CN108138121B - High-level production of long-chain dicarboxylic acids by microorganisms - Google Patents
High-level production of long-chain dicarboxylic acids by microorganisms Download PDFInfo
- Publication number
- CN108138121B CN108138121B CN201680055065.3A CN201680055065A CN108138121B CN 108138121 B CN108138121 B CN 108138121B CN 201680055065 A CN201680055065 A CN 201680055065A CN 108138121 B CN108138121 B CN 108138121B
- Authority
- CN
- China
- Prior art keywords
- lcda
- enzyme
- seq
- cell
- strain
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Active
Links
Images
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12P—FERMENTATION OR ENZYME-USING PROCESSES TO SYNTHESISE A DESIRED CHEMICAL COMPOUND OR COMPOSITION OR TO SEPARATE OPTICAL ISOMERS FROM A RACEMIC MIXTURE
- C12P7/00—Preparation of oxygen-containing organic compounds
- C12P7/64—Fats; Fatty oils; Ester-type waxes; Higher fatty acids, i.e. having at least seven carbon atoms in an unbroken chain bound to a carboxyl group; Oxidised oils or fats
- C12P7/6409—Fatty acids
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N9/00—Enzymes; Proenzymes; Compositions thereof; Processes for preparing, activating, inhibiting, separating or purifying enzymes
- C12N9/93—Ligases (6)
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Y—ENZYMES
- C12Y602/00—Ligases forming carbon-sulfur bonds (6.2)
- C12Y602/01—Acid-Thiol Ligases (6.2.1)
- C12Y602/01003—Long-chain-fatty-acid-CoA ligase (6.2.1.3)
Abstract
Disclosed are recombinant microbial cells comprising an engineered LCDA production pathway comprising at least one up-regulated long-chain acyl-coa synthetase (ACoS). These recombinant microbial cells are capable of producing one or more Long Chain Dicarboxylic Acid (LCDA) products from a substrate containing long chain fatty acids. Methods of producing LCDA using the recombinant microbial cells are also disclosed.
Description
This application claims the benefit of U.S. provisional application nos. 62/195,340 (filed 2016, 7, 22) and 62/195,338 (filed 2016, 7, 22), both of which are hereby incorporated by reference in their entirety.
Technical Field
The present disclosure is in the field of molecular biology. For example, the disclosure relates to microorganisms (e.g., yeast) that are genetically engineered to produce Long Chain Dicarboxylic Acids (LCDA) from fatty acid containing substrates.
Reference to electronically submitted sequence Listing
An official copy of this sequence listing was submitted electronically via EFS-Web as an ASCII formatted sequence listing with the file name "CL 6467WOPCT _ sequencing _ ST 25", created at 18 months 7 and 2016, and having a size of 480 kilobytes, and submitted concurrently with this specification. The sequence listing included in this ASCII formatted file is part of this specification and is incorporated herein by reference in its entirety.
Background
Dicarboxylic acids containing ten or more carbon atoms may be referred to as Long Chain Dicarboxylic Acids (LCDA). LCDA can be used as a constituent monomer for a variety of synthetic materials, such as polyamides (nylon), polyurethanes, and polyesters. Other uses of LCDA include, for example, the production of certain polycarbonates, powder coatings, fragrances, personal care products, food additives, solvents, cleaning additives, hot melt adhesives, lubricants, insecticides, and fungicides. For example, LCDA can also be used as a plasticizer for engineering plastics and as a corrosion inhibitor in metal working technology.
Many LCDAs suitable for performing commercial applications such as those described above are not typically found in nature. Certain LCDAs, such as dodecanedioic acid (DDDA), can be prepared via a variety of synthetic processes. However, biological processes (e.g., microbial fermentation) may also be used to produce LCDA. Feedstocks containing oil or free fatty acids, for example, may be suitable for use as substrates for fermenting the LCDA product. Efforts have been made to ferment LCDA with yeast biocatalysts (U.S. patent application publication nos. 2004/0146999, 2010/0041115, 2013/0267012, 2014/0228586).
Fatty acids can be activated in yeast for beta-oxidation and other downstream pathways, thereby carrying fatty acids away from the omega-oxidation pathway. Thus, some yeast biocatalysts have been modified to exhibit reduced fatty acid activation, for example by downregulating expression of long-chain fatty acyl-coa synthetases, thereby increasing fermentation of LCDA products via omega-oxidation (see, e.g., U.S. patent application publication nos. 2014/0228586 and 2013/0267012).
Despite the above disclosure, it has now been surprisingly found that increasing fatty acid activation in yeast by up-regulating long chain fatty acyl-coa synthetase allows for high LCDA production. Thus, engineering microbial biocatalysts for high level LCDA production is disclosed herein.
Disclosure of Invention
In one embodiment, the disclosure relates to a recombinant microbial cell comprising an engineered LCDA production pathway comprising upregulation of a polynucleotide sequence encoding a long-chain acyl-coa synthetase (ACoS enzyme), wherein the microbial cell is capable of producing one or more long-chain dicarboxylic acids (LCDA) from a substrate comprising a long-chain fatty acid.
Another embodiment relates to a recombinant microbial cell comprising:
(i) (ii) up-regulation of a polynucleotide sequence encoding a cytochrome P450 monooxygenase (CYP enzyme) and/or up-regulation of a polynucleotide sequence encoding a cytochrome P450 reductase (CPR enzyme),
(ii) up-regulation of a polynucleotide sequence encoding a long-chain acyl-CoA synthetase (ACoS enzyme), and
(iii) down-regulating an endogenous polynucleotide sequence encoding peroxisome biogenesis factor-3,
wherein the microbial cell is capable of producing one or more Long Chain Dicarboxylic Acid (LCDA) products from a substrate comprising long chain fatty acids.
Another embodiment relates to a process for producing Long Chain Dicarboxylic Acids (LCDA). The method comprises the following steps: a) contacting a recombinant microbial cell as disclosed herein with a substrate comprising a long chain fatty acid, wherein the microbial cell synthesizes LCDA from the substrate; and b) optionally recovering the LCDA of step (a).
Brief description of the figures and sequences
FIG. 1: lipid metabolic pathways are depicted, including fatty acid beta-oxidation and omega-oxidation aspects of lipid metabolism. The dashed line/arrow indicates low or weak activity of yarrowia lipolytica (y.
FIG. 2: strategies are shown for engineering yarrowia lipolytica to produce LCDA from oils, oil-derived fatty acids, and/or fatty acid esters.
FIG. 3: phylogenetic trees of candidate acyl-coa synthetases from saccharomyces cerevisiae (s. cerevisiae), yarrowia lipolytica (y. lipolytica) and candida tropicalis (c. tropicalis). Certain abbreviations used in the figure: FAA1 and FAA2 represent saccharomyces cerevisiae Faa1p and Faa2p, respectively. YA-1 represents YLFaa1 p. "YA-" means "YLACoS-". Reference is made to example 1.
FIG. 4: LCDA was produced in a flask test from strain D0145. DCA, dicarboxylic acid. Refer to example 2.
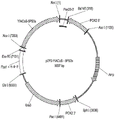
FIG. 5A: the plasmid construct pZP2-YLACoS-3Ps (SEQ ID NO: 63).
FIG. 5B: the plasmid construct pZP2-YLACoS-5Ps (SEQ ID NO: 64).
FIG. 5C: the plasmid construct pZP2-YLACoS-6Ps (SEQ ID NO: 65).
FIG. 5D: the plasmid construct pZP2-YLACoS-10Ps (SEQ ID NO: 66).
FIG. 5E: the plasmid construct pZKL7A-FYLFAAs (SEQ ID NO: 67).
FIG. 5F: the plasmid construct pZP2-YLACoS-5PS3s (SEQ ID NO: 68).
FIG. 6A: SDS-PAGE analysis of soluble and insoluble fractions of E.coli cells transformed to overexpress putative fatty acyl-CoA synthetases. Lanes 1,2, 3, 4, 5, 6: samples from E.coli cells overexpressing YLACoS-3P (SEQ ID NO: 39), YLACoS-5P (SEQ ID NO: 42), YLACoS-6P (SEQ ID NO: 44), YLACoS-10P (SEQ ID NO: 49), YLFAA (SEQ ID NO: 36), or YLACoS-5PS3(SEQ ID NO: 56), respectively. Lane C: samples from E.coli cells transformed with pET23d vector alone (negative control). Lane M: and (3) protein labeling. Refer to example 5.
FIG. 6B: SDS-PAGE of lysates of E.coli cells before and after IPTG-induced overexpression of putative fatty acyl-CoA synthetases. Lanes 1,2, 3, 4: samples from E.coli cells overexpressing YLACoS-3P (SEQ ID NO: 39), YLACoS-5P (SEQ ID NO: 42), YLACoS-6P (SEQ ID NO: 44), or YLACoS-10P (SEQ ID NO: 49), respectively. Lane C: samples from E.coli cells transformed with pET23d vector alone (negative control). Lane M: and (3) protein labeling. Refer to example 5.
FIG. 7A: a graph depicting the lineage of certain strains listed in table 7 is shown. Refer to example 6.
FIG. 7B: a graph depicting the lineage of certain strains disclosed herein is shown.
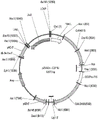
FIG. 8A: plasmid construct p12_3-B-Pex3del1(SEQ ID NO: 76).
FIG. 8B: plasmid construct p70_ Pox2: : leu2(SEQ ID NO: 77).
FIG. 9A: the plasmid construct pZKLY-FCtR17U (SEQ ID NO: 82).
FIG. 9B: the plasmid construct pZKADn-C2F1U (SEQ ID NO: 87).
FIG. 10: time course of LCDA production by Yarrowia (Yarrowia) strain D1308 in a 2-L fermentation experiment. Ethyl palmitate was used as substrate for LCDA production. Diamonds represent the total LCDA amount measured over the course of time, and squares represent C16: 0 LCDA. Refer to example 8.
FIG. 11A: plasmid construct pYRH213(SEQ ID NO: 92).
FIG. 11B: the plasmid construct pZSCpn-3FAOBU (SEQ ID NO: 98).
FIG. 12: time course of LCDA production by yarrowia strain D2300 in a 2-L fermentation experiment. Ethyl palmitate was used as substrate for LCDA production. Squares represent the total LCDA amount measured over the course of time, and circles represent C16: 0 LCDA. Refer to example 9.
FIG. 13. time course of LCDA production by yarrowia strain D3928 in a 5-L fed-batch fermentation experiment. Ethyl palmitate was used as substrate for LCDA production. Squares represent the total LCDA amount measured over the course of time, and diamonds represent C16: 0 LCDA. Refer to example 12.
TABLE 1 nucleic acids and proteins SEQ ID No
Detailed Description
The disclosures of all patent and non-patent documents cited herein are hereby incorporated by reference in their entirety.
The term "a" as used herein is intended to encompass one or more (i.e. at least one) of the referenced feature(s), unless otherwise disclosed.
Where present, all ranges are inclusive and combinable unless otherwise stated. For example, when a range of "1 to 5" is recited, the recited range should be interpreted to include ranges of "1 to 4", "1 to 3", "1 to 2 and 4 to 5", "1 to 3 and 5", and the like.
As used herein, the terms "long-chain acyl-CoA synthetase", "long-chain fatty acyl-co-enzymeEnzyme a synthetase "," long chain fatty acid coa ligase ", and the like are used interchangeably and may be abbreviated as" ACoS ". Using the energy provided by ATP, the ACoS enzymes herein with EC entry 6.2.1.3 can catalyze the activation of long fatty acid chains to fatty acyl-coa. Specifically, the reaction catalyzed by ACoS enzymes is as follows ("ACoS activity"): ATP + Long-chain carboxylate + CoA (coenzyme A) → AMP + diphosphate (PP)i) + acyl-coenzyme A. Typically, an ACoS enzyme is a peroxisomal protein in eukaryotic cells. Up-regulation of a polynucleotide sequence encoding an ACoS enzyme herein results in increased expression of the ACoS enzyme, which in turn can be used to activate increased amounts of long chain fatty acids to long chain acyl-coa. The ACoS enzyme herein is not a "fatty-acyl-coa synthase," with EC entry 2.3.1.86.
The terms "cytochrome P450 monooxygenase", "CYP enzyme", and the like are used interchangeably herein. CYP enzymes herein can catalyze diatomic oxygen (O)2) Is transferred to an organic substrate (typically resulting in an alcohol group), whereas the other oxygen atoms are reduced to water. CYP enzymes have Enzyme Commission (EC) entry 1.14.14.1. The CYP enzyme may be included in an omega-hydroxylase complex (below). CYP enzymes are generally classified herein as class II P450 enzymes, which utilize CPR enzymes for electron transfer. Typically, CYP enzymes are membrane bound. In general, CYP enzymes are described in Urlacher and Girhard (Cell [ Cell ]]30: 26-36) and van Bogaert et al (the european journal of the union of biochemistry (FEBS nuclear) 278: 206-221) incorporated herein by reference. Up-regulation of a polynucleotide sequence encoding a CYP enzyme herein results in increased amount of expression of the CYP enzyme, which in turn can be used to form increased amounts of the ω -hydroxylase complex.
The terms "cytochrome P450 reductase", "NADPH-cytochrome P450 reductase", "CPR enzyme", "NADPH-methemoglobin reductase (NADPH-ferribioprotein reductase)" and the like are used interchangeably herein. CPR enzyme, via FAD (flavin adenine dinucleotide) and FMN (flavin mononucleotide) redox cofactors, catalyzes the reduction of the heme-thiolate moiety in cytochrome P450 monooxygenase by transferring electrons thereto. CPR enzyme has EC entry 1.6.2.4. CPR enzymes may be included in the omega-hydroxylase complex (below). Typically, CPR enzymes are membrane bound. In general, CPR enzyme functions are described in Porter and Kasper (Biochemistry 25: 1682-. Up-regulation of the CPR enzyme-encoding polynucleotide sequence herein results in increased expression of CPR enzyme, which in turn can be used to form increased amounts of omega-hydroxylase complexes.
The terms "ω -hydroxylase complex", "enzyme complex of hydroxylase", "CPR-P450 system", and the like are used interchangeably herein. The omega-hydroxylase complex herein comprises CYP enzymes and CPR enzymes, and can perform omega-hydroxylation of certain organic substrates (e.g., alkanes, fatty alcohols, fatty aldehydes, and fatty acids). Typically, the ω -hydroxylase complex is membrane-bound. Omega-hydroxylation occurring in the Endoplasmic Reticulum (ER) membrane of yeast is typically the first step in omega-oxidation.
The terms "fatty alcohol oxidase" (FAO), "long chain fatty acid oxidase", "long chain alcohol oxidase", "FAO enzyme", and the like are used interchangeably herein. The FAO enzyme has EC entry 1.1.3.20. Herein, FAO enzymes can catalyze the following reactions: fatty alcohol + O2→ fatty aldehyde + H2O2Wherein the fatty alcohol is preferably an omega-hydroxy long chain fatty acid and the fatty aldehyde is preferably an omega-aldehyde long chain fatty acid, each having a carbon chain length of at least 10 (e.g., 10-24 carbons). Typically, the FAO enzyme is a peroxisomal protein in yeast cells.
The terms "fatty alcohol dehydrogenase" (FADH), "long chain fatty acid dehydrogenase", "ADH enzyme", "FADH enzyme", and the like are used interchangeably herein. The FADH enzyme has EC entry 1.1.1.1. The FADH enzymes herein can catalyze the following reactions: fatty alcohol + NAD+→ fatty aldehyde + NADH, wherein the fatty alcohol is preferably an omega-hydroxy long chain fatty acid and the fatty aldehyde is preferably an omega-aldehyde long chain fatty acid, each having a carbon chain length of at least 10 (e.g., 10-24 carbons). Typically, the FADH enzyme is an endoplasmic reticulum membrane protein in yeast cells. FADH enzymes typically use Zn2+Or Fe cation as adjuvantA factor.
The terms "fatty aldehyde dehydrogenase" (FALDH), "long chain aldehyde dehydrogenase", "FALDH enzyme", and the like are used interchangeably herein. The FALDH enzyme has EC entry 1.2.1.48. The FALDH enzyme herein may catalyze the following reaction: fatty aldehyde + NAD++H2O→LCDA+NADH+2H+Wherein the fatty aldehyde is preferably an omega-aldehyde long chain fatty acid having a carbon chain length of at least 10 (e.g., 10-24 carbons) (preferred LCDA are further disclosed herein). Typically, the FALDH enzyme is a peroxisomal protein and/or an endoplasmic reticulum membrane protein in the yeast cell.
The "engineered LCDA production pathway" herein may include, for example:
(i) upregulation of polynucleotide sequences encoding an ACoS enzyme, and
(ii) upregulation of polynucleotide sequences encoding CYP enzymes and/or CPR enzymes (i.e., upregulation of ω -hydroxylase). For example, such pathways can produce LCDA products from substrates containing long chain fatty acids.
As used herein, the term "omega-oxidation" refers to a fatty acid metabolic pathway in which an omega carbon (the carbon furthest from the carboxyl group of a fatty acid) is oxidized to a carboxyl group (see fig. 1). The first step of omega-oxidation is performed by an omega-hydroxylase complex, which catalyzes the addition of a hydroxyl (OH) group to the omega carbon, thereby producing an omega-hydroxy fatty acid. The next step of omega-oxidation involves oxidation of omega-hydroxyl groups to aldehyde (C ═ O) groups by a fatty alcohol oxidase (e.g., EC entry 1.1.3.20) or a fatty alcohol dehydrogenase (e.g., EC entry 1.1.1.66, 1.1.1.192), thereby producing omega-aldehyde fatty acids. The last step of omega-oxidation involves the oxidation of an aldehyde group to a Carboxyl (COOH) group (carboxylic acid group) by a fatty aldehyde dehydrogenase (e.g., EC entry 1.2.1.3, 1.2.1.48), thereby producing a dicarboxylic acid. The product of the omega-oxidation of long chain fatty acids is Long Chain Dicarboxylic Acid (LCDA).
The term "beta-oxidation" herein refers to a process in which fatty acids are catabolized by removing two carbons at a time from the carboxy terminus of the fatty acid. Beta-oxidation typically occurs only in the peroxisomes of the yeast. Peroxisomes are membrane-enclosed, cytoplasmic organelles containing multiple oxidoreductases. Blocking beta-oxidation of a fatty acid herein can be achieved, for example, by disrupting peroxisome development and/or down-regulating expression of one or more beta-oxidation pathway enzymes.
The terms "peroxisomal protein", "peroxisome-associated protein", and the like are used interchangeably herein. Peroxisomal proteins are proteins involved in peroxisome development and/or located in the peroxisome, where they are involved in maintaining peroxisome structure and/or metabolic function (e.g., β -oxidation pathway). Examples of peroxisome proteins herein include Pex protein and Pox protein.
The terms "peroxisome biosynthesis factor", "peroxisome biosynthesis factor protein", "peroxisome production protein (peroxin)", "Pex protein", and the like are used interchangeably herein and refer to proteins involved in peroxisome biosynthesis and/or in the process of inputting cellular proteins into the peroxisome. For example, the abbreviation of a polynucleotide sequence (e.g., a gene or open reading frame) encoding a Pex protein may be referred to as "Pex" or "Pex polynucleotide" or "Pex gene". The system of PEX sequence naming is described by Distel et al (J.cell Biol. [ J.Cell Biol. ] 135: 1-3). At least 32 different PEX sequences have been identified to date in a variety of eukaryotic organisms. Kiel et al (Traffic [ 7: 1291-1303) identified the following fungal Pex proteins: pex1p, Pex2p, Pex3p, Pex3Bp, Pex4p, Pex5p, Pex5Bp, Pex5Cp, Pex5/20p, Pex6p, Pex7p, Pex8p, Pex1Op, Pex12p, Pex13p, Pex14p, Pex15p, Pex16p, Pex17p, Pex14/17p, Pex18p, Pex19p, Pex20p, Pex21p, Pex21Bp, Pex22p, Pex22p and Pex26 p. Hong et al (U.S. patent application publication No. 2009/0117253) disclose that down-regulation of certain PEX sequences in yeast enhances the accumulation of lipids and fatty acids.
The term "PEX 3" as used herein refers to a polynucleotide sequence encoding peroxisome biogenesis factor-3 (PEX3 protein [ "PEX 3 p" ]). The Pex3 protein is a peroxisomal integral membrane protein, and is thought to play a role in peroxisomal membrane formation during peroxisomal biosynthesis (e.g., Baerends et al, J.biol.chem. [ J.Biochem ] 271: 8887-8894; Bascom et al, mol.biol.cell [ molecular biology of cells ] 14: 939-957).
The terms "peroxisomal acyl-coa oxidase", "Pox protein", "Aox protein", and the like are used interchangeably herein and refer to a protein involved in the β -oxidation pathway occurring in the peroxisome. Herein belonging to the EC entry EC: the Pox protein of 1.3.3.6 typically catalyzes the following reaction: fatty acyl coenzyme A + O2→ trans-2, 3-dehydroacyl coenzyme A + H2O2. Abbreviations for polynucleotide sequences (e.g., genes or open reading frames) encoding Pox proteins may be referred to, for example, as "Pox", "Pox polynucleotides", or "Pox genes" (e.g., Pox 4). Examples of Pox proteins are Pox-1, -2, -3, -4, -5 and-6.
The terms "diacylglycerol acyltransferase", "acyl-coa: diacylglycerol acyltransferase "," diacylglycerol O-acyltransferase "," DGAT "," DAGAT ", and the like are used interchangeably herein. DGAT enzymes have EC entry 2.3.1.20 and convert acyl-coa and 1, 2-Diacylglycerol (DAG) to Triacylglycerol (TAG) and coa (thereby participating in the final step of TAG biosynthesis). DGAT1 and DGAT2 are examples of DGATs herein. DGAT1 enzyme with acyl-coa: cholesterol acyltransferases share homology (Lardizabal et al, J.biol.chem. [ J.Biol.J. [ J.Biol ] 276: 38862-38869).
The terms "coumaroyl-coa synthetase", "4-coumarate-coa ligase", and the like are used interchangeably herein. A coumaroyl-coa synthetase having EC entry 6.2.1.12 herein can catalyze the following reaction ("coumaroyl-coa synthetase activity"): ATP + 4-coumaric acid + CoA → AMP + diphosphate + 4-coumaroyl-CoA.
As used herein, the term "long chain" refers to a straight chain of at least 10 carbon atoms, and typically up to 24 carbon atoms. For example, "long chain fatty acids" may have a length of 10 to 24A chain of carbon atoms. The number of carbon atoms in the carbon chain of a long chain fatty acid is determined by its aliphatic Carbon (CH)3-、-CH2-, and ═ CH- (if present)) and the carboxyl group Carbon (COOH).
The terms "long chain dicarboxylic acid" (LCDA), "long chain diacid," "long chain alpha, omega-dicarboxylic acid," "long chain fatty dicarboxylic acid," and the like are used interchangeably herein. LCDA results from the complete omega-oxidation of long chain fatty acids and thus has alpha and omega carboxylic acid groups (i.e., COOH at each end of the carbon chain). For example, the LCDA herein may have a chain length of 10 to 24 carbon atoms. The number of carbon atoms in the carbon chain of LCDA is determined by its aliphatic carbon (-CH)2-, and ═ CH- (if present)) and the carbon of the two carboxyl groups. For example, C18: 0 LCDA (18 carbon chain length, no double bonds) has 16 CH2And 2 carboxyl groups; and C18: 1 LCDA (18 carbon chain length, 1 double bond) has 14 CH 22 CH, and 2 carboxyl groups. The LCDA herein is preferably linear without any organic side chains of aliphatic carbon.
"Long-chain acyl-CoA" or "long-chain fatty acyl-CoA" herein refers to compounds in which the long-chain fatty acid is in a thioester bond with coenzyme A (CoA). Long chain acyl-coa is the product of long chain acyl-coa synthetase activity on long chain fatty acid substrates. Herein, "long chain fatty acid activation" refers to a process of converting a long chain fatty acid into a long chain acyl-coa in a cell via a long chain acyl-coa synthetase activity.
The terms "long-chain fatty acid-containing substrate" (long-chain fatty acid-containing substrate, substrate-containing a long-chain fatty acid), "long-chain fatty acid-containing feedstock" and the like are used interchangeably herein. Any substrate comprising long chain fatty acids obtained from a biological or biologically derived source herein can be characterized as "renewable" or "biorenewable" if desired. For example, a substrate containing long chain fatty acids may comprise "free long chain fatty acids" (e.g., non-esterified or non-amide linked long chain fatty acids) or "linked long chain fatty acids" (e.g., esterified or amide linked long chain fatty acids).
Herein, the COOH group of the free long chain fatty acid does not relate to a bond, such as an ester bond (i.e. the free long chain fatty acid is non-esterified) or an amide bond (i.e. the free long chain fatty acid is not amide-linked).
For example, the linked long chain fatty acid may be an "esterified long chain fatty acid" or an "amide linked long chain fatty acid".
The structure of the long chain fatty acid may be represented by "X: a simple notation system for Y "indicates where X is the total number of carbon (C) atoms in the fatty acid and Y is the number of double bonds, if any. Additional information regarding the differences between "saturated fatty acids" and "unsaturated fatty acids", "monounsaturated fatty acids" and "polyunsaturated fatty acids" (PUFAs), and "omega-6 fatty acids" and "omega-3 fatty acids" is provided, for example, in U.S. patent No. 7238482, which is incorporated herein by reference.
By "glyceride molecule" or "glyceride" is meant herein a mono-, di-and/or triglyceride comprising one, two, or three fatty acids, respectively, esterified with glycerol (alternatively referred to as monoacylglycerol, diacylglycerol, and/or triacylglycerol, respectively). Glyceride molecules are examples of neutral lipids.
Herein, "fatty acid alkyl ester" refers to an ester formed from an ester bond between a carboxyl group of a fatty acid and a hydroxyl group of an alkyl alcohol. For example, the fatty acid alkyl ester herein may be a fatty acid methyl ester, for example, produced by esterifying a fatty acid with methanol. Fatty acid alkyl esters are examples of fatty esters.
As used herein, "ester group" refers to an organic moiety having a carbonyl group (C ═ O) adjacent to an ether linkage. The general formula of the ester group is:
with respect to esterified long chain fatty acids, R in the above formula of the ester comprises a straight chain of aliphatic carbon atoms of the esterified fatty acid. For example, the R' group refers to an alkyl group, an aryl group, or other organic group. Examples of ester groups are found in mono-, di-, and triglycerides comprising one, two, or three fatty acids, respectively, esterified with glycerol. With respect to the above formula, the R' group of the monoglyceride will refer to the glycerol portion of the molecule; the R' group of a di-or triglyceride will refer to the glycerol moiety further linked to one or two other fatty acid esters, respectively.
As used herein, the term "lipid" refers to a molecule that is fat-soluble (i.e., lipophilic). A general overview of lipids is provided in U.S. patent application publication No. 2009/0093543 (see table 2 therein), which is incorporated herein by reference. Examples of lipids that may be used herein as substrates containing long chain fatty acids include glycerides (e.g., mono-, di-, and triacylglycerols); fatty acyl groups (e.g., fatty esters, fatty amides); glycerophospholipids (e.g., phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, phosphatidylinositol, phosphatidic acid); sphingolipids (e.g., ceramides, phospho-sphingolipids (e.g., sphingomyelins), glycosphingolipids (e.g., gangliosides and cerebrosides)); and glycolipids (compounds in which fatty acids are directly attached to the sugar backbone) (e.g., acylamino-sugars, acylamino-polysaccharides, acyltrehalose). The fatty acid-containing substrate can be characterized (if desired) as a fatty acid-containing lipid.
As used herein, the term "oil" refers to a lipid that is liquid at 25 ℃; the oil is hydrophobic and soluble in organic solvents. Oils typically consist primarily of triacylglycerols, but may also contain other neutral lipids, as well as phospholipids and free fatty acids.
As used herein, the terms "fatty acid distillate," "fatty acid distillate of an oil," and the like refer to a composition comprising fatty acids of a particular type of oil. For example, palm fatty acid distillate comprises fatty acids present in palm oil. Fatty acid distillates are typically by-products of vegetable oil refining processes.
The term "cell" herein refers to any type of cell, e.g., prokaryotic or eukaryotic cell. Eukaryotic cells have nuclei and other membrane-enclosed structures (organelles), while prokaryotic cells lack nuclei. "microbial cells" (microorganisms) herein may refer to, for example, fungal cells (e.g., yeast cells), prokaryotic cells, protist cells (e.g., algal cells), euglena cells, stramenopiles cells, or oomycete cells. Prokaryotic cells in this context typically refer to bacterial cells.
The term "yeast" herein refers to a fungal species that exists predominantly in the form of a single cell. Alternatively, yeast may be referred to as "yeast cells". The yeast herein can be characterized, for example, as a conventional yeast or a non-conventional yeast.
The term "conventional yeast" ("model yeast") herein generally refers to the Saccharomyces (Saccharomyces) or Schizosaccharomyces (Schizosaccharomyces) yeast species. In certain embodiments, the conventional yeast is one that prefers Homologous Recombination (HR) DNA repair over repair mediated by non-homologous end joining (NHEJ).
The term "non-conventional yeast" herein refers to any yeast that is not a species of yeast of the genus Saccharomyces or Schizosaccharomyces. Non-conventional yeasts are described inNon-Conventional Yeasts in Genetics,Biochemistry and Biotechnology:Practical Protocols[Non-conventional yeasts in genetics, biochemistry and biotechnology: fruit of Chinese wolfberry Plan for practicing](K.wolf, K.D.Breunig, G.Barth, eds., Springer-Verlag, Berlin, Germany [ Schpringer Press, Berlin, Germany ]]2003) and Spencer et al (appl. microbiol. biotechnol. [ application of microorganisms and biotechnology ]]58: 147-. Some strains of non-conventional yeast may additionally (or alternatively) be yeast that favor NHEJ DNA repair processes over HR-mediated repair processes. Following these principles, the definition of unconventional yeast-preference for NHEJ over HR-is further disclosed by Chen et al (PLoS ONE 8: e57952, which is incorporated herein by reference). Preferred non-conventional yeasts herein are those of the genus yarrowia (e.g., yarrowia lipolytica).
When used to describe the expression of a gene or polynucleotide sequence, the terms "downregulated," "disrupted," "inhibited," "inactivated," and "silenced," and the like are used interchangeably herein to refer to the situation when transcription of a polynucleotide sequence is reduced or eliminated. This results in a reduction or elimination of RNA transcripts from the polynucleotide sequence, which leads to a reduction or elimination of the expression of the protein derived from the polynucleotide sequence (if the gene comprises an ORF). Alternatively, downregulation may refer to a condition in which protein translation from a transcript produced from a polynucleotide sequence is reduced or eliminated. Alternatively, down-regulation may refer to the situation where a protein expressed by a polynucleotide sequence has reduced activity. The reduction of any of the above processes (transcription, translation, protein activity) in a cell can be at least about 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 100% relative to the corresponding process in a suitable control cell. For example, down-regulation may be caused by a targeted event (e.g., indels, knockouts, knockins) or by the use of antisense or RNAi technology.
The terms "targeting", "gene targeting", "DNA targeting", "editing", "gene editing" and "DNA editing" and the like are used interchangeably herein. DNA targeting herein may be the introduction of an indel, knockout or knock-in a particular DNA sequence (e.g. chromosome of a cell). Means of targeting in microbial cells, e.g., Homologous Recombination (HR), are known in the art and can be applied accordingly. Various HR procedures which may be carried out in yeast cells, e.g.as disclosed inDNA Recombination:Methods and Protocols:1st Edition[DNA recombination: methods and protocols (1 st edition)](H.Tsubauchi, eds., Springer-Verlag, New York Schpringer Press, N.Y.)]2011), which is hereby incorporated by reference. The HR process can be used to introduce, for example, indels, knockouts or knockins at DNA target sites.
The terms "knockout," "gene knockout," "genetic knockout," "disruption," and the like are used interchangeably herein. Knock-out means that the DNA sequence of the cells herein has been rendered partially or completely ineffective by DNA targeting; for example, such a DNA sequence may already encode an amino acid sequence prior to the knockout, or may already have a regulatory function (e.g., a promoter). For example, knock-out represents a particular means for providing a deletion of a DNA sequence. Knockouts can be generated, for example, by a mutagenesis process (e.g., resulting in the formation of indels) or by specifically removing sequences (e.g., by HR) and reducing or completely disrupting the function of a DNA sequence (e.g., a polynucleotide encoding a protein and/or its regulatory sequences). A knockout DNA polynucleotide sequence herein may also be characterized as partially or fully disrupted or partially or fully downregulated.
The terms "knock-in", "gene knock-in", "genetic knock-in", and the like are used interchangeably herein. Knock-in refers to the replacement or insertion of a DNA sequence on a specific DNA sequence in a cell by DNA targeting. Examples of knockins include the specific insertion of heterologous amino acid coding sequences into the protein coding region of a polynucleotide sequence and/or its regulatory sequences. For example, such insertions may result in down-regulation of the targeted sequence. For example, a knock-in may be created by specific insertion of a sequence (e.g., by HR).
The term "indel" as used herein refers to the insertion or deletion of one or more nucleotide bases in a target DNA sequence. Such insertions or deletions may be, for example, 1,2, 3, 4, 5, 6, 7, 8, 9, 10 or more bases. In certain embodiments, the indels may be even larger, at least about 20, 30, 40, 50, 60, 70, 80, 90, or 100 bases. If an indel is introduced in the Open Reading Frame (ORF) of a gene, it will usually disrupt the wild-type expression of the protein encoded by the ORF by generating a frame-shift mutation. For example, indels can be generated using a mutagenesis process.
The terms "volume percent" (percent by volume and volume percent), "vol%", "v/v%", and the like are used interchangeably herein. The volume percent of solute in the solution can be determined using the following equation: [ (solute volume)/(solution volume) ] × 100%.
The terms "weight percent" (percent by weight), "" weight percent (wt%) "," "weight-weight percent (% w/w)," and the like are used interchangeably herein. Weight percent refers to the percentage of material on a mass basis when included in a composition, mixture, or solution.
The terms "polynucleotide", "polynucleotide sequence", "nucleic acid sequence" and the like are used interchangeably herein. These terms encompass nucleotide sequences and the like. Polynucleotides may be polymers of DNA or RNA, single-or double-stranded, optionally comprising synthetic, non-natural or altered nucleotide bases. A polynucleotide may be composed of one or more segments of cDNA, genomic DNA, synthetic DNA, or a mixture thereof. Nucleotides (ribonucleotides or deoxyribonucleotides) may be abbreviated by the one-letter name as follows: "A" represents either adenylic or deoxyadenylic acid (for RNA or DNA, respectively), "C" represents cytidylic or deoxycytidylic acid (for RNA or DNA, respectively), "G" represents guanylic or deoxyguanylic acid (for RNA or DNA, respectively), "U" represents uridylic acid (for RNA), "T" represents deoxythymidylic acid (for DNA), "R" represents purine (A or G), "Y" represents pyrimidine (C or T), "K" represents G or T, "H" represents A or C or T, "I" represents inosine, "W" represents A or T, and "N" represents any nucleotide (e.g., N may be A, C, T or G if a DNA sequence is mentioned; N may be A, C, U or G if a RNA sequence is mentioned).
As used herein, the term "gene" refers to a DNA polynucleotide sequence that expresses RNA (which is transcribed from the DNA polynucleotide sequence) from a coding region, which RNA may be messenger RNA (coding for a protein) or non-protein coding RNA. A gene may refer to a separate coding region, or may include regulatory sequences upstream and/or downstream of the coding region (e.g., promoters, 5 '-untranslated regions, 3' -transcription terminator regions). Alternatively, the coding region encoding the protein may be referred to herein as an "open reading frame" (ORF). A "native" or "endogenous" gene refers to a gene found in nature with its own regulatory sequences; such genes are located in natural positions in the host cell genome. A "chimeric" gene refers to any gene that is not a native gene, which includes regulatory and coding sequences that are not found together in nature (i.e., the regulatory and coding regions are heterologous to one another). Thus, a chimeric gene may comprise regulatory sequences and coding sequences that are derived from different sources, or regulatory sequences and coding sequences derived from the same source but arranged in a manner different than that found in nature. A "foreign" or "heterologous" gene refers to a gene that is introduced into a host organism by gene transfer. The foreign/heterologous gene may comprise a native gene inserted into a non-native organism, a native gene introduced into a new location within a native host, or a chimeric gene. In certain embodiments the polynucleotide sequences disclosed herein are heterologous. A "transgene" is a gene that has been introduced into the genome by a gene delivery procedure (e.g., transformation). The codon usage frequency of the "codon optimized" open reading frame is designed to mimic the frequency of preferred codon usage of the host cell.
A "non-native" amino acid sequence or polynucleotide sequence as contained herein in a cell or organism does not occur in the natural (native) counterpart of such a cell or organism.
As used herein, "regulatory sequence" refers to a nucleotide sequence, 5 'untranslated region, intron, and 3' noncoding region that is located upstream from the transcriptional start site of a gene (e.g., a promoter), and that may affect transcription, processing or stability, and/or translation of RNA transcribed from the gene. Herein, regulatory sequences may include promoters, enhancers, silencers, 5' untranslated leader sequences, introns, polyadenylation recognition sequences, RNA processing sites, effector binding sites, stem-loop structures, and other elements involved in regulating gene expression. One or more regulatory elements herein may be heterologous to the coding region herein.
"promoter" as used herein refers to a DNA sequence capable of controlling the transcription of RNA from a gene. Typically, the promoter sequence is located upstream of the transcription start site of the gene. Promoters may be derived in their entirety from a native gene, or be composed of different elements derived from different promoters found in nature, or even comprise synthetic DNA segments. Promoters which in all cases cause expression of genes in cells at most times are commonly referred to as "constitutive promoters". One or more of the promoters herein may be heterologous to the coding regions herein.
As used herein, "inducible promoter" refers to a promoter that is capable of controlling transcription of RNA from a gene under certain specific conditions (i.e., by the presence or absence of biological or non-biological agents). These types of promoters typically have no or very low activity in the absence of inducing conditions.
As used herein, "strong promoter" refers to a promoter that can direct the initiation of a relatively large amount of productivity per unit time, and/or a promoter that drives a higher level of transcription of a gene than the average level of transcription of the gene in a cell.
As used herein, the terms "3' non-coding sequence", "transcription terminator" and "terminator" refer to a DNA sequence located downstream of a coding sequence. This includes polyadenylation recognition sequences and other sequences encoding regulatory signals capable of affecting mRNA processing or gene expression.
The terms "cassette", "expression cassette", "gene cassette" and the like are used interchangeably herein. A cassette may refer to a promoter operably linked to a DNA sequence encoding a protein-coding RNA or a non-protein-coding RNA. The cassette may optionally be operably linked to a 3' non-coding sequence. The structure of the cassette herein may optionally be represented by "X: : y: : z "is represented by a simple notation system. Specifically, X describes the promoter, Y describes the coding sequence, and Z describes the terminator (optional); x is operatively connected to Y, and Y is operatively connected to Z.
As used herein, the term "expression" refers to (i) transcription of RNA (e.g., mRNA or non-protein encoding RNA) from a coding region, and/or (ii) translation of a polypeptide from mRNA. In certain embodiments, the expression of the coding region of the polynucleotide sequence may be up-or down-regulated.
As used herein, the term "operably linked" refers to the association of two or more nucleic acid sequences such that the function of one nucleic acid sequence is affected by the other nucleic acid sequence. For example, a promoter is operably linked with a coding sequence when it is capable of affecting the expression of that coding sequence. That is, the coding sequence is under the transcriptional control of a promoter. For example, a coding sequence may be operably linked to one (e.g., a promoter) or more (e.g., a promoter and a terminator) regulatory sequences.
The term "recombinant" when used herein to characterize a DNA sequence, such as a plasmid, vector, or construct, refers to the artificial combination of two otherwise isolated sequence segments, for example, by chemical synthesis and/or by manipulating the isolated nucleic acid segments using genetic engineering techniques. The methods for preparing recombinant constructs/vectors herein may follow standard recombinant DNA and molecular cloning techniques, such as those described by j.sambrook and d.raroell (r) ((r))Molecular Cloning: a laboratory Manual [ molecular gram And (2) Longding: experimental manual]3 rd edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [ Cold Spring Harbor, N.Y. ]]2001); T.J.Silhavy et al (Experiments with Gene Fusions [ experiments using gene fusion]Cold spring harbor laboratory press: cold spring harbor, new york, 1984); and F.M.Ausubel et al (Short Protocols in Molecular Biology [ concise Protocols for Molecular Biology]5 th edition, Current Protocols [ Current Protocols ]]John Wiley and Sons, Inc., NY [ John Willi father, N.Y. ]]2002).
As used herein, the term "transformation" refers to the transfer of a nucleic acid molecule into a host organism or host cell by any method. The nucleic acid molecule that has been transformed into an organism/cell may be one that is autonomously replicated in the organism/cell, or integrated into the genome of the organism/cell, or transiently exists in the cell without replication or integration. Non-limiting examples of nucleic acid molecules suitable for transformation, such as plasmids and linear DNA molecules, are disclosed herein. A host organism/cell containing a transforming nucleic acid sequence herein may be referred to, for example, "transgenic", "recombinant", "transformed", "engineered", as "transformant", and/or as "modified for exogenous gene expression".
Constructs or vectors comprising the polynucleotides described herein can be introduced into a cell by any standard technique. These techniques include, for example, transformation (e.g., lithium acetate transformation [ Methods in Enzymology [ Methods of Enzymology ], 194: 186-187(1991) ]), gene gun impact, electroporation, and microinjection. As an example, U.S. Pat. Nos. 4880741 and 5071764, and Chen et al (1997, appl. Microbiol. Biotechnol. [ applied microbiology and biotechnology ] 48: 232-235), disclose integration of DNA-based linearized fragments into yarrowia lipolytica.
The terms "control cell" and "suitable control cell" are used interchangeably herein, and can be referenced with respect to a cell (i.e., "experimental cell") that has been specifically modified (e.g., overexpression of a polynucleotide, downregulation of a polynucleotide). The control cell may be any cell that does not have or does not express the particular modification of the test cell. Thus, the control cell may be an untransformed wild-type cell, or may be genetically transformed but not express a particular modification. For example, a control cell may be the immediate parent of a test cell, which does not have a particular modification in the test cell. Alternatively, the control cell may be the parent of the experimental cell removed by one or more generations. Still alternatively, the control cell may be a sibling of the test cell that does not include the particular modification present in the test cell. Control cells can optionally be characterized as cells that are present prior to modification into experimental cells.
As used herein, the term "sequence identity" or "identity" with respect to a polynucleotide or polypeptide sequence refers to the nucleic acid bases or amino acid residues in the two sequences that are identical when aligned for maximum correspondence over a specified comparison window. Thus, "percent sequence identity" or "percent identity" refers to the value determined by comparing two optimally aligned sequences over a comparison window, wherein the portion of the polynucleotide or polypeptide sequence in the comparison window may comprise additions or deletions (i.e., gaps) when the optimal alignment of the two sequences is compared to a reference sequence (which does not comprise additions or deletions). The percentage is calculated by: determining the number of positions at which the identical nucleic acid base or amino acid residue occurs in both sequences to yield the number of matched positions, dividing the number of matched positions by the total number of positions in the window of comparison, and then multiplying the result by 100 to yield the percentage of sequence identity. It will be understood that when calculating sequence identity between a DNA sequence and an RNA sequence, the T residues of the DNA sequence are aligned with the U residues of the RNA sequence and may be considered "identical" thereto. For the purpose of determining the percent complementarity of the first and second polynucleotides, it may be obtained by determining (i) the percent identity between complementary sequences of the first and second polynucleotides (or vice versa), for example and/or (ii) the percentage of bases between the first and second polynucleotides that will produce the canonical Watson and Crick base pairs.
The Basic Local Alignment Search Tool (BLAST) obtained online on the national center for biotechnology information (NCRI) website can be used, for example, to measure percent identity between two or more polynucleotide sequences (BLASTN algorithm) or polypeptide sequences (BLASTP algorithm). Alternatively, percent identity alignments between sequences can be made using a Clustal algorithm (e.g., ClustalW, ClustalV, or Clustal-omega). For multiple alignments using the Clustal alignment method, the default values may correspond to a gap penalty (GAP PENALTY) of 10 and a gap length penalty (GAP LENGTH PENALTY) of 10. Default parameters for calculation of percent identity for alignment-by-alignment pairs and protein sequences using the Clustal method may be KTUPLE-1, gap penalty-3, WINDOW-5, and stored diagonal (DIAGONALS SAVED) -5. For nucleic acids, these parameters may be KTUPLE 2, gap penalty 5, window 4, and stored diagonal 4. Still alternatively, percent identity between sequences may be performed using a BLOSUM matrix (e.g., BLOSUM62), using an EMBOSS algorithm (e.g., needle) with parameters such as GAP start (GAP OPEN) of 10, GAP extension (GAP extension) of 0.5, final GAP penalty (END GAP PENALTY) of error (false), END GAP start (END GAP OPEN) of 10, and final GAP extension (END GAP extension) of 0.5.
Herein, a first sequence that is "complementary" to a second sequence may alternatively be referred to as being in an "antisense" orientation to the second sequence.
As a feature of certain embodiments, various polypeptide amino acid sequences and polynucleotide sequences are disclosed herein. Variants of these sequences having at least about 70% -85%, 85% -90%, or 90% -95% identity to the sequences disclosed herein may be used or referenced. Alternatively, a variant amino acid sequence or polynucleotide sequence may be at least 70%, 71%, 72%, 73%, 74%, 75%, 76%, 77%, 78%, 79%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to a sequence disclosed herein. Variant amino acid sequences or polynucleotide sequences have the same function/activity of the disclosed sequences, or at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% of the function/activity of the disclosed sequences. Typically, any polypeptide amino acid sequence disclosed herein that does not start with a methionine may further comprise at least one initiating methionine at the N-terminus of the amino acid sequence.
All amino acid residues at each amino acid position of the proteins disclosed herein are examples. Given that certain amino acids share similar structural and/or charge characteristics (i.e., conserved) with one another, the amino acid at each position of the proteins herein can be provided in the disclosed sequences or substituted with conserved amino acid residues ("conservative amino acid substitutions") as follows:
1. the following small aliphatic, nonpolar or slightly polar residues may be substituted for one another: ala (A), Ser (S), Thr (T), Pro (P), Gly (G);
2. the following polar, negatively charged residues and their amides may be substituted for each other: asp (D), Asn (N), Glu (E), Gln (Q);
3. the following polar, positively charged residues may be substituted for each other: his (H), Arg (R), Lys (K);
4. the following aliphatic, nonpolar residues may be substituted for one another: ala (A), Leu (L), Ile (I), Val (V), Cys (C), Met (M); and
5. the following large aromatic residues may be substituted for each other: phe (F), Tyr (Y), Trp (W).
As used herein, the term "isolated" refers to a polynucleotide or polypeptide molecule that has been fully or partially purified from its natural source. In some cases, the isolated polynucleotide or polypeptide molecule is part of a larger composition, buffer system, or reagent mixture. For example, an isolated polynucleotide or polypeptide molecule can be contained in a cell or organism in a heterologous manner. Such cells or organisms containing heterologous components and/or one or more genetic deletions will not occur in nature. "isolated" herein may also characterize embodiments that are synthetic/man-made, and/or have non-naturally occurring properties.
The term "increase" as used herein may refer to an amount or activity that is at least about 1%, 2%, 3%, 4%, 5%, 6%, 7%, 8%, 9%, 10%, 11%, 12%, 13%, 14%, 15%, 16%, 17%, 18%, 19%, 20%, 50%, 100%, or 200% more than the amount or activity to which the increased amount or activity is compared. The terms "increased", "enhanced", "greater than", "improved", and the like are used interchangeably herein. For example, these terms may be used to characterize "overexpression" or "upregulation" of a polynucleotide encoding a protein.
New microbial biocatalysts with enhanced LCDA fermentation capacity are desirable. Accordingly, some embodiments disclosed herein relate to a recombinant microbial cell comprising an engineered LCDA production pathway comprising upregulation of a polynucleotide sequence encoding a long-chain acyl-coa synthetase (ACoS enzyme). Notably, such microbial cells can produce one or more Long Chain Dicarboxylic Acid (LCDA) products from a substrate containing long chain fatty acids.
Some embodiments disclosed herein relate to recombinant microbial cells (e.g., yeast cells) including:
(i) (ii) up-regulation of a polynucleotide sequence encoding a cytochrome P450 monooxygenase (CYP enzyme) and/or up-regulation of a polynucleotide sequence encoding a cytochrome P450 reductase (CPR enzyme),
(ii) up-regulation of a polynucleotide sequence encoding a long-chain acyl-CoA synthetase (ACoS enzyme), and
(iii) downregulating an endogenous polynucleotide sequence encoding a peroxisome biogenesis factor-3.
Notably, such microbial cells can produce one or more Long Chain Dicarboxylic Acid (LCDA) products from a substrate containing long chain fatty acids.
Upregulation of an ACoS enzyme in a recombinant cell herein by upregulation of a polynucleotide encoding the enzyme is believed to result in increased levels of long-chain acyl-coa in the cell. Such an increase in this metabolite reflects an increased level of long chain fatty acid activation in the cell.
In certain aspects herein, upregulation of an ACoS enzyme can be performed by upregulating a polynucleotide sequence encoding the ACoS enzyme. Such up-regulation, which results in overexpression of the ACoS enzyme, can be accomplished by one or more of a variety of methods. For example, a polynucleotide encoding an ACoS can be provided to a cell transiently or stably in multiple copies (such polynucleotide sequences are operably linked to a promoter sequence [ e.g., a heterologous promoter ]). Providing a polynucleotide sequence in multiple copies can be accomplished by providing one or more copies (e.g., at least 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, or 50 copies) of the polynucleotide to a cell. It will be appreciated that polynucleotide sequences provided in a stable manner typically have a lower copy number than polynucleotide sequences provided in a transient manner. As another example, a polynucleotide sequence encoding an ACoS may be upregulated by being operably linked to a constitutive promoter, a strong promoter, or an inducible promoter (any of which may be heterologous).
Upregulation (e.g., overexpression) of an ACoS enzyme in a cell herein relative to a suitable control cell can optionally be considered. For example, an increase in the level of an ACoS enzyme in a cell herein can be characterized as at least about 5%, 10%, 20%, 30%, 40%, 50%, 60%, 75%, 80%, 90%, 100%, 150%, 200%, 500%, or 1000% higher than the expression of an ACoS enzyme in a suitable control cell. An example of a suitable control cell is a cell (e.g., a parental cell) that existed before it was modified to have upregulated expression of the ACoS enzyme.
For example, the ACoS enzyme herein may be heterologous to the cell. An example of a heterologous ACoS enzyme may be one that is derived from a species or strain different from that of the cell in which the ACoS enzyme is upregulated.
Alternatively, the ACoS enzyme that is up-regulated in the cell may be native to the cell. For example, a native ACoS enzyme may be upregulated using any of the means disclosed above with respect to upregulation of a polynucleotide sequence. For example, a polynucleotide sequence encoding the enzyme that is native to the cell (operably linked to a promoter sequence [ e.g., a heterologous promoter ]) can be provided to the cell in a stable or transient manner (but the location of the polynucleotide sequence will be at a non-native site [ i.e., a heterologous site ]). As another example, a polynucleotide sequence encoding an ACoS enzyme that naturally occurs in the genome of a cell can be modified such that one or more native polynucleotide sequences are overexpressed. This can be achieved, for example, by modifying one or more regulatory elements (e.g., promoters) of a gene comprising a polynucleotide sequence encoding an ACoS enzyme.
One, two, three, four, or more ACoS enzymes may optionally be upregulated in the cells herein by providing two, three, four, or more sets (copies) of polynucleotide sequences encoding one or more ACoS enzymes. For example, an ACoS enzyme can be provided to a cell by introducing (i) multiple copies of a polynucleotide sequence encoding the same ACoS enzyme, and/or (ii) a polynucleotide sequence encoding a different ACoS enzyme (e.g., overexpressing both a saccharomyces ACoS and a yarrowia ACoS).
The ACoS enzyme herein may be derived from a eukaryote, such as, for example, any eukaryote disclosed as follows: the eukaryote herein may be an animal, a plant, a fungus, or a protist. The animal herein can be, for example, a mammal, bird, amphibian, reptile, fish, or invertebrate (e.g., insect, crustacean, mollusk, nematode). The mammal herein can be, for example, a human or a rodent (e.g., mouse, rat). The plant herein may be, for example, a monocotyledonous plant or a dicotyledonous plant. Examples of monocot plants herein include maize, rice, rye, sorghum, millet, wheat, sugarcane, oat, barley, and switchgrass. Examples of dicotyledonous plants herein include soybean, canola, alfalfa, tobacco, arabidopsis (e.g., arabidopsis thaliana (a. thaliana), arabidopsis thaliana (a. lyrata)), sunflower, cotton, peanut, tomato, potato, and green (e.g., arrow pea). The fungus herein may be, for example, a fungus of the class Basidiomycetes (Basidiomycetes), Zygomycetes (Zygomycetes), Chytridiomycetes (Chytridiomycetes) or Ascomycetes (Ascomycetes). In certain embodiments the fungus may be a yeast or filamentous fungus. Examples of yeasts include any of those species disclosed below (e.g., yarrowia species, such as yarrowia lipolytica; candida species, such as candida tropicalis; saccharomyces species, such as saccharomyces cerevisiae), which, in certain aspects herein, can be used to prepare recombinant yeast cells. Examples of filamentous fungi herein include those species of the genera: acremonium (Acremonium), Aspergillus (Aspergillus), Aureobasidium (Aureobasidium), Chrysosporium (Chrysosporium), Neurospora (Crypthecospiria), Cryptococcus (Cryptococcus), Neurospora (Filibasidium), Fusarium (Fusarium), Gibberella (Gibberella), Humicola (Humicola), Mucor (Mucor), Myceliophthora (Myceliophthora), Neurospora (Neurospora), Penicillium (Penicillium), Verbena (Piromyces), Acremonium (Scytalidium), Schizophyllum (Schizophyllum), Sporotrichum (Sporotrichum), Thielavia (Thielavia), Tolypocladium (Tolypocladium), and Trichoderma (Trichoderma). Examples of protists herein include algal cells (e.g., green algae, brown algae, red algae) and protists of the class Ciliata (cilata), the class flagellates (Mastigophora) subgenus (flagellates), the class Phytomastigophorea (Phytomastigophorea), the class Zoomastiophorea (Zoomastiophorea), the class Rhizopoda (Rhizopoda), the class Fopodophia (Lobosea), and the class Eumyceliophyceae (Eumycetozoea).
In certain embodiments, the ACoS enzyme may be derived from a prokaryote, such as, for example, any prokaryote disclosed as follows: the prokaryote herein may be, for example, a bacterium or archaea. Examples of bacteria include those that are gram negative and gram positive. Still other examples of bacteria include those of the genera: achromobacter (Achromobacter), Aminococcus (Aciaminococcus), Acinetobacter (Acinetobacter), Actinobacillus (Actinobacillus), Actinomyces (Actinomyces), Aerococcus (Aerococcus), Aeromonas (Aeromonas), Acropillum (Africaria), Agrobacterium (Agrobacterium), Alcaligenes (Alcaligenes), Cryptobacterium (Arcanobacter), Toxobacterium (Arcobacterium), Bacillus (Bacillus) (e.g., Bacillus subtilis, Bacillus megaterium), Bacteroides (Bacteroides), Bacteroides (Bartonella), Clostridium (Bordetella), Clostridium (Campylobacter), Bifidobacterium), Lactobacillus (Corynebacterium), Lactobacillus (Lactobacillus), Lactobacillus (Bilobatus), Lactobacillus (bilobacter), Lactobacillus (Brevibacterium), Lactobacillus (Campylobacter), and Bacillus (Campylobacter) bacteria (Corynebacterium), Corynebacterium (Corynebacterium) are used in the genus (Corynebacterium), Lactobacillus (Corynebacterium), Escherichia (Corynebacterium), Escherichia), and Bacillus (Corynebacterium) may be (Corynebacterium) may be used in the strain (Corynebacterium), Bacillus) may be used in the strain (Corynebacterium), Bacillus) may be used in the strain (Corynebacterium), Bacillus) may be used in the strain (Corynebacterium), Bacillus, Corynebacterium (Corynebacterium), Bacillus (Corynebacterium), Bacillus) may be strain (Corynebacterium), Bacillus) may be strain (Corynebacterium), Bacillus) may be used for producing strain (Corynebacterium), Bacillus) may be strain (Corynebacterium), Bacillus, Corynebacterium (Corynebacterium), Bacillus) may be used for example, Corynebacterium) may be used in the strain (Corynebacterium), Bacillus) may be strain (Corynebacterium), Bacillus) may be used for example, Corynebacterium (Corynebacterium) may be strain (Corynebacterium), Bacillus) may be strain (Corynebacterium) may be strain (Corynebacterium), Bacillus, Corynebacterium) may be strain (Corynebacterium), Bacillus) may be strain (Corynebacterium) may be strain (Corynebacterium) may be used for example, Corynebacterium (Corynebacterium) may be strain (Corynebacterium), Bacillus) may be used for example, Corynebacterium (Corynebacterium) may be used for example, Corynebacterium (Corynebacterium) may be used for example, Corynebacterium) may be strain (Corynebacterium) may be used for example, Corynebacterium) may be strain (Corynebacterium) may be used for example, Corynebacterium (Corynebacterium) for example, Corynebacterium (Corynebacterium), Clostridium (Clostridium), Comamonas (Comamonas), Coprococcus (Coprococcus), Cockerthus (Coxiella), Corynebacterium (Corynebacterium), Edwardsiella (Edwardsiella), Erichia (Ehrlichia), Eikeium (Eikeella), Enterobacter (Enterobacter), Enterococcus (Enterococcus), Erysipelothrix (Erysipelothrix), Escherichia (Escherichia) (e.g., Escherichia coli (E.coli)), Eubacterium (Eubacterium), Erwinia (Ewigella), Xanthomonas (Flavivionis), Flavobacterium (Flavobacterium), Francisella (Francisella), Fusobacter (Futerium), Gardnerella (Gardnerella), Geraniella (Klebsiella), Kluyveromyces (Kluyveromyces), Helicobacter (Kluyveromyces), Lactobacillus (Kluyveromyces), and Lactobacillus) Legionella (Legionella), Leptospira (Leptospira), cilium (Leptorickia), Leuconostoc (Leuconostoc), Listeria (Listeria), Macrococcus (Megasphaera), Mycobacterium (Mycobacterium), Micrococcus (Micrococcus), Micropolyspora (Micropolysporas), Flexibacter (Mobilucus), Moraxella (Moraxella), Morganella (Morganella), Mycoplasma (Mycoplasma), Neisseria (Neisseria), Nocardia (Norcardia), Nocardia (Norcarob), Nocardia (Norcaropsiopsis), Oligeella (Oligeella), Pasteurella (Pasteurella), Pediococcus (Pedicoccus), Peptococcus (Peptococcus), Streptococcus (Propionibacterium), Porphyromyces (Proptococcus), Porphyromonas (Porphyra), Porphyra (Porphyra) and/or Porphyra (P) and/or Porphyra (Porphyra) and/or P. sp Rhodococcus (Rhodococcus), Rickettsia (Rickettsia), Roshielima (Rochalimaea), Robinia (Rothia), Ruminococcus (Ruminococcus), Sarcinia, Salmonella (Salmonella), Shewanella (Shewanella), Shigella (Shigella), Serratia (Serratia), Spirobacterium (Spirillum), Staphylococcus (Staphylococcus), Oriococcus (Stomatococcus), Streptococcus (Streptococcus), Streptomyces (Streptomyces), Thermoactinomyces (Thermoactinomyces), Treponema (Treponema), Urenapma (Urepapmapmama), Veillonella (Veillonella), Vibrio (Vibrio), Williaceae (Wonella), Yersinia (Yersinia), or Yersinia (Yersinia).
In some embodiments, the ACoS enzyme can be characterized as being microbial (i.e., derived from a bacterial cell, a protist cell (e.g., an algal cell), a fungal cell (e.g., a yeast cell), an euglena cell, a stramenopile cell, or an oomycete cell).
The amino acid sequence of an ACoS enzyme herein may comprise, for example, any of the amino acid sequences disclosed in the following genbank accession numbers: XP _503862.1, XP _503608.1, XP _502959.1, AJT71734.1, NP _014962.3, AJU13255.1, NP _010931.3, EWG91402.1, EJT42092.1, NP _001153101.1, NP _001273637.1, XP _001146361.1, XP _003829365.1, XP _004033324.1, NP _001125625.1, XP _003266954.1, XP _001363547.2, XP _007422758.1, XP _002880290.1, NP _631034.1, O14975.2, CAH21295.1, CAL20709.1, AEV18827.1, CEM58466.1, CBA20954.1, BAK25224.1, AIU33175.1, CBJ51928.1, CAL 82 93650.1, CAL09544.1, CEE01548.1, GAE33988.1, AAY81441.1, BAH81064.1, CCA89166.1, EAWP _ 89166.1, EAZ 89166.1, EFH 89166.1, EFWP 89166.1, GAFF 89166.1, GAWP 89166.1, GCK 89166.1, EPG 89166.1, EPWP _ 89166.1, EPG _ 89166.1, EPWP _ 89166.1, EPG _ 363672, EPG _ 89166.1, EPG _ 36363672, EPG _ 363672, EPG _ 3636363672, EPG _ 363672, EPG _ 36363672, EPG _ 3636363672, EPG _ 89166.1, EPG _ 363636363636363672, EPG _ 363672, EPG _ 363636363672, EPW _ 36363672, EPW _ 89166.1, EPW _ 363672, EPW _ 89166.1, and EPW _ 89166.1, etc. Variants of any of these ACoS amino acid sequences can be used, but should have some (e.g., at least 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzymatic activity of the corresponding non-variant ACoS enzyme reference sequence (see above definitions). Such variant ACoS enzymes can comprise an amino acid sequence that is at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of a corresponding non-variant ACoS enzyme reference sequence.
In certain aspects herein, an ACoS enzyme may comprise SEQ ID NO: 44 (yarrowia lipolytica ACoS), SEQ ID NO: 49 (yarrowia lipolytica ACoS), SEQ ID NO: 36 (yarrowia lipolytica ACoS), SEQ ID NO: 33 (saccharomyces cerevisiae ACoS), or SEQ ID NO: 34 (saccharomyces cerevisiae ACoS). It is believed that in some other aspects, proteins comprising any one of the amino acid sequences listed in tables 2 and 3 (below) may be useful as ACoS enzymes. Alternatively, for example, an ACoS enzyme herein can comprise an amino acid sequence that is at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to any of the aforementioned ACoS enzyme amino acid sequences. Such variant ACoS enzymes should have some (e.g., at least about 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzymatic activity of the corresponding non-variant ACoS enzyme reference sequence (see definitions above). Methods available in the art (e.g., Galton and Fraser, Analytical Biochemistry 28: 59-64, incorporated herein by reference) or measuring ACoS enzyme activity as disclosed in example 5 below may be applied accordingly herein.
In certain embodiments, an ACoS enzyme herein has both long-chain acyl-coa synthetase activity and coumaroyl-coa synthetase activity. Examples of such ACoS enzymes as presently disclosed comprise a nucleotide sequence identical to SEQ ID NO: 44 or 49 has an amino acid sequence that is at least 90% identical.
The recombinant cells herein can optionally be characterized as comprising an engineered LCDA production pathway comprising at least one upregulated ACoS enzyme. In some aspects, further comprising: (i) up-regulation of a polynucleotide sequence encoding a cytochrome P450 monooxygenase (CYP enzyme), and/or (ii) up-regulation of an LCDA production pathway engineered by a polynucleotide sequence encoding a cytochrome P450 reductase (CPR enzyme). Either or both of these upregulations ([ i ] and/or [ ii ]) are expected to result in omega-hydroxylase upregulation. In some other embodiments, the engineered LCDA production pathway further comprises (optionally in addition to upregulating [ i ] and/or [ ii ]) at least one of: (iii) (iii) up-regulation of a polynucleotide sequence encoding a fatty alcohol oxidase (FAO enzyme), (iv) up-regulation of a polynucleotide sequence encoding a fatty alcohol dehydrogenase (FADH enzyme), and/or (v) up-regulation of a polynucleotide sequence encoding a fatty aldehyde dehydrogenase (FALDH enzyme).
For example, in certain embodiments recombinant cells may have both CYP enzymes and CPR enzymes up-regulated. Alternatively, CYP enzymes may be upregulated, or CPR enzymes may be upregulated. In embodiments where the CYP enzyme is up-regulated, but the CPR enzyme is expressed at wild-type levels, an up-regulated ω -hydroxylase complex may result from CYP enzyme up-regulation. In embodiments where CPR enzyme is up-regulated, but CYP enzyme is expressed at wild-type levels, an up-regulated ω -hydroxylase complex may result from CPR enzyme up-regulation.
In certain aspects herein, upregulation of CYP enzymes and/or CPR enzymes can be by upregulation of polynucleotide sequences encoding CYP enzymes and/or upregulation of polynucleotide sequences encoding CPR enzymes. Such up-regulation, which results in overexpression of CYP enzymes and/or CPR enzymes, can be accomplished by one or more of a variety of methods. For example, a polynucleotide encoding a CYP and/or a polynucleotide encoding a CYP enzyme can be transiently or stably provided to a cell in multiple copies (such polynucleotide sequences are operably linked to a promoter sequence [ e.g., a heterologous promoter ]). Providing a polynucleotide sequence in multiple copies can be accomplished by providing one or more copies (e.g., at least 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, or 50 copies) of the polynucleotide to a cell. It will be appreciated that polynucleotide sequences provided in a stable manner typically have a lower copy number than polynucleotide sequences provided in a transient manner. As another example, the polynucleotide sequence encoding a CYP enzyme and/or the polynucleotide encoding a CPR enzyme may be upregulated by being operably linked to a constitutive promoter, a strong promoter, or an inducible promoter (any of which may be heterologous).
In certain embodiments, both the polynucleotide sequence encoding the CYP enzyme and the polynucleotide sequence encoding the CPR enzyme are up-regulated; this upregulation can be performed, for example, following an overexpression strategy or a combination of overexpression strategies disclosed herein. For example, separate polynucleotides (e.g., vectors, such as plasmids) can be used — one encoding CYP enzymes and the other encoding CPR enzymes. As another example, a single polynucleotide (e.g., a vector, such as a plasmid) can be used, comprising each of the CYP and CPR encoding sequences; for example, each coding sequence may be contained in its own expression cassette (e.g., promoter-coding sequence-terminator) or in a bicistronic expression cassette.
Upregulation (e.g., overexpression) of CYP enzymes and/or CPR enzymes in the cell relative to a suitable control cell can optionally be considered. For example, an increase in the level of CYP enzymes and/or CPR enzymes in a cell herein can be characterized as at least about 5%, 10%, 20%, 30%, 40%, 50%, 60%, 75%, 80%, 90%, 100%, 150%, 200%, 500%, or 1000% higher than the expression of CYP enzymes and/or CPR enzymes in a suitable control cell. Examples of suitable control cells are cells (e.g., parental cells) that exist before they are modified to have up-regulated CYP enzyme and/or CPR enzyme expression.
For example, the CYP enzymes and/or CPR enzymes may be heterologous to the cell. An example of a heterologous CYP enzyme (and/or CPR enzyme) may be one that is derived from a species or strain different from that of the cell in which the CYP enzyme (and/or CPR enzyme) is upregulated. In certain aspects, both the CYP enzyme and CPR enzyme are heterologous to the cell. Heterologous expression of CYP enzymes and/or CPR enzymes in a cell can optionally be characterized as providing a heterologous omega-hydroxylase complex to the cell. The heterologous omega-hydroxylase complex comprises one or both of a heterologous CYP enzyme or CPR enzyme.
Alternatively, CYP enzymes and/or CPR enzymes that are up-regulated in the cell may be native to the cell. For example, the native CYP and/or CPR enzymes can be upregulated using any of the means disclosed above with respect to upregulation of polynucleotide sequences. For example, the respective polynucleotide sequences encoding these enzymes that are native to the cell (operably linked to a promoter sequence) can be provided to the cell in a stable or transient manner (although the location of one or more polynucleotide sequences will be at a non-native site [ i.e., a heterologous site ]). As another example, the respective polynucleotide sequences encoding the CYP enzymes and/or CPR enzymes as naturally occurring in the genome of a cell may be modified such that one or more of the native polynucleotide sequences is overexpressed. This may be achieved, for example, by modifying one or more regulatory elements (e.g., promoters) of one or more genes comprising a polynucleotide sequence encoding a CYP enzyme or a CPR enzyme.
Optionally, two, three, four, or more omega-hydroxylase complexes can be upregulated in the cells herein by providing two, three, four, or more (e.g., copies) sets of polynucleotide sequences encoding CYP and/or CPR enzymes, respectively. For example, a plurality of omega-hydroxylases may be provided to a cell by introducing (i) multiple copies of a polynucleotide sequence encoding a CYP and/or CPR enzyme (e.g., yeast cells transformed with two copies of a sequence encoding CYP/CPR) to overexpress the same omega-hydroxylase, and/or (ii) multiple sets of polynucleotide sequences encoding different omega-hydroxylases (e.g., overexpression of both murine and plant omega-hydroxylases). In some embodiments, the cells herein comprise two, or at least two, up-regulated polynucleotide sequences encoding CYP and CPR (e.g., VsCYP and VsCPR).
In embodiments in which both CYP and CPR enzymes are upregulated in the cells herein, the polynucleotide sequences encoding these enzymes may be derived from the same species/source. Alternatively, the polynucleotide sequences encoding these enzymes may be derived from different species/sources. An example is an embodiment wherein the CYP enzyme is encoded by a mammalian sequence and the CPR enzyme is encoded by a plant sequence. Another example is an embodiment in which one of the enzymes (e.g., CYP) may be heterologous to the cell and the other enzyme (e.g., CPR) is native to the cell. In these latter types of embodiments, in which the polynucleotide sequences encoding the CYP and CPR enzymes are derived from different species/sources, the resulting omega-hydroxylase (containing different sources of CYP and CPR enzyme components) can optionally be characterized as a chimeric omega-hydroxylase complex.
For example, the CYP enzymes and/or CPR enzymes herein can be derived from eukaryotes or prokaryotes. Examples of such eukaryotes and prokaryotes are disclosed above with respect to derivatives of the ACoS enzyme. In some aspects, the CYP enzymes having both CYP and CPR activity useful herein can be derived from prokaryotes. In some embodiments, the CYP enzymes and/or CPR enzymes may be characterized as being microbial (i.e., derived from a bacterial cell, a protist cell (e.g., an algal cell), a fungal cell (e.g., a yeast cell), an euglena cell, a stramenopile cell, or an oomycete cell).
In those embodiments in which the ω -hydroxylase complex has a CYP and CPR enzyme components derived from the same species or strain (e.g., any species/strain disclosed herein, e.g., mouse, rat, human, plant, Arabidopsis (Arabidopsis), nidus, yeast, candida), such ω -hydroxylase complex can optionally be characterized as being from that species or strain. For example, an omega-hydroxylase complex containing mouse CYP and CPR enzyme components can optionally be characterized as a mouse omega-hydroxylase complex. Likewise, certain omega-hydroxylase complexes herein can be characterized as, for example, rat, human, plant, arabidopsis, brassica, or candida omega-hydroxylase complexes, respectively.
In certain embodiments, the CYP enzymes may be from a particular subfamily of CYP enzymes. For example, CYP enzymes may be from the following subfamilies: CYP4 (e.g., mammalian CYP4 (e.g., CYP4a1 and CYP4a10)), CYP86 (e.g., plant CYP86), CYP94 (e.g., plant CYP94 (e.g., CYP94a1)), CYP96 (e.g., plant CYP96 (e.g., CYP96a4)), CYP52 (e.g., yeast CYP52 (e.g., CYP52a4 and CYP52a1)), or CYP102 (e.g., bacterial CYP 102).
The amino acid sequence of the CYP enzyme herein can comprise, for example, any of the CYP amino acid sequences disclosed in the following genbank accession numbers: BAA31435, BAA31437, BAA31439, P16496, P16141, Q12586, EEQ43763, P10615, P30609, P30610, AAO73952, AAO73953, AAO73954, AAO73955, AAO73958, AAO73959, NP _200694, NM _100042, NP _182121, DQ099538, AAD10204, P98188, Q9FMV7, Q9SMP5, Q9ZUX1, NP _200045, XP _002865907, NM _175837, P20816, NP _786936, AAH81771, NP _034141, and Q02928, which are incorporated herein by reference. Variants of any of these CYP amino acid sequences can be used, but should have some (e.g., at least 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzymatic activity of the corresponding non-variant CYP enzyme reference sequence (see above definition). Such variant CYP enzymes can comprise an amino acid sequence having at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity to the amino acid sequence of a corresponding non-variant CYP enzyme reference sequence.
In certain aspects herein, the CYP enzyme can comprise SEQ ID NO: 84 (candida tropicalis CYP) or SEQ ID NO: 94 (arrow pea CYP). Alternatively, for example, a CYP enzyme herein can comprise an amino acid sequence having at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity to any of the foregoing CYP enzyme amino acid sequences. Such variant CYP enzymes should have some (e.g., at least about 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzymatic activity of the corresponding non-variant CYP enzyme reference sequence (see above definition).
The amino acid sequence of CPR enzymes herein can comprise, for example, any of the CPR amino acid sequences disclosed in the following genbank accession numbers: x76226, P37201, X66016, X66017, NM _008898, M12516, and Z26252, which accession numbers are incorporated herein by reference. Variants of any of these CPR amino acid sequences can be used, but should have some (e.g., at least 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzymatic activity of the corresponding non-variant CPR enzyme reference sequence (see definition above). Such variant CPR enzymes can comprise an amino acid sequence that is at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of a corresponding non-variant CPR enzyme reference sequence.
In certain aspects herein, the CPR enzyme can comprise SEQ ID NO: 86 (candida tropicalis CPR) or SEQ ID NO: 96 (arrow pea CPR). Alternatively, for example, a CPR enzyme herein can comprise an amino acid sequence having at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity to any of the aforementioned CPR enzyme amino acid sequences. Such variant CPR enzymes should have some (e.g., at least 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzymatic activity of the corresponding non-variant CPR enzyme reference sequence (see definition above).
In some aspects herein, the recombinant cell can comprise (1) an upregulation of a fatty alcohol oxidase (FAO enzyme), and/or (2) an upregulation of a fatty alcohol dehydrogenase (FADH enzyme), and/or (3) an upregulation of a fatty aldehyde dehydrogenase (FALDH enzyme). Upregulation of FAO and/or FADH provides upregulation of omega-hydroxy fatty acids to omega-aldehyde fatty acids in the long chain fatty acid omega-oxidation pathway (fig. 1 and 2). Upregulation of FALDH provides upregulation of omega-aldehyde fatty acids to LCDA in the long-chain fatty acid omega-oxidation pathway (fig. 1 and 2).
Upregulation of FAO, FADH, and/or FALDH enzymes in recombinant cells herein may be, for example:
(i) at least one FAO enzyme is up-regulated,
(ii) at least one FADH enzyme is up-regulated,
(iii) at least one FALDH enzyme is up-regulated,
(iv) at least one FAO and at least one FADH enzyme are up-regulated,
(v) at least one FAO and at least one FALDH enzyme are up-regulated,
(vi) at least one FADH and at least one FALDH enzyme are up-regulated, or
(vii) At least one FAO, at least one FADH, and at least one FALDH enzyme are up-regulated.
In certain aspects herein, upregulation of FAO, FADH, and/or FALDH enzymes can be by (1) upregulation of a polynucleotide sequence encoding a FAO enzyme, (2) upregulation of a polynucleotide sequence encoding a FADH enzyme, and/or (3) upregulation of a polynucleotide sequence encoding a FALDH enzyme. Such up-regulation, which results in over-expression of FAO, FADH, and/or FALDH enzymes, can be accomplished by one or more of a variety of methods. For example, polynucleotides encoding FAO, FADH, and/or FALDH can be provided to a cell transiently or stably in multiple copies (such polynucleotide sequences operably linked to a promoter sequence [ e.g., a heterologous promoter ]). Providing a polynucleotide sequence in multiple copies can be accomplished by providing one or more copies (e.g., at least 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, or 50 copies) of the polynucleotide to a cell. As another example, polynucleotide sequences encoding FAO, FADH, and/or FALDH can be upregulated by being operably linked to a constitutive promoter or a strong promoter (any of which can be heterologous). The upregulation of any of the FAO, FADH and/or FALDH enzymes listed in (i) - (vii) above may be via upregulation of one or more polynucleotide sequences.
For example, up-regulation of a polynucleotide sequence may be performed following one or a combination of the overexpression strategies disclosed herein. For example, a separate polynucleotide (e.g., a vector, such as a plasmid) encoding FAO, FADH, or FALDH enzyme can be used. As another example, a single polynucleotide (e.g., a vector, such as a plasmid) comprising two or more FAO, FADH, or FALDH coding sequences can be used; for example, each coding sequence may be contained in its own expression cassette (e.g., promoter-coding sequence-terminator) or in a bicistronic expression cassette.
Upregulation (e.g., overexpression) of FAO, FADH, and/or FALDH enzymes in the cells herein relative to suitable control cells can optionally be considered. For example, an increase in the level of FAO, FADH, and/or FALDH enzyme in a cell herein can be characterized as at least about 5%, 10%, 20%, 30%, 40%, 50%, 60%, 75%, 80%, 90%, 100%, 150%, 200%, 500%, or 1000% higher than the expression of FAO, FADH, and/or FALDH enzyme in a suitable control cell. Examples of suitable control cells are cells (e.g., parental cells) that existed prior to their being modified to have up-regulated FAO, FADH, and/or FALDH enzyme expression.
For example, the FAO, FADH, and/or FALDH enzymes may be heterologous to the cell. An example of a heterologous FAO, FADH, or FALDH enzyme can be one that is derived from a species or strain different from that of the cell in which the FAO, FADH, and/or FALDH enzyme is upregulated. In certain aspects, at least one, two, or all of the FAO, FADH, and FALDH enzymes are heterologous to the cell (e.g., any of the upregulations listed in (i) - (vii) above).
Alternatively, the FAO, FADH, and FALDH enzymes that are up-regulated in the cell may be native to the cell. For example, native FAO, FADH, and FALDH enzymes can be upregulated using any of the means disclosed above for upregulation of polynucleotide sequences. For example, the respective polynucleotide sequences encoding these enzymes that are native to the cell (operably linked to a promoter sequence [ e.g., a heterologous promoter ]) can be provided to the cell in a stable or transient manner (but the location of one or more polynucleotide sequences will be at a non-native site [ i.e., a heterologous site ]). As another example, the respective polynucleotide sequences encoding the FAO, FADH, and/or FALDH enzymes naturally occurring in the genome of the cell can be modified such that one or more of the native polynucleotide sequences are overexpressed. This can be accomplished, for example, by modifying one or more regulatory elements (e.g., promoters) of one or more genes comprising polynucleotide sequences encoding FAO, FADH, and/or FALDH enzymes.
One, two, three, four, or more FAO, FADH, and/or FALDH enzymes can optionally be upregulated in the cells herein by providing one, two, three, four, or more (e.g., copies) of the FAO, FADH, and/or FALDH enzyme-encoding polynucleotide sequences, respectively. Multiple FAO, FADH, and/or FALDH enzymes can be provided to a cell, for example, by introducing (i) multiple copies of a polynucleotide sequence encoding a FAO, FADH, and/or FALDH enzyme (e.g., transforming a cell with two copies of a sequence encoding a FAO, FADH, and/or FALDH) to overexpress the same FAO, FADH, and/or FALDH enzyme, and/or (ii) multiple sets of polynucleotide sequences encoding different FAO, FADH, and/or FALDH enzymes (e.g., overexpression of both murine FAO and plant FAO). In some embodiments, the cells herein comprise three, or at least three, different up-regulated FAO-encoding polynucleotide sequences (e.g., CtFAO1M, CcFAO1, and CcFAO 2).
For example, the FAO, FADH, and/or FALDH enzymes herein can be derived from eukaryotes or prokaryotes. Examples of such eukaryotes and prokaryotes are disclosed above with respect to derivatives of the ACoS enzyme. In some embodiments, the FAO, FADH, and/or FALDH enzymes can be characterized as being microbial (i.e., derived from bacterial cells, protist cells (e.g., algal cells), fungal cells (e.g., yeast cells), euglena cells, stramenopile (stramenopile) cells, or oomycete cells).
The FAO, FADH, and/or FALDH enzymes may be from a particular enzyme family or sub-family. For example, the FAO enzyme may be FAO1, FAO2, FAO3, or FAO4 enzyme. For example, the FADH enzyme can be an ADH, ADH1, ADH2, ADH3, FADH1, FADH2, or FADH3 enzyme. For example, the FALDH enzyme may be a FALDH1, a FALDH2, a FALDH3, or a FALDH4 enzyme.
The amino acid sequence of the FAO enzyme herein comprises, for example, any of the amino acid sequences disclosed in the following genbank accession numbers: XP _001389382, XP _002867943, Q9ZWB9, CAA18625, AEE76762.1, AEE84174, AEE85508, XP _007158083, XP _007132926, XP _003540021, XP _003554295, XP _003534338, XP _009102621, EAK93199, CAB75351, CAB75352, XP _002422236, CCG23291, CCG23293, CCE42799, CCE42800, AAS46878, AAS46879, AAS46880, CAB75353, EGV61357, XP _459506, EFX04185, JX879776, XP _001525361, CAP15762.1, KEH23950, EGW33941, and XP _001386087, which are incorporated herein by reference. Variants of any of these FAO amino acid sequences can be used, but should have some (e.g., at least 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzymatic activity of the corresponding non-variant FAO enzyme reference sequence (see above definitions). Such variant FAO enzymes can comprise an amino acid sequence that is at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of a corresponding non-variant FAO enzyme reference sequence.
In certain aspects herein, the FAO enzyme may comprise SEQ ID NO: 100 (candida tropicalis FAO), SEQ ID NO: 102 (candida cloaca FAO), or SEQ ID NO: 104 (candida cloaca FAO). Alternatively, for example, a FAO enzyme herein may comprise an amino acid sequence having at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity to any of the aforementioned FAO enzyme amino acid sequences. Such variant FAO enzymes should have some (e.g., at least about 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzymatic activity of the corresponding non-variant FAO enzyme reference sequence (see definitions above).
The amino acid sequence of the fadh (adh) enzyme herein may comprise, for example, any of the amino acid sequences disclosed in the following genbank accession numbers: NP-982625, EEQ46516, EEQ42383, XM-712556, BAD12482, CD 36-07850, ABD60084, XP-002619012, ADM08005, ADM08008, XP-003870523, AFD29185, XP-006683745, XP-002546635, XP-002550829, GU056282, GU056283, GU056286, GU056287, XP-460537, WP-024173607, AHC53987, AAP51040, XP-001524974, AAP51047, AAP51048, AAP51049, XP-001485610, ESW95881, AFH35136, KGK40277, EJS44121, AAP51043, EHN00693, EJT43588, XP-007377163, AGO10074, CAA 7342, XP-003686595, XP-4629, XP-67884, XP-4143, XP-4642, XP-500087, XP-4643, and 4643. Variants of any of these amino acid sequences can be used, but should have some (e.g., at least 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzymatic activity of the corresponding non-variant fadh (adh) enzyme reference sequence (see definitions above). Such variant fadh (adh) enzymes may comprise an amino acid sequence having at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity to the amino acid sequence of a corresponding non-variant fadh (adh) enzyme reference sequence.
The amino acid sequence of the FALDH enzyme herein may comprise, for example, any of the amino acid sequences disclosed in the following genbank accession numbers: XP _719028, KGQ84508, KGQ98444, XP _002421401, EMG46594, EMG47675, XP _003868193, XP _002550173, XP _002550712, XP _505802, XP _500380, XP _503981, BAP82457, XP _500179, and CCH41136, which are incorporated herein by reference. Variants of any of these FALDH amino acid sequences can be used, but should have some (e.g., at least 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzymatic activity of the corresponding non-variant FALDH enzyme reference sequence (see definition above). Such variant FALDH enzymes may comprise an amino acid sequence that is at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of a corresponding non-variant FALDH enzyme reference sequence.
In certain aspects herein, the FALDH enzyme may comprise SEQ ID NO: 91 (candida tropicalis FALDH), or an amino acid sequence corresponding to SEQ ID NO: 91 has an amino acid sequence that is at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical. Such a variant FALDH enzyme should have the amino acid sequence of SEQ ID NO: 91 (e.g., at least about 30%, 40%, 50%, 60%, 70%, 80%, or 90%) or all of the enzyme activity of the FALDH enzyme (see definition above).
In some embodiments, the recombinant cell may comprise down-regulation of a peroxisome biogenesis factor (Pex protein). For example, the recombinant cell may comprise down-regulation of an endogenous polynucleotide sequence encoding peroxisome biogenesis factor-3 (Pex3 protein). While not intending to be bound by any particular theory or mechanism, it is expected that Pex protein down-regulation results in the level of β -oxidation being blocked or reduced in recombinant cells due to impairment of normal peroxisomal function (e.g., peroxisomal membrane function). It is expected that blocked or reduced levels of beta-oxidation result in the redirection of fatty acids to the omega-oxidation pathway, where the fatty acids serve as substrates for LCDA synthesis (see fig. 1 and 2). In certain embodiments, the expression of one or more of the following Pex proteins may be downregulated: pex1p, Pex2p, Pex3p, Pex3Bp, Pex4p, Pex5p, Pex5Bp, Pex5Cp, Pex5/20p, Pex6p, Pex7p, Pex8p, Pex10p, Pex12p, Pex13p, Pex14p, Pex15p, Pex16p, Pex17p, Pex14/17p, Pex18p, Pex19p, Pex20p, Pex21p, Pex21Bp, Pex22p, Pex22p, and Pex26 p.
Examples of Pex3 proteins that can be down-regulated, for example, by down-regulating polynucleotide sequences encoding such proteins, are disclosed in the following genbank accession numbers: CAG78565 (yarrowia lipolytica, also disclosed herein as SEQ ID NO: 107), NP-010616.3 (Saccharomyces cerevisiae S288), AHY75303.1 (Saccharomyces cerevisiae YJM993), EWH19033.1 (Saccharomyces cerevisiae P283), EWG96624.1 (Saccharomyces cerevisiae R103), EWG87344.1 (Saccharomyces cerevisiae R008), EGA75546.1 (Saccharomyces cerevisiae AWRI796), CAB10141 (Schizosaccharomyces pombe (S. pombe)), EKD00377.1 (Trichosporon atra), 49471 (Hansenula polymorpha)), Candida 569751.1 (Cryptococcus neoformans), XP _003193133.1 (Cryptococcus neoformans), Candida. degermatakii (Cryptococcus neoforma), Candida XP 3 (Candida albicans), Candida albicans (Candida utilis), Candida utilis (Candida utilis), Candida utilis (Candida utilis), Candida utilis (Candida utilis), Candida utilis (Candida utilis), Candida utilis (Candida utilis), Candida utilis (Candida utilis), Candida utilis (Candida utilis), Candida utilis) and Candida utilis (Candida utilis), Candida utilis (Candida utilis), Candida utilis (Candida utilis) and Candida utilis (Candida utilis) and Candida utilis), Candida utilis) and Candida utilis (Candida utilis), Candida utilis (Candida utilis, ABN67699.2 (Schiffersomyces stipitis), AAS52217.1 (Ashbya gossypii), and CCH44061.1(Wickerhamomyces ciferrii), which are hereby incorporated by reference. It will be appreciated that each of these Pex3 proteins will be targeted for down-regulation in the corresponding cell expressing the Pex3 protein (e.g. the s.cerevisiae Pex3 protein will be down-regulated in s.cerevisiae).
In cells of other embodiments, a Pex3 protein, for example, comprising an amino acid sequence at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to any of the foregoing Pex3 proteins can be downregulated. For example, a yarrowia cell expressing a Pex3 protein, or any other type of yeast cell herein, can be modified to have down-regulated expression of a Pex3 protein comprising an amino acid sequence identical to SEQ ID NO: 107, or a variant thereof, having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity.
In some embodiments, for example using yarrowia cells, the down-regulated endogenous polynucleotide sequence may encode a polypeptide comprising an amino acid sequence that is identical to SEQ ID NO: 107 a Pex3 protein having an amino acid sequence of at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity. In certain other embodiments, the downregulated endogenous polynucleotide sequence encoding a Pex3 protein comprises a nucleotide sequence identical to SEQ ID NO: 106 have at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity.
In certain aspects herein, down-regulation of an endogenous polynucleotide sequence encoding a Pex protein (e.g., Pex3) may be due to mutation of the polynucleotide sequence. Such mutations may be, for example, substitutions, deletions, or insertions.
For example, a deletion can remove (i) one or more nucleotides from the open reading frame encoding the Pex protein (i.e., the Pex open reading frame), and/or (ii) one or more nucleotides of the non-protein coding sequence within 500 or 1000 base pairs of the 5' -end of the open reading frame encoding the Pex protein. In certain embodiments, the insertion can occur in (i) an open reading frame encoding a Pex protein, or (ii) a non-protein coding sequence within 500 or 1000 base pairs of the 5' -end of the open reading frame encoding a Pex protein. Other types of mutations can also be used to down-regulate endogenous polynucleotide sequences encoding Pex proteins, if desired. For example, one or more point mutations that exchange a single nucleotide for another (i.e., a nucleotide substitution) can be used accordingly.
Example 6 discloses deletion of the endogenous polynucleotide sequence encoding Pex3 protein in yarrowia lipolytica. In one aspect of this work, the PEX3 open reading frame was removed by homologous recombination-based targeting and replaced with the appropriate donor DNA with the URA3 cassette. This substitution results in a down-regulated (disrupted, or knocked-out) polypeptide comprising SEQ ID NO: 71 comprising the 5 '-and 3' -non-coding PEX3 homology arm sequences (each 100-bp) parts flanking the LoxP-flanking URA3 box. Another aspect of this work involves removing the URA3 cassette by expressing Cre recombinase (stimulated recombination between LoxP sequences, leaving one LoxP sequence) to cause down-regulated (disrupted, or knocked-out) dna comprising SEQ ID NO: 72, in a sequence of seq id no. SEQ ID NO: 72 contains portions of the 5 '-and 3' -non-coding PEX3 homology arm sequences (each 100-bp) flanking one LoxP sequence. Thus, certain embodiments herein relate to a recombinant yarrowia yeast cell comprising a down-regulated endogenous polynucleotide sequence encoding a Pex3 protein, wherein the down-regulation is due to disruption (knock-out) of the endogenous polynucleotide sequence encoding the Pex3 protein; this disruption (knockout) comprises SEQ ID NO: 71 or 72, or a variant of SEQ ID NO: 71 or 72, or a nucleotide sequence having at least about 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity.
Mutations in codons that do not alter the PEX open reading frame of the amino acid encoded by the codon (i.e., silent mutations) are typically not mutations that down-regulate PEX polynucleotides as described herein. Typically, there is also no mutation that changes an amino acid encoded by a codon to a related amino acid that does not change the wild-type function of the Pex protein (e.g., a conservative mutation). In certain embodiments, related amino acids have pendant groups that share structure and/or charge, and may be grouped as follows: aliphatic (glycine, alanine, valine, leucine, isoleucine); aromatic (phenylalanine, tyrosine, tryptophan); hydroxyl-containing groups (serine, threonine); sulfur-containing groups (cysteine, methionine); carboxylic acid group-containing (aspartic acid, glutamic acid); amide-containing groups (asparagine, glutamine) and amino-containing groups (histidine, lysine, arginine). However, any of such mutations (silent or conservative) that down-regulate transcription and/or translation of a PEX polynucleotide (e.g., by inhibiting transactivating transcription and/or translation factors) is typically considered herein as a mutation that down-regulates a PEX polynucleotide.
It will be appreciated by those of ordinary skill in the art that any of the disclosed mutations to the endogenous polynucleotide sequence encoding a Pex protein can be determined to constitute down-regulated mutations by reference to the corresponding endogenous Pex protein coding sequence in a suitable control cell. For example, the PEX polynucleotide sequence in the modified cell can be compared to the endogenous corresponding PEX polynucleotide sequence of the corresponding cell (e.g., parent cell) from which the modified cell was derived.
In certain embodiments, the down-regulation of an endogenous polynucleotide sequence encoding a Pex protein is a reduction in transcription and/or translation of the endogenous polynucleotide sequence by at least about 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 100% relative to the transcription and/or translation of the corresponding polynucleotide sequence encoding the Pex protein in a suitable control cell (e.g., a parental cell). In other embodiments, the down-regulation of an endogenous polynucleotide sequence encoding a Pex protein is reflected by at least about a 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 100% reduction in the function (e.g., protein localization and/or activity) of the encoded Pex protein relative to the function of the corresponding Pex protein in a suitable control cell (e.g., a parent cell).
While not intending to be bound by any particular theory or mechanism, it is contemplated that downregulating a polynucleotide sequence encoding a Pex protein in a recombinant cell herein results in a blocked or reduced level of β -oxidation in the recombinant cell due to impairment of normal peroxisomal function (e.g., peroxisomal membrane function). For example, β -oxidation may be reduced by at least about 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 100% in a cell comprising a down-regulated polynucleotide sequence encoding a Pex protein, as compared to a suitable control cell (e.g., a parent cell that has not undergone down-regulation).
In certain aspects herein, downregulation of a polynucleotide encoding a Pex3 protein (e.g., SEQ ID NO: 107), but not a Pex10 protein (e.g., SEQ ID NO: 108) or a Pex16 protein (e.g., SEQ ID NO: 109) is suitable for use in preparing recombinant yeast cells (e.g., yarrowia lipolytica, reference example 14) that can produce one or more LCDA products from a substrate containing long chain fatty acids. Thus, in some embodiments, the yeast cell does not comprise a down-regulated polynucleotide encoding a Pex10 protein, a polynucleotide encoding a Pex16 protein, and/or a down-regulated polynucleotide encoding a Pex-1, -2, -4, -5, -6, -7, -8, -12, -13, -14, -15, -17, -18, -19, -20, -21, -22, or-26 protein. Examples of Pex10 protein or Pex16 protein herein comprise SEQ ID NO: 108 or SEQ ID NO: 109, or a sequence identical to SEQ ID NO: 108 or SEQ ID NO: 109 has an amino acid sequence that is at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical.
In some embodiments, downregulation of a polynucleotide sequence encoding a Pex3 protein may be the only modification of the polynucleotide sequence encoding a peroxisomal protein necessary for the production of an LCDA product by a recombinant yeast cell. Indeed, example 14 below demonstrates that recombinant yeast having only a downregulated PEX3 polynucleotide, but no downregulation of any other protein directly involved in peroxisomal function (e.g., peroxisomal development and/or maintenance; metabolic pathways, such as β -oxidation occurring in the peroxisome), are capable of producing LCDA from a fatty acid-containing substrate. Thus, certain embodiments disclosed herein relate to recombinant yeast cells in which a downregulated PEX3 polynucleotide is the only modification to a polynucleotide encoding a peroxisomal protein.
In certain aspects, the peroxisomal protein can be one that plays a role in development and/or maintenance of peroxisomal structure/function, such as a Pex protein (e.g., Pex-1, -2, -3, -4, -5, -6, -7, -8, -12, -13, -14, -15, -16, -17, -18, -19, -20, -21, -22, and/or-26 proteins). Another example of a peroxisomal protein herein is one that plays a role in the metabolic activities (e.g. beta-oxidation) carried out in the peroxisome. Examples of peroxisome proteins involved in β -oxidation include Pox proteins (e.g., Pox-1, -2, -3, -4, -5, -6). In some aspects herein, the yeast cell does not have down-regulated expression of Pex protein (other than Pex3), and/or down-regulated expression of Pox protein. In some other aspects, the yeast cell does not have down-regulated expression of: (i) pox-1, -2, -3, -4, -5, and-6 proteins; (ii) pox-1, -2, -3, -4, and-5 proteins; (iii) pox-2, -3, -4, and-5 proteins; (iv) pox-2, -3, and-5 proteins; or (v) Pox-4 and-5 proteins.
Although the Pex3 protein is expected to be the only Pex protein that is downregulated in the recombinant yeast cells herein, one or more additional Pex proteins may optionally be downregulated. Any of the Pex-proteins listed herein, for example, can be down-regulated; specific examples of such other Pex proteins are listed in table 4 of U.S. patent application publication No. 2009/0117253, which is incorporated herein by reference. For example, in addition to downregulating Pex3 protein, Pex10 and/or Pex16 protein may be downregulated.
In some embodiments, a recombinant cell as presently disclosed may include down-regulation of an endogenous polynucleotide sequence encoding a peroxisomal acyl-coa oxidase (Pox protein). For example, one or more of Pox-1, -2, -3, -4, -5, or-6 may be suitable for down-regulation. Any one, two, three, four, five or six, or any combination thereof, of these Pox proteins may be used down-regulated as desired. Examples of combinations of Pox proteins for down-regulation herein include: (i) pox-2, -3, -4; (ii) pox-2, -3, -4, -5; (iii) pox-1, -2, -3, -4, -5; (iv) pox-1, -2, -3, -4, -5, -6; (v) pox-1, -2, -3, -4; and (vi) Pox-2, -3, -4, -5, -6. As a further example, the recombinant cell may include down-regulation of acyl-CoA oxidase-2, -3, and/or-4 enzymes. The down-regulation of one or more Pox proteins herein can be performed using any of the strategies presently disclosed for down-regulating, for example, Pex3 protein expression (e.g., deletions, insertions, other types of mutations). Moreover, the level of such downregulation and the manner of determining downregulation may follow the relevant examples disclosed above with respect to downregulating Pex3 protein expression. In some aspects, the recombinant cell optionally does not comprise down-regulation of a Pox protein.
Any of the foregoing Pox proteins herein can be down-regulated, for example, by down-regulating one or more polynucleotide sequences encoding an endogenous Pox protein. In certain embodiments, the down-regulation of an endogenous polynucleotide sequence encoding a Pox protein is a reduction in transcription and/or translation of the endogenous polynucleotide sequence by at least about 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 100% relative to the transcription and/or translation of the corresponding polynucleotide sequence encoding a Pox protein in a suitable control cell (e.g., a parental cell). In other embodiments, the down-regulation of an endogenous polynucleotide sequence encoding a Pox protein is reflected by a reduction in the function (e.g., protein localization and/or activity) of the encoded Pox protein by at least about 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 100% relative to the function of the corresponding Pox protein in a suitable control cell (e.g., a parent cell).
Examples of Pox4 proteins that can be down-regulated herein (e.g., by down-regulating a polynucleotide sequence encoding such a protein) are disclosed in the following genbank accession numbers: CAG80078 (y. lipolytica), also disclosed herein as SEQ ID NO: 111), P06598 (Candida tropicalis), P05335 (Candida maltosa), KHC52040 (Candida albicans), EIF46613 (Brettanomyces bruxellensis), XP 007376225 (spathaspasalidarum), XP 001526373 (lodoromyces elongata), XP 001387042 (schersomonas stipitis), XP 011276972(Wickerhamomyces ciferrii), and ENH66703 (Fusarium oxysporum), which are incorporated herein by reference. It will be appreciated that each of these Pox4 proteins will be targeted for down-regulation in the corresponding cell expressing the Pox4 protein (e.g., the candida tropicalis Pox4 protein will be down-regulated in candida tropicalis).
In certain embodiments, a Pox4 protein comprising an amino acid sequence at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to any of the aforementioned Pox4 proteins and having Pox4 activity can be down-regulated in a cell. For example, a yarrowia cell or any other type of cell herein that expresses a Pox4 protein can be modified to have down-regulated expression of a Pox4 protein that comprises an amino acid sequence that is identical to SEQ ID NO: 111, or a variant thereof, and 111 has an amino acid sequence that is at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical.
Example 6 discloses deletion of the endogenous polynucleotide sequence encoding the Pox4 protein in yarrowia lipolytica. In one aspect of this work, the POX4 open reading frame was removed by targeting based on homologous recombination. This targeting results in down-regulated (disrupted, or knocked-out) expression of a polypeptide comprising SEQ ID NO: 74 comprising portions of the 5 'and 3' POX4 homology arm sequences. Specifically, SEQ ID NO: 74 with base positions 1-455 and 464-957 corresponding to certain 5 'and 3' POx4 gene sequences, respectively. Thus, certain embodiments herein relate to a recombinant yarrowia yeast cell comprising a down-regulated endogenous polynucleotide sequence encoding a Pox4 protein, wherein the down-regulation is due to disruption (knock-out) of the endogenous polynucleotide sequence encoding a Pox4 protein; this disruption (knockout) comprises SEQ ID NO: 74, or a variant of SEQ ID NO: 74 have a nucleotide sequence of at least about 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity.
Examples of Pox2 proteins that can be down-regulated herein (e.g., by down-regulating a polynucleotide sequence encoding such a protein) are disclosed in the following genbank accession numbers: q00468.1 (Candida maltosa), P11356.3 (Candida tropicalis), O74935.1 (yarrowia lipolytica (Y. lipolytica), also disclosed herein as SEQ ID NO: 79), CCA37459.1 (Saccharomyces pastorianus (Komagataella pastoris)), CAX42707.1 (Candida duchensis), and XP 721613.1 (Candida albicans), which are hereby incorporated by reference. It will be appreciated that each of these Pox2 proteins will be targeted for down-regulation in the corresponding cell expressing the Pox2 protein.
In certain embodiments, a Pox2 protein comprising an amino acid sequence at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to any of the aforementioned Pox2 proteins and having Pox2 activity can be down-regulated in a cell. For example, a yarrowia cell or any other type of cell herein that expresses a Pox2 protein can be modified to have down-regulated expression of a Pox2 protein that comprises an amino acid sequence that is identical to SEQ ID NO: 79, at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical.
Examples of Pox3 proteins that may be down-regulated herein (e.g., by down-regulating a polynucleotide sequence encoding such a protein) include sequences substantially identical to SEQ ID NO: 81 have an amino acid sequence that is at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical.
In certain aspects of the disclosure, the recombinant cell may have reduced lipid (oil) synthesis and/or storage capacity. For example, the lipid synthesis and/or storage capacity can be reduced by at least about 40%, 50%, 60%, 70%, 80%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% (as compared to a suitable control cell (e.g., a parent cell)). Reduced lipid synthesis and/or storage in cells can be determined using any number of means known in the art, such as chromatographic analysis of cellular lipid content (e.g., gas chromatography) and/or certain visual analysis (e.g., microscopic evaluation of liposomes).
Recombinant cells with reduced lipid synthesis and/or storage capacity can have, for example, less than about 50%, 25%, 10%, 5%, 4%, 3%, 2.5%, 2.0%, 1.5%, or 1.0% total lipid as measured as a percentage of Dry Cell Weight (DCW).
In some embodiments, the endogenous activity of converting Diacylglycerols (DAGs) to Triacylglycerols (TAGs) may be reduced to achieve a reduction in lipid synthesis and/or storage capacity. This reflects that TAG usually represents the major lipid storage molecule in the cell. An example of reducing TAG synthesis may be by downregulating at least one endogenous polynucleotide sequence encoding a diacylglycerol acyltransferase (DGAT). Examples of DGAT for down-regulation herein include DGAT1 and DGAT 2. In some aspects herein, either or both of DGAT1 and DGAT2 may be downregulated. Downregulation of DGAT1 and/or DGAT2 can be performed using any of the strategies disclosed herein (e.g., deletions, insertions, other types of mutations) that can be used to downregulate expression of the Pex3 protein. Moreover, the level of such downregulation and the manner of determining downregulation may follow the relevant examples disclosed above with respect to downregulating Pex3 protein expression.
Examples of DGAT1 enzymes that can be down-regulated herein are SEQ ID NOs: 113, which represents yarrowia lipolytica DGATl enzyme. Expression can be compared to SEQ ID NO: 113 a yarrowia cell of a DGAT1 enzyme having an amino acid sequence of at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity, or any other cell herein is modified to have down-regulated expression of such a DGAT1 enzyme. As another example, expression of a polypeptide having SEQ ID NO: 113 or any other cell herein modified to have down-regulated expression of such DGAT1 enzyme.
Examples of DGAT2 enzymes that can be down-regulated herein are SEQ ID NOs: 115, which represents the yarrowia lipolytica DGAT2 enzyme. Expression can be compared to SEQ ID NO: 115 or any other cell herein of DGAT2 having an amino acid sequence of at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity, is modified to have down-regulated expression of such DGAT2 enzyme. As another example, expression of a polypeptide having SEQ ID NO: yarrowia cells of an enzyme that is at least 80%, 90%, 95%, or 100% active of DGAT2 of 115, or any other cell herein, are modified to have down-regulated expression of such DGAT2 enzyme.
The DGAT enzymes herein may be down-regulated, for example, by down-regulating one or more polynucleotide sequences encoding endogenous DGAT. In certain embodiments, downregulation of an endogenous polynucleotide sequence encoding DGAT is at least about a 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 100% reduction in transcription and/or translation of the endogenous polynucleotide sequence relative to the transcription and/or translation of a corresponding polynucleotide sequence encoding DGAT in a suitable control cell (e.g., a parental cell). In other embodiments, the down-regulation of an endogenous polynucleotide sequence encoding DGAT is reflected by a reduction in the function (e.g., protein localization and/or activity) of the encoded DGAT by at least about 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 100% relative to the function of the corresponding DGAT in a suitable control cell (e.g., a parental cell).
If desired, other types of acyltransferases may be down-regulated in the recombinant cells herein to effect a reduction in lipid synthesis and/or storage capacity. Such downregulation may be independent of downregulation of DGAT1 and/or DGAT2 enzymes, or may be in addition to downregulation of DGAT1 and/or DGAT2 enzymes. Other acyltransferases that may optionally be targeted for down-regulation include lecithin-cholesterol acyltransferase (EC 2.3.1.43; also known as phosphatidylcholine-sterol O-acyltransferase) and phospholipids: diacylglycerol acyltransferase (PDAT, EC 2.3.1.158), both of which can generally catalyze the conversion of phospholipids and DAG to lysophospholipids and TAG.
Recombinant microbial cells herein can refer to, for example, fungal cells (e.g., yeast cells), prokaryotic cells, protist cells (e.g., algal cells), euglena cells, stramenopiles cells, or oomycete cells. Prokaryotic cells in this context may refer to, for example, bacterial cells or archaeal cells. The yeast cell can be any yeast as presently disclosed. For example, the yeast can be a yarrowia (e.g., yarrowia lipolytica), candida (e.g., candida tropicalis), debaryomyces (e.g., debaryomyces hansenii), saccharomyces (e.g., saccharomyces cerevisiae), schizosaccharomyces (e.g., schizosaccharomyces pombe (s.pombe)), or Pichia (e.g., Pichia pastoris)) yeast species.
The fungal cell herein may be a yeast (e.g., infra) or any other fungal type such as a filamentous fungus. For example, the fungus herein may be a Basidiomycetes (Basidiomycetes), Zygomycetes (zygomyycetes), chytriomycetes (chytriomycetes) or Ascomycetes (Ascomycetes). Examples of filamentous fungi herein include those of the genera: trichoderma (Trichoderma) (e.g., Trichoderma reesei)); chrysosporium (Chrysosporium); thielavia (Thielavia); neurospora (Neurospora) (e.g., Neurospora crassa (n.crassa), Neurospora eutropha (n.sitophila)); genus erythrocuprina (cryptonectria) (e.g., hypocrea castanea (c. parasitica)); aureobasidium (Aureobasidium) (e.g., Aureobasidium pullulans)); ustilago (Filibasidium); ruminal chytrium (Piromyces); cryptococcus (cryptococcus); acremonium (Acremonium); torticollis (Toypocladium); acremonium (Scytalidium); schizophyllum (Schizophyllum); sporotrichum (Sporotrichum); penicillium (Penicillium) (e.g., p.bilaiae, Penicillium salmonellae (p.camemberti), Penicillium albugineum (p.candidum), Penicillium chrysogenum (p.chrysogenum), Penicillium expansum (p.expansum), Penicillium funiculosum (p.funiculosum), Penicillium glaucum (p.glaucum), Penicillium marneffei (p.marneffei), Penicillium roqueforti (p.roqueforti), Penicillium verrucosum (p.verrucosum), fresh delicacium (p.glaucum), and mixtures thereofPenicillium viridans (p.viridicatum)); gibberella (e.g., g.acuminata, Gibberella avenae (g.avenaceae), g.baccata, g.circinata, g.cyanogena, Gibberella gambieri (g.fujikuroi), Gibberella zeae (g.internricans), Gibberella louse (g.pulicaris), g.stibroides, g.tricmcta, Gibberella zeae (g.zeae)); myceliophthora (mycestophthora); mucor (Mucor) (e.g., Mucor rucosa (m.rouxii), Mucor circinelloides (m.circinelloides), Aspergillus (e.g., Aspergillus niger (a.niger), Aspergillus oryzae (a.oryzae), Aspergillus nidulans (a.nidulans), Aspergillus flavus (a.flavus), Aspergillus lentus, Aspergillus terreus (a.terreus), Aspergillus clavus (a.clavatus), Aspergillus fumigatus (a.fumigatus)), Fusarium (Fusarium) (e.g., Fusarium graminearum, Fusarium oxysporum (f.oxydans), f.bubigensum, Fusarium solani (f.solanum), Fusarium oxysporum (f.oxysporum), f.oxydpora, Fusarium verticillium (f.verticillum), Fusarium laminaria solani), Fusarium (f.oxydiproducens), Fusarium solani (f.verticillium), and Fusarium solani (humuloides), and Fusarium solanum when necessaryIllustrated Genera of Imperfect Fungi, 3rd Edition,Burgess Publishing Company,1972[Illustrative genus of incomplete fungi (3 rd edition)Bogis publishing Co., 1972)]The genera and species of fungi herein are defined by morphology.
In certain aspects, the yeast herein can be an apomictic (anamorphic) or a sexual (sexual) yeast. Although the yeasts herein are typically present in single cell form, certain types of these yeasts may optionally be capable of forming pseudohyphae (strings of connected germ cells). In still further aspects, the yeast can be haploid or diploid, and/or can have the ability to exist in any of these ploidy forms.
Examples of yeast herein include conventional yeast and non-conventional yeast. For example, conventional yeast herein includes species of the genera: saccharomyces (Saccharomyces) (e.g., Saccharomyces cerevisiae, also known as Saccharomyces buddleyanus, Saccharomyces bakeri, and/or Saccharomyces cerevisiae; Saccharomyces bayanus; Saccharomyces boulardii; Saccharomyces cariocaiensis; Saccharomyces carhiocus; Saccharomyces chevalieri; Saccharomyces dawlensis; Saccharomyces cerevisiae; Schizosaccharomyces pombe; and Schizosaccharomyces pombe; also known as Schizosaccharomyces pombe).
The non-conventional yeast herein is not a conventional yeast, such as a saccharomyces (e.g., saccharomyces cerevisiae) or schizosaccharomyces (e.g., schizosaccharomyces pombe) species. The non-conventional yeast herein may be cultivated according to any means known in the art, such as described in the following references:Non-Conventional Yeasts in Genetics, Biochemistry and Biotechnology:Practical Protocols(K.Wolf,K.D.Breunig,G.Barth,Eds.,Springer-Verlag,Berlin,Germany,2003)[in genetics, biochemistry and biotechnology The non-conventional yeast of (1): practice plan(K.wolf, K.D.Breunig, G.Barth, eds., Berlin Schpringer, Germany, 2003)],Yeasts in Natural and Artificial Habitats(J.F.T.Spencer,D.M.Spencer,Eds.,Springer-Verlag,Berlin,Germany,1997)[Yeast in natural and artificial habitats(J.F.T.Spencer, D.M.Spencer, eds., Berlin Schpringer Press, Germany, 1997)]And/orYeast Biotechnology:Diversity and Applications(T.Satyanarayana,G.Kunze,Eds.,Springer,2009)[Yeast biotechnology: diversity and applications(T.Satyanarayana, G.Kunze, eds., Sppringe Press, 2009)]Which is hereby incorporated by reference in its entirety.
Non-limiting examples of non-conventional yeasts herein include yeasts of the genera: yarrowia (Yarrowia), Pichia (Pichia), Schwanniomyces (Schwanniomyces), Kluyveromyces (Kluyveromyces), arxua, trichomonas (trichosporin), Candida (Candida), Ustilago (Ustilago), Torulopsis (Torulopsis), Zygosaccharomyces (Zygosaccharomyces), Trigonopsis (Trigonopsis), Cryptococcus (Cryptococcus), Rhodotorula (Rhodotorula), favus (Phaffia), Sporobolomyces (spobolomyces), pachysolepsis (Pachysolen), and moniella (monilila). A suitable example of a Yarrowia species is Yarrowia lipolytica (y. Suitable examples of Pichia species (Pichia) include Pichia pastoris (p.pastoris) (also known as pasteurella foenum germania (Komagataella pastoris)), Pichia methanolica (p.methanolica), Pichia stipitis (p.stipitis), Pichia anomala (p.anomala), and Pichia angusta (p.angusta) (also known as Hansenula polymorpha). Suitable examples of Schwanniomyces species include s.castellii, s.aluvius, s.hominis, s.occidentalis, s.capriottii, s.etchellsii, Schwanniomyces polymorpha (s.polymorphus), Schwanniomyces pseudomorpha (s.pseudomorpholophorus), s.vanrijiae and s.yamadae. Suitable examples of Kluyveromyces species include Kluyveromyces lactis (k.lactis), Kluyveromyces marxianus (k.marxianus), Kluyveromyces fragilis (k.fragilis), k.drosophilum, Kluyveromyces thermotolerans (k.thermolerans), k.phaseolosporirus, k.vanudeni, k.waltii, Kluyveromyces africanus (k.africanus), and Kluyveromyces porosus (k.polysporaus). Suitable examples of Arxula species include a.adeninivorans and a.terrestre. Suitable examples of trichosporium (trichosporin) species include Trichosporon cutaneum (t. cutaneum), Trichosporon capitis (t. capitatum), Trichosporon chinese (t. incin) and t. beemeri. Suitable examples of Candida species (Candida) include Candida albicans (c.albicans), Candida acalaberianum, c.amphiaceae, Candida antarctica (c.antarctica), Candida beehives (c.apiicola), c.argentata, Candida atlantic (c.atlantic), Candida atmospherica (c.atmospherica), c.blakearia, c.bremeliacarum, c.carpophylaefm, c.carvajalis, c.cerambycidinium, c.chaulides, c.corydali, c.dosseyi, Candida duchenii (c.dubliniensis), c.ergeniensis, c.ergeniasicosis, Candida fruit (c.frutus), Candida glabrata (c.glacialis), Candida yeast (c.milfoil), Candida albicans (c.yeast), Candida rugosa, Candida rugosa, Candida rugosa, Candida rugosa, Candida rugosa, Candida rugosa, Candida rugosa, Candida rugosa, Candida Oleophila, c.oregon, candida parapsilosis (c.parapsilosis), candida citrinophyllata (c.quercitrirusa), candida rugosa (c.rugosa), candida sake (c.sake), candida sheu (c.lake), candida shehatae (c.sheharea), c.temnochiase, candida tenuis (c.tenuis), c.teae, c.tolerans, candida tropicalis (c.tropicalis), c.tsuchiyae, c.siloborantium, c.sojae, c.subhashii, candida virswanathii, candida utilis (c.utlubis), c.ubustensens and c.zelinina. Suitable examples of smut (Ustilago) species include Farinia avenae (U.avenae), Utilia sativa (U.esculenta), Utilia hordei (U.hordei), Utilia zeae (U.maydis), Utilia nuda (U.nuda) and Utilia tritici (U.tritici). Suitable examples of Torulopsis (Torulopsis) species include T.geochars, T.azyma, Torulopsis glabrata (T.glabrata) and Torulopsis albicans (T.Candida). Suitable examples of Zygosaccharomyces species include Zygosaccharomyces bayer (z. bailii), Zygosaccharomyces bisporus (z. bisporus), z.cidri, Zymobacter fermentati (z.fermentati), Z.florentinus, Z.kombuchaensis, Z.lentus, Z.mellis, Z.microbellissoids, Z.mrakii, Z.pseudorouzoxii and Zygosaccharomyces rouxii (z.rouxii). Suitable examples of Trigonopsis (Trigonopsis) species include variant Trigonopsis (t. Suitable examples of species of Cryptococcus (Cryptococcus) include Cryptococcus laurentii (c.laurentiii), Cryptococcus albidus (c.albicans), Cryptococcus neoformans (c.neoformans), Cryptococcus gattii (c.gattii), Cryptococcus monopharynensis (c.unlguttulatus), c.adensis, c.aereus, c.albicosimilis, c.antarcticaus, c.aquaticus, Cryptococcus nigricans (c.ater), c.bhutanensis, c.consortia, Cryptococcus curvatus (c.curvatus), c.phenocolicus, c.skinneri, Cryptococcus neoformans (c.terreus), and c.vishenniaci. Suitable examples of Rhodotorula species (Rhodotorula) include Rhodotorula gracilis (r. acheniorum), r.tula, r.acuta, Rhodotorula mays (r. americana), r.araucae, Rhodotorula arctica (r.arctica), r.armeniaca, Rhodotorula orangii (r.aurantiaca), r.auricularia, high yielding yeast strain (r.bacarum), Rhodotorula benthica (r.benthica), r.bioourcegi, r.goerinsis, Rhodotorula bronchiale (r.bronchus), r.bufelonii, r.caulyphylla, r.caulynensis, r.chunga, r.laurenciphenesis, r.sylvestris, Rhodotorula gracilis, r.laurencystis, r.gracilaria, r.sylla, Rhodotorula gracilis, r.balsamifera, r.rhodotorula, r.graciliaris, r.balsamifera, r.balsamia, r.gracilaria, r.balsamifera, r.sp A mineral, r.longissima, r.ludwigii, r.lisinophila, rhodotorula rubra (r.marina), r.martyniae-fragrantis, r.matritensiensis, r.melissa, rhodotorula gracilis (r.miniata), rhodotorula mucilaginosa (r.mucolanosa), r.nitens, r.nothofia, rhodotorula oryzae (r.oryza), rhodotorula pacifica (r.pacifica), rhodotorula rubra (r.pallida), r.peneaus, r.philiphylula, r.phylloplana, r.pilatii, rhodotorula rubra (r.pilula, rhodotorula rubra, rhodotorula rubrum, r.laurula, rhodotorula, r.rhodotorula, r.macrorrhula, r.yurea, r.rhodotorula, r.yuliu R.yunnanensis and r.zsolti. Suitable examples of Phaffia (Phaffia) species include Phaffia rhodozyma (P.rhodozyma). Suitable examples of saccharomyces sp (sporulosidia) species include s.alborubers, s.bannaensis, beijing saccharomyces sp (s.beiijingensis), s.bischofiae, s.clavatus, s.coproprosma, s.dipreniae, s.dracophylli, s.elongatus, s.gracilis, s.inostiphilus, s.johnsonii, s.koalae, s.magnisporus, s.novoalnidius, s.odorosporussp.sp.synucleus, s.pataginus, s.long-living saccharomyces sp.sp.roseus, r.roseosporussp.roseosporusus, s.biosporangiensis, s.microsporusususussp.syzygus, s.yunacanthusus sp.sp.sp.. Suitable examples of the ascomyces (Pachysolen) and aureobasidium (Moniliella) species include Pachysolen tannophilus (p.tannophilus) and aureobasidium pullulans (m.polisis), respectively. Other examples of non-conventional yeasts herein also include species of the genus Pseudozyma (e.g., s. antarctica), thodotoria (e.g., t. bogoriensis), Wickerhamiella (e.g., w. domercqi), Starmerella (e.g., s. bombicola), Debaryomyces (e.g., d. hansenii), Ogataea (e.g., o. angasta), and Ashbya (e.g., Ashbya gossypii (a. gossypii)).
In certain embodiments, the yeast is a yarrowia yeast (e.g., yarrowia lipolytica). Examples of suitable yarrowia lipolytica include the following isolates available from the American Type Culture Collection (ATCC, manassas, va): strain names ATCC #20362, #8862, #8661, #8662, #9773, #15586, #16617, #16618, #18942, #18943, #18944, #18945, #20114, #20177, #20182, #20225, #20226, #20228, #20327, #20255, # 20292, #20297, #20315, #20320, #2038, #20336, #20341, # 20320320320346, #20348, #20363, # 20764, #20372, #20373, #20383, #20390, #20400, #20460, #20461, #20462, #20496, #20510, # 20620620620620688, #2079, #20775, #20776, #2039, # 3263469, # 639, # 32469, #2079, # 639, #2079, # 639, #2079, #9, # 639, #9, # 5935, #9, # 593259469, #9, # 5935, #9, # 5935, #9, # 5935, #9, # 5935, #9, # 5935, #9, #, #46436, #60594, #62385, #64042, #74234, #76598, #76861, #76862, #76982, #90716, #90811, #90812, #90813, #90814, #90903, #90904, #90905, #96028, #201241, #201242, #201243, #201244, #201245, #201246, #201247, #201249, and/or # 201847.
In certain embodiments, the microbial cells are algal cells. For example, the algal cells may be from any of the following: chlorophyta (chlorella), Rhodophyta (Rhodophyta), Phaeophyceae (Phaeophyceae) (brown algae), Bacillariophyceae (diatom) and dinoflagellates (dinoflagellates). In other aspects, the algal cell can be a microalgae (e.g., phytoplankton, microsphyte, or planktonic algae) or a macroalgae (kelp, seaweed). As further examples, the algal cells herein may be of the following genera of species: chlamydomonas (e.g., Chlamydomonas reinhardtii); porphyra (Porphyra); the third carpal plate (Palmaria) (e.g., Palmaria (p. palmata) [ Palmaria palmata;)]) (ii) a Arthrospira (Arthrospira) (e.g., Arthrospira platensis (A. platensis) [ Spirulina (Spirulina)]) (ii) a Chlorella (Chlorella) (e.g., c. protothecoides, c. vulgaris)); chondrus (Chondrus) (e.g., Chondrus crispus (C. crispus) [ Ireland moss)]) (ii) a The genus Aphanizomenon (Aphanizomenon); sargassum (Sargassum); hai RongGenus (Cochayuyo); botryococcus (Botryococcus) (e.g. Botryococcus braunii (b.braunii)); dunaliella (Dunaliella) (e.g., Dunaliella salina (d.tertiolecta), Dunaliella salina (d.salina)); gracilaria (Gracilaria); pleurochrysis (e.g., p. carterae); fibrothrix (Ankistrodesmus); the genus Cyclotella (Cyclotella); genus Trapa (Hantzschia); micrococcus (Nannochloris); nannochloropsis (Nannochloropsis); genus Nitzschia (Nitzschia); phaeodactylum (Phaeodactylum) (e.g., Phaeodactylum tricornutum)); scenedesmus (e.g., Scenedesmus obliquus (s.obliquus)); schizophyllum (schizococcus); tetraselmis (Tetraselmis) (e.g., t. subeica); the genus Thalassiosira (Thalassiosira) (e.g., pseudothalassiosira (t. pseudonana)); crypthecodinium (cryptothecodinium) (e.g., Crypthecodinium cohnii); neochlorella (Neochloris) (e.g., oil-rich neochlorella (n. oleoakundans)); or schiiochytrium. May be as described, for example, in Thompson (R) ((R))Algal Cell Culture.Encvclopedia of Life Support System(EOLSS),Biotechnology Vol 1[Algae cell culture, life support system encyclopedia (EOLSS), Biotechnology, Vol.1]Net/sample-chapters internet website) (incorporated herein by reference) to culture and/or manipulate algal species.
In certain embodiments, the bacterial cell may be in the form of a coccus, bacillus, spirochete, spheroplast, protoplast, or the like. Still other non-limiting examples of bacteria include those in the genera: salmonella (Salmonella) (e.g., Salmonella typhi (s.typhi), Salmonella enteritidis (s.enteritidis)), Shigella (Shigella) (e.g., Shigella dysenteriae (s.dysenteriae)), escherichia (Fscherichia) (e.g., escherichia coli (e.coli)), Enterobacter (Enterobacter), Serratia (Serratia), Proteus (Proteus), Citrobacter (Citrobacter), Edwardsiella (edwards), Providencia (Providencia), Klebsiella (Klebsiella), Klebsiella (klisella), Hafnia (Hafnia), ehrlington (Ewingella), kluyveromyces (Kluyvera), Morganella (Morganella), coccus (paracoccus), Staphylococcus (Staphylococcus aureus (Staphylococcus) Haemophilus (Actinobacillus), Pasteurella (Pasteurella), Ureaplasma (Ureaplasma), Streptococcus (Streptococcus) (e.g. Streptococcus pyogenes), Streptococcus (s.mutans), Streptococcus pneumoniae (s.pneumoeae)), Enterococcus (Enterococcus) (e.g. Enterococcus faecalis (e.g. Enterococcus), Aerococcus (Aerococcus), facultative (gemellae), Lactococcus (e.g. Lactococcus lactis), leucococcus (leucococcus lactis), Corynebacterium (Leuconostoc) (e.g. leucococcus coli), Streptococcus (e.g. Leuconostoc) or Streptococcus sp (e.g. leucococcus lactis), Streptococcus (Leuconostoc) e.g. leucococcus (e.g. leucococcus), Bacillus (Corynebacterium) e.g. leucococcus (r. leucococcus), Bacillus (e.g. leucococcus), Bacillus (Corynebacterium) such as Bacillus cereus, Bacillus sp Actinomycetes (Actinomyces), Rhodococcus (Rhodococcus), Listeria (Listeria) (e.g. Listeria monocytogenes (l.monocytogenes)), erysiphe (Erysipelothrix), Gardnerella (Gardnerella), Campylobacter (Campylobacter), toxoplasma (arobacter), williamella (woliniella), Achromobacter (Achromobacter), Acinetobacter (Acinetobacter), Agrobacterium (agrobacter) (e.g. Agrobacterium tumefaciens (a.tumefaciens)), Alcaligenes (Alcaligenes), rhodomonas (chrysosporium), oligochaeta (amonas), ekalangium (Eikenella), Xanthomonas (flavomonas), Flavobacterium (Flavobacterium), Pseudomonas (Moraxella), Pseudomonas aeruginosa (Pseudomonas), Pseudomonas aeruginosa (Pseudomonas aeruginosa) The genus aphenanthe (Afipia), the genus Bartonella (Bartonella), the genus capsular (calomymatobacterium), the genus cardiobacter (Cardiobacterium), the genus streptomyces (streptococcus), the genus helicobacter (Spirillum), the genus Peptostreptococcus (Peptostreptococcus), the genus Peptococcus (Peptococcus), the genus Sarcinia (Coprococcus), the genus Ruminococcus (Ruminococcus), the genus Propionibacterium (Propionibacterium), the genus campylobacter (mobilus), the genus Bifidobacterium (bifidum), the genus Eubacterium (Eubacterium), the genus Lactobacillus (Lactobacillus) (e.g. Lactobacillus lactis), the genus acidophilus (l.acidophilus), the genus rhodinella (Rothia), the genus Clostridium (Clostridium) (e.g. Clostridium (Clostridium), the genus Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), Clostridium (Clostridium), megasphaera (Megasphaera), Vellonella (Veilonella), Nocardia (Norcardia), Actinomycera (Madura), Nocardiopsis (Norcadiopsis), Streptomyces (Streptomyces), Micropolyspora (Micropolyspora), Thermoactinomyces (Thermoactinomyces), Treponema (Treponema), Microspira (Leptospira), and Chlamydia (Chlamydia).
Recombinant cells herein can produce one or more LCDA products from a substrate containing long chain fatty acids. For example, the total amount of LCDA that can be produced by a cell in a volume of culture medium as presently disclosed can be about or at least about 10g/L, 20g/L, 30g/L, 40g/L, 50g/L, 60g/L, 70g/L, 80g/L, 90g/L, 100g/L, 110g/L, or 120g/L (or any integer between 5g/L and 120 g/L). Examples of recombinant cells of the disclosure can exhibit at least a 10-fold to 1000-fold increase in LCDA production when grown under the same fermentation conditions, as compared to a suitable control cell (e.g., a parental cell). For example, such an increase may be about or at least about 10-fold, 25-fold, 50-fold, 75-fold, 100-fold, 150-fold, 200-fold, 250-fold, 300-fold, 400-fold, 500-fold, 750-fold, or 1000-fold.
The degree of homogeneity or heterogeneity of the LCDA produced by the cells herein typically depends on the nature of the substrate containing long chain fatty acids supplied to the cells. For example, cells grown with a substrate comprising one type of long chain fatty acid (a substrate containing homogenous fatty acids) can typically produce an LCDA product that contains predominantly (e.g., at least 50 wt%, 55 wt%, 60 wt%, 65 wt%, 70 wt%, or 75 wt%) LCDA that has the same carbon chain length as the fatty acid in the substrate. For example, in some aspects, cells grown in media with substrates containing only palmitic acid (C16: 0) or oleic acid (C18: 1) can typically produce LCDA containing at least 50 wt% of LCDA products with carbon chain lengths of 16 or 18, respectively.
In some aspects, cells grown with substrates comprising more than one type of long chain fatty acid (substrates containing heterogeneous fatty acids) can typically produce LCDA product profiles having carbon chain lengths that are generally proportional to the corresponding carbon chain lengths of the fatty acids in the substrate. For example, cells grown with soybean oil, which typically comprises about 7% alpha-linolenic acid (C18: 3), about 55% linoleic acid (C18: 2), about 23% oleic acid (C18: 1), about 4% stearic acid (C18: 0), and about 11% palmitic acid (C16: 0) fatty acids (thus, about 89% of the fatty acids are C18, and about 11% are C16), herein can produce LCDA products having predominantly (e.g., at least 50 wt%, 55 wt%, 60 wt%, 65 wt%, 70 wt%, or 75 wt%) carbon chain lengths of 18.
For example, the LCDA herein may have a carbon chain length of 10 to 24. For example, the LCDA may be C10, C11, C12, C13, C14, C15, C16, C17, C18, C19, C20, C21, C22, C23, or C24 LCDA. In some embodiments, the LCDA can have a chain length of 10-22, 12-22, 14-22, 16-22, 18-22, 20-22, 16-18, 16-20, or 16-22 carbon atoms. In certain aspects, examples of LCDA products are saturated (their carbon chains do not contain any double bonds) and are listed in table a.
TABLE A
Examples of LCDA products
Still other examples of the LCDA products herein are unsaturated. The unsaturated LCDA may comprise, for example, an aliphatic carbon chain having 1,2, 3, 4, 5, or 6 double bonds. Examples of unsaturated LCDA herein include C16: 1. c16: 2. c18: 1. c18: 2. c18: 3. c18: 4. c20: 1. c20: 2. c20: 3. c20: 4. c20: 5. c22: 1. c22: 2. c22: 3. c22: 4. c22: 5 and C22: 6. any of the foregoing LCDAs can be produced, for example, by growing the presently disclosed recombinant cells with a substrate comprising fatty acids having the corresponding chain length and saturation/unsaturation profile. The position or positions of unsaturation in the carbon chain of the LCDA product may correspond, for example, to the position or positions of unsaturation in a fatty acid containing substrate used to prepare the LCDA.
For example, a long chain fatty acid as provided herein in a substrate containing a long chain fatty acid may have a carbon chain length of at least 10, or a length of 10 to 24 carbon atoms. The long chain fatty acid may be, for example, a C10, C11, C12, C13, C14, C15, C16, C17, C18, C19, C20, C21, C22, C23, or C24 long chain fatty acid. In some embodiments, the long chain fatty acid may have a chain length of 10-24, 12-24, 14-24, 16-24, 18-24, 20-24, 10-22, 12-22, 14-22, 16-22, 18-22, 20-22, 16-18, 16-20, or 16-22 carbon atoms. Although the presently disclosed substrates comprise fatty acids having a carbon chain length of at least 10 or in the range of 10 to 24 carbon atoms, other types of fatty acids may also be present in the substrate if desired. For example, the substrate may further comprise one or more types of fatty acids having a carbon chain length of less than 10.
The long chain fatty acids herein may be saturated or unsaturated. Examples of unsaturated long chain fatty acids are monounsaturated fatty acids (MUFA) if only one double bond is present in the fatty acid carbon chain, and polyunsaturated fatty acids (PUFA) if the fatty acid carbon chain has two or more double bonds. Examples of long chain fatty acids herein are provided in table B.
TABLE B
Examples of long chain fatty acids that may be included in the substrate
In some cases, the long chain fatty acid may be a substituted fatty acid, as long as the fatty acid is non-toxic or exhibits only low toxicity to the cell. For example, one OR more hydrogens in the aliphatic chain of a fatty acid may optionally be replaced by halogen, acetyl, OR, NR2Or SR groups, wherein R is independently H or a C1-C8 alkyl group. Certain examples of substituted fatty acids herein include fatty acids having an omega-alcohol or an omega-aldehyde group.
In some aspects herein, a substrate containing long chain fatty acids as presently disclosed may comprise free long chain fatty acids. Such fatty acids may optionally be characterized as non-esterified long chain fatty acids or non-linked long chain fatty acids. For example, any of the long chain fatty acids disclosed herein (e.g., as listed in table B) may be included in such a substrate. Other examples of substrates containing free long chain fatty acids include fatty acid distillates of oils. The fatty acid distillate may be any oil disclosed herein, such as a vegetable oil (e.g., palm oil fatty acid distillate [ PFAD ]).
In some aspects, a substrate containing long chain fatty acids as presently disclosed may comprise esterified long chain fatty acids. For example, any of the long chain fatty acids disclosed herein (e.g., as listed in table B) may be included in such a substrate. Some examples of long chain fatty acids esterified herein include long chain fatty acids included within a glyceride molecule or fatty acid alkyl ester.
The glyceride molecules herein may be mono-, di-, or triglycerides, or mixtures thereof. In those embodiments in which the substrate containing long chain fatty acids comprises di-and/or triglycerides, not all of the esterified fatty acids thereof need be long chain fatty acids. In some embodiments, the glyceride molecules herein are typically provided as an oil, although the molecules may also be provided as a fat. Thus, a substrate containing long chain fatty acids may optionally be characterized as containing one or more types of oils and/or fats.
Examples of oils (or fats) suitable for use herein may be derived from plants, microorganisms, yeasts, fungi, bacteria, algae, euglena, stramenopiles, animals, poultry, and fish. Examples of vegetable oils (vegetable oils) include canola oil, corn oil, palm kernel oil, cheru seed oil, wild almond oil, sesame oil, sorghum oil, soybean oil, rapeseed oil, tall oil, sunflower oil, hemp seed oil, olive oil, linseed oil, coconut oil, castor oil, peanut oil, palm oil, mustard oil, cottonseed oil, linseed oil, jatropha oil, and crambe oil. Other examples of oils and fats herein include refined fats and oils; restaurant grease; yellow and brown oils; waste industrial frying oil; tallow; lard oil; whale oil; fat in milk; fish oil; algae oil; a yeast oil; microbial oil; oils/fats from yeast biomass, microbial biomass, sludge; and phospholipids (e.g., as provided in the soap stock). Still other examples of oils that may be used herein include (i) fossil fuel-derived oils (e.g., oils from petroleum-based products), used and industrial oils and lubricants, coal-derived liquids; (ii) synthetic oils produced as by-products from petrochemical and chemical processes; and (iii) oil from industrial and/or agricultural waste.
For example, the fatty acid alkyl ester herein may include C1-C10Alkyl radicals are, for example, the methyl, ethyl, propyl, butyl, pentyl, hexyl, heptyl, octyl, nonyl or decyl radicals, respectively. Examples include fatty acid methyl esters and fatty acid ethyl esters. However, any of the long chain fatty acids disclosed herein may be included in the fatty acid alkyl esters, some examples include C16 (e.g., palmitic acid) andc18 (e.g., oleic acid) fatty acids. One or a mixture of fatty acid alkyl esters may be used with the cells herein for LCDA production. In some aspects, the fatty acid esters can be produced by chemically reacting any of the oils or fats (i.e., lipids) disclosed herein with an alcohol (e.g., methanol or ethanol) to provide a mixture of fatty acid alkyl esters using any suitable method known in the art. An example of such a mixture is biodiesel, which is typically derived from vegetable oils or animal fats and oils (e.g., tallow).
In some aspects, a substrate comprising a long chain fatty acid as presently disclosed may comprise an amide-linked long chain fatty acid. Examples of amide-linked long chain fatty acids herein include fatty amides, acylamino-sugars and acylamino-glycans. For example, any of the long chain fatty acids disclosed herein (e.g., as listed in table B) can be provided as amide-linked long chain fatty acids.
It is believed that the cells herein, although described as producing LCDA from substrates containing long chain fatty acids, are also capable of producing LCDA from other organic substrates (e.g., alkanes, fatty alcohols, and/or fatty aldehydes). Such other substrates may have the same carbon chain length as disclosed herein for substrates containing long chain fatty acids.
The disclosure also relates to methods of producing one or more Long Chain Dicarboxylic Acids (LCDA). This method includes contacting a recombinant cell (e.g., a microbial cell, such as a yeast cell) as disclosed herein with a substrate comprising a long chain fatty acid, wherein the cell synthesizes LCDA from the substrate. The method further includes the optional step of recovering the LCDA synthesized by the cells.
For example, the method can be practiced using any of the one or more features of the embodiments disclosed above or the examples below (e.g., cell type-related features; ACoS enzyme sequences; CYP and/or CPR enzyme sequences; FAO, FADH, and/or FALDH enzyme sequences; Pex3 protein sequences, etc.). Thus, any feature or any combination of features disclosed above or in the examples may be suitably used to characterize embodiments of the LCDA production methods herein. The following method features are additional examples.
The LCDA production method as presently disclosed includes the step of contacting a recombinant cell with a substrate comprising a long chain fatty acid, wherein the cell synthesizes LCDA from the substrate. Such contacting step can optionally be characterized as incubating, culturing, and/or growing the recombinant cell in a medium comprising a substrate comprising a fatty acid. This contacting step can also be characterized as a fermentation step (e.g., fermenting LCDA from a substrate containing long chain fatty acids) (e.g., an LCDA fermentation process), if desired.
For example, a suitable pH for fermenting the LCDA herein (e.g., the pH of the medium that contacts the cells with the substrate containing long chain fatty acids) is between about pH 4.0 and 9.0. A suitable pH within this range may be, for example, about 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, or 9.0. In some other aspects, a pH in the range of about pH 7.5 to 8.5 may be used. A pH of about 5.5 to 7.5 may sometimes be used for the initial growth conditions.
A suitable temperature for fermenting the LCDA herein (e.g., the temperature of the medium that contacts the cells with the substrate containing long chain fatty acids) can be a temperature at which the recombinant cells herein exhibit optimal growth. Examples of suitable temperatures include about 15 ℃, 16 ℃, 17 ℃, 18 ℃, 19 ℃, 20 ℃, 21 ℃,22 ℃, 23 ℃, 24 ℃, 25 ℃, 26 ℃, 27 ℃, 28 ℃, 29 ℃, 30 ℃, 31 ℃, 32 ℃,33 ℃,34 ℃, or 35 ℃. In some cases, suitable temperature ranges that may be used include 25 ℃ to 32 ℃, 28 ℃ to 32 ℃, and 28 ℃ to 30 ℃.
The amount of time for which recombinant cells are grown with one or more substrates containing long chain fatty acids for fermenting the one or more LCDAs can be about or at least about 36, 48, 60, 72, 84, 96, 108, 120, 132, 144, 156, 168, 180, 192, 204, 216, 228, or 240 hours. In certain other embodiments, the fermentation period may be about 3-7, 4-6, or 5 days. The cells may optionally be grown for about 12-24 hours prior to initial contact with one or more substrates containing long chain fatty acids.
In the medium in which the recombinant microbial cells herein are contacted with one or more such substrates, the concentration of the one or more substrates comprising long chain fatty acids can be, for example, about or at least about 1g/L, 3g/L, 5g/L, 10g/L, 15g/L, 20g/L, 25g/L, 30g/L, 35g/L, 40g/L, 45g/L, 50g/L, 55g/L, 60g/L, 65g/L, 70g/L, 75g/L, 80g/L, 85g/L, 90g/L, 95g/L, or 100g/L (or any integer between 1g/L and 100 g/L). In certain other embodiments, such a concentration may be from about 3g/L to 30g/L or from 5g/L to 20 g/L. Any of these concentrations may be an initial concentration (starting concentration) that is the concentration of substrate present immediately after addition to the medium in which the LCDA is fermented with the microbial cells. For example, the initial substrate concentration containing long chain fatty acids may optionally be characterized as the concentration at the beginning of a pulse feed or a continuous feed.
In some embodiments, the LCDA fermentation process can be performed using a batch, fed-batch, or continuous fermentation process. Batch fermentation processes typically include a closed system in which the medium (including the substrate containing long chain fatty acids) is fixed at the start of the process and no further additions/modifications are made other than those that may be required to maintain pH and/or oxygen levels during fermentation. The fed-batch process herein is similar to a batch process except that the process is subjected to one or more additions/modifications other than those that may be required to maintain pH and/or oxygen levels during fermentation. For example, a substrate containing a long chain fatty acid may be added to the system during the process; such additions may be staggered/periodic or continuous. Batch and fed-batch culture methods are known in the art (e.g., Brock,Biotechnology:A textbook of Industrial Microbiology [ Biotechnology: textbook for industrial microbiology]2 nd edition(Sinauer Association: Sandland, Mass, 1989; Deshpande, appl.Biochem.Biotechnol. [ application of biochemistry and Biotechnology ]]36: 227-234). The continuous fermentation process herein can generally be carried out by continuously adding defined medium to the fermentation vessel while removing an equal amount of culture volume for LCDA product recovery. Brock discloses a continuous fermentation process.
Other culture conditions may also optionally be applied to perform the LCDA production methods herein. For example, recombinant cells can be cultured under aerobic (e.g., micro-aerobic) or anaerobic conditions, with the former being preferred in some cases. Agitation in the form of shaking or rotation may optionally be applied to the culture, for example at a rate of about 100rpm, 150rpm, 200rpm, 300rpm, 500rpm, 800rpm, 1000rpm, 1200rpm, 1500rpm, 1800rpm, or 2000 rpm. In another example, a two-stage process may be used, where the first stage promotes cell proliferation and the second stage promotes LCDA production. In yet other examples, two, three, four or more different types of recombinant cells (preferably of the same species, genus or family) as presently disclosed may be used.
For example, the total amount of one or more LCDA produced in the LCDA production process as presently disclosed can be about or at least about 10g/L, 20g/L, 30g/L, 40g/L, 50g/L, 60g/L, 70g/L, 80g/L, 90g/L, 100g/L, 110g/L, or 120g/L (or any integer between 5g/L and 120 g/L). These concentrations can be as measured in the medium that the microbial cells herein are contacted with the substrate containing long chain fatty acids and at any of the growth stages disclosed above. The rate of LCDA production in certain LCDA production methods herein can be about or at least about 0.10 g/L/hr, 0.15 g/L/hr, 0.20 g/L/hr, 0.25 g/L/hr, 0.30 g/L/hr, 0.35 g/L/hr, 0.40 g/L/hr, 0.45 g/L/hr, 0.50 g/L/hr, 0.55 g/L/hr, 0.60 g/L/hr, 0.65 g/L/hr, 0.70 g/L/hr, 0.75 g/L/hr, 0.80 g/L/hr, 0.85 g/L/hr, 0.90 g/L/hr, 0.95 g/L/hr, 1.00 g/L/hr, 1.05 g/L/hr, or a combination thereof, 1.10 g/L/hr, 1.15 g/L/hr, or 1.20 g/L/hr. In certain aspects, the starting amount of microbial cells that results in these measurements of any LCDA output can be any of those tested in the examples below.
One or more LCDA products synthesized by the cells in the LCDA production methods herein can optionally be isolated. Any method known in the art for separating LCDA from a fermentation broth may be used, such as, for example, the methods disclosed in U.S. patent application publication nos. 2014/0228587 and 2012/0253069, which are incorporated herein by reference. Also, for example, any of the LCDA separation methods disclosed in the examples below may be used.
During the LCDA synthesis process herein, one or more omega-hydroxy long chain fatty acids and/or omega-aldehyde long chain fatty acids are produced as intermediates (see fig. 1 and 2). Thus, in certain alternative embodiments of the present disclosure, the method of synthesizing LCDA may optionally be characterized as a method of producing omega-hydroxy long chain fatty acids and/or omega-aldehyde long chain fatty acids. For example, such one or more LCDA metabolites may have a carbon number corresponding to any of the presently disclosed LCDA and long chain fatty acids.
Non-limiting examples of the compositions and methods disclosed herein include:
1. a recombinant microbial cell comprising an engineered LCDA production pathway comprising upregulation of a polynucleotide sequence encoding a long-chain acyl-coa synthetase (ACoS enzyme), wherein the microbial cell can produce one or more long-chain dicarboxylic acids (LCDA) from a substrate comprising a long-chain fatty acid.
2. The recombinant microbial cell of embodiment 1, wherein the ACoS enzyme comprises a sequence identical to SEQ ID NO: 44. 49, 36, 33, or 34, having at least 90% identity.
3. The recombinant microbial cell of example 1 or 2, wherein the ACoS enzyme has both long-chain acyl-coa synthetase activity and coumaroyl-coa synthetase activity.
4. The recombinant microbial cell of embodiment 3, wherein the ACoS enzyme comprises a nucleotide sequence identical to SEQ ID NO: 44 or 49 has an amino acid sequence that is at least 90% identical.
5. The recombinant microbial cell of example 1,2, 3, or 4, wherein the engineered LCDA production pathway further comprises one or more of the following features:
(i) upregulation of a polynucleotide sequence encoding a cytochrome P450 monooxygenase (CYP enzyme),
(ii) upregulation of polynucleotide sequences encoding cytochrome P450 reductase (CPR enzyme),
(iii) upregulation of a polynucleotide sequence encoding a fatty alcohol oxidase (FAO enzyme),
(iv) upregulation of a polynucleotide sequence encoding a fatty alcohol dehydrogenase (FADH enzyme), and/or
(v) Upregulation of a polynucleotide sequence encoding a fatty aldehyde dehydrogenase (FALDH enzyme).
6. The recombinant microbial cell of embodiment 5, wherein one or both of the polynucleotide sequence encoding the CYP enzyme and the polynucleotide sequence encoding the CPR enzyme are up-regulated.
7. The recombinant microbial cell of embodiment 1,2, 3, 4, 5, or 6, wherein the microbial cell further comprises down-regulation of an endogenous polynucleotide sequence encoding a peroxisome biosynthetic factor.
8. The recombinant microbial cell of embodiment 7, wherein the peroxisome biogenesis factor is peroxisome biogenesis factor-3.
9. The recombinant microbial cell of embodiment 1,2, 3, 4, 5, 6, 7, or 8, wherein the microbial cell further comprises down-regulation of an endogenous polynucleotide sequence encoding a peroxisomal acyl-coa oxidase.
10. The recombinant microbial cell of embodiment 9, wherein the peroxisomal acyl-coa oxidase is a peroxisomal acyl-coa oxidase-2, -3, and/or-4.
11. The recombinant microbial cell of embodiment 1,2, 3, 4, 5, 6, 7, 8, 9, or 10, wherein the microbial cell has reduced lipid synthesis and/or storage capacity.
12. The recombinant microbial cell of embodiment 11, wherein the decreased lipid synthesis and storage capacity is due to down-regulation of at least one endogenous polynucleotide sequence encoding a diacylglycerol acyltransferase (DGAT enzyme).
13. The recombinant microbial cell of embodiment 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 11, or 12, wherein the microbial cell is a yeast cell.
14. The recombinant microbial cell of embodiment 13, wherein the yeast cell is a yarrowia cell.
15. The recombinant microbial cell of example 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, or 14, wherein the LCDA product has a chain length of 10 to 24 carbon atoms and/or the substrate comprising long chain fatty acids comprises free long chain fatty acids or esterified long chain fatty acids.
16. A process for producing a Long Chain Dicarboxylic Acid (LCDA), the process comprising: a) contacting the recombinant microbial cell of example 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, or 15 with a substrate comprising a long chain fatty acid, wherein the microbial cell synthesizes LCDA from the substrate; and b) optionally recovering the LCDA of step (a).
17. The method of embodiment 16, wherein the microbial cell is a yeast cell, and optionally wherein the yeast cell is a yarrowia cell.
Examples of the invention
The present disclosure is further illustrated in the following examples. It should be understood that these examples, while indicating certain preferred aspects of the disclosure, are given by way of illustration only. From the above discussion and these examples, one skilled in the art can ascertain the essential characteristics of the disclosed embodiments, and without departing from the spirit and scope thereof, can make various changes and modifications to adapt the disclosed embodiments to various uses and conditions.
General procedure
Standard recombinant DNA and molecular cloning techniques used in the examples are well known in the art and are exemplified by: 1) sambrook and dMolecular Cloning:A Laboratory Manual [ molecular cloning: laboratory Handbook]3 rd edition, cold spring harbor laboratory press: cold spring harbor, new york, 2001); 2) T.J.Silhavy et al (Experiments with Gene Fusions [ Experiments using Gene Fusions ]]Cold spring harbor laboratory press: cold spring harbor, new york, 1984); and 3) F.M.Ausubel et al (Short Protocols in Molecular Biology [ conciseness Molecular biological assay protocol]5 th edition, Current Protocols [ Current Protocols ]Scheme testing method]John Wiley and Sons, Inc., NY [ John Willi father, N.Y. ]]2002) are described.
Materials and methods suitable for the maintenance and growth of microbial cultures are well known in the art. As in, for exampleManual of Methods for General Bacteriology [ Manual of Methods for General Bacteriology](P.Gerhardt, R.G.E.Murray, R.N.Costilow, E.W.Nester, W.A.Wood, N.R.Krieg, and G.B.Phillips, eds., American society for microbiology: Washington, 1994); and/or Thomas D.Brock, inBiotechnology:A Textbook of Industrial Microbiology [ biotechnology: textbook for industrial microbiology]2 nd edition(Sinauer Association: Sandland, Mass. (Sunderland, MA), 1989) may find suitable techniques for use in the following examples. Unless otherwise indicated, all reagents, restriction enzymes, and cell growth materials were obtained from either DIFCO laboratories (detroit, michigan), New England biology laboratories, Inc. (New England Biolabs, Inc.) (Bevery, Mass.), GIBCO/BRL (Gathersburg, Md.), or Sigma-Aldrich (Sigma-Aldrich) (St. Louis, Mo.). Coli strains were grown typically on Luria Bertani (LB) plates at 37 ℃.
General molecular cloning was performed according to standard methods (e.g., Sambrook and Russell). Oligonucleotides were synthesized by Sigma-Ginnos (Sigma-Genosys) (Spprine, Texas (Spring, TX)). Unless otherwise stated, separate PCR amplification reactions were performed in a total volume of 50- μ l, including: PCR buffer (containing 10mM KCl, 10mM (NH)4)2SO4、20mM Tris-HCl(pH 8.75)、2mM MgSO40.1% Triton X-100), 100. mu.g/mL BSA, 200. mu.M of each deoxyribonucleoside triphosphate, 10pmole of each primer, and 1. mu.l of Pfu DNA polymerase (Agilent Technologies, Santa Clara, Calif.). Site-directed mutagenesis was performed using the Agilent technologies kit according to the manufacturer's instructions. When PCR or site-directed mutagenesis involved subcloning, the constructs were sequenced to confirm that no mismatches were introduced into the sequences. Will be provided withCloning of the PCR product into-T simple vectors (Promega, Madison, Wis.) and/or4-In vectors (Invitrogen, carlsbad, ca). All codon optimized genes were synthesized by the firm kingsry (GenScript) (picscatavir, new jersey).
DNA sequences were generated on an ABI automated sequencer using dye terminator technology using a combination of vector-and insert-specific primers. Sequence editing and analysis was performed using sequence software (Gene coding Corporation (Gene Codes Corporation), ann arba, michigan). Comparison of gene sequences was achieved using DNASTAR software (DNA Star, Inc.). Alternatively, Vector NTI obtained from Life technologies, Inc. (Life technologies, Greenland, N.Y.) was usedThe program 10 enables the manipulation of gene sequences.
Results summarizing the alignment comparison of sequences with the greatest similarity to the query sequence are reported in terms of percent identity, percent similarity, and/or expected (E) values. The "expectation" estimates the statistical significance of the matches, specifying the number of matches, with a given score, which would be expected in an absolutely occasional search of a database of this size.
The meanings of certain abbreviations used herein are as follows: "sec" means seconds, "min" means minutes, "h" means hours, "d" means days, "μ L" means microliters, "mL" means milliliters, "L" means liters, "μ M" means micromoles, "mM" means millimoles, "M" means moles, "mmol" means millimoles, "μmole" micromoles, "g" means grams, "μ g" means micrograms, "ng" means nanograms, "U" means units, "bp" means base pairs, "kb" means kilobases, "DCW" means dry cell weight, and "TFAs" means total fatty acids.
Culture and transformation of yarrowia lipolytica
Yarrowia lipolytica strains ATCC # 20362 and # ATCC 90812 were purchased from the American type culture Collection (Rokville, Md.). Yarrowia lipolytica strains were routinely grown in several media at 28 ℃ -30 ℃ according to the formulations shown below. Agar plates were prepared by adding 20g/L agar to each liquid medium.
YPD agar Medium (per liter): 10g yeast extract (DIFCO), 20g BactoTMPeptone (DIFCO), 20g glucose.
Minimal Medium (MM) (per liter): 20g glucose, 1.7g yeast nitrogen source without amino acids, 1.0g proline, pH 6.1 (unadjusted).
Minimal medium + uracil (MM + uracil or MMU) (per liter): MM medium was prepared as above and 0.1g uracil and 0.1g uridine were added.
Minimal medium + uracil + sulfonylurea (MMU + SU) (per liter): MMU medium was prepared as above and 280mg sulfonylurea was added.
Minimal medium + leucine + lysine (MMLeuLys) (per liter): MM medium was prepared as above, and 0.1g leucine and 0.1g lysine were added.
Minimal Medium + 5-Fluorotic acid (MM +5-FOA) (Per liter): 20g glucose, 6.7g yeast nitrogen source, 75mg uracil, 75mg uridine and an appropriate amount of FOA (Zymo Research Corp., Orange, Calif.) based on FOA activity tested for a concentration range from 100mg/L to 1000mg/L (as variations occur within each batch received from the supplier).
MF medium (per liter):14.3g yeast extract, 7.15g peptone, 0.82g KH2PO4、16.37g K2HPO420g of grapeSugar, 1.2mL Trace Metal (100X), 3mL MgSO4(1M), 0.6mL thiamine HCl (1.5 g/L).
3 MF buffer 1 medium (per liter): 150g glucose, 100.12g KHCO, 4.29g urea.
YM medium: 0.5% peptone, 0.3% yeast extract, 0.3% maltose extract.
YNB medium (per liter):20g glucose, 1.7g yeast nitrogen source without amino acids, 20g agar, pH 6.1 (unadjusted).
YPD2-B Medium:10g yeast extract, 10g peptone, 20g glucose, 94mL K2HPO4(1M)、6mL KH2PO4(1M), 200. mu.L of trace metal (100X), 1mL of thiamine-HCl (75mg/mL), 1mL of MgSO4-7H2O(12.5g/100mL)。
YPD4-B Medium:10g yeast extract, 10g peptone, 40g glucose, 94ml K2HPO4(1M)、6mL KH2PO4(1M), 200. mu.L of trace metal (100X), 1mL of thiamine-HCl (75mg/mL), 1mL of MgSO4-7H2O(12.5g/100mL)。
Y2P1D2-B medium: 20g yeast extract, 10g peptone, 20g glucose, 94ml K2HPO4(1M)、6mL KH2PO4(1M), 200. mu.L of trace metal (100X), 1mL of thiamine-HCl (75mg/mL), 1mL of MgSO4-7H2O(12.5g/100mL)。
Trace metal formula (100X): 10.0g/L citric acid, 1.5g/L CaCl2·2H2O、10.0g/L FeSO4·7H2O、0.39g/L ZnSO4·7H2O、0.38g/L CuSO4·5H2O、0.20g/L CoCl2·6H2O、0.30g/L MnCl2·4H2O。
Yarrowia transformation:
Unless otherwise stated, the transformation of yarrowia lipolytica was carried out according to the method of Chen et al (applied. Microbiol. Biotechnol. [ applied microorganisms and Biotechnology ] 48: 232-235). Briefly, yarrowia was streaked onto YPD plates and grown at 30 ℃ for approximately 18 h. Several large ring numbers of cells were scraped from the plate and resuspended in 1mL of transformation buffer containing 2.25mL of 50% PEG (average MW 3350), 0.125mL of 2M lithium acetate, pH 6.0, and 0.125mL of 2M DTT. Then, approximately 500ng of linearized plasmid DNA was incubated in 100. mu.L of resuspended cells and maintained at 39 ℃ for 1h while vortex mixing at 15min intervals. These cells were seeded on selective media plates and maintained at 30 ℃ for 2 to 3 days.
Flask cultures for Long Chain Dicarboxylic Acid (LCDA) production:
a circular amount of freshly streaked yarrowia cells were inoculated in 15-mL FalconTMTubes were grown overnight (about 20 hours) in 3mL MM medium with shaking (250rpm) at 30 ℃. The overnight cultured cells were used to inoculate 50mL of liquid Y2P1D2-B medium in a 250-mL baffled flask and shaken at 250rpm at 30 ℃. After 24 hours, by adding 2.0mL of 1M NaHCO3And 1.0mL of glucose solution (200g/L) to adjust the culture to pH 8.0. Then, 1.5mL ethyl palmitate (substrate) was added directly to the medium to a final concentration of 23mg mL-1And the culture was shaken at 250rpm at 30 ℃ for 4 days. Samples of whole broth from each flask culture were subjected to LCDA analysis.
Micro-fermentation for LCDA production:
the strains used for the micro-fermenter analysis were grown as single colonies from the frozen stock on YPD agar plates. Single colonies were inoculated onto 3mL minimal medium in 15-mL FALCON tubes and grown overnight at 250rpm at 30 ℃. From these cultures, fermentation vials were constructed with 1mL seed culture and 1mL 50% glycerol stock and stored at-80 ℃. The fermentation vials were thawed and 200 μ Ι _ of culture were used to inoculate 4mL of MF medium in each well of the 24-well cassette. The micro-fermentor was operated at 700rpm at 30 ℃ with a DO of 20 for the first 24 hours and a DO of 75 for 72 hours of operation. MF buffer 1 medium was added to each well at 24 hours (200 μ L), 32 hours (150 μ L), 48 hours (150 μ L), 56 hours (150 μ L), and 72 hours (50 μ L). Ethyl palmitate substrate was added to each well at 24 hours (20 μ L), 32 hours (30 μ L), 48 hours (20 μ L), 56 hours (30 μ L), 72 hours (20 μ L), and 80 hours (30 μ L). The micro-fermentor cultures were harvested at 96 hours and aliquots were taken for LCDA analysis.
LCDA extraction and analysis from 250-mL flask cultures:
a sample of the whole fermentation broth (1.0mL) was harvested in a screw-top glass vial with TEFLON septum. The sample was acidified to pH 3.0 by addition of 1M HCl and then extracted once with 1.0mL t-butyl methyl ether (MTBE, Sigma-Aldrich) containing 5.0mg/mL myristic acid internal standard. The sample was vortexed, after which the aqueous and organic phases were separated by centrifugation at 4500rpm for 5-min. An aliquot (0.5mL) of the organic, MTBE phase (containing LCDA) was transferred to a new vial and H by adding 0.5mL of methanol2SO4(5% v/v) to perform derivatization of the LCDA product with methyl groups and heating at 80 ℃ for 1 hour. After derivatization, 1M NaCl in water (0.5mL) was added, the sample was vortexed, and the phases were separated while at rest. The upper MTBE organic layer containing the methyl-derived LCDA product was collected for analysis by Gas Chromatography (GC) and Flame Ionization Detector (FID). Compound residence time and mass spectral data were compared to those measured for methyl esters from commercial standards (Ultra Scientific, north kingston, rhode island). Using a device equipped withA320 fused silica capillary column (30m x 0.32mm x 0.25 μm) (Supelco, Bel Tont, Pa.) 7890GC (Agilent Technologies, Santa Clara, Calif.) was used for GC analysis. Hydrogen was used as a diluent at 5.5mL min-1A constant flow of gas carrier having a split ratio of 10: 1 and an inlet pressure of 18.0 psi. The oven temperature was initially programmed at 200 ℃ and then at 25 ℃ min-1Is immediately increased to 240 ℃; the detector was at 260 ℃.
LCDA extraction and analysis from 2-L fermentation samples:
the method involves transferring 100 μ L of a sample of whole fermentation broth into a reaction vial. The sample weight was measured using an analytical balance and recorded as ± 0.1 mg. Immediately after transfer, by adding 100. mu.L of 20mg/mL myristic acid internal standard (provided in toluene) and 2.0mL methanol in H2SO4(5% v/v) and heating the reaction vial at 80 ℃ for 1 hour to effect derivatization of the LCDA product with methyl groups. After derivatization, solvent extraction was performed by adding 2.0mL of 1M NaCl and 2.0mL of hexane in water to the reaction mixture. The upper hexane organic layer containing the derivatized product was collected for analysis by GC and FID. Compound residence time and mass spectral data were compared to those measured for methyl esters from commercial standards (Ultra Scientific, north kingston, rhode island). The concentration of the LCDA product in the sample was calculated with respect to the myristic acid internal standard. Using a device equipped withGC analysis was performed on a 320 fused silica capillary column (30m x 0.32 mm. times.0.25 μm) (Supelco) 6890GC (Agilent technologies). Helium was used at 2.8mL min-1A constant flow of the carrier gas, wherein the split ratio is 20: 1, and the inlet pressure is 18.0 psi. The oven temperature was initially programmed at 160 ℃ and then at 5 ℃ for min-1Immediately increasing to 200 deg.C for 10min-1Increase to 240 ℃ and keep for 4 min. The detector was at 260 ℃.
Strategy for engineering yarrowia to produce LCDA from vegetable oil-based substrates
Yarrowia lipolytica is an unconventional oleaginous yeast that produces lipids in excess of 25% Dry Cell Weight (DCW) when grown under nitrogen limited conditions with glucose as the carbon source. Because yarrowia lipolytica has a strong β -oxidizing ability, the yeast can readily use hydrophobic substrates (e.g., n-alkanes, oils, fats, and fatty acids) as the sole carbon source. When yarrowia lipolytica is fed with fatty acids or fatty acid esters, it can produce lipids in excess of 40% DCW. Most of the fatty acids and/or fatty acid esters supplied to yarrowia are stored in the form of triacylglycerols.
Figure 1 depicts lipid metabolic pathways that include aspects of fatty acid beta-oxidation and omega-oxidation. Yarrowia lipolytica has a very weak omega-oxidizing ability (indicated by the dashed line in FIG. 1). Because of this low activity, there is no detectable LCDA when the yeast (wild-type) is supplied with a vegetable oil, vegetable oil-derived fatty acid, or fatty acid ester. The strategy for engineering yarrowia lipolytica to convert vegetable oils, vegetable oil-derived fatty acids, and/or fatty acid esters to LCDA is illustrated in fig. 2 and comprises: (1) reducing storage lipids by knocking out genes encoding diacylglycerol acyltransferase 1(DGAT1), diacylglycerol acyltransferase 2(DGAT2), and Phospholipid Diacylglycerol Acyltransferase (PDAT); (2) reducing or eliminating beta-oxidation in peroxisomes by knocking out genes encoding one or more peroxisome biogenesis factor Proteins (PEXs); (3) omega-oxidation is enhanced by overexpressing cytochrome P450 monooxygenase (CYP) and Cytochrome P450 Reductase (CPR) genes.
In addition, as depicted in fig. 1 and 2, it is believed that the rate and extent of fatty acid transport across the cell membrane to the cytoplasm affects the production of LCDA in engineered yarrowia lipolytica cells due to fatty acid transporter and long chain fatty acyl coa synthetase activities. Indeed, as disclosed below, long-chain fatty acyl-coa synthetases were found to upregulate LCDA production in engineered yarrowia cells.
Example 1
Gene encoding putative long-chain fatty acyl-CoA synthetase from yarrowia lipolytica
This example describes the identification of candidate sequences for long-chain acyl-coa synthetases in yarrowia lipolytica for the production of long-chain acyl-coa metabolites in microorganisms.
Fatty acids must be activated by esterification to be transported into the cell. Enzymes of long-chain fatty acyl-coa synthetases catalyze this activation step by conjugating fatty acids to coa, thereby forming fatty acyl-coa. There are four genes (FAA-1, -2, -3, -4) encoding acyl-CoA synthetases specific for medium and long chain fatty acids in Saccharomyces cerevisiae. For example, FAA1 encodes the acyl-CoA synthetase ScFaalp (SEQ ID NO: 33) of fatty acids preferably having a chain length of C12 to C16, and FAA2 encodes the enzyme ScFaa2p (SEQ ID NO: 34) of fatty acids preferably having a chain length of C9 to C13 (J.CelBiol. [ J.Cell.Biobiol ] 127: 751 762; Biochim.Biophys.acta [ Proc.Biochem ] 1486: 18-27).
To identify FAA homologs in yarrowia lipolytica, the amino acid sequences encoded by the predicted Open Reading Frame (ORF) sequences in the yarrowia lipolytica genomic database (www.genolevures.org/yali. html) were aligned with the predicted amino acid sequences of Saccharomyces cerevisiae Faalp (SEQ ID NO: 33) and Faa2p (SEQ ID NO: 34). Fifteen yarrowia lipolytica ORFs were identified by these BLAST analyses (Table 2). Of the fifteen Faa1p and Faa2p homologs encoded by these ORFs, twelve of them are predicted to be peroxisomes (containing peroxisome localization signals), whereas three of them have unknown cellular localization information.
TABLE 2
Long-chain fatty acyl-CoA synthetase candidates in yarrowia lipolytica
Respectively, the Saccharomyces cerevisiae Faalp (SEQ ID NO: 33) and Faa2p (SEQ ID NO: 34) amino acid sequences were aligned with the amino acid sequence encoded by the Candida tropicalis genome (www.candidagenome.org/cgi-bin/computer/blast _ close. pl # Select _ Target _ Organisms). A total of six candidate ORFs were identified. Three of these ORFs encode amino acid sequences containing putative peroxisome localization signals, and are therefore predicted to encode peroxisome proteins. Table 3 lists each of these candidate sequences.
TABLE 3
Long-chain fatty acyl-CoA synthetase candidates in Candida tropicalis
Using the VECTOR NTI software, the amino acid sequences of Saccharomyces cerevisiae Faa1p (SEQ ID NO: 33) and Faa2p (SEQ ID NO: 34), fifteen yarrowia lipolytica long-chain acyl-CoA synthetase candidates, and six Candida tropicalis long-chain acyl-CoA synthetase candidates were aligned. The phylogenetic tree resulting from this alignment is shown in figure 3. Yarrowia candidates YLAcoS-2P (SEQ ID NO: 37), -3P (SEQ ID NO: 39), -4P (SEQ ID NO: 40), -5P (SEQ ID NO: 42), -6P (SEQ ID NO: 44), -7P (SEQ ID NO: 45), -9P (SEQ ID NO: 47), -10P (SEQ ID NO: 49), -11P (SEQ ID NO: 50) and-12P (SEQ ID NO: 51) were grouped together to form a group. All these sequences are predicted to be peroxisomal proteins. Six Candida long-chain acyl-CoA synthetase candidates and yarrowia long-chain acyl-CoA synthetase candidates YLFaa1(SEQ ID NO: 36), YLAcoS-8(SEQ ID NO: 46), -13P (SEQ ID NO: 52), -14(SEQ ID NO: 53), -15P (SEQ ID NO: 54) were pooled together with two Saccharomyces cerevisiae acyl-CoA synthetases. ScFaa1(SEQ ID NO: 33) was closely related to CA-1(SEQ ID NO: 57) and YLFaal (SEQ ID NO: 36, FIG. 3 "YA-1"). ScFaa2(SEQ ID NO: 34) and CA-2 to-6 form one group, and YLAcoS-8(SEQ ID NO: 46), -13P (SEQ ID NO: 52), -14(SEQ ID NO: 53) and-15P (SEQ ID NO: 54) form a third group.
Thus, the sequences of candidate long-chain fatty acyl-coa synthetases in yarrowia lipolytica were identified.
Example 2
Expression patterns of candidate long-chain fatty acyl-CoA synthetases in engineered yarrowia lipolytica cells
This example describes screening of yarrowia lipolytica long-chain acyl-coa synthetase candidates identified in example 1 by qRT-PCR to identify sequences induced under conditions where substrate is added to the culture medium. Any long-chain acyl-coa synthetase sequence whose expression is induced by a fatty acid-containing substrate may be a candidate enzyme for facilitating substrate import.
LCDA-producing yarrowia lipolytica strain D0145 (construction of this strain is described in example 13 below) was performed in triplicate in 50mL cultures in 250-mL flasks with Y2P2D2 growth medium (20g/L yeast extract; 20g/L BACTO-PEPTONE; 20g/L glucose) at 30 ℃ to start the OD600Growth was carried out at 0.15 with a shaking speed of 250 rpm. After 24 hours, 0.5mL and 1mL "day 0" culture samples were collected for RNA extraction and LCDA quantification, respectively. For the remaining culture, 1M NaHCO was added3The pH was adjusted to 8.0, after which ethyl palmitate substrate was added to a final concentration of 3%. 24 hours after substrate addition, 0.5mL and 1mL of "day 1" samples were collected for RNA extraction and LCDA quantification, respectively. Fig. 4 shows LCDA produced by strain D0145 at different time points. There was no LCDA production before the addition of ethyl palmitate to the medium, but there was production after the addition of substrate, which increased at a steady rate to about day 2 (figure 4).
To prepare RNA samples, 0.5-mL aliquots from each culture at day 0 and day 1 were harvested by centrifugation at 13,000x g for 1 min. The cell pellet was immediately frozen and stored at-80 ℃. Using TRIzolTMReagents (Life Technologies, Calsbad, Calif.) Total RNA was prepared from each cell pellet. Cell disruption was performed using MINI-BEADBEATER-8(BSP, Batterville, Oklahoma). Qiagen RNeasy was then usedTMThe kit purified the total RNA extracted from each sample.To remove any residual genomic DNA, 3. mu.g of total RNA was treated with DNase without RNase activity (Qiagen, Hilden, Germany). Then, the DNase was inactivated by adding 1mM EDTA and heating to 75 ℃ for 5 minutes. Using a high volume cDNA reverse transcription kit (Applied Biosystems, Foster City, Calif.), following the manufacturer's instructions, 1. mu.g of DNase-treated RNA was then converted to complementary DNA (cDNA). The cDNA was then expressed as 1: 10 dilutions were used in rnase-free water for quantitative pcr (qpcr) analysis.
qPCR was performed to detect the expression of the target genes listed in table 4. All PRIMERs listed in Table 4 were designed using PRIMER EXPRESS v 3.0.1 software (Applied Biosystems). Primer specificity was assessed by BLAST analysis against the yarrowia lipolytica genolevurs database (genolevurs. org/yali. html) and quantification was verified using genomic DNA (data not shown). Primers with PCR efficiencies between 0.85 and 1.15 were validated for quantification. Use ofGreen all qPCR reactions were performed in triplicate, andgreen was used for detection on an ABI 7900 SDS instrument (Applied Biosystems, Foster City, Calif.). Relative expression (RQ) was calculated using data-aided software v3.01 and the Δ Δ Ct method (applied biosystems, foster city, ca). The gene encoding 18S rRNA was identified by the software as the best endogenous control gene and used for data normalization. Then, the relative expression of each gene on day 1 was calculated by comparing the expression of each gene on day 1 with the expression on day 0, which was set to 1.0.
TABLE 4
Primers for qPCR analysis
Table 5 (below) shows the results of the qRT-PCR analysis. Expression measurements (SYBR) for each day 0 (D0) and day 1 (D1) sample were measurements relative to day 0-1 ('D0-1') for the sample set at 1.00. Each data point was run through three independent PCR reactions and normalized to yarrowia 18S rRNA expression. The "SYBR SD" value is the standard deviation for each three-pack (trio) of PCR reactions. Transcripts encoding YLAcoS-10P (SEQ ID NO: 49), YLAcoS-6P (SEQ ID NO: 44), and YLAcoS-3P (SEQ ID NO: 39) long-chain acyl-CoA synthetase exhibited more than a 4-fold increase in expression compared to day 0 (shown in grey boxes in Table 5).
Based on the data in Table 5, the expression of the putative long-chain acyl-CoA synthetases of YLAcoS-10P (SEQ ID NO: 49), YLAcoS-6P (SEQ ID NO: 44), and YLAcoS-3P (SEQ ID NO: 39) was induced in yarrowia lipolytica when treated with a substrate containing a long-chain fatty acid. These long-chain acyl-coa synthetases may therefore be useful for facilitating the import of substrates containing long-chain fatty acids.
Example 3
Codon optimization of a polynucleotide sequence encoding a putative long-chain acyl-coa synthetase for use in lipolytic enzymes
Expression in yarrowia
The DNA open reading frames encoding the long chain acyl-CoA synthetase candidates YLACoS-3P (SEQ ID NO: 39), YLACoS-5P (SEQ ID NO: 42), YLACoS-6P (SEQ ID NO: 44), YLACoS-10P (SEQ ID NO: 49), and YLFAA (SEQ ID NO: 36) were codon optimized for high expression in yarrowia lipolytica according to the methodology disclosed in U.S. Pat. No. 7125672, which is incorporated herein by reference. Thus, polynucleotide sequences YLACoS-3Ps (SEQ ID NO: 38), YLACoS-5Ps (SEQ ID NO: 41), YLACoS-6Ps (SEQ ID NO: 43), YLACoS-10Ps (SEQ ID NO: 48), and YLFAA1s (SEQ ID NO: 35) were prepared, which encode YLACoS-3P (SEQ ID NO: 39), YLACoS-5P (SEQ ID NO: 42), YLACoS-6P (SEQ ID NO: 44), YLACoS-10P (SEQ ID NO: 49), and YLFAA1(SEQ ID NO: 36), respectively. Each codon-optimized DNA sequence was individually synthesized and cloned into an expression vector by Kinsley (GenScript) (Piscataway, N.J.) to yield pZP2-YLACoS-3Ps (SEQ ID NO: 63), pZP2-YLACoS-5Ps (SEQ ID NO: 64), pZP2-YLACoS-6Ps (SEQ ID NO: 65), pZP2-YLACoS-10Ps (SEQ ID NO: 66), and pZKL7A-FYLFAAs (SEQ ID NO: 67) (respectively, in FIGS. 5A-E). Another vector, pZP2-YLACoS-5PS3s (SEQ ID NO: 68, FIG. 5F), was also prepared, which allows the expression of YLACoS-5PS3(SEQ ID NO: 56), a truncated version (six amino acid C-terminal truncation) of YLAcoS-5P (SEQ ID NO: 42).
The above constructs can be used to overexpress long-chain acyl-coa synthetase candidates in yarrowia.
Example 4
Expression of long-chain acyl-CoA synthetase candidates in E.coli
This example discloses overexpression of acyl-CoA synthetase candidates YLACoS-3P (SEQ ID NO: 39), YLACoS-5P (SEQ ID NO: 42), YLACoS-6P (SEQ ID NO: 44), YLACoS-10P (SEQ ID NO: 49), YLACoS-5PS3(SEQ ID NO: 56, a six amino acid C-terminally truncated version of YLACoS-5P), and YLFAA (SEQ ID NO: 36) under a T7 inducible promoter in E.coli (Escherichia coli).
First, polynucleotide sequences of YLACoS-3Ps (SEQ ID NO: 38), YLACoS-5Ps (SEQ ID NO: 41), YLACoS-6Ps (SEQ ID NO: 43), YLACoS-10Ps (SEQ ID NO: 48), YLACoS-5PS3s (SEQ ID NO: 55), and YLFAAs (SEQ ID NO: 35) (each codon-optimized for expression in yarrowia) were excised from pZP2-YLACoS-3Ps (SEQ ID NO: 63), pZP2-YLACoS-5Ps (SEQ ID NO: 64), pZP2-YLACoS-6Ps (SEQ ID NO: 65), pZP2-YLACoS-10Ps (SEQ ID NO: 66), YZP 2-389-5 Ps (SEQ ID NO: 64), pZP2-YLACoS-6Ps (SEQ ID NO: 65), pZP2-YLACoS-10Ps (SEQ ID NO: 66), YACoS-5 Ps (SEQ ID NO: 389) and pZP 3967, respectively, using NcoI/NotI restriction endonucleases, and separately ligated into pET23d vector (SEQ ID NO: 69) (Novagen, Madison, Wis.) at the NcoI/NotI endonuclease site. Restriction analysis was used to confirm each ligation (data not shown).
To overexpress each putative long-chain acyl-CoA synthetase, it was transformed with the appropriate pET23 d-based plasmid and placed in LBAMPAn 8-hour culture of E.coli BL (DE3) grown in medium (AMP: ampicillin, final concentration 100. mu.g/mL) was diluted 1: 50 in 100mL of the same medium in a 500-mL flask. Each culture was shaken at 37 ℃ until the optical density at 600nm reached 0.8-0.9, after which the flask was placed in an 18 ℃ incubator for about 20 minutes, and then isopropylthio-beta-D-galactoside (IPTG) was added to a final concentration of 100. mu.M. Then, each culture was shaken for another 10-12 hours at 18 ℃. Cells (approximately 100mg wet weight from 15mL culture) were collected by centrifugation, washed once with Phosphate Buffered Saline (PBS) (pH 7.4), then resuspended in 400 μ L of lysis buffer (BUGBUSTER HT, containing 25% glycerol, 0.5mg/mL lysozyme from Pierce and protease inhibitor mixture) and incubated on a shaking platform at room temperature for 20 minutes. Cell debris was removed by centrifugation at 12,000x g for 30 minutes at 4 ℃. To remove small molecules from the supernatant that could interfere with the enzyme assay described below, the supernatant was placed in a 10-kDa molecular weight cut-off (MWCO) centrifuge unit and centrifuged at 12,000x g for 30 minutes at 4 ℃. The retained protein solution (about 50. mu.L-100. mu.L) was resuspended in 400. mu.L (final volume) of buffer (0.1M KPi, 20% glycerol, pH 7.5) and concentrated once again by centrifugation at 12,000x g for 30 minutes at 4 ℃ on a MWCO apparatus. The concentrated protein solution was resuspended in 0.1M KPi, 20% glycerol, pH 7.5 in a final volume of about 200 μ L, transferred to a new centrifuge tube, and centrifuged briefly at maximum speed to remove any precipitated protein. Will be used for SDS-PAGE analysis, protein concentrationThe assay of (a), and the clear supernatant of the enzyme assay are stored at-80 ℃. As shown in FIGS. 6A and B, all six acyl-CoA synthetase candidates were successfully overexpressed in E.coli and were found in the soluble fraction of E.coli cell lysates, except for YLACoS-3P (SEQ ID NO: 39).
Example 5
Determination of specific Activity of Long-chain acyl-CoA synthetase candidates
This example discloses an assay of the specific activity of a long-chain acyl-coa synthetase candidate. Specifically, acyl-coa synthetase candidates present in soluble e.coli fractions (produced in example 4) were tested for activity using palmitic acid or p-coumaric acid as substrates.
The specific activity of each long-chain acyl-coa synthetase candidate for palmitic acid substrates was determined as follows. Formation of Adenosine Monophosphate (AMP) from the putative acyl-coa synthetase in the clear supernatant (example 4) in the presence of phosphoenolpyruvate (PEP), NADH, myokinase and pyruvate kinase coupled with oxidation of NADH by lactate dehydrogenase (monitored by absorbance at 340 nm), as depicted in the following scheme (1 → 4):
3.2 ADP +2 PEP → 2 ATP +2 pyruvate (pyruvate kinase catalyzed).
4.2 pyruvate +2 NADH → 2 lactate +2 NAD+(catalyzed by lactate dehydrogenase).
Specifically, each assay (300 μ Ι _ final volume) was performed at 30 ℃ and comprised: in 100mM Tris-Cl, 50mM NaCl, 10mM MgCl21mM Brown in pH7.2Palmitic acid (diluted from a10 mM stock solution prepared in DMSO), 4mM ATP, 1.5mM CoASH, 1mM PEP, 5 units pyruvate kinase, 5 units lactate dehydrogenase, 6 units myokinase. The reaction process was initiated by adding the appropriate amount of cell lysate containing the candidate long-chain fatty acyl-coa synthetase (example 4). After addition of the cell extract, oxidation of NADH (to NAD) at 340nm was monitored using Cary-100UV-Vis Spectrophotometer (Agilent)+) For 5 minutes. The initial slope was calculated by subtracting the background activity observed in the enzyme assay in which the palmitic acid substrate was replaced by DMSO.
The specific activities of the putative long-chain acyl-coa synthetases as measured above for palmitic acid substrates are summarized in table 6 below. Providing specific activity measurements in mU/mg, wherein one unit corresponds to the amount of enzyme producing 1.0. mU. mole of palmitoyl-CoA per minute at 30 ℃ and pH7.2 in the presence of 1mM palmitic acid, 4mM ATP and 1.5mM CoA; absorption coefficient of NADH 6,220M-1cm-1. No activity above background levels (indicated as "n.d." inTable 6) was detected in supernatants prepared from control cells (transformed with empty pET23d vector) and in supernatants prepared from cells expressing YLACoS-3P (SEQ ID NO: 39), YLACoS-5P (SEQ ID NO: 42) and YLACoS-5PS3(SEQ ID NO: 56).
Since the sequences associated with the acyl-CoA synthetase candidates YLACoS-3P (SEQ ID NO: 39), YLACoS-5P (SEQ ID NO: 42), and YLACoS-10P (SEQ ID NO: 49) were annotated in the NCBI Genbank database as putative 4-coumaric acid-CoA ligases, while YLFAA (SEQ ID NO: 36) showed 50% identity to Faa1P (SEQ ID NO: 33) (well-characterized long-chain fatty acyl-CoA synthetases from Saccharomyces cerevisiae, preferably C12: 0-C16: 0 fatty acids), the specific activities of the above-mentioned enzymes were also tested using P-coumaric acid (pCA) as a substrate. The specific activity of each long-chain acyl-coa synthetase candidate on the pCA substrate was determined as follows. Each assay (250 μ Ι final volume) was performed at 30 ℃ and contained: in 100mM Tris-C1, 50mM NaCl, 10mM MgCl21mM p-coumaric acid (diluted from a10 mM stock solution prepared in DMSO), 4mM ATP, 1.5mM CoASH in pH 7.2. By addingAn appropriate amount of cell lysate containing the candidate long-chain fatty acyl-coa synthetase (example 4) elicits the reaction. After addition of the cell extract, the increase in absorbance at 340nm (due to the formation of p-coumaroyl-CoA) was monitored for 10min using Cary-100UV-Vis Spectrophotometer (Agilent). The initial slope was calculated by subtracting the background activity observed in the enzyme assay in which the pCA was replaced by DMSO.
The putative specific activities of long-chain acyl-coa synthetases as measured above for the pCA substrates are summarized in table 6 below. Providing specific activity measurements in mU/mg, wherein one unit corresponds to the amount of enzyme producing 1.0. mU. mole of p-coumaroyl-CoA per minute in the presence of 1mM p-coumaric acid, 4mM ATP and 1.5mM coenzyme A at 30 ℃ and pH 7.2; the absorbance coefficient of coumaroyl-CoA was 21,000M-1cm-1. No activity above background levels (indicated as "n.d." inTable 6) was detected in supernatants prepared from control cells (transformed with empty pET23d vector) and in supernatants prepared from cells expressing YLACoS-3P (SEQ ID NO: 39), YLACoS-5P (SEQ ID NO: 42), YLACoS-5PS3(SEQ ID NO: 56) and YLFAA (SEQ ID NO: 36).
TABLE 6
Specific Activity of Long-chain acyl-CoA synthetase candidates on different substrates
an.a. (not detected).
These results support the insight that YLACoS-6P (SEQ ID NO: 44) and YLACoS-10P (SEQ ID NO: 49) can accept both aromatic carboxylic acids and long chain fatty acids as substrates. In contrast, YLFAA1(SEQ ID NO: 36) appears to be specific for palmitic acid. Neither YlACoS-3P (SEQ ID NO: 39) nor YlACoS-5P (SEQ ID NO: 42) showed activity on both substrates under the defined reaction conditions.
Example 6
Is generated fromSubstrate of vegetable oil production of LCDA A higher yarrowia lipolytica parent Strain
This example discloses yarrowia lipolytica strains suitable for additional genetic engineering to produce strains that produce large amounts of LCDA.
As noted above, it is expected that yarrowia lipolytica may need to be engineered to reduce or eliminate lipid storage and β -oxidation in order to efficiently produce LCDA from vegetable oils, vegetable oil-derived fatty acids or fatty acid esters. It is also possible that different genetic backgrounds may be beneficial for LCDA production. As shown in table 7, a series of yarrowia lipolytica strains were generated from wild type strains ATCC nos. 20362 and 90812. Some of these strains have reduced lipid storage capacity and reduced beta-oxidation function. Fig. 7A illustrates the lineage of some of these strains relative to each other.
TABLE 7
Yarrowia lipolytica parent strain for LCDA production
aEach of which is incorporated herein by reference.
Specifically, strain D0004 was produced by knocking out the PEX3 gene (encoding peroxisome biogenesis factor 3 protein [ PEX3p ]) in strain L183. Strain L183 (designated D0003) was transformed with AscI/SphI fragment containing URA3 of plasmid pY157(SEQ ID NO: 70, see FIG. 4A of U.S. Pat. No. 62/140,681) by homologous recombination to knock out the PEX3 gene. One of the transformants, designated strain T1876, was identified as pex3- (i.e., Δ pex3) by real-time PCR. The PEX3 knockout site of strain T1876 is expected to comprise the amino acid sequence of SEQ ID NO: 71 (in place of the wild-type PEX3 locus sequence) (see Table 1 for a description of SEQ ID NO: 71). Strain T1876 is transformed with plasmid pY117 (disclosed in table 20 of U.S. patent application publication No. 2012/0142082, which is incorporated herein by reference) to express Cre recombinase to excise the LoxP-flanked URA3 gene (introduced by a fragment of pY157 that knocks out PEX 3). The pY117 transformant failed to grow on MM, but could grow on MMU, indicating that the transformant lacks URA3 gene; this transformant was designated as strain D0004(dgat1-, dgat2-, pex3-, ura 3-). The PEX3 knock-out site of strain D0004 is expected to comprise SEQ ID NO: 72 (in place of the wild-type PEX3 locus sequence) (see Table 1 for a description of SEQ ID NO: 72).
Strain D0015 was produced from strain D0004 by the "pop-in/pop-out" method (see U.S. patent application publication No. 2014/0220645 for further details regarding this type of knockout strategy, which is incorporated herein by reference) by knocking out the POX4 gene (encoding peroxisomal acyl-coa oxidase-4 [ POX4 enzyme, genbank accession No. CAG80078 ]). Briefly, strain D0004 was transformed with XbaI digested plasmid pYRH146-Pox4KO (SEQ ID NO: 73, see FIG. 4C in U.S. patent application No. 62/140,681). A total of 28 transformants were grown on MM plates. Two transformants (#7 and #17) were tested by PCR analysis, where the first hybridization (spring-in) was between the native POX4 gene and the homologous 3' -arm sequence of construct pYRH146-Pox4 KO. The #7 transformant was picked, grown in liquid YPD medium, and then plated on FOA600 plates (to select for the ejection event that led to ura 3-). PCR analysis detected a second hybridization (between the corresponding 5' -homology arm sequences) in 13 of the 28 strains grown on the FOA600 plates. One of the 13 strains was designated as D0015, and the D0015 strain was identified as having a knock-out of the POX4 gene. D0015 has the following genotype: dgat1-, dgat2-, pex3-, pox4-, ura 3-. The POX4 knock-out site is expected to comprise the amino acid sequence of SEQ ID NO: 74 (in place of the wild-type POX4 locus sequence) (see Table 1 for a description of SEQ ID NO: 74).
Strain W101 was produced by transforming strain ATCC No. 90812 with the EcoRI/ClaI fragment of plasmid pYRH72 containing URA 3(SEQ ID NO: 75).
A diploid strain (1D2373) was produced by crossing W101 with D0004.
Strain 1D2373 was sporulated and one of its progeny (strain 2373I-6) was determined to be haploid with mating type B genotype by real-time PCR. Strain 2373I-6 was unable to grow on SC-1eu medium and was renamed to strain 1B 24791.
A diploid strain (2D2519) was produced by crossing 1B24791 with D0004.
Strain 2D2519 was sporulated and one of its progeny (strain 2519I-1) was determined by real-time PCR to be haploid with mating type B genotype. Strain 2519I-1 was unable to grow on SC-leu medium and was renamed to strain 2B 25831.
A diploid strain (3D2653) was produced by crossing 2B25831 with D0004.
Strain 3D2653 was sporulated and one of its progeny (strain 2653I-19) was determined by real-time PCR to be haploid with genotype dgat2-, MATB. Strain 2653I-19 was unable to grow on SC-leu medium and was renamed to strain 3B 27021.
Strain D0015 was crossed with strain 3B27021 to produce diploid strain 4D 2738.
Strain 4D2738 was sporulated and one of its progeny (strain 2738Y-14) was determined by real-time PCR to be haploid with genotypes dgat1-, dgat2-, pox4-, pex 3-and MATA. Strain 2738Y-14 was unable to grow on MM medium and was designated D0017.
Strain 4D2738 was sporulated and one of its progeny (strain 2738Y-45) was identified by real-time PCR as a haploid with genotypes dgat1-, dgat2-, pox 4-and pex 3-. Strain 2738Y-45 was unable to grow on SC-ura or SC-leu plates. Thus, strain 2738Y-45 has the genotype: MATA, dgat1-, dgat2-, pex3-, pox4-, ura3-, and leu 2-.
Strain 77T5-5 was produced by deleting the POX3 gene from 2738Y-45 via a one-step process. Strain 2738Y-45 was transformed with the AscI/SphI fragment of plasmid p12_3-B-Pex3del1 (FIG. 8A, SEQ ID NO: 76). One of the transformants was identified as pox 3-by real-time PCR. This transformant was named 77T5-5(MATA, dgat1-, dgat2-, leu2-, pex3-, pox3-, pox4-, Ura3 +).
Strain D0031 was produced by first deleting the POX2 gene from 77T5-5 via a one-step process. Strain 77T5-5 was transformed with plasmid p70_ Pox2: : leu (FIG. 8B, SEQ ID NO: 77). One of transformants 118T1-14 was identified as pox 2-by real-time PCR. Strain 118T1-14(MATA, dgat1-, dgat2-, Leu2+, Pex3-, pox2-, pox3-, pox4-, Ura3+) was in turn transformed with plasmid pY117 (disclosed in Table 20 of U.S. patent application publication No. 2012/0142082, which is incorporated herein by reference) to express Cre recombinase to excise the LoxP-flanked URA3 gene (introduced in a previous step by p12_3-B-Pex3del 1). One of the transformants, 118T1-14-7-1U, was unable to grow on MM but could grow on MMU, indicating that this transformant lacks the URA3 gene; this transformant was designated as strain D0031(MATA, dgat1-, dgat2-, Leu2+, pex3-, pox2-, pox3-, pox4-, ura 3-).
Thus, certain yarrowia lipolytica strains were generated, including some that lack functional PEX3(PEX3-), POX2(POX2-), POX3(POX3-), and POX4(POX4-) genes. These strains are amenable to additional genetic engineering to produce strains that can produce significant amounts of LCDA (examples below).
Example 7
Production of yarrowia lipolytica Strain D1017 for LCDA production by overexpression of CYP and CPR enzymesThis example discloses the construction of yarrowia strain D1017 by expressing codon-optimized sequences encoding candida tropicalis CYP and CPR enzymes in strain D0031. Strain D1017 is an intermediate strain used to develop strain D3928 (fig. 7B).
The construct pZKLY-FCtR17U (FIG. 9A, SEQ ID NO: 82) contained one copy each of the codon optimized CYP52A17(CtCYPA17s, GenBank accession No. AAO73958, SEQ ID NO: 83 encoding SEQ ID NO: 84) and CPR (CtCPRs, GenBank accession No. P37201, SEQ ID NO: 85 encoding SEQ ID NO: 86) coding sequences from Candida tropicalis. Each coding sequence is under the control of a heterologous promoter and 3' -termination sequence. The NcoI and NotI endonuclease sites were added near the translation start codon (ATG) and after the stop codon, respectively, of each codon optimized sequence encoding CtCYPA17 or CtCPR. The composition of the pZKLY-FCtR17U plasmid (SEQ ID NO: 82) is further described in Table 8.
TABLE 8
Description of plasmid pZKLY-FCtR17U (SEQ ID NO: 82)
Plasmid pZKLY-FCtR17U (SEQ ID NO: 82) was digested with AscI/SphI and then used to transform strain D0031 according to the general procedure. The transformant cells were seeded on MM plates and maintained at 30 ℃ for 2 days. Single colonies from each transformation were streaked again onto MM plates and then inoculated into liquid MM at 30 ℃ and shaken at 250rpm for 1 day. The overnight cultured cells were used to inoculate 25mL of liquid YPD4-B medium in 250-mL flasks, which were then shaken at 180rpm at 30 ℃. After 40 hours, 2.0mL of 1M NaHCO was added3These cultures were adjusted to pH 8.0, after which ethyl palmitate (W2450, Sigma Aldrich) was added directly to the medium to a final concentration of 8mg mL-1. These cultures were then shaken at 180rpm for an additional 4 days at 30 ℃ after which a sample of the whole fermentation broth from each flask culture was subjected to LCDA analysis according to the general method.
GC analysis showed no hexadecanedioic acid detected in the parent strain D0031 (C16: 0 LCDA). However, most transformants of the parent strain D0031 produced more than 8g/L C16: 0 LCDA. Transformants # 6, #8, #10 and #11 produced 9.5g/L, 12.1g/L and 9.1g/L C16, respectively: 0 LCDA. These four strains were designated strains D1015, D1016, D1017 and D1018, respectively.
Flask analyses of strains D1015, D1016 and D1017 were subsequently performed. Specifically, strains D1015, D1016 and D1017Each was placed in 50-mL cultures in 250-mL baffled flasks, to which ethyl palmitate was added to a final concentration of 16mg mL-1. These cultures were shaken at 180rpm for 4 days at 30 ℃. Strains D1015, D1016, and D1017 produced C16 at approximately 7.4g/L, 7.6g/L, and 9.3g/L, respectively: 0 LCDA.
Strain D1017 was also analyzed by micro-fermentation analysis. Whereas the control strain (D0285, data not shown) produced C16 at 6.4 g/L: 0 LCDA, strain D1017 produced C16 at about 7.4 g/L: 0 LCDA.
It should be noted that pZKLY-FCtR17U (SEQ ID NO: 82) DNA used to transform D0031 to produce strain D1017 and its cognate could potentially knock out the lipase Y locus (GenBank accession AJ 549519). However, such knockouts were not confirmed in these strains. D1017 and its cognate genotypes relative to wild-type yarrowia lipolytica ATCC # 20362 are dgat1-, dgat2-, Leu2+, pex3-, pox2-, pox3-, pox4-, Ura3+, to know 1-, FBA: : CtCPRs: : lip1, FBAINm 1: : CtCyPA17 s: : pex 20.
Thus, yarrowia strain D1017 was produced which can produce greater than 5g/L of LCDA product when fed with a substrate containing long chain fatty acids in a flask assay.
Example 8
Production of yarrowia lipolytica for LCDA production by overexpression of fatty alcohol oxidase and fatty aldehyde dehydrogenase
Strain D1308
This example discloses the construction of yarrowia strain D1308 by expressing codon-optimized sequences encoding candida cloacae Fatty Alcohol Oxidase (FAO) and candida tropicalis fatty aldehyde dehydrogenase (FALDH). Strain D1308 was an intermediate strain for the development of strain D3928 (fig. 7B).
First, strain D1017U was developed from strain D1017. Plasmid pY117 was used to transiently express Cre recombinase to excise the LoxP-flanked URA3 gene within strain D1017. The pY117 transformant failed to grow on MM, but could grow on MMU, indicating that the transformant lacks URA3 gene; this transformant was designated as strain D1017U.
Next, strain D1017U was transformed with the linearized plasmid construct pZKADn-C2F1U (FIG. 9B, SEQ ID NO: 87). This fragment contains two expression cassettes, one for overexpressing the codon-optimized FAO enzyme-encoding sequence (CcFAO1s, GenBank accession No. CAB75351, SEQ ID NO: 88 encoding SEQ ID NO: 89), and the other for overexpressing the codon-optimized FALDH enzyme-encoding sequence (CtFALDH2s, GenBank accession No. XP-002550712, SEQ ID NO: 90 encoding SEQ ID NO: 91). The composition of the pZKADn-C2F1U plasmid (SEQ ID NO: 87) is further described in Table 9.
TABLE 9
Description of plasmid pZKADn-C2F1U (SEO ID NO: 87)
Plasmid pZKADn-C2F1U (SEQ ID NO: 87) was digested with AscI and then used to transform strain D1017U according to the general method. The transformant cells were seeded on MM plates and maintained at 30 ℃ for 2 days. Individual colonies from each transformation were streaked again onto MM plates and then inoculated into liquid YPD2-B medium in 24-well blocks, which were then shaken at 375rpm for 20 hours at 30 ℃. 0.12mL of 1M NaHCO was added3These cultures were adjusted to pH 8.0, after which ethyl palmitate was added directly to the medium to a final concentration of 23mg mL-1. These cultures were then shaken at 375rpm for an additional 4 days at 30 ℃ after which a sample of the whole fermentation broth from each culture was subjected to LCDA analysis according to the general method.
GC analysis showed that three transformants of strain D1017U produced more than 10g/L C16: 0 LCDA. Specifically, transformants # 2, #5, and #10 produced 10.2g/L, 14.5g/L, and 10.8g/L C16, respectively: 0 LCDA. These three strains were designated as strains D1307, D1308, and D1309, respectively.
Strains D1307 and D1308 were also analyzed by micro-fermentation analysis. Whereas the control strain (D0285, data not shown) produced C16 at about 6.0 g/L: 0 LCDA, strains D1307 and D1308 produced C16 at approximately 9.7g/L and 10.8g/L, respectively: 0 LCDA.
Strain D1308 was further tested using a 2-L fermentation experiment. As shown in table 10 and figure 10, after 162 hours of fermentation, strain D1308 produced a total amount of LCDA of about 50.9g/L, where about 42.6g/L was C16: 0 LCDA.
Watch 10
LCDA produced by Strain D1308 grown in 2-L fermentation with Ethyl palmitate as substrate
It should be noted that the pZKADn-C2F1U (SEQ ID NO: 87) DNA used to transform D1017U to produce strain D1308 and its cognate can potentially knock out the alcohol dehydrogenase 3 locus (GenBank accession AF 175273). However, such knockouts were not confirmed in these strains. D1308 and its cognate genotype relative to wild-type yarrowia lipolytica ATCC # 20362 are dgat1-, dgat2-, Leu2+, pex3-, pox2-, pox3-, pox4-, Ura3+, unknown 1-, unknown 2-, FBA: : CtCPRs: : lip1, FBAINm 1: : CtCYPA17 s: : pex20, DG2 Pro-715: : CtALDH2 s: : lip1, FBAlL: : CcFAOls: : aco.
Thus, yarrowia strain D1308 was produced, which when fed with a substrate containing long chain fatty acids could produce more than 50g/L of LCDA product.
Example 9
Production of yarrowia lipolytica Strain D2300 for production of LCDA higher than 70g/L
This example discloses the construction of yarrowia strain D2300 by expressing codon-optimized sequences encoding the vetch CYP and CPR enzymes in strain D1308. Strain D2300 was an intermediate strain used to develop strain D3928 (fig. 7B).
First, strain D1308U was developed from strain D1308. Plasmid construct pY117 was used to transiently express Cre recombinase to excise the LoxP-flanked URA3 gene within strain D1308. The pY117 transformant failed to grow on MM, but could grow on MMU, indicating that the transformant lacks URA3 gene; this transformant was designated as strain D1308U.
Next, strain D1308U was transformed with the DNA fragment from plasmid construct pYRH213 (FIG. 11A, SEQ ID NO: 92). This fragment contains two expression cassettes, one for the overexpression of the codon-optimized sequence encoding the CYP enzyme (VsCYP94A1s, from Arrowia Ardisia, GenBank accession AAD10204, SEQ ID NO: 93 encoding SEQ ID NO: 94) and the other for the overexpression of the codon-optimized sequence encoding the CPRase enzyme (VsCPRs, from Arrowia Ardisia, GenBank accession Z26252, SEQ ID NO: 95 encoding SEQ ID NO: 96). Each coding sequence is under the control of a heterologous promoter and 3' -termination sequence. NcoI and NotI endonuclease sites were added near the translation start codon (ATG) and after the stop codon, respectively, of each codon optimized sequence encoding VsCYP or VsCPR. The composition of the pYRH213 plasmid (SEQ ID NO: 92) is further described in Table 11.
TABLE 11
Description of plasmid pYRH213(SEQ ID NO: 92)
Plasmid pYRH213(SEQ ID NO: 92) was digested with AscI/SphI and then used to transform strain D1308U according to the general method. The transformant cells were seeded on MM plates and maintained at 30 ℃ for 2 days. Single colonies from each transformation were streaked again onto MM plates. Both strains were directly analyzed for LCDA production using a flask assay. Specifically, single colonies were streaked again onto MM plates and then inoculated into 24 wellsLiquid YPD2-B medium in the block, which was then shaken at 375rpm for 20 hours at 30 ℃. 0.12mL of 1M NaHCO was added3These cultures were adjusted to pH 8.0, after which ethyl palmitate was added directly to the medium to a final concentration of 23mg mL-1. These cultures were then shaken at 375rpm for an additional 4 days at 30 ℃ after which a sample of the whole fermentation broth from each culture was subjected to LCDA analysis according to the general method.
GC analysis showed that the two transformants of strain D1308U each produced 8.2g/L and 12.6g/L C16: 0 LCDA. Production 12.6g/L C16: the strain with 0 LCDA was designated strain D2300.
Strain D2300 was further tested using a 2-L fermentation experiment. As shown in table 12 and fig. 12, strain D2300 produced a total amount of about 72.7g/L of LCDA 163 hours after fermentation, where about 64.6g/L was C16: 0 LCDA.
TABLE 12
LCDA produced by Strain D2300 grown in 2-L fermentation with Ethyl palmitate as substrate
It should be noted that the pYRH213(SEQ ID NO: 92) DNA used to transform D1308U to produce strain D2300 and its family could potentially knock out the lipase Y locus (GenBank accession AJ 549519). However, such knockouts were not confirmed in these strains. The genotype of strain D2300 and its cognate relative to wild-type yarrowia lipolytica ATCC # 20362 is dgat1-, dgat2-, Leu2+, pex3-, pox2-, pox3-, pox4-, Ura3+, unknown 1-, unknown 2-, unknown 3-, FBA: : CtCPRs: : lip1, FBA: : VsCPRs: : lip1, FBAINm 1: : CtCYPA17 s: : pex20, CPR 1: : VsCYP94A1 s: : pex20, DG2 Pro-715: : CtALDH2 s: : lip1, FBA 1L: : CcFAO1 s: : aco.
Thus, yarrowia strain D2300 was produced which could produce more than 70g/L of LCDA product when fed with a substrate containing long chain fatty acids.
Example 10
Production of yarrowia lipolytica Strain D2882 for LCDA production
This example discloses the construction of yarrowia strain D2882 by expressing three codon-optimized sequences encoding a Fatty Alcohol Oxidase (FAO) enzyme in strain D2300. Strain D2300 was an intermediate strain used to develop strain D3928 (fig. 7B).
First, strain D2300, which was Ura3+ as a result of transformation with pYRH213(SEQ ID NO: 92) (see example 9), was represented as Ura 3-. Specifically, D2300 was transformed with plasmid pZKUM to integrate the URA 3-mutant sequence into the complete URA3 sequence. The construction and use of plasmid pZKUM to obtain ura-yarrowia lipolytica cells has been described (U.S. patent application publication No. 2009/0093543, see Table 15 therein, which is incorporated herein by reference). Briefly, plasmid pZKUM was digested with SalI/PacI and then transformed into strain D2300 according to the general method. After transformation, cells were seeded on MM +5-FOA plates and maintained at 30 ℃ for 2-3 days. A total of 8 transformants grown on MM +5-FOA plates were picked and re-streaked onto MM plates and MM +5-FOA plates, respectively. All of these 8 transformants have the ura-phenotype (i.e., cells can grow on MM +5-FOA plates, but cannot grow on MM plates). Transformants # 1, #2, and #3 were designated as D2300U1, D2300U2, and D2300U3, which were collectively designated as D2300U 2300U.
To generate strain D2882, strain D2300U1 was transformed with a DNA fragment from the construct pZSCpn-3FAOBU (FIG. 11B, SEQ ID NO: 98) comprising three expression cassettes to overexpress codon optimized sequences encoding FAO enzymes (CtFAO1, CcFAO1, CcFAO 2). In particular, these expression cassettes comprise the following sequences: (i) CtFAO1Ms (SEQ ID NO: 99 encoding SEQ ID NO: 100, which is a mutant form of CtFAO1 of Genbank accession No. AAS 46878) (CtFAO 1M comprises a histidine residue at amino acid position 359 instead of a tyrosine residue as compared to wild-type CtFAO 1), (ii) CcFAO1s (SEQ ID NO: 101 encoding SEQ ID NO: 102), and (iii) CcFAO2s (SEQ ID NO: 103 encoding SEQ ID NO: 104). NcoI and NotI sites were added near the translation initiation codon (ATG) and after the stop codon, respectively, of each codon-optimized sequence encoding the aforementioned FAO enzyme. The composition of the pZSCpn-3FAOBU plasmid (SEQ ID NO: 98) is further described in Table 13.
Watch 13
Description of plasmid pZSCpn-3FAOBU (SEQ ID NO: 98)
Plasmid pZSCpn-3FAOBU (SEQ ID NO: 98) was digested with AscI/SphI and then used to transform strain D2300U1 according to the general method. The transformant cells were seeded on MM plates and maintained at 30 ℃ for 2 days. Individual colonies from each transformation were streaked again onto MM plates and then inoculated into liquid YPD2-B medium in 24-well blocks, which were then shaken at 375rpm for 20 hours at 30 ℃. Then, 0.12mL of 1M NaHCO was added3These cultures were adjusted to pH 8.0, after which ethyl palmitate was added directly to the medium to a final concentration of 23mg mL-1. These cultures were then shaken at 375rpm for an additional 4 days at 30 ℃ after which a sample of the whole fermentation broth from each culture was subjected to LCDA analysis according to the general method.
Twenty-four strains each causing transformation of strain D2300U1 with pZSCPn-3FAOBU (SEQ ID NO: 98) were cultured and analyzed by GC. Five of the twenty-four transformants produced C16 at more than 10.6 g/L: 0 LCDA. Specifically, transformants #11, #14, #18, and #21 produced C16 at 12.1g/L, 12.0g/L, 12.4g/L, and 10.6g/L, respectively: 0 LCDA. These four strains were designated as strains D2882, D2883, D2884, and D2885, respectively.
Strains D2882, D2883, D2884 and D2885 were also analyzed for LCDA production by flask assay according to the general method. As shown in Table 14, strains D2882, D2883, D2884, and D2885 produced C16 at approximately 15.1g/L, 13.2g/L, 15.0g/L, and 15.5g/L, respectively: 0 LCDA.
TABLE 14
LCDA production by strain D2882 and its homologues in a flask assay with ethyl palmitate as substrate
Strains D2882 and D2885 were further analyzed for LCDA production by micro-fermentation analysis according to the general procedure. As shown in Table 15, strains D2882 and D2885 produced C16 at approximately 23.4g/L and 21.0g/L, respectively: 0 LCDA.
Watch 15
LCDA production by strains D2882 and D2885 in a micro-fermentation assay with ethyl palmitate as substrate
It should be noted that the pZSCpn-3FAOBU (SEQ ID NO: 98) DNA used to transform D2300U1 to produce strain D2882 and its cognate could potentially knock out the yarrowia lipolytica SCP2 (sterol carrier protein) locus (Genbank accession number AJ431362, YALI0E01298 g). However, such knockouts were not confirmed in these strains. The genotype of strain D2882 and its cognates relative to wild-type yarrowia lipolytica ATCC # 20362 is dgat1-, dgat2-, Leu2+, pex3-, pox2-, pox3-, pox4-, Ura3+, unknown 1-, unknown 2-, unknown 3-, unknown 4-, FBA: : CtCPRs: : lip1, FBA: : VsCPRs: : lip1, FBAINm 1: : CtCYPA17 s: : pex20, CPR 1: : VsCYP94A1 s: : pex20, DG2 Pro-715: : CtALDH2 s: : lip1, FBA 1L: : CcFAO1 s: : aco; (ii) a YAT: : CtFAO1 sM: : pex20, FBA: : CcFAO1 s: : lip1, ALK2 LM-C: : CcFAO2 s: : aco 3.
Example 11
By passing through the tableProduction of yarrowia lipolytica Strain D3928 by Long-chain acyl-CoA synthetase
This example discloses the construction of yarrowia strain D3982 by expressing a codon-optimized sequence encoding a long-chain acyl-CoA synthetase (YLACOS-6P, SEQ ID NO: 44, see example 5). As shown in example 12, this strain was able to produce LCDA at over 100 g/L.
Strain D3928 was produced from strain D2882 as follows (fig. 7B).
First, strain D2882, which was Ura3+ due to transformation with pZSCpn-3FAOBU (SEQ ID NO: 98) DNA (see example 10), was represented as Ura 3-. Specifically, D2882 was transformed with plasmid pY117 for transient expression of Cre recombinase to excise the LoxP-flanked URA3 gene within strain D2882. The pY117 transformant failed to grow on MM, but could grow on MMU, indicating that the transformant lacks URA3 gene; this transformant was designated as strain D2882U.
To generate strain D3928, strain D2882U was transformed with a DNA fragment from construct pzP2-YLACoS-6Ps (FIG. 5C, SEQ ID NO: 65) comprising an expression cassette to overexpress a codon-optimized sequence encoding the enzyme YLACoS-6P (SEQ ID NO: 44). Specifically, the expression cassette comprises a nucleic acid sequence encoding SEQ ID NO: 44, long-chain acyl-CoA synthetase sequence YLACOS-6Ps (SEQ ID NO: 43). NcoI and NotI sites were added near the translation initiation codon (ATG) and after the stop codon, respectively, of the synthetic sequence encoding YLACOS-6P (SEQ ID NO: 44). The composition of the pZP2-YLACOS-6Ps plasmid (SEQ ID NO: 65) is further described in Table 16.
TABLE 16
Description of plasmid pZP2-YLACOS-6Ps (SEQ ID NO: 65)
Plasmid pZP2-YLACOS-6Ps (SEQ ID NO: 65) was digested with AscI/SphI and then used to transform strain D2882U according to the general method. The transformant cells were seeded on MM plates and maintained at 30 ℃ for 2 days. Will come fromIndividual colonies of each transformation were streaked again onto MM plates and then inoculated into liquid YPD2-B medium in 24-well blocks, which were then shaken at 375rpm for 20 hours at 30 ℃. Then, 0.12mL of 1M NaHCO was added3These cultures were adjusted to pH 8.0, after which ethyl palmitate was added directly to the medium to a final concentration of 23mg mL-1. These cultures were then shaken at 375rpm for an additional 4 days at 30 ℃ after which a sample of the whole fermentation broth from each culture was subjected to LCDA analysis according to the general method.
Twenty-four strains, each causing transformation of strain D2882U with pZP2-YLACOS-6Ps (SEQ ID NO: 65), were cultured and analyzed by GC. Nine of the twenty-four transformants produced C16 at more than 14.5 g/L: 0 LCDA. Specifically, transformants # 6, #7, #8, #9, #10, #11, #12, #13 and #20 produced C16 at 14.8g/L, 17.7g/L, 18.7g/L, 18.3g/L, 20.6g/L, 17.8g/L, 15.4g/L, 17.1g/L and 14.5g/L, respectively: 0 LCDA. These transformants were designated as strains D3924, D3925, D3926, D3927, D3928, D3929, D3930, D3931 and D3932, respectively.
The LCDA production of strains D3928, D3931 and D3932 was further analyzed by micro-fermentation analysis according to the general method. As shown in table 17, strains D3928, D3931, and D3932 produced C16 at approximately 23.0g/L, 21.2g/L, and 22.7g/L, respectively: 0 LCDA.
TABLE 17
LCDA by strains D3928, D3931 and D3932 in a micro-fermentation assay with ethyl palmitate as substrate
Production of
It should be noted that the pZP2-YLACoS-6Ps (SEQ ID NO: 65) DNA used to transform D2882U to produce strain D3928 and its cognate can potentially knock out the Pox2 gene (GenBank accession AJ 001300). However, such knockouts were not confirmed in these strains. The genotype of strain D3928 and its cognate relative to wild-type yarrowia lipolytica ATCC # 20362 is dgat1-, dgdt2-, Leu2+, pex3-, pox2-, pox3-, pox4-, Ura3+, unknown 1-, unknown 2-, unknown 3-, unknown 4-, unknown 5-, FBA: : CtCPRs: : lip1, FBA: : VsCPRs: : lip1, FBAINm 1: : CtCYPA17 s: : pex20, CPR 1: : VsCYP94A1 s: : pex20, DG2 Pro-715: : CtALDH2 s: : lip1, FBA 1L: : CcFAO1 s: : aco; YAT: : CtFAO1 sM: : pex20, FBA: : CcFAO1 s: : lip1, ALK2 LM-C: : CcFAO2 s: : aco3, FBAINm: : YLAcoS-6 Ps: : pex 20.
Thus, yarrowia strains are produced that overexpress long-chain acyl-coa synthetases and can synthesize significant amounts of LCDA product when fed with substrates containing long-chain fatty acids.
Example 12
LCDA production of yarrowia by overexpression of long-chain acyl-CoA synthetase under fed-batch fermentation conditions
Product produced by birth
This example discloses that yarrowia overexpressing long-chain acyl-coa synthetase can produce greater than 100g/L of LCDA product when grown in fed-batch fermentation. Specifically, after about 143 hours of fermentation, strain D3928 was able to produce C16 at 109 g/L: 0 LCDA and 119g/L total LCDA (Table 18, FIG. 13).
The seed culture scheme is as follows: engineered yarrowia strain D3928 stored at-80 ℃ was streaked onto YPD plates and incubated at 30 ℃ for about 24 hours. Single colonies were inoculated into 5mL of a complex medium (6.7g/L yeast nitrogen source without amino acids, 5g/L yeast extract, 20g/L D-glucose, 6g/L KH)2PO4、3.3g/L Na2HPO4·12H2O) 14-mL FALCON tubes (Corning, N.Y.). The test tube cultures were grown for about 24 hours at 30 ℃ with shaking at about 250 and 300 rpm. A portion of this culture (0.2-5.0mL) was transferred to a 250-mL flask containing 50mL of complex medium (described above) and incubated at 30 ℃ for an additional about 20 hours to OD600Is about 5.0-10.0. This culture was used as a seed culture to inoculate a 5-L fermentor at about 3% by volume.
5-L fermentation protocol: will be provided withThe shake flask seed culture prepared above was transferred to a 5-L fermenter (Sartorius BBI, biostate B plus) to start fermentation (t ═ 0 h). The fermentation medium contained 50g/L D-glucose, 6g/L KH2PO4、3.3g/L Na2HPO4·12H2O, 8mL/L trace metal (100X), 40g/L BactoTMYeast extract, 20g/L BactoTMPeptone, 20mM MgSO4, 6mg/L thiamine & HCl, and 15g/L (NH)4)2SO4. The trace metals (100X) are composed of 10g/L citric acid and 1.5g/L CaCl2·2H2O、10g/L FeSO4·7H2O、0.39g/L 10g/L ZnSO4·7H2O、0.38g/L CuSO4·5H2O、0.2g/L CoCl2·6H2O, and MnCl2·4H2And (C) O. The initial working volume was 3.0L. For the first 26 hours, the dissolved oxygen level (pO) was adjusted by cascading the stirring speed between 300rpm and 1200rpm2) Controlled at about 20% of air saturation. After 26h, the stirring speed was fixed at 1200rpm, and then pO was cascaded with pure oxygen supplement only2Control is at 60% of air saturation. Preparing a glucose feed comprising 700g/L glucose and 15-25g/L urea; when the initially added glucose was consumed, glucose feeding was started at about 18 hours. The glucose feed rate was initially as high as 20mL/hr and then gradually decreased to 10mL/hr at the end of fermentation (about 144 hours). Throughout the run, the aeration rate was controlled at 1.5-2.5L/min and the temperature was maintained at 30 ℃. The pH was controlled at 6.0 for the first 26 hours and then increased to 7.5 in the remainder of the run by KOH feeding. Starting from t ═ 28h, ethyl palmitate was fed to the fermentor to control the residual concentration within 1-20 g/L. Fermentation samples (approximately 25mL at each time point) were taken twice a day to analyze OD600Residual glucose in the fermentation medium, residual ethyl palmitate and LCDA.
5-L fermentation results: after 143.4 hours of fermentation, about 119g/L of LCDA was produced. Most of the LCDA products were hexadecanedioic acid (C16: 0 diacid) (table 18 and fig. 13).
Watch 18
LCDA produced by strain D3928 in a 5-L fed-batch fermentation with ethyl palmitate as substrate
Thus, yarrowia overexpressing long-chain acyl-coa synthetase can synthesize significant amounts of LCDA product when fed with substrates containing long-chain fatty acids.
Example 13
Yarrowia lipolytica strain D0145 was generated as a positive control for LCDA production
This example discloses the construction of various yarrowia strains by expressing codon-optimized sequences encoding certain Vicia sativa (nest) CYP and CPR enzymes. Most of these strains, including strain D0145, were able to produce LCDA.
Construct pZKLY-VsCPR & CYP (SEQ ID NO: 105) was generated to incorporate one copy of each of the codon optimized brassica CYP (VsCYP94A1s, from Arrowia Arrowiana, GenBank accession AAD10204, SEQ ID NO: 93 encoding SEQ ID NO: 94) and CPR (VsCPRs, from Arrowia Arrowiana, GenBank accession Z26252, SEQ ID NO: 95 encoding SEQ ID NO: 96) coding sequences. Each coding sequence is under the control of a heterologous promoter and 3' -termination sequence. NcoI and NotI endonuclease sites were added near the translation start codon (ATG) and after the stop codon, respectively, of each codon optimized sequence encoding VsCYP or VsCPR. The composition of the pZKLY-VsCPR & CYP (SEQ ID NO: 105) plasmid is further described in Table 19.
Watch 19
Plasmid pZKLY-VsCPR&Description of CYP (SEO ID NO: 105)
Digestion of plasmid pZKLY-VsCPR with AscI/SphI&CYP (SEQ ID NO: 105) and then used to transform strain D0004(dgat1-, dgat2-, pex3-, ura3-) (refer to Table 7) according to a general method. The transformant cells were seeded on MM plates and maintained at 30 ℃ for 2 days. Single colonies from each transformation were streaked again onto MM plates and then inoculated into liquid MM at 30 ℃ and shaken at 250rpm for 1 day. The overnight cultured cells were used to inoculate 50mL of liquid YPD2-B medium in a 250-mL baffled flask, then shaken at 250rpm at 30 ℃. After 24 hours, 2.0mL of 1M NaHCO was added3These cultures were adjusted to pH 8.0, after which ethyl palmitate was added directly to the medium to a final concentration of 16mg mL-1. These cultures were then shaken at 250rpm for an additional 4 days at 30 ℃ after which a sample of the whole fermentation broth from each flask culture was subjected to LCDA analysis according to the general method.
Each of forty-eight strains produced from parent strain D0004 transformed with pZKLY-VsCPR & CYP (SEQ ID NO: 105) was cultured and analyzed by GC. Almost all 48 strains produce C16 at over 3 g/L: 0 LCDA. For example, transformants #12, #15, #20, #23, #28, #29, #31, #37, #39, #44 and #48 produced C16 at 5.0g/L, 5.1g/L, 5.0g/L, 5.2g/L, 4.9g/L, 5.5g/L, 4.8g/L, 5.5g/L, 5.0g/L and 4.8g/L, respectively: 0 LCDA. These eleven transformants were designated as strains D0138, D0139, D0140, D0141, D0142, D0143, D0144, D0145, D0146, D0147 and D0148, respectively.
It should be noted that pZKLY-VsCPR & CYP (SEQ ID NO: 105) DNA used to transform D0004 to produce strain D0145 and its cognate could potentially knock out the lipase Y locus (GenBank accession AJ 549519). However, such knockouts were not confirmed in these strains. The genotype of strain D0145 and its cognate relative to wild-type yarrowia lipolytica ATCC # 20362 is Ura3+, dgat1-, dgat2-, pex3-, unknown 1-, FBA: : VsCPRs: : lip1, FBAINm: : VsCYP94A1 s: : pex 16.
Thus, a yeast (e.g., yarrowia) with up-regulated hydroxylase complex expression and down-regulated PEX3 expression can produce LCDA from a fatty acid-containing substrate.
Example 14
The pex 3-yarrowia genus can produce LCDA
This example discloses the construction of yarrowia strain D0101 by expression of codon-optimized sequences encoding candida tropicalis CYP and CPR enzymes. Moreover, this example discloses that the PEX 3-strain can produce LCDA, whereas the PEX3+ strain (e.g., a strain that does not have a PEX gene disruption, or that is PEX 10-or PEX 16-) does not have this ability.
The construct pZP2N-FCtA1R was generated to integrate one copy of each of the codon optimized CYP (CtALK1s, GenBank accession P10615) and CPR (CtCPRs, GenBank accession P37201) coding sequences from Candida tropicalis. Each coding sequence is under the control of a heterologous promoter and 3' -termination sequence. The NcoI and NotI endonuclease sites were added near the translation start codon (ATG) and after the stop codon, respectively, of each codon optimized sequence encoding ctak 1 or CtCPR. The composition of the pZP2N-FCtA1R plasmid is further described in Table 12.
Description of the plasmid pZP2N-FCtA1R
Plasmid pZP2N-FCtA1R was digested with AscI/SphI and then used to transform strains Y2224, D0003, D0004 and D0009 according to the general procedure. The transformant cells were seeded on MM plates and maintained at 30 ℃ for 2 days. Will be from each conversion of the sheetIndividual colonies were streaked again onto MM plates and then inoculated into liquid MM at 30 ℃ and shaken at 250rpm for 1 day. The overnight cultured cells were used to inoculate 25mL of liquid YPD4-B medium in 250-mL flasks, which were then shaken at 180rpm at 30 ℃. After 40 hours, 2.0mL of 1M NaHCO was added3These cultures were adjusted to pH 8.0 before ethyl palmitate (W2450, Sigma Aldrich) was added directly to the medium to a final concentration of 8mg mL-1. These cultures were then shaken at 180rpm for an additional 4 days at 30 ℃ after which a sample of the whole fermentation broth from each flask culture was subjected to LCDA analysis according to the general method.
Strains generated from each parent strain (Y2224, D0003, D0004, D0009) transformed with pZP2N-FCtA1R were analyzed by GC. No hexadecanedioic acid (C16: 0 LCDA) was detected in transformants of the parent strain Y2224, D0003, or D0009. However, transformants of parent strain D0004 produced more than 1g/L of C16: 0 LCDA. Production 1.24g/L C16: one D0004-transformant of 0 LCDA was designated strain D0101.
Flask analysis of strain D0101 was then performed. Specifically, D0101 was placed in a 25-mL culture in a 250-mL baffled flask, to which ethyl palmitate was added to a final concentration of 16mg mL-1. The culture was shaken at 180rpm for 4 days at 30 ℃. This culture produced C16 at about 5 g/L: 0 LCDA.
It should be noted that pZP2N-FCtA1R DNA used to transform D0004 to produce strain D0101 could potentially knock out the Pox2 gene (GenBank accession AJ 001300). However, such a knockout in D0101 was not confirmed. The genotype of strain D0101 relative to wild-type yarrowia lipolytica ATCC # 20362 is Ura3+, dgat1-, dgat2-, pex3-, unknown 1-, FBA 1: : CtALK1 s: : pex20, FBAINm: : CtCPRs: : pex 16.
It is notable that transformants of parent strain D0004(dgat1-, dgat2-, pex3-, ur α 3-) (e.g., strain D0101) produced LCDA, whereas transformants of parent strain D0009(dgat1-, dgat2-, pex10-, ura3-) did not have this ability. Although both types of transformants have (i) a down-regulated PEX gene (resulting in impaired peroxisome function and blocked beta-oxidation), and (ii) the same genotype under other conditions (including a down-regulated DGAT gene resulting in reduced oil storage), only yeasts with down-regulated PEX3 are able to produce LCDA. Similar to the pex 10-strain, the pex 16-strain also lacked the ability to produce LCDA (data not shown). Thus, blocking peroxisome function and β -oxidation has a significant impact on the production of LCDA from fatty acid containing substrates.
Thus, a yeast (e.g., yarrowia) with downregulated expression of PEX3 can produce LCDA from a fatty acid-containing substrate.
Sequence listing
<110> E.I. Moorel DuPont Co
Zhu, Quinn
<120> high-level production of long-chain dicarboxylic acid by microorganism
<130> CL6467
<150> US 62/195,340
<151> 2015-07-22
<150> US 62/195,338
<151> 2015-07-22
<160> 115
<170> PatentIn version 3.5
<210> 1
<211> 21
<212> DNA
<213> Artificial sequence
<220>
<223> primer 17864-900F
<400> 1
cacagaccgg cttctcaact t 21
<210> 2
<211> 22
<212> DNA
<213> Artificial sequence
<220>
<223> primer 17864-967R
<400> 2
aggtgaccat ctcgaacaca aa 22
<210> 3
<211> 21
<212> DNA
<213> Artificial sequence
<220>
<223> primer 5885-1034F
<400> 3
cttctccctg cgtcactctg t 21
<210> 4
<211> 20
<212> DNA
<213> Artificial sequence
<220>
<223> primer 5885-
<400> 4
<210> 5
<211> 18
<212> DNA
<213> Artificial sequence
<220>
<223> primer 14234-1341F
<400> 5
ggctccggct gagattga 18
<210> 6
<211> 23
<212> DNA
<213> Artificial sequence
<220>
<223> primer 14234-1404R
<400> 6
<210> 7
<211> 22
<212> DNA
<213> Artificial sequence
<220>
<223> primer 11979-
<400> 7
tcagctcaaa ctcgacgact tg 22
<210> 8
<211> 20
<212> DNA
<213> Artificial sequence
<220>
<223> primer 11979-
<400> 8
<210> 9
<211> 21
<212> DNA
<213> Artificial sequence
<220>
<223> primer 7755-
<400> 9
ttacagctcg ttgccctacc a 21
<210> 10
<211> 15
<212> DNA
<213> Artificial sequence
<220>
<223> primer 7755-
<400> 10
tggcgggcga aatgg 15
<210> 11
<211> 18
<212> DNA
<213> Artificial sequence
<220>
<223> primer 12419-
<400> 11
tgctggcatc gtggtgat 18
<210> 12
<211> 21
<212> DNA
<213> Artificial sequence
<220>
<223> primer 12419-
<400> 12
gcaacaatcg tcgcagaatc t 21
<210> 13
<211> 18
<212> DNA
<213> Artificial sequence
<220>
<223> primer 20405-626F
<400> 13
ccgtggagct cacccatt 18
<210> 14
<211> 24
<212> DNA
<213> Artificial sequence
<220>
<223> primer 20405-
<400> 14
ggttaggtgc attctttgct gtct 24
<210> 15
<211> 22
<212> DNA
<213> Artificial sequence
<220>
<223> primer 5456-
<400> 15
ctctgctgct atggttgtcg at 22
<210> 16
<211> 21
<212> DNA
<213> Artificial sequence
<220>
<223> primer 5456-
<400> 16
tgcaaccctc atcaccagtt c 21
<210> 17
<211> 17
<212> DNA
<213> Artificial sequence
<220>
<223> primer 15103-
<400> 17
caaggccgtg cgtgtca 17
<210> 18
<211> 20
<212> DNA
<213> Artificial sequence
<220>
<223> primer 15103-
<400> 18
<210> 19
<211> 19
<212> DNA
<213> Artificial sequence
<220>
<223> primer 5951-
<400> 19
gcattttgcc gcacttgat 19
<210> 20
<211> 18
<212> DNA
<213> Artificial sequence
<220>
<223> primer 5951-
<400> 20
gacgagctcc gccacagt 18
<210> 21
<211> 20
<212> DNA
<213> Artificial sequence
<220>
<223> primer 17314-47F
<400> 21
<210> 22
<211> 22
<212> DNA
<213> Artificial sequence
<220>
<223> primer 17314-
<400> 22
cacttgtttt ggagctcttg ga 22
<210> 23
<211> 20
<212> DNA
<213> Artificial sequence
<220>
<223> primer 6556-
<400> 23
<210> 24
<211> 20
<212> DNA
<213> Artificial sequence
<220>
<223> primer 6556-
<400> 24
<210> 25
<211> 23
<212> DNA
<213> Artificial sequence
<220>
<223> primer 12859-
<400> 25
ccagattctg ctgaacacaa aga 23
<210> 26
<211> 20
<212> DNA
<213> Artificial sequence
<220>
<223> primer 12859-1071
<400> 26
<210> 27
<211> 21
<212> DNA
<213> Artificial sequence
<220>
<223> primer 9284-
<400> 27
tctgcttgtt gacgaccgaa t 21
<210> 28
<211> 21
<212> DNA
<213> Artificial sequence
<220>
<223> primer 9284 and 995R
<400> 28
gggttgttca ccagcatgtt g 21
<210> 29
<211> 20
<212> DNA
<213> Artificial sequence
<220>
<223> primer 16016-
<400> 29
<210> 30
<211> 22
<212> DNA
<213> Artificial sequence
<220>
<223> primer/Probe 16016-1422T
<400> 30
catcctggcc acccgacaga cc 22
<210> 31
<211> 22
<212> DNA
<213> Artificial sequence
<220>
<223> primer YL-18S-329F
<400> 31
cctgagaaac ggctaccaca tc 22
<210> 32
<211> 21
<212> DNA
<213> Artificial sequence
<220>
<223> primer YL-18S-395R
<400> 32
ccctgtgtca ggattgggta a 21
<210> 33
<211> 700
<212> PRT
<213> Saccharomyces cerevisiae
<400> 33
Met Val Ala Gln Tyr Thr Val Pro Val Gly Lys Ala Ala Asn Glu His
1 5 10 15
Glu Thr Ala Pro Arg Arg Asn Tyr Gln Cys Arg Glu Lys Pro Leu Val
20 25 30
Arg Pro Pro Asn Thr Lys Cys Ser Thr Val Tyr Glu Phe Val Leu Glu
35 40 45
Cys Phe Gln Lys Asn Lys Asn Ser Asn Ala Met Gly Trp Arg Asp Val
50 55 60
Lys Glu Ile His Glu Glu Ser Lys Ser Val Met Lys Lys Val Asp Gly
65 70 75 80
Lys Glu Thr Ser Val Glu Lys Lys Trp Met Tyr Tyr Glu Leu Ser His
85 90 95
Tyr His Tyr Asn Ser Phe Asp Gln Leu Thr Asp Ile Met His Glu Ile
100 105 110
Gly Arg Gly Leu Val Lys Ile Gly Leu Lys Pro Asn Asp Asp Asp Lys
115 120 125
Leu His Leu Tyr Ala Ala Thr Ser His Lys Trp Met Lys Met Phe Leu
130 135 140
Gly Ala Gln Ser Gln Gly Ile Pro Val Val Thr Ala Tyr Asp Thr Leu
145 150 155 160
Gly Glu Lys Gly Leu Ile His Ser Leu Val Gln Thr Gly Ser Lys Ala
165 170 175
Ile Phe Thr Asp Asn Ser Leu Leu Pro Ser Leu Ile Lys Pro Val Gln
180 185 190
Ala Ala Gln Asp Val Lys Tyr Ile Ile His Phe Asp Ser Ile Ser Ser
195 200 205
Glu Asp Arg Arg Gln Ser Gly Lys Ile Tyr Gln Ser Ala His Asp Ala
210 215 220
Ile Asn Arg Ile Lys Glu Val Arg Pro Asp Ile Lys Thr Phe Ser Phe
225 230 235 240
Asp Asp Ile Leu Lys Leu Gly Lys Glu Ser Cys Asn Glu Ile Asp Val
245 250 255
His Pro Pro Gly Lys Asp Asp Leu Cys Cys Ile Met Tyr Thr Ser Gly
260 265 270
Ser Thr Gly Glu Pro Lys Gly Val Val Leu Lys His Ser Asn Val Val
275 280 285
Ala Gly Val Gly Gly Ala Ser Leu Asn Val Leu Lys Phe Val Gly Asn
290 295 300
Thr Asp Arg Val Ile Cys Phe Leu Pro Leu Ala His Ile Phe Glu Leu
305 310 315 320
Val Phe Glu Leu Leu Ser Phe Tyr Trp Gly Ala Cys Ile Gly Tyr Ala
325 330 335
Thr Val Lys Thr Leu Thr Ser Ser Ser Val Arg Asn Cys Gln Gly Asp
340 345 350
Leu Gln Glu Phe Lys Pro Thr Ile Met Val Gly Val Ala Ala Val Trp
355 360 365
Glu Thr Val Arg Lys Gly Ile Leu Asn Gln Ile Asp Asn Leu Pro Phe
370 375 380
Leu Thr Lys Lys Ile Phe Trp Thr Ala Tyr Asn Thr Lys Leu Asn Met
385 390 395 400
Gln Arg Leu His Ile Pro Gly Gly Gly Ala Leu Gly Asn Leu Val Phe
405 410 415
Lys Lys Ile Arg Thr Ala Thr Gly Gly Gln Leu Arg Tyr Leu Leu Asn
420 425 430
Gly Gly Ser Pro Ile Ser Arg Asp Ala Gln Glu Phe Ile Thr Asn Leu
435 440 445
Ile Cys Pro Met Leu Ile Gly Tyr Gly Leu Thr Glu Thr Cys Ala Ser
450 455 460
Thr Thr Ile Leu Asp Pro Ala Asn Phe Glu Leu Gly Val Ala Gly Asp
465 470 475 480
Leu Thr Gly Cys Val Thr Val Lys Leu Val Asp Val Glu Glu Leu Gly
485 490 495
Tyr Phe Ala Lys Asn Asn Gln Gly Glu Val Trp Ile Thr Gly Ala Asn
500 505 510
Val Thr Pro Glu Tyr Tyr Lys Asn Glu Glu Glu Thr Ser Gln Ala Leu
515 520 525
Thr Ser Asp Gly Trp Phe Lys Thr Gly Asp Ile Gly Glu Trp Glu Ala
530 535 540
Asn Gly His Leu Lys Ile Ile Asp Arg Lys Lys Asn Leu Val Lys Thr
545 550 555 560
Met Asn Gly Glu Tyr Ile Ala Leu Glu Lys Leu Glu Ser Val Tyr Arg
565 570 575
Ser Asn Glu Tyr Val Ala Asn Ile Cys Val Tyr Ala Asp Gln Ser Lys
580 585 590
Thr Lys Pro Val Gly Ile Ile Val Pro Asn His Ala Pro Leu Thr Lys
595 600 605
Leu Ala Lys Lys Leu Gly Ile Met Glu Gln Lys Asp Ser Ser Ile Asn
610 615 620
Ile Glu Asn Tyr Leu Glu Asp Ala Lys Leu Ile Lys Ala Val Tyr Ser
625 630 635 640
Asp Leu Leu Lys Thr Gly Lys Asp Gln Gly Leu Val Gly Ile Glu Leu
645 650 655
Leu Ala Gly Ile Val Phe Phe Asp Gly Glu Trp Thr Pro Gln Asn Gly
660 665 670
Phe Val Thr Ser Ala Gln Lys Leu Lys Arg Lys Asp Ile Leu Asn Ala
675 680 685
Val Lys Asp Lys Val Asp Ala Val Tyr Ser Ser Ser
690 695 700
<210> 34
<211> 744
<212> PRT
<213> Saccharomyces cerevisiae
<400> 34
Met Ala Ala Pro Asp Tyr Ala Leu Thr Asp Leu Ile Glu Ser Asp Pro
1 5 10 15
Arg Phe Glu Ser Leu Lys Thr Arg Leu Ala Gly Tyr Thr Lys Gly Ser
20 25 30
Asp Glu Tyr Ile Glu Glu Leu Tyr Ser Gln Leu Pro Leu Thr Ser Tyr
35 40 45
Pro Arg Tyr Lys Thr Phe Leu Lys Lys Gln Ala Val Ala Ile Ser Asn
50 55 60
Pro Asp Asn Glu Ala Gly Phe Ser Ser Ile Tyr Arg Ser Ser Leu Ser
65 70 75 80
Ser Glu Asn Leu Val Ser Cys Val Asp Lys Asn Leu Arg Thr Ala Tyr
85 90 95
Asp His Phe Met Phe Ser Ala Arg Arg Trp Pro Gln Arg Asp Cys Leu
100 105 110
Gly Ser Arg Pro Ile Asp Lys Ala Thr Gly Thr Trp Glu Glu Thr Phe
115 120 125
Arg Phe Glu Ser Tyr Ser Thr Val Ser Lys Arg Cys His Asn Ile Gly
130 135 140
Ser Gly Ile Leu Ser Leu Val Asn Thr Lys Arg Lys Arg Pro Leu Glu
145 150 155 160
Ala Asn Asp Phe Val Val Ala Ile Leu Ser His Asn Asn Pro Glu Trp
165 170 175
Ile Leu Thr Asp Leu Ala Cys Gln Ala Tyr Ser Leu Thr Asn Thr Ala
180 185 190
Leu Tyr Glu Thr Leu Gly Pro Asn Thr Ser Glu Tyr Ile Leu Asn Leu
195 200 205
Thr Glu Ala Pro Ile Leu Ile Phe Ala Lys Ser Asn Met Tyr His Val
210 215 220
Leu Lys Met Val Pro Asp Met Lys Phe Val Asn Thr Leu Val Cys Met
225 230 235 240
Asp Glu Leu Thr His Asp Glu Leu Arg Met Leu Asn Glu Ser Leu Leu
245 250 255
Pro Val Lys Cys Asn Ser Leu Asn Glu Lys Ile Thr Phe Phe Ser Leu
260 265 270
Glu Gln Val Glu Gln Val Gly Cys Phe Asn Lys Ile Pro Ala Ile Pro
275 280 285
Pro Thr Pro Asp Ser Leu Tyr Thr Ile Ser Phe Thr Ser Gly Thr Thr
290 295 300
Gly Leu Pro Lys Gly Val Glu Met Ser His Arg Asn Ile Ala Ser Gly
305 310 315 320
Ile Ala Phe Ala Phe Ser Thr Phe Arg Ile Pro Pro Asp Lys Arg Asn
325 330 335
Gln Gln Leu Tyr Asp Met Cys Phe Leu Pro Leu Ala His Ile Phe Glu
340 345 350
Arg Met Val Ile Ala Tyr Asp Leu Ala Ile Gly Phe Gly Ile Gly Phe
355 360 365
Leu His Lys Pro Asp Pro Thr Val Leu Val Glu Asp Leu Lys Ile Leu
370 375 380
Lys Pro Tyr Ala Val Ala Leu Val Pro Arg Ile Leu Thr Arg Phe Glu
385 390 395 400
Ala Gly Ile Lys Asn Ala Leu Asp Lys Ser Thr Val Gln Arg Asn Val
405 410 415
Ala Asn Thr Ile Leu Asp Ser Lys Ser Ala Arg Phe Thr Ala Arg Gly
420 425 430
Gly Pro Asp Lys Ser Ile Met Asn Phe Leu Val Tyr His Arg Val Leu
435 440 445
Ile Asp Lys Ile Arg Asp Ser Leu Gly Leu Ser Asn Asn Ser Phe Ile
450 455 460
Ile Thr Gly Ser Ala Pro Ile Ser Lys Asp Thr Leu Leu Phe Leu Arg
465 470 475 480
Ser Ala Leu Asp Ile Gly Ile Arg Gln Gly Tyr Gly Leu Thr Glu Thr
485 490 495
Phe Ala Gly Val Cys Leu Ser Glu Pro Phe Glu Lys Asp Val Gly Ser
500 505 510
Cys Gly Ala Ile Gly Ile Ser Ala Glu Cys Arg Leu Lys Ser Val Pro
515 520 525
Glu Met Gly Tyr His Ala Asp Lys Asp Leu Lys Gly Glu Leu Gln Ile
530 535 540
Arg Gly Pro Gln Val Phe Glu Arg Tyr Phe Lys Asn Pro Asn Glu Thr
545 550 555 560
Ser Lys Ala Val Asp Gln Asp Gly Trp Phe Ser Thr Gly Asp Val Ala
565 570 575
Phe Ile Asp Gly Lys Gly Arg Ile Ser Val Ile Asp Arg Val Lys Asn
580 585 590
Phe Phe Lys Leu Ala His Gly Glu Tyr Ile Ala Pro Glu Lys Ile Glu
595 600 605
Asn Ile Tyr Leu Ser Ser Cys Pro Tyr Ile Thr Gln Ile Phe Val Phe
610 615 620
Gly Asp Pro Leu Lys Thr Phe Leu Val Gly Ile Val Gly Val Asp Val
625 630 635 640
Asp Ala Ala Gln Pro Ile Leu Ala Ala Lys His Pro Glu Val Lys Thr
645 650 655
Trp Thr Lys Glu Val Leu Val Glu Asn Leu Asn Arg Asn Lys Lys Leu
660 665 670
Arg Lys Glu Phe Leu Asn Lys Ile Asn Lys Cys Thr Asp Gly Leu Gln
675 680 685
Gly Phe Glu Lys Leu His Asn Ile Lys Val Gly Leu Glu Pro Leu Thr
690 695 700
Leu Glu Asp Asp Val Val Thr Pro Thr Phe Lys Ile Lys Arg Ala Lys
705 710 715 720
Ala Ser Lys Phe Phe Lys Asp Thr Leu Asp Gln Leu Tyr Ala Glu Gly
725 730 735
Ser Leu Val Lys Thr Glu Lys Leu
740
<210> 35
<211> 2076
<212> DNA
<213> Artificial sequence
<220>
<223> YlFaa1
<400> 35
atggtcggat acaccatctc ctcgaagccc gtgtccgtcg aggttggccc cgccaagcct 60
ggcgagactg ctccccgacg gaacgtcatt gccaaggacg ctcctgtggt cttccccgac 120
aacgattcgt ccctcaccac tgtctacaag ctgttcaaaa agtacgccga gatcaactcc 180
gaacgaaagg ctatgggatg gcgagacacc atcgacattc acgtggagac caagcaggtc 240
acaaaggtgg tcgacggcgt ggagaagaaa gtgcccaagg aatggaagta cttcgagatg 300
ggtccttaca agtggctgtc ctacaaggag gccctcaagc tggttcacga ttatggagct 360
ggtcttcgac atctcggcat caagcccaaa gagaagatgc acatttacgc acagacctct 420
caccgatgga tgctttccgg actggcctct ctctcgcagg gcattcccat cgtcactgcc 480
tacgacaccc ttggagagga aggtctcaca cgatctctgc aggagaccaa ctccgtcatc 540
atgttcacgg acaaggctct tctgtcgtct ctcaaggtgt ccctcaaaaa gggcaccgat 600
ctgcgaatca ttatctacgg aggcgacctg actcccgatg acaagaaagc cggaaacacc 660
gagatcgacg ccatcaagga gattgttcca gacatgaaga tctacactat ggacgaggtt 720
gtcgctctcg gtcgagagca tcctcacccc gtggaagagg tcgactacga ggatctggcc 780
ttcatcatgt acacctctgg ctccacagga gttcccaagg gtgtcgtgct gcagcacaag 840
cagatcctcg cctctgtggc cggtgtcacc aagattatcg acagatccat tatcggcaat 900
acagatcgac tgctcaactt tcttcccctc gcacacatct tcgagtttgt gttcgagatg 960
gtcaccttct ggtggggtgc ctctctgggc tacggaactg tcaagaccat ttccgacctg 1020
tcgatgaaga actgcaaggg agacatccga gagctcaagc ccaccatcat ggtcggcgtt 1080
ccagctgtct gggaacccat gcggaagggt attcttggca aaatcaagga gctgtctcct 1140
ctcatgcagc gagtcttctg ggcctccttt gctgccaagc aacgtctcga cgagaacgga 1200
cttcccggtg gctctattct ggattcgctc atcttcaaga aagtcaagga cgccactgga 1260
ggctgtctcc gatacgtgtg caacggaggt gctccagttt ccgtcgacac ccagaagttc 1320
attactaccc ttatctgtcc catgctcatt ggatgcggtc tgaccgagac tacagccaac 1380
accactatca tgtctcccaa gtcctatgcc tttggcacca ttggagagcc tactgcagcc 1440
gtcaccctca agcttatcga cgtgcccgaa gctggctact tcgccgagaa caatcaggga 1500
gagctgtgca tcaagggcaa cgtggtcatg aaggagtatt acaagaacga ggaagagacc 1560
aagaaagcgt tctccgacga tggctacttt ctcaccggag acattgccga gtggactgcc 1620
aatggtcagc ttcgaattat cgacagacga aagaacctcg tcaagaccca gaacggagag 1680
tacattgctc tggagaagct cgaaacacag taccgatcgt cttcctacgt tgccaacctg 1740
tgcgtctacg ccgaccagaa ccgagtcaag cccatcgctc tggtcattcc caacgagggt 1800
cctaccaaaa agcttgccca gagcttgggc gtggattccg acgactggga tgccgtctgt 1860
tccaacaaga aagtggtcaa ggctgttctc aaggacatgc tggataccgg acgatctctc 1920
ggtctgtccg gcatcgagct gctgcaagga atcgtgttgc tgcctggcga gtggactccc 1980
cagaacagct acctcaccgc tgcccagaag ctcaaccgaa agaagattgt cgatgacaac 2040
aaaaaggaga tcgacgagtg ctacgagcag tcctaa 2076
<210> 36
<211> 691
<212> PRT
<213> yarrowia lipolytica
<400> 36
Met Val Gly Tyr Thr Ile Ser Ser Lys Pro Val Ser Val Glu Val Gly
1 5 10 15
Pro Ala Lys Pro Gly Glu Thr Ala Pro Arg Arg Asn Val Ile Ala Lys
20 25 30
Asp Ala Pro Val Val Phe Pro Asp Asn Asp Ser Ser Leu Thr Thr Val
35 40 45
Tyr Lys Leu Phe Lys Lys Tyr Ala Glu Ile Asn Ser Glu Arg Lys Ala
50 55 60
Met Gly Trp Arg Asp Thr Ile Asp Ile His Val Glu Thr Lys Gln Val
65 70 75 80
Thr Lys Val Val Asp Gly Val Glu Lys Lys Val Pro Lys Glu Trp Lys
85 90 95
Tyr Phe Glu Met Gly Pro Tyr Lys Trp Leu Ser Tyr Lys Glu Ala Leu
100 105 110
Lys Leu Val His Asp Tyr Gly Ala Gly Leu Arg His Leu Gly Ile Lys
115 120 125
Pro Lys Glu Lys Met His Ile Tyr Ala Gln Thr Ser His Arg Trp Met
130 135 140
Leu Ser Gly Leu Ala Ser Leu Ser Gln Gly Ile Pro Ile Val Thr Ala
145 150 155 160
Tyr Asp Thr Leu Gly Glu Glu Gly Leu Thr Arg Ser Leu Gln Glu Thr
165 170 175
Asn Ser Val Ile Met Phe Thr Asp Lys Ala Leu Leu Ser Ser Leu Lys
180 185 190
Val Ser Leu Lys Lys Gly Thr Asp Leu Arg Ile Ile Ile Tyr Gly Gly
195 200 205
Asp Leu Thr Pro Asp Asp Lys Lys Ala Gly Asn Thr Glu Ile Asp Ala
210 215 220
Ile Lys Glu Ile Val Pro Asp Met Lys Ile Tyr Thr Met Asp Glu Val
225 230 235 240
Val Ala Leu Gly Arg Glu His Pro His Pro Val Glu Glu Val Asp Tyr
245 250 255
Glu Asp Leu Ala Phe Ile Met Tyr Thr Ser Gly Ser Thr Gly Val Pro
260 265 270
Lys Gly Val Val Leu Gln His Lys Gln Ile Leu Ala Ser Val Ala Gly
275 280 285
Val Thr Lys Ile Ile Asp Arg Ser Ile Ile Gly Asn Thr Asp Arg Leu
290 295 300
Leu Asn Phe Leu Pro Leu Ala His Ile Phe Glu Phe Val Phe Glu Met
305 310 315 320
Val Thr Phe Trp Trp Gly Ala Ser Leu Gly Tyr Gly Thr Val Lys Thr
325 330 335
Ile Ser Asp Leu Ser Met Lys Asn Cys Lys Gly Asp Ile Arg Glu Leu
340 345 350
Lys Pro Thr Ile Met Val Gly Val Pro Ala Val Trp Glu Pro Met Arg
355 360 365
Lys Gly Ile Leu Gly Lys Ile Lys Glu Leu Ser Pro Leu Met Gln Arg
370 375 380
Val Phe Trp Ala Ser Phe Ala Ala Lys Gln Arg Leu Asp Glu Asn Gly
385 390 395 400
Leu Pro Gly Gly Ser Ile Leu Asp Ser Leu Ile Phe Lys Lys Val Lys
405 410 415
Asp Ala Thr Gly Gly Cys Leu Arg Tyr Val Cys Asn Gly Gly Ala Pro
420 425 430
Val Ser Val Asp Thr Gln Lys Phe Ile Thr Thr Leu Ile Cys Pro Met
435 440 445
Leu Ile Gly Cys Gly Leu Thr Glu Thr Thr Ala Asn Thr Thr Ile Met
450 455 460
Ser Pro Lys Ser Tyr Ala Phe Gly Thr Ile Gly Glu Pro Thr Ala Ala
465 470 475 480
Val Thr Leu Lys Leu Ile Asp Val Pro Glu Ala Gly Tyr Phe Ala Glu
485 490 495
Asn Asn Gln Gly Glu Leu Cys Ile Lys Gly Asn Val Val Met Lys Glu
500 505 510
Tyr Tyr Lys Asn Glu Glu Glu Thr Lys Lys Ala Phe Ser Asp Asp Gly
515 520 525
Tyr Phe Leu Thr Gly Asp Ile Ala Glu Trp Thr Ala Asn Gly Gln Leu
530 535 540
Arg Ile Ile Asp Arg Arg Lys Asn Leu Val Lys Thr Gln Asn Gly Glu
545 550 555 560
Tyr Ile Ala Leu Glu Lys Leu Glu Thr Gln Tyr Arg Ser Ser Ser Tyr
565 570 575
Val Ala Asn Leu Cys Val Tyr Ala Asp Gln Asn Arg Val Lys Pro Ile
580 585 590
Ala Leu Val Ile Pro Asn Glu Gly Pro Thr Lys Lys Leu Ala Gln Ser
595 600 605
Leu Gly Val Asp Ser Asp Asp Trp Asp Ala Val Cys Ser Asn Lys Lys
610 615 620
Val Val Lys Ala Val Leu Lys Asp Met Leu Asp Thr Gly Arg Ser Leu
625 630 635 640
Gly Leu Ser Gly Ile Glu Leu Leu Gln Gly Ile Val Leu Leu Pro Gly
645 650 655
Glu Trp Thr Pro Gln Asn Ser Tyr Leu Thr Ala Ala Gln Lys Leu Asn
660 665 670
Arg Lys Lys Ile Val Asp Asp Asn Lys Lys Glu Ile Asp Glu Cys Tyr
675 680 685
Glu Gln Ser
690
<210> 37
<211> 574
<212> PRT
<213> yarrowia lipolytica
<400> 37
Met Thr Thr Ile Ile His Lys Ser Thr Phe Pro Asp Ile Glu Leu Phe
1 5 10 15
Gln Gly Ser Ile Thr Asp Phe Ile Arg Thr Gly Ala Tyr Ala Glu Asp
20 25 30
Asp Tyr Lys Pro Cys Met Ile Asp Ala Glu Thr Gly Glu Gln Leu Thr
35 40 45
Gln Lys Gln Ile Leu Asp Cys Ala Asp Gln Phe Arg Ser Leu Leu Tyr
50 55 60
Gln His Gly Val Gln Lys Thr Pro Asn Arg Asp Glu Arg Ile Gly Asp
65 70 75 80
Val Val Ile Pro Phe Ile Asp Asn Asn Ile Tyr Leu Pro Ala Ile His
85 90 95
Tyr Ala Cys Leu Glu Leu Gly Cys Cys Met Asn Pro Ala Ser Thr Gln
100 105 110
Gln Thr Pro Leu Glu Leu Ser Lys Gln Ile Arg Val Thr Asp Pro Lys
115 120 125
Val Ile Ile Tyr Gln Arg Lys Tyr Arg Lys Thr Val Met His Ala Ile
130 135 140
Asp Leu Val Cys Tyr Thr Asn Phe Pro Ile Val Ile Glu Phe Glu Thr
145 150 155 160
Met Leu Phe Leu Arg Asn Ser Val Pro Pro Pro Pro Lys Lys Ala Lys
165 170 175
Phe His Ile Thr Ser Thr Glu Gln Ala Arg Lys Arg Ile Ala Tyr Leu
180 185 190
Gly Met Ser Ser Gly Thr Ser Gly Lys Ser Lys Ala Val Arg Leu Ser
195 200 205
His His Asn Ile Val Ala Cys Ser Gln Val Ser Gln Val Thr Phe Pro
210 215 220
Ala Leu Tyr Lys Ala Ser Asn Val Cys Val Ala Val Leu Pro Ser Cys
225 230 235 240
His Val Phe Gly Leu Tyr Ile Phe Phe Met Val Leu Pro Arg Ser Gly
245 250 255
Gly Thr Thr Ile Met His Thr Lys Phe Asp Leu Lys Gln Leu Leu Glu
260 265 270
Ser Gln Lys Lys Tyr Lys Ala Asn Phe Leu Pro Leu Val Pro Pro Ile
275 280 285
Ala Val Gln Leu Ala Lys Asn Pro Met Val Lys Asn Tyr Ala Asp Ser
290 295 300
Leu Lys Gln Val Lys Leu Ile Met Ser Ala Ala Ala Pro Leu Gly Ala
305 310 315 320
Glu Val Thr Gln Ser Leu Ile Lys Ala Ile Gly Pro Gln Val Arg Val
325 330 335
Val Gln Gly Tyr Gly Met Thr Glu Thr Ser Pro Cys Val Thr Leu Phe
340 345 350
Asp Pro Ala Asp Pro His Leu His Ile Lys Ala Cys Gly Lys Leu Val
355 360 365
Pro Asn Cys Glu Val Arg Ile Val Ala Asp Gly Val Asp Gln Pro Ala
370 375 380
Tyr Ser Gly Ser Val Ser Asp Val Ala Lys Asn Lys Thr Asp Asn Leu
385 390 395 400
Pro Val Gly Glu Ile Trp Val Arg Gly Pro Gln Val Met Asp Gly Tyr
405 410 415
His Lys Asn Lys Ser Ala Thr Ser Glu Ala Phe Val Glu Ala Asn Asp
420 425 430
Ser Ser Val Cys Tyr Asn Thr Lys Trp Leu Arg Thr Gly Asp Val Gly
435 440 445
Leu Val Asp Ser Leu Gly Arg Phe Met Ile Val Asp Arg Thr Lys Glu
450 455 460
Met Ile Lys Ser Met Ser Lys Gln Val Ala Pro Ala Glu Leu Glu Asp
465 470 475 480
Met Leu Leu Ala His Ala Asp Val Ser Asp Ala Ala Val Ile Gly Val
485 490 495
Glu Asn Glu Ala Lys Gly Thr Glu Gln Ile Arg Ala Phe Leu Val Leu
500 505 510
Lys Lys Gly Gly Asp Ala Leu Glu Val Lys Lys Trp Met Asp Ser Lys
515 520 525
Leu Pro Lys Tyr Lys Gln Leu His Gly Gly Val Val Val Ile Asp Gln
530 535 540
Ile Pro Lys Ser Gln Ala Gly Lys Ile Leu Arg Arg Met Leu Arg Leu
545 550 555 560
Arg Gln Asp Asp Val Val Leu Gly Thr Asp Gln Ala Lys Leu
565 570
<210> 38
<211> 1647
<212> DNA
<213> Artificial sequence
<220>
<223> YlACoS-3P
<400> 38
atggccatca tccactccac cggaactctg cccatcttca acggtaccgt caccgattac 60
ctgcgaacaa agccttctta ctcgtccaca gatccagcct acatcgacgt ggttacaggc 120
aactctatca gctactccga ggtctggaag cttgccgacc gactctcctc tgctctgtac 180
aacgactacg gactcaccga cgccaagccc gacgagaatg tgggtcctgt tgtcatgctg 240
cacgctgtca attcgcctct cctggcatct gttcactacg ctcttctgga tctgggcgtc 300
acaatcactc ccgcagctgc cacctacgag gctggcgatc tcgcacatca aatcaaggtg 360
tgctctccgt ccctggtcat ttgcaaccag cagttcgaac ccaaggtcaa atctgcctcc 420
agcaacacca agctcatttt catcgaggat ctgctcaaaa cccagtcgtc tgctccctgg 480
aaaaagttca ctacctccaa ccccaaccga gttgcctacc tgggcatgtc cagtggaacc 540
tctggtctcc ccaaggcggt tcaacagacc cacatcaaca tgtcgtcttc caccgaagcc 600
gtcatttcct ctcagaccat cttcagcgct cgaaagaacg tcaccgcagc cattgtgccc 660
atgactcatg tctacggact caccaagttt gttttccact ctgtcgcagg ctcaatgacc 720
accgttgtgt tccccaagtt ctccctggtc gacctcctgg aggcccagat caagtacaag 780
atcaacattc tgtatctggt tcctccagtg gtcttggctc tggccaagga ctctcgtgta 840
cagccctaca tcaagtccat ttgcgagctc accactctga ttgccactgg tgcggctccc 900
cttcctccca ctgcaggcga cgcccttctg gagcgactta cgggcaacaa agagggaaac 960
agagacaacg gtatggatcc cttggttctc atccagggct acggactcac agagactctc 1020
caggtgtctg tcttcaagcc agaggatccc gaacgagatc tcaagaccgt gggcaaactg 1080
cttcccaaca ccgaggttcg aattgtcggc gagaagggag atgttccgcg ttccaaatgg 1140
tcgtttgtca ctcctccaac cggcgaaatc tacattcgag gtccccacgt gactcctggt 1200
tacttcaaca acgactctgc caactctgag tcctttgacg gcgagtggct caagaccggc 1260
gatatcggat acatggacct ggaaggtcga ctcaccattg tggaccgaaa caaggagatg 1320
atcaaggtca acggacgtca ggttgctcct gccgagatcg aatctgtgct gctgggtcat 1380
cctatggtca aggatgtggc cgtcattgga gtcaccaatc ccgacagagg cacggagtct 1440
gctcgggcgt ttcttgttac tgaagctcga gctctccctg tcatcaagca gtggtttgac 1500
cgtcgagttc cctcctacaa gcgactttac ggaggcattg tggttgtcga tgccattccc 1560
aagtctgcct cgggcaagat tctgcgacgg gtcctcagag agcgaaaggg cgactccgtg 1620
tttggagagt atgtcgagga agtctaa 1647
<210> 39
<211> 548
<212> PRT
<213> Artificial sequence
<220>
<223> YLACoS-3P protein
<400> 39
Met Ala Ile Ile His Ser Thr Gly Thr Leu Pro Ile Phe Asn Gly Thr
1 5 10 15
Val Thr Asp Tyr Leu Arg Thr Lys Pro Ser Tyr Ser Ser Thr Asp Pro
20 25 30
Ala Tyr Ile Asp Val Val Thr Gly Asn Ser Ile Ser Tyr Ser Glu Val
35 40 45
Trp Lys Leu Ala Asp Arg Leu Ser Ser Ala Leu Tyr Asn Asp Tyr Gly
50 55 60
Leu Thr Asp Ala Lys Pro Asp Glu Asn Val Gly Pro Val Val Met Leu
65 70 75 80
His Ala Val Asn Ser Pro Leu Leu Ala Ser Val His Tyr Ala Leu Leu
85 90 95
Asp Leu Gly Val Thr Ile Thr Pro Ala Ala Ala Thr Tyr Glu Ala Gly
100 105 110
Asp Leu Ala His Gln Ile Lys Val Cys Ser Pro Ser Leu Val Ile Cys
115 120 125
Asn Gln Gln Phe Glu Pro Lys Val Lys Ser Ala Ser Ser Asn Thr Lys
130 135 140
Leu Ile Phe Ile Glu Asp Leu Leu Lys Thr Gln Ser Ser Ala Pro Trp
145 150 155 160
Lys Lys Phe Thr Thr Ser Asn Pro Asn Arg Val Ala Tyr Leu Gly Met
165 170 175
Ser Ser Gly Thr Ser Gly Leu Pro Lys Ala Val Gln Gln Thr His Ile
180 185 190
Asn Met Ser Ser Ser Thr Glu Ala Val Ile Ser Ser Gln Thr Ile Phe
195 200 205
Ser Ala Arg Lys Asn Val Thr Ala Ala Ile Val Pro Met Thr His Val
210 215 220
Tyr Gly Leu Thr Lys Phe Val Phe His Ser Val Ala Gly Ser Met Thr
225 230 235 240
Thr Val Val Phe Pro Lys Phe Ser Leu Val Asp Leu Leu Glu Ala Gln
245 250 255
Ile Lys Tyr Lys Ile Asn Ile Leu Tyr Leu Val Pro Pro Val Val Leu
260 265 270
Ala Leu Ala Lys Asp Ser Arg Val Gln Pro Tyr Ile Lys Ser Ile Cys
275 280 285
Glu Leu Thr Thr Leu Ile Ala Thr Gly Ala Ala Pro Leu Pro Pro Thr
290 295 300
Ala Gly Asp Ala Leu Leu Glu Arg Leu Thr Gly Asn Lys Glu Gly Asn
305 310 315 320
Arg Asp Asn Gly Met Asp Pro Leu Val Leu Ile Gln Gly Tyr Gly Leu
325 330 335
Thr Glu Thr Leu Gln Val Ser Val Phe Lys Pro Glu Asp Pro Glu Arg
340 345 350
Asp Leu Lys Thr Val Gly Lys Leu Leu Pro Asn Thr Glu Val Arg Ile
355 360 365
Val Gly Glu Lys Gly Asp Val Pro Arg Ser Lys Trp Ser Phe Val Thr
370 375 380
Pro Pro Thr Gly Glu Ile Tyr Ile Arg Gly Pro His Val Thr Pro Gly
385 390 395 400
Tyr Phe Asn Asn Asp Ser Ala Asn Ser Glu Ser Phe Asp Gly Glu Trp
405 410 415
Leu Lys Thr Gly Asp Ile Gly Tyr Met Asp Leu Glu Gly Arg Leu Thr
420 425 430
Ile Val Asp Arg Asn Lys Glu Met Ile Lys Val Asn Gly Arg Gln Val
435 440 445
Ala Pro Ala Glu Ile Glu Ser Val Leu Leu Gly His Pro Met Val Lys
450 455 460
Asp Val Ala Val Ile Gly Val Thr Asn Pro Asp Arg Gly Thr Glu Ser
465 470 475 480
Ala Arg Ala Phe Leu Val Thr Glu Ala Arg Ala Leu Pro Val Ile Lys
485 490 495
Gln Trp Phe Asp Arg Arg Val Pro Ser Tyr Lys Arg Leu Tyr Gly Gly
500 505 510
Ile Val Val Val Asp Ala Ile Pro Lys Ser Ala Ser Gly Lys Ile Leu
515 520 525
Arg Arg Val Leu Arg Glu Arg Lys Gly Asp Ser Val Phe Gly Glu Tyr
530 535 540
Val Glu Glu Val
545
<210> 40
<211> 616
<212> PRT
<213> yarrowia lipolytica
<400> 40
Met Pro Gln Ile Ile His Lys Ser Ala Trp Gly Asp Ile Pro Leu Ser
1 5 10 15
Thr Phe Phe Tyr Gly Asn Val Thr Asp Tyr Leu Arg Ser Lys Lys Ser
20 25 30
Phe Gly Ser Asp Lys Ile Gly Tyr Ile Asp Ala Glu Thr Gly Glu Gly
35 40 45
Ile Thr Tyr Lys Gln Leu Trp Lys Leu Ala Asn Gly Ile Ser Ala Val
50 55 60
Leu Tyr His His Tyr Gly Ile Gly His Ala Arg Ala Pro Val Ala Ser
65 70 75 80
Asp His Thr Leu Gly Asp Val Val Met Leu His Ala Pro Asn Ser Arg
85 90 95
Phe Phe Pro Ser Leu His Tyr Gly Met Leu Asp Met Gly Cys Thr Ile
100 105 110
Thr Ser Ala Ser Val Ser Tyr Asp Val Ala Asp Leu Ala His Gln Leu
115 120 125
Arg Val Thr Asp Ala Ser Leu Val Leu Cys Tyr Gln Glu Lys Glu Asn
130 135 140
Asn Val Arg Gln Ala Ile Lys Glu Ala Gln Lys Asp Ala Ala Phe Pro
145 150 155 160
Gly Ile Thr His Pro Val Arg Ile Leu Leu Ile Glu Asn Leu Leu Thr
165 170 175
Met Ala Cys Asn Ile Ser Glu Glu Lys Ile Asn Ser Ala Met Ala Arg
180 185 190
Lys Phe Glu Tyr Ser Pro Gln Glu Cys Thr Lys Arg Ile Ala Tyr Leu
195 200 205
Ser Met Ser Ser Gly Thr Thr Gly Gly Ile Pro Lys Ala Val Arg Leu
210 215 220
Thr His Phe Asn Met Ser Ser Cys Asp Thr Leu Gly Thr Leu Ser Thr
225 230 235 240
Pro Ser Phe Ser Thr Gly Asp Asp Ile Arg Val Ala Ala Ile Val Pro
245 250 255
Met Thr His Gln Tyr Gly Leu Thr Lys Phe Ile Phe Asn Met Cys Ser
260 265 270
Ser His Ala Thr Thr Val Val His Arg Gln Phe Asp Leu Val Lys Leu
275 280 285
Leu Glu Ser Gln Lys Lys Tyr Lys Leu Asn Arg Leu Met Leu Val Pro
290 295 300
Pro Val Ile Val Lys Met Ala Lys Asp Pro Ala Val Glu Pro Tyr Ile
305 310 315 320
Pro Ser Leu Tyr Glu His Val Asp Phe Ile Thr Thr Gly Ala Ala Pro
325 330 335
Leu Pro Gly Ser Ala Val Thr Asn Leu Leu Thr Arg Ile Thr Gly Asn
340 345 350
Pro Gln Gly Ile Arg His Ser Gln Ser Gly Arg Pro Pro Leu Thr Ile
355 360 365
Ser Gln Gly Tyr Gly Leu Thr Glu Thr Ser Pro Leu Cys Ala Val Phe
370 375 380
Asp Pro Leu Asp Pro Asp Val Asp Phe Arg Ser Ala Gly Lys Ala Thr
385 390 395 400
Ser His Val Glu Ile Arg Ile Val Ser Glu Asp Gly Val Asp Gln Pro
405 410 415
Gln Leu Lys Leu Asp Asp Leu Ser His Leu Asp Gly Met Leu Lys Arg
420 425 430
Asp Glu Pro Leu Pro Val Gly Glu Val Leu Ile Arg Gly Pro Met Ile
435 440 445
Met Asp Gly Tyr His Lys Asn Arg Gln Ser Ser Glu Glu Ser Phe Asp
450 455 460
Arg Ser Gln Glu Asp Pro Lys Thr Leu Ile His Trp Gln Asp Lys Trp
465 470 475 480
Leu Lys Thr Gly Asp Ile Gly Met Val Asp Gln Lys Gly Arg Leu Met
485 490 495
Ile Val Asp Arg Asn Lys Glu Met Ile Lys Ser Met Ser Lys Gln Val
500 505 510
Ala Pro Ala Glu Leu Glu Ser Leu Leu Leu Asn His Asp Gln Val Ile
515 520 525
Asp Cys Ala Val Ile Gly Val Asn Ser Glu Ala Lys Ala Thr Glu Ser
530 535 540
Ala Arg Ala Phe Leu Val Leu Lys Asp Pro Ser Tyr Asp Ala Val Lys
545 550 555 560
Ile Lys Ala Trp Leu Asp Gly Gln Val Pro Ser Tyr Lys Arg Leu Tyr
565 570 575
Gly Gly Val Val Val Leu Lys Asn Glu Gln Ile Pro Lys Asn Pro Ser
580 585 590
Gly Lys Ile Leu Arg Arg Ile Leu Arg Thr Arg Lys Asp Asp Phe Ile
595 600 605
Gln Gly Ile Asp Val Ser Lys Leu
610 615
<210> 41
<211> 1800
<212> DNA
<213> Artificial sequence
<220>
<223> YlACoS-5P
<400> 41
atggcctcaa tcattcacaa gtctcctgtg cccgacgttc agctgttcta cggttcctgg 60
ccagatctca tgcgaacctc tcctcatgcc cacaacgact ccaaacccgt ggtctttgac 120
ttcgatacca agcagcaact tacttggaag caggtctggc aactcagcgc tcgactcaga 180
gcccagctgt accacaagta cggaatcggc aaacccggtg ctcttgcacc tttccacaac 240
gatccctctc tcggagacgt ggtcatcttc tacactccca acacctacag ctcgttgccc 300
tatcatctgg ctcttcacga tctcggagcc accatttctc ctgcctccac atcttacgac 360
gtcaaggaca tttgccatca gatcgttact accgatgcgg tcgtggttgt cgctgcagcc 420
gagaaatccg agattgctcg agaggccgtt cagctgtctg gtcgagacgt cagagttgtg 480
gtcatggagg acctcatcaa caatgctccc accgttgcgc agaacgatat cgactcggca 540
cctcatgtgt ccctgtctcg ggaccaggct cgagccaaga ttgcatacct gggcatgtct 600
tccggtacgt ctggcggact tcccaaggct gttcgtctca ctcacttcaa cgttacctcg 660
aactgtctgc aggtctccgc tgccgcaccc aaccttgccc agaacgtggt tgccagcgcc 720
gtcattccaa ccactcacat ctacggtctc accatgtttc tgtcggttct tccctacaac 780
ggttccgtgg tcattcatca caagcaattc aacttgcgag atctgctcga ggctcagaag 840
acatacaagg tctctctgtg gattctcgtt cctcccgtca tcgtgcagct tgccaagaac 900
cctatggtcg acgagtacct ggactccatt cgagcccatg tgcggtgcat cgtctctgga 960
gctgctcctc tcggtggcaa tgtcgtggat caggtttcgg ttcgtcttac cggcaacaag 1020
gaaggcattc tgcccaacgg agacaagctc gtcattcatc aagcctacgg tcttaccgag 1080
tcctctccca tcgttggaat gctcgatcct ctgtcggacc acatcgacgt catgactgtg 1140
ggctgtctca tgcccaatac cgaggctcga attgtcgacg aagagggaaa cgatcagcca 1200
gcagtccacg ttaccgacac acgaggcatc ggtgccgctg tcaagcgagg cgagaagatt 1260
ccctccggag aactctggat tcgaggtcct cagatcatgg acggatacca caagaacccc 1320
gagtcgtctc gtgagtccct ggaacccagc acagagacct acggtctgca acatttccag 1380
gacagatggc ttcgaactgg agacgttgct gtcatcgaca ccttcggacg agtcatggtt 1440
gtggatcgaa ccaaggagct catcaagtcc atgtctcgac aggttgctcc tgccgagctc 1500
gaagctcttc tgctcaacca tccttccgtc aacgatgtgg ctgtcgttgg cgtccacaac 1560
gacgataatg gcacagagtc agcacgagcg tttgtcgttc ttcaaccagg cgacgcctgt 1620
gatcctacta ccatcaagca ctggatggac cagcaagttc cctcctacaa gcggctgtac 1680
ggaggcattg tggtcatcga cactgttccc aagaatgcct ctggcaagat tctgcgaaga 1740
ctgcttcgac agcggagaga cgatcgagtc tggggtctgg ccaaggttgc caagctctaa 1800
<210> 42
<211> 599
<212> PRT
<213> Artificial sequence
<220>
<223> YLACoS-5P protein
<400> 42
Met Ala Ser Ile Ile His Lys Ser Pro Val Pro Asp Val Gln Leu Phe
1 5 10 15
Tyr Gly Ser Trp Pro Asp Leu Met Arg Thr Ser Pro His Ala His Asn
20 25 30
Asp Ser Lys Pro Val Val Phe Asp Phe Asp Thr Lys Gln Gln Leu Thr
35 40 45
Trp Lys Gln Val Trp Gln Leu Ser Ala Arg Leu Arg Ala Gln Leu Tyr
50 55 60
His Lys Tyr Gly Ile Gly Lys Pro Gly Ala Leu Ala Pro Phe His Asn
65 70 75 80
Asp Pro Ser Leu Gly Asp Val Val Ile Phe Tyr Thr Pro Asn Thr Tyr
85 90 95
Ser Ser Leu Pro Tyr His Leu Ala Leu His Asp Leu Gly Ala Thr Ile
100 105 110
Ser Pro Ala Ser Thr Ser Tyr Asp Val Lys Asp Ile Cys His Gln Ile
115 120 125
Val Thr Thr Asp Ala Val Val Val Val Ala Ala Ala Glu Lys Ser Glu
130 135 140
Ile Ala Arg Glu Ala Val Gln Leu Ser Gly Arg Asp Val Arg Val Val
145 150 155 160
Val Met Glu Asp Leu Ile Asn Asn Ala Pro Thr Val Ala Gln Asn Asp
165 170 175
Ile Asp Ser Ala Pro His Val Ser Leu Ser Arg Asp Gln Ala Arg Ala
180 185 190
Lys Ile Ala Tyr Leu Gly Met Ser Ser Gly Thr Ser Gly Gly Leu Pro
195 200 205
Lys Ala Val Arg Leu Thr His Phe Asn Val Thr Ser Asn Cys Leu Gln
210 215 220
Val Ser Ala Ala Ala Pro Asn Leu Ala Gln Asn Val Val Ala Ser Ala
225 230 235 240
Val Ile Pro Thr Thr His Ile Tyr Gly Leu Thr Met Phe Leu Ser Val
245 250 255
Leu Pro Tyr Asn Gly Ser Val Val Ile His His Lys Gln Phe Asn Leu
260 265 270
Arg Asp Leu Leu Glu Ala Gln Lys Thr Tyr Lys Val Ser Leu Trp Ile
275 280 285
Leu Val Pro Pro Val Ile Val Gln Leu Ala Lys Asn Pro Met Val Asp
290 295 300
Glu Tyr Leu Asp Ser Ile Arg Ala His Val Arg Cys Ile Val Ser Gly
305 310 315 320
Ala Ala Pro Leu Gly Gly Asn Val Val Asp Gln Val Ser Val Arg Leu
325 330 335
Thr Gly Asn Lys Glu Gly Ile Leu Pro Asn Gly Asp Lys Leu Val Ile
340 345 350
His Gln Ala Tyr Gly Leu Thr Glu Ser Ser Pro Ile Val Gly Met Leu
355 360 365
Asp Pro Leu Ser Asp His Ile Asp Val Met Thr Val Gly Cys Leu Met
370 375 380
Pro Asn Thr Glu Ala Arg Ile Val Asp Glu Glu Gly Asn Asp Gln Pro
385 390 395 400
Ala Val His Val Thr Asp Thr Arg Gly Ile Gly Ala Ala Val Lys Arg
405 410 415
Gly Glu Lys Ile Pro Ser Gly Glu Leu Trp Ile Arg Gly Pro Gln Ile
420 425 430
Met Asp Gly Tyr His Lys Asn Pro Glu Ser Ser Arg Glu Ser Leu Glu
435 440 445
Pro Ser Thr Glu Thr Tyr Gly Leu Gln His Phe Gln Asp Arg Trp Leu
450 455 460
Arg Thr Gly Asp Val Ala Val Ile Asp Thr Phe Gly Arg Val Met Val
465 470 475 480
Val Asp Arg Thr Lys Glu Leu Ile Lys Ser Met Ser Arg Gln Val Ala
485 490 495
Pro Ala Glu Leu Glu Ala Leu Leu Leu Asn His Pro Ser Val Asn Asp
500 505 510
Val Ala Val Val Gly Val His Asn Asp Asp Asn Gly Thr Glu Ser Ala
515 520 525
Arg Ala Phe Val Val Leu Gln Pro Gly Asp Ala Cys Asp Pro Thr Thr
530 535 540
Ile Lys His Trp Met Asp Gln Gln Val Pro Ser Tyr Lys Arg Leu Tyr
545 550 555 560
Gly Gly Ile Val Val Ile Asp Thr Val Pro Lys Asn Ala Ser Gly Lys
565 570 575
Ile Leu Arg Arg Leu Leu Arg Gln Arg Arg Asp Asp Arg Val Trp Gly
580 585 590
Leu Ala Lys Val Ala Lys Leu
595
<210> 43
<211> 1788
<212> DNA
<213> Artificial sequence
<220>
<223> YlACoS-6P
<400> 43
atggccacac agattatcca caacgccacc atccccaata tccccgtcga ccagctctac 60
gacggcaaga tcaccgactt cattcgatcc ggaggccact ccaacgaaac caagccttct 120
gtcatcgacg ccaagacagg ccagactctc tcccaggcgg aaatgtggca gctgtcggac 180
aagtacgcgg cacttctcag ctctcagtac ggtctgtgcc gacacagaga caacgagctg 240
gacccatcta tgggagatgt gctcatcacc ttctttggaa acgttatcct cgctcctgtg 300
gtccattggg ctgccctcga cctcggagca accatttctc ctggatccac aggctactct 360
gcccaggatc tcgctcacca gttccgagtc accactccca aggtcgttgt gtacgccaag 420
gcgttcaagg atgtggtgga cgaggctacg aagctgtaca actccccaaa ccctccagca 480
cttgtcgagc tcgaggcgct ggacaagcag gcccgaatgg ttggaaacca caaggtcgaa 540
cacacccgaa agatcaagct ggctcctcac gagtcccgaa ctcggatcgc gtaccttggc 600
atgtcttcag gtacctccgg tggagtttca aaggctgtcc gactcaccca ttccaatctc 660
acgtcgtgtt ccgaaatctc gaacaaagcc tccgagtctc tcgcaactga ccagcagatc 720
gctgccgcca tcattcccgt gagtcatctg tttggactgt ccaagttcct cattggcaac 780
cctcacgccg gagccaccac tgtctatcac aatggcttcg atctgatcga ggtgctggag 840
gcacagaaga aatacaaagt caactcgtgg accctggttc ctcccatcat tgtcctgctc 900
accaaacacc ccattgtcga gaagtacatt ccttctctcc gtgcccacat gcgagccatc 960
ctctccggag ctgctcctct gggtgccaat gtcacagagg ctcttctcac ccgagtcact 1020
ggcaacaagt ttggcgagtc tcccgagggc ggtctgcgaa tcgttcaggg ctacggactt 1080
acagagacgt ctcccgttgc cactctgttt gaccccgaag acaaggaacg acacattcgg 1140
tcgtgtggaa agctcgtgcc caactctcag gttcgaattg tcaacgaaga cggcgtggat 1200
cagcctgcct acgatgtgga ccccaacgag ctggacgagg ccatcaaaca gggcactctg 1260
ccagtcggag agctttggat cagaggtccc caggttatgg acggctacca taacaacccc 1320
gaggccaacg aagcctgttt cgtcaaggct gacgatgctg aagcagatac tgcctactac 1380
aacagacact ggttccgaac cggagacgtt gctctggtcg acaagcaggg cagatacatg 1440
attgtggacc gaaccaagga gatgatcaag agtcagggta agcaggttgc tcctgccgag 1500
ctcgaagaca tgctcctggg acacgcacag gtggcagata ccgcagtcat cggtattcag 1560
gacgtggaga agggtaacga ggctcctcga gcttttgttg tgctcaagga cccgaagtac 1620
gacgctgtgg agatcaagac atggctggac aagcagcttc ccaagtacaa gcagcttcat 1680
gctggcatcg tggtcattga tgccattccc aagaacgcca gtggcaagat tctgcgacgt 1740
ctgttgcgtg ctagaaagga cgatgttgtt ctgggtctca acaagtaa 1788
<210> 44
<211> 595
<212> PRT
<213> Artificial sequence
<220>
<223> YLACoS-6P protein
<400> 44
Met Ala Thr Gln Ile Ile His Asn Ala Thr Ile Pro Asn Ile Pro Val
1 5 10 15
Asp Gln Leu Tyr Asp Gly Lys Ile Thr Asp Phe Ile Arg Ser Gly Gly
20 25 30
His Ser Asn Glu Thr Lys Pro Ser Val Ile Asp Ala Lys Thr Gly Gln
35 40 45
Thr Leu Ser Gln Ala Glu Met Trp Gln Leu Ser Asp Lys Tyr Ala Ala
50 55 60
Leu Leu Ser Ser Gln Tyr Gly Leu Cys Arg His Arg Asp Asn Glu Leu
65 70 75 80
Asp Pro Ser Met Gly Asp Val Leu Ile Thr Phe Phe Gly Asn Val Ile
85 90 95
Leu Ala Pro Val Val His Trp Ala Ala Leu Asp Leu Gly Ala Thr Ile
100 105 110
Ser Pro Gly Ser Thr Gly Tyr Ser Ala Gln Asp Leu Ala His Gln Phe
115 120 125
Arg Val Thr Thr Pro Lys Val Val Val Tyr Ala Lys Ala Phe Lys Asp
130 135 140
Val Val Asp Glu Ala Thr Lys Leu Tyr Asn Ser Pro Asn Pro Pro Ala
145 150 155 160
Leu Val Glu Leu Glu Ala Leu Asp Lys Gln Ala Arg Met Val Gly Asn
165 170 175
His Lys Val Glu His Thr Arg Lys Ile Lys Leu Ala Pro His Glu Ser
180 185 190
Arg Thr Arg Ile Ala Tyr Leu Gly Met Ser Ser Gly Thr Ser Gly Gly
195 200 205
Val Ser Lys Ala Val Arg Leu Thr His Ser Asn Leu Thr Ser Cys Ser
210 215 220
Glu Ile Ser Asn Lys Ala Ser Glu Ser Leu Ala Thr Asp Gln Gln Ile
225 230 235 240
Ala Ala Ala Ile Ile Pro Val Ser His Leu Phe Gly Leu Ser Lys Phe
245 250 255
Leu Ile Gly Asn Pro His Ala Gly Ala Thr Thr Val Tyr His Asn Gly
260 265 270
Phe Asp Leu Ile Glu Val Leu Glu Ala Gln Lys Lys Tyr Lys Val Asn
275 280 285
Ser Trp Thr Leu Val Pro Pro Ile Ile Val Leu Leu Thr Lys His Pro
290 295 300
Ile Val Glu Lys Tyr Ile Pro Ser Leu Arg Ala His Met Arg Ala Ile
305 310 315 320
Leu Ser Gly Ala Ala Pro Leu Gly Ala Asn Val Thr Glu Ala Leu Leu
325 330 335
Thr Arg Val Thr Gly Asn Lys Phe Gly Glu Ser Pro Glu Gly Gly Leu
340 345 350
Arg Ile Val Gln Gly Tyr Gly Leu Thr Glu Thr Ser Pro Val Ala Thr
355 360 365
Leu Phe Asp Pro Glu Asp Lys Glu Arg His Ile Arg Ser Cys Gly Lys
370 375 380
Leu Val Pro Asn Ser Gln Val Arg Ile Val Asn Glu Asp Gly Val Asp
385 390 395 400
Gln Pro Ala Tyr Asp Val Asp Pro Asn Glu Leu Asp Glu Ala Ile Lys
405 410 415
Gln Gly Thr Leu Pro Val Gly Glu Leu Trp Ile Arg Gly Pro Gln Val
420 425 430
Met Asp Gly Tyr His Asn Asn Pro Glu Ala Asn Glu Ala Cys Phe Val
435 440 445
Lys Ala Asp Asp Ala Glu Ala Asp Thr Ala Tyr Tyr Asn Arg His Trp
450 455 460
Phe Arg Thr Gly Asp Val Ala Leu Val Asp Lys Gln Gly Arg Tyr Met
465 470 475 480
Ile Val Asp Arg Thr Lys Glu Met Ile Lys Ser Gln Gly Lys Gln Val
485 490 495
Ala Pro Ala Glu Leu Glu Asp Met Leu Leu Gly His Ala Gln Val Ala
500 505 510
Asp Thr Ala Val Ile Gly Ile Gln Asp Val Glu Lys Gly Asn Glu Ala
515 520 525
Pro Arg Ala Phe Val Val Leu Lys Asp Pro Lys Tyr Asp Ala Val Glu
530 535 540
Ile Lys Thr Trp Leu Asp Lys Gln Leu Pro Lys Tyr Lys Gln Leu His
545 550 555 560
Ala Gly Ile Val Val Ile Asp Ala Ile Pro Lys Asn Ala Ser Gly Lys
565 570 575
Ile Leu Arg Arg Leu Leu Arg Ala Arg Lys Asp Asp Val Val Leu Gly
580 585 590
Leu Asn Lys
595
<210> 45
<211> 598
<212> PRT
<213> yarrowia lipolytica
<400> 45
Met Val Gln Ile Ile His Lys Ala Pro Leu Gly Asp Met Ala Glu Ser
1 5 10 15
Glu Leu Phe Tyr Gly Ser Ile Pro Asp Phe Met Arg Ser Ser Arg Phe
20 25 30
Ala Asp Asp Asp Thr Arg Ile Ser Val Val Asp Tyr Asp Thr Asp Lys
35 40 45
Ala Met Thr Leu Ala Arg Val Phe Lys Val Ser Gly Met Leu Arg Ala
50 55 60
Gln Phe Phe His Thr Tyr Asp Val Gly Lys Lys Lys Asp Gly Asp Ala
65 70 75 80
Asn Pro Lys Val Ile Phe Tyr Val Gly Asn Thr Ala Asp Asn Leu Ala
85 90 95
Cys His Ile Ala Leu His Asp Leu Gly Ala Ile Ile Ser Pro Ala Ser
100 105 110
Thr Ala Tyr Asp Val Asn Asp Leu Leu His Gln Ile Asn Val Val Asp
115 120 125
Ala Ala Leu Ile Val Ala Glu Ala Ala Arg Ala Asp Val Ala Arg Glu
130 135 140
Ala Val Ala Lys Ala Gly Asp Lys Phe Lys His Val Lys Val Val Val
145 150 155 160
Phe Glu Glu Leu Leu Glu Gln Asn Arg Arg Val Arg Pro Asn Leu Ile
165 170 175
Arg Val Ala Pro Ile Val His Leu Ser Lys Glu Gln Ala Tyr Thr Thr
180 185 190
Leu Ala Tyr Leu Gly Met Ser Ser Gly Thr Ser Gly Gly Val Pro Lys
195 200 205
Ala Val Glu Leu Thr His Phe Ala Met Thr Ser Asn Val Gln Gln Thr
210 215 220
Ala Lys Asn Ala Pro Asn Leu Val Asp Asp Asp Thr Val Cys Ser Ala
225 230 235 240
Val Ile Pro Thr Ser His Ile Tyr Gly Leu Ala Leu Phe Met Leu His
245 250 255
Met Pro Phe Leu Gly Ala Lys Val Val Tyr His Lys Lys Phe Asp Leu
260 265 270
Val Glu Met Leu Glu Gly Gln Lys Lys His Gly Val Asn Tyr Trp Val
275 280 285
Leu Val Pro Pro Ile Ile Val Ala Leu Ala Lys His Pro Ile Ile Asp
290 295 300
Arg Tyr Leu Asp Ser Ile Arg Ala Asn Leu Lys Thr Ile Thr Ser Gly
305 310 315 320
Ala Ala Pro Leu Gly Gly Asn Val Val Asp Ala Val Gln Thr Arg Phe
325 330 335
Thr Gly Asn Thr Arg Gly Thr Leu Pro Asn Asn Arg Arg Ile Val Ile
340 345 350
Tyr Gln Gly Tyr Gly Leu Thr Glu Thr Ala Pro Ile Ala Cys Leu Cys
355 360 365
Asp Pro Leu Trp Asp Asn Leu Asn Val Val Thr Val Gly Thr Leu Val
370 375 380
Pro Asn Thr Glu Ala Arg Ile Val Asp Glu Asn Gly Asp Asp Gln Pro
385 390 395 400
Ala Phe Glu Val Thr Asp Ala Arg Ala Leu Gly Asp Ala Val Arg Arg
405 410 415
Gly Asp Lys Ile Pro Ser Gly Glu Leu Tyr Leu Arg Gly Pro Gln Ile
420 425 430
Met Ser Gly Tyr His Lys Asn Pro Lys Ser Thr Glu Glu Ser Phe Glu
435 440 445
Tyr Val Asp Tyr Lys Ala Glu Gly Leu Arg His Tyr Gln Asp Arg Trp
450 455 460
Leu Lys Thr Gly Asp Val Ala Val Ile Asp Asn Phe Gly Arg Ile Gln
465 470 475 480
Ile Val Asp Arg Thr Lys Glu Leu Ile Lys Ser Met Ser Lys Gln Val
485 490 495
Ala Pro Ala Glu Leu Glu Ala Leu Leu Leu Ser His Pro Asp Val Val
500 505 510
Asp Val Ala Val Ile Gly Val Trp Gln Glu Glu Lys Ala Thr Glu Ser
515 520 525
Ala Arg Ala Phe Leu Val Val Arg Asp Pro Lys Val Asp Val Val Ala
530 535 540
Ile Lys Lys Trp Met Asp Glu Gln Val Pro Ser Tyr Lys Arg Leu Tyr
545 550 555 560
Gly Gly Val Val Val Ile Asp Ala Ile Pro Lys Asn Pro Ser Gly Lys
565 570 575
Ile Leu Arg Arg Leu Leu Arg Gln Arg Lys Asp Asp Val Val Gln Gly
580 585 590
Leu Asp Gln Ala Lys Leu
595
<210> 46
<211> 741
<212> PRT
<213> yarrowia lipolytica
<400> 46
Met Leu Arg Pro Arg Ala Pro Ser Thr Leu Arg Arg Ala Ser Thr Gln
1 5 10 15
Leu Ser Ala Arg Pro Gln Gly Ile Thr Gly Pro Gln Arg Ser Phe His
20 25 30
Leu Ala Cys Ser Arg Pro Thr Arg Ser Thr Thr Ser Glu Glu Asp Arg
35 40 45
Pro Lys Trp Leu Thr Pro Arg Asn Val Arg Leu Gly Ser Leu Pro Phe
50 55 60
Ser Arg Leu Leu Gln Gly His Ser Gln Arg Leu Tyr Ser Gly Leu Ser
65 70 75 80
Thr Pro Gly Ile Ser Glu Val Thr Gly Cys Ser Ser Pro Ala Leu Ile
85 90 95
Glu Ser His Ser Trp Asp Tyr Tyr Thr Thr Pro Ala Gln Arg Glu Leu
100 105 110
Trp Gly Ser Lys Gly Glu Tyr Pro Ala Leu Ile Ser Ala Tyr Gln Gln
115 120 125
Pro Asp Ile Ala Asn Ala Ile Leu Asp Arg Thr Ser Asp Tyr Thr Asn
130 135 140
Glu Thr His Leu Ser Phe Ser Glu Leu Leu Lys Leu Ser Asn Leu Phe
145 150 155 160
Ala Asp Ser Leu Tyr Ala His Ala Arg Glu Gln Gly Leu Val Phe Lys
165 170 175
Ser Gly Asp Ser Val Ala Val Cys Gly Gly Asn Val Trp Glu Tyr Thr
180 185 190
Ala Leu Gln Met Gly Leu Ser Lys Leu Gly Leu Val Leu Val Pro Leu
195 200 205
Asn Pro Ala Phe Thr Ala Asn Gln Phe Ala Ala Ala Leu Ala Ala Thr
210 215 220
Glu Ala Lys Ala Leu Ile Met Thr Ser His Leu Pro Gly Gly Lys Asp
225 230 235 240
Lys Ala Thr Gly Lys Met Thr Leu Lys Ser Ala Ala Pro Ile Cys Gln
245 250 255
Glu Val Ile Asp Asn Leu Asn Ala Ser Gly Lys Ser Lys Leu Lys Leu
260 265 270
Leu Ile Asn Leu Ala Ser Gly Glu Thr Pro Gly Ala Asp Thr Ile Lys
275 280 285
Asp Val Lys Phe Gln Gly Ser Gln Ser Asp Met His Glu Ile Val Phe
290 295 300
Gln His Lys Lys Ala Val Ala Asn Gly Thr Leu Pro Ala Ser Val Pro
305 310 315 320
Thr Glu Ile Arg Arg Leu Thr Ala Thr Val Asn Pro Asp Asp Ile Thr
325 330 335
Asn Met Gln Phe Thr Ser Gly Thr Thr Ser Gln Pro Lys Val Ser Cys
340 345 350
Leu Thr His Arg Asn Leu Leu Asn Asn Gly His Leu Ile Gly Thr Arg
355 360 365
Met Gly Leu Lys Pro Ala Thr Gly Pro Ala Val Asn Gly Ile Ala Pro
370 375 380
Asn Gln Asp Arg Leu Cys Ile Pro Val Pro Met Phe His Cys Phe Gly
385 390 395 400
Leu Val Leu Ser Asn Leu Ala Ala Leu Thr Thr Gly Ala Ala Leu Val
405 410 415
Tyr Pro Ser Glu Trp Phe Cys Ala Arg Ser Ala Ile Asp Asn Val Arg
420 425 430
Lys Tyr Lys Cys Thr Gly Leu His Gly Val Pro Thr Met Tyr Val Ala
435 440 445
Glu Leu Glu Tyr Leu Lys Asp Leu Glu Leu Lys Glu Ala Lys Ala Pro
450 455 460
Gly Gln Asn Phe Leu Pro Gly Phe Glu Leu Leu Arg Thr Gly Ile Ala
465 470 475 480
Ala Gly Ser Ala Val Pro Gly Glu Leu Met Thr Lys Leu Gly Gln Ser
485 490 495
Met Asn Leu Lys Ala Leu Thr Ile Cys Tyr Gly Met Thr Glu Thr Ala
500 505 510
Pro Val Thr Phe Met Thr Arg Pro Asp Asp Pro Val Glu Lys Arg Val
515 520 525
Glu Thr Val Gly Gln Ile Met Pro His Thr Ser Cys Arg Ile Ile Lys
530 535 540
Ser Gln Gln Glu Asp Leu Ser Glu Ser Glu Leu Asp Phe Thr Pro Leu
545 550 555 560
Ala Thr Gly Gln Lys Gly Glu Ile Ile Thr Ser Gly Tyr Ala Leu Gln
565 570 575
Lys Tyr Tyr Lys Asp Asp Pro Lys Lys Thr Ser Ala Ala Met Val Val
580 585 590
Asp Pro Ala Thr Gly Val Arg Trp Met Arg Thr Gly Asp Glu Gly Cys
595 600 605
Met Asp Asn Glu Gly Phe Leu Lys Val Thr Gly Arg Leu Lys Asp Leu
610 615 620
Ile Ile Arg Gly Gly Glu Asn Ile His Pro Leu Glu Ile Glu Asn Val
625 630 635 640
Leu Phe Ala His Asp Lys Ile Ala Gln Ala Ser Val Val Gly Val Lys
645 650 655
Asp Pro Lys Tyr Gly Glu Ala Val Cys Ala Phe Val Thr Pro His Ala
660 665 670
Phe Phe His Lys Gly His Gln His Val Lys His Asp Asp Ser Asp Lys
675 680 685
Leu Thr Ile Glu Gln Val Gln Glu Trp Val Arg Asn Lys Leu Gly His
690 695 700
Tyr Met Val Pro Lys Tyr Val Phe Phe Val Gly Asp Tyr Pro Lys Thr
705 710 715 720
Ala Ser Gly Lys Ile Arg Lys Val Asp Leu Arg Lys Thr Ala Glu Ser
725 730 735
Gln Leu Gly Leu Cys
740
<210> 47
<211> 554
<212> PRT
<213> yarrowia lipolytica
<400> 47
Met Ala Pro Ser Gln Gly Asp Lys Lys Met Phe Ile Ser Ala Glu Thr
1 5 10 15
Gly Glu Ser Leu Thr Thr Ala Gln Gln Trp Ala Thr Val Glu Leu Phe
20 25 30
Ala Ser Lys Leu Tyr Gln Leu Gly Ile Gly His Ser Leu Arg Pro Asn
35 40 45
Ser Asp Ala His Leu Gly Asp Val Val Leu Leu Tyr Val Lys Asn Ser
50 55 60
Ile Tyr Ile Pro Ala Ala His Trp Ala Leu Leu Asp Leu Gly Ala Thr
65 70 75 80
Val Ala Pro Ala Ala Ala Val Tyr Lys Ala Arg Asp Leu Val His Gln
85 90 95
Ile Glu Leu Val Lys Pro Lys Leu Ile Val Cys Asp Ala Asp Leu Lys
100 105 110
Ser Glu Ala Val Glu Ala Leu Lys Ile Leu Ser Lys Lys Met Pro Ile
115 120 125
Val Thr Met Glu Glu Leu Arg Gln Pro Val Lys Lys Leu Lys Gln Arg
130 135 140
Gln Arg Phe Arg Leu Ser Arg Pro Glu Ala Ala Lys Arg Val Ala Ala
145 150 155 160
Leu Val Met Ser Ser Gly Thr Ser Gly Gly Leu Pro Lys Ala Val Arg
165 170 175
Val Thr His His Val Val Thr Ser Asn Ala Gln Cys Ser Ala Ile Val
180 185 190
Ala Pro Asp Leu Phe Asp Asp Pro Thr Asn Val Ile Ser Ala Val Leu
195 200 205
Pro Met Ser His Ile Tyr Gly Tyr Phe Lys Phe Leu Phe Ala Cys Phe
210 215 220
Tyr Thr Gly Glu Thr Cys Val Val His Gln Ser Phe Asp Leu Lys Ala
225 230 235 240
Val Leu Asp Ala Gln Gln Lys Tyr Gly Ile Thr Ser Phe Phe Met Val
245 250 255
Pro Pro Ile Ile Ile Ala Leu Ala Lys Ser Pro Ile Val Asp Glu Tyr
260 265 270
Ile Pro Ser Leu Gln Lys Leu Arg Phe Ile Thr Ser Gly Ala Ala Pro
275 280 285
Leu Gly Gly Asn Val Ile Glu Asp Val Lys Arg Arg Leu Gly Ser His
290 295 300
Ile Ala Val Thr Gln Met Tyr Gly Met Thr Glu Ser Ile Leu Ser Thr
305 310 315 320
Cys Phe Asn Pro Ser Asp Ala Asp Val Ala Ser Arg Ser Val Gly Lys
325 330 335
Leu Cys Gly Asn Ile Glu Ala Arg Ile Val Gly His Asp Gly Val Asp
340 345 350
Gln Pro Ala Tyr Asn Glu Thr Asp Pro Asp Lys Ile Asp Ala Ala Phe
355 360 365
Lys Arg Gly Asp Ala Leu Pro Ser Gly Glu Leu Trp Leu Arg Gly Pro
370 375 380
Ala Ile Met Ala Gly Tyr His Gly Asn Cys Leu Ala Asn Glu Glu Ser
385 390 395 400
Phe Val Asp Ala Ser Asp Ala Ala Thr Val Pro His Tyr His Arg Lys
405 410 415
Trp Leu Arg Thr Gly Asp Val Ala Val Ile Asp Val Lys Gly Arg Ile
420 425 430
Val Ile Val Asp Arg Thr Lys Glu Met Ile Lys Ser Met Gly Arg Ala
435 440 445
Val Ala Pro Ala Glu Ile Glu Ala Leu Leu Leu Ser His Pro Gln Val
450 455 460
Met Asp Cys Ala Val Ile Gly Val His Val Pro Glu Lys Gly Thr Glu
465 470 475 480
Ala Ala Arg Ala Phe Leu Val Leu Arg Asp Ala Gln Ala Ser Val Ala
485 490 495
Arg Asp Val Ala Ala Trp Leu Asn Asp Gln Val Pro Ser Tyr Lys Arg
500 505 510
Leu His Gly Gly Val Val Val Phe Arg Gly Glu Val Ile Pro Lys Asn
515 520 525
Ala Ser Gly Lys Ile Leu Arg Arg Leu Leu Arg Gln Arg Lys Gly Asp
530 535 540
Glu Val Val Phe Pro Glu Arg Ala Lys Leu
545 550
<210> 48
<211> 1812
<212> DNA
<213> Artificial sequence
<220>
<223> YlACoS-10P
<400> 48
atggcctccg tcgctccatc ttccaacccc aatccgatcc accatctgtc gcgagtcgaa 60
gacgttcctc tctcccagac gttccgaggc aacattaccg actttgtgcg atctggaggc 120
tttgccgacg acgactccaa gccctgttgc atcgacgcga agactggcca acaacttaca 180
cagaagcaag tctgggacta cgccgacaag ttcagagcac tgctccatca cgacaacaat 240
ctgtgtcctt tcaatgccaa caccaccgat ccagctcttg gagacgtcat gatcaccctc 300
gtgcccaacc atctgttcat tacgtcgctg cactttgccg cactcgatct gggtgcgaca 360
gtttctcctg gctcggctgg atacactgtg gccgagctcg tcaaccagat caatcttacc 420
ggagcttctc tcatcgtgta cactcgaccc gtcttcaagg ttgtgcgaga ggcgcttgct 480
cagatcgtgg taccagtcaa gatcgtggag ttcgagggtc tcatcgaacg agccgagttt 540
gttcagagcc acaagattca gtccacaaag aaagtcacac tttctcctga ggagtcctac 600
tcgagaatcg cctacctggg catgtcttca ggcacctccg gaggtcttcc taaggccgtt 660
cgattgtcgc acttcaacat ggcgagttct gccgagctct ccaagcgagc tgcaccttcg 720
attgccggat ccgagcagat cgcaggtgcc attatccctg tcaaccatgt gtatggtctg 780
gccaagttcc tcattgccat gccaaagtcc ggagccacca cagtcttcca ctccaagttc 840
gacctcatcg agatcctcga ggctcaacag aagtacaagg tcaacatgta cgcccttgtt 900
cctcccatca ttgtcgttct ggccaagcat cctgctgttg agaagtacat cccttcgctg 960
cgagaacacc ttcgatacgt gtcctctgga gctgcacccc tgggtgccaa cgtcatcgag 1020
gcttgcaaca agcgtcttgc cggaactgct tctggcgaga acgagtttgg aggtctcaag 1080
attgttcagg gctacggtct cactgaaacc tcccctgtgg tctccacttt cgatcccaac 1140
gatcctgagc gacacgctcg gtcctgtggc aagctggttc ccaacaccca ggcacgaatc 1200
gtgtcggagg acggagtcga tcagcctgcc tacgagctca aggacctgtc tcagttggag 1260
gccgagctca aaaagggcaa ccttcccacc ggtgagttgt ggcttcgagg tccccagatc 1320
atggatggct accacaagaa cgacgaggcc aacgctgagt cgtttgtcga cgccactgac 1380
tacacttcca acatgccctt ctacatgaag cggtggttcc gaactggcga tgttgctctc 1440
gtcgatactc tgggcagata catgattgtc gatcgaacca aagagatgat caagagcatg 1500
agtaagcagg ttgctcctgc cgagctggag gacatcctgc ttggccatcc ccaggtagcc 1560
gatgctgctg tcatcggtgt tcagcaggtg gagaagggca ctgaggctcc ccgagcgttc 1620
gtggtgcttc gagatcccaa gttcgatgca gtggagatca aaaagtggat ggacgcccag 1680
gtgcccaagt acaaacaact tcatggaggt gtcgtggttc tggatgctgt tcccaagaat 1740
gccagcggca agattctcag acgactgctc cgtcagcgag agaatgacgt cgttcttgga 1800
ctcgacaagt aa 1812
<210> 49
<211> 603
<212> PRT
<213> Artificial sequence
<220>
<223> YLACoS-10P protein
<400> 49
Met Ala Ser Val Ala Pro Ser Ser Asn Pro Asn Pro Ile His His Leu
1 5 10 15
Ser Arg Val Glu Asp Val Pro Leu Ser Gln Thr Phe Arg Gly Asn Ile
20 25 30
Thr Asp Phe Val Arg Ser Gly Gly Phe Ala Asp Asp Asp Ser Lys Pro
35 40 45
Cys Cys Ile Asp Ala Lys Thr Gly Gln Gln Leu Thr Gln Lys Gln Val
50 55 60
Trp Asp Tyr Ala Asp Lys Phe Arg Ala Leu Leu His His Asp Asn Asn
65 70 75 80
Leu Cys Pro Phe Asn Ala Asn Thr Thr Asp Pro Ala Leu Gly Asp Val
85 90 95
Met Ile Thr Leu Val Pro Asn His Leu Phe Ile Thr Ser Leu His Phe
100 105 110
Ala Ala Leu Asp Leu Gly Ala Thr Val Ser Pro Gly Ser Ala Gly Tyr
115 120 125
Thr Val Ala Glu Leu Val Asn Gln Ile Asn Leu Thr Gly Ala Ser Leu
130 135 140
Ile Val Tyr Thr Arg Pro Val Phe Lys Val Val Arg Glu Ala Leu Ala
145 150 155 160
Gln Ile Val Val Pro Val Lys Ile Val Glu Phe Glu Gly Leu Ile Glu
165 170 175
Arg Ala Glu Phe Val Gln Ser His Lys Ile Gln Ser Thr Lys Lys Val
180 185 190
Thr Leu Ser Pro Glu Glu Ser Tyr Ser Arg Ile Ala Tyr Leu Gly Met
195 200 205
Ser Ser Gly Thr Ser Gly Gly Leu Pro Lys Ala Val Arg Leu Ser His
210 215 220
Phe Asn Met Ala Ser Ser Ala Glu Leu Ser Lys Arg Ala Ala Pro Ser
225 230 235 240
Ile Ala Gly Ser Glu Gln Ile Ala Gly Ala Ile Ile Pro Val Asn His
245 250 255
Val Tyr Gly Leu Ala Lys Phe Leu Ile Ala Met Pro Lys Ser Gly Ala
260 265 270
Thr Thr Val Phe His Ser Lys Phe Asp Leu Ile Glu Ile Leu Glu Ala
275 280 285
Gln Gln Lys Tyr Lys Val Asn Met Tyr Ala Leu Val Pro Pro Ile Ile
290 295 300
Val Val Leu Ala Lys His Pro Ala Val Glu Lys Tyr Ile Pro Ser Leu
305 310 315 320
Arg Glu His Leu Arg Tyr Val Ser Ser Gly Ala Ala Pro Leu Gly Ala
325 330 335
Asn Val Ile Glu Ala Cys Asn Lys Arg Leu Ala Gly Thr Ala Ser Gly
340 345 350
Glu Asn Glu Phe Gly Gly Leu Lys Ile Val Gln Gly Tyr Gly Leu Thr
355 360 365
Glu Thr Ser Pro Val Val Ser Thr Phe Asp Pro Asn Asp Pro Glu Arg
370 375 380
His Ala Arg Ser Cys Gly Lys Leu Val Pro Asn Thr Gln Ala Arg Ile
385 390 395 400
Val Ser Glu Asp Gly Val Asp Gln Pro Ala Tyr Glu Leu Lys Asp Leu
405 410 415
Ser Gln Leu Glu Ala Glu Leu Lys Lys Gly Asn Leu Pro Thr Gly Glu
420 425 430
Leu Trp Leu Arg Gly Pro Gln Ile Met Asp Gly Tyr His Lys Asn Asp
435 440 445
Glu Ala Asn Ala Glu Ser Phe Val Asp Ala Thr Asp Tyr Thr Ser Asn
450 455 460
Met Pro Phe Tyr Met Lys Arg Trp Phe Arg Thr Gly Asp Val Ala Leu
465 470 475 480
Val Asp Thr Leu Gly Arg Tyr Met Ile Val Asp Arg Thr Lys Glu Met
485 490 495
Ile Lys Ser Met Ser Lys Gln Val Ala Pro Ala Glu Leu Glu Asp Ile
500 505 510
Leu Leu Gly His Pro Gln Val Ala Asp Ala Ala Val Ile Gly Val Gln
515 520 525
Gln Val Glu Lys Gly Thr Glu Ala Pro Arg Ala Phe Val Val Leu Arg
530 535 540
Asp Pro Lys Phe Asp Ala Val Glu Ile Lys Lys Trp Met Asp Ala Gln
545 550 555 560
Val Pro Lys Tyr Lys Gln Leu His Gly Gly Val Val Val Leu Asp Ala
565 570 575
Val Pro Lys Asn Ala Ser Gly Lys Ile Leu Arg Arg Leu Leu Arg Gln
580 585 590
Arg Glu Asn Asp Val Val Leu Gly Leu Asp Lys
595 600
<210> 50
<211> 627
<212> PRT
<213> yarrowia lipolytica
<400> 50
Met Ile Ile His Thr Ser Asp Thr Ser Ser Ile Arg Val Asn Asp Leu
1 5 10 15
Phe Cys Gly Asn Ile Ala Asp Phe Ile Val Lys Gly Gly His Ser Lys
20 25 30
Ser Ser Lys Thr Ser Ala Ile Asp Ala Ala Thr Gly Glu Ser Leu Ser
35 40 45
His Val Asn Gln His Ile Leu Ser Arg Gln Ile Ala Ser Ile Leu Thr
50 55 60
Glu Ser Gly Tyr Glu Pro Asn Phe Asp Pro Lys Ser His Ile Gly Asp
65 70 75 80
Val Leu Val Thr Leu Phe Pro Asn Ser Ile Tyr Ser Ser Pro Val His
85 90 95
Trp Ala Ala Leu Ile Arg Gly Gly Thr Val Ser Pro Ala Ser Val Ser
100 105 110
Tyr Thr Leu Asn Glu Leu Ala His Gln Val Arg Thr Val Arg Pro Lys
115 120 125
Val Ile Val Ala Cys Lys Ser Lys Val Ser Leu Ala Lys Lys Ala Val
130 135 140
Leu Met Ala Arg Val Lys Thr Ala Val Leu Glu Leu Glu His Val Ile
145 150 155 160
Ser Asn Ala Pro Lys Tyr Pro Glu Ser Asp Ser Val Lys Phe Asn Lys
165 170 175
Asn Ser Gly Tyr Arg Arg Val Ala Tyr Leu Ala Met Ser Ser Gly Thr
180 185 190
Ser Gly Gly Ile Phe Lys Ala Val Lys Ile Thr His Phe Asn Ile Thr
195 200 205
Ser Cys Ile Gln Val Cys Gln Lys Ser Ala Pro Asn Arg Asp Thr Ala
210 215 220
Ser Gln Ile Ala Ser Ala Val Ile Pro Val Ser His Leu Tyr Gly Leu
225 230 235 240
Ser Lys Phe Leu Ile Met Ala Pro Tyr Val Gly Ser Thr Thr Val Phe
245 250 255
His Glu Lys Phe Glu Ile Lys Glu Phe Leu Glu Ala Gln Lys Gln Phe
260 265 270
Gln Val Asn Ser Trp Pro Ile Val Pro Pro Leu Val Val Leu Leu Thr
275 280 285
Asn His Pro Leu Val Lys Glu Phe Ser Glu Ser Leu Arg Ala His Leu
290 295 300
Arg Ile Val Cys Cys Gly Ala Ala Pro Leu Gly Glu Lys Ala Ala Arg
305 310 315 320
Asp Phe Leu Thr Ala Ile Thr Gly Ser Pro Asp Gly Ile Ile Gln Pro
325 330 335
Thr Ile Thr Ser Arg Asp Lys Ser Lys Ser Arg Asp Ser Gly Phe Phe
340 345 350
Ser Ser Ile Arg Ala His Val Ala Asp Pro Ala Ala Ala Gly Ile Thr
355 360 365
Ser Ala Asn Thr Ala Glu Ser Ala Gly Gln Ser Arg Asp Ala Pro Arg
370 375 380
Leu Gln Ile Ile Gln Gly Trp Gly Leu Thr Glu Thr Ser Pro Thr Cys
385 390 395 400
Thr Thr Phe Asp Pro Leu Asp Pro Asp Leu His Ile Lys Ala Cys Gly
405 410 415
Lys Ile Val Ala Asn Thr Glu Ile Arg Ile Arg Gly Gln Gly Gln Asp
420 425 430
Leu Gln Lys Ala Pro Ile Leu Ile Glu Asn Tyr Asp Ala Tyr Pro Ser
435 440 445
Lys Glu Thr Leu Pro Ile Gly Asp Ile Tyr Val Arg Gly Pro Gln Val
450 455 460
Thr Leu Gly Tyr Leu Asn Asp Asp His Ala Asp Ser Val Ser Phe Glu
465 470 475 480
Gln Cys Tyr Asp Pro His Val Pro Trp Phe His Leu Lys Trp Phe Lys
485 490 495
Thr Gly Asp Val Gly Phe Ile Asp Ala Lys Gly Arg Val Met Val Val
500 505 510
Asp Arg Thr Lys Glu Met Ile Lys Ser Met Gly Lys Gln Val Ala Pro
515 520 525
Ala Glu Ile Glu Asp Leu Leu Leu Ser His Glu Leu Val Ala Asp Ala
530 535 540
Ala Val Ile Gly Val Ser Asn Glu Lys Leu Gly Thr Glu Ser Pro Arg
545 550 555 560
Ala Phe Val Val Pro Lys Ser Gly Phe Lys Ala Ala Glu Leu Arg Ser
565 570 575
Trp Thr Asp Ser Gln Leu Pro Lys His Lys Gln Leu His Gly Gly Ile
580 585 590
Val Leu Val Asp Lys Val Pro Lys Asn Ala Ser Gly Lys Ile Leu Arg
595 600 605
Arg Val Leu Arg Glu Arg Arg Gly Asp Leu Val Glu Gly Val Lys Leu
610 615 620
Ser Lys Leu
625
<210> 51
<211> 593
<212> PRT
<213> yarrowia lipolytica
<400> 51
Met Ile Ile His Thr Ser Leu Asn Gln Pro Ile Asp Asp Arg Glu Phe
1 5 10 15
Phe Asp Gly Thr Ile Pro Asp Phe Ile Arg Thr Ser Pro Phe Val Asn
20 25 30
Asp Thr Thr Pro Phe Phe Ile Asp Ala Ala Thr Gly Glu Gln Leu Ser
35 40 45
Gln Arg Gln Leu Trp Lys Met Ser Asn Asp Phe Leu Thr Ile Phe Arg
50 55 60
Gln His Gly Leu Gly Asn Ala Arg Asp Asn Val Ser Asp Pro Ser Met
65 70 75 80
Gly Asp Val Phe Ile Thr Leu Phe Pro Asn Cys Ile Trp Ala Gly Pro
85 90 95
Val His Trp Ala Ala Leu Asp Ala Gly Ala Thr Leu Ser Pro Ala Ser
100 105 110
Cys Ser Tyr Thr Val Gln Glu Phe Ala His Gln Leu Gln Leu Val Val
115 120 125
Pro Lys Met Val Val Tyr Ser Glu Pro Phe Lys Gln Leu Leu Glu Asp
130 135 140
Ala Ile Ile Val Ser Lys Thr Asn Pro Thr Ile Leu Ser Leu Glu Gln
145 150 155 160
Leu Ile Glu Asp Ser Glu Arg Val Pro Leu Ala Gln Ala Asn Phe Gln
165 170 175
Phe Ala Asn Arg Leu Gln Leu Arg Pro Lys Glu Ser Ile Thr Arg Val
180 185 190
Ala Tyr Leu Ala Met Ser Ser Gly Thr Ser Gly Gly Leu Phe Lys Ala
195 200 205
Val Arg Ile Thr His Gly Asn Ile Thr Ser Asn Ala Ile Met Ser Thr
210 215 220
Lys Ser Ser Asn Ala Leu Leu Lys Thr Asn Gln Val Ala Ser Ala Ile
225 230 235 240
Ile Pro Val Ser His Leu Tyr Gly Leu Ala Gln Phe Leu Val Phe Gly
245 250 255
Val His Arg Gly Thr Ala Ala Val Phe His Lys Gly Phe Asp Phe Ile
260 265 270
Glu Phe Leu Asp Ala Ala Val Lys Tyr Lys Val Asn Ile Phe Pro Leu
275 280 285
Val Pro Pro Ile Ile Ile Leu Leu Ala Lys His Pro Phe Thr Gln Lys
290 295 300
Tyr Val Pro Asp Leu Lys Arg Asn Leu Thr Thr Val Leu Ser Gly Ala
305 310 315 320
Ala Pro Leu Gly Val Lys Ala Thr Glu Glu Phe Leu Glu Arg Ile Thr
325 330 335
Gly Arg Lys Asp Gly Val Ser Glu Tyr Gly Thr Leu Arg Val Ile Gln
340 345 350
Gly Trp Gly Met Thr Glu Thr Ser Pro Val Cys Thr Leu Phe Asp Pro
355 360 365
Glu Val Pro Val Ala His Ile Arg Ser Val Gly Lys Leu Val Ser Asn
370 375 380
Thr Glu Ala Arg Val Val Ser Glu Gly Val Asp Gln Pro Ala Cys Asp
385 390 395 400
Val Asp Pro Ala Ser Leu Asp Ala Ala Ile Lys Ala Gly Gly Leu Pro
405 410 415
Thr Gly Glu Ile Leu Ile Arg Gly Pro His Val Met Asp Gly Tyr His
420 425 430
Lys Asn Pro Ser Ala Asn Ala Asp Ala Phe Glu Glu Ala Ser Asp Trp
435 440 445
Thr Pro Asp Met Pro Trp Tyr Lys Lys Arg Trp Leu Arg Thr Gly Asp
450 455 460
Val Gly Phe Phe Asp Leu Gln Gly Arg Val Met Ile Val Asp Arg Thr
465 470 475 480
Lys Glu Leu Ile Lys Ser Met Gly Lys Gln Val Ala Pro Ala Glu Leu
485 490 495
Glu Asp Ala Leu Leu Ala Asn Pro Leu Val Ala Asp Cys Ala Val Ile
500 505 510
Gly Val Met Asp Val Asp Lys Gly Thr Glu Ser Pro Arg Ala Phe Val
515 520 525
Val Leu Arg Asp Pro Lys Ala Asp Ala Val Gly Ile Leu Lys Ser Leu
530 535 540
Asn Ser Gln Met Pro Lys Tyr Lys Asn Leu His Gly Gly Ile Val Val
545 550 555 560
Val Glu Ala Val Pro Arg Asn Pro Ser Gly Lys Val Leu Arg Arg Leu
565 570 575
Leu Arg Asp Arg Lys Asp Asp Val Val Leu Gly Leu Asp Val Ser Lys
580 585 590
Leu
<210> 52
<211> 583
<212> PRT
<213> yarrowia lipolytica
<400> 52
Met His Arg Arg Pro Asn Cys Pro Val Leu Phe Tyr Thr Ser Ser Ala
1 5 10 15
Ser Tyr Asp Ile Ala Leu Leu Val Leu Asn Thr Leu Ala Leu Pro Leu
20 25 30
Phe Leu Pro Gly Arg Thr Pro Leu Arg Cys Ile Val Phe Arg Leu Pro
35 40 45
Arg Asn Arg Ser Ala Phe Ile Thr His Leu Tyr Ile Thr Pro Leu Ser
50 55 60
Thr Pro Ser Ser His Asp Thr Thr Ser Ile His Thr Met Ala Thr Leu
65 70 75 80
Gln Lys Thr Ile Ser Lys Thr Gly Ala Gly Ile Phe Ile Pro Gly Ala
85 90 95
Gln Glu Leu Thr Tyr Ser Gln Phe Phe Asp Leu Ile Gly Asp Phe Gln
100 105 110
Lys Gln Leu Ala Gln Val Gly Leu Pro Pro Gln Ser Ala Val Ser Ile
115 120 125
Ala Ile Pro Asn Ser Leu Glu Phe Ala Val Thr Phe Leu Ala Val Thr
130 135 140
Phe Ser Arg Tyr Ile Ala Ala Pro Leu Asn Ser Ala Tyr Lys Lys Ser
145 150 155 160
Glu Phe Glu Phe Tyr Ile Asp Asp Leu Lys Ser Lys Leu Val Leu Val
165 170 175
Pro Lys Gly Ala Val Ala Gln Asn Leu Ala Ser Val Gln Ala Ala Arg
180 185 190
Thr Phe Asn Ala Ala Ile Ala Glu Val Tyr Trp Asp Asp Gln Lys Lys
195 200 205
Arg Ile Val Met Asp Ile Lys Glu Gly Pro Thr Asn Pro Pro Val Ala
210 215 220
Val Pro Thr Pro Asp Glu Val Ser Pro Glu Asp Val Ala Leu Val Leu
225 230 235 240
His Thr Ser Gly Thr Thr Gly Arg Pro Lys Ala Val Pro Leu Thr Gln
245 250 255
Arg Asn Leu Cys Arg Thr Met His Asn Ile Val Asp Thr Tyr Lys Leu
260 265 270
Thr Ser Lys Asp Thr Thr Tyr Leu Val Met Pro Leu Phe His Val His
275 280 285
Gly Leu Leu Cys Ala Phe Leu Ala Pro Leu Ala Ser Gly Gly Gly Ile
290 295 300
Val Ile Pro Ser Lys Phe Ser Ala Ser Gln Phe Trp Asp Asp Phe Val
305 310 315 320
Lys Tyr Lys Cys Asn Trp Tyr Thr Ala Val Pro Thr Ile His Gln Ile
325 330 335
Leu Leu Asn Thr Lys Ile Pro Gln Pro Leu Pro Glu Ile Arg Phe Ile
340 345 350
Arg Ser Cys Ser Ser Ala Leu Ala Pro Ala Thr Phe His Gln Ile Glu
355 360 365
Lys Ala Phe Lys Ala Pro Val Leu Glu Ala Tyr Ala Met Thr Glu Ala
370 375 380
Ala His Gln Met Thr Ser Asn Asn Leu Pro Pro Gly Gln Arg Lys Pro
385 390 395 400
Gly Thr Val Gly Val Gly Gln Gly Val Glu Val Ala Ile Leu Asp Asp
405 410 415
Asn Gly Asp Glu Val Pro Gln Gly Lys Ile Ala Glu Ile Cys Ile Arg
420 425 430
Gly Glu Asn Val Thr Lys Gly Tyr Ile Asn Asn Pro Glu Ala Asn Lys
435 440 445
Ser Ser Phe Thr Lys Ser Gly Phe Phe Arg Thr Gly Asp Gln Gly Phe
450 455 460
Leu Asp Lys Asp Gly Phe Val Asn Ile Thr Gly Arg Ile Lys Glu Leu
465 470 475 480
Ile Asn Arg Gly Gly Glu Lys Ile Ser Pro Ile Glu Leu Asp Gly Val
485 490 495
Met Leu Glu His Pro Ala Val Ala Glu Ala Val Cys Phe Gly Ala Pro
500 505 510
Asp Glu Met Tyr Gly Gln Gln Val Asn Ala Ala Ile Val Leu Lys Lys
515 520 525
Asp Ala Lys Ala Thr Glu Gln Asp Ile Lys Asp Phe Met Ala Asp Lys
530 535 540
Val Ala Lys Phe Lys Ile Pro Ala Arg Val Phe Phe Thr Asp Ile Met
545 550 555 560
Pro Lys Thr Ala Thr Gly Lys Ile Gln Arg Arg Phe Val Ala Gln Lys
565 570 575
Phe Leu Asp Lys Ala Lys Leu
580
<210> 53
<211> 585
<212> PRT
<213> yarrowia lipolytica
<400> 53
Met Ala Lys Gly Lys Leu Ser Ser Gly Gly Val Lys Ser Ser Val Ala
1 5 10 15
Asp Lys Thr Thr Ala Ala Ala Ile His Thr Leu Pro Arg Ile Gln Gly
20 25 30
Asp Asp Thr Val Tyr Val Pro Asp Lys Val Asn Arg His Asn Met Asn
35 40 45
Pro Thr Tyr Phe Leu Pro Arg Ala Ala Glu Ile Glu Pro Asn Ala Lys
50 55 60
Ala Tyr Ile His Lys Gly Ala Asp Gly Val Arg Val Glu Arg Thr Tyr
65 70 75 80
Gly Glu Met Ala Asp Arg Val Leu Gly Leu Ala Thr Tyr Phe Lys Ser
85 90 95
Lys Glu Phe Lys Arg Val Ala Ile Cys Gly Pro Asn Thr Pro Ala His
100 105 110
Leu Glu Thr Met Phe Gly Ala Val Ala Ala Gly Ala Tyr Val Leu Gly
115 120 125
Leu Asn Tyr Arg Leu Thr Met Gly Glu Ile Thr Tyr Lys Met Glu Leu
130 135 140
Gly Asp Ala Asp Cys Val Val Val Asp Arg Glu Phe Val His Leu Ile
145 150 155 160
Ser Pro Glu Ile Arg Ala Lys Cys Gln Val Ile Val Asp Asp Asp Val
165 170 175
Ser Gly Ala Ser Lys Pro Gln Gln Pro Gly Glu Ile Leu Tyr Ser Asn
180 185 190
Val Val Gln Gln Gly Met Gln Leu Ala Lys Glu Gln Lys Thr Thr Trp
195 200 205
Asp Asn Leu His Val Gln Asn Glu Asp Glu Asp Glu Ile Leu Gly Leu
210 215 220
Phe Tyr Thr Ser Gly Thr Thr Gly Lys Pro Lys Ala Val Glu Tyr Thr
225 230 235 240
His Arg Ser Val Tyr Leu Cys Ala Met Ser Asn Ile Ile Glu Ala Gly
245 250 255
Leu Asn Cys Glu Thr Val Asp Gly His Asn Arg Cys His Tyr Leu Trp
260 265 270
Thr Leu Pro Leu Phe His Ala Ala Gly Trp Thr Phe Pro Tyr Ser Val
275 280 285
Thr Ala Val Arg Gly Thr His Val Leu Leu Arg Lys Ile Glu Pro Asp
290 295 300
Tyr Ile Trp Asp Leu Leu Val Asp Asp Arg Ile Thr His Phe Asn Ala
305 310 315 320
Ala Pro Thr Val Asn Asn Met Leu Val Asn Asn Pro Lys Ala Arg Arg
325 330 335
Leu Pro Gln Thr Val Arg Val Thr Val Ala Ala Ser Pro Pro Ser Ala
340 345 350
Ala Leu Phe Asn Lys Met Phe Asp Met Asn Leu His Pro Val His Met
355 360 365
Tyr Gly Leu Thr Glu Ser Tyr Gly Pro Phe Val Arg Asn Tyr Phe Leu
370 375 380
Gln Asp Trp His Gly Ala Thr Gly Asp Glu Arg Tyr Ala Leu Met Ala
385 390 395 400
Arg Gln Gly Phe Ala Phe Val Gly Ser Gln Ser Val Gln Val Ile Ala
405 410 415
Asn Asn Asp Ile Asn Gln Pro Val Pro Arg Asn Gly Gln Glu Ile Gly
420 425 430
Glu Ile Val Cys Arg Gly Asn Ala Val Met Ala Arg Tyr His Lys Asp
435 440 445
Pro Glu Ala Thr Ala Lys Ala Phe Glu Gln Gly Trp Phe His Thr Gly
450 455 460
Asp Leu Ala Val Val Asn Pro Asp Gly Ser Ile Lys Ile Leu Asp Arg
465 470 475 480
Lys Lys Asp Ile Ile Ile Ser Gly Gly Glu Asn Ile Ser Ser Val Ala
485 490 495
Val Glu Gly Ile Ile Cys Lys Tyr Asp Asn Val Leu Glu Val Ala Val
500 505 510
Val Gly Ile Pro Asp Glu Lys Tyr Gly Glu Val Pro Lys Ala Phe Leu
515 520 525
Ile Leu Lys Asp Lys Ser Lys Pro Phe Asp Thr Asp Lys Met Ile Ala
530 535 540
Trp Met Arg Glu Arg Met Gly Ala Tyr Gln Ile Pro Arg Gln Val Ser
545 550 555 560
Val Val Asp Asp Leu Pro Arg Thr Ser Thr Gly Lys Ile Lys Lys Asn
565 570 575
Val Leu Arg Asp Ser Val Gln Ala Ala
580 585
<210> 54
<211> 712
<212> PRT
<213> yarrowia lipolytica
<400> 54
Met Lys Thr Ile Leu Lys Ile Thr Lys Ser Glu Asn Gln Asn Ala Leu
1 5 10 15
Phe Lys Asn Pro Ile Ser Pro Pro His Pro Pro Gln Thr Arg Thr Pro
20 25 30
Ser Leu Lys Ile Lys Val Gln Pro Gln Ile Pro His Phe Phe His Ala
35 40 45
Gly Pro Tyr Ile Asn Arg Gly Cys Pro Phe Leu Ser Pro Leu Leu His
50 55 60
Tyr His Leu Val Glu Ile Pro Thr Thr Met Thr Ala Gly Leu Val Ala
65 70 75 80
Ala Ala Ala Ile Gly Ala Ala Tyr Leu Glu Ala Lys Thr Leu Ile Ser
85 90 95
Glu Asp Ala Tyr Met Ile Arg Gly Ala Met Thr Asn Gly Leu Asp Phe
100 105 110
Phe Tyr Asn Ala Trp Lys Gly Arg Val Gln Tyr Trp Tyr Ala Phe Glu
115 120 125
Asp Ala Val Lys Lys Tyr Pro Asn Asn Pro Ala Ile Val Tyr Pro Lys
130 135 140
Pro Ile Glu Gly Lys Lys Pro Ser Gly Asp Ser Tyr Asp Asp Leu Phe
145 150 155 160
Asp Val Glu Thr Phe Thr Tyr Gln Gln Leu Tyr Asp Glu Val Leu Lys
165 170 175
Met Ser His Leu Leu Arg Asn Lys Tyr Gly Val Thr Ala Asn Asp Thr
180 185 190
Ile Ala Leu Asn Ala Met Asn Ser Pro Leu Phe Ile Ile Val Trp Phe
195 200 205
Ala Ile Trp Asn Leu Gly Ala Thr Pro Ala Phe Ile Asn Tyr Asn Leu
210 215 220
Ala Asp Lys Ser Leu Leu His Cys Leu Lys Val Gly His Ala Ser Ile
225 230 235 240
Met Phe Val Asp Thr Glu Val Glu Gly Asn Val Arg Pro Ser Leu Ala
245 250 255
Glu Ile Lys Ser Glu Ala Lys Cys Asp Thr Val Phe Met Asp Asp Asp
260 265 270
Phe Leu Ala Ala Tyr Ala Ala Ser Pro Ala Tyr Arg Ala Pro Asp Tyr
275 280 285
Glu Arg His Pro Glu Gln Lys Asp Tyr Asp Thr Ala Val Leu Ile Tyr
290 295 300
Thr Ser Gly Thr Thr Gly Leu Pro Lys Pro Ala Ile Met Ser Trp Lys
305 310 315 320
Lys Ala Lys Leu Met Ser Ser Leu Tyr Gly His Ser Ile Arg Leu Lys
325 330 335
Asn Asn Gly Val Val Tyr Ser Ala Met Pro Leu Tyr His Ser Thr Ala
340 345 350
Ala Ile Leu Gly Cys Leu Pro Cys Leu Asn Arg Gly Ala Ala Tyr Ala
355 360 365
Pro Gly Arg Lys Phe Ser Thr Thr Thr Phe Trp Thr Gln Ala Lys Leu
370 375 380
Thr Asn Ala Thr His Ile Gln Tyr Val Gly Glu Thr Cys Arg Tyr Leu
385 390 395 400
Ile Asn Ala Pro Pro Ser Pro Asp Glu Lys Ser His Gln Ile Lys Val
405 410 415
Ala Phe Gly Asn Gly Met Arg Arg Asp Ile Trp Val Lys Phe Lys Glu
420 425 430
Arg Phe Asn Ile Pro Ala Ile Gly Glu Phe Tyr Ala Ala Thr Glu Gly
435 440 445
Pro Leu Gly Thr Asn Asn Phe Gln Gln Gly Glu Ile Gly Ile Gly Ala
450 455 460
Met Gly Arg Tyr Gly Lys Leu Leu Ala Ala Ile Leu Ala Thr Arg Gln
465 470 475 480
Thr Ile Val Pro Val Asp Pro Glu Asp Glu Thr Glu Leu Trp Arg Asp
485 490 495
Pro Glu Thr Gly Phe Cys Arg Val Ala Gln Ser Asp Glu Pro Gly Glu
500 505 510
Phe Ile Gln Lys Ile Pro Asn Pro Glu Lys Val His Glu Thr Phe Gln
515 520 525
Gly Tyr Leu Gly Asn Asp Lys Ala Thr Asn Ser Lys Ile Met Arg Asp
530 535 540
Val Phe Lys Lys Gly Asp Ala Tyr Tyr Arg Thr Gly Asp Leu Val Arg
545 550 555 560
Leu Asn Asp Glu Gln Cys Tyr Tyr Phe Val Asp Arg Leu Gly Asp Thr
565 570 575
Phe Arg Trp Lys Ser Glu Asn Val Ser Thr Ser Glu Val Glu Glu His
580 585 590
Val Gly Ala Ser Asp Pro Asn Ile Glu Gln Val Val Cys Val Gly Val
595 600 605
Lys Val Pro Glu His Glu Gly Arg Ala Gly Phe Ala Val Val Lys Leu
610 615 620
Lys Asp Ala Ser Val Lys Pro Asn Leu Asp Gln Ile Ala Glu Tyr Ser
625 630 635 640
Leu Lys Gln Leu Pro Lys Tyr Ala Val Pro Leu Phe Ile Lys Phe Val
645 650 655
Asp Glu Ile Glu Arg Thr Gly Asn Asn Lys Val Gln Lys Val Lys Tyr
660 665 670
Lys Asn Gln Lys Met Pro His Glu Glu Gly Glu Ser Pro Ile Tyr Trp
675 680 685
Leu Lys Gly Asn Lys Tyr Val Glu Leu Asp Ala Gly Asp Trp Ala Ser
690 695 700
Leu Gly Ser Gly Lys Ile Lys Leu
705 710
<210> 55
<211> 1782
<212> DNA
<213> Artificial sequence
<220>
<223> YlACoS-5PS3
<400> 55
atggcctcaa tcattcacaa gtctcctgtg cccgacgttc agctgttcta cggttcctgg 60
ccagatctca tgcgaacctc tcctcatgcc cacaacgact ccaaacccgt ggtctttgac 120
ttcgatacca agcagcaact tacttggaag caggtctggc aactcagcgc tcgactcaga 180
gcccagctgt accacaagta cggaatcggc aaacccggtg ctcttgcacc tttccacaac 240
gatccctctc tcggagacgt ggtcatcttc tacactccca acacctacag ctcgttgccc 300
tatcatctgg ctcttcacga tctcggagcc accatttctc ctgcctccac atcttacgac 360
gtcaaggaca tttgccatca gatcgttact accgatgcgg tcgtggttgt cgctgcagcc 420
gagaaatccg agattgctcg agaggccgtt cagctgtctg gtcgagacgt cagagttgtg 480
gtcatggagg acctcatcaa caatgctccc accgttgcgc agaacgatat cgactcggca 540
cctcatgtgt ccctgtctcg ggaccaggct cgagccaaga ttgcatacct gggcatgtct 600
tccggtacgt ctggcggact tcccaaggct gttcgtctca ctcacttcaa cgttacctcg 660
aactgtctgc aggtctccgc tgccgcaccc aaccttgccc agaacgtggt tgccagcgcc 720
gtcattccaa ccactcacat ctacggtctc accatgtttc tgtcggttct tccctacaac 780
ggttccgtgg tcattcatca caagcaattc aacttgcgag atctgctcga ggctcagaag 840
acatacaagg tctctctgtg gattctcgtt cctcccgtca tcgtgcagct tgccaagaac 900
cctatggtcg acgagtacct ggactccatt cgagcccatg tgcggtgcat cgtctctgga 960
gctgctcctc tcggtggcaa tgtcgtggat caggtttcgg ttcgtcttac cggcaacaag 1020
gaaggcattc tgcccaacgg agacaagctc gtcattcatc aagcctacgg tcttaccgag 1080
tcctctccca tcgttggaat gctcgatcct ctgtcggacc acatcgacgt catgactgtg 1140
ggctgtctca tgcccaatac cgaggctcga attgtcgacg aagagggaaa cgatcagcca 1200
gcagtccacg ttaccgacac acgaggcatc ggtgccgctg tcaagcgagg cgagaagatt 1260
ccctccggag aactctggat tcgaggtcct cagatcatgg acggatacca caagaacccc 1320
gagtcgtctc gtgagtccct ggaacccagc acagagacct acggtctgca acatttccag 1380
gacagatggc ttcgaactgg agacgttgct gtcatcgaca ccttcggacg agtcatggtt 1440
gtggatcgaa ccaaggagct catcaagtcc atgtctcgac aggttgctcc tgccgagctc 1500
gaagctcttc tgctcaacca tccttccgtc aacgatgtgg ctgtcgttgg cgtccacaac 1560
gacgataatg gcacagagtc agcacgagcg tttgtcgttc ttcaaccagg cgacgcctgt 1620
gatcctacta ccatcaagca ctggatggac cagcaagttc cctcctacaa gcggctgtac 1680
ggaggcattg tggtcatcga cactgttccc aagaatgcct ctggcaagat tctgcgaaga 1740
ctgcttcgac agcggagaga cgatcgagtc tggggtctgt aa 1782
<210> 56
<211> 593
<212> PRT
<213> Artificial sequence
<220>
<223> YLACoS-5PS3 protein
<400> 56
Met Ala Ser Ile Ile His Lys Ser Pro Val Pro Asp Val Gln Leu Phe
1 5 10 15
Tyr Gly Ser Trp Pro Asp Leu Met Arg Thr Ser Pro His Ala His Asn
20 25 30
Asp Ser Lys Pro Val Val Phe Asp Phe Asp Thr Lys Gln Gln Leu Thr
35 40 45
Trp Lys Gln Val Trp Gln Leu Ser Ala Arg Leu Arg Ala Gln Leu Tyr
50 55 60
His Lys Tyr Gly Ile Gly Lys Pro Gly Ala Leu Ala Pro Phe His Asn
65 70 75 80
Asp Pro Ser Leu Gly Asp Val Val Ile Phe Tyr Thr Pro Asn Thr Tyr
85 90 95
Ser Ser Leu Pro Tyr His Leu Ala Leu His Asp Leu Gly Ala Thr Ile
100 105 110
Ser Pro Ala Ser Thr Ser Tyr Asp Val Lys Asp Ile Cys His Gln Ile
115 120 125
Val Thr Thr Asp Ala Val Val Val Val Ala Ala Ala Glu Lys Ser Glu
130 135 140
Ile Ala Arg Glu Ala Val Gln Leu Ser Gly Arg Asp Val Arg Val Val
145 150 155 160
Val Met Glu Asp Leu Ile Asn Asn Ala Pro Thr Val Ala Gln Asn Asp
165 170 175
Ile Asp Ser Ala Pro His Val Ser Leu Ser Arg Asp Gln Ala Arg Ala
180 185 190
Lys Ile Ala Tyr Leu Gly Met Ser Ser Gly Thr Ser Gly Gly Leu Pro
195 200 205
Lys Ala Val Arg Leu Thr His Phe Asn Val Thr Ser Asn Cys Leu Gln
210 215 220
Val Ser Ala Ala Ala Pro Asn Leu Ala Gln Asn Val Val Ala Ser Ala
225 230 235 240
Val Ile Pro Thr Thr His Ile Tyr Gly Leu Thr Met Phe Leu Ser Val
245 250 255
Leu Pro Tyr Asn Gly Ser Val Val Ile His His Lys Gln Phe Asn Leu
260 265 270
Arg Asp Leu Leu Glu Ala Gln Lys Thr Tyr Lys Val Ser Leu Trp Ile
275 280 285
Leu Val Pro Pro Val Ile Val Gln Leu Ala Lys Asn Pro Met Val Asp
290 295 300
Glu Tyr Leu Asp Ser Ile Arg Ala His Val Arg Cys Ile Val Ser Gly
305 310 315 320
Ala Ala Pro Leu Gly Gly Asn Val Val Asp Gln Val Ser Val Arg Leu
325 330 335
Thr Gly Asn Lys Glu Gly Ile Leu Pro Asn Gly Asp Lys Leu Val Ile
340 345 350
His Gln Ala Tyr Gly Leu Thr Glu Ser Ser Pro Ile Val Gly Met Leu
355 360 365
Asp Pro Leu Ser Asp His Ile Asp Val Met Thr Val Gly Cys Leu Met
370 375 380
Pro Asn Thr Glu Ala Arg Ile Val Asp Glu Glu Gly Asn Asp Gln Pro
385 390 395 400
Ala Val His Val Thr Asp Thr Arg Gly Ile Gly Ala Ala Val Lys Arg
405 410 415
Gly Glu Lys Ile Pro Ser Gly Glu Leu Trp Ile Arg Gly Pro Gln Ile
420 425 430
Met Asp Gly Tyr His Lys Asn Pro Glu Ser Ser Arg Glu Ser Leu Glu
435 440 445
Pro Ser Thr Glu Thr Tyr Gly Leu Gln His Phe Gln Asp Arg Trp Leu
450 455 460
Arg Thr Gly Asp Val Ala Val Ile Asp Thr Phe Gly Arg Val Met Val
465 470 475 480
Val Asp Arg Thr Lys Glu Leu Ile Lys Ser Met Ser Arg Gln Val Ala
485 490 495
Pro Ala Glu Leu Glu Ala Leu Leu Leu Asn His Pro Ser Val Asn Asp
500 505 510
Val Ala Val Val Gly Val His Asn Asp Asp Asn Gly Thr Glu Ser Ala
515 520 525
Arg Ala Phe Val Val Leu Gln Pro Gly Asp Ala Cys Asp Pro Thr Thr
530 535 540
Ile Lys His Trp Met Asp Gln Gln Val Pro Ser Tyr Lys Arg Leu Tyr
545 550 555 560
Gly Gly Ile Val Val Ile Asp Thr Val Pro Lys Asn Ala Ser Gly Lys
565 570 575
Ile Leu Arg Arg Leu Leu Arg Gln Arg Arg Asp Asp Arg Val Trp Gly
580 585 590
Leu
<210> 57
<211> 696
<212> PRT
<213> Candida tropicalis
<400> 57
Met Gly Ala Pro Leu Thr Val Ala Val Gly Glu Ala Lys Pro Gly Glu
1 5 10 15
Thr Ala Pro Arg Arg Lys Ala Ser Gln Lys Leu Ala Pro Val Glu Arg
20 25 30
Pro Thr Asp Ser Lys Ala Thr Thr Leu Pro Glu Phe Ile Glu Glu Cys
35 40 45
Phe Thr Arg Asn Gly Asn Arg Asp Ala Met Ala Trp Arg Asp Leu Leu
50 55 60
Glu Val His Val Glu Thr Lys Lys Val Thr Lys Ile Ile Asp Gly Glu
65 70 75 80
Gln Lys Lys Val Asp Lys Glu Trp Ile Tyr Tyr Glu Met Gly Pro Tyr
85 90 95
Asn Tyr Ile Ser Tyr Pro Lys Leu Leu Gln Leu Val Lys Asn Tyr Ser
100 105 110
Lys Gly Leu Leu Glu Leu Gly Leu Leu Pro Asp Gln Gln Ser Lys Leu
115 120 125
Met Ile Phe Ala Ser Thr Ser His Lys Trp Met Gln Thr Phe Leu Ala
130 135 140
Ser Ser Phe Gln Gly Ile Pro Val Val Thr Ala Tyr Asp Thr Leu Gly
145 150 155 160
Glu Ser Gly Leu Thr His Ser Leu Val Gln Thr Glu Ser Asp Ala Ile
165 170 175
Phe Thr Asp Asn Gln Leu Leu Gly Ser Leu Ile Arg Pro Leu Glu Lys
180 185 190
Ala Thr Ala Ile Lys Tyr Ile Ile His Gly Glu Lys Ile Asp Pro Asn
195 200 205
Asp Lys Arg Gln Gly Gly Lys Ile Tyr Gln Asp Ala Glu Lys Ala Lys
210 215 220
Glu Lys Ile Leu Gln Ile Arg Pro Asp Ile Lys Phe Ile Ser Tyr Asn
225 230 235 240
Glu Val Ile Ala Leu Gly Glu Lys Ser Ser Lys Glu Leu His Tyr Pro
245 250 255
Lys Pro Glu Asp Ser Ile Cys Ile Met Tyr Thr Ser Gly Ser Thr Gly
260 265 270
Asp Pro Lys Gly Val Val Ile Thr Asn His Asn Ile Val Ala Ala Val
275 280 285
Gly Gly Ile Ser Thr Asn Ala Thr Arg Asp Leu Val Arg Pro Ser Asp
290 295 300
Lys Val Val Ala Phe Leu Pro Leu Ala His Ile Phe Glu Leu Ala Phe
305 310 315 320
Glu Leu Ile Thr Phe Trp Trp Gly Ala Pro Leu Gly Tyr Ala Asn Val
325 330 335
Lys Thr Leu Thr Asp Ala Ser Cys Arg Asn Cys Gln Pro Asp Leu Ile
340 345 350
Glu Phe Lys Pro Thr Ile Met Val Gly Val Ala Ala Val Trp Glu Ser
355 360 365
Val Arg Lys Gly Val Leu Ser Lys Leu Lys Gln Ala Ser Pro Leu Gln
370 375 380
Gln Lys Ile Phe Trp Ala Ala Phe Lys Ala Lys Ser Thr Leu Asn His
385 390 395 400
Phe Gly Leu Pro Gly Gly Gly Met Phe Asp Val Ile Phe Lys Lys Val
405 410 415
Lys Ala Ala Thr Gly Gly Gln Leu Arg Tyr Val Leu Asn Gly Gly Ser
420 425 430
Pro Ile Ser Ile Asp Ala Gln Val Phe Ile Ser Thr Leu Leu Ala Pro
435 440 445
Met Leu Leu Gly Tyr Gly Leu Thr Glu Thr Cys Ala Asn Thr Thr Ile
450 455 460
Thr Glu His Thr Arg Phe Gln Ile Gly Thr Leu Gly Ala Leu Val Gly
465 470 475 480
Ser Val Thr Ala Lys Leu Val Asp Val Ala Asp Ala Gly Tyr Phe Ala
485 490 495
Lys Asn Asn Gln Gly Glu Ile Trp Leu Lys Gly Gly Pro Val Val Lys
500 505 510
Glu Tyr Tyr Lys Asn Glu Glu Glu Thr Lys Ala Ala Phe Thr Asp Asp
515 520 525
Gly Trp Phe Lys Thr Gly Asp Ile Gly Glu Trp Thr Ser Glu Gly Gly
530 535 540
Leu Asn Ile Ile Asp Arg Lys Lys Asn Leu Val Lys Thr Leu Asn Gly
545 550 555 560
Glu Tyr Ile Ala Leu Glu Lys Leu Glu Ser Val Tyr Arg Ser Asn His
565 570 575
Leu Val Met Asn Leu Cys Val Tyr Ala Asp Gln Thr Lys Val Lys Pro
580 585 590
Ile Ala Ile Val Leu Pro Ile Glu Ala Asn Leu Lys Thr Met Leu Lys
595 600 605
Asp Glu Lys Val Val Pro Asp Ala Asp Thr Gln Glu Leu Ser His Leu
610 615 620
Val His Asn Lys Lys Val Val Gln Ala Val Leu Arg His Leu Leu Gln
625 630 635 640
Thr Gly Lys Gln Gln Gly Leu Lys Gly Ile Glu Leu Leu Gln Asn Ile
645 650 655
Val Leu Leu Asp Glu Glu Trp Thr Pro Gln Asn Gly Phe Val Thr Ser
660 665 670
Ala Gln Lys Leu Gln Arg Lys Lys Ile Leu Glu Ser Cys Arg Lys Glu
675 680 685
Val Asp Glu Ala Tyr Lys Ser Ser
690 695
<210> 58
<211> 718
<212> PRT
<213> Candida tropicalis
<400> 58
Met Pro Ala Leu Phe Lys Glu Ser Ala Lys His Ile Leu Asp Thr Ile
1 5 10 15
Ala Ala Asp Leu Pro Leu Asp Gln Lys Leu Ala Ser Ile Ala Val Pro
20 25 30
Leu Glu Asn Ser Glu Glu Pro Gly Phe Ser Ala Ile Tyr Arg Asn Lys
35 40 45
Tyr Ser Leu Asp Lys Leu Ile Asp Thr Pro Tyr Pro Gly Val Asp Thr
50 55 60
Leu Tyr Lys Leu Phe Glu Val Ala Thr Glu Ala Tyr Gly Asp Lys Pro
65 70 75 80
Cys Leu Gly Ala Arg Val Lys Asn Gln Asp Gly Thr Phe Gly Glu Tyr
85 90 95
Lys Phe Gln Asp Tyr Asn Thr Ile Arg Gln Arg Arg Asn Asn Leu Gly
100 105 110
Ser Gly Ile Phe Phe Val Leu Gln Asn Asn Pro Tyr Lys Thr Ser Ser
115 120 125
Glu Ala His Ser Lys Leu Lys Tyr Asp Pro Ala Ser Lys Asp Ser Phe
130 135 140
Ile Leu Thr Ile Phe Ser His Asn Arg Pro Glu Trp Glu Leu Cys Asp
145 150 155 160
Leu Thr Ser Val Ala Tyr Ser Ile Thr Asn Thr Ala Leu Tyr Asp Thr
165 170 175
Leu Gly Pro Asp Thr Ser Lys Tyr Ile Leu Gly Leu Thr Glu Ser Pro
180 185 190
Ile Val Ile Cys Ser Lys Asp Lys Ile Lys Gly Leu Ile Asp Leu Lys
195 200 205
Lys Ser Asn Pro Glu Glu Leu Ser Asn Leu Ile Val Leu Val Ser Met
210 215 220
Asp Asp Leu Thr Thr Ala Asp Ser Ser Leu Lys Asn Tyr Gly His Glu
225 230 235 240
His Asn Val Thr Val Phe Asp Met Lys Gln Val Glu Lys Leu Gly Glu
245 250 255
Ile Asn Pro Leu Asp Pro Ile Glu Pro Thr Pro Asp Thr Asp Phe Thr
260 265 270
Ile Thr Phe Thr Ser Gly Thr Thr Gly Ala Asn Pro Lys Gly Val Val
275 280 285
Leu Asn His Arg Asn Ala Val Ala Gly Val Thr Phe Ile Leu Ser Arg
290 295 300
Tyr Asp Gly Lys Phe Asn Pro Arg Ala Tyr Ser Phe Leu Pro Leu Ala
305 310 315 320
His Ile Tyr Glu Arg Ala Ser Ile Gln Phe Ala Leu Ser Ile Gly Ser
325 330 335
Ala Ile Gly Phe Pro Gln Gly Pro Ser Pro Leu Thr Leu Leu Glu Asp
340 345 350
Val Lys Val Leu Gln Pro Asp Gly Leu Ala Leu Val Pro Arg Val Leu
355 360 365
Thr Lys Leu Glu Ala Ala Ile Arg Ser Gln Thr Ile Asn Asn Asp Glu
370 375 380
Lys Pro Leu Val Lys Ser Val Phe Ser Thr Val Ile Asn Ala Lys Met
385 390 395 400
Asp Leu Gln Thr Lys Asp Glu Asn Glu Asn Val Asn Pro Ser Leu Leu
405 410 415
Val Tyr Asp Arg Leu Leu Asn Met Leu Arg Lys Lys Ile Gly Met Gln
420 425 430
Asn Val Gln Tyr Ile Ser Thr Gly Ser Ala Pro Ile Ala Pro Ser Thr
435 440 445
Ile Gln Phe Leu Lys Ala Ala Leu Asn Val Gly Ile Met Gln Gly Tyr
450 455 460
Gly Leu Ser Glu Ser Phe Ala Gly Cys Met Ala Ser Ser Lys Phe Glu
465 470 475 480
Pro Glu Ala Ala Thr Cys Gly Ala Thr Ser Ile Thr Thr Glu Val Lys
485 490 495
Leu Lys Asp Leu Val Glu Met Gly Tyr Thr Ser Lys Asp Glu Gly Gly
500 505 510
Pro Arg Gly Glu Leu Leu Leu Arg Gly Pro Gln Ile Phe Arg Glu Tyr
515 520 525
Tyr Lys Asn Pro Glu Glu Thr Ala Lys Ala Ile Asp Glu Asp Gly Trp
530 535 540
Phe His Thr Gly Asp Val Ala Lys Ile Asn Ser Lys Gly Arg Ile Ser
545 550 555 560
Ile Ile Asp Arg Ala Lys Asn Phe Phe Lys Leu Ala Gln Gly Glu Tyr
565 570 575
Val Thr Pro Glu Lys Ile Glu Gly Leu Tyr Leu Ser Lys Phe Pro Tyr
580 585 590
Ile Ala Gln Leu Phe Val His Gly Asp Ser Lys Glu Ser Phe Leu Val
595 600 605
Ala Val Val Gly Leu Asp Pro Ile Ala Ala Lys Gln Tyr Met Glu His
610 615 620
Arg Phe His Asp Lys Ile Val Lys Glu Asp Asp Ile Val Glu Phe Phe
625 630 635 640
Lys Ser Pro Arg Asn Arg Lys Ile Leu Leu Gln Asp Met Asn Lys Ser
645 650 655
Ile Ala Asp Gln Leu Gln Gly Phe Glu Lys Leu His Asn Ile Tyr Val
660 665 670
Asp Phe Glu Pro Leu Thr Val Asp Arg Gly Val Ile Thr Pro Thr Met
675 680 685
Lys Ile Arg Arg Pro Ile Ala Val Lys Phe Phe Gln Asp Gln Ile Asp
690 695 700
Gly Met Tyr Asn Glu Gly Ser Leu Val Lys Asn Gly Ser Leu
705 710 715
<210> 59
<211> 718
<212> PRT
<213> Candida tropicalis
<400> 59
Met Pro Ala Leu Phe Lys Glu Ser Pro Gln Gln Ile Ser Gln Ser Leu
1 5 10 15
Lys Ala Lys Phe Ser Asp Pro Tyr Gln Phe Ala Thr Ser Val Pro Leu
20 25 30
Ser Asp Thr Lys Glu Pro Gly Tyr Ser His Ile Tyr Arg Asn Ser Tyr
35 40 45
Asp Pro Thr Thr Leu Ala Thr Cys Pro His Pro Glu Leu Asp Thr Leu
50 55 60
His Lys Ile Phe Glu Phe Ser Asn Thr Ile Tyr Ser Asp Ser Pro Phe
65 70 75 80
Leu Gly His Arg Val Lys Asn Pro Asp Gly Thr Phe Gly Glu Tyr Lys
85 90 95
Phe Gln Thr Tyr Arg Gln Ile Tyr Lys Arg Arg Asn Asp Phe Gly Ser
100 105 110
Gly Ile Tyr Tyr Val Leu Glu Asn Asn Pro Tyr Lys Thr Ser Ser Glu
115 120 125
Ala His Ser Lys Leu Lys Tyr Asp Pro Thr Asn Lys Asp Pro Phe Ile
130 135 140
Leu Ala Val Phe Ser His Asn Arg Pro Glu Trp Ala Leu Cys Asp Leu
145 150 155 160
Thr Thr Asn Ser Phe Gly Ile Ile Asn Thr Ala Leu Tyr Ser Thr Leu
165 170 175
Gly Pro Asp Thr Ser Arg Tyr Ile Leu Gly Val Thr Asp Cys Pro Ile
180 185 190
Val Val Thr Thr Lys Asp Lys Val Lys Gly Ile Ile Ser Leu Lys Asn
195 200 205
Ser Asn Gln Lys Glu Leu Ala Ser Leu Ile Thr Ile Val Ser Met Asp
210 215 220
Glu Leu Thr Glu Glu Asp Lys Glu Leu Arg Ser Phe Gly His Glu Asn
225 230 235 240
Asn Ile Thr Val Tyr Asp Ile Lys Glu Val Glu Asn Phe Gly Glu Lys
245 250 255
Asn Pro Leu Lys Pro Ile Glu Pro Thr Pro Asp Thr Ile Phe Thr Ile
260 265 270
Ser Phe Thr Ser Gly Thr Thr Gly Ala Ala Pro Lys Gly Val Val Leu
275 280 285
Thr Asn Arg Ile Leu Val Ser Gly Ile Thr Thr His Cys Thr Ile Leu
290 295 300
Ser Phe Gly Pro Glu Arg Val His Tyr Ser Phe Leu Pro Leu Ala His
305 310 315 320
Ile Tyr Glu Arg Met Leu Leu Gln Phe Gly Ile Leu Ala Gly Val Lys
325 330 335
Ile Gly Tyr Pro Gln Gly Pro Leu Pro Thr Thr Leu Phe Asp Asp Val
340 345 350
Lys Tyr Leu Gln Pro Thr Phe Leu Cys Leu Val Pro Arg Val Phe Thr
355 360 365
Lys Ile Glu Ala Ala Ile Lys Ala Gln Thr Val Glu Asn Asp Ala Asn
370 375 380
Pro Lys Ile Lys Thr Leu Phe Gln Asn Ile Val Asp Lys Lys Leu Lys
385 390 395 400
Leu Gln Gln Gln Glu Asp Phe Thr Asn Pro Ser Phe Pro Glu Gly Asp
405 410 415
Lys Val Leu Leu Gln Leu Arg Glu Lys Leu Gly Phe Gly Lys Ile Ala
420 425 430
Phe Met Asn Thr Gly Ser Ala Pro Leu Ser Glu Glu Thr Tyr Arg Phe
435 440 445
Leu Gln Ala Ile Leu Asn Leu Pro Asp Gly Phe Arg Ser Gly Tyr Gly
450 455 460
Leu Thr Glu Ser Ala Ser Gly Val Cys Ile Ser Pro Ala Tyr Ala Asn
465 470 475 480
Glu Phe Ser Cys Gly Pro Ile Ser Val Thr Thr Glu Phe Lys Leu Arg
485 490 495
Asp Ile Glu Glu Met Gly Tyr Thr Ser Ser Asp Lys Asp Gly Pro Arg
500 505 510
Gly Glu Leu Leu Leu Arg Gly Pro Gln Ile Phe Pro Tyr Tyr Tyr Lys
515 520 525
Asn Pro Glu Glu Thr Ala Lys Val Ile Asp Lys Asp Gly Trp Phe Tyr
530 535 540
Thr Gly Asp Val Ala Val Val Ser Pro Gln His Gly Asn Arg Leu Gln
545 550 555 560
Ile Ile Asp Arg Val Lys Asn Phe Phe Lys Leu Ser Gln Gly Glu Tyr
565 570 575
Val Ser Pro Glu Lys Ile Glu Asn Val Tyr Leu Ser Gln Phe Pro Tyr
580 585 590
Ile Ser Gln Leu Phe Ala His Gly Asp Ser Thr Glu Ser Tyr Leu Val
595 600 605
Gly Val Val Gly Ile Asp Lys Ala Ser Ile Asp Pro Tyr Leu Lys Lys
610 615 620
Arg Phe Asn Val Ser Ile Glu Lys Gln Ala Asp Ile Val Lys Tyr Phe
625 630 635 640
Glu Asn Pro Lys Asn Arg Arg Ala Leu Leu His Asp Met Asn Glu Ala
645 650 655
Ile Glu Gly Gln Leu Gln Gly Phe Glu Lys Leu His Asn Val Phe Val
660 665 670
Asp Phe Glu Pro Leu Thr Leu Glu Arg Glu Val Ile Thr Pro Thr Ile
675 680 685
Lys Ile Arg Arg Pro Val Ala Val Lys Phe Phe Lys Glu Gln Ile Glu
690 695 700
Asn Met Tyr Arg Glu Gly Ser Leu Ile Lys Gly Ser Asn Leu
705 710 715
<210> 60
<211> 741
<212> PRT
<213> Candida tropicalis
<400> 60
Met Thr Thr Leu Pro Ser Ile Ser Glu Thr Asp Ile Val Tyr Ala Thr
1 5 10 15
Asp Asp Lys Pro Tyr Val Phe Glu Asn Pro Asn Asp Leu Pro Val Glu
20 25 30
Thr Leu Val Asn His Ile Leu Pro Phe Pro Gln Glu Val Ala Gly Glu
35 40 45
Ser Ile Lys Ile Pro Gly Thr Ala Val Glu Gly Phe Ser Glu Ile Tyr
50 55 60
Arg Asn Ala Ala Thr Pro Asn Gly Ile Lys Ala Ser Leu Ile Lys Gly
65 70 75 80
Leu Asp Thr Tyr His Asp Ile Phe Glu Arg Ser Ala Asp Cys Tyr Ala
85 90 95
Asp Glu Pro Cys Leu Ala Phe His Glu Tyr Asp Tyr Glu Asn Ser Gln
100 105 110
His Leu Glu Arg Tyr Ala Thr Ile Ser Tyr Lys Glu Val Arg Gln Arg
115 120 125
Lys Asp Asp Phe Ala Ala Gly Leu Phe Phe Leu Leu Lys Ser Asn Pro
130 135 140
Phe Lys Asn Asn Ser Leu Glu Ser His Gln Lys Ile Asp Asn His Glu
145 150 155 160
Ala Asn Tyr Lys Ser Tyr Asn Ser Asp Asp Met Ser Phe Ile Ala Thr
165 170 175
Phe Tyr Ala Gly Asn Arg Val Glu Trp Ile Leu Ser Asp Leu Ala Cys
180 185 190
Ser Ser Asn Ser Ile Thr Ser Thr Ala Leu Tyr Asp Thr Leu Gly Pro
195 200 205
Glu Thr Ser Lys Tyr Ile Leu Glu Thr Thr Gln Ser Pro Val Ile Ile
210 215 220
Ser Ser Lys Asp His Ile Arg Gly Leu Ile Glu Leu Lys Arg Lys Tyr
225 230 235 240
Pro Glu Ala Leu Glu Ser Ile Ile Leu Ile Ile Ser Met Asp Pro Leu
245 250 255
Thr Lys Ser Asp Val Gly Leu Val Gln Leu Ala Glu Lys Ser Asn Ile
260 265 270
Lys Leu Tyr Asp Phe Ser Gln Val Glu Arg Thr Gly Ala Ile Phe Pro
275 280 285
His Glu Thr Asn Pro Pro Asn Ser Glu Thr Val Phe Thr Ile Thr Phe
290 295 300
Thr Ser Gly Thr Thr Gly Ala Asn Pro Lys Gly Val Val Leu Pro Gln
305 310 315 320
Arg Cys Ala Ala Ser Ala Met Leu Ala Tyr Ser Leu Leu Met Pro His
325 330 335
His Arg Gly Thr Arg Glu Phe Ala Phe Leu Pro Leu Ala His Ile Phe
340 345 350
Glu Arg Gln Met Val Ala Ser Met Phe Leu Phe Gly Gly Ser Ser Ala
355 360 365
Phe Pro Arg Leu Gly Gly Thr Pro Leu Thr Leu Val Glu Asp Leu Lys
370 375 380
Leu Trp Lys Pro Asn Phe Met Ala Asn Val Pro Arg Ile Phe Thr Lys
385 390 395 400
Ile Glu Ala Gly Ile Lys Ala Ser Thr Ile Asp Ser Thr Ser Gly Leu
405 410 415
Thr Arg Ser Leu Tyr Gly Arg Ala Ile Glu Ala Lys Arg Val Lys Gln
420 425 430
Ile Lys Asn Asp Asp Ser Gly Asp His Phe Ile Tyr Asp Lys Leu Leu
435 440 445
Ile Gln Lys Leu Arg Asn Ala Ile Gly Tyr Asp Lys Leu Glu Phe Cys
450 455 460
Val Thr Gly Ser Ala Pro Ile Ala Pro Glu Thr Ile Lys Phe Leu Lys
465 470 475 480
Ala Ser Leu Gly Ile Gly Phe Ala Gln Gly Tyr Gly Ser Ser Glu Ser
485 490 495
Phe Ala Gly Met Leu Phe Ala Leu Pro Phe Lys Thr Ser Ser Val Gly
500 505 510
Thr Cys Gly Val Ile Ala Pro Thr Met Glu Ala Arg Leu Arg Glu Leu
515 520 525
Pro Asp Met Gly Tyr Met Leu Asp Asp Ala Asn Gly Pro Arg Gly Glu
530 535 540
Leu Gln Ile Arg Gly Ala Gln Leu Phe Thr Lys Tyr Phe Lys Asn Asp
545 550 555 560
Glu Glu Thr Ala Lys Ser Ile Asp Glu Asp Gly Trp Phe Ser Thr Gly
565 570 575
Asp Val Ala Glu Ile Gly Ala Lys Asp Gly Tyr Phe Arg Ile Ile Asp
580 585 590
Arg Val Lys Asn Phe Tyr Lys Leu Ala Gln Gly Glu Tyr Val Ser Pro
595 600 605
Glu Lys Ile Glu Ser Leu Tyr Leu Ser Leu Asn Ser Thr Ile Ser Gln
610 615 620
Leu Phe Val His Gly Asp Ser Thr Lys Ser Tyr Leu Val Gly Val Val
625 630 635 640
Gly Leu Gln Pro Asp Val Ala Ser Lys Tyr Val Asp Leu Ser Ser Gly
645 650 655
Asp Lys Val Val Gln Glu Leu Asn Lys Pro Glu Leu Arg Lys Gln Ile
660 665 670
Leu Ser Asp Leu Asn Gly Lys Val Asn Gly Lys Leu Gln Gly Phe Glu
675 680 685
Lys Leu His Asn Ile Phe Ile Asp Ile Glu Pro Leu Thr Leu Glu Arg
690 695 700
Asn Val Val Thr Pro Thr Met Lys Leu Lys Arg His Phe Ala Ala Lys
705 710 715 720
Phe Phe Arg Ala Gln Ile Asp Ser Met Tyr Glu Glu Gly Ser Ile Val
725 730 735
Ala Asp Tyr Lys Leu
740
<210> 61
<211> 741
<212> PRT
<213> Candida tropicalis
<400> 61
Met Thr Thr Leu Pro Ser Ile Ser Glu Thr Asp Ile Val Tyr Ala Thr
1 5 10 15
Asp Asp Lys Pro Tyr Val Phe Glu Asn Pro Asn Asp Leu Pro Val Glu
20 25 30
Thr Leu Val Asn His Ile Leu Pro Phe Pro Gln Glu Val Ala Gly Glu
35 40 45
Ser Ile Lys Ile Pro Gly Thr Ala Val Glu Gly Phe Ser Glu Ile Tyr
50 55 60
Arg Asn Ala Ala Thr Pro Asn Gly Ile Lys Ala Ser Leu Ile Lys Gly
65 70 75 80
Leu Asp Thr Tyr His Asp Ile Phe Glu Arg Ser Ala Asp Cys Tyr Ala
85 90 95
Asp Glu Pro Cys Leu Ala Phe His Glu Tyr Asp Tyr Glu Asn Ser Gln
100 105 110
His Leu Glu Arg Tyr Ala Thr Ile Ser Tyr Lys Glu Val Arg Gln Arg
115 120 125
Lys Asp Asp Phe Ala Ala Gly Leu Phe Phe Leu Leu Lys Ser Asn Pro
130 135 140
Phe Lys Asn Asn Ser Leu Glu Ser His Gln Lys Ile Asp Asn His Glu
145 150 155 160
Ala Asn Tyr Lys Ser Tyr Asn Ser Asp Asp Met Ser Phe Ile Ala Thr
165 170 175
Phe Tyr Ala Gly Asn Arg Val Glu Trp Ile Leu Ser Asp Leu Ala Cys
180 185 190
Ser Ser Asn Ser Ile Thr Ser Thr Ala Leu Tyr Asp Thr Leu Gly Pro
195 200 205
Gly Thr Ser Lys Tyr Ile Leu Glu Ala Thr Gln Ser Pro Val Ile Ile
210 215 220
Thr Ser Lys Asp His Ile Lys Gly Leu Ile Glu Leu Lys Arg Lys Tyr
225 230 235 240
Pro Glu Ala Leu Glu Ser Ile Ile Leu Ile Ile Ser Met Asp Pro Leu
245 250 255
Thr Lys Ser Asp Val Gly Leu Val Gln Leu Ala Glu Asn Ser Asn Ile
260 265 270
Lys Leu Tyr Asp Phe Ser Gln Val Glu Arg Ala Gly Ala Ile Phe Pro
275 280 285
His Glu Thr Asn Pro Pro Asn Arg Glu Thr Val Phe Thr Ile Thr Phe
290 295 300
Thr Ser Gly Thr Thr Gly Ala Asn Pro Lys Gly Val Val Leu Ser Gln
305 310 315 320
Gly Ser Ala Ala Ser Ala Ser Phe Val Tyr Ser Leu Leu Met Pro His
325 330 335
Arg Arg Gly Ala Arg Asp Phe Ala Phe Leu Pro Leu Ala His Ile Phe
340 345 350
Gln Arg Gln Met Val Ala Ser Thr Leu Phe Phe Gly Gly Ser Ser Ala
355 360 365
Phe Pro Arg Leu Gly Gly Thr Pro Leu Thr Leu Val Glu Asp Leu Lys
370 375 380
Leu Trp Lys Pro Asn Phe Met Ala Asn Val Pro Arg Ile Phe Thr Lys
385 390 395 400
Ile Glu Ala Gly Ile Lys Ala Ser Thr Ile Asp Ser Thr Ser Gly Leu
405 410 415
Thr Arg Ser Leu Tyr Gly Arg Ala Ile Glu Ala Lys Arg Val Lys Gln
420 425 430
Ile Lys Asn Asp Asp Ser Gly Asp His Phe Ile Tyr Asp Lys Leu Leu
435 440 445
Ile Gln Lys Leu Arg Asn Ala Ile Gly Tyr Asp Lys Leu Glu Phe Leu
450 455 460
Leu Thr Gly Ser Ala Pro Ile Ser Pro Glu Thr Ile Lys Phe Leu Lys
465 470 475 480
Ser Ser Leu Gly Ile Gly Phe Gly Gln Gly Tyr Gly Ser Ser Glu Ser
485 490 495
Phe Gly Gly Ile Leu Phe Ala Leu Pro Phe Lys Asn Ser Ser Leu Gly
500 505 510
Thr Cys Gly Val Ile Ala Pro Thr Met Glu Ala Arg Leu Arg Glu Leu
515 520 525
Pro Asp Met Gly Tyr Met Leu Asp Asp Ala Asn Gly Pro Arg Gly Glu
530 535 540
Leu Gln Ile Arg Gly Ala Gln Leu Phe Ala Lys Tyr Phe Lys Asn Asp
545 550 555 560
Glu Glu Thr Ala Lys Ser Ile Asp Glu Asp Gly Trp Phe Ser Thr Gly
565 570 575
Asp Val Ala Glu Ile Gly Ala Lys Asp Gly Tyr Phe Arg Ile Ile Asp
580 585 590
Arg Val Lys Asn Phe Tyr Lys Leu Ala Gln Gly Glu Tyr Val Ser Pro
595 600 605
Glu Lys Ile Glu Asn Leu Tyr Leu Ser Leu Asn Ser Thr Ile Ser Gln
610 615 620
Leu Phe Ile His Gly Asp Ser Thr Lys Ser Tyr Leu Val Gly Val Val
625 630 635 640
Gly Leu Gln Pro Asp Val Ala Ser Lys Tyr Val Asp Leu Ser Ser Gly
645 650 655
Asp Lys Val Val Gln Glu Leu Asn Lys Pro Glu Leu Arg Lys Gln Ile
660 665 670
Leu Leu Asp Leu Asn Gly Lys Val Asn Gly Lys Leu Gln Gly Phe Glu
675 680 685
Lys Leu His Asn Ile Phe Ile Asp Ile Glu Pro Leu Thr Leu Glu Arg
690 695 700
Asn Val Val Thr Pro Thr Met Lys Leu Lys Arg His Phe Ala Ala Lys
705 710 715 720
Phe Phe Arg Ala Gln Ile Asp Ser Met Tyr Glu Glu Gly Ser Ile Ile
725 730 735
Ala Asp Tyr Lys Leu
740
<210> 62
<211> 749
<212> PRT
<213> Candida tropicalis
<400> 62
Met Ile Glu Ser Lys Ser Ile Phe Ser Gly Glu Lys Tyr Thr Lys Gln
1 5 10 15
Glu Ala Leu Ser Gln Leu Pro Phe Gly Ser Asp Val Glu Asn Ala Val
20 25 30
Met Ile Asp Glu Pro Val Thr Asn Val Lys Tyr Ser Pro Ile Phe Arg
35 40 45
Asn Lys Ala His Leu Asp Gly Leu Ile Gln Asn Val His Pro Asp Leu
50 55 60
Asn Thr His Tyr Lys Leu Phe Asn Asn Ala Ala Glu Met Tyr His Asp
65 70 75 80
Arg Pro Cys Leu Gly Lys Arg Pro Tyr Asn Tyr Thr Thr His Gln Ser
85 90 95
Asp Asp Tyr Phe Ser His Trp Thr Tyr Gly Glu Val Phe Thr Lys Lys
100 105 110
Asn Asn Leu Gly Ala Gly Phe Ile Arg Ala Leu Leu Glu Asn Pro Phe
115 120 125
Leu Asp Val Gln Leu Glu Ser His Arg Lys Val Val Asn His Leu Arg
130 135 140
Asp Trp Ser Asn Phe Gly Ile Asn Lys Leu Pro Arg Asp Asn Leu Asn
145 150 155 160
Cys Glu Ile Glu Lys Asn Cys Ser Phe Ile Leu Thr Ile Phe Ala Val
165 170 175
Asn Arg Ala Glu Trp Ile Leu Thr Asp Leu Ala Cys Ser Ser Tyr Gly
180 185 190
Ile Thr Asn Thr Ala Leu Tyr Asp Thr Leu Gly Pro Asp Val Ser Gln
195 200 205
Tyr Ile Leu Asn Leu Thr Glu Ser Pro Ile Val Val Cys Thr His Asp
210 215 220
Lys Ile Gln Val Leu Ile Asn Leu Lys Lys Lys Tyr Pro Gln Gln Thr
225 230 235 240
Lys Asn Leu Ile Ser Ile Val Ser Met Asp Pro Ile Asp Leu Val Thr
245 250 255
Gln Gly Thr Ile Glu Asp Ala Tyr Glu Leu Gly Ile Thr Ile Gln Gly
260 265 270
Leu Asn Gln Ile Glu Lys Ile Gly Ala Lys Asn Pro Ile His Gln Leu
275 280 285
Glu Thr Asn Pro Glu Ala Leu Phe Thr Ile Ser Phe Thr Ser Gly Thr
290 295 300
Thr Gly Ser Lys Pro Lys Gly Val Met Ile Ser Gln Gly Gly Ala Ala
305 310 315 320
Ala Tyr Ile Thr Tyr Leu Leu Cys Cys Glu Pro Gln Ala Lys Pro Gly
325 330 335
Asp Lys Ala Phe Ile Phe Leu Pro Leu Thr His Leu Tyr Glu Arg Gln
340 345 350
Thr Cys Gly Phe Ala Phe Ser Ser Gly Tyr Tyr Leu Gly Phe Pro Gln
355 360 365
Val Asn Leu Gly Lys Lys Lys Ile Asn Pro Phe Glu Asn Leu Leu Ala
370 375 380
Asp Leu Arg Ile Phe Lys Pro Thr Tyr Met Ser Met Val Pro Arg Leu
385 390 395 400
Leu Thr Arg Leu Glu Ala Leu Ile Lys Ser Lys Ile Lys Glu Leu Pro
405 410 415
Val Gln Glu Gln Glu Lys Val Asn Ser Ile Ile Glu Ala Lys Ile Lys
420 425 430
Lys Gln Ser Lys Gln Asp Gly Ser Thr Gly Phe Asp Ala Thr Leu Asp
435 440 445
Asn Asp Pro Thr Tyr Lys Ser Leu Ala Gln Phe Val Gly Tyr Asp Asn
450 455 460
Met Arg Trp Val Gln Thr Ala Ser Ala Pro Ile Ala Pro Thr Thr Leu
465 470 475 480
Ile Tyr Leu Lys Ala Ser Leu Asn Ile Gly Thr Arg Gln Gln Tyr Gly
485 490 495
Leu Thr Glu Ser Gly Ala Ala Ile Thr Ser Thr Gly Glu Tyr Glu Ala
500 505 510
Ser Pro Gly Ser Cys Gly Val Ile Leu Pro Thr Gly Gln Tyr Arg Leu
515 520 525
Tyr Ser Val Ser Glu Met Gly Tyr Asp Leu Asn Lys Leu Glu Gly Glu
530 535 540
Val Met Leu Gln Gly Pro Gln Met Phe Lys Gly Tyr Tyr Tyr Asn Tyr
545 550 555 560
Glu Glu Thr Ile Asn Ala Val Thr Glu Asp Gly Trp Phe His Ser Gly
565 570 575
Asp Ile Ala Arg Val Asp Ser Lys Thr Gly Arg Val Thr Ile Ile Asp
580 585 590
Arg Val Lys His Phe Phe Lys Leu Ala Gln Gly Glu Tyr Ile Ser Pro
595 600 605
Glu Arg Ile Glu Asn Arg Tyr Leu Ser Ser Asn Pro Asp Ile Cys Gln
610 615 620
Leu Trp Val His Gly Asp Ser Lys Glu His Tyr Leu Ile Gly Ile Val
625 630 635 640
Gly Val Glu Tyr Glu Lys Gly Leu Lys Phe Ile Asn Thr Glu Phe Gly
645 650 655
Tyr Asn Lys Ile Asp Met Pro Pro Gly Asp Leu Leu Asp Ile Leu Asn
660 665 670
Ser Pro Glu Val Lys Ser Lys Phe Leu Thr Lys Met Asn Gln Ser Val
675 680 685
Arg Asp Lys Leu Asn Gly Phe Glu Ile Leu His Asn Ile Phe Ile Glu
690 695 700
Phe Glu Pro Leu Thr Val Gln Arg Glu Val Val Thr Pro Thr Phe Lys
705 710 715 720
Ile Arg Arg Pro Ile Cys Arg Lys Phe Phe Lys Ser Gln Leu Asp Ala
725 730 735
Met Tyr Asn Glu Gly Ser Leu Ile Asn Asn Ala Lys Leu
740 745
<210> 63
<211> 8902
<212> DNA
<213> Artificial sequence
<220>
<223> plasmid pZP2-YLACoS-3Ps
<400> 63
ggccgcaagt gtggatgggg aagtgagtgc ccggttctgt gtgcacaatt ggcaatccaa 60
gatggatgga ttcaacacag ggatatagcg agctacgtgg tggtgcgagg atatagcaac 120
ggatatttat gtttgacact tgagaatgta cgatacaagc actgtccaag tacaatacta 180
aacatactgt acatactcat actcgtaccc gggcaacggt ttcacttgag tgcagtggct 240
agtgctctta ctcgtacagt gtgcaatact gcgtatcata gtctttgatg tatatcgtat 300
tcattcatgt tagttgcgta cgggcgtcgt tgcttgtgtg atttttgagg acccatccct 360
ttggtatata agtatactct ggggttaagg ttgcccgtgt agtctaggtt atagttttca 420
tgtgaaatac cgagagccga gggagaataa acgggggtat ttggacttgt ttttttcgcg 480
gaaaagcgtc gaatcaaccc tgcgggcctt gcaccatgtc cacgacgtgt ttctcgcccc 540
aattcgcccc ttgcacgtca aaattaggcc tccatctaga cccctccata acatgtgact 600
gtggggaaaa gtataaggga aaccatgcaa ccatagacga cgtgaaagac ggggaggaac 660
caatggaggc caaagaaatg gggtagcaac agtccaggag acagacaagg agacaaggag 720
agggcgcccg aaagatcgga aaaacaaaca tgtccaattg gggcagtgac ggaaacgaca 780
cggacacttc agtacaatgg accgaccatc tccaagccag ggttattccg gtatcacctt 840
ggccgtaacc tcccgctggt acctgatatt gtacacgttc acattcaata tactttcagc 900
tacaataaga gaggctgttt gtcgggcatg tgtgtccgtc gtatggggtg atgtccgagg 960
gcgaaattcg ctacaagctt aactctggcg cttgtccagt atgaatagac aagtcaagac 1020
cagtggtgcc atgattgaca gggaggtaca agacttcgat actcgagcat tactcggact 1080
tgtggcgatt gaacagacgg gcgatcgctt ctcccccgta ttgccggcgc gccagctgca 1140
ttaatgaatc ggccaacgcg cggggagagg cggtttgcgt attgggcgct cttccgcttc 1200
ctcgctcact gactcgctgc gctcggtcgt tcggctgcgg cgagcggtat cagctcactc 1260
aaaggcggta atacggttat ccacagaatc aggggataac gcaggaaaga acatgtgagc 1320
aaaaggccag caaaaggcca ggaaccgtaa aaaggccgcg ttgctggcgt ttttccatag 1380
gctccgcccc cctgacgagc atcacaaaaa tcgacgctca agtcagaggt ggcgaaaccc 1440
gacaggacta taaagatacc aggcgtttcc ccctggaagc tccctcgtgc gctctcctgt 1500
tccgaccctg ccgcttaccg gatacctgtc cgcctttctc ccttcgggaa gcgtggcgct 1560
ttctcatagc tcacgctgta ggtatctcag ttcggtgtag gtcgttcgct ccaagctggg 1620
ctgtgtgcac gaaccccccg ttcagcccga ccgctgcgcc ttatccggta actatcgtct 1680
tgagtccaac ccggtaagac acgacttatc gccactggca gcagccactg gtaacaggat 1740
tagcagagcg aggtatgtag gcggtgctac agagttcttg aagtggtggc ctaactacgg 1800
ctacactaga agaacagtat ttggtatctg cgctctgctg aagccagtta ccttcggaaa 1860
aagagttggt agctcttgat ccggcaaaca aaccaccgct ggtagcggtg gtttttttgt 1920
ttgcaagcag cagattacgc gcagaaaaaa aggatctcaa gaagatcctt tgatcttttc 1980
tacggggtct gacgctcagt ggaacgaaaa ctcacgttaa gggattttgg tcatgagatt 2040
atcaaaaagg atcttcacct agatcctttt aaattaaaaa tgaagtttta aatcaatcta 2100
aagtatatat gagtaaactt ggtctgacag ttaccaatgc ttaatcagtg aggcacctat 2160
ctcagcgatc tgtctatttc gttcatccat agttgcctga ctccccgtcg tgtagataac 2220
tacgatacgg gagggcttac catctggccc cagtgctgca atgataccgc gagacccacg 2280
ctcaccggct ccagatttat cagcaataaa ccagccagcc ggaagggccg agcgcagaag 2340
tggtcctgca actttatccg cctccatcca gtctattaat tgttgccggg aagctagagt 2400
aagtagttcg ccagttaata gtttgcgcaa cgttgttgcc attgctacag gcatcgtggt 2460
gtcacgctcg tcgtttggta tggcttcatt cagctccggt tcccaacgat caaggcgagt 2520
tacatgatcc cccatgttgt gcaaaaaagc ggttagctcc ttcggtcctc cgatcgttgt 2580
cagaagtaag ttggccgcag tgttatcact catggttatg gcagcactgc ataattctct 2640
tactgtcatg ccatccgtaa gatgcttttc tgtgactggt gagtactcaa ccaagtcatt 2700
ctgagaatag tgtatgcggc gaccgagttg ctcttgcccg gcgtcaatac gggataatac 2760
cgcgccacat agcagaactt taaaagtgct catcattgga aaacgttctt cggggcgaaa 2820
actctcaagg atcttaccgc tgttgagatc cagttcgatg taacccactc gtgcacccaa 2880
ctgatcttca gcatctttta ctttcaccag cgtttctggg tgagcaaaaa caggaaggca 2940
aaatgccgca aaaaagggaa taagggcgac acggaaatgt tgaatactca tactcttcct 3000
ttttcaatat tattgaagca tttatcaggg ttattgtctc atgagcggat acatatttga 3060
atgtatttag aaaaataaac aaataggggt tccgcgcaca tttccccgaa aagtgccacc 3120
tgatgcggtg tgaaataccg cacagatgcg taaggagaaa ataccgcatc aggaaattgt 3180
aagcgttaat attttgttaa aattcgcgtt aaatttttgt taaatcagct cattttttaa 3240
ccaataggcc gaaatcggca aaatccctta taaatcaaaa gaatagaccg agatagggtt 3300
gagtgttgtt ccagtttgga acaagagtcc actattaaag aacgtggact ccaacgtcaa 3360
agggcgaaaa accgtctatc agggcgatgg cccactacgt gaaccatcac cctaatcaag 3420
ttttttgggg tcgaggtgcc gtaaagcact aaatcggaac cctaaaggga gcccccgatt 3480
tagagcttga cggggaaagc cggcgaacgt ggcgagaaag gaagggaaga aagcgaaagg 3540
agcgggcgct agggcgctgg caagtgtagc ggtcacgctg cgcgtaacca ccacacccgc 3600
cgcgcttaat gcgccgctac agggcgcgtc cattcgccat tcaggctgcg caactgttgg 3660
gaagggcgat cggtgcgggc ctcttcgcta ttacgccagc tggcgaaagg gggatgtgct 3720
gcaaggcgat taagttgggt aacgccaggg ttttcccagt cacgacgttg taaaacgacg 3780
gccagtgaat tgtaatacga ctcactatag ggcgaattgg gcccgacgtc gcatgcgctg 3840
atgacacttt ggtctgaaag agatgcattt tgaatcccaa acttgcagtg cccaagtgac 3900
atacatctcc gcgttttgga aaatgttcag aaacagttga ttgtgttgga atggggaatg 3960
gggaatggaa aaatgactca agtatcaatt ccaaaaactt ctctggctgg cagtacctac 4020
tgtccatact actgcatttt ctccagtcag gccactctat actcgacgac acagtagtaa 4080
aacccagata atttcgacat aaacaagaaa acagacccaa taatatttat atatagtcag 4140
ccgtttgtcc agttcagact gtaatagccg aaaaaaaatc caaagtttct attctaggaa 4200
aatatattcc aatattttta attcttaatc tcatttattt tattctagcg aaatacattt 4260
cagctacttg agacatgtga tacccacaaa tcggattcgg actcggttgt tcagaagagc 4320
atatggcatt cgtgctcgct tgttcacgta ttcttcctgt tccatctctt ggccgacaat 4380
cacacaaaaa tggggttttt tttttaattc taatgattca ttacagcaaa attgagatat 4440
agcagaccac gtattccata atcaccaagg aagttcttgg gcgtcttaat taagtcatac 4500
acaagtcagc tttcttcgag cctcatataa gtataagtag ttcaacgtat tagcactgta 4560
cccagcatct ccgtatcgag aaacacaaca acatgcccca ttggacagat catgcggata 4620
cacaggttgt gcagtatcat acatactcga tcagacaggt cgtctgacca tcatacaagc 4680
tgaacaagcg ctccatactt gcacgctctc tatatacaca gttaaattac atatccatag 4740
tctaacctct aacagttaat cttctggtaa gcctcccagc cagccttctg gtatcgcttg 4800
gcctcctcaa taggatctcg gttctggccg tacagacctc ggccgacaat tatgatatcc 4860
gttccggtag acatgacatc ctcaacagtt cggtactgct gtccgagagc gtctcccttg 4920
tcgtcaagac ccaccccggg ggtcagaata agccagtcct cagagtcgcc cttaggtcgg 4980
ttctgggcaa tgaagccaac cacaaactcg gggtcggatc gggcaagctc aatggtctgc 5040
ttggagtact cgccagtggc cagagagccc ttgcaagaca gctcggccag catgagcaga 5100
cctctggcca gcttctcgtt gggagagggg actaggaact ccttgtactg ggagttctcg 5160
tagtcagaga cgtcctcctt cttctgttca gagacagttt cctcggcacc agctcgcagg 5220
ccagcaatga ttccggttcc gggtacaccg tgggcgttgg tgatatcgga ccactcggcg 5280
attcggtgac accggtactg gtgcttgaca gtgttgccaa tatctgcgaa ctttctgtcc 5340
tcgaacagga agaaaccgtg cttaagagca agttccttga gggggagcac agtgccggcg 5400
taggtgaagt cgtcaatgat gtcgatatgg gttttgatca tgcacacata aggtccgacc 5460
ttatcggcaa gctcaatgag ctccttggtg gtggtaacat ccagagaagc acacaggttg 5520
gttttcttgg ctgccacgag cttgagcact cgagcggcaa aggcggactt gtggacgtta 5580
gctcgagctt cgtaggaggg cattttggtg gtgaagagga gactgaaata aatttagtct 5640
gcagaacttt ttatcggaac cttatctggg gcagtgaagt atatgttatg gtaatagtta 5700
cgagttagtt gaacttatag atagactgga ctatacggct atcggtccaa attagaaaga 5760
acgtcaatgg ctctctgggc gtcgcctttg ccgacaaaaa tgtgatcatg atgaaagcca 5820
gcaatgacgt tgcagctgat attgttgtcg gccaaccgcg ccgaaaacgc agctgtcaga 5880
cccacagcct ccaacgaaga atgtatcgtc aaagtgatcc aagcacactc atagttggag 5940
tcgtactcca aaggcggcaa tgacgagtca gacagatact cgtcaaacgg taggttagtg 6000
cttggtatat gagttgtagg catgacaatt tggaaagggg tggactttgg gaatattgtg 6060
ggatttcaat accttagttt gtacagggta attgttacaa atgatacaaa gaactgtatt 6120
tcttttcatt tgttttaatt ggttgtatat caagtccgtt agacgagctc agtgccttgg 6180
cttttggcac tgtatttcat ttttagaggt acactacatt cagtgaggta tggtaaggtt 6240
gagggcataa tgaaggcacc ttgtactgac agtcacagac ctctcaccga gaattttatg 6300
agatatactc gggttcattt taggctcatc gattgccccg gagaagacgg ccaggccgcc 6360
tagatgacaa attcaacaac tcacagctga ctttctgcca ttgccactag gggggggcct 6420
ttttatatgg ccaagccaag ctctccacgt cggttgggct gcacccaaca ataaatgggt 6480
agggttgcac caacaaaggg atgggatggg gggtagaaga tacgaggata acggggctca 6540
atggcacaaa taagaacgaa tactgccatt aagactcgtg atccagcgac tgacaccatt 6600
gcatcatcta agggcctcaa aactacctcg gaactgctgc gctgatctgg acaccacaga 6660
ggttccgagc actttaggtt gcaccaaatg tcccaccagg tgcaggcaga aaacgctgga 6720
acagcgtgta cagtttgtct taacaaaaag tgagggcgct gaggtcgagc agggtggtgt 6780
gacttgttat agcctttaga gctgcgaaag cgcgtatgga tttggctcat caggccagat 6840
tgagggtctg tggacacatg tcatgttagt gtacttcaat cgccccctgg atatagcccc 6900
gacaataggc cgtggcctca tttttttgcc ttccgcacat ttccattgct cggtacccac 6960
accttgcttc tcctgcactt gccaacctta atactggttt acattgacca acatcttaca 7020
agcggggggc ttgtctaggg tatatataaa cagtggctct cccaatcggt tgccagtctc 7080
ttttttcctt tctttcccca cagattcgaa atctaaacta cacatcacag aattccgagc 7140
cgtgagtatc cacgacaaga tcagtgtcga gacgacgcgt tttgtgtaat gacacaatcc 7200
gaaagtcgct agcaacacac actctctaca caaactaacc cagctctggt accatggcca 7260
tcatccactc caccggaact ctgcccatct tcaacggtac cgtcaccgat tacctgcgaa 7320
caaagccttc ttactcgtcc acagatccag cctacatcga cgtggttaca ggcaactcta 7380
tcagctactc cgaggtctgg aagcttgccg accgactctc ctctgctctg tacaacgact 7440
acggactcac cgacgccaag cccgacgaga atgtgggtcc tgttgtcatg ctgcacgctg 7500
tcaattcgcc tctcctggca tctgttcact acgctcttct ggatctgggc gtcacaatca 7560
ctcccgcagc tgccacctac gaggctggcg atctcgcaca tcaaatcaag gtgtgctctc 7620
cgtccctggt catttgcaac cagcagttcg aacccaaggt caaatctgcc tccagcaaca 7680
ccaagctcat tttcatcgag gatctgctca aaacccagtc gtctgctccc tggaaaaagt 7740
tcactacctc caaccccaac cgagttgcct acctgggcat gtccagtgga acctctggtc 7800
tccccaaggc ggttcaacag acccacatca acatgtcgtc ttccaccgaa gccgtcattt 7860
cctctcagac catcttcagc gctcgaaaga acgtcaccgc agccattgtg cccatgactc 7920
atgtctacgg actcaccaag tttgttttcc actctgtcgc aggctcaatg accaccgttg 7980
tgttccccaa gttctccctg gtcgacctcc tggaggccca gatcaagtac aagatcaaca 8040
ttctgtatct ggttcctcca gtggtcttgg ctctggccaa ggactctcgt gtacagccct 8100
acatcaagtc catttgcgag ctcaccactc tgattgccac tggtgcggct ccccttcctc 8160
ccactgcagg cgacgccctt ctggagcgac ttacgggcaa caaagaggga aacagagaca 8220
acggtatgga tcccttggtt ctcatccagg gctacggact cacagagact ctccaggtgt 8280
ctgtcttcaa gccagaggat cccgaacgag atctcaagac cgtgggcaaa ctgcttccca 8340
acaccgaggt tcgaattgtc ggcgagaagg gagatgttcc gcgttccaaa tggtcgtttg 8400
tcactcctcc aaccggcgaa atctacattc gaggtcccca cgtgactcct ggttacttca 8460
acaacgactc tgccaactct gagtcctttg acggcgagtg gctcaagacc ggcgatatcg 8520
gatacatgga cctggaaggt cgactcacca ttgtggaccg aaacaaggag atgatcaagg 8580
tcaacggacg tcaggttgct cctgccgaga tcgaatctgt gctgctgggt catcctatgg 8640
tcaaggatgt ggccgtcatt ggagtcacca atcccgacag aggcacggag tctgctcggg 8700
cgtttcttgt tactgaagct cgagctctcc ctgtcatcaa gcagtggttt gaccgtcgag 8760
ttccctccta caagcgactt tacggaggca ttgtggttgt cgatgccatt cccaagtctg 8820
cctcgggcaa gattctgcga cgggtcctca gagagcgaaa gggcgactcc gtgtttggag 8880
agtatgtcga ggaagtctaa gc 8902
<210> 64
<211> 9055
<212> DNA
<213> Artificial sequence
<220>
<223> plasmid pZP2-YLACoS-5Ps
<400> 64
ggccgcaagt gtggatgggg aagtgagtgc ccggttctgt gtgcacaatt ggcaatccaa 60
gatggatgga ttcaacacag ggatatagcg agctacgtgg tggtgcgagg atatagcaac 120
ggatatttat gtttgacact tgagaatgta cgatacaagc actgtccaag tacaatacta 180
aacatactgt acatactcat actcgtaccc gggcaacggt ttcacttgag tgcagtggct 240
agtgctctta ctcgtacagt gtgcaatact gcgtatcata gtctttgatg tatatcgtat 300
tcattcatgt tagttgcgta cgggcgtcgt tgcttgtgtg atttttgagg acccatccct 360
ttggtatata agtatactct ggggttaagg ttgcccgtgt agtctaggtt atagttttca 420
tgtgaaatac cgagagccga gggagaataa acgggggtat ttggacttgt ttttttcgcg 480
gaaaagcgtc gaatcaaccc tgcgggcctt gcaccatgtc cacgacgtgt ttctcgcccc 540
aattcgcccc ttgcacgtca aaattaggcc tccatctaga cccctccata acatgtgact 600
gtggggaaaa gtataaggga aaccatgcaa ccatagacga cgtgaaagac ggggaggaac 660
caatggaggc caaagaaatg gggtagcaac agtccaggag acagacaagg agacaaggag 720
agggcgcccg aaagatcgga aaaacaaaca tgtccaattg gggcagtgac ggaaacgaca 780
cggacacttc agtacaatgg accgaccatc tccaagccag ggttattccg gtatcacctt 840
ggccgtaacc tcccgctggt acctgatatt gtacacgttc acattcaata tactttcagc 900
tacaataaga gaggctgttt gtcgggcatg tgtgtccgtc gtatggggtg atgtccgagg 960
gcgaaattcg ctacaagctt aactctggcg cttgtccagt atgaatagac aagtcaagac 1020
cagtggtgcc atgattgaca gggaggtaca agacttcgat actcgagcat tactcggact 1080
tgtggcgatt gaacagacgg gcgatcgctt ctcccccgta ttgccggcgc gccagctgca 1140
ttaatgaatc ggccaacgcg cggggagagg cggtttgcgt attgggcgct cttccgcttc 1200
ctcgctcact gactcgctgc gctcggtcgt tcggctgcgg cgagcggtat cagctcactc 1260
aaaggcggta atacggttat ccacagaatc aggggataac gcaggaaaga acatgtgagc 1320
aaaaggccag caaaaggcca ggaaccgtaa aaaggccgcg ttgctggcgt ttttccatag 1380
gctccgcccc cctgacgagc atcacaaaaa tcgacgctca agtcagaggt ggcgaaaccc 1440
gacaggacta taaagatacc aggcgtttcc ccctggaagc tccctcgtgc gctctcctgt 1500
tccgaccctg ccgcttaccg gatacctgtc cgcctttctc ccttcgggaa gcgtggcgct 1560
ttctcatagc tcacgctgta ggtatctcag ttcggtgtag gtcgttcgct ccaagctggg 1620
ctgtgtgcac gaaccccccg ttcagcccga ccgctgcgcc ttatccggta actatcgtct 1680
tgagtccaac ccggtaagac acgacttatc gccactggca gcagccactg gtaacaggat 1740
tagcagagcg aggtatgtag gcggtgctac agagttcttg aagtggtggc ctaactacgg 1800
ctacactaga agaacagtat ttggtatctg cgctctgctg aagccagtta ccttcggaaa 1860
aagagttggt agctcttgat ccggcaaaca aaccaccgct ggtagcggtg gtttttttgt 1920
ttgcaagcag cagattacgc gcagaaaaaa aggatctcaa gaagatcctt tgatcttttc 1980
tacggggtct gacgctcagt ggaacgaaaa ctcacgttaa gggattttgg tcatgagatt 2040
atcaaaaagg atcttcacct agatcctttt aaattaaaaa tgaagtttta aatcaatcta 2100
aagtatatat gagtaaactt ggtctgacag ttaccaatgc ttaatcagtg aggcacctat 2160
ctcagcgatc tgtctatttc gttcatccat agttgcctga ctccccgtcg tgtagataac 2220
tacgatacgg gagggcttac catctggccc cagtgctgca atgataccgc gagacccacg 2280
ctcaccggct ccagatttat cagcaataaa ccagccagcc ggaagggccg agcgcagaag 2340
tggtcctgca actttatccg cctccatcca gtctattaat tgttgccggg aagctagagt 2400
aagtagttcg ccagttaata gtttgcgcaa cgttgttgcc attgctacag gcatcgtggt 2460
gtcacgctcg tcgtttggta tggcttcatt cagctccggt tcccaacgat caaggcgagt 2520
tacatgatcc cccatgttgt gcaaaaaagc ggttagctcc ttcggtcctc cgatcgttgt 2580
cagaagtaag ttggccgcag tgttatcact catggttatg gcagcactgc ataattctct 2640
tactgtcatg ccatccgtaa gatgcttttc tgtgactggt gagtactcaa ccaagtcatt 2700
ctgagaatag tgtatgcggc gaccgagttg ctcttgcccg gcgtcaatac gggataatac 2760
cgcgccacat agcagaactt taaaagtgct catcattgga aaacgttctt cggggcgaaa 2820
actctcaagg atcttaccgc tgttgagatc cagttcgatg taacccactc gtgcacccaa 2880
ctgatcttca gcatctttta ctttcaccag cgtttctggg tgagcaaaaa caggaaggca 2940
aaatgccgca aaaaagggaa taagggcgac acggaaatgt tgaatactca tactcttcct 3000
ttttcaatat tattgaagca tttatcaggg ttattgtctc atgagcggat acatatttga 3060
atgtatttag aaaaataaac aaataggggt tccgcgcaca tttccccgaa aagtgccacc 3120
tgatgcggtg tgaaataccg cacagatgcg taaggagaaa ataccgcatc aggaaattgt 3180
aagcgttaat attttgttaa aattcgcgtt aaatttttgt taaatcagct cattttttaa 3240
ccaataggcc gaaatcggca aaatccctta taaatcaaaa gaatagaccg agatagggtt 3300
gagtgttgtt ccagtttgga acaagagtcc actattaaag aacgtggact ccaacgtcaa 3360
agggcgaaaa accgtctatc agggcgatgg cccactacgt gaaccatcac cctaatcaag 3420
ttttttgggg tcgaggtgcc gtaaagcact aaatcggaac cctaaaggga gcccccgatt 3480
tagagcttga cggggaaagc cggcgaacgt ggcgagaaag gaagggaaga aagcgaaagg 3540
agcgggcgct agggcgctgg caagtgtagc ggtcacgctg cgcgtaacca ccacacccgc 3600
cgcgcttaat gcgccgctac agggcgcgtc cattcgccat tcaggctgcg caactgttgg 3660
gaagggcgat cggtgcgggc ctcttcgcta ttacgccagc tggcgaaagg gggatgtgct 3720
gcaaggcgat taagttgggt aacgccaggg ttttcccagt cacgacgttg taaaacgacg 3780
gccagtgaat tgtaatacga ctcactatag ggcgaattgg gcccgacgtc gcatgcgctg 3840
atgacacttt ggtctgaaag agatgcattt tgaatcccaa acttgcagtg cccaagtgac 3900
atacatctcc gcgttttgga aaatgttcag aaacagttga ttgtgttgga atggggaatg 3960
gggaatggaa aaatgactca agtatcaatt ccaaaaactt ctctggctgg cagtacctac 4020
tgtccatact actgcatttt ctccagtcag gccactctat actcgacgac acagtagtaa 4080
aacccagata atttcgacat aaacaagaaa acagacccaa taatatttat atatagtcag 4140
ccgtttgtcc agttcagact gtaatagccg aaaaaaaatc caaagtttct attctaggaa 4200
aatatattcc aatattttta attcttaatc tcatttattt tattctagcg aaatacattt 4260
cagctacttg agacatgtga tacccacaaa tcggattcgg actcggttgt tcagaagagc 4320
atatggcatt cgtgctcgct tgttcacgta ttcttcctgt tccatctctt ggccgacaat 4380
cacacaaaaa tggggttttt tttttaattc taatgattca ttacagcaaa attgagatat 4440
agcagaccac gtattccata atcaccaagg aagttcttgg gcgtcttaat taagtcatac 4500
acaagtcagc tttcttcgag cctcatataa gtataagtag ttcaacgtat tagcactgta 4560
cccagcatct ccgtatcgag aaacacaaca acatgcccca ttggacagat catgcggata 4620
cacaggttgt gcagtatcat acatactcga tcagacaggt cgtctgacca tcatacaagc 4680
tgaacaagcg ctccatactt gcacgctctc tatatacaca gttaaattac atatccatag 4740
tctaacctct aacagttaat cttctggtaa gcctcccagc cagccttctg gtatcgcttg 4800
gcctcctcaa taggatctcg gttctggccg tacagacctc ggccgacaat tatgatatcc 4860
gttccggtag acatgacatc ctcaacagtt cggtactgct gtccgagagc gtctcccttg 4920
tcgtcaagac ccaccccggg ggtcagaata agccagtcct cagagtcgcc cttaggtcgg 4980
ttctgggcaa tgaagccaac cacaaactcg gggtcggatc gggcaagctc aatggtctgc 5040
ttggagtact cgccagtggc cagagagccc ttgcaagaca gctcggccag catgagcaga 5100
cctctggcca gcttctcgtt gggagagggg actaggaact ccttgtactg ggagttctcg 5160
tagtcagaga cgtcctcctt cttctgttca gagacagttt cctcggcacc agctcgcagg 5220
ccagcaatga ttccggttcc gggtacaccg tgggcgttgg tgatatcgga ccactcggcg 5280
attcggtgac accggtactg gtgcttgaca gtgttgccaa tatctgcgaa ctttctgtcc 5340
tcgaacagga agaaaccgtg cttaagagca agttccttga gggggagcac agtgccggcg 5400
taggtgaagt cgtcaatgat gtcgatatgg gttttgatca tgcacacata aggtccgacc 5460
ttatcggcaa gctcaatgag ctccttggtg gtggtaacat ccagagaagc acacaggttg 5520
gttttcttgg ctgccacgag cttgagcact cgagcggcaa aggcggactt gtggacgtta 5580
gctcgagctt cgtaggaggg cattttggtg gtgaagagga gactgaaata aatttagtct 5640
gcagaacttt ttatcggaac cttatctggg gcagtgaagt atatgttatg gtaatagtta 5700
cgagttagtt gaacttatag atagactgga ctatacggct atcggtccaa attagaaaga 5760
acgtcaatgg ctctctgggc gtcgcctttg ccgacaaaaa tgtgatcatg atgaaagcca 5820
gcaatgacgt tgcagctgat attgttgtcg gccaaccgcg ccgaaaacgc agctgtcaga 5880
cccacagcct ccaacgaaga atgtatcgtc aaagtgatcc aagcacactc atagttggag 5940
tcgtactcca aaggcggcaa tgacgagtca gacagatact cgtcaaacgg taggttagtg 6000
cttggtatat gagttgtagg catgacaatt tggaaagggg tggactttgg gaatattgtg 6060
ggatttcaat accttagttt gtacagggta attgttacaa atgatacaaa gaactgtatt 6120
tcttttcatt tgttttaatt ggttgtatat caagtccgtt agacgagctc agtgccttgg 6180
cttttggcac tgtatttcat ttttagaggt acactacatt cagtgaggta tggtaaggtt 6240
gagggcataa tgaaggcacc ttgtactgac agtcacagac ctctcaccga gaattttatg 6300
agatatactc gggttcattt taggctcatc gattgccccg gagaagacgg ccaggccgcc 6360
tagatgacaa attcaacaac tcacagctga ctttctgcca ttgccactag gggggggcct 6420
ttttatatgg ccaagccaag ctctccacgt cggttgggct gcacccaaca ataaatgggt 6480
agggttgcac caacaaaggg atgggatggg gggtagaaga tacgaggata acggggctca 6540
atggcacaaa taagaacgaa tactgccatt aagactcgtg atccagcgac tgacaccatt 6600
gcatcatcta agggcctcaa aactacctcg gaactgctgc gctgatctgg acaccacaga 6660
ggttccgagc actttaggtt gcaccaaatg tcccaccagg tgcaggcaga aaacgctgga 6720
acagcgtgta cagtttgtct taacaaaaag tgagggcgct gaggtcgagc agggtggtgt 6780
gacttgttat agcctttaga gctgcgaaag cgcgtatgga tttggctcat caggccagat 6840
tgagggtctg tggacacatg tcatgttagt gtacttcaat cgccccctgg atatagcccc 6900
gacaataggc cgtggcctca tttttttgcc ttccgcacat ttccattgct cggtacccac 6960
accttgcttc tcctgcactt gccaacctta atactggttt acattgacca acatcttaca 7020
agcggggggc ttgtctaggg tatatataaa cagtggctct cccaatcggt tgccagtctc 7080
ttttttcctt tctttcccca cagattcgaa atctaaacta cacatcacag aattccgagc 7140
cgtgagtatc cacgacaaga tcagtgtcga gacgacgcgt tttgtgtaat gacacaatcc 7200
gaaagtcgct agcaacacac actctctaca caaactaacc cagctctggt accatggcct 7260
caatcattca caagtctcct gtgcccgacg ttcagctgtt ctacggttcc tggccagatc 7320
tcatgcgaac ctctcctcat gcccacaacg actccaaacc cgtggtcttt gacttcgata 7380
ccaagcagca acttacttgg aagcaggtct ggcaactcag cgctcgactc agagcccagc 7440
tgtaccacaa gtacggaatc ggcaaacccg gtgctcttgc acctttccac aacgatccct 7500
ctctcggaga cgtggtcatc ttctacactc ccaacaccta cagctcgttg ccctatcatc 7560
tggctcttca cgatctcgga gccaccattt ctcctgcctc cacatcttac gacgtcaagg 7620
acatttgcca tcagatcgtt actaccgatg cggtcgtggt tgtcgctgca gccgagaaat 7680
ccgagattgc tcgagaggcc gttcagctgt ctggtcgaga cgtcagagtt gtggtcatgg 7740
aggacctcat caacaatgct cccaccgttg cgcagaacga tatcgactcg gcacctcatg 7800
tgtccctgtc tcgggaccag gctcgagcca agattgcata cctgggcatg tcttccggta 7860
cgtctggcgg acttcccaag gctgttcgtc tcactcactt caacgttacc tcgaactgtc 7920
tgcaggtctc cgctgccgca cccaaccttg cccagaacgt ggttgccagc gccgtcattc 7980
caaccactca catctacggt ctcaccatgt ttctgtcggt tcttccctac aacggttccg 8040
tggtcattca tcacaagcaa ttcaacttgc gagatctgct cgaggctcag aagacataca 8100
aggtctctct gtggattctc gttcctcccg tcatcgtgca gcttgccaag aaccctatgg 8160
tcgacgagta cctggactcc attcgagccc atgtgcggtg catcgtctct ggagctgctc 8220
ctctcggtgg caatgtcgtg gatcaggttt cggttcgtct taccggcaac aaggaaggca 8280
ttctgcccaa cggagacaag ctcgtcattc atcaagccta cggtcttacc gagtcctctc 8340
ccatcgttgg aatgctcgat cctctgtcgg accacatcga cgtcatgact gtgggctgtc 8400
tcatgcccaa taccgaggct cgaattgtcg acgaagaggg aaacgatcag ccagcagtcc 8460
acgttaccga cacacgaggc atcggtgccg ctgtcaagcg aggcgagaag attccctccg 8520
gagaactctg gattcgaggt cctcagatca tggacggata ccacaagaac cccgagtcgt 8580
ctcgtgagtc cctggaaccc agcacagaga cctacggtct gcaacatttc caggacagat 8640
ggcttcgaac tggagacgtt gctgtcatcg acaccttcgg acgagtcatg gttgtggatc 8700
gaaccaagga gctcatcaag tccatgtctc gacaggttgc tcctgccgag ctcgaagctc 8760
ttctgctcaa ccatccttcc gtcaacgatg tggctgtcgt tggcgtccac aacgacgata 8820
atggcacaga gtcagcacga gcgtttgtcg ttcttcaacc aggcgacgcc tgtgatccta 8880
ctaccatcaa gcactggatg gaccagcaag ttccctccta caagcggctg tacggaggca 8940
ttgtggtcat cgacactgtt cccaagaatg cctctggcaa gattctgcga agactgcttc 9000
gacagcggag agacgatcga gtctggggtc tggccaaggt tgccaagctc taagc 9055
<210> 65
<211> 9043
<212> DNA
<213> Artificial sequence
<220>
<223> plasmid pZP2-YLACoS-6Ps
<400> 65
ggccgcaagt gtggatgggg aagtgagtgc ccggttctgt gtgcacaatt ggcaatccaa 60
gatggatgga ttcaacacag ggatatagcg agctacgtgg tggtgcgagg atatagcaac 120
ggatatttat gtttgacact tgagaatgta cgatacaagc actgtccaag tacaatacta 180
aacatactgt acatactcat actcgtaccc gggcaacggt ttcacttgag tgcagtggct 240
agtgctctta ctcgtacagt gtgcaatact gcgtatcata gtctttgatg tatatcgtat 300
tcattcatgt tagttgcgta cgggcgtcgt tgcttgtgtg atttttgagg acccatccct 360
ttggtatata agtatactct ggggttaagg ttgcccgtgt agtctaggtt atagttttca 420
tgtgaaatac cgagagccga gggagaataa acgggggtat ttggacttgt ttttttcgcg 480
gaaaagcgtc gaatcaaccc tgcgggcctt gcaccatgtc cacgacgtgt ttctcgcccc 540
aattcgcccc ttgcacgtca aaattaggcc tccatctaga cccctccata acatgtgact 600
gtggggaaaa gtataaggga aaccatgcaa ccatagacga cgtgaaagac ggggaggaac 660
caatggaggc caaagaaatg gggtagcaac agtccaggag acagacaagg agacaaggag 720
agggcgcccg aaagatcgga aaaacaaaca tgtccaattg gggcagtgac ggaaacgaca 780
cggacacttc agtacaatgg accgaccatc tccaagccag ggttattccg gtatcacctt 840
ggccgtaacc tcccgctggt acctgatatt gtacacgttc acattcaata tactttcagc 900
tacaataaga gaggctgttt gtcgggcatg tgtgtccgtc gtatggggtg atgtccgagg 960
gcgaaattcg ctacaagctt aactctggcg cttgtccagt atgaatagac aagtcaagac 1020
cagtggtgcc atgattgaca gggaggtaca agacttcgat actcgagcat tactcggact 1080
tgtggcgatt gaacagacgg gcgatcgctt ctcccccgta ttgccggcgc gccagctgca 1140
ttaatgaatc ggccaacgcg cggggagagg cggtttgcgt attgggcgct cttccgcttc 1200
ctcgctcact gactcgctgc gctcggtcgt tcggctgcgg cgagcggtat cagctcactc 1260
aaaggcggta atacggttat ccacagaatc aggggataac gcaggaaaga acatgtgagc 1320
aaaaggccag caaaaggcca ggaaccgtaa aaaggccgcg ttgctggcgt ttttccatag 1380
gctccgcccc cctgacgagc atcacaaaaa tcgacgctca agtcagaggt ggcgaaaccc 1440
gacaggacta taaagatacc aggcgtttcc ccctggaagc tccctcgtgc gctctcctgt 1500
tccgaccctg ccgcttaccg gatacctgtc cgcctttctc ccttcgggaa gcgtggcgct 1560
ttctcatagc tcacgctgta ggtatctcag ttcggtgtag gtcgttcgct ccaagctggg 1620
ctgtgtgcac gaaccccccg ttcagcccga ccgctgcgcc ttatccggta actatcgtct 1680
tgagtccaac ccggtaagac acgacttatc gccactggca gcagccactg gtaacaggat 1740
tagcagagcg aggtatgtag gcggtgctac agagttcttg aagtggtggc ctaactacgg 1800
ctacactaga agaacagtat ttggtatctg cgctctgctg aagccagtta ccttcggaaa 1860
aagagttggt agctcttgat ccggcaaaca aaccaccgct ggtagcggtg gtttttttgt 1920
ttgcaagcag cagattacgc gcagaaaaaa aggatctcaa gaagatcctt tgatcttttc 1980
tacggggtct gacgctcagt ggaacgaaaa ctcacgttaa gggattttgg tcatgagatt 2040
atcaaaaagg atcttcacct agatcctttt aaattaaaaa tgaagtttta aatcaatcta 2100
aagtatatat gagtaaactt ggtctgacag ttaccaatgc ttaatcagtg aggcacctat 2160
ctcagcgatc tgtctatttc gttcatccat agttgcctga ctccccgtcg tgtagataac 2220
tacgatacgg gagggcttac catctggccc cagtgctgca atgataccgc gagacccacg 2280
ctcaccggct ccagatttat cagcaataaa ccagccagcc ggaagggccg agcgcagaag 2340
tggtcctgca actttatccg cctccatcca gtctattaat tgttgccggg aagctagagt 2400
aagtagttcg ccagttaata gtttgcgcaa cgttgttgcc attgctacag gcatcgtggt 2460
gtcacgctcg tcgtttggta tggcttcatt cagctccggt tcccaacgat caaggcgagt 2520
tacatgatcc cccatgttgt gcaaaaaagc ggttagctcc ttcggtcctc cgatcgttgt 2580
cagaagtaag ttggccgcag tgttatcact catggttatg gcagcactgc ataattctct 2640
tactgtcatg ccatccgtaa gatgcttttc tgtgactggt gagtactcaa ccaagtcatt 2700
ctgagaatag tgtatgcggc gaccgagttg ctcttgcccg gcgtcaatac gggataatac 2760
cgcgccacat agcagaactt taaaagtgct catcattgga aaacgttctt cggggcgaaa 2820
actctcaagg atcttaccgc tgttgagatc cagttcgatg taacccactc gtgcacccaa 2880
ctgatcttca gcatctttta ctttcaccag cgtttctggg tgagcaaaaa caggaaggca 2940
aaatgccgca aaaaagggaa taagggcgac acggaaatgt tgaatactca tactcttcct 3000
ttttcaatat tattgaagca tttatcaggg ttattgtctc atgagcggat acatatttga 3060
atgtatttag aaaaataaac aaataggggt tccgcgcaca tttccccgaa aagtgccacc 3120
tgatgcggtg tgaaataccg cacagatgcg taaggagaaa ataccgcatc aggaaattgt 3180
aagcgttaat attttgttaa aattcgcgtt aaatttttgt taaatcagct cattttttaa 3240
ccaataggcc gaaatcggca aaatccctta taaatcaaaa gaatagaccg agatagggtt 3300
gagtgttgtt ccagtttgga acaagagtcc actattaaag aacgtggact ccaacgtcaa 3360
agggcgaaaa accgtctatc agggcgatgg cccactacgt gaaccatcac cctaatcaag 3420
ttttttgggg tcgaggtgcc gtaaagcact aaatcggaac cctaaaggga gcccccgatt 3480
tagagcttga cggggaaagc cggcgaacgt ggcgagaaag gaagggaaga aagcgaaagg 3540
agcgggcgct agggcgctgg caagtgtagc ggtcacgctg cgcgtaacca ccacacccgc 3600
cgcgcttaat gcgccgctac agggcgcgtc cattcgccat tcaggctgcg caactgttgg 3660
gaagggcgat cggtgcgggc ctcttcgcta ttacgccagc tggcgaaagg gggatgtgct 3720
gcaaggcgat taagttgggt aacgccaggg ttttcccagt cacgacgttg taaaacgacg 3780
gccagtgaat tgtaatacga ctcactatag ggcgaattgg gcccgacgtc gcatgcgctg 3840
atgacacttt ggtctgaaag agatgcattt tgaatcccaa acttgcagtg cccaagtgac 3900
atacatctcc gcgttttgga aaatgttcag aaacagttga ttgtgttgga atggggaatg 3960
gggaatggaa aaatgactca agtatcaatt ccaaaaactt ctctggctgg cagtacctac 4020
tgtccatact actgcatttt ctccagtcag gccactctat actcgacgac acagtagtaa 4080
aacccagata atttcgacat aaacaagaaa acagacccaa taatatttat atatagtcag 4140
ccgtttgtcc agttcagact gtaatagccg aaaaaaaatc caaagtttct attctaggaa 4200
aatatattcc aatattttta attcttaatc tcatttattt tattctagcg aaatacattt 4260
cagctacttg agacatgtga tacccacaaa tcggattcgg actcggttgt tcagaagagc 4320
atatggcatt cgtgctcgct tgttcacgta ttcttcctgt tccatctctt ggccgacaat 4380
cacacaaaaa tggggttttt tttttaattc taatgattca ttacagcaaa attgagatat 4440
agcagaccac gtattccata atcaccaagg aagttcttgg gcgtcttaat taagtcatac 4500
acaagtcagc tttcttcgag cctcatataa gtataagtag ttcaacgtat tagcactgta 4560
cccagcatct ccgtatcgag aaacacaaca acatgcccca ttggacagat catgcggata 4620
cacaggttgt gcagtatcat acatactcga tcagacaggt cgtctgacca tcatacaagc 4680
tgaacaagcg ctccatactt gcacgctctc tatatacaca gttaaattac atatccatag 4740
tctaacctct aacagttaat cttctggtaa gcctcccagc cagccttctg gtatcgcttg 4800
gcctcctcaa taggatctcg gttctggccg tacagacctc ggccgacaat tatgatatcc 4860
gttccggtag acatgacatc ctcaacagtt cggtactgct gtccgagagc gtctcccttg 4920
tcgtcaagac ccaccccggg ggtcagaata agccagtcct cagagtcgcc cttaggtcgg 4980
ttctgggcaa tgaagccaac cacaaactcg gggtcggatc gggcaagctc aatggtctgc 5040
ttggagtact cgccagtggc cagagagccc ttgcaagaca gctcggccag catgagcaga 5100
cctctggcca gcttctcgtt gggagagggg actaggaact ccttgtactg ggagttctcg 5160
tagtcagaga cgtcctcctt cttctgttca gagacagttt cctcggcacc agctcgcagg 5220
ccagcaatga ttccggttcc gggtacaccg tgggcgttgg tgatatcgga ccactcggcg 5280
attcggtgac accggtactg gtgcttgaca gtgttgccaa tatctgcgaa ctttctgtcc 5340
tcgaacagga agaaaccgtg cttaagagca agttccttga gggggagcac agtgccggcg 5400
taggtgaagt cgtcaatgat gtcgatatgg gttttgatca tgcacacata aggtccgacc 5460
ttatcggcaa gctcaatgag ctccttggtg gtggtaacat ccagagaagc acacaggttg 5520
gttttcttgg ctgccacgag cttgagcact cgagcggcaa aggcggactt gtggacgtta 5580
gctcgagctt cgtaggaggg cattttggtg gtgaagagga gactgaaata aatttagtct 5640
gcagaacttt ttatcggaac cttatctggg gcagtgaagt atatgttatg gtaatagtta 5700
cgagttagtt gaacttatag atagactgga ctatacggct atcggtccaa attagaaaga 5760
acgtcaatgg ctctctgggc gtcgcctttg ccgacaaaaa tgtgatcatg atgaaagcca 5820
gcaatgacgt tgcagctgat attgttgtcg gccaaccgcg ccgaaaacgc agctgtcaga 5880
cccacagcct ccaacgaaga atgtatcgtc aaagtgatcc aagcacactc atagttggag 5940
tcgtactcca aaggcggcaa tgacgagtca gacagatact cgtcaaacgg taggttagtg 6000
cttggtatat gagttgtagg catgacaatt tggaaagggg tggactttgg gaatattgtg 6060
ggatttcaat accttagttt gtacagggta attgttacaa atgatacaaa gaactgtatt 6120
tcttttcatt tgttttaatt ggttgtatat caagtccgtt agacgagctc agtgccttgg 6180
cttttggcac tgtatttcat ttttagaggt acactacatt cagtgaggta tggtaaggtt 6240
gagggcataa tgaaggcacc ttgtactgac agtcacagac ctctcaccga gaattttatg 6300
agatatactc gggttcattt taggctcatc gattgccccg gagaagacgg ccaggccgcc 6360
tagatgacaa attcaacaac tcacagctga ctttctgcca ttgccactag gggggggcct 6420
ttttatatgg ccaagccaag ctctccacgt cggttgggct gcacccaaca ataaatgggt 6480
agggttgcac caacaaaggg atgggatggg gggtagaaga tacgaggata acggggctca 6540
atggcacaaa taagaacgaa tactgccatt aagactcgtg atccagcgac tgacaccatt 6600
gcatcatcta agggcctcaa aactacctcg gaactgctgc gctgatctgg acaccacaga 6660
ggttccgagc actttaggtt gcaccaaatg tcccaccagg tgcaggcaga aaacgctgga 6720
acagcgtgta cagtttgtct taacaaaaag tgagggcgct gaggtcgagc agggtggtgt 6780
gacttgttat agcctttaga gctgcgaaag cgcgtatgga tttggctcat caggccagat 6840
tgagggtctg tggacacatg tcatgttagt gtacttcaat cgccccctgg atatagcccc 6900
gacaataggc cgtggcctca tttttttgcc ttccgcacat ttccattgct cggtacccac 6960
accttgcttc tcctgcactt gccaacctta atactggttt acattgacca acatcttaca 7020
agcggggggc ttgtctaggg tatatataaa cagtggctct cccaatcggt tgccagtctc 7080
ttttttcctt tctttcccca cagattcgaa atctaaacta cacatcacag aattccgagc 7140
cgtgagtatc cacgacaaga tcagtgtcga gacgacgcgt tttgtgtaat gacacaatcc 7200
gaaagtcgct agcaacacac actctctaca caaactaacc cagctctggt accatggcca 7260
cacagattat ccacaacgcc accatcccca atatccccgt cgaccagctc tacgacggca 7320
agatcaccga cttcattcga tccggaggcc actccaacga aaccaagcct tctgtcatcg 7380
acgccaagac aggccagact ctctcccagg cggaaatgtg gcagctgtcg gacaagtacg 7440
cggcacttct cagctctcag tacggtctgt gccgacacag agacaacgag ctggacccat 7500
ctatgggaga tgtgctcatc accttctttg gaaacgttat cctcgctcct gtggtccatt 7560
gggctgccct cgacctcgga gcaaccattt ctcctggatc cacaggctac tctgcccagg 7620
atctcgctca ccagttccga gtcaccactc ccaaggtcgt tgtgtacgcc aaggcgttca 7680
aggatgtggt ggacgaggct acgaagctgt acaactcccc aaaccctcca gcacttgtcg 7740
agctcgaggc gctggacaag caggcccgaa tggttggaaa ccacaaggtc gaacacaccc 7800
gaaagatcaa gctggctcct cacgagtccc gaactcggat cgcgtacctt ggcatgtctt 7860
caggtacctc cggtggagtt tcaaaggctg tccgactcac ccattccaat ctcacgtcgt 7920
gttccgaaat ctcgaacaaa gcctccgagt ctctcgcaac tgaccagcag atcgctgccg 7980
ccatcattcc cgtgagtcat ctgtttggac tgtccaagtt cctcattggc aaccctcacg 8040
ccggagccac cactgtctat cacaatggct tcgatctgat cgaggtgctg gaggcacaga 8100
agaaatacaa agtcaactcg tggaccctgg ttcctcccat cattgtcctg ctcaccaaac 8160
accccattgt cgagaagtac attccttctc tccgtgccca catgcgagcc atcctctccg 8220
gagctgctcc tctgggtgcc aatgtcacag aggctcttct cacccgagtc actggcaaca 8280
agtttggcga gtctcccgag ggcggtctgc gaatcgttca gggctacgga cttacagaga 8340
cgtctcccgt tgccactctg tttgaccccg aagacaagga acgacacatt cggtcgtgtg 8400
gaaagctcgt gcccaactct caggttcgaa ttgtcaacga agacggcgtg gatcagcctg 8460
cctacgatgt ggaccccaac gagctggacg aggccatcaa acagggcact ctgccagtcg 8520
gagagctttg gatcagaggt ccccaggtta tggacggcta ccataacaac cccgaggcca 8580
acgaagcctg tttcgtcaag gctgacgatg ctgaagcaga tactgcctac tacaacagac 8640
actggttccg aaccggagac gttgctctgg tcgacaagca gggcagatac atgattgtgg 8700
accgaaccaa ggagatgatc aagagtcagg gtaagcaggt tgctcctgcc gagctcgaag 8760
acatgctcct gggacacgca caggtggcag ataccgcagt catcggtatt caggacgtgg 8820
agaagggtaa cgaggctcct cgagcttttg ttgtgctcaa ggacccgaag tacgacgctg 8880
tggagatcaa gacatggctg gacaagcagc ttcccaagta caagcagctt catgctggca 8940
tcgtggtcat tgatgccatt cccaagaacg ccagtggcaa gattctgcga cgtctgttgc 9000
gtgctagaaa ggacgatgtt gttctgggtc tcaacaagta agc 9043
<210> 66
<211> 9067
<212> DNA
<213> Artificial sequence
<220>
<223> plasmid pZP2-YLACoS-10Ps
<400> 66
ggccgcaagt gtggatgggg aagtgagtgc ccggttctgt gtgcacaatt ggcaatccaa 60
gatggatgga ttcaacacag ggatatagcg agctacgtgg tggtgcgagg atatagcaac 120
ggatatttat gtttgacact tgagaatgta cgatacaagc actgtccaag tacaatacta 180
aacatactgt acatactcat actcgtaccc gggcaacggt ttcacttgag tgcagtggct 240
agtgctctta ctcgtacagt gtgcaatact gcgtatcata gtctttgatg tatatcgtat 300
tcattcatgt tagttgcgta cgggcgtcgt tgcttgtgtg atttttgagg acccatccct 360
ttggtatata agtatactct ggggttaagg ttgcccgtgt agtctaggtt atagttttca 420
tgtgaaatac cgagagccga gggagaataa acgggggtat ttggacttgt ttttttcgcg 480
gaaaagcgtc gaatcaaccc tgcgggcctt gcaccatgtc cacgacgtgt ttctcgcccc 540
aattcgcccc ttgcacgtca aaattaggcc tccatctaga cccctccata acatgtgact 600
gtggggaaaa gtataaggga aaccatgcaa ccatagacga cgtgaaagac ggggaggaac 660
caatggaggc caaagaaatg gggtagcaac agtccaggag acagacaagg agacaaggag 720
agggcgcccg aaagatcgga aaaacaaaca tgtccaattg gggcagtgac ggaaacgaca 780
cggacacttc agtacaatgg accgaccatc tccaagccag ggttattccg gtatcacctt 840
ggccgtaacc tcccgctggt acctgatatt gtacacgttc acattcaata tactttcagc 900
tacaataaga gaggctgttt gtcgggcatg tgtgtccgtc gtatggggtg atgtccgagg 960
gcgaaattcg ctacaagctt aactctggcg cttgtccagt atgaatagac aagtcaagac 1020
cagtggtgcc atgattgaca gggaggtaca agacttcgat actcgagcat tactcggact 1080
tgtggcgatt gaacagacgg gcgatcgctt ctcccccgta ttgccggcgc gccagctgca 1140
ttaatgaatc ggccaacgcg cggggagagg cggtttgcgt attgggcgct cttccgcttc 1200
ctcgctcact gactcgctgc gctcggtcgt tcggctgcgg cgagcggtat cagctcactc 1260
aaaggcggta atacggttat ccacagaatc aggggataac gcaggaaaga acatgtgagc 1320
aaaaggccag caaaaggcca ggaaccgtaa aaaggccgcg ttgctggcgt ttttccatag 1380
gctccgcccc cctgacgagc atcacaaaaa tcgacgctca agtcagaggt ggcgaaaccc 1440
gacaggacta taaagatacc aggcgtttcc ccctggaagc tccctcgtgc gctctcctgt 1500
tccgaccctg ccgcttaccg gatacctgtc cgcctttctc ccttcgggaa gcgtggcgct 1560
ttctcatagc tcacgctgta ggtatctcag ttcggtgtag gtcgttcgct ccaagctggg 1620
ctgtgtgcac gaaccccccg ttcagcccga ccgctgcgcc ttatccggta actatcgtct 1680
tgagtccaac ccggtaagac acgacttatc gccactggca gcagccactg gtaacaggat 1740
tagcagagcg aggtatgtag gcggtgctac agagttcttg aagtggtggc ctaactacgg 1800
ctacactaga agaacagtat ttggtatctg cgctctgctg aagccagtta ccttcggaaa 1860
aagagttggt agctcttgat ccggcaaaca aaccaccgct ggtagcggtg gtttttttgt 1920
ttgcaagcag cagattacgc gcagaaaaaa aggatctcaa gaagatcctt tgatcttttc 1980
tacggggtct gacgctcagt ggaacgaaaa ctcacgttaa gggattttgg tcatgagatt 2040
atcaaaaagg atcttcacct agatcctttt aaattaaaaa tgaagtttta aatcaatcta 2100
aagtatatat gagtaaactt ggtctgacag ttaccaatgc ttaatcagtg aggcacctat 2160
ctcagcgatc tgtctatttc gttcatccat agttgcctga ctccccgtcg tgtagataac 2220
tacgatacgg gagggcttac catctggccc cagtgctgca atgataccgc gagacccacg 2280
ctcaccggct ccagatttat cagcaataaa ccagccagcc ggaagggccg agcgcagaag 2340
tggtcctgca actttatccg cctccatcca gtctattaat tgttgccggg aagctagagt 2400
aagtagttcg ccagttaata gtttgcgcaa cgttgttgcc attgctacag gcatcgtggt 2460
gtcacgctcg tcgtttggta tggcttcatt cagctccggt tcccaacgat caaggcgagt 2520
tacatgatcc cccatgttgt gcaaaaaagc ggttagctcc ttcggtcctc cgatcgttgt 2580
cagaagtaag ttggccgcag tgttatcact catggttatg gcagcactgc ataattctct 2640
tactgtcatg ccatccgtaa gatgcttttc tgtgactggt gagtactcaa ccaagtcatt 2700
ctgagaatag tgtatgcggc gaccgagttg ctcttgcccg gcgtcaatac gggataatac 2760
cgcgccacat agcagaactt taaaagtgct catcattgga aaacgttctt cggggcgaaa 2820
actctcaagg atcttaccgc tgttgagatc cagttcgatg taacccactc gtgcacccaa 2880
ctgatcttca gcatctttta ctttcaccag cgtttctggg tgagcaaaaa caggaaggca 2940
aaatgccgca aaaaagggaa taagggcgac acggaaatgt tgaatactca tactcttcct 3000
ttttcaatat tattgaagca tttatcaggg ttattgtctc atgagcggat acatatttga 3060
atgtatttag aaaaataaac aaataggggt tccgcgcaca tttccccgaa aagtgccacc 3120
tgatgcggtg tgaaataccg cacagatgcg taaggagaaa ataccgcatc aggaaattgt 3180
aagcgttaat attttgttaa aattcgcgtt aaatttttgt taaatcagct cattttttaa 3240
ccaataggcc gaaatcggca aaatccctta taaatcaaaa gaatagaccg agatagggtt 3300
gagtgttgtt ccagtttgga acaagagtcc actattaaag aacgtggact ccaacgtcaa 3360
agggcgaaaa accgtctatc agggcgatgg cccactacgt gaaccatcac cctaatcaag 3420
ttttttgggg tcgaggtgcc gtaaagcact aaatcggaac cctaaaggga gcccccgatt 3480
tagagcttga cggggaaagc cggcgaacgt ggcgagaaag gaagggaaga aagcgaaagg 3540
agcgggcgct agggcgctgg caagtgtagc ggtcacgctg cgcgtaacca ccacacccgc 3600
cgcgcttaat gcgccgctac agggcgcgtc cattcgccat tcaggctgcg caactgttgg 3660
gaagggcgat cggtgcgggc ctcttcgcta ttacgccagc tggcgaaagg gggatgtgct 3720
gcaaggcgat taagttgggt aacgccaggg ttttcccagt cacgacgttg taaaacgacg 3780
gccagtgaat tgtaatacga ctcactatag ggcgaattgg gcccgacgtc gcatgcgctg 3840
atgacacttt ggtctgaaag agatgcattt tgaatcccaa acttgcagtg cccaagtgac 3900
atacatctcc gcgttttgga aaatgttcag aaacagttga ttgtgttgga atggggaatg 3960
gggaatggaa aaatgactca agtatcaatt ccaaaaactt ctctggctgg cagtacctac 4020
tgtccatact actgcatttt ctccagtcag gccactctat actcgacgac acagtagtaa 4080
aacccagata atttcgacat aaacaagaaa acagacccaa taatatttat atatagtcag 4140
ccgtttgtcc agttcagact gtaatagccg aaaaaaaatc caaagtttct attctaggaa 4200
aatatattcc aatattttta attcttaatc tcatttattt tattctagcg aaatacattt 4260
cagctacttg agacatgtga tacccacaaa tcggattcgg actcggttgt tcagaagagc 4320
atatggcatt cgtgctcgct tgttcacgta ttcttcctgt tccatctctt ggccgacaat 4380
cacacaaaaa tggggttttt tttttaattc taatgattca ttacagcaaa attgagatat 4440
agcagaccac gtattccata atcaccaagg aagttcttgg gcgtcttaat taagtcatac 4500
acaagtcagc tttcttcgag cctcatataa gtataagtag ttcaacgtat tagcactgta 4560
cccagcatct ccgtatcgag aaacacaaca acatgcccca ttggacagat catgcggata 4620
cacaggttgt gcagtatcat acatactcga tcagacaggt cgtctgacca tcatacaagc 4680
tgaacaagcg ctccatactt gcacgctctc tatatacaca gttaaattac atatccatag 4740
tctaacctct aacagttaat cttctggtaa gcctcccagc cagccttctg gtatcgcttg 4800
gcctcctcaa taggatctcg gttctggccg tacagacctc ggccgacaat tatgatatcc 4860
gttccggtag acatgacatc ctcaacagtt cggtactgct gtccgagagc gtctcccttg 4920
tcgtcaagac ccaccccggg ggtcagaata agccagtcct cagagtcgcc cttaggtcgg 4980
ttctgggcaa tgaagccaac cacaaactcg gggtcggatc gggcaagctc aatggtctgc 5040
ttggagtact cgccagtggc cagagagccc ttgcaagaca gctcggccag catgagcaga 5100
cctctggcca gcttctcgtt gggagagggg actaggaact ccttgtactg ggagttctcg 5160
tagtcagaga cgtcctcctt cttctgttca gagacagttt cctcggcacc agctcgcagg 5220
ccagcaatga ttccggttcc gggtacaccg tgggcgttgg tgatatcgga ccactcggcg 5280
attcggtgac accggtactg gtgcttgaca gtgttgccaa tatctgcgaa ctttctgtcc 5340
tcgaacagga agaaaccgtg cttaagagca agttccttga gggggagcac agtgccggcg 5400
taggtgaagt cgtcaatgat gtcgatatgg gttttgatca tgcacacata aggtccgacc 5460
ttatcggcaa gctcaatgag ctccttggtg gtggtaacat ccagagaagc acacaggttg 5520
gttttcttgg ctgccacgag cttgagcact cgagcggcaa aggcggactt gtggacgtta 5580
gctcgagctt cgtaggaggg cattttggtg gtgaagagga gactgaaata aatttagtct 5640
gcagaacttt ttatcggaac cttatctggg gcagtgaagt atatgttatg gtaatagtta 5700
cgagttagtt gaacttatag atagactgga ctatacggct atcggtccaa attagaaaga 5760
acgtcaatgg ctctctgggc gtcgcctttg ccgacaaaaa tgtgatcatg atgaaagcca 5820
gcaatgacgt tgcagctgat attgttgtcg gccaaccgcg ccgaaaacgc agctgtcaga 5880
cccacagcct ccaacgaaga atgtatcgtc aaagtgatcc aagcacactc atagttggag 5940
tcgtactcca aaggcggcaa tgacgagtca gacagatact cgtcaaacgg taggttagtg 6000
cttggtatat gagttgtagg catgacaatt tggaaagggg tggactttgg gaatattgtg 6060
ggatttcaat accttagttt gtacagggta attgttacaa atgatacaaa gaactgtatt 6120
tcttttcatt tgttttaatt ggttgtatat caagtccgtt agacgagctc agtgccttgg 6180
cttttggcac tgtatttcat ttttagaggt acactacatt cagtgaggta tggtaaggtt 6240
gagggcataa tgaaggcacc ttgtactgac agtcacagac ctctcaccga gaattttatg 6300
agatatactc gggttcattt taggctcatc gattgccccg gagaagacgg ccaggccgcc 6360
tagatgacaa attcaacaac tcacagctga ctttctgcca ttgccactag gggggggcct 6420
ttttatatgg ccaagccaag ctctccacgt cggttgggct gcacccaaca ataaatgggt 6480
agggttgcac caacaaaggg atgggatggg gggtagaaga tacgaggata acggggctca 6540
atggcacaaa taagaacgaa tactgccatt aagactcgtg atccagcgac tgacaccatt 6600
gcatcatcta agggcctcaa aactacctcg gaactgctgc gctgatctgg acaccacaga 6660
ggttccgagc actttaggtt gcaccaaatg tcccaccagg tgcaggcaga aaacgctgga 6720
acagcgtgta cagtttgtct taacaaaaag tgagggcgct gaggtcgagc agggtggtgt 6780
gacttgttat agcctttaga gctgcgaaag cgcgtatgga tttggctcat caggccagat 6840
tgagggtctg tggacacatg tcatgttagt gtacttcaat cgccccctgg atatagcccc 6900
gacaataggc cgtggcctca tttttttgcc ttccgcacat ttccattgct cggtacccac 6960
accttgcttc tcctgcactt gccaacctta atactggttt acattgacca acatcttaca 7020
agcggggggc ttgtctaggg tatatataaa cagtggctct cccaatcggt tgccagtctc 7080
ttttttcctt tctttcccca cagattcgaa atctaaacta cacatcacag aattccgagc 7140
cgtgagtatc cacgacaaga tcagtgtcga gacgacgcgt tttgtgtaat gacacaatcc 7200
gaaagtcgct agcaacacac actctctaca caaactaacc cagctctggt accatggcct 7260
ccgtcgctcc atcttccaac cccaatccga tccaccatct gtcgcgagtc gaagacgttc 7320
ctctctccca gacgttccga ggcaacatta ccgactttgt gcgatctgga ggctttgccg 7380
acgacgactc caagccctgt tgcatcgacg cgaagactgg ccaacaactt acacagaagc 7440
aagtctggga ctacgccgac aagttcagag cactgctcca tcacgacaac aatctgtgtc 7500
ctttcaatgc caacaccacc gatccagctc ttggagacgt catgatcacc ctcgtgccca 7560
accatctgtt cattacgtcg ctgcactttg ccgcactcga tctgggtgcg acagtttctc 7620
ctggctcggc tggatacact gtggccgagc tcgtcaacca gatcaatctt accggagctt 7680
ctctcatcgt gtacactcga cccgtcttca aggttgtgcg agaggcgctt gctcagatcg 7740
tggtaccagt caagatcgtg gagttcgagg gtctcatcga acgagccgag tttgttcaga 7800
gccacaagat tcagtccaca aagaaagtca cactttctcc tgaggagtcc tactcgagaa 7860
tcgcctacct gggcatgtct tcaggcacct ccggaggtct tcctaaggcc gttcgattgt 7920
cgcacttcaa catggcgagt tctgccgagc tctccaagcg agctgcacct tcgattgccg 7980
gatccgagca gatcgcaggt gccattatcc ctgtcaacca tgtgtatggt ctggccaagt 8040
tcctcattgc catgccaaag tccggagcca ccacagtctt ccactccaag ttcgacctca 8100
tcgagatcct cgaggctcaa cagaagtaca aggtcaacat gtacgccctt gttcctccca 8160
tcattgtcgt tctggccaag catcctgctg ttgagaagta catcccttcg ctgcgagaac 8220
accttcgata cgtgtcctct ggagctgcac ccctgggtgc caacgtcatc gaggcttgca 8280
acaagcgtct tgccggaact gcttctggcg agaacgagtt tggaggtctc aagattgttc 8340
agggctacgg tctcactgaa acctcccctg tggtctccac tttcgatccc aacgatcctg 8400
agcgacacgc tcggtcctgt ggcaagctgg ttcccaacac ccaggcacga atcgtgtcgg 8460
aggacggagt cgatcagcct gcctacgagc tcaaggacct gtctcagttg gaggccgagc 8520
tcaaaaaggg caaccttccc accggtgagt tgtggcttcg aggtccccag atcatggatg 8580
gctaccacaa gaacgacgag gccaacgctg agtcgtttgt cgacgccact gactacactt 8640
ccaacatgcc cttctacatg aagcggtggt tccgaactgg cgatgttgct ctcgtcgata 8700
ctctgggcag atacatgatt gtcgatcgaa ccaaagagat gatcaagagc atgagtaagc 8760
aggttgctcc tgccgagctg gaggacatcc tgcttggcca tccccaggta gccgatgctg 8820
ctgtcatcgg tgttcagcag gtggagaagg gcactgaggc tccccgagcg ttcgtggtgc 8880
ttcgagatcc caagttcgat gcagtggaga tcaaaaagtg gatggacgcc caggtgccca 8940
agtacaaaca acttcatgga ggtgtcgtgg ttctggatgc tgttcccaag aatgccagcg 9000
gcaagattct cagacgactg ctccgtcagc gagagaatga cgtcgttctt ggactcgaca 9060
<210> 67
<211> 10109
<212> DNA
<213> Artificial sequence
<220>
<223> plasmid pZKL7A-FYLFAAs
<400> 67
ggccgcaagt gtggatgggg aagtgagtgc ccggttctgt gtgcacaatt ggcaatccaa 60
gatggatgga ttcaacacag ggatatagcg agctacgtgg tggtgcgagg atatagcaac 120
ggatatttat gtttgacact tgagaatgta cgatacaagc actgtccaag tacaatacta 180
aacatactgt acatactcat actcgtaccc gggcaacggt ttcacttgag tgcagtggct 240
agtgctctta ctcgtacagt gtgcaatact gcgtatcata gtctttgatg tatatcgtat 300
tcattcatgt tagttgattt aaatgtaacg aaactgaaca caagcttcca cacaacgtac 360
gatgtattta tacttgacag tgcgggtcgg aggaagttcg gttcgggcca ggtctgccaa 420
gttgactgtt actacgctaa tgcaatcata aggagaactg tagctaattc accgttgcag 480
ttagatctct cgaaggtgtc acgaaccatt atcatgtatg ctattgtaca ctgccctctg 540
tcctggtaag tgtgttattc caagtaagag ctcttcaatc atcctggaaa tgttcaaaga 600
agaaaaaacc ccgatatatt caactattct tgaaaggaaa tcatgaggga ggcaatgccc 660
actcgatttt gtgcttaaat gacagtgtcg tgcatcagtg tgatcctaac catatacagt 720
aattgtgtca agtgtcaggc ccctttcgtg cttgtacaag cctccaacag aagtaccgag 780
tcgcaaaatg gggtcaattg atgttctcac atgttggtcc tatgactctc ggtttcttca 840
acgagactac cctaagaagc gttacatgtg tggtcagggt ggtgtgtcat gtatagatac 900
agagcgagca cgactcacat ggagaacaac ttctgtaaga gcagagaaac cttaacagga 960
taataataaa taaatcgact cttcttgtga tgtcttttcg cttgttgtac cccacatttg 1020
tgatctgacg taaaagtggg cctgtgcttt gttatcgaga atcacatttc acactaatta 1080
gtgcacgact agtagcacgc agagttgcat gtcagtagtt attgtggatc ccgactactg 1140
tacggtatgt agttatgagc gacttatcat gaactagttt gctcgttatg gcgcgccagc 1200
tgcattaatg aatcggccaa cgcgcgggga gaggcggttt gcgtattggg cgctcttccg 1260
cttcctcgct cactgactcg ctgcgctcgg tcgttcggct gcggcgagcg gtatcagctc 1320
actcaaaggc ggtaatacgg ttatccacag aatcagggga taacgcagga aagaacatgt 1380
gagcaaaagg ccagcaaaag gccaggaacc gtaaaaaggc cgcgttgctg gcgtttttcc 1440
ataggctccg cccccctgac gagcatcaca aaaatcgacg ctcaagtcag aggtggcgaa 1500
acccgacagg actataaaga taccaggcgt ttccccctgg aagctccctc gtgcgctctc 1560
ctgttccgac cctgccgctt accggatacc tgtccgcctt tctcccttcg ggaagcgtgg 1620
cgctttctca tagctcacgc tgtaggtatc tcagttcggt gtaggtcgtt cgctccaagc 1680
tgggctgtgt gcacgaaccc cccgttcagc ccgaccgctg cgccttatcc ggtaactatc 1740
gtcttgagtc caacccggta agacacgact tatcgccact ggcagcagcc actggtaaca 1800
ggattagcag agcgaggtat gtaggcggtg ctacagagtt cttgaagtgg tggcctaact 1860
acggctacac tagaagaaca gtatttggta tctgcgctct gctgaagcca gttaccttcg 1920
gaaaaagagt tggtagctct tgatccggca aacaaaccac cgctggtagc ggtggttttt 1980
ttgtttgcaa gcagcagatt acgcgcagaa aaaaaggatc tcaagaagat cctttgatct 2040
tttctacggg gtctgacgct cagtggaacg aaaactcacg ttaagggatt ttggtcatga 2100
gattatcaaa aaggatcttc acctagatcc ttttaaatta aaaatgaagt tttaaatcaa 2160
tctaaagtat atatgagtaa acttggtctg acagttacca atgcttaatc agtgaggcac 2220
ctatctcagc gatctgtcta tttcgttcat ccatagttgc ctgactcccc gtcgtgtaga 2280
taactacgat acgggagggc ttaccatctg gccccagtgc tgcaatgata ccgcgagacc 2340
cacgctcacc ggctccagat ttatcagcaa taaaccagcc agccggaagg gccgagcgca 2400
gaagtggtcc tgcaacttta tccgcctcca tccagtctat taattgttgc cgggaagcta 2460
gagtaagtag ttcgccagtt aatagtttgc gcaacgttgt tgccattgct acaggcatcg 2520
tggtgtcacg ctcgtcgttt ggtatggctt cattcagctc cggttcccaa cgatcaaggc 2580
gagttacatg atcccccatg ttgtgcaaaa aagcggttag ctccttcggt cctccgatcg 2640
ttgtcagaag taagttggcc gcagtgttat cactcatggt tatggcagca ctgcataatt 2700
ctcttactgt catgccatcc gtaagatgct tttctgtgac tggtgagtac tcaaccaagt 2760
cattctgaga atagtgtatg cggcgaccga gttgctcttg cccggcgtca atacgggata 2820
ataccgcgcc acatagcaga actttaaaag tgctcatcat tggaaaacgt tcttcggggc 2880
gaaaactctc aaggatctta ccgctgttga gatccagttc gatgtaaccc actcgtgcac 2940
ccaactgatc ttcagcatct tttactttca ccagcgtttc tgggtgagca aaaacaggaa 3000
ggcaaaatgc cgcaaaaaag ggaataaggg cgacacggaa atgttgaata ctcatactct 3060
tcctttttca atattattga agcatttatc agggttattg tctcatgagc ggatacatat 3120
ttgaatgtat ttagaaaaat aaacaaatag gggttccgcg cacatttccc cgaaaagtgc 3180
cacctgatgc ggtgtgaaat accgcacaga tgcgtaagga gaaaataccg catcaggaaa 3240
ttgtaagcgt taatattttg ttaaaattcg cgttaaattt ttgttaaatc agctcatttt 3300
ttaaccaata ggccgaaatc ggcaaaatcc cttataaatc aaaagaatag accgagatag 3360
ggttgagtgt tgttccagtt tggaacaaga gtccactatt aaagaacgtg gactccaacg 3420
tcaaagggcg aaaaaccgtc tatcagggcg atggcccact acgtgaacca tcaccctaat 3480
caagtttttt ggggtcgagg tgccgtaaag cactaaatcg gaaccctaaa gggagccccc 3540
gatttagagc ttgacgggga aagccggcga acgtggcgag aaaggaaggg aagaaagcga 3600
aaggagcggg cgctagggcg ctggcaagtg tagcggtcac gctgcgcgta accaccacac 3660
ccgccgcgct taatgcgccg ctacagggcg cgtccattcg ccattcaggc tgcgcaactg 3720
ttgggaaggg cgatcggtgc gggcctcttc gctattacgc cagctggcga aagggggatg 3780
tgctgcaagg cgattaagtt gggtaacgcc agggttttcc cagtcacgac gttgtaaaac 3840
gacggccagt gaattgtaat acgactcact atagggcgaa ttgggcccga cgtcgcatgc 3900
tcaaatttca agactcatat cgagtctagt cggaacaggc gcgccagagt tgggttgggg 3960
acgatgtatg tccaggtacc ctgctcatac aagtacagta tatcctaggt gcaaaaaaag 4020
aaggtatttg tcttatatac cggtactagc acatgtgaat ctatgtttag tctcaagtat 4080
attcattaac atgccctcaa tcgtttctgt ttactttcag atactccttt ggtgcctatg 4140
gttccacaga atccacaatg atactgtaga aggggactac gtgaagtaga ataaaaccag 4200
cttttctgga aacttgtctg ttctatcacc tgttggtcat gtcagtttcc gcgttcttca 4260
cacccacccc cctaataata caacacaata aaatcattga aacaagagcg atgcgaccct 4320
cagtccagca aaccactctg gaaatattga gaaacaaaag acctacagta atacaattgt 4380
ggcatctata gaataggaaa tttaatatgt gcaactgtcc attctctcga cctgtacttc 4440
tacacgattg tcaaaagata ttatcatagt ctacagtaat ttatacagat tgaaagaggg 4500
tcattcaaat atggaagctg gtggtcaaag atcatcccca gtcactctct tctaccttct 4560
aaaagatggt ggaaccgttg gcaagacctc cgtagagcca agtaccttca aacttgcgcc 4620
ctctctccag atctcgacgt ccaatattga gagtgcataa agagatccag tcaaagtagg 4680
ccaggtggtt cttgacaatg tctaaccgag caagagcgtt aaagtccttt ctgtaacagc 4740
tcatactctt aattaagttg cgacacatgt cttgatagta tcttgaattc tctctcttga 4800
gcttttccat aacaagttct tctgcctcca ggaagtccat cggtggtttg atcatggttt 4860
tggtgtagtg gtagtgcagt ggtggtattg tgactgggga tgtagttgag aataagtcat 4920
acacaagtca gctttcttcg agcctcatat aagtataagt agttcaacgt attagcactg 4980
tacccagcat ctccgtatcg agaaacacaa caacatgccc cattggacag atcatgcgga 5040
tacacaggtt gtgcagtatc atacatactc gatcagacag gtcgtctgac catcatacaa 5100
gctgaacaag cgctccatac ttgcacgctc tctatataca cagttaaatt acatatccat 5160
agtctaacct ctaacagtta atcttctggt aagcctccca gccagccttc tggtatcgct 5220
tggcctcctc aataggatct cggttctggc cgtacagacc tcggccgaca attatgatat 5280
ccgttccggt agacatgaca tcctcaacag ttcggtactg ctgtccgaga gcgtctccct 5340
tgtcgtcaag acccaccccg ggggtcagaa taagccagtc ctcagagtcg cccttaggtc 5400
ggttctgggc aatgaagcca accacaaact cggggtcgga tcgggcaagc tcaatggtct 5460
gcttggagta ctcgccagtg gccagagagc ccttgcaaga cagctcggcc agcatgagca 5520
gacctctggc cagcttctcg ttgggagagg ggactaggaa ctccttgtac tgggagttct 5580
cgtagtcaga gacgtcctcc ttcttctgtt cagagacagt ttcctcggca ccagctcgca 5640
ggccagcaat gattccggtt ccgggtacac cgtgggcgtt ggtgatatcg gaccactcgg 5700
cgattcggtg acaccggtac tggtgcttga cagtgttgcc aatatctgcg aactttctgt 5760
cctcgaacag gaagaaaccg tgcttaagag caagttcctt gagggggagc acagtgccgg 5820
cgtaggtgaa gtcgtcaatg atgtcgatat gggttttgat catgcacaca taaggtccga 5880
ccttatcggc aagctcaatg agctccttgg tggtggtaac atccagagaa gcacacaggt 5940
tggttttctt ggctgccacg agcttgagca ctcgagcggc aaaggcggac ttgtggacgt 6000
tagctcgagc ttcgtaggag ggcattttgg tggtgaagag gagactgaaa taaatttagt 6060
ctgcagaact ttttatcgga accttatctg gggcagtgaa gtatatgtta tggtaatagt 6120
tacgagttag ttgaacttat agatagactg gactatacgg ctatcggtcc aaattagaaa 6180
gaacgtcaat ggctctctgg gcgtcgcctt tgccgacaaa aatgtgatca tgatgaaagc 6240
cagcaatgac gttgcagctg atattgttgt cggccaaccg cgccgaaaac gcagctgtca 6300
gacccacagc ctccaacgaa gaatgtatcg tcaaagtgat ccaagcacac tcatagttgg 6360
agtcgtactc caaaggcggc aatgacgagt cagacagata ctcgtcgacc ttttccttgg 6420
gaaccaccac cgtcagccct tctgactcac gtattgtagc caccgacaca ggcaacagtc 6480
cgtggatagc agaatatgtc ttgtcggtcc atttctcacc aactttaggc gtcaagtgaa 6540
tgttgcagaa gaagtatgtg ccttcattga gaatcggtgt tgctgatttc aataaagtct 6600
tgagatcagt ttggccagtc atgttgtggg gggtaattgg attgagttat cgcctacagt 6660
ctgtacaggt atactcgctg cccactttat actttttgat tccgctgcac ttgaagcaat 6720
gtcgtttacc aaaagtgaga atgctccaca gaacacaccc cagggtatgg ttgagcaaaa 6780
aataaacact ccgatacggg gaatcgaacc ccggtctcca cggttctcaa gaagtattct 6840
tgatgagagc gtatcgatga gcctaaaatg aacccgagta tatctcataa aattctcggt 6900
gagaggtctg tgactgtcag tacaaggtgc cttcattatg ccctcaacct taccatacct 6960
cactgaatgt agtgtacctc taaaaatgaa atacagtgcc aaaagccaag gcactgagct 7020
cgtctaacgg acttgatata caaccaatta aaacaaatga aaagaaatac agttctttgt 7080
atcatttgta acaattaccc tgtacaaact aaggtattga aatcccacaa tattcccaaa 7140
gtccacccct ttccaaattg tcatgcctac aactcatata ccaagcacta acctaccgtt 7200
taaacagtgt acgcagtact atagaggaac aattgccccg gagaagacgg ccaggccgcc 7260
tagatgacaa attcaacaac tcacagctga ctttctgcca ttgccactag gggggggcct 7320
ttttatatgg ccaagccaag ctctccacgt cggttgggct gcacccaaca ataaatgggt 7380
agggttgcac caacaaaggg atgggatggg gggtagaaga tacgaggata acggggctca 7440
atggcacaaa taagaacgaa tactgccatt aagactcgtg atccagcgac tgacaccatt 7500
gcatcatcta agggcctcaa aactacctcg gaactgctgc gctgatctgg acaccacaga 7560
ggttccgagc actttaggtt gcaccaaatg tcccaccagg tgcaggcaga aaacgctgga 7620
acagcgtgta cagtttgtct taacaaaaag tgagggcgct gaggtcgagc agggtggtgt 7680
gacttgttat agcctttaga gctgcgaaag cgcgtatgga tttggctcat caggccagat 7740
tgagggtctg tggacacatg tcatgttagt gtacttcaat cgccccctgg atatagcccc 7800
gacaataggc cgtggcctca tttttttgcc ttccgcacat ttccattgct cggtacccac 7860
accttgcttc tcctgcactt gccaacctta atactggttt acattgacca acatcttaca 7920
agcggggggc ttgtctaggg tatatataaa cagtggctct cccaatcggt tgccagtctc 7980
ttttttcctt tctttcccca cagattcgaa atctaaacta cacatcacac catggtcgga 8040
tacaccatct cctcgaagcc cgtgtccgtc gaggttggcc ccgccaagcc tggcgagact 8100
gctccccgac ggaacgtcat tgccaaggac gctcctgtgg tcttccccga caacgattcg 8160
tccctcacca ctgtctacaa gctgttcaaa aagtacgccg agatcaactc cgaacgaaag 8220
gctatgggat ggcgagacac catcgacatt cacgtggaga ccaagcaggt cacaaaggtg 8280
gtcgacggcg tggagaagaa agtgcccaag gaatggaagt acttcgagat gggtccttac 8340
aagtggctgt cctacaagga ggccctcaag ctggttcacg attatggagc tggtcttcga 8400
catctcggca tcaagcccaa agagaagatg cacatttacg cacagacctc tcaccgatgg 8460
atgctttccg gactggcctc tctctcgcag ggcattccca tcgtcactgc ctacgacacc 8520
cttggagagg aaggtctcac acgatctctg caggagacca actccgtcat catgttcacg 8580
gacaaggctc ttctgtcgtc tctcaaggtg tccctcaaaa agggcaccga tctgcgaatc 8640
attatctacg gaggcgacct gactcccgat gacaagaaag ccggaaacac cgagatcgac 8700
gccatcaagg agattgttcc agacatgaag atctacacta tggacgaggt tgtcgctctc 8760
ggtcgagagc atcctcaccc cgtggaagag gtcgactacg aggatctggc cttcatcatg 8820
tacacctctg gctccacagg agttcccaag ggtgtcgtgc tgcagcacaa gcagatcctc 8880
gcctctgtgg ccggtgtcac caagattatc gacagatcca ttatcggcaa tacagatcga 8940
ctgctcaact ttcttcccct cgcacacatc ttcgagtttg tgttcgagat ggtcaccttc 9000
tggtggggtg cctctctggg ctacggaact gtcaagacca tttccgacct gtcgatgaag 9060
aactgcaagg gagacatccg agagctcaag cccaccatca tggtcggcgt tccagctgtc 9120
tgggaaccca tgcggaaggg tattcttggc aaaatcaagg agctgtctcc tctcatgcag 9180
cgagtcttct gggcctcctt tgctgccaag caacgtctcg acgagaacgg acttcccggt 9240
ggctctattc tggattcgct catcttcaag aaagtcaagg acgccactgg aggctgtctc 9300
cgatacgtgt gcaacggagg tgctccagtt tccgtcgaca cccagaagtt cattactacc 9360
cttatctgtc ccatgctcat tggatgcggt ctgaccgaga ctacagccaa caccactatc 9420
atgtctccca agtcctatgc ctttggcacc attggagagc ctactgcagc cgtcaccctc 9480
aagcttatcg acgtgcccga agctggctac ttcgccgaga acaatcaggg agagctgtgc 9540
atcaagggca acgtggtcat gaaggagtat tacaagaacg aggaagagac caagaaagcg 9600
ttctccgacg atggctactt tctcaccgga gacattgccg agtggactgc caatggtcag 9660
cttcgaatta tcgacagacg aaagaacctc gtcaagaccc agaacggaga gtacattgct 9720
ctggagaagc tcgaaacaca gtaccgatcg tcttcctacg ttgccaacct gtgcgtctac 9780
gccgaccaga accgagtcaa gcccatcgct ctggtcattc ccaacgaggg tcctaccaaa 9840
aagcttgccc agagcttggg cgtggattcc gacgactggg atgccgtctg ttccaacaag 9900
aaagtggtca aggctgttct caaggacatg ctggataccg gacgatctct cggtctgtcc 9960
ggcatcgagc tgctgcaagg aatcgtgttg ctgcctggcg agtggactcc ccagaacagc 10020
tacctcaccg ctgcccagaa gctcaaccga aagaagattg tcgatgacaa caaaaaggag 10080
atcgacgagt gctacgagca gtcctaagc 10109
<210> 68
<211> 9037
<212> DNA
<213> Artificial sequence
<220>
<223> plasmid pZP2-YLACoS-5PS3s
<400> 68
ggccgcaagt gtggatgggg aagtgagtgc ccggttctgt gtgcacaatt ggcaatccaa 60
gatggatgga ttcaacacag ggatatagcg agctacgtgg tggtgcgagg atatagcaac 120
ggatatttat gtttgacact tgagaatgta cgatacaagc actgtccaag tacaatacta 180
aacatactgt acatactcat actcgtaccc gggcaacggt ttcacttgag tgcagtggct 240
agtgctctta ctcgtacagt gtgcaatact gcgtatcata gtctttgatg tatatcgtat 300
tcattcatgt tagttgcgta cgggcgtcgt tgcttgtgtg atttttgagg acccatccct 360
ttggtatata agtatactct ggggttaagg ttgcccgtgt agtctaggtt atagttttca 420
tgtgaaatac cgagagccga gggagaataa acgggggtat ttggacttgt ttttttcgcg 480
gaaaagcgtc gaatcaaccc tgcgggcctt gcaccatgtc cacgacgtgt ttctcgcccc 540
aattcgcccc ttgcacgtca aaattaggcc tccatctaga cccctccata acatgtgact 600
gtggggaaaa gtataaggga aaccatgcaa ccatagacga cgtgaaagac ggggaggaac 660
caatggaggc caaagaaatg gggtagcaac agtccaggag acagacaagg agacaaggag 720
agggcgcccg aaagatcgga aaaacaaaca tgtccaattg gggcagtgac ggaaacgaca 780
cggacacttc agtacaatgg accgaccatc tccaagccag ggttattccg gtatcacctt 840
ggccgtaacc tcccgctggt acctgatatt gtacacgttc acattcaata tactttcagc 900
tacaataaga gaggctgttt gtcgggcatg tgtgtccgtc gtatggggtg atgtccgagg 960
gcgaaattcg ctacaagctt aactctggcg cttgtccagt atgaatagac aagtcaagac 1020
cagtggtgcc atgattgaca gggaggtaca agacttcgat actcgagcat tactcggact 1080
tgtggcgatt gaacagacgg gcgatcgctt ctcccccgta ttgccggcgc gccagctgca 1140
ttaatgaatc ggccaacgcg cggggagagg cggtttgcgt attgggcgct cttccgcttc 1200
ctcgctcact gactcgctgc gctcggtcgt tcggctgcgg cgagcggtat cagctcactc 1260
aaaggcggta atacggttat ccacagaatc aggggataac gcaggaaaga acatgtgagc 1320
aaaaggccag caaaaggcca ggaaccgtaa aaaggccgcg ttgctggcgt ttttccatag 1380
gctccgcccc cctgacgagc atcacaaaaa tcgacgctca agtcagaggt ggcgaaaccc 1440
gacaggacta taaagatacc aggcgtttcc ccctggaagc tccctcgtgc gctctcctgt 1500
tccgaccctg ccgcttaccg gatacctgtc cgcctttctc ccttcgggaa gcgtggcgct 1560
ttctcatagc tcacgctgta ggtatctcag ttcggtgtag gtcgttcgct ccaagctggg 1620
ctgtgtgcac gaaccccccg ttcagcccga ccgctgcgcc ttatccggta actatcgtct 1680
tgagtccaac ccggtaagac acgacttatc gccactggca gcagccactg gtaacaggat 1740
tagcagagcg aggtatgtag gcggtgctac agagttcttg aagtggtggc ctaactacgg 1800
ctacactaga agaacagtat ttggtatctg cgctctgctg aagccagtta ccttcggaaa 1860
aagagttggt agctcttgat ccggcaaaca aaccaccgct ggtagcggtg gtttttttgt 1920
ttgcaagcag cagattacgc gcagaaaaaa aggatctcaa gaagatcctt tgatcttttc 1980
tacggggtct gacgctcagt ggaacgaaaa ctcacgttaa gggattttgg tcatgagatt 2040
atcaaaaagg atcttcacct agatcctttt aaattaaaaa tgaagtttta aatcaatcta 2100
aagtatatat gagtaaactt ggtctgacag ttaccaatgc ttaatcagtg aggcacctat 2160
ctcagcgatc tgtctatttc gttcatccat agttgcctga ctccccgtcg tgtagataac 2220
tacgatacgg gagggcttac catctggccc cagtgctgca atgataccgc gagacccacg 2280
ctcaccggct ccagatttat cagcaataaa ccagccagcc ggaagggccg agcgcagaag 2340
tggtcctgca actttatccg cctccatcca gtctattaat tgttgccggg aagctagagt 2400
aagtagttcg ccagttaata gtttgcgcaa cgttgttgcc attgctacag gcatcgtggt 2460
gtcacgctcg tcgtttggta tggcttcatt cagctccggt tcccaacgat caaggcgagt 2520
tacatgatcc cccatgttgt gcaaaaaagc ggttagctcc ttcggtcctc cgatcgttgt 2580
cagaagtaag ttggccgcag tgttatcact catggttatg gcagcactgc ataattctct 2640
tactgtcatg ccatccgtaa gatgcttttc tgtgactggt gagtactcaa ccaagtcatt 2700
ctgagaatag tgtatgcggc gaccgagttg ctcttgcccg gcgtcaatac gggataatac 2760
cgcgccacat agcagaactt taaaagtgct catcattgga aaacgttctt cggggcgaaa 2820
actctcaagg atcttaccgc tgttgagatc cagttcgatg taacccactc gtgcacccaa 2880
ctgatcttca gcatctttta ctttcaccag cgtttctggg tgagcaaaaa caggaaggca 2940
aaatgccgca aaaaagggaa taagggcgac acggaaatgt tgaatactca tactcttcct 3000
ttttcaatat tattgaagca tttatcaggg ttattgtctc atgagcggat acatatttga 3060
atgtatttag aaaaataaac aaataggggt tccgcgcaca tttccccgaa aagtgccacc 3120
tgatgcggtg tgaaataccg cacagatgcg taaggagaaa ataccgcatc aggaaattgt 3180
aagcgttaat attttgttaa aattcgcgtt aaatttttgt taaatcagct cattttttaa 3240
ccaataggcc gaaatcggca aaatccctta taaatcaaaa gaatagaccg agatagggtt 3300
gagtgttgtt ccagtttgga acaagagtcc actattaaag aacgtggact ccaacgtcaa 3360
agggcgaaaa accgtctatc agggcgatgg cccactacgt gaaccatcac cctaatcaag 3420
ttttttgggg tcgaggtgcc gtaaagcact aaatcggaac cctaaaggga gcccccgatt 3480
tagagcttga cggggaaagc cggcgaacgt ggcgagaaag gaagggaaga aagcgaaagg 3540
agcgggcgct agggcgctgg caagtgtagc ggtcacgctg cgcgtaacca ccacacccgc 3600
cgcgcttaat gcgccgctac agggcgcgtc cattcgccat tcaggctgcg caactgttgg 3660
gaagggcgat cggtgcgggc ctcttcgcta ttacgccagc tggcgaaagg gggatgtgct 3720
gcaaggcgat taagttgggt aacgccaggg ttttcccagt cacgacgttg taaaacgacg 3780
gccagtgaat tgtaatacga ctcactatag ggcgaattgg gcccgacgtc gcatgcgctg 3840
atgacacttt ggtctgaaag agatgcattt tgaatcccaa acttgcagtg cccaagtgac 3900
atacatctcc gcgttttgga aaatgttcag aaacagttga ttgtgttgga atggggaatg 3960
gggaatggaa aaatgactca agtatcaatt ccaaaaactt ctctggctgg cagtacctac 4020
tgtccatact actgcatttt ctccagtcag gccactctat actcgacgac acagtagtaa 4080
aacccagata atttcgacat aaacaagaaa acagacccaa taatatttat atatagtcag 4140
ccgtttgtcc agttcagact gtaatagccg aaaaaaaatc caaagtttct attctaggaa 4200
aatatattcc aatattttta attcttaatc tcatttattt tattctagcg aaatacattt 4260
cagctacttg agacatgtga tacccacaaa tcggattcgg actcggttgt tcagaagagc 4320
atatggcatt cgtgctcgct tgttcacgta ttcttcctgt tccatctctt ggccgacaat 4380
cacacaaaaa tggggttttt tttttaattc taatgattca ttacagcaaa attgagatat 4440
agcagaccac gtattccata atcaccaagg aagttcttgg gcgtcttaat taagtcatac 4500
acaagtcagc tttcttcgag cctcatataa gtataagtag ttcaacgtat tagcactgta 4560
cccagcatct ccgtatcgag aaacacaaca acatgcccca ttggacagat catgcggata 4620
cacaggttgt gcagtatcat acatactcga tcagacaggt cgtctgacca tcatacaagc 4680
tgaacaagcg ctccatactt gcacgctctc tatatacaca gttaaattac atatccatag 4740
tctaacctct aacagttaat cttctggtaa gcctcccagc cagccttctg gtatcgcttg 4800
gcctcctcaa taggatctcg gttctggccg tacagacctc ggccgacaat tatgatatcc 4860
gttccggtag acatgacatc ctcaacagtt cggtactgct gtccgagagc gtctcccttg 4920
tcgtcaagac ccaccccggg ggtcagaata agccagtcct cagagtcgcc cttaggtcgg 4980
ttctgggcaa tgaagccaac cacaaactcg gggtcggatc gggcaagctc aatggtctgc 5040
ttggagtact cgccagtggc cagagagccc ttgcaagaca gctcggccag catgagcaga 5100
cctctggcca gcttctcgtt gggagagggg actaggaact ccttgtactg ggagttctcg 5160
tagtcagaga cgtcctcctt cttctgttca gagacagttt cctcggcacc agctcgcagg 5220
ccagcaatga ttccggttcc gggtacaccg tgggcgttgg tgatatcgga ccactcggcg 5280
attcggtgac accggtactg gtgcttgaca gtgttgccaa tatctgcgaa ctttctgtcc 5340
tcgaacagga agaaaccgtg cttaagagca agttccttga gggggagcac agtgccggcg 5400
taggtgaagt cgtcaatgat gtcgatatgg gttttgatca tgcacacata aggtccgacc 5460
ttatcggcaa gctcaatgag ctccttggtg gtggtaacat ccagagaagc acacaggttg 5520
gttttcttgg ctgccacgag cttgagcact cgagcggcaa aggcggactt gtggacgtta 5580
gctcgagctt cgtaggaggg cattttggtg gtgaagagga gactgaaata aatttagtct 5640
gcagaacttt ttatcggaac cttatctggg gcagtgaagt atatgttatg gtaatagtta 5700
cgagttagtt gaacttatag atagactgga ctatacggct atcggtccaa attagaaaga 5760
acgtcaatgg ctctctgggc gtcgcctttg ccgacaaaaa tgtgatcatg atgaaagcca 5820
gcaatgacgt tgcagctgat attgttgtcg gccaaccgcg ccgaaaacgc agctgtcaga 5880
cccacagcct ccaacgaaga atgtatcgtc aaagtgatcc aagcacactc atagttggag 5940
tcgtactcca aaggcggcaa tgacgagtca gacagatact cgtcaaacgg taggttagtg 6000
cttggtatat gagttgtagg catgacaatt tggaaagggg tggactttgg gaatattgtg 6060
ggatttcaat accttagttt gtacagggta attgttacaa atgatacaaa gaactgtatt 6120
tcttttcatt tgttttaatt ggttgtatat caagtccgtt agacgagctc agtgccttgg 6180
cttttggcac tgtatttcat ttttagaggt acactacatt cagtgaggta tggtaaggtt 6240
gagggcataa tgaaggcacc ttgtactgac agtcacagac ctctcaccga gaattttatg 6300
agatatactc gggttcattt taggctcatc gattgccccg gagaagacgg ccaggccgcc 6360
tagatgacaa attcaacaac tcacagctga ctttctgcca ttgccactag gggggggcct 6420
ttttatatgg ccaagccaag ctctccacgt cggttgggct gcacccaaca ataaatgggt 6480
agggttgcac caacaaaggg atgggatggg gggtagaaga tacgaggata acggggctca 6540
atggcacaaa taagaacgaa tactgccatt aagactcgtg atccagcgac tgacaccatt 6600
gcatcatcta agggcctcaa aactacctcg gaactgctgc gctgatctgg acaccacaga 6660
ggttccgagc actttaggtt gcaccaaatg tcccaccagg tgcaggcaga aaacgctgga 6720
acagcgtgta cagtttgtct taacaaaaag tgagggcgct gaggtcgagc agggtggtgt 6780
gacttgttat agcctttaga gctgcgaaag cgcgtatgga tttggctcat caggccagat 6840
tgagggtctg tggacacatg tcatgttagt gtacttcaat cgccccctgg atatagcccc 6900
gacaataggc cgtggcctca tttttttgcc ttccgcacat ttccattgct cggtacccac 6960
accttgcttc tcctgcactt gccaacctta atactggttt acattgacca acatcttaca 7020
agcggggggc ttgtctaggg tatatataaa cagtggctct cccaatcggt tgccagtctc 7080
ttttttcctt tctttcccca cagattcgaa atctaaacta cacatcacag aattccgagc 7140
cgtgagtatc cacgacaaga tcagtgtcga gacgacgcgt tttgtgtaat gacacaatcc 7200
gaaagtcgct agcaacacac actctctaca caaactaacc cagctctggt accatggcct 7260
caatcattca caagtctcct gtgcccgacg ttcagctgtt ctacggttcc tggccagatc 7320
tcatgcgaac ctctcctcat gcccacaacg actccaaacc cgtggtcttt gacttcgata 7380
ccaagcagca acttacttgg aagcaggtct ggcaactcag cgctcgactc agagcccagc 7440
tgtaccacaa gtacggaatc ggcaaacccg gtgctcttgc acctttccac aacgatccct 7500
ctctcggaga cgtggtcatc ttctacactc ccaacaccta cagctcgttg ccctatcatc 7560
tggctcttca cgatctcgga gccaccattt ctcctgcctc cacatcttac gacgtcaagg 7620
acatttgcca tcagatcgtt actaccgatg cggtcgtggt tgtcgctgca gccgagaaat 7680
ccgagattgc tcgagaggcc gttcagctgt ctggtcgaga cgtcagagtt gtggtcatgg 7740
aggacctcat caacaatgct cccaccgttg cgcagaacga tatcgactcg gcacctcatg 7800
tgtccctgtc tcgggaccag gctcgagcca agattgcata cctgggcatg tcttccggta 7860
cgtctggcgg acttcccaag gctgttcgtc tcactcactt caacgttacc tcgaactgtc 7920
tgcaggtctc cgctgccgca cccaaccttg cccagaacgt ggttgccagc gccgtcattc 7980
caaccactca catctacggt ctcaccatgt ttctgtcggt tcttccctac aacggttccg 8040
tggtcattca tcacaagcaa ttcaacttgc gagatctgct cgaggctcag aagacataca 8100
aggtctctct gtggattctc gttcctcccg tcatcgtgca gcttgccaag aaccctatgg 8160
tcgacgagta cctggactcc attcgagccc atgtgcggtg catcgtctct ggagctgctc 8220
ctctcggtgg caatgtcgtg gatcaggttt cggttcgtct taccggcaac aaggaaggca 8280
ttctgcccaa cggagacaag ctcgtcattc atcaagccta cggtcttacc gagtcctctc 8340
ccatcgttgg aatgctcgat cctctgtcgg accacatcga cgtcatgact gtgggctgtc 8400
tcatgcccaa taccgaggct cgaattgtcg acgaagaggg aaacgatcag ccagcagtcc 8460
acgttaccga cacacgaggc atcggtgccg ctgtcaagcg aggcgagaag attccctccg 8520
gagaactctg gattcgaggt cctcagatca tggacggata ccacaagaac cccgagtcgt 8580
ctcgtgagtc cctggaaccc agcacagaga cctacggtct gcaacatttc caggacagat 8640
ggcttcgaac tggagacgtt gctgtcatcg acaccttcgg acgagtcatg gttgtggatc 8700
gaaccaagga gctcatcaag tccatgtctc gacaggttgc tcctgccgag ctcgaagctc 8760
ttctgctcaa ccatccttcc gtcaacgatg tggctgtcgt tggcgtccac aacgacgata 8820
atggcacaga gtcagcacga gcgtttgtcg ttcttcaacc aggcgacgcc tgtgatccta 8880
ctaccatcaa gcactggatg gaccagcaag ttccctccta caagcggctg tacggaggca 8940
ttgtggtcat cgacactgtt cccaagaatg cctctggcaa gattctgcga agactgcttc 9000
gacagcggag agacgatcga gtctggggtc tgtaagc 9037
<210> 69
<211> 3663
<212> DNA
<213> Artificial sequence
<220>
<223> pET23d plasmid
<400> 69
atccggatat agttcctcct ttcagcaaaa aacccctcaa gacccgttta gaggccccaa 60
ggggttatgc tagttattgc tcagcggtgg cagcagccaa ctcagcttcc tttcgggctt 120
tgttagcagc cggatctcag tggtggtggt ggtggtgctc gagtgcggcc gcaagcttgt 180
cgacggagct cgaattcgga tccgacccat ttgctgtcca ccagtcatgc tagccatggt 240
atatctcctt cttaaagtta aacaaaatta tttctagagg gaaaccgttg tggtctccct 300
atagtgagtc gtattaattt cgcgggatcg agatctcggg cagcgttggg tcctggccac 360
gggtgcgcat gatcgtgctc ctgtcgttga ggacccggct aggctggcgg ggttgcctta 420
ctggttagca gaatgaatca ccgatacgcg agcgaacgtg aagcgactgc tgctgcaaaa 480
cgtctgcgac ctgagcaaca acatgaatgg tcttcggttt ccgtgtttcg taaagtctgg 540
aaacgcggaa gtcagcgccc tgcaccatta tgttccggat ctgcatcgca ggatgctgct 600
ggctaccctg tggaacacct acatctgtat taacgaagcg ctggcattga ccctgagtga 660
tttttctctg gtcccgccgc atccataccg ccagttgttt accctcacaa cgttccagta 720
accgggcatg ttcatcatca gtaacccgta tcgtgagcat cctctctcgt ttcatcggta 780
tcattacccc catgaacaga aatccccctt acacggaggc atcagtgacc aaacaggaaa 840
aaaccgccct taacatggcc cgctttatca gaagccagac attaacgctt ctggagaaac 900
tcaacgagct ggacgcggat gaacaggcag acatctgtga atcgcttcac gaccacgctg 960
atgagcttta ccgcagctgc ctcgcgcgtt tcggtgatga cggtgaaaac ctctgacaca 1020
tgcagctccc ggagacggtc acagcttgtc tgtaagcgga tgccgggagc agacaagccc 1080
gtcagggcgc gtcagcgggt gttggcgggt gtcggggcgc agccatgacc cagtcacgta 1140
gcgatagcgg agtgtatact ggcttaacta tgcggcatca gagcagattg tactgagagt 1200
gcaccatata tgcggtgtga aataccgcac agatgcgtaa ggagaaaata ccgcatcagg 1260
cgctcttccg cttcctcgct cactgactcg ctgcgctcgg tcgttcggct gcggcgagcg 1320
gtatcagctc actcaaaggc ggtaatacgg ttatccacag aatcagggga taacgcagga 1380
aagaacatgt gagcaaaagg ccagcaaaag gccaggaacc gtaaaaaggc cgcgttgctg 1440
gcgtttttcc ataggctccg cccccctgac gagcatcaca aaaatcgacg ctcaagtcag 1500
aggtggcgaa acccgacagg actataaaga taccaggcgt ttccccctgg aagctccctc 1560
gtgcgctctc ctgttccgac cctgccgctt accggatacc tgtccgcctt tctcccttcg 1620
ggaagcgtgg cgctttctca tagctcacgc tgtaggtatc tcagttcggt gtaggtcgtt 1680
cgctccaagc tgggctgtgt gcacgaaccc cccgttcagc ccgaccgctg cgccttatcc 1740
ggtaactatc gtcttgagtc caacccggta agacacgact tatcgccact ggcagcagcc 1800
actggtaaca ggattagcag agcgaggtat gtaggcggtg ctacagagtt cttgaagtgg 1860
tggcctaact acggctacac tagaaggaca gtatttggta tctgcgctct gctgaagcca 1920
gttaccttcg gaaaaagagt tggtagctct tgatccggca aacaaaccac cgctggtagc 1980
ggtggttttt ttgtttgcaa gcagcagatt acgcgcagaa aaaaaggatc tcaagaagat 2040
cctttgatct tttctacggg gtctgacgct cagtggaacg aaaactcacg ttaagggatt 2100
ttggtcatga gattatcaaa aaggatcttc acctagatcc ttttaaatta aaaatgaagt 2160
tttaaatcaa tctaaagtat atatgagtaa acttggtctg acagttacca atgcttaatc 2220
agtgaggcac ctatctcagc gatctgtcta tttcgttcat ccatagttgc ctgactcccc 2280
gtcgtgtaga taactacgat acgggagggc ttaccatctg gccccagtgc tgcaatgata 2340
ccgcgagacc cacgctcacc ggctccagat ttatcagcaa taaaccagcc agccggaagg 2400
gccgagcgca gaagtggtcc tgcaacttta tccgcctcca tccagtctat taattgttgc 2460
cgggaagcta gagtaagtag ttcgccagtt aatagtttgc gcaacgttgt tgccattgct 2520
gcaggcatcg tggtgtcacg ctcgtcgttt ggtatggctt cattcagctc cggttcccaa 2580
cgatcaaggc gagttacatg atcccccatg ttgtgcaaaa aagcggttag ctccttcggt 2640
cctccgatcg ttgtcagaag taagttggcc gcagtgttat cactcatggt tatggcagca 2700
ctgcataatt ctcttactgt catgccatcc gtaagatgct tttctgtgac tggtgagtac 2760
tcaaccaagt cattctgaga atagtgtatg cggcgaccga gttgctcttg cccggcgtca 2820
atacgggata ataccgcgcc acatagcaga actttaaaag tgctcatcat tggaaaacgt 2880
tcttcggggc gaaaactctc aaggatctta ccgctgttga gatccagttc gatgtaaccc 2940
actcgtgcac ccaactgatc ttcagcatct tttactttca ccagcgtttc tgggtgagca 3000
aaaacaggaa ggcaaaatgc cgcaaaaaag ggaataaggg cgacacggaa atgttgaata 3060
ctcatactct tcctttttca atattattga agcatttatc agggttattg tctcatgagc 3120
ggatacatat ttgaatgtat ttagaaaaat aaacaaatag gggttccgcg cacatttccc 3180
cgaaaagtgc cacctgaaat tgtaaacgtt aatattttgt taaaattcgc gttaaatttt 3240
tgttaaatca gctcattttt taaccaatag gccgaaatcg gcaaaatccc ttataaatca 3300
aaagaataga ccgagatagg gttgagtgtt gttccagttt ggaacaagag tccactatta 3360
aagaacgtgg actccaacgt caaagggcga aaaaccgtct atcagggcga tggcccacta 3420
cgtgaaccat caccctaatc aagttttttg gggtcgaggt gccgtaaagc actaaatcgg 3480
aaccctaaag ggagcccccg atttagagct tgacggggaa agccggcgaa cgtggcgaga 3540
aaggaaggga agaaagcgaa aggagcgggc gctagggcgc tggcaagtgt agcggtcacg 3600
ctgcgcgtaa ccaccacacc cgccgcgctt aatgcgccgc tacagggcgc gtcccattcg 3660
cca 3663
<210> 70
<211> 12712
<212> DNA
<213> Artificial sequence
<220>
<223> pY157 plasmid
<400> 70
ttgagaagcc cattgtatat tattaggatc gtagcattat tgtggcaaaa aatattcaag 60
tgctcatgtg aattgacacg atcacgtaaa tacctggtga aattgctagt attcgtgatg 120
ttctaataca actctgttca atatttccgg cgctctcttg tatacaagag cacaagacat 180
gcaccccaca ttaaccgagg tcaagtgttt atgtatgaaa agtgacataa atcgtccaaa 240
aaaaagtagc acatagttgt atggctgtaa gttatgtgat tgtcagttct tcggccttcc 300
aactcctatg caccgtcttc aatcatctac ccccgtgccc cacaccccgc actattagag 360
tttatcacag tcagctaaac tgcttgcaca tctacacctc tgactacacc accatggatt 420
tcttcagacg gcaccagaaa aaggtgctgg cactggtagg tgtggcgctg agttcctacc 480
tgtttatcga ctatgtgaag aaaaagttct tcgagatcca gggtcgtttg agctcggagc 540
gaaccgctaa acagaatctc cggcgccgat ttgaacagaa ccagcaggat gcagatttta 600
caatcatggc tctgctatcc agcttgacga caccggtaat ggagcgttac cccgtcgacc 660
agatcaaggc agagttacag agcaagagac gccccacaga ccgggttttg gctctcgaga 720
gctccacctc gtcctcagct accgcacaaa ccgtgcccac catgacaagt ggcgccacag 780
aggagggcga gaagttaatt aactttggcc ggcctttacc tgcaggataa cttcgtataa 840
tgtatgctat acgaagttat gaattctctc tcttgagctt ttccataaca agttcttctg 900
cctccaggaa gtccatgggt ggtttgatca tggttttggt gtagtggtag tgcagtggtg 960
gtattgtgac tggggatgta gttgagaata agtcatacac aagtcagctt tcttcgagcc 1020
tcatataagt ataagtagtt caacgtatta gcactgtacc cagcatctcc gtatcgagaa 1080
acacaacaac atgccccatt ggacagatca tgcggataca caggttgtgc agtatcatac 1140
atactcgatc agacaggtcg tctgaccatc atacaagctg aacaagcgct ccatacttgc 1200
acgctctcta tatacacagt taaattacat atccatagtc taacctctaa cagttaatct 1260
tctggtaagc ctcccagcca gccttctggt atcgcttggc ctcctcaata ggatctcggt 1320
tctggccgta cagacctcgg ccgacaatta tgatatccgt tccggtagac atgacatcct 1380
caacagttcg gtactgctgt ccgagagcgt ctcccttgtc gtcaagaccc accccggggg 1440
tcagaataag ccagtcctca gagtcgccct taggtcggtt ctgggcaatg aagccaacca 1500
caaactcggg gtcggatcgg gcaagctcaa tggtctgctt ggagtactcg ccagtggcca 1560
gagagccctt gcaagacagc tcggccagca tgagcagacc tctggccagc ttctcgttgg 1620
gagaggggac taggaactcc ttgtactggg agttctcgta gtcagagacg tcctccttct 1680
tctgttcaga gacagtttcc tcggcaccag ctcgcaggcc agcaatgatt ccggttccgg 1740
gtacaccgtg ggcgttggtg atatcggacc actcggcgat tcggtgacac cggtactggt 1800
gcttgacagt gttgccaata tctgcgaact ttctgtcctc gaacaggaag aaaccgtgct 1860
taagagcaag ttccttgagg gggagcacag tgccggcgta ggtgaagtcg tcaatgatgt 1920
cgatatgggt tttgatcatg cacacataag gtccgacctt atcggcaagc tcaatgagct 1980
ccttggtggt ggtaacatcc agagaagcac acaggttggt tttcttggct gccacgagct 2040
tgagcactcg agcggcaaag gcggacttgt ggacgttagc tcgagcttcg taggagggca 2100
ttttggtggt gaagaggaga ctgaaataaa tttagtctgc agaacttttt atcggaacct 2160
tatctggggc agtgaagtat atgttatggt aatagttacg agttagttga acttatagat 2220
agactggact atacggctat cggtccaaat tagaaagaac gtcaatggct ctctgggcgt 2280
cgcctttgcc gacaaaaatg tgatcatgat gaaagccagc aatgacgttg cagctgatat 2340
tgttgtcggc caaccgcgcc gaaaacgcag ctgtcagacc cacagcctcc aacgaagaat 2400
gtatcgtcaa agtgatccaa gcacactcat agttggagtc gtactccaaa ggcggcaatg 2460
acgagtcaga cagatactcg tcgactcatc gatataactt cgtataatgt atgctatacg 2520
aagttatcct aggtatagat cttgcacttc ttattttctt cacgcgtttg cagctcaaca 2580
ttctaggacg acgaaactac gtcaacagtg ttgtcgctct ggcgcagcag ggccgagagg 2640
gtaatgccga gggtcgagtg gcgccctcgt ttggtgatct tgcagatatg ggctatttcg 2700
gcgacctttc aggctcgtcc agcttcggag aaactattgt cgatcccgat ctggacgaac 2760
agtaccttac cttttcgtgg tggctgctga acgagggatg ggtgtcgctg agcgagcgag 2820
tggaggaagc ggttcgtcga gtgtgggacc ccgtgtcacc caaggccgaa cttggatttg 2880
acgagttgtc ggaactcatt ggacgaacac agatgctcat tgatcgacct ctcaatccct 2940
cgtcgccact caactttctg agccagctgc tgccaccacg ggagcaggag gagtacgtgc 3000
ttgcccagaa ccccagcgat actgctgccc ccattgtagg acctaccctc cgacggcttc 3060
tggacgagac tgccgacttc atcgagtccc ctaatgccgc agaggtgatt gagcgacttg 3120
ttcactccgg tctctctgtg ttcatggaca agctggctgt cacgtttgga gccacacctg 3180
ctgattcggg ttcgccttat cctgtggtgc tgcctactgc aaaggtcaag ctgccctcca 3240
ttcttgccaa catggctcga caggctggag gcatggccca gggatcgccg ggcgtggaaa 3300
acgagtacat tgacgtgatg aaccaagtgc aggagctgac ctcctttagt gctgtggtct 3360
attcatcttt tgattgggct ctctagaggc tcattcacga aagacacgaa gaacgaagat 3420
ggggactgaa tacagcgctc tcatttgtac acaaatgatt tatgacagag taacttgtac 3480
atcatgtaga gcatacatac tgaaggtgtg atctcacggg atatcttgaa gaccactcgt 3540
agctggaggc ataggtagtg ctagtacgga tacttgcacc gtatccaaca taagtagagg 3600
agcctcctag tggctattgg tacaccgata aagatacaca tacatggcgc gccagctgca 3660
ttaatgaatc ggccaacgcg cggggagagg cggtttgcgt attgggcgct cttccgcttc 3720
ctcgctcact gactcgctgc gctcggtcgt tcggctgcgg cgagcggtat cagctcactc 3780
aaaggcggta atacggttat ccacagaatc aggggataac gcaggaaaga acatgtgagc 3840
aaaaggccag caaaaggcca ggaaccgtaa aaaggccgcg ttgctggcgt ttttccatag 3900
gctccgcccc cctgacgagc atcacaaaaa tcgacgctca agtcagaggt ggcgaaaccc 3960
gacaggacta taaagatacc aggcgtttcc ccctggaagc tccctcgtgc gctctcctgt 4020
tccgaccctg ccgcttaccg gatacctgtc cgcctttctc ccttcgggaa gcgtggcgct 4080
ttctcatagc tcacgctgta ggtatctcag ttcggtgtag gtcgttcgct ccaagctggg 4140
ctgtgtgcac gaaccccccg ttcagcccga ccgctgcgcc ttatccggta actatcgtct 4200
tgagtccaac ccggtaagac acgacttatc gccactggca gcagccactg gtaacaggat 4260
tagcagagcg aggtatgtag gcggtgctac agagttcttg aagtggtggc ctaactacgg 4320
ctacactaga agaacagtat ttggtatctg cgctctgctg aagccagtta ccttcggaaa 4380
aagagttggt agctcttgat ccggcaaaca aaccaccgct ggtagcggtg gtttttttgt 4440
ttgcaagcag cagattacgc gcagaaaaaa aggatctcaa gaagatcctt tgatcttttc 4500
tacggggtct gacgctcagt ggaacgaaaa ctcacgttaa gggattttgg tcatgagatt 4560
atcaaaaagg atcttcacct agatcctttt aaattaaaaa tgaagtttta aatcaatcta 4620
aagtatatat gagtaaactt ggtctgacag ttaccaatgc ttaatcagtg aggcacctat 4680
ctcagcgatc tgtctatttc gttcatccat agttgcctga ctccccgtcg tgtagataac 4740
tacgatacgg gagggcttac catctggccc cagtgctgca atgataccgc gagacccacg 4800
ctcaccggct ccagatttat cagcaataaa ccagccagcc ggaagggccg agcgcagaag 4860
tggtcctgca actttatccg cctccatcca gtctattaat tgttgccggg aagctagagt 4920
aagtagttcg ccagttaata gtttgcgcaa cgttgttgcc attgctacag gcatcgtggt 4980
gtcacgctcg tcgtttggta tggcttcatt cagctccggt tcccaacgat caaggcgagt 5040
tacatgatcc cccatgttgt gcaaaaaagc ggttagctcc ttcggtcctc cgatcgttgt 5100
cagaagtaag ttggccgcag tgttatcact catggttatg gcagcactgc ataattctct 5160
tactgtcatg ccatccgtaa gatgcttttc tgtgactggt gagtactcaa ccaagtcatt 5220
ctgagaatag tgtatgcggc gaccgagttg ctcttgcccg gcgtcaatac gggataatac 5280
cgcgccacat agcagaactt taaaagtgct catcattgga aaacgttctt cggggcgaaa 5340
actctcaagg atcttaccgc tgttgagatc cagttcgatg taacccactc gtgcacccaa 5400
ctgatcttca gcatctttta ctttcaccag cgtttctggg tgagcaaaaa caggaaggca 5460
aaatgccgca aaaaagggaa taagggcgac acggaaatgt tgaatactca tactcttcct 5520
ttttcaatat tattgaagca tttatcaggg ttattgtctc atgagcggat acatatttga 5580
atgtatttag aaaaataaac aaataggggt tccgcgcaca tttccccgaa aagtgccacc 5640
tgatgcggtg tgaaataccg cacagatgcg taaggagaaa ataccgcatc aggaaattgt 5700
aagcgttaat attttgttaa aattcgcgtt aaatttttgt taaatcagct cattttttaa 5760
ccaataggcc gaaatcggca aaatccctta taaatcaaaa gaatagaccg agatagggtt 5820
gagtgttgtt ccagtttgga acaagagtcc actattaaag aacgtggact ccaacgtcaa 5880
agggcgaaaa accgtctatc agggcgatgg cccactacgt gaaccatcac cctaatcaag 5940
ttttttgggg tcgaggtgcc gtaaagcact aaatcggaac cctaaaggga gcccccgatt 6000
tagagcttga cggggaaagc cggcgaacgt ggcgagaaag gaagggaaga aagcgaaagg 6060
agcgggcgct agggcgctgg caagtgtagc ggtcacgctg cgcgtaacca ccacacccgc 6120
cgcgcttaat gcgccgctac agggcgcgtc cattcgccat tcaggctgcg caactgttgg 6180
gaagggcgat cggtgcgggc ctcttcgcta ttacgccagc tggcgaaagg gggatgtgct 6240
gcaaggcgat taagttgggt aacgccaggg ttttcccagt cacgacgttg taaaacgacg 6300
gccagtgaat tgtaatacga ctcactatag ggcgaattgg gcccgacgtc gcatgcttga 6360
gaagcccatt gtatattatt aggatcgtag cattattgtg gcaaaaaata ttcaagtgct 6420
catgtgaatt gacacgatca cgtaaatacc tggtgaaatt gctagtattc gtgatgttct 6480
aatacaactc tgttcaatat ttccggcgct ctcttgtata caagagcaca agacatgcac 6540
cccacattaa ccgaggtcaa gtgtttatgt atgaaaagtg acataaatcg tccaaaaaaa 6600
agtagcacat agttgtatgg ctgtaagtta tgtgattgtc agttcttcgg ccttccaact 6660
cctatgcacc gtcttcaatc atctaccccc gtgccccaca ccccgcacta ttagagttta 6720
tcacagtcag ctaaactgct tgcacatcta cacctctgac tacaccacca tggatttctt 6780
cagacggcac cagaaaaagg tgctggcact ggtaggtgtg gcgctgagtt cctacctgtt 6840
tatcgactat gtgaagaaaa agttcttcga gatccagggt cgtttgagct cggagcgaac 6900
cgctaaacag aatctccggc gccgatttga acagaaccag caggatgcag attttacaat 6960
catggctctg ctatccagct tgacgacacc ggtaatggag cgttaccccg tcgaccagat 7020
caaggcagag ttacagagca agagacgccc cacagaccgg gttttggctc tcgagagctc 7080
cacctcgtcc tcagctaccg cacaaaccgt gcccaccatg acaagtggcg ccacagagga 7140
gggcgagaag ttaattaact ttggccggcc tttacctgca ggataacttc gtataatgta 7200
tgctatacga agttatgaat tctctctctt gagcttttcc ataacaagtt cttctgcctc 7260
caggaagtcc atgggtggtt tgatcatggt tttggtgtag tggtagtgca gtggtggtat 7320
tgtgactggg gatgtagttg agaataagtc atacacaagt cagctttctt cgagcctcat 7380
ataagtataa gtagttcaac gtattagcac tgtacccagc atctccgtat cgagaaacac 7440
aacaacatgc cccattggac agatcatgcg gatacacagg ttgtgcagta tcatacatac 7500
tcgatcagac aggtcgtctg accatcatac aagctgaaca agcgctccat acttgcacgc 7560
tctctatata cacagttaaa ttacatatcc atagtctaac ctctaacagt taatcttctg 7620
gtaagcctcc cagccagcct tctggtatcg cttggcctcc tcaataggat ctcggttctg 7680
gccgtacaga cctcggccga caattatgat atccgttccg gtagacatga catcctcaac 7740
agttcggtac tgctgtccga gagcgtctcc cttgtcgtca agacccaccc cgggggtcag 7800
aataagccag tcctcagagt cgcccttagg tcggttctgg gcaatgaagc caaccacaaa 7860
ctcggggtcg gatcgggcaa gctcaatggt ctgcttggag tactcgccag tggccagaga 7920
gcccttgcaa gacagctcgg ccagcatgag cagacctctg gccagcttct cgttgggaga 7980
ggggactagg aactccttgt actgggagtt ctcgtagtca gagacgtcct ccttcttctg 8040
ttcagagaca gtttcctcgg caccagctcg caggccagca atgattccgg ttccgggtac 8100
accgtgggcg ttggtgatat cggaccactc ggcgattcgg tgacaccggt actggtgctt 8160
gacagtgttg ccaatatctg cgaactttct gtcctcgaac aggaagaaac cgtgcttaag 8220
agcaagttcc ttgaggggga gcacagtgcc ggcgtaggtg aagtcgtcaa tgatgtcgat 8280
atgggttttg atcatgcaca cataaggtcc gaccttatcg gcaagctcaa tgagctcctt 8340
ggtggtggta acatccagag aagcacacag gttggttttc ttggctgcca cgagcttgag 8400
cactcgagcg gcaaaggcgg acttgtggac gttagctcga gcttcgtagg agggcatttt 8460
ggtggtgaag aggagactga aataaattta gtctgcagaa ctttttatcg gaaccttatc 8520
tggggcagtg aagtatatgt tatggtaata gttacgagtt agttgaactt atagatagac 8580
tggactatac ggctatcggt ccaaattaga aagaacgtca atggctctct gggcgtcgcc 8640
tttgccgaca aaaatgtgat catgatgaaa gccagcaatg acgttgcagc tgatattgtt 8700
gtcggccaac cgcgccgaaa acgcagctgt cagacccaca gcctccaacg aagaatgtat 8760
cgtcaaagtg atccaagcac actcatagtt ggagtcgtac tccaaaggcg gcaatgacga 8820
gtcagacaga tactcgtcga ctcatcgata taacttcgta taatgtatgc tatacgaagt 8880
tatcctaggt atagatcttg cacttcttat tttcttcacg cgtttgcagc tcaacattct 8940
aggacgacga aactacgtca acagtgttgt cgctctggcg cagcagggcc gagagggtaa 9000
tgccgagggt cgagtggcgc cctcgtttgg tgatcttgca gatatgggct atttcggcga 9060
cctttcaggc tcgtccagct tcggagaaac tattgtcgat cccgatctgg acgaacagta 9120
ccttaccttt tcgtggtggc tgctgaacga gggatgggtg tcgctgagcg agcgagtgga 9180
ggaagcggtt cgtcgagtgt gggaccccgt gtcacccaag gccgaacttg gatttgacga 9240
gttgtcggaa ctcattggac gaacacagat gctcattgat cgacctctca atccctcgtc 9300
gccactcaac tttctgagcc agctgctgcc accacgggag caggaggagt acgtgcttgc 9360
ccagaacccc agcgatactg ctgcccccat tgtaggacct accctccgac ggcttctgga 9420
cgagactgcc gacttcatcg agtcccctaa tgccgcagag gtgattgagc gacttgttca 9480
ctccggtctc tctgtgttca tggacaagct ggctgtcacg tttggagcca cacctgctga 9540
ttcgggttcg ccttatcctg tggtgctgcc tactgcaaag gtcaagctgc cctccattct 9600
tgccaacatg gctcgacagg ctggaggcat ggcccaggga tcgccgggcg tggaaaacga 9660
gtacattgac gtgatgaacc aagtgcagga gctgacctcc tttagtgctg tggtctattc 9720
atcttttgat tgggctctct agaggctcat tcacgaaaga cacgaagaac gaagatgggg 9780
actgaataca gcgctctcat ttgtacacaa atgatttatg acagagtaac ttgtacatca 9840
tgtagagcat acatactgaa ggtgtgatct cacgggatat cttgaagacc actcgtagct 9900
ggaggcatag gtagtgctag tacggatact tgcaccgtat ccaacataag tagaggagcc 9960
tcctagtggc tattggtaca ccgataaaga tacacataca tggcgcgcca gctgcattaa 10020
tgaatcggcc aacgcgcggg gagaggcggt ttgcgtattg ggcgctcttc cgcttcctcg 10080
ctcactgact cgctgcgctc ggtcgttcgg ctgcggcgag cggtatcagc tcactcaaag 10140
gcggtaatac ggttatccac agaatcaggg gataacgcag gaaagaacat gtgagcaaaa 10200
ggccagcaaa aggccaggaa ccgtaaaaag gccgcgttgc tggcgttttt ccataggctc 10260
cgcccccctg acgagcatca caaaaatcga cgctcaagtc agaggtggcg aaacccgaca 10320
ggactataaa gataccaggc gtttccccct ggaagctccc tcgtgcgctc tcctgttccg 10380
accctgccgc ttaccggata cctgtccgcc tttctccctt cgggaagcgt ggcgctttct 10440
catagctcac gctgtaggta tctcagttcg gtgtaggtcg ttcgctccaa gctgggctgt 10500
gtgcacgaac cccccgttca gcccgaccgc tgcgccttat ccggtaacta tcgtcttgag 10560
tccaacccgg taagacacga cttatcgcca ctggcagcag ccactggtaa caggattagc 10620
agagcgaggt atgtaggcgg tgctacagag ttcttgaagt ggtggcctaa ctacggctac 10680
actagaagaa cagtatttgg tatctgcgct ctgctgaagc cagttacctt cggaaaaaga 10740
gttggtagct cttgatccgg caaacaaacc accgctggta gcggtggttt ttttgtttgc 10800
aagcagcaga ttacgcgcag aaaaaaagga tctcaagaag atcctttgat cttttctacg 10860
gggtctgacg ctcagtggaa cgaaaactca cgttaaggga ttttggtcat gagattatca 10920
aaaaggatct tcacctagat ccttttaaat taaaaatgaa gttttaaatc aatctaaagt 10980
atatatgagt aaacttggtc tgacagttac caatgcttaa tcagtgaggc acctatctca 11040
gcgatctgtc tatttcgttc atccatagtt gcctgactcc ccgtcgtgta gataactacg 11100
atacgggagg gcttaccatc tggccccagt gctgcaatga taccgcgaga cccacgctca 11160
ccggctccag atttatcagc aataaaccag ccagccggaa gggccgagcg cagaagtggt 11220
cctgcaactt tatccgcctc catccagtct attaattgtt gccgggaagc tagagtaagt 11280
agttcgccag ttaatagttt gcgcaacgtt gttgccattg ctacaggcat cgtggtgtca 11340
cgctcgtcgt ttggtatggc ttcattcagc tccggttccc aacgatcaag gcgagttaca 11400
tgatccccca tgttgtgcaa aaaagcggtt agctccttcg gtcctccgat cgttgtcaga 11460
agtaagttgg ccgcagtgtt atcactcatg gttatggcag cactgcataa ttctcttact 11520
gtcatgccat ccgtaagatg cttttctgtg actggtgagt actcaaccaa gtcattctga 11580
gaatagtgta tgcggcgacc gagttgctct tgcccggcgt caatacggga taataccgcg 11640
ccacatagca gaactttaaa agtgctcatc attggaaaac gttcttcggg gcgaaaactc 11700
tcaaggatct taccgctgtt gagatccagt tcgatgtaac ccactcgtgc acccaactga 11760
tcttcagcat cttttacttt caccagcgtt tctgggtgag caaaaacagg aaggcaaaat 11820
gccgcaaaaa agggaataag ggcgacacgg aaatgttgaa tactcatact cttccttttt 11880
caatattatt gaagcattta tcagggttat tgtctcatga gcggatacat atttgaatgt 11940
atttagaaaa ataaacaaat aggggttccg cgcacatttc cccgaaaagt gccacctgat 12000
gcggtgtgaa ataccgcaca gatgcgtaag gagaaaatac cgcatcagga aattgtaagc 12060
gttaatattt tgttaaaatt cgcgttaaat ttttgttaaa tcagctcatt ttttaaccaa 12120
taggccgaaa tcggcaaaat cccttataaa tcaaaagaat agaccgagat agggttgagt 12180
gttgttccag tttggaacaa gagtccacta ttaaagaacg tggactccaa cgtcaaaggg 12240
cgaaaaaccg tctatcaggg cgatggccca ctacgtgaac catcacccta atcaagtttt 12300
ttggggtcga ggtgccgtaa agcactaaat cggaacccta aagggagccc ccgatttaga 12360
gcttgacggg gaaagccggc gaacgtggcg agaaaggaag ggaagaaagc gaaaggagcg 12420
ggcgctaggg cgctggcaag tgtagcggtc acgctgcgcg taaccaccac acccgccgcg 12480
cttaatgcgc cgctacaggg cgcgtccatt cgccattcag gctgcgcaac tgttgggaag 12540
ggcgatcggt gcgggcctct tcgctattac gccagctggc gaaaggggga tgtgctgcaa 12600
ggcgattaag ttgggtaacg ccagggtttt cccagtcacg acgttgtaaa acgacggcca 12660
gtgaattgta atacgactca ctatagggcg aattgggccc gacgtcgcat gc 12712
<210> 71
<211> 1947
<212> DNA
<213> Artificial sequence
<220>
<223> PEX3 knock-out site, including LoxP flanked URA3
<400> 71
acagaccggg ttttggctct cgagagctcc acctcgtcct cagctaccgc acaaaccgtg 60
cccaccatga caagtggcgc cacagaggag ggcgagaagt taattaactt tggccggcct 120
ttacctgcag gataacttcg tataatgtat gctatacgaa gttatgaatt ctctctcttg 180
agcttttcca taacaagttc ttctgcctcc aggaagtcca tgggtggttt gatcatggtt 240
ttggtgtagt ggtagtgcag tggtggtatt gtgactgggg atgtagttga gaataagtca 300
tacacaagtc agctttcttc gagcctcata taagtataag tagttcaacg tattagcact 360
gtacccagca tctccgtatc gagaaacaca acaacatgcc ccattggaca gatcatgcgg 420
atacacaggt tgtgcagtat catacatact cgatcagaca ggtcgtctga ccatcataca 480
agctgaacaa gcgctccata cttgcacgct ctctatatac acagttaaat tacatatcca 540
tagtctaacc tctaacagtt aatcttctgg taagcctccc agccagcctt ctggtatcgc 600
ttggcctcct caataggatc tcggttctgg ccgtacagac ctcggccgac aattatgata 660
tccgttccgg tagacatgac atcctcaaca gttcggtact gctgtccgag agcgtctccc 720
ttgtcgtcaa gacccacccc gggggtcaga ataagccagt cctcagagtc gcccttaggt 780
cggttctggg caatgaagcc aaccacaaac tcggggtcgg atcgggcaag ctcaatggtc 840
tgcttggagt actcgccagt ggccagagag cccttgcaag acagctcggc cagcatgagc 900
agacctctgg ccagcttctc gttgggagag gggactagga actccttgta ctgggagttc 960
tcgtagtcag agacgtcctc cttcttctgt tcagagacag tttcctcggc accagctcgc 1020
aggccagcaa tgattccggt tccgggtaca ccgtgggcgt tggtgatatc ggaccactcg 1080
gcgattcggt gacaccggta ctggtgcttg acagtgttgc caatatctgc gaactttctg 1140
tcctcgaaca ggaagaaacc gtgcttaaga gcaagttcct tgagggggag cacagtgccg 1200
gcgtaggtga agtcgtcaat gatgtcgata tgggttttga tcatgcacac ataaggtccg 1260
accttatcgg caagctcaat gagctccttg gtggtggtaa catccagaga agcacacagg 1320
ttggttttct tggctgccac gagcttgagc actcgagcgg caaaggcgga cttgtggacg 1380
ttagctcgag cttcgtagga gggcattttg gtggtgaaga ggagactgaa ataaatttag 1440
tctgcagaac tttttatcgg aaccttatct ggggcagtga agtatatgtt atggtaatag 1500
ttacgagtta gttgaactta tagatagact ggactatacg gctatcggtc caaattagaa 1560
agaacgtcaa tggctctctg ggcgtcgcct ttgccgacaa aaatgtgatc atgatgaaag 1620
ccagcaatga cgttgcagct gatattgttg tcggccaacc gcgccgaaaa cgcagctgtc 1680
agacccacag cctccaacga agaatgtatc gtcaaagtga tccaagcaca ctcatagttg 1740
gagtcgtact ccaaaggcgg caatgacgag tcagacagat actcgtcgac tcatcgatat 1800
aacttcgtat aatgtatgct atacgaagtt atcctaggta tagatcttgc acttcttatt 1860
ttcttcacgc gtttgcagct caacattcta ggacgacgaa actacgtcaa cagtgttgtc 1920
gctctggcgc agcagggccg agagggt 1947
<210> 72
<211> 280
<212> DNA
<213> Artificial sequence
<220>
<223> PEX3 knock-out site, comprising a single LoxP (URA3 removed)
<400> 72
acagaccggg ttttggctct cgagagctcc acctcgtcct cagctaccgc acaaaccgtg 60
cccaccatga caagtggcgc cacagaggag ggcgagaagt taattaactt tggccggcct 120
ttacctgcag gataacttcg tataatgtat gctatacgaa gttatcctag gtatagatct 180
tgcacttctt attttcttca cgcgtttgca gctcaacatt ctaggacgac gaaactacgt 240
caacagtgtt gtcgctctgg cgcagcaggg ccgagagggt 280
<210> 73
<211> 5164
<212> DNA
<213> Artificial sequence
<220>
<223> pYRH146-Pox4KO plasmid
<400> 73
cgcgccagct gcattaatga atcggccaac gcgcggggag aggcggtttg cgtattgggc 60
gctcttccgc ttcctcgctc actgactcgc tgcgctcggt cgttcggctg cggcgagcgg 120
tatcagctca ctcaaaggcg gtaatacggt tatccacaga atcaggggat aacgcaggaa 180
agaacatgtg agcaaaaggc cagcaaaagg ccaggaaccg taaaaaggcc gcgttgctgg 240
cgtttttcca taggctccgc ccccctgacg agcatcacaa aaatcgacgc tcaagtcaga 300
ggtggcgaaa cccgacagga ctataaagat accaggcgtt tccccctgga agctccctcg 360
tgcgctctcc tgttccgacc ctgccgctta ccggatacct gtccgccttt ctcccttcgg 420
gaagcgtggc gctttctcat agctcacgct gtaggtatct cagttcggtg taggtcgttc 480
gctccaagct gggctgtgtg cacgaacccc ccgttcagcc cgaccgctgc gccttatccg 540
gtaactatcg tcttgagtcc aacccggtaa gacacgactt atcgccactg gcagcagcca 600
ctggtaacag gattagcaga gcgaggtatg taggcggtgc tacagagttc ttgaagtggt 660
ggcctaacta cggctacact agaagaacag tatttggtat ctgcgctctg ctgaagccag 720
ttaccttcgg aaaaagagtt ggtagctctt gatccggcaa acaaaccacc gctggtagcg 780
gtggtttttt tgtttgcaag cagcagatta cgcgcagaaa aaaaggatct caagaagatc 840
ctttgatctt ttctacgggg tctgacgctc agtggaacga aaactcacgt taagggattt 900
tggtcatgag attatcaaaa aggatcttca cctagatcct tttaaattaa aaatgaagtt 960
ttaaatcaat ctaaagtata tatgagtaaa cttggtctga cagttaccaa tgcttaatca 1020
gtgaggcacc tatctcagcg atctgtctat ttcgttcatc catagttgcc tgactccccg 1080
tcgtgtagat aactacgata cgggagggct taccatctgg ccccagtgct gcaatgatac 1140
cgcgagaccc acgctcaccg gctccagatt tatcagcaat aaaccagcca gccggaaggg 1200
ccgagcgcag aagtggtcct gcaactttat ccgcctccat ccagtctatt aattgttgcc 1260
gggaagctag agtaagtagt tcgccagtta atagtttgcg caacgttgtt gccattgcta 1320
caggcatcgt ggtgtcacgc tcgtcgtttg gtatggcttc attcagctcc ggttcccaac 1380
gatcaaggcg agttacatga tcccccatgt tgtgcaaaaa agcggttagc tccttcggtc 1440
ctccgatcgt tgtcagaagt aagttggccg cagtgttatc actcatggtt atggcagcac 1500
tgcataattc tcttactgtc atgccatccg taagatgctt ttctgtgact ggtgagtact 1560
caaccaagtc attctgagaa tagtgtatgc ggcgaccgag ttgctcttgc ccggcgtcaa 1620
tacgggataa taccgcgcca catagcagaa ctttaaaagt gctcatcatt ggaaaacgtt 1680
cttcggggcg aaaactctca aggatcttac cgctgttgag atccagttcg atgtaaccca 1740
ctcgtgcacc caactgatct tcagcatctt ttactttcac cagcgtttct gggtgagcaa 1800
aaacaggaag gcaaaatgcc gcaaaaaagg gaataagggc gacacggaaa tgttgaatac 1860
tcatactctt cctttttcaa tattattgaa gcatttatca gggttattgt ctcatgagcg 1920
gatacatatt tgaatgtatt tagaaaaata aacaaatagg ggttccgcgc acatttcccc 1980
gaaaagtgcc acctgatgcg gtgtgaaata ccgcacagat gcgtaaggag aaaataccgc 2040
atcaggaaat tgtaagcgtt aatattttgt taaaattcgc gttaaatttt tgttaaatca 2100
gctcattttt taaccaatag gccgaaatcg gcaaaatccc ttataaatca aaagaataga 2160
ccgagatagg gttgagtgtt gttccagttt ggaacaagag tccactatta aagaacgtgg 2220
actccaacgt caaagggcga aaaaccgtct atcagggcga tggcccacta cgtgaaccat 2280
caccctaatc aagttttttg gggtcgaggt gccgtaaagc actaaatcgg aaccctaaag 2340
ggagcccccg atttagagct tgacggggaa agccggcgaa cgtggcgaga aaggaaggga 2400
agaaagcgaa aggagcgggc gctagggcgc tggcaagtgt agcggtcacg ctgcgcgtaa 2460
ccaccacacc cgccgcgctt aatgcgccgc tacagggcgc gtccattcgc cattcaggct 2520
gcgcaactgt tgggaagggc gatcggtgcg ggcctcttcg ctattacgcc agctggcgaa 2580
agggggatgt gctgcaaggc gattaagttg ggtaacgcca gggttttccc agtcacgacg 2640
ttgtaaaacg acggccagtg aattgtaata cgactcacta tagggcgaat tgggcccgac 2700
gtcgcgatcg cgagtatctg tctgactcgt cattgccgcc tttggagtac gactccaact 2760
atgagtgtgc ttggatcact ttgacgatac attcttcgtt ggaggctgtg ggtctgacag 2820
ctgcgttttc ggcgcggttg gccgacaaca atatcagctg caacgtcatt gctggctttc 2880
atcatgatca catttttgtc ggcaaaggcg acgcccagag agccattgac gttctttcta 2940
atttggaccg atagccgtat agtccagtct atctataagt tcaactaact cgtaactatt 3000
accataacat atacttcact gccccagata aggttccgat aaaaagttct gcagactaaa 3060
tttatttcag tctcctcttc accaccaaaa tgccctccta cgaagctcga gctaacgtcc 3120
acaagtccgc ctttgccgct cgagtgctca agctcgtggc agccaagaaa accaacctgt 3180
gtgcttctct ggatgttacc accaccaagg agctcattga gcttgccgat aaggtcggac 3240
cttatgtgtg catgatcaaa acccatatcg acatcattga cgacttcacc tacgccggca 3300
ctgtgctccc cctcaaggaa cttgctctta agcacggttt cttcctgttc gaggacagaa 3360
agttcgcaga tattggcaac actgtcaagc accagtaccg gtgtcaccga atcgccgagt 3420
ggtccgatat caccaacgcc cacggtgtac ccggaaccgg aatcattgct ggcctgcgag 3480
ctggtgccga ggaaactgtc tctgaacaga agaaggagga cgtctctgac tacgagaact 3540
cccagtacaa ggagttccta gtcccctctc ccaacgagaa gctggccaga ggtctgctca 3600
tgctggccga gctgtcttgc aagggctctc tggccactgg cgagtactcc aagcagacca 3660
ttgagcttgc ccgatccgac cccgagtttg tggttggctt cattgcccag aaccgaccta 3720
agggcgactc tgaggactgg cttattctga cccccggggt gggtcttgac gacaagggag 3780
acgctctcgg acagcagtac cgaactgttg aggatgtcat gtctaccgga acggatatca 3840
taattgtcgg ccgaggtctg tacggccaga accgagatcc tattgaggag gccaagcgat 3900
accagaaggc tggctgggag gcttaccaga agattaactg ttagaggtta gactatggat 3960
atgtaattta actgtgtata tagagagcgt gcaagtatgg agcgcttgtt cagcttgtat 4020
gatggtcaga cgacctgtct gatcgagtat gtatgatact gcacaacctg tgtatccgca 4080
tgatctgtcc aatggggcat gttgttgtgt ttctcgatac ggagatgctg ggtacagtgc 4140
taatacgttg aactacttat acttatatga ggctcgaaga aagctgactt gtgtatgacg 4200
catgcttgag cgattgggag agttggttgt gtacaattat tttaatacct cttctgattg 4260
ttttctattg ccttccattt ctatctttac ctgccatctc acgtcgtgtg taccatcccc 4320
acatacggaa ccagtaggtc ttttaggctc tgaacgtgca aatgagtttg gtggggtagg 4380
cagagatcgc atagagacgg gtagaatgag cagttaaaag ctgtgttgag tggtaaaaat 4440
ttacaataag tgttcctcaa ggcatcaagg agacgaaata agccattatg gacacgaacc 4500
aacagtccca ccacgttcta aacacattcc tccactgcca ctcccaaaca ccacgtccca 4560
cataaacttc taccccacat tttgacaagc ctattcgttt aataatcacc ccgaggagac 4620
agaaagccta acagctggag ccactatata gttgcagtgg ttaattaagg agttgtgtgt 4680
aacttgtaca ggtacaccta catactgtac tgtaggtcca aagataggta cactgtggca 4740
ataattatgc gagtacttgt accgtcatcg tagctgctgt aaagagatca gacacaggca 4800
cttttcccca ccatgagatc accactcgtc gtccgagtac ttctatggca cagccacaat 4860
cacatgtact tgtgcatgcc aatgtgtgac atcatcatct agagctatca tcatattccc 4920
gctgcaaatg gtctacgtat tactattaag cagggggggg ggaggaatta tgacgacatt 4980
gtacgtgtac tcgtaccggt acttgtagca cgccgaactg cggtattact gtgcactgta 5040
atttcggacc cctcttatag ccccaagttg gtctatacat ctgaaccggt gcagactcac 5100
tattaaaagt gcggcagcta attttgctga cacagccttg tcgataaaag tagctacttg 5160
tagg 5164
<210> 74
<211> 957
<212> DNA
<213> Artificial sequence
<220>
<223> POX4 knockout site
<400> 74
ttgagcgatt gggagagttg gttgtgtaca attattttaa tacctcttct gattgttttc 60
tattgccttc catttctatc tttacctgcc atctcacgtc gtgtgtacca tccccacata 120
cggaaccagt aggtctttta ggctctgaac gtgcaaatga gtttggtggg gtaggcagag 180
atcgcataga gacgggtaga atgagcagtt aaaagctgtg ttgagtggta aaaatttaca 240
ataagtgttc ctcaaggcat caaggagacg aaataagcca ttatggacac gaaccaacag 300
tcccaccacg ttctaaacac attcctccac tgccactccc aaacaccacg tcccacataa 360
acttctaccc cacattttga caagcctatt cgtttaataa tcaccccgag gagacagaaa 420
gcctaacagc tggagccact atatagttgc agtggttaat taaggagttg tgtgtaactt 480
gtacaggtac acctacatac tgtactgtag gtccaaagat aggtacactg tggcaataat 540
tatgcgagta cttgtaccgt catcgtagct gctgtaaaga gatcagacac aggcactttt 600
ccccaccatg agatcaccac tcgtcgtccg agtacttcta tggcacagcc acaatcacat 660
gtacttgtgc atgccaatgt gtgacatcat catctagagc tatcatcata ttcccgctgc 720
aaatggtcta cgtattacta ttaagcaggg gggggggagg aattatgacg acattgtacg 780
tgtactcgta ccggtacttg tagcacgccg aactgcggta ttactgtgca ctgtaatttc 840
ggacccctct tatagcccca agttggtcta tacatctgaa ccggtgcaga ctcactatta 900
aaagtgcggc agctaatttt gctgacacag ccttgtcgat aaaagtagct acttgta 957
<210> 75
<211> 6853
<212> DNA
<213> Artificial sequence
<220>
<223> pYRH72 plasmid
<400> 75
gcatgcgacg tcgggcccaa ttcgccctat agtgagtcgt attacaattc actggccgtc 60
gttttacaac gtcgtgactg ggaaaaccct ggcgttaccc aacttaatcg ccttgcagca 120
catccccctt tcgccagctg gcgtaatagc gaagaggccc gcaccgatcg cccttcccaa 180
cagttgcgca gcctgaatgg cgaatggacg cgccctgtag cggcgcatta agcgcggcgg 240
gtgtggtggt tacgcgcagc gtgaccgcta cacttgccag cgccctagcg cccgctcctt 300
tcgctttctt cccttccttt ctcgccacgt tcgccggctt tccccgtcaa gctctaaatc 360
gggggctccc tttagggttc cgatttagtg ctttacggca cctcgacccc aaaaaacttg 420
attagggtga tggttcacgt agtgggccat cgccctgata gacggttttt cgccctttga 480
cgttggagtc cacgttcttt aatagtggac tcttgttcca aactggaaca acactcaacc 540
ctatctcggt ctattctttt gatttataag ggattttgcc gatttcggcc tattggttaa 600
aaaatgagct gatttaacaa aaatttaacg cgaattttaa caaaatatta acgcttacaa 660
tttcctgatg cggtattttc tccttacgca tctgtgcggt atttcacacc gcatcaggtg 720
gcacttttcg gggaaatgtg cgcggaaccc ctatttgttt atttttctaa atacattcaa 780
atatgtatcc gctcatgaga caataaccct gataaatgct tcaataatat tgaaaaagga 840
agagtatgag tattcaacat ttccgtgtcg cccttattcc cttttttgcg gcattttgcc 900
ttcctgtttt tgctcaccca gaaacgctgg tgaaagtaaa agatgctgaa gatcagttgg 960
gtgcacgagt gggttacatc gaactggatc tcaacagcgg taagatcctt gagagttttc 1020
gccccgaaga acgttttcca atgatgagca cttttaaagt tctgctatgt ggcgcggtat 1080
tatcccgtat tgacgccggg caagagcaac tcggtcgccg catacactat tctcagaatg 1140
acttggttga gtactcacca gtcacagaaa agcatcttac ggatggcatg acagtaagag 1200
aattatgcag tgctgccata accatgagtg ataacactgc ggccaactta cttctgacaa 1260
cgatcggagg accgaaggag ctaaccgctt ttttgcacaa catgggggat catgtaactc 1320
gccttgatcg ttgggaaccg gagctgaatg aagccatacc aaacgacgag cgtgacacca 1380
cgatgcctgt agcaatggca acaacgttgc gcaaactatt aactggcgaa ctacttactc 1440
tagcttcccg gcaacaatta atagactgga tggaggcgga taaagttgca ggaccacttc 1500
tgcgctcggc ccttccggct ggctggttta ttgctgataa atctggagcc ggtgagcgtg 1560
ggtctcgcgg tatcattgca gcactggggc cagatggtaa gccctcccgt atcgtagtta 1620
tctacacgac ggggagtcag gcaactatgg atgaacgaaa tagacagatc gctgagatag 1680
gtgcctcact gattaagcat tggtaactgt cagaccaagt ttactcatat atactttaga 1740
ttgatttaaa acttcatttt taatttaaaa ggatctaggt gaagatcctt tttgataatc 1800
tcatgaccaa aatcccttaa cgtgagtttt cgttccactg agcgtcagac cccgtagaaa 1860
agatcaaagg atcttcttga gatccttttt ttctgcgcgt aatctgctgc ttgcaaacaa 1920
aaaaaccacc gctaccagcg gtggtttgtt tgccggatca agagctacca actctttttc 1980
cgaaggtaac tggcttcagc agagcgcaga taccaaatac tgttcttcta gtgtagccgt 2040
agttaggcca ccacttcaag aactctgtag caccgcctac atacctcgct ctgctaatcc 2100
tgttaccagt ggctgctgcc agtggcgata agtcgtgtct taccgggttg gactcaagac 2160
gatagttacc ggataaggcg cagcggtcgg gctgaacggg gggttcgtgc acacagccca 2220
gcttggagcg aacgacctac accgaactga gatacctaca gcgtgagcta tgagaaagcg 2280
ccacgcttcc cgaagggaga aaggcggaca ggtatccggt aagcggcagg gtcggaacag 2340
gagagcgcac gagggagctt ccagggggaa acgcctggta tctttatagt cctgtcgggt 2400
ttcgccacct ctgacttgag cgtcgatttt tgtgatgctc gtcagggggg cggagcctat 2460
ggaaaaacgc cagcaacgcg gcctttttac ggttcctggc cttttgctgg ccttttgctc 2520
acatgttctt tcctgcgtta tcccctgatt ctgtggataa ccgtattacc gcctttgagt 2580
gagctgatac cgctcgccgc agccgaacga ccgagcgcag cgagtcagtg agcgaggaag 2640
cggaagagcg cccaatacgc aaaccgcctc tccccgcgcg ttggccgatt cattaatgca 2700
gctggcgcgc caccatcatt gagaacctca gtaagctcaa gtgggataag accgccatcc 2760
acatccgtgc caccaaacag gcccatgctg gtgccattgt gcgtcatcca gacccgtcct 2820
tctatgaagg taagaccgtt gtcaagcact ggattgacaa caaggcccgt ctttgagagc 2880
agcctgactc ggcctttgta gagcaaggcc attattgatt tatagacatt ataccgggaa 2940
atactaaaca caagaacctt cggcgtgatt atctgcactt gtaccacgct gcagtaagtt 3000
agtaatcgga attcaactgc cattagacat tatttacctg cagacccaat tcgtccagca 3060
acggaggaat gaacgttggg ggttcgggtc taaagatgcg tcctgtaagt gggggcgttg 3120
gtctacgagt agggtcctgc tttagtgtgg aaaatttatc tctttatgtg tttccattgg 3180
ctggtgcgtt ttgttgtttt cctttattca gccaatcaga atgcgtgaga atacctgtta 3240
tgttttcctc acaatatttt tttccaccgc cattttggag agccgccgga aaccagagtg 3300
ccgaaaggct atcttttggg ctggaattat atatcctaag ccagacacaa tcctgactca 3360
ctaaccttgg agtagataga gaaagagcaa gaagctctgt gagtcagaga ggaagaggga 3420
ggcacataat gtgggagtta taggggtatc gtacgttgtg tggaagcttg tgagcggata 3480
acaatttcac acaggaaaca gctatgacca tgattacgcc aagctcgaaa ttaaccctca 3540
ctaaagggaa caaaagctgg agctccaccg cggacacaat atctggtcaa atttcagttt 3600
cgttacattt aaacggtagg ttagtgcttg gtatatgagt tgtaggcatg acaatttgga 3660
aaggggtgga ctttgggaat attgtgggat ttcaatacct tagtttgtac agggtaattg 3720
ttacaaatga tacaaagaac tgtatttctt ttcatttgtt ttaattggtt gtatatcaag 3780
tccgttagac gagctcagtg ccttggcttt tggcactgta tttcattttt agaggtacac 3840
tacattcagt gaggtatggt aaggttgagg gcataatgaa ggcaccttgt actgacagtc 3900
acagacctct caccgagaat tttatgagat atactcgggt tcattttagg ctcatcgata 3960
cgctctcatc aagaatactt cttgagaacc gtggagaccg gggttcgatt ccccgtatcg 4020
gagtgtttat tttttgctca accataccct ggggtgtgtt ctgtggagca ttctcacttt 4080
tggtaaacga cattgcttca agtgcagcgg aatcaaaaag tataaagtgg gcagcgagta 4140
tacctgtaca gactgtaggc gataactcaa tccaattacc ccccacaaca tgactggcca 4200
aactgatctc aagactttat tgaaatcagc aacaccgatt ctcaatgaag gcacatactt 4260
cttctgcaac attcacttga cgcctaaagt tggtgagaaa tggaccgaca agacatattc 4320
tgctatccac ggactgttgc ctgtgtcggt ggctacaata cgtgagtcag aagggctgac 4380
ggtggtggtt cccaaggaaa aggtcgacga gtatctgtct gactcgtcat tgccgccttt 4440
ggagtacgac tccaactatg agtgtgcttg gatcactttg acgatacatt cttcgttgga 4500
ggctgtgggt ctgacagctg cgttttcggc gcggttggcc gacaacaata tcagctgcaa 4560
cgtcattgct ggctttcatc atgatcacat ttttgtcggc aaaggcgacg cccagagagc 4620
cattgacgtt ctttctaatt tggaccgata gccgtatagt ccagtctatc tataagttca 4680
actaactcgt aactattacc ataacatata cttcactgcc ccagataagg ttccgataaa 4740
aagttctgca gactaaattt atttcagtct cctcttcacc accaaaatgc cctcctacga 4800
agctcgagct aacgtccaca agtccgcctt tgccgctcga gtgctcaagc tcgtggcagc 4860
caagaaaacc aacctgtgtg cttctctgga tgttaccacc accaaggagc tcattgagct 4920
tgccgataag gtcggacctt atgtgtgcat gatcaaaacc catatcgaca tcattgacga 4980
cttcacctac gccggcactg tgctccccct caaggaactt gctcttaagc acggtttctt 5040
cctgttcgag gacagaaagt tcgcagatat tggcaacact gtcaagcacc agtaccggtg 5100
tcaccgaatc gccgagtggt ccgatatcac caacgcccac ggtgtacccg gaaccggaat 5160
cattgctggc ctgcgagctg gtgccgagga aactgtctct gaacagaaga aggaggacgt 5220
ctctgactac gagaactccc agtacaagga gttcctagtc ccctctccca acgagaagct 5280
ggccagaggt ctgctcatgc tggccgagct gtcttgcaag ggctctctgg ccactggcga 5340
gtactccaag cagaccattg agcttgcccg atccgacccc gagtttgtgg ttggcttcat 5400
tgcccagaac cgacctaagg gcgactctga ggactggctt attctgaccc ccggggtggg 5460
tcttgacgac aagggagacg ctctcggaca gcagtaccga actgttgagg atgtcatgtc 5520
taccggaacg gatatcataa ttgtcggccg aggtctgtac ggccagaacc gagatcctat 5580
tgaggaggcc aagcgatacc agaaggctgg ctgggaggct taccagaaga ttaactgtta 5640
gaggttagac tatggatatg taatttaact gtgtatatag agagcgtgca agtatggagc 5700
gcttgttcag cttgtatgat ggtcagacga cctgtctgat cgagtatgta tgatactgca 5760
caacctgtgt atccgcatga tctgtccaat ggggcatgtt gttgtgtttc tcgatacgga 5820
gatgctgggt acagtgctaa tacgttgaac tacttatact tatatgaggc tcgaagaaag 5880
ctgacttgtg tatgacttat tctcaactac atccccagtc acaataccac cactgcacta 5940
ccactacacc aaaaccatga tcaaaccacc catggacttc ctggaggcag aagaacttgt 6000
tatggaaaag ctcaagagag agaattcaag atactatcaa gacatgtgtc gcaacttaat 6060
taatgtatct atacctaatc gacatggact gcaccgcatt cgttatctta tcgtagtcct 6120
gcttactttg atccgtatca cattcgtgta tcgcgataag aagtaaacat tgctgcaagt 6180
ataaatcagg tctcaaatct cttcttgaag atagttttaa gctcaccgtg aactgaacgt 6240
gaacactacg agtacgattt tcaatattat taccgtcgat ataacaacca tgcaatttct 6300
atatacatat atacattccc tcattcgtct gtgctgtttg gttcactgca gcaaatcaaa 6360
cagcttttcg tactttctgt gctgcgaatg tttgttacta tccgcgtgca ttctttgcag 6420
cttttgagat cccgtatagt ccttccacac gttggcactc gcctttttcg tcgtcgattg 6480
cagcttcata atcaacttgt cattgtccag cttggccccc tggacctcca tcatcacaat 6540
cttgtctctg tggtgtttga tagacgagtc gttcacttca caggtagcca caacgccgat 6600
gcaagcagct tttgtgcgct gatcatccac aatacgtgct ggtaacatgg gatttgccat 6660
gactccgtta cgagatgtgt gtagcgatcc cttggggaag taggacggtc cagccacggg 6720
tttcgtggat attgagtccc acggaccgaa tttttgtgtt gcagcgtact ctttggcaag 6780
ctggatgaga tactcgccgt ccagagcgct gcgcttgacg cctcggtcgt tgcaccactt 6840
gatgaaggcg taa 6853
<210> 76
<211> 6512
<212> DNA
<213> Artificial sequence
<220>
<223> p12_3-B-Pex3del1 plasmid
<400> 76
gtacgataac ttcgtatagc atacattata cgaagttatc gcgtcgacga gtatctgtct 60
gactcgtcat tgccgccttt ggagtacgac tccaactatg agtgtgcttg gatcactttg 120
acgatacatt cttcgttgga ggctgtgggt ctgacagctg cgttttcggc gcggttggcc 180
gacaacaata tcagctgcaa cgtcattgct ggctttcatc atgatcacat ttttgtcggc 240
aaaggcgacg cccagagagc cattgacgtt ctttctaatt tggaccgata gccgtatagt 300
ccagtctatc tataagttca actaactcgt aactattacc ataacatata cttcactgcc 360
ccagataagg ttccgataaa aagttctgca gactaaattt atttcagtct cctcttcacc 420
accaaaatgc cctcctacga agctcgagct aacgtccaca agtccgcctt tgccgctcga 480
gtgctcaagc tcgtggcagc caagaaaacc aacctgtgtg cttctctgga tgttaccacc 540
accaaggagc tcattgagct tgccgataag gtcggacctt atgtgtgcat gatcaaaacc 600
catatcgaca tcattgacga cttcacctac gccggcactg tgctccccct caaggaactt 660
gctcttaagc acggtttctt cctgttcgag gacagaaagt tcgcagatat tggcaacact 720
gtcaagcacc agtaccggtg tcaccgaatc gccgagtggt ccgatatcac caacgcccac 780
ggtgtacccg gaaccggaat cattgctggc ctgcgagctg gtgccgagga aactgtctct 840
gaacagaaga aggaggacgt ctctgactac gagaactccc agtacaagga gttcctagtc 900
ccctctccca acgagaagct ggccagaggt ctgctcatgc tggccgagct gtcttgcaag 960
ggctctctgg ccactggcga gtactccaag cagaccattg agcttgcccg atccgacccc 1020
gagtttgtgg ttggcttcat tgcccagaac cgacctaagg gcgactctga ggactggctt 1080
attctgaccc ccggggtggg tcttgacgac aagggagacg ctctcggaca gcagtaccga 1140
actgttgagg atgtcatgtc taccggaacg gatatcataa ttgtcggccg aggtctgtac 1200
ggccagaacc gagatcctat tgaggaggcc aagcgatacc agaaggctgg ctgggaggct 1260
taccagaaga ttaactgtta gaggttagac tatggatatg taatttaact gtgtatatag 1320
agagcgtgca agtatggagc gcttgttcag cttgtatgat ggtcagacga cctgtctgat 1380
cgagtatgta tgatactgca caacctgtgt atccgcatga tctgtccaat ggggcatgtt 1440
gttgtgtttc tcgatacgga gatgctgggt acagtgctaa tacgttgaac tacttatact 1500
tatatgaggc tcgaagaaag ctgacttgtg tatgacttat tctcaactac atccccagtc 1560
acaataccac cactgcacta ccactacacc aaaaccatga tcaaaccacc catggacttc 1620
ctggaggcag aagaacttgt tatggaaaag ctcaagagag agatcataac ttcgtatagc 1680
atacattata cgaagttatc ctgcaggtaa aggaattcag gagagaccgg gttggcggcg 1740
tatttgtgtc ccaaaaaaca gccccaattg ccccaattga ccccaaattg acccagtagc 1800
gggcccaacc ccggcgagag cccccttcac cccacatatc aaacctcccc cggttcccac 1860
acttgccgtt aagggcgtag ggtactgcag tctggaatct acgcttgttc agactttgta 1920
ctagtttctt tgtctggcca tccgggtaac ccatgccgga cgcaaaatag actactgaaa 1980
atttttttgc tttgtggttg ggactttagc caagggtata aaagaccacc gtccccgaat 2040
tacctttcct cttcttttct ctctctcctt gtcaactcac acccgaaatc gttaagcatt 2100
tccttctgag tataagaatc attcaccatg gacttcctgg aggcagaaga acttgttatg 2160
gaaaagctca agagagagaa gccaagatac tatcaagaca tgtgtcgcaa cttaattaag 2220
atgacgacat ttgcgagctg gacgaggaat agatggagcg tgtgttctga gtcgatgttt 2280
tctatggagt tgtgagtgtt agtagacatg atgggtttat atatgatgaa tgaatagatg 2340
tgattttgat ttgcacgatg gaattgagaa ctttgtaaac gtacatggga atgtatgaat 2400
gtgggggttt tgtgactgga taactgacgg tcagtggacg ccgttgttca aatatccaag 2460
agatgcgaga aactttgggt caagtgaaca tgtcctctct gttcaagtaa accatcaact 2520
atgggtagta tatttagtaa ggacaagagt tgagattctt tggagtccta gaaacgtatt 2580
ttcgcgttcc aagatcaaat tagtagagta atacgggcac gggaatccat tcatagtctc 2640
aattttccca taggtgtgct acaaggtgtt gagatgtggt acagtaccac catgattcga 2700
ggtaaagagc ccagaagtca ttgatgaggt caagaaatac acagatctac agctcaatac 2760
aatgaatatc ttctttcata ttcttcaggt gacaccaagg gtgtctattt tccccagaaa 2820
tgcgtgaaaa ggcgcgtgtg tagcgtggag tatgggttcg gttggcgtat ccttcatata 2880
tcgacgaaat agtagggcaa gagatgacaa aaagtatcta tatgtagaca gcgtagaata 2940
tggatttgat tggtataaat tcatttattg cgtgtctcac aaatactctc gataagttgg 3000
ggttaaactg gagatggaac aatgtcgata tctcgacgca tgcgacgtcg ggcccaattc 3060
gccctatagt gagtcgtatt acaattcact ggccgtcgtt ttacaacgtc gtgactggga 3120
aaaccctggc gttacccaac ttaatcgcct tgcagcacat ccccctttcg ccagctggcg 3180
taatagcgaa gaggcccgca ccgatcgccc ttcccaacag ttgcgcagcc tgaatggcga 3240
atggacgcgc cctgtagcgg cgcattaagc gcggcgggtg tggtggttac gcgcagcgtg 3300
accgctacac ttgccagcgc cctagcgccc gctcctttcg ctttcttccc ttcctttctc 3360
gccacgttcg ccggctttcc ccgtcaagct ctaaatcggg ggctcccttt agggttccga 3420
tttagtgctt tacggcacct cgaccccaaa aaacttgatt agggtgatgg ttcacgtagt 3480
gggccatcgc cctgatagac ggtttttcgc cctttgacgt tggagtccac gttctttaat 3540
agtggactct tgttccaaac tggaacaaca ctcaacccta tctcggtcta ttcttttgat 3600
ttataaggga ttttgccgat ttcggcctat tggttaaaaa atgagctgat ttaacaaaaa 3660
tttaacgcga attttaacaa aatattaacg cttacaattt cctgatgcgg tattttctcc 3720
ttacgcatct gtgcggtatt tcacaccgca tcaggtggca cttttcgggg aaatgtgcgc 3780
ggaaccccta tttgtttatt tttctaaata cattcaaata tgtatccgct catgagacaa 3840
taaccctgat aaatgcttca ataatattga aaaaggaaga gtatgagtat tcaacatttc 3900
cgtgtcgccc ttattccctt ttttgcggca ttttgccttc ctgtttttgc tcacccagaa 3960
acgctggtga aagtaaaaga tgctgaagat cagttgggtg cacgagtggg ttacatcgaa 4020
ctggatctca acagcggtaa gatccttgag agttttcgcc ccgaagaacg ttttccaatg 4080
atgagcactt ttaaagttct gctatgtggc gcggtattat cccgtattga cgccgggcaa 4140
gagcaactcg gtcgccgcat acactattct cagaatgact tggttgagta ctcaccagtc 4200
acagaaaagc atcttacgga tggcatgaca gtaagagaat tatgcagtgc tgccataacc 4260
atgagtgata acactgcggc caacttactt ctgacaacga tcggaggacc gaaggagcta 4320
accgcttttt tgcacaacat gggggatcat gtaactcgcc ttgatcgttg ggaaccggag 4380
ctgaatgaag ccataccaaa cgacgagcgt gacaccacga tgcctgtagc aatggcaaca 4440
acgttgcgca aactattaac tggcgaacta cttactctag cttcccggca acaattaata 4500
gactggatgg aggcggataa agttgcagga ccacttctgc gctcggccct tccggctggc 4560
tggtttattg ctgataaatc tggagccggt gagcgtgggt ctcgcggtat cattgcagca 4620
ctggggccag atggtaagcc ctcccgtatc gtagttatct acacgacggg gagtcaggca 4680
actatggatg aacgaaatag acagatcgct gagataggtg cctcactgat taagcattgg 4740
taactgtcag accaagttta ctcatatata ctttagattg atttaaaact tcatttttaa 4800
tttaaaagga tctaggtgaa gatccttttt gataatctca tgaccaaaat cccttaacgt 4860
gagttttcgt tccactgagc gtcagacccc gtagaaaaga tcaaaggatc ttcttgagat 4920
cctttttttc tgcgcgtaat ctgctgcttg caaacaaaaa aaccaccgct accagcggtg 4980
gtttgtttgc cggatcaaga gctaccaact ctttttccga aggtaactgg cttcagcaga 5040
gcgcagatac caaatactgt tcttctagtg tagccgtagt taggccacca cttcaagaac 5100
tctgtagcac cgcctacata cctcgctctg ctaatcctgt taccagtggc tgctgccagt 5160
ggcgataagt cgtgtcttac cgggttggac tcaagacgat agttaccgga taaggcgcag 5220
cggtcgggct gaacgggggg ttcgtgcaca cagcccagct tggagcgaac gacctacacc 5280
gaactgagat acctacagcg tgagctatga gaaagcgcca cgcttcccga agggagaaag 5340
gcggacaggt atccggtaag cggcagggtc ggaacaggag agcgcacgag ggagcttcca 5400
gggggaaacg cctggtatct ttatagtcct gtcgggtttc gccacctctg acttgagcgt 5460
cgatttttgt gatgctcgtc aggggggcgg agcctatgga aaaacgccag caacgcggcc 5520
tttttacggt tcctggcctt ttgctggcct tttgctcaca tgttctttcc tgcgttatcc 5580
cctgattctg tggataaccg tattaccgcc tttgagtgag ctgataccgc tcgccgcagc 5640
cgaacgaccg agcgcagcga gtcagtgagc gaggaagcgg aagagcgccc aatacgcaaa 5700
ccgcctctcc ccgcgcgttg gccgattcat taatgcagct ggcgcgccac caatcacaat 5760
tctgaaaagc acatcttgat ctcctcattg cggggagtcc aacggtggtc ttattccccc 5820
gaatttcccg ctcaatctcg ttccagaccg acccggacac agtgcttaac gccgttccga 5880
aactctaccg cagatatgct ccaacggact gggctgcata gatgtgatcc tcggcttgga 5940
gaaatggata aaagccggcc aaaaaaaaag cggaaaaaag cggaaaaaaa gagaaaaaaa 6000
atcgcaaaat ttgaaaaata gggggaaaag acgcaaaaac gcaaggaggg gggagtatat 6060
gacactgata agcaagctca caacggttcc tcttattttt ttcctcatct tctgcctagg 6120
ttcccaaaat cccagatgct tctctccagt gccaaaagta agtaccccac aggttttcgg 6180
ccgaaaattc cacgtgcagc aacgtcgtgt ggggtgttaa aatgtggggg gggggaacca 6240
ggacaagagg ctcttgtggg agccgaatga gagcacaaag cgggcgggtg tgataagggc 6300
atttttgccc attttccctt ctcctgtctc tccgacggtg atggcgttgt gcgtcctcta 6360
tttcttttta tttctttttg ttttatttct ctgactaccg atttggtttg atttcctcaa 6420
ccccacacaa ataagctcgg gccgaggaat atatatatac acggacacag tcgccctgtg 6480
gacaacacgt cactacctct acgatacaca cc 6512
<210> 77
<211> 6906
<212> DNA
<213> Artificial sequence
<220>
<223> p70_ Pox2: Leu2 plasmid
<400> 77
cgatagttgg agcaagggag aaatgtttgt acgggcgtcg ttgcttgtgt gatttttgag 60
gacccatccc tttggtatat aagtatactc tggggttaag gttgcccgtg tagtctaggt 120
tatagttttc atgtgaaata ccgagagccg agggagaata aacgggggta tttggacttg 180
tttttttcgc ggaaaagcgt cgaatcaacc ctgcgggcct tgcaccatgt ccacgacgtg 240
tttctcgccc caattcgccc cttgcacgtc aaaattaggc ctccatctag acccctccat 300
aacatgtgac tgtggggaaa agtataaggg aaaccatgca accatagacg acgtgaaaga 360
cggggaggaa ccaatggagg ccaaagaaat ggggtagcaa cagtccagga gacagacaag 420
gagacaagga gagggcgccc gaaagatcgg aaaaacaaac atgtccaatt ggggcagtga 480
cggaaacgac acggacactt cagtacaatg gaccgaccat ctccaagcca gggttattcc 540
ggtatcacct tggccgtaac ctcccgctgg tacctgatat tgtacacgtt cacattcaat 600
atactttcag ctacaataag agaggctgtt tgtcgggcat gtgtgtccgt cgtatggggt 660
gatgtccgag ggcgaaattc gctacaagct taactctggc gcttgtccag tatgaataga 720
caagtcaaga ccagtggtgc catgattgac agggaggtac aagacttcga tactcgagca 780
ttactcggac ttgtggcgat tgaacagacg ggcgatcgct tctcccccgt attgccggcg 840
cgccagctgc attaatgaat cggccaacgc gcggggagag gcggtttgcg tattgggcgc 900
tcttccgctt cctcgctcac tgactcgctg cgctcggtcg ttcggctgcg gcgagcggta 960
tcagctcact caaaggcggt aatacggtta tccacagaat caggggataa cgcaggaaag 1020
aacatgtgag caaaaggcca gcaaaaggcc aggaaccgta aaaaggccgc gttgctggcg 1080
tttttccata ggctccgccc ccctgacgag catcacaaaa atcgacgctc aagtcagagg 1140
tggcgaaacc cgacaggact ataaagatac caggcgtttc cccctggaag ctccctcgtg 1200
cgctctcctg ttccgaccct gccgcttacc ggatacctgt ccgcctttct cccttcggga 1260
agcgtggcgc tttctcatag ctcacgctgt aggtatctca gttcggtgta ggtcgttcgc 1320
tccaagctgg gctgtgtgca cgaacccccc gttcagcccg accgctgcgc cttatccggt 1380
aactatcgtc ttgagtccaa cccggtaaga cacgacttat cgccactggc agcagccact 1440
ggtaacagga ttagcagagc gaggtatgta ggcggtgcta cagagttctt gaagtggtgg 1500
cctaactacg gctacactag aagaacagta tttggtatct gcgctctgct gaagccagtt 1560
accttcggaa aaagagttgg tagctcttga tccggcaaac aaaccaccgc tggtagcggt 1620
ggtttttttg tttgcaagca gcagattacg cgcagaaaaa aaggatctca agaagatcct 1680
ttgatctttt ctacggggtc tgacgctcag tggaacgaaa actcacgtta agggattttg 1740
gtcatgagat tatcaaaaag gatcttcacc tagatccttt taaattaaaa atgaagtttt 1800
aaatcaatct aaagtatata tgagtaaact tggtctgaca gttaccaatg cttaatcagt 1860
gaggcaccta tctcagcgat ctgtctattt cgttcatcca tagttgcctg actccccgtc 1920
gtgtagataa ctacgatacg ggagggctta ccatctggcc ccagtgctgc aatgataccg 1980
cgagacccac gctcaccggc tccagattta tcagcaataa accagccagc cggaagggcc 2040
gagcgcagaa gtggtcctgc aactttatcc gcctccatcc agtctattaa ttgttgccgg 2100
gaagctagag taagtagttc gccagttaat agtttgcgca acgttgttgc cattgctaca 2160
ggcatcgtgg tgtcacgctc gtcgtttggt atggcttcat tcagctccgg ttcccaacga 2220
tcaaggcgag ttacatgatc ccccatgttg tgcaaaaaag cggttagctc cttcggtcct 2280
ccgatcgttg tcagaagtaa gttggccgca gtgttatcac tcatggttat ggcagcactg 2340
cataattctc ttactgtcat gccatccgta agatgctttt ctgtgactgg tgagtactca 2400
accaagtcat tctgagaata gtgtatgcgg cgaccgagtt gctcttgccc ggcgtcaata 2460
cgggataata ccgcgccaca tagcagaact ttaaaagtgc tcatcattgg aaaacgttct 2520
tcggggcgaa aactctcaag gatcttaccg ctgttgagat ccagttcgat gtaacccact 2580
cgtgcaccca actgatcttc agcatctttt actttcacca gcgtttctgg gtgagcaaaa 2640
acaggaaggc aaaatgccgc aaaaaaggga ataagggcga cacggaaatg ttgaatactc 2700
atactcttcc tttttcaata ttattgaagc atttatcagg gttattgtct catgagcgga 2760
tacatatttg aatgtattta gaaaaataaa caaatagggg ttccgcgcac atttccccga 2820
aaagtgccac ctgatgcggt gtgaaatacc gcacagatgc gtaaggagaa aataccgcat 2880
caggaaattg taagcgttaa tattttgtta aaattcgcgt taaatttttg ttaaatcagc 2940
tcatttttta accaataggc cgaaatcggc aaaatccctt ataaatcaaa agaatagacc 3000
gagatagggt tgagtgttgt tccagtttgg aacaagagtc cactattaaa gaacgtggac 3060
tccaacgtca aagggcgaaa aaccgtctat cagggcgatg gcccactacg tgaaccatca 3120
ccctaatcaa gttttttggg gtcgaggtgc cgtaaagcac taaatcggaa ccctaaaggg 3180
agcccccgat ttagagcttg acggggaaag ccggcgaacg tggcgagaaa ggaagggaag 3240
aaagcgaaag gagcgggcgc tagggcgctg gcaagtgtag cggtcacgct gcgcgtaacc 3300
accacacccg ccgcgcttaa tgcgccgcta cagggcgcgt ccattcgcca ttcaggctgc 3360
gcaactgttg ggaagggcga tcggtgcggg cctcttcgct attacgccag ctggcgaaag 3420
ggggatgtgc tgcaaggcga ttaagttggg taacgccagg gttttcccag tcacgacgtt 3480
gtaaaacgac ggccagtgaa ttgtaatacg actcactata gggcgaattg ggcccgacgt 3540
cgcatgcgct gatgacactt tggtctgaaa gagatgcatt ttgaatccca aacttgcagt 3600
gcccaagtga catacatctc cgcgttttgg aaaatgttca gaaacagttg attgtgttgg 3660
aatggggaat ggggaatgga aaaatgactc aagtatcaat tccaaaaact tctctggctg 3720
gcagtaccta ctgtccatac tactgcattt tctccagtca ggccactcta tactcgacga 3780
cacagtagta aaacccagat aatttcgaca taaacaagaa aacagaccca ataatattta 3840
tatatagtca gccgtttgtc cagttcagac tgtaatagcc gaaaaaaaat ccaaagtttc 3900
tattctagga aaatatattc caatattttt aattcttaat ctcatttatt ttattctagc 3960
gaaatacatt tcagctactt gagacatgtg atacccacaa atcggattcg gactcggttg 4020
ttcagaagag catatggcat tcgtgctcgc ttgttcacgt attcttcctg ttccatctct 4080
tggccgacaa tcacacaaaa atggggtttt ttttttaatt ctaatgattc attacagcaa 4140
aattgagata tagcagacca cgtattccat aatcaccaag gaagttcttg ggcgtcttaa 4200
ttaagttgcg acacatgtct tgatagtatc ttggcttctc tctcttgagc ttttccataa 4260
caagttcttc tgcctccagg aagtccatgg tgaatgattc ttatactcag aaggaaatgc 4320
ttaacgattt cgggtgtgag ttgacaagga gagagagaaa agaagaggaa aggtaattcg 4380
gggacggtgg tcttttatac ccttggctaa agtcccaacc acaaagcaaa aaaattttca 4440
gtagtctatt ttgcgtccgg catgggttac ccggatggcc agacaaagaa actagtacaa 4500
agtctgaaca agcgtagatt ccagactgca gtaccctacg cccttaacgg caagtgtggg 4560
aaccggggga ggtttgatat gtggggtgaa gggggctctc gccggggttg ggcccgctac 4620
tgggtcaatt tggggtcaat tggggcaatt ggggctgttt tttgggacac aaatacgccg 4680
ccaacccggt ctctcctgaa ttctgcagat gggctgcagg aattccgtcg tcgcctgagt 4740
cgacatcatt tatttaccag ttggccacaa acccttgacg atctcgtatg tcccctccga 4800
catactcccg gccggctggg gtacgttcga tagcgctatc ggcatcgaca aggtttgggt 4860
ccctagccga taccgcacta cctgagtcac aatcttcgga ggtttagtct tccacatagc 4920
acgggcaaaa gtgcgtatat atacaagagc gtttgccagc cacagatttt cactccacac 4980
accacatcac acatacaacc acacacatcc acaatggaac ccgaaactaa gaagaccaag 5040
actgactcca agaagattgt tcttctcggc ggcgacttct gtggccccga ggtgattgcc 5100
gaggccgtca aggtgctcaa gtctgttgct gaggcctccg gcaccgagtt tgtgtttgag 5160
gaccgactca ttggaggagc tgccattgag aaggagggcg agcccatcac cgacgctact 5220
ctcgacatct gccgaaaggc tgactctatt atgctcggtg ctgtcggagg cgctgccaac 5280
accgtatgga ccactcccga cggacgaacc gacgtgcgac ccgagcaggg tctcctcaag 5340
ctgcgaaagg acctgaacct gtacgccaac ctgcgaccct gccagctgct gtcgcccaag 5400
ctcgccgatc tctcccccat ccgaaacgtt gagggcaccg acttcatcat tgtccgagag 5460
ctcgtcggag gtatctactt tggagagcga aaggaggatg acggatctgg cgtcgcttcc 5520
gacaccgaga cctactccgt tcctgaggtt gagcgaattg cccgaatggc cgccttcctg 5580
gcccttcagc acaacccccc tcttcccgtg tggtctcttg acaaggccaa cgtgctggcc 5640
tcctctcgac tttggcgaaa gactgtcact cgagtcctca aggacgaatt cccccagctc 5700
gagctcaacc accagctgat cgactcggcc gccatgatcc tcatcaagca gccctccaag 5760
atgaatggta tcatcatcac caccaacatg tttggcgata tcatctccga cgaggcctcc 5820
gtcatccccg gttctctggg tctgctgccc tccgcctctc tggcttctct gcccgacacc 5880
aacgaggcgt tcggtctgta cgagccctgt cacggatctg cccccgatct cggcaagcag 5940
aaggtcaacc ccattgccac cattctgtct gccgccatga tgctcaagtt ctctcttaac 6000
atgaagcccg ccggtgacgc tgttgaggct gccgtcaagg agtccgtcga ggctggtatc 6060
actaccgccg atatcggagg ctcttcctcc acctccgagg tcggagactt gttgccaaca 6120
aggtcaagga gctgctcaag aaggagtaag tcgtttctac gacgcattga tggaaggagc 6180
aaactgacgc gcctgcgggt tggtctaccg gcagggtccg ctagtgtata agactctata 6240
aaaagggccc tgccctgcta atgaaatgat gatttataat ttaccggtgt agcaaccttg 6300
actagaagaa gcagattggg tgtgtttgta gtggaggaca gtggtacgtt ttggaaacag 6360
tcttcttgaa agtgtcttgt ctacagtata ttcactcata acctcaatag ccaagggtgt 6420
agtcggttta ttaaaggaag ggagttgtgg ctgatgtgga tagatatctt taagctggcg 6480
actgcaccca acgagtgtgg tggtagcttg ttagatctgt atattcggta agatatattt 6540
tgtggggttt tagtggtgtt taaacggtag gttagtgctt ggtatatgag ttgtaggcat 6600
gacaatttgg aaaggggtgg actttgggaa tattgtggga tttcaatacc ttagtttgta 6660
cagggtaatt gttacaaatg atacaaagaa ctgtatttct tttcatttgt tttaattggt 6720
tgtatatcaa gtccgttaga cgagctcagt gccttggctt ttggcactgt atttcatttt 6780
tagaggtaca ctacattcag tgaggtatgg taaggttgag ggcataatga aggcaccttg 6840
tactgacagt cacagacctc tcaccgagaa ttttatgaga tatactcggg ttcattttag 6900
<210> 78
<211> 2103
<212> DNA
<213> yarrowia lipolytica
<400> 78
atgaacccca acaacactgg caccattgaa atcaacggta aggagtacaa caccttcacc 60
gagccccccg tggccatggc tcaggagcga gccaagacct ccttccccgt gcgagagatg 120
acctacttcc tcgacggtgg cgagaagaac accctcaaaa acgagcagat catggaggag 180
attgagcgag accctctttt caacaacgac aactactacg atctcaacaa ggagcagatc 240
cgagagctca ccatggagcg agtcgccaag ctgtctctgt ttgtgcgtga tcagcccgag 300
gacgacatca agaagcgatt tgctctcatt ggtatcgccg atatgggaac ctacacccga 360
cttggtgtcc actacggcct cttctttggc gccgtccgag gtaccggaac tgccgagcag 420
tttggccact ggatctccaa gggagccgga gacctgcgaa agttctacgg atgtttctcc 480
atgaccgagc tgggccatgg ctccaacctg gctggtctcg agaccaccgc catctacgat 540
gaggagaccg acgagttcat catcaacacc cctcacattg ccgccaccaa gtggtggatt 600
ggaggagccg cccacaccgc cacccacact gtcgtgttcg cccgactcat tgtcaagggc 660
aaggactacg gtgtcaagac ctttgttgtc cagctgcgaa acatcaacga ccacagcctc 720
aaggtcggta tctctattgg tgatatcgga aagaagatgg gccgagacgg tatcgataac 780
ggatggatcc agttcaccaa cgtgcgaatc ccccgacaga acctgctcat gaagtacaca 840
aaggtcgacc gagagggtaa cgtgacccag cctcctctgg ctcagcttac ctacggttct 900
cttatcactg gtcgagtctc catggcctct gattctcacc aggtcggaaa gcgattcatc 960
accattgctc tgcgatacgc ctgcattcga cgacagttct ccaccacccc cggccagccc 1020
gagaccaaga tcatcgacta cccctaccat cagcgacgac ttctgcctct tctggcctat 1080
gtctatgctc ttaagatgac tgccgatgag gttggagctc tcttctcccg aaccatgctt 1140
aagatggacg acctcaagcc cgacgacaag gccggcctca atgaggttgt ttccgacgtc 1200
aaggagctct tctccgtctc cgccggtctc aaggccttct ccacctgggc ttgtgccgac 1260
gtcattgaca agacccgaca ggcttgcggt ggccacggtt actctggata caacggtttc 1320
ggccaggcct acgccgactg ggttgtccag tgcacctggg agggtgacaa caacattctc 1380
accctttctg ccggccgagc tcttatccag tctgccgttg ctctgcgaaa gggcgagcct 1440
gttggtaacg ccgtttctta cctgaagcga tacaaggatc tggccaacgc taagctcaat 1500
ggccgatctc tcaccgaccc caaggtcctc gtcgaggcct gggaggttgc tgccggtaac 1560
atcatcaacc gagccaccga ccagtacgag aagctcattg gcgagggtct taacgccgac 1620
caggcctttg aggttctgtc tcagcagcga ttccaggccg ccaaggtcca cacacgacga 1680
cacctcattg ccgctttctt ctcccgaatt gacaccgagg ctggcgaggc catcaagcag 1740
cccctgctta acctggctct gctgtttgcc ctgtggtcca tcgaagagga ctctggtctg 1800
ttcctgcgag agggcttcct cgagcccaag gatatcgaca ccgtcaccga gctcgtcaac 1860
aagtactgca ccactgtgcg agaggaggtc attggctaca ccgatgcctt caacctgtcc 1920
gactacttca tcaacgctcc tattggatgc tacgatggtg acgcttaccg acactacttc 1980
cagaaggtca acgagcagaa ccctgcccga gacccccgac ctccttacta cgcctctact 2040
ctcaagccct tccttttccg agaggaggag gatgatgaca tttgcgagct tgatgaggaa 2100
tag 2103
<210> 79
<211> 700
<212> PRT
<213> yarrowia lipolytica
<400> 79
Met Asn Pro Asn Asn Thr Gly Thr Ile Glu Ile Asn Gly Lys Glu Tyr
1 5 10 15
Asn Thr Phe Thr Glu Pro Pro Val Ala Met Ala Gln Glu Arg Ala Lys
20 25 30
Thr Ser Phe Pro Val Arg Glu Met Thr Tyr Phe Leu Asp Gly Gly Glu
35 40 45
Lys Asn Thr Leu Lys Asn Glu Gln Ile Met Glu Glu Ile Glu Arg Asp
50 55 60
Pro Leu Phe Asn Asn Asp Asn Tyr Tyr Asp Leu Asn Lys Glu Gln Ile
65 70 75 80
Arg Glu Leu Thr Met Glu Arg Val Ala Lys Leu Ser Leu Phe Val Arg
85 90 95
Asp Gln Pro Glu Asp Asp Ile Lys Lys Arg Phe Ala Leu Ile Gly Ile
100 105 110
Ala Asp Met Gly Thr Tyr Thr Arg Leu Gly Val His Tyr Gly Leu Phe
115 120 125
Phe Gly Ala Val Arg Gly Thr Gly Thr Ala Glu Gln Phe Gly His Trp
130 135 140
Ile Ser Lys Gly Ala Gly Asp Leu Arg Lys Phe Tyr Gly Cys Phe Ser
145 150 155 160
Met Thr Glu Leu Gly His Gly Ser Asn Leu Ala Gly Leu Glu Thr Thr
165 170 175
Ala Ile Tyr Asp Glu Glu Thr Asp Glu Phe Ile Ile Asn Thr Pro His
180 185 190
Ile Ala Ala Thr Lys Trp Trp Ile Gly Gly Ala Ala His Thr Ala Thr
195 200 205
His Thr Val Val Phe Ala Arg Leu Ile Val Lys Gly Lys Asp Tyr Gly
210 215 220
Val Lys Thr Phe Val Val Gln Leu Arg Asn Ile Asn Asp His Ser Leu
225 230 235 240
Lys Val Gly Ile Ser Ile Gly Asp Ile Gly Lys Lys Met Gly Arg Asp
245 250 255
Gly Ile Asp Asn Gly Trp Ile Gln Phe Thr Asn Val Arg Ile Pro Arg
260 265 270
Gln Asn Leu Leu Met Lys Tyr Thr Lys Val Asp Arg Glu Gly Asn Val
275 280 285
Thr Gln Pro Pro Leu Ala Gln Leu Thr Tyr Gly Ser Leu Ile Thr Gly
290 295 300
Arg Val Ser Met Ala Ser Asp Ser His Gln Val Gly Lys Arg Phe Ile
305 310 315 320
Thr Ile Ala Leu Arg Tyr Ala Cys Ile Arg Arg Gln Phe Ser Thr Thr
325 330 335
Pro Gly Gln Pro Glu Thr Lys Ile Ile Asp Tyr Pro Tyr His Gln Arg
340 345 350
Arg Leu Leu Pro Leu Leu Ala Tyr Val Tyr Ala Leu Lys Met Thr Ala
355 360 365
Asp Glu Val Gly Ala Leu Phe Ser Arg Thr Met Leu Lys Met Asp Asp
370 375 380
Leu Lys Pro Asp Asp Lys Ala Gly Leu Asn Glu Val Val Ser Asp Val
385 390 395 400
Lys Glu Leu Phe Ser Val Ser Ala Gly Leu Lys Ala Phe Ser Thr Trp
405 410 415
Ala Cys Ala Asp Val Ile Asp Lys Thr Arg Gln Ala Cys Gly Gly His
420 425 430
Gly Tyr Ser Gly Tyr Asn Gly Phe Gly Gln Ala Tyr Ala Asp Trp Val
435 440 445
Val Gln Cys Thr Trp Glu Gly Asp Asn Asn Ile Leu Thr Leu Ser Ala
450 455 460
Gly Arg Ala Leu Ile Gln Ser Ala Val Ala Leu Arg Lys Gly Glu Pro
465 470 475 480
Val Gly Asn Ala Val Ser Tyr Leu Lys Arg Tyr Lys Asp Leu Ala Asn
485 490 495
Ala Lys Leu Asn Gly Arg Ser Leu Thr Asp Pro Lys Val Leu Val Glu
500 505 510
Ala Trp Glu Val Ala Ala Gly Asn Ile Ile Asn Arg Ala Thr Asp Gln
515 520 525
Tyr Glu Lys Leu Ile Gly Glu Gly Leu Asn Ala Asp Gln Ala Phe Glu
530 535 540
Val Leu Ser Gln Gln Arg Phe Gln Ala Ala Lys Val His Thr Arg Arg
545 550 555 560
His Leu Ile Ala Ala Phe Phe Ser Arg Ile Asp Thr Glu Ala Gly Glu
565 570 575
Ala Ile Lys Gln Pro Leu Leu Asn Leu Ala Leu Leu Phe Ala Leu Trp
580 585 590
Ser Ile Glu Glu Asp Ser Gly Leu Phe Leu Arg Glu Gly Phe Leu Glu
595 600 605
Pro Lys Asp Ile Asp Thr Val Thr Glu Leu Val Asn Lys Tyr Cys Thr
610 615 620
Thr Val Arg Glu Glu Val Ile Gly Tyr Thr Asp Ala Phe Asn Leu Ser
625 630 635 640
Asp Tyr Phe Ile Asn Ala Pro Ile Gly Cys Tyr Asp Gly Asp Ala Tyr
645 650 655
Arg His Tyr Phe Gln Lys Val Asn Glu Gln Asn Pro Ala Arg Asp Pro
660 665 670
Arg Pro Pro Tyr Tyr Ala Ser Thr Leu Lys Pro Phe Leu Phe Arg Glu
675 680 685
Glu Glu Asp Asp Asp Ile Cys Glu Leu Asp Glu Glu
690 695 700
<210> 80
<211> 2103
<212> DNA
<213> yarrowia lipolytica
<400> 80
atgatctccc ccaacctcac agctaacgtc gagattgacg gcaagcagta caacaccttc 60
acagagccac ccaaggcgct cgccggcgag cgagccaagg tcaagttccc catcaaggac 120
atgacggagt ttctgcacgg tggcgaggag aacgtgacca tgatcgagcg actgatgacg 180
gagctcgagc gagaccccgt gctcaacgtg tcgggcgact acgacatgcc caaggagcag 240
ctgcgagaga cggccgtggc gcgaattgcg gcgctgtccg gccactggaa gaaggacaca 300
gaaaaggagg cgctgctgcg gtcccagctg cacggcattg tggacatggg cacccgaatc 360
cgactcggtg tgcacacggg cctgttcatg ggcgccatcc ggggttccgg caccaaggag 420
cagtacgact actgggtgcg aaagggcgcc gcggacgtca agggcttcta cggctgcttt 480
gctatgaccg agctgggcca tggctccaac gtggccggtc ttgagaccac cgccacctac 540
atccaggaca cggacgagtt catcatcaac acccccaaca ctggagccac caagtggtgg 600
attggaggag ccgcccactc ggccacccac accgcctgct ttgctcgtct gcttgtcgac 660
ggcaaggact acggcgtcaa gatctttgtt gtccagctgc gagacgtctc ttctcactct 720
ctcatgcccg gcatcgctct cggcgacatt ggaaagaaga tgggccgaga cgccatcgac 780
aacggctgga tccagttcac caatgtgcga atcccccgac agaacatgct catgaagtac 840
gccaaggtct cgtctaccgg caaggtgtcg cagcctcctc tggcccagct cacctacggc 900
gctctcattg gcggccgagt caccatgatt gccgactcct tctttgtctc ccagcgattc 960
atcaccattg ctctgcgata cgcctgtgtg cgacgacagt ttggcaccac ccccggccag 1020
cccgagacta agatcatcga ctacccctac catcagcgac gtctgctgcc tcttctggcc 1080
ttcacctacg ccatgaagat ggccgccgac cagtcccaga ttcagtacga tcagaccacc 1140
gatctgctgc agaccatcga ccctaaggac aagggcgctc tgggcaaggc cattgtcgac 1200
ctcaaggagc tgtttgcctc ttctgctggt ctcaaggcct tcaccacctg gacctgtgcc 1260
aacatcattg accagtgccg acaggcctgc ggtggccacg gctactctgg ctacaacggc 1320
tttggccagg cctacgccga ctgggttgtc cagtgcacct gggagggtga caacaacgtc 1380
ctgtgtctgt ccatgggccg aggtctcatc cagtcgtgtc tgggccaccg aaagggtaag 1440
cctctgggct cttctgtcgg ctacctggct aacaagggtc ttgagcaggc tactctgagc 1500
ggccgagacc tcaaggaccc caaggttctc atcgaggcct gggagaaggt cgccaacggc 1560
gccatccagc gggccactga caaatttgtc gagctcacca agggcggcct ctctcctgac 1620
caggcctttg aggagctgtc gcagcagcga ttccagtgtg ccaagatcca cacccgaaag 1680
cacctggtga ctgccttcta cgagcgaatc aacgcctctg cgaaggccga cgtcaagcct 1740
tacctcatca acctcgccaa cctcttcact ctgtggtcca ttgaggagga ctctggtctc 1800
ttcctgcgag agggtttcct gcagcccaag gacattgacc aggtgactga gctggtgaac 1860
cactactgca aggaggttcg agaccaggtt gccggctaca ccgatgcctt tggtctgtct 1920
gactggttca tcaacgctcc cattggaaac tacgatggtg acgtttacaa gcattacttt 1980
gccaaggtta accagcagaa ccctgctcag aacccccgac ctccttacta tgagagcact 2040
cttcgacctt tcctgttccg agaggatgag gatgacgaca tttgcgagct ggacgaggaa 2100
tag 2103
<210> 81
<211> 700
<212> PRT
<213> yarrowia lipolytica
<400> 81
Met Ile Ser Pro Asn Leu Thr Ala Asn Val Glu Ile Asp Gly Lys Gln
1 5 10 15
Tyr Asn Thr Phe Thr Glu Pro Pro Lys Ala Leu Ala Gly Glu Arg Ala
20 25 30
Lys Val Lys Phe Pro Ile Lys Asp Met Thr Glu Phe Leu His Gly Gly
35 40 45
Glu Glu Asn Val Thr Met Ile Glu Arg Leu Met Thr Glu Leu Glu Arg
50 55 60
Asp Pro Val Leu Asn Val Ser Gly Asp Tyr Asp Met Pro Lys Glu Gln
65 70 75 80
Leu Arg Glu Thr Ala Val Ala Arg Ile Ala Ala Leu Ser Gly His Trp
85 90 95
Lys Lys Asp Thr Glu Lys Glu Ala Leu Leu Arg Ser Gln Leu His Gly
100 105 110
Ile Val Asp Met Gly Thr Arg Ile Arg Leu Gly Val His Thr Gly Leu
115 120 125
Phe Met Gly Ala Ile Arg Gly Ser Gly Thr Lys Glu Gln Tyr Asp Tyr
130 135 140
Trp Val Arg Lys Gly Ala Ala Asp Val Lys Gly Phe Tyr Gly Cys Phe
145 150 155 160
Ala Met Thr Glu Leu Gly His Gly Ser Asn Val Ala Gly Leu Glu Thr
165 170 175
Thr Ala Thr Tyr Ile Gln Asp Thr Asp Glu Phe Ile Ile Asn Thr Pro
180 185 190
Asn Thr Gly Ala Thr Lys Trp Trp Ile Gly Gly Ala Ala His Ser Ala
195 200 205
Thr His Thr Ala Cys Phe Ala Arg Leu Leu Val Asp Gly Lys Asp Tyr
210 215 220
Gly Val Lys Ile Phe Val Val Gln Leu Arg Asp Val Ser Ser His Ser
225 230 235 240
Leu Met Pro Gly Ile Ala Leu Gly Asp Ile Gly Lys Lys Met Gly Arg
245 250 255
Asp Ala Ile Asp Asn Gly Trp Ile Gln Phe Thr Asn Val Arg Ile Pro
260 265 270
Arg Gln Asn Met Leu Met Lys Tyr Ala Lys Val Ser Ser Thr Gly Lys
275 280 285
Val Ser Gln Pro Pro Leu Ala Gln Leu Thr Tyr Gly Ala Leu Ile Gly
290 295 300
Gly Arg Val Thr Met Ile Ala Asp Ser Phe Phe Val Ser Gln Arg Phe
305 310 315 320
Ile Thr Ile Ala Leu Arg Tyr Ala Cys Val Arg Arg Gln Phe Gly Thr
325 330 335
Thr Pro Gly Gln Pro Glu Thr Lys Ile Ile Asp Tyr Pro Tyr His Gln
340 345 350
Arg Arg Leu Leu Pro Leu Leu Ala Phe Thr Tyr Ala Met Lys Met Ala
355 360 365
Ala Asp Gln Ser Gln Ile Gln Tyr Asp Gln Thr Thr Asp Leu Leu Gln
370 375 380
Thr Ile Asp Pro Lys Asp Lys Gly Ala Leu Gly Lys Ala Ile Val Asp
385 390 395 400
Leu Lys Glu Leu Phe Ala Ser Ser Ala Gly Leu Lys Ala Phe Thr Thr
405 410 415
Trp Thr Cys Ala Asn Ile Ile Asp Gln Cys Arg Gln Ala Cys Gly Gly
420 425 430
His Gly Tyr Ser Gly Tyr Asn Gly Phe Gly Gln Ala Tyr Ala Asp Trp
435 440 445
Val Val Gln Cys Thr Trp Glu Gly Asp Asn Asn Val Leu Cys Leu Ser
450 455 460
Met Gly Arg Gly Leu Ile Gln Ser Cys Leu Gly His Arg Lys Gly Lys
465 470 475 480
Pro Leu Gly Ser Ser Val Gly Tyr Leu Ala Asn Lys Gly Leu Glu Gln
485 490 495
Ala Thr Leu Ser Gly Arg Asp Leu Lys Asp Pro Lys Val Leu Ile Glu
500 505 510
Ala Trp Glu Lys Val Ala Asn Gly Ala Ile Gln Arg Ala Thr Asp Lys
515 520 525
Phe Val Glu Leu Thr Lys Gly Gly Leu Ser Pro Asp Gln Ala Phe Glu
530 535 540
Glu Leu Ser Gln Gln Arg Phe Gln Cys Ala Lys Ile His Thr Arg Lys
545 550 555 560
His Leu Val Thr Ala Phe Tyr Glu Arg Ile Asn Ala Ser Ala Lys Ala
565 570 575
Asp Val Lys Pro Tyr Leu Ile Asn Leu Ala Asn Leu Phe Thr Leu Trp
580 585 590
Ser Ile Glu Glu Asp Ser Gly Leu Phe Leu Arg Glu Gly Phe Leu Gln
595 600 605
Pro Lys Asp Ile Asp Gln Val Thr Glu Leu Val Asn His Tyr Cys Lys
610 615 620
Glu Val Arg Asp Gln Val Ala Gly Tyr Thr Asp Ala Phe Gly Leu Ser
625 630 635 640
Asp Trp Phe Ile Asn Ala Pro Ile Gly Asn Tyr Asp Gly Asp Val Tyr
645 650 655
Lys His Tyr Phe Ala Lys Val Asn Gln Gln Asn Pro Ala Gln Asn Pro
660 665 670
Arg Pro Pro Tyr Tyr Glu Ser Thr Leu Arg Pro Phe Leu Phe Arg Glu
675 680 685
Asp Glu Asp Asp Asp Ile Cys Glu Leu Asp Glu Glu
690 695 700
<210> 82
<211> 12355
<212> DNA
<213> Artificial sequence
<220>
<223> pZKLY-FCtR17U plasmid
<400> 82
cgatgagcct aaaatgaacc cgagtatatc tcataaaatt ctcggtgaga ggtctgtgac 60
tgtcagtaca aggtgccttc attatgccct caaccttacc atacctcact gaatgtagtg 120
tacctctaaa aatgaaatac agtgccaaaa gccaaggcac tgagctcgtc taacggactt 180
gatatacaac caattaaaac aaatgaaaag aaatacagtt ctttgtatca tttgtaacaa 240
ttaccctgta caaactaagg tattgaaatc ccacaatatt cccaaagtcc acccctttcc 300
aaattgtcat gcctacaact catataccaa gcactaacct accgtttaaa cagtgtacgc 360
agtactatag aggaacaatt gccccggaga agacggccag gccgcctaga tgacaaattc 420
aacaactcac agctgacttt ctgccattgc cactaggggg gggccttttt atatggccaa 480
gccaagctct ccacgtcggt tgggctgcac ccaacaataa atgggtaggg ttgcaccaac 540
aaagggatgg gatggggggt agaagatacg aggataacgg ggctcaatgg cacaaataag 600
aacgaatact gccattaaga ctcgtgatcc agcgactgac accattgcat catctaaggg 660
cctcaaaact acctcggaac tgctgcgctg atctggacac cacagaggtt ccgagcactt 720
taggttgcac caaatgtccc accaggtgca ggcagaaaac gctggaacag cgtgtacagt 780
ttgtcttaac aaaaagtgag ggcgctgagg tcgagcaggg tggtgtgact tgttatagcc 840
tttagagctg cgaaagcgcg tatggatttg gctcatcagg ccagattgag ggtctgtgga 900
cacatgtcat gttagtgtac ttcaatcgcc ccctggatat agccccgaca ataggccgtg 960
gcctcatttt tttgccttcc gcacatttcc attgctcggt acccacacct tgcttctcct 1020
gcacttgcca accttaatac tggtttacat tgaccaacat cttacaagcg gggggcttgt 1080
ctagggtata tataaacagt ggctctccca atcggttgcc agtctctttt ttcctttctt 1140
tccccacaga ttcgaaatct aaactacaca tcacacaatt ccgagccgtg agtatccacg 1200
acaagatcag tgtcgagacg acgcgttttg tgtaatgaca caatccgaaa gtcgctagca 1260
acacacactc tctacacaaa ctaacccagc tctccatggc catcgagcag ctgctcgagt 1320
actggtacgt cgttgtgccc gtcctgtaca ttatcaagca gctccttgcc tacaccaaga 1380
ctcgagtgct gatgaaaaag ctcggagccg ctcccgtcac caacaagctg tacgacaacg 1440
ccttcggtat cgtcaacggc tggaaggctc ttcagttcaa gaaagagggc cgagctcagg 1500
aatacaacga ctacaagttc gatcactcca agaatccttc tgtgggaacc tacgtctcca 1560
ttctgtttgg cactcgaatc gtggttacca aggatcccga gaacatcaag gccattctcg 1620
caacccagtt cggagacttt tcgctgggca agcgacacac tctcttcaag cccttgctgg 1680
gagacggtat cttcacactc gatggagaag gctggaagca ttccagagct atgctgcgac 1740
ctcagtttgc ccgagagcaa gttgctcacg tcaccagcct cgaaccacac ttccagctgc 1800
tcaagaaaca tatcctcaag cacaagggcg agtacttcga cattcaggag ctgttctttc 1860
gattcaccgt ggactctgcc acggagtttc tgttcggcga gtccgttcac tctctcaagg 1920
acgagtcgat tggaatcaac caggacgata tcgacttcgc tggtcggaag gactttgccg 1980
agtccttcaa caaggcacag gaatacttgg ccattcgaac tctggtgcag accttctact 2040
ggctcgtcaa caacaaggag tttcgagact gcaccaagct ggttcacaag ttcaccaact 2100
actatgtcca gaaggctctc gatgcatctc ccgaggaact tgagaagcaa agcggctacg 2160
tgttcctgta cgagcttgtc aagcagacca gagatcccaa cgtgctgcga gaccagtccc 2220
tcaacatctt gctggccgga cgagacacca ctgctggcct cctgtcgttt gcagtcttcg 2280
agttggctcg tcatcccgag atctgggcca agctgcgaga ggaaatcgag caacagttcg 2340
gacttggcga ggactctcgt gtcgaagaga ttaccttcga gagcctcaag cgatgcgagt 2400
acctcaaggc ctttctcaac gaaaccctgc ggatctaccc ttccgttcct cgaaacttca 2460
gaatcgctac caagaacaca acccttcccc gaggcggtgg atcggacggt acttctccaa 2520
tcctcattca gaagggcgag gccgtgtcct acggtatcaa ttctactcat ctggatcctg 2580
tctattacgg acccgacgct gccgagtttc gacccgagcg atggttcgaa ccttcgacca 2640
aaaagctcgg ctgggcctac cttcccttca acggaggtcc acgaatctgt ctgggccaac 2700
agtttgccct caccgaggct ggctacgtgc tggtcagact cgttcaggag ttctcccacg 2760
tccgatccga tcccgacgag gtgtaccctc ccaagcgact tacaaacctg accatgtgtc 2820
tccaggacgg tgccattgtc aagttcgact aagcggccgc aagtgtggat ggggaagtga 2880
gtgcccggtt ctgtgtgcac aattggcaat ccaagatgga tggattcaac acagggatat 2940
agcgagctac gtggtggtgc gaggatatag caacggatat ttatgtttga cacttgagaa 3000
tgtacgatac aagcactgtc caagtacaat actaaacata ctgtacatac tcatactcgt 3060
acccgggcaa cggtttcact tgagtgcagt ggctagtgct cttactcgta cagtgtgcaa 3120
tactgcgtat catagtcttt gatgtatatc gtattcattc atgttagttg atttaaacca 3180
tcatctaagg gcctcaaaac tacctcggaa ctgctgcgct gatctggaca ccacagaggt 3240
tccgagcact ttaggttgca ccaaatgtcc caccaggtgc aggcagaaaa cgctggaaca 3300
gcgtgtacag tttgtcttaa caaaaagtga gggcgctgag gtcgagcagg gtggtgtgac 3360
ttgttatagc ctttagagct gcgaaagcgc gtatggattt ggctcatcag gccagattga 3420
gggtctgtgg acacatgtca tgttagtgta cttcaatcgc cccctggata tagccccgac 3480
aataggccgt ggcctcattt ttttgccttc cgcacatttc cattgctcgg tacccacacc 3540
ttgcttctcc tgcacttgcc aaccttaata ctggtttaca ttgaccaaca tcttacaagc 3600
ggggggcttg tctagggtat atataaacag tggctctccc aatcggttgc cagtctcttt 3660
tttcctttct ttccccacag attcgaaatc taaactacac atcacaccat ggccctggac 3720
aagctcgacc tgtacgtcat cattaccctc gtggttgcta tcgccgctta cttcgccaag 3780
aaccagttcc tggatcagca acaggacact ggctttctca acaccgactc cggagatggc 3840
aactctcgag acatccttca ggctctcaag aaaaacaata agaacaccct gcttctcttc 3900
ggttcccaga ctggcacagc cgaggactac gccaacaagt tgtcgcgaga gctgcactcc 3960
cgatttggct tgaagactat ggtcgcagat ttcgccgatt acgacttcga gaactttgga 4020
gacattaccg aggacatcct cgtgttcttt atcgttgcta cctacggcga aggcgagccc 4080
accgacaatg ccgacgagtt tcacacttgg ctgaccgagg aagcagatac cctgtctaca 4140
ctcaagtata ccgtcttcgg tctgggtaac tccacctacg agtttttcaa cgccatcggt 4200
cgaaagttcg acagactgct cggagagaag ggtggcgacc gatttgccga gtacggcgaa 4260
ggagacgatg gtactggcac tctcgacgag gatttcctgg cttggaagga caacgtgttc 4320
gattctctca agaacgacct gaatttcgaa gagaaggagc tcaaatacga acccaacgtc 4380
aagttgaccg agcgggacga tctgtctggc aacgatcccg acgtttcgct gggcgagcca 4440
aacgtcaagt acatcaagtc cgagggtgtt gaccttacca agggaccttt cgatcacact 4500
catccctttc tggcccgaat cgtcaagacc aaagagctct ttacttccga ggacagacac 4560
tgcgtgcacg tcgagttcga cattagcgag tccaacctca agtatactac cggagatcat 4620
cttgctatct ggccctccaa ttcggacgag aacatcaagc agtttgccaa gtgctttggc 4680
ctggaggaca agctcgatac cgtcatcgag ctgaaggctc tcgattccac ttactccatt 4740
ccatttccca atccaatcac ctacggagcc gtcattcgtc accatttgga gatctctggt 4800
cctgtgtcgc gacagttctt tctgtccatt gccggatttg ctcccgacga agagaccaaa 4860
aagtccttca ctcgaatcgg tggcgacaag caagagttcg ccagcaaggt cacccgtcga 4920
aagttcaaca ttgccgatgc tcttctgttt gcctccaaca atcgaccctg gtccgacgtt 4980
cccttcgagt tccttatcga gaacgtccag catctcactc ctcggtacta ttccatttct 5040
tcgtcctctc tcagcgagaa gcagaccatc aacgttactg ctgtggtcga agccgaggaa 5100
gaggccgatg gacgacccgt tactggtgtc gttaccaacc tgctcaagaa catcgagatt 5160
gaacagaaca agactggcga gacaccaatg gtccactacg acctcaatgg tcccagaggc 5220
aagttctcca agtttcgact gcccgtgcac gtcagacgat ccaacttcaa acttcccaag 5280
aactctacta cccctgtcat cctgattggt ccaggcaccg gtgttgctcc cctgcgaggc 5340
tttgtccggg agcgagtgca gcaagtcaag aacggagtca acgttggtaa gactgtgctg 5400
ttctacggct gtcgaaattc cgaacaggac tttctctaca aacaggagtg gagcgagtat 5460
gcctccgtcc tgggagagaa cttcgaaatg ttcaacgcct tttctcgaca ggaccctacc 5520
aagaaagtgt acgttcaaga caagatcctc gagaactctg ctcttgtcga cgagctcctg 5580
tccagcggtg caattatcta cgtttgcgga gatgcctctc gaatggctcg agacgtgcag 5640
gctgcaattg ccaagatcgt tgccaagtcc cgagacatcc acgaggacaa ggctgccgag 5700
ctggtcaagt cttggaaggt gcagaaccga taccaggagg atgtctggta agcggccgca 5760
tgagaagata aatatataaa tacattgaga tattaaatgc gctagattag agagcctcat 5820
actgctcgga gagaagccaa gacgagtact caaaggggat tacaccatcc atatccacag 5880
acacaagctg gggaaaggtt ctatatacac tttccggaat accgtagttt ccgatgttat 5940
caatgggggc agccaggatt tcaggcactt cggtgtctcg gggtgaaatg gcgttcttgg 6000
cctccatcaa gtcgtaccat gtcttcattt gcctgtcaaa gtaaaacaga agcagatgaa 6060
gaatgaactt gaagtgaagg aatttaaatg taacgaaact gaaatttgac cagatattgt 6120
gtccgcggtg gagctccagc ttttgttccc tttagtgagg gttaatttcg agcttggcgt 6180
aatcatggtc atagctgttt cctgtgtgaa attgttatcc gctcacaagc ttccacacaa 6240
cgtacgttga ttgaggtgga gccagatggg ctattgtttc atatatagac tggcagccac 6300
ctctttggcc cagcatgttt gtatacctgg aagggaaaac taaagaagct ggctagttta 6360
gtttgattat tatagtagat gtcctaatca ctagagatta gaatgtcttg gcgatgatta 6420
gtcgtcgtcc cctgtatcat gtctagacca actgtgtcat gaagttggtg ctggtgtttt 6480
acctgtgtac tacaagtagg tgtcctagat ctagtgtaca gagccgttta gacccatgtg 6540
gacttcacca ttaacgatgg aaaatgttca ttatatgaca gtatattaca atggacttgc 6600
tccatttctt ccttgcatca catgttctcc acctccatag ttgatcaaca catcatagta 6660
gctaaggctg ctgctctccc actacagtcc accacaagtt aagtagcacc gtcagtacag 6720
ctaaaagtac acgtctagta cgtttcataa ctagtcaagt agcccctatt acagatatca 6780
gcactatcac gcacgagttt ttctctgtgc tatctaatca acttgccaag tattcggaga 6840
agatacactt tcttggcatc aggtatacga gggagcctat cagatgaaaa agggtatatt 6900
ggatccattc atatccacct acacgttgtc ataatctcct cattcacgtg attcatttcg 6960
tgacactagt ttctcacttt cccccccgca cctatagtca acttggcgga cacgctactt 7020
gtagctgacg ttgatttata gacccaatca aagcgggtta tcggtcaggt agcacttatc 7080
attcatcgtt catactacga tgagcaatct cgggcatgtc cggaaaagtg tcgggcgcgc 7140
cagctgcatt aatgaatcgg ccaacgcgcg gggagaggcg gtttgcgtat tgggcgctct 7200
tccgcttcct cgctcactga ctcgctgcgc tcggtcgttc ggctgcggcg agcggtatca 7260
gctcactcaa aggcggtaat acggttatcc acagaatcag gggataacgc aggaaagaac 7320
atgtgagcaa aaggccagca aaaggccagg aaccgtaaaa aggccgcgtt gctggcgttt 7380
ttccataggc tccgcccccc tgacgagcat cacaaaaatc gacgctcaag tcagaggtgg 7440
cgaaacccga caggactata aagataccag gcgtttcccc ctggaagctc cctcgtgcgc 7500
tctcctgttc cgaccctgcc gcttaccgga tacctgtccg cctttctccc ttcgggaagc 7560
gtggcgcttt ctcatagctc acgctgtagg tatctcagtt cggtgtaggt cgttcgctcc 7620
aagctgggct gtgtgcacga accccccgtt cagcccgacc gctgcgcctt atccggtaac 7680
tatcgtcttg agtccaaccc ggtaagacac gacttatcgc cactggcagc agccactggt 7740
aacaggatta gcagagcgag gtatgtaggc ggtgctacag agttcttgaa gtggtggcct 7800
aactacggct acactagaag aacagtattt ggtatctgcg ctctgctgaa gccagttacc 7860
ttcggaaaaa gagttggtag ctcttgatcc ggcaaacaaa ccaccgctgg tagcggtggt 7920
ttttttgttt gcaagcagca gattacgcgc agaaaaaaag gatctcaaga agatcctttg 7980
atcttttcta cggggtctga cgctcagtgg aacgaaaact cacgttaagg gattttggtc 8040
atgagattat caaaaaggat cttcacctag atccttttaa attaaaaatg aagttttaaa 8100
tcaatctaaa gtatatatga gtaaacttgg tctgacagtt accaatgctt aatcagtgag 8160
gcacctatct cagcgatctg tctatttcgt tcatccatag ttgcctgact ccccgtcgtg 8220
tagataacta cgatacggga gggcttacca tctggcccca gtgctgcaat gataccgcga 8280
gacccacgct caccggctcc agatttatca gcaataaacc agccagccgg aagggccgag 8340
cgcagaagtg gtcctgcaac tttatccgcc tccatccagt ctattaattg ttgccgggaa 8400
gctagagtaa gtagttcgcc agttaatagt ttgcgcaacg ttgttgccat tgctacaggc 8460
atcgtggtgt cacgctcgtc gtttggtatg gcttcattca gctccggttc ccaacgatca 8520
aggcgagtta catgatcccc catgttgtgc aaaaaagcgg ttagctcctt cggtcctccg 8580
atcgttgtca gaagtaagtt ggccgcagtg ttatcactca tggttatggc agcactgcat 8640
aattctctta ctgtcatgcc atccgtaaga tgcttttctg tgactggtga gtactcaacc 8700
aagtcattct gagaatagtg tatgcggcga ccgagttgct cttgcccggc gtcaatacgg 8760
gataataccg cgccacatag cagaacttta aaagtgctca tcattggaaa acgttcttcg 8820
gggcgaaaac tctcaaggat cttaccgctg ttgagatcca gttcgatgta acccactcgt 8880
gcacccaact gatcttcagc atcttttact ttcaccagcg tttctgggtg agcaaaaaca 8940
ggaaggcaaa atgccgcaaa aaagggaata agggcgacac ggaaatgttg aatactcata 9000
ctcttccttt ttcaatatta ttgaagcatt tatcagggtt attgtctcat gagcggatac 9060
atatttgaat gtatttagaa aaataaacaa ataggggttc cgcgcacatt tccccgaaaa 9120
gtgccacctg atgcggtgtg aaataccgca cagatgcgta aggagaaaat accgcatcag 9180
gaaattgtaa gcgttaatat tttgttaaaa ttcgcgttaa atttttgtta aatcagctca 9240
ttttttaacc aataggccga aatcggcaaa atcccttata aatcaaaaga atagaccgag 9300
atagggttga gtgttgttcc agtttggaac aagagtccac tattaaagaa cgtggactcc 9360
aacgtcaaag ggcgaaaaac cgtctatcag ggcgatggcc cactacgtga accatcaccc 9420
taatcaagtt ttttggggtc gaggtgccgt aaagcactaa atcggaaccc taaagggagc 9480
ccccgattta gagcttgacg gggaaagccg gcgaacgtgg cgagaaagga agggaagaaa 9540
gcgaaaggag cgggcgctag ggcgctggca agtgtagcgg tcacgctgcg cgtaaccacc 9600
acacccgccg cgcttaatgc gccgctacag ggcgcgtcca ttcgccattc aggctgcgca 9660
actgttggga agggcgatcg gtgcgggcct cttcgctatt acgccagctg gcgaaagggg 9720
gatgtgctgc aaggcgatta agttgggtaa cgccagggtt ttcccagtca cgacgttgta 9780
aaacgacggc cagtgaattg taatacgact cactataggg cgaattgggc ccgacgtcgc 9840
atgcattccg acagcagcga ctgggcacca tgatcaagcg aaacaccttc ccccagctgc 9900
cctggcaaac catcaagaac cctactttca tcaagtgcaa gaacggttct actcttctca 9960
cctccggtgt ctacggctgg tgccgaaagc ctaactacac cgctgatttc atcatgtgcc 10020
tcacctgggc tctcatgtgc ggtgttgctt ctcccctgcc ttacttctac ccggtcttct 10080
tcttcctggt gctcatccac cgagcttacc gagactttga gcgactggag cgaaagtacg 10140
gtgaggacta ccaggagttc aagcgacagg tcccttggat cttcatccct tatgttttct 10200
aaacgataag cttagtgagc gaatggtgag gttacttaat tgagtggcca gcctatggga 10260
ttgtataaca gacagtcaat atattactga aaagactgaa cagccagacg gagtgaggtt 10320
gtgagtgaat cgtagagggc ggctattaca gcaagtctac tctacagtgt actaacacag 10380
cagagaacaa atacaggtgt gcattcggct atctgagaat tagttggaga gctcgagacc 10440
ctcggcgata aactgctcct cggttttgtg tccatacttg tacggaccat tgtaatgggg 10500
caagtcgttg agttctcgtc gtccgacgtt cagagcacag aaaccaatgt aatcaatgta 10560
gcagagatgg ttctgcaaaa gattgatttg tgcgagcagg ttaattaact ttggccggaa 10620
ttcctttacc tgcaggataa cttcgtataa tgtatgctat acgaagttat gatctctctc 10680
ttgagctttt ccataacaag ttcttctgcc tccaggaagt ccatgggtgg tttgatcatg 10740
gttttggtgt agtggtagtg cagtggtggt attgtgactg gggatgtagt tgagaataag 10800
tcatacacaa gtcagctttc ttcgagcctc atataagtat aagtagttca acgtattagc 10860
actgtaccca gcatctccgt atcgagaaac acaacaacat gccccattgg acagatcatg 10920
cggatacaca ggttgtgcag tatcatacat actcgatcag acaggtcgtc tgaccatcat 10980
acaagctgaa caagcgctcc atacttgcac gctctctata tacacagtta aattacatat 11040
ccatagtcta acctctaaca gttaatcttc tggtaagcct cccagccagc cttctggtat 11100
cgcttggcct cctcaatagg atctcggttc tggccgtaca gacctcggcc gacaattatg 11160
atatccgttc cggtagacat gacatcctca acagttcggt actgctgtcc gagagcgtct 11220
cccttgtcgt caagacccac cccgggggtc agaataagcc agtcctcaga gtcgccctta 11280
ggtcggttct gggcaatgaa gccaaccaca aactcggggt cggatcgggc aagctcaatg 11340
gtctgcttgg agtactcgcc agtggccaga gagcccttgc aagacagctc ggccagcatg 11400
agcagacctc tggccagctt ctcgttggga gaggggacta ggaactcctt gtactgggag 11460
ttctcgtagt cagagacgtc ctccttcttc tgttcagaga cagtttcctc ggcaccagct 11520
cgcaggccag caatgattcc ggttccgggt acaccgtggg cgttggtgat atcggaccac 11580
tcggcgattc ggtgacaccg gtactggtgc ttgacagtgt tgccaatatc tgcgaacttt 11640
ctgtcctcga acaggaagaa accgtgctta agagcaagtt ccttgagggg gagcacagtg 11700
ccggcgtagg tgaagtcgtc aatgatgtcg atatgggttt tgatcatgca cacataaggt 11760
ccgaccttat cggcaagctc aatgagctcc ttggtggtgg taacatccag agaagcacac 11820
aggttggttt tcttggctgc cacgagcttg agcactcgag cggcaaaggc ggacttgtgg 11880
acgttagctc gagcttcgta ggagggcatt ttggtggtga agaggagact gaaataaatt 11940
tagtctgcag aactttttat cggaacctta tctggggcag tgaagtatat gttatggtaa 12000
tagttacgag ttagttgaac ttatagatag actggactat acggctatcg gtccaaatta 12060
gaaagaacgt caatggctct ctgggcgtcg cctttgccga caaaaatgtg atcatgatga 12120
aagccagcaa tgacgttgca gctgatattg ttgtcggcca accgcgccga aaacgcagct 12180
gtcagaccca cagcctccaa cgaagaatgt atcgtcaaag tgatccaagc acactcatag 12240
ttggagtcgt actccaaagg cggcaatgac gagtcagaca gatactcgtc gacgcgataa 12300
cttcgtataa tgtatgctat acgaagttat cgtacgatag ttagtagaca acaat 12355
<210> 83
<211> 1557
<212> DNA
<213> Artificial sequence
<220>
<223> CtCYPs(52A17)
<400> 83
atggccatcg agcagctgct cgagtactgg tacgtcgttg tgcccgtcct gtacattatc 60
aagcagctcc ttgcctacac caagactcga gtgctgatga aaaagctcgg agccgctccc 120
gtcaccaaca agctgtacga caacgccttc ggtatcgtca acggctggaa ggctcttcag 180
ttcaagaaag agggccgagc tcaggaatac aacgactaca agttcgatca ctccaagaat 240
ccttctgtgg gaacctacgt ctccattctg tttggcactc gaatcgtggt taccaaggat 300
cccgagaaca tcaaggccat tctcgcaacc cagttcggag acttttcgct gggcaagcga 360
cacactctct tcaagccctt gctgggagac ggtatcttca cactcgatgg agaaggctgg 420
aagcattcca gagctatgct gcgacctcag tttgcccgag agcaagttgc tcacgtcacc 480
agcctcgaac cacacttcca gctgctcaag aaacatatcc tcaagcacaa gggcgagtac 540
ttcgacattc aggagctgtt ctttcgattc accgtggact ctgccacgga gtttctgttc 600
ggcgagtccg ttcactctct caaggacgag tcgattggaa tcaaccagga cgatatcgac 660
ttcgctggtc ggaaggactt tgccgagtcc ttcaacaagg cacaggaata cttggccatt 720
cgaactctgg tgcagacctt ctactggctc gtcaacaaca aggagtttcg agactgcacc 780
aagctggttc acaagttcac caactactat gtccagaagg ctctcgatgc atctcccgag 840
gaacttgaga agcaaagcgg ctacgtgttc ctgtacgagc ttgtcaagca gaccagagat 900
cccaacgtgc tgcgagacca gtccctcaac atcttgctgg ccggacgaga caccactgct 960
ggcctcctgt cgtttgcagt cttcgagttg gctcgtcatc ccgagatctg ggccaagctg 1020
cgagaggaaa tcgagcaaca gttcggactt ggcgaggact ctcgtgtcga agagattacc 1080
ttcgagagcc tcaagcgatg cgagtacctc aaggcctttc tcaacgaaac cctgcggatc 1140
tacccttccg ttcctcgaaa cttcagaatc gctaccaaga acacaaccct tccccgaggc 1200
ggtggatcgg acggtacttc tccaatcctc attcagaagg gcgaggccgt gtcctacggt 1260
atcaattcta ctcatctgga tcctgtctat tacggacccg acgctgccga gtttcgaccc 1320
gagcgatggt tcgaaccttc gaccaaaaag ctcggctggg cctaccttcc cttcaacgga 1380
ggtccacgaa tctgtctggg ccaacagttt gccctcaccg aggctggcta cgtgctggtc 1440
agactcgttc aggagttctc ccacgtccga tccgatcccg acgaggtgta ccctcccaag 1500
cgacttacaa acctgaccat gtgtctccag gacggtgcca ttgtcaagtt cgactaa 1557
<210> 84
<211> 518
<212> PRT
<213> Artificial sequence
<220>
<223> CtCYPs (52A17) protein
<400> 84
Met Ala Ile Glu Gln Leu Leu Glu Tyr Trp Tyr Val Val Val Pro Val
1 5 10 15
Leu Tyr Ile Ile Lys Gln Leu Leu Ala Tyr Thr Lys Thr Arg Val Leu
20 25 30
Met Lys Lys Leu Gly Ala Ala Pro Val Thr Asn Lys Leu Tyr Asp Asn
35 40 45
Ala Phe Gly Ile Val Asn Gly Trp Lys Ala Leu Gln Phe Lys Lys Glu
50 55 60
Gly Arg Ala Gln Glu Tyr Asn Asp Tyr Lys Phe Asp His Ser Lys Asn
65 70 75 80
Pro Ser Val Gly Thr Tyr Val Ser Ile Leu Phe Gly Thr Arg Ile Val
85 90 95
Val Thr Lys Asp Pro Glu Asn Ile Lys Ala Ile Leu Ala Thr Gln Phe
100 105 110
Gly Asp Phe Ser Leu Gly Lys Arg His Thr Leu Phe Lys Pro Leu Leu
115 120 125
Gly Asp Gly Ile Phe Thr Leu Asp Gly Glu Gly Trp Lys His Ser Arg
130 135 140
Ala Met Leu Arg Pro Gln Phe Ala Arg Glu Gln Val Ala His Val Thr
145 150 155 160
Ser Leu Glu Pro His Phe Gln Leu Leu Lys Lys His Ile Leu Lys His
165 170 175
Lys Gly Glu Tyr Phe Asp Ile Gln Glu Leu Phe Phe Arg Phe Thr Val
180 185 190
Asp Ser Ala Thr Glu Phe Leu Phe Gly Glu Ser Val His Ser Leu Lys
195 200 205
Asp Glu Ser Ile Gly Ile Asn Gln Asp Asp Ile Asp Phe Ala Gly Arg
210 215 220
Lys Asp Phe Ala Glu Ser Phe Asn Lys Ala Gln Glu Tyr Leu Ala Ile
225 230 235 240
Arg Thr Leu Val Gln Thr Phe Tyr Trp Leu Val Asn Asn Lys Glu Phe
245 250 255
Arg Asp Cys Thr Lys Leu Val His Lys Phe Thr Asn Tyr Tyr Val Gln
260 265 270
Lys Ala Leu Asp Ala Ser Pro Glu Glu Leu Glu Lys Gln Ser Gly Tyr
275 280 285
Val Phe Leu Tyr Glu Leu Val Lys Gln Thr Arg Asp Pro Asn Val Leu
290 295 300
Arg Asp Gln Ser Leu Asn Ile Leu Leu Ala Gly Arg Asp Thr Thr Ala
305 310 315 320
Gly Leu Leu Ser Phe Ala Val Phe Glu Leu Ala Arg His Pro Glu Ile
325 330 335
Trp Ala Lys Leu Arg Glu Glu Ile Glu Gln Gln Phe Gly Leu Gly Glu
340 345 350
Asp Ser Arg Val Glu Glu Ile Thr Phe Glu Ser Leu Lys Arg Cys Glu
355 360 365
Tyr Leu Lys Ala Phe Leu Asn Glu Thr Leu Arg Ile Tyr Pro Ser Val
370 375 380
Pro Arg Asn Phe Arg Ile Ala Thr Lys Asn Thr Thr Leu Pro Arg Gly
385 390 395 400
Gly Gly Ser Asp Gly Thr Ser Pro Ile Leu Ile Gln Lys Gly Glu Ala
405 410 415
Val Ser Tyr Gly Ile Asn Ser Thr His Leu Asp Pro Val Tyr Tyr Gly
420 425 430
Pro Asp Ala Ala Glu Phe Arg Pro Glu Arg Trp Phe Glu Pro Ser Thr
435 440 445
Lys Lys Leu Gly Trp Ala Tyr Leu Pro Phe Asn Gly Gly Pro Arg Ile
450 455 460
Cys Leu Gly Gln Gln Phe Ala Leu Thr Glu Ala Gly Tyr Val Leu Val
465 470 475 480
Arg Leu Val Gln Glu Phe Ser His Val Arg Ser Asp Pro Asp Glu Val
485 490 495
Tyr Pro Pro Lys Arg Leu Thr Asn Leu Thr Met Cys Leu Gln Asp Gly
500 505 510
Ala Ile Val Lys Phe Asp
515
<210> 85
<211> 2043
<212> DNA
<213> Artificial sequence
<220>
<223> CtCPRs (CPR)
<400> 85
atggccctgg acaagctcga cctgtacgtc atcattaccc tcgtggttgc tatcgccgct 60
tacttcgcca agaaccagtt cctggatcag caacaggaca ctggctttct caacaccgac 120
tccggagatg gcaactctcg agacatcctt caggctctca agaaaaacaa taagaacacc 180
ctgcttctct tcggttccca gactggcaca gccgaggact acgccaacaa gttgtcgcga 240
gagctgcact cccgatttgg cttgaagact atggtcgcag atttcgccga ttacgacttc 300
gagaactttg gagacattac cgaggacatc ctcgtgttct ttatcgttgc tacctacggc 360
gaaggcgagc ccaccgacaa tgccgacgag tttcacactt ggctgaccga ggaagcagat 420
accctgtcta cactcaagta taccgtcttc ggtctgggta actccaccta cgagtttttc 480
aacgccatcg gtcgaaagtt cgacagactg ctcggagaga agggtggcga ccgatttgcc 540
gagtacggcg aaggagacga tggtactggc actctcgacg aggatttcct ggcttggaag 600
gacaacgtgt tcgattctct caagaacgac ctgaatttcg aagagaagga gctcaaatac 660
gaacccaacg tcaagttgac cgagcgggac gatctgtctg gcaacgatcc cgacgtttcg 720
ctgggcgagc caaacgtcaa gtacatcaag tccgagggtg ttgaccttac caagggacct 780
ttcgatcaca ctcatccctt tctggcccga atcgtcaaga ccaaagagct ctttacttcc 840
gaggacagac actgcgtgca cgtcgagttc gacattagcg agtccaacct caagtatact 900
accggagatc atcttgctat ctggccctcc aattcggacg agaacatcaa gcagtttgcc 960
aagtgctttg gcctggagga caagctcgat accgtcatcg agctgaaggc tctcgattcc 1020
acttactcca ttccatttcc caatccaatc acctacggag ccgtcattcg tcaccatttg 1080
gagatctctg gtcctgtgtc gcgacagttc tttctgtcca ttgccggatt tgctcccgac 1140
gaagagacca aaaagtcctt cactcgaatc ggtggcgaca agcaagagtt cgccagcaag 1200
gtcacccgtc gaaagttcaa cattgccgat gctcttctgt ttgcctccaa caatcgaccc 1260
tggtccgacg ttcccttcga gttccttatc gagaacgtcc agcatctcac tcctcggtac 1320
tattccattt cttcgtcctc tctcagcgag aagcagacca tcaacgttac tgctgtggtc 1380
gaagccgagg aagaggccga tggacgaccc gttactggtg tcgttaccaa cctgctcaag 1440
aacatcgaga ttgaacagaa caagactggc gagacaccaa tggtccacta cgacctcaat 1500
ggtcccagag gcaagttctc caagtttcga ctgcccgtgc acgtcagacg atccaacttc 1560
aaacttccca agaactctac tacccctgtc atcctgattg gtccaggcac cggtgttgct 1620
cccctgcgag gctttgtccg ggagcgagtg cagcaagtca agaacggagt caacgttggt 1680
aagactgtgc tgttctacgg ctgtcgaaat tccgaacagg actttctcta caaacaggag 1740
tggagcgagt atgcctccgt cctgggagag aacttcgaaa tgttcaacgc cttttctcga 1800
caggacccta ccaagaaagt gtacgttcaa gacaagatcc tcgagaactc tgctcttgtc 1860
gacgagctcc tgtccagcgg tgcaattatc tacgtttgcg gagatgcctc tcgaatggct 1920
cgagacgtgc aggctgcaat tgccaagatc gttgccaagt cccgagacat ccacgaggac 1980
aaggctgccg agctggtcaa gtcttggaag gtgcagaacc gataccagga ggatgtctgg 2040
taa 2043
<210> 86
<211> 680
<212> PRT
<213> Candida tropicalis
<400> 86
Met Ala Leu Asp Lys Leu Asp Leu Tyr Val Ile Ile Thr Leu Val Val
1 5 10 15
Ala Ile Ala Ala Tyr Phe Ala Lys Asn Gln Phe Leu Asp Gln Gln Gln
20 25 30
Asp Thr Gly Phe Leu Asn Thr Asp Ser Gly Asp Gly Asn Ser Arg Asp
35 40 45
Ile Leu Gln Ala Leu Lys Lys Asn Asn Lys Asn Thr Leu Leu Leu Phe
50 55 60
Gly Ser Gln Thr Gly Thr Ala Glu Asp Tyr Ala Asn Lys Leu Ser Arg
65 70 75 80
Glu Leu His Ser Arg Phe Gly Leu Lys Thr Met Val Ala Asp Phe Ala
85 90 95
Asp Tyr Asp Phe Glu Asn Phe Gly Asp Ile Thr Glu Asp Ile Leu Val
100 105 110
Phe Phe Ile Val Ala Thr Tyr Gly Glu Gly Glu Pro Thr Asp Asn Ala
115 120 125
Asp Glu Phe His Thr Trp Leu Thr Glu Glu Ala Asp Thr Leu Ser Thr
130 135 140
Leu Lys Tyr Thr Val Phe Gly Leu Gly Asn Ser Thr Tyr Glu Phe Phe
145 150 155 160
Asn Ala Ile Gly Arg Lys Phe Asp Arg Leu Leu Gly Glu Lys Gly Gly
165 170 175
Asp Arg Phe Ala Glu Tyr Gly Glu Gly Asp Asp Gly Thr Gly Thr Leu
180 185 190
Asp Glu Asp Phe Leu Ala Trp Lys Asp Asn Val Phe Asp Ser Leu Lys
195 200 205
Asn Asp Leu Asn Phe Glu Glu Lys Glu Leu Lys Tyr Glu Pro Asn Val
210 215 220
Lys Leu Thr Glu Arg Asp Asp Leu Ser Gly Asn Asp Pro Asp Val Ser
225 230 235 240
Leu Gly Glu Pro Asn Val Lys Tyr Ile Lys Ser Glu Gly Val Asp Leu
245 250 255
Thr Lys Gly Pro Phe Asp His Thr His Pro Phe Leu Ala Arg Ile Val
260 265 270
Lys Thr Lys Glu Leu Phe Thr Ser Glu Asp Arg His Cys Val His Val
275 280 285
Glu Phe Asp Ile Ser Glu Ser Asn Leu Lys Tyr Thr Thr Gly Asp His
290 295 300
Leu Ala Ile Trp Pro Ser Asn Ser Asp Glu Asn Ile Lys Gln Phe Ala
305 310 315 320
Lys Cys Phe Gly Leu Glu Asp Lys Leu Asp Thr Val Ile Glu Leu Lys
325 330 335
Ala Leu Asp Ser Thr Tyr Ser Ile Pro Phe Pro Asn Pro Ile Thr Tyr
340 345 350
Gly Ala Val Ile Arg His His Leu Glu Ile Ser Gly Pro Val Ser Arg
355 360 365
Gln Phe Phe Leu Ser Ile Ala Gly Phe Ala Pro Asp Glu Glu Thr Lys
370 375 380
Lys Ser Phe Thr Arg Ile Gly Gly Asp Lys Gln Glu Phe Ala Ser Lys
385 390 395 400
Val Thr Arg Arg Lys Phe Asn Ile Ala Asp Ala Leu Leu Phe Ala Ser
405 410 415
Asn Asn Arg Pro Trp Ser Asp Val Pro Phe Glu Phe Leu Ile Glu Asn
420 425 430
Val Gln His Leu Thr Pro Arg Tyr Tyr Ser Ile Ser Ser Ser Ser Leu
435 440 445
Ser Glu Lys Gln Thr Ile Asn Val Thr Ala Val Val Glu Ala Glu Glu
450 455 460
Glu Ala Asp Gly Arg Pro Val Thr Gly Val Val Thr Asn Leu Leu Lys
465 470 475 480
Asn Ile Glu Ile Glu Gln Asn Lys Thr Gly Glu Thr Pro Met Val His
485 490 495
Tyr Asp Leu Asn Gly Pro Arg Gly Lys Phe Ser Lys Phe Arg Leu Pro
500 505 510
Val His Val Arg Arg Ser Asn Phe Lys Leu Pro Lys Asn Ser Thr Thr
515 520 525
Pro Val Ile Leu Ile Gly Pro Gly Thr Gly Val Ala Pro Leu Arg Gly
530 535 540
Phe Val Arg Glu Arg Val Gln Gln Val Lys Asn Gly Val Asn Val Gly
545 550 555 560
Lys Thr Val Leu Phe Tyr Gly Cys Arg Asn Ser Glu Gln Asp Phe Leu
565 570 575
Tyr Lys Gln Glu Trp Ser Glu Tyr Ala Ser Val Leu Gly Glu Asn Phe
580 585 590
Glu Met Phe Asn Ala Phe Ser Arg Gln Asp Pro Thr Lys Lys Val Tyr
595 600 605
Val Gln Asp Lys Ile Leu Glu Asn Ser Ala Leu Val Asp Glu Leu Leu
610 615 620
Ser Ser Gly Ala Ile Ile Tyr Val Cys Gly Asp Ala Ser Arg Met Ala
625 630 635 640
Arg Asp Val Gln Ala Ala Ile Ala Lys Ile Val Ala Lys Ser Arg Asp
645 650 655
Ile His Glu Asp Lys Ala Ala Glu Leu Val Lys Ser Trp Lys Val Gln
660 665 670
Asn Arg Tyr Gln Glu Asp Val Trp
675 680
<210> 87
<211> 12573
<212> DNA
<213> Artificial sequence
<220>
<223> pZKADn-C2F1U plasmid
<400> 87
cgatagtgta cgcagtacta tagaggaaca attgccccgg agaagacggc caggccgcct 60
agatgacaaa ttcaacaact cacagctgac tttctgccat tgccactagg ggggggcctt 120
tttatatggc caagccaagc tctccacgtc ggttgggctg cacccaacaa taaatgggta 180
gggttgcacc aacaaaggga tgggatgggg ggtagaagat acgaggataa cggggctcaa 240
tggcacaaat aagaacgaat actgccatta agactcgtga tccagcgact gacaccattg 300
catcatctaa gggcctcaaa actacctcgg aactgctgcg ctgatctgga caccacagag 360
gttccgagca ctttaggttg caccaaatgt cccaccaggt gcaggcagaa aacgctggaa 420
cagcgtgtac agtttgtctt aacaaaaagt gagggcgctg aggtcgagca gggtggtgtg 480
acttgttata gcctttagag ctgcgaaagc gcgtatggat ttggctcatc aggccagatt 540
gagggtctgt ggacacatgt catgttagtg tacttcaatc gccccctgga tatagccccg 600
acaataggcc gtggcctcat ttttttgcct tccgcacatt tccattgctc ggtacccaca 660
ccttgcttct cctgcacttg ccaaccttaa tactggttta cattgaccaa catcttacaa 720
gcggggggct tgtctagggt atatataaac agtggctctc ccaatcggtt gccagtctct 780
tttttccttt ctttccccac agattcgaaa tctaaactac acatcacacc atggcctccc 840
accaggtcga ggaccacgat ctggacgtgt tctgcctcct ggccgacgct gttctccacg 900
agattcctcc ctccgaaatc gtcgagtacc ttcatcccga tttccccaag gacaagatcg 960
aagagtacct gaccggcttt tctcgaccct ccgccgttcc tcagttccga cagtgtgcca 1020
agaaactcat caaccgaggt tccgagctgt cgatcaagct cttcctttac ttgaccactg 1080
ctctggactc tcgaatcctt gcaccagccc tgaccaactc gctcactctg atcagagaca 1140
tggatctttc ccagcgagag gaactgttgc ggtcctggcg agattctcca ctgactgcca 1200
agcgaagact ctttcgagtc tacgcctcct ttaccctgtc tacattcaac aagctcggaa 1260
ccgacttgca cttcaaggcc ctgggctacc ctggtcgaga gctccggacc cagattcaag 1320
actacgaggt cgatcccttt cgatactcgt tcatggagaa gctcaaacac gagggacatg 1380
aactgttcct tcccgatatc gacgttctga tcattggctc tggatccggt gcaggcgtgg 1440
tcgctcagac tcttaccgag agcggactca agtctctggt tctcgagaag ggcaagtact 1500
ttgcctccga agagctgtgc atgaccgatc tcgacggaaa cgaggccctg ttcgaaagcg 1560
gtggcactat tccttccacc aatcaacagt tgttcatgat cgctggatct acttttggtg 1620
gaggctccac cgtcaactgg tctgcctgtc tcaagactcc cttcaaggtt cgaaaggagt 1680
ggtacgacga tttcggactg gactttgtgg ctacccagca atacgacgat tgcatggact 1740
acgtgtggaa gaaaatgggt gcctcgaccg agcacatcga gcattctgct gcaaatgccg 1800
tcatcatgga cggagctgcc aagcttggct acgctcaccg agccctcgag cagaacaccg 1860
gtggccatgt tcacgactgt ggaatgtgcc acctgggctg tcgattcggt atcaagcagg 1920
gaggcgtcaa ctgctggttt cgagaaccct ccgagaaggg ttccaagttc atggagcagg 1980
tcgttgtcga gaagattctg cagcacaagg gcaaggccac tggaattctc tgcagagata 2040
ccgagtctgg catcaagttc aagattactg gacccaagaa atacgtcgtg tccggtggct 2100
ctttgcagac ccctgttctc cttcagaagt ctggcttcaa gaacaagcac attggagcca 2160
acctcaagct gcatcccgtc tcggttgctc ttggcgactt tggtaacgag gtggacttcg 2220
aagcctacaa gcgacccctc atgaccgcag tctgcaatgc cgtggacgat ctggacggca 2280
aggctcacgg aacacgaatc gaggccattc tgcacgctcc ttacgtcact gctcccttct 2340
atccctggca gtccggtgcc caggctcgga agaacttgct caaatacaag cagaccgtgc 2400
ctctgctcct tctgtctcga gacacctcct cgggtaccgt tacatacgac aaacagaagc 2460
cagatgtctt ggtggtcgac tacactgtca acaagttcga tcgaaactcc atcctgcagg 2520
gatttctcgt tgcttccgac attttgtaca tcgagggtgc caaggagatt ctgtctcccc 2580
aggcttgggt gcccaccttc aagagcaaca agcccaagca cgccagatcc atcaaggacg 2640
aggactacgt caagtggcga gaaaccgtgg ccaagattcc ctttgattcc tacggctcgc 2700
cttacggttc tgctcatcag atgtcctcgt gtcgaatgtc tggcaaggga cccggatacg 2760
gtgcctgcga taccaaggga cgactcttcg agtgcaacaa tgtgtacgtt gcagacgcct 2820
ccgtcatgcc tactgcttct ggagtcaacc ccatgattac cacaatggcc tttgcacgac 2880
acgttgctct ctgtctggcc aaggaccttc aaccccagac caagctgtaa gcggccgcat 2940
ggagcgtgtg ttctgagtcg atgttttcta tggagttgtg agtgttagta gacatgatgg 3000
gtttatatat gatgaatgaa tagatgtgat tttgatttgc acgatggaat tgagaacttt 3060
gtaaacgtac atgggaatgt atgaatgtgg gggttttgtg actggataac tgacggtcag 3120
tggacgccgt tgttcaaata tccaagagat gcgagaaact ttgggtcaag tgaacatgtc 3180
ctctctgttc aagtaaacca tcaactatgg gtagtatatt tagtaaggac aagagttgag 3240
attctttgga gtcctagaaa cgtattttcg cgttccaaga tcaaattagt agagtaatac 3300
gggcacggga atccattcat agtctcaagt ttaaaccacg ccaactgata tccccttacg 3360
ttaccccctc atcacctggt gaggcaaaac tgtaaggtga aagctaaaaa tgacatctca 3420
gctgcacgaa ggaccggggc ttaaaagacg ggctggtgct tgtgatttaa aactggacaa 3480
atctcagctt gcttgaaatt ttggtctcca actgtttccg agcgaatcgc acacaaaccg 3540
ggcttctctc tgcagaccac gcccccgaaa ctctttctcc caccaccacc aacactccct 3600
ttccattccc acaccgttcc tctctcatcc ttgcgcaatc atcttcgtct gcgacatatt 3660
gtacgacata cagtaccacg gaacgtttca gaccgtcgac gtgaacacat cttaggaaca 3720
gcaacctgag ctacagaaat ctatctatag gcggataaaa aaacgcaccc actgctcgtc 3780
ctccttgctc ctcgaaaccg actcctctac acacgtcaaa tccgaggttg aaatcttccc 3840
cacatttggc agccaaacca gcacatccca gcaacctcgc acagcgccga aatcgacctg 3900
tcgacttggc cacaaaaaaa agcaccggct ctgcaacagt tctcacgacc aattacgtac 3960
aagtacgaaa tcgttcgtgg accgtgactg ataagctccc actttttctt ctaacaacag 4020
gcaacagaca agtcacacaa aacaaaagcc atggccaagt cctacaagct gcccaagcct 4080
tccaagatcg ctcccatcat tcgaggcaag acctctgcca agaccaaagg ctccactcag 4140
cctcccgagt ctccacctgc ctcggctaag atcacagctc cccagctcga acccgtcgag 4200
cccaccagcg actccgagat tccttctacc aaggtctccg ttcgacgtac atcgaccact 4260
tccagcaaga ccatcacgga cgattccatc tctgccactt ccaccgacca gatgaagtcg 4320
agcaccaacg aagccgagat tccaaacccc aagccagagt ccgtggttgc ccctatgacc 4380
aagcccgtcg aggacgataa actcgaggac cacaccaagc tggagactgc cgaatcgtac 4440
atcaacgttc agaaggaagc tgcaattcct ggcgagacca agagcgtcgt ttcctcgaag 4500
actgcttctg tgctcgagta cacacctctt tccgagatct ctggcggagt caagaaagtg 4560
gtcgacggtt tccacaccgg caagacgcat cccctggagt tcagactcaa gcagcttcga 4620
aacctgtact ttgctatgaa ggacaatcag gaggccatct gcgaagcgct tgccaaggac 4680
tttcaccgag ccccttccga gactcgaaac tacgagctgg tcacaggtct caacgagctg 4740
ctctacacca tgactcaact gcacaagtgg tccaagcccc ttcctgtgga cgcgctgccc 4800
atcaacctca agaccaatcc cgtctacatc gagcggattc cagtcggaac cgttctcgtc 4860
atttctgcct tcaactatcc cttctttgtc tccgtgtctc ccatcgcagg tgctattgcc 4920
gcaggcaact ccgtcgtgtt caagccgtcg gagcttacac cccactttac caagctgttc 4980
acagagttgc tcaccaaggc tctggatccc gagatcttct acgtggtcaa cggtgccgtt 5040
tccgagacta ccgaactgct caaccagaag ttcgacaaga tcgtctacac tggcagcgac 5100
attgtcggca agatcattgc caagaaagca gcggagaccc ttactccagt catcttggag 5160
ctcggtggca agtctcctgc tttcgtgctg gacgatgtct cggacaagga tcttcccgtc 5220
atcgctcgac gtatcgcctg gggacgatac gccaacgctg gtcaaacctg cattggcgtc 5280
gactacgttc tcgtggccga gtccaagcac gagaagttca ttcaggctct gcggaatgtc 5340
atcgaaaacg agttctttcc caacatcgac cagaactcca actttaccca catgatccac 5400
gagcgagcct tcctcaagat gaaaaagatc ctggatacca ctgccggaga gatcattatc 5460
ggtggcaagc tcgacagcga gtccaactac gtgtctccca ccgtcatcga caatgcttcg 5520
tgggacgatt cctcgatgaa ggaggaaatc ttcggtccta ttcttcccat cattacttac 5580
accgacctca agcaggcctg caacgaggtc atttctcatc atgacactcc ccttgctcag 5640
tacatcttca cgtctggctc cacctcgcga aagtacaact ctcagatcaa cacaatctcc 5700
accatgattc gatcgggtgg actggtcatc aacgacgttc tcatgcatat ctcccttcat 5760
aacgctccct tcggtggcgt gggaaagtcc ggctacggtg cctatcacgg agagttctcc 5820
tacagagcct ttacccacga gcgaaccgtc ctcgagcagc atctgtggaa cgattggatt 5880
atcaactctc ggtatcctcc ctactccaac aagaaagaac gactggtggc ctccagccag 5940
tccaactacg gtggcagagt ctggtttggt cgaaagggcg acgttcgaat cgagggaccc 6000
actaccttct tcagcgcctg gaccaacgtg ctcggcgttg ctgccgtcgt tcgagacttc 6060
atcggtgctt ccatgtaagc ggccgcatga gaagataaat atataaatac attgagatat 6120
taaatgcgct agattagaga gcctcatact gctcggagag aagccaagac gagtactcaa 6180
aggggattac accatccata tccacagaca caagctgggg aaaggttcta tatacacttt 6240
ccggaatacc gtagtttccg atgttatcaa tgggggcagc caggatttca ggcacttcgg 6300
tgtctcgggg tgaaatggcg ttcttggcct ccatcaagtc gtaccatgtc ttcatttgcc 6360
tgtcaaagta aaacagaagc agatgaagaa tgaacttgaa gtgaaggaat ttaaatgtaa 6420
cgaaactgaa atttgaccag atattgtgtc cgcggtggag ctccagcttt tgttcccttt 6480
agtgagggtt aatttcgagc ttggcgtaat catggtcata gctgtttcct gtgtgaaatt 6540
gttatccgct cacaagcttc cacacaacgt acgaacgcac ggtatcggag catcggatac 6600
cccacattga gccaacctac tttgtagtgt acatactgta gagaagaggg acgcttcgac 6660
atgattggcc gatgtgggca tgtagaaaca cgatatatgg tgcttactaa tggacacttg 6720
cacaaccatt tctcttctag ggtaacctcg acagtgacag ccgttttttc tgcgctagcg 6780
tgtcgtcgta tttttggttt cgacatgtta agatttgtgg ggcaatcgag cgacattaag 6840
gtgcatcgga tcatcggccc aagggagagt cactggagtc tcgtagggtg gaggaaaaga 6900
gcaatttggg acgatttggg gcgatttgaa gacggattgg ggcaggtgtt tgtcacgtga 6960
ctgtggtatt actattacta atcgtcattg ttcgaaagtc ctgtcaattg tatcactttg 7020
gtgggtctac caaaacactg gtcaaatcta cgccacatga aaatataaag tttcacatta 7080
gccacattga ggggtaccct tagttggaat ctacaaggag ggatgcagtg aaaaatgttc 7140
ctttgatcct tcagagatga aaatgccatt gaccaatcac agcgggttta aagagtggcg 7200
aaaagagccc cttttttgca ccggttggcc cagcagccac gtgactggcc ccttccccat 7260
cccactcaac tgttgaggag gtgggatgcc aagatgcacc gtcaatgtac ttccgtgtat 7320
ccttctgcaa ttgatccgag ataggcgcgc cagctgcatt aatgaatcgg ccaacgcgcg 7380
gggagaggcg gtttgcgtat tgggcgctct tccgcttcct cgctcactga ctcgctgcgc 7440
tcggtcgttc ggctgcggcg agcggtatca gctcactcaa aggcggtaat acggttatcc 7500
acagaatcag gggataacgc aggaaagaac atgtgagcaa aaggccagca aaaggccagg 7560
aaccgtaaaa aggccgcgtt gctggcgttt ttccataggc tccgcccccc tgacgagcat 7620
cacaaaaatc gacgctcaag tcagaggtgg cgaaacccga caggactata aagataccag 7680
gcgtttcccc ctggaagctc cctcgtgcgc tctcctgttc cgaccctgcc gcttaccgga 7740
tacctgtccg cctttctccc ttcgggaagc gtggcgcttt ctcatagctc acgctgtagg 7800
tatctcagtt cggtgtaggt cgttcgctcc aagctgggct gtgtgcacga accccccgtt 7860
cagcccgacc gctgcgcctt atccggtaac tatcgtcttg agtccaaccc ggtaagacac 7920
gacttatcgc cactggcagc agccactggt aacaggatta gcagagcgag gtatgtaggc 7980
ggtgctacag agttcttgaa gtggtggcct aactacggct acactagaag aacagtattt 8040
ggtatctgcg ctctgctgaa gccagttacc ttcggaaaaa gagttggtag ctcttgatcc 8100
ggcaaacaaa ccaccgctgg tagcggtggt ttttttgttt gcaagcagca gattacgcgc 8160
agaaaaaaag gatctcaaga agatcctttg atcttttcta cggggtctga cgctcagtgg 8220
aacgaaaact cacgttaagg gattttggtc atgagattat caaaaaggat cttcacctag 8280
atccttttaa attaaaaatg aagttttaaa tcaatctaaa gtatatatga gtaaacttgg 8340
tctgacagtt accaatgctt aatcagtgag gcacctatct cagcgatctg tctatttcgt 8400
tcatccatag ttgcctgact ccccgtcgtg tagataacta cgatacggga gggcttacca 8460
tctggcccca gtgctgcaat gataccgcga gacccacgct caccggctcc agatttatca 8520
gcaataaacc agccagccgg aagggccgag cgcagaagtg gtcctgcaac tttatccgcc 8580
tccatccagt ctattaattg ttgccgggaa gctagagtaa gtagttcgcc agttaatagt 8640
ttgcgcaacg ttgttgccat tgctacaggc atcgtggtgt cacgctcgtc gtttggtatg 8700
gcttcattca gctccggttc ccaacgatca aggcgagtta catgatcccc catgttgtgc 8760
aaaaaagcgg ttagctcctt cggtcctccg atcgttgtca gaagtaagtt ggccgcagtg 8820
ttatcactca tggttatggc agcactgcat aattctctta ctgtcatgcc atccgtaaga 8880
tgcttttctg tgactggtga gtactcaacc aagtcattct gagaatagtg tatgcggcga 8940
ccgagttgct cttgcccggc gtcaatacgg gataataccg cgccacatag cagaacttta 9000
aaagtgctca tcattggaaa acgttcttcg gggcgaaaac tctcaaggat cttaccgctg 9060
ttgagatcca gttcgatgta acccactcgt gcacccaact gatcttcagc atcttttact 9120
ttcaccagcg tttctgggtg agcaaaaaca ggaaggcaaa atgccgcaaa aaagggaata 9180
agggcgacac ggaaatgttg aatactcata ctcttccttt ttcaatatta ttgaagcatt 9240
tatcagggtt attgtctcat gagcggatac atatttgaat gtatttagaa aaataaacaa 9300
ataggggttc cgcgcacatt tccccgaaaa gtgccacctg atgcggtgtg aaataccgca 9360
cagatgcgta aggagaaaat accgcatcag gaaattgtaa gcgttaatat tttgttaaaa 9420
ttcgcgttaa atttttgtta aatcagctca ttttttaacc aataggccga aatcggcaaa 9480
atcccttata aatcaaaaga atagaccgag atagggttga gtgttgttcc agtttggaac 9540
aagagtccac tattaaagaa cgtggactcc aacgtcaaag ggcgaaaaac cgtctatcag 9600
ggcgatggcc cactacgtga accatcaccc taatcaagtt ttttggggtc gaggtgccgt 9660
aaagcactaa atcggaaccc taaagggagc ccccgattta gagcttgacg gggaaagccg 9720
gcgaacgtgg cgagaaagga agggaagaaa gcgaaaggag cgggcgctag ggcgctggca 9780
agtgtagcgg tcacgctgcg cgtaaccacc acacccgccg cgcttaatgc gccgctacag 9840
ggcgcgtcca ttcgccattc aggctgcgca actgttggga agggcgatcg gtgcgggcct 9900
cttcgctatt acgccagctg gcgaaagggg gatgtgctgc aaggcgatta agttgggtaa 9960
cgccagggtt ttcccagtca cgacgttgta aaacgacggc cagtgaattg taatacgact 10020
cactataggg cgaattgggc ccgacgtcgc atgctacaag tatcgcacca tacttttgct 10080
gacggcgcgc cttcttgcag tgatataatc ggtttcttgg agctgatggg gtgagcatca 10140
tacaagtatg agtacgagaa gtcgcacttg tactccaagt acaaatgccc ggaatggcag 10200
acacacaagt cctacgggtg ttcagagact actgactgga gattgcaact acaagtactg 10260
tacacacagt acaacacaca agttaactca tcattcataa ttatcataaa ctagacggcc 10320
aaaaagtcgt ggccgctcct cagcgtcaat agccgcgctt acttggagca gtccagaacg 10380
tatcgaccgg caatcttgcc ctcctccatg agcttgtaga cggattcgag ctcggagaga 10440
ccaacaataa tgatggggga cttgaccagt cctcgggcaa agaactcaat ggcctcctgg 10500
gagtcggctc ggtttccgac gtaagagccc ttgatctgaa tagatcgagc aacctgctgg 10560
aagatgggcg acttgcagac ggcaccggcg ggcagaccga ccagaacaac ggttcccagg 10620
gttcgcacgt actcaacaga ctggttgacg gcaaactcgg agacagacac gttgatgacg 10680
gcgtggggtc cgcccttggt ggcctcctgg acgtccttga ccagatcctt ggacttggca 10740
aagtcgatga agacctcggc gccgagctcc ttgcacatct tctccttgtc agcgccagtg 10800
tcaatggcca gcactcggtt aattaacttt ggccggaatt cctttacctg caggataact 10860
tcgtataatg tatgctatac gaagttatga tctctctctt gagcttttcc ataacaagtt 10920
cttctgcctc caggaagtcc atgggtggtt tgatcatggt tttggtgtag tggtagtgca 10980
gtggtggtat tgtgactggg gatgtagttg agaataagtc atacacaagt cagctttctt 11040
cgagcctcat ataagtataa gtagttcaac gtattagcac tgtacccagc atctccgtat 11100
cgagaaacac aacaacatgc cccattggac agatcatgcg gatacacagg ttgtgcagta 11160
tcatacatac tcgatcagac aggtcgtctg accatcatac aagctgaaca agcgctccat 11220
acttgcacgc tctctatata cacagttaaa ttacatatcc atagtctaac ctctaacagt 11280
taatcttctg gtaagcctcc cagccagcct tctggtatcg cttggcctcc tcaataggat 11340
ctcggttctg gccgtacaga cctcggccga caattatgat atccgttccg gtagacatga 11400
catcctcaac agttcggtac tgctgtccga gagcgtctcc cttgtcgtca agacccaccc 11460
cgggggtcag aataagccag tcctcagagt cgcccttagg tcggttctgg gcaatgaagc 11520
caaccacaaa ctcggggtcg gatcgggcaa gctcaatggt ctgcttggag tactcgccag 11580
tggccagaga gcccttgcaa gacagctcgg ccagcatgag cagacctctg gccagcttct 11640
cgttgggaga ggggactagg aactccttgt actgggagtt ctcgtagtca gagacgtcct 11700
ccttcttctg ttcagagaca gtttcctcgg caccagctcg caggccagca atgattccgg 11760
ttccgggtac accgtgggcg ttggtgatat cggaccactc ggcgattcgg tgacaccggt 11820
actggtgctt gacagtgttg ccaatatctg cgaactttct gtcctcgaac aggaagaaac 11880
cgtgcttaag agcaagttcc ttgaggggga gcacagtgcc ggcgtaggtg aagtcgtcaa 11940
tgatgtcgat atgggttttg atcatgcaca cataaggtcc gaccttatcg gcaagctcaa 12000
tgagctcctt ggtggtggta acatccagag aagcacacag gttggttttc ttggctgcca 12060
cgagcttgag cactcgagcg gcaaaggcgg acttgtggac gttagctcga gcttcgtagg 12120
agggcatttt ggtggtgaag aggagactga aataaattta gtctgcagaa ctttttatcg 12180
gaaccttatc tggggcagtg aagtatatgt tatggtaata gttacgagtt agttgaactt 12240
atagatagac tggactatac ggctatcggt ccaaattaga aagaacgtca atggctctct 12300
gggcgtcgcc tttgccgaca aaaatgtgat catgatgaaa gccagcaatg acgttgcagc 12360
tgatattgtt gtcggccaac cgcgccgaaa acgcagctgt cagacccaca gcctccaacg 12420
aagaatgtat cgtcaaagtg atccaagcac actcatagtt ggagtcgtac tccaaaggcg 12480
gcaatgacga gtcagacaga tactcgtcga cgcgataact tcgtataatg tatgctatac 12540
gaagttatcg tacgatagtt agtagacaac aat 12573
<210> 88
<211> 2100
<212> DNA
<213> Artificial sequence
<220>
<223> CcFAO1s (FAO)
<400> 88
atggcctccc accaggtcga ggaccacgat ctggacgtgt tctgcctcct ggccgacgct 60
gttctccacg agattcctcc ctccgaaatc gtcgagtacc ttcatcccga tttccccaag 120
gacaagatcg aagagtacct gaccggcttt tctcgaccct ccgccgttcc tcagttccga 180
cagtgtgcca agaaactcat caaccgaggt tccgagctgt cgatcaagct cttcctttac 240
ttgaccactg ctctggactc tcgaatcctt gcaccagccc tgaccaactc gctcactctg 300
atcagagaca tggatctttc ccagcgagag gaactgttgc ggtcctggcg agattctcca 360
ctgactgcca agcgaagact ctttcgagtc tacgcctcct ttaccctgtc tacattcaac 420
aagctcggaa ccgacttgca cttcaaggcc ctgggctacc ctggtcgaga gctccggacc 480
cagattcaag actacgaggt cgatcccttt cgatactcgt tcatggagaa gctcaaacac 540
gagggacatg aactgttcct tcccgatatc gacgttctga tcattggctc tggatccggt 600
gcaggcgtgg tcgctcagac tcttaccgag agcggactca agtctctggt tctcgagaag 660
ggcaagtact ttgcctccga agagctgtgc atgaccgatc tcgacggaaa cgaggccctg 720
ttcgaaagcg gtggcactat tccttccacc aatcaacagt tgttcatgat cgctggatct 780
acttttggtg gaggctccac cgtcaactgg tctgcctgtc tcaagactcc cttcaaggtt 840
cgaaaggagt ggtacgacga tttcggactg gactttgtgg ctacccagca atacgacgat 900
tgcatggact acgtgtggaa gaaaatgggt gcctcgaccg agcacatcga gcattctgct 960
gcaaatgccg tcatcatgga cggagctgcc aagcttggct acgctcaccg agccctcgag 1020
cagaacaccg gtggccatgt tcacgactgt ggaatgtgcc acctgggctg tcgattcggt 1080
atcaagcagg gaggcgtcaa ctgctggttt cgagaaccct ccgagaaggg ttccaagttc 1140
atggagcagg tcgttgtcga gaagattctg cagcacaagg gcaaggccac tggaattctc 1200
tgcagagata ccgagtctgg catcaagttc aagattactg gacccaagaa atacgtcgtg 1260
tccggtggct ctttgcagac ccctgttctc cttcagaagt ctggcttcaa gaacaagcac 1320
attggagcca acctcaagct gcatcccgtc tcggttgctc ttggcgactt tggtaacgag 1380
gtggacttcg aagcctacaa gcgacccctc atgaccgcag tctgcaatgc cgtggacgat 1440
ctggacggca aggctcacgg aacacgaatc gaggccattc tgcacgctcc ttacgtcact 1500
gctcccttct atccctggca gtccggtgcc caggctcgga agaacttgct caaatacaag 1560
cagaccgtgc ctctgctcct tctgtctcga gacacctcct cgggtaccgt tacatacgac 1620
aaacagaagc cagatgtctt ggtggtcgac tacactgtca acaagttcga tcgaaactcc 1680
atcctgcagg gatttctcgt tgcttccgac attttgtaca tcgagggtgc caaggagatt 1740
ctgtctcccc aggcttgggt gcccaccttc aagagcaaca agcccaagca cgccagatcc 1800
atcaaggacg aggactacgt caagtggcga gaaaccgtgg ccaagattcc ctttgattcc 1860
tacggctcgc cttacggttc tgctcatcag atgtcctcgt gtcgaatgtc tggcaaggga 1920
cccggatacg gtgcctgcga taccaaggga cgactcttcg agtgcaacaa tgtgtacgtt 1980
gcagacgcct ccgtcatgcc tactgcttct ggagtcaacc ccatgattac cacaatggcc 2040
tttgcacgac acgttgctct ctgtctggcc aaggaccttc aaccccagac caagctgtaa 2100
<210> 89
<211> 699
<212> PRT
<213> Artificial sequence
<220>
<223> CcFAO1s (FAO) protein
<400> 89
Met Ala Ser His Gln Val Glu Asp His Asp Leu Asp Val Phe Cys Leu
1 5 10 15
Leu Ala Asp Ala Val Leu His Glu Ile Pro Pro Ser Glu Ile Val Glu
20 25 30
Tyr Leu His Pro Asp Phe Pro Lys Asp Lys Ile Glu Glu Tyr Leu Thr
35 40 45
Gly Phe Ser Arg Pro Ser Ala Val Pro Gln Phe Arg Gln Cys Ala Lys
50 55 60
Lys Leu Ile Asn Arg Gly Ser Glu Leu Ser Ile Lys Leu Phe Leu Tyr
65 70 75 80
Leu Thr Thr Ala Leu Asp Ser Arg Ile Leu Ala Pro Ala Leu Thr Asn
85 90 95
Ser Leu Thr Leu Ile Arg Asp Met Asp Leu Ser Gln Arg Glu Glu Leu
100 105 110
Leu Arg Ser Trp Arg Asp Ser Pro Leu Thr Ala Lys Arg Arg Leu Phe
115 120 125
Arg Val Tyr Ala Ser Phe Thr Leu Ser Thr Phe Asn Lys Leu Gly Thr
130 135 140
Asp Leu His Phe Lys Ala Leu Gly Tyr Pro Gly Arg Glu Leu Arg Thr
145 150 155 160
Gln Ile Gln Asp Tyr Glu Val Asp Pro Phe Arg Tyr Ser Phe Met Glu
165 170 175
Lys Leu Lys His Glu Gly His Glu Leu Phe Leu Pro Asp Ile Asp Val
180 185 190
Leu Ile Ile Gly Ser Gly Ser Gly Ala Gly Val Val Ala Gln Thr Leu
195 200 205
Thr Glu Ser Gly Leu Lys Ser Leu Val Leu Glu Lys Gly Lys Tyr Phe
210 215 220
Ala Ser Glu Glu Leu Cys Met Thr Asp Leu Asp Gly Asn Glu Ala Leu
225 230 235 240
Phe Glu Ser Gly Gly Thr Ile Pro Ser Thr Asn Gln Gln Leu Phe Met
245 250 255
Ile Ala Gly Ser Thr Phe Gly Gly Gly Ser Thr Val Asn Trp Ser Ala
260 265 270
Cys Leu Lys Thr Pro Phe Lys Val Arg Lys Glu Trp Tyr Asp Asp Phe
275 280 285
Gly Leu Asp Phe Val Ala Thr Gln Gln Tyr Asp Asp Cys Met Asp Tyr
290 295 300
Val Trp Lys Lys Met Gly Ala Ser Thr Glu His Ile Glu His Ser Ala
305 310 315 320
Ala Asn Ala Val Ile Met Asp Gly Ala Ala Lys Leu Gly Tyr Ala His
325 330 335
Arg Ala Leu Glu Gln Asn Thr Gly Gly His Val His Asp Cys Gly Met
340 345 350
Cys His Leu Gly Cys Arg Phe Gly Ile Lys Gln Gly Gly Val Asn Cys
355 360 365
Trp Phe Arg Glu Pro Ser Glu Lys Gly Ser Lys Phe Met Glu Gln Val
370 375 380
Val Val Glu Lys Ile Leu Gln His Lys Gly Lys Ala Thr Gly Ile Leu
385 390 395 400
Cys Arg Asp Thr Glu Ser Gly Ile Lys Phe Lys Ile Thr Gly Pro Lys
405 410 415
Lys Tyr Val Val Ser Gly Gly Ser Leu Gln Thr Pro Val Leu Leu Gln
420 425 430
Lys Ser Gly Phe Lys Asn Lys His Ile Gly Ala Asn Leu Lys Leu His
435 440 445
Pro Val Ser Val Ala Leu Gly Asp Phe Gly Asn Glu Val Asp Phe Glu
450 455 460
Ala Tyr Lys Arg Pro Leu Met Thr Ala Val Cys Asn Ala Val Asp Asp
465 470 475 480
Leu Asp Gly Lys Ala His Gly Thr Arg Ile Glu Ala Ile Leu His Ala
485 490 495
Pro Tyr Val Thr Ala Pro Phe Tyr Pro Trp Gln Ser Gly Ala Gln Ala
500 505 510
Arg Lys Asn Leu Leu Lys Tyr Lys Gln Thr Val Pro Leu Leu Leu Leu
515 520 525
Ser Arg Asp Thr Ser Ser Gly Thr Val Thr Tyr Asp Lys Gln Lys Pro
530 535 540
Asp Val Leu Val Val Asp Tyr Thr Val Asn Lys Phe Asp Arg Asn Ser
545 550 555 560
Ile Leu Gln Gly Phe Leu Val Ala Ser Asp Ile Leu Tyr Ile Glu Gly
565 570 575
Ala Lys Glu Ile Leu Ser Pro Gln Ala Trp Val Pro Thr Phe Lys Ser
580 585 590
Asn Lys Pro Lys His Ala Arg Ser Ile Lys Asp Glu Asp Tyr Val Lys
595 600 605
Trp Arg Glu Thr Val Ala Lys Ile Pro Phe Asp Ser Tyr Gly Ser Pro
610 615 620
Tyr Gly Ser Ala His Gln Met Ser Ser Cys Arg Met Ser Gly Lys Gly
625 630 635 640
Pro Gly Tyr Gly Ala Cys Asp Thr Lys Gly Arg Leu Phe Glu Cys Asn
645 650 655
Asn Val Tyr Val Ala Asp Ala Ser Val Met Pro Thr Ala Ser Gly Val
660 665 670
Asn Pro Met Ile Thr Thr Met Ala Phe Ala Arg His Val Ala Leu Cys
675 680 685
Leu Ala Lys Asp Leu Gln Pro Gln Thr Lys Leu
690 695
<210> 90
<211> 2028
<212> DNA
<213> Artificial sequence
<220>
<223> CtFALDH2s (FALDH)
<400> 90
atggccaagt cctacaagct gcccaagcct tccaagatcg ctcccatcat tcgaggcaag 60
acctctgcca agaccaaagg ctccactcag cctcccgagt ctccacctgc ctcggctaag 120
atcacagctc cccagctcga acccgtcgag cccaccagcg actccgagat tccttctacc 180
aaggtctccg ttcgacgtac atcgaccact tccagcaaga ccatcacgga cgattccatc 240
tctgccactt ccaccgacca gatgaagtcg agcaccaacg aagccgagat tccaaacccc 300
aagccagagt ccgtggttgc ccctatgacc aagcccgtcg aggacgataa actcgaggac 360
cacaccaagc tggagactgc cgaatcgtac atcaacgttc agaaggaagc tgcaattcct 420
ggcgagacca agagcgtcgt ttcctcgaag actgcttctg tgctcgagta cacacctctt 480
tccgagatct ctggcggagt caagaaagtg gtcgacggtt tccacaccgg caagacgcat 540
cccctggagt tcagactcaa gcagcttcga aacctgtact ttgctatgaa ggacaatcag 600
gaggccatct gcgaagcgct tgccaaggac tttcaccgag ccccttccga gactcgaaac 660
tacgagctgg tcacaggtct caacgagctg ctctacacca tgactcaact gcacaagtgg 720
tccaagcccc ttcctgtgga cgcgctgccc atcaacctca agaccaatcc cgtctacatc 780
gagcggattc cagtcggaac cgttctcgtc atttctgcct tcaactatcc cttctttgtc 840
tccgtgtctc ccatcgcagg tgctattgcc gcaggcaact ccgtcgtgtt caagccgtcg 900
gagcttacac cccactttac caagctgttc acagagttgc tcaccaaggc tctggatccc 960
gagatcttct acgtggtcaa cggtgccgtt tccgagacta ccgaactgct caaccagaag 1020
ttcgacaaga tcgtctacac tggcagcgac attgtcggca agatcattgc caagaaagca 1080
gcggagaccc ttactccagt catcttggag ctcggtggca agtctcctgc tttcgtgctg 1140
gacgatgtct cggacaagga tcttcccgtc atcgctcgac gtatcgcctg gggacgatac 1200
gccaacgctg gtcaaacctg cattggcgtc gactacgttc tcgtggccga gtccaagcac 1260
gagaagttca ttcaggctct gcggaatgtc atcgaaaacg agttctttcc caacatcgac 1320
cagaactcca actttaccca catgatccac gagcgagcct tcctcaagat gaaaaagatc 1380
ctggatacca ctgccggaga gatcattatc ggtggcaagc tcgacagcga gtccaactac 1440
gtgtctccca ccgtcatcga caatgcttcg tgggacgatt cctcgatgaa ggaggaaatc 1500
ttcggtccta ttcttcccat cattacttac accgacctca agcaggcctg caacgaggtc 1560
atttctcatc atgacactcc ccttgctcag tacatcttca cgtctggctc cacctcgcga 1620
aagtacaact ctcagatcaa cacaatctcc accatgattc gatcgggtgg actggtcatc 1680
aacgacgttc tcatgcatat ctcccttcat aacgctccct tcggtggcgt gggaaagtcc 1740
ggctacggtg cctatcacgg agagttctcc tacagagcct ttacccacga gcgaaccgtc 1800
ctcgagcagc atctgtggaa cgattggatt atcaactctc ggtatcctcc ctactccaac 1860
aagaaagaac gactggtggc ctccagccag tccaactacg gtggcagagt ctggtttggt 1920
cgaaagggcg acgttcgaat cgagggaccc actaccttct tcagcgcctg gaccaacgtg 1980
ctcggcgttg ctgccgtcgt tcgagacttc atcggtgctt ccatgtaa 2028
<210> 91
<211> 675
<212> PRT
<213> Candida tropicalis
<400> 91
Met Ala Lys Ser Tyr Lys Leu Pro Lys Pro Ser Lys Ile Ala Pro Ile
1 5 10 15
Ile Arg Gly Lys Thr Ser Ala Lys Thr Lys Gly Ser Thr Gln Pro Pro
20 25 30
Glu Ser Pro Pro Ala Ser Ala Lys Ile Thr Ala Pro Gln Leu Glu Pro
35 40 45
Val Glu Pro Thr Ser Asp Ser Glu Ile Pro Ser Thr Lys Val Ser Val
50 55 60
Arg Arg Thr Ser Thr Thr Ser Ser Lys Thr Ile Thr Asp Asp Ser Ile
65 70 75 80
Ser Ala Thr Ser Thr Asp Gln Met Lys Ser Ser Thr Asn Glu Ala Glu
85 90 95
Ile Pro Asn Pro Lys Pro Glu Ser Val Val Ala Pro Met Thr Lys Pro
100 105 110
Val Glu Asp Asp Lys Leu Glu Asp His Thr Lys Leu Glu Thr Ala Glu
115 120 125
Ser Tyr Ile Asn Val Gln Lys Glu Ala Ala Ile Pro Gly Glu Thr Lys
130 135 140
Ser Val Val Ser Ser Lys Thr Ala Ser Val Leu Glu Tyr Thr Pro Leu
145 150 155 160
Ser Glu Ile Ser Gly Gly Val Lys Lys Val Val Asp Gly Phe His Thr
165 170 175
Gly Lys Thr His Pro Leu Glu Phe Arg Leu Lys Gln Leu Arg Asn Leu
180 185 190
Tyr Phe Ala Met Lys Asp Asn Gln Glu Ala Ile Cys Glu Ala Leu Ala
195 200 205
Lys Asp Phe His Arg Ala Pro Ser Glu Thr Arg Asn Tyr Glu Leu Val
210 215 220
Thr Gly Leu Asn Glu Leu Leu Tyr Thr Met Thr Gln Leu His Lys Trp
225 230 235 240
Ser Lys Pro Leu Pro Val Asp Ala Leu Pro Ile Asn Leu Lys Thr Asn
245 250 255
Pro Val Tyr Ile Glu Arg Ile Pro Val Gly Thr Val Leu Val Ile Ser
260 265 270
Ala Phe Asn Tyr Pro Phe Phe Val Ser Val Ser Pro Ile Ala Gly Ala
275 280 285
Ile Ala Ala Gly Asn Ser Val Val Phe Lys Pro Ser Glu Leu Thr Pro
290 295 300
His Phe Thr Lys Leu Phe Thr Glu Leu Leu Thr Lys Ala Leu Asp Pro
305 310 315 320
Glu Ile Phe Tyr Val Val Asn Gly Ala Val Ser Glu Thr Thr Glu Leu
325 330 335
Leu Asn Gln Lys Phe Asp Lys Ile Val Tyr Thr Gly Ser Asp Ile Val
340 345 350
Gly Lys Ile Ile Ala Lys Lys Ala Ala Glu Thr Leu Thr Pro Val Ile
355 360 365
Leu Glu Leu Gly Gly Lys Ser Pro Ala Phe Val Leu Asp Asp Val Ser
370 375 380
Asp Lys Asp Leu Pro Val Ile Ala Arg Arg Ile Ala Trp Gly Arg Tyr
385 390 395 400
Ala Asn Ala Gly Gln Thr Cys Ile Gly Val Asp Tyr Val Leu Val Ala
405 410 415
Glu Ser Lys His Glu Lys Phe Ile Gln Ala Leu Arg Asn Val Ile Glu
420 425 430
Asn Glu Phe Phe Pro Asn Ile Asp Gln Asn Ser Asn Phe Thr His Met
435 440 445
Ile His Glu Arg Ala Phe Leu Lys Met Lys Lys Ile Leu Asp Thr Thr
450 455 460
Ala Gly Glu Ile Ile Ile Gly Gly Lys Leu Asp Ser Glu Ser Asn Tyr
465 470 475 480
Val Ser Pro Thr Val Ile Asp Asn Ala Ser Trp Asp Asp Ser Ser Met
485 490 495
Lys Glu Glu Ile Phe Gly Pro Ile Leu Pro Ile Ile Thr Tyr Thr Asp
500 505 510
Leu Lys Gln Ala Cys Asn Glu Val Ile Ser His His Asp Thr Pro Leu
515 520 525
Ala Gln Tyr Ile Phe Thr Ser Gly Ser Thr Ser Arg Lys Tyr Asn Ser
530 535 540
Gln Ile Asn Thr Ile Ser Thr Met Ile Arg Ser Gly Gly Leu Val Ile
545 550 555 560
Asn Asp Val Leu Met His Ile Ser Leu His Asn Ala Pro Phe Gly Gly
565 570 575
Val Gly Lys Ser Gly Tyr Gly Ala Tyr His Gly Glu Phe Ser Tyr Arg
580 585 590
Ala Phe Thr His Glu Arg Thr Val Leu Glu Gln His Leu Trp Asn Asp
595 600 605
Trp Ile Ile Asn Ser Arg Tyr Pro Pro Tyr Ser Asn Lys Lys Glu Arg
610 615 620
Leu Val Ala Ser Ser Gln Ser Asn Tyr Gly Gly Arg Val Trp Phe Gly
625 630 635 640
Arg Lys Gly Asp Val Arg Ile Glu Gly Pro Thr Thr Phe Phe Ser Ala
645 650 655
Trp Thr Asn Val Leu Gly Val Ala Ala Val Val Arg Asp Phe Ile Gly
660 665 670
Ala Ser Met
675
<210> 92
<211> 12572
<212> DNA
<213> Artificial sequence
<220>
<223> pYRH213 plasmid
<400> 92
aaaccatcat ctaagggcct caaaactacc tcggaactgc tgcgctgatc tggacaccac 60
agaggttccg agcactttag gttgcaccaa atgtcccacc aggtgcaggc agaaaacgct 120
ggaacagcgt gtacagtttg tcttaacaaa aagtgagggc gctgaggtcg agcagggtgg 180
tgtgacttgt tatagccttt agagctgcga aagcgcgtat ggatttggct catcaggcca 240
gattgagggt ctgtggacac atgtcatgtt agtgtacttc aatcgccccc tggatatagc 300
cccgacaata ggccgtggcc tcattttttt gccttccgca catttccatt gctcggtacc 360
cacaccttgc ttctcctgca cttgccaacc ttaatactgg tttacattga ccaacatctt 420
acaagcgggg ggcttgtcta gggtatatat aaacagtggc tctcccaatc ggttgccagt 480
ctcttttttc ctttctttcc ccacagattc gaaatctaaa ctacacatca caccatggcc 540
acctcctcta actccgacct ggtccgaacc atcgagtccg ccctcggcat ttctctcggc 600
gacagcgtgt ccgattctgt cgttatcatt gccaccactt ctgctgccgt catcattgga 660
cttctggtct tcctctggcg aaagtctccc gacagatcgc gagagctgcg tcctgtcatt 720
gtgcccaagt ttaccgttaa gcacgaggac gatgaagtcg aggtggaccg aggcaaaacc 780
aaggttacag ttttctacgg aactcagacc ggtactgccg agggctttgc aaaggccctt 840
gcggaggaaa tcaaggccag atacgagaag gccgttgtca aggtggttga catggatgac 900
tacgctattg acgatgacca gtacgaggaa aagctcaaaa aggagaccct ggtcttcttt 960
atgcttgcca cctatggaga cggcgaaccc accgataacg ctgcacgatt ctacaagtgg 1020
tttaccgagg gcaaggagga acgaggaacc tggctgcagc aactcactta cggtgtgttc 1080
gccttgggca accgacagta cgagcatttc aacaagatcg gcaagattgt cgacgaggat 1140
cttaccgagc agggagccaa gcgtctggtt cccgtcggtc tcggcgacga tgaccagtcc 1200
atcgaggacg atttcaacgc ttggaaggaa accttgtggc cagagctgga ccaacttctc 1260
cgagacgagg atgacgtcaa cactgcttcg accccttaca ctgccgctat ctccgagtat 1320
cgtgtcgtta tccacgatcc taccgtgtct ccctcctacg agaaccattt caatgttgcc 1380
aacggtggag cagtgttcga cattcaccat ccctgtcgag tcaacgttgc cgtgcgacgg 1440
gaacttcaca agccccagtc cgaccgatct tgcattcacc tggagtttga tctctccggt 1500
actggcgtta catacgagac tggcgaccac gtcggagtgt acgccgagaa ctgcgacgaa 1560
actgtcgagg aagctggcaa gctgctcggt cagtcgctgg atcttctctt ttctctgcat 1620
accgacaagg aggatggcac aagccttggt ggatctctgc tccctccatt tcctggaccc 1680
tgtaccgttc gaactgccct cgcttgctac gccgaccttc ttaatcctcc acggaaagcc 1740
gctatcgtgg cacttgctgc ccatgcttcc gagcccagcg aggccgaacg actcaagttt 1800
ctttcttcgc ctcagggcaa ggacgagtac tccaagtggg tcgttggatc tcagcgatcg 1860
ctgctcgaag tcatggccga ttttccctcc gccaagcctc cactgggagt gttctttgct 1920
gccattgcac ctcgactgca gcctcgatac tattctatct cctcttcgcc cagaccagct 1980
ccccagcgag tgcacgttac ctgtgccctt gtcgagggac ccactcctac cggtcggatt 2040
cacaagggtg tgtgctccac ctggatgaag tctgctactc ccttggagaa gtctcacgac 2100
tgttcccgag cacctatctt cattcgaccc tccaacttca agcttcctgc cgaccattcg 2160
attcccatta tcatggtcgg acctggtaca ggtctggctc cctttcgagg attcctccag 2220
gaacgacttg ccctcaagga ggatggagtt cagcttggac ctgccctgct cttctttggt 2280
tgccgaaaca gacagatgga cttcatctac gaggacgaac tcaacaattt cgttcagcaa 2340
ggtgccattt ccgagctcat cgttgcgttt tctcgagagg gcccagaaaa ggagtacgtg 2400
cagcacaaga tgatggacaa ggccgagtat ctgtggtctc tcatttcgca gggaggctac 2460
ctgtacgtct gtggtgatgc caaaggcatg gctcgagacg tgcaccgatc ccttcatacc 2520
attgttcagc aacaggagaa cgcagattct tcgaaggctg aggccactgt caagaaactc 2580
cagatggacg gaagatacct gcgagacgtg tggtaagcgg ccgcatgaga agataaatat 2640
ataaatacat tgagatatta aatgcgctag attagagagc ctcatactgc tcggagagaa 2700
gccaagacga gtactcaaag gggattacac catccatatc cacagacaca agctggggaa 2760
aggttctata tacactttcc ggaataccgt agtttccgat gttatcaatg ggggcagcca 2820
ggatttcagg cacttcggtg tctcggggtg aaatggcgtt cttggcctcc atcaagtcgt 2880
accatgtctt catttgcctg tcaaagtaaa acagaagcag atgaagaatg aacttgaagt 2940
gaaggaattt aaatgtaacg aaactgaaat ttgaccagat attgtgtccg cggtggagct 3000
ccagcttttg ttccctttag tgagggttaa tttcgagctt ggcgtaatca tggtcatagc 3060
tgtttcctgt gtgaaattgt tatccgctca caagcttcca cacaacgtac gttgattgag 3120
gtggagccag atgggctatt gtttcatata tagactggca gccacctctt tggcccagca 3180
tgtttgtata cctggaaggg aaaactaaag aagctggcta gtttagtttg attattatag 3240
tagatgtcct aatcactaga gattagaatg tcttggcgat gattagtcgt cgtcccctgt 3300
atcatgtcta gaccaactgt gtcatgaagt tggtgctggt gttttacctg tgtactacaa 3360
gtaggtgtcc tagatctagt gtacagagcc gtttagaccc atgtggactt caccattaac 3420
gatggaaaat gttcattata tgacagtata ttacaatgga cttgctccat ttcttccttg 3480
catcacatgt tctccacctc catagttgat caacacatca tagtagctaa ggctgctgct 3540
ctcccactac agtccaccac aagttaagta gcaccgtcag tacagctaaa agtacacgtc 3600
tagtacgttt cataactagt caagtagccc ctattacaga tatcagcact atcacgcacg 3660
agtttttctc tgtgctatct aatcaacttg ccaagtattc ggagaagata cactttcttg 3720
gcatcaggta tacgagggag cctatcagat gaaaaagggt atattggatc cattcatatc 3780
cacctacacg ttgtcataat ctcctcattc acgtgattca tttcgtgaca ctagtttctc 3840
actttccccc ccgcacctat agtcaacttg gcggacacgc tacttgtagc tgacgttgat 3900
ttatagaccc aatcaaagcg ggttatcggt caggtagcac ttatcattca tcgttcatac 3960
tacgatgagc aatctcgggc atgtccggaa aagtgtcggg cgcgccagct gcattaatga 4020
atcggccaac gcgcggggag aggcggtttg cgtattgggc gctcttccgc ttcctcgctc 4080
actgactcgc tgcgctcggt cgttcggctg cggcgagcgg tatcagctca ctcaaaggcg 4140
gtaatacggt tatccacaga atcaggggat aacgcaggaa agaacatgtg agcaaaaggc 4200
cagcaaaagg ccaggaaccg taaaaaggcc gcgttgctgg cgtttttcca taggctccgc 4260
ccccctgacg agcatcacaa aaatcgacgc tcaagtcaga ggtggcgaaa cccgacagga 4320
ctataaagat accaggcgtt tccccctgga agctccctcg tgcgctctcc tgttccgacc 4380
ctgccgctta ccggatacct gtccgccttt ctcccttcgg gaagcgtggc gctttctcat 4440
agctcacgct gtaggtatct cagttcggtg taggtcgttc gctccaagct gggctgtgtg 4500
cacgaacccc ccgttcagcc cgaccgctgc gccttatccg gtaactatcg tcttgagtcc 4560
aacccggtaa gacacgactt atcgccactg gcagcagcca ctggtaacag gattagcaga 4620
gcgaggtatg taggcggtgc tacagagttc ttgaagtggt ggcctaacta cggctacact 4680
agaagaacag tatttggtat ctgcgctctg ctgaagccag ttaccttcgg aaaaagagtt 4740
ggtagctctt gatccggcaa acaaaccacc gctggtagcg gtggtttttt tgtttgcaag 4800
cagcagatta cgcgcagaaa aaaaggatct caagaagatc ctttgatctt ttctacgggg 4860
tctgacgctc agtggaacga aaactcacgt taagggattt tggtcatgag attatcaaaa 4920
aggatcttca cctagatcct tttaaattaa aaatgaagtt ttaaatcaat ctaaagtata 4980
tatgagtaaa cttggtctga cagttaccaa tgcttaatca gtgaggcacc tatctcagcg 5040
atctgtctat ttcgttcatc catagttgcc tgactccccg tcgtgtagat aactacgata 5100
cgggagggct taccatctgg ccccagtgct gcaatgatac cgcgagaccc acgctcaccg 5160
gctccagatt tatcagcaat aaaccagcca gccggaaggg ccgagcgcag aagtggtcct 5220
gcaactttat ccgcctccat ccagtctatt aattgttgcc gggaagctag agtaagtagt 5280
tcgccagtta atagtttgcg caacgttgtt gccattgcta caggcatcgt ggtgtcacgc 5340
tcgtcgtttg gtatggcttc attcagctcc ggttcccaac gatcaaggcg agttacatga 5400
tcccccatgt tgtgcaaaaa agcggttagc tccttcggtc ctccgatcgt tgtcagaagt 5460
aagttggccg cagtgttatc actcatggtt atggcagcac tgcataattc tcttactgtc 5520
atgccatccg taagatgctt ttctgtgact ggtgagtact caaccaagtc attctgagaa 5580
tagtgtatgc ggcgaccgag ttgctcttgc ccggcgtcaa tacgggataa taccgcgcca 5640
catagcagaa ctttaaaagt gctcatcatt ggaaaacgtt cttcggggcg aaaactctca 5700
aggatcttac cgctgttgag atccagttcg atgtaaccca ctcgtgcacc caactgatct 5760
tcagcatctt ttactttcac cagcgtttct gggtgagcaa aaacaggaag gcaaaatgcc 5820
gcaaaaaagg gaataagggc gacacggaaa tgttgaatac tcatactctt cctttttcaa 5880
tattattgaa gcatttatca gggttattgt ctcatgagcg gatacatatt tgaatgtatt 5940
tagaaaaata aacaaatagg ggttccgcgc acatttcccc gaaaagtgcc acctgatgcg 6000
gtgtgaaata ccgcacagat gcgtaaggag aaaataccgc atcaggaaat tgtaagcgtt 6060
aatattttgt taaaattcgc gttaaatttt tgttaaatca gctcattttt taaccaatag 6120
gccgaaatcg gcaaaatccc ttataaatca aaagaataga ccgagatagg gttgagtgtt 6180
gttccagttt ggaacaagag tccactatta aagaacgtgg actccaacgt caaagggcga 6240
aaaaccgtct atcagggcga tggcccacta cgtgaaccat caccctaatc aagttttttg 6300
gggtcgaggt gccgtaaagc actaaatcgg aaccctaaag ggagcccccg atttagagct 6360
tgacggggaa agccggcgaa cgtggcgaga aaggaaggga agaaagcgaa aggagcgggc 6420
gctagggcgc tggcaagtgt agcggtcacg ctgcgcgtaa ccaccacacc cgccgcgctt 6480
aatgcgccgc tacagggcgc gtccattcgc cattcaggct gcgcaactgt tgggaagggc 6540
gatcggtgcg ggcctcttcg ctattacgcc agctggcgaa agggggatgt gctgcaaggc 6600
gattaagttg ggtaacgcca gggttttccc agtcacgacg ttgtaaaacg acggccagtg 6660
aattgtaata cgactcacta tagggcgaat tgggcccgac gtcgcatgca ttccgacagc 6720
agcgactggg caccatgatc aagcgaaaca ccttccccca gctgccctgg caaaccatca 6780
agaaccctac tttcatcaag tgcaagaacg gttctactct tctcacctcc ggtgtctacg 6840
gctggtgccg aaagcctaac tacaccgctg atttcatcat gtgcctcacc tgggctctca 6900
tgtgcggtgt tgcttctccc ctgccttact tctacccggt cttcttcttc ctggtgctca 6960
tccaccgagc ttaccgagac tttgagcgac tggagcgaaa gtacggtgag gactaccagg 7020
agttcaagcg acaggtccct tggatcttca tcccttatgt tttctaaacg ataagcttag 7080
tgagcgaatg gtgaggttac ttaattgagt ggccagccta tgggattgta taacagacag 7140
tcaatatatt actgaaaaga ctgaacagcc agacggagtg aggttgtgag tgaatcgtag 7200
agggcggcta ttacagcaag tctactctac agtgtactaa cacagcagag aacaaataca 7260
ggtgtgcatt cggctatctg agaattagtt ggagagctcg agaccctcgg cgataaactg 7320
ctcctcggtt ttgtgtccat acttgtacgg accattgtaa tggggcaagt cgttgagttc 7380
tcgtcgtccg acgttcagag cacagaaacc aatgtaatca atgtagcaga gatggttctg 7440
caaaagattg atttgtgcga gcaggttaat taagttgcga cacatgtctt gatagtatct 7500
tgaattctct ctcttgagct tttccataac aagttcttct gcctccagga agtccatggg 7560
tggtttgatc atggttttgg tgtagtggta gtgcagtggt ggtattgtga ctggggatgt 7620
agttgagaat aagtcataca caagtcagct ttcttcgagc ctcatataag tataagtagt 7680
tcaacgtatt agcactgtac ccagcatctc cgtatcgaga aacacaacaa catgccccat 7740
tggacagatc atgcggatac acaggttgtg cagtatcata catactcgat cagacaggtc 7800
gtctgaccat catacaagct gaacaagcgc tccatacttg cacgctctct atatacacag 7860
ttaaattaca tatccatagt ctaacctcta acagttaatc ttctggtaag cctcccagcc 7920
agccttctgg tatcgcttgg cctcctcaat aggatctcgg ttctggccgt acagacctcg 7980
gccgacaatt atgatatccg ttccggtaga catgacatcc tcaacagttc ggtactgctg 8040
tccgagagcg tctcccttgt cgtcaagacc caccccgggg gtcagaataa gccagtcctc 8100
agagtcgccc ttaggtcggt tctgggcaat gaagccaacc acaaactcgg ggtcggatcg 8160
ggcaagctca atggtctgct tggagtactc gccagtggcc agagagccct tgcaagacag 8220
ctcggccagc atgagcagac ctctggccag cttctcgttg ggagagggga ctaggaactc 8280
cttgtactgg gagttctcgt agtcagagac gtcctccttc ttctgttcag agacagtttc 8340
ctcggcacca gctcgcaggc cagcaatgat tccggttccg ggtacaccgt gggcgttggt 8400
gatatcggac cactcggcga ttcggtgaca ccggtactgg tgcttgacag tgttgccaat 8460
atctgcgaac tttctgtcct cgaacaggaa gaaaccgtgc ttaagagcaa gttccttgag 8520
ggggagcaca gtgccggcgt aggtgaagtc gtcaatgatg tcgatatggg ttttgatcat 8580
gcacacataa ggtccgacct tatcggcaag ctcaatgagc tccttggtgg tggtaacatc 8640
cagagaagca cacaggttgg ttttcttggc tgccacgagc ttgagcactc gagcggcaaa 8700
ggcggacttg tggacgttag ctcgagcttc gtaggagggc attttggtgg tgaagaggag 8760
actgaaataa atttagtctg cagaactttt tatcggaacc ttatctgggg cagtgaagta 8820
tatgttatgg taatagttac gagttagttg aacttataga tagactggac tatacggcta 8880
tcggtccaaa ttagaaagaa cgtcaatggc tctctgggcg tcgcctttgc cgacaaaaat 8940
gtgatcatga tgaaagccag caatgacgtt gcagctgata ttgttgtcgg ccaaccgcgc 9000
cgaaaacgca gctgtcagac ccacagcctc caacgaagaa tgtatcgtca aagtgatcca 9060
agcacactca tagttggagt cgtactccaa aggcggcaat gacgagtcag acagatactc 9120
gtcgaccttt tccttgggaa ccaccaccgt cagcccttct gactcacgta ttgtagccac 9180
cgacacaggc aacagtccgt ggatagcaga atatgtcttg tcggtccatt tctcaccaac 9240
tttaggcgtc aagtgaatgt tgcagaagaa gtatgtgcct tcattgagaa tcggtgttgc 9300
tgatttcaat aaagtcttga gatcagtttg gccagtcatg ttgtgggggg taattggatt 9360
gagttatcgc ctacagtctg tacaggtata ctcgctgccc actttatact ttttgattcc 9420
gctgcacttg aagcaatgtc gtttaccaaa agtgagaatg ctccacagaa cacaccccag 9480
ggtatggttg agcaaaaaat aaacactccg atacggggaa tcgaaccccg gtctccacgg 9540
ttctcaagaa gtattcttga tgagagcgta tcgatgagcc taaaatgaac ccgagtatat 9600
ctcataaaat tctcggtgag aggtctgtga ctgtcagtac aaggtgcctt cattatgccc 9660
tcaaccttac catacctcac tgaatgtagt gtacctctaa aaatgaaata cagtgccaaa 9720
agccaaggca ctgagctcgt ctaacggact tgatatacaa ccaattaaaa caaatgaaaa 9780
gaaatacagt tctttgtatc atttgtaaca attaccctgt acaaactaag gtattgaaat 9840
cccacaatat tcccaaagtc cacccctttc caaattgtca tgcctacaac tcatatacca 9900
agcactaacc taccgtttaa acgagaatcg tacagagttg tttctgaacc atttcgaagc 9960
cgttcagagt cgtttaaccg cagtttgagt cgtttcagag cggttctcag tcgtttttga 10020
tccatctctg aatgattcag agccgcctaa ctccttgtga gccgttaaat cgcgaattga 10080
gccactctga gccgttgatt cttccatttg tggccccttc accatctcct catctccttc 10140
ctctcgtcgc catctccttt tctcctctcg cacctcgcac tactccaaac caactcactg 10200
actacatccc aacagcgatc taaccaacgc cgcgcaagca aacagacaca aaaacaacgt 10260
ccgcgccgcc gaccacgtca gcagatcccg ctctcctggg ttttgtcgtc gtttgccgcc 10320
ttttgccgcc gctatttgca acgccgccac actcaatggc cgagccatga tgccgtcgtc 10380
gtgtcttttc ccaaatagga aaccgggcac caaccctagc cccacaactg gagtactcaa 10440
ctcggcgaaa aatgggccaa tcgccgcggg agaaacgaca atcggcttgt tttgactcaa 10500
ttacctcacc aagcgcccct tcgtcgccgc catacctccg caacaccccc tcaccgtccc 10560
tccctccgcc cctctggaaa ctcatagaac cccaaacctt atttccgatg accgcaaact 10620
ttagactata caggcgaatc tgggtggtgg caccaaccct tcctcttctc catcaccccc 10680
ccctcaatct ctttttctca ccatggcctt ccagtttcac ctggaggtcc tcctgcccta 10740
cctccttcct ctgcttctgc tcatcctgcc caccactatc ttctttctca ccaagcccaa 10800
caataaggtg tcctctactt ccaccaacaa taacatcatt acactgccca agtcgtaccc 10860
tctcattggc tcctaccttt cgttccgaaa gaacctgcat cgacggatcc agtggctctc 10920
cgacattgtt cagatctctc cctccgctac cttccagctc gacggaaccc tgggcaagcg 10980
acagatcatt actggcaacc cttctaccgt ccagcacatt ctcaagaacc agttctccaa 11040
ctatcagaag ggcaccacat tcaccaacac tctgtccgac tttctcggaa caggcatctt 11100
caacaccaac ggtcccaact ggaagtttca acgacaggtt gcctctcacg agttcaacac 11160
caagtccatt cggaacttcg tcgagcacat cgtggatacc gaactcacca accgattgat 11220
tcccatcctc acttcgagca cccagacaaa caatatcctg gacttccagg atattctgca 11280
gcgatttacc ttcgacaaca tctgcaacat tgccttcgga tacgatcccg agtacctcac 11340
tccctcgacc aatcgttcca agttcgcgga ggcctacgaa gacgctaccg agatctccag 11400
caagcgattc agactgcctc ttcccatcat ttggaagatc aaaaagtact tcaacattgg 11460
ctccgagaag cgactcaagg aagccgtcac cgaggtccga tcctttgcca agaaactggt 11520
ccgagagaag aaacgggagc tcgaagagaa gtcttcgctg gagaccgaag acatgctttc 11580
tcgatttctg tccagcggtc actcggacga ggatttcgtt gccgacattg tcatctcctt 11640
cattctcgca ggcaaggaca ctacctctgc cgctcttacc tggtttttct ggctgctctg 11700
gaagaaccct cgagtggagg aagagatcgt caacgagctg tccaagaaat cggagcttat 11760
ggtgtacgac gaggtcaagg aaatggtcta cacccatgct gcgctgtccg agtcgatgag 11820
actctaccct cccgttccaa tggattccaa ggaggccgtc aacgacgatg tgctgcccga 11880
cggctgggtg gtcaagaaag gtacaatcgt cacctaccat gtctacgcta tgggtcgaat 11940
gaagtctctc tggggagacg attgggcaga gtttcgacca gaacggtggc tcgagaagga 12000
cgaggtcaac ggcaagtggg tgttcgtcgg acgagacagc tactcctatc ctgtgttcca 12060
ggctggtccc agagtctgcc tgggaaagga gatggccttc atgcagatga agcgaattgt 12120
ggctggcatc gtcggaaagt tcaaggtggt tcccgaagcc cacttggctc aggagccagg 12180
attcatttcc tttctgtcgt ctcagatgga gggtggattt cccgtcacta tccagaagcg 12240
agactcctaa gcggccgcaa gtgtggatgg ggaagtgagt gcccggttct gtgtgcacaa 12300
ttggcaatcc aagatggatg gattcaacac agggatatag cgagctacgt ggtggtgcga 12360
ggatatagca acggatattt atgtttgaca cttgagaatg tacgatacaa gcactgtcca 12420
agtacaatac taaacatact gtacatactc atactcgtac ccgggcaacg gtttcacttg 12480
agtgcagtgg ctagtgctct tactcgtaca gtgtgcaata ctgcgtatca tagtctttga 12540
tgtatatcgt attcattcat gttagttgat tt 12572
<210> 93
<211> 1548
<212> DNA
<213> Artificial sequence
<220>
<223> VsCYP94A1s (CYP)
<400> 93
atggccttcc agtttcacct ggaggtcctc ctgccctacc tccttcctct gcttctgctc 60
atcctgccca ccactatctt ctttctcacc aagcccaaca ataaggtgtc ctctacttcc 120
accaacaata acatcattac actgcccaag tcgtaccctc tcattggctc ctacctttcg 180
ttccgaaaga acctgcatcg acggatccag tggctctccg acattgttca gatctctccc 240
tccgctacct tccagctcga cggaaccctg ggcaagcgac agatcattac tggcaaccct 300
tctaccgtcc agcacattct caagaaccag ttctccaact atcagaaggg caccacattc 360
accaacactc tgtccgactt tctcggaaca ggcatcttca acaccaacgg tcccaactgg 420
aagtttcaac gacaggttgc ctctcacgag ttcaacacca agtccattcg gaacttcgtc 480
gagcacatcg tggataccga actcaccaac cgattgattc ccatcctcac ttcgagcacc 540
cagacaaaca atatcctgga cttccaggat attctgcagc gatttacctt cgacaacatc 600
tgcaacattg ccttcggata cgatcccgag tacctcactc cctcgaccaa tcgttccaag 660
ttcgcggagg cctacgaaga cgctaccgag atctccagca agcgattcag actgcctctt 720
cccatcattt ggaagatcaa aaagtacttc aacattggct ccgagaagcg actcaaggaa 780
gccgtcaccg aggtccgatc ctttgccaag aaactggtcc gagagaagaa acgggagctc 840
gaagagaagt cttcgctgga gaccgaagac atgctttctc gatttctgtc cagcggtcac 900
tcggacgagg atttcgttgc cgacattgtc atctccttca ttctcgcagg caaggacact 960
acctctgccg ctcttacctg gtttttctgg ctgctctgga agaaccctcg agtggaggaa 1020
gagatcgtca acgagctgtc caagaaatcg gagcttatgg tgtacgacga ggtcaaggaa 1080
atggtctaca cccatgctgc gctgtccgag tcgatgagac tctaccctcc cgttccaatg 1140
gattccaagg aggccgtcaa cgacgatgtg ctgcccgacg gctgggtggt caagaaaggt 1200
acaatcgtca cctaccatgt ctacgctatg ggtcgaatga agtctctctg gggagacgat 1260
tgggcagagt ttcgaccaga acggtggctc gagaaggacg aggtcaacgg caagtgggtg 1320
ttcgtcggac gagacagcta ctcctatcct gtgttccagg ctggtcccag agtctgcctg 1380
ggaaaggaga tggccttcat gcagatgaag cgaattgtgg ctggcatcgt cggaaagttc 1440
aaggtggttc ccgaagccca cttggctcag gagccaggat tcatttcctt tctgtcgtct 1500
cagatggagg gtggatttcc cgtcactatc cagaagcgag actcctaa 1548
<210> 94
<211> 515
<212> PRT
<213> Artificial sequence
<220>
<223> VsCYP94A1s (CYP) protein
<400> 94
Met Ala Phe Gln Phe His Leu Glu Val Leu Leu Pro Tyr Leu Leu Pro
1 5 10 15
Leu Leu Leu Leu Ile Leu Pro Thr Thr Ile Phe Phe Leu Thr Lys Pro
20 25 30
Asn Asn Lys Val Ser Ser Thr Ser Thr Asn Asn Asn Ile Ile Thr Leu
35 40 45
Pro Lys Ser Tyr Pro Leu Ile Gly Ser Tyr Leu Ser Phe Arg Lys Asn
50 55 60
Leu His Arg Arg Ile Gln Trp Leu Ser Asp Ile Val Gln Ile Ser Pro
65 70 75 80
Ser Ala Thr Phe Gln Leu Asp Gly Thr Leu Gly Lys Arg Gln Ile Ile
85 90 95
Thr Gly Asn Pro Ser Thr Val Gln His Ile Leu Lys Asn Gln Phe Ser
100 105 110
Asn Tyr Gln Lys Gly Thr Thr Phe Thr Asn Thr Leu Ser Asp Phe Leu
115 120 125
Gly Thr Gly Ile Phe Asn Thr Asn Gly Pro Asn Trp Lys Phe Gln Arg
130 135 140
Gln Val Ala Ser His Glu Phe Asn Thr Lys Ser Ile Arg Asn Phe Val
145 150 155 160
Glu His Ile Val Asp Thr Glu Leu Thr Asn Arg Leu Ile Pro Ile Leu
165 170 175
Thr Ser Ser Thr Gln Thr Asn Asn Ile Leu Asp Phe Gln Asp Ile Leu
180 185 190
Gln Arg Phe Thr Phe Asp Asn Ile Cys Asn Ile Ala Phe Gly Tyr Asp
195 200 205
Pro Glu Tyr Leu Thr Pro Ser Thr Asn Arg Ser Lys Phe Ala Glu Ala
210 215 220
Tyr Glu Asp Ala Thr Glu Ile Ser Ser Lys Arg Phe Arg Leu Pro Leu
225 230 235 240
Pro Ile Ile Trp Lys Ile Lys Lys Tyr Phe Asn Ile Gly Ser Glu Lys
245 250 255
Arg Leu Lys Glu Ala Val Thr Glu Val Arg Ser Phe Ala Lys Lys Leu
260 265 270
Val Arg Glu Lys Lys Arg Glu Leu Glu Glu Lys Ser Ser Leu Glu Thr
275 280 285
Glu Asp Met Leu Ser Arg Phe Leu Ser Ser Gly His Ser Asp Glu Asp
290 295 300
Phe Val Ala Asp Ile Val Ile Ser Phe Ile Leu Ala Gly Lys Asp Thr
305 310 315 320
Thr Ser Ala Ala Leu Thr Trp Phe Phe Trp Leu Leu Trp Lys Asn Pro
325 330 335
Arg Val Glu Glu Glu Ile Val Asn Glu Leu Ser Lys Lys Ser Glu Leu
340 345 350
Met Val Tyr Asp Glu Val Lys Glu Met Val Tyr Thr His Ala Ala Leu
355 360 365
Ser Glu Ser Met Arg Leu Tyr Pro Pro Val Pro Met Asp Ser Lys Glu
370 375 380
Ala Val Asn Asp Asp Val Leu Pro Asp Gly Trp Val Val Lys Lys Gly
385 390 395 400
Thr Ile Val Thr Tyr His Val Tyr Ala Met Gly Arg Met Lys Ser Leu
405 410 415
Trp Gly Asp Asp Trp Ala Glu Phe Arg Pro Glu Arg Trp Leu Glu Lys
420 425 430
Asp Glu Val Asn Gly Lys Trp Val Phe Val Gly Arg Asp Ser Tyr Ser
435 440 445
Tyr Pro Val Phe Gln Ala Gly Pro Arg Val Cys Leu Gly Lys Glu Met
450 455 460
Ala Phe Met Gln Met Lys Arg Ile Val Ala Gly Ile Val Gly Lys Phe
465 470 475 480
Lys Val Val Pro Glu Ala His Leu Ala Gln Glu Pro Gly Phe Ile Ser
485 490 495
Phe Leu Ser Ser Gln Met Glu Gly Gly Phe Pro Val Thr Ile Gln Lys
500 505 510
Arg Asp Ser
515
<210> 95
<211> 2082
<212> DNA
<213> Artificial sequence
<220>
<223> VsCPRs (CPR)
<400> 95
atggccacct cctctaactc cgacctggtc cgaaccatcg agtccgccct cggcatttct 60
ctcggcgaca gcgtgtccga ttctgtcgtt atcattgcca ccacttctgc tgccgtcatc 120
attggacttc tggtcttcct ctggcgaaag tctcccgaca gatcgcgaga gctgcgtcct 180
gtcattgtgc ccaagtttac cgttaagcac gaggacgatg aagtcgaggt ggaccgaggc 240
aaaaccaagg ttacagtttt ctacggaact cagaccggta ctgccgaggg ctttgcaaag 300
gcccttgcgg aggaaatcaa ggccagatac gagaaggccg ttgtcaaggt ggttgacatg 360
gatgactacg ctattgacga tgaccagtac gaggaaaagc tcaaaaagga gaccctggtc 420
ttctttatgc ttgccaccta tggagacggc gaacccaccg ataacgctgc acgattctac 480
aagtggttta ccgagggcaa ggaggaacga ggaacctggc tgcagcaact cacttacggt 540
gtgttcgcct tgggcaaccg acagtacgag catttcaaca agatcggcaa gattgtcgac 600
gaggatctta ccgagcaggg agccaagcgt ctggttcccg tcggtctcgg cgacgatgac 660
cagtccatcg aggacgattt caacgcttgg aaggaaacct tgtggccaga gctggaccaa 720
cttctccgag acgaggatga cgtcaacact gcttcgaccc cttacactgc cgctatctcc 780
gagtatcgtg tcgttatcca cgatcctacc gtgtctccct cctacgagaa ccatttcaat 840
gttgccaacg gtggagcagt gttcgacatt caccatccct gtcgagtcaa cgttgccgtg 900
cgacgggaac ttcacaagcc ccagtccgac cgatcttgca ttcacctgga gtttgatctc 960
tccggtactg gcgttacata cgagactggc gaccacgtcg gagtgtacgc cgagaactgc 1020
gacgaaactg tcgaggaagc tggcaagctg ctcggtcagt cgctggatct tctcttttct 1080
ctgcataccg acaaggagga tggcacaagc cttggtggat ctctgctccc tccatttcct 1140
ggaccctgta ccgttcgaac tgccctcgct tgctacgccg accttcttaa tcctccacgg 1200
aaagccgcta tcgtggcact tgctgcccat gcttccgagc ccagcgaggc cgaacgactc 1260
aagtttcttt cttcgcctca gggcaaggac gagtactcca agtgggtcgt tggatctcag 1320
cgatcgctgc tcgaagtcat ggccgatttt ccctccgcca agcctccact gggagtgttc 1380
tttgctgcca ttgcacctcg actgcagcct cgatactatt ctatctcctc ttcgcccaga 1440
ccagctcccc agcgagtgca cgttacctgt gcccttgtcg agggacccac tcctaccggt 1500
cggattcaca agggtgtgtg ctccacctgg atgaagtctg ctactccctt ggagaagtct 1560
cacgactgtt cccgagcacc tatcttcatt cgaccctcca acttcaagct tcctgccgac 1620
cattcgattc ccattatcat ggtcggacct ggtacaggtc tggctccctt tcgaggattc 1680
ctccaggaac gacttgccct caaggaggat ggagttcagc ttggacctgc cctgctcttc 1740
tttggttgcc gaaacagaca gatggacttc atctacgagg acgaactcaa caatttcgtt 1800
cagcaaggtg ccatttccga gctcatcgtt gcgttttctc gagagggccc agaaaaggag 1860
tacgtgcagc acaagatgat ggacaaggcc gagtatctgt ggtctctcat ttcgcaggga 1920
ggctacctgt acgtctgtgg tgatgccaaa ggcatggctc gagacgtgca ccgatccctt 1980
cataccattg ttcagcaaca ggagaacgca gattcttcga aggctgaggc cactgtcaag 2040
aaactccaga tggacggaag atacctgcga gacgtgtggt aa 2082
<210> 96
<211> 693
<212> PRT
<213> Artificial sequence
<220>
<223> VsCPRs (CPR) protein
<400> 96
Met Ala Thr Ser Ser Asn Ser Asp Leu Val Arg Thr Ile Glu Ser Ala
1 5 10 15
Leu Gly Ile Ser Leu Gly Asp Ser Val Ser Asp Ser Val Val Ile Ile
20 25 30
Ala Thr Thr Ser Ala Ala Val Ile Ile Gly Leu Leu Val Phe Leu Trp
35 40 45
Arg Lys Ser Pro Asp Arg Ser Arg Glu Leu Arg Pro Val Ile Val Pro
50 55 60
Lys Phe Thr Val Lys His Glu Asp Asp Glu Val Glu Val Asp Arg Gly
65 70 75 80
Lys Thr Lys Val Thr Val Phe Tyr Gly Thr Gln Thr Gly Thr Ala Glu
85 90 95
Gly Phe Ala Lys Ala Leu Ala Glu Glu Ile Lys Ala Arg Tyr Glu Lys
100 105 110
Ala Val Val Lys Val Val Asp Met Asp Asp Tyr Ala Ile Asp Asp Asp
115 120 125
Gln Tyr Glu Glu Lys Leu Lys Lys Glu Thr Leu Val Phe Phe Met Leu
130 135 140
Ala Thr Tyr Gly Asp Gly Glu Pro Thr Asp Asn Ala Ala Arg Phe Tyr
145 150 155 160
Lys Trp Phe Thr Glu Gly Lys Glu Glu Arg Gly Thr Trp Leu Gln Gln
165 170 175
Leu Thr Tyr Gly Val Phe Ala Leu Gly Asn Arg Gln Tyr Glu His Phe
180 185 190
Asn Lys Ile Gly Lys Ile Val Asp Glu Asp Leu Thr Glu Gln Gly Ala
195 200 205
Lys Arg Leu Val Pro Val Gly Leu Gly Asp Asp Asp Gln Ser Ile Glu
210 215 220
Asp Asp Phe Asn Ala Trp Lys Glu Thr Leu Trp Pro Glu Leu Asp Gln
225 230 235 240
Leu Leu Arg Asp Glu Asp Asp Val Asn Thr Ala Ser Thr Pro Tyr Thr
245 250 255
Ala Ala Ile Ser Glu Tyr Arg Val Val Ile His Asp Pro Thr Val Ser
260 265 270
Pro Ser Tyr Glu Asn His Phe Asn Val Ala Asn Gly Gly Ala Val Phe
275 280 285
Asp Ile His His Pro Cys Arg Val Asn Val Ala Val Arg Arg Glu Leu
290 295 300
His Lys Pro Gln Ser Asp Arg Ser Cys Ile His Leu Glu Phe Asp Leu
305 310 315 320
Ser Gly Thr Gly Val Thr Tyr Glu Thr Gly Asp His Val Gly Val Tyr
325 330 335
Ala Glu Asn Cys Asp Glu Thr Val Glu Glu Ala Gly Lys Leu Leu Gly
340 345 350
Gln Ser Leu Asp Leu Leu Phe Ser Leu His Thr Asp Lys Glu Asp Gly
355 360 365
Thr Ser Leu Gly Gly Ser Leu Leu Pro Pro Phe Pro Gly Pro Cys Thr
370 375 380
Val Arg Thr Ala Leu Ala Cys Tyr Ala Asp Leu Leu Asn Pro Pro Arg
385 390 395 400
Lys Ala Ala Ile Val Ala Leu Ala Ala His Ala Ser Glu Pro Ser Glu
405 410 415
Ala Glu Arg Leu Lys Phe Leu Ser Ser Pro Gln Gly Lys Asp Glu Tyr
420 425 430
Ser Lys Trp Val Val Gly Ser Gln Arg Ser Leu Leu Glu Val Met Ala
435 440 445
Asp Phe Pro Ser Ala Lys Pro Pro Leu Gly Val Phe Phe Ala Ala Ile
450 455 460
Ala Pro Arg Leu Gln Pro Arg Tyr Tyr Ser Ile Ser Ser Ser Pro Arg
465 470 475 480
Pro Ala Pro Gln Arg Val His Val Thr Cys Ala Leu Val Glu Gly Pro
485 490 495
Thr Pro Thr Gly Arg Ile His Lys Gly Val Cys Ser Thr Trp Met Lys
500 505 510
Ser Ala Thr Pro Leu Glu Lys Ser His Asp Cys Ser Arg Ala Pro Ile
515 520 525
Phe Ile Arg Pro Ser Asn Phe Lys Leu Pro Ala Asp His Ser Ile Pro
530 535 540
Ile Ile Met Val Gly Pro Gly Thr Gly Leu Ala Pro Phe Arg Gly Phe
545 550 555 560
Leu Gln Glu Arg Leu Ala Leu Lys Glu Asp Gly Val Gln Leu Gly Pro
565 570 575
Ala Leu Leu Phe Phe Gly Cys Arg Asn Arg Gln Met Asp Phe Ile Tyr
580 585 590
Glu Asp Glu Leu Asn Asn Phe Val Gln Gln Gly Ala Ile Ser Glu Leu
595 600 605
Ile Val Ala Phe Ser Arg Glu Gly Pro Glu Lys Glu Tyr Val Gln His
610 615 620
Lys Met Met Asp Lys Ala Glu Tyr Leu Trp Ser Leu Ile Ser Gln Gly
625 630 635 640
Gly Tyr Leu Tyr Val Cys Gly Asp Ala Lys Gly Met Ala Arg Asp Val
645 650 655
His Arg Ser Leu His Thr Ile Val Gln Gln Gln Glu Asn Ala Asp Ser
660 665 670
Ser Lys Ala Glu Ala Thr Val Lys Lys Leu Gln Met Asp Gly Arg Tyr
675 680 685
Leu Arg Asp Val Trp
690
<210> 97
<211> 783
<212> DNA
<213> yarrowia lipolytica
<400> 97
aaacgagaat cgtacagagt tgtttctgaa ccatttcgaa gccgttcaga gtcgtttaac 60
cgcagtttga gtcgtttcag agcggttctc agtcgttttt gatccatctc tgaatgattc 120
agagccgcct aactccttgt gagccgttaa atcgcgaatt gagccactct gagccgttga 180
ttcttccatt tgtggcccct tcaccatctc ctcatctcct tcctctcgtc gccatctcct 240
tttctcctct cgcacctcgc actactccaa accaactcac tgactacatc ccaacagcga 300
tctaaccaac gccgcgcaag caaacagaca caaaaacaac gtccgcgccg ccgaccacgt 360
cagcagatcc cgctctcctg ggttttgtcg tcgtttgccg ccttttgccg ccgctatttg 420
caacgccgcc acactcaatg gccgagccat gatgccgtcg tcgtgtcttt tcccaaatag 480
gaaaccgggc accaacccta gccccacaac tggagtactc aactcggcga aaaatgggcc 540
aatcgccgcg ggagaaacga caatcggctt gttttgactc aattacctca ccaagcgccc 600
cttcgtcgcc gccatacctc cgcaacaccc cctcaccgtc cctccctccg cccctctgga 660
aactcataga accccaaacc ttatttccga tgaccgcaaa ctttagacta tacaggcgaa 720
tctgggtggt ggcaccaacc cttcctcttc tccatcaccc ccccctcaat ctctttttct 780
cac 783
<210> 98
<211> 17083
<212> DNA
<213> Artificial sequence
<220>
<223> pZSCpn-3FAOBU plasmid
<400> 98
cgattcgaga ttttacagat atttctcgca gtttttcacg tccccttgtc cttgtcctat 60
tgtttcaaat aaactctcgt ctactgattt cacatggaac ctttgctatt tcggggataa 120
ccccctttgc cattgcacga tggacgtggc aaaagaaaga tcgccctgcg gggatactta 180
tcatgtggtc acatgctgtg attagaaata aagaaaaagg tgcttttttg gcgctgtgat 240
taacatctcg tctgccgtgc tctactagtc gcaatagcaa aaactcgctt aatagtgtgc 300
atagtgcggg gtagcaggat actgaactac agtacgattt gcttgctact gcttgtagca 360
attaccttta ctgtagggac cacacctcct ggtttcaatg tctttcctcg cctcgacaaa 420
gcaaaactgt cacccaatca caccttgttc atattcatta gtgcatccgt taaccttgac 480
atgacacttc tcatactagt gatagggctg tagttgagac aagttgattc acacggatac 540
atacaaagcc tcagagagca aatgttatat actcagggac cgaccaatca aaaaaacaca 600
ctcctaataa ccaccatttc catctacgcg tactcactct gtcagctgcc ccacattgcc 660
caatgcacaa tgcacaatga tgtgtgcaaa caacgcaatc aaaagtctat ggatgctgac 720
caaactctga tcaccaagtt gcgaacatga aaaagaagac ctgtgtatat ataagtaagg 780
gggagagccc taactagatc tttcgaaaac cccccgacct tcaccttcca caaccatggc 840
caaccccgtc gtggaggact cccatctgga cgtcttctgc ctccttgccg atgctgtggt 900
tcacgagatt cctccctccg agatcgtcga gtacctgcat cctgactttc ccaaggacaa 960
ggtcgaagag taccttgccg agttctctca tccctccgct attccagagt tccgagaggt 1020
tgccaagcga atcattaaca agggcaccgt gctgtcgatc aagctctttc tgctcttggc 1080
cactgctctg gattctcgaa tccttgctcc tgccttgacc aactccacta cactcatccg 1140
agacatggat ctttctcagc gggaggaact cctgagatcc tggcgagact ctcccttcac 1200
taccaaacga aagctgttcc gagtctacaa ctcgtttacc ctcaacgcct tcagcaagac 1260
tgctaccgac cttcacttca aggccctggg ataccctggt cgagagctcc gtactcagat 1320
tcaggactac gaggtcgatc cctttcgata cacgttcctg gagaagcctc agcaagacgg 1380
acaggagctc cactttcccg acattgatgt gctcattatc ggatctggct ccggtgcagg 1440
cgtcgttgct cagactcttt cggagaacgg actcaagtct ctggtgctcg agaagggcaa 1500
atacttttcc aacgacgagc tgaccatgaa cgacctcgaa ggttccgagg ccctgttcga 1560
aaacggaggt gctctctcct ctaccaatca acagatcttt atcattgccg gatcgacttt 1620
cggtggcgga tccacagtca actggtctgc ctgtctcaag actcccttca aggtgcgaaa 1680
ggagtggtac gacaactttg gactggattt cgttgctacc cagtattacg aggactgtat 1740
ggactacgtc tggaagaaaa tgggtgcctc caacgagaat atcgaccatt ctggagccaa 1800
ctcggtcatt ctggaaggtt ccaagaaact tggctaccct caccgtgccg tggaacagaa 1860
caatggaggc aagattcacg actgtggtat gtgccacctc ggatgtcgat ttggcatcaa 1920
gcagggatcg gtcaactgct ggtttcgtgg tccctccgag aacggctcga agttcatgca 1980
gcaagttctc gtggacaaga tcctgcagcg agatggcaag gctgtcggtg ttctctgtag 2040
agacgtggtt accggagtca agttcaagat cactggaccc aagaaaatcg tcgtgttctg 2100
gtggttcttt gccaactccg gatttgttac caagtctggt ttcaagaaca agcacatcgg 2160
tgcaaacctc aagctgcatc ccgtcagcct tacgctcggc gactttggta acaatgtgga 2220
tttcgaggcc taccgaaagc caatcatgac ctccatttgt aacaaggtcg aggacctgga 2280
tggaaaggct cacggcactc gaatcgaggc catgctcaat gctccctacg gtgttgctcc 2340
attctttccc tggaagtctg gcgcagagtc ccgaaaggac ctcctgcgat acaagcagac 2400
tgtgcccatt ctcctgcttt ccagagacac cacttctgga tccgtcacct acgacaaaca 2460
gaagcccgat gccttggtga tcgactacct gctcaacaag ttcgaccgaa actccatcct 2520
gcagggcttt ctcattgctt cggatcttct gtacatcgag ggtgccagcc gagaccatgt 2580
tacctacaag cttggatacc agtggttcaa gtcttccaag cccaagcacg ctcgatccat 2640
cgaagacgag gactacgtca actggagagc caaggttgca aagattccct ttgattccta 2700
tggatctcct tacggttcgg ctcaccagat gtccacttgc agaatgtctg gcaagggacc 2760
aggctacgga gcctgcgaca ccaagggcaa actcttcgag tgcagcaacg tgtacgtcgc 2820
cgatgcttcc actctgccca ccgcatctgg tgccaaccct atggtctcta ccatgtcctt 2880
tgcccgacac gtgtcgcttg gcatcgtcaa ggagctgcag caatccaagc tctaagcggc 2940
cgcatggagc gtgtgttctg agtcgatgtt ttctatggag ttgtgagtgt tagtagacat 3000
gatgggttta tatatgatga atgaatagat gtgattttga tttgcacgat ggaattgaga 3060
actttgtaaa cgtacatggg aatgtatgaa tgtgggggtt ttgtgactgg ataactgacg 3120
gtcagtggac gccgttgttc aaatatccaa gagatgcgag aaactttggg tcaagtgaac 3180
atgtcctctc tgttcaagta aaccatcaac tatgggtagt atatttagta aggacaagag 3240
ttgagattct ttggagtcct agaaacgtat tttcgcgttc caagatcaaa ttagtagagt 3300
aatacgggca cgggaatcca ttcatagtct caagtttaaa ccatcatcta agggcctcaa 3360
aactacctcg gaactgctgc gctgatctgg acaccacaga ggttccgagc actttaggtt 3420
gcaccaaatg tcccaccagg tgcaggcaga aaacgctgga acagcgtgta cagtttgtct 3480
taacaaaaag tgagggcgct gaggtcgagc agggtggtgt gacttgttat agcctttaga 3540
gctgcgaaag cgcgtatgga tttggctcat caggccagat tgagggtctg tggacacatg 3600
tcatgttagt gtacttcaat cgccccctgg atatagcccc gacaataggc cgtggcctca 3660
tttttttgcc ttccgcacat ttccattgct cggtacccac accttgcttc tcctgcactt 3720
gccaacctta atactggttt acattgacca acatcttaca agcggggggc ttgtctaggg 3780
tatatataaa cagtggctct cccaatcggt tgccagtctc ttttttcctt tctttcccca 3840
cagattcgaa atctaaacta cacatcacac catggcctcc caccaggtcg aggaccacga 3900
tctggacgtg ttctgcctcc tggccgacgc tgttctccac gagattcctc cctccgaaat 3960
cgtcgagtac cttcatcccg atttccccaa ggacaagatc gaagagtacc tgaccggctt 4020
ttctcgaccc tccgccgttc ctcagttccg acagtgtgcc aagaaactca tcaaccgagg 4080
ttccgagctg tcgatcaagc tcttccttta cttgaccact gctctggact ctcgaatcct 4140
tgcaccagcc ctgaccaact cgctcactct gatcagagac atggatcttt cccagcgaga 4200
ggaactgttg cggtcctggc gagattctcc actgactgcc aagcgaagac tctttcgagt 4260
ctacgcctcc tttaccctgt ctacattcaa caagctcgga accgacttgc acttcaaggc 4320
cctgggctac cctggtcgag agctccggac ccagattcaa gactacgagg tcgatccctt 4380
tcgatactcg ttcatggaga agctcaaaca cgagggacat gaactgttcc ttcccgatat 4440
cgacgttctg atcattggct ctggatccgg tgcaggcgtg gtcgctcaga ctcttaccga 4500
gagcggactc aagtctctgg ttctcgagaa gggcaagtac tttgcctccg aagagctgtg 4560
catgaccgat ctcgacggaa acgaggccct gttcgaaagc ggtggcacta ttccttccac 4620
caatcaacag ttgttcatga tcgctggatc tacttttggt ggaggctcca ccgtcaactg 4680
gtctgcctgt ctcaagactc ccttcaaggt tcgaaaggag tggtacgacg atttcggact 4740
ggactttgtg gctacccagc aatacgacga ttgcatggac tacgtgtgga agaaaatggg 4800
tgcctcgacc gagcacatcg agcattctgc tgcaaatgcc gtcatcatgg acggagctgc 4860
caagcttggc tacgctcacc gagccctcga gcagaacacc ggtggccatg ttcacgactg 4920
tggaatgtgc cacctgggct gtcgattcgg tatcaagcag ggaggcgtca actgctggtt 4980
tcgagaaccc tccgagaagg gttccaagtt catggagcag gtcgttgtcg agaagattct 5040
gcagcacaag ggcaaggcca ctggaattct ctgcagagat accgagtctg gcatcaagtt 5100
caagattact ggacccaaga aatacgtcgt gtccggtggc tctttgcaga cccctgttct 5160
ccttcagaag tctggcttca agaacaagca cattggagcc aacctcaagc tgcatcccgt 5220
ctcggttgct cttggcgact ttggtaacga ggtggacttc gaagcctaca agcgacccct 5280
catgaccgca gtctgcaatg ccgtggacga tctggacggc aaggctcacg gaacacgaat 5340
cgaggccatt ctgcacgctc cttacgtcac tgctcccttc tatccctggc agtccggtgc 5400
ccaggctcgg aagaacttgc tcaaatacaa gcagaccgtg cctctgctcc ttctgtctcg 5460
agacacctcc tcgggtaccg ttacatacga caaacagaag ccagatgtct tggtggtcga 5520
ctacactgtc aacaagttcg atcgaaactc catcctgcag ggatttctcg ttgcttccga 5580
cattttgtac atcgagggtg ccaaggagat tctgtctccc caggcttggg tgcccacctt 5640
caagagcaac aagcccaagc acgccagatc catcaaggac gaggactacg tcaagtggcg 5700
agaaaccgtg gccaagattc cctttgattc ctacggctcg ccttacggtt ctgctcatca 5760
gatgtcctcg tgtcgaatgt ctggcaaggg acccggatac ggtgcctgcg ataccaaggg 5820
acgactcttc gagtgcaaca atgtgtacgt tgcagacgcc tccgtcatgc ctactgcttc 5880
tggagtcaac cccatgatta ccacaatggc ctttgcacga cacgttgctc tctgtctggc 5940
caaggacctt caaccccaga ccaagctgta agcggccgca tgagaagata aatatataaa 6000
tacattgaga tattaaatgc gctagattag agagcctcat actgctcgga gagaagccaa 6060
gacgagtact caaaggggat tacaccatcc atatccacag acacaagctg gggaaaggtt 6120
ctatatacac tttccggaat accgtagttt ccgatgttat caatgggggc agccaggatt 6180
tcaggcactt cggtgtctcg gggtgaaatg gcgttcttgg cctccatcaa gtcgtaccat 6240
gtcttcattt gcctgtcaaa gtaaaacaga agcagatgaa gaatgaactt gaagtgaagg 6300
aatttaaata agtttgcaaa aagatcgtat tatagttgga gcaagggaga aatgtagagt 6360
gtgaaagact cactatggtc cgggcttatc tcgaccaata gccaaagtct ggagtttctg 6420
agagaaaaag gcaagatacg tatgtaacaa agcgacgcat ggtacaataa taccggaggc 6480
atgtatcata gagagttagt ggttcgatga tggcactggt gcctggtatg actttatacg 6540
gctgactaca tatttgtcct cagacataca attacagtca agcacttacc cttggacatc 6600
tgtaggtacc ccccggccaa gacgatctca gcgtgtcgta tgtcggattg gcgtagctcc 6660
ctcgctcgtc aattggctcc catctacttt cttctgcttg gctacaccca gcatgtctgc 6720
tatggctcgt tttcgtgcct tatctatcct cccagtatta ccaactctaa atgacatgat 6780
gtgattgggt ctacactttc atatcagaga taaggagtag cacagttgca taaaaagccc 6840
aactctaatc agcttcttcc tttcttgtaa ttagtacaaa ggtgattagc gaaatctgga 6900
agcttagttg gccctaaaaa aatcaaaaaa agcaaaaaac gaaaaacgaa aaaccacagt 6960
tttgagaaca gggaggtaac gaaggatcgt atatatatat atatatatat atacccacgg 7020
atcccgagac cggcctttga ttcttcccta caaccaacca ttctcaccac cctaattcac 7080
aaccatggct cccttcctgc ccgaccaggt cgactacaag cacgtcgata ccctcatgct 7140
gctgtgcgac ggcatcattc acgagactac cgtggacgag atcaaggatg tcattgctcc 7200
tgactttcca gccgacaagt acgaggaata cgttcgaacc ttcacaaagc cctccgagac 7260
tcccggtttc cgagagaccg tgtacaacac cgtcaatgcc aacactatgg atgccatcca 7320
tcagttcatt atcctgacca acgttctcgg atctcgagtc cttgctcctg ccctgaccaa 7380
ctccttgact cccatcaagg acatgtctct cgaagaccgg gagaagctgc ttgcctcgtg 7440
gcgagattct cccattgctg ccaagcggaa gctgttcaga ctcgtgtcca cgcttactct 7500
ggtcaccttt acacgacttg ccaacgagtt gcatctcaag gccattcact atccaggacg 7560
agaagaccga gagaaggctt acgagaccca ggagatcgac cccttcaagt accagtttct 7620
ggagaaaccc aagttctacg gcgcagagct gtacctccca gacattgatg tcatcattat 7680
cggatctggt gccggagctg gtgtcgttgc ccatactctc accaacgacg gcttcaagtc 7740
cctggttctc gaaaagggca gatactttag caactccgag ctcaacttcg acgataagga 7800
cggtgttcag gagctgtacc aatctggagg taccttgact accgtcaatc agcaactctt 7860
cgtgcttgct ggttccactt ttggaggtgg cactaccgtc aactggtctg cctgtctcaa 7920
gacgcccttc aaggtgcgga aggagtggta cgacgagttc ggcgtcgatt ttgctgccga 7980
cgaagcctac gacaaggcac aggattacgt gtggcagcaa atgggagcct cgaccgaagg 8040
catcactcac tccttggcca acgagatcat tatcgaaggt ggcaagaaac tcggatacaa 8100
ggccaaggtc ctggaccaga actctggtgg acatcctcat caccgatgcg gcttctgtca 8160
cctcggttgc aagcacggaa tcaagcaggg ctccgtcaac aattggtttc gagacgcagc 8220
tgcccacgga tcgcagttca tgcaacaggt gcgagttctg cagattctca acaagaaagg 8280
catcgcctac ggtatcttgt gcgaggatgt cgttaccgga gccaagttta ccattactgg 8340
tcccaaaaag ttcgtggtcg ctgcaggagc cctcaacact cccagcgtgc tggtcaactc 8400
cggattcaag aacaaaaaca ttggcaagaa ccttaccttg catcccgttt ctgtcgtgtt 8460
tggcgacttc ggaaaggacg tgcaggccga tcactttcac aattccatca tgactgctct 8520
gtgttcggaa gccgctgacc tcgacggcaa gggtcatgga tgccgaattg agaccatcct 8580
gaacgcaccc ttcattcagg cttcctttct tccttggcga ggttccaacg aggccagacg 8640
agacctcctg cgatacaaca atatggtcgc gatgctgctt ctctctcgag atacaacctc 8700
gggttccgtg tcttcccatc ccaccaaacc agaagccctg gttgtcgagt acgacgtcaa 8760
caagtttgat cgaaactcca tcttgcaggc cctgcttgtc actgcagacc tgctctacat 8820
tcagggagcc aagcgaatcc tttctcctca gccctgggtg ccaatcttcg agtccgacaa 8880
gcccaaggac aagcgatcta tcaaggacga ggattacgtc gaatggcgag ccaaggttgc 8940
caagattccc ttcgacacct acggctctcc ttatggttcg gctcaccaga tgtcttcctg 9000
tcgtatgagc ggcaagggtc ccaagtacgg agccgtcgat accgacggtc gactgtttga 9060
gtgctcgaac gtgtacgttg ccgacgcttc ccttctgccc actgctagcg gtgccaaccc 9120
tatggtcaca accatgactc tcgctcgaca cgttgccctc ggcttggcag actccctgaa 9180
gaccaaagcc aagctctaag cggccgcaag tgtggatggg gaagtgagtg cccggttctg 9240
tgtgcacaat tggcaatcca agatggatgg attcaacaca gggatatagc gagctacgtg 9300
gtggtgcgag gatatagcaa cggatattta tgtttgacac ttgagaatgt acgatacaag 9360
cactgtccaa gtacaatact aaacatactg tacatactca tactcgtacc cgggcaacgg 9420
tttcacttga gtgcagtggc tagtgctctt actcgtacag tgtgcaatac tgcgtatcat 9480
agtctttgat gtatatcgta ttcattcatg ttagttgcgt acgacccctc tcaggccaag 9540
cagaaggctg agtccatcaa gaaggccaac gctatcattg tcttcaacct caagaacaag 9600
gctggcaaga ccgagtcttg gtaccttgac ctcaagaacg acggtgacgt cggcaagggc 9660
aacaagtccc ccaagggtga tgctgacatc cagctcactc tctctgacga ccacttccag 9720
cagctcgttg agggtaaggc taacgcccag cgactcttca tgaccggcaa gctcaaggtt 9780
aagggcaacg tcatgaaggc tgccgccatt gagggtatcc tcaagaacgc tcagaacaac 9840
ctctaagcgc atcatttatt gattaattga tgatttacta tattgatttc gcaactgtag 9900
tgtgattgta tgtgatctgg ctcgtaggct tcagtaaata ctagacgggt atcctacgta 9960
gttgtatcat acatcgagcc tgtggttact tgtacaataa ttcgtaatgt agagataccc 10020
cttgatccat tgcctgtttc taacatacaa tgatctccac gcaataatcc cactcttgac 10080
taaaagttgc tactcttgca cggttacctc ggcatagtca cgcctctctt gtctcgtctc 10140
gaacgcacaa agtcaattga caacgccact cactcgagtg tgccccaaca gggcaccata 10200
tcgactaatt tgaggccaac tagggtgatt ttggatggaa tttgatcgga aaaaatagct 10260
gcagaaattc ctggagagaa aaattgaccg catccacatg gtttgaccaa aaaatcgtct 10320
ccatctctgt gctcaactct cctgacgaga tatgcgcgcg cacccccaca tgatgtgatt 10380
gatctcaaca aacttcaccc agacccttat ctttccggga aacttactgt ataagtggtc 10440
gtgcgaacag aaagtgtgcg cactttaggt gtctagatcc gattgttctc gttctgataa 10500
tgagccagcc ccgcgaggca atgtttttta caattgaaaa cttcgttaac cactcacatt 10560
accgtttttg ccccatattt accctctggt acactccctc ttgcatacac acacactgca 10620
gtgaaaatgc actccgttag caccgttgtg attggttcag ggcacgagtt tggtggttta 10680
aggcgcaact acatcaatat gaaaacagga gacgctgaaa aggggtaata tcggactgct 10740
gctatgttgt atgtactgca tgacgaattg gtgttattca agaccgtggc acaggttgct 10800
gcggtacgag acctggtagc ttctctaaac ggcatgtcta ggtggcgcgc cagctgcatt 10860
aatgaatcgg ccaacgcgcg gggagaggcg gtttgcgtat tgggcgctct tccgcttcct 10920
cgctcactga ctcgctgcgc tcggtcgttc ggctgcggcg agcggtatca gctcactcaa 10980
aggcggtaat acggttatcc acagaatcag gggataacgc aggaaagaac atgtgagcaa 11040
aaggccagca aaaggccagg aaccgtaaaa aggccgcgtt gctggcgttt ttccataggc 11100
tccgcccccc tgacgagcat cacaaaaatc gacgctcaag tcagaggtgg cgaaacccga 11160
caggactata aagataccag gcgtttcccc ctggaagctc cctcgtgcgc tctcctgttc 11220
cgaccctgcc gcttaccgga tacctgtccg cctttctccc ttcgggaagc gtggcgcttt 11280
ctcatagctc acgctgtagg tatctcagtt cggtgtaggt cgttcgctcc aagctgggct 11340
gtgtgcacga accccccgtt cagcccgacc gctgcgcctt atccggtaac tatcgtcttg 11400
agtccaaccc ggtaagacac gacttatcgc cactggcagc agccactggt aacaggatta 11460
gcagagcgag gtatgtaggc ggtgctacag agttcttgaa gtggtggcct aactacggct 11520
acactagaag aacagtattt ggtatctgcg ctctgctgaa gccagttacc ttcggaaaaa 11580
gagttggtag ctcttgatcc ggcaaacaaa ccaccgctgg tagcggtggt ttttttgttt 11640
gcaagcagca gattacgcgc agaaaaaaag gatctcaaga agatcctttg atcttttcta 11700
cggggtctga cgctcagtgg aacgaaaact cacgttaagg gattttggtc atgagattat 11760
caaaaaggat cttcacctag atccttttaa attaaaaatg aagttttaaa tcaatctaaa 11820
gtatatatga gtaaacttgg tctgacagtt accaatgctt aatcagtgag gcacctatct 11880
cagcgatctg tctatttcgt tcatccatag ttgcctgact ccccgtcgtg tagataacta 11940
cgatacggga gggcttacca tctggcccca gtgctgcaat gataccgcga gacccacgct 12000
caccggctcc agatttatca gcaataaacc agccagccgg aagggccgag cgcagaagtg 12060
gtcctgcaac tttatccgcc tccatccagt ctattaattg ttgccgggaa gctagagtaa 12120
gtagttcgcc agttaatagt ttgcgcaacg ttgttgccat tgctacaggc atcgtggtgt 12180
cacgctcgtc gtttggtatg gcttcattca gctccggttc ccaacgatca aggcgagtta 12240
catgatcccc catgttgtgc aaaaaagcgg ttagctcctt cggtcctccg atcgttgtca 12300
gaagtaagtt ggccgcagtg ttatcactca tggttatggc agcactgcat aattctctta 12360
ctgtcatgcc atccgtaaga tgcttttctg tgactggtga gtactcaacc aagtcattct 12420
gagaatagtg tatgcggcga ccgagttgct cttgcccggc gtcaatacgg gataataccg 12480
cgccacatag cagaacttta aaagtgctca tcattggaaa acgttcttcg gggcgaaaac 12540
tctcaaggat cttaccgctg ttgagatcca gttcgatgta acccactcgt gcacccaact 12600
gatcttcagc atcttttact ttcaccagcg tttctgggtg agcaaaaaca ggaaggcaaa 12660
atgccgcaaa aaagggaata agggcgacac ggaaatgttg aatactcata ctcttccttt 12720
ttcaatatta ttgaagcatt tatcagggtt attgtctcat gagcggatac atatttgaat 12780
gtatttagaa aaataaacaa ataggggttc cgcgcacatt tccccgaaaa gtgccacctg 12840
atgcggtgtg aaataccgca cagatgcgta aggagaaaat accgcatcag gaaattgtaa 12900
gcgttaatat tttgttaaaa ttcgcgttaa atttttgtta aatcagctca ttttttaacc 12960
aataggccga aatcggcaaa atcccttata aatcaaaaga atagaccgag atagggttga 13020
gtgttgttcc agtttggaac aagagtccac tattaaagaa cgtggactcc aacgtcaaag 13080
ggcgaaaaac cgtctatcag ggcgatggcc cactacgtga accatcaccc taatcaagtt 13140
ttttggggtc gaggtgccgt aaagcactaa atcggaaccc taaagggagc ccccgattta 13200
gagcttgacg gggaaagccg gcgaacgtgg cgagaaagga agggaagaaa gcgaaaggag 13260
cgggcgctag ggcgctggca agtgtagcgg tcacgctgcg cgtaaccacc acacccgccg 13320
cgcttaatgc gccgctacag ggcgcgtcca ttcgccattc aggctgcgca actgttggga 13380
agggcgatcg gtgcgggcct cttcgctatt acgccagctg gcgaaagggg gatgtgctgc 13440
aaggcgatta agttgggtaa cgccagggtt ttcccagtca cgacgttgta aaacgacggc 13500
cagtgaattg taatacgact cactataggg cgaattgggc ccgacgtcgc atgcgtcact 13560
aatcaaggat acctaccatg ccactatgat gtttgcagga ggtgtacctc ggcagtcatc 13620
aaaaaatgga actactggct ttagatcttg ttgtatggca tcgcgcctaa aaaagaaacc 13680
cccttccagc gagctactac aagtagttgt agttgcgggc gttggatacc gaaagtcaca 13740
agcacatgtc gaagctctca tctgaaacac cgacagtcgt ctgcaccccg caagtctcgg 13800
ttcgtaccag caccaatgtt aggcagaact atacacaaga gggcggacga tcacttcggc 13860
gttaggcaac tgaaggctat tttcggctgg tactgtaggg gacagaggaa acgcaagtga 13920
ttagtaaatc ggataatagg cctgttagtt taccgaaatg gtgggggagg ggttccgtgg 13980
atatcttgaa gttatggagg ctgatcgtta tttgtgggga tggatatcat tgtatggaca 14040
tactgtagct actgtataaa caacggatct tacacctgcc tcttgtatgc ccattgcttg 14100
atcatctatc gtgttactgt acatatacaa tagatatagg gaagaaaagc cggaagtaga 14160
gaccatagtc tggcagaagt aacggcctcg ggtcgagaga actataacaa agtccaacgg 14220
cgggtcttag aatagcccca aggatcacac agttccgcaa tccagtttca catgttccgt 14280
tgcatggact tttgcatgtc tactgttgct acgattcccc cattgcaacc acagtttggg 14340
gttaccccgc attatattag catgattacg aaagagataa gtatcatatg gaacatgtga 14400
agggtagtat gcaggtccgg cggagaaaga gaatgacgtt ttcattaagc gattcgcttg 14460
gcggcttgtg ggggatgtga cgatacttac ggtaaagacc ctgtgtgaga gctggtactc 14520
gctcgttact tcgctgatct gttgggccgt caatcgaatc tcgtggaact tgcattcttc 14580
ttaactgtgt ctatacaaga cacctaatga aacatacaag ctaccgaaat cattttactc 14640
gtactgaccg gtacggtact tgcacaagta gtgaaacttc cgaaaatagc cagcctcatg 14700
catcatcgct tcaccccttc tgttgacctc aaaagcattc caacggtaaa aaattataac 14760
gccgccaact ggatggttgt gacggcgttg accaccaatg tgtgggggct ggcggtagga 14820
ccgagcttat tcgtcccaat aagctctttg gatttgattc tttggggtgt gtggtaaaat 14880
tcacatgggg aagaacacgg tggcagtttg aggcagaggc ccagcgtgta gttcctaggg 14940
catgaatata ccgaactcat ggcgcagaat tgagctgaat gcgcaaaaag ctacaggatc 15000
aaccgcgtta gaaatgccgc aaatgtccac taattccccg gactgttcca aatgattctg 15060
tggggataaa tctcaaactg ggttaggctt tgtcacgttt ctttgtgtcg tgtcggttcg 15120
tccggggcaa tgtgcccacg cttggctgtc tccctacacc tcggtaaaaa ctatcacatg 15180
ctgcccctct cgagcaagca ttaaatgcat atagtcaatc taacgacata tatataggta 15240
gggtgcatcc tccggtttag ctccccagaa tatctcttat tcattacaca aaaacaacaa 15300
tgtctctcaa ggtcgacggc ttcacttctt aattaacttt ggccggaatt cctttacctg 15360
caggataact tcgtataatg tatgctatac gaagttatga tctctctctt gagcttttcc 15420
ataacaagtt cttctgcctc caggaagtcc atgggtggtt tgatcatggt tttggtgtag 15480
tggtagtgca gtggtggtat tgtgactggg gatgtagttg agaataagtc atacacaagt 15540
cagctttctt cgagcctcat ataagtataa gtagttcaac gtattagcac tgtacccagc 15600
atctccgtat cgagaaacac aacaacatgc cccattggac agatcatgcg gatacacagg 15660
ttgtgcagta tcatacatac tcgatcagac aggtcgtctg accatcatac aagctgaaca 15720
agcgctccat acttgcacgc tctctatata cacagttaaa ttacatatcc atagtctaac 15780
ctctaacagt taatcttctg gtaagcctcc cagccagcct tctggtatcg cttggcctcc 15840
tcaataggat ctcggttctg gccgtacaga cctcggccga caattatgat atccgttccg 15900
gtagacatga catcctcaac agttcggtac tgctgtccga gagcgtctcc cttgtcgtca 15960
agacccaccc cgggggtcag aataagccag tcctcagagt cgcccttagg tcggttctgg 16020
gcaatgaagc caaccacaaa ctcggggtcg gatcgggcaa gctcaatggt ctgcttggag 16080
tactcgccag tggccagaga gcccttgcaa gacagctcgg ccagcatgag cagacctctg 16140
gccagcttct cgttgggaga ggggactagg aactccttgt actgggagtt ctcgtagtca 16200
gagacgtcct ccttcttctg ttcagagaca gtttcctcgg caccagctcg caggccagca 16260
atgattccgg ttccgggtac accgtgggcg ttggtgatat cggaccactc ggcgattcgg 16320
tgacaccggt actggtgctt gacagtgttg ccaatatctg cgaactttct gtcctcgaac 16380
aggaagaaac cgtgcttaag agcaagttcc ttgaggggga gcacagtgcc ggcgtaggtg 16440
aagtcgtcaa tgatgtcgat atgggttttg atcatgcaca cataaggtcc gaccttatcg 16500
gcaagctcaa tgagctcctt ggtggtggta acatccagag aagcacacag gttggttttc 16560
ttggctgcca cgagcttgag cactcgagcg gcaaaggcgg acttgtggac gttagctcga 16620
gcttcgtagg agggcatttt ggtggtgaag aggagactga aataaattta gtctgcagaa 16680
ctttttatcg gaaccttatc tggggcagtg aagtatatgt tatggtaata gttacgagtt 16740
agttgaactt atagatagac tggactatac ggctatcggt ccaaattaga aagaacgtca 16800
atggctctct gggcgtcgcc tttgccgaca aaaatgtgat catgatgaaa gccagcaatg 16860
acgttgcagc tgatattgtt gtcggccaac cgcgccgaaa acgcagctgt cagacccaca 16920
gcctccaacg aagaatgtat cgtcaaagtg atccaagcac actcatagtt ggagtcgtac 16980
tccaaaggcg gcaatgacga gtcagacaga tactcgtcga cgcgataact tcgtataatg 17040
tatgctatac gaagttatcg tacgatagtt agtagacaac aat 17083
<210> 99
<211> 2115
<212> DNA
<213> Artificial sequence
<220>
<223> CtFAO1M (FAO)
<400> 99
atggctccct tcctgcccga ccaggtcgac tacaagcacg tcgataccct catgctgctg 60
tgcgacggca tcattcacga gactaccgtg gacgagatca aggatgtcat tgctcctgac 120
tttccagccg acaagtacga ggaatacgtt cgaaccttca caaagccctc cgagactccc 180
ggtttccgag agaccgtgta caacaccgtc aatgccaaca ctatggatgc catccatcag 240
ttcattatcc tgaccaacgt tctcggatct cgagtccttg ctcctgccct gaccaactcc 300
ttgactccca tcaaggacat gtctctcgaa gaccgggaga agctgcttgc ctcgtggcga 360
gattctccca ttgctgccaa gcggaagctg ttcagactcg tgtccacgct tactctggtc 420
acctttacac gacttgccaa cgagttgcat ctcaaggcca ttcactatcc aggacgagaa 480
gaccgagaga aggcttacga gacccaggag atcgacccct tcaagtacca gtttctggag 540
aaacccaagt tctacggcgc agagctgtac ctcccagaca ttgatgtcat cattatcgga 600
tctggtgccg gagctggtgt cgttgcccat actctcacca acgacggctt caagtccctg 660
gttctcgaaa agggcagata ctttagcaac tccgagctca acttcgacga taaggacggt 720
gttcaggagc tgtaccaatc tggaggtacc ttgactaccg tcaatcagca actcttcgtg 780
cttgctggtt ccacttttgg aggtggcact accgtcaact ggtctgcctg tctcaagacg 840
cccttcaagg tgcggaagga gtggtacgac gagttcggcg tcgattttgc tgccgacgaa 900
gcctacgaca aggcacagga ttacgtgtgg cagcaaatgg gagcctcgac cgaaggcatc 960
actcactcct tggccaacga gatcattatc gaaggtggca agaaactcgg atacaaggcc 1020
aaggtcctgg accagaactc tggtggacat cctcatcacc gatgcggctt ctgtcacctc 1080
ggttgcaagc acggaatcaa gcagggctcc gtcaacaatt ggtttcgaga cgcagctgcc 1140
cacggatcgc agttcatgca acaggtgcga gttctgcaga ttctcaacaa gaaaggcatc 1200
gcctacggta tcttgtgcga ggatgtcgtt accggagcca agtttaccat tactggtccc 1260
aaaaagttcg tggtcgctgc aggagccctc aacactccca gcgtgctggt caactccgga 1320
ttcaagaaca aaaacattgg caagaacctt accttgcatc ccgtttctgt cgtgtttggc 1380
gacttcggaa aggacgtgca ggccgatcac tttcacaatt ccatcatgac tgctctgtgt 1440
tcggaagccg ctgacctcga cggcaagggt catggatgcc gaattgagac catcctgaac 1500
gcacccttca ttcaggcttc ctttcttcct tggcgaggtt ccaacgaggc cagacgagac 1560
ctcctgcgat acaacaatat ggtcgcgatg ctgcttctct ctcgagatac aacctcgggt 1620
tccgtgtctt cccatcccac caaaccagaa gccctggttg tcgagtacga cgtcaacaag 1680
tttgatcgaa actccatctt gcaggccctg cttgtcactg cagacctgct ctacattcag 1740
ggagccaagc gaatcctttc tcctcagccc tgggtgccaa tcttcgagtc cgacaagccc 1800
aaggacaagc gatctatcaa ggacgaggat tacgtcgaat ggcgagccaa ggttgccaag 1860
attcccttcg acacctacgg ctctccttat ggttcggctc accagatgtc ttcctgtcgt 1920
atgagcggca agggtcccaa gtacggagcc gtcgataccg acggtcgact gtttgagtgc 1980
tcgaacgtgt acgttgccga cgcttccctt ctgcccactg ctagcggtgc caaccctatg 2040
gtcacaacca tgactctcgc tcgacacgtt gccctcggct tggcagactc cctgaagacc 2100
aaagccaagc tctaa 2115
<210> 100
<211> 704
<212> PRT
<213> Artificial sequence
<220>
<223> CtFAO1M (FAO) protein
<400> 100
Met Ala Pro Phe Leu Pro Asp Gln Val Asp Tyr Lys His Val Asp Thr
1 5 10 15
Leu Met Leu Leu Cys Asp Gly Ile Ile His Glu Thr Thr Val Asp Glu
20 25 30
Ile Lys Asp Val Ile Ala Pro Asp Phe Pro Ala Asp Lys Tyr Glu Glu
35 40 45
Tyr Val Arg Thr Phe Thr Lys Pro Ser Glu Thr Pro Gly Phe Arg Glu
50 55 60
Thr Val Tyr Asn Thr Val Asn Ala Asn Thr Met Asp Ala Ile His Gln
65 70 75 80
Phe Ile Ile Leu Thr Asn Val Leu Gly Ser Arg Val Leu Ala Pro Ala
85 90 95
Leu Thr Asn Ser Leu Thr Pro Ile Lys Asp Met Ser Leu Glu Asp Arg
100 105 110
Glu Lys Leu Leu Ala Ser Trp Arg Asp Ser Pro Ile Ala Ala Lys Arg
115 120 125
Lys Leu Phe Arg Leu Val Ser Thr Leu Thr Leu Val Thr Phe Thr Arg
130 135 140
Leu Ala Asn Glu Leu His Leu Lys Ala Ile His Tyr Pro Gly Arg Glu
145 150 155 160
Asp Arg Glu Lys Ala Tyr Glu Thr Gln Glu Ile Asp Pro Phe Lys Tyr
165 170 175
Gln Phe Leu Glu Lys Pro Lys Phe Tyr Gly Ala Glu Leu Tyr Leu Pro
180 185 190
Asp Ile Asp Val Ile Ile Ile Gly Ser Gly Ala Gly Ala Gly Val Val
195 200 205
Ala His Thr Leu Thr Asn Asp Gly Phe Lys Ser Leu Val Leu Glu Lys
210 215 220
Gly Arg Tyr Phe Ser Asn Ser Glu Leu Asn Phe Asp Asp Lys Asp Gly
225 230 235 240
Val Gln Glu Leu Tyr Gln Ser Gly Gly Thr Leu Thr Thr Val Asn Gln
245 250 255
Gln Leu Phe Val Leu Ala Gly Ser Thr Phe Gly Gly Gly Thr Thr Val
260 265 270
Asn Trp Ser Ala Cys Leu Lys Thr Pro Phe Lys Val Arg Lys Glu Trp
275 280 285
Tyr Asp Glu Phe Gly Val Asp Phe Ala Ala Asp Glu Ala Tyr Asp Lys
290 295 300
Ala Gln Asp Tyr Val Trp Gln Gln Met Gly Ala Ser Thr Glu Gly Ile
305 310 315 320
Thr His Ser Leu Ala Asn Glu Ile Ile Ile Glu Gly Gly Lys Lys Leu
325 330 335
Gly Tyr Lys Ala Lys Val Leu Asp Gln Asn Ser Gly Gly His Pro His
340 345 350
His Arg Cys Gly Phe Cys His Leu Gly Cys Lys His Gly Ile Lys Gln
355 360 365
Gly Ser Val Asn Asn Trp Phe Arg Asp Ala Ala Ala His Gly Ser Gln
370 375 380
Phe Met Gln Gln Val Arg Val Leu Gln Ile Leu Asn Lys Lys Gly Ile
385 390 395 400
Ala Tyr Gly Ile Leu Cys Glu Asp Val Val Thr Gly Ala Lys Phe Thr
405 410 415
Ile Thr Gly Pro Lys Lys Phe Val Val Ala Ala Gly Ala Leu Asn Thr
420 425 430
Pro Ser Val Leu Val Asn Ser Gly Phe Lys Asn Lys Asn Ile Gly Lys
435 440 445
Asn Leu Thr Leu His Pro Val Ser Val Val Phe Gly Asp Phe Gly Lys
450 455 460
Asp Val Gln Ala Asp His Phe His Asn Ser Ile Met Thr Ala Leu Cys
465 470 475 480
Ser Glu Ala Ala Asp Leu Asp Gly Lys Gly His Gly Cys Arg Ile Glu
485 490 495
Thr Ile Leu Asn Ala Pro Phe Ile Gln Ala Ser Phe Leu Pro Trp Arg
500 505 510
Gly Ser Asn Glu Ala Arg Arg Asp Leu Leu Arg Tyr Asn Asn Met Val
515 520 525
Ala Met Leu Leu Leu Ser Arg Asp Thr Thr Ser Gly Ser Val Ser Ser
530 535 540
His Pro Thr Lys Pro Glu Ala Leu Val Val Glu Tyr Asp Val Asn Lys
545 550 555 560
Phe Asp Arg Asn Ser Ile Leu Gln Ala Leu Leu Val Thr Ala Asp Leu
565 570 575
Leu Tyr Ile Gln Gly Ala Lys Arg Ile Leu Ser Pro Gln Pro Trp Val
580 585 590
Pro Ile Phe Glu Ser Asp Lys Pro Lys Asp Lys Arg Ser Ile Lys Asp
595 600 605
Glu Asp Tyr Val Glu Trp Arg Ala Lys Val Ala Lys Ile Pro Phe Asp
610 615 620
Thr Tyr Gly Ser Pro Tyr Gly Ser Ala His Gln Met Ser Ser Cys Arg
625 630 635 640
Met Ser Gly Lys Gly Pro Lys Tyr Gly Ala Val Asp Thr Asp Gly Arg
645 650 655
Leu Phe Glu Cys Ser Asn Val Tyr Val Ala Asp Ala Ser Leu Leu Pro
660 665 670
Thr Ala Ser Gly Ala Asn Pro Met Val Thr Thr Met Thr Leu Ala Arg
675 680 685
His Val Ala Leu Gly Leu Ala Asp Ser Leu Lys Thr Lys Ala Lys Leu
690 695 700
<210> 101
<211> 2100
<212> DNA
<213> Artificial sequence
<220>
<223> CcFAO1 (FAO)
<400> 101
atggcctccc accaggtcga ggaccacgat ctggacgtgt tctgcctcct ggccgacgct 60
gttctccacg agattcctcc ctccgaaatc gtcgagtacc ttcatcccga tttccccaag 120
gacaagatcg aagagtacct gaccggcttt tctcgaccct ccgccgttcc tcagttccga 180
cagtgtgcca agaaactcat caaccgaggt tccgagctgt cgatcaagct cttcctttac 240
ttgaccactg ctctggactc tcgaatcctt gcaccagccc tgaccaactc gctcactctg 300
atcagagaca tggatctttc ccagcgagag gaactgttgc ggtcctggcg agattctcca 360
ctgactgcca agcgaagact ctttcgagtc tacgcctcct ttaccctgtc tacattcaac 420
aagctcggaa ccgacttgca cttcaaggcc ctgggctacc ctggtcgaga gctccggacc 480
cagattcaag actacgaggt cgatcccttt cgatactcgt tcatggagaa gctcaaacac 540
gagggacatg aactgttcct tcccgatatc gacgttctga tcattggctc tggatccggt 600
gcaggcgtgg tcgctcagac tcttaccgag agcggactca agtctctggt tctcgagaag 660
ggcaagtact ttgcctccga agagctgtgc atgaccgatc tcgacggaaa cgaggccctg 720
ttcgaaagcg gtggcactat tccttccacc aatcaacagt tgttcatgat cgctggatct 780
acttttggtg gaggctccac cgtcaactgg tctgcctgtc tcaagactcc cttcaaggtt 840
cgaaaggagt ggtacgacga tttcggactg gactttgtgg ctacccagca atacgacgat 900
tgcatggact acgtgtggaa gaaaatgggt gcctcgaccg agcacatcga gcattctgct 960
gcaaatgccg tcatcatgga cggagctgcc aagcttggct acgctcaccg agccctcgag 1020
cagaacaccg gtggccatgt tcacgactgt ggaatgtgcc acctgggctg tcgattcggt 1080
atcaagcagg gaggcgtcaa ctgctggttt cgagaaccct ccgagaaggg ttccaagttc 1140
atggagcagg tcgttgtcga gaagattctg cagcacaagg gcaaggccac tggaattctc 1200
tgcagagata ccgagtctgg catcaagttc aagattactg gacccaagaa atacgtcgtg 1260
tccggtggct ctttgcagac ccctgttctc cttcagaagt ctggcttcaa gaacaagcac 1320
attggagcca acctcaagct gcatcccgtc tcggttgctc ttggcgactt tggtaacgag 1380
gtggacttcg aagcctacaa gcgacccctc atgaccgcag tctgcaatgc cgtggacgat 1440
ctggacggca aggctcacgg aacacgaatc gaggccattc tgcacgctcc ttacgtcact 1500
gctcccttct atccctggca gtccggtgcc caggctcgga agaacttgct caaatacaag 1560
cagaccgtgc ctctgctcct tctgtctcga gacacctcct cgggtaccgt tacatacgac 1620
aaacagaagc cagatgtctt ggtggtcgac tacactgtca acaagttcga tcgaaactcc 1680
atcctgcagg gatttctcgt tgcttccgac attttgtaca tcgagggtgc caaggagatt 1740
ctgtctcccc aggcttgggt gcccaccttc aagagcaaca agcccaagca cgccagatcc 1800
atcaaggacg aggactacgt caagtggcga gaaaccgtgg ccaagattcc ctttgattcc 1860
tacggctcgc cttacggttc tgctcatcag atgtcctcgt gtcgaatgtc tggcaaggga 1920
cccggatacg gtgcctgcga taccaaggga cgactcttcg agtgcaacaa tgtgtacgtt 1980
gcagacgcct ccgtcatgcc tactgcttct ggagtcaacc ccatgattac cacaatggcc 2040
tttgcacgac acgttgctct ctgtctggcc aaggaccttc aaccccagac caagctgtaa 2100
<210> 102
<211> 699
<212> PRT
<213> Artificial sequence
<220>
<223> CcFAO1 (FAO) protein
<400> 102
Met Ala Ser His Gln Val Glu Asp His Asp Leu Asp Val Phe Cys Leu
1 5 10 15
Leu Ala Asp Ala Val Leu His Glu Ile Pro Pro Ser Glu Ile Val Glu
20 25 30
Tyr Leu His Pro Asp Phe Pro Lys Asp Lys Ile Glu Glu Tyr Leu Thr
35 40 45
Gly Phe Ser Arg Pro Ser Ala Val Pro Gln Phe Arg Gln Cys Ala Lys
50 55 60
Lys Leu Ile Asn Arg Gly Ser Glu Leu Ser Ile Lys Leu Phe Leu Tyr
65 70 75 80
Leu Thr Thr Ala Leu Asp Ser Arg Ile Leu Ala Pro Ala Leu Thr Asn
85 90 95
Ser Leu Thr Leu Ile Arg Asp Met Asp Leu Ser Gln Arg Glu Glu Leu
100 105 110
Leu Arg Ser Trp Arg Asp Ser Pro Leu Thr Ala Lys Arg Arg Leu Phe
115 120 125
Arg Val Tyr Ala Ser Phe Thr Leu Ser Thr Phe Asn Lys Leu Gly Thr
130 135 140
Asp Leu His Phe Lys Ala Leu Gly Tyr Pro Gly Arg Glu Leu Arg Thr
145 150 155 160
Gln Ile Gln Asp Tyr Glu Val Asp Pro Phe Arg Tyr Ser Phe Met Glu
165 170 175
Lys Leu Lys His Glu Gly His Glu Leu Phe Leu Pro Asp Ile Asp Val
180 185 190
Leu Ile Ile Gly Ser Gly Ser Gly Ala Gly Val Val Ala Gln Thr Leu
195 200 205
Thr Glu Ser Gly Leu Lys Ser Leu Val Leu Glu Lys Gly Lys Tyr Phe
210 215 220
Ala Ser Glu Glu Leu Cys Met Thr Asp Leu Asp Gly Asn Glu Ala Leu
225 230 235 240
Phe Glu Ser Gly Gly Thr Ile Pro Ser Thr Asn Gln Gln Leu Phe Met
245 250 255
Ile Ala Gly Ser Thr Phe Gly Gly Gly Ser Thr Val Asn Trp Ser Ala
260 265 270
Cys Leu Lys Thr Pro Phe Lys Val Arg Lys Glu Trp Tyr Asp Asp Phe
275 280 285
Gly Leu Asp Phe Val Ala Thr Gln Gln Tyr Asp Asp Cys Met Asp Tyr
290 295 300
Val Trp Lys Lys Met Gly Ala Ser Thr Glu His Ile Glu His Ser Ala
305 310 315 320
Ala Asn Ala Val Ile Met Asp Gly Ala Ala Lys Leu Gly Tyr Ala His
325 330 335
Arg Ala Leu Glu Gln Asn Thr Gly Gly His Val His Asp Cys Gly Met
340 345 350
Cys His Leu Gly Cys Arg Phe Gly Ile Lys Gln Gly Gly Val Asn Cys
355 360 365
Trp Phe Arg Glu Pro Ser Glu Lys Gly Ser Lys Phe Met Glu Gln Val
370 375 380
Val Val Glu Lys Ile Leu Gln His Lys Gly Lys Ala Thr Gly Ile Leu
385 390 395 400
Cys Arg Asp Thr Glu Ser Gly Ile Lys Phe Lys Ile Thr Gly Pro Lys
405 410 415
Lys Tyr Val Val Ser Gly Gly Ser Leu Gln Thr Pro Val Leu Leu Gln
420 425 430
Lys Ser Gly Phe Lys Asn Lys His Ile Gly Ala Asn Leu Lys Leu His
435 440 445
Pro Val Ser Val Ala Leu Gly Asp Phe Gly Asn Glu Val Asp Phe Glu
450 455 460
Ala Tyr Lys Arg Pro Leu Met Thr Ala Val Cys Asn Ala Val Asp Asp
465 470 475 480
Leu Asp Gly Lys Ala His Gly Thr Arg Ile Glu Ala Ile Leu His Ala
485 490 495
Pro Tyr Val Thr Ala Pro Phe Tyr Pro Trp Gln Ser Gly Ala Gln Ala
500 505 510
Arg Lys Asn Leu Leu Lys Tyr Lys Gln Thr Val Pro Leu Leu Leu Leu
515 520 525
Ser Arg Asp Thr Ser Ser Gly Thr Val Thr Tyr Asp Lys Gln Lys Pro
530 535 540
Asp Val Leu Val Val Asp Tyr Thr Val Asn Lys Phe Asp Arg Asn Ser
545 550 555 560
Ile Leu Gln Gly Phe Leu Val Ala Ser Asp Ile Leu Tyr Ile Glu Gly
565 570 575
Ala Lys Glu Ile Leu Ser Pro Gln Ala Trp Val Pro Thr Phe Lys Ser
580 585 590
Asn Lys Pro Lys His Ala Arg Ser Ile Lys Asp Glu Asp Tyr Val Lys
595 600 605
Trp Arg Glu Thr Val Ala Lys Ile Pro Phe Asp Ser Tyr Gly Ser Pro
610 615 620
Tyr Gly Ser Ala His Gln Met Ser Ser Cys Arg Met Ser Gly Lys Gly
625 630 635 640
Pro Gly Tyr Gly Ala Cys Asp Thr Lys Gly Arg Leu Phe Glu Cys Asn
645 650 655
Asn Val Tyr Val Ala Asp Ala Ser Val Met Pro Thr Ala Ser Gly Val
660 665 670
Asn Pro Met Ile Thr Thr Met Ala Phe Ala Arg His Val Ala Leu Cys
675 680 685
Leu Ala Lys Asp Leu Gln Pro Gln Thr Lys Leu
690 695
<210> 103
<211> 2100
<212> DNA
<213> Artificial sequence
<220>
<223> CcFAO2 (FAO)
<400> 103
atggccaacc ccgtcgtgga ggactcccat ctggacgtct tctgcctcct tgccgatgct 60
gtggttcacg agattcctcc ctccgagatc gtcgagtacc tgcatcctga ctttcccaag 120
gacaaggtcg aagagtacct tgccgagttc tctcatccct ccgctattcc agagttccga 180
gaggttgcca agcgaatcat taacaagggc accgtgctgt cgatcaagct ctttctgctc 240
ttggccactg ctctggattc tcgaatcctt gctcctgcct tgaccaactc cactacactc 300
atccgagaca tggatctttc tcagcgggag gaactcctga gatcctggcg agactctccc 360
ttcactacca aacgaaagct gttccgagtc tacaactcgt ttaccctcaa cgccttcagc 420
aagactgcta ccgaccttca cttcaaggcc ctgggatacc ctggtcgaga gctccgtact 480
cagattcagg actacgaggt cgatcccttt cgatacacgt tcctggagaa gcctcagcaa 540
gacggacagg agctccactt tcccgacatt gatgtgctca ttatcggatc tggctccggt 600
gcaggcgtcg ttgctcagac tctttcggag aacggactca agtctctggt gctcgagaag 660
ggcaaatact tttccaacga cgagctgacc atgaacgacc tcgaaggttc cgaggccctg 720
ttcgaaaacg gaggtgctct ctcctctacc aatcaacaga tctttatcat tgccggatcg 780
actttcggtg gcggatccac agtcaactgg tctgcctgtc tcaagactcc cttcaaggtg 840
cgaaaggagt ggtacgacaa ctttggactg gatttcgttg ctacccagta ttacgaggac 900
tgtatggact acgtctggaa gaaaatgggt gcctccaacg agaatatcga ccattctgga 960
gccaactcgg tcattctgga aggttccaag aaacttggct accctcaccg tgccgtggaa 1020
cagaacaatg gaggcaagat tcacgactgt ggtatgtgcc acctcggatg tcgatttggc 1080
atcaagcagg gatcggtcaa ctgctggttt cgtggtccct ccgagaacgg ctcgaagttc 1140
atgcagcaag ttctcgtgga caagatcctg cagcgagatg gcaaggctgt cggtgttctc 1200
tgtagagacg tggttaccgg agtcaagttc aagatcactg gacccaagaa aatcgtcgtg 1260
ttctggtggt tctttgccaa ctccggattt gttaccaagt ctggtttcaa gaacaagcac 1320
atcggtgcaa acctcaagct gcatcccgtc agccttacgc tcggcgactt tggtaacaat 1380
gtggatttcg aggcctaccg aaagccaatc atgacctcca tttgtaacaa ggtcgaggac 1440
ctggatggaa aggctcacgg cactcgaatc gaggccatgc tcaatgctcc ctacggtgtt 1500
gctccattct ttccctggaa gtctggcgca gagtcccgaa aggacctcct gcgatacaag 1560
cagactgtgc ccattctcct gctttccaga gacaccactt ctggatccgt cacctacgac 1620
aaacagaagc ccgatgcctt ggtgatcgac tacctgctca acaagttcga ccgaaactcc 1680
atcctgcagg gctttctcat tgcttcggat cttctgtaca tcgagggtgc cagccgagac 1740
catgttacct acaagcttgg ataccagtgg ttcaagtctt ccaagcccaa gcacgctcga 1800
tccatcgaag acgaggacta cgtcaactgg agagccaagg ttgcaaagat tccctttgat 1860
tcctatggat ctccttacgg ttcggctcac cagatgtcca cttgcagaat gtctggcaag 1920
ggaccaggct acggagcctg cgacaccaag ggcaaactct tcgagtgcag caacgtgtac 1980
gtcgccgatg cttccactct gcccaccgca tctggtgcca accctatggt ctctaccatg 2040
tcctttgccc gacacgtgtc gcttggcatc gtcaaggagc tgcagcaatc caagctctaa 2100
<210> 104
<211> 699
<212> PRT
<213> Artificial sequence
<220>
<223> CcFAO2 (FAO) protein
<400> 104
Met Ala Asn Pro Val Val Glu Asp Ser His Leu Asp Val Phe Cys Leu
1 5 10 15
Leu Ala Asp Ala Val Val His Glu Ile Pro Pro Ser Glu Ile Val Glu
20 25 30
Tyr Leu His Pro Asp Phe Pro Lys Asp Lys Val Glu Glu Tyr Leu Ala
35 40 45
Glu Phe Ser His Pro Ser Ala Ile Pro Glu Phe Arg Glu Val Ala Lys
50 55 60
Arg Ile Ile Asn Lys Gly Thr Val Leu Ser Ile Lys Leu Phe Leu Leu
65 70 75 80
Leu Ala Thr Ala Leu Asp Ser Arg Ile Leu Ala Pro Ala Leu Thr Asn
85 90 95
Ser Thr Thr Leu Ile Arg Asp Met Asp Leu Ser Gln Arg Glu Glu Leu
100 105 110
Leu Arg Ser Trp Arg Asp Ser Pro Phe Thr Thr Lys Arg Lys Leu Phe
115 120 125
Arg Val Tyr Asn Ser Phe Thr Leu Asn Ala Phe Ser Lys Thr Ala Thr
130 135 140
Asp Leu His Phe Lys Ala Leu Gly Tyr Pro Gly Arg Glu Leu Arg Thr
145 150 155 160
Gln Ile Gln Asp Tyr Glu Val Asp Pro Phe Arg Tyr Thr Phe Leu Glu
165 170 175
Lys Pro Gln Gln Asp Gly Gln Glu Leu His Phe Pro Asp Ile Asp Val
180 185 190
Leu Ile Ile Gly Ser Gly Ser Gly Ala Gly Val Val Ala Gln Thr Leu
195 200 205
Ser Glu Asn Gly Leu Lys Ser Leu Val Leu Glu Lys Gly Lys Tyr Phe
210 215 220
Ser Asn Asp Glu Leu Thr Met Asn Asp Leu Glu Gly Ser Glu Ala Leu
225 230 235 240
Phe Glu Asn Gly Gly Ala Leu Ser Ser Thr Asn Gln Gln Ile Phe Ile
245 250 255
Ile Ala Gly Ser Thr Phe Gly Gly Gly Ser Thr Val Asn Trp Ser Ala
260 265 270
Cys Leu Lys Thr Pro Phe Lys Val Arg Lys Glu Trp Tyr Asp Asn Phe
275 280 285
Gly Leu Asp Phe Val Ala Thr Gln Tyr Tyr Glu Asp Cys Met Asp Tyr
290 295 300
Val Trp Lys Lys Met Gly Ala Ser Asn Glu Asn Ile Asp His Ser Gly
305 310 315 320
Ala Asn Ser Val Ile Leu Glu Gly Ser Lys Lys Leu Gly Tyr Pro His
325 330 335
Arg Ala Val Glu Gln Asn Asn Gly Gly Lys Ile His Asp Cys Gly Met
340 345 350
Cys His Leu Gly Cys Arg Phe Gly Ile Lys Gln Gly Ser Val Asn Cys
355 360 365
Trp Phe Arg Gly Pro Ser Glu Asn Gly Ser Lys Phe Met Gln Gln Val
370 375 380
Leu Val Asp Lys Ile Leu Gln Arg Asp Gly Lys Ala Val Gly Val Leu
385 390 395 400
Cys Arg Asp Val Val Thr Gly Val Lys Phe Lys Ile Thr Gly Pro Lys
405 410 415
Lys Ile Val Val Phe Trp Trp Phe Phe Ala Asn Ser Gly Phe Val Thr
420 425 430
Lys Ser Gly Phe Lys Asn Lys His Ile Gly Ala Asn Leu Lys Leu His
435 440 445
Pro Val Ser Leu Thr Leu Gly Asp Phe Gly Asn Asn Val Asp Phe Glu
450 455 460
Ala Tyr Arg Lys Pro Ile Met Thr Ser Ile Cys Asn Lys Val Glu Asp
465 470 475 480
Leu Asp Gly Lys Ala His Gly Thr Arg Ile Glu Ala Met Leu Asn Ala
485 490 495
Pro Tyr Gly Val Ala Pro Phe Phe Pro Trp Lys Ser Gly Ala Glu Ser
500 505 510
Arg Lys Asp Leu Leu Arg Tyr Lys Gln Thr Val Pro Ile Leu Leu Leu
515 520 525
Ser Arg Asp Thr Thr Ser Gly Ser Val Thr Tyr Asp Lys Gln Lys Pro
530 535 540
Asp Ala Leu Val Ile Asp Tyr Leu Leu Asn Lys Phe Asp Arg Asn Ser
545 550 555 560
Ile Leu Gln Gly Phe Leu Ile Ala Ser Asp Leu Leu Tyr Ile Glu Gly
565 570 575
Ala Ser Arg Asp His Val Thr Tyr Lys Leu Gly Tyr Gln Trp Phe Lys
580 585 590
Ser Ser Lys Pro Lys His Ala Arg Ser Ile Glu Asp Glu Asp Tyr Val
595 600 605
Asn Trp Arg Ala Lys Val Ala Lys Ile Pro Phe Asp Ser Tyr Gly Ser
610 615 620
Pro Tyr Gly Ser Ala His Gln Met Ser Thr Cys Arg Met Ser Gly Lys
625 630 635 640
Gly Pro Gly Tyr Gly Ala Cys Asp Thr Lys Gly Lys Leu Phe Glu Cys
645 650 655
Ser Asn Val Tyr Val Ala Asp Ala Ser Thr Leu Pro Thr Ala Ser Gly
660 665 670
Ala Asn Pro Met Val Ser Thr Met Ser Phe Ala Arg His Val Ser Leu
675 680 685
Gly Ile Val Lys Glu Leu Gln Gln Ser Lys Leu
690 695
<210> 105
<211> 12358
<212> DNA
<213> Artificial sequence
<220>
<223> pZKLY-VsCPR & CYP plasmid
<400> 105
aaaccatcat ctaagggcct caaaactacc tcggaactgc tgcgctgatc tggacaccac 60
agaggttccg agcactttag gttgcaccaa atgtcccacc aggtgcaggc agaaaacgct 120
ggaacagcgt gtacagtttg tcttaacaaa aagtgagggc gctgaggtcg agcagggtgg 180
tgtgacttgt tatagccttt agagctgcga aagcgcgtat ggatttggct catcaggcca 240
gattgagggt ctgtggacac atgtcatgtt agtgtacttc aatcgccccc tggatatagc 300
cccgacaata ggccgtggcc tcattttttt gccttccgca catttccatt gctcggtacc 360
cacaccttgc ttctcctgca cttgccaacc ttaatactgg tttacattga ccaacatctt 420
acaagcgggg ggcttgtcta gggtatatat aaacagtggc tctcccaatc ggttgccagt 480
ctcttttttc ctttctttcc ccacagattc gaaatctaaa ctacacatca caccatggcc 540
acctcctcta actccgacct ggtccgaacc atcgagtccg ccctcggcat ttctctcggc 600
gacagcgtgt ccgattctgt cgttatcatt gccaccactt ctgctgccgt catcattgga 660
cttctggtct tcctctggcg aaagtctccc gacagatcgc gagagctgcg tcctgtcatt 720
gtgcccaagt ttaccgttaa gcacgaggac gatgaagtcg aggtggaccg aggcaaaacc 780
aaggttacag ttttctacgg aactcagacc ggtactgccg agggctttgc aaaggccctt 840
gcggaggaaa tcaaggccag atacgagaag gccgttgtca aggtggttga catggatgac 900
tacgctattg acgatgacca gtacgaggaa aagctcaaaa aggagaccct ggtcttcttt 960
atgcttgcca cctatggaga cggcgaaccc accgataacg ctgcacgatt ctacaagtgg 1020
tttaccgagg gcaaggagga acgaggaacc tggctgcagc aactcactta cggtgtgttc 1080
gccttgggca accgacagta cgagcatttc aacaagatcg gcaagattgt cgacgaggat 1140
cttaccgagc agggagccaa gcgtctggtt cccgtcggtc tcggcgacga tgaccagtcc 1200
atcgaggacg atttcaacgc ttggaaggaa accttgtggc cagagctgga ccaacttctc 1260
cgagacgagg atgacgtcaa cactgcttcg accccttaca ctgccgctat ctccgagtat 1320
cgtgtcgtta tccacgatcc taccgtgtct ccctcctacg agaaccattt caatgttgcc 1380
aacggtggag cagtgttcga cattcaccat ccctgtcgag tcaacgttgc cgtgcgacgg 1440
gaacttcaca agccccagtc cgaccgatct tgcattcacc tggagtttga tctctccggt 1500
actggcgtta catacgagac tggcgaccac gtcggagtgt acgccgagaa ctgcgacgaa 1560
actgtcgagg aagctggcaa gctgctcggt cagtcgctgg atcttctctt ttctctgcat 1620
accgacaagg aggatggcac aagccttggt ggatctctgc tccctccatt tcctggaccc 1680
tgtaccgttc gaactgccct cgcttgctac gccgaccttc ttaatcctcc acggaaagcc 1740
gctatcgtgg cacttgctgc ccatgcttcc gagcccagcg aggccgaacg actcaagttt 1800
ctttcttcgc ctcagggcaa ggacgagtac tccaagtggg tcgttggatc tcagcgatcg 1860
ctgctcgaag tcatggccga ttttccctcc gccaagcctc cactgggagt gttctttgct 1920
gccattgcac ctcgactgca gcctcgatac tattctatct cctcttcgcc cagaccagct 1980
ccccagcgag tgcacgttac ctgtgccctt gtcgagggac ccactcctac cggtcggatt 2040
cacaagggtg tgtgctccac ctggatgaag tctgctactc ccttggagaa gtctcacgac 2100
tgttcccgag cacctatctt cattcgaccc tccaacttca agcttcctgc cgaccattcg 2160
attcccatta tcatggtcgg acctggtaca ggtctggctc cctttcgagg attcctccag 2220
gaacgacttg ccctcaagga ggatggagtt cagcttggac ctgccctgct cttctttggt 2280
tgccgaaaca gacagatgga cttcatctac gaggacgaac tcaacaattt cgttcagcaa 2340
ggtgccattt ccgagctcat cgttgcgttt tctcgagagg gcccagaaaa ggagtacgtg 2400
cagcacaaga tgatggacaa ggccgagtat ctgtggtctc tcatttcgca gggaggctac 2460
ctgtacgtct gtggtgatgc caaaggcatg gctcgagacg tgcaccgatc ccttcatacc 2520
attgttcagc aacaggagaa cgcagattct tcgaaggctg aggccactgt caagaaactc 2580
cagatggacg gaagatacct gcgagacgtg tggtaagcgg ccgcatgaga agataaatat 2640
ataaatacat tgagatatta aatgcgctag attagagagc ctcatactgc tcggagagaa 2700
gccaagacga gtactcaaag gggattacac catccatatc cacagacaca agctggggaa 2760
aggttctata tacactttcc ggaataccgt agtttccgat gttatcaatg ggggcagcca 2820
ggatttcagg cacttcggtg tctcggggtg aaatggcgtt cttggcctcc atcaagtcgt 2880
accatgtctt catttgcctg tcaaagtaaa acagaagcag atgaagaatg aacttgaagt 2940
gaaggaattt aaatgtaacg aaactgaaat ttgaccagat attgtgtccg cggtggagct 3000
ccagcttttg ttccctttag tgagggttaa tttcgagctt ggcgtaatca tggtcatagc 3060
tgtttcctgt gtgaaattgt tatccgctca caagcttcca cacaacgtac gttgattgag 3120
gtggagccag atgggctatt gtttcatata tagactggca gccacctctt tggcccagca 3180
tgtttgtata cctggaaggg aaaactaaag aagctggcta gtttagtttg attattatag 3240
tagatgtcct aatcactaga gattagaatg tcttggcgat gattagtcgt cgtcccctgt 3300
atcatgtcta gaccaactgt gtcatgaagt tggtgctggt gttttacctg tgtactacaa 3360
gtaggtgtcc tagatctagt gtacagagcc gtttagaccc atgtggactt caccattaac 3420
gatggaaaat gttcattata tgacagtata ttacaatgga cttgctccat ttcttccttg 3480
catcacatgt tctccacctc catagttgat caacacatca tagtagctaa ggctgctgct 3540
ctcccactac agtccaccac aagttaagta gcaccgtcag tacagctaaa agtacacgtc 3600
tagtacgttt cataactagt caagtagccc ctattacaga tatcagcact atcacgcacg 3660
agtttttctc tgtgctatct aatcaacttg ccaagtattc ggagaagata cactttcttg 3720
gcatcaggta tacgagggag cctatcagat gaaaaagggt atattggatc cattcatatc 3780
cacctacacg ttgtcataat ctcctcattc acgtgattca tttcgtgaca ctagtttctc 3840
actttccccc ccgcacctat agtcaacttg gcggacacgc tacttgtagc tgacgttgat 3900
ttatagaccc aatcaaagcg ggttatcggt caggtagcac ttatcattca tcgttcatac 3960
tacgatgagc aatctcgggc atgtccggaa aagtgtcggg cgcgccagct gcattaatga 4020
atcggccaac gcgcggggag aggcggtttg cgtattgggc gctcttccgc ttcctcgctc 4080
actgactcgc tgcgctcggt cgttcggctg cggcgagcgg tatcagctca ctcaaaggcg 4140
gtaatacggt tatccacaga atcaggggat aacgcaggaa agaacatgtg agcaaaaggc 4200
cagcaaaagg ccaggaaccg taaaaaggcc gcgttgctgg cgtttttcca taggctccgc 4260
ccccctgacg agcatcacaa aaatcgacgc tcaagtcaga ggtggcgaaa cccgacagga 4320
ctataaagat accaggcgtt tccccctgga agctccctcg tgcgctctcc tgttccgacc 4380
ctgccgctta ccggatacct gtccgccttt ctcccttcgg gaagcgtggc gctttctcat 4440
agctcacgct gtaggtatct cagttcggtg taggtcgttc gctccaagct gggctgtgtg 4500
cacgaacccc ccgttcagcc cgaccgctgc gccttatccg gtaactatcg tcttgagtcc 4560
aacccggtaa gacacgactt atcgccactg gcagcagcca ctggtaacag gattagcaga 4620
gcgaggtatg taggcggtgc tacagagttc ttgaagtggt ggcctaacta cggctacact 4680
agaagaacag tatttggtat ctgcgctctg ctgaagccag ttaccttcgg aaaaagagtt 4740
ggtagctctt gatccggcaa acaaaccacc gctggtagcg gtggtttttt tgtttgcaag 4800
cagcagatta cgcgcagaaa aaaaggatct caagaagatc ctttgatctt ttctacgggg 4860
tctgacgctc agtggaacga aaactcacgt taagggattt tggtcatgag attatcaaaa 4920
aggatcttca cctagatcct tttaaattaa aaatgaagtt ttaaatcaat ctaaagtata 4980
tatgagtaaa cttggtctga cagttaccaa tgcttaatca gtgaggcacc tatctcagcg 5040
atctgtctat ttcgttcatc catagttgcc tgactccccg tcgtgtagat aactacgata 5100
cgggagggct taccatctgg ccccagtgct gcaatgatac cgcgagaccc acgctcaccg 5160
gctccagatt tatcagcaat aaaccagcca gccggaaggg ccgagcgcag aagtggtcct 5220
gcaactttat ccgcctccat ccagtctatt aattgttgcc gggaagctag agtaagtagt 5280
tcgccagtta atagtttgcg caacgttgtt gccattgcta caggcatcgt ggtgtcacgc 5340
tcgtcgtttg gtatggcttc attcagctcc ggttcccaac gatcaaggcg agttacatga 5400
tcccccatgt tgtgcaaaaa agcggttagc tccttcggtc ctccgatcgt tgtcagaagt 5460
aagttggccg cagtgttatc actcatggtt atggcagcac tgcataattc tcttactgtc 5520
atgccatccg taagatgctt ttctgtgact ggtgagtact caaccaagtc attctgagaa 5580
tagtgtatgc ggcgaccgag ttgctcttgc ccggcgtcaa tacgggataa taccgcgcca 5640
catagcagaa ctttaaaagt gctcatcatt ggaaaacgtt cttcggggcg aaaactctca 5700
aggatcttac cgctgttgag atccagttcg atgtaaccca ctcgtgcacc caactgatct 5760
tcagcatctt ttactttcac cagcgtttct gggtgagcaa aaacaggaag gcaaaatgcc 5820
gcaaaaaagg gaataagggc gacacggaaa tgttgaatac tcatactctt cctttttcaa 5880
tattattgaa gcatttatca gggttattgt ctcatgagcg gatacatatt tgaatgtatt 5940
tagaaaaata aacaaatagg ggttccgcgc acatttcccc gaaaagtgcc acctgatgcg 6000
gtgtgaaata ccgcacagat gcgtaaggag aaaataccgc atcaggaaat tgtaagcgtt 6060
aatattttgt taaaattcgc gttaaatttt tgttaaatca gctcattttt taaccaatag 6120
gccgaaatcg gcaaaatccc ttataaatca aaagaataga ccgagatagg gttgagtgtt 6180
gttccagttt ggaacaagag tccactatta aagaacgtgg actccaacgt caaagggcga 6240
aaaaccgtct atcagggcga tggcccacta cgtgaaccat caccctaatc aagttttttg 6300
gggtcgaggt gccgtaaagc actaaatcgg aaccctaaag ggagcccccg atttagagct 6360
tgacggggaa agccggcgaa cgtggcgaga aaggaaggga agaaagcgaa aggagcgggc 6420
gctagggcgc tggcaagtgt agcggtcacg ctgcgcgtaa ccaccacacc cgccgcgctt 6480
aatgcgccgc tacagggcgc gtccattcgc cattcaggct gcgcaactgt tgggaagggc 6540
gatcggtgcg ggcctcttcg ctattacgcc agctggcgaa agggggatgt gctgcaaggc 6600
gattaagttg ggtaacgcca gggttttccc agtcacgacg ttgtaaaacg acggccagtg 6660
aattgtaata cgactcacta tagggcgaat tgggcccgac gtcgcatgca ttccgacagc 6720
agcgactggg caccatgatc aagcgaaaca ccttccccca gctgccctgg caaaccatca 6780
agaaccctac tttcatcaag tgcaagaacg gttctactct tctcacctcc ggtgtctacg 6840
gctggtgccg aaagcctaac tacaccgctg atttcatcat gtgcctcacc tgggctctca 6900
tgtgcggtgt tgcttctccc ctgccttact tctacccggt cttcttcttc ctggtgctca 6960
tccaccgagc ttaccgagac tttgagcgac tggagcgaaa gtacggtgag gactaccagg 7020
agttcaagcg acaggtccct tggatcttca tcccttatgt tttctaaacg ataagcttag 7080
tgagcgaatg gtgaggttac ttaattgagt ggccagccta tgggattgta taacagacag 7140
tcaatatatt actgaaaaga ctgaacagcc agacggagtg aggttgtgag tgaatcgtag 7200
agggcggcta ttacagcaag tctactctac agtgtactaa cacagcagag aacaaataca 7260
ggtgtgcatt cggctatctg agaattagtt ggagagctcg agaccctcgg cgataaactg 7320
ctcctcggtt ttgtgtccat acttgtacgg accattgtaa tggggcaagt cgttgagttc 7380
tcgtcgtccg acgttcagag cacagaaacc aatgtaatca atgtagcaga gatggttctg 7440
caaaagattg atttgtgcga gcaggttaat taagttgcga cacatgtctt gatagtatct 7500
tgaattctct ctcttgagct tttccataac aagttcttct gcctccagga agtccatggg 7560
tggtttgatc atggttttgg tgtagtggta gtgcagtggt ggtattgtga ctggggatgt 7620
agttgagaat aagtcataca caagtcagct ttcttcgagc ctcatataag tataagtagt 7680
tcaacgtatt agcactgtac ccagcatctc cgtatcgaga aacacaacaa catgccccat 7740
tggacagatc atgcggatac acaggttgtg cagtatcata catactcgat cagacaggtc 7800
gtctgaccat catacaagct gaacaagcgc tccatacttg cacgctctct atatacacag 7860
ttaaattaca tatccatagt ctaacctcta acagttaatc ttctggtaag cctcccagcc 7920
agccttctgg tatcgcttgg cctcctcaat aggatctcgg ttctggccgt acagacctcg 7980
gccgacaatt atgatatccg ttccggtaga catgacatcc tcaacagttc ggtactgctg 8040
tccgagagcg tctcccttgt cgtcaagacc caccccgggg gtcagaataa gccagtcctc 8100
agagtcgccc ttaggtcggt tctgggcaat gaagccaacc acaaactcgg ggtcggatcg 8160
ggcaagctca atggtctgct tggagtactc gccagtggcc agagagccct tgcaagacag 8220
ctcggccagc atgagcagac ctctggccag cttctcgttg ggagagggga ctaggaactc 8280
cttgtactgg gagttctcgt agtcagagac gtcctccttc ttctgttcag agacagtttc 8340
ctcggcacca gctcgcaggc cagcaatgat tccggttccg ggtacaccgt gggcgttggt 8400
gatatcggac cactcggcga ttcggtgaca ccggtactgg tgcttgacag tgttgccaat 8460
atctgcgaac tttctgtcct cgaacaggaa gaaaccgtgc ttaagagcaa gttccttgag 8520
ggggagcaca gtgccggcgt aggtgaagtc gtcaatgatg tcgatatggg ttttgatcat 8580
gcacacataa ggtccgacct tatcggcaag ctcaatgagc tccttggtgg tggtaacatc 8640
cagagaagca cacaggttgg ttttcttggc tgccacgagc ttgagcactc gagcggcaaa 8700
ggcggacttg tggacgttag ctcgagcttc gtaggagggc attttggtgg tgaagaggag 8760
actgaaataa atttagtctg cagaactttt tatcggaacc ttatctgggg cagtgaagta 8820
tatgttatgg taatagttac gagttagttg aacttataga tagactggac tatacggcta 8880
tcggtccaaa ttagaaagaa cgtcaatggc tctctgggcg tcgcctttgc cgacaaaaat 8940
gtgatcatga tgaaagccag caatgacgtt gcagctgata ttgttgtcgg ccaaccgcgc 9000
cgaaaacgca gctgtcagac ccacagcctc caacgaagaa tgtatcgtca aagtgatcca 9060
agcacactca tagttggagt cgtactccaa aggcggcaat gacgagtcag acagatactc 9120
gtcgaccttt tccttgggaa ccaccaccgt cagcccttct gactcacgta ttgtagccac 9180
cgacacaggc aacagtccgt ggatagcaga atatgtcttg tcggtccatt tctcaccaac 9240
tttaggcgtc aagtgaatgt tgcagaagaa gtatgtgcct tcattgagaa tcggtgttgc 9300
tgatttcaat aaagtcttga gatcagtttg gccagtcatg ttgtgggggg taattggatt 9360
gagttatcgc ctacagtctg tacaggtata ctcgctgccc actttatact ttttgattcc 9420
gctgcacttg aagcaatgtc gtttaccaaa agtgagaatg ctccacagaa cacaccccag 9480
ggtatggttg agcaaaaaat aaacactccg atacggggaa tcgaaccccg gtctccacgg 9540
ttctcaagaa gtattcttga tgagagcgta tcgattgccc cggagaagac ggccaggccg 9600
cctagatgac aaattcaaca actcacagct gactttctgc cattgccact aggggggggc 9660
ctttttatat ggccaagcca agctctccac gtcggttggg ctgcacccaa caataaatgg 9720
gtagggttgc accaacaaag ggatgggatg gggggtagaa gatacgagga taacggggct 9780
caatggcaca aataagaacg aatactgcca ttaagactcg tgatccagcg actgacacca 9840
ttgcatcatc taagggcctc aaaactacct cggaactgct gcgctgatct ggacaccaca 9900
gaggttccga gcactttagg ttgcaccaaa tgtcccacca ggtgcaggca gaaaacgctg 9960
gaacagcgtg tacagtttgt cttaacaaaa agtgagggcg ctgaggtcga gcagggtggt 10020
gtgacttgtt atagccttta gagctgcgaa agcgcgtatg gatttggctc atcaggccag 10080
attgagggtc tgtggacaca tgtcatgtta gtgtacttca atcgccccct ggatatagcc 10140
ccgacaatag gccgtggcct catttttttg ccttccgcac atttccattg ctcggtaccc 10200
acaccttgct tctcctgcac ttgccaacct taatactggt ttacattgac caacatctta 10260
caagcggggg gcttgtctag ggtatatata aacagtggct ctcccaatcg gttgccagtc 10320
tcttttttcc tttctttccc cacagattcg aaatctaaac tacacatcac agaattccga 10380
gccgtgagta tccacgacaa gatcagtgtc gagacgacgc gttttgtgta atgacacaat 10440
ccgaaagtcg ctagcaacac acactctcta cacaaactaa cccagctctc catggccttc 10500
cagtttcacc tggaggtcct cctgccctac ctccttcctc tgcttctgct catcctgccc 10560
accactatct tctttctcac caagcccaac aataaggtgt cctctacttc caccaacaat 10620
aacatcatta cactgcccaa gtcgtaccct ctcattggct cctacctttc gttccgaaag 10680
aacctgcatc gacggatcca gtggctctcc gacattgttc agatctctcc ctccgctacc 10740
ttccagctcg acggaaccct gggcaagcga cagatcatta ctggcaaccc ttctaccgtc 10800
cagcacattc tcaagaacca gttctccaac tatcagaagg gcaccacatt caccaacact 10860
ctgtccgact ttctcggaac aggcatcttc aacaccaacg gtcccaactg gaagtttcaa 10920
cgacaggttg cctctcacga gttcaacacc aagtccattc ggaacttcgt cgagcacatc 10980
gtggataccg aactcaccaa ccgattgatt cccatcctca cttcgagcac ccagacaaac 11040
aatatcctgg acttccagga tattctgcag cgatttacct tcgacaacat ctgcaacatt 11100
gccttcggat acgatcccga gtacctcact ccctcgacca atcgttccaa gttcgcggag 11160
gcctacgaag acgctaccga gatctccagc aagcgattca gactgcctct tcccatcatt 11220
tggaagatca aaaagtactt caacattggc tccgagaagc gactcaagga agccgtcacc 11280
gaggtccgat cctttgccaa gaaactggtc cgagagaaga aacgggagct cgaagagaag 11340
tcttcgctgg agaccgaaga catgctttct cgatttctgt ccagcggtca ctcggacgag 11400
gatttcgttg ccgacattgt catctccttc attctcgcag gcaaggacac tacctctgcc 11460
gctcttacct ggtttttctg gctgctctgg aagaaccctc gagtggagga agagatcgtc 11520
aacgagctgt ccaagaaatc ggagcttatg gtgtacgacg aggtcaagga aatggtctac 11580
acccatgctg cgctgtccga gtcgatgaga ctctaccctc ccgttccaat ggattccaag 11640
gaggccgtca acgacgatgt gctgcccgac ggctgggtgg tcaagaaagg tacaatcgtc 11700
acctaccatg tctacgctat gggtcgaatg aagtctctct ggggagacga ttgggcagag 11760
tttcgaccag aacggtggct cgagaaggac gaggtcaacg gcaagtgggt gttcgtcgga 11820
cgagacagct actcctatcc tgtgttccag gctggtccca gagtctgcct gggaaaggag 11880
atggccttca tgcagatgaa gcgaattgtg gctggcatcg tcggaaagtt caaggtggtt 11940
cccgaagccc acttggctca ggagccagga ttcatttcct ttctgtcgtc tcagatggag 12000
ggtggatttc ccgtcactat ccagaagcga gactcctaag cggccgcatt gatgattgga 12060
aacacacaca tgggttatat ctaggtgaga gttagttgga cagttatata ttaaatcagc 12120
tatgccaacg gtaacttcat tcatgtcaac gaggaaccag tgactgcaag taatatagaa 12180
tttgaccacc ttgccattct cttgcactcc tttactatat ctcatttatt tcttatatac 12240
aaatcacttc ttcttcccag catcgagctc ggaaacctca tgagcaataa catcgtggat 12300
ctcgtcaata gagggctttt tggactcctt gctgttggcc accttgtcct tgctgttt 12358
<210> 106
<211> 1296
<212> DNA
<213> yarrowia lipolytica
<400> 106
atggatttct tcagacggca ccagaaaaag gtgctggcac tggtaggtgt ggcgctgagt 60
tcctacctgt ttatcgacta tgtgaagaaa aagttcttcg agatccaggg tcgtttgagc 120
tcggagcgaa ccgctaaaca gaatctccgg cgccgatttg aacagaacca gcaggatgca 180
gattttacaa tcatggctct gctatccagc ttgacgacac cggtaatgga gcgttacccc 240
gtcgaccaga tcaaggcaga gttacagagc aagagacgcc ccacagaccg ggttttggct 300
ctcgagagct ccacctcgtc ctcagctacc gcacaaaccg tgcccaccat gacaagtggc 360
gccacagagg agggcgagaa gtcgaaaaca cagttgtggc aggatctcaa gcgaacgacc 420
atttcccgag cgttttctct tgtctatgca gatgcacttc ttattttctt cacgcgtttg 480
cagctcaaca ttctaggacg acgaaactac gtcaacagtg ttgtcgctct ggcgcagcag 540
ggccgagagg gtaatgccga gggtcgagtg gcgccctcgt ttggtgatct tgcagatatg 600
ggctatttcg gcgacctttc aggctcgtcc agcttcggag aaactattgt cgatcccgat 660
ctggacgaac agtaccttac cttttcgtgg tggctgctga acgagggatg ggtgtcgctg 720
agcgagcgag tggaggaagc ggttcgtcga gtgtgggacc ccgtgtcacc caaggccgaa 780
cttggatttg acgagttgtc ggaactcatt ggacgaacac agatgctcat tgatcgacct 840
ctcaatccct cgtcgccact caactttctg agccagctgc tgccaccacg ggagcaggag 900
gagtacgtgc ttgcccagaa ccccagcgat actgctgccc ccattgtagg acctaccctc 960
cgacggcttc tggacgagac tgccgacttc atcgagtccc ctaatgccgc agaggtgatt 1020
gagcgacttg ttcactccgg tctctctgtg ttcatggaca agctggctgt cacgtttgga 1080
gccacacctg ctgattcggg ttcgccttat cctgtggtgc tgcctactgc aaaggtcaag 1140
ctgccctcca ttcttgccaa catggctcga caggctggag gcatggccca gggatcgccg 1200
ggcgtggaaa acgagtacat tgacgtgatg aaccaagtgc aggagctgac ctcctttagt 1260
gctgtggtct attcatcttt tgattgggct ctctag 1296
<210> 107
<211> 431
<212> PRT
<213> yarrowia lipolytica
<400> 107
Met Asp Phe Phe Arg Arg His Gln Lys Lys Val Leu Ala Leu Val Gly
1 5 10 15
Val Ala Leu Ser Ser Tyr Leu Phe Ile Asp Tyr Val Lys Lys Lys Phe
20 25 30
Phe Glu Ile Gln Gly Arg Leu Ser Ser Glu Arg Thr Ala Lys Gln Asn
35 40 45
Leu Arg Arg Arg Phe Glu Gln Asn Gln Gln Asp Ala Asp Phe Thr Ile
50 55 60
Met Ala Leu Leu Ser Ser Leu Thr Thr Pro Val Met Glu Arg Tyr Pro
65 70 75 80
Val Asp Gln Ile Lys Ala Glu Leu Gln Ser Lys Arg Arg Pro Thr Asp
85 90 95
Arg Val Leu Ala Leu Glu Ser Ser Thr Ser Ser Ser Ala Thr Ala Gln
100 105 110
Thr Val Pro Thr Met Thr Ser Gly Ala Thr Glu Glu Gly Glu Lys Ser
115 120 125
Lys Thr Gln Leu Trp Gln Asp Leu Lys Arg Thr Thr Ile Ser Arg Ala
130 135 140
Phe Ser Leu Val Tyr Ala Asp Ala Leu Leu Ile Phe Phe Thr Arg Leu
145 150 155 160
Gln Leu Asn Ile Leu Gly Arg Arg Asn Tyr Val Asn Ser Val Val Ala
165 170 175
Leu Ala Gln Gln Gly Arg Glu Gly Asn Ala Glu Gly Arg Val Ala Pro
180 185 190
Ser Phe Gly Asp Leu Ala Asp Met Gly Tyr Phe Gly Asp Leu Ser Gly
195 200 205
Ser Ser Ser Phe Gly Glu Thr Ile Val Asp Pro Asp Leu Asp Glu Gln
210 215 220
Tyr Leu Thr Phe Ser Trp Trp Leu Leu Asn Glu Gly Trp Val Ser Leu
225 230 235 240
Ser Glu Arg Val Glu Glu Ala Val Arg Arg Val Trp Asp Pro Val Ser
245 250 255
Pro Lys Ala Glu Leu Gly Phe Asp Glu Leu Ser Glu Leu Ile Gly Arg
260 265 270
Thr Gln Met Leu Ile Asp Arg Pro Leu Asn Pro Ser Ser Pro Leu Asn
275 280 285
Phe Leu Ser Gln Leu Leu Pro Pro Arg Glu Gln Glu Glu Tyr Val Leu
290 295 300
Ala Gln Asn Pro Ser Asp Thr Ala Ala Pro Ile Val Gly Pro Thr Leu
305 310 315 320
Arg Arg Leu Leu Asp Glu Thr Ala Asp Phe Ile Glu Ser Pro Asn Ala
325 330 335
Ala Glu Val Ile Glu Arg Leu Val His Ser Gly Leu Ser Val Phe Met
340 345 350
Asp Lys Leu Ala Val Thr Phe Gly Ala Thr Pro Ala Asp Ser Gly Ser
355 360 365
Pro Tyr Pro Val Val Leu Pro Thr Ala Lys Val Lys Leu Pro Ser Ile
370 375 380
Leu Ala Asn Met Ala Arg Gln Ala Gly Gly Met Ala Gln Gly Ser Pro
385 390 395 400
Gly Val Glu Asn Glu Tyr Ile Asp Val Met Asn Gln Val Gln Glu Leu
405 410 415
Thr Ser Phe Ser Ala Val Val Tyr Ser Ser Phe Asp Trp Ala Leu
420 425 430
<210> 108
<211> 377
<212> PRT
<213> yarrowia lipolytica
<400> 108
Met Trp Gly Ser Ser His Ala Phe Ala Gly Glu Ser Asp Leu Thr Leu
1 5 10 15
Gln Leu His Thr Arg Ser Asn Met Ser Asp Asn Thr Thr Ile Lys Lys
20 25 30
Pro Ile Arg Pro Lys Pro Ile Arg Thr Glu Arg Leu Pro Tyr Ala Gly
35 40 45
Ala Ala Glu Ile Ile Arg Ala Asn Gln Lys Asp His Tyr Phe Glu Ser
50 55 60
Val Leu Glu Gln His Leu Val Thr Phe Leu Gln Lys Trp Lys Gly Val
65 70 75 80
Arg Phe Ile His Gln Tyr Lys Glu Glu Leu Glu Thr Ala Ser Lys Phe
85 90 95
Ala Tyr Leu Gly Leu Cys Thr Leu Val Gly Ser Lys Thr Leu Gly Glu
100 105 110
Glu Tyr Thr Asn Leu Met Tyr Thr Ile Arg Asp Arg Thr Ala Leu Pro
115 120 125
Gly Val Val Arg Arg Phe Gly Tyr Val Leu Ser Asn Thr Leu Phe Pro
130 135 140
Tyr Leu Phe Val Arg Tyr Met Gly Lys Leu Arg Ala Lys Leu Met Arg
145 150 155 160
Glu Tyr Pro His Leu Val Glu Tyr Asp Glu Asp Glu Pro Val Pro Ser
165 170 175
Pro Glu Thr Trp Lys Glu Arg Val Ile Lys Thr Phe Val Asn Lys Phe
180 185 190
Asp Lys Phe Thr Ala Leu Glu Gly Phe Thr Ala Ile His Leu Ala Ile
195 200 205
Phe Tyr Val Tyr Gly Ser Tyr Tyr Gln Leu Ser Lys Arg Ile Trp Gly
210 215 220
Met Arg Tyr Val Phe Gly His Arg Leu Asp Lys Asn Glu Pro Arg Ile
225 230 235 240
Gly Tyr Glu Met Leu Gly Leu Leu Ile Phe Ala Arg Phe Ala Thr Ser
245 250 255
Phe Val Gln Thr Gly Arg Glu Tyr Leu Gly Ala Leu Leu Glu Lys Ser
260 265 270
Val Glu Lys Glu Ala Gly Glu Lys Glu Asp Glu Lys Glu Ala Val Val
275 280 285
Pro Lys Lys Lys Ser Ser Ile Pro Phe Ile Glu Asp Thr Glu Gly Glu
290 295 300
Thr Glu Asp Lys Ile Asp Leu Glu Asp Pro Arg Gln Leu Lys Phe Ile
305 310 315 320
Pro Glu Ala Ser Arg Ala Cys Thr Leu Cys Leu Ser Tyr Ile Ser Ala
325 330 335
Pro Ala Cys Thr Pro Cys Gly His Phe Phe Cys Trp Asp Cys Ile Ser
340 345 350
Glu Trp Val Arg Glu Lys Pro Glu Cys Pro Leu Cys Arg Gln Gly Val
355 360 365
Arg Glu Gln Asn Leu Leu Pro Ile Arg
370 375
<210> 109
<211> 391
<212> PRT
<213> yarrowia lipolytica
<400> 109
Met Thr Asp Lys Leu Val Lys Val Met Gln Lys Lys Lys Ser Ala Pro
1 5 10 15
Gln Thr Trp Leu Asp Ser Tyr Asp Lys Phe Leu Val Arg Asn Ala Ala
20 25 30
Ser Ile Gly Ser Ile Glu Ser Thr Leu Arg Thr Val Ser Tyr Val Leu
35 40 45
Pro Gly Arg Phe Asn Asp Val Glu Ile Ala Thr Glu Thr Leu Tyr Ala
50 55 60
Val Leu Asn Val Leu Gly Leu Tyr His Asp Thr Ile Ile Ala Arg Ala
65 70 75 80
Val Ala Ala Ser Pro Asn Ala Ala Ala Val Tyr Arg Pro Ser Pro His
85 90 95
Asn Arg Tyr Thr Asp Trp Phe Ile Lys Asn Arg Lys Gly Tyr Lys Tyr
100 105 110
Ala Ser Arg Ala Val Thr Phe Val Lys Phe Gly Glu Leu Val Ala Glu
115 120 125
Met Val Ala Lys Lys Asn Gly Gly Glu Met Ala Arg Trp Lys Cys Ile
130 135 140
Ile Gly Ile Glu Gly Ile Lys Ala Gly Leu Arg Ile Tyr Met Leu Gly
145 150 155 160
Ser Thr Leu Tyr Gln Pro Leu Cys Thr Thr Pro Tyr Pro Asp Arg Glu
165 170 175
Val Thr Gly Glu Leu Leu Glu Thr Ile Cys Arg Asp Glu Gly Glu Leu
180 185 190
Asp Ile Glu Lys Gly Leu Met Asp Pro Gln Trp Lys Met Pro Arg Thr
195 200 205
Gly Arg Thr Ile Pro Glu Ile Ala Pro Thr Asn Val Glu Gly Tyr Leu
210 215 220
Leu Thr Lys Val Leu Arg Ser Glu Asp Val Asp Arg Pro Tyr Asn Leu
225 230 235 240
Leu Ser Arg Leu Asp Asn Trp Gly Val Val Ala Glu Leu Leu Ser Ile
245 250 255
Leu Arg Pro Leu Ile Tyr Ala Cys Leu Leu Phe Arg Gln His Val Asn
260 265 270
Lys Thr Val Pro Ala Ser Thr Lys Ser Lys Phe Pro Phe Leu Asn Ser
275 280 285
Pro Trp Ala Pro Trp Ile Ile Gly Leu Val Ile Glu Ala Leu Ser Arg
290 295 300
Lys Met Met Gly Ser Trp Leu Leu Arg Gln Arg Gln Ser Gly Lys Thr
305 310 315 320
Pro Thr Ala Leu Asp Gln Met Glu Val Lys Gly Arg Thr Asn Leu Leu
325 330 335
Gly Trp Trp Leu Phe Arg Gly Glu Phe Tyr Gln Ala Tyr Thr Arg Pro
340 345 350
Leu Leu Tyr Ser Ile Val Ala Arg Leu Glu Lys Ile Pro Gly Leu Gly
355 360 365
Leu Phe Gly Ala Leu Ile Ser Asp Tyr Leu Tyr Leu Phe Asp Arg Tyr
370 375 380
Tyr Phe Thr Ala Ser Thr Leu
385 390
<210> 110
<211> 2106
<212> DNA
<213> yarrowia lipolytica
<400> 110
atgatcaccc caaaccccgc taacgacatt gtccatgacg gcaagctcta cgacaccttc 60
actgagcccc ccaagctgat ggctcaggag cgagctcagc tggacttcga ccctagagac 120
atcacctact ttctggatgg ctctaaggag gagaccgagc tgctggagtc gctcatgctc 180
atgtacgagc gagaccctct cttcaacaac cagaacgagt acgatgaatc gtttgaaaca 240
ctgcgagagc gatctgtgaa gcgaattttc cagctgtcca agtccatcgc catggacccc 300
gagcccatgt ctttccgaaa gattgggttc ctgggtattc ttgacatggg aacgtatgct 360
cgactgggag tccactacgc gctcttctgt aactccatcc ggggccaggg aacccccgat 420
cagctcatgt actggctgga ccagggagcc atggtcatca agggcttcta cggctgtttt 480
gccatgaccg aaatgggcca tggatctaac ctgtcgcgtc tggaaaccat cgccactttc 540
gacaaagaga ccgacgaatt tatcattaac acgccccacg ttggagccac aaagtggtgg 600
attggaggag ccgcccacac tgctactcac acacttgcct ttgcccgtct tcaagtagac 660
ggaaaggact acggtgtgaa atcgtttgtc gtacctctcc gaaacctgga cgaccattcg 720
ctgcgtcctg gaatcgccac aggtgatatt ggtaagaaga tgggtcgaga tgccgttgac 780
aacggctgga ttcagttcac caacgtccga gtgccccgaa actacatgct catgaagcat 840
accaaggttc ttcgagacgg taccgtcaag cagccgcctt tggcccaact gacttacgga 900
tctctcatca ctggacgagt ccagatgacc actgactctc acaatgtgtc caaaaagttc 960
ctcaccattg ccctgagata cgccaccatc cgacgacagt tctcgtcaac tccaggagag 1020
cccgaaaccc gactaattga ctacctgtac caccaaagac gactcctgcc tcttatggct 1080
tactcttacg ccatgaaact agctggagat cacgtccgag agctgttctt tgcatcccag 1140
gagaaggctg agagcctcaa ggaggacgac aaagccggag ttgagtctta cgtccaggat 1200
atcaaggagc tcttctctgt ttctgctggt ctcaaggctg ccactacatg ggcttgtgct 1260
gacatcattg acaaggcccg acaggcgtgt ggaggccacg gatactctgc ctacaacggc 1320
tttggacagg ccttccagga ctgggttgtc cagtgcactt gggagggtga caatactgtt 1380
ctgactctat ctgccggccg agctctgatc caatctgctc tcgtctaccg aaaggagggc 1440
aaactaggta acgccacgaa gtacctctct cggtccaagg agcttgccaa cgccaagaga 1500
aacggacgat ccctggaaga ccccaagctg ctcgtggagg catgggaggc tgtctctgcc 1560
ggtgctatca acgctgctac tgacgcttac gaggagctct ccaagcaggg agtttctgtt 1620
gacgagtgct ttgagcaggt gtcccaggag cgattccagg ctgcccgaat ccacactcga 1680
cgagctctta tcgaggcctt ctactcacga atcgccactg ctgatgagaa ggtgaagcct 1740
catctgatcc ctctggccaa cctgtttgcc ctgtggtcca ttgaggagga ctctgctctg 1800
ttcctggctg agggctactt tgagcctgag gatatcattg aggtgacttc tcttgtcaac 1860
aagtactgcg gaattgttcg aaagaacgtt attggataca ccgatgcctt caacctgtcc 1920
gactacttca tcaacgctgc cattggacga tacgacggag acgtgtacaa gaactacttt 1980
gagaaggtca aacagcagta ccctcctgag ggtggcaagc ctcactacta cgaggatgtc 2040
atgaagccct tcctgcatcg agagcgaatt cccgatgtcc ccatggagcc cgaggatatt 2100
cagtaa 2106
<210> 111
<211> 701
<212> PRT
<213> yarrowia lipolytica
<400> 111
Met Ile Thr Pro Asn Pro Ala Asn Asp Ile Val His Asp Gly Lys Leu
1 5 10 15
Tyr Asp Thr Phe Thr Glu Pro Pro Lys Leu Met Ala Gln Glu Arg Ala
20 25 30
Gln Leu Asp Phe Asp Pro Arg Asp Ile Thr Tyr Phe Leu Asp Gly Ser
35 40 45
Lys Glu Glu Thr Glu Leu Leu Glu Ser Leu Met Leu Met Tyr Glu Arg
50 55 60
Asp Pro Leu Phe Asn Asn Gln Asn Glu Tyr Asp Glu Ser Phe Glu Thr
65 70 75 80
Leu Arg Glu Arg Ser Val Lys Arg Ile Phe Gln Leu Ser Lys Ser Ile
85 90 95
Ala Met Asp Pro Glu Pro Met Ser Phe Arg Lys Ile Gly Phe Leu Gly
100 105 110
Ile Leu Asp Met Gly Thr Tyr Ala Arg Leu Gly Val His Tyr Ala Leu
115 120 125
Phe Cys Asn Ser Ile Arg Gly Gln Gly Thr Pro Asp Gln Leu Met Tyr
130 135 140
Trp Leu Asp Gln Gly Ala Met Val Ile Lys Gly Phe Tyr Gly Cys Phe
145 150 155 160
Ala Met Thr Glu Met Gly His Gly Ser Asn Leu Ser Arg Leu Glu Thr
165 170 175
Ile Ala Thr Phe Asp Lys Glu Thr Asp Glu Phe Ile Ile Asn Thr Pro
180 185 190
His Val Gly Ala Thr Lys Trp Trp Ile Gly Gly Ala Ala His Thr Ala
195 200 205
Thr His Thr Leu Ala Phe Ala Arg Leu Gln Val Asp Gly Lys Asp Tyr
210 215 220
Gly Val Lys Ser Phe Val Val Pro Leu Arg Asn Leu Asp Asp His Ser
225 230 235 240
Leu Arg Pro Gly Ile Ala Thr Gly Asp Ile Gly Lys Lys Met Gly Arg
245 250 255
Asp Ala Val Asp Asn Gly Trp Ile Gln Phe Thr Asn Val Arg Val Pro
260 265 270
Arg Asn Tyr Met Leu Met Lys His Thr Lys Val Leu Arg Asp Gly Thr
275 280 285
Val Lys Gln Pro Pro Leu Ala Gln Leu Thr Tyr Gly Ser Leu Ile Thr
290 295 300
Gly Arg Val Gln Met Thr Thr Asp Ser His Asn Val Ser Lys Lys Phe
305 310 315 320
Leu Thr Ile Ala Leu Arg Tyr Ala Thr Ile Arg Arg Gln Phe Ser Ser
325 330 335
Thr Pro Gly Glu Pro Glu Thr Arg Leu Ile Asp Tyr Leu Tyr His Gln
340 345 350
Arg Arg Leu Leu Pro Leu Met Ala Tyr Ser Tyr Ala Met Lys Leu Ala
355 360 365
Gly Asp His Val Arg Glu Leu Phe Phe Ala Ser Gln Glu Lys Ala Glu
370 375 380
Ser Leu Lys Glu Asp Asp Lys Ala Gly Val Glu Ser Tyr Val Gln Asp
385 390 395 400
Ile Lys Glu Leu Phe Ser Val Ser Ala Gly Leu Lys Ala Ala Thr Thr
405 410 415
Trp Ala Cys Ala Asp Ile Ile Asp Lys Ala Arg Gln Ala Cys Gly Gly
420 425 430
His Gly Tyr Ser Ala Tyr Asn Gly Phe Gly Gln Ala Phe Gln Asp Trp
435 440 445
Val Val Gln Cys Thr Trp Glu Gly Asp Asn Thr Val Leu Thr Leu Ser
450 455 460
Ala Gly Arg Ala Leu Ile Gln Ser Ala Leu Val Tyr Arg Lys Glu Gly
465 470 475 480
Lys Leu Gly Asn Ala Thr Lys Tyr Leu Ser Arg Ser Lys Glu Leu Ala
485 490 495
Asn Ala Lys Arg Asn Gly Arg Ser Leu Glu Asp Pro Lys Leu Leu Val
500 505 510
Glu Ala Trp Glu Ala Val Ser Ala Gly Ala Ile Asn Ala Ala Thr Asp
515 520 525
Ala Tyr Glu Glu Leu Ser Lys Gln Gly Val Ser Val Asp Glu Cys Phe
530 535 540
Glu Gln Val Ser Gln Glu Arg Phe Gln Ala Ala Arg Ile His Thr Arg
545 550 555 560
Arg Ala Leu Ile Glu Ala Phe Tyr Ser Arg Ile Ala Thr Ala Asp Glu
565 570 575
Lys Val Lys Pro His Leu Ile Pro Leu Ala Asn Leu Phe Ala Leu Trp
580 585 590
Ser Ile Glu Glu Asp Ser Ala Leu Phe Leu Ala Glu Gly Tyr Phe Glu
595 600 605
Pro Glu Asp Ile Ile Glu Val Thr Ser Leu Val Asn Lys Tyr Cys Gly
610 615 620
Ile Val Arg Lys Asn Val Ile Gly Tyr Thr Asp Ala Phe Asn Leu Ser
625 630 635 640
Asp Tyr Phe Ile Asn Ala Ala Ile Gly Arg Tyr Asp Gly Asp Val Tyr
645 650 655
Lys Asn Tyr Phe Glu Lys Val Lys Gln Gln Tyr Pro Pro Glu Gly Gly
660 665 670
Lys Pro His Tyr Tyr Glu Asp Val Met Lys Pro Phe Leu His Arg Glu
675 680 685
Arg Ile Pro Asp Val Pro Met Glu Pro Glu Asp Ile Gln
690 695 700
<210> 112
<211> 1581
<212> DNA
<213> yarrowia lipolytica
<400> 112
atggaagtcc gacgacgaaa aatcgacgtg ctcaaggccc agaaaaacgg ctacgaatcg 60
ggcccaccat ctcgacaatc gtcgcagccc tcctcaagag catcgtccag aacccgcaac 120
aaacactcct cgtccaccct gtcgctcagc ggactgacca tgaaagtcca gaagaaacct 180
gcgggacccc cggcgaactc caaaacgcca ttcctacaca tcaagcccgt gcacacgtgc 240
tgctccacat caatgctttc gcgcgattat gacggctcca accccagctt caagggcttc 300
aaaaacatcg gcatgatcat tctcattgtg ggaaatctac ggctcgcatt cgaaaactac 360
ctcaaatacg gcatttccaa cccgttcttc gaccccaaaa ttactccttc cgagtggcag 420
ctctcaggct tgctcatagt cgtggcctac gcacatatcc tcatggccta cgctattgag 480
agcgctgcca agctgctgtt cctctctagc aaacaccact acatggccgt ggggcttctg 540
cataccatga acactttgtc gtccatctcg ttgctgtcct acgtcgtcta ctactacctg 600
cccaaccccg tggcaggcac aatagtcgag tttgtggccg ttattctgtc tctcaaactc 660
gcctcatacg ccctcactaa ctcggatctc cgaaaagccg caattcatgc ccagaagctc 720
gacaagacgc aagacgataa cgaaaaggaa tccacctcgt cttcctcttc ttcagatgac 780
gcagagactt tggcagacat tgacgtcatt cctgcatact acgcacagct gccctacccc 840
cagaatgtga cgctgtcgaa cctgctgtac ttctggtttg ctcccacact ggtctaccag 900
cccgtgtacc ccaagacgga gcgtattcga cccaagcacg tgatccgaaa cctgtttgag 960
ctcgtctctc tgtgcatgct tattcagttt ctcatcttcc agtacgccta ccccatcatg 1020
cagtcgtgtc tggctctgtt cttccagccc aagctcgatt atgccaacat ctccgagcgc 1080
ctcatgaagt tggcctccgt gtctatgatg gtctggctca ttggattcta cgctttcttc 1140
cagaacggtc tcaatcttat tgccgagctc acctgttttg gaaacagaac cttctaccag 1200
cagtggtgga attcccgctc cattggccag tactggactc tatggaacaa gccagtcaac 1260
cagtacttta gacaccacgt ctacgtgcct cttctcgctc ggggcatgtc gcggttcaat 1320
gcgtcggtgg tggttttctt tttctccgcc gtcatccatg aactgcttgt cggcatcccc 1380
actcacaaca tcatcggagc cgccttcttc ggcatgatgt cgcaggtgcc tctgatcatg 1440
gctactgaga accttcagca tattaactcc tctctgggcc ccttccttgg caactgtgca 1500
ttctggttca cctttttcct gggacaaccc acttgtgcat tcctttatta tctggcttac 1560
aactacaagc agaaccagta g 1581
<210> 113
<211> 526
<212> PRT
<213> yarrowia lipolytica
<400> 113
Met Glu Val Arg Arg Arg Lys Ile Asp Val Leu Lys Ala Gln Lys Asn
1 5 10 15
Gly Tyr Glu Ser Gly Pro Pro Ser Arg Gln Ser Ser Gln Pro Ser Ser
20 25 30
Arg Ala Ser Ser Arg Thr Arg Asn Lys His Ser Ser Ser Thr Leu Ser
35 40 45
Leu Ser Gly Leu Thr Met Lys Val Gln Lys Lys Pro Ala Gly Pro Pro
50 55 60
Ala Asn Ser Lys Thr Pro Phe Leu His Ile Lys Pro Val His Thr Cys
65 70 75 80
Cys Ser Thr Ser Met Leu Ser Arg Asp Tyr Asp Gly Ser Asn Pro Ser
85 90 95
Phe Lys Gly Phe Lys Asn Ile Gly Met Ile Ile Leu Ile Val Gly Asn
100 105 110
Leu Arg Leu Ala Phe Glu Asn Tyr Leu Lys Tyr Gly Ile Ser Asn Pro
115 120 125
Phe Phe Asp Pro Lys Ile Thr Pro Ser Glu Trp Gln Leu Ser Gly Leu
130 135 140
Leu Ile Val Val Ala Tyr Ala His Ile Leu Met Ala Tyr Ala Ile Glu
145 150 155 160
Ser Ala Ala Lys Leu Leu Phe Leu Ser Ser Lys His His Tyr Met Ala
165 170 175
Val Gly Leu Leu His Thr Met Asn Thr Leu Ser Ser Ile Ser Leu Leu
180 185 190
Ser Tyr Val Val Tyr Tyr Tyr Leu Pro Asn Pro Val Ala Gly Thr Ile
195 200 205
Val Glu Phe Val Ala Val Ile Leu Ser Leu Lys Leu Ala Ser Tyr Ala
210 215 220
Leu Thr Asn Ser Asp Leu Arg Lys Ala Ala Ile His Ala Gln Lys Leu
225 230 235 240
Asp Lys Thr Gln Asp Asp Asn Glu Lys Glu Ser Thr Ser Ser Ser Ser
245 250 255
Ser Ser Asp Asp Ala Glu Thr Leu Ala Asp Ile Asp Val Ile Pro Ala
260 265 270
Tyr Tyr Ala Gln Leu Pro Tyr Pro Gln Asn Val Thr Leu Ser Asn Leu
275 280 285
Leu Tyr Phe Trp Phe Ala Pro Thr Leu Val Tyr Gln Pro Val Tyr Pro
290 295 300
Lys Thr Glu Arg Ile Arg Pro Lys His Val Ile Arg Asn Leu Phe Glu
305 310 315 320
Leu Val Ser Leu Cys Met Leu Ile Gln Phe Leu Ile Phe Gln Tyr Ala
325 330 335
Tyr Pro Ile Met Gln Ser Cys Leu Ala Leu Phe Phe Gln Pro Lys Leu
340 345 350
Asp Tyr Ala Asn Ile Ser Glu Arg Leu Met Lys Leu Ala Ser Val Ser
355 360 365
Met Met Val Trp Leu Ile Gly Phe Tyr Ala Phe Phe Gln Asn Gly Leu
370 375 380
Asn Leu Ile Ala Glu Leu Thr Cys Phe Gly Asn Arg Thr Phe Tyr Gln
385 390 395 400
Gln Trp Trp Asn Ser Arg Ser Ile Gly Gln Tyr Trp Thr Leu Trp Asn
405 410 415
Lys Pro Val Asn Gln Tyr Phe Arg His His Val Tyr Val Pro Leu Leu
420 425 430
Ala Arg Gly Met Ser Arg Phe Asn Ala Ser Val Val Val Phe Phe Phe
435 440 445
Ser Ala Val Ile His Glu Leu Leu Val Gly Ile Pro Thr His Asn Ile
450 455 460
Ile Gly Ala Ala Phe Phe Gly Met Met Ser Gln Val Pro Leu Ile Met
465 470 475 480
Ala Thr Glu Asn Leu Gln His Ile Asn Ser Ser Leu Gly Pro Phe Leu
485 490 495
Gly Asn Cys Ala Phe Trp Phe Thr Phe Phe Leu Gly Gln Pro Thr Cys
500 505 510
Ala Phe Leu Tyr Tyr Leu Ala Tyr Asn Tyr Lys Gln Asn Gln
515 520 525
<210> 114
<211> 1545
<212> DNA
<213> yarrowia lipolytica
<400> 114
atgactatcg actcacaata ctacaagtcg cgagacaaaa acgacacggc acccaaaatc 60
gcgggaatcc gatatgcccc gctatcgaca ccattactca accgatgtga gaccttctct 120
ctggtctggc acattttcag cattcccact ttcctcacaa ttttcatgct atgctgcgca 180
attccactgc tctggccatt tgtgattgcg tatgtagtgt acgctgttaa agacgactcc 240
ccgtccaacg gaggagtggt caagcgatac tcgcctattt caagaaactt cttcatctgg 300
aagctctttg gccgctactt ccccataact ctgcacaaga cggtggatct ggagcccacg 360
cacacatact accctctgga cgtccaggag tatcacctga ttgctgagag atactggccg 420
cagaacaagt acctccgagc aatcatctcc accatcgagt actttctgcc cgccttcatg 480
aaacggtctc tttctatcaa cgagcaggag cagcctgccg agcgagatcc tctcctgtct 540
cccgtttctc ccagctctcc gggttctcaa cctgacaagt ggattaacca cgacagcaga 600
tatagccgtg gagaatcatc tggctccaac ggccacgcct cgggctccga acttaacggc 660
aacggcaaca atggcaccac taaccgacga cctttgtcgt ccgcctctgc tggctccact 720
gcatctgatt ccacgcttct taacgggtcc ctcaactcct acgccaacca gatcattggc 780
gaaaacgacc cacagctgtc gcccacaaaa ctcaagccca ctggcagaaa atacatcttc 840
ggctaccacc cccacggcat tatcggcatg ggagcctttg gtggaattgc caccgaggga 900
gctggatggt ccaagctctt tccgggcatc cctgtttctc ttatgactct caccaacaac 960
ttccgagtgc ctctctacag agagtacctc atgagtctgg gagtcgcttc tgtctccaag 1020
aagtcctgca aggccctcct caagcgaaac cagtctatct gcattgtcgt tggtggagca 1080
caggaaagtc ttctggccag acccggtgtc atggacctgg tgctactcaa gcgaaagggt 1140
tttgttcgac ttggtatgga ggtcggaaat gtcgcccttg ttcccatcat ggcctttggt 1200
gagaacgacc tctatgacca ggttagcaac gacaagtcgt ccaagctgta ccgattccag 1260
cagtttgtca agaacttcct tggattcacc cttcctttga tgcatgcccg aggcgtcttc 1320
aactacgatg tcggtcttgt cccctacagg cgacccgtca acattgtggt tggttccccc 1380
attgacttgc cttatctccc acaccccacc gacgaagaag tgtccgaata ccacgaccga 1440
tacatcgccg agctgcagcg aatctacaac gagcacaagg atgaatattt catcgattgg 1500
accgaggagg gcaaaggagc cccagagttc cgaatgattg agtaa 1545
<210> 115
<211> 514
<212> PRT
<213> yarrowia lipolytica
<400> 115
Met Thr Ile Asp Ser Gln Tyr Tyr Lys Ser Arg Asp Lys Asn Asp Thr
1 5 10 15
Ala Pro Lys Ile Ala Gly Ile Arg Tyr Ala Pro Leu Ser Thr Pro Leu
20 25 30
Leu Asn Arg Cys Glu Thr Phe Ser Leu Val Trp His Ile Phe Ser Ile
35 40 45
Pro Thr Phe Leu Thr Ile Phe Met Leu Cys Cys Ala Ile Pro Leu Leu
50 55 60
Trp Pro Phe Val Ile Ala Tyr Val Val Tyr Ala Val Lys Asp Asp Ser
65 70 75 80
Pro Ser Asn Gly Gly Val Val Lys Arg Tyr Ser Pro Ile Ser Arg Asn
85 90 95
Phe Phe Ile Trp Lys Leu Phe Gly Arg Tyr Phe Pro Ile Thr Leu His
100 105 110
Lys Thr Val Asp Leu Glu Pro Thr His Thr Tyr Tyr Pro Leu Asp Val
115 120 125
Gln Glu Tyr His Leu Ile Ala Glu Arg Tyr Trp Pro Gln Asn Lys Tyr
130 135 140
Leu Arg Ala Ile Ile Ser Thr Ile Glu Tyr Phe Leu Pro Ala Phe Met
145 150 155 160
Lys Arg Ser Leu Ser Ile Asn Glu Gln Glu Gln Pro Ala Glu Arg Asp
165 170 175
Pro Leu Leu Ser Pro Val Ser Pro Ser Ser Pro Gly Ser Gln Pro Asp
180 185 190
Lys Trp Ile Asn His Asp Ser Arg Tyr Ser Arg Gly Glu Ser Ser Gly
195 200 205
Ser Asn Gly His Ala Ser Gly Ser Glu Leu Asn Gly Asn Gly Asn Asn
210 215 220
Gly Thr Thr Asn Arg Arg Pro Leu Ser Ser Ala Ser Ala Gly Ser Thr
225 230 235 240
Ala Ser Asp Ser Thr Leu Leu Asn Gly Ser Leu Asn Ser Tyr Ala Asn
245 250 255
Gln Ile Ile Gly Glu Asn Asp Pro Gln Leu Ser Pro Thr Lys Leu Lys
260 265 270
Pro Thr Gly Arg Lys Tyr Ile Phe Gly Tyr His Pro His Gly Ile Ile
275 280 285
Gly Met Gly Ala Phe Gly Gly Ile Ala Thr Glu Gly Ala Gly Trp Ser
290 295 300
Lys Leu Phe Pro Gly Ile Pro Val Ser Leu Met Thr Leu Thr Asn Asn
305 310 315 320
Phe Arg Val Pro Leu Tyr Arg Glu Tyr Leu Met Ser Leu Gly Val Ala
325 330 335
Ser Val Ser Lys Lys Ser Cys Lys Ala Leu Leu Lys Arg Asn Gln Ser
340 345 350
Ile Cys Ile Val Val Gly Gly Ala Gln Glu Ser Leu Leu Ala Arg Pro
355 360 365
Gly Val Met Asp Leu Val Leu Leu Lys Arg Lys Gly Phe Val Arg Leu
370 375 380
Gly Met Glu Val Gly Asn Val Ala Leu Val Pro Ile Met Ala Phe Gly
385 390 395 400
Glu Asn Asp Leu Tyr Asp Gln Val Ser Asn Asp Lys Ser Ser Lys Leu
405 410 415
Tyr Arg Phe Gln Gln Phe Val Lys Asn Phe Leu Gly Phe Thr Leu Pro
420 425 430
Leu Met His Ala Arg Gly Val Phe Asn Tyr Asp Val Gly Leu Val Pro
435 440 445
Tyr Arg Arg Pro Val Asn Ile Val Val Gly Ser Pro Ile Asp Leu Pro
450 455 460
Tyr Leu Pro His Pro Thr Asp Glu Glu Val Ser Glu Tyr His Asp Arg
465 470 475 480
Tyr Ile Ala Glu Leu Gln Arg Ile Tyr Asn Glu His Lys Asp Glu Tyr
485 490 495
Phe Ile Asp Trp Thr Glu Glu Gly Lys Gly Ala Pro Glu Phe Arg Met
500 505 510
Ile Glu
Claims (12)
1. A recombinant microbial cell comprising an engineered LCDA production pathway comprising upregulation of a polynucleotide sequence encoding a long-chain acyl-CoA synthetase (ACoS enzyme) represented by the amino acid sequence of SEQ ID NO: 44 or 49,
wherein the microbial cells can produce one or more Long Chain Dicarboxylic Acid (LCDA) products from a substrate comprising long chain fatty acids,
wherein the microbial cells are of the genus yarrowia (Yersinia) ((R))Yarrowia) A cell.
2. The recombinant microbial cell of claim 1, wherein said ACoS enzyme has both long-chain acyl-coa synthetase activity and coumaroyl-coa synthetase activity.
3. The recombinant microbial cell of claim 1, wherein the engineered LCDA production pathway further comprises one or more of the following features:
(i) up-regulation of a polynucleotide sequence encoding a cytochrome P450 monooxygenase (CYP enzyme)
(ii) Upregulation of polynucleotide sequences encoding cytochrome P450 reductase (CPR enzyme),
(iii) upregulation of a polynucleotide sequence encoding a fatty alcohol oxidase (FAO enzyme),
(iv) upregulation of a polynucleotide sequence encoding a fatty alcohol dehydrogenase (FADH enzyme), and/or
(v) Upregulation of a polynucleotide sequence encoding a fatty aldehyde dehydrogenase (FALDH enzyme).
4. The recombinant microbial cell of claim 3, wherein one or both of the polynucleotide sequence encoding the CYP enzyme and the polynucleotide sequence encoding the CPR enzyme are up-regulated.
5. The recombinant microbial cell of claim 1, wherein the microbial cell further comprises down-regulation of an endogenous polynucleotide sequence encoding a peroxisome biogenesis factor.
6. The recombinant microbial cell of claim 5, wherein the peroxisome biogenesis factor is peroxisome biogenesis factor-3.
7. The recombinant microbial cell of claim 1, wherein the microbial cell further comprises down-regulation of an endogenous polynucleotide sequence encoding a peroxisomal acyl-coa oxidase.
8. The recombinant microbial cell of claim 7, wherein the peroxisomal acyl-CoA oxidase is peroxisomal acyl-CoA oxidase-2, -3, and/or-4.
9. The recombinant microbial cell of claim 1, wherein the microbial cell has reduced lipid synthesis and/or storage capacity.
10. The recombinant microbial cell of claim 9, wherein said decreased lipid synthesis and storage capacity is due to down-regulation of at least one endogenous polynucleotide sequence encoding a diacylglycerol acyltransferase (DGAT enzyme).
11. The recombinant microbial cell of claim 1, wherein:
the LCDA product has a chain length of 10 to 24 carbon atoms, and/or
The long chain fatty acid-containing substrate comprises a free long chain fatty acid or an esterified long chain fatty acid.
12. A method of producing a Long Chain Dicarboxylic Acid (LCDA), the method comprising:
a) contacting the recombinant microbial cell of claim 1 with a substrate comprising a long chain fatty acid, wherein said microbial cell synthesizes LCDA from said substrate; and is
b) Optionally recovering the LCDA of step (a), wherein the microbial cells are yarrowia cells.
Applications Claiming Priority (5)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US201562195340P | 2015-07-22 | 2015-07-22 | |
| US201562195338P | 2015-07-22 | 2015-07-22 | |
| US62/195340 | 2015-07-22 | ||
| US62/195338 | 2015-07-22 | ||
| PCT/US2016/043133 WO2017015368A1 (en) | 2015-07-22 | 2016-07-20 | High level production of long-chain dicarboxylic acids with microbes |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| CN108138121A CN108138121A (en) | 2018-06-08 |
| CN108138121B true CN108138121B (en) | 2022-04-19 |
Family
ID=57834583
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| CN201680055065.3A Active CN108138121B (en) | 2015-07-22 | 2016-07-20 | High-level production of long-chain dicarboxylic acids by microorganisms |
Country Status (6)
| Country | Link |
|---|---|
| US (1) | US10626424B2 (en) |
| EP (1) | EP3325607A1 (en) |
| JP (1) | JP6898915B2 (en) |
| CN (1) | CN108138121B (en) |
| MY (1) | MY191314A (en) |
| WO (1) | WO2017015368A1 (en) |
Families Citing this family (13)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN107400675B (en) * | 2017-08-29 | 2021-01-26 | 海南省农业科学院植物保护研究所 | Cloning and sequence analysis method of anti-melon wilt active product gene |
| US11136596B2 (en) * | 2018-07-06 | 2021-10-05 | Cibt America Inc. | Long-chain dibasic acid with low content of hydroxyl acid impurity and production method thereof |
| CN110684676B (en) * | 2018-07-06 | 2023-08-08 | 上海凯赛生物技术股份有限公司 | Long-chain dibasic acid with low content of hydroxy acid impurities and production method thereof |
| CN111394400B (en) * | 2019-01-03 | 2022-08-19 | 上海凯赛生物技术股份有限公司 | Application of SCT1 gene in production of long-chain dicarboxylic acid |
| CN111394399B (en) * | 2019-01-03 | 2022-06-28 | 上海凯赛生物技术股份有限公司 | Method for reducing content of acylglycerol ester impurities in long-chain dibasic acid |
| CN112301066B (en) * | 2019-07-31 | 2022-08-02 | 上海凯赛生物技术股份有限公司 | Bacterial strain for producing long-chain dicarboxylic acid by fermentation and preparation method and application thereof |
| KR102212882B1 (en) * | 2019-09-25 | 2021-02-08 | 한국원자력연구원 | Chlamydomonas sp. Microalgae with increased fatty acid productivity and method for increasing fatty acid productivity of Chlamydomonas sp. Microalgae |
| KR102343479B1 (en) * | 2020-06-23 | 2021-12-27 | 한국생명공학연구원 | Pichia ciferrii mutant strains Having Enhanced Spingoid Base and Sphingolipid Production And Preparation Method of the same |
| JP2024517798A (en) | 2021-05-04 | 2024-04-23 | ニュートリション・アンド・バイオサイエンシーズ・ユーエスエー・フォー,インコーポレイテッド | Compositions Comprising Insoluble Alpha-Glucans |
| CN115710584A (en) * | 2021-08-18 | 2023-02-24 | 上海凯赛生物技术股份有限公司 | Application of superoxide dismutase in preparing LCDA (LCDA) and genetically engineered bacterium for over-expressing superoxide dismutase |
| CN113943760B (en) * | 2021-12-02 | 2023-09-12 | 王婷 | Method for producing long-chain dibasic acid by fermentation of candida viscidosa, product and strain |
| WO2023137258A1 (en) | 2022-01-12 | 2023-07-20 | Nutrition & Biosciences USA 4, Inc. | Coating compositions comprising rubber and insoluble alpha-glucan |
| CN116656505B (en) * | 2023-04-23 | 2024-04-16 | 四川大学 | High-yield alcohol dehydrogenase and acyltransferase strain, and culture method and application thereof |
Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN101228282A (en) * | 2004-12-15 | 2008-07-23 | 法国石油公司 | Production d'acides dicarboxyliques par des souches mutantes ameliorees de yarrowia lipolytica |
| CN104321424A (en) * | 2012-04-03 | 2015-01-28 | 纳幕尔杜邦公司 | Expression of cytosolic malic enzyme in transgenic yarrowia to increase lipid production |
| CN104781411A (en) * | 2012-11-09 | 2015-07-15 | 凯利斯塔公司 | Compositions and methods for biological production of fatty acid derivatives |
Family Cites Families (14)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US4880741A (en) | 1983-10-06 | 1989-11-14 | Pfizer Inc. | Process for transformation of Yarrowia lipolytica |
| US5071764A (en) | 1983-10-06 | 1991-12-10 | Pfizer Inc. | Process for integrative transformation of yarrowia lipolytica |
| US20040146999A1 (en) | 1997-07-21 | 2004-07-29 | E.I. Du Pont De Nemours And Company | Transformed yeast strains and their use for the production of monoterminal and diterminal aliphatic carboxylates |
| US7238482B2 (en) | 2003-05-07 | 2007-07-03 | E. I. Du Pont De Nemours And Company | Production of polyunsaturated fatty acids in oleaginous yeasts |
| US7125672B2 (en) | 2003-05-07 | 2006-10-24 | E. I. Du Pont De Nemours And Company | Codon-optimized genes for the production of polyunsaturated fatty acids in oleaginous yeasts |
| US8846374B2 (en) | 2006-12-12 | 2014-09-30 | E I Du Pont De Nemours And Company | Carotenoid production in a recombinant oleaginous yeast |
| CN101970638B (en) | 2007-10-03 | 2015-02-11 | 纳幕尔杜邦公司 | Optimized strains of yarrowia lipolytica for high eicosapentaenoic acid production |
| WO2012071439A1 (en) | 2010-11-22 | 2012-05-31 | The Regents Of The University Of California | Host cells and methods for producing diacid compounds |
| US8729298B2 (en) | 2011-03-28 | 2014-05-20 | Cathay Industrial Biotech Ltd. | Method for separation and purification of long-chain diacids |
| MY165893A (en) * | 2011-07-06 | 2018-05-18 | Verdezyne Inc | Biological methods for preparing a fatty dicarboxylic acid |
| WO2013082186A2 (en) * | 2011-11-28 | 2013-06-06 | Solazyme, Inc. | Genetically engineered microbial strains including prototheca lipid pathway genes |
| EP2935563B1 (en) * | 2012-12-19 | 2020-11-04 | Corvay Bioproducts GmbH | Biological methods for preparing a fatty dicarboxylic acid |
| JP2016502851A (en) | 2012-12-21 | 2016-02-01 | イー・アイ・デュポン・ドウ・ヌムール・アンド・カンパニーE.I.Du Pont De Nemours And Company | Down-regulation of polynucleotides encoding SOU2 sorbitol-utilizing proteins to modify lipogenesis in microbial cells |
| EP2843043A1 (en) * | 2013-08-27 | 2015-03-04 | Evonik Industries AG | A method for producing acyl amino acids |
-
2016
- 2016-07-20 JP JP2018502751A patent/JP6898915B2/en active Active
- 2016-07-20 WO PCT/US2016/043133 patent/WO2017015368A1/en active Application Filing
- 2016-07-20 CN CN201680055065.3A patent/CN108138121B/en active Active
- 2016-07-20 EP EP16747664.7A patent/EP3325607A1/en active Pending
- 2016-07-20 MY MYPI2018700157A patent/MY191314A/en unknown
- 2016-07-20 US US15/746,094 patent/US10626424B2/en active Active
Patent Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN101228282A (en) * | 2004-12-15 | 2008-07-23 | 法国石油公司 | Production d'acides dicarboxyliques par des souches mutantes ameliorees de yarrowia lipolytica |
| CN104321424A (en) * | 2012-04-03 | 2015-01-28 | 纳幕尔杜邦公司 | Expression of cytosolic malic enzyme in transgenic yarrowia to increase lipid production |
| CN104781411A (en) * | 2012-11-09 | 2015-07-15 | 凯利斯塔公司 | Compositions and methods for biological production of fatty acid derivatives |
Non-Patent Citations (2)
| Title |
|---|
| Involvement of acyl-CoA synthetase genes in n-alkane assimilation and fatty acid utilization in yeast Yarrowia lipolytica;Tenagy等;《FEMS Yeast Research》;20150630;第15卷(第4期);第4页"Orthologs of ACS genes in Y.lipolytica" * |
| YALI0E12419p [Yarrowia lipolytica CLIB122];NCBI;《NCBI》;20150227;ORIGIN部分 * |
Also Published As
| Publication number | Publication date |
|---|---|
| JP2018519843A (en) | 2018-07-26 |
| CN108138121A (en) | 2018-06-08 |
| US10626424B2 (en) | 2020-04-21 |
| US20190144897A1 (en) | 2019-05-16 |
| WO2017015368A1 (en) | 2017-01-26 |
| JP6898915B2 (en) | 2021-07-07 |
| EP3325607A1 (en) | 2018-05-30 |
| MY191314A (en) | 2022-06-15 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| CN108138121B (en) | High-level production of long-chain dicarboxylic acids by microorganisms | |
| AU2023226754A1 (en) | Compositions and methods for modifying genomes | |
| DK2664670T3 (en) | perhydrolase | |
| CN101939434B (en) | Dgat genes from yarrowia lipolytica for increased seed storage lipid production and altered fatty acid profiles in soybean | |
| CN101365788B (en) | Delta-9 elongases and their use in making polyunsaturated fatty acids | |
| DK2087106T3 (en) | MUTATING DELTA8 DESATURATION GENES CONSTRUCTED BY TARGETED MUTAGENES AND USE THEREOF IN THE MANUFACTURE OF MULTI-Saturated FAT ACIDS | |
| DK2087105T3 (en) | DELTA 17 DESATURASE AND ITS USE IN THE MANUFACTURE OF MULTI-Saturated FAT ACIDS | |
| DK2443248T3 (en) | IMPROVEMENT OF LONG-CHAIN POLYUM Saturated OMEGA-3 AND OMEGA-6 FATTY ACID BIOS SYNTHESIS BY EXPRESSION OF ACYL-CoA LYSOPHOSPHOLIPID ACYL TRANSFERASES | |
| KR20140092759A (en) | Host cells and methods for production of isobutanol | |
| AU2018220469A1 (en) | Method and cell line for production of phytocannabinoids and phytocannabinoid analogues in yeast | |
| KR20130032897A (en) | Production of alcohol esters and in situ product removal during alcohol fermentation | |
| KR20190002470A (en) | Improved method for modification of target nucleic acid | |
| KR20140099224A (en) | Keto-isovalerate decarboxylase enzymes and methods of use thereof | |
| EP2181195A2 (en) | Fermentative production of acetone from renewable resources by means of novel metabolic pathway | |
| KR20070085669A (en) | High arachidonic acid producing strains of yarrowia lipolytica | |
| KR20120099509A (en) | Expression of hexose kinase in recombinant host cells | |
| BRPI0806354A2 (en) | transgender oilseeds, seeds, oils, food or food analogues, medicinal food products or medicinal food analogues, pharmaceuticals, beverage formulas for babies, nutritional supplements, pet food, aquaculture feed, animal feed, whole seed products , mixed oil products, partially processed products, by-products and by-products | |
| KR20150014953A (en) | Ketol-acid reductoisomerase enzymes and methods of use | |
| KR20180053684A (en) | Manufacture of FDCA fungi | |
| KR20100037031A (en) | Gene knockout mesophilic and thermophilic organisms, and methods of use thereof | |
| CN101646766B (en) | Detal 17 desaturases and use thereof in making polyunsaturated fatty acids | |
| KR20220012327A (en) | Methods and cells for production of phytocannabinoids and phytocannabinoid precursors | |
| CN108779480A (en) | The method for producing sphingosine and sphingolipid | |
| KR20180084135A (en) | Methods for producing proteins from filamentous fungi with reduced CLR2 activity | |
| CN108473968A (en) | The method for producing phytosphingosine or dihydrosphingosine |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| PB01 | Publication | ||
| PB01 | Publication | ||
| SE01 | Entry into force of request for substantive examination | ||
| SE01 | Entry into force of request for substantive examination | ||
| GR01 | Patent grant | ||
| GR01 | Patent grant |