with a target domain complementary to a site in the target gene's terminal exon and a 'Ul domain' that binds to the Ul smaller nuclear RNA component of the Ul snRNP (Goraczniak, et al., 2008, Nature Biotechnology, 27(3), 257-263). Ul snRNP is a ribonucleoprotein complex that functions primarily to direct early steps in spliceosome formation by binding to the pre-mRNA exon- intron boundary (Brown and Simpson, 1998, Annu Rev Plant Physiol Plant Mol Biol 49:77-95). Nucleotides 2-11 of the 5'end of Ul snRNA base pair bind with the 5'ss of the pre mRNA. In one embodiment, oligonucleotides are Ul adaptors. In one embodiment, the Ul adaptor can be administered in combination with at least one other iRNA agent.
- In other different embodiments, the invention is directed to a method for administering a therapeutic agent to a patient in need thereof, the method comprising preparing or providing any of the foregoing LNPs and/or administering a composition comprising the same to the patient. In some embodiments, the therapeutic agent is effective to treat the disease.
- For the purposes of administration, the lipid nanoparticles of embodiments of the present invention may be administered alone or may be formulated as pharmaceutical compositions. Pharmaceutical compositions of certain embodiments comprise a lipid nanoparticle according to any of the foregoing embodiments and one or more pharmaceutically acceptable carrier, diluent or excipient. The lipid nanoparticle may be present in an amount which is effective to deliver the therapeutic agent, e.g., for treating a particular disease or condition of interest. Appropriate concentrations and dosages can be readily determined by one skilled in the art.
- Administration of the lipid nanoparticles of some embodiments can be carried out via any of the accepted modes of administration of agents for serving similar utilities. The pharmaceutical compositions of some embodiments may be formulated into preparations in solid, semi-solid, liquid or gaseous forms, such as tablets, capsules, powders, granules, ointments, solutions, suspensions, suppositories, injections, inhalants, gels, microspheres, and aerosols. Typical routes of administering such pharmaceutical compositions include, without limitation, oral, topical, transdermal, inhalation, parenteral, sublingual, buccal, rectal, vaginal, and intranasal. The term parenteral as used herein includes subcutaneous injections, intravenous, intramuscular, intradermal, intrasternal injection or infusion techniques. Pharmaceutical compositions of some embodiments are formulated so as to allow the active ingredients contained therein to be bioavailable upon administration of the composition to a patient. Compositions that may be administered to a subject or patient may take the form of one or more dosage units, where for example, a tablet may be a single dosage unit, and a container comprising LNPs in aerosol form may hold a plurality of dosage units. Actual methods of preparing such dosage forms are known, or will be apparent, to those skilled in this art; for example, see Remington: The Science and Practice of Pharmacy, 20th Edition (Philadelphia College of Pharmacy and Science, 2000). The composition to be administered will typically contain a therapeutically effective amount of a lipid nanoparticle of any of the embodiments disclosed herein, comprising a therapeutic agent, or a pharmaceutically acceptable salt thereof, for treatment of a disease or condition of interest.
- A pharmaceutical composition of some embodiments may be in the form of a solid or liquid. In one aspect, the carrier(s) are particulate, so that the compositions are, for example, in tablet or powder form. The carrier(s) may be liquid, with the compositions being, for example, an oral syrup, injectable liquid or an aerosol, which is useful in, for example, inhalatory administration.
- When intended for oral administration, the pharmaceutical composition is preferably in either solid or liquid form, where semi-solid, semi-liquid, suspension and gel forms are included within the forms considered herein as either solid or liquid.
- As a solid composition for oral administration, the pharmaceutical composition may be formulated into a powder, granule, compressed tablet, pill, capsule, chewing gum, wafer or the like form. Such a solid composition will typically contain one or more inert diluents or edible carriers. In addition, one or more of the following may be present: binders such as carboxymethylcellulose, ethyl cellulose, microcrystalline cellulose, gum tragacanth or gelatin; excipients such as starch, lactose or dextrins, disintegrating agents such as alginic acid, sodium alginate, Primogel, corn starch and the like; lubricants such as magnesium stearate or Sterotex; glidants such as colloidal silicon dioxide; sweetening agents such as sucrose or saccharin; a flavoring agent such as peppermint, methyl salicylate or orange flavoring; and a coloring agent.
- When the pharmaceutical composition is in the form of a capsule, for example, a gelatin capsule, it may contain, in addition to materials of the above type, a liquid carrier such as polyethylene glycol or oil.
- The pharmaceutical composition may be in the form of a liquid, for example, an elixir, syrup, solution, emulsion or suspension. The liquid may be for oral administration or for delivery by injection, as two examples. When intended for oral administration, preferred composition contain, in addition to the present compounds, one or more of a sweetening agent, preservatives, dye/colorant and flavor enhancer. In a composition intended to be administered by injection, one or more of a surfactant, preservative, wetting agent, dispersing agent, suspending agent, buffer, stabilizer and isotonic agent may be included.
- The liquid pharmaceutical compositions of some embodiments, whether they be solutions, suspensions or other like form, may include one or more of the following adjuvants: sterile diluents such as water for injection, saline solution, preferably physiological saline, Ringer's solution, isotonic sodium chloride, fixed oils such as synthetic mono or diglycerides which may serve as the solvent or suspending medium, polyethylene glycols, glycerin, propylene glycol or other solvents; antibacterial agents such as benzyl alcohol or methyl paraben; antioxidants such as ascorbic acid or sodium bisulfite; chelating agents such as ethylenediaminetetraacetic acid; buffers such as acetates, citrates or phosphates and agents for the adjustment of tonicity such as sodium chloride or dextrose; agents to act as cryoprotectants such as sucrose or trehalose. The parenteral preparation can be enclosed in ampoules, disposable syringes or multiple dose vials made of glass or plastic. Physiological saline is a preferred adjuvant. An injectable pharmaceutical composition is preferably sterile.
- A liquid pharmaceutical composition of certain embodiments intended for either parenteral or oral administration should contain an amount of a lipid nanoparticle of the invention such that a suitable dosage will be obtained.
- The pharmaceutical composition of embodiments of the invention may be intended for topical administration, in which case the carrier may suitably comprise a solution, emulsion, ointment or gel base. The base, for example, may comprise one or more of the following: petrolatum, lanolin, polyethylene glycols, bee wax, mineral oil, diluents such as water and alcohol, and emulsifiers and stabilizers. Thickening agents may be present in a pharmaceutical composition for topical administration. If intended for transdermal administration, the composition may include a transdermal patch or iontophoresis device.
- The pharmaceutical composition of some embodiments may be intended for rectal administration, in the form, for example, of a suppository, which will melt in the rectum and release the drug. The composition for rectal administration may contain an oleaginous base as a suitable nonirritating excipient. Such bases include, without limitation, lanolin, cocoa butter and polyethylene glycol.
- The pharmaceutical composition of other embodiments may include various materials, which modify the physical form of a solid or liquid dosage unit. For example, the composition may include materials that form a coating shell around the active ingredients. The materials that form the coating shell are typically inert, and may be selected from, for example, sugar, shellac, and other enteric coating agents. Alternatively, the active ingredients may be encased in a gelatin capsule.
- The pharmaceutical composition of embodiments in solid or liquid form may include an agent that binds to the LNP or therapeutic agent, and thereby assists in the delivery of the LNP or therapeutic agent. Suitable agents that may act in this capacity include a monoclonal or polyclonal antibody, or a protein.
- In other embodiments, the pharmaceutical composition may comprise or consist of dosage units that can be administered as an aerosol. The term aerosol is used to denote a variety of systems ranging from those of colloidal nature to systems consisting of pressurized packages. Delivery may be by a liquefied or compressed gas or by a suitable pump system that dispenses the active ingredients. Aerosols of compounds of the invention may be delivered in single phase, bi-phasic, or tri-phasic systems in order to deliver the active ingredient(s). Delivery of the aerosol includes the necessary container, activators, valves, subcontainers, and the like, which together may form a kit. One skilled in the art, without undue experimentation may determine preferred aerosols.
- In some embodiments, the pharmaceutical compositions may be prepared by methodology well known in the pharmaceutical art. For example, a pharmaceutical composition intended to be administered by injection can be prepared by combining the lipid nanoparticles of the invention with sterile, distilled water or other carrier so as to form a solution. A surfactant may be added to facilitate the formation of a homogeneous solution or suspension. Surfactants are compounds that non-covalently interact with the compound of the invention so as to facilitate dissolution or homogeneous suspension of the compound in the aqueous delivery system.
- The pharmaceutical compositions of some embodiments are administered in a therapeutically effective amount, which will vary depending upon a variety of factors including the activity of the specific therapeutic agent employed; the metabolic stability and length of action of the therapeutic agent; the age, body weight, general health, sex, and diet of the patient; the mode and time of administration; the rate of excretion; the drug combination; the severity of the particular disorder or condition; and the subject undergoing therapy.
- The pharmaceutical compositions of various embodiments may also be administered simultaneously with, prior to, or after administration of one or more other therapeutic agents. Such combination therapy includes administration of a single pharmaceutical dosage formulation of a composition of the invention and one or more additional active agents, as well as administration of the composition of the invention and each active agent in its own separate pharmaceutical dosage formulation. For example, a pharmaceutical composition of one embodiments and the other active agent can be administered to the patient together in a single oral dosage composition such as a tablet or capsule, or each agent administered in separate oral dosage formulations. Where separate dosage formulations are used, the compounds of the invention and one or more additional active agents can be administered at essentially the same time, i.e., concurrently, or at separately staggered times, i.e., sequentially; combination therapy is understood to include all these regimens.
- The following examples are provided for purpose of illustration and not limitation.
- Cationic lipids and polymer conjugated lipids (PEG-lipid) were prepared and tested according to the general procedures described in
PCT Pub. Nos. WO 2020/061426 ,WO 2015/199952 ,WO 2017/004143 ,WO 2017/075531 andWO 2017/117528 , or were prepared as described herein. LNPs were prepared according to the following exemplary procedure. - The indicated cationic lipid (e.g. III-45), DSPC, cholesterol and PEG-lipid were solubilized in ethanol at the indicated molar ratio e.g. 47.5:10:40.7:1.8. Lipid nanoparticles (LNP) were prepared at a total lipid to mRNA weight ratio of approximately 10:1 to 30:1. Briefly, the mRNA was diluted to 0.2 mg/mL in 10 to 50 mM citrate or acetate buffer,
pH 4 topH 6. Syringe pumps or piston pumps were used to mix the ethanolic lipid solution with the mRNA aqueous solution at a ratio of about 1:5 to 1:3 (vol/vol) with total flow rates above 15 ml/min, for example 20 ml/min to 40 ml/min or above 100 ml/min or about 500 ml/min or about 1 000 ml/min. The ethanol was then removed and the external buffer replaced with PBS by dialysis. Finally, the lipid nanoparticles were filtered through a 0.2 µm pore sterile filter. Lipid nanoparticle particle size was approximately 45-105 nm, 55-95 nm diameter, 50-65 nm, 65-80 nm and in some instances approximately 70-90 nm diameter as determined by quasi-elastic light scattering using a Malvern Zetasizer Nano ZS (Malvern, UK) (not according to the invention unless embraced by the claims). - Luciferase mRNA in vivo evaluation studies are performed in 6-8 week old female C57BL/6 mice (Charles River) 8-10 week old CD-1 (Harlan) mice (Charles River) according to guidelines established by an institutional animal care committee (ACC) and the Canadian Council on Animal Care (CCAC). Varying doses of mRNA-lipid nanoparticle are systemically administered by tail vein injection and animals euthanized at a specific time point (e.g., 4 hours) post-administration. Liver and spleen are collected in pre-weighed tubes, weights determined, immediately snap frozen in liquid nitrogen and stored at -80 °C until processing for analysis.
- For liver, approximately 50 mg is dissected for analyses in a 2 mL FastPrep tubes (MP Biomedicals, Solon OH). ¼" ceramic sphere (MP Biomedicals) is added to each tube and 500 µL of Glo Lysis Buffer - GLB (Promega, Madison WI) equilibrated to room temperature is added to liver tissue. Liver tissues are homogenized with the FastPrep24 instrument (MP Biomedicals) at 2 × 6.0 m/s for 15 seconds. Homogenate is incubated at room temperature for 5 minutes prior to a 1:4 dilution in GLB and assessed using SteadyGlo Luciferase assay system (Promega). Specifically, 50 µL of diluted tissue homogenate is reacted with 50 µL of SteadyGlo substrate, shaken for 10 seconds followed by 5 minute incubation and then quantitated using a CentroXS3 LB 960 luminometer (Berthold Technologies, Germany). The amount of protein assayed is determined by using the BCA protein assay kit (Pierce, Rockford IL). Relative luminescence units (RLU) are then normalized to total µg protein assayed. To convert RLU to ng luciferase a standard curve is generated with QuantiLum Recombinant Luciferase (Promega). For a representative formulation, a four-hour time point is chosen for an efficacy evaluation of the lipid formulation.
- The FLuc mRNA (e.g L-6107 or L-7202 from Trilink Biotechnologies) will express a luciferase protein, originally isolated from the firefly, photimus pyralis. FLuc is commonly used in mammalian cell culture to measure both gene expression and cell viability. It emits bioluminescence in the presence of the substrate, luciferin. This capped and polyadenylated mRNA is fully substituted with 5-methylcytidine and pseudouridine (L-6107) or 5-methoxyuridine (L-7202).
- Expression of the exogenous protein, luciferase in a murine small animal model was evaluated as a function of the acyl chain length of the PEG lipid component of the liquid nanoparticle (LNP) formulation.
- Briefly, mice were given a tail vein injection of lipid nanoparticle formulation comprising either 1.5% or 2.5% PEG polymer lipid and containing an mRNA expression vector for the luciferase enzyme. The molar ratios of the cationic lipid (compound I-6), DSPC, Cholesterol and pegylated lipid were 50:10:38.5:1.5 and 50:10:37.5:2.5, respectively. LNPs were formulated according to standard methods as described herein in Example 1, using PEG lipids with varied lengths of their acyl chains; namely di-C12, di-C13, di-C14, C12/14 (asymmetric tail combination) and di-C15. Animals were dosed at 0.3 or 0.5 mg/kg RNA and quantitation of luciferase expression in the liver was accomplished using standard methods known to those of ordinary skill in the art or as described herein in Example 2.
- The pegylated lipid used in the studies described herein was a compound having the following structure:
- The luciferase expression data for the various LNP formulations presented in
Figures 1 and 2 is shown as a ratio relative to the quantity observed for the LNP having a 1.5%, di-C14 PEG lipid formulation. Luciferase expression was highest for C14 length acyl chains and embodiments containing 2.5% PEG polymer lipid demonstrated reduced or equivalent expression of the enzyme. - A related murine study investigated additional LNP embodiments wherein PEG lipid quantities were varied from 0.5 to 5.0%. The luciferase expression data for the various LNP formulations presented in
Figures 3 and 4 is shown as a ratio relative to the quantity observed for the LNP having a 1.5%, di-C14 PEG lipid formulation. No significant benefit is observed for LNP embodiments wherein the PEG lipid quantity is greater than 1.5% and the trends in mice generally indicate improved performance for lower LNPs with lower pegylated lipid concentrations. - Experimentally naive male cynomolgus monkeys (Macaca fascicularis, macaque) were given control (saline) or test doses of LNP formulations via a 1-hour intravenous (IV) infusion in groups of three animals. The LNP formulation contained an expression vector for human immunoglobulin G, type 1 (IgG1). LNPs were synthesized according to standard methods known to those skilled in the art, or as described herein in Example 1, using cationic lipid III-45 and PEG lipids with varied lengths of their di-acyl chains; namely C12, C13, C14, C15 and C16 as described above. An additional LNP test group included a di-C14 formulation having smaller LNP diameter (~60nm). Non-control animals were dosed at 1.0 mg/kg RNA with a dose volume of 5 mL/kg. One control and seven test groups were used.
- Pharmacodynamic samples to evaluate plasma concentrations of IgG1 were obtained by blood draw (K3EDTA, 0.5 mL) prior to infusion; 3 and 9 hours post infusion and on
days Figure 5 shows IgG1 plasma concentration levels determined onDay 2, demonstrating that IgG1 was expressed greatest for LNP embodiments with PEG lipids containing di-acyl chains shorter than C14. - Plasma amino lipid levels were checked by blood draw (K3EDTA, 1 mL) at the end of infusion (EOI) and at
hours Figure 6 plots the plasma concentration of compound III-45 as a function of time for certain LNP embodiments. Results of this analysis, shown as a maximum average concentration (Cmax, ug/mL), are given in Table 12 (control group not shown).Table 12: Amino lipid concentrations in blood plasma No. Di-Acyl chain length Particle Diameter Cmax (ug/mL) 1 C12 77 187 ± 87 2 C13 68 152 ± 56 3 C14 71 300 ± 57 4 C15 77 532 ± 85 5 C16 79 541 ± 26 6 C14 (small) 61 230 ± 28 - In the present study, the lowest Cmax levels of amino lipids corresponded to LNPs having PEG lipids comprising shorter acyl chains (di-C12 and di-C 13). Without wishing to be bound by theory, applicants believe the specific lipophilic qualities imparted by the di-C12 and di-C13 acyl chains promote their distribution out of the LNP at a rate that enables delivery of the LNP to target tissues in a primate in a way that is not indicated by the analogous data in a murine model.
- Additionally, comparison of the di-C14 embodiments of
entries - Again, without wishing to be bound by theory, applicant believes the smaller LNP size for the 60 nm C14 formulation affords more rapid clearance from the blood and into hepatocytes, relative to the standard di-C14 preparation, promoting delivery of the LNP to target tissues and resulting in greater expression. Consequently, a synergistic increase in delivery of LNPs may be realized by combining short di-acyl chain PEG lipids with LNP sizes of approximately 60 nm.
- Liver amino lipid levels were checked by obtaining a liver sample via liver biopsy at 4, 12 and 24 hours post EOI. Higher relative levels of amino lipids in liver tissue is an indicator that the LNPs have accumulated in this tissue of interest.
Figure 7 plots the liver tissue concentration of compound III-45 as a function of time for certain LNP embodiments. Results of this analysis, shown as a maximum average concentration (Cmax, ng/g) are given in Table 13 (control group not shown).Table 13: Amino lipid concentrations in liver tissue No. Acyl chain length Particle Diameter Cmax (ug/mL) 1 C12 77 352 2 C13 68 300 3 C14 71 246 4 C15 77 260 5 C16 79 177 6 C14 (small) 61 370 - For LNPs comprising differences only from the length of the di-acyl chain (No 1-5 in Table 13), the highest Cmax levels of amino lipids observed in liver tissue corresponded to the embodiment having PEG lipids with a di-C12 acyl chain. Without wishing to be bound by theory, applicants believe the specific lipophilic qualities imparted by the shorter di-acyl chain promotes enhanced accumulation of the LNP in liver tissue of primates. This increased accumulation promotes increased relative expression of the encapsulated mRNA resulting in the higher IgG1 concentrations observed above (
Figure 5 ). - Additionally, comparison of the di-C14 embodiments of
entries - Further, plasma cytokine levels for the LNPs of Example 4 were determined as shown in
Figure 12 . Quantitation of cytokines in blood plasma was accomplished using standard methods known to those of ordinary skill in the art. Measurements were made pre-dose, EOI, and 6 and 24 hours post EOI. These data show lower peak induction (i.e. at 6 hours) of IL-6, MCP-1 and MIP-1a for the embodiment with a smaller diameter LNP (60 nm, entry 6) formulation compared to formulations 1-5 shown in Table 13 which have diameters of -70-80 nm. - Experimentally naive male cynomolgus monkeys (Macaca fascicularis, macaque) were given control (saline) or test doses of LNP formulations via a 1-hour intravenous (IV) infusion in groups of three animals. Liver biopsy samples were collected at 4 hours and 12 hours post end-of-infusion. The samples were flash frozen and stored until histological analysis could be performed. Additional details regarding experimental protocol for the NHP study are in Example 4.
- Samples of macaque liver were sliced thin for histological analysis and in situ hybridization analysis was performed according to standard methods known to those skilled in the art.
- RNA of the target sequence can be identified as darkened punctate spots within the hepatocytes and as broad regions of dark color within the sinusoidal space.
-
Figures 8 and 9 are provided to demonstrate the differential in distribution of LNP over time in the hepatocytes and sinusoidal space for different size LNP (60 nm vs. 70-80 nm) of the same composition. Both particles show significant distribution into hepatocytes at 4 hours as well as significant accumulation in the sinusoidal spaces. At 12 hours, both LNP show relatively little mRNA within hepatocytes, which is consistent with the timeframe for uptake, expression and natural degradation of the mRNA within the cells. However, the larger size LNP still shows relatively high signal in the sinusoidal spaces (Figure 9 ) whereas mRNA is relatively absent from the sinusoidal spaces for the small LNP at 12 hours (Figure 8 ). Without wishing to bound by theory, the higher expression of the smaller LNP is consistent with larger LNP being prevented from accessing hepatocytes to be productively expressed while smaller LNP cross the sinusoidal wall more readily for fast uptake into the hepatocytes. -
Figures 10 and 11 provide an expanded view of the 12 hour tissue sample, better demonstrating the difference in LNP density in the sinusoidal space. - Without wishing to be bound by theory, Applicant believes the smaller diameter (60nm) lipid nanoparticles allow for increased uptake into hepatocytes, thus resulting in decreased incidence of the LNP in the sinusoidal space at the 12 hour time point. An increase of LNP uptake into hepatocytes promotes concomitant increases in expression of the delivered payload.
- Experimentally naive male and female cynomolgus monkeys (Macaca fascicularis, macaque) are given control (saline) or test doses of LNP compositions via a 1-hour intravenous (IV) infusion in groups of three. The test LNP compositions are made up of five groups; four of these use a LNP formulation comprising a PEG lipid with a di-C12 acyl chain, with each group using a different proportion of said lipid (1.8%, 2.3%, 2.5% and 2.8% respectively). The fifth group uses a LNP formulation comprising a PEG lipid with a di-C13 acyl chain. All test LNP formulations contain an expression vector for human immunoglobulin G, type 1 (IgG1). LNPs were formulated according to standard methods as described herein in Example 1. Control subjects receive a 5 mL/kg saline injection. Non-control animals are nominally dosed at 1.0 mg/kg RNA with a dose volume of 5 mL/kg.
- Pharmacodynamic samples to evaluate plasma concentrations of IgG1 are obtained by blood draw (K3EDTA, 0.5 mL) prior to infusion; 6 hours post infusion and on
days - Plasma amino lipid levels are checked by blood draw (K3EDTA, 1 mL) at the end of infusion (EOI) and at
hours - Liver amino lipid levels are checked by obtaining a liver sample via liver biopsy at 4 hours post EOI. Higher relative levels of amino lipids in liver issue is an indicator that the LNPs have accumulated in this regions of interest.
- Experimentally naive male cynomolgus monkeys (Macaca fascicularis, macaque) were given control (saline) or test doses of LNP formulations via a 1-hour intravenous (IV) infusion in groups of four animals. The LNP formulation contained an mRNA expression vector for human immunoglobulin G, type 1 (IgG1). LNPs were synthesized according to standard methods known to those skilled in the art, or as described herein in Example 1, using cationic lipd III-45 and PEG lipid with C14 di-acyl chains as described above and size of 70 nm (LNP 8-1). Another LNP test group had the same composition but smaller LNP diameter of 52 nm (LNP 8-2). Non-control animals were dosed at 1.0 mg/kg RNA with a dose volume of 5 mL/kg.
- Pharmacodynamic samples to evaluate plasma concentrations of IgG1 were obtained by blood draw (K3EDTA, 0.5 mL) prior to infusion; 6 hours post infusion and on
days Figure 13 shows IgG1 plasma concentration levels demonstrating that IgG1 was expressed greatest for LNP embodiments with size ~50 nm (LNP8-2) than size ~70 nm (LNP 8-1). The same preparations were administered in a murine model as described in Example 1 and the results are provided inFigure 14 . These data demonstrate the smaller 50 nm LNP formulation (LNP 8-2) performs less well compared to the larger 70 nm formulation (LNP 8-1), which is in stark contrast the results in NHP. - Plasma cytokine levels were determined as shown in
Figure 15 . Quantitation of cytokines in blood plasma was accomplished using standard methods known to those of ordinary skill in the art. Measurements were made pre-dose, EOI, and 6 and 24 hours post EOI. These data show lower peak induction (i.e. at 6 hours) of IL-6 and MCP-1 at 6 hours post EOI for the embodiment with a smaller 50 nm diameter LNP formulation (LNP8-2) compared to the larger 70 nm formulation (LNP 8-1). - The distribution of the LNP to hepatocytes was characterized by In situ hybridization as described in Example 5.
Figures 16A and 16B are provided to demonstrate the differential in distribution at 4 hours post administration of LNP in the hepatocytes and sinusoidal space for different size LNP (~50 nm vs. ~70 nm) of the same composition. The smaller ~50 nm LNP show greater distribution into hepatocytes at 4 hours as well as less accumulation in the sinusoidal spaces than the larger ~70 nm LNP. Without wishing to bound by theory, the higher expression of the smaller LNP is consistent with larger LNP being prevented from accessing hepatocytes to be productively expressed while smaller LNP cross the sinusoidal wall more readily for fast uptake into the hepatocytes. - Experimentally naive male cynomolgus monkeys (Macaca fascicularis, macaque) were given control (saline) or test doses of LNP formulations via a 1-hour intravenous (IV) infusion in groups of four animals. The LNP formulation contained an mRNA expression vector for human immunoglobulin G, type 1 (IgG1). LNPs were synthesized according to standard methods known to those skilled in the art, or as described herein in Example 1, using cationic lipd III-45 and PEG lipid with C14 di-acyl chains as described above and size of 70 nm (LNP 9-1). Another LNP test group had the same composition but smaller LNP diameter of 54 nm (LNP 9-2). Non-control animals were dosed at 0.5 mg/kg or 2.0 mg/kg RNA with a dose volume of 5 mL/kg.
- Pharmacodynamic samples to evaluate plasma concentrations of IgG1 were obtained by blood draw (K3EDTA, 0.5 mL) prior to infusion; 6 hours post infusion and on
days Figure 17 shows IgG1 plasma concentration levels demonstrating that IgG1 was expressed greatest for LNP embodiments with size ~54 nm (LNP 9-2) than size ~70 nm (LNP 9-1) in the NHP. The same preparations were administered in a murine model as described in Example 1 and the results are provided inFigure 18 . These data demonstrate the smaller 54 nm LNP formulation (LNP 9-2) performs less well compared to the larger 70 nm formulation (LNP 9-1), which is in stark contrast the results in NHP. - Experimentally naive male cynomolgus monkeys (Macaca fascicularis, macaque) were given control (saline) or test doses of LNP formulations via a 1-hour intravenous (IV) infusion in groups of three animals. The LNP formulation contained an mRNA expression vector for human immunoglobulin G, type 1 (IgG1). LNPs were synthesized according to standard methods known to those skilled in the art, or as described herein in Example 1, using cationic lipd II-15 and PEG lipid with C14 di-acyl chains as described above and size of 67nm (LNP 10-1). Another LNP test group had the same composition but smaller LNP diameter of 59 nm (LNP 10-2). Non-control animals were dosed at 3.0 mg/kg RNA with a dose volume of 5 mL/kg.
- Pharmacodynamic samples to evaluate plasma concentrations of IgG1 were obtained by blood draw (K3EDTA, 0.5 mL) prior to infusion; 6 hours post infusion and on
days Figure 19 shows IgG1 plasma concentration levels demonstrating that IgG1 was expressed greatest for LNP embodiments with size ~59 nm (LNP 10-2) than size ~67 nm (LNP 10-1).
Claims (15)
- Lipid nanoparticles (LNPs) for use in a method of treating or preventing a disease in a primate in need thereof, wherein the method comprises administering the LNPs to the primate, each of the LNPs comprising:i) a nucleic acid, or a pharmaceutically acceptable salt thereof, encapsulated within the LNP;ii) a cationic lipid;iii) a neutral lipid;iv) a steroid; andv) a polymer-conjugated lipid,wherein a mean particle diameter of the LNPs ranges from 40 nm to 68 nm as determined by quasi-elastic light scattering.
- The LNPs for use according to claim 1, wherein the mean particle diameter ranges from 50 nm to 68 nm, from 55 nm to 65 nm, from 50 nm to 60 nm or from 60 nm to 68 nm.
- The LNPs for use according to claim 1, wherein the mean particle diameter is about 47 nm, about 48 nm, about 49 nm, about 50 nm, about 51 nm, about 52 nm, about 53 nm, about 54 nm, about 55 nm, about 56 nm, about 57 nm, about 58 nm, about 59 nm, about 60 nm, about 61 nm, about 62 nm, about 63 nm, about 64 nm or about 65 nm.
- The LNPs for use according to any one of claims 1-3, wherein:i) the cationic lipid has a structure of Formula (I):
- The LNPs for use according to any one of claims 1-5, wherein the molar ratio of cationic lipid to neutral lipid ranges from about 2:1 to about 8:1.
- The LNPs for use according to any one of claims 1-6, wherein the neutral lipid is distearoylphosphatidylcholine (DSPC), dioleoylphosphatidylcholine (DOPC), dipalmitoylphosphatidylcholine (DPPC), dioleoylphosphatidylglycerol (DOPG), dipalmitoylphosphatidylglycerol (DPPG), dioleoyl-phosphatidylethanolamine (DOPE), palmitoyloleoylphosphatidylcholine (POPC), palmitoyloleoyl-phosphatidylethanolamine (POPE) and dioleoyl-phosphatidylethanolamine 4-(N-maleimidomethyl)-cyclohexane-1 carboxylate (DOPE-mal), dipalmitoyl phosphatidyl ethanolamine (DPPE), dimyristoylphosphoethanolamine (DMPE), distearoyl-phosphatidylethanolamine (DSPE), 16-O-monomethyl PE, 16-O-dimethyl PE, 18-1-trans PE, 1-stearioyl-2-oleoylphosphatidyethanol amine (SOPE) or 1,2-dielaidoyl-sn-glycero-3-phophoethanolamine (transDOPE).
- The LNPs for use according to any one of claims 1-6, wherein the neutral lipid is DSPC, DPPC, DMPC, DOPC, POPC, DOPE or SM.
- The LNPs for use according to any one of claims 1-6, wherein the neutral lipid is DSPC.
- The LNPs for use according to any one of claims 1-9, wherein the steroid is cholesterol.
- The LNPs for use according to any one of claims 1-10, wherein the molar ratio of cationic lipid to steroid ranges from 5:1 to 1:1.
- The LNPs for use according to any one of claims 1-11, wherein the molar ratio of cationic lipid to polymer conjugated lipid ranges from about 100:1 to about 20:1.
- The LNPs for use according to any one of claims 1-12, wherein the nucleic acid is selected from antisense and messenger RNA.
- The LNPs for use according to any one of claims 1-13, wherein the nucleic acid comprises an mRNA capable of translating an immunogenic protein.
- The LNPs for use according to any one of claims 1-14, wherein the administering comprises intraveneously administering.
Priority Applications (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| SI202030493T SI4013385T1 (en) | 2019-08-14 | 2020-08-14 | Improved lipid nanoparticles for delivery of nucleic acids |
| EP24185708.5A EP4454640A2 (en) | 2019-08-14 | 2020-08-14 | Improved lipid nanoparticles for delivery of nucleic acids |
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US201962886894P | 2019-08-14 | 2019-08-14 | |
| PCT/US2020/046407 WO2021030701A1 (en) | 2019-08-14 | 2020-08-14 | Improved lipid nanoparticles for delivery of nucleic acids |
Related Child Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| EP24185708.5A Division EP4454640A2 (en) | 2019-08-14 | 2020-08-14 | Improved lipid nanoparticles for delivery of nucleic acids |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| EP4013385A1 EP4013385A1 (en) | 2022-06-22 |
| EP4013385B1 true EP4013385B1 (en) | 2024-07-03 |
Family
ID=72322523
Family Applications (2)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| EP20765121.7A Active EP4013385B1 (en) | 2019-08-14 | 2020-08-14 | Improved lipid nanoparticles for delivery of nucleic acids |
| EP24185708.5A Pending EP4454640A2 (en) | 2019-08-14 | 2020-08-14 | Improved lipid nanoparticles for delivery of nucleic acids |
Family Applications After (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| EP24185708.5A Pending EP4454640A2 (en) | 2019-08-14 | 2020-08-14 | Improved lipid nanoparticles for delivery of nucleic acids |
Country Status (26)
| Country | Link |
|---|---|
| US (1) | US20230097090A1 (en) |
| EP (2) | EP4013385B1 (en) |
| JP (1) | JP2022544652A (en) |
| KR (1) | KR20220053599A (en) |
| CN (1) | CN114901253A (en) |
| AU (1) | AU2020328596A1 (en) |
| BR (1) | BR112022002708A2 (en) |
| CA (1) | CA3150458A1 (en) |
| CL (1) | CL2022000351A1 (en) |
| CO (1) | CO2022002685A2 (en) |
| CR (1) | CR20220108A (en) |
| DE (1) | DE112020003843T5 (en) |
| DK (1) | DK4013385T3 (en) |
| DO (1) | DOP2022000038A (en) |
| EC (1) | ECSP22018209A (en) |
| ES (1) | ES2918001A2 (en) |
| FI (1) | FI4013385T3 (en) |
| GB (1) | GB2600859B (en) |
| IL (1) | IL290477A (en) |
| JO (1) | JOP20220037A1 (en) |
| LT (1) | LT4013385T (en) |
| MX (1) | MX2022001720A (en) |
| PE (1) | PE20220968A1 (en) |
| PT (1) | PT4013385T (en) |
| SI (1) | SI4013385T1 (en) |
| WO (1) | WO2021030701A1 (en) |
Families Citing this family (46)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| FI3313829T3 (en) | 2015-06-29 | 2024-07-01 | Acuitas Therapeutics Inc | Lipids and lipid nanoparticle formulations for delivery of nucleic acids |
| US11357856B2 (en) | 2017-04-13 | 2022-06-14 | Acuitas Therapeutics, Inc. | Lipids for delivery of active agents |
| JP7355731B2 (en) | 2017-08-16 | 2023-10-03 | アクイタス セラピューティクス インコーポレイテッド | Lipids for use in lipid nanoparticle formulations |
| WO2019036030A1 (en) | 2017-08-17 | 2019-02-21 | Acuitas Therapeutics, Inc. | Lipids for use in lipid nanoparticle formulations |
| AU2019359299B2 (en) | 2018-10-09 | 2022-04-21 | The University Of British Columbia | Compositions and systems comprising transfection-competent vesicles free of organic-solvents and detergents and methods related thereto |
| US20240277830A1 (en) | 2020-02-04 | 2024-08-22 | CureVac SE | Coronavirus vaccine |
| JP2021185136A (en) | 2020-04-22 | 2021-12-09 | ビオエンテッヒ・アールエヌエイ・ファーマシューティカルズ・ゲゼルシャフト・ミット・ベシュレンクテル・ハフツング | Coronavirus vaccine |
| BR112023000327A2 (en) | 2020-07-16 | 2023-01-31 | Acuitas Therapeutics Inc | CATION LIPIDS FOR USE IN LIPID NANOPARTICLES |
| WO2022023284A1 (en) | 2020-07-27 | 2022-02-03 | Anjarium Biosciences Ag | Compositions of dna molecules, methods of making therefor, and methods of use thereof |
| WO2022115645A1 (en) | 2020-11-25 | 2022-06-02 | Akagera Medicines, Inc. | Lipid nanoparticles for delivery of nucleic acids, and related methods of use |
| KR20230164648A (en) | 2020-12-22 | 2023-12-04 | 큐어백 에스이 | RNA vaccines against SARS-CoV-2 variants |
| CN113402404B (en) * | 2021-04-30 | 2022-03-11 | 苏州科锐迈德生物医药科技有限公司 | Lipid compound, lipid carrier containing same, nucleic acid lipid nanoparticle composition and pharmaceutical preparation |
| CN115385820A (en) * | 2021-05-09 | 2022-11-25 | 英维沃生物科技(苏州)有限公司 | Cationic lipids and uses thereof |
| WO2022247801A1 (en) * | 2021-05-28 | 2022-12-01 | 北京启辰生生物科技有限公司 | Lipid compound and use thereof in delivery of nucleic acid |
| CN113403313B (en) * | 2021-06-23 | 2022-08-19 | 北京理工大学 | sgRNA, plasmid and nano-composite for specifically recognizing human PLK1 locus and application |
| WO2023273364A1 (en) * | 2021-06-30 | 2023-01-05 | 天津键凯科技有限公司 | Polyethylene glycol lipid and use thereof |
| CN115403761A (en) * | 2021-07-23 | 2022-11-29 | 天津键凯科技有限公司 | Polyglycol modified lipid compound and preparation method and application thereof |
| CN115710191A (en) * | 2021-08-23 | 2023-02-24 | 广州谷森制药有限公司 | Novel cationic lipid compounds |
| WO2023031394A1 (en) | 2021-09-03 | 2023-03-09 | CureVac SE | Novel lipid nanoparticles for delivery of nucleic acids |
| CN115869262A (en) * | 2021-09-27 | 2023-03-31 | 广州谷森制药有限公司 | Novel PEG lipid compound, preparation method, composition and application thereof |
| AR127312A1 (en) * | 2021-10-08 | 2024-01-10 | Suzhou Abogen Biosciences Co Ltd | LIPID COMPOUNDS AND LIPID NANOPARTICLE COMPOSITIONS |
| US20240335384A1 (en) * | 2021-10-15 | 2024-10-10 | Xiamen Sinopeg Biotech Co., Ltd. | Nitrogen-containing cationic lipid and application thereof |
| WO2023073228A1 (en) | 2021-10-29 | 2023-05-04 | CureVac SE | Improved circular rna for expressing therapeutic proteins |
| KR20240123832A (en) | 2021-12-16 | 2024-08-14 | 아퀴타스 테라퓨틱스 인크. | Lipids for use in lipid nanoparticle formulations |
| CN114044741B (en) * | 2022-01-13 | 2022-04-15 | 北京悦康科创医药科技股份有限公司 | Cationic lipid compound, composition containing same and application |
| WO2023144330A1 (en) | 2022-01-28 | 2023-08-03 | CureVac SE | Nucleic acid encoded transcription factor inhibitors |
| CN116969851A (en) * | 2022-04-29 | 2023-10-31 | 北京剂泰医药科技有限公司 | Ionizable lipid compounds |
| WO2023227608A1 (en) | 2022-05-25 | 2023-11-30 | Glaxosmithkline Biologicals Sa | Nucleic acid based vaccine encoding an escherichia coli fimh antigenic polypeptide |
| AU2023275780A1 (en) | 2022-05-25 | 2024-12-05 | Akagera Medicines, Inc. | Lipid nanoparticles for delivery of nucleic acids and methods of use thereof |
| CN117263818A (en) * | 2022-06-14 | 2023-12-22 | 杭州高田生物医药有限公司 | Cationic lipid compound, and preparation method and application thereof |
| CN114989182B (en) * | 2022-06-23 | 2023-06-23 | 尧唐(上海)生物科技有限公司 | Lipid compound, composition containing lipid compound and application of lipid compound |
| US11878055B1 (en) | 2022-06-26 | 2024-01-23 | BioNTech SE | Coronavirus vaccine |
| CN117417264A (en) * | 2022-07-19 | 2024-01-19 | 深圳深信生物科技有限公司 | Amino lipid compound, preparation method and application thereof |
| WO2024022263A1 (en) * | 2022-07-25 | 2024-02-01 | 苏州艾博生物科技有限公司 | Lipid compound and lipid nanoparticle composition |
| WO2024031051A1 (en) * | 2022-08-05 | 2024-02-08 | Life Technologies Corporation | Lipids for nucleic acid delivery |
| US20240270679A1 (en) * | 2022-09-07 | 2024-08-15 | Acuitas Therapeutics, Inc. | Lipids for use in lipid nanoparticle formulations |
| CN118265692A (en) * | 2022-09-09 | 2024-06-28 | 厦门赛诺邦格生物科技股份有限公司 | Carbon nucleus cationic lipid |
| WO2024089638A1 (en) | 2022-10-28 | 2024-05-02 | Glaxosmithkline Biologicals Sa | Nucleic acid based vaccine |
| CN115417779B (en) * | 2022-11-03 | 2023-06-16 | 北京华芢生物技术有限公司 | Ionizable cationic lipid C6-A1 and nano liposome particles composed of same |
| CN115417780B (en) * | 2022-11-04 | 2023-06-20 | 北京华芢生物技术有限公司 | Ionizable cationic lipid C5-A2 and nano liposome particles composed of same |
| WO2024119098A1 (en) * | 2022-12-02 | 2024-06-06 | Prime Medicine, Inc. | Lipid nanoparticle (lnp) delivery systems and formulations |
| WO2024131810A1 (en) * | 2022-12-21 | 2024-06-27 | Suzhou Abogen Biosciences Co., Ltd. | Lipid nanoparticles comprising sterol-modified phospholipids |
| WO2024184500A1 (en) | 2023-03-08 | 2024-09-12 | CureVac SE | Novel lipid nanoparticle formulations for delivery of nucleic acids |
| CN118666726A (en) * | 2023-03-17 | 2024-09-20 | 尧唐(上海)生物科技有限公司 | Lipid compounds for delivering therapeutic agents, methods of making and uses thereof |
| CN116891423B (en) * | 2023-07-07 | 2024-03-01 | 荣灿生物医药技术(上海)有限公司 | Lipid compound, composition, preparation method and application thereof |
| CN118319880A (en) * | 2024-04-15 | 2024-07-12 | 荣灿生物医药技术(上海)有限公司 | Lipid nanoparticle for targeted lung efficient delivery of nucleic acid, inhalation formulation and application |
Family Cites Families (82)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US5453566A (en) | 1986-03-28 | 1995-09-26 | Calgene, Inc. | Antisense regulation of gene expression in plant/cells |
| US4987071A (en) | 1986-12-03 | 1991-01-22 | University Patents, Inc. | RNA ribozyme polymerases, dephosphorylases, restriction endoribonucleases and methods |
| CA1340323C (en) | 1988-09-20 | 1999-01-19 | Arnold E. Hampel | Rna catalyst for cleaving specific rna sequences |
| GB8822492D0 (en) | 1988-09-24 | 1988-10-26 | Considine J | Apparatus for removing tumours from hollow organs of body |
| CA2039718C (en) | 1989-08-31 | 2003-02-25 | John J. Rossi | Chimeric dna-rna catalytic sequences |
| US6365730B1 (en) | 1990-06-19 | 2002-04-02 | Gene Shears Pty. Limited | DNA-Armed ribozymes and minizymes |
| US5789573A (en) | 1990-08-14 | 1998-08-04 | Isis Pharmaceuticals, Inc. | Antisense inhibition of ICAM-1, E-selectin, and CMV IE1/IE2 |
| WO1992007065A1 (en) | 1990-10-12 | 1992-04-30 | MAX-PLANCK-Gesellschaft zur Förderung der Wissenschaften e.V. | Modified ribozymes |
| DE4216134A1 (en) | 1991-06-20 | 1992-12-24 | Europ Lab Molekularbiolog | SYNTHETIC CATALYTIC OLIGONUCLEOTIDE STRUCTURES |
| US5652094A (en) | 1992-01-31 | 1997-07-29 | University Of Montreal | Nucleozymes |
| WO1993023569A1 (en) | 1992-05-11 | 1993-11-25 | Ribozyme Pharmaceuticals, Inc. | Method and reagent for inhibiting viral replication |
| EP0786522A2 (en) | 1992-07-17 | 1997-07-30 | Ribozyme Pharmaceuticals, Inc. | Enzymatic RNA molecules for treatment of stenotic conditions |
| IL108367A0 (en) | 1993-01-27 | 1994-04-12 | Hektoen Inst For Medical Resea | Antisense polynzcleotide inhibition of human growth factor-sensitive cancer cells |
| US5801154A (en) | 1993-10-18 | 1998-09-01 | Isis Pharmaceuticals, Inc. | Antisense oligonucleotide modulation of multidrug resistance-associated protein |
| US5591317A (en) | 1994-02-16 | 1997-01-07 | Pitts, Jr.; M. Michael | Electrostatic device for water treatment |
| US5631359A (en) | 1994-10-11 | 1997-05-20 | Ribozyme Pharmaceuticals, Inc. | Hairpin ribozymes |
| US6197553B1 (en) | 1994-07-15 | 2001-03-06 | Merck & Co., Inc. | Method for large scale plasmid purification |
| US5783683A (en) | 1995-01-10 | 1998-07-21 | Genta Inc. | Antisense oligonucleotides which reduce expression of the FGFRI gene |
| US5795587A (en) | 1995-01-23 | 1998-08-18 | University Of Pittsburgh | Stable lipid-comprising drug delivery complexes and methods for their production |
| US5747470A (en) | 1995-06-07 | 1998-05-05 | Gen-Probe Incorporated | Method for inhibiting cellular proliferation using antisense oligonucleotides to gp130 mRNA |
| US5739119A (en) | 1996-11-15 | 1998-04-14 | Galli; Rachel L. | Antisense oligonucleotides specific for the muscarinic type 2 acetylcholine receptor MRNA |
| US5965542A (en) | 1997-03-18 | 1999-10-12 | Inex Pharmaceuticals Corp. | Use of temperature to control the size of cationic liposome/plasmid DNA complexes |
| US6441111B1 (en) | 1997-08-15 | 2002-08-27 | Chisso Corporation | Polydisperse propylene polymer and process for producing the same |
| AU749881B2 (en) | 1998-02-03 | 2002-07-04 | Inex Pharmaceuticals Corporation | Systemic delivery of serum stable plasmid lipid particles for cancer therapy |
| US6410328B1 (en) | 1998-02-03 | 2002-06-25 | Protiva Biotherapeutics Inc. | Sensitizing cells to compounds using lipid-mediated gene and compound delivery |
| US6211140B1 (en) | 1999-07-26 | 2001-04-03 | The Procter & Gamble Company | Cationic charge boosting systems |
| AU2002319668A1 (en) | 2001-07-27 | 2003-02-17 | President And Fellows Of Harvard College | Laminar mixing apparatus and methods |
| US20050222064A1 (en) | 2002-02-20 | 2005-10-06 | Sirna Therapeutics, Inc. | Polycationic compositions for cellular delivery of polynucleotides |
| WO2004002453A1 (en) | 2002-06-28 | 2004-01-08 | Protiva Biotherapeutics Ltd. | Method and apparatus for producing liposomes |
| CA2518475C (en) | 2003-03-07 | 2014-12-23 | Alnylam Pharmaceuticals, Inc. | Irna agents comprising asymmetrical modifications |
| EP2567693B1 (en) | 2003-07-16 | 2015-10-21 | Protiva Biotherapeutics Inc. | Lipid encapsulated interfering RNA |
| US6927663B2 (en) | 2003-07-23 | 2005-08-09 | Cardiac Pacemakers, Inc. | Flyback transformer wire attach method to printed circuit board |
| KR101164256B1 (en) | 2003-09-15 | 2012-07-10 | 프로티바 바이오쎄라퓨틱스, 인코포레이티드 | Polyethyleneglycol-modified lipid compounds and uses thereof |
| US7745651B2 (en) | 2004-06-07 | 2010-06-29 | Protiva Biotherapeutics, Inc. | Cationic lipids and methods of use |
| WO2005121348A1 (en) | 2004-06-07 | 2005-12-22 | Protiva Biotherapeutics, Inc. | Lipid encapsulated interfering rna |
| WO2007012191A1 (en) | 2005-07-27 | 2007-02-01 | Protiva Biotherapeutics, Inc. | Systems and methods for manufacturing liposomes |
| US20070213292A1 (en) | 2005-08-10 | 2007-09-13 | The Rockefeller University | Chemically modified oligonucleotides for use in modulating micro RNA and uses thereof |
| US20070123482A1 (en) | 2005-08-10 | 2007-05-31 | Markus Stoffel | Chemically modified oligonucleotides for use in modulating micro RNA and uses thereof |
| US20100184209A1 (en) | 2006-02-17 | 2010-07-22 | Dharmacon, Inc. | Compositions and methods for inhibiting gene silencing by rna interference |
| JP5352462B2 (en) | 2006-09-22 | 2013-11-27 | ダーマコン, インコーポレイテッド | Double-stranded oligonucleotide complex, gene silencing method by RNA interference, and pharmaceutical composition |
| US20100015218A1 (en) | 2007-02-16 | 2010-01-21 | Vasant Jadhav | Compositions and methods for potentiated activity of biologically active molecules |
| CA2709875C (en) | 2008-01-02 | 2019-07-16 | Tekmira Pharmaceuticals Corporation | Improved compositions and methods for the delivery of nucleic acids |
| ES2638448T3 (en) | 2008-04-15 | 2017-10-20 | Protiva Biotherapeutics Inc. | Novel lipid formulations for nucleic acid administration |
| DK2355851T3 (en) | 2008-11-10 | 2018-06-25 | Arbutus Biopharma Corp | Newly known lipids and compositions for release of therapeutic agents |
| KR101987962B1 (en) | 2009-06-10 | 2019-06-11 | 알닐람 파마슈티칼스 인코포레이티드 | Improved lipid formulation |
| US8283333B2 (en) | 2009-07-01 | 2012-10-09 | Protiva Biotherapeutics, Inc. | Lipid formulations for nucleic acid delivery |
| US8569256B2 (en) | 2009-07-01 | 2013-10-29 | Protiva Biotherapeutics, Inc. | Cationic lipids and methods for the delivery of therapeutic agents |
| CA2816925C (en) | 2009-11-04 | 2023-01-10 | The University Of British Columbia | Nucleic acid-containing lipid particles and related methods |
| WO2011066651A1 (en) | 2009-12-01 | 2011-06-09 | Protiva Biotherapeutics, Inc. | Snalp formulations containing antioxidants |
| EP2509636B1 (en) | 2009-12-07 | 2017-07-19 | Arbutus Biopharma Corporation | Compositions for nucleic acid delivery |
| US20130017223A1 (en) | 2009-12-18 | 2013-01-17 | The University Of British Columbia | Methods and compositions for delivery of nucleic acids |
| EP3391877A1 (en) | 2010-04-08 | 2018-10-24 | The Trustees of Princeton University | Preparation of lipid nanoparticles |
| US20130156845A1 (en) | 2010-04-29 | 2013-06-20 | Alnylam Pharmaceuticals, Inc. | Lipid formulated single stranded rna |
| WO2011141705A1 (en) | 2010-05-12 | 2011-11-17 | Protiva Biotherapeutics, Inc. | Novel cationic lipids and methods of use thereof |
| WO2011149733A2 (en) | 2010-05-24 | 2011-12-01 | Merck Sharp & Dohme Corp. | Novel amino alcohol cationic lipids for oligonucleotide delivery |
| JP5893611B2 (en) | 2010-06-03 | 2016-03-23 | アルニラム・ファーマシューティカルズ・インコーポレーテッド | Biodegradable lipids for delivery of active agents |
| WO2012016184A2 (en) | 2010-07-30 | 2012-02-02 | Alnylam Pharmaceuticals, Inc. | Methods and compositions for delivery of active agents |
| KR101761388B1 (en) | 2010-07-30 | 2017-07-25 | 큐어백 아게 | Complexation of nucleic acids with disulfide-crosslinked cationic components for transfection and immunostimulation |
| CA2808901A1 (en) | 2010-08-20 | 2012-02-23 | Cerulean Pharma Inc. | Conjugates, particles, compositions, and related methods |
| US8466122B2 (en) | 2010-09-17 | 2013-06-18 | Protiva Biotherapeutics, Inc. | Trialkyl cationic lipids and methods of use thereof |
| US9999673B2 (en) | 2011-01-11 | 2018-06-19 | Alnylam Pharmaceuticals, Inc. | PEGylated lipids and their use for drug delivery |
| US8710200B2 (en) | 2011-03-31 | 2014-04-29 | Moderna Therapeutics, Inc. | Engineered nucleic acids encoding a modified erythropoietin and their expression |
| US8691750B2 (en) | 2011-05-17 | 2014-04-08 | Axolabs Gmbh | Lipids and compositions for intracellular delivery of biologically active compounds |
| WO2013016058A1 (en) | 2011-07-22 | 2013-01-31 | Merck Sharp & Dohme Corp. | Novel bis-nitrogen containing cationic lipids for oligonucleotide delivery |
| EP2760477B1 (en) * | 2011-09-27 | 2018-08-08 | Alnylam Pharmaceuticals, Inc. | Di-aliphatic substituted pegylated lipids |
| US8762704B2 (en) | 2011-09-29 | 2014-06-24 | Apple Inc. | Customized content for electronic devices |
| AU2012330819B2 (en) | 2011-11-04 | 2017-08-31 | Nitto Denko Corporation | Single use system for sterilely producing lipid-nucleic acid particles |
| WO2013086322A1 (en) | 2011-12-07 | 2013-06-13 | Alnylam Pharmaceuticals, Inc. | Branched alkyl and cycloalkyl terminated biodegradable lipids for the delivery of active agents |
| EP3988537A1 (en) | 2011-12-07 | 2022-04-27 | Alnylam Pharmaceuticals, Inc. | Biodegradable lipids for the delivery of active agents |
| WO2013086373A1 (en) | 2011-12-07 | 2013-06-13 | Alnylam Pharmaceuticals, Inc. | Lipids for the delivery of active agents |
| KR20140102759A (en) | 2011-12-16 | 2014-08-22 | 모더나 세라퓨틱스, 인코포레이티드 | Modified nucleoside, nucleotide, and nucleic acid compositions |
| WO2014008334A1 (en) | 2012-07-06 | 2014-01-09 | Alnylam Pharmaceuticals, Inc. | Stable non-aggregating nucleic acid lipid particle formulations |
| EP3033325B1 (en) * | 2013-07-23 | 2019-12-04 | Arbutus Biopharma Corporation | Compositions and methods for delivering messenger rna |
| LT3766916T (en) | 2014-06-25 | 2023-01-10 | Acuitas Therapeutics Inc. | Novel lipids and lipid nanoparticle formulations for delivery of nucleic acids |
| FI3313829T3 (en) | 2015-06-29 | 2024-07-01 | Acuitas Therapeutics Inc | Lipids and lipid nanoparticle formulations for delivery of nucleic acids |
| CA3003055C (en) | 2015-10-28 | 2023-08-01 | Acuitas Therapeutics, Inc. | Lipids and lipid nanoparticle formulations for delivery of nucleic acids |
| WO2018081480A1 (en) * | 2016-10-26 | 2018-05-03 | Acuitas Therapeutics, Inc. | Lipid nanoparticle formulations |
| EP3397613A1 (en) | 2015-12-30 | 2018-11-07 | Acuitas Therapeutics Inc. | Lipids and lipid nanoparticle formulations for delivery of nucleic acids |
| CA3045122A1 (en) * | 2016-12-09 | 2018-06-14 | Sangamo Therapeutics, Inc. | Delivery of target specific nucleases |
| WO2019089828A1 (en) * | 2017-10-31 | 2019-05-09 | Acuitas Therapeutics, Inc. | Lamellar lipid nanoparticles |
| WO2020061367A1 (en) | 2018-09-19 | 2020-03-26 | Modernatx, Inc. | Compounds and compositions for intracellular delivery of therapeutic agents |
| WO2020061426A2 (en) | 2018-09-21 | 2020-03-26 | Acuitas Therapeutics, Inc. | Systems and methods for manufacturing lipid nanoparticles and liposomes |
-
2020
- 2020-08-14 CN CN202080072049.1A patent/CN114901253A/en active Pending
- 2020-08-14 LT LTEPPCT/US2020/046407T patent/LT4013385T/en unknown
- 2020-08-14 GB GB2201685.1A patent/GB2600859B/en active Active
- 2020-08-14 EP EP20765121.7A patent/EP4013385B1/en active Active
- 2020-08-14 DE DE112020003843.2T patent/DE112020003843T5/en active Pending
- 2020-08-14 EP EP24185708.5A patent/EP4454640A2/en active Pending
- 2020-08-14 CA CA3150458A patent/CA3150458A1/en active Pending
- 2020-08-14 PE PE2022000237A patent/PE20220968A1/en unknown
- 2020-08-14 FI FIEP20765121.7T patent/FI4013385T3/en active
- 2020-08-14 ES ES202290010A patent/ES2918001A2/en active Pending
- 2020-08-14 WO PCT/US2020/046407 patent/WO2021030701A1/en active Application Filing
- 2020-08-14 PT PT207651217T patent/PT4013385T/en unknown
- 2020-08-14 JP JP2022508754A patent/JP2022544652A/en active Pending
- 2020-08-14 US US17/634,516 patent/US20230097090A1/en active Pending
- 2020-08-14 CR CR20220108A patent/CR20220108A/en unknown
- 2020-08-14 SI SI202030493T patent/SI4013385T1/en unknown
- 2020-08-14 DK DK20765121.7T patent/DK4013385T3/en active
- 2020-08-14 KR KR1020227008228A patent/KR20220053599A/en unknown
- 2020-08-14 AU AU2020328596A patent/AU2020328596A1/en active Pending
- 2020-08-14 JO JOP/2022/0037A patent/JOP20220037A1/en unknown
- 2020-08-14 MX MX2022001720A patent/MX2022001720A/en unknown
- 2020-08-14 BR BR112022002708A patent/BR112022002708A2/en unknown
-
2022
- 2022-02-09 IL IL290477A patent/IL290477A/en unknown
- 2022-02-11 CL CL2022000351A patent/CL2022000351A1/en unknown
- 2022-02-14 DO DO2022000038A patent/DOP2022000038A/en unknown
- 2022-03-07 CO CONC2022/0002685A patent/CO2022002685A2/en unknown
- 2022-03-11 EC ECSENADI202218209A patent/ECSP22018209A/en unknown
Also Published As
| Publication number | Publication date |
|---|---|
| FI4013385T3 (en) | 2024-09-27 |
| WO2021030701A1 (en) | 2021-02-18 |
| KR20220053599A (en) | 2022-04-29 |
| DOP2022000038A (en) | 2023-01-31 |
| IL290477A (en) | 2022-04-01 |
| US20230097090A1 (en) | 2023-03-30 |
| ES2918001A2 (en) | 2022-07-13 |
| CA3150458A1 (en) | 2021-02-18 |
| EP4013385A1 (en) | 2022-06-22 |
| CO2022002685A2 (en) | 2022-04-19 |
| JP2022544652A (en) | 2022-10-20 |
| LT4013385T (en) | 2024-09-25 |
| GB2600859B (en) | 2024-04-03 |
| EP4454640A2 (en) | 2024-10-30 |
| PE20220968A1 (en) | 2022-06-10 |
| CR20220108A (en) | 2022-05-27 |
| CL2022000351A1 (en) | 2023-02-03 |
| MX2022001720A (en) | 2022-03-11 |
| SI4013385T1 (en) | 2024-10-30 |
| DK4013385T3 (en) | 2024-09-02 |
| GB2600859A (en) | 2022-05-11 |
| CN114901253A (en) | 2022-08-12 |
| BR112022002708A2 (en) | 2022-05-31 |
| AU2020328596A1 (en) | 2022-03-31 |
| DE112020003843T5 (en) | 2022-05-19 |
| JOP20220037A1 (en) | 2023-01-30 |
| PT4013385T (en) | 2024-10-01 |
| ECSP22018209A (en) | 2022-07-29 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| EP4013385B1 (en) | Improved lipid nanoparticles for delivery of nucleic acids | |
| JP7086870B2 (en) | Compositions and Methods for Delivering Messenger RNA | |
| EP2281041B1 (en) | Silencing of csn5 gene expression using interfering rna | |
| EP3201338B1 (en) | Compositions and methods for silencing hepatitis b virus gene expression | |
| JP2022009505A (en) | Compositions and Methods for Delivering Messenger RNA | |
| US9222086B2 (en) | Compositions and methods for silencing genes expressed in cancer | |
| JP5766188B2 (en) | Lipid formulations for delivering therapeutic agents to solid tumors | |
| JP2022530018A (en) | Lipid nanoparticles | |
| US20160115483A1 (en) | Silencing of polo-like kinase expression using interfering rna | |
| JP2014530602A (en) | Compositions and methods for silencing aldehyde dehydrogenase | |
| US20220273567A1 (en) | Systems and methods for manufacturing lipid nanoparticles and liposomes | |
| TR2022001658T2 (en) | Developed lipid nanoparticles for the delivery of nucleic acids. | |
| WO2023205628A1 (en) | Lipid nanoparticles, nucleic acids, and methods of use |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| STAA | Information on the status of an ep patent application or granted ep patent |
Free format text: STATUS: UNKNOWN |
|
| STAA | Information on the status of an ep patent application or granted ep patent |
Free format text: STATUS: THE INTERNATIONAL PUBLICATION HAS BEEN MADE |
|
| PUAI | Public reference made under article 153(3) epc to a published international application that has entered the european phase |
Free format text: ORIGINAL CODE: 0009012 |
|
| STAA | Information on the status of an ep patent application or granted ep patent |
Free format text: STATUS: REQUEST FOR EXAMINATION WAS MADE |
|
| 17P | Request for examination filed |
Effective date: 20220303 |
|
| AK | Designated contracting states |
Kind code of ref document: A1 Designated state(s): AL AT BE BG CH CY CZ DE DK EE ES FI FR GB GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL PT RO RS SE SI SK SM TR |
|
| STAA | Information on the status of an ep patent application or granted ep patent |
Free format text: STATUS: EXAMINATION IS IN PROGRESS |
|
| 17Q | First examination report despatched |
Effective date: 20230116 |
|
| REG | Reference to a national code |
Ref country code: HK Ref legal event code: DE Ref document number: 40076667 Country of ref document: HK |
|
| P01 | Opt-out of the competence of the unified patent court (upc) registered |
Effective date: 20230515 |
|
| GRAP | Despatch of communication of intention to grant a patent |
Free format text: ORIGINAL CODE: EPIDOSNIGR1 |
|
| STAA | Information on the status of an ep patent application or granted ep patent |
Free format text: STATUS: GRANT OF PATENT IS INTENDED |
|
| INTG | Intention to grant announced |
Effective date: 20240201 |
|
| GRAS | Grant fee paid |
Free format text: ORIGINAL CODE: EPIDOSNIGR3 |
|
| GRAA | (expected) grant |
Free format text: ORIGINAL CODE: 0009210 |
|
| STAA | Information on the status of an ep patent application or granted ep patent |
Free format text: STATUS: THE PATENT HAS BEEN GRANTED |
|
| AK | Designated contracting states |
Kind code of ref document: B1 Designated state(s): AL AT BE BG CH CY CZ DE DK EE ES FI FR GB GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL PT RO RS SE SI SK SM TR |
|
| REG | Reference to a national code |
Ref country code: CH Ref legal event code: EP |
|
| REG | Reference to a national code |
Ref country code: DE Ref legal event code: R096 Ref document number: 602020033395 Country of ref document: DE |
|
| REG | Reference to a national code |
Ref country code: DK Ref legal event code: T3 Effective date: 20240826 |
|
| REG | Reference to a national code |
Ref country code: NL Ref legal event code: FP |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: LU Payment date: 20240827 Year of fee payment: 5 |
|
| REG | Reference to a national code |
Ref country code: SE Ref legal event code: TRGR |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: NL Payment date: 20240826 Year of fee payment: 5 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: IS Payment date: 20240822 Year of fee payment: 5 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: BG Payment date: 20240822 Year of fee payment: 5 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: DE Payment date: 20240828 Year of fee payment: 5 Ref country code: LT Payment date: 20240820 Year of fee payment: 5 Ref country code: FI Payment date: 20240826 Year of fee payment: 5 Ref country code: IE Payment date: 20240827 Year of fee payment: 5 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: GR Payment date: 20240904 Year of fee payment: 5 Ref country code: DK Payment date: 20240826 Year of fee payment: 5 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: GB Payment date: 20240827 Year of fee payment: 5 Ref country code: PT Payment date: 20240929 Year of fee payment: 5 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: BE Payment date: 20240827 Year of fee payment: 5 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: FR Payment date: 20240826 Year of fee payment: 5 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: ES Payment date: 20240902 Year of fee payment: 5 Ref country code: CH Payment date: 20240903 Year of fee payment: 5 |
|
| REG | Reference to a national code |
Ref country code: GR Ref legal event code: EP Ref document number: 20240402049 Country of ref document: GR Effective date: 20241007 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: SI Payment date: 20240819 Year of fee payment: 5 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: LV Payment date: 20240820 Year of fee payment: 5 |
|
| PGFP | Annual fee paid to national office [announced via postgrant information from national office to epo] |
Ref country code: RO Payment date: 20240829 Year of fee payment: 5 Ref country code: NO Payment date: 20240828 Year of fee payment: 5 Ref country code: SE Payment date: 20240827 Year of fee payment: 5 |