WO2018164573A1 - Viral polypeptide inhibitors - Google Patents
Viral polypeptide inhibitors Download PDFInfo
- Publication number
- WO2018164573A1 WO2018164573A1 PCT/NL2018/050137 NL2018050137W WO2018164573A1 WO 2018164573 A1 WO2018164573 A1 WO 2018164573A1 NL 2018050137 W NL2018050137 W NL 2018050137W WO 2018164573 A1 WO2018164573 A1 WO 2018164573A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- seq
- amino acid
- region
- amino acids
- inhibitor
- Prior art date
Links
Classifications
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K38/00—Medicinal preparations containing peptides
- A61K38/16—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- A61K38/55—Protease inhibitors
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
- A61P31/12—Antivirals
- A61P31/14—Antivirals for RNA viruses
Definitions
- the disclosure provides polypeptide inhibitors of viral proteins, useful in the treatment and prevention of viral infection.
- the viral inhibitors are particularly useful for treating Middle East respiratory syndrome coronaviral (MERS-CoV) infection, Crimean-Congo hemorrhagic fever viral infection, or the symptoms of infection.
- MERS-CoV Middle East respiratory syndrome coronaviral
- One object of the invention is to provide viral inhibitors with high specificity for their respective target.
- a further object of the invention is to provide viral inhibitors with low toxicity.
- One aspect of the invention provides an inhibitor comprising a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l, wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and wherein said substitutions include an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1.
- the inhibitor has an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1.
- the inhibitor has an amino acid substitution of V to I at the amino acid position corresponding to 70 of SEQ ID NO: 1.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5;
- region 2 has an ammo acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5;
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5.
- region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5; SEQ ID NO: 12, or SEQ ID NO: 14;
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, or SEQ ID NO: 13; and/or
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 13, or SEQ ID NO: 15.
- region 1 is linked to region 2 by a polypeptide comprising amino acids 15-41 of SEQ ID NO: 1 or having at least 80% identity to amino acids 15-41 of SEQ ID NO: 1
- region 2 is linked to region 3 by a polypeptide comprising amino acids 50-61 of SEQ ID NO: 1 or having at least 80% identity to amino acids 50-61 of SEQ ID NO: 1.
- the inhibitor comprises an amino acid sequence selected from SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, and SEQ ID NO: 5. It is clear from the sequences disclosed herein that SEQ ID Nos: 12, 13, and 15 share the same amino acids at positions 2-14; SEQ ID Nos:5, 12, 14, and 15 share the same amino acids at positions 42-49; and SEQ ID Nos: 5, 12, and 14 share the same amino acids at positions 62-78.
- the invention further provides nucleic acid molecules encoding the inhibitor as well as recombinant expression vector expressing said nucleic acid molecules.
- the invention further provides cell lines, non- human cells and non-human organisms comprising said inhibitors, said nucleic acid molecules, or said recombinant expression vectors.
- the invention further provides pharmaceutical compositions comprising said inhibitors, said nucleic acid molecules, or said recombinant expression vectors.
- the invention further provides the use of said inhibitor or a nucleic acid molecule encoding said an inhibitor, or pharmaceutical compositions thereof, for inhibiting the biological activity of a viral protein.
- the use is for inhibiting the proteolytic cleavage activity of a viral protein.
- the use if for inhibiting the polyprotein processing activity of a viral protein.
- the inhibitor inhibits the biological activity of MERS-CoV PLP 10 domain.
- the inhibitor or a nucleic acid molecule encoding said inhibitor is provided for use in therapy.
- the use is for the treatment and/or prevention of Middle East respiratory syndrome coronaviral (MERS-CoV) infection and/or the symptoms thereof.
- MERS-CoV Middle East respiratory syndrome coronaviral
- One aspect of the invention provides an inhibitor comprising a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l, wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and wherein said substitutions include an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10;
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10;
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, SEQ ID NO: 10 or SEQ ID NO: 16..
- region 1 is linked to region 2 by a polypeptide comprising amino acids 15-41 of SEQ ID NO: 1 or having at least 80% identity to amino acids 15-41 of SEQ ID NO: 1 and where in region 2 is linked to region 3 by a polypeptide comprising amino acids 50-61 of SEQ ID NO: 1 or having at least 80% identity to amino acids 50- 61 of SEQ ID NO: 1.
- the inhibitor comprises an amino acid sequence selected from SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, SEQ ID NO: 10 or SEQ ID NO: 16.
- the invention further provides nucleic acid molecules encoding the inhibitor as well as recombinant expression vector expressing said nucleic acid molecules.
- the invention further provides cell lines, non-human cells and non-human organisms comprising said inhibitors, said nucleic acid molecules, or said recombinant expression vectors.
- the invention further provides pharmaceutical compositions comprising said inhibitors, said nucleic acid molecules, or said recombinant expression vectors.
- the invention further provides the use of said inhibitor or a nucleic acid molecule encoding said an inhibitor, or pharmaceutical compositions thereof, for inhibiting the biological activity of a viral protein.
- the use is for inhibiting the proteolytic cleavage activity of a viral protein.
- the inhibitor inhibits the biological activity of CCHFV OTU domain.
- the inhibitor or a nucleic acid molecule encoding said inhibitor is provided for use in therapy.
- the use is for the treatment and/or prevention of Crimean-Congo hemorrhagic fever viral infection and/or the symptoms thereof.
- One aspect of the invention provides methods for screening and identifying polypeptides that inhibit viral proteins, preferably viral proteins that bind to or interact with ubiquitin and/or ubiquitin-like proteins (i.e., viral ubiquitin binding partners), said method comprising providing a library of polypeptides and screening said library against a viral ubiquitin binding partner in order to identify inhibitors that bind to said viral ubiquitin binding partner, wherein said polypeptide library comprises at least 1000 different polypeptides, wherein each polypeptide comprises a beta-grasp fold comprising region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l and comprises at least one amino acid mutation in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l.
- FIG. 1 Inhibitors inhibit activity of MERS-CoV PLpro and CCHFV OTU in vitro.
- A Sequences of inhibitors that bind MERS-CoV or CCHFV vDUBs. Only regions subjected to diversification relative to Ub.wt in the phage-displayed library are shown. Amino acids discussed in the text are highlighted.
- B The binding specificities of phage-displayed inhibitors (y-axis) are shown across a group of 12 DUBs (x-axis), as assessed by phage ELISA. Sub-saturating concentrations of phage were added to immobilized proteins as indicated.
- Bound phages were detected by the addition of anti-M13-HRP and colorimetric development of TMB peroxidase substrate. The mean value of absorbance at 450 nm is shaded in a black-red-yellow gradient.
- C Inhibition of MERS-CoV PLpro (solid lines) or CCHFV OTU (dashed lines) by the cognate inhibitors shown as dose-response curves using Ub-AMC (left) or ISG15-AMC (right) as a substrate.
- the IC50 value was determined as the concentration of inhibitor that reduced proteolytic activity by 50% (Table 2).
- the wt Ub data obtained in the delSGylation assay cannot be fitted by GraphPad Prism so no lines are shown.
- FIG. 3 Structural basis for inhibition of MERS-CoV PLpro.
- A Crystal structure of the MERS-CoV PLpro-ME.4 complex
- B MERS-CoV PLpro-ME.2 complex
- C MERS-CoV PLpro-Ub.wt complex
- PLpro domains are shown as surface representations, and coloured in wheat, gray and chartreuse for the PLpro- ME.4, -ME.2 and -Ub complexes, respectively.
- ME.4, ME.2 and Ub are shown as tubes and coloured in marine, red and orange, respectively.
- FIG. 3 Structural basis for inhibition of CCHFV OTU.
- A Crystal structure of the CCHFV OTU-CC.2 complex
- B CCHFV OTU-CC.4 complex
- C CCHFV OTU- Ub.wt complex
- OTU domains are shown as surface representations, and coloured in cyan, light cyan and slate for the OTU-CC.2, -CC.4 and -Ub.wt complexes, respectively.
- CC.2, CC.4 and Ub.wt are shown as tubes and coloured in yellow, magenta and orange, respectively.
- Figure 4 Inhibition of proteolytic activity of MERS-CoV PLpro in cell culture and affect MERS-CoV replication.
- A The effects of inhibitors on the DUB activity of MERS-CoV PLpro was determined by co-transfecting HEK293T cells with plasmids encoding HA-Ub, MERS-CoV PLpro-V5 (wild type or the active site mutant C1592A annotated as "C" throughout the rest of the figure), FLAG-ME- inhibitor as indicated (in increasing dose) and GFP (as a transfection control). Cells were lysed 18 hours post transfection and expressed proteins were analyzed by western blotting.
- mitochondrial antiviral signaling protein MAVS
- MERS-CoV PLpro-V5 wild type or the active site mutant C
- FLAG-tagged inhibitors in increasing dose.
- Cells were lysed 16 hours post transfection and both firefly and Renilla luciferase activities were measured. Shown results represent at least three independent experiments.
- MERS-CoV titers of collected supernatants from lentivirus transduced and, subsequently, MERS- CoV infected MRC5 cells were transduced with lentiviruses encoding FLAG-inhibitors, FLAG-Ub.AA or GFP (latter two as controls) and, either 32 hours or 48 hours post-transduction, the cells were infected with MERS-CoV at a multiplicity of infection of 0.01.
- MERS-CoV titers were determined by plaque assays on Vero cells. Significant difference relative to MERS-CoV titers from lentivirus transduced MRC5 cells expressing Ub.AA is indicated: * p ⁇ 0.05. Bars represent mean and error bars represent S.D.
- FIG. 5 Inhibitors bound with high affinity to MERS-CoV PLpro and CCHFV OTU.
- A Binding curves of inhibitors to the cognate viral proteases (left panel: MERS-CoV PLpro; right panel: CCHFV OTU), measured by ELISA. The half maximal binding concentrations (EC50) of inhibitors to indicated vDUBs were determined by established methods [2] and are listed in Table 2. Viral proteases (1 ⁇ ) were immobilized in microtiter plates. Serial dilutions of FLAG-tagged inhibitor or Ub (up to 4 ⁇ , 24 points) were added and incubated for 20 min at room temperature.
- MERS-CoV PLpro and CCHFV OTU are inhibited by inhibitors in vitro.
- A- B Inhibition of MERS-CoV PLpro (left) or CCHFV OTU (right) by the cognate inhibitors shown as dose-response curves using Ub-AMC (A) or ISG15-AMC (B) as a substrate.
- the IC50 values were determined as the concentrations of inhibitors that reduced deubiquitination or delSGylation activity by 50% (Table 2).
- the wt Ub data obtained in the delSGylation assay can not be fitted by GraphPad Prism so no lines were shown.
- CCHFV OTU-Ub.wt, -CC.2 and CC.4 complexes CCHFV OTU is displayed as ribbons, and coloured in slate, cyan and pale cyan in the CCHFV OTU-Ub.wt, -CC.2 and -CC.4 structures, respectively.
- the Ub and inhibitors structures are displayed as tubes, and coloured in orange, yellow and magenta in the CCHFV OTU-Ub.wt, -CC.2 and -CC.4 structures, respectively. Structures were aligned within PyMOL [18].
- FIG. 8 Comparison of the C-terminal regions of ME.2 and ME.4 in the active site of MERS-CoV PLpro.
- A Superposition of the C-terminal regions of the MERS-CoV PLpro-ME.2 and -ME.4 structures.
- PLpro is coloured in gray and wheat in the MERS-CoV PLpro-ME.2 and -ME.4 structures, and ME.2 and ME.4 are coloured in red and marine, respectively.
- PLpro active site residues Hisl759 and Cysl592 are shown as sticks, along with additional PLpro, ME.2 and ME.4 residues involved in binding.
- FIG. 9 Residues in the N-terminal ⁇ -hairpin of ME.4 and ME.2 are disordered.
- A Cartoon representation of ME.4 (marine). Dashed line indicates missing residues 8-10 which were not resolved in the electron density maps. A 2Fo-Fc electron density map is displayed as blue mesh and contoured at 1.0 RMSD.
- B Cartoon representation of ME.2 (red). Dashed line indicates missing residues 7-10. Figure generated with PyMOL [18].
- FIG. 10 Proteolytic activity of MERS-CoV PLpro is inhibited by inhibitors.
- A Inhibition of MERS-CoV PLpro DUB activity by ME. l, ME.2 and ME.3 was determined by expressing HA-Ub, MERS-CoV PLpro-V5 (wild type or the active site mutant C1592A designated as C), FLAG-ME- inhibitor (500, 750 or 1000 ng of the appropriate plasmid) and GFP (as a transfection control) in HEK293T cells. After obtaining protein lysates the expressed proteins were separated on a SDS-PAGE gel, blotted and visualized after antibody incubations.
- MERS-CoV PLpro Proteolytic cleavage capability of MERS-CoV PLpro was assessed in the presence of the inhibitors. N- terminally HA-tagged and C-terminally V5-tagged nsp3C-4 (a polyprotein fragment excluding PLpro) was co-expressed with MERS-CoV PLpro-V5 (wild type or the active site mutant C), FLAG-ME-inhibitors (at increasing concentrations) and GFP (as a transfection control). Cells were lysed 18 h post-transfection and expressed proteins were analyzed by Western blotting. Figure 11. MERS-CoV- directed inhibitors do not inhibit the DUB activity of SARS- CoV PLpro.
- SARS-CoV PLpro's DUB activity in the presence of inhibitors was determined by co-transfecting HEK293T cells with plasmids encoding HA-Ub, SARS- CoV PLpro-V5 (wild type or the active site mutant C1651A designated as C), FLAG- ME-inhibitor (1000 ng) and GFP (as a transfection control). 18 h post-transfection cells were lysed and deconjugation of HA-tagged Ub by SARS-CoV PLpro was visualized via Western blotting.
- HEK293T cells were transfected with plasmids encoding firefly luciferase reporter gene under control of the IFN-B promoter, Renilla luciferase, MAVS, SARS-CoV PLpro-V5 (wild type or the active site mutant C; 100 ng) and FLAG-tagged inhibitors (750 ng).
- FIG. 12 Structural model of the SARS-CoV PLpro domain bound to the MERS-CoV PLpro-specific ME.4.
- A The SARS-CoV PLpro domain is shown as a cartoon representation (yellow-orange). ME.4 and Ub.wt are shown in tubes (marine and orange, respectively). The ME.4 structure determined herein was superposed over Ub bound to the SARS-CoV PLpro domain (4M0W [19])
- FIG. 13 Analysis of lentivirus transduction of MRC5 and HuH-7 cells.
- A, B Western blot analysis of transduced MRC5 and HuH-7 cells with lentiviruses encoding GFP (A) or FLAG-ME. l (B) both 32 h and 48 h pt. As a control cells were mock transduced (designated as M). Relative expression of GFP and FLAG-ME. l was quantified and normalized to actin and expression levels in MRC5 cells 48 h pt were set at 100%.
- C GFP transduced MRC5 and HuH-7 cells were fixed 32 h or 48 h pt and nuclear DNA was stained using Hoechst.
- FIG. 14 Western blot analysis of MERS-CoV infection on transduced MRC5 cells shows decreased viral protein production as a result of inhibitor expression.
- protein lysates were obtained. Expression of two viral proteins was analyzed by Western blotting, MERS-CoV nsp4 (using cross-reacting SARS-CoV nsp4 antiserum), and MERS-CoV ORF4B. Lentivirus-induced expression of FLAG-inhibitors or GFP was confirmed and actin was used as a loading control.
- Representative Western blots are shown for transduced MRC5 cells that were infected with MERS-CoV at a multiplicity of infection of 0.01 either 32 h pt (A, B) or 48 h pt (C, D).
- FIG. 15 Titers of MERS-CoV progeny decreased upon infection of HuH-7 cells expressing inhibitors.
- HuH-7 cells were transduced with lentiviruses encoding
- FIG. 16 Exemplary beta-GF containing scaffold of the ubiquitin like superfamily. Diagonal shading in 16A (slanting upwards to right) and 16B indicate the beta-GF. 16C depicts the location of region 1 (cross-hatching), region 2 (stippling), and region 3 (slanting downwards to right).
- FIG. 17 Titers of MERS-CoV progeny decreased upon infection of cells expressing inhibitors.
- MRC5 cells were transduced with lentiviruses encoding FLAG-inhibitors or GFP (as control) respectively and these cells were infected with MERS-CoV.
- Culture supernatants were collected 32 h (A) or 48h (B) post MERS-CoV infection and infectious progeny titers were determined by plaque assays.
- C HuH-7 cells were transduced with lentiviruses encoding FLAG-inhibitors or GFP (as control) respectively and these cells were infected with MERS-CoV.
- Culture supernatants were collected or 48h post MERS-CoV infection and infectious progeny titers were determined by plaque assays.
- FIG. 18 A) Binding curves of inhibitors to MERS-CoV PLpro; right panel were measured by ELISA. The half maximal binding concentrations (EC50) of inhibitors were determined as described in Figure 5A and are listed in Table 3.
- B-C Inhibition of MERS-CoV PLpro by inhibitors shown as dose-response curves using Ub-AMC (B) or ISG15-AMC (C) as a substrate. The IC50 values were determined as the
- FIG. 19 Overview of sequences of MERS-CoV-specific inhibitors. Changes compared to inhibitors of Figure 1A are highlighted and critical amino acid residues that are important for the strong binding to MERS-CoV PLpro are indicated at positions 46, 64, and 74.
- ME.1.1 and ME.3.1 were generated to determine the role of V70I on binding affinity. Region 1 of variant ME.4 was changed to region 1 of Ub WT (ME.4.2) or region 1 of ME. l (ME.4.4) to determine if stability could be increased.
- Figure 20 Variants ME. l and ME.3 are more thermostable than ME.2 and ME.4. DSC curves are shown for (A) Ub, (B) ME.2, (C) ME.3, (D) ME.4 and (E) Ub. Tm values are indicated.
- FIG. 21 Mutation RIOG stabilizes ME.4. Fluorescence melt curves are shown for (A) ME.2, (B) ME.4 and (C) ME.4R10G. Assays were performed in triplicate, and representative curves for each variant are reported, with the Tm and standard deviations indicated.
- Figure 22 The expression levels of the inhibitors were determined by transfecting HEK293T cells with plasmids encoding FLAG-ME- Inh as indicated. Cells were lysed 18 hours post transfection and expressed proteins were analyzed on a Western blot.
- Protein levels of ME.2, ME.4 and ME.4.1 were lower compared to ME. l, ME.1.1, ME.3 and ME.3.1. Changing region 1 of ME.4 either to region 1 of Ub WT or region 1 of ME. l results in higher protein levels (ME.4.2, ME.4.3 and ME4.4).
- FIG. 23 A. Relative FLAG-Inh protein levels were determined by quantification of Western blots. Protein bands were quantified of least three independent experiments and error bars indicate the standard error of the mean. ME.2, ME.4, ME.4.1 and ME.4.5 have the lowest relative protein levels whereas ME. l, ME.1.1, ME.3, ME.3.1 as well as stabilized variants of ME.4 (ME.4.2, ME.4.3 and ME4.4) are higher expressed.
- mRNA levels of inhibitors were determined by RT-qPCR. mRNA levels of the inhibitors were normalized to the transfection efficiency (neomycin) and to the normalization gene (actin). Error bars show the standard deviation. Although the protein levels of ME. l, ME.3, ME.4.2, ME.4.3 and ME4.4 are higher when compared to ME.2, ME.4, ME.4.1 their relative mRNA levels are lower.
- A. ME.4, ME.4.2 and ME4.4 bind equally strong to MERS-CoV PLpro in the binding assay. Mutating amino acid residues that are important for the binding to MERS-CoV PLpro to Ub WT residues severely reduced the binding affinity (ME.4.5, ME.4.2.1 and ME.4.4.1). Also Q48 and V62 in ME.4.2 contribute to the high binding affinity of this variant because these residues were mutated to Ub WT residues in ME.4.3 and this resulted in a lower affinity of this variant compared to ME.4.2.
- Figure 25 Binding assays comparing affinities between variants (A and B).
- Figure 26 Bindings assay with CCHFV inhibitors.
- viruses including MERS-CoV and the Crime an- Congo hemorrhagic fever virus (CCHFV) encode deubiquitinating (DUB) enzymes that are critical for viral replication and pathogenicity. They bind and remove ubiquitin (Ub) and the Ub-like protein interferon stimulated gene 15 (ISG15) from cellular proteins to suppress host antiviral innate immune responses.
- Ub ubiquitin
- ISG15 Ub-like protein interferon stimulated gene 15
- vDUBs including the MERS-CoV papain-like protease, are responsible for cleaving the viral replicase polyproteins during replication, and are thereby critical components of the viral replication cycle.
- the present disclosure provides highly selective protein -based inhibitors. Unlike small chemical molecules which bind their target over a small surface area, the protein-based inhibitors disclosed herein are relatively large molecules and bind their target over a larger surface area. This has several advantages including high target selectivity, low cross-reactivity to host proteins, and low toxicity. In addition, the protein-based inhibitors disclosed herein share homology with a host protein (i.e., ubiquitin). Thus the host cells are less likely to view the inhibitors as "foreign" as compared to small chemical molecules. In addition, the protein-based inhibitors disclosed herein are very soluble.
- a host protein i.e., ubiquitin
- the present disclosure demonstrates that these inhibitors bind vDUBs with high affinity and specificity to inhibit deubiquitination and delSGylation.
- the disclosure demonstrates that polypeptides having a beta-grasp fold and an F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or a Y at the amino acid position corresponding to 64 of SEQ ID NO: 1 effectively inhibit a viral protease.
- the present disclosure also provides that inhibitors further having a substitution of V to I at the amino acid position corresponding to 70 of SEQ ID NO: 1 are effective.
- the disclosure provides an inhibitor comprising a beta- grasp fold, wherein said fold comprises region 1 (amino acids 2-14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l, wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, wherein said substitutions include an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1.
- the inhibitor comprises between 5 to 20 amino acid substitutions, more preferably between 10-15, most preferably between 10-13 amino acid substitutions, as compared to the sequence of regions 1, 2, and 3 of SEQ ID NO: 1.
- said substitutions further include an amino acid substitution of V to I at the amino acid position corresponding to 70 of SEQ ID NO: 1.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5;
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5; and/or wherein region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5; SEQ ID NO: 12, or SEQ ID NO: 14; b) wherein region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, or SEQ ID NO: 13; and/or c) wherein region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 13, SEQ ID NO: 15.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 2
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2.
- region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 3
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 3
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 3.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 4
- region 2 has an amino acid sequence
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 5
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 5
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 5.
- region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 12
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 12
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 12.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 13
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 13
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 13.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 14
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 14
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 14.
- region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO:
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO:
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 15.
- the inhibitors comprise an amino acid comprising SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5.
- the inhibitors comprise an ammo acid comprising SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 12, SEQ ID NO: 13, SEQ ID NO: 14, or SEQ ID NO: 15. It is clear to a skilled person that additional tags or sequences may be added to the inhibitors.
- ME inhibitors having an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1 are referred to herein as "ME inhibitors". Most preferred inhibitors are depicted in figure 1A.
- the disclosure further demonstrates that polypeptides having a beta-grasp fold and a G at the amino acid position corresponding to 74 of SEQ ID NO: 1 effectively inhibit a viral protease.
- the disclosure provides an inhibitor comprising a beta- grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l, wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and wherein the inhibitor has an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1.
- the inhibitor comprises between 5 to 20 amino acid substitutions, more preferably between 7-9 amino acid substitutions, as compared to the sequence of regions 1, 2, and 3 of SEQ ID NO: 1.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 6
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 6
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 6.
- region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 7
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 7
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 7.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 8
- region 2 has an amino acid sequence
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 9
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 9
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 9.
- region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 16
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 16
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 16.
- region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 10
- region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 10
- region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 10.
- Inhibitors having an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1 are referred to herein as "CC inhibitors". Most preferred inhibitors are depicted in figure 1A.
- the inhibitors disclosed herein are fusion molecules.
- the inhibitor is covalently linked with a label or tag allowing the detection and/or isolation of the fusion molecule.
- the fusion molecule further comprises one or more modifications increasing the stability of the fusion molecule and/or extending the serum half- life of the fusion molecule.
- the inhibitor may be conjugated to a polyalkylene glycol molecule.
- Beta-GF beta -grasp fold
- a-GF refers to a common folding structure comprising a beta(2)-alpha-beta(2) motif, where the alpha helix is situated against a 4-stranded mixed beta sheet.
- Beta-GF are present in, e.g., IF3-N, archaeo- eukaryotic RNA poly beta-sub unit, Ymll08w, BofC, and POZ.
- a variation of a 4- stranded beta-GF domain is found in Nudix, whereas a "barrelizing version is present in L25, Fasciclin, and phosphoribosyl AMP cyclohdrolase.
- Beta-grasp folds are reviewed in Burroughs et al. 2012 Methods in Molecular Biology vol 832, Chapter 2, which is hereby incorporated by reference.
- Preferred beta-grasp fold containing scaffolds comprise an additional beta strand.
- Proteins comprising 5-stranded beta-grasp folds include molybdopterin- dependent oxidoreductase, SLBB, 2Fe-2S ferredoxm, L-proDH alpha, AOR-N, MoaD, TGS, ThiS, TmoB, superantigen toxins, streptokinase, and S4.
- Most preferred proteins having this structure are members of the ubiquitin-like superfamily which include proteins such as Urml, Apgl2, NeddS, ISG15 and SUMO.
- Such preferred scaffolds have the features referred to in Burroughs et al. 2012 for "Classic UB-like" scaffolds, namely, a loop connecting strands beta2 and beta3 termed a "lateral shelf; and a at the loop connecting strands beta4 and beta5 termed a "connector arm”.
- Figure 16 depicts an exemplary beta-GF containing scaffold of the ubiquitin like superfamily. As depicted in Figure 16, region 1 forms strands betal and beta2, region 2 forms beta3, and region 3 forms beta5. Preferred inhibitors share this three dimensional structure and orientation of regions 1, 2, and 3.
- any beta-GF may be used as a scaffold for presenting regions 1, 2, and 3 as defined herein. While not wishing to be bound by theory, the inventors propose that while the alpha helix of the beta-GF provides structure, it does not bind to the viral proteins. Thus, any similar alpha helix structure can be used (or rather, the primary sequence is not relevant).
- the scaffold is from ubiquitin or a ubiquitin-like protein. As is known to a skilled person, ubiquitin from different species is extremely homologous. The term
- ubiquitin or "Ub” as used herein refers to ubiquitin from any species or source and includes the full-length protein as well as fragments or portions of the protein.
- Human ubiquitin has the amino acid sequence as shown in SEQ ID NO: 1.
- the scaffold is from ubiquitin, more preferably from human ubiquitin.
- the beta-GF fold is provided by linking region 1 to region 2 by a
- polypeptide comprising amino acids 15-41 of SEQ ID NO: 1 or having at least 80% identity to amino acids 15-41 of SEQ ID NO: 1.
- the polypeptide has at least 90%, or at least 95% identity to amino acids 15-41 of SEQ ID NO: 1.
- This linkage provides the alpha helix of the beta-GF.
- the beta-GF is also provided by linking region 2 to region 3 by a polypeptide comprising amino acids 50-61 of SEQ ID NO: 1 or having at least 80% identity to amino acids 50-61 of SEQ ID NO: 1.
- the polypeptide has at least 90%, or at least 95% identity to amino acids 50-61 of SEQ ID NO: 1. As shown in Figure 2, the linkage of region 2 to region 3 does not form part of the beta-GF fold.
- linkages having at least 80% identify to SEQ ID NO: l differ by
- one amino acid residue is replaced with another amino acid residue without altering the tertiary structure of the resulting protein.
- a hydrophobic residue such as glycine can be substituted for another hydrophobic residue such as alanine.
- An alanine residue may be substituted with a more hydrophobic residue such as leucine, valine or isoleucine.
- a negatively charged amino acid such as aspartic acid may be substituted for glutamic acid.
- a positively charged amino acid such as lysine may be substituted for another positively charged amino acid such as arginine.
- Conservative substitutions typically include substitutions within the following groups: glycine, alanine; valine, isoleucine, leucine: aspartic acid, glutamic acid, asparagine, glutamine; serine, threonine; lysine, arginine; and phenylalanine, tyrosine.
- conserveed amino acid substitutions involve replacing one or more amino acids of the polypeptides of the disclosure with amino acids of similar charge, size, and/or hydrophobicity
- the inhibitor comprises an amino acid sequence selected from SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, SEQ ID NO: 10, SEQ ID NO: 12, SEQ ID NO: 13, SEQ ID NO: 14, SEQ ID NO: 15 or SEQ ID NO: 16.
- the disclosure provides nucleic acid molecules encoding said inhibitors.
- a skilled person can determine the nucleic acid sequences which encode the polypeptide inhibitors disclosed herein. Based on the degeneracy of the genetic code, sixty-four codons may be used to encode twenty amino acids and translation termination signal. As is known to a skilled person, codon usage bias in different organisms can effect gene expression level. Various computational tools are available to the skilled person in order to optimize codon usage depending on which organism the desired nucleic acid will be expressed.
- the polypeptide based inhibitors disclosed herein can be produced by any method known to a skilled person. In some embodiments, the inhibitors are chemically synthesized. The inhibitors can also be produced using molecular genetic techniques, such as by inserting a nucleic acid into an expression vector, introducing the expression vector into a host cell, and isolating the polypeptide.
- the disclosure provides vectors which comprise said nucleic acid molecules.
- Said vectors and plasmids are useful, e.g., for generating transgenic organisms or for expressing said polypeptide inhibitors.
- a "vector” is a recombinant nucleic acid construct, such as plasmid, phase genome, virus genome, cosmid, or artificial chromosome, to which another DNA segment may be attached.
- vector includes both viral and non-viral means for introducing the nucleic acid into a cell in vitro, ex vivo or in vivo.
- Preferred vectors are expression vectors. It is within the purview of a skilled person to prepare suitable expression vectors for expressing the inhibitors disclosed hereon. Suitable regulatory sequences including enhancers, promoters, translation initiation signals, and polyadenylation signals may be included. Additionally, depending on the host cell chosen and the vector employed, other sequences, such as an origin of replication, additional DNA restriction sites, enhancers, and sequences conferring inducibility of transcription may be incorporated into the expression vector.
- the expression vectors may also contain a selectable marker gene which facilitates the selection of host cells transformed or transfected. Examples of selectable marker genes are genes encoding a protein such as G418 and hygromycin which confer resistance to certain drugs, 6- galactosidase,
- chloramphenicol acetyl transferase and firefly luciferase.
- Viral vectors include lentivirus, retrovirus, adeno-associated virus (AAV), poxvirus (including MVA), baculovirus, , herpes simplex, Epstein-Barr and adenovirus vectors.
- Vector sequences may also contain one or more regulatory regions, and/or selectable markers useful in selecting, measuring, and monitoring nucleic acid transfer results (transfer to which tissues, duration of expression, etc.). Lentiviruses have been previously described for transgene delivery to the hippocampus (van Hooijdonk BMC Neuroscience 2009, 10:2).
- nucleic acids there are a variety of techniques available for introducing nucleic acids into viable cells.
- the techniques vary depending upon whether the nucleic acid is transferred into cultured cells in vitro, or in vivo in the cells of the intended host or animal model.
- Techniques suitable for the transfer of nucleic acid into mammalian cells in vitro include the use of liposomes, electroporation, microinjection, cell fusion, DEAE- dextran, the calcium phosphate precipitation method, etc.
- the currently preferred in vivo gene transfer techniques include transfection with viral (typically retroviral) vectors and viral coat protein-liposome mediated transfection (Dzau et al., Trends in Biotechnology 11:205-210 (1993)).
- the nucleic acids may stably integrate into the genome of the host cell (for example, with retroviral introduction, outlined below), or may exist either transiently or stably in the cytoplasm (i.e. through the use of traditional plasmids, utilizing standard regulatory sequences, selection markers, etc.). Such cells are useful for producing isolated polypeptides, which may be used in the methods described herein.
- the examples disclosed herein provide exemplary methods and vectors for expressing nucleic acids. Accordingly, the disclosure provides cells and organisms comprising the nucleic acids or vectors comprising said nucleic acids as disclosed herein. Preferably, said cells and organisms express the respective polypeptide inhibitor as disclosed herein.
- Such cells include bacteria, e.g., E. coli, as well as filamentous fungi, yeast, plant, mammalian and insect cells.
- the cells and organisms are from animals (in particular mammals; e.g., mice, rats, rabbits, bovine, Camelidae family members)
- animals in particular mammals; e.g., mice, rats, rabbits, bovine, Camelidae family members
- the animal is a non-human transgenic animal.
- the cells are in vitro or in vivo non-human cells.
- Preferred host cells for expression include MRC5 cells (human cell line derived from lung tissue), HuH7 cells (human liver cell line), CHO-cells (Chinese Hamster Ovary), COS-cells (derived from monkey kidney (African green monkey), Vero-cells (kidney epithelial cells extracted from African green monkey), Hela-cells (human cell line), BHK-cells (baby hamster kidney cells, HEK-cells (Human Embryonic Kidney), NSO-cells (Murine myeloma cell line), C127-cells (nontumorigenic mouse cell line), and PerC6®-cells (human cell line, Crucell).
- MRC5 cells human cell line derived from lung tissue
- HuH7 cells human liver cell line
- CHO-cells Choinese Hamster Ovary
- COS-cells derived from monkey kidney (African green monkey)
- Vero-cells kidney epithelial cells extracted from African green monkey
- the disclosed inhibitors are useful in therapy.
- compositions comprising one or more of the polypeptide inhibitors disclosed herein, one or more of the nucleic acid molecules encoding said inhibitors, or one or more vectors disclosed herein.
- Such pharmaceutical compositions are useful in therapy and for inhibiting viral proteins, as disclosed herein.
- Actual dosage levels of the pharmaceutical preparations described herein may be varied so as to obtain an amount of the active ingredient which is effective to achieve the desired therapeutic response for a particular patient, composition, and mode of administration, without being toxic to the patient.
- the selected dosage level will depend upon a variety of factors including the activity of the particular compound, the route of administration, the time of administration, the rate of excretion of the particular compound being employed, the duration of the treatment, other drugs, compounds and/or materials used in combination, the age, sex, weight, condition, general health and prior medical history of the patient being treated, and like factors well known in the medical arts.
- a physician or veterinarian having ordinary skill in the art can readily determine and prescribe the effective amount of the
- the disclosure provides the use of the inhibitors disclosed herein and nucleic acids encoding said inhibitors for inhibiting the biological activity of a viral protein as well as providing methods for screening for new inhibitors.
- viral proteins refers to viral ubiquitin binding protein partners, or rather, proteins that bind/interact with ubiquitin or ubiquitin-like proteins.
- Such proteins include viral proteases with deubiquitinating activity (vDUBs), viral E2 conjugating enzymes, viral E3 ubiquitin ligases, viral El enzymes, and other viral proteins containing ubiquitin binding domains (UBD).
- the viral protein is a vDUB.
- the inhibitors inhibit the proteolytic cleavage activity of said vDUB.
- Ubiquitination is a post-translational modification mediated by an enzyme cascade that results in the conjugation of ubiquitin (Ub) to cellular proteins [1, 2] .
- This process is regulated in part through activity of cellular deubiquitinating enzymes (DUBs), which remove Ub from cellular proteins [1, 2].
- DUBs cellular deubiquitinating enzymes
- virus-encoded DUBs reverse the ubiquitination process to alter host signaling pathways critical to the induction of cellular antiviral and pro-inflammatory innate immune responses [3].
- vDUBs In addition to removing Ub molecules from host proteins, many viral DUBs (vDUBs) also remove the Ub-like protein interferon- stimulated gene 15 (ISG15) to further suppress antiviral responses [4, 5]. Importantly, a number of vDUBs also play an essential role in viral replication [4-6]. Together, the replicative and/or deubiquitinating activities of viral proteases contribute directly to pathogenesis during viral infection in vivo [7], making them ideal antiviral drug targets.
- the viral protein is from a coronavirus.
- Coronaviruses initially express their non-structural proteins (nsps) as large viral polyproteins, which are processed into functional domains by proteases encoded within the polyproteins to establish a viral replication-transcriptase complex.
- SARS- and MERS-CoV release nspl-3 through the activity of a papain-like protease (PLt jro ) domain situated within nsp3, in a process that is indispensable for replication [4] ,
- 3CLP ro corresponding to nsp5
- coronaviral PLt jro s also act as vDUBs to suppress host antiviral innate immune responses by targeting cellular Ub- conjugated substrates [9- 14].
- the ME inhibitors disclosed herein inhibit the biological activity of the MERS-CoV papain-like protease (PLf ,ro ) domain.
- the inhibitors inhibit the proteolytic cleavage activity of the PLr >n> domain.
- the inhibitors inhibit the polyprotein processing activity of the PL/" "0 domain.
- expression of the inhibitors during MERS-CoV infection reduces infectious progeny titer.
- the ME inhibitors disclosed herein are useful in therapy.
- MERS-CoV Middle East respiratory syndrome coronaviral
- the inhibitor comprises a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l,
- the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and
- inhibitor has an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1.
- Additional ME inhibitors as disclosed herein may also be used in the treatment method, in particular inhibitors comprising an amino acid sequence selected from SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, , SEQ ID NO: 5, as well as SEQ ID NO: 12, SEQ ID NO: 13, SEQ ID NO: 14, and SEQ ID NO: 15.
- an individual infected with MERS-CoV is an animal such as a member of the Camelidae family.
- Treatment of reservoir species can reduce transmission to humans.
- the individual is a human.
- said treatment reduces the severity and/or duration of the viral infection and/or reduces the severity and/or duration of symptoms (such as respiratory illness, fever, dyspnea, and myalgia).
- the viral protein is from the nairovirus Crimean-Congo hemorrhagic fever virus (CCHFV).
- CCHFV Crimean-Congo hemorrhagic fever virus
- This pathogenic virus also encodes a vDUB.
- the CCHFV vDUB domain is located within the large (L) segment of the genome, and has also been explicitly implicated in the evasion of host Ub- and ISG 15- dependent innate immune responses [17]. This domain is also referred to as the CCHFV OTU domain (ovarian tumor (OTU) protease domain ).
- the CC inhibitors disclosed herein inhibit the biological activity of the CCHFV vDUB domain.
- the inhibitors inhibit the proteolytic cleavage activity of said domain.
- the CE inhibitors disclosed herein are useful in therapy.
- methods are provided for the treatment and/or prevention of Crimean-CongO hemorrhagic fever viral infection and/or the symptoms thereof, the method comprising administering to an individual in need thereof a therapeutically effective amount of an inhibitor or a nucleic acid molecule encoding said inhibitor, wherein the inhibitor comprising a beta- grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l, wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and wherein the inhibitor has an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1.
- Additional CC inhibitors as disclosed herein may also be used in the treatment method, in particular inhibitors comprising an amino acid sequence selected from SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10 as well as SEQ ID NO: 16.
- an individual infected with CCHFV is an animal such as ruminants and ostriches.
- Treatment of reservoir species can reduce transmission to humans.
- the individual is a human.
- said treatment reduces the severity and/or duration of the viral infection and/or reduces the severity and/or duration of symptoms (such as flu-like symptoms, hemorrhage, respiratory distress).
- the disclosure provides methods of identifying an inhibitor or a nucleic acid molecule encoding said inhibitor that inhibits the biological activity of a viral protein, the method comprising providing a library of polypeptides and screening said library against a viral ubiquitin binding partner in order to identify inhibitors that bind to said viral ubiquitin binding partner, wherein said polypeptide library comprises at least 1000 distinct polypeptides (or rather each polypeptide varies from every other polypeptide by at least one amino acid residue), wherein each polypeptide comprises a beta-grasp fold comprising region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l and comprises at least one amino acid mutation in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l.
- each polypeptide of the library is a distinct ubiquitin polypeptide having one ore more amino acid substitutions in region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and/or region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l.
- the polypeptide has at least 2, at least 4, at least 8, at least 10, at least 15, or at least 20 amino acid substitutions.
- the polypeptides comprise two additional amino acids at the C-terminus.
- the library comprises at least 10 6 , 10 8 , or 10 9 distinct polypeptides.
- the library can be screened for inhibition of a viral protein, for example, by phage display, mRNA display, ribosome display, yeast display or other similar technologies to determine the inhibition of biological activity compared to a control.
- the control is a different protein to test for specificity.
- Example 1 provides an exemplary embodiment of methods to screen for candidate inhibitors.
- the methods comprise purifying the candidate inhibitors and testing them in additional in vitro and/or in vivo assays in order to determine their effect on viral infection.
- the viral ubiquitin binding partner is a viral proteases with
- vDUBs deubiquitinating activity
- UBD viral proteins containing ubiquitin binding domains
- polypeptide libraries such as those described in WO2012/020289 and Ernst et al. can be used to rapidly identify viral inhibitors.
- the present disclosure provides vDUBs as a preferred target for such libraries.
- vDUBs generally have a low affinity for ubiquitin.
- the present disclosure demonstrates that high affinity polypeptide inhibitors can be identified using the disclosed methods.
- the large binding surface of the candidate polypeptide inhibitors offers the potential to target a diverse range of vDUBs.
- screening methods are advantageous over screening small chemical compound libraries.
- the probability of identifying a compound that specifically binds it's target is quite small.
- Candidates that bind a target with low or moderate affinity may be identified. These candidates must be further chemically optimized in order to produce a compound with the potential to bind with high affinity to its target.
- the screening methods disclosed herein can identify highly specific inhibitors without the need for further optimization.
- the disclosed screening methods are faster and more efficient than small chemical compound screening.
- to comprise and its conjugations is used in its non-limiting sense to mean that items following the word are included, but items not specifically mentioned are not excluded.
- verb "to consist” may be replaced by "to consist essentially of meaning that a compound or adjunct compound as defined herein may comprise additional component(s) than the ones specifically identified, said additional component(s) not altering the unique characteristic of the invention.
- treatment refers to reversing, alleviating, delaying the onset of, or inhibiting the progress of a disease or disorder, or one or more symptoms thereof, as described herein.
- treatment may be administered after one or more symptoms have developed.
- treatment may be administered in the absence of symptoms.
- treatment may be administered to a susceptible individual prior to the onset of symptoms (e.g., in light of a history of symptoms and/or in light of genetic or other susceptibility factors). Treatment may also be continued after symptoms have resolved, for example to prevent or delay their recurrence.
- a polypeptide library [25] was screened against the MERS-CoV PLpro domain (MERS-CoV PLpro) and the CCHFV OTU domain (CCHFV OTU). Inhibitors were identified that bound with high affinity to either MERS-CoV PLpro (ME. l to ME.4) or CCHFV OTU (CC. l to CC.5) (Fig. 1A). To confirm the specificity of the inhibitors towards their cognate vDUBs, the phage-displayed inhibitors were challenged against a diverse panel of 11 DUBs from several species representing distinct DUB families (USP, OTU, and ubiquitin C-terminal hydrolases (UCH)).
- each inhibitor also potently inhibited the deubiquitinating and delSGylating activities of MERS-CoV PLpro or CCHFV OTU as measured using the fluorogenic substrates Ub-AMC or ISG15-AMC, respectively (Fig. 1C and Fig. 6).
- the most potent inhibitors of MERS-CoV PLpro and CCHFV OTU were ME.4
- Inhibitor residue He 70 extends further into a hydrophobic pocket of PLpro formed by residues Thrl730* and Vall691* (asterisks denote amino acid numbering of MERS-CoV polyprotein), in comparison to Ub.wt residue Val70 (Fig. 2D).
- ME.2 and ME.4 residue Phe46 inserts into a hydrophobic pocket formed by PLpro residues Trpl668*,
- CCHFV OTU crystal structures of CCHFV OTU were determined bound to CC.2 and to CC.4 to gain insight into how they selectively block the DUB and delSGylating activities of this viral enzyme (Fig. 3A,B and Table 1).
- CC.2 and CC.4 were found to bind in the same orientation as Ub.wt with similar buried surface areas of -1000 A2 (Fig. 3C and Fig. 7B) [20, 21].
- substitutions in these inhibitors were concentrated in the C-terminal region (Fig. 1A) and only substitutions at position 68 and downstream were found to interact with the enzyme.
- Tyr68 improves hydrophobic packing with ThrlO*, Vall2* and Vall8* (asterisks denote amino acid numbering of the large segment-encoded protein of CCHFV), relative to His68 in Ub.wt (Fig. 3D).
- residue Leu70 projects further into a hydrophobic cavity of CCHFV OTU formed by residues Vall2*, He 14*, Vall8* and He 131*, than the equivalent Ub.wt residue Val70 (Fig. 3E).
- the conformational freedom of a Gly substitution at position 74 enables the C-terminal tail of each inhibitor to form numerous favorable interactions with the enzyme.
- Trp76 in both CC.2 and CC.4 does not interact with CCHFV OTU, but instead packs into a hydrophobic cavity within each inhibitor (Fig. 3E,F).
- Example 3 Inhibition of MERS-CoV PLpro activity in cell culture-based assays To explore the effects of inhibitors on MERS-CoV PLpro activity in cells,
- deubiquitination assays were performed by transfecting cells with combinations of plasmids encoding the following proteins: HA-tagged Ub , MERS-CoV PLpro, and inhibitors. Unlike Ub which becomes conjugated to cellular proteins by its C-terminal glycine residue, the inhibitors have substitutions in the C-terminal di-Gly motif.
- the inhibitors cannot interfere with the natural ubiquitin regulatory machinery, further avoiding toxicity/adverse effects to the cell.
- ME.2 and ME.4 were more potent than ME. l and ME.3 at blocking the ability of MERS-CoV PLpro to suppress IFN- ⁇ promoter activation, whereas none of the inhibitors were able to block suppression of the IFN- ⁇ promoter activity by SARS-CoV PLpro (Fig. 11B).
- the inhibitors thus prevented MERS-CoV PLpro-mediated suppression of cellular antiviral innate immune responses, and in a remarkably selective, virus-specific manner.
- a critical step in the replication cycle of MERS-CoV is the processing of viral polyproteins into functional non-structural proteins (nsps) that is accomplished in part by the protease activity of PLpro, which cleaves the nsp lj,2, nsp2
- nsps functional non-structural proteins
- FLAG-tagged inhibitors and V5-tagged MERS-CoV PLpro were co-expressed with N-terminally HA-tagged and C-terminally V5-tagged nsp3C-4 (HA-nsp3C-4-V5), a fragment of the viral polyprotein encompassing the C-terminal part of nsp3 (excluding the PLpro domain) and nsp4.
- HA-nsp3C-4-V5 N-terminally HA-tagged and C-terminally V5-tagged nsp3C-4
- nsp4 a fragment of the viral polyprotein encompassing the C-terminal part of nsp3 (excluding the PLpro domain) and nsp4.
- MERS-CoV PLpro efficiently cleaved HA-nsp3C-4-V5 into HA-nsp3C and nsp4-V5 products, whereas the active site mutant did not (Fig.
- Example 4 Inhibitors block MERS-CoV replication in cells
- MERS- CoV PLpro-specific inhibitors were ectopically expressed in cell culture, and cells were subsequently infected with MERS-CoV.
- MRC5 and HuH-7 cell lines were transduced with lentiviruses encoding FLAG-tagged inhibitors, Ub.AA, or GFP.
- Efficient expression of FLAG-ME. l and GFP in these cells was confirmed by fluorescence microscopy and by western blotting (Fig. 13). Either 32 or 48 hours post-transduction, cells were infected with MERS-CoV at a multiplicity of infection of 0.01, and MERS- CoV titers were determined from supernatants harvested 32 hours post infection (Fig.
- MRC5 cells In MRC5 cells, ME. l and ME.4 expression resulted in significantly lower virus titers as these dropped from 5 x 105 plaque forming units (PFU)/ml recovered from control cells to 1,000 or 10 PFU/ml, respectively, when the MERS-CoV infection was started 32 hours post-transduction (Fig. 4D).
- the effect of the inhibitors was even more pronounced in MRC5 cells that were infected with MERS-CoV 48 hours post- transduction, as virus titers dropped below 10 PFU/ml upon expression of ME.4, which represented a reduction in infectious progeny titers of more than four orders of magnitude (Fig. 4D) and correlated with higher expression of the inhibitors at this time point (Fig. 13 and 14).
- ME.4 was mutated at position 46 from F to A, at position 64 from Y to E, and at position 70 from I to V to generate ME.4.1 (SEQ ID NO: 11).
- ME4.1 was expressed in MRC5 or HuH-7 cells using the lentivirus -based expression system and infected the transduced cells with MERS-CoV.
- ME.4.1 was less effective in inhibiting MERS-CoV replication, highlighting the importance of the F46, Y64 and 170 residues in ME.4 for the inhibitory effect (Figure 17).
- IC50 and EC50 values (Table 3) were also determined as described herein (see Figure 18).
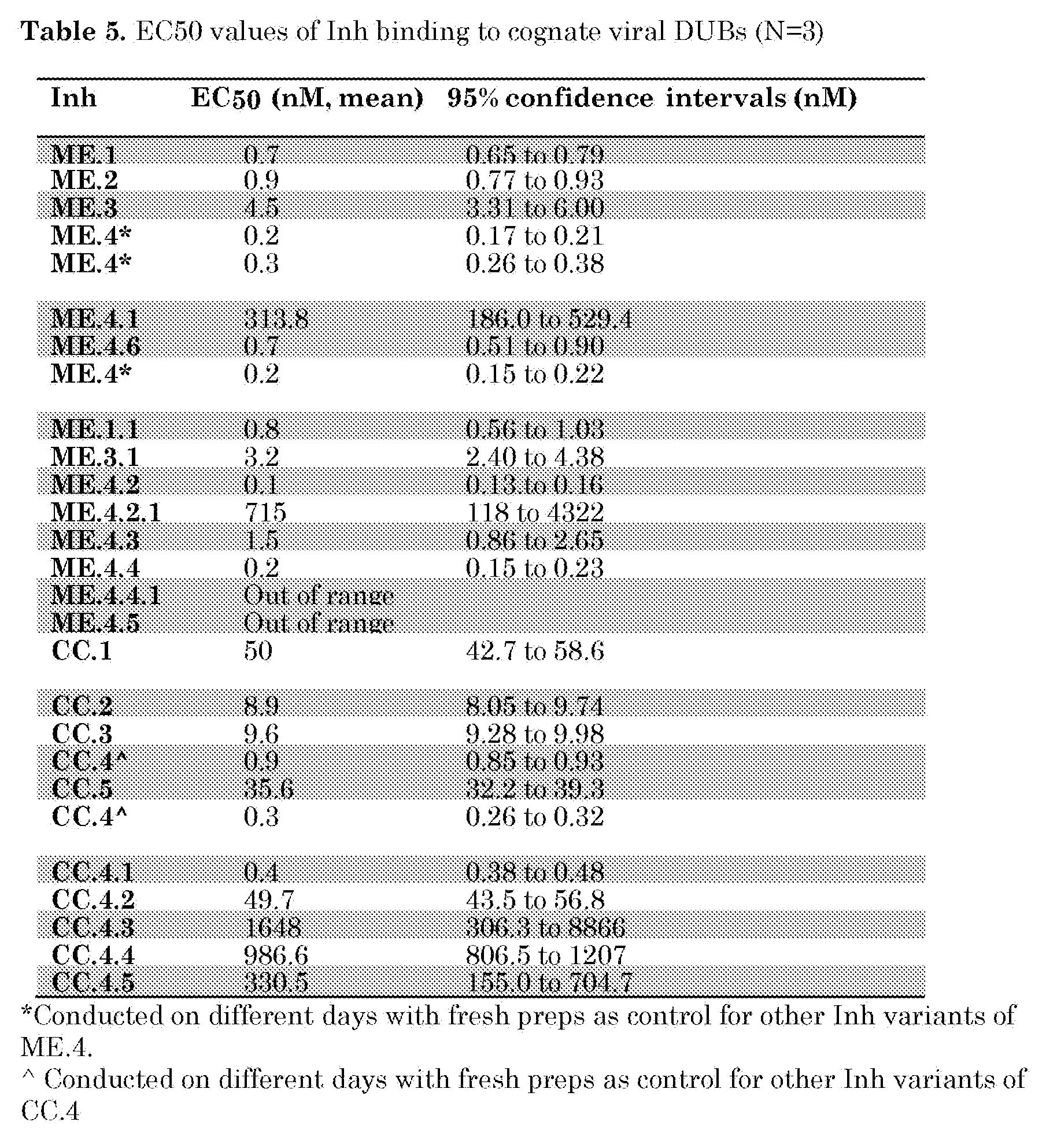
- CC.4 was mutated to remove 1 to 4 amino acids of the C-terminus (CC.4.1-CC.4.4) or to mutate position 75 from V to G (CC.4.5). Binding curves of these inhibitors to CCHFV OTU are shown in Figure 26 and EC50 values were determined and are listed in Table 5. The results demonstrate that residues V75, P76 and W77 in the tail region of CC.4 play a role in the high binding affinity to CCHFV OTU.
- Example 7 Modification of ME. l and ME.3 inhibitors
- ME.1.1 and ME.3.1 were generated to determine whether the binding affinity of ME. l and ME.3 can be increased by introducing the V70I substitution. As shown in Table 5, this mutation does not increase the binding affinity to MERS-CoV PLpro compared to their original variants, ME. l and ME.3.
- the phage-display library used in this study was re-amplified from Library 2 as previously described [25] . Protein immobilization and the following inhibitor selections were done according to established protocols [26, 45]. Briefly, purified viral proteases were coated on 96-well MaxiSorp plates (Thermo Scientific 12565135) by adding 100 ⁇ of 1 ⁇ proteins and incubating overnight at 4°C.
- phage-display library five rounds of selections using the phage-display library were performed against immobilized proteins including the following steps: (a) Each phage particle in the library pool displays a unique candidate inhibitor and encapsulates the encoding DNA; (b) Binding phages are captured with an immobilized protein; (c) Non-binding phages are washed away; and (d) Bound phages are amplified by infection of bacteria. The enriched phage pool is cycled through additional rounds of selection to further enrich for protein-binding candidate inhibitors. After the fifth round of binding selections individual phages with improved binding properties were identified by phage ELISA using established techniques and subjected to DNA sequencing of the phagemids to obtain candidate inhibitor sequences [26, 45] .
- MERS-CoV PLpro-inhibitor complexes To form the non-covalent MERS-CoV PLpro-inhibitor complexes, a 4-fold molar excess of ME.4 or ME.2 was incubated with MERS-CoV PLpro overnight at 4°C. The excess, unbound inhibitors were removed from the sample using a Superdex 75 size exclusion column and fractions containing the MERS-CoV PLpro- inhibitor complex were pooled and concentrated to 10 mg/mL.
- the MERS-CoV PLpro-ME.4 complex was found to crystallize under similar conditions to those previously reported for the MERS-CoV PLpro-Ub complex [9], with optimal crystals appearing in 0.1 M trisodium citrate pH 5.6, 20% (w/v) polyethylene glycol (PEG) 4000 and 20% (v/v) isopropanol. Crystals of the MERS-CoV PLpro-ME.2 complex were grown in 0.1 M trisodium citrate pH 5.6, 19% (w/v) PEG 4000 and 19% (v/v) 1,2-isopropanediol.
- Crystals were grown by mixing PLpro- inhibitor (10 mg/niL and 9 mg/mL for PLpro-ME.4 and PLpro-ME.2, respectively) with crystallization solution at a 1: 1 volumetric ratio (2 ⁇ MERS-CoV PLpro-inhibitor + 2 ⁇ well solution). Immediately prior to mixing, 1 M DTT was added to the MERS-CoV PLpro- inhibitor complexes to a final concentration of 10 mM to prevent oxidation of the sample.
- Purified CCHFV OTU was pooled with 2-3-fold molar excess of purified inhibitor and dialyzed overnight against 50 mM Tris pH 8.0, 150 mM NaCl and 2 mM DTT. Protein complexes were concentrated and loaded onto a Superdex 75 size exclusion column and eluted in 50 mM Tris, 150 mM NaCl and 2 mM DTT. For all samples, a single peak corresponding to the respective complex was observed in the gel filtration profile and two bands corresponding to the CCHFV OTU and respective inhibitor were observed by SDS-PAGE, indicating the high purity of the complexes.
- the CCHFV OTU169-CC.2 complex was concentrated to 12 mg/ml for crystallization trials, and initial crystals and crystalline material obtained from preliminary screens were used to prepare seed stocks for microseed matrix screening [46, 47], which was set up for the hanging drop vapor diffusion method in 48-well VDX plates (Hampton Research) and carried out using conventional screens (Qiagen) at 4°C, with and without heterogeneous nucleation using 0.3-0.4 cm strands of human hair [48] . Total drop volume was 2 ⁇ containing equal volumes of the protein complex and the well solution. Crystals of the CCHFV OTU169-CC.2 complex were grown in 30% (w/v) PEG 4000, 0.2 M CaC12 and 0.1 M HEPES pH 7.5 and appeared after 5-8 days.
- the CCHFV OTU185-CC.4 complex was concentrated to 23 mg/ml and initial leads were observed with a combinatorial approach using microseed matrix screening with crystallization screens (Qiagen) along with the Silver Bullets screen (Hampton Research) and micro-seeding using crystals of a CCHFV OTU185-CC.5 (a weaker binding variant selected by phage display) complex.
- crystallization screens Qiagen
- Silver Bullets screen Hampton Research
- screens were set up at 20°C with a reservoir volume of 150 ⁇ , and a drop size of 3.5 ⁇ , which comprised of 1.5 ⁇ of the protein complex, 1 ⁇ of the reservoir solution and 1 ⁇ of Silver Bullet additive, added in this order.
- Crystals were obtained with 25% (w/v) PEG 3350, 0.1 M Tris pH 7.0 and 0.2 M sodium chloride.
- the Silver Bullet formulation in the drop was as follows: 0.16% (w/v) each of 5-Sulfosalicylic acid dehydrate, dodecanedioic acid, hippuric acid, mellitic acid, oxalacetic acid, suberic acid and 0.02 M HEPES sodium pH 6.8.
- HEK293T cells (Virgin lab, Washington University School of Medicine) grown in a T175 flask were transfected with packaging vectors
- MRC5 cells (CCL-171; American Type Culture Collection) grown in a 12-wells plate were transduced with lentiviruses encoding GFP or FLAG- ME.
- l diluted in DMEM containing 2% FCS and 8 ⁇ g/ml Polybrene (Sigma Aldrich).
- Medium was replaced 24 h post transduction (pt), and 32 h or 48 h pt protein lysates were obtained by adding 250 ⁇ 2xLSB containing 25 mM NEM to each well while cells grown on coverslips were fixed with 3%) paraformaldehyde (PFA) in PBS.
- PFA paraformaldehyde
- GFP and FLAG-ME. l expression were analyzed by Western blotting as described in the supporting information.
- HuH-7 or MRC5 cells were transduced with lentiviruses encoding GFP, FLAG-Ub.AA, FLAG-ME. l or FLAG-ME.4.
- Cells were infected with MERS-CoV with a multiplicity of infection of 0.01 32 h or 48 h pt.
- MERS-CoV mocula were prepared in PBS containing 50 ⁇ g/ml DEAE-dextran and 2% FCS. Cells were inoculated for 1 h at 37°C and the inoculum was replaced with EMEM containing 2% FCS. Supernatants were harvested 32 h post MERS-CoV infection and
- MERS-CoV titers were determined by plaque assays on Vero cells (Department of Viroscience, Erasmus Medical Center) as described by van den Worm et al. [50]. MERS-CoV infection experiments were performed at least twice and plaque assays were performed in duplicate in order to determine MERS-CoV titers. Work with MERS- CoV was performed inside biosafety cabinets in Biosafety Level 3 facilities at Leiden University Medical Center.
- Mielech AM Kilianski A
- Baez-Santos YM Mesecar AD
- Baker SC MERS-CoV papaindike protease has delSGylating and deubiquitinating activities. Virology.
- PubMed PMID 22412934; PubMed Central PMCID: PMC3296753.
- Binding- constants KD were obtained by fitting the response wavelength shifts in the steady- state regions using single -site binding system (Eq. 1) shown below.
- Req is the value of the response shift in the steady-state region in each sensorgram curve
- [C] is the titrant concentration
- Rmax is the maximal response in the steady-state region
- KD is the binding constant for the single-site binding system.
- Rmax and KD values are unknown and the Levenberg-Marquardt algorithm was used to perform iterative non-linear least squares curve fitting in Profit 6.2 (QuantumSoft) to obtain the fitted Rmax and KD.
- deconjugation substrates were performed as described before [1, 3] . Experiments were performed in assay buffer (50 niM HE PES, pH 7.5, 0.01% Tween 20, 1 mM
- DTT dithiothreitol
- vDUBs dithiothreitol
- 12 serial dilutions of inhibitor were mixed in assay buffer as indicated and incubated at room temperature for 2 min prior to the addition of Ub- AMC. All serial dilutions were performed in 96-well plates and subsequently transferred to 384-well black plates (Thermo Scientific) for making measurements. Deconjugation activity was measured by monitoring the increase of AMC fluorescence emission at 460 nm (excitation at 360 nm) for 30 min using a BioTek Synergy2 plate reader (BioTek Instruments, Winooski, VT). IC50 values were calculated using the GraphPad Prism software with the built-in equation formula (non-linear regression curve).
- Plasmids named pET53-ME.2, and -ME.4 were transformed into CaC12-competent Escherichia coli BL21 (DE3) Gold cells (Agilent) to allow for T7 polymerase- driven expression of N-terminally His6-tagged ME.2 and ME.4 respectively.
- Cells were grown at 37°C in the presence of 150 ⁇ g/mL ampicillin to an optical density (OD600) of 0.6 and then induced at 16°C by addition of Isopropyl B-D- l-thiogalactopyranoside (IPTG; final concentration 1 mM).
- lysis buffer 500 mM NaCl, 50 mM Tris pH 8.0
- the cell lysate was clarified by centrifugation at 17,211 x g at 4°C for 30 min, then incubated with 2 niL Ni-NTA Superflow resin (Qiagen) at 4°C for 30 min, and poured into a gravity column. The column was washed with 50 niL of lysis buffer, followed by 50 mL of lysis buffer containing 50 mM imidazole. Protein was eluted in lysis buffer containing 250 mM imidazole. Following affinity purification, inhibitors were further purified using a Superdex 75 size exclusion column (GE Healthcare), eluting in 20 mM Tris pH 8.5, 150 mM NaCl and 2 mM DTT.
- GE Healthcare Superdex 75 size exclusion column
- Plasmids encoding CC.2 and CC.4, named pET53-CC2 and -CC.4 respectively, were transformed into CaC12-competent E. coli BL21 (DE3) Gold cells, and grown in Luria- Bertani media supplemented with 150 pg/ml Ampicillin at 37°C with shaking to an OD600 of 0.8-1.5. Protein expression was induced by the addition of a final
- CC.2 was eluted in 20 mM Tris pH 7.4, 150 mM NaCl and 1 mM DTT, and CC.4 was eluted in 50 mM Tris pH 8.0, 150 mM NaCl and 2 mM DTT.
- the MERS-CoV PLpro domain was expressed and purified as described previously [4] . Briefly, E. coli BL21 (DE3) Gold cells transformed with plasmid pE-SUMO PLpro were grown to an OD600 ⁇ of 0.6-0.8 at 37°C in the presence of 35 ⁇ g/mL kanamycin.
- Protein expression was induced by the addition of a final concentration of 1 mM IPTG and overnight incubation at 16C Cells were resuspended in lysis buffer (150 mM Tris, pH 8.5, 1 M NaCl, 2 mM DTT), lysed via French press and clarified via centrifugation at 17,21 lx g Clarified lysate was loaded onto a Ni-NTA gravity column and washed with lysis buffer, followed by lysis buffer supplemented with 25 mM imidazole and subsequent elution in lysis buffer supplemented with 250 mM
- lysis buffer 150 mM Tris, pH 8.5, 1 M NaCl, 2 mM DTT
- the SUMO-PLpro -1-1 fusion was cleaved overnight in 150 mM NaCl, 50 mM Tris, pH 8.0, 1 mM DTT in the presence of Ulp l SUMO protease. Cleaved, tagless MERS-CoV PLpro was subsequently passed through a second Ni-NTA column, and further purified on a Superdex 75 size exclusion column equilibrated in 20 mM Tris, pH 8.5, 150 mM NaCl, 2 mM DTT.
- Plasmids encoding the CCHFV OTU domain residues 1-169 (pGEX-CCHFV OTU169) and residues 1-185 (pET49b-CCHFV OTU 185) fused with a GST tag and an HRV3c protease cleavage site were used for the expression and purification of the CCHFV OTU domain as described previously [5, 6] .
- E. coli BL21-Gold (DE3) cells transformed with either of the plasmids were grown to an OD600 of 0.9-1.0 at 37°C with shaking, and protein expression was induced with a final concentration of 1 mM IPTG at 30°C for 19-21 hrs.
- lysis buffer 50mM Tris-Cl pH 7.2, 200 mM NaCl, 5 mM EDTA, 5 mM DTT
- the lysate was clarified by centrifugation at 48,298 x g for 30-40 min.
- Cleaved CCHF- vOTU proteins were collected as flow-through by passing the digest through a recharged GST-Bind column, concentrated and loaded onto the Superdex-75 column (GE Healthcare). Purified proteins, eluted in 20 mM Tris pH 7.2, 150 mM NaCl and ImM DTT, were then used for further analyses.
- Crystals were harvested by sweeping through a cryoprotectant solution containing
- PLpro- ME.4 0.1M trisodium citrate pH 5.6, 22% (w/v) PEG 4000, 21% (v/v) 1,2-propanediol (PLpro- ME.4), or harvested directly from the crystallization solution (PLpro-ME.2) and flash- cooled in liquid nitrogen.
- X-ray diffraction data for PLpro-ME.4 crystals were collected using a Rigaku 007HF MicroFocus X-ray generator and R-AXIS IV++ detector.
- PLpro- ME.2 data collection was carried out at the Canadian Light Source on beamline 08B1-
- Diffraction data for the PLpro-ME.4 and -ME.2 crystals were integrated and scaled using XDS [7], followed by merging using Aimless within the CCP4 software suite.
- Plasmids used for cell culture work The following plasmids were described elsewhere or provided by others: pcDNA3.1- MERS-CoV-PLpro WT and active site mutant C1592A [12], pCAGGS-HA-nsp3C-4-V5 [12], pcDNA-eGFP [13], pCAGGS- MAVS (provided by N. Frias-Staheli), pLuc-IFN-B [14] and pRL-TK (Promega).
- pcDNA3-HA-Ub was generated by cloning PCR-amplified Ub (using pCMV-FLAG-Ub [15] as a template) in pcDNA3.1(-) (Invitrogen) in frame with an N-terminal HA tag.
- Codon optimized SARS-CoV-PLpro amino acids 1541- 1855 of the SARS-CoV ppla/pp lab (NCBI ID: AY291315.1)
- IDT polyadenylation signals
- HEK293T were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS; Bodinco BV), 100 units/ml penicillin, 100 units/ml streptomycin and 2 niM L-glutamine.
- DMEM Dulbecco's modified Eagle's medium
- FCS fetal calf serum
- Vero cells Erasmus Medical Center
- MRC5 cells ATCC CCL- 171
- HuH-7 cells were maintained in DMEM containing 8% FCS, antibiotics and non-essential amino acids.
- DMEM, EMEM and supplements were obtained from Lonza.
- Proteins on Western blot were visualized using the following primary antibodies: mouse anti-FLAG (F3165; Sigma-Aldrich), mouse anti-V5 (37-7500; Invitrogen), mouse anti-HA (ab 18181; Abeam), rabbit anti-GFP [13], rabbit anti-SARS-CoV nsp4 [16], rabbit anti-MERS-CoV p4b [17] and mouse anti-actin (A5316; Sigma-Aldrich). Primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (P0447 and P0217; Dako).
- primary antibody mouse anti-FLAG F3165; Sigma-Aldrich
- secondary antibody Alexa488-conjugated goat anti-mouse immunoglobulin G (IgG) antibody A- 11001; Thermo Fisher Scientific
- the in trans cleavage activity of MERS-CoV PLpro in the presence of inhibitors was determined by co-expressing HA-nsp3C-4-V5 (0.2 ⁇ g), MERS-CoV-PLpro-V5 (0.15 ⁇ g), FLAG-tagged inhibitors (0.5; 0.75 or 1 ⁇ & and GFP (0.25 ⁇ g) in HEK293T cells.
- Empty pcDNA vector was added to supplement to a total of 2 ⁇ g of plasmid DNA transfected per well of a 12-wells cluster.
- LSB Laemmli sample buffer
- NEM N-ethylmaleimide
- Proteins were separated in an SDS-PAGE gel, blotted onto Hybond-P (0,45 ⁇ pore size, GE-Heathcare) and visualized after antibody incubation steps using Pierce ECL 2 Western blotting substrate (Thermo Fisher Scientific). To visualize FLAG-tagged inhibitors the proteins separated in an SDS-page gel were blotted onto 0,2 ⁇ PVDF membranes (GE-Heathcare). The membranes were blocked with dried milk powder in PBS containing 0,05% Tween-20 followed by antibody incubation steps.
- HEK293T cells grown to 80% confluency in a 24-wells plate were co-transfected with a combination of plasmids encoding the firefly luciferase reporter gene under control of the IFN- ⁇ promoter (25 ng), Renilla luciferase (5 ng), innate immune inducer mitochondrial antiviral signalling protein (MAVS; 25 ng), MERS-CoV PLpro-V5 (250 ng) and FLAG-tagged inhibitors (250, 500, 750 ng).
- SARS-CoV PLpro- V5 100 ng
- FLAG-tagged inhibitors 750 ng
- Firefly and Renilla luciferase activities were measured in triplicate and assays were repeated independently at least three times.
- pET53-ME.4R10G was generated via around-the-horn PGR [ Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989; 17:6545-51.
- E. coli BL21 (DE3) Gold cells harbouring inhibitors were started by inoculating 3 mL LB supplemented with 150 ⁇ g/mL ampicillin (amp) with a glycerol stock scraping, and were incubated overnight at 37°C with aeration. The next morning, 60 ⁇ of overnight culture were subcultured into 6 mL LB + 150 ⁇ g/mL amp and grown at 37°C to an OD600 of 0.8. Inhibitor expression was induced overnight at 16°C with the addition IPTG to a final concentration of 1 mM. Cell pellets were frozen at -80°C until use.
- Cell pellets were resuspended in 1 mL [50 mM Tris pH 8.5, 500 mM NaCl, 100 ⁇ PMSF, 5 ⁇ g/mL DNase] and lysed by sonication. Cell lysates were clarified by centrilugation at 16, 100 rcf, and 200 ⁇ of His Mag Sepharose Ni resin (GE Healthcare) was added to the clarified lysate supernatant, and incubated at room temperature for 30 min with end- over-end mixing.
- DSC curves were collected on a Nano DSC (TA Instruments) with a scan rate of l°C/min at 3 atm, using protein concentrations of 2-3.25 mg/mL. Data analysis was performed using NanoAnalyze, and DSC curves were fit to a two-state scaled model for melting temperature (Tm) determination.
- Thermal shift assay reactions were performed using 4 ⁇ g inhibitor, and a final concentration of 5x SYPRO orange (Sigma). Assays were performed in 96-well format using an StepOnePlus Real-Time PCR System (Applied Biosystems), with a scan rate of l°C/min. Fluorescence melt curves were fit to a Boltzmann equation with Tm values corresponding to the midpoint of the transition curve using Protein Thermal Shift Software Version 1.1 (Thermofisher). Tm values are reported as an average of 3 replicates.
- Inhibitor-containing pDONR-221 vectors were previously described [Zhang et al.]. By classical cloning with Xhol and Pvul, region 1 of Ub WT or ME. l was cloned in place of region 1 of ME.4 in DONR-221-FLAG-ME.4 to generate pDONR-221-FLAG-ME.4.2 and -ME.4.4 respectively. Single amino acid substitutions were introduced using the QuikChangeTM strategy with pDONR-221- FLAG-Inh as template to generate pDONR-221-FLAG-ME. l. l, -ME.3.1, -ME.4.1, - ME4.3 and -ME.4.5. The newly generated FLAG-Inh were cloned in destination vectors pcDNA3.1-DEST or pLenti6.3/TO/V5-DEST (Thermo Fisher Scientific) using the Gateway technology (Thermo Fisher Scientific).
- HEK293T cells grown to 80% confluency in a 12-well plate were translected with 1 ⁇ g pcDNA3.1-DEST-FLAG-Inh using the calcium phosphate transfection method [ Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456-67.].
- LSB Laemmli sample buffer
- NAM N-ethylmaleimide
- RA1 buffer supplemented with ⁇ - mercaptoethanol for total RNA isolation using the NucleoSpin RNA II kit (Machery- Nagel).
- Protein samples were loaded on SDS-polyacrylamide gels which were blotted onto PVDF membranes (GE-Heatheare) using the semi- dry blotting system (Trans- Blot turbo Tranfer System, BioRad). Membranes were blocked with 5% dried milk powder in PBS containing 0,05% Tween-20, followed by antibody incubation steps.
- qRT real-time quantitative reverse-transcriptase
- iTaq SYBR Green Supermix BioRad
- BioRad CFX384 TouchTM Real-Time PGR Detection System
Landscapes
- Health & Medical Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Animal Behavior & Ethology (AREA)
- Virology (AREA)
- Chemical & Material Sciences (AREA)
- Veterinary Medicine (AREA)
- Public Health (AREA)
- Medicinal Chemistry (AREA)
- Pharmacology & Pharmacy (AREA)
- General Health & Medical Sciences (AREA)
- Immunology (AREA)
- Communicable Diseases (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Engineering & Computer Science (AREA)
- Molecular Biology (AREA)
- Gastroenterology & Hepatology (AREA)
- Epidemiology (AREA)
- Oncology (AREA)
- Chemical Kinetics & Catalysis (AREA)
- General Chemical & Material Sciences (AREA)
- Nuclear Medicine, Radiotherapy & Molecular Imaging (AREA)
- Organic Chemistry (AREA)
- Medicines That Contain Protein Lipid Enzymes And Other Medicines (AREA)
Abstract
The disclosure provides polypeptide inhibitors of viral proteins, useful in the treatment and prevention of viral infection. The viral inhibitors are particularly useful for treating Middle East respiratory syndrome coronaviral (MERS-CoV) infection, Crimean-Congo hemorrhagic fever viral infection, or the symptoms of infection.
Description
Title: Viral polypeptide inhibitors FIELD OF THE INVENTION
The disclosure provides polypeptide inhibitors of viral proteins, useful in the treatment and prevention of viral infection. The viral inhibitors are particularly useful for treating Middle East respiratory syndrome coronaviral (MERS-CoV) infection, Crimean-Congo hemorrhagic fever viral infection, or the symptoms of infection.
BACKGROUND OF THE INVENTION
Emerging viruses pose a tremendous challenge to both human and animal health. While vaccine-based approaches are desirable in terms of infection prevention in the longer term, alternative antiviral strategies are needed, especially when providing treatment options for infected patients during acute outbreaks.
Various strategies for treating and preventing viral infection have been developed. One approach has been to strengthen the host's immune system against viral infection. Another has been to target the specific virus itself. For example, virally encoded enzymes have been used as putative targets. For some viruses, viral proteases are essential for viral replication and various protease inhibitors have been developed. However, effective viral protease inhibitors are lacking for many viral infections. The recent Middle East respiratory syndrome coronavirus (MERS-CoV), Ebola and Zika virus outbreaks exemplify the continued threat of (re-)emerging viruses to human health, and our inability to rapidly develop effective therapeutic countermeasures. In some cases the development of such inhibitors is hampered by the structural similarity between potential viral targets and the corresponding host targets. This leads to potential toxicity issues. Highly specific inhibitors are thus in need.
One object of the invention is to provide viral inhibitors with high specificity for their respective target. A further object of the invention is to provide viral inhibitors with low toxicity. SUMMARY OF THE INVENTION
One aspect of the invention provides an inhibitor comprising a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l, wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and wherein said substitutions include an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1.
Preferably, wherein the inhibitor has an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1. Preferably, wherein the inhibitor has an amino acid substitution of V to I at the amino acid position corresponding to 70 of SEQ ID NO: 1. Preferably, wherein a) region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5; b) region 2 has an ammo acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5; and/or c) region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5. Preferably, a) wherein region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5; SEQ ID NO: 12, or SEQ ID NO: 14;
b) wherein region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, or SEQ ID NO: 13; and/or
c) wherein region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 13, or SEQ ID NO: 15. Preferably, wherein region 1 is linked to region 2 by a polypeptide comprising amino acids 15-41 of SEQ ID NO: 1 or having at least 80% identity to
amino acids 15-41 of SEQ ID NO: 1 and where in region 2 is linked to region 3 by a polypeptide comprising amino acids 50-61 of SEQ ID NO: 1 or having at least 80% identity to amino acids 50-61 of SEQ ID NO: 1. Preferably, wherein the inhibitor comprises an amino acid sequence selected from SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, and SEQ ID NO: 5. It is clear from the sequences disclosed herein that SEQ ID Nos: 12, 13, and 15 share the same amino acids at positions 2-14; SEQ ID Nos:5, 12, 14, and 15 share the same amino acids at positions 42-49; and SEQ ID Nos: 5, 12, and 14 share the same amino acids at positions 62-78. The invention further provides nucleic acid molecules encoding the inhibitor as well as recombinant expression vector expressing said nucleic acid molecules. The invention further provides cell lines, non- human cells and non-human organisms comprising said inhibitors, said nucleic acid molecules, or said recombinant expression vectors. The invention further provides pharmaceutical compositions comprising said inhibitors, said nucleic acid molecules, or said recombinant expression vectors. The invention further provides the use of said inhibitor or a nucleic acid molecule encoding said an inhibitor, or pharmaceutical compositions thereof, for inhibiting the biological activity of a viral protein.
Preferably, the use is for inhibiting the proteolytic cleavage activity of a viral protein. Preferably, the use if for inhibiting the polyprotein processing activity of a viral protein. Preferably, the inhibitor inhibits the biological activity of MERS-CoV PLP10 domain. Preferably, the inhibitor or a nucleic acid molecule encoding said inhibitor is provided for use in therapy. Preferably the use is for the treatment and/or prevention of Middle East respiratory syndrome coronaviral (MERS-CoV) infection and/or the symptoms thereof. One aspect of the invention provides an inhibitor comprising a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l, wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and wherein said substitutions include an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1. Preferably, a) wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10; b) wherein region 2 has
an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10; and/or c) wherein region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, SEQ ID NO: 10 or SEQ ID NO: 16.. Preferably, wherein region 1 is linked to region 2 by a polypeptide comprising amino acids 15-41 of SEQ ID NO: 1 or having at least 80% identity to amino acids 15-41 of SEQ ID NO: 1 and where in region 2 is linked to region 3 by a polypeptide comprising amino acids 50-61 of SEQ ID NO: 1 or having at least 80% identity to amino acids 50- 61 of SEQ ID NO: 1. Preferably, wherein the inhibitor comprises an amino acid sequence selected from SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, SEQ ID NO: 10 or SEQ ID NO: 16.The invention further provides nucleic acid molecules encoding the inhibitor as well as recombinant expression vector expressing said nucleic acid molecules. The invention further provides cell lines, non-human cells and non-human organisms comprising said inhibitors, said nucleic acid molecules, or said recombinant expression vectors. The invention further provides pharmaceutical compositions comprising said inhibitors, said nucleic acid molecules, or said recombinant expression vectors. The invention further provides the use of said inhibitor or a nucleic acid molecule encoding said an inhibitor, or pharmaceutical compositions thereof, for inhibiting the biological activity of a viral protein.
Preferably, the use is for inhibiting the proteolytic cleavage activity of a viral protein. Preferably, the inhibitor inhibits the biological activity of CCHFV OTU domain.
Preferably, the inhibitor or a nucleic acid molecule encoding said inhibitor is provided for use in therapy. Preferably the use is for the treatment and/or prevention of Crimean-Congo hemorrhagic fever viral infection and/or the symptoms thereof.
One aspect of the invention provides methods for screening and identifying polypeptides that inhibit viral proteins, preferably viral proteins that bind to or interact with ubiquitin and/or ubiquitin-like proteins (i.e., viral ubiquitin binding partners), said method comprising providing a library of polypeptides and screening said library against a viral ubiquitin binding partner in order to identify inhibitors that bind to said viral ubiquitin binding partner, wherein said polypeptide library comprises at least 1000 different polypeptides, wherein each polypeptide comprises a beta-grasp fold comprising region 1 (amino acids 2- 14), region 2 (amino acids 42-49),
and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l and comprises at least one amino acid mutation in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l. BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1. Inhibitors inhibit activity of MERS-CoV PLpro and CCHFV OTU in vitro. (A) Sequences of inhibitors that bind MERS-CoV or CCHFV vDUBs. Only regions subjected to diversification relative to Ub.wt in the phage-displayed library are shown. Amino acids discussed in the text are highlighted. (B) The binding specificities of phage-displayed inhibitors (y-axis) are shown across a group of 12 DUBs (x-axis), as assessed by phage ELISA. Sub-saturating concentrations of phage were added to immobilized proteins as indicated. Bound phages were detected by the addition of anti-M13-HRP and colorimetric development of TMB peroxidase substrate. The mean value of absorbance at 450 nm is shaded in a black-red-yellow gradient. (C) Inhibition of MERS-CoV PLpro (solid lines) or CCHFV OTU (dashed lines) by the cognate inhibitors shown as dose-response curves using Ub-AMC (left) or ISG15-AMC (right) as a substrate. The IC50 value was determined as the concentration of inhibitor that reduced proteolytic activity by 50% (Table 2). The wt Ub data obtained in the delSGylation assay cannot be fitted by GraphPad Prism so no lines are shown. (D) Effects of inhibitors on vDUB activity against K48/K63 tetra-Ub substrates. Purified MERS-CoV PLpro (top panels) or CCHFV OTU (bottom panels) was incubated with the indicated inhibitors or Ub.wt (negative control) and biotinylated tetra-Ub at 37°C for a time course of 30 minutes. Western blots were probed with ExtrAvidin-HRP (EA- HRP) to detect biotin-Ub. Inhibition of proteolysis was shown by a delay of
appearance of the digestion products tri-Ub (Ub3), di-Ub (Ub2) and mono-Ub (Ubl). Figure 2. Structural basis for inhibition of MERS-CoV PLpro. (A) Crystal structure of the MERS-CoV PLpro-ME.4 complex (B) MERS-CoV PLpro-ME.2 complex and (C) MERS-CoV PLpro-Ub.wt complex (PDB ID: 4RF0). PLpro domains are shown as surface representations, and coloured in wheat, gray and chartreuse for the PLpro- ME.4, -ME.2 and -Ub complexes, respectively. ME.4, ME.2 and Ub are shown as tubes and coloured in marine, red and orange, respectively. (D) Close up of a superposition of the MERS-CoV PLpro-ME.4, -ME.2 complexes (left panel) showing detailed interactions between PLpro and residue He 70 of ME.4 or ME.2, and a
comparison (right panel) of the previously determined PLpro-Ub.wt complex and homologous residue Val70. PLpro residues are shown as sticks and labeled with asterisks in italics. (E) Close up of the MERS-CoV PLpro-ME.4 and -ME.2 complexes (left panel) showing detailed interactions between PLpro and residue Phe46 of ME.4 or ME.2, and a comparison (right panel) PLpro-Ub.wt and homologous residue Ala46. (F) Close up of the MERS-CoV PLpro-ME.4 and -ME.2 complexes (left panel) showing detailed interactions between PLpro and residue Tyr64 of ME.4 or ME.2, and a comparison (right panel) PLpro-Ub.wt and homologous residue Glu64. (G) Close up of ME.4 residue Asn74 bound near the active site of PLpro. Hydrogen bonds are represented by dashed black lines. (H) Close up of the C-terminus of Ub.wt covalently bound in the active site of PLpro. (I) Close up of ME.2 residue Pro 74 bound near the active site of PLpro. Figure generated using PyMOL [51].
Figure 3. Structural basis for inhibition of CCHFV OTU. (A) Crystal structure of the CCHFV OTU-CC.2 complex (B) CCHFV OTU-CC.4 complex and (C) CCHFV OTU- Ub.wt complex (PDB ID: 3PT2). OTU domains are shown as surface representations, and coloured in cyan, light cyan and slate for the OTU-CC.2, -CC.4 and -Ub.wt complexes, respectively. CC.2, CC.4 and Ub.wt are shown as tubes and coloured in yellow, magenta and orange, respectively. (D) Overlay of the CCHFV OTU-CC.2, CC.4 and -Ub.wt structures demonstrating interactions occurring between CC.2 or CC.4 residue Tyr68 and CCHFV OTU, compared to the homologous Ub.wt residue His68. Inhibitor and Ub.wt residues are shown as sticks and labeled in regular font. CCHFV OTU residues are shown as sticks and labeled in with asterisks in italics. (E) Detailed interactions occurring between the C-terminus of CC.2 and CCHFV OTU. (F) Detailed interactions occurring between the C-terminus of CC.4 and CCHFV OTU. (G) The C- terminus of Ub.wt bound in the active site of CCHFV OTU. Figure generated using PyMOL [51].
Figure 4. Inhibition of proteolytic activity of MERS-CoV PLpro in cell culture and affect MERS-CoV replication. (A) The effects of inhibitors on the DUB activity of MERS-CoV PLpro was determined by co-transfecting HEK293T cells with plasmids encoding HA-Ub, MERS-CoV PLpro-V5 (wild type or the active site mutant C1592A annotated as "C" throughout the rest of the figure), FLAG-ME- inhibitor as indicated (in increasing dose) and GFP (as a transfection control). Cells were lysed 18 hours post transfection and expressed proteins were analyzed by western blotting. DUB
activity of MERS- Co V PLpro was visualized by the deconjugation of HA-Ub from cellular proteins. (B) Assessment of the inhibitory effects of inhibitors on the suppression of the IFN- β promoter by MERS-CoV PLpro. HEK293T cells were transfected with plasmids encoding firefly luciferase reporter gene under control of the IFN-β promoter, Renilla luciferase, innate immune response inducer
mitochondrial antiviral signaling protein (MAVS), MERS-CoV PLpro-V5 (wild type or the active site mutant C) and FLAG-tagged inhibitors (in increasing dose). Cells were lysed 16 hours post transfection and both firefly and Renilla luciferase activities were measured. Shown results represent at least three independent experiments.
Significance relative to wild- type without expression of an inhibitor was calculated using an unpaired two-tailed Student's t test and significant values were indicated: ** p < 0.01. Bars represent mean and error bars represent S.D. (C) Proteolytic cleavage capability of MERS-CoV PLpro was assessed in the presence of the inhibitors. N- terminally HA-tagged and C-terminally V5-tagged nsp3C-4 (excluding the PLpro domain) was co-expressed with V5-tagged MERS-CoV PLpro-V5 (wild type or the active site mutant C), FLAG-ME-inhibitor (with increasing doses) and GFP (as a transfection control). Cells were lysed 18 hours post transfection and proteolytic cleavage activity was assessed by western blotting to detect generation of N-terminal HA-tagged nsp3C and C-terminal V5-tagged nsp4 cleavage products. (D) MERS-CoV titers of collected supernatants from lentivirus transduced and, subsequently, MERS- CoV infected MRC5 cells. MRC-5 cells were transduced with lentiviruses encoding FLAG-inhibitors, FLAG-Ub.AA or GFP (latter two as controls) and, either 32 hours or 48 hours post-transduction, the cells were infected with MERS-CoV at a multiplicity of infection of 0.01. After another 32 hours, culture supernatants were harvested and MERS-CoV titers were determined by plaque assays on Vero cells. Significant difference relative to MERS-CoV titers from lentivirus transduced MRC5 cells expressing Ub.AA is indicated: * p < 0.05. Bars represent mean and error bars represent S.D.
Figure 5. Inhibitors bound with high affinity to MERS-CoV PLpro and CCHFV OTU. (A) Binding curves of inhibitors to the cognate viral proteases (left panel: MERS-CoV PLpro; right panel: CCHFV OTU), measured by ELISA. The half maximal binding concentrations (EC50) of inhibitors to indicated vDUBs were determined by established methods [2] and are listed in Table 2. Viral proteases (1 μΜ) were
immobilized in microtiter plates. Serial dilutions of FLAG-tagged inhibitor or Ub (up to 4 μΜ, 24 points) were added and incubated for 20 min at room temperature. Wells were washed and bound Inh/Ub was detected by anti-FLAG-HRP conjugate antibody and colorimetric development of TMB peroxidase substrate. The absorbance at 450 nm (y-axis) was plotted against Log (Inh/Ub concentration, nM) (x-axis). Data were presented as the mean ± SD (N = 3). (B) Binding curves of wild type Ub (Ub.wt) to MERS-CoV PLpro (left) and CCHFV OTU (right). Experiments were performed as described in (A) except the concentration of Ub was increased (up to 10 mM, 24 serial dilutions). Data were presented as the mean ± SD (N = 3).
Figure 6. MERS-CoV PLpro and CCHFV OTU are inhibited by inhibitors in vitro. (A- B) Inhibition of MERS-CoV PLpro (left) or CCHFV OTU (right) by the cognate inhibitors shown as dose-response curves using Ub-AMC (A) or ISG15-AMC (B) as a substrate. The IC50 values were determined as the concentrations of inhibitors that reduced deubiquitination or delSGylation activity by 50% (Table 2). The wt Ub data obtained in the delSGylation assay can not be fitted by GraphPad Prism so no lines were shown.
Figure 7. MERS-CoV- and CCHFV-specific inhibitors bind their cognate DUBs in comparable orientations to Ub.wt. (A) Superposition of the MERS-CoV PLpro-Ub.wt, - ME.2 and -ME.4 complexes. PLpro is displayed as ribbons, and coloured in
chartreuse, gray and wheat in the PLpro-Ub.wt, -ME.2 and -ME.4 structures, respectively. The Ub and inhibitor structures are displayed as tubes, and coloured in orange, red and marine in the PLpro-Ub.wt, -ME.2 and -ME.4 structures,
respectively. (B) Superposition of the CCHFV OTU-Ub.wt, -CC.2 and CC.4 complexes. CCHFV OTU is displayed as ribbons, and coloured in slate, cyan and pale cyan in the CCHFV OTU-Ub.wt, -CC.2 and -CC.4 structures, respectively. The Ub and inhibitors structures are displayed as tubes, and coloured in orange, yellow and magenta in the CCHFV OTU-Ub.wt, -CC.2 and -CC.4 structures, respectively. Structures were aligned within PyMOL [18].
Figure 8. Comparison of the C-terminal regions of ME.2 and ME.4 in the active site of MERS-CoV PLpro. (A) Superposition of the C-terminal regions of the MERS-CoV PLpro-ME.2 and -ME.4 structures. PLpro is coloured in gray and wheat in the MERS-CoV PLpro-ME.2 and -ME.4 structures, and ME.2 and ME.4 are coloured in red and marine, respectively. PLpro active site residues Hisl759 and Cysl592 are
shown as sticks, along with additional PLpro, ME.2 and ME.4 residues involved in binding. (B) Close up of the C-terminus of ME.4 in the MERS-CoV PLpro ME.4 complex, with PLpro depicted in a surface representation. (C) Close up of the C- terminus of ME.2 in the MERS-CoV PLpro ME.2 complex, with PLpro depicted in a surface representation.
Figure 9. Residues in the N-terminal β-hairpin of ME.4 and ME.2 are disordered. (A) Cartoon representation of ME.4 (marine). Dashed line indicates missing residues 8-10 which were not resolved in the electron density maps. A 2Fo-Fc electron density map is displayed as blue mesh and contoured at 1.0 RMSD. (B) Cartoon representation of ME.2 (red). Dashed line indicates missing residues 7-10. Figure generated with PyMOL [18].
Figure 10. Proteolytic activity of MERS-CoV PLpro is inhibited by inhibitors. (A) Inhibition of MERS-CoV PLpro DUB activity by ME. l, ME.2 and ME.3 was determined by expressing HA-Ub, MERS-CoV PLpro-V5 (wild type or the active site mutant C1592A designated as C), FLAG-ME- inhibitor (500, 750 or 1000 ng of the appropriate plasmid) and GFP (as a transfection control) in HEK293T cells. After obtaining protein lysates the expressed proteins were separated on a SDS-PAGE gel, blotted and visualized after antibody incubations. (B) Suppression of the IFN-β promoter activity by MERS-CoV PLpro in the presence of inhibitors was assessed by transfecting plasmids encoding firefly luciferase reporter gene under control of the IFN-6 promoter, Renilla luciferase, MAVS, MERS-CoV PLpro-V5 (wild type or the active site mutant C) and FLAG-tagged inhibitors (250, 500 or 750 ng). Firefly and Renilla luciferase activities were measured 16 h post transfection and significance relative to wild-type without expression of an inhibitor was calculated using an unpaired two-tailed Student's t test. Significant values were indicated: ** p < 0.01. Bars represent mean and error bars represent S.D (N=3). (C) Proteolytic cleavage capability of MERS-CoV PLpro was assessed in the presence of the inhibitors. N- terminally HA-tagged and C-terminally V5-tagged nsp3C-4 (a polyprotein fragment excluding PLpro) was co-expressed with MERS-CoV PLpro-V5 (wild type or the active site mutant C), FLAG-ME-inhibitors (at increasing concentrations) and GFP (as a transfection control). Cells were lysed 18 h post-transfection and expressed proteins were analyzed by Western blotting.
Figure 11. MERS-CoV- directed inhibitors do not inhibit the DUB activity of SARS- CoV PLpro. (A) SARS-CoV PLpro's DUB activity in the presence of inhibitors was determined by co-transfecting HEK293T cells with plasmids encoding HA-Ub, SARS- CoV PLpro-V5 (wild type or the active site mutant C1651A designated as C), FLAG- ME-inhibitor (1000 ng) and GFP (as a transfection control). 18 h post-transfection cells were lysed and deconjugation of HA-tagged Ub by SARS-CoV PLpro was visualized via Western blotting. (B) HEK293T cells were transfected with plasmids encoding firefly luciferase reporter gene under control of the IFN-B promoter, Renilla luciferase, MAVS, SARS-CoV PLpro-V5 (wild type or the active site mutant C; 100 ng) and FLAG-tagged inhibitors (750 ng). Cells were lysed 16 h post-transfection and both firefly and Renilla luciferase activities were measured. Significance relative to wild- type without expression of a inhibitor was measured using an unpaired two-tailed Student's t test; significant values were indicated: ** p < 0.01. Bars represent mean and error bars represent S.D (N=3).
Figure 12. Structural model of the SARS-CoV PLpro domain bound to the MERS-CoV PLpro-specific ME.4. (A) The SARS-CoV PLpro domain is shown as a cartoon representation (yellow-orange). ME.4 and Ub.wt are shown in tubes (marine and orange, respectively). The ME.4 structure determined herein was superposed over Ub bound to the SARS-CoV PLpro domain (4M0W [19]) (B) Close-up of residue clashes occurring between SARS-CoV PLpro and ME.4. Residues are shown as spheres, with SARS-CoV PLpro residues indicated with asterisks and in italics. SARS-CoV PLpro residue M209 clashed with ME.4 residue 170, compared with Ub residue V70 (C). (D) SARS-CoV PLpro residues E204 and V188 clash with ME.4 residues Q48 and V188, respectively, compared to Ub.wt residues K48 and A46 (E). Figure generated in PyMOL [18].
Figure 13. Analysis of lentivirus transduction of MRC5 and HuH-7 cells. (A, B) Western blot analysis of transduced MRC5 and HuH-7 cells with lentiviruses encoding GFP (A) or FLAG-ME. l (B) both 32 h and 48 h pt. As a control cells were mock transduced (designated as M). Relative expression of GFP and FLAG-ME. l was quantified and normalized to actin and expression levels in MRC5 cells 48 h pt were set at 100%. (C) GFP transduced MRC5 and HuH-7 cells were fixed 32 h or 48 h pt and nuclear DNA was stained using Hoechst. Images were taken using fixed exposure times for both the GFP and Hoechst signal. (D) Immunofluorescence assay of FLAG-
ME. l-transduced MRC5 and HuH-7 cells that were fixed 32 h or 48 h pt. Cells were labelled with a mouse monoclonal antibody recognizing FLAG followed by labelling with a secondary Alexa488-conjugated goat anti-mouse antibody. Exposure times were kept the same for each image.
Figure 14. Western blot analysis of MERS-CoV infection on transduced MRC5 cells shows decreased viral protein production as a result of inhibitor expression. Upon collection of supernatants after MERS-CoV infection of transduced MRC5 cells protein lysates were obtained. Expression of two viral proteins was analyzed by Western blotting, MERS-CoV nsp4 (using cross-reacting SARS-CoV nsp4 antiserum), and MERS-CoV ORF4B. Lentivirus-induced expression of FLAG-inhibitors or GFP was confirmed and actin was used as a loading control. Representative Western blots are shown for transduced MRC5 cells that were infected with MERS-CoV at a multiplicity of infection of 0.01 either 32 h pt (A, B) or 48 h pt (C, D).
Figure 15. Titers of MERS-CoV progeny decreased upon infection of HuH-7 cells expressing inhibitors. (A) HuH-7 cells were transduced with lentiviruses encoding
FLAG-inhibitors or GFP (as control) respectively and 48 h pt these cells were infected with MERS-CoV at a multiplicity of infection of 0.01. Culture supernatants were collected 32 h post MERS-CoV infection and infectious progeny titers were determined by plaque assays. (B) Lentivirus transduced and MERS-CoV infected HuH-7 cells were 32 h post MERS-CoV infection lysed and expression of MERS-CoV nsp4, MERS- CoV ORF4B as well as expression of FLAG-inhibitors or GFP was visualized via Western blotting.
Figure 16. Exemplary beta-GF containing scaffold of the ubiquitin like superfamily. Diagonal shading in 16A (slanting upwards to right) and 16B indicate the beta-GF. 16C depicts the location of region 1 (cross-hatching), region 2 (stippling), and region 3 (slanting downwards to right).
Figure 17. Titers of MERS-CoV progeny decreased upon infection of cells expressing inhibitors. MRC5 cells were transduced with lentiviruses encoding FLAG-inhibitors or GFP (as control) respectively and these cells were infected with MERS-CoV. Culture supernatants were collected 32 h (A) or 48h (B) post MERS-CoV infection and infectious progeny titers were determined by plaque assays. (C) . HuH-7 cells were transduced with lentiviruses encoding FLAG-inhibitors or GFP (as control) respectively and these cells were infected with MERS-CoV. Culture supernatants
were collected or 48h post MERS-CoV infection and infectious progeny titers were determined by plaque assays.
Figure 18. A) Binding curves of inhibitors to MERS-CoV PLpro; right panel were measured by ELISA. The half maximal binding concentrations (EC50) of inhibitors were determined as described in Figure 5A and are listed in Table 3. (B-C) Inhibition of MERS-CoV PLpro by inhibitors shown as dose-response curves using Ub-AMC (B) or ISG15-AMC (C) as a substrate. The IC50 values were determined as the
concentrations of inhibitors that reduced deubiquitination or delSGylation activity by 50% (Table 3).
Figure 19. Overview of sequences of MERS-CoV-specific inhibitors. Changes compared to inhibitors of Figure 1A are highlighted and critical amino acid residues that are important for the strong binding to MERS-CoV PLpro are indicated at positions 46, 64, and 74. ME.1.1 and ME.3.1 were generated to determine the role of V70I on binding affinity. Region 1 of variant ME.4 was changed to region 1 of Ub WT (ME.4.2) or region 1 of ME. l (ME.4.4) to determine if stability could be increased. Figure 20. Variants ME. l and ME.3 are more thermostable than ME.2 and ME.4. DSC curves are shown for (A) Ub, (B) ME.2, (C) ME.3, (D) ME.4 and (E) Ub. Tm values are indicated.
Figure 21. Mutation RIOG stabilizes ME.4. Fluorescence melt curves are shown for (A) ME.2, (B) ME.4 and (C) ME.4R10G. Assays were performed in triplicate, and representative curves for each variant are reported, with the Tm and standard deviations indicated.
Figure 22. The expression levels of the inhibitors were determined by transfecting HEK293T cells with plasmids encoding FLAG-ME- Inh as indicated. Cells were lysed 18 hours post transfection and expressed proteins were analyzed on a Western blot.
Protein levels of ME.2, ME.4 and ME.4.1 were lower compared to ME. l, ME.1.1, ME.3 and ME.3.1. Changing region 1 of ME.4 either to region 1 of Ub WT or region 1 of ME. l results in higher protein levels (ME.4.2, ME.4.3 and ME4.4).
Figure 23. A. Relative FLAG-Inh protein levels were determined by quantification of Western blots. Protein bands were quantified of least three independent experiments and error bars indicate the standard error of the mean. ME.2, ME.4, ME.4.1 and ME.4.5 have the lowest relative protein levels whereas ME. l, ME.1.1, ME.3, ME.3.1
as well as stabilized variants of ME.4 (ME.4.2, ME.4.3 and ME4.4) are higher expressed.
B. Relative mRNA levels of inhibitors were determined by RT-qPCR. mRNA levels of the inhibitors were normalized to the transfection efficiency (neomycin) and to the normalization gene (actin). Error bars show the standard deviation. Although the protein levels of ME. l, ME.3, ME.4.2, ME.4.3 and ME4.4 are higher when compared to ME.2, ME.4, ME.4.1 their relative mRNA levels are lower.
Figure 24. Binding assays
A. ME.4, ME.4.2 and ME4.4 bind equally strong to MERS-CoV PLpro in the binding assay. Mutating amino acid residues that are important for the binding to MERS-CoV PLpro to Ub WT residues severely reduced the binding affinity (ME.4.5, ME.4.2.1 and ME.4.4.1). Also Q48 and V62 in ME.4.2 contribute to the high binding affinity of this variant because these residues were mutated to Ub WT residues in ME.4.3 and this resulted in a lower affinity of this variant compared to ME.4.2.
B. Mutating critical amino acid residues that are important for the strong binding to MERS-CoV PLpro to Ub WT residues reduced the binding affinity.
Figure 25. Binding assays comparing affinities between variants (A and B).
Figure 26. Bindings assay with CCHFV inhibitors.
DETAILED DESCRIPTION OF THE DISCLOSED EMBODIMENTS
Many viruses, including MERS-CoV and the Crime an- Congo hemorrhagic fever virus (CCHFV) encode deubiquitinating (DUB) enzymes that are critical for viral replication and pathogenicity. They bind and remove ubiquitin (Ub) and the Ub-like protein interferon stimulated gene 15 (ISG15) from cellular proteins to suppress host antiviral innate immune responses. A variety of viral DUBs (vDUBs), including the MERS-CoV papain-like protease, are responsible for cleaving the viral replicase polyproteins during replication, and are thereby critical components of the viral replication cycle.
Together, this makes vDUBs highly attractive antiviral drug targets. However, structural similarity between the catalytic cores of vDUBs and human DUBs complicates the development of selective small molecule vDUB inhibitors. Despite intensive efforts, only a handful of inhibitors targeting vDUB proteases have been reported, and none have been approved for clinical use [24].
The present disclosure provides highly selective protein -based inhibitors. Unlike small chemical molecules which bind their target over a small surface area, the protein-based inhibitors disclosed herein are relatively large molecules and bind their target over a larger surface area. This has several advantages including high target selectivity, low cross-reactivity to host proteins, and low toxicity. In addition, the protein-based inhibitors disclosed herein share homology with a host protein (i.e., ubiquitin). Thus the host cells are less likely to view the inhibitors as "foreign" as compared to small chemical molecules. In addition, the protein-based inhibitors disclosed herein are very soluble.
The present disclosure demonstrates that these inhibitors bind vDUBs with high affinity and specificity to inhibit deubiquitination and delSGylation. The disclosure demonstrates that polypeptides having a beta-grasp fold and an F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or a Y at the amino acid position corresponding to 64 of SEQ ID NO: 1 effectively inhibit a viral protease. The present disclosure also provides that inhibitors further having a substitution of V to I at the amino acid position corresponding to 70 of SEQ ID NO: 1 are effective. Although this
amino acid substitution does not appear essential, and while not wishing to be bound by theory, we hypothesize that an He residue at position 70 extends further into a hydrophobic pocket of the vDUB target in comparison to Val and thereby promotes a more favourable hydrophobic interaction.
Accordingly, in one aspect, the disclosure provides an inhibitor comprising a beta- grasp fold, wherein said fold comprises region 1 (amino acids 2-14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l, wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, wherein said substitutions include an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1. Preferably, the inhibitor comprises between 5 to 20 amino acid substitutions, more preferably between 10-15, most preferably between 10-13 amino acid substitutions, as compared to the sequence of regions 1, 2, and 3 of SEQ ID NO: 1. In preferred embodiments, said substitutions further include an amino acid substitution of V to I at the amino acid position corresponding to 70 of SEQ ID NO: 1. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5;
wherein region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5; and/or wherein region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5. Preferably, a) wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5; SEQ ID NO: 12, or SEQ ID NO: 14; b) wherein region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, or SEQ ID NO: 13; and/or c) wherein region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 13, SEQ ID NO: 15.
Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 2, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 3, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 3, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 3. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 4, region 2 has an amino acid sequence
corresponding to amino acids 42-49 of SEQ ID NO: 4, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 4. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 5, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 5, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 5. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 12, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 12, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 12.
Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 13, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 13, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 13. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 14, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 14, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 14. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 15, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 15, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 15.
Preferably, the inhibitors comprise an amino acid comprising SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, or SEQ ID NO: 5. Preferably, the inhibitors comprise an ammo acid comprising SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 12, SEQ ID NO: 13, SEQ ID NO: 14, or SEQ ID NO: 15. It is clear to a
skilled person that additional tags or sequences may be added to the inhibitors. For example Inhibitors having an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1 are referred to herein as "ME inhibitors". Most preferred inhibitors are depicted in figure 1A.
The disclosure further demonstrates that polypeptides having a beta-grasp fold and a G at the amino acid position corresponding to 74 of SEQ ID NO: 1 effectively inhibit a viral protease.
Accordingly, in one aspect, the disclosure provides an inhibitor comprising a beta- grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l, wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and wherein the inhibitor has an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1. Preferably, the inhibitor comprises between 5 to 20 amino acid substitutions, more preferably between 7-9 amino acid substitutions, as compared to the sequence of regions 1, 2, and 3 of SEQ ID NO: 1.
Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10; region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10; and/or region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 6, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 6, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 6. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 7, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 7, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID
NO: 7. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 8, region 2 has an amino acid sequence
corresponding to amino acids 42-49 of SEQ ID NO: 8, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 8. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 9, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 9, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 9. Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 16, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 16, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 16.
Preferably, wherein region 1 has an amino acid sequence corresponding to amino acids 2-14 of SEQ ID NO: 10, region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 10, and region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 10. Inhibitors having an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1 are referred to herein as "CC inhibitors". Most preferred inhibitors are depicted in figure 1A. In some embodiments, the inhibitors disclosed herein are fusion molecules. In a preferred embodiment, the inhibitor is covalently linked with a label or tag allowing the detection and/or isolation of the fusion molecule. Such tags include affinity tags such as a myc-tag, FLAG-tag, His6-tag or HA-tag. In a preferred embodiment, the fusion molecule further comprises one or more modifications increasing the stability of the fusion molecule and/or extending the serum half- life of the fusion molecule. For example, the inhibitor may be conjugated to a polyalkylene glycol molecule.
As used herein, the term "beta -grasp fold" (beta-GF) refers to a common folding structure comprising a beta(2)-alpha-beta(2) motif, where the alpha helix is situated against a 4-stranded mixed beta sheet. Beta-GF are present in, e.g., IF3-N, archaeo- eukaryotic RNA poly beta-sub unit, Ymll08w, BofC, and POZ. A variation of a 4- stranded beta-GF domain is found in Nudix, whereas a "barrelizing version is present in L25, Fasciclin, and phosphoribosyl AMP cyclohdrolase. Beta-grasp folds are
reviewed in Burroughs et al. 2012 Methods in Molecular Biology vol 832, Chapter 2, which is hereby incorporated by reference.
Preferred beta-grasp fold containing scaffolds comprise an additional beta strand. Proteins comprising 5-stranded beta-grasp folds include molybdopterin- dependent oxidoreductase, SLBB, 2Fe-2S ferredoxm, L-proDH alpha, AOR-N, MoaD, TGS, ThiS, TmoB, superantigen toxins, streptokinase, and S4. Most preferred proteins having this structure are members of the ubiquitin-like superfamily which include proteins such as Urml, Apgl2, NeddS, ISG15 and SUMO. Such preferred scaffolds have the features referred to in Burroughs et al. 2012 for "Classic UB-like" scaffolds, namely, a loop connecting strands beta2 and beta3 termed a "lateral shelf; and a at the loop connecting strands beta4 and beta5 termed a "connector arm".
Figure 16 depicts an exemplary beta-GF containing scaffold of the ubiquitin like superfamily. As depicted in Figure 16, region 1 forms strands betal and beta2, region 2 forms beta3, and region 3 forms beta5. Preferred inhibitors share this three dimensional structure and orientation of regions 1, 2, and 3.
It is clear to a skilled person that any beta-GF may be used as a scaffold for presenting regions 1, 2, and 3 as defined herein. While not wishing to be bound by theory, the inventors propose that while the alpha helix of the beta-GF provides structure, it does not bind to the viral proteins. Thus, any similar alpha helix structure can be used (or rather, the primary sequence is not relevant). Preferably, the scaffold is from ubiquitin or a ubiquitin-like protein. As is known to a skilled person, ubiquitin from different species is extremely homologous. The term
"ubiquitin" or "Ub" as used herein refers to ubiquitin from any species or source and includes the full-length protein as well as fragments or portions of the protein.
Human ubiquitin has the amino acid sequence as shown in SEQ ID NO: 1. Preferably, the scaffold is from ubiquitin, more preferably from human ubiquitin.
Preferably, the beta-GF fold is provided by linking region 1 to region 2 by a
polypeptide comprising amino acids 15-41 of SEQ ID NO: 1 or having at least 80% identity to amino acids 15-41 of SEQ ID NO: 1. Preferably, the polypeptide has at
least 90%, or at least 95% identity to amino acids 15-41 of SEQ ID NO: 1. This linkage provides the alpha helix of the beta-GF. Preferably, the beta-GF is also provided by linking region 2 to region 3 by a polypeptide comprising amino acids 50-61 of SEQ ID NO: 1 or having at least 80% identity to amino acids 50-61 of SEQ ID NO: 1.
Preferably, the polypeptide has at least 90%, or at least 95% identity to amino acids 50-61 of SEQ ID NO: 1. As shown in Figure 2, the linkage of region 2 to region 3 does not form part of the beta-GF fold.
Preferably, linkages having at least 80% identify to SEQ ID NO: l differ by
conservative amino acid substitutions. Or rather, one amino acid residue is replaced with another amino acid residue without altering the tertiary structure of the resulting protein. For example, a hydrophobic residue, such as glycine can be substituted for another hydrophobic residue such as alanine. An alanine residue may be substituted with a more hydrophobic residue such as leucine, valine or isoleucine. A negatively charged amino acid such as aspartic acid may be substituted for glutamic acid. A positively charged amino acid such as lysine may be substituted for another positively charged amino acid such as arginine. Conservative substitutions typically include substitutions within the following groups: glycine, alanine; valine, isoleucine, leucine: aspartic acid, glutamic acid, asparagine, glutamine; serine, threonine; lysine, arginine; and phenylalanine, tyrosine. Conserved amino acid substitutions involve replacing one or more amino acids of the polypeptides of the disclosure with amino acids of similar charge, size, and/or hydrophobicity
characteristics. When only conserved substitutions are made, the resulting molecule should retain the same tertiary structure. Preferably the inhibitor comprises an amino acid sequence selected from SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, SEQ ID NO: 10, SEQ ID NO: 12, SEQ ID NO: 13, SEQ ID NO: 14, SEQ ID NO: 15 or SEQ ID NO: 16.
In a further aspect, the disclosure provides nucleic acid molecules encoding said inhibitors. Based on the genetic code, a skilled person can determine the nucleic acid sequences which encode the polypeptide inhibitors disclosed herein. Based on the degeneracy of the genetic code, sixty-four codons may be used to encode twenty amino acids and translation termination signal. As is known to a skilled person, codon usage
bias in different organisms can effect gene expression level. Various computational tools are available to the skilled person in order to optimize codon usage depending on which organism the desired nucleic acid will be expressed. The polypeptide based inhibitors disclosed herein can be produced by any method known to a skilled person. In some embodiments, the inhibitors are chemically synthesized. The inhibitors can also be produced using molecular genetic techniques, such as by inserting a nucleic acid into an expression vector, introducing the expression vector into a host cell, and isolating the polypeptide.
In a further aspect, the disclosure provides vectors which comprise said nucleic acid molecules. Said vectors and plasmids are useful, e.g., for generating transgenic organisms or for expressing said polypeptide inhibitors. A "vector" is a recombinant nucleic acid construct, such as plasmid, phase genome, virus genome, cosmid, or artificial chromosome, to which another DNA segment may be attached. The term
"vector" includes both viral and non-viral means for introducing the nucleic acid into a cell in vitro, ex vivo or in vivo. Preferred vectors are expression vectors. It is within the purview of a skilled person to prepare suitable expression vectors for expressing the inhibitors disclosed hereon. Suitable regulatory sequences including enhancers, promoters, translation initiation signals, and polyadenylation signals may be included. Additionally, depending on the host cell chosen and the vector employed, other sequences, such as an origin of replication, additional DNA restriction sites, enhancers, and sequences conferring inducibility of transcription may be incorporated into the expression vector. The expression vectors may also contain a selectable marker gene which facilitates the selection of host cells transformed or transfected. Examples of selectable marker genes are genes encoding a protein such as G418 and hygromycin which confer resistance to certain drugs, 6- galactosidase,
chloramphenicol acetyl transferase, and firefly luciferase.
Viral vectors include lentivirus, retrovirus, adeno-associated virus (AAV), poxvirus (including MVA), baculovirus, , herpes simplex, Epstein-Barr and adenovirus vectors. Vector sequences may also contain one or more regulatory regions, and/or selectable markers useful in selecting, measuring, and monitoring nucleic acid transfer results (transfer to which tissues, duration of expression, etc.). Lentiviruses have been
previously described for transgene delivery to the hippocampus (van Hooijdonk BMC Neuroscience 2009, 10:2).
There are a variety of techniques available for introducing nucleic acids into viable cells. The techniques vary depending upon whether the nucleic acid is transferred into cultured cells in vitro, or in vivo in the cells of the intended host or animal model. Techniques suitable for the transfer of nucleic acid into mammalian cells in vitro include the use of liposomes, electroporation, microinjection, cell fusion, DEAE- dextran, the calcium phosphate precipitation method, etc. The currently preferred in vivo gene transfer techniques include transfection with viral (typically retroviral) vectors and viral coat protein-liposome mediated transfection (Dzau et al., Trends in Biotechnology 11:205-210 (1993)). The nucleic acids may stably integrate into the genome of the host cell (for example, with retroviral introduction, outlined below), or may exist either transiently or stably in the cytoplasm (i.e. through the use of traditional plasmids, utilizing standard regulatory sequences, selection markers, etc.). Such cells are useful for producing isolated polypeptides, which may be used in the methods described herein. The examples disclosed herein provide exemplary methods and vectors for expressing nucleic acids. Accordingly, the disclosure provides cells and organisms comprising the nucleic acids or vectors comprising said nucleic acids as disclosed herein. Preferably, said cells and organisms express the respective polypeptide inhibitor as disclosed herein. A skilled person can select suitable host cells for cloning and/or expressing the inhibitors. Such cells include bacteria, e.g., E. coli, as well as filamentous fungi, yeast, plant, mammalian and insect cells.
In some embodiments, the cells and organisms are from animals (in particular mammals; e.g., mice, rats, rabbits, bovine, Camelidae family members) In some embodiments the animal is a non-human transgenic animal. In some embodiments, the cells are in vitro or in vivo non-human cells. Preferred host cells for expression include MRC5 cells (human cell line derived from lung tissue), HuH7 cells (human liver cell line), CHO-cells (Chinese Hamster Ovary), COS-cells (derived from monkey kidney (African green monkey), Vero-cells (kidney epithelial cells extracted from
African green monkey), Hela-cells (human cell line), BHK-cells (baby hamster kidney cells, HEK-cells (Human Embryonic Kidney), NSO-cells (Murine myeloma cell line), C127-cells (nontumorigenic mouse cell line), and PerC6®-cells (human cell line, Crucell).
As further described herein, the disclosed inhibitors are useful in therapy.
Accordingly, the disclosure provides pharmaceutical compositions comprising one or more of the polypeptide inhibitors disclosed herein, one or more of the nucleic acid molecules encoding said inhibitors, or one or more vectors disclosed herein. Such pharmaceutical compositions are useful in therapy and for inhibiting viral proteins, as disclosed herein.
Actual dosage levels of the pharmaceutical preparations described herein may be varied so as to obtain an amount of the active ingredient which is effective to achieve the desired therapeutic response for a particular patient, composition, and mode of administration, without being toxic to the patient. The selected dosage level will depend upon a variety of factors including the activity of the particular compound, the route of administration, the time of administration, the rate of excretion of the particular compound being employed, the duration of the treatment, other drugs, compounds and/or materials used in combination, the age, sex, weight, condition, general health and prior medical history of the patient being treated, and like factors well known in the medical arts. A physician or veterinarian having ordinary skill in the art can readily determine and prescribe the effective amount of the
pharmaceutical composition required.
In a further aspect, the disclosure provides the use of the inhibitors disclosed herein and nucleic acids encoding said inhibitors for inhibiting the biological activity of a viral protein as well as providing methods for screening for new inhibitors. Preferably, "viral proteins" as used herein refers to viral ubiquitin binding protein partners, or rather, proteins that bind/interact with ubiquitin or ubiquitin-like proteins. Such proteins include viral proteases with deubiquitinating activity (vDUBs), viral E2 conjugating enzymes, viral E3 ubiquitin ligases, viral El enzymes, and other viral proteins containing ubiquitin binding domains (UBD). Preferably, the viral protein is
a vDUB. Preferably, the inhibitors inhibit the proteolytic cleavage activity of said vDUB.
Ubiquitination is a post-translational modification mediated by an enzyme cascade that results in the conjugation of ubiquitin (Ub) to cellular proteins [1, 2] . This process is regulated in part through activity of cellular deubiquitinating enzymes (DUBs), which remove Ub from cellular proteins [1, 2]. Given the essential role of the Ub system in regulating a large number of critical cellular processes, it is not surprising that viruses have acquired the means to modulate this system in order to promote infection and replication in host cells [3]. In particular, virus-encoded DUBs reverse the ubiquitination process to alter host signaling pathways critical to the induction of cellular antiviral and pro-inflammatory innate immune responses [3]. In addition to removing Ub molecules from host proteins, many viral DUBs (vDUBs) also remove the Ub-like protein interferon- stimulated gene 15 (ISG15) to further suppress antiviral responses [4, 5]. Importantly, a number of vDUBs also play an essential role in viral replication [4-6]. Together, the replicative and/or deubiquitinating activities of viral proteases contribute directly to pathogenesis during viral infection in vivo [7], making them ideal antiviral drug targets. Preferably, the viral protein is from a coronavirus. Coronaviruses initially express their non-structural proteins (nsps) as large viral polyproteins, which are processed into functional domains by proteases encoded within the polyproteins to establish a viral replication-transcriptase complex. SARS- and MERS-CoV release nspl-3 through the activity of a papain-like protease (PLtjro) domain situated within nsp3, in a process that is indispensable for replication [4] , The chymotryp sin- like protease
(3CLPro), corresponding to nsp5, is responsible for cleaving the remaining part of the polyproteins, releasing mature nsps [8]. Strikingly, coronaviral PLtjros also act as vDUBs to suppress host antiviral innate immune responses by targeting cellular Ub- conjugated substrates [9- 14].
Preferably, the ME inhibitors disclosed herein inhibit the biological activity of the MERS-CoV papain-like protease (PLf,ro) domain. Preferably, the inhibitors inhibit the proteolytic cleavage activity of the PLr>n> domain. Preferably, the inhibitors inhibit the
polyprotein processing activity of the PL/""0 domain. Preferably, expression of the inhibitors during MERS-CoV infection reduces infectious progeny titer. Preferably, the ME inhibitors disclosed herein are useful in therapy. In particular, methods are provided for the treatment and/or prevention of Middle East respiratory syndrome coronaviral (MERS-CoV) infection and/or the symptoms thereof, the method comprising administering to an individual in need thereof a therapeutically effective amount of an inhibitor or a nucleic acid molecule encoding said inhibitor,
wherein the inhibitor comprises a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l,
wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and
wherein the inhibitor has an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1. Additional ME inhibitors as disclosed herein may also be used in the treatment method, in particular inhibitors comprising an amino acid sequence selected from SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, , SEQ ID NO: 5, as well as SEQ ID NO: 12, SEQ ID NO: 13, SEQ ID NO: 14, and SEQ ID NO: 15.
In some embodiments, an individual infected with MERS-CoV is an animal such as a member of the Camelidae family. Treatment of reservoir species can reduce transmission to humans. Preferably, the individual is a human. Preferably, said treatment reduces the severity and/or duration of the viral infection and/or reduces the severity and/or duration of symptoms (such as respiratory illness, fever, dyspnea, and myalgia).
Preferably, the viral protein is from the nairovirus Crimean-Congo hemorrhagic fever virus (CCHFV). This pathogenic virus also encodes a vDUB. The CCHFV vDUB domain is located within the large (L) segment of the genome, and has also been explicitly implicated in the evasion of host Ub- and ISG 15- dependent innate immune responses [17]. This domain is also referred to as the CCHFV OTU domain (ovarian tumor (OTU) protease domain ). Preferably, the CC inhibitors disclosed herein inhibit
the biological activity of the CCHFV vDUB domain. Preferably, the inhibitors inhibit the proteolytic cleavage activity of said domain. Preferably, expression of the inhibitors during CCHFV infection reduces infectious progeny titer. Preferably, the CE inhibitors disclosed herein are useful in therapy. In particular, methods are provided for the treatment and/or prevention of Crimean-CongO hemorrhagic fever viral infection and/or the symptoms thereof, the method comprising administering to an individual in need thereof a therapeutically effective amount of an inhibitor or a nucleic acid molecule encoding said inhibitor, wherein the inhibitor comprising a beta- grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l, wherein the inhibitor comprises one or more amino acid mutations in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and wherein the inhibitor has an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1. Additional CC inhibitors as disclosed herein may also be used in the treatment method, in particular inhibitors comprising an amino acid sequence selected from SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10 as well as SEQ ID NO: 16.
In some embodiments, an individual infected with CCHFV is an animal such as ruminants and ostriches. Treatment of reservoir species can reduce transmission to humans. Preferably, the individual is a human. Preferably, said treatment reduces the severity and/or duration of the viral infection and/or reduces the severity and/or duration of symptoms (such as flu-like symptoms, hemorrhage, respiratory distress). In a further aspect, the disclosure provides methods of identifying an inhibitor or a nucleic acid molecule encoding said inhibitor that inhibits the biological activity of a viral protein, the method comprising providing a library of polypeptides and screening said library against a viral ubiquitin binding partner in order to identify inhibitors that bind to said viral ubiquitin binding partner, wherein said polypeptide library comprises at least 1000 distinct polypeptides (or rather each polypeptide varies from every other polypeptide by at least one amino acid residue), wherein each polypeptide comprises a beta-grasp fold comprising region 1 (amino acids 2- 14), region 2
(amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l and comprises at least one amino acid mutation in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l. Such libraries have been previously described in WO 2012/020289 and Ernst et al. Science 2013;339:590-595; which are hereby incorporated by reference in their entirety. As described in these references, each polypeptide of the library is a distinct ubiquitin polypeptide having one ore more amino acid substitutions in region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and/or region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l. In some embodiments, the polypeptide has at least 2, at least 4, at least 8, at least 10, at least 15, or at least 20 amino acid substitutions. Preferably, between 5-15 amino acid substitutions, more preferably between 7- 13 substitutions. Preferably, the polypeptides comprise two additional amino acids at the C-terminus. Preferably, the library comprises at least 106, 108, or 109 distinct polypeptides.
The library can be screened for inhibition of a viral protein, for example, by phage display, mRNA display, ribosome display, yeast display or other similar technologies to determine the inhibition of biological activity compared to a control. In one embodiment, the control is a different protein to test for specificity. Example 1 provides an exemplary embodiment of methods to screen for candidate inhibitors.
Preferably, the methods comprise purifying the candidate inhibitors and testing them in additional in vitro and/or in vivo assays in order to determine their effect on viral infection.
Preferably, the viral ubiquitin binding partner is a viral proteases with
deubiquitinating activity (vDUBs), a viral E2 conjugating enzyme, a viral E3 ubiquitin ligase, a viral El enzyme, or other viral proteins containing ubiquitin binding domains (UBD). Preferred viral ubiquitin binding partner are discussed in Wimmer and Schreiner, Viruses 2015 7:4854-4872, which is hereby incorporated by reference.
The present disclosure demonstrates that polypeptide libraries, such as those described in WO2012/020289 and Ernst et al. can be used to rapidly identify viral inhibitors. The present disclosure provides vDUBs as a preferred target for such libraries. vDUBs generally have a low affinity for ubiquitin. Surprisingly, the present disclosure demonstrates that high affinity polypeptide inhibitors can be identified using the disclosed methods. In addition, the large binding surface of the candidate polypeptide inhibitors offers the potential to target a diverse range of vDUBs.
Such screening methods are advantageous over screening small chemical compound libraries. In a typical small chemical compound library screen, the probability of identifying a compound that specifically binds it's target is quite small. Candidates that bind a target with low or moderate affinity may be identified. These candidates must be further chemically optimized in order to produce a compound with the potential to bind with high affinity to its target. In contrast, the screening methods disclosed herein can identify highly specific inhibitors without the need for further optimization. Thus the disclosed screening methods are faster and more efficient than small chemical compound screening. A skilled person will appreciate that with the continual emergence of new viruses in the human population, rapid methods for identifying new viral inhibitors is crucial.
Sequence Listing
SEQ ID NO: 1 wild- type ubiquitin
MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLS DYNIQKESTLHLVLRLRGG SEQ ID NO: 2 ME.1
MHILVKTHTGNTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFFGSQLEDGRTLS
DYNINKYSTLHLVLRLNGRKF
SEQ ID NO: 3 ME.2
MRIFVETLRRLTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFFGQQLEDGRTLS DYNI VKYSTLH LI LRLPR WN SEQ ID NO: 4 ME.3
MHILVKTHTGNTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFFGSQLEDGRTLS DYNINKYSTLHLVLRLNGWNV
SEQ ID NO: 5 ME.4
MRIFVETLRRLTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFFGQQLEDGRTLS
DYNIVKYSTLHLILRLNSWY
SEQ ID NO: 6 CC. l
MQIFVKTLKGHVITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFALQQLEDGRTLS DYNIQKESNLHLVLRLGGGRR SEQ ID NO: 7 CC.2
MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLS DYNIRKQSNLYLLWRLGGWKK SEQ ID NO: 8 CC.3
MYIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLS
DYNIHNESPLRLLWRLGGFRR
SEQ ID NO: 9 CC.4
MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLS DYNIQKLSLLYLVWRLGVPWV SEQ ID NO: 10 CC.5
MQIYVKTFKGTTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAFKQLEDGRTLS
DYNIDKESTLQLVLRLGGWAS
SEQ ID NO: 11 ME4.1
MRIFVETLRRLTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGQQLEDGRTLS DYNIVKESTLHLVLRLNSWY SEQ ID NO: 12 ME.4.2:
MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFFGQQLEDGRTLS DYNIVKYSTLHLILRLNSWY SEQ ID NO: 13 ME.4.3:
MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFFGKQLEDGRTLS
DYNIQKYSTLHLILRLNSWY
SEQ ID NO: 14 ME.4.4:
MHILVKTHTGNTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFFGQQLEDGRTLS DYNIVKYSTLHLILRLNSWY SEQ ID NO: 15 ME.4.6:
MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFFGQQLEDGRTLS DYNIVKYSTLHLILRLRGG
SEQ ID NO: 16 CC.4.1
MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLS DYNIQKLSLLYLVWRLGVPW Definitions
As used herein, "to comprise" and its conjugations is used in its non-limiting sense to mean that items following the word are included, but items not specifically mentioned are not excluded. In addition the verb "to consist" may be replaced by "to consist essentially of meaning that a compound or adjunct compound as defined herein may comprise additional component(s) than the ones specifically identified, said additional component(s) not altering the unique characteristic of the invention.
The articles "a" and "an" are used herein to refer to one or to more than one (i.e., to at least one) of the grammatical object of the article. By way of example, "an element" means one element or more than one element.
As used herein, the terms "treatment," "treat," and "treating" refer to reversing, alleviating, delaying the onset of, or inhibiting the progress of a disease or disorder, or one or more symptoms thereof, as described herein. In some embodiments, treatment may be administered after one or more symptoms have developed. In other
embodiments, treatment may be administered in the absence of symptoms. For example, treatment may be administered to a susceptible individual prior to the onset of symptoms (e.g., in light of a history of symptoms and/or in light of genetic or other susceptibility factors). Treatment may also be continued after symptoms have resolved, for example to prevent or delay their recurrence.
All patent and literature references cited in the present specification are hereby incorporated by reference in their entirety.
The invention is further explained in the following examples. These examples do not limit the scope of the invention, but merely serve to clarify the invention.
EXAMPLES
Example 1. Potent and selective inhibitors of MERS-CoV and CCHFV vDUBs
A polypeptide library [25] was screened against the MERS-CoV PLpro domain (MERS-CoV PLpro) and the CCHFV OTU domain (CCHFV OTU). Inhibitors were
identified that bound with high affinity to either MERS-CoV PLpro (ME. l to ME.4) or CCHFV OTU (CC. l to CC.5) (Fig. 1A). To confirm the specificity of the inhibitors towards their cognate vDUBs, the phage-displayed inhibitors were challenged against a diverse panel of 11 DUBs from several species representing distinct DUB families (USP, OTU, and ubiquitin C-terminal hydrolases (UCH)). Indeed, all inhibitors bound only to their cognate viral proteins but not to the 11 additional DUBs tested (Fig. IB). To determine the binding affinities of each identified inhibitor, phage enzyme-linked immunosorbent assays (Fig. 5) and Bio-Layer Interferometry (BLI) measurements were performed (Table 4). Each inhibitor was found to bind its cognate vDUB with affinities in the low to sub -nanomolar range, whereas wild-type Ub (Ub.wt) showed binding to MERS-CoV or CCHFV vDUBs only in the high micromolar range (Fig. 5B and Table 2). Consistent with the high affinities observed for the inhibitors toward their respective vDUBs, each inhibitor also potently inhibited the deubiquitinating and delSGylating activities of MERS-CoV PLpro or CCHFV OTU as measured using the fluorogenic substrates Ub-AMC or ISG15-AMC, respectively (Fig. 1C and Fig. 6). The most potent inhibitors of MERS-CoV PLpro and CCHFV OTU were ME.4
(deubiquitination IC50 = 0.8 nM and delSGlyation IC50 = 1.2 nM) and CC.4
(deubiquitination IC50 = 3.3 nM and delSGlyation IC50 = 11 nM), respectively (Table 2). Furthermore, the inhibitors were confirmed to inhibit processing of K48- and K63- linked tetra-Ub substrates by their respective vDUBs (Fig. ID).
Example 2. Structural basis for vDUB inhibition
To reveal the molecular basis for the inhibition of MERS-CoV PLpro by inhibitors, crystal structures of the enzyme were determined bound to ME.2 or ME.4 (Fig. 2A,B and Table 1). Both inhibitors bound in nearly identical orientations as Ub.wt (Fig. 2C and Fig. 7A) with interface surface areas of -1000 A2 [9, 27]. Substitutions common to both ME.4 and ME.2 at positions 46, 64 and 70 (Fig. 1A) were found to promote more favorable hydrophobic interactions with the enzyme relative to Ub.wt. Inhibitor residue He 70 extends further into a hydrophobic pocket of PLpro formed by residues Thrl730* and Vall691* (asterisks denote amino acid numbering of MERS-CoV polyprotein), in comparison to Ub.wt residue Val70 (Fig. 2D). ME.2 and ME.4 residue Phe46 inserts into a hydrophobic pocket formed by PLpro residues Trpl668*,
Glul670*, Vall680*, Leul682*, Tyrl690* and Tyrl705*. and forms a cation-π
interaction with Argl715* (Fig. 2E). These extensive interactions are not formed at the PLpro-Ub.wt interface with the homologous residue Ala46. In addition, ME.2 and ME.4 residue Tyr64 undergoes more extensive hydrophobic interaction with Vall706* and Glyl71()*, compared to Ub.wt residue Glu64 (Fig. 2F). The inhibitors also differ in their C-terminal residues at positions 74, 75 and 77 (Fig. 1A). In ME.4, Asn74 extends into the active site and forms a hydrogen bonding network with Asp 1645* and
Glyl758* (Fig. 2G), which mimics hydrogen bonds formed by the same region of MERS-CoV PLpro with Ub.wt residues Arg74, Gly75 and Gly76 (Fig. 2H). In ME.2, Pro74 cannot form an analogous hydrogen bonding network, which likely contributes to the decreased inhibitory potency of ME.2 compared to ME.4 (Fig. ID and Fig.
ΙΟΑ,Β). Instead, Pro74 and the C-terminal tail are excluded from the active site and the main-chains interact through a hydrogen bond formed between the main-chain amide nitrogen of Arg75 and the backbone carbonyl of Glul754* (Fig. 21). A structural comparison of the C-terminal regions of ME.2 and ME.4 near the PLpro active site is shown in Fig. 8. Additional substitutions in the N-terminal region of ME.4 and ME.2 (Fig. 1A, region 1) do not make favourable contacts with MERS-CoV PLpro, and in fact, residues 8-10 and 7- 10 of ME.4 and ME.2, respectively, failed to resolve in electron density maps (Fig. 9). Taken together, our structural analyses revealed that a small subset of substitutions is sufficient to enhance hydrophobic packing and hydrogen bond networks to endow ME.4 and ME.2 with highly potent and specific inhibitory activity against MERS-CoV PLpro.
Similarly, crystal structures of CCHFV OTU were determined bound to CC.2 and to CC.4 to gain insight into how they selectively block the DUB and delSGylating activities of this viral enzyme (Fig. 3A,B and Table 1). CC.2 and CC.4 were found to bind in the same orientation as Ub.wt with similar buried surface areas of -1000 A2 (Fig. 3C and Fig. 7B) [20, 21]. Interestingly, substitutions in these inhibitors were concentrated in the C-terminal region (Fig. 1A) and only substitutions at position 68 and downstream were found to interact with the enzyme. In both inhibitors, Tyr68 improves hydrophobic packing with ThrlO*, Vall2* and Vall8* (asterisks denote amino acid numbering of the large segment-encoded protein of CCHFV), relative to His68 in Ub.wt (Fig. 3D). In CC.2, residue Leu70 projects further into a hydrophobic cavity of CCHFV OTU formed by residues Vall2*, He 14*, Vall8* and He 131*, than
the equivalent Ub.wt residue Val70 (Fig. 3E). The conformational freedom of a Gly substitution at position 74 enables the C-terminal tail of each inhibitor to form numerous favorable interactions with the enzyme. Residues Gly75 and Val75 of CC.2 and CC.4, respectively, occupy space on the CCHFV OTU surface, which in the case of Ub.wt is occupied by the side-chain of Arg74 (Fig. 3 E-G). This alternative
conformation permitted by Gly74 in CC.2 and CC.4 allows the side-chain of Trp76 or Trp77 of CC.2 or CC.4, respectively, to pack within different adjacent grooves near the CCHFV OTU active site, with Trp76 of CC.2 forming a cation-π interaction with Arg92* (Fig. 3E), and the amide nitrogen group of CC.4 residue Trp77 forming a hydrogen bond with the side chain of Gin 149* (Fig. 3F). Conversely, Trp71 in both CC.2 and CC.4 does not interact with CCHFV OTU, but instead packs into a hydrophobic cavity within each inhibitor (Fig. 3E,F). Differences between the orientations of the C-terminal tails of CC.2 and CC.4 arise from the variation at positions 75 and 76. In CC.4, residue Val75 packs against CCHFV OTU residue Trp99*, and Pro 76 appears to restrict conformational freedom and enable hydrophobic interactions with Trp99* and Thrl50* (Fig. 3F). These additional contacts, together with the unique packing orientation of Trp77, likely account for the remarkably high binding affinity of CC.4 for CCHFV OTU (Table 4). See example 6 for a description of mutations in the tail region which effect binding.
Example 3. Inhibition of MERS-CoV PLpro activity in cell culture-based assays To explore the effects of inhibitors on MERS-CoV PLpro activity in cells,
deubiquitination assays were performed by transfecting cells with combinations of plasmids encoding the following proteins: HA-tagged Ub , MERS-CoV PLpro, and inhibitors. Unlike Ub which becomes conjugated to cellular proteins by its C-terminal glycine residue, the inhibitors have substitutions in the C-terminal di-Gly motif.
Thus, the inhibitors cannot interfere with the natural ubiquitin regulatory machinery, further avoiding toxicity/adverse effects to the cell.
A clear decrease of cellular HA-Ub conjugates was observed during co-expression of MERS-CoV PLpro, while there was no effect upon expression of a catalytically inactive mutant (Fig. 4A, compare lanes 3 and 4). The co-expression of increasing doses of different inhibitors attenuated MERS-CoV PLpro DUB activity to varying
degrees (Fig. 4A and Fig. 10A). In a dose- dependent manner, ME.4 co-expression resulted in severe inhibition of HA-Ub deconjugation mediated by MERS-CoV PLpro, whereas co-expression of an unconjugatable form of Ub.wt as a negative control (Ub.AA, which contains Gly75Ala and Gly76Ala substitutions) had no effect on the DUB activity of MERS-CoV PLpro at any dose (Fig. 4A, compare lanes 5-7 to 8- 10). Like ME.4, ME.2 had a strong effect resulting in near complete inhibition of MERS- CoV PLpro DUB activity at the lowest inhibitor dose, whereas the inhibitory effect of ME. l and ME.3 was explicit only at higher doses (Fig. 10A). Notably, none of the inhibitors inhibited the DUB activity of the closely related SARS-CoV PLpro, highlighting their specificity for MERS-CoV PLpro (Fig. 11A). A superposition of
ME.4 onto the Ub domain of a previously determined SARS-CoV-Ub complex suggests that Phe46 and Ile70 of ME.4 clash with Vall88* and Met209* of SARS-CoV PLpro, respectively, and offers a plausible explanation for the specificity of ME.2 and ME.4 toward MERS-COV PLpro (Fig. 12).
We previously found that the DUB activity of MERS-CoV PLpro suppresses IFN-β promoter activity upon activation of cellular innate immune signaling [9]. In a luciferase-based IFN-β reporter assay we show that ectopically expressed inhibitors competed with endogenous Ub for binding to MERS-CoV PLpro resulting in an alleviated suppression of the IFN-6 promoter activity (Fig. 4B and Fig. 10B).
Consistent with described binding and inhibition data, ME.2 and ME.4 were more potent than ME. l and ME.3 at blocking the ability of MERS-CoV PLpro to suppress IFN-β promoter activation, whereas none of the inhibitors were able to block suppression of the IFN-β promoter activity by SARS-CoV PLpro (Fig. 11B). The inhibitors thus prevented MERS-CoV PLpro-mediated suppression of cellular antiviral innate immune responses, and in a remarkably selective, virus-specific manner.
A critical step in the replication cycle of MERS-CoV is the processing of viral polyproteins into functional non-structural proteins (nsps) that is accomplished in part by the protease activity of PLpro, which cleaves the nsp lj,2, nsp2|3, and nsp3|4 junctions. In order to assess the ability of inhibitors to inhibit MERS-CoV PLpro- mediated polyprotein processing activity, an in trans cleavage assay was performed [9]. FLAG-tagged inhibitors and V5-tagged MERS-CoV PLpro were co-expressed with
N-terminally HA-tagged and C-terminally V5-tagged nsp3C-4 (HA-nsp3C-4-V5), a fragment of the viral polyprotein encompassing the C-terminal part of nsp3 (excluding the PLpro domain) and nsp4. In trans cleavage of the nsp3|4 junction is indicative of proteolytic cleavage activity of PLpro during infection [9]. MERS-CoV PLpro efficiently cleaved HA-nsp3C-4-V5 into HA-nsp3C and nsp4-V5 products, whereas the active site mutant did not (Fig. 4C, compare lanes 3 and 4). The cleavage of the nsp3J,4 site was not affected upon expression of the negative control Ub.AA, whereas only a fraction of HA-nsp3C-4-V5 was cleaved upon expression of ME.4 or ME.2 at the lowest dose, and cleavage was completely blocked at higher doses (Fig. 4C compare lanes 5-7 to 8- 10, Fig. IOC). Increasing doses of ME. l and ME.3 also resulted in reduced cleavage as gradually more HA-nsp3C-4-V5 precursor was observed (Fig. IOC).
Example 4. Inhibitors block MERS-CoV replication in cells
To directly assess the ability of inhibitors to inhibit MERS-CoV replication, MERS- CoV PLpro-specific inhibitors were ectopically expressed in cell culture, and cells were subsequently infected with MERS-CoV. MRC5 and HuH-7 cell lines were transduced with lentiviruses encoding FLAG-tagged inhibitors, Ub.AA, or GFP. Efficient expression of FLAG-ME. l and GFP in these cells was confirmed by fluorescence microscopy and by western blotting (Fig. 13). Either 32 or 48 hours post-transduction, cells were infected with MERS-CoV at a multiplicity of infection of 0.01, and MERS- CoV titers were determined from supernatants harvested 32 hours post infection (Fig. 4D). In MRC5 cells, ME. l and ME.4 expression resulted in significantly lower virus titers as these dropped from 5 x 105 plaque forming units (PFU)/ml recovered from control cells to 1,000 or 10 PFU/ml, respectively, when the MERS-CoV infection was started 32 hours post-transduction (Fig. 4D). The effect of the inhibitors was even more pronounced in MRC5 cells that were infected with MERS-CoV 48 hours post- transduction, as virus titers dropped below 10 PFU/ml upon expression of ME.4, which represented a reduction in infectious progeny titers of more than four orders of magnitude (Fig. 4D) and correlated with higher expression of the inhibitors at this time point (Fig. 13 and 14). In HuH-7 cells the expression of GFP or Ub.AA did not affect MERS-CoV titers compared to the non- transduced cells, whereas ME. l expression led to a two orders of magnitude reduction in virus titer, and an even
greater reduction of more than three orders of magnitude was observed upon ME.4 expression (Fig. 15). The effect of inhibitors on MERS-CoV progeny titers was more severe in MRC5 cells compared to HuH-7 cells, which might be due to generally higher expression of inhibitors in MRC5 cells than in HuH-7 cells (Fig. 3D, 13B and 15). Taken together, these studies show that the inhibitors readily inhibit the proteolytic activities of MERS-CoV PLpro in cells and provide extremely effective protection from MERS-CoV infection.
Example 5. Modification of ME.4 inhibitor
ME.4 was mutated at position 46 from F to A, at position 64 from Y to E, and at position 70 from I to V to generate ME.4.1 (SEQ ID NO: 11). We expressed ME4.1 in MRC5 or HuH-7 cells using the lentivirus -based expression system and infected the transduced cells with MERS-CoV. Compared to ME.4, ME.4.1 was less effective in inhibiting MERS-CoV replication, highlighting the importance of the F46, Y64 and 170 residues in ME.4 for the inhibitory effect (Figure 17). IC50 and EC50 values (Table 3) were also determined as described herein (see Figure 18).
Example 6. Modification of CC.4 inhibitor
CC.4 was mutated to remove 1 to 4 amino acids of the C-terminus (CC.4.1-CC.4.4) or to mutate position 75 from V to G (CC.4.5). Binding curves of these inhibitors to CCHFV OTU are shown in Figure 26 and EC50 values were determined and are listed in Table 5. The results demonstrate that residues V75, P76 and W77 in the tail region of CC.4 play a role in the high binding affinity to CCHFV OTU. Example 7 Modification of ME. l and ME.3 inhibitors
ME.1.1 and ME.3.1 were generated to determine whether the binding affinity of ME. l and ME.3 can be increased by introducing the V70I substitution. As shown in Table 5, this mutation does not increase the binding affinity to MERS-CoV PLpro compared to their original variants, ME. l and ME.3.
Example 8. Inhibitors with increased stability
Upon expression in E. coli, it was apparent the ME. l and ME.3 were significantly more soluble that ME.2 and ME.4. Further preliminary experiments using differential
scanning calorimetry (DSC) determined that ME. l and ME.3 exhibited higher Tm values (83.7°C and 83.9°C respectively) than ME.2 and ME.4 (47.5°C and 46.6°C respectively) (Fig. 20). Analysis of the primary sequences of ME.2 and ME.4 showed that a number of mutations which are unique to these inhibitors near the N-terminus appeared to be uninvolved in PLpro binding ( Zhang, W., Bailey-Elkin, B. A., Knaap, R. C. M., Khare, B., Dalebout, T. J., Johnson, G. G., van Kasteren, P. B., McLeish, N. J., Gu, J., He, W., Kikkert, M., Mark, B. L., and Sidhu, S. S. (2017) Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants. PLOS Pathogens 13, e 1006372), indicating that mutations at these positions may impart destabilizing effects. Specifically, Ub residue GlylO has been mutated to arginine in ME.2 and ME.4, and this residue is known to be critical for the formation of a Gl-type β-bulge, which is formed at the juncture of strands 61 and 62 of Ub (Chen, P. Y., Gopalacushina, B. G., Yang, C. C., Chan, S. I., and Evans, P. A. (2001) The role of a beta-bulge in the folding of the beta-hairpin structure in ubiquitin. Protein Sci. 10, 2063-2074). The crystal structures of ME.2 and ME.4 are lacking electron density in the region representing the Ub 6-bulge, indicating that this region may be
unstructured in the inhibitors. As a final indication that this 6-bulge region may be implicated in the destabilization of ME.2 and ME.4, previous work on the folding dynamics of Ub have suggested that the hairpin structure formed by strands 61 and 62 form a folding nucleus, representing an ensemble of high energy folding
intermediates which must be passed through in order to reach the folded tertiary Ub structure ( Piana, S., Lindorff-Larsen, K., and Shaw, D. E. (2013) Atomic-level description of ubiquitin folding. Proceedings of the National Academy of Sciences of the United States of America 110, 5915-5920). These observations together suggest that the stability of the first- generation inhibitors can be improved.
In order to assess the role of position 10 in stability, an ArglOGly mutation was introduced into the highest affinity ME.4 (ME.4R10G), reverting the position to wild- type with respect to Ub. In order to assess the Tm of ME.4R10G, thermal shift assays were performed in the presence of Sypro® orange dye, which fluoresces upon interaction with the hydrophobic core of unfolded proteins ( Pantoliano, M. W.,
Petrella, E. C, Kwasnoski, J. D., Lobanov, V. S., Myslik, J., Graf, E., Carver, T., Asel, E., Springer, B. A., Lane, P., and Salemme, F. R. (2001) High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen 6, 429-
440). The measured Tm values for ME.2 and ME.4 using this method were 53.3 ± 0.2°C and 51.4 ± 0.2°C, respectively, consistent with the DSC data (Fig. 21). Thermal shift analysis of ME.4R10G determined a Tm of 71.6 ± 0.1°C, demonstrating that the introduction of an ArglOGly mutation at position 10 of ME.4 stabilized the inhibitor, increasing the Tm by ~20°C.
Materials and Methods
Selection of inhibitors
The phage-display library used in this study was re-amplified from Library 2 as previously described [25] . Protein immobilization and the following inhibitor selections were done according to established protocols [26, 45]. Briefly, purified viral proteases were coated on 96-well MaxiSorp plates (Thermo Scientific 12565135) by adding 100 μΕ of 1 μΜ proteins and incubating overnight at 4°C. Afterwards, five rounds of selections using the phage-display library were performed against immobilized proteins including the following steps: (a) Each phage particle in the library pool displays a unique candidate inhibitor and encapsulates the encoding DNA; (b) Binding phages are captured with an immobilized protein; (c) Non-binding phages are washed away; and (d) Bound phages are amplified by infection of bacteria. The enriched phage pool is cycled through additional rounds of selection to further enrich for protein-binding candidate inhibitors. After the fifth round of binding selections individual phages with improved binding properties were identified by phage ELISA using established techniques and subjected to DNA sequencing of the phagemids to obtain candidate inhibitor sequences [26, 45] .
Protein crystallization
MERS-CoV PLpro-ME.2 and -ME.4 complexes
To form the non-covalent MERS-CoV PLpro-inhibitor complexes, a 4-fold molar excess of ME.4 or ME.2 was incubated with MERS-CoV PLpro overnight at 4°C. The excess, unbound inhibitors were removed from the sample using a Superdex 75 size exclusion column and fractions containing the MERS-CoV PLpro- inhibitor complex were pooled and concentrated to 10 mg/mL. The MERS-CoV PLpro-ME.4 complex was found to crystallize under similar conditions to those previously reported for the MERS-CoV PLpro-Ub complex [9], with optimal crystals appearing in 0.1 M trisodium citrate pH 5.6, 20% (w/v) polyethylene glycol (PEG) 4000 and 20% (v/v) isopropanol. Crystals of
the MERS-CoV PLpro-ME.2 complex were grown in 0.1 M trisodium citrate pH 5.6, 19% (w/v) PEG 4000 and 19% (v/v) 1,2-isopropanediol. Crystals were grown by mixing PLpro- inhibitor (10 mg/niL and 9 mg/mL for PLpro-ME.4 and PLpro-ME.2, respectively) with crystallization solution at a 1: 1 volumetric ratio (2 μΕ MERS-CoV PLpro-inhibitor + 2 μΕ well solution). Immediately prior to mixing, 1 M DTT was added to the MERS-CoV PLpro- inhibitor complexes to a final concentration of 10 mM to prevent oxidation of the sample.
CCHFV OTU169-CC.2 and CCHFV OTU185-CC.4 complexes
Purified CCHFV OTU was pooled with 2-3-fold molar excess of purified inhibitor and dialyzed overnight against 50 mM Tris pH 8.0, 150 mM NaCl and 2 mM DTT. Protein complexes were concentrated and loaded onto a Superdex 75 size exclusion column and eluted in 50 mM Tris, 150 mM NaCl and 2 mM DTT. For all samples, a single peak corresponding to the respective complex was observed in the gel filtration profile and two bands corresponding to the CCHFV OTU and respective inhibitor were observed by SDS-PAGE, indicating the high purity of the complexes. The CCHFV OTU169-CC.2 complex was concentrated to 12 mg/ml for crystallization trials, and initial crystals and crystalline material obtained from preliminary screens were used to prepare seed stocks for microseed matrix screening [46, 47], which was set up for the hanging drop vapor diffusion method in 48-well VDX plates (Hampton Research) and carried out using conventional screens (Qiagen) at 4°C, with and without heterogeneous nucleation using 0.3-0.4 cm strands of human hair [48] . Total drop volume was 2 μΐ containing equal volumes of the protein complex and the well solution. Crystals of the CCHFV OTU169-CC.2 complex were grown in 30% (w/v) PEG 4000, 0.2 M CaC12 and 0.1 M HEPES pH 7.5 and appeared after 5-8 days.
The CCHFV OTU185-CC.4 complex was concentrated to 23 mg/ml and initial leads were observed with a combinatorial approach using microseed matrix screening with crystallization screens (Qiagen) along with the Silver Bullets screen (Hampton Research) and micro-seeding using crystals of a CCHFV OTU185-CC.5 (a weaker binding variant selected by phage display) complex. Using the hanging drop vapor diffusion method and 48-well VDX trays (Hampton Research), screens were set up at 20°C with a reservoir volume of 150 μΐ, and a drop size of 3.5 μΐ, which comprised of 1.5 μΐ of the protein complex, 1 μΐ of the reservoir solution and 1 μΐ of Silver Bullet additive, added in this order. Crystals were obtained with 25% (w/v) PEG 3350, 0.1 M
Tris pH 7.0 and 0.2 M sodium chloride. The Silver Bullet formulation in the drop was as follows: 0.16% (w/v) each of 5-Sulfosalicylic acid dehydrate, dodecanedioic acid, hippuric acid, mellitic acid, oxalacetic acid, suberic acid and 0.02 M HEPES sodium pH 6.8.
Data collection procedures are described in Supporting Information.
Lentivirus transduction and MERS-CoV infections
To produce lentiviruses, HEK293T cells (Virgin lab, Washington University School of Medicine) grown in a T175 flask were transfected with packaging vectors
pMDLg/pRRE and pRSV-REV, envelope protein-expressing vector pCMV-VSVG [49] and the transfer vector (pLenti6.3/TC)/V5-DEST containing GFP or FLAG- inhibitors) using polyethylenimine (PEI; Polysciences Inc.). Medium was replaced 24 h post transfection and 48 h and 72 h post transfection supernatant was collected, centrifuged (1000 x g for 10 min) and filtered through a 0.45 μηι filter before storage at -80°C. Titers of the lentivirus particles were determined by p24 antigen enzyme- linked immunosorbent assay (ELISA; ZeptoMetrix). HuH-7 (Bartenschlager lab,
Heidelberg University) and MRC5 cells (CCL-171; American Type Culture Collection) grown in a 12-wells plate were transduced with lentiviruses encoding GFP or FLAG- ME. l diluted in DMEM containing 2% FCS and 8 μg/ml Polybrene (Sigma Aldrich). Medium was replaced 24 h post transduction (pt), and 32 h or 48 h pt protein lysates were obtained by adding 250 μΐ 2xLSB containing 25 mM NEM to each well while cells grown on coverslips were fixed with 3%) paraformaldehyde (PFA) in PBS. GFP and FLAG-ME. l expression were analyzed by Western blotting as described in the supporting information. Fixed cells that were grown on coverslips were permeabilized with 0.1%) Triton X-100 in PBS and subsequently indirect immunofluorescence assays were carried out. Primary and secondary antibodies were diluted in PBS containing 5% FCS and Hoechst 33258 was added to the secondary antibody dilution to stain nuclear DNA. Coverslips were analyzed with a Zeiss Axioskop 2 fluorescence microscope with an Axiocam HRc camera. To obtain a cell population in which >90% of the cells expressed GFP, the amount of lentivirus yielding 120 ng of p24 was required to transduce 1x105 HuH-7 cells and 40 ng of p24/lxl()5 MRC5 cells was required. HuH-7 or MRC5 cells (lxl05/12-well) were transduced with lentiviruses encoding GFP, FLAG-Ub.AA, FLAG-ME. l or FLAG-ME.4. Cells were infected with MERS-CoV with a multiplicity of infection of 0.01 32 h or 48 h pt. MERS-CoV mocula
were prepared in PBS containing 50 μg/ml DEAE-dextran and 2% FCS. Cells were inoculated for 1 h at 37°C and the inoculum was replaced with EMEM containing 2% FCS. Supernatants were harvested 32 h post MERS-CoV infection and
simultaneously cells were lysed in 4x LSB for Western blotting analysis. MERS-CoV titers were determined by plaque assays on Vero cells (Department of Viroscience, Erasmus Medical Center) as described by van den Worm et al. [50]. MERS-CoV infection experiments were performed at least twice and plaque assays were performed in duplicate in order to determine MERS-CoV titers. Work with MERS- CoV was performed inside biosafety cabinets in Biosafety Level 3 facilities at Leiden University Medical Center.
References
1. Komander D, Rape M. The ubiquitin code. Annual review of biochemistry. 2012;81:203-29. Epub 2012/04/25. doi: 10.1146/annurev-biochem-060310- 170328. PubMed PMID: 22524316.
2. Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016; 18(6):579-86. doi: 10.1038/ncb3358. PubMed PMID: 27230526.
3. Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nature reviews Immunology. 2012; 12(l):35-48. doi: 10.1038/nri3111. PubMed PMID: 22158412; PubMed Central PMCID: PMC3864900.
4. Mielech AM, Chen Y, Mesecar AD, Baker SC. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and delSGylating activities. Virus Res. 2014; 194: 184-90. doi: 10.1016/j.virusres.2014.01.025. PubMed PMID: 24512893; PubMed Central PMCID: PMCPMC4125544.
5. Morales DJ, Lenschow DJ. The antiviral activities of ISG15. Journal of molecular biology. 2013;425(24):4995-5008. doi: 10.1016/j.jmb.2013.09.041. PubMed PMID: 24095857; PubMed Central PMCID: PMCPMC4090058.
6. Chenon M, Camborde L, Cheminant S, Jupin I. A viral deubiquitylating enzyme targets viral RN A- dependent RNA polymerase and affects viral infectivity. The EMBO journal. 2012;31(3):741-53. doi: 10.1038/emboj.2011.424. PubMed PMID: 22117220; PubMed Central PMCID: PMCPMC3273391.
7. Isaacson MK, Ploegh HL. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell host & microbe. 2009;5(6):559-70. Epub 2009/06/17. doi: 10.1016/j.chom.2009.05.012. PubMed PMID: 19527883.
8. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nature Reviews Microbiology.
2016; 14:523-34. doi: doi: 10.1038/nrmicro.2016.81.
9. Bailey-Elkin BA, Knaap RC, Johnson GG, Dalebout TJ, Ninaber DK, van Kasteren PB, et al. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papaindike protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. The Journal of biological chemistry. 2014;289(50):34667-82. doi:
10.1074/jbc.M114.609644. PubMed PMID: 25320088; PubMed Central PMCID:
PMCPMC4263872.
10. Mielech AM, Kilianski A, Baez-Santos YM, Mesecar AD, Baker SC. MERS-CoV papaindike protease has delSGylating and deubiquitinating activities. Virology.
2014;450-451:64-70. doi: 10.1016/j.virol.2013.11.040. PubMed PMID: 24503068;
PubMed Central PMCID: PMCPMC3957432.
11. Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, et al.
Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84(9):4619-29. doi: 10.1128/ JVI.02406-09. PubMed PMID: 20181693; PubMed Central PMCID: PMC2863753.
12. Xing Y, Chen J, Tu J, Zhang B, Chen X, Shi H, et al. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. The Journal of general virology. 2013;94(Pt 7): 1554- 67. doi: 10.1099/vir.0.051169-0. PubMed PMID: 23596270.
13. Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. 2007;282(44):32208-21. doi: 10.1074/jbc.M704870200. PubMed PMID: 17761676; PubMed Central PMCID:
PMCPMC2756044.
14. Zheng D, Chen G, Guo B, Cheng G, Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008; 18(11): 1105-13. doi: 10.1038/cr.2008.294. PubMed PMID: 18957937.
15. Baez-Santos YM, St John SE, Mesecar AD. The SARS -coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds.
Antiviral Res. 2015; 115:21-38. doi: 10.1016/j.antiviral.2014.12.015. PubMed PMID: 25554382.
16. Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085-96. doi:
10.1111/febs.12936. PubMed PMID: 25039866.
17. Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG 15- dependent innate immune responses. Cell Host Microbe. 2()07;2(6):404- 16. doi: 10.1016/j.chom.2007.09.014. PubMed PMID: 18078692; PubMed Central PMCID: PMCPMC2184509.
18. Lei J, Mesters JR, Drosten C, Anemuller S, Ma Q, Hilgenfeld R. Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features. Antiviral research. 2014; 109:72-82. doi: 10.1016/j.antiviral.2014.06.()l l. PubMed PMID: 24992731.
19. Ratia K, Saikatendu KS, Santarsiero BD, Barretto N, Baker SC, Stevens RC, et al. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2006; 103(15):5717-22. doi: 10.1073/pnas.0510851103. PubMed PMID: 16581910; PubMed Central PMCID: PMC1458639.
20. James TW, Frias-Staheli N, Bacik JP, Levingston Macleod JM, Khajehpour M, Garcia-Sastre A, et al. Structural basis for the removal of ubiquitin and interferon- stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proceedings of the National Academy of Sciences of the United States of America.
2011; 108(6):2222-7. doi: 10.1073/pnas.1013388108. PubMed PMID: 21245344;
PubMed Central PMCID: PMCPMC3038750.
21. Akutsu M, Ye Y, Virdee S, Chin JW, Komander D. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proceedings of the National Academy of Sciences of the United States of America. 2011; 108(6):2228- 33. doi: 10.1073/pnas.1015287108. PubMed PMID: 21266548; PubMed Central PMCID: PMCPMC3038727.
22. Calistri A, Munegato D, Carli I, Parolin C, Palu G. The ubiquitin-conjugating system: multiple roles in viral replication and infection. Cells. 2014;3(2):386-417. doi: 10.3390/cells3020386. PubMed PMID: 24805990; PubMed Central PMCID:
PMCPMC4092849.
23. Heideker J, Wertz IE. DUBs, the regulation of cell identity and disease.
Biochem J. 2015;465(l):l-26. doi: 10.1042/BJ20140496. PubMed PMID: 25631680. 24. Kemp M. Recent Advances in the Discovery of Deubiquitinating Enzyme Inhibitors. Prog Med Chem. 2016;55: 149-92. doi: 10.1016/bs.pmch.2015.10.002.
PubMed PMID: 26852935.
25. Ernst A, Avvakumov G, Tong J, Fan Y, Zhao Y, Alberts P, et al. A strategy for modulation of enzymes in the ubiquitin system. Science. 2013;339(6119):590-5. Epub 2013/01/05. doi: 10.1126/science.1230161. PubMed PMID: 23287719.
26. Zhang W, Wu KP, Sartori MA, Kamadurai HB, Ordureau A, Jiang C, et al. System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes. Molecular cell. 2016;62(l): 121-36. doi: 10.1016/j.molcel.2016.02.005. PubMed PMID: 26949039.
27. Krissinel E, Henrick K. Inference of macromolecular assemblies from
crystalline state. Journal of molecular biology. 2007;372(3):774-97. doi:
10.1016/j.jmb.2007.05.022. PubMed PMID: 17681537.
28. Haagmans BL, van den Brand JM, Raj VS, Volz A, Wohlsein P, Smits SL, et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science. 2016;351(6268):77-81. doi: 10.1126/science.aadl283.
PubMed PMID: 26678878.
29. Inn KS, Lee SH, Rathbun JY, Wong LY, Toth Z, Machida K, et al. Inhibition of RIG-I-mediated signaling by Kaposi's sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J Virol. 2011;85(20): 10899-904. doi: 10.1128/JVI.00690-11. PubMed PMID: 21835791; PubMed Central PMCID: PMCPMC3187500.
30. Wang S, Wang K, Li J, Zheng C. Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J Virol. 2013;87(21): 11851-60. doi: 10.1128/JVI.01211-13. PubMed PMID: 23986588; PubMed Central PMCID: PMCPMC3807349.
31. Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. The papain-like protease of severe acute respiratory syndrome coronavirus has
deubiquitinating activity. J Virol. 2005;79(24): 15189-98. doi: 10.1128/JVI.79.24.15189- 15198.2005. PubMed PMID: 16306590; PubMed Central PMCID: PMCPMC1316023.
32. Chen Z, Wang Y, Ratia K, Mesecar AD, Wilkinson KD, Baker SC. Proteolytic processing and deubiquitinating activity of papain-like proteases of human
coronavirus NL63. J Virol. 2007;81(11):6007- 18. doi: 10.1128/JVL02747-06. PubMed PMID: 17392370; PubMed Central PMCID: PMCPMC 1900296.
33. Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Menard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol. 2005;79(24): 15199-208. doi:
10.1128/JVI.79.24.15199-15208.2005. PubMed PMID: 16306591; PubMed Central PMCID: PMCPMC1316033.
34. Ziebuhr J, Schelle B, Karl N, Minskaia E, Bayer S, Siddell SG, et al. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J Virol. 2007;81(8):3922-32. doi: 10.1128/JVI.02091-06. PubMed PMID: 17251282; PubMed Central PMCID: PMCPMC 1866161.
35. Bergeron E, Albarino CG, Khristova ML, Nichol ST. Crimean-Congo
hemorrhagic fever virus-encoded ovarian tumor protease activity is dispensable for virus RNA polymerase function. J Virol. 2010;84(l):216-26. doi: 10.1128/JVI.01859-09. PubMed PMID: 19864393; PubMed Central PMCID: PMCPMC2798392.
36. Lombardi C, Ayach M, Beaurepaire L, Chenon M, Andreani J, Guerois R, et al. A compact viral processing proteinase/ubiquitin hydrolase from the OTU family. PLoS Pathog. 2013;9(8):el003560. doi: 10.1371/journal.ppat.1003560. PubMed PMID:
23966860; PubMed Central PMCID: PMCPMC3744425.
37. Miao X. Recent advances in the development of new transgenic animal technology. Cellular and molecular life sciences : CMLS. 2()13;70(5):815-28. doi:
10.1007/800018-012- 1081-7. PubMed PMID: 22833168.
38. Auger A, Park M, Nitschke F, Minassian LM, Beilhartz GL, Mmassian BA, et al. Efficient Delivery of Structurally Diverse Protein Cargo into Mammalian Cells by a Bacterial Toxin. Mol Pharm. 2015; 12(8):2962-71. doi:
10.1021/acs.molpharmaceut.5b()0233. PubMed PMID: 26103531.
39. D Astolfo DS, Pagliero RJ, Pras A, Karthaus WR, Clevers H, Prasad V, et al. Efficient intracellular delivery of native proteins. Cell. 2015;161(3):674-90. doi:
10.1016/j.cell.2015.03.028. PubMed PMID: 25910214.
40. Gaj T, Liu J, Anderson KE, Sirk SJ, Barbas CF, 3rd. Protein delivery using Cys2-His2 zinc-finger domains. ACS Chem Biol. 2014;9(8): 1662-7. doi:
10.1021/cb500282g. PubMed PMID: 24936957; PubMed Central PMCID:
PMCPMC4519095.
41. Schott JW, Morgan M, Galla M, Schambach A. Viral and Synthetic RNA Vector Technologies and Applications. Mol Ther. 2016. doi: 10.1038/mt.2016.143. PubMed PMID: 27377044.
42. Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004; 10(3):31()-5. doi: 10.1038/nm996. PubMed PMID: 14770178.
43. van Doremalen N, Munster VJ. Animal models of Middle East respiratory syndrome coronavirus infection. Antiviral Res. 2015;122:28-38. doi:
10.1016/j.antiviral.2015.()7.005. PubMed PMID: 26192750; PubMed Central PMCID: PMCPMC4561025.
44. Cockrell AS, Yount BL, Scobey T, Jensen K, Douglas M, Beall A, et al. A mouse model for MERS coronavirus -induced acute respiratory distress syndrome. Nat Microbiol. 2016;2: 16226. doi: 10.1038/nmicrobiol.2016.226. PubMed PMID: 27892925.
45. Tonikian R, Zhang Y, Boone C, Sidhu SS. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nature protocols. 2007;2(6): 1368-86. Epub 2007/06/05. doi: 10.1038/nprot.2007.151. PubMed PMID: 17545975.
46. D'Arcy A, Villard F, Marsh M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 4):550-4. doi: 10.1107/S0907444907007652. PubMed PMID: 17372361.
47. Ireton GC, Stoddard BL. Microseed matrix screening to improve crystals of yeast cytosine deaminase. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 3):601-5. doi: 10.1107/S0907444903029664. PubMed PMID: 14993707.
48. Georgieva DC, Kuil ME, Oosterkamp TH, Zandbergen HW, Abrahams JP.
Heterogeneous nucleation of three-dimensional protein nanocrystals. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 5):564-70. doi: 10.1107/S0907444907007810. PubMed
PMID: 17452781.
49. Carlotti F, Bazuine M, Kekarainen T, Seppen J, Pognonec P, Maassen JA, et al. Lentiviral vectors efficiently transduce quiescent mature 3T3-L1 adipocytes. Mol Ther. 2004;9(2):209- 17. doi: 10.1016/j.ymthe.2003.11.021. PubMed PMID: 14759805.
50. van den Worm SH, Eriksson KK, Zevenhoven JC, Weber F, Zust R, Kuri T, et al. Reverse genetics of SARS-related coronavirus using vaccinia virus-based recombination. PloS one. 2012;7(3):e32857. doi: 10.1371/journal.pone.0032857.
PubMed PMID: 22412934; PubMed Central PMCID: PMC3296753.
51. Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015. Supplemental methods
ELISA assays to evaluate binding and specificity
Proteins under study were immobilized on 384-well MaxiSorp plates (Thermo
Scientific 12665347) by adding 30 μΐ^ of 1 μΜ proteins for overnight incubation at 4 °C. Phage and protein ELISA against immobilized proteins was performed as previously described [1, 2]. Binding of phage was detected using anti-M13-HRP antibody (GE Healthcare 27942101) and binding of FLAG-tagged inhibitors was detected using anti-FLAG-HRP antibody (Sigma-Aldrich A8592). To measure the half maximal binding concentration (EC50) of inhibitors binding to viral proteases, the concentration of inhibitors or wild type Ub was varied from 0 to 4 μΜ (24 points, 1:2 dilution), while the concentration of target proteins immobilized on the plate remained at 1 μΜ. EC50 values were calculated using the GraphPad Prism software with the built-in equation formula (non-linear regression curve).
Octet Bio- Layer Interferometry (BLI)
Experiments were performed as previously described [2]. Concentrated analyte and ligand proteins were diluted into BLI reaction buffer (25 mM HEPES pH 7.0, 150 mM NaCl, 0.1 mg/ml bovine serum albumin and 0.01% tween 20). Experiments of BLI were carried out on Octet RED96 system (ForteBio) using anti-GST antibody biosensors for GST-tagged ligands and His-tagged analytes at 25°C. 7-9 concentration points of analytes covering a wide titration range were applied for BLI experiments. Sensorgram raw data was processed and extracted by Octet Analysis 9.0. Binding- constants KD were obtained by fitting the response wavelength shifts in the steady- state regions using single -site binding system (Eq. 1) shown below.
where Req is the value of the response shift in the steady-state region in each sensorgram curve, [C] is the titrant concentration, Rmax is the maximal response in the steady-state region, KD is the binding constant for the single-site binding system. In both equations, Rmax and KD values are unknown and the Levenberg-Marquardt algorithm was used to perform iterative non-linear least squares curve fitting in Profit 6.2 (QuantumSoft) to obtain the fitted Rmax and KD.
Deconjugation assays
Ub/ISG15-amido-4-methylcoumarin (AMC)
Inhibition assays using Ub-AMC or ISG15-AMC (both Boston Biochem) as
deconjugation substrates were performed as described before [1, 3] . Experiments were performed in assay buffer (50 niM HE PES, pH 7.5, 0.01% Tween 20, 1 mM
dithiothreitol (DTT)) containing 1 μΜ Ub-AMC substrate, 10 nM viral DUBs (vDUBs) and 12 serial dilutions of inhibitor. vDUBs and inhibitors were mixed in assay buffer as indicated and incubated at room temperature for 2 min prior to the addition of Ub- AMC. All serial dilutions were performed in 96-well plates and subsequently transferred to 384-well black plates (Thermo Scientific) for making measurements. Deconjugation activity was measured by monitoring the increase of AMC fluorescence emission at 460 nm (excitation at 360 nm) for 30 min using a BioTek Synergy2 plate reader (BioTek Instruments, Winooski, VT). IC50 values were calculated using the GraphPad Prism software with the built-in equation formula (non-linear regression curve).
K48/K63 tetra-Ub
Inhibition assays using Biotin Tetra-Ub/Ub4 WT Chains (K48- or K63-linked, Boston Biochem) as deubiquitination substrates were performed in assay buffer (50 mM
HEPES, pH 7.5, 100 mM NaCl, 1 mM DTT) containing 1 μΜ substrate, 1 μΜ vDUB and 10 μΜ inhibitor as indicated. After incubation at 37°C for the indicated times, reactions were stopped by the addition of 10 mM EDTA and SDS-PAGE sample buffer and resolved using 4-20% gradient gels (Bio-Rad). The cleavage of K48/K63 tetra Ub chains was evaluated by Western blotting, probed with ExtrAvidinlC-Peroxidase (Sigma).
Protein expression and purification
ME.2 and ME.4
Plasmids named pET53-ME.2, and -ME.4 were transformed into CaC12-competent Escherichia coli BL21 (DE3) Gold cells (Agilent) to allow for T7 polymerase- driven expression of N-terminally His6-tagged ME.2 and ME.4 respectively. Cells were grown at 37°C in the presence of 150 μg/mL ampicillin to an optical density (OD600) of 0.6 and then induced at 16°C by addition of Isopropyl B-D- l-thiogalactopyranoside (IPTG; final concentration 1 mM). After overnight incubation, cells were resuspended in lysis buffer (500 mM NaCl, 50 mM Tris pH 8.0) and lysed via French press. The cell lysate was clarified by centrifugation at 17,211 x g at 4°C for 30 min, then incubated with 2 niL Ni-NTA Superflow resin (Qiagen) at 4°C for 30 min, and poured into a gravity column. The column was washed with 50 niL of lysis buffer, followed by 50 mL of lysis buffer containing 50 mM imidazole. Protein was eluted in lysis buffer containing 250 mM imidazole. Following affinity purification, inhibitors were further purified using a Superdex 75 size exclusion column (GE Healthcare), eluting in 20 mM Tris pH 8.5, 150 mM NaCl and 2 mM DTT.
CC.2 and CC.4
Plasmids encoding CC.2 and CC.4, named pET53-CC2 and -CC.4 respectively, were transformed into CaC12-competent E. coli BL21 (DE3) Gold cells, and grown in Luria- Bertani media supplemented with 150 pg/ml Ampicillin at 37°C with shaking to an OD600 of 0.8-1.5. Protein expression was induced by the addition of a final
concentration of 1 mM IPTG for 19-21 hours at 28'C. Cells were resuspended in lysis buffer (for CC.2: 50 mM Tris pH 7.2, 150 mM NaCl; for CC.4: 50 mM Tris pH 8.0, 150 mM NaCl) and lysed using either French Press or a freeze-thaw cycle that was performed three times. For the latter, cells were frozen at -80'C after addition of 150 μΐ of 20 mg/ml lysozyme followed by incubation on ice for 20 min, and thawed at room temperature. Lysis was followed by addition of 100-300 μΐ of DNase I (10 mg/ml) while mixing on a magnetic stirrer. Lysates from French Press or the freeze-thaw procedure were clarified by centrifugation at 48,298 x g for 30-40 min. Supernatants were loaded onto a gravity column containing Ni-NTA Superflow resin (Qiagen) that had been pre- equilibrated with Buffer A (50 mM Tris pH 8.0, 150 mM NaCl). The column was washed with 2-3 column volumes (CV) of Buffer A supplemented with 20 mM imidazole and eluted in a step-wise manner with Buffer A containing 500 mM imidazole. Fractions of interest, as determined from SDS-PAGE, were pooled and
dialyzed twice against Buffer A. The proteins were concentrated and loaded onto the Superdex 75 size exclusion column. CC.2 was eluted in 20 mM Tris pH 7.4, 150 mM NaCl and 1 mM DTT, and CC.4 was eluted in 50 mM Tris pH 8.0, 150 mM NaCl and 2 mM DTT.
MERS-CoV PLpro
The MERS-CoV PLpro domain was expressed and purified as described previously [4] . Briefly, E. coli BL21 (DE3) Gold cells transformed with plasmid pE-SUMO PLpro were grown to an OD600 ^of 0.6-0.8 at 37°C in the presence of 35 μg/mL kanamycin. Protein expression was induced by the addition of a final concentration of 1 mM IPTG and overnight incubation at 16C Cells were resuspended in lysis buffer (150 mM Tris, pH 8.5, 1 M NaCl, 2 mM DTT), lysed via French press and clarified via centrifugation at 17,21 lx g Clarified lysate was loaded onto a Ni-NTA gravity column and washed with lysis buffer, followed by lysis buffer supplemented with 25 mM imidazole and subsequent elution in lysis buffer supplemented with 250 mM
imidazole. The SUMO-PLpro-1-1 fusion was cleaved overnight in 150 mM NaCl, 50 mM Tris, pH 8.0, 1 mM DTT in the presence of Ulp l SUMO protease. Cleaved, tagless MERS-CoV PLpro was subsequently passed through a second Ni-NTA column, and further purified on a Superdex 75 size exclusion column equilibrated in 20 mM Tris, pH 8.5, 150 mM NaCl, 2 mM DTT.
CCHFV OTU169 and CCHFV OTU185
Plasmids encoding the CCHFV OTU domain residues 1-169 (pGEX-CCHFV OTU169) and residues 1-185 (pET49b-CCHFV OTU 185) fused with a GST tag and an HRV3c protease cleavage site were used for the expression and purification of the CCHFV OTU domain as described previously [5, 6] . Briefly, E. coli BL21-Gold (DE3) cells transformed with either of the plasmids were grown to an OD600 of 0.9-1.0 at 37°C with shaking, and protein expression was induced with a final concentration of 1 mM IPTG at 30°C for 19-21 hrs. Cells were resuspended in 25 ml of lysis buffer (50mM Tris-Cl pH 7.2, 200 mM NaCl, 5 mM EDTA, 5 mM DTT) and lysed via French Press. The lysate was clarified by centrifugation at 48,298 x g for 30-40 min. The
supernatant was loaded onto GST-Bind resin (VWR) in a gravity column pre- equilibrated with the lysis buffer, and proteins were eluted in 50 mM Tris-Cl pH 7.2, 500 mM NaCl, 5 mM EDTA, 5 mM DTT and 15 mM reduced glutathione. The GST tag was cleaved by addition of GST-tagged HRV 3c protease during overnight dialysis
against 50 niM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 niM DTT. Cleaved CCHF- vOTU proteins were collected as flow-through by passing the digest through a recharged GST-Bind column, concentrated and loaded onto the Superdex-75 column (GE Healthcare). Purified proteins, eluted in 20 mM Tris pH 7.2, 150 mM NaCl and ImM DTT, were then used for further analyses.
Protein crystallization data collection
MERS-CoV PLpro-ME.2 and -ME.4 complexes
Crystals were harvested by sweeping through a cryoprotectant solution containing
0.1M trisodium citrate pH 5.6, 22% (w/v) PEG 4000, 21% (v/v) 1,2-propanediol (PLpro- ME.4), or harvested directly from the crystallization solution (PLpro-ME.2) and flash- cooled in liquid nitrogen. X-ray diffraction data for PLpro-ME.4 crystals were collected using a Rigaku 007HF MicroFocus X-ray generator and R-AXIS IV++ detector. PLpro- ME.2 data collection was carried out at the Canadian Light Source on beamline 08B1-
1. Diffraction data for the PLpro-ME.4 and -ME.2 crystals were integrated and scaled using XDS [7], followed by merging using Aimless within the CCP4 software suite.
Initial phase estimates were determined via molecular replacement within
phenix.phaser using the previously determined MERS-CoV PLpro domain (PDB ID: 4RF0) and a polyAla Ub model (PDB ID: 4RF0) as independent search models [8]. Model building and refinement were performed using Coot and phenix.refine, respectively [9, 10] .
CCHFV OTU169-CC.2 and CCHFV OTU185-CC.4 complexes
Single crystals of the CCHFV OTU169-CC.2 and CCHFV OTU185-CC.4 complexes were swept through cryoprotectant containing well solution supplemented with 20- 30% ethylene glycol, and X-ray diffraction data were collected at the CLS on beamline 08ID-1 at 100K. X-ray diffraction data were indexed and scaled using XDS [7] and merged with Aimless [11]. Initial phases for the CCHFV OTU169-CC.2 and CCHFV OTU185-CC.4 complexes were determined by molecular replacement within phenix.phaser [8] using the previously reported CCHFV OTU domain bound to Ub (PDB 3PT2) [5] as a search model. The structures of CC.2 and CC.4 bound to CCHFV OTU were subsequently modelled and refined into the MR maps using Coot [9] and phenix.refine, respectively [10].
Plasmids used for cell culture work
The following plasmids were described elsewhere or provided by others: pcDNA3.1- MERS-CoV-PLpro WT and active site mutant C1592A [12], pCAGGS-HA-nsp3C-4-V5 [12], pcDNA-eGFP [13], pCAGGS- MAVS (provided by N. Frias-Staheli), pLuc-IFN-B [14] and pRL-TK (Promega). pcDNA3-HA-Ub was generated by cloning PCR-amplified Ub (using pCMV-FLAG-Ub [15] as a template) in pcDNA3.1(-) (Invitrogen) in frame with an N-terminal HA tag. Codon optimized SARS-CoV-PLpro (amino acids 1541- 1855 of the SARS-CoV ppla/pp lab (NCBI ID: AY291315.1)) with removed potential splice sites and polyadenylation signals (IDT) was cloned into pcDNA3.1(-) in frame with a C-terminal V5-tag generating pcDNA3.1(-)-SARS-CoV-PLpro-V5. pcDNA3.1(-)- SARS-CoV-PLpro-V5 was used as template for site-directed mutagenesis using the
QuikChangeTM strategy (Stratagene using Accuzyme DNA polymerase from Bioline) to mutate the active site cysteine to alanine (C1651A).
In the inhibitor -containing pDONR-221 vectors (Thermo Fisher Scientific) a methionine was introduced before the already present N-terminal FLAG tag using the QuikChangeTM strategy. In pDONR-221-FLAG-Ub substitutions G75A+G76A were introduced in Ub to generate pDONR-221-FLAG-Ub.AA. FLAG- inhibitors were cloned in destination vectors pcDNA3.1-DEST or pLenti6.3/TO/V5-DEST (Thermo Fisher Scientific) using the Gateway technology (Thermo Fisher Scientific). Primer sequences are available upon request.
(Jell culture and antibodies
HEK293T were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS; Bodinco BV), 100 units/ml penicillin, 100 units/ml streptomycin and 2 niM L-glutamine. Vero cells (Erasmus Medical Center) and MRC5 cells (ATCC CCL- 171) were cultured in Eagle's minimum essential medium (EMEM) with 8% FCS, 100 units/ml penicillin and streptomycin, 2 niM L-glutamine and nonessential amino acids (PAA). HuH-7 cells were maintained in DMEM containing 8% FCS, antibiotics and non-essential amino acids. DMEM, EMEM and supplements were obtained from Lonza.
Proteins on Western blot were visualized using the following primary antibodies: mouse anti-FLAG (F3165; Sigma-Aldrich), mouse anti-V5 (37-7500; Invitrogen), mouse anti-HA (ab 18181; Abeam), rabbit anti-GFP [13], rabbit anti-SARS-CoV nsp4 [16], rabbit anti-MERS-CoV p4b [17] and mouse anti-actin (A5316; Sigma-Aldrich). Primary antibodies were detected with horseradish peroxidase-conjugated secondary
antibodies (P0447 and P0217; Dako). In an indirect immunofluorescence assay primary antibody mouse anti-FLAG (F3165; Sigma-Aldrich) and secondary antibody Alexa488-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (A- 11001; Thermo Fisher Scientific) were used.
Protease activity assays in cell culture
Protease activity assays as described in Bailey-Elkin et al. [12] were performed with slight modifications. Briefly, to assess the DUB activity of MERS-CoV PLpro in the presence of inhibitors, HEK293T cells were co-transfected using the calcium
phosphate transfection method with plasmids encoding HA-tagged Ub (0.25 μg), MERS-CoV-PLpro-V5 (0.2 μg), FLAG-tagged inhibitors (0.5; 0.75 or 1 μg, as indicated) and GFP (0.25 ^ig). In order to establish whether the MERS-CoV-directed inhibitors target SARS-CoV PLpro, DUB assays were also performed with SARS-CoV- PLpro- V5 (0.2 μg) instead of MERS-CoV-FLpro-V5. The in trans cleavage activity of MERS-CoV PLpro in the presence of inhibitors was determined by co-expressing HA-nsp3C-4-V5 (0.2 μg), MERS-CoV-PLpro-V5 (0.15 μg), FLAG-tagged inhibitors (0.5; 0.75 or 1 μ& and GFP (0.25 μg) in HEK293T cells. Empty pcDNA vector was added to supplement to a total of 2 μg of plasmid DNA transfected per well of a 12-wells cluster. At 18 h post transfection, cells were lysed in 2x Laemmli sample buffer (LSB) containing 25 niM N-ethylmaleimide (NEM; Sigma). Proteins were separated in an SDS-PAGE gel, blotted onto Hybond-P (0,45μηι pore size, GE-Heathcare) and visualized after antibody incubation steps using Pierce ECL 2 Western blotting substrate (Thermo Fisher Scientific). To visualize FLAG-tagged inhibitors the proteins separated in an SDS-page gel were blotted onto 0,2 μιη PVDF membranes (GE-Heathcare). The membranes were blocked with dried milk powder in PBS containing 0,05% Tween-20 followed by antibody incubation steps.
Luciferase-based IFN-6 reporter assay
In a luciferase-based IFN-β reporter assay the effect of the inhibitors on the suppression of IFN- B promoter activity by viral PLpros was investigated. HEK293T cells grown to 80% confluency in a 24-wells plate were co-transfected with a combination of plasmids encoding the firefly luciferase reporter gene under control of the IFN- β promoter (25 ng), Renilla luciferase (5 ng), innate immune inducer mitochondrial antiviral signalling protein (MAVS; 25 ng), MERS-CoV PLpro-V5 (250 ng) and FLAG-tagged inhibitors (250, 500, 750 ng). Alternatively, SARS-CoV PLpro-
V5 (100 ng) and FLAG-tagged inhibitors (750 ng) were co-expressed with firefly luciferase under control of the IFN-β promoter, Renilla luciferase and MAVS. Total amounts of transfected DNA were equalized by the addition of empty pcDNA vector. Both firefly and Renilla luciferase activities were measured (Mithras LB 940 multimode reader; Berthold Technologies) using the Dual-Luciferase reporter assay system (Promega) 16 h post transfection. After normalizing the firefly luciferase activity to Renilla luciferase activity, an unpaired two-tailed Student's t test was performed and p values <0.05 were considered statistically significant. Firefly and Renilla luciferase activities were measured in triplicate and assays were repeated independently at least three times.
Methods for further variants
Expression plasmids and host strains
pET53-ME.4R10G was generated via around-the-horn PGR [ Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989; 17:6545-51.
[3] Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456-67.
Protein expression and purification
For protein thermal shift assays, overnight cultures of E. coli BL21 (DE3) Gold cells harbouring inhibitors (Inh) were started by inoculating 3 mL LB supplemented with 150 μg/mL ampicillin (amp) with a glycerol stock scraping, and were incubated overnight at 37°C with aeration. The next morning, 60 μΕ of overnight culture were subcultured into 6 mL LB + 150 μg/mL amp and grown at 37°C to an OD600 of 0.8. Inhibitor expression was induced overnight at 16°C with the addition IPTG to a final concentration of 1 mM. Cell pellets were frozen at -80°C until use. Cell pellets were resuspended in 1 mL [50 mM Tris pH 8.5, 500 mM NaCl, 100 μΜ PMSF, 5 μg/mL DNase] and lysed by sonication. Cell lysates were clarified by centrilugation at 16, 100 rcf, and 200 μΥ· of His Mag Sepharose Ni resin (GE Healthcare) was added to the clarified lysate supernatant, and incubated at room temperature for 30 min with end- over-end mixing. His Mag resin was washed with 2 mL [50 mM Tris pH 8.5, 500 mM NaCl], followed by 2 mL [50 mM Tris pH 8.5, 500 mM NaCl, 25 mM imidazole], followed by 2 mL [50 mM Tris pH 8.5, 500 mM NaCl, 50 mM imidazole] and eluted in [50 mM Tris pH 8.5, 500 mM NaCl, 250-500 mM imidazole] .
Protein purification for DSC assays were performed as described in [Zhang et al.], and Inhibitors were either flash-frozen in LN2 or held at 4°C until use. Inhibitors were dialyzed into PBS pH 7.4 prior to analysis. Bovine Ub was purchased from Sigma. Differential scanning calorimetry analysis of Inhibitors
DSC curves were collected on a Nano DSC (TA Instruments) with a scan rate of l°C/min at 3 atm, using protein concentrations of 2-3.25 mg/mL. Data analysis was performed using NanoAnalyze, and DSC curves were fit to a two-state scaled model for melting temperature (Tm) determination.
Protein thermal shift analysis of Inhibitors
Thermal shift assay reactions were performed using 4 μg inhibitor, and a final concentration of 5x SYPRO orange (Sigma). Assays were performed in 96-well format using an StepOnePlus Real-Time PCR System (Applied Biosystems), with a scan rate of l°C/min. Fluorescence melt curves were fit to a Boltzmann equation with Tm values corresponding to the midpoint of the transition curve using Protein Thermal Shift Software Version 1.1 (Thermofisher). Tm values are reported as an average of 3 replicates.
Plasmids used for cell culture work
Inhibitor-containing pDONR-221 vectors (Thermo Fisher Scientific) were previously described [Zhang et al.]. By classical cloning with Xhol and Pvul, region 1 of Ub WT or ME. l was cloned in place of region 1 of ME.4 in DONR-221-FLAG-ME.4 to generate pDONR-221-FLAG-ME.4.2 and -ME.4.4 respectively. Single amino acid substitutions were introduced using the QuikChangeTM strategy with pDONR-221- FLAG-Inh as template to generate pDONR-221-FLAG-ME. l. l, -ME.3.1, -ME.4.1, - ME4.3 and -ME.4.5. The newly generated FLAG-Inh were cloned in destination vectors pcDNA3.1-DEST or pLenti6.3/TO/V5-DEST (Thermo Fisher Scientific) using the Gateway technology (Thermo Fisher Scientific).
Determination of Inh mRNA and protein levels
HEK293T cells grown to 80% confluency in a 12-well plate were translected with 1 μg pcDNA3.1-DEST-FLAG-Inh using the calcium phosphate transfection method [ Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456-67.]. At 18 h post transfection, cells were lysed either in 2x Laemmli sample buffer (LSB) containing 25 mM N-ethylmaleimide (NEM; Sigma) to obtain protein samples or in RA1 buffer supplemented with β-
mercaptoethanol for total RNA isolation using the NucleoSpin RNA II kit (Machery- Nagel). Protein samples were loaded on SDS-polyacrylamide gels which were blotted onto PVDF membranes (GE-Heatheare) using the semi- dry blotting system (Trans- Blot turbo Tranfer System, BioRad). Membranes were blocked with 5% dried milk powder in PBS containing 0,05% Tween-20, followed by antibody incubation steps. After antibody incubation, the protein bands were visualized using Amersham ECL Plus detection reagent (GE Healthcare). Protein quantification was performed using ImageJ on Western blots of at least three independent experiments [ Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671-5.]. FLAG-lnh protein levels were normalized to actin that was used as loading control and Ub AA was set to 100%. Isolated RNA was treated with DNase (TURBO DNA-free™ Kit, Thermo Fisher Scientific) to ensure removal of DNA contaminants before it was used as a template for cDNA synthesis using RevertAid H minus reverse transcriptase (ThermoFisher) and oligo(dT)20 primer. Samples were analysed in triplicate by real-time quantitative reverse-transcriptase (qRT) PGR using iTaq SYBR Green Supermix (BioRad) on a CFX384 Touch™ Real-Time PGR Detection System (BioRad). Primers targeting mRNAs encoding human 6-actin (Forward (Fwd): 5'AGGCACCAGGGCGTGAT-3'; Reverse (Rev):
5' GC C C AC AT AGGAATC CTTCT GAG - 3 ') , neomycin (Fwd:
5' CT C CTGC C GAGAAAGT ATC C - 3' , Rev: 5'GCTCTTCGTCCAGATCATCC-3') and Ub(V) (Fwd: 5'CAGCTGGAAGATGGACGTAC-3', Rev:
5'CCAGTGTGGTGGAATTCTGC-3') were used. In all PGR runs a standard dilution series was included and melting-curve analysis was done. PGR efficiencies were determined which allowed quantification using the standard curve method. For relative quantification of Inh expression levels, the results were normalized to the transfection efficiency (neomycin) and to the normalization gene (actin). The normalization gene in the sample set was determined using NormFinder software [ Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription- PGR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer research. 2004;64:5245-50.].
Claims
1. Use of an inhibitor or a nucleic acid molecule encoding said inhibitor for inhibiting the biological activity of a viral protein, preferably a viral protease having
deubiquitination activity,
wherein the inhibitor comprises a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l,
wherein the inhibitor comprises one or more amino acid substitutions in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and
wherein said substitutions include an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1.
2. The use according to claim 1,
a) wherein region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5; SEQ ID NO: 12, or
SEQ ID NO: 14;
b) wherein region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, or SEQ ID NO: 13; and/or
c) wherein region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 13, SEQ ID NO: 15;
preferably wherein the inhibitor comprises an amino acid sequence selected from SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 12, SEQ ID NO: 13, SEQ ID NO: 14, and SEQ ID NO: 15.
3. Use of an inhibitor or a nucleic acid molecule encoding said an inhibitor for inhibiting the biological activity of a viral protein, preferably a viral protease having deubiquitination activity,
wherein the inhibitor comprises a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l,
wherein the inhibitor comprises one or more amino acid substitutions in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and
wherein said substitution include an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1.
4. The use according to claim 3,
a) wherein region 1 has an amino acid sequence corresponding to amino acids 2- 14 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10; b) wherein region 2 has an amino acid sequence corresponding to amino acids 42-49 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10; and/or
c) wherein region 3 has an amino acid sequence corresponding to amino acids 62-78 of SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10, preferably wherein the inhibitor comprises an amino acid sequence selected from SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, or SEQ ID NO: 10.
5. The use according to any one of the preceding claims wherein the use is for inhibiting the proteolytic cleavage activity of a viral protein.
6. The use according to any one of the preceding claims, wherein region 1 is linked to region 2 by a polypeptide comprising amino acids 15-41 of SEQ ID NO: 1 or having at least 80% identity to amino acids 15-41 of SEQ ID NO: 1 and where in region 2 is linked to region 3 by a polypeptide comprising amino acids 50-61 of SEQ ID NO: 1 or having at least 80% identity to amino acids 50-61 of SEQ ID NO: 1.
7. An inhibitor comprising a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l,
wherein the inhibitor comprises one or more amino acid substitutions in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and
wherein said substitutions include an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1.
8. An inhibitor comprising a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l,
wherein the inhibitor comprises one or more amino acid substitutions in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and
wherein said substitution include an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1.
9. A nucleic acid molecule encoding the inhibitor of claims 7 or 8.
10. A recombinant expression vector expressing the nucleic acid molecule of claim 9.
11. An in vitro cell line, or a non-human cell or non-human organism comprising the inhibitor of claims 7 or 8, the nucleic acid molecule of claim 9, or the recombinant expression vector of claim 10.
12. A pharmaceutical composition comprising the inhibitor of claims 7 or 8, the nucleic acid molecule of claim 9, or the recombinant expression vector of claim 10.
13. An inhibitor or a nucleic acid molecule encoding said inhibitor for use in therapy, wherein the inhibitor comprises a beta-grasp fold, wherein said fold comprises region
1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l,
wherein the inhibitor comprises one or more amino acid substitutions in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and
wherein said substitutions include an amino acid substitution of A to F at the amino acid position corresponding to 46 of SEQ ID NO: 1 and/or an amino acid substitution of E to Y at the amino acid position corresponding to 64 of SEQ ID NO: 1, preferably
wherein the use is for the treatment and/or prevention of Middle East respiratory syndrome coronaviral (MERS-CoV) infection and/or the symptoms thereof.
14. An inhibitor or a nucleic acid molecule encoding said an inhibitor for use in therapy,
wherein the inhibitor comprising a beta-grasp fold, wherein said fold comprises region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l,
wherein the inhibitor comprises one or more amino acid substitutions, in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l, and
wherein said substitution include an amino acid substitution of R to G at the amino acid position corresponding to 74 of SEQ ID NO: 1, preferably wherein the use is for the treatment and/or prevention of Crimean-Congo hemorrhagic fever viral infection and/or the symptoms thereof.
15. A method of identifying an inhibitor or a nucleic acid molecule encoding said inhibitor that inhibits the biological activity of a viral ubiquitin binding protein partner, the method comprising providing a library of polypeptides and screening said library against a viral ubiquitin binding partner in order to identify inhibitors that bind to said viral ubiquitin binding partner, wherein said polypeptide library comprises at least 1000 different polypeptides, wherein each polypeptide comprises a beta-grasp fold comprising region 1 (amino acids 2- 14), region 2 (amino acids 42-49), and region 3 (amino acids 62-76) of the amino acid sequence set forth in SEQ ID NO: l and comprises at least one amino acid mutation in said regions as compared to the amino acid sequence set forth in SEQ ID NO: l.
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| EP17159420 | 2017-03-06 | ||
| EP17159420.3 | 2017-03-06 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2018164573A1 true WO2018164573A1 (en) | 2018-09-13 |
Family
ID=58264426
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/NL2018/050137 WO2018164573A1 (en) | 2017-03-06 | 2018-03-06 | Viral polypeptide inhibitors |
Country Status (1)
| Country | Link |
|---|---|
| WO (1) | WO2018164573A1 (en) |
Citations (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2012020289A2 (en) | 2010-08-10 | 2012-02-16 | The Governing Council Of The University Of Toronto | Specific active site inhibitors of enzymes or substrate binding partners and methods of producing same |
-
2018
- 2018-03-06 WO PCT/NL2018/050137 patent/WO2018164573A1/en active Application Filing
Patent Citations (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2012020289A2 (en) | 2010-08-10 | 2012-02-16 | The Governing Council Of The University Of Toronto | Specific active site inhibitors of enzymes or substrate binding partners and methods of producing same |
Non-Patent Citations (65)
| Title |
|---|
| "PROGRESS IN MEDICINAL CHEMISTRY.", vol. 55, 1 January 2016, ELSEVIER, AMSTERDAM., NL, ISSN: 0079-6468, article MARK KEMP ET AL: "Recent Advances in the Discovery of Deubiquitinating Enzyme Inhibitors", pages: 149 - 192, XP055333853, DOI: 10.1016/bs.pmch.2015.10.002 * |
| AKUTSU M; YE Y; VIRDEE S; CHIN JW; KOMANDER D: "Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains", PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA, vol. 108, 2011, pages 2228 - 33, XP055249441, DOI: doi:10.1073/pnas.1015287108 |
| AUGER A; PARK M; NITSCHKE F; MINASSIAN LM; BEILHARTZ GL; MINASSIAN BA ET AL.: "Efficient Delivery of Structurally Diverse Protein Cargo into Mammalian Cells by a Bacterial Toxin", MOL PHARM., vol. 12, no. 8, 2015, pages 2962 - 71, XP055332136, DOI: doi:10.1021/acs.molpharmaceut.5b00233 |
| BAEZ-SANTOS YM; ST JOHN SE; MESECAR AD: "The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds", ANTIVIRAL RES., vol. 115, 2015, pages 21 - 38, XP029136855, DOI: doi:10.1016/j.antiviral.2014.12.015 |
| BAILEY-ELKIN BA; KNAAP RC; JOHNSON GG; DALEBOUT TJ; NINABER DK; VAN KASTEREN PB ET AL.: "Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression", THE JOURNAL OF BIOLOGICAL CHEMISTRY, vol. 289, no. 50, 2014, pages 34667 - 82 |
| BARRETTO N; JUKNELIENE D; RATIA K; CHEN Z; MESECAR AD; BAKER SC: "The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity", J VIROL., vol. 79, no. 24, 2005, pages 15189 - 98 |
| BERGERON E; ALBARINO CG; KHRISTOVA ML; NICHOL ST: "Crimean-Congo hemorrhagic fever virus-encoded ovarian tumor protease activity is dispensable for virus RNA polymerase function", J VIROL., vol. 84, no. 1, 2010, pages 216 - 26, XP055249446, DOI: doi:10.1128/JVI.01859-09 |
| BURROUGHS ET AL.: "Methods in Molecular Biology", vol. 832, 2012 |
| CALISTRI A; MUNEGATO D; CARLI I; PAROLIN C; PALU G: "The ubiquitin-conjugating system: multiple roles in viral replication and infection", CELLS, vol. 3, no. 2, 2014, pages 386 - 417 |
| CANCER RESEARCH, vol. 64, 2004, pages 5245 - 50 |
| CARLOTTI F; BAZUINE M; KEKARAINEN T; SEPPEN J; POGNONEC P; MAASSEN JA ET AL.: "Lentiviral vectors efficiently transduce quiescent mature 3T3-L1 adipocytes", MOL THER., vol. 9, no. 2, 2004, pages 209 - 17 |
| CHEN Z; WANG Y; RATIA K; MESECAR AD; WILKINSON KD; BAKER SC: "Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63", J VIROL., vol. 81, no. 11, 2007, pages 6007 - 18 |
| CHEN, P. Y.; GOPALACUSHINA, B. G.; YANG, C. C.; CHAN, S. I.; EVANS, P. A.: "The role of a beta-bulge in the folding of the beta-hairpin structure in ubiquitin", PROTEIN SCI., vol. 10, 2001, pages 2063 - 2074 |
| CHENON M; CAMBORDE L; CHEMINANT S; JUPIN I: "A viral deubiquitylating enzyme targets viral RNA-dependent RNA polymerase and affects viral infectivity", THE EMBO JOURNAL, vol. 31, no. 3, 2012, pages 741 - 53 |
| CLEMENTZ MA; CHEN Z; BANACH BS; WANG Y; SUN L; RATIA K ET AL.: "Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases", J VIROL., vol. 84, no. 9, 2010, pages 4619 - 29 |
| COCKRELL AS; YOUNT BL; SCOBEY T; JENSEN K; DOUGLAS M; BEALL A ET AL.: "A mouse model for MERS coronavirus-induced acute respiratory distress syndrome", NAT MICROBIOL, vol. 2, 2016, pages 16226 |
| D'ARCY A; VILLARD F; MARSH M: "An automated microseed matrix-screening method for protein crystallization", ACTA CRYSTALLOGR D BIOL CRYSTALLOGR, vol. 63, no. 4, 2007, pages 550 - 4 |
| D'ASTOLFO DS; PAGLIERO RJ; PRAS A; KARTHAUS WR; CLEVERS H; PRASAD V ET AL.: "Efficient intracellular delivery of native proteins", CELL, vol. 161, no. 3, 2015, pages 674 - 90, XP055338589, DOI: doi:10.1016/j.cell.2015.03.028 |
| DE WIT E; VAN DOREMALEN N; FALZARANO D; MUNSTER VJ: "SARS and MERS: recent insights into emerging coronaviruses", NATURE REVIEWS MICROBIOLOGY, vol. 14, 2016, pages 523 - 34, XP055369190, DOI: doi:10.1038/nrmicro.2016.81 |
| DEVARAJ SG; WANG N; CHEN Z; CHEN Z; TSENG M; BARRETTO N ET AL.: "Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus", J BIOL CHEM., vol. 282, no. 44, 2007, pages 32208 - 21 |
| DZAU ET AL., TRENDS IN BIOTECHNOLOGY, vol. 11, 1993, pages 205 - 210 |
| ERNST A; AVVAKUMOV G; TONG J; FAN Y; ZHAO Y; ALBERTS P ET AL.: "A strategy for modulation of enzymes in the ubiquitin system", SCIENCE, vol. 339, no. 6119, 2013, pages 590 - 5 |
| ERNST ET AL., SCIENCE, vol. 339, 2013, pages 590 - 595 |
| FRIAS-STAHELI N; GIANNAKOPOULOS NV; KIKKERT M; TAYLOR SL; BRIDGEN A; PARAGAS J ET AL.: "Ovarian tumor domain-containing viral proteases evade ubiquitin-and ISG15-dependent innate immune responses", CELL HOST MICROBE, vol. 2, no. 6, 2007, pages 404 - 16, XP055049785, DOI: doi:10.1016/j.chom.2007.09.014 |
| GAJ T; LIU J; ANDERSON KE; SIRK SJ; BARBAS CF: "3rd. Protein delivery using Cys2-His2 zinc-finger domains", ACS CHEM BIOL., vol. 9, no. 8, 2014, pages 1662 - 7 |
| GEORGIEVA DG; KUIL ME; OOSTERKAMP TH; ZANDBERGEN HW; ABRAHAMS JP: "Heterogeneous nucleation of three-dimensional protein nanocrystals", ACTA CRYSTALLOGR D BIOL CRYSTALLOGR., vol. 63, no. 5, 2007, pages 564 - 70 |
| GRAHAM FL; VAN DER EB AJ: "A new technique for the assay of infectivity of human adenovirus 5 DNA", VIROLOGY, vol. 52, 1973, pages 456 - 67, XP023052128, DOI: doi:10.1016/0042-6822(73)90341-3 |
| HAAGMANS BL; VAN DEN BRAND JM; RAJ VS; VOLZ A; WOHLSEIN P; SMITS SL ET AL.: "An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels", SCIENCE, vol. 351, no. 6268, 2016, pages 77 - 81, XP055265765, DOI: doi:10.1126/science.aad1283 |
| HEIDEKER J; WERTZ IE: "DUBs, the regulation of cell identity and disease", BIOCHEM J., vol. 465, no. 1, 2015, pages 1 - 26 |
| HEMSLEY A; ARNHEIM N, TONEY MD; CORTOPASSI G; GALAS DJ: "A simple method for site-directed mutagenesis using the polymerase chain reaction", NUCLEIC ACIDS RES., vol. 17, 1989, pages 6545 - 51, XP002027312 |
| HILGENFELD R: "From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design", FEBS J., vol. 281, no. 18, 2014, pages 4085 - 96 |
| INN KS; LEE SH; RATHBUN JY; WONG LY; TOTH Z; MACHIDA K ET AL.: "Inhibition of RIG-I-mediated signaling by Kaposi's sarcoma-associated herpesvirus-encoded deubiquitinase ORF64", J VIROL., vol. 85, no. 20, 2011, pages 10899 - 904 |
| IRETON GC; STODDARD BL: "Microseed matrix screening to improve crystals of yeast cytosine deaminase", ACTA CRYSTALLOGR D BIOL CRYSTALLOGR, vol. 60, no. 3, 2004, pages 601 - 5 |
| ISAACSON MK; PLOEGH HL: "Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection", CELL HOST & MICROBE, vol. 5, no. 6, 2009, pages 559 - 70 |
| JAMES TW; FRIAS-STAHELI N; BACIK JP; LEVINGSTON MACLEOD JM; KHAJEHPOUR M; GARCIA-SASTRE A ET AL.: "Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease", PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA, vol. 108, 2011, pages 2222 - 7, XP055049784, DOI: doi:10.1073/pnas.1013388108 |
| JIANG X; CHEN ZJ: "The role of ubiquitylation in immune defence and pathogen evasion", NATURE REVIEWS IMMUNOLOGY, vol. 12, no. 1, 2012, pages 35 - 48 |
| KEMP M.: "Recent Advances in the Discovery of Deubiquitinating Enzyme Inhibitors", PROG MED CHEM., vol. 55, 2016, pages 149 - 92, XP055333853, DOI: doi:10.1016/bs.pmch.2015.10.002 |
| KOMANDER D; RAPE M, THE UBIQUITIN CODE. ANNUAL REVIEW OF BIOCHEMISTRY, vol. 81, 2012, pages 203 - 29 |
| KRISSINEL E; HENRICK K: "Inference of macromolecular assemblies from crystalline state", JOURNAL OF MOLECULAR BIOLOGY, vol. 372, no. 3, 2007, pages 774 - 97, XP022220069, DOI: doi:10.1016/j.jmb.2007.05.022 |
| LEI J; MESTERS JR; DROSTEN C; ANEMULLER S; MA Q; HILGENFELD R: "Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features", ANTIVIRAL RESEARCH., vol. 109, 2014, pages 72 - 82, XP029045736, DOI: doi:10.1016/j.antiviral.2014.06.011 |
| LINDNER HA; FOTOUHI-ARDAKANI N; LYTVYN V; LACHANCE P; SULEA T; MENARD R: "The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme", J VIROL., vol. 79, no. 24, 2005, pages 15199 - 208 |
| LOMBARDI C; AYACH M; BEAUREPAIRE L; CHENON M; ANDREANI J; GUEROIS R ET AL.: "A compact viral processing proteinase/ubiquitin hydrolase from the OTU family", PLOS PATHOG., vol. 9, no. 8, 2013, pages elO03560 |
| MIAO X.: "Recent advances in the development of new transgenic animal technology", CELLULAR AND MOLECULAR LIFE SCIENCES : CMLS, vol. 70, no. 5, 2013, pages 815 - 28, XP055073911, DOI: doi:10.1007/s00018-012-1081-7 |
| MIELECH AM; CHEN Y; MESECAR AD; BAKER SC: "Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities", VIRUS RES., vol. 194, 2014, pages 184 - 90, XP029105621, DOI: doi:10.1016/j.virusres.2014.01.025 |
| MIELECH AM; KILIANSKI A; BAEZ-SANTOS YM; MESECAR AD; BAKER SC: "MERS-CoV papain-like protease has deISGylating and deubiquitinating activities", VIROLOGY, vol. 450-451, 2014, pages 64 - 70 |
| MORALES DJ; LENSCHOW DJ: "The antiviral activities of ISG15", JOURNAL OF MOLECULAR BIOLOGY, vol. 425, no. 24, 2013, pages 4995 - 5008, XP028775405, DOI: doi:10.1016/j.jmb.2013.09.041 |
| PANTOLIANO, M. W.; PETRELLA, E. C.; KWASNOSKI, J. D.; LOBANOV, V. S.; MYSLIK, J.; GRAF, E.; CARVER, T.; ASEL, E.; SPRINGER, B. A.;: "High-density miniaturized thermal shift assays as a general strategy for drug discovery", J BIOMOL SCREEN, vol. 6, 2001, pages 429 - 440, XP055027496, DOI: doi:10.1177/108705710100600609 |
| PIANA, S.; LINDORFF-LARSEN, K.; SHAW, D. E.: "Atomic-level description of ubiquitin folding", vol. 110, 2013, PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES, pages: 5915 - 5920 |
| RATIA K; SAIKATENDU KS; SANTARSIERO BD; BARRETTO N; BAKER SC; STEVENS RC ET AL.: "Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme", PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA, vol. 103, 2006, pages 5717 - 22 |
| SCHNEIDER CA; RASBAND WS; ELICEIRI KW: "NIH Image to ImageJ: 25 years of image analysis", NATURE METHODS, vol. 9, 2012, pages 671 - 5, XP055403257 |
| SCHOTT JW; MORGAN M; GALLA M; SCHAMBACH A: "Viral and Synthetic RNA Vector Technologies and Applications", MOL THER., 2016 |
| SCHRODINGER, LLC, THE PYMOL MOLECULAR GRAPHICS SYSTEM, 2015 |
| TONIKIAN R; ZHANG Y; BOONE C; SIDHU SS: "Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries", NATURE PROTOCOLS, vol. 2, no. 6, 2007, pages 1368 - 86, XP008167972, DOI: doi:10.1038/nprot.2007.151 |
| VAN DEN WORM SH; ERIKSSON KK; ZEVENHOVEN JC; WEBER F; ZUST R; KURI T ET AL.: "Reverse genetics of SARS-related coronavirus using vaccinia virus-based recombination", PLOS ONE, vol. 7, no. 3, 2012, pages e32857 |
| VAN DOREMALEN N; MUNSTER VJ: "Animal models of Middle East respiratory syndrome coronavirus infection", ANTIVIRAL RES., vol. 122, 2015, pages 28 - 38, XP029262472, DOI: doi:10.1016/j.antiviral.2015.07.005 |
| VAN HOOIJDONK BMC NEUROSCIENCE, vol. 10, 2009, pages 2 |
| WADIA JS; STAN RV; DOWDY SF: "Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis", NAT MED., vol. 10, no. 3, 2004, pages 310 - 5, XP002477563, DOI: doi:10.1038/nm996 |
| WANG S; WANG K; LI J; ZHENG C: "Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3", J VIROL., vol. 87, no. 21, 2013, pages 11851 - 60 |
| WIMMER; SCHREINER, VIRUSES, vol. 7, 2015, pages 4854 - 4872 |
| XING Y; CHEN J; TU J; ZHANG B; CHEN X; SHI H ET AL.: "The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase", THE JOURNAL OF GENERAL VIROLOGY, vol. 94, no. 7, 2013, pages 1554 - 67 |
| YAU R; RAPE M: "The increasing complexity of the ubiquitin code", NAT CELL BIOL., vol. 18, no. 6, 2016, pages 579 - 86 |
| ZHANG W; WU KP; SARTORI MA; KAMADURAI HB; ORDUREAU A; JIANG C ET AL.: "System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes", MOLECULAR CELL, vol. 62, no. 1, 2016, pages 121 - 36, XP029496716, DOI: doi:10.1016/j.molcel.2016.02.005 |
| ZHANG, W.; BAILEY-ELKIN, B. A.; KNAAP, R. C. M.; KHARE, B.; DALEBOUT, T. J.; JOHNSON, G. G.; VAN KASTEREN, P. B.; MCLEISH, N. J.;: "Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants", PLOS PATHOGENS, vol. 13, 2017, pages el006372 |
| ZHENG D; CHEN G; GUO B; CHENG G; TANG H: "PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production", CELL RES., vol. 18, no. 11, 2008, pages 1105 - 13 |
| ZIEBUHR J; SCHELLE B; KARL N; MINSKAIA E; BAYER S; SIDDELL SG ET AL.: "Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication", J VIROL., vol. 81, no. 8, 2007, pages 3922 - 32 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| Cong et al. | Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle | |
| Zhang et al. | Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants | |
| Richard et al. | RSV hijacks cellular protein phosphatase 1 to regulate M2-1 phosphorylation and viral transcription | |
| Christensen et al. | PDZ domains as drug targets | |
| Schulte et al. | Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication | |
| Huthoff et al. | RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1 | |
| Matreyek et al. | Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity | |
| Arold et al. | The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling | |
| Gogrefe et al. | Structure of a functional cap-binding domain in Rift Valley fever virus L protein | |
| US10815476B2 (en) | Methods and compositions for synthetic RNA endonucleases | |
| Patzina et al. | Structural requirements for the antiviral activity of the human MxA protein against Thogoto and influenza A virus | |
| Yu et al. | Accelerating PERx reaction enables covalent nanobodies for potent neutralization of SARS-CoV-2 and variants | |
| van Vliet et al. | Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site | |
| Qiu et al. | N-substituted pyrrole derivative 12m inhibits HIV-1 entry by targeting Gp41 of HIV-1 envelope glycoprotein | |
| Dutta et al. | Structural and functional characterization of human SGT and its interaction with Vpu of the human immunodeficiency virus type 1 | |
| Yao et al. | Structural basis of potent and broad HIV-1 fusion inhibitor CP32M | |
| Lamb et al. | Charge-surrounded pockets and electrostatic interactions with small ions modulate the activity of retroviral fusion proteins | |
| Tran et al. | Insights into human Lck SH3 domain binding specificity: different binding modes of artificial and native ligands | |
| Singh et al. | A novel dimer-tetramer transition captured by the crystal structure of the HIV-1 Nef | |
| Lee et al. | Multimerization potential of the cytoplasmic domain of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 | |
| Nielsen et al. | Structure of TSA2 reveals novel features of the active-site loop of peroxiredoxins | |
| Noad et al. | Sequences of avian reovirus M1, M2 and M3 genes and predicted structure/function of the encoded μ proteins | |
| Cross et al. | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin α‐dependent nuclear localization signals | |
| Pascoe et al. | Yeast two-hybrid analysis for ubiquitin variant inhibitors of human deubiquitinases | |
| Meier et al. | Peptides presenting the binding site of human CD4 for the HIV-1 envelope glycoprotein gp120 |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 18710569 Country of ref document: EP Kind code of ref document: A1 |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| 122 | Ep: pct application non-entry in european phase |
Ref document number: 18710569 Country of ref document: EP Kind code of ref document: A1 |