WO1999027084A1 - Novel pectate lyases - Google Patents
Novel pectate lyases Download PDFInfo
- Publication number
- WO1999027084A1 WO1999027084A1 PCT/DK1998/000515 DK9800515W WO9927084A1 WO 1999027084 A1 WO1999027084 A1 WO 1999027084A1 DK 9800515 W DK9800515 W DK 9800515W WO 9927084 A1 WO9927084 A1 WO 9927084A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- polypeptide
- amino acid
- seq
- pectate lyase
- nucleotide
- Prior art date
Links
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N9/00—Enzymes; Proenzymes; Compositions thereof; Processes for preparing, activating, inhibiting, separating or purifying enzymes
- C12N9/14—Hydrolases (3)
- C12N9/24—Hydrolases (3) acting on glycosyl compounds (3.2)
- C12N9/2402—Hydrolases (3) acting on glycosyl compounds (3.2) hydrolysing O- and S- glycosyl compounds (3.2.1)
- C12N9/2465—Hydrolases (3) acting on glycosyl compounds (3.2) hydrolysing O- and S- glycosyl compounds (3.2.1) acting on alpha-galactose-glycoside bonds, e.g. alpha-galactosidase (3.2.1.22)
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N9/00—Enzymes; Proenzymes; Compositions thereof; Processes for preparing, activating, inhibiting, separating or purifying enzymes
- C12N9/88—Lyases (4.)
-
- D—TEXTILES; PAPER
- D06—TREATMENT OF TEXTILES OR THE LIKE; LAUNDERING; FLEXIBLE MATERIALS NOT OTHERWISE PROVIDED FOR
- D06L—DRY-CLEANING, WASHING OR BLEACHING FIBRES, FILAMENTS, THREADS, YARNS, FABRICS, FEATHERS OR MADE-UP FIBROUS GOODS; BLEACHING LEATHER OR FURS
- D06L4/00—Bleaching fibres, filaments, threads, yarns, fabrics, feathers or made-up fibrous goods; Bleaching leather or furs
- D06L4/40—Bleaching fibres, filaments, threads, yarns, fabrics, feathers or made-up fibrous goods; Bleaching leather or furs using enzymes
Definitions
- the present invention relates to microbial pectate lyases, more specifically to microbial enzymes exhibiting pectate lyase activity as their major enzymatic activity in the neutral and alkaline pH ranges; to a method of producing such enzymes; and to methods for using such enzymes in the textile, detergent and cellulose fiber processing industries.
- Pectin polymers are important constituents of plant cell walls.
- Pectin is a hetero-polysaccharide with a backbone composed of alternating homogalacturonan (smooth regions) and rhamnogalacturonan (hairy regions) .
- the smooth regions are linear polymers of 1,4 -linked alpha-D-galacturonic acid.
- the galacturonic acid residues can be methyl-esterified on the carboxyl group to a varying degree, usually in a non-random fashion with blocks of polygalacturonic acid being completely methyl-esterified.
- Pectinases can be classified according to their preferential substrate, highly methyl-esterified pectin or low methyl-esterified pectin and polygalacturonic acid (pectate) , and their reaction mechanism, beta-elimination or hydrolysis. Pectinases can be mainly endo-acting, cutting the polymer at random sites within the chain to give a mixture of oligomers, or they may be exo-acting, attacking from one end of the polymer and producing monomers or dimers .
- pectinase activities acting on the smooth regions of pectin are included in the classification of enzymes provided by the Enzyme Nomenclature (1992) such as pectate lyase (EC 4.2.2.2), pectin lyase (EC 4.2.2.10), polygalacturonase (EC 3.2.1.15), exo- polygalacturonase (EC 3.2.1.67), exo-polygalacturonate lyase (EC 4.2.2.9) and exo-poly-alpha-galacturonosidase (EC 3.2.1.82).
- pectate lyase EC 4.2.2.2
- pectin lyase EC 4.2.2.10
- polygalacturonase EC 3.2.1.15
- exo- polygalacturonase EC 3.2.1.67
- exo-polygalacturonate lyase EC 4.2.2.9
- Pectate lyases have been cloned from different bacterial genera such as Erwinia , Pseudomonas , Klebsiella and Xanthomonas . Also from Bacillus subtilis (Nasser et al . (1993) FEBS 335:319- 326) and Bacillus sp . YA-14 (Kim et al . (1994) Biosci . Biotech. Biochem. 58:947-949) cloning of a pectate lyase has been described. Purification of pectate lyases with maximum activity in the pH range of 8-10 produced by Bacillus pumilus (Dave and Vaughn (1971) J. Bacteriol .
- WO 98/45393 discloses detergent compositions containing protopectinase with remarkable detergency agains muddy soilings.
- pectinase producing microorganisms exhibit a broad range of pectin degrading or modifying enzymes.
- the microorganisms also produce cellulases and/or hemicellulases and complex multi-component enzyme preparations from such microorganisms may be difficult to optimise for various applications, they even may contain enzymes with detrimental effect.
- this invention relates to a pectate lyase comprising a first amino acid sequence consisting of seven (7) amino acid residues having the following sequence: Asn Leu Asn Ser Arg Val Pro (NLNSRVP) .
- the pectate lyase may additionally hold a second amino acid sequence consisting of six (6) amino acid residues selected from the group consisting of the sequences Trp Val Asp His Asn Glu (WVDHNE) and Trp He Asp His Asn Glu (WIDHNE) ; and optionally also a third amino acid sequence consisting of three (3) amino acid residues having the following sequence: Ser Trp Asn (SWN) .
- the DNA sequences of five pectate lyases of the invention are listed in the sequence listing as SEQ ID No. 1, 3, 5, 7 and 9, respectively, and the deduced amino acid sequences are listed in the sequence listing as SEQ ID No. 2, 4, 6, 8 and 10, 5 respectively. It is believed that the novel enzyme will be classified according to the Enzyme Nomenclature in the Enzyme Class EC 4.2.2.2. However, it should be noted that the enzyme of the invention also exhibits catalytic activity on pectin (which may be esterified) besides the activity on pectate and 0 polygalacturonides conventionally attributed to enzymes belonging to EC 4.2.2.2.
- the present invention relates to a pectate lyase which is i) a polypeptide produced by Bacillus agaradhaerens, NCIMB 40482 or DSM 8721, or ii) a polypeptide 5 comprising an amino acid sequence as shown in positions 27-359 of SEQ ID NO: 2, or iii) an analogue of the polypeptide defined in i) or ii) which is at least 45% homologous with said polypeptide, or iv) is derived from said polypeptide by substitution, deletion or addition of one or several amino 0 acids, provided that the arginine in position 240, and optionally also the arginine in position 245, is conserved and the derived polypeptide is at least 42% homologous with said polypeptide, or v) is immunologically reactive with a polyclonal antibody raised against said polypeptide in purified form.
- the present invention provides an isolated polynucleotide molecule selected from the group consisting of (a) polynucleotide molecules encoding a polypeptide having pectate lyase activity and comprising a sequence of nucleotides as shown in SEQ ID NO : 1 from nucleotide 30 79 to nucleotide 1077; (b) species homologs of (a); (c) polynucleotide molecules that encode a polypeptide having pectate lyase activity that is at least 45% identical to the amino acid sequence of SEQ ID NO: 2 from amino acid residue 27 to amino acid residue 359; (d) molecules complementary to (a), (b) 35 or (c) ; and (e) degenerate nucleotide sequences of (a) , (b) , (c) or (d) .
- the plasmid pSJ1678 comprising the polynucleotide molecule (the DNA sequence) encoding a pectate lyase of the present invention has been transformed into a strain of the Escherichia coli which was deposited by the inventors according to the Budapest Treaty on the International Recognition of the Deposit of Microorganisms for the Purposes of Patent Procedure at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Mascheroder Weg lb, D-38124 Braunschweig, Federal Republic of Germany, on 25 September 1997 under the deposition number DSM 11788.
- the present invention relates to a pectate lyase which is i) a polypeptide produced by Bacillus licheniformis, ATCC 14580, or ii) a polypeptide comprising an amino acid sequence as shown in positions 28-341 of SEQ ID NO:4, or iii) an analogue of the polypeptide defined in i) or ii) which is at least 45% homologous with said polypeptide, or iv) is derived from said polypeptide by substitution, deletion or addition of one or several amino acids, provided that the arginine in position 233, and optionally also the arginine in position 238, is conserved and the derived polypeptide is at least 42% homologous with said polypeptide, or v) is immunologically reactive with a polyclonal antibody raised against said polypeptide in purified form.
- the present invention provides an isolated polynucleotide molecule selected from the group consisting of (a) polynucleotide molecules encoding a polypeptide having pectate lyase activity and comprising a sequence of nucleotides as shown in SEQ ID NO : 3 from nucleotide 82 to nucleotide 1026; (b) species homologs of (a); (c) polynucleotide molecules that encode a polypeptide having pectate lyase activity that is at least 45% identical to the amino acid sequence of SEQ ID NO : 4 from amino acid residue 28 to amino acid residue 341; (d) molecules complementary to (a), (b) or (c) ; and (e) degenerate nucleotide sequences of (a) , (b) , (c) or (d) .
- the plasmid pSJ1678 comprising the polynucleotide molecule (the DNA sequence) encoding a pectate lyase of the present invention has been transformed into a strain of the Escherichia coli which was deposited by the inventors according to the Budapest Treaty on the International Recognition of the Deposit of Microorganisms for the Purposes of Patent Procedure at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Mascheroder Weg lb, D-38124 Braunschweig, Federal Republic of Germany, on 25 September 1997 under the deposition number DSM 11789.
- the present invention relates to a pectate lyase which is a polypeptide produced by a Bacillus species having the 16S rDNA sequence of SEQ ID NO: 14 or by a Bacillus species having a 16S rDNA sequence homology to SEQ ID NO:14 higher than 97.3%; ii) a polypeptide comprising an amino acid sequence as shown in positions 181-509 of SEQ ID NO : 6 , or iii) an analogue of the polypeptide defined in i) or ii) which is at least 50% homologous with said polypeptide, or iv) is derived from said polypeptide by substitution, deletion or addition of one or several amino acids, provided that the arginine in position 390, and optionally also the arginine in position 395, is conserved and the derived polypeptide is at least 44% homologous with said polypeptide, or v) is immunologically reactive with a polyclonal antibody raised against said polypeptid
- the present invention provides an isolated polynucleotide molecule selected from the group consisting of (a) polynucleotide molecules encoding a polypeptide having pectate lyase activity and comprising a sequence of nucleotides as shown in SEQ ID NO : 5 from nucleotide 541 to nucleotide 1530; (b) species homologs of (a); (c) polynucleotide molecules that encode a polypeptide having pectate lyase activity that is at least 50% identical to the amino acid sequence of SEQ ID NO : 6 from amino acid residue 181 to amino acid residue 509; (d) molecules complementary to (a),
- the plasmid pSJ1678 comprising the polynucleotide molecule (the DNA sequence) encoding a pectate lyase of the present invention has been transformed into a strain of the Escherichia coli which was deposited by the inventors according to the Budapest Treaty on the International Recognition of the Deposit of Microorganisms for the Purposes of Patent Procedure at the D
- the present invention relates to a pectate lyase which is i) a polypeptide produced by a strain of the species Bacillus halodurans, preferably the species Bacillus sp .
- Bacillus sp . KJ59 which is believed to be a strain belonging to or at least very closely related to the known species Bacillus halodurans was deposited by the inventors according to the Budapest Treaty on the International Recognition of the Deposit of Microorganisms for the Purposes of Patent Procedure at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Mascheroder Weg lb, D-38124 Braunschweig, Federal Republic of Germany, on 21 September 1998 under the deposition number DSM 12419.
- the present invention provides an isolated polynucleotide molecule selected from the group consisting of (a) polynucleotide molecules encoding a polypeptide having pectate lyase activity and comprising a sequence of nucleotides as shown in SEQ ID NO : 7 from nucleotide 124 to nucleotide 1047; (b) species homologs of (a); (c) polynucleotide molecules that encode a polypeptide having pectate lyase activity that is at least 45% identical to the amino acid sequence of SEQ ID NO : 8 from amino acid residue 42 to amino acid residue 348; (d) molecules complementary to (a), (b) or (c) ; and (e) degenerate nucleotide sequences of (a) , (b) , (c) or (d) .
- the present invention relates to a pectate lyase which is a polypeptide produced by a Bacillus species having the 16S rDNA sequence of SEQ ID NO: 13 or by a Bacillus species having a 16S rDNA sequence homology to SEQ ID NO: 13 higher than 98.1%; ii) a polypeptide comprising an amino acid sequence as shown in positions 25-335 of SEQ ID NO: 10, or iii) an analogue of the polypeptide defined in i) or ii) which is at least 45% homologous with said polypeptide, or iv) is derived from said polypeptide by substitution, deletion or addition of one or several amino acids, provided that the arginine in position 227, and optionally also the argininge in position 232, is conserved and the derived polypeptide is at least 41% homologous with said polypeptide, or v) is immunologically reactive with a polyclonal antibody raised against said polypeptide in
- the present invention provides an isolated polynucleotide molecule selected from the group consisting of (a) polynucleotide molecules encoding a polypeptide having pectate lyase activity and comprising a sequence of nucleotides as shown in SEQ ID NO : 9 from nucleotide 73 to nucleotide 1008; (b) species homologs of (a); (c) polynucleotide molecules that encode a polypeptide having pectate lyase activity that is at least 45% identical to the amino acid sequence of SEQ ID NO: 10 from amino acid residue 25 to amino acid residue 335; (d) molecules complementary to (a),
- the plasmid pSJ1678 comprising the polynucleotide molecule (the DNA sequence) encoding a pectate lyase of the present invention has been transformed into a strain of the Escheri chia coli which was deposited by the inventors according to the Budapest Treaty on the International Recognition of the Deposit of Microorganisms for the Purposes of Patent Procedure at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Mascheroder Weg lb, D-38124 Braunschweig, Federal Republic of Germany, on 8 September 1998 under the deposition number DSM 12404.
- an expression vector comprising the following operably linked elements: a transcription promoter; a DNA segment selected from the group consisting of a) polynucleotide molecules encoding a polypeptide having pectate lyase activity comprising a nucleotide sequence as shown in SEQ ID NO : 1 from nucleotide 79 to nucleotide 1077, in SEQ ID NO : 3 from nucleotide 82 to nucleotide 1026, in SEQ ID NO : 5 from nucleotide 541 to nucleotide 1530, in SEQ ID NO : 7 from nucleotide 124 to nucleotide 1047 or as shown in SEQ ID NO : 9 from nucleotide 73 to nucleotide 1008, b) polynucleotide molecules encoding a polypeptide having pectate lyase activity that is at least 50% identical to the amino acid sequence of SEQ ID NO : 2 from amino
- a cultured cell into which has been introduced an expression vector as disclosed above, wherein said cell expresses the polypeptide encoded by the DNA segment .
- a further aspect of the present invention provides an isolated polypeptide having pectate lyase activity selected from the group consisting of a) polypeptide molecules having pectate lyase activity and comprising an amino acid sequence as shown in SEQ ID NO: 2 from residue 27 to residue 359; b) polypeptide molecules having pectate lyase activity and which are at least 45% identical to the amino acids of SEQ ID NO : 2 from amino acid residue 27 to amino acid residue 359; c) polypeptide molecules having pectate lyase activity and comprising an amino acid sequence as shown in SEQ ID NO : 4 from residue 28 to residue 241; d) polypeptide molecules having pectate lyase activity and which are at least 45% identical to the amino acids of SEQ ID NO : 4 from amino acid residue 28 to amino acid residue 341; e) polypeptide molecules having pectate lyase activity and comprising an amino acid sequence as shown in SEQ ID NO : 6 from residue 181 to residue 509;
- a polypeptide according to the invention comprising culturing a cell into which has been introduced an expression vector as disclosed above, whereby said cell expresses a polypeptide encoded by the DNA segment and recovering the polypeptide.
- the novel pectate lyase enzymes of the present invention are useful for the treatment of cellulosic material, especially cellulose-containing fiber, yarn, woven or non-woven fabric, treatment of mechanical paper-making pulps or recycled waste paper, and for retting of fibres.
- the treatment can be carried 30 out during the processing of cellulosic material into a material ready for garment manufacture or fabric manufacture, e.g. in the desizing or scouring step; or during industrial or household laundering of such fabric or garment .
- the present invention 35 relates to a detergent composition comprising an enzyme having substantial pectate lyase activity; and to use of the enzyme of the invention for the treatment of cellulose-containing fibers, yarn, woven or non-woven fabric.
- pectate lyases of the invention are very effective for OJ J to h- 1 on o n o on o On
- polypeptide or protein obtained from a given species that has homology to a distinct polypeptide or protein from that same species.
- expression vector denotes a DNA molecule, linear or circular, that comprises a segment encoding a polypeptide of interest operably linked to additional segments that provide for its transcription. Such additional segments may include promoter and terminator sequences, and may optionally include one or more origins of replication, one or more selectable markers, an enhancer, a polyadenylation signal, and the like. Expression vectors are generally derived from plasmid or viral DNA, or may contain elements of both.
- the expression vector of the invention may be any expression vector that is conveniently subjected to recombinant DNA procedures, and the choice of vector will often depend on the host cell into which the vector it is to be introduced. Thus, the vector may be an autonomously replicating vector, i.e.
- a vector which exists as an extra- chromosomal entity, the replication of which is independent of chromosomal replication e.g. a plasmid.
- the vector may be one which, when introduced into a host cell, is integrated into the host cell genome and replicated together with the chromosome (s) into which it has been integrated.
- the term "recombinant expressed” or “recombinantly expressed” used herein in connection with expression of a polypeptide or protein is defined according to the standard definition in the art. Recombinantly expression of a protein is generally performed by using an expression vector as described immediately above.
- isolated when applied to a polynucleotide molecule, denotes that the polynucleotide has been removed from its natural genetic milieu and is thus free of other extraneous or unwanted coding sequences, and is in a form suitable for use within genetically engineered protein production systems.
- isolated molecules are those that are separated from their natural environment and include cDNA and genomic clones.
- Isolated DNA molecules of the present invention are free of other genes with which they are ordinarily associated, but may include naturally occurring 5 ' and 3 ' untranslated regions such as promoters and terminators. The identification of associated regions will be evident to one of ordinary skill in the art (see for example, Dynan and Tijan, Nature 316 :774-78, 1985).
- the term "an isolated polynucleotide” may alternatively be termed "a cloned polynucleotide” .
- the term "isolated” indicates that the protein is found in a condition other than its native environment.
- the isolated protein is substantially free of other proteins, particularly other homologous proteins (i.e. "homologous impurities” (see below) ) .
- homologous impurities i.e. "homologous impurities” (see below)
- isolated protein/polypeptide may alternatively be termed “purified protein/polypeptide” .
- homologous impurities means any impurity (e.g. another polypeptide than the polypeptide of the invention) which originate from the homologous cell where the polypeptide of the invention is originally obtained from.
- the term "obtained from” as used herein in connection with a specific microbial source means that the polynucleotide and/or polypeptide produced by the specific source, or by a cell in which a gene from the source have been inserted.
- endogeneous to means that a polypeptide is produced by the specific source due to the presence in the source of a native gene, ie a gene which has not been recombinantly inserted into a cell of the source but is naturally occurring.
- operably linked when referring to DNA segments, denotes that the segments are arranged so that they function in concert for their intended purposes, e.g. transcription initiates in the promoter and proceeds through the coding segment to the terminator n n
- PGL enzyme which catalyzes the random cleavage of alpha- 1,4- glycosidic linkages in pectic acid also called polygalacturonic acid by transelimination
- PGL enzyme class polygalacturonate lyase
- PGL enzyme class polygalacturonate lyase
- poly (1, 4-alpha-D-galacturonide) lyase also known as pectate lyase .
- the disclosed sequence information herein relating to a polynucleotide sequence encoding a pectate lyase of the invention can be used as a tool to identify other homologous pectate lyases.
- polymerase chain reaction PCR
- PCR polymerase chain reaction
- an isolated polynucleotide of the invention will hybridize to similar sized regions of SEQ ID No. 1, 3, 5, 7 or 9, respectively, or a sequence complementary thereto, under at least medium stringency conditions .
- polynucleotides of the invention will hybridize to a denatured double- stranded DNA probe comprising either the full sequence (encoding for the mature part of the polypeptide) shown in positions 79-1077 of SEQ ID NO:l, in positions 82-1026 of SEQ ID NO : 3 , in positions 541-1530 of SEQ ID NO:5, in positions 124-1047 of SEQ ID NO : 7 or in positions 73-1008 of SEQ ID NO : 9 , or any probe comprising a subsequence of SEQ ID NO : 1 , 3, 5, 7 or 9 , respectively, or any probe comprising a subsequence of SEQ ID NO : 1 , 3, 5, 7 or 9 having a length of at least about 100 base pairs under at least medium stringency conditions, but preferably at high stringency conditions as described in detail below.
- Suitable experimental conditions for determining hybridization at medium, or high stringency between a nucleotide probe and a homologous DNA or RNA sequence involves presoaking of the filter containing the DNA fragments or RNA to hybridize in 5 x SSC (Sodium chloride/Sodium citrate, Sambrook o o on rt TJ ⁇ to cr ⁇ ⁇ Z ⁇ 0 TJ to Xi SU 3 LQ D TJ ⁇ ⁇ _ ⁇ ⁇ . P- rt td ⁇ rt to CQ ⁇ tr 0 tr ⁇ >- X P- 0 rt ⁇ rt o 0 ⁇ ⁇ a ii ⁇ ⁇ ! o in ⁇ CO tr P- SD tr TJ 0 r
- a DNA can also be cloned using the polymerase chain reaction, or PCR (Mullis, U.S. Patent 4,683,202), using primers designed from the sequences disclosed herein.
- the DNA library can be used to transform or transfect host cells, and expression of the DNA of interest can be detected with an antibody (mono-clonal or polyclonal) raised against the pectate lyase cloned from B . licheniformis, ATCC 14580, the pectate lyase cloned from B .
- the polypeptide encoding part of the DNA sequence cloned into plasmid pSJ1678 present in Escherichia coli DSM 11789 and/or an analogue DNA sequence of the invention may be cloned from a strain of the bacterial species Bacillus licheniformis, preferably the strain ATCC 14580, producing the pectate lyase enzyme, or another or related organism as described herein.
- polypeptide encoding part of the DNA sequence cloned into plasmid pSJ1678 present in Escherichia coli DSM 11789 and/or an analogue DNA sequence of the invention may be cloned from a strain of the bacterial species Bacillus agaradhaerens as represented by the type strain DSM 8721, producing the pectate lyase enzyme, or another or related organism as described herein.
- polypeptide encoding part of the DNA sequence cloned into plasmid pSJ1678 present in Escherichia coli DSM 12403 and 12404, respectively, and/or an analogue DNA sequence of the invention may be cloned from a strain of Bacillus sp . AAI12, Bacillus sp . KJ59, DSM 12419, or Bacillus sp . 1534 producing the pectate lyase enzyme, or another or related organism as described herein.
- analogous sequence may be constructed on t
- the polynucleotide molecule of the invention may be isolated from Escherichia coli , DSM 11788, DSM 11789, DSM 12403 or DSM 12404, in which the each of the plasmids obtained by cloning such as described above is deposited. Also, the present invention relates to an isolated substantially pure 0 biological culture of each of the strains Escherichia coli , DSM 11788, DSM 11789, DSM 12403 and DSM 12404, respectively.
- sequence of amino acids no. 27-359 of SEQ ID No 2 is a
- positions 32-86 is a first lectin domain
- positions 87- 134 is a second lectin domain
- positions 135-180 is a third lectin domain.
- the sequence of amino acids no. 42-348 of SEQ ID No 8 is a mature pectate lyase sequence; positions 1-41 is a propeptide.
- pectate lyases of the invention belongs to family 1 of polysaccharide lyases.
- the present invention also provides pectate lyase polypeptides that are substantially homologous to the mature
- the term "substantially homologous” is used herein to denote polypeptides having at least 45% preferably at least 50%, preferably at least 60%, more preferably at least 70%, more preferably at least 85%, and even
- sequence identity to the sequences shown in SEQ ID NO : 2 , 4, 6, 8 and 10, or their orthologs or paralogs.
- polypeptides will more preferably be at least 95% identical, and most preferably 98% or more identical to the sequences shown in SEQ ID NO: 2, 4, 6, 8 and 10, or their orthologs or paralogs.
- Percent sequence identity is determined by conventional methods, by means of computer programs known in the art such as GAP provided in the GCG program package (Program Manual for the Wisconsin Package, Version 8, August 1994, Genetics Computer Group, 575 Science Drive, Madison, Wisconsin, USA 53711) as disclosed in Needleman, S.B.
- GAP is used with the following settings for polypeptide sequence comparison: GAP creation penalty of 3.0 and GAP extension penalty of 0.1.
- Sequence identity of polynucleotide molecules is determined by similar methods using GAP with the following settings for DNA sequence comparison: GAP creation penalty of 5.0 and GAP extension penalty of 0.3.
- the pectate lyases of the invention comprising the unique first amino acid sequence NLNSRVP, which is believed to be unique and thereby sufficient for identifying any new pectate lyase belonging to this novel group of pectate lyase of the present invention having excellent performance in industrial processes such as textile treatment and laundering, are preferably derived from a microorganism, preferably from a bacterium, an archea or a fungus, especially from a bacterium such as a bacterium belonging to Bacillus , preferably to an alkalophilic Bacillus strain which may be selected from the group consisting of the species Bacillus licheniformis, Bacillus agaradhaerens , Bacillus halodurans and the Bacillus species 1534 and AAI12 identified by the 16S rDNA sequence listed as SEQ ID Nos: 13 or 14, respectively, and other Bacillus species which are highly related to any of these species based on aligned 16S rDNA sequences as explained below, preferably
- Sequence similarities were established using the Phylip Distance Matrix option (with default settings, i.e. no corrections) integrated in the ARB program package, by converting the distances to per cent sequence similarity.
- B . licheniformis ATCC 14580
- B . subtilis the species most closely related to Bacillus sp .
- KJ59, DSM 12419, is B . halodurans, DSM 8718 (16S data X76442) which is so close that the strain KJ59 is believed to be a strain of this species; the species most closely related to Bacillus sp .
- AAI12 is B .
- DSM 485 (16S data X76436) which shows a 16S homology of 97.3%; and the species most closely related to Bacillus sp . 1534 is Bacillus sp . PN1 , DSM 8714 (16S data X76438) which shows a 16S homology of 98.1%.
- Phylogenetic trees were calculated using the ARB program by the maximum likelihood method (FastDnaML algorithm from G. J. Olsen, H. Matsuda, R. Hagstrom, and R. Overbeek. fastDNAml : a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput .Appl .Biosci .
- Substantially homologous proteins and polypeptides are characterized as having one or more amino acid substitutions, deletions or additions. These changes are preferably of a minor nature, that is conservative amino acid substitutions (see Table 2) and other substitutions that do not significantly affect the folding or activity of the protein or polypeptide; small deletions, typically of one to about 30 amino acids; and small amino- or carboxyl -terminal extensions, such as an amino- terminal methionine residue, a small linker peptide of up to about 20-25 residues, or a small extension that facilitates purification (an affinity tag), such as a poly-histidine tract, protein A (Nilsson et al .
- an affinity tag such as a poly-histidine tract, protein A (Nilsson et al .
- non-standard ammo acids such as 4-hydroxyprolme, 6 -N-methyl lysine, 2- ammoisobuty ⁇ c acid, isovalme and a-methyl serine
- a limited number of non-conservative ammo acids, ammo acids that are not encoded by the genetic code, and unnatural ammo acids may be substituted for ammo acid residues.
- "Unnatural ammo acids” have been modified after protein synthesis, and/or have a chemical structure their side chain (s) different from that of the standard amino acids.
- Unnatural amino acids can be chemically synthesized, or preferably, are commercially available, and include pipecolic acid, thiazolidine carboxylic acid, dehydroproline, 3- and 4- 5 methylproline, and 3 , 3-dimethylproline .
- Essential amino acids in the pectate lyase polypeptides of the present invention can be identified according to procedures known in the art, such as site-directed mutagenesis or alanine- scanning mutagenesis (Cunningham and Wells, Science 244 : 1081-
- the active site of the enzyme or other biological interaction can also be determined by physical analysis of structure, as determined by such techniques as nuclear magnetic resonance, crystallography, electron diffraction or photoaffinity labeling, in conjunction
- Mutagenesis/shuffling methods as disclosed above can be combined with high-throughput , automated screening methods to detect activity of cloned, mutagenized polypeptides in host cells.
- Mutagenized DNA molecules that encode active polypeptides can be recovered from the host cells and rapidly sequenced using modern equipment. These methods allow the rapid determination of the importance of individual amino acid residues in a polypeptide of interest, and can be applied to polypeptides of unknown structure .
- polypeptides that are substantially homologous to residues 27 to 359 of SEQ ID NO:2, to residues 28 to 341 of SEQ ID NO : 4 , to residues 181 to 509 of SEQ ID NO: 6, to residues 42 to 348 of SEQ ID NO : 8 and to residues 25 to 335 of SEQ ID NO: 10 and retain the pectate lyase activity of the wild-type protein.
- Such variants of the invention are pectate lyases having, in position 223 relative to the numbering in the sequence alignment of Figure 1, the amino acid residue arginine.
- such variant also holds a conserved arginine in position 228 relative to the numbering in the sequence alignment of Figure 1.
- the present invention relates to pectate lyases having an amino acid sequence which is derived from any of the amino acid sequences SEQ ID No: 2, 4, 6, 8 and 10 by deletion, replacement or addition of one or more amino acid residues (hereinafter referred to as mutation) provided that the pectate lyase activity is not deactivated and the mutation conserves arginine at the 223rd position and optionally also arginine at the 228th position of the sequence numbering in the alignment of Figure 1.
- positions corresponds to arginine (R) at the 240th position and at the 245th position in SEQ ID No: 2, to positions 233 and 238 in SEQ ID NO : 4 , to positions 390 and 395 in SEQ ID NO: 6, to positions 240 and 245 in SEQ ID NO : 8 , and to positions 227 and 232 in SEQ ID NO: 10.
- the mutation preferably conserves aspartic acid (D) at the 169th position and/or aspartic acid (D) at the 173rd position and/or lysine (K) at the 193rd position of the sequence numbering in the alignment of Figure 1.
- positions corresponds to aspartic acid (D) at the at the 186th position and at the 190th position and lysine (K) at the 210th position in SEQ ID NO : 2 ; to positions D180, D184 and K204 in SEQ ID NO : 4 ; to positions D336, D340 and K360 in SEQ ID NO : 6 ; to positions D187, D191 and K211 in SEQ ID NO : 8 , and to positions D174, D178 and K198 in SEQ ID NO: 10.
- D aspartic acid
- K lysine
- the degree of mutation is not particularly limited provided that the above described arginine in the 223rd position position is conserved.
- 40% or higher homology exists between such mutation variants of the native or parent pectate lyase enzyme, calculated on the any of the sequence SEQ ID Nos: 2, 4, 6, 8 and 10: 42% or higher homology exists between amino acid positions 39 and 359 of SEQ ID NO : 2 and amino acid positions 46 and 341 of SEQ ID NO: 4; 44% or higher homology exists between amino acid positions 187 and 509 of SEQ ID NO : 6 ; 40% or higher homology exists between amino acid positions 50 and 348 of SEQ ID NO : 8 ; and 41% or higher homology exists between amino acid positions 40 and 335 ofSEQ ID NO: 10.
- the homology is at least 45%, preferably at least 50%, more preferably at least 55%, more preferably at least 60%, even more preferably at least 70%, even more preferably at least 75%, even more preferably at least 80%, even more preferably at least 85%, even more preferably at least 90%, even more preferably at least 95%, especially at least 98%.
- the pectate lyase of the invention may, in addition to the enzyme core comprising the catalytically domain, also comprise a cellulose binding domain (CBD) , the cellulose binding domain and enzyme core (the catalytically active domain) of the enzyme being operably linked.
- the cellulose binding domain (CBD) may exist as an integral part of the encoded enzyme, or a CBD from another origin may be introduced into the pectin degrading enzyme thus creating an enzyme hybrid.
- the term "cellulose-binding domain” is intended to be understood as defined by Peter Tomme et al . "Cellulose-Binding Domains: Classification and Properties" in "Enzymatic Degradation of Insoluble Carbohydrates", John N. Saddler and Michael H.
- CBDs are found in various enzymes such as cellulases, xylanases, mannanases, arabinofuranosidases, acetyl esterases and chitinases. CBDs have also been found in algae, e.g. the red alga Porphyra purpurea as a non-hydrolytic polysaccharide-binding protein, see Tomme et al . , op . ci t .
- Enzyme hybrids are known in the art, see e.g. WO 90/00609 and WO 95/16782, and may be prepared by transforming into a host cell a DNA construct comprising at least a fragment of DNA encoding the cellulose-binding domain ligated, with or without a linker, to a DNA sequence encoding the pectin degrading enzyme and growing the host cell to express the fused gene. Enzyme hybrids may be described by the following formula:
- CBD is the N-terminal or the C-terminal region of an amino acid sequence corresponding to at least the cellulose- binding domain
- MR is the middle region (the linker) , and may be a bond, or a short linking group preferably of from about 2 to about 100 carbon atoms, more preferably of from 2 to 40 carbon atoms; or is preferably from about 2 to to about 100 amino acids, more preferably of from 2 to 40 amino acids
- X is an N-terminal or C-terminal region of the pectin degrading enzyme of the invention.
- the enzyme of the present invention has its maximum catalytic activity at a pH of at least 8, more preferably higher than 8.5, more preferably higher than 9, more preferably higher than 9.5, more preferably higher than 10, even more preferably higher than 10.5, especially higher than 11; and preferably the maximum activity of the enzyme is obtained at a temperature of at least 50°C, more preferably of at least 55°C.
- polypeptides of the present invention can be produced in genetically engineered host cells according to conventional techniques.
- Suitable host cells are those cell types that can be transformed or transfected with exogenous DNA and grown in culture, and include bacteria, fungal cells, and cultured higher eukaryotic cells.
- Bacterial cells, particularly cultured cells of gram-positive organisms, are preferred.
- Gram- positive cells from the genus of Bacillus are especially preferred, such as Bacillus subtilis, Bacillus lentus, Bacillus brevis, Bacillus stearothermophilus , Bacillus alkalophilus, Bacillus amyloliquefaciens, Bacillus coagulans, Bacillus cir- culans , Bacillus lautus, Bacillus thuringiensis , Bacillus agaradhaerens or Bacillus licheniformis .
- Techniques for manipulating cloned DNA molecules and introducing exogenous DNA into a variety of host cells are disclosed by Sambrook et al . , Molecular Cloning: A Laboratory Manual , 2nd ed.
- a DNA sequence encoding a pectate lyase of the present invention is operably linked to other genetic elements required for its expression, generally including a transcription promoter and terminator within an expression vector.
- the vector will also commonly contain one or more selectable markers and one or more origins of replication, although those skilled in the art will recognize that within certain systems selectable markers may be provided on separate vectors, and replication of the exogenous DNA may be provided by integration into the host cell genome. Selection of promoters, terminators, selectable markers, vectors and other elements is a matter of routine design within the level of ordinary skill in the art. Many such elements are described in the literature and are available through commercial suppliers.
- a secretory signal sequence (also known as a leader sequence, prepro sequence or pre sequence) is provided in the expression vector.
- the secretory signal sequence may be that of the polypeptide, or may be derived from another secreted protein or synthesized de novo . Numerous suitable secretory signal sequences are known in the art and reference is made to
- secretory signal sequences especially for secretion in a Bacillus host cell.
- the secretory signal sequence is joined to the DNA sequence in the correct reading frame.
- Secretory signal sequences are commonly positioned 5 ' to the DNA sequence encoding the polypeptide of interest, although certain signal sequences may be positioned elsewhere in the DNA sequence of interest (see, e.g., Welch et al . , U.S. Patent No. 5,037,743; Holland et al . , U.S. Patent No. 5,143,830).
- Transformed or transfected host cells are cultured according to conventional procedures in a culture medium containing nutrients and other components required for the growth of the chosen host cells.
- suitable media including defined media and complex media, are known in the art and generally include a carbon source, a nitrogen source, essential amino acids, vitamins and minerals. Media may also contain such components as growth factors or serum, as required.
- the growth medium will generally select for cells containing the exogenously added DNA by, for example, drug selection or deficiency in an essential nutrient which is complemented by the selectable marker carried on the expression vector or co- transfected into the host cell.
- the polypeptides of the present invention may also be produced by fermenting a wildtype strain belonging to the genus Bacillus, preferably a strain which may be selected from the group consisting of the species Bacillus licheniformis, Bacillus agaradhaerens and highly related Bacillus species in which all species are at least 95% homologous to Bacillus licheniformis based on aligned 16S rDNA sequences.
- a wildtype strain belonging to the genus Bacillus preferably a strain which may be selected from the group consisting of the species Bacillus licheniformis, Bacillus agaradhaerens and highly related Bacillus species in which all species are at least 95% homologous to Bacillus licheniformis based on aligned 16S rDNA sequences.
- Specific and highly preferred examples are Bacillus licheniformis, ATCC 14580, and Bacillus agaradhaerens, DSM 8721.
- polypeptides of the present invention may be produced by fermenting a mutant or a variant derived from the above mentioned strain.
- a mutant may be obtained by using conventional mutagenesis by subjecting the strain in question to treatment with a mutagen (eg NTG (n-methyl-N-nitro-N- nitrosoguanidine) ) or to ultraviolet radiation, eg as described in Manual of methods for General Bacteriology; ASM 1981, Chapter 13.
- a mutagen eg NTG (n-methyl-N-nitro-N- nitrosoguanidine)
- ultraviolet radiation eg as described in Manual of methods for General Bacteriology; ASM 1981, Chapter 13.
- This mutagenesis is performed to stimulate mutation of the strains.
- a screening for mutants giving higher pectinase yields aer possible using conventional plate assays or liquid assays.
- the fermentation may be carried out by cultivation of the strain under aerobic conditions in a nutrient medium containing carbon and nitrogen sources together with other essential nutrients, the medium being composed in accordance with the principles of the known art .
- the medium may be a complex rich medium or a minimal medium.
- the nitrogen source may be of inorganic and/or organic nature. Suitable inorganic nitrogen sources are nitrates and ammonium salts. Among the organic nitrogen sources quite a number are used regularly in fermentations. Examples are soybean meal, casein, corn, corn steep liquor, yeast extract, urea and albumin. Suitable carbon sources are carbohydrates or carbohydrate containing materials.
- the nutrient medium contains pectate, polygalacturonic acid and/or pectin esterified to a higher or lower degree as carbon source and/or inducer of pectinase production.
- the medium contains a pectin rich material such as soybean meal, apple pulp or citrus peel. Since the Bacillus species of this invention are alkalophilic the cultivation is preferably conducted at alkaline pH values such as at least pH 8 or at least pH 9, which can be obtained by addition of suitable buffers such as sodium carbonate or mixtures of sodium carbonate and sodium bicarbonate after sterilisation of the growth medium.
- Selection of a particular method is a matter of routine design and is determined in part by the properties of the chosen support. See, for example, Affinity Chromatography: Principles & Methods . Pharmacia LKB Biotechnology, Uppsala, Sweden, 1988.
- Polypeptides of the invention or fragments thereof may also be prepared through chemical synthesis.
- Polypeptides of the invention may be monomers or multimers; glycosylated or non- glycosylated; pegylated or non-pegylated; and may or may not include an initial methionine amino acid residue.
- the present invention also relates to a transgenic plant, plant part or plant cell which has been transformed with a DNA sequence encoding the pectin degrading enzyme of the invention so as to express and produce this enzyme in recoverable quantities.
- the enzyme may be recovered from the plant or plant part.
- the plant or plant part containing the recombinant enzyme may be used as such.
- the transgenic plant can be dicotyledonous or monocotyledonous, for short a dicot or a monocot .
- monocot plants are grasses, such as meadow grass (blue grass, Poa) , forage grass such as festuca, lolium, temperate grass, such as Agrostis, and cereals, e.g. wheat, oats, rye, barley, rice, sorghum and maize (corn) .
- dicot plants are tobacco, legumes, such as lupins, potato, sugar beet, pea, bean and soybean, and cruciferous (family Brassicaceae) , such as cauliflower, oil seed rape and the closely related model organism Arabidopsis thaliana .
- plant parts are stem, callus, leaves, root, fruits, seeds, and tubers.
- plant tissues such as chloroplast, apoplast, mitochondria, vacuole, peroxisomes and cytoplasm are considered to be a plant part.
- any plant cell whatever the tissue origin, is considered to be a plant part .
- progeny of such plants, plant parts and plant cells are also included within the scope of the invention.
- the transgenic plant or plant cell expressing the enzyme of the invention may be constructed in accordance with methods known in the art.
- the plant or plant cell is constructed by incorporating one or more expression constructs encoding the enzyme of the invention into the plant host genome and propagating the resulting modified plant or plant cell into a transgenic plant or plant cell .
- the expression construct is a DNA construct which comprises a gene encoding the enzyme of the invention in operable association with appropriate regulatory sequences required for expression of the gene in the plant or plant part of choice.
- the expression construct may comprise a selectable marker useful for identifying host cells into which the expression construct has been integrated and DNA sequences necessary for introduction of the construct into the plant in question (the latter depends on the DNA introduction method to be used) .
- regulatory sequences such as promoter and terminator sequences and optionally signal or transit sequences is determined, eg on the basis of when, where and how the enzyme is desired to be expressed.
- the expression of the gene encoding the enzyme of the invention may be constitutive or inducible, or may be developmental, stage or tissue specific, and the gene product may be targeted to a specific tissue or plant part such as seeds or leaves.
- Regulatory sequences are eg described by Tague et al , Plant, Phys . , 86, 506, 1988.
- the 35S-CaMV promoter may be used (Franck et al . , 1980. Cell 21: 285-294).
- Organ-specific promoters may eg be a promoter from storage sink tissues such as seeds, potato tubers, and fruits (Edwards & Coruzzi, 1990. Annu. Rev. Genet. 24: 275-303), or from metabolic sink tissues such as meristems (Ito et al . , 1994. Plant Mol . Biol. 24: 863-878), a seed specific promoter such as the glutelin, prolamin, globulin or albumin promoter from rice (Wu et al . , Plant and Cell Physiology Vol. 39, No. 8 pp.
- Vicia faba promoter from the legumin B4 and the unknown seed protein gene from Vicia faba described by Conrad U. et al , Journal of Plant Physiology Vol. 152, No. 6 pp. 708-711 (1998), a promotter from a seed oil body protein (Chen et al . , Plant and cell physiology vol. 39, No. 9 pp. 935-941 (1998), the storage protein napA promoter from Brassica napus , or any other seed specific promoter known in the art, eg as described in WO 91/14772.
- the promoter may be a leaf specific promoter such as the rbcs promoter from rice or tomato (Kyozuka et al . , Plant Physiology Vol. 102, No. 3 pp. 991-1000 (1993), the chlorella virus adenine methyltransferase gene promoter (Mitra, A. and Higgins, DW, Plant Molecular Biology Vol. 26, No. 1 pp. 85-93 (1994) , or the aldP gene promoter from rice (Kagaya et al . , Molecular and General Genetics Vol. 248, No. 6 pp .
- a promoter enhancer element may be used to achieve higher expression of the enzyme in the plant.
- the promoter enhancer element may be an intron which is placed between the promoter and the nucleotide sequence encoding the enzyme.
- Xu et al . op ci t disclose the use of the first intron of the rice actin 1 gene to enhance expression.
- the selectable marker gene and any other parts of the expression construct may be chosen from those available in the art .
- the DNA construct is incorporated into the plant genome according to conventional techniques known in the art, including Agrobacterium-mediated transformation, virus-mediated transformation, micro • injection, particle bombardment, biolistic transformation, and electroporation (Gasser et al , Science, 244, 1293; Potrykus, Bio/Techn. 8, 535, 1990; Shimamoto et al , Nature , 338 , 274 , 1989 ) .
- transgenic dicots for review Hooykas & Schilperoort , 1992. Plant Mol . Biol. 19: 15-38
- the method of choice for generating transgenic monocots is particle bombardment (microscopic gold or tungsten particles coated with the transforming DNA) of embryonic calli or developing embryos (Christou, 1992. Plant J. 2: 275-281; Shimamoto, 1994. Curr. Opin. Biotechnol . 5: 158-162; Vasil et al . , 1992.
- transformants having incorporated the expression construct are selected and regenerated into whole plants according to methods well-known in the art .
- enzyme preparation is intended to mean either be a conventional enzymatic fermentation product, possibly isolated and purified, from a single species of a microorganism, such preparation usually comprising a number of different enzymatic activities; or a mixture of monocomponent enzymes, preferably enzymes derived from bacterial or fungal species by using conventional recombinant techniques, which enzymes have been fermented and possibly isolated and purified separately and which may originate from different species, preferably fungal or bacterial species; or the fermentation product of a microorganism which acts as a host cell for expression of a recombinant pectate lyase, but which microorganism simultaneously produces other enzymes, e.g.
- pectin lyases being naturally occurring fermentation products of the microorganism, i.e. the enzyme complex conventionally produced by the corresponding naturally occurring microorganism.
- the pectate lyase preparation of the invention may further comprise one or more enzymes selected from the group consisting of proteases, cellulases (endo- ⁇ -1 , 4-glucanases) , ⁇ -glucanases (endo- ⁇ -1, 3 (4) -glucanases) , lipases, cutinases, peroxidases, laccases, amylases, glucoamylases, pectinases, reductases, oxidases, phenoloxidases , ligninases, pullulanases, arabinanases, hemicellulases , mannanases, xyloglucanases, xylanases, pectin acetyl esterases, rhamnogalac
- one or more or all enzymes in the preparation is produced by using recombinant techniques, i.e. the enzyme (s) is/are mono-component enzyme (s) which is/are mixed with the other enzyme (s) to form an enzyme preparation with the desired enzyme blend.
- Polyclonal antibodies (which are monospecific for a given enzyme protein) to be used in determining immunological cross- reactivity may be prepared by use of a purified pectate lyase enzyme. More specifically, antiserum against the pectate lyase of the invention may be raised by immunizing rabbits (or other rodents) according to the procedure described by N. Axelsen et al . in: A Manual of Quantitative Immunoelectrophoresis , Blackwell Scientific Publications, 1973, Chapter 23, or A. Johnstone and R. Thorpe, Immunochemistry in Practice, Blackwell Scientific Publications, 1982 (more specifically p. 27-31).
- Purified immunoglobulins may be obtained from the antisera, for example by salt precipitation ((NH 4 ) 2 S0 4 ) , followed by dialysis and ion exchange chromatography, e . g. on DEAE-Sephadex.
- Immunochemical characterization of proteins may be done either by Outcherlony double-diffusion analysis (0. Ouchterlony in: Handbook of Experimental Immunology (D.M. Weir, Ed.), Blackwell Scientific Publications, 1967, pp. 655-706), by crossed immunoelectrophoresis (N. Axelsen et al . , supra . Chapters 3 and 4), or by rocket immunoelectrophoresis (N. Axelsen et al . , Chapter 2) .
- TJ P- i ⁇ - rt 0 ⁇ ⁇ Pi ⁇ SU ⁇ TJ LQ P- LQ 3 ⁇ o P- SD SD 3 rt
- compositions of the invention preferably contain a surfactant and preferably other detergent compounds selected from organic polymeric compounds, suds enhancing agents, group II metal ions, solvents, hydrotropes and additional enzymes.
- compositions suitable for use in a laundry machine washing method preferably contain both a surfactant and a builder compound and additionally one or more detergent components preferably selected from organic polymeric compounds, bleaching agents, additional enzymes, suds suppressors, dispersants, lime- soap dispersants, soil suspension and anti-redeposition agents and corrosion inhibitors.
- Laundry compositions can also contain softening agents, as additional detergent components.
- Such compositions containing carbohydrase can provide fabric cleaning, stain removal, whiteness maintenance, softening, colour appearance, dye transfer inhibition and sanitization when formulated as laundry detergent compositions.
- compositions of the invention can also be used as detergent additive products in solid or liquid form.
- Such additive products are intended to supplement or boost the performance of conventional detergent compositions and can be added at any stage of the cleaning process.
- the density of the laundry detergent compositions herein ranges from 400 to 1200 g/litre, preferably 500 to 950 g/litre of composition measured at 20°C.
- compositions herein are best reflected by density and, in terms of composition, by the amount of inorganic filler salt; inorganic filler salts are conventional ingredients of detergent compositions in powder form; in conventional detergent compositions, the filler salts are present in substantial amounts, typically 17-35% by weight of the total composition. In the compact compositions, the filler salt is present in amounts not exceeding 15% of the total composition, preferably not exceeding 10%, most preferably not exceeding 5% by weight of the composition.
- the inorganic filler salts, such as meant in the present compositions are selected from the alkali and alkaline-earth-metal salts of sulphates and chlorides. A preferred filler salt is sodium sulphate.
- Liquid detergent compositions according to the present invention can also be in a "concentrated form", in such case, the liquid detergent compositions according the present invention will contain a lower amount of water, compared to conventional liquid detergents.
- the water content of the concentrated liquid detergent is preferably less than 40%, more preferably less than 30%, most preferably less than 20% by weight of the detergent composition.
- Suitable specific detergent compounds for use herein are selected from the group consisting of the specific compounds as described in WO 97/01629 which is hereby incorporated by reference in its entirety.
- Mannanase may be incorporated into the cleaning compositions in accordance with the invention preferably at a level of from 0.0001% to 2%, more preferably from 0.0005% to 0.5%, most preferred from 0.001% to 0.1% pure enzyme by weight of the composition.
- the cellulases usable in the present invention include both bacterial or fungal cellulases. Preferably, they will have a pH optimum of between 5 and 12 and a specific activity above 50 CEVU/mg (Cellulose Viscosity Unit) .
- Suitable cellulases are disclosed in U.S. Patent 4,435,307, J61078384 and WO96/02653 which discloses fungal cellulase produced from Humicola insolens, Trichoderma, Thielavia and Sporotrichum, respectively.
- EP 739 982 describes cellulases isolated from novel Bacillus species. Suitable cellulases are also disclosed in GB-A-2075028 ; GB-A-2095275; DE-OS-22 47 832 and W095/26398.
- cellulases examples include cellulases produced by a strain of Humicola insolens (Humicola grisea var. thermoidea) , particularly the strain Humicola insolens , DSM 1800.
- Other suitable cellulases are cellulases originated from Humicola insolens having a molecular weight of about 50kD, an isoelectric point of 5.5 and containing 415 amino acids; and a ⁇ 43kD endo- beta-1 , 4-glucanase derived from Humicola insolens , DSM 1800; a preferred cellulase has the amino acid sequence disclosed in PCT Patent Application No. WO 91/17243.
- suitable cellulases are the EGIII cellulases from Trichoderma longibrachiatum described in WO94/21801. Especially suitable cellulases are the cellulases 5 having color care benefits. Examples of such cellulases are the cellulases described in W096/29397, EP-A-0495257 , WO 91/17243, W091/17244 and WO91/21801. Other suitable cellulases for fabric care and/or cleaning properties are described in WO96/34092, W096/17994 and W095/24471.
- Said cellulases are normally incorporated in the detergent composition at levels from 0.0001% to 2% of pure enzyme by weight of the detergent composition.
- Preferred cellulases for the purpose of the present invention are alkaline cellulases, i.e. enzyme having at least
- a preferred alkaline cellulase is the cellulase sold under the tradename Carezyme ® by Novo Nordisk A/S.
- Amylases ( ⁇ and/or ⁇ ) can be included for removal of carbohydrate-based stains.
- WO94/02597 Novo Nordisk A/S published February 03, 1994, describes cleaning compositions which incorporate mutant amylases. See also WO95/10603, Novo Nordisk A/S, published April 20, 1995.
- Other amylases known for
- 25 use in cleaning compositions include both ⁇ - and ⁇ -amylases.
- ⁇ - Amylases are known in the art and include those disclosed in US Pat. no. 5,003,257; EP 252,666; WO/91/00353 ; FR 2,676,456; EP 285,123; EP 525,610; EP 368,341; and British Patent specification no. 1,296,839 (Novo) .
- Other suitable amylases are examples of suitable amylases.
- amylases described in W094/18314, published August 18, 1994 and WO96/05295, Genencor, published February 22, 1996 and amylase variants having additional modification in the immediate parent available from Novo Nordisk A/S, disclosed in WO 95/10603, published April 95. Also suitable are amylases
- ⁇ -amylases characterised by having a specific activity at least 25% higher than the specific activity of Termamyl® at a temperature range of 25°C to 55°C and at a pH value in the range of 8 to 10, measured by the Phadebas
- ® ⁇ -amylase activity assay Suitable are variants of the above enzymes, described in W096/23873 (Novo Nordisk) .
- Other amylolytic enzymes with improved properties with respect to the activity level and the combination of thermostability and a higher activity level are described in W095/35382.
- Preferred amylases for the purpose of the present invention are the amylases sold under the tradename Termamyl, Duramyl and Maxamyl and or the ⁇ -amylase variant demonstrating increased thermostability disclosed as SEQ ID No. 2 in W096/23873.
- Preferred amylases for specific applications are alkaline amylases, ie enzymes having an enzymatic activity of at least 10%, preferably at least 25%, more preferably at least 40% of their maximum activity at a pH ranging from 7 to 12. More preferred amylases are enzymes having their maximum activity at a pH ranging from 7 to 12.
- amylolytic enzymes are incorporated in the detergent compositions of the present invention a level of from 0.0001% to 2%, preferably from 0.00018% to 0.06%, more preferably from 0.00024% to 0.048% pure enzyme by weight of the composition.
- xyloglucanase encompasses the family of enzymes described by Vincken and Voragen at Wageningen University [Vincken et al (1994) Plant Physiol . , 104, 99-107] and are able to degrade xyloglucans as described in Hayashi et al (1989) Plant. Physiol. Plant Mol . Biol., 40, 139-168.
- Vincken et al demonstrated the removal of xyloglucan coating from cellulase of the isolated apple cell wall by a xyloglucanase purified from Trichoderma viride (endo-IV-glucanase) .
- This enzyme enhances the enzymatic degradation of cell wall -embedded cellulose and work in synergy with pectic enzymes.
- Rapidase LIQ+ from Gist-Brocades contains an xyloglucanase activity.
- This xyloglucanase is incorporated into the cleaning compositions in accordance with the invention preferably at a level of from 0.0001% to 2%, more preferably from ⁇ .0005% to 0.5%, most preferred from 0.001% toO .1 % pure enzyme by weight of the composition.
- Preferred xyloglucanases for specific applications are alkaline xyloglucanases, ie enzymes having an enzymatic activity of at least 10%, preferably at least 25%, more preferably at least 40% of their maximum activity at a pH ranging from 7 to 12. More preferred xyloglucanases are enzymes having their maximum activity at a pH ranging from 7 to 12.
- the above-mentioned enzymes may be of any suitable origin, such as vegetable, animal, bacterial, fungal and yeast origin. Origin can further be mesophilic or extremophilic (psychrophilic, psychrotrophic, thermophilic, barophilic, alkalophilic, acidophilic, halophilic, etc.). Purified or non- purified forms of these enzymes may be used.
- the variants may be designed such that the compatibility of the enzyme to commonly encountered ingredients of such compositions is increased.
- the variant may be designed such that the optimal pH, bleach or chelant stability, catalytic activity and the like, of the enzyme variant is tailored to suit the particular cleaning application.
- the isoelectric point of such enzymes may be modified by the substitution of some charged amino acids, e.g. an increase in isoelectric point may help to improve compatibility with anionic surfactants.
- the stability of the enzymes may be further enhanced by the creation of e.g. additional salt bridges and enforcing metal binding sites to increase chelant stability.
- the pectate lyase of the present invention can be used in combination with other carbohydrate degrading enzymes (for instance arabinanase, xyloglucanase, pectinase) for biopreparation of fibers or for cleaning of fibers in combination with detergents.
- Cotton fibers consist of a primary cell wall layer containing pectin and a secondary layer containing mainly cellulose. Under cotton preparation or cotton refining part of the primary cell wall will be removed.
- the present invention relates to either help during cotton refining by removal of the primary cell wall. Or during cleaning of the cotton to remove residual pectic substances and prevent graying of the textile.
- cellulosic material is intended to mean fibers, sewn and unsewn fabrics, including knits, wovens, denims, yarns, and toweling, made from cotton, cotton blends or natural or manmade cellulosics (e.g. originating from xylan-containing cellulose fibers such as from wood pulp) or blends thereof.
- blends are blends of cotton or rayon/viscose with one or more companion material such as wool, synthetic fibers (e.g.
- polyamide fibers acrylic fibers, polyester fibers, polyvinyl alcohol fibers, polyvinyl chloride fibers, polyvinylidene chloride fibers, polyurethane fibers, polyurea fibers, aramid fibers), and cellulose-containing fibers (e.g. rayon/viscose, ramie, hemp, flax/linen, jute, cellulose acetate fibers, lyocell) .
- cellulose-containing fibers e.g. rayon/viscose, ramie, hemp, flax/linen, jute, cellulose acetate fibers, lyocell
- the preparation of the present invention is useful in the cellulosic fiber processing industry for the pretreatment or retting of fibers from hemp, flax or linen.
- the processing of cellulosic material for the textile industry, as for example cotton fiber, into a material ready for garment manufacture involves several steps : spinning of the fiber into a yarn; construction of woven or knit fabric from the yarn and subsequent preparation, dyeing and finishing operations.
- Woven goods are constructed by weaving a filling yarn between a series of warp yarns; the yarns could be two different types.
- Knitted goods are constructed by forming a network of interlocking loops from one continuous length of yarn.
- the cellulosic fibers can also be used for non-woven fabric.
- the preparation process prepares the textile for the proper response in dyeing operations.
- the sub-steps involved in preparation are a. Desizing (for woven goods) using polymeric size like co ⁇ to en o C ⁇ o en o C ⁇
- woven cotton fabrics are desized by a combination of hot water, the enzyme ⁇ -amylase to hydrolyze the starch and a wetting agent or surfactant.
- the cellulosic material is allowed to stand with the desizing chemicals for a "holding period" sufficiently long to accomplish the desizing.
- the holding period is dependent upon the type of processing regime and the temperature and can vary from 15 minutes to 2 hours, or in some cases, several days.
- the desizing chemicals are applied in a saturator bath which generally ranges from about 15°C to about 55°C.
- the fabric is then held in equipment such as a "J-box" which provides sufficient heat, usually between about 55°C and about 100°C, to enhance the activity of the desizing agents.
- the chemicals, including the removed sizing agents are washed away from the fabric after the termination of the holding period.
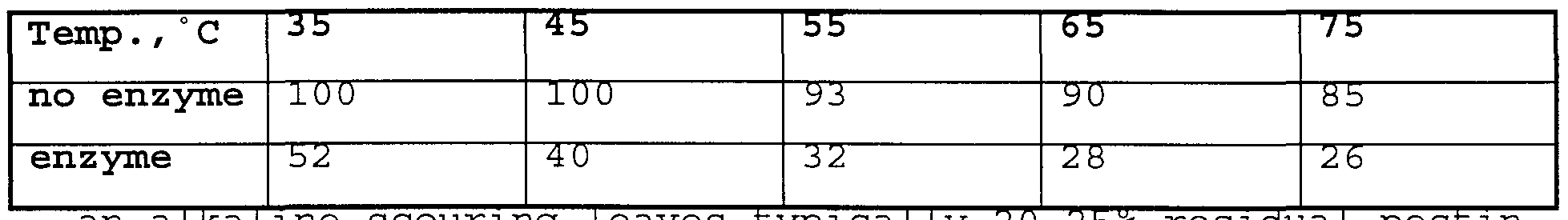
- the scouring process removes much of the non-cellulosic compounds naturally found in cotton. In addition to the natural non-cellulosic impurities, scouring can remove dirt, soils and residual manufacturing introduced materials such as spinning, coning or slashing lubricants.
- the scouring process employs sodium hydroxide or related causticizing agents such as sodium carbonate, potassium hydroxide or mixtures thereof. Generally an alkali stable surfactant is added to the process to enhance solubilization of hydrophobic compounds and/or prevent their redeposition back on the fabric.
- the treatment is generally at a high temperature, 80°C - 100°C, employing strongly alkaline solutions, pH 13-14, of the scouring agent.
- the softness of the cellulosic fabric is a function of residual natural cotton waxes.
- the non-specific nature of the high temperature strongly alkaline scouring process cannot discriminate between the desirable natural cotton lubricants and the manufacturing introduced lubricants.
- the conventional scouring process can cause environmental problems due to the highly alkaline effluent from these processes.
- the scouring stage prepares the fabric for the optimal response in bleaching. An inadequately scoured fabric will need a higher level of bleach chemical in the subsequent bleaching stages.
- the bleaching step decolorizes the natural cotton pigments and removes any residual natural woody cotton trash components not completely removed during ginning, carding or scouring.
- the main process in use today is an alkaline hydrogen peroxide bleach. In many cases, especially when a very high whiteness is not needed, bleaching can be combined with scouring.
- the scouring step can be carried out using the pectate lyase or pectate lyase preparation of the present invention a temperature of about 50°C - 80°C and a pH of about 7-11, thus substituting or supplementing the highly causticizing agents.
- An optimized enzymatic process ensures a high pectin removal and full wettability.
- the enzyme or enzyme preparation according to the invention is preferably used as an agent for degradation or modification of plant cell walls or any pectin-containing material originating from plant cells walls due to the high plant cell wall degrading activity of the pectate lyase of the invention.
- pectate lyase of the present invention may be used alone or together with other enzymes like glucanases, pectinases and/or hemicellulases to improve the extraction of oil from oil -richêt _,
- the pectate lyase of the present invention may be used for separation of components of plant cell materials.
- the separation of sugar or starch rich plant material into components of considerable commercial interest like sucrose from sugar beet or starch from potato
- components of low interest like pulp or hull fractions
- the separation of protein-rich or oil-rich crops into valuable protein and oil and invaluable hull fractions The separation process may be performed by use of methods known in the art .
- the pectate lyase of the invention may also be used in the preparation of fruit or vegetable juice in order to increase yield, and in the enzymatic hydrolysis of various plant cell wall -derived materials or waste materials, e.g. from wine or juice production, or agricultural residues such as vegetable hulls, bean hulls, sugar beet pulp, olive pulp, potato pulp, and the like.
- the plant material may be degraded in order to improve different kinds of processing, facilitate purification or extraction of other component than the galactans like purification of pectins from citrus, improve the feed value, decrease the water binding capacity, improve the degradability in waste water plants, improve the conversion of plant material to ensilage, etc .
- an enzyme preparation of the invention it is possible to regulate the consistency and appearence of processed fruit or vegetables .
- the consistency and appearence has been shown to be a product of the actual combination of enzymes used for processing, i.e. the specificity of the enzymes with which the pectate lyase of the invention is combined. Examples include the production of clear juice e.g. from apples, pears or berries; cloud stable juice e.g. from apples, pears, berries, citrus or tomatoes; and purees e.g. from carrots and tomatoes.
- the pectate lyase of the invention may be used in modifying the viscosity of plant cell wall derived material.
- the pectate lyase may be used to reduce the viscosity of feed ⁇ M c ⁇ o C ⁇ o en o C ⁇
- the GrafPad Prism program using a non linear fit with a one phase exponential decay with a plateau, was used for calculations.
- the plateau plus span is the mV obtained without enzyme.
- the plateau is the mV of more than 100 APSU and the half reduction of viscosity in both examples was found to be 12 APSU units with a standard error of 1.5 APSU.
- the lyase assay (at 235 mti) For determination of the ⁇ -elimination an assay measuring the increase in absorbance at 235 nm was carried out using the substrate 0.1% polygalacturonic acid sodium salt (Sigma P-1879) solubilised in 0.1 M Glycin buffer pH 10. For calculation of the catalytic rate an increase of 5.2 Absorbency at 235 units per min corresponds to formation of 1 ⁇ mol of unsaturated product (Nasuna and Starr (1966) J. Biol. Chem. Vol 241 page 5298-5306; and Bartling, egener and Olsen (1995) Microbiology Vol 141 page 873-881) .
- Pectate lyase activity can be measured by applying a test solution to 4 mm holes punched out in agar plates (such as, for example, LB agar), containing 0.7% w/v sodium polygalacturonate (Sigma P 1879) . The plates are then incubated for 6 h at a particular temperature (such as, e.g., 75°C) . The plates are then soaked in either (i) 1M CaC12 for 0.5h or (ii) 1% mixed alkyl trimethylammonium Br (MTAB, Sigma M-7635) for 1 h. Both of these procedures cause the precipitation of polygalacturonate within the agar.
- agar plates such as, for example, LB agar
- MTAB mixed alkyl trimethylammonium Br
- Pectate lyase activity can be detected by the appearance of clear zones within a background of precipitated polygalacturonate. Sensitivity of the assay is calibrated using dilutions of a standard preparation of pectate lyase.
- the substrate and enzyme is incubated for 20 min at 37°C followed by measurement at 235 nm of the formation of double bounds. Finally, the rate of the degradation is calculated based on the molar extinction coefficient in terms of Trans Units .
- Substrate Polygalactoronic acid from Sigma P-1879 lot 77H3784
- Buffer 2x 0.1M Glycin pH 10 + 2.0 mmol CaCl 2
- Bacillus agaradhaerens comprises the pectate lyase encoding DNA sequence presented in SEQ ID NO: 1.
- Bacillus sp . AAI12 comprises the pectate lyase encoding DNA sequence presented in SEQ ID NO : 5.
- Bacillus sp . KJ59, DSM 12419 comprises the pectate lyase encoding DNA sequence presented in SEQ ID NO: 7.
- Bacillus sp . 1534 comprises the pectate lyase encoding DNA sequence presented in SEQ ID NO : 9.
- E. coli DSM 12403 comprises the plasmid containing the pectate lyase encoding DNA sequence of the invention presented in SEQ ID NO: 5.
- E. coli DSM 12404 comprises the plasmid containing the pectate lyase encoding DNA sequence of the invention presented in SEQ ID NO: 9.
- E. coli DSM 11789 comprises the plasmid containing the pectate lyase encoding DNA sequence of the invention presented in SEQ ID NO: 3.
- E. coli DSM 11788 comprises the plasmid containing the pectate lyase encoding DNA sequence of the invention presented in SEQ ID NO: 1.
- This strain is the B. subtilis DN1885 with disrupted apr and npr genes (Diderichsen, B., Wedsted, U. , Hedegaard, L., Jensen, B. R. , Sj ⁇ holm, C. (1990) Cloning of aldB, which encodes alpha-acetolactate decarboxylase, an exoenzyme from Bacillus brevis. J. Bacteriol . , 172, 4315-4321) disrupted in the transcriptional unit of the known Bacillus subtili s cellulase gene, resulting in cellulase negative cells. The disruption was performed essentially as described in ( Eds. A.L.
- Competent cells were prepared and transformed as described by Yasbin, R.E., Wilson, G.A. and Young, F.E. (1975) Transformation and transfection in lysogenic strains of Bacillus subtilis : evidence for selective induction of prophage in competent cells. J. Bacteriol, 121:296-304.
- E. coli SJ2 (Diderichsen, B., Wedsted, U., Hedegaard, L., .- ,
- Plasmids pBK-CAMV (Stratagene inc., La Jolla Ca.) pSJ1678 (see WO 94/19454 which is hereby incorporated by reference in its entirety) .
- pMOL944 :
- This plasmid is a pUBllO derivative essentially containing elements making the plasmid propagatable in Bacillus subtilis, kanamycin resistance gene and having a strong promoter and signal peptide cloned from the amyL gene of B . licheniformis ATCC14580.
- the signal peptide contains a SacII site making it convenient to clone the DNA encoding the mature part of a protein in-fusion with the signal peptide. This results in the expression of a Pre-protein which is directed towards the exterior of the cell .
- the plasmid was constructed by means of conventional genetic engineering techniques which are briefly described in the following.
- the two PCR primers used have the following sequences:
- the primer #LWN5494 inserts a NotI site in the plasmid.
- the plasmid pSJ2624 was then digested with Sad and NotI and a new PCR fragment amplified on amyL promoter encoded on the ,_ _
- pDN1981 was digested with Sad and NotI and this DNA fragment was inserted in the SacI-NotI digested pSJ2624 to give the plasmid pSJ2670.
- This cloning replaces the first amyL promoter cloning with the same promoter but in the opposite direction.
- the two primers used for PCR amplification have the following sequences:
- the plasmid pSJ2670 was digested with the restriction enzymes PstI and Bell and a PCR fragment amplified from a cloned DNA sequence encoding the alkaline amylase SP722 (disclosed in the International Patent Application published as W095/26397 which is hereby incorporated by reference in its entirety) was digested with PstI and Bell and inserted to give the plasmid pMOL944.
- the two primers used for PCR amplification have the following sequence: #LWN7864 5" -AACAGCTGATCACGACTGATCTTTTAGCTTGGCAC-3 '
- the primer #LWN7901 inserts a SacII site in the plasmid.
- Enzymes for DNA manipulations were used according to the specifications of the suppliers ⁇ e . g. restriction endonucleases, ligases etc. are obtainable from New England Biolabs, Inc.) .
- the strain Bacillus licheniformis ATCC 14580 was propagated in liquid medium 3 as specified by ATCC (American Type Culture Collection, USA) . After 18 hours incubation at 37°C and 300 rpm, the cells were harvested, and genomic DNA was isolated by the method described below. Bacillus agaradherens NCIMB No. 40482, Bacillus sp . AAI12, Bacillus sp . KJ59, DSM 12419, and the Bacillus sp. 1534 were all grown in TY with pH adjusted to approximately pH 9.7 by the addition of 50 ml of 1M Sodium-Sesquicarbonat per 500 ml TY. After 24 hours incubation at 30°C and 300 rpm, the cells were harvested, and genomic DNA was isolated by the method described below.
- the Bacillus sp . strains described above as donor organisms were propagated in liquid media as described above.
- the cells were harvested, and genomic DNA was isolated by the method described by Pi tcher et al . [Pi tcher, D . G. , Saunders , N. A . , Owen, R . J; Rapid extraction of bacterial genomic DNA with guanidium thiocyanate; Lett Appl Microbiol 1989 8 151-156] .
- LambdaZAP express cloning kit with BamHI digested and dephosphorylated arms from Stratagene.
- Bacillus DNA was isolated by the method of Pitcher et al . , 1989) .
- Isolated DNA was partially digested with Sau3A and size fractionated on a 1% DNA agarose gel.
- DNA was excised from the agarose gel between 2 and 6 Kb and purified using Qiaspin DNA fragment purification procedure (Qiagen GmBH) .
- Qiaspin DNA fragment purification procedure Qiagen GmBH
- lOOng of purified, fractionated DNA was ligated with 1 ug of BamHI dephosphorylated ZAPexpress vector arms (4 degrees overnight) .
- Ligation reaction was packaged directly with GigaPacklll Gold according to the manufacturers instructions (Stratagene) . Phage libraries were titered with XLlblue mrf- (Stratagene) .
- XLl-blue cells (Stratagene, La Jolla Ca.) were prepared and resuspended in lOmM MgS04 as recommended in the mass excission protocol in the Stratagene ZAPexpress handbook. 40,000 plaque forming units from each library were placed in Falcon 2059 tubes. Samples were incubated with 400uls of XLl-blue cells and > 10 10 pfus/ml EXassist M13 helper phage (Stratagene) at 37 C for 15 minutes. Six mis of NZY broth was added to each tube and then the tubes were agitated at 37 C for 2.5 hours. Samples were then heated at 65 C for 20 minutes to kill E.
- XLOLR cells had been prepared as described in the Stratagene ZAPexpress manual (Stratagene inc., La Jolla CA) .
- Standard screening for pectinases on LB agar plates was performed as follows: Plasmid libraries in XLOLR E. coli cells were plated on LB kanamycin plates at a density of 5000 colony forming units per 140mm diamater petri plates. Plates were incubated overnight at 37 C then overlayed with 1% pectin DE 35% or DE 75% and 1% agarose. Plates were incubated overnight at 37 C before being overlayed with 1% MTAB solution. After two hours ,_ ,- o the MTAB was poured off and the positive recombinant clones identified by a clearing zone around the colony. Positive isolates were reconfirmed by streaking on fresh LB media and testing again.
- LBPG is LB agar supplemented with 0.5% Glucose and 0.05 M potassium phosphate, pH 7.0 BPX media is described in EP 0 506 780 (WO 91/09129) .
- ID No.l and encoding the pectate lyase of the invention can be obtained from the deposited organism E. coli , DSM 11788, by extraction of plasmid DNA by methods known in the art (Sambrook et al . (1989) Molecular cloning: A laboratory manual, Cold
- Genomic library construction of Bacillus agaradhaerens Genomic DNA of Bacillus agaradhaerens NCIMB 40482 was partially digested with restriction enzyme Sau3A, and size- fractionated by electrophoresis on a 0.7 % agarose gel. Fragments between 2 and 7 kb in size were isolated by electrophoresis onto DEAE-cellulose paper (Dretzen, G., Bellard, M., Sassone-Corsi, P., Chambon, P. (1981) A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal. Biochem. , 112, 295-298).
- Isolated DNA fragments were ligated to BamHI digested pSJ1678 plasmid DNA, and the ligation mixture was used to transform E. coli SJ2.
- Transformed cells from the Genomic library of Bacillus agaradhaerens NCIMB 40482 were plated on LB-agar plates containing 10 ⁇ g/ml of Chloramphenicol and 0.7%, Sodium Polygalacturonate (SIGMA P-1879) . The plated cells were incubated 16 hours at 37°C.
- the colonies were replica plated onto fresh LB-agar plates containing 10 ⁇ g/ml of Chloramphenicol and 0.7% , Sodium Polygalacturonate (SIGMA P-1879) these plates were incubated 8 hours at 37°C.
- the original master plates were flooded with 5 ml 1 M CaCl2, after 5 to 30 min distinct cloudy halos appeared around putative Sodium Polygalacturonate degrading clones. The corresponding masterplate clones were picked for further characterisation.
- clones were further characterized by preparing plasmid DNA from overnight 30°C liquid TY cultures of the E.coli clones and preparing plasmid DNA using Qiagen Qiaspin Prep Kit as according to manufacturer (Qiagen, Germany) .
- the pectate lyase positive clone of Bacillus agaradhaerens NCIMB 40482 Gene library was deposited as DSM 11788. After primer walking on the plasmid of the E. coli DSM 11788 the SEQ ID NO : 1 of the pectate lyase encoding DNA from Bacillus agaradhaerens NCIMB No. 40482 was identified.
- pectinase positive clones were obtained as single colonies, and plasmids were extracted using Qiagen Plasmid Prep as indicated by the manufacturer (Qiagen, Germany) . Phenotypes were confirmed by retransformation of E . coli SJ2 , and. plasmids characterized by restriction di- gests.
- the transformed cells were plated on LB agar plates containing 6 mg/ml Chloramphenicol, 0.4% glucose, 10 mM KH2P04, and incubated at 37°C for 18 hours. Pectate lyase positive colonies were identified as done above with E. coli .
- Each of the positive transformants were inoculated in 10 ml TY-medium containing 6 mg/ml Chloramphenicol . After 1 day of incubation at 37°C and shaking at 250rpm, 50 ⁇ l supernatant was removed. The pectate lyase activity was identified by adding 10 ⁇ l supernatant to holes punched in the agar of LB agar plates containing 0.7 % Sodium Polypectate (Sigma, US).
- the cells were removed by centrifugation and the remaining supernatant was used as source for purifying the pectate lyase.
- the B . subtilis transformant obtained as described above was incubated in 100 ml of TY containing 6 mg/ml
- the fermentation medium was adjusted to pH 7.5 with NaOH and flocculated using cationic flocculation agent C521 (10% solution) and 0.1% solution of anionic agent A130: To 6500 ml of fermentation medium was added 306 ml of C521 (10%) simultaneous with 608 ml of A130 under stirring at room temperature. The flocculated material was separated by centrifugation using a Sorval RC 3B centrifuge at 10,000 rpm for 30 minutes. The supernatant was clarified using Whatman glass filter number F. In total was obtained 7200 ml of clear solution.
- the liquid was concentrated into 2 portions of 500 ml and 840 ml, respectively, using filtron ultrafiltration with a MW cut off of 10 kDa.
- the pH was adjusted to 5.3 using acetic acid, and the concentrate was applied to a 200 ml S-Sepharose column equilibrated with 50 mM sodium acetate buffer, pH 5.3.
- the pectate lyase of the invention (B. agaradhaerens) eluted using a linear gradient of 2 1 with 0.5 M NaCl as final concentration.
- Pectate lyase from Bacillus subtilis will also bind to Sepharose at this pH but it has a higher pi (7.6 versus 6.0) .
- pectate lyase of the invention elutes first, fractions were analyzed for APSU units and for reaction with antiserum raised against Bacillus subtilis pectate lyase.
- the Bacillus agaradhaerens pectate lyase was concentrated using an Amicon ultrafiltration cell with a GR61 membrane with a cut off of 20 kDa.
- a total of 90,000 APSU units was obtained. This sample was free of protease and of the Bacillus subtili s pectate lyase determined using antiserum raised against Bacillus subtilis pectate lyase .
- the pectate lyase enzyme of the invention could be easily seen in SDS-PAGE as a band with a MW of 36 kDa. After electroblotting of this band the N-terminal was determined as: Ser-Asn-Gly-Pro-Gln-Gly-Tyr-Ala-Ser-Met-Asn-Gly-Gly-Thr
- pH profile was determined using the following buffers: pH 6.0: Na-MES 0.1M pH 6.5, 7.0 and 7.5: Na-MOPS 0.1M pH 8.0 & 8.5: Tris 0.1M pH 9.0, 9.5, 10.0 and 10.5: Na-glycine 0.1M pH 11-11.5: Na-Carbonate 0.1M
- MES is 2 [N-Morpholino] ethanesulfonicAcid (SIGMA, No. M-8250) .
- MOPS is 3- [N-Morpholino]propanesulfonic Acid (SIGMA, No. M-1254]
- Tris (Merck No. 1.08382 ).
- the pectate lyase encoding DNA sequence of the invention (SEQ ID No:l) was PCR amplified using the PCR primer set consisting of these two oligo nucleotides:
- Chromosomal DNA isolated from B . agaradhaerens NCIMB 40482 as described above was used as template in a PCR reaction using Amplitaq DNA Polymerase (Perkin Elmer) according to manufacturers instructions.
- the PCR reaction was set up in PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KC1, 1.5 mM MgCl 2 , 0.01 % (w/v) gelatin) containing 200 ⁇ M of each dNTP, 2.5 units of AmpliTaq polymerase (Perkin-Elmer, Cetus, USA) and 100 pmol of each primer.
- the PCR reaction was performed using a DNA thermal cycler (Landgraf , Germany) . One incubation at 94°C for 1 min followed by thirty cycles of PCR performed using a cycle profile of denaturation at 94°C for 30 sec, annealing at 60°C for 1 min, and extension at 72°C for 2 min. Five- ⁇ l aliquots of the amplification product was analysed by electrophoresis in 0.7 % agarose gels (NuSieve, FMC) . The appearance of a DNA fragment size 1.0 kb indicated proper amplification of the gene segment. Subcloning of PCR fragment
- the isolated PCR DNA fragment was then ligated to the SacII-NotI digested and purified pMOL944. The ligation was performed overnight at 16°C using 0.5 ⁇ g of each DNA fragment, 1 U of T4 DNA ligase and T4 ligase buffer (Boehringer Mannheim, Germany) .
- the ligation mixture was used to transform competent B. subtilis PL2306.
- the transformed cells were plated onto LBPG- 10 ⁇ g/ml of Kanamycin plates. After 18 hours incubation at 37°C several clones were restreaked on fresh agar plates and also grown in liquid TY cultures with 10 ⁇ g/ ml kanamycin and incubated overnight at 37°C. Next day 1 ml of cells were used to isolate plasmid from the cells using the Qiaprep Spin Plasmid Miniprep Kit #27106 according to the manufacturers recommendations for B. subtilis plasmid preparations. This plasmid DNA was used as template for DNA sequencing.
- the DNA corresponding to the mature part of the pectate lyase was characterised by DNA sequencing by primerwalking, using the Taq deoxy-terminal cycle sequencing kit (Perkin-Elmer, USA) , fluorescent labelled terminators and appropriate oligonucleotides as primers.
- the pure enzyme was dialysed against EDTA at pH 8.0 (20 mM tris pH 8.0), and at pH 10 (20 mM Glycine pH 10) and was analysed in Circular dichroism: No differences were seen in the spectra with and with out EDTA.
- Differential Scanning Calorimetry DSC of the 4 samples showed that the enzyme was most stable at pH 8.0 with a melting temperature around 61°C in Tris pH 8.0 and 62 °C after dialysis against EDTA. At pH 10 the enzyme melted at 59°C with and without EDTA.
- the catalytic activity is inhibited by the presence of EDTA during incubation with substrate but the enzyme dialysed against EDTA was still active if EDTA was omitted during incubation with substrate.
- Divalent cations like Fe++, Li++, Mg++, Cu++, Mn++ have no effect on the catalytic activity.
- the enzyme was active in the commercially available European powder laundry detergent Ariel FuturTM with 37% relative activity, in the commercially available US powder laundry detergent TideTM with 58% relative activity and in the commercially available US liquid detergent TideTM with 37% relative activity to the activity measured in Glycine buffer.
- the detergent concentration was equal to the concentration recommended on the detergent packages for household use, and the used water tap water had 18 degrees German hardness (European detergent/European conditions) and 9 degrees German hardness (US detergents/US conditions) .
- rabbit polyclonal monospecific serum was raised against the highly purified pectate lyase prepared as described above using conventional techniques.
- the serum formed a nice single precipitate in agarose gels with the B. agarahaerens pectate lyase of the invention.
- Genomic library construction of Bacillus licheniformis . ATCC 14580 was carried out as described m example 1 for B. agaradhaerens .
- the pectate lyase positive clone of Bacillus licheniformis ATCC 14580 Gene library was deposited as DSM 11789. After primer walking on the plasmid of the E. coli DSM 11789 the SEQ ID No : 3 of the pectate lyase encoding DNA from Bacillus licheniformis ATCC 14580 was identified.
- the pectate lyase represented by ammo acid sequence SEQ ID NO:4j encoding DNA sequence of the invention was PCR amplified using the PCR primer set consisting of these two oligo nucleotides:
- the DNA corresponding to the mature part of the pectate lyase was characterised by DNA sequencing by primerwalking, using the Taq deoxy-terminal cycle sequencing kit (Perkin-Elmer, USA) , fluorescent labelled terminators and appropriate oligonucleotides as primers.
- MB541 was grown in 25 x 200 ml BPX media with 10 ⁇ g/ml of Kanamycin in 500 ml two baffled shakeflasks for 5 days at 37°C at 300 rpm, whereby 3500 ml of culture broth was obtained.
- the pH was adjusted to 5.0 using acetic acid and 100 ml of cationic agent (C521) and 200 ml of anionic agent (A130) was added during agitation for flocculation.
- the flocculated material was separated by centrifugation using a Sorval RC 3B centrifuge at 10000 rpm for 30 min at 6°C.
- the resulting supernatant contained 370 APSU per ml in a total volume of 3600 ml.
- the supernatant was clarified using Whatman glass filters GF/D and C and finally concentrated on a filtron UF membrane with a cut off of 10 kDa.
- the total volume of 2000 ml was adjusted to pH 8.5.
- 50 gram of DEAE A-50 Sephadex (Pharmacia) was swelled in 2000 ml 50 mM Tris pH 8.5.
- Excess buffer was discarded and the clear concentrated enzyme solution was mixed with the slurry for 15 min.
- the enzyme was separated from the ion-exchange material by suction on a Buchner funnel.
- the resulting solution was concentrated on a filtron with a cut off of 10 kDa to a final volume of 800 ml.
- the protein concentration was determined using a molar extinction coefficient of 57750 (based on the amino acid composition deducted from the sequence) .
- 57750 based on the amino acid composition deducted from the sequence
- the pure enzyme was dialysed against EDTA at pH 8.0 (20 mM tris pH 8.0, and at pH 10 (20 mM Glycine pH 10) and the enzyme analysed in Circular dichroism, no differences was seen in the spectra with and with out EDTA.
- the catalytic activity of the pectate lyase is inhibited by the presence of EDTA during incubation with substrate but the enzyme dialysed against EDTA was still active if EDTA was omitted during incubation with substrate.
- Divalent cation like Fe ++ ,
- Li++, Mg++, Cu++, Mn++ has no effect on the catalytic activity.
- the ⁇ -transelimination activity (using the lyase assay at
- MES From SIGMA number M-8250 (2 [N-Morpholino] ethane sulfonic acid) .
- MOPS From SIGMA number M-1254 (3- [N-Morpholino] propane sulfonic acid) .
- the resulting B. subtilis clone expressing the Pectate lyase was termed MB888.
- the cloned DNA sequence was expressed in B. subtilis and the protein that appeared in the supernatant corresponded to the mature protein represented in SEQ ID NO : 8 mature protein.
- Bacillus subtilis transformed with this plasmid was grown PS medium containing Kanamycin.
- Flocculation was done using cationic flocculation agent C521 (10% solution) and 0.1% solution of anionic agent A130: To 2000 ml of fermentation medium was added 2000 ml ion free water and it had pH 6.0 then 80 ml of C521 (10%) simultaneous with 40 ml of A130 was added under stirring at room temperature. The flocculated material was separated by centrifugation using a Sorval RC 3B centrifuge at 10,000 rpm for 30 minutes. The supernatant was clarified using Whatman glass filter number F. In total was obtained 40000 ml of clear solution containing 204,000 Trans Units.
- the liquid was concentrated into 500 ml using filtron ultrafiltration with a MW cut off of 10 kDa.
- the Concentrate was treated with 5 gram of DEAE A-50 Sephadex equilibrated in 25 mM Tris pH 8.0 for 30 minutes and the pectate lyase did not bind and was filtrated free of the ionexchange material.
- the filtrate was adjusted to pH 5.0 using HCl and applied to S-Sepharose column equilibrated with 25 mM Sodium acetate pH 5.0. The pectate lyase bound and was eluted as a pure protein using a NaCl gradient.
- the pure enzyme has a MW of 36 kDa and a pi of 8.98.
- the temperature optimum at pH 10 is about 65 °C.
- the relative activity is higher than 50% active at 40 °C between pH 9 and 11.
- the pectate lyase has a melting temperature of 74°C measured using DSC in 0.1 M sodium acetate pH 6.0.
- B. subtilis clone expressing the Pectate lyase was termed MB746.
- the cloned DNA sequence was expressed in B. subtilis and the protein that appeared in the supernatant corresponded to the mature protein represented in SEQ ID NO: 10 mature protein.
- Bacillus subtilis transformed with this plasmid was grown in PS medium containing Kanamycin.
- Flocculation was done using cationic flocculation agent C521 (10% solution) and 0.1% solution of anionic agent A130: To 3700 ml of fermentation medium was adjusted to pH 5.5 using HCl and then added 37 ml of C521 (10%) simultaneous with 75 ml of A130 under stirring at room temperature. The flocculated material was separated by centrifugation using a Sorval RC 3B centrifuge at 10,000 rpm for 30 minutes. The supernatant was clarified using Whatman glass filter number F. In total was obtained 40000 ml of clear solution containing 1,044,000 Trans Units . The liquid was concentrated into 550 ml using filtron ultrafiltration with a MW cut off of 10 kDa. This product was used for application trials after stabilization using 50% MPG (batch #9831) .
- the pure enzyme has a MW of 35 kDa and an isoelectrial point (pi) of 6.2.
- the temperature optimum at pH 10 is above 70 °C.
- the relative activity is more than 50% between pH 9 and 11 at a temperature of 40°C.
- the N-terminal of the purified pectate lyase has the amino acid sequence KGESDSTMNA starting at position 25 of the amino acid sequence SEQ ID NO: 10.
- Bacillus subtilis transformed with this plasmid (MB644) was grown in PS medium containing Kanamycin. The purification
- this enzyme contains 3 lectin binding domains at the N-terminal.
- the percentage of residual pectin was calculated using the dye uptake of the starting material as 100% residual pectin and that of fully chemically scoured and bleached fabric as 0%. Results are shown in Table 1. Further, the wettability (drop test - measuring the time in seconds for a drop of water to become absorbed by the fabric) was measured and compared to a no enzyme control. Results are shown in Table 2.
- the pH op imum is ound. to ioe at app . .5, but a good activity is demonstrated in a very broad alkaline interval,
- the purified enzyme obtained as described in example 2 (batch 9751) showed improved cleaning performance when tested at a level of 1 ppm in a miniwash test using a conventional commercial liquid detergent. The test was carried out under conventional North American wash conditions.
- the stained cotton textile was washed in the commercial liquid detergent brand Ariel Futur Liquid under European wash conditions, with an addition of 0.1 ppm, 0.2ppm, lppm and 10 ppm, respectively, of the pectate lyase of example 2 to the detergent liquid and an addition of 10 ppm of the pectate lyase of example 1. The test was repeated.
- Ariel liquid % removal of the banana stains (100% is total removal of stain)
- the CBD encoding DNA sequence of the CipB gene from Clostridium thermocellum strain YS (Poole D M; Morag E; Lamed R; Bayer EA; Hazlewood GP; Gilbert HJ (1992) Identification of the cellulose-binding domain of the cellulosome subunit SI from Clostridium thermocellum YS, Fems Microbiology Letters Vol. 78 , No. 2-3 pp. 181-186 was PCR amplified using the PCR primer set consisting of the following two oligo nucleotides:
- Chromosomal DNA encoding the CBD can be obtained as described in Poole DM; Morag E; Lamed R; Bayer EA; Hazlewood GP Gilbert HJ (1992) Identification of the cellulose-binding domain of the cellulosome subunit SI from Clostridium thermocellum YS, Fems Microbiology Letters Vol. 78 , No. 2-3 pp.
- the subcloning was carried out as described in example 1 except that the purified PCR fragment was digested with Sail and NotI .
- Several clones were analyzed by isolating plasmid DNA from overnight culture broth.
- One such positive clone was restreaked several times on agar plates as used above, this clone was called MB914.
- the clone MB914 was grown overnight in TY-lO ⁇ g/ml Kanamycin at 37°C, and next day 1 ml of cells were used to isolate plasmid from the cells using the Qiaprep Spin Plasmid Miniprep Kit #27106 according to the manufacturers recommendations for B. subtilis plasmid preparations.
- This DNA was DNA sequenced and revealed the DNA sequence corresponding to the fusionprotein of: Pectate lyase-linker-cbd as represented in SEQ ID NO: 11 and in the appended protein sequence SEQ ID NO: 12.
- MB914 was incubated for 20 hours in TY-medium at 37°C and
- Eijuipment A Labomat (Mathis, Switzerland) was used at a liquor ratio of 12.5:1 (12 g fabric in 150 ml buffer/enzyme solution) .
- Pectate lyase In Experiment 1, a pectate lyase corresponding to SEQ ID NO: 10 was used, formulated in a solution containing 0.02 M phosphate buffer and 0.4 g/L non- ionic surfactant (Tergitol 15-S-12 from Union Carbide) . In Experiment 2, the pectate lyase of SEQ ID NO: 4 was used, formulated in a solution containing 0.05 M phosphate/borate buffer, in 2.0 g/L non-ionic surfactant (Tergitol 15-S-12 from Union carbide) , and 1.0 g/L wetter (Dioctyl sulfosuccinate) .
- test fabrics were contacted with the aqueous solution containing the pectate lyase for 15 minutes at temperatures ranging between 60-80°C and pHs ranging between 7-11, after which residual pectin was quantified.
- the Figure 3 below shows a contour plot of the % residual pectin as a function of both pH and temperature
- Figure 3 shows the % residual pectin as a function of the enzyme dosage.
- the pH optimum for pectin removal was 9.2 and the temperature optimum was above 80°C.
- the test fabrics were contacted with the aqueous solution containing the pectate lyase at 600APSU/kg cotton, squeezed in a roller system to give a solution pickup of 85%, and incubated for 60 minutes at temperatures between 40-70°C, after which residual pectin was quantified.
- the % residual pectin as a function of temperature is shown in the Table below.
Landscapes
- Health & Medical Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Chemical & Material Sciences (AREA)
- Engineering & Computer Science (AREA)
- Genetics & Genomics (AREA)
- Organic Chemistry (AREA)
- Zoology (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Wood Science & Technology (AREA)
- Medicinal Chemistry (AREA)
- General Health & Medical Sciences (AREA)
- Microbiology (AREA)
- Biomedical Technology (AREA)
- Molecular Biology (AREA)
- Biochemistry (AREA)
- General Engineering & Computer Science (AREA)
- Biotechnology (AREA)
- Textile Engineering (AREA)
- Enzymes And Modification Thereof (AREA)
- Micro-Organisms Or Cultivation Processes Thereof (AREA)
- Detergent Compositions (AREA)
- Immobilizing And Processing Of Enzymes And Microorganisms (AREA)
- Chemical Or Physical Treatment Of Fibers (AREA)
Abstract
Description
Claims
Priority Applications (6)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| AU14825/99A AU1482599A (en) | 1997-11-24 | 1998-11-24 | Novel pectate lyases |
| JP2000522226A JP4246386B2 (en) | 1997-11-24 | 1998-11-24 | A new pectate lyase |
| KR1020007005621A KR20010032382A (en) | 1997-11-24 | 1998-11-24 | Novel pectate lyases |
| EP98958820A EP1032658B1 (en) | 1997-11-24 | 1998-11-24 | Pectate lyases |
| CA2310562A CA2310562C (en) | 1997-11-24 | 1998-11-24 | Novel pectate lyases |
| BR9815007-3A BR9815007A (en) | 1997-11-24 | 1998-11-24 | Pectate lyase, isolated polynucleotide molecule encoding a polypeptide, expression vector, cultured cell into which an expression vector was introduced, isolated and fused polypeptides, enzyme preparation, processes for producing a polypeptide showing pectate lyase activity, for the cleaning of a hard surface, for the treatment of fabrics by machine, to improve the properties of cellulosic fibers, yarn, woven or nonwoven fabric, for the degradation or modification of plant material, for preparing animal food, and for processing wine or juice, isolated enzyme showing pectate lyase activity, and detergent composition |
Applications Claiming Priority (8)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| DK134397 | 1997-11-24 | ||
| DK134497 | 1997-11-24 | ||
| DK1343/97 | 1997-11-24 | ||
| DK1344/97 | 1997-11-24 | ||
| US09/073,684 | 1998-05-06 | ||
| US09/073,684 US6124127A (en) | 1997-11-24 | 1998-05-06 | Pectate lyase |
| US09/184,217 US6258590B1 (en) | 1998-11-02 | 1998-11-02 | Biopreparation of textiles at high temperatures |
| US09/184,217 | 1998-11-02 |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| WO1999027084A1 true WO1999027084A1 (en) | 1999-06-03 |
| WO1999027084A9 WO1999027084A9 (en) | 1999-08-12 |
Family
ID=27439469
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/DK1998/000515 WO1999027084A1 (en) | 1997-11-24 | 1998-11-24 | Novel pectate lyases |
Country Status (10)
| Country | Link |
|---|---|
| EP (1) | EP1032658B1 (en) |
| JP (1) | JP4246386B2 (en) |
| KR (1) | KR20010032382A (en) |
| CN (1) | CN1244695C (en) |
| AU (1) | AU1482599A (en) |
| BR (1) | BR9815007A (en) |
| CA (1) | CA2310562C (en) |
| PL (1) | PL343254A1 (en) |
| TR (1) | TR200001489T2 (en) |
| WO (1) | WO1999027084A1 (en) |
Cited By (89)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2000026464A2 (en) * | 1998-11-02 | 2000-05-11 | Novozymes North America Inc. | Enzymatic treatment of textiles at high temperatures |
| WO2000055309A1 (en) * | 1999-03-16 | 2000-09-21 | Novozymes A/S | Novel pectate lyases |
| WO2001079440A2 (en) * | 2000-04-13 | 2001-10-25 | Novozymes A/S | Extracellular expression of pectate lyase using bacillus or escherichia coli |
| US6399351B1 (en) | 1999-03-16 | 2002-06-04 | Novozymes A/S | Pectate lyases |
| WO2002006442A3 (en) * | 2000-07-19 | 2002-09-26 | Novozymes As | Cell-wall degrading enzyme variants |
| WO2004053039A2 (en) | 2002-12-11 | 2004-06-24 | Novozymes A/S | Detergent composition comprising endo-glucanase |
| WO2008021761A2 (en) | 2006-08-11 | 2008-02-21 | Novozymes Biologicals, Inc. | Bacteria cultures and compositions comprising bacteria cultures |
| EP1975229A2 (en) | 2000-10-13 | 2008-10-01 | Novozymes A/S | Alpha-amylase variant with altered properties |
| WO2008118749A2 (en) | 2007-03-23 | 2008-10-02 | Novozymes Biologicals, Inc. | Preventing and reducing biofilm formation and planktonic proliferation |
| US7601529B2 (en) | 2002-05-14 | 2009-10-13 | Novozymes A/S | Pectate lyase variants |
| US7611882B2 (en) | 2001-05-14 | 2009-11-03 | Novozymes A/S | Detergent compositions comprising Bacillus subtilis pectate lyases |
| EP2135934A1 (en) | 2008-06-16 | 2009-12-23 | Unilever PLC | Use of a laundry detergent composition |
| EP2149786A1 (en) | 2008-08-01 | 2010-02-03 | Unilever PLC | Improvements relating to detergent analysis |
| EP2202290A1 (en) | 2008-12-23 | 2010-06-30 | Unilever PLC | A flowable laundry composition and packaging therefor |
| WO2011080267A2 (en) | 2009-12-29 | 2011-07-07 | Novozymes A/S | Polypetides having detergency enhancing effect |
| WO2011104339A1 (en) | 2010-02-25 | 2011-09-01 | Novozymes A/S | Variants of a lysozyme and polynucleotides encoding same |
| WO2011134809A1 (en) | 2010-04-26 | 2011-11-03 | Novozymes A/S | Enzyme granules |
| DE212009000119U1 (en) | 2008-09-12 | 2011-12-30 | Unilever N.V. | Dispenser and pretreatment agent for viscous liquids |
| WO2012028483A1 (en) | 2010-08-30 | 2012-03-08 | Novozymes A/S | A concentrated soak wash |
| WO2012028482A1 (en) | 2010-08-30 | 2012-03-08 | Novozymes A/S | A two-soak wash |
| WO2012035103A1 (en) | 2010-09-16 | 2012-03-22 | Novozymes A/S | Lysozymes |
| WO2012038144A1 (en) | 2010-09-20 | 2012-03-29 | Unilever Plc | Fabric treatment compositions comprising target benefit agents |
| WO2012047430A1 (en) | 2010-10-07 | 2012-04-12 | Danisco Us Inc. | Processing of palm kernel waste using mannanase and pectinase |
| WO2012052306A1 (en) | 2010-10-22 | 2012-04-26 | Unilever Plc | Externally structured aqueous detergent liquid |
| EP2476743A1 (en) | 2011-04-04 | 2012-07-18 | Unilever Plc, A Company Registered In England And Wales under company no. 41424 of Unilever House | Method of laundering fabric |
| WO2012104159A1 (en) | 2011-01-31 | 2012-08-09 | Unilever Plc | Alkaline liquid detergent compositions |
| WO2012112718A1 (en) | 2011-02-15 | 2012-08-23 | Novozymes Biologicals, Inc. | Mitigation of odor in cleaning machines and cleaning processes |
| EP2522714A1 (en) | 2011-05-13 | 2012-11-14 | Unilever Plc, A Company Registered In England And Wales under company no. 41424 of Unilever House | Aqueous concentrated laundry detergent compositions |
| EP2522715A1 (en) | 2011-05-13 | 2012-11-14 | Unilever Plc, A Company Registered In England And Wales under company no. 41424 of Unilever House | Aqueous concentrated laundry detergent compositions |
| WO2012156250A1 (en) | 2011-05-13 | 2012-11-22 | Unilever Plc | Aqueous concentrated laundry detergent compositions |
| EP2537918A1 (en) | 2011-06-20 | 2012-12-26 | The Procter & Gamble Company | Consumer products with lipase comprising coated particles |
| WO2012175401A2 (en) | 2011-06-20 | 2012-12-27 | Novozymes A/S | Particulate composition |
| WO2013004636A1 (en) | 2011-07-01 | 2013-01-10 | Novozymes A/S | Stabilized subtilisin composition |
| WO2013004635A1 (en) | 2011-07-01 | 2013-01-10 | Novozymes A/S | Liquid detergent composition |
| WO2013007594A1 (en) | 2011-07-12 | 2013-01-17 | Novozymes A/S | Storage-stable enzyme granules |
| WO2013092052A1 (en) | 2011-12-20 | 2013-06-27 | Unilever Plc | Isotropic liquid detergents comprising soil release polymer |
| WO2013160025A1 (en) | 2012-04-23 | 2013-10-31 | Unilever Plc | Structured aqueous liquid detergent |
| WO2014068083A1 (en) | 2012-11-01 | 2014-05-08 | Novozymes A/S | Method for removal of dna |
| WO2014086659A2 (en) | 2012-12-06 | 2014-06-12 | Ahmedabad Textile Industry's Research Association | Method for enzymatical preparation of textiles |
| WO2014191170A1 (en) | 2013-05-30 | 2014-12-04 | Novozymes A/S | Particulate enzyme composition |
| WO2014198840A1 (en) | 2013-06-12 | 2014-12-18 | Earth Alive Clean Technologies Inc. | Dust suppressant |
| WO2016001319A1 (en) | 2014-07-03 | 2016-01-07 | Novozymes A/S | Improved stabilization of non-protease enzyme |
| WO2016155993A1 (en) | 2015-04-02 | 2016-10-06 | Unilever Plc | Composition |
| EP3101108A1 (en) | 2015-06-04 | 2016-12-07 | The Procter and Gamble Company | Hand dishwashing liquid detergent composition |
| EP3101109A1 (en) | 2015-06-04 | 2016-12-07 | The Procter and Gamble Company | Hand dishwashing liquid detergent composition |
| WO2017036916A1 (en) | 2015-08-28 | 2017-03-09 | Unilever N.V. | Process to manufacture cross-linked enzyme aggregates |
| WO2017117089A1 (en) | 2015-12-28 | 2017-07-06 | Novozymes Bioag A/S | Heat priming of bacterial spores |
| WO2017133879A1 (en) | 2016-02-04 | 2017-08-10 | Unilever Plc | Detergent liquid |
| WO2017210188A1 (en) | 2016-05-31 | 2017-12-07 | Novozymes A/S | Stabilized liquid peroxide compositions |
| WO2017211697A1 (en) | 2016-06-09 | 2017-12-14 | Unilever Plc | Laundry products |
| EP3284805A1 (en) | 2016-08-17 | 2018-02-21 | The Procter & Gamble Company | Cleaning composition comprising enzymes |
| WO2018060475A1 (en) | 2016-09-29 | 2018-04-05 | Novozymes A/S | Spore containing granule |
| WO2018083093A1 (en) | 2016-11-01 | 2018-05-11 | Novozymes A/S | Multi-core granules |
| WO2018099762A1 (en) | 2016-12-01 | 2018-06-07 | Basf Se | Stabilization of enzymes in compositions |
| WO2019002356A1 (en) | 2017-06-30 | 2019-01-03 | Novozymes A/S | Enzyme slurry composition |
| WO2019038187A1 (en) | 2017-08-24 | 2019-02-28 | Unilever Plc | Improvements relating to fabric cleaning |
| WO2019038186A1 (en) | 2017-08-24 | 2019-02-28 | Unilever Plc | Improvements relating to fabric cleaning |
| EP3461881A1 (en) | 2013-05-03 | 2019-04-03 | Novozymes A/S | Microencapsulation of detergent enzymes |
| WO2019063499A1 (en) | 2017-09-27 | 2019-04-04 | Novozymes A/S | Lipase variants and microcapsule compositions comprising such lipase variants |
| WO2019076833A1 (en) | 2017-10-16 | 2019-04-25 | Novozymes A/S | Low dusting granules |
| WO2019076834A1 (en) | 2017-10-16 | 2019-04-25 | Novozymes A/S | Low dusting granules |
| WO2019091822A1 (en) | 2017-11-09 | 2019-05-16 | Basf Se | Coatings of enzyme particles comprising organic white pigments |
| WO2019154951A1 (en) | 2018-02-08 | 2019-08-15 | Novozymes A/S | Lipases, lipase variants and compositions thereof |
| WO2019154955A1 (en) | 2018-02-08 | 2019-08-15 | Novozymes A/S | Lipase variants and compositions thereof |
| WO2019175240A1 (en) | 2018-03-13 | 2019-09-19 | Novozymes A/S | Microencapsulation using amino sugar oligomers |
| WO2019206994A1 (en) | 2018-04-26 | 2019-10-31 | Basf Se | Lipase enzymes |
| WO2019211143A1 (en) | 2018-05-03 | 2019-11-07 | Basf Se | Amylase enzymes |
| EP3569611A1 (en) | 2013-04-23 | 2019-11-20 | Novozymes A/S | Liquid automatic dish washing detergent compositions with stabilised subtilisin |
| WO2019238761A1 (en) | 2018-06-15 | 2019-12-19 | Basf Se | Water soluble multilayer films containing wash active chemicals and enzymes |
| US10655093B2 (en) | 2015-06-05 | 2020-05-19 | The Procter & Gamble Company | Compacted liquid laundry detergent composition |
| US10662417B2 (en) | 2016-07-05 | 2020-05-26 | Novozymes A/S | Pectate lyase variants and polynucleotides encoding same |
| US10683474B2 (en) | 2015-06-05 | 2020-06-16 | The Procter & Gamble Company | Compacted liquid laundry detergent composition |
| US10711225B2 (en) | 2015-06-05 | 2020-07-14 | The Procter & Gamble Company | Compacted liquid laundry detergent composition |
| WO2020169564A1 (en) | 2019-02-20 | 2020-08-27 | Basf Se | Industrial fermentation process for bacillus using defined medium and trace element feed |
| WO2020169563A1 (en) | 2019-02-20 | 2020-08-27 | Basf Se | Industrial fermentation process for bacillus using defined medium and magnesium feed |
| WO2020193534A2 (en) | 2019-03-25 | 2020-10-01 | Basf Se | Amylase enzymes |
| WO2020193535A2 (en) | 2019-03-25 | 2020-10-01 | Basf Se | Amylase enzymes |
| WO2020193532A1 (en) | 2019-03-25 | 2020-10-01 | Basf Se | Cleaning composition having amylase enzymes |
| WO2020249546A1 (en) | 2019-06-13 | 2020-12-17 | Basf Se | Method of recovering a protein from fermentation broth using a divalent cation |
| WO2021001400A1 (en) | 2019-07-02 | 2021-01-07 | Novozymes A/S | Lipase variants and compositions thereof |
| WO2021001244A1 (en) | 2019-07-01 | 2021-01-07 | Basf Se | Peptide acetals for stabilising enzymes |
| WO2021004830A1 (en) | 2019-07-05 | 2021-01-14 | Basf Se | Industrial fermentation process for microbial cells using a fed-batch pre-culture |
| WO2021032881A1 (en) | 2019-08-22 | 2021-02-25 | Basf Se | Amylase variants |
| WO2021105336A1 (en) | 2019-11-29 | 2021-06-03 | Basf Se | Compositions comprising polymer and enzyme |
| EP3907271A1 (en) | 2020-05-07 | 2021-11-10 | Novozymes A/S | Cleaning composition, use and method of cleaning |
| WO2022023250A1 (en) | 2020-07-27 | 2022-02-03 | Unilever Ip Holdings B.V. | Use of an enzyme and surfactant for inhibiting microorganisms |
| WO2022063698A1 (en) | 2020-09-22 | 2022-03-31 | Basf Se | Liquid composition comprising peptide aldehyde |
| WO2022063699A1 (en) | 2020-09-22 | 2022-03-31 | Basf Se | Improved combination of protease and protease inhibitor with secondary enzyme |
| WO2023227356A1 (en) | 2022-05-27 | 2023-11-30 | Unilever Ip Holdings B.V. | Composition containing enzyme |
Families Citing this family (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2017102542A1 (en) * | 2015-12-15 | 2017-06-22 | Metgen Oy | Method for producing mechanical pulp from a biomass comprising lignocellulosic material |
| CN108004821B (en) * | 2017-11-28 | 2020-05-19 | 嘉兴温华环保科技有限公司 | Waste paper papermaking process |
Citations (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| EP0870834A1 (en) * | 1997-04-09 | 1998-10-14 | Kao Corporation | Pectic acid lyase |
-
1998
- 1998-11-24 CA CA2310562A patent/CA2310562C/en not_active Expired - Fee Related
- 1998-11-24 TR TR2000/01489T patent/TR200001489T2/en unknown
- 1998-11-24 KR KR1020007005621A patent/KR20010032382A/en not_active Application Discontinuation
- 1998-11-24 PL PL98343254A patent/PL343254A1/en unknown
- 1998-11-24 WO PCT/DK1998/000515 patent/WO1999027084A1/en not_active Application Discontinuation
- 1998-11-24 EP EP98958820A patent/EP1032658B1/en not_active Expired - Lifetime
- 1998-11-24 AU AU14825/99A patent/AU1482599A/en not_active Abandoned
- 1998-11-24 CN CNB988128012A patent/CN1244695C/en not_active Expired - Fee Related
- 1998-11-24 JP JP2000522226A patent/JP4246386B2/en not_active Expired - Fee Related
- 1998-11-24 BR BR9815007-3A patent/BR9815007A/en not_active Application Discontinuation
Patent Citations (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| EP0870834A1 (en) * | 1997-04-09 | 1998-10-14 | Kao Corporation | Pectic acid lyase |
Non-Patent Citations (7)
| Title |
|---|
| AGRIC. BIOL. CHEM., Volume 55, No. 1, 1991, YOSHIMITSU MIYAZAKI, "Purification and Characterization of Endo-Pectate Lyase from Bacillus Macerans", pages 25-30. * |
| CANADIAN JOURNAL OF MICROBIOLOGY, Volume 24, 1979, CATHERINE T. KELLY et al., "Production and Properties of Polygalacturonate Lyase by an Alkalophilic Microorganism Bacillus sp. RK9", pages 1164-1172. * |
| EMBL, Database GenBank/DDBJ, Accession No. L41673, ID PFPELYB, LIAO C.H. et al., "Cloning of E Pectate Lyase Gene from Xanthomonas Campestris pv. Malvacearum and Comparison of Its Sequence Relationship with Pel Genes of Soft-Rot Erwinia and Pseudomonas"; & MEDLINE, 96143678, 19 Feb. 1996. * |
| FILE WPI, Derwent Accession No. 81-54142D, AGENCY OF IND SCI AND TECHNOLOGY, "Pectate-Lyase Prepn. - Using Basophilic Bacillus Strain"; & JP,A,56 068 393 (09-06-81) DW8130. * |
| FILE WPI, Derwent Accession No. 88-209811, HOHNEN OIL KK, "Pectic Acid Lyase Mfr. - by Cloning Pectic Acid Lyase Gene Using E. Coli by Recombinant Deoxyribonucleic Acid Technique, Culturing E. Coli and Sepg. the Lyase"; & JP,A,63 146 790 (18-06-88) DW8830. * |
| FILE WPI, Derwent Accession No. 90-285860, OJI PAPER CO., "Protein and Region Relating to its Expression and Secretion - Where DNA Sequence is Used to Increase Prodn. Yield of Pectic Acid Lyase by Microorganism"; & JP,A,02 200 191 (08-08-90) DW9038. * |
| STN INTERNATIONAL, File CA, CHEMICAL ABSTRACTS, Volume 118, No. 25, 21 June 1993, (Columbus, Ohio, US), HAN HYE JEONG et al., "Characterization of Pectate Lyase from Alkalitorerant Bacillus sp. YA-14: Its Action Pattern and Active Center", Abstract No. 250320; & J. MICROBIOL. BIOTECHNOL., (1992), 2(4), 260-7. * |
Cited By (114)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2000026464A3 (en) * | 1998-11-02 | 2000-08-10 | Novo Nordisk Biochem Inc | Enzymatic treatment of textiles at high temperatures |
| US6258590B1 (en) | 1998-11-02 | 2001-07-10 | Novozymes A/S | Biopreparation of textiles at high temperatures |
| US6630342B2 (en) | 1998-11-02 | 2003-10-07 | Novozymes A/S | Biopreparation of textiles at high temperatures |
| WO2000026464A2 (en) * | 1998-11-02 | 2000-05-11 | Novozymes North America Inc. | Enzymatic treatment of textiles at high temperatures |
| WO2000055309A1 (en) * | 1999-03-16 | 2000-09-21 | Novozymes A/S | Novel pectate lyases |
| US6399351B1 (en) | 1999-03-16 | 2002-06-04 | Novozymes A/S | Pectate lyases |
| WO2001079440A2 (en) * | 2000-04-13 | 2001-10-25 | Novozymes A/S | Extracellular expression of pectate lyase using bacillus or escherichia coli |
| WO2001079440A3 (en) * | 2000-04-13 | 2002-01-24 | Novozymes As | Extracellular expression of pectate lyase using bacillus or escherichia coli |
| WO2002006442A3 (en) * | 2000-07-19 | 2002-09-26 | Novozymes As | Cell-wall degrading enzyme variants |
| EP1975229A2 (en) | 2000-10-13 | 2008-10-01 | Novozymes A/S | Alpha-amylase variant with altered properties |
| US7611882B2 (en) | 2001-05-14 | 2009-11-03 | Novozymes A/S | Detergent compositions comprising Bacillus subtilis pectate lyases |
| US7601529B2 (en) | 2002-05-14 | 2009-10-13 | Novozymes A/S | Pectate lyase variants |
| US8288144B2 (en) | 2002-05-14 | 2012-10-16 | Novozymes A/S | Pectate lyase variants |
| US8563290B2 (en) | 2002-05-14 | 2013-10-22 | Novozymes A/S | Pectate lyase variants |
| US9005950B2 (en) | 2002-05-14 | 2015-04-14 | Novozymes A/S | Pectate lyase variants |
| WO2004053039A2 (en) | 2002-12-11 | 2004-06-24 | Novozymes A/S | Detergent composition comprising endo-glucanase |
| EP2311941A1 (en) | 2002-12-11 | 2011-04-20 | Novozymes A/S | Detergent composition comprising endo-glucanase |
| WO2008021761A2 (en) | 2006-08-11 | 2008-02-21 | Novozymes Biologicals, Inc. | Bacteria cultures and compositions comprising bacteria cultures |
| WO2008118749A2 (en) | 2007-03-23 | 2008-10-02 | Novozymes Biologicals, Inc. | Preventing and reducing biofilm formation and planktonic proliferation |
| EP2500325A1 (en) | 2007-03-23 | 2012-09-19 | Novozymes Biologicals, Inc. | Preventing and Reducing Biofilm Formation and Planktonic Proliferation |
| EP2135934A1 (en) | 2008-06-16 | 2009-12-23 | Unilever PLC | Use of a laundry detergent composition |
| EP2149786A1 (en) | 2008-08-01 | 2010-02-03 | Unilever PLC | Improvements relating to detergent analysis |
| DE212009000119U1 (en) | 2008-09-12 | 2011-12-30 | Unilever N.V. | Dispenser and pretreatment agent for viscous liquids |
| EP2202290A1 (en) | 2008-12-23 | 2010-06-30 | Unilever PLC | A flowable laundry composition and packaging therefor |
| WO2011080267A2 (en) | 2009-12-29 | 2011-07-07 | Novozymes A/S | Polypetides having detergency enhancing effect |
| WO2011104339A1 (en) | 2010-02-25 | 2011-09-01 | Novozymes A/S | Variants of a lysozyme and polynucleotides encoding same |
| WO2011134809A1 (en) | 2010-04-26 | 2011-11-03 | Novozymes A/S | Enzyme granules |
| EP2840134A1 (en) | 2010-04-26 | 2015-02-25 | Novozymes A/S | Enzyme granules |
| WO2012028483A1 (en) | 2010-08-30 | 2012-03-08 | Novozymes A/S | A concentrated soak wash |
| WO2012028482A1 (en) | 2010-08-30 | 2012-03-08 | Novozymes A/S | A two-soak wash |
| WO2012035103A1 (en) | 2010-09-16 | 2012-03-22 | Novozymes A/S | Lysozymes |
| WO2012038144A1 (en) | 2010-09-20 | 2012-03-29 | Unilever Plc | Fabric treatment compositions comprising target benefit agents |
| WO2012047430A1 (en) | 2010-10-07 | 2012-04-12 | Danisco Us Inc. | Processing of palm kernel waste using mannanase and pectinase |
| WO2012052306A1 (en) | 2010-10-22 | 2012-04-26 | Unilever Plc | Externally structured aqueous detergent liquid |
| WO2012104159A1 (en) | 2011-01-31 | 2012-08-09 | Unilever Plc | Alkaline liquid detergent compositions |
| WO2012112718A1 (en) | 2011-02-15 | 2012-08-23 | Novozymes Biologicals, Inc. | Mitigation of odor in cleaning machines and cleaning processes |
| EP3431581A2 (en) | 2011-02-15 | 2019-01-23 | Novozymes Biologicals, Inc. | Mitigation of odor in cleaning machines and cleaning processes |
| EP2476743A1 (en) | 2011-04-04 | 2012-07-18 | Unilever Plc, A Company Registered In England And Wales under company no. 41424 of Unilever House | Method of laundering fabric |
| WO2012136427A1 (en) | 2011-04-04 | 2012-10-11 | Unilever Plc | Method of laundering fabric |
| EP2522714A1 (en) | 2011-05-13 | 2012-11-14 | Unilever Plc, A Company Registered In England And Wales under company no. 41424 of Unilever House | Aqueous concentrated laundry detergent compositions |
| EP2522715A1 (en) | 2011-05-13 | 2012-11-14 | Unilever Plc, A Company Registered In England And Wales under company no. 41424 of Unilever House | Aqueous concentrated laundry detergent compositions |
| WO2012156250A1 (en) | 2011-05-13 | 2012-11-22 | Unilever Plc | Aqueous concentrated laundry detergent compositions |
| WO2013003025A1 (en) | 2011-06-20 | 2013-01-03 | The Procter & Gamble Company | Consumer products with lipase comprising coated particles |
| EP2537918A1 (en) | 2011-06-20 | 2012-12-26 | The Procter & Gamble Company | Consumer products with lipase comprising coated particles |
| WO2012175401A2 (en) | 2011-06-20 | 2012-12-27 | Novozymes A/S | Particulate composition |
| WO2013004636A1 (en) | 2011-07-01 | 2013-01-10 | Novozymes A/S | Stabilized subtilisin composition |
| WO2013004635A1 (en) | 2011-07-01 | 2013-01-10 | Novozymes A/S | Liquid detergent composition |
| WO2013007594A1 (en) | 2011-07-12 | 2013-01-17 | Novozymes A/S | Storage-stable enzyme granules |
| WO2013092052A1 (en) | 2011-12-20 | 2013-06-27 | Unilever Plc | Isotropic liquid detergents comprising soil release polymer |
| WO2013160025A1 (en) | 2012-04-23 | 2013-10-31 | Unilever Plc | Structured aqueous liquid detergent |
| WO2014068083A1 (en) | 2012-11-01 | 2014-05-08 | Novozymes A/S | Method for removal of dna |
| WO2014086659A2 (en) | 2012-12-06 | 2014-06-12 | Ahmedabad Textile Industry's Research Association | Method for enzymatical preparation of textiles |
| EP3569611A1 (en) | 2013-04-23 | 2019-11-20 | Novozymes A/S | Liquid automatic dish washing detergent compositions with stabilised subtilisin |
| EP3461881A1 (en) | 2013-05-03 | 2019-04-03 | Novozymes A/S | Microencapsulation of detergent enzymes |
| WO2014191170A1 (en) | 2013-05-30 | 2014-12-04 | Novozymes A/S | Particulate enzyme composition |
| WO2014198840A1 (en) | 2013-06-12 | 2014-12-18 | Earth Alive Clean Technologies Inc. | Dust suppressant |
| WO2016001319A1 (en) | 2014-07-03 | 2016-01-07 | Novozymes A/S | Improved stabilization of non-protease enzyme |
| WO2016155993A1 (en) | 2015-04-02 | 2016-10-06 | Unilever Plc | Composition |
| EP3101109A1 (en) | 2015-06-04 | 2016-12-07 | The Procter and Gamble Company | Hand dishwashing liquid detergent composition |
| EP3101108A1 (en) | 2015-06-04 | 2016-12-07 | The Procter and Gamble Company | Hand dishwashing liquid detergent composition |
| EP3287513A1 (en) | 2015-06-04 | 2018-02-28 | The Procter & Gamble Company | Hand dishwashing liquid detergent composition |
| WO2016196874A1 (en) | 2015-06-04 | 2016-12-08 | The Procter & Gamble Company | Hand dishwashing liquid detergent composition |
| WO2016196872A1 (en) | 2015-06-04 | 2016-12-08 | The Procter & Gamble Company | Hand dishwashing liquid detergent composition |
| US10377974B2 (en) | 2015-06-04 | 2019-08-13 | The Procter & Gamble Company | Hand dishwashing liquid detergent composition |
| US10377973B2 (en) | 2015-06-04 | 2019-08-13 | The Procter & Gamble Company | Hand dishwashing liquid detergent composition |
| EP3284811A1 (en) | 2015-06-04 | 2018-02-21 | The Procter & Gamble Company | Hand dishwashing liquid detergent composition |
| US10711225B2 (en) | 2015-06-05 | 2020-07-14 | The Procter & Gamble Company | Compacted liquid laundry detergent composition |
| US10683474B2 (en) | 2015-06-05 | 2020-06-16 | The Procter & Gamble Company | Compacted liquid laundry detergent composition |
| US10655093B2 (en) | 2015-06-05 | 2020-05-19 | The Procter & Gamble Company | Compacted liquid laundry detergent composition |
| WO2017036916A1 (en) | 2015-08-28 | 2017-03-09 | Unilever N.V. | Process to manufacture cross-linked enzyme aggregates |
| WO2017036915A1 (en) | 2015-08-28 | 2017-03-09 | Unilever N.V. | Liquid detergency composition comprising protease and non-protease enzyme |
| WO2017036917A1 (en) | 2015-08-28 | 2017-03-09 | Unilever N.V. | Liquid detergency composition comprising lipase and protease |
| WO2017117089A1 (en) | 2015-12-28 | 2017-07-06 | Novozymes Bioag A/S | Heat priming of bacterial spores |
| WO2017133879A1 (en) | 2016-02-04 | 2017-08-10 | Unilever Plc | Detergent liquid |
| WO2017210188A1 (en) | 2016-05-31 | 2017-12-07 | Novozymes A/S | Stabilized liquid peroxide compositions |
| WO2017211697A1 (en) | 2016-06-09 | 2017-12-14 | Unilever Plc | Laundry products |
| US10662417B2 (en) | 2016-07-05 | 2020-05-26 | Novozymes A/S | Pectate lyase variants and polynucleotides encoding same |
| EP3284805A1 (en) | 2016-08-17 | 2018-02-21 | The Procter & Gamble Company | Cleaning composition comprising enzymes |
| WO2018034842A1 (en) | 2016-08-17 | 2018-02-22 | The Procter & Gamble Company | Cleaning composition comprising enzymes |
| WO2018060475A1 (en) | 2016-09-29 | 2018-04-05 | Novozymes A/S | Spore containing granule |
| WO2018083093A1 (en) | 2016-11-01 | 2018-05-11 | Novozymes A/S | Multi-core granules |
| WO2018099762A1 (en) | 2016-12-01 | 2018-06-07 | Basf Se | Stabilization of enzymes in compositions |
| WO2019002356A1 (en) | 2017-06-30 | 2019-01-03 | Novozymes A/S | Enzyme slurry composition |
| WO2019038186A1 (en) | 2017-08-24 | 2019-02-28 | Unilever Plc | Improvements relating to fabric cleaning |
| WO2019038187A1 (en) | 2017-08-24 | 2019-02-28 | Unilever Plc | Improvements relating to fabric cleaning |
| WO2019063499A1 (en) | 2017-09-27 | 2019-04-04 | Novozymes A/S | Lipase variants and microcapsule compositions comprising such lipase variants |
| WO2019076833A1 (en) | 2017-10-16 | 2019-04-25 | Novozymes A/S | Low dusting granules |
| WO2019076834A1 (en) | 2017-10-16 | 2019-04-25 | Novozymes A/S | Low dusting granules |
| WO2019091822A1 (en) | 2017-11-09 | 2019-05-16 | Basf Se | Coatings of enzyme particles comprising organic white pigments |
| WO2019154955A1 (en) | 2018-02-08 | 2019-08-15 | Novozymes A/S | Lipase variants and compositions thereof |
| WO2019154952A1 (en) | 2018-02-08 | 2019-08-15 | Novozymes A/S | Lipase variants and compositions thereof |
| WO2019154954A1 (en) | 2018-02-08 | 2019-08-15 | Novozymes A/S | Lipase variants and compositions thereof |
| WO2019154951A1 (en) | 2018-02-08 | 2019-08-15 | Novozymes A/S | Lipases, lipase variants and compositions thereof |
| WO2019175240A1 (en) | 2018-03-13 | 2019-09-19 | Novozymes A/S | Microencapsulation using amino sugar oligomers |
| WO2019206994A1 (en) | 2018-04-26 | 2019-10-31 | Basf Se | Lipase enzymes |
| WO2019211143A1 (en) | 2018-05-03 | 2019-11-07 | Basf Se | Amylase enzymes |
| WO2019238761A1 (en) | 2018-06-15 | 2019-12-19 | Basf Se | Water soluble multilayer films containing wash active chemicals and enzymes |
| WO2020169564A1 (en) | 2019-02-20 | 2020-08-27 | Basf Se | Industrial fermentation process for bacillus using defined medium and trace element feed |
| WO2020169563A1 (en) | 2019-02-20 | 2020-08-27 | Basf Se | Industrial fermentation process for bacillus using defined medium and magnesium feed |
| WO2020193534A2 (en) | 2019-03-25 | 2020-10-01 | Basf Se | Amylase enzymes |
| WO2020193535A2 (en) | 2019-03-25 | 2020-10-01 | Basf Se | Amylase enzymes |
| WO2020193532A1 (en) | 2019-03-25 | 2020-10-01 | Basf Se | Cleaning composition having amylase enzymes |
| WO2020249546A1 (en) | 2019-06-13 | 2020-12-17 | Basf Se | Method of recovering a protein from fermentation broth using a divalent cation |
| WO2021001244A1 (en) | 2019-07-01 | 2021-01-07 | Basf Se | Peptide acetals for stabilising enzymes |
| WO2021001400A1 (en) | 2019-07-02 | 2021-01-07 | Novozymes A/S | Lipase variants and compositions thereof |
| WO2021004830A1 (en) | 2019-07-05 | 2021-01-14 | Basf Se | Industrial fermentation process for microbial cells using a fed-batch pre-culture |
| WO2021032881A1 (en) | 2019-08-22 | 2021-02-25 | Basf Se | Amylase variants |
| WO2021105336A1 (en) | 2019-11-29 | 2021-06-03 | Basf Se | Compositions comprising polymer and enzyme |
| EP3907271A1 (en) | 2020-05-07 | 2021-11-10 | Novozymes A/S | Cleaning composition, use and method of cleaning |
| WO2021224389A1 (en) | 2020-05-07 | 2021-11-11 | Novozymes A/S | Medical cleaning composition, use and method of cleaning |
| WO2022023250A1 (en) | 2020-07-27 | 2022-02-03 | Unilever Ip Holdings B.V. | Use of an enzyme and surfactant for inhibiting microorganisms |
| WO2022063698A1 (en) | 2020-09-22 | 2022-03-31 | Basf Se | Liquid composition comprising peptide aldehyde |
| WO2022063699A1 (en) | 2020-09-22 | 2022-03-31 | Basf Se | Improved combination of protease and protease inhibitor with secondary enzyme |
| WO2023227356A1 (en) | 2022-05-27 | 2023-11-30 | Unilever Ip Holdings B.V. | Composition containing enzyme |
Also Published As
| Publication number | Publication date |
|---|---|
| KR20010032382A (en) | 2001-04-16 |
| EP1032658B1 (en) | 2012-06-27 |
| CN1244695C (en) | 2006-03-08 |
| PL343254A1 (en) | 2001-07-30 |
| CA2310562A1 (en) | 1999-06-03 |
| BR9815007A (en) | 2000-10-03 |
| AU1482599A (en) | 1999-06-15 |
| TR200001489T2 (en) | 2000-11-21 |
| CN1285870A (en) | 2001-02-28 |
| CA2310562C (en) | 2011-01-11 |
| EP1032658A1 (en) | 2000-09-06 |
| JP2001526022A (en) | 2001-12-18 |
| WO1999027084A9 (en) | 1999-08-12 |
| JP4246386B2 (en) | 2009-04-02 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| EP1032658B1 (en) | Pectate lyases | |
| US7273745B2 (en) | Pectate lyases | |
| US6280995B1 (en) | Pectate lyases | |
| WO1999027083A1 (en) | PECTIN DEGRADING ENZYMES FROM $i(BACILLUS LICHENIFORMIS) | |
| EP1305408B1 (en) | Cell-wall degrading enzyme variants | |
| WO2001062903A1 (en) | Family 44 xyloglucanases | |
| US6808915B2 (en) | Cell-wall degrading enzyme variants | |
| US6429000B1 (en) | Pectin degrading enzymes from Bacillus licheniformis | |
| JP4837855B2 (en) | Novel endo-β-1,4-glucanase | |
| US6399351B1 (en) | Pectate lyases | |
| WO2000055309A1 (en) | Novel pectate lyases | |
| EP1261698A1 (en) | Family 5 xyloglucanases | |
| CA2310806C (en) | Pectin degrading enzymes from bacillus licheniformis | |
| US7256030B1 (en) | Family 9 endo-β-1,4-glucanases | |
| MXPA00005108A (en) | Novel pectate lyases | |
| MXPA00005110A (en) | Pectin degrading enzymes from bacillus licheniformis |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| WWE | Wipo information: entry into national phase |
Ref document number: 98812801.2 Country of ref document: CN |
|
| AK | Designated states |
Kind code of ref document: A1 Designated state(s): AL AM AT AU AZ BA BB BG BR BY CA CH CN CU CZ DE DK EE ES FI GB GD GE GH GM HR HU ID IL IS JP KE KG KP KR KZ LC LK LR LS LT LU LV MD MG MK MN MW MX NO NZ PL PT RO RU SD SE SG SI SK SL TJ TM TR TT UA UG UZ VN YU ZW |
|
| AL | Designated countries for regional patents |
Kind code of ref document: A1 Designated state(s): GH GM KE LS MW SD SZ UG ZW AM AZ BY KG KZ MD RU TJ TM AT BE CH CY DE DK ES FI FR GB GR IE IT LU MC NL PT SE BF BJ CF CG CI CM GA GN GW ML MR NE SN TD TG |
|
| DFPE | Request for preliminary examination filed prior to expiration of 19th month from priority date (pct application filed before 20040101) | ||
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application | ||
| AK | Designated states |
Kind code of ref document: C2 Designated state(s): AL AM AT AU AZ BA BB BG BR BY CA CH CN CU CZ DE DK EE ES FI GB GD GE GH GM HR HU ID IL IS JP KE KG KP KR KZ LC LK LR LS LT LU LV MD MG MK MN MW MX NO NZ PL PT RO RU SD SE SG SI SK SL TJ TM TR TT UA UG UZ VN YU ZW |
|
| AL | Designated countries for regional patents |
Kind code of ref document: C2 Designated state(s): GH GM KE LS MW SD SZ UG ZW AM AZ BY KG KZ MD RU TJ TM AT BE CH CY DE DK ES FI FR GB GR IE IT LU MC NL PT SE BF BJ CF CG CI CM GA GN GW ML MR NE SN TD TG |
|
| COP | Corrected version of pamphlet |
Free format text: PAGES 1-16, SEQUENCE LISTING, ADDED |
|
| REG | Reference to national code |
Ref country code: DE Ref legal event code: 8642 |
|
| ENP | Entry into the national phase |
Ref document number: 2310562 Country of ref document: CA Ref document number: 2310562 Country of ref document: CA Kind code of ref document: A |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 1020007005621 Country of ref document: KR |
|
| WWE | Wipo information: entry into national phase |
Ref document number: PA/a/2000/005108 Country of ref document: MX Ref document number: 2000/01489 Country of ref document: TR |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 1998958820 Country of ref document: EP |
|
| WWP | Wipo information: published in national office |
Ref document number: 1998958820 Country of ref document: EP |
|
| WWP | Wipo information: published in national office |
Ref document number: 1020007005621 Country of ref document: KR |
|
| WWW | Wipo information: withdrawn in national office |
Ref document number: 1020007005621 Country of ref document: KR |